Abstract
Aberrant activation of the TAL1 is associated with up to 60% of T-ALL cases and is involved in CTCF-mediated genome organization within the TAL1 locus, suggesting that CTCF boundary plays a pathogenic role in T-ALL. Here, we show that −31-Kb CTCF binding site (−31CBS) serves as chromatin boundary that defines topologically associating domain (TAD) and enhancer/promoter interaction required for TAL1 activation. Deleted or inverted −31CBS impairs TAL1 expression in a context-dependent manner. Deletion of −31CBS reduces chromatin accessibility and blocks long-range interaction between the +51 erythroid enhancer and TAL1 promoter-1 leading to inhibition of TAL1 expression in erythroid cells, but not T-ALL cells. However, in TAL1-expressing T-ALL cells, the leukemia-prone TAL1 promoter-IV specifically interacts with the +19 stem cell enhancer located 19 Kb downstream of TAL1 and this interaction is disrupted by the −31CBS inversion in T-ALL cells. Inversion of −31CBS in Jurkat cells alters chromatin accessibility, histone modifications and CTCF-mediated TAD leading to inhibition of TAL1 expression and TAL1-driven leukemogenesis. Thus, our data reveal that −31CBS acts as critical regulator to define +19-enhancer and the leukemic prone promoter IV interaction for TAL1 activation in T-ALL. Manipulation of CTCF boundary can alter TAL1 TAD and oncogenic transcription networks in leukemogenesis.
INTRODUCTION
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive fatal disease that affects both children and adults. Approximately 30% of T-ALL cases relapse within the first 2 years following diagnosis (1–4). The poor prognosis is a consequence of insufficient knowledge of molecular mechanisms underlying T-ALL pathogenesis. Better understanding of the molecular changes associated with T-ALL biology will lead to development of novel diagnostic and therapeutic strategies.
Activation of TAL1, a basic helix-loop-helix (bHLH) transcription factor, is the most frequent gain-of-function mutation observed in T-ALL patients and is found in 40–60% of T-ALL cases resulted from chromosomal translocation (4–5%), interstitial chromosome deletion (25–30%), or an undefined mechanism (60%) (5–7). Upregulation of Tal1 in T-cells also led to leukemia in mice (8,9). Deletion of TAL1 in T-ALL eliminated the leukemic phenotype and induced apoptosis (10–12), implicating one important role of TAL1 in T-cell neoplastic disease. Despite having identified several enhancers in TAL1 regulation, it remains largely unknown how these enhancers are differentially utilized and whether they are involved in TAL1 aberration in T-ALL.
TAL1 is required for HSC self-renewal (13) and the commitment of hematopoietic lineages (14,15). Deletion of Tal1 in mice leads to embryonic lethality at embryonic day 9.5 (E9.5) due to a complete loss of hematopoietic cells (16,17). Further, Tal1-null embryonic stem cells (ESCs) are unable to generate both primitive and definitive erythroid cells in vitro and do not contribute to hematopoiesis in vivo in a chimeric mouse (18,19). These results demonstrate that TAL1 acts as a master regulator of hematopoiesis. Because of its relevance to normal hematopoiesis and T-ALL, transcriptional regulation of the TAL1 becomes a fundamental issue for controlling normal and malignant hematopoiesis.
The human TAL1 gene is located on chromosome 1p32 and is tightly regulated by various cis regulatory elements (20–23). The organization of this 199-Kb gene dense region are conserved among chicken, mouse, and human genomes (21). Several studies including transgenic reporter and knock-in mouse, DNase I hypersensitive assay, and ChIP on chip assays have revealed that expression of the TAL1 during hematopoiesis is controlled by distinct promoters and enhancers. Some of these enhancers are located far away from the transcription start site (TSS) of the TAL1 gene (23–26,20). However, the detailed mechanisms governing differential enhancer/promoter actions that selectively activate TAL1 in different stages of hematopoiesis and leukemogenesis remain unclear and must still be illustrated.
Genome-wide studies of K562 cells (27) and CD4+ T cells (28) revealed that there are four CTCF binding sites (CBSs) in the TAL1 locus bound by CTCF to separate the STIL, TAL1 and MAP17 genes (27,29). CTCF, as enhancer-blocking insulator, prevents enhancer and promoter interactions when placed between them. CTCF also acts as chromatin boundary to play a critical role in defining topologically associating domains (TAD) and chromatin signature within the TAD (30,31). CTCF is a highly conserved zinc-finger protein involved in transcription activation/repression, insulation, imprinting and X chromosome inactivation (32–35). Recent studies implicated that CTCF regulates intra- and interchromosomal contacts within the nucleus at several developmentally regulated genomic loci (36,37) and suggested a primary function for CTCF in global organization of chromatin architecture (32,38). It is conceivable that altered CTCF defined boundary might result in inappropriate enhancer/promoter interactions leading to changes in transcription of oncogene or tumor suppressor. We and others showed that the −31Kb CBS (−31CBS) plays a critical role in organizing enhancer/promoter communications to activate TAL1 (39,40). However, it remains to be determined whether CTCF is directly involved in enhancer/promoter interactions for TAL1 activation. Furthermore, whether and how aberrant TAL1 activation is depended on CTCF defined chromatin neighborhood within the TAL1 locus. Defining molecular mechanisms that are involved in differential activation of TAL1 are critical for understanding its role in the pathogenesis of T-ALL for potential target therapy.
Here, we demonstrated that inversion of the −31CBS orientation alters TAL1 three-dimensional genome organization and chromatin signature in the TAL1 locus that results in inhibition of the TAL1-driven oncogenic transcription program and T-cell leukemogenesis. Thus, targeting the CTCF-mediated chromatin neighborhood provides an opportunity to correct the aberrant oncogene transcription program and to develop new molecular therapy for acute leukemia.
MATERIALS AND METHODS
Patient samples and cell lines
Primary T-ALL patient samples including TAL1-positive (08H125) and TAL1-negative (08H028) blasts were obtained from the Quebec Leukemia Cell Bank (41) and expanded in non-obese diabetic (NOD)/LtSz-severe combined immunodeficiency (SCID) IL2Rγcnull (NSG) female mice as previously described (42). All experiments were approved by The Ottawa Health Science Network Research Ethics Board (2009009-01H). K562 and Jurkat cells were cultured in RPMI1640 supplemented with 10% fetal bovine serum as described before (43). All cell lines were verified by short tandem repeat analysis and tested for mycoplasma contamination.
T-ALL xenograft model
Wild-type (WT) control, −31CBS-KO or −31CBS-INV Jurkat cells were transduced with lentiviral particles expressing the mCHERRY fluorescent protein and luciferase (FUW-CHERRY-puro-LUC) (44). The viability and transduction efficiency of cells was measured by flow cytometry and was equivalent for all conditions. NSG mice (6–8 weeks old, The Jackson Laboratory) were maintained under sterile conditions. Mice were sublethally irradiated at 3-Gy total body irradiation and anesthetized with a solution of 10% ketamine and 5% xylasine. NGS mice were injected with WT control, −31CBS-KO or −31CBS-INV Jurkat cells at a dose of 50 000 cells per mouse by intrafemoral injection. Leukemia progression for each condition (control, −31CBS-KO or −31CBS-INV) was then measured 1, 3, 7, 10 and 13 days after injection by invivo bio-imaging using the in Vivo Imaging System (IVIS, Xenogen). Briefly, mice were anesthetized and given an intraperitoneal injection of 100 mg/kg D-luciferin (Perkin Elmer #122799). After 5–10 min. luciferase activity was measured by IVIS. Animals were used in accordance with a protocol approved by the Institutional Animal Care and use committees of the Ottawa Hospital Research Institute and the Pennsylvania State University.
RNA extraction, qRT-PCR and RNA-seq
Total RNA was extracted using TRIzol Reagent (Invitrogen). A total of 1 μg of total RNA was subjected to reverse transcription with M-MLV Reverse Transcriptase (New England Biolabs) and analyzed by the CFX96™ Real-Time System (Bio-Rad). RNA-seq library was prepared by ‘TruSeq Stranded mRNA Library Prep Kit’ (Illumina, #RS-122-2101), and paired-end RNA-seq was performed by the Penn State University Genome Sciences and Bioinformatics Facility using the Hiseq2500 (100PE) platform with 40 M reads per sample according to standard protocols. The primers are listed in the Supplementary Table S1.
ChIP, ChIP-seq and ATAC-seq
ChIP and ChIP-seq were performed as previously described (30,45–46). Briefly, cells were harvested and fixed with 1% formaldehyde. Chromatin was fragmented using a Bioruptor™ UCD200 sonicator (Diagenode SA, Belgium) on high power at 4°C and precipitated using antibodies against CTCF (Millipore, #07-729), H3K4me3 (Millipore, #04-745), H3K4me2 (Millipore, 07-030), H3K27Ac (Abcam, #ab4729) and H3K9/14ac (Diagenode, #C15410005). ChIP-DNA was recovered using phenol/chloroform/isoamyl alcohol and assayed by quantitative polymerase chain reaction (qPCR). ChIP primers are listed in the Supplementary Table S1. For ChIP-seq, the ChIP DNA library was prepared by ‘TruSeq ChIP Sample Preparation Kit’ (Illumina, #IP-202-1024). Assay for transposase-accessible chromatin using sequencing (ATAC-seq) was performed as previously described (30). Briefly, 5 × 104 cells were harvested and lysed in lysis buffer on ice. Nuclei were extracted and processed by ‘Nextera DNA Library Preparation Kit’ (Illumina). Libraries were purified with the Qiagen MinElute PCR Purification Kit. ChIP-seq and ATAC-seq libraries were sequenced using the Hiseq2500 (100PE) platform with 25M reads per sample.
Sequencing data analysis
RNA-seq, ChIP-seq and ATAC-seq analyses were performed as previously described (30). In brief, sequencing reads were aligned to human genome (hg19) using TopHat (version 2.0) and Bowtie2 (47–49). A detailed data analysis protocol is provided in Supplementary Materials and Methods. Sequence reads have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) under accession number GSE135320.
Chromosome conformation capture (3C), circular chromosome conformation capture (4C) and Hi-C
Chromosome conformation capture (3C) assay was performed as described previously with minor modifications (40). In brief, 2 × 106 cells were cross-linked with 2% formaldehyde. Nuclei were extracted and digested with 800U of BamH1 or DpnII (New England Biolabs) at 37°C overnight with shaking. The digested chromatin was treated with 400U of T4 DNA ligase (New England Biolabs) to ligate interacting DNA fragments at 16°C for 3 days. The ligated chromatin was reverse-crosslinked, followed by phenol/chloroform extraction to purify 3C DNA. Purified 3C ligated DNA was amplified using PCR, the products were visualized on a 2% agarose gel with SYBR safe (Life Technologies). The PCR products were cloned into pGEM-T Easy vector (Promega) for Sanger sequencing (Genewiz). Relative crosslinking frequencies were calculated and plotted after normalization to the loading control. The 3C primers are listed in the Supplementary Table S1.
The circular chromosome conformation capture (4C) assay was performed as described previously (30,50) with minor modifications. Briefly, 1 × 106 cells were cross-linked with 2% formaldehyde and digested with restriction enzyme overnight. Circular ligated chromatin was reversely cross-linked. Then, 4C DNA library was prepared by ‘TruSeq ChIP Sample Preparation Kit’ (Illumina, #IP-202-1024) for further sequencing.
Hi-C assay was performed to generate a genome-wide interaction as described previously with Arima-HiC Kit (Cat: A410030) (https://arimagenomics.com/) with minor modifications. In brief, 2 × 106 cells were collected and cross-linked in 10 ml of phosphate-buffered saline (PBS) buffer containing 1% formaldehyde at room temperature for 10 min. The reaction was quenched by 0.125 M glycine solution. Cross-linked cell pellet was washed in 1× PBS buffer and collected. Cross-linked cell pellet was treated with lysis Buffer in a tube and incubated at 4°C for 15 min, and then added conditioning solution to incubate at 62°C for 10 min. Add stop solution and incubate at 37°C for 15 min. Cell pellet was digested with reaction buffer and restriction enzyme cocktail to digest chromatin overnight or for at least 2 h at 37°C with rotation. Digested DNA was purified with DNA purification beads (AMPure XP Beads), and then quantified its concentration with Qubit. A total of 750 ng of DNA per sample was sheared through sonication (Bioruptor) with default parameters (30 s ON, 30 s OFF pulse intervals). Fragmented DNA must then be size-selected to have a size distribution between 200 and 600 bp. 250ng of size-selected DNA was used to generate sequence library with KAPA Hyper Prep Kit (Catalog # KK8500, KK4824 and KK8502). Final libraries were submitted to paired-end sequencing of 100 bp length on an Illumina HiSeq 2500. The detailed Hi-C data analysis protocol is provided in the Supplementary Materials and Methods. Sequence reads have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) under accession number GSE135320.
CRISPR/Cas9 for genome editing
We generated −31CBS deletion and inversion cell lines by CRISPR/Cas9 technology. Single-guide RNA (sgRNA) oligos (Sigma) were designed using the CRISPR Design Tool (crispr.mit.edu) according to the target genome sequence and cloned into a pLKO5.sgRNA.EFS.tRFP (Addgene, # 57823) or pL-CRISPR.EFS.GFP (Addgene, # 57818) lentiviral vector. After lentiviral infection, GFP+/RFP+ cells were sorted by flow cytometry. Single cell clones were then selected, and −31CBS deletion and inversion clones were verified by genotyping PCR and Sanger sequencing. The genotyping primers are listed in the Supplementary Table S1.
RESULTS
−31CBS is critical for TAL1 expression in erythroid cells, but not in TAL1-expressing T-ALL cells
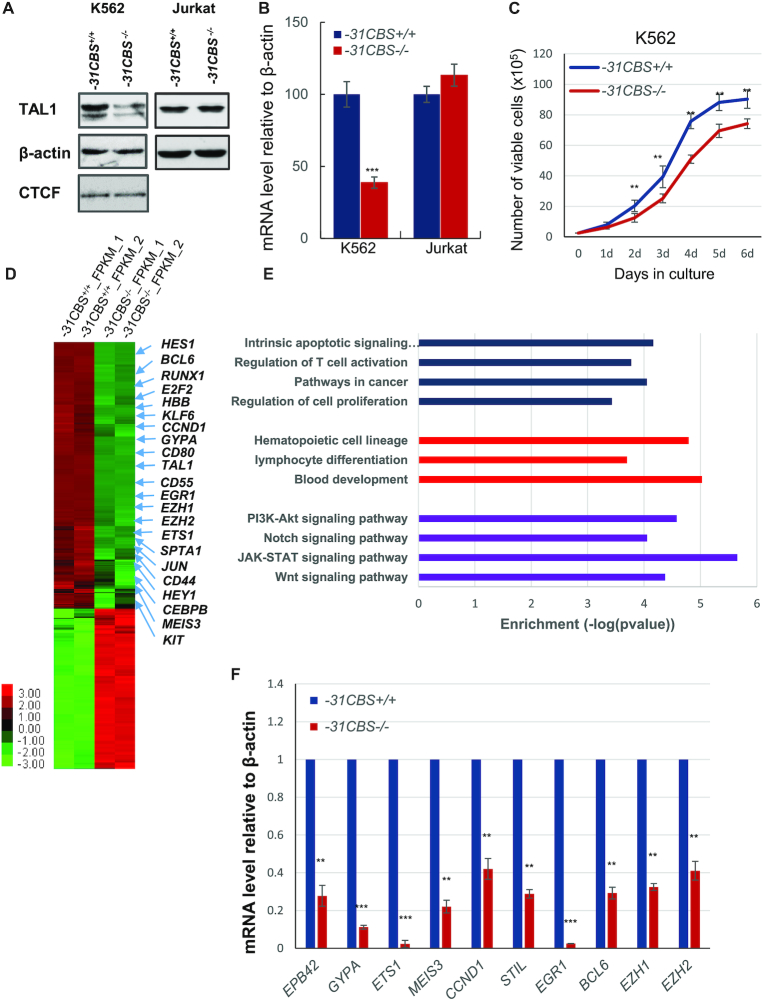
CTCF ChIP-seq study in different hematopoietic cells has established that there are four CBSs flanking TAL1 gene, +57CBS, +53CBS, +40CBS and −31CBS relative to TAL1 TSS. Computational analysis revealed that orientations of all four CBSs are point toward TAL1 gene (51). Recently, we and others employed extensive 3C analyses to demonstrate that −31CBS functions as a nucleation center and recruits +53CBS, +57CBS, +51 erythroid enhancer and promoter-I to form an active chromatin domain in much higher frequencies in erythroid cells than in T-ALL cells (39,40). To characterize the direct role of −31CBS in chromatin domain organization and TAL1 transcription, we employed CRISPR-Cas9 to delete the core CTCF motif of the −31CBS in the TAL1-expressed erythroleukemia K562 and T-ALL Jurkat cells (Supplementary Figure S1A). Deletion of −31CBS (−31CBS−/−) exhibited very distinct effects on TAL1 expression between K562 and Jurkat cells (Figure 1A and B). −31CBS−/− significantly decreased TAL1 transcript and protein levels in K562 cells, but not in Jurkat cells (Figure 1A and B; Supplementary Figure S1B and C). −31CBS−/− resulted in decreased cell growth of erythroid K562 cells (Figure 1C), while there is no negative growth effect on T-ALL Jurkat cells (data not shown). Given that TAL1 is required for both hematopoietic and leukemia cell survival, we further examined if −31CBS−/− affected cell cycle progression of K562 cells. −31CBS−/− in K562 cells resulted in increased Sub-G1 apoptotic cells and reduction of the G2 phase (Supplementary Figure S1D).
Figure 1.
Deletion of −31CBS inhibits TAL1 expression in K562 cells, but not T-ALL cells. (A) Western blot analysis of TAL1 protein levels in the −31CBS deletion clone in K562 (Clone D10) or Jurkat cells compared to WT control. β-actin is served as a loading control. (B) RT-qPCR analysis of TAL1 mRNA levels in the −31CBS deleted K562 (Clone D10) or Jurkat cells compared to WT control. (C) Proliferation curves of the WT control and −31CBS−/− K562 cells (Clone D10) were assessed by counting cell viability for 6 days. (D) Heat map of RNA-seq analysis shows upregulated and downregulated genes upon −31CBS deletion in K562 cells. (E) Downregulated genes upon the −31CBS deletion were analyzed and annotated by Gene Ontology analysis. (F) RT-qPCR analysis and confirmation of representative downregulated genes identified in the RNA-seq analysis. The data are presented as mean ± SD from three independent experiments, *P < 0.05, **P < 0.01, ***P < 0.001 by student’s t-test.
To explore the mechanism underlying the effect of the −31CBS−/−, we performed RNA-seq analysis comparing WT and the two different −31CBS−/− K562 cell clones. A total of 1568 genes exhibited more than 2-fold decreases in mRNA levels, whereas 939 genes had increased expression upon −31CBS deletion (Figures 1D and Supplementary Table S2). Gene ontology (GO) analysis revealed that pathways involved in hematopoietic cell lineage, lymphocyte differentiation, blood development, T-cell activation, Notch signaling, JAK-STAT signaling and Wnt signaling, etc. were affected by −31CBS−/− (Figure 1E). Among them, TAL1, HBB, GYPA, RUNX1, SPTA1, E2F2, CCND1 and BCL6 were significantly downregulated (Figure 1D). These genes play an important role in hematopoietic/erythroid differentiation and cell cycle regulation. A subset of downregulated genes involved in erythropoiesis and cell cycle were further verified by RT-qPCR analysis (Figure 1F). Perturbation of the cell-cycle pathway by GO term analysis is consistent with the observation that −31CBS−/− blocks cell cycle progression leading to apoptosis (Figure 1C and Supplementary Figure S1D).). However, given that −31CBS is overlapped with the STIL coding exon, the changes in STIL gene expression may be directly resulted from disruption of this exon rather than altered genome organization (Figure 1F).
−31CBS is required for formation of active TAL1 chromatin domain in erythroid cells
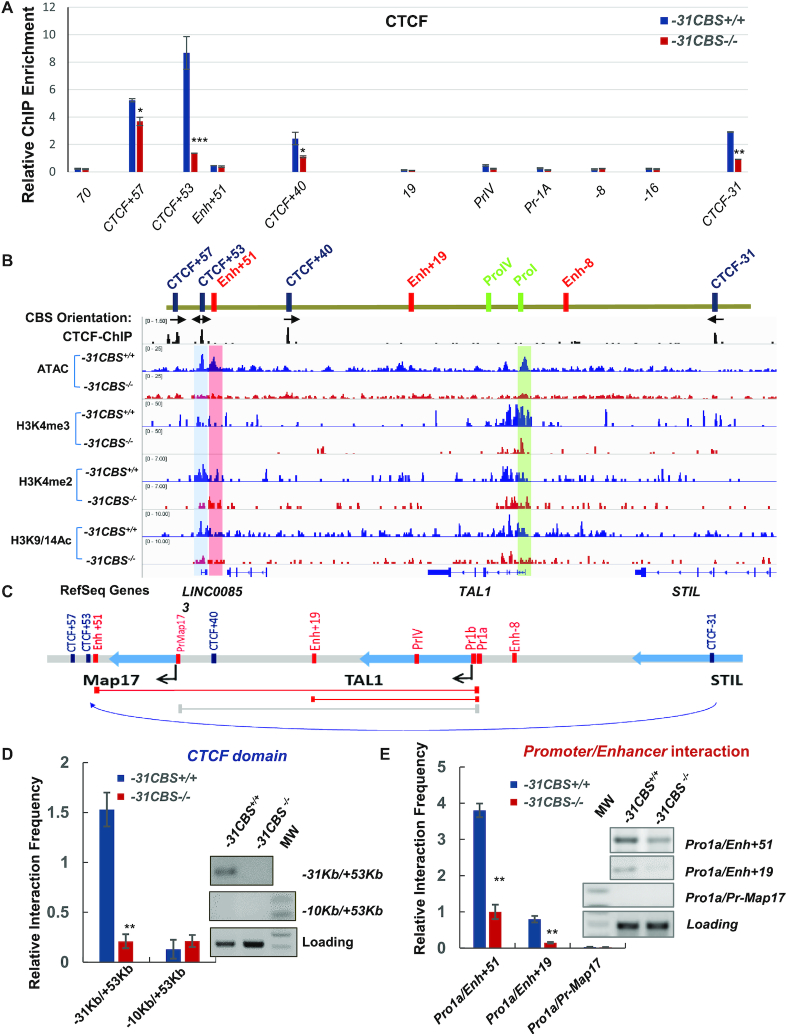
It was reported that interaction between −31CBS and +53/+57CBS facilitates the +51 enhancer to activate the TAL1 promoter-I in erythroid cells (40). Thus, it is likely that −31CBS−/− will interfere with chromatin signatures in the TAL1 locus required for TAL1 expression. To test this possibility, we performed ChIP to investigate the effect of the −31CBS−/− on CTCF binding in the TAL1 locus. There are four CBSs in the TAL1 locus located at −31Kb, +40Kb, +53Kb and +57Kb relative to the TAL1 TSS (27,29). While −31CBS−/− only reduced CTCF binding to the −31CBS in Jurkat cells (data not shown), −31CBS−/− in K562 cells significantly inhibited CTCF binding to all CBSs in the TAL1 locus (Figure 2A). As a control, −31CBS−/− did not alter CTCF protein levels in K562 cells (Figure 1A). The data indicate that −31CBS may play an important role in organizing the erythroid specific chromatin domain to support TAL1 transcription in K562 cells.
Figure 2.
−31CBS loss alters CTCF defined chromatin domain and looped enhancer/promoter interaction in the TAL1 locus in erythroid cells. (A) ChIP analysis of CTCF binding across the TAL1 locus compared the WT control and −31CBS−/− cells. (B) CTCF binding and CTCF binding site orientations in the TAL1 locus were indicated. The abundance of ATAC-seq peak as well as enrichment of H3K4me3, H3K4me2 and H3K9/14Ac patterns at the TAL1 locus analyzed by ATAC-seq and ChIP-seq compared WT control and −31CBS−/− K562 cells, respectively. (C) Schematic representation of the 3C fragments after restriction digestion in the human TAL1 locus. Red lines: the primer used to detect known 3C interactions. Grey line: negative control primers. (D) PCR analysis of 3C products examining the −31CBS mediated DNA looping in the TAL1 locus compared WT control and −31CBS−/− K562 cells. Relative interaction frequency is quantitated in triplicate experiments. (E) PCR assay examining the enhancer and promoter interactions in the TAL1 locus upon −31CBS deletion in K562 cells. Relative interaction frequency is quantitated in triplicate experiments. The data from the panel A, D and E are presented as mean ± SD from three independent experiments, *P < 0.05, **P < 0.01, ***P < 0.001 by student’s t-test.
Next, we examined the effect of −31CBS−/− on chromatin accessibility and histone modifications over the TAL1 locus in K562 cells by ATAC-seq and ChIP-seq analyses. Wild-type cells exhibit highly accessible chromatin regions at the +53CBS, +51 enhancer and promoter-I that are correlated with enriched H3K4me2, H3K4me3 and H3K9/K14ac modifications (Figure 2B). −31CBS−/− resulted in decreases of enhancer/promoter accessibility and H3K4me3, H3K4me2 and H3K9/K14ac modifications, especially in the +53CBS, +51 enhancer and the TAL1 promoter-I (Figure 2B). The ChIP-qPCR confirmed that −31CBS−/− (two clones) leads to diminishment of active chromatin signature in the TAL1 locus in K562 cells (Supplementary Figures S2A and B). Furthermore, H3K4me3 enrichment was markedly reduced in the promoters of a subset of TAL1 target genes identified by RNA-seq (Supplementary Figure S2C), perhaps, due to the reduction of TAL1 expression by −31CBS deletion.
Given that CTCF-mediated chromatin organization plays an important role in regulating enhancer/promoter interactions, we carried out 3C analyses to examine whether −31CBS−/−affects −31CBS loop interactions and TAL1 enhancer/promoter interaction (Figure 2C). Consistent with previous reports (40), −31CBS interacts with +53 CTCF site to form an erythroid-specific CTCF loop in WT K562 cells (Figure 2D). As a result of the −31CBS/+53CBS CTCF organization, the +51 erythroid enhancer also strongly interacts with the TAL1 promoter-I to activate TAL1 gene (Figure 2E). Interestingly, −31CBS−/− abolished the formation of +53/−31CBS chromatin loop (Figure 2D), and significantly reduced the interaction frequencies of the TAL1 promoter-I with the +51 erythroid enhancer or the +19 enhancer (Figure 2E) demonstrating that −31CBS plays a direct role in organizing erythroid genome to allow enhancer/promoter interactions and transcription of TAL1.
The T-ALL-prone TAL1 promoter-IV differentially interacts with stem cell enhancer in T-ALL cells
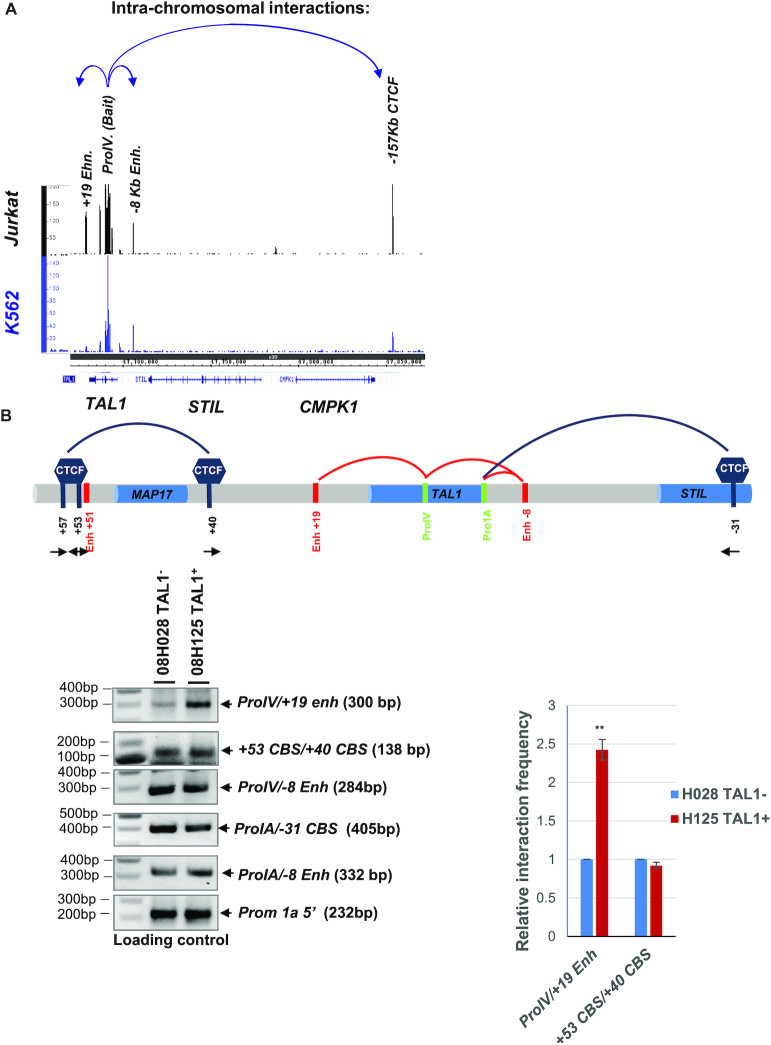
In addition to the TAL1 promoter-I, promoter-IV of the TAL1 gene is also highly active in T-ALL (52,53). Interestingly, all cases of the chromosomal translocation involving the TAL1 gene are located in the promoter-IV region (53–56,53–56). Thus, we reason that the promoter-IV may play a differential role in TAL1 transcription in T-ALL cells. To test this hypothesis, we performed the 4C-seq using the TAL1 promoter-IV as a viewpoint, comparing differential long-range interactions associated with the promoter-IV of T-ALL Jurkat and erythroid K562 cells. When we compared the cumulative normalized intensities of the TAL1 promoter-IV interactome in Jurkat cells with internal control or the interactions in K562 cells, we found that the TAL1 promoter-IV-mediated loops were very distinct from that of the TAL1 promoter-I. The TAL1 promoter-IV interacts with the +19 stem cell enhancer and the -8 T-ALL super-enhancer in Jurkat cells, while K562 cells show no interaction between promoter-IV and the +19 enhancer (Figure 3A). It was previously reported that +40CBS predominantly interacted with +53CBS in T-ALL cell line and patient samples, while −31CBS associated with +53CBS in erythroid cells (40). This CTCF organization may allow TAL1 promoter to be activated by enhancers located within or outside the TAL1 locus (40). When we examine the interaction between the TAL1 promoter-IV and +19 enhancer in a TAL1-negative primary T-ALL patient sample (08H028) and a TAL1-positive patient sample (08H125), the TAL1 promoter-IV only weakly interacts with the +19 enhancer in the TAL1-negative patient (08H028) (Figure 3B). In contrast, the TAL1 promoter-IV strongly interacts with +19 enhancer in the TAL1-positive T-ALL patient (08H125) (Figure 3B). Thus, our data reveal that the +19 stem cell enhancer located 19 Kb downstream of the TAL1 gene may also drive TAL1 expression in T-ALL leukemia.
Figure 3.
The TAL1 promoter IV interacts with +19 stem cell enhancer in T-ALL cells. (A) Long-range intra-chromosomal interactions in the TAL1 locus determined by 4C-seq analysis using the TAL1 Promoter IV as a viewpoint compared K562 to Jurkat cells. The blue arrows indicate long-range interactions detected by 4C-seq. (B) 3C analysis confirms the interaction between the TAL1 promoter IV and +19 stem cell enhancer in the TAL1-negative (08H028) or TAL1-positive (08H125) primary T-ALL patient samples. Relative interaction frequency is quantitated from triplicate experiments. The data are presented as mean ± SD, **P < 0.01 by student’s t-test.
Inversion of −31CBS inhibits TAL1 expression and TAL1-mediated T-ALL leukemogenesis
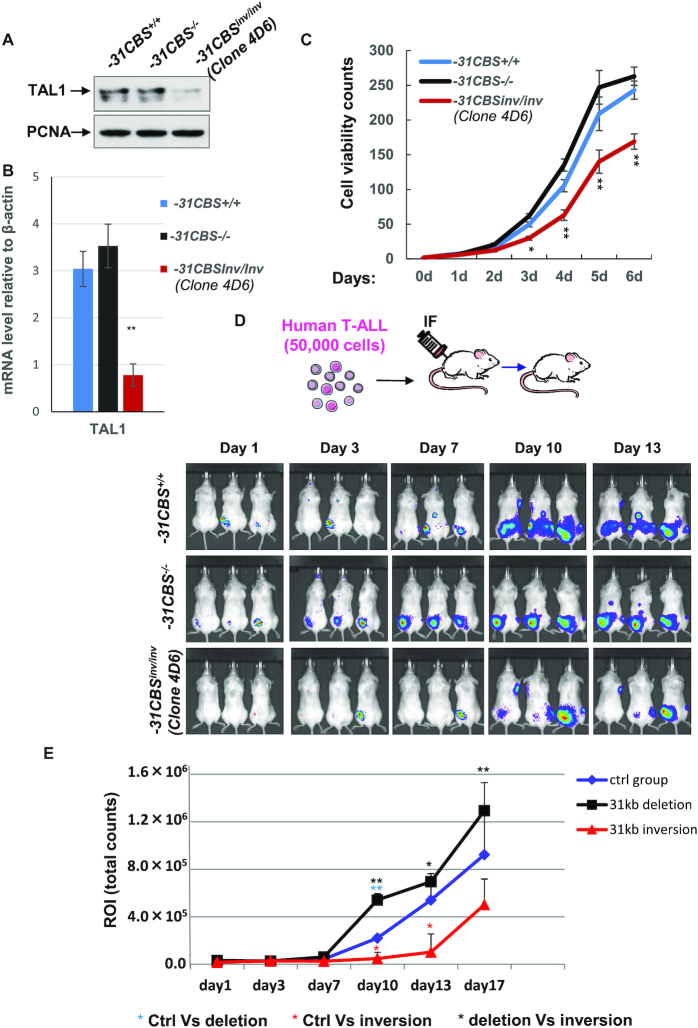
It was recently reported that altered CBS orientation reshapes the genome organization and leads to changes in CTCF protected gene expression (57,58) suggesting that CBS polarity determines the status of gene activity. Convergent orientation of each pair of CBSs flanking TAL1 gene (Figure 2B) may establish TAL1 TAD that is required for oncogenic TAL1 expression. We thus examined whether changing the −31CBS orientation would alter genome topology within the CTCF-defined chromatin neighborhood and prevent ectopic activation of TAL1 in T-ALL cells. Using 2-sgRNA-mediated CRISPR-Cas9 editing, we screened the −31CBS-KO Jurkat cell pools for single clones containing −31CBS inversion (−31CBSinv/inv). Two clones that exhibited −31CBSinv/inv were obtained (Supplementary Figure S3A). −31CBSinv/inv significantly inhibits TAL1 transcription and protein levels (Figure 4A and B; Supplementary Figure S3B and C). However, −31CBSinv/inv does not affect CTCF binding (Supplementary Figure S3D), suggesting that inversion may only alter CBS polarity-mediated genome topology, but not CTCF binding per se. Given that TAL1 is required for leukemic cell survival (10–12), we further test if −31CBSinv/inv affects T-ALL cell proliferation and leukemogenesis in vitro and in vivo as compared to the vector control and the −31CBS−/− clone. −31CBSinv/inv significantly inhibited proliferation of Jurkat cells (Figure 4C and Supplementary Figure S3E) and colony growth in soft agar (Supplementary Figure S3F), presumably by inhibiting TAL1 expression. Interestingly, −31CBS−/−, which slightly increases TAL1 transcription, indeed enhances cell proliferation (Figure 4C).
Figure 4.
Inversion of −31CBS inhibits T-ALL cell proliferation invitro and leukemogenesis invivo. (A) Western blot analysis of TAL1 protein levels in the −31CBS−/− and −31CBSinv/inv (Clone 4D6) Jurkat cells as compared to the WT control cells. PCNA serves as a loading control. (B) RT-qPCR analysis of TAL1 RNA expression in the −31CBS−/− and −31CBSinv/inv (Clone 4D6) Jurkat cells as compared to the WT control. (C) Proliferation curves of WT control, −31CBS−/− and −31CBSinv/inv (Clone 4D6) Jurkat cells were assessed by counting cell viability. (D) A total of 50 000 mCherry-Luc-Jurkat cells are intrafemorally injected into mice. Bioluminescent imaging of tumor burden from mice injected with WT control, −31CBS−/− and −31CBSinv/inv mCherry-Luc-Jurkat cells at day 0, 3, 7, 9 or 13. (E) Total flux from the respective regions of interest (ROI) (photons/sec) is evaluated in mice injected with WT control, −31CBS−/− and −31CBSinv/inv Jurkat cells at day 1, 3, 7, 10, 13 or 17. *P < 0.05, **P < 0.01 by student t test.
To examine whether −31CBSinv/inv affects T-ALL leukemogenesis in vivo, we employed a T-ALL xenograft mouse model (42) by transplanting 50 000 vector control, −31CBS−/− and −31CBSinv/inv Jurkat cells into irradiated NSG mice (Figure 4D, top). All mice transplanted with WT or −31CBS−/− Jurkat cells developed leukemia 7 days after transplantation. However, the −31CBSinv/inv cells showed significant delays and inhibition of T-ALL disease progression (Figure 4D and E). Interestingly, consistent with slightly increased growth of −31CBS−/− Jurkat cells in vitro, −31CBS−/− resulted in more aggressive and faster engraftment of leukemia cells in vivo (Figure 4D and E).
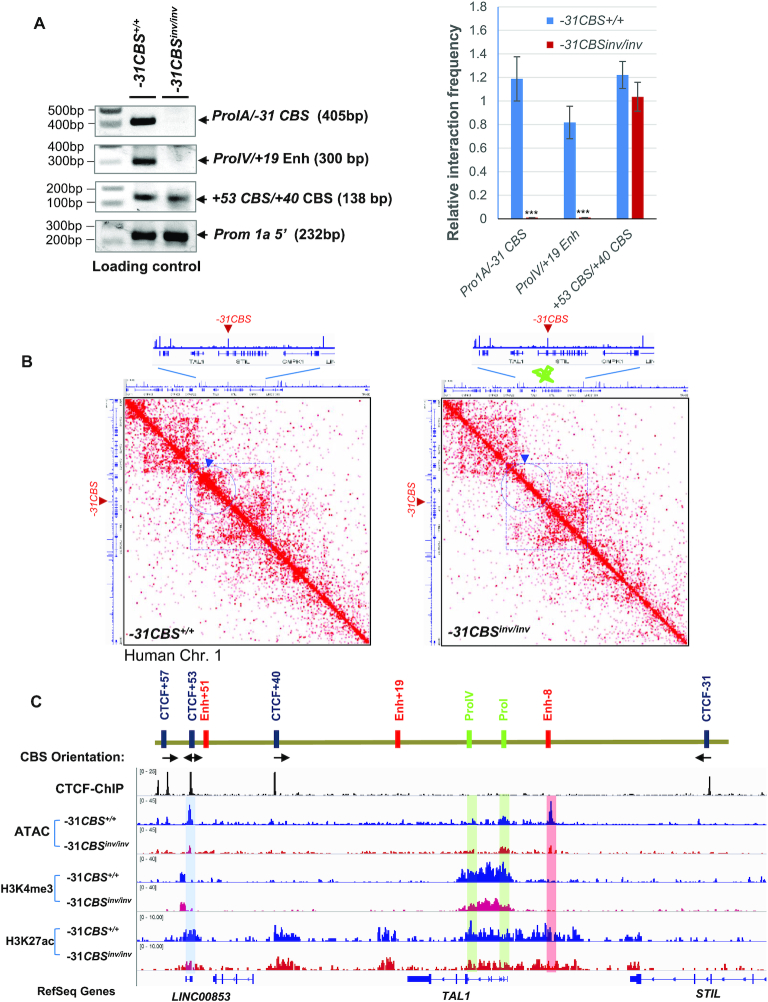
Inversion of −31CBS alters TAL1 genome topology in T-ALL cells
Next, we employed 3C analysis to examine whether −31CBSinv/inv alters genome topology. Although both promoter-I and promoter-IV interact with −8 superenhancer in the TAL1-negative and TAL1-positive T-ALL patient cells (Figure 3B), −31CBSinv/inv does not prevent the −8 super-enhancer to interact with the TAL1 promoters (Supplementary Figure S4A). In contrast, inversion resulted in complete loss of interactions of −31CBS and the TAL1 promoter-1a, and disruption of proximity between the TAL1 promoter-IV and +19 enhancer (Figure 5A and Supplementary Figure S4B). The interactions of the TAL1 promoter-IV/+19 enhancer, as well as the −31CBS/TAL1 promoter 1A was further validated by sequencing analysis as the fusion molecules constituting two genomic regions were confirmed by Sanger sequencing (Supplementary Figure S4C). We further investigated whether −31CBSinv/inv alters T-ALL genome organization by Hi-C analysis, comparing vector control and the −31CBSinv/inv Jurkat cells (Figure 5B, Supplementary Table S3). Inversion of −31CBS has not or very little effects on global chromosome organization (Supplementary Figure S5A-S5B), rather it affects chromatin organization at the TAL1 locus (Figure 5B). In control cells, there is predominant small TAD formed between +53CBS and +40CBS (blue arrowhead), and a weak TAD between +53CBS and −31CBS (broken blue cycle dash line) (Figure 5B, left). Interestingly, +53CBS also interact with −157CBS to form larger TAD (Figure 5B, broken purple square dash line). −31CBSinv/inv disrupts the TAD formation in the TAL1 locus, especially the smaller TAD between +53/57CBS and +40CBS (blue arrowhead), as well as the larger TAD between +53CBS and −157CBS (broken purple square dash line) encompassing the whole TAL1 locus (Figure 5B, right). Thus, the data demonstrate that −31CBS plays critical role in organizing the 3D TAL1 genome to support TAL1 oncogenic transcription program.
Figure 5.
Inversion of −31CBS inhibits TAL1 genome topology. (A) PCR results of 3C assays examining DNA looping at the TAL1 locus in WT control and −31CBSinv/inv (Clone #1–8) Jurkat cells (left). Relative interaction frequency is quantitated in biological triplicate experiments (right). (B) Hi-C analysis of genome organization compared between WT control and −31CBSinv/inv (Clone #1–8) Jurkat cells. (C) ATAC-seq and ChIP-seq analysis of chromatin accessibility as well as enrichment of H3K4me3, H3K4me2 and H3K27Ac patterns at the TAL1 locus compared between WT control and −31CBSinv/inv (Clone #1–8) Jurkat cells. ***P < 0.001 by student’s t-test.
−31CBSinv/inv remodels chromatin signature leading to perturbation of TAL1-driven leukemic transcription program
The CTCF boundary defines the chromatin neighborhood and genome topology. Altering CBS may change chromatin modifications and accessibility. To address this, we assessed the effects of −31CBSinv/inv on enhancer/promoter chromatin accessibility and histone modifications by ATAC-seq and ChIP-seq assays, comparing vector control and −31CBSinv/inv Jurkat cells. −31CBSinv/inv leads to decreases in chromatin accessibility in −31CBS, -8 Jurkat super-enhancer, TAL1 promoter-I and +53CBS. Consistent with loss of chromatin accessibility, H3K4me3, a promoter mark, is significantly decreased in the TAL1 promoter regions, while enhancer mark H3K27ac is reduced in the -8 super-enhancer, promoter-IV, +19 enhancer and +53CBS (Figure 5C). Effects of −31CBSinv/inv on histone modifications in the TAL1 locus was further validated in two independent −31CBS inverted Jurkat clones (Supplementary Figure S5C and D). This suggests that −31CBSinv/inv perturbs the CTCF-defined leukemic chromatin neighborhood required for the TAL1 oncogene activation.
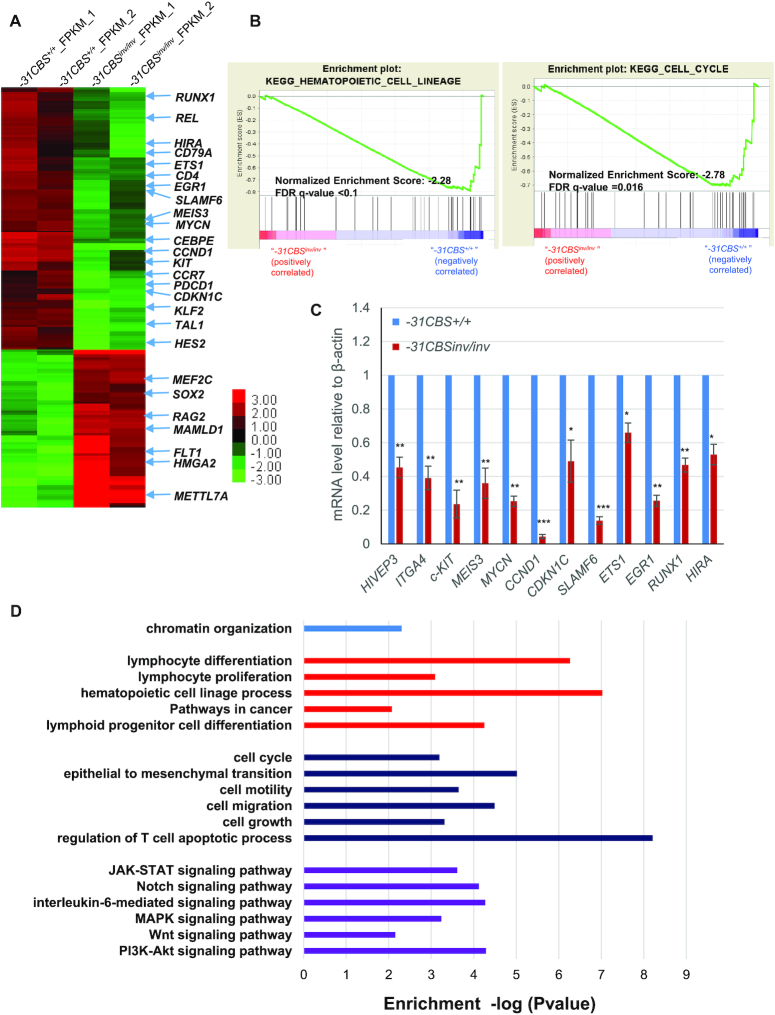
To investigate whether −31CBSinv/inv perturbs the TAL1-driven leukemic transcription program, we performed RNA-seq analysis comparing the −31CBSinv/inv Jurkat cells to the vector control. A total of 189 genes exhibited greater than 2-fold decreases in mRNA levels, while 70 genes had increased expression upon −31CBSinv/inv (Figure 6A and Supplementary Table S4). −31CBSinv/inv significantly impairs the transcription of many genes important for hematopoiesis, HSC function, cell cycle and leukemogenesis (Figure 6A and B). Many known TAL1 target genes (∼19%) are affected by −31CBSinv/inv (Supplementary Figure S6A). A subset of known TAL1 target genes important for T-ALL leukemogenesis were confirmed by RT-qPCR (Figure 6C). Together, the data suggest that −31CBSinv/inv alters the TAL1-driven transcription program by remodeling genome topology and chromatin accessibility/modifications. GO analysis further revealed that many of the pathways affected by −31CBSinv/inv are involved in cell cycle, apoptosis, hematopoiesis, lymphoid progenitor/cell differentiation, JAK-STAT signaling, Notch signaling, IL-6 signaling, Wnt signaling and regulation of cell growth (Figure 6D and Supplementary Figure S6B). These pathways play critical roles in hematopoiesis, lymphoid development and leukemogenesis.
Figure 6.
Inversion of −31CBS inhibits TAL1-mediated leukemic transcription program. (A) Heat map of RNA-seq analysis shows upregulated and downregulated genes upon the −31CBS inversion (Clone #1–8) in Jurkat cells as compared to the WT control. (B) Enrichment plots by GSEA of regulated target genes involved in cell cycle and hematopoietic cell lineage in the −31CBSinv/inv (Clone #1–8) Jurkat cells as compared to the WT control. (C) RT-qPCR analysis and confirmation of representative downregulated genes identified in the RNA-seq analysis. (D) Gene Ontology analysis-identified signaling pathways differently regulated by the WT control and −31CBSinv/inv (Clone #1–8) Jurkat cells. *P < 0.05, **P < 0.01, ***P < 0.001 by student’s t-test.
DISCUSSION
We have identified −31CBS as a critical CTCF boundary that differentially regulates the chromatin topology, allowing for differential expression of TAL1 in erythroid cells and T-ALL cells. However, in TAL1-positive T-ALL cells, inversion of −31CBS blocks enhancer/promoter communication and alters chromatin signature, including histone modification and chromatin accessibility, leading to inhibition of TAL1 expression and T-ALL leukemogenesis. Chromatin boundaries, CTCF binding sites in many cases, play a critical role in defining TADs and chromatin signature within the TAD (30,31). CTCF boundary mediated loop formation is proposed to create an insulated neighborhood that organizes genes and enhancers in close proximity to determine a lineage-specific transcription program and to define cell identity (59,60). The chromatin boundary in the genome may not be permanent depending in looped interactions (61). It was proposed that TADs are formed by loop extrusion model (62). In this model, the progressive and dynamic movement of loops driven by cohesion are stalled at chromatin boundaries that required convergent orientation to form TADs by interacting with boundary proteins including CTCF (62). It is particularly interested that RNAs are involved in CTCF-mediated genome organization (63,64). Since that −31CBS inversion does not affect the CTCF binding to the TAL1 locus (Supplementary Figure S3D), it is also possible that changes in convergent boundary orientation alters CTCF association with other boundary factors or lncRNAs leading to altered boundary activity and ability to recruit target genes to its proximity. Nevertheless, change of chromatin insulator polarity or orientation may potentially facilitate or stall the extrusion of loops. Such action may also affect CTCF-driven TAD resulting in disruption of TAD-mediated enhancer/promoter communications and the oncogenic transcription program, such as that of the TAL1 oncogene in T-ALL, eventually blocking leukemogenesis.
TAL1 is critically required for HSC function and erythroid differentiation (65). Aberrant activation of TAL1 in T-lymphocytes leads to leukemic transformation in the majority of childhood T-ALL (6). Molecular mechanisms controlling TAL1 transcription in normal hematopoiesis and leukemic T-cells remains unclear. It is also largely unknown which of TAL1′s enhancers differentially regulate its transcription in normal and malignant hematopoiesis, and how they might do so. We found that TAL1 expression in erythroid cells is, in part, controlled by a long-range intrachromatin loop that brings the +51 enhancer into close proximity of the TAL1 promoter-I. This interaction is specific to erythroid cells and is absent in T-ALL cells (40). CTCF boundary mediates different chromatin loops of the TAL1 locus in erythroid and T-ALL cells, thereby providing another layer of regulation to ensure proper TAL1 expression in distinct cell lineages or differentiation stages (39,40). It is proposed that −31CBS plays a crucial role in determining looping interactions at the TAL1 locus (39). In order to test this notion, we deleted −31CBS and showed that −31CBS−/− impaired the enhancer and promoter interaction at the TAL1 locus in erythroid cells, but not affecting TAL1 transcription in T-ALL cells, suggesting that −31CBS may regulate enhancer/promoter interaction in cell context-dependent manner.
However, inversion of −31CBS reshapes the genome topology resulting in following consequences (Figure 5). First, disruption of the TAD between +53/57CBS and −157CBS that protect the whole TAL1 locus from surrounding chromatin environment. Second, impairment of the association of −31CBS with the TAL1 promoter-I. Third, decreased the looped interaction between the +19 enhancer and TAL1 promoter-IV (Figure 5). These data support that −31CBS acts as a nucleation site crucial in orchestrating looping interactions and defining TAL1 gene expression. Interestingly, previous studies have shown that the enhancer/promoter loop interactions required looping factor LDB1 and TAL1/GATA/LMO2 transcription super complexes that are present in both normal hematopoiesis and T-ALL leukemia (39,66–68). Thus, it remains to be determined whether and how the TAL1 super transcription complexes including TAL1, GATA factors, LMO2 and LDB1 cooperate with the CTCF boundary to determine enhancer/promoter communication and TAL1 transcription activity.
The +19 enhancer in the TAL1 locus is active in HSCs and progenitor cells, but not in mature erythroid cells (69). Thus, it is not surprising that this enhancer is required for development of hematopoietic stem/progenitor cells, but not necessary for mature hematopoietic lineage (69). Here, we demonstrated that the TAL1 +19Kb enhancer interacts and activates the TAL1 promoter-IV in TAL1 positive T-ALL cells, but not in K562 erythroid cells (Figure 3). Moreover, the TAL1 positive T-ALL patient sample exhibited a very strong looped interaction between the +19Kb enhancer and the TAL1 promoter-IV, while the TAL1 negative T-ALL exhibits a very weak looped interaction (Figure 3). It is very interesting that the TAL1 promoter-IV is specifically active in human T-cell leukemia (53). Thus, our data reveal that the TAL1 +19 enhancer acts not only as stem cell enhancer, but also as a leukemia specific enhancer to activate the TAL1 in T-ALL.
Recently, it has been shown that CBS orientation determines the genome topology and chromatin looping, which potentially affects promoter function and gene expression (57,58). This may provide a potential strategy to reshape genome organization, especially in the oncogenic loci, to prevent or disrupt the oncogene transcription program for cancer therapy. We reported recently that disruption of a critical CTCF boundary located at the HOXA gene locus blocks the oncogenic transcription program and prolongs survival of transplanted AML mice (30). Consistent with this notion, CRISPR-mediated inversion of −31CBS disrupts an existing TAL1 promoter/enhancer interaction and inhibits TAL1 expression in T-ALL cells leading to interfering leukemia progression. Although −31CBS is far away from any known enhancer in the TAL1 locus, it is likely that change in its orientation alters polarity of boundary and reshapes the TAD organization. In supporting this claim, our Hi-C analysis revealed that altered −31CBS orientation disrupts the normal TAL1 TAD/Sub-TADs, which usually prevents influence signals from the surrounding chromatin environment. It is conceivable that altering CTCF boundary by enhancing or impairment of CTCF binding to boundary can dramatically change TAD structure and oncogenic transcription program to affect oncogenic/leukemogenic processes. Thus, CTCF boundaries that are involved in determining specific topology of leukemic genome and gene expression programs can potentially be targeted against pathogenesis of leukemia.
DATA AVAILABILITY
Sequence reads have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) under accession number GSE135320.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Penn State College of Medicine Genome Science Facility for Illumina sequencing. We are grateful for helpful discussions from members of the Brand and Huang laboratories. We also thank Rachael Mills for editing the manuscript. We thank Adolfo Ferrando and Andrew Kung from the Columbia University for providing the FUW-Luc-Cherry-puro lentiviral vector.
Author contributions’: M.B., K Z. and S.H. conceived and designed experiments. Y.L., Z.L., A.B., Y.K. Q.L. I.C. performed experiments. H.L. H.L. and I.C. performed bioinformatics and statistical analysis. A.B. and M.B. established transplantation T-ALL mouse model and performed invivo assays. Y.L. S.D. K.Z. and M.B. provide critical reagents and human patient sample analysis. S.H. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01DK110108, R01CA204044 to S.H.]; Four Diamonds Endowed Epigenetic and Gene Regulation Research Program (to S.H.); Canadian Institutes of Health Research [MOP-343603 to M.B.]; Intramural Research Program, the National Heart, Lung and Blood Institute, NIH (to K.Z.). Funding for open access charge: NIH [R01CA204044].
Conflict of interest statement. None declared.
REFERENCES
- 1. Pui C.H., Relling M.V., Downing J.R.. Acute lymphoblastic leukemia. N. Engl. J. Med. 2004; 350:1535–1548. [DOI] [PubMed] [Google Scholar]
- 2. van Grotel M., Meijerink J.P., Beverloo H.B., Langerak A.W., Buys-Gladdines J.G., Schneider P., Poulsen T.S., den Boer M.L., Horstmann M., Kamps W.A. et al.. The outcome of molecular-cytogenetic subgroups in pediatric T-cell acute lymphoblastic leukemia: a retrospective study of patients treated according to DCOG or COALL protocols. Haematologica. 2006; 91:1212–1221. [PubMed] [Google Scholar]
- 3. Iacobucci I., Papayannidis C., Lonetti A., Ferrari A., Baccarani M., Martinelli G.. Cytogenetic and molecular predictors of outcome in acute lymphocytic leukemia: recent developments. Curr. Hematol. Malig. Rep. 2012; 7:133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kraszewska M.D., Dawidowska M., Szczepanski T., Witt M.. T-cell acute lymphoblastic leukaemia: recent molecular biology findings. Br. J. Haematol. 2012; 156:303–315. [DOI] [PubMed] [Google Scholar]
- 5. Brown L., Cheng J.T., Chen Q., Siciliano M.J., Crist W., Buchanan G., Baer R.. Site-specific recombination of the tal-1 gene is a common occurrence in human T cell leukemia. EMBO J. 1990; 9:3343–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bash R.O., Hall S., Timmons C.F., Crist W.M., Amylon M., Smith R.G., Baer R.. Does activation of the TAL1 gene occur in a majority of patients with T-cell acute lymphoblastic leukemia? A pediatric oncology group study. Blood. 1995; 86:666–676. [PubMed] [Google Scholar]
- 7. Carroll A.J., Crist W.M., Link M.P., Amylon M.D., Pullen D.J., Ragab A.H., Buchanan G.R., Wimmer R.S., Vietti T.J.. The t(1;14)(p34;q11) is nonrandom and restricted to T-cell acute lymphoblastic leukemia: a Pediatric Oncology Group study. Blood. 1990; 76:1220–1224. [PubMed] [Google Scholar]
- 8. Condorelli G.L., Facchiano F., Valtieri M., Proietti E., Vitelli L., Lulli V., Huebner K., Peschle C., Croce C.M.. T-cell-directed TAL-1 expression induces T-cell malignancies in transgenic mice. Cancer Res. 1996; 56:5113–5119. [PubMed] [Google Scholar]
- 9. Kelliher M.A., Seldin D.C., Leder P.. Tal-1 induces T cell acute lymphoblastic leukemia accelerated by casein kinase IIalpha. EMBO J. 1996; 15:5160–5166. [PMC free article] [PubMed] [Google Scholar]
- 10. Palii C.G., Perez-Iratxeta C., Yao Z., Cao Y., Dai F., Davison J., Atkins H., Allan D., Dilworth F.J., Gentleman R. et al.. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. EMBO J. 2011; 30:494–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palomero T., Odom D.T., O’Neil J., Ferrando A.A., Margolin A., Neuberg D.S., Winter S.S., Larson R.S., Li W., Liu X.S. et al.. Transcriptional regulatory networks downstream of TAL1/SCL in T-cell acute lymphoblastic leukemia. Blood. 2006; 108:986–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O’Neil J., Shank J., Cusson N., Murre C., Kelliher M.. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell. 2004; 5:587–596. [DOI] [PubMed] [Google Scholar]
- 13. Reynaud D., Ravet E., Titeux M., Mazurier F., Renia L., Dubart-Kupperschmitt A., Romeo P.H., Pflumio F.. SCL/TAL1 expression level regulates human hematopoietic stem cell self-renewal and engraftment. Blood. 2005; 106:2318–2328. [DOI] [PubMed] [Google Scholar]
- 14. D'Souza S.L., Elefanty A.G., Keller G.. SCL/Tal-1 is essential for hematopoietic commitment of the hemangioblast but not for its development. Blood. 2005; 105:3862–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y., Payne K.J., Zhu Y., Price M.A., Parrish Y.K., Zielinska E., Barsky L.W., Crooks G.M.. SCL expression at critical points in human hematopoietic lineage commitment. Stem Cells. 2005; 23:852–860. [DOI] [PubMed] [Google Scholar]
- 16. Robb L., Lyons I., Li R., Hartley L., Kontgen F., Harvey R.P., Metcalf D., Begley C.G.. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:7075–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shivdasani R.A., Mayer E.L., Orkin S.H.. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995; 373:432–434. [DOI] [PubMed] [Google Scholar]
- 18. Porcher C., Swat W., Rockwell K., Fujiwara Y., Alt F.W., Orkin S.H.. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996; 86:47–57. [DOI] [PubMed] [Google Scholar]
- 19. Robb L., Elwood N.J., Elefanty A.G., Kontgen F., Li R., Barnett L.D., Begley C.G.. The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J. 1996; 15:4123–4129. [PMC free article] [PubMed] [Google Scholar]
- 20. Delabesse E., Ogilvy S., Chapman M.A., Piltz S.G., Gottgens B., Green A.R.. Transcriptional regulation of the SCL locus: identification of an enhancer that targets the primitive erythroid lineage in vivo. Mol. Cell Biol. 2005; 25:5215–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gottgens B., Barton L.M., Gilbert J.G., Bench A.J., Sanchez M.J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M. et al.. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 2000; 18:181–186. [DOI] [PubMed] [Google Scholar]
- 22. Gottgens B., Barton L.M., Chapman M.A., Sinclair A.M., Knudsen B., Grafham D., Gilbert J.G., Rogers J., Bentley D.R., Green A.R.. Transcriptional regulation of the stem cell leukemia gene (SCL)–comparative analysis of five vertebrate SCL loci. Genome Res. 2002; 12:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gothert J.R., Gustin S.E., Hall M.A., Green A.R., Gottgens B., Izon D.J., Begley C.G.. In vivo fate-tracing studies using the Scl stem cell enhancer: embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood. 2005; 105:2724–2732. [DOI] [PubMed] [Google Scholar]
- 24. Follows G.A., Dhami P., Gottgens B., Bruce A.W., Campbell P.J., Dillon S.C., Smith A.M., Koch C., Donaldson I.J., Scott M.A. et al.. Identifying gene regulatory elements by genomic microarray mapping of DNaseI hypersensitive sites. Genome Res. 2006; 16:1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Silberstein L., Sanchez M.J., Socolovsky M., Liu Y., Hoffman G., Kinston S., Piltz S., Bowen M., Gambardella L., Green A.R. et al.. Transgenic analysis of the stem cell leukemia +19 stem cell enhancer in adult and embryonic hematopoietic and endothelial cells. Stem Cells. 2005; 23:1378–1388. [DOI] [PubMed] [Google Scholar]
- 26. Gottgens B., Gilbert J.G., Barton L.M., Grafham D., Rogers J., Bentley D.R., Green A.R.. Long-range comparison of human and mouse SCL loci: localized regions of sensitivity to restriction endonucleases correspond precisely with peaks of conserved noncoding sequences. Genome Res. 2001; 11:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dhami P., Bruce A.W., Jim J.H., Dillon S.C., Hall A., Cooper J.L., Bonhoure N., Chiang K., Ellis P.D., Langford C. et al.. Genomic approaches uncover increasing complexities in the regulatory landscape at the human SCL (TAL1) locus. PLoS One. 2010; 5:e9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K.. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837. [DOI] [PubMed] [Google Scholar]
- 29. Follows G.A., Ferreira R., Janes M.E., Spensberger D., Cambuli F., Chaney A.F., Kinston S.J., Landry J.R., Green A.R., Gottgens B.. Mapping and functional characterisation of a CTCF-dependent insulator element at the 3′ border of the murine Scl transcriptional domain. PLoS One. 2012; 7:e31484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luo H., Wang F., Zha J., Li H., Yan B., Du Q., Yang F., Sobh A., Vulpe C., Drusbosky L. et al.. CTCF boundary remodels chromatin domain and drives aberrant HOX gene transcription in acute myeloid leukemia. Blood. 2018; 132:837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O., Reinberg D.. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015; 347:1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Phillips J.E., Corces V.G.. CTCF: master weaver of the genome. Cell. 2009; 137:1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bushey A.M., Dorman E.R., Corces V.G.. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol. Cell. 2008; 32:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gaszner M., Felsenfeld G.. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006; 7:703–713. [DOI] [PubMed] [Google Scholar]
- 35. West A.G., Gaszner M., Felsenfeld G.. Insulators: many functions, many mechanisms. Genes Dev. 2002; 16:271–288. [DOI] [PubMed] [Google Scholar]
- 36. Handoko L., Xu H., Li G., Ngan C.Y., Chew E., Schnapp M., Lee C.W., Ye C., Ping J.L., Mulawadi F. et al.. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 2011; 43:630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao Z., Tavoosidana G., Sjolinder M., Gondor A., Mariano P., Wang S., Kanduri C., Lezcano M., Sandhu K.S., Singh U. et al.. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006; 38:1341–1347. [DOI] [PubMed] [Google Scholar]
- 38. Wallace J.A., Felsenfeld G.. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 2007; 17:400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou Y., Kurukuti S., Saffrey P., Vukovic M., Michie A.M., Strogantsev R., West A.G., Vetrie D.. Chromatin looping defines expression of TAL1, its flanking genes, and regulation in T-ALL. Blood. 2013; 122:4199–4209. [DOI] [PubMed] [Google Scholar]
- 40. Patel B., Kang Y., Cui K., Litt M., Riberio M.S., Deng C., Salz T., Casada S., Fu X., Qiu Y. et al.. Aberrant TAL1 activation is mediated by an interchromosomal interaction in human T-cell acute lymphoblastic leukemia. Leukemia. 2014; 28:349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simon C., Chagraoui J., Krosl J., Gendron P., Wilhelm B., Lemieux S., Boucher G., Chagnon P., Drouin S., Lambert R. et al.. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012; 26:651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Benyoucef A., Palii C.G., Wang C., Porter C.J., Chu A., Dai F., Tremblay V., Rakopoulos P., Singh K., Huang S. et al.. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev. 2016; 30:508–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu X., Li X., Valverde K., Fu X., Noguchi C., Qiu Y., Huang S.. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. PNAS. 2009; 106:10141–10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kimbrel E.A., Davis T.N., Bradner J.E., Kung A.L.. In vivo pharmacodynamic imaging of proteasome inhibition. Mol. Imaging. 2009; 8:140–147. [PubMed] [Google Scholar]
- 45. Li X., Wang S., Li Y., Deng C., Steiner L.A., Xiao H., Wu C., Bungert J., Gallagher P.G., Felsenfeld G. et al.. Chromatin boundaries require functional collaboration between the hSET1 and NURF complexes. Blood. 2011; 118:1386–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y., Schulz V.P., Deng C., Li G., Shen Y., Tusi B.K., Ma G., Stees J., Qiu Y., Steiner L.A. et al.. Setd1a and NURF mediate chromatin dynamics and gene regulation during erythroid lineage commitment and differentiation. Nucleic Acids Res. 2016; 44:7173–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Trapnell C., Pachter L., Salzberg S.L.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van de Werken H.J., Landan G., Holwerda S.J., Hoichman M., Klous P., Chachik R., Splinter E., Valdes-Quezada C., Oz Y., Bouwman B.A. et al.. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat. Methods. 2012; 9:969–972. [DOI] [PubMed] [Google Scholar]
- 51. Rivera-Reyes A., Hayer K.E., Bassing C.H.. Genomic alterations of non-coding regions underlie human cancer: Lessons from T-ALL. Trends Mol. Med. 2016; 22:1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Courtes C., Lecointe N., Le Cam L., Baudoin F., Sardet C., Mathieu-Mahul D.. Erythroid-specific inhibition of the tal-1 intragenic promoter is due to binding of a repressor to a novel silencer. J. Biol. Chem. 2000; 275:949–958. [DOI] [PubMed] [Google Scholar]
- 53. Bernard O., Azogui O., Lecointe N., Mugneret F., Berger R., Larsen C.J., Mathieu-Mahul D.. A third tal-1 promoter is specifically used in human T cell leukemias. J. Exp. Med. 1992; 176:919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Aplan P.D., Raimondi S.C., Kirsch I.R.. Disruption of the SCL gene by a t(1;3) translocation in a patient with T cell acute lymphoblastic leukemia. J. Exp. Med. 1992; 176:1303–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bernard O., Barin C., Charrin C., Mathieu-Mahul D., Berger R.. Characterization of translocation t(1;14)(p32;q11) in a T and in a B acute leukemia. Leukemia. 1993; 7:1509–1513. [PubMed] [Google Scholar]
- 56. Fitzgerald T.J., Neale G.A., Raimondi S.C., Goorha R.M.. c-tal, a helix-loop-helix protein, is juxtaposed to the T-cell receptor-beta chain gene by a reciprocal chromosomal translocation: t(1;7)(p32;q35). Blood. 1991; 78:2686–2695. [PubMed] [Google Scholar]
- 57. Guo Y., Xu Q., Canzio D., Shou J., Li J., Gorkin D.U., Jung I., Wu H., Zhai Y., Tang Y. et al.. CRISPR Inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015; 162:900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. de Wit E., Vos E.S., Holwerda S.J., Valdes-Quezada C., Verstegen M.J., Teunissen H., Splinter E., Wijchers P.J., Krijger P.H., de Laat W.. CTCF binding polarity determines chromatin looping. Mol. Cell. 2015; 60:676–684. [DOI] [PubMed] [Google Scholar]
- 59. Dowen J.M., Fan Z.P., Hnisz D., Ren G., Abraham B.J., Zhang L.N., Weintraub A.S., Schujiers J., Lee T.I., Zhao K. et al.. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014; 159:374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hnisz D., Day D.S., Young R.A.. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell. 2016; 167:1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. et al.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N., Mirny L.A.. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016; 15:2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yao H., Brick K., Evrard Y., Xiao T., Camerini-Otero R.D., Felsenfeld G.. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010; 24:2543–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Luo H., Zhu G., Xu J., Lai Q., Yan B., Guo Y., Fung T.K., Zeisig B.B., Cui Y., Zha J. et al.. HOTTIP lncRNA promotes hematopoietic stem cell Self-Renewal leading to AML-like disease in mice. Cancer Cell. 2019; 36:645–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hu X., Ybarra R., Qiu Y., Bungert J., Huang S.. Transcriptional regulation by TAL1: a link between epigenetic modifications and erythropoiesis. Epigenetics. 2009; 4:357–361. [DOI] [PubMed] [Google Scholar]
- 66. Song S.H., Hou C., Dean A.. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol. Cell. 2007; 28:810–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xu Z., Huang S., Chang L.S., Agulnick A.D., Brandt S.J.. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol. Cell Biol. 2003; 23:7585–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wadman I.A., Osada H., Grutz G.G., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H.. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997; 16:3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Spensberger D., Kotsopoulou E., Ferreira R., Broccardo C., Scott L.M., Fourouclas N., Ottersbach K., Green A.R., Gottgens B.. Deletion of the Scl +19 enhancer increases the blood stem cell compartment without affecting the formation of mature blood lineages. Exp. Hematol. 2012; 40:588–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence reads have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) under accession number GSE135320.