Abstract
Genomic instability is an underlying hallmark of cancer and is closely associated with defects in DNA damage repair (DDR). Chromatin relaxation is a prerequisite for DDR, but how chromatin accessibility is regulated remains elusive. Here we report that the histone deacetylase SIRT6 coordinates with the chromatin remodeler CHD4 to promote chromatin relaxation in response to DNA damage. Upon DNA damage, SIRT6 rapidly translocates to DNA damage sites, where it interacts with and recruits CHD4. Once at the damage sites, CHD4 displaces heterochromatin protein 1 (HP1) from histone H3 lysine 9 trimethylation (H3K9me3). Notably, loss of SIRT6 or CHD4 leads to impaired chromatin relaxation and disrupted DNA repair protein recruitment. These molecular changes, in-turn, lead to defective homologous recombination (HR) and cancer cell hypersensitivity to DNA damaging agents. Furthermore, we show that SIRT6-mediated CHD4 recruitment has a specific role in DDR within compacted chromatin by HR in G2 phase, which is an ataxia telangiectasia mutated (ATM)-dependent process. Taken together, our results identify a novel function for SIRT6 in recruiting CHD4 onto DNA double-strand breaks. This newly identified novel molecular mechanism involves CHD4-dependent chromatin relaxation and competitive release of HP1 from H3K9me3 within the damaged chromatin, which are both essential for accurate HR.
INTRODUCTION
DNA damage repair (DDR) defects are a pervasive hallmark of cancer cells; as such, the processes that drive DDR provide opportunities for therapeutic intervention (1,2). Genomic DNA is under constant threat from replication stress, endogenous metabolites and environmental stress factors, such as ultraviolet (UV) and ionizing radiation (IR) (3), which can elicit different types of DNA damage (4). DNA double-strand breaks (DSBs) are a particularly harmful type of DNA damage and have thus been widely studied (5). To limit genomic instability and ensure complete and accurate DNA-mediated processes, cells have evolved mechanisms to respond to DNA damage by activating complex DNA repair signaling networks (6,7).
Chromatin is the primary DDR substrate, but DNA wrapping into chromatin limits the access of repair proteins to DNA damage sites (8,9), to overcome this barrier, heterochromatin must be relaxed (10–12). Heterochromatin is packed and maintained via heterochromatin protein 1 (HP1) binding to histone H3 lysine 9 trimethylation (H3K9me3) and suppressor of variegation 3–9 homolog 1 (SUV39H1), which trimethylates H3K9 (13). In response to DNA damage, casein kinase 2 (CK2) phosphorylates HP1β and disrupts the HP1β interaction with H3K9me3 to induce transient heterochromatin relaxation (14). In addition, upon sensing DSBs, KRAB-associated protein 1 (KAP-1) phosphorylation mediated by ataxia telangiectasia mutated (ATM) and checkpoint kinase 2 (Chk2) promotes HP1β mobilization from heterochromatin and induces chromatin relaxation (15,16). Moreover, HP1 release from H3K9me3 is reportedly necessary for the Tip60 histone acetyltransferase binding to H3K9me3 and Tip60 activation, thus inducing chromatin decondensation and ATM signaling (17).
Many chromatin remodelers help open chromatin during DDR, such as INOsitol requiring 80 (INO80), the SWItch/Sucrose Non-Fermentable (SWI–SNF) complex, the histone acetyltransferase p300 and the mammalian nucleosome remodeling and histone deacetylase (NuRD) complex (18–20). Chromodomain helicase DNA-binding protein 4 (CHD4) is a core subunit of the NuRD complex (21), and a number of studies have demonstrated a role for CHD4 in mediating the DNA damage response. CHD4 moves to DNA damage sites and promotes DNA repair through various pathways (22–26). For example, CHD4 recruits BRCT- repeat inhibitor of hTERT expression (BRIT1) to influence replication protein A (RPA) and breast cancer susceptibility gene 1 (BRCA1) loading on DNA damage sites (27), and also interacts with ring finger protein 8 (RNF8) to relax chromatin (28). CHD4 depletion impairs DSB repair efficiency and sensitizes cancer cells to IR, DSB-inducing agents and Poly (ADP-ribose) polymerase 1 (PARP1) inhibitors (22,27,29,30). The mechanisms underlying CHD4 recruitment to DNA damage sites, however, are unclear and its function in DDR needs further mechanistic clarification.
Sirtuin 6 (SIRT6) has a key role in DNA repair and chromatin relaxation. SIRT6 is one of the seven mammalian sirtuins and can catalyse deacetylation, defatty-acylation and mono-ADP ribosylation (31–37). SIRT6 is responsible for robust DSB repair across rodent species and its activity in stimulating DSB repair coevolves with longevity (38). SIRT6 knock-out mice display increased genomic instability and SIRT6-deficient cells are more sensitive to IR than wild-type cells (39). A recent study implied that lamin A, a protein of nuclear lamina, is an endogenous SIRT6 activator that facilitates SIRT6 localization to chromatin upon sensing DNA damage (40). Once at DNA damage sites, SIRT6 catalyzes and activates PARP1 to promote DNA repair (37). SIRT6 also has a critical role in regulating SNF2H-dependent chromatin accessibility and DNA repair (41).
Because both SIRT6 and CHD4 are key chromatin regulators that can promote chromatin remodeling upon DNA damage, we hypothesized that these two proteins might regulate chromatin accessibility in response to DNA damage in a coordinated manner. Here, we show that SIRT6 interacts with CHD4 and is required for recruiting CHD4 to DNA damage sites. Once recruited, CHD4 competes with HP1 to bind H3K9me3, excluding HP1 from DNA damage sites and facilitating chromatin relaxation to permit proper homologous recombination (HR). Specifically, SIRT6-dependent CHD4 recruitment participates in compacted DSB repair by HR in G2 phase that requires ATM activity. These data reveal a novel link between SIRT6 and CHD4 in modulating chromatin relaxation and provide new mechanistic insight into CHD4-dependent chromatin relaxation in HR.
MATERIALS AND METHODS
Cells and reagents
Human cervical cancer HeLa cells, human embryonic kidney HEK293T cells, human osteosarcoma U2OS cells, human lung cancer A549 cells, human colon cancer HCT116 cells and LoVo cells were obtained from ATCC (American Type Culture Collection). DR-U2OS and EJ5-U2OS cells were gifts from Dr. Xingzhi Xu (Shenzhen University, China). These cells were maintained in McCoy’s 5A (M&C, 10051) medium or DMEM (M&C, 15019) supplemented with 10% FBS (Hyclone, SV30087.02) in a humidified incubator containing 5% CO2. AO3_1 cells were provided by Dr. Qinong Ye (Beijing Institute of Biotechnology, China) and maintained in Ham's F12 (M&C, 10081) medium. Etoposide (VP16) (Sigma, E1383) and doxorubicin (DOX) (Sigma, D1515) were stored in DMSO at 80 mM and 10 mM stock concentrations, respectively. Cisplatin was purchased from TRC and ATMi (KU55933) was purchased from Selleck.
Antibodies
CHD4 (ab72418), H3 (ab1791), H3K9me3 (ab8898), H3K9Ac (ab4441) antibodies were purchased from Abcam. HDAC1 (sc-8410), MTA2 (H-170), Tubulin (sc8035), RPA (sc53496), 53BP1 (sc22760), IgG (sc2025) and Actin (C-11) antibodies were purchased from Santa Cruz Biotechnology. HP1α (3584), HP1γ (05690) and HP1β (3448) antibodies were purchased from Millipore. HA (m180-3), HIS (PM032) and GFP (D153-3) antibodies were purchased from MBL. SIRT6 (2590S), γH2AX (80312 and 9718) and p-ATM S1981 (5883) antibodies were purchased from Cell Signalling Technology. KAP1 (66630-1-Ig) and SNF2H (13066-1-AP) antibodies were purchased from Proteintech. Other antibodies used in this study were: GST (Applygen, C1303), Flag (sigma, F1804) and BRCA1 (Bethy, A301-378a).
Plasmids
SIRT6 full length (FL) or fragments were amplified by polymerase chain reaction (PCR) from a human cell line and cloned into 3XFlag-CMV-10, pGEX-4T-3 or pET-28b vectors. CHD4 FL and fragments were provided by Dr. Shiaw-Yih Lin (The University of Texas, USA). GST-CHD4 constructs were generated based on the HA fragments. The I-SceI expression construct was a gift from Dr. Xingzhi Xu. The Lac-re-EGFP plasmid into which SIRT6 was cloned, was provided by Dr. Pingkun Zhou (Beijing Institute of Radiation Medicine, China).
RNA interference
All RNAi oligonucleotides were purchased from Shanghai GenePharma Company. Cells were transfected with siRNA duplexes using Lipofectamine (Invitrogen, 11668-019), following the manufacturer's instructions. The siRNA sequences were as follows: SIRT6: 5′-AAGAATGTGCCAAGTGTAAGA-3′ and 5′-CACGGGAACATGTTTGTGGAA-3′; CHD4: 5′-CCCAGAAGAGGAUUUGUCA-3′ and 5′-GGUGUUAUGUCUUUGAUUC-3′; HP1α:5′-CCTGAGAAAAACTTGGATT-3′; HP1β:5′-AGGAATATGTGGTGGAAAA-3′; HP1γ:5′-AGGTCTTGATCCTGAAAGA-3′; HDAC1: 5′-UGGCCAUCCUGGAACUGCUAAAGUA-3′; BRCA1: 5′-AAAUGUCACUCUGAGAGGAUAGCCC-3′ and 5′-UUCUAACACAGCUUCUAGUUCAGCC-3′; 53BP1: 5′-CACACAGAUUGAGGAUACG-3′; KAP1: 5′-CAGUGCUGCACUAGCUGUGAGGAUA-3′ and 5′-GCAUGAACCCCUUGUGCUGUUUUGU-3′; SNF2H: 5′-CCGGGCAAAUAGAUUCGAGUAUUUA-3′ and 5′-CAGGGAAGCUCUUCGUGUUAGUGAA-3′; and nonspecific siRNA: 5′-UUCUCCGAACGUGUCACGU-3′.
Establishment of stable cell lines
An shRNA construct targeting SIRT6 or a scrambled shRNA control was transfected into HeLa cells using Lipofectamine (Invitrogen, 11668-019), according to the manufacturer's instructions. After 48 h, the cells were cultured in the presence of G418 to select for stable cell lines. For rescue experiments, the indicated RNAi-resistant plasmids were transfected into shSIRT6 cells with the re-SIRT6 sequence, AAGAATGTGCCTAGTGTAAGA. Bold indicates the mutation sites in the re-SIRT6 plasmid.
Chromatin purification and cell fractionation
Cells were washed and harvested in phosphate-buffered saline (PBS). The cell pellet was re-suspended in buffer 1 (150 mM NaCl, 50 mM Hepes 7.5, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.1% Triton X-100, protease inhibitor cocktail [Roche]) for 3 min on ice and the detergent extractable (Dt) supernatant was collected. The insoluble pellet was washed twice in Buffer 1 without Triton X-100. The remaining pellet chromatin sample (Chr) was re-suspended in sodium dodecyl sulphate (SDS) loading buffer, boiled and sonicated prior to SDS-polyacrylamide gel electrophoresis (PAGE) and western blotting.
Chromatin immunoprecipitation (ChIP) and sequential ChIP-reChIP
Chromatin immunoprecipitation (ChIP) was performed as previously described (42). Briefly, cells were cross-linked in formaldehyde, re-suspended in lysis buffer and fragmented by sonication. The soluble chromatin was diluted and immunoprecipitated with the indicated antibodies and precipitated with Protein A/G Sepharose beads, washed sequentially with low-salt, high-salt, LiCl and TE buffer before elution in elution buffer (1% SDS and 0.1 M NaHCO3). The DNA was purified with a DNA extraction kit (Qiagen, 28106). For Sequential ChIP-reChIP, the primary immunoprecipitated complexes were eluted with buffer (10 mM DTT, 500 mM NaCl and 0.1% SDS) at 37°C instead of elution buffer (1% SDS and 0.1 M NaHCO3). Samples were diluted 1:50 in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, and 20 mM Tris–HCl pH 8.1) and immunoprecipitated with the second antibody. ChIP PCR primers for the HR assay were as follows: HRChIP.S: 5′-TCTTCTTCAAGGACGACGGCAACT-3′ and HRChIP.R: 5′-TTGTAGTTGTACTCCAGCTTGTGC-3′.
Co-immunoprecipitation (Co-IP) and nuclear IP
Cells were washed and harvested in PBS. The cell pellet was re-suspended in lysis buffer (20 mM Tris-HCl pH 8.0, 137 mM NaCl, 10% glycerol, 1% Nonidet P-40 (NP-40), 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride and cocktail protease inhibitors) and incubated with primary antibodies or normal IgG overnight at 4°C. Protein A/G Sepharose beads (GE Healthcare) were added and incubated with the samples for 2 h at 4°C. The beads were washed with lysis buffer and boiled in SDS loading buffer prior to SDS-PAGE and western blotting. A nuclear IP was performed using a Nuclear Complex Co-IP Kit (54001; Active motif), according to the manufacturer's instructions.
GST pull-down assay
GST or GST-fusion proteins were expressed in E. coli induced with isopropyl-D-thio-galactoside (IPTG) and purified using glutathione-Sepharose 4B beads (GE Healthcare) and then washed with TEN buffer (20 mM Tris-HCl pH 7.4, 0.1 mM EDTA, and 100 mM NaCl). Recombinant HIS-tagged proteins were expressed in and purified from bacteria by Ni (ii)-Sepharose affinity (GE healthcare). Proteins were incubated at 4°C overnight before washing the beads three times with TEN buffer and eluted by boiling in 2x SDS loading buffer. The proteins were analyzed by western blotting with an anti-HIS or anti-GST antibody.
Nucleosome solubilisation and Co-IP
This assay was adapted from Goodarzi et al. (43). Briefly, the cells were washed with PBS and low-salt buffer (LSB: 10 mM HEPES pH 7.4, 25 mM KCl, 10 mM NaCl, 1 mM MgCl2, 0.1 mM EDTA). Pelleted cells were re-suspended in LSB with 0.1 mM MC-LR and protease inhibitor cocktail and then snap frozen in liquid nitrogen. Cells were quickly thawed and immediately centrifuged at 9391 g for 10 min. The resulting supernatant was discarded and the pellet was re-suspended in nuclease buffer (10mM HEPES pH 7.9, 10 mM KCl, 1.0 mM CaCl2, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 0.1% [v/v] Triton X-100) containing 100 U/ml MNase. Samples were incubated at 37°C for 45 min before adding an equal volume of solubilisation buffer (nuclease buffer + 2% [v/v] NP-40, 2% [v/v] Triton X-100, 600 mM NaCl). The samples were then sonicated briefly and centrifuged at 9391 g for 10 min. The resulting supernatant, containing solubilized nucleosomes, was incubated with primary antibody at 4°C overnight before Co-IP.
MNase assay qPCR
Cells were collected and re-suspended in buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% Glycerol, 1 mM DTT, 0.1% Triton X-100 and protease inhibitor cocktail) for 8 min on ice. Lysates were pelleted at 1,400 g for 5 min and washed twice with free-detergent buffer A, and then the pellet was lysed in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT and protease inhibitor cocktail) for 30 min on ice followed by centrifugation at 1,700 g for 5 min. The pellet was washed twice with PBS, and then re-suspended in MNase buffer (200 mM Tris-HCl pH 8.0, 50 mM NaCl, 25 mM CaCl2) with 10 U MNase for 2 min at 25°C. The reaction was stopped by adding 0.5 M EDTA. RNase A and proteinase K were added and then DNA was extracted by phenol-chloroform method. The purified DNA was then separated on a 1.2% gel and analyzed using Quantity One software (Bio-Rad). For qPCR analysis, DR-U2OS cells were infected with a retrovirus carrying I-SceI. Cells were harvested and the purified DNA was separated as described above. The DNA bands were then isolated and purified using a Gel Extraction Kit (AxyGEN), and the extracted DNA was used in qPCR reactions as indicated.
Real-time RT-PCR assay
Total RNA was extracted by TRIzol (Invitrogen) method and precipitated in ethanol. cDNA was then synthesized using a Rever Tra Ace qPCR RT Master Kit (TOYOBO). The relative expression of α-Sat or Sat-2 and GAPDH were measured by real-time PCR with the following primers: α-Sat: 5′-CTGCACTACCTGAAGAGGAC-3′ (sense), 5′-GATGGTTCAACACTCTTACA-3′ (anti); Sat-2: 5′-CATCGAATGGAAATGAAAG GAGTC-3′ (sense), 5′-ACCATTGGATGATTGCAGTCAA-3′ (anti); GAPDH: 5′-GGAGCGAGATCCCTCCAAAAT-3′ (sense), 5′-GGCTGTTGTCATACTTCTCATGG-3′ (anti).
Laser micro-irradiation
Laser micro-irradiation (micro-IR) was performed as previously reported (44). Briefly, cells were grown on a glass-bottomed dish and irradiated with a 365 nm pulsed nitrogen UV laser (16 Hz pulse, 41% laser output) generated using a Micropoint System (Andor). The cells were cultured at 37°C for the indicated times and then fixed with 4% paraformaldehyde.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and permeabilized with methanol. The dishes were incubated with blocking solution (0.8% BSA in PBS) and exposed overnight to primary antibody at 4°C. After being washed three times with blocking buffer, the cells were exposed to a secondary FITC/TRITC-conjugated antibody. For the pre-extraction of soluble proteins, the cells were incubated with pre-extraction buffer (0.5% triton-100, 50 mM Hepes pH 7, 150 mM NaCl, 10 mM EGTA, 2 mM MgCl2) for 30 sec on ice prior to fixation. Morphological alterations in the cells were visualized under an Olympus BX-51 confocal microscope.
Analysis of DSB repair at compacted regions
Cells were arrested in G1 or G2 phase, irradiated, fixed and immunostained with an anti-γH2AX antibody, before the γH2AX foci were quantified. For cell enrichment in G1 phase, A549 cells were synchronized by 48 h serum starvation as described previously (45). For cell enrichment in G2 phase, HeLa cells were first synchronized by double thymidine arrest (12 h incubation with 2 mM thymidine, 10 h release, 12 h incubation with 2 mM thymidine), and then 6 h after release, the cells were treated with 5 μM RO-3306 (RO) to arrest the cells at the G2/M boundary. The cell-cycle status of the cells was verified by flow cytometry (FACS) analysis.
DSB repair analysis
DR-U2OS or EJ5-U2OS cells were transfected with the indicated plasmids or siRNAs for 24 h, and then transfected with the I-SceI expression plasmid or infected with the retrovirus expressing I-SceI. After 48 h recovery, the cells were analyzed by FACS. At least three biological repeats were performed.
Colony formation assay
Colony formation assay was performed as previously described (46). Briefly, the cells were treated with VP16 for 2 h and then seeded in 6-well plates for 24 h. The cells were then washed five times with PBS and re-cultured in fresh medium. For IR, the cells were irradiated by X-ray and then seeded in 6-well plates. All cells were cultured for 2 weeks under normal conditions, stained with crystal violet and the number of colonies consisting of >50 cells were counted.
Statistical analyses
All data were evaluated by Student's t-test. A P < 0.05 was considered statistically significant (NS, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001). At least three independent experiments were performed in all cases.
RESULTS
The interaction between SIRT6 and CHD4 markedly increases in response to DNA damage
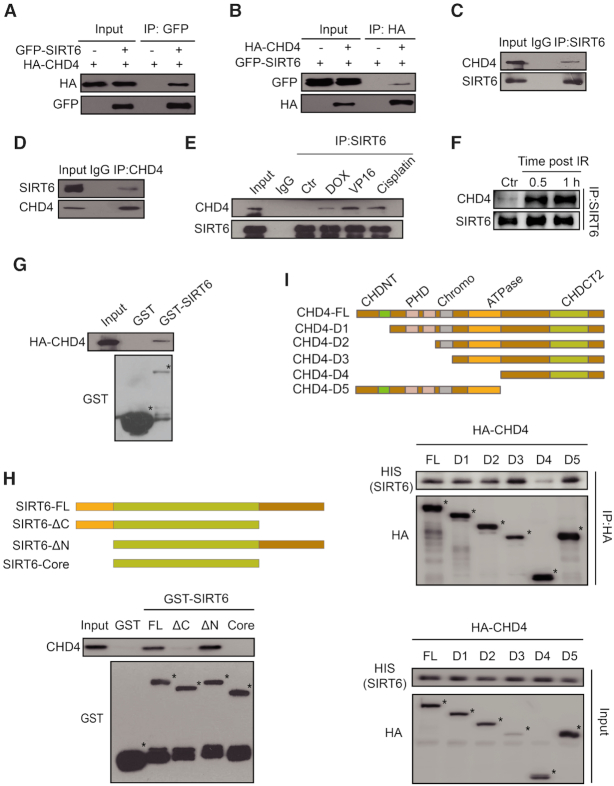
To investigate whether there is a physiological interaction between SIRT6 and CHD4, we first performed a Co-IP assay in HEK293T cells over-expressing GFP-SIRT6 and HA-CHD4. Here, we detected a notable interaction between SIRT6 and CHD4 (Figure 1A and B). In addition, we found that endogenous SIRT6 and CHD4 could mutually precipitate each other in HCT116 cells (Figure 1C and D). This endogenous interaction also occurred in human cervical cancer HeLa cells and human colon cancer LoVo cells (Supplementary Figure S1A and B), indicating that the interaction between SIRT6 and CHD4 is universal. Importantly, the interaction between SIRT6 and CHD4 markedly increased after HeLa or HCT116 cells were exposed to various DNA damaging agents, including doxorubicin (DOX), etoposide (VP16) and cisplatin (Figure 1E and Supplementary Figure S1C).
Figure 1.
The interaction between SIRT6 and CHD4 markedly increases in response to DNA damage. (A) Whole-cell lysates of HEK293T cells transfected with HA-CHD4 with or without GFP-SIRT6 transfection were precipitated with an anti-GFP antibody and analyzed by western blotting, as indicated. (B) Whole-cell lysates of HEK293T cells transfected with GFP-SIRT6 with or without HA-CHD4 transfection were precipitated with an anti-HA antibody and analyzed by western blotting, as indicated. (C andD) Nuclear proteins from HCT116 cells were extracted and immunoprecipitated using an anti-SIRT6 (C) or an anti-CHD4 (D) antibody. Rabbit IgG was used as a negative control. Western blotting was performed with the indicated antibodies. (E) HCT116 cells were treated with 1 μM doxorubicin (DOX) /40 μM etoposide (VP16)/10 μM cisplatin for 1 h and cell extracts were then precipitated with an anti-SIRT6 antibody before western blotting with the indicated antibodies. (F) HCT116 cells were exposed to 10 Gy IR and released for 0.5 or 1 h. The cell extracts were then precipitated with an anti-SIRT6 antibody and western blotting was performed with the indicated antibodies. (G) GST-SIRT6 proteins were purified and incubated with HCT116 whole-cell lysates that expressed HA-CHD4, and analyzed by western blotting. (H) GST-fusion proteins of full length (FL) SIRT6 and SIRT6 fragments were purified and incubated with HCT116 whole cell lysates, and analyzed by western blotting. (I) HA-fragments of CHD4 were infected into HCT116 cells. The cell extracts were incubated with HIS-SIRT6 proteins and then analyzed by western blotting.
To verify the existence of this interaction upon DNA damage, we performed a Co-IP assay in which protein extracts from HCT116 cells irradiated (or not) with 10 Gy were precipitated with an anti-SIRT6 antibody and blotted with an anti-CHD4 antibody. The interaction between SIRT6 and CHD4 was dramatically enhanced from 0.5 h to 1 h post-irradiation (IR) as compared to the control cells (Figure 1F). Interestingly, other components of the NuRD complex, including histone deacetylase 1 (HDAC1), histone deacetylase 2 (HDAC2) and metastasis-associated protein 2 (MTA2), also interacted with SIRT6, and their interaction with SIRT6 also markedly increased in response to IR (Supplementary Figure S1D). Moreover, CHD4 depletion impaired the increased interaction between SIRT6 and HDAC1 as well as MTA2 after DNA damage (Supplementary Figure S1E), suggesting that CHD4 is the main component of the NuRD complex for interacting with SIRT6 in response to DNA damage. These data indicate that SIRT6 interacts with CHD4 in physiological settings and that this interaction is enhanced in response to DNA damage.
SIRT6 binds the CHD4 ATPase domain via its C-terminal domain
To determine whether SIRT6 and CHD4 directly interact, we performed a GST pull-down assay by incubating purified GST-SIRT6 or GST (as a negative control) with HCT116 whole-cell lysates. We found that HA-CHD4 interacted with GST-SIRT6 but not GST alone (Figure 1G). We then mapped the mutual interacting domains between SIRT6 and CHD4. To do so, we constructed and purified full length (FL) GST-SIRT6 (FL, 1–355 aa) and several fragments (C-terminal deleted fragment ΔC, 1–271 aa; N-terminal deleted fragment ΔN, 35–355 aa; core catalytic domain fragment, 35–271 aa), and repeated the GST pull-down assay. Here, CHD4 could only bind to SIRT6 FL and the ΔN fragment (Figure 1H). This finding was also confirmed by Co-IP assay in HCT116 cells, whereby CHD4 only precipitated with SIRT6 FL or the ΔN fragment (Supplementary Figure S1F). These data imply that SIRT6 interacts with CHD4 via the C-terminal domain.
Using a similar approach, we next mapped the region(s) of CHD4 required for SIRT6 binding using a series of CHD4 deletion mutants (Figure 1I). SIRT6 specifically interacted with the FL and adenosine triphosphate (ATP)-helicase domain-containing fragments of CHD4 (Figure 1I). A GST pull-down assay, in which we incubated purified HIS-SIRT6 with the purified GST, FL or fragments of GST-CHD4 confirmed that SIRT6 directly associated with the CHD4 ATPase domain-containing fragments, but not with the ATPase domain-deleted fragment D4 or GST alone (Supplementary Figure S1G). These data indicate that the CHD4 ATPase domain is required for interacting with SIRT6. In summary, we found that SIRT6 directly interacts with the CHD4 ATPase domain via its C-terminal domain.
SIRT6 is required to recruit CHD4 to chromatin following DNA damage
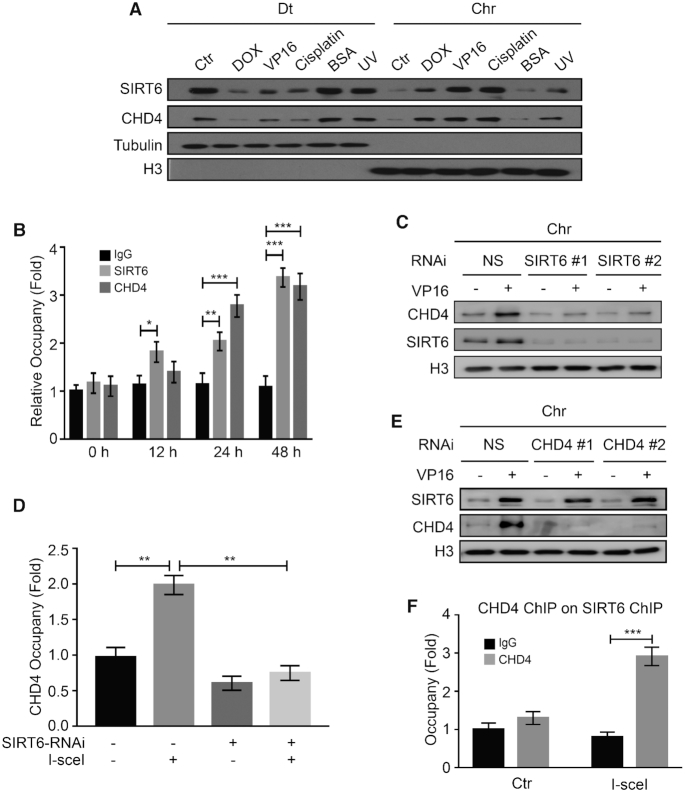
SIRT6 and CHD4 rapidly arrive at sites of DNA damage and are both implicated in DNA repair (22,41). We thus proposed that SIRT6 and CHD4 may coordinate their action in DDR. To verify this hypothesis, we first examined SIRT6 and CHD4 recruitment onto DNA break sites. Here, we found that both SIRT6 and CHD4 localized onto the nuclear chromatin (Chr) following cellular exposure to the DNA-damaging agents DOX, VP16, cisplatin or UV radiation, but not in response to non-genotoxic BSA treatment (Figure 2A). These findings suggest that the noted movement of SIRT6 and CHD4 to chromatin might be a universal response to DSBs. To measure the dynamics of SIRT6 and CHD4 recruitment to DNA damage sites, we established a cellular assay using the DR-U2OS reporter system. This reporter system contains two separate stable GFP genes and can be specifically induced to form a DSB site upon endonuclease I-SceI over-expression. We performed ChIP assays in DR-U2OS cells to detect the enrichment of DSB-responsive proteins in response to a DSB. After I-SceI over-expression, both SIRT6 and CHD4 were recruited to the DSB sites (up to ∼3-fold enrichment) in a time-dependent manner (Figure 2B). These data indicate that indeed, SIRT6 and CHD4 localize to DSB sites during the DNA damage response.
Figure 2.
SIRT6 is required to recruit CHD4 to chromatin following DNA damage. (A) HCT116 cells were treated with 1 μM DOX/40 μM VP16 /10 μM cisplatin/0.5% BSA for 0.5 h or treated with 200 J/m2 UV. The cells were fractionated (Dt/Chr) and analyzed by western blotting. (B) DR-U2OS cells were transfected with an I-SceI expression plasmid for up to 48 h. ChIP experiments were performed using an anti-SIRT6 or an anti-CHD4 antibody. SIRT6 and CHD4 binding to the DNA sequences flanking the I-SceI sites were measured. IgG was used as a negative control. Data are normalized to the control (no I-SceI) samples. (C) HCT116 cells were transfected with NS (non-specific) or two independent SIRT6 specific siRNAs for 48 h in the presence or absence of 40 μM VP16. Chromatin fractions were extracted and analyzed by western blotting. (D) DR-U2OS cells were transfected with NS or SIRT6-specific siRNA for 48 h; then, the cells were transfected with or without an I-SceI expression plasmid for 24 h. ChIP experiments were performed using an anti-CHD4 antibody. CHD4 binding to the DNA sequences flanking the I-SceI sites was measured. (E) HCT116 cells were transfected with NS or two independent CHD4-specific siRNAs for 48 h in the presence or absence of 40 μM VP16. Chromatin fractions were extracted and analyzed by western blotting. (F) DR-U2OS cells were transfected with or without the I-SceI expression plasmid for 48 h. Sequential ChIP-reChIP was performed on the I-SceI DNA damage sites by immunoprecipitation with an anti-SIRT6 antibody followed by immunoprecipitation with an anti-CHD4 antibody. The data represent the means ± SEM (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001).
We next explored the functional significance of the SIRT6 and CHD4 interaction in cells exposed to DNA-damaging agents. We first found that CHD4 proteins levels were unchanged in SIRT6-depleted cells (Supplementary Figure S2A). We then purified the chromatin-enriched fraction of these depleted cells to test whether SIRT6 is required for recruiting CHD4 onto chromatin. Indeed, we found that CHD4 was efficiently recruited onto chromatin after VP16 treatment in control siRNA cells, but this recruitment was largely abrogated in SIRT6 siRNA-depleted cells (Figure 2C). We obtained similar results when using HeLa cells (Supplementary Figure S2B). In addition, CHD4 recruitment to the DSB sites was impaired upon I-SceI expression in SIRT6-depleted cells (Figure 2D), suggesting that SIRT6 is required for CHD4 recruitment to DSBs. Conversely, depleting CHD4 with a CHD4 siRNA did not affect SIRT6 recruitment to chromatin upon DNA damage (Figure 2E) or SIRT6 total protein levels (Supplementary Figure S2C). Similar results were also obtained in HeLa cells (Supplementary Figure S2D), suggesting that SIRT6 recruitment onto chromatin after DNA damage is independent of CHD4.
By performing a sequential ChIP-reChIP assay, we further confirmed the role of SIRT6 in CHD4 recruitment to DSB sites. CHD4 and SIRT6 chromatin occupancy significantly increased at DSB sites after induction of I-SceI (Figure 2F). Moreover, other components of the NuRD complex, including HDAC1 and MTA2, also localized onto chromatin in response to DNA damage (Supplementary Figure S2E), and depletion of either SIRT6 or CHD4 clearly suppressed their localization onto chromatin (Supplementary Figure S2E). HDAC1 knockdown, however, did not affect SIRT6 and CHD4 recruitment onto chromatin after DNA damage (Supplementary Figure S2F). Taken together, these data indicate that SIRT6 is required for efficient CHD4 recruitment to DNA damage sites.
The SIRT6 C-terminal domain and SIRT6 enzymatic activity are required to recruit CHD4 to chromatin upon DNA damage
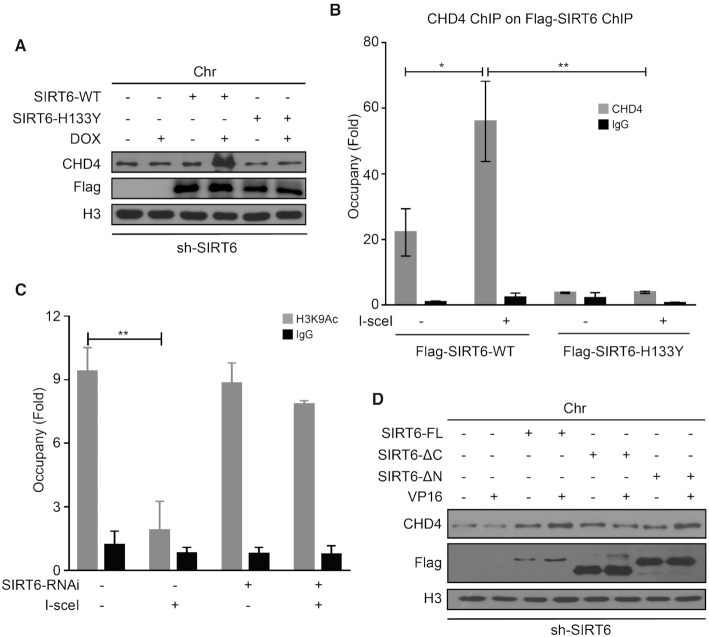
SIRT6 is well characterized as a deacetylase implicated in DDR (41,47). To test whether SIRT6 enzymatic activity is also required for CHD4 recruitment in response to DNA damage, we overexpressed an empty vector, wild-type SIRT6 (SIRT6-WT) or a SIRT6 catalytic mutant (SIRT6-H133Y) in SIRT6-depleted cells and then monitored CHD4 recruitment to chromatin. Reduced CHD4 movement onto chromatin in SIRT6-depleted cells significantly increased upon SIRT6-WT reintroduction (Figure 3A); however, SIRT6-H133Y failed to rescue the impaired CHD4 enrichment onto chromatin after DNA damage (Figure 3A). CHD4 total protein levels were unchanged in response to SIRT6-WT or SIRT6-H133Y over-expression (Supplementary Figure S3A).
Figure 3.
The SIRT6 C-terminal domain and SIRT6 enzymatic activity are required to recruit CHD4 to chromatin upon DNA damage. (A) SIRT6-depleted HeLa cells were transfected with an empty plasmid, a plasmid expressing SIRT6-WT, or a plasmid expressing SIRT6-H133Y. At 48 h after transfection, the cells were treated with or without 1 μM DOX for 1 h. Chromatin fractions were then extracted and analyzed by western blotting. (B) DR-U2OS cells were transfected with a plasmid expressing SIRT6-WT, or a plasmid expressing SIRT6-H133Y for 48 h, and then the cells were transfected with or without an I-SceI expression plasmid for 24 h. Sequential ChIP-reChIP was performed on the I-SceI-induced DSB sites by immunoprecipitation with an anti-Flag antibody followed by immunoprecipitation with an anti-CHD4 antibody. (C) DR-U2OS cells were transfected with NS (non-specific) or SIRT6 specific siRNA for 48 h; then, the cells were transfected with or without an I-SceI expression plasmid for 24 h. ChIP experiments were performed using an anti-H3K9Ac antibody. (D) An empty plasmid, a Flag-SIRT6-FL, a Flag-SIRT6-ΔC mutant or a Flag-SIRT6-ΔN mutant plasmid was transfected into shSIRT6 HeLa cells. After 36 h, the cells were treated with or without 40 μM VP16 for 1 h. Chromatin fractions were extracted and analyzed by western blotting. The data represent the means ± SEM (n = 3, *P < 0.05, **P < 0.01).
To confirm that SIRT6 enzymatic activity is required to recruit CHD4 to DSBs, we performed a sequential ChIP-reChIP assay after I-SceI break-site induction. We found significant CHD4 enrichment on SIRT6-WT-chipped chromatin at I-SceI break sites (Figure 3B). This finding was not observed on SIRT6-H133Y-chipped chromatin (Figure 3B). These data again support that SIRT6 enzymatic activity is required for CHD4 recruitment to DSBs.
SIRT6 is required for global H3K9 deacetylation in response to DNA damage (47). We therefore tested whether SIRT6 functions as a H3K9 deacetylase at DSBs by ChIP assay in DR-U2OS cells. H3K9 acetylation (H3K9Ac) was substantially decreased at damaged sites following I-SceI induction, which was not prevented by HDAC1 knockdown (Supplementary Figure S3B). However, we observed no difference in H3K9Ac levels in SIRT6-depleted cells with or without I-SceI transfection (Figure 3C), indicating that SIRT6 might deacetylate H3K9 in the regions where it associates with CHD4 independently of HDAC1. Notably, SIRT6-ΔC (containing the core catalytic domain but without the C-terminal domain) over-expression failed to rescue impaired recruitment of CHD4 in SIRT6-depleted cells (Figure 3D). By contrast, we could rescue impaired CHD4 loading onto chromatin upon DNA damage by over-expressing either SIRT6-FL or SIRT6-ΔN (containing both the core catalytic domain and the C-terminal domain) in SIRT6-depleted cells (Figure 3D). Collectively, these studies demonstrate that both the C-terminal domain (responsible for binding CHD4) and SIRT6 enzymatic activity (responsible for H3K9 deacetylation) are responsible for modulating CHD4 subcellular localization in response to DNA damage.
SIRT6 promotes chromatin relaxation and DSB repair through CHD4
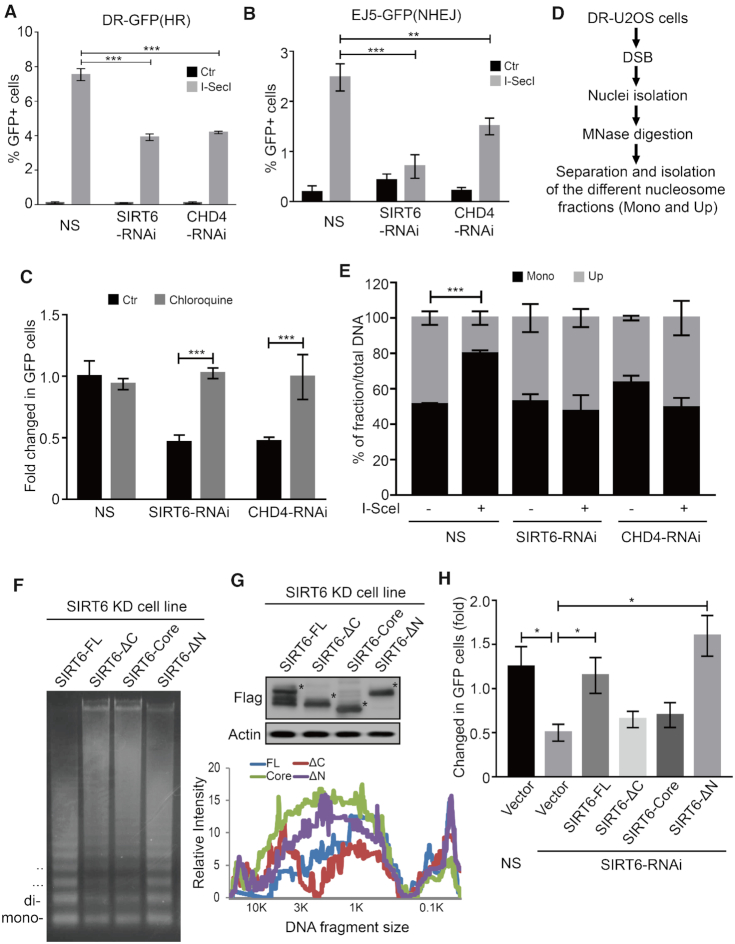
Two major DNA repair pathways are involved in repairing DSBs in mammalian cells: HR and nonhomologous end-joining (NHEJ) (48,49). To confirm the role of the CHD4–SIRT6 in DSB repair, we next analyzed DNA repair efficiency in DR-U2OS or EJ5-U2OS cells that provide readouts for HR or NHEJ, respectively. Here, we found that SIRT6 or CHD4 depletion significantly decreased HR (Figure 4A and Supplementary Figure S4A) and NHEJ (Figure 4B) efficiency compared to BRCA1 and p53 binding-protein 1 (53BP1) that were used as a positive control in each assay (Supplementary Figure S4B and C). This finding might be because SIRT6 and CHD4 are required for both HR-mediated and NHEJ-mediated DNA repair.
Figure 4.
SIRT6 promotes chromatin relaxation and DSB repair through CHD4. (A and B) NS (non-specific), CHD4 or SIRT6-specific siRNA were infected into DR-U2OS (A) or EJ5-U2OS (B) cells for 24 h. Then, the cells were transfected with or without an I-SceI expression plasmid for 48 h. HR (A) and NHEJ (B) efficiency were determined by FACS. The data represent the means ± s.d. (n = 3, **P < 0.01, ***P < 0.001). (C) DR-U2OS cells were transfected with NS, SIRT6 or CHD4-specific siRNA for 24 h. Then, the cells were transfected with an I-SceI expression plasmid for 48 h with or without chloroquine treatment, and HR efficiency was determined by FACS. The data represent the means ± s.d. (n = 3, ***P < 0.001). (D and E) DR-U2OS cells were transfected with NS, SIRT6 or CHD4-specific siRNA for 24 h. Then, the cells were transfected with or without an I-SceI expression plasmid for 48 h. The nuclei were isolated and digested with MNase. Different nucleosomal fractions (mono and upper) were separated on a 1.2% agarose gel. I-SceI site enrichment in each nucleosomal fraction was quantified by qPCR using specific primers. The data represent the means ± s.d. (n = 3, ***P < 0.001). (F) A Flag-SIRT6-FL, Flag-SIRT6-ΔC mutant, Flag-SIRT6-Core mutant, or Flag-SIRT6-ΔN mutant plasmid was transfected into SIRT6 knock-down HeLa cells. After 36 h, the cells were treated with 40 μM VP16 for 1 h. Different nucleosomal fractions were separated on a 1.2% gel after MNase digestion. (G) Whole cell lysates from (F) were extracted and analyzed by western blotting as indicated. The intensity of each lane was consecutively quantified using Quantity One software. (H) DR-U2OS cells were transfected with NS or SIRT6 specific siRNA for 24 h. An empty plasmid, Flag-SIRT6-FL, Flag-SIRT6-ΔC, Flag-SIRT6-Core or Flag-SIRT6-ΔN mutant was transfected into SIRT6 knockdown cells, and then the cells were transfected with an I-SceI expression plasmid for 48 h. HR efficiency was determined by FACS. The data represent the means ± SEM (n = 3, *P < 0.05).
Because chromatin relaxation is necessary for optimal repair and CHD4 is critical for DDR (22,28,41), we hypothesized that SIRT6-dependent CHD4 recruitment might also be required for chromatin decondensation and thus contribute to efficient DNA repair. Strikingly, we could rescue SIRT6 or CHD4 depletion-induced defects in HR with the chromatin relaxation agent chloroquine (Figure 4C). These data suggest that SIRT6 coordinates with CHD4 to modulate HR through chromatin relaxation at DNA damage sites, and that forced chromatin relaxation by chromatin opening agents (i.e. chloroquine) might be sufficient for reversing the effects of SIRT6 or CHD4 depletion on HR.
We next performed a MNase qPCR assay to assess the level of chromatin accessibility at the I-SceI break site in DR-U2OS cells (41). After I-SceI break-site induction, we digested DNA with MNase and then separated the nucleosomal fragments by electrophoresis. We then measured I-SceI break-site enrichment in each nucleosome fraction (mononucleosomes and upper nucleosomes, upper nucleosomes = total DNA-mononucleosomes) by qPCR (Figure 4D). The amount of DSBs in mononucleosomes markedly increased in control cells, whereas its enrichment in the upper fraction significantly decreased (Figure 4E), indicating a marked increase in chromatin relaxation around DSB sites following I-SceI induction in these cells. This effect was markedly abrogated in SIRT6-depleted or CHD4-depleted DR-U2OS cells (Figure 4E), indicating that SIRT6 and CHD4 are required for opening up chromatin at DSB sites. The MNase sensitivity assay also showed that SIRT6 and CHD4 have a role in increasing chromatin accessibility in response to a DSB (Supplementary Figure S4D and E).
We also used a CHO cell line, AO3_1, to further confirm the role of SIRT6 in chromatin decondensation: these cells contain a lac repressor and produce a 90-Mb heterochromatic region (Green) (50). SIRT6 over-expressing cells showed a larger sub-nuclear structure than control cells, suggesting that SIRT6 induces large-scale chromatin decondensation (Supplementary Figure S4F). SIRT6-FL or SIRT6-ΔN over-expression, but not SIRT6-ΔC or SIRT6-core over-expression in SIRT6-depleted cells, was sufficient to rescue chromatin relaxation and HR (Figure 4F-H), suggesting that the SIRT6 C-terminal domain is necessary for chromatin remodeling and HR. In addition, expression of the acetyl mimetic H3K9Q, impaired HR repair efficiency compared to WT H3K9-transfected cells (Supplementary Figure S4G). Together, these results support the notion that SIRT6-dependent CHD4 recruitment to DSBs is required for efficient HR via inducing chromatin relaxation.
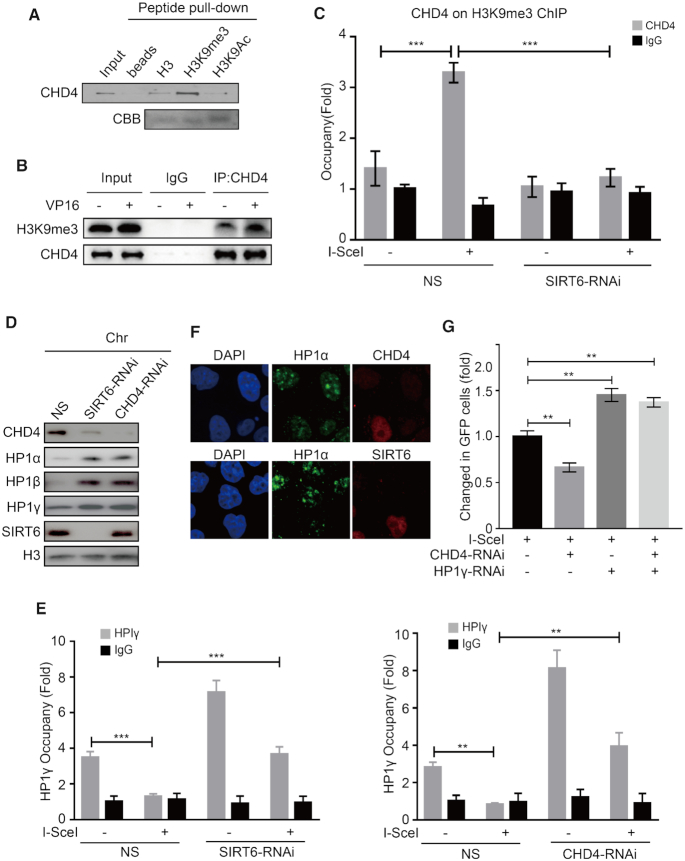
CHD4 competes with HP1 for interacting with H3K9me3
CHD4 has tandem plant homeodomain 1/2 (PHD1/2) fingers that recognize methylated histones with high affinity (51). HP1 is a conserved chromosomal protein that binds H3K9me and forms silent heterochromatin (52). We thus hypothesized that CHD4 might compete with HP1 to bind H3K9me3. To test this hypothesis, we first performed a peptide pull-down assay and found that CHD4 indeed specifically bound to H3K9me3 (Figure 5A). In addition, the interaction between CHD4 and H3K9me3 increased in HCT116 cells upon DNA damage (Figure 5B and Supplementary Figure S5A). To confirm the interaction between CHD4 and H3K9me3 at DSBs, we next performed a sequential ChIP-reChIP assay by first immunoprecipitating the cell lysates with an anti-H3K9me3 antibody and then with an anti-CHD4 antibody. Notably, after I-SceI induction, we readily detected CHD4 enrichment onto H3K9me3 at DSBs in DR-U2OS cells (Figure 5C). Strikingly, this enrichment was completely abolished in SIRT6 knockdown cells (Figure 5C). Together, these data reveal an enhanced SIRT6-dependent interaction between CHD4 and H3K9me3 at DNA damage sites.
Figure 5.
CHD4 competes with HP1 for interacting with H3K9me3. (A) HeLa cell lysates were extracted and incubated with biotin-tagged H3, H3K9me3 or H3K9Ac histone peptides, and a peptide pull down assay was performed and analyzed by western blotting. (B) HCT116 cells were treated with 40 μM VP16 for 30 min. CHD4 was immunoprecipitated from soluble nucleosome extract, washed with solubilization buffer and analyzed by western blotting. Rabbit IgG was used as a negative control. (C) DR-U2OS cells were transfected with NS (non-specific) or SIRT6-specific siRNA for 24 h, and then the cells were transfected with or without an I-SceI expression plasmid for 24 h. Sequential ChIP-reChIP was performed on the I-SceI DNA damage sites by immunoprecipitation with an anti-H3K9me3 antibody followed by immunoprecipitation with an anti-CHD4 antibody. The data represent the means ± SEM (n = 3, ***P < 0.001). (D) HeLa cells were transfected with NS, CHD4 or SIRT6-specific siRNA for 48 h, and the chromatin fractions were extracted and analyzed by western blotting. (E) DR-U2OS cells were transfected with NS, CHD4 or SIRT6-specific siRNA for 48 h, and then the cells were transfected with or without an I-SceI expression plasmid for 24 h. ChIP experiments were performed using an anti-HP1γ antibody, and HP1γ binding to the DNA sequences flanking the I-SceI sites was measured. The data represent the means ± SEM (n = 3, **P < 0.01, ***P < 0.001). (F) HeLa cells were transfected with CHD4 or SIRT6-specific siRNA for 48 h. Cells were exposed to 10 Gy IR, fixed after 1 h, and immunostained with the anti-SIRT6 or anti-CHD4 (right) and anti-HP1α (middle) antibodies and DAPI, and then observed under a confocal microscope. (G) NS, CHD4 or HP1γ-specific siRNA or combined siRNA duplexes targeted to CHD4 and HP1γ were infected into DR-U2OS cells for 24 h. The cells were then transfected with or without an I-SceI expression plasmid for 48 h. HR efficiency was determined by FACS. The data represent the means ± s.d. (n = 3, **P < 0.01).
We next investigated the effects of CHD4 on HP1 binding with H3K9me3. As expected, CHD4 or SIRT6 knockdown in HeLa cells increased HP1α, HP1β or HP1γ retention on chromatin (Figure 5D), but did not affect their total protein levels (Supplementary Figure S5B). In addition, we found that HP1γ recruitment to I-SceI break sites was severely impaired in control DR-U2OS cells but significantly increased in SIRT6-knockdown or CHD4-knockdown cells (Figure 5E). H3K9me3 levels in the I-SceI region were not markedly downregulated by SIRT6 or CHD4 knockdown (Supplementary Figure S5C), indicating that HP1 dispersal from H3K9me3 is not caused by a decrease in H3K9me3 levels, but by SIRT6-depdendent CHD4 recruitment. Immunofluorescence analysis also demonstrated that CHD4 or SIRT6 knockdown induced a substantial increase of HP1 retention to chromatin (Figure 5F; Supplementary Figure S5D and E). Interestingly, we found that the interaction between CHD4 and H3K9me3 was markedly enhanced following siRNA-mediated HP1 knockdown in HCT116 cells (Supplementary Figure S5F). Importantly, HP1 depletion rescued or at least alleviated the HR defect conferred by CHD4 knockdown (Figure 5G; Supplementary Figure S5G and H). Taken together, these data support that CHD4 associates with H3K9me3 in a SIRT6-dependent manner and competitively displaces HP1. We presume that such a mechanism might facilitate chromatin relaxation to permit efficient HR.
SIRT6 and CHD4 enable chromatin relaxation and DSB repair within compacted regions
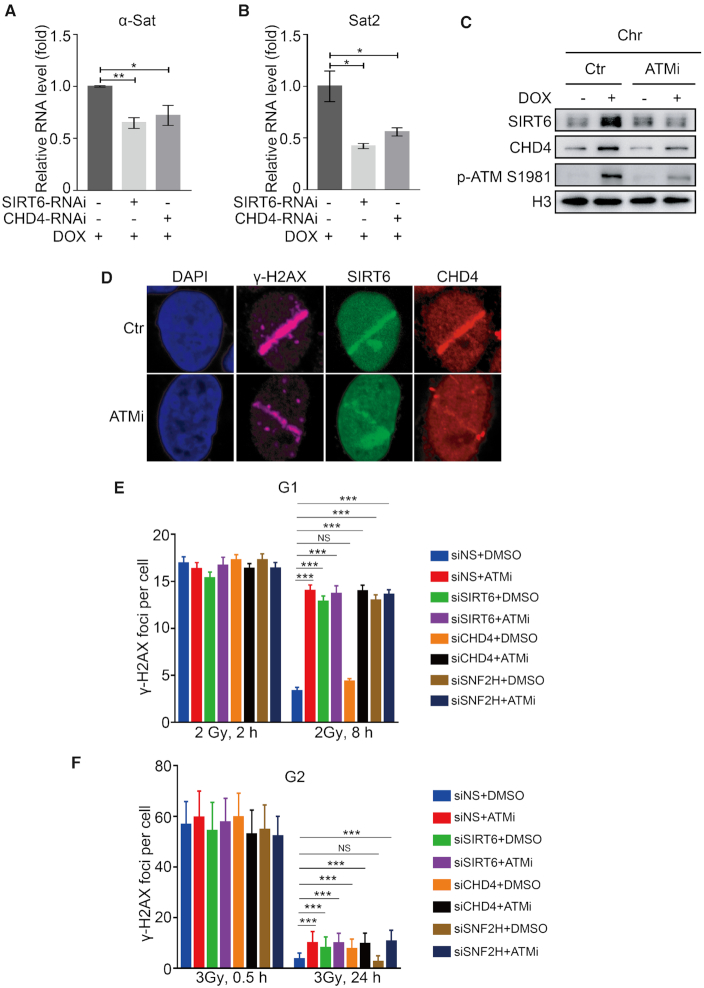
Based on the above observations, we hypothesized that SIRT6 and CHD4 enable compacted regions relaxation in response to DNA damage. To verify this hypothesis, we evaluated the expression levels of the centromeric satellite repeats, α-satellite (α-Sat) and satellite 2 (Sat2), as markers for compacted chromatin. α-Sat and Sat2 are transcriptionally repressed and are only transcribed at a low level under basal conditions; however, when the centromeric region is relaxed, their transcripts are increased (53). Here, we found that the relative expression of α-Sat and Sat2 was significantly elevated in response to DOX treatment (Supplementary Figure S6A and B). SIRT6 or CHD4 depletion compromised this DOX-induced expression of both α-Sat and Sat2 (Figure 6A and B). These data indicate that SIRT6 and CHD4 participate in DNA damage-induced chromatin relaxation.
Figure 6.
SIRT6 and CHD4 enable chromatin relaxation and DSB repair within compacted regions. (A and B) HeLa cells were transfected with NS, SIRT6 or CHD4-specific siRNA for 48 h in the presence of DOX. The relative expression levels of α-Sat (A) and Sat2 (B) were measured by real-time PCR. The data represent the means ± SEM (n = 3, ∗P < 0.05, **P < 0.01). (C) HeLa cells were pre-treated for 30 min with or without 20 μM ATM inhibitor (ATMi, Ku55933), followed by treatment with 1 μM DOX. Chromatin fractions were extracted and analyzed by western blotting. (D) HeLa cells were pre-treated for 30 min with or without 20 μM ATMi before micro-irradiation (micro-IR), fixed after 3 min and immunostained with the indicated antibodies. (E) A549 cells were transfected with NS, SIRT6, CHD4 or SNF2H-specific siRNA and arrested at G1 by serum starvation. Cells were then irradiated with 2 Gy IR with or without ATMi, harvested 2 or 8 h later and stained for γH2AX. Average γH2AX foci numbers per cell were quantified. Background foci numbers were subtracted. The data represent the means ± SEM (n = 40-50, NS, P > 0.05, ***P < 0.001). (F) HeLa cells were synchronized at G2 with double thymidine block and release, and kept with the Cdk1 inhibitor RO-3306. Cells were transfected with NS, SIRT6, CHD4 or SNF2H-specific siRNA, irradiated with 3 Gy IR with or without ATMi, harvested 0.5 or 24 h later. Cells were stained and quantified as in E. The data represent the means ± s.d. (n = 50-100, NS, P > 0.05, ***P < 0.001).
Because DSBs within compacted regions are repaired by an ATM-dependent slow component (43), we tested for the potential contribution of ATM to SIRT6 and CHD4 recruitment. We found that ATM inhibitor (ATMi, KU55933) pre-treatment did not affect the interaction between SIRT6 and CHD4 (Supplementary Figure S6C) but did diminish SIRT6/CHD4 recruitment onto chromatin and to sites of micro-IR (Figure 6C and D). These data imply that ATM recruits SIRT6/CHD4 to sites of DNA damage. Next, we investigated whether SIRT6 and CHD4 are involved in ATM-dependent slow-repair signaling. ATM-dependent compacted repair is mainly mediated by NHEJ in G0/G1 phase and by HR in G2 phase (43,54); here, ATM phosphorylates KAP1 to allow chromatin relaxation and KAP1 knockdown relieves the requirement for ATM in compacted DSB repair (43,54). To address whether SIRT6 has a role in these processes, we transfected KAP1 or SIRT6 siRNAs into DR-U2OS cells. Interestingly, KAP1 knockdown alleviated the HR defects induced by SIRT6 depletion (Supplementary Figure S6D), indicating that SIRT6 participates in ATM-dependent signaling.
Next, we enumerated γH2AX foci in cells arrested in G1 or G2 phase (Supplementary Figure S6E and F) to monitor repair progression (43,54). ATMi-treated cells displayed repair defects and lesions (γ-H2AX foci enumeration) that persisted 8 h after IR in G1 phase and 24 h after IR in G2 (Figure 6E and F). Interestingly, SIRT6 depletion also produced a comparable repair defect to that induced by ATMi in both G1 and G2 cells (Figure 6E and F), indicating that SIRT6 functions downstream of ATM in DSB repair within compacted regions by both NHEJ (in G1) and HR (in G2). SNF2H is a known chromatin remodeler functioning within ATM-dependent slow DSB repair component and a known downstream of SIRT6 (41,55). In our control assay, SNF2H depletion did not affect the SIRT6–CHD4 interaction (Supplementary Figure S6G), suggesting that SNF2H does not compete with CHD4 for binding SIRT6 and that CHD4 and SNF2H likely have distinct functions in SIRT6-dependent DNA repair. Interestingly, SNF2H-depleted cells showed a specific repair defect at G1 (Figure 6E and F), whereas CHD4-depleted cells displayed a repair defect and the persistence of lesions in G2 phase (at 24 h after IR) (Figure 6F), but not in G1 phase (at 8 h after IR) (Figure 6E). These data indicate that SIRT6 is required for DSB repair within compacted regions—a process that is regulated by ATM. More specifically, SIRT6/SNF2H is indispensable for NHEJ at compacted DSBs in G1 phase, whereas SIRT6/CHD4 is required for DSB repair within compacted regions by HR in G2 phase.
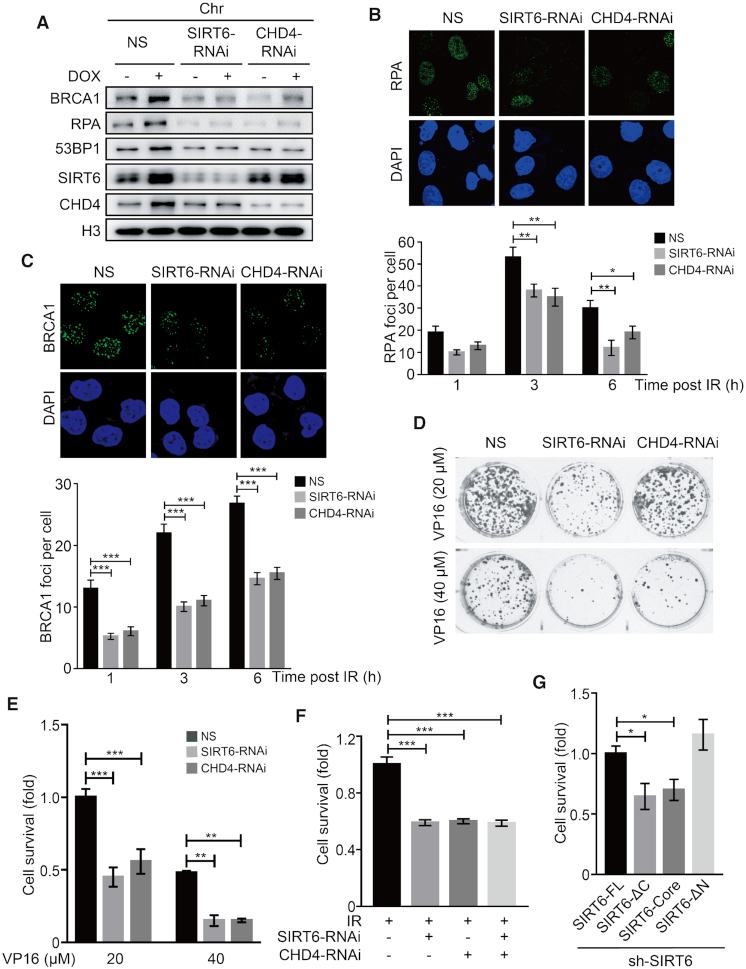
SIRT6 or CHD4 depletion impairs repair protein loading and compromises cell survival
In our final analyses, we aimed to reveal the physiological significance of the coordinated interaction between SIRT6 and CHD4 in DDR biology. First, we analyzed the effects of SIRT6 and CHD4 depletion on downstream repair protein recruitment and the survival of target cells following DNA damage. We found that SIRT6 or CHD4 depletion impaired RPA, 53BP1 and BRCA1 recruitment onto chromatin and repair-foci formation in response to DOX-induced or IR-induced DNA damage (Figure 7A–C and Supplementary Figure S7A–B). Depletion of SIRT6, but not CHD4, significantly impaired γ-H2AX foci formation (1 h after IR exposure)(Supplementary Figure S7C). ATM phosphorylation, however, was unaffected by SIRT6 depletion (Supplementary Figure S7D).
Figure 7.
SIRT6 or CHD4 depletion impairs repair protein loading and compromises cell survival. (A) HeLa cells were transfected with NS (non-specific), SIRT6 or CHD4-specific siRNA for 48 h in the presence or absence of DOX. Chromatin fractions were extracted and analyzed by western blotting. (B and C) HeLa cells were transfected with NS, CHD4 or SIRT6-specific siRNA for 48 h. Cells were exposed to 10 Gy irradiation (IR), and then fixed at 1, 3 or 6 h post-IR and observed under a confocal microscope. The numbers of PRA (B) and BRCA1 (C) foci per cell were quantified. The data represent the means ± SEM (n = 100, *P < 0.05, **P < 0.01, ***P < 0.001). (D and E) HeLa cells stably expressing NS, SIRT6 or CHD4 specific siRNA were treated with 20 or 40 μM VP16 for 2 h and then washed free of the drug. The cells were then cultured in a six-well plate for 2 weeks, and clone formation was analyzed by crystal violet staining. (F) HeLa cells stably expressing NS, SIRT6 or CHD4-specific siRNA were exposed to 3 Gy IR. The cells were then cultured in a 6-well plate for 2 weeks, and clone formation was analyzed by crystal violet staining. (G) A Flag-SIRT6-FL, Flag-SIRT6-ΔC mutant, Flag-SIRT6-Core or Flag-SIRT6-ΔN mutant plasmid was transfected into shSIRT6 HeLa cells. The cells were then treated with 40 μM VP16 for 2 h and washed free of the drug. Then the cells were cultured in a 6-cm plate for 2 weeks and clone formation was analyzed by crystal violet staining. The data represent the means ± SEM (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001).
SIRT6 or CHD4 depletion also compromised clonogenic cell survival after VP16 treatment (Figure 7D and E) or IR exposure (Figure 7F and Supplementary Figure S7E). Interestingly, simultaneous knockdown of SIRT6 and CHD4, did not further compromise cell survival after IR exposure (Figure 7F and Supplementary Figure S7E), supporting the cooperation of these two molecules within the same pathway. To validate these findings, we next performed a rescue experiment in shSIRT6 cells and analyzed the status of cell survival: SIRT6-ΔC or SIRT6-core over-expression failed to increase cell survival as compared to shSIRT6 cells transfected with SIRT6-FL or SIRT6-ΔN (Figure 7G and Supplementary Figure S7F). These findings indicate that both the SIRT6 C-terminus, which facilitates the interaction between SIRT6 and CHD4, and the core catalytic domain are critical for cell survival after exposure to DNA damaging agents.
DISCUSSION
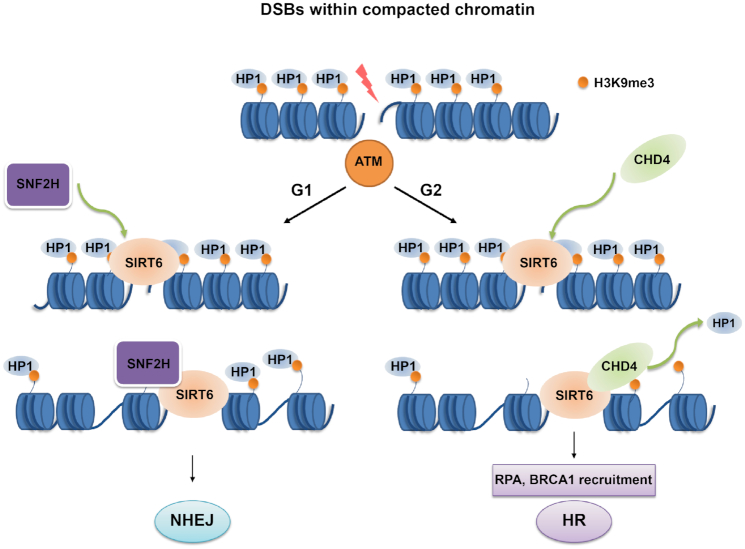
This study demonstrates that SIRT6 is required for CHD4 localization at DSBs and that defective CHD4 recruitment compromises cell survival by impairing DSB repair. We provide evidence to establish that following SIRT6-dependent recruitment of CHD4 to the chromatin, CHD4 competitively binds H3K9me3 at DSBs and excludes HP1 from the chromatin, leading to chromatin relaxation, increased accessibility for downstream repair factors and proper HR. We also show that SIRT6 is required for compacted DSB repair involving ATM-dependent signaling—a process representing NHEJ in G1 phase and HR in G2 phase. Specifically, SIRT6/SNF2H enables DSB repair by NHEJ in G1 phase, whereas SIRT6-dependent CHD4 recruitment enables DSB repair by HR in G2 phase. We have thus established that SIRT6 is upstream of CHD4 for directing it to DSBs and have delineated a novel mechanism for CHD4-dependent chromatin relaxation (Figure 8).
Figure 8.
SIRT6 coordinates with CHD4 to regulate chromatin relaxation and DNA repair. In response to DNA damage, SIRT6 functions downstream of ATM in DSB repair within compacted chromatin. In G1 phase, SIRT6 recruits SNF2H to promote chromatin relaxation and DSB repair by NHEJ within compacted regions. In G2 phase, SIRT6 recruits CHD4 to DNA damage sites, where CHD4 competitively binds H3K9me3 to exclude the retention of HP1, leading to increased chromatin accessibility, repair-factor loading and efficient DNA repair by HR.
CHD4 rapidly moves to chromatin as an early sensor of DNA damage (56). Previous studies have established that CHD4 recruitment onto DNA lesions occurs in a PARP-dependent manner (22,23), and the bromodomain protein zinc finger and MYND [myeloid, Nervy and DEAF-1] domain containing 8 (ZMYND8) recruits CHD4 to DNA-damage sites in a PAR-dependent manner (57). These studies have emphasized that PAR likely acts upstream of CHD4 recruitment in response to DNA damage. Although the CHD4 N-terminal region is reportedly important for binding PAR and redirecting it to DSB sites, it does not contain any structurally characterized PAR binding domain (22,58). These studies suggest that CHD4 recruitment to chromatin during DDR might be more complex than originally thought. In this context, RNF8-mediated CHD4 recruitment to DSBs follows a distinct mechanism from the action of PARP (28). Our study has defined a novel mechanism by which SIRT6 directly binds to and recruits CHD4 to sites of DNA damage. Although ATM inhibition did not affect the interaction between SIRT6 and CHD4, it clearly diminished SIRT6 and CHD4 recruitment to DSB sites. Consistently, CHD4 is phosphorylated by ATM at S1349 in response to DNA damage, and this event is required for CHD4 recruitment to chromatin (59). Conversely, another study reported that ATM catalyzes CHD4 phosphorylation at S1346 and that ATM activity is unnecessary for CHD4 recruitment to DSB sites (22). These paradoxical situations might be explained by the expression of different CHD4 isoforms and the dynamic kinetics of CHD4 recruitment.
SIRT6-dependent CHD4 recruitment to chromatin is required to subsequently recruit downstream repair factors and ensure proper DNA repair. SIRT6 or CHD4 depletion impaired RPA, BRCA1 and 53BP1 loading (27,29,41). Consistent with our findings, SIRT6 and SNF2H were shown to be necessary for 53BP1 recruitment in both U2OS cells and primary brain cultures from SIRT6 KO mice, by stabilizing H2AX (41,60). Another study, however, reported no substantial impact of SNF2H depletion on γ-H2AX/53BP1 formation (55); this discrepancy might be attributed to the complex roles of SNF2H in the WSTF-ISWI chromatin remodeling (WICH) complex and different knockdown efficiencies between experiments (by siRNA or shRNA).
In this study, we found that SIRT6 but not CHD4 depletion impaired γ-H2AX foci formation, suggesting that 53BP1 is recruited by SIRT6/CHD4 through other mechanisms. It is well accepted that H4K16 deacetylation by HDAC1/2 increases the binding affinity of 53BP1 for dimethylated H4K20 (H4K20me2) and promotes 53BP1 recruitment following DSB induction (61). In addition, HDAC1/2 particularly function in NHEJ and regulate the acetylation status of H3K56Ac, which is inversely correlated with 53BP1 (62). In this study, we showed that SIRT6 also bound HDAC1/2 through CHD4 and that this interaction markedly increased after DNA damage. These data support the possibility that SIRT6/CHD4 might modulate NHEJ and 53BP1 recruitment to DSBs through HDAC1/2. In addition, we can’t preclude the possibility that SIRT6/CHD4 contributes to NHEJ though HP1 release and chromatin remodeling. SIRT6 and CHD4 showed comparable effects on HR repair, whereas SIRT6 was more efficient in NHEJ than CHD4. These findings imply that SIRT6 has distinct functions from CHD4 in NHEJ. Indeed, in addition to CHD4 recruitment, SIRT6 is involved in other activities that are critical for NHEJ, including recruiting SNF2H (41), stimulating PARP1 and stabilizing DNA-dependent protein kinases (DNA-PKs) (37,47). In support of these findings, CHD4 is dispensable for SIRT6-dependent compacted DSB repair by NHEJ in G1 phase, whereas SNF2H has a key role in this process (55). Future studies are needed to confirm whether and how CHD4 functions in NHEJ outside the compacted regions in cell-cycle progression.
SIRT6 consists of a conserved central ‘sirtuin domain’ that is flanked by N-terminal and C-terminal extensions. The C-terminal extension contributes to proper nuclear localization but is dispensable for SIRT6 enzymatic activity (63). In this study, we showed that the C-terminal, but not the enzymatic core domain or the N-terminal, was essential for binding CHD4. We also showed, however, that both the SIRT6 C-terminus and SIRT6 enzymatic activity were required to recruit CHD4 to DSBs and contributed to cell survival in response to DNA damage. Moreover, SIRT6 modulated H3K9 deacetylation at DSB sites, which is key for efficient DNA repair.
It should be noted that in addition to CHD4, SIRT6 also interacts with and recruits other components of the NuRD complex to chromatin, including HDAC1/2, which also exhibit H3K9 deacetylase activity. HDAC1 depletion did not prevent I-SceI-induced H3K9 deacetylation at DSB breaks, nor did it markedly affect SIRT6 or CHD4 recruitment onto chromatin in response to DNA damage. Consistently, a previous study reported that CHD4 has a key role in mediating NuRD complex recruitment after DNA damage (22). Together, these findings emphasize the importance of both the C terminus and enzymatic activity of SIRT6 in CHD4 recruitment in response to DNA damage. Similarly, SIRT6 recruits SNF2H through its C-terminus and deacetylates H3K56 to open up the condensed chromatin and permit downstream DDR signaling events (41). Together with this previous report, it seems that SIRT6-dependent histone deacetylation and the C terminus of SIRT6 recruit chromatin remodelers to DSB sites and ensure DNA repair procession. Further work is now necessary to identify other SIRT6 C-terminus binding partners and to subsequently explore their roles in regulating DNA repair. This mechanism may then be exploited to improve chemotherapeutic sensitivity.
SIRT6-dependent CHD4 recruitment enables chromatin relaxation and DSB repair within compacted regions under the control of the ATM-dependent signaling. CHD4 and SNF2H are both chromatin remodelers that are implicated in DNA repair, but because SNF2H did not compete with CHD4 to bind SIRT6, it is reasonable to speculate that the two proteins might have distinct functions in SIRT6-dependent DNA repair. ATM-dependent KAP1 phosphorylation are essential for compacted DSB repair, which represents NHEJ in G0/G1 and HR in G2 phase (43,54,55,64–66). Interestingly, our study demonstrates that SIRT6 has a key role in compacted DSB repair by both NHEJ (in G1) and HR (G2), and that its depletion compromises compacted chromatin relaxation and repair. In these processes, CHD4 specifically participates in chromatin relaxation and DSB repair within compacted regions by HR in G2 phase, whereas SNF2H is only essential for DSB repair by NHEJ in G1 phase (55). Together, CHD4 and SNF2H both act downstream of SIRT6 in DNA repair.
Analysis of compacted DSB repair using γH2AX enumeration is best carried out in hTERT immortalized cells (54,55). In this study, A549 cells were arrested in G1 by serum starvation and analyzed as previously reported (45). Double thymidine block and RO-3306 treatment are well established methods for cell synchronization, including for HeLa cells (45,66–71). Here, HeLa cells were efficiently synchronized in G2, and most of the cells remained in G2 after 24 h post-IR with RO-3306 incubation. Although thymidine (2.5 mM for 24 h) treatment induces moderate DNA breaks in HeLa cells (72), ∼20% HCT116 cells exhibite low levels of γH2AX foci (1–10 foci/cell) after treatment with 2 mM thymidine for 24 h, and only ∼5% cells have high levels of foci (>10 foci/cell) (73). In our hands, we observed that there were ∼5–10% cells with >10 γH2AX foci after double thymidine arrest and ∼11% cells (with >10 γH2AX foci) after 24 h in NS siRNA-transfected cells (data not shown). RO-3306 is a selective inhibitor for CDK1 (71), which is required for end resection and HR in budding yeast (74). In mammalian cells, treatment with RO-3306 diminishes the formation of BRCA1 foci and reduces the phosphorylation of Chk1, Chk2 and SMC1, but without markedly affecting RPA foci (75). Another study showed that when HeLa cells were treated with RO-3306 for G2 arrest, the level of T13/S14 doubly phosphorylated Rad51 increased, which is responsible for binding NBS1 (70). In addition, this study showed that increased HR events were detected after overexpression of WT-Rad51, whereas no increase was found with Rad51 variants at S14 and T13 (S14A, T13A). Similar phenotypes were also observed even after BRCA2 downregulation. Interestingly, overexpression of S14D supported modest HR recovery in BRCA2-depleted cells (70). Based on these studies, the DSBs in G2 can be repaired by HR even with RO-3306 incubation. In addition, ATMi pre-treatment and SNF2H knockdown are well established factors within the slow DSB repair process (43,54,55,65), and they worked well in our hands as positive controls in the present study, further supporting that these analyses are, in principle, reasonable.
Histone modification and HP1 release from chromatin are critical for DNA repair (14,43,53,76). In this study, we confirmed that CHD4 specifically bound H3K9me3 rather than H3K9Ac or unmodified H3. We also observed an enhanced interaction between CHD4 and H3K9me3 at DNA damage sites in a SIRT6-dependent manner, leading to decreased HP1 retention at DSB sites. Interestingly, HP1 inhibition could rescue or alleviate CHD4 depletion-induced defects in HR. Based on these results, we propose that in response to DNA damage, CHD4 recruitment by SIRT6 to damaged sites facilitates chromatin opening and promotes HR by competitively inhibiting the association between HP1 and H3K9me3. In support of this concept, HP1 is released from chromatin at DNA damage sites (14,16,77), and HP1 knockdown alleviates the DSB repair defect within compacted chromatin by ATM inhibition (43); however, persistent HP1 retention on chromatin results in inefficient HR (77).
In summary, our findings provide novel mechanistic insights into the process of CHD4 recruitment to DNA damage sites and the combined roles of SIRT6 and CHD4 in chromatin relaxation, efficient HR and cell survival. These findings broaden our understanding of chromatin remodeling in DNA repair and may provide new targets for cancer therapy in the future.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Xingzhi Xu (Shenzhen University, China) and Dr. Qinong Ye (Beijing Institute of Biotechnology, China) for kindly providing the DR-U2OS/EJ5-U2OS cells and the AO3_1 cells. The authors also appreciate Dr. Shiaw-Yih Lin (The University of Texas, USA) and Dr. Pingkun Zhou (Beijing Institute of Radiation Medicine, China) for sharing the CHD4 and Lac-re-EGFP plasmids. At last, the authors thank Dr. Jessica Tamanini (Shenzhen University) for proofreading the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2017YFA0503900]; National Natural Science Foundation of China [81720108027, 81530074, 81621063, 31570812, 81802803]; Science and Technology Program of Guangdong Province in China [2017B030301016]; Shenzhen Municipal Commission of Science and Technology Innovation [JCYJ20160427104855100, JCYJ20170818092450901]; Discipline Construction Funding of Shenzhen [20160422092356147, 20170815170915596, (2016)1452]; Shanghai Sailing Program [17YF1415800]. Funding for open access charge: National Key R&D Program of China [2017YFA0503900].
Conflict of interest statement. None declared.
REFERENCES
- 1. O’Connor M.J. Targeting the DNA damage response in cancer. Mol. Cell. 2015; 60:547–560. [DOI] [PubMed] [Google Scholar]
- 2. Pearl L.H., Schierz A.C., Ward S.E., Al-Lazikani B., Pearl F.M.G.. Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer. 2015; 15:166–180. [DOI] [PubMed] [Google Scholar]
- 3. Hoeijmakers J.H. DNA damage, aging, and cancer. N. Engl. J. Med. 2009; 361:1475–1485. [DOI] [PubMed] [Google Scholar]
- 4. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khanna K.K., Jackson S.P.. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001; 27:247–254. [DOI] [PubMed] [Google Scholar]
- 6. Negrini S., Gorgoulis V.G., Halazonetis T.D.. Genomic instability–an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010; 11:220–228. [DOI] [PubMed] [Google Scholar]
- 7. Seviour E.G., Lin S.Y.. The DNA damage response: balancing the scale between cancer and ageing. Aging. 2010; 2:900–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jeggo P.A., Downs J.A.. Roles of chromatin remodellers in DNA double strand break repair. Exp. Cell Res. 2014; 329:69–77. [DOI] [PubMed] [Google Scholar]
- 9. Price B.D., D’Andrea A.D.. Chromatin remodeling at DNA double-strand breaks. Cell. 2013; 152:1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cann K.L., Dellaire G.. Heterochromatin and the DNA damage response: the need to relax. Biochem. Cell Biol. 2011; 89:45–60. [DOI] [PubMed] [Google Scholar]
- 11. Goodarzi A.A., Jeggo P.A.. The heterochromatic barrier to DNA double strand break Repair: How to get the entry visa. Int. J. Mol. Sci. 2012; 13:11844–11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feng Y.L., Xiang J.F., Kong N., Cai X.J., Xie A.Y.. Buried territories: heterochromatic response to DNA double-strand breaks. Acta Biochim. Biophys. Sin. (Shanghai). 2016; 48:594–602. [DOI] [PubMed] [Google Scholar]
- 13. Maison C., Almouzni G.. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 2004; 5:296–304. [DOI] [PubMed] [Google Scholar]
- 14. Ayoub N., Jeyasekharan A.D., Bernal J.A., Venkitaraman A.R.. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008; 453:682–686. [DOI] [PubMed] [Google Scholar]
- 15. Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D.C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y.. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 2006; 8:870–876. [DOI] [PubMed] [Google Scholar]
- 16. Bolderson E., Savage K.I., Mahen R., Pisupati V., Graham M.E., Richard D.J., Robinson P.J., Venkitaraman A.R., Khanna K.K.. Kruppel-associated Box (KRAB)-associated co-repressor (KAP-1) Ser-473 phosphorylation regulates heterochromatin protein 1beta (HP1-beta) mobilization and DNA repair in heterochromatin. J. Biol. Chem. 2012; 287:28122–28131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun Y., Jiang X., Xu Y., Ayrapetov M.K., Moreau L.A., Whetstine J.R., Price B.D.. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat. Cell Biol. 2009; 11:1376–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morrison A.J., Highland J., Krogan N.J., Arbel-Eden A., Greenblatt J.F., Haber J.E., Shen X.. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004; 119:767–775. [DOI] [PubMed] [Google Scholar]
- 19. Das C., Lucia M.S., Hansen K.C., Tyler J.K.. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009; 459:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stanley F.K.T., Moore S., Goodarzi A.A.. CHD chromatin remodelling enzymes and the DNA damage response. Mutat. Res. Fund. Mol. M. 2013; 750:31–44. [DOI] [PubMed] [Google Scholar]
- 21. Lai A.Y., Wade P.A.. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer. 2011; 11:588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Polo S.E., Kaidi A., Baskcomb L., Galanty Y., Jackson S.P.. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010; 29:3130–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiacovo M.P., Elledge S.J.. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:18475–18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qi W., Chen H., Xiao T., Wang R., Li T., Han L., Zeng X.. Acetyltransferase p300 collaborates with chromodomain helicase DNA-binding protein 4 (CHD4) to facilitate DNA double-strand break repair. Mutagenesis. 2016; 31:193–203. [DOI] [PubMed] [Google Scholar]
- 25. Sperlazza J., Rahmani M., Beckta J., Aust M., Hawkins E., Wang S.Z., Zu Zhu S., Podder S., Dumur C., Archer K. et al.. Depletion of the chromatin remodeler CHD4 sensitizes AML blasts to genotoxic agents and reduces tumor formation. Blood. 2015; 126:1462–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gong F., Chiu L.Y., Cox B., Aymard F., Clouaire T., Leung J.W., Cammarata M., Perez M., Agarwal P., Brodbelt J.S. et al.. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015; 29:197–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pan M.R., Hsieh H.J., Dai H., Hung W.C., Li K., Peng G., Lin S.Y.. Chromodomain helicase DNA-binding protein 4 (CHD4) regulates homologous recombination DNA repair, and its deficiency sensitizes cells to poly(ADP-ribose) polymerase (PARP) inhibitor treatment. J. Biol. Chem. 2012; 287:6764–6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luijsterburg M.S., Acs K., Ackermann L., Wiegant W.W., Bekker-Jensen S., Larsen D.H., Khanna K.K., van Attikum H., Mailand N., Dantuma N.P.. A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO J. 2012; 31:2511–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Larsen D.H., Poinsignon C., Gudjonsson T., Dinant C., Payne M.R., Hari F.J., Rendtlew Danielsen J.M., Menard P., Sand J.C., Stucki M. et al.. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J. Cell Biol. 2010; 190:731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smeenk G., Wiegant W.W., Vrolijk H., Solari A.P., Pastink A., van Attikum H.. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 2010; 190:741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frye R.A. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 1999; 260:273–279. [DOI] [PubMed] [Google Scholar]
- 32. Frye R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000; 273:793–798. [DOI] [PubMed] [Google Scholar]
- 33. Kawahara T.L., Michishita E., Adler A.S., Damian M., Berber E., Lin M., McCord R.A., Ongaigui K.C., Boxer L.D., Chang H.Y. et al.. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009; 136:62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sebastian C., Zwaans B.M., Silberman D.M., Gymrek M., Goren A., Zhong L., Ram O., Truelove J., Guimaraes A.R., Toiber D. et al.. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012; 151:1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang P., Tu B., Wang H., Cao Z., Tang M., Zhang C., Gu B., Li Z., Wang L., Yang Y. et al.. Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:10684–10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jiang H., Khan S., Wang Y., Charron G., He B., Sebastian C., Du J., Kim R., Ge E., Mostoslavsky R. et al.. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013; 496:110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., Seluanov A., Gorbunova V.. SIRT6 promotes DNA repair under stress by activating PARP1. Science (New York, N.Y.). 2011; 332:1443–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tian X., Firsanov D., Zhang Z.H., Cheng Y., Luo L.F., Tombline G., Tan R.Y., Simon M., Henderson S., Steffan J. et al.. SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell. 2019; 177:622–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mostoslavsky R., Chua K.F., Lombard D.B., Pang W.W., Fischer M.R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M.M. et al.. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006; 124:315–329. [DOI] [PubMed] [Google Scholar]
- 40. Ghosh S., Liu B., Wang Y., Hao Q., Zhou Z.. Lamin a is an endogenous SIRT6 activator and promotes SIRT6-mediated DNA repair. Cell Rep. 2015; 13:1396–1406. [DOI] [PubMed] [Google Scholar]
- 41. Toiber D., Erdel F., Bouazoune K., Silberman D.M., Zhong L., Mulligan P., Sebastian C., Cosentino C., Martinez-Pastor B., Giacosa S. et al.. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell. 2013; 51:454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li M., Hou T., Gao T., Lu X., Yang Q., Zhu Q., Li Z., Liu C., Mu G., Liu G. et al.. p53 cooperates with SIRT6 to regulate cardiolipin de novo biosynthesis. Cell Death. Dis. 2018; 9:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goodarzi A.A., Noon A.T., Deckbar D., Ziv Y., Shiloh Y., Lobrich M., Jeggo P.A.. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008; 31:167–177. [DOI] [PubMed] [Google Scholar]
- 44. Li Y., Li Z., Dong L., Tang M., Zhang P., Zhang C., Cao Z., Zhu Q., Chen Y., Wang H. et al.. Histone H1 acetylation at lysine 85 regulates chromatin condensation and genome stability upon DNA damage. Nucleic Acids Res. 2018; 46:7716–7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Geuting V., Reul C., Lobrich M.. ATM release at resected double-strand breaks provides heterochromatin reconstitution to facilitate homologous recombination. PLos Genet. 2013; 9:e1003667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tang M., Li Z., Zhang C., Lu X., Tu B., Cao Z., Li Y., Chen Y., Jiang L., Wang H. et al.. SIRT7-mediated ATM deacetylation is essential for its deactivation and DNA damage repair. Sci. Adv. 2019; 5:eaav1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McCord R.A., Michishita E., Hong T., Berber E., Boxer L.D., Kusumoto R., Guan S.H., Shi X.B., Gozani O., Burlingame A.L. et al.. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging Us. 2009; 1:109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chapman J.R., Taylor M.R., Boulton S.J.. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012; 47:497–510. [DOI] [PubMed] [Google Scholar]
- 49. Sung P., Klein H.. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006; 7:739–750. [DOI] [PubMed] [Google Scholar]
- 50. Ye Q., Hu Y.F., Zhong H., Nye A.C., Belmont A.S., Li R.. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J. Cell Biol. 2001; 155:911–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Musselman C.A., Ramirez J., Sims J.K., Mansfield R.E., Oliver S.S., Denu J.M., Mackay J.P., Wade P.A., Hagman J., Kutateladze T.G.. Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nielsen A.L., Oulad-Abdelghani M., Ortiz J.A., Remboutsika E., Chambon P., Losson R.. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell. 2001; 7:729–739. [DOI] [PubMed] [Google Scholar]
- 53. Wang D., Zhou J., Liu X., Lu D., Shen C., Du Y., Wei F.Z., Song B., Lu X., Yu Y. et al.. Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:5516–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Beucher A., Birraux J., Tchouandong L., Barton O., Shibata A., Conrad S., Goodarzi A.A., Krempler A., Jeggo P.A., Lobrich M.. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009; 28:3413–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Klement K., Luijsterburg M.S., Pinder J.B., Cena C.S., Del Nero V., Wintersinger C.M., Dellaire G., van Attikum H., Goodarzi A.A.. Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J. Cell Biol. 2014; 207:717–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. O'Shaughnessy A., Hendrich B.. CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochem. Soc. Trans. 2013; 41:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Spruijt C.G., Luijsterburg M.S., Menafra R., Lindeboom R.G.H., Jansen P.W.T.C., Edupuganti R.R., Baltissen M.P., Wiegant W.W., Voelker-Albert M.C., Matarese F. et al.. ZMYND8 co-localizes with NuRD on target genes and regulates Poly(ADP-Ribose)-Dependent recruitment of GATAD2A/NuRD to sites of DNA damage. Cell Rep. 2016; 17:783–798. [DOI] [PubMed] [Google Scholar]
- 58. Silva A.P.G., Ryan D.P., Galanty Y., Low J.K.K., Vandevenne M., Jackson S.P., Mackay J.P.. The N-terminal region of chromodomain helicase DNA-binding protein 4 (CHD4) is essential for activity and contains a high mobility group (HMG) Box-like-domain that can bind Poly(ADP-ribose). J. Biol. Chem. 2016; 291:924–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Urquhart A.J., Gatei M., Richard D.J., Khanna K.K.. ATM mediated phosphorylation of CHD4 contributes to genome maintenance. Genome Integrity. 2011; 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Atsumi Y., Minakawa Y., Ono M., Dobashi S., Shinohe K., Shinohara A., Takeda S., Takagi M., Takamatsu N., Nakagama H. et al.. ATM and SIRT6/SNF2H Mediate Transient H2AX Stabilization When DSBs Form by Blocking HUWE1 to Allow Efficient gammaH2AX Foci Formation. Cell Rep. 2015; 13:2728–2740. [DOI] [PubMed] [Google Scholar]
- 61. Panier S., Boulton S.J.. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Bio. 2014; 15:7–18. [DOI] [PubMed] [Google Scholar]
- 62. Miller K.M., Tjeertes J.V., Coates J., Legube G., Polo S.E., Britton S., Jackson S.P.. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010; 17:1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tennen R.I., Berber E., Chua K.F.. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech. Ageing Dev. 2010; 131:185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Goodarzi A.A., Kurka T., Jeggo P.A.. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 2011; 18:831–839. [DOI] [PubMed] [Google Scholar]
- 65. Shibata A., Conrad S., Birraux J., Geuting V., Barton O., Ismail A., Kakarougkas A., Meek K., Taucher-Scholz G., Lobrich M. et al.. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011; 30:1079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kakarougkas A., Ismail A., Klement K., Goodarzi A.A., Conrad S., Freire R., Shibata A., Lobrich M., Jeggo P.A.. Opposing roles for 53BP1 during homologous recombination. Nucleic Acids Res. 2013; 41:9719–9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lee M., Daniels M.J., Venkitararnan A.R.. Phosphorylation of BRCA2 by the Polo-like kinase Plk1 is regulated by DNA damage and mitotic progression. Oncogene. 2004; 23:865–872. [DOI] [PubMed] [Google Scholar]
- 68. Hockemeyer D., Sfeir A.J., Shay J.W., Wright W.E., de Lange T.. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005; 24:2667–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Esashi F., Christ N., Gannon J., Liu Y.L., Hunt T., Jasin M., West S.C.. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005; 434:598–604. [DOI] [PubMed] [Google Scholar]
- 70. Yata K., Lloyd J., Maslen S., Bleuyard J.Y., Skehel M., Smerdon S.J., Esashi F.. Plk1 and CK2 Act in Concert to Regulate Rad51 during DNA Double Strand Break Repair. Mol. Cell. 2012; 45:371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vassilev L.T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D.C., Chen L.. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:10660–10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Szuts D., Krude T.. Cell cycle arrest at the initiation step of human chromosomal DNA replication causes DNA damage. J. Cell Sci. 2004; 117:4897–4908. [DOI] [PubMed] [Google Scholar]
- 73. Gagou M.E., Zuazua-Villar P., Meuth M.. Enhanced H2AX phosphorylation, DNA replication fork arrest, and cell death in the absence of Chk1. Mol. Biol. Cell. 2010; 21:739–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L.H., Hollingsworth N.M. et al.. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004; 431:1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Johnson N., Cai D.P., Kennedy R.D., Pathania S., Arora M., Li Y.C., D’Andrea A.D., Parvin J.D., Shapiro G.I.. Cdk1 participates in BRCA1-Dependent S phase checkpoint control in response to DNA damage. Mol. Cell. 2009; 35:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li Z., Li Y., Tang M., Peng B., Lu X., Yang Q., Zhu Q., Hou T., Li M., Liu C. et al.. Destabilization of linker histone H1.2 is essential for ATM activation and DNA damage repair. Cell Res. 2018; 28:756–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kalousi A., Hoffbeck A.S., Selemenakis P.N., Pinder J., Savage K.I., Khanna K.K., Brino L., Dellaire G., Gorgoulis V.G., Soutoglou E.. The nuclear oncogene SET controls DNA repair by KAP1 and HP1 retention to chromatin. Cell Rep. 2015; 11:149–163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.