Highlights
-
•
Major reassortment and transmission features of H7N9 were summarized.
-
•
Structural bases of interspecies transmission/drug resistance of H7N9 were proposed.
-
•
We summarized the major immunological characteristics of H7N9 infection.
-
•
The major strategies for vaccine development were proposed.
-
•
The disease burden of H7N9 infection was calculated.
Abstract
Human infections by the newly reassorted avian influenza A (H7N9) virus were reported for the first time in early 2013, and the virus was confirmed to be a low pathogenic avian influenza virus in poultry. Because continuously reported cases have been increasing since the summer of 2013, this novel virus poses a potential threat to public health in China and is attracting broad attention worldwide. In this review, we summarize and discuss the characteristics of the H7N9 virus revealed by the recent timely studies from the perspectives of epidemiology, host preference, clinical manifestations, immunopathogenesis, drug resistance, vaccine development, and burden of disease. This knowledge about the novel avian-origin H7N9 virus will provide a useful reference for clinical interventions of human infections and help to rapidly pave the way to develop an efficient and safe vaccine.
Current Opinion in Virology 2014, 5:91–97
This review comes from a themed issue on Emerging viruses
Edited by Christopher F Basler and Patrick CY Woo
For a complete overview see the Issue and the Editorial
Available online 3rd April 2014
1879-6257/$ – see front matter, © 2014 Elsevier B.V. All rights reserved.
Introduction
Cases of laboratory-confirmed human infections with avian-origin influenza A (H7N9) virus in the Yangtze River Delta region of China date back to February 2013 [1••]. In the subsequent two months, the number of laboratory-confirmed H7N9 infections soon grew to >130, with an apparent case fatality rate of ∼30% and cases occurring in additional locations throughout China [2]. Most patients had a history of recent exposure to poultry, generally at live poultry markets (LPMs), suggesting direct transmission of the virus from poultry to humans [3•, 4•, 5•]. Because human H7N9 infections have continued to be reported and H7N9 virus could be detected in LPMs during the 2013–2014 flu season, the virus still poses a threat to public health.
Before 2013, direct transmission of avian influenza viruses from domestic poultry to humans had been documented only for the H5N1, H7N2, H7N3, H7N7, H9N2, and H10N7 subtypes [6, 7, 8, 9]. Generally, H7N2, H7N3, H9N2, and H10N7 are low pathogenic avian influenza A viruses (LPAIVs) that are usually associated with mild disease in poultry. It has been reported that human infections with these LPAIVs cause upper respiratory tract illness varying from mild (conjunctivitis or uncomplicated influenza-like illness) to moderate in severity. Whereas, highly pathogenic avian influenza viruses (HPAIVs) can cause severe illness and high mortality in poultry. Most human infections with HPAIVs result in conjunctivitis or uncomplicated influenza illness, except for the H5N1 subtype (which is associated with >50% mortality [10]) and the H7N7 subtype (which caused one death during an outbreak in the Netherlands [9]). The recent H7N9 outbreak represents the first time ever that this LPAIV has infected humans and caused disease. Herein, we review the characteristics of this novel avian influenza virus and the disease that it causes, from the perspectives of epidemiology, clinical features, immunology, receptor binding, drug resistance, and vaccine development.
Origin and epidemiology
Virological surveillance and phylogenetic analyses confirm the transmission of the H7N9 infection from poultry to humans (Figure 1 ) [1••, 3•, 11]. Direct evidence was revealed by an infection case in Jiangsu Province, China, where a female butcher was infected, and a phylogenetically related H7N9 virus was isolated from a chicken cage 1 m away [5•]. Thus, there was an urgent call to control exposure at poultry farms and LPMs [12, 13], as many LPMs are still open or have re-opened after a short closure. A recent ecological study has assured the effectiveness of the closure of LPMs since April 2013 [14].
Figure 1.
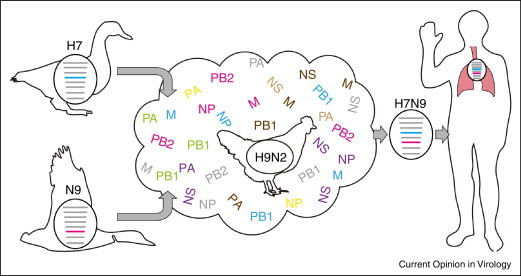
Illustration of the transmission routes of the novel H7N9 virus. H7 and N9 subtypes of undefined avian influenza viruses were transmitted into the poultry population, where H9N2 influenza viruses were circulating, and resulted in the varied genotypes of H7N9 reassortant viruses. Later on, humans were infected through the contact with virus-carrying poultry.
The novel H7N9 viruses appear to be reassortments with segments from at least four different origins [15••]: the hemagglutinin (HA) gene from duck H7 viruses; the neuraminidase (NA) gene from wild birds in Korea; and internal genes from two distinct lineages of poultry H9N2 viruses. Our most recent work [16•, 17] demonstrates that within the first H7N9 influenza infection wave, a total of 27 genotypes were observed, with diverse internal genes from varied H9N2 lineages but conserved HA and NA genes. Because the H7N9 viruses originated from poultry through genetic reassortment, it is not a surprise that the H7N9 virus could easily reassort with poultry H9N2 viruses (Figure 1). Current studies suggest that none of the H7N9 virus genotypes has gained an overwhelming fitness advantage and that they are still undergoing dynamic reassortment within poultry H9N2 gene pools [16•, 17]. Whether a ‘super’ reassortant fitting the mammalian hosts would appear is unpredictable because genetic reassortment often has major effects on influenza virus replication within cells. Therefore, extensive surveillance and control of poultry production and transportation is an urgent priority.
Clinical features and immunology
The clinical characteristics of the human LPAIV H7N9 virus infections are somewhat comparable to infections with HPAIV H5N1 and severe acute respiratory syndrome coronavirus (SARS-CoV) [4•]. Acute H7N9 infection causes severe illness, that is, respiratory failure and acute respiratory distress syndrome (ARDS), with high rates of ICU admission [2, 18]. Multivariate analyses revealed that the presence of an underlying medical condition is an independent risk factor for ARDS [19•]. Together with the existence of the mild-to-moderate infection cases detected by China's national sentinel surveillance system for influenza-like illness [20], the H7N9 virus infections seem to be less serious than initially thought at the beginning of the outbreak.
Similar to what has been observed for H5N1 infections, immunopathogenetic analysis of H7N9 infection samples reveals high levels of plasma proinflammatory cytokines and chemokines, termed hypercytokinemia, in the early stage of the infection [21, 22]. The concentrations of several cytokines/chemokines, including MIP-1β, IL-6, and IL-8, are even higher than those of previously reported H5N1 infection cases [23]. Furthermore, the hypercytokinemia is correlated with disease severity for H7N9 infection. Higher levels of IL-6, IL-8, IL-10, and MIP-1β are detected in fatal cases compared to survivors from H7N9 infections [24•]. Our data indicated that plasma levels of angiotensin II are correlated with disease severity and predict fatal outcomes in H7N9-infected patients [25•]. A recent study elucidated that the human single nucleotide polymorphism (SNP) rs12252-C, which is associated with dysfunctional interferon induced transmembrane protein-3 (IFITM3), plays a crucial genetic role in early hypercytokinemia development in H7N9-infected patients [24•]. Despite this progress, it remains important to profile the sequential connections of these cytokines/chemokines and clarify the key factors in the infection process, particularly in the local mucosal tissues of the lungs of H7N9 patients.
Most H7N9 patients display a robust serological response after the onset of the disease, as revealed by antibody reactivity kinetics [26, 27]. Detectable H7N9-specific antibodies appear in the sera of survivors earlier than in fatal cases. In addition, the highest antibody titers in fatal cases are also lower than in survivors [28]. Among the survivors of H7N9 infections, the group with higher antibody titers experiences a shorter time from the onset of the disease to recovery [26, 27]. These data indicate that an earlier and robust H7N9-specific humoral response provides a protective effect and improves clinical outcomes in patients.
Data on T-cell responses in H7N9-infected patients are still lacking. However, immunoinformatics analysis of H7N9 proteins indicates that conservation of T-cell epitopes with other recently circulating strains of influenza is very limited [29]. Approximately 60% of the previously well-defined major histocompatibility complex class I (MHC I)-restricted T-cell epitopes have substitutions in the corresponding sequences in H7N9 as revealed by the comparison of H7N9 with 2009 pH1N1 (Figure 2 ). Furthermore, a study of human leukocyte antigen (HLA)-typed healthy subjects demonstrates that seasonal influenza virus-specific T-cells poorly cross-react to individual H7N9 variant epitopes, apart from the recognition of the whole virus [30]. It is also indicated that cross-CD8+ T-cell immunity to the H7N9 influenza A virus varies across different ethnic groups based on the HLA background [31]. Thus, further investigations into the features of the H7N9-specific T-cell responses in patients and a comparison of CD8+ and CD4+ T-cell-related immunogenic variance of H7N9 to other currently circulating influenza viruses, for example, 2009 pH1N1, should be performed.
Figure 2.
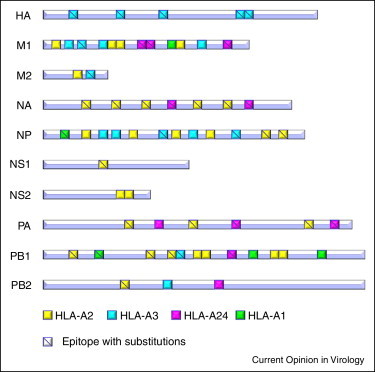
The T-cell immunogenic variation of H7N9 as compared with 2009 pH1N1. The previously well-defined antigenic epitopes of influenza virus (colored squares) are distributed in all of the major proteins (gray stripes, 10 well-defined proteins as illustrated herein) of the virus. The peptides have different HLA-restrictions (yellow for HLA-A2, pink for HLA-A24, green for HLA-A1, and cyan for HLA alleles from A3 supertype), which may cover a large majority of the population. The peptides with mutated sites when compared between H7N9 and 2009 pH1N1 are marked with a bias line in the square.
Molecular basis of the ‘host jump’
Influenza A viruses enter host cells through endocytosis after binding to cell-surface receptors, which are sialic acids (SAs) linked to cell-surface glycolipids or glycoproteins [32, 33, 34]. The SA-binding protein HA is a major determinant of the virus ‘host jump’. It is known that three secondary elements and one base element in the HA receptor binding pocket are directly involved in SA binding. For the shallow receptor binding cavity, the three secondary elements forming the edge are the 130-loop, the 190-helix, and the 220-loop (in H3 numbering, respectively), and the base element is mainly composed of four conserved residues (Y98, W153, H183, and Y195).
The molecular bases of the receptor binding shift of H7N9 were explored in several studies by different groups, including our own, which elucidated the basis for the avian-to-human host jump by the H7N9 influenza virus [35, 36, 37, 38, 39••]. In the beginning of the H7N9 outbreak, A/Anhui/1/2013 (Anhui-H7N9) and A/Shanghai/1/2013 (Shanghai-H7N9) viruses were two of the earliest strains. Shanghai-H7N9 preferentially binds to the avian receptor analog, whereas Anhui-H7N9 recognizes both avian and human receptor analogs. In contrast to the H5N1 influenza [40, 41], the human-signature substitution Q226L is not solely responsible for binding of the H7N9 influenza virus HA to the human receptor. Rather, other amino acids around the receptor binding site (RBS) are also very important. Four residues (S138A, G186V, T221P, and Q226L) in the RBS that differ between Shanghai-H7N9 and Anhui-H7N9 are able to create a larger hydrophobic region on the left side of the RBS of Anhui-H7N9, which allows Anhui-H7N9 to easily bind the human receptor analog [39••].
Features and the molecular basis of drug resistance
Available flu-specific anti-virus drugs applied in clinical treatment mainly focus on two influenza proteins: firstly, M2 is targeted by amantadine/rimantadine and finally, NA is targeted by four types of inhibitors, those are, oseltamivir, zanamivir, peramivir, and laninamivir [42]. The M2 proteins derived from all the H7N9 reported in 2013 contain the S31N substitution, indicating resistance to amantadine/rimantadine [1••]. For the other target, NA inhibitors are commonly used in the clinic and are the most effective drugs against influenza. However, some H7N9 virus strains also contain substitutions in NA that are associated with drug resistance, raising serious concerns [43•].
For N9, the A/Anhui/1/2013 and A/Shanghai/1/2013 viruses, which have distinct drug resistant features, exhibit differences at two residues (40 and 294, N9 numbering) [43•]. Specifically, N9 in A/Shanghai/1/2013 contains K294, while residue 294 in A/Anhui/1/2013 is an Arg. Residue 294 is one of the three key arginines at the active site (the other two are R118 and R371) that directly interacts with the substrate/inhibitor carboxylate [44]. The decreased interaction between K294 and the inhibitor carboxylate group plays an essential role in reducing drug sensitivity (Figure 3 ). Furthermore, K294 has an effect on the rotation of the E276 side chain, which is in an unfavorable position to accommodate the oseltamivir hydrophobic pentyloxy group, thereby significantly reducing oseltamivir sensitivity. However, in vitro assays demonstrate that R294K impairs NA activity, which is not advantageous for virus replication [43•]. This is good news for the treatment of the H7N9-infected patients.
Figure 3.
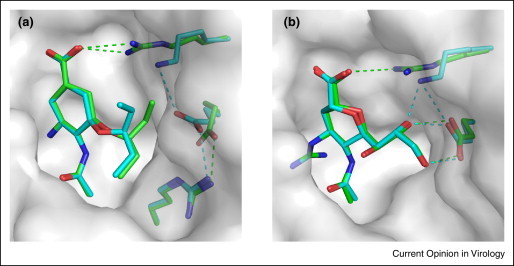
The molecular basis of the drug resistance of N9. Structural analysis of the oselatmivir carboxylate and zanamivir binding models in Anhui N9 and Shanghai N9. (a) Comparison of the oseltamivir binding model in Shanghai N9 (cyan) and Anhui N9 (green). (b) Comparison of the zanamivir binding models in Shanghai N9 (cyan) and Anhui N9 (green). The inhibitors and the surrounding residues are shown in sticks with hydrogen bonds and salt bridges indicated by dotted lines. The structure of Shanghai N9 is also displayed in surface representation. The binding models of peramivir and laninamivir with Shanghai N9 and Anhui N9 are quite similar to zanamivir.
Vaccine development
Several different approaches, including using live recombinant viral vector-based vaccines, live attenuated influenza vaccines, universal DNA vaccines, and virus-like particles (VLPs), have been explored to meet the urgent challenge of vaccine development to combat H7N9 infection [45, 46, 47, 48, 49, 50, 51, 52]. One vaccine containing recombinant Newcastle disease virus expressing HA from H7N3 and two live attenuated vaccines with H7N3 and H7N7 were investigated for their cross-reactivity with the H7N9 virus in May 2013 [45, 46, 47]. Subsequently, universal DNA vaccines based on antibodies targeting the HA stalk region were examined as a possible candidate to prevent H7N9 infection. The H3 stalk-based vaccine is able to elicit antibodies reactive with H7 proteins and also protects mice from H7N9 challenge. In addition, insect cell-derived VLPs containing HA and NA from H7N9 display promising protection in mice against H7N9 [50, 51]. Indeed, shortly after these animal experiments, enrolment of 284 adults was initiated for a phase 1 clinical trial of a monovalent VLP (H7N9) vaccine candidate with and without adjuvant [52]. The clinical trial suggests that VLPs in the presence of adjuvant induce an immune response at a low dosage. Recent data also indicates that antigen preparations from divergent H7 strains are able to induce protective immunity against H7N9 infection [53]. All of these efforts to develop effective H7N9 influenza vaccines with traditional or new techniques are attempting to shorten production cycles and meet the manufacturing requirements for rapid deployment.
Lessons learned and the burden of the disease
The outbreak of H7N9 infections on mainland, as well as in Taiwan and Hong Kong, China, has posed a great burden for the society. On the basis of the most recent data [54•], the total direct medical cost of H7N9 as of May 31, 2013 is 16 422 535 CNY (2 627 606 USD). For the mean cost of each patient, the cost per day increase correspondingly as the severity of the illness increases. The total cost of disability-adjusted life years (DALYs) is 17 356 561 CNY (2 777 050 USD). Further, the poultry industry losses amount to 7.75 billion CNY (1.24 billion USD) in 10 affected provinces in China and 3.68 billion CNY (0.59 billion USD) in eight non-affected adjacent provinces. The huge poultry industry losses resulted from LPM closings and poultry culling in some areas. Compared to the poultry-industry losses, direct medical losses and DALY losses have been relatively small. Closure of LPMs reduced the mean daily number of infections by 99% in Shanghai and Hangzhou and by 97% in Huzhou and Nanjing based on a Bayesian statistical model for the time series of the onset of illness [14]. However, the medical costs per case are extremely high (particularly for addressing the use of modern medical devices). A cost-effectiveness assessment for disease intervention has yet to be determined.
Conclusions and perspectives
The outbreak of the LPAIV H7N9 virus in early 2013 generated tremendous concern, and recent studies reveal the novel properties of the virus. Epidemical and genetic analysis illustrates the novel reassortment events that apparently facilitate the transmission from poultry to humans (the so called ‘host jump’). This was facilitated by the unconventional features of the HA receptor binding pocket. Further, the preliminary understanding of the clinical manifestations and the immunopathogenetic properties of H7N9 infections indicates a complicated interaction between the virus and host. Knowledge gained regarding this novel avian virus may help to pave a way to rapidly conquer it, including approaches to develop an efficient and safe vaccine. However, considering the continuing and unexpected emergence of human infections by novel avian influenza viruses (for example, H6N1 and H10N8) [55, 56], it is necessary to devote more attention to surveillance for interspecies transmission of influenza viruses and toward the development of universal vaccines.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC, Nos. 81021003, 31030030, and 81030032/H19); Grand S&T project of China Health and Family Planning Commission (Nos. 2013ZX10004608-002, 2012ZX10004201-009, and 2013ZX10001004); and intramural special grant for influenza virus research from the Chinese Academy of Sciences (KJZD-EW-L09). GFG is a leading principal investigator of the Innovative Research Group of the National Natural Science Foundation of China (No. 81321063).
Contributor Information
Jun Liu, Email: liuj333@gmail.com.
George F Gao, Email: gaofu@chinacdc.cn.
References
- 1••.Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W., Chen J., Jie Z., Qiu H., Xu K. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]; This is the first report of the human infection of the H7N9 virus, characterized the etiology, clinical features, and gene properties.
- 2.Yu H., Cowling B.J., Feng L., Lau E.H., Liao Q., Tsang T.K., Peng Z., Wu P., Liu F., Fang V.J. Human infection with avian influenza A H7N9 virus: an assessment of clinical severity. Lancet. 2013;382:138–145. doi: 10.1016/S0140-6736(13)61207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Li J., Yu X., Pu X., Xie L., Sun Y., Xiao H., Wang F., Din H., Wu Y., Liu D. Environmental connections of novel avian-origin H7N9 influenza virus infection and virus adaptation to the human. Sci China Life Sci. 2013;56:485–492. doi: 10.1007/s11427-013-4491-3. [DOI] [PubMed] [Google Scholar]; This paper proposed a possible transmission route of H7N9 from poultry to human, through analyzing the virus isolations from both patients and wet markets.
- 4•.Chen Y., Liang W., Yang S., Wu N., Gao H., Sheng J., Yao H., Wo J., Fang Q., Cui D. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013;381:1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref. [3•].
- 5•.Bao C.J., Cui L.B., Zhou M.H., Hong L., Gao G.F., Wang H. Live-animal markets and influenza A (H7N9) virus infection. N Engl J Med. 2013;368:2337–2339. doi: 10.1056/NEJMc1306100. [DOI] [PubMed] [Google Scholar]; See annotation to Ref. [3•].
- 6.Arzey G.G., Kirkland P.D., Arzey K.E., Frost M., Maywood P., Conaty S., Hurt A.C., Deng Y.M., Iannello P., Barr I. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg Infect Dis. 2012;18:814–816. doi: 10.3201/eid1805.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirst M., Astell C.R., Griffith M., Coughlin S.M., Moksa M., Zeng T., Smailus D.E., Holt R.A., Jones S., Marra M.A. Novel avian influenza H7N3 strain outbreak, British Columbia. Emerg Infect Dis. 2004;10:2192–2195. doi: 10.3201/eid1012.040743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avian influenza A/(H7N2) outbreak in the United Kingdom. Euro Surveill. 2007;12:E070531–E070532. [PubMed] [Google Scholar]
- 9.Fouchier R.A., Schneeberger P.M., Rozendaal F.W., Broekman J.M., Kemink S.A., Munster V., Kuiken T., Rimmelzwaan G.F., Schutten M., Van Doornum G.J. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuen K.Y., Chan P.K., Peiris M., Tsang D.N., Que T.L., Shortridge K.F., Cheung P.T., To W.K., Ho E.T., Sung R. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 11.Shi J., Deng G., Liu P., Zhou J., Guan L., Li W., Li X., Guo J., Wang G., Fan J. Isolation and characterization of H7N9 viruses from live poultry markets — implication of the source of current H7N9 infection in humans. Chin Sci Bull. 2013;58:7. [Google Scholar]
- 12.Xu J., Lu S., Wang H., Chen C. Reducing exposure to avian influenza H7N9. Lancet. 2013;381:1815–1816. doi: 10.1016/S0140-6736(13)60950-2. [DOI] [PubMed] [Google Scholar]
- 13.Wu Y., Gao G.F. Lessons learnt from the human infections of avian-origin influenza A H7N9 virus: live free markets and human health. Sci China Life Sci. 2013;56:493–494. doi: 10.1007/s11427-013-4496-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu H., Wu J.T., Cowling B.J., Liao Q., Fang V.J., Zhou S., Wu P., Zhou H., Lau E.H., Guo D. Effect of closure of live poultry markets on poultry-to-person transmission of avian influenza A H7N9 virus: an ecological study. Lancet. 2014;383:541–548. doi: 10.1016/S0140-6736(13)61904-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Liu D., Shi W., Shi Y., Wang D., Xiao H., Li W., Bi Y., Wu Y., Li X., Yan J. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]; Using phylogenetic, structural and coalescent analysis, the authors proposed a four-origin reassortment model and the diversity of H7N9 virus. This is the first study to systematically analyse the H7N9 origin.
- 16•.Cui L., Liu D., Shi W., Pan J., Qi X., Li X., Guo X., Zhou M., Li W., Li J. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun. 2014;5:3142. doi: 10.1038/ncomms4142. [DOI] [PubMed] [Google Scholar]; In this study, using a broader dataset of H7N9 virus genomes, the authors comprehensively studied the distribution, increment, and migration of virus genotypes, and proposed a dynamic reassortment model of H7N9 within poultry H9N2 gene pools.
- 17.Liu D., Shi W., Gao G.F. Poultry carrying H9N2 act as incubators for novel human avian influenza viruses. Lancet. 2014 doi: 10.1016/S0140-6736(14)60386-X. [DOI] [PubMed] [Google Scholar]
- 18.Yu L., Wang Z., Chen Y., Ding W., Jia H., Chan J.F., To K.K., Chen H., Yang Y., Liang W. Clinical, virological, and histopathological manifestations of fatal human infections by avian influenza A(H7N9) virus. Clin Infect Dis. 2013;57:1449–1457. doi: 10.1093/cid/cit541. [DOI] [PubMed] [Google Scholar]
- 19•.Gao H.N., Lu H.Z., Cao B., Du B., Shang H., Gan J.H., Lu S.H., Yang Y.D., Fang Q., Shen Y.Z. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013;368:2277–2285. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]; This paper described the clinical features of H7N9 and analyzed the potential independent risk factor for ARDS based on a systematical integration of the clinical data from most of the H7N9 cases available in China thus far.
- 20.Ip D.K., Liao Q., Wu P., Gao Z., Cao B., Feng L., Xu X., Jiang H., Li M., Bao J. Detection of mild to moderate influenza A/H7N9 infection by China's national sentinel surveillance system for influenza-like illness: case series. BMJ. 2013;346:f3693. doi: 10.1136/bmj.f3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Z., Chen Z., Li X., Xu L., Guan W., Cao Y., Hu Y., Zhang J. Host immunological response and factors associated with clinical outcome in patients with the novel influenza A H7N9 infection. Clin Microbiol Infect. 2013 doi: 10.1111/1469-0691.12505. [DOI] [PubMed] [Google Scholar]
- 22.Chi Y., Zhu Y., Wen T., Cui L., Ge Y., Jiao Y., Wu T., Ge A., Ji H., Xu K. Cytokine and chemokine levels in patients infected with the novel avian influenza A (H7N9) virus in China. J Infect Dis. 2013;208:1962–1967. doi: 10.1093/infdis/jit440. [DOI] [PubMed] [Google Scholar]
- 23.Zhou J., Wang D., Gao R., Zhao B., Song J., Qi X., Zhang Y., Shi Y., Yang L., Zhu W. Biological features of novel avian influenza A (H7N9) virus. Nature. 2013;499:500–503. doi: 10.1038/nature12379. [DOI] [PubMed] [Google Scholar]
- 24•.Wang Z., Zhang A., Wan Y., Liu X., Qiu C., Xi X., Ren Y., Wang J., Dong Y., Bao M. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc Natl Acad Sci U S A. 2014;111:769–774. doi: 10.1073/pnas.1321748111. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study indicated the correlation of the high cytokine/chemokine production with the outcome of the H7N9 infection and fingered out the risk factors, for example, genetic background of the patients in the infection.
- 25•.Huang F., Guo J., Zou Z., Liu J., Cao B., Zhang D S., Li H., Wang W., Sheng M., Liu S. Angiotensin II plasma levels are linked to disease severity and predict fatal outcomes in H7N9-infected patients. Nat Commun. 2014 doi: 10.1038/ncomms4595. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]; This study evaluated the predictive value of angiotensin II for disease severity and fatal outcomes in H7N9-infected patients and indicated that angiotensin II is a biomarker for lethality in flu infections.
- 26.Yang S., Chen Y., Cui D., Yao H., Lou J., Huo Z., Xie G., Yu F., Zheng S., Yang Y. Avian-origin influenza A(H7N9) infection in influenza A(H7N9)-affected areas of China: a serological study. J Infect Dis. 2014;209:265–269. doi: 10.1093/infdis/jit430. [DOI] [PubMed] [Google Scholar]
- 27.Zhang A., Huang Y., Tian D., Lau E.H., Wan Y., Liu X., Dong Y., Song Z., Zhang X., Zhang J. Kinetics of serological responses in influenza A(H7N9)-infected patients correlate with clinical outcome in China. Euro Surveill. 2013;18:20657. doi: 10.2807/1560-7917.es2013.18.50.20657. [DOI] [PubMed] [Google Scholar]
- 28.Yu L., Wang Z.M., Chen Y., Ding W., Jia H.Y., Chan J.F.W., To K.K.W., Chen H.L., Yang Y.D., Liang W.F. Clinical virological, and histopathological manifestations of fatal human infections by avian influenza A(H7N9) virus. Clin Infect Dis. 2013;57:1449–1457. doi: 10.1093/cid/cit541. [DOI] [PubMed] [Google Scholar]
- 29.De Groot A.S., Ardito M., Terry F., Levitz L., Ross T., Moise L., Martin W. Low immunogenicity predicted for emerging avian-origin H7N9: implication for influenza vaccine design. Hum Vaccin Immunother. 2013;9:950–956. doi: 10.4161/hv.24939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van de Sandt C.E., Kreijtz J.H., de Mutsert G., Geelhoed-Mieras M.M., Hillaire M.L., Vogelzang-van Trierum S.E., Osterhaus A.D., Fouchier R.A., Rimmelzwaan G.F. Human cytotoxic T lymphocytes directed to seasonal influenza A viruses cross-react with the newly emerging H7N9 virus. J Virol. 2014;88:1684–1693. doi: 10.1128/JVI.02843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinones-Parra S., Grant E., Loh L., Nguyen T.H., Campbell K.A., Tong S.Y., Miller A., Doherty P.C., Vijaykrishna D., Rossjohn J. Preexisting CD8+ T-cell immunity to the H7N9 influenza A virus varies across ethnicities. Proc Natl Acad Sci U S A. 2014;111:1049–1054. doi: 10.1073/pnas.1322229111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauter N.K., Bednarski M.D., Wurzburg B.A., Hanson J.E., Whitesides G.M., Skehel J.J., Wiley D.C. Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: a 500-MHz proton nuclear magnetic resonance study. Biochemistry. 1989;28:8388–8396. doi: 10.1021/bi00447a018. [DOI] [PubMed] [Google Scholar]
- 33.Takemoto D.K., Skehel J.J., Wiley D.C. A surface plasmon resonance assay for the binding of influenza virus hemagglutinin to its sialic acid receptor. Virology. 1996;217:452–458. doi: 10.1006/viro.1996.0139. [DOI] [PubMed] [Google Scholar]
- 34.Gambaryan A.S., Tuzikov A.B., Piskarev V.E., Yamnikova S.S., Lvov D.K., Robertson J.S., Bovin N.V., Matrosovich M.N. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6′-sialyl(N-acetyllactosamine) Virology. 1997;232:345–350. doi: 10.1006/viro.1997.8572. [DOI] [PubMed] [Google Scholar]
- 35.Yang H., Carney P.J., Chang J.C., Villanueva J.M., Stevens J. Structural analysis of the hemagglutinin from the recent 2013 H7N9 influenza virus. J Virol. 2013;87:12433–12446. doi: 10.1128/JVI.01854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu R., de Vries R.P., Zhu X., Nycholat C.M., McBride R., Yu W., Paulson J.C., Wilson I.A. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science. 2013;342:1230–1235. doi: 10.1126/science.1243761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong X., Martin S.R., Haire L.F., Wharton S.A., Daniels R.S., Bennett M.S., McCauley J.W., Collins P.J., Walker P.A., Skehel J.J. Receptor binding by an H7N9 influenza virus from humans. Nature. 2013;499:496–499. doi: 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
- 38.Tharakaraman K., Jayaraman A., Raman R., Viswanathan K., Stebbins N.W., Johnson D., Shriver Z., Sasisekharan V., Sasisekharan R. Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell. 2013;153:1486–1493. doi: 10.1016/j.cell.2013.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Shi Y., Zhang W., Wang F., Qi J., Wu Y., Song H., Gao F., Bi Y., Zhang Y., Fan Z. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science. 2013;342:243–247. doi: 10.1126/science.1242917. [DOI] [PubMed] [Google Scholar]; The host receptor binding features and the corresponding molecular bases of H7N9 HA were determined, which may shed light on the host jump of the virus.
- 40.Lu X., Shi Y., Zhang W., Zhang Y., Qi J., Gao G.F. Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut) Protein Cell. 2013;4:502–511. doi: 10.1007/s13238-013-3906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W., Shi Y., Lu X., Shu Y., Qi J., Gao G.F. An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level. Science. 2013;340:1463–1467. doi: 10.1126/science.1236787. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki Y. Sialobiology of influenza: molecular mechanism of host range variation of influenza viruses. Biol Pharm Bull. 2005;28:399–408. doi: 10.1248/bpb.28.399. [DOI] [PubMed] [Google Scholar]
- 43•.Wu Y., Bi Y., Vavricka C.J., Sun X., Zhang Y., Gao F., Zhao M., Xiao H., Qin C., He J. Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses. Cell Res. 2013;23:1347–1355. doi: 10.1038/cr.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors compared the drug resistance and virus fitness of two distinct NAs from prototypes of the H7N9 viruses, that is, A/Anhui/1/2013 and A/Shanghai/1/2013, and elucidated the molecular basis.
- 44.von Itzstein M. The war against influenza: discovery and development of sialidase inhibitors. Nat Rev Drug Discov. 2007;6:967–974. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- 45.Goff P.H., Krammer F., Hai R., Seibert C.W., Margine I., Garcia-Sastre A., Palese P. Induction of cross-reactive antibodies to novel H7N9 influenza virus by recombinant Newcastle disease virus expressing a North American lineage H7 subtype hemagglutinin. J Virol. 2013;87:8235–8240. doi: 10.1128/JVI.01085-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudenko L., Isakova-Sivak I., Donina S. H7N3 live attenuated influenza vaccine has a potential to protect against new H7N9 avian influenza virus. Vaccine. 2013;31:4702–4705. doi: 10.1016/j.vaccine.2013.08.040. [DOI] [PubMed] [Google Scholar]
- 47.Xu Q., Chen Z., Cheng X., Xu L., Jin H. Evaluation of live attenuated H7N3 and H7N7 vaccine viruses for their receptor binding preferences, immunogenicity in ferrets and cross reactivity to the novel H7N9 virus. PLoS ONE. 2013;8:7688–7694. doi: 10.1371/journal.pone.0076884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krammer F., Margine I., Hai R., Flood A., Hirsh A., Tsvetnitsky V., Chen D., Palese P. H3 stalk-based chimeric hemagglutinin influenza virus constructs protect mice from H7N9 challenge. J Virol. 2014;88:2340–2343. doi: 10.1128/JVI.03183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Margine I., Krammer F., Hai R., Heaton N.S., Tan G.S., Andrews S.A., Runstadler J.A., Wilson P.C., Albrecht R.A., Garcia-Sastre A. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J Virol. 2013;87:10435–10446. doi: 10.1128/JVI.01715-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith G.E., Flyer D.C., Raghunandan R., Liu Y., Wei Z., Wu Y., Kpamegan E., Courbron D., Fries L.F., 3rd, Glenn G.M. Development of influenza H7N9 virus like particle (VLP) vaccine: homologous A/Anhui/1/2013 (H7N9) protection and heterologous A/chicken/Jalisco/CPA1/2012 (H7N3) cross-protection in vaccinated mice challenged with H7N9 virus. Vaccine. 2013;31:4305–4313. doi: 10.1016/j.vaccine.2013.07.043. [DOI] [PubMed] [Google Scholar]
- 51.Klausberger M., Wilde M., Palmberger D., Hai R., Albrecht R.A., Margine I., Hirsh A., Garcia-Sastre A., Grabherr R., Krammer F. One-shot vaccination with an insect cell-derived low-dose influenza A H7 virus-like particle preparation protects mice against H7N9 challenge. Vaccine. 2014;32:355–362. doi: 10.1016/j.vaccine.2013.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fries L.F., Smith G.E., Glenn G.M. A recombinant viruslike particle influenza A (H7N9) vaccine. N Engl J Med. 2013;369:2564–2566. doi: 10.1056/NEJMc1313186. [DOI] [PubMed] [Google Scholar]
- 53.Krammer F., Albrecht R.A., Tan G.S., Margine I., Hai R., Schmolke M., Runstadler J., Andrews S.F., Wilson P.C., Cox R.J. Divergent H7 immunogens offer protection from H7N9 challenge. J Virol. 2014 doi: 10.1128/JVI.03095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54•.Qi X., Jiang D., Wang H., Zhuang D., Ma J., Fu J., Qu J., Sun Y., Yu S., Meng Y. Calculating the burden of disease of avian-origin H7N9 infections in China. BMJ Open. 2014;4:e890041. doi: 10.1136/bmjopen-2013-004189. [DOI] [PMC free article] [PubMed] [Google Scholar]; The overall burden of H7N9 cases in China was calculated, which provides an example for the comprehensive burden of disease in the 21st century from an acute animal-borne emerging infectious disease.
- 55.Chen H., Yuan H., Gao R., Zhang J., Wang D., Xiong Y., Fan G., Yang F., Li X., Zhou J. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 56.Wei S.H., Yang J.R., Wu H.S., Chang M.C., Lin J.S., Lin C.Y., Liu Y.L., Lo Y.C., Yang C.H., Chuang J.H. Human infection with avian influenza A H6N1 virus: an epidemiological analysis. Lancet Respir Med. 2013;1:771–778. doi: 10.1016/S2213-2600(13)70221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


