Abstract
Cyclic diadenylate (c-di-AMP) is a widespread second messenger in bacteria and archaea that is involved in the maintenance of osmotic pressure, response to DNA damage, and control of central metabolism, biofilm formation, acid stress resistance, and other functions. The primary importance of c-di AMP stems from its essentiality for many bacteria under standard growth conditions and the ability of several eukaryotic proteins to sense its presence in the cell cytoplasm and trigger an immune response by the host cells. We review here the tertiary structures of the domains that regulate c-di-AMP synthesis and signaling, and the mechanisms of c-di-AMP binding, including the principal conformations of c-di-AMP, observed in various crystal structures. We discuss how these c-di-AMP molecules are bound to the protein and riboswitch receptors and what kinds of interactions account for the specific high-affinity binding of the c-di-AMP ligand. We describe seven kinds of non-covalent–π interactions between c-di-AMP and its receptor proteins, including π–π, C–H–π, cation–π, polar–π, hydrophobic–π, anion–π and the lone pair–π interactions. We also compare the mechanisms of c-di-AMP and c-di-GMP binding by the respective receptors that allow these two cyclic dinucleotides to control very different biological functions.
INTRODUCTION
Cyclic bis(3′→5′) dimeric adenosine monophosphate (cyclic di-AMP or c-di-AMP) is a dinucleotide second messenger that is widespread in bacteria and archaea. It was initially synthesized in 1985 as a potential inhibitor of RNA polymerase (1); five years later, it was tested for its (in)ability to replace the closely related cyclic diguanylate (c-di-GMP) as an activator of the bacterial cellulose synthase (2). The discovery of c-di-AMP in a biological system happened much later, in 2008, when it was serendipitously found in the crystal structure of the DNA integrity scanning protein DisA from Thermotoga maritima, whose close homolog in Bacillus subtilis is a sporulation checkpoint protein that senses DNA double-strand breaks (3,4). The N-terminal domain of DisA, DisA_N, has been identified as a diadenylate cyclase (DAC), responsible for producing c-di-AMP from two molecules of ATP; this activity is suppressed when DisA encounters branched DNA structures of stalled replication forks (3,4). The DisA_N domain [Pfam database entry PF02457 and COG database entries COG1623 and COG1624 (5,6)] is often referred to as the DAC domain (3,7). Although it is generally not a good idea to name a protein domain after a specific enzymatic activity (which may be lacking or altered), we are using the same name here, as all DisA_N domains characterized so far exhibited the DAC activity.
Like its better-studied sibling c-di-GMP, c-di-AMP consists of two nucleotide moieties bound by 3′→5′ phosphodiester bridges that form a 12-atom central ribose-phosphate ring. Both molecules can be seen in a wide range of conformations, from two nucleobases located side-by-side (Figure 1A, B) to fully stretched conformations where the bases are far apart (Figure 1C, D). However, while c-di-GMP is typically found in a dimer form with four guanine bases forming a parallel stack (Figure 1E), c-di-AMP is almost always seen in a monomer form with its adenine bases either parallel (Figure 1B) or arranged at an angle with each other (Figure 1F). The c-di-GMP conformations and binding mechanisms have been analyzed in detail (8) and those of c-di-AMP are discussed below.
Figure 1.
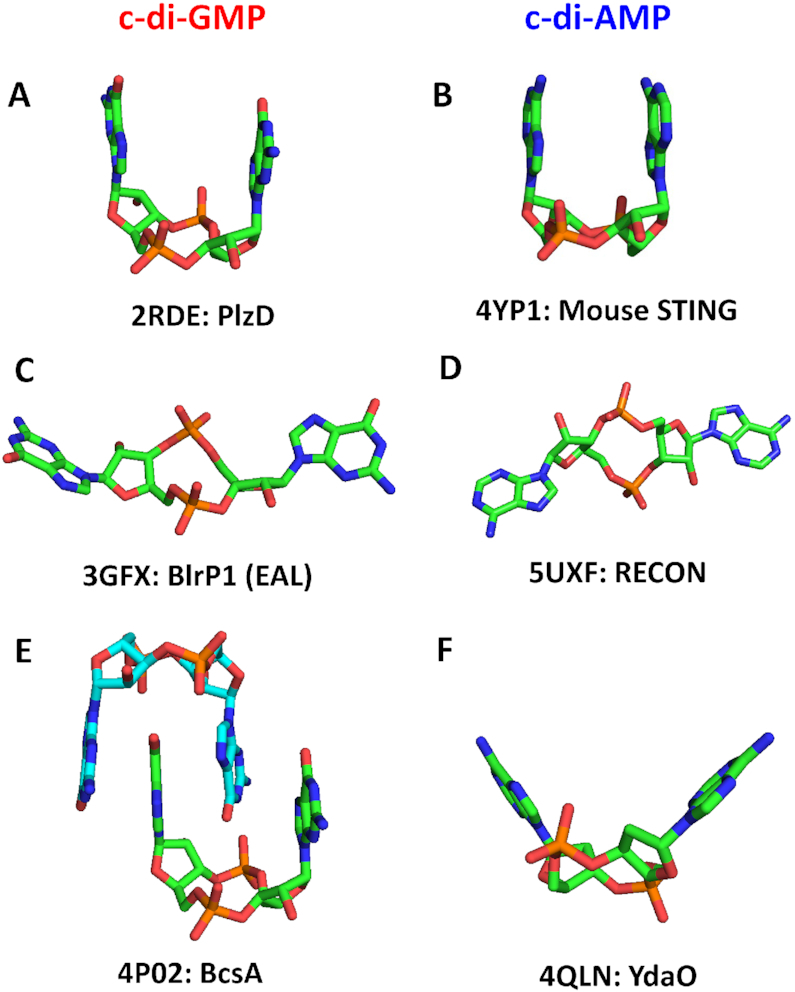
Structures of c-di-GMP and c-di-AMP molecules. (A, B) U-type conformations of c-di-GMP from the PDB entry 2RDE (in complex with the V. cholerae protein PlzD) (A), and c-di-AMP from the PDB entry 4YP1 (in complex with the cation-proton antiporter CpaA from S. aureus) (B); the two bases are oriented in an almost parallel fashion. (C, D) Extended conformations of c-di-GMP from its complex with the EAL domain of Klebsiella pneumoniae photoreceptor BlrP1, PDB: 3GFX (C), and c-di-AMP from its complex with the RECON protein, PDB: 5UXF (D); the two bases are wide apart. (E, F) Widespread conformations of c-di-GMP, a dimer from its complex with the cellulose synthase from Rhodobacter sphaeroides, PDB: 4P02 (E), and c-di-AMP, a V-type monomer from its complex with the YdaO riboswitch, PDB: 4QLN, with two adenine bases pointed in the same direction but inclined by ∼45° from vertical (F). The carbon atoms are in green (in panel E, also in light blue), N atoms are in blue, O in red and P in orange.
The two second messengers are also similar in that they are both synthesized by dedicated (but distinct) nucleotidyl cyclase domains (DAC for c-di-AMP and GGDEF for c-di-GMP) and both can be hydrolyzed by two different enzymes. For c-di-GMP, those hydrolases are the EAL and HD-GYP domains, whereas c-di-AMP hydrolysis can be catalyzed by two distinct classes of phosphodiesterases, GdpP/DhhP, which have the DHH/DHHA1 domain combination, and PgpH, which has an HD-type phosphohydrolase domain; see Table 1 for a comparison and references (9–17) for reviews. A survey of the phylogenetic distribution of the respective domains identified the likely c-di-AMP turnover machinery in most bacterial phyla and in the archaeal phylum Euryarchaeota; see Supplementary Table S1 or the respective entries in the Pfam and COG databases (5,6). Remarkably, the major groups of bacteria that do not produce c-di-AMP are alpha-, beta- and gamma subdivisions of Proteobacteria (Supplementary Table S1), the same ones that encode multiple c-di-GMP-related enzymes and where the c-di-GMP signaling has been studied in most detail (11,12). The mechanisms of c-di-AMP synthesis and hydrolysis and its physiological effects have been investigated and reviewed in detail (13–20). This review is centered on the structural aspects of c-di-AMP signaling and the specific interactions between c-di-AMP and its protein and riboswitch receptors, something that has not been comprehensively addressed until now.
Table 1.
Comparison of c-di-AMP- and c-di-GMP-mediated signaling
| c-di-GMP | c-di-AMP | |
|---|---|---|
| Discovery year | 1987 | 2008 |
| Chemical properties | ||
| Structural formula | C20H24N10O14P2 | C20H24N10O12P2 |
| Molecular weight | 690.4 Da | 658.4 Da |
| PDB structure code | C2E (95 structures) | 2BA (33 structures) |
| Oligomeric structure | Monomer, dimer (tetramer) | Monomer (dimer) |
| Intracellular levelsa | 0.1–10 μM | 0.5–18.8 μM |
| Turnover enzymes: gene and domain namesb, structures | ||
| Synthetase(cyclase) : | DgcA, AdrA GGDEF domain (PF00990, PDB: 1W25b) |
DisA, CdaA, CdaS, CdaM, CdaZ DisA_N (DAC) domain (PF02457, PDB: 3C1Y) |
| Hydrolase class I | YhjH, YkuI, BlrP1 EAL domain (PF00563, PDB: 2R6O) |
GdpP/PdeA, DhhP, DHH and DHHA1 domains (PF01388/PF02272, PDB: 5XSN) |
| Hydrolase class II | RpfG, PmGH, TM0186 HD-GYP domain (COG2206, PDB: 4R8Z) |
PgpH HDc domain (PF01966, PDB: 4S1B) |
| Receptor types | ||
| Riboswitches | Class I (PDB: 3IRW), Class II (PDB:3Q3Z) | YdaO (PDB: 4QLN) |
| Bacterial proteinsa | ||
| Widespread c-di-NMP-binding domains | PilZ domain (PF07238, PDB: 1YWU) | RCK_C domain (PF02080, PDB: 4XTT) |
| MshEN domain (PDB: 5HTL) | CBS domain pair (PDB: 5KS7) | |
| Histidine kinases | CckA, HATPase domain (PDB: 5IDM) | KdpD, USP-like domain (PDB: 2R8R) |
| Transcriptional regulators | BldD (PDB: 5KHD); FleQ (PDB: 5EXX) | BusR (RCK_C domain); DarR (COG4977) |
| Eukaryotic proteins | STING (PDB: 4EMT) | STING (PDB: 6CFF) |
| DDX41 (PDB: 5GVR) | RECON (PDB: 5UXF) | |
| Phylogenetic distribution | All bacterial phyla, Dictyostelium | Most bacterial phyla, Euryarchaeota |
| Regulated processes | Transcription, protein and polysaccharide secretion, motility, cell development, biofilm formation | DNA repair, sporulation, K+ transport, osmotic homeostasis, central metabolism, biofilm formation |
SECOND MESSENGER: SIGNAL TRANSDUCTION BY CYCLIC DI-AMP
As a second messenger, c-di-AMP serves as a signaling molecule that is being synthesized (or not synthesized) in response to certain extra- and intracellular signals and transmits this information by binding to certain cellular receptors, RNA (riboswitches) or protein molecules. As it is, unfortunately, still the case with many other signaling systems (21), the signals controlling c-di-AMP turnover remain poorly understood.
Regulation of diadenylate cyclases
All DACs characterized to date share the same DisA_N domain (DAC, Pfam: PF02457), which consists of seven β-strands forming a central β-sheet that is flanked on both sides by five α-helices (3,22). However, they differ in the associated domains (Supplementary Figure S1A), which appear to play regulatory roles (13,17). In this section, we briefly review what is known - and unknown - about these regulatory domains.
The most widespread version of DACs, exemplified by the CdaA proteins of B. subtilis and Listeria monocytogenes, consists of a DAC domain (22) that is anchored in the membrane by three N-terminal transmembrane (TM) helices, which carry several conserved residues (Supplementary Figure S1B). In these two organisms, and apparently many others, the DAC activity of CdaA is controlled by the interaction of its TM segments with the N-terminal TM fragment of the membrane-anchored extracytoplasmic protein CdaR (formerly YbbR, PF07946 and COG4856), which is usually encoded in the same conserved cdaA-cdaR operon – or in a longer cdaA-cdaR-glmM operon (23,24). The interaction with CdaR dramatically increased the activity of CdaA from B. subtilis (23) but inhibited the enzyme from L. monocytogenes (24). In some organisms, CdaA and CdaR are fused on a single polypeptide chain, forming a single protein consisting of the DAC domain that is followed by one, two, three, or four YbbR domains. High-resolution crystal structures of the ∼100-aa YbbR domain [Protein Data Bank (PDB) entries 3LYW and 5HQH] show that it consists of 7 β-strands, sometimes with an additional helix in the middle, arranged in an elongated slightly bent barrel (Supplementary Figure S1C); solution NMR structures (PDB: 2L3U, 2L5N) show much shorter β-strands (25), suggesting possible conformational flexibility of the YbbR domain. The conserved cdaA-cdaR operon is found in the members of the Firmicutes and in certain Gram-negative bacteria from Acidobacteria, Chloroflexi, Deinococcus-Thermus, Spirochetes, Synergistes, and some other phyla (compare COG1624 and COG4856, Supplementary Table S1). It remains unknown whether the YbbR domain binds peptidoglycan or any other ligand and if so, how this signal is transmitted to its N-terminal TM segment. The product of the third gene of the cdaA-cdaR-glmM operon, phosphoglucosamine mutase GlmM, has been shown to interact with CdaA and inhibit its DAC activity in B. subtilis, Lactococcus lactis and Staphylococcus aureus, apparently by masking the active site of the cyclase and preventing the formation of the catalytically active dimers (26–28). While GlmM is an essential enzyme of the peptidoglycan biosynthesis and is found in almost every bacterium, CdaA has a more limited distribution (Supplementary Table S1). Accordingly, CdaA from S.aureus has been shown to interact with its cognate GlmM but not with GlmMs from E. coli or Pseudomonas aeruginosa, which do not encode CdaA or any other DACs (28). A crystal structure of the CdaA-GlmM complex from B. subtilis should help to understand the mechanisms of such selectivity.
The DNA integrity scanning protein DisA, the first experimentally characterized form of DAC, is the only one where the signal controlling c-di-AMP synthesis has been identified. DisA combines the DAC domain with a long linker domain and a C-terminal DNA-binding helix-hairpin-helix domain that binds to branched DNA structures such as Holliday junctions and stalled replication forks, which inhibits c-di-AMP synthesis (3,4). The DisA-encoding gene is often located in the conserved radA-disA operon, and the RadA-type DNA repair ATPases from B. subtilis, Bacillus thuringiensis, Mycobacterium smegmatis, Mycobacterium tuberculosis and Streptomyces coelicolor have been shown to interact with the respective DisA proteins, forming potential tripartite complexes with branched DNA that suppressed their DAC activity (29,30). Thus, DisA interactions with branched DNA and/or RadA control the cellular levels of c-di-AMP, which therefore reflect the integrity of the chromosomal DNA and provide a sporulation checkpoint for B. subtilis. In addition to the Firmicutes, DisA-type DACs are found in the members of Actinobacteria, Thermotogae, and Dictyoglomi.
Certain spore-forming members of the Firmicutes, including B. subtilis, encode yet another form of DAC, CdaS, that combines the enzymatic DAC domain with an N-terminal YojJ domain (Pfam: PF10372), which consists of two long α-helices (32 and 28 aa, Supplementary Figure S1D) and is responsible for the formation of CdaS hexamers (31,32). Studies of this enzyme in B. subtilis and B. thuringiensis brought conflicting results. In B. subtilis, CdaS was only expressed in the forespore compartment and deletion of the YojJ domain dramatically stimulated the DAC activity of CdaS, prompting labeling it an ‘autoinhibitory domain’ (31). In B. thuringiensis, CdaS was expressed in stationary-phase cells, stimulating sporulation and parasporal crystal formation, while the YojJ domain was required for the DAC activity (32). B. subtilis spores were suggested to contain some ligand that could interact with the YojJ domain to allow the formation of the catalytically active CdaS dimers and thereby stimulate c-di-AMP production in the germinating spore (31) but the nature of that ligand remains unknown.
An experimentally characterized DAC from Mycoplasma pneumoniae (33), referred to as CdaM, has a single N-terminal TM segment (an uncleavable signal peptide) that anchors the enzyme in the membrane. Although it is tempting to suggest that this TM segment allows CdaM activity to be regulated by the status of the membrane (e.g. its turgor), there is no data to support or disprove this idea.
Many members of the archaeal phylum Euryarchaeota encode CdaZ/DacZ-type DACs that combine the DAC domain with a specific version of the PK_C (Pfam: PF02887) domain (Supplementary Figure S1E), which serves as the ligand-binding domain for allosteric regulators of pyruvate kinase activity, such as pyruvate and glucose 6-phosphate (34–36). This 110-aa α/β domain (Supplementary Figure S1F) has also been seen binding such osmolytes as proline and glycerol (34), which would be in line with the role of c-di-AMP in maintaining the osmotic balance in bacteria (see below). However, the residues that bind these osmolytes are not conserved in the CdaZ version of the PK_C domain (Supplementary Figure S1E). While the DAC activity of CdaZ-type enzymes from Methanocaldococcus jannaschii and Haloferax volcanii has been experimentally demonstrated (37,38), the ligands sensed by their N-terminal PK_C domains remain to be identified. It is also not known if ligand binding activates or inhibits the DAC activity of these enzymes.
Certain members of the Planctomycetes, Verrucomicrobiae, and Spirochetes phyla encode DACs that combine the DacZ architecture with yet another N-terminal regulatory domain (Supplementary Figure S1A), the 150-aa EIIA-like domain (Pfam: PF00359) of the phosphoenolpyruvate:sugar phosphotransferase system (PTS), which contains a conserved His residue (Supplementary Figure S1G,H) that can be phosphorylated in a PEP-dependent reaction via the Enzyme I – HPr protein cascade of the PTS (39). None of these DACs have been characterized so far, and the regulation, if any, imposed by the EIIA-like domains remains obscure.
Finally, as noted back in 2008 (3,7), some archaeal DACs contain N-terminal PAS domains. These enzymes, found in the euryarchaeal order Methanomicrobiales, have the REC–PAS1–2–DAC domain architecture, combining the ligand-binding PAS domain with the phosphoacceptor receiver (REC) domain (Supplementary Figure S1A). This puts the DAC activity of these enzymes under the control of the two-component signal transduction systems, which are particularly complex in the organisms of this group (40).
There are also other forms of DACs with additional domain architectures, such as DAC–TPR5 in Actinosynnema mirum protein Amir_4183 (Supplementary Figure S1A), which might be regulated by protein-protein interactions, but their partners remain to be identified.
Regulation of c-di-AMP phosphodiesterases
Two major classes of c-di-AMP-hydrolyzing phosphodiesterases (PDEs) have been described, those containing the DHH/DHHA1 domain combination (GdpP and DhhP families) and those containing the HD phosphohydrolase domain (26,41–45).
Historically, the first characterized PDE was the GdpP (formerly YybT) enzyme from B. subtilis, a multidomain protein that, in addition to the C-terminal DHH and DHHA1 enzymatic domains, contains two N-terminal TM segments, followed by a ligand-binding PAS domain and a degenerate GGDEF (xGGDEF) domain (41). The construct consisting solely of the DHH/DHHA1 domain combination (which is similar to the bifunctional oligoribonuclease/pAp phosphatase NrnA of B. subtilis, PDB: 5IPP) was catalytically active, hydrolyzing c-di-AMP with the Km of 1.3 μM and Kcat of 0.55 s−1 (41). The PAS domain was shown to play a regulatory role: it binds b-type heme, which in turn can bind NO and CO; binding of NO, but not CO, stimulated the PDE activity of GdpP (46). The role of the xGGDEF domain remains obscure; it was shown to possess some ATPase activity (41) but whether it contributes to the regulation of the PDE activity remains unknown. GdpP can also interact with the stringent response factor (p)ppGpp, which serves as a competitive inhibitor of its PDE activity (41). The phylogenetic distribution of GdpP-type PDEs is limited to the Firmicutes and Tenericutes (Mollicutes), see Supplementary Table S1 or COG3887 in the COG database (6).
The second type of c-di-AMP-specific PDE, PgpH, has been initially characterized in B. subtilis and L. monocytogenes (26,42) and subsequently described in the cyanobacterium Synechocystis sp. PCC 6803 (47). This protein consists of three distinct domains: an extracellular 7TMR-HDED domain preceded by a signal peptide, an integral membrane domain with 7 TM helices (7TM-7TMR-HD), and a cytoplasmic HD-type phosphodiesterase domain (14). The extracellular ligands sensed by the 7TMR-HDED domain and presumably controlling the PDE activity of PgpH still remain unknown. PgpH plays an important role in cold shock response of L. monocytogenes: a pgpH mutant was cold-sensitive, while cold-adapted strains showed a 9- to 76-fold increase in the expression of the pgpH gene (48,49). This protein has a wide phylogenetic distribution, being found, besides Firmicutes, in members of Bacteroidetes, Chlamydiae, Chloroflexi, Cyanobacteria, Fusobacteria, Planctomycetes, Spirochaetes, and other bacterial phyla, as well as in the delta subdivision of Proteobacteria, see Supplementary Table S1 or COG1480 in the COG database (6).
The observation that the DHH/DHHA1 domain combination of GdpP was capable of hydrolyzing c-di-AMP (41) suggested that the DhhP-type enzymes (COG0618), which consist solely of these two domains, should also function as c-di-AMP-specific PDEs. This suggestion proved to be partly correct, as this activity has been verified in several members of the DhhP family from Borrelia burgdorferi, M. smegmatis, M. tuberculosis, M. pneumoniae, S. aureus, Streptococcus pneumoniae and T. maritima (33,43,50–54). However, other members of the same family have been shown to also – or instead – catalyze the second step of c-di-AMP degradation, hydrolysis of pApA to AMP (33,51,53). Indeed, Supplementary Table S1 shows that many bacteria encode two paralogous members of this family, often along with either GdpP (Mollicutes, Negativicutes) or PgpH (Bacteroidetes, Chloroflexi, Cyanobacteria, Planctomycetes, Spirochaetes, Thermotogae). However, in archaea, most members of Actinobacteria and in some members of Bacteroidetes, GdpP and PgpH are both missing and the DhhP family enzymes are the only c-di-AMP hydrolases (51,52). Further, Chlamydia trachomatis, Chlamydia avium and Chlamydia muridarum encode a CdaA-type DAC but do not encode any of the known c-di-AMP PDEs (Supplementary Table S1); the mechanisms of c-di-AMP hydrolysis in such organisms remain enigmatic.
Signal transduction by c-di-AMP
The above sections show that the cellular levels of c-di-AMP can be regulated both by affecting its synthesis and hydrolysis. Studies of the bacteria with elevated or decreased c-di-AMP levels revealed its involvement in the following processes:
Finally, c-di-AMP contributes to virulence of several bacterial pathogens (50,53,74–79).
All these responses are mediated by c-di-AMP binding to its RNA and protein receptors, which come from several different protein families (Table 2). The principal c-di-AMP-binding domains are as follows.
Table 2.
Regulatory domains of c-di-AMP receptorsa
| Receptor typeb | Characterized examples | |||
|---|---|---|---|---|
| Organismc, accession numbers | Structure, PDB coded | Estimated Kd, μMe | References | |
| Riboswitch | ||||
| ydaO (potE) | Bacillus subtilis | 4W90 | <0.1 nM, 0.7 nM | (100,116) |
| Bacillus subtilis | 7.4 ± 2.2 nM | (117) | ||
| Nostoc punctiforme | – | 30 nM | (100) | |
| Syntrophus aciditrophicus | – | 0.55 nM | (100) | |
| Clostridium acetobutylicum | – | 1 nM | (100) | |
| Thermovirga lienii | 4QK9 | 300 ± 166 nM | (117) | |
| Thermoanaerobacter pseudethanolicus | 4QK8 | – | (117) | |
| Caldanaerobacter subterraneus | 4QLN | – | (118) | |
| Protein (domain) name | ||||
| RCK_C (TrkA_C) domain, PF02080 | ||||
| K+ uptake protein TrkA, COG0569 | S. aureus (ABD22474) | 4XTT | 0.043 ± 0.016; 0.064 ± 0.003; 0.369 ± 0.044 | (57,111) |
| B. subtilis (CAB15087, BAA07051) | – | – | (66) | |
| L. monocytogenes (CAC99101) | – | 1.2 ± 0.2 | (62) | |
| M. pneumoniae (AAB96028) | – | – | (33) | |
| S. agalactiae (CAD47298, CAD47337) | – | – | (88) | |
| S. mutans (AAN59208, AAN59343) | – | 7.8 ± 0.3 | (67) | |
| S. pneumoniae (ABJ54144) | 0.15 | (58) | ||
| Na+/H+ and K+/H+ antiporter CpaA, COG0475 | S. aureus (ABD21176) | 5F29 | 9.0 | (57,99) |
| B. subtilis (CAB13021) | – | – | (66) | |
| K+/H+ antiporter subunit KhtT, COG0490 | B. subtilis (CAB12826, CAB14723) | – | – | (66) |
| Transcriptional regulator BusR | L. lactis R (CAL97640) | – | 10 | (65) |
| S. agalactiae (CAD46860) | (88) | |||
| CBS pair (PF00571) | ||||
| Carnitine transporter subunit OpuCA, COG1125 | L. monocytogenes (CAC99506) | 5KS7 | 1.2 | (63) |
| E. faecalis (AAO80496) | – | 6 | (63) | |
| S. aureus (AAW38543) | – | 2.46 ± 0.14 | (64) | |
| S. agalactiae (CAD45880) | – | – | (88) | |
| B. subtilis (CAB15388) | – | – | (66) | |
| Mg2+ transporter MgtE, COG2239 | B. subtilis (CAB13187) | – | – | (66) |
| Stand-alone CBS domain, COG0517 | B. subtilis (CAB13286) | – | 1.8 ± 0.2 | (66) |
| L. monocytogenes (CAC98632, CAC99087) | – | 2.2 ± 0.4 | (69) | |
| USP domain | ||||
| K+-sensing histidine kinase KdpD, COG2205 | S. aureus (AAW37032, CAG39096) | – | 2.01 ± 0.18 | (57,59) |
| L. monocytogenes (CAD00892) | – | – | (62,63) | |
| S. elongatus (ABB57759) | – | – | (72) | |
| K+ uptake protein KimA, COG0531, PF13520 | B. subtilis (CAB12239) | – | – | (60,66) |
| L. monocytogenes (CAD00208) | – | – | (62) | |
| S. aureus (AAW38535) | – | – | (66) | |
| KupAC domain | ||||
| K+ transporter Kup/TrkD, COG3158, PF02705 | L. lactis (AAK04721, AAK04722, CAL97188) | – | – | (65,82) |
| Other receptors | ||||
| PII-like protein DarA/PstA, COG3870, PF06153 | L. monocytogenes (CAD00905) | 4RWW | 1.4 | (69,86) |
| B. subtilis (CAB11805) | 4RLE | 0.64, 1.3 ± 0.07 | (84) | |
| S. aureus (AAW37644) | 4WK1, 4D3H | 0.109 | (85,87) | |
| Pyruvate carboxylase, COG1038 | L. monocytogenes (CAC99150), | 4QSH | 8 ± 0.2 | (69) |
| L. lactis (AAK04767) | 5VYZ | – | (139) | |
| E. faecalis (AAO82174) | – | – | (69) | |
| Transcriptional regulators | ||||
| M. smegmatis DarR (ABK70852) | – | 2.3 ± 0.5 | (71) | |
| L. monocytogenes NrdR (CAC99640) | – | – | (69) | |
| Eukaryotic proteins | ||||
| STING | Sea anemone | 5CFN | – | (113) |
| Mouse | 4YP1 | 1.85 | (99) | |
| Pig | 6IYF | – | (112) | |
| Human | 6CFF | – | (140) | |
| DDX41 | Mouse | – | – | (103) |
| RECON | Mouse | 5UXF | 0.087 | (104) |
| ERAdP | Mouse | – | 0.076 | (105) |
aThe L. monocytogenes c-di-AMP phosphodiesterases PdeA (lmo0052) and PgpH (lmo1466), identified as c-di-AMP binders structurally, by binding to the c-di-AMP-linked Sepharose beads, and in the DRaCALA assay (42,69) are omitted here but listed in Table 1. The ClpC (lmo0232), CtaP (lmo0135), OppA (lmo2196), Ndh (lmo2638) and Adh (lmo1634) proteins, identified as potential c-di-AMP binders in the DRaCALA assay (69) are not listed as their interaction with c-di-AMP is likely indirect.
bThe protein and domain names are taken from Pfam (5) and/or the COG database (6). Sequence alignments and sequence logos of the newly described KimAC and KupAC domains are provided in Figures S3 and S4 in the Supplementary Materials.
cFor the full names of organisms, see the text or the respective GenBank or UniProt entries.
dWhere available, protein structures listed are those in complex with bound c-di-AMP.
e K d values are in nM for the riboswitches and in μM for protein receptors.
The RCK_C (regulator of conductance of K+C-terminal) domain, referred to as the TrkA_C (PF02080) domain in Pfam (5). Four distinct classes of c-di-AMP receptors containing this domain have been characterized; in two cases, the structures of c-di-AMP–RCK_C complexes have been solved (Table 2). So far, most experimentally studied RCK_C domains have been found to bind c-di-AMP (66), although they are also widespread in alpha-, beta- and gamma-proteobacteria that do not produce c-di-AMP. Several widespread RCK_C-containing domain architectures, including its associations with voltage-gated chloride channel (PF00654), sodium-sulfate symporter (PF00939), citrate transporter (PF03600) and aspartate-alanine exchanger (PF06826), remain to be experimentally characterized.
Pairs of the CBS (cystathionine beta-synthase) domain (Pfam: PF00571), also referred to as the Bateman domain (80,81), bind a variety of adenine derivatives and are found in a wide variety of domain architectures. Two of these domain combinations, along with the stand-alone CBS domain pairs, have been shown to bind c-di-AMP (Table 2). By contrast, the majority of B. subtilis CBS-containing proteins do not bind c-di-AMP (66). Accordingly, only a relatively small fraction of the widespread CBS domains (Supplementary Table S1) can be expected to function as c-di-AMP receptors; those associated with the voltage-gated chloride channel (PF00654) and magnesium transporters of the CNNM (cyclin M, PF01595) family look like plausible candidates.
K+ transport-regulating histidine kinases KdpD from several bacteria have been shown to bind c-di-AMP (Table 2) and the binding site has been localized to the USP-like (universal stress protein, Pfam: PF00582) domain of this protein (59). In addition, binding of c-di-AMP by the K+ uptake protein KimA has been localized to its C-terminal cytoplasmic domain (62), which also belongs to the USP superfamily (Supplementary Figure S2).
Two closely related c-di-AMP-regulated K+ uptake proteins from L. lactis, KupA and KupB (82), combine a 12-TM membrane transporter domain with a previously uncharacterized C-terminal cytoplasmic domain. This domain, which we refer to as the KupAC domain (Supplementary Figure S3), can be predicted to serve as yet another receptor of c-di-AMP.
Several more proteins, listed in Table 2, do not appear to have dedicated c-di-AMP-binding domains. Instead, c-di-AMP binding occurs at allosteric sites on the protein surface. One of such c-di-AMP binding proteins is structurally related to the family of PII-like signal transduction proteins that are involved in many pathways, often associated with nitrogen metabolism. It was originally named PstA (PII-like signal transduction protein) (57), which caused certain confusion, as that name had been previously used for the widespread membrane permease subunit of the bacterial ABC-type phosphate transporter (83). In B. subtilis, this protein has been renamed from YaaQ to DarA (c-di-AMP receptor A) (84), which was not ideal either because the darA symbol is also used for the genes for darobactin family peptide antibiotic and the phage defense against restriction protein A. Since both DarA and PstA designations are used in the literature (69,84–87), we refer to this protein here as DarA/PstA.
Other c-di-AMP receptors without dedicated binding domains but with experimentally characterized c-di-AMP binding sites include pyruvate carboxylase and the eukaryotic STING and RECON proteins.
Physiological responses to c-di-AMP have been described in detail in several comprehensive reviews (13–20), which is why in subsequent sections, we discuss primarily the mechanistic aspects of c-di-AMP-binding.
PRIMARY IMPORTANCE: ESSENTIALITY OF CYCLIC DI-AMP
Uniquely among known second messengers, c-di-AMP plays important roles in representatives of all three domains of life. Most bacteria and some archaea can produce c-di-AMP and it has been found to be essential for the growth of bacteria (23,26,33,56) and, recently, for an archaeon (38). In eukaryotic cells, c-di-AMP is not synthesized; its presence in cell cytoplasm is sensed by such proteins as STING, RECON, DDX41 and ERAdP and is perceived as a sign of an infection and a signal to trigger an immune response by the host cells.
Essentiality of c-di-AMP in bacteria
The very first attempts to prepare deletion mutants lacking the DAC activity showed that c-di-AMP was essential for such bacteria as B. subtilis (23,56), S. aureus (55), L. monocytogenes (77), S. pneumoniae (53), Streptococcus agalactiae (88) and other species. The essentiality of c-di-AMP was also observed for the spirochete Borrelia burgdorferi (50), the cell wall-less mollicute M. pneumoniae (33), and the archaeon H. volcanii (38), showing that this phenomenon is not limited to the members of Firmicutes or just Gram-positive bacteria. In contrast, a disA deletion mutant of M. tuberculosis was viable; it showed no c-di-AMP production and decreased virulence but no slowdown of growth (79). A viable c-di-AMP-less mutant has also been constructed in another actinobacterium, Streptomyces venezuelae, albeit not in S. coelicolor (89). Likewise, in the cyanobacterium Synechococcus elongatus, a cdaA transposon mutant with a dramatically decreased c-di-AMP level was viable but showed a disrupted light-dark cycle (72), whereas a cdaA mutant of Synechocystis could not be obtained (47). On the other hand, c-di-AMP proved to be non-essential for L. monocytogenes and S. aureus cells grown in chemically defined minimal media (90,91). In B. subtilis, a c-di-AMP-lacking mutant could also grow on minimal media, but only at low (0.1 mM) levels of K+ ions (60). These observations indicated that the essentiality of c-di-AMP was dependent on the growth conditions, or, more generally, on its distinct roles as a second messenger.
Judging from the complementation studies, properties of the DAC-missing suppressor mutants, and the functions of c-di-AMP-binding proteins, of all the c-di-AMP-mediated processes mentioned above, the most universal – and the most likely to make it essential for a variety of diverse organisms—is maintaining the osmotic homeostasis through regulation of K+ uptake and, as a result, of the cellular level of this cation (58–62,65,92,93). K+ plays several important roles in bacterial, archaeal and eukaryotic cells, both as a major cation and as a component of the ribosome and a cofactor for some key cellular enzymes that cannot be replaced, e.g. by Na+ ions (94,95). It has been proposed that the first live cells originated in a K+-rich environment (96) and only later, after the acquisition of sophisticated ion pumps, ventured into the habitats with the prevalence of Na+ over K+ (95). Some of the ancestral and most widespread ATPases appear to be K+-dependent (97). However, most environments contain far less K+ than Na+. Accordingly, most organisms possess a variety of K+ uptake systems that allow them to maintain the K+ gradient with [K+]in >> [K+]out (98). Obviously, these systems are not foolproof and need to be regulated to prevent the unlimited accumulation of K+ ions, particularly when the cells grow in the media with relatively high K+ levels. In all cases where it has been studied, c-di-AMP was found to do exactly that, by inhibiting K+ uptake via both high- and low-affinity transporters and by stimulating K+ exit via the K+/H+ antiporter CpaA (99).
The essentiality of c-di-AMP is probably enhanced by its other regulatory roles, such as the interaction of the c-di-AMP riboswitch YdaO with its multiple targets (100) and its involvement in controlling central metabolism (69,70,90) and aerobic respiration (91) of bacterial cells.
c-di-AMP, an alarmone in eukaryotes
Along with other cyclic nucleotide second messengers, c-di-AMP serves as a signaling molecule that can be recognized by eukaryotic receptors and trigger an immune response in the host cells (101). The importance of this response is such that it uses several redundant mechanisms and relies on c-di-AMP binding by at least four distinct receptors. These include STING (stimulator of interferon genes, formerly Tmem173), DDX41 (DEAD-box helicase 41), RECON (reductase controlling NF-κB, also known as aldo-keto reductase family 1 member C13, encoded by Akr1c13), ERAdP (CTD nuclear envelope phosphatase 1 regulatory subunit 1 or C16orf69, encoded by Cnep1r1, formerly Tmem188), and possibly NLRP3 (C1orf7) inflammasome (102–106). STING and DDX41 trigger innate immunity response by activating the production of interferon, RECON and ERAdP help in the activation of a pro-inflammatory antibacterial state, whereas NLRP3 stimulates secretion of interleukin-1β. In addition to c-di-AMP, STING binds other cyclic dinucleotides, including c-di-GMP and c-GMP-AMP (3′,3′-cGAMP) synthesized by bacterial pathogens, as well as the 2′,3′-cGAMP that is synthesized by the host cGMP-AMP synthetase (cGAS), activated by the presence of foreign DNA (102,107). RECON binds c-di-AMP and 3′,3′-cGAMP but not c-di-GMP or 2′,3′-cGAMP (104). We discuss the detailed mechanisms of c-di-AMP binding by STING and RECON in the next section.
The ability of the eukaryotic cells to sense c-di-AMP raises several interesting questions. For example, is c-di-AMP production beneficial or detrimental for the host-associated bacteria? Group B streptococci actively degrade secreted c-di-AMP, ostensibly to decrease the inflammatory response by the host (108). In contrast, c-di-AMP secretion by L. monocytogenes enhanced the cell-to-cell spread of this organism between different hepatocytes, suggesting that it could benefit the pathogen (109). In any case, intracytoplasmic surveillance for foreign nucleic acids and individual nucleotides is an important phenomenon that deserves further study and promises a better insight into the immune mechanisms of the eukaryotic cell.
TERTIARY STRUCTURE: MECHANISMS OF CYCLIC DI-AMP BINDING
Diversity of c-di-AMP conformations
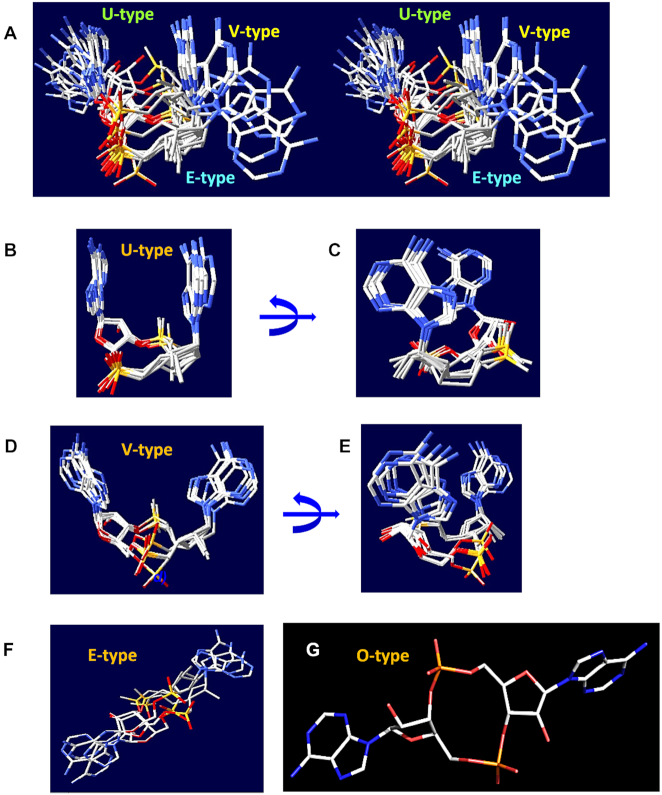
As discussed above, c-di-AMP regulates a wide range of functions in bacteria. This variety is partly due to the diversity of conformations that c-di-AMP adopts when binding to diverse cellular receptors. A comparison of the crystal structures of the c-di-AMP-protein and c-di-AMP-riboswitch complexes available in the PDB reveals a variety of conformations of this interesting molecule (Figures 1 and 2). We could identify at least four principal conformation types, two of which have been discussed before (99,110), while the other two have not.
Figure 2.
Diversity of c-di-AMP conformations. (A) Stereo view of three main conformations of c-di-AMP: the U-, V- and E-types. (B, C) Superposition of U-type c-di-AMP molecules from PDB entries 4QSH1, 4XTT, 4YP1, 5CFN, and 5F29 displayed from two different angles. (D,E) Superposition of V-type c-di-AMP molecules from PDB entries 4RLE, 4RWW, 4WK1, 4D3H and 4S1B displayed in two different angles. (F) E-type c-di-AMP molecules from PDB entries 5XSN, 5UXF and 4QSH2. (G) The O-type c-di-AMP from PDB entry 5KS7.
U-type
The most widespread is the ‘U-type’ conformation of c-di-AMP, in which the two adenine bases are oriented in an almost parallel fashion (Figures 1B and 2B). This conformation was first seen by Witte and colleagues in the structure of the T. maritima protein DisA (PDB: 3C1Y) that led to the initial discovery of c-di-AMP in a biological system (3). After that, a very similar U-type conformation has been observed in the structures of S. aureus K+ transport proteins KtrA (PDB: 4XTT) and CpaA (PDB: 5F29), which both bind c-di-AMP via their C-terminal RCK_C (PF02080) domains (99,111). This conformation of c-di-AMP was also seen in the structure of the pyruvate carboxylase PycA from L. monocytogenes (PDB: 4QSH), which binds c-di-AMP at the interface between the enzyme monomers (69). In one of the crystal forms of PycA, however, there is also another c-di-AMP molecule that bridges two c-di-AMPs from two different PycA monomers; each of its adenine bases fits into the cavity between the adenines of the U-shaped c-di-AMP (Supplementary Figure S4). The U-type conformation of c-di-AMP has also been seen in its complexes with the murine (PDB: 4YP1, Supplementary Figure S4C) and porcine (PDF: 6IYF) molecules of the STING protein (112). One more STING structure, the one from the sea anemone Nematostella vectensis (PDB: 5CFN), contains two U-shaped c-di-AMP molecules (Supplementary Figure S4D), whose adenine bases are stacked in a pairwise manner (113). A similar U-shaped conformation is also adopted by 3′,3′-cGAMP in its complexes with its synthase cGAS (PDB: 5VDT) and its riboswitch (PDB: 4YB1) (114,115).
V-type
The structure of the c-di-AMP-specific PDE PgpH (42) from L. monocytogenes (PDB: 4S1B) revealed a slightly different conformation, in which the adenine bases point in the same direction but the respective glycosidic bonds are inclined by approximately 45°, leaving more open space between the adenines (Figures 1F and 2D). This conformation, referred to as ‘V-type’ (99), has been also observed in the c-di-AMP-riboswitch complexes from B. subtilis (PDB: 4W90) and three thermophilic bacteria (PDB: 4QLN, 4QK8 and 4QK9) (116–118). Essentially the same V-type conformation is seen in the structures of the c-di-AMP receptor DarA/PstA from S. aureus (PDB: 4D3H and 4WK1), B. subtilis (PDB: 4RLE), and L. monocytogenes (PDB: 4RWW) (84–87). The c-di-AMP molecule bound to the pyruvate carboxylase from L. lactis (PDB: 5VYZ) also displays a V-type conformation (Supplementary Figure S4A), in contrast to the U-shaped one in L. monocytogenes (PDB: 4QSH) (Supplementary Figure S4B). One could speculate that at higher concentrations of c-di-AMP, the second c-di-AMP molecule binds to first one, inserting one of its bases between the two adenines of the first molecule and converting it from the V-shape into a U-shape (Supplementary Figure S4C, D).
E-type
The third type of c-di-AMP conformation, seen in its complex with the murine RECON protein (PDB: 5UXF), is almost flat (Figures 1D and 2F) with the two adenine bases looking in the opposite directions (104). We refer to this extended conformation of c-di-AMP as the ‘E-type’.
O-type
Finally, c-di-AMP bound to the c-di-AMP-specific PDE GdpP from S. aureus (PDB: 5XSN) displays another type of extended conformation with one adenine base almost orthogonal to the other one (45). We refer to this conformation as ‘O-type’ (Figure 2G). The same conformation is adopted by c-di-AMP in its complex with the CBS domain dimer (PF00571) in the carnitine transporter subunit OpuCA from L. monocytogenes (PDB: 5KS7) (63) and by the second c-di-AMP molecule in the pyruvate carboxylase PycA structure from L. monocytogenes (the upper one in Supplementary Figure S4B) (69).
Obviously, as has been previously observed for c-di-GMP (8), the distinct shapes of the c-di-AMP molecule allow distinct modes of its interactions with diverse protein and RNA targets.
Non-covalent interactions involved in c-di-NMP binding
Analysis of the c-di-AMP- and c-di-GMP-binding proteins and riboswitches identified the following principal types of interactions that contribute to the tight binding of these cyclic dinucleotides.
Hydrogen bonds
In our earlier review of c-di-GMP binding mechanisms, we noted the nearly universal presence of Arg residues in all kinds of c-di-GMP-binding receptors (8). In most cases, these Arg residues formed hydrogen bonds with O6 and N7 atoms of the guanine Hoogsteen edge, whereas Asp and Glu residues often formed hydrogen bonds with N1 and N(2) atoms of the Watson-Crick edge of the guanine base (8); see reference (119) for the nomenclature. In contrast, Arg residues are rarely involved in c-di-AMP binding. Instead, there is a wide variety of hydrogen bonds formed between N1, N3, N6 (amino group), and N7 atoms of c-di-AMP adenines and assorted residues of the receptor proteins, either through their side-chain groups (Asn, Thr or Glu) or backbone atoms (see below). Binding of c-di-AMP by its YdaO riboswitch is also supported by multiple hydrogen bonds (Figure 3). In most cases, these hydrogen bonds are formed by adenine-specific nitrogen atoms and contribute to the specificity of the c-di-AMP binding.
Figure 3.
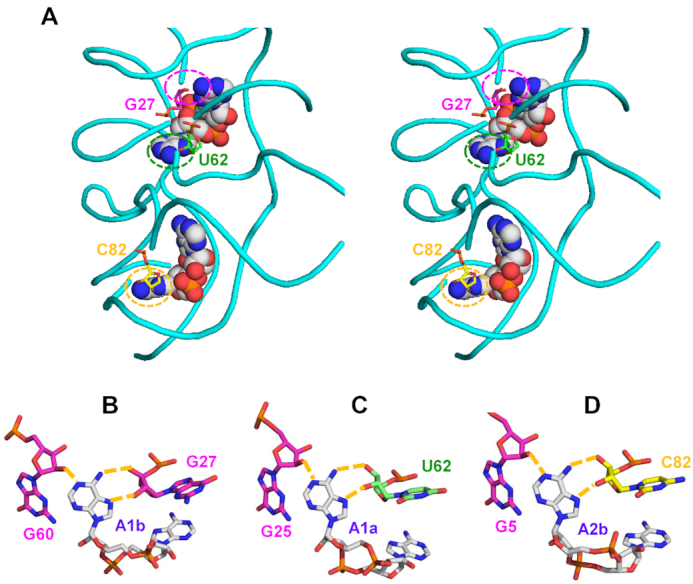
Binding of c-di-AMP to the YdaO riboswitch. (A) Stereo view of two c-di-AMP molecules (shown as spheres) bound to the YdaO riboswitch from B. subtilis (PDB: 4W90), whose sugar-phosphate backbone is shown in cyan (116). C-di-AMP-binding ribose moieties of G27, U62, and C82 are shown in stick representation and indicated by dashed circles. (B–D) Role of the ribose hydroxyl groups of the riboswitch in binding c-di-AMP (carbon atoms in grey). Hoogsteen side hydrogen bonds between N6 and N7 atoms of adenine bases of c-di-AMP and the ribose 2′- and 3′-hydroxyls from riboswitch bases G27 (B), U62 (C), and C82 (D) are coupled with adenine-N1 binding by 2′-hydroxyl groups of G60 (B), G25 (C) and G5(D). The abundance of hydrogen bonds explains the much higher affinity of c-di-AMP binding by the riboswitch than by protein receptors.
π–π interaction
Base-base stacking has been found to be very important for stabilizing the DNA duplex structure. Accordingly, Phe and Tyr (and sometimes Trp) residues can stack the adenine base of c-di-AMP with their side-chain aromatic groups. Such an interaction has also been seen in the case of c-di-GMP binding to its receptors (8).
Cation–π interaction (mainly by the Arg guanidino cation)
Cation–π interaction has been found to provide a significant contribution to the stabilization of protein–cofactor and enzyme–substrate interactions (120–123). In c-di-GMP-binding proteins, Arg residues exhibited two distinct types of interaction by their side-chain guanidino group: (i) binding the Hoogsteen side of the guanine base and (ii) planar stacking against the guanine base, causing significant stabilization of the c-di-GMP/receptor complexes (8). Although the Arg guanidino group cannot bind adenine base from its Hoogsteen side (99), stacking with the adenine base is still possible, so the cation (guanidino group)–π interaction plays an important role in c-di-AMP–receptor interactions.
Polar–π interaction (by the backbone peptide bond or from Asn or Gln side-chain amide bond)
Since the peptide bond is both planar and polar, it has been shown to stack with a guanine base of c-di-GMP bound to the allosteric inhibitory site (I-site) of the GGDEF domain that suppresses its diguanylate cyclase activity (124). In a similar fashion, c-di-AMP can interact with a protein receptor through the wide upper region of its V-shape (the U-type conformation is too narrow to accommodate a peptide segment, see below). Since Asn and Gln side-chain amide groups are also planar, they can also stack with the adenine base.
C-H bond–π interaction
Although the C–H bond has no charge or polarity, it can still interact with an aromatic group when it is situated perpendicularly to an aromatic ring (122,125–127). Such a special interaction mode is present in c-di-GMP–receptor complexes (8) and can also be seen in the complex of c-di-AMP with the carnitine transporter OpuCA (64).
Hydrophobic–π interaction
The previous type of interaction depended on perpendicularity in forming the C–H–π bond. However, even when the C–H bond is not located directly above the aromatic ring, it can still contribute to the hydrophobic–aromatic group interaction. For example, a triple Leu–π interaction was shown to be crucial for c-di-GMP binding to the MshEN domain, where replacement of any of those Leu residues reduced the binding affinity at least 10-fold (128). However, two nearby hydrophobic groups should be enough for binding and long and bulky hydrophobic side chains of Leu, Ile, Phe or Val can all participate in such a ‘hydrophobic–π’ interaction.
Anion–π interaction
While no anion–π interaction has been seen in the c-di-GMP receptor complexes, such an interaction is theoretically possible (122,129,130). Indeed, the planar carboxylate group of Glu has been found to stack upon the adenine base of c-di-AMP in its complex with the RECON protein (109), see below.
Lone pair–π interaction
In addition to a cation, neutral C-H bond, and an anion that all interact with π systems, a lone pair of electrons of the H2O molecule can do that as well (122,131–133). Such an interaction has been found in the crystal structures of c-di-AMP-bound RCK_C domains of CpaA and KtrA proteins (99,111). Here, a water molecule is situated on top of an adenine base and coordinates with three surrounding chemical moieties (see below). A similar interaction can also be seen in the recently reported complex (PDB: 6PFJ) of the S. venezuelae sigma factor σWhiG with an anti-sigma RsiG and two c-di-GMP molecules (134,135).
c-di-AMP binding by the YdaO riboswitch
Like its sibling c-di-GMP, c-di-AMP has a specific riboswitch that binds it with a very high affinity (Kd in sub-nanomolar to nanomolar range), see Table 1 and reference (100). While the structures of these riboswitches are not related, changing just two bases in the c-di-GMP-binding riboswitch GEMM was enough to convert it into one with a 4-fold preference for c-di-AMP over c-di-GMP (136,137). Surprisingly, this apparently never happened in the course of the riboswitch evolution and the structure of the c-di-AMP-specific riboswitch YdaO is dramatically different from both class I and class II c-di-GMP-binding riboswitches. The riboswitch GEMM-I from Geobacter sulfurreducens has been shown to bind c-di-GMP and 3′,3′-cGAMP, but not c-di-AMP or 2′,3′-cGAMP (138).
Structures of the c-di-AMP-riboswitch complexes from B. subtilis (PDB: 4W90) and the thermophilic bacteria Caldanaerobacter subterraneus (PDB: 4QLN), Thermoanaerobacter pseudethanolicus (PDB: 4QK8) and Thermovirga lienii (PDB: 4QK9) revealed very similar binding modes that combined partial stacking of the adenine bases of c-di-AMP by the adenine bases of the riboswitch with multiple hydrogen bonds between the atoms of the ligand and the riboswitch (116–118). As already noted, the planes of these adenine bases are not parallel, they form a 10–30° angle (117). However, Hoogsteen edge atoms of c-di-AMP adenines form hydrogen bonds with the ribose hydroxyls of the sugar-phosphate backbone of the riboswitch, particularly those of U62, G27, and C82, as shown in Figure 3. In addition, the 2′-hydroxyls of c-di-AMP interact with several bases of the riboswitch, whereas the central ribose-phosphate ring of c-di-AMP (which is also present in c-di-GMP) does not seem to form any bonds, ensuring binding specificity and the absence of competition by c-di-GMP. Thus, several components of the c-di-AMP ligand participate in interactions with the riboswitch, which probably accounts for the extremely tight binding (116–118).
Binding mechanisms in c-di-AMP–protein complexes
Crystal structures of several c-di-AMP–protein complexes have been solved (Table 2), which allows taking a close look at the c-di-AMP binding mechanisms.
c-di-AMP–RCK_C domain complex
The c-di-AMP-binding RCK_C domain is a Rossmann-fold domain that is found at the C-termini of several distinct potassium transporters, including the K+ uptake protein KtrA, K+/H+ antiporter subunit KhtT, and Na+/H+ and K+/H+ antiporter CpaA; it is also found in the transcriptional regulator BusR (Table 2). In its complex with a CpaA dimer from S. aureus (PDB: 5F29), the U-shaped c-di-AMP is coordinated by lone-pair electrons from two water molecules located between the adenine bases (Figure 4). Each of these water molecules is coordinated by the N7 atom of the adenine base and a phosphate oxygen atom from c-di-AMP, as well as the nitrogen atom from the side chain of His184, leaving a pair of electrons to form a lone pair–adenine interaction (indicated by a white arrow in Figure 4B). Indeed, replacement of this His184 with Ala resulted in a >300-fold weaker binding of c-di-AMP (99). In addition, N1 and N6 atoms of each adenine base of c-di-AMP form hydrogen bonds with the backbone nitrogen and carbonyl oxygen atoms, respectively, of the Phe171 residues of the CpaA dimer, while the oxygen atoms of the central ribose-phosphate ring form hydrogen bonds with the backbone nitrogen atom of Gly185. With the exception of the last one, all these interactions are specific for the adenine base and account for the specificity of the c-di-AMP binding by the RCK_C domain.
Figure 4.
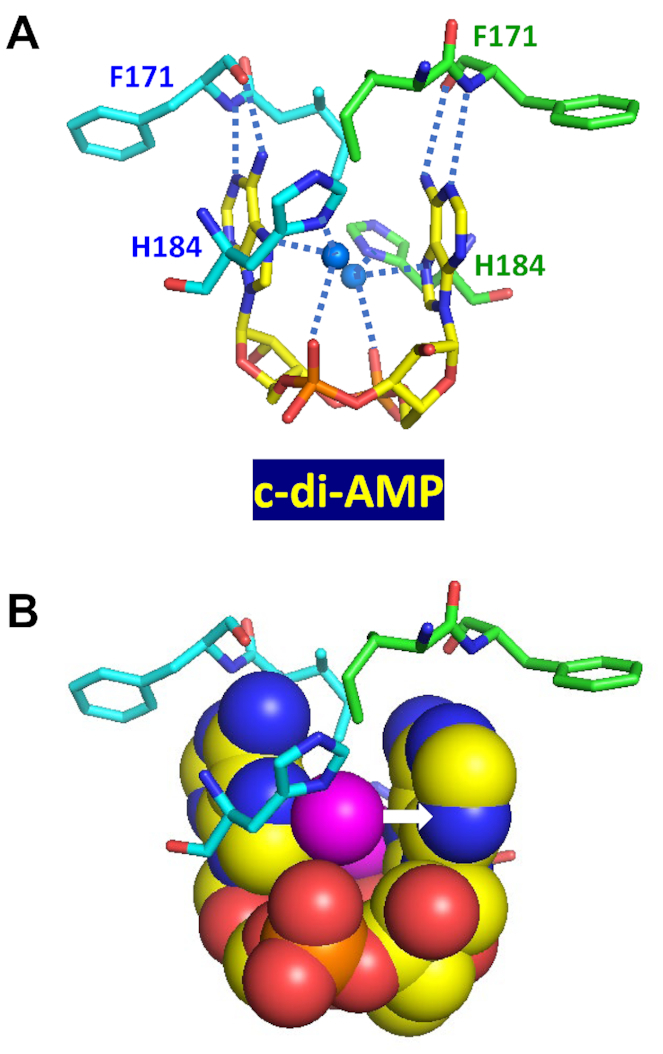
Binding of c-di-AMP to CpaA. Complex of U-shaped c-di-AMP with two RCK_C domains of S. aureus K+/H+ antiporter CpaA (PDB: 5F29) (99) shows each water molecule (solid blue circles in (A) and magenta spheres in (B)) coordinated by the N7 atom and phosphate oxygen atom of c-di-AMP, as well as the N atom from the side chain of His184. The lone pair electrons of oxygen atom form a water-mediated lone pair–adenine interaction (indicated by a white arrow in B). The N1 and N6 atoms of c-di-AMP adenines bind the backbone nitrogen and carbonyl oxygen atoms of Phe171, respectively, of both RCK_C domains.
A similar lone pair–adenine interaction is seen in the c-di-AMP complex with the RCK_C domain of the S. aureus KtrA receptor (PDB: 4XTT). Here, a water molecule is coordinated by the guanidino nitrogen atom of one of the Arg169 residues and the phosphate oxygen atom, leaving one lone pair of electrons to interact with the adenine base (Figure 5). However, in this case, the binding mode is asymmetric, and the side chain guanidino group of the Arg169 from the other RCK_C monomer (marked by a blue arrow in Figure 5A) is turned sideways to prevent a steric clash with the water oxygen atom. Thus, there is only one H2O lone pair–adenine interaction in KtrA (indicated by a white arrow in Figure 5B). In addition, the c-di-AMP molecule forms hydrogen bonds with the backbone nitrogen and carbonyl oxygen atoms, respectively, of the Asn175 residue. Thus, the same U-shaped molecule of c-di-AMP is bound by the two RCK_C domains by the same types of interactions, albeit provided by totally different residues.
Figure 5.
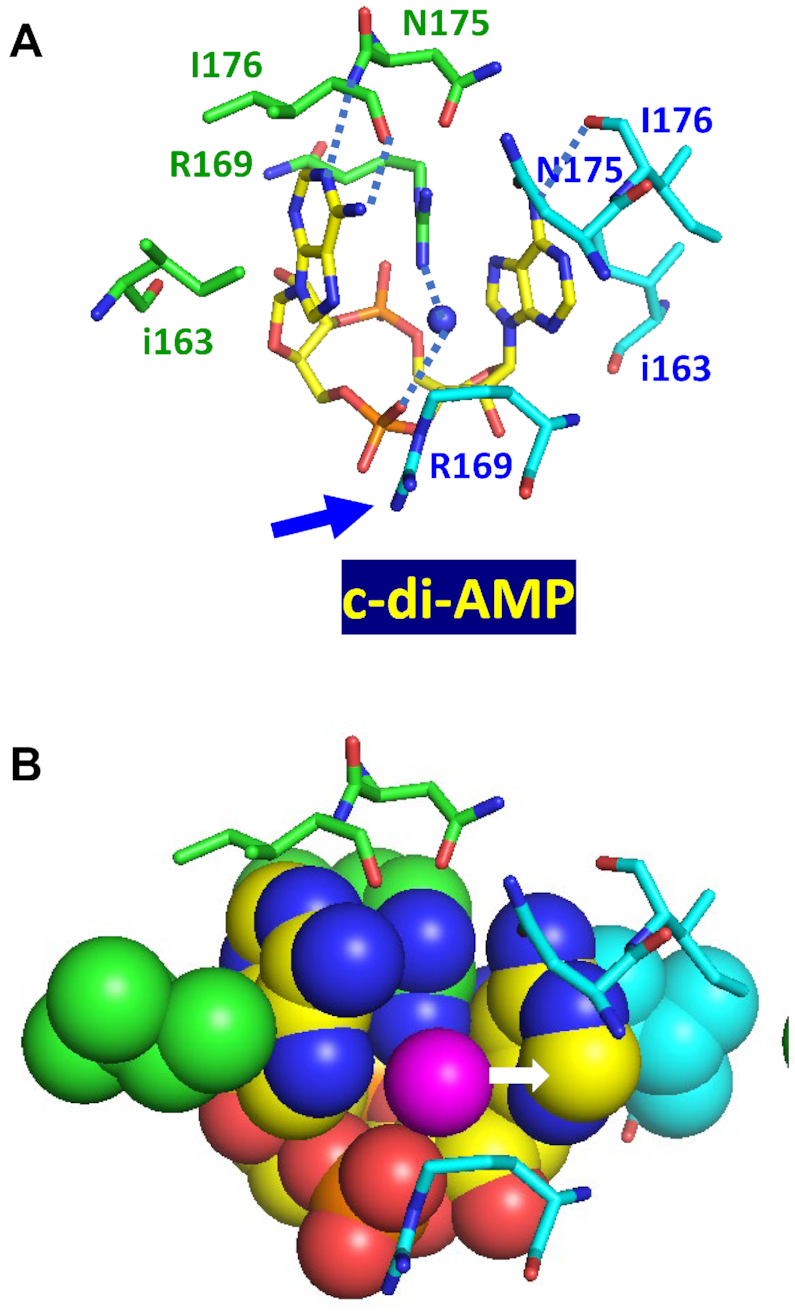
Binding of c-di-AMP to KtrA. (A) Complex of U-shaped c-di-AMP with two RCK_C domains of S. aureus K+ transporter KtrA (PDB: 4XTT) (111) shows an H2O lone pair–adenine interaction, in which a water molecule is coordinated by the guanidino nitrogen atom of Arg169 and the phosphate oxygen atom, with its lone pair electrons interacting with the adenine base, as shown in (B). Here, the binding mode is asymmetric and the side-chain guanidino group of Arg169 from the other RCK_C monomer (marked by a blue arrow) is turned sideways to prevent a steric clash with the water oxygen atom.
c-di-AMP–CBS domain complex
Interaction of c-di-AMP with the carnitine transporter subunit OpuCA, demonstrated in B. subtilis, L. monocytogenes, S. aureus and S. agalactiae (Table 2), is important for controlling cellular turgor for normal bacterial growth and survival (63,64,66,88). The OpuCA subunit is an ATPase that binds c-di-AMP via the tandem of cystathionine beta-synthase domains of the βαββα structure (CBS pair), located at its C-terminus. In its complex with an OpuCA dimer from L. monocytogenes (PDB: 5KS7) (64), c-di-AMP adopts an extended orthogonal (O-type) conformation, with the one adenine base turned almost 90° relative to the other (Figure 6). In this complex, both adenine bases of c-di-AMP are involved in extensive C–H bond–π interactions, sandwiched between the C–H bonds of Val280 on one side and of Ile355 on the other side, from each OpuCA monomer. The side-chain hydroxyl group of Thr282 forms two hydrogen bonds with the N6 and N7 atoms of each adenine base and contributes to the specific recognition of c-di-AMP by OpuCA. In addition, oxygen atoms of the central ribose-phosphate ring form hydrogen bonds with Arg358 residues of both monomers, one of those cases where Arg residues are involved in c-di-AMP binding.
Figure 6.
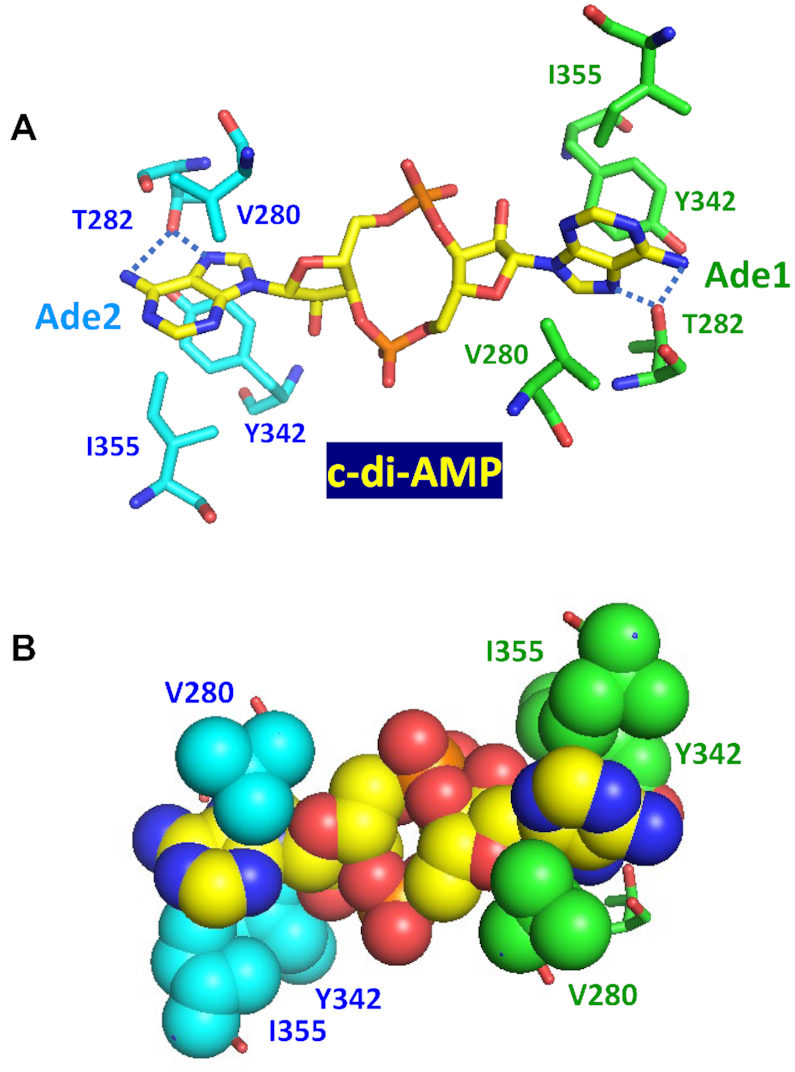
Binding of c-di-AMP to the CBS domains of the carnitine transporter OpuCA. In this structure (PDB: 5KS7), c-di-AMP is in an O-type conformation and both adenine bases are sandwiched between Val280 on one side and Ile355 and Tyr342 on the other. In (A), blue dotted lines indicate the hydrogen bonds between the side-chain hydroxyl group of Thr282 and the N6 and N7 atoms of each adenine base. The bottom part (B) shows this binding site in a space-filling model.
c-di-AMP–DarA/PstA complexes
The DarA/PstA protein has a ferredoxin-like fold and is structurally related to the family of PII-like signal transduction proteins that are involved in many pathways, often associated with nitrogen metabolism. There are already four separately solved structures of c-di-AMP–DarA/PstA complexes from three organisms: S. aureus (PDB: 4D3H and 4WK1), L. monocytogenes (PDB: 4RWW), and B. subtilis (PDB: 4RLE) (84–87). The top row in Figure 7 shows that when bound to B. subtilis DarA, c-di-AMP adopts a V-type conformation, which allows a peptide fragment to pass through the upper region of the V-shape. This Arg26-Val27-Thr28 tripeptide is important for binding c-di-AMP. Arg26 plays a dual role in c-di-AMP binding: its terminal guanidino group forms a cation–π interaction with the Ade1 base (indicated by an orange arrow), while the planar peptide bond between Arg26 and Val27 stacks with the Ade2 base to form a polar–π interaction. These stacking interactions are clearly seen in the space-filling model on the right (indicated by a blue arrow). In addition, the side-chain hydroxyl group of Thr28 forms two hydrogen bonds (indicated by blue dotted lines): with the N3 atom of the Ade2 base and with the 2′-OH group of the Ade1 adenosine ribose, thus contributing to the specificity of the interaction. This is different from what was seen above for OpuCA, where Thr282 also forms two hydrogen bonds, but with the N6 and N7 atoms of the adenine base.
Figure 7.
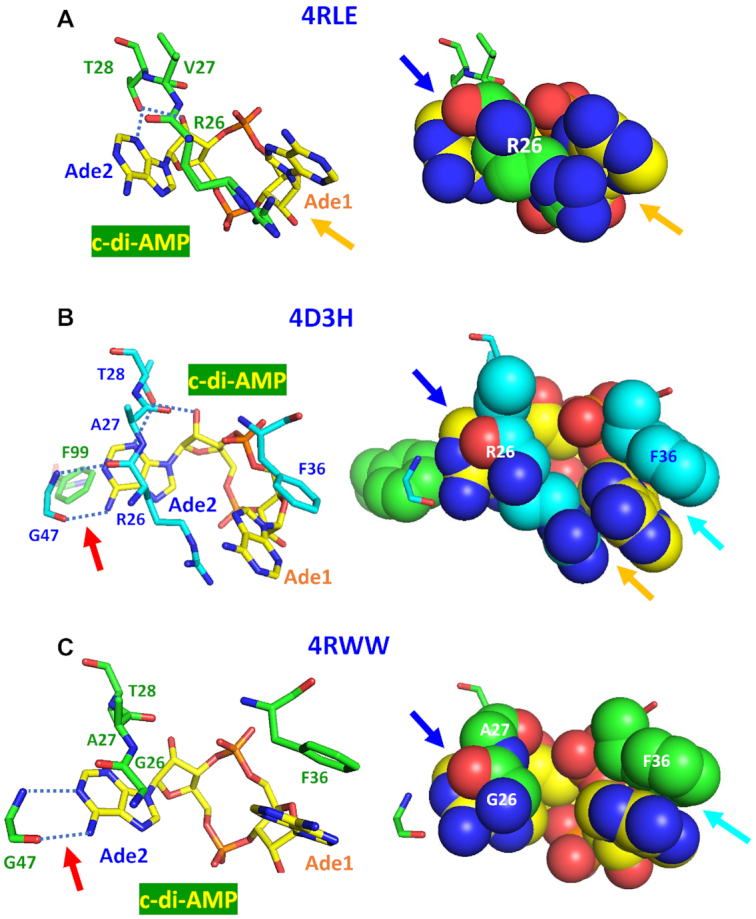
Binding of c-di-AMP to DarA/PstA proteins. Mechanisms of c-di-AMP binding to the DarA/PstA proteins from B. subtilis (PDB: 4RLE, top row, A), S. aureus (PDB: 4D3H, middle row, B), and L. monocytogenes (PDB: 4RWW, bottom row, C) (84–86) are shown in the sticks view on the left and as space-filling models on the right. The c-di-AMP molecules in V-type conformation are shown with carbon atoms in yellow. In (A), the guanidino group of Arg26 stacks with Ade1, while the peptide bond linking it to Val27 stacks with Ade2 (indicated by an orange arrow and a blue arrow, respectively). The dotted blue lines indicate hydrogen bonds between the β-OH group of Thr28 and N3 atom of Ade2 and 2′-OH of its ribose. In (B), the guanidino group and peptide bond of Arg26 also stack with the Ade1 and Ade2 and the β-OH group of Thr28 also forms a hydrogen bond with N3 atom of Ade2. The hydrogen bonds between the N and O atoms of the peptide bond of Gly47 and N1 and N6 atoms of Ade2 are indicated by a red arrow on the left panel. Additional stacking of Ade1 by Phe36 is indicated by a cyan arrow on the right. In (C), the Arg residue of the tripeptide Arg26-Ala/Val27-Thr28 is replaced by Gly, resulting in the loss of cation–π stacking of the guanidino group with Ade1 base. However, this complex retains the polar-π interaction of the peptide bond with Ade2 (indicated by a blue arrow), the hydrogen bonds between backbone atoms of Gly47 and nitrogen atoms of Ade2 (indicated by a red arrow), and the stacking of Ade1 by Phe36 (indicated by a cyan arrow).
The middle row of Figure 7 shows c-di-AMP binding by the DarA/PstA from S. aureus, which involves a similar tripeptide Arg26-Ala27-Thr28. The binding mechanism is essentially the same as in B. subtilis protein with additional stacking interactions of the phenyl groups of Phe36 on the Ade1 base and of Phe99 on the Ade2 base. Also, in addition to the hydrogen bonds between the side-chain hydroxyl group of Thr28 and the N3 atom of Ade2 and the 2′-OH of ribose (marked by blue dotted lines), the backbone nitrogen and carbonyl oxygen atoms of the nearby Gly47 residue form two hydrogen bonds with the N1 and N6 atoms of Ade2 (marked by blue dotted lines and a red arrow). Like the previous one, this structure forms a homo-trimer, with O1P and O2P oxygen atoms of the c-di-AMP central ring each forming two hydrogen bonds with the backbone nitrogen atoms of Phe36 and Leu37 residues of the neighboring subunit. Here, c-di-AMP shows a more complex interaction with the DarA/PstA protein than in the case of B. subtilis, which is consistent with an at least 10-fold higher binding affinity of S. aureus protein (see Table 2).
The bottom row in Figure 7 shows c-di-AMP binding to the DarA/PstA from L. monocytogenes [PDB: 4RWW, (139)]. The c-di-AMP in this complex is also in the V-type conformation but the tripeptide motif is now Gly26-Ala27-Thr28, losing the contribution of the side chain of Arg26 that was seen in the two above-described DarA/PstA complexes. Further, there is only one Phe residue (F36) that stacks upon the Ade1 base from above (indicated by a cyan arrow). However, the backbone atoms of Gly47 again form two hydrogen bonds with the N1 and N6 atoms of Ade2. This structure also forms a homo-trimer, with the O1P oxygen atom of the c-di-AMP central ring forming a hydrogen bond with the backbone nitrogen atom of Phe36. The other oxygen atom the c-di-AMP central ring, O2P, forms two hydrogen bonds with the backbone nitrogen atom of Gly94 and the NE2 atom of His108 from a neighboring subunit. Accordingly, the Kd for c-di-AMP binding by this protein is intermediate between the previous two (Table 2).
These three cases illustrate the flexibility of the interactions between c-di-AMP and DarA/PstA. The stacking partners can be either an aromatic group or a guanidino group, and all three cases reveal a good stacking between the polar peptide bond with an adenine base. Additional recognition of the adenine base is achieved by the side chain hydroxyl group of Thr or the backbone atoms of Gly.
c-di-AMP–PycA complexes
Pyruvate carboxylase PycA belongs to the aldolase superfamily of TIM barrel proteins. In its complexes with pyruvate carboxylases from L. monocytogenes (LmPycA, PDB: 4QSH) and L. lactis (LlPycA, PDB: 5VYZ), c-di-AMP adopts slightly different shapes. In the former, c-di-AMP is in a typical U-shape (69) that is similar to the ones in its complex with the riboswitch and the RCK_C domain. In the L. lactis structure, the adenine bases are a bit shifted and located at an angle, closer to the V-shape (139). The binding mechanism is essentially the same in both complexes, with Tyr722 of LmPycA (Tyr715 in LlPycA) of two PycA monomers providing π-π stacking interaction with the two adenines of c-di-AMP (Figure 8A). This stack is further strengthened by the insertion of an adenine base from the second, bridging c-di-AMP molecule that creates a five-member Tyr/Ade1/Ade3/Ade2/Tyr stack (Figure 8A). The importance of this π–π stacking was highlighted by the loss of the c-di-AMP effect on the PycA activity (likely due to the loss of c-di-AMP binding) caused by the Tyr715Thr substitution in LlPycA (139). C-di-AMP binding is further stabilized by the hydrogen bonds to the oxygen atoms of its central ribose-phosphate ring (Gln749 of LmPycA and Ser756 of LlPycA) and the ribose hydroxyl group (Tyr749 of LlPycA) (139).
Figure 8.
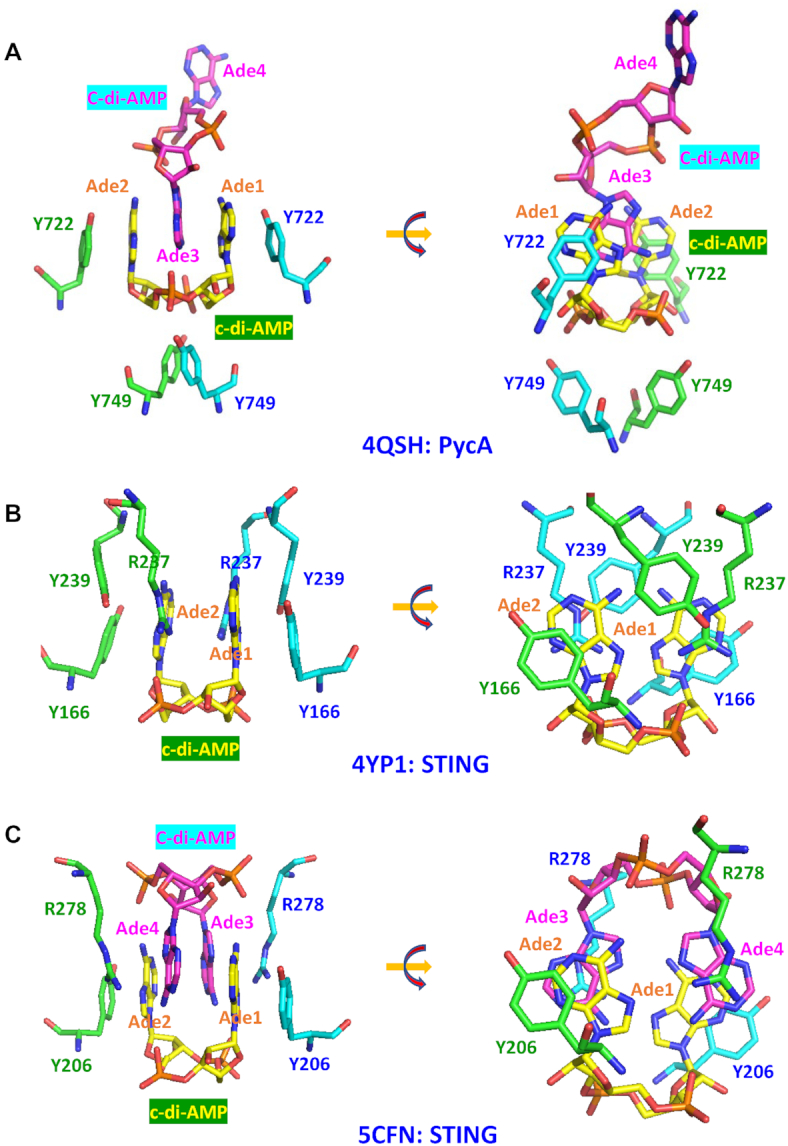
Binding of U-shaped c-di-AMP to the pyruvate carboxylase and STING molecules. (A) In the complex of U-shaped c-di-AMP with pyruvate carboxylase (PycA) dimer from L. monocytogenes (PDB: 4QSH) (69), adenine bases Ade1 and Ade2 form π–π stacking interactions with Tyr722 residues of the two PycA monomers (shown in green and cyan, respectively). This stack also contains an adenine base Ade3 from another c-di-AMP molecule (shown in magenta) that is inserted between Ade1 and Ade2. In addition, oxygen atoms of the central ribose-phosphate ring of c-di-AMP form hydrogen bonds with Tyr749 of both PycA monomers (69). (B) In the mouse STING complex (PDB: 4YP1), a single c-di-AMP molecule (carbon atoms in yellow) is present in the dimeric interface to stabilize the STING dimer formation, with Ade1 stacking with Tyr166 from one subunit (carbon atoms in cyan) and with Arg237 and Tyr239 from another subunit (carbon atoms in green), while Ade2 is stacking with Tyr166 from the second subunit (carbon atoms in green) and Arg237 and Tyr239 from the first subunit (carbon atoms in cyan). The rotated view on the right provides a better view of these four-layer Y166/Ade/R237/Y239 π-π-cation-π stacks. (C) In the STING complex from the sea anemone Nematostella vectensis (PDB: 5CFN), an additional c-di-AMP molecule (carbon atoms in magenta) forms a stack with the first c-di-AMP (carbon atoms in yellow), and, as shown on the right, the four-layer stacks are now formed by R278/Ade4/Ade1/Y206 or R278/Ade3/Ade2/Y206 cation-π-π-π stacking.
c-di-AMP–STING complexes
As described above, STING is a crucial membrane protein located in the endoplasmic reticulum that interacts with c-di-GMP, c-di-AMP, cGAMP and even 2′,3′-cGAMP to elicit a strong immune response (102,113). Several c-di-GMP–STING or cGAMP–STING complex structures have been solved (see (112) and references therein), but only four structures of c-di-AMP–STING complexes are available at this time; the mammalian (murine, PDB: 4YP1; porcine, PDB: 6IYF and human, PDB: 6CFF) structures (99,112,140) show substantial differences from the one from sea anemone [PDB: 5CFN (113), compare panels B and C on Figure 8]. In mammalian structures, the c-di-AMP molecule links two STING monomers, with each adenine base stacking against the aromatic group of Tyr166 in one monomer and the guanidino group of Arg237 in another monomer (Figure 8B). Thus, two different types of stacking are involved here: π–π and cation–π interactions. The partial stacking of Tyr237 completes an array of four planar moieties (Y166/Ade2/R237/Y239 and Y239/R237/Ade1/Y166) that are involved in stabilizing the STING dimer to activate its downstream reactions. There are two hydrogen bonds from the Arg237 side-chain amide nitrogen atom to the O2P atom of the ribose-phosphate central ring of c-di-AMP, c-di-GMP and cGAMP, which contribute to the recognition by STING of all these cyclic dinucleotides. In addition, there are two weaker hydrogen bonds from the backbone oxygen atoms of Val238 to the adenine base N6 atom. STING complexes with other cyclic dinucleotide messenger molecules show essentially the same binding mechanism.
The structure of the c-di-AMP–STING complex from the sea anemone N. vectensis (Figure 8C) presents a somewhat different story. In this structure, adenine bases from two molecules of c-di-AMP stack against each other and each of them further stacks with Arg278 of one STING monomer and Tyr206 of the other monomer to form two stacks consisting of Y206/Ade2/Ade3/R278 and R278/Ade4/Ade1/Y206, respectively. Thus, in all STING–c-di-AMP complexes, the quanidino group of Arg participates in a four-layer stack with an adenine base and a Tyr residue. In all these cases, both the π–π interaction and cation–π interaction modes are used to stabilize the dimeric structure of STING protein. Further, in this structure, two c-di-AMP molecules mutually stack to each other, with each of the c-di-AMP N6 atom forming a hydrogen bond with its opposite O2P oxygen atom of the central ring (four in total). The terminal guanidino group nitrogens of Arg237 are no longer involved in hydrogen bonds but form a cation-π interaction with the adenine base.
c-di-AMP–RECON complex
The oxidoreductase RECON has a typical TIM barrel fold and is involved in activating NF-κB to induce a proinflammatory antibacterial state in Mus musculus (104). In its complex with RECON (PDB: 5UXF), c-di-AMP adopts an extended (E-type) conformation (Figure 2C) with both adenine bases stacked by the residues coming from a single RECON molecule. As shown on Figure 9, base Ade1 is sandwiched between an aromatic ring of Tyr24 from below and a C–H–π stacking Leu306 from above, while base Ade2 is stacked by a hydrophobic–π interaction (Ala253 and Leu219) from above and an anion–π interaction with the carboxylate anion of Glu276 from below (indicated by a red arrow at the bottom of Figure 9). As mentioned above, this anion–π interaction has not been seen before, including any of the c-di-GMP protein complexes. Another unusual feature is that the side-chain amide group of Asn280 forms two hydrogen bonds with Ade2: between its N atom and N7 of Ade2 and between its carbonyl oxygen and the N6 (amino group) atom of Ade2. This N6 atom is also linked by a hydrogen bond to Glu279 (Figure 9). Like the Thr residue described above, the Asn280 residue recognizes the Hoogsteen edge of the adenine base. All these bonds go to adenine-specific atoms and account for binding of c-di-AMP and 3′,3′-cGAMP, as well as the recently described cyclic dinucleotide cUMP-AMP and the trinucleotide cAMP–AMP–GMP, but not c-di-GMP (104,141). Binding of these ligands is additionally supported by the hydrogen bonds between oxygen atoms in the central ribose-phosphate ring with the backbone nitrogen atoms of Gly217, Leu219 and Gln270 of RECON.
Figure 9.
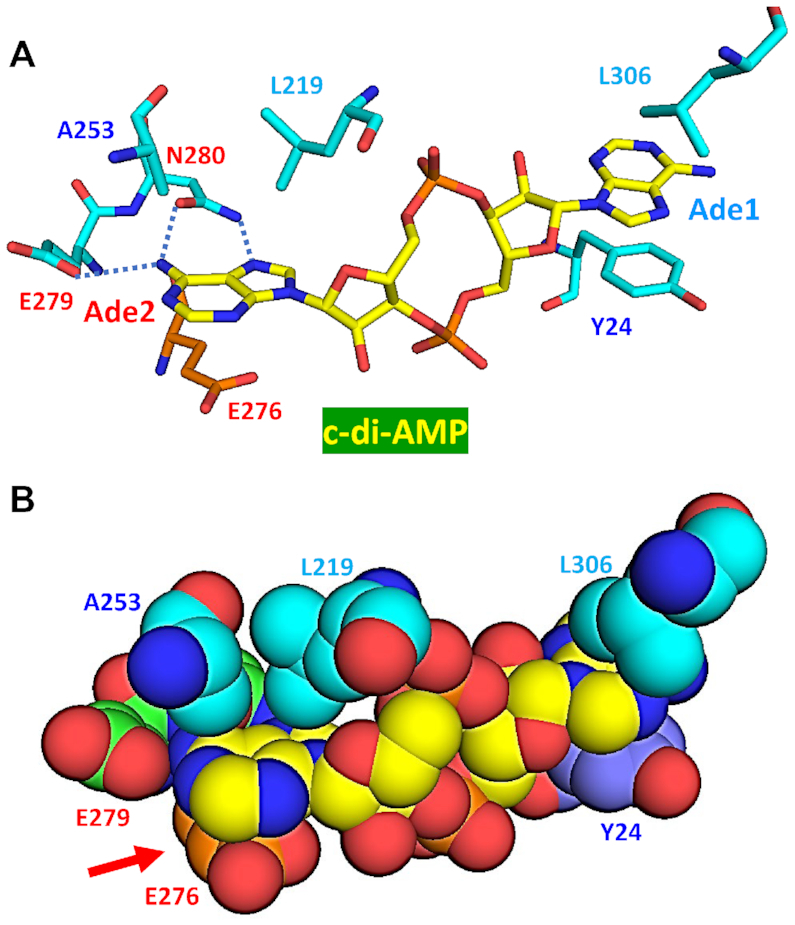
Binding of c-di-AMP to the RECON protein. In its complex with RECON (PDB: 5UXF), c-di-AMP adopts an E-type conformation, with both adenine bases stacked from both sides. Base Ade1 is held by a π–π stacking with Tyr24 from below and a C–H–π stacking with Leu306 from above. Base Ade2 is held by a triple hydrophobic–π interaction with Ala253 and Leu219 from above and an anion–π interaction of the carboxylate of Glu276 from below (indicated by the red arrow in panel B). In (A), dotted blue lines indicate the hydrogen bonds between N and O atoms of the Asn280 side-chain amide group and the N7 and N6 atoms of Ade2, respectively, and between Glu279 and the N6 atom of Ade2.
General trends in c-di-NMP binding modes
As discussed above, binding of c-di-AMP to its receptors involves a plethora of distinct binding modes, some of which have not been seen elsewhere and many of which may remain unknown. Recently, various non-covalent π interactions, including π–π, X–H–π (where X = C, N, O), cation–π, anion–π, and lone pair–π interactions in many different chemical environments have been reviewed in the context of catalyst design (122). It is therefore interesting to note that binding of a single c-di-NMP molecule to its various receptors can utilize almost all such non-covalent π interactions to ensure sufficiently high binding affinity and specificity to control many crucial physiological functions in bacteria and eukaryotes. Binding mechanisms of c-di-GMP (8,135) and c-di-AMP by various receptors seem to utilize similarly wide sets of non-covalent interactions, although the anion–π interaction has not yet been seen used for c-di-GMP binding.
Most of the c-di-GMP binding motifs contain at least one, often two Arg residues that either participate in the cation–π interactions or specifically interact with the guanine base via its Hoogsteen edge (8). In contrast, in c-di-AMP receptors, very few specific binding motifs could be defined. The [R/G]xT motif seen in three DarA/PstA structures (Figure 7) could serve as a signature motif to cross-link the two adenine bases of V-shaped c-di-AMP. Another potential motif is ExxEN, with the planar carboxylate of first Glu stacking the adenine base and the side-chain amide group of Asn forming hydrogen bonds with two nitrogen atoms of the same adenine base (Figure 9). Most c-di-AMP receptors just use backbone atoms to bind with the Hoogsteen edge of the adenine base with little, if any, contribution from the side chains (Figures 4 and 5). For example, in the complex of c-di-AMP with the central metabolic enzyme pyruvate carboxylase from L. monocytogenes (69), its adenine bases are only engaged in a π–π interaction with a nearby Tyr722 residue; the adenine edge atoms are not bound by any specific residue. This picture is dramatically different from what has been seen in the c-di-GMP receptors (8) and could be due to the fact that the Hoogsteen edge of a guanine base comprises only hydrogen-bond acceptors (O6 and N7 atoms), which can form bonds with the protons of arginine terminal guanidino group NH1, NH2 or NϵH, which are all hydrogen bond donors (99). In contrast, in an adenine base, the Hoogsteen edge comprises an N6 amino group and an N7 atom, which are a hydrogen bond donor and acceptor, respectively. Therefore, an adenine base cannot form stable bonds with the guanidino group of Arg. However, this restriction can be loosened by the interaction of the adenine base of c-di-AMP with either the 2′- and 3′-OH groups of ribose from the riboswitch, as seen in the c-di-AMP/riboswitch interaction (Figure 3) or with the water lone-pair electrons and oxygen atoms as shown in Figures 4 and 5.
It is worth mentioning that, depending on whether c-di-AMP is bound within a monomeric binding site, in a dimeric interface (e.g. in STING), or in a trimeric interface (e.g. in DarA/PstA), the protein can adopt different symmetrical elements. In the monomeric case, c-di-AMP is mostly symmetrical, but the binding site is usually not; in the dimeric interface, both the c-di-AMP and binding sites are symmetrical with a C2 axis; in the trimeric case, the protein adopts a C3 axis with the c-di-AMP exhibiting a local pseudo-C2 symmetry axis.
Another interesting distinction between c-di-GMP and c-di-AMP is that the former binds its receptor proteins in its monomer, dimer, or even tetramer states (8). In contrast, c-di-AMP is almost always found bound as a monomer. Exceptions, as shown in Supplementary Figure S4, include the c-di-AMP complexes with STING from the sea anemone (PDB: 5CFN, Figure 3B) and with the pyruvate carboxylase from L. monocytogenes (PDB: 4QSH) (69). The reasons why c-di-AMP does not form more dimers remain unclear.
CONCLUSIONS AND OUTLOOK
This Survey and Summary describes the structural aspects of c-di-AMP-mediated signaling (summarized in Figure 10) with occasional references to the better-studied second messenger c-di-GMP (8). Use of these c-di-NMPs as second messengers requires specific recognition of these molecules by their receptors against >1000-fold excess of the respective nucleoside triphosphates, ATP and GTP. The biochemical mechanisms that underlie this very specific and selective binding are still only partly understood; we hope that this review would attract the attention of a wider community and provide a much-needed boost to the studies of such mechanisms.
Figure 10.
Cyclic di-AMP signaling in bacteria and eukaryotes. (A) In bacteria and archaea, c-di-AMP (magenta double dots) is synthesized by five kinds of diadenylate cyclases (green ovals at the top) and hydrolyzed by three kinds of phosphodiesterases (pink round rectangles on the right). C-di-AMP receptors include, among others, a specific riboswitch that controls expression of a number of K+ uptake proteins, RCK_C domains of K+ transporters KtrA and CpaA, the USP domain of the osmosensitive histidine kinase KdpD that regulates high-affinity K+ uptake system KdpABC, CBS domains of the OpuCA subunit of the carnitine transporter OpuC, K+ transporters KupA and KupB, pyruvate carboxylase PycA, PII-like signal transduction protein DarA (also referred to as PstA), and BusR, transcriptional regulator of osmolyte uptake. (B) In eukaryotic cells, five types of c-di-AMP receptors have been characterized: STING, DDX41, RECON, ERAdP, and NLRP3. Signaling by STING and DDX41 involves protein kinases TBK1 and IKK; TBK1 phosphorylates interferon regulatory factor IRF3 and promotes the production of type I interferons, whereas IKK phosphorylates IκB to release NF-κB, which leads to the production of pro-inflammatory cytokines (interleukin-6, TNF-α and others). RECON and ERAdP modulate the NF-κB to stimulate the production of pro-inflammatory cytokines (interleukin-6, TNF-α and others), while NLRP3 regulates the production of interleukin-1β. The dashed arrows represent steps that remain to be clarified. See the text for references and further details.
Analysis of the available crystal structures shows that tight binding of c-di-AMP and c-di-GMP is usually accomplished through a combination of Hoogsteen-side hydrogen bonds, which partly define the selectivity with respect to the particular nucleobase, and various stacking interactions, which include π–π, C–H–π, cation–π, polar–π, hydrophobic–π, anion–π and the lone pair–π interactions. Still, it appears that c-di-AMP and c-di-GMP receptors have evolved substantially distinct mechanisms to bind their respective ligands, consistent with the fact that the two cyclic dinucleotides control very different biological functions.
The case of the c-di-AMP–DarA/PstA complexes, with four independently solved crystal structures from three different organisms (Figure 7) and experimentally determined Kd values (Table 2), nicely illustrates the correlation between the binding affinity and the complexity of interactions between the receptor and the c-di-AMP ligand: the availability of additional bonds between c-di-AMP and DarA/PstA from S. aureus results in a 10-fold higher binding affinity than in the same complex from B. subtilis.
On a more practical note, structural studies of the c-di-AMP synthases and hydrolases and the c-di-AMP-receptor complexes are contributing to a better understanding of the mechanisms of c-di-AMP-mediated signaling and promise eventual use of the c-di-AMP-related machinery as a potential drug target (18,21,142). In addition, tracing the bonds between the c-di-AMP ligand and its receptors offers a way to rationalize why certain CBS and USP-like domains function as efficient c-di-AMP receptors while the others do not. That said, signals that control c-di-AMP turnover remain poorly understood, as are mechanisms of how these ligand-domain interactions affect the respective enzyme activities.
It is worth mentioning that the world of cyclic nucleotide second messenger keeps expanding: the discovery of cAMP in 1958 was followed by cGMP, then c-di-GMP and c-di-AMP and, later, 3′,3′- and 2′,3′-cGMP-AMP (cGAMP) (143–145). In the past two years, this list was further enriched to include cyclic oligoadenylates (146,147) and, more recently, uridine-containing cyclic dinucleotides and cyclic trinucleotides (141,148). 3′,3′-cGAMP and cyclic triadenylate have been identified as components of the bacterial cyclic-oligonucleotide-based anti-phage signaling systems (CBASS), which are encoded in a wide variety of bacterial genomes (148–151).
Cyclic triadenylate and higher oligoadenylates, including cyclic hexaadenylate, have been shown to be produced by the Palm domains of the Cas10 enzymes of type III CRISPR–Cas systems and to serve as messengers controlling the nuclease activity of these systems (146,147). Upon binding to the CARF (CRISPR-associated Rossmann fold) domain of the Csm6 protein, they activate the nuclease activity of Csm6 that helps it intercept and degrade invader RNAs (146,147). Cyclic oligoadenylate binding by Csm6 has been documented in three distantly related bacteria and in the archaeon Methanothermobacter thermoautotrophicus (146,147), indicating that this type of signaling is widespread in the microbial world. Obviously, hydrolysis of viral RNA is essential for anti-phage immunity but, if insufficiently specific, it could hurt cellular mRNAs and lead to dormancy or even programmed cell death (152), so the signaling by cyclic oligoadenylates is critical for cell survival. Analysis of the structure of the CARF domain (PDB: 5FSH) and structure-guided mutagenesis revealed a putative ligand-binding cleft in the CARF dimer that was proposed as the potential cyclic oligoadenylate binding site [see Figure 3 and S15 in (146)]. Judging from the recently solved structures of Csm6 in complex with cyclic triadenylate and cyclic tetraadenylate [PDB: 6O78 and 6O7B, (153)], binding of these ligands involves the same kinds of interactions, stacking and hydrogen bonding, as described above for c-di-AMP. Importantly, not all CARF domains are associated with the nuclease domains of the CRISPR-Cas systems: many of them are found in association with the DNA-binding helix-turn-helix domains, suggesting a possible role in transcriptional regulation (154,155). In their comment on the cyclic oligoadenylate story, Johnson and Bailey stated ‘The case of the prokaryotic secondary messengers has been cracked’ (156). It is probably safe to say that there is still a long way from cracking this case to fully wrapping it up. Anyway, cyclic oligoadenylates represent yet another widespread and important group of prokaryotic second messengers.
The list of cyclic nucleotide second messengers was further expanded last year, when nucleotidyltransferases of the cGAS/DncV family were shown to produce a wide variety of cyclic di- and trinucleotides (141). Two of those, cyclic dinucleotide cUMP-AMP and the trinucleotide cAAG, were found to bind RECON and modulate its catalytic activity, potentially regulating host innate immunity (141). Previously, uracil and cytosine were judged to have limited capacity for hydrogen bonding and base stacking interactions and therefore considered less favorable ligands for the receptors that regulate signaling networks (157). However, the crystal structure of cGAS-like nucleotidyltransferase CdnE (PDB: 6E0L) in complex with non-hydrolyzable analogs of ATP and UTP (141) revealed a binding mode supported by a set of non-covalent interactions that was very similar to those described above. These included (i) hydrogen bonds between the Watson-Crick edge atoms of uridine and the side-chain amide group of Asn166; (ii) stacking of adenine by Tyr213, and (iii) stacking of Asn166-uridine area by the hydrophobic side-chains of Phe155 and Val164 (Supplementary Figure S5). This set of interactions could potentially support binding of other pyrimidine-containing nucleotides beyond cUMP-AMP, suggesting that (a) other uridine-containing cyclic dinucleotides described by Whiteley and colleagues could use the same binding mechanisms; (b) even more such cyclic nucleotide second messengers could be discovered in the future (148) and (c) the interaction mechanisms covered in this review are likely to apply in those cases as well.
In conclusion, while cyclic dinucleotide second messengers appear to be ancient molecules, widely believed to be the last vestiges of the primordial RNA World (157), they play key roles in signaling in all three domains of life. While c-di-GMP is often said to guide the choice between motility and sessility (11,12), c-di-AMP deals with even more critical matters of bacterial life (and death). Following Patrick Henry's maxim, the choice for bacteria is rather ‘Give me liberty (from viruses, osmotic shock, DNA damage) or give me death’, and c-di-AMP is a major factor in making that choice.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health Intramural Research Program at the National Library of Medicine (M.Y.G); National Natural Science Foundation of China [31770087, 31970074, and 31270105 to J.H.]; Fundamental Research Funds for the Central Universities [2662017PY112 and 2662015PY175 to J.H.]; China Postdoctoral Science Foundation [2018M630872 to W.Y.]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hsu C.-Y.J., Dennis D., Oger J.. Synthesis and physical characterization of bis 3′→5′ cyclic dinucleotides (NpNp): RNA polymerase inhibitors. Nucleosides Nucleotides. 1985; 4:377–389. [Google Scholar]
- 2. Ross P., Mayer R., Weinhouse H., Amikam D., Huggirat Y., Benziman M., de Vroom E., Fidder A., de Paus P., Sliedregt L.A. et al.. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 1990; 265:18933–18943. [PubMed] [Google Scholar]
- 3. Witte G., Hartung S., Buttner K., Hopfner K.P.. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell. 2008; 30:167–178. [DOI] [PubMed] [Google Scholar]
- 4. Oppenheimer-Shaanan Y., Wexselblatt E., Katzhendler J., Yavin E., Ben-Yehuda S.. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011; 12:594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A. et al.. The Pfam protein families database in 2019. Nucleic Acids Res. 2019; 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Galperin M.Y., Makarova K.S., Wolf Y.I., Koonin E.V.. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015; 43:D261–D269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Römling U. Great times for small molecules: c-di-AMP, a second messenger candidate in Bacteria and Archaea. Sci. Signal. 2008; 1:e39. [DOI] [PubMed] [Google Scholar]
- 8. Chou S.H., Galperin M.Y.. Diversity of cyclic di-GMP-binding proteins and mechanisms. J. Bacteriol. 2016; 198:32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schirmer T., Jenal U.. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 2009; 7:724–735. [DOI] [PubMed] [Google Scholar]
- 10. Hengge R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009; 7:263–273. [DOI] [PubMed] [Google Scholar]
- 11. Römling U., Galperin M.Y., Gomelsky M.. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013; 77:1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jenal U., Reinders A., Lori C.. Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 2017; 15:271–284. [DOI] [PubMed] [Google Scholar]
- 13. Corrigan R.M., Gründling A.. Cyclic di-AMP: another second messenger enters the fray. Nat. Rev. Microbiol. 2013; 11:513–524. [DOI] [PubMed] [Google Scholar]
- 14. Huynh T.N., Woodward J.J.. Too much of a good thing: regulated depletion of c-di-AMP in the bacterial cytoplasm. Curr. Opin. Microbiol. 2016; 30:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Commichau F.M., Dickmanns A., Gundlach J., Ficner R., Stülke J.. A jack of all trades: the multiple roles of the unique essential second messenger cyclic di-AMP. Mol. Microbiol. 2015; 97:189–204. [DOI] [PubMed] [Google Scholar]
- 16. Commichau F.M., Gibhardt J., Halbedel S., Gundlach J., Stülke J.. A delicate connection: c-di-AMP affects cell integrity by controlling osmolyte transport. Trends Microbiol. 2018; 26:175–185. [DOI] [PubMed] [Google Scholar]
- 17. Commichau F.M., Heidemann J.L., Ficner R., Stülke J.. Making and breaking of an essential poison: the cyclases and phosphodiesterases that produce and degrade the essential second messenger cyclic di-AMP in bacteria. J. Bacteriol. 2019; 201:e00462-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fahmi T., Port G.C., Cho K.H.. c-di-AMP: An essential molecule in the signaling pathways that regulate the viability and virulence of Gram-positive bacteria. Genes (Basel). 2017; 8:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gundlach J., Commichau F.M., Stülke J.. Perspective of ions and messengers: an intricate link between potassium, glutamate, and cyclic di-AMP. Curr. Genet. 2018; 64:191–195. [DOI] [PubMed] [Google Scholar]
- 20. Devaux L., Kaminski P.A., Trieu-Cuot P., Firon A.. Cyclic di-AMP in host-pathogen interactions. Curr. Opin. Microbiol. 2018; 41:21–28. [DOI] [PubMed] [Google Scholar]
- 21. Galperin M.Y. What bacteria want. Environ. Microbiol. 2018; 20:4221–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosenberg J., Dickmanns A., Neumann P., Gunka K., Arens J., Kaever V., Stülke J., Ficner R., Commichau F.M.. Structural and biochemical analysis of the essential diadenylate cyclase CdaA from Listeria monocytogenes. J. Biol. Chem. 2015; 290:6596–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mehne F.M., Gunka K., Eilers H., Herzberg C., Kaever V., Stülke J.. Cyclic di-AMP homeostasis in Bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J. Biol. Chem. 2013; 288:2004–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rismondo J., Gibhardt J., Rosenberg J., Kaever V., Halbedel S., Commichau F.M.. Phenotypes associated with the essential diadenylate cyclase CdaA and its potential regulator CdaR in the human pathogen Listeria monocytogenes. J. Bacteriol. 2016; 198:416–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barb A.W., Cort J.R., Seetharaman J., Lew S., Lee H.W., Acton T., Xiao R., Kennedy M.A., Tong L., Montelione G.T. et al.. Structures of domains I and IV from YbbR are representative of a widely distributed protein family. Protein Sci. 2011; 20:396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gundlach J., Mehne F.M., Herzberg C., Kampf J., Valerius O., Kaever V., Stülke J.. An essential poison: synthesis and degradation of cyclic di-AMP in Bacillus subtilis. J. Bacteriol. 2015; 197:3265–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu Y., Pham T.H., Nhiep T.H., Vu N.M., Marcellin E., Chakrabortti A., Wang Y., Waanders J., Lo R., Huston W.M. et al.. Cyclic-di-AMP synthesis by the diadenylate cyclase CdaA is modulated by the peptidoglycan biosynthesis enzyme GlmM in Lactococcus lactis. Mol. Microbiol. 2016; 99:1015–1027. [DOI] [PubMed] [Google Scholar]
- 28. Tosi T., Hoshiga F., Millership C., Singh R., Eldrid C., Patin D., Mengin-Lecreulx D., Thalassinos K., Freemont P., Gründling A.. Inhibition of the Staphylococcus aureus c-di-AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM. PLoS Pathog. 2019; 15:e1007537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang L., He Z.G.. Radiation-sensitive gene A (RadA) targets DisA, DNA integrity scanning protein A, to negatively affect cyclic di-AMP synthesis activity in Mycobacterium smegmatis. J. Biol. Chem. 2013; 288:22426–22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Torres R., Carrasco B., Gandara C., Baidya A.K., Ben-Yehuda S., Alonso J.C.. Bacillus subtilis DisA regulates RecA-mediated DNA strand exchange. Nucleic Acids Res. 2019; 47:5141–5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mehne F.M., Schroder-Tittmann K., Eijlander R.T., Herzberg C., Hewitt L., Kaever V., Lewis R.J., Kuipers O.P., Tittmann K., Stülke J.. Control of the diadenylate cyclase CdaS in Bacillus subtilis: an autoinhibitory domain limits cyclic di-AMP production. J. Biol. Chem. 2014; 289:21098–21107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng C., Ma Y., Wang X., Xie Y., Ali M.K., He J.. Functional analysis of the sporulation-specific diadenylate cyclase CdaS in Bacillus thuringiensis. Front. Microbiol. 2015; 6:908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blötz C., Treffon K., Kaever V., Schwede F., Hammer E., Stülke J.. Identification of the components involved in cyclic di-AMP signaling in Mycoplasma pneumoniae. Front. Microbiol. 2017; 8:1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fenton A.W., Johnson T.A., Holyoak T.. The pyruvate kinase model system, a cautionary tale for the use of osmolyte perturbations to support conformational equilibria in allostery. Protein Sci. 2010; 19:1796–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ishwar A., Tang Q., Fenton A.W.. Distinguishing the interactions in the fructose 1,6-bisphosphate binding site of human liver pyruvate kinase that contribute to allostery. Biochemistry. 2015; 54:1516–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abdelhamid Y., Brear P., Greenhalgh J., Chee X., Rahman T., Welch M.. Evolutionary plasticity in the allosteric regulator-binding site of pyruvate kinase isoform PykA from Pseudomonas aeruginosa. J. Biol. Chem. 2019; 294:15505–15516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kellenberger C.A., Chen C., Whiteley A.T., Portnoy D.A., Hammond M.C.. RNA-based fluorescent biosensors for live cell imaging of second messenger cyclic di-AMP. J. Am. Chem. Soc. 2015; 137:6432–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Braun F., Thomalla L., van der Does C., Quax T.E.F., Allers T., Kaever V., Albers S.V.. Cyclic nucleotides in archaea: cyclic di-AMP in the archaeon Haloferax volcanii and its putative role. MicrobiologyOpen. 2019; 8:e829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bordo D., van Monfort R.L., Pijning T., Kalk K.H., Reizer J., Saier M.H. Jr, Dijkstra B.W.. The three-dimensional structure of the nitrogen regulatory protein IIANtr from Escherichia coli. J. Mol. Biol. 1998; 279:245–255. [DOI] [PubMed] [Google Scholar]
- 40. Galperin M.Y., Makarova K.S., Wolf Y.I., Koonin E.V.. Phyletic distribution and lineage-specific domain architectures of archaeal two-component signal transduction systems. J. Bacteriol. 2018; 200:e00681–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rao F., See R.Y., Zhang D., Toh D.C., Ji Q., Liang Z.X.. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 2010; 285:473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huynh T.N., Luo S., Pensinger D., Sauer J.D., Tong L., Woodward J.J.. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E747–E756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bowman L., Zeden M.S., Schuster C.F., Kaever V., Gründling A.. New insights into the cyclic di-adenosine monophosphate (c-di-AMP) degradation pathway and the requirement of the cyclic dinucleotide for acid stress resistance in Staphylococcus aureus. J. Biol. Chem. 2016; 291:26970–26986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Konno H., Yoshida Y., Nagano K., Takebe J., Hasegawa Y.. Biological and biochemical roles of two distinct cyclic dimeric adenosine 3′,5′-monophosphate- associated phosphodiesterases in Streptococcus mutans. Front. Microbiol. 2018; 9:2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang F., He Q., Su K., Wei T., Xu S., Gu L.. Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase. Biochem. J. 2018; 475:191–205. [DOI] [PubMed] [Google Scholar]
- 46. Rao F., Ji Q., Soehano I., Liang Z.X.. Unusual heme-binding PAS domain from YybT family proteins. J. Bacteriol. 2011; 193:1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Agostoni M., Logan-Jackson A.R., Heinz E.R., Severin G.B., Bruger E.L., Waters C.M., Montgomery B.L.. Homeostasis of second messenger cyclic-di-AMP is critical for cyanobacterial fitness and acclimation to abiotic stress. Front. Microbiol. 2018; 9:1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu S., Bayles D.O., Mason T.M., Wilkinson B.J.. A cold-sensitive Listeria monocytogenes mutant has a transposon insertion in a gene encoding a putative membrane protein and shows altered (p)ppGpp levels. Appl. Environ. Microbiol. 2006; 72:3955–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Arguedas-Villa C., Stephan R., Tasara T.. Evaluation of cold growth and related gene transcription responses associated with Listeria monocytogenes strains of different origins. Food Microbiol. 2010; 27:653–660. [DOI] [PubMed] [Google Scholar]
- 50. Ye M., Zhang J.J., Fang X., Lawlis G.B., Troxell B., Zhou Y., Gomelsky M., Lou Y., Yang X.F.. DhhP, a cyclic di-AMP phosphodiesterase of Borrelia burgdorferi, is essential for cell growth and virulence. Infect. Immun. 2014; 82:1840–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tang Q., Luo Y., Zheng C., Yin K., Ali M.K., Li X., He J.. Functional analysis of a c-di-AMP-specific phosphodiesterase MsPDE from Mycobacterium smegmatis. Int. J. Biol. Sci. 2015; 11:813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Manikandan K., Sabareesh V., Singh N., Saigal K., Mechold U., Sinha K.M.. Two-step synthesis and hydrolysis of cyclic di-AMP in Mycobacterium tuberculosis. PLoS One. 2014; 9:e86096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bai Y., Yang J., Eisele L.E., Underwood A.J., Koestler B.J., Waters C.M., Metzger D.W., Bai G.. Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence. J. Bacteriol. 2013; 195:5123–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Drexler D.J., Müller M., Rojas-Cordova C.A., Bandera A.M., Witte G.. Structural and biophysical analysis of the soluble DHH/DHHA1-type phosphodiesterase TM1595 from Thermotoga maritima. Structure. 2017; 25:1887–1897. [DOI] [PubMed] [Google Scholar]
- 55. Corrigan R.M., Abbott J.C., Burhenne H., Kaever V., Gründling A.. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011; 7:e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Luo Y., Helmann J.D.. Analysis of the role of Bacillus subtilis σM in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol. Microbiol. 2012; 83:623–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Corrigan R.M., Campeotto I., Jeganathan T., Roelofs K.G., Lee V.T., Gründling A.. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:9084–9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bai Y., Yang J., Zarrella T.M., Zhang Y., Metzger D.W., Bai G.. Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J. Bacteriol. 2014; 196:614–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Moscoso J.A., Schramke H., Zhang Y., Tosi T., Dehbi A., Jung K., Gründling A.. Binding of cyclic di-AMP to the Staphylococcus aureus sensor kinase KdpD occurs via the Universal Stress Protein domain and downregulates the expression of the Kdp potassium transporter. J. Bacteriol. 2016; 198:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gundlach J., Herzberg C., Kaever V., Gunka K., Hoffmann T., Weiss M., Gibhardt J., Thurmer A., Hertel D., Daniel R. et al.. Control of potassium homeostasis is an essential function of the second messenger cyclic di-AMP in Bacillus subtilis. Sci. Signal. 2017; 10:eaal3011. [DOI] [PubMed] [Google Scholar]
- 61. Commichau F.M., Stülke J.. Coping with an essential poison: a genetic suppressor analysis corroborates a key function of c-di-AMP in controlling potassium ion homeostasis in Gram-positive bacteria. J. Bacteriol. 2018; 200:e00166–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gibhardt J., Hoffmann G., Turdiev A., Wang M., Lee V.T., Commichau F.M.. c-di-AMP assists osmoadaptation by regulating the Listeria monocytogenes potassium transporters KimA and KtrCD. J. Biol. Chem. 2019; 294:16020–16033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Huynh T.N., Choi P.H., Sureka K., Ledvina H.E., Campillo J., Tong L., Woodward J.J.. Cyclic di-AMP targets the cystathionine beta-synthase domain of the osmolyte transporter OpuC. Mol. Microbiol. 2016; 102:233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schuster C.F., Bellows L.E., Tosi T., Campeotto I., Corrigan R.M., Freemont P., Gründling A.. The second messenger c-di-AMP inhibits the osmolyte uptake system OpuC in Staphylococcus aureus. Sci. Signal. 2016; 9:ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pham H.T., Nhiep N.T.H., Vu T.N.M., Huynh T.N., Zhu Y., Huynh A.L.D., Chakrabortti A., Marcellin E., Lo R., Howard C.B. et al.. Enhanced uptake of potassium or glycine betaine or export of cyclic-di-AMP restores osmoresistance in a high cyclic-di-AMP Lactococcus lactis mutant. PLoS Genet. 2018; 14:e1007574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gundlach J., Kruger L., Herzberg C., Turdiev A., Poehlein A., Tascon I., Weiss M., Hertel D., Daniel R., Hanelt I. et al.. Sustained sensing in potassium homeostasis: Cyclic di-AMP controls potassium uptake by KimA at the levels of expression and activity. J. Biol. Chem. 2019; 294:9605–9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Peng X., Zhang Y., Bai G., Zhou X., Wu H.. Cyclic di-AMP mediates biofilm formation. Mol. Microbiol. 2016; 99:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gundlach J., Rath H., Herzberg C., Mäder U., Stülke J.. Second messenger signaling in Bacillus subtilis: Accumulation of cyclic di-AMP inhibits biofilm formation. Front. Microbiol. 2016; 7:804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sureka K., Choi P.H., Precit M., Delince M., Pensinger D.A., Huynh T.N., Jurado A.R., Goo Y.A., Sadilek M., Iavarone A.T. et al.. The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell. 2014; 158:1389–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Whiteley A.T., Garelis N.E., Peterson B.N., Choi P.H., Tong L., Woodward J.J., Portnoy D.A.. c-di-AMP modulates Listeria monocytogenes central metabolism to regulate growth, antibiotic resistance and osmoregulation. Mol. Microbiol. 2017; 104:212–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang L., Li W., He Z.G.. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J. Biol. Chem. 2013; 288:3085–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rubin B.E., Huynh T.N., Welkie D.G., Diamond S., Simkovsky R., Pierce E.C., Taton A., Lowe L.C., Lee J.J., Rifkin S.A. et al.. High-throughput interaction screens illuminate the role of c-di-AMP in cyanobacterial nighttime survival. PLoS Genet. 2018; 14:e1007301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zarrella T.M., Yang J., Metzger D.W., Bai G.. The bacterial second messenger cyclic di-AMP modulates the competence state in Streptococcus pneumoniae. J. Bacteriol. 2020; 202:e00691-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Block K.F., Hammond M.C., Breaker R.R.. Evidence for widespread gene control function by the ydaO riboswitch candidate. J. Bacteriol. 2010; 192:3983–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cron L.E., Stol K., Burghout P., van Selm S., Simonetti E.R., Bootsma H.J., Hermans P.W.. Two DHH subfamily 1 proteins contribute to pneumococcal virulence and confer protection against pneumococcal disease. Infect. Immun. 2011; 79:3697–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cho K.H., Kang S.O.. Streptococcus pyogenes c-di-AMP phosphodiesterase, GdpP, influences SpeB processing and virulence. PLoS One. 2013; 8:e69425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Witte C.E., Whiteley A.T., Burke T.P., Sauer J.D., Portnoy D.A., Woodward J.J.. Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. mBio. 2013; 4:e00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Du B., Ji W., An H., Shi Y., Huang Q., Cheng Y., Fu Q., Wang H., Yan Y., Sun J.. Functional analysis of c-di-AMP phosphodiesterase, GdpP, in Streptococcus suis serotype 2. Microbiol. Res. 2014; 169:749–758. [DOI] [PubMed] [Google Scholar]
- 79. Yang J., Bai Y., Zhang Y., Gabrielle V.D., Jin L., Bai G.. Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol. Microbiol. 2014; 93:65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 1997; 22:12–13. [DOI] [PubMed] [Google Scholar]
- 81. Kemp B.E. Bateman domains and adenosine derivatives form a binding contract. J. Clin. Invest. 2004; 113:182–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Quintana I.M., Gibhardt J., Turdiev A., Hammer E., Commichau F.M., Lee V.T., Magni C., Stülke J.. The KupA and KupB proteins of Lactococcus lactis IL1403 are novel c-di-AMP receptor proteins responsible for potassium uptake. J. Bacteriol. 2019; 201:e00028-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cox G.B., Rosenberg H., Downie J.A., Silver S.. Genetic analysis of mutants affected in the Pst inorganic phosphate transport system. J. Bacteriol. 1981; 148:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gundlach J., Dickmanns A., Schroder-Tittmann K., Neumann P., Kaesler J., Kampf J., Herzberg C., Hammer E., Schwede F., Kaever V. et al.. Identification, characterization, and structure analysis of the cyclic di-AMP-binding PII-like signal transduction protein DarA. J. Biol. Chem. 2015; 290:3069–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Campeotto I., Zhang Y., Mladenov M.G., Freemont P.S., Gründling A.. Complex structure and biochemical characterization of the Staphylococcus aureus cyclic diadenylate monophosphate (c-di-AMP)-binding protein PstA, the founding member of a new signal transduction protein family. J. Biol. Chem. 2015; 290:2888–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Choi P.H., Sureka K., Woodward J.J., Tong L.. Molecular basis for the recognition of cyclic-di-AMP by PstA, a PII-like signal transduction protein. Microbiol. Open. 2015; 4:361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Müller M., Hopfner K.P., Witte G.. c-di-AMP recognition by Staphylococcus aureus PstA. FEBS Lett. 2015; 589:45–51. [DOI] [PubMed] [Google Scholar]
- 88. Devaux L., Sleiman D., Mazzuoli M.V., Gominet M., Lanotte P., Trieu-Cuot P., Kaminski P.A., Firon A.. Cyclic di-AMP regulation of osmotic homeostasis is essential in Group B Streptococcus. PLoS Genet. 2018; 14:e1007342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. St-Onge R.J., Haiser H.J., Yousef M.R., Sherwood E., Tschowri N., Al-Bassam M., Elliot M.A.. Nucleotide second messenger-mediated regulation of a muralytic enzyme in Streptomyces. Mol. Microbiol. 2015; 96:779–795. [DOI] [PubMed] [Google Scholar]
- 90. Whiteley A.T., Pollock A.J., Portnoy D.A.. The PAMP c-di-AMP Is essential for Listeria monocytogenes growth in rich but not minimal media due to a toxic increase in (p)ppGpp. Cell Host Microbe. 2015; 17:788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zeden M.S., Schuster C.F., Bowman L., Zhong Q., Williams H.D., Gründling A.. Cyclic di-adenosine monophosphate (c-di-AMP) is required for osmotic regulation in Staphylococcus aureus but dispensable for viability in anaerobic conditions. J. Biol. Chem. 2018; 293:3180–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zarrella T.M., Metzger D.W., Bai G.. Stress suppressor screening leads to detection of regulation of cyclic di-AMP homeostasis by a Trk family effector protein in Streptococcus pneumoniae. J. Bacteriol. 2018; 200:e00045-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang X., Cai X., Ma H., Yin W., Zhu L., Li X., Lim H.M., Chou S.H., He J.. A c-di-AMP riboswitch controlling kdpFABC operon transcription regulates the potassium transporter system in Bacillus thuringiensis. Commun. Biol. 2019; 2:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Epstein W. The roles and regulation of potassium in bacteria. Prog. Nucleic Acid. Res. Mol. Biol. 2003; 75:293–320. [DOI] [PubMed] [Google Scholar]
- 95. Dibrova D.V., Galperin M.Y., Koonin E.V., Mulkidjanian A.Y.. Ancient systems of sodium/potassium homeostasis as predecessors of membrane bioenergetics. Biochemistry (Mosc.). 2015; 80:495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mulkidjanian A.Y., Bychkov A.Y., Dibrova D.V., Galperin M.Y., Koonin E.V.. Origin of first cells at terrestrial, anoxic geothermal fields. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E821–E830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shalaeva D.N., Cherepanov D.A., Galperin M.Y., Golovin A.V., Mulkidjanian A.Y.. Evolution of cation binding in the active sites of P-loop nucleoside triphosphatases in relation to the basic catalytic mechanism. Elife. 2018; 7:e37373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Brown I. I., Galperin M.Y., Glagolev A.N., Skulachev V.P.. Utilization of energy stored in the form of Na+ and K+ ion gradients by bacterial cells. Eur. J. Biochem. 1983; 134:345–349. [DOI] [PubMed] [Google Scholar]
- 99. Chin K.H., Liang J.M., Yang J.G., Shih M.S., Tu Z.L., Wang Y.C., Sun X.H., Hu N.J., Liang Z.X., Dow J.M. et al.. Structural insights into the distinct binding mode of cyclic di-AMP with SaCpaA_RCK. Biochemistry. 2015; 54:4936–4951. [DOI] [PubMed] [Google Scholar]
- 100. Nelson J.W., Sudarsan N., Furukawa K., Weinberg Z., Wang J.X., Breaker R.R.. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat. Chem. Biol. 2013; 9:834–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Danilchanka O., Mekalanos J.J.. Cyclic dinucleotides and the innate immune response. Cell. 2013; 154:962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Burdette D.L., Monroe K.M., Sotelo-Troha K., Iwig J.S., Eckert B., Hyodo M., Hayakawa Y., Vance R.E.. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011; 478:515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Parvatiyar K., Zhang Z., Teles R.M., Ouyang S., Jiang Y., Iyer S.S., Zaver S.A., Schenk M., Zeng S., Zhong W. et al.. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 2012; 13:1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. McFarland A.P., Luo S., Ahmed-Qadri F., Zuck M., Thayer E.F., Goo Y.A., Hybiske K., Tong L., Woodward J.J.. Sensing of bacterial cyclic dinucleotides by the oxidoreductase RECON promotes NF-κB activation and shapes a proinflammatory antibacterial state. Immunity. 2017; 46:433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Xia P., Wang S., Xiong Z., Zhu X., Ye B., Du Y., Meng S., Qu Y., Liu J., Gao G. et al.. The ER membrane adaptor ERAdP senses the bacterial second messenger c-di-AMP and initiates anti-bacterial immunity. Nat. Immunol. 2018; 19:141–150. [DOI] [PubMed] [Google Scholar]
- 106. Abdul-Sater A.A., Tattoli I., Jin L., Grajkowski A., Levi A., Koller B.H., Allen I.C., Beaucage S.L., Fitzgerald K.A., Ting J.P. et al.. Cyclic-di-GMP and cyclic-di-AMP activate the NLRP3 inflammasome. EMBO Rep. 2013; 14:900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Barker J.R., Koestler B.J., Carpenter V.K., Burdette D.L., Waters C.M., Vance R.E., Valdivia R.H.. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. mBio. 2013; 4:e00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Andrade W.A., Firon A., Schmidt T., Hornung V., Fitzgerald K.A., Kurt-Jones E.A., Trieu-Cuot P., Golenbock D.T., Kaminski P.A.. Group B Streptococcus degrades cyclic-di-AMP to modulate STING-dependent type I interferon production. Cell Host Microbe. 2016; 20:49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. McFarland A.P., Burke T.P., Carletti A.A., Glover R.C., Tabakh H., Welch M.D., Woodward J.J.. RECON-dependent inflammation in hepatocytes enhances Listeria monocytogenes cell-to-cell spread. mBio. 2018; 9:e00526-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Meehan R.E., Torgerson C.D., Gaffney B.L., Jones R.A., Strobel S.A.. Nuclease-resistant c-di-AMP derivatives that differentially recognize RNA and protein receptors. Biochemistry. 2016; 55:837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kim H., Youn S.-J., Kim S.O., Ko J., Lee J.-O., Choi B.-S.. Structural studies of potassium transport protein KtrA Regulator of Conductance of K+ (RCK) C domain in complex with cyclic diadenosine monophosphate (c-di-AMP). J. Biol. Chem. 2015; 290:16393–16402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Cong X., Yuan Z., Du Y., Wu B., Lu D., Wu X., Zhang Y., Li F., Wei B., Li J. et al.. Crystal structures of porcine STING(CBD)-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins. J. Biol. Chem. 2019; 294:11420–11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kranzusch P.J., Wilson S.C., Lee A.S., Berger J.M., Doudna J.A., Vance R.E.. Ancient origin of cGAS-STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol. Cell. 2015; 59:891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ren A., Wang X.C., Kellenberger C.A., Rajashankar K.R., Jones R.A., Hammond M.C., Patel D.J.. Structural basis for molecular discrimination by a 3′,3′-cGAMP sensing riboswitch. Cell Rep. 2015; 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hall J., Ralph E.C., Shanker S., Wang H., Byrnes L.J., Horst R., Wong J., Brault A., Dumlao D., Smith J.F. et al.. The catalytic mechanism of cyclic GMP-AMP synthase (cGAS) and implications for innate immunity and inhibition. Protein Sci. 2017; 26:2367–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Jones C.P., Ferré-D’Amaré A.R.. Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA. EMBO J. 2014; 33:2692–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Gao A., Serganov A.. Structural insights into recognition of c-di-AMP by the ydaO riboswitch. Nat. Chem. Biol. 2014; 10:787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ren A., Patel D.J.. c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets. Nat. Chem. Biol. 2014; 10:780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Leontis N.B., Westhof E.. Geometric nomenclature and classification of RNA base pairs. RNA. 2001; 7:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Dougherty D.A. The cation–π interaction. Acc. Chem. Res. 2013; 46:885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gallivan J.P., Dougherty D.A.. Cation–π interactions in structural biology. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:9459–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Neel A.J., Hilton M.J., Sigman M.S., Toste F.D.. Exploiting non-covalent π interactions for catalyst design. Nature. 2017; 543:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wang Q.Y., Lu J., Liao S.M., Du Q.S., Huang R.B.. Unconventional interaction forces in protein and protein-ligand systems and their impacts to drug design. Curr. Top. Med. Chem. 2013; 13:1141–1151. [DOI] [PubMed] [Google Scholar]
- 124. Yang C.Y., Chin K.H., Chuah M.L., Liang Z.X., Wang A.H., Chou S.H.. The structure and inhibition of a GGDEF diguanylate cyclase complexed with (c-di-GMP)2 at the active site. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:997–1008. [DOI] [PubMed] [Google Scholar]
- 125. Chakrabarti P., Samanta U.. CH/π interaction in the packing of the adenine ring in protein structures. J. Mol. Biol. 1995; 251:9–14. [DOI] [PubMed] [Google Scholar]
- 126. Brandl M., Weiss M.S., Jabs A., Suhnel J., Hilgenfeld R.. C-H… π -interactions in proteins. J. Mol. Biol. 2001; 307:357–377. [DOI] [PubMed] [Google Scholar]
- 127. Nishio M., Umezawa Y., Fantini J., Weiss M.S., Chakrabarti P.. CH–π hydrogen bonds in biological macromolecules. Phys. Chem. Chem. Phys. 2014; 16:12648–12683. [DOI] [PubMed] [Google Scholar]
- 128. Wang Y.C., Chin K.H., Tu Z.L., He J., Jones C.J., Sanchez D.Z., Yildiz F.H., Galperin M.Y., Chou S.H.. Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain. Nat. Commun. 2016; 7:12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cotelle Y., Lebrun V., Sakai N., Ward T.R., Matile S.. Anion–π enzymes. ACS Cent. Sci. 2016; 2:388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zhao Y., Cotelle Y., Liu L., Lopez-Andarias J., Bornhof A.B., Akamatsu M., Sakai N., Matile S.. The emergence of anion–π catalysis. Acc. Chem. Res. 2018; 51:2255–2263. [DOI] [PubMed] [Google Scholar]
- 131. Sarkhel S., Rich A., Egli M.. Water-nucleobase “stacking”: H–π and lone pair–π interactions in the atomic resolution crystal structure of an RNA pseudoknot. J. Am. Chem. Soc. 2003; 125:8998–8999. [DOI] [PubMed] [Google Scholar]
- 132. Jain A., Ramanathan V., Sankararamakrishnan R.. Lone pair … π interactions between water oxygens and aromatic residues: quantum chemical studies based on high-resolution protein structures and model compounds. Protein Sci. 2009; 18:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Kozelka J. Lone pair–π interactions in biological systems: occurrence, function, and physical origin. Eur. Biophys. J. 2017; 46:729–737. [DOI] [PubMed] [Google Scholar]
- 134. Gallagher K.A., Schumacher M.A., Bush M.J., Bibb M.J., Chandra G., Holmes N.A., Zeng W., Henderson M., Zhang H., Findlay K.C. et al.. c-di-GMP arms an anti-σ to control progression of multicellular differentiation in Streptomyces. Mol. Cell. 2020; 77:586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Chou S.H., Galperin M.Y.. Cyclic di-GMP in streptomycetes: a new conformation, new binding mode, new receptor, and a new mechanism to control cell development. Mol. Cell. 2020; 77:443–445. [DOI] [PubMed] [Google Scholar]
- 136. Smith K.D., Lipchock S.V., Ames T.D., Wang J., Breaker R.R., Strobel S.A.. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat. Struct. Mol. Biol. 2009; 16:1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Smith K.D., Lipchock S.V., Livingston A.L., Shanahan C.A., Strobel S.A.. Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch. Biochemistry. 2010; 49:7351–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Kellenberger C.A., Wilson S.C., Hickey S.F., Gonzalez T.L., Su Y., Hallberg Z.F., Brewer T.F., Iavarone A.T., Carlson H.K., Hsieh Y.F. et al.. GEMM-I riboswitches from Geobacter sense the bacterial second messenger cyclic AMP-GMP. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:5383–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Choi P.H., Vu T.M.N., Pham H.T., Woodward J.J., Turner M.S., Tong L.. Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E7226–E7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Ergun S.L., Fernandez D., Weiss T.M., Li L.. STING polymer structure reveals mechanisms for activation, hyperactivation, and inhibition. Cell. 2019; 178:290–301. [DOI] [PubMed] [Google Scholar]
- 141. Whiteley A.T., Eaglesham J.B., de Oliveira Mann C.C., Morehouse B.R., Lowey B., Nieminen E.A., Danilchanka O., King D.S., Lee A.S.Y., Mekalanos J.J. et al.. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature. 2019; 567:194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Heidemann J.L., Neumann P., Dickmanns A., Ficner R.. Crystal structures of the c-di-AMP-synthesizing enzyme CdaA. J. Biol. Chem. 2019; 294:10463–10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Gomelsky M. cAMP, c-di-GMP, c-di-AMP and now cGMP: bacteria use them all. Mol. Microbiol. 2011; 79:562–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Davies B.W., Bogard R.W., Young T.S., Mekalanos J.J.. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012; 149:358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Gao J., Tao J., Liang W., Jiang Z.. Cyclic (di)nucleotides: the common language shared by microbe and host. Curr. Opin. Microbiol. 2016; 30:79–87. [DOI] [PubMed] [Google Scholar]
- 146. Kazlauskiene M., Kostiuk G., Venclovas C., Tamulaitis G., Siksnys V.. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017; 357:605–609. [DOI] [PubMed] [Google Scholar]
- 147. Niewoehner O., Garcia-Doval C., Rostøl J.T., Berk C., Schwede F., Bigler L., Hall J., Marraffini L.A., Jinek M.. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017; 548:543–548. [DOI] [PubMed] [Google Scholar]
- 148. Severin G.B., Waters C.M.. Pyrimidines and cyclic trinucleotides join the second messenger symphony. Cell Host Microbe. 2019; 25:471–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Cohen D., Melamed S., Millman A., Shulman G., Oppenheimer-Shaanan Y., Kacen A., Doron S., Amitai G., Sorek R.. Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature. 2019; 574:691–695. [DOI] [PubMed] [Google Scholar]
- 150. Ye Q., Lau R.K., Mathews I.T., Birkholz E.A., Watrous J.D., Azimi C.S., Pogliano J., Jain M., Corbett K.D.. HORMA domain proteins and a Trip13-like ATPase regulate bacterial cGAS-like enzymes to mediate bacteriophage immunity. Mol. Cell. 2020; 77:709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Lau R.K., Ye Q., Birkholz E.A., Berg K.R., Patel L., Mathews I.T., Watrous J.D., Ego K., Whiteley A.T., Lowey B.. Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Mol. Cell. 2020; 77:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Koonin E.V., Zhang F.. Coupling immunity and programmed cell suicide in prokaryotes: Life-or-death choices. Bioessays. 2017; 39:1–9. [DOI] [PubMed] [Google Scholar]
- 153. Jia N., Jones R., Yang G., Ouerfelli O., Patel D.J.. CRISPR-Cas III-A Csm6 CARF domain is a ring nuclease triggering stepwise cA4 cleavage with ApA>p formation terminating RNase activity. Mol. Cell. 2019; 75:944–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Makarova K.S., Anantharaman V., Grishin N.V., Koonin E.V., Aravind L.. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front. Genet. 2014; 5:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Koonin E.V., Makarova K.S.. Discovery of oligonucleotide signaling mediated by CRISPR-associated polymerases solves two puzzles but leaves an enigma. ACS Chem. Biol. 2018; 13:309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Johnson K., Bailey S.. Microbiology: the case of the mysterious messenger. Nature. 2017; 548:527–528. [DOI] [PubMed] [Google Scholar]
- 157. Nelson J.W., Breaker R.R.. The lost language of the RNA world. Sci. Signal. 2017; 10:eaam8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.