Abstract
Background:
Abnormal DNA methylation leading to altered transcription of certain genes occurs frequently in colorectal cancer (CRC). As with protein-coding genes, microRNAs (miRNAs) may be targeted for methylation in CRC; however, the methylation state of miRNA genes in CRC, especially in primary lesions, has not yet been completely elucidated. To understand the impact of DNA methylation on the miR-200c/141 cluster promoter, we investigated the methylation and expression of miR-141 in precancerous lesions and colorectal cancer.
Methods:
In this cross-sectional study, 208 colorectal tissue samples, including 34 tumor tissue samples, 60 precancerous lesions with matched normal adjacent tissues, and 20 normal tissue samples, were collected. Promoter methylation of the miR-200c/141 cluster was studied using methylation-specific PCR. MiR-141 expression was examined using quantitative real-time PCR.
Results:
Our findings showed that the miR-200c/141 cluster promoter region was most frequently hypermethylated in colorectal tumors and adenomatous polyps, but unmethylated in hyperplastic polyp tissues (P < 0.001). DNA methylation of the miR-200c/141 cluster and the tumor stage were significantly correlated (P = 0.002); however, miR-141 expression difference between the tumor and polyp samples was not significant (p = 0.6).
Conclusion:
The DNA methylation status of the miR-200c/141 cluster could serve as a progression marker from benign polyps to colorectal cancer.
Key Words: Colonic Polyps, Colorectal Cancer, DNA methylation, MiR-141
Introduction
Colorectal cancer (CRC) is the most common malignant neoplasm of the gastrointestinal tract (1). This type of cancer is the second and third most common malignancy diagnosed in women and men, respectively. It accounts for more than 1.2 million new cases each year (2) and 10 percent of cancer-related deaths worldwide (3).
Pre-existing colonic polyps are the main source for colorectal cancer generation;however, data regarding epigenetic changes related to colorectal polyps are limited (4, 5). Understanding the prevalence and epigenetic mechanism of colorectal polyps would help clarify the efficacy of a CRC screening program. Therefore, updating the current knowledge in the scope of colorectal polyps and CRC is essential.
MicroRNAs (miRNAs) serve as particular targets of genomic lesions that are associated with the silencing of tumor suppressor genes or activation of specific oncogenes (6, 7). Both loss and gain of miRNA function have been found to contribute to cancer development via different mechanisms (8). It has been well documented that miRNAs are involved in the development of gastrointestinal tumors, such as colorectal cancer (9-11).
The miR-200 family members are grouped into potent tumor-suppressive miRNAs (12). MiR-141,an miR-200 family member, is located with miR200c on chromosome 12 (13). MiR-141 has been identified to be associated with the organization of cancer stem cells (CSCs) and epithelialmesenchymal transition (EMT) regulation, although some researchers have shown that miR141 plays a dual role in the tumorigenicity of gastrointestinal cancers, such as CRC (14-16). It has also become apparent that several miRNAs are controlled through epigenetic mechanisms, such as DNA methylation (17).Previous reports indicate that the miR-200 family is linked to aberrant DNA methylation patterns in multiple human malignancies (18,19).This family is silenced by aberrant CpG methylation of its promoter in a large percentage of bladder tumors (20).Epigenetic silencing of the miR-200 family, including miR200c, in gastric cancer cells has been shown through experimental data (19).
Evidence suggests that miR-200 family dysregulation induces EMT during carcinogenesis through direct regulation of ZEB1 and ZEB2, and downregulation of the miR-200 family in some malignancies promotes progression and metastasis (21). Although miR-200 family members have been shown to both promote proliferation and suppress metastasis during colorectal tumorigenesis, the precise mechanism of action of the miR-200 family in different tumor types and stages is not clear (21,22).
Early detection and clinical management of precancerous polyps in the large bowel are essential for CRC prevention and treatment. Here, we examined the tissue-dependent methylation status of the miR-200c/141 cluster promoter and miR-141 expression in normal tissue, colorectal polyps, and malignancies.
Materials and Methods
Study Population
In this cross-sectional study, colorectal samples (n=208) were collected from individuals referred to Taleghani Hospital, Tehran, Iran in 2015 and 2016. Informed consent was given by the participants and the demographic and clinical data, including age, gender, BMI, diabetes state, smoking status, inflammatory bowel disease (IBD), blood pressure, and family history were collected using a standard questionnaire. The questionnaire was completed by a trained research scientist through an interview. All subjects also received colonoscopies. All the tissue samples were histologically examined by a pathologist and divided into groups including adenomatous polyps, hyperplastic polyps, and tumor specimens. Tumor tissues were categorized at stages I, II, and III using the Tumor-Node-Metastasis criteria from the American Joint Committee for Cancer. The characteristic and clinicopathologic parameters are summarized in Table I. This research project was performed under the approval of the Ethics Committee of the Research Center for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran (ethics code: IR.SBMU.RIGLD.REC.1395.925).
DNA Extraction and Methylation-Specific PCR (MSP)
The specimens were snap-frozen in liquid nitrogen and stored at −70 °C until used. Genomic DNAs were extracted from the frozen tissues with QIAamp DNA Mini kit (Qiagen,Germany) according to the manufacturer’s protocol and then stored. The promoter methylation states of miR200c-141 cluster were determined by MSP.
Methylation-specific PCR primer sequences were 5'-TTCGGGAGTAGTTCGGTTC-3' (forward) and 5'AATTAAACTATACCGCCCCG-3'(reverse)for methylated miR-141 and 5'GGTTTGGGAGTAGTTTGGTTT-3' (forward) and 5'-AAAT TAAACTATACCACCCCAC-3'(reverse) for the unmethylated form (18).
Aliquots of the extracted DNA were converted by the Qiagen EpiTect Bisulfite kit (Qiagen, Hilden, Germany) following the manufacturer's guidelines. Then, PCR amplification with MSP primers was performed using 11 µl of master mix and 1.5 µl of bisulfite-converted DNA in a final reaction mixture of 12.5 µL. The master mix included 0.5 µL of the forward primer (10 pmol concentration), 0.5 µl of the reverse primer (10 pmol concentration), 1.25 µl of 10x MSP PCR buffer, 0.25 µL of 10 mM dNTP mix, 0.5 µL of Mg, 2.5 µL of Q PCR buffer, 7.4 µl of nuclease-free H2O, and 0.1 µL of HotStart Taq DNA Polymerase (Qiagene, Germany).
For each DNA sample, the PCR was performed with both methylated and unmethylated primer pairs. The PCR conditions were: a preliminary denaturation at 94 °C for 7 min, then 36 cycles of 94 °C for 40 s (denaturation), 58.5 °C for 40 s (annealing), 72 °C for 40 s (extension), and the final elongation for 10 min. The MSP products were confirmed with 2% agarose gel electrophoresis, stained with a green viewer, and visualized with a UV transilluminator. A human control DNA set containing both bisulfite-converted methylated and unmethylated DNAs and an unconverted unmethylated DNA (Qiagen, Germany) were used to detect methylated and unmethylated sequences. Several examples of methylated and unmethylated samples (tumor and polyp tissue) are shown in Figure 1A.
RNA Isolation and Real-time PCR analysis
The total RNAs were isolated from human colorectal tissues using the miRNeasy Mini Kit(Qiagen, Hilden, Germany). RNA concentration and quality were determined by the NanoDrop system (NanoDrop Technologies). The purified total RNA was stored at -70 °C for subsequent use.The cDNA was synthesized using 5 µg of total RNA with RB cDNA Synthesis kit (RNA biotechnology Co, Iran). The cDNA fragments were used as the templates to amplify miR-141 using a specific miR-141 primer (RNA biotechnology Co, Iran) and 2X RB sybr master mix (RNA biotechnology Co, Iran) according to the manufacturer’s guidelines. The qPCR was performed on a 7500 Real-Time PCR System(Applied Biosystem, USA). Real time PCR amplifications were performed at 94 °C for 4 min,followed by 40 cycles at 95 °C for 30 s, 58 °C for 40 s, and 72 °C for 40 s. The analysis was performed using U6 as an endogenous control and results were calculated by the 2-ΔΔCT method.
Statistical Analyses
Statistics were analyzed with IBM SPSS statistical software, version 19 (SPSS Inc., Chicago, IL).Quantitative data was described as the mean ± standard deviation (SD). Correlations between clinicopathological factors and methylation status of the miR-200c/141 cluster promoter were compared by Chi-square (X2) or Fisher’s exact tests and one-way ANOVA. GraphPad Prism software version 7(Graph Pad Inc., USA) was used for statistical analysis of miR-141 expression. The t-test was used to examine the miR-141 expression levels between groups. P values < 0.05 were considered statistically significant.
Results
General Statistical Information
This study was performed on 208 fresh/frozen colorectal tissues and adjacent samples from 58 male and 56 female subjects. The samples were 14.9, 36.8, 30.7, and 17.5 percent hyperplastic polyps, adenomas, colorectal tumors, and normal tissues, respectively.
The mean ages of the patients with polyps,tumors, and normal tissues were 58.6 ±12.7,61±13.7, and 57.3±9.2 years, respectively.Demographic and clinicopathological characteristics are shown in Table 1.
Table 1.
Demographic and clinicopathological characteristics of the study subjects
| Variables | Polyp(N=60) | Tumor(N=34) | Normal(N=20) | P value |
|---|---|---|---|---|
| Age(years; x ± SD) ** | 58.6±12.7 | 61±13.7 | 57.3±9.2 | 0.54 |
| BMI (Kg/m2; x± SD) ** | 25.2±2.8 | 24.9±2.9 | 26.3±3.8 | 0.26 |
| Sex,n(%)* | 0.19 | |||
| Female | 27(45.0%) | 21(61.8%) | 8 (40.0%) | |
| Male | 33(55.0%) | 13(38.2%) | 12(60.0%) | |
| FH n(%)* | 0.50 | |||
| No | 47(78.3%) | 28(82.4%) | 18(90.0%) | |
| Yes | 13(21.7%) | 6(17.6%) | 2(10.0%) | |
| Smoking n(%)* | 0.10 | |||
| No | 54(90.0%) | 25(73.5%) | 17(85.0%) | |
| Yes | 6(10.0%) | 9(26.5%) | 3(15.0%) | |
| Location n(%)* | 0.008 | |||
| Colon | 51(85.0%) | 19(55.9%) | 14(70.0%) | |
| Rectum | 9(15.0%) | 15(44.1%) | 6(30.0%) | |
| IBD n (%)* | 0.14 | |||
| No | 53(88.3%) | 28(82.4%) | 20(100.0%) | |
| Yes | 7(11.7%) | 6(17.6%) | 0(0.0%) | |
| HBn(%)* | 0.57 | |||
| No | 48(80.0%) | 30(88.2%) | 16(80.0%) | |
| Yes | 12 (20.0%) | 4(11.8%) | 4 (20.0%) | |
| Diabetes n(%)* | 0.74 | |||
| No | 51(85.0%) | 28(82.4%) | 18(90.0%) | |
| Yes | 9(15.0%) | 6(17.6%) | 2(10.0%) |
Data are presented as number (%), x± SD, and P value;BMI:Body Mass Index; FH:Family History; IBD: Inflammatory Bowel Disease; HB: High Blood Pressure;
X2 test;
One-way ANOVA
Correlations between the methylation of the miR-200c/141 cluster promoter and Clinical Characteristics
No significant correlations were found between the miR-200c/141 cluster promoter DNA methylation state and patient demographic data, including BMI, family history (FH), diabetes, high blood pressure (HBP),and inflammatory bowel disease (IBD)(Table 2).
Table 2.
Correlation between miR-200c/141 cluster promoter methylation status and clinicopathological properties of the study subjects.
| Methylation status | P value | |||
|---|---|---|---|---|
| Variables | M | U | X2 test | Fisher's exact test |
| Age(year) | 0.45 | 0.57 | ||
| Under 50 | 12(18.2%) | 7 (25.0%) | ||
| Upper 50 | 54(81.8%) | 21(75.0%) | ||
| Sex(n%) | 0.55 | 0.65 | ||
| Female | 35(53.0%) | 13(46.4%) | ||
| Male | 31(47.0%) | 15(53.6%) | ||
| BMI(Kg/m2) | 0.61 | |||
| 18-23.9 | 15(22.7%) | 8(28.6%) | ||
| 24-29.9 | 42(63.6%) | 18 (64.3%) | ||
| 30-35.9 | 9(13.6%) | 2(7.1%) | ||
| FH (n%) | 0.35 | 0.41 | ||
| No 51 | 51(77.3%) | 24(85.7%) | ||
| Yes | 15(22.7%) | 4(14.3%) | ||
| Diabetes(n%) | 0.36 | 0.54 | ||
| No | 54(81.8%) | 25(89.3%) | ||
| Yes | 12(18.2%) | 3(10.7%) | ||
| HB(n%) | 0.64 | 0.77 | ||
| No | 54(81.8%) | 24(85.7%) | ||
| Yes | 12(18.2%) | 4(14.3%) | ||
| Smoking(n%) | 0.34 | 0.36 | ||
| No | 57(86.4%) | 22 (78.6%) | ||
| Yes | 9(13.6%) | 6(21.4%) | ||
| Position(n%) | 0.14 | 0.19 | ||
| Colon | 52(78.8%) | 18(64.3% | ||
| Rectum | 14 (21.2%) | 10 (35.7%) | ||
| IBD (n%) | 0.22 | 0.33 | ||
| No | 55 (83.3%) | 26 (92.9%) | ||
| Yes | 11(16.7%) | 2(7.1%) | ||
| Stage (n%) | 0.002 | 0.007 | ||
| Stage I,II | 23(85.2%) | 2(28.6%) | ||
| Stage III | 4(14.8%) | 5(71.4%) | ||
M and U indicate Methylated and Unmethylated statuses, respectively; P value of less than 0.05 is considered significant. Data are presented as number (%), x± SD, and P value; BMI: Body Mass Index; FH: Family History; IBD: Inflammatory Bowel Disease; HB: High Blood Pressure
Of the 34 tumor tissue samples obtained from colorectal cancer patients, 8 (23.5%), 17 (50%),and 9 (26.5%) were in stages I, II, and III,respectively. We found that stages I and II samples were mostly methylated, while those in stage III were mostly unmethylated.This difference was statistically significant (P =0.002, Table 2).
Correlations between miR-200c/141 cluster methylation and colorectal specimen type
The miR-200c/141 cluster promoter was methylated in approximately 80% of the adenocarcinoma specimens, while it was mostly unmethylated in the hyperplastic samples. The methylation status of miR-200c/141 cluster correlated significantly with the tissue sample type P < 0.001, Table 3).
Table 3.
miR-200c/141 cluster methylation and colorectal specimen types
| Type | Methylation status | P value | |
|---|---|---|---|
| M | U | P<0.001* | |
| Normal | 5(25.0%) | 15(75.0%) | |
| Adenoma | 32(76.2%) | 10(23.8%) | |
| Hyperplastic | 6(35.3%) | 11(64.7%) | |
| Tumor | 28(80.0%) | 7(20.0%) | |
According to X2 test
Expression of miR-141 in the Samples of Patients
No significant difference in miR-141 expression was found between polyps and control tissues (P= 0.07, Fig. 1B) or between tumor and control tissues (P = 0.19, Fig. 1C).
Fig.1.
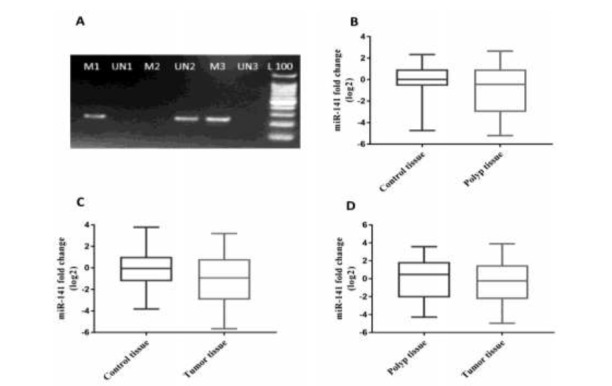
A)representative 2% agarose gel electrophoresis for identification of miR-200c/141 cluster methylation status in colorectal specimens, M and UN indicate methylated and unmethylated states, number 1 represents a tumor tissue, number 2 showed a hyperplastic polyp tissue and number 3 showed adenoma polyp tissue, respectively. L: 100 bp ladder, B) miR-141 expression in polyp and adjacent tissues C) miR-141 expression in tumors and adjacent controls D) miR-141 expression in tumor and polyp tissues.
miR-141 expression was 0.77-fold less in tumors than in polyps; however, this difference was not significant (P = 0.63,Fig.1D).
Discussion
DNA methylation is an important mechanism of inactivating tumor suppressor genes such as miR-141 (23, 24). miR-141 methylation may participate in CRC progression. Colorectal cancer can arise from polyps; therefore, the identification of mechanisms that correlate with CRC generation it is critical (25).
In the current study, the miR-200c/141 cluster promoter methylation profile in colon cancer tissues was characterized in an Iranian population.We found the miR-200c/141 cluster promoter was hypermethylated in colorectal tumors and adenomatous polyps, but not in hyperplastic polyps or normal tissues.
We also found that miR-141 expression was less in tumors than in polyp but the difference was not statistically significant.
Previous studies have demonstrated this cellular plasticity is controlled by the miR-200 family,which plays a fundamental role in EMT suppression by directly silencing ZEB transcription factors (26,27).
The evidence supports the idea that the imbalanced expression between ZEB transcription factors and the miR-200 family could promote EMT and development of epithelial cancers (28).Also, reports have suggested that cellular plasticity may be associated with epigenetic modifications such as DNA methylation (29).
The miR-200 family is epigenetically silenced by methylation of promoters in lung (30) and bladder (31) cancers and glioma (18).
miR-141 functions as a tumor suppressor in gastric adenocarcinoma (16), renal cell carcinoma(26), and pancreatic cancer (32). MiR-200c/141locus methylation correlates with miRNA downregulation in endometrial carcinosarcoma(21). Also, miR-200c-141 cluster promoter methylation has been correlated with aggressive phenotypes such as poor differentiation and high proliferation in carcinoma cell lines (33,34).
Neves and colleagues demonstrated that the miR-200c-141 cluster promoter is silenced by hypermethylation, resulting in decreased miR-141 expression (35). Their silencing states in prostate cancer cells also occur with the aberrant DNA methylation patterns (33,36). While silencing of miR-141/200c is associated with tumorigenesis, an association has been shown between their increased expression and hepatic metastatic cancer (21).
We found that miR-200c-141 cluster methylation was greater in adenomatous than in hyperplastic polyps. Because these adenomatous polyps are more likely to become malignant, the methylation state can be used as an epigenetic marker in the prognosis of colorectal cancer.
Accordingly, hypermethylation of the miR-200c-141 cluster promoter leads to altered miR-141 and miR200c expression, which can potentially affect a number of downstream targets.However, abnormal methylation of this locus may be correlated with disease pathogenesis.
Although our results revealed significant differences between the methylation states of tumor tissues and precancerous lesions, the establishment of this correlation needs further studies. Furthermore, the role of the miR-200c-141 cluster and those of other miRNAs in the regulation of genes involved in CRC progression should be determined to validate their use as appropriate markers for the prediction of CRC.
Our results provide further insight into the role of epigenetic changes underpinning colorectal tumor progression from colorectal polyps. To our knowledge, this is the first report describing the methylation state of miR-200c-141 cluster in colorectal polyps and the association between its methylation state and various CRC stages.
Acknowledgements
We thank all the patients who participated in this study. The project is supported by Research Center for Gastroenterology and Liver Diseases, grant No:925. This article resulted from Zahra Taheri PhD thesis project. All authors declare that there is no conflict of interest related to this study.
References
- 1.Riihimäki M, Hemminki A, Sundquist J, Hemminki K. Patterns of metastasis in colon and rectal cancer. Sci Rep. 2016;6:29765. doi: 10.1038/srep29765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010:1055–9965. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 4.Chan AT, Giovannucci EL. Primary prevention of colorectal cancer. Gastroenterology. 2010;138(6):2029–43. e10. doi: 10.1053/j.gastro.2010.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tänzer M, Balluff B, Distler J, Hale K, Leodolter A, Röcken C, et al. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One. 2010;5(2):e9061. doi: 10.1371/journal.pone.0009061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang H, Mendell J. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94(6):776–80. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira HJ, Davalos V, de Moura MC, Soler M, Perez-Salvia M, Bueno-Costa A, et al. Circular RNA CpG island hypermethylationassociated silencing in human cancer. Oncotarget. 2018;9(49):29208–19. doi: 10.18632/oncotarget.25673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6(6):590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schetter AJ, Okayama H, Harris CC. The role of microRNAs in colorectal cancer. Cancer J. 2012;18(3):244–52. doi: 10.1097/PPO.0b013e318258b78f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen J, KK Wu W, X Ren S, Zhang L, LY Chan R, CM Wong C, et al. miRNAs in gastrointestinal and liver cancers: Their perspectives and clinical applications. Curr Pharm Des. 2013;19(7):1301–10. doi: 10.2174/138161213804805720. [DOI] [PubMed] [Google Scholar]
- 11.Stiegelbauer V, Perakis S, Deutsch A, Ling H, Gerger A, Pichler M. MicroRNAs as novel predictive biomarkers and therapeutic targets in colorectal cancer. World Journal of Gastroenterology: WJG. 2014;20(33):11727–35. doi: 10.3748/wjg.v20.i33.11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mongroo PS, Rustgi AK. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol Ther. 2010;10(3):219–22. doi: 10.4161/cbt.10.6312548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu L, Li Q, Xu D, Wang Q, An Y, Du Q, et al. hsa-miR-141 downregulates TM4SF1 to inhibit pancreatic cancer cell invasion and migration. Int J Oncolon. 2014;44(2):459–66. doi: 10.3892/ijo.2013.2189. [DOI] [PubMed] [Google Scholar]
- 14.Park S-M, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the Ecadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu M, Xia M, Chen X, Lin Z, Xu Y, Ma Y, et al. MicroRNA-141 regulates Smad interacting protein 1 (SIP1) and inhibits migration and invasion of colorectal cancer cells. Dig Dis Sci. 2010;55(8):2365–72. doi: 10.1007/s10620-009-1008-9. [DOI] [PubMed] [Google Scholar]
- 16.Zuo Q, Zhang R, Li B, Lin Z, Zhao Y, Zhuang Y, Yu T, et al. MicroRNA-141 inhibits tumor growth and metastasis in gastric cancer by directly targeting transcriptional co-activator with PDZbinding motif, TAZ. Cell Death Dis. 2015;6(1):e1623. doi: 10.1038/cddis.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73(2):473–7. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- 18.Bian E-B, Ma C-C, He X-J, Wang C, Zong G, Wang H-L, et al. Epigenetic modification of miR-141 regulates SKA2 by an endogenous‘sponge’HOTAIR in glioma. Oncotarget. 2016;7(21):30610–25. doi: 10.18632/oncotarget.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Nie Y, Tu S, Wang H, Zhou Y, Du Y, et al. Epigenetically deregulated miR-200c is involved in a negative feedback loop with DNMT3a in gastric cancer cells. Oncol Rep. 2016;36(4):2108–16. doi: 10.3892/or.2016.4996. [DOI] [PubMed] [Google Scholar]
- 20.Shindo T, Niinuma T, Nishiyama N, Shinkai N, Kitajima H, Kai M, et al. Epigenetic silencing of miR-200b is associated with cisplatin resistance in bladder cancer. Oncotarget. 2018;9(36):24457–69. doi: 10.18632/oncotarget.25326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hur K, Toiyama M, Takahashi M, Toiyama M, Balaguer F, Nagasaka T, Koike J, et al. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2012:1315–26. doi: 10.1136/gutjnl-2011-301846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan Y, Liang H, Kosta K, Chen W, Zhang H, Wang N, Wang F, et al. microRNA-200b and microRNA-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with Kazal motifs. RNA Biol. 2015;12(3):276–89. doi: 10.1080/15476286.2015.1017208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, Wu W, Claret FX. Mutual regulation of microRNAs and DNA methylation in human cancers. Epigenetics. 2017;12(3):187–97. doi: 10.1080/15592294.2016.1273308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu S, He X, Wang L. Correlation analysis of miR-200b, miR-200c, and miR-141 with liver metastases in colorectal cancer patients. Eur Rev Med Pharmacol Sci. 2017;21:2357–63. [PubMed] [Google Scholar]
- 25.Øines M, Helsingen LM, Bretthauer M, Emilsson L. Epidemiology and risk factors of colorectal polyps. Best Pract Res Clin Gastroenterol. 2017;31(4):419–24. doi: 10.1016/j.bpg.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Yu X-y, Zhang Z, Liu J, Zhan B, Kong C-z. MicroRNA-141 is downregulated in human renal cell carcinoma and regulates cell survival by targeting CDC25B. Onco Targets Ther. 2013;6:349–54. doi: 10.2147/OTT.S41343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knezevic J, Pfefferle AD, Petrovic I, Greene SB, Perou CM, Rosen JM. Expression of miR-200c in claudin-low breast cancer alters stem cell functionality, enhances chemosensitivity and reduces metastatic potential. Oncogene. 2015;34(49):5997–6006. doi: 10.1038/onc.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamagawa S, Beder LB, Hotomi M, Gunduz M, Yata K, Grenman R, et al. Role of miR-200c/miR-141 in the regulation of epithelialmesenchymal transition and migration in head and neck squamous cell carcinoma. Int J Mol Med. 2014;33(4):879–86. doi: 10.3892/ijmm.2014.1625. [DOI] [PubMed] [Google Scholar]
- 29.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19(11):1438–49. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Humphries B, Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6(9):6472–98. doi: 10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braicu C, Cojocneanu-Petric R, Martin J, Chira S, Truta A, Floares A, Petrut B, et al. Clinical and pathological implications of miRNA in bladder cancer. Int J Nanomedicine. 2015;10:791–800. doi: 10.2147/IJN.S72904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao. G, Wang B, Liu Y, Zhang J-g, Deng Sc, Qin Q, et al. miRNA-141, downregulated in pancreatic cancer, inhibits cell proliferation and invasion by directly targeting MAP4K4. Mol Cancer Ther. 2013;12:2569–80. doi: 10.1158/1535-7163.MCT-13-0296. [DOI] [PubMed] [Google Scholar]
- 33.Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5(1):e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiklund ED, Bramsen JB, Hulf T, Dyrskjøt L, Ramanathan R, Hansen TB, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2011;128(6):1327–34. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- 35.Neves R, Scheel C, Weinhold S, Honisch E, Iwaniuk KM, Trompeter H-I, et al. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res Notes. 2010;3(1):219. doi: 10.1186/1756-0500-3-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynch SM, O'neill KM, McKenna MM, Walsh CP, McKenna DJ. Regulation of miR‐200c and miR‐141 by Methylation in Prostate Cancer. The Prostate. 2016;76(13):1146–59. doi: 10.1002/pros.23201. [DOI] [PMC free article] [PubMed] [Google Scholar]


