Abstract
In vitro assembly of alphavirus nucleocapsid cores, called core-like particles (CLPs), requires a polyanionic cargo. There are no sequence or structure requirements to encapsidate single-stranded nucleic acid cargo. In this work, we wanted to determine how the length of the cargo impacts the stability and structure of the assembled CLPs. We hypothesized that cargo neutralizes the basic region of the alphavirus capsid protein and if the cargo is long enough, it will also act to scaffold the CP monomers together. Experimentally we found that CLPs encapsidating short 27mer oligonucleotides were less stable than CLPs encapsidating 48mer or 90mer oligonucleotides under different chemical and thermal conditions. Furthermore, cryo-EM studies showed there were structural differences between CLPs assembled with 27mer and 48mer cargo. To mimic the role of the cargo in CLP assembly we made a mutant (4D) where we substituted a cluster of four Lys residues in the CP with four Asp residues. We found that these few amino acid substitutions were enough to initiate CLP assembly in the absence of cargo. The cargo-free 4D CLPs show higher resistance to ionic strength and increased temperature compared to wild-type cargo containing CLPs suggesting their CLP assembly mechanism might also be different.
Keywords: self-assembly, protein, nucleic acid, virus capsid
Introduction
A virus capsid has dual roles in a viral infection. First, during extracellular transmission and intracellular transport, the capsid protects the viral genome from being released prematurely or being targeted for degradation. Second, in response to an environmental trigger such as a change in pH, the capsid releases its genome and viral replication begins inside the cell [1]. A capsid that is misassembled may not disassemble at the right time or location during an infection, and subsequently the infection will be attenuated. Understanding the mechanisms and physical basis behind capsid assembly will identify targets for anti-viral therapeutics and provide insight for the design of capsid-based nanoparticles [2–4].
Capsids have traditionally been characterized as closed, spherical shells held together by intersubunit contacts [5–7]. Interactions between either capsid proteins (CP) or nucleic acid and CP contribute to capsid stability, but their exact stoichiometric composition varies for each virus. Individual subunit contacts may be weak but as an entity, capsids are extremely stable. As a result of complete capsid assembly, there may be conformational changes of subunits and hysteresis or a high barrier to subunit dissociation from the capsid [8–10].
The requirements for nucleic acid to promote in vitro self-assembly vary. Some viruses like MS2 require a stem loop-containing cargo for assembly to occur [11, 12]. Other viruses like the immature HIV capsid and alphavirus nucleocapsid core require a polyanionic cargo but there is no sequence or structure requirement in vitro [13–16]. In contrast, Cowpea chlorotic mottle virus and brome mosaic virus can assemble in the absence of nucleic acid but if nucleic acid is present, regardless of its sequence, it will be packaged [17, 18]. In vivo, viruses must package their genomic material to propagate an infection. Many CP contain a domain rich in basic residues (e.g. arginine-rich motifs) which binds to the negatively-charged viral genome. Theoretical studies have shown that a charge ratio (negative charges from cargo to positive charges from CP) ranging between ~1 and 2 is optimal for assembly of a closed capsid; capsids have a preference for being ‘overcharged’ or containing more negative charges [19–22]. However, experimental studies have shown that particle assembly can occur with a charge ratio as broad as 0.3 and 9 [23–27].
Alphaviruses are enveloped viruses with an icosahedral nucleocapsid core [28]. Self-assembly of the core can be recapitulated in vitro to form nucleocapsid core-like particles (CLPs), but only in the presence of anionic cargo [13, 16]. The cargo requirement can be fulfilled by single-stranded DNA or RNA oligos of at least 14 bases in length, heparan sulfate, or gold nanoparticles coated with carboxylated PEG. CLP assembly has been demonstrated with CP from several alphaviruses including Ross River (RRV), Sindbis, and Western equine encephalitis [16, 29–34]. Alphavirus CP has a conserved N-terminal domain (residues 1–120 RRV numbering) enriched in basic residues (~30 out of 120 residues), and is thought to mediate electrostatic interactions between the CP and viral genome during core assembly [35–37]. There is no atomic structure of the alphavirus N-terminus domain. The C-terminus of alphavirus CP ~121–270 (RRV numbering) has a conserved chymotrypsin-like domain and folds independently of any CP-cargo interactions [38, 39]. In this work we use RRV CP.
By negative stain transmission electron microscopy (TEM), alphavirus CLPs are spherical with a diameter between 36–40 nm, consistent with the size of the core in the virion. Early cryo-EM reconstructions of CLPs showed the CP were arranged as a series of pentamers and hexamers in T = 4 icosahedral symmetry [32]. The resolution of the CLP structures was limited to ~25 Å suggesting heterogeneity between particles. More recently, Wang et al, used a 27mer oligo and RRV CP and determined that these CLPs did not undergo hysteresis to dissociation as a result of changing ionic strength, one possibility being incomplete capsids were being formed [24]. Cryo-EM studies showed a prevalence of regions of discontinuity in the CP layer confirming the presence of incomplete shells.
From the results from Wang et al [24], we hypothesized that cargo length might impact alphavirus CLP assembly. We wanted to determine if cargo was important for neutralizing the basic N-terminus of CP, acting as a scaffold to localize CP, or both. We demonstrated that RRV CLPs assembled with oligos longer than 27 bases were more chemically and thermally stable than CLPs made with the 27mer. However, despite differences in stability, the CLPs with longer cargo still had incomplete capsid shells. To test the importance of electrostatics-mediated CP scaffolding, we mutated the CP and introduced a cluster of anionic residues in the N-terminus of RRV CP. We demonstrated that by providing intrinsic anionic charge, CP–CP scaffolding and CLP self-assembly can be promoted without exogenous cargo.
Results
CLPs assembled with longer oligo nucleotides are more stable than those assembled with the shorter 27mer
In our previous work we used a 27mer oligo to drive RRV CP assembly into CLPs [24]. From our results we concluded that the role of the 27mer oligo was to neutralize the basic residues of the CP to allow CLP assembly. In this current work we hypothesized that longer cargo would both neutralize and scaffold interactions between CP monomers in a CLP. Furthermore, the CLPs encapsidating longer cargo would be more stable compared to those containing the shorter 27mer. To test this, we monitored CLP formation using four different length oligos. Two cargos, the 27mer and 48hp were used in previous work [16, 24, 32]. The 27mer was predicted to be linear and the 48hp was predicted to have a hairpin structure. In addition, we also used a 48mer with no predicted secondary structure (48lin), and a 90mer with minimal predicted secondary structure (90mer) (figure 1(A)) as assembly substrates. Dynamic light scattering (DLS) and TEM (figure S1 (stacks.iop.org/JPhysCM/29/484003/mmedia)) were used to measure particle size, and light scattering intensity was used to determine the relative amount of CLPs assembled with each oligo. CLPs were assembled at a charge ratio of 1:1 cargo anion to CP cation.
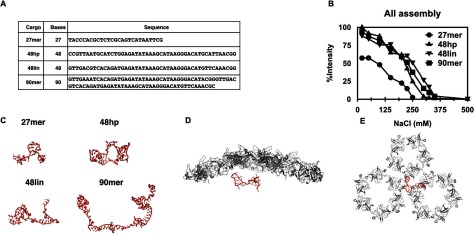
Figure 1.
Cargo length influences CLP sensitivity to ionic strength. (A) Nucleotide sequences of the four oligo cargo used in this study. (B) DLS of CLP assembly reactions using 27mer, 48hp, 48lin, and 90mer cargo. The values plotted are the average of at least three independent protein preparations and salt titrations. Determination of derived count rates and analysis of data are described in materials and methods. (C) 3D models of the different oligo cargo built using 3dRNA. 48hp is the only cargo predicted to have a hairpin motif, the others are primarily linear in structure. Details of parameters used are in materials and methods.(D) and (E) CP docked into cryo-EM virion structure (EMD 1121, PBD 1z8y) with the model of 27mer. One hexamer-pentamer-hexamer is shown. (D) is the side view and (E) is a view from the interior of the core.
Modest differences in particle size on DLS and EM suggest that changes in light scattering intensity indicate that similar amounts of CLPs were formed using 48hp, 48lin, and 90mer oligos than with the shorter 27mer (figure 1(B)). The ionic strength at which half of the total CP assembles into CLPs (50% intensity or CLP50) was used to determine the relative stability of CLPs assembled with different length oligos. The CLP50 for the 27mer encapsidated CLPs required ~125 mM NaCl while the CLP50 concentration for 48hp, 48lin, and 90mer fell between ~240–250 mM NaCl (figure 1(B)). The particle size, however, was about ~40 nm in diameter independent of the ionic strength of the reaction (data not shown), which is similar to the size of the nucleocapsid core in the virion [24, 28]. Our results show that CLPs containing longer oligos are more stable and require increased ionic strength to disassemble. These results are consistent with our hypothesis that the length of the oligo in the CLP also dictates the function of the oligo. For example, shorter oligos, like the 27mer, are long enough to neutralize the charge of the basic region of the N-terminus of the CP. However, longer oligos may not only neutralize the charge but act as a scaffold and bring CP monomers together. In addition to the cargo having a role as scaffold, it is entropically less advantageous to encapsidate many short cargo segments than a few longer ones. Thus the increase in CLPs made with longer oligos could be from both the cargo length and cargo function.
Because the different lengths of cargo gave such different results for ionic strength sensitivity, we calculated the representative oligo structures to determine if there was a correlation between cargo structure and its potential function. The surface area of the oligo that potentially contacts the CP is predicted to affect CLP assembly. We built a 3D model using 3dRNA [40, 41]. We performed ten independent molecular dynamics simulations on each oligo, at low and high salt concentrations, for a cumulative of 480–1000 ns each, the results are shown in figure 1(C). We concluded that our oligo sequences had increasing footprints based on length and presence of secondary structure. We found that only the 48hp contained a hairpin, consistent with our initial design. The presence of the hairpin made 48hp have a smaller radius of gyration than 48lin (figure 1(C), table S1). Furthermore, we determined that all oligos could collapse into a more compact structure under high-salt conditions, presumably because the salt ions screen and attenuate the Coulomb interactions of the negative charges on the oligo (table S1). To help understand the relationship between the cargo and the CPs, we show the 27mer in the presence of the CP (residues 120–264 of Sindbis) arranged as seen in the mature virion [42] (figures 1(D) and (E)).
CLPs containing 48hp and 48lin cargos are not complete, closed capsids
From our ionic strength stability results (figure 1(B)), we postulated that longer cargo that functioned to both neutralize charge and scaffold CP would favor the formation of complete, closed, spherical cores. As a result the CPs may undergo a conformational change when forming CLPs. If so, the disassembly of CLPs made with these longer cargo would require higher ionic strength compared to its assembly. Experimentally, the CLP50 for CLP disassembly would be higher than the CLP50 determined during assembly reactions. We compared assembly (figure 1(B)) and disassembly of CLPs in the presence of increasing ionic strength (figure 2(A)). For disassembly measurements we took preassembled CLPs made in low salt, titrated increasing amounts of NaCl, and monitored light scattering intensity for decreasing amounts of CLP. While there was a small shift in CLP50 for the disassembly of 48hp CLPs compared to the assembly, no detectable hysteresis was observed in CLPs made with 27mer, 48lin, or 90mer, suggesting that these CLPs are not closed capsids and/or the CP does not undergo a major conformational change when forming a CLP.
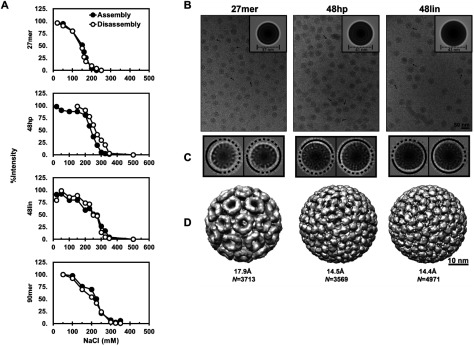
Figure 2.
CLPs have incomplete capsid shells independent of cargo length. (A) The amount of CLPs present during assembly and disassembly at increasing NaCl concentrations based on light scattering intensity. (B) Images of CLPs used for cryo image reconstruction. The arrows indicate non-spherical particles. Inset, rotationally and translationally averaged particle images showing different concentric densities between the 27mer CLP and the 48hp/48lin CLPs. (C) 2D images and class-averaging of 27mer, 48hp, and 48lin CLPs shows regions of discontinuity in the CP shell. (D) cryo-EM reconstructions imposing icosahedral symmetry show that 27mer CLPs have different surface arrangement of CP compared to CLPs assembled with 48hp and 48lin oligos.
From figure 2(A), the properties of 48lin and 90mer CLPs were similar to each other yet different when compared to the CLPs containing 27mer, and slightly different when compared to CLPs containing 48hp. We examined CLPs assembled with 27mer, 48hp, and 48lin by cryo-EM. We had three questions: Are the CLPs made with longer cargo closed capsids? Are the structures of CLPs made with longer cargos (48hp and 48lin) different than the structure of the 27mer CLP possibly a consequence of cargo length? Is there a difference between the structures of the CLPs made with 48hp and 48lin as suggested in the hysteresis experiments (figure 2(A))? Cryo-EM images of 48hp and 48lin CLPs showed that, like the 27mer CLPs, a majority of the particles were spherical (figure 2(B), top panel) but we also observed some individual particles with non-uniform density distribution at the surface (figure 2(B), arrow); particle diameters did not vary by more than 4% from the average size. We also observed very low amounts of multiplets in all CLP samples. When processing images, we determined that there were major structural differences between CLPs assembled with the 27mer compared to those assembled with longer cargo. The 48hp CLPs and 48lin CLPs were larger, with average diameters of 43 nm and 41 nm respectively, compared to 27mer CLPs which were 37 nm in diameter (figure 2(B), insets). Rotationally and translationally averaged particle images showed that both 48hp and 48lin CLPs have three concentric rings surrounding a dense center; whereas the 27mer CLP has only two rings around its center (figure 2(B), insets). Reference-free 2D classification was used to assess the structural features from the cryo-EM images (figure 2(C)). The resulting class averages revealed partially ordered density at the same positions as the three rings seen in the rotationally and translationally averaged particles (figure 2(B), inset). However, all of the class averages showed the CLPs with longer cargo were not complete, closed capsids (figure 2(C)).
To determine the arrangement of CP in the CLPs, the 3D structure of each CLP was determined imposing icosahedral symmetry. Applying icosahedral symmetry will allow resolution of the overall surface topology and arrangement of the CLPs while minimizing the effects of gaps or defects in a largely icosahedral CP lattice. If we imposed icosahedral symmetry on completely asymmetric particles we would expect to see a smooth surface. We determined the structure of 48hp and 48lin CLPs to ~14.5 Å resolution using 4971 and 3569 particles respectively (figure 2(D)). These CLP structures were compared to our previously published 27mer CLP structure. All CLPs had T = 4 symmetry but their surface arrangements were not the same (figure 2(D)). When comparing the CLPs made with the different oligos we saw that the orientation corresponding to the CP density was rotated between each structure. Taken together, our DLS and cryo-EM data show that CLPs assembled with 27mer cargo are structurally different than CLPs assembled with the longer cargos, possibly contributing to their differences in stability. The 48hp CLPs showed a slight shift in CLP50 between assembly/disassembly compared to 48lin CLPs, however, at our resolution, CLPs assembled with both 48hp and 48lin oligos have incomplete CP shells.
Mutant CP self-assembles into CLPs independent of cargo
Previous work has shown that Alphavirus CP does not self-assemble in the absence of polyanionic cargo [13, 16]. Our current results indicate that the cargo is important for both charge neutralization and scaffolding multiple CP monomers. We hypothesized that we could fulfill the function of the cargo by introducing polyanionic charge into the N-terminus of the CP itself. To test this hypothesis we made a 4D mutant where we mutated four consecutive Lys residues (104–107, RRV CP numbering) to Asp (figure 3(A)) thus substituting four basic amino acids with four acidic ones. We would predict cargo-free assembly to occur with the 4D mutant because the 4D could either neutralize charges on the same CP monomer, similar to a 27mer oligo, or neutralize charges on an adjacent CP monomer. In the process of neutralizing charges on an adjacent monomer, the N-terminus would be fulfilling the scaffolding role of the cargo as well. We further predicted that the 4D mutant would be more sensitive to ionic strength because there would be fewer electrostatic interactions between CP monomers were present, similar to CLPs made with shorter oligos.
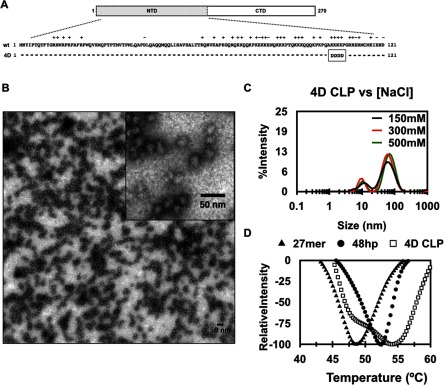
Figure 3.
RRV 4D CP self-assembles into CLPs in the absence of cargo. (A) Schematic showing the four lysine residues, amino acids 104–107, in the N-terminus of RRV CP that were mutated to aspartic acid in the 4D mutant. (B) TEM micrograph showing 4D cargo-free CLPs. (C) DLS spectra of 4D cargo-free CLPs in the presence of increasing ionic strength (black = 150 mM, red = 300 mM, and green = 500 mM NaCl). One representative result from three experiments that were performed is shown. (D) Thermal denaturation of 4D cargo-free CLPs, and 27mer and 48hp containing CLPs. The average of three experimental runs is shown.
We found that 4D CP does indeed self-assemble into CLPs in the absence of cargo (figures 3(B)–(D)). These cargo-free CLPs have different properties than cargo-containing CLPs made with wild-type CP. The 4D particles are spherical and have diameters of ~33–37 nm by TEM and ~43–58 nm by DLS (figures 3(B) and (C)). In contrast to CLPs assembled with the four different oligos, the total amount of cargo-free 4D CLPs formed was lower than the amount of CLPs formed using wild-type CP, even at the highest 4D CP concentration used.
Unlike CLPs assembled with oligos and WT CP, when the ionic strength of the reaction mixture was increased, the amount of cargo-free 4D CLPs formed did not decrease (figure 3(C)). This was surprising since the mutations in 4D CP were expected to promote electrostatic-mediated scaffolding between adjacent N-termini of CP monomers. In the 4D cargo-free samples we also observed a small peak corresponding to about 10 nm that was also present in low and high ionic strength buffer. We speculate this smaller peak corresponds to unassembled proteins but do not know their oligomeric state. The differences in particle diameter and resistance to ionic strength suggested that 4D CP in CLPs interacted differently than WT CP in CLPs assembled with oligo. To explore this possibility, we used differential scanning fluorometry to compare the thermal stability of cargo-free 4D CLPs to CLPs assembled with WT CP and 27mer and 48hp cargos (figure 3(D)) [43, 44]. Based on the stability of the 4D CLPs under increased ionic strength, we expected 4D CLPs to be more stable to thermal denaturation than WT CLPs assembled in the presence of cargo: that the CP–CP interactions in 4D CLPs would be stronger than the cargo-CP scaffolding interactions in WT, cargo-containing CLPs. The inflection point for 27mer and 48hp CLPs were ~49 °C and ~52 °C respectively, consistent with our previous findings that 48hp CLPs were more stable than 27mer CLPs. These monophasic melting curves suggest that CLP and CP melting are simultaneous. We found that 4D CLPs produced a biphasic melt-curve with inflection points of ~48 °C and 54 °C (figure 3(D)). These results indicate that 4D CLPs have two classes of interactions that combine to stabilize CP. We propose that the first inflection point may correspond to the electrostatic interactions between Asp rich region of one CP monomer with the basic rich region of adjacent CP monomer. The second inflection point may correspond to new hydrophobic or protein–protein contacts between the CP monomers that are present in the 4D CLPs but not in the WT+cargo CLPs.
Most surprising from this work is that CP self-assembly can be promoted in the absence of cargo by introducing as few as 4 anionic charges in the N-terminus of RRV CP. While 4D CLPs share some similarities with CLPs assembled from WT CP and different oligomer cargos, the differences in particle diameter, as well as chemical and thermal sensitivity, indicate that CLPs formed with and without cargo have different assembly properties and are most likely forming different structures.
Discussion
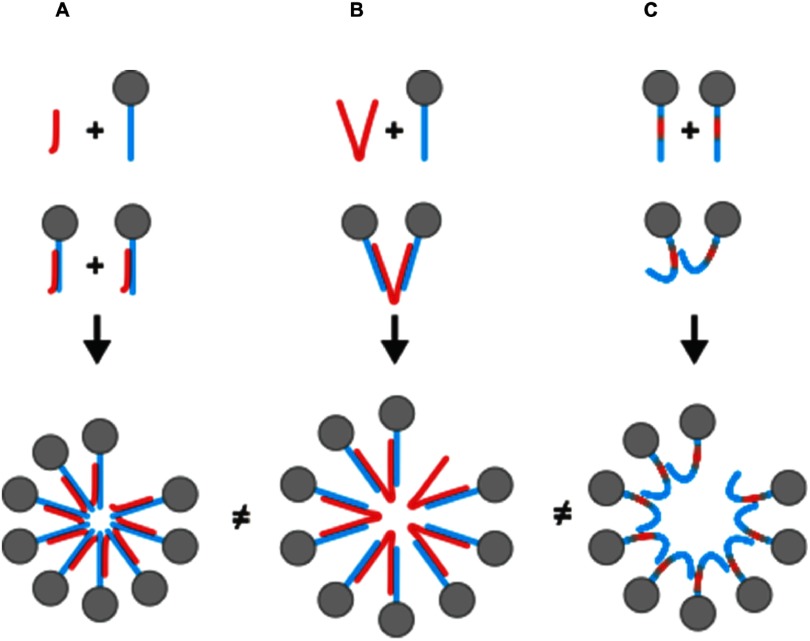
Taking our results and previously published work, we propose that cargo has two functions in alphavirus CLP assembly (figure 4). First, the anionic charges neutralize basic residues on the CP N-terminus, preventing electrostatic repulsion between CP monomers and possibly inducing structural changes in CP that confer assembly-competence. Second, cargo acts as a molecular scaffold upon which multiple CP monomers can adhere and interact. Short-range CP–CP interactions that drive assembly are promoted when CP monomers are brought within close proximity to each other via molecular scaffolding. The capacity of cargo to scaffold multiple CP monomers over long intermolecular distances increases with cargo length and the availability of anionic charge. Therefore non-adjacent CP monomers too distant to interact directly via CP–CP interactions can still be templated for CLP assembly. As cargo length decreases, so does the available anionic charge to neutralize basic residues on multiple CP monomers and to scaffold CP over long distances. Charge neutralization is favored over long-distance scaffolding when cargo is short, and assembly is driven primarily by CP–CP interactions between adjacent CP monomers [24]. However when longer cargo is present, both neutralization and scaffolding of multiple CP monomers is possible, allowing for a greater number of intermolecular contacts between CP and cargo alike. We suggest that CLPs formed with longer cargo are more stable to increasing ionic strength and thermal denaturation because of increased intermolecular interactions between CP and cargo conferred with an increase in available anionic charge and scaffold length. One could also speculate that with longer cargo, the assembly competent CP is in a different conformation compared to CP bound to shorter cargo.
Figure 4.
Model for how cargo functions to neutralize and scaffold CP depending on its length. (A) The short length of the 27mer (red) will neutralize the CP N-terminus (blue) but limits cargo-mediated scaffolding of CP monomers and subunits. (B) Cargo 48 nucleotides and longer neutralize and scaffold CP monomers that potentially stabilize the assembled CLP. (C) Mutagenesis of the N-terminus of CP can fulfill the role of neutralization and all for CLP assembly as observed with 4D cargo-free CLPs. The CLPs formed via the three pathways have different sensitivities to ionic strength and heat suggesting, consistent with them being structurally different.
In the 4D mutant we set out to combine the electrostatic and scaffolding roles of the cargo into the N-terminus of the CP (figure 3). By replacing four basic residues with four acidic residues, the N-terminus of 4D CP could function both to neutralize and scaffold CP monomers. The cargo-free CLPs (4D CLPs) had very different properties compared to cargo-containing CLPs. The 4D CLPs were smaller than cargo-containing CLPs and were not sensitive to ionic strength, suggesting that CP–CP interactions between adjacent monomers are not primarily electrostatic.
In addition, the thermal stability of the 4D CLPs showed a biphasic profile in contrast to single peaks seen in the cargo-containing CLPs suggesting at least two types of contacts—contacts between adjacent N-termini in addition to different contacts between CP monomers—are occurring in the 4D mutant (figure 4). One explanation for this phenotype is the mutations have minimized any repulsion that may be present between the more positively charged N-terminal domains of CP. Now, the N-termini of CP can come in closer contact and new CP–CP interactions are formed. When the N-terminus arginine-rich RNA binding domain from Cowpea chlorotic mottle virus is removed, T = 1 particles are formed presumably because the repulsion between basic residues was removed. With HBV, CP protein missing the arginine-rich domain can self-assemble but when this domain is present either nucleic acid or phosphorylation of the CP is necessary, implying that electrostatic repulsion between arginine-rich domains must be neutralized before assembly can occur, ([45, 46], personal communication M Comos Garcia and W Gelbart).
Virus capsids have usually been shown to be spherical, closed shells. Our results showed the contrary. We found that regardless of the length of cargo used, the alphavirus CLPs still had incomplete CP shells, as seen either by cryo-EM reconstructions or testing for hysteresis (figures 2(A) and (B)). Upon closer analysis, we found that CLPs assembled with the 27mer and 48mer cargo show variances in their structure and, perhaps as a consequence, explain the differences in sensitivity to ionic strength and temperature (figures 2(A) and 3(D)). There was no obvious difference between CLPs made with 48mer and 90mer in our assays. One possible explanation for the retention of defects is that we are observing kinetic traps, often caused by relatively strong interactions between subunits [7, 8]. If this is the case for RRV CP, it would not be surprising that assembly around a longer cargo would allow for differential orientation of CP in a CLP compared to the CP arrangement in a CLP made with a shorter cargo.
We see that CLPs assembled with a 27mer DNA oligo are structurally distinct from CLPs assembled with longer cargo of 48 or 90 bases, specifically in the interior region of the CLP where CP interacts with cargo (figure 2(B)). From our cryo-EM 2D classification we see multiple layers of density in particles encapsidating the longer cargo (longer than 27mer) suggesting increased order in the CP or additional contacts made because of the scaffolding function of the cargo. The C-terminal domain of the atomic structure of RRV CP fit well in the cryo density of the CLPs suggesting the differences in topology and inner density is due to rotation/translation and N-terminal CP conformation, all dependent on the length of the encapsidated cargo. We also speculate that the different numbers of concentric rings of density implies different N-terminal conformations in CP and thus a structural difference between CLPs containing 27mer compared to CLPs with 48mer (figure 2(B)). This model agrees well with the role of cargo as a scaffold or bridge between CP monomers, orienting them into different conformation, and provides an explanation for why cargo is required for assembly. This model would also be consistent with previous work that showed crosslinking retroviral Gag proteins, with a nucleic acid or by inserting a coiled -coil domain, led to enhanced assembly [47, 48]; an effect readily explained by increasing the valency of the assembly competent CP [6, 49].
How do our observations compare to previous work on alphavirus core assembly and assembly models using different spherical viruses? Using full length RRV CP and 27mer as cargo, Wang et al proposed that an assembly competent CP subunit consists of one 27mer bound to the N-terminus of one CP [24]. Going from subunit to CLP assembly, the authors put forth a model where strong CP–CP subunit interactions trap structural defects, leading to incomplete CP shells. This model provides an explanation for the structural defects observed in the cryo-EM classification, but does not account for the possible role of longer cargo in scaffolding or bridging between CP monomers to promote CLP assembly. Reports using CP peptides and longer cargo (48–132 nucleotides in length) have proposed that CP-nucleic acid binding promotes the stabilization of a CP dimer bound to nucleic acid. In this model, CP dimers function as the assembly competent subunit, and multiple dimers then oligomerize to form CLPs [29, 33, 34]. Additional interactions between adjacent helical N-terminal domains [30, 31], and/or interactions between adjacent C-terminal domains [50] were thought to follow during CLP assembly. Whether CLP assembly occurs with subunits of monomeric or multiple CPs bound to cargo, or additional interactions between purported helical domains of CP occur subsequent to cargo-binding, is not directly addressed in our experiments. However, our results suggest a direct correlation between the length of cargo and its ability to interact with the N-terminus of CP, leading to altered CP conformations in an assembled CLP and differential CLP stability.
Studies with MS2 bacteriophage [51–53], Hepatitis B virus [45], and Cowpea chlorotic mottle virus [25, 26, 54, 55] suggest that initial interactions between cargo and CP may be strongly influenced by cargo length and structure [49, 51, 52], and that a series of coordinated cargo-CP and CP–CP interactions are required for assembly. These studies suggest that specific RNA-CP interactions occur and then subsequently the CP–CP interactions drive assembly around the viral RNA. It has also been proposed that very weak CP–CP interactions do not promote assembly but too strong CP–CP interactions lead to kinetic traps [55].
Assembly models that favor strong CP–CP interactions have also been observed. SV40 favors stepwise assembly of the capsid that occur concomitantly with nucleic acid binding [56]. Consistent with our results, SV40 assembly has shown tertiary structure of the cargo and length of cargo influence the assembly of the virion [57]. Computational models show that weak CP–CP interactions favor initial formation of a disordered CP-nucleic acid oligomer that then assembles, while strong interactions favor co-assembly [19, 20, 21].
Our experimental results are consistent with the observations of Perlmutter et al [20] and work by van der Schoot and Zandi et al [58–60] that assembled capsid tend to be ‘overcharged’ with excess genomic material. Through computational simulations Perlmutter and Hagan [20] determined that the most stable capsids have regions of the genome that are not directly binding to CP but are bridging adjacent CP monomers or subunits, acting as a scaffold or linker. In our model, we propose that overcharging occurs in the CLPs as the length of encapsidated cargo and the amount of available anionic charge increases over the number of basic residues to neutralize the N-terminus of CP. It is important to note that in these studies we do not purify CLPs or subsequently quantify the amount of encapsidated cargo or extent of overcharging.
The results presented in this work emphasizes that in order to fully understand how capsid assembly occurs—in the presence or absence of cargo—we must investigate the subunit interactions that occur. All CLPs may have similar structures but the chemical properties of the subunits, or complexes of subunits, alter their physical interactions and drastically modify the properties of the CLP.
Materials and methods
Ross River capsid protein purification
Ross River virus capsid protein (RRV CP) was purified as described previously [24] except the cell pellet was resuspended into Buffer A (20 mM HEPES, pH 6.8, 250 mM NaCl, 5 mM EDTA, 5% Glycerol, and 1 mM DTT) with a protease inhibitor cocktail (1 Roche™ complete mini protease tablet/gram of cell pellet, 1 µg ml−1 Leupeptin, 1 µg ml−1 Aprotinin, 1 mM PMSF). A ratio of 10 ml buffer to 0.6–0.8 g of cell pellet was used. Full length CP was buffer exchanged into Storage Buffer (20 mM HEPES, pH 6.8, 150 mM NaCl, 0.1 mM EDTA) using Vivaspin 6 10 kDa filters (GE Healthcare) at 5000 × g. Protein concentration was determined by measuring absorbance at 280 nm using an extinction coefficient of 39 670 M−1 cm−1.
Oligonucleotides used for cargo
Oligos for the assembly reactions were purchased from IDT (Coralville, IA), resuspended in water. The nucleotide sequences are TACCCACGCTCTCGCAGTCATAATTCG (27mer), CCG TTAATGCATGTCGAGATATAAAGCATAAGGGACATGCATTAACGG (48hp), GTTGACGTCACAGATGAGATATAAAGCATAAGGGACATGTTCAAACGG (48lin), and GTTGAAATCACAGATGAGATATAAAGCATAAGGGACATACGGGTTGACGTCACAGAT- GAGATATAAAGCATAAGGGACATGTTCAAACGC (90mer).
3D model generation and simulations of oligo
The 3D model of the different oligo cargo was built using 3dRNA [40, 41]. The generated RNA model was transformed into DNA by changing uracil to thymine. The cargo models (27mer, 48hp, 48lin, and 90mer) were similarly solvated in a sufficiently large water box at low and high salt concentrations. Ten independent equilibrium molecular dynamics simulations were performed for a cumulative of ~480–1000 ns for each system. To quantify the size of the cargo, the average radius of gyration was calculated based on the final structures of the ten independent simulations.
Dynamic light scattering (DLS)
CLP assembly was monitored using DLS. For assembly reactions, CLPs were assembled by adding cargo and NaCl in Buffer C (20 mM HEPES, 0.1 mM EDTA at pH 6.8) to the indicated concentration. Assembly was initiated by adding CP to obtain a final concentration of 3 µM in a final volume of 30 µl. Measurements were taken on a Malvern Zetasizer™ Nano Z with the attenuator set to 8 and the measurement window at 4.2 mm. Each measurement consisted of at least three runs, each run averaged over 10 s. A peak corresponding to the hydrodynamic diameter of the core like particles (~48 nm) was observed. The data was plotted with the percent derived count rate on the Y-axis and the salt concentration on the X-axis. For disassembly reactions CLPs were pre-assembled in 20 mM salt and diluted into Buffer C containing indicated amounts of NaCl. The final concentration of CP was 2.5 µM in a volume of 30 µl for each reaction. The data was visualized using the Zetasizer software from Malvern and analysis was done using Microsoft Excel 2013 and GraphPad Prism.
DSF
CLP stability was monitored by the increase in the fluorescence of the SYPRO Orange dye. CLPs were prepared at a charge ratio of 1:1 and diluted in 20 mM Phosphate, pH 6.8, 50 mM NaCl, 0.1 mM EDTA to yield a final protein concentration of 2.5 µM in a final volume of 30 µl. Phosphate buffer was used instead of HEPES because it has a low temperature dependence (ΔpH/ΔT = −0.0028). For all samples 3 ul of SYPRO Orange was added, yielding a final concentration of 2.5× (the absolute concentration is not disclosed by the manufacturer). MicroAmp optical adherent film was used to cover the microplate during the heating process. Buffer and water were used as negative controls and CP without the cargo was used as the positive control. The CLPs were incubated in a StepOnePlus™ Real-Time PCR instrument. Samples were treated from 25 °C to 99.9 °C, in increments of 0.26° steps per 20 s and each temperature was maintained for 80 s. The fluorescence intensity was measured every 20 s. The negative first derivatives of fluorescence intensities were plotted as a function of temperature by using the internally developed software package. At least two measurements were performed. The data was analyzed using the StepOnePlus™ software and analyzed using Microsoft Excel 2013 and GraphPad Prism.
Negative stain TEM
For TEM, 3–5 µl of a CLP reaction mix was applied to a Formvar and carbon-coated 400-mesh copper grid (Electron Microscopy Sciences, Hatfield, PA) and stained with 1% uranyl acetate. The stained grids were analyzed using a JEOL 1010 transmission electron microscope (Tokyo, Japan) operating at 80 kV. Images were recorded using a Gatan UltraScan 4000 charge-coupled-device camera (Pleasanton, CA).
Cryo-electron microscopy of CLPs
CLPs were prepared using a charge ratio of 1 in Buffer C (20 mM HEPES pH 6.8, 0.1 mM EDTA, 50 mM NaCl) with a final protein concentration of 3 µM. Samples were prepared and data collected as described previously [24]. In total 104 and 195 cryo-EM images were collected for CLP 48lin and CLP 48hp, respectively. Image processing was performed as described previously [24]. A total of 3929 particles were selected for 48lin dataset and 6282 particles were selected for 48hp dataset. For 3D reconstruction, the initial model was generated from a cryo-EM model of Sindbis virus, in which the glycoprotein shell and the encpasidated RNA core were both removed. Structure determination and 3D reconstruction were carried out by applied icosahedral symmetry using auto3dem v4.05 [61]. The final resolution was estimated to 14.5 Å for 48lin (3569 particles) and 14.4 Å for 48hp (4971 particles) based on a Fourier shell correlation cutoff at 0.5. The 3D models were visualized using UCSF Chimera [62].
Supplementary Material
Funding
We thank the IU Nanoscale Characterization Facility for access to the instrumentation and members of the Mukhopadhyay and Zlotnick lab for scientific input. This work was supported by NIH Training Grant T32 GM0007757 (AM), Institute for Advanced Study at IU (SM), NIH 9P41GM104601 (BCG/JRP), and NIH R01 AI067417 (AZ). Molecular dynamics simulations were performed on the Blue Waters Supercomputers, financed by the National Science Foundation (OCI-0725070 and ACI-1238993) under Petascale Computational Resource (PRAC) grant ACI-1440026.
References
- 1.Enquist L W, Shalka A M, Flint S J, Racaniello V R, Flint S J. In: Principles of Virology: Molecular Biology, Pathogenesis, and Control of Animal Viruses. FLint S J, editor. Washington DC: American Society for Microbiology; 2003. [Google Scholar]
- 2.Czapar A E, Steinmetz N F. Plant viruses and bacteriophages for drug delivery in medicine and biotechnology. Curr. Opin. Chem. Biol. 2017;38:108–16. doi: 10.1016/j.cbpa.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen A M, Steinmetz N F. Design of virus-based nanomaterials for medicine, biotechnology, and energy. Chem. Soc. Rev. 2016;45:4074–126. doi: 10.1039/C5CS00287G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwarz B, Uchida M, Douglas T. Biomedical and catalytic opportunities of virus-like particles in nanotechnology. Adv. Virus Res. 2017;97:1–60. doi: 10.1016/bs.aivir.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perlmutter J D, Hagan M F. Mechanisms of virus assembly. Annu. Rev. Phys. Chem. 2015;66:217–39. doi: 10.1146/annurev-physchem-040214-121637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zlotnick A. Are weak protein-protein interactions the general rule in capsid assembly? Virology. 2003;315:269–74. doi: 10.1016/S0042-6822(03)00586-5. [DOI] [PubMed] [Google Scholar]
- 7.Katen S, Zlotnick A. The thermodynamics of virus capsid assembly. Methods Enzymol. 2009;455:395–417. doi: 10.1016/S0076-6879(08)04214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh S, Zlotnick A. Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. J. Biol. Chem. 2003;278:18249–55. doi: 10.1074/jbc.M211408200. [DOI] [PubMed] [Google Scholar]
- 9.Hagan M F, Chandler D. Dynamic pathways for viral capsid assembly. Biophys. J. 2006;91:42–54. doi: 10.1529/biophysj.105.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Schoot P, Zandi R. Kinetic theory of virus capsid assembly. Phys. Biol. 2007;4:296–304. doi: 10.1088/1478-3975/4/4/006. [DOI] [PubMed] [Google Scholar]
- 11.Toropova K, et al. The three-dimensional structure of genomic RNA in bacteriophage MS2: implications for assembly. J. Mol. Biol. 2008;375:824–36. doi: 10.1016/j.jmb.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 12.Witherell G W, Gott J M, Uhlenbeck O C. Specific interaction between RNA phage coat proteins and RNA. Prog. Nucl. Acid Res. Mol. Biol. 1991;40:185–220. doi: 10.1016/s0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]
- 13.Cheng F, et al. The packaging of different cargo into enveloped viral nanoparticles. Mol. Pharm. 2013;10:51–8. doi: 10.1021/mp3002667. [DOI] [PubMed] [Google Scholar]
- 14.Feng Y X, et al. Reversible binding of recombinant human immunodeficiency virus type 1 gag protein to nucleic acids in virus-like particle assembly in vitro. J. Virol. 2002;76:11757–62. doi: 10.1128/JVI.76.22.11757-11762.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 1999;73:2270–9. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tellinghuisen T L, et al. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J. Virol. 1999;73:5309–19. doi: 10.1128/jvi.73.7.5309-5319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfeiffer P, Hirth L. Aggregation states of brome mosaic virus protein. Virology. 1974;61:160–7. doi: 10.1016/0042-6822(74)90251-7. [DOI] [PubMed] [Google Scholar]
- 18.Pfeiffer P, Hirth L. Formation of artificial top component from brome mosaic virus at high salt concentrations. Virology. 1974;58:362–8. doi: 10.1016/0042-6822(74)90071-3. [DOI] [PubMed] [Google Scholar]
- 19.Perlmutter J D, Perkett M R, Hagan M F. Pathways for virus assembly around nucleic acids. J. Mol. Biol. 2014;426:3148–65. doi: 10.1016/j.jmb.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perlmutter J D, Qiao C, Hagan M F. Viral genome structures are optimal for capsid assembly. Elife. 2013;2:e00632. doi: 10.7554/eLife.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zlotnick A, Porterfield J Z, Wang J C. To build a virus on a nucleic acid substrate. J. Biophys. 2013;104:1595. doi: 10.1016/j.bpj.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belyi V A, Muthukumar M. Electrostatic origin of the genome packing in viruses. Proc. Natl Acad. Sci. USA. 2006;103:17174–8. doi: 10.1073/pnas.0608311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni P, et al. An examination of the electrostatic interactions between the N-terminal tail of the brome mosaic virus coat protein and encapsidated RNAs. J. Mol. Biol. 2012;419:284–300. doi: 10.1016/j.jmb.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J C, et al. Self-assembly of an alphavirus core-like particle is distinguished by strong intersubunit association energy and structural defects. ACS Nano. 2015;9:8898–906. doi: 10.1021/acsnano.5b02632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cadena-Nava R D, et al. Self-assembly of viral capsid protein and RNA molecules of different sizes: requirement for a specific high protein/RNA mass ratio. J. Virol. 2012;86:3318–26. doi: 10.1128/JVI.06566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garmann R F, et al. Role of electrostatics in the assembly pathway of a single-stranded RNA virus. J. Virol. 2014;88:10472–9. doi: 10.1128/JVI.01044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Y, et al. Packaging of a polymer by a viral capsid: the interplay between polymer length and capsid size. Biophys. J. 2008;94:1428–36. doi: 10.1529/biophysj.107.117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng R H, et al. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–30. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tellinghuisen T L, Perera R, Kuhn R J. In vitro assembly of Sindbis virus core-like particles from cross-linked dimers of truncated and mutant capsid proteins. J. Virol. 2001;75:2810–7. doi: 10.1128/JVI.75.6.2810-2817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perera R, et al. Alphavirus nucleocapsid protein contains a putative coiled coil alpha-helix important for core assembly. J. Virol. 2001;75:1–10. doi: 10.1128/JVI.75.1.1-10.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong E M, Perera R, Kuhn R J. Alphavirus capsid protein helix I controls a checkpoint in nucleocapsid core assembly. J. Virol. 2006;80:8848–55. doi: 10.1128/JVI.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukhopadhyay S, et al. In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J. Virol. 2002;76:11128–32. doi: 10.1128/JVI.76.21.11128-11132.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warrier R, et al. Role of sindbis virus capsid protein region II in nucleocapsid core assembly and encapsidation of genomic RNA. J. Virol. 2008;82:4461–70. doi: 10.1128/JVI.01936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linger B R, et al. Sindbis virus nucleocapsid assembly: RNA folding promotes capsid protein dimerization. RNA. 2004;10:128–38. doi: 10.1261/rna.5127104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garoff H, et al. The capsid protein of Semliki Forest virus has clusters of basic amino acids and prolines in its amino-terminal region. Proc. Natl Acad. Sci. USA. 1980;77:6376–80. doi: 10.1073/pnas.77.11.6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bell J R, Strauss E G, Strauss J H. Purification and amino acid compositions of the structural proteins of sindbis virus. Virology. 1979;97:287–94. doi: 10.1016/0042-6822(79)90340-4. [DOI] [PubMed] [Google Scholar]
- 37.Bell J R, et al. Structural proteins of Western equine encephalitis virus: amino acid compositions and N-terminal sequences. J. Virol. 1983;45:708–14. doi: 10.1128/jvi.45.2.708-714.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi H K, et al. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 39.Choi H K, et al. Structural analysis of Sindbis virus capsid mutants involving assembly and catalysis. J. Mol. Biol. 1996;262:151–67. doi: 10.1006/jmbi.1996.0505. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Xiao Y. Using 3dRNA for RNA 3D structure prediction and evaluation. Curr. Protoc. Bioinform. 2017;57:5.9.1–12. doi: 10.1002/cpbi.21. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y, et al. Automated and fast building of three-dimensional RNA structures. Sci. Rep. 2012;2:734. doi: 10.1038/srep00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukhopadhyay S, et al. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure. 2006;14:63–73. doi: 10.1016/j.str.2005.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rayaprolu V, et al. Comparative analysis of adeno-associated virus capsid stability and dynamics. J. Virol. 2013;87:13150–60. doi: 10.1128/JVI.01415-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rayaprolu V, et al. Fluorometric estimation of viral thermal stability. Bio Protoc. 2014;4:e1199. doi: 10.21769/BioProtoc.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel N, et al. HBV RNA pre-genome encodes specific motifs that mediate interactions with the viral core protein that promote nucleocapsid assembly. Nat. Microbiol. 2017;2:17098. doi: 10.1038/nmicrobiol.2017.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selzer L, Kant R, Wang J C, Bothner B, Zlotnick A. Hepatitis B virus core protein phosphorylation sites affect capsid stability and transient exposure of the C-terminal Domain. J. Biol. Chem. 2015;290:28584. doi: 10.1074/jbc.M115.678441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Datta S A, et al. Dimerization of the SP1 region of HIV-1 gag induces a helical conformation and association into helical bundles: implications for particle assembly. J. Virol. 2015;90:1773–87. doi: 10.1128/JVI.02061-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson M C, et al. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 2002;76:11177–85. doi: 10.1128/JVI.76.22.11177-11185.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zlotnick A, Stray S J. How does your virus grow? Understanding and interfering with virus assembly. Trends Biotechnol. 2003;21:536–42. doi: 10.1016/j.tibtech.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 50.Snyder J E, et al. Probing the early temporal and spatial interaction of the Sindbis virus capsid and E2 proteins with reverse genetics. J. Virol. 2012;86:12372–83. doi: 10.1128/JVI.01220-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stockley P G, et al. Bacteriophage MS2 genomic RNA encodes an assembly instruction manual for its capsid. Bacteriophage. 2016;6:e1157666. doi: 10.1080/21597081.2016.1157666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rolfsson O, et al. Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J. Mol. Biol. 2016;428:431–48. doi: 10.1016/j.jmb.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borodavka A, Tuma R, Stockley P G. Evidence that viral RNAs have evolved for efficient, two-stage packaging. Proc. Natl Acad. Sci. USA. 2012;109:15769–74. doi: 10.1073/pnas.1204357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garmann R F, et al. Physical principles in the self-assembly of a simple spherical virus. Acc. Chem. Res. 2016;49:48–55. doi: 10.1021/acs.accounts.5b00350. [DOI] [PubMed] [Google Scholar]
- 55.Garmann R F, et al. The assembly pathway of an icosahedral single-stranded RNA virus depends on the strength of inter-subunit attractions. J. Mol. Biol. 2014;426:1050–60. doi: 10.1016/j.jmb.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kler S, et al. RNA encapsidation by SV40-derived nanoparticles follows a rapid two-state mechanism. J. Am. Chem. Soc. 2012;134:8823–30. doi: 10.1021/ja2110703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li C, et al. Single particle observation of SV40 VP1 polyanion-induced assembly shows that substrate size and structure modulate capsid geometry. ACS Chem. Biol. 2017;12:1327–34. doi: 10.1021/acschembio.6b01066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Erdemci-Tandogan G, et al. RNA topology remolds electrostatic stabilization of viruses. Phys. Rev. E. 2014;89:032707. doi: 10.1103/PhysRevE.89.032707. [DOI] [PubMed] [Google Scholar]
- 59.Erdemci-Tandogan G, et al. Effects of RNA branching on the electrostatic stabilization of viruses. Phys. Rev. E. 2016;94:022408. doi: 10.1103/PhysRevE.94.022408. [DOI] [PubMed] [Google Scholar]
- 60.Li S, et al. Impact of a nonuniform charge distribution on virus assembly. Phys. Rev. E. 2017;96:022401. doi: 10.1103/PhysRevE.96.022401. [DOI] [PubMed] [Google Scholar]
- 61.Yan X, Sinkovits R S, Baker T S. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pettersen E F, et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.