Abstract
During oocyte maturation, changes in gene expression depend exclusively on translation and degradation of maternal mRNAs rather than transcription. Execution of this translation program is essential for assembling the molecular machinery required for meiotic progression, fertilization, and embryo development. With the present study, we used a RiboTag/RNA-Seq approach to explore the timing of maternal mRNA translation in quiescent oocytes as well as in oocytes progressing through the first meiotic division. This genome-wide analysis reveals a global switch in maternal mRNA translation coinciding with oocyte re-entry into the meiotic cell cycle. Messenger RNAs whose translation is highly active in quiescent oocytes invariably become repressed during meiotic re-entry, whereas transcripts repressed in quiescent oocytes become activated. Experimentally, we have defined the exact timing of the switch and the repressive function of CPE elements, and identified a novel role for CPEB1 in maintaining constitutive translation of a large group of maternal mRNAs during maturation.
INTRODUCTION
Cell development relies on elaborate changes in gene expression in order to transition through different phenotypic and functional stages that ultimately lead to terminal differentiation. Changes in gene expression are achieved through transcriptional and post-transcriptional regulations. Although transcriptional regulation is understood in considerable detail (1,2), much less is known about the molecular machinery involved in translation regulation.
Large oligomeric complexes involving proteins and non-coding RNAs are assembled on the mRNA (3) to regulate its interaction with ribosomes, its translation rate, and its stability (4,5). In somatic cells, numerous observations indicate that translation is intimately coupled with degradation of mRNAs (5,6). Proteins recruited to the mRNA interact with elements located throughout the length of the transcript (3,7). However, complexes nucleated around the 5′ and 3′ untranslated regions (UTRs) play a predominant role in translation and stabilization, often by controlling the length of the poly(A) tail, which is present in most mRNAs (4,8). Particularly, in gametes and embryos the poly(A) tail determines the translation rate and stability of the mRNA (9–14).
Germ cells are unique in their properties as they progressively acquire specialized functions during development (14). At the same time, they maintain traits that allow for rapid transition to totipotency (15). Throughout development, germ cells often rely on unique post-transcriptional regulations rather than on transcription itself (14,16). Striking examples of this property are the growth and maturation stages of an oocyte and its transition to zygote and early embryo (13,14). During the growth phase, oocytes amass a large number of maternal mRNAs through high transcriptional activity. These mRNAs are either used immediately to synthesize proteins involved in growth or are stored for future use. Indeed in all species studied, transcription ceases when an oocyte is fully grown and resumes only in the embryo. Thus, critical steps in oocyte maturation and early embryo development rely exclusively on a program of maternal mRNA translation.
Some properties of the molecular machinery involved in maternal mRNA translation repression or activation have been elucidated in model organisms (13,16,17). In frogs, the cytoplasmic polyadenylation element-binding protein (CPEB) is considered a master regulator of polyadenylation and translation (18,19). Much less is known about the role of CPEB in mammalian oocytes. Here, we have used a genome-wide approach to investigate the role of this RNA-binding protein (RBP) during the transition from quiescence to re-entry into meiosis. Through a detailed time course, we have investigated the temporal association between maternal mRNA translation and the different steps involved in oocyte re-entry into and progression through meiosis. Using a RiboTag/RNA-Seq strategy, we describe a genome-wide switch in the translation program of maternal mRNAs, and define new, critical functions of CPEB in the control of this switch.
MATERIALS AND METHODS
Animals
All experimental procedures involving mice were approved by the University of California, San Francisco Institutional Animal Care and Use Committee (Approval #AN163021-03C). Animal care and use were performed according to relevant guidelines and regulations. All animals used were of the C57BL/6J inbred strain. C57BL/6-Zp3cre-Rpl22tm1.1Psam (Zp3-CreT RiboTagF/F) mice were obtained from Jackson Laboratories and bred as previously described (20). Cpeb1-targeted mice were a gift from Raúl Méndez et al. (21) and bred in our laboratory.
Oocyte isolation and culture
Three-week old female mice were injected with 5 I.U. PMSG to induce superovulation. Forty-four hours after injection, the mice were euthanized and the ovaries dissected into media containing HEPES and 1 μM cilostamide (Millipore, 231085) (HC media). Antral follicles were punctured, allowing release of cumulus-oocyte complexes (COCs). Repeated aspiration through a glass pipette allowed for removal of the surrounding cumulus cells. Denuded oocytes were maintained at prophase I arrest (Pro I) in MEM Alpha (Gibco, 12561-056) supplemented with sodium pyruvate (Gibco, 11360070) and penicillin-streptomycin (Gibco, 15140122) in addition to 1 μM cilostamide (αC media) at 37°C under 5% CO2. If indicated, oocytes were transferred to cilostamide-free MEM Alpha (α media), allowed to mature, and collected at various time points. Where specified, oocytes were treated with 5 μM dinaciclib (Selleckchem, SCH727965), 10 μM Ro-3306 (Selleckchem, S7747), or 10 mM Rp-cAMPS.
Immunofluorescence staining and confocal microscopy
Oocytes were collected at various time points and fixed in DPBS (GE Healthcare, SH30264.02) supplemented with 0.1% Triton X-100 (Sigma, X-100) and 2% formaldehyde (ThermoFisher, 28908) for 30 min. After washing in blocking buffer (1× DPBS, 0.3% BSA and 0.01% Tween), the oocytes were incubated in blocking buffer for 16 h and permeabilized for 15 min in DPBS supplemented with 0.3% BSA and 0.1% Triton X-100. Samples were washed and then incubated for 1 h with primary antibody diluted in blocking buffer. The antibodies used were: 1:100 β-tubulin (9F3) rabbit mAb (Cell Signaling Technology, 3623); 1:200 human antibody against centromere (ImmunoVision, HCT-0100). After another round of washing, samples were incubated for 1 h with the corresponding secondary antibody, 1:500 goat anti-rabbit IgG, Alexa Fluor 488 (ThermoFisher, A-11008) or 1:500 goat anti-human IgG, Alexa Fluor 568 (ThermoFisher, A-21090). Oocytes were washed again and then mounted with VECTASHIELD® Antifade Mounting Medium with DAPI (Vector, H-1200). All washes were done three times each round in blocking buffer for 10 min each wash. Images were captured with a confocal Nikon C1SI equipped with ×60 oil immersion lens and processed with ImageJ (22).
RiboTag-immunoprecipitation (RiboTag-IP)
Only Zp3-CreT RiboTagF/F female mice were used for RiboTag-immunoprecipitation. Oocytes were collected in 5 μl 0.1% polyvinylpyrrolidone (PVP; Sigma, P0930) in 1x PBS (Invitrogen, AM9625), flash frozen in liquid nitrogen, and stored at −80°C.
The appropriate volume (50 μl per sample) of Dynabeads™ Protein G (Invitrogen, 10004D) was washed three times in 500 μl homogenization buffer (HB: 50 mM Tris–HCl pH 7.4, 100 mM KCl, 12 mM MgCl2 and 1% NP-40) on a rotor at 4°C for 5 min per wash. Two additional washes were performed with 500 μl supplemented HB (sHB: HB supplemented with 1 mM DTT, 1× protease inhibitors, 200 units/ml RNaseOUT, 100 μg/ml cycloheximide and 1 mg/ml heparin) on a rotor at 4°C for 10 min per wash. The final wash solution was removed and the beads were eluted in the original volume of sHB. Samples were thawed, randomly pooled to yield a total of 200 oocytes per time point per replicate, and 300 μl sHB was added to each pooled sample. To lyse the cells, samples were vortexed for 30 s, flash frozen in liquid nitrogen, and thawed at room temperature (RT); this process was repeated twice. Finally, the homogenates were centrifuged for 10 min at maximum speed and 4°C and the supernatants were transferred to new tubes. To pre-clear the samples, 20 μl washed beads was added to each supernatant and samples were incubated on a rotor at 4°C for 1 h. A magnetic rack was used to remove the beads and 15 μl of each pre-cleared lysate was collected and added to 250 μl RLT buffer per sample (Qiagen, 74034) to serve as the input samples. The input samples were frozen and kept at −80°C until RNA extraction. Three μl (3 μg) anti-HA.11 epitope tag antibody (BioLegend, 901501) was added to each of the extracts and all samples were incubated on a rotor at 4°C for 4 h. Thirty microliter washed beads was then added to the samples and incubated overnight on a rotor at 4°C. The beads (now bound by HA-tagged ribosomes and the associated mRNAs) were washed five times in 1 ml urea wash buffer (uWB: 50 mM Tris–HCl pH 7.4, 150 mM KCl, 12 mM MgCl2, 1% NP-40, 1× protease inhibitors, 1 mM DTT, 40 U RNaseOUT, 1 mg/ml heparin, 100 μg/ml cycloheximide, and 1 M urea) on a rotor at 4°C for 10 min per wash. The beads were then pelleted via a magnetic rack and the uWB removed. Two-hundred and fifty microliters RLT buffer was added to each sample and samples were vortexed for 30 s. RNA extraction was performed using the Rneasy Plus Micro Kit (Qiagen, 74034). Samples were eluted in 10 μl of RNase-free water and used downstream for RNA-Seq or RT-qPCR analysis.
RNA-Seq
Library preparation and sequencing
RiboTag RNA samples were sent to the Gladstone Institutes Genomics Core for quality control using Bioanalyzer (Agilent) and cDNA library preparation. cDNA was prepared with the Ovation RNA-Seq System V2 Kit (NuGEN, 7102), which uses a proprietary combination of enzymes and primers to preferentially prime non-rRNA sequences. First-strand cDNA synthesis is achieved with random hexamer primers and chimeric poly(T) primers, which bind to the region spanning the end of the 3′UTR and the start of the poly(A) tail. Because stable mRNAs have at least 20 adenosine residues, use of these chimeric poly(T) primers allows for preferential priming of adenylated messages (mRNAs) with minimal bias for messages with longer poly(A) tails. RNA-Seq libraries were constructed using the Ovation Ultralow System V2 (NuGEN, 0344) and analyzed by Bioanalyzer, quantified by qPCR (Kapa Biosystems, KR0405), and sequenced using the HiSeq 4000 system (Illumina).
Sequence quality assessment and trimming
The quality of the raw sequence data was checked via FASTQC. The sequence files were then trimmed using tools in Trimmomatic-0.36 (23). The following were removed: Illumina TruSeq3 single-ended adapter sequences, bases with a quality score lower than three at the start and end of a read, bases that had an average quality per base of <15 calculated using a sliding window to average four bases, and any reads that were shorter than 36 bases. The reads were single-ended and qualities were ASCII characters equal to the Phred quality plus 33.
Mapping and counting reads
HiSat2 (24) was used to build indexes from the Reference Consortium Mouse Build 38 (mm10) and to align sequence reads to the genome. The resulting .bam files were sorted and indexed with SAMtools (25). Count files for each group were created with HTSeq using the Mouse GENCODE Gene set release M11. The inputted data were .bam files, the data were not from a strand-specific assay and the feature type used was ‘gene’.
Differential expression (DE) analysis
The Bioconductor packages edgeR (26) and limma (27) were used for statistical analyses. Only reads with greater or equal to 10 counts per million (CPM) in at least two samples were kept. Trimmed mean of M-values (TMM) normalization, which accounts for compositional differences among the libraries, was then performed on HA reads and input reads, separately. Using the raw counts, dispersion, and design matrix, the negative binomial generalized linear model was fitted for each gene. Finally, pairwise likelihood ratio tests for 2, 4, 6 and 8 h versus 0 h were conducted.
Gene ontology (GO) analysis
Gene lists were uploaded to DAVID 6.8 (28,29) and processed with the Functional Annotation Tool.
Analysis of 3′UTR sequences
The 3′UTR sequences of the genes of interest (including known mRNA isoforms) were downloaded using the Table Browser (UCSC Genes track) provided by GBShape (30). The locations of putative PAS (AATAAA, ATTAAA and AAGAAA) (31) and CPE (TTTTAT, TTTTAAT, TTTTACT, TTTTAAAT, TTTTAAGT and TTTTCAT) (32) sequences were determined via the Find Individual Motif Occurrences tool (33) that is part of the MEME Suite (34); only exact matches were used for downstream analysis. Python scripts were written to calculate the distance of each CPE from each PAS for individual 3′UTRs.
Data visualization
ggplot2 (35) was used to create the multi-dimensional scaling (MDS) plots used to visualize sample-to-sample distances, and translation activation and repression time course plots. All other data were plotted using GraphPad Prism 8.
RNA immunoprecipitation (RNA-IP)
The appropriate volume (50 μl per sample) of Dynabeads™ Protein G (Invitrogen, 10004D) was washed twice in 250 ul incomplete lysis buffer (iLB: 15 mM Tris–HCl pH 7.4, 75 mM NaCl, 5 mM MgCl2, 0.25% NP-40, 0.125 mM Na3VO4 and 5 mM β-glycerophosphate) on a rotor at 4°C for 5 min. Two additional washes were performed with 250 μl complete LB (cLB: iLB supplemented with protease inhibitor, DTT, RNAseOUT, ribonucleoside vanadyl complex and cycloheximide) on a rotor at 4°C for 5 min per wash. The final wash solution was removed and the beads were eluted in the original volume of cLB. Oocytes were isolated as described and kept arrested at Pro I. Two hundred oocytes were collected in 5 μl 0.1% PVP in PBS, flash frozen in liquid nitrogen, and stored at −80°C. To homogenize the cells, 250 μl of cLB was added to the samples, samples were vortexed for 30 secs, and then incubated on ice for 10 min. The homogenates were then centrifuged for 10 mins at maximum speed at 4°C and the supernatants were transferred to new tubes. Fifteen μl of each supernatant was saved in 250 μl RLT buffer as input samples and the rest were equally aliquoted for the CPEB1-IP and the IgG-IP (control). The volume of each sample was increased to 300 μl with cLB, 2 μl (2 μg) of the appropriate antibody (anti-CPEB: Abcam, ab73287 and control IgG: Abcam, ab172730) was added to each tube, and samples were incubated on a rotor for 2 h at 4°C. Thirty microliter washed beads were then added to each tube and samples were incubated on a rotor overnight at 4°C. The beads of each sample were pelleted using a magnetic rack and washed 5 times on a rotor at 4°C with 750 ml wash buffer (WB: 30 mM Tris–HCl pH 7.4, 200 mM NaCl, 10 mM MgCl2, 0.5% NP-40, 0.25 mM Na3VO4, 10 mM β-glycerophosphate, 1× protease inhibitor, 1 mM DTT, 1 M urea and 1× RNase out) for 10 min each wash. The beads were then pelleted and the WB removed. Two hundred and fifty microliters RLT buffer was added to each sample and the samples were vortexed for 30 s. RNA extraction was performed following the Rneasy Plus Micro Kit protocol. Samples were eluted in 10 μl of RNase-free water and used for downstream RT-qPCR analysis.
Real-time quantitative PCR (RT-qPCR)
Extracted RNA was reverse-transcribed using the SuperScript™ III First-Strand Synthesis System with random hexamer primers (Invitrogen, 18080051) and the resulting cDNA was diluted with RNase-free water. Gene expression was measured using TaqMan Assays™ and TaqMan™ Fast Advanced Master Mix (ThermoFisher, 4444557). The assays used were: Astl (Mm00553165_m1), Bcl2l10 (Mm00478988_m1), Ccnb1 (Mm03053893_gH), Cdk8 (Mm01223097_m1), Depdc7 (Mm00522683_m1) Dnmt (Mm01151063_m1), Dppa3 (Mm01184198_g1), Ewsr1 (Mm01191469_g1), Ing3 (Mm00458324_m1), Mos (Mm01700521_g1), Nlrp5 (Mm01143609_m1), Obox5 (Mm00773197_gH), Oosp1 (Mm00504796_m1), Oosp2 (Mm03015599_m1), Padi6 (Mm00462201_m1), Smc4 (Mm00713073_m1), Tcl1 (Mm00493475_m1), Tiparp (Mm00724822_m1), Zp1 (Mm00494367_m1), Zp2 (Mm00442173_m1) and Zp3 (Mm00442176_m1). Ten microliters reactions were run on the QuantStudio 6 Flex Real-time PCR System (Applied Biosystems). Gene expression was quantified via the 2−ΔΔCt method and statistical analysis was performed via GraphPad Prism 8.
Construction of florescent protein reporters
The 3′ UTR sequences of Ccnb1, Ccnb2 short, Ewsr1, Mos, Oosp1, Oosp2, Smc4 and Zp2 were retrieved from the RNA-Seq .bam files using the Integrative Genomics Viewer (Table S1). Primers were used to amplify the target 3′ UTRs from oocyte cDNA and portions of YPet-containing vector. Using the Choo-Choo Cloning™ Kit (MCLAB, CCK-20), the PCR fragments were fused together and transfected into competent 5-α Escherichia coli cells. The DNA plasmids of ampicillin-resistant bacteria were extracted using the QIAprep Spin Miniprep Kit (Qiagen, 27106). The sequences were then confirmed via DNA sequencing. The extracted plasmid was linearized using a forward primer upstream of the YPet sequence and a reverse primer with 20 additional thymine residues. The PCR product was then transcribed in vitro using the mMESSAGE mMACHINE T7 Transcription Kit (Invitrogen, AM1344) and the resulting cRNA was purified using the MEGAclear™ Transcription Clean-Up Kit (Invitrogen, AM1908); cRNA were eluted in RNase-free water and kept at −80°C. The mCherry reporter was similarly produced. However, the transcripts contained no 3′UTR, and instead was polyadenylated (150–200 nts) using the Poly(A) Tailing Kit (Invitrogen, AM1350).
Oocyte microinjection
Oocytes were collected as described and allowed to recover in αC media for 2 h, after which they were transferred into HC media for microinjection. Oocytes were injected with 5–10 pl of a 12.5 ng/μl solution of the YPet reporter of interest mixed with mCherry mRNA and allowed to recover in αC media for the specified amount of time before live cell imaging.
Live cell imaging and fluorescence microscopy
Live cell imaging experiments were performed using a Nikon Eclipse T2000-E equipped with mobile stage and environmental chamber at 37°C and 5% CO2. Filter set: dichroic mirror YFP/CFP/mCherry 69008BS; YFP channel (Ex: S500/20 × 49057; Em: D535/30 m 47281), mCherry channel (Ex: 580/25 × 49829; Em: 632/60 m). Images were processed and fluorescence was quantified using MetaMorph (Molecular Devices).
Poly(A) tail length (PAT) assay
This assay was performed as previously described (36).
Histology
Ovaries were dissected from 8-week-old female mice, fixed in Bouin's solution, and preserved in 70% ethanol. Tissues were then processed, cut at 8 μm, and stained (H&E) by the Cancer Center Tissue Core at UCSF.
Western blot
Oocytes were collected in 0.1% PVP in DPBS and boiled for 3 min at 95°C in 1× Laemmli Sample Buffer (Bio-Rad, 161-0747) supplemented with β-mercaptoethanol, proteinase inhibitor and phosphatase inhibitor. Samples were resolved on a 10% Laemmli gel and transferred onto supported nitrocellulose membranes, 0.2 μm (Bio-Rad, 1620097). Membranes were incubated in 5% blocking buffer for 1 h then incubated for 18 h in primary antibody at 4°C. The antibodies used: 1:1000 rabbit anti-CPEB1 (Abcam, ab73287), 1:1000 rabbit anti-CDC27 (Abcam, ab129085), 1:1000 mouse anti-CCNB1 (Abcam, ab72), 1:1000 mouse anti-STAT3 (Cell signaling, #3139), 1:1000 goat anti-CCNB2 (R&D Systems, AF6004), 1:10 000 rabbit anti-CPEB1 (Abcam, ab181051) and 1:1000 rabbit anti-ZAR1 (Bioss, bs-13549R). The membrane was then washed in 1× TBST, incubated in the appropriate secondary antibodies, 1:10 000 anti-rabbit IgG (GE Healthcare, NA934V), 1:10 000 anti-mouse IgG (GE Healthcare, NA931V), and 1:10 000 anti-goat IgG (TermoFisher, 31400) for 2 h, and washed again in 1× TBST. Clarity Western ECL substrate (Bio-Rad, 1705061) was then used to develop the membrane. All washes were done four times each round in TBST for 10 min each wash.
RESULTS
Re-entry into meiosis coincides with rapid translational changes of stable mRNAs
We have used a RiboTag/RNA-Seq strategy to characterize mRNA translation in oocytes arrested at prophase I (Pro I), as well as those undergoing meiotic maturation (Figure 1A). This strategy has been previously validated by us using RiboTag/qPCR, luciferase reporter assays, and Western blots for proteins encoded by the regulated mRNAs (20,36–38). Additional quality controls are included here (Supplementary Figure S1A–D). Upon meiotic resumption, we detect both progressive increases and decreases in ribosome loading on maternal mRNAs (Figure 1B). By metaphase I (Met I) and using a pairwise comparison with Pro I, we identified three functionally distinct translation patterns: mRNAs constitutively translated (n = 4284, CONSTITUTIVE), translationally repressed (n = 1722, DOWN), or translationally activated (n = 1537, UP) (FDR ≤ 0.05, Supplementary Figure S1E). Nearly identical results were obtained when the statistical analysis was applied to the entire time course (ANOVA f-test with five time points in duplicate) (FDR < 0.05, Supplementary Figure S1G). Changes in translation for 34 mRNAs are consistent with protein accumulation during meiosis monitored by published (Supplementary Figure S2A) as well as our western blots (Supplementary Figure S2B). Confirming the multidimensional scaling plots (Supplementary Figure S1A and B), total mRNA levels (input) remained stable up to late pro-metaphase I and significant destabilization was detectable only for 3% of maternal mRNAs at Met I (Figure 1B and Supplementary Figure S1G–H). Comparison of changes in total mRNA levels (transcriptome) to changes in ribosome-associated mRNA levels (translatome) confirms this late MI destabilization (Figure 1C). Changes in translation that initiate during MI were extended into and amplified at MII (Figure 1D); similarly, the pool of destabilized and repressed mRNA becomes prominent at the MII arrest (Figure 1D). Yet, a subset of mRNAs remained stable even if their translation were repressed (Figure 1D), indicating additional delayed waves of destabilization. Furthermore, we found a robust, positive correlation between translational changes at late MI and MII (Supplementary Figure S1G). Therefore, the patterns of differential ribosome loading are consistent across multiple in vitro and in vivo biological replicates and across two distinct detection platforms.
Figure 1.
The translational program during oocyte meiotic cell cycle involves both translational repression and activation. (A) Spindle and chromatin conformation in the oocyte during meiosis. Oocytes were matured in vitro and fixed at 0, 2, 4, 6 and 8 h post-meiotic resumption. Immunofluorescence staining for tubulin (green), kinetochores (red) and chromatin (blue) was performed. Maturing oocytes either presented chromosome condensation, but no spindle assembly (2 h, early pro-metaphase I), visible initial spindle formation (4 h, mid pro-metaphase I), progressive spindle formation with kinetochore attachment (6 h, late pro-metaphase I), or a fully attached, Met I-bipolar spindle (8 h, Met I). (B) Total mRNA levels and differential ribosome loading during meiotic progression as compared to Pro I. Oocytes were matured in vitro and collected at 0, 2, 4, 6 and 8 h post-meiotic resumption. Total RNA samples were collected prior to RiboTag-IP for each time point. cDNA libraries were prepared from total and ribosome-bound RNA samples, RNA-Seq was performed, and the data processed and analyzed as described in ‘Materials and Methods.’ The data are presented as volcano plots with log2(fold change) (LFC) CPM at each time point compared to 0 h and plotted against false discovery rate (FDR). Statistically significant increased (red) and decreased (blue) (FDR ≤ 0.05) genes are reported as well as non-significant changes (gray). –1 ≥ LFC ≥ 1 are considered biologically significant and are marked by dashed lines. Two biological replicates of 200 oocytes per time point were used for this experiment. (C) Changes in the transcriptome and translatome during meiosis. Met I data are derived from the experiment described in (B), Met II translation data are from a deposited dataset generated from oocytes matured in vivo followed by polysome fractionation/microarray (polysome array) (49,65), and Pro I-to-Met II total mRNA data were from a deposited dataset (66). The data are reported as scatterplots with LFC in total mRNA CPM at either Met I or Met II compared to Pro I versus the LFC of ribosome-bound mRNA CPM at the same time points. We identified four groups of messages: transcripts that showed significant changes only in translation (purple), significant changes only in total message levels (orange), no significant changes in translation nor in total transcript levels (gray), and significant changes in both translation and total transcript levels (black); significant changes are defined as FDR ≤ 0.05. Two biological replicates of 200 oocytes per time point were used to generate the RNA-Seq data, while six biological replicates of 500 oocytes per time point were used to generate the polysome array data. (D) Overlap of translatome changes between Met I and Met II. Both DOWN (blue) and UP (red) genes were analyzed. The data were collected as described in (C). –1 ≥ LFC ≥ 1 with FDR ≤ 0.05 are considered statistically significant.
It is widely accepted that destabilization of maternal mRNAs is an obligatory step in the reprogramming of gene expression at the oocyte-to-zygote transition (13). Although methylation of RNA had been known for five decades (39–41), only recently the functional significance of these modifications has come into focus (42). Methylation of mRNA at specific sites plays a role in pre-mRNA splicing, mRNA export, mRNA translation and stability (42). Genetic inactivation of the m6-methyl reader Ythdf2 in both zebrafish and mice causes female infertility (43,44). It has been proposed that defective destabilization of maternal mRNAs is the cause of the phenotype. To determine the mechanisms underlying the wave of destabilization detected at 8 hrs, we compared our set of destabilized mRNAs with the transcriptomic signatures associated with deletion of Ydhtf2 in mouse oocytes (43). We found minimal overlap, suggesting that the two processes are unrelated (Supplementary Figure S3A and B). Additional components involved in maternal mRNA destabilization include CNot6l, a component of the CCR4 complex (45), and Btg4, a member of the TOB family of proteins that interacts with CNOT7/8 and is required for mRNA destabilization in MI (46–48). Whereas little similarity was found with Cnot6l targets (Supplementary Figure S3C and D), we detect a significant overlap between the mRNAs destabilized at 8 hrs and those mRNAs stabilized in MII in the Btg4−/− mouse (Supplementary Figure S3E and F). Thus, CNOT7/CNOT8-mediated deadenylation is a likely cause of the decreased stability of the mRNA we detect at the end of Met I.
Divergent mechanisms control gene expression during mitosis and meiosis
Repression of maternal mRNA translation during meiosis is associated predominantly with mitochondrial and ribosomal biogenesis, while translationally activated mRNAs code for proteins with functions related to cell cycle and embryo development (49). Gene ontology (GO) analysis of UP and DOWN transcripts at Met I reinforces this association (Figure 2A). Genome-wide comparison of our RNA-Seq data to those available for translation and transcription during mitosis in HeLa cells synchronized at S or M phase (50) did not reveal any significant correlation (Figure 2B), suggesting profound differences in gene expression regulation during these processes. When the comparison between mitosis and meiosis is restricted to genes specific to cell cycle function, only six mRNAs overlap in translation activation between mitosis and meiosis (Figure 2C). However, a sizable group of mRNAs whose translation is activated during meiosis instead is activated transcriptionally at the S-to-M-phase transition. Although limited, overlap is also detected when translation repression during meiosis is compared with changes in translation during mitosis. Manual curation of the data from multiple datasets confirms that decreased translation of Cdk1 and increased translation of Bub1b occur during meiosis (Supplementary Figure S4).
Figure 2.
Cell cycle components are regulated via translation in meiosis, but via transcription in mitosis (A) Gene ontology analysis of DOWN and UP genes. DOWN (blue) and UP (red) mRNAs significantly changed from 0 h (Pro I) to 8 h (Met I) post-meiotic resumption were used (–1 ≥ LFC ≥ 1 and FDR ≤ 0.05). Only terms with FDR ≤ 0.05 were considered. (B) Pairwise comparison of translation during meiosis and transcription during mitosis. FCs in translational efficiency (TE) from 0 h (Pro I) to 8 h (Met I) in our RNA-Seq dataset are plotted against FCs in RNA levels from S-phase to M-phase of a deposited dataset (50). (C) Heat maps comparing fold changes in translation of cell cycle components during meiosis in oocytes and fold changes in transcription or translation during the mitotic cell cycle. The data were collected as described in (B). Genes involved in the cell cycle are defined under GO:0007049.
A switch in the pattern of maternal mRNA translation is detected at the time of meiotic re-entry
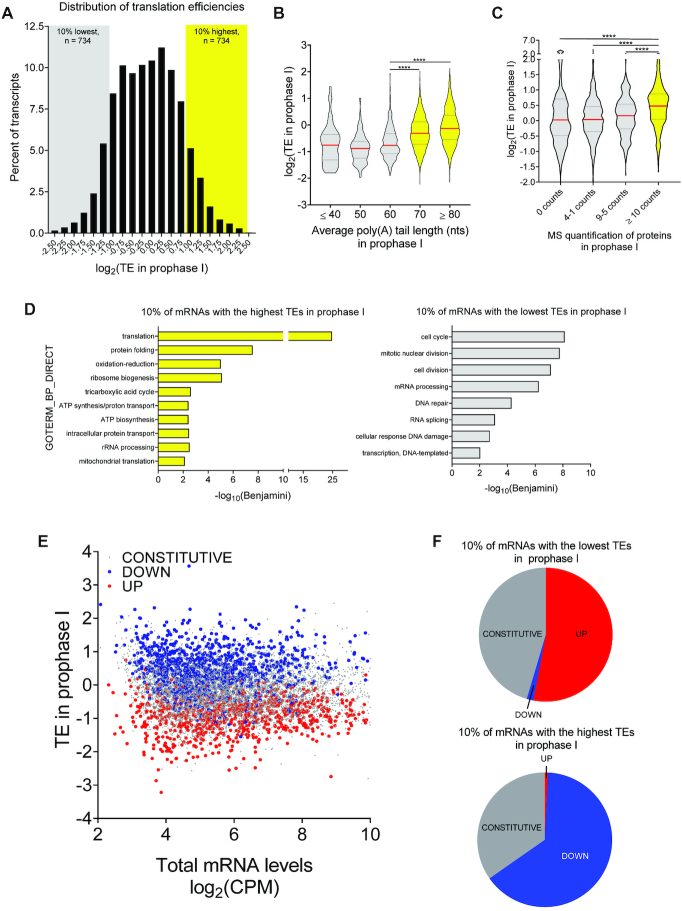
During Pro I, maternal messages display a broad spectrum of translation efficiencies (TEs) (Figure 3A), defined as the ratio between ribosome-bound and total mRNA levels (51); there is a seven-fold difference when comparing the average TE of the 10% of mRNAs with the highest TEs (high-TEs) to that of the 10% with the lowest TEs (low-TEs). To validate that TE reflects rate of translation, we related these values to other available measurements in Pro I-arrested oocytes (52,53). High-TE messages have significantly longer poly(A) tails (≥70 nts) (Figure 3B) and are associated with increased protein accumulation as assessed by mass spectrometry (Figure 3C).
Figure 3.
Genome-wide analysis of translation reveals a switch in the translation program at the quiescence-to-meiotic cell cycle re-entry transition (A) Histogram of translational efficiencies in Pro I-arrested oocytes. Translation efficiency (TE) for individual mRNAs was calculated as the ratio between ribosome-associated and total mRNA CPMs. Plotted is the distribution of maternal mRNA TEs during Pro I. The 10% of mRNAs with the lowest TEs are designated as low-TE mRNAs (n = 734, gray box) and the 10% of mRNAs with the highest TEs as high-TE mRNAs (n = 734, yellow box); this definition is used for all subsequent comparisons. (B) Genome-wide relationship between TE and poly(A) tail length in Pro I-arrested oocytes. TE was calculated for individual mRNAs as described in (A). Deposited TAIL-Seq data on poly(A) tail length of maternal mRNAs during Pro I (52) were associated with TEs during this time. Median values are represented by red lines and the 25% and 75% quartiles are represented by black, dashed lines. Statistical significance was evaluated by unpaired, two-tailed t-tests. ****P < 0.0001. (C) Genome-wide relationship between TE and protein levels in Pro I-arrested oocytes. TE was calculated for individual mRNAs as described in (A). Deposited data on protein levels in Pro I-arrested oocytes as quantified by mass spectrometry (53) were compared to the TEs of maternal mRNAs. Median values are represented by red lines and the 25% and 75% quartiles are represented by black, dashed lines. Statistical significance was evaluated by unpaired, two-tailed t-test; ****P < 0.0001. (D) Gene ontology analysis of low- and high-TE maternal mRNAs in Pro I-arrested oocytes. Only terms with a Benjamini coefficient ≤0.05 were considered. (E) Genome-wide relationship between TE in Pro I and translation pattern during meiotic resumption of oocyte maternal mRNAs. TE was calculated for individual mRNAs as described in (A). The data are presented as a scatterplot of total mRNA CPMs compared to TE values in Pro I-arrested oocytes. Transcripts were then categorized as CONSTITUTIVE (gray), DOWN (blue) or UP (red) according to their translation pattern during maturation to Met I (–1 ≥ LFC ≥ 1 and FDR ≤ 0.05). (F) Detailed analysis of the relationship between TE in Pro I and translation pattern during meiotic resumption of low- and high-TE mRNAs. Pie charts report the percentage of low- or high-TE mRNAs in Pro I-arrested oocytes that are UP, DOWN or CONSTITUTIVE during meiotic maturation. Ninety-nine percent of low-TE mRNAs are either UP (53%) or CONSTITUTIVE (46%) and all the high-TE mRNAs are either DOWN (65%) or CONSTITUTIVE (35%).
GO analysis of low- and high-TE messages during Pro I reveals associations antithetical to those found during meiotic maturation (Figure 2A). Functions important for oocyte growth are significantly enriched for high-TE mRNAs, while functions important during oocyte maturation are enriched in low-TE mRNAs (Figure 3D). Thus, we hypothesize that, upon meiotic resumption, a switch in the translation program occurs in order for the oocyte to progress through meiosis and prepare for embryogenesis. Indeed, transcripts with greater TEs become translationally downregulated (DOWN), while transcripts with lower TEs become activated (UP) during meiotic maturation (Figure 3E). More quantitative analysis reveals that translation of 99% of low-TE mRNAs is either constitutive or upregulated during meiotic maturation, while translation of virtually all of high-TE mRNAs is either constitutive or repressed (Figure 3F).
Unique mRNA features are associated with the opposing translation patterns in prophase I-arrested oocytes
To understand how such a broad array of TEs is established in Pro I-arrested oocytes, we performed a genome-wide correlation between these data and various mRNA features (Figure 4A). In the 5′UTR, ATG density, GC content, and UTR length are inversely related with TE. As for the 3′UTR, polyadenylation signal (PAS) density and GC content are positively correlated, whereas DAZL-binding element density, UTR length, and cytoplasmic polyadenylation element (CPE) density are all inversely correlated with TE. Detailed analysis confirms this significant inverse relationship between CPE density and TE, as 87% of low-TE mRNAs contain putative CPEs in the 3′UTR, while this is only true for 57% of high-TE mRNAs (Figure 4B). The presence of CPEs in mRNAs translated at high rates might be due to their position relative to the PAS in the 3′-UTR. Indeed, CPEs present 200–300 nucleotides away from the PAS are associated with higher TEs, while CPEs near the PAS are associated with lower TEs (Figure 4C and D).
Figure 4.
Features associated with maternal mRNAs translated with high or low efficiency in prophase I-arrested oocytes (A) Genome-wide correlation between mRNA features with TE in Pro I-arrested oocytes. PAS density in the 3′UTR, GC content in the 3′ and 5′UTRs, ATG density in the 5′UTR, DAZL and CPEB cis-acting element densities in the 3′UTR, and 3′UTR and 5′UTR lengths were calculated as detailed in the ‘Methods and Materials.’ These data were then correlated with TEs during Pro I and Spearman correlation coefficients were calculated for every comparison; P < 0.0001 for all pairs. In mRNAs with higher TEs, the reduced number of 3′UTR cis-acting elements is not due to shorter 3′UTR length, as element number was normalized for 3′UTR length when calculating densities. (B) Detailed analysis of the relationship between TE in Pro I and presence of CPEs in the 3′UTR of low- and high-TE mRNAs. Pie charts report the percentage of low- or high-TE mRNAs in Pro I-arrested oocytes where a CPE could be identified. Scanning for CPE in the 3′ UTRs was performed as detailed in the ‘Materials and Methods.’ (C) Genome-wide relationship between TE and number of CPEs found within 100 nts of the PAS in Pro I-arrested oocytes. Median values are represented by red lines and the 25% and 75% quartiles are represented by black, dashed lines. Statistical significance was evaluated by unpaired, two-tailed t-tests; *P = 0.0313; ****P < 0.0001. (D) Detailed analysis of the relationship between TE in Pro I and the distance of the closest CPE to the PAS. Median values are represented by red lines and the 25% and 75% quartiles are represented by black, dashed lines. (E) Enrichment of low-TE mRNAs bound to CPEB1 in Pro I-arrested oocytes. Pro I-arrested oocytes were collected and RNA-IP followed by RT-qPCR was performed as described in the ‘Materials and Methods.’ Nlrp5 was used as a reference gene as it is known to not bind to CPEB1. Three biological replicates of 200 oocytes per time point were used and RT-qPCR reactions were run in triplicate. Data are presented as fold difference in mRNA levels in CPEB1-IP as compared to the IgG-IP. The bars represent the mean ± SEM of three experiments. TE and the number of putative CPEs for each gene are reported. *The Mos 3′UTR has a single embryonic CPE (67).
This correlation was confirmed by CPEB1 RNA-immunoprecipitation (RNA-IP) followed by RT-qPCR (Figure 4E) of selected candidates. While only two of the eight candidate high-TE mRNAs (Cdk8 and Dnmt1) were immunoprecipitated above background levels, both of which have ≥1 CPEs in the 3′UTR, all low-TE transcripts had CPEs in the 3′UTR and were efficiently recovered in the CPEB1-IP pellet.
Binding of CPEB1 to CPE recruits a complex that represses translation in prophase I
To elucidate the mechanisms controlling translation during Pro I, we focused on members of the oocyte-secreted protein (OOSP) cluster (Oosp1, 2 and 3) (54). Oosp1 (red symbols) and Oosp3 (black symbols) are translationally repressed in Pro I and activated after meiotic resumption, while Oosp2 (blue symbols) is highly translated in Pro I and its translation becomes repressed during meiotic maturation (Figure 5A). While 60% of Oosp2 transcripts have poly(A) tails with ≥80 nts, Oosp1 (21%) and Oosp3 (6%) do not show this bias (Figure 5B). The 3′UTRs of Oosp1 and Oosp3 have two and four putative CPEs upstream of the PAS, respectively, while Oosp2 has no obvious CPE. Oosp1 and Oosp2 were selected as candidates to further investigate the mechanisms of translation control.
Figure 5.
The presence of CPEs in the 3′UTR is associated with translational repression in prophase I-arrested oocytes. (A) TE values of members of the Oosp cluster during meiosis. The average and range of TEs are reported. (B) Polyadenylation state of members of the Oosp cluster in Pro I-arrested oocytes. Data were from a published TAIL-Seq study (52). (C) Accumulation of YPet reporters for Oosp1 and Oosp2 3′UTRs during meiotic maturation. Pro I-arrested oocytes were collected and microinjected with oligoadenylated YPet-Oosp1 (red) or YPet-Oosp2 (blue) mRNA along with polyadenylated mCherry mRNA. Oocytes were allowed to recover for 16 h after microinjection, released from cilostamide, and imaged for 16 h with a sampling frequency of 15 min. Each point is the mean ± SEM of individual oocyte traces obtained in three separate experiments. The total number of oocytes analyzed is in parentheses. (D) YPet reporters for Oosp1 and Oosp2 3′UTRs. 3′UTRs expressed in the oocytes were cloned downstream of the YPet ORF (yellow box). CPEs (gray ovals) and PASes (green hexagons) are shown along with nucleotide positions relative to the start of the 3′UTR. (E) Accumulation of Oosp1 and Oosp2 3′UTR YPet reporters in Pro I-arrested oocytes. Pro I-arrested oocytes were collected and microinjected with oligoadenylated YPet-Oosp1 (red) or YPet-Oosp2 (blue) mRNA along with polyadenylated mCherry mRNA. Oocytes were allowed to recover for 2.5 hrs after microinjection, maintained in Pro I, and imaged for 9 h with a sampling frequency of 15 min. Each point is the mean ± SEM of individual oocyte traces obtained in three separate experiments. The total number of oocytes analyzed is in parentheses. (F) Translation rates of the Oosp1 and Oosp2 YPet reporters in Pro I-arrested oocytes. The translation rate for each oocyte was calculated by linear regression of the reporter data (E) between 6 and 9 h. Mean ± SEM is reported. Statistical significance was evaluated by Mann–Whitney test; ****P < 0.0001. (G) Translation rates of oligoadenylated and polyadenylated Oosp1 YPet reporters in Pro I-arrested oocytes. Experimental conditions were as described in (E). The data were collected from two independent experiments (Supplementary Figure S5C) and the total number of oocytes analyzed and mean ± SEM are reported. Statistical significance was evaluated by Kruskal?–Wallis test; ****P < 0.0001 and ns: not significant. (H) Translation rates of oligoadenylated or polyadenylated Oosp2 3′UTR YPet reporter in Pro I-arrested oocytes. Pro I-arrested oocytes were collected and microinjected with either YPet-Oosp2-oligo(A) or YPet-Oosp2-poly(A) mRNA along with polyadenylated mCherry mRNA. Experimental conditions were as described in (E). The translation rate for each oocyte was calculated by linear regression of the reporter data (Supplementary Figure S5D) between 0 and 3 h or 6 and 9 h. The data were collected from two independent experiments and the total number of oocytes analyzed and mean ± SEM are reported. Statistical significance was evaluated by Kruskal–Wallis test; ****P < 0.0001 and ns: not significant. (I) Mutations of CPE(s) in the Oosp1 YPet reporter. The proximal site is designated as CPE1 and the distal as CPE2. CPE1 (TTTTAAATaaa) was mutated to ‘CGACAAATaaa,’, preserving the downstream, overlapping PAS, while CPE2 (TTTTAAT) was mutated to ‘CGACTCC’ as previously described (36). (J) Accumulation of wild type Oosp1, wild type Oosp2, and mutant Oosp1 reporters in Pro I-arrested oocytes. Pro I-arrested oocytes were collected and microinjected with oligoadenylated YPet-Oosp1 (red circle), YPet-Oosp2 (blue circle), YPet-Oosp1(ΔCPE1) (red square), YPet-Oosp1(ΔCPE2) (red triangle) or YPet-Oosp1(ΔCPE1+2) (red diamond) mRNA along with polyadenylated mCherry mRNA. Experimental conditions were as described in (E). Each point is the mean ± SEM of individual oocyte traces obtained in two separate experiments. The total number of oocytes analyzed is in parentheses. (K) Translation rates of wild type Oosp1, wild type Oosp2 and mutant Oosp1 reporters in Pro I-arrested oocytes. The translation rate for each oocyte was calculated by linear regression of the reporter data (J) between 6 and 9 h. Mean ± SEM is reported. Statistical significance was evaluated by Kruskal–Wallis test; ****P < 0.0001 and ns: not significant. (L) Accumulation of YPet-Oosp1 in Pro I-arrested CPEB1+/+, CPEB1+/− and CPEB1−/− oocytes. Oocytes were collected from hormone-primed wild type, Zp3-CreTCpeb1F/+, and Zp3-CreTCpeb1F/F mice. Experimental conditions were as described in (E). Each point is the mean ± SEM of individual oocyte traces obtained in two separate experiments. The total number of oocytes analyzed is in parentheses. (M) Translation rates of YPet-Oosp1 in Pro I-arrested CPEB1+/+, CPEB1+/− and CPEB1−/− oocytes. The translation rate for each oocyte was calculated by linear regression of the reporter data (L) between 6 and 9 h. Mean ± SEM is reported. Statistical significance was evaluated by Kruskal–Wallis test; **P = 0.0043 and ns: not significant.
To test whether the 3′UTR of the two mRNAs by itself is sufficient to reproduce the opposing translation patterns, we constructed reporters with the YPet fluorescent protein fused to the 3′UTRs of the two mRNAs. Using this approach, we can manipulate the 3′UTR to characterize the specific cis-acting elements functioning to control translation. Either YPet-Oosp1 or YPet-Oosp2 mRNAs (Figure 5C and D) were injected into oocytes along with mCherry mRNA and the translation of YPet was monitored via quantitative live cell imaging (Supplementary Figure S5A). The patterns of YPet accumulation for the Oosp1 and Oosp2 reporters recapitulate the mRNA translation during meiotic maturation predicted by Ribotag/RNA-Seq (Figure 5C). During Pro I arrest, YPet-Oosp1 was translated at a significantly lower rate than YPet-Oosp2 (Figure 5E and F). However, we noted that the translation rate of YPet-Oosp2 significantly increased during the incubation, suggesting that modifications of the reporter develop within this timeframe. It is established that rates of mRNA translation in oocytes are dependent on the length of poly(A), and macromolecular complexes control repression by recruiting deadenylases to the mRNA (9). Thus, we surmised that changes in polyadenylation might be responsible for the increased translation of Oosp2 but not Oosp1 reporter. To test this possibility, we compared the rate of translation in Pro I of reporters with a short poly(A) (oligo-adenylated) or long poly(A) (poly-adenylated) tails to gain insight into the balance between adenylases/deadenylases impinging on the two reporters (Supplementary Figure S5B). Unlike the oligo-adenylated counterpart, the translation rate of the poly-adenylated Oosp1 reporter is initially increased; however, translation rates rapidly decrease to levels identical to the oligo-adenylated probe (Figure 5G and Supplementary Figure S5C). Conversely, the translation of the poly-adenylated Oosp2 reporter is high and remains steady throughout the incubation (Figure 5H and Supplementary Figure S5D). These findings strongly suggest that deadenylation predominates for the Oosp1 reporter while the 3′ UTR of the Oosp2 reporter promotes poly-adenylation and high translation rates. Furthermore, regardless of its initial polyadenylation state, a reporter will eventually be translated at a rate dictated by the 3′UTR (Supplementary Figure S5E).
It is established that CPEB1 plays a central role in assembling repressive complexes on a mRNA in frog oocytes (9). Although the components identified in different studies are often inconsistent, it is generally accepted that this repressive complex maintains low translation rate by skewing the balance of adenylase/deadenylases toward deadenylation. To test whether this is true for Oosp1 repression, we mutated the two CPE elements found in the 3′UTR of this mRNA. In the YPet-Oosp1 reporter, single as well as combined mutations of CPE1 and CPE2 in the 3′UTR (Figure 5I) resulted in de-repressed translation to levels similar to those of YPet-Oosp2 (Figure 5J and K). Using a complementary approach, we determined whether depletion of CPEB1 in the oocytes causes de-repression of the Oosp1 reporter. To this aim, we used oocytes derived from Zp3-Cre^T Cpeb1^F/F mice. The phenotype of these mice has been characterized (Supplementary Figure S6.) While no significant difference in Ypet-Oosp1 translation was detected between CPEB1+/+ and CPEB1+/− oocytes, translation in CPEB1−/− oocytes was significantly de-repressed (Figure 5L and M).
To conclusively confirm the repressive role of CPEB1 using a gain-of-function approach, we inserted two CPE elements in 3'UTR of Oosp2 or in a synthetic 3′UTR that contains only PAS element. While repression could be induced in the synthetic 3′UTR, no significant effect on translation was detected when two CPEs are inserted in the Oosp2 3′UTR (Supplementary Figure S7). This gain-of-function approach suggests that two CPE elements are sufficient to induce repression but that the context of the 3′UTR plays an important role in translational regulation as well.
Translation repression during meiotic maturation is dependent on mRNA deadenylation and is dissociated from destabilization
From Pro I to Met I, ribosome loading for 1722 transcripts is decreased (DOWN, FDR ≤ 0.05). Translation repression is observed from early pro-metaphase I, but also occurs later on during meiosis (Figure 1B and Supplementary Figures S8 and S9). For 92% of DOWN transcripts, repression does not coincide with message destabilization, indicating that these two processes are mechanistically decoupled. Several DOWN candidates with stable mRNA levels in meiosis I were chosen for further investigation, including Oosp2 and mRNAs that code for components of the zona pellucida (Zp1, Zp2 and Zp3) and the chromosome condensin complex (Smc4) (Figure 6A). RT-qPCR confirms the stability of these mRNAs, with all levels remaining constant up to 8 hrs and most up to 16 hrs into meiotic maturation (Met II) (Figure 6B). Poly(A) tail length (PAT) assays confirm that Smc4 and Zp2 are polyadenylated in Pro I (Supplementary Figure S10A) and, by 2 h, their poly(A) tails were significantly shortened (Figure 6C), indicating that repression and deadenylation are associated.
Figure 6.
Translational repression during oocyte re-entry into the cell cycle is dissociated from destabilization and requires deadenylation. (A) Time course of ribosome loading onto repressed candidate mRNAs (DOWN) during meiotic maturation. Values are from our RiboTag/RNA-Seq dataset and the mean and range of duplicate biological replicates are plotted. (B) Translational repression of endogenous mRNAs is dissociated from destabilization. Oocytes were matured in vitro up to Met II and samples were collected at different times during maturation. RNA was extracted from the oocytes, reverse transcribed, and used for RT-qPCR. Bcl2l10 was used as a reference gene as its levels are known to be stable during this time. Data are represented as fold changes in mRNA levels as compared to 0 hrs. Three biological replicates of 30 oocytes per time point were used and RT-qPCR reactions were run in triplicate. The bars represent the mean ± SEM of three experiments. Statistical significance was evaluated by Friedman tests; *P < 0.05. (C) Translational repression of endogenous Smc4 and Zp2 is associated with message deadenylation. Oocytes were either maintained in Pro I (0 h) or allowed to mature for 2 or 8 h. At the end of the incubation, RNA was extracted and used for PAT assays with anchored oligo-dT primers. A representative experiment of the three performed is reported.
A CPE in close proximity of the PAS is required to maintain translation during meiotic maturation
The YPet-Zp2 reporter is translated at relatively high and steady rates during Pro I whereas translation is repressed shortly after germinal vesicle breakdown GVBD (Figure 7A and B). This pattern of translation is in agreement with the RiboTag/RNA-Seq data (Figure 6A) and PAT assay (Figure 6C). Similar results were obtained with YPet-Smc4 (Supplementary Figure S10B and C) and Ypet-Oosp2 (see below). Oocytes released from cilostamide and simultaneously treated with the CDK1 inhibitor dinaciclib did not show differences in YPet-Zp2 translation rates when compared to oocytes maintained in cilostamide, suggesting that CDK1 activation and GVBD are required for translation repression (Figure 7C). PKA inhibitor treatment (Rp-cAMPS), used to block cAMP signaling, again resulted in no repression, indicating that cAMP is not a signal directly involved in translation repression. However, treatment of oocytes with dinaciclib after GVBD (2 h) resulted in decreased translation repression as compared to control oocytes. Therefore, CDK1 activation and GVBD are events necessary to trigger translational repression of Zp2 upon meiotic resumption while the decrease in cAMP levels, by itself, is not sufficient to signal repression when activation of CDK1 is also inhibited.
Figure 7.
Translational repression during meiotic maturation is recapitulated by the 3′UTR of DOWN mRNAs, requires CDK1 activation, and is prevented by the presence of a CPE in close proximity of the PAS. (A) The 3′UTR of Zp2 (high-TE in Pro I and DOWN transcript) recapitulates the rapid translation repression post-GVBD. Oocytes were injected with an oligoadenylated YPet-Zp2 mRNA together with a polyadenylated mCherry mRNA. Oocytes were then either maintained in Pro I with cilostamide treatment (empty circles) or allowed to mature (solid circles) and imaged for 10 h with a sampling frequency of 30 min. Data are reported as the fold change of the YPet/mCherry ratios as compared to 0 h. Each point is the mean ± SEM of individual oocyte traces obtained in two separate experiments. The total number of oocytes analyzed is in parentheses. (B) Translation rates of the YPet-Zp2 reporter in Pro I-arrested or maturing oocytes. The translation rate for each oocyte was calculated by linear regression of the reporter data (A) between 3 and 6 h. Mean ± SEM are reported. Statistical significance was evaluated by unpaired, two-tailed t-test; ****P < 0.0001. (C) Translation repression of the YPet-Zp2 reporter during meiosis resumption requires GVBD and CDK1 activation but not PKA activity. After microinjection of the YPet-Zp2 reporter, oocytes were released in cilostamide-free medium and incubated with a CDK1 inhibitor (5 μM dinaciclib) or a combination of CDK1 and PKA inhibitors (Rp-cAMPS) from the time of release (0 h). The translation rate for each oocyte was calculated by linear regression of the reporter data between 3 and 6 h. In another group, dinaciclib was added after GVBD at 2 h into incubation. Statistical significance was evaluated by unpaired, two-tailed t-tests; ns: not significant; **P = 0.0053. (D) Deletion mutagenesis of the Ccnb2 3′UTR. Pro I-arrested oocytes were collected, microinjected with oligoadenylated YPet-CcnB2 short or reporters fused to Ccnb2 3′UTRs with progressive deletions along with a polyadenylated mCherry reporter. Sixteen hours after microinjection, oocytes were either maintained in Pro I with cilostamide (empty circles) or allowed to mature (solid circles) and imaged for 6 hrs with a sampling frequency of 15 min. Data are reported as the fold change of the YPet/mCherry ratios as compared to 0 h. Each point is the mean ± SEM of individual oocyte traces obtained in two separate experiments. The total number of oocytes analyzed is in parentheses. (E) Insertion of a CPE in the 3′ UTR of Zp2 (DOWN), prevents repression during meiotic maturation. Pro I-arrested oocytes were collected, microinjected with oligoadenylated YPet-Zp2 +CPE mRNA together with a polyadenylated mCherry mRNA. Experimental conditions were as described in (A). Data are reported as the fold change of the YPet/mCherry ratios as compared to 0 h. Each point is the mean ± SEM of individual oocyte traces obtained in two separate experiments. The total number of oocytes analyzed is in parentheses. (F) Insertion of a CPE in close proximity of the PAS in the Oosp2 3′UTR prevents translational repression during meiotic maturation. Pro I-arrested oocytes were collected, microinjected with oligoadenylated YPet-Oosp2 or a reporter with a CPE inserted in the Oosp2 3′UTR along with a polyadenylated mCherry reporter. Oocytes were incubated for 16 hrs after microinjection, allowed to mature, and imaged for 16 h with a sampling frequency of 15 min. Data are reported as the fold change of the YPet/mCherry ratios as compared to 0 h. Each point is the mean ± SEM of individual oocyte traces obtained in three separate experiments. The total number of oocytes analyzed is in parentheses. (G) Detailed analysis of the relationship between translation patterns during meiotic resumption and the presence of CPEs in the 3′UTR. Pie charts report the percentage of CONSTITUTIVE or DOWN mRNAs in Pro I-arrested oocytes that have or lack CPEs in the 3′UTR.
To elucidate the mechanisms of constitutive or repressed translation during meiotic maturation, we monitored the translation of the Ccnb2 reporter, which is constitutively translated before and after GVBD. Progressive deletions of the Ccnb2 short 3′UTR revealed that a reporter retaining only the PAS sequence (YPet-Ccnb2 short (102–118)) is translated like a prototypical DOWN gene; it is highly translated during Pro I and translation became repressed after GVBD (Figure 7D). Presence of the first 42 nts of the 3′UTR did not significantly affect the translation pattern (Supplementary Figure S11A). If the 3′UTR included a CPE (YPet-Ccnb2 short (Δ49–102), translation of the reporter was no longer repressed post-GVBD (Figure 7D) and resembled the pattern of a prototypical CONSTITUTIVE mRNA. Therefore, the presence a CPE is critical for an mRNA to evade translation repression during meiotic maturation. This was also confirmed by a gain-of-function experiment using a repressed mRNA, where insertion of a CPE was sufficient to maintain the high, Pro I translation rate of YPet-Zp2 after meiotic resumption (Figure 7E).
We then tested whether CPE position in relation to the PAS is important for the maintenance of translation post-GVBD by using the Oosp2 3′UTR. When a CPE was added 22 nts upstream of the PAS, reporter translation was no longer repressed upon meiotic resumption, (Figure 7F and Supplementary Figure S11B). However, if the same CPE was added 111 nts upstream of the PAS, translation was still repressed post-GVBD and the pattern did not differ from that of YPet-Oosp2 (Figure 7F and Supplementary Figure S11B). Therefore, during meiotic maturation, inclusion of a CPE proximal to the PAS in the 3′UTR of a translationally repressed mRNA changed its translation pattern to that of a constitutively translated mRNA (gain-of-function). Conversely, removal of a CPE proximal to the PAS of a constitutively translated mRNA switched its translation pattern to that of a repressed mRNA (loss-of-function).
Confirming the observations with candidate 3′UTRs, genome-wide analysis shows that 82% of CONSTITUTIVE mRNAs have at least ≥1 CPEs in the 3′UTR, while this is true for only 47% of DOWN mRNAs (Figure 7G). Further analysis of these classes reveals a bias towards the presence of CPEs within 50 nts upstream of the PAS in constitutively translated mRNAs (Supplementary Figure S12).
Activation of translation during meiotic maturation is dependent on CDK1 activation and GVBD
Concurrent with translation repression, progression through meiosis is associated with significant increases in the translation of 1537 maternal mRNAs (UP, FDR ≤ 0.05) (Supplementary Figure S1E). These UP mRNAs show both early and late increased translation (Supplementary Figure S13). To investigate the mechanisms underlying this activation, we chose candidates with some of the highest fold-changes in ribosome loading from Pro I to Met I, Tcl1, Oosp1, Obox5, Ccnb1 and Ewsr1 (Figure 8A). RiboTag/RT-qPCR confirmed the translation pattern of these UP transcripts (Figure 8B). To investigate the link between cell cycle and the translation program during meiosis, we used a YPet reporter fused with the Ccnb1 3′UTR, a transcript whose translation activation after GVBD has been shown to be CDK1-dependent (38). When CDK1 activity was inhibited immediately after release from PDE inhibition, GVBD did not occur and translation was maintained at levels prior to cilostamide release (Figure 8C). Oocytes treated with dinaciclib after GVBD (2 hrs) eventually regained a nuclear membrane, indicating effective CDK1 inhibition, and translation of Ypet-Ccnb1 was reduced. Similar results were obtain with a second small molecule CDK1 inhibitor, Ro-3306 (Supplementary Figure S14A-B). Experiments with dinaciclib were also performed using YPet-Ewsr1, YPet-Oosp1, and YPet-Mos (Supplementary Figure S14C–E). While dinaciclib treatment of Pro I-arrested oocytes completely inhibited translation of all the reporters, CDK1 inhibition after GVBD only decreased the translation of YPet-Ccnb1 and Ypet-Ewsr1; the translation of YPet-Oosp1 and YPet-Mos was unaffected (Figure 8D). Therefore, early CDK1 activity responsible for GVBD is required for translation activation of these candidates. The variable effects of dinaciclib may be due to subtle differences in the timing and mechanisms of translation activation. Indeed, more detailed analysis of the time course of Mos and CcnB1 reporter activation shows that translational activation of the Mos reporter precedes significantly that of CcnB1 (Supplementary Figure S14A–E and F–G). These differences were present on a background of identical GVBD times (Supplementary Figure S14F–G).
Figure 8.
CPEB binding to mRNAs activated during maturation is necessary, but not sufficient, for full translational activation (A) Pattern of ribosome loading onto UP mRNAs during meiotic maturation. mRNAs whose translation increased by at least 3-fold from Pro I to Met I in our RiboTag/RNA-Seq dataset are shown. Traces of the 149 mRNAs with the highest activation are in grey and transcripts recovered in the pellet of RNA-IP/RT-qPCR with CPEB1 antibody are in black. * denotes transcripts that are also immunoprecipitated by DAZL antibodies (data under review). (B) RiboTag-IP/RT-qPCR validation of ribosome loading for selected UP candidates. Zp3-CreTRiboTagF/F mice were hormone primed and the oocytes isolated. Oocytes were either maintained in Pro I or matured in vitro for 8 h and collected for downstream RiboTag-IP/RT-qPCR analysis. We quantified several candidates with some of the greatest fold changes in ribosome loading from Pro I to Met I. Dppa3 was used as a reference gene as it is known to be constitutively translated during this time. Data are represented as fold changes in message levels as compared to 0 h. Three biological replicates of 200 oocytes per time point were used and RT-qPCR reactions were run in triplicate. The bars represent the mean ± SEM of three experiments. Statistical significance was evaluated by unpaired, two-tailed t-tests; ****P < 0.0001. (C) The effect of CDK1 inhibition on the translation of Ccnb1 mRNA (UP). Pro I-arrested oocytes were collected and microinjected with oligoadenylated YPet-Ccnb1 3′UTR mRNA along with polyadenylated mCherry mRNA. Oocytes were incubated for 16 h then two groups of oocytes were maintained in Pro I with either cilostamide (empty, black circle) or dinaciclib without cilostamide (blue circle). Another two groups of oocytes were either matured without (solid, black circle) or with dinaciclib added at 2 h after release (red circle). Imaging started 2 h after cilostamide release and lasted for 10 h with a sampling frequency of 15 min. Each point is the mean ± SEM of individual oocyte traces obtained in three separate experiments. The total number of oocytes analyzed is in parentheses. (D) Translation rates of YPet-CcnB1 and YPet-Ewsr1 are affected by CDK1 inhibition during meiotic maturation. The translation rate for each oocyte was calculated by linear regression of the reporter data (C and Supplementary Figure S14C and D) between 8 and 12 h. Mean ± SEM is reported. Statistical significance was evaluated by Kruskal–Wallis test; ns: not significant; ****P < 0.0001. (E) Detailed analysis of the relationship between mRNAs that are translationally activated during meiotic resumption and the presence of CPEs in the 3′UTR. Pie charts report the percentage of UP mRNAs in Pro I-arrested oocytes that have or lack CPEs in the 3′UTR. (F) CPEB1 is required for efficient translational activation of CcnB1. CPEB1+/+ (black), CPEB1+/− (light red) and CPEB1−/− (red) oocytes were collected, maintained in Pro I, and microinjected with oligoadenylated YPet-CcnB1 mRNA along with polyadenylated mCherry mRNA. After 2.5 h incubation, oocytes were matured and imaged for 10 h with a sampling frequency of 15 min. Each point is the mean ± SEM of individual oocyte traces obtained in two separate experiments. The total number of oocytes analyzed is in parentheses. (G) Translation rates of the YPet-CcnB1 reporter during oocyte maturation in CPEB1+/+, CPEB1+/− and CPEB1−/− oocytes. The translation rate for each oocyte was calculated by linear regression of the reporter data (F) between 0 and 2 h or 6 and 10 h. Mean ± SEM is reported. Statistical significance was evaluated by Kruskal–Wallis test; ns: not significant; ****P < 0.0001. (H) Accumulation of wild type Oosp1 and mutant Oosp1 YPet reporters during meiotic maturation. Pro I-arrested oocytes were collected and microinjected with oligoadenylated YPet-Oosp1 (circle), YPet-Oosp1(ΔCPE1) (square), YPet-Oosp1(ΔCPE2) (triangle) or YPet-Oosp1(ΔCPE1+2) (diamond) mRNA along with polyadenylated mCherry mRNA. After 16 h of recovery after microinjection, oocytes were allowed to mature, and imaged for 10 h with a sampling frequency of 15 min. Each point is the mean ± SEM of individual oocyte traces obtained in two separate experiments. The total number of oocytes analyzed is in parentheses. (I) Translation rates of wild type Oosp1 and mutant Oosp1 YPet reporters during meiotic maturation. The translation rate for each oocyte was calculated by linear regression of the reporter data (H) between 0 and 2 h or 6 and 10 h (post-GVBD). Mean ± SEM is reported. Statistical significance was evaluated by Kruskal–Wallis test; ns: not significant; ****P < 0.0001.
Regulation of mRNA translation by CDK1 is thought to be mediated by phosphorylation of CPEB1 (38,55). Genome-wide analysis reveals that 95% of UP transcripts have ≥ 1 CPEs in the 3′UTR (Figure 8E). Using CPEB1−/− oocytes, we investigated the role of CPEB1 in the regulation of Ccnb1 translation. CPEB1−/− oocytes showed significantly decreased, but not abolished, translation rates as compared to wild type oocytes (Figure 8F-G). Moreover, depletion of CPEB1 resulted in higher translation rates prior to GVBD (Figure 8G), confirming the role of this RBP in translation repression during Pro I. Single mutations of CPE1 and CPE2 in Oosp1 resulted in a significant increase in initial reporter translation, but there were no effects on translation activation post-GVBD (Figure 8H and I). Mutation of both CPEs did not further de-repress translation before GVBD, but completely abolished translation activation post-GVBD (Figure 8H and I), suggesting that translation activation is not simply due to de-repression and that the two processes are dissociated.
DISCUSSION
Female gamete development is driven by transcription of maternal mRNAs essential for the expansive growth of the oocyte, but also mRNAs needed for the synthesis of proteins that will be used later on during meiotic progression and embryo development. To accomplish this elaborate program of gene expression, not all mRNAs are translated immediately after synthesis (14). Instead, some are stored in repressive complexes throughout the growth phase and unmasked for translation after oocyte re-entry into the cell cycle. Here, we have taken advantage of a genome-wide approach to explore the translation patterns of maternal mRNAs during mouse oocyte meiosis I. We establish that re-entry into meiosis is associated with termination of the translation program associated with growth and activation of a meiotic program of mRNA translation. This switch in translation is dependent on GVBD and CDK1 activation and involves widespread CPEB1-dependent regulations. The extensive genome-wide data on ribosome loading as well as the candidate approach provide clues on the code of regulatory elements in 3′UTR involved in this developmental switch. Moreover, our data show that the rapid repression of translation at nuclear envelope breakdown is dissociated from mRNA destabilization, as the first wave of mRNA degradation is detected at the exit from Met I.
Our genome wide analysis demonstrates a global switch in translation pattern around the exit from prophase I, at the time of GVBD. During this transition, maternal mRNAs required for oocyte growth, which are translated at high rates in Pro I, become repressed, whereas mRNAs coding for cell cycle and for the components of the transcriptional and epigenetic machinery, which were mostly repressed during growth, become translationally activated. The existence of this switch is confirmed with the analysis of the translational efficiency (TE), the changes in polyadenylation state of the mRNAs, and with the translation pattern of reporters for candidate 3′UTRs. The timing of this switch generally coincides with CDK1 activation and GVBD. A causative link between CDK1 activation and increased translation could be established for some candidate mRNAs including Ccnb1 and Ewsr1. In other cases, we have not been able to dissociate GVBD from CDK1 activation and the translational control, in part because of the timeframe of the mouse experimental model. In mouse oocytes, CDK1 activation and GVBD occurs within 90 minutes after relief from cAMP-mediated suppression. Conversely, in frog oocytes, activation of translation of some mRNAs including CcnB1 and Mos precedes the prophase-to metaphase transition. One hypothesis to explore is the presence of critical translational component(s) sequestered in the nucleus that, only upon GVBD, are released into the cytoplasm and function to promote translation. Given the observation that several transcripts are detected in the large nucleus (GV) of the oocyte, it is also possible that nuclear envelope breakdown may release and expose mRNAs to the translation machinery (56).
The RiboTag/RNA-Seq data indicate that the time at which translation repression initiates is variable. These variable times are recapitulated by experiments using fluorescent protein reporters fused to 3′UTRs. Similar to ribosome loading, repression of the Zp2 and Smc4 reporters occur ∼1.5–2 h after cilostamide release in coincidence with GVBD, whereas repression of Oosp2 occurs at ∼4 h. Thus, the timing of repression is ‘encoded’ by the 3′UTR of any given mRNA. In frog oocytes, it has been proposed that the deadenylase PARN is released at GVBD, activating the default process of widespread deadenylation (57,58). This mechanism may apply to only a minority of mRNAs in mouse oocytes, as 77% of the maternal mRNAs become repressed at later times. Thus, these variable time courses imply that multiple mechanisms of repression are operating during meiosis in the mouse.
Our genome-wide analysis demonstrates that the environment of stable maternal mRNAs established during oocyte growth extends well into the late stage of meiosis I. We have identified a first wave of mRNA destabilization at the Met I-to-anaphase transition; most mRNAs that are translationally activated along with a subset of translationally repressed mRNAs continue to remain stable at this stage. Thus, additional waves of mRNA destabilization must take place up to the time of zygote genome activation. Recently, mRNA methylation has emerged as a key regulator of mRNA stability and the m6A-methyl reader YTHDF2 has been implicated in message destabilization in the oocyte (43,59). However, we found minimal overlap between mRNAs destabilized in meiosis I and those stabilized by Ythdf2 loss-of-function. Similarly, marginal overlap was found between mRNAs destabilized at the end of MI and mRNAs stabilized after Cnot6 inactivation. Conversely, we found significant overlap with mRNAs stabilized after Btg4 ablation. Thus, it is likely that the CCR4/CNOT complex that includes CNOT7/8 and BTG4 is responsible for destabilization of a subset of repressed mRNAs (46–48). It should be also noted that the ribosome loading data indicate that the machinery required for mRNA destabilization/degradation is synthesized late during oocyte maturation due to delayed translation of mRNAs including Btg4, Cnot7 and Dcp1a. This late translation coincides with destabilization of a subset of mRNAs at the end of MI. The coincidence of these two events suggests that destabilization of mRNAs is encoded in the translation program itself.
Similar to translation repression, the timing of translation activation during meiotic maturation is characteristic of each individual maternal mRNA. As an example, translation of Mos mRNA is activated early after GVBD, while translation of the Btg4 mRNA is delayed by several hours. Of note, we find significant differences in the timing of Mos and CcnB1 reporter translation. Although requiring further investigation, subtle differences in the mechanisms of translation activation may explain the different time courses as well as the CDK inhibitor sensitivity we have observed. This finding is not unique to the mouse, as differences in translation timing and underlying mechanisms for Mos and Ccnb1 have been documented in frog oocytes (60,61). A combinatorial code of different cis-acting elements is likely responsible for these divergent time courses in this species (32,62).
CPEB1 is considered a master regulator of translation during oocyte meiosis. Our findings are consistent with this tenet, as 95% of mRNAs significantly activated from prophase I to metaphase I have at least one CPE in the 3′UTR and all the translationally activated candidates we tested interact with CPEB1. The role of this RBP in translational activation is further supported by the mutagenesis of the cis-acting elements that interact with CPEB. Removal of these interaction sites prevent translational activation. However, we have identified an additional central function of this RBP during oocyte maturation: CPEB1 is required for maintaining translation of thousands of maternal mRNAs by preventing their deadenylation. Our genome-wide data provide evidence that a CPE in close proximity of the PAS is required for this widespread translation of the majority of maternal mRNAs. As mentioned above, studies in frog oocytes have revealed the presence of a combinatorial code of CPEs enforcing repression and early and late activation (32). Our global analysis documents that CPE elements cluster in the vicinity of the PAS (< 100 nts), and that both repression in prophase I and activation in metaphase I are associated with the presence of more than one CPE element in the 3′UTR. The location of a CPE in the proximity of PAS elements has been associated with repression (63). We show that a CPE located <100 nucleotides from the PAS is critical to maintain translation of reporters that would be otherwise repressed. Using Oosp1 mRNA as prototypic mRNA repressed in prophase I and activated in MI, we demonstrated that two CPEs are required for repression, but one CPE is sufficient for activation. These findings are consistent with some of the rules of the combinatorial code established by Pique et al. (32) but inconsistent with the requirement of a single CPE necessary for repression proposed by Dai et al. (63). To reconcile these discordant results, we propose that more complex rules govern the functions of CPEs in oocyte translation. We believe that the 3′UTR context in which a CPE is located plays a central role in setting the repression or activation as well as the quantitative aspects and the timing of translational regulation.
Our experiments shed light on the mechanisms controlling the translation of mRNAs coding for cyclin B1 and B2, proteins that play a pivotal role in the progression through the meiotic cell cycle (37,64). Although CcnB1 and CcnB2 mRNA levels are present at comparable levels in fully-grown oocytes, their translation diverges substantially in GV oocytes. CcnB2 mRNA is translated at sustained rates while CcnB1 is one of the most repressed mRNAs in prophase I. During maturation, the translation of CcnB1 mRNA increases four-fold, while little change in the translation of CcnB2 mRNA is detected. The functional significance of these divergent patterns of translation is verified by genetic studies in the mouse, where depletion of CCNB2 results in defective meiosis re-entry (37), while CCNB1-depleted oocytes are able to resume meiosis, but are unable to enter meiosis II (64). We have reported that the CcnB1 mRNA in the oocyte is present with three different 3′UTRs and at least four functional CPE elements that contribute to the translational repression in GV and activation after GVBD (36). Conversely and although present with two distinct 3′UTRs, the CcnB2 mRNA contains a single CPE. We document that this CPE plays a critical role in maintaining constant translation during maturation and in protecting the mRNA from deadenylation and repression. These findings further confirm the critical role of CPEs in shaping translation in oocytes during meiosis.
In summary, our genome-wide approach provides a novel perspective on the dynamics of the translational program in mouse oocytes and its interdependence on the cell cycle. In addition to providing a comprehensive genome-wide view of changes in translation during re-entry into the meiotic cell cycle, we have developed novel strategies to measure the dynamic aspects of translation in an intact cell. These powerful approaches are of wide applicability affording quantitative measurement of translational changes in a wide variety of biological systems. The in-depth quantification of a reporter accumulation allows to measure steady state translation rates at any given time during progression through meiosis. Such single cell measurements provide insight into the behavior of different cell populations adding power to the analysis. Moreover, the time-lapse based strategy we developed allows to pinpoint changes in translation rates during biologically relevant cellular transitions. Our manipulations of the length of the poly(a) tail of the reporters also provides information on the endogenous activities of adenylases/deadenylases targeting a given 3′UTR in a cell. All these tools we have developed can be readily adapted to any cell to precisely measure translation rates associated with different functional transitions.
DATA AVAILABILITY
Raw sequences and TMM-normalized CPM values from the RiboTag/RNA-Seq experiment are available on NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE135525 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135525). Scripts used for statistical analysis of the RiboTag/RNA-Seq data and those used to calculate CPE and PAS distances are available on UCSF Box: https://ucsf.app.box.com/s/k474fl8cvkfql5l8m4o2mxq4dg0kgb9g.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to John C. Wright (University of California, Berkeley) for assistance with bioinformatics analyses, as well as Elena Gochez (San Francisco VA Health Care System) and Emily Miller (Stanford University) for assistance with some experiments. We thank Dr Raúl Méndez and Dr Gonzalo Fernandez-Miranda (The Barcelona Institute of Science and Technology) for sharing the Cpeb1-targeted mice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01 GM116926, P50 HD055764 to M.C.]; X.G.L. was supported by 5T32HD7263-35, and E.M.D. by the Lalor Foundation. Funding for open access charge: NIH [P50 HD055764].
Conflict of interest statement. None declared.
REFERENCES
- 1. Chen T.P., Dent S.Y.R.. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat. Rev. Genet. 2014; 15:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klemm S.L., Shipony Z., Greenleaf W.J.. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019; 20:207–220. [DOI] [PubMed] [Google Scholar]
- 3. Rissland O.S. The organization and regulation of mRNA-protein complexes. Wiley Interdiscipl. Rev.-RNA. 2017; 8:doi:10.1002/wrna.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rissland O.S., Subtelny A.O., Wang M., Lugowski A., Nicholson B., Laver J.D., Sidhu S.S., Smibert C.A., Lipshitz H.D., Bartel D.P.. The influence of microRNAs and poly(A) tail length on endogenous mRNA-protein complexes. Genome Biol. 2017; 18:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu X.Y., Brewer G.. The regulation of mRNA stability in mammalian cells: 2.0. Gene. 2012; 500:10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jonas S., Izaurralde E.. NON-CODING RNA towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015; 16:421–433. [DOI] [PubMed] [Google Scholar]
- 7. Cheng J., Maier K.C., Avsec Z., Rus P., Gagneur J.. Cis-regulatory elements explain most of the mRNA stability variation across genes in yeast. RNA. 2017; 23:1648–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jacobson A., Peltz S.W.. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 1996; 65:693–739. [DOI] [PubMed] [Google Scholar]
- 9. Radford H.E., Meijer H.A., de Moor C.H.. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim. Biophys. Acta-Gene Regul. Mech. 2008; 1779:217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tay J., Hodgman R., Richter J.D.. The control of cyclin B1 mRNA translation during mouse oocyte maturation. Dev. Biol. 2000; 221:1–9. [DOI] [PubMed] [Google Scholar]
- 11. Subtelny A.O., Eichhorn S.W., Chen G.R., Sive H., Bartel D.P.. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014; 508:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Richter J.D., Lasko P.. Translational control in oocyte development. Cold Spring Harb. Perspect. Biol. 2011; 3:a002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yartseva V., Giraldez A.J.. The Maternal-to-Zygotic transition during vertebrate development: a model for reprogramming. Maternal-to-Zygotic Transition. 2015; 113:191–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clarke H.J. Post-transcriptional control of gene expression during mouse oogenesis. Results Probl. Cell Differ. 2012; 55:1–21. [DOI] [PubMed] [Google Scholar]
- 15. Seydoux G., Braun R.E.. Pathway to totipotency: lessons from germ cells. Cell. 2006; 127:891–904. [DOI] [PubMed] [Google Scholar]
- 16. Kimble J., Crittenden S.L.. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 2007; 23:405–433. [DOI] [PubMed] [Google Scholar]
- 17. Tadros W., Lipshitz H.D.. Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev. Dyn. 2005; 232:593–608. [DOI] [PubMed] [Google Scholar]
- 18. Mendez R., Richter J.D.. Translational control by CPEB: A means to the end. Nat. Rev. Mol. Cell Biol. 2001; 2:521–529. [DOI] [PubMed] [Google Scholar]
- 19. Richter J.D. CPEB: a life in translation. Trends Biochem. Sci. 2007; 32:279–285. [DOI] [PubMed] [Google Scholar]
- 20. Martins J.P.S., Liu X., Oke A., Arora R., Franciosi F., Viville S., Laird D.J., Fung J.C., Conti M.. DAZL and CPEB1 regulate mRNA translation synergistically during oocyte maturation. J. Cell Sci. 2016; 129:1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Calderone V., Gallego J., Fernandez-Miranda G., Garcia-Pras E., Maillo C., Berzigotti A., Mejias M., Bava F.A., Angulo-Urarte A., Graupera M. et al.. Sequential functions of CPEB1 and CPEB4 regulate pathologic expression of vascular endothelial growth factor and angiogenesis in chronic liver disease. Gastroenterology. 2016; 150:982–997. [DOI] [PubMed] [Google Scholar]
- 22. Rueden C.T., Schindelin J., Hiner M.C., DeZonia B.E., Walter A.E., Arena E.T., Eliceiri K.W.. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017; 18:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim D., Landmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–U121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data, P . The sequence alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ritchie M.E., Phipson B., Wu D., Hu Y.F., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang D.W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 29. Huang D.W., Sherman B.T., Lempicki R.A.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chiu T.P., Yang L., Zhou T.Y., Main B.J., Parker S.C.J., Nuzhdin S.V., Tullius T.D., Rohs R.. GBshape: a genome browser database for DNA shape annotations. Nucleic Acids Res. 2015; 43:D103–D109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beaudoing E., Freier S., Wyatt J.R., Claverie J.M., Gautheret D.. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000; 10:1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pique M., Lopez J.M., Foissac S., Guigo R., Mendez R.. A combinatorial code for CPE-mediated translational control. Cell. 2008; 132:434–448. [DOI] [PubMed] [Google Scholar]
- 33. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J.Y., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wickham H. ggplot2 Elegant graphics for data analysis introduction. Ggplot2. 2009; Springer. [Google Scholar]
- 36. Yang Y., Yang C.-R., Han S.J., Daldello E.M., Cho A., Martins J.P.S., Xia G., Conti M.. Maternal mRNAs with distinct 3' UTRs define the temporal pattern of Ccnb1 synthesis during mouse oocyte meiotic maturation. Genes Dev. 2017; 31:1302–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Daldello E.M., Luong X.G., Yang C.R., Kuhn J., Conti M.. Cyclin B2 is required for progression through meiosis in mouse oocytes. Development. 2019; 146:dev172734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Han S.J., Martins J.P.S., Yang Y., Kang M.K., Daldello E.M., Conti M.. The translation of cyclin B1 and B2 is differentially regulated during mouse oocyte reentry into the meiotic cell cycle. Sci. Rep. 2017; 7:14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Desrosiers R., Friderici K., Rottman F.. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Adams J.M., Cory S.. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature. 1975; 255:28–33. [DOI] [PubMed] [Google Scholar]
- 41. Dubin D.T., Taylor R.H.. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975; 2:1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nachtergaele S., He C.. Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 2018; 52:349–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ivanova I., Much C., Di Giacomo M., Azzi C., Morgan M., Moreira P.N., Monahan J., Carrieri C., Enright A.J., O’Carroll D.. The RNA m(6)a reader YTHDF2 is essential for the Post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol. Cell. 2017; 67:1059–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao B.S., Wang X., Beadell A.V., Lu Z., Shi H., Kuuspalu A., Ho R.K., He C.. m. Nature. 2017; 542:475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Horvat F., Fulka H., Jankele R., Malik R., Jun M., Solcova K., Sedlacek R., Vlahovicek K., Schultz R.M., Svoboda P.. Role of Cnot6l in maternal mRNA turnover. Life Sci. Alliance. 2018; 1:e201800084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pasternak M., Pfender S., Santhanam B., Schuh M.. The BTG4 and CAF1 complex prevents the spontaneous activation of eggs by deadenylating maternal mRNAs. Open Biol. 2016; 6: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu C., Ji S.Y., Sha Q.Q., Dang Y.J., Zhou J.J., Zhang Y.L., Liu Y., Wang Z.W., Hu B.Q., Sun Q.Y. et al.. BTG4 is a meiotic cell cycle-coupled maternal-zygotic transition licensing factor in oocytes. Nat. Struct. Mol. Biol. 2016; 23:387–394. [DOI] [PubMed] [Google Scholar]
- 48. Liu Y.S., Lu X.K., Shi J.C., Yu X.J., Zhang X.X., Zhu K., Yi Z.H., Duan E.K., Li L.. BTG4 is a key regulator for maternal mRNA clearance during mouse early embryogenesis. J. Mol. Cell Biol. 2016; 8:366–368. [DOI] [PubMed] [Google Scholar]
- 49. Conti M., Franciosi F.. Acquisition of oocyte competence to develop as an embryo: integrated nuclear and cytoplasmic events. Hum. Reprod. Update. 2018; 24:245–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Park J.E., Yi H., Kim Y., Chang H., Kim V.N.. Regulation of poly(A) tail and translation during the somatic cell cycle. Mol. Cell. 2016; 62:462–471. [DOI] [PubMed] [Google Scholar]
- 51. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morgan M., Much C., DiGiacomo M., Azzi C., Ivanova I., Vitsios D.M., Pistolic J., Collier P., Moreira P.N., Benes V. et al.. mRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature. 2017; 548:347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang S.F., Kou Z.H., Jing Z.Y., Zhang Y., Guo X.Z., Dong M.Q., Wilmut I., Gao S.R.. Proteome of mouse oocytes at different developmental stages. PNAS. 2010; 107:17639–17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Paillisson A., Dade S., Callebaut I., Bontoux M., Dalbies-Tran R., Vaiman D., Monget P.. Identification, characterization and metagenome analysis of oocyte-specific genes organized in clusters in the mouse genome. BMC Genomics. 2005; 6:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ballantyne S., Daniel D.L., Wickens M.. A dependent pathway of cytoplasmic polyadenylation reactions linked to cell cycle control by c-mos and CDK1 activation. Mol. Biol. Cell. 1997; 8:1633–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Susor A., Jansova D., Cerna R., Danylevska A., Anger M., Toralova T., Malik R., Supolikova J., Cook M.S., Oh J.S. et al.. Temporal and spatial regulation of translation in the mammalian oocyte via the mTOR-eIF4F pathway. Nat. Commun. 2015; 6:6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Copeland P.R., Wormington M.. The mechanism and regulation of deadenylation: Identification and characterization of Xenopus PARN. RNA-a Publ. RNA Soc. 2001; 7:875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim J.H., Richter J.D.. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006; 24:173–183. [DOI] [PubMed] [Google Scholar]
- 59. Wang X., Lu Z.K., Gomez A., Hon G.C., Yue Y.N., Han D.L., Fu Y., Parisien M., Dai Q., Jia G.F. et al.. N-6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mendez R., Barnard D., Richter J.D.. Differential rnRNA translation and rneiotic progression require Cdc2-mediated CPEB destruction. EMBO J. 2002; 21:1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sheets M.D., Fox C.A., Hunt T., Vandewoude G., Wickens M.. The 3′-Untranslated regions of C-Mos and cyclin messenger-rnas stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994; 8:926–938. [DOI] [PubMed] [Google Scholar]
- 62. Charlesworth A., Wilczynska A., Thampi P., Cox L.L., MacNicol A.M.. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006; 25:2792–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dai X.X., Jiang J.C., Sha Q.Q., Jiang Y., Ou X.H., Fan H.Y.. A combinatorial code for mRNA 3-UTR-mediated translational control in the mouse oocyte. Nucleic Acids Res. 2019; 47:328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li J., Tang J.X., Cheng J.M., Hu B., Wang Y.Q., Aalia B., Li X.Y., Jin C., Wang X.X., Deng S.L. et al.. Cyclin B2 can compensate for Cyclin B1 in oocyte meiosis I. J. Cell Biol. 2018; 217:3901–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen J., Melton C., Suh N., Oh J.S., Horner K., Xie F., Sette C., Blelloch R., Conti M.. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011; 25:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Su Y.Q., Sugiura K., Woo Y., Wigglesworth K., Kamdar S., Affourtit J., Eppig J.J.. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev. Biol. 2007; 302:104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Simon R., Richter J.D.. Further analysis of cytoplasmic polyadenylation in xenopus embryos and identification of embryonic cytoplasmic polyadenylation element-binding proteins. Mol. Cell. Biol. 1994; 14:7867–7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences and TMM-normalized CPM values from the RiboTag/RNA-Seq experiment are available on NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE135525 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135525). Scripts used for statistical analysis of the RiboTag/RNA-Seq data and those used to calculate CPE and PAS distances are available on UCSF Box: https://ucsf.app.box.com/s/k474fl8cvkfql5l8m4o2mxq4dg0kgb9g.