Abstract
Three-dimensional structures have been solved for several naturally occurring RNA triple helices, although all are limited to six or fewer consecutive base triples, hindering accurate estimation of global and local structural parameters. We present an X-ray crystal structure of a right-handed, U•A-U-rich RNA triple helix with 11 continuous base triples. Due to helical unwinding, the RNA triple helix spans an average of 12 base triples per turn. The double helix portion of the RNA triple helix is more similar to both the helical and base step structural parameters of A′-RNA rather than A-RNA. Its most striking features are its wide and deep major groove, a smaller inclination angle and all three strands favoring a C3′-endo sugar pucker. Despite the presence of a third strand, the diameter of an RNA triple helix remains nearly identical to those of DNA and RNA double helices. Contrary to our previous modeling predictions, this structure demonstrates that an RNA triple helix is not limited in length to six consecutive base triples and that longer RNA triple helices may exist in nature. Our structure provides a starting point to establish structural parameters of the so-called ‘ideal’ RNA triple helix, analogous to A-RNA and B-DNA double helices.
INTRODUCTION
The formation of an RNA triple helix, generated by mixing together poly(U) and poly(A) RNA strands at a 2:1 ratio, was first reported in 1957 (1). Subsequent characterization of this structure revealed stacks of U•A-U base triples in which one strand (U) establishes hydrogen bonds via Hoogsteen interactions (•) along the major groove of a Watson-Crick RNA duplex (A-U) (2–4). Since then, major-groove RNA triple helices composed of three or more consecutive base triples have been structurally validated by nuclear magnetic resonance (NMR), X-ray crystallography or cryo-EM methods for several natural RNAs, such as telomerase, group II introns and U2/U6 spliceosomal RNAs, various classes of ligand-bound riboswitches and RNA stability elements for nuclear expression from viral and cellular long noncoding RNAs (5–17). Although each RNA triple helix is structurally unique, the majority contain three to five consecutive base triples, which are most commonly a combination of the canonical U•A-U and C•G-C base triples and non-canonical base triples. Interestingly, none of the natural RNA triple helices discovered to date have more than six consecutive base triples. Modeling studies using the MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) RNA triple helix indicated that this length limitation is to avoid a putative steric clash between the 2′-hydroxyl of the Hoogsteen strand and phosphate oxygen of the purine-rich strand in the Watson–Crick duplex (12). If this model were true, then a structural distortion, such as an extrahelical nucleotide in the Hoogsteen strand or subtle backbone rearrangements, would be expected in an RNA triple helix with the potential to form more than six consecutive base triples, such as a poly(U•A-U) RNA triple helix. In the case of the MALAT1 triple helix having ten base triples, an intervening C-G doublet was proposed to reset the helical axis after six consecutive base triples, avoiding a steric clash (Supplementary Figure S1A) (12).
Thus far, there is no high-resolution structure of an RNA triple helix with the potential to form more than six consecutive base triples. The earliest structural model of a poly(U•A-U) RNA triple helix was proposed based on X-ray fiber diffraction data and molecular modeling (3,4). This model is suboptimal from a stereochemical evaluation; therefore, an alternative model was generated using the linked atom least squares program (18). Most recently, X-ray fiber diffraction data of a poly(U•A-U) RNA triple helix were refined using the continuous X-ray intensity data on layer lines (19). None of these structural models indicate a structural distortion to accommodate triple helices more than six consecutive base triples. Surprisingly, the sugar pucker, which could affect the putative steric clash limiting consecutive base triples, of a poly(U•A-U) RNA triple helix has not been resolved. Structural models and infrared (IR) data have not converged on the sugar pucker, although the C3′-endo conformation seems to be favored in structures of both poly(U•A-U) and natural RNA triple helices (3–17). From these studies, there is a clear need for an accurate structure of an RNA triple helix.
We present here the crystal structure analysis of a U•A-U-rich RNA triple helix that forms 11 consecutive base triples. This structural model allows us to establish the structural parameters of an RNA triple helix forming nearly one complete helical turn. Comparing it to A-RNA, A′-RNA and B-DNA helices, we show that the RNA triple helix is an A′-RNA conformer with all three strands favoring the C3′-endo conformation. Moreover, the stereochemistry of the RNA triple helix accommodates all three strands, allowing formation of a continuous stack of 11 consecutive base triples.
MATERIALS AND METHODS
RNA preparation
The pHDV-MALAT1_th11 plasmid (insert 5′-GGAAGGTTTTTCTTTTTCCTGAAGGCGAAAGTCTTCAGGTTTTTCTTTTTGGCCTTTCTTAAAAAAAAAAAAAGAAAAA-3′) was created by site-directed mutagenesis to modify the previously described pHDV-MALAT1_Core plasmid (referred to as pHDV-MALAT1 ENE+A core in reference (12)). Two guanosines were included at the 5′ end to ensure high transcription efficiency. The HDV ribozyme was used to generate homogeneous 3′ ends (20). To prepare RNA, in vitro transcription reactions were performed, with final gel-purified RNA in crystallization buffer (5 mM sodium cacodylate pH 6.5, 50 mM KCl, 1 mM MgCl2 and 0.1 mM ethylenediaminetetraacetic acid) as described previously (12).
Crystallization of MALAT1_th11 RNA and diffraction data collection
The 79-nt MALAT1_th11 RNA solution (11 mg/ml) was heated at 95°C for 3 min, snap-cooled on ice for 10 min and allowed to equilibrate at room temperature for at least 1 h prior to setting up crystal tray. The crystallization drop included 1.5 μl of MALAT1_th11 RNA and 1 μl reservoir solution containing 50 mM sodium cacodylate pH 6.0–6.3, 200 mM calcium acetate and 2.5 M sodium chloride. Crystals grew at 19°C using the hanging-drop method. Crystals (approximate dimensions: 0.1 × 0.5 × 0.02 mm) appeared after ∼3 days on a dust particle in the drop. Crystals were cryoprotected by stepwise addition of crystallization solution supplemented with glycerol until the final concentration of 20% glycerol was reached. Crystals were flash frozen in liquid nitrogen and stored until data collection. Diffraction data were collected at 100 K on a single crystal at the Advanced Photon Source, Argonne National Laboratory, on the SER-CAT 22-ID beamline. Data collection and processing statistics are summarized in Table 1.
Table 1.
Diffraction data and refinement statistics for the MALAT1_th11 model
| Data collection | |||
| Wavelength (Å) | 1.000 | ||
| Space group | P21 | ||
| Unit cell parameters | |||
| a, b, c (Å) | 54.7, 78.1, 84.0 | ||
| β (°) | 104.2 | ||
| Resolution (Å)a | 40.7–2.5 | 40.7–8.3 | 2.7–2.5 |
| Unique reflectionsa | 13040 | 648 | 664 |
| Multiplicity a | 3.9 | 3.5 | 4.5 |
| Ellipsoidal completeness (%)a | 90.8 | 95.7 | 83.8 |
| Spherical completeness (%)a | 54.4 | 95.7 | 13.1 |
| R meas (%)a | 4.5 | 2.9 | 166 |
| <I/σ(I)> a | 17.9 | 40.9 | 1.1 |
| CC(1/2) a | 0.999 | 0.998 | 0.405 |
| Refinement | |||
| R free reflections | 633 | ||
| No. of atoms (non-H) | 3397 | ||
| RNA | 3372 | ||
| Ca2+ | 8 | ||
| glycerol | 12 | ||
| solvent | 5 | ||
| R work/Rfree (%) | 16.9/21.3 | ||
| RMSD from ideal geometry | |||
| bond lengths (Å) | 0.005 | ||
| bond angles (o) | 0.9 | ||
| Average B-factors molecules A/B (Å2) | 76.0/108.1 | ||
| PDB code | 6svs |
aData processing statistics are given separately for all reflections (left column), inner shell (middle column) and outer shell (right column).
Determination and refinement of the crystal structure
Data were indexed and processed using XDS (21) and corrected for anisotropy using the STARANISO server (http://staraniso.globalphasing.org/cgi-bin/staraniso.cgi), which resulted in gradual omission of weak reflections between 4.2 and 2.5 Å resolution. The crystal structure of MALAT1_th11 was solved by molecular replacement in Phaser (22) using the X-ray crystal structure of the MALAT1_Core (94% identity, PDB ID: 4plx (12)) as the initial model. Structural models for two RNA molecules were built in the asymmetric unit. Manual fitting in the electron density maps was performed in Coot (23) with multiple rounds of model refinement in Phenix.refine (24). The refinement statistics are shown in Table 1. Three TLS (25) groups were added for each molecule. Our final model includes nucleotides 1–79 in both molecules. Four calcium cations and one glycerol molecule interact with each molecule.
Determination of structural parameters
Except for diameter, all structural parameters of the MALAT1_th11 model were computed by the online versions of CURVES+ Version 3.0nc (26,27) and 3DNA DSSR v1.9.6 (28,29) accessed from January-September 2019 (Supplementary Table S1). Due to strand order bias for CURVES+ calculations of triple helix structure, we determined helical and base pairs parameters by analyzing the Watson-Crick (WC) helix (input: 5′ 40:50 3′/3′ 79:69 5′) and Hoogsteen-Watson (HW) helix (input: 5′ 7:17 3′/5′ 69:79 3′). Groove measurements determined by CURVES+ (26,27) are calculated using a vectors-based approach as follows: width is measured between spline curves passing through phosphorus atoms and depth is measured as a distance from a groove width vector to the long axis of base pair. Structural parameters for both RNA molecules are presented in Supplementary Table S1. Differences between molecules include major-groove width, inclination angle and select nucleotides exhibiting C2′-endo sugar puckers. To calculate diameter, we measured the radius of the helix, which is the average distance between the helical axis and the outermost atoms (O2′ for triple helix, A-RNA, A′-RNA; OP1 for B-DNA). Radii measurements are as follows: 10.6 (RMSD 0.8) Å for MALAT1_th11, 10.3 (RMSD 0.0) Å for ideal A-RNA, 10.1 (RMSD 0.9) Å for A-RNA, 10.6 (RMSD 0.8) Å for A′-RNA, 10.5 (RMSD 0.0) Å for ideal B-DNA and 10.5 (RMSD 0.5) Å for B-DNA. An additional 1.4 Å was added to account for van der Waals radius of oxygen atom.
RESULTS AND DISCUSSION
Determination of MALAT1_th11 structure
The largest naturally occurring RNA triple helix that has been structurally validated to date is the one at the 3′ end of the MALAT1 long noncoding RNA (12). This triple helix is composed of ten major-groove base triples interrupted by a C-G doublet (Supplementary Figure S1A). Because this RNA triple helix readily crystallizes, we generated a variant of the previously determined X-ray crystal structure of the MALAT1 triple helix core (Supplementary Figure S1B). This variant of the MALAT1 triple helix, which we refer to as MALAT1_th11, has the internal C-G doublet replaced with a canonical U•A-U base triple so that there is the potential to form 11 consecutive base triples (Figure 1A and Supplementary Figure S1C). Additionally, insertion of an A-U base pair in stem II of MALAT1_th11 facilitated formation of crystal contacts to produce crystals that diffracted up to 2.5 Å (Supplementary Figure S2). Due to anisotropic diffraction, the high-resolution cutoff was set to 2.5 Å in the best direction and 4.2 Å in the worst direction (Table 1). Despite the anisotropy, high completeness (>94%) of the data below 4.2 Å allowed us to solve the structure of MALAT1_th11 by molecular replacement using Phaser (22), whereby the X-ray crystal structure of the MALAT1 triple helix core (PDB ID: 4plx) was the initial model (12). Electron density is visible for all 79 nts of MALAT1_th11 and structural models were built for two RNA molecules, which we refer to as A and B, in the asymmetric unit (Supplementary Figure S2). Superposition of molecule A with B established RMSD values of 1.6 Å for the entire model and 1 Å for only the triple helix (Supplementary Figure S2B and D). Additionally, inserting the model for molecule A into the electron density map for molecule B showed a good fit between the two models (Supplementary Figure S2E). Herein, data analyses and figures, unless indicated otherwise, are based on molecule A because of its better electron density maps and a lower average B-factor of ∼76 Å2 compared to ∼108 Å2 for molecule B ( Figure 1, Supplementary Figure S2 and Table 1).
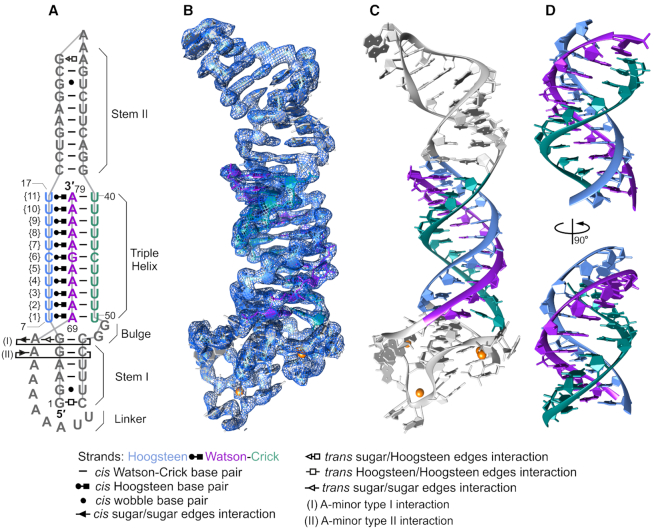
Figure 1.
Structure of MALAT1_th11. (A) A schematic diagram shows the secondary and tertiary structures observed in MALAT1_th11. Interactions between nucleotides are represented using Leontis–Westhof notation in all figures (36). Numbering of nucleotides, triples and structural elements of MALAT1_th11 are presented. (B) Crystal structure of MALAT1_th11 (stick representation) and 2Fo - Fc electron density map (blue mesh) of RNA molecule A contoured at 1.5 σ level is shown. (C) Crystal structure of MALAT1_th11 (ribbon representation). The major-groove triple helix is highlighted by blue (Hoogsteen strand), purple (Watson strand) and green (Crick strand). (D) Two different views of major-groove triple helix rotated by 90°, starting from orientation presented in panel C. Orange spheres in panels B and C represent calcium ions.
RNA triple helix of MALAT1_th11 forms 11 consecutive base triples
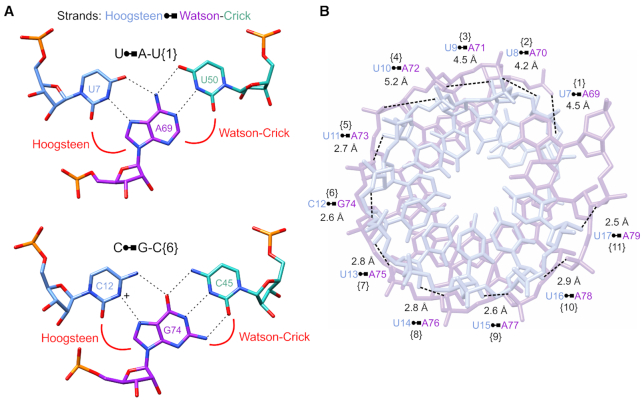
In the MALAT1_th11 structure, the 3′-terminal A-rich tract (nts 69–79, Watson strand) interacts with the U-rich internal stem-loop (nts 7–17, Hoogsteen strand and nts 40–50, Crick strand) to form a continuous RNA triple helix of 11 canonical major-groove base triples: ten U•A-U triples interrupted by one C•G-C triple (Figures 1 and 2A; Supplementary Table S2 and Movie S1). All bases of these 11 triples are within hydrogen-bonding distance (2.5–3.3 Å) along the Watson–Crick and the Hoogsteen faces except for three weak interactions along the Hoogsteen–Watson face of U•A-U base triples {8} (3.7 Å), {9} (3.8 Å) and {11} (3.6 Å) (Supplementary Figure S3). Moreover, the ribose moieties of the Hoogsteen strand are stabilized by a hydrogen bond between the 2′-hydroxyl group of the Hoogsteen strand and phosphate backbone of the Watson strand (2′-OH…OP2) (Figure 2B) (12,30–31). The 2′-OH…OP2 hydrogen bonds observed for U•A-U{5–11} likely stabilize the highly electronegative RNA triple helix, minimizing possible clashes between the O2′ and O4′ of the Hoogsteen strand and phosphate of the Watson strand. Thus, the triple helix of MALAT1_th11 is stabilized by both inter- and intrastrand hydrogen bonds.
Figure 2.
Nucleotide interactions in MALAT1_th11 triple helix. (A) Base-pairing interactions of U•A-U and C•G-C base triples (stick representation) are shown. Tricolor code indicates nucleotides located in Hoogsteen (blue), Watson (purple) and Crick (green) strands of triple helix. The C•G-C triple requires protonation (+) of the cytosine N3 to form the same number of Hoogsteen hydrogen bonds as U•A. Dashed lines represent hydrogen-bonding interactions. (B) The hydrogen-bonding pattern and distances between the 2′-OH of the Hoogsteen strand (light blue) and phosphate oxygen of the Watson strand (light purple) are shown; numbers in brackets correspond to base triple numbering scheme in Figure 1A.
The major-groove triple helix is adjacent to a GG dinucleotide bulge and a double-helical stem engaged in two A-minor interactions (Figure 1A). The G5-C54 and G6-C53 base pairs in stem I interact with A67 and A68, respectively, to form type II and I A-minor interactions, a common tertiary motif in RNA structures (Supplementary Figure S4A-B and Table S2) (32,33). These two consecutive A-minor interactions consequently form a ribose zipper motif (Supplementary Figure S4C), which is another structural element that stabilizes the tertiary fold of MALAT1_th11 (34,35). Both the A-minor interactions and ribose zipper are present in the MALAT1_Core (Supplementary Figure S1B) (12). Additional non-canonical nucleotide interactions present in MALAT1_th11 are as follows: (i) GTP1-C58 in stem I interact via their Hoogsteen edges in a trans orientation (tHH in Supplementary Figure S4D), (ii) G2-U57 in stem I (Supplementary Figure S4E) and G25-U32 in stem II (not shown) form cis Watson–Crick (cWC) wobble pairs and (iii) G27-A30 in stem II interact via their sugar/Hoogsteen edges in a trans conformation (tSH in Supplementary Figure S4F and Table S2) (36). Overall, most nucleotide interactions in MALAT1_th11 are similar to those observed previously in the MALAT1_Core structure (Supplementary Figure S1B and C) (12).
Structural parameters of RNA triple helix resemble A′-RNA
A range of conformations for nucleic acid double helices have been observed, with there being two predominant forms: A for RNA and B for DNA (37). These two forms are characterized by multiple parameters, such as helical twist, rise, pitch, number of base pairs per turn, base pair geometry, sugar pucker and phosphate backbone geometry. Therefore, we used CURVES+ web server to quantitatively examine the conformation of nucleic acid structures and to compute these structural parameters for the MALAT1_th11 structure, analyzing both the Watson–Crick (WC-helix) and Hoogsteen–Watson (HW-helix) strands of the RNA triple helix (26,27). The RNA triple helix of MALAT1_th11 is a right-handed helix, whose helical twist is ∼30° (WC-helix: 29.7°; HW-helix: 30.3°), helical rise is 2.9 Å (WC-helix: 2.9 Å; HW-helix: 2.9 Å), pitch is 35.1 Å and X displacement is −5 Å (Table 2; Supplementary Tables S1, 3 and 4). For one full 360° rotation of the triple helix, there are 12 base triples; therefore, the MALAT1_th11 structure captures, for the first time, nearly one complete helical turn of a continuous RNA triple helix. These structural parameters, along with other parameters described below (e.g. C3′-endo sugar pucker assignments), suggested that the MALAT1_th11 triple helix belongs to an A-family conformation.
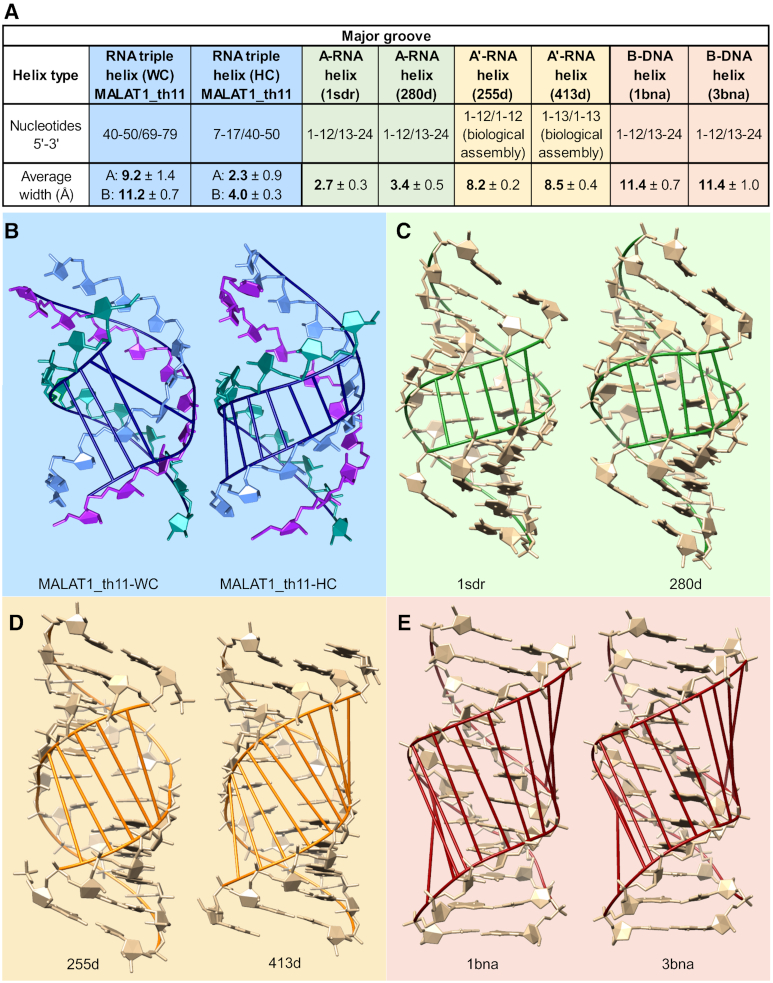
Table 2.
Structural parameters for the MALAT1_th11 RNA triple helix and various double helices
| RNA triple helixa | Ideal A-RNA double helixb | A-RNA double helix (1sdr, 280d) | A′-RNA double helix (255d, 413d) | Ideal B-DNA double helixb | B-DNA double helix (1bna, 3bna) | |
|---|---|---|---|---|---|---|
| Helical sense | Right-handed | Right-handed | Right-handed | Right-handed | Right-handed | Right-handed |
| Helical twist | 30° ± 5.3 | 32.7° | 33.7° ± 4.7 | 31.4° ± 8.1 | 36° | 36.2° ± 3.9 |
| Helix rise per base pair/triple | 2.9 Å ± 0.2 | 2.8 Å | 2.6 Å ± 0.3 | 2.9 Å ± 0.1 | 3.4 Å | 3.4 Å ± 0.3 |
| Helix pitch | 35.1 Å | 31 Å | 27.5 Å | 32.8 Å | 34 Å | 33.7 Å |
| X displacement | −5 Å ± 1 | −4.3 Å | −3.9 Å ± 0.6 | −4.9 Å ± 1 | 0.6 Å | 0.3 Å ± 0.6 |
| Base pairs/base triples per helical turn | 12 | 11 | 10.7 | 11.5 | 10 | 10 |
| Inclination | A: 8.4° ± 2.6c | 11.9° | 17.6° ± 2.2 | 13.6° ± 3.4 | 2.9° | -0.5 ° ± 4.2 |
| B: 6.3° ± 5.7 | ||||||
| Major-groove | widec | narrow (4 Å) | narrow (3.1 Å ± 0.5) | wide (8.4 Å ± 0.3) | wide (11.4 Å) | wide (11.4 Å ± 0.8) |
| dimensions | (A: 9.2 Å ± 1.4) | |||||
| (B: 11.2 Å ± 0.7) | ||||||
| deep (11.0 Å ± 1.2) | deep (8.9 Å) | deep (9.2 Å ± 0.7) | deep (9.3 Å ± 1.6) | shallow (3.8 Å) | shallow (4.5 Å ± 1.2) | |
| Minor-groove | wide (9 Å ± 0.5) | wide (11 Å) | wide (9.9 Å ± 0.9) | wide (9.7 ± 0.7) | narrow (5.9 Å) | narrow (4.9 Å ± 1.4) |
| dimensions | shallow (1.5 Å ± 0.4) | shallow (−0.8 Åd) | shallow (0.5 Å ± 0.5) | shallow (0.5 Å ± 1.3) | shallow (5.7 Å) | shallow (5.3 Å ± 0.3) |
| Diameter | 24 Å (RMSD 1.6) | 23.3 Å | 22.9 Å (RMSD 1.7) | 24 Å (RMSD 1.6) | 23.8 Å | 23.9 Å (RMSD 1.0) |
| External atom of helix | O2′ | O2′ | O2′ | O2′ | OP1 | OP1 |
| Sugar pucker | C3′-endo | C3′-endo | C3′-endo | C3′-endo | C2′-endo | C2′-endo |
All parameters, except for diameter (see ‘Materials and Methods’ section), were calculated using CURVES+ web server (26,27). Values represent an average ± standard deviation (or RMSD) for all base pairs/triples. By definition, idealized structures have a standard deviation of 0. PDB IDs of analyzed structures are shown in column headers.
aAnalysis was performed on molecule A (see ‘Materials and Methods’ section—Determination of structural parameters).
bThe ideal A-RNA helix (UUUUUCUUUUU/AAAAAGAAAAA) and ideal B-DNA (TTTTTCTTTTT/AAAAAGAAAAA) were generated using Coot software (23) and then both were analyzed using CURVES+ web server (26,27).
cDimensions for inclination angle and major groove of WC-helix are presented for both molecules A and B.
dA negative value implies a convex surface rather than a groove.
Next, we used CURVES+ to analyze previously determined X-ray crystal structures for A-RNA (PDB IDs: 1sdr and 280d), A′-RNA (PDB IDs: 255d and 413d) and B-DNA (PDB IDs: 1bna and 3bna) double helices to directly compare with the structural parameters of the RNA triple helix (26–27,38–43). MALAT1_th11 appears to be an A-family conformer; therefore, we considered the two characterized A-family conformations: A-RNA and A′-RNA. X-ray fiber diffraction data and CURVES+ analyses of X-ray crystal structures have shown that both A-RNA and A′-RNA conformations are similar except for quantitative differences in helical twist, rise, pitch, number of base pairs per turn, inclination angle and major-groove width (Table 2) (40,43–45). Because nucleotide sequence can affect structural parameters, we also used Coot to cross-validate our observations against idealized A-RNA and B-DNA helices composed of the same sequence found in the Watson–Crick strands of the MALAT1_th11 triple helix (Figure 1A) and then analyzed these idealized structures using CURVES+ (23,26–27,46). Our comparative analysis revealed that the overall helical geometry of the RNA triple helix more closely resembles A′-RNA, which has 11.5 bp per turn and a helical rise of 2.9 Å, than A-RNA, which has 10.7–11 bp per turn and a helical rise of 2.6–2.8 Å (Table 2; Supplementary Tables S3 and 4) (43). Despite being a three-stranded structure, a diameter of 24 Å for the MALAT1_th11 triple helix is nearly identical (within RMSD) to the diameters of both DNA and RNA double-stranded helices, which range from 22.9–24 Å (Table 2). This finding confirms early structural characterizations of RNA triple helices supporting an A-helix derivative (1,3).
In addition to the helical geometric parameters, we also examined base step and base pair geometrical parameters generated by CURVES+ (26,27). Some parameters have relatively high standard deviation values so only general conclusions can be drawn from our comparative analysis (Supplementary Tables S3 and 5). In general, the average slide, tilt, roll, twist, propeller, inclination and X displacement values for the WC-helix of the MALAT1_th11 triple helix were more consistent with A′-RNA than A-RNA (Supplementary Tables S3 and 5). Notably, the average inclination angle of base pairs at 8.4° was 9.2° and 5.2° lower than A-RNA and A′-RNA, respectively (Supplementary Table S5). Such a decrease suggests the WC-helix major groove of the RNA triple helix may be wider, as observed previously for A′-RNA (43). From our analysis of multiple geometric parameters, we conclude that the RNA triple helix of MALAT1_th11 is A′ form.
Double helix of MALAT1_th11 triple helix has a wide and deep major groove
A-RNA and B-DNA are also distinguished by the dimensions of their major and minor grooves. Therefore, we used CURVES+ to determine the groove dimensions of the WC-helix of the MALAT1_th11 triple helix for both molecules A and B (26,27). We examined both molecules because the C2′-endo sugar pucker of A71 may alter groove dimensions in molecule A (read next section for details). For both molecules A and B, the major groove is wider (9.2–11.2 Å) and deeper (11 Å) compared to the relatively narrow (3.1–4 Å) and deep (8.9–9.2 Å) major groove of A-RNA (Table 2 and Figure 3). However, A′-RNA has a widened major groove at 8.4 Å (Table 2 and Figure 3). When the Hoogsteen strand is considered, the average major-groove width of the MALAT1_th11 triple helix is 2.3–4.0 Å, which is more similar to A-RNA. Widening the major groove to accommodate the Hoogsteen strand does not affect the dimensions of the major groove in stem II, for it is narrow (3–3.2 Å) like A-RNA (Supplementary Figure S5). Likewise, the minor groove of the WC-helix of the RNA triple helix in MALAT1_th11 is similar to that of A-RNA and A′-RNA, i.e. wide and shallow (Table 2). The unique major-groove dimensions of the RNA triple helix and RNA double helices provide a structural rationale for proteins to discriminate between different conformational forms of nucleic acids (43,47).
Figure 3.
Dimensions of major grooves in various helices. (A) Table summarizing major-groove width calculations determined using CURVES+ (26,27). Major grooves are highlighted in (B) WC-helix from MALAT1_th11triple helix (left) and Hoogsteen–Crick (HC) groove of MALAT1_th11triple helix (right), with both structures being molecule A,(C) two A-RNA helices, (D) two A′-RNA helices and (E) two B-DNA helices. PDB IDs of structures are included in each panel.
A wider major groove of the WC-helix in the MALAT1_th11 triple helix is consistent with lower average inclination angles of 8.4° for molecule A and 6.3° for molecule B (broad/inconsistent distribution, see Supplementary Table S1) in comparison to inclination angles of 17.6° in A-RNA and 13.6° in A′-RNA (Table 2 and Supplementary Table S5). When the inclination angle becomes smaller, the backbone of one strand moves upward while the backbone of the complementary strand goes downward, thereby causing the major groove to widen. Both the inclination angle and major-groove dimensions are two structural hallmarks of A′-RNA and both are observed in the WC-helix of the RNA triple helix in MALAT1_th11 (43).
Triple helix of MALAT1_th11 favors a C3′-endo sugar pucker and maintains regular phosphate backbone geometry except for Watson strand
The sugar-phosphate backbone contributes to the conformation of nucleic acids. The sugar-phosphate backbone can be defined by its sugar pucker, glycosidic bond orientation and torsion angles. In general, an A-RNA double helix adopts a C3′-endo ribose conformation. However, Fourier-transform infrared spectroscopy (FTIR), UV and Raman spectroscopy and NMR structural studies of poly(U•A-U) triple helices have not converged on the sugar pucker in an RNA triple helix (3,19,31,48–49). Our structural model of an 11-base-triple-long RNA triple helix and those of natural triple helices (5–17) favor riboses in a C3′-endo conformation and glycosidic bonds in an anti-conformation for all three strands (Supplementary Table S6). These observations are in agreement with those presenting molecular models of poly(U•A-U) triple helices derived from an oriented noncrystalline fiber (3), FTIR (49) and NMR studies (31). However, some poly(U•A-U) models, based on Raman spectroscopy and computational refinement of an X-ray fiber diffraction model, propose that one or two strands of the triple helix adopt a C2′-endo conformation (19,48). Raman spectroscopic analysis of a poly(U•A-U) triple helix detected a Raman band at 863 cm−1, which was assigned to be the Hoogsteen strand in a C2′-endo conformation (48). Computational models of eight different combinations of C2′- and C3′-endo sugar puckers indicated that a poly(U•A-U) triple helix was most favorable when the Watson and Hoogsteen strands adopt a C2′-endo conformation (19). Interestingly, the MALAT1_th11 RNA triple helix does have two nucleotides in a C2′-endo conformation (Supplementary Table S6 and Figure S6). First, the ribose of U50 is in a C2′-endo conformation in both molecules A and B. This C2′-endo conformation arises from an interaction between the OP1 phosphate of G52 and a calcium ion, which is also coordinated by GTP1 (Supplementary Figure S6A). This coordination arrangement circumvents a steric clash between OP2 of G52 and the 2′-hydroxyl group of U50 in a C3′-endo conformation. Second, A71 adopts a C2′-endo sugar pucker in molecule A but not B, which suggests the C2′-endo sugar pucker is not structurally required at this position in the triple helix (Supplementary Figure S6B). This local C2′-endo conformation of A71 disrupts the 2′-OH…OP2 hydrogen bond at U•A-U{3&4} (Figure 2B) and instead creates a different zipper-like hydrogen bonding pattern. Here, the OP2 of A70 and A71 interact with the 2′-hydroxyl group of the bases one step above, U9 and U10, respectively, to stabilize the Hoogsteen face of the triple helix along with the 2′-hydroxyl group of A71 forming hydrogen bonds with O2 of U10 and O4′ of U11 (Supplementary Figure S7). Thus, it is possible that a local C2′-endo ribose conformation in the Watson strand can stabilize an RNA triple helix, although the C3′-endo ribose conformation facilitating the 2′-OH…OP2 hydrogen bond seems to be preferred.
To fully describe the backbone conformation of a nucleotide, we used CURVES+ to measure all six torsion angles (α, β, γ, δ, ϵ, ζ) of phosphate backbone as well as the glycosidic bond (χ, which connects the base to the sugar moiety) of the MALAT1_th11 triple helix (Supplementary Figure S8A) (26,27). The most accurate backbone conformations of nucleic acids have been extrapolated from X-ray crystallographic datasets at 1.9-Å resolution or better; therefore, we can draw only general conclusions from the torsion angles of our MALAT1_th11 structure (Table 3; Supplementary Table S1 and Figure S8) (50–52). The torsion angles β, δ, ϵ, ζ and χ are within the range typically observed for nucleic acids (Table 3 and Supplementary Figure S8B-C). The torsion angle δ depends on sugar pucker so U50 and A71 have δ values for C2′-endo sugar pucker (universal: 147.3 ± 4.9°; observed average: 145.2 ± 2.8°) while all other nucleotides within the triple helix have a δ value characteristic of C3′-endo sugar pucker (universal: 81 ± 4.4°; observed average: 79.6 ± 6.1°) (Table 3) (51). The values for χ support an anti conformation for both purines and pyrimidines in C2′-endo and C3′-endo sugar puckers (Table 3 and Supplementary Table S6) except for two outliers in the Watson strand (A76 and A78; average χ: 179.5°). However, both the α and γ values deviate from the universal values. A strand-by-strand analysis of torsion angles at the individual nucleotide level showed that the more negative values of α and γ occur predominantly in the Watson strand of the MALAT1_th11 triple helix but also the β values of the Watson strand vary more than those from the Hoogsteen and Crick strands (Supplementary Figure S8B and C). Such a widespread distribution of α, β and γ torsion angles in the Watson strand is generally not observed in A-RNA nor A′-RNA helices (Supplementary Figure S8B and C). Thus, the Watson strand may need to make subtle backbone rearrangements to maintain hydrogen-bonding interactions among the three bases and/or to optimize the number of 2′-OH…OP2 hydrogen bonds (Figure 2 and Supplementary Figure S3). Overall, the sugar-phosphate backbone geometry of the RNA triple helix largely follows A-RNA except for the Watson strand, whose geometry is perturbed slightly to mediate optimal interactions among all three strands of the triple helix.
Table 3.
Universal torsion angles for nucleic acids and torsion angles in RNA triple helix of MALAT1_th11
| Torsion angles (°) | ||||
|---|---|---|---|---|
| Universala | MALAT1_th11b | |||
| Alpha (α) | −74.7 ± 9.8 | −149.7 ± 18.3 [22] | ||
| 81 ± 12.1 | −61.2 ± 13.2 [5] | |||
| 171 ± 15 | 147.6 ± 17.9 [6] | |||
| Beta (β) | −173.5 ± 13 | −157.4 ± 12.1 [8] | ||
| 163.8 ± 23 | 159.6 ± 16.2 [25] | |||
| Gamma (γ) | −67.1 ± 12.3 | −167.5 ± 3.9 [3] | ||
| 52.5 ± 5.7 | 56.8 ± 10.5 [21] | |||
| 179.4 ± 6.4 | 149 ± 20.4 [9] | |||
| Delta (δ) | C2′-endo | C3′-endo | C2′-endo | C3′-endo |
| 147.3 ± 4.9 (RNA) | 81 ± 4.4 (RNA) | 145.2 ± 2.8 [2] | 79.6 ± 6.1 [31] | |
| 145.2 ± 4 (DNA) | 84.8 ± 4.5 (DNA) | |||
| Epsilon (ϵ) | −146 ± 8.6 | −144.8 ± 17.9 [32] | ||
| Zeta (ζ) | −70.8 ± 4.8 | −75.6 ± 20.1 [32] | ||
| 80.7 ± 14.3 | ||||
| 163.1 ± 0.6 | ||||
| Chi (χ) | C2′-endo purines | C3′-endo purines | C2′-endo purines | C3′-endo purinesc |
| −123 ± 24.3 {anti} | −166.7 ± 14 {anti} | −126 {anti} [1] | −172.1 ± 5.6 {anti} [8] | |
| 58.6 ± 12 {syn} | 35.6 {syn} | |||
| pyrimidines | pyrimidines | pyrimidines | pyrimidines | |
| −130.2 ± 18.4 {anti} | −164.3 ± 6.6 {anti} | −120.6 {anti} [1] | −164.2 ± 6.7 {anti} [21] | |
| 63.4 ± 10.2 {syn} | ||||
All values represent average ± standard deviation. Torsion angles are defined in Supplementary Figure S8A.
aUniversal torsion angles were obtained from references (50–52).
bAll torsion angles were calculated for molecule A using CURVES+ (26,27). The number in brackets indicates number of nucleotides included in calculation.
cThis value excludes two outliers, A76 and A78, which have an average χ value of 179.5°.
Comparative analysis of MALAT1_th11 triple helix with previous poly(U•A-U) models
The global features of our U•A-U-rich triple helix are largely consistent with poly(U•A-U) structures modeled using X-ray fiber diffraction data, i.e. canonical base triple interactions, helical twist of 30–32.7°, helical rise of ∼3.1 Å, pitch of 33.4–36.5 Å, up to 12 base triples per turn, double helix portion classified as A-RNA/A′-RNA, diameter of ∼25 Å and a major groove occupied by the Hoogsteen strand (Figures 1 and 2, Table 2 and Supplementary Tables S3–5) (3–4,19). However, there were several notable differences. The MALAT1_th11 structure shows that an all-C3′-endo triple helix is favorable, in contrast to a previous structural model having both the Hoogsteen and Watson strands in a C2′-endo ribose conformation (19). Moreover, our model demonstrates that having a C3′-endo sugar pucker in a 12-base-triple-per-turn triple helix is stereochemically possible. Previous studies indicated that modeling all three strands of a 12-base-triple-per-turn poly(U•A-U) triple helix with C3′-endo sugar pucker was suboptimal due to a possible distortion of the sugar ring, which may explain (i) the isolated instances of C2′-endo sugar puckers observed in the MALAT1_th11 structure (U50 and A71, Supplementary Figure S6) and natural triple helices as well as (ii) torsion angles that deviate from universal angles (Supplementary Figure S8) (6,8–9,13,16,19). Another important factor for triple helix formation is widening of the major groove. Our A′-RNA-like double helix has a wide major groove of 9.2–11.2 Å due to a low inclination angle of 6.3–8.4° whereas a poly(U•A-U) structural model has a reported major-groove width of 8.5 Å and an A-RNA-like inclination angle of 17.2° (Figure 3, Table 2 and Supplementary Table S5) (19). The MALAT1_th11 triple helix structure establishes the structural parameters for an RNA triple helix and provides direct evidence that an RNA triple helix can exhibit A-RNA character while maintaining proper stereochemistry through subtle changes to phosphate backbone torsion angles.
RNA triple helix is not limited to six consecutive base triples
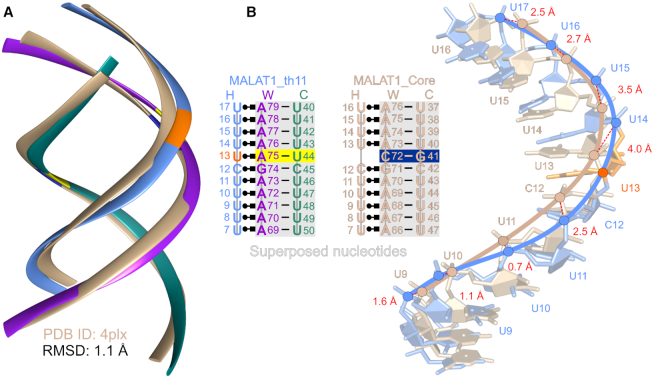
It has been postulated that natural RNA triple helices are restricted to six consecutive base triples (12). Previously, our computational modeling analysis suggested that extension of the MALAT1_Core triple helix would lead to a clash between the 2′-hydroxyl of the Hoogsteen strand and phosphate oxygen of the Watson strand if the stack of base triples was greater than six (12). Here, our structure of MALAT1_th11 shows that this triple helix of 11 consecutive base triples is intact with no major structural distortions nor suboptimal stereochemistry, demonstrating that a triple helix is not limited to six consecutive base triples (Figure 1 and Supplementary Figure S3). However, the length of this triple helix, which resides within an internal loop, may be topologically constrained to one helical turn due to the flanking RNA duplexes. To identify structural differences, we first superposed each of the three strands from the MALAT1_th11 triple helix with the corresponding strands from the MALAT1_Core triple helix. Most structural differences occur in the Hoogsteen strand (RMSD 1.6 Å for all corresponding atom pairs) rather than the WC-helix portion (RMSD 1 Å for all corresponding atom pairs in each strand) (Supplementary Figure S9). By superposing the WC-helix portions of MALAT1_th11 and MALAT1_Core, we determined an RMSD value of 1.1 Å over 424 atom pairs (Figure 4A). Importantly, MALAT1_th11 avoids the 2′-OH…OP2 steric clash because the phosphate backbone near U13 of Hoogsteen strand, namely C12, U14 and U15, are shifted 2.5–4 Å away from the corresponding positions, i.e. C12, U13 and U14, in the superposed MALAT1_Core triple helix (Figure 4B). This expansion by the Hoogsteen strand accommodates all nucleotides, including U13, in the Hoogsteen strand to form a continuous RNA triple helix (Figure 4 and Supplementary Figure S9). However, subtle irregularities do occur, such as the lack of 2′-OH…OP2 hydrogen bonds for U•A-U{1–4} (Figure 2B), the base pair parameters for U•A-U{3–4} deviate from average most frequently among all triples (Supplementary Tables S3–5) and backbone torsion angles of Watson strand differ the most from universal values (Table 3 and Supplementary Figure S8). Currently, it is not clear if these irregularities represent a mild helical reset or if they are artifacts specific to this RNA triple helix and its crystal packing (e.g. C2′-endo sugar pucker for A71 in molecule A). More recently, it was discovered that a protein, methyltransferase-like protein 16, interacts with the MALAT1 triple helix in vivo (53,54). Thus, the C-G doublet that is present in the MALAT1 triple helix may serve a critical role in protein recognition rather than resetting the helical axis to prevent a steric clash. Regardless, the MALAT1_th11 RNA triple helix is not a rigid structure and steric rearrangements in the three strands allow formation of triple helices up to 11 triples; therefore, RNA triple helices with stacks of more than six consecutive base triples may exist in nature.
Figure 4.
Superposition of MALAT1_th11 and MALAT1_Core RNA triple helices. (A) WC-helix of MALAT1_th11 triple helix (tricolor ribbon representation as described in Figure 1) superposed with WC-helix of MALAT1_Core triple helix (tan ribbon representation). RMSD is calculated over 424 atom pairs of WC-helix. (B) Shift of Hoogsteen strand in MALAT1_th11 triple helix (blue stick representation) relative to that in MALAT1_Core (tan stick representation) is observed in superposition analysis. The distances in red font between corresponding P atoms (blue and tan circles) are shown by dashed red lines. Nucleotide numbering is shown in schematics for each triple helix. The C72-G41 base pair in MALAT1_Core is marked by navy blue box and corresponding A75-U44 pair in MALAT_th11 is marked by yellow box while U13 in Hoogsteen strand is orange.
CONCLUSION
In this work, we report the X-ray crystal structure of a U•A-U-rich triple helix spanning 11 consecutive base triples, which is nearly one complete helical turn. By analyzing the structural parameters of this RNA triple helix, we discovered that its Watson–Crick helix is an A′-RNA conformer, with its defining features being a wide and deep major-groove and a decreased inclination angle. One helical twist accommodates 12 base triples per turn. Importantly, the sugar-phosphate backbone, with C3′-endo sugar puckers and glycosidic bonds in the anti conformation for all three strands, shows normal stereochemistry so that the RNA triple helix is not limited to six consecutive base triples, as previously speculated. On the contrary, an RNA triple helix with at least 11 consecutive base triples can form and its solvent-exposed surface presents a unique interface for RNA–protein interactions. Like the high-resolution structural studies of B-DNA in the early 1980s, we have established the structural parameters of one RNA triple helix. It remains to be seen if these parameters are also applicable to other RNA triple helices, such as a C+•G-C-rich triple helix or to a DNA triple helix.
DATA AVAILABILITY
Coordinates and experimental data of X-ray crystal structure have been deposited in PDB (PDB ID: 6svs).
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Suzanne DeGregorio (Yale University) for preparing plasmid, to SER-CAT beamline 22-ID (Argonne National Laboratory) during data collection and to all members of the Brown Laboratory for their helpful discussions and critical review of the manuscript. The authors are also grateful to Neocles B. Leontis (Bowling Green State University) and to Richard Lavery (Institute of Biology and Protein Chemistry) for their kind assistance during data analysis.
Author contributions: A.R. and J.A.B. designed experiments and supervised the work; A.R. and J.P.H. performed the experiments; A.R., M.R., Z.D. and J.A.B. processed the experimental data and performed the analysis; A.R. and J.A.B. wrote the manuscript.
Notes
Present address: Agnieszka Ruszkowska and Milosz Ruszkowski, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznan, Poland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R00GM111430 to J.A.B.]; Clare Boothe Luce Program of the Henry Luce Foundation; University of Notre Dame, Startup Funds (to J.A.B.); Intramural Research Program of the National Cancer Institute, Center for Cancer Research (in part); U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract [W-31-109-Eng-38]. Funding for open access charge: University of Notre Dame, Startup Funds (to J.A.B.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Felsenfeld G., Davies D.R., Rich A.. Formation of a 3-stranded polynucleotide molecule. J. Am. Chem. Soc. 1957; 79:2023–2024. [Google Scholar]
- 2. Hoogsteen K. The crystal and molecular structure of a hydrogen-bonded complex between 1-methylthymine and 9-methyladenine. Acta Crystallogr. 1959; 12:822–823. [Google Scholar]
- 3. Arnott S., Bond P.J.. Structures for poly(U)-poly(A)-poly(U) triple stranded polynucleotides. Nat. N. Biol. 1973; 244:99–101. [DOI] [PubMed] [Google Scholar]
- 4. Arnott S., Bond P.J., Selsing E., Smith P.J.. Models of triple-stranded polynucleotides with optimised stereochemistry. Nucleic Acids Res. 1976; 3:2459–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Theimer C.A., Blois C.A., Feigon J.. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005; 17:671–682. [DOI] [PubMed] [Google Scholar]
- 6. Gilbert S.D., Rambo R.P., Van Tyne D., Batey R.T.. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat. Struct. Mol. Biol. 2008; 15:177–182. [DOI] [PubMed] [Google Scholar]
- 7. Toor N., Keating K.S., Taylor S.D., Pyle A.M.. Crystal structure of a self-spliced group II intron. Science. 2008; 320:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mitton-Fry R.M., DeGregorio S.J., Wang J., Steitz T.A., Steitz J.A.. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010; 330:1244–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith K.D., Shanahan C.A., Moore E.L., Simon A.C., Strobel S.A.. Structural basis of differential ligand recognition by two classes of bis-(3′-5′)-cyclic dimeric guanosine monophosphate-binding riboswitches. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:7757–7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cash D.D., Cohen-Zontag O., Kim N.K., Shefer K., Brown Y., Ulyanov N.B., Tzfati Y., Feigon J.. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:10970–10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liberman J.A., Salim M., Krucinska J., Wedekind J.E.. Structure of a class II preQ1 riboswitch reveals ligand recognition by a new fold. Nat. Chem. Biol. 2013; 9:353–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown J.A., Bulkley D., Wang J., Valenstein M.L., Yario T.A., Steitz T.A., Steitz J.A.. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 2014; 21:633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kang M., Eichhorn C.D., Feigon J.. Structural determinants for ligand capture by a class II preQ1 riboswitch. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E663–E671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liberman J.A., Suddala K.C., Aytenfisu A., Chan D., Belashov I.A., Salim M., Mathews D.H., Spitale R.C., Walter N.G., Wedekind J.E.. Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3485–E3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yan C., Hang J., Wan R., Huang M., Wong C.C., Shi Y.. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015; 349:1182–1191. [DOI] [PubMed] [Google Scholar]
- 16. Huang L., Wang J., Wilson T.J., Lilley D.M.J.. Structure of the guanidine III riboswitch. Cell Chem. Biol. 2017; 24:1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang L., Lilley D.M.J.. Structure and ligand binding of the SAM-V riboswitch. Nucleic Acids Res. 2018; 46:6869–6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Raghunathan G., Miles H.T., Sasisekharan V.. Symmetry and structure of RNA and DNA triple helices. Biopolymers. 1995; 36:333–343. [DOI] [PubMed] [Google Scholar]
- 19. Chandrasekaran R., Giacometti A., Arnott S.. Structure of poly(U)-poly(A)-poly(U). J. Biomol. Struct. Dyn. 2000; 17:1023–1034. [DOI] [PubMed] [Google Scholar]
- 20. Walker S.C., Avis J.M., Conn G.L.. General plasmids for producing RNA in vitro transcripts with homogeneous ends. Nucleic Acids Res. 2003; 31:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kabsch W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 25. Winn M.D., Isupov M.N., Murshudov G.N.. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D. Biol. Crystallogr. 2001; 57:122–133. [DOI] [PubMed] [Google Scholar]
- 26. Lavery R., Moakher M., Maddocks J.H., Petkeviciute D., Zakrzewska K.. Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res. 2009; 37:5917–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blanchet C., Pasi M., Zakrzewska K., Lavery R.. CURVES+ web server for analyzing and visualizing the helical, backbone and groove parameters of nucleic acid structures. Nucleic Acids Res. 2011; 39:W68–W73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu X.J., Olson W.K.. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003; 31:5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu X.J., Bussemaker H.J., Olson W.K.. DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res. 2015; 43:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roberts R.W., Crothers D.M.. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992; 258:1463–1466. [DOI] [PubMed] [Google Scholar]
- 31. Holland J.A., Hoffman D.W.. Structural features and stability of an RNA triple helix in solution. Nucleic Acids Res. 1996; 24:2841–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doherty E.A., Batey R.T., Masquida B., Doudna J.A.. A universal mode of helix packing in RNA. Nat. Struct. Biol. 2001; 8:339–343. [DOI] [PubMed] [Google Scholar]
- 33. Nissen P., Ippolito J.A., Ban N., Moore P.B., Steitz T.A.. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cate J.H., Gooding A.R., Podell E., Zhou K., Golden B.L., Kundrot C.E., Cech T.R., Doudna J.A.. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996; 273:1678–1685. [DOI] [PubMed] [Google Scholar]
- 35. Tamura M., Holbrook S.R.. Sequence and structural conservation in RNA ribose zippers. J. Mol. Biol. 2002; 320:455–474. [DOI] [PubMed] [Google Scholar]
- 36. Leontis N.B., Westhof E.. Geometric nomenclature and classification of RNA base pairs. RNA. 2001; 7:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ghosh A., Bansal M.. A glossary of DNA structures from A to Z. Acta Crystallogr. D. Biol. Crystallogr. 2003; 59:620–626. [DOI] [PubMed] [Google Scholar]
- 38. Drew H.R., Wing R.M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R.E.. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:2179–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fratini A.V., Kopka M.L., Drew H.R., Dickerson R.E.. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J. Biol. Chem. 1982; 257:14686–14707. [PubMed] [Google Scholar]
- 40. Holbrook S.R., Cheong C., Tinoco I. Jr., Kim S.H.. Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature. 1991; 353:579–581. [DOI] [PubMed] [Google Scholar]
- 41. Schindelin H., Zhang M., Bald R., Furste J.P., Erdmann V.A., Heinemann U.. Crystal structure of an RNA dodecamer containing the Escherichia coli Shine-Dalgarno sequence. J. Mol. Biol. 1995; 249:595–603. [DOI] [PubMed] [Google Scholar]
- 42. Lietzke S.E., Barnes C.L., Berglund J.A., Kundrot C.E.. The structure of an RNA dodecamer shows how tandem U-U base pairs increase the range of stable RNA structures and the diversity of recognition sites. Structure. 1996; 4:917–930. [DOI] [PubMed] [Google Scholar]
- 43. Tanaka Y., Fujii S., Hiroaki H., Sakata T., Tanaka T., Uesugi S., Tomita K., Kyogoku Y.. A′-form RNA double helix in the single crystal structure of r(UGAGCUUCGGCUC). Nucleic Acids Res. 1999; 27:949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arnott S., Fuller W., Hodgson A., Prutton I.. Molecular conformations and structure transitions of RNA complementary helices and their possible biological significance. Nature. 1968; 220:561–564. [DOI] [PubMed] [Google Scholar]
- 45. Arnott S., Hukins D.W.L., Dover S.D., Fuller W., Hodgson A.R.. Structures of synthetic polynucleotides in the A-RNA and A′-RNA conformations: X-ray diffraction analyses of the molecular conformations of polyadenylic acid•polyuridylic acid and polyinosinic acid•polycytidylic acid. J. Mol. Biol. 1973; 81:107–122. [DOI] [PubMed] [Google Scholar]
- 46. Yanagi K., Prive G.G., Dickerson R.E.. Analysis of local helix geometry in three B-DNA decamers and eight dodecamers. J. Mol. Biol. 1991; 217:201–214. [DOI] [PubMed] [Google Scholar]
- 47. Zhang X., Zheng Q.C.. Exploring the influence of hyperthermophilic protein Ssh10b on the stability and conformation of RNA by molecular dynamics simulation. Biopolymers. 2018; 109:e23068. [DOI] [PubMed] [Google Scholar]
- 48. O’Connor T., Bina M.. The structure of triple helical poly(U)-poly(A)-poly(U) studied by Raman spectroscopy. J. Biomol. Struct. Dyn. 1984; 2:615–625. [DOI] [PubMed] [Google Scholar]
- 49. Liquier J., Taillandier E., Klinck R., Guittet E., Gouyette C., Huynh-Dinh T.. Spectroscopic studies of chimeric DNA-RNA and RNA 29-base intramolecular triple helices. Nucleic Acids Res. 1995; 23:1722–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gelbin A., Schneider B., Clowney L., Hsieh S.H., Olson W.K., Berman H.M.. Geometric parameters in nucleic acids: sugar and phosphate constituents. J. Am. Chem. Soc. 1996; 118:519–529. [Google Scholar]
- 51. Parkinson G., Vojtechovsky J., Clowney L., Brunger A.T., Berman H.M.. New parameters for the refinement of nucleic acid-containing structures. Acta Crystallogr. D. Biol. Crystallogr. 1996; 52:57–64. [DOI] [PubMed] [Google Scholar]
- 52. Kowiel M., Brzezinski D., Jaskolski M.. Conformation-dependent restraints for polynucleotides: I. Clustering of the geometry of the phosphodiester group. Nucleic Acids Res. 2016; 44:8479–8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brown J.A., Kinzig C.G., DeGregorio S.J., Steitz J.A.. Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:14013–14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Warda A.S., Kretschmer J., Hackert P., Lenz C., Urlaub H., Hobartner C., Sloan K.E., Bohnsack M.T.. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017; 18:2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and experimental data of X-ray crystal structure have been deposited in PDB (PDB ID: 6svs).