Abstract
Inhalation of carbon black nanoparticles (CBNPs) causes pulmonary inflammation; however, time course data to evaluate the detailed evolution of lung inflammatory responses are lacking. Here we establish a time-series of lung inflammatory response to CBNPs. Female C57BL/6 mice were intratracheally instilled with 162 μg CBNPs alongside vehicle controls. Lung tissues were examined 3 h, and 1, 2, 3, 4, 5, 14, and 42 days (d) post-exposure. Global gene expression and pulmonary inflammation were assessed. DNA damage was evaluated in bronchoalveolar lavage (BAL) cells and lung tissue using the comet assay. Increased neutrophil influx was observed at all time-points. DNA strand breaks were increased in BAL cells 3 h post-exposure, and in lung tissues 2–5 d post-exposure. Approximately 2600 genes were differentially expressed (± 1.5 fold; p ≤ 0.05) across all time-points in the lungs of exposed mice. Altered transcript levels were associated with immune-inflammatory response and acute phase response pathways, consistent with the BAL profiles and expression changes found in common respiratory infectious diseases. Genes involved in DNA repair, apoptosis, cell cycle regulation, and muscle contraction were also differentially expressed. Gene expression changes associated with inflammatory response followed a biphasic pattern, with initial changes at 3 h post-exposure declining to base-levels by 3 d, increasing again at 14 d, and then persisting to 42 d post-exposure. Thus, this single CBNP exposure that was equivalent to nine 8-h working days at the current Danish occupational exposure limit induced biphasic inflammatory response in gene expression that lasted until 42 d post-exposure, raising concern over the chronic effects of CBNP exposure.
Keywords: Carbon black nanoparticles, Biphasic response, Inflammation, DNA repair, Cell cycle regulation, Muscle contraction
Highlights
-
•
A single exposure to CBNPs induced expression changes in over 2600 genes in mouse lungs.
-
•
Altered genes were associated with immune-inflammatory and acute phase responses.
-
•
Several genes were involved in DNA repair, apoptosis, and muscle contraction.
-
•
Effects of a single exposure to CBNPs lasted until 42 d post-exposure.
-
•
A single exposure to CBNPs induced a biphasic inflammatory response in gene expression.
1. Introduction
Carbon black particles with diameters less than 100 nm are referred to as ultrafine powder or carbon black nanoparticles (CBNPs) and are used as reference material for diesel exhaust particles stripped of their adsorbed compounds in toxicological studies (Levy et al., 2012, Kyjovska et al., 2015b). CBNPs are one of the most widely produced and used nanomaterial. Approximately 70% of CBNPs are used by the tire industry, with the remaining 30% used for other rubber products and some non-rubber applications (e.g., as pigments and in printing ink) (IARC, 2010). Given the widespread production of CBNPs, a broad understanding of their toxicity and potential long-term health effects is required.
Pulmonary oxidative stress and inflammation are known responses to CBNPs. Substantive dose- and time-dependent neutrophil influxes have been observed in mice following intratracheal instillation and inhalation of CBNPs (Saber et al., 2009, Bourdon et al., 2012b, Jackson et al., 2012a, Saber et al., 2013, Kyjovska et al., 2015a). Generation of reactive oxygen species and induction of oxidatively damaged DNA has also been documented in cellular and acellular systems following exposure to CBNPs (Jacobsen et al., 2008). Increased oxidative DNA damage has been observed in mouse lungs following intratracheal instillation of CBNPs (Bourdon et al., 2012c); CBNPs have been shown to induce oxidative DNA damage, DNA strand breaks, and mutations both in vivo and in vitro, and in offspring following prenatal exposure (Driscoll et al., 1996, Saber et al., 2005, Jacobsen et al., 2008, Jacobsen et al., 2009, Saber et al., 2009, Jackson et al., 2012a). The mutation spectrum supports the notion that oxidative DNA damage as a result of CBNP-induced generation of reactive oxygen species may be the cause of this mutagenicity (Jacobsen et al., 2011). Furthermore, pulmonary exposure to CBNPs induces a pulmonary acute phase response in mice leading to alterations in cholesterol homeostasis (Bourdon et al., 2012a), which may increase the risk of cardiovascular effects (Saber et al., 2013, Saber et al., 2014).
In a previous study we showed that intratracheal instillation of 162 μg CBNPs (a dose corresponding to pulmonary deposition during nine 8-h working days at the current Danish occupational exposure limit for carbon black) resulted in increased neutrophil influx, changes in the expression of genes associated with inflammation, and genotoxicity in the lungs of C57BL/6 mice that lasted for 28 d following a single exposure (Bourdon et al., 2012a, Bourdon et al., 2012c). However, it remains unclear whether these inflammatory responses persist at time-points beyond 28 d, and how these inflammatory responses evolve over time. In this study, we investigate lung neutrophil influx, global gene expression changes, and genotoxicity at multiple post-exposure time-points following exposure to 162 μg CBNPs via intratracheal instillation. We used intratracheal instillation in order to have an exact time point following which to measure the onset of the pulmonary response to CBNP exposure to increase precision in measurement of temporal changes in gene expression. In addition, although inhalation is a more physiologically relevant exposure model, the generation of aerosol for the desired dose of nanomaterials is complex, expensive, time-consuming, and poses hazards to laboratory personnel (Brain et al., 1976). Moreover, inhalation methodologies require high concentrations of experimental dose and it can be difficult to deliver the desired dose to the experimental animal (Brain et al., 1976). Intratracheal instillation enables the delivery of the desired dose and provides a fairly uniform distribution of the experimental materials throughout the lungs of the exposed mice (Brain et al., 1976, Mikkelsen et al., 2011). Overall, our experiment examines the effects of CBNP exposure at early (3 h), intermediate (1, 2, 3, 4, 5, and 14 d), and late (42 d) time-points to understand the implications of CBNP exposure.
2. Materials and methods
2.1. Animals
Six-to-seven week old female C57BL/6 mice were purchased from Taconic (Ry, Denmark) and were allowed 1–2 weeks to acclimate. A total of 112 mice were divided into 8 experimental (N = 8 per group) and 8 control groups (N = 6 per group), and were maintained in polypropylene cages with sawdust bedding and enrichment, at 20–22 °C temperature and relative humidity of 40–60% with a 12 h light-to-dark cycle. All mice received food and water ad libitum during the period of the whole experiment. All animal experiments were approved by the Danish “Animal Experiments Inspectorate” and performed according to their guidelines for ethical conduct and care for animals in research (The Danish Ministry of Justice, Animal Experiments Inspectorate, permission 2010/561-1779).
2.2. Carbon black nanoparticle characterization
CBNPs were a kind gift from Evonik/Degussa (Frankfurt, Germany) and were extensively characterized in previous studies (Jacobsen et al., 2007, Bourdon et al., 2012c, Saber et al., 2012). Dynamic Light Scattering (DLS) was employed to determine the hydrodynamic particle size distributions in the exposure media, using a Malvern Zetasizer Nano ZS (Malvern, UK). Transmission electron microscopy (TEM) and scanning electron microscopy (SEM) were used to determine the aggregation levels of CBNPs.
2.3. Mouse exposures and tissue collection
Preparation of exposure stock, exposures, and tissue collection were described previously (Bourdon et al., 2012a, Bourdon et al., 2012c, Jackson et al., 2012a, Jackson et al., 2012b, Kyjovska et al., 2015a). Briefly, CBNPs were suspended in 0.2 μm filtered, ɤ-irradiated Nanopure Diamond UV water (Pyrogens: <0.001 EU/ml, total organic carbon: <3.0 ppb), and subjected to sonication using a Branson Sonifier S-450D (Branson Ultrasonics Corp., Danbury, CT, USA) equipped with a disruptor horn (model number 101–147-037) as described (Jackson et al., 2012a, Jackson et al., 2012b, Kyjovska et al., 2015a). Total sonication time was 16 min without pause; samples were kept cool on ice during the sonication procedure. Vehicle control solutions contained only Nanopure Diamond UV water and were also sonicated as described above.
Prior to intratracheal instillation, mice were anesthetized with 4% Isoflurane (Jackson et al., 2011). Mice from the experimental groups received a single dose of 162 μg in a 50 μl volume (162 μg per mouse) of CBNPs suspension, followed by 200 μl air to ensure proper dispersion and deposition of the instilled suspension. Mice from matching control groups received 50 μl of Nanopure water followed by 200 μl air. At the time of the necropsy, mice were anesthetized with a subcutaneous injection of Hypnorm® (fentanyl citrate 0.315 mg/ml and fluanisone 10 mg/ml from Janssen Pharma) and Dormicum® (Midazolam 5 mg/mL from Roche). The lungs were flushed twice with 0.8 ml sterile 0.9% NaCl through the trachea to obtain BAL fluid. BAL fluids were stored on ice until centrifugation at 400 × g for 10 min at 4 °C. The BAL cells were re-suspended in 100 μl medium (HAM F-12 with 1% penicillin/streptomycin and 10% fetal bovine serum). Acellular BAL fluid was recovered and stored at − 80 °C. The total number of living and dead cells in BAL was determined by NucleoCounter NC-200TM (Chemometec, Denmark) from diluted cell suspension following the manufacturer's protocol, the total cell counts were determined for each mouse. Lung and liver tissue were collected following BAL collection, snap frozen in liquid nitrogen, and stored at − 80 °C until analyzed.
2.4. Inflammatory cell counts and protein content in BAL
Differential cell count in BAL was conducted as described previously (Bourdon et al., 2012c). Briefly, cells from 40 μl of BAL fluid were collected on microscope slides by centrifugation at 10,000 rpm for 4 min in a Cytofuge 2 (StatSpin, TRIOLAB, Rødovre, Denmark). Cells were fixed for 5 min in 96% ethanol and stained with May-Grünwald-Giemsa stain. Cellular composition of BAL fluid was determined by differentiation of cell types in 200 cells from each slide, under light microscope with 100 × magnification (immersion oil). The total amount of cells of each type was recalculated using the total number of BAL cells. Protein content in BAL was determined as previously described (Kyjovska et al., 2015a).
2.5. Comet assay
The comet assay was performed to determine DNA strand breaks; frozen BAL cell suspension, lung (3 × 3 mm of right lobe), and liver (3 × 3 mm of median lobe) tissue from control and CBNP exposed mice were used as samples. The detailed experimental protocol for sample preparation and comet analysis was described previously (Jackson et al., 2013, Kyjovska et al., 2015a). Briefly, cells from BAL, lung, and liver tissue were suspended and embedded in 0.7% agarose gel on Trevigen 20-Well CometSlides™. Slides were submerged into lysis solution and stored overnight at 4 °C. The next day, samples were subjected to alkaline electrophoresis following alkaline treatment; electrophoresis was conducted in ice cold circulating electrophoresis solution for 25 min at 1.15 V/cm and pH > 13. Alkaline electrophoresis was followed by sample neutralization, fixing, and staining with SYBRGreen®. Comets were scored using the PathFinder™ system (IMSTAR, France). Up to 1500 nucleoids were scored for each sample, and DNA strand breaks were quantified as % DNA in the comet tail (%TDNA) and the comet tail length (TL). A549 cells exposed to phosphate buffered saline (PBS) and 60 μM H2O2 were used as negative and positive controls, respectively.
2.6. Statistical analysis of BAL and comet assay data
The BAL data were normalized by ranking and assessed by two-way ANOVA (dose and time were used as categorical variables) with a post-hoc Tukey-type multiple comparison test. Comet assay data and protein concentrations were assessed by two-way ANOVA (dose and time were used as categorical variables) with a post-hoc Tukey-type multiple comparison test. Statistical significance was tested at the p < 0.05 level and reported as p < 0.05, p < 0.01 or p < 0.001. The statistical analyses were performed using SAS version 9.2 (SAS Institute Inc., Cary, NC, USA).
2.7. Total RNA extraction and purification
Total RNA was isolated from random sections of lung tissues from 80 mice in total (n = 5 per exposed and control group) using TRIzol (Invitrogen, Carlsbad, CA, USA) and purified using RNeasy Plus Mini Kit (Qiagen, Mississauga, ON, Canada) according to the manufacturer's instructions. Total RNA concentration was measured using NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA), and RNA quality and integrity were assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Mississauga, ON, Canada) according to the manufacturer's instruction. All RNA samples showed A260/280 ratios between 2.0 and 2.15, and RNA integrity numbers (RIN) of 7 and above, and were used to conduct microarray and quantitative real-time RT-PCR (RT-qPCR) experiments. Total RNA samples were stored at − 80 °C until analysis.
2.8. Microarray hybridization
Complementary DNA (cDNA) and cyanine labeled complementary RNA (cRNA) were synthesized from 200 ng of total RNA from individual mice of the CBNP exposed and control groups (5 mice per group), and universal reference total RNA (Agilent Technologies, Inc., Mississauga, ON, Canada), using Low Input Quick Amp Labeling Kit (Agilent Technologies Inc., Mississauga, ON, Canada) according to the manufacturer's instruction. Cyanine-labeled cRNAs were synthesized using T7 RNA polymerase in vitro transcription kits (Agilent Technologies Inc., Mississauga, ON, Canada) and later purified using RNeasy Mini Kits (Qiagen, Mississauga, ON, Canada); experimental and control cRNAs were labeled with Cyanine 5-CTP, and reference RNAs were labeled with Cyanine 3-CTP. Each experimental or control Cy-5 labeled cRNA sample (300 ng) was mixed with equal amounts of Cy-3 labeled reference cRNA and hybridized to Agilent SurePrint G3 Mouse GE 8 × 60K oligonucleotide microarrays (Agilent Technologies Inc., Mississauga, ON, Canada) for 17 h in a hybridization chamber at 65 °C with a rotation speed of 10 rpm. Microarray slides were washed twice according to the manufacturer's instructions (1 × with wash buffer 1 and 1 × with wash buffer 2; Agilent Technologies Inc., Mississauga, ON, Canada), and were scanned on an Agilent G2505C scanner at 3 μm resolution. Data from the scanned images were extracted using Agilent Feature Extraction software version 11.0.1.1 (Agilent Technologies Inc., Mississauga, ON, Canada).
2.9. Statistical analysis of microarray data
The quality of each array was determined using Agilent Feature Extraction software version 11.0.1.1; a total of four arrays (each from experimental 1, 2, and 3 d, and one control 42 d) did not satisfy the quality criteria and were removed from the analysis. Statistical analysis was performed using a reference randomized block design (Kerr and Churchill, 2007) to analyze gene expression microarray data. Data were normalized using the LOcally WEighted Scatterplot Smoothing (LOWESS) (Cleveland, 1979) regression modeling method, and statistical analysis to identify differentially expressed genes was done using MicroArray ANalysis of VAriance (MAANOVA) (Wu et al., 2003) in R statistical software (http://www.r-project.org). The Fs statistic (Cui et al., 2005), a shrinkage estimator for the gene-specific variance components, was used to test for treatment effects. The p-values for statistical tests were estimated using the permutation method with residual shuffling (30,000 permutations). Fold change calculations were based on the least-square means; genes with expression changes of at least 1.5 fold (up- or down-regulated) compared to time-matched controls and that had p-values less than or equal to 0.05 (i.e., FC ± 1.5; p ≤ 0.05) were considered significantly differentially expressed. All microarray data from this study are available at the National Center for Biotechnology Information (NCBI) and Gene Expression Omnibus (GEO) databases, accession number: GSE68036.
2.10. Functional and pathway analysis of differentially expressed genes
Differentially expressed genes from each time-point were subjected to gene ontology (GO), biological network, and pathway analysis. GO analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.7 (Huang da et al., 2009), and GO processes with a Fisher's exact p-value p ≤ 0.05 (Benjamini-Hochberg corrected) were considered significantly enriched (i.e., over-represented). Specific biological networks and pathways associated with the differentially expressed genes were identified using the Metacore data-mining and pathway analysis tool (Thomson Reuters Scientific LLC, PA, USA); pathways and networks with a Fisher's exact p-value p ≤ 0.05 (FDR corrected) were considered significantly enriched.
2.11. Quantitative real-time RT-PCR (RT-qPCR)
Approximately 250 ng of total RNA (N = 3 per group) from each mouse of the experimental (CBNP exposed) and control groups was reverse transcribed using iScript™ Advanced cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA, USA). Real-time qPCR was done using SsoAdvanced™ Universal SYBRR Green Supermix and 96-well custom PrimePCR™ plates for mouse genes (Bio-Rad Laboratories, Hercules, CA, USA), in a CFX96 Real-Time System (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer's instruction. Threshold cycle (Ct) values were normalized using Hprt, Rpl13a, and Gapdh as internal control (housekeeping) genes, and relative expression of the differentially expressed genes was determined using Bio-Rad CFX Manager 3.0 software (Bio-Rad Laboratories, Hercules, CA, USA).
2.12. Public database mining and disease prediction
Differentially expressed genes for each time-point were used for mining against available genomic data repositories in NextBio (http://nextbio.com). Disease prediction analysis was conducted for CBNP datasets from each time-point in order to identify potential disease outcomes following CBNP exposure. NextBio scores for each disease were calculated using pairwise gene signature correlations and rank-based enrichment statistics. The highest ranking disease (i.e., most correlated expression pattern) relative CBNP exposed groups was given a score of 100; the rest of the results were normalized accordingly (Kupershmidt et al., 2010). A meta-analysis was also conducted using differentially expressed gene-sets from all time-points to identify the most consistently and highly disregulated genes from various studies available in the NexBio repository. Ranking and scoring of common genes and biogroups among the studies was conducted according to the method described by Kupershmidt et al. (2010).
3. Results
3.1. Characterization of CBNPs (Printex 90)
Detailed characterization of the CBNPs was reported by Jacobsen et al. (2007), Bourdon et al. (2012a, 2012b), Saber et al. (2012) and Kyjovska et al. (2015a). Briefly, the primary particle size reported by the manufacturer was 14 nm. The estimated specific surface area was 295–338 m2/g. The total carbon content was greater than 99%, with 0.82% nitrogen and 0.01% hydrogen by weight. The content of organic impurity was less than 1% and included very low levels of polycyclic aromatic hydrocarbon (74.2 ng/g) and endotoxin (142 EU/g). The size distribution of CBNP particles suspended in Nanopure water determined by dynamic light scattering (DLS) has been reported previously to be dominated by agglomerates in the size range of 50–100 nm (Jackson et al., 2012a, Jackson et al., 2012b, Kyjovska et al., 2015a).
3.2. Influx of inflammatory cells in BAL fluid
BAL fluid was collected 3 h and 1, 2, 3, 4, 5, 14, and 42 d post-exposure from mice exposed to 162 μg of CBNPs by intratracheal instillation, alongside concurrent vehicle controls. Total numbers of neutrophils, macrophages, eosinophils, lymphocytes, and epithelial cells were counted in BAL fluid. Significant increases in neutrophil number were observed at all post-exposure time-points and persisted until 42 d post-exposure, with the largest influx observed 1 d post-exposure (Table 1 ). The net increase in total BAL cells in exposed mice relative to the controls was mainly due to large influxes of neutrophils. Thus, the inflammatory response in the lungs was predominantly driven by neutrophils. However, we observed significantly increased counts of macrophages on 4, 5, and 42 d; lymphocytes on 4 and 14 d; epithelial cells after 3 h, and eosinophils on 4 d (Table 1). Successful pulmonary deposition of CBNPs was confirmed by the presence of particles observed by light microscopy at all assessed post-exposure days (data not shown). The amount of free particles decreased over time; however, macrophages containing black particles were present at all post-exposure time-points for all CBNP exposed mice (data not shown). Additionally, protein content in BAL fluid was determined as a marker of cell membrane integrity; significant increases in protein level were observed in CBNP exposed mice compared to vehicle controls at all time-points except 3 h and 14 d (Fig. S1).
Table 1.
Inflammatory cell counts in bronchoalveolar lavage (BAL) fluid of C57BL/6 mice exposed to 162 μg of CBNPs at eight different post-exposure time-points. The number of cells is expressed as ± standard error of the mean (SEM) × 103.
| Time-points | Cell types | Number of cells × 103 |
|
|---|---|---|---|
| Control | CBNPs | ||
| 3 h | Neutrophils | 3.9± 1.2 | 26.7 ± 5.5⁎⁎ |
| Macrophages | 38.2 ± 8.3 | 46.7 ± 3.1 | |
| Eosinophils | 0.7 ± 0.2 | 0.6 ± 0.2 | |
| Lymphocytes | 0.4 ± 0.2 | 0.1 ± 0.1 | |
| Epithelial | 16.6 ± 2.7 | 42.6 ± 3.6⁎⁎ | |
| Total BAL cells | 59.7 ± 11.4 | 116.8 ± 7.7⁎⁎ | |
| 1 d | Neutrophils | 4.1 ± 1.0 | 123.4 ± 8.4⁎⁎⁎ |
| Macrophages | 39.8 ± 5.4 | 44.2 ± 8.4 | |
| Eosinophils | 1.9 ± 1.2 | 7.1 ± 2.4 | |
| Lymphocytes | 0.6 ± 0.2 | 1.0 ± 0.6 | |
| Epithelial | 11.0 ± 3.1 | 14.2 ± 2.2 | |
| Total BAL cells | 57.3 ± 3.9 | 189.8 ± 12.1⁎⁎⁎ | |
| 2 d | Neutrophils | 4.0 ± 2.3 | 97.4 ± 6.4⁎⁎⁎ |
| Macrophages | 56.8 ± 8.3 | 50.8 ± 5.0 | |
| Eosinophils | 2.3 ± 0.9 | 2.5 ± 1.0 | |
| Lymphocytes | 1.3 ± 0.4 | 1.6 ± 0.4 | |
| Epithelial | 22.4 ± 4.3 | 16.0 ± 1.9 | |
| Total BAL cells | 86.9 ± 11.4 | 168.4 ± 9.8⁎⁎ | |
| 3 d | Neutrophils | 2.0 ± 0.8 | 84.5 ± 6.0⁎⁎⁎ |
| Macrophages | 48.9 ± 13.5 | 65.0 ± 5.3 | |
| Eosinophils | 1.3 ± 0.4 | 3.7 ± 1.9 | |
| Lymphocytes | 0.6 ± 0.2 | 3.3 ± 1.1 | |
| Epithelial | 13.0 ± 3.3 | 14.5 ± 2.7 | |
| Total BAL cells | 65.9 ± 17.5 | 171.1 ± 7.1⁎⁎⁎ | |
| 4 d | Neutrophils | 0.4 ± 0.2 | 80.0 ± 9.0⁎⁎⁎ |
| Macrophages | 38.6 ± 8.2 | 62.5 ± 7.2⁎ | |
| Eosinophils | 0.8 ± 0.7 | 11.0 ± 2.9⁎ | |
| Lymphocytes | 0.2 ± 0.1 | 8.1 ± 1.4⁎⁎⁎ | |
| Epithelial | 9.3 ± 1.0 | 15.1 ± 4.3 | |
| Total BAL cells | 49.2 ± 9.3 | 176.8 ± 13.1⁎⁎⁎ | |
| 5 d | Neutrophils | 0.2 ± 0.1 | 90.6 ± 12.9⁎⁎⁎ |
| Macrophages | 40.0 ± 6.0 | 89.6 ± 6.5⁎⁎⁎ | |
| Eosinophils | 9.1 ± 5.6 | 20.4 ± 9.4 | |
| Lymphocytes | 1.5 ± 0.7 | 16.9 ± 6.6⁎ | |
| Epithelial | 17.4 ± 1.7 | 16.8 ± 4.7 | |
| Total BAL cells | 68.2 ± 10.6 | 234.3 ± 18.5⁎⁎⁎ | |
| 14 d | Neutrophils | 1.1 ± 0.4 | 29.2 ± 5.4⁎⁎⁎ |
| Macrophages | 53.4 ± 11.6 | 59.1 ± 9.5 | |
| Eosinophils | 0.4 ± 0.3 | 0.9 ± 0.3 | |
| Lymphocytes | 0.2 ± 0.1 | 1.6 ± 0.4⁎ | |
| Epithelial | 27.3 ± 6.7 | 26.3 ± 1.7 | |
| Total BAL cells | 98.8 ± 8.2 | 117.1 ± 15.4 | |
| 42 d | Neutrophils | 0.7 ± 0.3 | 30.5 ± 4.9⁎⁎⁎ |
| Macrophages | 37.5 ± 2.5 | 97.1 ± 10.8⁎⁎⁎ | |
| Eosinophils | 0.5 ± 0.2 | 3.8 ± 1.3 | |
| Lymphocytes | 0.0 ± 0.0 | 0.8 ± 0.7 | |
| Epithelial | 17.4 ± 2.2 | 30.3 ± 4.7 | |
| Total BAL cells | 56.1 ± 4.5 | 162.5 ± 19.7⁎⁎⁎ | |
h — hour; d — day.
p < 0.05.
p < 0.01.
p < 0.001.
3.3. DNA strand breaks
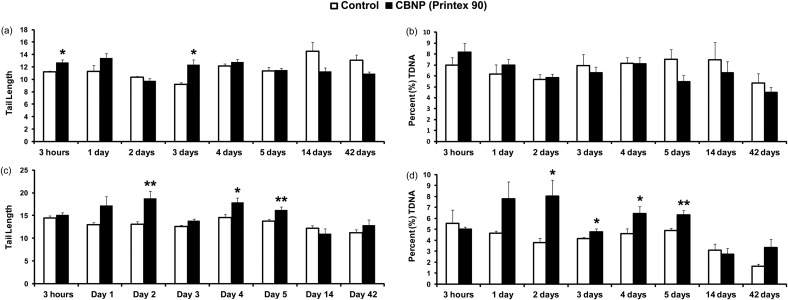
DNA strand break levels were assessed in BAL cells, lung, and liver tissue using the comet assay, and quantified by TL and %TDNA. Statistically significant increased levels of DNA strand breaks were observed in BAL cells 3 h and 3 d post-exposure, expressed as TL (Fig. 1a); %TDNA showed the same trend at 3 h without statistical significance (Fig. 1b). In the lungs, DNA strand break levels, expressed as TL, showed statistically significant increased levels 2, 4, and 5 d post-exposure (Fig. 1c), whereas, %TDNA had significant increases 2, 3, 4, and 5 d post-exposure (Fig. 1d). Increased levels of DNA strand breaks were also observed in liver tissue, expressed both as TL (3 h and 3 d) and %TDNA (3 h, 3 and 4 d) respectively (Fig. S2).
Fig. 1.
DNA strand break determined using the comet assay in BAL cells and lung tissues. DNA strand breaks were quantified as TL and %TDNA in BAL (a) and (b), and in lung tissue (c) and (d) respectively. Statistical significance compared to vehicle controls is indicated by *p < 0.05 and **p < 0.01, respectively.
3.4. Gene expression analysis
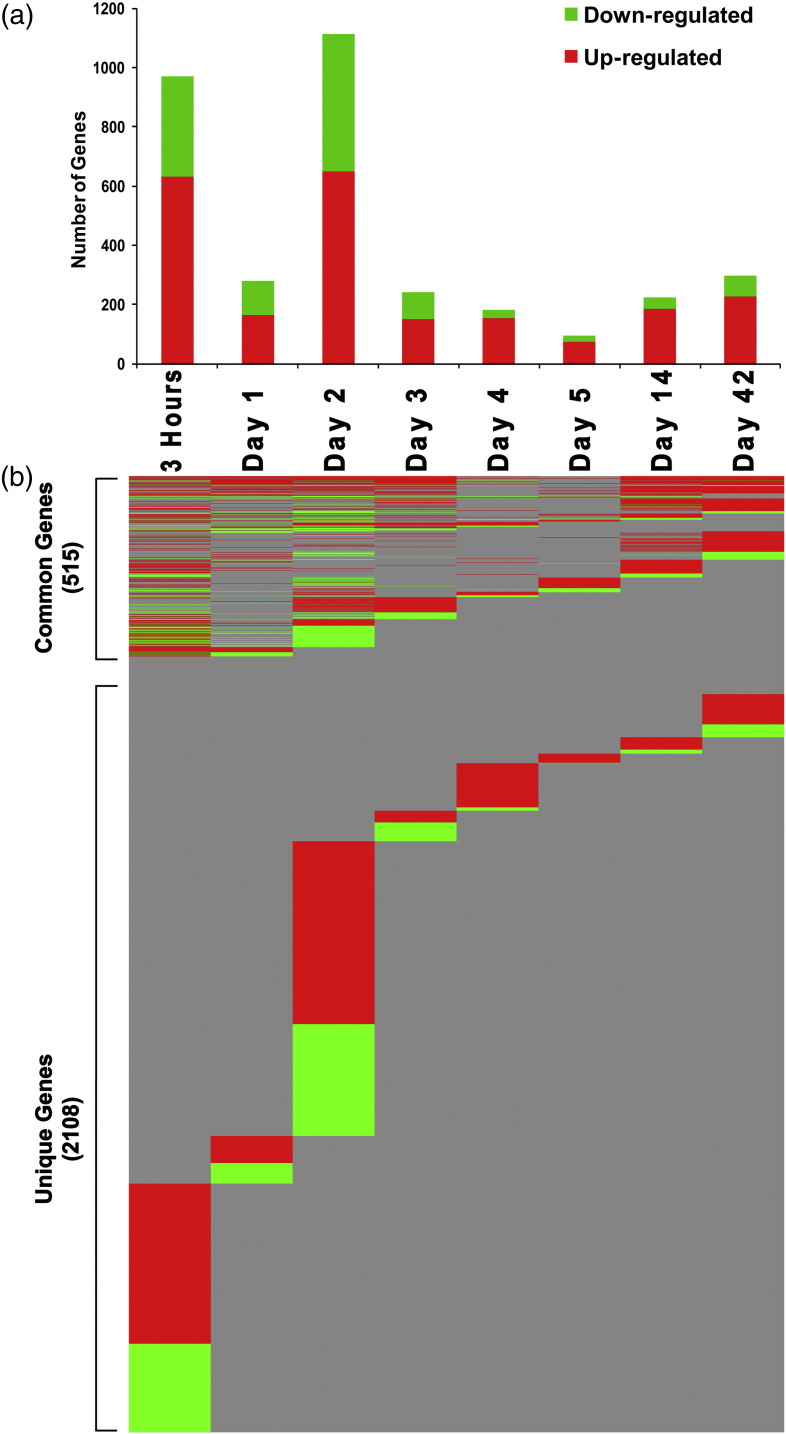
Gene expression analysis identified a total of 2623 unique genes significantly (p ≤ 0.05) differentially expressed (at least ± 1.5 fold) across all time-points that were represented by 3019 probes. Detailed analysis revealed that CBNP exposure induced changes in the greatest number of genes 3 h and 2 d post-exposure, with a total of 970 genes differentially expressed 3 h and 1112 genes 2 d post-exposure (Fig. 2a). In addition, 281 genes were differentially expressed 1 d, 242 genes 3 d, 183 genes 4 d, 94 genes 5 d, 223 genes 14 d, and 297 genes 42 d post-exposure; the majority of the differentially expressed genes were up-regulated (Fig. 2a and Table S1).
Fig. 2.
Genes significantly differentially expressed in response to CBNP exposure. (a) Total number of genes differentially expressed at each post-exposure time-point. (b) Common and unique genes from each post-exposure time-point. Red bars/lines indicate up-regulated and green bars/lines indicate down-regulated genes.
Gene expression signatures were time-point specific, with relatively few genes being differentially expressed consistently across time-points (Fig. 2b). Of the total 2623 differentially expressed genes, 515 genes were commonly expressed (overlapping) across more than one time-point (Fig. 2b and Table S2): 13 genes were in common across 6 time-points, 16 genes were in common across 5 time-points (Table 2 ), and 35 genes were in common across 4 time-points (Table 3 ). The rest of the 451 genes were commonly expressed across 3 and 2 time-points (Table S2). The majority of these common genes belong to pro-inflammatory chemokine (C–C and C–X–C motifs) and acute phase response groups; a few notable common genes include Complement factor 3 (C3), CD14 antigen (Cd14), Chemokine (C–C motif) ligand 2 (Ccl2), Chemokine (C–C motif) ligand 7 (Ccl7), Chemokine (C–C motif) ligand 12 (Ccl12), C-type lectin domain family 4, member D (Clec4d), Colony stimulating factor 2 (granulocyte-macrophage) (Csf2), Chemokine (C–X–C motif) ligand 5 (Cxcl5), Chemokine (C–X–C motif) ligand 10 (Cxcl10), Interleukin 33 (Il33), Serum amyloid A 1 (Saa1), Serum amyloid A 3 (Saa3), Tissue inhibitor of metalloproteinase 1 (Timp1), Vanin 1 (Vnn1), and Vanin 3 (Vnn3). No single gene was differentially expressed across all time-points. The rest of the 2108 genes were uniquely expressed at individual time-points (Fig. 2b and Fig. 3 ). Exposure to CBNPs also induced differential expression of long intergenic noncoding RNAs (lincRNAs) in the lungs of the CBNP exposed mice. From the total 2623 differentially expressed (1.5 fold; p ≤ 0.05) transcripts, 359 were lincRNAs (Table S3).
Table 2.
Fold changes of genes that were differentially expressed across either six or five post-exposure time-points in CBNP exposed versus control mice.
| Gene name | Gene symbol | 3 h | 1 d | 2 d | 3 d | 4 d | 5 d | 14 d | 42 d |
|---|---|---|---|---|---|---|---|---|---|
| Common to 6 post-exposure time-points | |||||||||
| CD14 antigen | Cd14 | 1.7 | 1.6 | 2.3 | 1.7 | – | – | 1.8 | 2.4 |
| Chemokine (C–C motif) ligand 12 | Ccl12 | 1.7 | 1.8 | 1.5 | 1.7 | – | – | 1.8 | 1.6 |
| Chemokine (C–C motif) ligand 2 | Ccl2 | 3.3 | 2.0 | 3.0 | 2.1 | – | – | 2.1 | 2.4 |
| Chemokine (C–C motif) ligand 7 | Ccl7 | 3.2 | 2.4 | 3.0 | 2.4 | – | – | 2.4 | 3.0 |
| Chemokine (C–X–C motif) ligand 1 | Cxcl1 | 9.3 | 2.1 | 2.8 | 2.2 | – | – | 3.0 | 2.7 |
| Chemokine (C–X–C motif) ligand 5 | Cxcl5 | 8.2 | 4.2 | 11.7 | 5.2 | – | – | 6.0 | 13.8 |
| Chemokine (C–X–C motif) receptor 1 | Cxcr1 | – | – | 1.9 | 1.6 | 1.8 | 1.5 | 2.3 | 2.7 |
| D site albumin promoter binding protein | Dbp | − 3.6 | 2.9 | − 1.7 | 2.2 | − 1.9 | – | – | − 3.0 |
| Expressed sequence AA467197 | AA467197 | 2.5 | 2.0 | 2.3 | 1.8 | – | – | 3.8 | 4.7 |
| Leukocyte immunoglobulin-like receptor, subfamily B, member 4 | Lilrb4 | 1.6 | 1.8 | 1.7 | 1.5 | – | – | 2.1 | 2.3 |
| Lymphocyte antigen 6 complex, locus F | Ly6f | – | 2.9 | 3.3 | 3.3 | 1.6 | – | 8.0 | 9.0 |
| Serum amyloid A 1 | Saa1 | 3.8 | 6.4 | 3.7 | 2.3 | – | – | 2.2 | 2.4 |
| Serum amyloid A 3 | Saa3 | 7.1 | 30.4 | 12.4 | 8.0 | – | – | 9.7 | 9.1 |
| Common to 5 post-exposure time-points | |||||||||
| Aryl hydrocarbon receptor nuclear translocator-like | Arntl | 1.8 | − 1.6 | – | − 1.6 | 1.6 | – | – | 1.7 |
| Brevican | Bcan | – | − 1.7 | − 1.6 | − 1.5 | – | – | − 1.6 | − 2.1 |
| Cholesterol 25-hydroxylase | Ch25h | 4.0 | 1.6 | – | 1.7 | – | – | 2.9 | 2.7 |
| Colony stimulating factor 2 (granulocyte-macrophage) | Csf2 | 1.6 | 2.2 | – | 1.8 | – | – | 1.7 | 1.6 |
| Chemokine (C–X–C motif) ligand 10 | Cxcl10 | 3.9 | 2.3 | 2.1 | – | – | 1.7 | – | 1.9 |
| Glutathione S-transferase, alpha 2 (Yc2) | Gsta2 | – | − 1.8 | − 1.6 | – | − 1.5 | – | − 1.5 | − 1.6 |
| Interleukin 33 | Il33 | − 1.7 | 1.6 | 2.0 | 1.7 | – | – | – | 1.7 |
| Lipocalin 2 | Lcn2 | – | 3.1 | 2.9 | 2.5 | – | – | 3.3 | 3.5 |
| Lymphocyte antigen 6 complex, locus I | Ly6i | – | 2.8 | 3.2 | 3.2 | – | – | 8.0 | 9.3 |
| NADPH oxidase activator 1 | Noxa1 | – | 1.8 | 1.8 | 1.6 | – | – | 3.3 | 2.0 |
| NADPH oxidase organizer 1 | Noxo1 | – | 2.2 | 2.6 | 2.7 | – | – | 3.8 | 3.9 |
| Orosomucoid 2 | Orm2 | – | 3.7 | 1.7 | 1.6 | – | – | 3.0 | 1.7 |
| Polymeric immunoglobulin receptor | Pigr | – | 1.9 | 2.2 | 2.4 | – | – | 2.4 | 2.8 |
| STEAP family member 4 | Steap4 | 1.7 | 1.7 | – | 1.5 | – | – | 1.9 | 2.1 |
| Tissue inhibitor of metalloproteinase 1 | Timp1 | 2.6 | 3.8 | 3.1 | 2.1 | – | – | 1.7 | – |
| WAP four-disulfide core domain 17 | Wfdc17 | – | 2.4 | – | 1.9 | 1.7 | – | 4.0 | 5.1 |
All genes had a p < 0.05; “–” indicates that gene in that specific time-point did not pass the selection criteria. h — hour; d — day.
Table 3.
Fold changes of genes that were differentially expressed across four post-exposure time-points in CBNP exposed versus control mice.
| Gene name | Gene symbol | 3 h | 1 d | 2 d | 3 d | 4 d | 5 d | 14 d | 42 d |
|---|---|---|---|---|---|---|---|---|---|
| Amiloride binding protein 1 | Abp1 | − 1.7 | – | − 1.9 | 1.8 | − 1.6 | – | – | – |
| Angiopoietin-like 4 | Angptl4 | 5.2 | − 2.1 | 1.7 | – | 2.2 | – | – | – |
| Asialoglycoprotein receptor 1 | Asgr1 | – | − 2.2 | − 1.8 | – | − 1.8 | – | − 2.2 | – |
| Cathepsin K | Ctsk | – | – | 1.9 | 2.0 | – | – | 2.6 | 2.5 |
| Cell adhesion molecule with homology to L1CAM | Chl1 | – | 1.5 | 1.6 | 1.6 | – | – | – | 1.5 |
| Ceruloplasmin | Cp | 1.5 | – | 1.9 | 1.7 | – | – | – | 1.5 |
| Chemokine (C-C motif) receptor-like 1 | Ccrl1 | − 2.0 | − 1.9 | − 1.6 | − 1.8 | – | – | – | – |
| Complement component 3 | C3 | – | 1.5 | 1.6 | – | – | – | 1.8 | 1.8 |
| C-type lectin domain family 4, member d | Clec4d | – | 1.9 | 1.9 | – | – | – | 2.1 | 2.4 |
| Cytidine 5′-triphosphate synthase | Ctps | 1.9 | 1.9 | 1.6 | – | – | – | 1.5 | – |
| Cytochrome P450, family 2, subfamily a, polypeptide 4 | Cyp2a4 | – | – | − 1.9 | – | − 2.3 | − 2.3 | − 2.2 | – |
| Eosinophil-associated, ribonuclease A family, member 6 | Ear6 | – | – | 1.6 | 1.7 | – | – | 2.3 | 1.7 |
| Glycoprotein (transmembrane) nmb | Gpnmb | – | – | 2.0 | 2.0 | – | – | 2.3 | 3.4 |
| Growth differentiation factor 15 | Gdf15 | 1.7 | – | 1.6 | – | – | – | 1.7 | 1.9 |
| Hemopexin | Hpx | 2.7 | 2.6 | 1.9 | – | – | – | 1.6 | – |
| Immunoresponsive gene 1 | Irg1 | – | 1.9 | 3.2 | – | – | – | 1.8 | 2.7 |
| Inter alpha-trypsin inhibitor, heavy chain 4 | Itih4 | – | 1.5 | – | 1.7 | – | – | 1.9 | 2.4 |
| Interleukin 1 receptor, type II | Il1r2 | 2.7 | 2.0 | 1.5 | – | – | – | 1.6 | – |
| Interleukin 4 induced 1 | Il4i1 | – | 1.6 | 1.7 | – | – | – | 2.0 | 2.2 |
| LincRNA:chr1:134867906-134885481_R | – | 1.8 | – | – | – | – | 1.6 | 1.8 | 2.6 |
| LincRNA:chr10:93774693-93893616_F | – | − 1.5 | – | − 1.7 | − 1.5 | – | 1.6 | – | – |
| LincRNA:chr12:5303823-5379098_R | – | 1.9 | − 1.7 | – | – | – | 1.7 | 1.9 | – |
| LincRNA:chr18:35967933-35986564_F | – | 1.7 | – | – | – | – | 1.8 | 1.9 | 1.8 |
| LincRNA:chr18:63525907-63526504_R | – | − 1.5 | – | − 1.8 | – | – | − 1.6 | − 1.5 | – |
| LincRNA:chr8:80581177-80621652_F | – | 1.6 | – | – | – | – | 1.5 | 1.6 | 1.5 |
| Mab-21-like 3 (C. elegans) | Mab21l3 | – | 1.6 | 1.9 | – | – | – | 1.8 | 2.0 |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | Nfkbia | 2.8 | – | – | – | – | 1.5 | 1.5 | 1.6 |
| Palmdelphin | Palmd | − 1.8 | – | − 1.7 | − 1.6 | – | 1.5 | – | – |
| Predicted gene 3161 | Gm3161 | − 1.6 | – | – | 1.6 | – | − 1.6 | – | − 1.5 |
| Protein Z, vitamin K-dependent plasma glycoprotein | Proz | 1.6 | 1.8 | – | – | – | – | 2.2 | 3.6 |
| Serine peptidase inhibitor, Kazal type 5 | Spink5 | 1.5 | 2.0 | – | 1.5 | – | – | 1.8 | – |
| Solute carrier family 26, member 4 | Slc26a4 | – | – | 2.3 | 3.0 | – | – | 9.3 | 8.2 |
| Tumor necrosis factor receptor superfamily, member 9 | Tnfrsf9 | – | 1.7 | – | 1.7 | – | – | 1.6 | 1.6 |
| Vanin 1 | Vnn1 | – | 1.9 | – | 1.6 | – | – | 2.9 | 2.3 |
| Vanin 3 | Vnn3 | – | 2.2 | – | 1.6 | – | – | 2.3 | 1.7 |
All genes had p < 0.05; “–” indicates that gene in that specific time-point did not pass the selection criteria. h — hour; d — day.
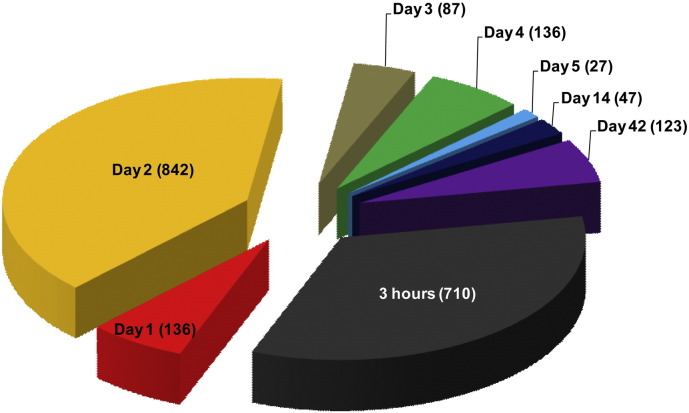
Fig. 3.
The number of unique genes differentially expressed at each time-point following CBNP exposure.
3.5. Functional classification of differentially expressed genes
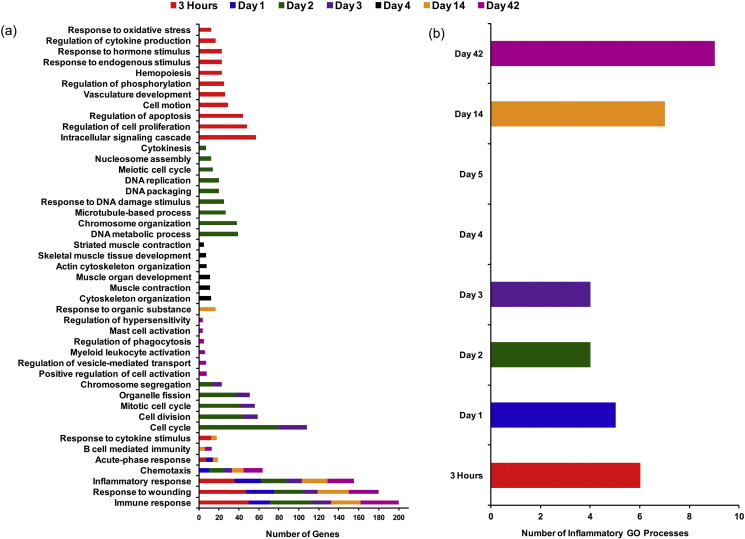
Enrichment of a number of immune-inflammatory GO processes was observed at six of the eight time-points, except 4 and 5 d. Three biological processes, immune response [GO:0006955], inflammatory response [GO:0006954], and response to wounding [GO:0009611] were significantly enriched at six time-points including 3 h, 1, 2, 3, 14, and 42 d post-exposure (Fig. 4a); whereas, chemotaxis [GO:0006935] was significantly enriched at 5 of the 8 time-points, except 3 h, and 4 and 5 d post-exposure (Fig. 4a). A detailed analysis revealed that a group of 165 genes was involved in these four immune-inflammatory response processes (Table S4). Additionally, we observed enrichment of the acute phase response [GO:0006953] GO process at 3 h, 1 d, and 14 d post-exposure. Genes involved in this process from these three time-points are summarized in Table 4 . Enrichment of these immune-inflammatory GO processes 3 h, 1, 2, 3, 14, and 42 d post-exposure suggests a biphasic inflammatory response (i.e., a bimodal distribution in the number of inflammatory genes differentially expressed and corresponding inflammatory biological processes enriched across time-points), which was activated almost immediately following CBNP exposure at 3 h, and gradually declined to 4 d; this response was detected again on 14 d and continued until 42 d post-exposure (Fig. 4b).
Fig. 4.
(a) Common and unique GO biological processes that were significantly enriched at each CBNP post-exposure time-point. (b) Enrichment of immune-inflammatory GO processes follows a biphasic expression pattern; all of these processes were enriched 3 h, and 1, 2, 3, 14, and 42 d post-exposure. No immune-inflammatory GO processes were enriched 4 or 5 d post-exposure.
Table 4.
Genes involved in acute phase response GO processes that were differentially expressed 3 h, 1 d, and 14 d post-exposure to CBNPs.
| Genbank accession | Gene name |
Gene symbol | 3 h |
1 d |
14 d |
|||
|---|---|---|---|---|---|---|---|---|
| P-value | Fold change | P-value | Fold change | P-value | Fold change | |||
| NM_013465 | Alpha-2-hs-glycoprotein | Ahsg | < 0.001 | 1.8 | – | – | – | – |
| NM_031168 | Interleukin 6 | Il6 | < 0.001 | 4.4 | – | – | – | – |
| NM_008768 | Orosomucoid 1 | Orm1 | – | – | < 0.001 | 2.7 | < 0.001 | 2.1 |
| NM_011016 | Orosomucoid 2 | Orm2 | – | – | < 0.001 | 3.7 | < 0.001 | 3.0 |
| NM_011260 | Regenerating islet-derived 3 gamma | Reg3g | – | – | 0.02 | − 6.7 | – | – |
| NM_009117 | Serum amyloid a 1 | Saa1 | < 0.001 | 3.8 | < 0.001 | 6.4 | < 0.001 | 2.2 |
| NM_011314 | Serum amyloid a 2 | Saa2 | < 0.001 | 6.6 | < 0.001 | 3.9 | – | – |
| NM_011315 | Serum amyloid a 3 | Saa3 | < 0.001 | 7.1 | < 0.001 | 30.4 | < 0.001 | 9.7 |
| NM_009243 | Serine (or cysteine) peptidase inhibitor, clade a, member 1a | Serpina1a | < 0.001 | 2.5 | – | – | – | – |
| NM_009252 | Serine (or cysteine) peptidase inhibitor, clade a, member 3n | Serpina3n | < 0.001 | 2.2 | < 0.001 | 2.2 | 0.01 | 1.6 |
“–” indicates that gene in that specific time-point did not pass the selection criteria. h — hour; d — day.
The rest of the enriched GO processes were unique for each time-point, indicating very specific time-dependent transitions in the genes that were differentially expressed. With over 900 differentially expressed genes, the 3 h post-exposure time-point exhibited more than ten uniquely enriched GO processes, including intracellular signaling cascade [GO:0007242], regulation of cell proliferation [GO:0042127], regulation of apoptosis [GO:0042981], and response to oxidative stress [GO:0006979] (Fig. 4a). Enrichment of these GO processes indicates early defense responses that are occurring in response to the initial particle instillation and damage inflicted by high levels of particles in the respiratory system within 3 h of the CBNPs exposure.
The 2 and 3 d post-exposure time-points had five common GO processes significantly enriched: cell cycle [GO:0007049], cell division [GO:0051301], mitotic cell cycle [GO:0000278], organelle fission [GO:0048285], and chromosome segregation [GO:0007059] (Fig. 4a). Additionally, there were a number of perturbed processes that were unique to 2 d post-exposure including: DNA metabolic process [GO:0006259], chromosome organization [GO:0051276], and response to DNA damage stimulus [GO:0006974].
The 4 d time-point exhibited the most unique profile in terms of perturbed biological functions. All of the GO processes that were altered at this time-point were related to muscle contraction and development (Fig. 4a), indicating an alteration in lung muscle regulation. Finally, a number of unique GO processes were significantly enriched at the 42 d post-exposure time-point including positive regulation of cell activation [GO:0050867], regulation of vesicle-mediated transport [GO:0060627], myeloid leukocyte activation [GO:0002274], and regulation of phagocytosis [GO:0050764]. These enriched processes suggest re-activation of immune-inflammatory response after the 14 d time-point. Moreover, a few other GO processes that were enriched at this time-point, for example mast cell activation [GO:0045576] and regulation of hypersensitivity [GO:0002883], suggest possible onset of allergic airway inflammation at 42 d after CBNP exposure (Fig. 4a).
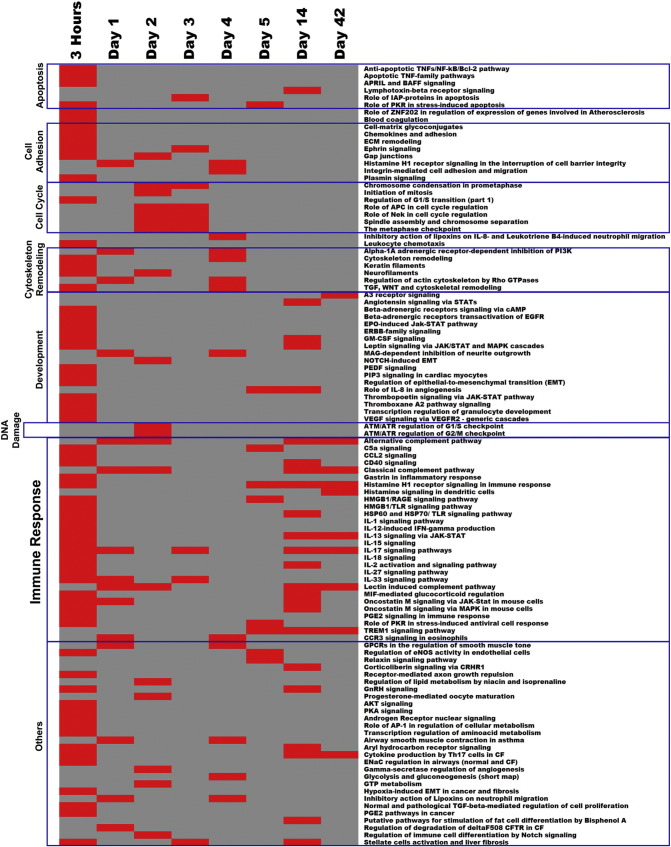
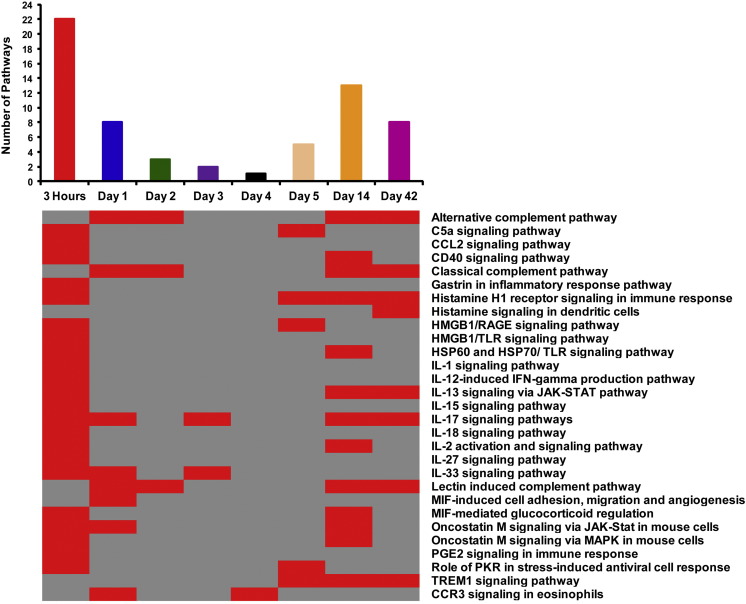
3.6. Biological pathways perturbed in response to CBNP exposure
Out of a total of 109 significantly enriched pathways, 29 were related to immune-inflammatory response (Fig. 5 ). Among the 29 immune-inflammatory pathways, 22 were enriched 3 h post-exposure, with the number of enriched pathways decreasing up to the 4 d post-exposure time-point, where only one immune-inflammatory response pathway was enriched (CCR3 signaling in eosinophils pathway). Beyond 5 d, the number of enriched immune-responsive pathways then began to increase again, and many enriched pathways were found 42 d post-exposure (Fig. 6 ). Perturbation of these pathways is consistent with our GO analysis and further strengthens the argument of induction of a biphasic inflammatory response following CBNP exposure (Fig. 6). Additionally, we observed enrichment of pathways related to apoptosis and survival, cell adhesion, and development at 3 h post-exposure; pathways related to cell cycle were perturbed 2 and 3 d post-exposure, whereas two pathways related to DNA damage were perturbed only 2 d post-exposure (Fig. 5). Pathway data were also suggestive of a possible interruption of cell cycle and DNA damage at intermediate (2 and 3 d) post-exposure time-points.
Fig. 5.
Biological pathways that were significantly enriched at each CBNP post-exposure time-point.
Fig. 6.
Biphasic enrichment pattern of immune-inflammatory pathways following CBNP exposure in the lungs of C57BL/6 mice. The majority of these inflammatory pathways were enriched 3 h, 1, 2, 14, and 42 d post-exposure.
3.7. Validation microarray data by RT-qPCR
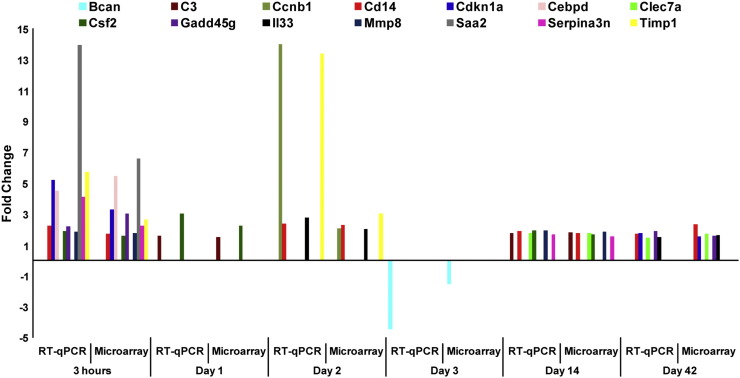
A total of 18 representative genes (Atp2a1, Bcan, C3, Ccnb1, Cd14, Cdkn1a, Cebpd, Clec7a, Csf2, Gadd45g, Il1b, Il33, Mmp8, Myh2, Nfkbia (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha), Saa2, Serpina3n, Timp1) from the microarray experiment were selected for validation by RT-qPCR. We primarily focused on the biological functions and pathways significantly enriched in response to CBNP exposure in the selection of genes for validation. Out of 18 genes, 14 genes (Bcan, C3, Ccnb1, Cd14, Cdkn1a, Cebpd, Clec7a, Csf2, Gadd45g, Il33, Mmp8, Saa2, Serpina3n, Timp1) had the same directional fold changes by RT-qPCR as microarrays (1.5 fold; p < 0.05; Fig. 7 and Table S5). Genes that were selected from 4 and 5 d post-exposure time-points (Atp2a1, Il1b, Myh2, and Nfkbia) did not achieve 1.5 fold and p < 0.05 values; lack of consistency across the platforms may be due to differences in the probe versus primer sequences, quantification approaches, and data analysis for microarrays versus RT-qPCR.
Fig. 7.
Genes identified as significantly differentially expressed using DNA microarrays validated by RT-qPCR.
3.8. Dataset and disease prediction by NextBio analysis
We examined the top 50 datasets that were the most highly correlated with CBNP exposed mouse lungs from a NextBio meta-analysis. Interestingly, the datasets with the highest scores (an indication of high correlation) were from studies related to mouse lung tissues exposed either to cigarette smoke or viruses (e.g., influenza or SARS virus; Table S6). Gene sets that were differentially expressed in response to cigarette smoke or viruses were highly correlated with CBNP-induced profiles at all time-points except 4 and 5 d. Select representative genes from this association are summarized in Table 5 .
Table 5.
Genes differentially expressed in response to CBNP exposure that were similarly altered in response to cigarette smoke or viral exposure.
References related to cigarette smoke (Thomson et al., 2013, Tilton et al., 2013) and virus response studies (Josset et al., 2012).
In order to predict the potential relevance of CBNP exposures to human diseases, we used the NextBio Human Disease Atlas. This analysis revealed that severe acute respiratory syndrome (SARS) was the disease profile with the strongest correlation with gene expression profiles of CBNP exposed mice across all time-points (Table S7). Additionally, other viral and bacterial infectious diseases, parasitic diseases, and respiratory disorders scored high in this analysis at all time-points, indicating that gene expression following CBNP exposure exhibits some similarity to gene expression patterns induced during viral or bacterial infectious diseases.
4. Discussion
Previously we reported that pulmonary CBNP exposure induces increased neutrophil influx, DNA strand breaks, and oxidative stress related DNA lesions in lung tissue (Saber et al., 2005, Jacobsen et al., 2009, Bourdon et al., 2012a, Bourdon et al., 2012c, Kyjovska et al., 2015b). The inflammatory response following intratracheal instillation of CBNPs was dominated by neutrophils, and gene expression analysis revealed substantive immune, inflammatory, and acute phase responses, as well as pathways signaling oxidative stress and DNA damage that persisted up to 28 d following exposure of mice to 162 μg CBNPs. The results of these studies (Bourdon et al., 2012a, Bourdon et al., 2012c) suggested significant changes in the pulmonary response over time and especially in the intermediate time window (within 3 d). In the present study we provide more detailed insight into the temporal sequence of pulmonary gene expression changes that occur following CBNP exposure and the persistence of these effects. The use of intratracheal instillation allowed the delivery of one bolus dose for resolution of a highly time-dependent pulmonary response. We demonstrate significant changes in the pulmonary response over time, with major events occurring immediately post-exposure (3 h), but with some molecular changes persisting up to 42 d post-exposure following a single intratracheal instillation. Our findings support immediate early initiation of oxidative stress, DNA damage, and apoptosis following CBNP exposure, leading to changes in cellular division/replication, consistent with mounting an inflammatory response, and prolonged inflammation to 42 d post-exposure. We observed a biphasic response in gene expression changes, but not BAL cell counts, which was also observed to a lesser extent in our previous investigations (Saber et al., 2006, Bourdon et al., 2012a). Our exposure (162 μg CBNPs/mouse) is equivalent to the cumulative dose expected in nine 8-h working days at the current Danish occupational exposure limit based on observed particle size distribution during aerosolization of CBNPs (Jackson et al., 2012a). A recent study on workers exposed to CBNPs reported exposure levels of 14.90 mg/m3 in the inhalation zone (Zhang et al., 2014); the dose used in the present study corresponds roughly to the pulmonary deposition during two 8-h working days at 15 mg/m3.
The inflammatory BAL profiles observed in this study were indicative of both acute and chronic inflammation, and were mainly neutrophil driven. Inflammatory BAL cell count showed that neutrophil infiltration was initiated as soon as 3 h post-exposure and remained elevated until 42 d post-exposure. Elevation in neutrophil count was accompanied by an increase in macrophage counts on 4, 5, and 42 d in the CBNP exposed mice, with the highest numbers observed 42 d post-exposure; furthermore, a brief surge in lymphocytes was observed 4 and 14 d post-exposure. Our gene expression, GO, and pathway data showed significant enrichment of acute phase response, and several immune-inflammatory GO processes and pathways at early and late time-points that suggest the presence of a biphasic inflammatory response that was initiated almost immediately following CBNP exposure. We observed significant up-regulation of genes associated with chemokines and their receptors, cytokines and their receptors, complement factors, acute phase genes, TNF superfamily members, and other pro-inflammatory genes (e.g., Csf2, Orm2 (Orosomucoid 2), Timp1, Nfkbia, Vnn1, Vnn3) at multiple post-exposure time-points. Serum amyloid A (Saa1, Saa2, and Saa3), macrophage inflammatory protein-2 (Mip-2, also known as Cxcl2), interleukin-6 (Il-6), and macrophages/monocyte chemoattractant protein-1 (Mcp-1, also known as Ccl2) are biomarkers of chemotactic recruitment of inflammatory cells including neutrophils and induction of inflammation following CBNP exposure (Jacobsen et al., 2009, Saber et al., 2014). In a previous study, Jacobsen et al. (2009) reported significant up-regulation of Il6, Mip-2 (Cxcl2), and Mcp-1 (Ccl2) 3 h and 1 d post-intratracheal instillation of CBNPs in ApoE−/− and C57BL/6 mice. In this study, we observed significant up-regulation of Il6 (4.4 fold; p < 0.0001) at 3 h, Cxcl2 (2.0 fold; p < 0.005) at 1 d, and Ccl2 at 3 h (3.3 fold; p < 0.0001) and 1 d (2.0 fold; p < 0.05) post-exposure. Additionally, significant up-regulation of Ccl2 was observed 2 d (3.0 fold; p < 0.001), 3 d (2.0 fold; p < 0.05), 14 d (2.0 fold; p < 0.01), and 42 d (2.0 fold; p < 0.01) post-exposure. Increased expression of several neutrophil chemoattractants (e.g., Saa1, Saa3, Il6 and Ccl2) in our study is in agreement with the previous study, and may explain the recruitment and presence of pro-inflammatory cells in the lung of CBNP exposed mice.
Although acute inflammation and neutrophil influx are considered to be protective, uncontrolled or chronic neutrophil influx caused by persistent exposure to toxicants, such as insoluble particles, can lead to pulmonary injury and may result in acute lung injury and acute respiratory distress syndrome, two life-threatening conditions (Moldoveanu et al., 2009, Konrad and Reutershan, 2012). Tissue injury following excessive neutrophil activation is well documented in major organs including the respiratory, cardiovascular, gastrointestinal, genitourinary, and musculoskeletal systems, among which the respiratory system is the most susceptible, and may lead to the development of chronic obstructive pulmonary disease (COPD), asthma, or cystic fibrosis (Tintinger et al., 2005). The acute phase response, which is shown to occur here after pulmonary exposure to different types of particles and nanomaterials (Bourdon et al., 2012a, Saber et al., 2013, Saber et al., 2014, Kyjovska et al., 2015a, Kyjovska et al., 2015b), is the likely cause of this intense neutrophil recruitment and is implicated in health effects that extend beyond the respiratory system, such as adverse cardiovascular effects including systemic inflammation and alterations in cholesterol homeostasis, and increased plaque progression (Mikkelsen et al., 2011, Bourdon et al., 2012a, Saber et al., 2013, Saber et al., 2014, Poulsen et al., 2015). We note that increased levels of acute phase proteins including SAA are associated with risk of cardiovascular disease in prospective epidemiological studies (Ridker et al., 2000). Thus, the data from our study and from previous work suggest long-term systemic complications following CBNP exposure.
Chronic inflammation can occur when there is an incomplete resolution of acute inflammation caused by excessive tissue damage or particles that cannot be removed from the lungs. The major cells involved in this process are macrophages and lymphocytes (Moldoveanu et al., 2009). Alveolar macrophages are the resident macrophages in the alveoli that account for 95% of the macrophage population and provide protection against pathogens and exogenous materials by phagocytosis; however, prolonged surges in macrophages are indicative of loss of pulmonary homeostasis and may be implicated in multiple chronic pulmonary diseases including asthma and chronic COPD (Moldoveanu et al., 2009, Scherbart et al., 2011, Duan et al., 2012). We observed a significant surge in macrophage counts starting from 4 d post-exposure and continuing until 42 d in our study, and elevation of lymphocyte counts 14 d post-exposure, which are in agreement with established mechanisms for chronic inflammation (Moldoveanu et al., 2009). A number of previous studies have suggested that exposure to nanoparticles results in internalization and deposition of the particles deeper into the lung tissue that may induce prolonged inflammatory response. Results from a recent study by Zhang et al. (2014) on workers exposed to carbon black, which was validated by a mouse model study, showed that CBNPs are deposited in lung tissue following inhalation exposure and may contribute to long-term inflammation and lung diseases (Zhang et al., 2014). In another study using nano-scale hyperspectral microscope, intratracheally instilled titanium dioxide nanoparticles (nano-TiO2) were shown to evade particle clearing mechanisms and deposit in the lung, thus inducing long-lasting pulmonary inflammation and translocation of nanoparticles to the blood, heart, and liver (Husain et al., 2013, Husain et al., 2015). In fact, it has been established in several studies that nanosized particles are not readily recognized by macrophages, and thus their clearance is hampered (Ferin et al., 1992, Katsnelson et al., 2010). This may lead to accumulation of CBNPs within the lung, sustained inflammation, development of granulomas, tissue damage, and eventually pulmonary disease (Heinrich et al., 1995, Driscoll et al., 1996, Bermudez et al., 2002, Bermudez et al., 2004, Carter et al., 2006). At the later time-points in this study, we observed black matter in macrophages, but there was no evidence of free particles in the BAL fluid, which indicates that CBNPs were likely engulfed by macrophages at these time-points (data not shown). However, we did not quantify CBNP retention in the lungs. In previous studies we assessed lung burden following inhalation exposure to 42 mg/m3 of nano-TiO2 (primary size 21 nm) for 1 h daily for 11 days, which resulted in a similar estimated pulmonary deposited dose as the present study (Hougaard et al., 2010, Hougaard et al., 2011). Five days following exposure, 24% of the estimated deposited dose was still present in the lungs, indicating some particle clearance; whereas, 21% of the estimated deposited dose was still present 26–27 d post-exposure, indicating prolonged pulmonary particle retention (Hougaard et al., 2010, Hougaard et al., 2011). The prolonged particle retention was accompanied by increased neutrophil influx at 26–27 d post-exposure (Hougaard et al., 2010, Hougaard et al., 2011).
One interesting observation from our gene expression data was the absence of inflammatory response 4 d post-exposure. Functional analysis of gene expression changes revealed that inflammation-associated gene expression changes were resolved by 3 d post-exposure, with little to no inflammatory response 4 d post-exposure observed by gene expression profiling despite the BAL cell counts showing significant increases in neutrophil, macrophages, eosinophils, and lymphocyte numbers. Inflammatory gene expression increased again at later time-points (e.g., 14 and 42 d post-exposure). Genes affected 4 d post-exposure were mainly involved in muscle regulatory processes; several muscle contraction and regulation related GO processes and pathways were perturbed at this time-point, which indicate a possible change in the smooth muscle regulatory process in the lungs. CBNPs are known to affect lung structure and can cause destruction of lung tissue. In a study on Balb/c mice that were exposed to 30 to 50 nm carbon black particles by inhalation, Zhang et al. (2014) reported histopathological changes in the lungs of exposed mice. Seven and 14 d following exposure, these authors observed thickening of the alveolar wall, disarrangement, and swelling of some cells with vacuolar degeneration in the experimental groups compared to the control groups (Zhang et al., 2014). Our GO and pathway data from the 4 d post-exposure time-point indicate similar changes that may lead to cytoskeleton remodeling and alteration in the alveolar architecture.
Exposure to CBNPs is known to cause oxidative stress and cell death, which is supported by a number of in vitro studies (Hussain et al., 2010, Reisetter et al., 2011) and is evident from gene expression profiles at the earliest time-point in the present study. Cell death may occur in several ways (e.g., necrosis, apoptosis, and pyroptosis). Apoptosis plays an important role in many cellular processes, and is mainly characterized by the shrinking of cells (Hussain et al., 2010, Reisetter et al., 2011). Decreased mitochondrial membrane potential, increased production of reactive oxygen species (ROS), and DNA strand breaks have been implicated in CBNP-induced apoptosis (Hussain et al., 2010). Although apoptosis is necessary for controlling inflammation and clearing inflammatory cells, brief or incomplete apoptosis, which could occur due to abnormal functioning of inducer or suppressor genes, may lead to prolonged infiltration of inflammatory cells in the lungs and may play a central role in various chronic lung inflammatory conditions, including asthma and acute respiratory distress syndrome (Moldoveanu et al., 2009, Hussain et al., 2010, Reisetter et al., 2011). Pro-inflammatory cytokines (e.g., GM–CSF) may be responsible for delaying the process of apoptosis (Moldoveanu et al., 2009). Exposure to CBNPs causes an inflammasome-dependent form of cell death known as pyroptosis in macrophages (Reisetter et al., 2011). Pyroptosis shares characteristics with both necrosis and apoptosis; however, inflammatory in nature and unlike apoptosis, it causes swelling of cells by fluid influx that leads to loss of membrane integrity and results in the release of inflammatory contents of the damaged cells (Reisetter et al., 2011). Interestingly, our GO and pathway analysis indicates significant enrichment of oxidative stress, cell proliferation, and apoptosis regulatory processes mainly at the 3 h post-exposure time-point. Differential expression of numerous genes, including several mitochondrial genes, were consistent with this: Thioredoxin interacting protein (Txnip), Glutamate–cysteine ligase, catalytic subunit (Gclc), Hypoxia inducible factor 1, alpha subunit (Hif1a), Uncoupling protein 3 (mitochondrial, proton carrier) (Ucp3), Uncoupling protein 2 (mitochondrial, proton carrier) (Ucp2), Prostaglandin-endoperoxide synthase 2 (Ptgs2), Clusterin (Clu), Isocitrate dehydrogenase 1 (NADP +) soluble (Idh1), Thioredoxin reductase 1 (Txnrd1), Cytoglobin (Cygb), Tyrosine aminotransferase (Tat), and Sulfiredoxin 1 homolog (Srxn1). We also observed transcriptional changes in several pro-inflammatory genes (colony stimulating factor 2, granulocyte-macrophage) and pro-apoptotic genes (Casp3, Casp4, and Casp8). Therefore, it is reasonable to argue that our gene expression data provide evidence of oxidative stress, cell death, and imbalance of systemic homeostasis in the lungs of C57BL/6 mice following this single exposure to CBNPs that resulted in the observed prolonged pulmonary inflammation.
CBNP exposure is known to cause DNA damage. In our previous studies, increased DNA strand breaks were observed in the lungs of C57BL/6 mice following 0.7, 2, 6, 18, 54, and 162 μg CBNP exposure at various time-points relative to control lungs; although we consistently observed 50–100% increases in %TDNA, no dose–response relationship was observed (Bourdon et al., 2012c, Kyjovska et al., 2015a). In this study, we found significantly increased levels of DNA strand breaks in BAL cells after 3 h, and in lung tissues 2–5 d post-exposure. Interestingly, we also observed increased levels of DNA strand breaks in liver tissue 3 h post-exposure; this phenomenon could be due to translocation of CBNPs from the lungs to other secondary organs. We recently reported translocation of low levels of nano-TiO2 from the lungs to the blood, heart, and liver 24 h after the exposure (Husain et al., 2015); such observations support the possibility of translocation of CBNPs from lung to liver. Alternatively, DNA damage in the liver could result from circulating second messengers (e.g., acute phase proteins, cytokines, and chemokines) that were differentially expressed in the lungs at those respective time-points (Jackson et al., 2012a).
We previously showed that CBNPs induce ROS formation both in acellular and cellular assays, and generate oxidative lesions in DNA in vitro and in lung tissue in vivo; the spectrum of the induced mutations indicate ROS as a possible effector (Jacobsen et al., 2007, Jacobsen et al., 2008, Jacobsen et al., 2011, Bourdon et al., 2012c). Gene expression data in the present study suggest that CBNP exposure induces oxidative stress and causes DNA strand breaks at the early post-exposure time-points, which may then lead to inhibition of cell growth relatively early after the exposure in the lungs of C57BL/6 mice. CBNP-induced gene expression profiles were highly correlated with expression profiles derived from tissues and cell culture models treated with a cyclin-dependent kinase inhibitor (CDKI), cigarette smoke, formaldehyde, and tumor necrosis factor (TNFα), in particular from cultured epithelial cells and ApoE−/− mouse tissues (Thomson et al., 2013) (Table S6). Several networks and pathways, including pulmonary inflammation, cell cycle arrest, necroptosis, apoptosis, and DNA damage that were perturbed in response to CDKI, cigarette smoke, and formaldehyde exposure have been identified (Thomson et al., 2013). The high similarity of our expression profiles with these published data supports the notion that CBNP exposure may induce long-term detrimental effects in the pulmonary system, similar to effects caused by smoke or formaldehyde inhalation.
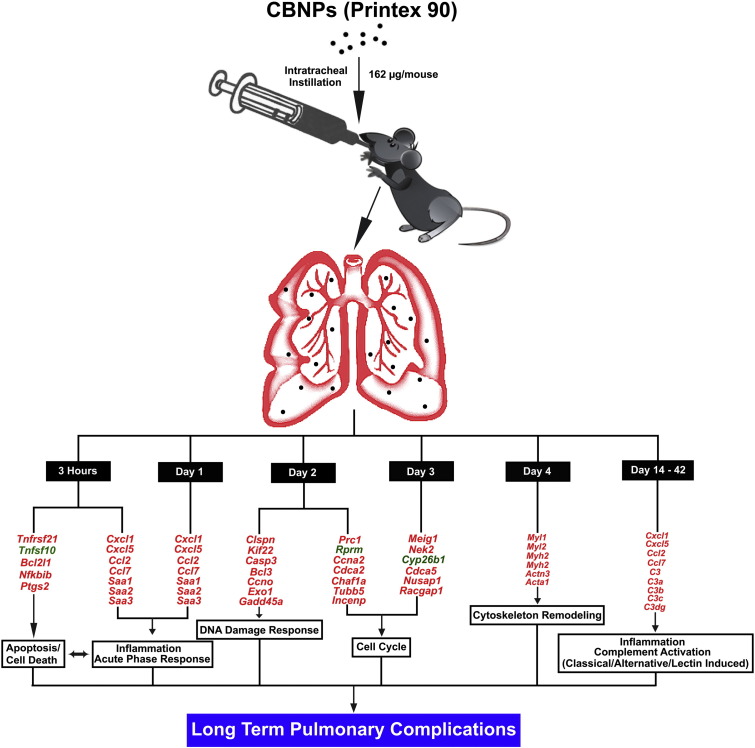
The biphasic nature of the response that we observed suggests that the possible consequences of CBNP pulmonary exposure are multifold. The first level of response is an acute inflammatory response, driven by neutrophil influx and acute phase responses, which is immediately followed by the induction of oxidative stress, DNA strand breaks, cell cycle arrest, and cell death (apoptosis, necrosis, and pyroptosis). These immediate responses in the initial post-exposure days may then facilitate the worsening of pulmonary homeostasis (Fig. 8 ). We speculate that all of these dysfunctions modulate the development of a secondary inflammatory response at the later time-points, which may ultimately lead to multiple chronic pulmonary inflammatory processes. Thus, the results support a pivotal transitional time period around 4–5 d post-exposure, during which there is a shift between the immediate responses, and sustained effects resulting from particle retention and pathological changes. Instillation or aspiration of a single dose in toxicological studies is considered more economic and safer alternatives to 28 or 90-day inhalation studies. Our time series study suggests that the biological responses following pulmonary particle exposures evolve over time, and can be effectively tracked using single-exposure study designs. During the continuous exposures, the transcriptional responses will likely be a mixture of these time-dependent responses. The present time-series study suggests that for early time-points, the transcriptional responses following exposure to a single dose will likely differ significantly from those observed following continuous exposures, whereas the long-term responses are more likely to be similar, since the transcriptional responses at 14 and 42 d following a single exposure were somewhat similar (Fig. 5, Fig. 6).
Fig. 8.
Schematic representation of genes and biological processes/pathways induced by CBNP exposure at various post-exposure time-points, and their possible long-term consequences. Genes represented in red were up-regulated and those in green were down-regulated.
The primary goal of this study was to assess the temporal series of molecular events leading to sustained effects following acute pulmonary exposure to nanoparticles. Overall, the results provide a thorough description of the inflammatory, DNA damaging, and transcriptional events that occur over post-CBNP exposure times. The data clearly demonstrate a biphasic response in gene expression changes, and persistence of these effects up to 42 d post-exposure in the lungs of C57BL/6 mice. The results of the current study on female mice agree well with previously published data on inflammation performed only in male mice (Zhang et al., 2014). The experimental design used addresses the need for temporal data to produce adverse outcome pathways linking diverse molecular perturbations to pulmonary diseases driven by sustained inflammation.
Funding
This work was supported by Health Canada's Genomics Research and Development Initiative, the Chemicals Management Plan-Nano, and the Danish Working Environment Fund (Danish Centre for Nanosafety, grant# 20110092173-3).
Conflicts of interest
All authors, except Dr. Ulla Vogel and Professor Håkan Wallin, disclose that there is no actual or potential conflict of interests. Dr. Ulla Vogel (Danish Centre for Nanosafety, grant# 20110092173-3) and Professor Håkan Wallin disclose that they received grants from the Danish Work Environment Fund and that Professor Wallin is an employee of the National Research Centre for the Working Environment, Copenhagen, Denmark (Institute under the Ministry of Employment Danish Government.
The following are the supplementary data related to this article.
Fig. S1 Total protein level in BAL fluid following CBNP exposure. Statistical significance compared to vehicle controls is indicated by **p < 0.01 and ***p < 0.001, respectively.
Fig. S2 DNA strand break in the liver expressed as tail length (TL) and %TDNA. Statistical significance compared to vehicle controls is indicated by *p < 0.05 and **p < 0.01, respectively.
Genes differentially expressed in response to CBNP exposure.
Genes commonly expressed across the time-points.
Table S3 List of lincRNAs differentially expressed in response to CBNP exposure.
Table S4 Genes involved in immune-inflammatory response GO processes.
Table S5 Genes from the microarray experiment selected for validation by RT-qPCR.
Commonality between CBNP datasets and datasets in the NextBio repository. Top 50 datasets correlated with data from CBNP post-exposure time-points.
Table S7 Disease prediction from NextBio repository.
Transparency document
Transparency document
Acknowledgements
The authors thank Dr. Azam F. Tayabali and Dr. Hongyan Dong for their valuable comments on this manuscript.
Footnotes
The Transparency document associated with this article can be found, in the online version.
References
- Bermudez E., Mangum J.B., Asgharian B., Wong B.A., Reverdy E.E., Janszen D.B., Hext P.M., Warheit D.B., Everitt J.I. Long-term pulmonary responses of three laboratory rodent species to subchronic inhalation of pigmentary titanium dioxide particles. Toxicol. Sci. 2002;70:86–97. doi: 10.1093/toxsci/70.1.86. [DOI] [PubMed] [Google Scholar]
- Bermudez E., Mangum J.B., Wong B.A., Asgharian B., Hext P.M., Warheit D.B., Everitt J.I. Pulmonary responses of mice, rats, and hamsters to subchronic inhalation of ultrafine titanium dioxide particles. Toxicol. Sci. 2004;77:347–357. doi: 10.1093/toxsci/kfh019. [DOI] [PubMed] [Google Scholar]
- Bourdon J.A., Halappanavar S., Saber A.T., Jacobsen N.R., Williams A., Wallin H., Vogel U., Yauk C.L. Hepatic and pulmonary toxicogenomic profiles in mice intratracheally instilled with carbon black nanoparticles reveal pulmonary inflammation, acute phase response, and alterations in lipid homeostasis. Toxicol. Sci. 2012;127:474–484. doi: 10.1093/toxsci/kfs119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon J.A., Saber A.T., Halappanavar S., Jackson P.A., Wu D., Hougaard K.S., Jacobsen N.R., Williams A., Vogel U., Wallin H., Yauk C.L. Carbon black nanoparticle intratracheal installation results in large and sustained changes in the expression of miR-135b in mouse lung. Environ. Mol. Mutagen. 2012;53:462–468. doi: 10.1002/em.21706. [DOI] [PubMed] [Google Scholar]
- Bourdon, J. A., Saber, A. T., Jacobsen, N. R., Jensen, K. A., Madsen, A. M., Lamson, J. S., Wallin, H., Moller, P., Loft, S., Yauk, C. L., and Vogel, U. B. (2012c). Carbon black nanoparticle instillation induces sustained inflammation and genotoxicity in mouse lung and liver. Part. Fibre Toxicol. 9, 5-8977-8979-8975. [DOI] [PMC free article] [PubMed]
- Brain J.D., Knudson D.E., Sorokin S.P., Davis M.A. Pulmonary distribution of particles given by intratracheal instillation or by aerosol inhalation. Environ. Res. 1976;11:13–33. doi: 10.1016/0013-9351(76)90107-9. [DOI] [PubMed] [Google Scholar]
- Carter J.M., Corson N., Driscoll K.E., Elder A., Finkelstein J.N., Harkema J.N., Gelein R., Wade-Mercer P., Nguyen K., Oberdorster G. A comparative dose-related response of several key pro- and antiinflammatory mediators in the lungs of rats, mice, and hamsters after subchronic inhalation of carbon black. J. Occup. Environ. Med. 2006;48:1265–1278. doi: 10.1097/01.jom.0000230489.06025.14. [DOI] [PubMed] [Google Scholar]
- Cleveland W.S. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 1979;74:829–836. [Google Scholar]
- Cui X., Qiu J.T.H.J., Blades N.J., Churchill G.A. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6:59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- Driscoll K.E., Carter J.M., Howard B.W., Hassenbein D.G., Pepelko W., Baggs R.B., Oberdorster G. Pulmonary inflammatory, chemokine, and mutagenic responses in rats after subchronic inhalation of carbon black. Toxicol. Appl. Pharmacol. 1996;136:372–380. doi: 10.1006/taap.1996.0045. [DOI] [PubMed] [Google Scholar]
- Duan M., Li W.C., Vlahos R., Maxwell M.J., Anderson G.P., Hibbs M.L. Distinct macrophage subpopulations characterize acute infection and chronic inflammatory lung disease. J. Immunol. 2012;189:946–955. doi: 10.4049/jimmunol.1200660. [DOI] [PubMed] [Google Scholar]
- Ferin J., Oberdorster G., Penney D.P. Pulmonary retention of ultrafine and fine particles in rats. Am. J. Respir. Cell Mol. Biol. 1992;6:535–542. doi: 10.1165/ajrcmb/6.5.535. [DOI] [PubMed] [Google Scholar]
- Heinrich U., Fuhst R., Rittinghausen S., Creutzenberg O., Bellmann B., Koch W., Levsen K. Chronic inhalation exposure of wistar rats and two different strains of mice to diesel engine exhaust, carbon black, and titanium dioxide. Inhal. Toxicol. 1995;7:533–556. [Google Scholar]
- Hougaard K.S., Jackson P., Jensen K.A., Sloth J.J., Loschner K., Larsen E.H., Birkedal R.K., Vibenholt A., Boisen A.M., Wallin H., Vogel U. Effects of prenatal exposure to surface-coated nanosized titanium dioxide (UV-Titan). A study in mice. Part. Fibre Toxicol. 2010;7(16) doi: 10.1186/1743-8977-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hougaard K.S., Jackson P., Jensen K.A., Sloth J.J., Löschner K., Larsen E.H., Birkedal R.K., Vibenholt A., Boisen A.Z., Wallin H., Vogel U. Correction: effects of prenatal exposure to surface-coated nanosized titanium dioxide (UV-Titan). A study in mice. Part. Fibre Toxicol. 2011;8(14) doi: 10.1186/1743-8977-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Husain M., Saber A.T., Guo C., Jacobsen N.R., Jensen K.A., Yauk C.L., Williams A., Vogel U., Wallin H., Halappanavar S. Pulmonary instillation of low doses of titanium dioxide nanoparticles in mice leads to particle retention and gene expression changes in the absence of inflammation. Toxicol. Appl. Pharmacol. 2013;269:250–262. doi: 10.1016/j.taap.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Husain M., Wu D., Saber A.T., Decan N., Jacobsen N.R., Williams A., Yauk C.L., Wallin H., Vogel U., Halappanavar S. Intratracheally instilled titanium dioxide nanoparticles translocate to heart and liver and activate complement cascade in the heart of C57BL/6 mice. Nanotoxicology. 2015;1-10 doi: 10.3109/17435390.2014.996192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S., Thomassen L.C.J., Ferecatu I., Borot M.C., Andreau K., Martens J.A., Fleury J., Baeza-Squiban A., Marano F., Boland S. Carbon black and titanium dioxide nanoparticles elicit distinct apoptotic pathways in bronchial epithelial cells. Part. Fibre Toxicol. 2010;7 doi: 10.1186/1743-8977-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans/World Health Organization, International Agency for Research on Cancer. Vol. 93. 2010. IARC monographs on the evaluation of carcinogenic risks to humans: carbon black, titanium dioxide, and talc; pp. 1–413. [PMC free article] [PubMed] [Google Scholar]
- Jackson P., Lund S.P., Kristiansen G., Andersen O., Vogel U., Wallin H., Hougaard K.S. An experimental protocol for maternal pulmonary exposure in developmental toxicology. Basic Clin. Pharmacol. Toxicol. 2011;108:202–207. doi: 10.1111/j.1742-7843.2010.00644.x. [DOI] [PubMed] [Google Scholar]
- Jackson P., Hougaard K.S., Boisen A.M., Jacobsen N.R., Jensen K.A., Moller P., Brunborg G., Gutzkow K.B., Andersen O., Loft S., Vogel U., Wallin H. Pulmonary exposure to carbon black by inhalation or instillation in pregnant mice: effects on liver DNA strand breaks in dams and offspring. Nanotoxicology. 2012;6:486–500. doi: 10.3109/17435390.2011.587902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P., Hougaard K.S., Vogel U., Wu D., Casavant L., Williams A., Wade M., Yauk C.L., Wallin H., Halappanavar S. Exposure of pregnant mice to carbon black by intratracheal instillation: toxicogenomic effects in dams and offspring. Mutat. Res. 2012;745:73–83. doi: 10.1016/j.mrgentox.2011.09.018. [DOI] [PubMed] [Google Scholar]
- Jackson P., Pedersen L.M., Kyjovska Z.O., Jacobsen N.R., Saber A.T., Hougaard K.S., Vogel U., Wallin H. Validation of freezing tissues and cells for analysis of DNA strand break levels by comet assay. Mutagenesis. 2013;28:699–707. doi: 10.1093/mutage/get049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen N.R., Saber A.T., White P., Moller P., Pojana G., Vogel U., Loft S., Gingerich J., Soper L., Douglas G.R., Wallin H. Increased mutant frequency by carbon black, but not quartz, in the lacZ and cII transgenes of muta mouse lung epithelial cells. Environ. Mol. Mutagen. 2007;48:451–461. doi: 10.1002/em.20300. [DOI] [PubMed] [Google Scholar]
- Jacobsen N.R., Pojana G., White P., Moller P., Cohn C.A., Korsholm K.S., Vogel U., Marcomini A., Loft S., Wallin H. Genotoxicity, cytotoxicity, and reactive oxygen species induced by single-walled carbon nanotubes and C(60) fullerenes in the FE1-mutatrade markMouse lung epithelial cells. Environ. Mol. Mutagen. 2008;49:476–487. doi: 10.1002/em.20406. [DOI] [PubMed] [Google Scholar]
- Jacobsen N.R., Moller P., Jensen K.A., Vogel U., Ladefoged O., Loft S., Wallin H. Lung inflammation and genotoxicity following pulmonary exposure to nanoparticles in ApoE −/− mice. Part. Fibre Toxicol. 2009;6:2. doi: 10.1186/1743-8977-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen N.R., White P.A., Gingerich J., Moller P., Saber A.T., Douglas G.R., Vogel U., Wallin H. Mutation spectrum in FE1-MUTA(TM) mouse lung epithelial cells exposed to nanoparticulate carbon black. Environ. Mol. Mutagen. 2011;52:331–337. doi: 10.1002/em.20629. [DOI] [PubMed] [Google Scholar]
- Josset L., Belser J.A., Pantin-Jackwood M.J., Chang J.H., Chang S.T., Belisle S.E., Tumpey T.M., Katze M.G. Implication of inflammatory macrophages, nuclear receptors, and interferon regulatory factors in increased virulence of pandemic 2009 H1N1 influenza A virus after host adaptation. J. Virol. 2012;86:7192–7206. doi: 10.1128/JVI.00563-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsnelson B., Privalova L.I., Kuzmin S.V., Degtyareva T.D., Sutunkova M.P., Yeremenko O.S., Minigalieva I.A., Kireyeva E.P., Khodos M.Y., Kozitsina A.N., Malakhova N.A., Glazyrina J.A., Shur V.Y., Shishkin E.I., Nikolaeva E.V. Some peculiarities of pulmonary clearance mechanisms in rats after intratracheal instillation of magnetite (Fe3O4) suspensions with different particle sizes in the nanometer and micrometer ranges: are we defenseless against nanoparticles? Int. J. Occup. Environ. Health. 2010;16:508–524. doi: 10.1179/107735210799160011. [DOI] [PubMed] [Google Scholar]
- Kerr M.K., Churchill G.A. Statistical design and the analysis of gene expression microarray data. Genet. Res. 2007;89:509–514. doi: 10.1017/S0016672308009713. [DOI] [PubMed] [Google Scholar]
- Konrad F.M., Reutershan J. CXCR2 in acute lung injury. Mediat. Inflamm. 2012;2012:740987. doi: 10.1155/2012/740987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupershmidt I., Su Q.J., Grewal A., Sundaresh S., Halperin I., Flynn J., Shekar M., Wang H., Park J., Cui W., Wall G.D., Wisotzkey R., Alag S., Akhtari S., Ronaghi M. Ontology-based meta-analysis of global collections of high-throughput public data. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyjovska Z.O., Jacobsen N.R., Saber A.T., Bengtson S., Jackson P., Wallin H., Vogel U. DNA damage following pulmonary exposure by instillation to low doses of carbon black (Printex 90) nanoparticles in mice. Environ. Mol. Mutagen. 2015;56:41–49. doi: 10.1002/em.21888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyjovska Z.O., Jacobsen N.R., Saber A.T., Bengtson S., Jackson P., Wallin H., Vogel U. DNA strand breaks, acute phase response and inflammation following pulmonary exposure by instillation to the diesel exhaust particle NIST1650b in mice. Mutagenesis. 2015;30(4):499–507. doi: 10.1093/mutage/gev009. [DOI] [PubMed] [Google Scholar]
- Levy L., Chaudhuri I.S., Krueger N., McCunney R.J. Does carbon black disaggregate in lung fluid? A critical assessment. Chem. Res. Toxicol. 2012;25:2001–2006. doi: 10.1021/tx300160z. [DOI] [PubMed] [Google Scholar]
- Mikkelsen L., Sheykhzade M., Jensen K.A., Saber A.T., Jacobsen N.R., Vogel U., Wallin H., Loft S., Moller P. Modest effect on plaque progression and vasodilatory function in atherosclerosis-prone mice exposed to nanosized TiO(2) Part. Fibre Toxicol. 2011;8:32. doi: 10.1186/1743-8977-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldoveanu B., Otmishi P., Jani P., Walker J., Sarmiento X., Guardiola J., Saad M., Yu J. Inflammatory mechanisms in the lung. J. Inflamm. Res. 2009;2:1–11. [PMC free article] [PubMed] [Google Scholar]
- Poulsen S.S., Saber A.T., Mortensen A., Szarek J., Wu D., Williams A., Andersen O., Jacobsen N.R., Yauk C.L., Wallin H., Halappanavar S., Vogel U. Changes in cholesterol homeostasis and acute phase response link pulmonary exposure to multi-walled carbon nanotubes to risk of cardiovascular disease. Toxicol. Appl. Pharmacol. 2015;283:210–222. doi: 10.1016/j.taap.2015.01.011. [DOI] [PubMed] [Google Scholar]
- Reisetter A.C., Stebounova L.V., Baltrusaitis J., Powers L., Gupta A., Grassian V.H., Monick M.M. Induction of inflammasome-dependent pyroptosis by carbon black nanoparticles. J. Biol. Chem. 2011;286:21844–21852. doi: 10.1074/jbc.M111.238519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker P.M., Hennekens C.H., Buring J.E., Rifai N. C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N. Engl. J. Med. 2000;342:836–843. doi: 10.1056/NEJM200003233421202. [DOI] [PubMed] [Google Scholar]
- Saber A.T., Bornholdt J., Dybdahl M., Sharma A.K., Loft S., Vogel U., Wallin H. Tumor necrosis factor is not required for particle-induced genotoxicity and pulmonary inflammation. Arch. Toxicol. 2005;79:177–182. doi: 10.1007/s00204-004-0613-9. [DOI] [PubMed] [Google Scholar]
- Saber A.T., Jacobsen N.R., Bornholdt J., Kjaer S.L., Dybdahl M., Risom L., Loft S., Vogel U., Wallin H. Cytokine expression in mice exposed to diesel exhaust particles by inhalation. Role of tumor necrosis factor. Part. Fibre Toxicol. 2006;3:4. doi: 10.1186/1743-8977-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saber A.T., Halappanavar S., Folkmann J.K., Bornholdt J., Boisen A.M., Moller P., Williams A., Yauk C., Vogel U., Loft S., Wallin H. Lack of acute phase response in the livers of mice exposed to diesel exhaust particles or carbon black by inhalation. Part. Fibre Toxicol. 2009;6:12. doi: 10.1186/1743-8977-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saber A.T., Jensen K.A., Jacobsen N.R., Birkedal R., Mikkelsen L., Moller P., Loft S., Wallin H., Vogel U. Inflammatory and genotoxic effects of nanoparticles designed for inclusion in paints and lacquers. Nanotoxicology. 2012;6:453–471. doi: 10.3109/17435390.2011.587900. [DOI] [PubMed] [Google Scholar]
- Saber A.T., Lamson J.S., Jacobsen N.R., Ravn-Haren G., Hougaard K.S., Nyendi A.N., Wahlberg P., Madsen A.M., Jackson P., Wallin H., Vogel U. Particle-induced pulmonary acute phase response correlates with neutrophil influx linking inhaled particles and cardiovascular risk. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saber A.T., Jacobsen N.R., Jackson P., Poulsen S.S., Kyjovska Z.O., Halappanavar S., Yauk C.L., Wallin H., Vogel U. Particle-induced pulmonary acute phase response may be the causal link between particle inhalation and cardiovascular disease. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2014;6:517–531. doi: 10.1002/wnan.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherbart A.M., Langer J., Bushmelev A., van Berlo D., Haberzettl P., van Schooten F.J., Schmidt A.M., Rose C.R., Schins R.P., Albrecht C. Contrasting macrophage activation by fine and ultrafine titanium dioxide particles is associated with different uptake mechanisms. Part. Fibre Toxicol. 2011;8:31. doi: 10.1186/1743-8977-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T.M., Sewer A., Martin F., Belcastro V., Frushour B.P., Gebel S., Park J., Schlage W.K., Talikka M., Vasilyev D.M., Westra J.W., Hoeng J., Peitsch M.C. Quantitative assessment of biological impact using transcriptomic data and mechanistic network models. Toxicol. Appl. Pharmacol. 2013;272:863–878. doi: 10.1016/j.taap.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Tilton S.C., Karin N.J., Webb-Robertson B.J., Waters K.M., Mikheev V., Lee K.M., Corley R.A., Pounds J.G., Bigelow D.J. Impaired transcriptional response of the murine heart to cigarette smoke in the setting of high fat diet and obesity. Chem. Res. Toxicol. 2013;26:1034–1042. doi: 10.1021/tx400078b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintinger G., Steel H.C., Anderson R. Taming the neutrophil: calcium clearance and influx mechanisms as novel targets for pharmacological control. Clin. Exp. Immunol. 2005;141:191–200. doi: 10.1111/j.1365-2249.2005.02800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Kerr K.M., Cui X., Churchill G.A. 2003. MAANOVA: A Software Package for the Analysis of Spotted cDNA Microarray Experiments. Statistics for Biology and Health: The Analysis of Gene Expression Data; pp. 313–341. [Google Scholar]
- Zhang R., Dai Y., Zhang X., Niu Y., Meng T., Li Y., Duan H., Bin P., Ye M., Jia X., Shen M., Yu S., Yang X., Gao W., Zheng Y. Reduced pulmonary function and increased pro-inflammatory cytokines in nanoscale carbon black-exposed workers. Part. Fibre Toxicol. 2014;11:73. doi: 10.1186/s12989-014-0073-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Total protein level in BAL fluid following CBNP exposure. Statistical significance compared to vehicle controls is indicated by **p < 0.01 and ***p < 0.001, respectively.
Fig. S2 DNA strand break in the liver expressed as tail length (TL) and %TDNA. Statistical significance compared to vehicle controls is indicated by *p < 0.05 and **p < 0.01, respectively.
Genes differentially expressed in response to CBNP exposure.
Genes commonly expressed across the time-points.
Table S3 List of lincRNAs differentially expressed in response to CBNP exposure.
Table S4 Genes involved in immune-inflammatory response GO processes.
Table S5 Genes from the microarray experiment selected for validation by RT-qPCR.
Commonality between CBNP datasets and datasets in the NextBio repository. Top 50 datasets correlated with data from CBNP post-exposure time-points.
Table S7 Disease prediction from NextBio repository.
Transparency document