Abstract
SGTA is a co-chaperone that, in collaboration with the complex of BAG6/UBL4A/TRC35, facilitates the biogenesis and quality control of hydrophobic proteins, protecting them from the aqueous cytosolic environment. This work includes targeting tail-anchored proteins to their resident membranes, sorting of membrane and secretory proteins that mislocalize to the cytoplasm and endoplasmic reticulum-associated degradation of misfolded proteins. Since these functions are all vital for the cell's continued proteostasis, their disruption poses a threat to the cell, with a particular risk of protein aggregation, a phenomenon that underpins many diseases. Although the specific disease implications of machinery involved in quality control of hydrophobic substrates are poorly understood, here we summarize much of the available information on this topic.
Keywords: SGTA, BAG6, UBL4A, TRC35, Chaperones, Co-chaperones, Quality control, Proteostasis, Tail-anchored proteins, Hydrophobic proteins
1. Introduction
The cytoplasm of cells is a bustling, crowded environment which requires layers of management to prevent descent into chaos. This chapter will address some of the machinery that maintains the cellular environment and what happens when it breaks down or gets out of control. In particular we will consider the case of stray hydrophobic proteins that become exposed to the aqueous cytosol. These pose an aggregation risk and, as relevant, must be sent to a membrane, refolded to tuck the hydrophobic parts back into the safety of the core or labeled for disposal with ubiquitin tags and then degraded by the proteasome. Since these processes are crucial for maintaining cellular proteostasis, it is inevitable that failures in them, which, among other things, can be caused by absence or disruption of any of their machine components, may result in disease. At present, the precise roles of these proteins in disease are not well understood. This review will consider some of the major known constituents of the mammalian quality control machinery for hydrophobic proteins exposed to the cytoplasm, SGTA, BAG6, TRC35 and UBL4A, and catalog some of the current disease connections that exist in today's literature. It is likely that these references barely scratch the surface of the disease relevance and therapeutic potential of these proteins and that this field will continue burgeoning in the coming years.
2. Hydrophobic parts of proteins exposed to the cytoplasm pose an aggregation risk
There are many scenarios in which hydrophobic areas of proteins become aberrantly exposed to the aqueous cell cytoplasm. Large proteins often reach their native folded state via a number of intermediate states. During these folding pathways there is the potential for protein misfolding, which can result in additional deleterious functions that can lead to a range of diseases (Hartl & Hayer-Hartl, 2009). The more complex the protein, the higher the chance it will misfold. During the folding pathway there has evolved a need for a quality control network. As proteins pass through intermediate states where they are incompletely folded, they can expose buried regions to the solvent which can lead to undesired interactions; the exposure of these regions can be an indicator of protein misfolding (Buchberger, Bukau, & Sommer, 2010). It's the role of molecular chaperones and co-chaperones of the quality control network to protect these exposed regions and thus aid either the refolding of the protein, further prevention of misfolding or transport of the protein to a desired safe site (Buchberger et al., 2010). While chaperones do not increase the rate of folding, they do increase its efficiency by reducing the chances of undesired reactions such as aggregation (Fernandez-Fernandez & Valpuesta, 2018). Hence molecular chaperones and co-chaperones play a role in the essential protein quality control network that deals with unfolded and misfolded proteins.
3. The specific case of tail-anchored proteins
Tail-anchored (TA) proteins represent a widespread but distinct case of hydrophobic regions requiring protection from the aqueous cytosol (Rabu, Schmid, Schwappach, & High, 2009). They are a class of membrane proteins that make up 3–5% of all membrane proteins including SNAREs (involved in vesicular trafficking); signaling proteins; endoplasmic reticulum translocon components and various enzymes that are spatially restricted (Borgese & Fasana, 2011). They therefore have critical roles in cell biology in membrane biogenesis, apoptosis, and protein degradation among many others. The specific localization of TA proteins within membranes is crucial to their task and, while their functions vary greatly, they are united in a common topology, based on the positioning of their single transmembrane helix (Kalbfleisch, Cambon, & Wattenberg, 2007). Newly synthesized TA proteins cannot be co-translationally directed to the endoplasmic reticulum (ER) for insertion, as is traditional for membrane proteins, as their single membrane-spanning helix is at the extreme C-terminus and therefore inaccessible to the signal recognition particle during translation (Borgese & Fasana, 2011). This is where quality control proteins come into play, they are nature's solution to the insertion of the TA proteins into the ER membrane and this process shares common sorting and targeting machinery with the MLPs described above (Shao, Rodrigo-Brenni, Kivlen, & Hegde, 2017).
4. Triage and fate of hydrophobic substrates in the cytoplasm
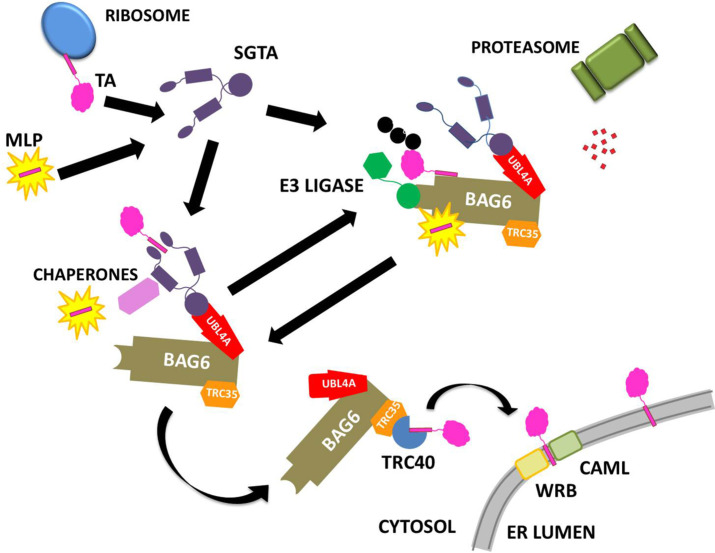
The details of these processes are still being established but many advances have been made in recent years including structure solution of some of the machinery for sorting and delivery (which will be discussed in the individual sections for each protein) and mechanistic understanding from cell biology and isolated systems (Shao et al., 2017). Our current understanding is summarized in Fig. 1 . It is thought that SGTA can catch the TMDs of newly translated TA proteins or exposed hydrophobic patches on mislocalised membrane and secretory proteins (MLP), which bind to its C-terminal domain. In collaboration with the heterotrimeric BAG6 complex, which comprises BAG6, TRC35 and UBL4A, SGTA determines the fate of these proteins. Hydrophobic substrates bound to the BAG6 complex can be ubiquitinated by the actions of the E3 ligase RNF126 (Krysztofinska et al., 2016; Rodrigo-Brenni, Gutierrez, & Hegde, 2014) and thus targeted for proteasomal degradation. SGTA can interact with the RPN13 subunit of the 19S regulatory particle of the proteasome through its TPR domain (Leznicki et al., 2015; Thapaliya et al., 2016), which has led to the proposal of an SGTA/BAG6 cycle operating at the proteasome (Leznicki & High, 2012). SGTA hands tail-anchored (TA) proteins over to TRC40 facilitating their post-translational integration into the ER by means of the transmembrane proteins WRB and CAML (Vilardi, Stephan, Clancy, Janshoff, & Schwappach, 2014). Furthermore, SGTA has been implicated in hormone receptor signaling and has been associated with viral lifecycles (Philp et al., 2013). SGTA's interactions with Hsp70/Hsp90 chaperones via its TPR domain likely provide substrate access to additional branches of the global cellular quality control network (Yin et al., 2006).
Fig. 1.
Current ideas on quality control pathways for hydrophobic proteins exposed to the aqueous cytoplasm. SGTA can catch the TMDs of newly translated TA proteins or exposed hydrophobic patches on MLPs, which bind to its C-terminal domain. In collaboration with the heterotrimeric BAG6 complex, which comprises BAG6, TRC35 and UBL4A, SGTA determines the fate of these proteins. Hydrophobic substrates bound to the BAG6 complex can be ubiquitinated by the actions of the E3 ligase RNF126 and thus targeted for proteasomal degradation. SGTA can interact with the RPN13 subunit of the 19S regulatory particle of the proteasome through its TPR domain, which has led to the proposal of an SGTA/BAG6 cycle operating at the proteasome. SGTA hands tail-anchored (TA) proteins over to TRC40 facilitating their post-translational integration into the ER by means of the transmembrane proteins WRB and CAML. Furthermore, SGTA has been implicated in hormone receptor signaling and has been associated with viral lifecycles. SGTA's interactions with Hsp70/Hsp90 chaperones via its TPR domain likely provide substrate access to additional branches of the global cellular quality control network.
5. SGTA
The focus of the first section of this chapter will be the molecular co-chaperone known as the small glutamine-rich tetratricopeptide repeat-containing protein alpha (SGTA) (Wunderley, Leznicki, Payapilly, & High, 2014). A major known function of SGTA is to aid in determining the fate of secretory and membrane proteins that have mislocalized to the cytosol (Leznicki et al., 2013). SGTA is also thought to be the first port of call for tail-anchored proteins upon termination of translation when the two ribosomal subunits separate (Philp et al., 2013). On many occasions SGTA works in concert with the BAG6 complex, a heterotrimeric complex comprising stoichiometric volumes of BAG6 (BCL2-associated athanogene 6, formerly known as BAT3 or Scythe), UBL4A (ubiquitin-like protein 4A, sometimes known as GDX) and TRC35 (transmembrane domain recognition complex 35), that is also involved in the protein quality control network (Xu, Cai, Yang, Huang, & Ye, 2012) and these three proteins will be addressed in detail later on in this chapter. It's been suggested that one of the roles of the SGTA/BAG6 combination is in facilitation of the ER-associated degradation pathway (ERAD) in which poorly folded proteins are retro-translocated through the ER membrane (Hampton & Sommer, 2012). SGTA has also been shown to rescue proteins from degradation by reversing the ubiquitination caused by BAG6 probably via recruitment of an, as yet unidentified, DUB or deubiquitinating enzyme (Leznicki & High, 2012). Disruption or incorrect use of these functions can lead to diseases such as cancer of the prostate, ovary, liver, polycystic ovary syndrome, Alzheimer's and many more (Philp et al., 2016). Here we will examine some of the current disease implications of SGTA.
6. SGTA structure
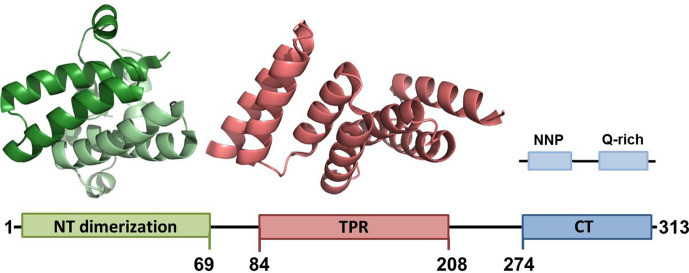
SGTA (shown in Fig. 2 ) is a homodimeric protein with each monomer composed of 313 amino acids encompassing 3 domains connected by flexible linkers (Roberts, Thapaliya, Martinez-Lumbreras, Krysztofinska, & Isaacson, 2015). First is the N-terminal domain spanning the first 69 residues (Chartron, VanderVelde, & Clemons, 2012; Simon, Simpson, Goldstone, et al., 2013), and then following a 14 residue linker is the central tetratricopeptide repeat domain (TPR) (from which part of the name originates) which spans residues 86–208 (Dutta & Tan, 2008). Finally, from residues 211 to 313 is the C-terminal domain that contains the glutamine-rich region from residues 274 to 313 (Liou & Wang, 2005).
Fig. 2.
Schematic representation of SGTA showing domain boundaries and solved structures. N-Terminal dimerization domain (monomer chains in two shades of green) residues 1–69 from PDB: 4CPG (Darby et al., 2014); TPR domain (red) residues 84–210 from PDB: 2VYI (Dutta & Tan, 2008). There is currently no solved structure for the C-terminal domain (blue) which encompasses NNP and Q-rich regions (Martinez-Lumbreras et al., 2018).
SGTA belongs to a family of co-chaperones that are categorized with a “nPR” nomenclature system. The “n” represents the varying number of residues in a single repeat, and thus SGTA belongs to the canonical 34PR motif (Thapaliya et al., 2016) as each repeat, made of 2 helices, consists of 34 amino acids; tetratrico means 34 in Greek.
SGTA is highly conserved throughout mammals and higher eukaryotes with the TPR domain sharing the greatest similarity across the different species (Martinez-Lumbreras et al., 2018). The N-terminal domain of SGTA forms a tight dimer with a negatively charged patch on its surface which has been shown to bind with positively charged UBL domains from both BAG6 and UBL4A domain through electrostatic interactions (Darby et al., 2014). The trade-off between binding ability of SGTA and these two UBLs is thought to be behind the sorting mechanism of the protein quality control network (Darby et al., 2014; Shao et al., 2017).
As previously discussed, it's the existence of the TPR domain that places SGTA in the 34PR family (Nguyen et al., 2017). Typically TPR domains are made up of between 3 and 16 tandem repeats—SGTA contains 3 repeats with each repeat containing 2 almost identically structured antiparallel folded alpha helices (Dutta & Tan, 2008; Krysztofinska et al., 2017). There is additionally a seventh alpha helix known as the C-terminal capping helix that aligns against the second helix of the third repeat, helix 6. The three repeats are structured in a parallel arrangement which creates a super helical structure containing a surface groove. This groove has been shown to play a role in intra-molecular interactions (Liou & Wang, 2005). The TPR domain is responsible for SGTA binding to Hsp70 and Hsp90, along with other varying receptors and proteins (Philp et al., 2013; Roberts et al., 2015). Due to this, the TPR domain has been linked with SGTA's role in diseases such as HIV (Dutta & Tan, 2008) and the parvovirus (Cziepluch et al., 1998).
Out of the three domains, the least is known about the C-terminal domain with no high resolution structure currently available. It has been shown that it contains an important glutamine-rich region between residues 274 and 313 where 13 glutamine residues are located. This region has been implicated as responsible for the C-terminal's ability to interact with hydrophobic amino acids within polypeptides (Liou & Wang, 2005). Removal of this region resulted in no interactions with hydrophobic substrates (Liou & Wang, 2005). This allows for SGTA to bind to and then shield improperly or incompletely folded proteins with exposed hydrophobic regions, and thus provide a vital role within the protein quality control network. Very recently another important motif has been identified in the C-terminus called the NNP motif consisting of three copies of asparagine-asparagine-proline (Martinez-Lumbreras et al., 2018). The C-terminal domain has also been found to transiently dimerize, probably around hydrophobic substrates in the manner of a pair of tweezers (Martinez-Lumbreras et al., 2018).
7. Regulation of cytosolic quality control by SGTA
As previously stated, exposed hydrophobic residues on proteins can form inappropriate interactions within the cytosol (Buchberger et al., 2010). Typically, when membrane proteins with exposed hydrophobic residues enter the secretory pathway, they are met and transported by signal recognition particle dependent delivery to their desired location. This process prevents interactions between the cytosol and the exposed hydrophobic residues (Cross, Sinning, Luirink, & High, 2009). However sometimes membrane and secretory proteins end up mislocalized which can lead to undesired interactions with the cytosol; it's the responsibility of the protein quality control network to deal with this (Ast, Cohen, & Schuldiner, 2013). SGTA, alongside the BAG6 complex, plays a role in responding to MLPs while encouraging the transportation of tail-anchored proteins into the ER membrane (Hessa et al., 2011). Studies have suggested that while BAG6 promotes the polyubiquitylation of mislocalized hydrophobic substrates, SGTA can antagonize this process by activating substrate deubiquitylation (Leznicki & High, 2012). Both complexes also play a role in the ERAD pathway for irregular proteins (Xu et al., 2012).
Studies have shown that SGTA binds directly to hydrophobic regions of MLPs both in vitro and in vivo, which highlights their importance in signaling for degradation. Results also suggest that SGTA promotes substrate deubiquitylation. Overexpression of SGTA is shown to inhibit proteasomal degradation of irregular proteins while also stabilizing MLPs; this links to undesirable results such as prolonging the lifespan of aberrant proteins, and several forms of cancer. Overall SGTA plays an important role in cytosolic quality control; at the ER it encourages the biogenesis of certain hydrophobic precursors while additionally promoting the removal of irregular and misfolded proteins (Shao et al., 2017). Without the presence of SGTA, not only would the lifespan of potentially dangerous proteins be extended, but undesirable interactions between proteins and the cytosol could lead to aggregation, a suggested cause of some neurodegenerative diseases (Chakrabarti & Hegde, 2009; Wunderley et al., 2014).
8. Role of SGTA in neurodegenerative diseases
Protein aggregation plays an important role in neurodegenerative diseases, such as Alzheimer's disease, Parkinson's disease, Huntington's disease, amyotrophic lateral sclerosis and prion diseases (Cox, Raeburn, Sui, & Hatters, 2018; Ross & Poirier, 2004). If proteins with exposed hydrophobic regions are allowed to interact with the cytosol they can form aggregates and inclusion bodies (Ross & Poirier, 2004). In Huntington's disorder, for example, inclusions have been found in regions of the brain that degenerate (Vonsattel et al., 1985). Another example is Alzheimer's disease, which is shown to be caused by two types of protein aggregates: extracellular and intracellular (Cox et al., 2018). Interestingly, both increased and decreased expression of SGTA can lead to the formation of aggregates which has been correlated with potentially fatal neurodegenerative diseases (Wunderley et al., 2014).
9. The androgen signaling pathway and SGTA
Steroid hormone receptors (SHRs) follow an incredibly complex and labile biogenesis which is aided by a number of different chaperones along the way (Echeverria & Picard, 2010). One such SHR, the androgen receptor (AR), is a testosterone-dependent, specific transcription factor (Cutress, Whitaker, Mills, Stewart, & Neal, 2008). Located in the cytoplasm, upon binding of the ligand to the AR, interactions take place within its N- and C-terminal domains that result in activation and transfer to the nucleus to activate transcription of specific genes (Cutress et al., 2008). ARs play an important role in locations such as the prostate gland; although their primary role in this organ is found during the growth of the gland, they still function after maturity (Brooke & Bevan, 2009).
SGTA comes into play as a chaperone that stabilizes the unliganded form of the AR within the cytoplasm by binding to the AR's hinge region with its TPR domain; after the ligand binds to AR, the receptor dissociates from the chaperone (Trotta et al., 2012). SGTA activity has been implicated in androgen regulation as a downregulator of the receptor by promoting retention within the cytoplasm (Buchanan et al., 2007). Studies suggest that this downregulation is caused by controlling the sensitivity to the AR ligand, which leads to retention within the cytoplasm (Trotta et al., 2013). However, one study showed that SGTA's role in this downregulation could be during the folding of the receptor, with SGTA affecting the ubiquitination and recycling of the receptor during the early folding stages (Paul et al., 2014). This links into its role in cytosolic quality control which supports this theory (Wunderley et al., 2014). The same study also suggests a link between SGTA and other SHRs, the glucocorticoid and progesterone receptor activity suggesting that SGTA's role in hormone signaling extends beyond the androgen receptors (Paul et al., 2014).
Improper regulation of AR signaling has been related to prostate cancer, polycystic ovary syndrome and breast cancer thus it's suggested that irregular activity of SGTA could be a factor in these diseases (Goodarzi et al., 2008; Trotta et al., 2013).
10. The role of SGTA in prostate cancer
Prostate cancer is one of the most common forms of cancer in males, and is considered one of the deadliest cancers. Usually, initial tumor growth is androgen dependent and driven by the AR activity within the cells (Brooke & Bevan, 2009). Androgen dependent prostate cancer involves the AR stimulating proliferation while inhibiting apoptosis of cells, thus ensuring the survival of cancerous cells (Brooke & Bevan, 2009). This form of the cancer is normally treated with androgen ablation and while most tumors respond to this treatment, some evolve into the androgen independent form which has been largely considered incurable (Cattrini et al., 2017). Androgen independent cancer evolves from genetic mutations within the cell that allow it to grow independent of AR activity; instead it follows pathways involving overexpression of BCL2 or inactivation of tumor suppressor genes among many others (Brooke & Bevan, 2009; Cattrini et al., 2017). These pathways have also been linked to other hormone independent cancers such as breast cancer (Bullock, 2016).
One such cause of androgen independent prostate cancer has been linked to SGTA; it has been suggested that SGTA is a negative regulator of AR transport into the nucleus by promoting AR retention within the cytoplasm, as previously stated (Kato et al., 2017). One theory as to the mechanism behind this involves Hsp70, which is suggested to provide the energy needed by chaperones that are responsible for association and dissociation of proteins. SGTA interactions with Hsp70 increase protein affinity and result in more efficient protein folding; in yeast cells that lack SGTA there is a decrease in Hsp70 activity, and thus it's suggested that SGTA can indirectly affect the ligand binding capacity of the AR via this mechanism (Buchanan et al., 2007). Since it has separately been suggested that SGTA directly binds to the AR's hinge region, it correlates that SGTA overexpression directly inhibits the AR by reducing its ability to bind with ligands (Trotta et al., 2012). Despite conflicting ideas behind the mechanism of which SGTA affects AR activity, there has been evidence to suggest that overexpression leads to decreased activity, while underexpression leads to increased activity (Buchanan et al., 2007). Thus its widely accepted SGTA plays a role in prostate cancer development; however the mechanism by which this occurs is still not fully understood.
11. The role of SGTA in polycystic ovary syndrome
Polycystic ovary syndrome (PCOS) is a common genetic disorder that affects between 6% and 8% of women of reproductive age. Common symptoms include infertility, insulin resistance, obesity and high cardiovascular risk factors (Azziz et al., 2004). While there is still much debate as to the cause of PCOS, with suggestions such as improper insulin signaling, genetic mutations and irregular androgen signaling; most patients present with hyperandrogenism (Goodarzi et al., 2008).
It has been suggested that SGTA's role in this disease is much the same as its role in prostate cancer, promoting retention of the AR within the cytoplasm and thus preventing further function (Buchanan et al., 2007). Overexpression of SGTA is implied to inhibit AR function by causing said retention in the cytoplasm, whereas knockdown of SGTA is said to result in increased AR activity and promiscuous activation of the receptor by ligands other than testosterone, such as progesterone (Buchanan et al., 2007; Goodarzi et al., 2008). Asides from affecting androgen signaling, SGTA has been linked to PCOS via its role in apoptosis, promoting cell death when both over- and under-expressed (Yin et al., 2006). A further role of SGTA within PCOS is insulin signaling. A study has shown that the SGTA gene is related to insulin resistance; however more research is needed to determine how and to what extent (Goodarzi et al., 2008). Overall it's shown that there is a probable role of SGTA in the cause of PCOS through multiple pathways, making it a possible target for therapy of this disease.
12. The role of SGTA in breast cancer
Another disease linked to androgen signaling is breast cancer, one of the most common cancers among women. A 2014 study (Zhu et al., 2014) first examined the relationship between SGTA and breast cancer cells and found that SGTA was overexpressed in breast cancer cells compared to normal tissue. They also found a positive correlation between SGTA concentration and breast cancer cell proliferation following SGTA knockout experiments. These results suggest that SGTA levels could be a potential indicator of breast cancer progression as concentration was found to increase at a higher histological grade. Additionally, SGTA was recognized as a potential prognosis indicator (Zhu et al., 2014). This leaves SGTA as an interesting potential treatment target for breast cancer and thus more research should be done into this relationship (Zhu et al., 2014).
13. SGTA in the cell cycle
SGTA is required for the cell to successfully complete the cell division pathway (Winnefeld, Rommelaere, & Cziepluch, 2004). Reduced levels of SGTA results in reduced proliferation through an arrest in mitosis, which is subsequently followed by cell death. Studies have shown that SGTA depleted cells mostly arrested in their metaphase stage, suggesting that SGTA was limiting the cells' exit from the metaphase; however some cells were able to progress through to the G1 stage before cell death took place (Winnefeld et al., 2004).
On the other hand, overexpression of SGTA seems to be just as disastrous for the cell lifespan. There is a correlation between SGTA and apoptosis with a proposal that the TPR domain was responsible for this function (Wang et al., 2005). The role of SGTA is also linked to that of Hsp90s (highly expressed protein chaperones that assist in protein degradation), suggesting that while they usually bind in the cytoplasm, during cell apoptosis SGTA dissociates and then accumulates in the nucleus, although much more research is needed to suggest how this further links to apoptosis (Yin et al., 2006). The importance of SGTA within the cell cycle is another likely contributor to its role in tumorigenesis.
14. The role of SGTA in lung cancer
Non-small-cell lung cancer (NSCLC) is the most common form of lung cancer, and a leading cause of cancer deaths worldwide. The prognosis is generally poor with 5 year survival rates less than 15%; this is mostly due to poor diagnostic rates and difficulty in treatment (Ji et al., 2011). There is a known relationship between NSCLC and SGTA expression; SGTA is positively correlated with cellular proliferation and was frequently highly expressed in NSCLC cells. A link is suggested between SGTA and the chemoresistance of NSCLC cells by SGTA suppressing cisplatin-induced apoptosis. This suggests SGTA as a potential target for NSCLC treatments especially due to its role in chemoresistance (Xue et al., 2013).
15. The role of SGTA in esophageal cancer
Like NSCLC, esophageal squamous cell carcinoma (ESCC) reports a poor prognosis. The etiology is complex and involves multiple genes; therefore important diagnostic indicators are needed to help with early stage diagnosis, which, like in NSCLC, is a reason for the poor prognosis (Talukdar et al., 2018). One study suggested SGTA contributed to the malignant progression of ESCC and thus could be used a prognostic biomarker (Yang et al., 2014). Much like the NSCLC study, overexpression of SGTA was shown to play a role in the development and progression of tumors, although more research is still needed to decipher the mechanisms behind these observations to further understand SGTA's role (Yang et al., 2014).
16. Early identifications of SGTA in viral infections
Early in the literature, SGTA was discovered in complex with the parvovirus H-1, specifically NS1, a non-structural protein of H-1 that is essential for DNA replication (Cziepluch et al., 1998). Another example of SGTA in complex with a virus involves Vpu, a viral core protein precursor from HIV-1 (Dutta & Tan, 2008). It was suggested that in the absence of Vpu, overexpression of SGTA leads to interactions with HIV-1 Gag, another HIV-1 protein, and the resulting complex inhibits HIV-1 release. However, in the presence of Vpu stable interactions form between SGTA and Vpu that prevent SGTA-Gag interactions (Waheed et al., 2016).
Another viral protein interacting with SGTA is protein 7a from the severe acute respiratory syndrome coronavirus (SARS-CoV), a member of the Coronaviridae family, the largest known RNA viruses (Fielding et al., 2006; Philp et al., 2013). SGTA is thought to play a role in virus assembly or release from the cell but more research is needed to fully understand the implications of SGTA's involvement with SARS-CoV and the mechanisms behind this (Fielding et al., 2006).
17. SGTA and the simian virus 40
A study into SGTA's relationship with the simian virus 40 (SV40) suggested that SGTA facilitates transport of the virus between the endoplasmic reticulum (ER) and the cytosol (Walczak, Ravindran, Inoue, & Tsai, 2014).
SV40 is a non-enveloped polyomavirus that has to travel through biological membranes to infect cells. As it lacks a surrounding lipid bilayer it cannot infect cells in the same way as enveloped viruses, which usually proceed via membrane fusion. Thus it needs to find a different path for entry to the cell; this is usually taken through virus-induced pore formation or disruption of the cell membrane's integrity (Wiethoff, Wodrich, Gerace, & Nemerow, 2005). Polyomaviruses are unique in that they travel past the endosomal system and reach the ER for membrane penetration; SV40 is known to infect cells via engaging the ganglioside receptor GM1 at the cell surface. Endocytosis then brings SV40 molecules complexed with lipid rafts into the cell where they proceed through endosomes before reaching the ER. After this SV40 has to exit through the ER membrane and cross the cytosol to reach the nucleus where it subsequently activates transcription and replication of the viral genome; thus cellular transformation is caused (Wiethoff et al., 2005). It is thought that SGTA associates with a B14 and B12 complex at the ER membrane and, during infection, SGTA binds directly to SV40 and thus promotes its ER membrane penetration. During this penetration, SGTA dissociates from the B14 and B12 complex and hence it is implied that SGTA is directly responsible for SV40’s transport between the ER and the nucleus (Walczak et al., 2014).
A further study in 2017 showed that two of SGTA's domains are responsible for this function (Dupzyk, Williams, Bagchi, Inoue, & Tsai, 2017). First it linked the TPR domain's ability to interact with Hsc70; this aids penetration of the ER membrane and subsequent transport to the cytosol. Then it linked the N-terminal domain with the infection of SV40 into the nucleus; although the exact role the N-terminal plays is uncertain, results showed that the N-terminal was crucial for further infection after SV40 has entered the cytosol. The study suggested that this could be via the N-terminal being directly involved in transport to the nucleus, or the N-terminal could recruit cellular components that subsequently transport the virus to the nucleus. Although it has been seen in other studies that SGTA can localize to the nucleus, and this suggests the direct involvement of the N-terminal in SV40 infection, more research is needed to further understand the role of SGTA in virus infection, and how its domains affect each stage (Dupzyk et al., 2017).
18. BAG6
The human BAG family consists of six members, the largest being the BAG6 protein constituting of 1132 amino acids in the major isoform. It is situated in the gene cluster of the human major histocompatibility (MHC) class lll on chromosome 6 which contains many genes responsible for immune function (Banerji, Sands, Strominger, & Spies, 1990). The highly conserved BAG6 protein is flanked by a ubiquitin-like (UBL) domain on the N-terminus and a BAG domain on the C-terminus. The central region consists of a disordered proline-rich region, zinc-finger like and Nuclear Localization Signal (NLS) domain. BAG6 was designated a member of the BAG family due to its observed sequence homology to the BAG domain and its collaboration with Heat Shock Protein 70 (Hsp70) family possessing chaperoning activity (Kuwabara et al., 2015).
BAG6 is an abundant cellular protein expressed ubiquitously in higher eukaryotes. Some tissues in multi-cellular organisms, such as testis and brain, express relatively higher levels of BAG6. This suggests a potential function in spermatogenesis and brain development respectively. Numerous other functions associated with BAG6 have been identified, including the regulation of apoptosis, T-cell response and protein homeostasis (Kuwabara et al., 2015; Lee & Ye, 2013). Due to the diversity of its functions, the significance of BAG6 in disease and therapy is of particular interest. As previously mentioned, a heterotrimeric complex (known as the BAG6 complex) is formed between BAG6, UBL4A and TRC35, which together determine the fate of hydrophobic nascent polypeptides. Hence, BAG6 has been identified as a multi-functional protein that plays a critical role in the quality control of hydrophobic polypeptides whereby it is at the interface of protein synthesis and degradation.
Fundamentally, the BAG6 complex can function in several cell conditions and carry out essential processes depending on individual cell requirements. Therefore, dysfunctions in the BAG6 complex cause various cellular abnormalities which lead to both physiological and pathological diseases (Lee & Ye, 2013). An overview of BAG6 functions and its association with disease are given in the following sections.
19. BAG6 structure
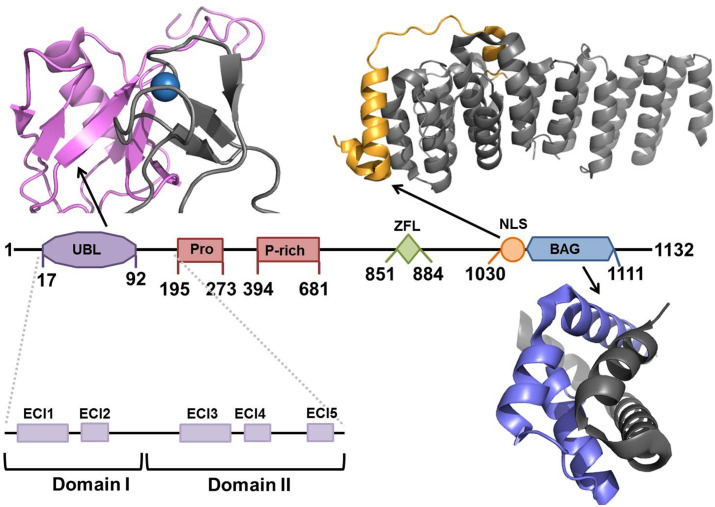
The primary sequence of BAG6 can be subdivided into three distinct domains: the N-terminal UBL-containing region spanning amino acids 1–474; a central proline-rich domain constituting amino acids 475–1029, a possible site for binding of hydrophobic substrates according to in vitro and in vivo studies (Leznicki et al., 2013); and a C-terminal NLS and designated unconventional BAG domain constituting amino acids 1030–1132. The small sections of BAG6 whose structures have been solved are shown in Fig. 3 but the vast majority of the protein still remains structurally undetermined. How BAG6 contributes to causing disease may be somewhat understood from understanding its primary sequence and the functions of the key domains illustrated in Fig. 3.
Fig. 3.
Schematic representation of BAG6 showing domain boundaries and solved structures. UBL domain (residues 17–92, purple) in complex with E3 ligase RNF126 (residues 1–40, gray) from PDB: 2N9P (Krysztofinska et al., 2016); NLS (residues 1008–1050, orange) and TRC35 (residues 23–305, gray) from PDB: 6AU8 (Mock, Xu, Ye, & Clemons, 2017); BAG domain (blue) in complex with UBL4A (residues 95–147, gray), from PDB: 4X86 (Kuwabara et al., 2015).
20. BAG6 structure: N-Terminal UBL domain
Multiple alignments of the UBL-containing domain, responsible for recruiting ubiquitination machinery, revealed further subdivision of this region into two smaller domains: Domain I and Domain II, both containing designated Evolutionarily Conserved Island (ECI) regions (shown in Fig. 3). Hessa et al. (2011) discovered ECI1 to be the UBL domain which is directly linked to protein quality control, specifically ER substrates. This was experimentally found in in vitro experiments whereby absence of ECI1 failed to successfully ubiquitinate mislocalized proteins. Further experimental evidence showed an E3 ubiquitin ligase needed to interact with ECI1 to permit the degradation of ER substrates (Payapilly & High, 2014).
The binding of hydrophobic stretches of protein onto BAG6 is imperative for substrate discrimination. Domain I contains ECI1 and ECI2, both of which are capable of binding hydrophobic substrates, but cannot necessarily maintain adequate hydrophobic stabilizing interactions. Domain II contains further ECI with UBL sequence similarity, and therefore can potentially bind hydrophobic substrates. Therefore, collectively, the UBL-containing domain is capable of directly recognizing and binding hydrophobic substrates (Kawahara, Minami, & Yokota, 2013). Further structural understanding is required to gain a mechanistic understanding of substrate binding to the UBL domain.
21. BAG6 structure: C-Terminal BAG domain
A highly conserved BAG domain exists at the C-terminal region of BAG6. It is involved in direct interaction with the molecular chaperone family Hsp70/Hsc70 via the ATPase domain. Mechanistically, the interaction between Hsp70 and C-terminal BAG domain commences with Hsp70 in an ADP-bound state possessing a vacant nucleotide binding domain. This allows the binding of the BAG6 domain and Hsp70 via electrostatic and hydrophobic interactions. Thereafter, phosphorylation of ADP to ATP results in the simultaneous release of the BAG domain due to conformational changes to the nucleotide binding region. Regeneration of ADP-bound Hsp70 allows this cycle to continue and supports the recruitment of hydrophobic proteins by acting as a critical checkpoint in determining the fate of polypeptides.
Interestingly, multiple alignment of BAG domains from several species of the BAG family suggested that the BAG domain of BAG6 is atypical. In vivo binding assays have revealed the co-precipitation of the BAG6 and Hsp70, reinforcing that BAG6 is a Hsp70-binding motif. However, differences in the residual interaction between the BAG domain and Hsp70 across the six members of BAG proteins rationalizes why in vivo studies showed reduced affinity of BAG6 to Hsp70 (Thress, Henzel, Shillinglaw, & Kornbluth, 1998). Moreover, due to evolutionary pressure on the BAG domain, it has undergone phylogenetic divergence to accommodate for more complex demands in mammals. This indicates that on a functional and sequence level, the BAG6 C-terminal BAG domain is unique (Kuwabara et al., 2015; Mock et al., 2017).
Although Hsp70 participates in recognizing and binding hydrophobic polypeptides, BAG6 itself can also bind these nascent polypeptides independently. In cytosolic conditions, several chaperones are required to hinder undesired interactions leading to aggregate formation. This suggests that BAG6 exhibits critical chaperoning characteristics, supported by studies which shows preferential binding of hydrophobic Tail-Anchored proteins via the TMD which allows sufficient time frame for the fate of polypeptides to be decided (Slavotinek & Biesecker, 2001). BAG6 behaves as a special transporter which exploits its “holdase” activity to sort aggregation prone substrates depositing them into their desired functional system. Essentially, cofactors of BAG6 complex as well as the interplays between the BAG6 complex and nascent polypeptides determine its destination.
22. BAG6 structure: NLS domain
The significance of BAG6 protein lies in its position as a central hub mediating various cellular processes, involving but not limited to apoptosis, proteostasis and immunoregulation. A bipartite NLS exists in BAG6 adjacent to the N-terminal side of BAG domain in the primary sequence. The NLS signal in BAG6 is masked by TRC35-bound BAG6 promoting BAG6 nuclear translocation to the cytosol, indicative of a functional link between transmembrane protein biosynthesis and the NLS (Kawahara et al., 2013). BAG6 variants lacking the NLS due to alternative splicing have shown preferential localization at the cytosol. Although BAG6 is primarily localized in the cytosol, the NLS domain indicates that its presence in the nucleus is important too, and functions of BAG6 as a quality control site have been displayed in both localities (Mock et al., 2017). However, some cases show that increased nuclear levels of BAG6 can be associated with tumorigenesis, for example, in osteosarcoma tissue (Tsukahara et al., 2009).
Some functions of nuclear BAG6 have been proposed including DNA damage regulation via tempering of post-translational modification of nuclear factors such as histones. BAG6 regulates chromatin structure in testes as well as gene expression via employment of histone modifiers. During genotoxic stress in the testes, BAG6 transactivates tumor suppressor p53 target genes by enhancing p53 acetylation. This is aided by a direct stabilizing interaction between p53 gene and p300 acetyltransferase complex (Kawahara et al., 2013; Lee & Ye, 2013; Luce, Akpawu, Tucunduva, Mason, & Scott, 2016). Immunocytochemical analysis demonstrated that point mutations of the NLS domain led to the absence of BAG6 in the nucleus (Kawahara et al., 2013). This signifies the importance of the conserved NLS domain for BAG6 localization in the nucleus. Moreover, the studies emphasized the importance of regulated levels of nuclear localization of BAG6, which if overexpressed or underexpressed leads to disease.
23. BAG6 is an essential apoptotic regulator of nascent polypeptides
The earliest cellular function associated with BAG6 was its role as an apoptosis regulator. Reaper, an apoptosis-inducer protein in Drosophila, binds its C-terminal 50 amino acids to BAG6 (previously called Scythe) which inhibits apoptosis. This is possible due to Reaper having the ability to bind directly to ribosomes, thus inhibiting protein synthesis. This is the case for several cell systems including HeLa human cancerous cells, indicating possible health conditions that may be caused in humans due to unregulated apoptosis (Mock et al., 2015). In vitro assays revealed that Reaper induces rapid cytochrome c release during mitochondria-mediated apoptosis and therefore is BAG6 dependent (Thress et al., 1998). Mechanistically, Reaper works by disrupting the interaction between BAG6 and UBL4A which leads to BAG6 sequestering pro-apoptotic Reaper. Therefore, BAG6 serves as an anti-apoptotic regulator. An example of unregulated apoptosis causing health defects is in mice, whereby because of in vivo genetic ablation of BAG6, embryonic lethality is caused due to amplified levels of apoptosis and cell proliferation (Desmots, Russell, Lee, Boyd, & McKinnon, 2005). This concludes that functional BAG6 has a direct effect on regulating developmental defects. Although deficiency of BAG6 increases apoptosis which negatively impacts tissues, chemicals which affect calcium flow via ER stress are more resistant against cortical neurons deficient of BAG6 (Desmots et al., 2005; Kawahara et al., 2013; Tsukahara et al., 2009). This deduces the complex roles of BAG6 with the ability to adapt its consequences on its clients requirements.
Moreover, BAG6 is deemed compulsory for the operation of Reaper-induced apoptosis. This is deduced from cases of depleted endogenous BAG6 whereby the caspase activation of Reaper is hindered suggesting the need of functional BAG6 to act as an anti-apoptotic regulator (Thress et al., 1998). Surprisingly, BAG6 suppressed Reaper-induced apoptosis upon overexpression of wild-type BAG6 which is functionally beneficial for cells (Thress et al., 1998).
Despite reports of BAG6 being an apoptotic regulator, conflicting results show otherwise regarding BAG6 proteins role in apoptosis and viability. For that reason, during development, BAG6 is proposed to play ambiguous yet case-dependent roles. Several studies support this hypothesis whereby the absence of the gene encoding BAG6 in mice resulted in neonatal lethality due to extensive abnormal apoptosis.
Conversely, underexpression of BAG6 in mice with ICR background was viable. Strikingly, apoptosis was impaired in ICR mice upon deletion of BAG6 as a result of genotoxic stress. Other independent studies demonstrate that slight underexpression of BAG6 causes mitotic and G1/S phase arrest in mislocalized chromosomes and in mammalian in vitro cells respectively, as well as enhanced apoptotic activity in HCT116 cells, thus contributing negatively to health conditions (Kawahara et al., 2013). This portrays the differential roles of BAG6 in precise cellular contexts as well as its contribution to cells with varying cellular levels.
24. BAG6 involved in the triage system during protein quality control
As has already been discussed, BAG6 serves as a dynamic platform in determining the fate of post-translational polypeptides with exposed hydrophobicity. Importantly, BAG6 associates itself with nascent polypeptides and evidence supports this experimentally as, on co-treatment of eukaryotic cells with protein synthesis inhibitor, polyubiquitinated associated BAG6 proteins disappeared (Minami et al., 2010). There are three common pathways coupled to BAG6 to maintain quality control within the cytoplasm upon recognizing exposed hydrophobic regions of polypeptides.
25. BAG6 coupled degradation of defective nascent polypeptides
When membrane protein insertion fails, BAG6 triages uninserted substrates to the degradation pathway via the proteasome upon being ubiquitinated or attempts are made to correctly refold the protein and for reinsertion in the ER. Furthermore, during degradation deficiency, a ubiquitin-positive aggresome forms and BAG6 along with the hydrophobic substrates deposits into an aggregate (Kawahara et al., 2013).
One of the BAG6 quality control processes is the degradation of defective nascent polypeptides in the cytosol. The breakdown of BAG6 anatomy found the N-terminal UBL domain to be associated with the ubiquitin proteasome system (UPS). BAG6 associates with several protein binding partners when carrying out quality control functions. BAG6 also binds to polyubiquitinated substrates showing that it is interacting multivalently with the proteasome for degradation (Lee & Ye, 2013; Payapilly & High, 2014). This is also the case for Defective Ribosome Products (DRiP) which behave using a similar mechanism.
BAG6 can detect nascent defective polypeptides and subsequently target it to polyubiquitination by E3 ubiquitin ligase. This targets the substrates to the proteasome where it undergoes degradation (Lee & Ye, 2013). It was experimentally deduced that BAG6 heterotrimeric complex is involved in the direct interaction of the proteasome-bound substrate whereby BAG6 plays a key role in the interaction. Therefore, depletion of BAG6 prevented turnover as degradation at the proteasome was inhibited by the absence of BAG6. The turnover of DRiPs is proposed to create MHC class I antigenic peptides, which, in the case of BAG6 depletion, have limited cell surface presentation (Minami et al., 2010). Antigen production results due to degradation of misfolded and mislocalized proteins. The importance of BAG6 here lies in its underexpression causing limited suppression of cell surface presentation in MHC class I which may be a basis of peptide ligands. Alongside this, BAG6 is a possible participant in transmembrane protein assembly, vital for antigen presentation (Kawahara et al., 2013).
Initial studies proposed BAG6 to be a protein associated with the proteasome. However, newer research via MS analysis elucidated extremely high sequence similarity between the 26S proteasome subunits and BAG6 immunoprecipitates as well as BAG6’s collaboration with interferon-γ (IFN-γ)-induced immunoproteasomes, which are typically favored due to their ability to produce peptides satisfactory to MHC class I molecules (Minami et al., 2010). Therefore, in controlling antigen presentation, IFN-γ exposure increased BAG6 expression which can be regulated in health conditions. These results conclude that BAG6 is a functional collaborator of the proteasome.
BAG6 also assists mislocalized protein degradation by BAG6 heterotrimeric complex. Nascent polypeptides existing the ribosomal tunnel are targeted by BAG6 and “sorted” to its appropriate destination. Issues associated with MLPs are that their hydrophobic region is exposed to the aqueous cytosolic environment; thus they are inclined to form aggregates. This poses challenges for cells experiencing ER-stress as membrane integration efficiency is further reduced. Ideally, TA proteins captured by BAG6 are transferred to TRC40 for ER membrane targeting. However, this pathway is not always efficient which can contribute to the build-up of mislocalized proteins in the cytosol. For some mislocalized transmembrane proteins, the TMD is targeted to polyubiquitination by E3 ligase and thus undergoes degradation at the proteasome (Amm, Sommer, & Wolf, 2014). Interestingly, BAG6 may behave in a similar manner to the ER lumen timer which controls the degradation of misfolded protein whereby only in the case of mislocalized proteins will BAG6 triage them to degradation at the proteasome (Lee & Ye, 2013).
A third major evolutionary conserved quality control process is ERAD of hydrophobic proteins which fail to be successfully inserted into the ER lumen or fail to obtain its native conformation. Typically, the efficiency of ER-targeting is relatively low depending on the strength of the ER signal. However, to enhance ERAD efficiency and, in turn, maintain homeostasis of both transmembrane and secretory proteins which are disassembled or misfolded, sophisticated chaperones are used to retro-translocate them to the proteasome for degradation (Payapilly & High, 2014). Sequentially, membrane-associated E3 ubiquitin ligase including gp78 targets retro-translocated proteins for polyubiquitination. An ATPase associated multiprotein complex then extracts the ubiquitinated protein from the membrane and BAG6 is recruited. This suggests preferential chaperoning activity of BAG6 as opposed to the assisting multiprotein complex which shuttles the substrates to the proteasome for degradation. However, whether BAG6 directly interacts with the proteasome or is facilitated by an adaptor remains elusive (Lee & Ye, 2013; Payapilly & High, 2014).
At a structural level, interaction between BAG6 and ERAD de-glycosylated intermediates was observed as well as the association of the BAG6 heterotrimeric complex with retro-translocated membrane targeted complex involving gp78 and p97, as revealed by affinity chromatography (Wang, Whynot, Tung, & Denic, 2011). This implies that BAG6 works as a p97-mediated downstream retro-translocation of ERAD. Additionally, soluble and membrane proteins are stabilized in the case of BAG6 depletion. Notably, some polyubiquitin conjugates remain which can potentially form aggregates, and thus become unaffected by proteasome degradation as a result of its inhibitory effect (Wang, Whynot, et al., 2011). This concludes that the absence of BAG6 prevents its detrimental role in maintaining client ERAD protein solubility and escorting of substrates to the proteasome for degradation.
The heterotrimeric BAG6 complex elevates the quality control of nascent polypeptides, particularly ERAD substrates. This has been observed in studies where underexpression of BAG6 impaired the degradation of ERAD substrate T-cell receptor, TRCα, a single-spanning transmembrane helix. On the other hand, non-ERAD substrates such as cytosolic Ub-V-GFP protein remained unaffected in degradation (Wang, Whynot, et al., 2011). This exemplifies the strong inhibitory effect of absent BAG6 on ERAD degradation. Although BAG6 can independently maintain retro-translocated substrates in a soluble, unfolded state, underexpression of its heterotrimeric complex counterparts TRC35 and UBL4A allowed ERAD substrates to remain stabilized. This is because BAG6 is the main chaperone protein involved in ERAD but requires its counterparts for increased ERAD efficiency (Wang, Whynot, et al., 2011).
26. BAG6 complex—Structural interaction with UBL4A
To successfully achieve protein homeostasis through ER-associated degradation and mislocalized protein pathways, the formation of the BAG6 complex with TRC35 and UBL4A is vital. Notably, the BAG domain of BAG6 is necessary and can bind UBL4A self-sufficiently which plays a pivotal role in the correct assembly of TA proteins (Kuwabara et al., 2015). The precise role of UBL4A is obscure; however it is proposed that it recruits additional chaperone machinery and ubiquitination enzymes through the UBL domains to module protein quality control. Thus, the essential interaction between the two binding partners was explored in depth. GST pull-down analysis on Escherichia coli presented the co-precipitation of both GST-BAG domain and UBL4A, indicating that the domains interact directly. Interestingly, the BAG domain of BAG6 preferentially binds with UBL4A relative to other members of the BAG family, supporting that its domain is distinct from the other canonical BAG family members and direct binding with UBL4A is optimized (Kuwabara et al., 2015).
To explore the key residues involved in the sufficient binding of BAG6 and UBL4a, the interactions between the BAG domain and evolutionarily conserved C-terminal half of UBL4A were closely examined. The secondary structure reveals overlapping of three α-helices from each domain. Across the dimer, several key hydrophobic and electrostatic interactions were observed playing a vital role in its strong binding character.
Characterization of the binding interface by point mutation allowed the most critical binding residues to be identified. Constructed combination of BAG6 mutations highlighted the significance of hydrophobic interactions. On single substitution of a BAG6 residue, no apparent effect was observed on the dimer interaction. However, double and triple substitution abolished the interaction altogether, particularly simultaneous Val-1068 and Leu-1086, establishing these residues as the most critical for the BAG6-UBL4A interaction. Fascinatingly, out of the four interactions illustrated, point mutations on UBL4A His-106 and Phe-107 significantly lowered the binding affinity of the BAG6-UBL4A complex indicating the importance of both hydrophobic and electrostatic interactions in stabilizing the dimer. However, point mutations of UBL4a Val-02 and Tyr-123 had a lesser effect on the binding stabilization as the former residues are located on the linker region which may be critical for complex formation (Kuwabara et al., 2015).
Importantly, the heterotrimeric complex of BAG6 effectively shields long hydrophobic exposed regions on the client proteins, thus preventing aggregates forming. Essentially, undesired interactions with client proteins are avoided before being triaged to the correct destination. Within mislocalized proteins, BAG6 can bind to broader specific hydrophobic stretches including GPI-anchors and signal sequences. An example of the necessary requirement of BAG6 in ERAD is demonstrated by interaction with Derlin2, an ER dislocon protein. BAG6-Derlin2 interaction facilitates the recruitment of BAG6 to the mislocalized protein site as well as retro-translocation of substrates back to the cytosol. Importantly, BAG6 is also essential during the ER exit of substrates and in the prevention of hydrophobic domains forming a blockade (Payapilly & High, 2014; Wang, Whynot, et al., 2011). Therefore, the heterotrimeric complex with BAG6 plays a crucial quality control role by functioning at the interface of protein biosynthesis and degradation. This regulatory mechanism needs the use of special “holdase” enzyme activity which sorts and channels hydrophobic polypeptides to its required destination (Lee & Ye, 2013).
27. BAG6 associations with disease
It is a logical corollary that dysfunctions in the BAG6 system can cause severe cellular abnormalities that may be associated with pathological conditions. Due to the diverse functions of the BAG6 complex in mammals, it is not surprising that deficiency in BAG6 may cause several developmental consequences and human diseases. Several roles of BAG6 in disease have already been identified and will be discussed below. However, it is likely that this review will barely scratch the surface of future disease implications of BAG6 that remain to be discovered and understood.
28. The role of BAG6 in male infertility
Consistent with the view that BAG6 binds to the family of heat shock proteins, detailed studies on the interaction between BAG6 and Hsp2A, a testis-enriched member of the Hsp70 family, have revealed a crucial regulatory role of BAG6 in human spermatozoa (Bromfield, Aitken, & Nixon, 2015). Extensive studies into the primary role of BAG6 localized in human testes have been implicated to protect Hsp2A from polyubiquitination and subsequent degradation, thereby maintaining its stability in germ cells. It has been established that hydrophobic and electrostatic interactions govern the binding between BAG6 and Hsp2A. The release of BAG6 from Hsp2A is promoted by these intermolecular interactions.
Recent studies have revealed Hsp2A as a regulator of zona pellucida-receptor complex whereby in infertile male subjects, this complex formation is perturbed due to a lack of Hsp2A expression due to a knock-on effect from the lack of BAG6. Ultimately, BAG6 plays a role in male infertility by stabilizing Hsp2A by a similar manner of Hsp70-BAG6 interaction mechanism. Therefore, BAG6 may be a crucial candidate in future understanding of male idiopathic infertility due to its suggested underexpression in zona pellucida-spermatozoa binding deficiency (Bromfield et al., 2015).
29. The role of BAG6 in lung cancer
As mentioned earlier, the major known roles of BAG6 in disease are associated with underexpression or overexpression of BAG6; however single nucleotide polymorphisms (SNPs) are also a factor. The genotype and alleles of BAG6 are prognostic for non-small cell lung cancer in Norwegian-Croatian subjects, as implicated by logistic regression analyses. It was observed that the C allele of BAG6 rs3117582 SNP posed a gene-dosage increased risk of lung cancer whereby subjects carrying a single C allele had 1.7 fold increased risk and an even greater 7-fold risk for subjects carrying two C alleles (Etokebe et al., 2015; Rosenberger et al., 2017).
Typically, mutations in the promoter-region of genes have an impact on expression and when the specific rs3117582 SNP is situated in the promoter-region of BAG6, it may perturb BAG6 expression. As a result, normal functions of BAG6 in regulating cellular processes are disturbed. The significantly increased susceptibility to lung cancer with SNPs in the BAG6 gene leads to consideration of these SNPs as potential therapeutic targets (Etokebe et al., 2015; Zhao, Wang, Hu, & Jin, 2014).
30. The role of BAG6 in osteoarthritis
Additionally, the influence of BAG6 expression as a result of the BAG6 rs3117582 SNP located in the promoter region effects the risk of osteoarthritis (Mock et al., 2017). The normal major allele homozygote was associated with reduced risk to osteoarthritis while the major-minor allele heterozygote contributed to the increased susceptibility, irrespective of anatomical site and gender in the Croatian population. Results from the study indicate that variations in alleles and genotype of BAG6 affect the etiology of osteoarthritis due to dysfunctions in BAG6 protein regulatory roles (Wang et al., 2016).
31. The role of BAG6 in autoimmune disease
BAG6 polymorphism is associated with increased incidence of several autoimmune diseases such as Kawasaki syndrome and type I diabetes (Hsieh et al., 2011; Piras et al., 2014). As BAG6 influences numerous substrate proteins essential to immune regulations, it suggests a functional link between BAG6 and autoimmune disorders. This is demonstrated by examination of single-spanning transmembrane protein T-cell immunoglobulin and mucin domain-containing molecule-3 (Tim-3), an inhibitory type I membrane receptor. In cases of tumor-bearing subjects, exhausted CD4 + T-cells show increased expression of Tim-3 which results in negative-regulation of immune functions as the T-cell response is repressed. Consequently, exhausted T-cells diminish effector functions including cytokine secretion and cytotoxicity in response to antigen stimulation. However, BAG6 plays a role in binding the tail of Tim-3 and galectin-9 facilitates the disruption of the binding by mediating phosphorylation at the Tim-3 tail on residues Tyr-256 and Tyr-263. Subsequently, BAG6 is released which permits the transduction of inhibitory signals (Binici et al., 2013). Therefore, when BAG6 is overexpressed, levels of Tim-3 is reduced which results in earlier onset of autoimmune development by promoting helper T-cell response. This is supported experimentally whereby increased levels of Tim-3 and low frequency of IFN-γ-production were observed in BAG6-deficient T-cells. This is suggestive of BAG6’s behavior as a repressor of Tim-3 activity which functions by protecting helper T-cells from galectin-9-mediated death, thereby promoting pro-inflammatory cytokine production and proliferation (Binici & Koch, 2014; Kawahara et al., 2013; Rangachari et al., 2012). Therefore, manipulations to the BAG6/Tim-3 interaction may be a potential molecular target in treatment of several chronic infections and autoimmune disorders.
32. BAG6: future prospects
As outlined in this section, BAG6 is a crucial multi-functional protein contributing to several diseases. It is involved in several unrelated cellular pathways and plays a pivotal role by functioning at the interface of protein synthesis and quality control of nascent polypeptides. This is governed by the unique interaction of BAG6 with the two-opposing cellular machinery, the ribosome and proteasome. As BAG6 is involved in many key cellular processes in healthy mammalian cells, it makes sense that defects in such processes will lead to unwanted cellular functions, leading to disease.
The role of BAG6 in immune response regulation allows the multi-functional protein to behave as a promising diagnostic, more so a prognostic marker protein, whereby this unravels promising new perspectives for conventional therapies, for example, in autoimmune disease. However, to fully understand the functions of BAG6 in such diseases, a range of patient cohorts need to be investigated, not limiting to certain geographical regions to understand the full potential of BAG6 as a target, thus allowing elucidation of specific diseases that may benefit from BAG6 counteractions.
Further exploring BAG6 mechanisms into how it distinguishes and targets different substrates to its correct fate remains unsolved. It has been postulated that hydrophobic interactions between BAG6 and substrates govern substrate recognition. Furthermore, the BAG6 cofactors contribute to recognizing and determining the fate of hydrophobic substrates but the mechanism it follows is yet to be fully understood. This requires full mapping and structural characterization of substrate-BAG6 interactions and upon discovery, manipulations can be made helping combat disease.
A number of answers regarding BAG6’s role in health and disease are yet to be discovered. Although BAG6 binds its substrates with high affinity, it is unknown how BAG6 is able to release individual substrates upon reaching their final destination. Additionally, how BAG6 interacts with accessory factors helps determine the fate of polypeptides. Understanding these questions would allow the function of BAG6 to be exploited and manipulated for treatments in disease.
33. Other members of the BAG6 complex and machinery for TA protein insertion
Understanding the targeting mechanism of the TA proteins is expected to have far reaching conjectures on molecular biology, and much work has been carried out to identify the components of the insertion system (Simpson, Schwappach, Dohlman, & Isaacson, 2010; Yamamoto & Sakisaka, 2012). Initial biochemical analyses between 1995 and 2005 (mostly yeast based studies) suggested that TA proteins were inserted via an energy dependent process, and an ATP requirement was confirmed. The process was defined in yeast systems as the guided entry of TA proteins (GET) pathway (Denic, 2012) or transmembrane domain recognition complex (TRC) pathway in mammalian systems (Johnson, Powis, & High, 2013). The studies made evident that TA protein targeting and insertion can be regulated, particularly in mammalian systems.
The challenges involved in identifying the components in the TRC pathway were likely a result of heterogeneity of insertion assays measuring binding of TA proteins (Stefanovic & Hegde, 2007). Using a new protease protection assay a key component of the pathway essential for membrane insertion was found: the ATPase TRC40 (also known as ASNA1, yeast homolog termed Get3), trailblazing the proximate identification of the other components in further studies using genetic and biochemical approaches.
Get3 forms a complex with Get1 and Get2, and these proteins provide a route for insertion of the TA proteins into the ER membrane by Get3 (TRC40 in mammalian constructs) (Schuldiner et al., 2008). WRB (Tryptophan-rich basic protein) is now thought to be the Get1 mammalian analogue (Vilardi, Lorenz, & Dobberstein, 2011), and CAML (calcium-modulating cyclophilin ligand) the Get2 mammalian analogue (Vilardi et al., 2014; Yamamoto & Sakisaka, 2012). Using the endogenous sensor of yeast cells (unfolded protein response), Jonikas et al. (2009) identified two new components of the TA protein biogenesis machinery: Get4 and Get5, these were expected to form a complex and capture nascent proteins to be delivered to Get3 (Jonikas et al., 2009). The mammalian analogues were discovered by Mariappan et al. (2010) to be TRC35 and UBL4A, respectively, forming a complex with an additional protein BAG6 (Bat3) (Mariappan et al., 2010). In large scale yeast genetic interaction profiles Sgt2 was found as another essential factor in protein targeting by virtue of its strong functional relationship with the GET pathway (Chang et al., 2010). The human homolog is known to be SGTA and has been found to interact with BAG6 (Darby et al., 2014).
Since discovery a complex model of the TA protein membrane insertion has been formulated with the most up-to-date configuration simulated by a fully constructed mammalian system proposed by Shao et al. (2017). They have conglomerated the investigations of the well-defined GET pathway in yeast and the evolutionarily conserved mammalian homologues to suggest the chaperones involved act as a molecular triage system. BAG6, SGTA and TRC40 are the three proteins that can recognize and shield the transmembrane domain (TMD) of a TA protein (Leznicki et al., 2013). The C-terminal 110 residues of BAG6 form a complex with two tightly associated subunits UBL4A and TRC35 (collectively known as the cBag6 complex) (Mock et al., 2015), while the N-terminal part of BAG6 contains a UBL domain (nBag6) (Tanaka et al., 2016). As mentioned earlier, SGTA is the most competitive binder to exposed hydrophobic stretches of proteins, and in the first step the proteins undergo binding and shielding by SGTA, following which the proteins have one of two fates. The first involves the targeting module which consists of SGTA and TRC40 bridged by the cBag6 complex, where the TA protein binds to the TRC40 and is delivered to the ER for insertion. The second involves the quality control module exclusive to mammalian systems comprised of SGTA, nBag6 and a newly defined protein RNF126 (Krysztofinska et al., 2016) where the TA protein binds to nBag6 and is targeted for ubiquitination. These events are depicted in Fig. 1.
This quality control pathway is well-balanced allowing nascent proteins to mature but forcing degradation of poorly folded proteins over time. Accurate triage of degradation versus biosynthesis is essential for maintaining the desired quantity of TA proteins in the membrane, and ensuring the quality of proteins meets the requirements. Upregulation or inhibition of these proteins is postulated to lead to cancer or disease. Important reviews (Denic, Dotsch, & Sinning, 2013; Hegde & Keenan, 2011; Shao & Hegde, 2016; Simpson et al., 2010) have focused much on the GET system and on the known structures and interactions of the protein chaperones.
34. Formation of the Get4/5 and cBag6 complex
The year 2010 was revolutionary to the understanding of much of the GET pathway, and the structure of Get4 and Get5 was determined and their interactions with the other proteins in the pathway better understood. Chang et al. (2010) obtained the first crystal structure of Get4 in a complex with a fragment of Get5 obtained by co-expression in E. coli. Almost simultaneously Bozkurt et al. (2010) published the structure of Get4, and Chartron, Suloway, Zaslaver, and Clemons (2010) further defined the structural elements of the Get4/5 complex. The first structural analyses of Get5 were the crystal structure of just the Get5-N domain in complex with Get4, followed by determination of the C-terminal Get5 (Get5-C) structure by X-ray and SAXS by Chartron et al. (2012). The full Get5 protein structure was only later published in complexes with Get3 and Get4 by Gristick et al. (2014). While the interactions of the mammalian analogues of Get4 and Get5 were well studied in relation to the TA protein insertion, their molecular structures were not elucidated until more recently. The UBL4A crystal structure was elucidated by Kuwabara et al. (2015), and the TRC35 structure was recently published by Mock et al. (2017).
The standalone Get4 structure had significant distortions compared to the structure of Get4 in complex with Get5-N, suggestive of a tightly bound complex of Get4/5. Based on immunoprecipitation results where both Get4 and Get5 were completely immunoprecipitated from yeast cytosol by anti-Get5 this tight Get4/5 complex was verified. It was reported that the full length Get4/5 complex exists as a dimer, mediated by the C-terminal homodimerization domain of Get5, resulting in a heterotetramer. Prior to the structure elucidation, sequencing alignment experiments suggested that UBL4A in higher eukaryotes lacks the Get5-N domain, and TRC35 does not have a β-loop, both of which interact in yeast to form the Get4/5 complex. BAG6 was found to bridge UBL4A and TRC35 and form a similar construct (cBag6). Knockdown experiments using siRNA demonstrated that knocking down UBL4A or TRC35 alone has negligible effect on BAG6 presence; however simultaneous knockdown results in a threefold reduction of BAG6. When BAG6 was knocked down the levels of UBL4A and TRC35 were greatly reduced, but PCR analysis demonstrated that the mRNA levels remain almost constant, and BAG6 is needed to maintain TRC35 and UBL4A at the protein level. This experiment further proved that Bag6 can functionally substitute the Get5-N domain.
35. Structures and binding domains of proteins in the Get4/5 and the cBag6 complexes
Get4 was found to contain 15 α-helices, composed of several helix-turn-helix motifs in an α-2-solenoid fold as well as an antiparallel β-sheet inserted between helices α11 and α12 only allowing packing of α13-α14 to the side leading to an empty helix binding site. The N-terminal domain (NTD) consists of the first seven helices, they do not contain an obvious internal consensus however they do exhibit some curvature. The C-terminal domain (CTD) continues with right handed helical coils, more varied in length. Bar the absent β-hairpin, TRC35 was found by Mock et al. (2017) to have the same overall conserved construct as Get4, with the first 14 α-helices forming the α-solenoid fold comprised of the NTD (RMSD = 1.38 Å) and CTD (RMSD = 3.43 Å). As a result of the absent β-hairpin α11, α13 and α15 are shifted compared to Get4 resulting in the greater RMSD of the CTD. Get4 was found to physically interact with Get3 as well as Get5, the mammalian homolog TRC35 interacts with TRC40 as well as BAG6 (mediator of UBL4A interaction), and the residues of TRC35 at both the TRC40 and BAG6 binding interfaces are well conserved from the yeast homologs. The convex surface of Get4 has conserved regions in the C-domain as well as helices α1-α4, suggesting a usefulness of the convex surface for protein interactions. A cleft between α2 and α4 with conserved positive charges is proposed to be the N-terminal binding site to Get3 and similarly in TRC35 the binding site of TRC40.
The first Get4-Get5 binding interface is between α12 and α13, the cleft formed by empty helix binding site with conserved hydrophobic interactions, in which the Get5-N α1-helix lies (Bozkurt et al., 2010; Chang et al., 2010; Chartron et al., 2010). Similarly α1 and α2 of BAG6 (W1004, V1008 and W1012) bind within the cleft formed by α12 and α13 of TRC35 (V254, V257, F242, L258 and Y262) (Mock et al., 2017). An extended, highly ordered loop 1 following the α1-helix of Get5-N lies along the helices α9, α10, α11 of Get4 which form a well-conserved pronounced hydrophobic cleft, a result of an insertion in the loop connecting α6 and α7 and subsequent deviation of the α7 and α8 from the helical arrangement (Chang et al., 2010). This interface is also conserved in mammalian systems and the BA G6 α3 (L1032, Y1036 and M1040) interacts along the helices α9, α10, α11 of TRC35 (F195 and M271) (Mock et al., 2017). The loop 2 of Get5-N fits into the concavity on the surface of Get4 comprised of helices α8, α9, α10 and α14 via backbone contacts; however the BAG6 extended domain (second loop) wraps around the TRC35 at the concave side of the α-solenoid scaffold (Bozkurt et al., 2010; Chartron et al., 2010).
The binding interface between the C-terminal heterodimerization domains of BAG6 and UBL4A (BAG6-C and UBL4A-C) was structurally solved by Mock et al. (2015) and Kuwabara et al. (2015) verifying that BAG6 mediates the indirect interaction between TRC35 and UBL4A. UBL4A-C domain was found to contain three helices, helices 1 and 2 form the dimer interface with conserved hydrophobic residues, and helix 3 wraps around BAG6-C (Kuwabara et al., 2015). The UBL4A heterodimerization domain is identical to the Get5-C homodimerization domain, and the hydrophobic residues (W179, I182, L186, F190, V200, L204, W208) of the Get5-C core are conserved in UBL4A (W96, I99, L103, F107, V115, L119, Y123) (Mock et al., 2015). The central ubiquitin-like domain of Get5 (Get5-UBL) domain was found to form a complex with Sgt2, and structural information was elucidated by Chartron, Gonzalez, and Clemons (2011) via small angle X-ray scattering, then by Simon, Simpson, Hawthorne, et al. (2013) via NMR. Crystal structures of the Get5-UBL domain were published shortly thereafter by Chartron et al. (2012), Simon, Simpson, Goldstone, et al. (2013) and Tung, Li, Lin, and Hsiao (2013), and consist of an α-helix: two sets of antiparallel β-strands and two 3/10 (η) helices that form a UBL beta-grasp fold. Compared to the solution NMR structure of the UBL domain of UBL4A (UBL4A-UBL; PDB ID: 2DZI), the Get5-UBL domain is very similar with an average RMSD of 1.23 Å, the few differences due to small sequence insertions, and the β-sheet surface elements are conserved between the two proteins (Chartron et al., 2012). Using NMR, Xu et al. (2012) determined that the UBL site on UBL4A is a non-conventional type IIb class, unrecognizable by UBD and CUE. This Get5/UBL4a-UBL domain has been demonstrated to be unique due to a charged polar residue near the binding site, and is clearly distinguished from other UBLs resulting in specific and favored interactions (Xu et al., 2012).
36. Protein-protein interactions of the Get4/5 (cBag6) complex in the GET/TRC pathway
The cBag6 complex in mammals acts as a pre-targeting step for insertion of TA proteins, and it is a crucial part of the TRC pathway mediating the transfer of proteins from SGTA (Sgt2 in yeast) to TRC40 (Get3 in yeast). The interaction of the complex with these two proteins provides insight into how priority and time are encoded within the sorting system for different hydrophobic substrates (Shao et al., 2017).
37. Interaction of Get4/5 (cBag6) complex with Sgt2 (SGTA)
Sgt2 is a small glutamine-rich protein, and like its mammalian homolog, SGTA, it comprises an N-terminal homodimerization domain; a tetratrico-repeat (TPR) domain; and a glutamine-rich C-terminal domain. Early links were found between Sgt2, Get5, Get4 and Get3 in large scale yeast two-hybrid yeast two-hybrid and tandem-affinity assays, as well as mass spectrometry analyses of protein complexes (Gavin et al., 2006; Ito, Chiba, & Yoshida, 2001; Uetz et al., 2000). In a large scale genetic yeast study using small molecules, Get4 and Get5 were found to have highest correlation scores as interactors of Sgt2 based on homozygous co-sensitivity for yeast deletion strains (Hillenmeyer et al., 2008). In vitro immunoprecipitation results showed Sgt2 in varying (almost invariably lower) concentrations compared to the identical amounts of Get4 and Get5, suggestive of a transient association of Sgt2 with the Get4/5 complex, therefore a dynamic stoichiometry (Chang et al., 2010). It was postulated following these studies that Sgt2 mediates the transfer to Get3 via the Get4/5 complex especially as Sgt2 was found to be the first point of contact for TA membrane proteins in yeast following release from the ribosome (Brodsky, 2010). The mammalian homologs SGTA, BAG6, UBL4A, TRC35 and TRC40 were found via proteomic analyses to interact with one another, indicative of similar TA protein transfer between the chaperones (Sowa, Bennett, Gygi, & Harper, 2009).
Liou, Cheng, and Wang (2007) used recombinant protein assays to identify the N-terminal region of Sgt2 (Sgt2-NT) to mediate the physical interaction between Get4/5 and Sgt2. Later structural studies and proteolysis assays indicated that it interacts with Get5-UBL forming a canonical binding interface facilitating the indirect interaction with Get4 (Chang et al., 2010; Chartron et al., 2011). NMR chemical shift experiments indicated that the UBL4A-UBL is sequentially and structurally highly conserved, as is its interaction surface with SGTA's N-terminal domain (SGTA-NT) (Simon, Simpson, Goldstone, et al., 2013). Xu et al. (2012) confirmed the direct interaction with mass spectrometry; co-immunoprecipitation and immunoblotting experiments. The results indicated that UBL4A promotes an interaction between SGTA and cBag6 complex.
The MALLS molecular weight of a complex of Sgt2 and Get5 purified by SEC was consistent with a 1:1 binding ratio, confirmed by SAXS, indicating that the Sgt2 dimer binds to just one Get5 (Chartron et al., 2012). This was confirmed in computer docking, ITC and NMR relaxation experiments (Chartron et al., 2012; Simon, Simpson, Goldstone, et al., 2013). ITC and microscale thermophoresis (MST) were used by Darby et al. (2014) to demonstrate a corresponding 1:1 stoichiometry between the mammalian homologs SGTA and UBL4A. Simon, Simpson, Goldstone, et al. (2013) analyzed the interface between constructs covering Sgt2-NT (residues 1–78) and that make up Get5-UBL (residues 70–152). Following X-ray crystallographic structure identification, reciprocal chemical shift perturbation and site-directed mutagenesis experiments were used to map the binding interactions. A well conserved helix in the Sgt2-NT helical dimer interface with an adjoining negatively charged surface (D28, D31, D38, E42) forms strong electrostatic interactions with the highly positively charged binding patch in the Get5-UBL fragment (K85, K79, K122, K124). The two pairs of antiparallel β-sheets of Get5-UBL provide the main interactions, specifically β1 and β4 and the loops connecting β1-β2 and β3-β4 (Simon, Simpson, Goldstone, et al., 2013). Exposed hydrophobic residues of Sgt2-NT (V35, C39) were also found to be crucial for the binding interface, and interact with compatible residues of Get5-UBL (I81, L120, V125, G143, M147) (Chartron et al., 2012). A salt concentration dependency in the interaction of SGTA-NT with UBL4A-UBL was observed by Leznicki et al. (2013), indicating that the strong electrostatic binding is conserved in mammalian analogues. Darby et al. (2014) biochemically characterized the interaction using NMR perturbation shifts followed by computer docking analysis and confirmed the electrostatic binding. The negatively charged residues of SGTA-NT at the binding interface (D27, E30, E33, E40) are highly conserved, interacting with conserved positively charged residues of UBL4A-UBL (K6, K66, K46, K48). The greatest difference between the interactions of mammalian and yeast complexes is the 45° rotation of the mammalian system due to the shift in the charge distribution of key residues on the surface of each interacting partner.
38. Interaction of Get4/5 (cBag6) complex with Get3 (TRC40)
Get3 binding by a hydrophobic substrate is thought to be the point at which it is committed to the TA protein insertion route. Get3 is an ATPase, it forms a “closed” configuration when bound to ATP (and in some cases ADP); in this form a hydrophobic nucleotide-binding groove is available, thereby increasing its affinity for TA proteins (Mateja et al., 2009). Early analyses demonstrated that the Get4/5 complex functions upstream of Get3 in the GET targeting pathway enabling transfer of TA proteins from the ribosome to Get3, a direct, but transient, physical interaction was expected (Hu, Li, Qian, Denic, & Sha, 2009; Jonikas et al., 2009). Crystal structure analysis confirmed this reaction, and computational docking experiments performed found that the conserved patch of positive residues at the Get4 N-terminal face acts as the binding interface suggestive of complementary electrostatic interactions with Get3 justifying the salt-sensitivity of the interaction observed in size-exclusion chromatography (SEC) (Chang et al., 2010). A 1:1 stoichiometry was confirmed by SAXS, wherein the Get3 exists as a dimer binding to two Get4-N termini of the Get4/5 dimer complex (Chang et al., 2010).
Rome, Rao, Clemons, and Shan (2013) challenged the model of the assumed passive bridging complex Get4/5, and suggested that it drives the transfer of TA proteins to Get3 via an active process. Using ATPase assays they found that binding of Get4/5 to Get3 reduces the ATPase activity of Get3 10-fold, and inhibits Get3 tetramerization indicating that Get4/5 induces tighter ATP binding by Get3, this demonstrates that ATP-bound “closed” Get3 binds with greater affinity than the apo-Get3 to Get4/5 (Rome et al., 2013). Gel filtration chromatography further established this relationship since a complex was only formed between Get4/5 and apo-Get3 with saturating ATP present. SEC, pull-down and ITC experiments carried out by Gristick et al. (2014) confirmed a preference of Get4/5 for the “closed” ATP-bound Get3 form. Get4/5 was verified to be actively involved in TA protein loading rather than just acting as convenient bridge bringing Get3 in closer proximity with the ribosome. It regulates the Get3 protein by induced ATP-hydrolysis delay, stabilizing it in a conformation favored for efficient substrate capture.
Mariappan et al. (2010) found that the Get4/5 mammalian analog, the cBag6 complex, regulates the nucleotide cycle. They observed that absence of cBag6 slows down the activity of TRC40, but in purified systems TRC40 can still operate suggesting that it is not essential, but increases efficiency and fidelity. It has since been proposed that it actively regulates the conformation of TRC40 to facilitate substrate loading, and perhaps as with Get3 preferentially binds open conformations.
In vitro isothermal titration calorimetry experiments (ITC) with purified wild type and mutant Get4/5-N and Get3 components together with the crystal structures of the Get3-Get4/5 complex found two functionally distinct binding interfaces between Get3 and Get4 (Chang et al., 2010; Chartron et al., 2010; Gristick et al., 2014). The first is the anchoring interface responsible for binding specificity, and it involves α2 (Y30, R37, R42) of Get4 forming hydrophobic and electrostatic contacts within the groove created by α10 and α11 of Get 3 (F246, Y250, E253, Q257, E258, E304, D308). α1 of Get4 containing conserved charged residues (K15, R19) is tilted toward α10 of Get3. The second interface is responsible for ATPase regulation, located on the opposite monomer, and involves conserved complementary charged residue interactions, and the C-terminal end of α4 (D74, E1) of Get4 lies in the post-α3 loop (K69, K72, R75) of Get3. The regulatory interface was found by mutating residues on the expected regulatory surface: K69D on Get3 and D74K on Get4; Mock et al. (2017) demonstrated that the corresponding regulatory mutations: K86D in Trc40 and D84K in TRC35 resulted in a similar response observing a loss if ATPase inhibition and reduction of TA insertion. Gristick et al. (2014) refined the model for Get3-Get4/5 complex formation, using pre-steady state kinetics. They found that an intermediate is formed by initial electrostatic interactions with a single Get4/5 heterotetramer bound to one monomer of a Get3 dimer; this then rearranges in a second step to a stable final complex dominated by hydrophobic interactions. Upon binding of the TA protein ATP hydrolysis is promoted and Get3 is released from the Get4/5 complex. Mock et al. demonstrate that as expected when histidine tagged SGTA (hSGTA) was incubated with TRC40 in the presence of the cBag6 complex a resultant ATP-dependent increase in the rate of transfer of a TA protein was observed compared to its absence (Mock et al., 2015). In yeast the increased rate in transfer is not just a result of proximity, rather a regulatory role carried out by the Get4/5 complex; Mock et al. (2015) have illustrated that the cBag6 complex performs a similar regulatory role to the Get4/5 complex as originally postulated.
39. Protein interactions in the TRC(GET) pathway
In yeast systems the Get4/5 complex is associated with the ribosome via the interaction of Get5 with Sgt2, and Get4/5 interacts with Get3 targeting protein by the Get4. Get4/5 has been indisputably proven to use a dual mechanism to aid TA protein insertion, a first passive mode involves binding Get3 and tethering it near TA proteins held by Sgt2, acting simply as a bridge. The second mechanism involves the assembly of the Get3 membrane-targeting complex, and binding of Get3 to ATP shifts the protein to the closed conformation, the only form recognized by the Get4/5 heterotetramer. Get4/5 binding promotes inhibition of ATPase activity, enabling the Get3 in the tetrameric complex to bind a TA substrate transferred from Sgt2 (Wang, Brown, Mak, Zhuang, & Denic, 2010).
This has been related to mammalian systems as the TRC pathway, the picture that has emerged is of initial hydrophobic protein capture at the ribosome, then assembly into highly dynamic sorting complex containing many potential binding partners. The pathway protects against high levels of aggregation, and it allows a rapid and private bridging mechanism of transfer from SGTA to TRC40 using the Get4/5 complex equivalent cBag6 (Wang et al., 2010). These specific interactions: of SGTA with UBL4A and of TRC40 with TRC35 confirm the role of the cBag6 complex as an active tethering device, it brings TRC40 into proximity with its substrates and activates it for TMD recognition serving as more than just an adapter function or bridge (Hegde & Keenan, 2011). If any proteins in Get4/5 or cBag6 complexes are absent, then bridging is impaired and so is the capture of the protein by TRC40/Get3. It is clear therefore that the protein chaperones involved in bridging are crucial for delivery of TA proteins, and their up/downregulation will likely affect subsequent processes and lead to disease.
40. Disease related to this quality control system
Hu, Potthoff, Hollenberg, and Ramezani-Rad (2006) demonstrated that cells lacking Get4 or Get5 show identical phenotypes as expected in co-fitness chemical genomic assays, deletion of Get4 and Get5 showed reduction in mating efficiency in yeast cells. Get4 and Get5 were found to be closely related in a physical and functional sense, allowing the transfer of TA proteins from Sgt2 at the ribosome to the targeting Get3 (Chang et al., 2010). Mutagenesis experiments suggest that loss of regulation of Get3 by Get4 leads to growth defects in vivo, and knock out yeast strains of Get4 and Get5 show defects in TA protein biogenesis (Bozkurt et al., 2010; Gristick et al., 2014).
Following these observed phenotypes in yeast Get4 and Get5 deletion assays it appeared likely that the deletion of UBL4A and TRC35 which are closely related in sequence and function will have similar effects in mammalian constructs. As yet, there is no published report as to the effects of deletion of these crucial bridging proteins in relation to the delivery of TA proteins, and the consequences of reduced delivery. However, as will be discussed at length below, the BAG6 complex is found to be a multiprotein “holdase” complex having crucial other roles in cells; in particular the protein quality control processes that are the subject of this chapter. The holdase activity has been found to play a role in retro-translocation of ERAD (endoplasmic reticulum assisted degradation) substrates to the proteasome; mediation of DNA damage response signaling (DDR); degradation of mislocalized membrane proteins, as well as the above discussed chaperoning of nascent TA proteins to the ER. The effect of TRC35 and UBL4A in the role of some of these activities has been studied and it is likely the observations can be correlated in future experiments to their role in TA protein membrane insertion.
In the study by Wang, Song, Brancati, and Segatori (2011) to understand ERAD and how misfolded proteins are directed for proteasomal degradation, the BAG6 complex was found to act as a “holdase.” They experimentally identified cBag6 complex as a key mediator in the chaperoning step in ERAD. The cBag6 complex maintains the solubility of retro-translocated polypeptides, facilitating movement of the misfolded proteins across the ER membrane for targeting to the cytosolic proteasome before aggregation. This action promotes cell survival under stress conditions. BAG6 is a nuclear protein, but was found to be present in the cytosol and in an ER-containing membrane fraction. For engagement in ERAD there must be a sufficient presence of BAG6 in cytosol; TRC35 maintains a cytosolic pool of BAG6 (Xu et al., 2012). TRC35 is also involved in the holdase activity of BAG6 in the cell (Mock et al., 2017). UBL4A function is still unclear. Although knockout experiments of UBL4A greatly reduced ERAD, in TA protein transfer UBL4A regulates substrate handoff to TRC40, so it may have a similar function in substrate transfer to the proteasome.
Krenciute et al. (2013) demonstrated that cBag6 complex mediates DNA damage response (DDR) signaling and damage-induced cell death. All three components of the cBag6 complex are essential for DDR signaling since they regulate the recruitment of BRCA1 (breast cancer protein) to sites of DNA damage, and depletion of the complex compromises the role of BRCA1 in maintaining genome integrity. Knockdown experiments using siRNA demonstrated that knocking down UBL4A or TRC40 alone has negligible effect on Bag6 presence and therefore cell death, knockdown of both simultaneously results in a threefold reduction of bag6 and cells are more resistant to death. UBL4A knockdown largely reduces BAG6 phosphorylation, while TRC40 had no obvious effect, suggesting that UBL4A may play a more direct role in DDR (Krenciute et al., 2013).
These effects indicate that cBag6 complex regulates cellular protein quality control in the cytoplasm as well as the nucleus by targeting multiple substrates for a variety of cellular functions. The cBag6 complex plays a similar “holdase” role in all its seemingly unrelated functions, by shielding hydrophobic or transmembrane domain (TMD) sequences in a variety of substrates, thereby maintaining solubility and preventing aggregation. The model proposed by Krenciute et al. (2013) suggests that UBL4A and TRC35 can interact independently of one another with BAG6, and both can act individually as a stabilizing partner; therefore only knockdown of both will lead to total resistance to cell death. It is interesting to note that in ERAD TRC35 plays the major role while in DDR UBL4A plays the major role. This may be linked to their roles in the GET/TRC pathways in which specifically Get4/TRC35 enables Get3/TRC40 binding, and Get5/UBL4A enables Sgt2/SGTA binding.
41. Disease seemingly unrelated to the chaperone system
UBL4A deficiency has been found to play roles in disease beyond the cBag6 complex. It was found by Zhao et al. (2015) to be critical for insulin-induced plasma membrane translocation, it interacts via Arp2/3 promoting Arp2/3 mediated actin branching, and therefore it was predicted that it regulates other cellular activities related to actin cytoskeletal reorganization dependent on Arp2/3. Recently new results published by Zhao et al. (2017) found that indeed UBL4A is crucial for other cellular activities that depend on Arp2/3, and found it critical for migration of fibroblasts and macrophages. Mice studies found that neutrophils lacking UBL4A showed significant directionality defects during chemotaxis, and loss of UBL4A delayed wound closure by fibroblasts.
STAT3 activation occurs via phosphorylation of tyrosine reside Y705; its activated form is widely accepted as a hallmark in a wide range of cancers such as leukemia or solid tumors. Failure of the balance of STAT3 activation results in tumorigenesis. Wang et al. (2014) revealed that UBL4A bridges an association between TC45 and STAT3, mediating efficient phosphorylation by TC45, and blocking p-STAT3-dependent cancer cell growth. The authors found that tumor growth rate and tumor size were both greatly reduced in B16 murine melanoma cells in vivo when transfected with UBL4A, and it does so by targeting p-STAT3. The results suggest that UBL4A may function as a tumor suppressor, further implied when the deletion of UBL4A facilitated MEF cell proliferation, potentially providing a new strategy for tumor therapy.
Liang et al. (2018) recently identified UBL4A as being a novel regulator in bone development. GdX is the UBL4A gene; GdX knockout mice indicate a critical role of UBL4A in skeletal biology; at the embryonic stage they have decreased bone mineral density, and later demonstrate a blended spine and reduced growth. UBL4A mutations may be a potential cause for skeletal dysplasias and other bone disorders via repressed osteoblast formation and increased chrondrocyte differentiation; however this is yet to be proven in human systems.
42. Conclusion
This chapter has described some of the core machinery involved in quality control of hydrophobic proteins exposed to the aqueous cytoplasm of cells. This field is still rapidly evolving and there are many outstanding questions, in particular with regard to understanding the roles of this machinery in disease states. It is likely that one or more of the proteins discussed here is already under investigation as a drug target and that many of the interaction partners for these proteins have yet to be identified. In the future these ambiguities will become clearer but for now, we have outlined the currently suggested connections between SGTA, BAG6, UBL4A and TRC35 and a range of diseases.
References
- Amm I., Sommer T., Wolf D.H. Protein quality control and elimination of protein waste: The role of the ubiquitin-proteasome system. Biochimica et Biophysica Acta. 2014;1843(1):182–196. doi: 10.1016/j.bbamcr.2013.06.031. [DOI] [PubMed] [Google Scholar]
- Ast T., Cohen G., Schuldiner M. A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell. 2013;152(5):1134–1145. doi: 10.1016/j.cell.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Azziz R., Woods K.S., Reyna R., Key T.J., Knochenhauer E.S., Yildiz B.O. The prevalence and features of the polycystic ovary syndrome in an unselected population. The Journal of Clinical Endocrinology and Metabolism. 2004;89(6):2745–2749. doi: 10.1210/jc.2003-032046. [DOI] [PubMed] [Google Scholar]
- Banerji J., Sands J., Strominger J.L., Spies T. A gene pair from the human major histocompatibility complex encodes large proline-rich proteins with multiple repeated motifs and a single ubiquitin-like domain. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(6):2374–2378. doi: 10.1073/pnas.87.6.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binici J., Hartmann J., Herrmann J., Schreiber C., Beyer S., Guler G. A soluble fragment of the tumor antigen BCL2-associated athanogene 6 (BAG-6) is essential and sufficient for inhibition of NKp30 receptor-dependent cytotoxicity of natural killer cells. The Journal of Biological Chemistry. 2013;288(48):34295–34303. doi: 10.1074/jbc.M113.483602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binici J., Koch J. BAG-6, a jack of all trades in health and disease. Cellular and Molecular Life Sciences. 2014;71(10):1829–1837. doi: 10.1007/s00018-013-1522-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N., Fasana E. Targeting pathways of C-tail-anchored proteins. Biochimica et Biophysica Acta. 2011;1808(3):937–946. doi: 10.1016/j.bbamem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Bozkurt G., Wild K., Amlacher S., Hurt E., Dobberstein B., Sinning I. The structure of Get4 reveals an alpha-solenoid fold adapted for multiple interactions in tail-anchored protein biogenesis. FEBS Letters. 2010;584(8):1509–1514. doi: 10.1016/j.febslet.2010.02.070. [DOI] [PubMed] [Google Scholar]
- Brodsky J.L. The special delivery of a tail-anchored protein: Why it pays to use a dedicated courier. Molecular Cell. 2010;40(1):5–7. doi: 10.1016/j.molcel.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromfield E., Aitken R.J., Nixon B. Novel characterization of the HSPA2-stabilizing protein BAG6 in human spermatozoa. Molecular Human Reproduction. 2015;21(10):755–769. doi: 10.1093/molehr/gav041. [DOI] [PubMed] [Google Scholar]
- Brooke G.N., Bevan C.L. The role of androgen receptor mutations in prostate cancer progression. Current Genomics. 2009;10(1):18–25. doi: 10.2174/138920209787581307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan G., Ricciardelli C., Harris J.M., Prescott J., Yu Z.C., Jia L. Control of androgen receptor signaling in prostate cancer by the cochaperone small glutamine rich tetratricopeptide repeat containing protein alpha. Cancer Research. 2007;67(20):10087–10096. doi: 10.1158/0008-5472.CAN-07-1646. [DOI] [PubMed] [Google Scholar]
- Buchberger A., Bukau B., Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: Brothers in arms. Molecular Cell. 2010;40(2):238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Bullock M. FOXO factors and breast cancer: Outfoxing endocrine resistance. Endocrine-Related Cancer. 2016;23(2):R113–R130. doi: 10.1530/ERC-15-0461. [DOI] [PubMed] [Google Scholar]
- Cattrini C., Zanardi E., Vallome G., Cavo A., Cerbone L., Di Meglio A. Targeting androgen-independent pathways: New chances for patients with prostate cancer? Critical Reviews in Oncology/Hematology. 2017;118:42–53. doi: 10.1016/j.critrevonc.2017.08.009. [DOI] [PubMed] [Google Scholar]
- Chakrabarti O., Hegde R.S. Functional depletion of mahogunin by cytosolically exposed prion protein contributes to neurodegeneration. Cell. 2009;137(6):1136–1147. doi: 10.1016/j.cell.2009.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.W., Chuang Y.C., Ho Y.C., Cheng M.Y., Sun Y.J., Hsiao C.D. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. The Journal of Biological Chemistry. 2010;285(13):9962–9970. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron J.W., Gonzalez G.M., Clemons W.M., Jr. A structural model of the Sgt2 protein and its interactions with chaperones and the Get4/Get5 complex. The Journal of Biological Chemistry. 2011;286(39):34325–34334. doi: 10.1074/jbc.M111.277798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron J.W., Suloway C.J., Zaslaver M., Clemons W.M., Jr. Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(27):12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron J.W., VanderVelde D.G., Clemons W.M., Jr. Structures of the Sgt2/SGTA dimerization domain with the Get5/UBL4A UBL domain reveal an interaction that forms a conserved dynamic interface. Cell Reports. 2012;2(6):1620–1632. doi: 10.1016/j.celrep.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D., Raeburn C., Sui X., Hatters D.M. Protein aggregation in cell biology: An aggregomics perspective of health and disease. Seminars in Cell & Developmental Biology. 2018 doi: 10.1016/j.semcdb.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Cross B.C., Sinning I., Luirink J., High S. Delivering proteins for export from the cytosol. Nature Reviews. Molecular Cell Biology. 2009;10(4):255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- Cutress M.L., Whitaker H.C., Mills I.G., Stewart M., Neal D.E. Structural basis for the nuclear import of the human androgen receptor. Journal of Cell Science. 2008;121(Pt. 7):957–968. doi: 10.1242/jcs.022103. [DOI] [PubMed] [Google Scholar]
- Cziepluch C., Kordes E., Poirey R., Grewenig A., Rommelaere J., Jauniaux J.C. Identification of a novel cellular TPR-containing protein, SGT, that interacts with the nonstructural protein NS1 of parvovirus H-1. Journal of Virology. 1998;72(5):4149–4156. doi: 10.1128/jvi.72.5.4149-4156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby J.F., Krysztofinska E.M., Simpson P.J., Simon A.C., Leznicki P., Sriskandarajah N. Solution structure of the SGTA dimerisation domain and investigation of its interactions with the ubiquitin-like domains of BAG6 and UBL4A. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0113281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V. A portrait of the GET pathway as a surprisingly complicated young man. Trends in Biochemical Sciences. 2012;37(10):411–417. doi: 10.1016/j.tibs.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V., Dotsch V., Sinning I. Endoplasmic reticulum targeting and insertion of tail-anchored membrane proteins by the GET pathway. Cold Spring Harbor Perspectives in Biology. 2013;5(8):a013334. doi: 10.1101/cshperspect.a013334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmots F., Russell H.R., Lee Y., Boyd K., McKinnon P.J. The reaper-binding protein scythe modulates apoptosis and proliferation during mammalian development. Molecular and Cellular Biology. 2005;25(23):10329–10337. doi: 10.1128/MCB.25.23.10329-10337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupzyk A., Williams J.M., Bagchi P., Inoue T., Tsai B. SGTA-dependent regulation of Hsc70 promotes cytosol entry of simian virus 40 from the endoplasmic reticulum. Journal of Virology. 2017;91(12) doi: 10.1128/JVI.00232-17. e00232-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S., Tan Y.J. Structural and functional characterization of human SGT and its interaction with Vpu of the human immunodeficiency virus type 1. Biochemistry. 2008;47(38):10123–10131. doi: 10.1021/bi800758a. [DOI] [PubMed] [Google Scholar]
- Echeverria P.C., Picard D. Molecular chaperones, essential partners of steroid hormone receptors for activity and mobility. Biochimica et Biophysica Acta. 2010;1803(6):641–649. doi: 10.1016/j.bbamcr.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Etokebe G.E., Zienolddiny S., Kupanovac Z., Enersen M., Balen S., Flego V. Association of the FAM46A gene VNTRs and BAG6 rs3117582 SNP with non small cell lung cancer (NSCLC) in Croatian and Norwegian populations. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0122651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Fernandez M.R., Valpuesta J.M. Hsp70 chaperone: A master player in protein homeostasis. F1000Research. 2018;7:1497. doi: 10.12688/f1000research.15528.1. [version 1; referees: 3 approved] (F1000 Faculty Rev) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding B.C., Gunalan V., Tan T.H., Chou C.F., Shen S., Khan S. Severe acute respiratory syndrome coronavirus protein 7a interacts with hSGT. Biochemical and Biophysical Research Communications. 2006;343(4):1201–1208. doi: 10.1016/j.bbrc.2006.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440(7084):631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Goodarzi M.O., Xu N., Cui J., Guo X., Chen Y.I., Azziz R. Small glutamine-rich tetratricopeptide repeat-containing protein alpha (SGTA), a candidate gene for polycystic ovary syndrome. Human Reproduction. 2008;23(5):1214–1219. doi: 10.1093/humrep/den065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gristick H.B., Rao M., Chartron J.W., Rome M.E., Shan S.O., Clemons W.M., Jr. Crystal structure of ATP-bound Get3-Get4-Get5 complex reveals regulation of Get3 by Get4. Nature Structural & Molecular Biology. 2014;21:437–442. doi: 10.1038/nsmb.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R.Y., Sommer T. Finding the will and the way of ERAD substrate retrotranslocation. Current Opinion in Cell Biology. 2012;24(4):460–466. doi: 10.1016/j.ceb.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nature Structural & Molecular Biology. 2009;16(6):574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Hegde R.S., Keenan R.J. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nature Reviews. Molecular Cell Biology. 2011;12(12):787–798. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa T., Sharma A., Mariappan M., Eshleman H.D., Gutierrez E., Hegde R.S. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature. 2011;475(7356):394–397. doi: 10.1038/nature10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer M.E., Fung E., Wildenhain J., Pierce S.E., Hoon S., Lee W. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science. 2008;320(5874):362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y.Y., Chang C.C., Hsu C.M., Chen S.Y., Lin W.H., Tsai F.J. Major histocompatibility complex class I chain-related gene polymorphisms: Associated with susceptibility to Kawasaki disease and coronary artery aneurysms. Genetic Testing and Molecular Biomarkers. 2011;15(11):755–763. doi: 10.1089/gtmb.2011.0001. [DOI] [PubMed] [Google Scholar]
- Hu J., Li J., Qian X., Denic V., Sha B. The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS One. 2009;4(11) doi: 10.1371/journal.pone.0008061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Potthoff B., Hollenberg C.P., Ramezani-Rad M. Mdy2, a ubiquitin-like (UBL)-domain protein, is required for efficient mating in Saccharomyces cerevisiae. Journal of Cell Science. 2006;119(Pt. 2):326–338. doi: 10.1242/jcs.02754. [DOI] [PubMed] [Google Scholar]
- Ito T., Chiba T., Yoshida M. Exploring the protein interactome using comprehensive two-hybrid projects. Trends in Biotechnology. 2001;19(10 Suppl):S23–S27. doi: 10.1016/S0167-7799(01)01790-5. [DOI] [PubMed] [Google Scholar]
- Ji X.D., Li G., Feng Y.X., Zhao J.S., Li J.J., Sun Z.J. EphB3 is overexpressed in non-small-cell lung cancer and promotes tumor metastasis by enhancing cell survival and migration. Cancer Research. 2011;71(3):1156–1166. doi: 10.1158/0008-5472.CAN-10-0717. [DOI] [PubMed] [Google Scholar]
- Johnson N., Powis K., High S. Post-translational translocation into the endoplasmic reticulum. Biochimica et Biophysica Acta. 2013;1833(11):2403–2409. doi: 10.1016/j.bbamcr.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Jonikas M.C., Collins S.R., Denic V., Oh E., Quan E.M., Schmid V. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323(5922):1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbfleisch T., Cambon A., Wattenberg B.W. A bioinformatics approach to identifying tail-anchored proteins in the human genome. Traffic. 2007;8(12):1687–1694. doi: 10.1111/j.1600-0854.2007.00661.x. [DOI] [PubMed] [Google Scholar]
- Kato Y., Ochiai K., Kawakami S., Nakao N., Azakami D., Bonkobara M. Canine REIC/Dkk-3 interacts with SGTA and restores androgen receptor signalling in androgen-independent prostate cancer cell lines. BMC Veterinary Research. 2017;13(1):170. doi: 10.1186/s12917-017-1094-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara H., Minami R., Yokota N. BAG6/BAT3: Emerging roles in quality control for nascent polypeptides. Journal of Biochemistry. 2013;153(2):147–160. doi: 10.1093/jb/mvs149. [DOI] [PubMed] [Google Scholar]
- Krenciute G., Liu S., Yucer N., Shi Y., Ortiz P., Liu Q. Nuclear BAG6-UBL4A-GET4 complex mediates DNA damage signaling and cell death. The Journal of Biological Chemistry. 2013;288(28):20547–20557. doi: 10.1074/jbc.M112.443416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysztofinska E.M., Evans N.J., Thapaliya A., Murray J.W., Morgan R.M.L., Martinez-Lumbreras S. Structure and interactions of the TPR domain of Sgt2 with yeast chaperones and Ybr137wp. Frontiers in Molecular Biosciences. 2017;4:68. doi: 10.3389/fmolb.2017.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysztofinska E.M., Martinez-Lumbreras S., Thapaliya A., Evans N.J., High S., Isaacson R.L. Structural and functional insights into the E3 ligase, RNF126. Scientific Reports. 2016;6 doi: 10.1038/srep26433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara N., Minami R., Yokota N., Matsumoto H., Senda T., Kawahara H. Structure of a BAG6 (Bcl-2-associated athanogene 6)-UBL4a (ubiquitin-like protein 4a) complex reveals a novel binding interface that functions in tail-anchored protein biogenesis. The Journal of Biological Chemistry. 2015;290(15):9387–9398. doi: 10.1074/jbc.M114.631804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.G., Ye Y. Bag6/Bat3/Scythe: A novel chaperone activity with diverse regulatory functions in protein biogenesis and degradation. BioEssays. 2013;35(4):377–385. doi: 10.1002/bies.201200159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P., High S. SGTA antagonizes BAG6-mediated protein triage. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(47):19214–19219. doi: 10.1073/pnas.1209997109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P., Korac-Prlic J., Kliza K., Husnjak K., Nyathi Y., Dikic I. Binding of SGTA to Rpn13 selectively modulates protein quality control. Journal of Cell Science. 2015;128(17):3187–3196. doi: 10.1242/jcs.165209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P., Roebuck Q.P., Wunderley L., Clancy A., Krysztofinska E.M., Isaacson R.L. The association of BAG6 with SGTA and tail-anchored proteins. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0059590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Li J., Fu Y., Ren F., Xu J., Zhou M. GdX/UBL4A null mice exhibit mild kyphosis and scoliosis accompanied by dysregulation of osteoblastogenesis and chondrogenesis. Cell Biochemistry and Function. 2018;36(3):129–136. doi: 10.1002/cbf.3324. [DOI] [PubMed] [Google Scholar]
- Liou S.T., Cheng M.Y., Wang C. SGT2 and MDY2 interact with molecular chaperone YDJ1 in Saccharomyces cerevisiae. Cell Stress & Chaperones. 2007;12(1):59–70. doi: 10.1379/CSC-220R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou S.T., Wang C. Small glutamine-rich tetratricopeptide repeat-containing protein is composed of three structural units with distinct functions. Archives of Biochemistry and Biophysics. 2005;435(2):253–263. doi: 10.1016/j.abb.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Luce M.J., Akpawu A.A., Tucunduva D.C., Mason S., Scott M.S. Extent of pre-translational regulation for the control of nucleocytoplasmic protein localization. BMC Genomics. 2016;17:472. doi: 10.1186/s12864-016-2854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan M., Li X., Stefanovic S., Sharma A., Mateja A., Keenan R.J. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466(7310):1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lumbreras S., Alfano C., Evans N.J., Collins K.M., Flanagan K.A., Atkinson R.A. Structural and functional insights into Bacillus subtilis sigma factor inhibitor, CsfB. Structure. 2018;26(4):640–648.e5. doi: 10.1016/j.str.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja A., Szlachcic A., Downing M.E., Dobosz M., Mariappan M., Hegde R.S. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461(7262):361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami R., Hayakawa A., Kagawa H., Yanagi Y., Yokosawa H., Kawahara H. BAG-6 is essential for selective elimination of defective proteasomal substrates. The Journal of Cell Biology. 2010;190(4):637–650. doi: 10.1083/jcb.200908092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock J.Y., Chartron J.W., Zaslaver M., Xu Y., Ye Y., Clemons W.M., Jr. Bag6 complex contains a minimal tail-anchor-targeting module and a mock BAG domain. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(1):106–111. doi: 10.1073/pnas.1402745112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock J.Y., Xu Y., Ye Y., Clemons W.M., Jr. Structural basis for regulation of the nucleo-cytoplasmic distribution of Bag6 by TRC35. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(44):11679–11684. doi: 10.1073/pnas.1702940114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.Q., Chenon M., Vilela F., Velours C., Aumont-Nicaise M., Andreani J. Structural plasticity of the N-terminal capping helix of the TPR domain of kinesin light chain. PLoS One. 2017;12(10) doi: 10.1371/journal.pone.0186354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A., Garcia Y.A., Zierer B., Patwardhan C., Gutierrez O., Hildenbrand Z. The cochaperone SGTA (small glutamine-rich tetratricopeptide repeat-containing protein alpha) demonstrates regulatory specificity for the androgen, glucocorticoid, and progesterone receptors. The Journal of Biological Chemistry. 2014;289(22):15297–15308. doi: 10.1074/jbc.M113.535229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payapilly A., High S. BAG6 regulates the quality control of a polytopic ERAD substrate. Journal of Cell Science. 2014;127(Pt. 13):2898–2909. doi: 10.1242/jcs.145565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philp L.K., Butler M.S., Hickey T.E., Butler L.M., Tilley W.D., Day T.K. SGTA: A new player in the molecular co-chaperone game. Hormones & Cancer. 2013;4(6):343–357. doi: 10.1007/s12672-013-0151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philp L.K., Day T.K., Butler M.S., Laven-Law G., Jindal S., Hickey T.E. Small glutamine-rich tetratricopeptide repeat-containing protein alpha (SGTA) ablation limits offspring viability and growth in mice. Scientific Reports. 2016;6:28950. doi: 10.1038/srep28950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras I.S., Angius A., Andreani M., Testi M., Lucarelli G., Floris M. BAT2 and BAT3 polymorphisms as novel genetic risk factors for rejection after HLA-related SCT. Bone Marrow Transplantation. 2014;49(11):1400–1404. doi: 10.1038/bmt.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabu C., Schmid V., Schwappach B., High S. Biogenesis of tail-anchored proteins: The beginning for the end? Journal of Cell Science. 2009;122(Pt. 20):3605–3612. doi: 10.1242/jcs.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangachari M., Zhu C., Sakuishi K., Xiao S., Karman J., Chen A. Bat3 promotes T cell responses and autoimmunity by repressing Tim-3-mediated cell death and exhaustion. Nature Medicine. 2012;18(9):1394–1400. doi: 10.1038/nm.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J.D., Thapaliya A., Martinez-Lumbreras S., Krysztofinska E.M., Isaacson R.L. Structural and functional insights into small, glutamine-rich, tetratricopeptide repeat protein alpha. Frontiers in Molecular Biosciences. 2015;2:71. doi: 10.3389/fmolb.2015.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni M.C., Gutierrez E., Hegde R.S. Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Molecular Cell. 2014;55(2):227–237. doi: 10.1016/j.molcel.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rome M.E., Rao M., Clemons W.M., Shan S.O. Precise timing of ATPase activation drives targeting of tail-anchored proteins. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(19):7666–7671. doi: 10.1073/pnas.1222054110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger A., Sohns M., Friedrichs S., Hung R.J., Fehringer G., McLaughlin J. Gene-set meta-analysis of lung cancer identifies pathway related to systemic lupus erythematosus. PLoS One. 2017;12(3) doi: 10.1371/journal.pone.0173339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nature Medicine. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Metz J., Schmid V., Denic V., Rakwalska M., Schmitt H.D. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134(4):634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S., Hegde R.S. Target selection during protein quality control. Trends in Biochemical Sciences. 2016;41(2):124–137. doi: 10.1016/j.tibs.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Shao S., Rodrigo-Brenni M.C., Kivlen M.H., Hegde R.S. Mechanistic basis for a molecular triage reaction. Science. 2017;355(6322):298–302. doi: 10.1126/science.aah6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A.C., Simpson P.J., Goldstone R.M., Krysztofinska E.M., Murray J.W., High S. Structure of the Sgt2/Get5 complex provides insights into GET-mediated targeting of tail-anchored membrane proteins. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(4):1327–1332. doi: 10.1073/pnas.1207518110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A.C., Simpson P.J., Hawthorne W., Hale L.R., Goldstone R.M., Isaacson R.L. 1H, 13C and 15N assignments of Sgt2 N-terminal dimerisation domain and its binding partner, Get5 ubiquitin-like domain. Biomolecular NMR Assignments. 2013;7(2):271–274. doi: 10.1007/s12104-012-9425-7. [DOI] [PubMed] [Google Scholar]
- Simpson P.J., Schwappach B., Dohlman H.G., Isaacson R.L. Structures of Get3, Get4, and Get5 provide new models for TA membrane protein targeting. Structure. 2010;18(8):897–902. doi: 10.1016/j.str.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek A.M., Biesecker L.G. Unfolding the role of chaperones and chaperonins in human disease. Trends in Genetics. 2001;17(9):528–535. doi: 10.1016/s0168-9525(01)02413-1. [DOI] [PubMed] [Google Scholar]
- Sowa M.E., Bennett E.J., Gygi S.P., Harper J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S., Hegde R.S. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128(6):1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Talukdar F.R., di Pietro M., Secrier M., Moehler M., Goepfert K., Lima S.S.C. Molecular landscape of esophageal cancer: Implications for early detection and personalized therapy. Annals of the New York Academy of Sciences. 2018 doi: 10.1111/nyas.13876. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Tanaka H., Takahashi T., Xie Y., Minami R., Yanagi Y., Hayashishita M. A conserved island of BAG6/Scythe is related to ubiquitin domains and participates in short hydrophobicity recognition. The FEBS Journal. 2016;283(4):662–677. doi: 10.1111/febs.13618. [DOI] [PubMed] [Google Scholar]
- Thapaliya A., Nyathi Y., Martinez-Lumbreras S., Krysztofinska E.M., Evans N.J., Terry I.L. SGTA interacts with the proteasomal ubiquitin receptor Rpn13 via a carboxylate clamp mechanism. Scientific Reports. 2016;6:36622. doi: 10.1038/srep36622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thress K., Henzel W., Shillinglaw W., Kornbluth S. Scythe: A novel reaper-binding apoptotic regulator. The EMBO Journal. 1998;17(21):6135–6143. doi: 10.1093/emboj/17.21.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta A.P., Need E.F., Butler L.M., Selth L.A., O'Loughlin M.A., Coetzee G.A. Subdomain structure of the co-chaperone SGTA and activity of its androgen receptor client. Journal of Molecular Endocrinology. 2012;49(2):57–68. doi: 10.1530/JME-11-0152. [DOI] [PubMed] [Google Scholar]
- Trotta A.P., Need E.F., Selth L.A., Chopra S., Pinnock C.B., Leach D.A. Knockdown of the cochaperone SGTA results in the suppression of androgen and PI3K/Akt signaling and inhibition of prostate cancer cell proliferation. International Journal of Cancer. 2013;133(12):2812–2823. doi: 10.1002/ijc.28310. [DOI] [PubMed] [Google Scholar]
- Tsukahara T., Kimura S., Ichimiya S., Torigoe T., Kawaguchi S., Wada T. Scythe/BAT3 regulates apoptotic cell death induced by papillomavirus binding factor in human osteosarcoma. Cancer Science. 2009;100(1):47–53. doi: 10.1111/j.1349-7006.2008.00991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J.Y., Li Y.C., Lin T.W., Hsiao C.D. Structure of the Sgt2 dimerization domain complexed with the Get5 UBL domain involved in the targeting of tail-anchored membrane proteins to the endoplasmic reticulum. Acta Crystallographica. Section D, Biological Crystallography. 2013;69(Pt. 10):2081–2090. doi: 10.1107/S0907444913019379. [DOI] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403(6770):623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Vilardi F., Lorenz H., Dobberstein B. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. Journal of Cell Science. 2011;124(Pt. 8):1301–1307. doi: 10.1242/jcs.084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardi F., Stephan M., Clancy A., Janshoff A., Schwappach B. WRB and CAML are necessary and sufficient to mediate tail-anchored protein targeting to the ER membrane. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0085033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., Richardson E.P., Jr. Neuropathological classification of Huntington's disease. Journal of Neuropathology and Experimental Neurology. 1985;44(6):559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- Waheed A.A., MacDonald S., Khan M., Mounts M., Swiderski M., Xu Y. The Vpu-interacting protein SGTA regulates expression of a non-glycosylated tetherin species. Scientific Reports. 2016;6 doi: 10.1038/srep24934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C.P., Ravindran M.S., Inoue T., Tsai B. A cytosolic chaperone complexes with dynamic membrane J-proteins and mobilizes a nonenveloped virus out of the endoplasmic reticulum. PLoS Pathogens. 2014;10(3) doi: 10.1371/journal.ppat.1004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Brown E.C., Mak G., Zhuang J., Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Molecular Cell. 2010;40(1):159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Liang Y., Li H., Li H., He Q., Xue Y. Single nucleotide polymorphisms and osteoarthritis: An overview and a meta-analysis. Medicine (Baltimore) 2016;95(7) doi: 10.1097/MD.0000000000002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ning H., Ren F., Zhang Y., Rong Y., Wang Y. GdX/UBL4A specifically stabilizes the TC45/STAT3 association and promotes dephosphorylation of STAT3 to repress tumorigenesis. Molecular Cell. 2014;53(5):752–765. doi: 10.1016/j.molcel.2014.01.020. [DOI] [PubMed] [Google Scholar]
- Wang H., Shen H., Wang Y., Li Z., Yin H., Zong H. Overexpression of small glutamine-rich TPR-containing protein promotes apoptosis in 7721 cells. FEBS Letters. 2005;579(5):1279–1284. doi: 10.1016/j.febslet.2004.12.092. [DOI] [PubMed] [Google Scholar]
- Wang F., Song W., Brancati G., Segatori L. Inhibition of endoplasmic reticulum-associated degradation rescues native folding in loss of function protein misfolding diseases. The Journal of Biological Chemistry. 2011;286(50):43454–43464. doi: 10.1074/jbc.M111.274332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Whynot A., Tung M., Denic V. The mechanism of tail-anchored protein insertion into the ER membrane. Molecular Cell. 2011;43(5):738–750. doi: 10.1016/j.molcel.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiethoff C.M., Wodrich H., Gerace L., Nemerow G.R. Adenovirus protein VI mediates membrane disruption following capsid disassembly. Journal of Virology. 2005;79(4):1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnefeld M., Rommelaere J., Cziepluch C. The human small glutamine-rich TPR-containing protein is required for progress through cell division. Experimental Cell Research. 2004;293(1):43–57. doi: 10.1016/j.yexcr.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Wunderley L., Leznicki P., Payapilly A., High S. SGTA regulates the cytosolic quality control of hydrophobic substrates. Journal of Cell Science. 2014;127(Pt. 21):4728–4739. doi: 10.1242/jcs.155648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Cai M., Yang Y., Huang L., Ye Y. SGTA recognizes a noncanonical ubiquitin-like domain in the Bag6-UBL4A-Trc35 complex to promote endoplasmic reticulum-associated degradation. Cell Reports. 2012;2(6):1633–1644. doi: 10.1016/j.celrep.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Q., Lv L., Wan C., Chen B., Li M., Ni T. Expression and clinical role of small glutamine-rich tetratricopeptide repeat (TPR)-containing protein alpha (SGTA) as a novel cell cycle protein in NSCLC. Journal of Cancer Research and Clinical Oncology. 2013;139(9):1539–1549. doi: 10.1007/s00432-013-1474-5. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Sakisaka T. Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Molecular Cell. 2012;48(3):387–397. doi: 10.1016/j.molcel.2012.08.028. [DOI] [PubMed] [Google Scholar]
- Yang X., Cheng L., Li M., Shi H., Ren H., Ding Z. High expression of SGTA in esophageal squamous cell carcinoma correlates with proliferation and poor prognosis. Journal of Cellular Biochemistry. 2014;115(1):141–150. doi: 10.1002/jcb.24641. [DOI] [PubMed] [Google Scholar]
- Yin H., Wang H., Zong H., Chen X., Wang Y., Yun X. SGT, a Hsp90beta binding partner, is accumulated in the nucleus during cell apoptosis. Biochemical and Biophysical Research Communications. 2006;343(4):1153–1158. doi: 10.1016/j.bbrc.2006.03.090. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Lin Y., Zhang H., Manas A., Tang W., Zhang Y. UBL4A is required for insulin-induced Akt plasma membrane translocation through promotion of Arp2/3-dependent actin branching. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(31):9644–9649. doi: 10.1073/pnas.1508647112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Wang H., Hu W., Jin Y. Effect of HLA-B-associated transcript 3 polymorphisms on lung cancer risk: A meta-analysis. Medical Science Monitor. 2014;20:2461–2465. doi: 10.12659/MSM.891141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhang H., Affonso C., Bonomo R., Manas A., Xiang J. Deficiency in ubiquitin-like protein UBL4A impairs migration of fibroblasts and macrophages. Biochemical and Biophysical Research Communications. 2017;483(1):617–623. doi: 10.1016/j.bbrc.2016.12.094. [DOI] [PubMed] [Google Scholar]
- Zhu T., Ji Z., Xu C., Peng Z., Gu L., Zhang R. Expression and prognostic role of SGTA in human breast carcinoma correlates with tumor cell proliferation. Journal of Molecular Histology. 2014;45(6):665–677. doi: 10.1007/s10735-014-9586-z. [DOI] [PubMed] [Google Scholar]