Abstract
Non-alcoholic fatty liver disease (NAFLD) has a developmental origin and is influenced in utero. We aimed to evaluate if maternal diet intervention before pregnancy would be beneficial to reduce the risk of offspring NAFLD. In our study, female mice were either on a normal-fat diet (NF group), or a high-fat diet for 12 weeks and continued on this diet throughout pregnancy and lactation (HF group), or switched from HF to NF diet 1 week (H1N group), or 9 weeks (H9N group) before pregnancy. Compared to the NF offspring the H1N and HF, but not the H9N offspring, displayed more severe hepatic steatosis and glucose intolerance. More specifically, an abnormal blood lipid panel was shown in the H1N offspring and abnormal hepatic free fatty acid composition was present in both the HF and H1N offspring, while the H9N offspring displayed both at normal levels. These physiological changes were associated with desensitized hepatic insulin/AKT signaling, increased expression of genes and proteins for de novo lipogenesis and cholesterol synthesis, decreased expression of genes and proteins for fatty acid oxidation, increased Pcsk9 expression, and hypoactivation of AMPK signaling in the HF and H1N offspring. However, these effects were completely or partially rescued in the H9N offspring. In summary, we found that early maternal diet intervention is effective in reducing the risk of offspring NAFLD caused by maternal HF diet. These findings provide significant support for promoting the development of effective diet intervention strategies, policy for prevention of obesity and NAFLD, and improvement of health outcomes for mothers and children.
Introduction
The rapid rise in obesity and associated diseases throughout the world has major negative impacts on human health and healthcare resources. According to data from the National Center for Health Statistics, 71.6% of the adult population in the United states from 2015 to 2016 was overweight or obese in the United States (1). An estimated 18.5% of adolescents and US children were obese and nearly one-half of childbearing age women were overweight or obese (2, 3). Non-alcoholic fatty liver disease (NAFLD), regarded as the hepatic manifestation of metabolic syndrome, affects 10% to 24% of the general population in various countries. The prevalence of NAFLD is up to 75% in obese people (4). In recent years, the population of NAFLD patients has increased and is becoming younger, perhaps due to changes in diet structure and decreased physical activity (5). The American Heart Association Council on Epidemiology and Prevention states that obesity among girls and women of childbearing age is trans-generational, which may fuel the obesity epidemic for decades to come especially among children.
Recent study efforts have been put on investigating maternal over-nutrition to reflect the dietary habits of Western society. In both human and animal models, embryos exposed to over-nutrition during gestation have increased risks for obesity, diabetes, and other complications including NAFLD attributed to catch-up growth, increased adiposity, impaired glucose tolerance, impaired insulin sensitivity, and abnormal liver function in offspring (6–10). Thus, it has been suggested that prevention of obesity and its related diseases may need to begin before pregnancy (11–18). However, previous studies that evaluated pre-pregnancy dietary interventions composed of a balanced diet and regular physical activity only focus on the short-term effects on pregnancy outcomes, failing to look at the long-term effects of maternal diet on offspring (19–22).
Previously, we conducted a mouse study to evaluate if the transition of maternal diet from high-fat (HF) to normal-fat (NF) before pregnancy remediates the obesogenic effects of maternal HF diet on offspring 12-weeks post-weaning. We reported that neither a short (1-week) nor a medium (5-week), but a long-term (9-week) diet transition, effectively avoided the effects of maternal HF diet on exacerbating offspring obesity, glucose intolerance, adiposity and adipose tissue inflammation (23–25). Additionally, we found a sex-specific phenotype wherein male offspring from a dam with HF-to-NF transition one week prior to pregnancy had more severe hepatic steatosis than male offspring exposed to a continuous maternal HF diet (25). These results suggested that a proper maternal adaptation period before pregnancy is important to “re-program” offspring energy metabolism, especially fatty acid metabolism in the liver. We hypothesize that starting a maternal diet transition early enough would be beneficial in reducing NAFLD in male offspring. Thus, the aim of this study was to investigate if a maternal diet intervention could release the “priming” effects of maternal HF diet on NAFLD observed in male offspring and to understand the underlying mechanisms.
Materials and Methods
Experimental design (Table 1)
Table 1.
Study Design for maternal diet transition from HF to NF diet before pregnancy.
| Maternal Diet | Offspring Diet | |||
|---|---|---|---|---|
| Pre-pregnancy | Transition | Pregnancy/Lactation | After Weaning | |
| REF (n=15) | CD | - | CD | CD |
| NF (n=15) | NF | - | NF | HF |
| HF (n=15) | HF | - | HF | HF |
| H1N (n=15) | HF | NF (1 week) | NF | HF |
| H9N (n=15) | HF | NF (9 weeks) | NF | HF |
Diet description:
REF Rodent diet: normal chow diet (CD, 4% kcal. from fat)
NF Rodent diet: 10% kcal from fat.
HF Rodent diet: 60% kcal from fat.
Four-week-old female mice (mixed background) were fed either a normal fat diet (10% kcal from fat, NF group) or a high fat diet (60% kcal from fat, HF) for 12 weeks. The female mice on HF diet either continued a HF diet through gestation and lactation (HF group), or were transferred to a NF diet at either one-week (H1N group) or nine-weeks (H9N group) prior to pregnancy. To ensure different diet transition periods, female mice were housed together with breeding males for 24 hours, at the end of diet transition period (1 week or 9 weeks). The vaginal plug was also checked and recorded early the next morning to confirm if timely mating occurred. The breeding pairs were then separated and the female mouse was single housed until the end of lactation stage. The H1N and the H9N mothers remained on the NF diet through gestation and lactation. Our previous study has demonstrated a sex-dependent phenotype of fatty liver disease in offspring mice, with female offspring only displaying very mild fat deposition regardless of treatment groups (24, 25). Thus, three male offspring were randomly selected per litter from five different litters for a total of 15 (average litter size = 8) for each group and were given a HF diet for 12 weeks post-weaning prior to sacrifice. A separate group, birthed from the breeders who adhered to NF diet, was fed NF diet for 12 weeks and utilized as a reference control group (REF group).
Mouse experiments were conducted according to a protocol reviewed and approved by the Institutional Animal Care and Use Committee of the University of North Dakota and Texas A&M University, in compliance with the USA Public Health Service Policy on Humane Care and Use of Laboratory Animals.
Diet Composition
Diets were purchased from Research Diets, LLC (New Brunswick, NJ). The normal fat diet (Cat#D12450B) had an energy density of 3.771 kcal/g (10% fat energy, 70% carbohydrate energy, and 20% protein energy). The HF diet (Cat#D12492) had an energy density of 5.157 kcal/g (60% fat energy, 20% carbohydrate energy, and 20% protein energy). The fat source is composed of 92% lard and 8% soybean oil.
Antibodies
Antibodies against IRS1, P-IRS1–612, AKT, p-AKT-473, PPAR-α, Srebp1, AMPK, p-AMPK-α were purchased from Cell Signaling Technology (Cell signaling Technology, Danvers, MA).
Western Blots
About 100 mg of frozen tissue was homogenized in 1 ml of lysis buffer (10 mL of RIPA, 1 complete ULTRA Tablet, 1 tablet of Phosphatase inhibitor, and 100 μL of 200 mM PMSF). The protein concentration was determined using a BCA protein concentration assay. Aliquots of 150 μg of total protein were run through routine western blots.
Realtime-PCR (RT-PCR)
Total RNA was extracted from visceral adipose tissue using Trizol reagent (Invitrogen). cDNA was converted using Ready Script cDNA synthesis mix (Sigma Aldrich, St. Louis, MO). Primers used are listed in Table 2. RT-PCR was performed using a POWER SYBER Green PCR Master Mix from Applied Biosystems on a CFX384 Touch Real-Time PCR Detection System (BioRad Laboratories, Hercules CA). Results were analyzed using the ΔΔCT method with GAPDH as the normalization control (26).
Table 2.
Primers used for mRNA quantification by real-time PCR
| Gene | Forward primer 5′–3′ | Reverse primer 5′–3′ |
|---|---|---|
| Srebf1 | TGACCCGGCTATTCCGTGA | CTGGGCTGAGCAATACAGTTC |
| Fasn | GGAGGTGGTGATAGCCGGTAT | TGGGTAATCCATAGAGCCCAG |
| Acc1 | ATCCAGGCCATGTTGAGACG | AGATGTGCTGGGTCATGTGG |
| Acc2 | CGCTCACCAACAGTAAGGTGG | GCTTGGCAGGGAGTTCCTC |
| Elovl6 | GAAAAGCAGTTCAACGAGAACG | AGATGCCGACCACCAAAGATA |
| Scd1 | TTCTTGCGATACACTCTGGTGC | CGGGATTGAATGTTCTTGTCGT |
| Cd36 | GAACCAAACTGAGGAATGGATCT | GAACCAAACTGAGGAATGGATCT |
| Srebf2 | GCAGCAACGGGACCATTCT | CCCCATGACTAAGTCCTTCAACT |
| Cyp51 | AACGAAGACCTGAATGCAGAAG | GTGGGCTATGTTAAGGCCACT |
| Hmgcr | AGCTTGCCCGAATTGTATGTG | TCTGTTGTGAACCATGTGACTTC |
| Hmgcs1 | AAATGCCAGACCTACAGGTGG | ATGCTGCATGTGTGTCCCA |
| Hmgcs2 | GAAGAGAGCGATGCAGGAAAC | GTCCACATATTGGGCTGGAAA |
| Abca1 | GCTTGTTGGCCTCAGTTAAGG | GTAGCTCAGGCGTACAGAGAT |
| Ldlr | AGAGCCTGTGCCGAGATGT | TGGTCTGAGTAGATCCAGGAGT |
| Pcsk9 | TTGCCCCATGTGGAGTACATT | GGGAGCGGTCTTCCTCTGT |
| Igf1 | CTGGACCAGAGACCCTTTGC | GGACGGGGACTTCTGAGTCTT |
| Insig1 | CTAGTGCTCTTCTCATTTGGCG | AGGGATACAGTAAACCGACAACA |
| Insig2a | GGAGTCACCTCGGCCTAAAAA | CAAGTTCAACACTAATGCCAGGA |
| Insr | TCAAGACCAGACCCGAAGATT | TCTCGAAGATAACCAGGGCATAG |
| Lepr | ATGTGCCCTTCCGATATACAACC | CGTGTCATCCACTAATCTTCTGG |
| Adipor1 | AGACAACGACTACCTGCTACA | GTGGATGCGGAAGATGCTCT |
| Adipor2 | GGAGTGTTCGTGGGCTTAGG | GCAGCTCCGGTGATATAGAGG |
| Cpt1a | CTCCGCCTGAGCCATGAAG | CACCAGTGATGATGCCATTCT |
| Cpt1b | GCACACCAGGCAGTAGCTTT | CAGGAGTTGATTCCAGACAGGTA |
| Cpt2 | CAAAAGACTCATCCGCTTTGTTC | CATCACGACTGGGTTTGGGTA |
| Acsl1 | CGCACCCTTCCAACCAACA | CGCTATTTCCACTGACTGCAT |
| Mlycd | GCACGTCCGGGAAATGAAC | GCCTCACACTCGCTGATCTT |
| B2m* | TTCTGGTGCTTGTCTCACTGA | CAGTATGTTCGGCTTCCCATTC |
B2m was selected as the internal control.
Intraperitoneal Glucose Tolerance Test (IPGTT)(25)
Offspring from each experimental group were fasted overnight and were subsequently subjected to an IPGTT the following morning. Glucose tolerance tests were performed as previously reported (25, 27). Glucose tolerance was assessed by area under the curve (AUC) analysis.
Mass spectrometry (MS) for hepatic free fatty acid, triglyceride and cholesterol esters
Liver samples were pulverized under liquid nitrogen conditions to a fine homogeneous powder and about 10 mg was extracted according to the Folch procedure (28). Fatty acids (FA) internal standards were added before extraction. A one hundred microliter aliquot was subjected to saponification to release FA before UPLC-MS analysis. Fatty acids were resolved on the ultra-performance liquid chromatography mass spectrometer (UPLC-MS) column and quantified against FA stable isotope labeled internal standards using a quadrupole time-of-flight mass spectrometer as previously described (29, 30). Briefly, the liquid chromatography (LC) system consisted of a Waters ACQUITY UPLC pump with a well-plate autosampler (Waters, Milford, MA) equipped with an ACQUITY UPLC HSS T3 column (1.8 μM, 100 A pore diameter, 2.1×150 mm, Waters) and an ACQUITY UPLC HSS T3 Vanguard precolumn (1.8 μM, 100 A pore diameter 2.1 × 5 mm, Waters). The column temperature was 55 °C and the autosampler temperature was 8 °C. The flow rate was 0.3 mL/min. Solvent A consisted of acetonitrile : water (40 : 60) with 10 μM ammonium acetate and 0.025% acetic acid. Solvent B was acetonitrile:2-propanol (10 : 90) containing 10 μM ammonium acetate and 0.02% acetic acid. Solvent B was initially held at 35% for 0.1 minutes and was then increased to 99% over 20 minutes using a linear gradient. Solvent B was held at 99% for 8 min before returning to initial conditions over 0.5 minutes. The column was equilibrated for 2.5 min between injections. The Q-TOF (Synapt G2-S, Waters) with electrospray ionization in negative ion mode was used for quantification. The cone voltage was 20 V and the capillary voltage was 1.51 kV. The source and desolvation temperatures were 110 °C and 350 °C respectively. The analyzer was operated with an extended dynamic range at 10,000 resolution (fwhm at m/z 554) with an acquisition time of 0.1s. MSE mode was used to collect data with the T-wave element alternating between a low energy of 2V and a high energy state where the transfer T-wave element voltage was 10–25 V (29). The cone gas flow rate was 10 L/h and the desolvation gas flow was 1,000 L/h. Leucine enkephalin (400 pg/μL, ACN:water, 50 : 50 by volume) was infused at a rate of 10 μL/min for mass correction. MassLynx V4.1 software (Waters) was used for instrument control, acquisition, and sample analysis. Saturated long chain FA was quantified against 16:0-[U-13C], medium chain FA against 14:0-d27, short chain – against 10:0-d19, mono-unsaturated – against 18:1-d17, di-unsaturated – against 18:2-[U-13C], and polyunsaturated – against 20:4-d8.
L-Carnitine detection
About 100 mg (±2 mg) of frozen liver tissue was homogenized after adding 4 mL methanol and 0.85 mL water per gram tissue. The methanol-chloroform method was used to separate the polar metabolites (upper methanol/water phase, contains L-Carnitine) and the lipophilic compounds (lower chloroform phase). Upper layer solvents were removed using a speed vacuum concentrator and under a stream of nitrogen. The samples and standard (Acetyl-d3-L-carnitine, CDN isotopes Quebec) were analyzed on a Waters ACQUITY UPLC TQD MS system (Waters Corp., Milford, MA) equipped with a column heater, sample manager, binary solvent manager, photodiode array eλ (PDA) detector, and ESI source. Samples were separated on an Acquity UPLC BEH HILIC column (100 mm × 2.1 mm (i.d), 1.7 um particle size). The internal standards were detected in positive ion mode with source temperature at 150 °C, desolvation temperature at 400 °C, desolvation gas (N2) flow rate at 700 L/h, and cone gas flow rate at 50 L/h. Capillary voltage was set at 3.5 kV and collision gas flow (Ar) at 0.15 L/h. Dwell time was set at 0.05 s with a span of 0.1 Da. The Cone and collision energy voltage, and MRM mass transitions are summarized in Table 3.
Table 3.
Cone and collision energy voltage and MRM mass transitions used for L-Carnitine detection
| Analyte | Cone voltage (V) | Collision energy (V) | Precursor ion (m/z) | Product ion (m/z) | Retention time (min) |
|---|---|---|---|---|---|
| L-carnitine | 30 | 20 | 162 | 85 | 4.3 |
Hepatic triglyceride (TG) measurement
Blood lipid levels were measured using an Adipogenesis Detection Kit (Abcam Inc, #ab102513, Cambridge, UK) according to the manufacturer’s instructions and as reported previously (25).
Blood Lipid Panel
The mouse serum total cholesterol, free cholesterol, HDL and LDL/VLDL cholesterol levels were detected using an HDL and LDL/VLDL cholesterol assay kit (Abcam, Cambridge, UK). Following the manufacturer’s instructions, the cholesterol standard curve was prepared by diluting the provided cholesterol standard. The concentration of lipid was calculated based on the standard curve generated.
Blood C-peptide content
Blood C-peptide level was detected using the ALPCO Mouse C-peptide ELISA kit (ALPCO, Salem, NH) according to the user’s manual.
RNA-Seq and data analysis
Illumina next-generation sequencing was performed at University of Chicago Genomics Core (Chicago, USA) using Illumina HiSeq2000 instruments. The RNA-Seq data analysis was performed as published previously (31). Briefly, RNA-Seq short reads were mapped to mouse reference genome GRCm38 using a Bowtie2 package. The gene expression level was then quantified by a Cufflink package. The hierarchical clustering analysis (HCA) was used to assess the global gene-expression variations. Regression analysis was used to identify the genes that were correlated with the different maternal diet. Genes that showed significant correlation were selected by controlling the false discovery rate at 10%. The activated gene pathways were analyzed using a hypergeometric distribution-based method, which was developed in house using R programing language. The biochemical pathway information was obtained from KEGG (www.genome.jp/kegg).
Statistical Analysis
Measurements for single time points were analyzed by Fisher’s least significant difference test to take into account the multiple comparisons between groups REF, NF, HF, H1N, and H9N. Fisher’s least significant difference test was performed by first carrying out a one-way analysis of variance for all treatment groups. For the longitudinal data such as body weight, a linear mixed model was used for the analysis of repeated measures with each individual mouse as a random effect. A p-value less than 0.05 is considered a significant difference, while a p-value less than 0.1 is considered a marginal significance. All analyses were carried out using SAS JMP software (SAS Institute Inc., Cary, NC, USA). Data is presented as Mean ± SE.
Results
The H1N diet resulted in rapid body weight gain and glucose intolerance in male offspring, while the H9N diet only caused moderate body weight gain
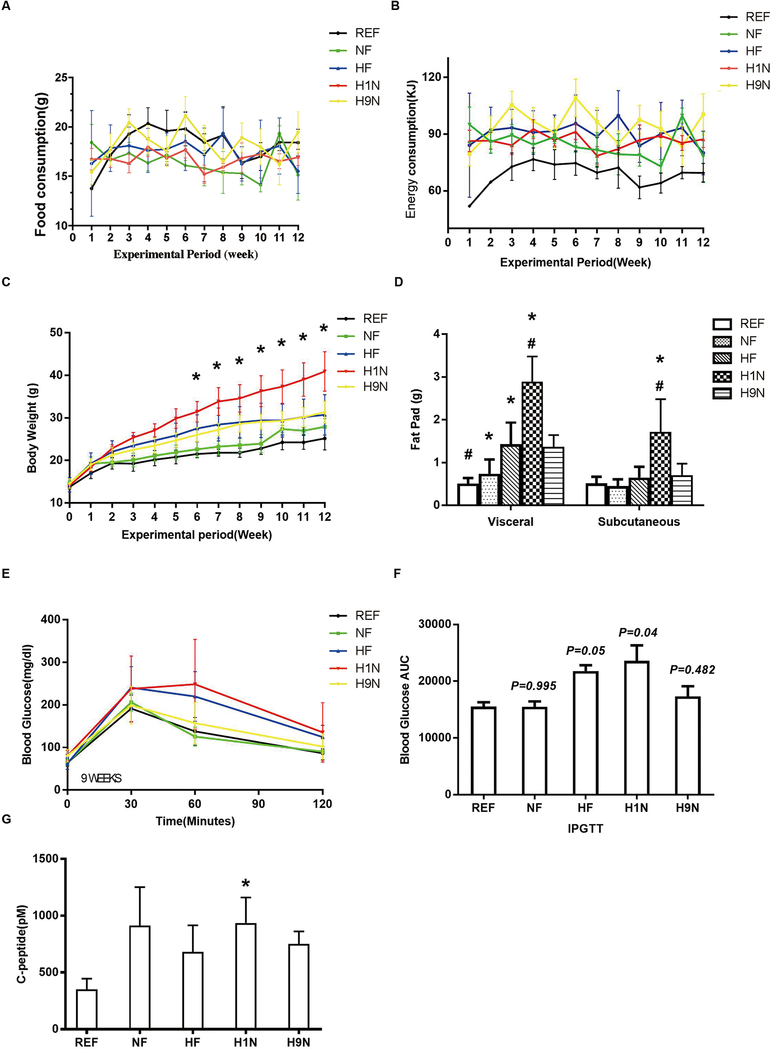
The weekly food consumption (Fig 1A) of male offspring mice was recorded for calculating the weekly energy consumption via diet (Fig 1B). Food consumption was the same among all groups; however, the energy consumption of the REF group on post-weaning NF diet was significantly lower than the other four groups on post-weaning HF-diet (Figs 1A and B). The energy consumption among NF, HF, H1N and H9N groups were not different (Fig 1B). The offspring mice were weaned at similar body weights. On the post-weaning HF-diet, all offspring constantly gained weight during the 12-week experimental period, with H1N offspring gaining the fastest. Longitudinal analysis of repeated measurements showed that H1N offspring gained significantly more weight than the REF or NF offspring. Interestingly, both the HF offspring and the H9N offspring weighed marginally more than the REF offspring (Fig 1C, P = 0.093 for the HF offspring and P = 0.071 for the H9N offspring). We further weighed the posterior subcutaneous fat tissue and the gonadal adipose tissue of the offspring mice. The results showed significantly more visceral fat in the NF, HF and H1N offspring compared to the REF offspring, with the H1N offspring having the largest amount (Fig 1D). The amount of subcutaneous fat pad from the highest to the lowest rank is H1N > H9N ≈ HF ≈ NF ≈ REF (Fig 1D).
Figure1. The H1N diet resulted in rapid body weight gain and glucose intolerance in male offspring, while the H9N diet only caused moderate body weight gain.
1A. Food consumption of male offspring mice was recorded weekly since post-weaning for 12 weeks.
1B. Energy consumption was calculated weekly since post-weaning for 12 weeks.
1C. Body weight of male offspring was recorded weekly since post-weaning for 12 weeks.
1D. Posterior subcutaneous and gonadal adipose tissue of male offspring mice were collected and weighed.
1E. IPGTT was conducted on male offspring mice at the end of 9 weeks of post-weaning HF diet.
1F. Area under the curve (AUC) was calculated for the results of IPGTT in male offspring mice.
1G. Blood C-Peptide was measured in male offspring mice.
Data is presented as Mean ± SE, n=15 from five litters. * P<0.05 vs. REF group. # P<0.05 vs. NF group.
The IPGTT was performed on the offspring at the end of week 9. At this time point, all offspring had similar fasting glucose levels (Fig 1E). The NF offspring were glucose tolerant at checkpoint; however, neither the HF nor the H1N offspring were glucose tolerant (Figs 1E and F). In contrast, the H9N offspring were glucose tolerant (Figs 1E and F). Similarly, the H1N offspring, but not the H9N nor HF offspring, had higher levels of blood C-Peptide than that of the REF offspring. (Fig 1G).
The maternal HF and H1N diets resulted in hepatic steatosis and a disrupted lipid panel in male offspring, while the maternal H9N diet avoided such effects
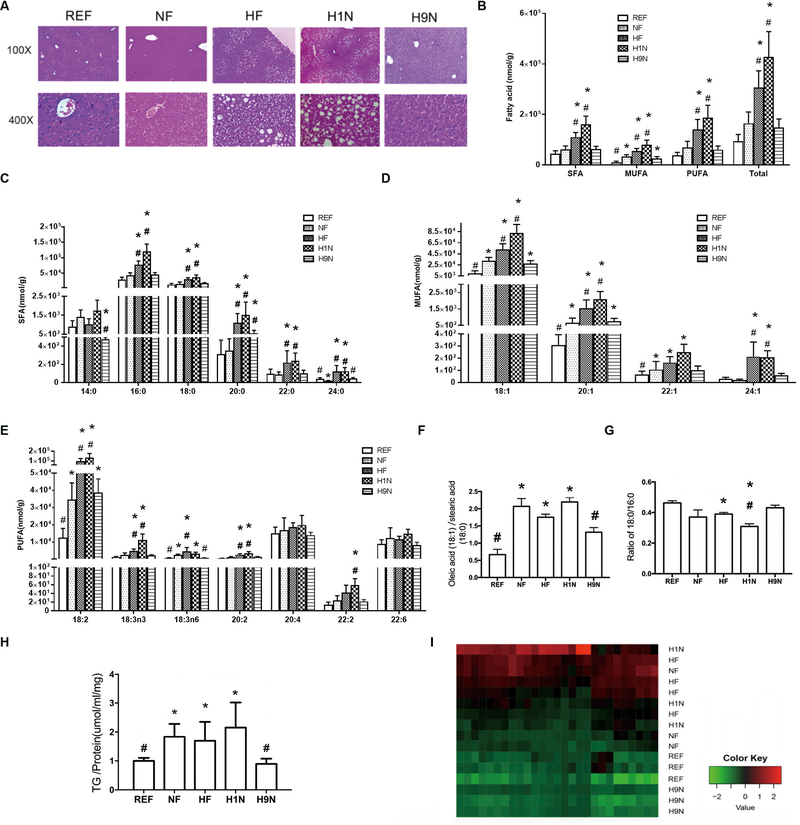
We next evaluated if lipid metabolism in the liver was affected by different maternal diet transitions before pregnancy. Histology showed that the NF offspring only had microvesicular fat in hepatocytes instead of visible ballooning cells, in which the nuclei remain centrally located and appeared to be indented by the small fat droplets (Fig 2A). However, hepatic steatosis was observed in both HF and H1N offspring along with ballooning cells that indicated intracellular lipid accumulation. In contrast, fatty acid accumulation was not visible in hepatocytes of the H9N offspring.
Figure2. The maternal HF and H1N diets resulted in hepatic steatosis and a disrupted lipid panel of male offspring, while the maternal H9N diet avoided such effects.
2A. HE staining was performed on the liver tissue of male offspring mice.
2B-E. The major hepatic long or very long chain free fatty acid (L/VLCFFA) panel (chain lengths of 12–24 carbons) were measured via high-resolution UPLC-MS/MS method after Folch extraction.
2F. The relative amount of oleic acid (18:1) on the stearic acid (18:0) was calculated by dividing the amount of oleic acid with the amount of stearic acid.
2G. The ratio of stearic acid (18:0) to the palmitic acid (16:0) was calculated by dividing the amount of stearic acid with the amount of palmitic acid.
2H. Hepatic TG content was detected in male offspring mice.
2I. A heatmap for the types of TG detected via high-resolution UPLC-MS/MS method was generated by cluster analysis.
Data is presented as Mean ± SE, n = 5 from 5 litters. * P<0.05 vs. REF group. # P<0.05 vs. NF group.
Using a high-resolution UPLC-MS/MS method, we determined whether the major hepatic long or very long chain free fatty acid (FFA) panel (chain lengths of 12–24 carbons) was affected by different maternal diet interventions before pregnancy (Figs 2B–H). The total FFA was significantly greater in H1N and HF offspring than in the NF offspring (Fig 2B). In addition, the saturated FFA (SFA) in the liver of H1N and HF offspring was higher than that of NF offspring (Fig 2B). Of note, the H1N and HF offspring displayed higher amounts of monounsaturated FFA (MUFA) and polyunsaturated FFA (PUFA). Generally, the HF and H1N diets increased most of the major SFA (Fig 2C), MUFA (Fig 2D) and PUFA (Fig 2E), while the H9N diet avoided such effects in the FFA panel in hepatocytes. Importantly, offspring of NF, HF, and H1N groups significantly enhanced the ratio of oleic acid (18:1) to stearic acid (18:0) compared to REF offspring whereas H9N offspring had the level recovered (Fig 2F). In addition, the HF diet significantly decreased the ratio of C18:0 to C16:0. Compared to the NF offspring, the level of C18:0 to C16:0 in HF offspring was similar, but this ratio was much lower in H1N offspring when compared to NF offspring. However, H9N offspring had no changes in this ratio (Fig 2G). These results suggest that maternal diet differentially affects the hepatic FFA panel in offspring, not only in quantity but also in FFA subtype.
With a disruption of the hepatic FFA panel, we asked if the hepatic TG content was also altered by different maternal diet interventions. The NF offspring had higher TG content in the liver, showing a positive effect of post-weaning HF diet (Fig 2H). Both the HF and H1N offspring had increased TG content. Surprisingly H9N offspring had lower TG content than NF offspring, which made it a level similar to REF offspring (Fig 2H). We further measured several major types of TG and found that the NF, HF, and H1N groups were unable to be clustered, which were distinct from the cluster of H9N and REF offspring (Fig 2I).
The H1N offspring had disrupted blood lipids, which were normal in the H9N offspring
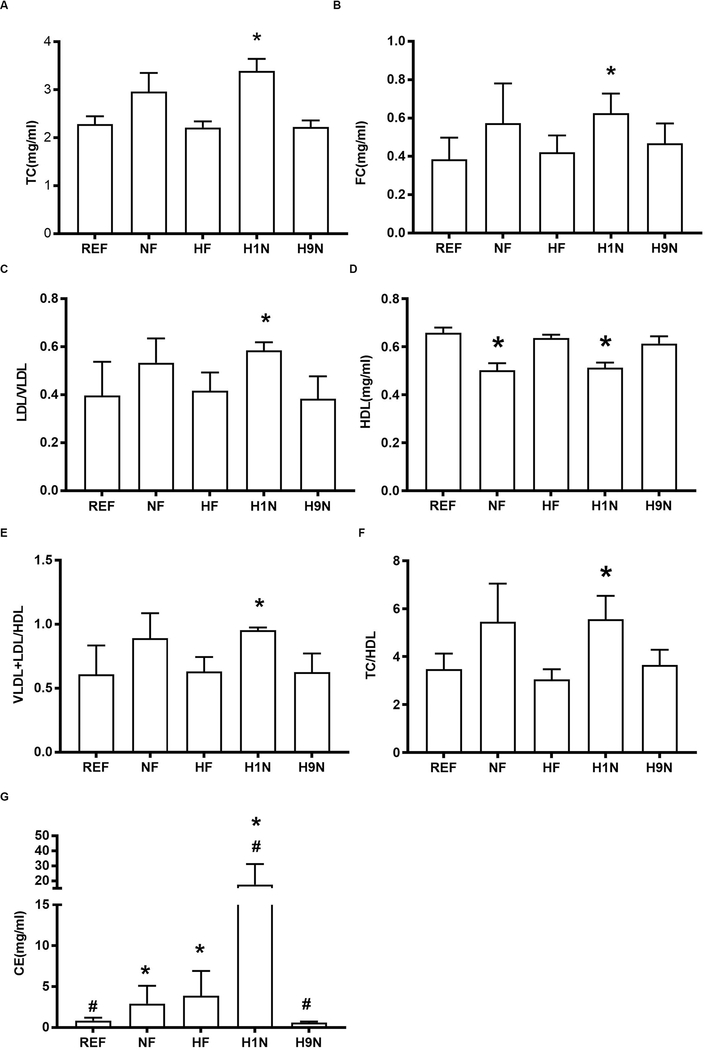
We wondered if the disrupted hepatic FFA and TG panel in HF and H1N offspring was associated with disrupted blood lipid levels. The post-weaning HF diet (NF vs. REF) did not affect the total cholesterol (TC) (Fig 3A), total amount of free cholesterol (FC) (Fig 3B) or blood VLDL and LDL content (VLDL+LDL) (Fig 3C), but significantly decreased HDL content (Fig 3D). The ratio of TC/HDL and (LDL+ VLDL)/HDL were known to be associated with a higher risk of cardiovascular disease (32). The maternal HF diet did not seem to change the blood lipid panel. However, H1N offspring had all six biochemical parameters significantly increased. In contrast, H9N offspring had an entirely normal lipid panel (Figs 3A–F). With an overall similar trend to the blood lipid panel, the hepatic cholesterol ester content from the highest to the lowest was H1N > HF ≈ NF > REF ≈ H9N (Fig 3G).
Figure3. The H1N offspring had disrupted blood lipids, which were normal in the H9N offspring.
3A-D. The blood lipid panel of male offspring mice including the amount of total cholesterol (TC), free cholesterol (FC), LDL/VLDL, and HDL was detected using a cholesterol assay kit from Abcam.
3E. The ratio of (VLDL+LDL)/HDL was calculated by dividing the sum of VLDL and LDL with the amount of HDL.
3F. The ratio of TC/HDL was calculated by dividing the amount of TC with the amount of HDL.
3G. The hepatic cholesterol of male offspring mice was detected using a cholesterol detection kit from Sigma.
Data is presented as Mean ± SE, n=5 from 5 litters. * P<0.05 vs. REF group. # P<0.05 vs. NF group.
RNA-Seq analysis identified significant KEGG pathways involving lipid metabolism by different maternal diet interventions
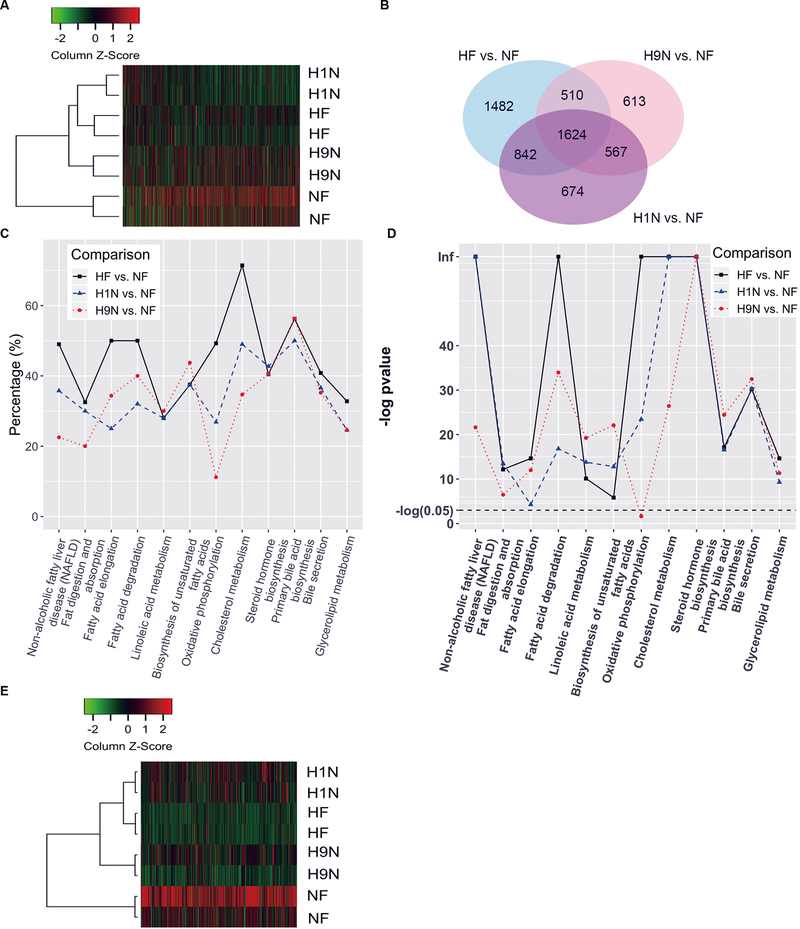
Next, we investigated the molecular mechanism contributing to the phenotypical differences between H1N and the H9N offspring. For each sample, 20–25 million short reads were generated using Illumina HiSeq 2000, and >90% of short reads were mapped to the mouse reference genome GRCm38. Hierarchical clustering analysis of the 2,000 genes with the largest variance across the groups showed that HF and H1N had similar transcript profiles. The H9N offspring clustered with the H1N and the HF groups, which are distinct from the NF offspring (Fig 4A). We found 4458, 3314, and 3707 differentially expressed genes (DEGs) for HF, H1N and H9N respectively when compared with NF offspring using a false discovery rate 0.05 as the cutoff. There were 2,466 DEGs overlapping between the comparisons of HF and H1N versus NF, as shown by Venn diagram. There were 2,191 DEGs overlapping between H9N and H1N versus NF. The number of overlapped DEGs between the comparisons of H9N and HF versus NF was 2,134. There were 1,624 overlapping DEGs among all three comparisons (Fig 4B).
Figure4. RNA-Seq analysis identified significant KEGG pathways involving lipid metabolism by different maternal diet interventions.
4A. RNA-seq analysis using Illumina HiSeq 2000 were performed on NF, HF, H1N and H9N offspring livers. Hierarchical clustering analysis was performed on the top 2,000 DEGs.
4B. DEGs for HF, H1N and H9N when compared with NF offspring using false discovery rate 0.05 as the cutoff. DEGs overlapping among the comparisons of HF, H1N and H9N versus NF was shown by Venn diagram.
4C. Gene ontology (GO) analysis identified distinct KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways by HF, H1N, and H9N diets. The percentile of DEGs in each pathway are plotted.
4D.The –Log (p-value) of the altered KEGG signaling pathway are plotted.
4E. DEGs involved in the lipid metabolism signaling pathways were analyzed by Hierarchical clustering analysis.
In addition, gene ontology (GO) analysis identified distinct KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways following HF, H1N, and H9N diets. We noticed several significant KEGG pathways were involved in lipid metabolism and homeostasis, including NAFLD, fat digestion and absorption, fatty acid elongation, fatty acid degradation, linoleic acid metabolism, biosynthesis of unsaturated fatty acids, oxidative phosphorylation, cholesterol metabolism, steroid hormone biosynthesis, primary bile acid synthesis, bile secretion, and glycerolipid metabolism (Fig 4C). Overall, HF offspring had more DEGs of the significant pathways than H1N offspring, which had more DEGs than H9N offspring. Compared to the NF offspring, three pathways (NAFLD, oxidative phosphorylation and cholesterol metabolism) had more than or close to 50% of DEGs in HF offspring, while the number decreased in H9N offspring to half of that of HF offspring. The H1N offspring had the lowest percentile of DEGs in the pathways for fatty acid elongation and fatty acid degradation. There were also pathways with similar or slightly lower percentage of DEGs among HF, H1N, and H9N offspring including linoleic acid metabolism, biosynthesis of unsaturated fatty acid, steroid hormone biosynthesis, primary bile acid biosynthesis, and bile secretion. Among all these pathways, only the pathway for oxidative phosphorylation was insignificantly associated with the H9N diet (Fig 4D). We further performed a hierarchical clustering analysis to show that the DEGs, involved in the mentioned signaling pathways for lipid metabolism, of H1N offspring clustered together with HF offspring, which further grouped with those of H9N offspring (Fig 4E). The DEGs of NF offspring was distinct from the hierarchy of HF, H1N, and H9N. These results suggest that different maternal diets differentially affected the lipid metabolism at the mRNA level.
The maternal HF and H1N diets increased the gene expression for fatty acid and cholesterol biosynthesis while the H9N diet had no such effects
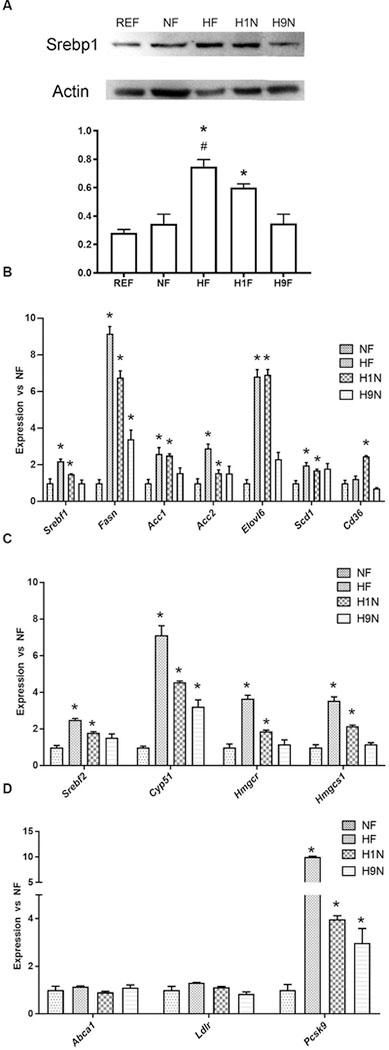
To confirm that different maternal diet transition strategies affect lipid metabolism differently at the gene expression level, we first measured the expression of key lipogenesis genes. Insulin promotes lipogenesis by activation of sterol regulatory element binding protein 1c (SREBP1c), a transcription factor that promotes the expression of lipogenic genes including Fasn and Acc (33). The HF and H1N offspring had significantly higher levels of SREBP1c than REF offspring, and its level in HF offspring was higher than that of NF offspring. However, the H9N offspring had a similar amount of SREBP1c as the NF and REF offspring (Fig 5A). The changes of Srebp1c found at the RNA level were similar to levels measured at the protein level (Fig 5B, Srebf1). Consistently, we observed significantly enhanced Fasn, Acc1, and Acc2 expression in the HF and H1N hepatocytes versus the NF hepatocytes, whereas these gene expression changes were either completely (Acc1 and Acc2) or partially (Fasn) returned to normal in H9N offspring hepatocytes (Fig 5B). We further measured two important genes, one for fatty acid elongation (Elovl6) and the other for fatty acid desaturation (Scd1). The Elovl6 expression was remarkably elevated in HF and H1N offspring versus NF offspring, while completely recovered in H9N offspring (Fig. 5B). Similarly, HF and H1N offspring had higher expression of Scd1 than NF offspring, while H9N offspring expressed a similar level of Scd1 as NF offspring. In addition, Cd36, an important gene encoding fatty acid translocase for selective cholesteryl ester and fatty acid uptake, was upregulated in H1N offspring compared to NF offspring, while HF and H9N offspring had similar expression levels (Fig 5B).
Figure 5. The maternal HF and H1N diets enhanced expression of genes in fatty acids and cholesterol biosynthesis while the H9N diet avoided such effects.
5A. The expression of SREBP1c in liver tissue was measured by western blots of male offspring mice.
5B. The hepatic expression of Srebf1, Fasn, Acc1, Acc2, Elovl6, Scd1, and Cd36 of male offspring mice was measured by RT-PCR.
5C. The hepatic expression of Srebf2, Cyp51, Hmgcr, and Hmgcs1 of male offspring mice was measured by RT-PCR.
5D. The hepatic expression of Abca1, Ldlr and Pcsk9 of male offspring mice was measured by RT-PCR.
Data is presented as Mean ± SE, n = 5 from 5 litters. * P < 0.05 vs. REF group. # P < 0.05 vs. NF group.
Srebp-2 is an important transcription factor that regulates cholesterol metabolism (34). The Srebp-2 expression was higher in the HF and H1N offspring compared to NF offspring liver while H9N offspring had expression levels similar to NF offspring (Fig 5C). We further assessed the expression of key genes encoding the rate-limiting enzyme for de novo cholesterol biosynthesis, Hmgcr and Hmgcs1. Clearly, both the HF and H1N diet significantly enhanced the expression of both genes, while the H9N diet avoided such effects (Fig 5C). Cyp51a1 is the only cytochrome P450 enzyme involved in de novo cholesterol biosynthesis (35). Expression of Cyp51a1 was significantly higher in HF and H1N offspring compared to NF offspring, while Cyp51a1 expression was partially retracted in H9N offspring. (Fig 5C HF vs. H9N: 7.129 ± 0.509 vs. 3.230 ± 0.363, P < 0.05). These results suggested an increase in de novo cholesterol biosynthesis in HF and H1N offspring.
Furthermore, we observed a 9-fold increase in Pcsk9 expression in HF (10.04±0.12) and 3-fold increase in H1N offspring (3.97±0.15) compared to NF offspring (1.00±0.23). In contrast, only a 2-fold increase was measured in H9N offspring (2.98±0.61) (Fig 5D). We did not detect expression differences of Abca1 and Ldlr among the groups.
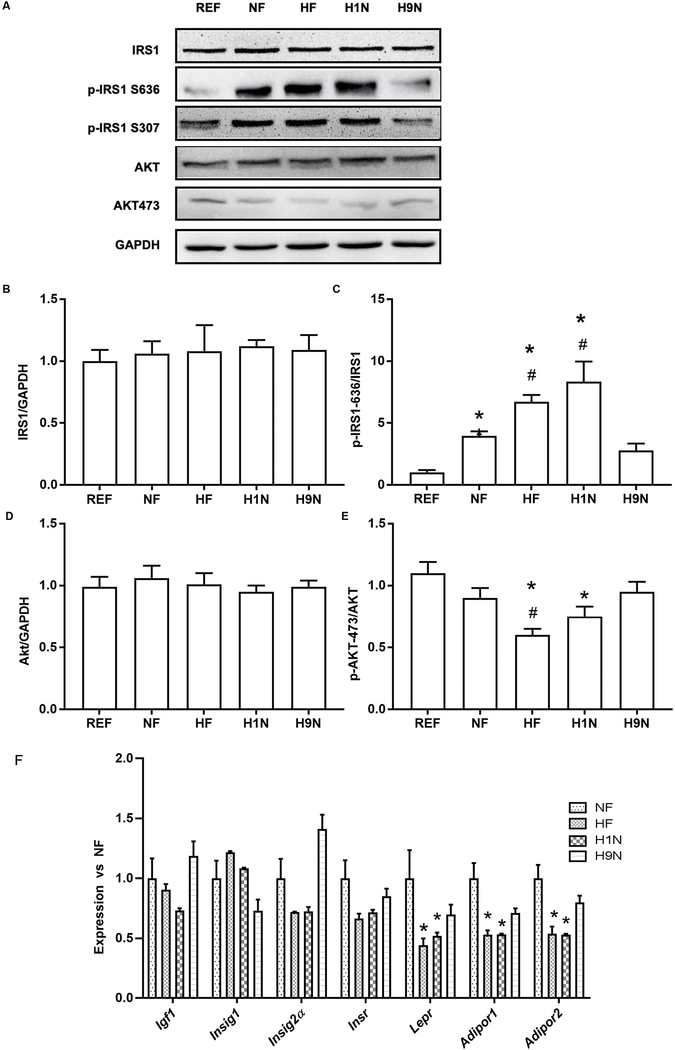
The HF and H1N diet desensitized the Insulin/AKT signaling in liver, while the H9N diet avoided such effects
In hepatocytes, insulin sensitivity was disrupted by the post-weaning HF diet, evidenced by over-phosphorylation of IRS1 at Ser636 (p-IRS1636) in the NF offspring. The maternal HF and H1N diets further over-phosphorylated IRS1 at Ser636 (Figs6 A–C) and thus resulted in the inhibition of insulin signaling that dephosphorylates its downstream p-AKT at Ser473 (Figs 6A and E). In contrast, the maternal H9N diet avoided such effects by normalizing the phosphorylation of IRS1 and its downstream AKT. Furthermore, we measured several key genes whose hepatic expression reflects insulin sensitivity including Igf1, Insig1, Insig2a, Insr, Lepr, Adipor1, and Adipor2. Both the maternal HF and the H1N diet significantly decreased the expression of Lepr, Adipor1, and Adipor2 in offspring liver (Fig 6F). However, their expressions were recovered in H9N offspring (Fig 6F). Although there was a decreasing trend for the expression of Igf1, Insig1, Insig2a, and Insr in the HF and H1N offspring versus the NF offspring, the changes were not significant. To be noted, the expression of Insig2a (0.722 ± 0.039 vs. 1.412 ± 0.118, p < 0.05 compared to H1N group) and Igf1 (0.731 ± 0.020 vs. 1.188 ± 0.122, p < 0.05 compared to H1N group) in H9N offspring was significantly higher than H1N offspring and was similar to the REF offspring.
Figure6. The HF and H1N diet desensitized Insulin/AKT signaling in liver disrupted by post-weaning HF diet, while the H9N diet avoided such effects.
6A. The expression of IRS-1, phosphorylation of IRS-1 at Ser636, the expression of Akt, phosphorylation of Akt at Ser473 was measured by western blots in male offspring mice.
6B-E. Relative gene expressions are shown as ratios of IRS1/GAPDH, Phospho-IRS-1-Ser636/ IRS-1, Akt/GAPDH, Phospho-Akt (Ser473)/ Akt.
6F. The expression of Igf1, Insig1, Insig2a, Insr, Lepr, Adipor1, and Adipor2 was measured by RT-PCR in the liver of male offspring mice.
Mice were sacrificed under a fasting status, thus the Insulin and AKT signaling is measured at basal level. Data is presented as Mean ± SE, n = 5 from 5 litters. * P < 0.05 vs. REF group. # P < 0.05 vs. NF group.
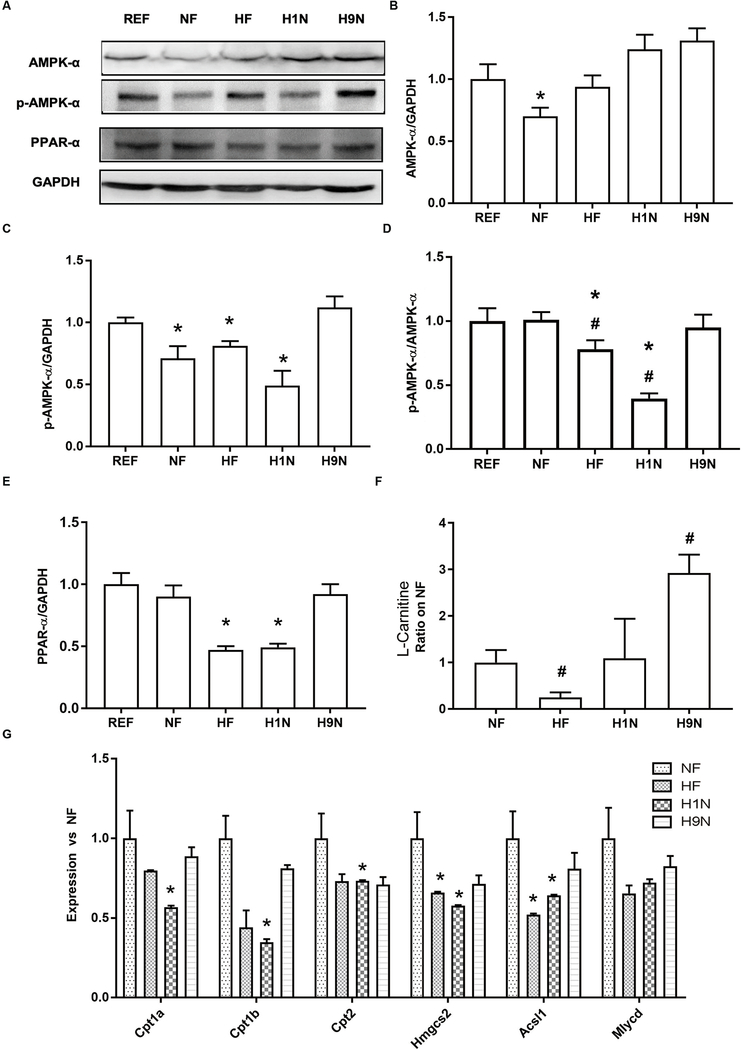
The maternal HF and H1N diets inhibited hepatic AMPK signaling activity and decreased expression of β-oxidation related genes, while the H9N diet avoided such effects
We measured the activity of AMPK signaling to evaluate if changes in lipid metabolism in HF, H1N, and H9N offspring were associated with its deactivation or overactivation. We observed decreased AMPK expression in the NF offspring, but not the other groups (Figs 7A and B). The results showed a decreased ratio of p-AMPK-α/GAPDH, in the NF, HF, and H1N groups compared to the REF offspring (Figs 7A and C). The levels of p-AMPK-α/AMPK-α was significantly lower in HF and H1N offspring than NF or REF offspring (Figs 7A and D). However, these ratios in H9N offspring were not different from REF or NF offspring (Figs 7D and E). These results suggested that the HF and H1N diet, but not the H9N diet, deactivated AMPK signaling.
Figure7. The maternal HF and H1N diets inhibited the hepatic AMPK signaling activity and overexpression of β-oxidation related genes, while the H9N diet avoided such effects.
7A. The expression of AMPK-α, phosphorylation of AMPK-α, and PPAR-α were detected by western blots in male offspring mice.
7B-E. Relative expressions were expressed as ratios of AMPK-α/GAPDH (B), Phospho- AMPK-α/GAPDH (C), Phospho- AMPK-α/ AMPK-α (D) and PPAR-α/ GAPDH(E).
7F. L-Carnitine content was detected via high-resolution UPLC-MS/MS method.
7G. The hepatic expression of key genes for fatty acid degradation and β-oxidation, including Cpt1a, Cpt1b, Cpt2, Hmgcs2, Mlycd, and Acsl1 of male offspring mice was measured by RT-PCR.
Data is presented as Mean ± SE, n = 5 from 5 litters. * P < 0.05 vs. REF group. # P < 0.05 vs. NF group.
Next, we evaluated the level of β-oxidation in the liver by measuring the expression of key modulators and genes involved. Compared to NF offspring, both HF and H1N offspring decreased expression of PPAR-α protein in the liver, while H9N offspring had a similar expression of PPAR-α (Fig 7E). Consistently, L-Carnitine content in the liver significantly decreased in the HF offspring while significantly increased in the H9N offspring compared to NF offspring (Fig 7F). In addition, we measured the expression of key genes for fatty acid degradation and β-oxidation, which include Cpt1a, Cpt1b, Cpt2, Hmgcs2, Acsl1, and Mlycd. Significantly lower expressions of Hmgcs2 and Acsl1 were observed in HF offspring compared to NF offspring. In addition to Hmgcs2 and Acsl1 downregulation, H1N offspring also had decreased hepatic expressions for Cpt1a, Cpt1b, and Cpt2 (Fig 7G). Importantly, the expression of all these genes in H9N offspring were not different from NF offspring. These results suggest an inhibition of β-oxidation in the HF and H1N offspring liver, which was not observed in the H9N offspring.
Discussion
Previous animal studies have shown that western-style obesogenic diets prime NAFLD in offspring adulthood (24, 25, 36–43), providing strong evidence that greater focus is needed on maternal metabolic health prior to and during pregnancy. Thus, it is important to know the long-term effectiveness and etiology of maternal pre-pregnancy dietary intervention on mitigating the deleterious effects of maternal HF diet on offspring NAFLD risk. This new knowledge will promote the development of effective diet intervention strategies and policy for prevention of obesity and NAFLD and improvement of health outcomes for mothers and children. In the current study, we showed that the HF and H1N offspring had more severe hepatic steatosis than the NF and H9N offspring, suggesting that simply switching to a NF diet before pregnancy and not allowing a proper transition period was not beneficial in reducing the risk for offspring NAFLD. In our model, the H1N diet causes an even more severe phenotype of metabolic dysregulation than the HF and NF offspring, evidenced by more body weight gain, glucose intolerance, hepatic steatosis, and hyperlipidemia. However, the H9N diet has a beneficial effect indicated by glucose tolerance, no NAFLD, and normal blood lipid panel in offspring. In humans, the diagnosis of NAFLD is strongly predictive of insulin resistance (44, 45). Similarly, the HF and H1N hepatocytes were less sensitive to insulin signaling due to inhibited Insulin/Akt signaling and lower expression of genes responsible for insulin sensitivity. In contrast to the HF and H1N diet, the H9N diet did not cause these damaging effects of maternal HF diet by promoting normal Insulin/Akt signaling and a normal expression of genes involved in insulin signaling.
We found that different maternal diet interventions differentially affected the offspring lipid panel. Abnormal cellular lipid composition is lipotoxic as it can lead to toxic lipid accumulation, organelle dysfunction, cell injury, and chronic inflammation (46, 47). Taking into account that TG is an inert form of lipid storage that protects against lipotoxicity (48), changes in hepatic FFAs of offspring is critical for understanding how different maternal diet interventions led to different phenotypes of NAFLD. The significantly elevated total amount of FFAs in offspring liver by the HF diet and H1N diet, and the normal amount of FFAs by the H9N diet suggest that different maternal diets before pregnancy prime the quantity of hepatic FFAs in offspring. In addition to quantity, the composition of FFAs is various among groups treated with different maternal diet interventions. In our model, increased MUFAs and PUFAs in HF and H1N offspring versus NF offspring, suggests that maternal diets prime the desaturation of SFAs. It is not clear if the increasing MUFAs and PUFAs are simply due to overall higher amount of SFAs as substrates, or due to enhanced desaturation activity in these offspring. We also reported two associations: one between increased C18:1/C18:0 ratio and increased expression of Scd1, and the other between decreased C18:1/C18:0 ratio and increased expression of Elovl6. Both trends were seen in HF and H1N offspring, but not among H9N offspring. The association between C18:1/C18:0 ratio and Elovl6 expression might suggest and be explained by a deactivation of Elovl6 enzyme in the HF and H1N offspring which was averted in H9N offspring. Actually, while Elovl6 gene overexpression promotes steatohepatitis (49), increased C16:0 from C14:0 with decreased activity of Elovl6 enzyme is recently reported in the liver of NASH patients (50). Noteworthily, previous human studies of NAFLD patients reported increased SFAs, MUFAs, C18:1/C18:0 ratio, and decreased C18:0/C16:0 ratio associated with the severity of NAFLD (50–52). Our HF and H1N offspring displayed changes of the mentioned biomarkers in parallel with more severe hepatic steatosis than H9N and NF offspring, confirming this association reported in human studies. This suggests new biomarkers for the progression of NAFLD.
Our study discloses the various mechanisms through which different maternal diets prime lipid metabolism in offspring liver. A previous study has shown that hepatic de novo lipogenesis (DNL) is marginal in fetal liver and the expression of genes involved in DNL is not significantly changed with over-nutrition exposure in utero (53). In our study, the HF and H1N offspring had desensitized insulin signaling, higher expression of genes involved in DNL and de novo cholesterol synthesis, and increased Srebp-1 expression under post-weaning HF diet compared to the NF offspring. This result is consistent with multiple studies showing that in utero exposure to HF diet primes hepatic lipid metabolism under malnutrition, such that it worsens the NAFLD phenotype in offspring fed on post-weaning HF diet (40, 41, 54–56). To be noted, the H9N diet but not the H1N diet avoided the adverse effects of maternal HF diet to alter the molecular markers of lipogenesis. These results support the concept that maternal diet during the gestational and pre-pregnancy stage potentiates postnatal metabolic liver disease via modification of hepatic lipogenesis. We also identified the deactivation of FA oxidation to be the cause of hepatic lipid accumulation seen in HF and H1N offspring. Others have demonstrated that adult mice previously exposed to overnutrition in utero display decreased FA oxidation under postnatal HF diet (55, 57). In our study, the deactivation of AMPK signaling, decreased expression of PPAR-α and genes involved in FA oxidation in HF and H1N offspring, suggesting an inability to sufficiently increase β-oxidation in response to over-nutrition. These molecular changes were not noted in the H9N offspring liver. Thus, modification of the capability for FA oxidation could be another molecular mechanism that potentiates postnatal NAFLD in offspring of maternal overnutrition. Interestingly, most of the lipogenesis genes showed higher expressions in the HF offspring than the H1N offspring, while the expression level of genes involved in β-oxidation was more down-regulated in H1N offspring. These results suggest that maternal HF or H1N diet have different modes of pathological action, either by downregulating fatty acid oxidation more (H1N diet), or by inducing lipogenesis more (HF diet). Similar discrepancy between H1N and HF offspring was also noted in the molecular events involved in β-oxidation. Such that, the HF diet yielded lower content of L-carnitine, while the H1N diet inhibited expression of genes involved in β-oxidation.
In addition, different maternal diets seemed to result in different capacities for hepatic lipoprotein metabolism. Enhanced expression of Pcsk9 was found in HF, H1N, and H9N offspring with the lowest increase seen in H9N offspring. PCSK9 binds to the receptor for low-density lipoprotein particles (LDLR), which blocks its binding with the LDL-particles, preventing hepatic uptake (45). Thus, these results suggest a higher potential for NF offspring to process lipoprotein than those offspring (HF, H1N, and H9N) from mothers exposed to HF diet previously. Indeed, H1N offspring had an abnormal blood lipid panel with hepatic steatosis. The HF offspring had hepatic steatosis, while NF and H9N offspring had generally normal lipid panel and no hepatic steatosis. In H9N offspring, lipid metabolism was balanced with DNL, FA oxidation level, and Pcsk9 expression partially recovered. This potential mechanism needs to be confirmed by future studies focused on evaluating the effects of different maternal diets on lipoprotein metabolism. Nonetheless, our results suggest that different maternal diets prime offspring lipid metabolism differently.
In summary, our study indicates that an early maternal diet intervention is effective in reducing the risk for offspring NAFLD caused by maternal HF diet. Clinically, this encouraging discovery may provide valuable data-supported evidence for designing clinical trials to evaluate urgently required intervention strategies to minimize the vicious cycle of obesity and its related metabolic complications for future generations.
Acknowledgement
This project is supported by grants from the National Institutes of Health (NIDDK 1R01DK112368-01 to Drs. Xie and Zhang). This work is supported by the USDA National Institute of Food and Agriculture, [Hatch] project [1010406] to Dr. Xie. Portions of these studies were supported by a VA Merit Award, 4I01BX000574 to Dr. Alpini from the United States Department of Veteran’s affairs, the Hickam Endowed Chair Funds from Indiana University and the Strategic Research Initiative (SRI) from Indiana University Health, 41-855-96. The content is the responsibility of the author(s) alone and does not necessarily reflect the views or policies of the Department of Veterans Affairs or the United States Government.
Reference
- 1.Hales CM, Fryar CD, Carroll MD, Freedman DS, Aoki Y, Ogden CL. Differences in Obesity Prevalence by Demographic Characteristics and Urbanization Level Among Adults in the United States, 2013–2016. JAMA. 2018;319(23):2419–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hales CM, Fryar CD, Carroll MD, Freedman DS, Ogden CL. Trends in Obesity and Severe Obesity Prevalence in US Youth and Adults by Sex and Age, 2007–2008 to 2015–2016. JAMA. 2018;319(16):1723–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogden CL, Fryar CD, Hales CM, Carroll MD, Aoki Y, Freedman DS. Differences in Obesity Prevalence by Demographics and Urbanization in US Children and Adolescents, 2013–2016. JAMA. 2018;319(23):2410–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miele L, Forgione A, Hernandez AP, Gabrieli ML, Vero V, Di Rocco P, et al. The natural history and risk factors for progression of non-alcoholic fatty liver disease and steatohepatitis. Eur Rev Med Pharmacol Sci. 2005;9(5):273–7. [PubMed] [Google Scholar]
- 5.Charlton M Nonalcoholic fatty liver disease: a review of current understanding and future impact. Clin Gastroenterol Hepatol. 2004;2(12):1048–58. [DOI] [PubMed] [Google Scholar]
- 6.Ainge H, Thompson C, Ozanne SE, Rooney KB. A systematic review on animal models of maternal high fat feeding and offspring glycaemic control. Int J Obes (Lond). 2011;35(3):325–35. [DOI] [PubMed] [Google Scholar]
- 7.Alfaradhi MZ, Ozanne SE. Developmental programming in response to maternal overnutrition. Frontiers in genetics. 2011;2:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams L, Seki Y, Vuguin PM, Charron MJ. Animal models of in utero exposure to a high fat diet: a review. Biochim Biophys Acta. 2014;1842(3):507–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuyama H, Hiramatsu Y. Effects of a high-fat diet exposure in utero on the metabolic syndrome-like phenomenon in mouse offspring through epigenetic changes in adipocytokine gene expression. Endocrinology. 2012;153(6):2823–30. [DOI] [PubMed] [Google Scholar]
- 10.Ornellas F, Souza-Mello V, Mandarim-de-Lacerda CA, Aguila MB. Programming of obesity and comorbidities in the progeny: lessons from a model of diet-induced obese parents. PLoS One. 2015;10(4):e0124737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stettler N, Kumanyika SK, Katz SH, Zemel BS, Stallings VA. Rapid weight gain during infancy and obesity in young adulthood in a cohort of African Americans. Am J Clin Nutr. 2003;77(6):1374–8. [DOI] [PubMed] [Google Scholar]
- 12.Magarey AM, Daniels LA, Boulton TJ, Cockington RA. Predicting obesity in early adulthood from childhood and parental obesity. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 2003;27(4):505–13. [DOI] [PubMed] [Google Scholar]
- 13.Danielzik S, Czerwinski-Mast M, Langnase K, Dilba B, Muller MJ. Parental overweight, socioeconomic status and high birth weight are the major determinants of overweight and obesity in 5–7 y-old children: baseline data of the Kiel Obesity Prevention Study (KOPS). International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 2004;28(11):1494–502. [DOI] [PubMed] [Google Scholar]
- 14.Salsberry PJ, Reagan PB. Dynamics of early childhood overweight. Pediatrics. 2005;116(6):1329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravelli GP, Stein ZA, Susser MW. Obesity in young men after famine exposure in utero and early infancy. The New England journal of medicine. 1976;295(7):349–53. [DOI] [PubMed] [Google Scholar]
- 16.Ravelli AC, van Der Meulen JH, Osmond C, Barker DJ, Bleker OP. Obesity at the age of 50 y in men and women exposed to famine prenatally. Am J Clin Nutr. 1999;70(5):811–6. [DOI] [PubMed] [Google Scholar]
- 17.Law CM, Barker DJ, Osmond C, Fall CH, Simmonds SJ. Early growth and abdominal fatness in adult life. Journal of epidemiology and community health. 1992;46(3):184–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips DI, Walker BR, Reynolds RM, Flanagan DE, Wood PJ, Osmond C, et al. Low birth weight predicts elevated plasma cortisol concentrations in adults from 3 populations. Hypertension. 2000;35(6):1301–6. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt SF, Jorgensen M, Chen Y, Nielsen R, Sandelin A, Mandrup S. Cross species comparison of C/EBPalpha and PPARgamma profiles in mouse and human adipocytes reveals interdependent retention of binding sites. BMC genomics. 2011;12:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villamor E, Cnattingius S. Interpregnancy weight change and risk of adverse pregnancy outcomes: a population-based study. Lancet. 2006;368(9542):1164–70. [DOI] [PubMed] [Google Scholar]
- 21.Bogaerts A, Van den Bergh BR, Ameye L, Witters I, Martens E, Timmerman D, et al. Interpregnancy weight change and risk for adverse perinatal outcome. Obstet Gynecol. 2013;122(5):999–1009. [DOI] [PubMed] [Google Scholar]
- 22.Viana LV, Gross JL, Azevedo MJ. Dietary intervention in patients with gestational diabetes mellitus: a systematic review and meta-analysis of randomized clinical trials on maternal and newborn outcomes. Diabetes Care. 2014;37(12):3345–55. [DOI] [PubMed] [Google Scholar]
- 23.Summerfield M, Zhou Y, Zhou T, Wu C, Alpini G, Zhang KK, et al. A long-term maternal diet transition from high-fat diet to normal fat diet during pre-pregnancy avoids adipose tissue inflammation in next generation. PLoS One. 2018;13(12):e0209053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu H, Fu Q, Zhou Y, Xue C, Olson P, Lynch EC, et al. A long-term maternal diet intervention is necessary to avoid the obesogenic effect of maternal high-fat diet in the offspring. J Nutr Biochem. 2018;62:210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Q, Olson P, Rasmussen D, Keith B, Williamson M, Zhang KK, et al. A short-term transition from a high-fat diet to a normal-fat diet before pregnancy exacerbates female mouse offspring obesity. Int J Obes (Lond). 2016;40(4):564–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8. [DOI] [PubMed] [Google Scholar]
- 27.Fu Q, Olson P, Rasmussen D, Keith B, Williamson M, Zhang KK, et al. A short-term transition from a high-fat diet to a normal-fat diet before pregnancy exacerbates female mouse offspring obesity. Int J Obes (Lond). 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Folch J, Lees M, Stanley GHS. A SIMPLE METHOD FOR THE ISOLATION AND PURIFICATION OF TOTAL LIPIDES FROM ANIMAL TISSUES. Journal of Biological Chemistry. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 29.Brose S, Baker A, Golovko M. A Fast One-Step Extraction and UPLC–MS/MS Analysis for E2/D2 Series Prostaglandins and Isoprostanes. Lipids. 2013;48(4):411–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Golovko SA, Golovko MY. Plasma Unesterified Fatty-Acid Profile Is Dramatically and Acutely Changed under Ischemic Stroke in the Mouse Model. Lipids. 2018;53(6):641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang KK, Xiang M, Zhou L, Liu J, Curry N, Heine Suner D, et al. Gene network and familial analyses uncover a gene network involving Tbx5/Osr1/Pcsk6 interaction in the second heart field for atrial septation. Hum Mol Genet. 2016;25(6):1140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosmas CE, Martinez I, Sourlas A, Bouza KV, Campos FN, Torres V, et al. High-density lipoprotein (HDL) functionality and its relevance to atherosclerotic cardiovascular disease. Drugs Context. 2018;7:212525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang P, Li X, Xu C, Eckert RL, Reece EA, Zielke HR, et al. Maternal hyperglycemia activates an ASK1-FoxO3a-caspase 8 pathway that leads to embryonic neural tube defects. Science signaling. 2013;6(290):ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89(3):331–40. [DOI] [PubMed] [Google Scholar]
- 35.Pikuleva IA. Cholesterol-metabolizing cytochromes P450: implications for cholesterol lowering. Expert Opin Drug Metab Toxicol. 2008;4(11):1403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinclair KJ, Friesen-Waldner LJ, McCurdy CM, Wiens CN, Wade TP, de Vrijer B, et al. Quantification of fetal organ volume and fat deposition following in utero exposure to maternal Western Diet using MRI. PLoS One. 2018;13(2):e0192900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brumbaugh DE, Friedman JE. Developmental origins of nonalcoholic fatty liver disease. Pediatr Res. 2014;75(1–2):140–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hintze KJ, Benninghoff AD, Cho CE, Ward RE. Modeling the Western Diet for Preclinical Investigations. Adv Nutr. 2018;9(3):263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Itoh H, Kanayama N. Developmental Origins of Nonalcoholic Fatty Liver Disease (NAFLD). Adv Exp Med Biol. 2018;1012:29–39. [DOI] [PubMed] [Google Scholar]
- 40.Musial B, Vaughan OR, Fernandez-Twinn DS, Voshol P, Ozanne SE, Fowden AL, et al. A Western-style obesogenic diet alters maternal metabolic physiology with consequences for fetal nutrient acquisition in mice. J Physiol. 2017;595(14):4875–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruis MG, Lendvai A, Bloks VW, Zwier MV, Baller JF, de Bruin A, et al. Maternal western diet primes non-alcoholic fatty liver disease in adult mouse offspring. Acta Physiol (Oxf). 2014;210(1):215–27. [DOI] [PubMed] [Google Scholar]
- 42.Steegenga WT, Mischke M, Lute C, Boekschoten MV, Lendvai A, Pruis MG, et al. Maternal exposure to a Western-style diet causes differences in intestinal microbiota composition and gene expression of suckling mouse pups. Mol Nutr Food Res. 2017;61(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trevino LS, Katz TA. Endocrine Disruptors and Developmental Origins of Nonalcoholic Fatty Liver Disease. Endocrinology. 2018;159(1):20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sundaram SS, Zeitler P, Nadeau K. The metabolic syndrome and nonalcoholic fatty liver disease in children. Curr Opin Pediatr. 2009;21(4):529–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinreich M, Frishman WH. Antihyperlipidemic therapies targeting PCSK9. Cardiol Rev. 2014;22(3):140–6. [DOI] [PubMed] [Google Scholar]
- 46.Musso G, Cassader M, Gambino R. Non-alcoholic steatohepatitis: emerging molecular targets and therapeutic strategies. Nat Rev Drug Discov. 2016;15(4):249–74. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi K, Yang L, McCall S, Huang J, Yu XX, Pandey SK, et al. Inhibiting triglyceride synthesis improves hepatic steatosis but exacerbates liver damage and fibrosis in obese mice with nonalcoholic steatohepatitis. Hepatology. 2007;45(6):1366–74. [DOI] [PubMed] [Google Scholar]
- 48.Musso G, Cassader M, Paschetta E, Gambino R. Bioactive Lipid Species and Metabolic Pathways in Progression and Resolution of Nonalcoholic Steatohepatitis. Gastroenterology. 2018;155(2):282–302 e8. [DOI] [PubMed] [Google Scholar]
- 49.Matsuzaka T, Atsumi A, Matsumori R, Nie T, Shinozaki H, Suzuki-Kemuriyama N, et al. Elovl6 promotes nonalcoholic steatohepatitis. Hepatology. 2012;56(6):2199–208. [DOI] [PubMed] [Google Scholar]
- 50.Chiappini F, Coilly A, Kadar H, Gual P, Tran A, Desterke C, et al. Metabolism dysregulation induces a specific lipid signature of nonalcoholic steatohepatitis in patients. Sci Rep. 2017;7:46658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiappini F, Desterke C, Bertrand-Michel J, Guettier C, Le Naour F. Hepatic and serum lipid signatures specific to nonalcoholic steatohepatitis in murine models. Sci Rep. 2016;6:31587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Y, Chen J, Huang C, Li N, Zou L, Chia SE, et al. Comparison of hepatic and serum lipid signatures in hepatocellular carcinoma patients leads to the discovery of diagnostic and prognostic biomarkers. Oncotarget. 2018;9(4):5032–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCurdy CE, Bishop JM, Williams SM, Grayson BE, Smith MS, Friedman JE, et al. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest. 2009;119(2):323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bayol SA, Simbi BH, Fowkes RC, Stickland NC. A maternal “junk food” diet in pregnancy and lactation promotes nonalcoholic Fatty liver disease in rat offspring. Endocrinology. 2010;151(4):1451–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruce KD, Cagampang FR, Argenton M, Zhang J, Ethirajan PL, Burdge GC, et al. Maternal high-fat feeding primes steatohepatitis in adult mice offspring, involving mitochondrial dysfunction and altered lipogenesis gene expression. Hepatology. 2009;50(6):1796–808. [DOI] [PubMed] [Google Scholar]
- 56.Gregorio BM, Souza-Mello V, Carvalho JJ, Mandarim-de-Lacerda CA, Aguila MB. Maternal high-fat intake predisposes nonalcoholic fatty liver disease in C57BL/6 offspring. Am J Obstet Gynecol. 2010;203(5):495 e1–8. [DOI] [PubMed] [Google Scholar]
- 57.Borengasser SJ, Lau F, Kang P, Blackburn ML, Ronis MJ, Badger TM, et al. Maternal obesity during gestation impairs fatty acid oxidation and mitochondrial SIRT3 expression in rat offspring at weaning. PLoS One. 2011;6(8):e24068. [DOI] [PMC free article] [PubMed] [Google Scholar]