Abstract
The hexametric T7 helicase (gp4) adopts a spiral lock-washer form and encircles a coil-like DNA (tracking) strand with two nucleotides bound to each subunit. However, the chemo-mechanical coupling mechanism in unwinding has yet to be elucidated. Here, we utilized nanotensioner-enhanced Förster resonance energy transfer with one nucleotide precision to investigate gp4-induced unwinding of DNA that contains an abasic lesion. We observed that the DNA unwinding activity of gp4 is hindered but not completely blocked by abasic lesions. Gp4 moves back and forth repeatedly when it encounters an abasic lesion, whereas it steps back only occasionally when it unwinds normal DNA. We further observed that gp4 translocates on the tracking strand in step sizes of one to four nucleotides. We propose that a hypothetical intermediate conformation of the gp4–DNA complex during DNA unwinding can help explain how gp4 molecules pass lesions, providing insights into the unwinding dynamics of gp4.
INTRODUCTION
Helicases are central to all replisomes, playing crucial roles in unwinding DNA duplexes and coupling DNA unwinding with subsequent leading- and lagging-strand DNA syntheses (1,2). DNA replisomes inevitably encounter a variety of DNA lesions. According to current replisome models (3), replicative helicases function as vanguards of DNA replication and, therefore, would be the first to encounter potential DNA lesions. It follows that DNA lesions that can prevent DNA helicases from unwinding would have an impact on the overall replication process (3). How replicative helicases overcome these obstacles is an outstanding question, as the process is important for the maintenance of genomic stability and healthy life process (4,5). An apurinic/apyrimidinic or ‘abasic’ site is one of the most common DNA lesions produced by both endogenous and exogenous sources (6,7). Previous studies have revealed that the replicative helicase of Escherichia coli (DnaB) and the bacteriophage T4 were not impeded by abasic lesions in tracking DNA templates (8–11). The unwinding rate of DnaB from Thermus aquaticus was reportedly only inhibited 13-fold using forked DNA with a loading tail of 30 abasic sites (12). Beyond these valuable reports, little is currently known about the dynamic process of helicase passing an abasic lesion.
The bacteriophage T7 helicase (gp4) often serves as a model of hexameric helicase and is, therefore, one of the most intensively studied helicases (13,14). The first insights into its translocation on ssDNA were derived from the crystal structure of gp4 without a DNA substrate (15) (Supplementary Figure S1A). Positions of loops protruding into the central pore are reportedly asymmetric and considered responsible for sequential hydrolysis around the ring formed by the six subunits of gp4 (15). More recently, three distinct gp4–DNA complex conformations have been observed by cryo-EM (16) (Supplementary Figure S1B). The six subunits of gp4 adopt a spiral lock-washer form that encircles a coil-like DNA (tracking) strand with 2 nucleotides (nt) bound to each subunit. Upon deoxythymidine triphosphate (dTTP) hydrolysis, the sixth subunit at the 5′ end trailing behind the fifth subunit (conformation I) is moved to the 3′ end of the strand ahead of the first subunit (conformation III) via an intermediate state of translocation (conformation II) (16). The process occurs in a hand-over-hand fashion, resulting in translocation of the helicase in the 5′–3′ direction. The mechanism predicts that gp4 translocates along the tracking strand in 2 nt steps. However, a variety of average step sizes ranging from 1 to 4 nt in the tracking strand have been reported (17–19). In addition, occasional backward movements of gp4 during DNA unwinding were observed at low dTTP concentrations (19), but the available static structures can neither explain why nor answer whether the backward movements correspond to a certain special state. Furthermore, whether a certain lesion (e.g. abasic lesion) would affect the unwinding of gp4 is unknown. If that is the case, the dynamic process should be explored.
Using a nanotensioner that we recently developed to enhance the precision of single-molecule Förster resonance energy transfer (FRET) up to 1 nt (20), we investigated the dynamic stepping mechanism of gp4 unwinding dsDNA that contains an abasic site. Surprisingly, a single abasic lesion impeded the unwinding activity of gp4. While a majority of the helicases were not blocked, a certain number of them were stalled or completely blocked by the lesion. The translocating step sizes were random and distributed widely among 1, 2, 3 and 4 nt per step. Our dynamic structural studies also revealed a chemo-mechanical coupling mechanism for gp4 that could not be deduced from the available static structures.
MATERIALS AND METHODS
Protein expression and purification
T7 helicase gene 4 protein wild-type (gp4) and a catalytically dead mutant of gp4 (gp4-D237A) were expressed and purified as previously described (21–23).
DNA constructs and annealing procedures
All oligonucleotides required to prepare the DNA substrates were purchased from Sangon Biotech Co., Ltd (Shanghai, China). The sequences of each DNA template are described in Supplementary Figure S2 and Supplementary Table S1. DNA was annealed by incubating the mixture at 95°C for 5 min and then cooling it to room temperature for ∼7 h. The annealing was carried out with a buffer containing 50 mM NaCl and 25 mM Tris–HCl (pH 7.5 at 25°C) (20). For magnetic tweezer assays, the DNA substrate was constructed as described previously (24).
Buffers
At conditions without helicase, a buffer containing 20 mM Tris–HCl (pH 8.0 at 25°C) and 50 mM NaCl was used. The gp4 helicase unwinding buffer contained 20 mM Tris–HCl (pH 8.0 at 25°C), 50 mM NaCl, 10 mM MgCl2 and 5 mM dithiothreitol. In single-molecule FRET measurements, an oxygen-scavenging system containing 0.8% (w/w) d-glucose, 1 mg/ml glucose oxidase (266.6 units/mg, Sigma), 0.4 mg/ml catalase (2000–5000 units/mg, Sigma), and 1 mM Trolox was added to the T7 helicase unwinding buffer. Final concentrations were used in all cases.
Single-molecule magnetic tweezer experiments
A flow chamber was assembled with two coverslips placed on an inverted microscope (IX71, Olympus). The coverslips were cleaned and modified with anti-digoxigenin proteins. A magnetic bead was tethered to the modified coverslip through a single DNA substrate. An external magnetic force was applied to the magnetic bead by placing a permanent magnet above the chamber. The DNA extension was monitored by analyzing the shape of the diffraction rings of the magnetic beads (25), which depended on the distance between the bead and the focal plane of the objective (100×, N.A. 1.45, Olympus). A stack of calibration images that recorded the shape of the diffraction rings versus distance was obtained by stepping the focal plane through a series of positions. After checking the state of the DNA–magnetic bead connection, the proteins (20 nM) were added to the chamber and incubated for 5 min, followed by the addition of dTTP (0.25 or 2 mM) to initiate unwinding. The DNA unwinding events were monitored by recording the distance between the magnetic bead and the surface (the DNA extension) as a function of time. The time resolution was 10 ms. The DNA extension change at a certain force can be converted into the number of base pairs unwound by using the force-extension curve of single-stranded DNA, which is well characterized by the freely jointed chain model at low forces (26). All magnetic tweezer (MT) assays were performed at a constant force of ∼8 pN.
Single-molecule FRET (smFRET) setup
The smFRET study was carried out with a home-built objective-type total internal reflection fluorescence microscopy (27). Cy3 was excited by a 532 nm sapphire laser (Coherent Inc., USA). An oil immersion objective (100×, N.A. 1.49) was used to generate an evanescent field of illumination. The fluorescence signals from Cy3 and Cy5 were split by a dichroic mirror and collected by an electron-multiplying charge-coupled-device camera (iXON, Andor Technology, South Windsor, CT, USA). The fluorescence imaging process was controlled and recorded by MetaMorph (Molecular Devices, CA, USA).
Single-molecule FRET sample preparation and data acquisition
The coverslips (Fisher Scientific, USA) and slides were cleaned thoroughly by rinsing with acetone, methanol, a mixture of sulfuric acid and hydrogen peroxide (7:3, v/v), and sodium ethoxide. The surfaces of coverslip were coated with a mixture of 99% mPEG (methoxy-PEG-5,000, Laysan Bio, Inc.) and 1% biotin-PEG (biotin-PEG-5,000, Laysan Bio, Inc.). Streptavidin was added to the microfluidic chamber and incubated for 5 min. The chamber was made of the PEG-coated coverslip. After washing with buffer, ∼100 pM DNA was added to the chamber and immobilized for 5 min. Free DNA molecules were removed by washing the chamber. After the proteins (50 nM) were incubated in the chamber for 5 min, 0.03–1 mM dTTP was added to initiate the unwinding. The unbound helicases were flushed out. The time resolution was 30 ms. All the experiments were carried out at 25°C.
Data analysis
In the single-molecule FRET measurements, the FRET efficiency was calculated by using IA/(ID + IA), where ID and IA represent the intensities of the donor and acceptor, respectively. We used a step-finding algorithm developed by Kerssemakers et al. (28) to analyze the steps. Briefly, we fitted a single large step to the data, finding the size and location based on a calculation of a chi-square. We then searched for new steps by fitting them to the plateaus from the previous cycles, each time selecting the most prominent one. This eventually led to a series of ‘best’ fits that differed only by a single-step. We then compared each best fit in the series to a ‘counter’ fit that had an equal number of steps as the original one but with step locations displaced randomly. A ‘step-indicator’ S defined as the ratio between the chi-square of the counter fit and the chi-square of the best fit was then used to evaluate the quality of the step fits. S was large when the number of steps in the best fit was close to the real number of steps. After fitting the original FRET data, the FRET of the pauses was determined, and hence the change of FRET (ΔFRET) between two adjacent pauses was calculated.
RESULTS
An abasic site in a tracking strand impedes gp4 from effectively unwinding DNA
We first used MT to measure the gp4-induced unwinding of a 120-bp dsDNA hairpin containing an abasic lesion positioned at the 51st base in the tracking strand (Figure 1A). Unwinding bursts were observed upon the addition of 20 nM gp4 and 0.25 mM dTTP. Time intervals between adjacent bursts were much longer than the bursts themselves (Supplementary Figure S3), indicating that each unwinding burst was produced by a single helicase. The unwinding can be sorted into three patterns (Figure 1B): (i) gp4 was blocked and dissociated from the substrate at the lesion to allow the hairpin to restore, ∼10%; (ii) gp4 was stalled at the lesion for a while, and then either passed the lesion or dissociated from the substrate, ∼30% and (iii) gp4 was unblocked by the lesion as if the lesion did not exist, ∼60%. The distribution of the stalling times showed an exponential function with a characteristic decay time 0.25 ± 0.02 s at 0.25 mM dTTP (Figure 1C), suggesting that the escape of gp4 from the stalled state is a simple Poisson random process. The bursts observed in the unblocked pattern were not distinguishable from those using a DNA without the lesion (Supplementary Figure S3). Similar results were also observed at 2 mM dTTP (Supplementary Figure S4).
Figure 1.
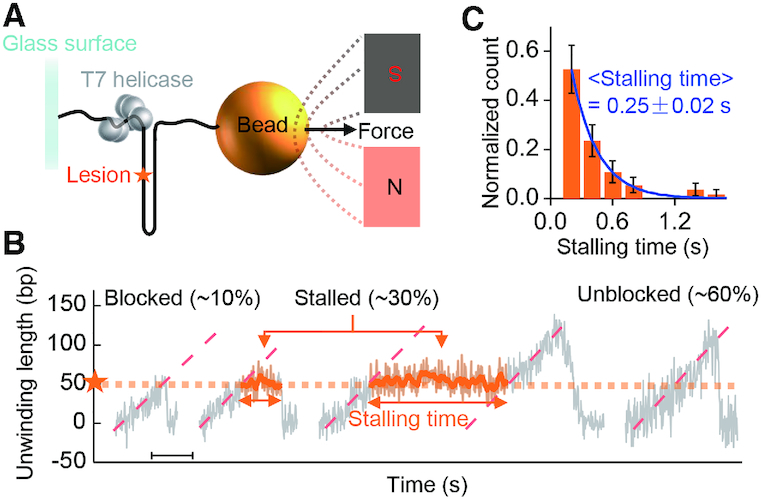
Magnetic tweezer assays of gp4 unwinding dsDNA containing an abasic lesion. (A) The experimental setup. DNA with random sequences (no more than three contiguous G/C) was used in the assays. The abasic lesion is marked by an orange asterisk. (B) Three types of unwinding patterns were observed. Scale bar, 1 s. (C) Histogram of stalling time and corresponding exponential fitting (blue line). Data were collected at 0.25 mM dTTP. The error bars are inversely proportional to the square root of the number of points for each bin. Stalling data were from 55 unwinding curves.
Gp4 moves back and forth repetitively before an abasic lesion
The dynamics of gp4 unwinding dsDNA containing an abasic lesion was further investigated using the nanotensioner-enhanced FRET (20). In the nanotensioner, a DNA duplex was bent to stretch the much softer ssDNA overhangs of a forked DNA (∼6 pN, left panel in Figure 2A) (20). Compared with the DNA fork substrates (19), the distance between the two fluorescent dyes (Cy3 and Cy5) in the nanotensioner stabilized and increased due to the tension on the overhangs. As a result, the precision of the FRET improved to 1 nt for dsDNA unwinding (20), making it ideal to reveal the distinct steps in gp4 unwinding. No unwinding signal was observed in the absence of either dTTP or MgCl2 (Supplementary Figure S5). A catalytically dead mutant of gp4 (gp4-D237A) was able to bind the DNA but could not unwind it (Supplementary Figure S6). Typical unwinding bursts for wild-type gp4 are displayed in Figure 2 for a forked DNA with an abasic lesion. We adopted an unbiased step-finding algorithm to find the unwinding steps (19,20,28,29). The configuration of gp4 having unwound all the base pairs before the lesion corresponds to a FRET value of ∼0.4, which was calibrated by a binding assay (Supplementary Figure S7). In such a configuration, two strands before the lesion were not base paired so that the lesion was at the fork (see Supplementary Figure S7).
Figure 2.
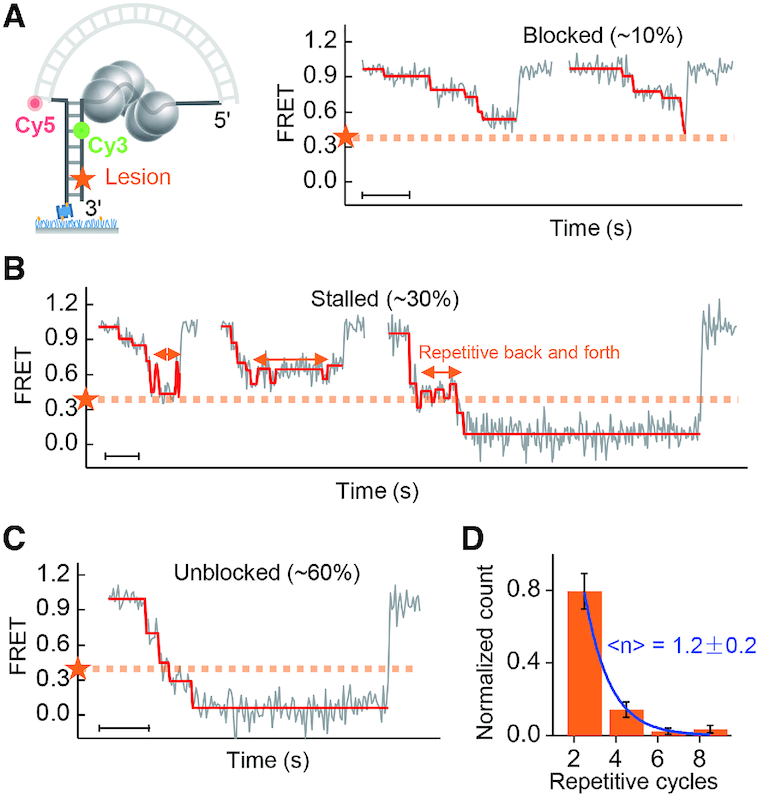
Single-molecule FRET assays of gp4 unwinding a DNA containing an abasic lesion. (A–C) DNA unwinding patterns at 0.1 mM dTTP. Scale bars, 1 s. (D) Histogram of the repetitive cycles. Each repetitive cycle includes one backward and one forward movement of gp4 in front of the lesion. The error bars are inversely proportional to the square root of the number of points for each bin. Data of repetitive cycles were from 83 unwinding curves.
Consistent with MT results, gp4 was blocked by the lesion (∼10%), stalled at the lesion (∼30%) for a while, or unblocked by the lesion (∼60%). A significant fraction of the helicases did not reach the position marked by the orange dashed lines (representing the position of the lesion) before they dissociated from the substrate or slid back to the original position. In the stalled pattern (Figure 2B), the helicase moved back and forth for a few cycles before the lesion. A histogram of the repetitive cycles (each cycle consisting of a back and forth movement of the gp4) showed an exponential decay with a characteristic cycle number of 1.2 ± 0.2 (Figure 2D). The average duration of the repetitive back and forth movements was ∼0.5 s at 0.1 mM dTTP, which was calculated by multiplying the average duration of the repetitive cycles by the average cycle number (Figure 5C). In the unblocked pattern (Figure 2C), the unwinding bursts were similar to those for normal DNA without a lesion (Supplementary Figure S3). These three unwinding patterns were also observed in measurements at different dTTP concentrations (Supplementary Figures S8 and S9).
Figure 5.
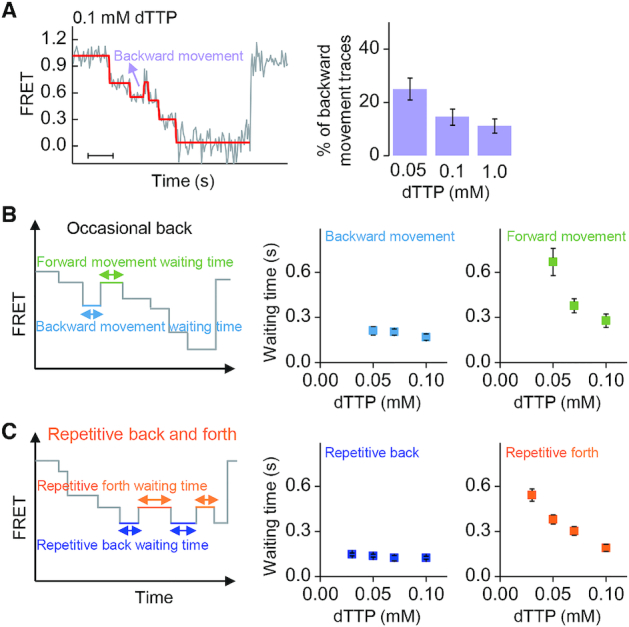
Backward movement during unwinding. (A) An example of occasional backward movement when gp4 unwound normal DNA (left panel). Scale bar, 1 s. The frequency of such single-step backward movement decreases with increasing dTTP concentration (right panel). (B) Waiting times of the occasional backward and forward movements at a normal site. (C) Waiting times of the backward and forward movements at an abasic lesion. The error bars in (A) are inversely proportional to the square root of the number of points in the bins. The error bars in (B) and (C) represent standard error of the mean.
A single lesion affects the unwinding activity of gp4 1 to 4 bp ahead of the lesion
We compared the stepping kinetics of gp4 in three different situations: when gp4 unwound the DNA with a lesion, with a mismatched base, and without any defect (Figure 3). We converted the FRET value at each step to the Cy3–Cy5 distance. The Cy3–Cy5 distance was 17 nt at FRET ≈ 0.4 (Figure 3A, Supplementary Figure S7). We constrained the analysis in the most sensitive range of FRET (0.2–0.8) and then calculated the average waiting time at each step. The averaged waiting times for the unwinding of 1–4 bp before the lesion were shorter than those for the base pairs after the lesion (Figure 3B). When the abasic site was replaced by a mismatched base, the position-specific waiting times showed a similar feature (Figure 3C). Shortened waiting time, therefore, may be attributed to the absence of Watson–Crick base pairing at the defects. In the negative control, the abasic site was replaced by a complementary base, and the average waiting times were homogenous along the DNA (Figure 3D). Our results indicated that a single defect in DNA can affect gp4 unwinding of the 1–4 preceding base pairs. Similar results were obtained in experiments at a different dTTP concentration (Supplementary Figure S10).
Figure 3.
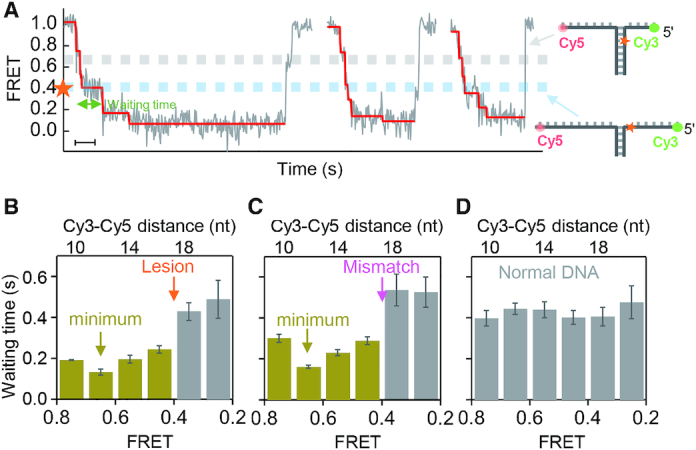
Position-specific waiting times during stepwise unwinding. (A) Typical unwinding bursts in the unblocked pattern of gp4 unwinding a lesion-containing DNA at 0.1 mM dTTP. The Cy3-Cy5 distance is 17 nt at FRET ≈ 0.4 as calibrated in Supplementary Figure S7. Scale bar, 1 s. (B–D) Distributions of the waiting times of the unblocked pattern along the DNA with a lesion (B), with a mismatched base pair (C) and without any defect (D), respectively. The histograms were built from 192, 198 and 195 unwinding curves, respectively. The error bars represent standard error of the mean.
Gp4 steps unevenly during DNA unwinding
To better understand the mechanism of gp4 passing an abasic lesion, we analyzed the stepping of gp4 when it unwound normal DNA without a lesion (Figure 4A, same sequence as in Figure 3D). The histogram of step sizes (ΔFRET) showed four major peaks that can be attributed to increases in the Cy3-Cy5 distances by 1, 2, 3 and 4 nt (Figure 4B). Similar results were obtained at different dTTP concentrations and on DNA constructions in which the dyes were labeled at different positions (see Supplementary Figure S11). The probability of missing steps was too low to explain the four major peaks (see Supplementary Figure S12). Although each unwinding burst displayed three steps on average, we found that gp4 paused at 12 positions in the range of the FRET signal from 0.2 to 0.8 when all traces were taken into account. Each pause is represented by a peak in the histogram of the pause positions in Figure 4C. The average distance between the adjacent peaks is ΔFRET ≈ 0.05, corresponding to a step size of 1 nt (20). A roughly linear relationship between the FRET value of each pause and the Cy3–Cy5 distance (in units of nt) enabled us to correlate each pause with a specific number of nucleotides translocated by the helicase, assuming that gp4 unwinding N bp would result in an increase in the Cy3–Cy5 distance of 2N nt (20,29). The possibility that the helicase could statistically visit every nucleotide during DNA unwinding was consistent with the observation that the step sizes of gp4 were uneven.
Figure 4.
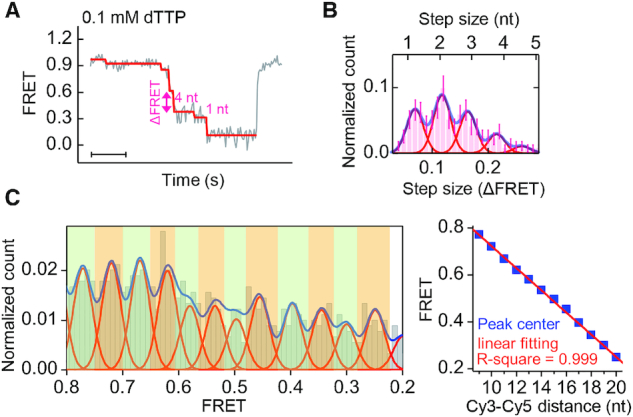
Statistical analysis of step positions and step sizes. (A) Random steps in a typical unwinding burst for a normal DNA measured at 0.1 mM dTTP. Scale bar, 1 s. (B) Histogram of step sizes (ΔFRET) with four major peaks. (C) Distribution of the FRET values of pauses after making ensemble average of 216 unwinding curves (left panel). An approximately linear relationship (with a slope of −0.047 ± 0.001) is observed between the Cy3–Cy5 distance (nt) and the FRET value of the corresponding peak (right panel). The error bars are inversely proportional to the square root of the number of points in the bins.
Gp4 occasionally moves a step back on normal DNA
We found that gp4 might occasionally move a step back even when it unwound normal dsDNA (Figure 5A). Such backward movements have been reported previously (19). About 15% of the unwinding bursts displayed a single-step backward movement at 0.1 mM dTTP (Figure 5A, right panel). The percentage, which depended on dTTP concentrations, was lower at higher dTTP concentrations (Figure 5A and Supplementary Figure S13A). The occasional backward movement occurred at random positions, in contrast with that when gp4 unwound lesion-containing DNA (Supplementary Figure S13B and C). The step size of a backward movement was equal to that of the forward movement right before the backward movement, implying that the backward movement was just the reverse of the forward movement.
We further compared the kinetics of the repetitive movements of gp4 at different dTTP concentrations (Figure 5B and C). Similar features were found for the occasional backward movements and the lesion-induced backward movements. The average waiting times of the backward movements were independent of dTTP concentrations, with an average of ∼200 ms at a normal site and ∼130 ms at the lesion site. In contrast, waiting times of the forward movements in both cases increased when dTTP concentrations decreased. The results in Figure 5B and C suggest that the forward movement was correlated with dTTP binding, and the backward movement was independent of dTTP binding (30,31).
DISCUSSION
The excluded 3′ tail is released from the surface of gp4 in a delay mode
Because gp4 is basically a passive helicase, a less stable sequence of a DNA duplex would enhance its unwinding rate (32). Our results showed that a single lesion or mismatch can affect gp4’s unwinding activity in a region that precedes the defect by 1–4 bp (Figure 3). A straightforward interpretation would be that a lesion/mismatch site destabilizes the base pairs in this region. We observed that ∼20% of forked DNA molecules can open spontaneously when only 3 bp are sandwiched between the fork and the lesion/mismatch site, even in the absence of the helicase (Supplementary Figure S14A and B), whereas forked DNA without a lesion was relatively stable (Supplementary Figure S14C). This implies that the nearer the lesion to the fork, the less stable the base pairs are between the fork and the lesion. The most unstable site should, therefore, be just 1 bp before the lesion/mismatch, where the waiting time would be the shortest. On the contrary, we observed that the shortest waiting time was on average 2.5 bp before the lesion/mismatch site (Figure 3B, C and Supplementary Figure S10). That is, it shifted by ∼1.5 bp to a higher FRET value. Therefore, it is unfeasible to interpret our data by simply assuming that a lesion/mismatch would destabilize a few base pairs surrounding it.
In general, the opening of N bp by the helicase would generate N nt in the 3′ strand and N nt in the 5′ strand. The dyes were labeled on the ssDNA overhangs in our smFRET measurements. Therefore, the observed step sizes should be even numbers of nucleotides if the newly generated nucleotides on both strands were released simultaneously after the duplex unwound. However, if the helicase released only one strand at a time, the observed step sizes could be odd numbers of nucleotides. In the hand-over-hand translocation model, gp4 unwinds DNA by encircling and translocating along the 5′ strand and partitioning the 3′ strand to the outside of the ring (‘steric exclusion’) (17–19,33). The model predicts that the nascent nucleotides in the 5′ strand would be sent immediately to the central channel of the helicase, increasing the Cy3-Cy5 distance by N nt (4,16,34–39). An odd number of nucleotides implies that gp4 sequesters the nascent nucleotides on the 3′ strand and releases them after a short delay (20,40). This view is supported by the results shown in Figure 3. If we assume that the 3′ tail was released immediately after the base pairs were uncoupled, the position with the shortest waiting time should be only 1 bp, rather than ∼2.5 bp (on average) before the lesion/mismatch. Only when the release of the newly generated 3′ tail was delayed could one observe the shift of the position with the shortest waiting time.
Gp4 passes abasic lesions by uneven stepping along the tracking strand
A previous study revealed that a cyclobutane pyrimidine dimer lesion at the tracking strand does not stall gp4 unwinding (41). Here by using single-molecule methods (MT and FRET), we observed that a single abasic lesion hinders gp4 from effectively unwinding DNA. However, the abasic lesion does not block the helicase completely. One would argue that, because the helicase binds ssDNA in a 2 nt per subunit increment, it would randomly start to unwind from either the even or the odd register where it binds the DNA substrate initially. In the presence of an abasic site, there would be two potential outcomes for the helicase. As a result, 50% of the helicases would be blocked by the lesion if the helicase initiates only from the abasic lesion, whereas the other 50% would pass the site smoothly. The value is close to the percentage observed in the experiments that ∼60% of the helicases were unblocked by the lesion. However, we performed an experiment using a DNA substrate containing two consecutive abasic lesions on the tracking strand. We expected that all helicases would be blocked. On the contrary, the experiments showed that nearly 30% of the helicases could still pass the lesion as if no lesion existed (Supplementary Figure S15). Based on the observations depicted in Figure 4, we propose that gp4 can translocate along the tracking strand in random step sizes (1, 2, 3 and 4 nt), giving the helicase chances to pass the lesion if the step size is sufficient to skip over the lesion. Accordingly, we estimated the relative probability of an unblocked pattern at the abasic site in Supplementary Figure S16. The estimate is in agreement with our experimental results. Moreover, we observed that, in a single unwinding burst, the step sizes remained unchanged when gp4 moved back and forth repetitively at the lesion (Figure 2 and Supplementary Figure S9) until it dissociated from the substrate or passed the lesion when its step size changed to a different value. Our analysis further showed that, on average, gp4 changed its step sizes after 1.2 ± 0.2 struggling cycles (Figure 2).
A recent cryo-EM study of gp4–DNA complexes revealed that 2 nt bind to each monomer of the helicase via two DNA-binding loops (16). In general, a 3D cryo-EM structure is obtained by averaging tens of thousands of particles (42), suggesting that only stable configurations can be reconstructed. However, two cases that deviate from the static structures may exist because of thermal fluctuations at room temperature at which our single molecule experiments were performed. First, only 1 nt interacts with one of the two loops of a subunit, leaving the other loop free of any nucleotide. Second, 2 nt interact with the two loops of a subunit, but with one or two extra nucleotides extruding out of the two loops with a random configuration. The first case results in 1 nt stepping, whereas the second case leads to 3 or 4 nt stepping in our single-molecule observations. The two cases do not contradict the cryo-EM results. The defect in the first case is simply overlooked, whereas the extruding nucleotides in the second case are blurred because of the randomness and are therefore difficult to see in the cryo-EM structures.
Dynamic structure and chemo-mechanical coupling of gp4
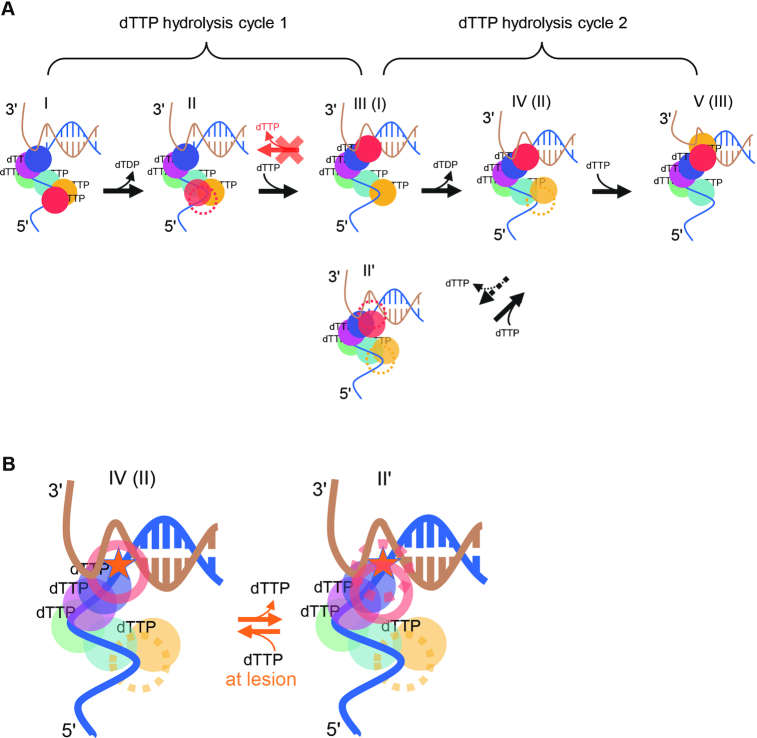
The cryo-EM structures of gp4–DNA complexes (16) (Supplementary Figure S1B) support the hypothesis that the hydrolysis of dTTP at the interface between the sixth subunit (HelF, red sphere in Figure 6) and the fifth subunit (HelE, yellow sphere) propels HelF from the conformation I to the conformation II in Figure 6, during which the FRET signal remains unchanged because the position of the front-most subunit (HelA, blue sphere) remains unchanged. When a random number of base pairs melt at the ssDNA–dsDNA junction, HelF (red sphere in the conformation II) may move to the position ahead of HelA and form a new interface with HelA (blue) to which a fresh dTTP will bind. The FRET value falls when the complex transforms from conformation II to conformation III. The process is repeated in a hand-over-hand fashion, resulting in translocation of the helicase in the 5′–3′ direction. In Figure 6, conformation III is equivalent to conformation I, whereas conformation IV is equivalent to the conformation II, except that the helicase has moved one step forward along the tracking strand.
Figure 6.
Chemo-mechanical coupling of gp4. (A) In the first hydrolysis cycle,hydrolysis of the dTTP between the red and the yellow subunits in conformation I propels the red subunit to an intermediate position in conformation II. The red subunit then moves to the front of the helicase upon the binding of a new dTTP in conformation III. The helicase moves a step forward when it changes from conformation II to conformation III. The same occurs in the second hydrolysis cycle. Gp4 may occasionally move a step back in normal DNA from conformation IV (II) to conformation II'. (B) When the leading subunit (red) steps on the abasic lesion (asterisk), gp4 may transfer between conformation IV (II) and conformation II' for a few cycles.
We then introduced an abasic site in DNA and probed the mechanism of gp4 unwinding through it. Previous studies have shown that all six subunits are required for gp4 to translocate along ssDNA (15). Our data indicate that the helicase moves a step back when the front-most subunit encounters the lesion. We also noticed that gp4 can occasionally move one step back, even when gp4 unwinds normal DNA. Because we observed very similar dTTP-dependent backward and forward movements of gp4 at the lesion and at a normal site (Figure 5B and C), we expect that the mechanisms underlying the backward movements at the lesion and at a normal site would be similar. A simple model for this would be that gp4 can step back from conformation III to II, or equivalently from conformation V to IV in Figure 6A. This would occur when the leading subunit (HelF, red in the conformation III) disengages from the lesion site and returns to its original position in conformation II, allowing the just-unwound strands to rewind. As for the normal DNA, it is possible that the leading subunit occasionally disengages from the base of a normal nucleotide by thermal fluctuation alone.
However, the aforementioned simple model cannot explain the kinetics of the occasional backward movement. The frequency of backward movements of gp4 on normal DNA decreased with increasing dTTP concentrations (Figure 5A). When gp4 is in conformation III, it has five dTTP molecules on it (16). Thus, the transition from conformation III to conformation II would not involve dTTP binding and, therefore, should not depend on dTTP concentration. We, therefore, have to build an alternative model. We argue that if gp4 is in conformation IV, it can move a step forward to conformation V by binding a new dTTP element to the leading subunit (yellow) (Figure 6A). When it is waiting for this new dTTP, the dTTP that has been at the blue-red interface may be released so that the HelF subunit (red) in conformation IV can detach from the ssDNA–dsDNA junction to allow for the unwound base pairs to re-anneal. This may result in a new conformation (conformation II′ in Figure 6) that has only three dTTP molecules on it. When the dTTP concentration falls further, gp4 may stay in conformation IV for a longer time before it transfers to conformation V. Therefore, gp4 may have more chances to transform from conformation IV to conformation II′ at lower dTTP concentrations, explaining the tendency in the right panel of Figure 5A.
This raises the question of whether the backward movement induced by the lesion follows the III to II pathway or the IV-to-II′ pathway. The average waiting time for backward movement was approximately 200 ms (Figure 5B and C). When gp4 is in conformation III, releasing a dTTP from conformation III would not take such a long time. But it may take only a few milliseconds because the binding-to-releasing cycle of dTTP by gp4 can be as fast as ∼160 dTTP/s at high dTTP concentrations (32). Alternatively, when gp4 is in conformation IV, the release of dTTP from the blue-red interface may be hindered because the yellow subunit, which is at the intermediate position of the spiral lock-washer form of gp4, can sterically hinder the motion of the red subunit. We, therefore, suspect that gp4 moves back at the lesion through the IV-to-II′ pathway (Figure 6B). Both the red sphere and the yellow sphere disengage from the tracking strand in this hypothetical gp4–DNA-II′ complex. However, this complex was not observed in the cryo-EM structures probably because the frequency of occurrence of such a conformation is low when normal DNA is used in a cryo-EM study. Our findings shed new insights into the unwinding dynamics of gp4 beyond the available static structures and suggest that new high-resolution structural studies are required to better understand the chemo-mechanical coupling mechanism of gp4 at atomic levels.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation of China [11674382, 11834018, 91753104, 11574381]; CAS Key Research Program of Frontier Sciences [QYZDJ-SSW-SYS014]; China Key Research and Development Program [2017YFC1002002]; Fundamental Research Funds for the Central Universities; Y.L. is supported by the Youth Innovation Promotion Association of CAS [2017015]. Funding for open access charge: National Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kim S.S., Dallmann H.G., McHenry C.S., Marians K.J.. Coupling of a replicative polymerase and helicase: a tau–DnaB interaction mediates papid replication fork movement. Cell. 1996; 84:643–650. [DOI] [PubMed] [Google Scholar]
- 2. Sun J.C., Shi Y., Georgescu R.E., Yuan Z.N., Chait B.T., Li H.L., O’Donnell M.E.. The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol. 2015; 22:976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yong Y., Romano L.J.. Benzo[a]pyrene-DNA adducts inhibit the DNA helicase activity of the bacteriophage T7 gene 4 protein. Chem. Res. Toxicol. 1996; 9:179–187. [DOI] [PubMed] [Google Scholar]
- 4. O’Donnell M.E., Li H.L.. The ring-shaped hexameric helicases that function at DNA replication forks. Nat. Struct. Mol. Biol. 2018; 25:122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sparks J.L., Chistol G., Gao A.O., Räschle M., Larsen N.B., Mann M., Duxin J.P., Walter J.C.. The CMG helicase bypasses DNA-protein cross-links to facilitate their repair. Cell. 2019; 176:167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kozmin S.G., Sedletska Y., Reynaud-Angelin A., Gasparutto D., Sage E.. The formation of double-strand breaks at multiply damaged sites is driven by the kinetics of excision/incision at base damage in eukaryotic cells. Nucleic Acids Res. 2009; 37:1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao B., Xie Z.W., Shen H.Y., Wang Z.G.. Role of DNA polymerase in eta in the bypass of abasic sites in yeast cells. Nucleic Acids Res. 2004; 32:3984–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Higuchi K., Katayama T., Iwai S., Hidaka M., Horiuchi T., Maki H.. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells. 2003; 8:437–449. [DOI] [PubMed] [Google Scholar]
- 9. McInerney P., O’Donnell M.. Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J. Biol. Chem. 2004; 279:21543–21551. [DOI] [PubMed] [Google Scholar]
- 10. Nelson S.W., Benkovic S.J.. Response of the bacteriophage T4 replisome to noncoding lesions and regression of a stalled replication fork. J. Mol. Biol. 2010; 401:743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marians K.J. Lesion bypass and the reactivation of stalled replication forks. Annu. Rev. Biochem. 2018; 87:217–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaplan D.L. The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J. Mol. Biol. 2000; 301:285–299. [DOI] [PubMed] [Google Scholar]
- 13. Richardson C.C. Bacteriophage T7: minimal requirements for the replication of a duplex DNA molecule. Cell. 1983; 33:315–317. [DOI] [PubMed] [Google Scholar]
- 14. Patel S.S., Hingorani M.M.. Oligomeric structure of bacteriophage T7 DNA helicase/primase proteins. J. Biol. Chem. 1993; 268:10668–10675. [PubMed] [Google Scholar]
- 15. Singleton M.R., Sawaya M.R., Ellenberger T., Wigley D.B.. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000; 101:589–600. [DOI] [PubMed] [Google Scholar]
- 16. Gao Y., Cui Y.X., Fox T., Lin S.Q., Wang H.B., de Val N., Zhou Z.H., Yang W.. Structures and operating principles of the replisome. Science. 2019; 363:eaav7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim D.E., Narayan M., Patel S.S.. T7 DNA helicase: a molecular motor that processively and unidirectionally translocates along single-stranded DNA. J. Mol. Biol. 2002; 321:807–819. [DOI] [PubMed] [Google Scholar]
- 18. Donmez I., Patel S.S.. Coupling of DNA unwinding to nucleotide hydrolysis in a ring-shaped helicase. EMBO J. 2008; 27:1718–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Syed S., Pandey M., Patel S.S., Ha T.J.. Single-molecule fluorescence reveals the unwinding stepping mechanism of replicative helicase. Cell Rep. 2014; 6:1037–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin W.X., Ma J.B., Nong D.G., Xu C.H., Zhang B., Li J.H., Jia Q., Dou S.X., Ye F.F., Xi X.G. et al.. Helicase stepping investigated with one-nucleotide resolution fluorescence resonance energy transfer. Phys. Rev. Lett. 2017; 119:138102. [DOI] [PubMed] [Google Scholar]
- 21. Zhang H.D., Lee S.J., Zhu B., Tran N.Q., Tabor S., Richardson C.C.. Helicase-DNA polymerase interaction is critical to initiate leading-strand DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:9372–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang H.D., Tang Y., Lee S.J., Wei Z.L., Cao J., Richardson C.C.. Binding affinities among DNA helicase-primase, DNA polymerase, and replication intermediates in the replisome of bacteriophage T7. J. Biol. Chem. 2016; 291:1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee S.J., Richardson C.C.. Acidic residues in the nucleotide-binding site of the bacteriophage T7 DNA primase. J. Biol. Chem. 2005; 280:26984–26991. [DOI] [PubMed] [Google Scholar]
- 24. Wang S., Qin W., Li J.H., Lu Y., Lu K.Y., Nong D.G., Dou S.X., Xu C.H., Xi X.G., Li M.. Unwinding forward and sliding back: an intermittent unwinding mode of the BLM helicase. Nucleic Acids Res. 2015; 43:3736–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Strick T.R., Allemand J.F., Bensimon D., Bensimon A., Croquette V.. The elasticity of a single supercoiled DNA molecule. Science. 1996; 271:1835–1837. [DOI] [PubMed] [Google Scholar]
- 26. Smith S.B., Cui Y.J., Bustamante C.. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996; 271:795–799. [DOI] [PubMed] [Google Scholar]
- 27. Myong S., Rasnik I., Joo C., Lohman T.M., Ha T.J.. Repetitive shuttling of a motor protein on DNA. Nature. 2005; 437:1321–1325. [DOI] [PubMed] [Google Scholar]
- 28. Kerssemakers J.W.J., Munteanu E.L., Laan L., Noetzel T.L., Janson M.E., Dogterom M.. Assembly dynamics of microtubules at molecular resolution. Nature. 2006; 442:709–712. [DOI] [PubMed] [Google Scholar]
- 29. Blosser T.R., Yang J.G., Stone M.D., Narlikar G.J., Zhuang X.W.. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009; 462:1022–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang J.C., Zhou R.B., Inoue J., Mikawa T., Ha T.J.. Single molecule analysis of Thermus thermophilus SSB protein dynamics on single-stranded DNA. Nucleic Acids Res. 2014; 42:3821–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu H.P., Xun L.Y., Xie X.S.. Single-molecule enzymatic dynamics. Science. 1998; 282:1877–1882. [DOI] [PubMed] [Google Scholar]
- 32. Johnson D.S., Bai L., Smith B.Y., Patel S.S., Wang M.D.. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007; 129:1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Satapathy A.K., Kulczyk A.W., Ghosh S., van Oijen A.M., Richardson C.C.. Coupling dTTP hydrolysis with DNA unwinding by the DNA helicase of bacteriophage T7. J. Biol. Chem. 2011; 286:34468–34478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Enemark E.J., Joshua-Tor L.. On helicases and other motor proteins. Curr. Opin. Struct. Biol. 2008; 18:243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lyubimov A.Y., Strycharska M., Berger J.M.. The nuts and bolts of ring-translocase structure and mechanism. Curr. Opin. Struct. Biol. 2011; 21:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Itsathitphaisarn O., Wing R.A., Eliason W.K., Wang J.M., Steitz T.A.. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell. 2012; 151:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Enemark E.J., Joshua-Tor L.. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006; 442:270–275. [DOI] [PubMed] [Google Scholar]
- 38. Thomsen N.D., Berger J.M.. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009; 139:523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun B., Johnson D.S., Patel G., Smith B.Y., Pandey M., Patel S.S., Wang M.D.. ATP-induced helicase slippage reveals highly coordinated subunits. Nature. 2011; 478:132–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cheng W., Arunajadai S.G., Moffitt J.R., Tinoco I.J., Bustamante C.. Single-base pair unwinding and asynchronous RNA release by the hepatitis C virus NS3 helicase. Science. 2011; 333:1746–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun B., Pandey M., Inman J.T., Yang Y., Kashlev M., Patel S.S., Wang M.D.. T7 replisome directly overcomes DNA damage. Nat. Commun. 2015; 6:10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li M.J., Xia X., Tian Y.Y., Jia Q., Liu X.Y., Lu Y., Li M., Li X.M., Chen Z.C.. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature. 2019; 567:409–413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.