Abstract
Long non-coding RNAs (lncRNAs) have emerged as important biological tuners. Here, we reveal the role of an uncharacterized lncRNA we call SENEBLOC that is expressed by both normal and transformed cells under homeostatic conditions. SENEBLOC was shown to block the induction of cellular senescence through dual mechanisms that converge to repress the expression of p21. SENEBLOC facilitates the association of p53 with MDM2 by acting as a scaffold to promote p53 turnover and decrease p21 transactivation. Alternatively, SENEBLOC was shown to affect epigenetic silencing of the p21 gene promoter through regulation of HDAC5. Thus SENEBLOC drives both p53-dependent and p53-independent mechanisms that contribute to p21 repression. Moreover, SENEBLOC was shown to be involved in both oncogenic and replicative senescence, and from the perspective of senolytic agents we show that the antagonistic actions of rapamycin on senescence are dependent on SENEBLOC expression.
INTRODUCTION
Cell senescence was described by Hayflick as a concept accounting for the finite lifespan of non-transformed fibroblasts (1). Senescence involves cells entering an essentially irreversible non-proliferative but nonetheless viable state. Characteristics of senescent cells include an enlarged size (1), resistance to apoptosis (2), changes in metabolic phenotype (3) the acquisition of senescence-associated heterochromatin foci (SAHF) (4), senescence-associated β-galactosidase (SA-β-gal) activity (5) and the senescence-associated secretory phenotype (SASP) (6). Senescence is proposed as a defense mechanism to mitigate cancer development through preventing the replication of damaged genomes (7,8). Senescence also contributes to the age-related decline in organ function through the loss of progenitors and the accumulation of senescent cells (9,10).
Broadly, there is replicative senescence (RS) involving the telomere length-dependent limit of cell divisions or stress-induced premature senescence which occurs independently of telomere erosion (11,12). Nevertheless, both forms involve sustained repression of pro-proliferative genes regulated through the retinoblastoma (Rb) pocket proteins to induce cell-cycle arrest. Senescence programming is principally achieved by activation of tumor suppressor networks encompassing p53/p21CIP1 and p16INK4a/ARF and is typified by increased levels of cyclin-dependent kinase inhibitors, p21 and p16 (8,10). Moreover, radiation and chemotherapy induce senescence in tumors, indicative that cancer cells possess the latent ability to undergo senescence (13,14). Of interest, the inactivation of c-Myc in cancer cells can also trigger senescence (15) and in melanoma, c-Myc can suppress oncogene-induced senescence (OIS) induced by activated forms of Braf and N-Ras (16).
Although drivers of senescence are well accepted, the underlying control mechanisms are not fully understood. It has recently emerged that long non-coding RNAs (lncRNAs) play important regulatory roles (17,18). For example, the lncRNA PANDA is co-induced with p21 by DNA damage and serves to prevent transactivation of proliferative genes during senescence (19). The lncRNA HOTAIR increases during replicative and irradiation-induced senescence (20) and reducing the levels of lncRNA MALAT in cycling cells also induces senescence, an effect attributed in part to p53 activation (21). Thus, lncRNAs play positive and negative roles in senescence.
The role of senescence in aging has given rise to the notion of senolytics, therapeutics that selectively remove senescent cells to prevent or reverse organ deterioration (9,14). Indeed such agents can re-sensitize senescent cells to apoptosis for example, the tyrosine kinase inhibitor, dasatinib can induce apoptosis in senescent adipocytes but not their non-senescent counterparts (22). The activation of mTOR signaling during senescence has been shown to promote the SASP and this is counteracted by rapamycin (23,24). Nevertheless, the mechanistic actions of rapamycin appear multifactorial (25).
Here we describe SENEBLOC, a lncRNA that maintains normal and transformed cells in the non-senescent state. Under steady state conditions, SENEBLOC acts pleiotropically to repress p21 expression through both p53-dependent and independent mechanisms. SENEBLOC serves as a scaffold to facilitate p53-MDM2 association which decreases p53-dependent transactivation of p21. Alternatively, SENEBLOC acts as a decoy to sequester miR-3175 and prevent HDAC5 mRNA turnover which also contributes to p21 repression. Additionally, we show that the antagonistic actions of rapamycin on p21 expression are dependent on SENEBLOC. Moreover, we show that manipulating SENEBLOC in cancer cells has a profound growth effect.
MATERIALS AND METHODS
Cell culture
HCT116, A549, IMR90, HAFF, 293T and P493-6 cells carrying a c-Myc tet-off system were maintained as previously described including mycoplasma testing and cell line authentication (26).
Antibodies and reagents
Western blotting
Equal amounts of protein or IP eluates were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes before immunodetection using ECL as previously described (26).
RNAi
Lentiviral supernatants were prepared in HEK293T cells after transfecting with shRNAs (cloned in PLKO.1; Supplementary Tables S5 and 6), gag/pol, rev and vsvg plasmids at the ratio of 2:2:2:1. Cell free culture supernatants were used to infect cells for 24 h before selection with puromycin (8 μg/ml).
PCR
One microgram of total RNA isolated using TRIzol reagent (Invitrogen) was used to synthesize cDNA using the PrimeScript RT Reagent Kit (Takara). Quantitative polymerase chain reaction (qPCR) was performed using SYBR Green real‐time PCR analysis (Takara) with the specified primers (Supplementary Table S7). PCR results, recorded as cycle threshold (Ct), were normalized against an internal control (β‐actin). Alternatively, RT-PCR was performed using Taq DNA polymerase (Vazyme). Standard curves assays were utilized where absolute quantitation of RNA expression was required. Plasmids containing the target cDNA of interest were used to construct standard curves by plotting Ct values against copy number as determined from the adjusted final concentration of plasmids. RNA was extracted from a fixed number of cells and cDNA equivalents from 2000 cells used in each qPCR reaction. Average RNA copy numbers/cell were calculated from the sample Ct values referenced against the linear portion of the standard curve.
SA-β-gal and SASP assays
SA-β-gal staining along with quantitation was performed as previously described (26). Human IL-6 and IL-8 were measured by ELISA according to the manufacturer's recommendations (Thermo Fisher #88-7066 and 88-8066, respectively).
Cell proliferation, cell cycle and colony formation assays
Cells adhered to glass coverslips were pulsed for 30 min with 50 μM EdU before fixation and staining for EdU incorporation. Cell nuclei were counterstained with Hoechst and samples observed using epifluorescence microscopy. Quantitation was performed using ImageJ using the Analyze Particles module. For cell-cycle analysis, cells were fixed with 70% ethanol were stained with 20 μg per ml propidium iodide in the presence of 10 μg per ml RNase A before data acquisition by flow cytometry (GE). Data were analysed using FlowJo with the cell-cycle analysis module. Colony formation assays were conducted as previously described (26) after 14 days of culture.
Cell fractionation
Nuclear and cytoplasmic cell fractions were obtained as previously described (26).
Fluorescence insitu hybridization (ISH) and immunofluorescence (IF) staining
In vitro transcribed antisense probes targeting SBLC (Supplementary Table S8) were fluorescently labeled and hybridized with fixed cells as previously described (26). For IF, cells permeabilized in 0.2% Triton X-100 and incubated overnight at 4°C with primary antibodies and detection performed with either Alexa Fluor-488 or -568 secondary antibodies against rabbit or mouse IgG, respectively.
Tumor xenograft model
HCT116 cells expressing the control or SBLC shRNA were subcutaneously injected into the dorsal flanks of 4-week-old male nude mice (Beijing Vital River Laboratory Animal Technology Co., Ltd.). Mice were humanely euthanized and tumors dissected and weighed before analysis. To prepare RNA, tumor samples were homogenized in TRIZOL Reagent (Invitrogen) using a power homogenizer prior to the RNA extraction protocol and qPCR analysis. Alternatively for Western blotting, tumor samples were freeze-dried in liquid nitrogen, vibrated into powder, and this dissolved with RIPA buffer containing protease inhibitors (Beyotime). Studies on animals were conducted in accordance with relevant guidelines and regulations and were approved by the Animal Research Ethics Committee of the University of Science & Technology of China.
GFP reconstitution assay
The predicted IRES of SBLC (258–379 bp) was inserted into the pCirc-GFP-IRES circRNA translation reporter containing a split GFP system using EcoRI and EcoRV (Supplementary Table S7).
Luciferase reporter assays
Assays were performed as described previously (26) using vectors listed in Supplementary Table S5.
Immunoprecipitation, RNA immunoprecipitation and RNA pull-down assays
IP and RNA IP assays were performed as previously described (26). For RNA pull-down, cell lysates were incubated with streptavidin beads (Invitrogen) coated with labeled antisense or sense probes (3 μg) at 4°C for 3 h to overnight. After washing the beads, samples were eluted in Laemmli buffer prior to further analysis.
Chromatin immunoprecipitation (ChIP) assays
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (26) with analysis by using semi-quantitative PCR or qPCR as indicated with the primers shown in Supplementary Table S7.
Northern blot
Equal amounts of total RNA were resolved on 1% agarose gels before transfer to Hybond-N membranes (GE) and UV-crosslinking. Digoxin-labeled oligonucleotide probes were prepared using the DIG Northern Starter Kit (Roche) according to the manufacturer's instructions. HRP-conjugated anti-digoxin antibody was used after the hybridization. Visualized images were obtained using Image Quant LAS-4000 mini (GE Fujifilm).
Electrophoretic mobility shift assay
EMSAs were performed using the EMSA/gel shift kit (Beyotime, China) according to the manufacturers protocol. Flag-tagged p53 and MDM2 proteins purified from 293T cells were used in combination with invitro transcribed biotin-labeled SBLC prepared using the T7-Flash Biotin RNA Transcription Kit (Epicentre). Reactions were resolved on native PAGE gels before transfer to nylon membranes with results visualized using SA-HRP and ECL to detect biotin-labeled SBLC.
Statistical analysis
Statistical significance analyses were performed by two-tailed Student's t-test using Microsoft Excel.
RESULTS
Identification of SENEBLOC as a c-Myc responsive lncRNA involved in senescence
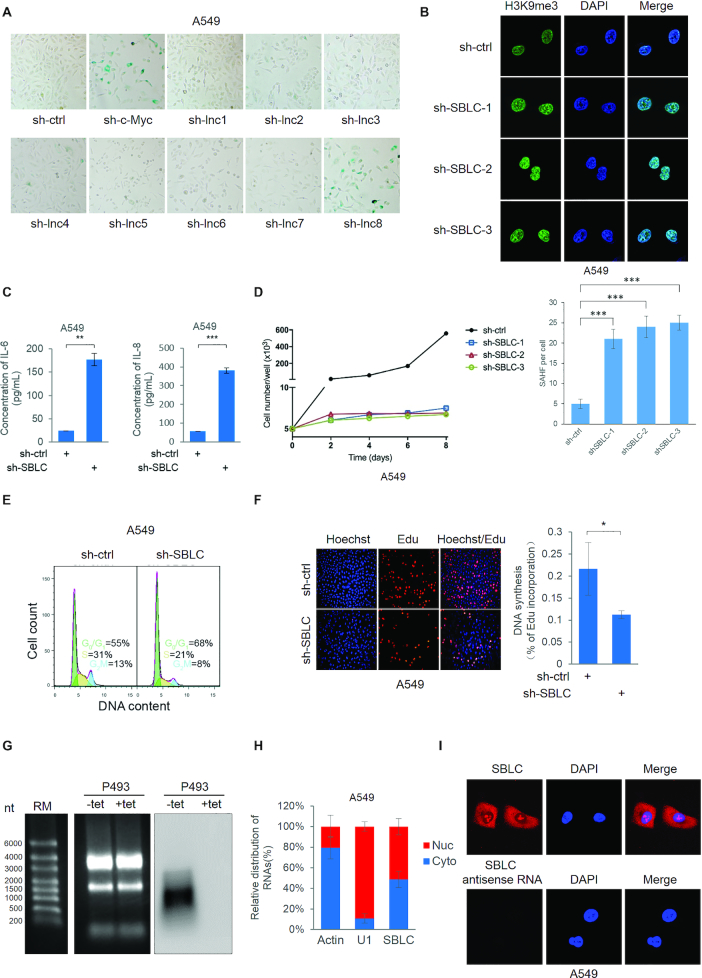
To identify c-Myc responsive lncRNAs that regulate senescence we took advantage of array data generated in P493-6 cells bearing Tet-Off c-Myc expression (26). Eight c-Myc responsive lncRNAs (Supplementary Table S1) were validated using qPCR along with the previously characterized Glut1 gene (Supplementary Figure S1A). Thereafter we depleted each lncRNA in A549 lung carcinoma cells and examined SA-β-gal activity (Figure 1A). Notably knockdown of lnc8 but not others promoted increased β-galactosidase staining, comparable to the effects of depleting c-Myc. On the basis of further investigations we ascribed the name SENEBLOC (SBLC) to lnc8 given its blocking action against senescence.
Figure 1.
Identification of SBLC, a c-Myc responsive lncRNA involved in evasion of senescence. (A) Senescence-associated (SA) β-galactosidase staining in A549 cells after 48 h transduction with either control (sh-ctrl), c-Myc or lentiviruses targeting c-Myc responsive lncRNAs1–8 identified by screening in Supplementary Figure S1A. (B) SAHF in A549 cells after silencing of lnc8 (SBLC) using independent shRNAs or shRNA control (sh-ctrl) for 48h. (Upper) IF staining with anti-H3K9me3 decorating SAHF with nuclear counterstaining using DAPI. (Lower) quantitation of SAHF/cell. (C) IL-6 and IL-8 secretion by A549 cells using ELISA. (D) shRNA-mediated silencing of SBLC in A549 cells inhibited proliferation, (E) entry into the cell cycle as determined by flow cytometry and (F) DNA synthesis as measured by EdU incorporation. (G) Northern blot analysis of SBLC in P493 cells. (H) SBLC distribution in nuclear and cytoplasmic compartments in A549 cells using subcellular fractionation and (I) FISH. (A–I) Results are representative of three independent experiments. Values are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t-test.
In addition to the SA-β-gal assays, depleting SBLC expression in A549 cells using independent shRNAs (Supplementary Figure S1B) resulted in an increased number of foci stained for H3K9me3 (SAHF; Figure 1B), these being representative of heterochromatin regions contributing to gene silencing in senescent cells (4). Additionally, IL-6 and IL-8 cytokine secretion also increased following sh-SBLC treatment (Figure 1C), indicative of the SASP (6). Thus, three independent characteristics of senescent cells were increased following SBLC silencing. Extending analyses to untransformed cells, SBLC silencing in human foetal lung myofibroblasts (IMR-90) and adult foreskin fibroblasts (HAFFs) also caused senescence (Supplementary Figure S1F). Thus SBLC appears broadly relevant to transformed and untransformed cells.
Depletion of SBLC in A549 cells also retarded cell growth (Figure 1D) with accompanying decreases in S/G2/M phases (Figure 1E) and DNA synthesis (Figure 1F). Growth inhibition was also recapitulated in colony formation using HCT116 colorectal carcinoma cells (Supplementary Figure S1C). Given the observed growth suppression, we considered the impact of SBLC on tumorigenicity. Indeed HCT116 xenografts bearing sh-SBLC showed a marked retardation of tumor growth (Supplementary Figure S1D,E). We next sought to better characterize SBLC.
Characterization of SENEBLOC
SBLC (AL161785.2) is located on chromosome 9 (132,020,633-132,022,125) with the annotated transcript (RP11-344B5.4) comprised of three exons (966 bp; Supplementary Figure S1G). Notably, SBLC does not bear significant homology to sequences within the mouse genome. Consistent with this annotation, Northern blotting analysis conducted in P493 Tet-Off cells (Supplementary Figure S1A) predominantly detected transcripts resolving at ∼1 kb which were diminished when c-MYC expression was inhibited (Figure 1G). Examining the SBLC sequence uncovered the presence of an internal ribosome entry site (IRES) with a high probability score along with several potential open reading frames (27). To examine whether SBLC RNA was translatable as has been reported with some lncRNAs (28), we cloned the IRES region into a split GFP system (29). This analysis showed the predicted IRES was unable to mediate the translation of GFP (Supplementary Figure S1H), confirming that SBLC is a bona fide lncRNA. Finally, we examined the distribution of SBLC within cells. Subcellular fractionation analyses revealed pools of SBLC within the nucleus and cytoplasm (Figure 1H), consistent with in situ localization patterns observed by FISH (Figure 1I).
To supplement these data, we conducted an assessment of SBLC expression in a cohort of 22 pairs of colon cancer versus normal tissues. This analysis showed that the average SBLC levels were significantly higher in colon tumors compared to normal adjacent tissues (Supplementary Figure S2A) and pairwise analysis confirmed that the majority of cases exhibited increased SBLC as a result of transformation (Supplementary Figure S2B). Thus at least from the perspective of colon cancers, increased expression of SBLC is evident in tumors.
SBLC expression is controlled by c-Myc
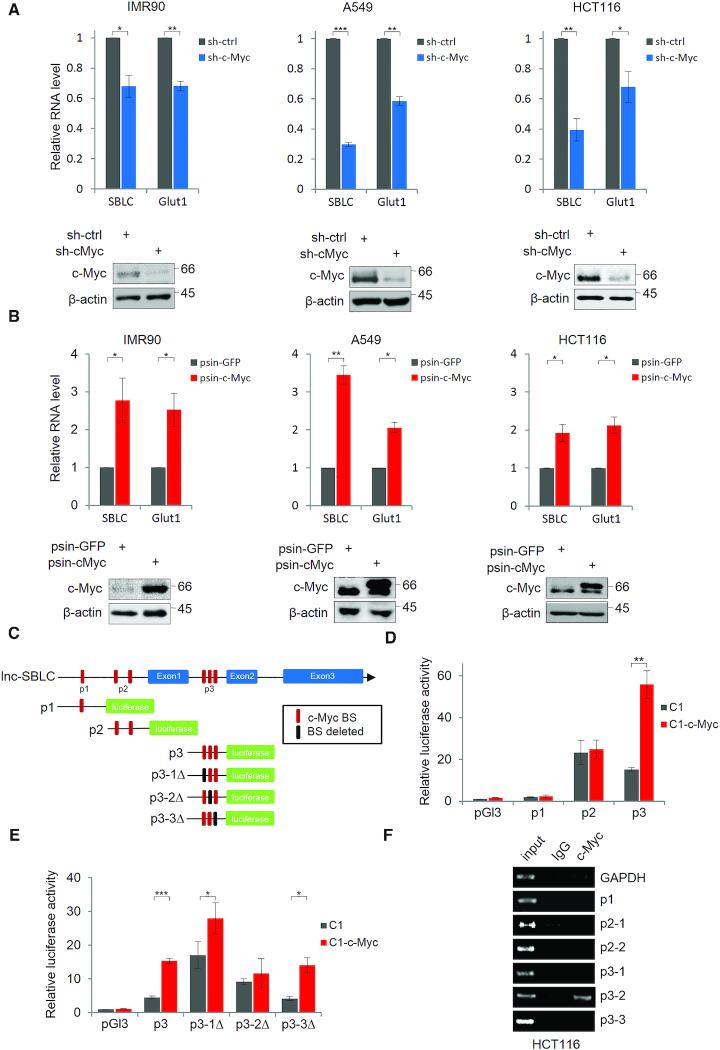
We sought to understand the relationship between c-Myc and SBLC by expanding experiments in additional cell lines. Depletion of c-Myc in IMR90, A549 and HCT116 cells resulted in decreased levels of SBLC (Figure 2A). Conversely, ectopic c-Myc expression increased SBLC levels (Figure 2B). We then considered if c-Myc was involved in SBLC transcriptional regulation.
Figure 2.
LncRNA SBLC is directly regulated by c-Myc. (A) Depletion of endogenous c-Myc expression decreases SBLC levels in IMR90, A549 and HCT116 cells. RNA and protein levels measured using qPCR and western blot, respectively with Glut-1 and β-actin serving as controls. (B) The experiment from (A) was repeated after transfection with Flag-tagged GFP or c-Myc. (C) Designated c-Myc binding sequences (p1, p2 and p3) in the proximal promoter/intronic region of SBLC. pGL3 luciferase reporter plasmids were constructed as illustrated. (D) Reporter assays in 293T cells using control, p1, p2 or p3 pGL3 reporter plasmids. (E) Individual c-Myc BS in the p3 region were mutated and the experiment from (D) repeated. (F) ChIP assays conducted on HCT116 cell lysates using anti-c-Myc or isotype-matched antibodies. GAPDH target sequence was employed as negative control. (A–F) Results are representative of three independent experiments. Values are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t-test.
Interrogation of SBLC identified multiple c-Myc consensus binding sites (c-Myc-BS) in its proximal promoter and between exons 1 and 2 (Supplementary Figure S2C). Luciferase reporters designed to test these BSs (Figure 2C) showed that only the region designated p3 was responsive to c-Myc (Figure 2D). Mapping the three consensus motifs within p3 (Figure 2C) showed that p3-1 and p3-3 mutants remained responsive to c-Myc overexpression whereas p3-2 was not (Figure 2E). Thus, p3-2 is required for c-Myc-mediated transcription of SBLC. Consistently, ChIP assays directed against endogenous c-Myc captured only p3-2 (Figure 2F). Collectively, these data support the transactivation of SBLC by c-Myc.
SBLC modulates p21 expression via p53-mediated transcription
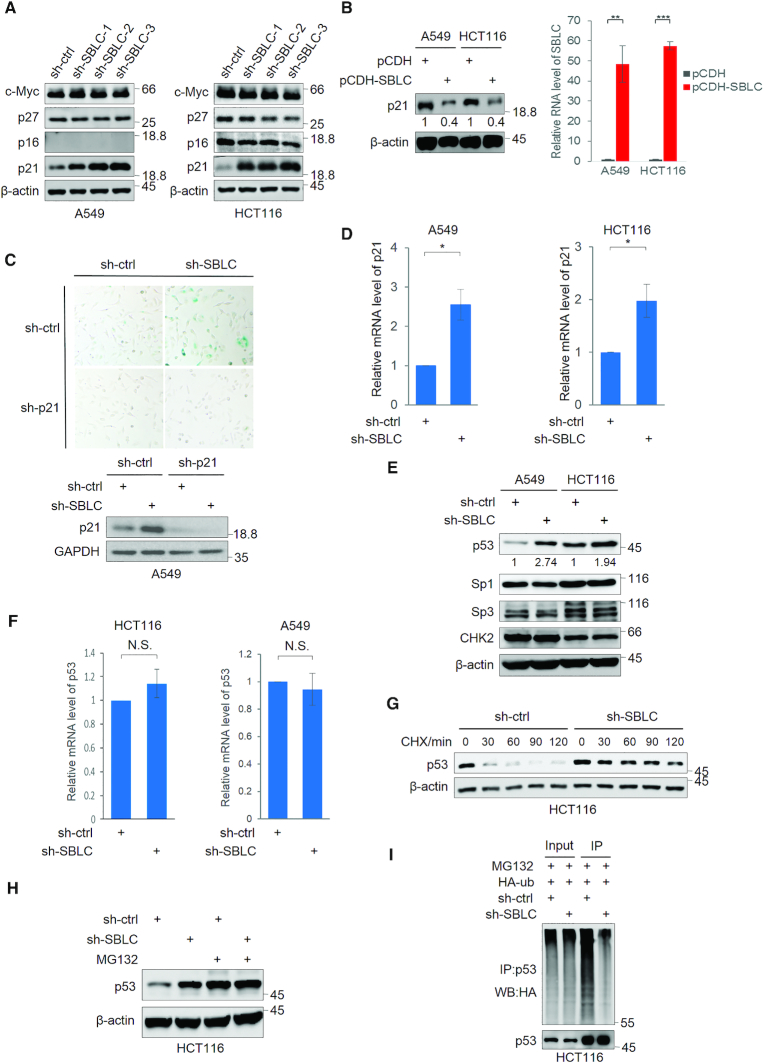
To understand how antagonizing SBLC induces senescence, we examined effectors of senescence, namely p27 (30), p16 and p21. Instructively, only p21 was induced following SBLC depletion (Figure 3A). Notably SBLC itself does not appear to impart regulatory effects on c-Myc levels. Conversely, enforced expression of SBLC caused reduction in the levels of p21 protein (Figure 3B). Similarly p21 protein increased in IMR90 and HAFF cells after SBLC depletion (Supplementary Figure S3A) as was the levels of p21 mRNA and protein measured ex vivo in HCT116 sh-SBLC tumors (Supplementary Figure S3B). Together these data suggested that SBLC acts to restrains p21-mediated senescence. Confirming this, silencing of p21 in A549 cells was effective in preventing senescence induction following SBLC depletion (Figure 3C).
Figure 3.
SBLC acts to repress p21 expression via transcriptional control via p53. Western blotting for c-Myc, p27, p16 and p21 in A549 and HCT116 cells after transduction with sh-ctrl or independent shRNAs against SBLC (A) or p21 levels after transfection of SBLC (pCDH-SBLC) or empty vector (pCDH). Overexpression efficiency was assessed by qPCR (B). β-actin served as a loading control throughout. (C) SA-β-galactosidase staining (top) and western blotting for p21/GAPDH (bottom) in A549 cells after transduction with sh-ctrl or sh-SBLC#3 in combination with p21 shRNA (sh-p21) for 24 h. (D) qPCR measurement of p21 mRNA in A549 cells. (E) Western blotting for p53, Sp3, Sp1 and Chk2 in A549 and HCT116 cells after transduction with sh-ctrl or sh-SBLC. Relative p53 levels were estimated using densitometry. (F) qPCR analysis of p53 mRNA levels in A549 and HCT116 cells. (G) Western blot against p21 in A549 cells treated with cycloheximide. (H) Western blot against p53 in HCT116 cells following 6 h pre-treatment with 20 μM MG132. (I) Western blotting against HA in cell lysates and p53 IPs from HCT116 transfected with HA-ubiquitin. (A–I) Results are representative of three independent experiments. Values are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t-test.
Determining how p21 was regulated, there was a significant increase in p21 mRNA after SBLC silencing (Figure 3D). Moreover, while p21 protein levels were increased, its degradation rate appeared similar (Supplementary Figure S3C). Additionally, analysis of p21 mRNA levels after actinomycin D treatment indicated that its stability was unchanged after SBLC depletion (Supplementary Figure S3D). These data imply that the increased p21 observed after SBLC silencing predominantly results from transcriptional regulation.
We then examined the transcription factors p53, Sp1, Sp3 and also CHK2, regulators of p21 (31,32). Indeed, p53 levels but not others were increased following depletion of SBLC (Figure 3E), thus pointing to p53 as a likely effector. While the levels of p53 mRNA did not significantly change after sh-SBLC treatment (Figure 3F), the increase in p53 resulted from a marked increase in protein half-life (Figure 3G). Moreover, similar levels of p53 were observed in both sh-ctrl and sh-SBLC after treating cells with MG132 (Figure 3H), indicative that the increased levels of p53 after SBLC silencing resulted from inhibiting proteasomal activity. Consistently, the polyubiquitination of p53 decreased following SBLC depletion (Figure 3I). Notably, although p53 was induced after sh-SBLC treatment, activation of PARP and caspase 3 did not occur, indicating no induction of apoptosis (Supplementary Figure S3E). Together these data suggest that SBLC largely acts post-translationally to influence proteasomal mediated degradation and turnover of p53.
SBLC facilitates a ternary complex between p53 and MDM2 that promotes p53 degradation
We next interrogated the levels of MDM2, Pirh2 and COP1, E3-ubiquitin ligases involved in the proteasomal degradation of p53 (33). Western blotting showed that the respective levels of MDM2 increased between sh-ctrl and sh-SBLC treated cells whereas the levels of other E3 ligases appeared similar (Figure 4A). The changes in MDM2 levels inferred that loss of SBLC may increase the p53-dependent transcription of MDM2.
Figure 4.
SBLC enhances proteasomal degradation of p53 through facultative assembly of the p53-MDM2 complex. (A) Western blot analysis for MDM2, Pirh2 and COP1 in A549 and HCT116 cells transduced with sh-ctrl or sh-SBLC#3. β-actin served as a loading control throughout. (B) RNA pulldown assays using biotin-labeled sense/antisense SBLC DNA probes conducted against A549 and HCT116 cell lysates before Western blotting. SBLC enrichment efficiency was assessed by RT-PCR. (C) RIP assay in HCT116 cells against p53 or MDM2 assessed by RT-PCR (top) and Western blotting (bottom). (D) HCT116 cells were transfected with Flag-control or Flag-p53 vectors before sequential IP. First phase Flag IPs were eluted with FLAG peptides and the eluates subjected to secondary IP with MDM2 antibodies or isotype-matched IgG. Samples were subjected to western blotting and RT-PCR analysis for Flag/MDM2 and SBLC, respectively. (E) SBLC localization in A549 cells decorated with RNA FISH and MDM2 or p53 by immunofluorescence. Nuclear staining with DAPI. (F) HCT116 cells expressing sh-ctrl or sh-SBLC#3 were pre-treated with 20μM MG132 before lysis and immunoprecipitation with anti-p53 antibodies and analysis by Western blot. (G) In vitro binding assays were conducted using recombinant sense or antisense SBLC, Flag-p53 and HA-MDM2 as indicated. Analyses were performed on Flag IPs using Western blotting against HA and Flag, respectively while inputs for recombinant proteins and SBLC were subjected to Coomassie blue staining and RT-PCR, respectively. (A–G) Results are representative of three independent experiments. Values are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t-test.
We considered that SBLC might act via binding to p53 and/or MDM2. Indeed, RNA pulldown assays revealed that both p53 and MDM2 were selectively recovered with SBLC (Figure 4B). In support, the association of SBLC with p53 and MDM2 was confirmed using p53 and MDM2 immunoprecipitation (RIP) assays (Figure 4C). Thus SBLC appears to interact with both p53 and MDM2 in the endogenous context, perhaps as a complex. To test this, sequential immunoprecipitation (IP) studies were conducted in cells transfected with Flag-p53. In the first phase IP, Flag-p53 was recovered along with enrichment of MDM2 and SBLC (Figure 4D, left). After elution using FLAG-peptides, the second phase IP against MDM2 recovered Flag-p53 along with SBLC (Figure 4D, right), indicative of a ternary complex formed between SBLC, p53 and MDM2. Consistently, visualizing the cellular distribution of p53 or MDM2 in comparison to SBLC indicated similar patterns of localization particularly in the cytoplasm (Figure 4E). Notably the amount of co-associated MDM2 within p53 immunoprecipitates was substantially reduced when SBLC was silenced (Figure 4F) and similarly, the amount of MDM2 co-precipitating with p53 was reduced when samples were pretreated with RNAse (Supplementary Figure S4A). Supporting this finding, SBLC but not antisense SBLC increased the association between recombinant p53 and MDM2 proteins in an in vitro binding assay (Figure 4G). Thus SBLC influences the association between p53 and MDM2.
To reveal the structural basis for these interactions, domain truncation constructs of p53 and MDM2 were used in binding assays against SBLC. Using this approach, SBLC was shown to bind to p53 through its C-terminal regulatory domain (Supplementary Figure S4B) whereas SBLC binds to the central acidic region of MDM2 (Supplementary Figure S4C). In support, in vitro gel shift assays showed SBLC caused size-shifted proteins bands with purified p53 and MDM2 and all purified domain truncation constructs containing the p53 C-terminal domain or MDM2 central acidic region (Supplementary Figure S4D and S4E, respectively).
Collectively these findings propose that SBLC drives the association between p53 and MDM2 which serves to facilitate p53 degradation.
p53-independent regulation of p21 by SBLC occurs via regulatory effects on HDAC5
As part of our investigations we considered if p53 was an absolute requirement for downstream actions of SBLC. Experiments performed in the p53 replete (+/+) and null (−/−) variants of HCT116 cells showed that SBLC knockdown increased the levels of p21 protein in the absence of p53, albeit to a lesser extent than in cells with p53 expression (Figure 5A). Similarly, silencing of SBLC in MD-MBA-468 cells expressing the tumor-associated p53 mutant R273H (34) also induced p21 expression, resulting in increased cell senescence (Supplementary Figure S5A). Together these data indicate a p53-independent component to SBLC-mediated suppression of p21. To examine the contribution of epigenetic mechanisms we treated HCT116 (p53−/−) cells with chaetocin and trichostatin A (TSA), inhibitors of methyltransferases and histone deacetylases, respectively. Both agents increased p21 levels whereas only the combination of TSA treatment with SBLC silencing failed to further increase p21 levels (Supplementary Figure S5B). Hence, p53-independent effects of SBLC on p21 expression may result from antagonism of histone acetylation.
Figure 5.
SBLC facilitates p53-independent regulation of p21 through miR-3175-dependent effects on HDAC5. (A) p53+/+ and p53−/− HCT116 cells were transduced with sh-ctrl or sh-SBLC#2 and #3 before analysis of p53, p21 and GAPDH levels by western blot.(B) Schematic illustrating ChIP target sequences designed across the p21/WAF1 gene (top). HCT116 p53−/− cells transduced with control sh-ctrl or sh-SBLC were subjected to ChIP assays using anti-H3K9ac or H4K5ac antibodies along with normal rabbit IgG. (C) Western blot of Class I and II HDAC levels in HCT116 (p53+/+) cells after transduction with sh-ctrl and sh-SBLC#3. The levels of HDAC5 and p21 were measured in HCT116 p53−/− cells after knockdown (D) or overexpression of Flag-HDAC5 (E) using western blotting. (F) HDAC5 and p21 levels were measured in HCT116 p53−/− cells following overexpression of Flag-HDAC5 alone and in combination with sh-SBLC#3 treatment to deplete SBLC. Senescence phenotypes were assessed by SA-β-galactosidase staining assay. (G) Western blot of p21 in HCT116 cells transfected with NC controls or the indicated miR mimics for 48 h. (H) The experiment in (G) was repeated with miR-3175 inhibitors. (I) Schematic illustrating pSI-CHECK2-based luciferase constructs incorporating miR-3175 binding sites (BSs) from the HDAC5 3′UTR region (top). HCT116 cells transfected with either control or miR-3175 mimics for 8 h before transfection with the indicated pSI-CHECK2 plasmids for a further 24 h before conducting reporter assays. (J) Schematic representing psiCHECK2-based reporter plasmids incorporating wild-type and mutant SBLC sequences (left). Reporter assays conducted as per (I) (right). (A–J) Results are representative of three independent experiments. Values are mean ± SEM. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t-test.
To evaluate this, ChIP assays were conducted across the p21 gene (Figure 5B, top). We first determined that the p21 promoter was responsive to TSA and indeed increased H3 and H4 histone acetylation was observed (Supplementary Figure S5C). Instructively, silencing of SBLC in HCT116 (p53−/−) cells increased the recovery of ChIP fragments by H3K9ac and H4K5ac antibodies (Figure 5B, bottom). As hyperacetylated H3 and H4 histones are generally indicative of increased gene transcription (35), this reconciles with the increased expression of p21 after sh-SBLC and TSA (Figure 5A and Supplementary Figure S5B). This implies that p53-independent regulation of p21 by SBLC results from maintaining p21 in a repressed state.
We sought to resolve whether specific histone deacetylases were influenced by SBLC. Analysis of candidate HDACs in HCT116 p53−/− cells after depletion of SBLC showed that only HDAC5 levels were decreased (Figure 5C). Indeed, ChIP analysis after knockdown of HDAC5 confirmed that hyperacetylation of H3 and H4 histones were increased in the p21 proximal promoter (Supplementary Figure S5D), supporting the role of HDAC5 in p21 regulation. Consistently, manipulating HDAC5 expression in HCT116 (p53−/−) cells by either knockdown or transfection with Flag-tagged HDAC5 resulted in increased or diminished p21 expression levels, respectively (Figure 5D and E). Bringing together these findings, overexpression of HDAC5 prevented the increase in p21 expression observed after SBLC silencing along with preventing senescence induction (Figure 5F, top and bottom, respectively). Thus the actions of SBLC on p21 expression appears to involve HDAC5.
Instructively there were no changes in HDAC5 mRNA after sh-SBLC (Supplementary Figure S5E) and furthermore since proteasomal inhibition failed to stabilize HDAC5 protein levels after SBLC silencing (Supplementary Figure S5E and F), it appeared likely that SBLC affects HDAC5 though translational efficiency. This also suggested that SBLC might act as a competing endogenous RNA for microRNAs (miRNAs). To evaluate this, we identified four complementary miRs (miR-1321, -3175, -4756 and -4739) that potentially bind to both the 3′-UTR of HDAC5 mRNA and SBLC (Supplementary Table S2). We observed that increasing miR-3175 expression using miR mimics selectively diminished HDAC5 expression along with increased p21 levels (Figure 5G). Conversely, transfection of miR-3175 inhibitors increased HDAC5 while decreasing p21 levels, respectively (Figure 5H). Instructively, miR-3175 inhibitors also prevented the induction of senescence caused by sh-SBLC treatment (Supplementary Figure S5G). Thus manipulating miR-3175 phenocopied the effects of SBLC.
To confirm the actions of miR-3175 we turned to the pSICHECK2 system, dividing the HDAC5 3′UTR into constructs containing two putative miR-3175 binding sites (Figure 5I, left). Only the reporter activity of the 3175-BS-1 construct was inhibited by miR-3175 mimics and such inhibition was completely abrogated when the binding sequence was mutated (Figure 5I, right). We then considered the structural basis for the interaction between miR-3175 and SBLC.
Bioinformatics examination revealed complementarity between SBLC and the seed region of miR-3175 (Supplementary Table S2). Pulldown assays demonstrated miR-3175 was recovered along with SBLC (Supplementary Figure S5H), indicating that SBLC binds to miR-3175 in the endogenous state. Moreover, the activity of a SBLC psiCHECK2 reporter containing three putative miR-3175 binding sites (Figure 5J, left) was inhibited by miR-3175 mimics but inhibition was alleviated if the motifs were disrupted by mutagenesis (Figure 5J, right). This implies that SBLC acts as a decoy to sequester miR-3175 which prevents inhibition of HDAC5 translation. Estimating the absolute cellular levels of SBLC versus miR-3175 in three cell lines showed the copy numbers/cell of miR-3175 was around six-ten fold higher than SBLC while HDAC5 levels were more abundant (Supplementary Figure S5I), similar to the patterns of expression seen in a paired liver versus tumor sample (Supplementary Figure S5J). On the basis that SBLC containing four miR-3175 binding sites (Supplementary Table S2), the proposed sequestration of miR-3175 by SBLC appears plausible.
Rapamycin promotes SBLC transcription through effects on E2F1
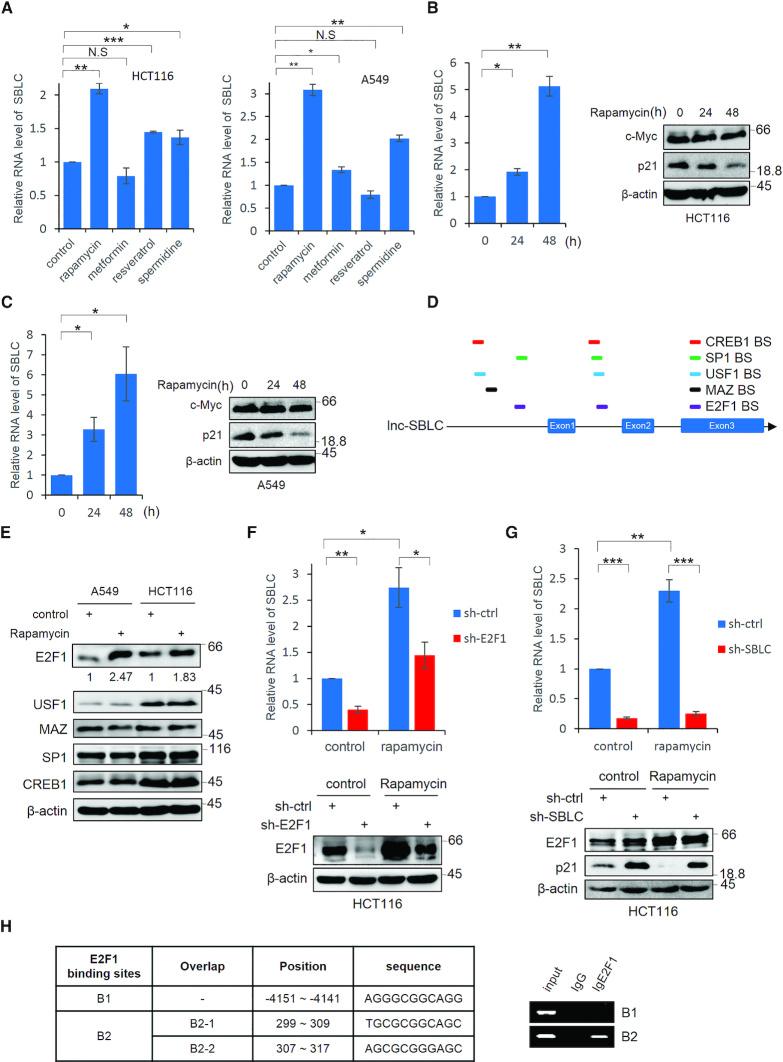
As a corollary it was important to establish the relevance of SBLC to the actions of senolytic drugs. Among rapamycin, metformin, resveratrol and spermidine treatments, rapamycin was most effective in inducing SBLC (Figure 6A). We therefore focused on understanding whether SBLC played a mechanistic role in the senolytic activity of rapamycin.
Figure 6.
Rapamycin promotes SBLC transcription through effects on E2F1. (A) HCT116 and A549 cells were treated with rapamycin, metformin, resveratrol and spermidine (1 μM, 1 mM, 25 μM and 0.1 μM, respectively) for 24 h before measuring the SBLC levels by qPCR. (B) SBLC levels in HCT116 cells measured by qPCR (left) along with the levels of c-Myc and p21 by blotting (right) after addition of 1 μM rapamycin. β-actin was included throughout as a loading control. (C) The experiment in (B) was repeated using A549 cells. (D) Schematic of predicted TF binding sites for E2F1, USF-1, MAZ, SP1 and CREB in SBLC. (E) A549 and HCT116 cells were treated with 1 μM rapamycin and the levels of TFs in (D) were measured by western blot. Relative levels of E2F1 were estimated using densitometric analysis. (F) Rapamycin was added to HCT116 cells for 24 h in which E2F1 had been depleted by shRNA along with respective controls. SBLC levels measured by qPCR (top) along with E2F1 by blotting (bottom). (G) The experiment in (F) was repeated on HCT116 cells after treatment with sh-SBLC. (H) Localization of the three intergenic E2F1 binding sites in SBLC (left). ChIP using E2F1 or IgG control assessed by RT-PCR analysis against B1 and B2 regions (right). (A–H) Results are representative of three independent experiments. Values are mean ± SEM. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t-test.
Initial investigations showed there was a time dependent increase in SBLC in response to rapamycin (Figure 6B and C, respectively) but the resulting changes were not caused by c-Myc. In addition to the presence of binding motifs for c-Myc (Figure 2C), there are potential binding motifs for a number of transcription factors within the SBLC promoter including E2F1, USF1, MAZ, SP1 and CREB1 (Figure 6D). Examination of these indicated that only E2F1 levels were responsive to rapamycin (Figure 6E). Moreover, depletion of E2F1 by shRNA significantly decreased SBLC expression in the context of both vehicle and rapamycin-treated cells (Figure 6F).
Treating cells with rapamycin served to diminish p21 to below basal levels; however, rapamycin did not antagonize the increased p21 expression resulting from SBLC silencing, implying that the presence of SBLC is required for the activity of rapamycin to suppress p21 (Figure 6G). Depleting SBLC had no effect on E2F1 levels indicative that there was no feedback loop, similar to the regulatory relationship observed between SBLC and c-Myc. Supporting the involvement of E2F1 as a transcriptional driver contributing to SBLC expression after rapamycin treatment, ChIP analysis confirmed that E2F1 could bind to SBLC through motifs in the intergenic region between exons 1 and 2 (Figure 6H).
Role of SBLC in OIS and RS
At last, we considered whether SBLC played roles in either oncogene-induced or RS. We first confirmed that transfection of the oncogenic V12 mutant of H-Ras in IMR90 cells resulted in premature senescence (Supplementary Figure S6A). Notably after H-RasV12 overexpression there was decreased SBLC along with increased p21 and p53 expression (Supplementary Figure S6B and C). Moreover, E2F1 and HDAC5 levels were decreased whereas no changes occurred in c-Myc expression.
We next assessed the expression and function of SBLC during RS in HAFF fibroblasts (Supplementary Figure S6D). Examination at 10 doubling intervals indicated that SBLC expression gradually decreased by ∼45% from 10 to 50 doublings whereas the levels of p21 and p53 progressively increased (Supplementary Figure S6E). In concert with these experiments, SBLC was transfected in HAFF cells to determine effects on RS. Maintaining high (70- to160-fold) ectopic SBLC expression throughout the experiment resulted in delayed induction of p21 expression but nevertheless similar levels of p21 were eventually evident at 50 cell doublings in both control and SBLC-overexpressing populations (Supplementary Figure S6F and G).
DISCUSSION
Senescence pathways converge on Rb family pocket proteins (36,37) and notwithstanding their importance, p21 lies directly upstream of Rb signaling and functions as a key determinant of senescence programming (9,10). Here we show lncRNA SENEBLOC functions to repress p21 gene expression through both p53-dependent and independent mechanisms. SENEBLOC promotes the assembly of p53 and MDM2 as a ternary complex, leading to ubiquitin-mediated degradation and low steady state p53 levels. In contrast, depletion of SENEBLOC profoundly increased cellular p53 levels and this was associated with the induction of senescence involving p21. While p21 is a classic p53-dependent mRNA, transcriptional upregulation of p21 by p53 was only partly responsible for increased p21 expression since epigenetic regulation was also identified. SENEBLOC acts as a decoy RNA to sponge miR-3175 that would otherwise bind HDAC5 mRNA 3′-UTR and reduce its translational efficiency. Higher levels of HDAC5 were shown to directly impact the chromatin structure of the p21 gene promoter, keeping it in a repressed transcriptional state whereas depleting SENEBLOC reversed this inhibition. It is presently unclear how frequently lncRNA functions converge on a single target or cellular process and indeed, such reported instances in the literature are few (26,38).
It is notable that HDAC5 decreased in response to SENEBLOC depletion. Histone deacetylase (HDAC) inhibitors (HDACi) can induce p21 expression in transformed cells (39) but the selective actions of HDAC5 in this context has not been reported. A prior study also utilising HCT116 cells determined that HDAC5 acted to exert effects on p21, not through epigenetic changes, but through effects on p53 activity and localization (40). They showed during early responses to genotoxic stress, HDAC5 bound to p53 and inhibited K120 acetylation, leading to p53-mediated transcription of p21. Unlike our observations, these authors did not find p21 levels were responsive to genotoxic stress in HCT116 p53−/− cells, perhaps reflecting different experimental contexts.
The cellular pools of SENEBLOC are divided into nuclear and cytoplasmic compartments which appear consistent with its dual modes of action. Complexes between p53/MDM2 are initiated in the nucleus before subsequent export and degradation (33) while competing endogenous RNA functions are typically played out in the cytosol (41). In the context of cancer lines, homeostatic levels of SENEBLOC appeared to be maintained via c-Myc transcription, whereas the enforced loss of SENEBLOC resulted in senescence, phenocopying the knockdown of c-Myc. Nevertheless, given the broad transcriptional program elicited by c-Myc (42) it remains a possibility that indirect mechanisms involving c-Myc target genes also contribute to SENEBLOC regulation.
It has been previously established that down-regulation of c-Myc can initiate senescence in a range of tumor types through a p53-dependent mechanism, resulting in tumor regression in vivo (15). Similarly our study defined a requirement for p53 along with the identification of an additional epigenetic mechanism as illustrated in our working model (Supplementary Figure S6H). One of the striking observations in the cancer context was the profound inhibition of tumor growth observed when SENEBLOC expression was inhibited. Conceptually, this proposes that maintaining SBLC expression would benefit tumor progression and accordingly, there were increases in the expression of SBLC in colon tumors compared to normal tissues. Loss or amplification of the SBLC locus has not been previously highlighted and whether gene amplification underlies the expression increases in colon cancers is not fully clear. Nevertheless, as with any proposed cancer biomarker, findings with cell lines need to be considered in each disease context. While senescence is often discussed in terms of its protective role in preventing cancer development, there is growing appreciation that the senescent state results in a proinflammatory microenvironment which may itself promote cancer formation (8,14). Our data propose the possible significance of SBLC to cancer eitiology but more work is needed to more precisely decipher its contribution.
We also found that irrespective of whether cells were transformed or not, reducing SENEBLOC expression also induced cellular senescence. Eliciting OIS caused reduced expression of SENEBLOC along with increased p53 and p21 levels as well as downregulation of HDAC5, consistent with the epigenetic regulatory mechanism. A progressive reduction in SENEBLOC levels also occurred during RS and SENEBLOC overexpression delayed p21 induction at early cell doublings. Thus SENEBLOC fulfils important roles during stress-associated senescence triggered by either loss of c-Myc or ectopic expression of oncogenes along with a contribution to RS. Nevertheless, as multiple mechanisms contribute to senescence induction and maintenance, it is likely that the relative importance of SENEBLOC is highly dependent on the cell/tissue type and the nature of the stimulus involved.
At last, our results with senolytic drugs produced intriguing findings. Rapamycin inhibits cell senescence through a multiplicity of mechanisms including suppression of p21 (25,43). Consistently, rapamycin decreased p21 expression along with augmenting SENEBLOC levels by ∼2.5-fold. Notably the increased SENEBLOC expression showed dependency on E2F1 expression but not c-Myc, thus proposing that steady state regulation of SENEBLOC requires c-Myc whereas E2F1-dependent regulation underpins its transactivation following rapamycin treatment. Depletion of SENEBLOC was sufficient to counteract the inhibitory effects of rapamycin on p21 levels with the clear implication that increased levels of SENEBLOC underpin the actions rapamycin to antagonize p21. As such the identification of SENEBLOC serves as exemplary case involving lncRNA-mediated regulation of an important anti-aging drug.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Zefeng Wang for the GFP translation reporter plasmid, Prof. Ping Gao for providing P493–6 cells carrying a c-Myc tet-off system and Prof. Yide Mei for the pRK5 constructs.
Author Contributions’: C.L.X., B.S., X.D.Z., R.F.T., G.Z.L., L.X.L. and M.W. designed the research. C.L.X., B.S. and J.M.L. performed the experiments and data analysis. R.F.T. and M.W. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology, China [2018YFA0107100]; National Natural Science Foundation of China [81820108021, 31871437, 81970153]; National Health and Medical Research Council of Australia (NHMRC) [1147271]. Funding for open access charge: Ministry of Science and Technology, China [2018YFA0107100].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 1965; 37:614–636. [DOI] [PubMed] [Google Scholar]
- 2. Wang E. Senescent human fibroblasts resist programmed cell death, and failure to suppress bcl2 is involved. Cancer Res. 1995; 55:2284–2292. [PubMed] [Google Scholar]
- 3. Wiley C.D., Campisi J.. From ancient pathways to aging cells-connecting metabolism and cellular senescence. Cell Metab. 2016; 23:1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Narita M., Nunez S., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W.. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003; 113:703–716. [DOI] [PubMed] [Google Scholar]
- 5. Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O. et al.. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coppe J.P., Patil C.K., Rodier F., Sun Y., Munoz D.P., Goldstein J., Nelson P.S., Desprez P.Y., Campisi J.. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008; 6:2853–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campisi J. Cancer, aging and cellular senescence. In vivo (Athens, Greece). 2000; 14:183–188. [PubMed] [Google Scholar]
- 8. Rodier F., Campisi J.. Four faces of cellular senescence. J. Cell Biol. 2011; 192:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McHugh D., Gil J.. Senescence and aging: causes, consequences, and therapeutic avenues. J. Cell Biol. 2018; 217:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Childs B.G., Durik M., Baker D.J., van Deursen J.M.. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat. Med. 2015; 21:1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ishikawa F. Cellular senescence, an unpopular yet trustworthy tumor suppressor mechanism. Cancer Sci. 2003; 94:944–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sharpless N.E., DePinho R.A.. Telomeres, stem cells, senescence, and cancer. J. Clin. Invest. 2004; 113:160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collado M., Serrano M.. Senescence in tumours: evidence from mice and humans. Nat. Rev. Cancer. 2010; 10:51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schosserer M., Grillari J., Breitenbach M.. The dual role of cellular senescence in developing tumors and their response to cancer therapy. Front. Oncol. 2017; 7:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu C.H., van Riggelen J., Yetil A., Fan A.C., Bachireddy P., Felsher D.W.. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:13028–13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhuang D., Mannava S., Grachtchouk V., Tang W.H., Patil S., Wawrzyniak J.A., Berman A.E., Giordano T.J., Prochownik E.V., Soengas M.S. et al.. C-MYC overexpression is required for continuous suppression of oncogene-induced senescence in melanoma cells. Oncogene. 2008; 27:6623–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ghanam A.R., Xu Q., Ke S., Azhar M., Cheng Q., Song X.. Shining the light on senescence associated LncRNAs. Aging Dis. 2017; 8:149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Montes M., Lund A.H.. Emerging roles of lncRNAs in senescence. FEBS J. 2016; 283:2414–2426. [DOI] [PubMed] [Google Scholar]
- 19. Puvvula P.K., Desetty R.D., Pineau P., Marchio A., Moon A., Dejean A., Bischof O.. Long noncoding RNA PANDA and scaffold-attachment-factor SAFA control senescence entry and exit. Nat. Commun. 2014; 5:5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yoon J.H., Abdelmohsen K., Kim J., Yang X., Martindale J.L., Tominaga-Yamanaka K., White E.J., Orjalo A.V., Rinn J.L., Kreft S.G. et al.. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 2013; 4:2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tripathi V., Shen Z., Chakraborty A., Giri S., Freier S.M., Wu X., Zhang Y., Gorospe M., Prasanth S.G., Lal A. et al.. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013; 9:e1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu Y., Tchkonia T., Fuhrmann-Stroissnigg H., Dai H.M., Ling Y.Y., Stout M.B., Pirtskhalava T., Giorgadze N., Johnson K.O., Giles C.B. et al.. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell. 2016; 15:428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herranz N., Gallage S., Mellone M., Wuestefeld T., Klotz S., Hanley C.J., Raguz S., Acosta J.C., Innes A.J., Banito A. et al.. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat. Cell Biol. 2015; 17:1205–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laberge R.M., Sun Y., Orjalo A.V., Patil C.K., Freund A., Zhou L., Curran S.C., Davalos A.R., Wilson-Edell K.A., Liu S. et al.. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat. Cell Biol. 2015; 17:1049–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang R., Yu Z., Sunchu B., Shoaf J., Dang I., Zhao S., Caples K., Bradley L., Beaver L.M., Ho E. et al.. Rapamycin inhibits the secretory phenotype of senescent cells by a Nrf2-independent mechanism. Aging Cell. 2017; 16:564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sang B., Zhang Y.Y., Guo S.T., Kong L.F., Cheng Q., Liu G.Z., Thorne R.F., Zhang X.D., Jin L., Wu M.. Dual functions for OVAAL in initiation of RAF/MEK/ERK prosurvival signals and evasion of p27-mediated cellular senescence. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E11661–E11670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen X., Han P., Zhou T., Guo X., Song X., Li Y.. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 2016; 6:34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ji Z., Song R., Regev A., Struhl K.. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015; 4:e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y. et al.. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017; 27:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alexander K., Hinds P.W.. Requirement for p27(KIP1) in retinoblastoma protein-mediated senescence. Mol. Cell. Biol. 2001; 21:3616–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gartel A.L., Radhakrishnan S.K.. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005; 65:3980–3985. [DOI] [PubMed] [Google Scholar]
- 32. Kastan M.B., Bartek J.. Cell-cycle checkpoints and cancer. Nature. 2004; 432:316–323. [DOI] [PubMed] [Google Scholar]
- 33. Nag S., Qin J., Srivenugopal K.S., Wang M., Zhang R.. The MDM2-p53 pathway revisited. J. Biomed. Res. 2013; 27:254–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Willis A., Jung E.J., Wakefield T., Chen X.. Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene. 2004; 23:2330–2338. [DOI] [PubMed] [Google Scholar]
- 35. Clayton A.L., Hazzalin C.A., Mahadevan L.C.. Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell. 2006; 23:289–296. [DOI] [PubMed] [Google Scholar]
- 36. Dannenberg J.H., van Rossum A., Schuijff L., te Riele H.. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000; 14:3051–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sage J., Mulligan G.J., Attardi L.D., Miller A., Chen S., Williams B., Theodorou E., Jacks T.. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000; 14:3037–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu W.L., Jin L., Xu A., Wang Y.F., Thorne R.F., Zhang X.D., Wu M.. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat. Cell Biol. 2018; 20:492–502. [DOI] [PubMed] [Google Scholar]
- 39. Gui C.Y., Ngo L., Xu W.S., Richon V.M., Marks P.A.. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sen N., Kumari R., Singh M.I., Das S.. HDAC5, a key component in temporal regulation of p53-mediated transactivation in response to genotoxic stress. Mol. Cell. 2013; 52:406–420. [DOI] [PubMed] [Google Scholar]
- 41. Rashid F., Shah A., Shan G.. Long Non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics. 2016; 14:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dang C.V., O’Donnell K.A., Zeller K.I., Nguyen T., Osthus R.C., Li F.. The c-Myc target gene network. Semin. Cancer Biol. 2006; 16:253–264. [DOI] [PubMed] [Google Scholar]
- 43. Llanos S., Garcia-Pedrero J.M., Morgado-Palacin L., Rodrigo J.P., Serrano M.. Stabilization of p21 by mTORC1/4E-BP1 predicts clinical outcome of head and neck cancers. Nat. Commun. 2016; 7:10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.