Abstract
Human CLEC4G (previously named LSECtin), DC-SIGN, and L-SIGN are three important C-type lectins capable of mediating viral and bacterial pathogen recognitions. These three genes, together with CD23, form a lectin gene cluster at chromosome 19p13.3. In this study, we have experimentally identified the cDNA and the gene encoding porcine CLEC4G (pCLEC4G). Full-length pCLEC4G cDNA encodes a type II transmembrane protein of 290 amino acids. pCLEC4G gene has the same gene structure as the human and the predicted bovine, canis, mouse and rat CLEC4G genes with nine exons. A multi-species-conserved site at the extreme 3′-untranslated region of CLEC4G mRNAs was predicted to be targeted by microRNA miR-350 in domesticated animals and by miR-145 in primates, respectively. We detected pCLEC4G mRNA expression in liver, lymph node and spleen tissues. We also identified a series of sequential intermediate products of pCLEC4G pre-mRNA during splicing from pig liver. The previously unidentified porcine CD23 cDNA containing the complete coding region was subsequently cloned and found to express in spleen, thymus and lymph node. Furthermore, we compared the chromosomal regions syntenic to the human cluster of genes CD23/CLEC4G/DC-SIGN/L-SIGN in representative mammalian species including primates, domesticated animal, rodents and opossum. The L-SIGN homologues do not exist in non-primates mammals. The evolutionary processes of the gene cluster, from marsupials to primates, were proposed based upon their genomic structures and phylogenetic relationships.
Keywords: C-type lectins, CLEC4G (LSECtin), CD23, DC-SIGN (CD209), L-SIGN (CD209L), Pig, MicroRNA, Pre-mRNA splicing, Mammals
1. Introduction
The C-type lectin receptor (CLR) family includes a large number of proteins that perform protein–carbohydrate interactions by binding to the polysaccharide chains present on the glycoprotein ligands in a calcium-dependent manner [1]. Numerous CLRs belong to pattern recognition receptors (PRRs) on the surface of antigen-presenting cells (APCs) that recognize foreign pathogens, and play key roles in host immune responses [1], [2]. The type II CLRs are classified as their cytoplasmic tail (CT) located in the NH2 terminus domain. Other type II CLRs domains include the transmembrane domain (TMD) following the CT, a single carbohydrate recognition domain (CRD) exposed extracellularly at the carboxyl terminus, and the neck domain between the TMD and CRD [1], [2].
A human gene cluster of type II CLRs, CD23/CLEC4G/DC-SIGN/L-SIGN, which is localized in human chromosome 19p13.3, has received increasing interest during the past few years. Human CD23 (FCER2) is a low affinity IgE receptor that plays an important role in cell–cell adhesions, B cells survival and antigen presentation [3]. Human DC-SIGN (hDC-SIGN, CD209) was initially identified as an ICAM-3 binding protein mediating dendritic cells (DCs) and T cell interaction, an ICAM-2 binding protein regulating chemokine-induced trafficking of DCs across both resting and activated endothelium, and a HIV-1 gp120 receptor mediating transmission of HIV-1 to susceptible cells in trans [4], [5], [6]. The second hDC-SIGN homologue, hL-SIGN (CD209L), was subsequently shown to have similar but subtly distinct property of pathogen recognition to hDC-SIGN [7]. Both hDC-SIGN and hL-SIGN bind to asparagines-linked high-mannose glycans present on a broad spectrum of enveloped viruses [8]. Human CLEC4G (hCLEC4G, previously named LSECtin), co-expressed with hL-SIGN on liver and lymph node sinusoidal endothelial cell (LSECs), is the third hDC-SIGN-related CLR recently identified [9], [10]. hCLEC4G was found to interact with the surface glycoproteins of Ebola virus and severe acute respiratory syndrome coronavirus (SARS-CoV) [11]. However, unlike hDC-SIGN and hL-SIGN, the hCLEC4G selectively binds to glycoproteins terminating in a disaccharide, GlcNAcβ1-2Man, resulting in its interacting with truncated glycans on glycoprotein of Ebola virus [12]. Since these four CLRs form a tight gene cluster and share overall protein domain structure, similar genomic organization and possible analogous function, it has been proposed that they are derived from a common ancestor [9].
DC-SIGN homologues have been experimentally identified from other mammalian species including non-human primates and mouse [13], [14], [15]. Interestingly, there exist seven mouse DC-SIGN paralogues, SIGNRs 1–5 as well as SIGNRs 7–8, on mouse chromosome 8A1.1, indicating widely divergent biochemical and probably physiological properties of DC-SIGN-related proteins in mouse [14]. However, none of these SIGNR molecules were shown to be the functional orthologue to hDC-SIGN. Recently, DC-SIGN homologues from domesticated animal species such as dog, cattle and horse have also been predicted from the completed genome projects. We recently reported the molecular cloning and characterization of the full-length cDNA and gene of porcine DC-SIGN (pDC-SIGN) in the absence of computer-based screening of DC-SIGN homologues in pig genome database [16]. Phylogenetic analysis revealed that pDC-SIGN, together with the putative bovine, canis and equine DC-SIGN, are more closely related to mouse SIGNR7 and SIGNR8 than to human DC-SIGN or other mouse SIGNR homologues and form a separate clade, indicating a distinct evolutionary pathway [16]. Since no other DC-SIGN sequences were detected in the bovine and canis genomes, the pDC-SIGN likely exists as a single gene analogously, although the relevant porcine genomic region has not been identified. Moreover, we observed that L-SIGN homologues only exist in human and non-human primates but not in other non-primate mammalian species [16]. Indeed, a previous study showed that the current L-SIGN gene, presented in apes such as chimpanzee and human but not in Old World monkeys (OWM) such as rhesus macaque, was newly duplicated from the ancestral DC-SIGN, whereas the older duplicator, CD209L2, was lost in human but still retained in OWM and apes [13]. The study indicated that the DC-SIGN/L-SIGN gene family in primates has undergone duplications and deletions during recent evolutionary processes [13].
Besides hCLEC4G, CLEC4G homologues in other mammalian species have not been experimentally identified although the gene information could be searched using computer programs from the genome databases. We report here the cloning and phylogenetic analysis of pCLEC4G cDNA and gene, its order of intron removal during pre-mRNA splicing, and the tissue distribution. In addition, the complete coding region of porcine CD23 (pCD23) cDNA was also determined. Furthermore, with the available information of CD23, DC-SIGN and CLEC4G genes obtained from the completed genome projects, we compare the chromosomal regions syntenic to the cluster of human genes CD23/CLEC4G/DC-SIGN/L-SIGN in the representative mammalian species including chimpanzee (Pan troglodytes), rhesus macaque (Macaca mulatta), cattle (Bos taurus), dog (Canis lupus), horse (Equus caballus), sheep (Ovis aries), pig (Sus scrofa), mouse (Mus musculus), rat (Rattus norvegicus), and opossum (Monodelphis domestica). Finally, the evolutionary processes of the gene cluster, from marsupials to primates, were proposed based upon their genomic structures and phylogenetic relationships.
2. Materials and methods
2.1. RNA extraction and reverse transcription PCR (RT-PCR)
Healthy crossbred conventional pigs of 7 weeks of age were used for the collection of tissue samples. Pigs were maintained in an isolated room under experimental conditions. Total RNA was isolated from homogenized pig liver using the RNeasy mini kit (Qiagen Inc.) followed by an RNase-free DNase I treatment. First-strand cDNA was synthesized from total RNA with SuperScript II reverse transcriptase (Invitrogen) using oligo-dT (Promega) as the reverse primer. A pair of gene-specific primers, PLST-F (5′-TATGCCCAGAGCAGGGCACC-3′) and PLST-R (5′-GGGCTAGGTCAGCAGTTGTGC-3′), was designed for the amplification of the complete coding region of pCLEC4G cDNA according to a porcine cDNA sequence with the GenBank accession number AK232603. PCR was performed in 50 μL reaction with an Advantage 2 PCR kit (Clontech, Palo Alto, CA) using the following PCR parameters: 94 °C for 2 min, 30 cycles of 94 °C for 15 s, 60.0 °C for 30 s and 72 °C for 90 s, and a final incubation at 72 °C for 3 min. For the amplification of the complete coding region of pCD23 cDNA, pig spleen tissue as well as primers PCD23-F (5′-GCGCTCCCATGGAGGAAAGTTTATACTC-3′) and PCD23-R (5′-TGAACAGATGCTCAGCAAGTGGCCA-3′) was used. The primer sequences were based on the S. scrofa chromosome 2 clone CH242-334A8 (working draft sequence; GenBank accession number CU929919) that was released most recently on September 24, 2008. The obtained PCR products were individually excised, purified, and subsequently cloned into a pCR2.1 vector (Invitrogen) by TA cloning strategy followed by DNA sequencing.
2.2. Genomic PCR and gene sequencing
The same primers PLST-F and PLST-R were used for one-step genomic PCR, which was performed with a Platinum PCR HiFi Supermix kit (Invitrogen) using 150 ng of the pig genomic DNA (Novagen) in a total volume of 50 μL. The PCR condition was 35 cycles of 94 °C for 30 s, 68 °C for 4 min with an initial denaturation step at 94 °C for 2 min. The resulting fragment was cloned into a pCR2.1 vector by the TA cloning strategy. The M13 forward and reverse primers together with a gene-specific primer PLST-E3F (5′-CAGGATCTACTGAGGACAAACG-3′) were used for DNA sequencing.
2.3. Tissue distribution of porcine CLEC4G and CD23 gene expression detected by RT-PCR
Total RNA was isolated from ten homogenized pig tissues including spleen, duodenum, thymus, kidney, lung, lymph node, heart, bone marrow, liver and muscles using the RNeasy mini kit (Qiagen) followed by an RNase-free DNase I treatment, and cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen) using oligo-dT (Promega) as the reverse primer. To avoid the contamination of genomic DNA, PCR was performed in 50 μL reactions with Clontech's Advantage 2 PCR kit using primer PLST-E67F (5′-GAGAGTCCGGTTCCAGAACAGCTCCT-3′) spanning the boundary between exons 6 and 7, and primer PLST-E89R (5′-TCCCCCAGATTCCAGTGGCTGAAG-3′) spanning the boundary of exons 8 and 9 of pCLEC4G gene sequence that had been determined by genomic sequencing. For the detection of pCD23 mRNA expression, primers PCD23-E89F (5′-CTACACGAGTCCAACGGCTCCGTG-3′) and PCD23-E1011R (5′-CGGGCTGCCAGTTGCTATAGTCCAG-3′) were used. The PCR parameters include 30 cycles of 95 °C for 20 s, 68 °C for 1 min with an initial denaturation step for 2 min. The house keeping gene, porcine glyceraldehyde 3-phosphate dehydrogenase (GAPDH), was also amplified using primers GAPDH5 (5′-GCTGAGTATGTCGTGGAGTC-3′) and GAPDH3 (5′-CTTCTGGGTGGCAGTGAT-3′) by PCR (95 °C for 1 min, 30 cycles of 95 °C for 20 s, 55 °C for 20 s and 68 °C for 40 s, and 72 °C for 3 min).
2.4. Sequence and phylogenetic analyses
Analyses and alignment of DNA and amino acid sequences were performed using Lasergene package (DNASTAR Inc., Madison, WI). The sequences of the human, chimpanzee, rhesus macaque, cattle, dog, horse, sheep, pig, mouse, rat and opossum CD23/CLEC4G/DC-SIGN genomic loci were retrieved from the public draft assemblies available at the NCBI Map viewer (http://www.ncbi.nlm.nih.gov/mapview/) released in March 2008 (human Build 36.3), March 2006 (chimpanzee Build 2.1), February 2006 (rhesus macaque Build 1.1), August 2006 (cattle Build 3.1), March 2005 (dog Build 2.1), September 2007 (horse EquCab2.0), December 2006 (Sheep SM4.7), February 2008 (pig Sscrofa5), May 2006 (mouse Build 37.1), July 2006 (rat RGSC v3.4) and October 2006 (opossum MonDom5). The following coordinates were used to obtain the sequences used in Fig. 8a: human chromosome 19, 7659662–7761897; chimpanzee chromosome 19, 7904925–8008046; rhesus macaque chromosome 19, 7653843–7776647; cattle chromosome 7, 14121567–14185245; dog chromosome 20, 55452895–55590209; horse chromosome 7, 1635615–1825170; mouse chromosome 8, 3681737–4208046; rat chromosome 12, 1657463–2469678; opossum chromosome 3, 462891145–463096613.
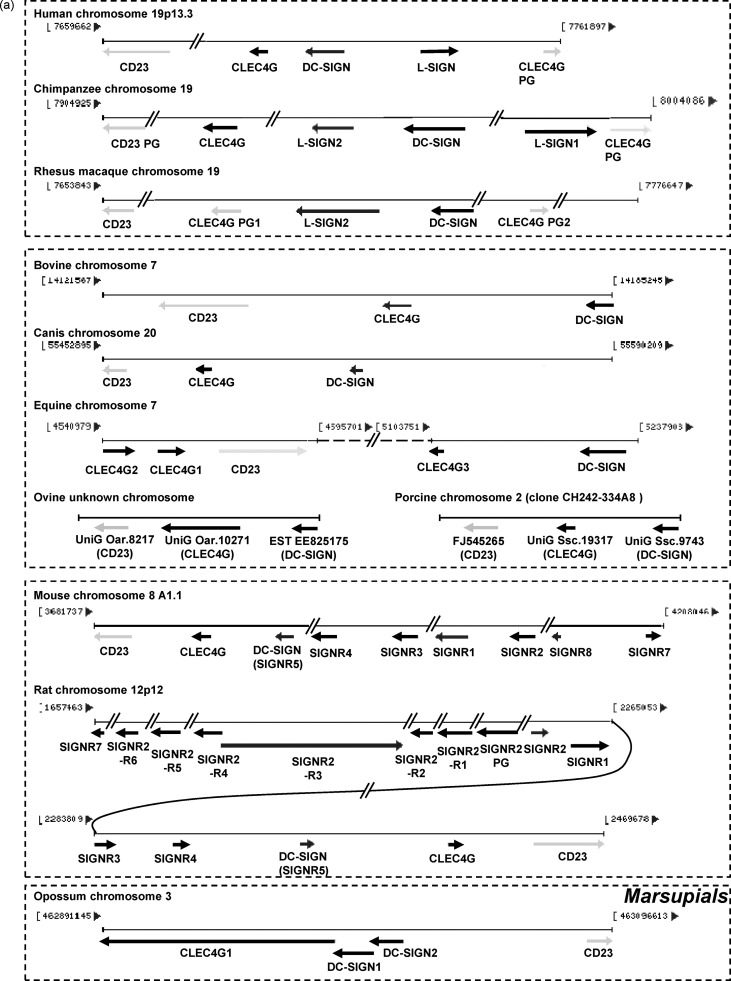
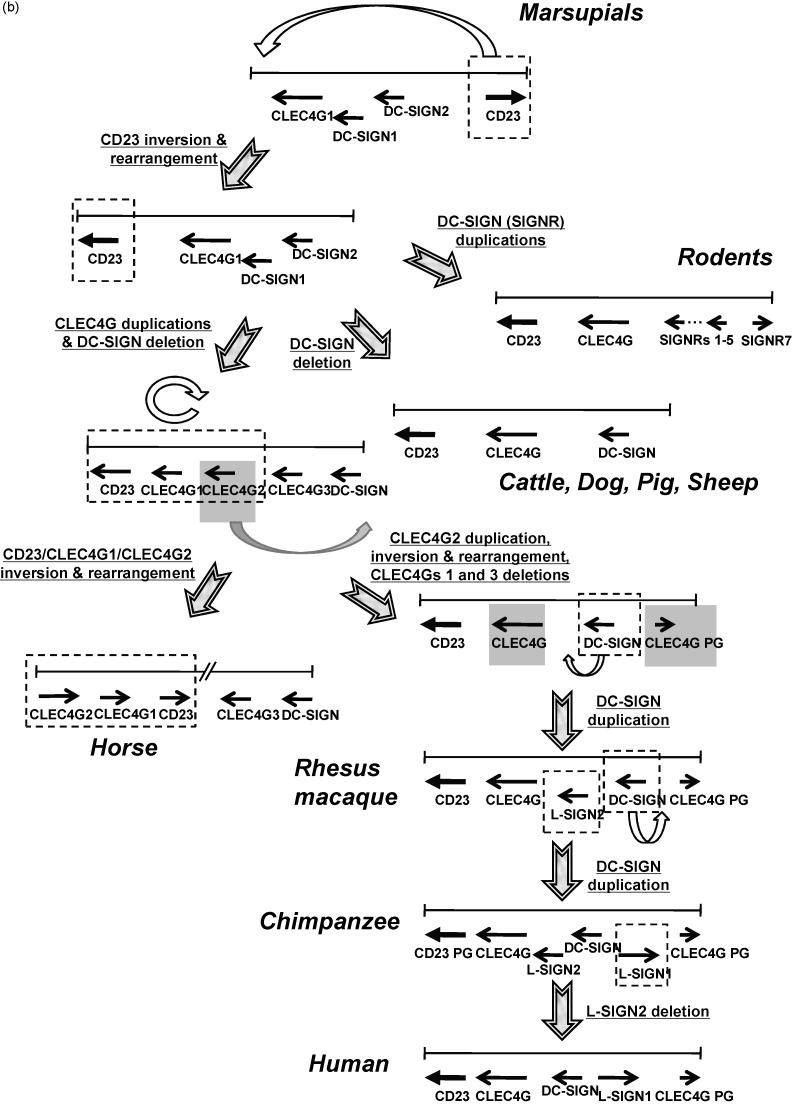
Fig. 8.
(a) Schematic representations of the clusters of genes CD23/CLEC4G/DC-SIGN in human, chimpanzee, rhesus macaque, bovine, canis, equine, mouse, rat and opossum. The relative size and orientation of CLEC4G, DC-SIGN, L-SIGN and SIGNR genes on their respective chromosome were shown as black arrows, whereas the CD23 and pseudogenes (PG) were indicted as light gray arrows. The genomic loci and coordinates were described in Section 2. (b) Proposed evolutionary processes of the cluster of genes CD23/CLEC4G/DC-SIGN from marsupials to primates based on their genomic structures and phylogenetic relationships. Only the orientations and arrangement of the genes were shown as black arrows, whereas the relative size of the genes and genomic coordinates were not shown. Notched arrows indicated the proposed key processes. Possible gene mutation events including inversion, rearrangement, duplication and deletion in each process were described with underlines adjacent to the corresponding notched arrow.
The putative cDNA and genes of CD23, CLEC4G and DC-SIGN-like homologues from the above vertebrate species and their corresponding UniGene numbers (or symbols) and GenBank accession numbers used for the alignment and comparison were summarized in Table 1 .
Table 1.
Summary of the putative genes and mRNAs of CD23, LSECtin and DC-SIGN-like homologues among mammalian species and their corresponding UniGene numbers (or symbols) and GenBank accession numbers used for the alignment and comparison.
| Species | Gene |
|||||||
|---|---|---|---|---|---|---|---|---|
| CD23 (FCER2) |
LSECtin (CLEC4G) |
DC-SIGN (CD209) |
L-SIGN (CD209L) |
|||||
| UniGene # or symbol | GenBank accession # | UniGene # or symbol | GenBank accession # | UniGene # or symbol | GenBank accession # | UniGene # or symbol | GenBank accession # | |
| Human (Homo sapiens) | Hs.465778 | NC_000019 (gene), NM_002002 (mRNA) | Hs.220649, Hs.568222 (pseudogene) | NC_000019 (gene), NM_198492 (mRNA), NR_002931 (non-coding RNA from pseudogene) | Hs.278694 | NC_000019 (Gene), NM_021155 (mRNA) | Hs.421437 | NC_000019 (Gene), NM_014257 (Isoform1 mRNA) |
| Chimpanzee (Pan troglodytes) | LOC456434 | NC_006486 (pseudogene), XR_023699 (non-coding RNA from pseudogene) | CLEC4G | NC_006486 (gene), XM_001154312 (mRNA), XR_023719 (non-coding RNA from pseudogene) | CD209 | NC_006486 (Gene), NM_001009064 (mRNA) | CD209L1, CD209L2 | NC_006486 (Genes), XM_512333 (CD209L1 mRNA), XM_001146279 (CD209L2 mRNA) |
| Rhesus macaque (Macaca mulatta) | FCER2 | NC_007876 (gene), XM_001097471 (Incomplete mRNA) | LOC704872 (pseudogene1), LOC705334 (pseudogene2) | NC_007876 (gene), XR_011056 and XR_011166 (non-coding RNAs from pseudogenes 1 and 2) | Mmu.3596 | NC_007876 (Gene), NM_001032870 (Isoform1 mRNA) | Mmu.3664 | NC_007876 (Gene), NM_001032951 (CD209L2 mRNA) |
| Cattle (Bos taurus) | Bt.29723 | NC_007305 (gene), XM_592551 (mRNA) | Bt.22180 | NC_007305 (gene), XM_583924 (mRNA) | Bt.9532 | NC_007305 (Gene), XM_590928 (mRNA) | None | None |
| Dog (Canis lupus) | Cfa.41676 | NC_006602 (gene), XM_542116 (mRNA) | Cfa.41675 | NC_006602 (gene), XM_542117 (mRNA) | Cfa.14490 | NC_006602 (Gene), XM_542118 (mRNA) | None | None |
| Horse (Equus caballus) | Eca.13012 | NC_009150 (gene), NM_001081807 (mRNA) | LOC100066617 (CLEC4G1), Eca.14020 (CLEC4G2), LOC100066686 (CLEC4G3) | NC_009150 (genes), XM_001496835, XM_001496849, XM_001496883 (CLEC4Gs 1-3 mRNAs) | LOC100066738 | NC_009150 (Genes), XM_001496929 (mRNA) | None | None |
| Pig (Sus scrofa) | Unassigned | CU929919 (gene), FJ545265 (mRNA) | Ssc.19137 | CU929919 or EU814899 (gene), AK232603 (mRNA) | Ssc.9743 | CU929919 (Gene), NM_001129972 or EU684956 (mRNA) | None | None |
| Sheep (Ovis aries) | Oar.8217 | EE816942 (incomplete mRNA) | Oar.10271 | EE825489 and EE789989 overlapping (complete mRNA) | Unassigned | EE825175 (mRNA) | None | None |
| Opossum (Monodelphis domestica) | LOC100026894 | NC_008803 (gene), XM_001377323 (mRNA) | LOC100026825 (CLEC4G1), LOC100013537 (CLEC4G2) | NC_008803 (genes), XM_001377275 and XM_001367882 (CLEC4Gs 1 and 2 mRNAs) | LOC100026844 (DC-SIGN1), LOC100026862 (DC-SIGN2) | NC_008803 (Genes), XM_001377290 and XM_001377303 (DC-SIGNs 1 and 2 mRNAs) | None | None |
| Duck-billed platypus (Ornithorhynchus anatinus) | Unknown | Unknown | LOC100081235 (CLEC4G1), LOC100081208 (CLEC4G2) | NW_001765451 (gene), XM_001512010 and XM_001511987 (CLEC4Gs 1 and 2 mRNAs) | Unknown | Unknown | Unknown | Unknown |
| Mouse (Mus musculus) | Mm.1233 | NC_000074 (gene), NM_013517 (mRNA) | Mm.109183 | NC_000074 (gene), NM_029465 (mRNA) | SIGNR (CD209) | |||
| UniGene # or symbol | GenBank accession # | |||||||
| Mm.175163 (SIGNR1), Mm.215729 (SIGNR2), Mm.111026 (SIGNR3), Mm.52281 (SIGNR4), Mm.32510 (SIGNR5), Mm.390006 (SIGNR7), Mm.171061 (SIGNR8) | NC_000074 (Genes), mRNAs: NM_026972 (SIGNR1), NM_130903 (SIGNR2), NM_130904 (SIGNR3), NM_130905 (SIGNR4), NM_133238 (SIGNR5), XM_284376 (SIGNR7), XM_284386 (SIGNR8) | |||||||
| Rat (Rattus norvegicus) | Rn.10326 | NC_005111 (gene), NM_001033924 (mRNA) | Clec4g | NC_005111 (gene), XM_001069149 (mRNA) | Rn.138846 (SIGNR1), Cd209c (SIGNR2), Rn.218629 (SIGNR3), Rn.220862 (SIGNR4), Rn.218460 (SIGNR5), Rn.198282 (SIGNR2-R1), LOC688858 (SIGNR2-R2), Rn.214149 (SIGNR2-R3), RGD1561234 (SIGNR2-R4), RGD1564563 (SIGNR2-R5), RGD1564571 (SIGNR2-R6), Rn.137828 (SIGNR7) | NC_005111 (Genes), mRNAs: XM_213687 (SIGNR1), XM_001068950 (SIGNR2), XM_344064 (SIGN3), XM_577206 (SIGN4), XM_221778 (SIGNR5), XM_001068850 (SIGNR2-R1), XM_001068599 (SIGNR2-R2), XM_221789 (SIGNR2-R3), XM_221790 (SIGNR2-R4), XM_577205 (SIGNR2-R5), XM_221808 (SIGNR2-R6), XM_001068161 (SIGNR7) | ||
Phylogenetic tree was constructed by the neighbor-joining method in the PAUP 4.0 program (David Swofford, Smithsonian Institute, Washington, DC, distributed by Sinauer Associate Inc.) based upon the available complete amino acid coding sequences of CD23, CLEC4G and DC-SIGN family proteins. Pairwise comparison of the porcine CLEC4G gene with the bovine, canis, equine, human, chimpanzee, rhesus macaque, mouse, rat, opossum and duck-billed platypus CLEC4G genes and porcine DC-SIGN was accomplished with the mVISTA program (http://genome.lbl.gov/vista/mvista/submit.shtml).
Prediction of targets of microRNA (miRNA) by searching for the presence of conserved 8-mer and 7-mer sites that match the seed region of each miRNA was performed by the TargetScan program (release 4.2, April 2008; http://www.targetscan.org/). The sequences of the predicted mature miRNAs were confirmed in the miRNA database miRBase (http://microrna.sanger.ac.uk/; release 12.0, September 2008).
3. Results and discussion
3.1. Molecular cloning and the structure of porcine CLEC4G cDNA and gene
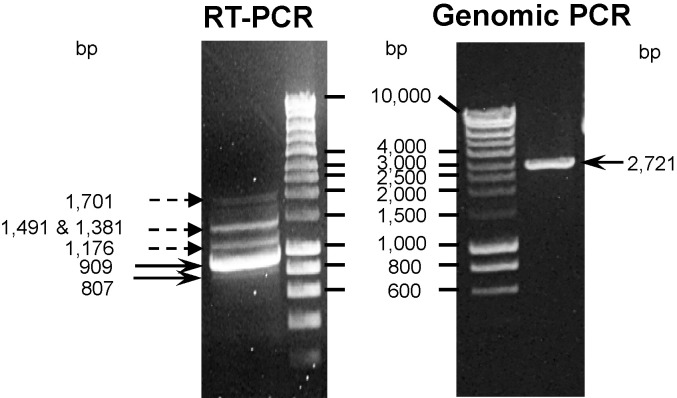
To identify the porcine homologue of human CLEC4G, a series of sequence similarity searches in the GenBank database were performed. A porcine cDNA sequence with the GenBank accession number AK232603 that shared significant sequence homology with human CLEC4G cDNA was identified. Based on this sequence, we designed gene-specific primers and then successfully amplified a 909-bp fragment containing the complete coding region of pCLEC4G cDNA from pig liver by RT-PCR (Fig. 1 ). In addition, a smaller fragment representing a pCLEC4G isoform lacking the transmembrane domain (807 bp) was also identified (Fig. 1). Besides these two isoforms, a series of higher-molecular-weight bands were amplified (Fig. 1), which are recognized as the intermediate products of pCLEC4G pre-mRNA during splicing (see discussion below). The pCLEC4G gene (2721 bp), which was not available on the public draft assembly of pig genome project in the early 2008, was also cloned by using the genomic PCR with the same pair of PCR primers (Fig. 1).
Fig. 1.
Amplification of the intermediate and mature products (isoforms) of pCLEC4G pre-mRNA during splicing from pig liver by RT-PCR and amplification of pCLEC4G gene from pig genomic DNA by genomic PCR. Dashed-line arrows showed the spliced intermediate products and solid-line arrows indicated the isoforms and pCLEC4G gene.
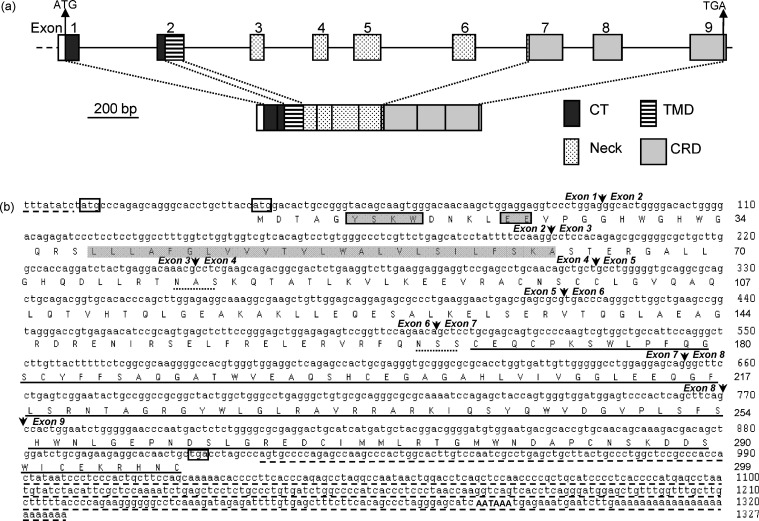
As illustrated in Fig. 2a, the pCLEC4G gene is encoded by nine exons spanning the complete coding region of the gene in which the exact sizes of the exons 1 and 9 are not yet known. The sequence of all the nine exons was identical to that of the cloned 909-bp cDNA as well as to the pCLEC4G EST, indicating the authenticity of the cloned gene. The sizes of eight introns vary from 110 to 320 bp, and the sequences of all acceptors and donors on the introns conform to the GT-AG rule. Like other type II C-type lectins, the putative coding region of pCLEC4G encodes four domains, CT, TMD, neck and CRD, from the amino- to the carboxyl-terminus (Fig. 2b). The 3′-end of exon 1 and the 5′-end of exon 2 encode the CT. The remaining part of exon 2 encodes the TMD. The neck region spans the entire exons 3–6 and the first 5 nucleotides (nt) of exon 7. The rest of the exon 7, the entire exon 8 and the 5′-end of the exon 9 encode the CRD (Fig. 2a).
Fig. 2.
(a) Gene structure of pCLEC4G gene. The upper row displayed the exon allocation of domains. The lower row represented the domain structure of the putative pCLEC4G coding region. CT: cytoplasmic tail; TMD: transmembrane domain; CRD: carbohydrate recognition domain. Un-translated regions in exons 1 and 9 were shown as open boxes. (b) Complete nucleotide sequence of pCLEC4G cDNA and its deduced amino acid sequence. Extra nucleotide sequences at both termini in the non-coding region of pCLEC4G cDNA that were not determined in this study were included with dashed underlines (available from nucleotide sequence with GenBank accession number AK232603). The two in-frame initiation codons and the stop codon were boxed. Two potential internalization motifs, YSKW and EE in the CT, were indicated by gray boxes. The putative TMD was indicated by a gray background and the carbohydrate recognition domain (CRD) is underlined. Two predicted glycosylation sites in the neck region were indicated by dotted underlines. The polyadenylation signal (AATAAA) was indicated by capitals. Arrows show the boundary of exons.
During the preparation of this manuscript, a S. scrofa chromosome 2 clone CH242-334A8 (working draft sequence; GenBank accession number CU929919) without any assigned gene loci was released on September 24, 2008. Our cloned pCLEC4G gene sequence is nearly identical to the sequence between nucleotides 148,895 and 146,200 (minus strand) on this clone. However, there exists a gap with unknown length in this region of the clone corresponding to the 3′-partial intron 7, the complete exon 8 and the 5′-partial intron 8, which has determined in our cloned pCLEC4G gene (data not shown).
3.2. Comparison of the pCLEC4G gene with other putative mammalian CLEC4G homologues available from the genome databases
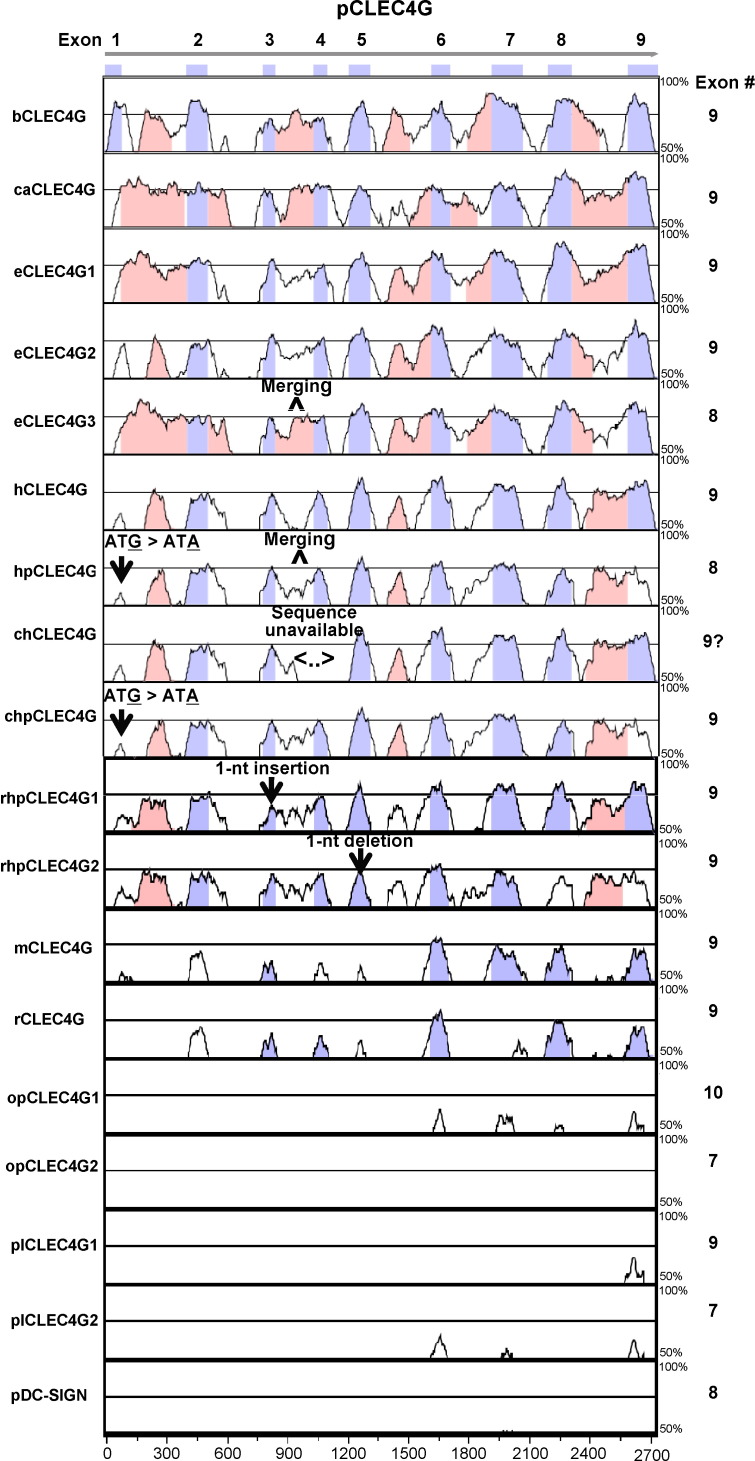
The pCLEC4G gene shares a similar structure and size of nine exons including the localization of the four domains to the corresponding exons of the human as well as the predicted bovine, canis, mouse and rat CLEC4G genes (Fig. 3 ). The exon 4 sequence of chimpanzee CLEC4G gene that is assumed to contain nine exons has not been available thus far. Three CLEC4G gene homologues, named as equine CLEC4Gs 1–3 in this study, were found in horse genome database. The equine CLEC4G1 and CLEC4G2 also have 9 exons with the same gene structure whereas equine CLEC4G3 only contains 8 exons. The missing of one exon in equine CLEC4G3 is caused by the fusion of two neck-domain-encoding exons (exons 3 and 4 corresponding to pCLEC4G gene) of the canonical 9-exon-containing CLEC4G gene into one. Similarly, the merging of exons 3 and 4 of the canonical 9-exon-containing CLEC4G gene into one exon results in a total of eight exons for the putative human CLEC4G pseudogene. However, the loss of protein-coding ability of the human pseudogene, along with the chimpanzee CLEC4G pseudogene, is due to a point mutation (G to A) at the proposed ATG start codon (Fig. 3). Two rhesus macaque CLEC4G homologues, CLEC4G1 and CLEC4G2, were predicted based upon the genomic sequencing data. However, in spite of the existence of nine exons, both homologues encode a carboxyl terminus-truncated protein product with CRD deletion due to a 1-nt insertion on exon 3 (for CLEC4G1) or a 1-nt deletion on exon 5 (for CLEC4G2), which are also recognized as the pseudogenes (Fig. 3).
Fig. 3.
Comparison of the gene sequences and numbers of exons of pCLEC4G gene with other CLEC4G homologues as well as with pDC-SIGN gene generated by the mVISTA program. Conserved regions between pairs of sequences are displayed as peaks of similarity (Y axis) relative to the positions of the gene sequence of pCLEC4G (X axis). The blue-violet boxes above the plots represent the nine exons of the pCLEC4G gene. The peaks in the same color indicate conserved regions within exons while the peaks in pink color denote conserved regions within introns. The cutoff value of percent identity is set to 70%. The human and chimpanzee CLEC4G pseudogenes lost their protein-coding ability due to a point mutation (G to A) at the proposed start codon. The two rhesus macaque CLEC4G pseudogenes are unable to encode functional CLEC4G proteins due to a 1-nt insertion or a 1-nt deletion leading to the frame shift. The exon 4 sequence of chimpanzee CLEC4G gene is not available thus far. Abbreviations: porcine CLEC4G (pCLEC4G), bovine CLEC4G (bCLEC4G), canis CLEC4G (caCLEC4G), equine CLEC4G (eCLEC4G), human CLEC4G (hCLEC4G), human CLEC4G pseudogene (hpCLEC4G), chimpanzee CLEC4G (chCLEC4G), chimpanzee CLEC4G pseudogene (chpCLEC4G), rhesus macaque CLEC4G pseudogene (rhpCLEC4G), mouse CLEC4G (mCLEC4G), rat CLEC4G (rCLEC4G), opossum CLEC4G (opCLEC4G), platypus CLEC4G (plCLEC4G), and porcine DC-SIGN (pDC-SIGN).(For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Two CLEC4G homologues were identified in both opossum and duck-billed platypus (Ornithorhynchus anatinus) that represents the non-placental and ancestral model animal of the mammalian species. The putative opossum CLEC4G1 contains 10 exons. The first 9 exons have a similar structure with those from the other 9-exon-containing CLEC4G genes, whereas the tenth exon encodes an extra 119-aa tail at the carboxyl-terminus. The putative opossum CLEC4G2 gene, containing 7 exons, encodes a soluble protein due to lack of the deduced TMD. The platypus CLEC4G1 is a canonical 9-exon-containing gene, whereas the platypus CLEC4G2 gene has 7 exons in which only two exons (exons 3 and 4) encode the neck domain. Except for the two rhesus macaque CLEC4G pseudogenes, all identified CLEC4G homologues share an important structure feature in which the CRD always spans the last three exons. The DC-SIGN homologues in mammals identified thus far share the same feature [16].
Pairwise comparison of the genomic sequences of pCLEC4G with bovine, canis, equine, human, chimpanzee, rhesus macaque, mouse, rat, opossum or platypus using mVISTA program revealed that significant conservations in both exons (especially the last three exons encoding the CRD) and intron (especially introns 1, 3, 5, 6 and 8) sequences exist between pCLEC4G and CLEC4G homologues from domesticated animals and primates (Fig. 3). Less conservation, mainly in exon sequences, was identified between pCLEC4G and rodent CLEC4G homologues, whereas the opossum and platypus CLEC4Gs have the least conservation in exon sequences. No significant sequence identity was found between pCLEC4G and pDC-SIGN (Fig. 3).
3.3. Sequence analysis of pCLEC4G encoding product and prediction of a multi-species-conserved microRNA target sequence at the 3′-untranslated region (3′-UTR) of CLEC4G mRNAs
The 1327-bp pCLEC4G cDNA has two in-frame ATG start codons at nt position 10 or 37, respectively (Fig. 2b). Compared to other CLEC4G homologues without the first ATG at the corresponding position, the deduced pCLEC4G protein is predicted to start at the second in-frame ATG and encompasses an ORF of 873-nt encoding a protein of 290 amino acids (Fig. 2b). The pCLEC4G protein is a putative type II transmembrane protein beginning from a 28-aa cytoplasmic tail followed by a predicted 22-aa TMD. The extracellular domain consists of a 111-aa neck region followed by a 129-aa CRD (Fig. 2b). Two potential internalization motifs, YSKW and EE at aa position 6–9 and 14–15, respectively, were found within the CT, which are conserved in human, chimpanzee, bovine, ovine and canis CLEC4Gs. Mutagenesis analysis has shown that the internalization ability of the hCLEC4G is dependent on the integrity of both motifs [10]. Equine CLEC4Gs 1 and 3, mouse, rat and two platypus CLEC4G homologues also harbor the tyrosine-based motif as the potential internalization signal (data not shown). The neck region of hCLEC4G contains two potential N-linked glycosylation sites and has a typical heptad repeat pattern that is expected to form α-helix coiled-coil structures [12]. A recent study also revealed that hCLEC4G exists as a disulfide-lined dimmer by two cysteine residues in the neck region [12]. All these features are identical in pCLEC4G as well as in other mammal CLEC4Gs except for equine CLEC4G3, opossum and platypus CLEC4Gs (data not shown). We previously found that human DC-SIGN, L-SIGN, non-human primate DC-SIGN and mouse SIGNR1 contains variable repeated sequence within the neck region, whereas the remaining mouse SIGNR members (except for SIGNR2 and SIGNR6) together with porcine, bovine, ovine, canis and equine DC-SIGNs do not have the repeated sequence [16]. These data suggested that the evolution of the neck region of CLEC4G family members is less divergent than that of the DC-SIGN family members.
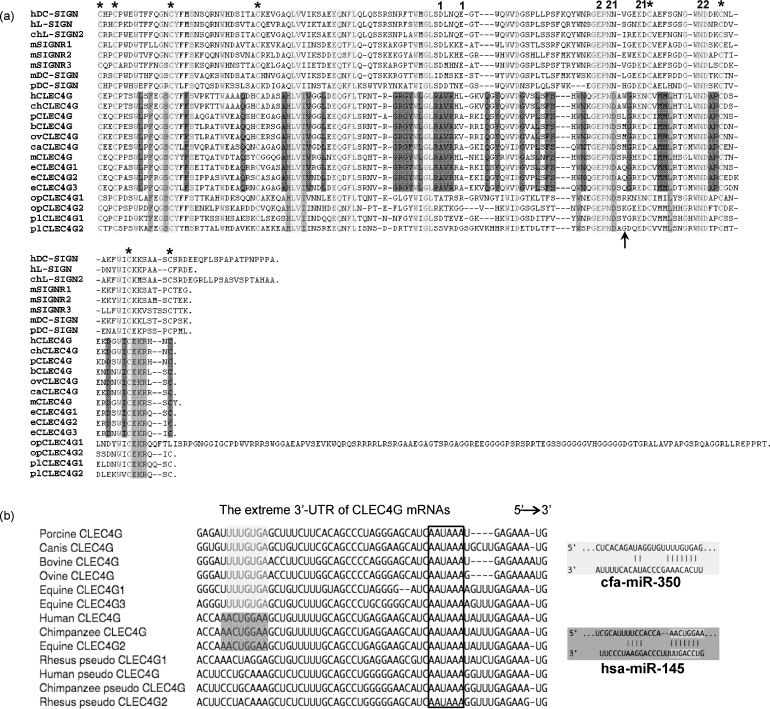
The CRD of pCLEC4G was the most conserved region shared by porcine and all the other CLEC4G homologue proteins, containing the key residues that form Ca2+- and carbohydrate-binding sites (Fig. 4a). Eight conserved cysteines predicted to form disulfide bonds were identified in the CRD of all CLEC4G homologues except for opossun CLEC4G1that contains an extra 119-aa tail. All CLEC4G as well as DC-SIGN family members possess five conserved amino acid residues, Glu260, Asn262, Asn268, Asn280 and Asp281 (aa position corresponding to pCLEC4G), for calcium-binding site 2 and the common Glu-Pro-Asn sequence (EPN sequences; aa position 260–262) that are critical for binding mannose-, fucose- or galactose-containing oligosaccharides. However, three of the four residues (aa positions 233, 237, 263 and 269) forming calcium-binding site 1 are unique in CLEC4G family members. All placental mammalian CLEC4Gs share a unique Ala residue distinct from DC-SIGNs at aa 233, whereas the residue at aa 237 is variable among CLEC4G homologues. All CLEC4G members share the Asp, residue instead of the conserved Asn residue, at aa 263. The conserved Asp269 of C-type lectins is identical in most of the CLEC4G proteins but is substituted by an Asn residue in human and chimpanzee CLEC4G as well as in opossum CLEC4G1. In addition, there are 9 and 29 unique residues in the CRD shared by all mammalian CLEC4G proteins and placental mammalian CLEC4Gs, respectively (Fig. 4a). These unique substitutions suggested that the CLEC4G family members would be expected to have different sugar-binding abilities.
Fig. 4.
(a) Alignment of amino acid sequences of the CRD of pCLEC4G and other CLEC4G as well as DC-SIGN homologues among various vertebrate species. Amino acid residues that form Ca2+-binding site 1 are indicated by “1”, residues that form Ca2+-binding site 2 are indicated by “2”, and conserved cysteine residues involved in disulfide bond formations are indicated by “*”. An arrow indicated the aa position 265 following the EPN motif where Trp residue of hCLEC4G had been proposed to interact with the GlcNAc residue of a disaccharide GlcNAcβ1-2Man. Alignment gaps are indicated by dashes. The consensus residues with identical amino acids among all placental CLEC4G members, all mammalian CLEC4G members, and both DC-SIGN and CLEC4G members are highlighted in dark gray, middle gray and light gray, respectively. Abbreviation: ovine CLEC4G (ovCLEC4G). Other abbreviations are the same as those in Fig. 3. (b) Prediction of a multi-species-conserved microRNA (miRNA) target sequence at the three prime untranslated regions (3′-UTR) of domesticated animal and primate CLEC4G mRNAs by the TargetScan program. Only one common region (nt position 1247–1254 corresponding to pCLEC4G, see Fig. 2b) that was targeted by miR-350 in domesticated animal CLEC4G members except equine CLEC4G2 (highlighted in light gray) and by miR-145 in human and chimpanzee CLEC4Gs as well as equine CLEC4G2 (highlighted in dark gray), respectively, was identified at the extreme 3′UTR. The polyadenylation signals (AAUAAA) were indicated by boxed. The respective predicted pairing of target region and miRNA was shown on the right.
Recently, human CLEC4G was shown to bind to a novel disaccharide, GlcNAcβ1-2Man, through the EPN motif and the two nearby residues Gly259 and Trp265 [12]. The contact between the GlcNAc residue and the side chain of Trp265 was predicted to be mediated by the packing of the indole ring of tryptophan against the methyl group of the N-acetyl substituent of GlcNAc [12]. However, although the Gly259 is conserved in all CLEC4G proteins, only chimpanzee CLEC4G shares the Trp265 residue with the hCLEC4G (Fig. 4a). The residue at this position is variable among other CLEC4G proteins: Leu in pCLEC4G, Met in bovine, ovine and canis CLEC4Gs, and Gln in three equine CLEC4Gs. It will be interesting to see if pCLEC4G and others have the analogous ability to bind GlcNAcβ1-2Man or other truncated glycans.
A recent study showed that the hCLEC4G is the putative receptor for Nipah virus surface glycoprotein protein (NiV-G) [17]. The interaction was mediated by the GlcNAcβ1-2Man terminal structures in NiV-G. The envelope protein of Ebola virus as well as the spike protein of SARS-CoV also bear these carbohydrate motifs and thus are uniquely recognized by hCLEC4G [11]. Nipah virus is a zoonotic Paramyxovirus transmitted from pig to human with high mortality rates. If pCLEC4G shares the same carbohydrate–protein interaction pattern with hCLEC4G, it will be very interesting to see whether pCLEC4G can serve as a pathogen recognition receptor to trigger the host innate immune responses and facilitate the transmission and spread of Nipah virus or other pathogenic porcine enveloped viruses during infections in pigs.
MicroRNAs (miRNAs) are a class of small (∼22 nt long) endogenous non-coding RNAs that bind to imperfectly complementary sites in the 3′-UTR of target mRNAs and thus repress mRNA expression [18], [19]. Thousands of different miRNAs from multicellular organisms and some viruses have been identified and shown to have both tissue-specific and development-stage-specific expression, which are thought to regulate almost every biological process. It has been estimated that more than one-third of human genes could be controlled by microRNAs [19]. MiRNA-mediated repression often requires perfect base pairing of the miRNA seed region (nt 2–7 from the miRNA 5′-end) to the 3′-UTR of an mRNA target sequence [18], [19]. Both miRNAs and their 3′-UTR binding sites are evolutionarily conserved in many cases. Thus far, functional C-type lectin expression has not been linked to miRNA regulation. With the available information of 3′-UTR sequences from different mammalian CLEC4G genes, we are interested to see if there exist miRNA target sequences that are conserved across multiple mammalian species.
Using the TargetScan program, we identified a unique site located 27-nt upstream of the polyadenylation signal (AAUAAA) in the putative canis CLEC4G mRNA that was predicted to be the target of a dog miRNA cfa-miR-350 (Fig. 4b). The 7-nt sequence UUUGUGA on this site was fully conserved among porcine, bovine and ovine CLEC4Gs as well as two equine homologues CLEC4G1 and 3 (Fig. 4b). Other conserved sequences at the 3′-UTR are less than 6-nt (data not shown), which did not fulfill the proposed perfect base pairing of the miRNA seed region. Although the miR-350 homologues in pig, cattle, sheep and horse have not been available from miRBase, they likely will have the identical sequence due to the evolutionary conservation. Interestingly, an 8-nt sequence AACUGGAA at the same position in hCLEC4G and chimpanzee CLEC4G mRNA was also targeted by a human miRNA hsa-miR-145 and a chimpanzee ptr-miR-145, respectively. This unique sequence was shared by equine CLEC4G2 but not the other primate pseudogenes (Fig. 4b). We did not identify any specific miRNAs recognizing the same site in rodent or non-placental CLEC4G members, probably due to the limiting data of miRNAs for these species available from miRBase (data not shown). The computer-based identification of a position-conserved and multi-species-conserved miRNA target sequence in CLEC4G members from domesticated animals and primates should provide some insights for the study of potential miRNA-mediated regulation of CLEC4G in the future.
3.4. Tissue distribution of pCLEC4G gene expression
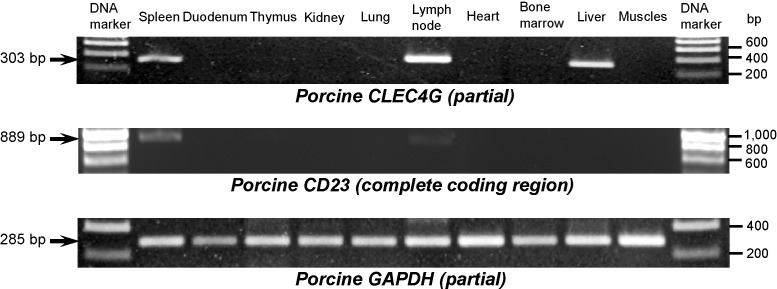
Expression of pCLEC4G mRNA was detected in spleen, lymph node and liver tissues but not in duodenum, thymus, kidney, lung, heart, bone marrow or skeletal muscle tissues of pig as determined by RT-PCR (Fig. 5 ). The pCLEC4G mRNA expression level in lymph node is the highest. It has been reported that hCLEC4G is expressed not only on LSECs but also on monocyte-derived macrophages and dendritic cells [9], [10], lymph node and bone marrow sinusoids [25]. However, the hCLEC4G expression was not found on peripheral blood lymphocytes, NK cells, CD34+-derived endothelial-like cells [10], liver Kupffer cells, thymus or placenta [25]. The pCLEC4G expression shares an analogous pattern with hCLEC4G mRNA expression.
Fig. 5.
Detection of pCLEC4G and pCD23 mRNA expression in selected pig tissues by RT-PCR. Pig tissue cDNAs were used as templates in PCR reactions with primers PLST-E67F/PLST-E89R, PCD23-F/PCD23-R and porcine GAPDH-specific primers, respectively. Arrows indicate the sizes of expected PCR products.
3.5. Cloning of the complete coding region of pCD23 cDNA and detection of its tissue expression distribution
The newly released porcine genomic clone CH242-334A8 also contains the putative pCD23 gene (12,500 bp) at nt positions 127,986–115,487 (minus strand) when compared with the bovine CD23 homologue. The pCD23 gene was predicted to encompass 11 exons, which shares the same genomic structure with bovine CD23 but not with human, mouse, rat, canis and equine CD23 genes consisting of 10, 12, 12, 9 and 12 exons, respectively. The deduced start codon of pCD23 is located on the exon 2 whereas the stop codon is on the exon 11. Based on the sequence information, we designed a pair of gene-specific primers (PCD23-F/PCD23-R) containing the putative start and the stop codons to amplify the complete coding region of pCD23 cDNA using the pig spleen tissue by RT-PCR (Fig. 5). Sequence analysis showed the expected size (889 bp) and nucleotide sequence identical to the putative 10 exons (exons 2–11) of pCD23 in the clone CH242-334A8 (data not shown), which indeed encodes a C-type lectin homologous to bovine and human CD23. The lymph node and thymus also had a very weak pCD23 mRNA expression that was not easily visible in the gel picture (Fig. 5). The PCR amplification signal in the three tissues can be strengthened (as strong bands) using a pair of primers (PCD23-E89F/PCD23-E1011R) specific to amplify the CRD sequence from the last three exons (data not shown). Other seven selected tissues did not have the pCD23 mRNA expression (Fig. 5).
3.6. Phylogenetic analysis of the pCLEC4G and pCD23 encoding protein products
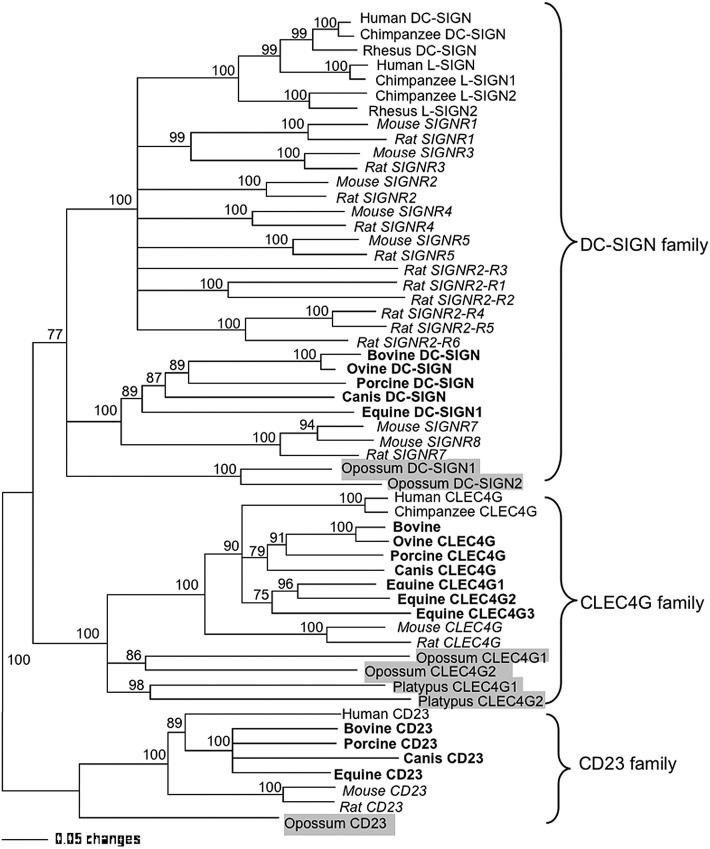
Phylogenetic analyses of the full-length encoding protein of all the available DC-SIGN, CLEC4G and CD23 family members in mammalian species were performed to determine their evolution relationship. As expected, three distinct clusters were identified in the phylogenetic tree, in which the CLEC4G family is more closely related to the DC-SIGN family than to the CD23 family (Fig. 6 ). In each family cluster, non-placental mammalian (opossum and duck-billed platypus) homologues form separate clades different from the clades containing placental mammalian homologues, indicating that they are the ancestral mammalian genes (“ancestral” is defined as the phylogenetically most divergent gene). The domesticated animal including porcine, bovine, ovine, equine and canis CLEC4Gs were clustered together, which is similar to the evolutionary relationships of their DC-SIGN and CD23 proteins. Interestingly, a phylogenetic tree generated on the basis of the 3′-UTR sequence of CLEC4G members showed that the equine CLEC4G2 was clustered into the clade containing primate homologues instead of homologues from other domesticated animals such as equine CLEC4G1 and CLEC4G3 (data not shown), which is consistent with the predicted miRNA target results that equine CLEC4G2, human and chimpanzee CLEC4Gs are uniquely recognized by miR-145. Mouse and rat CLEC4Gs are phylogenetically more divergent than other placental mammalian homologues.
Fig. 6.
Phylogenetic tree constructed by the neighbor-joining method based upon the amino acid sequences of DC-SIGN family, CLEC4G family and CD23 family proteins. Bootstrap values are indicated for each node from 1000 re-sampling. Proteins from rodents (mouse and rat), domesticated animals and non-placentals (opossum and platypus) were highlighted in italic, bold and highlighted gray, respectively.
We previously reported that mouse SIGNRs 7 and 8 along with DC-SIGN homologues from domesticated animal species form a divergent evolution pathway distinct from mouse SIGNRs 1–5 and primates [16]. In this study, we included rat SIGNR homologues, which has not been reported but are available from the rat genomic database, for the phylogenetic analysis (Fig. 6). Thirteen rat SIGNR genes related to mouse SIGNRs 1–7 were found on the rat chromosome 12p12 (see discussion below), indicating that the rat SIGNR homologues are more widely presented than mouse SIGNRs. However, the pre-assumed rat SIGNR8 does not exist. Instead, six SIGNR2-related homologues, designated here as SIGNR2-R1 to -R6, were identified. Although the phylogenetic tree based upon the alignment of DC-SIGN, CLEC4G and CD23 family members did not show the evolutionary relations of these new SIGNR-related homologues, another tree generated with only mouse and rat SIGNR molecules clearly indicated that they are probably derived from SIGNR2 progressively, from SIGNR-R1 to SIGNR-R6, and that rat SIGNR7 is more close-related to SIGNR-R6 (data not shown).
3.7. Identification of splicing intermediate products and proposed order of intron removal of pCLEC4G pre-mRNA
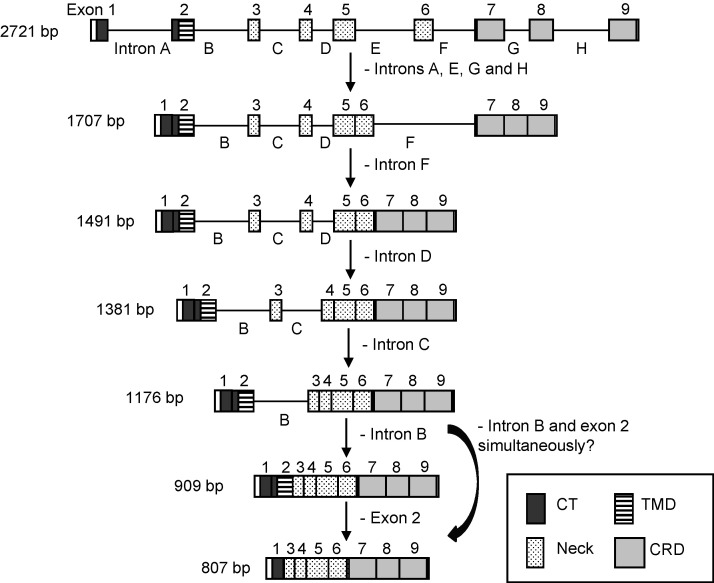
When the coding region of pCLEC4G cDNA from pig liver was amplified, we observed the possible processing of pCLEC4G pre-mRNA characterized by the appearance of a series of high-molecular-weight PCR fragments in addition to the expected mature mRNA (Fig. 1a). Each of these high-molecular weight PCR fragments was excised and cloned to determine the respective sequence. The sequencing results showed that these transcripts were actually splicing intermediate products of pCLEC4G mRNA precursors with the sizes of 1707, 1491, 1381 and 1176 bp, respectively. These intermediate splicing products retained various introns compared to the pCLEC4G gene (2721 bp). Based on the intron removals of these splicing intermediate products, we proposed a temporal order of the splicing pathway of pCLEC4G pre-mRNA (Fig. 7 ). First, introns A, E, G and H are to be removed either simultaneously or in an order that could not be predicted from the RT-PCR result to yield an intermediate product of 1707 bp, leading to the integrities of the CRD encoded by the last three exons as well as the CT encoded by the first two exons. Further processing of the 1707-bp intermediate results in a 1491-bp pre-mRNA by splicing of the intron F. Subsequent removal of the intron D yields an mRNA that is 1381 bp. Intron C appears to be spliced at this point to yield the 1176-bp pre-mRNA that retains only the intron B. The mature pCLEC4G mRNA, detected as a 909-bp product, is produced by the removal of the intron B. The processing of pCLEC4G exons encoding the neck region appears to follow a strictly temporal and positional order by splicing of the relevant introns one-by-one, from the 3 prime exon E to the 5 prime exon B. Furthermore, exon 2 is removed from the mature mRNA product to yield an 807-bp isoform lacking the TMD. Alternatively, since the exon 2 and intron B are linked together, the isoform may be generated by simultaneous splicing of them from the 1176-bp pre-mRNA (Fig. 7).
Fig. 7.
Proposed order of intron removal from porcine CLEC4G pre-mRNA. Boxes with numbers 1–9 represent the nine pCLEC4G exon sequences. The eight intron sequences, letters A to H, are indicated by the black lines between the exons. The arrows show the splicing pathway. The gene, splicing intermediate products and isoforms detected by RT-PCR, are indicated with their respective sizes shown on the left.
We proposed this temporal order of the splicing pathway based upon the fact that the amounts of the splicing intermediate products reached to the level that could be detectable by RT-PCR, thus indicating that they represent the majority in all the intermediate products. It should be noted that the RT-PCR detection was done in liver, which may accurately reflect the processing pathways of these pre-mRNAs in vivo. It will be interesting to see whether the processing of the exons encoding CRD and CT domain prior to the TMD and neck region observed here could also be present in other C-type lectins such as DC-SIGN and L-SIGN. The known DC-SIGN/L-SIGN mRNA as well as the hCLEC4G isoforms identified so far exist as TMD-lacking or partial-tandem-neck-repeats-lacking variants due to the skipping of the exon encoding the TMD and/or the presence of cryptic splicing sites on the exon encoding the neck region [10], [20]. This may be linked to the temporal order of the splicing in that different patterns of aberrant splicing occur during the later splicing events. Moreover, the variant neck-region tandem repeats of L-SIGN have been associated with the susceptibility of several infectious diseases such as SARS-CoV, HIV-1, hepatitis C virus (HCV) and Mycobacterium tuberculosis [21]. Other factors including the quality of the donor and acceptor sites, splice enhancers or suppressors, the RNA secondary structures or the size of introns and exons may also contribute to controlling the order of intron removal [22], [23], [24]. The identification of sequential splicing intermediate products of pCLEC4G pre-mRNA in vivo may provide a good model to study how the splicing machinery selects the correct pairs of splice sites to ensure the orderly intron removal in C-type lectins, and whether these could be linked to the interactions with the pathogens.
3.8. Comparative genomic analysis of the cluster of genes CD23/CLEC4G/DC-SIGN among mammalian species: the L-SIGN homologues do not exist in non-primates mammals
Thus far, the sequence information and genomic loci of CD23, DC-SIGN and CLEC4G genes from human, chimpanzee, rhesus macaque, cattle, dog, horse, mouse, rat and opossum have been available from their completed genome projects in respective species. The newly released pig chromosome 2 clone CH242-334A8 (working draft sequence) also contains necessary sequence information of the three genes and contexts according to our analysis (data not shown). For sheep, although the sequences of the genes and their contexts have not been released, the UniGene numbers and positions could be retrieved by comparison with the syntenic chromosome region of the cattle genome (data not shown).
It is well known that genes CD23, CLEC4G, DC-SIGN and L-SIGN are arranged in tandem, forming a tight gene cluster on chromosome 19p13.3. Comparison of the gene clusters among the three primate species revealed that there exists a CLEC4G pseudogene (designated here as pseudogene 2 in rhesus macaque) downstream of the gene cluster in each species (box of primates, Fig. 8a). The position of rhesus macaque CLEC4G pseudogene 1 corresponds to the functional human and chimpanzee CLEC4G genes. In addition, chimpanzee CD23 homologue is encoded by a pseudogene. Bashirova et al. [13] had reported that the L-SIGN genes emerged from a duplication event in the common DC-SIGN ancestor of anthropoids and were designated as CD209L2 (L-SIGN2) in OWM such as rhesus macaque [13]. A DC-SIGN-duplication event subsequently occurred, which resulted in the existence of L-SIGN1 in apes such as chimpanzee [13]. The L-SIGN2 was deleted and led to the proposed four-gene cluster CD23/CLEC4G/DC-SIGN/L-SIGN in human (Fig. 8b) [13].
The L-SIGN homologues do not exist in domesticated mammalian species as shown on the bovine, canis and porcine genomic regions where the C-type lectins arrange as a three-gene cluster CD23/CLEC4G/DC-SIGN instead of a four-gene cluster for primates (Fig. 8a). Either DC-SIGN or CLEC4G exists as a single gene in each species due to the evolutionary pathway that is distinct from that in primates. For sheep, previous phylogenetic analysis and comparison of gene organization indicated that its DC-SIGN and CLEC4G homologues are highly related to cattle, pig and dog and hence share similar characteristics (box of domesticated animals, Fig. 8a).
There is also no existence of L-SIGN in horse since equine DC-SIGN is a single gene. However, horse has three CLEC4G homologous genes and the arrangement of them along with CD23 and DC-SIGN is significantly different from that of other domesticated mammals and primates. CLEC4G2, CLEC4G1 and CD23 are arrayed as a three-gene cluster in the same orientation whereas CLEC4G3 and DC-SIGN are linked together in opposition to them. The two gene clusters are separated by a 508-kb region consisting of many equine genes on the same chromosome (Fig. 8a).
Based on the phylogenetic analysis, we found that, in addition to the known SIGNRs 1–7 homologues similar to mouse, rat has six SIGNR2-related homologues, SIGNR2-R1 to -R6 (Fig. 6), which are absent in the corresponding genomic region of mouse (Fig. 8a). These SIGNR2-related homologues, along with a SIGNR2 pseudogene, situate between the SIGNR2 and SIGNR7 genes. Since the SIGNR2-related homologues may be the sequential derivates from SIGNR2, and since SIGNR-R6 is closely related to SIGNR7, we speculated that they probably belong to the remnants of the intermediate products during the evolution from rat SIGNR2 to SIGNR7, which would bridge the relationship between SIGNR2 and SIGNR7 that are phylogenetically distinct in mouse [14]. The SIGNR-R1 to -R6 homologues have probably been deleted in mouse during the evolutionary process.
The gene cluster arranged as CLEC4G1/DC-SIGN1/DC-SIGN2/CD23 is shown on opossum chromosome 3, in which the orientation of the CD23 is opposite to CLEC4G1, DC-SIGN1 and DC-SIGN2 (Fig. 8a). The position of opossum CLEC4G2 is far from this cluster on the same chromosome (data not shown). Again, no L-SIGN homologue is present since both DC-SIGN1 and DC-SIGN2 are phylogenetically unrelated to L-SIGN (Fig. 6).
Proposed evolutionary processes of the cluster of genes CD23/CLEC4G/DC-SIGN from marsupials to primates based on the genomic structures and phylogenetic relationships.
First, the CD23 of ancestral opossum underwent gene inversion and was rearranged in the upstream of CLEC4G1 so that all the four genes had the same orientation (Fig. 8b). The ancestral opossum may not have two DC-SIGN genes and one may be a duplicate from another. Nevertheless, the cluster of genes CD23/CLEC4G/DC-SIGN with the same gene orientation is the “consensus” arrangement in all identified placental mammalian species. Domesticated animals except for horse retain this structure without other gene duplications, whereas multiple duplications of DC-SIGN (SIGNR) genes occurred in rodents, indicating two distinct evolutionary pathways (Fig. 8b). The third pathway with two CLEC4G duplications and/or DC-SIGN deletion followed by the inversion and rearrangement of the three-gene cluster CD23/CLEC4G1/CLEC4G2 may lead to the formation of the current arrangement of these genes in horse (Fig. 8b).
An equine gene remnant (GenBank accession number XM_001496908) homologous to primate DC-SIGN was found on horse chromosome 7 (data not shown). In addition, the 3′-UTR of equine CLEC4G2 was shown to be phylogenetically more closely related to primates as mentioned before. Taken together, we speculated that primates and horses probably share an ancestral gene cluster. The later process of evolving to OWM may include the events of CLEC4G2 duplication, inversion and rearrangement, which resulted in the arising of the ancestral CLEC4G pseudogene downstream of DC-SIGN, and the events of the deletions of ancestral CLEC4Gs 1 and 3. The L-SIGN2 duplicated from the ancestral OWM DC-SIGN may emerge subsequently (Fig. 8b). The subsequent evolutionary processes in apes and human had been described.
A thorough illumination of these processes, which is not the scope of the present study, requires more genomic information available from other mammals in the future, especially those model animals representing the intermediate stages. Detailed events of gene mutations in each process may be modified in further when more genomic information becomes available. Nevertheless, the model proposed herein provides an overall evolutionary outline of the cluster of genes CD23/CLEC4G/DC-SIGN in mammalian species, which will help better understand the biological roles of DC-SIGN and CLEC4G family in innate immunity during the evolution.
Acknowledgements
This project is funded in part by a grant from Fort Dodge Animal Health Inc. We thank Ms. Barbara Dryman for her assistance in plasmid extraction, and Dr. Stephen Wu for his encouragement and support.
Footnotes
References
- 1.Cambi A., Koopman M., Figdor C.G. How C-type lectins detect pathogens. Cell Microbiol. 2005;7(4):481–488. doi: 10.1111/j.1462-5822.2005.00506.x. [DOI] [PubMed] [Google Scholar]
- 2.Weis W.I., Taylor M.E., Drickamer K. The C-type lectin superfamily in the immune system. Immunol Rev. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 3.Conrad D.H., Ford J.W., Sturgill J.L., Gibb D.R. CD23: an overlooked regulator of allergic disease. Curr Allergy Asthma Rep. 2007;7(5):331–337. doi: 10.1007/s11882-007-0050-y. [DOI] [PubMed] [Google Scholar]
- 4.Geijtenbeek T.B., Krooshoop D.J., Bleijs D.A. DC-SIGN–ICAM-2 interaction mediates dendritic cell trafficking. Nat Immunol. 2000;1(4):353–357. doi: 10.1038/79815. [DOI] [PubMed] [Google Scholar]
- 5.Geijtenbeek T.B., Torensma R., van Vliet S.J., van Duijnhoven G.C., Adema G.J., van Kooyk Y., Figdor C.G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100(5):575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 6.Geijtenbeek T.B., Kwon D.S., Torensma R. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100(5):587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 7.Bashirova A.A., Geijtenbeek T.B., van Duijnhoven G.C. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J Exp Med. 2001;193(6):671–678. doi: 10.1084/jem.193.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lozach P.Y., Burleigh L., Staropoli I., Amara A. The C type lectins DC-SIGN and L-SIGN: receptors for viral glycoproteins. Methods Mol Biol. 2007;379:51–68. doi: 10.1007/978-1-59745-393-6_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W., Tang L., Zhang G. Characterization of a novel C-type lectin-like gene, LSECtin: demonstration of carbohydrate binding and expression in sinusoidal endothelial cells of liver and lymph node. J Biol Chem. 2004;279(18):18748–18758. doi: 10.1074/jbc.M311227200. [DOI] [PubMed] [Google Scholar]
- 10.Dominguez-Soto A., Aragoneses-Fenoll L., Martin-Gayo E. The DC-SIGN-related lectin LSECtin mediates antigen capture and pathogen binding by human myeloid cells. Blood. 2007;109(12):5337–5345. doi: 10.1182/blood-2006-09-048058. [DOI] [PubMed] [Google Scholar]
- 11.Gramberg T., Hofmann H., Moller P. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology. 2005;340(2):224–236. doi: 10.1016/j.virol.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powlesland A.S., Fisch T., Taylor M.E., Smith D.F., Tissot B., Dell A., Pohlmann S., Drickamer K. A novel mechanism for LSECtin binding to Ebola virus surface glycoprotein through truncated glycans. J Biol Chem. 2008;283(1):593–602. doi: 10.1074/jbc.M706292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bashirova A.A., Wu L., Cheng J. Novel member of the CD209 (DC-SIGN) gene family in primates. J Virol. 2003;77(1):217–227. doi: 10.1128/JVI.77.1.217-227.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powlesland A.S., Ward E.M., Sadhu S.K., Guo Y., Taylor M.E., Drickamer K. Widely divergent biochemical properties of the complete set of mouse DC-SIGN-related proteins. J Biol Chem. 2006;281(29):20440–20449. doi: 10.1074/jbc.M601925200. [DOI] [PubMed] [Google Scholar]
- 15.Yamakawa Y., Pennelegion C., Willcocks S., Stalker A., Machugh N., Burt D., Coffey T.J., Werling D. Identification and functional characterization of bovine orthologue to DC-SIGN. J Leukoc Biol. 2008;83(6):1396–1403. doi: 10.1189/jlb.0807523. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y.W., Dryman B.A., Li W., Meng X.J. Porcine DC-SIGN: molecular cloning, gene structure, tissue distribution and binding characteristics. Dev Comp Immunol. 2009;33(4):464–480. doi: 10.1016/j.dci.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowden T.A., Crispin M., Harvey D.J., Aricescu A.R., Grimes J.M., Jones E.Y., Stuart D.I. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: a template for antiviral and vaccine design. J Virol. 2008;82(23):11628–11636. doi: 10.1128/JVI.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Cullen B.R. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16(6):861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Mummidi S., Catano G., Lam L. Extensive repertoire of membrane-bound and soluble dendritic cell-specific ICAM-3-grabbing nonintegrin 1 (DC-SIGN1) and DC-SIGN2 isoforms. Inter-individual variation in expression of DC-SIGN transcripts. J Biol Chem. 2001;276(35):33196–33212. doi: 10.1074/jbc.M009807200. [DOI] [PubMed] [Google Scholar]
- 21.Khoo U.S., Chan K.Y., Chan V.S., Lin C.L. DC-SIGN and L-SIGN: the SIGNs for infection. J Mol Med. 2008;86(8):861–874. doi: 10.1007/s00109-008-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lear A.L., Eperon L.P., Wheatley I.M., Eperon I.C. Hierarchy for 5′ splice site preference determined in vivo. J Mol Biol. 1990;211(1):103–115. doi: 10.1016/0022-2836(90)90014-D. [DOI] [PubMed] [Google Scholar]
- 23.Robberson B.L., Cote G.J., Berget S.M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCullough A.J., Berget S.M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17(8):4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gramberg T., Soilleux E., Fisch T. Interactions of LSECtin and DC-SIGN/DC-SIGNR with viral ligands: differential pH dependence, internalization and virion binding. Virology. 2008;373(1):189–201. doi: 10.1016/j.virol.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]