Abstract
Induced pluripotent stem cells (iPSCs) were first generated from mouse embryonic fibroblasts in the year 2006. These cells resemble the typical morphology of embryonic stem cells, express pluripotency markers, and are able to transmit through germlines. To date, iPSCs of many species have been generated, whereas generation of bat iPSCs (biPSCs) has not been reported. To facilitate in-depth study of bats at the molecular and cellular levels, we describe the successful derivation of biPSCs with a piggyBac (PB) vector that contains eight reprogramming factors Oct4, Sox2, Klf4, Nanog, cMyc, Lin28, Nr5a2, and miR302/367. These biPSCs were cultured in media containing leukemia inhibitory factor and three small molecule inhibitors (CHIR99021, PD0325901, and A8301). They retained normal karyotype, displayed alkaline phosphatase activity, and expressed pluripotency markers Oct4, Sox2, Nanog, TBX3, and TRA-1-60. They could differentiate in vitro to form embryoid bodies and in vivo to form teratomas that contained tissue cells of all three germ layers. Generation of biPSCs will facilitate future studies on the mechanisms of antiviral immunity and longevity of bats at the cellular level.
Keywords: Induced pluripotent stem cells, Pluripotency, Reprogramming, Embryonic stem cells, Bat
1. Introduction
Embryonic stem cells (ESCs) can proliferate indefinitely without differentiation and are capable of chimera formation and germline transmission [2]. Currently, ESCs with such properties have only been derived from the mouse and rat [1]. Mouse ESCs were established from the inner cell mass of mouse embryos in the year 1981 [3], [4]. They can differentiate into multiple tissue cells [5] or form chimeric mice [6]. Rat ESCs were successfully isolated and propagated in the presence of small molecules in the year 2008 [7]. The derived rat ESCs also contribute to germline chimeric rats [7], [8].
In the year 2006, Yamanaka [9] used four pluripotency factors Oct4, Sox2, cMyc, and Klf4 to reprogram mouse fibroblasts into mouse-induced pluripotent stem cells (miPSCs) via retroviral infection. Human-induced pluripotent stem cells (hiPSCs) were generated successfully in the year 2007 [10], [11]. Subsequently, iPSCs were generated from monkeys [12], rats [13], [14], pigs [15], [16], [17], rabbits [18], bovines [19], [20], dogs [21], horses [22], buffalo [23], and sheep [24], [25], [26]. This opened up a new era of disease modeling, animal traits improvement, and translational medicine.
Induced pluripotent stem cells have similar biological properties to ESCs, such as infinite proliferation, expression of pluripotency markers, and the ability to differentiate into three germ layers, as well as germline transmission in vivo [9]. Induced pluripotent stem cell technology has been widely used to create disease models and to generate individual-specific hiPSCs that can differentiate into various cell types [27], [28], [29]. For livestock, iPSCs can be used to create gene-modified animals through nuclear transfer or generating chimeras. Gene targeting has been successfully performed with both miPSCs [28] and hiPSCs [30]. Chimeras of miPSCs [31] and rat-induced pluripotent stem cells (riPSCs) [14] were also generated successfully. As a result, iPSCs are a good substitute for both ESCs and somatic cells for researches and applications in other mammalian species [32].
As an ancient mammal, bats are the second largest group of mammals and live all over the world. Compared with other mammalian species of similar body size, the lifespan of bats is much longer [33], [34]. It is thought that hibernation, high body metabolic rates, low radical generation, and low reproductive rates are the causes of the longevity of bats [33], [35]. DNA mismatch repair systems, microsatellite instability, and antioxidant activity may be associated with bat longevity at the molecular level [36]. Sequencing analysis of the bat Myotis brandtii genome and transcriptome discovered that altered growth hormone/insulin-like growth factor-1 axis may have effect on longevity [37].
Bats are considered as natural virus reservoirs and support the replication of high titers of viruses in vivo without any clinical signs [38]. In the year 2013, severe acute respiratory syndrome coronavirus (SARS-CoV) ancestor was isolated and characterized in Chinese bats [39]. Whole-genome sequencing and comparative analyses of two distantly related bat species have been reported recently [40]. All these findings indicate that bats possess unique physiological characteristics and immune systems that can be utilized in biomedical research.
Because bats are not suitable for artificial rearing and bat embryonic fibroblasts (BEFs) can only be proliferated with limited passages, the successful generation of bat-induced pluripotent stem cells (biPSCs) would provide ideal and original materials to facilitate bat-related research at the cellular and molecular levels.
In this study, we have successfully derived biPSCs with a piggyBac (PB)-based vector, pSTEM-h103, containing eight stem cell factors. These cells have typical characteristics of high-quality ESCs or iPSCs. They should have broad applications for bat-related researches.
2. Materials and methods
2.1. Cell culture
Bat embryonic fibroblasts isolated from bat of Myotis lucifugus were obtained as a gift from Dr Mario Capecchi. They were cultured at 37 °C with 5% CO2 in mouse embryonic fibroblast (MEF) medium. Mouse embryonic fibroblast medium consisted of Dulbecco’s modified eagle medium, 15% fetal bovine serum, 0.5% GlutaMax, 1.0% non-essential amino acid, 1 mM sodium pyruvate, and 0.5% penicillin and streptomycin. Embryonic stem cell medium consisted of Dulbecco’s modified eagle medium, 15% ESC grade fetal bovine serum, 0.5% GlutaMax, 1.0% non-essential amino acid, 1 mM sodium pyruvate, 0.5% penicillin and streptomycin, 1000 units/mL mouse leukemia inhibitory factor (LIF, Chemicon); 0.1 mM β-mercaptoethanol, and 50 μg/mL vitamin C. The 3i medium consisted of equal volume of Neurobasal medium and Dulbecco’s modified eagle medium/F12, 0.5% N2 supplement, 1.0% B27 supplement, 0.5% penicillin and streptomycin, 0.1 mM β-mercaptoethanol, 1 mM PD0325901 (Selleck); 3 mM CHIR99021 (Selleck); 0.5 μM A8301 (Tocris Biosciences), and 1000 units/mL mouse LIF. The concentration of G418 sulfate (Calbiochem) for BEFs after nucleofection was 500 μg/mL. The final concentration of Fialuridine (FIAU, Moravek) was used at 0.3 μM. All reagents used above were purchased from Gibco, unless otherwise noted.
2.2. Generation of biPSC lines
Vector construction. To construct PB-based pSTEM-h103, human cDNA for OCT4, KLF4, SOX2, cMYC, NR5A2, NANOG, LIN28, and bat-specific miR302/367 gene cluster were assembled onto the pSP72 (Promega) plasmid backbone using polymerase chain reaction (PCR), restriction enzyme–based cloning, and recombining. The CAG promoter-driven PBase was from the CAG-PBase vector, and the PB 5′ and 3′ terminals were from the ZGs vector. pCAG-PBase vector that expresses the PB transposase can insert the PB sequences into the biPSC genome [41]. Detailed maps and sequences are available on request.
The BEF culture was replenished with MEF medium every 24 hours until about 75% confluency. Lonza Amaxa Nucelofector II device electroporation program A024 for fibroblasts was used to deliver 4 μg of the inducing vector pSTEM-h103 and 2 μg PBase expressing vector pCAG-PBase simultaneously (Fig. 1 A) into 105 BEFs. After transfections, BEFs were spread on 10-cm cell culture dishes with MEF feeder layers. The cells were cultured with ESC medium with G418 selection for 6 days. From the seventh day onward, the medium was substituted with 3i medium without G418. The cells were fed with fresh medium every 24 hours until iPSC colonies appeared (Fig. 1B). The colonies were picked with Pasteur pipettes under dissection microscope. Colonies were subsequently dissociated with 0.1% trypsin and passaged onto 24-well plates. When the cells reached about 60% to 75% confluency, they were individually passaged onto new six-well dishes and later expanded to 10-cm dishes.
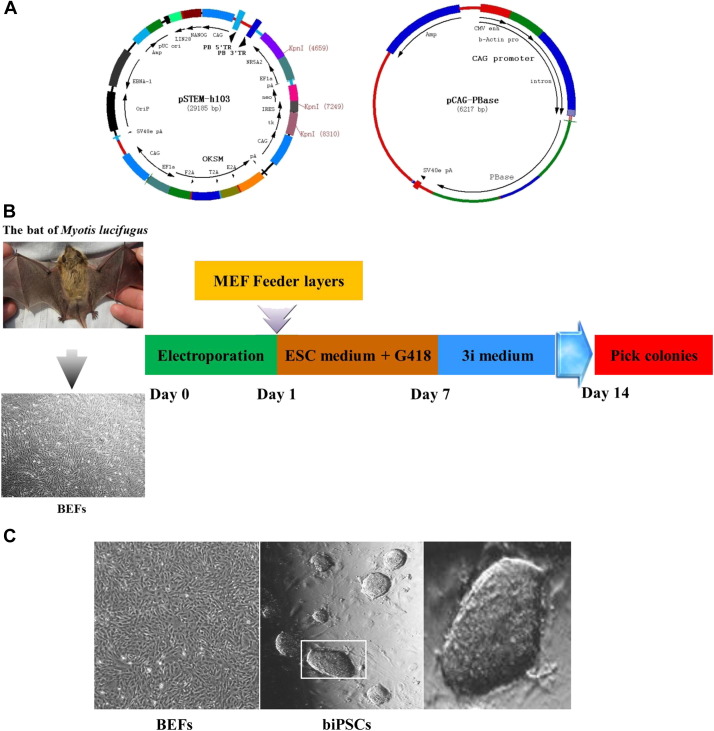
Fig. 1.
Generation of biPSCs. (A) Maps of plasmid vectors pSTEM-h103 and pCAG-PBase. The pSTEM-h103 contains eight reprogramming factors, PB sequences, and drug screening genes Neo and TK. The pCAG-PBase contains CAG-promoted PBase that can effect transposition of PB sequences into cell genome. (B) Schematic diagram of the reprogramming protocols used in the experiment. Bat embryonic fibroblasts isolated from bats of Myotis lucifugus were transfected with pSTEM-h103 and pCAG-PBase simultaneously and cultured on feeder layers with MEF medium. After 1 day, the medium was changed into ESC medium with G418. On Day 7, the medium was replaced with 3i medium, which was beneficial for the generation of iPSCs of high quality. The colonies could be picked from Day 14. (C) Bat embryonic fibroblasts derived from little brown bats of Myotis lucifugus (left). A representative biPSC colony with mouse ESC-like morphology at passage 45 is shown at a low magnification (middle) and at a higher magnification (right). biPSC, bat-induced pluripotent stem cell; MEF, mouse embryonic fibroblast; ESC, embryonic stem cell (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
2.3. Optimization of electroporation conditions for biPSCs
Bat-induced pluripotent cells were cultured on MEF feeder layers and passaged by trypsinization. About 1 × 105 iPSCs were suspended by Lonza electroporation buffer and transfected with 2 μg pMax-GFP (Fig. 2 A) with Lonza Amaxa Nucelofector II device. We chose nine programs (A012, A013, A023, A024, A030, B016, T013, T016, and U013) to test electroporation efficiency for biPSCs.
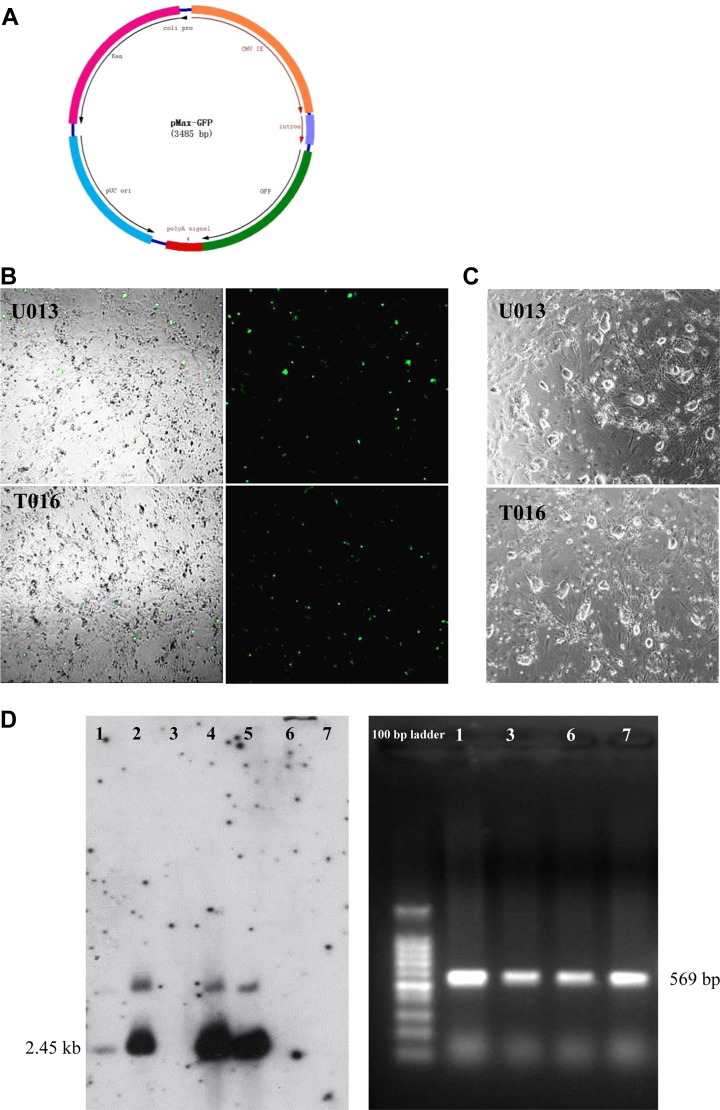
Fig. 2.
Removal of PB sequences from biPSCs. (A) The map of plasmid vector pMax-GFP. (B) Two optimal transfection programs U013 and T016 of biPSCs in Lonza nucleofection system were selected by GFP fluorescence intensity after electroporation. (C) Morphology of the pMax-GFP transfected biPSC colonies after 4 days' culture. (D) Southern blot analysis of the biPSC lines obtained from transfecting biPSCs with pCAG-PBase vector and subsequent 5 days FIAU screening (left), and PCR validating of the seemingly “transgene-free” clones from Southern blot analysis (right), the serial number labeled is consistent with that of Southern blot analysis. biPSC, bat-induced pluripotent stem cell; GFP, green fluorescent protein; FIAU, Fialuridine; PCR, polymerase chain reaction (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
2.4. Removal of PB sequences from biPSCs
pCAG-PBase vector was used to remove exogenous insertional sequences from the biPSC genome [41]. Bat-induced pluripotent cell lines were electroporated with pCAG-PBase vector and cultured with 3i medium supplemented with FIAU as negative screening drug. After 7 days, the survived cells formed colonies that could be picked and proliferated into dozens of cell lines. Southern blot analysis and PCR were performed to examine the removal effect.
2.5. Southern blot analysis and PCR
Southern blot analysis was performed with a PB probe using a protocol described earlier [40]. The genomic DNAs were digested with KpnI (NEB). The probe was synthesized by PCR with DIG-labeled dNTPs.
The colonies that were detected as negative in Southern blots were further confirmed by PCR with GoTaq polymerase using primers specific for the Neo gene of the pSTEM-h103 vector, as it is more sensitive than the Southern blot.
2.6. Karyotype analysis
On the day of karyotyping, the biPSC culture was replenished with fresh 3i medium containing 1% colcemid and incubated at 37 °C for 1 hour. The cells were then trypsinized, pelleted by centrifugation at 200 g for 5 minutes, gently resuspended in 1 mL of 0.75 M KCl, and incubated for 15 minutes at room temperature (RT). After another centrifugation process, the supernatant was discarded and replaced with ice-cold fixative solution mixture of acetic acid and methanol (1:3) to a final volume of 5 mL. Cells were dropped on cold slides, stained by Giemsa dye, and analyzed under a Nikon microscope.
2.7. Alkaline phosphatase staining
Bat-induced pluripotent cell colonies were fixed with 4% paraformaldehyde on the 10-cm dish for 30 minutes after PBS wash. At the end of fixation, the colonies were completely washed with PBS for three times, followed by staining with BM-purple (Millipore) for 1 to 2 hours in dark.
2.8. Teratoma formation and hematoxylin–eosin staining
The biPSCs suspended with PBS at a cell density of 1 × 107/mL were injected intramuscularly into severe combined immune deficient (SCID) mice. Mice with PBS injection were used as controls. Tumors developed at the injection sites were harvested, fixed, and processed for paraffin sections, and subsequently hematoxylin–eosin (HE) stained for histological analysis. Genomic DNAs of teratomas and SCID mice were extracted using DNeasy Blood & Tissue Kit (Qiagen). Polymerase chain reaction with GoTaq polymerase using bat-specific primers was performed to identify the origin of species of teratomas.
2.9. Embryoid body formation
The biPSCs were cultured with ESC medium without LIF at a cell density of 5 × 104/mL. Droplets of the mixture for 20 μL each were dropped gently on the inner wall of 10-cm dishes filled with PBS to keep moisture. After 3 days, the droplets were transferred to 0.1% gelatin-coated 96-well plates, to 48-well plates when overgrown, and finally to six-well plates. The embryoid body (EB) colonies were observed with a Nikon microscope.
2.10. Quantitative real-time PCR
Total RNA of biPSCs and BEFs (control) was extracted by RNeasy mini kit (Qiagen) and reverse transcribed to synthesize first-strand DNA by QuantiTect Reverse Transcription Kit (Qiagen). The cDNA was used as a template for real-time PCR. Real-time PCR was performed with LightCycler 480 by SYBR Green I Master mix (Roche). The cDNA of bat Myotis Oct4 (bOct4), Sox2 (bSox2), Klf4 (bKlf4), and cMyc (bcMyc) were obtained by BLAT analysis at www.genome.ucsc.edu/. Primers for real-time PCR are listed in Appendix I.
In this experiment, β-actin was used to calibrate gene expression of bOct4, bSox2, bKlf4, and bcMyc between biPSCs and BEFs. Data for real-time PCR were analyzed using the 2−ΔΔCt method, which were normalized to β-actin mRNA, and were presented as fold-changes relative to the control values. The standard deviation (SD) (n = 3) was used to quantify variability.
2.11. Immunofluorescence staining
Cells were fixed as described in AP staining section. For nuclear antigen staining, the fixed colonies were treated with 0.2% Triton X-100 (Sigma) for 15 minutes. For both nuclear and cell-surface staining, colonies were exposed to blocking solution containing 5% goat serum (Sigma) and 1% BSA (Sigma) in PBS for 1 hour. The colonies were incubated overnight in blocking buffer with primary antibodies at 4 °C. After that, they were washed with PBS completely and incubated in blocking buffer with secondary antibodies for 30 minutes at RT. After incubation, they were washed with PBS and incubated with 4,6-diamidino-2-phenylindole (1:10,000) (Roche) diluted with PBS for 1 to 2 minutes. Finally, 4,6-diamidino-2-phenylindole was washed off with PBS and the colonies were observed under a Nikon fluorescence microscope.
The primary antibodies used were anti-Oct4 (C-10) (1:100, Santa Cruz), anti-Sox2 (1:400, Novus), anti-Nanog (1:400, Abcam), anti-TBX3 (1:200, Santa Cruz), and anti-TRA-1-60 (1:250, Millipore). The secondary antibodies (Invitrogen) used at 1:500 were Alexa-Fluor-488 goat anti-mouse IgM (μ-chain), Alexa-Fluor-488 goat anti-rabbit IgG (H + L), and Alexa-Fluor-594 goat anti-mouse IgG (H + L).
2.12. Western blot analysis
Western blot was performed as previously described [9]. The secondary antibodies were detected using the electrochemiluminescence chemiluminescent color reagent (Beyotime). Antibodies used for Western blotting were mouse anti-Oct4 (C-10) (1:500; Santa Cruz), rabbit anti-Sox2 (1:2000; Millipore), rabbit anti-β-actin (1:500; Santa Cruz), anti-rabbit IgG-HRP (1:10,000; Santa Cruz), and anti-mouse IgG-HRP (1:10,000; Santa Cruz). The primary antibodies anti-β-actin, anti-Oct4, and anti-Sox2 used in the study have cross-reactivity to mouse, bat, and human proteins.
3. Results
3.1. Generating biPSC lines
To derive biPSCs, BEFs were nucleofected with the PB transposon vector, pSTEM-h103, by the Lonza Amaxa Nucelofector II device. After about 14 days of culture, biPSCs colonies began to appear (Fig. 1B). There are about 100 colonies per dish with 104 nucleofected BEFs. Colonies that were compact, shiny, and with clear edges and with 3D structure were manually picked and expanded.
Some of the picked cell lines were passaged for over 45 times in 3i medium. This confirmed their ability of unlimited proliferation without differentiation. Bat-induced pluripotent stem cells at passage 45 with mouse ESC-like morphology are shown in Figure 1C. When biPSCs were cultured in medium without LIF, they could not maintain the typical ESC morphology and differentiated (Supplementary Fig. 1). This indicated that biPSCs were more similar to the mouse ESCs.
3.2. Optimization of electroporation conditions for biPSCs
Electroporation conditions are critical for gene manipulations. To find the optimal electroporation condition for biPSCs with the Lonza Amaxa Nucleofector II device, we utilized the manufacturer-suggested pMax-GFP plasmid. As a transient and episomal vector, it expressed green fluorescent protein (GFP) after being delivered into biPSCs using different programs. The best program was judged by GFP fluorescence intensity and survival rate of electroporated biPSCs.
Twenty-four hours after electroporation of pMax-GFP, the GFP signals could be observed. Among the nine programs tested, the efficiency of two programs T016 and U013 was much higher than that of the rest (Fig. 2B). However, after 4 days of culture, we observed that biPSCs morphology, growth, and survival rates of U013 electroporated cells was slightly better than that of T016 (Fig. 2C), suggesting that U013 was more suitable for biPSCs electroporation.
3.3. Removal of PB sequences in biPSCs
It is a consensus that iPSCs from species other than the mouse and rat are usually incompletely reprogrammed, relying on the expression of exogenous factors for maintenance. Bat-induced pluripotent cells without exogenous genes would allow broader uses of these cells, such as differentiation-related experiments or gene targeting. Furthermore, successful generation of insertion-free biPSCs could provide important clues for derivation of transgene-free iPSCs for other species, such as farm animals.
Southern blot analysis was used to initially screen for exogenous sequence-free clones and PCR was used to further validate the clones. Southern blot probe was designed according to the Neo sequences of vector pSTEM-h103. As shown in Figure 2D (left), it appeared that PB insertions were removed in some lines. Because PCR is a more sensitive method than the Southern blot, we further performed PCR validation (Fig. 2D, right). The clones that were seemingly “transgene-free” (Nos. 1, 3, 6, and 7) turned out to be positive for exogenous genes, suggesting that exogenous sequences were required for the biPSCs to maintain an undifferentiated state, and that future work would be needed to obtain completely transgene-free biPSCs.
3.4. Characterization of the biPSCs
Karyotype abnormality is often associated with low-quality iPSCs, and affects further uses of the cells. We karyotyped our biPSC lines with mouse ESC-like morphology. The results showed that biPSC line No. 4 exhibited a normal karyotype of bat of 42 + XY (Fig. 3 A).
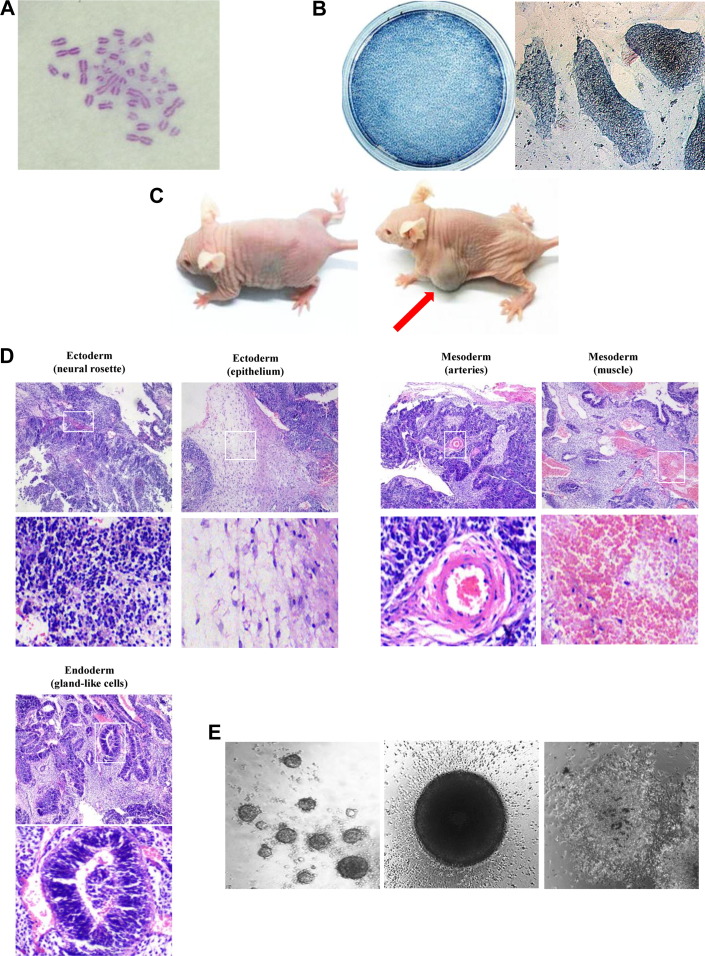
Fig. 3.
Characterization of the biPSCs. (A) The karyotype of the biPSC line at the 45 generation was 42 + XY analyzed under a Nikon microscope. (B) Alkaline phosphatase staining on the 10-cm dish (left) and analyzed under a Nikon microscope (right). (C) Teratomas formed by subcutaneous injection of biPSCs into SCID mice (right) compared with the mouse injected with PBS (left). (D) Hematoxylin–eosin staining analysis of teratomas. They contained cell types of all three germ layers (low magnification, above; higher magnification, below). (E) In vitro differentiated biPSCs could form embryoid bodies (low magnification, left; higher magnification, middle). Embryoid bodies could further differentiate by adherent culture (right). biPSC, bat-induced pluripotent stem cell; SCID, severe combined immune deficient.
Alkaline phosphatase staining is a rapid and effective preliminary method to test the pluripotency of iPSCs. It is also an important method to determine whether the iPSCs have differentiated. Bat-induced pluripotent cells were nearly 100% positive for AP staining, which demonstrated that the biPSC lines were pluripotent and had no signs of differentiation (Fig. 3B).
Teratomas-related experiments were widely used to determine pluripotency and the in vivo differentiation ability of iPSCs [9]. Various amounts of biPSCs were subcutaneously injected into SCID mice, and tumors (teratomas) of different sizes were observed after approximately 4 to 5 weeks (Fig. 3C, Supplementary Fig. 2). Hematoxylin–eosin staining confirmed that the teratomas contained various types of tissues representing all three germ layers, including ectoderm (neural rosettes and epithelium), mesoderm (arteries and muscles), and endoderm (gland-like cells), indicating that biPSCs had a high in vivo pluripotency (Fig. 3D). In addition, the origin of these teratoma tissues was confirmed to be from biPSCs as shown by PCR using biPSC-specific primers (Supplementary Fig. 3).
Embryoid body formation is an important criterion to determine the in vitro differentiation ability of iPSCs. Similar to miPSCs, biPSCs formed ball-shaped EBs effectively after 1 to 2 weeks in culture without LIF or bFGF and then differentiated into cells with different lineages after an additional 1 to 2 weeks of adherent culture (Fig. 3E). These data indicated that our biPSCs were pluripotent and could be differentiated into different lineages both in vivo and in vitro.
3.5. Expression of pluripotency markers in biPSCs
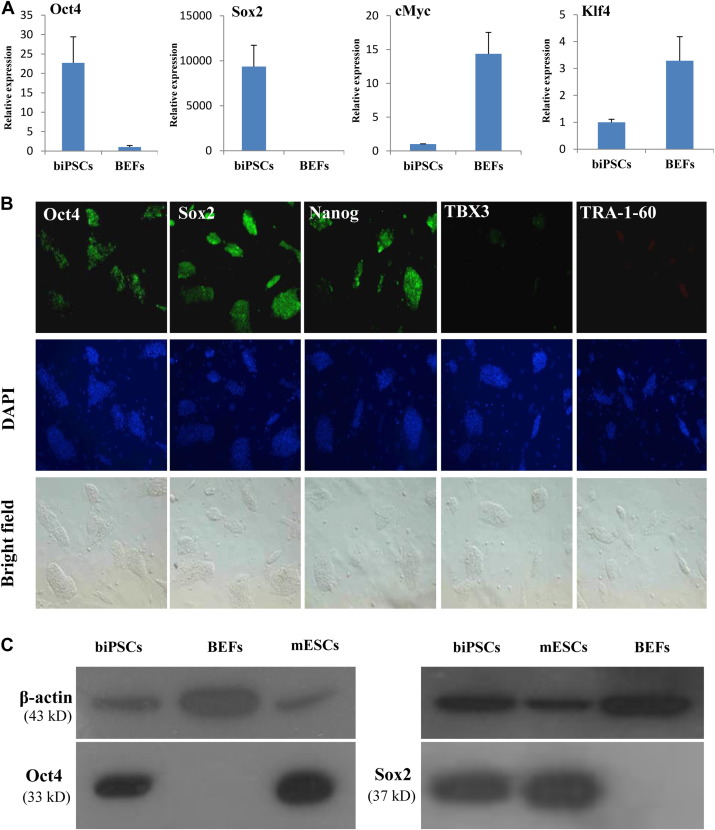
Real-time quantitative PCR was used to compare expression of pluripotency genes between biPSCs and BEFs. Relative gene expression of the four pluripotency genes Oct4, Sox2, Klf4, and cMyc was analyzed (Fig. 4 A). Compared with BEFs, expression of Oct4 and Sox2 was increased dramatically for biPSCs. However, expression of cMyc and Klf4 was decreased. When biPSCs were differentiated, the expression of core pluripotency genes Oct4 and Sox2 was decreased (Supplementary Fig. 4).
Fig. 4.
Expression of pluripotency factors in biPSCs. (A) Real-time PCR analysis of the relative RNA expression levels of pluripotency genes (Oct4, Sox2, cMyc, and Klf4) in biPSCs versus that in BEFs. Data were normalized to β-actin mRNA expression levels and error bars were calculated by standard deviation (SD), n = 3. (B) The immunofluorescence staining of pluripotency markers Oct4, Sox2, Nanog, TBX3, and TRA-1-60 of biPSC colonies are shown (fluorescence, upper; 4,6-diamidino-2-phenylindole, middle; bright field, bottom). (C) Western blot analysis of Oct4 and Sox2 proteins. BiPSCs protein expression levels of BEFs and mouse ESCs were used as negative and positive controls, respectively. β-Actin was used as the loading control. biPSC, bat-induced pluripotent stem cell; PCR, polymerase chain reaction; BEF, bat embryonic fibroblast; ESC, embryonic stem cell.
Immunofluorescence staining is widely used to detect the protein expression of pluripotency genes of iPSCs. Results of immunostaining also confirmed the pluripotency of our biPSCs at the protein level (Fig. 4B). Bat-induced pluripotent cell colonies stained positive for antibodies against nuclear markers Oct4, Sox2, Nanog, and TBX3. The colonies were also slightly positive for the cell-surface marker TRA-1-60, showing that pluripotency proteins were highly expressed in biPSCs.
To further detect the protein expression of pluripotency factors of iPSCs, we performed Western blot analysis. Western blot analysis of Oct4 and Sox2 (Fig. 4C) showed that the total protein expression levels of the two genes in biPSCs were similar to those in mouse ESCs, which further confirmed the pluripotency of biPSCs.
4. Discussion
In this study, biPSC lines were established with the pSTEM-h103 PB vector and a suitable cell culture system. Our biPSCs exhibited ESC-specific characteristics, such as mouse ESC-like morphology, AP staining, expression of pluripotency markers, and the ability to differentiate both in vivo and in vitro.
Previously reported iPSC lines were generated by four to six reprogramming factors including Oct4, Sox2, cMyc, Klf4, Nanog, and Lin28. Expression of the miR302/367 cluster could rapidly reprogram mice or human somatic cells into pluripotent stem cells even without the requirement of other exogenous pluripotency factors [42]. As an important nuclear receptor, Nr5a2 can replace exogenous Oct4 and work in combination with Sox2 and Klf4 to maintain the pluripotency of iPSCs [43]. To reprogram BEFs, we constructed eight reprogramming factors in pSTEM-h103, because of the use of all these factors in pSTEM-h103, the efficiency of biPSC generation has improved significantly.
Transfection methods also affect iPSC generation and cell quality. Previously, retroviral and lentiviral vectors were widely used to deliver reprogramming factors. However, they tend to randomly and stably integrate into the chromosomes that could be risky of tumorigenesis [44]. Later, recombinant adenoviral [45] and Sendai viral delivery vectors [46] were used to reduce the integration rates into the genomes and yet the efficiency of iPSC generation was very low [47]. Protein transduction method was also attempted to generate iPSCs [48]. However, it is costly, time-consuming, and low in efficiency, preventing its wide applications. PB system has many advantages over the viral systems, such as high safety, convenient operation, easy integration into the genomes, and high expression efficiency of exogenous genes [49], [50]. Most importantly, the inserted PB fragments could be excised without leaving a footprint.
Growth factors are essential to maintain the pluripotent status of ESCs and iPSCs. Epiblast-derived stem cells from humans depend on bFGF for self-renewal [51]. Mouse ESCs need to be maintained in medium containing LIF [52]. Our biPSCs were LIF-dependent, not bFGF-dependent.
Three small molecule compounds PD0325901, CHIR99021, and A8301 were supplemented into the medium as well. They are inhibitors of MAP kinase/ERK kinase, glycogen synthase kinase-3, and transforming growth factor-β/activin receptors pathways [53], [54]. Only iPSCs of high quality could survive and proliferate without differentiation when cultured in 3i medium [53]. The generated biPSCs could maintain undifferentiated status, typical ESC morphology, and proliferated indefinitely in 3i medium. As vitamin C has been reported to suppress cell senescence [55], it was added together with LIF into the ESC medium to enhance the biPSC generation.
Teratoma formation demonstrated that the reprogrammed biPSC lines had the developmental potential to give rise to tissues of all three primary germ layers. Bat-induced pluripotent stem cells also had the ability to form ball-shaped EBs without LIF. This confirmed that biPSCs had the pluripotent potential to differentiate both in vitro and in vivo.
Induced pluripotent stem cells of high quality should have similar features to ESCs. In this experiment, Oct4 and Sox2 expression was upregulated greatly compared with BEFs, showing that the biPSC lines stably maintained an ESC status. The decreased expression of Klf4 and cMyc explained that they were not essential for the maintenance of the ESC status of the biPSCs. Klf4 only plays a role in the early stage of the cellular reprogramming [56], and cMyc as an oncogene is not essential for the generation and maintenance of the iPSCs [57]. High-level expression of cMyc has negative effect on the pluripotency of iPSCs [58]. Low-level expression of cMyc suggested that the biPSCs were impossible to be cancerous cells. Both immunostaining and western blot analyses demonstrated that pluripotency factors could be expressed at the protein level in biPSCs, which further confirmed the pluripotent status of biPSCs. Our biPSCs could be used as cells for antiviral or other stress-related study purposes. Various types of tissue cells generated from biPSCs could be used for further exploring the mechanisms of bat's antiviral immunity and longevity that would greatly benefit the mankind.
Compared with the generation of riPSCs and miPSCs, the generation of biPSCs needs further researches in the future. In particular, more efforts are needed to derive completely insertion-free biPSCs. Culture conditions function as a driving force to promote complete reprogramming of iPSCs [32]. In this research, cell culture medium components have been improved and optimized from a variety of aspects to fit for the growth and proliferation of biPSCs. Encouraged by Hou's [59] success in using small molecules to reprogram mouse fibroblasts, large-scale screening of small molecule compounds is being carried out to further improve the quality of biPSCs.
4.1. Conclusion
According to our knowledge, this is the first report about derivation of bat iPSCs. It will provide an important reference for the generation of iPSCs of other species and offers clues to improve the iPSCs quality of species that have already been successfully generated. This experiment also proved that the pSTEM-h103 vector, in combination with the 3i medium, could generate biPSCs efficiently. The generated biPSCs may be important tools for cell biology researches on bats.
Acknowledgments
We are grateful to the Capecchi Lab at the University of Utah for gift of BEFs from the little brown bats of Myotis lucifugus. We thank members of the Wu laboratory for their help. This work was supported in part by the New Techniques and New Methods on Transgenic Research (grant no. 2013ZX08010001).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.theriogenology.2014.04.001.
Appendix I.
Primers used in this study.
| PCR target region | Sequences (5′–3′) | Applications |
|---|---|---|
| pSTEM-h103 PB probe | F: TGACGTACGTTAAAGATAATCATGC | Southern blot (340 bp) |
| R: GCTTCGATTAATAAGTATAATTTGTTTCT | ||
| pSTEM-h103 Neo-IRES | F: GGACAGGTCGGTCTTGACAAAAAG | GoTaq PCR (569 bp) |
| R: TCTGTTGAATGTCGTGAAGGAAGC | ||
| b-βactin | F: ACTGGGACGACATGGAGAAG | Real-time PCR as reference genes |
| R: AGAGGCGTACAGGGACAGC | ||
| bOct4 | F: GGTACACCCAGGCCGATGT | Real-time PCR for pluripotent genes expression |
| R: GATGGTCGTTTGGCTGAACA | ||
| bSox2 | F: CTGCGAGCGCTGCACAT | |
| R: TCATGAGCGTCTTGGTTTTCC | ||
| bKlf4 | F: CGAACCCACACAGGTGAGAAA | |
| R: CTGAGCGGGCAAACTTCCA | ||
| bcMyc | F: ACGTCAGCTTCGCCAACAG | |
| R: GTTCTCTTCCTCGTCGCAGAA | ||
| Bat genomic Oct4 | F: ATACTTCCTCAAGGCTCCCAGGAC | GoTaq PCR (281 bp) |
| R: GGGTGCATTTCCCACGTAAAGATA | ||
| Bat genomic Hprt | F: TACTGCAGGAGGTCAGCGCTACTAA | GoTaq PCR (262 bp) |
| R: ATCACAACACTGGGGCTGTGAGTC |
Appendix II.
PCR system with GoTaq polymerase.
| Template DNA | 1.5 μL |
| 5x buffer | 2.5 μL |
| Mg2+ | 1.0 μL |
| Primer F | 0.5 μL |
| Primer R | 0.5 μL |
| Hotshot neutralization buffer | 1.5 μL |
| dNTPs (40 mM) | 0.25 μL |
| GoTaq polymerase | 0.1 μL |
| ddH2O | To 12.5 μL |
| Total volume | 12.5 μL |
Appendix A. Supplementary data
LIF dependency of biPSCs. (A) BiPSCs cultured in 3i medium supplemented with LIF showed undifferentiated morphology. (B, C) BiPSCs cultured in media without LIF differentiated.
Supplementary Figure 2. SCID mice injected with different amount of biPSCs produced teratomas of different sizes. The cell density of biPSCs was 107/ml.
Supplementary Figure 3. PCR results of genomic DNAs isolated from teratomas of SCID mice injected with biPSCs. Two specific primers of bat genomic DNAs (bat Oct4 and bat Hprt) were used. Sizes of PCR products of target sequences were 281 bp (bat Oct4) and 262 bp (bat Hprt).
Supplementary Figure 4. Comparison of Oct4 and Sox2 expression between undifferentiated biPSCs and differentiated biPSCs.
References
- 1.Gafni O., Weinberger L., Mansour A.A., Manor Y.S., Chomsky E., Ben-Yosef D. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 2.Hanna J.H., Saha K., Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 4.Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibel M., Richter J., Schrenk K., Tucker K.L., Staiger V., Korte M. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- 6.Brook F., Gardner R. The origin and efficient derivation of embryonic stem cells in the mouse. Proc Natl Acad Sci U S A. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li P., Tong C., Mehrian-Shai R., Jia L., Wu N., Yan Y. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Men H., Bauer B.A., Bryda E.C. Germline transmission of a novel rat embryonic stem cell line derived from transgenic rats. Stem Cells Dev. 2012;21:2606–2612. doi: 10.1089/scd.2012.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 12.Liu H., Zhu F., Yong J., Zhang P., Hou P., Li H. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–590. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Liao J., Cui C., Chen S., Ren J., Chen J., Gao Y. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell. 2009;4:11–15. doi: 10.1016/j.stem.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Li W., Wei W., Zhu S., Zhu J., Shi Y., Lin T. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–19. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Esteban M.A., Xu J., Yang J., Peng M., Qin D., Li W. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J Biol Chem. 2009;284:17634–17640. doi: 10.1074/jbc.M109.008938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ezashi T., Telugu B.P., Alexenko A.P., Sachdev S., Sinha S., Roberts R.M. Derivation of induced pluripotent stem cells from pig somatic cells. Proc Natl Acad Sci U S A. 2009;106:10993–10998. doi: 10.1073/pnas.0905284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Z., Chen J., Ren J., Bao L., Liao J., Cui C. Generation of pig induced pluripotent stem cells with a drug-inducible system. J Mol Cell Biol. 2009;1:46–54. doi: 10.1093/jmcb/mjp003. [DOI] [PubMed] [Google Scholar]
- 18.Honda A., Hirose M., Hatori M., Matoba S., Miyoshi H., Inoue K. Generation of induced pluripotent stem cells in rabbits: potential experimental models for human regenerative medicine. J Biol Chem. 2010;285:31362–31369. doi: 10.1074/jbc.M110.150540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang B., Li T., Alonso-Gonzalez L., Gorre R., Keatley S., Green A. A virus-free poly-promoter vector induces pluripotency in quiescent bovine cells under chemically defined conditions of dual kinase inhibition. PloS One. 2011;6:e24501. doi: 10.1371/journal.pone.0024501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han X., Han J., Ding F., Cao S., Lim S.S., Dai Y. Generation of induced pluripotent stem cells from bovine embryonic fibroblast cells. Cell Res. 2011;21:1509–1512. doi: 10.1038/cr.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo J., Suhr S.T., Chang E.A., Wang K., Ross P.J., Nelson L.L. Generation of leukemia inhibitory factor and basic fibroblast growth factor-dependent induced pluripotent stem cells from canine adult somatic cells. Stem Cells Dev. 2011;20:1669–1678. doi: 10.1089/scd.2011.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy K., Sung H.K., Zhang P., Laflamme S., Vincent P., Agha-Mohammadi S. Induced pluripotent stem cell lines derived from equine fibroblasts. Stem Cell Rev. 2011;7:693–702. doi: 10.1007/s12015-011-9239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng Y., Liu Q., Luo C., Chen S., Li X., Wang C. Generation of induced pluripotent stem cells from buffalo (Bubalus bubalis) fetal fibroblasts with buffalo defined factors. Stem Cells Dev. 2012;21:2485–2494. doi: 10.1089/scd.2012.0018. [DOI] [PubMed] [Google Scholar]
- 24.Bao L., He L., Chen J., Wu Z., Liao J., Rao L. Reprogramming of ovine adult fibroblasts to pluripotency via drug-inducible expression of defined factors. Cell Res. 2011;21:600–608. doi: 10.1038/cr.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Cang M., Lee A.S., Zhang K., Liu D. Reprogramming of sheep fibroblasts into pluripotency under a drug-inducible expression of mouse-derived defined factors. PloS One. 2011;6:e15947. doi: 10.1371/journal.pone.0015947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J., Balehosur D., Murray B., Kelly J.M., Sumer H., Verma P.J. Generation and characterization of reprogrammed sheep induced pluripotent stem cells. Theriogenology. 2012;77:338–346.e1. doi: 10.1016/j.theriogenology.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y.-C., Tsai K.-L., Hung C.-W., Ding D.-C., Chen L.-H., Chang Y.-L. Induced pluripotent stem cells and regenerative medicine. J Clin Gerontol Geriatr. 2011;2:1–6. [Google Scholar]
- 28.Hanna J., Wernig M., Markoulaki S., Sun C.W., Meissner A., Cassady J.P. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 29.Xu D., Alipio Z., Fink L.M., Adcock D.M., Yang J., Ward D.C. Phenotypic correction of murine hemophilia A using an iPS cell-based therapy. Proc Natl Acad Sci U S A. 2009;106:808–813. doi: 10.1073/pnas.0812090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou J., Maeder M.L., Mali P., Pruett-Miller S.M., Thibodeau-Beganny S., Chou B.K. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 32.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson G.S., South J.M. Life history, ecology and longevity in bats. Aging Cell. 2002;1:124–131. doi: 10.1046/j.1474-9728.2002.00020.x. [DOI] [PubMed] [Google Scholar]
- 34.Brunet-Rossinni A.K., Austad S.N. Ageing studies on bats: a review. Biogerontology. 2004;5:211–222. doi: 10.1023/B:BGEN.0000038022.65024.d8. [DOI] [PubMed] [Google Scholar]
- 35.Brunet-Rossinni A.K. Reduced free-radical production and extreme longevity in the little brown bat (Myotis lucifugus) versus two non-flying mammals. Mech Ageing Dev. 2004;125:11–20. doi: 10.1016/j.mad.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Conde-Pérezprina J.C., Luna-López A., González-Puertos V.Y., Zenteno-Savín T., León-Galván M.Á., Königsberg M. DNA MMR systems, microsatellite instability and antioxidant activity variations in two species of wild bats: Myotis velifer and Desmodus rotundus, as possible factors associated with longevity. Age. 2012;34:1473–1492. doi: 10.1007/s11357-012-9399-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seim I., Fang X., Xiong Z., Lobanov A.V., Huang Z., Ma S. Genome analysis reveals insights into physiology and longevity of the Brandt's bat Myotis brandtii. Nat Commun. 2013;4 doi: 10.1038/ncomms3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omatsu T., Watanabe S., Akashi H., Yoshikawa Y. Biological characters of bats in relation to natural reservoir of emerging viruses. Comp Immun Microbiol Infect Dis. 2007;30:357–374. doi: 10.1016/j.cimid.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge X.-Y., Li J.-L., Yang X.-L., Chmura A.A., Zhu G., Epstein J.H. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013 doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang G., Cowled C., Shi Z., Huang Z., Bishop-Lilly K.A., Fang X. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S., Ying G., Wu Q., Capecchi M.R. Toward simpler and faster genome-wide mutagenesis in mice. Nat Genet. 2007;39:922–930. doi: 10.1038/ng2060. [DOI] [PubMed] [Google Scholar]
- 42.Anokye-Danso F., Trivedi C.M., Juhr D., Gupta M., Cui Z., Tian Y. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heng J.-C.D., Feng B., Han J., Jiang J., Kraus P., Ng J.-H. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 44.Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 45.Stadtfeld M., Nagaya M., Utikal J., Weir G., Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ban H., Nishishita N., Fusaki N., Tabata T., Saeki K., Shikamura M. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci U S A. 2011;108:14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simara P., Motl J.A., Kaufman D.S. Pluripotent stem cells and gene therapy. Trans Res J Lab Clin Med. 2013;161:284–292. doi: 10.1016/j.trsl.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D., Kim C.H., Moon J.I., Chung Y.G., Chang M.Y., Han B.S. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaji K., Norrby K., Paca A., Mileikovsky M., Mohseni P., Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yusa K., Rad R., Takeda J., Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amit M., Carpenter M.K., Inokuma M.S., Chiu C.P., Harris C.P., Waknitz M.A. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol. 2000;227:271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- 52.Matsuda T., Nakamura T., Nakao K., Arai T., Katsuki M., Heike T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu B., Li W., Wang L., Liu Z.H., Zhao X.Y. Stem cells and small molecule screening: haploid embryonic stem cells as a new tool. Acta Pharmacol Sin. 2013;34:725–731. doi: 10.1038/aps.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esteban M.A., Wang T., Qin B., Yang J., Qin D., Cai J. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 56.Chen J., Liu J., Chen Y., Yang J., Chen J., Liu H. Rational optimization of reprogramming culture conditions for the generation of induced pluripotent stem cells with ultra-high efficiency and fast kinetics. Cell Res. 2011;21:884–894. doi: 10.1038/cr.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wernig M., Meissner A., Cassady J.P., Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 58.Habib O., Habib G., Choi H.W., Hong K.S., Tae Do J., Moon S.H. An improved method for the derivation of high quality iPSCs in the absence of c-Myc. Exp Cell Res. 2013;319:3190–3200. doi: 10.1016/j.yexcr.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 59.Hou P., Li Y., Zhang X., Liu C., Guan J., Li H. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LIF dependency of biPSCs. (A) BiPSCs cultured in 3i medium supplemented with LIF showed undifferentiated morphology. (B, C) BiPSCs cultured in media without LIF differentiated.
Supplementary Figure 2. SCID mice injected with different amount of biPSCs produced teratomas of different sizes. The cell density of biPSCs was 107/ml.
Supplementary Figure 3. PCR results of genomic DNAs isolated from teratomas of SCID mice injected with biPSCs. Two specific primers of bat genomic DNAs (bat Oct4 and bat Hprt) were used. Sizes of PCR products of target sequences were 281 bp (bat Oct4) and 262 bp (bat Hprt).
Supplementary Figure 4. Comparison of Oct4 and Sox2 expression between undifferentiated biPSCs and differentiated biPSCs.