Abstract
Methylation of nucleotides in ribosomal RNAs (rRNAs) is a ubiquitous feature that occurs in all living organisms. The formation of methylated nucleotides is performed by a variety of RNA-methyltransferases. Chloroplasts of plant cells result from an endosymbiotic event and possess their own genome and ribosomes. However, enzymes responsible for rRNA methylation and the function of modified nucleotides in chloroplasts remain to be determined. Here, we identified an rRNA methyltransferase, CMAL (Chloroplast MraW-Like), in the Arabidopsis chloroplast and investigated its function. CMAL is the Arabidopsis ortholog of bacterial MraW/ RsmH proteins and accounts to the N4-methylation of C1352 in chloroplast 16S rRNA, indicating that CMAL orthologs and this methyl-modification nucleotide is conserved between bacteria and the endosymbiont-derived eukaryotic organelle. The knockout of CMAL in Arabidopsis impairs the chloroplast ribosome accumulation and accordingly reduced the efficiency of mRNA translation. Interestingly, the loss of CMAL leads not only to defects in chloroplast function, but also to abnormal leaf and root development and overall plant morphology. Further investigation showed that CMAL is involved in the plant development probably by modulating auxin derived signaling pathways. This study uncovered the important role of 16S rRNA methylation mediated by CMAL in chloroplast ribosome biogenesis and plant development.
INTRODUCTION
Ribosomes are essential ribonucleoprotein complexes engaged in protein translation in cells. The plastid of plant and algal cells is thought to result from an endosymbiotic event where an early eukaryotic cell engulfed a photosynthetic cyanobacterium (1). As such, plastids possess their own genome, as well as a translational apparatus, the plastid ribosome, which is related to that of bacteria, but has adopted novel mechanisms in order to execute the specific roles that this organelle performs within a eukaryotic cell. For instance, four plastid-specific ribosomal proteins (PSRP2/cS22, PSRP3/cS23, PSRP5/cL37 and PSRP6/cL38) have been implicated to play an important role in the regulation of translation and stability of the ribosome (2–7). Interestingly, in addition to their housekeeping functions in plastid protein biosynthesis, there is mounting genetic evidence to suggest that defects in plastid ribosome function impair plant development (8). However, the underlying mechanism(s) still remain largely unknown.
The plastid ribosome is a bacterial-type 70S ribosome composed of a large (50S) and a small (30S) subunit. The ribosomal subunits consist of a few ribosomal RNA (rRNA) species and a set of ribosomal proteins. The biosynthesis of mature rRNAs requires a complex series of post-transcriptional processing steps including nucleotide modifications, some of which take place during or immediately after transcription, while others occur in a ribosome assembly-assisted manner (9,10). The methylation of rRNA is ubiquitous among all living organisms (9,10), but its extent and complexity varies between species. The process of rRNA methylation appears to have evolved to refine the structure of rRNA to optimize ribosomal function (11). The Escherichia coli rRNA contains at least 24 methylated residues, and >20 site-specific methyltransferases responsible for these modifications have been described (9). Although the catalytic mechanism of rRNA methylation is well understood, its physiological significance in vivo is still an open question; all of the rRNA methylation sites studied so far in E. coli are dispensable for cell survival, suggesting that they are not vital for the core translation cycle reactions in this bacterium (12–14). The m2G966/m5C967 methylations of the 16S rRNA may shape bacterial fitness by modulating the initiation of translation (15); however, it is currently unclear whether this function can be attributed to the majority of modified rRNA residues. To date, three different forms of nucleotides methylation have been experimentally identified in plant chloroplast RNAs (16–18). However, methyltransferases responsible for these modifications have not been described yet.
RNA methylation is catalyzed by a variety of RNA-methyltransferases which include four superfamilies. The largest number of known RNA-methyltransferases is the Rossmann-fold methyltransferase superfamily, whose name is derived from the universal structural motif known for binding of adenosine-containing cofactors (19,20). This superfamily and all other methyltransferases, with a single exception, utilize S-adenosyl-methionine (SAM) to supply the methyl group (19,20). The second largest group is made up of the SPOUT superfamily, which is named after two evolutionarily-related RNA methyltransferases, SpoU and TrmD (21,22). All known SPOUT methyltransferases function as dimers, with the catalytic site located at the interface of two monomers. Members of the third family belong to the radical SAM-dependent methyltransferase family, contain [4Fe-4S] clusters and generate radicals as part of the reaction mechanism (23). Up to now, the fourth family contains only one RNA-methyltransferase: the folate/FAD-dependent RNA methyltransferase (20). Compared to cytoplasmic RNA-methyltransferases, less information is available for organelle RNA-methyltransferases. Surprisingly, two mitochondrial proteins, h-mtTFB1 and h-mtTFB2, which share significant similarity to bacterial methyltransferases of the KsgA family, can also stimulate transcriptional activity of mitochondrial genes (24,25), suggesting that novel functions might have been acquired for organelle RNA-methyltransferases during evolution.
Here, we uncovered the role of a chloroplast-localized rRNA methyltransferase, CMAL (Chloroplast MraW-Like), in Arabidopsis. We found that CMAL accounts for the N4-methylation of C1352 in chloroplast 16S rRNA, which is indispensable for the accumulation of chloroplast ribosomes. Interestingly, the loss of CMAL leads not only to defects in chloroplast function, but also to abnormal leaf and root development and overall plant morphology, suggesting a critical role of CMAL in plant development. We further showed that CMAL is involved in the plant development probably by modulating auxin-signaling pathways.
MATERIALS AND METHODS
Plant materials and growth conditions
All Arabidopsis strains used in this study had the Columbia-0 (Col-0) background. The mutant CS823952 (cmal) was obtained from the Arabidopsis Biological Resource Center (ABRC, http://abrc.osu.edu/), and was genotyped using PCR or genomic DNA sequencing.
The homozygous cmal mutant was crossed to wild type (WT) Arabidopsis expressing DII-VENUS, PIN1:PIN1-GFP and PIN2:PIN2-GFP reporter (Stocks CS799175, CS799173 and CS23889 from ABRC) and homozygotes were recovered from F2 populations. To produce the ProCMAL:GUS construct, a 1.0-kb DNA fragment upstream of the CMAL start codon was amplified from WT genomic DNA using PCR, then inserted into the binary vector pCAMBIA-1305 and transformed into the WT. The ProDR5:GUS construct was a kind gift from Dr Tom J. Guilfoyle (26), and was transformed into both the WT and cmal mutant plants. For the complementation experiments, the full-length coding sequence of CMAL was amplified from the WT using PCR and cloned into the pCAMBIA1305 binary vector under the control of the CaMV 35S promoter. All of the constructs were transformed into Arabidopsis using the floral dip method.
The Arabidopsis seeds were surface-sterilized, cold-stratified in the dark for three days, and sown onto a half-strength MS medium. The plants were grown in a growth chamber at 22°C under a 12-h light/12-h dark cycle, with a light intensity of 120 μmol m–2 s–1 provided by cool-white fluorescent bulbs.
RT-PCR, RNA blotting and polysome profiling
Frozen leaves from 14-day-old plants were ground in liquid nitrogen and their RNA was extracted using Trizol reagent (Thermo Fisher Scientific). The RNA samples were treated with DNase I for 30 min at 37°C, then transcribed using SuperScript II Reverse Transcriptase with reduced RNase H activity (Thermo Fisher Scientific) and random primers, according to the manufacturer's instructions. The resulting cDNA samples were used for a PCR analysis with gene-specific primers. The TUBULIN6 gene was amplified as an internal control to quantify the relative cDNA abundance of the target sequences.
For the RNA blot analysis, 10 μg total RNA was loaded onto a 1.5% formaldehyde agarose gel and transferred onto nylon membranes using capillary blotting. The hybridization probes for the chloroplast genes were generated in a PCR amplification with gene-specific primers, purified using agarose gel electrophoresis, and labeled with [α32P]-dCTP. The polysomes were isolated from the leaf extracts following the method described by Barkan (27). Polysomal aliquots (0.5 ml) were layered on 4.4-ml 15–55% sucrose gradients and centrifuged for 65 min at 45 000 rpm at 4°C, after which 0.4-ml fractions were collected. The RNA in each fraction was isolated, separated, transferred onto nylon membranes and subjected to RNA blotting.
Subcellular localization assay
To determine the subcellular localization of the CMAL protein, the open reading frames of full-length CMAL was PCR-amplified and subcloned into the pUC18-35S-sGFP vector to generate their respective fusion proteins with GFP at the C-terminus. The control constructs for the subcellular localization assay were generated as described previously (28).
Protein immunoblotting
Total protein was isolated from Arabidopsis seedlings, according to the method described previously (29). Briefly, 0.05 g of Arabidopsis leaves were ground in extraction buffer (125 mM Tris, 1% SDS (w/v), 10% glycerol (v/v) and 50 mM Na2S2O5) and subsequently centrifuged at 13 000 g for 10 min. The total proteins in the supernatant were quantified using the DC Protein Assay Kit, according to the manufacturer's instructions (Bio-Rad Laboratories). The proteins were resolved using 10% and 15% SDS-PAGE and were transferred onto nitrocellulose membranes. The immunoblots were performed using specific primary antibodies, and the signals from the secondary conjugated antibodies were detected using the enhanced chemiluminescence method. Antibodies used for protein immunoblotting were described previously (30,31).
Microscopy
Embryos of WT and heterozygous plants were dissected from the ovules, fixed, and cleared as previously described (32). The embryos were then examined with Nomarski optics using a Leica DMRB microscope (Leica Microsystems) equipped with a video camera (Hitachi, HV-C20A). Samples of three-week-old WT and cmal rosette leaves were prepared for transmission electron microscopy and were imaged using a transmission electron microscope (JEM-1230; JEOL). The measurement of root epidermal cells was performed as described in Han et al. (33).
For visualization of starch in roots, tissues were decolorized in hot 80% (v/v) ethanol, rinsed in water, stained in Lugol's iodine solution. Stained sections were photographed with a stereomicroscope equipped with a CCD camera.
For GUS staining, the seedlings were fixed for 20 min in ice-cold 90% (v/v) acetone, washed three times (5 min per wash) with ice-cold phosphate buffer [100 mM sodium phosphate (pH 7)], 10 μM EDTA, 0.1% Triton X-100 and 1 mM potassium ferricyanide, and stained with X-gluc (5- bromo-4-chloro-3-indolyl-β-d-glucuronic acid) solution (2 mM X-gluc in the same phosphate buffer) for 4 h at 37°C. Pictures of individual representative seedlings were taken using a stereomicroscope equipped with a CCD camera.
GFP fluorescence was visualized using confocal microscopy (Leica TCS SP5) at an excitation wavelength of 488 and 647 nm, respectively. The autofluorescence of chlorophyll was captured between 662 and 721 nm.
Methylation analysis by bisulfite treatment
The level of RNA methylation was assessed using bisulfite-sequencing, performed according to the method described by Schaefer et al. (34). Total RNA isolated from Arabidopsis seedlings was digested with DNase I (Promega) then treated with the EpiTect Bisulfite Kit (Qiagen). The conversions were carried out in a 70-ml reaction volume containing 1 μg total RNA in 20 μl RNase-free water, 42.55 ml bisulfite mix and 17.5 ml DNA protect buffer (both provided with the kit). The RNA was denatured at 70°C for 5–10 min, which was followed by a 1-h reaction period at 60°C. The RNA was isolated from the bisulfite reaction mix then subjected to RT-PCR to amplify the target sequence. The PCR products were cleaned using a DNA Clean Kit (Qiagen) and cloned into the pGEM-T vector (Promega) for sequencing.
RNA sequencing analysis of the WT and cmal mutant
The RNA sequencing and data analysis were performed by BGI Tech Solutions, as described previously (35). Poly(A) mRNA was isolated from total RNAs of two-week-old Arabidopsis seedlings using oligo(dT) beads. The first-strand cDNA was generated using random hexamer-primed reverse transcription, after which the second-strand cDNA was synthesized using RNase H and DNA polymerase I. The RNA sequencing libraries were prepared following Illumina's protocols and were sequenced using the Illumina GA II platform. A gene expression profiling analysis was performed based on the number of tags matching the exon regions, and the number of reads per kilobase of exon model per million mapped reads (RPKMs) were calculated to evaluate the expressed value and quantify transcript levels. Gene expression differences were evaluated using a chi-square test and the false discovery rate (FDR) was also controlled. Genes that had an FDR < 0.001 and for which the FPKM estimate was 2-fold higher than that of the lowest one were identified as differentially expressed genes (DEGs). GO enrichment annotations of DEGs were calculated using the GO:TermFinder software (version v0.86). A corrected P-value ≤0.05 or a Q-value ≤0.05 was used as a threshold for ‘enriched’ DEGs. The Pathfinder Internal software was used for analysis of statistical enrichment of DEGs in KEGG pathways. Three biological replicates each with three technical replicates were performed.
Free IAA measurement
Free IAA content was measured in seedlings using ultraperformance liquid chromatography–tandem mass spectrometer (UPLC–MS/MS) as described previously (36).
RESULTS
The cmal mutant has multiple developmental defects
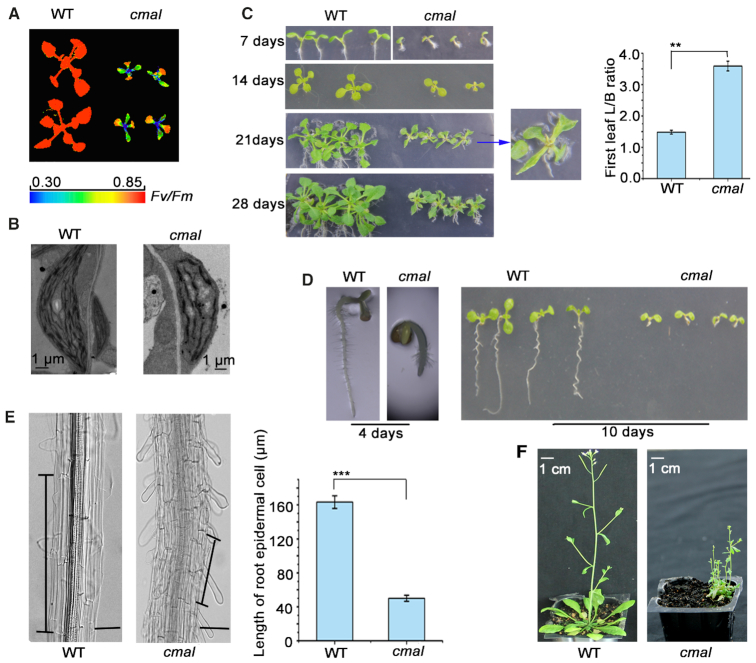
The cmal mutant was identified in a genetic screen of Arabidopsis thaliana mutants with dysfunctional chloroplasts (37). The cmal plants display a high chlorophyll fluorescence phenotype. The Fv/Fm ratio (indicating the maximum potential capacity of the photochemical reactions of photosystem II) of the newly emerging true leaves in cmal was significantly lower than that of the WT plants (Figure 1A), suggesting that photosynthetic performance is affected in cmal. A transmission electron microscopy analysis revealed that thylakoid development was also impaired in cmal (Figure 1B).
Figure 1.
The cmal mutant displays aberrant development. (A) The cmal mutant has a high chlorophyll fluorescence phenotype. The cmal and WT plants were grown on half-strength MS medium for three weeks and the chlorophyll fluorescence was measured using a CF Imager (Technologica). The Fv/Fm values are indicated in the pseudocolor index. (B) Transmission electron microscope images of the chloroplast thylakoid membranes of three-week-old leaves of WT and cmal plants. Bars = 1 μm. (C) Phenotype of cmal and WT plants grown on half-strength MS medium for 1–4 weeks. The image of one cmal seedling grown for 3 weeks was enlarged to display the leaf shape. The length:breadth ratios (L/B) of the first true leaves of 4-week-old seedlings are shown. The data represent the mean ± SD of three independent experiments, each containing ten plants per line. Statistical analysis was performed using the Student's t-test (**P < 0.01). (D) Primary roots of cmal and WT plants grown on half-strength MS medium for four and 10 days. (E) Epidermal cells of WT and cmal roots grown for 28 days. Black vertical bars mark a representative epidermal cell. The epidermal cell lengths of the WT and cmal plants are shown on the right. The data represent the mean ± SD of three independent experiments, each containing ten plants per line. Statistical analysis was performed using the Student's t-test (***P < 0.001). Bars = 20 μM. (F) Representative mature WT (6-week-old) and cmal plants (12-week-old) grown on soil. The WT plants grown for 12 weeks were already senescent and are not showed here.
In addition to the defects in chloroplast function, the cmal plants displayed several developmental disorders rarely observed in other mutants with dysfunctional chloroplasts. First, the leaf blades of cmal were significantly narrower than those of the WT (Figure 1C), which was particularly apparent in the first true leaves of cmal grown on Murashige and Skoog (MS) medium. In addition, the growth of the primary roots was significantly retarded in cmal (Figure 1D), reaching <10% of the length of the WT roots when grown for 10 days on MS medium under constant light. The root epidermal cells of cmal were shorter than those of the WT, suggesting that cell expansion is impaired in cmal roots (Figure 1E). In addition, light microscopy of iodine-stained root tips showed that levels of starch were reduced in cmal (Supplemental Figure S1), suggesting that the development of amyloplasts was affected as well in cmal. Interestingly, exogenously supplied sucrose in the MS medium was unable to relieve the development defects of roots (Supplemental Figure S2), suggesting that these defects did not merely result from the photosynthetic malfunction occurring in cmal. In conclusion, these data also indicate that CMAL acts not only in chloroplasts but also in other plastid types.
When transferred from the MS medium to soil, the cmal seedlings grew extremely slowly and their flowering time was delayed accordingly. At maturity, the cmal seedlings grown on soil had significantly more branches and displayed a bushier phenotype than the WT due to their reduced apical dominance (Figure 1F). These developmental defects are reminiscent of defects in auxin signaling (38,39), suggesting that the auxin-signaling pathway may be affected in cmal.
CMAL encodes an RNA methyltransferase localized in chloroplasts
A thermal asymmetric interlaced PCR was used to identify a T-DNA insert in the second intron of the AT5G10910 gene in cmal, which resulted in the loss of AT5G10910 mRNA in these plants (Supplemental Figure S3A). When AT5G10910 was expressed in the cmal mutants under the control of the cauliflower mosaic virus (CaMV) 35S promoter, the WT phenotype was fully restored (Supplemental Figure S3B). These results indicate that AT5G10910 is indeed responsible for the cmal mutant phenotype, and this gene is therefore referred to as CMAL hereafter.
The CMAL protein consists of 434 amino acids and shares a significant sequence identity with a SAM-dependent methyltransferase, MraW, in eubacteria (40–42). Two chloroplast-targeting prediction algorithms (TargetP, and Predotar) predicted a chloroplast transit peptide at the N terminus of the CMAL protein sequence (Supplemental Figure S4). Using a CMAL-GFP fusion protein, we showed that CMAL was indeed localized in chloroplasts (Figure 2A). It should be noted that the fluorescence signals generated with CMAL-GFP displayed a punctuated pattern resembling the appearance of nucleoids (43), suggesting that CMAL is a nucleoid-associated protein. We then further examined whether CMAL-GFP was colocalized with red fluorescent protein (RFP) fused with pTAC5, a well-known nucleoid-localized protein (44). The merged fluorescence signal of CMAL-GFP and pTAC5-RFP confirmed that CMAL and pTAC5 colocalized in chloroplast nucleoids (Figure 2A). We also constructed several transgenic lines expressing CMAL-FLAG fusion proteins and performed immunoblotting with anti-FLAG antibody to investigate the subcellular localization of CMAL. In accordance with the result of the GFP assay, we detected the CMAL-FLAG protein in chloroplasts but not in nuclei or mitochondria (Figure 2B). In addition, a proteomic assay confirmed that CMAL was localized in the chloroplast (48).
Figure 2.
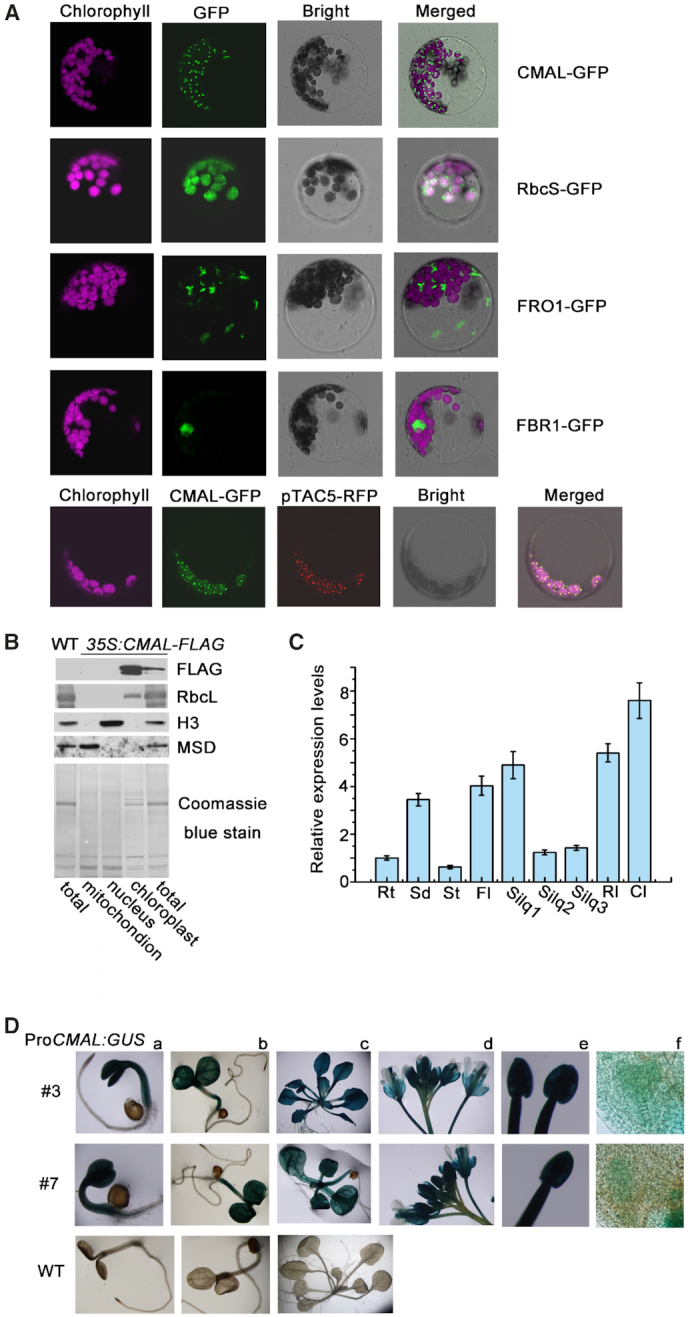
The subcellular localization and expression pattern of CMAL. (A) Subcellular localization of the CMAL protein. The CMAL-GFP fusion proteins were transiently expressed under the control of CaMV 35S promoter in Arabidopsis protoplasts and visualized with confocal laser-scanning microscopy. CMAL-GFP, signals from the CMAL-GFP fusion protein; FBR1-GFP, control for nuclear localization using fibrillarin 1 (45); FRO1-GFP, control for mitochondrial localization using FERRIC REDUCTION OXIDASE 1 (FRO1) (46); RbcS-GFP, control for chloroplast localization using the transit peptide of the ribulose bisphosphate carboxylase small subunit. Magenta signals indicate the chloroplast autofluorescence. The bottom panel indicates the colocalization of CMAL-GFP with the pTAC5-dsRED protein in the chloroplast nucleoids. The variation of chloroplast content in mesophyll protoplasts shown in this image might be due to the different developmental stages and/or cell types of the mesophyll tissue (47). (B) Immunoblot analysis of CMAL subcellular localization. Total protein, chloroplast protein, mitochondrial protein and nucleoprotein preparations from WT and transgenic plants expressing CMAL-FLAG fusion proteins were analyzed using immunoblot analysis with specific antisera against FLAG, H3, mitochondrial Mn-superoxide dismutase (MSD1), and ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (RbcL). (C) qRT-PCR analysis of CMAL mRNA in different Arabidopsis tissues and organs. Rt, roots; Sd, seedlings of 1-week-old; St, stems; Fl, flower buds; Silq1, siliques from the zygote to globular stages; Silq2, siliques from the globular to heart stages; Silq3, siliques from the heart to cotyledon stages; RL, rosette leaves; Cl, cauline leaves. Different tissues and organs were harvested from 6-week-old Arabidopsis seedlings. Expression levels of CMAL mRNAs in different tissues were normalized (expression in roots was set to 1). (D) Assay of the CMAL promoter activity. CMAL promoter-driven GUS constructs (ProCMAL:GUS) were transformed into WT plants and a histochemical analysis of the GUS activity was performed in three-day-old transgenic seedlings (a), 6-day-old seedlings (b), three-week-old seedlings (c), inflorescences (d), anthers (e), and developing seeds at the heart stage (f). Results from two independent lines (# 3 and # 7) are shown. WT plants were used as negative controls.
CMAL was differentially expressed in different Arabidopsis tissues as revealed by qRT-PCR assays (Figure 2C). To further investigate the expression profile of CMAL, the amplified promoter DNA sequence of CMAL was fused to the GUS reporter gene. The histochemical assays from two independent ProCMAL:GUS transgenic lines showed that CMAL was broadly expressed in a variety of plant tissues and organs (Figure 2D).
CMAL is involved in the N4-methylation of C1352 in chloroplast 16S rRNA
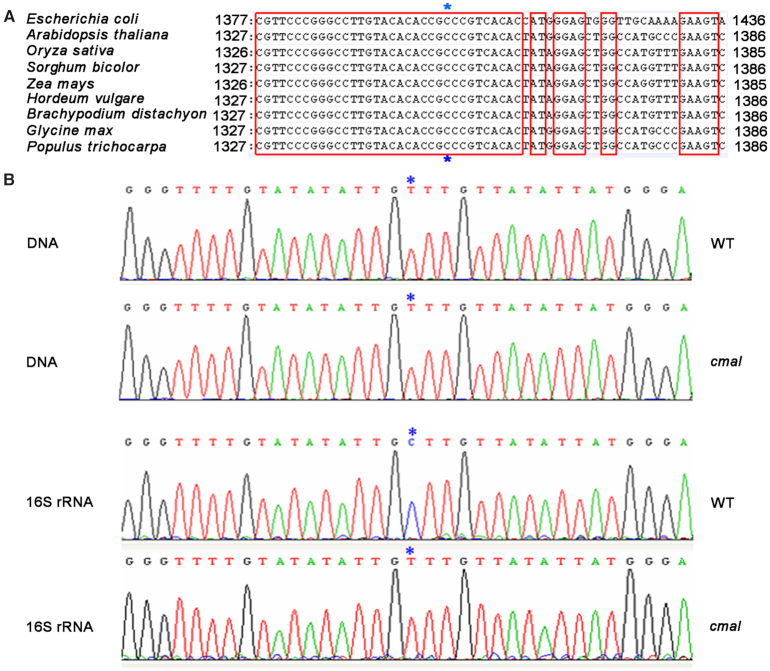
MraW uses SAM as a methyl donor and catalyzes the methyltransferase reaction to various RNA nucleotides (41). The E. coli MraW protein, RsmH, is responsible for N4-methylation at the C1402 position of 16S rRNA, which is required for the fine-tuning of the ribosomal decoding center (41). Chloroplast rRNAs are most closely related to eubacterial rRNAs, and typically share over 70% of their primary sequence identity with the E. coli 16S rRNA (49,50). In addition, the secondary structure of plastid 16S rRNA resembles that of bacteria except for the shortening of helices H6, H10, and H17 in the plastid 16S rRNA, leading to a truncated plastid 30S ribosome (4,6,7). We found that the E. coli C1402 position corresponds to C1352 of Arabidopsis 16S rRNA, and is well conserved among the 16S rRNA sequences of several plant species (Figure 3A). Based on these results, we suspected that the methylation of this nucleotide in E. coli might also occur in chloroplasts, and that CMAL might participate in its N4-methylation.
Figure 3.
CMAL is involved in the N4-methylation of C1352 in the chloroplast 16S rRNA. (A) Alignment of 16S rDNA sequences from different species. The conserved nucleotides are enclosed in red lines. Blue asterisks indicate the C1352 site of the Arabidopsis 16S rDNA. (B) Methylation analysis of the C1352 site of the 16S rRNA. Total RNA from the WT and cmal was treated with sodium bisulfite and then subjected to RT-PCR with 16S-rRNA-specific oligonucleotides. The RT-PCR products were subcloned into T-vectors for sequencing. The PCR products generated using chloroplast genomic DNAs treated with sodium bisulfite were used as controls. The sequences were assayed using the Chrosome software. Blue asterisks indicate the C1352 site.
To address this possibility, we investigated the methylation of C1352 in Arabidopsis 16S rRNA using bisulfite-sequencing in both cmal and WT plants. In this assay, the incubation of nucleotides with sodium bisulfite causes unmethylated cytosine residues to be converted into uracil, but methylated cytosines remains unchanged, thereby giving rise to different DNA sequences from methylated and unmethylated RNAs during reverse transcription (51). Our sequencing results showed that the genomic nucleotide C1352 was sequenced as T1352 following the reverse transcription of cmal 16S rRNA that had been incubated with sodium bisulfite; however, this nucleotide difference between genomic and reverse transcription products of the 16S rRNA was not detected in the WT (Figures 3A and B). As a control, the 16S rDNA from both the WT and the mutant was also subjected to bisulfite sequencing, revealing the presence of T1352 in both genotypes and indicating that no methylation was present on this nucleotide in the chloroplast genomic DNA (Figure 3B). These results clearly indicate that the methylation of C1352 in 16S rRNA occurs in the WT plants but is blocked in the cmal mutant.
In addition to C1402, E. coli rRNAs contain several other methylcytosines, which can also be detected by the RNA bisulfite sequencing (34). Thus, we investigated whether CMAL is also responsible or the methylation of other cytosines of chloroplast rRNAs using the same bisulfite sequencing as done for C1352. In addition to C1352, we identified three methylcytosine in chloroplast rRNAs from WT: C897 of the 16S rRNA, C1977 and C1940 of the 23S rRNA. Nevertheless, none of these methylation sites was affected in cmal (Supplemental Figure S5).
Chloroplast ribosome accumulation is altered in cmal
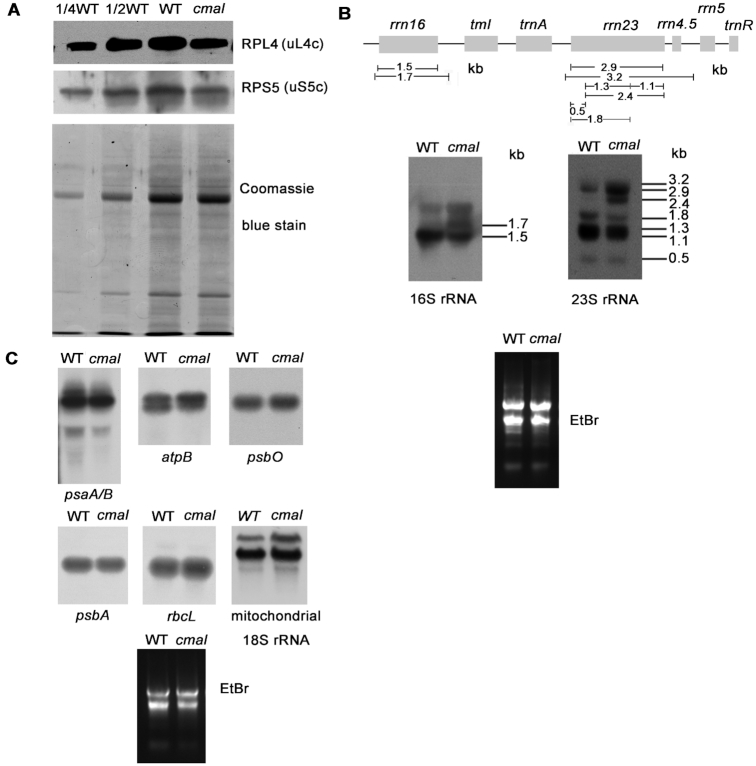
To address the possible impacts of the methylation of C1352 in the 16S rRNA on the integrity and function of the chloroplast ribosomes, we first examined the levels of chloroplast ribosomal proteins in cmal and the WT plants using immunoblot analyses. The levels of RPS5 (SMALL RIBOSOMAL SUBUNIT 5, uS5c) and RPL4 (LARGE RIBOSOMAL SUBUNIT 4, uL4c), which represent components of the 30S and 50S ribosomal subunits, respectively (2,52), were both reduced in the cmal mutant (Figure 4A). The levels of chloroplast rRNA were investigated using RNA blot analyses, which revealed that the abundance of the mature form of the 16S rRNA (1.5-kb molecule) was dramatically decreased in cmal relative to the WT, whereas the level of the precursor rRNA (1.7-kb) molecule was increased (Figure 4B). Levels of the three 23S rRNA species of 3.2, 2.9, and 2.4 kb were increased in cmal, while four shorter forms of 0.5, 1.1, 1.3 and 1.8 kb were decreased (Figure 4B). The accumulation of the 1.7 and 3.2 kb RNA species of the 16S and 23S rRNA, respectively, suggests that chloroplast rRNA processing and/or ribosome assembly may be impaired in the cmal mutant. Our analysis of the levels and patterns of representative chloroplast mRNAs revealed no significant differences between cmal and WT plants (Figure 4C). In contrast to the change of chloroplast rRNAs, we found that the abundance of mitochondrial 18S rRNA was not reduced but rather slightly increased (Figure 4C), suggesting that the mitochondrial ribosome accumulation was virtually not affected in cmal.
Figure 4.
Accumulation of chloroplast ribosomes in cmal. (A) Immunoblot analysis of chloroplast ribosomal proteins of 3-week-old cmal and WT plants. Total leaf protein (20 μg or the indicated dilution of WT samples) was analyzed by probing the immmunoblots with antibodies against RPL4 and RPS5. The Coomassie blue-stained blot at the bottom serves as a loading control. RPS5 and RPL4 are also renamed as uS5c and uL4c, respectively, according to the new nomenclature of ribosomal proteins (53) (https://bangroup.ethz.ch/research/nomenclature-of-ribosomal-proteins.html). (B) Levels of 16S and 23S rRNAs in 4-week-old cmal and WT plants. Total RNA from Arabidopsis leaves was analyzed using an RNA gel-blot hybridization with specific probes for 16S and 23S rRNA. The diagram of the rRNA operon and locations and sizes of distinct forms of rRNA species observed by RNA gel blotting are shown on the top. The gel was stained with ethidium bromide (EtBr) to visualize the rRNA, and was used as a loading control. (C) The abundance of chloroplast-encoded mRNAs and mitochondrial 18S rRNA was assayed using RNA gel-blot hybridizations as performed in (B).
The translation activity was reduced in cmal chloroplasts
Given that cmal contained significantly fewer mature ribosomal RNA molecules than the WT, and that the mutant accumulated fewer chloroplast-encoded proteins (Figure 5A), we next prepared leaf extracts fractionated in sucrose gradients under conditions that maintained intact polysomes and investigated the distribution of polysomes and free ribosomes reflecting the translation efficiency in chloroplasts. We found that the 16S rRNA was clearly shifted towards lower sucrose concentration in the mutant in comparison with the WT, suggesting that cmal contains fewer polysomes (Figure 5B). We therefore concluded that a general translation defect in cmal due to the absence of C1352 methylation in the 16S rRNA. In agreement with this hypothesis, the polysomal loading of several chloroplast mRNAs was also reduced in cmal mutant (Figure 5B).
Figure 5.
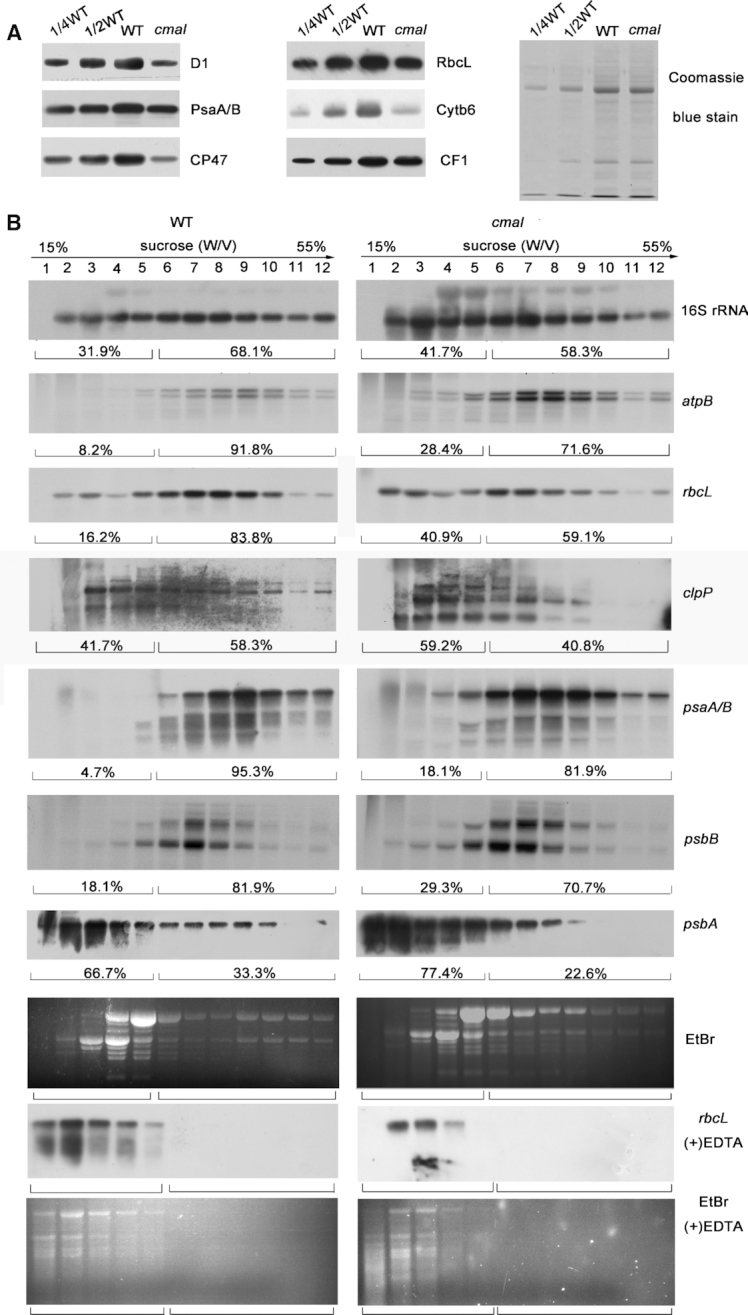
The translation efficiency of chloroplast mRNAs was reduced in cmal. (A) Immunoblot analysis of chloroplast-encoded proteins in cmal and WT plants. Total protein (10 μg) was extracted from four-week-old WT and cmal plants, separated using SDS-PAGE, and probed with different antibodies for specific proteins. The Coomassie blue-stained blot at the bottom serves as a loading control. (B) Polysome profiles of 4-week-old WT and cmal leaves. Twelve fractions of equal volume were collected from the top to bottom of 15% to 55% sucrose gradients. Equal proportions of the RNA purified from each fraction were analyzed using gel-blot hybridizations with the different probes. As a control, polysomes were isolated in the presence of EDTA which can disrupt polysome association, blotted, and hybridized to the rbcL probe. Signals of the polysomal fractions (fractions 1–5) and monosomes/free RNA fractions (fractions 6–12) were quantified with Image J respectively as described in Schult et al. (54). For the analysis of the rRNA distribution, an ethidium bromide-stained agarose gel prior to blotting is shown.
Auxin signaling is altered in cmal
To further elucidate the roles of CMAL in developmental regulation, we examined global gene expression in the WT and cmal plants using RNA sequencing. We identified 1,688 differently expressed genes (DEGs) between the cmal and WT plants (FDR < 0.001 and absolute values of log2(ratio) ≥ 1). Only 15% of the DEG products were predicted to be localized within chloroplasts (Supplemental Figure 6A). Many DEG products were predicted to be associated with the cell wall, endoplasmic reticulum, vacuole, peroxisome, cytoskeleton, Golgi apparatus, mitochondria, or the nucleus, suggesting that some biological processes outside of the chloroplast might also be affected in cmal. To explore the biological processes in which CMAL is involved, the DEGs were annotated with the reference pathway categories in the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp). Identified DEGs with highest P-values are involved in several biological pathways, including plant-pathogen interactions, plant hormone signal transduction, and stilbenoid biosynthesis (Supplemental Figure S6B). Considering the obvious developmental defects occurring in the cmal mutant, the pathway of plant hormone signal transduction came to our attention.
We found that ∼40% of the hormone-related genes belonged to the auxin pathway (Table 1), which is agreement with the fact that the developmental defects of cmal are reminiscent of defects in auxin signaling. The expression change of these auxin-related genes was further verified by qRT-PCR assays (Supplemental Figure S7). Three well-known groups of early auxin-responsive genes, the auxin/indole-3-acetic acid (Aux/IAA), small auxin-up RNA (SAUR), and GH3 gene families (55), were remarkably enriched among the genes repressed in cmal (5, 14 and 4 genes, respectively) (Table 1). In addition to the early auxin-responsive genes, several genes involved in auxin biosynthesis were down-regulated, including YUCCA8 (YUC8) encoding a putative flavin monooxygenase-like enzyme (56,57), CYP71A27 and CYP79B3 encoding two putative P450 monooxygenases (58). Interestingly, a myb-like transcription factor RVE1 that regulates free auxin levels by directly controlling the expression of YUCCA8 (59) was also down-regulated in cmal mutant (Table 1). In contrast to the change of auxin biosynthetic genes, the expression of two genes involved in auxin homeostasis ILL6 and IAR3 encoding two IAA amino acid conjugate hydrolases (60) was clearly increased in cmal (Table 1).
Table 1.
Expression change of auxin-related genes in cmal mutant
| Gene ID | Gene name | P Value | Fold changea |
|---|---|---|---|
| Early auxin-responsive genes | |||
| AT4G14560 | IAA1 | 9.82E-09 | −1.04 |
| AT1G04240 | IAA3 | 1.14E-16 | −1.17 |
| AT3G15540 | IAA19 | 1.70E-16 | −1.35 |
| AT4G32280 | IAA29 | 6.36E-20 | −1.78 |
| AT5G13370 | GH3L | 2.69E-09 | 1.03 |
| AT1G48690 | GH3 | 1.29E-06 | −2.49 |
| AT4G37390 | GH3.2 | 4.68E-08 | −1.63 |
| AT4G03400 | GH3.10 | 1.87E-06 | −1.23 |
| AT4G34770 | SAUR1 | 1.14E-19 | −2.12 |
| AT2G18010 | SAUR10 | 1.27E-05 | −2.45 |
| AT4G38850 | SAUR15 | 6.92E-11 | −2.79 |
| AT4G38860 | SAUR16 | 7.14E-14 | −1.17 |
| AT3G03840 | SAUR27 | 7.68E-06 | −1.59 |
| AT4G00880 | SAUR31 | 9.34E-08 | −1.58 |
| AT2G46690 | SAUR32 | 1.01E-16 | −1.63 |
| AT2G45210 | SAUR36 | 4.09E-07 | −1.95 |
| AT4G31320 | SAUR37 | 5.44E-10 | −3.83 |
| AT4G34760 | SAUR50 | 6.13E-14 | −1.39 |
| AT5G50760 | SAUR55 | 4.37E-17 | −2.27 |
| AT1G29440 | SAUR63 | 9.56E-07 | −1.86 |
| AT1G29450 | SAUR64 | 4.24E-05 | −1.70 |
| AT5G20820 | SAUR76 | 3.97E-06 | −1.92 |
| Auxin metabolism genes | |||
| AT5G17300 | RVE1(REVEILLE1) | 1.31E-06 | −1.08 |
| AT3G07390 | AIR12(AUXIN-INDUCED IN ROOT CULTURES 12) | 2.61E-24 | 1.27 |
| AT4G31500 | SUR2 (SUPERROOT 2) | 1.59E-57 | 1.14 |
| AT1G44350 | ILL6 [IAA-LEUCINE RESISTANT (ILR)-LIKE GENE 6] | 9.57E-65 | 2.49 |
| AT1G51760 | IAR3 (IAA-ALANINE RESISTANT3) | 5.48E-46 | 1.61 |
| AT4G28720 | YUCCA8 b | 1.94E-09 | −1.32 |
| AT4G20240 | CYP71A27 (CYTOCHROME P450, FAMILY 71) | 5.61E-06 | −1.85 |
| AT2G22330 | CYP79B3 (CYTOCHROME P450, FAMILY 79) | 3.75E-39 | −2.01 |
aLog2 ratios (cmal/WT).
bA flavin-monooxygenase in auxin biosynthesis.
Transcriptional reprogramming of early auxin-responsive genes can be triggered precisely and rapidly by the change of auxin levels in planta (39). In addition, the expression of auxin biosynthetic and homeostasis genes was altered in cmal. Based on these two lines of evidence, we suspected that the auxin level might be affected in cmal. Indeed, the free IAA content was reduced significantly in cmal compared to that of WT whereas the zeatin content was not changed as a control (Figure 6A). Therefore, the reduced auxin level in cmal might account for the down-regulation of early auxin-responsive gene expression.
Figure 6.
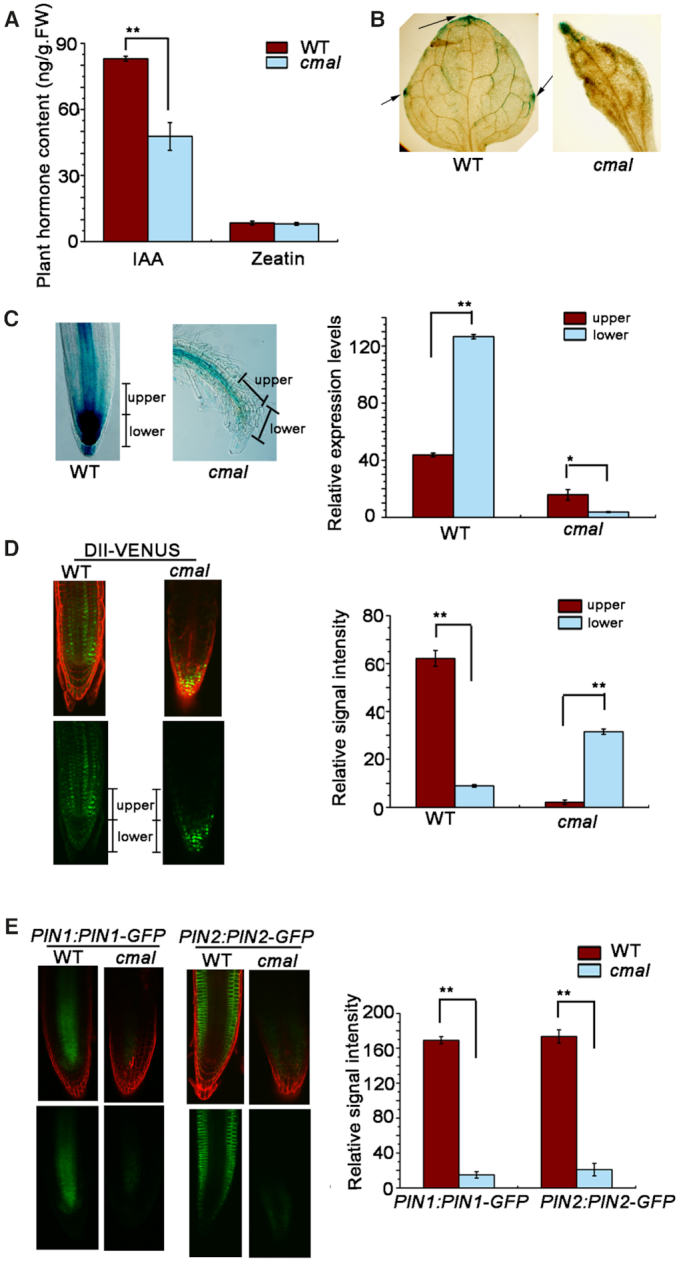
The auxin signaling and transport is affected in cmal. (A) Free IAA contents in two-week-old seedlings. Data are represented as mean ± SD (n = 5). ** P < 0.01 by Student's t test. The content of zeatin was used as a control. (B) Expression of the ProDR5:GUS reporter gene in leaves of WT and cmal plants. Arrows indicate GUS signals in the leaf margins. (C) Expression of the ProDR5:GUS reporter gene in roots of the WT and cmal plants. The ratio of GUS signals between the upper and lower sides (root tips) of WT and cmal roots. (D) Distribution of auxin indicated by DII-VENUS fluorescence in the roots of cmal and WT plants. Green channel, DII-VENUS; red channel, cell walls stained with propidium iodide. The ratio of DII-VENUS fluorescence between the upper and lower sides of WT and cmal roots is shown. (E) Expression of PIN1::PIN1-GFP and PIN2::PIN2-GFP reporters in the roots of WT and cmal plants. The green and red channels represent the GFP signal and the cell walls stained with propidium iodide, respectively. From (C) to (E), signal intensities are quantified by the Image J software and displayed in arbitrary units. Error bars represent SDs from 20 seedlings for each genotype and results were analyzed using the Student's t test (**P < 0.01).
We further examined auxin signaling by expressing the synthetic auxin-inducible reporter ProDR5:GUS (26) in transgenic cmal and WT plants (Figure 6B). We detected ProDR5:GUS staining in the leaf margins of WT. The strongest signals were observed in the leaf tip and two horizontal ends of the leaf blade, suggesting the local auxin maximum in these regions. However, the local auxin maximum was only observed in leaf tips of cmal. This apparent altered auxin distribution is in agreement with the needle-like leaf shape observed in the mutant background. In the root of WT, the strong GUS signals were observed in the root tip (root cap and meristems) and the relative weak signal along the stele of the primary root. However, signals at root tips were barely detectable and only weak signals were observed along the stele of primary roots in cmal (Figure 6B). Taken together, these findings suggest that not only the auxin level but also the auxin distribution was affected in the leaves and roots of cmal.
To further verify the alteration of auxin distribution, we used DII VENUS transgenic plants in which the fluorescence intensity of VENUS is inversely related to the auxin level (61). The fluorescence signals were less intense in the lower side of WT roots (root tips) but higher in those regions of cmal root (Figure 6C), which is in accordance with the results of the ProDR5:GUS assay.
The alternation of auxin distribution might be the consequence of defects in auxin transport (62). To test this hypothesis, we analyzed the polar distribution of the PIN-FORMED auxin efflux carriers PIN1 and PIN2 in the cmal mutant background. As shown in Figure 6D, the polar distribution pattern of the PIN1:PIN1-GFP and PIN2:PIN2-GFP markers was similar in cmal and WT plants; however, both PIN1-GFP and PIN2-GFP fluorescence signals were significantly reduced in the cmal mutant background. These results suggested that alterations of polar auxin transport might be responsible for the reduction of auxin levels in the root tips of cmal.
DISCUSSION
CMAL is critical for chloroplast ribosome biogenesis
MraW/RsmH is conserved in almost all species of eubacteria including cyanobacteria but not in archaeal species. In eukaryotes, RsmH orthologs exist in almost all plant species, several algae (red and green algae) and vertebrates (Supplemental Figure S8). Here, we revealed that the RsmH ortholog in higher plants encodes a chloroplast methyltransferse responsible for the C1352 methylation of 16S rRNA, suggesting that the role of the RsmH orthologs is conserved between eubacteria and the endosymbiont-derived chloroplast. Interestingly, m4C was identified at position 841 in the hamster mitochondrial 12S rRNA, which is equivalent to the position of C1402 in the E. coli 16S rRNA (63). This finding suggests that the animal orthologs of RsmH seem to encode a mitochondrial N4-methyltransferase that forms m4C in the 12S rRNA. Our GFP and immunoblotting assays showed that CMAL is not localized in mitochondria (Figure 2A). In addition, the accumulation of mitochondrial 18S rRNA is not affected in cmal (Figure 4C), which is distinct from the decrease of chloroplast rRNAs. These results indicate that CMAL does not participate in the methylation of mitochondrial rRNA.
It has been proposed that the E. coli Mraw/RsmH plays a role in fine-tuning the conformation and function of the ribosome P site, thus increasing decoding fidelity (41). Unfortunately, it is not feasible to assay the translation fidelity of plastids yet. Considering the conservation of helix 44 between plastid and eubacterial 30S ribosomes (4,6,7), it is likely that CMAL has the same function as RsmH in the translation fidelity. Nevertheless, knock-outs of rsmH in E. coli only result in a mild reduction in cellular growth (41). In contrast, the knock-out of CMAL in Arabidopsis resulted in strong defects in plant growth and development. Our studies showed that the loss of CMAL function led to a decrease of the mature form of the chloroplast 16S rRNAs and an increase of precursor rRNA levels (Figure 4B), suggesting that C1352 methylation of 16S rRNA is important for efficient biogenesis of the 30S ribosomal subunit in chloroplasts. The assembly of ribosomes is guided by RNA folding, wherein proteins are used to ‘lock in’ productive RNA folding and drive the structure towards its mature conformation (64). CMAL-mediated methylation may facilitate the structural rearrangements of helix 44 where C1352 is located in the 16S rRNA to establish a functionally optimal conformation during 30S ribosome assembly. In yeast, it has been reported that loss of rRNA modifications in the decoding center can affect the biogenesis of the small subunit as well as its translation activity (65). This study together with our finding suggests that a subset of rRNA modifications can influence both ribosome synthesis and function in synergistic ways.
Our results clearly indicate that the accumulation of 50S as well as 30S ribosomal subunits is altered in the cmal mutant (Figure 4A and B). CMAL might also methylate rRNAs of the 50S subunit, which could potentially affect its biogenesis. However, the methylation of the 23S rRNA was not affected as revealed by RNA-bisulfite sequencing. We cannot exclude completely that CMAL is also involved in the methylation of mRNAs. However, to data, all known rRNA methyltransferases of E. coli are specific for their substrates except RsmA/KsgA, which is responsible for the methylation of two nucleotides of the 16S rRNA (66). The crystal structure of RsmH also showed that not only C1402 itself but also the nucleotides neighboring C1402 may be necessary to trigger catalysis (42). Considering these findings together with our bisulfite sequencing data, we proposed that CMAL is specific for C1352 of the 16S rRNA rather than a promiscuous RNA methyltransferase. Indeed, 50S ribosome accumulation defects are often observed in mutants that impair 30S ribosome assembly or lack 30S ribosomal subunits in chloroplasts of higher plants (67–69), suggesting the interdependence of the biogenesis of the two ribosomal subunits. Taken together, we propose that the defect in 50S ribosome accumulation reflects an indirect consequence of 30S ribosome deficiency in cmal rather than a direct role of CMAL in 50S ribosome biogenesis.
In bacteria, C1402 of the 16S rRNA is subjected to dimethyl modification. Another methyltransferase Yral/RsmI is responsible for 2′-O-methylations of C1402 (41). The closely related Arabidopsis AT1G45110 protein showed 63% similarity to RsmI of E. coli, suggesting that AT1G45110 could be the Arabidopsis ortholog of RsmI (Supplemental Figure S9). Nevertheless, several algorithms used could not unequivocally predict the presence of an N-terminal signal peptide within this protein (http://aramemnon.uni-koeln.de). In addition, no proteome data revealed that this protein is localized in plastids yet. Thus, it remains to be clarified whether AT1G45110 is responsible for a potential 2′-O-methylation of C1352 of the plastid 16S rRNA. Some methyltransferases are highly conserved across various organisms which probably represent the core of methylations for the proper function of rRNA whereas others are not conserved (70). In agreement with this, many methylated nucleotides of rRNAs are conserved throughout prokaryotes and eukaryotes while some are unique to one or few kingdoms of life (71).
Mechanisms of CMAL involved in plant development
Several studies have suggested that plastid ribosomes or translation have important roles in particular developmental events. The formation of the palisade parenchyma is impaired in several plastid translation mutants, including those with mutations in ribosome components (72), factors involved in ribosome biogenesis (73), and plastid translation factors (74–76). Developmental changes that are not typically associated with chloroplast function, such as aberrant leaf shapes or plant architectures, can also occur in some plastid ribosome mutants (77,78). It is very interesting that the rpl36 knock-out in tobacco also leads to a similar needle-like leaf shape and reduced apical dominance as cmal (77). However, it is still unknown whether rpl36 knock-outs in Arabidopsis also lead to such developmental defects. Mutations in RFC3, a gene for the plastid ribosomal S6-like protein, in Arabidopsis result in abnormal lateral roots in which stem cell pattering of the root apical meristem is disrupted (79). Although the precise role of RFC3 in plastid ribosome biogenesis and/or translation has not been determined yet, it established a possible link between plastid ribosome and root development. In addition to genes encoding plastid ribosomal subunits or associated factors, a surprisingly large number of genes encoding for enzymes or proteins with functions in basic cellular processes such as transcription, post-transcriptional modification and translation have been found to be involved in plant organogenesis (80). This raises the still open question how organellar processes mediated by housekeeping genes are translated into a specific cellular behavior leading to defined organogenesis.
In cmal mutant, the free IAA content was reduced significantly (Figure 6A). In addition, the RNA-sequencing and ProDR5:GUS assays showed that the transcriptional response of auxin was impaired in cmal (Table 1 and Figure 6B). This clearly links chloroplast ribosome function to auxin singling pathways in regulating plant development. As the key auxin in most plants, IAA is synthesized from tryptophan (Trp) through four proposed routes according to their key intermediates, namely indole-3-acetaldoxime (IAOx), indole-3-pyruvic acid (IPA), indole-3-acetamide (IAM), and tryptamine (TAM) (81). Thus, it is possible that the reduced levels of chloroplast translation in cmal affect the chloroplast-localized enzymes involved in Trp biosynthesis, which might impact the IAA biosynthesis in cmal. This could occur by a plastome-encoded gene product influencing the activity or turnover of the (nuclear-encoded) enzymes involved in Trp metabolism. In this respect, one candidate is the CLPP1 protein, which is a subunit of the 350-kDa Clp protease complex encoded by the plastid genome (82). The polysomal loading assay showed that the translational activity of the clpP mRNA was clearly reduced in cmal (Figure 5B). It is possible that the CMAL mutation impairs the translation of the CLPP1 protein, which functions on the activity and/or stability of Trp-related enzyme(s) at the post-translation level. Interestingly, similar to cmal, disruption of CLPP1 in tobacco resulted in a slender leaf shape (83).
We found that the expression of the probable indole-3-pyruvate monooxygenase YUC8, which directly converts IPA into IAA, was reduced significantly in cmal (Table 1). Given that the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA)/YUCCA (YUC) linear pathway has been considered as a predominant Trp-dependent auxin biosynthetic pathway (84,85), it is likely that the decrease of IAA is caused by the down-regulation of YUC8. This could occur via a retrograde signaling pathway emanating from chloroplast gene expression. Interestingly, the expression of two IAA-amino acid conjugate hydrolase genes was up-regulated in cmal. This could reflect a compensatory response to the decline of free IAA levels because this enzyme can hydrolyze amide-linked conjugates to free IAA (60). The relationship between chloroplast function and auxin metabolism is even more complex because chloroplast redox homeostasis and many secondary factors affect auxin biosynthesis in plant (86).
It remains an open question how the biosynthesis and signaling pathways of auxin are affected by chloroplast translation. Impaired auxin biosynthesis/signaling and developmental defects in cmal might be related to retrograde signals which had been discovered nearly 40 ago in the barley albostrians mutant which lacks chloroplast ribosomes (87). Thereafter, many investigations confirmed the impact of chloroplast translation on nuclear gene expression and several signaling pathways linking these two processes have been proposed (88–90). It is likely that chloroplast ribosome-mediated retrograde signaling regulates the expression of auxin biosynthesis genes (for instance, YUC8) thus affecting auxin-derived signals to regulate plant development. Such signals from dysfunctional mitochondrial ribosomes have been reported to be crucial for cell proliferation in mammals (91).
In addition to free IAA levels, the auxin distribution was also affected in cmal, which might result from the impaired auxin polar transport mediated by PIN proteins. This impairment might reflect a secondary effect of auxin decline because auxin can regulate its own transport by affecting PIN polar localization and PIN protein abundance in the cell via a feedback mechanism (62). It remains possible that the putative retrograde signal from chloroplast ribosomes directly regulates the expression of PIN protein. Although the PIN1 and PIN2 mRNA levels were not changed in cmal, this signal might act on the post-transcriptional level, such as the phosphorylation of PIN proteins, which plays a central role in PIN-mediated auxin transport (62).
Interestingly, impairment of cytosolic ribosome function also affects specific development processes of plant leaves and roots (92,93). In addition, some cytosolic ribosomal proteins directly control developmental programs through translational regulation of auxin response factors (94). The phenotypic comparability between mutants defective in chloroplast and cytosolic ribosome related genes raises the interesting question whether overlapping signaling components exist. A recent study suggested a mechanism involved in balancing cytosolic and chloroplast translation programs during chloroplast biogenesis (95). It remains an outstanding challenge to study the functional interplay between cytosolic and chloroplast ribosomes in plant development.
DATA AVAILABILITY
Sequence data from this article can be found in the GenBank/EMBL library under the following accession numbers: CMAL (AT5G10910). The RNA-sequencing datasets reported in this paper have been deposited in the ArrayExpress database (accession no.E-MTAB-7904.)
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jean-David Rochaix of University of Geneva, Reimo Zoschke of Max Planck Institute of Molecular Plant Physiology in Golm and Zhongnan Yang of Shanghai Normal University for critical reading of this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDINGS
Strategic Priority Research Program of CAS [XDB17030100 to W.C.]; Ministry of Agriculture of China [2016ZX08009003-005 to W.C.]; German Science Foundation [TRR 175, project A03 to J.M.]. Funding for open access charge: Strategic Priority Research Program of CAS.
Conflict of interest statement. None declared.
REFERENCES
- 1. Reyes-Prieto A., Weber A.P., Bhattacharya D.. The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 2007; 41:147–168. [DOI] [PubMed] [Google Scholar]
- 2. Yamaguchi K., Subramanian A.R.. Proteomic identification of all plastid-specific ribosomal proteins in higher plant chloroplast 30S ribosomal subunit. Eur. J. Biochem. 2003; 270:190–205. [DOI] [PubMed] [Google Scholar]
- 3. Manuell A.L., Quispe J., Mayfield S.P.. Structure of the chloroplast ribosome: novel domains for translation regulation. PLoS Biol. 2007; 5:e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharma M.R., Wilson D.N., Datta P.P., Barat C., Schluenzen F., Fucini P., Agrawal R.K.. Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:19315–19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahmed T., Shi J., Bhushan S.. Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation. Nucleic Acids Res. 2017; 45:8581–8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bieri P., Leibundgut M., Saurer M., Boehringer D., Ban N.. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 2017; 36:475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graf M., Arenz S., Huter P., Donhofer A., Novacek J., Wilson D.N.. Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions. Nucleic Acids Res. 2017; 45:2887–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tiller N., Bock R.. The translational apparatus of plastids and its role in plant development. Mol. Plant. 2014; 7:1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Motorin Y., Helm M.. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA. 2011; 2:611–631. [DOI] [PubMed] [Google Scholar]
- 10. Nachtergaele S., He C.. The emerging biology of RNA post-transcriptional modifications. RNA Biol. 2017; 14:156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Decatur W.A., Fournier M.J.. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002; 27:344–351. [DOI] [PubMed] [Google Scholar]
- 12. Lesnyak D.V., Osipiuk J., Skarina T., Sergiev P.V., Bogdanov A.A., Edwards A., Savchenko A., Joachimiak A., Dontsova O.A.. Methyltransferase that modifies guanine 966 of the 16 S rRNA: functional identification and tertiary structure. J. Biol. Chem. 2007; 282:5880–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sergiev P.V., Serebryakova M.V., Bogdanov A.A., Dontsova O.A.. The ybiN gene of Escherichia coli encodes adenine-N6 methyltransferase specific for modification of A1618 of 23 S ribosomal RNA, a methylated residue located close to the ribosomal exit tunnel. J. Mol. Biol. 2008; 375:291–300. [DOI] [PubMed] [Google Scholar]
- 14. Sergiev P.V., Golovina A.Y., Sergeeva O.V., Osterman I.A., Nesterchuk M.V., Bogdanov A.A., Dontsova O.A.. How much can we learn about the function of bacterial rRNA modification by mining large-scale experimental datasets. Nucleic Acids Res. 2012; 40:5694–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burakovsky D.E., Prokhorova I.V., Sergiev P.V., Milon P., Sergeeva O.V., Bogdanov A.A., Rodnina M.V., Dontsova O.A.. Impact of methylations of m2G966/m5C967 in 16S rRNA on bacterial fitness and translation initiation. Nucleic Acids Res. 2012; 40:7885–7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trempe M.R., Glitz D.G.. Chloroplast ribosome structure. Electron microscopy of ribosomal subunits and localization of N6,N6-dimethyladenosine by immunoelectronmicroscopy. J. Biol. Chem. 1981; 256:11873–11879. [PubMed] [Google Scholar]
- 17. Burgess A.L., David R., Searle I.R.. Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol. 2015; 15:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Z., Tang K., Zhang D., Wan Y., Wen Y., Lu Q., Wang L.. High-throughput m6A-seq reveals RNA m6A methylation patterns in the chloroplast and mitochondria transcriptomes of Arabidopsis thaliana. PLoS One. 2017; 12:e0185612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Delk A.S., Rabinowitz J.C.. Biosynthesis of ribosylthymine in the transfer RNA of Streptococcus faecalis: a folate-dependent methylation not involving S-adenosylmethionine. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:528–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Urbonavicius J., Skouloubris S., Myllykallio H., Grosjean H.. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bac–ria–evolutionary implications. Nucleic Acids Res. 2005; 33:3955–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anantharaman V., Koonin E.V., Aravind L.. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 2002; 4:71–75. [PubMed] [Google Scholar]
- 22. Tkaczuk K.L., Dunin-Horkawicz S., Purta E., Bujnicki J.M.. Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinformatics. 2007; 8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Atta M., Mulliez E., Arragain S., Forouhar F., Hunt J.F., Fontecave M.. S-Adenosylmethionine-dependent radical-based modification of biological macromolecules. Curr. Opin. Struct. Biol. 2010; 20:684–692. [DOI] [PubMed] [Google Scholar]
- 24. Shadel G.S. Coupling the mitochondrial transcription machinery to human disease. Trends Genet. 2004; 20:513–519. [DOI] [PubMed] [Google Scholar]
- 25. Cotney J., McKay S.E., Shadel G.S.. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum. Mol. Genet. 2009; 18:2670–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J.. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997; 9:1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barkan A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 1988; 7:2637–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cai W., Ji D., Peng L., Guo J., Ma J., Zou M., Lu C., Zhang L.. LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol. 2009; 150:1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martinez-Garcia J.F., Monte E., Quail P.H.. A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 1999; 20:251–257. [DOI] [PubMed] [Google Scholar]
- 30. Zhang L., Paakkarinen V., van Wijk K.J., Aro E.M.. Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem. 1999; 274:16062–16067. [DOI] [PubMed] [Google Scholar]
- 31. Jiang J., Chai X., Manavski N., Williams-Carrier R., He B., Brachmann A., Ji D., Ouyang M., Liu Y., Barkan A. et al.. An RNA chaperone-Like protein plays critical roles in chloroplast mRNA stability and translation in Arabidopsis and Maize. Plant Cell. 2019; 31:1308–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu C.M., Meinke D.W.. The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 1998; 16:21–31. [DOI] [PubMed] [Google Scholar]
- 33. Han S., Fang L., Ren X., Wang W., Jiang J.. MPK6 controls H2 O2-induced root elongation by mediating Ca2+ influx across the plasma membrane of root cells in Arabidopsis seedlings. New Phytol. 2015; 205:695–706. [DOI] [PubMed] [Google Scholar]
- 34. Schaefer M., Pollex T., Hanna K., Lyko F.. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009; 37:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chi W., Li J., He B., Chai X., Xu X., Sun X., Jiang J., Feng P., Zuo J., Lin R. et al.. DEG9, a serine protease, modulates cytokinin and light signaling by regulating the level of ARABIDOPSIS RESPONSE REGULATOR 4. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E3568–E3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun X., Ouyang Y., Chu J., Yan J., Yu Y., Li X., Yang J., Yan C.. An in-advance stable isotope labeling strategy for relative analysis of multiple acidic plant hormones in sub-milligram Arabidopsis thaliana seedling and a single seed. J. Chromatogr. A. 2014; 1338:67–76. [DOI] [PubMed] [Google Scholar]
- 37. Chi W., Ma J., Zhang D., Guo J., Chen F., Lu C., Zhang L.. The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol. 2008; 147:573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vanneste S., Friml J.. Auxin: a trigger for change in plant development. Cell. 2009; 136:1005–1016. [DOI] [PubMed] [Google Scholar]
- 39. Weijers D., Wagner D.. Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 2016; 67:539–574. [DOI] [PubMed] [Google Scholar]
- 40. Carrion M., Gomez M.J., Merchante-Schubert R., Dongarra S., Ayala J.A.. mraW, an essential gene at the dcw cluster of Escherichia coli codes for a cytoplasmic protein with methyltransferase activity. Biochimie. 1999; 81:879–888. [DOI] [PubMed] [Google Scholar]
- 41. Kimura S., Suzuki T.. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2010; 38:1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wei Y., Zhang H., Gao Z.Q., Wang W.J., Shtykova E.V., Xu J.H., Liu Q.S., Dong Y.H.. Crystal and solution structures of methyltransferase RsmH provide basis for methylation of C1402 in 16S rRNA. J. Struct. Biol. 2012; 179:29–40. [DOI] [PubMed] [Google Scholar]
- 43. Powikrowska M., Oetke S., Jensen P.E., Krupinska K.. Dynamic composition, shaping and organization of plastid nucleoids. Front. Plant Sci. 2014; 5:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pfalz J., Liere K., Kandlbinder A., Dietz K.J., Oelmuller R.. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006; 18:176–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pih K.T., Yi M.J., Liang Y.S., Shin B.J., Cho M.J., Hwang I., Son D.. Molecular cloning and targeting of a fibrillarin homolog from Arabidopsis. Plant Physiol. 2000; 123:51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee B.H., Lee H., Xiong L., Zhu J.K.. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell. 2002; 14:1235–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lundquist P.K., Rosar C., Brautigam A., Weber A.P.. Plastid signals and the bundle sheath: mesophyll development in reticulate mutants. Mol. plant. 2014; 7:14–29. [DOI] [PubMed] [Google Scholar]
- 48. Dell’Aglio E., Giustini C., Salvi D., Brugiere S., Delpierre F., Moyet L., Baudet M., Seigneurin-Berny D., Matringe M., Ferro M. et al.. Complementary biochemical approaches applied to the identification of plastidial calmodulin-binding proteins. Mol. Biosyst. 2013; 9:1234–1248. [DOI] [PubMed] [Google Scholar]
- 49. Graf L., Roux E., Stutz E., Kossel H.. Nucleotide sequence of a Euglena gracilis chloroplast gene coding for the 16S rRNA: homologies to E. coli and Zea mays chloroplast 16S rRNA. Nucleic Acids Res. 1982; 10:6369–6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harris E.H., Boynton J.E., Gillham N.W.. Chloroplast ribosomes and protein synthesis. Microbiol. Rev. 1994; 58:700–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schaefer M. RNA 5-methylcytosine analysis by bisulfite sequencing. Methods Enzymol. 2015; 560:297–329. [DOI] [PubMed] [Google Scholar]
- 52. Yamaguchi K., Subramanian A.R.. The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 2000; 275:28466–28482. [DOI] [PubMed] [Google Scholar]
- 53. Ban N., Beckmann R., Cate J.H., Dinman J.D., Dragon F., Ellis S.R., Lafontaine D.L., Lindahl L., Liljas A., Lipton J.M. et al.. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014; 24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schult K., Meierhoff K., Paradies S., Toller T., Wolff P., Westhoff P.. The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell. 2007; 19:1329–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hagen G., Guilfoyle T.. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 2002; 49:373–385. [PubMed] [Google Scholar]
- 56. Sun J., Qi L., Li Y., Chu J., Li C.. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 2012; 8:e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hentrich M., Bottcher C., Duchting P., Cheng Y., Zhao Y., Berkowitz O., Masle J., Medina J., Pollmann S.. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 2013; 74:626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhao Y., Hull A.K., Gupta N.R., Goss K.A., Alonso J., Ecker J.R., Normanly J., Chory J., Celenza J.L.. Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002; 16:3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rawat R., Schwartz J., Jones M.A., Sairanen I., Cheng Y., Andersson C.R., Zhao Y., Ljung K., Harmer S.L.. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:16883–16888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Widemann E., Miesch L., Lugan R., Holder E., Heinrich C., Aubert Y., Miesch M., Pinot F., Heitz T.. The amidohydrolases IAR3 and ILL6 contribute to jasmonoyl-isoleucine hormone turnover and generate 12-hydroxyjasmonic acid upon wounding in Arabidopsis leaves. J. Biol. Chem. 2013; 288:31701–31714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brunoud G., Wells D.M., Oliva M., Larrieu A., Mirabet V., Burrow A.H., Beeckman T., Kepinski S., Traas J., Bennett M.J. et al.. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012; 482:103–106. [DOI] [PubMed] [Google Scholar]
- 62. Adamowski M., Friml J.. PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell. 2015; 27:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dubin D.T., Taylor R.H., Davenport L.W.. Methylation status of 13S ribosomal RNA from hamster mitochondria: the presence of a novel riboside, N4-methylcytidine. Nucleic Acids Res. 1978; 5:4385–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shajani Z., Sykes M.T., Williamson J.R.. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 2011; 80:501–526. [DOI] [PubMed] [Google Scholar]
- 65. Liang X.H., Liu Q., Fournier M.J.. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007; 28:965–977. [DOI] [PubMed] [Google Scholar]
- 66. Sergeeva O.V., Bogdanov A.A., Sergiev P.V.. What do we know about ribosomal RNA methylation in Escherichia coli. Biochimie. 2015; 117:110–118. [DOI] [PubMed] [Google Scholar]
- 67. Fristedt R., Scharff L.B., Clarke C.A., Wang Q., Lin C., Merchant S.S., Bock R.. RBF1, a plant homolog of the bacterial ribosome-binding factor RbfA, acts in processing of the chloroplast 16S ribosomal RNA. Plant Physiol. 2014; 164:201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Komatsu T., Kawaide H., Saito C., Yamagami A., Shimada S., Nakazawa M., Matsui M., Nakano A., Tsujimoto M., Natsume M. et al.. The chloroplast protein BPG2 functions in brassinosteroid-mediated post-transcriptional accumulation of chloroplast rRNA. Plant J. 2010; 61:409–422. [DOI] [PubMed] [Google Scholar]
- 69. Ehrnthaler M., Scharff L.B., Fleischmann T.T., Hasse C., Ruf S., Bock R.. Synthetic lethality in the tobacco plastid ribosome and its rescue at elevated growth temperatures. Plant Cell. 2014; 26:765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mosquera-Rendon J., Cardenas-Brito S., Pineda J.D., Corredor M., Benitez-Paez A.. Evolutionary and sequence-based relationships in bacterial AdoMet-dependent non-coding RNA methyltransferases. BMC Res. Notes. 2014; 7:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M. et al.. MODOMICS: a database of RNA modification pat–ays–2013 update. Nucleic Acids Res. 2013; 41:D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tiller N., Weingartner M., Thiele W., Maximova E., Schottler M.A., Bock R.. The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J. 2012; 69:302–316. [DOI] [PubMed] [Google Scholar]
- 73. Bang W.Y., Chen J., Jeong I.S., Kim S.W., Kim C.W., Jung H.S., Lee K.H., Kweon H.S., Yoko I., Shiina T. et al.. Functional characterization of ObgC in ribosome biogenesis during chloroplast development. Plant J. 2012; 71:122–134. [DOI] [PubMed] [Google Scholar]
- 74. Ruppel N.J., Hangarter R.P.. Mutations in a plastid-localized elongation factor G alter early stages of plastid development in Arabidopsis thaliana. BMC Plant Biol. 2007; 7:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang L., Ouyang M., Li Q., Zou M., Guo J., Ma J., Lu C., Zhang L.. The Arabidopsis chloroplast ribosome recycling factor is essential for embryogenesis and chloroplast biogenesis. Plant Mol. Biol. 2010; 74:47–59. [DOI] [PubMed] [Google Scholar]
- 76. Zheng M., Liu X., Liang S., Fu S., Qi Y., Zhao J., Shao J., An L., Yu F.. Chloroplast translation initiation factors regulate leaf variegation and development. Plant Physiol. 2016; 172:1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fleischmann T.T., Scharff L.B., Alkatib S., Hasdorf S., Schottler M.A., Bock R.. Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell. 2011; 23:3137–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mateo-Bonmati E., Casanova-Saez R., Quesada V., Hricova A., Candela H., Micol J.L.. Plastid control of abaxial-adaxial patterning. Sci. Rep. 2015; 5:15975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Horiguchi G., Kodama H., Iba K.. Mutations in a gene for plastid ribosomal protein S6-like protein reveal a novel developmental process required for the correct organization of lateral root meristem in Arabidopsis. Plant J. 2003; 33:521–529. [DOI] [PubMed] [Google Scholar]
- 80. Tsukaya H., Byrne M.E., Horiguchi G., Sugiyama M., Van Lijsebettens M., Lenhard M.. How do ‘housekeeping’ genes control organogenesis?–Unexpected new findings on the role of housekeeping genes in cell and organ differentiation. J. Plant Res. 2013; 126:3–15. [DOI] [PubMed] [Google Scholar]
- 81. Mano Y., Nemoto K.. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012; 63:2853–2872. [DOI] [PubMed] [Google Scholar]
- 82. Peltier J.B., Ytterberg J., Liberles D.A., Roepstorff P., van Wijk K.J.. Identification of a 350-kDa ClpP protease complex with 10 different Clp isoforms in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 2001; 276:16318–16327. [DOI] [PubMed] [Google Scholar]
- 83. Shikanai T., Shimizu K., Ueda K., Nishimura Y., Kuroiwa T., Hashimoto T.. The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 2001; 42:264–273. [DOI] [PubMed] [Google Scholar]
- 84. Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., Hanada A., Yaeno T., Shirasu K., Yao H. et al.. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2011; 108:18512–18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., Kasahara H., Kamiya Y., Chory J., Zhao Y.. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:18518–18523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lepisto A., Kangasjarvi S., Luomala E.M., Brader G., Sipari N., Keranen M., Keinanen M., Rintamaki E.. Chloroplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis. Plant Physiol. 2009; 149:1261–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bradbeer J.W., Atkinson Y.E., Borner T., Hagemann R.. Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesized RNA. Nature. 1979; 279:816–817. [Google Scholar]
- 88. de Souza A., Wang J.Z., Dehesh K.. Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu. Rev. Plant Biol. 2017; 68:85–108. [DOI] [PubMed] [Google Scholar]
- 89. Chan K.X., Phua S.Y., Crisp P., McQuinn R., Pogson B.J.. Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 2016; 67:25–53. [DOI] [PubMed] [Google Scholar]
- 90. Kleine T., Leister D.. Retrograde signaling: organelles go networking. Biochim. Biophys. Acta. 2016; 1857:1313–1325. [DOI] [PubMed] [Google Scholar]
- 91. Battersby B.J., Richter U.. Why translation counts for mitochon–ria - retrograde signalling links mitochondrial protein synthesis to mitochondrial biogenesis and cell proliferation. J. Cell Sci. 2013; 126:4331–4338. [DOI] [PubMed] [Google Scholar]
- 92. Byrne M.E. A role for the ribosome in development. Trends Plant Sci. 2009; 14:512–519. [DOI] [PubMed] [Google Scholar]
- 93. Horiguchi G., Van Lijsebettens M., Candela H., Micol J.L., Tsukaya H.. Ribosomes and translation in plant developmental control. Plant Sci. 2012; 191–192:24–34. [DOI] [PubMed] [Google Scholar]
- 94. Rosado A., Li R., van de Ven W., Hsu E., Raikhel N.V.. Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:19537–19544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang R., Zhao J.. Balance between cytosolic and chloroplast translation affects leaf variegation. Plant Physiol. 2018; 176:804–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data from this article can be found in the GenBank/EMBL library under the following accession numbers: CMAL (AT5G10910). The RNA-sequencing datasets reported in this paper have been deposited in the ArrayExpress database (accession no.E-MTAB-7904.)