Abstract
Background:
Tuberculosis (TB) still remains endemic worldwide making epidemiological studies essential to mitigating efforts implicated in identifying its source, controlling, and preventing the spread of dangerous strains amongst humans such as Mycobacterium tuberculosis (Mtb).
Methods:
In this study, we sought to determine the 6 Mycobacterial Interspersed Repetitive Unit-Variable-Number Tandem Repeat (MIRU-VNTR) loci with high discriminatory powers for Mtb genotyping as well as the loci with the highest and the lowest discriminatory powers for MIRU-VNTR. To conduct our search, we used several databases such as science direct, Embase (Elsevier), Web of Science, Scopus and Medline via PubMed. Searches were performed using key words including: Mycobacterium tuberculosis, MIRU–VNTR, Allele diversity, Genetic diversity and human patient. Finally, 56 articles were selected after filtering out titles, abstracts and full texts.
Results:
Loci with high discriminatory powers included MIRU10 and MIRU26, while MIRU2, MIRU20, MIRU24 and ETRD had poor discriminatory powers. According to previous data in the literature, the loci MIRU10, MIRU26, MIRU40, QUB 26, QUB 11b and Mtub21 have high discriminatory powers.
Conclusion:
Therefore, these loci recommended for genotyping Mtb to save time and cost and to ensure the production of reliable results.
Key Words: Discriminatory power, Genotyping, MIRU-VNTR, Mycobacterium tuberculosis
Introduction
In spite of recent efforts to control and eliminate TB, this highly infectious disease still remains the second leading cause of death worldwide (1, 2). In 2018, WHO predicted that 10 million patients (ranging from 9 to 11.1 million) were stricken by TB. Mtb is a member of the TB complex which causes TB and can be transmitted via aerosolization of bodily fluids from coughing, sneezing or speaking (2).
To effectively control TB, preventive policies based on transmission routes are required. Molecular epidemiology of Mtb involves monitoring special strains like multi-drug-resistant (MDR) Mtb during periods of increasing prevalence, inspecting regions of latest and potential outbreaks, locating the source and route of transmission and transmission gene sequence, discovering hidden strains and tracking circulating immigrant strains (3).
Various molecular methods are used in epidemiological studies about Mtb strains. Each method has a particular specificity and sensitivity. Some of these approaches include IS6110, IS6110-restriction fragment length polymorphism (RFLP), MIRU-VNTR, Spoligotyping, whole genome sequencing (WGS), Random Amplification of Polymorphic DNA PCR (RAPD_PCR), Repetitive element sequence-based PCR (rep-PCR), Pulsed-field gel electrophoresis (PFGE), Next Generation Sequencing (NGS) and finally, a combination of two or more techniques listed above will be applied. Among current typing methods, a test has to be chosen according to its feasibility, cost-benefit and discriminatory powers (4-7)
In a survey comparing PFGE, 24 locus MIRU-VNTR and IS6110-RFLP, it was revealed that the 24 locus MIRU-VNTR method was the preferred method due to a high power of discrimination and time management during epidemiological investigations (7).
This study was performed to review published applications of the Mycobacterial Interspersed Repetitive Unit-Variable-Number Tandem Repeat (MIRU-VNTR) method in Mtb genotyping and to introduce the best 6 loci for MIRU-VNTR in typing Mtb isolated from human patients along with determining the loci with highest and lowest discriminatory powers for MIRU- VNTR.
Materials and Methods
To identify the best loci of the 6 loci in MIRU-VNTR method for Mtb genotyping, the literature search was performed using several databases including science direct, Embase (Elsevier), Web of Science, Scopus, ISC and Medline via PubMed. Chosen keywords were: Mtb, MIRU–VNTR, Allele diversity, genetic diversity and human patient. Inclusion and exclusion criteria were determined by the following:
1. Isolation of Mtb from human patients.
2. Investigating the genetic diversity among Mtb just based on MIRU-VNTR.
3. Excluding the studies where lineage determination was based on MIRU-VNTR where Allele diversity “h” was not measured for each locus.
4- Excluding the studies in which MIRU-VNTR ability in cluster analysis and Hunter-Gaston discriminatory index (HGDI) for this method was investigated and h was not estimated for each of their locus.
Several articles were excluded from our study since allele diversity (h) was not mentioned for each locus. Screening the articles was done in 3 steps: 1. Title screening, 2. Abstract evaluation, 3. Full text evaluation based on these criteria.
Results
A total number of 228 articles were found collectively amongst the databases. As the title screening was performed, 90 articles were removed. Abstract screening resulted in 82 more studies to be omitted during the search. Finally, after full text screening, 56 articles remained. In the remaining articles, genotyping for Mtb using MIRU-VNTR was investigated from 2002 to 2019. Allele diversity (h) was evaluated amongst the 56 articles for each locus separately (Table 1).
Table 1.
Fifty-six studies after full-text evaluation that MIRU-VNTR were used.
| No. | Authors reference | year | Geographical region | Continent | Locus of MIRU-VNTR | Numbers of locus | High power | Low power |
|---|---|---|---|---|---|---|---|---|
| 1 | Cowan et al (8) | 2002 | United States (Michigan) | America | ETR D,E MIRU2,10,16,20,23,24,26,27,39,40 | 12 | MIRU40 | MIRU2,20 |
| 2 | Sola et al (9) | 2003 | Different regions such as; USA, Thailand, Sicily, Guadeloupe and Russia | ETR A-E MIRU2,10,16,20,23,24,26,27,39,40 | 15 | ETRA, MIRU40 ,26,10 | MIRU2,20, 27 | |
| 3 | Sun et al (10) | 2004 | Singapore | Asia | ETR D,E MIRU2,10,16,20,23,24,26,27,39,40 | 12 | MIRU26,10, ETRE | MIRU2,20 |
| 4 | Kremer et al (11) | 2005 | China | Asia | ETR A-E MIRU10,16,26,39,40 QUB 11a,11b,26,1895 | 14 | QUB 11b,11a MIRU10 | MIRU16 ETRC |
| 5 | Kovalev et al (12) | 2005 | Russian Federation | Asia | ETR D,E MIRU2,10,16,20,23,24,26,27,39,40 | 12 | MIRU26, ETRE | MIRU24,27 |
| 6 | Asgarzade et al (13) | 2007 | Iran | Asia | ETR D,E MIRU2,10,16,20,23,24,26,27,39,40 | 12 | MIRU26, 40 | MIRU16,39 |
| 7 | Çavuşoğlu et al (14) | 2007 | Turkey | Asia | ETR D,E MIRU2,10,16,20,23,24,26,27,39,40 | 12 | MIRU16,40,26 | MIRU24,27 |
| 8 | Maes et al (15) | 2008 | Venezuela | America | ETR A-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26,4156 | 24 | MIRU40,26, ETRB | MIRU20,2, 24 |
| 9 | Alonso-Rodríguez et al (16) | 2008 | Spain | Europe | ETRA,C-E MIRU10,16,26,40 Mtub4,21,30,39 QUB 11b,26,4156 | 15 | QUB, 26,11b, MIRU40, 10 | ETRD |
| 10 | Yun et al (17) | 2009 | Korea | Asia | ETRA-F MIRU2,10,16,20,23,24,26,27,39,40 | 6 | MIRU26, ETRE,F | MIRU24,20 ETRD |
| 11 | Stavrum et al (18) | 2009 | South Africa | Africa | ETRA,C-E MIRU10,16,26,40 Mtub4,21,30,39 QUB 11b,26,4156 | 15 | QUB 11b | ETRD |
| 12 | Shamputa et al (19) | 2010 | South Korea | Asia | ETR A-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26,4156 | 24 | QUB 11b,26, Mtub4 | MIRU2,24 ,23 ETRB,D |
| 13 | Noguti et al (20) | 2010 | Brazil | America | ETR D,E MIRU2,10,16,20,23,24,26,27,39,40 | 12 | MIRU40,23,10 | MIRU24,39 ETRD |
| 14 | Jafarian et al (21) | 2010 | Iran | Asia | ETR D,E MIRU2,10,16,20,23,24,26,27,39,40 | 12 | MIRU 26, 10,16 | MIRU2, 24 ETRD |
| 15 | Zhang et al (22) | 2011 | Cambodia | Asia | ETR A-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26,4156 | 24 | ETRD,Mtub 39, QUB26 | Mtub 34, MIRU2,20 |
| 16 | Asgarzad et al (23) | 2011 | Iran | Asia | ETR A-E MIRU2,10,16,20,23,24,26,27,39,40 | 15 | MIRU10,26,40 | MIRU39,2,4 |
| 17 | Bidovec-Stojkovi et al (24) | 2011 | Slovenia | Europe | ETR A-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26,4156 | 24 | QUB 26, 11b, MIRU40 ,10 | MIRU24,39 |
| 18 | Cerezo et al (25) | 2012 | Colombia | America | ETR D,E MIRU2,10,16,20,23,24,26,27,39,40 | 12 | MIRU10, 40 | MIRU24,39 |
| 19 | Chatterjee et al (26) | 2013 | India | Asia | ETR D,E MIRU2,10,16,20,23,24,26,27,39,40 | 12 | MIRU26, 10 | MIRU2,20 |
| 20 | Zamani et al (27) | 2013 | Iran | Asia | ETRA,C-E MIRU10,16,26,40 Mtub4,21,30,39 QUB 11b,26,4156 | 15 | MIRU16 ETRA | MIRU26, Mtub 21, 30, 39 QUB4156 |
| 21 | Joseph et al (28) | 2013 | India | Asia | ETRA,C-E MIRU10,16,26,40 Mtub4,21,30,39 QUB 11b,26,4156 | 15 | ETRA,B MIRU40 | MIRU2,20 |
| 21 | Yasmin et al (29) | 2014 | Pakistan | Asia | ETR A-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26,4156 | 24 | MIRU26 QUB26 MIRU10 | MIRU2,20,27,24 |
| 23 | Ali et al (30) | 2014 | Pakistan | Asia | ETRA,C-E MIRU10,16,26,40 Mtub4,21,30,39 QUB 11b,26,4156 | 15 | QUb26 ,MIRU10,26 | ETRD |
| 24 | Chaoui et al (31) | 2014 | Morocco | Africa | ETR D,E MIRU2,10,16,20,23,24,26,27,39,40 | 12 | MIRU40,23,10 | MIRU24,39 |
| 25 | Zheng et al (32) | 2014 | China | Asia | ETR A-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26,4156 | 24 | QUB11b,26 Mtub 21 MIRU26 | MIRU24 Mtub 34 |
| 26 | Vasconcellos et al (33) | 2014 | Brazil | America | ETR A-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26,4156 | 24 | QUB 4156, 11b MIRU10 | MIRU24,39 |
| 27 | Rindi et al (34) | 2014 | Italy | Europe | ETRA,D,E MIRU10,16,26,40 Mtub21 QUB 11b, 26 VNTR 42,43,47,52,53 | 15 | QUB 26 MIRU 40 QUB 11b | MIRU 04 ETRE |
| 28 | Bouklata et al (35) | 2015 | Morocco | Africa | ETR A-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26,4156 | 24 | QUB 26, RU40,26 | MIRU20,27 |
| 29 | Devi et al (36) | 2015 | India | Asia | ETR A-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26,4156 | 24 | Qub26, 11b MIRU10 QUB 4156 MIRU26 Mtub 21 | MIRU2,20, 27 |
| 30 | Zamani et al (37) | 2016 | Iran | Asia | ETRA,C-E MIRU10,16,26,40 Mtub4,21,30,39 QUB 11b,26,4156 | 15 | MIRU 40 | MIRU 10 QUB 4156 |
| 31 | Hoza et al (38) | 2016 | Tanzania | Africa | ETRA,C-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,39 QUB 11b,26,4156 | 22 | MIRU26,10,16 ETRAE QUB 26, 4156 | MIRU27,2, 20 |
| 32 | Cheng et al (39) | 2016 | China | Asia | ETR A-E MIRU10,16,20,26,27,39,40 Mtub31 QUB 11a,11b | 15 | QUB11a | ETRB,C |
| 33 | Bhembe et al (40) | 2017 | South Africa (Eastern Cape) | Africa | ETR D,E MIRU2,10,16,20,23,24,26,27,39,40 | 12 | ETRE, MIRU27,24 | MIRU40 |
| 34 | Zhang et al (41) | 2017 | China | Asia | ETR A-E MIRU 10,16,23,26,27,39,40 Mtub21,30,39 | 15 | Mtub 21, MIRU26,10 | ETRC,B |
| 35 | Liu et al (42) | 2017 | China | Asia | ETRA-E MIRU10,16,23,26,27,39,40 Mtub21,30,39 | 15 | MIRU26 Mtub 21 | MIRU27,23 ETRD |
| 36 | Pasechnik et al (43) | 2017 | West Siberia | Asia | ETRA-D MIRU2,10,16,20,23,24,26,27,39,40 | 15 | MIRU26 | MIRU24 ETRB |
| 37 | Pan et al (44) | 2017 | China | Asia | ETRA,D,E MIRU10,16,26,39,40 Mtub4,21,24,30,39 QUB11a,11b,18,26,3232,1895,4156 VNTR3820, 4120 | 22 | VNTR3820 QUB3232 | QUB4156 Mtub 24 |
| 38 | Khosravi et al (45) | 2017 | Iran | Asia | ETR D,E MIRU2,10,16,20,23,24,26,27,39,40 | 12 | MIRU 10, 26 | MIRU2,20 |
| 39 | Ravansalar et al (46) | 2017 | Iran | Asia | ETRA-F MIRU10,16,26,39,40 QUB 11b | 12 | MIRU10, 26 ETRF | ETRD |
| 40 | Shah et al (47) | 2017 | Nepal | Asia | ETRA-F MIRU2,10,16,20,23,24,26,27,39,40 Mtub21,30,39 QUB 11b,11b, 26, 4156 | 24 | QUB 26 MIRU10 | ETR B,C |
| 41 | Rasoaha et al (48) | 2017 | Madagascar | Africa | ETRA,C-E MIRU10,16,26,40 Mtub4,21,30,39 QUB 11b,26,4156 | 15 | MIRU26 QUB 11b Mtub 21 QUB 26 | ETRD,C Mtub4 |
| 42 | Chen et al (49) | 2017 | Asian countries (Cambodia, Singapore and Taiwan) | Asia | ETRA-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26, 4156 | 24 | Mtub 21 QUB 11b | MIRU2 QUB 4156 |
| 43 | Li et al (50) | 2018 | China | Asia | ETRA-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26, 4156 | 24 | Mtub4 MIRU40,10 | Mtub21 MIRU27 |
| 44 | Xu et al (51) | 2018 | China | Asia | ETRA,C-E MIRU10,16,26,40 Mtub4,21,30,39 QUB 11b,26,4156 | 15 | QUB 11b Mtub21 MIRU26 | ETRC MIRU16 QUB 4156 |
| 45 | Esteves et al (52) | 2018 | Brazil | America | ETRA-E MIRU2,10,16,20,23,24,26,27,39,40 QUB11,26 VNTR42,1955,47,52,53,49 | 24 | QUB26 QUB11 VNTR42 | MIRU39,24 ETRD |
| 46 | Augusto et al (6) | 2018 | Brazil | America | ETRD,E MIRU2,10,16,20,23,24,26,27,39,40 | 12 | MIRU16, 10,26 | MIRU20,24 |
| 47 | Riyahi Zaniani et al (53) | 2018 | Iran | Asia | ETRA-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26, 4156 | 24 | MIRU 10 QUB 26 Mtub 4,34 | ETRD, MIRU 20, Mtub 29 |
| 48 | Azimi et al (54) | 2018 | Iran | Asia | ETRA,C-E MIRU10,16,26,40 Mtub4,21,30,39 QUB11b,26,4156 | 15 | MIRU26, 10 Mtub 21 QUB 26 | ETR D,E |
| 49 | Lil et al (3) | 2018 | China | Asia | ETRA-E MIRU16,23,26,27,39,40 Mtub21,30,39 | 15 | ETRE, MIRU10, 26,39,40 Mtub 21 | ETRB, C MIRU16,23 |
| 50 | Mansoori et al (55) | 2018 | Iran | Asia | ETRA-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26, 4156 | 24 | MIRU 10, 16, 26, | MIRU 2, 20 ETRD |
| 51 | Chawla et al (56) | 2018 | India | Asia | ETRD,E MIRU2,10,16,20,23,24,26,27,39 40 | 12 | MIRU 39,10,26 | MIRU 2,20 |
| 52 | Shi et al (57) | 2018 | China | Asia | ETRA-F MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,38,39 QUB 11b,26, 4156 | 26 | QUB 11b Mtub 21 MIRU26 | MIRU24,2, 20 |
| 53 | Ei et al (58) | 2018 | Myanmar | Asia | ETRA-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26, 4156 | 24 | QUB 26 QUB 11b MIRU26 | Mtu34 MIRU20 |
| 54 | Weerasekera et al (59) | 2019 | Sri Lanka | Asia | ETRA-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26, 4156 | 24 | Mtub 21,39 QUB 11b | MIRU20,2 ,16 |
| 55 | Zarzour et al (60) | 2019 | Syria | Asia | ETRA-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26, 4156 | 24 | QUB 26 MIRU10,26 | MIRU24,20, Mtub 29 |
| 56 | Li et al (61) | 2019 | China | Asia | ETRA-E MIRU2,10,16,20,23,24,26,27,39,40 Mtub4,21,29,30,34,39 QUB 11b,26, 4156 | 24 | QUB11b MIRU 26 Mtub21 | MIRU2, 20,24 |
Mycobacterium tuberculosis genotyping was performed on 56 studies using the MIRU-VNTR technique; 39 of which were conducted in Asia, seven in America, six in Africa, three in Europe and one in a different country. The location and number of studies are shown in Figure 1. Each individual study employed a different number of loci. The results revealed that MIRU10 and MIRU26 had the highest discriminatory powers while MIRU2, MIRU20, MIRU24 and ETRD had the lowest discriminatory powers, respectively.
Figure 1.
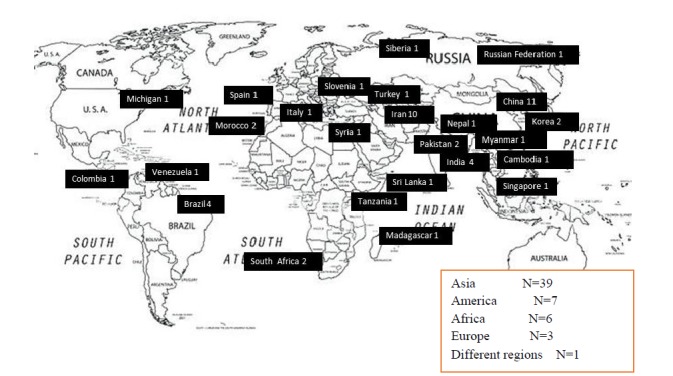
Location and numbers of studies was shown in the world from 2002 to 2019. Two studies were performed in different regions: Studies No. 2 and 42
Table 2 shows both the number of studies in which MIRU10, MIRU26, QUB26, MIRU40, QUB11b and Mtub21 was reported to be the loci with the highest discriminatory powers (h>0.6), including the range of h for the remaining loci.
Table 2.
Six MIRU-VNTR loci for Mycobacterium tuberculosis based on the number of studies and the reported allele diversity (h) range for each locus.
| locus | Number of studies | h range |
|---|---|---|
| MIRU10 | 28 | 0.61 to 0.82 |
| MIRU26 | 32 | 0.61 to 0.81 |
| QUB26 | 18 | 0.604 to 0.89 |
| MIRU40 | 17 | 0.604 to 0.76 |
| QUB11b | 17 | 0.72 to 0.84 |
| Mtub21 | 12 | 0.64 to 0.83 |
MIRU2 and MIRU20 (each in 21 studies), MIRU24 (17 studies) and ETRD (13 studies) were suggested as the loci with the lowest discriminatory powers (h<0.3).
Discussion
An alarming 62% of emerging TB cases occurred in the South-East Asia and Western Pacific regions, followed by Africa, which accounts for 25% of new cases. Countries such as India, China, Indonesia, the Philippines, Pakistan, Bangladesh, Nigeria and South Africa hold a prevalence rate greater than 60% for TB. It is believed that the surge in TB research in Asia could be that TB is highly prevalent in this continent based on 2018 WHO report (2). Of the 56 studies extracted, 39 studies were conducted within Asia. More specifically, 11 were performed in China, ten in Iran, four in India, two in Pakistan, two in Korea and only one in Nepal, Singapore, Myanmar, Turkey, Cambodia, Sri Lanka, Syria, Russia, Siberia, Taiwan, and Cambodia. Among the 39 studies conducted in Asia, MIRU26 and MIRU10 was reported having high discriminatory power loci. Among the 7 studies conducted in the Western Hemisphere, four studies were carried out in Brazil, and one study was performed in Michigan, Venezuela and Colombia. Both MIRU40 and MIRU10 had high power discriminatory power loci in these ancients.
Among the six studies performed in Africa, four studies were conducted in South Africa and Morocco, respectively. One study, performed in Madagascar and Tanzania, reported that QUB26 and MIRU26 had high discriminatory power loci in this continent. Several investigations in this study used PCR-based techniques such as spacer oligonucleotide typing (spoligotyping) and mycobacterial interspersed repetitive units–variable-number of tandem repeats (MIRU-VNTR) analyses. Although spoligotyping benefits from genetic diversity, it can minimize Mtb clonal diversity. This method uses one direct repeat (DR) which incorporates identical alternate and variable spacers and the results are represented as a single digit pattern. The basis of MIRU-VNTR depends on the (variable) number of tandem repeat elements called mycobacterial interspersed repetitive units (MIRU). The results are represented as a code, and offers a low discriminatory power when used alone. The MIRU-VNTR method analyzes DNA segments which involves tandem repeat sequences and the copy number which differs amongst various strains. This method depends on PCR efficiency, specifically the quantity of repeats which is based on the size of the amplified product. The results are illustrated as characters that range from 15 –24 characters, in which each character represents the number of repeats at a single locus (3, 62).
These results are evaluated in comparison to a strain database on the web-based tool MIRU-VNTR plus. Therefore, MIRU-VNTR typing is considered the gold-standard for genotypic analysis of Mtb (7, 9). The discriminatory power of these methods was found to be different which was determined by the Hunter and Gaston Index factor. Several studies reported different Hunter-Gaston Index for MIRU-VNTR ranging from 0.951 to 0.999 (10, 13, 45, 47, 48, 53-56).
The allele diversity index (h) is used to describe the discriminatory power of MIRU-VNTR loci. If the index is greater than 0.6 (h>0.6), the discriminatory power of the locus is high. If the index lies between 0.3 and 0.6 (0.3<h <0.6), the locus has medium discriminatory power, however, if the index is less than 0.3 (0.3>h), the discriminatory power is considered weak. Our results suggest that among the 24 defined loci for MIRU-VNTR introduced by the MIRU-VNTR plus database, the MIRU26, MIRU10, MIRU40, QUB26, QUB11b and Mtub21 loci had the highest discriminatory powers, in contrast to the MIRU2, MIRU20, MIRU24 and ETRD loci which yielded low discriminatory powers.
Mycobacterium bovis (M. bovis) is the causative agent of TB in humans. When comparing the discriminatory powers of loci between Mtb and M. bovis, the loci QUB 11b and QUB 3232 have the highest discriminatory powers (for M. bovis), whereas ETRD (for both strains) and MIRU10 (for M. bovis) had the lowest discriminatory powers. Therefore, it can be concluded from our study that QUB 11b in both Mtb and M. bovis has a high discriminatory power while ETRD for both strains is considered a low power locus. However, a difference exists between the two strains in the MIRU10 locus which has a high discriminatory power in Mtb but low in M. bovis (63).
While the 24 loci MIRU-VNTR has been introduced as the best typing method for Mtb based on multiple comparisons between various molecular techniques, it is, therefore, highly recommended to include the following loci, QUB26, QUB11b, MIRU10, MIRU26, MIRU40 and Mtub21, provided that MIRU-VNTR is done with less than 24 loci, in order to obtain the best results when genotyping Mtb isolates. In this regard, data extraction becomes inexpensive and time efficient.
Among typing methods, MIRU-VNTR is considered to be one of the best. Further, the 6 loci including MIRU10, MIRU26, MIRU40, QUB 26, QUB 11b and Mtub21, all of which have high discriminatory powers, are recommended in Mtb genotyping to save time and cost. Indeed, epidemiological studies are critical for disease surveillance of TB as they enable us to track circulating strains on a global scale, as well as for identifying important risk factors necessary for implementing control measures in vulnerable populations worldwide.
Acknowledgements
We thank Antimicrobial Resistance Research Center and Mashhad University of Medical Sciences The authors of this study declare no conflicts of interest.
References
- 1.Kaufmann SH, Schaible UE. 100th anniversary of Robert Koch's Nobel Prize for the discovery of the tubercle bacillus. Trends in microbiology. . 2005;13(10):469–75. doi: 10.1016/j.tim.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Global tuberculosis report 2018. World Health Organization Geneva. 2018.
- 3.Lil B, Ma Y, Liu H, Shenl X, Ma B, Jiang M, et al. Molecular typing of Mycobacterium tuberculosis isolates from Qinghai province of northwest china by spoligotyping and 15-locus MIRUVNTR. . Journal of Microbiology and Biotechnology Reports. . 2018;2(1):1–4. [Google Scholar]
- 4.Ravansalar H, Tadayon K, Ghazvini K. Molecular typing methods used in studies of Mycobacterium tuberculosis in Iran: a systematic review. . Iranian journal of microbiology. 2016;8(5):338. [PMC free article] [PubMed] [Google Scholar]
- 5.Ei PW, Aung WW, Lee JS, Choi G-E, Chang CL. Molecular strain typing of Mycobacterium tuberculosis: a review of frequently used methods. . Journal of Korean medical science. 2016;31(11):1673, 83. doi: 10.3346/jkms.2016.31.11.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Augusto CJ, Carvalho WdS, Almeida INd, Figueiredo LJdA, Dantas NGT, Suffys PN, et al. Comparative study of RFLP-IS6110 and MIRU-VNTR from Mycobacterium tuberculosis isolated in the state of Minas Gerais, Brazil. brazilian journal of microbiology. 2018;49(3):641–6. doi: 10.1016/j.bjm.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeon S, Lim N, Park S, Park M, Kim S. Comparison of PFGE, IS6110-RFLP, and 24-Locus MIRU-VNTR for Molecular Epidemiologic Typing of Mycobacterium tuberculosis Isolates with Known Epidemic Connections. Journal of microbiology and biotechnology. 2018;28(2):338–46. doi: 10.4014/jmb.1704.04042. [DOI] [PubMed] [Google Scholar]
- 8.Cowan LS, Mosher L, Diem L, Massey JP, Crawford JT. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. Journal of clinical microbiology. 2002;40(5):1592–602. doi: 10.1128/JCM.40.5.1592-1602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sola C, Filliol I, Legrand E, Lesjean S, Locht C, Supply P, et al. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infection, Genetics and Evolution. 2003;3(2):125–33. doi: 10.1016/s1567-1348(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 10.Sun Y-J, Bellamy R, Lee AS, Ng ST, Ravindran S, Wong S-Y, et al. Use of mycobacterial interspersed repetitive unit-variable-number tandem repeat typing to examine genetic diversity of Mycobacterium tuberculosis in Singapore. Journal of clinical microbiology. 2004;42(5):1986–93. doi: 10.1128/JCM.42.5.1986-1993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kremer K, Arnold C, Cataldi A, Gutiérrez MC, Haas WH, Panaiotov S, et al. Discriminatory power and reproducibility of novel DNA typing methods for Mycobacterium tuberculosis complex strains. Journal of clinical microbiology. 2005;43(11):5628–38. doi: 10.1128/JCM.43.11.5628-5638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovalev S, Kamaev E, Kravchenko M, Kurepina N, Skorniakov S. Genetic analysis of Mycobacterium tuberculosis strains isolated in Ural region, Russian Federation, by MIRU-VNTR genotyping. The International Journal of Tuberculosis and Lung Disease. 2005;9(7):746–52. [PubMed] [Google Scholar]
- 13.Asgharzadeh M, Khakpour M, Salehi TZ, Kafil HS. Use of mycobacterial interspersed repetitive unit-variable-number tandem repeat typing to study Mycobacterium tuberculosis isolates from East Azarbaijan province of Iran. . Pakistan journal of biological sciences. 2007;10(21):3769–77. doi: 10.3923/pjbs.2007.3769.3777. [DOI] [PubMed] [Google Scholar]
- 14.Cavuşoğlu C, Karataş E, Soeyler I. Genotyping of rifampin-resistant Mycobacterium tuberculosis isolates by mycobacterial interspersed repetitive unit-variable number tandem repeat (MIRU-VNTR) analysis. Mikrobiyoloji bulteni. 2007;41(3):385–93. [PubMed] [Google Scholar]
- 15.Maes M, Kremer K, van Soolingen D, Takiff H, de waeerd JH. 24-locus MIRU-VNTR genotyping is a useful tool to study the molecular epidemiology of tuberculosis among Warao Amerindians in Venezuela. Tuberculosis (Edinburgh, Scotland). . 2008;88(5):490–4. doi: 10.1016/j.tube.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Alonso-Rodríguez N, Martínez-Lirola M, Herránz M, Sanchez-Benitez M, Barroso P, Bouza E, et al. Evaluation of the new advanced 15-loci MIRU-VNTR genotyping tool in Mycobacterium tuberculosis molecular epidemiology studies. BMC microbiology. 2008;8(34):1–9. doi: 10.1186/1471-2180-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yun KW, Song EJ, Choi GE, Hwang IK, Lee EY, Chang CL. Strain typing of Mycobacterium tuberculosis isolates from Korea by mycobacterial interspersed repetitive units-variable number of tandem repeats. The Korean journal of laboratory medicine. 2009;29(4):314–9. doi: 10.3343/kjlm.2009.29.4.314. [DOI] [PubMed] [Google Scholar]
- 18.Stavrum R, Mphahlele M, Øvreås K, Muthivhi T, Fourie PB, Weyer K. High diversity of Mycobacterium tuberculosis genotypes in South Africa and preponderance of mixed infections among ST53 isolates. Journal of clinical microbiology. 2009;47(6):1848–56. doi: 10.1128/JCM.02167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shamputa IC, Lee J, Allix-Béguec C, Cho E-J, Rajan V, Lee EG, et al. Genetic diversity of Mycobacterium tuberculosis isolates from a tertiary care tuberculosis hospital in South Korea. Journal of clinical microbiology. 2010;48(2):387–94. doi: 10.1128/JCM.02167-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noguti EN, Leite CQF, Malaspina AC, Santos ACB, Hirata RDC, Hirata MH, et al. Genotyping of Mycobacterium tuberculosis isolates from alow-endemic setting in northwestern state of Paraná in Southern Brazil. Memorias do Instituto Oswaldo Cruz. 2010;105(6):779–85. doi: 10.1590/s0074-02762010000600008. [DOI] [PubMed] [Google Scholar]
- 21.Jafarian M, Aghali-Merza M, Farnia P, Ahmadi M, Masjedi MR, Velayati AA. Synchronous comparison of Mycobacterium tuberculosis epidemiology strains by “MIRU-VNTR” and “MIRU-VNTR and spoligotyping” technique. . Avicenna journal of medical biotechnology. . 2010;2(3):145–52. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Heng S, Le Moullec S, Refregier G, Gicquel B, Sola C. A first assessment of the genetic diversity of Mycobacterium tuberculosis complex in Cambodia. . BMC infectious diseases. 2011;11(42):1–7. doi: 10.1186/1471-2334-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asgharzadeh M, Kafil HS, Roudsary AA, Hanifi GR. Tuberculosis transmission in Northwest of Iran: using MIRU-VNTR, ETR-VNTR and IS6110-RFLP methods. . Infection, Genetics and Evolution. 2011;11(1):124–31. doi: 10.1016/j.meegid.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Bidovec-Stojkovic U, Zolnir-Dovc M, Supply P. One year nationwide evaluation of 24-locus MIRU-VNTR genotyping on Slovenian Mycobacterium tuberculosis isolates. Respiratory medicine. 2011;105:S67–S73. doi: 10.1016/S0954-6111(11)70014-2. [DOI] [PubMed] [Google Scholar]
- 25.Cerezo I, Jiménez Y, Hernandez J, Zozio T, Murcia MI, Rastogi N. A first insight on the population structure of Mycobacterium tuberculosis complex as studied by spoligotyping and MIRU-VNTRs in Bogotá, Colombia. Infection, Genetics and Evolution. 2012;12(4):657–63. doi: 10.1016/j.meegid.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Chatterjee A, Mistry N. MIRU–VNTR profiles of three major Mycobacterium tuberculosis spoligotypes found in western India. Tuberculosis. 2013;93(2):250–6. doi: 10.1016/j.tube.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Zamani S, Aflaki M, Fooladi AAI, Darban-Sarokhalil D, Bameri Z, Khazaee S, et al. MIRU-VNTR analysis of the Mycobacterium tuberculosis isolates from three provinces of Iran. . Scandinavian journal of infectious diseases. 2013;45(2):124–30. doi: 10.3109/00365548.2012.717233. [DOI] [PubMed] [Google Scholar]
- 28.Joseph BV, Soman S, Hill V, Rastogi N, Mundayoor S. Efficient discrimination by MIRU-VNTRs of Mycobacterium tuberculosis clinical isolates belonging to the predominant SIT11/EAI3-IND ancestral genotypic lineage in Kerala, India. . International journal of mycobacteriology. 2013;2(4):244–7. doi: 10.1016/j.ijmyco.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Yasmin M, Gomgnimbou MK, Siddiqui RT, Refrégier G, Sola C. Multi-drug resistant Mycobacterium tuberculosis complex genetic diversity and clues on recent transmission in Punjab, Pakistan. Infection, Genetics and Evolution. 2014;27:6–14. doi: 10.1016/j.meegid.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 30.Ali A, Hasan Z, Jafri S, Inayat R, Hasan R. Mycobacterium tuberculosis Central Asian Strain (CAS) lineage strains in Pakistan reveal lower diversity of MIRU loci than other strains. International journal of mycobacteriology. 2014;3(2):108–16. doi: 10.1016/j.ijmyco.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Chaoui I, Zozio T, Lahlou O, Sabouni R, Abid M, El Aouad R, et al. Contribution of spoligotyping and MIRU-VNTRs to characterize prevalent Mycobacterium tuberculosis genotypes infecting tuberculosis patients in Morocco. . Infection, Genetics and Evolution. 2014;21(463):71. doi: 10.1016/j.meegid.2013.05.023. [DOI] [PubMed] [Google Scholar]
- 32.Zheng C, Zhao Y, Zhu G, Li S, Sun H, Feng Q, et al. Suitability of IS6110-RFLP and MIRU-VNTR for differentiating spoligotyped drug-resistant Mycobacterium tuberculosis clinical isolates from Sichuan in China. BioMed research international. . 2014;2014:1–13. doi: 10.1155/2014/763204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vasconcellos S, Acosta C, Gomes L, Conceiçao E, Lima K. Strain Classification of Mycobacterium tuberculosis Isolates in Brazil Based on Genotypes Obtained by Spoligotyping, Mycobacterial Interspersed Repetitive Unit Typing and the Presence of Large Sequence and Single Nucleotide Polymorphism. PloS one. 2014;9(10):e107747. doi: 10.1371/journal.pone.0107747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rindi L, Medici C, Bimbi N, Buzzigoli A, Lari N, Garzelli C. Genomic Variability of Mycobacterium tuberculosis Strains of the Euro-American Lineage Based on Large Sequence Deletions and 15-Locus MIRU-VNTR Polymorphism. PloS one. 2014;9(11):e114676. doi: 10.1371/journal.pone.0107150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouklata N, Supply P, Jaouhari S, Charof R, Seghrouchni F, Sadki K ea. Molecular Typing of Mycobacterium tuberculosis complex by 24-locus based MIRU-VNTR typing in conjunction with spoligotyping to assess genetic diversity of strains circulating in Morocco. PloS one. 2015;10(8):e0135695. doi: 10.1371/journal.pone.0135695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devi KR, Bhutia R, Bhowmick S, Mukherjee K, Mahanta J, Narain K. Genetic diversity of Mycobacterium tuberculosis isolates from Assam, India: dominance of Beijing family and discovery of two new clades related to CAS1_Delhi and EAI family based on spoligotyping and MIRU-VNTR typing. PloS one. 2015;10(12):e0145860. doi: 10.1371/journal.pone.0145860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zamani S, Haeili M, Nasiri MJ, Fooladi I, Ali A, Javadpour S, et al. Genotyping of Mycobacterium tuberculosis isolates from Hormozgan province of Iran based on 15-locus MIRU-VNTR and spoligotyping. International journal of bacteriology. . 2016;2016:1–8. doi: 10.1155/2016/7146470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoza A, Mfinanga S, Moser I, König B. Molecular characterization of Mycobacterium tuberculosis isolates from Tanga, Tanzania: First insight of MIRU-VNTR and microarray-based spoligotyping in a high burden country. . Tuberculosis. 2016;98:116–24. doi: 10.1016/j.tube.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Cheng X-f, Jiang C, Zhang M, Xia D, Chu L-l, Wen Y-f, et al. Mycobacterial interspersed repetitive unit can predict drug resistance of Mycobacterium tuberculosis in China. Frontiers in Microbiology. 2016;7:378. doi: 10.3389/fmicb.2016.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhembe NL, Nwodo UU, Okoh AI, Obi CL, Mabinya LV, Green E. Clonality and genetic profiles of drug-resistant Mycobacterium tuberculosis in the Eastern Cape Province, South Africa. MicrobiologyOpen. 2017:1–10. doi: 10.1002/mbo3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Huang H, Liu C, Jia T, Zhang L, Zhou D, et al. Genotyping and drug-resistance epidemiology of mycobacterium tuberculosis in Xuzhou, China. International Journal of Clinical and Experimental Pathology. . 2017;10(9):9675–86. [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Li J, Liu J, Zhao X, Lian L, Liu H, et al. Genotypic diversity of mycobacterium tuberculosis clinical isolates in the multiethnic area of the Xinjiang Uygur autonomous region in China. BioMed research international. 2017;2017:1–8. doi: 10.1155/2017/3179535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasechnik O, Dymova MA, Stasenko VL, Blokh AI, Tatarintseva MP, Kolesnikova LP, et al. Molecular & genetic characteristics of Mycobacterium tuberculosis strains circulating in the southern part of West Siberia. The Indian journal of medical research. 2017;146:49–55. doi: 10.4103/ijmr.IJMR_162_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan X-L, Zhang C-L, Nakajima C, Fu J, Shao C-X, Zhao L-N, et al. A quantitative and efficient approach to select MIRU–VNTR loci based on accumulation of the percentage differences of strains for discriminating divergent Mycobacterium tuberculosis sublineages. . Emerging microbes & infections. 2017;6(7):e68. doi: 10.1038/emi.2017.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khosravi AD, Shahraki AH, Dezfuli SK, Hashemzadeh M, Goodarzi H, Mohajeri P, et al. Genetic diversity of multidrug-resistant Mycobacterium tuberculosis strains isolated from tuberculosis patients in Iran using MIRU-VNTR technique. . Kaohsiung Journal Medical Science. 2017;33(11):550–7. doi: 10.1016/j.kjms.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 46.Ravansalar H, Tadayon K, Mosavari N, Derakhshan M, Ghazvini k. Genetic Diversity of Mycobacterium tuberculosis Complex Isolated from Patients in the Northeast of Iran by MIRU-VNTR and Spoligotyping. Jundishapur Journal of Microbiology. 2017;10(4):e39568. [Google Scholar]
- 47.Shaha Y, Maharjana B, Thapaa J, Poudelc A, Diabd H. High diversity of multidrug-resistant Mycobacterium tuberculosis Central Asian Strain isolates in Nepal. International Journal of Infectious Diseases. . 2017;63:13–20. doi: 10.1016/j.ijid.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Rasoahanitralisoa R, Rakotosamimanana N, Stucki D, Sola C, Gagneux S, Razanamparany VR. Evaluation of spoligotyping, SNPs and customised MIRU-VNTR combination for genotyping Mycobacterium tuberculosis clinical isolates in Madagascar. . PloS one. 2017;12(10):e0186088. doi: 10.1371/journal.pone.0186088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen Y-Y, Chang J-R, Huang W-F, Hsu C-H, Cheng H-Y, Sun J-R, et al. Genetic diversity of the Mycobacterium tuberculosis East African–Indian family in three tropical Asian countries. Journal of Microbiology, Immunology and Infection. 2017;50(6):886–92. doi: 10.1016/j.jmii.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Hu Y, Mansjö M, Zhao Q, Jiang W, Ghebremichael S, et al. The Epidemiological Significance and Temporal Stability of Mycobacterial Interspersed Repetitive Units-Variable Number of Tandem Repeats-Based Method Applied to Mycobacterium tuberculosis in China. International journal of environmental research and public health. 2018;15(4):782–93. doi: 10.3390/ijerph15040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu C, Mao X, Wang J, Pan H. Clustering and recent transmission of Mycobacterium tuberculosis in a Chinese population. Infection and drug resistance. 2018;11:323–30. doi: 10.2147/IDR.S156534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esteves LS, Dalla Costa ER, Vasconcellos SEG, Vargas A, Junior SLMF, Halon ML, et al. Genetic diversity of Mycobacterium tuberculosis isoniazid monoresistant and multidrug-resistant in Rio Grande do Sul, a tuberculosis high-burden state in Brazil. . Tuberculosis. 2018;110:36–43. doi: 10.1016/j.tube.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 53.Zaniani FR, Moghim S, Esfahani BN. Genetic Diversity of Drug-resistant Mycobacterium tuberculosis Isolates in Isfahan Province of Iran. . Advanced Biomedical Research. 2018;7(23):1–6. doi: 10.4103/2277-9175.225594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Azimi T, Nasiri MJ, Zamani S, Hashemi A, Goudarzi H, Fooladi AAI, et al. High genetic diversity among Mycobacterium tuberculosis strains in Tehran, Iran. . Journal of Clinical Tuberculosis and Other Mycobacterial Diseases. 2018;11:1–6. doi: 10.1016/j.jctube.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mansoori N, Yaseri M, Vaziri F, Douraghi M. Genetic diversity of Mycobacterium tuberculosis complex isolates circulating in an area with high tuberculosis incidence: Using 24-locus MIRU-VNTR method. Tuberculosis. 2018;112:89–97. doi: 10.1016/j.tube.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Chawla K, Kumar A, Shenoy VP, Chauhan D, Sharma P, Feng Q. Strain diversity and relative transmission of Mycobacterium tuberculosis in south coastal Karnataka, India. The International Journal of Tuberculosis and Lung Disease. 2018;22(8):878–83. doi: 10.5588/ijtld.17.0732. [DOI] [PubMed] [Google Scholar]
- 57.Shi J, Zheng D, Zhu Y, Ma X, Wang S, Li H, et al. Role of MIRU-VNTR and spoligotyping in assessing the genetic diversity of Mycobacterium tuberculosis in Henan Province, China. BMC infectious diseases. 2018;18(447):1–12. doi: 10.1186/s12879-018-3351-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ei PW, Aung WW, Nyunt WW, Swe TL, Aung ST, Htwe MM, et al. Evaluation Of Mycobacterial Interspersed Repetitive Unit-Variable Number Tandem Repeat Typing To Discriminate Mycobacterium Tuberculosis Strains From Myanma. . The Southeast Asian journal of tropical medicine and public health. 2018;49(2):276–84. [Google Scholar]
- 59.Weerasekera D, Pathirane H, Madegedara D, Dissanayake N, Thevanesam V, Magana-Arachchi DN. Evaluation of the 15 and 24-loci MIRU-VNTR genotyping tools with spoligotyping in the identification of Mycobacterium tuberculosis strains and their genetic diversity in molecular epidemiology studies. Infectious Diseases. 2019:1–10. doi: 10.1080/23744235.2018.1551619. [DOI] [PubMed] [Google Scholar]
- 60.Zarzour H, Madania A, Ghoury I, Habous M. High-resolution genotyping of Mycobacterium tuberculosis isolates from syria using mycobacterial interspersed repetitive unit-variable-number tandem repeat. Biomedical and Biotechnology Research Journal (BBRJ). . 2019;3(1):1–8. [Google Scholar]
- 61.Li D, Song Y, Yang P, Li X, Zhang AM, Xia X. Genetic diversity and drug resistance of Mycobacterium tuberculosis in Yunnan, China. . Journal of clinical laboratory analysis. 2019:e22884. doi: 10.1002/jcla.22884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jagielski T, Minias A, Van Ingen J, Rastogi N, Brzostek A, Żaczek A, et al. Methodological and clinical aspects of the molecular epidemiology of Mycobacterium tuberculosis and other mycobacteria. Clinical microbiology reviews. 2016;29(2):239–90. doi: 10.1128/CMR.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghavidel M, Mansury D, Nourian K, Ghazvini K. The most common spoligotype of Mycobacterium bovis isolated in the world and the recommended loci for VNTR typing; A systematic review. Microbial pathogenesis. 2018;118:310–5. doi: 10.1016/j.micpath.2018.03.036. [DOI] [PubMed] [Google Scholar]


