Abstract
Telomere anchoring to nuclear envelope (NE) is a key feature of nuclear genome architecture. Peripheral localization of telomeres is important for chromatin silencing, telomere replication and for the control of inappropriate recombination. Here, we report that fission yeast quiescent cells harbor predominantly a single telomeric cluster anchored to the NE. Telomere cluster association to the NE relies on Rap1–Bqt4 interaction, which is impacted by the length of telomeric sequences. In quiescent cells, reducing telomere length or deleting bqt4, both result in an increase in transcription of the telomeric repeat-containing RNA (TERRA). In the absence of Bqt4, telomere shortening leads to deep increase in TERRA level and the concomitant formation of subtelomeric rearrangements (STEEx) that accumulate massively in quiescent cells. Taken together, our data demonstrate that Rap1–Bqt4-dependent telomere association to NE preserves telomere integrity in post-mitotic cells, preventing telomeric transcription and recombination. This defines the nuclear periphery as an area where recombination is restricted, creating a safe zone for telomeres of post-mitotic cells.
INTRODUCTION
Eukaryotic chromosome ends or telomeres fulfill important functions to preserve the stability of the genome. They consist in G-rich repetitive sequences ending in a 3′ single-stranded overhang (G-tail) that are specifically bound by telomeric proteins. This multiprotein complex, named shelterin, protects chromosome extremities from degradation, end-to-end fusion and recombination, and also ensures the recruitment of telomerase which utilizes its RNA subunit as a template for de novo telomeric repeat DNA addition (1–5). In mammals, telomeres localize all over the nucleus and are attached to the nuclear matrix through shelterin and lamins (6,7), and only a subset of telomeres are found at the nuclear periphery (8). Irrespective of their localization within the nucleus, mammalian telomeres are heterochromatinized regions enriched with repressive histone modifications (9), associated with proper Telomere repeat-containing RNA (TERRA) expression (10,11). In fission yeast, the repressive chromatin mark H3K9me2 is restricted to a 10–20 kb subtelomeric region (12). The repressed-state at subtelomeres is maintained by the Fun30 (Fft3) chromatin remodeler that binds to long-terminal repeat (LTR) elements and associates with the inner nuclear membrane (INM) protein Man1 (13,14). The interaction between Man1 and Fft3 creates domain boundaries and anchors subtelomeric chromatin to the nuclear envelope (NE), a peripheral localization that is important for transcriptional silencing. In addition, telomeric repeated sequences are tethered to the nuclear periphery by the INM Bqt3–Bqt4 complex. Indeed, Bqt4 is able to bind directly to double-stranded DNA (15), and this primes its association with the telomeric protein Rap1 (16). Rap1 interacts directly with Taz1, the main component of the shelterin that directly binds to telomeric tracks. Telomeres are released from the NE when the interaction between Rap1 and Bqt4 is perturbed (e.g. in bqt4 deleted cells or when Rap1 is phosphorylated) (16,17). In summary, both Fft3–Man1 and Bqt4–Rap1 interactions ensure chromosome ends localization to the nuclear periphery (Figure 1A). When Bqt4 is absent, in addition to Fft3, the entire telomeres can move away from the NE toward the interior of the nucleus (13).
Figure 1.
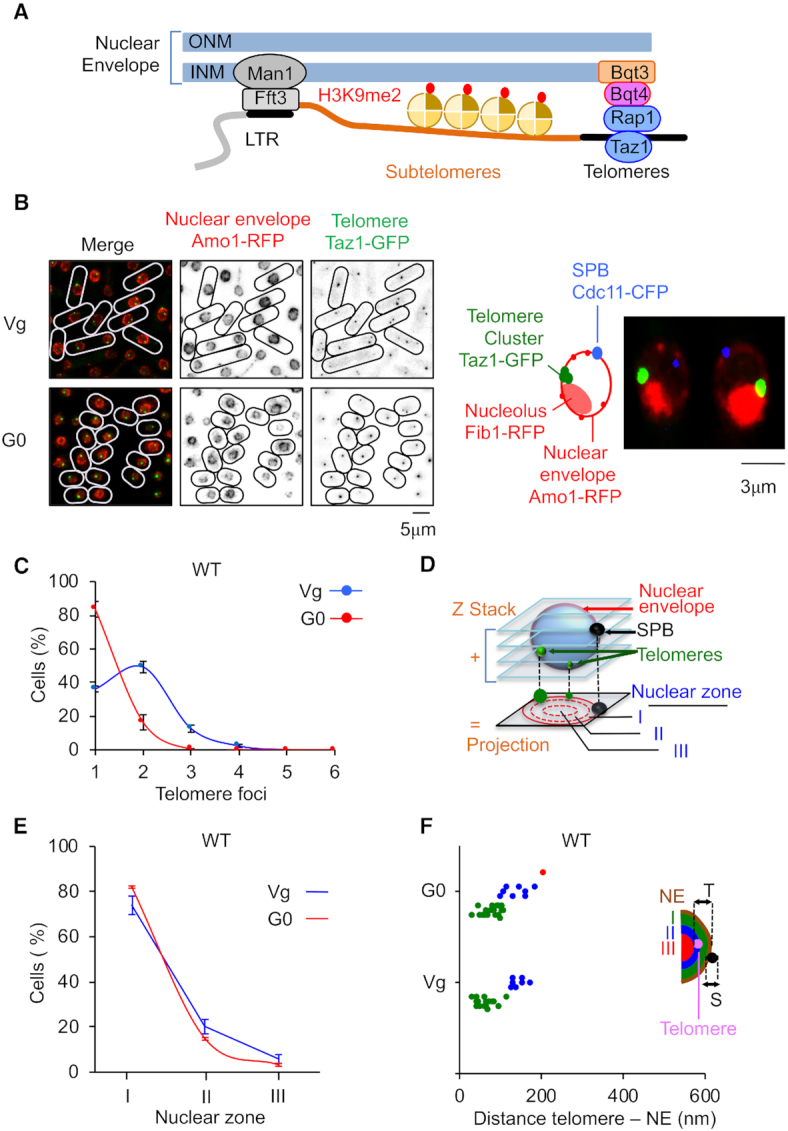
Telomeres hyperclusterize and anchor to NE in quiescent cells. (A) Schematic representation of nuclear envelope (NE) anchoring of telomeres in fission yeast. (B) Visualization of telomeric foci and NE by live microscopy in vegetative (Vg) and quiescent (G0) cells. Left panel: Taz1-GFP (green) and Amo1-RFP (red) mark the telomeres and the NE, respectively. Right panel: Image of Taz1-GFP (green), Amo1-RFP (red), Fib1-RFP (red) illustrating the Rabl conformation of telomere cluster located close to the nucleolus at the opposite of spindle pole body. (C) Percentage of cells that display one or several telomeric foci. (D) Z-stacks images of living cells and scoring of the position of the Taz1-GFP hypercluster in one of the three equal concentric zones of the nucleus with respect to Amo1-RFP in the focal plane of the GFP focus. (E) Taz1-GFP hypercluster localization relative to the NE (Amo1-RFP) in the three zones. (F) Distance to NE of the telomeric foci in Vg and G0 cells. This experiment was repeated in triplicate and for each experiment >100 nuclei with a hypercluster were analyzed. Error bars represent SEM.
In interphase, telomeres form two to three clusters at the NE (16,18), and this localization persists when telomeres are replicated (19). Tethering of telomeres to NE by Bqt4 stabilizes the subtelomeric heterochromatin and is thought to create a specialized environment that prevents collisions between transcription and replication machineries (19). The peripheral localization of telomeres is therefore important for genome organization, chromatin silencing and telomere replication. Interestingly, studies in budding yeast also revealed that the INM suppresses recombination at telomeres (20,21).
While the mechanisms of telomere maintenance have been investigated in vegetative cells, little is known about the stability of telomeres in quiescent cells. We recently examined the stability of telomeres in fission yeast cells that were maintained in quiescence by nitrogen starvation (G0) (22). We discovered that eroded telomeres were highly rearranged during quiescence in the absence of telomerase. These rearrangements correspond to duplication of a subtelomeric region (STE1) adjacent to telomeric repeats, named STEEx for STE1-Expansion. These telomeric rearrangements depend on homologous recombination (HR) and correlate with a higher transcription of TERRA (23). Because localization of telomeres at nuclear periphery is important for heterochromatin establishment and possibly for the control of recombination, we investigated the localization of telomeres in quiescent S. pombe cells.
In this study, we show that quiescent fission yeast cells mainly display a single telomeric cluster anchored to the NE. In quiescent cells, when telomeres shorten or in the absence of Bqt4, telomeres detach from nuclear periphery and this change in their localization within the nucleus is associated with an increase of TERRA transcription. When telomere shortening occurs in the absence of Bqt4, TERRA level deeply increases and subtelomeric rearrangements (STEEx) massively accumulate in these quiescent cells. This change in telomere structure prevents cells to exit properly from quiescence. Our results reveal the importance of telomere positioning at the NE during quiescence, representing a safe area in which repressed transcription limits the recombination caused by telomere attrition.
MATERIALS AND METHODS
Strains and growth conditions
All yeast strains used in this study are prototrophic (leu1+, ura4+, his+ and ade+) unless indicated in the strain list (Table 1). Most of the deleted or epitope tagged strains were produced by homologous recombination using modified pFA6a series of plasmids, carrying hygromycin resistance gene (hyg in strain list) (24) or previously described pFA6a series of plasmids carrying kanamycin resistance gene (kanR in strain list) (25). This includes taz1-gfp (a gift from J. Cooper, Center for Cancer Research, NIH), amo1-rfp (a gift from P. Nurse, Cancer Research UK, London), mis6-2rfp (a gift from T. Toda, Cancer Research UK, London), ndc80-gfp cdc11-cfp (26), swi6-gfp (a gift from J.P Javerzat, Bordeaux, France). Visualization of the nucleolus was achieved by the expression of the plasmid pREP41x/fib1-mRFP (27). Yeast transformations were performed by electroporation in the presence of thiamine (repressive condition). Fib1-mRFP protein was induced by growing the cells in the absence of thiamine for 24 h.
Table 1.
Strains used in this study
| Strain number | Genotype |
|---|---|
| CS158 | taz1-GFP:kanR cdc11-CFP:kanR amo1-RFP:kanR |
| ST1837 | taz1-GFP:kanR cdc11-CFP:kanR amo1-GFP:kanR h+ |
| CS165 | taz1-GFP:kanR cdc11-CFP:kanR amo1-RFP :kanR + pREP41x/fib1-mRFP:LEU2 |
| LM1317 | swi6-GFP:kanR amo1-RFP:kanR cdc11-CFP:kanR h- |
| ST2166 | bqt4::ura4+ taz1-GFP:kanR cdc11-CFP:kanR amo1-GFP:kanR |
| SC1404 | bqt4::hphR swi6-GFP:kanR amo1-RFP:kanR cdc11-CFP:kanR h- |
| LM1224 | bqt4::ura4+ h- |
| SC1406 | bqt4::ura4+ cid14 ::natR |
Telomerase was deleted by substituting the ter1 gene by kanamycin cassette by one-step homologous insertion using FwTer1275 and RevTer1276 primers (FwTer1275 5′-AACGCAACGCCCATGCTTAGAAGGTTGACAAGGAAAATTAATCAAACGGT-3′ and RevTer1276 5′-TTCATCTCTTCTAGTACGCAAATAAATACATTAAATTTATTTTACATTAT-3′) (25). Prior starvation, strains were grown in minimal medium MM (28), at 32 or 25°C to a density of 6 × 106 cells/ml, washed twice in MM deprived from nitrogen (MM-N), and re-suspended in MM-N at a density of 2 × 106 cells per ml at 32 or 25°C, reaching after two rounds of cell division 8 × 106 cells/ml (29). This density of cell bodies is stable for several weeks and was used as a source of G0 cells. Genome data and annotations were obtained from Pombase2018 (30,31).
Liquid senescence assay
After micromanipulation or transformation, cells were grown in 25 ml of YES medium at 32°C and the starting cell density was set to 106 cell/ml each day. Population doublings were calculated each day as the log2(cell density/106), where the cell density is measured after 24 h.
Telomere analysis and spotting assay
Genomic DNA was prepared from 2 × 108 cells according to standard protocols and digested with the indicated restriction enzymes (New England Biolabs). The digested DNA was resolved in a 1.2% agarose gel and blotted onto a Hybond-XL membrane (GE Healthcare). After transfer, the membrane was cross-linked with UV and hybridized with different probes. 32P labeling of DNA probes was performed by random priming using Klenow fragment exonuclease- (New England Biolabs), in presence of [α-32P]CTP and hybridizations were performed in Church buffer at 65°C for Telo/STE1 and STE1 (STE1 = Subtelomeric Element 1) probes and 55°C for chromosomal probe. For spotting assays, genomic DNA were directly spotted onto a Hybond-XL membrane (GE Healthcare). Radioactive signal was detected using a Biorad molecular imager FX.
Fluorescence microscopy
For the fluorescence microscopy experiments, cells were grown at 25°C and fixed in 3.7% formaldehyde for 7 min at room temperature. Fixed cells were washed twice in phosphate-buffered saline (PBS) and observed after staining with DAPI (Vectashield). Acquisition of Z stacks (maximum twelve stacks of 0.3 μm steps) were taken. Exposure times were 300–500 ms with a Sola source (Lumencor, France) reduced to 30% to avoid photobleaching. Images were visualized with a Princeton CCD CoolSNAP HQ2 camera (Roper Scientific, Evry, France) fitted to a Leica DM6000 upright microscope (Leica Microsystems, Rueil-Malmaison, France) with a 100× (1.4 NA) objective and SEMROCK filters, and were recorded using the MetaMorph software package (Molecular Devices France, St. Gregoire, France). Intensity adjustments were made using the MetaMorph, ImageJ, and Adobe Photoshop packages (Adobe Systems France, Paris, France).
As previously reported in budding yeast (32), we analyzed the position of the foci relative to the nuclear envelope by defining three zones of similar surfaces within a normalized nucleus (zone I, II or III). In this analysis, for simplicity, we always assume that the nucleus is a sphere. However, this assumption is not always true and envelope invaginations occur. Thus, to determine the telomere to NE distance (center of the telomere to center of the NE signal), the image with the best focal plane was used. When an invagination of the envelope is present, the telomere to envelope distance can be small, still, the telomere appears to be in zone III. Of note, when analyzing the telomere to envelope distance, we have to keep in mind that the theoretical resolution of our microscope is approximately of 200 nm. To quantify the number of telomere foci projected images were used.
RNA isolation and quantitative PCR
RNA isolation and quantitative PCR analysis were performed as described previously by Maestroni et al. (22).
Cell viability at exit of quiescence
Exit of quiescence was monitored by micromanipulation of single-quiescent cells on rich-medium YES plates. After 3 days of incubation at 32°C plates were observed under microscope and colonies and microcolonies were counted.
RESULTS
Telomeres hyperclusterize in quiescent cells
In vegetative Schizosaccharomyces pombe cells, telomeres form two to three clusters that are localized at the nuclear periphery (16,18,33). We tested whether this genome organization feature is conserved in G0 cells. We analyzed telomere foci organization at the nuclear envelope in vegetative cells (Vg) or in quiescent cells (G0) (Figure 1B). Taz1 and Amo1 proteins were used as markers of telomeres and NE, respectively. We confirmed that Vg cells display from one to three foci (Figure 1C–F), mainly concentrated into the peripheral zone (Figure 1E). When cells were starved from nitrogen source they enter into quiescence and become round and short (Figure 1B) (34). The attachment to NE was conserved in G0 cells (Figure 1E and F), however these cells predominantly exhibited a unique bright focus (Figure 1B and C). We concluded that telomeres in post-mitotic cells hyperclusterize and anchor to the NE, as described for budding yeast (35).
When the spindle pole body (SPB) and the nucleolus were marked in addition to telomeres and NE, we observed that the bright telomeric cluster was located close to the nucleolus facing the SPB (Figure 1B, right panel). This indicated that post-mitotic cells exhibit a Rabl organization in which telomeres form a unique cluster. Noteworthy, we observed that the average size of nucleus in wild-type (and mutants used in this study) was significantly reduced in G0 cells (Supplementary Figure S1). This might indicate that telomere hyperclusterization is also caused by nuclear size reduction.
The telomere hypercluster persists when telomeres are eroded
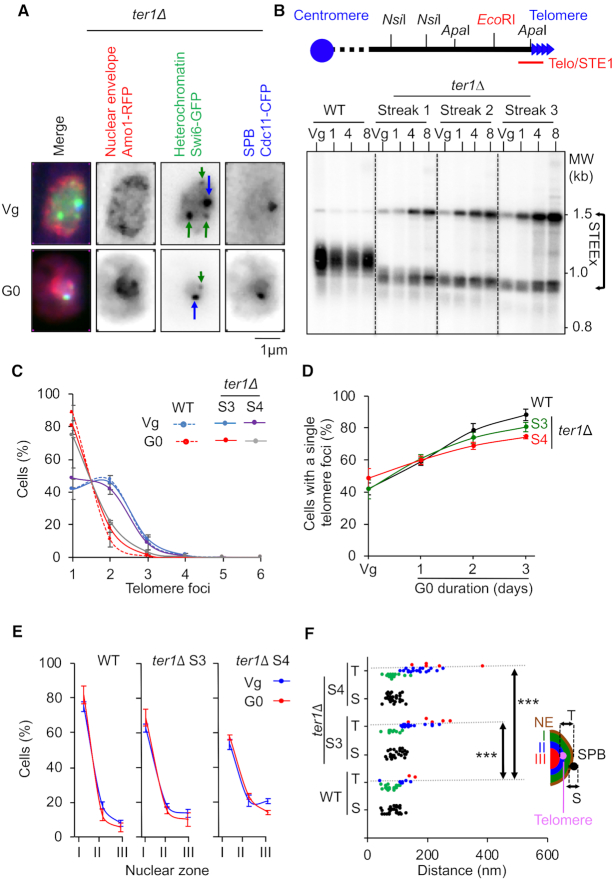
We previously showed that eroded telomeres undergo subtelomeric rearrangements in quiescent cells in the absence of telomerase activity (ter1Δ cells) (22). We wondered if telomere attrition together with the associated recombination of subtelomeric regions would modify telomere localization. In the absence of telomerase, since the Taz1-GFP signal rapidly faded, we decided to use Swi6-GFP to monitor subtelomere/telomere localization (Figure 2). These cells also expressed Amo1-RFP (NE marker) and Cdc11-CFP (SPB marker) that enables us to distinguish between centromeres (at the SPBs) or telomeres foci that are both marked with Swi6-GFP (Figure 2A).
Figure 2.
Telomere localization in absence of telomerase activity in quiescent cells. Telomerase positive (WT, ter1+) and negative (ter1Δ) strains were starved from nitrogen. (A) Visualization of telomeric foci, the nuclear envelope (NE) and the spindle pole body (SPB) by live microscopy in vegetative (Vg) and quiescent (G0) ter1Δ cells. Swi6-GFP (green), Amo1-RFP (red) and Cdc11-GFP (green) mark the telomeres and centromeres (heterochromatin), the NE and the SPBs, respectively. Green arrows and blue arrows indicate telomeres and SPB, respectively. (B) Top, relative position of the restriction sites in the telomeric and subtelomeric regions of S. pombe chromosomes based on pNSU70. The Telo/STE1 probe used for southern blot reveals telomeric and subtelomeric signals. Bottom, Genomic DNA from WT and ter1Δ cells was digested with EcoRI and southern blotted. The membrane was hybridized with a Telo/STE1 probe. Vg, replicative sample; 1, 4, 8 correspond to the number of days in quiescence. (C) Percentage of cells that display one or several telomeric foci after 3 days in G0. For ter1Δ cells, imaging was processed from streaks 3 and 4. (D) Percentage of cells that display one telomeric focus with time in quiescence monitored from streak 3 and streak 4 for ter1Δ cells. (E) Percentage of cells that display a unique telomeric hypercluster and its localization relative to the NE in the three equal concentric zones of the nucleus. (F) Distance to NE of the telomeric foci in Vg cells. p-values are indicated. This experiment was repeated in triplicate and for each experiment >100 nuclei with a hyper-cluster were analyzed (***P < 0.0005). Error bars represent SEM.
Deletion of ter1 was carried out in vegetative cells and the progressive shortening of telomeres was monitored by southern blot after several streaks on YES agar plates (Vg streak 1 to streak 3) (Figure 2B). Vegetative ter1Δ cells with different settings of telomere length (streaks 1–3) were taken from the YES agar plates and placed in medium depleted for nitrogen to induce quiescence. Cells were harvested at different times during quiescence and telomere structure was analyzed (Figure 2B). As expected, STEEx accumulated with time in quiescence and these rearrangements correlated with the shortening of telomeres (Figure 2B). Next, we determined the number of foci per cell in Vg and G0 cells (Figure 2C) and the percentage of cells that contain a unique telomeric cluster in G0 after streaks 3 and 4 (Figure 2D). We found that telomere attrition observed in the absence of telomerase did not significantly impair telomere hyperclusterization in quiescence. However, telomere clusterization did not reach WT level in ter1Δ cells after 3 days in G0. The Figure 2E shows that telomere clusters slightly move from zone 1 to zones 2 and 3 when telomeres are eroded. More significantly, the distance of telomere clusters to the NE increased with telomere shortening in Vg cells (Figure 2F). From these observations, we conclude that telomere shortening does not markedly change telomere organization in foci but causes a slight detachment of telomeres from the NE.
Telomeres detach from NE in bqt4Δ quiescent cells
Next, we asked if telomere detachment from NE that we observed when telomeric tracts shorten might impact the formation of subtelomeric rearrangements in G0. We used bqt4Δ cells in which telomeres are released from the NE in replicative cells (16). We first examined telomeres localization in quiescent bqt4Δ cells by following Taz1-GFP foci in a strain expressing Cdc11-CFP (SPB) and Amo1-GFP (NE) (Figure 3A). The distribution of foci number per cells was not significantly different between WT and bqt4Δ in Vg and G0 cells (Figure 3B) although in several experiments, we observed that the kinetic of appearance of this cluster over time was slightly accelerated in bqt4Δ cells (Figure 3C). We confirmed that telomere foci moved from nuclear periphery to a more central area (zone 1 to zone 2 or 3) in bqt4Δ Vg cells (Figure 3D). This was also observed in bqt4Δ G0 cells in which telomeric hypercluster seemed to accumulate in the central area (zone 3) in more that 20% of cells. The change in telomere positioning observed in bqt4Δ G0 cells was confirmed by measuring the telomeric to NE distance (the centromeric foci located at SPBs are used here as an internal control) (Figure 3E). Thus, we concluded that the distance between telomeres and the NE is increased in the absence of Bqt4 in vegetative cells and that this change in telomere positioning is exacerbated in quiescence.
Figure 3.
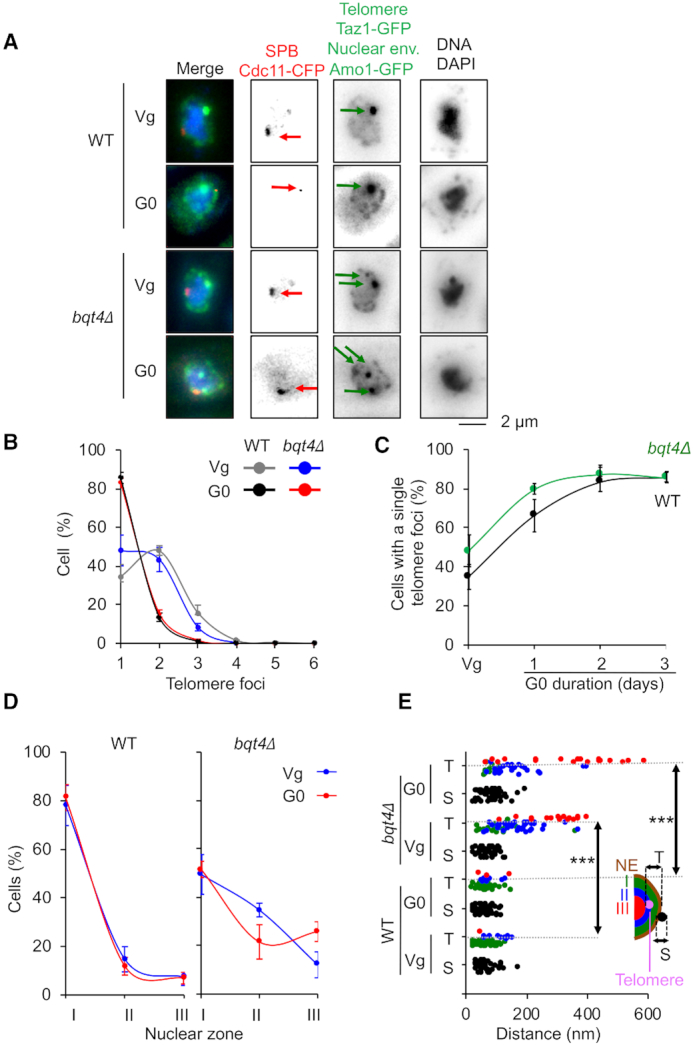
Telomeres detach from NE in bqt4Δ quiescent cells. Visualization of telomeric foci, spindle pole body (SPB) and the nuclear envelope (NE) by live microscopy in vegetative (Vg) and quiescent (G0) cells for the WT and bqt4Δ strains. (A) Taz1-GFP (green), Cdc11-CFP (red) and Amo1-GFP (green) are used to visualize the telomeres, the SPBs and the NE, respectively. Green arrows and red arrows indicate telomeres and SPB, respectively. (B) Percentage of cells that display one or several telomeric foci. (C) Percentage of WT and bqt4Δ cells that display a unique telomeric focus with time in quiescence. (D) Taz1-GFP hypercluster localization relative to the NE (Amo1-GFP). Z-stacks images of living cells and scoring of the position of the Taz1-GFP hypercluster in one of the three equal concentric zones of the nucleus with respect to Amo1-GFP in the focal plane of the GFP focus. (E) Distance to NE of the telomeric foci in Vg and G0 cells. P-values are indicated. This experiment was repeated in triplicate and for each experiment >100 nuclei with a hyper-cluster were analyzed (***P < 0.0005). Error bars represent SEM.
NE attachment limits STEEx formation
We next sought to determine the impact of bqt4 deletion in quiescent cells harboring short telomeres. In particular, we asked whether the defect in telomere positioning observed in bqt4Δ G0 cells could affect STEEx formation. We induced STEEx as previously described (22) by first performing daily dilution of ter1Δ spore colonies in vegetative cells and monitoring their growth (Figure 4A and Supplementary Figure S2) and telomere shortening (Figure 4B). In contrast to ter1Δ cells in which the loss of growth capacity was progressive, the growth of bqt4Δ ter1Δ cells was severely impaired (Figure 4A and Supplementary Figure S2). Accordingly, shortening of telomeres was much more severe in the absence of Bqt4, however cell death is unlikely caused by telomere loss (see discussion). Next, we monitored simultaneously telomere and SPB foci with the nuclear envelope in bqt4Δ ter1Δ strain (Supplementary Figure S3). Microscopic observations revealed that the double mutant rapidly undergoes mitotic catastrophe, exhibiting in many cells a ‘cut’ phenotype where the septum physically divides the nucleus in two parts (Supplementary Figure S4). The distribution of foci number per cells was similar between WT, bqt4Δ and bqt4Δ ter1Δ in Vg and G0 cells (Supplementary Figure S3B) but the kinetic of appearance of this cluster over time was compromised in ter1Δ bqt4Δ cells (Supplementary Figure S3C). Noticeably, the zoning of telomere foci within the nuclear envelope was severely impaired in bqt4Δ ter1Δ for vegetative and quiescent cells (Supplementary Figure S3D and E). These severe positioning defects of eroded telomeres correlated with growth defect of bqt4Δ ter1Δ cells. These results suggest that Bqt4 and likely NE attachment fulfill important function in replicative senescence. After 1, 3 and 5 days of senescence (D1, D3, D5 in Figure 4A), cells with different settings of telomere length were starved from nitrogen and maintained in quiescence for a total of 4 days. In the case of bqt4Δ ter1Δ cells, we only starved cells from D1 and D3 of the senescence kinetics because of the extremely low number of cells alive at day 5. Cells were collected at different times in quiescence and the formation of STEEx was monitored by southern blot (Figure 5A). bqt4Δ telomerase positive cells exhibited wild-type telomeres that were stable in post-mitotic cells (Figure 5A, middle panel). EcoRI digestion and Telo/STE1 probe also revealed a 1500 bp DNA fragment that corresponds to subtelomeric regions. This subtelomeric band can be detected in some specific genetic background in which subtelomeric regions are homogenous at each chromosome ends (22). Nevertheless, subtelomeres were stable in quiescent bqt4Δ cells (Figure 5A). In contrast, in ter1Δ cells, three bands that correspond to the amplification of subtelomeric regions appeared and gradually accumulated with time in quiescence when telomeres shorten (D3 and D5) (Figure 5A, middle panel). In bqt4Δ ter1Δ cells, STEEx were readily detected as two bands at 1500 and 900 bp, the highest one being prevalent (Figure 5A, right panel). Strikingly, we observed a massive accumulation of STEEx in quiescent bqt4Δ ter1Δ cells at early time points of quiescence (Figure 5A). This was obvious when comparing D5 of ter1Δ with D3 of bqt4Δ ter1Δ for which the size of telomeres is comparable at these time points of senescence. This strong amplification of subtelomeric regions was also confirmed by spotting assay quantification (Figure 5B and C). We concluded that STEEx appearance and accumulation were exacerbated in the absence of Bqt4, thereby correlating defects in telomeres anchoring at NE with STEEx formation. Taken together, these results suggest that telomere anchoring at the NE inhibits telomere recombination in quiescent cells.
Figure 4.
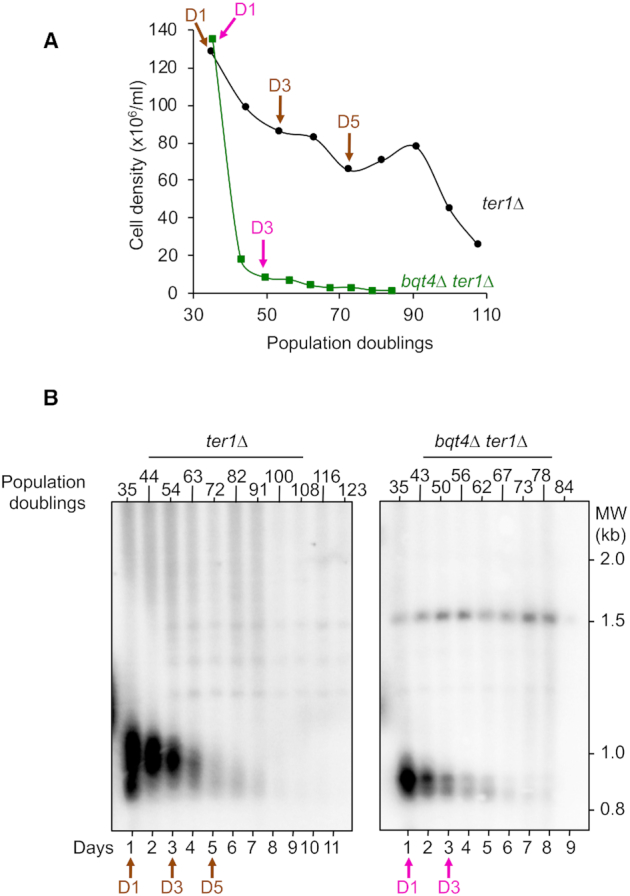
Telomere shortening in WT and bqt4Δ telomerase negative cells. (A) Freshly deleted ter1 colonies from WT and bqt4Δ strains were grown in liquid YES medium for several days by serial dilutions. Population doublings were monitored by cell counting. (B) Genomic DNA was digested with EcoRI and southern blotted. The membrane was hybridized with a Telo/STE1 probe. Arrows indicate the time points (Day 1, Day 3 and Day 5) of the replicative senescence kinetics that have been shifted to nitrogen-deprived medium (Day 1 = 34 pds, Day 3 = 54 pds and Day 5 = 72 for ter1Δ; Day 1 = 35 pds and Day 3 = 49 pds for ter1Δ bqt4Δ).
Figure 5.
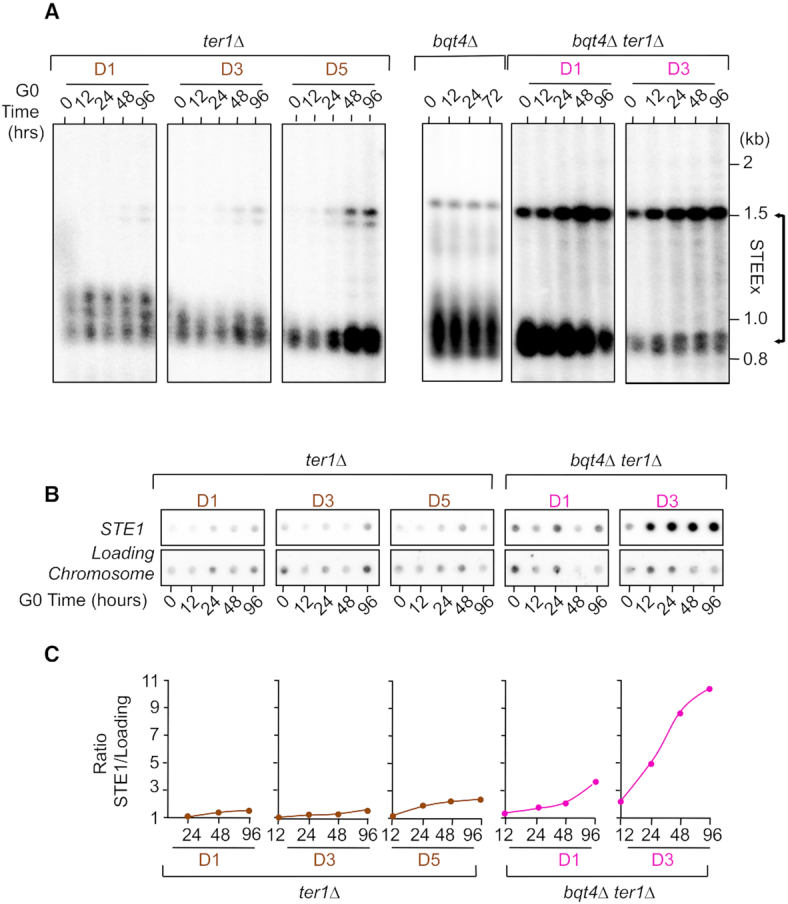
NE attachment of telomeres limits STEEx formation. (A) Cells were maintained in quiescence for 4 days and collected at different time points. Genomic DNA was digested by EcoRI and southern blotted. Membrane was hybridized with a Telo/STE1 probe. D1, D3 and D5 indicate the days during the senescence kinetics at which cells were starved from nitrogen. Results are presented in separated patterns but are issued from the same membrane. (B) Genomic DNA from quiescent ter1Δ and bqt4Δ ter1Δ cells was spotted onto Hybond-XL membrane and hybridized with a subtelomeric probe (STE1) and a chromosomic probe (Chromosome) as a loading control. (C) Signal quantification has been performed with ‘Quantity one’ software and STE1/Loading control ratio was determined.
bqt4Δ and short telomeres alleviate TERRA repression in quiescence
Since STEEx formation is promoted by transcription of telomeres, we investigated TERRA level in bqt4Δ cells. Strikingly, TERRA level was higher in bqt4Δ than WT in vegetative cells and this difference was substantially intensified after 48H in quiescence (Figure 6A), suggesting that telomeres association to NE through Bqt4 interaction participates in the repression of transcription at telomeres. We previously showed that the level of TERRA in G0 cells increased when telomeres are eroded (22) and this was confirmed here (Figure 6B). When ter1+ gene was deleted in bqt4Δ cells, we observed that the combination of telomere erosion and NE dissociation provokes a massive accumulation of TERRA in Vg cells and this robust increase in transcription is even stronger after 48H in quiescence (Figure 6B). We further determined that in bqt4Δ telomerase minus cells, accumulation of TERRA depends on Cid14, a RNA poly adenyl-transferase, (Supplementary Figure S5). This indicates that polyadenylation regulates TERRA stability in bqt4Δ quiescent cells. This seems to differ from what was previously observed by Bah and colleagues (36), who showed that Cid14 destabilizes TERRA in WT (bqt4+) context. Despite the destabilization by lack of Cid14, TERRA transcription seems highly induced in bqt4Δ ter1Δ cells. Collectively our results revealed a critical role of Bqt4 in repressing TERRA in Vg cells that is exacerbated in quiescent cells. When telomeres are short, the lack of repression is strongly intensified leading to an extremely high level of TERRA. Interestingly, STEEx formation correlates with TERRA level, suggesting that dissociation of telomeres from the NE exacerbates subtelomeric rearrangements by increasing transcription. Our results strengthen the concept that the nuclear periphery creates a safe zone that represses telomeric transcription and protects telomere from recombination in post-mitotic cells.
Figure 6.
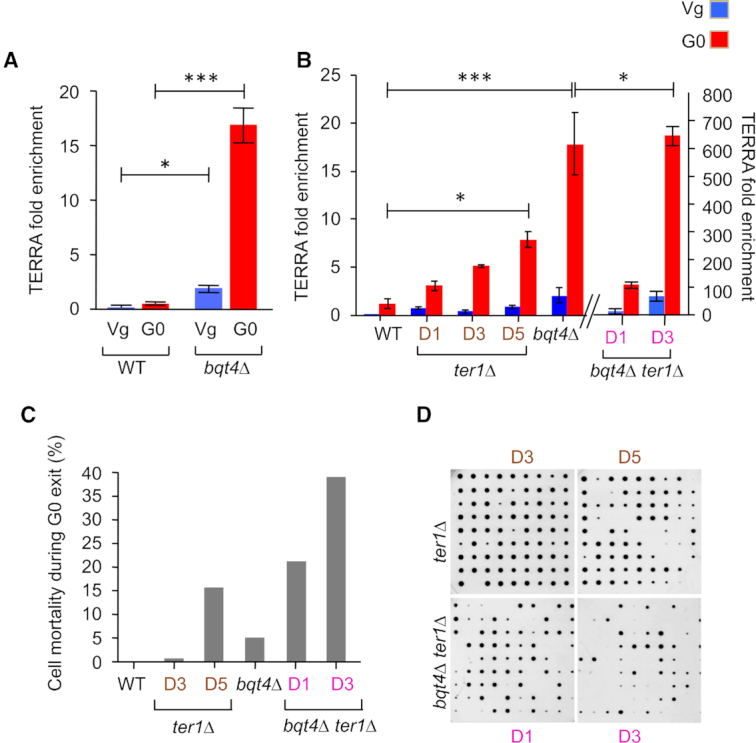
TERRA accumulates in bqt4Δ cells. (A and B) RNA from telomerase positive and telomerase negative cells (ter1Δ) were extracted in vegetative (Vg) and in quiescent (G0 after 48 h) cells from WT and bqt4Δ strains. RNA levels were determined by RT-qPCR using a random hexanucleotide primer followed by qPCR with specific oligonucleotides for TERRA and control Fcp1. TERRA fold enrichment was determined as the ratio of TERRA over Fcp1 RNA levels. Error bars represent SEM (***P < 0.0005; *P < 0.05). (C and D) Telomerase positive and negative WT and bqt4Δ single cells were maintained in quiescence for 2 days, micromanipulated and plated onto YES plates allowing them to exit quiescence. The percentage of cells that were not able to form a colony was plotted.
Our previous results suggest that STEEx formation correlates with defective exit from quiescence (22). We thus analyzed cell viability by micromanipulating ter1Δ and bqt4Δ ter1Δ cells onto rich media and monitored the proportion of cells forming colonies (CFC). Figure 6C and 6D show the percentage of cells that are unable to form a visible colony when cells re-enter the cell cycle after 48H of quiescence in different genetic backgrounds. As previously shown, we observed that telomere erosion and STEEx formation in ter1Δ cells correlates with defects to exit properly from G0 (22). Indeed, in bqt4Δ ter1Δ cells the percentage of cells that are unable to form a colony increased in correlation with the massive accumulation of STEEx at D1 and D3 of senescence. This is particularly clear at D1 of senescence where bqt4Δ ter1Δ mutant shows no growth defect in vegetative state (Figure 4A and Supplementary Figure S2) but is not able to exit properly from G0.
Collectively, our results suggest that in bqt4Δ ter1Δ cells, NE dissociation and erosion of telomeres cause both, high level of transcription and accumulation of STEEx, which in turn limit the capacity of cells to exit from quiescence (see the model in Figure 7).
Figure 7.
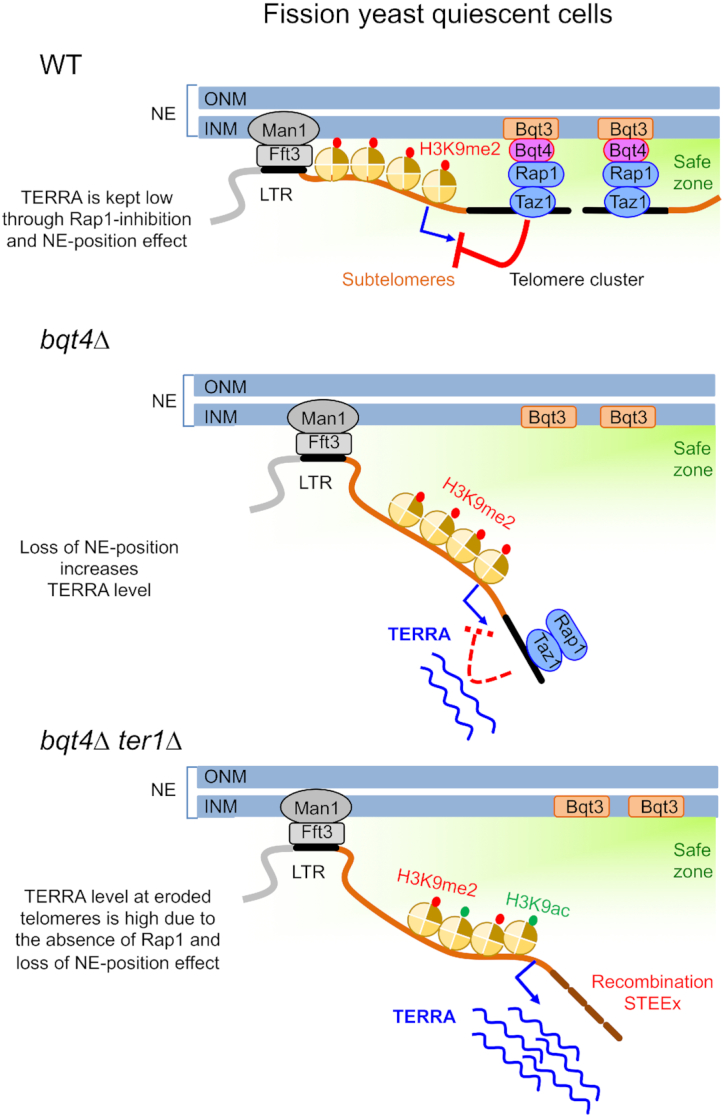
Model of telomere cluster anchoring in arrested cells. For the sake of clarity, only two over the six fission yeast telomeres are represented in the cluster that associates to the INM through Rap1–Bqt4 interaction. Anchoring of chromosome ends to the nuclear periphery is ensured by Man1–Fft3 interaction that creates domain boundaries within subtelomeric regions (100 kb away from terminal sequences) and by Bqt4–Rap1 interaction that tethers telomeric repeats to the INM. The nuclear periphery represents a safe zone that protects telomeres and keeps the transcription low. In this environment, TERRA transcription is down-regulated in a Rap1-dependent manner, named here the NE-position effect. In absence of Bqt4, telomeres dissociate from the NE and exit from the safe zone, leading to an increase of TERRA level in a Rap1-independent manner. At eroded telomeres, the loss of Rap1 exacerbates the NE-position effect and provokes a substantial increase of TERRA that may be controlled by a change of heterochromatin marks, such as higher level of H3K9ac. The erosion of telomeres and high transcription leads to the massive accumulation of STEEx.
DISCUSSION
In this study, we report that telomeres hyperclusterize in quiescent cells forming a unique focus in the nucleus, consistent with a recent report (37). We also observe that this unique cluster localizes to the nuclear periphery and that this positioning relies on the interaction between Rap1 and the INM protein Bqt4. In those cells, the Rabl organization is maintained since we observed that the telomeric cluster faces the SPB and intriguingly it contacts the nucleolus, which seems to be a particularity of fission yeast (Figure 1B). In budding yeast, this hyperclusterization has also been observed in long-lived quiescent cells (35,38,39) suggesting that this common feature might reflect a need to compartmentalize their genome when growth is stopped. This spatial re-organization during quiescence is not restricted to telomeres (39,40) and likely uncovers a specific feature of the nuclear periphery in arrested cells.
Here, we also report that eroded telomeres dissociate from the nuclear periphery in vegetative and quiescent cells (Figure 2E and F). We explain this change of telomere positioning by decreased Rap1 levels at eroded telomeres that in turn would lower their interaction with Bqt4 and promote the dissociation of short telomeres from the NE. In addition, it has been previously shown that Rap1 is a negative regulator of transcription at telomeres (41). This supports the model that the progressive shortening of telomeres and the associated loss of Rap1 account for the increase of TERRA level. Here we show that a substantial increase of TERRA transcription is also observed in absence of Bqt4, in both vegetative and arrested cells (Figure 6). In this case, the increase in transcription is caused by the dissociation of telomeres from the NE in a Rap1-independent manner. These results indicate that the nuclear periphery participates in the repression of telomeric transcription (Figure 7). Because Fft3 and Bqt4 cooperate in anchoring chromosome ends to the nuclear envelope, it would be interesting to also investigate the impact of fft3 deletion in quiescent cells, alone and in combination with bqt4Δ. This will allow to determine first the impact of Fft3-LTR anchoring mode on telomere positioning and telomeric transcription, and second to establish the functional links between these two modes of anchoring.
How the NE-position affects TERRA transcription remains elusive. The methylation of H3K9 does not seem to be involved in TERRA up-regulation since only a subtle increase of H3K9me2 level at telomeres was detected in bqt4Δ cells (19). Moreover, TERRA remains repressed in the absence of the methyl transferase Clr4 that abolishes all forms of H3K9 methylation (42). Noteworthy, it was shown that TERRA levels increase along with acetylation of H3K9 in telomerase negative cells displaying short telomeres (43). Thus, repression of TERRA transcription at the nuclear periphery likely occurs independently of H3K9 methylation but possibly in association with a higher level of H3K9 acetylation in vegetative cells. Interestingly, a recent work showed that within subtelomeric regions, H3K9me2 levels decreases rapidly upon nitrogen removal (44) and we previously established that TERRA is up-regulated in quiescence (22). In contrast to cycling cells, this suggests that in arrested cells the partial loss of the H3K9me2, within subtelomeric regions, might participate to the positive regulation of telomere transcription. Further investigations will be necessary to determine if the reduction of H3K9me2 occurs at the expense of H3K9ac in quiescence which would explain the up-regulation of transcription.
In our previous work, we showed that telomeric transcription at eroded telomeres correlates with subtelomeric rearrangements (STEEx) in quiescent cells (22). Importantly, the absence of RNaseH1 exacerbated STEEx formation, indicating a causal connection between telomeric transcription and STEEx formation in quiescent fission yeast cells. In the present study, we further show a correlative effect of the NE positioning of telomeres and TERRA transcription in quiescence. This relies on the observations that either the lack of Bqt4 or telomere erosion provokes the detachment of telomeres from NE. When both are combined, dissociation of telomeres from the NE is exacerbated and causes a substantial increase of TERRA transcripts and a massive accumulation of STEEx (Figure 7, lower panel). This supports a model in which telomere anchoring participates to the regulation of transcription at telomeres and telomere stability. Altogether, this defines the nuclear periphery as an area where recombination, that is likely dependent on transcription, is restricted, creating a safe zone for telomeres of post-mitotic cells.
Strikingly, we further observed that in vegetative cells, the lack of Bqt4 is deleterious in the absence of telomerase (Figure 4A and Supplementary Figure S3). Premature cell death of bqt4Δ ter1Δ mutant is unlikely to be caused by telomere loss as telomeric signal is still present (Figure 4B), suggesting that Bqt4 fulfills important functions during replicative senescence. Telomeres are known as difficult region to replicate and this is mostly revealed in the absence of telomerase activity (45). It has been recently reported that Bqt4-dependent NE-anchoring facilitates telomere replication by preventing telomeric transcription and avoiding conflicts between transcription and replication (19). Thus, it is likely that the lack of Bqt4 certainly exacerbates replicative stress at telomeres that may result in unreplicated DNA and in the entanglement of telomeres causing the death of telomerase minus cells. This is supported by the fact that a significant percentage of arrested cells with ‘cut’ phenotype is observed in bqt4Δ ter1Δ. This indicates that cells may undergo catastrophic mitosis with entangled telomeres leading to their death, as it was observed for taz1Δ cells when grown at low temperature (46). Taken together, these results suggest that nuclear periphery is the area of the nucleus that ensures faithful replication of telomeres. In budding yeast, eroded telomeres are targeted to the nuclear pore complex (47,48). Such mechanism remains to be uncovered in fission yeast.
In conclusion, in telomerase negative cells, either vegetative or arrested, Bqt4 preserves telomere integrity through anchoring telomeres to the nuclear periphery. In vegetative cells, defect in NE-position causes an abrupt loss of telomeres while in arrested cells it provokes a massive accumulation of STEEx that prevents cells to exit properly from quiescence.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Cooper, J. Kanoh, C. Azzalin, S. Braun, P. Nurse, T. Toda and JP. Javerzat for sharing strains.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale de la Recherche [ANR-QuiescenceDNA SVSE8 and ANR-16-CE12-0015 TeloMito]; l’Association de la Recherche sur le Cancer (ARC), fonctionnement, 2016; L.M. is supported by the ANR-TeloMito; S.C. is supported by ‘Projet Fondation ARC’ (Association pour la Recherche contre le Cancer) and Cancéropôle-PACA (Emergence); V.G. laboratory is supported by the ‘Ligue Nationale Contre le Cancer’ (LNCC) (Equipe labélisée). Funding for open access charge: Agence Nationale de la Recherche.
Conflict of interest statement. None declared.
REFERENCES
- 1. Arnoult N., Karlseder J.. Complex interactions between the DNA-damage response and mammalian telomeres. Nat. Struct. Mol. Biol. 2015; 22:859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nandakumar J., Cech T.R.. Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 2013; 14:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Verdun R.E., Karlseder J.. Replication and protection of telomeres. Nature. 2007; 447:924–931. [DOI] [PubMed] [Google Scholar]
- 4. Gilson E., Géli V.. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 2007; 8:825–838. [DOI] [PubMed] [Google Scholar]
- 5. de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005; 19:2100–2110. [DOI] [PubMed] [Google Scholar]
- 6. Ludérus M.E., van Steensel B., Chong L., Sibon O.C., Cremers F.F., de Lange T.. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J. Cell Biol. 1996; 135:867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaminker P.G., Kim S.-H., Desprez P.-Y., Campisi J.. A novel form of the telomere-associated protein TIN2 localizes to the nuclear matrix. Cell Cycle. 2009; 8:931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arnoult N., Schluth-Bolard C., Letessier A., Drascovic I., Bouarich-Bourimi R., Campisi J., Kim S.-H., Boussouar A., Ottaviani A., Magdinier F. et al.. Replication timing of human telomeres is chromosome arm-specific, influenced by subtelomeric structures and connected to nuclear localization. PLos Genet. 2010; 6:e1000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nishibuchi G., Déjardin J.. The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals. Chromosome Res. 2017; 25:77–87. [DOI] [PubMed] [Google Scholar]
- 10. Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J.. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007; 318:798–801. [DOI] [PubMed] [Google Scholar]
- 11. Schoeftner S., Blasco M.A.. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nature. 2007; 10:228–236. [DOI] [PubMed] [Google Scholar]
- 12. Steglich B., Sazer S., Ekwall K.. Transcriptional regulation at the yeast nuclear envelope. Nucleus. 2013; 4:379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Steglich B., Strålfors A., Khorosjutina O., Persson J., Smialowska A., Javerzat J.-P., Ekwall K.. The Fun30 chromatin remodeler Fft3 controls nuclear organization and chromatin structure of insulators and subtelomeres in fission yeast. PLos Genet. 2015; 11:e1005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Strålfors A., Walfridsson J., Bhuiyan H., Ekwall K.. The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLoS Genet. 2011; 7:e1001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu C., Inoue H., Sun W., Takeshita Y., Huang Y., Xu Y., Kanoh J., Chen Y.. The inner nuclear membrane protein Bqt4 in fission yeast contains a DNA-Binding domain essential for telomere association with the nuclear envelope. Structure. 2018; 27:335–343. [DOI] [PubMed] [Google Scholar]
- 16. Chikashige Y., Yamane M., Okamasa K., Tsutsumi C., Kojidani T., Sato M., Haraguchi T., Hiraoka Y.. Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J. Cell Biol. 2009; 187:413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fujita I., Nishihara Y., Tanaka M., Tsujii H., Chikashige Y., Watanabe Y., Saito M., Ishikawa F., Hiraoka Y., Kanoh J.. Telomere-nuclear envelope dissociation promoted by Rap1 phosphorylation ensures faithful chromosome segregation. Curr. Biol. 2012; 22:1932–1937. [DOI] [PubMed] [Google Scholar]
- 18. Funabiki H., Hagan I., Uzawa S., Yanagida M.. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 1993; 121:961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ebrahimi H., Masuda H., Jain D., Cooper J.P.. Distinct ‘safe zones’ at the nuclear envelope ensure robust replication of heterochromatic chromosome regions. eLife. 2018; 7:914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oza P., Jaspersen S.L., Miele A., Dekker J., Peterson C.L.. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009; 23:912–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schober H., Ferreira H., Kalck V., Gehlen L.R., Gasser S.M.. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009; 23:928–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maestroni L., Audry J., Matmati S., Arcangioli B., Géli V., Coulon S.. Eroded telomeres are rearranged in quiescent fission yeast cells through duplications of subtelomeric sequences. Nat. Commun. 2017; 8:1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maestroni L., Géli V., Coulon S.. STEEx, a boundary between the world of quiescence and the vegetative cycle. Curr. Genet. 2018; 17:3107–3105. [DOI] [PubMed] [Google Scholar]
- 24. Hentges P., Van Driessche B., Tafforeau L., Vandenhaute J., Carr A.M.. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 2005; 22:1013–1019. [DOI] [PubMed] [Google Scholar]
- 25. Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., Steever A.B., Wach A., Philippsen P., Pringle J.R.. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998; 14:943–951. [DOI] [PubMed] [Google Scholar]
- 26. Tournier S., Gachet Y., Buck V., Hyams J.S., Millar J.B.A.. Disruption of astral microtubule contact with the cell cortex activates a Bub1, Bub3, and Mad3-dependent checkpoint in fission yeast. Mol. Biol. Cell. 2004; 15:3345–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beauregard P.B., Guérin R., Turcotte C., Lindquist S., Rokeach L.A.. A nucleolar protein allows viability in the absence of the essential ER-residing molecular chaperone calnexin. J. Cell Sci. 2009; 122:1342–1351. [DOI] [PubMed] [Google Scholar]
- 28. Moreno S., Klar A., Nurse P.. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991; 194:795–823. [DOI] [PubMed] [Google Scholar]
- 29. Ben Hassine S., Arcangioli B.. Tdp1 protects against oxidative DNA damage in non-dividing fission yeast. EMBO J. 2009; 28:632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lock A., Rutherford K., Harris M.A., Hayles J., Oliver S.G., Bähler J., Wood V.. PomBase 2018: user-driven reimplementation of the fission yeast database provides rapid and intuitive access to diverse, interconnected information. Nucleic Acids Res. 2019; 47:D821–D827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wood V., Gwilliam R., Rajandream M.-A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S. et al.. The genome sequence of Schizosaccharomyces pombe. Nature. 2002; 415:871–880. [DOI] [PubMed] [Google Scholar]
- 32. Hediger F., Berthiau A.-S., van Houwe G., Gilson E., Gasser S.M.. Subtelomeric factors antagonize telomere anchoring and Tel1-independent telomere length regulation. EMBO J. 2006; 25:857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reyes C., Serrurier C., Gauthier T., Gachet Y., Tournier S.. Aurora B prevents chromosome arm separation defects by promoting telomere dispersion and disjunction. J. Cell Biol. 2015; 208:713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yanagida M. Cellular quiescence: are controlling genes conserved. Trends Cell Biol. 2009; 19:705–715. [DOI] [PubMed] [Google Scholar]
- 35. Laporte D., Courtout F., Tollis S., Sagot I.. Quiescent Saccharomyces cerevisiae forms telomere hyperclusters at the nuclear membrane vicinity through a multifaceted mechanism involving Esc1, the Sir complex, and chromatin condensation. Mol. Biol. Cell. 2016; 27:1875–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bah A., Wischnewski H., Shchepachev V., Azzalin C.M.. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 2012; 40:2995–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Swygert S.G., Kim S., Wu X., Fu T., Hsieh T.-H., Rando O.J., Eisenman R.N., Shendure J., McKnight J.N., Tsukiyama T.. Condensin-dependent chromatin compaction represses transcription globally during quiescence. Mol. Cell. 2019; 73:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guidi M., Ruault M., Marbouty M., Loïodice I., Cournac A., Billaudeau C., Hocher A., Mozziconacci J., Koszul R., Taddei A.. Spatial reorganization of telomeres in long-lived quiescent cells. Genome Biol. 2015; 16:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rutledge M.T., Russo M., Belton J.-M., Dekker J., Broach J.R.. The yeast genome undergoes significant topological reorganization in quiescence. Nucleic Acids Res. 2015; 43:8299–8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Laporte D., Sagot I.. Microtubules move the nucleus to quiescence. Nucleus. 2014; 5:113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bah A., Azzalin C.M.. The telomeric transcriptome: from fission yeast to mammals. Int. J. Biochem. Cell Biol. 2012; 44:1055–1059. [DOI] [PubMed] [Google Scholar]
- 42. Greenwood J., Cooper J.P.. Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res. 2012; 40:2956–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moravec M., Wischnewski H., Bah A., Hu Y., Liu N., Lafranchi L., King M.C., Azzalin C.M.. TERRA promotes telomerase‐mediated telomere elongation in Schizosaccharomyces pombe. EMBO Rep. 2016; 17:999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oya E., Durand-Dubief M., Cohen A., Maksimov V., Schurra C., Nakayama J.-I., Weisman R., Arcangioli B., Ekwall K.. Leo1 is essential for the dynamic regulation of heterochromatin and gene expression during cellular quiescence. Epigenet. Chromatin. 2019; 12:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Simon M.N., Churikov D., Géli V.. Replication stress as a source of telomere recombination during replicative senescence in Saccharomyces cerevisiae. FEMS Yeast Res. 2016; 16:fow085. [DOI] [PubMed] [Google Scholar]
- 46. Miller K.M., Cooper J.P.. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol. Cell. 2003; 11:303–313. [DOI] [PubMed] [Google Scholar]
- 47. Khadaroo B., Teixeira M.T., Luciano P., Eckert-Boulet N., Germann S.M., Simon M.N., Gallina I., Abdallah P., Gilson E., Géli V. et al.. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nature. 2009; 11:980–987. [DOI] [PubMed] [Google Scholar]
- 48. Churikov D., Charifi F., Eckert-Boulet N., Silva S., Simon M.N., Lisby M., Géli V.. SUMO-dependent relocalization of eroded telomeres to nuclear pore complexes controls telomere recombination. Cell Rep. 2016; 15:1242–1253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.