Highlights
-
•
There are two calnexin genes in the trout genome.
-
•
There are two different calnexin proteins.
-
•
Only one of the calnexin proteins has an ER retention motif.
-
•
The smaller form of calnexin is inducible, the larger is not.
Keywords: Chaperone, ER stress, Calcium ionophore, Major histocompatibility, MHC class I, Antigen presentation
Abstract
Calnexin (IP90/P88) is an integral membrane protein of the endoplasmic reticulum that binds newly synthesized N-linked glycoproteins during their folding in the ER including MHC class I molecule. This manuscript reports the identification of two unique cDNA clones of calnexin in rainbow trout. Both encode putative mature proteins of 579 and 592 aa respectively in addition to a 24 aa signal peptide. Sequence analysis revealed that only one of the two cDNA clones encodes a putative ER retention signal, K/QEDDL, followed by a serine phosphorylation site conserved with mammalian homologs. Amino acid sequence alignment illustrated conservation of the calnexin luminal domain, which consists of a globular and a P domain, in both copies. Southern blotting revealed that there are at least two copies of the calnexin gene in the trout genome and northern blotting showed a wide tissue distribution of an estimated 3 kbp calnexin transcript with an additional minor transcript of 2.3 kbp expressed only in head kidney, spleen PBLs and strongly in RTS11. Importantly, the smaller transcript was predominantly upregulated in RTS11 after a 24 h treatment with the calcium ionophore A23187. In western blots, calnexin was detected primarily as a 120 kDa protein and upon A23187 treatment; a 100 kDa band was most prominently expressed. These results suggest that in salmonids there are two differentiated versions of the calnexin gene which encode proteins that may have diverged to perform unique biological functions.
1. Introduction
Calnexin (IP90/P88) is an integral membrane protein of the endoplasmic reticulum that binds to monoglycosylated oligosaccharides. It was first identified as a protein that associates with partly folded MHC class I molecules, T-cell receptors and membrane immunoglobulins (Degen and Williams, 1991, Ahluwalia et al., 1992, Hochstenbach et al., 1992). Together with calreticulin, its soluble homologue, it participates in the calnexin/calreticulin cycle responsible for folding and quality control of newly-synthesized glycoproteins before their export from the ER (Ou et al., 1993, Hammond et al., 1994, Ellgaard and Helenius, 2003). Calnexin has also been shown to play a role in ER calcium regulation (Wada et al., 1991, Roderick et al., 2000), phagocytosis (Müller-Taubenberger et al., 2001) and cell sensitivity to apoptosis (Guérin et al., 2008, Takizawa et al., 2004, Zuppini et al., 2002, Delom et al., 2007). Two distinct luminal domains of calnexin were revealed by crystallization (Schrag et al., 2001): the globular domain that contains the lectin binding site and an extended arm domain, containing two tandem proline rich motifs, which binds to ERp57 (Leach et al., 2002). The C-terminal domain is highly acidic with a protein kinase-dependent phosphorylation site that plays a possible role regulating the chaperone function (Ou et al., 1992, Chevet et al., 1999). One well investigated function of calnexin is stabilizing both major histocompatibility MHC class I and class II molecules (Anderson and Cresswell, 1994, Carreno et al., 1995). In humans, newly translated MHC class I heavy chain associates with calnexin rapidly, but it is released and replaced by calreticulin upon assembly of heavy chain with beta2 microglobulin. However, in murine cells either calnexin or calreticulin may associate with beta2 microglobulin-heavy chain dimers. Interestingly, experiments assessing the function of calnexin in the biogenesis of class I molecules showed contradicting results. Co-expression of calnexin or calreticulin with MHC class I heavy chain in Drosophila melanogaster cells demonstrated fivefold enhanced assembly with the beta2 microglobulin, while treatment with ER glucosidase I and II, inhibitors of monoglycosylated oligosaccharide formation, reduced the assembly and surface expression of MHC class I (Vassilakos et al., 1996). However, other experiments using calnexin human deficient cell lines showed no effect at all on class I assembly, transport or peptide loading (Sadasivan et al., 1995, Scott and Dawson, 1995). These experimental discrepancies may be either explained by the different model systems used or by the redundancy of molecular chaperones that can functionally replace calnexin in the ER such as calreticulin (Zhang and Williams, 2006).
Co-immunoprecipitation studies have also demonstrated that calnexin can function as chaperone for some viral glycoproteins such as the G protein of vesicular stomatitis virus (VSV) and more recently the S glycoprotein of severe acute respiratory syndrome coronavirus by assisting in their folding and full maturation (Hammond and Helenius, 1994, Fukushi et al., 2012). Calnexin homologues have been identified in many eukaryotes such as plants, yeast, Xenopus (Huang et al., 1993, Parlati et al., 1995, Yamamoto and Nakamura, 1996) several fish species [Fuller et al., 2004, Danio rerio NM_213448.1, Takifugu rubripes XM_003978229 and XM_003978694] and remarkably in Dictyostelium (Müller-Taubenberger et al., 2001), which implies possible conserved functions for this molecule. To date, calnexin’s role in teleost fish has only been investigated in channel catfish where it was shown to associate both with the non-glycosylated MHC class II alpha chain and the glycosylated beta chain (Fuller et al., 2004). However, no studies have been carried out to assess its role during MHC class I folding, MHC class I receptor assembly in the ER or its possible association with viral glycoproteins mainly due to the lack of available antibodies. In rainbow trout, besides calnexin, most of the genes encoding molecular chaperones involved in this pathway have been fully characterised such as tapasin (Landis et al., 2006), calreticulin (Kales et al., 2004) and recently ERp57 (Sever et al., 2013). In this work two unique cDNA clones for calnexin from trout peripheral blood leukocytes are reported and their regulation under ER stress induced conditions is described.
2. Materials and methods
2.1. Fish
Rainbow trout were obtained from Silver Creek Aquaculture (Erin, ON) and kept at 13 °C in 200 L fresh-water flow-through tanks at the University of Waterloo. Animals were kept using CACC guidelines under a permit from the University of Waterloo animal care committee. Adult fish (∼800 g) were anesthetized in 1 ml/L of 2-phenoxyethanol (Sigma Aldrich St. Louis, MO). Blood was drawn from the caudal sinus as previously described (Sever et al., 2013) and tissues samples were collected in RNA later (2.5 mM Na citrate, 5.3 M (NH4)2SO4, 0.01 M EDTA, pH 5.2).
2.2. Rainbow trout cell lines
The rainbow trout cell lines utilized were the monocyte/macrophage cell line, RTS11 (Ganassin and Bols, 1998), and the epithelial-like cell lines: RTL-W1 from a normal liver (Lee et al., 1993), RTgill-W1 from the gill (Bols et al., 1990) and RTovarian fluid [TK Vo and Bols, unpublished]. RTS11 was grown at 20 °C in L-15 media with 15% FBS and other cell lines were grown in 10% FBS as described by Kawano et al. (2010). The epithelial cell lines maintained epithelial-like morphology during the course of this study.
2.2.1. RTS11 stimulation with A23187 calcium ionophore and poly I:C
Cultures were seeded to 2 × 106 in a 25 cm2 flasks containing L-15 media supplemented with 150 U/ml of penicillin and 150 mg/ml streptomycin in 15% fetal bovine serum (ThermoFisher Scientific, Nepean, ON) Treatment included either 2 μM of A23187 dissolved in DMSO or 50 μg/ml of polyinosinic-polycytidylic acid (poly I:C) in PBS (Sigma Aldrich, St. Louis, MO). The same volume of vehicle was added to the control.
2.3. Cloning of rainbow trout calnexin cDNA
Degenerate primers were designed based on conserved regions identified by alignment of the following sequences from GenBank: Homo sapiens [NP_001019820.1], Mus musculus [NP_031623.1], Canis familirais [NP_001003232.1], Rattus norvegicus [NP_742005.1], D. rerio [XP_002665576.1] and Ictalurus punctatus [AAQ18011.1]. Total RNA was extracted from 2 × 106 RTS11 cells using Qiagen RNeasy extraction kit according to manufacturer’s instructions (Qiagen, Mississauga, ON), followed by single strand cDNA synthesis using a Fermentas RevertAid™ First Strand cDNA Synthesis Kit with 1 μg of total RNA (ThermoFisher Scientific,Nepean, ON). PCR reactions of 25 μl included 1× PCR buffer, 200 μM dNTP mix, 2 mM MgCl2, 10 μM each of the forward 5AARTAYGAYGGNAARTGG3′ and reverse primer 5′TCTTCTTTCCAGTGCAGCAGA3′, 1 μl of RTS11 cDNA and 1 U of Taq Polymerase (MP Biomedical, Solon, OH). The reaction parameters were 95 °C for 5 min followed by 30 cycles of (95 °C 40 s, 50 °C 30 s, 72 °C 2 min) with a final 15 min 72 °C using a BioRad DNA Engine thermocycler (BioRad, Mississauga Ontario). An estimated 1200 bp PCR product was purified using a Qiagen gel extraction kit (Mississauga, ON), then ligated into pGEM-T Easy vector, subcloned into XL1-Blue MRF′ E. coli competent cells and sequenced at The Centre for Applied Genomics (Toronto, ON, Canada as described in Sever et al. (2013).
2.3.1. Generation of full length cDNA
The full 5′ end of the calnexin transcript was obtained using RACE adapted from Frohman et al. (1988) and described in detail in Sever et al. (2013). Briefly, cDNA synthesis used four μg of total RNA derived from peripheral blood leukocytes and gene specific primer1 (GSP1) 5′ CATGTCCTCCACCTCCCA 3′. Reverse transcription was performed for 60 min at 42 °C, 5 min at 45 °C, 5 min at 50 °C and 10 min at 70 °C followed by addition of RNAseH (Invitrogen, Carlsbad, CA), dATP and terminal deoxynucleotidyl transferase (Invitrogen, Carlsbad, CA) as described (Sever et al., 2013). The PCR reaction was performed using 10 μM gene specific primer sense primer 2 (GSP2) 5′CGGGCGCTTTGTAGGTCACTT3′ combined with the RACE primer 5′CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTTTTTTTTTTTT3′ and 2 μl of template obtained from the 5′cDNA pool. The PCR reaction for the first cycle was performed under the following conditions: 95 °C for 5 min, 48 °C for 2 min and 72 °C for 15 min with additional 30 cycles of 94 °C for 15 s, 52 °C for 30 s and 72 °C for 3 min.
Sequence alignment of the 3′UTR region of both I. punctatus NM_001200180 and D. rerio NM_213448 revealed a conserved region of 18 nucleotides which facilitated the design for a PCR primer to amplify this end of the cDNA. The primers that were used in this PCR reaction were as follows: 5′ATGGAGTTGAATGTGAGGTGTG3‘ and anti-sense primer 5′AAGTCCATCAGTCCTTTCT3′. The PCR reaction to amplify the 3′ UTR was performed under the following conditions: 95 °C for 5 min, 53 °C for 30 s and 72 °C for 4 min followed by with additional 30 cycles of 94 °C for 3 min, 53 °C for 30 s and 72 °C for 3 min.
2.4. Southern blotting analysis
Genomic DNA obtained from rainbow trout peripheral blood leukocytes was extracted using a Wizard Genomic DNA purification kit (Promega, Madison, USA). Briefly, 10 μg of genomic DNA was completely digested for 4 h at 37 °C using Fast digest enzymes EcoRV, HindIII, KpnI and PstI and BamHI (ThermoFisher Scientific, Nepean, ON), separated on a 1% agarose gel and transferred to a positively charged membrane (Roche, Manheim, Germany), followed by a UV cross linking. Membranes were washed and hybridized with DIG labelled probes as previously described (Sever et al., 2013). Two distinct probes were generated either to the proximal or distal end of calnexin. The primers used to generate the 477 bp proximal probe were as follows: forward 5′AGGAGGACATTGATGAGGATATTGC3′ and reverse 5′GACCCACCTTTACACTCGGTGGTGAACCCAG3′. For the distal 540 bp probe primers were as follows: forward 5′TGGATGATCAGCCAGAGTACA3′ and reverse 5′TGCAGAAGGAAGACAATGATGAGGA3′. The bands were detected using anti-DIG antibody conjugated to alkaline phosphatase and CDP star (Roche, Mannheim, Germany).
2.5. Northern blotting analysis
RNA was isolated from both rainbow trout tissues and the RTS11 cell line using Trizol according to manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). Briefly, 8 μg of RNA from tissues and 6 μg RNA obtained from RTS11 cells were separated on a 1% agarose-formaldehyde gel and transferred to a positively charged membrane (Roche, Manheim, Germany), followed by a UV crosslinking. The membrane was washed, hybridized and detected as previously described (Sever et al., 2013). The primers used to generate the 728 bp probe were as follows: forward 5′TGGTGGTGAACCCAGAC3′ and reverse 5′CCAGGGACGCTCGTCTGCTG3′.
2.6. Phylogenetic tree construction
Nucleotide sequences were obtained from the GenBank database (accession No.): H. sapiens CANX (NM_001024649.1), Pan troglodytes CANX (XM_003310996.2), Bos taurus CANX (NM_001105612.1), CANX (NM_001003232.1), Mus musculus CANX (NM_007597.3), R. norvegicus CANX (NM_172008.2), Gallus gallus CANX (NM_001030620.1), Danio rerio CANX (NM_213448.1), Caenorhabditis elegans CANX (NM_066775.3), Oryza sativa Os04g0402100 (NM_001059227.1), Arabidopsis thaliana CNX1 (NM_125573.3), Xenopus laevis CANX-a (NM_001086857), Xenopus tropicalis CANX (NM_001005668), D. melanogaster CNX99A (NM_170407.3), T. rubripes LOC10107085 (XM_003978229), T. rubripes LOC101069714 (XM_003978694), Oryzias latipes LOC101165199 (XM_004084923), O. latipes LOC101159642 (XM_004065645), I. punctatus LOC100304606 (NM_001200180.1) and Oreochromis niloticus LOC100701646 (XM_003447644). The alignment was generated using Muscle (Edgar, 2004) and was used to construct a phylogenetic tree using the neighbour-joining method (Saitou and Nei, 1987) with the Jukes and Cantor correction (Jukes and Cantor, 1969) for 1000 bootstrap replications. Numbers above the tree lines indicate the per cent bootstrap confidence values.
2.7. Sequence analysis
SignalP 3.0 server was used to identify the signal peptide (Bendtsen et al., 2004), while TMHMM Server v. 2.0 was used to identify transmembrane regions (Krogh et al., 2001), NetPhos 2.0 Server was used to identify possible phosphorylation sites (Blom et al., 1999), palmitolytion sites were predicted by CSS-Palm 3.0 (Ren et al., 2008) and both InterProScan (EBI) (Zdobnov and Apweiler, 2001) and the NCBI conserved domain (CDD) platform (Marchler-Bauer et al., 2005) were used to identify protein conserved regions.
2.8. Western blot analysis
Cell pellets were lysed in 1% NP-40 lysis buffer containing 150 mM NaCl and 50 mM Tris [pH 8.0] supplemented with 1× protease inhibitor cocktail (Roche). Thirty micrograms of protein samples were separated on a 10% acrylamide gel and transferred to nitrocellulose membranes (Bio-Rad, Mississauga, ON) overnight. Membranes were blocked with 5% skim milk in TBS-T (136 mM NaCl, 2 mM KCl, 2.4 mM Tris, 0.05% Tween 20) and probed with 1:500 rabbit polyclonal antibody for human calnexin GTX101676 (GeneTex Irvine, CA) followed by 1:30,000 goat anti-rabbit alkaline phosphatase (Sigma Aldrich St. Louis, MO). Bands were detected using NBT/BCIP (Roche, Manheim, Germany) according to manufacturer’s instructions.
2.8.1. Isolation of peripheral blood leukocytes
Trout peripheral blood was centrifuged at 400g for 10 min, a buffy coat was obtained and diluted 1:10 with L-15 media (Sigma Aldrich St. Louis, MO). Diluted Fractions were layered on the top of 3 ml histopaque 1077 (Sigma–Aldrich, St. Louis, MO) and centrifuged for 30 min at 400g. White blood cells were collected, centrifuged and washed once with 1× phosphate buffered saline (PBS) for 5 min. Pellets were stored in −80 °C until later use.
2.9. Immunofluorescence
RTS11 cells were seeded to 2 × 105 cells per well in 4 chamber slides containing 2% FBS 150 U/ml of penicillin and 150 mg/ml streptomycin and grown overnight. All steps were conducted at 20 °C. Cells were washed once with PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4·7H2O, 1.4 mM KH2PO4; pH 7.4) and fixed for 15 min with 4% paraformaldehyde (Thermo Fisher Scientific, Nepean, ON) followed by two washes with PBS. Cells were permeabilized by incubation for 10 min with PBS containing 0.25% Triton X-100 (PBST), followed by 3 washes with PBS of 5 min each. Cells were incubated with 1% BSA in PBST containing 0.1% (v/v) of Triton X-100 for 30 min and then probed with 1:100 of polyclonal rabbit anti-human calnexin (GTX101676) in 1% BSA for 1 h. After three washes for 5 min each in PBS, cells were incubated for 1 h with 1:3000 of goat anti-rabbit Alexa Fluor 488 (Invitrogen, Carlsbad, CA) in 1% BSA followed by 3 washes with PBST of 5 min each. Cells were dried for 5 min in room temperature, mounted with Fluoroshield containing DAPI (Sigma Aldrich St. Louis, MO), coverslips were attached and the slides were stored at 4 °C. Slides were examined by laser scanning confocal microscopy using a Zeiss Axiovert 200 microscope and ZEN 2009 software (Carl Zeiss, Mississauga, ON).
3. Results
3.1. Sequence analysis
An estimated 1230 bp amplicon was isolated from RTS11 cDNA which showed 83% nucleotide identity with I. punctatus calnexin (NM_001200180.1). In order to obtain the 5′end and 3′end of the trout calnexin cDNA, PCR primers were used for 5′ RACE and for a conserved region in the 3′UTR using a PBL cDNA template. The corresponding PCR fragments were 380 bp and 320 bp respectively. PCR amplification of the full open reading frame of rainbow trout calnexin was then conducted using 5′ and 3′ gene specific primers using PBL cDNA template. Interestingly, two unique cDNA clones similar in length were obtained.
The first calnexin cDNA clone contains 1812 nt in its coding region and 94 nt of 3′UTR corresponding to putative 603 aa protein of 68 kDa. The second cDNA clone contains slightly longer sequence of 1851 bp with additional 23 bp of 3′UTR (117 bp total) which encodes a 616 aa protein with predicted molecular weight of 70 kDa.
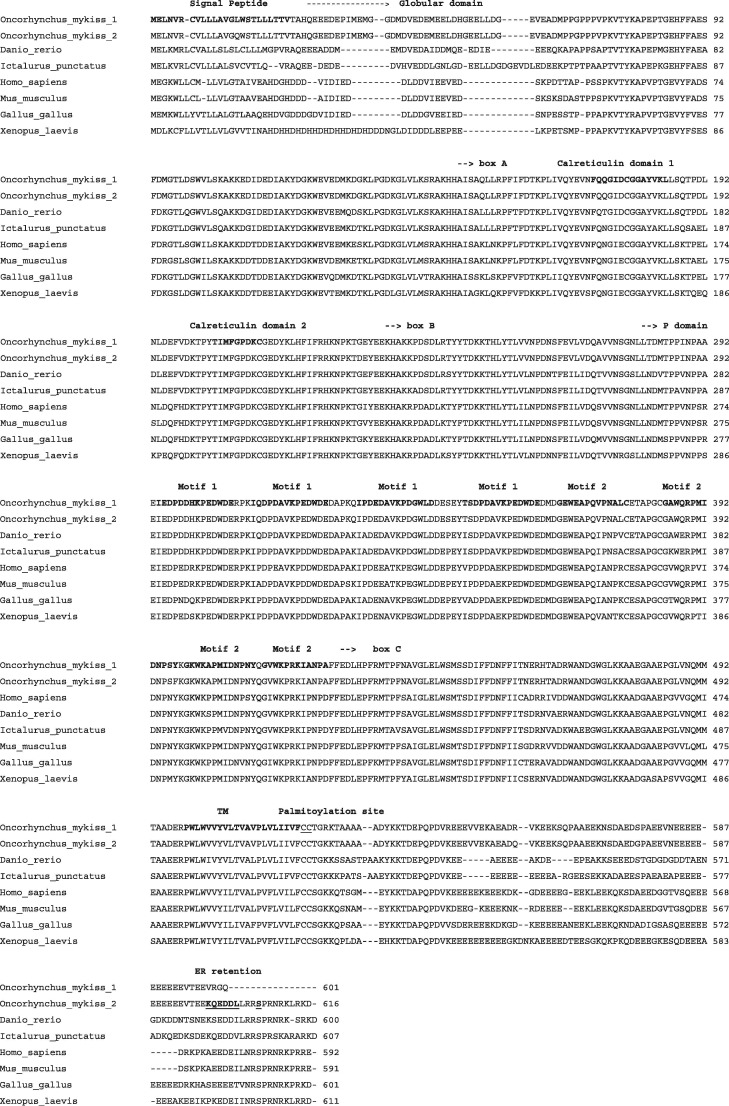
Both cDNAs showed 73% and 79% nucleotide identity with H. sapiens and I. punctatus Calnexin sequences, respectively with 72% and 75% sequence identity to their encoded proteins. The main characteristic of the rainbow trout calnexin proteins are illustrated in Fig. 1 . The N terminal domain includes a 24 aa signal peptide, followed by a luminal domain which contains a globular domain and an extended arm domain with a central segment, the P domain. This segment contains two proline motifs: M1 and M2 repeated in tandem 4 times each with the consensus sequences of M1: I-DP(D/E)A-KPEDWD(D/E) and M2: G-W-P-IN-P-Y. Moreover, there are 3 regions that have high sequence identity with human calnexin that are named box a, b and c. The box a segment is located upstream to calreticulin domain 1 whereas box b and c flank the repeated motifs. There is also a transmembrane domain of 21 aa that was predicted by TMHMM Server. Interestingly, there are two pairs of cysteines predicted to be palmitoylated by CSS-Palm 3.0 immediately after the transmembrane domain. These cysteines are found to be conserved across many species and when palmitoylated were shown to be critical for the association of calnexin with the ribosome translocon complex, allowing calnexin to bind to the N-linked glycans as they exit from the translocon (Lakkaraju et al., 2012). Another shared feature with mammalian calnexin is the presence of an acidic cytoplasmic tail enriched with glutamic acid and ER retention signal KQEDDL, which appears only in one of the clones and interestingly does not appear in the channel catfish sequence (Fuller et al., 2004). There are 5 phosphorylation sites predicted by NetPhos 2.0 Server for both trout clones, however a unique serine phosphorylation site downstream of the ER retention signal known to regulate calnexin activation (Chevet et al., 2010) appears only in calnexin cDNA sequence containing the ER retention motif.
Fig. 1.
An alignment of rainbow trout calnexin protein sequences with other known vertebrate sequences obtained from the GenBank (see Section 2 for specific accession numbers). Numbers on the right indicate amino acid positions. The conserved domains are labelled in bold above the sequences. The palmitoylation site in the juxta-membranal region is underlined. Both the putative ER retention signals and the serine phosphorylation site are labelled in bold and underlined.
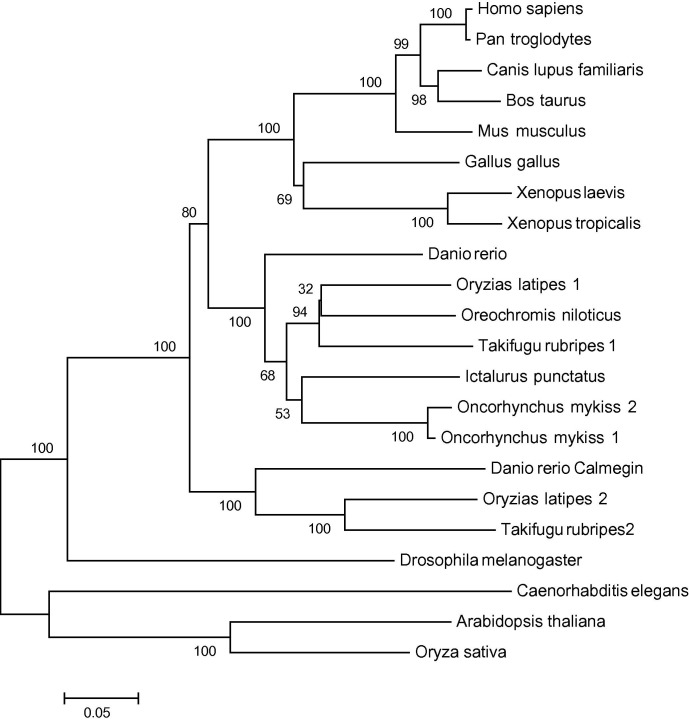
3.2. Phylogenetic tree
In order to further examine the evolutionary relationship between the unique trout clones and other calnexin nucleotide sequences, a phylogenetic tree were constructed (Fig. 2 ). As expected, strong distinct clusters can be seen for the tetrapod, teleost fish and plant sequences. Both trout sequences clustered together with a short genetic distance suggesting a recent gene duplication event, which is common in salmonids (Schmidtke and Kandt, 1981). Interestingly a more ancient duplication event also seems to have occurred in teleost fish, as both pufferfish, and rice fish showed a second sequence which grouped with zebrafish calmegin outside the main cluster of fish, frog and mammalian calnexin sequences, but still within a vertebrate cluster. This suggests a very early duplication event to produce this testis-specific ER chaperone homologous to calnexin.
Fig. 2.
A phylogenetic tree constructed using known nucleotide coding sequences of calnexin homologues in GenBank. The sequences were aligned with Muscle and Mega4 using neighbour joining with the Jukes and Cantor correction and bootstrap values determined through 1000 replications. The bar at the bottom indicates Nei’s genetic distance.
3.3. Trout calnexin gene copy number
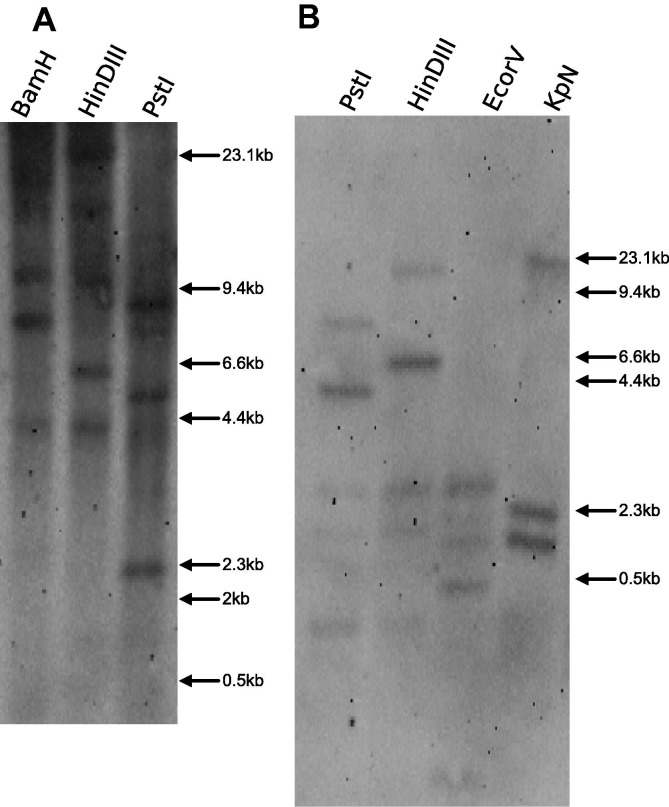
To further examine the possibility that the calnexin gene might be duplicated in trout, two separate probes were designed to proximal and distal segments of the calnexin gene. When the proximal probe was used, between 3 to 4 bands were observed in each digest (Fig 3 A), The distal probe showed similar results showing two dark bands and either one to two faint bands (Fig 3B). These results suggest that the calnexin gene is present in at least two copies in trout, unlike its soluble paralog calreticulin which appears in rainbow trout as single copy gene (Kales et al., 2004).
Fig. 3.
Southern blots of the rainbow trout calnexin gene. Left panel, labelled (A) shows the results obtained with a probe spanning the proximal domain of calnexin, right panel (B) using a probe for the distal coding domain of calnexin. The position of the size markers are shown by arrows on the right margin.
3.4. Calnexin tissue expression
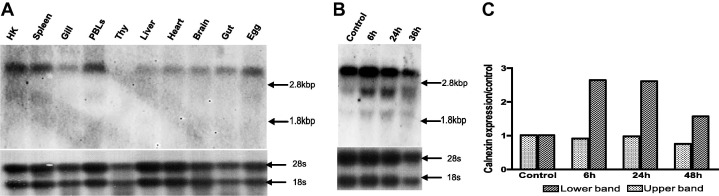
Northern blotting shows wide tissue distribution of the calnexin mRNA in trout tissues with the most prominent message being an estimated 3 kb transcript band (Fig. 4 A), but with a faint lower band detected in spleen, head kidney and PBLs. This lower band is strongly expressed in RTS11 (Fig 4B). The highest expression of that transcript was observed in spleen, head kidney PBLs (Fig 4A) and in the cell line RTS11 (Fig 4B). In order to determine if both bands were similarly regulated, RTS11 cells were treated with A23187, which is known to induce ER stress and upregulate other molecular chaperones in vitro such as ERp57 (Sever et al., 2013). Importantly, the lower calnexin transcript was upregulated up to 2.6-fold in 24 h followed by a decreased expression at 36 h. Interestingly, the upper band expression remained relatively constant (Fig. 4B and C). This result points out to the possibility that trout may have two unique transcripts responding differentially to ER induced stress due to unique modes of regulation.
Fig. 4.
Northern blots of rainbow trout calnexin showing tissue distribution of calnexin mRNA in normal rainbow trout tissues (4A) and in RTS11 upon ER stress stimulation using 2 μM Calcium ionophore A23187 for 36 h (4B). Densitometry analysis of the RTS11 experiment was conducted using image J software (4C). The estimated size of the 28s and 18s ribosomal bands is shown by the arrows on the right margin.
3.5. Calnexin protein expression under normal and induced ER stress conditions
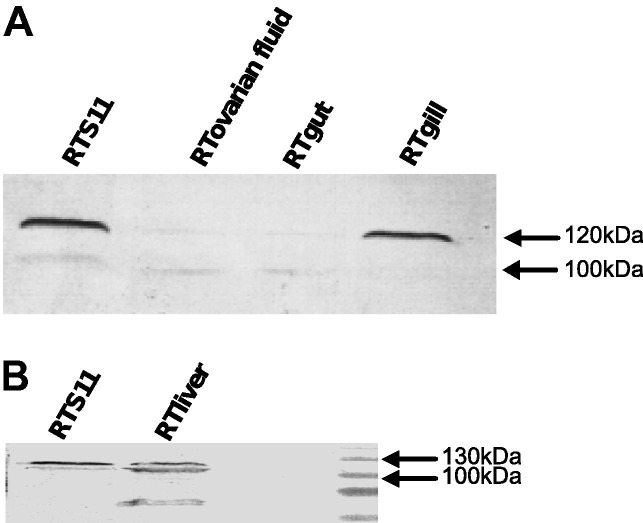
In order to further explore the possible role of calnexin during ER stress, an anti-human calnexin antibody (GTX101676) was used. This antibody was generated against the recombinant P domain of human calnexin, which shares 81% identity with the trout calnexin sequence. As opposed to its 70 KDa predicted molecular weight, trout calnexin protein was detected as a 120 kDa band in RTS11, RTgill, RTgut, RTovarian fluid and RTliver (Fig. 5 A and B). Interestingly, a lower fainter band around 100 KDa was also detected in all cell lines tested. A doublet band around 55 kDa was observed in RT liver, probably due to antibody binding to calreticulin, known to migrate as a doublet band in trout (Kales et al., 2007). In channel catfish calnexin is detected as a single band around 100 kDa compared to its 69 kDa molecular weight (Fuller et al., 2004). This discrepancy is common in many species and might be explained by the abundance of acidic amino acids which interferes with SDS binding and reduce the mobility on SDS–PAGE, and post-translational modifications. Importantly, induction of RTS11 with A23187 in a set of three repeats showed detection of the lower band in the treated cells but not in the control (Fig. 6 ). RTS11 treated with poly IC did not show any induction of the lower band (data not shown) which suggests that the expression of the smaller version of calnexin is modified specifically under ER stress conditions.
Fig. 5.
Distribution of calnexin protein in trout cell lines using an anti-human calnexin antibody. Lysates were separated on a 10% (Fig. 6A) or 12% (Fig. 6B) SDS PAGE gels. The position of the size marker is shown on the right margin.
Fig. 6.
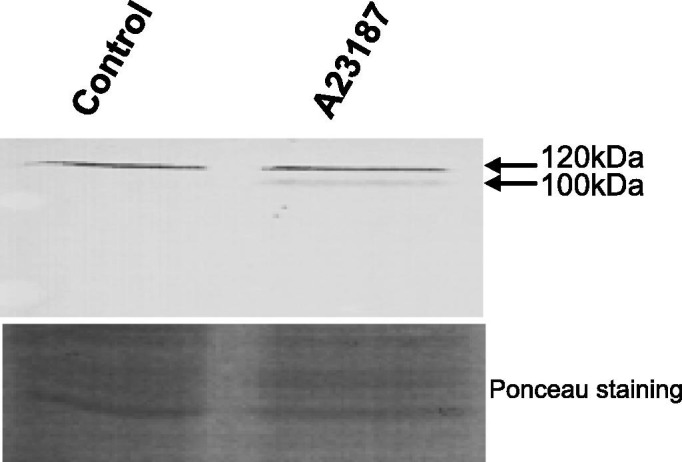
Upregulation of Calnexin following in vitro stimulation of RTS11 with Calcium Ionophore A23187 for 24 h. This experiment is representative of the results in a three replicate set.
3.6. Calnexin localisation in RTS11 cells
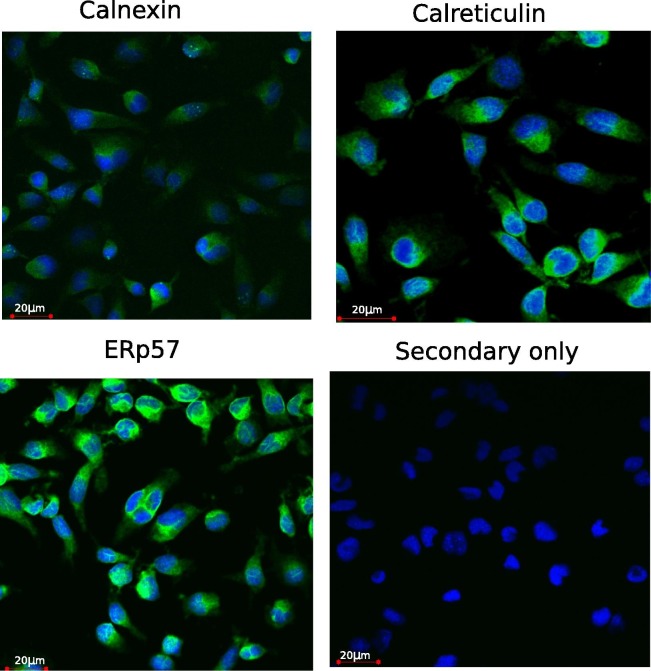
In order to further validate the specificity of the GTX101676 antibody, RTS11 were stained with anti-calnexin antibody (GTX101676) and examined under the confocal microscope (Fig. 7 ). The staining revealed a meshwork structure within the cells, with additional staining observed surrounding the nucleus. A similar and stronger staining pattern was observed for other ER resident proteins, trout ERp57 and calreticulin the latter of which contains a putative ER retention signal (Kales et al., 2004). These findings further support the localisation of calnexin to the ER membrane.
Fig. 7.
Immunofluorescence detection of calnexin, ERp57 and calreticulin in RTS11 using anti human calnexin GTX 10167, and antibodies generated against trout ERp57 and calreticulin proteins. Alexa Fluor 488 was used as anti-rabbit secondary antibody. Fluoroshield containing DAPI was used for nuclear staining. The negative control was stained with secondary antibody alone.
4. Discussion
In this study, rainbow trout calnexin was characterised and its expression was investigated to examine the regulation of a possible peptide loading complex in fish using rainbow as a model organism. Two unique cDNA clones encoding trout calnexin which share 99% sequence identity in their coding region were identified. Intriguingly, only one cDNA version contains a unique c-terminal with putative ER retention signal and conserved serine phosphorylation site which activates calnexin when protein misfolding occurs and is important for the recruitment of calnexin to ribosome translocons (Cameron et al., 2009, Chevet et al., 1999, Lakkaraju et al., 2012). Sequence alignment with the human calnexin protein reveals 72% identity and implies structural and functional conservation of calnexin as an ER lectin chaperone, calcium binding protein and as a protein indirectly assisting in oxidative folding. The luminal domain of calnexin contains the two conserved domains: the globular and the arm domain. The lectin binding site was found in a pocket on the surface of the globular domain (Schrag et al., 2001) whereas the tip of the arm domain contains the docking site that recruits ERp57 (Leach et al., 2002) and therefore helps to support oxidative folding in the ER. The P domain in the central arm segment contains two conserved motifs repeated four times each which contain low capacity and high affinity calcium binding sites, while the c-terminal domain is highly enriched with acidic amino acids shown to be as high capacity low affinity calcium binding sites (Michalak et al., 1992). All of these features are evident in the trout calnexin sequences, suggesting that many of its roles are conserved in trout, however the differences in the C terminal domains between the calnexin cDNA sequences might suggest of unique mode of regulation and perhaps differential subcellular localisation.
A phylogenetic tree constructed with calnexin nucleotide coding sequences indicates that the two copies arose in a recent gene duplication event within trout, or more probably salmonids (Schmidtke and Kandt, 1981) similar to that seen for trout ERp57 (Sever et al., 2013), suggesting that both are bone fide calnexin homologues. A duplication of calnexin like sequences of pufferfish and rice fish obtained from different loci may be more ancient as these sequences cluster outside all of the vertebrate calnexin sequences. Sequence alignment of their corresponding proteins reveals sequence identity of 73% and 84% respectively to zebrafish calmegin, a calnexin homologue in the testis which is expressed exclusively during spermatogenesis both in mouse and human calnexin and was suggested to bind sperm surface proteins important for egg-sperm interaction (Watanabe et al., 1994, Tanaka et al., 1997, Ikawa et al., 1997).
In order to investigate the regulation of calnexin during ER stress, cells were treated with calcium ionophore A23187 which has been previously shown to upregulate calnexin transcript expression by up to 2.5-fold in S. pombe and appeared as a single band in a northern blot (Parlati et al., 1995). Interestingly, in trout two possible transcript bands for calnexin were observed that are expressed only in immune system organs. Surprisingly, ER stress induction in RTS11 using A23187 showed that the smaller, less abundant transcript band was induced in contrast to the main transcript which remained unchanged. Similar results under the same stimulating conditions were obtained at the protein level showing the induction of a less abundant 100 kDa band with no change in expression of the higher molecular weight protein band. These results support the presence of two trout genes for calnexin that are regulated differently under ER stress conditions and perhaps performing unique biological functions. Importantly, according to their predicted cDNA sequences their difference in size might be more due to post translational modification rather than simply amino acid differences.
In contrast to what was seen here with trout calnexin, trout calreticulin, the soluble homologue of calnexin known to perform similar functions in the ER, was identified as a single copy gene in trout and failed to upregulate under several ER stress conditions (Kales et al., 2007). This suggests that fish calnexin may perform functions during ER stress that calreticulin cannot. Under normal conditions, trout calreticulin expression was found to be highest in the liver (Kales et al., 2007) while trout calnexin seems to be more abundant in immune system organs. Interestingly, while calnexin deficient mice display impaired motor function they are not embryonic lethal (Denzel et al., 2002) while calreticulin deficient mice exhibit an embryonic lethal phenotype due to a defect in heart development (Michalak et al., 2002). In addition, it has been recently demonstrated that both genes are important for generation of neuromasts during lateral line development in zebrafish using calnexin and calreticulin knockout morpholinos, however the calnexin knockout had a more profound effect on this development (Hung et al., 2013). These results suggest that both in mammals and in fish calreticulin and calnexin play a role during embryonic development, which suggest these two proteins may have distinctive roles outside of their chaperone function. Monitoring the expression of these two calnexin genes during stages of embryogenesis in rainbow trout could shed a light on their possible unique functions during development.
The immunocytochemistry of calnexin in RTS11 suggests that calnexin is localised to the ER and possibly resides close to the nucleus, as in some cells it showed staining near the nucleus. Interestingly, calnexin has been found as the major calcium binding protein in rat hepatic nuclear membranes although appeared as a minor member of the total nuclear protein (Gilchrist and Pierce, 1993). In addition, immunostaining of calnexin in rat epithelial cells has demonstrated a reticular network structure and nuclear envelope staining (Wada et al., 1991) supporting our findings. It may be that the trout calnexin without the ER retention signal can escape the ER and perhaps be localised in the nuclear periphery where it can regulate calcium levels in this region. However, further experimentation with monoclonal antibodies to each specific form is required to see which form(s) is localised to the nuclear region.
The role of calnexin as a chaperone has been widely investigated in mammals, however some research conducted in yeast and the microorganism Dictyostelium shed a light on the other important cellular functions of calnexin such as during apoptosis and phagocytosis. The first report linking calnexin to apoptosis was conducted in an S. pombe yeast two hybrid system demonstrating that calnexin is required for cell death mediated by the interaction of its cytosolic tail with the a pro-apoptotic protein BAK (Torgler et al., 1997). Later studies in mammals using calnexin-deficient cells further demonstrated its possible role in mediating apoptosis as these cells showed resistance to ER stress induced apoptosis (Zuppini et al., 2002, Groenendyk et al., 2006). Another surprising function for calnexin came from a study with Dictyostelium, the only microorganism containing both calnexin and calreticulin proteins. This study showed that co-disruption of calnexin and calreticulin results in severely impaired phagocytosis and compromised growth of the phagocytic cups (Müller-Taubenberger et al., 2001), pointing to a possible primary function for calnexin in this organism. This highlights the significance of studying the evolution of this molecule.
This study reports the first identification of calnexin sequences in a salmonid species, shows that this gene is duplicated in the trout genome and shows for the first time the possible differential regulation of the two forms under ER stress stimulating conditions, which was not seen for its soluble homologue trout calreticulin. Further research will focus on its role in the MH class I assembly and its possible interaction with fish viral glycoproteins.
Acknowledgements
The authors would like to thank Prof. John J. Heikkila and Prof. David Williams for valuable discussions. The research was supported by NSERC Discovery Grant number 217529-2008 to B.D.
References
- Ahluwalia N., Bergeron J.J., Wada I., Degen E., Williams D.B. The p88 molecular chaperone is identical to the endoplasmic reticulum membrane protein, calnexin. J. Biol. Chem. 1992;267:10914–10918. [PubMed] [Google Scholar]
- Anderson K.S., Cresswell P. A role for calnexin (IP90) in the assembly of class II MHC molecules. EMBO J. 1994;13(3):675–682. doi: 10.1002/j.1460-2075.1994.tb06306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294(5):1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Bols N.C., Barlian A., Chirino-Trejo M., Caldwell S.J., Goegan P., Lee L.E.J. Development of a cell line from primary cultures of rainbow trout, Oncorhynchus mykiss (Walbaum), gills. J. Fish Dis. 1990;17:601–611. [Google Scholar]
- Cameron P.H., Chevet E., Pluquet O., Thomas D.Y., Bergeron J.J. Calnexin phosphorylation attenuates the release of partially misfolded alpha1-antitrypsin to the secretory pathway. J. Biol. Chem. 2009;284:34570–34579. doi: 10.1074/jbc.M109.053165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno B.M., Schreiber K.L., McKean D.J., Stroynowski I., Hansen T.H. A glycosylated and phosphatidylinositol-anchored MHC class I molecules are associated with calnexin. J. Immunol. 1995;154:5173–5180. [PubMed] [Google Scholar]
- Chevet E., Wong H.N., Gerber D., Cochet C., Fazel A., Cameron P.H., Gushue J.N., Thomas D.Y., Bergeron J.J. Phosphorylation by CK2 and MAPK enhances calnexin association with ribosomes. EMBO J. 1999;18:3655–3666. doi: 10.1093/emboj/18.13.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevet E., Wong H.N., Gerber D., Cochet C., Fazel A., Cameron P.H., Gushue J.N., Thomas D.Y., Bergeron J.J. Phosphorylation by CK2 and MAPK enhances calnexin association with ribosome. EMBO J. 1999;18:3655–3666. doi: 10.1093/emboj/18.13.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevet E., Smirle J., Cameron P.H., Thomas D.Y., Bergeron J.J. Calnexin phosphorylation: linking cytoplasmic signalling to endoplasmic reticulum lumenal functions. Semin Cell Dev. Biol. 2010;21:486–490. doi: 10.1016/j.semcdb.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Degen E., Williams D.B. Participation of a novel 88-kD protein in the biogenesis of murine class I histocompatibility molecules. J. Cell Biol. 1991;112:1099–1115. doi: 10.1083/jcb.112.6.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delom F., Fessart D., Chevet E. Regulation of calnexin sub-cellular localization modulates endoplasmic reticulum stress-induced apoptosis in MCF-7 cells. Apoptosis. 2007;12:293–305. doi: 10.1007/s10495-006-0625-4. [DOI] [PubMed] [Google Scholar]
- Denzel A., Molinari M., Trigueros C., Martin J.E., Velmurgan S., Brown S., Stamp G., Owen M.J. Early postnatal death and motor disorders in mice congenitally deficient in calnexin expression. Mol. Cell. Biol. 2002;22:7398–7404. doi: 10.1128/MCB.22.21.7398-7404.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;19(5):113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Frohman M.A., Dush M.K., Martin G.R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi M., Yoshinaka Y., Matsuoka Y., Hatakeyama S., Ishizaka Y., Kirikae T., Sasazuki T. Monitoring of S protein maturation in the endoplasmic reticulum by calnexin is important for the infectivity of severe acute respiratory syndrome coronavirus. J. Virol. 2012;86:11745–11753. doi: 10.1128/JVI.01250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller J.R., Pitzer J.E., Godwin U., Albertino M., Machon B.D., Kearse K.P., McConnell T.J. Characterization of the molecular chaperone calnexin in the channel catfish, Ictalurus punctatus, and its association with MHC class II molecules. Dev. Comp. Immunol. 2004;28:603–617. doi: 10.1016/j.dci.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Ganassin R.C., Bols N.C. Development from the rainbow trout spleen of a cell line, RTS11, in the monocyte/macrophage lineage. Fish Shellfish Immunol. 1998:457–476. [Google Scholar]
- Gilchrist J.S., Pierce G.N. Identification and purification of a calcium-binding protein in hepatic nuclear membranes. J. Biol. Chem. 1993;268:4291–4299. [PubMed] [Google Scholar]
- Groenendyk J., Zuppini A., Shore G., Opas M., Bleackley R.C. Caspase 12 in calnexin-deficient cells. Biochemistry. 2006;45:13219–13226. doi: 10.1021/bi061428z. [DOI] [PubMed] [Google Scholar]
- Guérin R., Arseneault G., Dumont S., Rokeach L.A. Calnexin is involved in apoptosis induced by endoplasmic reticulum stress in the fission yeast. Mol. Biol. Cell. 2008;19:4404–4420. doi: 10.1091/mbc.E08-02-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C., Helenius A. Folding of VSV G protein: sequential interaction with BiP and calnexin. Science. 1994;266:456–458. doi: 10.1126/science.7939687. [DOI] [PubMed] [Google Scholar]
- Hammond C., Braakman I., Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstenbach F., David V., Watkins S., Brenner M.B. Endoplasmic reticulum resident protein of 90 kilodaltons associates with the T- and B-cell antigen receptors and major histocompatibility complex antigens during their assembly. Proc. Natl. Acad. Sci. USA. 1992;89:4734–4738. doi: 10.1073/pnas.89.10.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Franklin A.E., Hoffman N.E. Primary structure and characterization of an Arabidopsis thaliana calnexin-like protein. J. Biol. Chem. 1993;268:6560–6566. [PubMed] [Google Scholar]
- Hung I.C., Cherng B.W., Hsu W.M., Lee S.J. Calnexin is required for zebrafish posterior lateral line development. Int. J. Dev. Biol. 2013;57:427–438. doi: 10.1387/ijdb.120166sl. [DOI] [PubMed] [Google Scholar]
- Ikawa M., Wada I., Kominami K., Watanabe D., Toshimori K., Nishimune Y., Okabe M. The putative chaperone calmegin is required for sperm fertility. Nature. 1997;387:607–611. doi: 10.1038/42484. [DOI] [PubMed] [Google Scholar]
- Jukes T.H., Cantor C.R. Evolution of Protein Molecules. In: Munro H.N., editor. Mammalian Protein Metabolism. Academic Press; New York: 1969. pp. 21–132. [Google Scholar]
- Kales S., Fujiki K., Dixon B. Molecular cloning and characterization of calreticulin from rainbow trout (Oncorhynchus mykiss) Immunogenetics. 2004;55:717–723. doi: 10.1007/s00251-003-0631-4. [DOI] [PubMed] [Google Scholar]
- Kales S.C., Bols N.C., Dixon B. Calreticulin in rainbow trout: a limited response to endoplasmic reticulum (ER) stress. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 2007;147:607–615. doi: 10.1016/j.cbpb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Kawano A., Kales S.C., Fujiki K., DeWitte-Orr S.J., Dixon B., Lee L.E.J., Bols N.C. A comparison of rainbow32 trout cell lines for their expression of the Major Histocompatibility Complex genes and the induction beta 2-microglobulin by dsRNA. Fish Shellfish Immunol. 2010;29:312–318. doi: 10.1016/j.fsi.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lakkaraju A.K., Abrami L., Lemmin T., Blaskovic S., Kunz B., Kihara A., Dal Peraro M., van der Goot F.G. Palmitoylated calnexin is a key component of the ribosome–translocon complex. EMBO J. 2012;31:823–835. doi: 10.1038/emboj.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis E.D., Palti Y., Dekoning J., Drew R., Phillips R.B., Hansen J.D. Identification and regulatory analysis of rainbow trout tapasin and tapasin-related genes. Immunogenetics. 2006;58:56–69. doi: 10.1007/s00251-005-0070-5. [DOI] [PubMed] [Google Scholar]
- Leach M.R., Cohen-Doyle M.F., Thomas D.Y., Williams D.B. Localization of the lectin, ERp57 binding, and polypeptide binding sites of calnexin and calreticulin. J. Biol. Chem. 2002;277:29686–29697. doi: 10.1074/jbc.M202405200. [DOI] [PubMed] [Google Scholar]
- Lee L.E.J., Clemons J.H., Bechtel D.G., Caldwell S., Han K.B., Pasitschniak M., Mosser D.D., Bols N.C. Development and characterization of a rainbow trout liver cell line expressing cytochrome P450-dependent monooxygenase activity. Cell Biol. Toxicol. 1993;9:279–294. doi: 10.1007/BF00755606. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J.B., Cherukuri P.F., DeWeese-Scott C., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., Lanczycki C.J., Liebert C.A., Liu C., Lu F., Marchler G.H., Mullokandov M., Shoemaker B.A., Simonyan V., Song J.S., Thiessen P.A., Yamashita R.A., Yin J.J., Zhang D., Bryant S.H. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 2005;33:192–196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak M., Milner R.E., Burns K., Opas M. Calreticulin. Biochem. J. 1992;285:681–692. doi: 10.1042/bj2850681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak M., Lynch J., Groenendyk J., Guo L., Robert Parker J.M., Opas M. Calreticulin in cardiac development and pathology. Biochim. Biophys. Acta. 2002;1600:32–37. doi: 10.1016/s1570-9639(02)00441-7. [DOI] [PubMed] [Google Scholar]
- Müller-Taubenberger A., Lupas A.N., Li H., Ecke M., Simmeth E., Gerisch G. Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO J. 2001;20:6772–6782. doi: 10.1093/emboj/20.23.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou W.J., Thomas D.Y., Bell A.W., Bergeron J.J. Casein kinase II phosphorylation of signal sequence receptor alpha and the associated membrane chaperone calnexin. J. Biol. Chem. 1992;267:23789–23796. [PubMed] [Google Scholar]
- Ou W.J., Cameron P.H., Thomas D.Y., Bergeron J.J. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature. 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- Parlati F., Dominguez M., Bergeron J.J., Thomas D.Y. Saccharomyces cerevisiae CNE1 encodes an endoplasmic reticulum (ER) membrane protein with sequence similarity to calnexin and calreticulin and functions as a constituent of the ER quality control apparatus. J. Biol. Chem. 1995;270:244–253. doi: 10.1074/jbc.270.1.244. [DOI] [PubMed] [Google Scholar]
- Parlati F., Dignard D., Bergeron J.J., Thomas D.Y. The calnexin homologue cnx1+ in Schizosaccharomyces pombe, is an essential gene which can be complemented by its soluble ER domain. EMBO J. 1995;14:3064–3072. doi: 10.1002/j.1460-2075.1995.tb07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderick H.L., Lechleiter J.D., Camacho P. Cytosolic phosphorylation of calnexin controls intracellular Ca(2+) oscillations via an interaction with SERCA2b. J. Cell Biol. 2000;149:1235–1248. doi: 10.1083/jcb.149.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivan B.K., Cariappa A., Waneck G.L., Cresswell P. Assembly, peptide loading, and transport of MHC class I molecules in a calnexin-negative cell line. Cold Spring Harb. Symp. Quant. Biol. 1995;60:267–275. doi: 10.1101/sqb.1995.060.01.031. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schmidtke J., Kandt I. Single-copy DNA relationships between diploid and tetraploid teleostean fish species. Chromosoma. 1981;83:191–197. doi: 10.1007/BF00286788. [DOI] [PubMed] [Google Scholar]
- Schrag J.D., Bergeron J.J., Li Y., Borisova S., Hahn M., Thomas D.Y., Cygler M. The Structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol. Cell. 2001;8:633–644. doi: 10.1016/s1097-2765(01)00318-5. [DOI] [PubMed] [Google Scholar]
- Scott J.E., Dawson J.R. MHC class I expression and transport in a calnexin-deficient cell line. J. Immunol. 1995;155:143–148. [PubMed] [Google Scholar]
- Sever L., Bols N.C., Dixon B. The cloning and inducible expression of the rainbow trout ERp57 gene. Fish Shellfish Immunol. 2013;34:410–419. doi: 10.1016/j.fsi.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Takizawa T., Tatematsu C., Watanabe K., Kato K., Nakanishi Y. Cleavage of calnexin caused by apoptotic stimuli: implication for the regulation of apoptosis. J. Biochem. 2004;136:399–405. doi: 10.1093/jb/mvh133. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Ikawa M., Tsuchida J., Nozaki M., Suzuki M., Fujiwara T., Okabe M., Nishimune Y. Cloning and characterization of the human calmegin gene encoding putative testis-specific chaperone. Gene. 1997;204:159–163. doi: 10.1016/s0378-1119(97)00537-4. [DOI] [PubMed] [Google Scholar]
- Torgler C.N., de Tiani M., Raven T., Aubry J.P., Brown R. Expression of bak in S. pombe results in a lethality mediated through interaction with the calnexin homologue Cnx1. Cell Death Differ. 1997;4:263–271. doi: 10.1038/sj.cdd.4400239. [DOI] [PubMed] [Google Scholar]
- Vassilakos A., Cohen-Doyle M.F., Peterson P.A., Jackson M.R., Williams D.B. The molecular chaperone calnexin facilitates folding and assembly of class I histocompatibility molecules. EMBO J. 1996;15:1495–1506. [PMC free article] [PubMed] [Google Scholar]
- Wada I., Rindress D., Cameron P.H., Ou W.J., Doherty J.J., 2nd, Louvard D., Bell A.W., Dignard D., Thomas D.Y., Bergeron J.J. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J. Biol. Chem. 1991;266:19599–19610. [PubMed] [Google Scholar]
- Watanabe D., Yamada K., Nishina Y., Tajima Y., Koshimizu U., Nagata A., Nishimune Y. Molecular cloning of a novel Ca(2+)-binding protein (calmegin) specifically expressed during male meiotic germ cell development. J. Biol. Chem. 1994;269(10):7744–7749. [PubMed] [Google Scholar]
- Yamamoto S., Nakamura M. Calnexin: its molecular cloning and expression in the liver of the frog, Rana rugosa. FEBS Lett. 1996;387:27–32. doi: 10.1016/0014-5793(96)00443-7. [DOI] [PubMed] [Google Scholar]
- Zdobnov E.M., Apweiler R. InterProScan – an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Williams D.B. Assembly of MHC class I molecules within the endoplasmic reticulum. Immunol. Res. 2006;35:151–162. doi: 10.1385/IR:35:1:151. [DOI] [PubMed] [Google Scholar]
- Zuppini A., Groenendyk J., Cormack L.A., Shore G., Opas M., Bleackley R.C., Michalak M. Calnexin deficiency and endoplasmic reticulum stress-induced apoptosis. Biochemistry. 2002;41:2850–2858. doi: 10.1021/bi015967+. [DOI] [PubMed] [Google Scholar]