Abstract
OBJECTIVE
Heterozygous loss-of-function mutations in HNF1A cause maturity-onset diabetes of the young (MODY). Affected individuals can be treated with low-dose sulfonylureas. Individuals with homozygous HNF1A mutations causing MODY have not been reported.
RESEARCH DESIGN AND METHODS
We phenotyped a kindred with young-onset diabetes and performed molecular genetic testing, a mixed meal tolerance test, a sulfonylurea challenge, and in vitro assays to assess variant protein function.
RESULTS
A homozygous HNF1A variant (p.A251T) was identified in three insulin-treated family members diagnosed with diabetes before 20 years of age. Those with the homozygous variant had low hs-CRP levels (0.2–0.8 mg/L), and those tested demonstrated sensitivity to sulfonylurea given at a low dose, completely transitioning off insulin. In silico modeling predicted a variant of unknown significance; however, in vitro studies supported a modest reduction in transactivation potential (79% of that for the wild type; P < 0.05) in the absence of endogenous HNF1A.
CONCLUSIONS
Homozygous hypomorphic HNF1A variants are a cause of HNF1A-MODY. We thus expand the allelic spectrum of variants in dominant genes causing diabetes.
Introduction
Heterozygous loss-of-function mutations in the hepatocyte nuclear factor 1-α gene (HNF1A) cause maturity-onset diabetes of the young (MODY) (1). Young age at onset, autosomal dominant inheritance, and an excellent glycemic response to sulfonylurea monotherapy characterize HNF1A-MODY (2). A spectrum of heterozygous HNF1A variants have been identified—from classical MODY-causing mutations that impair insulin secretion (3,4), to milder alleles that increase the risk of type 2 diabetes. Homozygous loss-of-function germline mutations are thought to be embryonically lethal (5). Establishing the pathogenicity of missense variants is challenging (6,7): hypomorphic variants in the heterozygous state may be dismissed as benign, but when inherited recessively, they may mimic loss-of-function heterozygous mutations (8).
We describe an insulin-treated individual in whom a homozygous HNF1A missense variant, p.A251T (c.751G>A), was identified. We metabolically characterized both heterozygous carriers of the p.A251T variant and individuals homozygous for the p.A251T variant, performed in vitro functional studies to test our hypothesis that p.A251T is a hypomorphic HNF1A mutation, and assessed the transition to sulfonylurea therapy.
Research Design and Methods
In the proband, recruited to the MYDIABETES (MODY in Young-Onset Diabetes in Different Ethnicities) study (clinical trial no. NCT02082132, ClinicalTrials.gov), we sequenced HNF1A promoters, exons, and key flanking intronic sequences, along with 22 genes implicated in monogenic diabetes (9), and we assessed copy number variation using multiplex ligation-dependent probe amplification (9).
In Silico Modeling of Pathogenicity
We assessed predicted pathogenicity using established in silico tools, in line with the American College of Medical Genetics classification (10) and as previously described (11). For structural analysis, we used FoldX (12), introducing variants p.A251T and a nearby MODY-causing mutation, p.V246 L (13), into the structure of the HNF1A DNA binding domain complexed with DNA (Research Collaboratory for Structural Bioinformatics Protein Data Bank identifier 1IC8) (14).
Clinical Studies
We assessed clinical features and hs-CRP, a marker of HNF1A protein function (15), in the proband and his family. We also assessed in the proband β-cell function and glucose response to a mixed meal tolerance test (MMTT) (16), and his response to a 2.5-mg glibenclamide challenge. The proband underwent continuous glucose monitoring for 7-days before and after the switch to the sulfonylurea.
In Vitro Functional Characterization
The impact of p.A251T on HNF1A function was assessed by using a suite of in vitro assays, as described previously (11). This impact on function was compared with that of wild-type HNF1A and two DNA binding domain mutations that cause MODY.
Results
Clinical Characteristics of p.A251T Variant Carriers
Young-onset diabetes cosegregated with the homozygous, but not heterozygous, HNF1A p.A251T carriers; three homozygous individuals (proband, his identical twin, and a sister) presented with type 1 diabetes as teenagers and were treated with insulin (Supplementary Fig. 1 and Supplementary Table 1). The father fulfilled criteria for prediabetes and the mother developed gestational diabetes during her third pregnancy at age 29 years; her diabetes persisted postpartum.
In the proband, no additional mutations or variants were detected, and anti-GAD65 and IA2 antibodies were negative. Although consanguinity was not reported, both parents were from the same region in Eastern Europe.
Levels of hs-CRP were low (≤0.2–0.8 mg/L) in all those who were homozygous for p.A251T and in the heterozygous mother (see Supplementary Table 1). The heterozygous father displayed a higher value (1.4 mg/L).
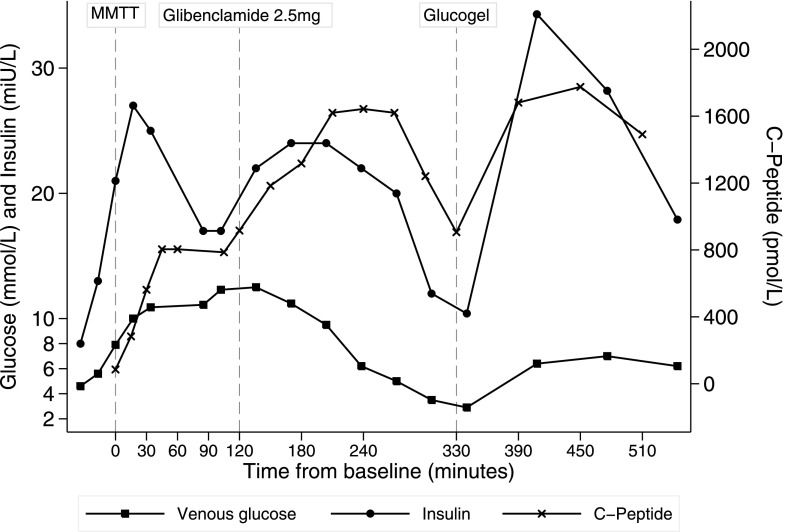
During the MMTT (see Fig. 1), blood glucose plateaued at 12.5 mmol/L; this was accompanied by a rise in C-peptide. After the proband ingested glibenclamide, C-peptide increased further, beyond the levels measured during the MMTT. This increase was sustained and decreased only after glucose levels began to fall. Glucose dropped beyond fasting levels (to 2.9 mmol/L), however, causing hypoglycemia. Oral glucose was administered, resulting in another, higher C-peptide increase that restored normoglycemia. The proband was discharged with a prescription for 1.25-mg glibenclamide once daily (which was subsequently increased to twice daily); insulin was stopped. Continuous glucose monitoring performed before and after the treatment change demonstrated the resolution of hypoglycemia and maintenance of glucose levels (Supplementary Fig. 2), and HbA1c was reduced to 6.9% (52 mmol/mol). The proband’s identical twin also transitioned from insulin to sulfonylurea therapy.
Figure 1.
Composite graph depicting results of the MMTT and the sulfonylurea challenge in the proband after morning premixed insulin was omitted. Glucose, C-peptide, and insulin during an MMTT, and the glibenclamide test dose, at indicated time points are displayed. Glucogel 25 g, an oral glucose preparation B (BI Healthcare, Bridgend, U.K.), was administered sublingually.
In Silico Characterization of the p.A251T Variant
The p.A251T variant is located in a region of HNF1A that encodes a highly conserved POU homeodomain DNA binding domain, which is crucial for transcription factor function (14). Neither p.A251T nor other rare variants at this position were present in the Genome Aggregation Database (examined July 2019). A variant of uncertain significance was predicted by in silico approaches (see Supplementary Table 2). Detailed structural analysis of p.A251T compared with that of a MODY-causing mutation (p.V246 L) at a similar location revealed the formation of a novel hydrogen bond (Supplementary Fig. 3).
In Vitro Functional Assessment
In vitro functional assays demonstrated a modest reduction in transactivation activity and DNA binding for p.A251T-HNF1A, which is less severe than bona fide mutations (p.P112L and p.R203H) that cause early-onset diabetes (Supplementary Fig. 4).
Conclusions
We report a homozygous hypomorphic HNF1A mutation in a family with young-onset diabetes. The clinical phenotype is consistent with classical heterozygous HNF1A-MODY: individuals have low hs-CRP and demonstrate sensitivity to sulfonylureas, enabling a transition off insulin. Importantly, young-onset diabetes cosegregated with homozygous individuals, not heterozygous carriers. In vitro studies support p.A251T as being a hypomorphic variant with modest transactivation defects only in the absence of endogenous HNF1A. This defect is likely driven by low DNA-binding potential. The defects observed, however, were less pronounced than those of reported bona fide heterozygous MODY-causing mutations in a similar location.
Our in silico studies provide a putative explanation for why p.A251T could have modest structural effects despite being located in the POU homeodomain. We showed that p.A251T effects are attenuated by the predicted formation of a novel hydrogen bond and that structural changes are less deleterious than those of a similarly located MODY-causing mutation. The hypomorphic status of this variant is further supported by the phenotype observed in heterozygous carriers, which is atypical for heterozygous loss-of-function HNF1A mutations.
The p.A251T HNF1A variant has been reported in two studies describing heterozygous carriers (17,18), bringing the total number of reported heterozygous carriers to eight. In the study by Thanabalasingham et al. (17), the proband, who presented with diabetes at the age of 43 years, was managed with metformin and sulfonylurea therapy for 25 years. A heterozygous relative of the proband developed diabetes later in life, at age 80 years. Although hs-CRP levels were low in the three carriers studied, the level of fucosylated glycan GP30 (low levels of which are predictive of HNF1A mutations) was above the cutoff in two of the three (18). In another study, a heterozygous proband received a diagnosis at age 39 years and had a BMI of 27 kg/m2. The proband’s father, who was diagnosed at age 50 years, and a sister, diagnosed at age 53 years, had only impaired fasting glycemia when studied (11,18). Thus the age of heterozygous carriers at diagnosis has been markedly older (11) than that of individuals who are homozygous (12, 15, and 16 years). This is consistent with incomplete penetrance, and reported data suggest that a high BMI might be required to unmask the effect of the variant in the heterozygous state.
In our study, two homozygous individuals had hs-CRP levels ≤0.2 mg/L, whereas the heterozygous father had a level of 1.4 mg/L—within the range observed for individuals with type 2 diabetes and those without diabetes (15,19). This higher level contrasts data from previous reports in which hs-CRP levels were all <0.3 mg/L in the heterozygous state (17), but it could be accounted for by his age and BMI.
The most compelling evidence favoring pathogenicity of the biallelically inherited p.A251T variant is the successful transition from insulin to low-dose sulfonylurea therapy after >10 years of diabetes. It would not be expected that alternative diabetes types could be effectively managed with low-dose glibenclamide replicating the observed responses to a sulfonylurea in individuals with typical heterozygous HNF1A mutations (20).
Conclusion
We conclude that biallelic hypomorphic variants of HNF1A may present with a typical heterozygous MODY phenotype. Hypomorphic alleles causing MODY may be encountered more frequently as diverse populations, particularly those with autozygosity, are studied using whole-exome or whole-genome approaches.
Supplementary Material
Article Information
Funding. This work was undertaken with funds from the Diabetes Research & Wellness Foundation (through a Sutherland-Earl Fellowship 2013–2016) and the Imperial College Healthcare Charity, and with infrastructure support from the National Institute for Health Research (NIHR) Imperial Biomedical Research Centre (BRC), Imperial Clinical Research Facility, and Clinical Research Network. S.M. is currently supported by a Future Leaders Mentorship Award from the European Association for the Study of Diabetes. A.J. was a Diabetes UK George Alberti Clinical Research Fellow when contributing to this study. S.E. received a Senior Investigator Award from Wellcome Trust. A.L.G. is a Wellcome Senior Fellow in Basic Biomedical Science. Part of this work was funded in Oxford by the Wellcome Trust (grants 095101 and 200837 [both to A.L.G.]). The research was also funded by the NIHR Oxford and BRC (to A.L.G.).
The views expressed are those of the author(s) and not necessarily those of the funding bodies or oganizations involved in this research.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. S.M. undertook patient studies and in vitro experiments and wrote and edited the manuscript. N.H., A.J.B., and A.J. contributed to in vitro experiments and reviewed the manuscript. R.C., K.C., and S.E. contributed to genetic analyses and reviewed the manuscript. S.M., J.V., N.S.O., and A.L.G. contributed to the discussion and reviewed and edited the manuscript. All authors approved the manuscript. N.S.O. and A.L.G. are the guarantors of this work and, as such, had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Part of this work was presented as a poster at the 76th Scientific Sessions of the American Diabetes Association, New Orleans, LA, 10–14 June 2016.
Footnotes
N.S.O. and A.L.G. jointly supervised this work.
This article contains Supplementary Data online at https://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc19-1843/-/DC1.
References
- 1.Frayling TM, Evans JC, Bulman MP, et al. beta-cell genes and diabetes: molecular and clinical characterization of mutations in transcription factors. Diabetes 2001;50(Suppl. 1):S94–S100 [DOI] [PubMed] [Google Scholar]
- 2.Shepherd MH, Shields BM, Hudson M, et al.; UNITED study . A UK nationwide prospective study of treatment change in MODY: genetic subtype and clinical characteristics predict optimal glycaemic control after discontinuing insulin and metformin. Diabetologia 2018;61:2520–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjørkhaug L, Sagen JV, Thorsby P, Søvik O, Molven A, Njølstad PR. Hepatocyte nuclear factor-1 alpha gene mutations and diabetes in Norway. J Clin Endocrinol Metab 2003;88:920–931 [DOI] [PubMed] [Google Scholar]
- 4.Galán M, García-Herrero C-M, Azriel S, et al. Differential effects of HNF-1α mutations associated with familial young-onset diabetes on target gene regulation. Mol Med 2011;17:256–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harries LW, Brown JE, Gloyn AL. Species-specific differences in the expression of the HNF1A, HNF1B and HNF4A genes. PLoS One 2009;4:e7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Althari S, Gloyn AL. When is it MODY? Challenges in the interpretation of sequence variants in MODY genes. Rev Diabet Stud 2015;12:330–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sagen JV, Bjørkhaug L, Haukanes BI, et al. The HNF1A mutant Ala180Val: clinical challenges in determining causality of a rare HNF1A variant in familial diabetes. Diabetes Res Clin Pract 2017;133:142–149 [DOI] [PubMed] [Google Scholar]
- 8.Monies D, Maddirevula S, Kurdi W, et al. Autozygosity reveals recessive mutations and novel mechanisms in dominant genes: implications in variant interpretation. Genet Med 2017;19:1144–1150 [DOI] [PubMed] [Google Scholar]
- 9.Ellard S, Lango Allen H, De Franco E, et al. Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia 2013;56:1958–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards S, Aziz N, Bale S, et al.; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juszczak A, Pavić T, Vučković F, et al. Plasma fucosylated glycans and C-reactive protein as biomarkers of HNF1A-MODY in young adult-onset nonautoimmune diabetes. Diabetes Care 2019;42:17–26 [DOI] [PubMed] [Google Scholar]
- 12.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res 2005;33:W382–W388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellard S, Colclough K. Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha (HNF1A) and 4 alpha (HNF4A) in maturity-onset diabetes of the young. Hum Mutat 2006;27:854–869 [DOI] [PubMed] [Google Scholar]
- 14.Chi Y-I, Frantz JD, Oh B-C, Hansen L, Dhe-Paganon S, Shoelson SE. Diabetes mutations delineate an atypical POU domain in HNF-1alpha. Mol Cell 2002;10:1129–1137 [DOI] [PubMed] [Google Scholar]
- 15.Owen KR, Thanabalasingham G, James TJ, et al. Assessment of high-sensitivity C-reactive protein levels as diagnostic discriminator of maturity-onset diabetes of the young due to HNF1A mutations. Diabetes Care 2010;33:1919–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oram RA, Jones AG, Besser REJ, et al. The majority of patients with long-duration type 1 diabetes are insulin microsecretors and have functioning beta cells. Diabetologia 2014;57:187–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thanabalasingham G, Huffman JE, Kattla JJ, et al. Mutations in HNF1A result in marked alterations of plasma glycan profile. Diabetes 2013;62:1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavić T, Juszczak A, Pape Medvidović E, et al. Maturity onset diabetes of the young due to HNF1A variants in Croatia. Biochem Med (Zagreb) 2018;28:020703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald TJ, Shields BM, Lawry J, et al. High-sensitivity CRP discriminates HNF1A-MODY from other subtypes of diabetes. Diabetes Care 2011;34:1860–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson ER, Flechtner I, Njølstad PR, et al.; Neonatal Diabetes International Collaborative Group . Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. N Engl J Med 2006;355:467–477 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.