Abstract
WDR5 is a highly-conserved nuclear protein that performs multiple scaffolding functions in the context of chromatin. WDR5 is also a promising target for pharmacological inhibition in cancer, with small molecule inhibitors of an arginine-binding pocket of WDR5 (the ‘WIN’ site) showing efficacy against a range of cancer cell lines in vitro. Efforts to understand WDR5, or establish the mechanism of action of WIN site inhibitors, however, are stymied by its many functions in the nucleus, and a lack of knowledge of the conserved gene networks—if any—that are under its control. Here, we have performed comparative genomic analyses to identify the conserved sites of WDR5 binding to chromatin, and the conserved genes regulated by WDR5, across a diverse panel of cancer cell lines. We show that a specific cohort of protein synthesis genes (PSGs) are invariantly bound by WDR5, demonstrate that the WIN site anchors WDR5 to chromatin at these sites, and establish that PSGs are bona fide, acute, and persistent targets of WIN site blockade. Together, these data reveal that WDR5 plays a predominant transcriptional role in biomass accumulation and provide further evidence that WIN site inhibitors act to repress gene networks linked to protein synthesis homeostasis.
INTRODUCTION
WDR5 is a highly-conserved WD40-repeat protein that participates in numerous chromatin-centric processes. Its most prominent role is in scaffolding the assembly of epigenetic ‘writer’ complexes, including the MLL/SET histone methyltransferases (HMTs) that catalyze histone H3 lysine 4 (H3K4) di- and tri-methylation (me2,3), and the non-specific lethal (NSL) and Ada2-containing (ATAC) histone acetyltransferase (HAT) complexes that acetylate histone H4 at lysine 16 (H4K16) (1). But WDR5 also functions as an epigenetic ‘reader’, recognizing specific post-translational modifications on histone H3 (2,3), it features in the genetic compensation response (4), and it serves as a critical co-factor for the retinoic acid receptor (5) and for the MYC family of oncoprotein transcription factors (6–9). The many and disparate functions of WDR5 in the nucleus clearly establish its moonlighting capabilities (10), but also hamper a fundamental prehension of the protein, making it difficult to establish the predominant biological setting in which WDR5 operates.
Difficulties in pinpointing a clear biologic role for WDR5 are not just conceptual, as WDR5 has emerged as a promising target for anti-cancer therapies (11). The motivation for targeting WDR5 is based on its overexpression in multiple cancers (12–20), its involvement in malignant processes such as the epithelial to mesenchymal transition (21) and cell motility (22), and its ties to oncogenic drivers such as MLL-fusion oncoproteins (23) and MYC (6–9). To date, drug discovery efforts have centered on targeting the ‘WIN’ site of WDR5 (10), a deep pocket that binds arginine-containing motifs (consensus ‘ARA’) present in a number of key WDR5-interaction partners, including histone H3 (24), KANSL1 [NSL complex; (25)], and all five MLL/SET family members (24). With regard to the latter, the actions of WIN site inhibitors against cancer cell lines have generally been attributed to their ability to inhibit H3K4 methylation via MLL1-containing HMT complexes, which in turn drives decreases in the expression of linked genes (23,26,27). But with the relatively long time-frame over which mechanism of action studies have been performed, skepticism as to whether changes in H3K4 methylation would even cause changes in transcription (28), and the absence of a coherent biological framework for understanding WDR5, the true mode of action of WIN site inhibitors in cancer cells is uncertain.
Recently, we discovered potent small molecule inhibitors of the WIN site of WDR5 and determined their mechanism of cell inhibition in the human Mixed Lineage Leukemia (MLL) cell line MV4:11, which carries the MLL–AF4 fusion oncogene (29,30). This study produced several surprising findings. First, despite the general association of H3K4me3 with transcriptionally active chromatin (28), WDR5 binds to just ∼160 sites on chromatin in this setting, as measured by chromatin immunoprecipitation-sequencing (ChIP-Seq). Although the instances of WDR5 binding are small, the sites involved are strongly enriched in genes connected to protein synthesis (protein synthesis genes; PSGs), and encompass half of the ribosome protein genes (RPGs), as well as translation initiation factors and nucleolar components. Second, WIN site inhibitors act rapidly to displace WDR5 from chromatin and diminish transcription from WDR5-bound PSGs, and do so before changes in H3K4me3 status are detected. And third, WIN site inhibitors decrease the protein synthesis capacity of MV4:11 cells, induce nucleolar stress, and activate p53-mediated apoptosis. From these data, we proposed that WDR5 is tethered to chromatin at PSGs via the WIN site, and that WIN site inhibitors act through a primary non-epigenetic mechanism to trigger an ultimately lethal nucleolar stress response in sensitive cell lines.
Although empirical tests of cancer cell sensitivity can be informative, a detailed knowledge of the conserved gene networks controlled by WDR5, and the role of the WIN site in this context, is needed to meaningfully evaluate the potential of WDR5 inhibition as an anti-cancer strategy. It is thus important to establish whether the behavior of WDR5 in MV4:11 cells is atypical, or reflects more general unappreciated aspects of the role WDR5 plays in cells. In this study, we compare the location of WDR5 on chromatin in a panel of mouse and human cells lines of diverse cancer types, and use a WIN site inhibitor to interrogate how WDR5 is tethered to chromatin in different contexts and to reveal the gene networks under the influence of WDR5. We find that the number and location of WDR5 binding sites on chromatin across different cell lines is variable, but that there is a conserved cohort of PSGs that are invariantly bound by WDR5 via the WIN site and transcriptionally repressed by WIN site inhibition. These studies define WDR5 as a conserved regulator of gene networks that promote biomass accumulation, and support the notion that—if a therapeutic window can be established—drug-like WIN site inhibitors could have utility as triggers of nucleolar surveillance and ribosome quality control pathways in cancer cells.
MATERIALS AND METHODS
Cell lines
MV4:11 (male), K562 (female), Be(2)C (male), CHP-134 (male), IMR-32 (male), SK-N-SH (female) and SK-N-AS (female) cell lines were cultured in RPMI-1640 media with 10% FBS. MC-38 (female) and NIH3T3 (male) cell lines were maintained in DMEM with 10% Bovine Serum. All media was supplemented with 100 IU/ml Penicillin and 100 μg/ml streptomycin. To generate p53 knock-down CHP-134 cells, HEK293 (female) cells—growing in DMEM supplemented with 10% FBS and 1% Penicillin/Streptomycin—were transfected with the appropriate shRNA expression construct, the pMD2.G envelope expressing plasmid, and the psPAX2 packaging plasmid, which were gifts from Didier Trono (Addgene plasmid #12259 and #12260, respectively). pLKO-p53-shRNA-941 was a gift from Todd Waldman [Addgene plasmid #25637 (31)]. Scrambled shRNA pLKO.1 plasmid was a gift from David Sabatini [Addgene plasmid # 1864; (32)]. Viral supernatant from transfected HEK293 cells was collected in normal CHP-134 maintenance media, and used to infect CHP-134 cells for 2 days, followed by 1 day of recovery. Stable cells were selected using 1 μg/ml of puromycin.
Antibodies
Antibodies used in this study were: (i) IgG (2729, Cell Signaling), (ii) α-WDR5 (D9E1I, Cell Signaling), (iii) α-p53 (sc-126, Santa Cruz Biotechnology), (iv) α-MYCN (51705, Cell Signaling), (v) α-p21 (mAb#2947, Cell Signaling Technology), (vi) α-RPL5 (51345, Cell Signaling), (vii) α-GAPDH–HRP (MA5-15738-HRP, ThermoFisher), (viii) α-HA-HRP (clone 3F10, Roche), (ix) α-GAPDH–HRP (D16H11, Cell Signaling Technology) and (x) goat α-rabbit IgG Fc secondary antibody (31463, ThermoFisher).
Immunoblotting
Cell lysates were prepared in Kischkel buffer (50 mM Tris pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, PMSF, Na3VO4, and Complete Protease Inhibitor Cocktail (Roche)) as previously described (29). Proteins in the lysate was resolved by SDS-PAGE, transferred to PVDF membrane (PerkinElmer), and antigens detected using the specified antibodies in conjunction with the Supersignal West Pico PLUS reagent (Pierce).
Proliferation assays
White, opaque, flat-bottomed 96-well plates were used, seeded with 2,000–5,000 cells per well. Cells were treated with 0.1% DMSO vehicle only and five 2-fold dilutions of WDR5 inhibitor. For neuroblastoma cell lines, 4-fold dilutions were used. Final DMSO concentration was 0.1% in all compound treatment experiments. Each concentration of inhibitor was tested in triplicate wells and at least two biological replicates were performed. Plates were incubated at 37°C for 3–7 days, depending on doubling time of the cells, and cells quantified using the CellTiter-Glo Luminescent Kit (Promega) according to the manufacturer's instructions. Raw luminescence values were normalized to the DMSO vehicle only wells and PRISM software was used to generate GI50 values. Error bars on proliferation curves represent standard errors of the mean.
Flow cytometry
For cell cycle analysis, treated cells were fixed in ice-cold 70% ethanol for at least 4 h (–20°C), washed in PBS, and resuspended in Propidium iodide (PI) staining buffer (PBS containing 10 μg/ml PI [Sigma], 100 μg/ml RNAse [Roche], and 2mM MgCl2) for overnight staining at 4°C. Samples were filtered through 35 μm nylon mesh and submitted to the Vanderbilt University Medical Center Flow Cytometry Shared Resource for distribution of cell cycle phases using a Becton Dickinson LSRFortessa instrument. For each sample, at least 10 000 cells were counted using forward and side scatter to select single cells. For quantification of apoptosis, cells were collected and fixed as described, and TUNEL staining was performed using the APO-BrdU TUNEL assay kit (Invitrogen) with Alexa-BrdU 488 according to manufacturer's protocol. After staining, samples were filtered through 35 μm nylon mesh and submitted to the Vanderbilt University Medical Center Flow Cytometry Shared Resource for analysis using a Becton Dickinson LSRFortessa instrument. For each sample, at least 10 000 cells were counted using forward and side scatter to select single cells. Each sample was gated against their own individual unstained control. For quantification of protein synthesis, the Click-iT™ Plus OPP Alexa Fluor™ 488 Protein Synthesis Assay Kit (Thermo Fisher C10456) was used. Briefly, CHP-134 cells treated with either DMSO or 5 μM C6 were pulsed with O-propargyl-puromycin (OPP) for 1 h at 37°C. Following incubation with OPP, cells were collected into PBS, fixed in ice-cold 70% ethanol, and stored at −20°C until staining. As a positive control for inhibition of protein synthesis, 50 μg/ml cycloheximide was added to DMSO-treated cells and incubated at 37°C for 30 min prior to addition of OPP. To control for background staining, a sample of DMSO and C6-treated cells were subject to the same staining procedure but no OPP was added. For staining, fixed cells were washed twice with PBS and then the The Click-iT reaction kit was used to conjugate Alexa Flour488-Azide to OPP following the manufacturer's instructions. Mean Alexa488 fluorescence was quantified using a Becton Dickinson LSRFortessa instrument. Single cells were selected using forward and side scatter. Signal from autofluorescence was excluded by gating against a unstained channel (APC) and selecting for cells that were APC negative but Alexa-488 positive.At least 10 000 events were recorded per sample.
dTAG
To tag endogenous WDR5 with the FKBP12(F36V)-HA module (33), a chemically-modified synthetic guide RNA (sequence: CTCTCGCGGGCAGGAGCAAAGGG) was synthesized by Synthego; this gRNA directs cutting of the WDR5 locus 8 bp downstream of the stop codon. Backbone targeting vectors pAW62.YY1.FKBP.knock-in.mCherry and pAW63.YY1.FKBP.knock-in.BFP were a gift from Richard Young (Addgene plasmid #104370 and #104371, respectively). pAW62.YY1.FKBP.knock-in.mCherry carries the FKBP12(F36V)-2xHA-P2A-mCherry cassette; pAW63.YY1.FKBP.knock-in.BFP carries the FKBP12(F36V)-2xHA-P2A-BFP cassette. These vectors were modified by Gibson cloning to include asymmetric homology arms for WDR5: 200 bp 5′ (up to the stop codon) and 800 bp 3′ (starting 17 bp after the stop codon to ensure deletion of PAM sequence). After construction, plasmids were purified with the QIAGEN Midi-prep kit and their integrity confirmed by DNA sequencing. Targeting vectors and Cas9-sgRNA ribonucleoprotein complexes were introduced to cells using the Neon Electroporation Transfection System (Invitrogen). Three reactions of Cas9-sgRNA ribonucleoprotein complexes (RNPs) were formed at a time by combining 10 pmol recombinant Cas9 (Synthego) and 30 pmol sgRNA (Synthego) in Neon buffer R (Invitrogen) and incubating at room temperature for 10 min. CHP-134 cells were counted and resuspended in Neon Buffer R. 600 000 CHP-134 cells and 12.5 μg targeting vectors (1:1 BFP:mCherry) were added to each RNP triplicate reaction and volume brought to 35 μl with Neon Buffer R. The Neon 10 μl reaction delivery tips were used for electroporating with the conditions of 1200 V, 20 ms pulse width and three pulses. After electroporating, cells were immediately placed into warm antibiotic-free RPMI supplemented with 10% FBS and allowed to recover for 2 days before replacing with media containing penicillin and streptomycin and expanding. After growing for 7 days, cells were counter-stained for 15–30 min with 1:2000 Zombie NIR viability dye (BioLegend #423105) in PBS, washed in PBS, and resuspended in 0.5% BSA in PBS. Cells were then analyzed by the Vanderbilt University Flow Cytometry Shared Resource using a BD LSRII Fortessa (BD Biosciences-US) instrument for expression of BFP and mCherry fluorescent markers as a proxy for proper integration. After confirmation of BFP/mCherry positive cells, a population of double positive cells was sorted using a BD FACSAria™ III (BD Biosciences-US), expanded, and analyzed by immunoblotting. For lysate analysis, cells were plated and allowed to adhere. One or two days later the media was replaced with media containing 500 nM dTAG-47 or DMSO vehicle control. Cells were cultured under treatment for the indicated timepoints and collected by scraping into PBS. Cell pellets were lysed in RIPA buffer (10 mM Tris pH 8.0, 0.5 mM EDTA, 1% NP-40, 0.1% deoxycholate, 0.1% SDS, 140 mM NaCl) supplemented fresh with with Roche Complete Protease Inhibitor Cocktail, 1 mM PMSF, and Roche PhosSTOP inhibitor and taken forward for analysis by immunoblotting.
Chromatin immunoprecipitation (ChIP) and ChIP-Sequencing (ChIP-Seq)
For ChIP experiments involving Be(2)C, LoVo, MC-38, and NIH3T3 cells, ChIP assays were performed using a published protocol (34). Briefly, cross-linking was carried out with 0.75% formaldehyde (Sigma-Aldrich; in PBS pH 7.4) at room temperature for 10 min. The reaction was quenched with 125 mM glycine for 10 min at room temperature, after which cells were washed twice with ice-cold PBS. Nuclei were prepared by incubating cells in Nuclear Lysis Buffer A (1 M HEPES pH 7.9, 1 M KCl, 0.5 M EDTA, 0.4% NP-40, PMSF, Na3VO4 and Complete Protease Inhibitor Cocktail) for 5 min on ice, after which they were collected by centrifugation. Nuclei were then lysed in Formaldehyde Lysis Buffer (50 mM HEPES pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% SDS, PMSF, Na3VO4 and Complete Protease Inhibitor Cocktail) for 15 min on ice. Chromatin was sheared by 17–24 min sonication (BioRuptor UCD-200, Diagenode; on highest setting, alternating between 30 s on and 30 s off) to yield a mean chromatin size of ∼250 bp. After sonication, debris was cleared by centrifugation, and sheared chromatin diluted 10-fold in Formaldehyde Lysis Buffer without SDS. Chromatin from 107 cells was used per immunoprecipitation reaction. Immunoprecipitation reactions were performed with either 0.8 μl of IgG or 4.0 μl of α-WDR5 antibody as indicated. Immune complexes were recovered on Protein A agarose beads (Roche) and washed sequentially with Low Salt Wash buffer (20 mM Tris pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, PMSF, and Complete Protease Inhibitor Cocktail), High Salt Wash Buffer (20 mM Tris pH 8.0, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100, PMSF, and Complete Protease Inhibitor Cocktail), LiCl Wash buffer (10 mM Tris pH 8.9, 250 mM LiCl, 1 mM EDTA, 1% Triton X-100, PMSF, and Complete Protease Inhibitor Cocktail) and TE (10 mM Tris pH 8.0, 1 mM EDTA, supplemented with PMSF and Complete Protease Inhibitor Cocktail). Protein–DNA complexes were de-crosslinked overnight at 65°C in 50 μl Elution Buffer (10 mM Tris pH 8.0, 1 mM EDTA, 200 mM NaCl, 0.1% SDS and 20 μg Proteinase K (740506, Clontech), and DNA prepared as described below, depending on whether ChIP DNA was interrogated by quantitative PCR (Q-PCR) or next-generation sequencing (NGS)). ChIP experiments in K562 cells were performed exactly as described (29).
For analysis of ChIP-DNA by Q-PCR, proteinase K was heat-inactivated for 20 min 95°C, and samples diluted by addition of 150 μl of TE. Samples were then analyzed, in technical triplicate, by Q-PCR using primers described in Supplementary Table S1. ChIP signals were calculated as percent input. ChIP experiments were completed in biological quadruplicate. For analysis of ChIP DNA by NGS (ChIP-Seq), de-crosslinked DNA samples from three ChIP reactions were pooled (a total of 3 × 107 cellular equivalents per sample), and DNA purified by phenol-chloroform extraction and recovered by ethanol precipitation. Indexed libraries were then made using the DNA Ultra II Library Prep Kit for Illumina (E7645, New England Biolabs). Library quality was assessed using the 2100 Bioanalyzer (Agilent) and libraries quantitated using KAPA Library Quantification Kits (KAPA Biosystems). Pooled libraries were subject to 75 bp single-end sequencing according to the manufacturer's protocol (Illumina NextSeq500). Sequencing was performed by the Vanderbilt Technologies for Advanced Genomics (VANTAGE) Shared Resource. Bcl2fastq2 Conversion Software (Illumina) was used to generate de-multiplexed Fastq files.
RNA analyses
Cells were lysed in Trizol (ThermoFisher), after which RNA was isolated using the Direct-zol RNA MiniPrep Kit (Zymo Research) with on-column DNase digestion, following the manufacturer's instructions. For transcript-specific analysis by reverse-transcription and Q-PCR (RT-Q-PCR), RNA was reverse-transcribed with M-MLV Reverse Transcriptase (Promega) to generate cDNA prior to analysis. Q-PCR primers are described in Supplementary Table S1. RNA-Seq analysis (with rRNA reduction) was performed at GENEWIZ or VANTAGE. Library preparation with rRNA depletion and paired-end 150 base pair sequencing was performed by GENEWIZ (Illumina HiSeq) or VANTAGE (Illumina NovaSeq6000 (S4)). RNA-Seq for K562 cells was completed with five biological replicates (GENEWIZ); for CHP-134 cells treated with C6, three biological replicates were obtained (GENEWIZ); for CHP-134 cell dTAG experiments, three biological replicates were obtained (VANTAGE).
SLAM-Seq
Nascent RNA was labeled using the SLAMseq Kinetics Kit–Anabolic Kinetics Module (061, Lexogen). Cells were pretreated with C6, C6nc or 0.1% DMSO prior to a 3-h labeling with 1 mM 4-thiouridine (S4U). Following the manufacturer's instructions, total RNA was isolated, alkylated with iodoacetamide, flash frozen, and shipped to Lexogen for analysis. After quality control analyses, libraries were prepared (250 ng RNA per sample) using Lexogen's QuantSeq 3′ mRNA-Seq Library Prep Kit FWD for Illumina, following the User Guide (015UG009V0251) recommendations. Sequencing was performed by Lexogen on an Illumina NextSeq 500 system, using the SR75 High Output Kit. The SLAMdunk analysis pipeline (35) was used to analyze SLAMseq sequencing data. SLAM-Seq completed with three biological replicates.
ChIP-Seq analysis
ChIP-Seq reads were aligned to the genome hg19 or mm10 using Bowtie2 (36) after adaptor trimming by Cutadapt [DOI:10.14806/ej.17.1.200]. Peaks were called using MACS2 with a q value of 0.01 (37). Peak sets overlapping and quantification were determined by Diffbind [Stark R, Brown G (2011). DiffBind: differential binding analysis of ChIP-Seq peak data. http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf]. Peaks were annotated using Homer (38) command annotatePeaks, and enriched motifs were identified by Homer command findMotifsGenome with the default region size and the motif length (http://homer.ucsd.edu/homer/). DAVID (39) was used for all reported Functional GO analyses. Gene Set Enrichment Analysis (GSEA) (40) was performed to evaluate the enrichment of WDR5 binding genes in the repressed genes in response to 2 μM C6 treatment (RNA-Seq) in K562.
RNA-Seq analysis
After adapter trimming by Cutadapt (41), RNA-Seq reads were aligned to the human reference genome using STAR (42), and quantified by featureCounts (43). Read counts were normalized by the Relative Log Expression (RLE) method. Differential analysis were performed by DESeq2 (44), which determined the log2 fold changes, Wald test P-values and adjusted P-value (FDR) by the Benjamini–Hochberg procedure. Significantly changed genes were assessed with an FDR <0.05 and a |log2FC| > 1.
RESULTS
Comparison of the WDR5–chromatin association in disparate cell types
To begin to understand the chromatin context in which WDR5 operates, we compared the genomic location of WDR5, using ChIP-Seq, in six different cell lines: MV4:11, from our previous study (29), K562 (human chronic myelogenous leukemia), Be(2)C (human neuroblastoma), LoVo (human colorectal adenocarcinoma), MC-38 (mouse colon adenocarcinoma), and NIH3T3 (immortal mouse fibroblast). Fortunately, mouse and human WDR5 are identical, and we were able to use the same α-WDR5 antibody for this analysis, allowing for direct comparison of our results.
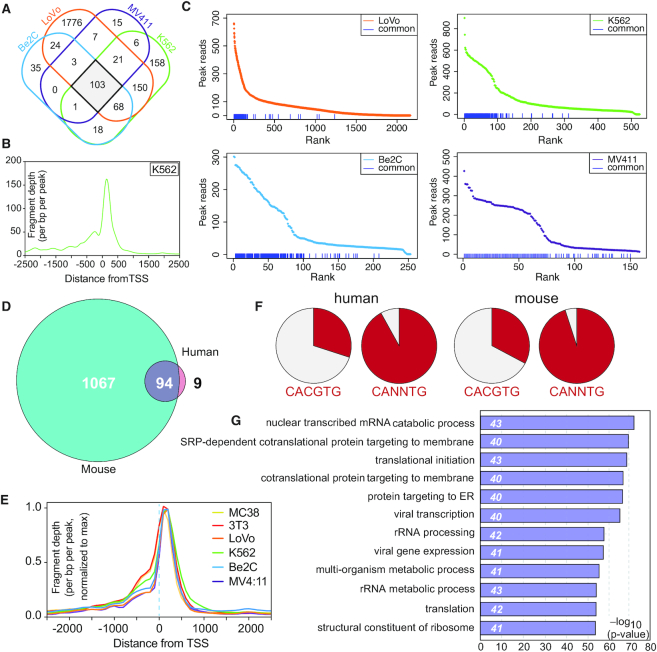
First, we compared the number and location of WDR5 binding sites in each cell type. Across the six cell lines, the total number of WDR5 peaks differs by over an order of magnitude (Figure 1A, B), ranging from ∼2900 in MC-38 to ∼160 in MV4:11 cells. The number of genes associated with these peaks, as defined by all genes within 2 kb upstream of the transcription start sites (TSS) or within the transcription unit, reflects a similar distribution (Figure 1A). The large differential between the number of WDR5 binding sites measured across the different lines does not appear to be due to differences in ChIP-Seq efficiencies or peak-calling issues, as evidenced by example browser images shown in Supplementary Figure S1A, and by quantitative PCR (qPCR) analysis of WDR5 ChIP at select loci in human (Supplementary Figure S1B) and mouse (Supplementary Figure S1C) cell lines. Interestingly, peak tally does not correlate with steady-state levels of WDR5 (Figure 1C), and indeed there appears to be a trend toward anti-correlation—with MV4:11 cells having the highest, and MC-38 the lowest, levels of WDR5 protein. Peak tally does, however, correlate with the distribution of WDR5 relative to TSS. In cell lines with fewer WDR5 binding events (K562, Be(2)C and MV4:11), the majority of WDR5 binding is promoter–proximal (Figure 1D, Supplementary Figure S2A), and is distributed equally between 500 bp upstream and 500 bp downstream of the TSS. In cell lines with more WDR5 binding sites (MC-38, NIH3T3 and LoVo), peaks are equally distributed between those close to the TSS (promoter proximal) and those >5 kb away. Despite the differing number and location of WDR5 binding sites in these different contexts, three themes are apparent. First, relative to the TSS, there is a bimodal distribution of WDR5 in all lines, with distinct peaks at both 300–500 bp upstream, and 50–100 bp downstream, of the TSS (Figure 1E). This distribution results from genes having either one upstream or one downstream WDR5 site; few genes show a WDR5 peak at both locations. Second, WDR5 binding sites in all lines are strongly enriched in consensus motifs for sequence-specific DNA binding proteins (Supplementary Figure S2B), the most conspicuous of which is the E-box motif (CACGTG; Figure 1F) that is bound by MYC and a half-dozen other transcription factors. And third, WDR5-bound genes are highly-significantly enriched in Gene Ontology (GO) terms connected to protein synthesis (Supplementary Figure S2C), with sites in all six lines sharing the overlapping GO categories of peptide biosynthetic processes, translation, ribosome, structural constituent of ribosome, and cytosolic large ribosomal subunit (Figure 1G).
Figure 1.
Survey of WDR5 binding sites in six different cell lines. (A) Results of ChIP-Seq analyses of WDR5 in six cell lines, showing (top row) the number of WDR5 peaks, and (bottom row) the number of genes assigned to those peaks in each line. (B) Heat maps of WDR5 ChIP-Seq peak intensity in each line. The figure represents the combined average of normalized peak intensity in 100-bp bins ± 2 kb around the center of peaks. For mouse cells, the heatmap presents the combined peaks in the two mouse cell lines; for human cells, the heatmap presents the combined peaks in the four human cell lines. Peaks are ranked based on MC-38 for mouse cells and LoVo for human cells. (C) Immunoblotting of steady-state WDR5 levels in the indicated cell lines. GAPDH is a loading control. (D) Distribution of WDR5 binding sites, relative to annotated TSS. ChIP-Seq peaks for all six cell lines are plotted according to distance from nearest TSS, binned by region: ±0–500, 500–1000, 1000–5000 and >5000 bp away from TSS. (E) Averaged TSS-proximal ChIP-Seq peak shape and distribution in all six cell lines. Fragment depth is normalized to the maximum peak read for each cell line. (F) Results of known-motif analysis of WDR5 ChIP-Seq peaks, showing the top-ranked E-box motif for each line. p-value, motif name, and percent of target sequences containing the motif are listed beneath each motif sequence. (G) GO enrichment analysis of WDR5-bound genes in each cell line. The top five conserved GO categories for each line are shown; numbers to the right of each bar show the number of genes in each category.
From this analysis, we conclude that there are substantial differences in the number of sites on chromatin that are bound by WDR5 in different contexts, that these differences are not driven by variations in WDR5 expression, and that as the number of WDR5 binding sites increases so does the chance that these will occur in non-promoter (intergenic) regions. We also conclude that WDR5 binding sites are replete with DNA-binding motifs for sequence-specific transcription factors, and that WDR5-bound genes have a strong and recurring tendency to be connected to protein synthesis.
A conserved set of genes are bound by WDR5 in all cell types
Having established that many of the characteristics of WDR5 binding to chromatin are common across all six cell types, we next asked whether these common characteristics derive from a conserved set of WDR5-bound loci. Because of genome sequence differences, we first compared WDR5 peaks in human and mouse data individually, and then overlaid their shared gene assignments. We identified ∼100 common WDR5 peaks in the four human cell lines (Figure 2A), and ∼1200 in the two mouse lines (Supplementary Figure S3A). Across cell lines and species, the general profiles of these common peaks are similar (Figure 2B, Supplementary Figure S3B), with particularly sharp definition at sites immediately downstream of the TSS, indicating an almost invariant positioning of WDR5 at its 3′ TSS-proximal locations. More variation is apparent at 5′ TSS-proximal sites. Comparing the intensity of these common peaks among all sites bound by WDR5, we observed that, in human cells, conserved WDR5 peaks generally have the strongest WDR5 signal (Figure 2C), whereas in the two mouse lines there is no correlation between conservation and intensity (Supplementary Figure S3C), as to be expected from the similar number of WDR5 binding events in these lines (Figure 1A).
Figure 2.
Identification and characterization of conserved WDR5-bound genes. (A) Venn diagram, showing the overlap of WDR5 peaks in ChIP-Seq data from all four human cell lines. (B) Averaged ChIP-Seq peak distribution for WDR5 at the 103 conserved sites in human cells (corresponds to 103 genes). Data from K562 cells is represented here; data from other cell lines are shown in Supplementary Figure S3B. (C) WDR5 peaks in all four human cell lines were ranked according to peak intensity; the blue lines at the bottom of each graph denote the 103 conserved human WDR5 peaks. (D) Venn diagram, showing the overlap of genes bound by WDR5 in human and mouse cells. (E) Averaged ChIP-Seq peak distribution for WDR5 at the 94 genes bound by WDR5 in all six cell lines, relative to the TSS. Peak signal was normalized to the maximum peak signal per cell line. (F) Representation of perfect (CACGTG) or imperfect (CANNTG) E-box motifs in the 74 human, or 76 mouse WDR5 binding sites corresponding to the 94 common genes. (G) GO enrichment analysis of the 94 conserved WDR5-bound genes. The top 12 GO categories are shown; numbers in italics show the number of genes in each category.
Overlaying all six sets of gene assignments, we identified 94 common WDR5-bound genes (Figure 2D, Supplementary Table S2) which due to multiple gene assignments, corresponded to 74 unique human, and 76 unique mouse, WDR5-binding sites. At these common genes, WDR5 binding occurs just downstream of the TSS (Figure 2E). There is no common DNA motif shared among these conserved WDR5 binding sites, and enrichments detected by known motif analysis using HOMER (38) were modest, likely due to the small number of sequences involved. We did manually observe, however, that 68 of the 74 human sites carried an E-box motif (CANNTG), 22 of which were perfect (CACGTG; Figure 2F). In mice, 72 of the 76 sites carried an E-box motif (CANNTG), 25 of which were perfect (CACGTG; Figure 2F). Interestingly, these 94 shared genes are sites of high WDR5 signal in mouse (Supplementary Figure S3C, red lines) as well as human (Figure 2C) cells. GO analysis reveals that these common genes are highly enriched in those connected to protein synthesis (Figure 2G), and we find that half of the common WDR5 binding sites occur within the same subset of small and large subunit RPGs (Supplementary Figure S3D); these RPGs are also bound by WDR5 in published ChIP-Seq data from prostate (12) and gastric (45) cancer cell lines. Taken together, these data identify a set of ∼75 broadly-conserved WDR5 binding sites, and reveal that they are typically high intensity sites located proximal to the TSS (downstream) in genes that are overtly linked to protein synthesis.
Binding of WDR5 to conserved genes is WIN site-dependent
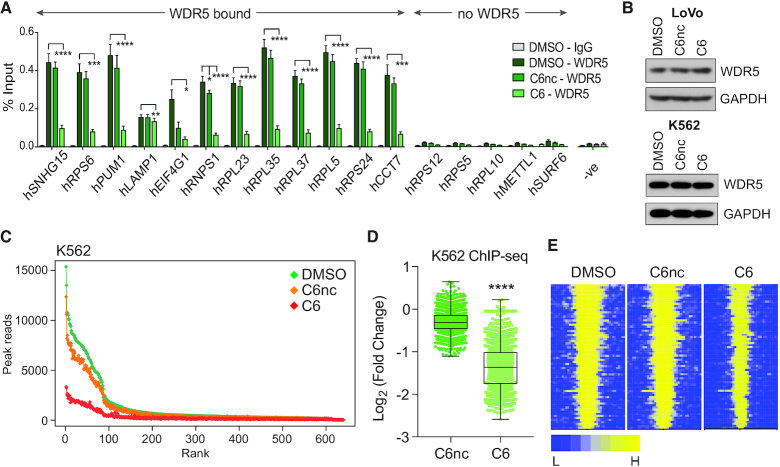
Having identified a set of conserved WDR5-bound loci, we next asked whether the interaction of WDR5 with these conserved genes is WIN site-dependent. First, we used gene-specific ChIP in LoVo cells, and found that overnight treatment of LoVo with C6, but not its isomeric negative control compound C6nc (29), reduces the interaction of WDR5 with chromatin at all 12 loci tested by gene-specific ChIP (Figure 3A). These changes in chromatin association are not accompanied by changes in the steady-state levels of WDR5 (Figure 3B). To extend these findings globally, we treated K562 cells with C6 (or C6nC) for 4 h and performed ChIP-Seq. Here, the results were comparable to our MV4:11 (29) and LoVo cell findings. There is no change in the steady state levels of WDR5 in K562 cells upon inhibitor treatment (Figure 3B), but we observed a widespread reduction in the interaction of WDR5 with chromatin (Figure 3C and Supplementary Figure S4), the average magnitude of which was ∼2.5-fold (Figure 3D). Notably, the impact of C6 treatment is most pronounced at high intensity WDR5-binding sites (Figure 3C), which correspond to the 94 conserved WDR5-bound genes (Figure 3E). We conclude from this analysis that the WIN site plays a general role in linking WDR5 to chromatin, especially at the conserved sites of WDR5 binding, and by extension that WIN site inhibitors function broadly to displace WDR5 from conserved target genes.
Figure 3.
The WIN site links WDR5 to chromatin at conserved target genes. (A) ChIP for WDR5 was performed in LoVo cells treated with DMSO, or 25 μM C6nc or C6, for 16 h. Co-precipitating DNA was detected by Q-PCR and expressed as a percentage of signal in input chromatin. Genes are grouped according to whether or not they are bound by WDR5 in our LoVo ChIP-Seq data. ‘-ve’ corresponds to a primer set within the SNHG15 gene body that does not bind WDR5. Data are presented as mean ± SEM, n = 4 independent ChIP experiments. One-Way ANOVA followed by Dunnett's Post-Hoc Test was performed on data from each gene to determine the statistical significance of WDR5 displacement upon C6/C6nc vs DMSO treatment. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (B) Immunoblotting of steady-state WDR5 levels in the LoVO cells treated with DMSO, or 25 μM C6nc or C6, for 16 h (top) or K562 cells treated with DMSO, or 2 μM C6nc or C6, for 4 h (bottom). GAPDH is a loading control. (C) Scatterplot of normalized average read counts for WDR5 binding peaks in K562 cells treated for 4 h with DMSO, 2 μM C6nc, or 2 μM C6, as determined by ChIP-Seq. Peaks are ranked based on read counts in DMSO-treated cells. (D) Box and whisker plot, showing the log2-fold change in WDR5 ChIP-Seq peak intensity in K562 cells, comparing C6nc and C6 treatments. The difference in signal for each peak is represented as a dot in the scatter plot. The box extends from the 25th to the 75th percentile, with the median marked by the middle line; whiskers extend from minimum to maximum points. Wilcoxon test shows a significant difference in the fold change of C6nc/DMSO versus C6/DMSO, ****P< 0.0001. (E) Heatmap of WDR5 ChIP-Seq peak intensity in K562 cells treated for 4 h with DMSO, C6nc or C6, representing the combined average of normalized peak intensity in 100-bp bins ±2 kb around the center of peaks, for the 94 conserved WDR5-bound genes. Genes ranked based on DMSO-treated cells.
Conserved WDR5-bound protein synthesis genes are inhibited by WIN site blockade
Next, we used WIN site inhibitor to ask whether any of the 94 conserved WDR5-bound genes are transcriptionally regulated by WDR5 binding. To avoid transcriptional complications from induction of cell death, we began by looking in cells that are not phenotypically sensitive to WIN site inhibitor treatment: LoVo (GI50 > 25 μM; Supplementary Figure S5A) and K562 [GI50 ∼25 μM (29)].
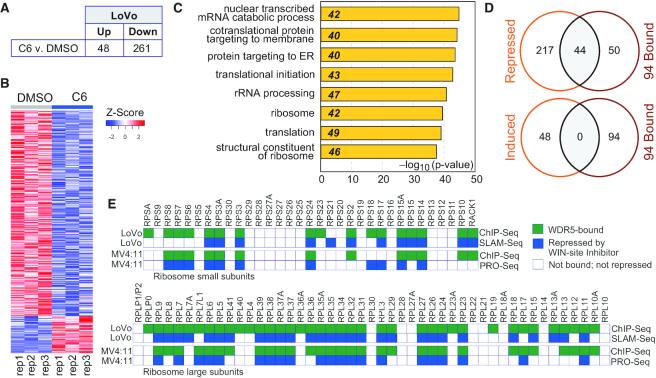
To examine immediate-early transcriptional responses, we coupled C6 treatment with Thiol (SH)-Linked Alkylation for the Metabolic Sequencing of RNA—SLAM-Seq (46)—an approach that allows quantification of changes in newly-synthesized mRNAs after exposure to inhibitor. LoVo cells were pre-treated with DMSO or C6 for 1 h, after which new mRNAs were labeled with 4-thiouridine (4sU) for 3 h before harvest and completion of the SLAM-Seq pipeline. In this timeframe, we expect to see both direct as well as early secondary effects of WIN site blockade. Using this approach, we identified ∼50 genes that are induced, and ∼260 genes that are repressed, by C6 treatment (Figure 4A, B). Genes induced by C6 in LoVo cells show weak clustering in fairly uninformative GO categories (Supplementary Figure S5B), whereas C6-repressed genes cluster strongly in GO terms connected to protein synthesis (Figure 4C). Overlaying WDR5 ChIP-Seq data onto these mRNA changes to separate direct from secondary effects, a number of themes emerge. First, WDR5-bound genes that respond to WIN site inhibition in this timeframe are generally repressed. Seventy (26%) of the genes repressed by C6 treatment in SLAM-Seq are bound by WDR5, compared to just seven (15%) of the genes that are induced by C6 exposure (Supplementary Figure S5C). Importantly, genes bound by WDR5 in LoVo cells are more likely to be repressed by C6 treatment than those that are unbound in this cell line (Supplementary Figure S5D). If we focus on the 94 conserved WDR5-bound genes (Figure 2D), this relationship becomes clearer; almost half (44) of the conserved WDR5-linked genes are repressed by C6 in LoVo cells—none are induced (Figure 4D). Second, WDR5-bound and C6-repressed genes are strongly enriched in those connected to protein synthesis (Supplementary Figure S5E), including a specific subset of small and large subunit RPGs. Indeed, approximately 40 of the C6-repressed genes are WDR5-bound RPGs, and a majority of these are also directly repressed by C6 treatment of MV4:11 cells (Figure 4E), as determined by PRO-Seq (29). And finally, early secondary effects of WIN site inhibition (i.e. at non-WDR5-bound genes) are generally not as tightly biologically clustered as the primary effects, although among the repressed genes there is modest enrichment in GO categories relating to autophagy and protein synthesis (Supplementary Figure S5F).
Figure 4.
Immediate-early transcriptional consequences of WIN site inhibition. (A) Results of SLAM-Seq analysis. Table shows the number of transcripts significantly (FDR < 0.05) altered by 4 h of treatment of LoVo cells with 25 μM C6, compared to DMSO control. (B) Heatmap, displaying z-transformed gene expression for significantly changed genes in C6 versus DMSO (FDR < 0.05) for all three replicates (rep1–rep3) of SLAM-Seq, examining the impact of 4 h of DMSO or 25 μM C6 treatment of LoVo cells. (C) GO enrichment clusters for gene transcripts significantly repressed by C6 treatment of LoVo cells, as determined by SLAM-Seq. Numbers in italics represent the number of repressed genes in each category. (D) Venn diagrams, showing overlap of genes repressed (top) or induced (bottom) by C6 treatment of LoVo cells with the 94 genes bound by WDR5 across all six cell types. (E) Ribosomogram, showing small (top) and large (bottom) ribosome subunit RPGs; a green box indicates whether WDR5 is bound to each RPG in the indicated cell type, a blue box indicates whether the gene is repressed by WIN site inhibition. LoVo cell data are from this study. MV4:11 data are taken from (29).
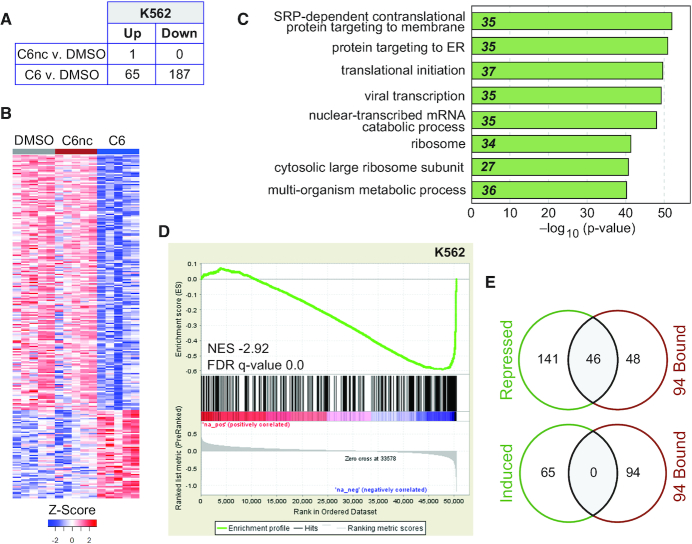
To examine persistent transcriptional changes induced by WIN site inhibition, K562 cells were treated with C6 (or C6nc) for 3 days and RNA-Seq analysis performed (Figure 5A, B). The negative control compound C6nc altered the expression of just one gene. The active compound, in contrast, had broader transcriptional impact; 65 genes were induced, and ∼190 repressed, by C6 treatment (Figure 5A). In general, the long-term transcriptional changes we observed in K562 cells mirrored the shorter term study in LoVo cells. In K562 cells, genes induced by C6 are not strongly clustered (Supplementary Figure S6A), but repressed genes are strongly enriched in GO terms connected to protein synthesis (Figure 5C). There is a highly significant tendency for WDR5-bound (C6-displaced) genes to be transcriptionally modulated by C6 in K562 cells (Figure 5D). The predominant effect of C6 on WDR5-bound genes is repression—52 of the repressed genes are bound by WDR5 in K562 cells, compared to just three of the induced genes (Supplementary Figure S6B). In total, half of the 94 conserved WDR5-bound genes are repressed by C6 in K562 cells (Figure 5E); none are induced. Again, genes bound by WDR5 in K562 cells are more likely to be repressed by C6 treatment than those that are unbound in this cell line (Supplementary Figure S6C). And again, we observe a very specific pattern of WDR5-bound RPGs that are repressed by WIN site blockade in this context, which overlaps strongly with those found to be repressed by C6 in MV4:11 and LoVo cells (Supplementary Figure S6D).
Figure 5.
Prolonged transcriptional consequences of WIN site blockade. (A) Table shows the number of transcripts significantly (FDR < 0.05) altered (in RNA-Seq analysis) by 3 days of treatment of K562 cells with 2 μM C6 or C6nc, compared to DMSO control. (B) Heatmap, displaying z-transformed gene expression for significantly changed genes in C6 or C6nc versus DMSO (FDR < 0.05) for five biological replicates of RNA-Seq, examining the impact of 3 days of DMSO, 2 μM C6nc, or 2 μM C6 treatment of K562 cells. (C) GO enrichment clusters for gene transcripts significantly repressed by C6 treatment of K562 cells, as determined by RNA-Seq. Numbers in italics represent the number of repressed genes in each category. (D) Gene set enrichment analysis showing the distribution of K562 genes repressed in response to 2 μM C6 treatment (RNA-Seq) against the list of all WDR5-bound genes in K562 cells (ChIP-Seq). FDR q = 0.0, genes ranked by log2-fold change. (E) Venn diagrams, showing overlap of genes repressed (top) or induced (bottom) by C6 treatment of K562 cells with the 94 genes bound by WDR5 across all six cell types.
Taken together, these data support the notion that there is a conserved cohort of protein synthesis genes, mostly encoding large and small subunit ribosomal proteins, that are directly and positively regulated by WIN site-dependent binding of WDR5 to chromatin.
WIN site inhibitor represses PSG expression and induces p53 in neuroblastoma cells
In our study of C6 in MLL-fusion cells, we showed that transcriptional changes associated with WIN site inhibition lead to a modest decrease in protein synthesis capacity, induction of nucleolar stress, and activation of p53-mediated apoptosis (29). We suggested that sensitivity of MLL-fusion cells to WIN site inhibition may result from two factors: (i) the tendency of MLL-fusion oncoproteins to drive ectopic protein synthesis as part of their tumorigenic program and (ii) the tendency of MLL-fusion leukemias to retain p53. If this suggestion is correct—and if WIN inhibition consistently represses PSG expression—we predict that additional cancer cells with similar characteristics will be sensitive to WIN site blockade. Because MYC oncogenes function in part through stimulation of biomass accumulation (47), and because p53 status is commonly retained in N-MYC-driven neuroblastoma (48), we tested the prediction that C6 will inhibit MYCN amplified, p53 wild-type, cancer cell lines.
We paneled C6 against five different neuroblastoma lines: (i) CHP-134 (N-MYC amplified, wild-type p53), (ii) IMR32 (N-MYC amplified, wild-type p53), (iii) Be(2)C (N-MYC amplified, mutant p53), (iv) SK-N-SH (non N-MYC amplified, wild-type p53) and (v) SK-N-AS (non N-MYC amplified, mutant p53) (49). To allow direct comparison, treatment times were adjusted for cell doubling time. Interestingly, the only two neuroblastoma lines that are sensitive to C6 are CHP-134 and IMR32 (Figure 6A), both of which are N-MYC amplified and p53 wild-type, and both of which are as sensitive to C6 as MV4:11 cells. The GI50 of C6 in CHP-134 cells is 3.9 μM, in IMR32 cells the GI50 is 2.3 μM, and in MV4:11 cells the GI50 is 3.0 μM (29). Measurable GI50 values were not obtained in the single-copy N-MYC or mutant p53 cell lines. Thus, consistent with our prediction, C6 WIN site inhibitor is also active against cancer cell lines driven by oncogenic lesions other than MLL-fusions.
Figure 6.
WIN site inhibitor is active against N-MYC amplified neuroblastoma cells with wild-type p53. (A) Dose response of neuroblastoma cell lines to C6. CHP-134 and Be(2)C cells were treated with compound for 4 days, the rest of the cell lines for seven days. The blue and red dotted lines indicate 100% and 50% of the DMSO levels, respectively. Data are presented as mean ± SEM, n = 3. (B) Table shows the number of transcripts significantly (FDR < 0.05) altered (in RNA-Seq analysis) by 1 day of treatment of CHP-134 cells with 5 μM C6, compared to DMSO control. (C) CHP-134 cells were treated with DMSO, or 5 μM C6, and counted on the indicated days post-treatment. Fold-change was calculated based on the number of total cells at each time point over the number of cells plated. For the 4 and 7 day time points, cells were replated at the starting concentration with fresh C6 on day three. Data are presented as mean ± SEM, n = 3. (D) GO enrichment clusters for gene transcripts significantly repressed by C6 treatment of CHP-134 cells, as determined by RNA-Seq. Numbers in italics represent the number of repressed genes in each category. (E) Venn diagrams, showing overlap of genes repressed (top) or induced (bottom) by C6 treatment of CHP-134 cells with the 94 genes bound by WDR5 across all six cell types. (F) Bar graph depicting mean (± SEM) Alexa488 labeled-OPP incorporation into nascent polypeptides of CHP-134 cells treated with DMSO or 5 μM C6 for one or 2 days. As a positive control for protein translation inhibition, 50 μg/ml of cycloheximide (CHX) was added to a sample of DMSO treated cells for 30 min prior to addition of OPP. No OPP was added to a sample of DMSO- or C6-treated cells to control for background staining. Mean Alexa488 fluorescence measurements were normalized to DMSO treated samples at each time point. **P = 0.003 by Student's t-test. n = 3. (G) Immunoblotting of steady-state levels of the indicated proteins in CHP-134 cells that were treated with DMSO, or 5 μM C6, for 2 days. GAPDH is a loading control. (H) RT-Q-PCR analysis of RNA extracted from CHP-134 cells treated for four days with 5 μM C6, or DMSO. Relative mRNA levels are normalized against GAPDH in each sample, setting the DMSO sample for each primer set to 1. Data are presented as mean ± SEM, n = 3. (I) CHP-134 cells were transduced to express an shRNA against p53 (sh-p53), or a scrambled control (scr). They were then treated with increasing concentrations of C6, as indicated, for 4 days. Cell number at each dose at day four is presented relative to the DMSO-treated cell numbers. Data are presented as mean ± SEM, n = 3. Inset: Immunoblot (IB) of p53 and GAPDH (loading control) in the indicated cells treated with DMSO, or 5 μM C6, for 2 days. n = 3. (J) Distribution of cell cycle phases as determined by flow cytometry for CHP-134 cells treated as in (C) for 2 or 7 days. Data are presented as mean, error bars are standard error. *P = 0.04, **P = 0.003 by Student's t-test. n = 3. (K) TUNEL staining of CHP-134 cells treated as in (C) for 7 days. Results are expressed as percent of cells with TUNEL staining. Data are presented as mean ± SEM, **P = 0.007 by Student's t-test. n = 3.
To determine if recurring WDR5-bound genes are transcriptionally impacted by WIN inhibition in neuroblastoma cells, we treated CHP-134 cells with 5 μM of C6 for 24 h and performed RNA-Seq analysis (Figure 6B, Supplementary Figure S7A). At this time, C6 has no measurable impact on cell viability (Figure 6C), allowing us to focus on the transcriptional changes that occur before an overt cellular response. Under these conditions, ∼1000 genes are induced, and ∼300 repressed by C6. As predicted, among the repressed genes we observed strong enrichment in GO terms connected to protein synthesis (Figure 6D), including the recurring subset of WDR5-bound RPGs (Supplementary Figure S7B), and 56 of the 94 conserved WDR5-bound genes (Figure 6E). None of the 94 conserved genes are induced by the WIN site inhibitor. Further interrogating gene expression changes by gene set enrichment analysis [GSEA; (40)], the only significant category for repressed genes was myogenesis (Supplementary Figure S7C). Compared to the insensitive cell lines previously profiled, the number of genes induced by C6 in CHP-134 cells is larger, and the genes are more enriched in specific GO categories (Supplementary Figure S7D), including those connected to nitrogen metabolism, the cell cycle, and neurogenesis. By GSEA, we also observed significant induction of genes connected to protein secretion, K-RAS signaling, and the G2M checkpoint (Supplementary Figure S7E). From this analysis we conclude that WIN site inhibitor C6 represses the same set of recurring WDR5-target genes in CHP-134 cells as it does in other settings, and—even before changes in cellular viability are observed—this is associated with transcriptional alterations in genes linked to critical cellular pathways.
Lastly, we asked if C6 elicits the same phenotypic response in CHP-134 cells as we reported for MLL-fusion cells: diminution of protein synthesis, induction of p53, and initiation of apoptosis (29). To measure protein synthesis, we treated CHP-134 cells with C6 for 1 or 2 days, pulsed with O-propargyl-puromycin (OPP) to label nascent polypeptide chains (50), and quantified OPP incorporation by fluorescent OPP tagging and flow cytometry (51). We observed a ∼17% reduction in protein synthesis capacity of CHP-134 at 2 days post-treatment (Figure 6F), which is comparable to the ∼25% reduction we reported in MLL-fusion cells over a similar time frame (29). We also observed that p53 is induced by 2 days of C6 treatment of CHP-134 cells (Figure 6G), along with its key target genes CDNK1A (p21) (Figure 6G), and BAX, NOXA, and PUMA (Figure 6H). As we saw in MLL-fusion cells, shRNA-mediated knockdown of p53 blunted the response to C6 (Figure 6I), confirming that at least part of the CHP-134 cell sensitivity to WIN site blockade is p53 dependent. Further, and as in our previous MLL-fusion cell study, we did not detect changes in cell cycle distribution of CHP-134 cells 2 days after treatment (Figure 6J), but by day 7 there were significantly fewer cells in G1 and significantly more in the sub-G1 population, consistent with induction of apoptosis; which we confirmed by TUNEL staining (Figure 6K). Thus, despite the different oncogenic drivers at play, the WIN site inhibitor C6 induces changes in CHP-134 neuroblastoma cells that are similar to those observed in MV4:11 MLL-fusion cells.
WDR5 depletion represses PSG expression and induces p53 in neuroblastoma cells
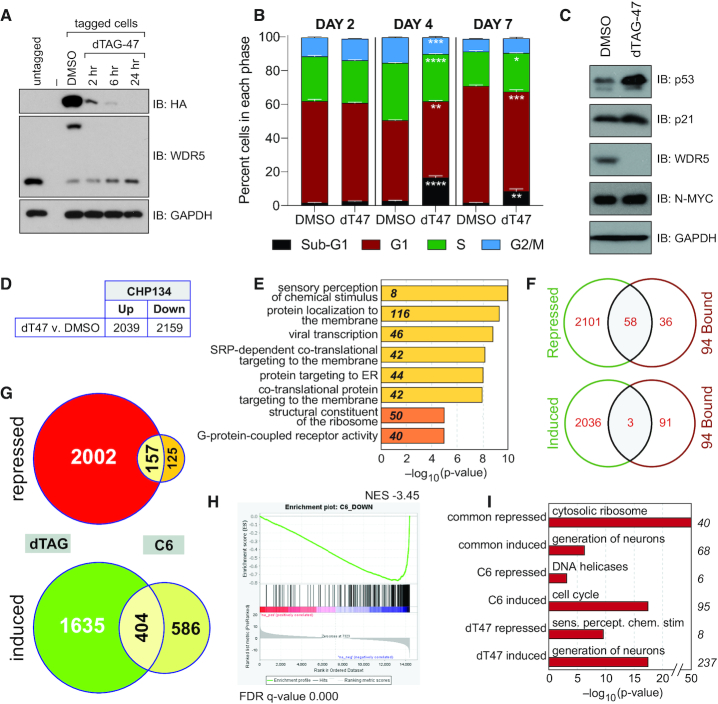
To determine if C6 acts via inhibition of WDR5, and to expose the extent to which the WIN site participates in the totality of WDR5 function, we next examined the phenotypic consequences of WDR5 depletion in CHP-134 cells. We created CHP-134 cells in which endogenous WDR5 is tagged with an FKBP12(F36V)–HA module, permitting acute degradation of WDR5 via the dTAG approach (33). CRISPR-mediated genome engineering was used to knock in the dTAG module, together with fluorescent protein cassettes that permit sorting of tagged cell populations (52). Using this approach, we tagged the majority of WDR5 in the sorted population (Figure 7A), and showed that WDR5 levels decrease quickly (within 2 h) and persistently (at 24 h) after addition of the dTAG-47 ligand (Figure 7A). At 2 days, depletion of WDR5 had little effect on CHP-134 cell proliferation (Supplementary Figure S8A) or cell cycle distribution (Figure 7B). As we saw with WIN site inhibitor C6, however, extended depletion of WDR5 results in decreased proliferation (Supplementary Figure S8A) and alterations to the cell cycle profile, specifically an increase in cells with sub-G1 content, which appear to arise at the expense of the G1, S, and G2/M populations (Figure 7B). Most notably, and like C6, 2-day depletion of WDR5 leads to an increase in the levels of both p53 and p21 (Figure 7C), suggesting that the induction of these proteins by WIN site inhibitor at day 2 (Figure 6G) is an authentic consequence of WDR5 inhibition.
Figure 7.
Depletion of WDR5 phenocopies WIN site inhibition. (A) Immunoblotting for WDR5 in CHP-134 cells in which endogenous WDR5 is tagged with an FKBP12(F36V)–HA cassette. The blot for WDR5 shows that its molecular weight is shifted upon introduction of the cassette in tagged cell populations, compared to untagged CHP-134 cells (middle panel). Addition of 500 nM of dTAG-47 ligand induces disappearance of tagged WDR5, as judged by both the total WDR5 blot and the anti-HA blot (top panel), which visualizes only tagged WDR5. GAPDH is a loading control. (B) Distribution of cell cycle phases as determined by flow cytometry of tagged CHP-134 cells treated with DMSO or 500 nM dTAG-47 (dT47) for 2, 4 or 7 days. Data are presented as mean, error bars are standard error. *P = 0.01, **P = 0.005, ***P = 0.0003, ****P < 0.0001 by Student's t-test. n = 3. (C) Immunoblotting of steady-state levels of the indicated proteins in tagged CHP-134 cells that were treated with DMSO or 500 nM dTAG-47 for 2 days. GAPDH is a loading control. (D) Table shows the number of transcripts significantly (FDR < 0.05) altered (in RNA-Seq analysis) by one day of treatment of tagged CHP-134 cells with 500 nM dTAG-47, compared to DMSO control. (E) GO enrichment clusters for gene transcripts significantly repressed by dTAG-47 in modified CHP-134 cells, as determined by RNA-Seq. Numbers in italics represent the number of repressed genes in each category. Categories in yellow are Biological Process; categories in orange are Molecular Function. (F) Venn diagrams, showing overlap of genes repressed (top) or induced (bottom) by degradation of WDR5 in CHP-134 cells with the 94 genes bound by WDR5 across all six cell types. (G) Venn diagrams, showing the overlap of gene expression changes (FDR < 0.05) induced by degradation of WDR5 (dTAG) or chemical blockade of the WIN site (C6) in CHP-134 cells. Diagrams are broken down into repressed or induced genes. (H) GSEA, comparing genes repressed by dTAG-mediated degradation of WDR5 in CHP-134 cells versus C6-treatment of CHP-134 cells. (I) Top GO enrichment categories for genes repressed by both C6 and dTAG-47 (common repressed), induced by both C6 and dTAG-47 (common induced), repressed by C6 but not dTAG-47 (C6 repressed), induced by C6 but not dTAG-47 (C6 induced), repressed by dTAG-47 but bot C6 (dT47 repressed), and induced by dTAG-47 but bot C6 (dT47 induced). Numbers to the right of the graph indicate the number of genes in each category.
Next, we performed RNA-Seq on our modified CHP-134 cells, 24 h after addition of dTAG-47; the same time point as used for analysis of C6 (Figure 6). In total, depletion of WDR5 by this approach induced ∼2000 and repressed ∼2200 genes (Figure 7D, Supplementary Figure S8B). For repressed genes, GO analysis (Figure 7E) revealed enrichment in terms related to principally to protein synthesis, but we also observed two overlapping categories that are related to sensory perception and G-protein coupled receptor activity. For induced genes, we observed particularly strong enrichment in terms connected to neurogenesis and neural differentiation (Supplementary Figure S8C), which likely reflects the cell of origin of these lines and the known ability of p53 to induce differentiation in neuronal cells (53). Gene set enrichment analysis showed that repressed genes were modestly enriched in those connected to myogenesis (Supplementary Figure S8D), whereas induced genes showed enrichment in several categories including hedgehog and tumor necrosis factor α signaling, the androgen response, and genes connected to the mitotic spindle (Supplementary Figure S8E). Looking at the 94 conserved WDR5-bound genes, 58 are repressed by WDR5 depletion, compared to three that are induced (Figure 7F), again reinforcing the notion that—among the common WDR5-bound genes—the predominant role of WDR5 is to positively regulate their expression.
Finally, we compared the RNA-Seq datasets for WDR5 degradation and WIN site inhibition in CHP-134 cells. Depletion of WDR5 produced a more extensive range of gene expression changes than WIN site blockade, and there was a greater number of induced genes shared between the two approaches than repressed genes (Figure 7G). Importantly, there is a highly significant tendency for genes repressed by WIN site inhibition to be repressed by WDR5 depletion (Figure 7H; and vice versa (Supplementary Figure S8F). Indeed more than 40% of the gene expression changes induced by C6 are also induced by depletion of WDR5 (561/1272 changes in the C6-treated samples). Importantly, every RPG that is repressed by WIN site blockade in CHP-134 cells is also repressed by WDR5 depletion (Supplementary Figure S8G). As expected, there are also common GO term enrichments between the two data sets, notably protein synthesis for repressed genes and neurogenesis for induced genes (Figure 7I). But it also clear that there are biologically clustered differences between the two methods of WDR5 perturbation; WDR5 degradation has more extensive effects on neuronal differentiation (induced) and sensory perception (repressed) genes, while WIN site inhibition has broader effects on the expression of genes linked to the cell cycle (induced; Figure 7I). Based on this comparison, we conclude that WIN site inhibition disrupts a large portion of the direct and indirect transcriptional functions of WDR5. We further conclude, based on the sum of our studies, that protein synthesis genes—most importantly a specific subset of RPGs—are direct transcriptional targets of WDR5, and that the action of WIN site inhibitors on these genes is via a WDR5-targeted mechanism. We also conclude, based on the differences described here, that not all functions of WDR5 are likely disabled by WIN site blockade.
DISCUSSION
Despite intense interest in WDR5, a fundamental understanding of the biological framework in which WDR5 acts has been lacking. Here, through comparative genomics and use of a potent WIN site inhibitor tool compound, we demonstrate that the predominant and conserved biological context in which WDR5 functions is in regulation of genes connected to protein synthesis. We identify a set of 94 genes that are bound by WDR5 in all cell types examined, and show that a majority of these encode proteins that function in translation, including a discrete collection of small and large subunit RPGs. We show that the WIN site tethers WDR5 to chromatin at these sites, and demonstrate that a defined subset of these are recurrently repressed by WIN site inhibition. Finally, we demonstrate that WIN site inhibitors induce p53 and inhibit N-MYC amplified (p53 wild-type) cancer cells, forecasting that they could have anti-tumor utility beyond the established niche of heme malignancies.
Side-by-side comparison of WDR5 in multiple cancer cell lines reveals several important commonalities—and a notable disparity—in how WDR5 behaves in different settings. In terms of disparities, we observed more than a 10-fold range in the number of sites bound by WDR5 across the six cell lines. In cells with fewer WDR5 binding events, most of the binding is promoter proximal. In cells with more WDR5 binding sites, the absolute number of promoter-proximal sites is generally preserved, with additional sites largely appearing in gene-distal (intergenic) regions, which are possibly enhancers. This phenomenon is reminiscent of ‘enhancer invasion’ by MYC (54), a process in which MYC can be thought of as essentially saturating promoter proximal sites at normal levels of expression, and then invading enhancers to control new gene expression patterns as its expression passes an oncogenic threshold. In the case of WDR5, however, although its transcriptional influence could be magnified by increased enhancer binding, the number of WDR5 binding sites is disconnected from the average amount of WDR5 protein. This observation is interesting because it illustrates that there must be some other determinant—extrinsic from WDR5—that dictates its gene-distal and cell type-specific chromatin binding patterns. It is also provocative in light of studies showing that WDR5 is overexpressed in a variety of cancers (12–20), where its overexpression often correlates with poor clinical outcomes. If levels of WDR5 do not predict either the number of sites bound by WDR5, nor the relative level of WDR5 at those sites, the impact of enhanced WDR5 expression in cancer cells is unlikely to manifest itself at WDR5-bound target genes. Perhaps WDR5 overexpression in cancer is irrelevant to the malignant state, or perhaps its impact is on events that do not require stable association of WDR5 with chromatin—such as H3K4 methylation (29) or non-transcriptional ‘moonlighting’ processes (10).
Turning to the commonalities, our study allows us to paint a portrait of a prototypical ‘universal’ WDR5-bound gene. Key characteristics of this prototype are high WDR5 signal by ChIP, localization of WDR5 to sequences immediately downstream of the TSS, and a frank connection to protein synthesis, most commonly by encoding a subunit of the ribosome. It is impossible to know if the high ChIP signal of WDR5 at these genes reflects increased occupancy, increased stoichiometry, or increased epitope accessibility. But it is interesting to note that in LoVo and K562 cells, and in our previous work in MV4:11 (29) cells, WIN site inhibitor C6 is unable to completely displace WDR5 from chromatin at the conditions used, even at high concentrations. For the prototypical WDR5-bound gene, therefore, there are likely two modes of association of WDR5 with chromatin; one that is sensitive to C6, and another that is not. One possibility is that there are two distinct anchoring mechanisms for WDR5 on chromatin, only one of which is WIN site-mediated. Alternatively, it is possible all recruitment of WDR5 to chromatin is WIN site dependent, but that there are some high affinity WIN site recruiters that are refractory to competition with C6. We do not know the molecular mechanism of WDR5 recruitment to its target genes, but the precise and conserved registration of WDR5 at prototype genes and the enrichment of DNA-motifs at these sites suggests that there may be a WIN-motif containing DNA-binding protein that recruits WDR5 to chromatin. This protein is unlikely to be MYC, as the WDR5–chromatin interaction is insensitive to disruption of the MYC–WDR5 interaction (7,9), but it could be any one of a number of E-box binding proteins, at least for the universal WDR5 target genes. By extension, it is easy to imagine how additional WIN motif-carrying DNA-binding proteins—perhaps expressed in a cell type-specific manner—could drive the cell type-specific differences in distribution of WDR5 that we observe at non-universal sites. A deeper understanding of the biochemical context in which WDR5 associates with chromatin is needed to address this issue.
The links between WDR5 and protein synthesis genes, particularly RPGs, are profound and intriguing. One particularly curious feature is the location of WDR5 binding at these sites: The region bound by WDR5 is downstream of the TSS, situated within the crucial +1 nucleosome (55)—the dynamics and composition of which can profoundly impact transcriptional processes (56)—and contained entirely within transcribed intronic sequences. Our results with C6 show that displacement of WDR5 from chromatin results, on average, in less than a 2-fold decrease in transcription at its target genes: The transcriptional effects of WDR5 depletion are equally modest. The implication of this finding is that the function of WDR5 is not to turn these genes on or off, but to modulate their expression within a fairly narrow window. The location and behavior of WDR5 at these sites is thus akin to enhancer function, leading us to speculate that these elements may be intronic enhancers (57). Indeed, several intronic enhancers have been described within mammalian RPGs (58–61), the boundaries of which encompass the conserved WDR5 binding sites defined here. One possibility is that WDR5 acts as part of a rheostat at these intronic enhancers, fine-tuning the transcription of select RPGs according to cellular needs. Why some RPGs would use WDR5 for this purpose, and others not, is unclear, but it could reflect specific aspects of the post-transcriptional control of individual subunits (e.g. mRNA and protein synthesis/stability) to balance subunit protein levels, or be related to non-canonical ribosome protein functions, such as extra-ribosomal activities (62) and specialized ribosome formation (63), which impose unique demands for transcriptional control of specific ribosome protein subunits.
Only about half of the prototypical WDR5-bound genes are transcriptionally-repressed by WIN site inhibitor, although WDR5 is displaced from chromatin by C6 at all locations. Indeed, the transcriptional effects of WIN site inhibition at WDR5-bound loci are confined mostly to RPGs. This finding implies that the function of WDR5 is either not the same at all its target genes, or there is some aspect of finer transcriptional regulation by WDR5 that is not apparent under the conditions used in our experiments, such as cell cycle regulation of protein synthesis capacity (64) or during periods of altered mTOR signaling. Regardless, for the WDR5-bound genes that do transcriptionally respond to WIN inhibitor, the pattern of response is highly consistent, allowing us to forecast with high confidence precisely which RPGs will be repressed by a WIN inhibitor in any cellular setting. Moreover, because these same WDR5-bound RPGs are also repressed by WDR5 depletion, we can confidently state that these group of genes are repressed via on-target actions of WIN site inhibitors.
The ability to predict which genes will be bound by WDR5 in any cell type, and which will respond to WIN site blockade, has important ramifications for the implementation of WIN site inhibition as an anti-cancer therapy. Our comparison of the impact of WIN site inhibition with WDR5 depletion in CHP-134 neuroblastoma cells, together with our other studies, clearly demonstrates that changes in RPG expression are predictable, direct, and bona fide consequences of WIN site blockade, and thus likely to happen in every cell treated with WIN site inhibitors. In one sense, therefore, WIN site inhibitors could be thought of as agents that act to decrease the supply of ribosomal components, much in line with the action of RNA polymerase I inhibitors that are gaining momentum as potential chemotherapeutic agents (65). Thinking of WIN site inhibitors in this context frees them from historical links to MLL-fusion oncoproteins (23) and implies broader utility, especially considering that aberrant protein synthesis is a recurring and characteristic feature of malignancy (66). The analogy with RNA polymerase I poisons is strengthened by a recent study showing that one such poison, CX-5461 (67), induces p53 in MYCN-amplified neuroblastoma cell lines and shows a near identical pattern of response in neuroblastoma cell lines (68) to WIN site inhibitor C6. We imagine that WIN site inhibitors could have a similar anti-tumor spectrum, and therapeutic window, to other nucleolar-targeted therapies. Importantly, however, because WIN site inhibitors act through a fundamentally different mechanism to rRNA poisons, they will likely have different on-target toxicities and a different spectrum of drug synergies. We do not yet know whether an effective therapeutic window can be established with WIN site inhibitors, nor do we know if they will have anti-cancer activity in vivo. Development of more potent small molecule WIN site inhibitors with true drug-like properties is needed to fully understand the therapeutic potential of this new class of compounds.
DATA AVAILABILITY
All genomic data have been deposited at GEO (https://www.ncbi.nlm.nih.gov/geo/, accession codes GSE136451 and GSE115377).
Supplementary Material
ACKNOWLEDGEMENTS
For reagents we thank Scott Hiebert, David Sabatini, Didier Trono, Todd Waldman, and Richard Young. We thank Kristy Stengel and Monica Bomber for advice and discussions about using the dTAG system. We thank the Vanderbilt University Chemical Synthesis Core for synthesis of the dTAG-47 molecule.
Notes
Present address: Eric. R. Rellinger, Division of Pediatric General and Thoracic Surgery, Cincinnati Children's Hospital Medical Center, Cincinnati, OH 45229, USA.
Present address: Dai H. Chung, Department of Surgery, Children's Medical Center Dallas, UT Southwestern Medical Center, Dallas, TX 75235, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [CA200709 to W.P.T., Chemical Biology Consortium Contract No. HHSN261200800001E to S.W.F., CA119925 to E.R.A., W.P.T., CA210429 to A.F.B, CA225065 to A.D.G, GM008554 to A.F.B., CA009582 to A.D.G., CA236733 to W.P.T., S.F.W., CA095103 to W.P.T.]; Robert J. Kleberg, Jr., and Helen C. Kleberg Foundation [to W.P.T. and S.W.F.]; The TJ Martell Foundation [to W.P.T. and S.W.F.]; Edward P. Evans Foundation [to W.P.T.]; Rally Foundation for Childhood Cancer Research Fellowship [to A.M.W.]; Open Hands Overflowing Hearts co-funded research fellowship [to A.M.W.]; American Association for Cancer Research Basic Cancer Research Fellowship [to A.M.W.]; The VANTAGE Shared Resource is supported by the National Institutes of Health [CA068485, EY08126, RR030956]; The Vanderbilt University Medical Center Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center [CA68485]; Vanderbilt Digestive Disease Research Center [DK058404]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. S.W.F., S.R. Stauffer, W.P.T., E.T. Olejniczak, J. Phan, F. Wang, K. Jeon and R.D. Gogliotti were granted US Patent 10,160,763 ‘WDR5 Inhibitors and Modulators,’ on December 25, 2018. S.W.F., S.R. Stauffer, J.M. Salovich, W.P.T, F. Wang, J. Phan, and E.T. Olejniczak were granted US Patent 10,501,466 ‘WDR5 Inhibitors and Modulators,’ on December 10, 2019.
REFERENCES
- 1. Wu M., Shu H.B.. MLL1/WDR5 complex in leukemogenesis and epigenetic regulation. Chin. J. Cancer. 2011; 30:240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D.. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005; 121:859–872. [DOI] [PubMed] [Google Scholar]
- 3. Migliori V., Muller J., Phalke S., Low D., Bezzi M., Mok W.C., Sahu S.K., Gunaratne J., Capasso P., Bassi C. et al.. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat. Struct. Mol. Biol. 2012; 19:136–144. [DOI] [PubMed] [Google Scholar]
- 4. Ma Z., Zhu P., Shi H., Guo L., Zhang Q., Chen Y., Chen S., Zhang Z., Peng J., Chen J.. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature. 2019; 568:259–263. [DOI] [PubMed] [Google Scholar]
- 5. Vilhais-Neto G.C., Fournier M., Plassat J.L., Sardiu M.E., Saraf A., Garnier J.M., Maruhashi M., Florens L., Washburn M.P., Pourquie O.. The WHHERE coactivator complex is required for retinoic acid-dependent regulation of embryonic symmetry. Nat. Commun. 2017; 8:728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun Y., Bell J.L., Carter D.R., Gherardi S., Poulos R.C., Milazzo G., Wong J.W., Al-Awar R., Tee A.E., Liu P.Y. et al.. WDR5 supports an N-Myc transcriptional complex that drives a pro-tumorigenic gene expression signature in neuroblastoma. Cancer Res. 2015; 75:5143–5154. [DOI] [PubMed] [Google Scholar]
- 7. Thomas L.R., Wang Q., Grieb B.C., Phan J., Foshage A.M., Sun Q., Olejniczak E.T., Clark T., Dey S., Lorey S. et al.. Interaction with WDR5 promotes target gene recognition and tumorigenesis by MYC. Mol. Cell. 2015; 58:440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carugo A., Genovese G., Seth S., Nezi L., Rose J.L., Bossi D., Cicalese A., Shah P.K., Viale A., Pettazzoni P.F. et al.. In vivo functional platform targeting patient-derived xenografts identifies WDR5-Myc association as a critical determinant of pancreatic cancer. Cell Rep. 2016; 16:133–147. [DOI] [PubMed] [Google Scholar]
- 9. Thomas L.R., Adams C.M., Wang J., Weissmiller A.M., Creighton J., Lorey S.L., Liu Q., Fesik S.W., Eischen C.M., Tansey W.P.. Interaction of the oncoprotein transcription factor MYC with its chromatin cofactor WDR5 is essential for tumor maintenance. Proc. Natl Acad. Sci. U.S.A. 2019; 116:25260–25268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guarnaccia A.D., Tansey W.P.. Moonlighting with WDR5: A cellular multitasker. J. Clin. Med. 2018; 7:E21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu K., Tao H., Si X., Chen Q.. The histone H3 lysine 4 presenter WDR5 as an oncogenic protein and novel epigenetic target in cancer. Front. Oncol. 2018; 8:502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim J.Y., Banerjee T., Vinckevicius A., Luo Q., Parker J.B., Baker M.R., Radhakrishnan I., Wei J.J., Barish G.D., Chakravarti D.. A role for WDR5 in integrating threonine 11 phosphorylation to lysine 4 methylation on histone H3 during androgen signaling and in prostate cancer. Mol. Cell. 2014; 54:613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen X., Gu P., Li K., Xie W., Chen C., Lin T., Huang J.. Gene expression profiling of WDR5 regulated genes in bladder cancer. Genom Data. 2015; 5:27–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen X., Xie W., Gu P., Cai Q., Wang B., Xie Y., Dong W., He W., Zhong G., Lin T. et al.. Upregulated WDR5 promotes proliferation, self-renewal and chemoresistance in bladder cancer via mediating H3K4 trimethylation. Sci. Rep. 2015; 5:8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dai X., Guo W., Zhan C., Liu X., Bai Z., Yang Y.. WDR5 expression is prognostic of breast cancer outcome. PLoS One. 2015; 10:e0124964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ge Z., Song E.J., Imamura Kawasawa Y., Li J., Dovat S., Song C.. WDR5 high expression and its effect on tumorigenesis in leukemia. Oncotarget. 2016; 7:37740–37754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan X., Chen S., Wu J., Lin J., Pan C., Ying X., Pan Z., Qiu L., Liu R., Geng R. et al.. PI3K/AKT-mediated upregulation of WDR5 promotes colorectal cancer metastasis by directly targeting ZNF407. Cell Death. Dis. 2017; 8:e2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cui Z., Li H., Liang F., Mu C., Mu Y., Zhang X., Liu J.. Effect of high WDR5 expression on the hepatocellular carcinoma prognosis. Oncol. Lett. 2018; 15:7864–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun W., Guo F., Liu M.. Up-regulated WDR5 promotes gastric cancer formation by induced cyclin D1 expression. J. Cell. Biochem. 2018; 119:3304–3316. [DOI] [PubMed] [Google Scholar]
- 20. Wu Y., Diao P., Li Z., Zhang W., Wang D., Wang Y., Cheng J.. Overexpression of WD repeat domain 5 associates with aggressive clinicopathological features and unfavorable prognosis in head neck squamous cell carcinoma. J. Oral Pathol. Med. 2018; 47:502–510. [DOI] [PubMed] [Google Scholar]
- 21. Wu M.Z., Tsai Y.P., Yang M.H., Huang C.H., Chang S.Y., Chang C.C., Teng S.C., Wu K.J.. Interplay between HDAC3 and WDR5 is essential for hypoxia-induced epithelial-mesenchymal transition. Mol. Cell. 2011; 43:811–822. [DOI] [PubMed] [Google Scholar]
- 22. Wang P., Dreger M., Madrazo E., Williams C.J., Samaniego R., Hodson N.W., Monroy F., Baena E., Sanchez-Mateos P., Hurlstone A. et al.. WDR5 modulates cell motility and morphology and controls nuclear changes induced by a 3D environment. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:8581–8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cao F., Townsend E.C., Karatas H., Xu J., Li L., Lee S., Liu L., Chen Y., Ouillette P., Zhu J. et al.. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol. Cell. 2014; 53:247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang P., Lee H., Brunzelle J.S., Couture J.F.. The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases. Nucleic Acids Res. 2012; 40:4237–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dias J., Van Nguyen N., Georgiev P., Gaub A., Brettschneider J., Cusack S., Kadlec J., Akhtar A.. Structural analysis of the KANSL1/WDR5/KANSL2 complex reveals that WDR5 is required for efficient assembly and chromatin targeting of the NSL complex. Genes Dev. 2014; 28:929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grebien F., Vedadi M., Getlik M., Giambruno R., Grover A., Avellino R., Skucha A., Vittori S., Kuznetsova E., Smil D. et al.. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPalpha N-terminal leukemia. Nat. Chem. Biol. 2015; 11:571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu J., Sammons M.A., Donahue G., Dou Z., Vedadi M., Getlik M., Barsyte-Lovejoy D., Al-awar R., Katona B.W., Shilatifard A. et al.. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015; 525:206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Howe F.S., Fischl H., Murray S.C., Mellor J.. Is H3K4me3 instructive for transcription activation. Bioessays. 2017; 39:1–12. [DOI] [PubMed] [Google Scholar]
- 29. Aho E.R., Wang J., Gogliotti R.D., Howard G.C., Phan J., Acharya P., Macdonald J.D., Cheng K., Lorey S.L., Lu B. et al.. Displacement of WDR5 from Chromatin by a WIN Site Inhibitor with Picomolar Affinity. Cell Reports. 2019; 26:2916–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aho E.R., Weissmiller A.M., Fesik S.W., Tansey W.P.. Targeting WDR5: A WINning Anti-Cancer strategy. Epigenet Insights. 2019; 12:2516865719865282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim J.S., Lee C., Bonifant C.L., Ressom H., Waldman T.. Activation of p53-dependent growth suppression in human cells by mutations in PTEN or PIK3CA. Mol. Cell Biol. 2007; 27:662–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sarbassov D.D., Guertin D.A., Ali S.M., Sabatini D.M.. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005; 307:1098–1101. [DOI] [PubMed] [Google Scholar]
- 33. Nabet B., Roberts J.M., Buckley D.L., Paulk J., Dastjerdi S., Yang A., Leggett A.L., Erb M.A., Lawlor M.A., Souza A. et al.. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 2018; 14:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weissmiller A.M., Wang J., Lorey S.L., Howard G.C., Martinez E., Liu Q., Tansey W.P.. Inhibition of MYC by the SMARCB1 tumor suppressor. Nat. Commun. 2019; 10:2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Neumann T., Herzog V.A., Muhar M., von Haeseler A., Zuber J., Ameres S.L., Rescheneder P.. Quantification of experimentally induced nucleotide conversions in high-throughput sequencing datasets. BMC Bioinformatics. 2019; 20:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feng J., Liu T., Qin B., Zhang Y., Liu X.S.. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012; 7:1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 40. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011; 17:10–12. [Google Scholar]
- 42. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 44. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun T.T., He J., Liang Q., Ren L.L., Yan T.T., Yu T.C., Tang J.Y., Bao Y.J., Hu Y., Lin Y. et al.. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016; 6:784–801. [DOI] [PubMed] [Google Scholar]
- 46. Muhar M., Ebert A., Neumann T., Umkehrer C., Jude J., Wieshofer C., Rescheneder P., Lipp J.J., Herzog V.A., Reichholf B. et al.. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science. 2018; 360:800–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tansey W.P. Mammalian MYC proteins and cancer. New J.Sci. 2014; 2014:757534. [Google Scholar]
- 48. Carr-Wilkinson J., O’Toole K., Wood K.M., Challen C.C., Baker A.G., Board J.R., Evans L., Cole M., Cheung N.K., Boos J. et al.. High frequency of p53/MDM2/p14ARF pathway abnormalities in relapsed neuroblastoma. Clin. Cancer Res. 2010; 16:1108–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Van Maerken T., Rihani A., Dreidax D., De Clercq S., Yigit N., Marine J.C., Westermann F., De Paepe A., Vandesompele J., Speleman F.. Functional analysis of the p53 pathway in neuroblastoma cells using the small-molecule MDM2 antagonist nutlin-3. Mol. Cancer Ther. 2011; 10:983–993. [DOI] [PubMed] [Google Scholar]
- 50. Liu J., Xu Y., Stoleru D., Salic A.. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Signer R.A., Magee J.A., Salic A., Morrison S.J.. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014; 509:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Weintraub A.S., Li C.H., Zamudio A.V., Sigova A.A., Hannett N.M., Day D.S., Abraham B.J., Cohen M.A., Nabet B., Buckley D.L. et al.. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017; 171:1573–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tedeschi A., Di Giovanni S.. The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO Rep. 2009; 10:576–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zeid R., Lawlor M.A., Poon E., Reyes J.M., Fulciniti M., Lopez M.A., Scott T.G., Nabet B., Erb M.A., Winter G.E. et al.. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat. Genet. 2018; 50:515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schones D.E., Cui K., Cuddapah S., Roh T.Y., Barski A., Wang Z., Wei G., Zhao K.. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008; 132:887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weber C.M., Ramachandran S., Henikoff S.. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol. Cell. 2014; 53:819–830. [DOI] [PubMed] [Google Scholar]
- 57. Chorev M., Carmel L.. The function of introns. Front Genet. 2012; 3:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chung S., Perry R.P.. Importance of introns for expression of mouse ribosomal protein gene rpL32. Mol. Cell Biol. 1989; 9:2075–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hariharan N., Kelley D.E., Perry R.P.. Equipotent mouse ribosomal protein promoters have a similar architecture that includes internal sequence elements. Genes Dev. 1989; 3:1789–1800. [DOI] [PubMed] [Google Scholar]
- 60. Chung S., Perry R.P.. The importance of downstream delta-factor binding elements for the activity of the rpL32 promoter. Nucleic Acids Res. 1993; 21:3301–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Antoine M., Kiefer P.. Functional characterization of transcriptional regulatory elements in the upstream region and intron 1 of the human S6 ribosomal protein gene. Biochem. J. 1998; 336:327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhou X., Liao W.J., Liao J.M., Liao P., Lu H.. Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol. 2015; 7:92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xue S., Barna M.. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012; 13:355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stumpf C.R., Moreno M.V., Olshen A.B., Taylor B.S., Ruggero D.. The translational landscape of the mammalian cell cycle. Mol. Cell. 2013; 52:574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Penzo M., Montanaro L., Trere D., Derenzini M.. The Ribosome biogenesis-cancer connection. Cells. 2019; 8:E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vaklavas C., Blume S.W., Grizzle W.E.. Translational dysregulation in cancer: molecular insights and potential clinical applications in biomarker development. Front. Oncol. 2017; 7:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Drygin D., Lin A., Bliesath J., Ho C.B., O’Brien S.E., Proffitt C., Omori M., Haddach M., Schwaebe M.K., Siddiqui-Jain A. et al.. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011; 71:1418–1430. [DOI] [PubMed] [Google Scholar]
- 68. Hald O.H., Olsen L., Gallo-Oller G., Elfman L.H.M., Lokke C., Kogner P., Sveinbjornsson B., Flaegstad T., Johnsen J.I., Einvik C.. Inhibitors of ribosome biogenesis repress the growth of MYCN-amplified neuroblastoma. Oncogene. 2019; 38:2800–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic data have been deposited at GEO (https://www.ncbi.nlm.nih.gov/geo/, accession codes GSE136451 and GSE115377).