Abstract
NAD+-dependent SIRT7 deacylase plays essential roles in ribosome biogenesis, stress response, genome integrity, metabolism and aging, while how it is transcriptionally regulated is still largely unclear. TGF-β signaling is highly conserved in multicellular organisms, regulating cell growth, cancer stemness, migration and invasion. Here, we demonstrate that histone deacetylase HDAC8 forms complex with SMAD3/4 heterotrimer and occupies SIRT7 promoter, wherein it deacetylates H4 and thus suppresses SIRT7 transcription. Treatment with HDAC8 inhibitor compromises TGF-β signaling via SIRT7-SMAD4 axis and consequently, inhibits lung metastasis and improves chemotherapy efficacy in breast cancer. Our data establish a regulatory feedback loop of TGF-β signaling, wherein HDAC8 as a novel cofactor of SMAD3/4 complex, transcriptionally suppresses SIRT7 via local chromatin remodeling and thus further activates TGF-β signaling. Targeting HDAC8 exhibits therapeutic potential for TGF-β signaling related diseases.
INTRODUCTION
SIRT7 belongs to NAD+-dependent sirtuin family, which predominantly localizes to nucleolus and widely expressed in various organs and tissues (1,2). SIRT7 interacts with mTOR and GTF3C1, thus regulating protein synthesis (3), and cooperates with Myc to suppress ribosome biogenesis (4). SIRT7 regulates DNA damage response and DNA repair via facilitating recruitment of 53BP1 and inhibition of ATM deacetylation (5,6). 5-Fluorouracil (5-FU)-induced SIRT7 reduction promotes radiosensitivity in colorectal cancer (7). Sirt7–/– mice suffer from genomic instability, cardiomyopathy, hepatic steatosis as well as early death (8,9). Bone marrow and liver carry high level of SIRT7, which regulates hematopoietic stem cell aging and hepatic lipid metabolism (4,10,11). Upon energetic stress, SIRT7 is released from nucleolus and degraded by REGγ (12). SIRT7 is progressively downregulated and thus activates TGF-β signaling during breast cancer metastasis (13). MicroRNAs, such as miR-125b, miR-125a-5p, hsa-miR-125b, miR-93 and miR-3666, negatively regulate SIRT7 expression in various cancers and adiposity (14–17). Albeit advances affirming pivotal function and post-transcriptional regulation of SIRT7, how SIRT7 is transcriptionally regulated is still elusive.
TGF-β signaling is highly conserved in multicellular organisms, involved in multiple cellular processes, such as cell growth, stemness, migration and invasion, epithelial–mesenchymal transition (EMT), extracellular matrix (ECM) remodeling and immune regulation (18). The canonical TGF-β signaling is initially transduced through the formation of a heterotetrametric receptor complex composed of TGF-β type I (TβRI) and type II (TβRII) receptors. Activated TβRI phosphorylates SMAD2 and SMAD3 (R-SMADs) at C-terminal serine residues, allowing them to assemble into homomeric complex and then to form heterotrimeric complex with SMAD4 (Co-SMAD). These complexes translocate to the nucleus, wherein regulating the transcription of dozens of genes (19,20). In certain case, SMAD3 recruits histone deacetylases HDAC4/5 to achieve transcriptional repression of Runx2 via local chromatin condensation (21). So far, however, the evidences that HDACs cooperate with SMADs to modify histone acetylation are still few. Their target genes and physiological roles need to be further explored.
HDAC8 is a class I HDAC that deacetylates histone H3 and H4 at nonspecific lysines (22,23). HDAC8 coordinates with DEC1 to suppress the transcription of TAp73 and DeltaNp73; HDAC8/YY1 signals suppress mutant p53 transcription in triple negative breast cancer (TNBC) cells (24,25). So far, however identified HDAC8 target genes are few, which restrains its mechanism clarification. Here, we reveal a feedback loop that regulates TGF-β signaling–HDAC8 forms complex with SMAD3/4 heterotrimers and represses SIRT7 transcription via local chromatin remodeling; reduction of SIRT7 further activates TGF-β signaling. The data highlight that manipulating level of SIRT7 or HDAC8 has great therapeutic potential for TGF-β signaling-related diseases.
MATERIALS AND METHODS
Cell lines and chemicals
The breast cancer cell lines 4T1, MDA-MB-231, BT549 and HEK 293 human kidney cells were obtained from the American Type Culture Collection (ATCC®). 4T1, MDA-MB-231 and HEK 293 cells were cultured in high glucose DMEM (Gibco®) supplemented with 10% FBS (Gibco®). BT549 cells were cultured in RPMI-1640 (Gibco®) supplemented with 10% FBS. All cells were maintained at 37°C in a humidified 5% CO2 atmosphere. The ALK5 inhibitor A8301, HDAC8 inhibitor PCI-34051 and paclitaxel were obtained from MedChemExpress (MCE®). Other HDAC inhibitors mentioned in manuscript were from Selleck.
Cell transfection
Cells were transfected with plasmids or siRNAs using either Polyethyleneimine (PEI) or Lipofectamine®3000 (Thermo Fisher), following the manufacturer's instructions. Oligo siRNAs or shRNAs were obtained from GenePharma Company (Shanghai). The siRNA sequences are listed in Supplementary Table S2.
Lentivirus package and stable cell line selection
Lentiviral constructs with shRNA, pSPAX2 and pMD2G (1:1:0.5) were co-transfected into HEK293 cells using Lipofectamine®3000. After 48 h, the supernatant was collected and filtered through a 0.22 μm membrane (Millipore). The virus titer was measured before infection. 4T1 or MDA-MB-231 cells were infected with shRNA lentivirus and selected with puromycin (Sigma) to obtain stable Sirt7 knockdown. The oligo shRNA sequences used are listed in Supplementary Table S2.
RNA isolation, qRT-PCR and chromatin immunoprecipitation assays (ChIP)
Cells were lysed in Trizol reagent (RNAiso Plus, Takara) and the total RNA was isolated by standard protocol before transcribing into cDNA using 5 × Primescript® RT Master Mix (Takara), according to the manufacturer's instructions. qRT-PCR analysis of gene expression was performed using 2 × SYBR® Green Mix (Takara) on a Bio-Rad detection system. Fast ChIP assays were performed according to previously reported protocols (26). All primer sequences and ChIP antibodies are listed in Supplementary Tables S2 and S3.
Luciferase reporter assay
The SIRT7 promoter sequence predicated at –1930/+54 (S7Pro) was cloned into a pGL4.17 vector (Promega). To monitor promoter activity, the luciferase plasmid and the Renilla luciferase (pRL-CMV) internal control plasmid were co-transfected into breast cancer cells. The transcriptional activity was determined by Dual-Luciferase Reporter Assay System (Promega), performed according to the manufacturer's instructions. The SMAD3/SMAD4 binding sites on the SIRT7 promoter were identified using the rVista 2.0 online tool (https://rvista.dcode.org/). To mutate the SMAD3/4 binding site on the SIRT7 promoter, site direction mutagenesis was performed by KOD-Plus-Neo (TOYOBO).
Transwell migration assay
For Transwell migration, 5 × 104 cells suspended in 200 μl FBS-free medium were placed in the upper chamber (8 μm pore size, Corning). The insert was incubated in a 24-well-plate supplemented with 500 μl 10% FBS medium as a chemoattractant. The top membrane was swiped with cotton swabs to remove the non-migrated cells, and the migrated cells were stained with crystal violet before the cell number was counted to evaluate the migration ability.
Protein extraction and western blotting
Total cell lysate was prepared following RIPA buffer extraction (Thermo Fisher). Protein extracts were resolved in 5× Laemmli sample buffer (100 mM Tris–HCl [pH6.8], 4% SDS, 20% glycerol and 1 mM DTT), separated by SDS-PAGE, transferred to a PVDF membrane (Millipore), blocked with 5% non-fat milk and probed with the respective primary antibodies. Immunoblotting images were collected on a Bio-Rad system after incubation with HRP-conjugated secondary antibodies. The antibodies listed in Supplementary Table S3.
Immunoprecipitation
The indicated cells were first lysed in IP lysis buffer [200 mM NaCl, 20 mM Tris–HCl (pH 7.9), 5 mM MgCl2, 10% glycerol, 0.2 mM EDTA and 0.1% NP-40] supplemented with protease inhibitors (Roche Complete). After sonication and centrifugation, the cleared cell supernatant was collected and incubated with the respective antibodies for 4 h or overnight at 4°C. The immunoprecipitates were washed with IP lysis buffer and eluted in Laemmli sample buffer for immunoblotting.
In vivo xenograft model of breast cancer
Pathogen-free female BALB/c mice were purchased from the Shanghai Laboratory Animal Center, CAS (Shanghai). Then, 4T1 cells (5 × 105) were injected into the fourth mammary fat pad of virgin BALB/c mice. After 7 days of inoculation, tumor-bearing mice were randomized for treatment with saline, PCI-34051 (10 mg/kg, i.p.) or paclitaxel (10 mg/kg, i.p.) as indicated. The tumor diameters were measured and the tumor volume (mm3) was calculated by the formula: volume = 0.5 × length × width2. The lung metastatic nodules were counted after H&E staining of the whole lung. Experimental animals were housed and handled in accordance with protocols approved by the Committee on the Use of Live Animals in Teaching and Research of Shenzhen University.
Statistical analyses
All statistical analyses were performed using GraphPad® Prism 5 or Microsoft excel. The data obtained by at least three independent experiments are represented as the means ± s.e.m. Normally distributed data were analyzed by Student's t-test. Non-normally distributed data were analyzed by non-parametric Mann–Whitney U-tests. A P < 0.05 was considered statistically significant.
RESULTS
TGF-β signaling suppresses SIRT7 transcription
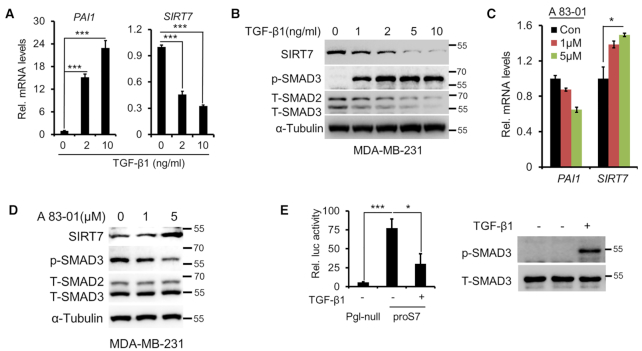
SIRT7 antagonizes TGF-β signaling via promoting SMAD4 degradation (13). Based on the intrinsic nature of TGF-β regulatory feedback loop (27), we examined whether TGF-β signaling in turn represses SIRT7 expression. We treated MDA-MB-231 cells with TGF-β1 and found that mRNA and protein levels of SIRT7 were both inhibited in a dose-dependent manner (Figure 1A, B). This finding was confirmed in two other cell lines (Supplementary Figure S1A–C). Of note, phosphorylated SMAD3 and upregulated PAI1 indicated activated TGF-β signaling (28). We next exposed the cells to TGFβRI inhibitor A83–01. As shown, blocking TGF-β signaling resulted in significant SIRT7 upregulation (Figure 1C, D). Similarly, A83-01 treatment abolished the TGF-β1-induced downregulation of SIRT7 in three other cell lines (Supplementary Figure S1D). To further confirm that TGF-β1 represses SIRT7 transcription, we cloned SIRT7 promoter (proS7) into a PGL-4.17 luciferase reporter system (Supplementary Figure S1E). The luciferase activity was significantly inhibited by TGF-β1 (Figure 1E), suggestive of transcriptional repression of SIRT7.
Figure 1.
TGF-β suppresses SIRT7 transcription. (A, B) qRT-PCR (A) and Immunoblotting (B) analysis of SIRT7 levels in MDA-MB-231 cells following TGF-β1 treatment. PAI1 upregulation indicates TGF-β signaling activation. Data are shown as means ± s.e.m. n = 3, ***P < 0.0001; Student's t-test. (C, D) MDA-MB-231 cells were incubated with the TβRI inhibitor A83-01 (1, 5 μM). qRT-PCR (C) and Immunoblotting (D) analysis of SIRT7 levels in MDA-MB-231 cells. The data represent the means ± s.e.m. n = 3, *P < 0.05; Student's t-test. (E) Luciferase reporter assay of SIRT7 promoter activity in MDA-MB-231 cells treated or untreated with TGF-β1 (5 ng/ml) (right). Pgl-null, PGL.417 empty vector with minimal promoter activity; proS7, PGL.417 cloned with the promoter of SIRT7. Immunoblot analysis of p-SMAD3 upon TGF-β1 treatment (left). The data represent the means ± s.e.m. n = 3, *P < 0.05 and ***P < 0.0001; Student's t-test.
The effect of TGFβ signaling highly relies on tissue or cellular context (29). We thus explored whether TGF-β-mediated suppression of SIRT7 expression is a general effect. As shown, SIRT7 expression was downregulated upon activation of TGF-β signaling in various tissue-derived cells, including human skin fibroblast F2S cells, human skin keratinocyte HaCat cells and lung cancer A549 cells (Supplementary Figure S2A). Of note, colorectal cancer Caco2 cells were insensitive to TGFβ signaling and failed to repress SIRT7 expression even at high dosage, indicating that the full activation of TGFβ is requisite for SIRT7 suppression. Further, based on public data (http://rna.sysu.edu.cn/chipbase/) (30), SIRT7 expression was negatively correlated to TGFBR1, TGFBR2 or SMAD3 in multiple tissues and cancers (Supplementary Tables S4–S9). Together, these data implicate that TGF-β signaling suppresses SIRT7 expression.
SMAD3/4 underlines TGF-β-induced SIRT7 suppression
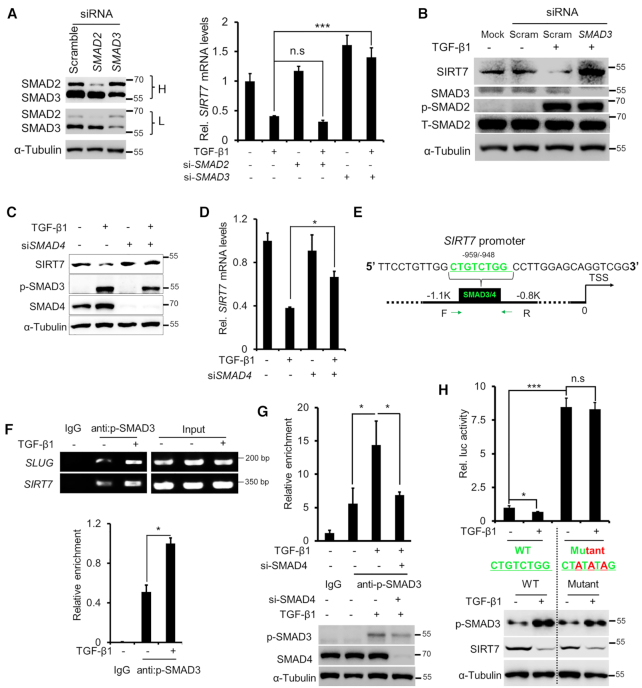
SMAD2/3 are the major TGF-β signaling modulators (28). We knocked down SMAD2 or SMAD3 via siRNA and monitored SIRT7 expression induced by TGF-β1. SMAD2 and SMAD3 knockdown (KD) attenuated PAI1 transcription (Supplementary Figure S2B). Interestingly, while SMAD3 KD blocked TGF-β1-induced SIRT7 repression, SMAD2 KD had little such effect (Figure 2A, B). Consistently, p-SMAD3 levels were reversely correlated with SIRT7 expression in series of breast cancer cell lines (Supplementary Figure S2C). As SMAD3 and SMAD4 heterotrimerize to regulate transcription (20), we asked whether SMAD4 is required for SMAD3-mediated SIRT7 transcriptional suppression. We knocked down SMAD4 in MDA-MB-231 cells and confirmed TGF-β1 inhibition by PAI1 reduction (Supplementary Figure S2D, E). As shown, SMAD4 KD significantly attenuated TGF-β1-induced SIRT7 downregulation (Figure 2C, D). These results suggest that TGF-β signaling suppresses SIRT7 transcription via SMAD3/4.
Figure 2.
SMAD3/SMAD4 mediates TGF-β-induced SIRT7 suppression. (A) qRT-PCR analysis of SIRT7 mRNA levels following TGF-β1 (5 ng/ml) treatment and SMAD2 and SMAD3 knockdown. Immunoblots showing SMAD2 and SMAD3 levels in MDA-MB-231 cells treated with SMAD2, SMAD3 siRNAs or scrambled siRNA. L, low exposure; H, high exposure. n.s., non-significant. The data represent the means ± s.e.m. n = 3, ***P < 0.001; Student's t-test. (B) Immunoblotting analysis of SIRT7 levels in cells treated with TGF-β1 (5 ng/ml) or SMAD3 siRNA. Increased p-SMAD2 levels indicate TGF-β activation. (C, D) Immunoblotting (C) and qRT-PCR (D) analysis of SIRT7 levels treated with TGF-β1 (5 ng/ml) and/or SMAD4 siRNA. The data represent the means ± s.e.m. n = 3, *P < 0.05; Student's t-test. (E) Predicated SMAD3/SMAD4 binding sites (SBEs) on the SIRT7 promoter by the online tool rVISTA 2.0 (31). The green color highlights the conserved SBE sequence (upper). (F) ChIP PCR using a p-SMAD3 antibody in breast cancer cells. SLUG serves as a positive control (middle); Fold change of p-SMAD3 enrichment on the SIRT7 promoter by ChIP-qPCR (below). The data represent the means ± s.e.m. n = 3, *P < 0.05; Student's t-test. (G) Fold change of p-SMAD3 enrichment on the SIRT7 promoter region following SMAD4 siRNA or TGF-β1 (5 ng/ml) treatment in breast cancer cells, determined by ChIP-qPCR (upper). Immunoblotting (below) of SMAD4 knockdown and TGF-β1 signaling activation. The data represent the means ± s.e.m. n = 3, *P < 0.05; Student's t-test. (H) Luciferase reporter assay (upper) of SIRT7 promoter activity with/without TGF-β1 (5 ng/ml) treatment. Immunoblotting (below) of endogenous SIRT7 and p-SMAD3 levels under the indicated treatments. n.s, non-significant. The data represent the means ± s.e.m. n = 3, *P < 0.05, ***P < 0.001; Student's t-test.
The SMAD3/4 complex recognizes conserved GTCT motif on target gene promoter, known as SMAD binding element (SBE) (19). We searched for putative SBE motifs on SIRT7 promoter region using an rVista 2.0 online tool (31). A highly conserved SBE (–959 to –948 bp) was predicted (Figure 2E). To gain experimental evidence, anti p-SMAD3 chromatin immunoprecipitation (ChIP) was conducted. Indeed, p-SMAD3 was significantly enriched on SIRT7 promoter, which was enhanced by TGF-β1 (Figure 2F). SLUG, a known transcription target of SMAD3/4 complex, served as a positive control (28). Further, SMAD4 KD significantly attenuated TGF-β1-induced SMAD3 enrichment on SIRT7 promoter (Figure 2G). To test the binding specificity, we mutated the core binding motif TGTCTGG (WT) to TATATAG (Mutant) (19) and performed dual-luciferase reporter assay. As shown, the Mutant promoter exhibited much higher luciferase activity compared to WT. TGF-β1 treatment inhibited WT promoter activity but hardly affected the Mutant (Figure 2H), implicating that the SBE motif is critical for SIRT7 transcriptional inhibition. Of note, TGF-β1 treatment significantly inhibited the expression of endogenous SIRT7 (Figure 2H, below), wherein the SBE motif is intact. Together, the findings suggest that SMAD3/4 complex recognizes the SIRT7 promoter to suppress transcription in response to TGF-β signaling.
HDAC8 is required for SIRT7 transcriptional repression
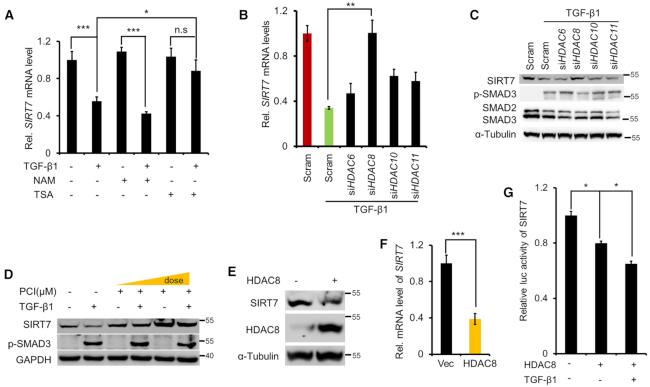
Direct transcriptional repression by SMAD3/4 complex is rare (20). Instead, SMAD recruits deacetylase HDACs to repress transcription by chromatin remodeling (19). To test whether HDACs are involved in TGF-β-induced SIRT7 transcriptional repression, we manipulated the enzyme activity of various HDACs by using Trichostatin A (TSA) and nicotinamide (NAM), which are specific class I/II and class III HDAC inhibitors respectively. TSA but not NAM treatment rescued TGF-β1-mediated transcriptional suppression of SIRT7 (Figure 3A), suggesting the involvement of class I/II HDACs in the SIRT7 transcriptional regulation. PAI1 is transcriptionally regulated by HDACs (13,32). We found increase of PAI1 in response to NAM and TSA (Supplementary Figure S3A). Using selective HDAC inhibitors, we further narrowed down the candidates that likely suppress SIRT7 transcription to HDAC6, 8, 10 and 11 (Supplementary Figure S3B, C and Supplementary Table S1). We then knocked down each of these candidates in MDA-MB-231 cells by siRNA (Supplementary Figure S3D). Only HDAC8 KD restored SIRT7 expression, which otherwise was suppressed by TGF-β1 (Figure 3B, C). Moreover, treatment with HDAC8 inhibitor PCI-34051 (33) restored SIRT7 levels in a dose-dependent manner (Figure 3D). On the other front, overexpression of ectopic HDAC8 downregulated SIRT7 expression and suppressed its promoter activity (Figure 3E–G). Together, we reason that TGF-β signaling-mediated SIRT7 repression is dependent on HDAC8.
Figure 3.
HDAC8 is required for SIRT7 transcriptional repression. (A) qRT-PCR analysis of SIRT7 in MDA-MB-231 cells treated with TGF-β1, NAM and TSA as indicated for 24 h. n.s, non-significant. The data represent the means ± s.e.m. n = 3, *P < 0.05, ***P < 0.001; Student's t-test. (B, C) SIRT7 levels were measured by qRT-PCR (B) and Immunoblotting (C) in HDAC6, HDAC8, HDAC10 and HDAC11 knocked down cells. The data represent the means ± s.e.m. n = 3, **P < 0.01; Student's t-test. (D) Immunoblotting analysis of SIRT7 level in cells incubated with the HDAC8 inhibitor PCI-34051 (PCI), in the presence or absence of TGF-β1 (5 ng/ml). (E, F) Immunoblotting (E) and qRT-PCR analysis (F) of SIRT7 expression in cells with ectopic HDAC8 over-expression. The data represent the means ± s.e.m. n = 3, ***P < 0.001; Student's t-test. (G) Luciferase assay of SIRT7 promoter activity with/without ectopic HDAC8 in the absence or presence of TGF-β1 (5 ng/ml). The data represent the means ± s.e.m.*P < 0.05; Student's t-test.
HDAC8 induces repressive chromatin remodeling of SIRT7 promoter
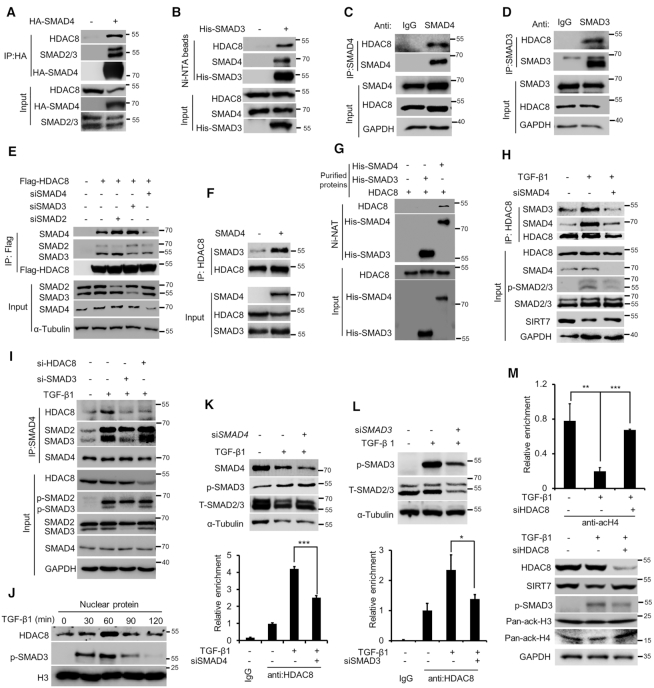
We hypothesized that SMAD3/4 might directly recruit HDAC8 to the SIRT7 promoter. To address this, we did Co-IP and found that HDAC8 was immunoprecipitated with His-SMAD3 and HA-SMAD4 (Figure 4A, B). Physical interaction of HDAC8-SMAD4 and HDAC8-SMAD3 was evidenced by immunoprecipitation of endogenous SMAD4 or SMAD3 (Figure 4C, D). While SMAD3 KD had little effect on the HDAC8–SMAD4 interaction, SMAD4 KD markedly jeopardized the HDAC8–SMAD3 interaction (Figure 4E). On the other hand, forced expression of SMAD4 significantly enhanced the SMAD3–HDAC8 interaction (Figure 4F). Furthermore, HDAC8 detectably interacted with SMAD4 but not SMAD3 based on pulldown assay in the test tube (Figure 4G). As determined by domain mapping, HDAC8 interacted with the MH2 domain of SMAD4, which is well dictated as an interface for binding transcriptional cofactors (34) (Supplementary Figure S4A). These data suggest that SMAD4 directly interacts with HDAC8 and acts as a linker in the SMAD3/4/HDAC8 complex.
Figure 4.
HDAC8 regulates chromatin remodeling of the SIRT7 promoter. (A) Immunoblotting analysis of HDAC8 and SMAD2/3 expression in anti-HA immunoprecipitates in HEK293 cells overexpressing ectopic HA-SMAD4. (B) Immunoblotting analysis of HDAC8 and SMAD4 levels in Ni-NAT-based pulldown from HEK293 cells overexpressing ectopic His-SMAD3. (C, D) IP of endogenous SMAD3 and SMAD4 in breast cancer MDA-MB-231 cells and immunoblotting analysis of HDAC8. (E) Immunoblotting analysis of SMAD2, SMAD3 and SMAD4 expression in anti-FLAG immunoprecipitates in HEK293 cells overexpressing FLAG-HDAC8 treated with indicated siRNAs. (F) Immunoblotting analysis of SMAD3 expression in anti-HDAC8 immunoprecipitates in HEK293 cells with or without ectopic SMAD4 overexpression. (G) In vitro Ni-NAT pulldown of purified His-SMAD3 and His-SMAD4 followed by immunoblotting of HDAC8. (H) Immunoblotting analysis of endogenous SMAD3 and SMAD4 expression in anti-HDAC8 immunoprecipitates in MDA-MB-231 breast cancer cells with indicated treatments. (I) Immunoblotting analysis of endogenous HDAC8 and SMAD3 expression in anti-SMAD4 immunoprecipitates with indicated treatments. (J) Immunoblotting analysis of nuclear HDAC8 in MDA-MB-231 breast cancer cells after TGF-β1 (5 ng/ml) treatment for indicated time. Histone H3 served as the loading control for nuclear proteins. (K, L) ChIP-qPCR analysis of HDAC8 enrichment on the SIRT7 promoter region in MDA-MB-231 breast cancer cells (below). Immunoblotting analysis (upper) of SMAD protein expression. The data represent the means ± s.e.m. n = 3, *P < 0.05, ***P < 0.001; Student's t-test. (M) ChIP-qPCR analysis of histone H4 acetylation levels at the SIRT7 promoter under the indicated treatments (upper). Immunoblotting analysis (below) of HDAC8, p-SMAD3 and total histone H3 and H4 acetylation levels. The data represent the means ± s.e.m. n = 3, **P < 0.01, ***P < 0.001; Student's t-test.
TGFβ induces SMAD3/4 oligomerization, which recruits cofactors to regulate gene transcription. Particularly, revealed by crystallographic data, SMADs oligomers were important for transcriptional complex stabilization (35,36). We explored how TGFβ signaling regulates the SMAD3/4/HDAC8 complex formation. As shown, TGF-β1 treatment significantly enhanced the binding of HDAC8 to SMAD3/4, which was inhibited by SMAD3 KD or SMAD4 KD (Figure 4H, I). By contrast, HDAC8 KD hardly affected SMAD3/4 oligomerization. Consistent with the nuclear trans-localization of SMAD3/4 complex, TGF-β1 treatment also enhanced nuclear HDAC8 level (Figure 4J). Collectively, the data suggest that TGFβ signaling induces SMAD3/4 oligomerization, which in turn enhances HDAC8 recruitment and stabilizes the SMAD3/4/HDAC8 complex.
We next employed ChIP to assess whether HDAC8 is recruited to the SBE motif on SIRT7 promoter. As shown, HDAC8 was markedly enriched on the SIRT7 promoter and TGF-β1 treatment significantly increased HDAC8 occupation, whereas SMAD3 or SMAD4 KD profoundly weakened it (Figure 4K, L). HDAC8 deacetylates histones H3 and H4 (37,38). We used ChIP to examine histone H3 and H4 acetylation levels. As shown, decreased histone H4 but not H3 acetylation level on the SIRT7 promoter was observed upon TGF-β1 treatment (Supplementary Figure S4B). We then knocked down HDAC8 via siRNA and again performed ChIP based on H4 acetylation. Significantly, HDAC8 KD restored H4 acetylation level (Figure 4M). Overall, these findings suggest that HDAC8 regulates local chromatin remodeling of the SIRT7 promoter in response to TGF-β signaling by deacetylating H4.
HDAC8 hyperactivates TGF-β signaling via SIRT7-SMAD4 axis
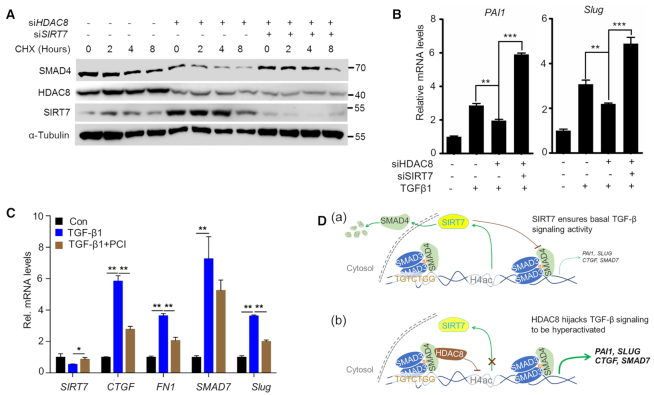
SIRT7 promotes SMAD4 degradation and attenuates TGF-β signaling (13), we next examined whether HDAC8 is involved in SMAD4 regulation. We established CHX chase assay and found that HDAC8 KD accelerated SMAD4 protein degradation in a SIRT7-dependent manner (Figure 5A), without affecting its mRNA (Supplementary Figure S5A). HDAC8 blockade by siRNA or PCI-34051 significantly inhibited TGF-β signaling, indicated by the expression of the downstream gene PAI1, SLUG, CTGF and FN1 (Figure 5B, C). All together, we propose that SIRT7 deacetylates SMAD4, leading to its degradation and ensuring basal activity of TGF-β signaling (13) (Figure 5D(a)); HDAC8 cooperates with SMAD3/4 heterotrimer to suppress SIRT7 transcription, causing hyperactivation of TGF-β signaling (Figure 5D(b)).
Figure 5.
HDAC8 inhibition attenuates TGF-β signaling. (A) SMAD4 protein levels in MDA-MB-231 breast cancer cells exposed to the indicated treatments. CHX, cycloheximide (50 μg/ml). (B, C) qRT-PCR analysis of TGF-β downstream genes in MDA-MB-231 breast cancer cells. The data represent the means ± s.e.m. n = 3, *P < 0.05, **P < 0.01, ***P < 0.001; Student's t-test. (D) A schematic model: (a) SIRT7 deacetylates SMAD4, leading to its degradation and ensuring basal activity of TGF-β signaling; (b) HDAC8 cooperates with SMAD3/4 heterotrimer to suppress SIRT7 transcription, causing hyperactivation of TGF-β signaling.
Targeting HDAC8 suppresses cancer metastasis and chemotherapy resistance
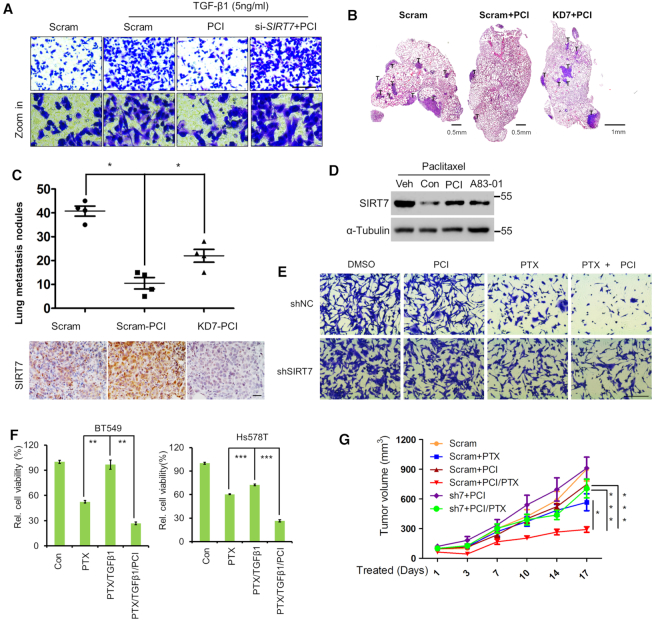
TGF-β signaling promotes breast cancer metastasis (39,40). Inhibition of HDAC8 via PCI-34051 markedly attenuated breast cancer cell migration induced by TGF-β1; SIRT7 KD completely abolished this effect (Figure 6A and Supplementary Figure S5B). We examined whether inhibiting HDAC8 prevents breast cancer metastasis. We employed 4T1 murine breast cancer cells that can readily develop into lung metastases (41). As shown, PCI-34051 treatment (10 mg/Kg/mouse) markedly decreased lung metastatic nodules compared to the vehicle control, while simultaneous SIRT7 KD largely attenuated the effect (Figure 6B. C and Supplementary Figure S5C). We noticed increased Sirt7 protein level in 4T1 tumors exposed to PCI-34051 (Figure 6C, below). By contrast, PCI-34051 treatment merely affected the acetylation levels of H3K18 and SMAD4, substrates of SIRT7, indicating that PCI-34051 less likely inhibits SIRT7 deacetylase activity (Supplementary Figure S5E. F). These findings implicate that HDAC8 inhibition suppresses breast cancer metastasis via SIRT7.
Figure 6.
HDAC8 inhibition suppresses cancer metastasis and attenuates chemotherapy resistance. (A) Transwell assay of MDA-MB-231 breast cancer cell migratory capacity after incubation with TGF-β1 or PCI-34051 (PCI), as indicated. Scale bar, 100 μm. (B) Representative H&E staining to detect lung metastasis of 4T1 breast cancer cells. Scram, mice inoculated with 4T1 cells treated with scrambled shRNA; Scram+PCI, mice inoculated with cells treated with scrambled shRNA and PCI-34051 (10 mg/kg body weight); KD7+PCI, mice inoculated with cells treated with SIRT7 shRNA and PCI-34051. Scale bar, 0.5 mm. (C) Scatter plots (upper) of the number of lung metastatic nodules in individual mice in (B), n = 4 mice per group. IHC analysis of Sirt7 expression in lung metastasis sections (lower). *P < 0.05; non-parametric Mann–Whitney U-test. Scale bar, 100 μm. (D) Immunoblotting analysis of SIRT7 levels in breast cancer cells treated with paclitaxel (PTX, 10 nM), PCI-34051 (PCI, 10 μM) or TβR1 inhibitor A83–01 (5 μM). (E) Crystal violet staining of control (shNC) and SIRT7 knockdown (shSIRT7) cells with the indicated treatments. Scale bar, 200 μm. (F) CCK8 assay of TNBC BT549 and Hs578T cell viability under the indicated treatments. The data represent the means ± s.e.m. n = 3, **P < 0.01, ***P < 0.001; Student's t-test. (G) Tumor volumes at the indicated time points in BALB/c mice treated with PTX (10 mg/kg), PCI (10 mg/kg) or PCI (10 mg/kg) + PTX (10 mg/kg). Mice were inoculated with 4T1 cells (5 × 105) in the mammary fat pad 2 weeks before the treatment. *P < 0.05 and ***P < 0.001; one-way ANOVA.
TGF-β signaling activation renders breast cancers insensitive to chemotherapy (42). SIRT7 is also downregulated by chemotherapeutic agents (7), and SIRT7 KD induces chemoresistance to paclitaxel in breast cancer (43). We asked whether HDAC8 is involved in breast cancer chemoresistance via SIRT7-SMAD4 axis. Interestingly, the paclitaxel-induced downregulation of SIRT7 was rescued by A-8301 and PCI-34501 treatment in MDA-MB-231 cells (Figure 6D and Supplementary Figure S5D). It suggests that HDAC8 underlies SIRT7 reduction induced by paclitaxel. We asked whether blocking HDAC8 sensitizes breast cancer cells to paclitaxel. As shown, the combined treatment with paclitaxel and PCI-34051 eliminated cancer cells more effectively than just one treatment alone (Figure 6E, upper). SIRT7 KD largely abrogated the effect of PCI-34051 (Figure 6E, lower). Likewise, TGF-β1-induced paclitaxel resistance was also attenuated by combined PCI-34051 treatment in BT549 and Hs578T breast cancer cells (Figure 6F). Importantly, PCI-34051 (10 mg/kg body weight) combined with paclitaxel (10 mg/kg body weight) greatly suppressed breast cancer growth in xenograft mouse model (Figure 6G). SIRT7 KD reversed tumor growth inhibition induced by the combined treatment. Based on the survival analysis tool (http://kmplot.com/) (44), high HDAC8 expression predicated poor prognosis in breast cancer patients (Supplementary Figure S7, left). Particularly, patients received chemotherapy with high HDAC8 had much worse prognosis, further indicating a drug resistance role of HDAC8 (Supplementary Figure S7, right). These data suggest that HDAC8 blockade suppresses breast cancer lung metastasis and attenuates paclitaxel resistance.
DISCUSSION
We propose that TGF-β1 activates the SMAD3/4/HDAC8 complex, which translocate into the nucleus and suppresses SIRT7 transcription via local H4 deacetylation. As a feedback, SIRT7 reduction activates TGF-β signaling and promotes related cellular functions, such as cell migration and cancer chemoresistance. HDAC8 inhibition represses TGF-β signaling, breast cancer metastasis and chemotherapy resistance.
SIRT7 has critical roles in ribosome biogenesis (45,46), cellular stress responses (4,10,47), genome stability (5,48), metabolic regulation (11,49), aging (8,9) and cancers (50). However, how SIRT7 is regulated at upstream is elusive. We have previously shown that SIRT7 deacetylates and destabilizes SMAD4, thus antagonizing TGF-β signaling (13). Here, we identified SIRT7 as a new target that is transcriptionally repressed by SMAD3/4 complex in collaboration with HDAC8, which mediates H4 deacetylation at local chromatin.
The association of SMADs with co-activators, such as CBP/p300, leads to chromatin remodeling via histone acetylation and modulates gene transcription (19). Conversely, transcriptional repression mediated by deacetylation may occur by recruiting HDACs to the SMAD binding sites (20). For example, SMAD3 recruits HDAC4/5 to mediate transcriptional repression of Runx2 (21). Other than that, SMADs-mediated HDACs recruitment in suppressing transcription is rare. HDAC8 is the first human HDACs to be crystalized, however its roles are still obscure as few downstream target is found (51). We are first to show that HDAC8 is involved in TGF-β signaling, and SIRT7 is a bona fide transcriptional repression target of the SMAD3/4/HDAC8 complex. On the other front, acetylation/deacetylation of SMADs is also critical for TGFβ signaling. For instance, p300/CBP mediates SMAD3 acetylation and Sirt1/7 induces SMAD4 deacetylation (13,52,53). Albeit HDACs are frequently identified as SMAD-recruited components, they mainly regulate local chromatin remodeling rather than deacetylate SMADs (34). Herein, HDAC8 less likely deacetylates SMAD3 or SMAD4 (see Supplementary Figure S6A, B). Previous studies suggest that HDAC8 could be a potential therapeutic target as it enhances breast cancer stemness and invasion. This is consistent with its effect on SIRT7 repression, as SIRT7 inhibits breast cancer stemness and metastasis (13,54). HDAC3 is the closest HDAC8 human homolog (55). Interestingly, HDAC3 represses SIRT7 expression in hepatocellular carcinoma via C/EBPα (56). However, such a suppressive effect was merely observed in TGF-β signaling. The diversified effects suggest that the regulation of SIRT7 might be determined by specific physiological context. Phenotypical studies suggest SIRT7 as an aging-related gene and its level is significantly decreased during aging (9,10). As such, it would be interesting to explore whether HDAC8 is involved in age-related diseases, such as tissue fibrosis and cardiovascular dysfunction.
The role of SIRT7 in cancers is hotly debated. SIRT7 is aberrantly increased in CRC, HCC, early stage breast cancers and thyroid cancers (57). Many studies have shown that SIRT7 promotes tumor cell growth, emphasizing an oncogenic action. Seemingly contradictory, SIRT7 inhibits metastasis in breast cancers and oral squamous cell carcinomas by antagonizing TGF-β signaling (13,58). Our finding that SIRT7 is downregulated by TGF-β provides a reasonable explanation for this effect; furthermore, it is well known that TGF-β is a ‘double-edged sword’ in the cancer context (59). During the pre-malignant stage, TGF-β has a tumor suppressive role by inducing cell-cycle arrest and apoptosis (59). Malignant cancers often bypass this unfavorable effect, e.g. mutations in TβRII, SMAD2 and SMAD4 are common in HCC, CRC, pancreatic cancer and lung cancer (39), which obtain a growth advantage, consistent with the oncogenic role of SIRT7. Conversely, for breast cancers, TGF-β signaling is intact, and is instead engaged to promote cancer-cell EMT, stemness and metastasis. SIRT7 promotes SMAD4 degradation and antagonizes TGF-β signaling (13). Our finding that TGF-β transcriptionally represses SIRT7 partially explains low SIRT7 expression in breast cancer lung metastases. As an attractive target for drug development, due to the aforementioned complicated roles, however, it is critical to consider the functional diversities of SIRT7 and select patients who could obtain the best benefit.
TNBCs are the most malignant breast cancer subtype, and are characterized by high metastasis and poor prognosis (60). To date, TNBCs still lack well-defined molecular targets: traditional chemotherapy (such as paclitaxel) and surgery are most widely applied (61). While initial paclitaxel treatment is effective for some patients, many patients later develop resistance (62) that is largely attributed to TGF-β activation (42). We found that HDAC8 enhances cell migration and boosts cancer stemness in TNBCs and PCI-34051 abrogates breast cancer lung metastasis and TGF-β-induced paclitaxel resistance by repressing SIRT7, suggesting that HDAC8 or its targets has therapeutic potential (24,63,64).
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr Jessica Tamanini (Shenzhen University and ETediting) for editing the manuscript prior to submission.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2017YFA0503900]; National Natural Science Foundation of China [91849208, 81702909, 81972602, 81871114, 81571374, 91949124, 8187051935]; Science and Technology Program of Guangdong Province in China [2017B030301016, 2019B030301009, 2019A151510472]; Shenzhen Municipal Commission of Science and Technology Innovation [ZDSYS20190902093401689, JCYJ20160226191451487, KQJSCX20180328093403969, JCYJ20180507182044945]; China Postdoctoral Science Foundation [2015M582419]. Funding for open access charge: National Key R&D Program of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Michishita E., Park J.Y., Burneskis J.M., Barrett J.C., Horikawa I.. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005; 16:4623–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kiran S., Anwar T., Kiran M., Ramakrishna G.. Sirtuin 7 in cell proliferation, stress and disease: rise of the seventh Sirtuin. Cell Signal. 2015; 27:673–682. [DOI] [PubMed] [Google Scholar]
- 3. Tsai Y.C., Greco T.M., Cristea I.M.. Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. Mol. Cell. Proteomics. 2014; 13:73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shin J., He M., Liu Y., Paredes S., Villanova L., Brown K., Qiu X., Nabavi N., Mohrin M., Wojnoonski K. et al.. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep. 2013; 5:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vazquez B.N., Thackray J.K., Simonet N.G., Kane-Goldsmith N., Martinez-Redondo P., Nguyen T., Bunting S., Vaquero A., Tischfield J.A., Serrano L.. SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J. 2016; 35:1488–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tang M., Li Z., Zhang C., Lu X., Tu B., Cao Z., Li Y., Chen Y., Jiang L., Wang H. et al.. SIRT7-mediated ATM deacetylation is essential for its deactivation and DNA damage repair. Sci. Adv. 2019; 5:eaav1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tang M., Lu X., Zhang C., Du C., Cao L., Hou T., Li Z., Tu B., Cao Z., Li Y. et al.. Downregulation of SIRT7 by 5-fluorouracil induces radiosensitivity in human colorectal cancer. Theranostics. 2017; 7:1346–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghiraldini F.G., Crispim A.C., Mello M.L.. Effects of hyperglycemia and aging on nuclear sirtuins and DNA damage of mouse hepatocytes. Mol. Biol. Cell. 2013; 24:2467–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wronska A., Lawniczak A., Wierzbicki P.M., Kmiec Z.. Age-related changes in Sirtuin 7 expression in calorie-restricted and Refed rats. Gerontology. 2016; 62:304–310. [DOI] [PubMed] [Google Scholar]
- 10. Mohrin M., Shin J., Liu Y., Brown K., Luo H., Xi Y., Haynes C.M., Chen D.. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science. 2015; 347:1374–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshizawa T., Karim M.F., Sato Y., Senokuchi T., Miyata K., Fukuda T., Go C., Tasaki M., Uchimura K., Kadomatsu T. et al.. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab. 2014; 19:712–721. [DOI] [PubMed] [Google Scholar]
- 12. Sun L., Fan G., Shan P., Qiu X., Dong S., Liao L., Yu C., Wang T., Gu X., Li Q. et al.. Regulation of energy homeostasis by the ubiquitin-independent REGgamma proteasome. Nat. Commun. 2016; 7:12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang X., Shi L., Xie N., Liu Z., Qian M., Meng F., Xu Q., Zhou M., Cao X., Zhu W.G. et al.. SIRT7 antagonizes TGF-beta signaling and inhibits breast cancer metastasis. Nat. Commun. 2017; 8:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi H., Ji Y., Zhang D., Liu Y., Fang P.. MicroRNA-3666-induced suppression of SIRT7 inhibits the growth of non-small cell lung cancer cells. Oncol. Rep. 2016; 36:3051–3057. [DOI] [PubMed] [Google Scholar]
- 15. Cioffi M., Vallespinos-Serrano M., Trabulo S.M., Fernandez-Marcos P.J., Firment A.N., Vazquez B.N., Vieira C.R., Mulero F., Camara J.A., Cronin U.P. et al.. MiR-93 controls adiposity via inhibition of Sirt7 and Tbx3. Cell Rep. 2015; 12:1594–1605. [DOI] [PubMed] [Google Scholar]
- 16. Kim J.K., Noh J.H., Jung K.H., Eun J.W., Bae H.J., Kim M.G., Chang Y.G., Shen Q., Park W.S., Lee J.Y. et al.. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013; 57:1055–1067. [DOI] [PubMed] [Google Scholar]
- 17. Han Y., Liu Y., Zhang H., Wang T., Diao R., Jiang Z., Gui Y., Cai Z.. Hsa-miR-125b suppresses bladder cancer development by down-regulating oncogene SIRT7 and oncogenic long noncoding RNA MALAT1. FEBS Lett. 2013; 587:3875–3882. [PubMed] [Google Scholar]
- 18. Akhurst R.J., Hata A.. Targeting the TGFbeta signalling pathway in disease. Nat. Rev. Drug Discov. 2012; 11:790–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hill C.S. Transcriptional control by the SMADs. Cold Spring Harb. Perspect. Biol. 2016; 8:a022079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moustakas A., Souchelnytskyi S., Heldin C.H.. Smad regulation in TGF-beta signal transduction. J. Cell Sci. 2001; 114:4359–4369. [DOI] [PubMed] [Google Scholar]
- 21. Kang J.S., Alliston T., Delston R., Derynck R.. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005; 24:2543–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buggy J.J., Sideris M.L., Mak P., Lorimer D.D., McIntosh B., Clark J.M.. Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem. J. 2000; 350 Pt 1:199–205. [PMC free article] [PubMed] [Google Scholar]
- 23. Hu E., Chen Z., Fredrickson T., Zhu Y., Kirkpatrick R., Zhang G.F., Johanson K., Sung C.M., Liu R., Winkler J.. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J. Biol. Chem. 2000; 275:15254–15264. [DOI] [PubMed] [Google Scholar]
- 24. Wang Z.T., Chen Z.J., Jiang G.M., Wu Y.M., Liu T., Yi Y.M., Zeng J., Du J., Wang H.S.. Histone deacetylase inhibitors suppress mutant p53 transcription via HDAC8/YY1 signals in triple negative breast cancer cells. Cell Signal. 2016; 28:506–515. [DOI] [PubMed] [Google Scholar]
- 25. Qian Y., Zhang J., Jung Y.S., Chen X.. DEC1 coordinates with HDAC8 to differentially regulate TAp73 and DeltaNp73 expression. PLoS One. 2014; 9:e84015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nelson J.D., Denisenko O., Bomsztyk K.. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006; 1:179–185. [DOI] [PubMed] [Google Scholar]
- 27. Palumbo-Zerr K., Zerr P., Distler A., Fliehr J., Mancuso R., Huang J., Mielenz D., Tomcik M., Furnrohr B.G., Scholtysek C. et al.. Orphan nuclear receptor NR4A1 regulates transforming growth factor-beta signaling and fibrosis. Nat. Med. 2015; 21:150–158. [DOI] [PubMed] [Google Scholar]
- 28. Xue J., Lin X., Chiu W.T., Chen Y.H., Yu G., Liu M., Feng X.H., Sawaya R., Medema R.H., Hung M.C. et al.. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-beta-dependent cancer metastasis. J. Clin. Invest. 2014; 124:564–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Massague J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012; 13:616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang J.H., Li J.H., Jiang S., Zhou H., Qu L.H.. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res. 2013; 41:D177–D187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loots G.G., Ovcharenko I.. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004; 32:W217–W221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kutz S.M., Higgins C.E., Higgins P.J.. Novel combinatorial therapeutic targeting of PAI-1 (SERPINE1) gene expression in Alzheimer's disease. Mol. Med. Ther. 2012; 1:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Balasubramanian S., Ramos J., Luo W., Sirisawad M., Verner E., Buggy J.J.. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia. 2008; 22:1026–1034. [DOI] [PubMed] [Google Scholar]
- 34. Massague J., Seoane J., Wotton D.. Smad transcription factors. Genes Dev. 2005; 19:2783–2810. [DOI] [PubMed] [Google Scholar]
- 35. Chacko B.M., Qin B.Y., Tiwari A., Shi G., Lam S., Hayward L.J., De Caestecker M., Lin K.. Structural basis of heteromeric smad protein assembly in TGF-beta signaling. Mol. Cell. 2004; 15:813–823. [DOI] [PubMed] [Google Scholar]
- 36. Wu J.W., Fairman R., Penry J., Shi Y.. Formation of a stable heterodimer between Smad2 and Smad4. J. Biol. Chem. 2001; 276:20688–20694. [DOI] [PubMed] [Google Scholar]
- 37. Somoza J.R., Skene R.J., Katz B.A., Mol C., Ho J.D., Jennings A.J., Luong C., Arvai A., Buggy J.J., Chi E. et al.. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004; 12:1325–1334. [DOI] [PubMed] [Google Scholar]
- 38. Vannini A., Volpari C., Filocamo G., Casavola E.C., Brunetti M., Renzoni D., Chakravarty P., Paolini C., De Francesco R., Gallinari P. et al.. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:15064–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Colak S., Dijke Ten. Targeting TGF-beta signaling in cancer. Trends Cancer. 2017; 3:56–71. [DOI] [PubMed] [Google Scholar]
- 40. Muraoka R.S., Dumont N., Ritter C.A., Dugger T.C., Brantley D.M., Chen J., Easterly E., Roebuck L.R., Ryan S., Gotwals P.J. et al.. Blockade of TGF-beta inhibits mammary tumor cell viability, migration, and metastases. J. Clin. Invest. 2002; 109:1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pulaski B.A., Ostrand-Rosenberg S.. Mouse 4T1 breast tumor model. Curr. Protoc. Immunol. 2001; doi:10.1002/0471142735.im2002s39. [DOI] [PubMed] [Google Scholar]
- 42. Bhola N.E., Balko J.M., Dugger T.C., Kuba M.G., Sanchez V., Sanders M., Stanford J., Cook R.S., Arteaga C.L.. TGF-beta inhibition enhances chemotherapy action against triple-negative breast cancer. J. Clin. Invest. 2013; 123:1348–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu J., Qin B., Wu F., Qin S., Nowsheen S., Shan S., Zayas J., Pei H., Lou Z., Wang L.. Regulation of serine-threonine kinase Akt activation by NAD(+)-dependent deacetylase SIRT7. Cell Rep. 2017; 18:1229–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gyorffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., Szallasi Z.. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010; 123:725–731. [DOI] [PubMed] [Google Scholar]
- 45. Ford E., Voit R., Liszt G., Magin C., Grummt I., Guarente L.. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006; 20:1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grob A., Roussel P., Wright J.E., McStay B., Hernandez-Verdun D., Sirri V.. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J. Cell Sci. 2009; 122:489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen S., Seiler J., Santiago-Reichelt M., Felbel K., Grummt I., Voit R.. Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol. Cell. 2013; 52:303–313. [DOI] [PubMed] [Google Scholar]
- 48. Tang M., Li Z., Zhang C., Lu X., Tu B., Cao Z., Li Y., Chen Y., Jiang L., Wang H. et al.. SIRT7-mediated ATM deacetylation is essential for its deactivation and DNA damage repair. Sci. Adv. 2019; 5:eaav1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tang B.L. SIRT7 and hepatic lipid metabolism. Front. Cell Dev. Biol. 2015; 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Blank M.F., Grummt I.. The seven faces of SIRT7. Transcription. 2017; 8:67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Delcuve G.P., Khan D.H., Davie J.R.. Targeting class I histone deacetylases in cancer therapy. Expert Opin. Ther. Targets. 2013; 17:29–41. [DOI] [PubMed] [Google Scholar]
- 52. Inoue Y., Itoh Y., Abe K., Okamoto T., Daitoku H., Fukamizu A., Onozaki K., Hayashi H.. Smad3 is acetylated by p300/CBP to regulate its transactivation activity. Oncogene. 2007; 26:500–508. [DOI] [PubMed] [Google Scholar]
- 53. Simic P., Williams E.O., Bell E.L., Gong J.J., Bonkowski M., Guarente L.. SIRT1 suppresses the epithelial-to-mesenchymal transition in cancer metastasis and organ fibrosis. Cell Rep. 2013; 3:1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shi L., Tang X., Qian M., Liu Z., Meng F., Fu L., Wang Z., Zhu W.G., Huang J.D., Zhou Z. et al.. A SIRT1-centered circuitry regulates breast cancer stemness and metastasis. Oncogene. 2018; 37:6299–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gregoretti I.V., Lee Y.M., Goodson H.V.. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 2004; 338:17–31. [DOI] [PubMed] [Google Scholar]
- 56. Liu G.F., Lu J.Y., Zhang Y.J., Zhang L.X., Lu G.D., Xie Z.J., Cheng M.B., Shen Y.F., Zhang Y.. C/EBPalpha negatively regulates SIRT7 expression via recruiting HDAC3 to the upstream-promoter of hepatocellular carcinoma cells. Biochim. Biophys. Acta. 2016; 1859:348–354. [DOI] [PubMed] [Google Scholar]
- 57. Wu D., Li Y., Zhu K.S., Wang H., Zhu W.G.. Advances in cellular characterization of the sirtuin isoform, SIRT7. Front. Endocrinol. 2018; 9:652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li W., Zhu D., Qin S.. SIRT7 suppresses the epithelial-to-mesenchymal transition in oral squamous cell carcinoma metastasis by promoting SMAD4 deacetylation. J. Exp. Clin. Cancer Res. 2018; 37:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Akhurst R.J., Derynck R.. TGF-beta signaling in cancer–a double-edged sword. Trends Cell Biol. 2001; 11:S44–S51. [DOI] [PubMed] [Google Scholar]
- 60. Dent R., Trudeau M., Pritchard K.I., Hanna W.M., Kahn H.K., Sawka C.A., Lickley L.A., Rawlinson E., Sun P., Narod S.A.. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 2007; 13:4429–4434. [DOI] [PubMed] [Google Scholar]
- 61. Liedtke C., Mazouni C., Hess K.R., Andre F., Tordai A., Mejia J.A., Symmans W.F., Gonzalez-Angulo A.M., Hennessy B., Green M. et al.. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008; 26:1275–1281. [DOI] [PubMed] [Google Scholar]
- 62. Li X., Lewis M.T., Huang J., Gutierrez C., Osborne C.K., Wu M.F., Hilsenbeck S.G., Pavlick A., Zhang X., Chamness G.C. et al.. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J. Natl. Cancer Inst. 2008; 100:672–679. [DOI] [PubMed] [Google Scholar]
- 63. An P., Li J., Lu L., Wu Y., Ling Y., Du J., Chen Z., Wang H.. Histone deacetylase 8 triggers the migration of triple negative breast cancer cells via regulation of YAP signals. Eur. J. Pharmacol. 2019; 845:16–23. [DOI] [PubMed] [Google Scholar]
- 64. Chao M.W., Chu P.C., Chuang H.C., Shen F.H., Chou C.C., Hsu E.C., Himmel L.E., Huang H.L., Tu H.J., Kulp S.K. et al.. Non-epigenetic function of HDAC8 in regulating breast cancer stem cells by maintaining Notch1 protein stability. Oncotarget. 2016; 7:1796–1807. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.