Abstract
The NEIL3 DNA glycosylase is a base excision repair enzyme that excises bulky base lesions from DNA. Although NEIL3 has been shown to unhook interstrand crosslinks (ICL) in Xenopus extracts, how NEIL3 participants in ICL repair in human cells and its corporation with the canonical Fanconi anemia (FA)/BRCA pathway remain unclear. Here we show that the NEIL3 and the FA/BRCA pathways are non-epistatic in psoralen-ICL repair. The NEIL3 pathway is the major pathway for repairing psoralen-ICL, and the FA/BRCA pathway is only activated when NEIL3 is not present. Mechanistically, NEIL3 is recruited to psoralen-ICL in a rapid, PARP-dependent manner. Importantly, the NEIL3 pathway repairs psoralen-ICLs without generating double-strand breaks (DSBs), unlike the FA/BRCA pathway. In addition, we found that the RUVBL1/2 complex physically interact with NEIL3 and function within the NEIL3 pathway in psoralen-ICL repair. Moreover, TRAIP is important for the recruitment of NEIL3 but not FANCD2, and knockdown of TRAIP promotes FA/BRCA pathway activation. Interestingly, TRAIP is non-epistatic with both NEIL3 and FA pathways in psoralen-ICL repair, suggesting that TRAIP may function upstream of the two pathways. Taken together, the NEIL3 pathway is the major pathway to repair psoralen-ICL through a unique DSB-free mechanism in human cells.
INTRODUCTION
DNA interstrand cross-links (ICLs) are toxic lesions that prevent DNA replication and transcription by blocking DNA strand separation, and unrepaired ICLs lead to apoptosis and cell death (1). The Fanconi anemia (FA) pathway is essential for the repair of DNA-ICLs, and defects in the FA pathway result in Fanconi anemia, a chromosomal instability disorder characterized by congenital abnormalities, progressive bone marrow failure, and cancer predisposition (2). The FA proteins function in a multistep pathway required for the repair of endogenous and exogenous ICLs, such as ICLs induced by the therapeutic agent Mitomycin C (MMC). To date, 23 FA genes have been identified, which are grouped into three categories: the FA core complex (an E3 ligase complex), the FANCI/FANCD2 (ID) complex, and the downstream effector proteins, such as structure-specific nuclease and double-strand break (DSB) repair proteins (3,4). When DNA replication is blocked by an ICL, the FA core complex (containing FANCA, B, C, E, F, G, L and M) monoubiquitinates the FANCI–FANCD2 complex (ID2), a pivotal step in the FA pathway (5). The FA core complex is recruited to a stalled replication fork by an ICL via the anchoring complex containing FANCM subunit, along with Fanconi-associated proteins (FAAPs). Nucleolytic processing of the ICLs, which involves nucleases recruited by the SLX4/FANCP scaffold protein (6,7), generates DSBs that can be repaired by multiple downstream repair pathways (8).
Base excision repair (BER) is the major pathway for repairing DNA base damage and single strand breaks. If left unrepaired, these lesions can be mutagenic, blocking replication fork (9), or even perturbing epigenetic marks (10,11). The first and most critical step of BER is the searching and excision of damaged bases, a step that is carried out by DNA glycosylases. NEIL3 (DNA Endonuclease VIII-like 3) is a member of the Fpg/Nei glycosylase family (12,13), also including NEIL1 (14) and NEIL2 (15). Like other members of the Fpg/Nei family, NEIL3 contains a DNA glycosylase activity that excises damaged bases and an AP (apurinic/apyrimidinic site) lyase activity that cleaves the DNA backbone at an AP site, thus generating a single-strand break (SSB) (13,16). NEIL3 is distinguished from the other NEILs by its long C-terminal domain (CTD) (13). The glycosylase domain of NEIL3 prefers base lesions in single-stranded DNA (ssDNA) or ssDNA-containing structures (i.e. fork DNA) (13,16). NEIL3 also possesses the unique activity of removing damaged bases from G-quadruplex DNA (17–19).
The biochemical features of NEIL3 have been well characterized in the past decade, although the cellular function of NEIL3 has just begun to be understood. NEIL3 repairs telomere damage and protects telomeres during S phase to ensure accurate segregation of chromosome during mitosis (20). NEIL3 also plays a critical role in preventing autoimmunity, and its glycosylase activity is required for this function (21). NEIL3 appears to be important for cell proliferation, as evidenced by its role in regulating proliferation of cardiac fibroblasts in the heart and neural progenitor cells in the human brain (22,23). Its expression is increased in highly replicative tissues such as bone marrow (12,24) and in cancerous tissues (24). These studies demonstrate that NEIL3 is a versatile DNA glycosylase that functions outside of BER.
A new role for NEIL3 in ICL repair has recently been discovered. NEIL3 directly unhooks psoralen- and AP-ICLs during DNA replication in Xenopus egg extracts (25). This function of NEIL3 appears to be the first choice for the repair of a psoralen-ICL or AP-ICL (formed by AP site and an adenosine on opposite strands), and its failure activates the Fanconi anemia pathway. Purified NEIL3 and NEIL1 excises psoralen-induced DNA–DNA cross-links in vitro (26,27). Moreover, NEIL3 unhooks ICLs between glycosidic bonds and does not nick the DNA backbone, and accordingly does not generate DSBs (25,26). The ability of NEIL3 to accommodate those bulky lesions is supported by its crystal structure, which features a large lesion binding pocket of its glycosylase domain (28). In addition, mouse embryonic fibroblasts (MEFs) derived from Neil3-/- mice are sensitive to ICL-inducing agents such as cisplatin (29). These studies show that NEIL3 plays an important role in ICL repair.
Additional players in ICL repair have been recently discovered. The RING-Type E3 ubiquitin ligase TRAIP (TRAF Interacting Protein) for example is essential for cell proliferation and replication fork progression through DNA damage, and TRAIP mutations cause microcephalic primordial dwarfism (30,31). TRAIP is required for optimal phosphorylation of H2AX and RPA2 during S-phase after UV-C irradiation (31), and knockdown of TRAIP sensitizes cells to MMC (32). More recently, TRAIP was found to operate a switch between the NEIL3 and FA pathways in ICL repair by controlling the length of ubiquitin chain on CMG complex (33). In addition, the AAA+ ATPase family proteins RuvB Like 1 (RUVBL1) and its paralog RUVBL2 are DNA-dependent ATPases/helicases that are involved in diverse cellular activities. The RUVBL1/2 complex facilitates homologous recombination (34) and ICL repair (35,36). The RUVBL1/2 complex also regulates the abundance of FA core complex proteins, and depletion of RUVBL1/2 causes FA phenotypes in human cells, including defective in FA pathway activation, chromosomal instability and sensitivity to MMC (36).
Although NEIL3 was shown to act on ICLs biochemically in the Xenopus egg extracts, little is known about how NEIL3 contributes to ICL repair in human cells, and how the NEIL3 and FA pathways interact. Here we show that the NEIL3 pathway is the major pathway that repairs psoralen-induced ICLs (psoralen-ICLs), while the FA pathway is mostly responsible for repairing MMC- and cisplatin- induced ICLs. NEIL3 provides a strong response to psoralen-ICLs, resulting in a rapid, PARP-dependent recruitment to psoralen-ICL sites and protein accumulation/stabilization. Unlike the canonical FA pathway, NEIL3 unhooks psoralen-ICLs without creating DSBs in human cells. TRAIP appears to function upstream of the NEIL3 and the FA pathways, while RUVBL1/2 binds directly to NEIL3 and functions within the NEIL3 pathway. Collectively, we show the NEIL3 pathway is prioritized in psoralen-ICL repair through a unique mechanism in human cells.
MATERIALS AND METHODS
Cell lines and culture conditions
Human FA fibroblasts including PD20 (FANCD2-/-), and PD326 (FANCG-/-) were derived from FA patients as previously described (37). All human FA fibroblasts, U2OS and HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum (Thermo Fisher Scientific) and 1% penicillin/streptomycin (Thermo Fisher Scientific) in a 5% CO2 incubator at 37°C. The primary wild-type and Nelil3-/- mouse embryonic fibroblasts (MEFs) were gifts from Magnar Bjørås. The MEFs were immortalized with SV40 antigen according to the protocol described previously (38). The immortalized MEFs were grown in minimum essential medium supplemented with 10% (v/v) fetal bovine serum (Thermo Fisher Scientific), 2 mM l-glutamine and 1% penicillin/streptomycin (Thermo Fisher Scientific) in a 5% CO2 incubator at 37°C.
Plasmids, siRNA, and transfection
The EGFP-FANCD2 and NEIL3-HA plasmids were described previously (20,39). The RUVBL1-Flag (#OHu26261) and RUVBL2-Flag (#OHu25367) plasmids were purchased from GenScript. Plasmid was transfected using Lipofectamine LTX Reagent (Thermo Fisher Scientific). For siRNA treatment, cells were transfected with specific siRNA (Dharmacon and Qiagen; see Supplementary Table S1) using Lipofectamine RNAiMAX Reagent (Thermo Fisher Scientific).
Generation of NEIL3-/-, FANCA-/- and FANCD2-/- cells
HeLa-FANCA-/- and U2OS-FANCD2-/- cell lines were generated using the CRISPR-Cas9 system.To generate HeLa-NEIL3-/- and U2OS-NEIL3-/- cell lines, HeLa- and U2OS-CAS9 stable cell lines were firstly generated by transfection with the Cas9 plasmid and selected by blasticidin (Thermo Fisher Scientific). Oligonucleotides encoding guide RNAs targeting NEIL3 (5′-ATTCGCGCGCGGGTGCTCCC-3′) was cloned into the lentiGuide-Puro vector, a gift from Feng Zhang (Addgene plasmid # 52963). To produce lentivirus, HEK293T cells were seeded at 60% confluence in 60-mm dishes 24 h before transfection. The gRNA-lentiGuide-Puro construct was transfected together with the packaging plasmids (psPAX2 and PMD2.G) using LipoLTX reagent (Thermo Fisher Scientific). Virus was harvested 48 h after transfection, filtered through a 0.45 mm low-protein-binding membrane (Millipore), and then placed on the HeLa- or U2OS-CAS9 cells with a density of 10%. Mono-clone was collected after 2–3 weeks of selection by puromycin, and knockout clones were identified by western blotting.
PUVA (psoralen plus ultraviolet A) treatment
To induce psoralen-ICLs, the cells were washed using 1× PBS and were treated with the indicated concentrations of psoralen (Sigma-Aldrich #P8399) in 1× Hank's Balanced Salt Solution (HBSS) containing 2% FBS. After incubation for 30 min in the dark, cells were exposed to ultraviolet A (UVA) light (365 nm) at a dose of 0.8 J/cm2 (40,41). Following treatment, cells were allowed to recover at 37°C in complete medium.
CellTiter-Glo cell viability assay
The CellTiter-Glo luminescent cell viability assay kit (Promega) was used to measure cell viability after drug treatment. The CellTiter-Glo kit determines the number of viable cells in culture based on quantitation of the ATP present, an indicator of metabolically active cells Generally, to determine the drug sensitivity of NEIL3-/- cells, 2000 cells (HeLa, U2OS or MEF) per well were plated in a 96-well plate. The sources and doses of DNA damaging agents were as following: camptothecin (Selleckchem #S1288: 0, 0.625, 1.25, 2.5, 5, 10 nM), mitomycin C (MMC, Sigma Aldrich #M0503: 0, 1.25, 2.5, 5, 10, 20 ng/ml), cisplatin (Sigma Aldrich #P4394: 0, 0.0625, 0.125, 0.25, 0.5, 1 μM), Hydroxyurea (HU, Sigma Aldrich #H8627: 0, 0.03125, 0.0625, 0.125, 0.25, 0.5 mM), psoralen (Sigma-Aldrich #P8399, 0, 0.03125, 0.0625, 0.125, 0.25, 0.5 μM), X-ray irradiation (IR: 0, 1, 2, 5, 7.5, 10 Gy), PUVA (0, 0.015625, 0.03125, 0.0625, 0.125 or 0.25 μM psoralen incubation for 30 min followed by exposure to 0.8 J/cm2 UVA), UVA (0, 0.0625, 0.125, 0.25, 0.5, 1.0 J/cm2), Ultraviolet B (UVB: 0, 50, 100, 200, 400, 800 J/m2), or Ultraviolet C (UVC: 0, 2, 5, 10, 20, 40 J/m2). Cells were cultured in fresh medium for 4–5 days after treatment, by then cell viability was measured. To test knockdown of NEIL3, FANCA, FANCD2 and TRAIP (Supplementary Table S1), indicated cells were transfected with control or specific siRNA (Final concentration of 20 nM siRNA) in 6-well plate and the cells were seeded into 96-well plates next day prior to treatment with DNA damage agents, using the condition stated above. To test knockdown of RUVBL1 and RUVBL2 (Supplementary Table S1), cells were transfected with control or specific siRNA (Final concentration of 10 nM siRNA) in 96-well plate (5000 cells/well). 24 h after transfection, the cells were treated with PUVA (0, 0.03125, 0.0625, 0.125, 0.25 or 0.5 μM psoralen incubation for 30 min followed by exposure to 0.8 J/cm2 UVA) or MMC (0, 1.25, 2.5, 5, 10, 20 ng/ml) and were subsequently incubated for a further 48 h to measure the cell viability. To test PARP inhibitor, cells were pre-treated with 10 μM olaparib (Selleckchem #AZD8821) for 2 h and then treated with indicated concentrations of psoralen (0, 0.015625, 0.03125, 0.0625, 0.125 or 0.25 μM) in HBSS containing 2% FBS and 10 μM olaparib for 30 min. After exposed with 0.8 J/cm2 UVA, cells were cultured in fresh medium with olaparib (0.5 μM) for 4–5 days before measuring cell viability.
Immunofluorescence
Cells were seeded on cover slide in 12-well plate and then were treated with PUVA (50 nM psoralen for 30 min and then irradiation with 0.8 J/cm2 UVA) or MMC (5 ng/ml). The γH2AX foci was determined at 0, 1, 6 and 20 h, respectively, after releasing into fresh medium. Briefly, cells were washed by 1× PBS and fixed using 4% paraformaldehyde at room temperature for 20 min. After washing with 1× PBS three times and blocking (blocking buffer, 1× PBS/5% goat serum/0.3% Triton X-100) for 1h, cells were incubated with anti-γH2AX (Ser139) (Cell Signaling Technology #2577) at 4°C overnight. γH2AX foci were generated after incubation with goat anti-rabbit IgG (H+L) secondary antibody (Life Technologies #A32731) for 1 h and detected using fluorescence microscope. For siRNA, 48 h after transfection, cells were then treated with PUVA and followed with above protocols to detect the γH2AX foci formation.
Neutral comet assay
Cells were seeded in six-well plate and exposed to PUVA (10 μM psoralen for 1 h followed by 1.0 J/cm2 UVA). After 4 h, the neutral comet assay was performed to detect DSBs, using a commercial kit (Trevigen, # 4250-050-K) according the manufacturer's protocol. In siRNA transfected groups, cells were treated with PUVA (10 μM psoralen for 1 h followed by 1.0 J/cm2 UVA) 72 h after siRNA transfection and were subjected to the neutral comet assay 4 h after PUVA. Quantification of the comet length was performed using Image J v.1.8.0_112 software.
Psoralen/UVA laser striping
To test the recruitment of NEIL3, the U2OS cells were seeded in glass bottomed dishes and transfected indicated siRNAs. Forty eight hours after transfection, NEIL3-EGFP plasmid was transfection into the cells. Next day the cells were incubated with psoralen (10 μM) for 1 h. A Zeiss PALM microdissection microscope equipped with a 360 nm UV laser at the 34% energy dose was then used to observe the recruitment.
To test the recruitment of FANCD2, we generated the U2OS stable cell line using the EGFP-FANCD2 plasmid by selection with puromycin. Cells were seeded in glass bottomed dishes and treated with indicated siRNAs. Seventy two hours after transfection, cells were incubated with psoralen (10 μM) in culture medium for 1 h and then used to perform the laser irradiation.
To test the PARPi, NEIL3-EGFP transfection or EFFP-FANCD2 stable U2OS cells were pre-treated with DMSO or 10 μM olaparib for 3 h, and then were used to perform the laser irradiation described as above.
Cell synchronization and Cell cycle analysis
HeLa cells were double blocked with 2 mM thymidine (arrested for 16 h, released for 8 h, and arrested for another 16 h), and then the cells were harvested for cell cycle and NEIL3 expression analysis at indicated time point. Cells were collected by digestion using trypsin and were washed twice with 1× cold-PBS. Cells were then fixed in 70% ethanol at −20°C overnight and stained with propidium iodide for 20 min. Cell cycle distribution was analyzed using a FACS Canto machine. The data was analyzed by FlowJo software v10.07 (BD Biosciences) or Modfit LT software v5.0 (Verity Software House).
Mass spectrometry
HEK293T cells were seeded in 15 cm dishes and transfected with NEIL3-EGFP or empty vector. Twenty four hours after transfection, cells were treated with 10 μM psoralen plus UVA irradiation (0.8 J/cm2) and released to culture for indicated time. Cells were then washed with pre-chilled PBS three times and lysed into the lysis buffer (20 mM HEPES-K+, pH 7.6, 0.1 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 10% glycerol, 0.01% NP-40, 1 mM DTT, 0.2 mM PMSF, 0.5 mM benzamidine, 1 ug/ml each of leupeptin, aprotinin and pepstatin A) for 30 min at 4°C. Cell lysates were incubated with GFP-Trap resin (ChromoTek) at 4°C overnight and the resin was washed with cold lysis buffer four times. The immunoprecipitated NEIL3-eGFP and its binding partners were eluted with SDS elution buffer (2% SDS, 50 mM Tris–Cl, pH 6.8). The samples were prepared by trichloroacetic acid (TCA) precipitation as previously described (42), and sent to for mass spec analysis.
Immunoprecipitation (IP)
At 48 h after plasmid transfection in 10 cm dish, HEK293T cells were washed twice with ice-cold phosphate-buffered saline. Cells were scraped from dish using 1 ml cold PBS and then were spun at 500 g for 3 min at 4°C. After removal of the supernatant, the cell pellet were lysed in 600 μl IP lysis buffer (250 mM NaCl, 50 mM pH 7.5 Tris–HCl, 1 mM EDTA, 10% glycerol, 0.5% NP40) containing EDTA-free protease inhibitor (Roche) at 4°C for 30 min. Centrifuge cell lysate at 20 000 g for 10 min at 4°C and transfer supernatant to a pre-cooled tube. The FLAG® M2 magnetic beads (Sigma-Aldrich) or GFP-Trap resin (ChromoTek) were washed three times using the IP lysis buffer and 25 μl beads were added to each supernatant tube. After incubating for 4 h at 4°C, the beads were washed three times with the IP lysis buffer and then were boiled with 100 μl 2× SDS-sample buffer for 10 min at 95°C to dissociate immunocomplexes.
Immunoblotting and antibodies
To prepare whole-cell extracts, cells were washed once with ice-cold PBS and lysed with SDS lysis buffer (100 Mm pH 6.8 Tris–HCl, 20% glycerol, 1% SDS). Equal amounts of samples were subjected to 3–8% Tris-acetate gel (for FANCD2-ubiquitination detection) or 4–12% Bis–tris gel (Thermo Fisher Scientific). The protein was transferred to a PVDF membrane. Membranes were blocked in 2% BSA in TBS-T for 1 h and then incubated overnight at 4°C with the following antibodies: anti-FLAG M2 antibody (Sigma-Aldrich #F1804), anti-HA-tag (Cell Signaling Technology #3724), anti-GFP (Cell Signaling Technology #2555), anti-beta-actin (Cell Signaling Technology #4967), anti-GAPDH (Cell Signaling Technology #2118), anti-NEIL3 (Proteintech, 11621-1-AP), anti-FANCA (Bethyl Laboratories #A301-980), anti-FANCD2 (Novus Biologicals #NB100-182), anti-FANCI (Santa Cruz Biotechnology #sc-271316), anti-γH2A.X (Ser139) (Cell Signaling Technology #2577), anti-PARP1 (Cell Signaling Technology #9542), anti-PARP3 (Novus Biologicals #NBP1-31415), anti-MCM7 (Cell Signaling Technology #3735), anti-RUVBL1 (Abcam #ab51500) and anti-RUVBL2 (BD Biosciences #612482). Proteins were detected using a chemiluminescence system with a horseradish peroxidase conjugated secondary antibody.
Quantitative RT-PCR
Total RNA was extracted applying a RNeasy Mini kit (Qiagen), and cDNA was obtained using the SuperScript™ III First-Strand Synthesis kit (Thermo Fisher Scientific). The following primers were used for qPCR. NEIL3: forward, 5′-GTCCTAATTGTGGTCAGTGCC-3′; reverse, 5′-TCCAGTGCTCTTCCGACTTC-3′; TRAIP: forward, 5′-CTAAAAGAGGCACGGAAGGC-3′; reverse, 5′- TTTCCTGCAGCATCGTTAGC-3′; GAPDH: forward, 5′-CCATCTTCCAGGAGCGAGAT-3′; reverse, 5′-TGCTGATGATCTTGAGGCTG-3′: NEIL1: forward, 5′-CTCGCCCTATGTTTCGTGGA -3′; reverse, 5′-TTCAGCCGGTACAGGATCTC-3′.
RESULTS
NEIL3 plays a major role in psoralen-ICL repair independent of the FA/BRCA pathway.
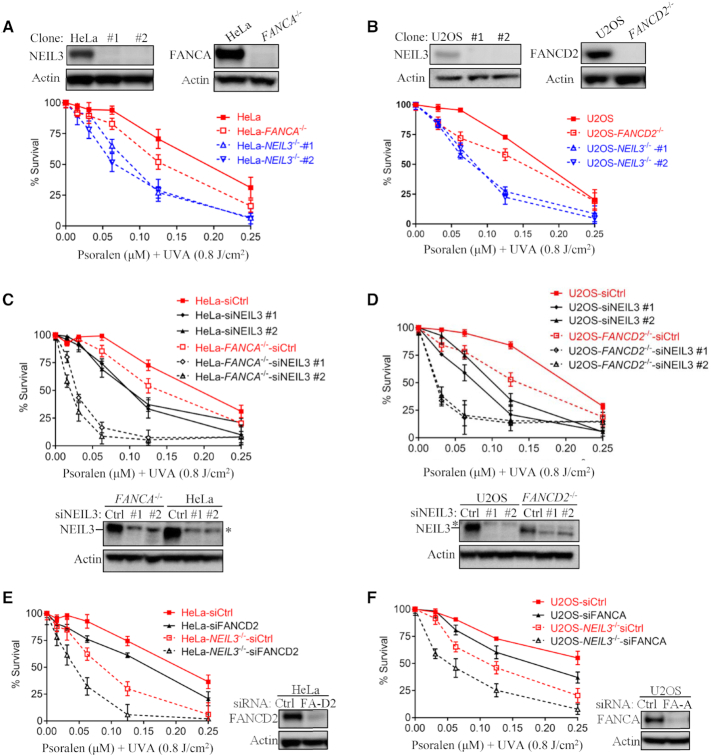
The biochemical role of NEIL3 in psoralen-ICL repair has been demonstrated biochemically in vitro (26) and in the Xenopus egg extract system (25). Whether NEIL3 plays a role in psoralen-ICL repair or in other types of DNA damage (i.e. double-strand break) repair in human cells remains unclear. We thus generated NEIL3-/- human cells and asked whether these cells are sensitive to any DNA damaging agents. NEIL3-/- HeLa and U2OS cells were hypersensitive to PUVA (psoralen plus UVA irradiation) (Figure 1A, B and Supplementary Figure S1A), while treatment with psoralen or UVA alone had no impact on cell growth (Supplementary Figure S1B). Neil3-/- MEFs were also sensitive to PUVA (but not UVA or psoralen) when compared to WT control (Supplementary Figure S1C). NEIL3-/- HeLa cells were also sensitive to ICL-inducing agents MMC and cisplatin, but to a lesser extent than PUVA. (Supplementary Figure S1A). NEIL3-/- HeLa also showed mild sensitivity to HU and no sensitivity to camptothecin, IR, UVB or UVC (Supplementary Figure S1A). Taken together, the sensitivity profile results suggest that NEIL3 plays a major role in psoralen-ICL repair.
Figure 1.
NEIL3 pathway plays a dominant role in psoralen-ICL repair in human cells. (A, B) Single cell HeLa-NEIL3-/-, U2OS-NEIL3-/-, HeLa-FANCA-/- and U2OS-FANCD2-/- clones were validated by Western blot. Cell viability was measured by CellTiter-Glo assays (Promega). PUVA, psoralen plus UVA. knockout cell lines were generated by CRISPR-Cas9. Guide RNA sequences and method used are shown in Materials and Methods. (C, D) Cell viability after PUVA treatment was measured after NEIL3 knockdown in HeLa WT versus HeLa-FANCA-/- and U2OS WT versus U2OS-FANCD2-/- cells. Western blots show that NEIL3 was knocked down. *Asterisk indicates a non-specific band above the NEIL3 band detected by the NEIL3 antibody. (E, F) PUVA sensitivity was determined after knockdown of FANCA or FANCD2 in U2OS-NEIL3-/- and HeLa-NEIL3-/- cells, respectively. Western blots show successful knockdown of FANCD2 and FANCA. Data are mean ± standard error of the mean (s.e.m.) from three independent experiments.
The FA/BRCA pathway has been extensively studied in MMC- and cisplatin-ICL repair, and it was recently implicated in psoralen-ICL repair (25,33). To study the relative contribution of NEIL3 and FA/BRCA pathways to the repair of those three types of ICLs, the sensitivities of NEIL3-deficient and FA-deficient were compared. Although both FANCA-/- and FANCD2-/- cells were sensitive to PUVA, NEIL3-/- cells showed much more sensitivity to PUVA, suggesting that NEIL3 is the major player in psoralen-ICL repair in HeLa and U2OS cells (Figure 1A and B). Conversely, NEIL3 pathway plays a less important role than the FA/BRCA pathway in MMC- and cisplatin-ICL repair, as demonstrated by the lesser sensitivity of HeLa-NEIL3-/- to those two agents (Supplementary Figure S1D).
To determine the relationship between the NEIL3 and FA pathways in psoralen-ICL repair, we knocked down NEIL3 in FA/BRCA-deficient cells and examined their PUVA sensitivity. siRNA knockdown of NEIL3 in FA/BRCA-deficient HeLa and U2OS cells led to additional sensitivity of those cells to PUVA (Figure 1C and D). Similar results were observed in FANCD2-deficient cells (PD20), which are SV40-transformed fibroblast cells derived from a FANCD2-deficient FA patient. We observed additional PUVA sensitivity when NEIL3 was knocked down in PD20 cells (Supplementary Figure S1E). Consistently, depletion of FANCA or FANCD2 by siRNA in NEIL3-/- cells also led to an additional sensitivity to PUVA (Figure 1E and F). These PUVA sensitivity data indicates that the NEIL3 pathway and the FA/BRCA pathway are functionally non-epistatic in psoralen-ICL repair. In contrast, HeLa-NEIL3-/- cells showed no additional sensitivity to MMC or cisplatin after FANCD2 knockdown (Supplementary Figure S1F), indicating that NEIL3 participates in MMC- and cisplatin-ICLs repair in a FA/BRCA pathway dependent manner. To investigate whether other NEIL DNA glycosylases is involved in psoralen-ICL repair, we knocked down NEIL1 in HeLa cells and tested their sensitivity to PUVA. NEIL1 knockdown resulted in resistance to PUVA in HeLa cells, where NEIL3 was concurrently upregulated (Supplementary Figure S1G). These results were consistent with a previous study (43) and supported the idea that NEIL3 is the major enzyme to repair psoralen-ICL. Taken together, NEIL3 plays a major role in psoralen-ICL repair in human cells, which is independent of the FA/BRCA pathway. NEIL3 is also involved in MMC- and cisplatin-ICL repair in human cells, which is governed by the FA/BRCA pathway.
NEIL3 knockdown activates the FA/BRCA pathway in psoralen-ICL repair
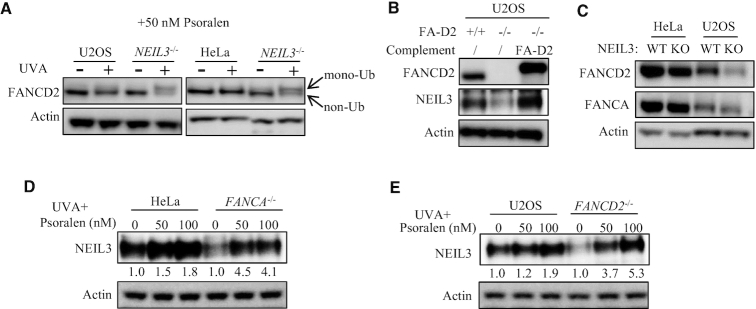
In Xenopus egg extracts, psoralen-ICLs are repaired via the FA/BRCA pathway when the NEIL3 pathway is blocked (25). We asked whether this mechanism is also found in human cells. We first monitored FA/BRCA pathway activation after PUVA by detecting FANCD2 mono-ubiquitination. FANCD2 mono-ubiquitination was not detected in U2OS or HeLa parental cells treated with 50 nM psoralen plus 0.8 J/cm2 UVA, when NEIL3 was present. In contrast, mono-ubiquitination of FANCD2 was clearly observed in NEIL3-/- HeLa and in NEIL3-/- U2OS cells under the same dose of PUVA treatment (Figure 2A). This is consistent with our observation that NEIL3 plays a dominant role in psoralen-ICL repair in human cells (Figure 1), and with the observation in Xenopus that the FA pathway is activated in the absence of NEIL3. These results suggest that the NEIL3 pathway is the major and prioritized pathway for repairing psoralen-ICLs in human cells. Only when the psoralen-ICLs exceed the repair capability of NEIL3 does the FA/BRCA pathway become activated as a backup repair mechanism.
Figure 2.
The relationship between the NEIL3 and the FA/BRCA pathways in psoralen-ICL repair. (A) WT and NEIL3-/- cells were treated with PUVA (pre-incubated with 50 nM psoralen for 30 min and then irradiation with 0.8 J/cm2 UVA). 24 h after PUVA, the level of FANCD2 mono-ubiquitination was detected by WB to determine activation of the FA/BRCA pathway. (B) U2OS-FANCD2-/- cells were complemented with EGFP-FANCD2 and NEIL3 expression level was determined by western blot in those cells. (C) The FANCA and FANCD2 expression levels were determined in wild-type and NEIL3 deficiency cells. (D, E) The NEIL3 expression level was determined in wild-type and FANC gene deficiency cells, 24 h after being treated with indicated dose of psoralen plus UVA (0.8 J/cm2) irradiation. Quantification of the Western blots was performed by ImageJ software and data were normalized to no psoralen.
Expression of NEIL3 and FA genes were interdependent at protein level
Since both NEIL3 and FA pathways contribute to the repair of PUVA-ICLs, we expected a compensatory relationship between the NEIL3 and FA pathway proteins. To our surprise, NEIL3 expression was down-regulated at the protein level in U2OS-FANCD2-/- and HeLa-FANCA-/- cells, compared to their WT counterparts (Figure 2B and Supplementary Figure S2A). Complementation with EGFP-FANCD2 in the U2OS-FANCD2-/- cells rescued the expression of NEIL3 (Figure 2B), and similar rescue results could be achieved in FANCD2 deficient (PD20) or FANCG deficient (PD326) FA patient fibroblasts (Supplementary Figure S2B). We excluded the possibility of robust regulation at NEIL3 mRNA level in FA-deficient cells, since NEIL3 mRNA level was not changed in those cells (Supplementary Figure S2C). We also excluded that the decreased level of NEIL3 was a result of cell cycle alteration. Consistent with our previous report (20), NEIL3 expression increases during S phase and reached maximal levels in G2/M (Supplementary Figure S2D). However, there was no significant change in the cell cycle distribution of NEIL3 in U2OS-FANCD2-/- and HeLa-FANCA-/- cells (Supplementary Figure S2E and F). Next, we studied how NEIL3 deficiency affected FA genes. FANCA and FANCD2 expression levels were decreased in NEIL3-/- HeLa and U2OS cells (Figure 2C). Further investigation showed the proteasome inhibitor MG-132 was able to stabilize NEIL3 in FANCD2 cells and FANCD2 in NEIL3-/- cells, suggesting NEIL3 and FANC protein stabilize each other by reducing proteasome degradation (Supplementary Figure S2G). The mutually dependent expression levels led us to speculate that NEIL3 might form a complex with FANC proteins. However, we were not able to detect an interaction between NEIL3 and the FA proteins after many attempts of co-immunoprecipitation experiments, and the FA proteins were not detected in the NEIL3 immunoprecipitant in our IP-MS experiment (Supplementary Figure S2H).
Response of NEIL3 to psoralen-ICLs was enhanced in FA/BRCA-deficient cells
We next studied how NEIL3 responses to PUVA treatment. Upon PUVA treatment, rather than UVA irradiation or psoralen treatment, the NEIL3 expression was up-regulated in both HeLa and U2OS cells, suggesting that NEIL3 plays an active role in the DNA damage response (Figure 2D and E, Supplementary Figure S2A). Interestingly, NEIL3 protein was strongly induced after PUVA exposure in these FA-deficient cells (more than 4-fold), despite a low starting level of NEIL3 protein in these cells (Figure 2D and E). This increase is not likely an effect of cell cycle, since the S and G/M populations combined showed no difference in WT and FANCA-/- HeLa cells after PUVA treatment (Supplementary Figure S2I and J). These results demonstrated an active response of NEIL3 to PUVA, and also explained why FA/BRCA-deficient cells were only mildly sensitive to PUVA.
The NEIL3 pathway repairs psoralen-ICLs without generating double-strand breaks
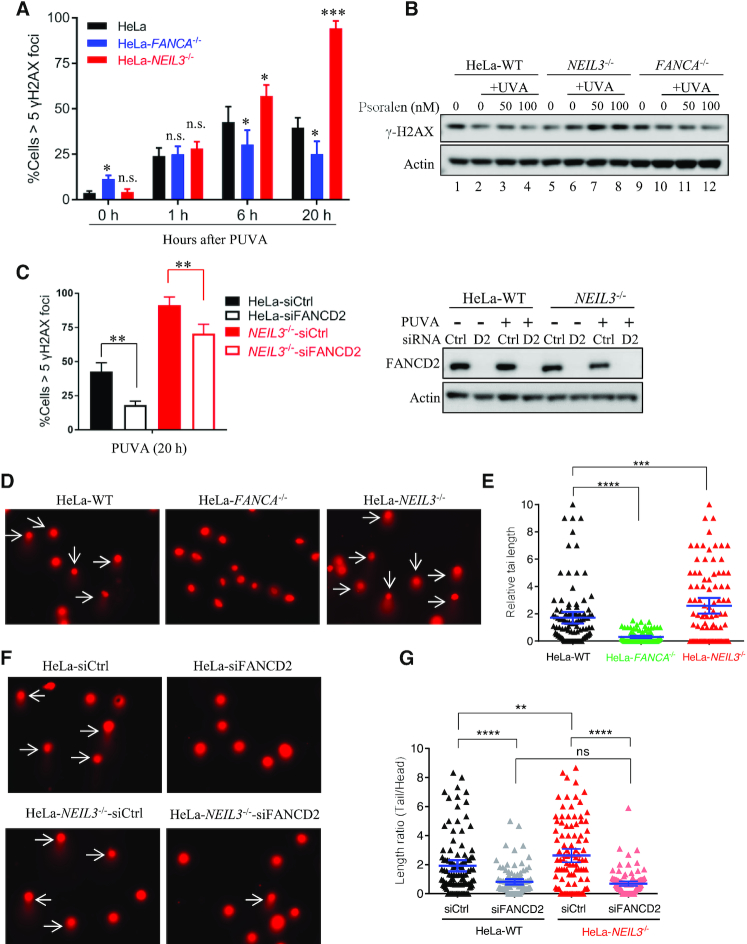
Previous studies in Xenopus egg extracts revealed that NEIL3 unhooks DNA psoralen-ICLs without generating double-strand breaks (DSBs) (25). We next determined whether a similar mechanism was present in human cells. We investigated DSB generation after PUVA treatment in HeLa cells that were defective in either the NEIL3 pathway or the FA/BRCA pathway, using γH2AX foci as markers. Following PUVA treatment, WT HeLa cells exhibited a slight increase of γH2AX foci compared to the untreated cells (0 h). FANCA-/- cells, which only retained the NEIL3 pathway, had a lower level of γH2AX foci compared to WT HeLa cells. Conversely, NEIL3-/- cells, which only have the FA/BRCA pathway for repair of the psoralen-ICLs, contained an increased number of γH2AX foci compared to WT HeLa cells (at 6 and 20 h) (Figure 3A and Supplementary Figure S3A). Consistent with the IF results, the γH2AX level was also increased in the NEIL3-/- cells after PUVA treatment as determined by western blot (Figure 3B, lanes 5–8). Moreover, depletion of FANCD2 decreased γH2AX foci formation in both WT and NEIL3 HeLa cells (Figure 3C and Supplementary Figure S3B), indicating the FA pathway is responsible for the DSBs generated after PUVA. Since γH2AX is not a perfect marker for DSBs, we evaluated cellular DSB levels after PUVA treatment using the neutral comet assay, which detects DSBs directly at DNA level. Similar to the γH2AX foci and Western blot data, we observed that there were more DSBs in NEIL3-/- HeLa cells compared to WT counterparts after PUVA, as indicated in the increased tail length of the comets. Consistently, much shorter comet tails were observed in FANCA-/- cells (Figure 3D-E). In addition, depletion of FANCD2 decreased the tail length of comets in both HeLa WT and NEIL3 cells (Figure 3F, G), indicating that DSB generation after PUVA is dependent on the FA pathway but not the NEIL3 pathway. To note, the elevated γH2AX foci in NEIL3-/- cells were completely rescued by siFANCD2. In addition, we did not observe a difference in γH2AX foci formation in FANCA-/- and NEIL3-/- cells following MMC treatment (Supplementary Figure S3C-D), suggesting again that NEIL3 shares the same downstream factors with FA/BRCA pathway in MMC-ICLs repair. Taken together, our data demonstrated that, unlike the FA/BRCA pathway, the unique psoralen-ICLs unhooking mechanism of NEIL3 is not associated with DSB formation in human cells.
Figure 3.
NEIL3 -/- cells show increased DSBs compared to WT cells after PUVA treatment. (A) Quantification of the γH2AX foci at 0, 1, 6 and 20 h after PUVA. Cells with more than five γH2AX foci were counted and quantified, from IF image shown in Figure S3. Data are mean ± s.e.m. from three independent experiments. (B) γH2AX level was determined in WT, FANCA-/- and NEIL3-/- HeLa cells after treatment. (C) Quantification and statistical analysis of the γH2AX foci 20 h after PUVA in WT and NEIL3−/- HeLa cells after FANCD2 knockdown. Data are mean ± s.e.m. from three independent experiments. Western blot on the right shows the knockdown of FANCD2. PUVA, 50 nM psoralen + 0.8 J/cm2 UVA. **P < 0.01 (paired t test). (D) Representative images of comets, showing tail formation (arrows) 4 h after PUVA in WT, FANCA-/- and NEIL3-/- HeLa cells. (E) Quantification of the comet length at 4 h after PUVA shown in (D). At least 100 cells were analyzed for each group. (F) Representative images of comets showing tail formation in WT and NEIL3-/- HeLa cells after FANCD2 knockdown and PUVA. 72 h after siRNA transfection, cells were treated with PUVA and released for 4 h before comet assays. PUVA, 10 μM psoralen plus 1.0 J/cm2 UVA; White arrows, cells with comet tails. (G) Quantification and statistical analysis of comet lengths from experiments shown in (F). ns, not significant, **P < 0.01, ****P < 0.0001 (paired t test).
PARP is required for the recruitment of NEIL3 and FANCD2-Ub to DNA damage
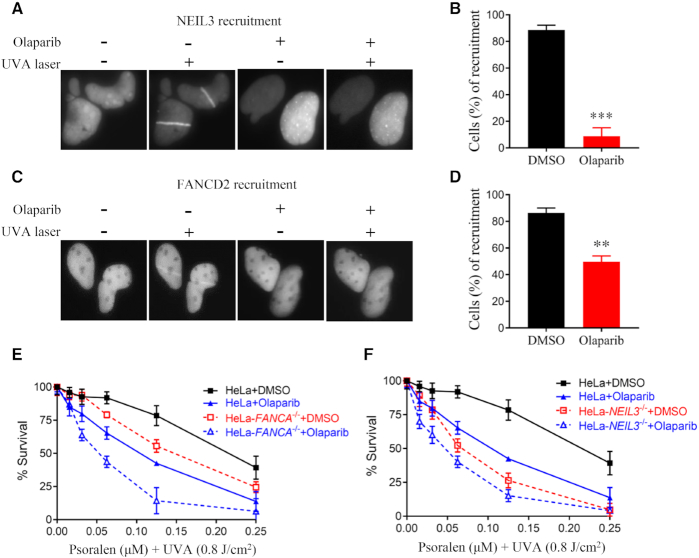
We next examined how NEIL3 is recruited to psoralen-ICL sites. Cells were pre-treated with psoralen and irradiated with a UVA laser (357 nm), and the NEIL3 recruitment was monitored by a GFP tag (Supplementary Figure S4A). In the absence of psoralen, UVA laser alone did not trigger the recruitment of NEIL3-EGFP (Figure S4B). The recruitment of NEIL3-EGFP to the PUVA laser stripe was very efficient; 90% of the cells showed positive NEIL3-EGFP recruitment to the laser track. The recruitment was also rapid, which occurs within one minute (Figure 4A, B). To determine whether NEIL3 recruitment to psoralen-ICL is dependent on any known DNA damage response pathways during psoralen-ICLs repair, we tested inhibitors of ATM, ATR, DNA-PK and PARP. We found only the PARP inhibitor (olaparib) strongly blocked NEIL3 recruitment to psoralen-ICLs (Figure 4A, B), while ATM, ATR or DNA-PKcs inhibitors did not affect NEIL3 recruitment (Supplementary Figure S4C). Note that olaparib almost completely blocked the recruitment of NEIL3, indicating this event is highly PARP dependent. In contrast, the recruitment of EGFP-FANCD2 was much weaker and less efficient, and the recruitment could only be clearly observed five minutes after PUVA laser irradiation. PARPi olaparib only partially suppressed FANCD2 recruitment (Figure 4C, D and Supplementary Figure S4D). The PARP-dependent recruitment of NEIL3 and FANCD2 (to a lesser extent) was confirmed in PUVA laser irradiation experiments with PARP knockdown (Supplementary Figure S4E–G). Interestingly, the recruitment of NEIL3 and FANCD2 was mediated by different PARP family members. Knockdown of PARP1 blocked NEIL3 recruitment, while depletion of PARP3 blocked FANCD2 recruitment (Supplementary Figure S4E–G). In addition, depletion of NEIL3 had no impact on FANCD2 recruitment, and depletion of FANCD2 did not affect NEIL3 recruitment (Supplementary Figure S4H and I). These results are consistent with the idea that the NEIL3 pathway is promptly activated upon PUVA exposure, while the FA pathway is delayed.
Figure 4.
PARP is essential for NEIL3 recruitment to psoralen-ICLs and facilitates FANCD2 recruitment. (A) Recruitment of NEIL3 to PUVA laser tracts. U2OS cells expressing NEIL3-GFP were pre-incubated with 10 μM psoralen and the recruitment of NEIL3 was visualized 1 min after exposure to UVA laser. PARP inhibitor (olaparib) was added 1 hour before laser striping. (B) Quantification of the NEIL3 recruitment to psoralene-ICL laser tracts after treatment with DMSO or olaparib. A total of 100 cells were quantified for each group. (C) U2OS cells stably expressing EGFP-FANCD2 were pre-treated with 10 μM psoralen for 1 h before laser striping. Recruitment efficiency was determined 10 min after exposure to UVA laser. Olaparib or DMSO was added 1 h before laser striping. (D) Quantification of the recruitment efficiency of EGFP-FANCD2 after treatment with DMSO or olaparib, as shown in (C). A total of 100 cells were counted for each group. (E, F) PUVA sensitivity was determined in WT, FANCA-/- and NEIL3-/- HeLa cells after DMSO or olaparib treatment. CellTiter-Glo cell viability assays were performed. Mean and s.e.m. of three independent experiments are shown. **P < 0.01; ***P < 0.001 (paired t test).
So far, we showed that NEIL3 was critical for psoralen-ICL repair and PARP was indispensable for NEIL3 recruitment to psoralen ICL. We asked whether treating cells with a PARP inhibitor would have same effect as depleting NEIL3 in PUVA sensitivity assays. Consistently, olaparib treatment sensitized HeLa cells to PUVA and caused an additional sensitivity of FANCA-/- cells to PUVA (Figure 4E), in agreement with the PUVA laser striping data (Figure 4A, B) and PUVA sensitivity data with NEIL3 depletion (Figure 1C, D). On the other hand, olaparib caused a mild increase in PUVA sensitivity in NEIL3-/- cells (Figure 4F), which may be due to a less efficient inhibition of FANCD2 by olaparib. These results show that NEIL3 repairs PUVA-ICLs in a strictly PARP-dependent manner.
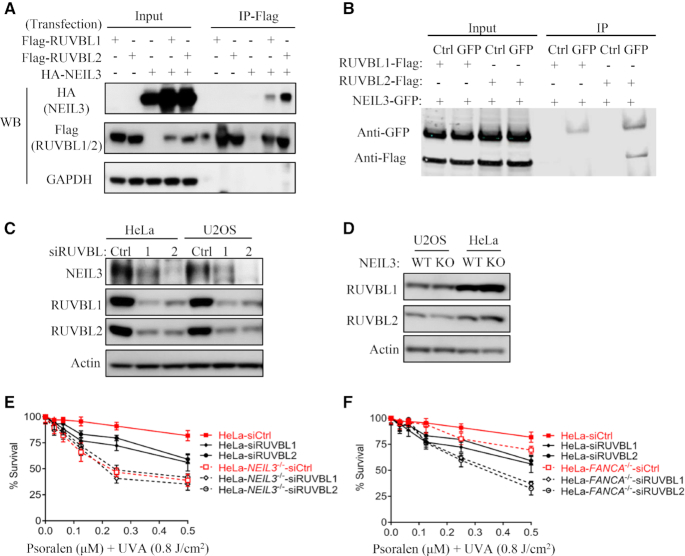
NEIL3 and RUVBL1/2 interact and function in the same pathway
To better understand the function of NEIL3 in psoralen-ICL repair, we performed mass spectrometry to identify novel NEIL3 binding partners upon PUVA treatment. NEIL3-GFP was expressed in HEK293T cells and immunoprecipitated by the GFP tag. The quality of immunoprecipitation was accessed by silver staining, in which both the bait NEIL3-GFP and the binding partners were evident (Supplementary Figure S5A). A list of the top interactors is shown in Supplementary Figure S5B. Consistent with the PARP-dependent recruitment of NEIL3, PARP1 was detected by the mass spectrometry. To our interest, RUVBL1 and RUVBL2, known to form a heterodimeric complex (44), were among top hits. The RUVBL1/2 complex has been reported to associate with FA/BRCA pathway proteins and its knockdown sensitizes cells to mitomycin C (MMC) (36), a phenotype of FA/BRCA pathway deficient cells. We therefore investigated whether NEIL3 functions in concert with the RUVBL1/RUVBL2 complex. To confirm the interaction between NEIL3 and RUVBL2 and/or RUVBL1, we co-transfected the NEIL3-HA and the RUVBL1-Flag or RUVBL2-Flag plasmids Supplementary Figure S5C), and co-IP experiments were carried out. RUVBL2, and RUVBL1 to a lesser extent, pulled down NEIL3 (Figure 5A), while NEIL3 pulled down RUVBL2 but not RUVBL1 (Figure 5B). These data suggest NEIL3 primarily binds to RUVBL2 of the RUVBL1/2 complex. Depletion of RUVBL1 or RUVBL2 had no impact on the recruitment of NEIL3 or FANCD2 to PUVA laser stripes (Supplementary Figure S5D and E). However, a decreased NEIL3 protein level was observed when RUVBL1 or RUVBL2 was knocked down (Figure 5C). A similar reduction of FANCD2 and FANCI levels was observedSupplementary Figure S5F), which was consistent with previous work (36). Since NEIL3 and RUVBL1/2 form a complex in cells, the down-regulation of NEIL3 might be due to its instability resulting from knockdown of RUVBL1/RUVBL2. On the opposite, RUVBL1 and RUVBL2 levels were not changed significantly in NEIL3-/- and FANCA-/- cells (Figure 5D and Supplementary Figure S5G). In addition, the cell cycle profiles of RUVBL1-depleted HeLa cells were not altered (Supplementary Figure S5H and (36)). These results show that the RUVBL1/2 complex either stabilize NEIL3 in a complex or regulate NEIL3 through transcription.
Figure 5.
The RUVBL1/RUVBL2 complex is involved in psoralen-ICLs repair via the NEIL3 pathway. (A) 293T cells were transiently transfected with indicated plasmids, and co-IP were performed using anti-Flag antibody. WB of the inputs and IPs are shown to detect interactions. (B) 293T cells were transiently transfected with indicated plasmids, and co-IP were performed using normal IgG or anti-GFP antibody. WB of the inputs and IPs are shown to detect interactions. (C) NEIL3 protein levels were analyzed after knockdown of RUVBL1 or RUVBL2 in HeLa or U2OS cells. (D) RUVBL1 and RUVBL2 protein levels were analyzed by WB in WT and NEIL3-/- cells. (E, F) Cell viability was measured and compared after knockdown of RUVBL1 or RUVBL2 and PUVA treatment in WT, NEIL3-/- (E) and FANCA-/- (F) HeLa cells. CellTiter-Glo cell viability assays were performed.
We next studied the contribution of the RUVBL1/2 complex to PUVA-ICL repair and its relationship with NEIL3 in this process. Depletion of either RUVBL1 or RUVBL2 led to mild PUVA sensitivity in WT HeLa cells, while NEIL3-/- cells were very sensitive to PUVA. We did not observe further PUVA sensitivity after RUVBL1/2 knockdown in NEIL3-/- cells, indicating NEIL3 and RUVBL1/2 have an epistatic function in PUVA-ICL repair (Figure 5E). On the other hand, FANCA and RUVBL1/2 showed a non-epistatic relationship in PUVA repair (Figure 5F). Given that the role of RUVBL1/RUVBL2 complex in MMC-ICL repair was dependent on the FA/BRCA pathway (36), which was distinct from psoralen-ICL repair, we next examined MMC sensitivity in FANCA-/- and NEIL3-/- cells after depletion of RUVBL1/RUVBL2. We did not observe any additional MMC sensitivity after knocking down of RUVBL1 or RUVBL2 in either HeLa-NEIL3-/- or HeLa-FANCA-/- (Supplementary Figure S5I and J). These data suggest that the RUVBL1/RUVBL2 complex are epistatic with both NEIL3 and FA pathways in MMC-ICL repair. Taken together, these data suggest that RUVBL1/2 and NEIL3 function in the same pathway for psoralen-ICL repair, and this pathway is distinct from the FA/BRCA pathway, which is majorly responsible for MMC-ICL repair.
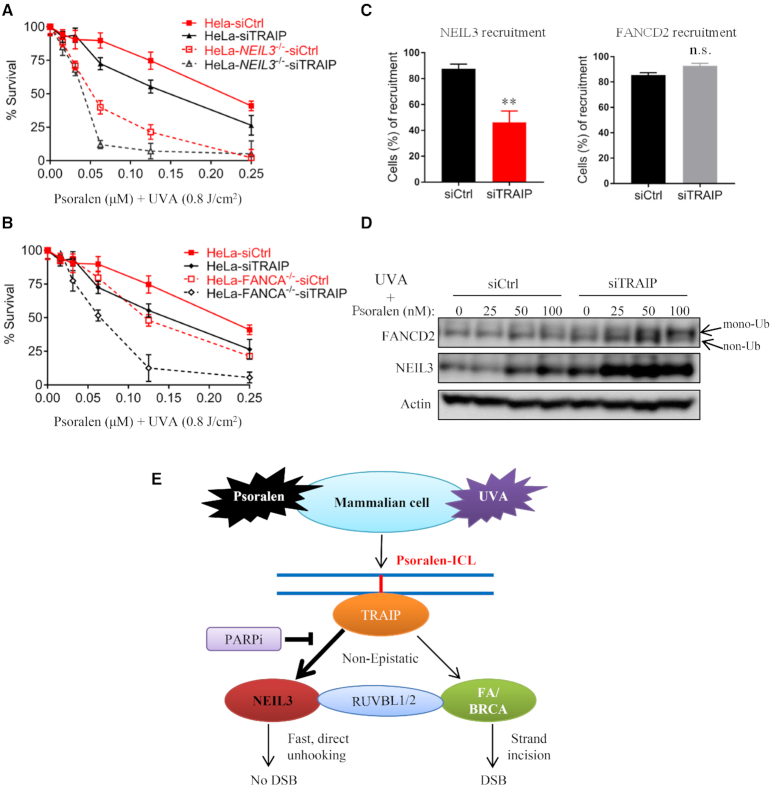
TRAIP is required for both NEIL3 and FA/BRCA pathways in psoralen-ICLs repair
Recently, TRAIP was identified as a master regulator of psoralen-ICL repair in both the NEIL3 and FA/BRCA pathways in Xenopus egg extracts (33). To investigate the role of TRAIP in psoralen-ICLs repair in human cells, we investigated PUVA sensitivity after TRAIP depletion. Lacking a good TRAIP antibody, the efficiency of siRNA-mediated depletion of TRAIP was evaluated by qPCR and Western blotting of exogenously expressed TRAIP-myc (Supplementary Figure S6A and B). Knockdown of TRAIP caused PUVA sensitivity, and more importantly, TRAIP knockdown presented additional PUVA sensitivity in both FANCA-/- and NEIL3-/- HeLa cells (Figure 6A and B), indicating that TRAIP is non-epistatic with both the FA/BRCA and the NEIL3 pathways in psoralen-ICL repair. In addition, depletion of TRAIP partially impeded NEIL3 recruitment to psoralen-ICLs, but it did not affect FANCD2 recruitment (Figure 6C and Supplementary Figure S6D and E). These results are consistent with a model in which ubiquitylation of the CMG complex by TRAIP is required for NEIL3 recruitment to chromatin in Xenopus egg extracts (33). Although MCM7 ubiquitylation has been identified as a key event that is required for NEIL3 recruitment, we only observed a weak impact of MCM7 knockdown in NEIL3 recruitment (Supplementary Figure S6C). Intriguingly, depletion of TRAIP increased the expression of NEIL3 and FANCD2 proteins, and it also increased the mono-ubiquitylation of FANCD2 (Figure 6D). We speculate that this elevated activation of NEIL3 and FA pathways might be a compensatory response to their nonfunctional recruitment in the absence of TRAIP. In conclusion, these data show TRAIP is non-epistatic with both the FA/BRCA and the NEIL3 pathways in psoralen-ICL repair, and suggest that TRAIP may function upstream of both pathways.
Figure 6.
TRAIP show synthetic lethality with both the NEIL3 pathway and the FA pathway upon PUVA treatment. (A, B) Cell viability was measured after knockdown of TRAIP and PUVA treatment in WT, FANCA-/- and NEIL3-/- HeLa cells. (C) PUVA laser striping was used to analyze the recruitment efficiency of NEIL3-EGFP and EGFP-FANCD2 in U2OS cells after siRNA knockdown of TRAIP. A total of 100 cells were counted in each group. **P < 0.01, n.s., no significance (paired t test). (D) FANCD2 and NEIL3 were analyzed by Western blotting in cell lysates from HeLa cells, which were transfected with control or TRAIP siRNA and treated with PUVA. (E) A model showing the NEIL3 pathway in psoralen-ICL repair in the context of the FA/BRCA pathway.
DISCUSSION
In this study, we investigated the role of NEIL3 in the repair of psoralen-induced ICLs. We show that NEIL3 plays a major role in psoralen-ICL repair while the FA pathway is primarily responsible for the repair of MMC- and cisplatin-induced ICLs. The two pathways are non-epistatic in psoralen-ICL repair. Unlike the FA pathway, NEIL3 repair mechanism is PARP dependent and does not introduce DSBs. On the one hand, RUVBL1/2 forms a complex with NEIL3 and is functionally epistatic with NEIL3. On the other hand, TRAIP is functionally non-epistatic with both NEIL3 and FA/BRCA pathways, indicating that TRAIP may function upstream of the two pathways. A model is provided to summarize our findings (Figure 6E)
The hypersensitivity of NEIL3 depleted cells to psoralen-ICLs in multiple human cell lines demonstrates that NEIL3 is the major pathway to repair psoralen-ICLs, and that the NEIL3 pathway is prioritized over the FA pathway in human cells (Figure 1). Based on in vitro studies, NEIL3 is also important for repair of AP-ICLs in human cells (25,26). We also tested the sensitivity of NEIL3-/- cells to other types of ICLs – namely, cisplatin- and MMC-induced ICLs. Although NEIL3-/- cells were sensitive to these ICLs, they were more sensitive to psoralen-ICL, and the repair of cisplatin-ICLs and MMC-ICLs was primarily governed by FA pathway (Supplementary Figure S1). These results are consistent with previous studies indicating that NEIL3 does not process cisplatin-ICL (25,33). It will be interesting to determine how cells choose a pathway upon different ICL exposures. The rapid recruitment kinetics of NEIL3 and relatively slow kinetics of FANCD2 suggest that NEIL3 is the first line of defense. Cells will proceed with the mechanistically simpler NEIL3 pathway when the ICL substrate can be directly unhooked by NEIL3 (i.e. psoralen-ICL) and activate the more complex FA pathway when the ICL cannot be unhooked by NEIL3.
Wu et al. showed that both the FA and NEIL3 pathways must to be eliminated in order to generate cellular hypersensitivity to psoralen-ICLs in HAP1 cells (33). Synthetic lethality between FA and NEIL3 pathway was also evident in our study. However, we observed that NEIL3 depletion alone, either by siRNA or CRISPR-Cas9 knockout, led to hypersensitivity of HeLa and U2OS cells to psoralen-ICLs (PUVA), emphasizing that the NEIL3 pathway is the more efficient pathway in psoralen-ICL repair. This discrepancy may be a result of different cell lines being used. One question was whether the NEIL3 pathway is prioritized in human cells, similar to the observation in Xenopus extracts. Our data in human cell lines support this hypothesis. First, NEIL3 is recruited to PUVA-induced DNA damages faster than the FA protein FANCD2 (Figure 4). In our hands, strong NEIL3 recruitment could be observed within one minute, while FANCD2 recruitment was only evident after 5 minutes of PUVA treatment. Secondly, FANCD2 is not mono-ubiquitinated when NEIL3 is present in WT HeLa and U2OS cells, while, in response to PUVA, NEIL3 protein levels are elevated regardless of FA pathway availability (Figure 2). Thirdly, PUVA does not trigger significant phosphorylation of H2AX in WT cells, a sign that NEIL3-mediated repair occurs in the normal setting, while γH2AX foci levels are higher after PUVA treatment of NEIL3-/- cells, indicating that the FA pathway is activated as a backup pathway (Figure 3). In summary, our data show that NEIL3 pathway is prioritized in PUVA repair in human cells.
The Fanconi Anemia pathway plays an essential role in ICL repair during DNA replication (3). One of the key steps of this pathway is nucleolytic incision of DNA strands flanking the ICL (45), which generates a DSB that need further processing. Both purified NEIL3 and NEIL1 remove psoralen-induced DNA–DNA cross-links in vitro, the former does it without nicking DNA backbone (26), which is consistent with results in Xenopus extracts (25). Our data in human cells are consistent with those in vitro observations. When NEIL3 was deleted in HeLa cells, a strong accumulation of γH2AX was observed after PUVA treatment, and this γH2AX accumulation was attenuated by FANCD2 depletion (Figure 4). Our data demonstrate that the direct unhooking mechanism utilized by NEIL3, which occurs without the generation of DSBs, is conserved in human cells.
Replication fork stalling at a DNA–protein crosslink leads to TRAIP-dependent ubiquitylation and proteasome degradation (46). Wu et al. show that CMG ubiquitylation by TRAIP is required to recruit NEIL3 for ICL unhooking. TRAIP associates with the replisome and non-specifically ubiquitylates lysine residues on bulky protein obstacles. The generation of a short ubiquitin chain can recruit NEIL3 (33). Here, we show that NEIL3 is rapidly recruited to PUVA laser tracts in a PARP-dependent manner. The PARP inhibitor olaparib, or knockdown of PARP1, strongly blocks NEIL3 recruitment (Figure 4). TRAIP, on the other hand, only weakly blocks NEIL3 recruitment (Figure 6). Since we looked at an unsynchronized cell population while the Xenopus extracts represent S phase, it is possible that the recruitment of NEIL3 relies more on TRAIP during S phase as opposed to other phases of the cell cycle. TRAIP itself is recruited to chromatin following ICL exposure during replication (47). BRCA-related FA proteins (BRCA2, FANCJ/BACH1 and FANCN/PALB2) requires replication for crosslink association, unlike other FA core and I/D2 complexes (48). In agreement with this study, we did not see reduction of FANCD2 recruitment after TRAIP knockdown. Our results suggest that psoralen-ICL repair by NEIL3 is not solely governed by TRAIP, and that it may also function outside of S phase.
Another interesting observation is that the protein levels of NEIL3 and FANC proteins are inter-dependent. We observed FANCD2 and FANCA proteins were decreased in NEIL3-/- cells and NEIL3 protein was decreased in the FA cells. Transcription regulation did not appear to be involved, since NEIL3 mRNA levels in FANCD2-/- or FANCA-/- cells were similar to the WT counterparts. Cell cycle alteration cannot explain the inter-dependency, since cell cycle distribution did not change in those FA cells. A previous study demonstrated that the NEIL1 level is dependent on the presence of an intact FA pathway and NEIL1 undergoes proteolytic degradation in FA cells (49). After treatment with the proteasome inhibitor MG-132, NEIL3 in U2OS-FANCD2-/- cells was increased to a similar level as in WT cells. Also, FANCD2 was increased after MG-132 treatment in NEIL3-/- HeLa cells, although the increase was not as prominent. There data suggest NEIL3 and FANC protein stabilize each other by reducing proteasome-mediated protein degradation.
The interdependency between NEIL3 and FANC proteins suggested that they may share a physical interaction. However, we failed to detect a specific protein-protein interaction between NEIL3 and FANC proteins using co-immunoprecipitation. The interaction may be transient or weak in nature and cannot be captured by co-immunoprecipitation. Alternatively, the stabilization between NEIL3 and FANC proteins may not be mediated by a direct interaction; rather, it may be mediated by a shared interaction with additional factors such as the RUVBL complex. We have shown that NEIL3 interacts with RUVBL2 and the interaction between FANC protein and RUVBL1/2 complex has been previously reported (36).
A potential role of NEIL1 in ICL repair has also been recently described. Macé-Aimé et al. demonstrated that the NEIL1 protein level is dependent on an intact FA pathway, suggesting downregulation of NEIL1 may contribute to hypersensitivity of FA cells to ICLs (49). However, McNeill et al. later showed that NEIL1 is recruited to psoralen-ICL, but it interferes and delays psoralen-ICL repair (43). Our results were consistent with the later study. NEIL1 knockdown cells were mildly resistant to psoralen-ICL. Interestingly, NEIL3 was concurrently upregulated in the NEIL1-depleted cells, providing a mechanism for the resistance of NEIL1-deficient cells to psoralen-ICL. Taken together, these results further suggest that NEIL3, but not the other NEIL glycosylases, is the major enzyme to repair psoralen-ICL.
In summary, our study shows that NEIL3 plays a major role in psoralen-ICL repair, which is non-epistatic with the FA pathway, a pathway which is primarily responsible for MMC- and cisplatin-induced ICLs. The psoralen-ICL repair mechanism of NEIL3 is PARP-dependent and does not introduce DSBs.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (50) with the dataset identifier PXD016256.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Magnar Bjørås at the Norwegian University of Science and Technology for providing the primary wild-type and Nelil3-/- MEF cells. We thank Dr Ross Tomaino of the Taplin Biological Mass Spectrometry Facility in Harvard Medical School, for his help in mass spectrometry sample handling and data analysis. The authors also thank members of the D’Andrea lab for technical support and critical discussions.
Author contributions: N.L., J.Z. and A.D.D. designed the experiments. N.L. and J.Z. carried out the experiments and analyzed the data. N.L., J.Z., S.S.W. and A.D.D. wrote and edited the manuscript. A.D.D., J.Z., J.W. and J.C. supervised this work. J.Z. and A.D.D. conceived the original idea of this study. S.S.W. provided critical reagents. All authors were involved in critical discussions of this project.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. National Institutes of Health [R37HL052725, P01HL048546]; U.S. Department of Defense [BC151331P1]; Breast Cancer Research Foundation; Fanconi Anemia Research Fund (to A.D.D.); National Natural Science Foundation of China [81601869 to N.L.]. Funding for open access charge: U.S. National Institutes of Health [R37HL052725, P01HL048546].
Conflict of interest statement. None declared.
REFERENCES
- 1. Deans A.J., West S.C.. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011; 11:467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rodriguez A., D’Andrea A.. Fanconi anemia pathway. Curr. Biol. 2017; 27:R986–R988. [DOI] [PubMed] [Google Scholar]
- 3. Ceccaldi R., Sarangi P., D’Andrea A.D.. The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol. 2016; 17:337–349. [DOI] [PubMed] [Google Scholar]
- 4. Niraj J., Farkkila A., D’Andrea A.D.. The fanconi anemia pathway in cancer. Annu Rev Cancer Biol. 2019; 3:457–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garner E., Smogorzewska A.. Ubiquitylation and the Fanconi anemia pathway. FEBS Lett. 2011; 585:2853–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crossan G.P., van der Weyden L., Rosado I.V., Langevin F., Gaillard P.H., McIntyre R.E., Sanger Mouse Genetics P., Gallagher F., Kettunen M.I., Lewis D.Y. et al.. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat. Genet. 2011; 43:147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim Y., Lach F.P., Desetty R., Hanenberg H., Auerbach A.D., Smogorzewska A.. Mutations of the SLX4 gene in Fanconi anemia. Nat. Genet. 2011; 43:142–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moldovan G.L., D’Andrea A.D.. How the fanconi anemia pathway guards the genome. Annu. Rev. Genet. 2009; 43:223–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barnes D.E., Lindahl T.. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004; 38:445–476. [DOI] [PubMed] [Google Scholar]
- 10. He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L. et al.. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011; 333:1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fleming A.M., Ding Y., Burrows C.J.. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. PNAS. 2017; 114:2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morland I., Rolseth V., Luna L., Rognes T., Bjoras M., Seeberg E.. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002; 30:4926–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu M., Bandaru V., Bond J.P., Jaruga P., Zhao X., Christov P.P., Burrows C.J., Rizzo C.J., Dizdaroglu M., Wallace S.S.. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. PNAS. 2010; 107:4925–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hazra T.K., Izumi T., Boldogh I., Imhoff B., Kow Y.W., Jaruga P., Dizdaroglu M., Mitra S.. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. PNAS. 2002; 99:3523–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hazra T.K., Kow Y.W., Hatahet Z., Imhoff B., Boldogh I., Mokkapati S.K., Mitra S., Izumi T.. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 2002; 277:30417–30420. [DOI] [PubMed] [Google Scholar]
- 16. Krokeide S.Z., Laerdahl J.K., Salah M., Luna L., Cederkvist F.H., Fleming A.M., Burrows C.J., Dalhus B., Bjoras M.. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst.). 2013; 12:1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou J., Liu M., Fleming A.M., Burrows C.J., Wallace S.S.. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J. Biol. Chem. 2013; 288:27263–27272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou J., Fleming A.M., Averill A.M., Burrows C.J., Wallace S.S.. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015; 43:4039–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fleming A.M., Zhou J., Wallace S.S., Burrows C.J.. A role for the fifth G-Track in G-Quadruplex forming oncogene promoter sequences during oxidative stress: Do these ‘Spare Tires’ have an evolved function?. ACS Cent. Sci. 2015; 1:226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou J., Chan J., Lambele M., Yusufzai T., Stumpff J., Opresko P.L., Thali M., Wallace S.S.. NEIL3 Repairs telomere damage during S Phase to secure chromosome segregation at mitosis. Cell Rep. 2017; 20:2044–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Massaad M.J., Zhou J., Tsuchimoto D., Chou J., Jabara H., Janssen E., Glauzy S., Olson B.G., Morbach H., Ohsumi T.K. et al.. Deficiency of base excision repair enzyme NEIL3 drives increased predisposition to autoimmunity. J. Clin. Invest. 2016; 126:4219–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jalland C.M., Scheffler K., Benestad S.L., Moldal T., Ersdal C., Gunnes G., Suganthan R., Bjoras M., Tranulis M.A.. Neil3 induced neurogenesis protects against prion disease during the clinical phase. Sci. Rep. 2016; 6:37844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Regnell C.E., Hildrestrand G.A., Sejersted Y., Medin T., Moldestad O., Rolseth V., Krokeide S.Z., Suganthan R., Luna L., Bjoras M. et al.. Hippocampal adult neurogenesis is maintained by Neil3-dependent repair of oxidative DNA lesions in neural progenitor cells. Cell Rep. 2012; 2:503–510. [DOI] [PubMed] [Google Scholar]
- 24. Hildrestrand G.A., Neurauter C.G., Diep D.B., Castellanos C.G., Krauss S., Bjoras M., Luna L.. Expression patterns of Neil3 during embryonic brain development and neoplasia. BMC Neurosci. 2009; 10:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Semlow D.R., Zhang J., Budzowska M., Drohat A.C., Walter J.C.. Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell. 2016; 167:498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin P.R., Couvé S., Zutterling C., Albelazi M.S., Groisman R., Matkarimov B.T., Parsons J.L., Elder R.H., Saparbaev M.K.. The human DNA glycosylases NEIL1 and NEIL3 excise Psoralen-induced DNA-DNA cross-links in a four-stranded DNA structure. Sci. Rep. 2017; 7:17438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang Z., Nejad M.I., Varela J.G., Price N.E., Wang Y., Gates K.S.. A role for the base excision repair enzyme NEIL3 in replication-dependent repair of interstrand DNA cross-links derived from psoralen and abasic sites. DNA Repair (Amst.). 2017; 52:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu M., Imamura K., Averill A.M., Wallace S.S., Doublie S.. Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure. 2013; 21:247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rolseth V., Krokeide S.Z., Kunke D., Neurauter C.G., Suganthan R., Sejersted Y., Hildrestrand G.A., Bjoras M., Luna L.. Loss of Neil3, the major DNA glycosylase activity for removal of hydantoins in single stranded DNA, reduces cellular proliferation and sensitizes cells to genotoxic stress. Biochim. Biophys. Acta. 2013; 1833:1157–1164. [DOI] [PubMed] [Google Scholar]
- 30. Chapard C., Hohl D., Huber M.. The role of the TRAF-interacting protein in proliferation and differentiation. Exp. Dermatol. 2012; 21:321–326. [DOI] [PubMed] [Google Scholar]
- 31. Harley M.E., Murina O., Leitch A., Higgs M.R., Bicknell L.S., Yigit G., Blackford A.N., Zlatanou A., Mackenzie K.J., Reddy K. et al.. TRAIP promotes DNA damage response during genome replication and is mutated in primordial dwarfism. Nat. Genet. 2016; 48:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoffmann S., Smedegaard S., Nakamura K., Mortuza G.B., Raschle M., Ibanez de Opakua A., Oka Y., Feng Y., Blanco F.J., Mann M. et al.. TRAIP is a PCNA-binding ubiquitin ligase that protects genome stability after replication stress. J. Cell Biol. 2016; 212:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu R.A., Semlow D.R., Kamimae-Lanning A.N., Kochenova O.V., Chistol G., Hodskinson M.R., Amunugama R., Sparks J.L., Wang M., Deng L. et al.. TRAIP is a master regulator of DNA interstrand crosslink repair. Nature. 2019; 567:267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clarke T.L., Sanchez-Bailon M.P., Chiang K., Reynolds J.J., Herrero-Ruiz J., Bandeiras T.M., Matias P.M., Maslen S.L., Skehel J.M., Stewart G.S. et al.. PRMT5-dependent methylation of the TIP60 coactivator RUVBL1 is a key regulator of homologous recombination. Mol. Cell. 2017; 65:900–916.e907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Runge J.S., Raab J.R., Magnuson T.. Epigenetic regulation by ATP-dependent chromatin-remodeling enzymes: SNF-ing out crosstalk. Curr. Top. Dev. Biol. 2016; 117:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rajendra E., Garaycoechea J.I., Patel K.J., Passmore L.A.. Abundance of the Fanconi anaemia core complex is regulated by the RuvBL1 and RuvBL2 AAA+ ATPases. Nucleic Acids Res. 2014; 42:13736–13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li N., Ding L., Li B., Wang J., D’Andrea A.D., Chen J.. Functional analysis of Fanconi anemia mutations in China. Exp. Hematol. 2018; 66:32–41.e38. [DOI] [PubMed] [Google Scholar]
- 38. Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol. 2005; doi:10.1002/0471142727.mb2801s70. [DOI] [PubMed] [Google Scholar]
- 39. Kim H., Yang K., Dejsuphong D., D’Andrea A.D.. Regulation of Rev1 by the Fanconi anemia core complex. Nat. Struct. Mol. Biol. 2012; 19:164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muniandy P.A., Thapa D., Thazhathveetil A.K., Liu S.T., Seidman M.M.. Repair of laser-localized DNA interstrand cross-links in G1 phase mammalian cells. J. Biol. Chem. 2009; 284:27908–27917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chowdhari S., Saini N.. hsa-miR-4516 mediated downregulation of STAT3/CDK6/UBE2N plays a role in PUVA induced apoptosis in keratinocytes. J. Cell Physiol. 2014; 229:1630–1638. [DOI] [PubMed] [Google Scholar]
- 42. Koontz L. TCA precipitation. Methods Enzymol. 2014; 541:3–10. [DOI] [PubMed] [Google Scholar]
- 43. McNeill D.R., Paramasivam M., Baldwin J., Huang J., Vyjayanti V.N., Seidman M.M., Wilson D.M. 3rd. NEIL1 responds and binds to psoralen-induced DNA interstrand crosslinks. J. Biol. Chem. 2013; 288:12426–12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gorynia S., Bandeiras T.M., Pinho F.G., McVey C.E., Vonrhein C., Round A., Svergun D.I., Donner P., Matias P.M., Carrondo M.A.. Structural and functional insights into a dodecameric molecular machine - the RuvBL1/RuvBL2 complex. J. Struct. Biol. 2011; 176:279–291. [DOI] [PubMed] [Google Scholar]
- 45. Kim H., D’Andrea A.D.. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012; 26:1393–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Larsen N.B., Gao A.O., Sparks J.L., Gallina I., Wu R.A., Mann M., Raschle M., Walter J.C., Duxin J.P.. Replication-coupled DNA-protein crosslink repair by SPRTN and the proteasome in xenopus egg extracts. Mol. Cell. 2019; 73:574–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Raschle M., Smeenk G., Hansen R.K., Temu T., Oka Y., Hein M.Y., Nagaraj N., Long D.T., Walter J.C., Hofmann K. et al.. DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science. 2015; 348:1253671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shen X., Do H., Li Y., Chung W.H., Tomasz M., de Winter J.P., Xia B., Elledge S.J., Wang W., Li L.. Recruitment of Fanconi anemia and breast cancer proteins to DNA damage sites is differentially governed by replication. Mol. Cell. 2009; 35:716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mace-Aime G., Couve S., Khassenov B., Rosselli F., Saparbaev M.K.. The Fanconi anemia pathway promotes DNA glycosylase-dependent excision of interstrand DNA crosslinks. Environ. Mol. Mutagen. 2010; 51:508–519. [DOI] [PubMed] [Google Scholar]
- 50. Vizcaino J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al.. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (50) with the dataset identifier PXD016256.