Abstract
Alternatives to efficient viral vectors in gene therapy are desired because of their poor safety profiles. Chitosan is a promising non-viral nucleotide delivery vector because of its biocompatibility, biodegradability, low immunogenicity and ease of manufacturing. Since the transfection efficiency of chitosan polyplexes is relatively low compared to viral counterparts, there is an impetus to gain a better understanding of the structure–performance relationship. Recent progress in preparation and characterisation has enabled coupling analysis of chitosans structural parameters that has led to increased TE by tailoring of chitosan's structure. In this review, we summarize the recent advances that have lead to a more rational design of chitosan polyplexes. We present an integrated review of all major areas of chitosan-based transfection, including preparation, chitosan and polyplexes physicochemical characterisation, in vitro and in vivo assessment. In each, we present the obstacles to efficient transfection and the strategies adopted over time to surmount these impediments.
Keywords: Chitosans, Gene delivery, RNAi, Nanotechnology, Nucleic acid, Polysaccharide
Graphical abstract
1. Introduction
The concept of genes as therapeutics was published as early as 1972 in Science [1]. The initial focus was for genetic defects where the insertion of exogenous DNA into cells using attenuated viruses replaced defective DNA. Although viral vectors still currently dominate the field of gene transfer because of their high efficiency, there is an increasing interest in the development of non-viral gene delivery systems to overcome viral safety issues such as immunogenicity and insertional mutagenesis [2], [3]. Non-viral nucleic acid (NA) delivery systems are also generally endowed with inherent formulation flexibility and lower manufacturing cost [4]. Moreover, interest in the field of non-viral vectors has gained new momentum recently due to the need for effective delivery systems for small interfering RNA (siRNA), which are 21–23 nucleotide duplexes that induce specific mRNA cleavage via RNA interference (Nobel Prize 2006 [5]) and promise to be a new major class of pharmaceutical therapeutics [6].
In 1979, Mulligan published in Nature the use of a non-viral technique (calcium phosphate) to introduce foreign DNA into mammalian cells [7]. Non-viral vectors are now broadly classified into two categories (excluding vector free systems such as electroporation and calcium phosphate): i) cationic lipids such as 1,2-dioleyloxy-3-trimethylammonium propane (DOTAP) and N-[1-(2,3-dioleyloxypropyl]-N,N,N-trimethylammonium chloride (DOTMA); and ii) cationic polymers such as polyethylenimine (PEI), poly(l-lysine) (PLL), and chitosan (CS). Cationic lipids and polymers are designed to form liposomes or nanocomplexes, respectively, with negatively charged NAs by means of electrostatic complex formation. Although cationic lipids are the most studied non-viral systems to date, they are associated with significant in vitro and in vivo toxicity [8], [9]. Cationic polymers have been shown to be particularly promising among non-viral vectors with successful nucleotide delivery both in vitro and in vivo [10], [11], [12], [13], [14]. Cationic polymers display less toxicity associated with cytokine induction compared to their cationic lipid counterparts [15]. Among these, chitosan-derived vectors have garnered increasing interest as chitosan is a naturally-derived, biocompatible, biodegradable, mucoadhesive and nontoxic polymer [16], [17]. Chitosan is composed of glucosamine and N-acetyl-glucosamine units and has been approved for use in several medical applications as a wound dressing (Tegasorb® by 3M) or hemostatic patch (Hem-con® by HemCon). Please see Table 2 of reference [18] for an exhaustive list of chitin or chitosan-based products that are used for wound dressing. A chitosan-based biomaterial, discovered and developed in part by our group, was shown to improve cartilage repair in animal models [19], [20], [21], [22], [23], [24]. This product has since completed clinical trial evaluation by Piramal Healthcare Canada who received European regulatory approval in 2012 for this chitosan-based cartilage repair product (BST-CarGel) [25].
Improved understanding of structure–function relationships in chitosan based delivery systems in the past decade has enabled a significant optimization of this family of delivery systems [14], [26], [27], [28]. Chitosan is one of the most studied polymers for nucleotide delivery, with more than 215 citations in 2011 and about 164 in the first half of 2012, up from a mere 22 a decade ago in 2002 (data obtained from Thompson Reuter, search defined as: Topic = chitosan AND [gene delivery OR siRNA]). Thus chitosan based nucleotide delivery has been the subject of many reviews, each with a specific focus such as targeted delivery of pDNA [29] and siRNA [30], and formulation influence on in vitro efficiency [31]. Based on these significant advancements in chitosan-based polynucleotide delivery, one may expect it to emerge shortly from a successful clinical trial. The aim of this review is to highlight recent developments that have improved understanding of the chitosan-based NA systems and to identify current limitations encountered in the field. We produce here an integrated review of the main aspects of chitosan-based polynucleotide delivery, including sourcing, preparation and characterization, in vitro and in vivo assessment, all with an eye towards achieving clinical applications.
2. Chitosan production and characterization for gene delivery
2.1. Origin and processing to obtain chitosans with specific properties
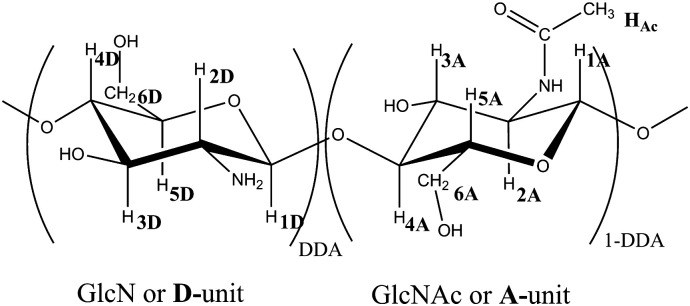
Chitin and its derivate chitosan are the second most abundant polysaccharides on Earth after cellulose. These natural linear copolymers, composed of 2-amino-2-deoxy-d-glucopyranose (noted GlcN or D unit) and 2-acetamido-2-deoxy-d-glucopyranose (noted GlcAc or A unit) units linked by β(1 → 4) glycosidic bonds (Fig. 1 ) are present in several species and characterized by the degree of deacetylation (DDA) which correspond to the molar fraction of 2-amino-2-deoxy-d-glucopyranose units: DDA = (D / (D + A) × 100).
Fig. 1.
Chemical structure of chitosan. DDA is the degree of deacetylation of chitosan. 1D, 2D, 3D, 4D, 5D, 6D correspond to the 2-amino-2-deoxy-d-glucopyranose in NMR and 1A, 2A, 3A, 4A, 5A, 6A and Ac to the 2-acetamido-2-deoxy-d-glucopyranose.
2.1.1. Sources of chitin
Although Braconnot discovered chitin from the cell wall of mushrooms in 1811 [32] and Rouget [33] extracted chitosan from insect chitin in 1859, these polysaccharides were first industrially extracted from marine organisms to eliminate fisheries wastes. In coastal regions, waste quickly leads to decreased oxygen content in water [34] and on land and is a source of environmental and public health concern [35]. Knowledge of the source of chitin is critical to ensure repeatability between different batches for medical applications. Brück et al. [36] have reviewed the preparation of chitin from marine organisms. They computed the variation of chitin content in different species and concluded, based on Kurita [37], that exoskeletons are composed of 15–40% chitin, 20–40% protein and 20–50% calcium carbonate. It is necessary to specify the source of chitin and whether or not these sources are a mixture of different species. Although several reviews affirmed seasonal variation of chitin content in exoskeletons [38], Rødde et al. [39] observed no change in chitin content over different seasons for deep water shrimp (Pandalus borealis). Seasonal variations may depend on species.
Methods have also been developed to extract chitin from terrestrial organisms such as insects and mushrooms. Nwe et al. [38] has reviewed these different terrestrial sources of chitin which are primarily insects (mosquitoes, cockroaches, honeybees, silkworms…). As in the case of marine organisms, the industrial production of chitin involves the use of insect wastes. For example, Nemtsev et al. [40] showed that honeybees are composed of 23–42% chitin, 35–45% proteins, 30–40% melanin and 3% minerals. Paulino et al. [41] also extracted chitin from dead silkworm larvae bodies. The presence of melanin covalently bonded to this chitin is a drawback given the difficulty of dissociation of the two products [38]. In the case of Nemtsev, 5% of degraded melanin residues are still present in the chitin.
Extraction of chitin from fungi has also been developed. With mushrooms, it is possible to grow a single species throughout the year. However the low chitin content is a challenge for industrial production of chitin. Knezevic-Jugovic et al. [42] concluded, based on several studies, that the content of chitin and chitosan in dry mycelia is 2% to 60%. This percentage of chitin in the dry biomass depends on the nature of the culture media as demonstrated by Chatterjee et al. [43] with Mucor rouxii or Stamford et al. [44] with Cunninghamella elegans mushrooms. Chatterjee also observed variation of crystallinity with the variation of the culture media used to grow these species. A particular issue with chitosan from mushrooms is that glucan chains, are covalently bonded to the chitin and remain difficult to eliminate [45], [46]. Given multiple possible origins of chitin, it is evident that a detailed description of location and species be given as well as an appreciation of potential side products or contaminants (melanin, glucan chains, etc.) and their absence verified for biomedical applications. Moreover, depending on the origin of chitin, its structure can vary, demanding changes in specific extraction protocols.
2.1.2. Structures of chitin
Chitin is a semicrystalline polymer, mainly in crystalline form of α and β allomorphs, but with an amorphous component between different crystalline regions. The most common α-form occurs in marine and terrestrial living organisms and in fungal cell walls. The β-form is less common and only found in certain species. In a recent review, Rinaudo [47] summarized the different species that produce the β-form: squid pens [48] and tubes synthesized by pogonophoran and vestimetiferan worms [49], [50]. A third allophorm, the γ-form is only present in cocoon fibers of the Ptinus beetle and the stomach of Loligo [51], [52] and seems to be a variant of the α-form [53].
N-acetyl-d-glucosamine is chiral and since the β(1 → 4) glycosidic bond links all units, the chitin chain can also be considered chiral. In both α and β forms, chains are organized into sheets where —C O⋯HN— intra-sheet hydrogen bonds bind chains. The crystalline forms specific to the three allomorphs depend on how the chains are stacked. In the α-form, adjacent chains are oriented in opposite directions, allowing strong intra- and inter-sheet hydrogen bonds with the hydroxymethyl group, leading to a strong crystal form where solvent has difficulty to penetrate [54]. In the β-form, chains are oriented in the same direction with only weak intra-sheet hydrogen bonds which result in higher affinity for solvents. Upon dissolution in strong acid media (HNO3 [55] or HCl 6–8 M [56], [57]), β-chitin converts itself invariably, and irreversibly, to α-chitin. The above different allomorphs are in part responsible for variability of chitin properties and impose certain restrictions on the extraction process of the chitin from the raw materials, described further below.
2.1.3. Extraction of chitin
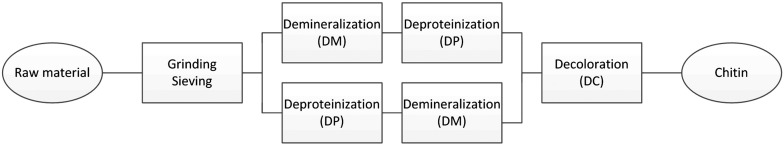
As shown in Fig. 2 , four steps are required to extract chitin from raw materials (marine exoskeletons, insect or fungi). The first step, grinding and sieving, is necessary to maximize chitin extraction and remove unwanted products. Depending upon the nature of raw materials [38] and the targeted molar mass and degree of deacetylation (DDA) of the chitin, subsequent steps need to be adjusted. The second step is usually deproteinization (DP) and then demineralization (DM) [36], [52], although DP and DM may also be reversed [58] for protein recovery. Before drying or further treatment, chitin may be decolored (DC). DP is carried out by treatment in mild alkali solutions (NaOH, KOH, Na2CO3,K2CO3…) [36], [52] where NaOH is most often used. The time and temperature of treatment depend on the origin of raw material (chitin from insect or fungi are covalently bonded to protein and require stronger treatment), the crystalline form of chitin and the targeted DDA since alkali conditions may deacetylate chitin [59], [60]. Minerals (mainly CaCO3 and Ca2PO4 but also other mineral salts) are removed using acids during the DM step. Acids used are HCl, HNO3, H2SO3, CH3COOH, and HCOOH with HCl as the preferred reagent [36]. Hydrolysis of the polymer chain may occur during DM if treatment is too strong. Percot et al. [58] optimized this step for α-chitin from shells of marine shrimp Parapenaeopsis stylifera and found that HCl at 0.25 M at room temperature for 15 min was sufficient to remove minerals. Krepets et al. [61] succeeded to extract chitin from raw materials without DM by applying an electrical current during the DP step.
Fig. 2.
Chitin manufacturing process. All step needed to obtain chitin from marine wastes.
At this level of the chitin extraction process, the resulting product is chitin, a brown to brown-white product due to the remaining carotenoids and derivates. Oxidants like KMnO4, H2O2 or NaOCl can be use for decoloration (DC or bleaching) [52] and may subsequently be removed by extraction with ethanol or acetone. The extraction also allows the removal of residual lipids [36]. The DC step is important for biomedical application where extraction with ethanol or acetone is preferred to ensure maximum removal of the side products. The DP, DM and DC steps use a large amount of water (to neutralize acid and base), energy (need to heat) and produce corrosive wastes. Alternatives have been developed using biological process. Jo et al. [62] has reviewed the different microorganisms or enzymes used for DM and DP. DM is done by fermentation using microorganisms whereas DP is done by bacterial fermentation or proteolytic enzymes. Pacheco et al. [63] compared chemical versus biological (use of Lactobacillus plantarum) DM and DP. In both cases, the DA of the resulting chitin is similar but weight average molar mass (Mw) is higher in the case of biological cleaning. Extracted proteins can be recovered during the DP step for other applications (i.e. cosmetics) and “biologically” extracted chitin contains more protein and ash than the “chemically” extracted chitin. In some cases DM and DP may be performed in one step. Rao et al. [64] used L. plantarum 541 bacteria and compared with a chemical process to show the advantage of the bacterial process with a drastic reduction of chemical use, but still requiring acid and alkali treatment to remove proteins and minerals remaining after fermentation. Biological production of chitin eliminates some pollution and depolymerization problems but the main drawbacks are time consumption, lower efficiency than chemical extraction and of particular importance for biomedical use, some undesirable residual protein in the chitin.
2.1.4. Preparation of chitosan
Chitosans are prepared from chitin by deacetylation. Several definitions of chitosan versus chitin exist. Roberts [52] assumed that chitosan is the soluble fraction of polymer soluble in dilute aqueous acid solutions. For Kumar et al. [65], chitosans are polysaccharides that have, at the elemental analysis, nitrogen content higher than 7 wt.% and degree of acetylation lower than 0.4, corresponding to a DDA higher than 60%. These two definitions are not always compatible since solubility not only depends on DDA but also on concentration and molar mass. Furthermore nitrogen content by elemental analysis can be affected by residual protein content. A third definition given by Rinaudo [47] where chitin with a DDA higher than 50% is called chitosan, seems to be appropriate for biomedical chitosan. In most of cases in gene delivery, chitosans employed are typically with DDA higher than 80% [26], [28], [66], [67].
2.1.5. Deacetylation of chitin
Chitosan can be obtained by deacetylation of chitin in different processes with resulting properties that vary according to the method used. Indeed, the means by which chitin is deacetylated will affect the distribution of N-acetyl and amine groups along the chitosan chain. Homogeneously reacetylated chitosans display different enzymatic degradation properties compared to heterogeneously prepared chitosans, and are typically less degradable [68], due to the random distribution of acetyl groups on the polymer backbone [69].
Chitosan is usually obtained by alkaline deacetylation of chitin, where particulate chitin is suspended in a hot alkaline solution (typically 40–50%, w/w, NaOH). Adjusting time and temperature will allow a wide range of DDA to be produced. Acetyl content can be targeted by alkaline deacetylation, and this heterogeneous reaction is thought to result in a block distribution of acetyl groups [70], [71]. Using 1H NMR, Vårum et al. [72], [73] confirmed that heterogeneously prepared chitosans have a slightly more blockwise and less random distribution than homogenous deacetylation described below. Lamarque et al. [74], [75] studied the role of the allomorph during deacetylation using X-ray diffraction and 1H NMR spectroscopy and concluded that crystalline areas of chitin need to first become amorphous in order to be deacetylated. In case of β-chitin, the deacetylation is fast due to its weak hydrogen bonding and the resulting acetyl distribution is random. In case of α-chitin, crystalline domains are not initially available to NaOH, needing more time, inert atmosphere and higher temperature, thus resulting in a more block acetyl distribution and decreasing final molar mass of chitosan. To avoid these phenomena, Lamarque et al. [76] developed a method of freeze–pump out–thaw cycle to improve deacetylation and reduce chain cleavage. The resulting chitosan has a higher molar mass and a random distribution of amine groups along the chain. A random distribution of amines along the chain can also be achieved by dissolving chitin in a 40% w/w NaOH solution and leaving it several hours stirring below 0 °C [71], [77], [78]. A third method to obtain random acetyl distribution is by reacetylation of fully deacetylated chitosan in 80% water–methanol (v/v) with a stoichiometric amount of acetic anhydride for the desired DDA [79], [80], [81], [82].
As for extraction of chitin from raw materials, several biological processes have also been developed using enzymatic pathways to deacetylate chitin. Zhao has summarized all the enzymatic modifications of chitin and chitosan [83]. The enzymes used for deacetylation are part of the carbohydrate esterase family 4 (CE-4s) as defined in the CAZy database (http://www.cazy.org/): chitin deacetylase (CDA, EC 3.5.1.41), NodB protein (EC 3.5.1.–) [84], peptidoglycan deacetylase (EC 3.1.1.–) [85] and an enzyme from Colletotrichum lindemuthianum, a Deuteromycetes (ATeC 56676) [86]. The main challenge for the enzymatic deacetylation is to succeed to deacetylate the crystalline form of the chitin since only 1% of crystalline chitin may be deacetylated via this procedure [87].
2.1.6. Depolymerization of chitosan
Several studies have highlighted that the length of chitosan chains plays a crucial role in gene delivery [88], [89]. To modulate the size of the chains and thus the molar mass of chitosan, the glycosidic linkage between saccharide units needs to be cleaved. This cleavage, also called depolymerization can be achieved by several means. Chitosans were first depolymerized using concentrated HCl by Falk et al. [90] who found that in concentrated HCl, hydrolysis of glycosidic bonds is faster than deacetylation. These results were confirmed by Rupley [91] and this method was employed by several groups [92], [93], [94], [95]. However, deacetylation inevitably occurs with HCl, leading to an increase of DDA. The enzymatic hydrolysis of chitin and chitosan has been proposed as an alternative method for the production of chitin/chitosan oligomers but at elevated cost. Chitosanases are enzymes that can depolymerize chitosan chains and many studies have been done to identify their mechanism of action [96], [97], [98], [99]. The efficiency of the enzyme will depend on the distribution of A and D units along the polymer chain. Two kinds of chitosanases exist: exo-chitosanases which cleave D units continuously from the non-reducing end of the substrate and endo-chitosanases (EC 3.2.1.132), more specifically family 5, 7, 8, 46, 75 and 80 glycoside hydrolases as defined in the CAZy database (http://www.cazy.org/) [85]. Fukamizo et al. [96] classified these endo-chitosanases into three subclasses depending their substrate specificity: Chitosanases in subclass I can hydrolyze A–D and D–D linkages, subclass II enzymes can hydrolyze D–D linkages only, whereas subclass III enzymes can hydrolyze D–A and D–D linkages.
Currently the most cost-effective method of depolymerizing chitosan is a second chemical method involving the use of nitrous acid (HONO) proposed by Allan et al. [100], [101], [102]. In this selective depolymerization, HONO attacks only amine groups without any deacetylation, is rapid and is easily controlled [26], [103]. The second advantage of this reaction is that one end of the cleaved chain is an aldehyde which can be exploited to make gels or graft oligomers to chitosan [104]. In the Tommeraas studies [103], chitosan was depolymerized to oligomers with low DP (2 to 5), allowing 1H NMR to observe the reducing end of the chain as gem diol or Schiff base rather than aldehyde in certain conditions (basic or acid conditions).
Multiple steps are required from the starting material to obtain a final chitosan appropriate for biomedical applications. Specific processes need to be chosen depending on the structure of the original raw material and of the desired product. Rigorous and precise chemical analysis is then required to characterize the resulting chitosans since their specific characteristics greatly affect experimental findings in vitro and in vivo) as described further below.
2.2. Analysis of chitosans
Several parameters need to be precisely determined for the use of chitosan in biomedical applications and more particularly for gene delivery. In 2011, chitosan was added in the list of excipients in the USP34-NF29 [105] and the American Society for Testing and Materials (ASTM) also published a “Standard Guide for Characterization and Testing of Chitosan Salts as Starting Materials Intended for Use in Biomedical and Tissue-Engineered Medical Product Applications” [106].
2.2.1. Identification
Both standards suggest the use of infrared spectroscopy (IR) or Fourier Transform Infrared Spectroscopy (FTIR) to identify the polymer. An infrared spectrum shows absorption bands relating to bending or stretching of unique bond and the resulting spectrum is a fingerprint of the product. Chitosan can be identified by several absorption bands with specific wavenumbers (Table 1 ). 1H and 13C NMR can also be used to identify chitosan via particular chemical shift of protons and carbons [75], [109] (Table 2 ).
Table 1.
Frequencies of infrared bands of chitosan, their intensities from ASTM edition 2011, test F2103-11 and attribution from Kasaai et al. [110].
| Bond | Frequency (cm− 1) |
Intensity | |
|---|---|---|---|
| Chitosan base (as acetate) | Chitosan Chloride | ||
| Intermolecular hydrogen bonding | 3362 | 3344 | Broad |
| NH2 band | 1605 | ||
| Amide II band | 1556 | ||
| NH3+ | 1513 | ||
| CH2 bending | 1406 | ||
| C—H bending | 1379 | ||
| C—O—C bridge | 1153 | 1154 | Sharp |
| C—O—C stretching | 1083 | 1086 | Sharp |
Table 2.
| Specie | Nucleus | Chemical Shift (ppm) |
|---|---|---|
| HAc | 1H | 2.36 |
| H2D | 1H | 3.52 |
| H2A, H3, H4, H5, H6 | 1H | 3.9–4.2 |
| H1A | 1H | 4.92 |
| H1D | 1H | 5.21 |
| C1A | 13C | 103 |
| C1D | 13C | 100 |
| C2 | 13C | 58 |
| C3 | 13C | 73–75 |
| C4 | 13C | 80–82 |
| C5 | 13C | 77 |
| C6 | 13C | 63 |
| C acetyl | 13C | 25 |
2.2.2. Characterization
2.2.2.1. DDA
The degree of deacetylation is one of the most important parameters characterizing chitosan, especially in gene delivery. Indeed, DDA determination allows the knowledge of the average amount of amine available to interact with NAs and strongly influences degradability [108], inflammation [24] and immune modulation [109]. In 2009, Kasaai reviewed different methods used for DDA determination [110] which we summarize below. Moreover, in the USP, 1H NMR is recommended for DDA determination whereas in the ASTM edition 2011, tests F2103-11, both 1H NMR and UV spectroscopy are advised.
2.2.2.2. DDA by UV
In the ASTM F2103-11, determination of the DDA by UV spectroscopy is based on Muzzarelli et al. studies [111]. Contrary to what is stated in the standard, this is a quantitative measure of the number of acetylated groups in the polymer at a wavelength of 202 nm and not the number of amine groups that allows the determination of the DDA. The DDA is calculated by building a calibration curve with various concentrations of N-acetyl glucosamine monomer. This method is very sensitive but the chitosan sample needs to be absolutely dry to determine an accurate concentration of polymer, and solutions need to be at a minimum concentration of 0.1 M N-acetyl glucosamine monomer for good accuracy. The presence of residual proteins may affect the measurement. In gene delivery, chitosans often have high DDA and can thus be problematic for this method that quantifies acetyl groups. For these reasons 1H NMR is recommended for determination of the DDA.
2.2.2.3. DDA by 1H NMR spectroscopy
Use of 1H NMR to measure chitosan DDA is a very fast method, does not need a calibration curve or standards and is not affected by water content in contrast to the UV method. USP34-NF29 recommends dissolving chitosan in deuterated formic acid and using the average area of the region containing all non-acetylated protons between 3 and 6 ppm (Int3–6) and the area of the methylene group of N-acetyl to calculate DDA according to the formula DDA = [1 − [(7 × Int3–6) / (3 × HAc)] × 100]. Even if chitosan is completely dry and deuterated formic acid is fresh, the HDO peak at 4.7 ppm chemical shift relative to the sodium 3-(trimethylsilyl)propanesulfonate standard can result in overestimation of the integration between 3 and 6 ppm, leading to errors. The following alternative calculation methods can be more precise for determination of DDA with 1H NMR [72], [107], [112], [113]. A specific ASTM standard, ASTM F2260-03R08 has been published for DDA determination by 1H NMR. This method is based on Vårum [72] and chitosans need to be first depolymerized to a degree of depolymerization (DP) between 15 and 30 by nitrous acid prior to analysis to enhance the quality of spectra (especially chitosan with low DDA which can precipitate at low temperature). The calculation for DDA here is done following Eq. (1) where H2–H6 are integrations of the protons H2 to H6 of both D and A monomers between 3.5 and 4 ppm, HAc, is the integration of the three protons of the acetyl group at 2.04 ppm, H1D is the integration of the anomeric proton of the D monomer at 4.85 ppm and H1A is the integration of the anomeric proton of A monomer between 4.55 and 4.65 ppm.
| (1) |
The drawback of this method is the HONO step to prepare the samples, needing more time and preparation, and the use of protons H2 to H6 since this region of the spectra can be affected in case of modification of chitosan (for example when poly(ethylene glycol) is grafted to chitosan [114]). As a recommended alternative we developed an accurate method to determine DDA using H1D and HAc [107], which can be used for very high DDA (the HAc peak corresponds to three protons) and with certain modified chitosans via the following.
| (2) |
Others techniques (acid or base and potentiometric titration, conductometry, 13C and 15N NMR, colloidal titration ninhydrin assay or elemental analysis) are also summarized in Kasaai [110] but are less accurate, need treatments prior to analysis and may be sensitive to side-products. Infrared is especially not recommended for DDA because the IR bands are strongly affected by the presence of water and impurities.
2.2.3. Pattern of acetylation
The distribution of the acetyl and amine groups along the chitosan chain will slightly depend on the mode of deacetylation and can be characterized by 1H 13C NMR. Vårum et al. [72], [73] and Weinhold et al. [115] concluded that most chitosans are close to a random distribution of acetyl and amine groups in heterogeneously deacetylated chitosan and perhaps it is for this reason that such an analysis is not mentioned in the standards.
2.2.4. Molar mass
Molar mass is another critically important characteristic of chitosan as it permits an assessment of the average number of monomers per chain and the dispersity which is related to the actual distribution of chain lengths in the sample. As for 1H NMR, a specific ASTM standard: F2602-08e1 “Standard Test Method for Determining the Molar Mass of Chitosan and Chitosan Salts by Size Exclusion Chromatography (SEC) with Multi angle Light Scattering Detection (SEC MALS)” has been published for chitosan molar mass determination [116]. A section treating molar mass determination is also present in USP34-NF29 but the suggested method is not accurate, as specified in ASTM F2103-11. Indeed, the use of a calibration curve with PEG or pullulan standards is not recommended since chitosan, a polyelectrolyte, has a hydrodynamic radius larger than these non-charged polymers [117], [118] and at same molar mass, its exclusion volume will not be the same. The use of the SEC coupled with MALS to measure absolute mass-average molar mass is necessary to obtain chitosan molar mass. Number and mass average molar mass of chitosan (Eqs. (3) and (4)), dispersity (Eq. (5)), radius of gyration rg and the second Virial coefficient A2 (Eqs. (6) and (7)) can be determined by the use of SEC with Multi Angle Light Scattering (MALS) detection coupled with refractometer detectors:
| (3) |
| (4) |
| (5) |
In Eqs. (3) and (4), ci is the concentration of the polymer measured by the refractometer, ni the number of molecules having a molar mass Mi and w i the mass of the molecules having a specific molar mass Mi. The use of MALS involves static light scattering theory for determination of Mw, rg and A2 (Eq. (6)) [119]. Nevertheless, for gene delivery, the number average molar mass Mn is recommended because it allows a better understanding of the number of amines available per chain.
The Rayleigh–Gans–Debye (RGD) approximation is a powerful generalization of light scattering theory that is applicable for particles much smaller than the wavelength of the light and the measurement of scattered light depends on several factors as explicated in Eq. (6). In this equation, the excess Rayleigh ratio (R θ) is a ratio of the scattered and incident light intensities that takes into account these different factors including the angle, distance from detector to scattering volume, incident light intensity, and the volume of sample illuminated.
| (6) |
In Eq. (6), c is the mass concentration of the solute (g/mL), Mw is the weight average molar mass (g/mol), A2 is the second Virial coefficient (mol mL/g2), θ is scattering angle (angle between incident beam and scattered signal), P(θ) is the form factor or “scattering function” that relates the angular variation in scattering intensity to the root mean square radius (Rg) of the particle and K* is the following optical constant,
| (7) |
In Eq. ((7), n0 is the refractive index of the solvent at the incident wavelength, λ0 is the incident wavelength, expressed in nm, NA is Avogadro's number and dn/dc is the refractive index increment of the solvent–solute solution with respect to a change in solute concentration, expressed in mL/g. dn/dc must be measured independently using a refractometer detector. In SEC MALS, the computed molar mass depends only upon dn/dc to the first order because of the use of an on-line refractive index detector to determine the polymer concentration, whereas in a batch type experiment, in which the concentration of the polymer is determined independently, the molar mass depends on the square dn/dc. This parameter is specific to each polymer and also depends on the mobile phase employed and the wavelength of incident light. Table 3 summarizes protocols and chitosans employed in several SEC studies [115], [118], [120], [121], [122], [123], [124], [125], [126], [127], [128], [129], [130], [131], [132], [133], [134]. Surprisingly, despite only slight changes in mobile phase composition and wavelength, a wide range of refractive index increments (dn/dc) for chitosan have been calculated. Some studies [120], [127], [128], [129], [130] found a dependence of dn/dc with DDA whereas others [134], [135] used an average value since the coefficient of variation was low. For good accuracy, it is recommended to measure the dn/dc specific to the chitosan and mobile phase used in the SEC system, prior to analysis.
Table 3.
Current mobile phases for molar mass determination: composition, dn/dc used, wavelength, DDA and molar mass of chitosan analyzed.
| Mobile phase | pH | λ (nm) | dn/dc | Mw (kg/mol) | DDA (%) | Ref |
|---|---|---|---|---|---|---|
| 0.2 M AcOH/0.1 M AcONa | 436 | 0.175–0.208 | 194–2510 | 69–100 | [120] | |
| 0.3 M AcOH/0.2 M AcONa | – | 0.163 | 200–290 | 79–98 | [121] | |
| 0.190 | 60–260 | 40–100 | [135] | |||
| 0.2 M AcONH4 | 4.5 | – | 0.150 | 16–2610 | 40–85 | [118] |
| 0.2 M AcONH4 | 4.5 | – | 0.162a | 22–720 | 40–100 | [122] |
| 0.2 M AcONH4 | 4.5 | 633 | 0.162 | 550 | [122] | |
| 0.02 M AcONa + 0.1 M NaCl | 4.5 | 633 | 0.203 | 31–57 (Mn) | 75–93 | [123], [126] |
| 0.2 M AcOH/0.1 M AcONa | – | 633 | 0.189 | 78–2770 | 83 | [124] |
| 0.25 M AcOH/0.25 M AcONa | 4.7 | 633 | 0.158 | 38–2260 | 75–80 | [125] |
| 0.1 M AcOH/0.2 M NaCl | – | 633 | 0.180–0.201 | 170–1200 | 58–85 | [423], [424] |
| 0.2 M AcONa | 4.3 | 690 | 0.201–0.056(1-DDA)a | 42–207 | 70–96 | [127] |
| 0.2 M AcOH/0.15 M AcONH4 | 4.5 | 633 | 0.154–0.195 | 360–400 | 30–95 | [130] |
| 0.2 M AcOH/0.15 M AcONa | 4.5 | 633 | 0.154–0.198 | 130–270 | 30–99 | [128], [129] |
| 0.2 M AcOH/0.1 M AcONH4 | 4.5 | 633 | 0.203b | 130–600 | 30–99 | [131] |
| 0.5 M AcOH/0.1 M NaNO3 | 690 | 0.203b | 24–416 | 53–99.1 | [132] | |
| 0.2 M AcONH4 | 4.5 | 633 | 0.142c | 7–360 | 45–99.5 | [133] |
| 0.3 M ACOH/0.3 M AcONa + 1% ethylene glycol | 4.5 | 670 | 0.163 d | 9.7–491 | 52–99.8 | [115] |
| 0.15 M AcOH/0.1 AcONa | 4.5 | 690 | 0.192 | 10–205 (Mn) | 77–91.7 | [134] |
AcOH = acetic acid, AcONa = sodium acetate, AcONH4 = ammonium acetate, NaNO3 = sodium nitrate.
(Mn): Authors only provided number average molar mass.
0.02 M AcOH/0.02 M AcONa, 0.1 M NaCl, pH = 4.5, λ = 633 nm by Anthonsen et al. [136].
Used Schatz et al. [129] values.
Expressed in chitosan acetate concentration otherwise change with DDA.
Used Rinaudo et al. [121] values.
Several mobile phases have been also proposed for SEC, with many composed of acetic acid (AcOH) and sodium acetate (AcONa), but also ammonium acetate (AcONH4). Although ammonium acetate is recommended in the ASTM, F2602-08E01, Christensen et al. [133], observed instability attributed to the volatility of AcONH4 in the RI signal. Thus AcONa is advised for mobile phase preparation. Anthonsen et al. [136] observed a “high molecular weight” component which depended on the concentration of polymer. Several subsequent studies [122], [129], [130] confirmed these phenomena. Schatz et al. [129] found aggregates and suggested that acetyl groups drove the formation of aggregates whereas Sorlier et al. [130] concluded that the degree of ionization (α) also played a role in these phenomena. More recently, Popa-Nita et al. [137] suggested hydrophobic interactions as explanation. To avoid the presence of aggregates, several mobile phases have been proposed. Nguyen et al. [134] compared the two principal compositions (0.3 M AcOH/0.2 M AcONa and 0.15 M AcOH/0.1 M AcONa) and concluded that the latter allows better molar mass determination without observation of aggregates. Although the standard ASTM, F2602-08E01 recommends “Dissolve the chitosan base in 1% acetic acid to a 1% solution by shaking at about 100 min− 1 overnight at cool temperature (3 to 8 °C)” with a 0.2 mol/L ammonium acetate solution, an alternative is to directly solubilize the chitosans in the mobile phase overnight. Using the Mark–Houwink equation ([η] = KMa), the measurement of intrinsic viscosity and knowledge of K and a, the Mark–Houwink parameters for the chitosan in the studied solvent, provide the viscometric-average molar masses (Mv). However, this formula is empirical and the calculated mass does not have the same rigor as the molar masses determined by light scattering and should be avoided. The use of the SEC coupled with MALS and refractometer is a minimum to measure absolute molar masses and polydispersity of chitosan.
2.2.5. Dry matter content
As Brugnerotto et al. [138] observed by IR, chitosan is a hygroscopic polymer. Determination of water content or dry mass must be done to prepare solutions with precise concentration. ASTM F2103-11 as USP34/NF29 refers to the USP 34/NF29 test < 731 > where 1 to 2 g of chitosan is dried in an oven at 105° for 5 h and weighed. Water content should be lower than 5% of the mass.
2.2.6. Additional required characterization
2.2.6.1. Insolubles
Characterization of insolubles is required by the ASTM F2103-11and allows the determination of insoluble impurities by filtering a solution of chitosan in dilute acetic acid and weighing the remaining chitosan and mass of insolubles collected on the filter. As specified in the ASTM F2103-11, the content should be as low as possible for biomedical applications.
2.2.6.2. Ash content or residue on ignition
Residue on ignition of a sample represents the quantity of inorganic matter in the sample. ASTM F2103-11 as USP34/NF use the test < 281 > and the value must be lower than 1%.
2.2.6.3. Heavy metal content
As chitosan can form complexes with metal ions [52], [139], determination of the heavy metal content is essential. The determination of some specific heavy metal content is necessary as stipulated in USP34/NF29 and ASTM-2103-11. Metals to be quantified are Pb, Hg, Cr, Ni, Cd, As, and Fe using inductively coupled plasma (ICP) and following Method III of USP < 231 >. Maximal values are summarized in Table 4 .
Table 4.
Maximum permitted concentration of heavy metals in chitosan (chitosan monograph NF29 [105]).
| Element | Maximal concentration (ppm) |
|---|---|
| Pb | 0.5 |
| Hg | 0.2 |
| Cr | 1.0 |
| Ni | 1.0 |
| Cd | 0.2 |
| As | 0.5 |
| Fe | 10.0 |
2.2.6.4. Endotoxin content
Endotoxins are pyrogenic high molar mass lipopolysaccharide complexes associated with the cell wall of Gram-negative bacteria. The surface of the LPS molecule carries a negative charge owing to phosphate, pyrophosphate, and carboxylic groups located predominantly in lipid A and the inner part of core [140]. This toxin is a structural molecule of bacteria that is recognized by the immune system. In pharmaceutical production, it is necessary to remove all traces of endotoxin. As explained in a patent application [141], [142], the DP step (treatment with NaOH) allows endotoxin removal but an exact final quantification of endotoxin content is needed. Based on FDA specifications, the maximum allowable dose is 5 EU/kg for non-intrathecal administration routes. Measurements are made using the Limulus amebocyte lysate (LAL) tests according to USP < 85 >.
2.3. Modification of chitosan
Chitosan has been modified for several applications such as water cleansing (ion removal for example) [47], fiber formation (enhance solubility of chitosan for processing) [143], cosmetics [144], and drug delivery [145]. In gene delivery, chemical modifications have been done to address certain issues. Chitosans were modified to enhance their solubility at physiological pH [114], [146], [147], [148], [149], [150]. Increased chitosan solubility can enhance the colloidal stability of the chitosan/NA particles allowing for longer circulating time and can be achieved by grafting certain polymers (PEG, dextran) to chitosan [13], [27], [104], [114], [149], [150], [151], [152], [153], [154], [155], [156], [157], [158], [159], [160], [161]. Specific cells can also be targeted by grafting appropriate ligands (like galactose for liver cells) on chitosan [152], [153], [154], [155], [157], [162], [163], [164]. Chitosans were also modified to increase proton sponge capacity using arginine, histidine, urocanic acid, imidazole or PEI to facilitate endosomal escape [152], [154], [165], [166], [167], [168], [169], [170], [171], [172]. Functional assessments of these modifications are reviewed in subsequent sections.
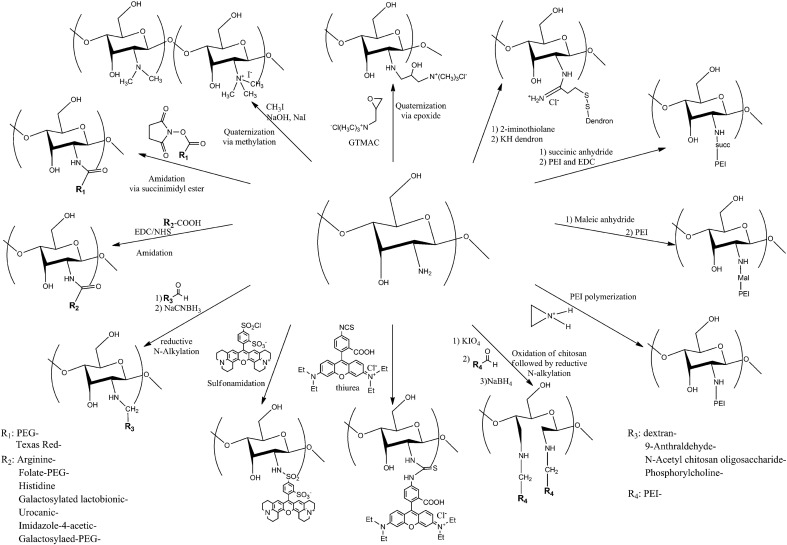
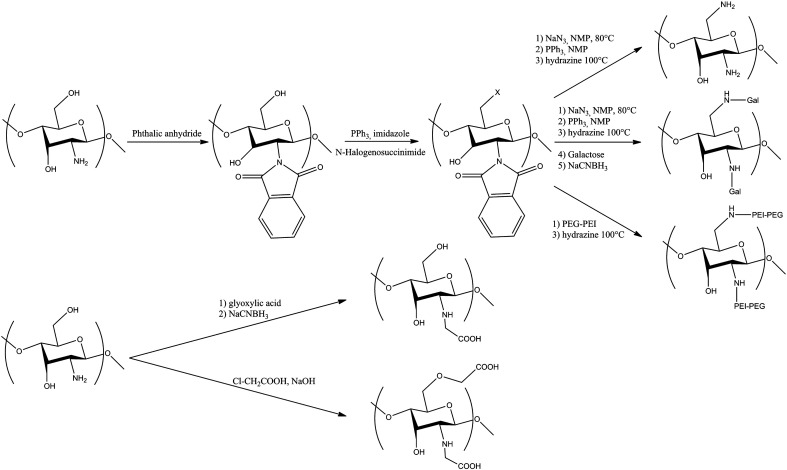
An essential consideration in the interpretation of data obtained using modified chitosans is the nature and extent of characterization of the final products. 1H NMR is a powerful tool for chemical quantification and should be preferentially chosen for determination of the degree of substitution. Nevertheless, 1H NMR spectroscopy only typically quantifies protons and is not always accurate in bond identification. The presence of a new bond that confirms the grafting to the chitosan can be obtained by infrared spectroscopy [173]. If newly created bond(s) are still difficult to observe, a comparison with the physical mixture at the same ratio should be done as in Darras et al. [174] for the grafting of a ligand to the amine function of chitosan. By comparing the grafted polymer and the physical mixture with the same amount of ligand, the change in the amide II band due to the grafting of the molecule was evident. Chitosan can be modified by chemical grafting of molecules or polymers on the C2 amine, the C6 hydroxyl function or both. The hydroxyl on C3 is less reactive and more difficult to use [175]. Table 5, Table 6 along with Fig. 3, Fig. 4 summarize the different modifications of chitosan and chemical synthesis performed to date.
Table 5.
Modifications on the amine functional group of C2 of chitosan.
| Grafted compound | Type of modification | Objective | Analysis | Ref |
|---|---|---|---|---|
| GTMAC | Epoxide opening | Solubility | IR, 1H 13C NMR | [181] |
| GTMAC | Epoxide opening | Solubility | IR, 1H NMR | [182] |
| CH3I | Methylation | Solubility | IR, 1H 13C NMR, SEC | [176], [177], [178], [179] |
| [180] | ||||
| CH3I | Methylation | Solubility | 1H NMR, SEC | [114] |
| PEG-succinimidyl ester | Amidation | Enhance pharmacokinetic | [151] | |
| CH3I | Methylation | Solubility | 1H NMR, SEC | [152] |
| Arginine | Amidation | Enhance uptake | ||
| Folate–PEG–COOH | Amidation | Enhance pharmacokinetics and folate for cancer cell recognition | ||
| CH3I | Methylation | Solubility | 1H NMR, SEC | [152] |
| Histidine | Amidation | Proton sponge effect | ||
| Folate–PEG–COOH | Amidation | Enhance pharmacokinetics and folate for cancer cell recognition | ||
| Galactosylated lactobionic acid | Amidation | Target liver cell | 1H NMR, IR | [153] |
| Dextran-aldehyde | Reductive N-alkylation | Enhance complexes stability | ||
| Galactosylated lactobionic acid | Amidation | Target liver cell | 1H NMR, SEC | [155] |
| PEG-succinimidyl ester | Amidation | Enhance pharmacokinetic | ||
| Galactosylated lactobionic acid | Amidation | Target liver cell | IR, SEC | [162] |
| Urocanic acid | Amidation | Endosmal escape | 1H NMR | [165] |
| Imidazole-4-acetic acid | Amidation | Endosmal escape | IR, 1H NMR | [166] |
| PEG-succinimidyl ester | Amidation | Solubilization | IR, 1H NMR, SEC | [156] |
| Texas Red Succinimidyl Ester | Amidation | Fluorescent chitosan | 1H NMR | [185] |
| 9-Anthraldehyde | Reductive N-alkylation | Fluorescent chitosan | 1H NMR | [186] |
| N-acetyl chitosan oligosaccharide aldehyde terminated | Reductive N-alkylation | Colloidal stability | 1H NMR, SEC | [13], [27], [104], [150], [160], [161] |
| Phosphorylcholine-glyceraldehyde | Reductive N-alkylation | Solubility | 1H NMR, SEC | [147], [148] |
| Sulforhodamine 101 | Sulfonamide | Fluorescent chitosan | [11] | |
| Rhodamine isothiocyanate | Thiourethane | Fluorescent chitosan | SEC | [188] |
| PEI | Reductive N-alkylation to oxidized chitosan | Reduce cytotoxicity, increase transfection efficiency | 1H NMR, SEC | [167] |
| PEI | Reductive N-alkylation to oxidized chitosan | Reduce cytotoxicity, increase transfection efficiency | 1H NMR, SEC | [163] |
| Galactosylated lactobionic acid | Amidation | Target liver cell | ||
| PEI | Reductive N-alkylation to oxidized chitosan | Reduce cytotoxicity, increase transfection efficiency | 1H NMR, SEC | [157] |
| Galactosylated-PEG-acid | Amidation | Target liver cell | ||
| PEI | Polymerization on Amine group | Reduce cytotoxicity, increase transfection efficiency | 13C NMR, SEC | [168] |
| PEI | Amidation on N-Maleated chitosan | Reduce cytotoxicity, increase transfection efficiency | 1H NMR, SEC | [169] |
| PEI | Amidation on N-Succinylated chitosan | Reduce cytotoxicity, increase transfection efficiency | IR, 1H NMR, SEC | [170] |
| 2-Iminothiolane and then thiolated histidine dendron | Disulfide bond | Endosomal escape | 1H NMR | [171] |
Table 6.
Modifications on the alcohol functional group of the C6 of chitosan and on both the amine and alcohol functional groups.
| Compound graft | Type of modification | Objective | Analysis | Ref |
|---|---|---|---|---|
| Modification of alcohol on C6 to an amine | Amination | Colloidal stability | IR, 13C NMR | [158], [159] |
| PEG–PEI | Addition to 6-iodide-6-deoxy-chitosan | Endosomal escape | IR, 1H NMR, | [172] |
| ClCH2COOH (glyoxylic acid) | O,N and N-Carboxymethylation of chitosan | Solubility | IR, 1H 13C NMR | [146] |
| Galactolysation | Reductive N and O-Alkylation to 6-amino-6-deoxy-chitosan | Target liver cell | 1H NMR, SEC | [164] |
Fig. 3.
Modifications on the amine functional group of chitosan (see Table 5). The scheme summarized all modifications done on the amine function of the chitosan.
Fig. 4.
Modifications on the C6 alcohol functional group of chitosan and on both amine and C6 alcohol (see Table 2). The scheme summarized all modifications done on the C6 alcohol or on the amine and the C6 alcohol function of the chitosan.
2.3.1. Modification on the C2 amine
The principal drawback in the use of chitosan's amine functional groups for modification is that these groups when ionized are the basis for electrostatic complexation with the anionic phosphate groups of NAs. Modifications may generate steric hindrance and decrease the number of ionizable amines that bind to NAs. The nonbonding pair of electrons on the primary amine of the chitosan is nonetheless a good candidate for nucleophilic attack and allows several reactions (Table 5 and Fig. 3).
2.3.1.1. Quaternization
Quaternization of chitosan has been performed to improve solubility at physiological pH. Two methods of quaternization have been performed: N-trialkylation using halogenoalkanes (usually CH3I) [176], [177], [178], [179], [180] and a reaction with a quaternized epoxide [181], [182], [183]. For modification with halogenoalkanes, precise characterization is required since Sieval et al. [176] demonstrated that, depending on the method of synthesis, N-dimethylalkylation can be obtained and Snyman et al. [178] also observed O-alkylation on the C6 hydroxyl.
2.3.1.2. Amidation
Amidation is one of the most common techniques to graft molecules and polymers to the amine of chitosan. It can be achieved using carboxylic compounds with carbodiimides such as EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) or DCC (dicyclohexyl carbodiimide) coupled with NHS (N-hydroxysulfosuccinimide) to accelerate the reaction [184]. Several species have been grafted including amino acids (arginine [152] or histidine [154], galactosylated lactobionic acid [153], [155], [162], [163], imidazole derivated acids [165], [166] and PEG [152], [154], [157]. Amidation can also be carried out with succinimidyl ester moieties which directly react with the primary amine of chitosan. PEG is often grafted by this method [114], [151], [156] as are some fluorescent probes like Texas Red [185]. Characterization should be rigorous to clearly demonstrate that the amide bond is formed by IR as in Darras et al. [174] since the carboxylate form of the species can bind electrostatically with the protonated amines of chitosan.
2.3.1.3. Reductive N-alkylation
Reductive N-alkylation is a method to graft an aldehyde derivative on the amine of chitosan. First, a Schiff base is formed between the amine and the aldehyde and then the resulting imine is reduced by the addition of a reducing agent (NaCNBH3 or NaBH4). Grafted species include fluorescent probes (as 9-anthraldehyde [186]), phosphorylcholine [147], [148], oligosaccharides (dimer from chitosan [13], [27], [104], [149], [150], [160] or dimer to hexamer [161]) and polymers such as dextran [153]. Reductive N-alkylation was also performed on an oxidized chitosan. First chitosan is oxidized by KIO4 and the resulting product with modified D-units where the sugar ring is opened between C2 and C3 and two aldehydes are formed on C2 and C3. The two aldehydes then react with the primary amine of PEI [157], [163], [167], [185]. The drawback of this method is the oxidizing step where Vold et al. [187] observed reduction of the molar mass of chitosan as the concentration of aldehyde increases in the chitosan. Accurate measurements of molar mass are required.
2.3.1.4. Sulfonamidation, thiurea, polymerization and sulfur bonds
Additional methods have been used to modify chitosan. Grafting of fluorescent probes was achieved by Ishii et al. [11] with Sulforhodamine 101 grafted to chitosan whereas Ma et al. [188] grafted Rhodamine B thioisocyanate. To bind PEI to chitosan, Wong et al. [168] directly polymerized PEI on the amine of chitosan via cationic ring opening polymerization of azaridine in acidic conditions. Lu et al. attached PEI to chitosan by first transforming chitosan via a maleic anhydride [169] or succinic anhydride [170] and then adding PEI to the modified polysaccharide. More recently, Chang et al. [171] succeeded in grafting a histidine dendrimer to chitosan via a disulfide bond. Finally, Morris et al. [152], [154] trimethylated chitosan, then grafted an amino acid and added galactosylated PEG.
2.3.2. Modification on the C6 hydroxyl
The C6 hydroxyl group can also be modified but, due to its high reactivity, the amine groups must be first protected (Table 6 and Fig. 4). For chitosan, amine protection is a challenge since this group needs to be protonated for solubility. The most commonly employed protecting group is the phthalic anhydride [189], [190]. More recently, Cai et al. [191] protected the amine by electrostatic interaction with sodium dodecylsulfate (SDS) but the subsequent steps then need to be in organic media otherwise the protection may be compromised. After protection, several modifications can be carried out: 6-amination [158], [159] to increase positive charge available for complexes formation with NAs, 6-amination followed by galactosylation [164] or 6-amination and PEI–PEG addition. Lastly, Rinaudo et al. [146] synthesized O and N-carboxylmethylated chitosan by reaction with chloroacetic acid.
All of the above modified polymers have been characterized and authors have proven by direct or indirect methods that chitosans were modified. In most studies, cytotoxicity and transfection efficiency were studied. In most cases, modifications did not affect cell viability whereas modifications can have more impact on size of the complexes and the transfection efficiency as will be detailed in subsequent sections. In the case of chitosans modified to increase proton sponge capacity, effects of adding proton sponge species will also be discussed below.
3. Preparation and physicochemical assessment of chitosan–polynucleotide nanoparticles
3.1. Polyelectrolyte complex overview
As for most polycations used to complex and deliver NA, chitosan–NA particles are generally prepared by fast addition and mixing of chitosan to NA [270], [66], [192], [193] solutions under dilute conditions (~ 0.1 mg/ml or below). Chitosan–NA particles prepared in this manner constitute a particular type of polyelectrolyte complexes (PECs). Prior to treating the details of preparation and properties of chitosan–NA PECs, it is useful to summarize the general properties of PECS and their associated characterization methods (Table 7 ).
Table 7.
Characterization techniques of chitosan–nucleotide particles.
| Parameter | Method |
|---|---|
| Size | Dynamic light scattering (DLS) |
| Atomic force microscopy | |
| Nanoparticle tracking analysis (NTA) [252] | |
| Electronic microscopy (EM) | |
| Asymmetrical flow field-flow fractionation (AF4) coupled with LS [229] | |
| MW | Static light scattering (SLS) |
| NTA (if stoiochiometry is known) | |
| Stoichiometry | Orange II dye depletion assay [425] |
| AF4 | |
| Binding affinity | Isothermal titration calorimetry (ITC) [256] |
| Stability | Polyanion competition assay |
| EtBr displacement assay [28] | |
| Gel retardation assay | |
| ITC | |
| Colloidal stability | DLS vs time |
| Surface charge | Laser Doppler velocimetry |
| Concentration | NTA |
| Morphology | AFM, EM |
| Protection against endonuclease | Agarose gel electrophoresis |
| Buffering capacity | Potentiometric Titration |
Mixing of oppositely charged polyelectrolytes (PELs) results in the spontaneous formation of polyelectrolyte complexes (PECs), via a process mainly driven by strong electrostatic attractions [194]. Non-viral gene delivery mainly relies on the use of cationic species (polycations or cationic lipids) that form complexes with NA (DNA, RNA), referred to respectively as polyplexes or lipoplexes. These systems have been the object of intense research since their first use to deliver DNA in 1987 [195], [196]. Depending on the nature of the PEL and on the mixing conditions such as media composition, mixing stoichiometry and PEL concentrations, water-soluble on a molecular level [194], [195] or colloidal PECs (i.e. comprising many chains of each species in a particle) [199] may be formed in dilute conditions (~≤ 0.1 mg/ml [200], [201]). The limiting cases for the resulting structures of soluble and colloidal polyelectrolyte complexes are modeled as ladder-like structures where complexes are formed on a molecular scale by conformational adaptation and scrambled-egg structures that contain a large number of PELs per particle, respectively [199], [202]. The production of soluble PECs requires special preparation conditions that were identified in the pioneering work of Kabanov [197] and Tsuchida [203]. Soluble PECs form when polyions with significantly different molecular weights bearing weak ionic groups are mixed at non-stoichiometric ratios in the presence of a minimal amount of salt. In most practical applications, colloidal PECs form in dilute conditions and to the best of our knowledge, the formation of soluble PECs between chitosan and NA hasn't been reported in the literature. Studies of colloidal PEC formation indicate that these PECs are mainly spherical and consist of a neutral stoichiometric core surrounded by a charged stabilizing shell composed of the excess polyion [199], [201], [204]. However, polyplexes formed between a polycation and a plasmid often consist of a mixture of globular, toroidal and rod-like structure [205].
The properties of PECs are influenced by several parameters such as PEL structural properties of length and charge spacing [201], mixing regime [206], [207], molar mixing ratio [201], medium pH [204] (i.e. polymer ionization) and ionic strength [200], [206]. Salt screening of electrostatic interactions in solution enables rearrangement processes [202]; it has been shown in some cases that a small amount of salt can significantly reduce the aggregation numbers of PECs (i.e. the number of chains of each species comprised in a particle), constituting a means to control size and mass of PECs [200] aside from varying PEL concentration. Reducing the ionization degree of the PEL by changing pH reduces the binding affinity and influences the structure of PECs [28], [204].
It is worth mentioning that in order to be able to elucidate the influence of mixing conditions and PEL characteristics on PEC properties, the mixing process must be performed using very dilute solutions. In many practical cases where mixing is not performed in a very dilute regime, PEC structure formation is mostly determined by the fast kinetics of the association process masking the influence of the other parameters such as polyelectrolyte structure, ionization degree and ionic strength of the solutions [202]. Some contradictory results reported in the literature regarding the influence of formulation parameters like molecular weight of chitosan and N:P ratio on particles size could possibly be explained by differences in the mixing concentrations, an aspect that will be further discussed in Section 3.4.1.
3.2. Preparation techniques for chitosan nanoparticles
Apart from direct mixing of a chitosan solution with a NA solution [26], [27], [66], [193], [208], [209], [210] (sometimes referred to as complex coacervation method), other preparation methods of chitosan-based nanoparticles for the delivery of NA have been proposed in the literature. Among these methods are: i) an ionic gelation method that relies on the inclusion of a small polyion such as tripolyphospate (TPP) in the NA solution that acts as an ionic cross-linker upon mixing with chitosan [211], [212], [213]. Note that some authors [211], [214] have mixed preformed chitosan/TPP particles with NAs that are adsorbed to the surface of particles, but this method raises concerns regarding NA protection against degradation [213]. ii) The inclusion of chitosan in the matrix of nanoparticles such as poly (d,l-lactide-co-glycolide) (PLGA) nanoparticles [215], [216] to bind NAs by virtue of their positive charge. iii) The use of self-aggregating hydrophobically modified chitosans forming nanoparticles that can adsorb NA [217], [218].
Each of the above methods has its pros and cons and produces particles that have different physico-chemical properties, but there is no clear indication to date that any one of these techniques produces chitosan-based nanoparticles that are superior in terms of their delivery efficiency or transfection efficiency. The direct mixing of a chitosan solution with a NA solution is the simplest method and is most often used to produce chitosan-based nanoparticles for the delivery of NAs; the following will focus mainly on this preparation method. It is worth mentioning that most of the systems using modified chitosans are also prepared by simple mixing of the two oppositely charged polyelectrolyte solutions.
Even for the apparently simple particle preparation method such as the direct mixing of chitosan and a NA solutions, several different procedures have been proposed in the literature where the following parameters have been varied: chitosan and NA have been mixed in the presence [66], [219] or absence [27], [208] of salt or buffer; in a very dilute regime [27], [220] or concentrated regime [210], [221], [222], [223]; mixing has been performed mainly at room temperature although sometimes at 50–55 °C [66], [166], [224]; equal volumes of NA and chitosan solutions [169], [192], [225] or unequal volumes [208], [210], [219], [221] have been mixed; most authors report rapid mixing of the solutions, while some have added one solution to the other in a dropwise fashion [12], [211], [226], [227]. Note that an excess of chitosan has been used to confer a positive charge to the polyplex in all formulations that have successfully transfected cells in vitro, whatever the mixing conditions used. Among these numerous preparation methods, a method including sodium sulfate in DNA solution where mixing is performed at 50–55 °C based on the method proposed by Leong et al. 1998 [224] appears frequently. There is however little evidence in the literature that heating the solutions and including sodium sulfate in the DNA solution results in polyplexes having specific physico-chemical properties conferring superior transfection efficiency. The use of many different mixing conditions listed above where ionic strength, pH, temperature, buffer, method of mixing and mixing concentration are varied renders difficult comparison and reconciliation of the sometimes contradictory reported influences of formulation parameters like molecular weight of chitosan and N:P ratio on polyplex physicochemical and structural properties such as size and morphology. There are also a significant number of papers presenting an incomplete description of the polyplex preparation method, a situation that limits or simply precludes comparison with other studies as well. There is however one mixing parameter that has a clear influence on the resulting polyplexes properties, whatever the preparation conditions used, namely the mixing concentration of NA where size of polyplexes increase as the concentration of NA concentration increases, discussed further below.
Most studies of chitosan–NA polyplexes focus on their biological activities with few dedicated to a detailed analysis of the actual formation of polyplex and the influence of chitosan structural parameters on their properties [28], [225], [228], [229], [230]. To the best of our knowledge, no systematic study of chitosan–NA polyplex physico-chemical properties (size, mass, density, stoichiometry, morphology) as a function of mixing parameters (concentration, ionic strength and pH of medium, N:P ratio and mixing regime) has yet been reported in the literature. This situation is not specific to chitosan as it is recognized that a finer characterization of polyplexes and a more complete theoretical underpinning of their mechanisms of formation are greatly needed in general.
3.3. Current limitations of preparation techniques of chitosan polynucleotide particles
Apart from the requirement for a better characterization and understanding of the influence of mixing parameters on resulting chitosan–nucleotide particles, there are other shortcomings associated with the current preparation of chitosan–nucleotide particles that must be overcome to generate pre-clinical data that can translate to clinical application: i) chitosan particles are currently prepared by manual mixing of small volumes of chitosan and NA solutions and then used within a few hours, raising concerns about process reproducibility and seriously limiting the quantity and applicability of these products and ii) the mixing process must be performed in dilute conditions in order to obtain a stable colloidal suspension of small and relatively homogeneous nanoparticles — another drawback that limits the deliverable dose for these systems and that requires the development of concentration methods post-mixing. Note that this shortcoming is also not specific to chitosan-based systems as it is generally recognized that preparations of non-viral gene delivery systems necessitate dilute conditions of preparation near or below 0.1 mg/mL of NA to obtain small and relatively homogeneous nanoparticles [231]. To the best of our knowledge, these shortcomings for chitosan-based systems have not been directly addressed in the literature. However, there are clear avenues for progress towards solutions that have been developed in studies for other systems. Adapting these approaches for chitosan–NA polyplexes would be beneficial to the field of chitosan-based gene delivery systems.
3.3.1. In-line mixing for the production of scaled-up homogenous chitosan–NA polyplexes
The kinetics of polyplex formation is fast (formation time < 5 μs has been reported [232]) and a rapid addition of a polycation solution to an NA solution or the reverse has practical limitations regarding scalability, reproducibility and homogeneity. In order to overcome these limitations, mixing techniques or devices have been proposed in the literature to prepare large volumes of polyplexes [233], [234]. These techniques rely on in-line mixing of the two PEL solutions via a T connector [233] or in a static mixer coupled to a dual syringe [234]. By varying mixing speeds, Kasper et al. [233] showed that small and homogeneous PEI [235]–pDNA polyplexes could be prepared and that size and polydispersity determined by dynamic light scattering (DLS) increased at higher pDNA concentration. Davies et al. [234] also obtained small and homogeneous PEI–pDNA polyplexes using a static mixer. PECs of two synthetic polymers have also been prepared by jet-mixing [236] where smaller PECs were obtained as compared to those prepared by polyelectrolyte titration (i.e. slow addition of a polyelectrolyte solution to the other).
3.3.2. Concentration of particles using freeze-drying and tangential flow filtration
One means to increase concentration of polyplexes is freeze-drying followed by rehydration using reduced volumes [231]. Successful freeze-drying of polyplexes has been reported in a limited number of studies, with [237] or without [233], [238], [239] concentration at rehydration. A certain amount of excipient is required for freeze-drying and concentration can therefore be limited by isotonicity requirements. Freeze-drying of chitosan–NA polyplexes have been reported in the literature [66], [240] but no concentration at rehydration has been reported in these studies. The use of vacuum centrifugation has also been reported as a means to concentrate chitosan–NA [89], [220].
Tangential flow filtration (TFF) can also be used to concentrate chitosan–nucleotide particles. TFF overcomes the main limitations of direct flow filtration, such as rapid flux decrease over time and is particularly indicated for applications where concentration of a product is required or when diafiltration is required for buffer exchange or desalting [241]. As compared to concentration upon rehydration after freeze-drying, TFF offers the advantage of not concentrating small solutes so that isotonicity requirement can be theoretically easily fulfilled. TFF has been used for nanoparticle purification [242], [243] and concentration purposes [244] or both [245].
3.4. Physico-chemical properties and characterization methods of chitosan particles
As pointed out above, a fine and complete characterization of chitosan–NA polyplexes is greatly needed to better understand their mechanism of formation as well as the influence of their properties on transfection efficiency. Chitosan–NA particles have been characterized for their physico-chemical properties to various extents in many studies since their first use for delivery of NA was reported by Mumper et al. in 1995 [246]. An overview of the reported physico-chemical properties of chitosan–nucleotide particles and the influence of formulation parameters on these properties as well as the characterization techniques is given below. Particular differences between plasmid DNA and siRNA are discussed. Table 1 provides a list of techniques that have been used or that could be used to characterize chitosan–NA particles.
3.4.1. Size
Generally, it is believed that smaller particles are more efficient at transfecting cells [247], [248]. Small particles are also required to reach various organs (e.g., particles below ~ 100 nm are required to cross liver fenestra and reach the hepatocytes). In addition to the surface properties of particles, their size might also determine the composition of protein corona that surrounds the particle and this could be a determinant in particle biological fate [249]. The pK a of chitosan is about 6.5 [8] and the polymer is therefore only weakly charged and barely soluble at neutral pH or above. Chitosan–NA polyplex surface charge as well as colloidal stability is therefore reduced at physiological pH and the particles are prone to aggregation under such conditions. It is therefore important to develop methods to control the size and mass of colloidal PECs at the time of their formation and to limit their aggregation once placed in protein containing physiological solutions, especially for their systemic delivery via intravenous injection.
Typically, in any study reporting the use of chitosan for gene delivery applications, chitosan–nucleotide particles are minimally characterized for their size by dynamic light scattering (DLS). DLS analysis of chitosan–NA polyplexes typically reveals relatively polydisperse nanoparticles. Size values (usually z-average diameter) reported in the literature for chitosan–NA polyplexes vary from a few tens of nm [27], [160] to a few hundred nm [66], [210], [250], depending on the preparation method (concentration, pH, volume mixed, etc.), the properties of chitosan and NA and the suspension medium used for analysis. Note that diameters as high as 5 to 8 μm have also been reported [11], but particles in these cases were analyzed in conditions where they were colloidally unstable and were undergoing aggregation. Size has also been measured by electron microscopy and atomic force microscopy. Size reported by microscopy is typically smaller than values obtained by DLS for polydisperse particles since size distribution measured in DLS is weighted to the scattered light intensity which is proportional to the square of molecular mass. Some authors have however attributed increased size up to 100% in DLS vs TEM or SEM as due to the fact that particles are fully hydrated when characterized by DLS [251] or that they aren't spherical [227]. Empirically, volume-weighted size distribution in DLS and size measured directly by microscopy are close to each other but conversion of DLS diameters from intensity (volume squared)- to volume- or number-weighted distributions must be used with great care since accurate values of the refractive index of particles is required and this parameter is not precisely known for chitosan–NA particles. Another technique available recently, Nanoparticle Tracking Analysis (NTA), is based on tracking Brownian motion of single particles that overcomes the sensitivity bias toward larger particles of DLS due the intensity weighted DLS measurement (see Filipe et al. [252] for evaluation and comparison of the technique with DLS). The use of NTA has not yet been reported for chitosan particles but this technique would complement DLS by providing a number-weighted distribution (i.e. that measured in microscopy) in addition to being able to estimate concentration of particles.
3.4.1.1. Influence of mixing concentration
Probably the most important experimental parameter to control size at the time of polyplex formation is the concentration of polyelectrolyte solutions to be mixed (NA). This parameter is however often overlooked in comparison to other formulation parameters such as structural parameters of chitosan or the N:P ratio. Comparison of the results of two studies where mixing concentration differ by a factor of more than 1000 provides a clear illustration of concentration influence on size of particles: Malmo et al. [27] prepared chitosan–siRNA particles using NA concentration of 1.2 μg/ml and report sizes ranging from ~ 50 to 60 nm at N:P = 30, while Nielsen et al. [221] prepared particles using a siRNA concentration of ~ 3.5 mg/ml and obtained polydisperse particles with much larger size of about 300 nm at N:P = 23. Other studies have also reported an increase of polyplexes size with increasing NA mixing concentration [208], [225].
3.4.1.2. Influence of chitosan structural parameters
Many studies report a chitosan–pDNA polyplex size increase as chitosan molecular weight increases. Mumper et al. [246] and MacLaughlin et al. [208] both report a significant increase of polyplex size with increasing molecular weight of chitosan. They found that size increases from about 100 nm to 500 nm when chitosan's molecular weight passes from a few kDa to ~ 500 kDa. These changes in size were attributed to reduced solubility and mobility of longer chitosan chains. Koping-Hoggard et al. [89] observed a size increase from 68 to 174 nm when chitosan molecular weight increases from ~ 2 to 7 kDa for chitosan–pDNA polyplexes prepared using an N:P ratio of 60. Much less marked size increase with chitosan molecular weight has been reported [160], [193], [220], [229]. On the other hand, some studies report a decrease in size of polyplex with increased chitosan molecular weight [219] or no significant influence of this parameter on polyplexes size [225]. Although the exact reason for these contradictory results remains unclear, they could possibly be attributed to differences in the polyplex preparation method. For instance, in the Huang et al. study [219], polyplexes were prepared in a medium with relatively high ionic strength (i.e. HBSS-MES buffer at pH 6.2) as compared to most other studies where polyplexes were prepared in water or in presence of a limited amount of salt. It has been reported elsewhere that mixing of pDNA and low molecular weight chitosans in the presence of a buffer with moderate ionic strength (~ 20–25 mM) results in the formation of large aggregated structures [253] or particles with reduced colloidal stability as compared to those prepared with high molecular weight chitosan [28]. It appears that the explanation for the apparent contradictory results regarding the influence of chitosan molecular weight on polyplex size could lie in the reduced colloidal stability of polyplex prepared with low molecular weight chitosan that are stable in water but aggregate in the presence of salt. We have also observed this phenomenon and attributed this size increase to the potential inability of low molecular weight chitosans to fully condense plasmid DNA [26]. However, in this study, polyplexes were suspended in PBS and it is most likely that the presence of salt and close to neutral pH of the resuspension medium favored particle aggregation. To the best of our knowledge, no explanation of the influence of chitosan molar mass on polyplex colloidal stability has been proposed in the literature. It could be that high molecular weight chitosans have dangling ends at the polyplex surface that sterically stabilize the particles, a phenomenon that is not observed for polyplex formed with shorter chains that lack protruding ends.
Only a few studies report the influence of chitosan's molecular weight on size of chitosan–siRNA polyplexes. Malmo et al. [27] and Holzerny et al. [226] both report a slight increase of chitosan–siRNA size with increasing chitosan molecular weight while Mittnacht et al. [254] report the reverse. On the other hand, Liu et al. [255] observed no influence of chitosan molecular weight on polyplex size, except for a 9 kDa chitosan that formed particles of a few microns. It is worth mentioning that particles in this case were prepared in a medium having a significant ionic strength and that probably limit polyplex colloidal stability for this 9 kDa chitosan, as discussed previously. It should also be pointed out that the polyplex preparation conditions used in each study reporting the influence of chitosan's molar mass on size of chitosan–siRNA polyplexes are significantly different and could explain some contradictory results regarding the influence of chitosan's molecular weight on size of chitosan–siRNA polyplexes.
A few studies report the influence of chitosan DDA on polyplex size. Size measurements by DLS revealed that chitosan–pDNA polyplex size slightly increases as DDA decreases [28], [219], [220], [225]. This has been attributed to weaker binding of chitosan to NA and to stiffening of the polymer chain as DDA decreases [225]. Another explanation is that the actual amount of chitosan chains bound in polyplexes increases as the DDA decreases to achieve a given N:P ratio in the polyplexes [28]. This is also consistent with findings of Ma et al. [256], according to which polyplexes charge stoichiometry (i.e. N:P ratio within the polyplex) is independent of DDA. It should be noted that these results were obtained for polyplexes prepared by dropwise addition of chitosan to pDNA, but it is reasonable to assume that similar results would be observed using a flash mixing preparation method.
3.4.1.3. Influence of N:P ratio
The influence of N:P ratio on polyplex size has been examined in numerous studies [14], [27], [66], [150], [193], [208], [225]. In most cases, an increase of the relative excess of chitosan vs NA results in the formation of polyplexes with slightly larger sizes [27], [150], [193], [208]. Mao et al. [66] reported no significant influence of N:P ratio on size but they tested a somewhat limited range of N:P ratios as compared to those tested in other studies. At least two studies report a decrease of polyplex size with N:P ratio [225] and observed this decrease in size by comparing a N:P ratio of 0.5 to a N:P ratio of 5. The pDNA is probably not completely complexed and compacted by chitosan for a N:P ratio below 1 which could explain these results. Howard et al. [14] observed a significant decrease in polyplex size with an ~ 10 × increase in N:P but this result is most probably due to a concentration effect since the mixing chitosan concentration was held constant and NA concentration was therefore about ten times lower for the highest N:P ratio.
3.4.1.4. DNA vs siRNA
siRNA are a few hundred times smaller than pDNA and can theoretically form smaller polyplexes. This is experimentally verified as long as polyplex sizes are compared for similar preparation conditions, more specifically if NA mixing concentrations are comparable. For instance, the hydrodynamic diameter of chitosan–siRNA prepared in a diluted regime using a chitosan of 16 kDa at N:P 10 was found to be 45 nm [27] while it was 70 nm [160] for chitosan–pDNA polyplexes prepared in similar conditions (chitosan of 14 kDa and N:P of 5). The mixing method also influences size and a dropwise addition of chitosan to a diluted siRNA solution led to the formation of larger polyplexes [211], most probably because aggregation is favored by a gradual traversing of the polyplex neutralization point (i.e. when the N:P ratio is 1). It is worth mentioning that larger chitosan–siRNA polyplexes with hydrodynamic diameter of a few hundred nanometers have been reported in many studies where particles were prepared using relatively concentrated NA solutions and/or where mixing was performed at either high pH or high ionic strength.
3.4.1.5. Overview of the influence of chitosan modifications
As discussed earlier in Section 2.3, there is an increasing number of publications reporting the use of modified chitosans for gene delivery applications. Since these modifications differ in nature, they are expected to have a variable influence on polyplex size at formation. Among the few papers where the size of polyplexes prepared with unmodified and modified chitosans was compared, particular modifications were reported to increase [257], decrease [13] or not significantly alter polyplex size [258]. Actually, many of these modifications were proposed as means to increase solubility of chitosan as well as polyplex colloidal stability and have a clearly defined influence on particle size and stability in physiological media. This particular topic is discussed in more detail in Section 3.4.7 below.
3.4.2. Zeta potential
High surface charge density of chitosan–NA polyplexes determines the colloidal stability of non-sterically stabilized formulations and correlates with higher cell uptake and transfection efficiency in vitro [150], [222], [259], [260]. Zeta potential provides an indirect measurement of particle surface charge density and has been measured by laser Doppler velocimetry in numerous studies. Values reported for polyplexes prepared using an excess of chitosan vs NA range from a few millivolts to a few tens of mV, depending mostly on pH and ionic strength of measurement media as well as on N:P ratio and to a lesser extent on structural parameters of chitosan. As discussed previously, since the pK a of chitosan is about 6.5, zeta potential is strongly influenced by pH changes near this value. For example, reduction of zeta potential from about 20 to 0 mV when pH passes from ~ 6 to 7 has been reported [66], [222]. Zeta potential typically rapidly increases from 0 to a few tens of mV when N:P passes from 1 to ~ 2 [28] and only slightly increases as N:P is increased above ~ 2 [28], [66], [150], the latter reflecting the mostly constant polyplex stoichiometry as N:P is increased above ~ 2. Slight zeta potential increases with increasing chitosan molar mass [28], [193], [255] or DDA [28], [255] have been reported, influences that are expected at least for DDA since it determines chitosan charge density. Chitosan modifications often rely on reaction with chitosan amino groups and are therefore expected to reduce zeta potential, unless substituents are also positively charged (e.g. PEI). One should expect a more marked decrease for bulky hydrophilic substituents such as PEG since they should move the slip plane further away from the polyplex surface. Finally, the nature of the NA (pDNA or siRNA) hasn't been reported to significantly change the effects of formulation parameters or medium properties on zeta potential discussed above.
3.4.3. Morphology
The morphology of chitosan–pDNA complexes have been examined by electron microscopy and/or by atomic force microscopy in numerous studies. In most cases, a mixture of toroids, rod-like and globular particles have been observed [205], [208], [220], [261] but some authors have reported mostly spherical particles [66], [219], [262]. The differences in the morphology reported in these studies could be related to the use of different mixing conditions and/or to the use of chitosans with different structural properties. Chain stiffness is theoretically predicted to determine the morphology of collapsed structures [263] and chitosans with different structural properties could result in the formation of polyplexes with different morphology by modifying DNA chain stiffness to various extents upon binding [205]. It seems that the inclusion in the polyplex mixing solutions of another polyanion such as tripolyphosphate (TPP) [213], and hyaluronic acid (HA) [264] favors formation of chitosan–pDNA polyplexes with a more homogeneous and mostly spherical morphology. The use of modified chitosans has also been reported to favor formation of spherical chitosan–pDNA polyplexes [265], [266], [267].
Studies where chitosan–siRNA polyplex morphology was examined report mostly spherical particles [14], [254] but the presence of an increasing proportion of rod-like particles for chitosan with higher molecular weight has been reported [226]. Theoretically, the formation of toroidal structures requires NA chain length significantly higher than ~ 21 bp [268] and this would explain the absence of this type of structure for chitosan–siRNA polyplexes. As for pDNA polyplexes, the inclusion of another polyanion [269] or the use of modified chitosans [[214], [267], [270]] appears to result in the formation of spherical particles.
3.4.4. Binding affinity/stability
The binding affinity of chitosan for NA determines chitosan's ability to protect the NA as well as polyplex stability, which in turn greatly determines polyplex transfection efficiency [26], [89], [160]. The gel retardation assay is a means to qualitatively assess the ability of chitosan to complex pDNA and the polyplex stability that has been reported in many studies. Polyplexes have been incubated in various conditions (e.g. in presence of competing polyanion such as heparin, at various pH, in transfection medium, etc.) prior to being submitted to this electrophoresis-based assay to monitor DNA dissociation from the polyplex. This type of experiment reveals that polyplexes formed with chitosan with low DDA [220], low Mn [89], [193] or low N:P [89], [193] are more easily dissociated and therefore less stable. Koping-Hoggard et al. showed that formulations that are very stable (i.e. high Mn chitosan not dissociated by heparin) are much less efficient at transfecting cells than their less stable counterparts [89]. The ethidium bromide displacement assay is another technique that has been used to analyze somewhat more quantitatively the chitosan–pDNA polyplex stability [28]. In addition to the influence of N:P and chitosan structural properties discussed above, polyplexes were found to be less stable at high pH and high salt content. Gel retardation and ethidium bromide displacement assays only qualitatively or indirectly assess the chitosan to DNA binding affinity. In contrast, isothermal titration calorimetry (ITC) combined with the single set of identical sites model (SSIS) can quantify binding constants of chitosans to pDNA [256]. Depending on pH, ionic strength and chitosan structure properties, binding constants ranging from ~ 1 to 15 nM− 1 were found. In agreement with results discussed above, the binding constant was found to increase as pH or ionic strength decrease and as chitosan DDA or molar mass increase.
Similar ITC experiments were performed to determine chitosan to siRNA binding affinity [226]. siRNA was added to chitosan solution at pH 5 in low salt conditions and binding constants ten times lower than those found for pDNA in equivalent conditions were determined (~ 1–2 nM), without any significant influence of chitosan molar mass. This relative insensitivity of binding affinity towards chitosan molar mass contrasts with results obtained for pDNA. This could be explained by the fact that the guest–host roles are reversed for siRNA, the NA acting here as a guest so that DDA is the only chitosan property expected to influence binding affinity. This lower binding affinity of chitosan to siRNA and its relative insensitivity to molar mass have led to the use of very large N:P ratios in a few studies where formulations were selected on the basis of their stability evaluated through the gel retardation assay [14], [210], [255], [271]. However, most of these gel retardation assays were performed in a buffer at a higher than physiological pH (~ 8), a condition where chitosan is barely charged and polyplexes are therefore easily dissociated particularly with small siRNA. Reduction of the pH in gel retardation assay revealed much more stable polyplexes and successful silencing using much lower N:P ratios [27], [226], [272].
Finally, since most chitosan modifications create steric hindrance and reduce chitosan charge density as they usually rely on reactions with the amino groups, they are expected to reduce chitosan binding affinity to NA. The resulting reduction in stability was revealed by the gel retardation assay in a few studies ([13], [150], [267]. As expected, this reduction was not observed for the case where a positively charged substituent, namely PEI, was used [267]).
3.4.5. Stoiochiometry
As discussed above, chitosan–NA polyplexes formulated for in vitro transfection are prepared using an excess of chitosan vs NA (i.e. N:P ratio above one). Using asymmetrical flow field-flow fractionation (AF4, see these references [273], [274] detailed discussion of AF4 theory) coupled with UV spectroscopy and light scattering, it was determined that a significant proportion of chitosan is unbound (~ 75%) in a polyplex dispersion formed using an N:P mixing ratio of 5 [230]. This AF4 technique offers the advantage of providing three important physicochemical properties of polyplex, namely, 1) the proportion of free chitosan 2) the polyplex hydrodynamic size and 3) their size distribution. In a second study, the influence of N:P mixing ratio was examined AF4 [229]. It was shown that the polyplex stoichiometry (i.e. N:P ratio within the polyplex) ranges from ~ 1.3 to 1.6 (or equivalently, that free chitosan ranges from ~ 53 to 92%) for polyplex prepared using an N:P mixing ratio ranging from 3 to 15 [229]. Orange II dye depletion method constitutes a relatively simple means to evaluate the proportion of unbound chitosan in polyplex suspensions and confirmed AF4 results [229]. It is worth mentioning that since the calculation of the amount of bound chitosan relies on the determination of free chitosan (for both AF4 and Orange II methods), the accuracy of this calculation decreases somewhat as the mixing N:P ratio increases.
The presence of unbound chitosan is required to efficiently transfect cells in vitro, a topic that will be discussed in more detail in Section 4.3.2. However, despite this requirement for a significant fraction of unbound polycation in chitosan–NA polyplex formulations and even if chitosan is recognized as non-toxic, the use of very high N:P values reported in a few studies raises concerns about possible side effects or off-target effects of these formulations that consist mostly of unbound chitosan.
3.4.6. Buffering capacity
It has been proposed that PEI transfection efficiency lies in its high buffering capacity in the endosomal pH range, ~ 4.5 to ~ 7, that can mediate escape from the endosome by the proton-sponge effect [10]. In this proposed endo-lysosomal escape mechanism, the acidification of the endosome or lysosome by the additional pumping of protons into the endosome, along with the concurrent influx of chloride ions to maintain charge neutrality causes an increase in osmotic pressure and the vesicle is ruptured by direct membrane perturbation or a mechanical swelling process. In order to elucidate the transfection mechanism of chitosan-based polyplex, the polycation buffering capacity was measured by simple titration and compared to PEI in numerous studies [66], [220], [265], [266]. These studies concluded that chitosan had significantly lower buffering capacity as compared to PEI and therefore it is unlikely that chitosan could escape the endosome/lysosome via the proton sponge effect. However, the experiments that led to the conclusion that chitosan has a lower buffering capacity vs PEI compared their buffering capacity on a mass concentration basis instead of molar charge concentration basis. PEI, with a mass of 43 g per mol of ionizable amine, has ~ 4 times more charge per gram than chitosan with 161 g per mol of ionizable sites for DDA = 100%. Both of these polyelectrolyte complexes (PEI and chitosan) form on a charge-to-charge basis (N:P) so that the particles involved in endolysosomal escape will have similar charges of chitosan and PEI rather than similar mass, not accounting for the unbound components that are not associated to the particles. Comparison on a molar charge basis reveals that chitosan actually has a larger buffering capacity than PEI in the endosomal/lysosomal relevant pH range [275]. Implications of these results regarding the transfection mechanism of chitosan–NA polyplex are further discussed in Section 4.3.2.
3.4.7. Colloidal stability
As pointed out previously, one drawback of chitosan is its poor solubility at neutral pH which translates into colloidally unstable polyplexes in physiological conditions. Colloidal stability of polyplexes is usually assessed by monitoring their hydrodynamic size vs time in medium mimicking physiological conditions (e.g. transfection medium, PBS, 10% serum + 150 mM NaCl, etc.) and size of chitosan–NA polyplexes was shown to increase from ~ 100 nm to more than a micron after 1 h incubation in PBS [150]. The colloidal instability of chitosan–NA polyplexes in physiological media may influence their behavior in vivo, for example in systemic administration via intravenous injection. Several chitosan modifications have been proposed in order to improve its solubility and polyplex colloidal stability. Among the strategies proposed are PEGylation [267], [276], [277], [278], quaternization [179], [180], [279] and glycolyzation [13], [150]. Most of these modifications were shown to improve chitosan solubility and polyplex colloidal stability in vitro to some extent. To the best of our knowledge, apart from the study of Lee et al. [280] where NPs formed with hydrophobically modified glycol chitosans have been shown to possess a relatively long-circulating time, the colloidal stability and circulation lifetime these modified polyplexes after systemic administration by intravenous injection have not been characterized to date and it remains to be seen if these modifications are only appropriate for applications where local administration is sufficient. It is however unlikely that any modification, such as chitosan quaternization, that doesn't reduce or shield surface charge in order to limit the strong interactions with blood components [281], [282], [283] will be efficient at increasing polyplex colloidal stability and circulation lifetime in vivo.
PEGylation has been shown to increase circulation lifetime and to limit aggregation of particles [281], [284], [285], and is therefore a most promising strategy to sterically shield nanoparticles from blood components and increase colloidal stability. PEGylation of chitosan nanoparticle surface has been reported [286], [66], but in most studies reporting PEGylation of chitosan, the hydrophilic PEG is grafted to chitosan backbone prior to form the polyplex [267], [276], [278] and the presence of this bulky substituent could limit polycation ability to complex NA [287]. The synthesis of a chitosan–PEG block copolymer (PEG-b–chitosan) is another avenue yet to be explored that could potentially circumvent this issue. The synthesis could be achieved via the reactive 2,5-anhydro-d-mannose unit at the new reducing of chitosan depolymerized with nitrous acid [101], [103]. Developing sterically stabilized chitosan–NA formulations with increased colloidal stability and circulation lifetime would be highly beneficial to the field of chitosan-based gene delivery systems.
3.4.8. Molecular weight and degree of aggregation
Aggregation number for polyplexes can be measured by static light scattering and possibly by NTA if polyplex stoichiometry is known since NTA measures particle concentration. This parameter is rarely measured although static light scattering measurements on polylysine–pDNA polyplexes revealed that colloidal and compact PEC with radius of gyration smaller than 120 nm can contain more than 1000 pDNA copies [288]. To the best of our knowledge, aggregation number hasn't been measured for chitosan–NA polyplexes. Characterization of chitosan–NA polyplex molecular weight as a function of formulation parameters and preparation method would improve understanding of their mechanisms of formation.
4. In vitro biological evaluation of chitosan-based polynucleotide delivery
4.1. Transfection efficiency
Transfection efficiency (TE) refers broadly to a pDNA carrier's ability to induce transgene expression. TE can be affected by multiple experimental parameters and a robust detection assay is required to determine how these factors influence transfection. Traceable reporter genes such as GFP, luciferase and galactosidase are frequently employed in a variety of gene delivery strategies [289]. Only fluorescence transgenes such as GFP allow lysis-free flow cytometry based quantitative assessment. Both GFP and galactosidase enable qualitative measurements via fluorescence and colorimetric microscopy of cell cultures. Most published measurements of TE use GFP and/or luciferase, two complementary quantitative methods [290]. GFP provides the percentage of cells transiently expressing the transgene while luciferase determines the level of transgene expression. To date, the multiplicity of methods of analysis and data presentation of TE renders comparison of one study to another quite challenging. The GFP-based parameter of percent cells positive for transgene expression provides a good reference point for inter-study comparison, and should be used alongside relative light units (RLU) from luciferase, since RLU varies with instrument type and settings such as excitation/emission filters, detector gains and reading speed. As such, it provides relative expression levels rather than absolute luciferase concentration. Absolute luciferase expression quantification obtained with standards of purified luciferase (Quantilum recombinant luciferase, Cat nb. E1071, Promega) can be used to facilitate inter-study comparison when only luciferase based transgene is assessed without GFP. Established positive controls such as Lipofectamine or PEI, in addition to proper negative controls such as carrier (chitosan) alone, DNA or siRNA alone and non-targeting siRNA, should be used and are important to facilitate data interpretation and comparison to literature [291].
4.1.1. Influence of Mn and DDA on transfection efficiency
The first studies of chitosan as a carrier for pDNA by Rolland et al. revealed an effect of Mn, where high Mn (from 100 to 500 kDa) resulted in higher TE in serum, but with expression levels that were 250-fold less than Lipofectamine controls [208], [246] and thus similarly low compared to what is now being achieved with chitosans [107], [150], [160], [260]. Since commercially available bulk chitosans were typically high molecular weight (150–500 kDa), subsequent studies used chitosans that were mostly of the high Mn type without depolymerization and often without appropriate characterization of Mn and DDA that would be required to understand their influence on carrier performance [224], [227], [292]. TE in these latter studies was still generally low but these studies nonetheless constitute the foundation of chitosan-based transfection and created the impetus to further optimize the system. Numerous methods to depolymerize chitosan were subsequently developed (reviewed above), with nitrous acid depolymerization the favored technique, to allow more reliable structure–function screening that accounts for Mn and DDA. Such screening generally contradicted Rolland's et al. original work and suggested lower Mn (< 150 kDa) chitosans to increase TE compared to higher Mn [12], [95], [208], [220], [293]. Since DDA is the second primary structural parameter of chitosan (along with Mn), studies also examined its influence on TE and suggested higher DDA (> 80%) were more efficient [28], [219], [225], except for studies using very large (390 kDa) chitosans, which suggested that lower DDA facilitates release of DNA [294].
These two critical parameters Mn and DDA were then screened in the high DDA low Mn range with accurate preparation and characterization techniques (SEC MALS for Mn and NMR for DDA), enabling a thorough structure–biological performance screening [26]. This systematic screening revealed significant coupling of Mn and DDA to determine TE, thereby establishing a rational structure–function relationship where both Mn and DDA modulate the binding affinity of chitosan to pDNA to obtain a fine intermediate balance between high and low affinity for DNA to produce efficient transfection, at levels similar to Lipofectamine or Fugene. Physicochemical studies subsequently confirmed that the chitosans with high TE did in fact possess an intermediate binding affinity [28], [256].
These pioneering studies resolved some of the many inconsistencies in the literature concerning the effectiveness of chitosan systems for gene transfer, via a precise accounting of the effects of Mn and DDA and identification of the central importance of intermediate binding affinity that leads to efficient systems in vitro. For example a chitosan with high DDA = 92% and low Mn = 10 kDa was found to be most efficient while simply increasing Mn to 40 kDa resulted in a large ~ 10 × reduction in TE by creating a more tightly bound polyplex. Simply reducing DDA to 80% from 92%, thereby reducing linear charge density on the chitosan, at constant Mn = 40 kDa then recovered most of the lost TE leading to similar efficiencies for 92–10 (DDA-Mn) and 80–40 chitosans [26]. These results highlight the need for precise preparation and characterization of chitosans as gene delivery vehicles, using the methods described above in Section 2. In addition to finely controlling Mn and DDA to optimize TE, tailoring chitosans by co-complexation with alginate [295] or more recently by substitution with trisaccharides [160] and self-branching [149], [150] also affect binding affinity and can improve transfection or silencing efficiency [27]. Although Mn and DDA are still important parameters affecting siRNA silencing performance, it seems that the small size of the oligonucleotide make it less susceptible to changes in chitosan Mn and DDA. Holzerny et al. [226] have found that at NP 5, chitosan of Mn 44, 63 and 93 performed equally well at DDAs in the range of 80–85% with only Mn 143 resulting in lower silencing. Using ITC, no significant differences between the binding affinities of those chitosans to siRNA were found. Moreover, Malmo et al. [27] reported similar efficiencies for Mn in the range 18–50 kDa (DPn 150–320) with fully deacetylated chitosans. Interestingly, using conditions similar to Holzerny, in serum at pH 6.5 but with a different cell type (HEK 293 vs. H1299 pGL3 human lung epithelial cells), we have shown optimal silencing with a smaller but more deacetylated chitosan (92–10) [209], [272]. At first glance, this recalls a similar relationship of chitosan oligonucleotide binding affinity and efficiency, as with plasmid. Data from Holzerny et al. and Malmo et al. combined with preliminary data from our lab showing effective silencing efficiencies for chitosans of increasing Mn at 92% DDA suggest a threshold where sufficient condensation is achieved and over which any increase in either DDA or Mn does not affect performance. Possibly because of its small size, siRNA can't be strongly bound and highly stabilized by chitosan architecture, compared to plasmids. Thorough screenings of chitosans–siRNA formulations for both Mn and DDA similar to Lavertu et al. for plasmid would provide a more complete understanding of chitosan–siRNA polyplexes.
4.1.2. Other formulation parameters affecting TE
4.1.2.1. Serum
The high transfection efficiency of chitosan in serum is a key advantage compared to other non-viral vectors since in vitro studies can be performed in conditions that are physiologically relevant for clinical translation. Transfection in serum also indirectly assesses polyplex stability, as positive uptake and TE imply nuclease protection of cargo polynucleotide. Early studies were quick to point out this advantage of chitosan over other non-viral vectors such as PEI [227] and liposomes [222]. Erbacher et al. were the first to find no inhibitory effect of serum with chitosan [227] while subsequent studies mostly underscored that serum enhances efficacy of chitosan-based polyplexes [11], [222], [260]. Despite these observations and the added value of using serum, a recent study using glycosylated chitosan did not use serum, preferring Opti-MEM serum free [160] even if a prior study demonstrated serum compatibility of glycosylated chitosan and similar transfection for linear chitosan [150]. Subsequent studies with siRNA from this group were again conducted in serum-free media [27].
It is still not entirely clear why serum increases TE with chitosan. Some have suggested better cell function [11], including cell division and endocytosis [260]. Nimesh et al. have shown that the level of polyplex uptake at pH 6.5, 7.1 and 7.4 can be upregulated by the presence of serum during the first 24 h [260] corresponding to increased luciferase levels. This increased uptake by serum was shown to be even more accentuated for chitosan only, possibly due to the formation of condensed particles of chitosan and serum protein. Thus, increased uptake of the free fraction of chitosan at N:P = 5 by serum addition might facilitate endosomal release [296].
Characterization in near physiological conditions would allow a better estimate of the type of polyplexes that are actually presented to cells in vitro and gain understanding of serum-mediated effects. However, measurement methods for nanosized particle are difficult in biological media and prone to significant background signal [260]; therefore such analyses are often overlooked in the development of new vectors. One potential future avenue we are following for improvement of polyplex size measurement in serum is nanoparticle tracking [297]. Other methods such as single particle tracking [298] and flow cytometry-based size determination [299] have shown promises for the size measurement of nano-complexes in serum. Such techniques could improve the predictive value of physicochemical and size characterization for both in vitro and in vivo efficiencies of chitosan-based polyplexes.
4.1.2.2. pH
The cationic charge density of chitosan is pH-dependent. Low charge density at physiological pH leads to low solubility, aggregation and poor stability of chitosan-based formulations [26], [28], [66], [222], [227], [260]. Hence, polyplex preparations are usually made in acidic conditions to optimize complexation with DNA and to provide colloidal stability, while addition to cell cultures imposes near physiological pH and ionic strength. Evaluation of the above literature reveals that pH between 6.5 and 7 is frequently used with success, and is attributed primarily to 1) positive zeta potential favoring cell contact and interparticle electrostatic repulsion (colloidal stability), and 2) an optimal physical stability of the polyplexes, providing protection from nucleases in the serum of transfection media. Nimesh et al. showed a strong correlation between uptake and TE at pH 6.5, 7.1 and 7.4 with no uptake and no TE at 7.4 and subsequent increases with decreasing pH. Size measurement showed an increase in size with pH and with ionic strength, with indications of aggregates at higher pH, correlated with microscopy images [260]. This is supported by previous observations of aggregates at neutral pH in isoosmolar conditions [28], [151], [227]. To circumvent this problem, many types of chitosan derivatives have been synthesized with the goal of improving solubility and resulting colloidal stability of the polyplexes in physiological media. These include PEGylation [114], [151], [172], glycosylation [13], [150], [300] and trimethylation [114], [151], [179], [180], [183]. These modified chitosans have shown improvements in DNA protection and/or TE in vitro at physiological pH, motivating in vivo studies to ascertain the advantages of such chitosans.
4.1.2.3. N:P ratio
As reviewed above in Section 3.4, up to a certain level, the N:P ratio determines the stoichiometry of the polyplexes which affects both colloidal and physical stability, especially in high ionic strength media. Mumper was the first to provide evidence of the importance of N:P to achieve stability and TE [246], which was followed by the others [222]. It is now established that, as with all polycation-based gene delivery, the most efficient N:P ratios for chitosan gene expression usually involve N:P ratios of at least 3–5 and sometimes of up to 30 [10], [26], [109], [160], [220], [301], [302], where a significant amount of the excess polycation remains free and soluble in solution without being physically bound to chitosan–DNA polyplexes [198], [230], [303]. These efficient N:P ratios with excess polycation have always been empirically identified but the means by which excess polycation improves TE was not identified until recently. A recent study revealed for the first time that the mechanism by which excess free polycation induces efficient transfection mainly involves promoting the release of the chitosan–DNA polyplexes from the lysosomes into the cytoplasm, an important step in nucleotide delivery as discussed further below [296].
This finding also partly explains the surprisingly high N:P ratio used in some studies of chitosan–siRNA delivery, ranging from 50 to 200 [14], [211], [255], [304]. In contrast, others have shown good silencing with N:P ratios of 5 [209], [226], [272] and N:P 10 [27]. Malmo et al. [27] have found that with fully deacetylated chitosans, only low Mn (10 kDa) required N:P up to 60 for efficient silencing while at higher Mn, all 3 tested ratios (10, 30, 60) performed equally with about 80% silencing at pH 7.2 in serum-free media, highlighting the stabilizing effect of increased N:P. At pH 6.5 in serum, we have shown that 92–10 performed best (80% silencing) at N:P 5 similar to Holzerny et al. [226] with other chitosans. Again, discrepancies among studies make overall comparisons difficult but it is our opinion that extremely high N:P ratios limit dosage of nucleotide and are difficult to translate to clinical application.
4.2. Cytotoxicity
Chitosan is often described as biodegradable, non-toxic and safe, and the FDA has approved a chitosan-based wound dressing biomaterial [305]. The biodegradability of chitosan by a number of physiological enzymes present in human such as lysozyme, di-N-acetylchitobiase, N-acetyl-beta-d-glucosaminidase and chitiotriosidase, has been well documented [17], [108], [306], [307]. Perhaps for these reasons, cytotoxic effects of chitosan have not been the focus of most studies in either gene or drug delivery. Yet, most of these studies report a slowed degradation of chitosan at high DDA, which could therefore present a risk of accumulation in the tissues over long period of administration [308]. This lowered biodegradability concurs with Huang et al. [309] finding of increased cytotoxicity of polyplexes prepared with increasing DDA. In recent reviews on chitosan's broad use in nanomedicine, Kean et al. and Garcia-Fuentes et al. also argue that toxicity appears with increasing charge density, and that at high DDA only, toxicity becomes Mn-dependent [[16], [17]]. Most studies from which this trend is observed are not in polynucleotide delivery. However, toxicity at high DDA compounded by high Mn follows general observations in our lab of decreased cell density and rounded morphology for high degrees of deacetylation or charge density (near 100% DDA at pH6.5) that could imply cytotoxicity or a cytostatic effect, however extensive cell viability and cytotoxicity testing has not yet been performed for all of these chitosan types [26]. To date we have only provided evidence that 92% DDA at 10 kDa Mn is non-toxic [260], [296]. Cytotoxicity of high DDA chitosan (85–99%) has also been found to be dose-dependence in human intestinal epithelial Caco-2 cells [310]. Garcia-Fuentes cites an IC50 from 0.2 to 2 mg/ml in most cell models, varying with DDA and Mn [16]. Moreover, some have described chitosan as a macrophage activator [311], but Guzman-Morales et al. [312] showed that while chitosan can concentrate and attract macrophages [24], it does not directly activate marrow-derived macrophages towards either the alternative nor the classical pathway, but rather through neutrophils in a yet to be determined cascade [312].
Interestingly, the group of Varum and Strand et al. consistently work with fully deacetylated chitosan with no adverse effects on cell health according to the MTT [150], Alamar Blue [149] and for siRNA studies, a judicious mix of both MTT and lactate dehydrogenase leakage assay (LDH) [27]. It is of note that assays whose detection rely on live cells such as MTT, based on the dehydrogenase activity of healthy mitochondria, have superior sensitivity than LDH that detects cell death [313]. Strand et al. have argued that the low Mn of their oligomers increase their solubilization at normal pH, particularly when glycosylated [149], which helps prevent cytotoxicity often associated with high charge density at acidic pH (high DDA, high Mn in acidic pH). Lower toxicity for low Mn (≤ 10 kDa) for any DDA is reported elsewhere as well [17]. The short 4 h incubation time with polyplexes might also limit cytotoxicity, particularly with fully deacetylated chitosan chains of Mn above 50 with siRNA [27]. MTT was performed at both 4 and 48 h post-transfection, while only at 48 h for LDH when the half-life of the leaked LDH is reported to be around 9 h by the manufacturer [314]. Despite the high number of published reports and patents highlighting the potential of chitosan for nanomedicine, chitosan is not yet approved by the FDA for drug or gene delivery, and incomplete cytotoxicity profiles possibly account for the slow translational progress, even if, by and large, cytotoxicity of chitosan is insignificant as compared to other synthetic vectors [17]. Proper and thorough cytotoxicity effect screening in relation to its structure and charge density with cells relevant to the application, including primary cells, has to be conducted (in addition to in vivo) for the translation to clinical application, and particularly for application requiring IV administration.
4.3. Intracellular trafficking
Once in proximity to cells, polynucleotide delivery systems need to overcome several obstacles to achieve expression, including: cell binding and internalization, escape from endo-lysosomal vesicles, polyplex unpacking/dissociation and, for plasmid DNA, nuclear translocation. One limitation with most studies investigating chitosan–polynucleotide delivery systems is that most rely entirely on end-point quantification of reporter gene expression to assess efficacy and potential formulation improvements. A more informative approach to study transfection mechanisms and resulting efficiency of carriers is to track their intracellular processing by microscopy using fluorescently labeled NAs and carriers in order to observe and potentially quantify the interactions of the components with the cell substructures at each of the major barriers. This approach can significantly contribute to a rational design of gene-carriers.
4.3.1. Uptake
The nature of the relationship between cellular uptake and physicochemical properties of chitosan has been recently reviewed [31]. The effects of intracellular and extracellular environments, as well as particle size, shape and surface charge characteristics on uptake were discussed in detail. In brief, some early studies found strong correlation of uptake with size [31]. However, in the presence of serum in acidic pH (6.5–7), size as measured in DLS has been shown to exert little influence on uptake [219]. This is consistent with Rolland's original work showing that particle size did not influence transgene expression when serum was present [208], [246]. The correlation of size as measured in DLS and TE in complex media is still not clear, especially in serum, were negatively charged proteins are known to change the morphology of polyplexes, sometimes forming protein coronas, as well as causing aggregation [17].
Uptake has been shown in many cell lines not to be a bottleneck [26], [160], [259], [309], [315]. These studies have shown that variations in uptake between chitosan formulations do not necessarily translate into variations in TE, and that for internalized polyplexes, important barriers still must be overcome. This is especially true for particles that are too stable for effective intracellular dissociation, as these particles usually have a positive zeta potential, bind to negatively charged cell membranes and are easily internalized [160], [259]. Evidently, cell membrane binding and permeation is the precondition of transfection, but once this is attained to a certain level, it has been shown that lysosomal escape and unpacking of DNA from its chitosan carriers may be an even more important factor in determining the transfection efficiency [160], [259], [296].
As with all polyplexes, chitosan TE is cell-line dependent. Many have demonstrated high transfection efficiency in established cell lines such as HEK 293 [26], [160], [259], [260] while moderate TE was observed in more challenging cell lines such as HELA, MDCK and COS-7 cells [224], [315], [316]. A chitosan–PEI graft was shown to increase transfection in HELA and HepG2 cells [167] but uptake was not assessed. Cell-line dependency of TE has been attributed to variations in membrane phenotype, types of receptors and variation in the most active endocytic pathways in each cell types [315]. For instance, Douglas et al. [315] show that the cell line dependency of TE is mainly due to the preferred endocytic pathway by which chitosan polyplexes are internalized and not to the level of uptake per se. In their study, cells favoring clathrin mediated pathway such as in HEK293 and COS7 promoted transfection, as opposed to CHO cells that prefer caveolin mediated endocytosis for reasons that are still unclear.
TE challenges are even more pronounced with primary cells, which are generally quiescent while most cell lines divide within 24 h. Cell division facilitates access of transgene to the nucleus while for quiescent cells, active nuclear translocation via nuclear pores is essential [317]. Even for primary dividing cell types such as mesenchymal stem cells, early attempts to transfect them proved unsuccessful [262]. Screening in various cell lines as well as in non-dividing cells would improve the predictive values of in vitro testing.
4.3.2. Endolysosomal transit & release mechanism
4.3.2.1. Transit
Once endocytosed, endosome acidification and pathways that lead to lysosomes are often regarded as a major barrier to non-viral based gene transfer [318], [319]. As compared to PEI, few studies have examined the trafficking of chitosan polyplexes through the endo-lysosomal pathway [219], [309], [320], [321]. These studies, exclusively using Lysotracker as a lysosome stain to assess colocalization with polyplexes to track their intracellular distribution, appeared to suggest that chitosan polyplexes do not transit through lysosomes at all [320] or only for short periods (within 2–4 h of transfection) [219], [321]. This suggestion is in direct contradiction with a more recent study using pulse-chase fluorescent dextran lysosomal staining that revealed that all chitosan-based polyplexes, independently of their structure, do transit through lysosomes, with nearly 80% colocalization at 12 h (4 h post-transfection with 8 h incubation time), followed by a gradual release to about 50–60% at 48 h, the moment when transfection peaks [259].
The lack of consensus between the above studies could stem in part from the use of different methodologies for transfection and lysosomal staining, including different cell types, transfection pH or the use of an acidophilic Lysotracker dye versus dextran. In this context, it is important to note that inhibition of lysosomal acidification will abolish Lysotracker fluorescence and acidophilic partitioning [322]. Therefore, this agent may not be an accurate marker of lysosomes when loaded with polycations endowed with a proton buffering capacity such as PEI [10] and chitosan [11] that can prevent lysosomal acidification.
As noted elsewhere using electron microscopy [220], [227], lysosomal sequestration appears to account for a later onset of gene expression, as compared to other polycations such as PEI, where vesicular escape is observed within the first 4 h after transfection and with significant gene expression detected as early as 5–6 h post-transfection [323], [324]. Thus lysosomal sequestration is a rate limiting step for chitosan carriers that is independent of chitosan Mn and DDA, and delays gene expression to that currently observed and possibly the attained number of transfected cells to ~ 40–50% of all cells.
4.3.2.2. Escape mechanism
The speed and high effectiveness of polymers such as PEI have generally been attributed to an ability to rapidly escape endo-lysosomes by means of its high buffering capacity by a mechanism known as the proton sponge effect. First described by Behr's group [10], [325] following the observation that lysosomotropic agents' chloroquine did not increase TE of PEI, the proton sponge hypothesis describes the ability of a polycation to swell and rupture vesicles by raising intra-vesicular osmotic pressure through proton buffering coupled to a concomitant influx of negatively charged ions such as Cl− [319]. Studies comparing PEI and chitosan buffering capacity suggested that PEI possesses a higher buffer capacity in a broader pH interval than chitosan as determined by acid/base titration [66], [220]. For these reasons, combined with the slow transfection kinetics as noted above, the endosomal escape mechanism of chitosan systems has usually been attributed to either direct hydrostatic membrane destabilization [326] or through a rise in vesicular osmolarity by lysosomal enzyme-induced degradation products of the polycation [89], [108], although neither of these hypothesis have been supported by data.
However, as mentioned above in Section 3, Richard et al. recently addressed a technical flaw found in the literature, namely that the comparison of buffering capacity of chitosan versus PEI was performed on a mass basis, without accounting for the ratio of protonation sites to mass [66], [170], [220], [266]. Richard et al. [275] recently found that chitosan buffering capacity was superior to PEI at equivalent charge ratios, and at pH relevant to vesicular trafficking (4.5–7). These findings are in agreement with the generalized absence of any beneficial effect of completely preventing endosomal fusion to lysosomes, hence inhibiting the lysosomal transit of polyplexes, as occurs when chloroquine is added at the start of transfection [224], [296], similar to that found with PEI. The reason for chitosan's long lysosomal transit time compared to PEI remains to be determined but could be due to a lower linear charge density and weaker membrane disrupting ability for chitosan. To this effect, it is worth mentioning that the importance and validity of the proton sponge hypothesis to induce escape and improve TE has been challenged [327], [328], [329]. Also, when complexed to a polynucleotide, the protonable amines of chitosan become more ionized [256], possibly reducing buffering capacity. The latter may also account for slower transfection kinetics and highlights the important role of excess DNA-free chitosan in promoting release in vitro [296]. In view of this, chitosan may also benefit from intravesicular degradation to increase osmolarity, possibly necessitating longer processing time inside lysosomes. On the other hand, PEI and its higher charge density might be less masked by binding to the polynucleotide [275] and may be more active in destabilizing vesicular membranes, for which its cytotoxicity is partly blamed [330].
Despite the slow transit kinetics of chitosan–NA polyplexes and a reasonable buffering capacity, chitosan has been mixed or functionalized with buffering moieties in attempts to increase TE. Mixing with PEI [331] or polypropyl acrylic acid [294] and grafting of PEI [266], [267], histidine [171] and other imidazole-containing components [166], [300] constitute some of the recent strategies to overcome any endo-lysosomal barrier.
4.3.3. Polyplexes dissociation
Timely intracellular DNA unpacking and release is another important rate-limiting step for chitosan [28], [89], [150], [160] and condensing polycations in general [332]. Despite the importance of the coupling of Mn and DDA towards optimal binding for high transfection efficiency, few studies have assessed how these structural parameters of chitosan influence the state of polyplex condensation inside cells. Particle stability is difficult to monitor intracellularly in part because it pertains to distances below (1–10 nm) the resolution limit of light microscopes (~ 250 nm) such that techniques such as fluorescent resonant energy transfer (FRET) are important tools to monitor the condensation state of the polyplexes intracellularly [259], [320], [333].
The first studies to use FRET to assess polyplex stability mostly compared one chitosan type to other polycations [320], [333] without accounting for the strong influence of DDA and Mn on the transfection process. Quantum dots were used, as they are efficient and flexible for FRET studies and provided cues that dissociation is an important factor for TE. However, with QD–chitosan complexes, it was shown that TE was nearly completely abolished [333], possibly due to the size of the QD label. By following chitosan of different formulations using organic dyes (rhodamine B for donor on chitosan, Cy5 for acceptor on pDNA), we revealed that the kinetics of polyplex unpacking or decondensation was the most critical formulation-dependent intracellular process and could account for the important relationship between TE and chitosan structure, validating for the first time the DDA–Mn coupling effect for optimal binding and resulting fine-balance [259].
The exact mechanism that promotes dissociation of self-assembled polyplexes is not yet understood. Many have suggested competition with cytoplasmic polyanions including proteins and RNA [334]. In this case, partial degradation of enzyme-labile biodegradable chitosan [69], [108] in lysosomes may facilitate downstream cytoplasmic decomplexation. Coincubation with chitosanase [335] was suggested to potentiate lysosomal-induced degradation, escape and to increase the TE of condensed polyplexes. The importance of dissociation is also evidenced in the addition of labile bonds to chitosans in attempts to facilitate intracellular decomplexation and increase TE [266], [163], [320]. To the best of our knowledge, no study to date has examined with tools such as FRET or live-cell lysosomal stain the intracellular dissociation dynamics and lysosomal colocalization, respectively, of chitosan–siRNA polyplexes.
5. In vivo application of chitosan-based nanoparticles for gene therapy
5.1. Chitosan-based plasmid DNA nanoparticles
Viral vectors have been proven to be efficient in gene delivery, however their clinical application is limited due to safety risks and unexpected adverse effects [336], [337]. In contrast, non-viral vectors, such as liposomes and polycation complexes, have been increasingly developed in recent years because of their low immunogenicity, ease of production and modification under controllable conditions.
5.1.1. Vaccination
The need for effective humoral and cellular responses induced by genetic immunization requires a delivery vehicle that will lead to improved presentation of transgene products to antigen-presenting cells (APCs) [338]. Chitosan packaging of DNA can improve cell uptake, as it is an adhesive biomaterial that extends the period of contact between the active agent and targeted cells and tissues. Also, chitosan has an innate ability to open tight junctions between cells, thereby, increasing the uptake of carried agents into these cells and increase membrane permeability [339], [340]. The choice of an adequate route of administration for a DNA vaccine is an important decision towards clinical application. Administration can be directly to a target tissue or through systemic delivery. The potential for transfection of the appropriate cell types in vivo that are able to elicit subsequent immunity to the DNA encoded antigens is key to successfully induce immunity. The encoded protein must be presented on the surface of appropriate cells to the responding T and B cells. This presentation takes place through transfection of somatic cells (e.g. myocytes, fibroblasts), or APCs, leading to uptake of the expressed antigen by professional APCs and presentation through cross-priming pathways [338].
The route of mucosal vaccination (oral and intranasal), a common route for pathogen entry to the body, is preferred as it can generate a local immune protection in addition to a systemic immune response. Roy et al. used chitosan–pDNA nanoparticles, of 150–300 nm in size, for oral immunization against the dominant peanut allergen gene (Arah2); immunized mice showed substantial reduction in allergen-induced anaphylaxis associated with reduced levels of IgE, plasma histamine and vascular leakage [341]. Also, the immunized mice produced secretory IgA and plasma IgG2 proving that chitosan–pDNA nanoparticles reached the target of mucosal-associated lymphoid tissue of the Peyer's patches in the gut and respiratory tract, and M cells where the presence of APCs such macrophages process the recombinant antigen leading to the activation of T cells and B cells (see Table 8 for a summary of oral vaccination studies).
Table 8.
Selected studies using chitosan-based pDNA nanoparticles for gene delivery in animal models.
| Animal model | Delivery system | Route of administration | Gene expression | Disease | Comments | References |
|---|---|---|---|---|---|---|
| Vaccine | ||||||
| BALB/c mice | Biotinylated chitosan nanoparticles is appended with bifunctional fusion protein (bfFp) for DEC-205 restricted DC targeting. | Intranasal Intramuscular |
Nucleocapsid (N) protein of severe acute respiratory syndrome coronavirus (SARS-CoV) | Acute respiratory syndrome coronavirus (SARS-CoV) | Nanoparticles administrated intranasally with a DC maturation stimuli increase the production of IgA and IgG against N protein. Intramuscular administration lead to higher induction of systemic IgG responses compared to intranasal route of administration. | [399] |
| BALB/c mice | Chitosan (390 kDa) | Oral | pDer p 2 | Allergy Dermatophagoides pteronyssinus | Oral chitosan pDer p 2 nanoparticles initiate Th1 immune responses and express pDer p 2 gene in epithelial cells of small intestine and stomach. Chitosan pDer p 2 effective for gene delivery for Th1-immunity. | [379] |
| BALB/c mice | Chitosan 92-10-5, 80-10-10 and 80-80-5 (DDA-number average molecular weight or Mn in kDa). | IM and SC | FGF-2 and PDGF-BB | High DDA — low molecular weight chitosans are efficient protein expressors with minimal or no generation of neutralizing antibodies, while lowering DDA resulted in greater antibody levels and correspondingly lower levels of detected recombinant protein. | [109] | |
| Fish — Japanese flounder | Chitosan (MW 1080 kDa; DDA 80%) | Oral | Major capsid protein (MCP) of lymphocystis disease virus (LCDV) | Lymphocystis disease | pDNA-loaded chitosan microspheres expressed MCP in tissues of fish 10–90 days after oral administration. Positive immune response. | [384] |
| BALB/c mice | Chitosan (MW 400 kDa; DDA 85%) | Intranasal | HBsAg | Hepatitis B | Chitosan nanoparticles produced humoral and cellular immune responses when administered intranasally. Chitosan effective nontoxic carrier of pDNA through nasal mucosal delivery. | [342] |
| BALB/c mice | Chitosan (MW 390 kDa; DDA 83.5%). Chitosan–DNA nanoparticles embedded in jelly |
Oral | House dust mite allergen (Der p 1) |
Allergy | Oral administration of chitosan–DNA nanoparticles primes an antigen-specific immune responses to Der p 1 and Der p 1-derived peptides. IgG2a was preferentially induced and was detected from day 56 after oral administration. | [407] |
| AKR/J mice | Chitosan (MW 390,000 kDa) | Oral | Dominant peanut allergen gene (Arah2) | Food Allergy | Oral immunization of mice with chitosan-based nanoparticles resulted in a considerable reduction of allergen-induced anaphylaxis with lower levels of IgE, plasma histamine and vascular leakage. | [341] |
| Km mice | Chitosan (MW 390 kDa; DDA 83.5%) | Oral | Zona pellucida (ZP) | Contraceptive vaccine | Transcription and expression of DNA vaccines after 5 days of feeding. | [381] |
| BALB/c mice | Chitosan oligomers (3.6–7 kDa) substituted by reductive N-alkylation with the trimer 2-acetamido-2-deoxy-Dglucopyranosyl-β-(1–4)-2-acetamido-2-deoxy-d-glucopyranosyl-β-(1–4)-2,5-anhydro-d-mannofuranose (A-A-M) | Intranasal | CTL epitope from the M2 protein of the Respiratory syncytial virus (RSV) | Respiratory syncytial virus (RSV) | Intrasal and intradermal nanoparticles administration induced peptide- and virus-specific CTL responses in mice. | [426] |
| Recombinant protein | ||||||
| Ascites tumor-bearing imprinting control region mice | CPT: Cancer-targeted specific peptide (FQHPSF sequence) linked with chitosan-linked polyethylenimine (CP). Chitosan (MW 100 kDa) | Intraperitoneal | IL-12 | Cancer | CPT/DNA complexes allow delivery of interleukin-12 which significantly enhanced the antitumor effect. | [427] |
| C.B-17/Icr-scid-bg mice bearing HeLa tumors. | Chitosan (MW 15.5 kDa; DDA 75–85%) | Intratumoral and followed by UltraSound exposure | Reporte gene encoding luciferase protein | Cancer | Tumor transfected with chitosan lead to higher (2 ×) luciferase expression compare to tumor transfected without chitosan. | [428] |
| Balb/c Mice | N-linoleyl LMWC N-oleyl LMWC Chitosan (MW 50 kDa, DDA 85%) |
Intramuscular | Interleukin-4 (IL-4) and Interleukin-10 (IL-10) | Type 1 diabetes | Both nanomicelles-based polyplexes administration can increase IL-4, and IL-10 expression, lower the blood glucose, TNF-α and IFN-γ, and thereby enhance pancreatic beta cells protection from inflammation and insulitis. | [389] |
| BALB/c mice bearing C26 tumors (model of murine liver metastasis of colon cancer). | Galactosylated Chitosan (GC) Chitosan (MW 300 kDa; DDA 84%) |
Intravenous | Interleukin 21 (IL-21) and granulocyte-macrophage colony-stimulating factor (GM-CSF) | Liver and colon cancer | Nanoparticles improved the transfection efficiency, exhibited specificity for hepatocytes and tumor and activated the function of cytolytic T lymphocytes (CTL) and natural killer (NK) cells. | [377] |
| Bama miniature pigs | Chitosan chloride (TMC) complexes loaded into a bilayer porous collagen chitosan/silicone membrane dermal equivalents (BDEs). Chitosan (MW 6 K, DDA 90%) for the TMC synthesis. Chitosan (DDA75–85%, Mn: 1.0 × 105–1.7 × 105) for the scaffold fabrication |
Skin implantation | Vascular Endothelial Growth Factor-165 (VEGF-165) | Burn injures | Nanoparticles use lead to a higher expression of VEGF, CD31 and alpha-SMA proteins involved in angiogenesis and dermal regeneration. | [429] |
| Lewis rats and AIA rat model. | Folic acid- chitosan (50 kDa; DDA 96%) (Ch-Fa) | Intravenous-hydrodynamic injection and normal femoral intravenous injection. | Reporter gene: Fluorescence emitting proteins (DesRed) and β-galactosidase Therapeutic protein: IL-1Ra |
Adjuvant induced Arthritis | Chitosan-folate-based nanovectors efficient hydrodynamic delivery of β-gal DNA nanocarriers. Hydrodynamic delivery of plasmid IL-1Ra DNA and chitosan-DNA-based nanocarriers in limb controlled progression of inflammation in an AIA rat model. | [398] |
| Sprague–Dawley rats | NOVAFECT O 15 (MW 5.7 kDa; DDA 99%) | Injection into the corneas | Reporter gene: luciferase and GFP | Corneal diseases | Compared to polyethylenimine-DNA nanoparticles, the chitosan-DNA nanoparticles increased 5.4 times the luciferase gene expression when injected into the corneal stroma. | [193] |
| ZDF rat | Chitosan 92-10-5, 80-10-10 and 80-80-5 (DDA-number average molecular weight or Mn in kDa N:P ratio). | Intramuscular and intravenous | GLP-1 | Type 2 Diabetes | Animal injected with chitosan 92-10-5/GLP-1 increase GLP-1 plasma levels and plasma insulin concentration and normalization of blood glucose level up to 24 days after treatment. | [235] |
| Nu/nu mice with C6 xenograft tumors | PEI–PEG–chitosan copolymer coated iron oxide nanoparticles (NP-CP-PEI) | Intravenous | GFP | Cancer | NP-CP-PEI was able to deliver intact DNA for transgene expression into tumors of various sizes. | [430] |
| FVIII exon 16 knock-out mice. | Chitosan (MW 390 kDa; DDA 84% and 70%) | Oral | Factor VIII | Hemophilia A | FVIII was detected in both local and systemic tissues reaching a peak level 22 days post administration. Treatment with of FVIII DNA-chitosan nanoparticles resulted in phenotypic correction in 13/20 mice compared to 1/13 mice treated with naked FVIII DNA and 0/6 in untreated mice. | [250] |
| BALB/c nude mice bearing SaOS-2 tumors | Low viscous chitosan ([2-amino-2-deoxy-(144)-b-d-glucopyranan]) | Injection through the cortex of the anterior tuberosity of the tibiae | Epithelium-derived factor (PEDF) | Osteosarcoma | pPEDF microparticles treatment resulted in a decrease in primary tumor growth, reduction of bone lysis and reduction of lung metastases. | [431] |
| BALB/c mice | Mannosylated chitosan | Intratumoral | IL-12 | Cancer | The complexes enhance the production of IL-12 p70 and IFN-γ and thereby suppressed tumor growth. | [432] |
| ICR mice | Chitosan (300 kDa) | Oral | LacZ gene and Erythropoietin | Anemia | Mice fed with chitosan-mEpo nanoparticles showed rapid enhancement of hematocrit that went up to 65% after a second administration. | [380] |
| C57BL mice | N-acetylated chitosan (MW 9.5 × 105; DDA 100%) | Oral | LacZ gene and IL-10 | Vaccin for Gastrointestinal diseases | Effective for transgene delivery to the gastrointestinal track. | [314] |
Intranasal administration of chitosan–pDNA nanoparticles is a second principal route of mucosal immunization for which several authors speculated that chitosan–pDNA nanoparticles may pass the mucosal membrane and directly transfect the APCs or be taken up first by the M cell-like cells (as for oral administration) in nasal associated lymphoid tissue and then presented to APCs. Khatri et al. used intranasal administration of chitosan (85% DDA and 400 kDa Mn) pDNA nanoparticles expressing the surface antigen of hepatitis B virus (HBsAg) for immunization against hepatitis B in Balb/c mice model showing successful generation of a systemic immune response (humoral and cellular) [342]. These spherical chitosan–pDNA nanoparticles with diameter of ~ 350 nm were able to elicit a systemic immune response with IgG levels (anti-HBsAg) above the clinical protective level of 10 mIU/ml. In addition, this intranasal vaccination elicited a mucosal immune response with the production of IgA secretory antibodies (see Table 8 for a summary of intranasal vaccination studies).
Systemic vaccination routes of chitosan–pDNA based vaccines such as intravenous, intramuscular, subcutaneous and intraperitoneal are considered for targeting APCs and macrophages. However in comparison with other DNA vaccine nanocarriers, fewer conclusive studies have been performed to date with chitosan–pDNA nanoparticles for systemic vaccination. Jean et al. showed that injection of Balb/c mice with chitosan–pDNA (expressing human FGF-2 and PDGF-BB) nanoparticles by a subcutaneous (SC) route led to enhanced levels of antigen-specific antibodies (anti-FGF-2 and anti-PDGF-BB) as compared with intramuscular (IM) injection [109]. Interestingly, the specific immune response obtained depended intimately on the specific chitosan that was used as a complexing agent for pDNA. The lower DDA (80%) and higher Mn (80 kDa) chitosans induced the highest levels of antigen-specific antibodies and was associated with a sustained influx of host cells associated with degradation of this chitosan. Both SC and IM routes of administration produce Th2-type response but act in different manners in terms of raising specific antibodies [343], [344], [345]. As the prevalence of APCs is different in muscle than in SC tissues, it is likely that the network of APCs residing in the target tissue act as a decision-maker of the extent of the humoral response versus accumulation of recombinant protein elicited by these two administration routes for chitosan–pDNA expressing vectors. The differences in the amplitude and in the quality of the immune responses suggest that the APCs transfected in IM and SC locations are functionally distinct and therefore prime the immune response uniquely [346], [347]. Following IM injection, DNA is taken up by myocytes [348]. However, myocytes do not normally express MHC II or costimulatory molecules and cannot prime T cells, hence the need for APC/macrophage intervention. However, APC transfection may be present during SC injection, as has been demonstrated after intradermal DNA injection [349].
Other studies examining immune modulating responses to chitosan have provided variable results, possibly due to different chitosan types (Mn and DDA) and preparation methods used in these studies [309], [350], [351]. Additionally, the recombinant products were detectable at the injection site and surrounding tissues several weeks after administration and were not broken down rapidly, permitting APCs/macrophages to process the antigens and eventually activate the B cells and the T cells [109], [235], [342], [352], [353].
5.1.2. Expression of therapeutic proteins
It is now known that the transfection efficiency of chitosan-based nanoparticles depends on several factors such as chemical structure of the specific chitosan (DDA, Mn, chitosan modification), size and charge and composition of polyplexes (including type, form and size of the DNA/RNA cargo), interaction between polyplexes and cells (endocytosis or pinocytosis, cytoplasm trafficking and crossing the nuclear pore complex) and the cell type. A number of studies showing successful delivery of recombinant DNA with sustained transgene expression in local, systemic and targeted sites were performed in animal models with a myriad of chitosan parameters (DDA, Mn, NP ratios, nanoparticle size and intrinsic and extrinsic modifications) (Table 9 ). Among these studies, effective oral delivery of chitosan (84% and 70% DDA with 390 kDa Mn) as therapeutic gene carrier was demonstrated by Bowman et al. by successfully inducing factor VIII synthesis in various organs of a hemophilia A murine model (FVIII exon 16 knock-out mice) [250]. Transgene DNA was detected in both local and systemic tissues resulting, after one month, in hemophilia A phenotype being correlated to 13/20 mice treated with chitosan–pDNA FVIII (250–600 μg of DNA) nanoparticles, compared to just 1/3 mice treated with naked FVIII plasmid DNA and 0/6 untreated mice. However no explanation was offered for the presence of chitosan–pDNA nanoparticles in the liver and spleen and the mechanism of transport from the intestinal lumen to the hepatic circulation. The success of chitosan as gene carrier was corroborated by Jean et al., who used low molecular weight chitosans (92% DDA and 80% DDA with 10 kDa Mn) to deliver, intramuscularly and subcutaneously, recombinant pVAX-GLP-1 plasmid expressing glucagon-like peptide 1 (GLP-1) for the control of type 2 diabetes in the Zucker Diabetic fatty (ZDF) rat model [235]. The animals treated with chitosan-pVAX-GLP-1 saw an increase of plasma concentration levels of GLP-1 and insulin associated with normalization of blood glucose which persisted for 24 days after the last injection, compared to a half life of just several minutes for the injected peptide. Also, Klausner et al. demonstrated that the injection, into the stroma of rat corneas, of a formulation of oligomeric chitosan–pDNA (99% DDA and 5.7 kDa Mn) expressing luciferase or green fluorescent protein (GFP) led to a gene expression 5.4 × greater than the injection of PEI–pDNA nanoparticles [193].
Table 9.
Studies using chitosan-based siRNA nanoparticles delivery system for gene silencing in animal models.
| Animal model | Delivery system | Route of administration | Silenced gene | Disease/objective | Comments | References |
|---|---|---|---|---|---|---|
| Female athymic nude mice (NCr-nu). | Chitosan (MW 50–190 kDa), sodium tripolyphosphate (TPP) | Intravenous | Zeste homolog 2 gene | Breast cancer | Significant reduction in tumor growth. | [433] |
| Sprague–Dawley female rats bearing breast tumors. | Chitosan (MW 75 kDa; DDA 75–85%) | Intratumoral | Vascular endothelial growth factor (VEGF-A) and VEGF receptor (VEGFR1, VEGFR2) and co-receptors for VEGF; the neuropilin-1 (NRP-1) | Cancer | Nanoplexes containing chitosan and mixture of siRNAs (siVEGF-A, siVEGFR-1, siVEGFR-2, and siNRP-1) have a suppressive effect on VEGF expression and tumor volume. | [364] |
| Female athymic nude mice (NCr-nu) bearing IGROV-AF1 or SKOV3TRip2 tumors. | Chitosan (MW 50–190 kDa) | Intravenous | Jagged1 | Ovarian cancer | Treatment with anti-human Jagged1 siRNA-CH and/or anti-murine Jagged1 siRNA-CH in two different orthotopic ovarian cancer models, treatment resulted in significantly reduced tumor weight up to 93%. | [434] |
| Female athymic nude mice (NCr-nu) bearing SKOV3ip1 or HeyA8tumors. | Chitosan (MW 50–190 kDa), sodium tripolyphosphate (TPP) Chitosan: TPP (3:1) |
Intravenous | Zeste homologue 2 (EZH2) Also used: Alexa555-labeled siRNA and Cy 5.5 labeled siRNA |
Ovarian cancer | After nanoparticles injection, siRNAs were delivered to the tumor and to various organs, such as kidney, liver, lung, and spleen. EZH2 gene silencing lead to a decrease of tumor burden (reduction up to 83%) and an inhibition of angiogenesis mediated by reactivation of vasohibin1 (VASH1). | [435] |
| Female athymic nude mice (NCr-nu) bearing melanoma (A375SM) and breast (MDA-MD231) cancer. | Chitosan hydrogel (CH-HG) chitosan (161 kDa; DDA 80%) | Intratumoral | Tissue Transglutaminase (TG2) Also used: Alexa555-labeled siRNA |
Cancer | Higher localization into tumor cells for siRNA CH-HG compared to siRNA alone. TG2 expression level was suppressed by up to 80% at 2 days and up to 50% until 10 days after a single injection of TG2 siRNA/CH-HG compared to control siRNA/CH-HG. Result in significant inhibition of tumor growth up to 92%. | [365] |
| Female athymic nude mice (NCr-nu) bearing SKOV3ip1, HeyA8 or HeyA8-MDR tumors. | Chitosan-tripolyphosphate (TPP) | Intravenous | Src and Fgr (Src family kinases (SFKs)) | Ovarian cancer | Tumor growth was significantly reduced up to 84%. Tumor associated microvessel density significantly reduced up to 75%. Dual silencing resulted in significantly increased tumor cell apoptosis by more than 2-fold compared to all other groups. | [366] |
| BALB/C and C57BL/6J mice | Chitosan (MW 130 kDa; DDA 86%) Imidazole-modified chitosan (chitosan-IAA) |
Intranasal | GAPDH | Non-specific disease | Significant silencing was seen in the lungs with chitosan and chitosan-IAA nanoparticles. | [436] |
| PEG–chitosan (MW 130 kDa; DDA 86%) PEG–chitosan-IAA |
Intravenous | GAPDH and apolipoprotein B (ApoB) | PEG–chitosan-IAA nanoparticles delivery demonstrated significant knockdown in both lung and liver. | |||
| Female athymic nude mice (NCr-nu) bearing SKOV3ip1, HeyA8, and A2780 tumors. | Arg-Gly-Asp peptide-labeled chitosan nanoparticles (RGD-CHNP) CH (MW 50–190 kDa) |
Intravenous | Periostin (POSTN) Focal Adhesion Kinase (FAK) Plexin domain-containing protein 1 (PLXDC1) Also used: Alexa555 siRNA |
Ovarian Cancer | The targeted delivery-mediated gene silencing significantly enhanced tumor localization, downregulation of specific gene and anti-tumor therapeutic efficacy compared to a non-targeted delivery system in ovarian cancer models. | [437] |
| Sprague–Dawley rats | Chitosan (Low MW; DDA 75–85%) | Intrathecal | Muscarinic acetylcholine receptors (mAChR) subtypes: M2, M3 and M4. Also used: Alexa Fluor 488-labeled chitosan-siRNA nanoparticles |
Investigation to determine each subtype's role in controlling nociception at the spinal level | Chitosan nanoparticles can be used for efficient delivery of siRNA for silencing in neuronal tissues in vivo. By selective knockdown authors demonstrated that M2 and M4, but not M3, contribute to nociceptive regulation by mAChRs at the spinal level. |
[438] |
| Female nu/nu mice bearing B16F10-red fluorescent protein (RFP) tumors. | Chitosan–Poly-l-arginine (PLR)–PEG Chitosan (MW 50–150 kDa; DDA 87% |
Intratumoral | Red fluorescent proteins (RFP) | Cancer | siRFP complexed to CS–PLR or to PEG–CS–PLR produced a significant reduction in RFP fluorescence 16.9 ± 1.7% and 10.4 ± 4.5% respectively compared to untreated tumor tissues. | [257] |
| Nude male mice bearing LNCaP or PC3 tumors. | Chitosan-TPP | Intratumoral | RLN family peptide receptor 1 (RXFP1) | Prostate cancer | Treatment leads to a downregulation of RXFP1 receptor expression and an important reduction in tumor growth. | [439] |
| Balb/c nude (nu/nu) mice bearing RFP/B16F10 tumors. | Glycol chitosan–polyethylenimine (GC–PEI) GC: MW 250 kDa; DDA 82.7%), |
Intravenous | Red fluorescent proteins (RFP) Also used: Cy5.5 labeled siRNA |
Cancer | siRNA–GC–PEI NPs presented higher tumor-targeting ability and a significant inhibition of gene expression in tumor. | [373] |
| EGFP-transgenic mice (C57BL/6-Tg) (ACTb-EGFP). | Chitosan (CS) (MW 50 kDa; DDA 92%) Guanidinylated chitosan (GCS) Guanidinylated chitosan with β(2)-adrenoceptor agonist (SGCS) |
Endotracheal | Green fluorescent protein (GFP) | Respiratory diseases | Guanidinylation of chitosan improve cellular internalization of siRNA nanoparticles, reduce nanoparticle cytotoxicity and thereby increase downregulation of specific gene in the bronchial epithelial cells in vivo compared to unmodified chitosan. | [386] |
| Mice bearing PC-3 tumor. Mice bearing SCC-7 tumor. Mice bearing RFP-B16F10 tumor. |
Poly-siRNA/thiolated glycol chitosan nanoparticles (psi-TGC) | Intravenous | Vascular endothelial growth factor (VEGF) Also used: FPR675-labeled poly-siRNA Reed fluorescent protein (RFP) |
Cancer | Chitosan nanoparticles (psi-TGC) provided sufficient in vivo stability for systemic delivery of siRNAs. Knockdown of genes coding for tumor antigens by psi-TGC resulted in a reduction in tumor size (by 80%) and vascularization. psi-TGC were mainly co-localized with tumor cells in animal model. RFP signal in mice RFP-B16F10 tumor clearly diminished to about 17% after psi(RFP)-TGC administration. |
[280] |
| C57BL/6J mice Collagen type II DBA/I arthritic mice. |
Chitosan (114 kDa; DDA 84%; N:P ratio 63) | Intraperitoneal | Tumor necrosis factor-alpha (TNF-α) Also used: Cy3-labeled siRNA |
Rheumatoid arthritis | Treatment lead to TNF-α knockdown in peritoneal macrophages, minimal cartilage destruction and inflammatory cell infiltration in collagen-induced arthritic (CIA) mice. | [210] |
| EGFP-transgenic mice (C57BL/6-Yg(ACTbEGFP)1Osb/J) |
Chitosan (MW 114 kDa; DDA 84%) | Intranasal | Enhanced green fluorescent protein (EGFP) | Systemic and mucosal disease | Chitosan-based systems to mediate EGFP knockdown in intact epithelial bronchiole. | [14] |
| Nude mice bearing RP tumor. | Chitosan-coated poly(isobutylcyanoacrylate) (Chitosan: 20 kDa) |
Intratumoral | Ret tyrosine kinase domain juxtaposed with H4 gene (Ret/PTC1) | Papillary thyroid carcinoma | Chitosan nanoparticles allowed down-regulation of the expression of ret/PTC1 gene and significant tumor growth inhibition. | [440] |
| Balb/c mice | Chitosan (MW 160 kDa; DDA 80%) | Intravenous and Intraperitoneal | No target | Chemical Modification and nanoparticle formulation to improve the systemic delivery of siRNA | Chitosan substantially improved the stability and biodistribution of siRNA. High siRNA concentration within the kidney was observed 24 h post IV injection and in peritoneal fluid 30 min post IP injection. | [371] |
| Female athymic nude bearing MDA-MB-231 tumors. | Chitosan-coated polyisohexylcyanoacrylate (PIHCA) | Intravenous | Ras homologous A (RhoA) | Aggressive breast cancer | Treatment inhibited angiogenesis and the growth of tumors (more than 90%) in absence of toxicity. | [441] |
| Urethane-induced lung cancer model A/J mouse | Folate–chitosan-graft-polyethylenimine (FC-g-PEI) | Intranasal-aerosol | Akt1, aserine/threonine-protein kinase | Lung cancer | Authors used plasmids encoding Akt1 shRNA. Aerosol delivery of FC-g-PEI/Akt1 shRNA complexes suppressed lung tumorigenesis through the Akt signaling pathway. |
[388] |
| Fischer 344 rats | High MW chitosan | Intranasal | NS1 gene of respiratory syncytial virus (RSV) | Respiratory syncytial virus infection | Authors used plasmids encoding siNS1. Rats treated with Chitosan-siNS1 prior to RSV exposure showed a switch from a Th2 to a Th1 phenotype and a reduction of CD4 + T cells and thereby lead to effective lung viral clearance without damaging airway tissues. |
[387] |
Chitosan and chitosan derivative formulations have been increasingly studied recently for delivery of therapeutic protein expressing plasmids in animal models for variety of applications including cancer, diabetes, anemia, hemophilia, osteosarcoma, arthritis, corneal disease and burn injuries using intratumoral, intraveneous, oral, intramuscular, subcutaneous, intraperitoneal, intratracheal, intrastroma and topical route of administration. A summary of these studies is provided in Table 8.
5.2. Chitosan-based siRNA nanoparticles
RNA interference (RNAi) is an innate mechanism whereby a small double-stranded RNA (dsRNA) can lead to sequence-specific gene silencing [5], [354]. RNAi has provided a potential new class of therapeutics [6] effective in mammalian cells [6], [355], [356], [357], [358] and has reached clinical trials [359], [360], [361]. However, direct delivery of siRNA to achieve RNAi continues to be problematic owing to their rapid extra/intra-cellular degradation by ubiquitous nucleases, limited blood stability, poor cellular uptake as well as off-target effects [211], [362], [363]. Despite the progress achieved through chemical modification to increase siRNA half-life, transfection efficiency, cellular targeting, and uptake remain as obstacles to effective delivery. Therefore, packaging systems which can both transport and protect, from ubiquitous nucleases, chemically unmodified/modified siRNA to target cells are required.
5.2.1. Therapeutic fields for siRNA
Chitosans have been increasingly studied for siRNA delivery in animal models in the fields of cancer, bone repair, rheumatoid arthritis, inflammatory diseases and infectious diseases (Table 9). In the field of cancer, the most important proposed application of gene therapy, Salva et al., targeted the vascular endothelial growth factor gene (VEGF) in a rat breast cancer model and found that chitosan (75 kDa, 75–85% DDA)–VEGF siRNA nanocomplexes had a remarkable suppressive effect on VEGF expression and tumor volume [364]. Han et al. targeted the transglutaminase 2 gene (TG2) in melanoma and breast cancer bearing nude mice and demonstrated that chitosan hydrogel nanocomplexes (CH-HG) (80% DDA, 161 kDa) TG2/siRNA (150 μg-siRNA/kg) delivered intratumorally permitted a higher localization of siRNA into the tumor; the incorporation of siRNA into CH-HG was indeed required for effective delivery and target modulation [365]. A decrease in tumor cell proliferation and angiogenesis and an increase of tumor cell apoptosis were observed by Kim et al., in an ovarian carcinoma nude mouse model following intravenous administration of chitosan-TPP-siRNA silencing Scr and Fgr family kinases twice a week at a dose of 150 μg-siRNA/kg [366]. Lee et al. administered poly-siRNA/glycol chitosan nanoparticles (psi-TGC) targeting VEGF specific siRNAs intravenously in a prostate cancer xenograft murine model; they found that chitosan nanoparticles provided sufficient in vivo stability for systemic delivery of siRNAs resulting in a knockdown of VEGF-specific mRNA (by 35.9%) associated with reduction in tumor size (by 80%) and vascularization [280]. This indicates that the inhibition of angiogenesis originated from VEGF-specific gene silencing. Howard et al. downregulated TNF-α expression in rheumatoid arthritis in collagen-induced arthritic (CIA) mice model by intra-peritoneal administration of chitosan (84% DDA, N:P ratio 63)–siTNF-α nanoparticles and found that downregulation of TNF-α-induced inflammatory responses arrested joint swelling [210]. However, despite these attempts to identify optimal physicochemical parameters for siRNA delivery [255], inconclusive results have been observed in the literature due to experimental discrepancies [14], [211]. For example, it was reported that intermediate DDA (80%) and high Mn (64–170 kDa) chitosans were more efficient than low Mn chitosans (10 kDa) in delivering siRNA [211], [255]. Yet high Mn chitosans were found to be cytotoxic [95], [309], [367], thus potentially limiting their use. Additionally, most of the reports evaluating the physicochemical parameters of chitosan/siRNA nanoparticles were performed at high N:P ratios (N:P > 25) [14], [211], [255]. Such formulations possess significant practical problems including limited dosing due to aggregation and the nonspecific effects of large quantities of soluble chitosan [230].
It has been demonstrated that maximization of in vitro transfection efficiency for the delivery of pDNA depends on a fine balance between these tunable parameters of chitosan [26], [259], [260]: maximum transgene expression was found for DDA:Mn values that run along a diagonal from high DDA/low Mn to low DDA/high Mn [26]. It has also been demonstrated that specific chitosan formulations [DDA, Mw, and N:P ratio] are able to efficiently express transgene in vivo [109], [235] and/or trigger an anti-transgene immune response [109]; therefore, chitosan/polynucleotide nanoparticles must be designed based on the fine-tuning of chitosan parameters for application-specific purposes such as gene therapy or genetic vaccination.
5.3. Biodegradation and biodistribution of chitosan–polynucleotide nanoparticles
The extent of chitosan biodegradability depends on Mn and DDA and needs to be considered when new chitosan formulations are proposed for gene therapy, along with the fact that the degradation pathways of chitosans and chitosan derivatives in vivo have not been fully elucidated. Chitosan is degraded in vertebrates by enzymes such lysozymes and by bacterial enzymes in the colon which hydrolyze glucosamine–glucosamine, glucosamine–N-acetyl-glucosamine and N-acetyl-glucosamine–N-acetyl-glucosamine linkages [17], [368], [369], [370].
5.3.1. Nanoparticle biodistribution after intravenous administration
Richardson et al. followed the biodistribution of chitosan in rat after IV injection of radioiodinated (125I) chitosans at different Mn with a DDA around 60% [95]. Five and sixty minutes after administration, 125I-labeled chitosans recovered from the organs varied significantly, the majority was found in blood, liver and lung, while small doses were found in spleen, kidney, thyroid and heart. More specifically for 125I-labeled chitosan of Mn > 5 kDa, after 5 min, less than 15% of the injected fraction was recovered from blood with more than 50% already in the liver. The chitosan with Mn < 5 kDa was cleared from the circulation more slowly, with 30% of injection fraction present in the blood, and less than 30% in the liver, at 60 min post-administration. Gao et al. show that chitosan, liposomal and JetPEI based formulations all greatly improved the stability and biodistribution of siRNA [371]. However, a high accumulation of the chitosan-based siRNA nanocomplexes was observed within the kidney 24 h post-administration. This observation was confirmed be Lee et al. when monitoring by fluorescent imaging, the biodistribution of multimeric (polymerized) siRNA (poly-siRNA-FPR675-labeled)/glycol chitosan nanoparticle (psi-TGC), intravenously administrated into prostate tumor xenograft mice [280]. This significant retention of chitosan nanoparticles in the kidneys, 3 days post-injection, may originate from their longer blood circulation time compared to free siRNAs. Lee et al. suggested that the retention of poly-siRNA/glycol chitosan nanoparticles (psi-TGC) in the kidneys does not represent a health risk since their excretion from the body via the kidney takes place several days post-injection. Gao et al. suggested that nanoparticle accumulation may reflect the mucoadhesion of chitosan to the mucosal epithelium lining the kidney and could be exploited for siRNA-based treatments for renal diseases [371]. Yan et al. evaluated the pharmacokinetic profiles of 5-fluorouracil-loaded N-succinyl–chitosan nanoparticles (without NA) in tail vein injected rats and demonstrated that nanoparticles accumulate mainly in the tumor [372] while Gao et al. and Mao et al. showed an accumulation in the kidneys of chitosan/siRNA and chitosan/pDNA nanoparticles [66], [371].
Time-dependent biodistribution studies were also performed by Huh et al. on siRNA–Glycol chitosan–PEI nanoparticle using Cy5.5-siRNA [373]. Cancer bearing mice injected intravenously with nanoparticles showed a strong fluorescence signal in tumor tissue within 1 h of injection. Nanoparticles preferentially accumulated in tumors for up to 2 days, indicating their higher tumor-targeting ability. This supports the work of Lee et al. [280] showing that nanoparticles could preferentially deliver siRNAs to targeting tumor tissue for at least 3 days. The prolonged circulation time of polymeric nanoparticles allows them to extravasate and accumulate in tumors tissue, due to the disorganized and defective vascular architecture in tumors, an effect referred to as the enhanced permeability and retention (EPR) effect in tumor tissue [280], [374], [375]. These results indicate the stability of nanoparticles in the bloodstream until they reach the tumor and are supported by the group of Torchilin et al. who showed that nanoparticles with size ranges of 100–500 nm accumulate efficiently at tumor sites through the EPR effect in vivo [376]. In contrast, Lee et al. [280] showed that tumor-bearing mice injected with the poly-siRNA only or psi-PEIs did not show any significant increase of fluorescence signal at the tumor site during the first 3 days post-injection and most of psi-PEIs were found in the liver. As expected, among all excised organs, tumor tissue of psi-Thiolate Glycol Chitosan (TGC) treated mice showed the strongest fluorescence signals. These observations of nanoparticle biodistribution are also supported by Cheng et al. following intravenous injection of galactosylated chitosan–pDNA nanoparticles expressing mGM-SCF and mIL21 transgenes in an animal model bearing colon and liver tumors [377]. The expression levels of the transgenes increased significantly in colon cancer and liver compared to those in spleen, kidney, lung, muscle, and heart.
5.3.2. Nanoparticle biodistribution after oral administration
Despite the harsh environment of the gastrointestinal tract, the effective in vivo delivery of pDNA was clearly demonstrated by oral administration of chitosan-based nanoparticles. Oral administration of chitosan and chitosan derivatives pDNA nanoparticles led to efficient gene transduction in the intestinal epithelium [314], [341], [378], [379], [380], [381], stomach [378], [379], [380] and gastric and upper intestinal mucosa [382]. It is well established that chitosan and chitosan derivatives, depending on their DDA and Mn are able to protect the complexed NA against nuclease degradation. Chitosan and chitosan derivatives, positively charged at gastric and duodenal pH (4.0–6.5) and neutral at intestinal alkaline pH (7.0–8.5), are able to increase trans- and para-cellular intestinal permeability. Gene expression may be triggered after the solubility/degradation of the chitosan-based nanocomplexes in acidic pH environment and/or by microflora of the colon [383].
Kai and Ochiya used chitosan alone (DDA 100%, MW 9.5 × 105) and N-acetylated chitosan to deliver the β Gal reporter gene to intestine by oral administration confirmed gene expression for both chitosans at the duodenum, jejunum, ileum, and colon, 5 days post-administration [314]. Bowman et al., using chitosan (84% DDA)–Factor VIII pDNA nanoparticles mixed with gelatin for oral administration to FVIII exon 16 knock-out mice, showed the presence of plasmid DNA in Peyer's patch but also in stomach, ileum, liver, and spleen 72 h post-administration [250]. The finding of plasmid DNA in non-gastrointestinal tissues indicates that pDNA was able to cross the intestinal barrier to reach either the blood or lymph circulation. It is noteworthy that plasmid biodistribution was determined by qualitative assays and confirmed by real-time PCR where none of real-time PCR results achieve statistical significance, due to the high variability. Zheng et al., using quaternized chitosan–trimethylated chitosan oligomer (60% DDA and 221 kDa MW) plasmid DNA expressing a GFP reporter to orally feed nude mice and showed GFP expression in the stomach, duodenum, jejunum, ileum, and large intestine [382]. To date, few investigations related to time-dependent biodistribution of orally administrated nanoparticles have been performed. Tian et al. [384] encapsulated pDNA (encodes for Major Capsid Protein, MCP, of LCD virus and GFP) in chitosan (80% DDA, 1080 kDa MW) microspheres through an emulsion-based methodology in orally vaccinated fish, and detected MCP recombinant mRNA in gill, intestine, spleen and kidney 10–90 days post-vaccination. These results were confirmed by a significant enhancement of green fluorescent protein production in the same tissues 2–90 days post-vaccination.
5.3.3. Nanoparticle biodistribution after mucosal administration
Intranasal administration of chitosan-based pDNA/siRNA nanoparticles has been mainly for nasal vaccination to target the nasal-associated lymphoid tissue of the pharynx [385]. Biodistribution of chitosan–siRNA nanoparticles following intranasal and intratracheal administration was studied in EGFP-transgenic mice by monitoring EGFP gene silencing [14], [386]. Chitosan (84% DDA, 114 kDa Mn) siRNA–EGFP nanoparticles, administrated intranasally over a 5-day period achieved a reduction up to 43% of the EGFP expression in bronchiolar epithelia both in lower and upper regions of the mouse lung [14]. Moreover, a higher EGFP gene silencing was achieved in the bronchial epithelial cells of transgenic mice when using guanidinylated chitosan GFP–siRNA or salbutamol modified guanidinylated chitosan GFP–siRNA nanoparticles instead of unmodified chitosan (92% DDA and 50 kDa Mn) GFP–siRNA nanoparticles [386]. Similar results were obtained by Kong et al. with intranasal administration of high Mn chitosan-based nanoparticles to deliver plasmid DNA encoding for siNS1 for the control of viral load in an RSV infected mice lung [387]. Hiang et al. succeeded in delivering, by aerosol, plasmid DNA producing shRNA, complexed to folate–chitosan-graft-PEI, to specifically silence the kinase AKT1 in the targeted lung tumor cells [388].
Csaba et al. showed that intratracheal administration of low Mn pentasodium tripolyphosphate chitosan (Cs/TPP) pCMV-XGal nanoparticles loaded with pCMV-XGal mediated a strong gene expression in the mouse lungs 72 h post-administration in contrast to high Mn CS/TPP nanoparticles and naked pDNA [213]. However Koping-Hoggard et al. [89] monitored luciferase expression in mouse lung, 24 h after intratracheal administration of nanoparticles, and found a 120- to 260-fold increase in luciferase gene expression with low Mn compared to high Mn chitosan. Later this group found that luciferase expression in the mouse lung was 4-fold higher with trisaccharide-substituted chitosan oligomers than with the corresponding linear chitosan oligomers [13]. These results emphasize the importance of using well-defined low Mn nanoparticles for intratracheal route of administration.
5.3.4. Nanoparticle biodistribution after intramuscular and subcutaneous administration
Fewer studies address the biodistrution of chitosan-based pDNA/siRNA nanoparticles following intramuscular or subcutaneous administration. Jean et al. [109] used different chitosan formulations to deliver, IM or SC, plasmid DNA encoding for FGF-2 and PDGF-BB recombinant proteins, and showed the efficient expression of the transgenes in muscle cells and in surrounding tissues. In addition, the recombinant protein levels in Balb/c mice muscle were significantly higher when the pDNA were complexed to low Mn chitosans (92% and 80% DDA and 10 kDa MnW) compared to high Mn chitosans (80% DDA and 80 kDa Mn). Mandke and Singh demonstrated that the IM administration into the mice anterior tibialis muscle of a bicistronic plasmid, encoding for both IL-4 and IL-10, complexed to N-linoleyl low Mn chitosan or N-oleyl low Mn chitosan (85% DDA, 50 kDa MW) led to a sustained expression of IL-4 and IL-10 with a maximum 1 week following the injection of a single dose of nanoparticles [389]. The expression of the transgene gradually decreased until the end of 6 week period, probably due to the deactivation of the promoter and/or turnover of muscle fibers [390]. The recombinant proteins reached the blood circulation and a therapeutic effect was confirmed by the control of the blood glucose levels of a Diabetes type 1 induced murine model. Jean et al. also succeeded to efficiently deliver IM and SC low Mn chitosan (92% DDA and 10 kDa MW) with pDNA encoding for GLP-1 nanoparticles. Their therapeutic effect was confirmed in type 2 diabetes rat model, ZDF by dosing GLP-1 plasma levels (fivefold higher than in non-treated animals) and by an increase in plasma insulin concentration [235].
5.4. Potential clinical applications of chitosan–NA polyplexes
5.4.1. Clinical trials with chitosan
Since the first gene therapy clinical trial initiated by Rosenberg in 1980 [391] over 1840 gene therapy clinical trials worldwide have been approved [392]. Adenovirus and retrovirus are the most widely used vectors. More than 60% of gene therapy clinical trials are for cancer treatment [392]. Liposomes and polymers formulated as nanocarriers for DNA and siRNA are also currently under clinical investigation. For example, Liposome (DOPA) [393], cationic lipid Allovectin-7® [394] and PEI [395] have been applied in human trials for pDNA delivery via intranasal, intralesional and intravesical administration to deliver, respectively, the cystic fibrosis transmembrane regulator gene (CFTR), for patients with cystic fibrosis, leukocyte antigen-B7 and beta-2 microglobulin for patients with advanced metastatic melanoma and H19 gene (non-protein coding RNA gene) regulatory sequences in patients with superficial bladder cancer. Also, systemically administrated nanoparticles formed of linear cyclodextrin-based polymer (CDP), a human transferrin protein (TF) targeting ligand, polyethylene glycol (PEG) and siRNA-RRM2 CDP-PEG-Tf [396] have been evaluated for the silencing of the M2 subunit of ribonucleotide reductase in patients with solid cancers. To date no gene therapy clinical trials have been reported using chitosan-based pDNA or siRNA nanoparticles. However clinical trials involving chitosan are in progress including the Norwalk VLP vaccine, a virus-like particle (VLP) vaccine adjuvanted with monophosphoryl lipid A and chitosan. In this study, chitosan is used as a mucoadhesive to nasal mucus and epithelial cells to prolong antigen adherence [397]. Table 10 summarizes the most significant human studies and clinical trials using chitosan over the last decade.
Table 10.
Summary of the most significant human clinical trials using chitosan over the last decade.
| Study title | Product information | Purpose | Conclusion | Location | Phase | References |
|---|---|---|---|---|---|---|
| Use of chitosan for Femoral (USF) haemostasis after percutaneous procedures. (NCT00716365) | HemCon® containging chitosan. HemCon® is used by the US army to control traumatic bleeding. |
Control of vascular access site bleeding and return to haemostasis following percutaneous coronary angiography. | The HemCon® pad decreases time-to-haemostasis when compared to the regular pad and an incidence of haematoma of 6% when compared to the regular pad (14.8%) group. | Israel | 4 | [442], [443] |
| Hemostasis of oral surgery wounds with the HemCon Dental Dressing. (NCT00707486) | HemCon® Dental Dressing (HDD) manufactured from freeze dried chitosan. | Control of bleeding during dental surgeries. | HDD significantly shortened bleeding time in patients undergoing oral surgery procedures including those taking oral anticoagulants. HDD also improved surgical wound healing compared to the control groups. | United States | 3 | [443], [444] |
| BST-DERMON versus standard of care in the treatment of diabetic foot ulcers. (NCT00434538) | BST-DermOn is a wound dressing of an aqueous mixture of chitosan, hydrochloric acid (HCl) and disodium beta-glycerol phosphate. | Evaluation of the safety and efficacy of BST-DermOn in the repair of diabetic foot ulcers. | This study was stopped forfinancial reasons. | Canada | 3 | [443] |
| BST-CarGel versus microfracture in the repair of articular cartilage lesions in the knee. (NCT00314236) |
BST-CarGel is a chitosan glycerol phosphate solution BST-CarGel® is mixed with whole blood of patient, to provide a polymer-reinforced blood clot for improved cartilage repair. |
Treatment of damaged cartilage in the knee with BST-CarGel with microfracture compared microfracture alone. Evaluation of amount and quality of cartilage repair tissue. | BST-CarGel treatment resulted in cartilage repair of higher quantity and quality with a similar safety profile to microfracture, known as orthopedic standard of care | Canada Korea Spain |
3 | [443] |
| Efficacy and safety of chitosan HEP-40 in the management of hypercholesterolemia: a randomized, multicenter, placebo-controlled trial. (NCT00454831) | HEP-40 chitosan (enzymatic polychitosamine hydrolysate) is low-MW chitosan ~ 40 kDa; DDA 93%. | Efficacy and safety of HEP-40 chitosan as sole treatment to lower LDL-cholesterol levels in patients with mild to moderate levels of cholesterol | As sole treatment, HEP-40 chitosan was effective and safe in lowering LDL-C concentrations in patients with low-to-moderate hypercholesterolemia but not as effective as statins. HEP-40 chitosan was also associated with significant reduction of total cholesterol starting four weeks post treatment. | Canada | 2 | [443], [445] |
| A study of HS219 in chronic kidney disease patients on hemodialysis with hyperphosphatemia. (NCT01039428) | HS219 is a chitosan-loaded chewing gum. | Efficacy and safety of HS219, three times a day for 3 weeks as regimen, to patients with hyperphosphatemia and undergoing hemodialysis. The patients included in this study have serum inorganic phosphorus that is not well controlled with calcium carbonate or sevelamer hydrogen chloride. | Information not available. | Japan | 2 | [443] |
| Norwalk vaccine study. (NCT00806962) |
Virus-like particle (VLP) vaccine adjuvanted with monophosphoryl lipid A (MPL) and chitosan. | Safety and immunogenicity of Norwalk VLP vaccine. | Adjuvanted Norwalk VLP vaccine administrated intranasally was highly immunogenic while second dose of vaccine significantly increased the level of specific antibodies in serum antibody responses. | United States | 1 | [397], [443] |
| A poloxamer/chitosan in situ forming gel with prolonged retention time for ocular delivery. | Gel combination of a thermosetting polymer, poly (ethylene oxide)–poly (propylene oxide)–poly (ethylene oxide) (PEO–PPO–PEO, poloxamer), with chitosan (MW 190–310 kDa; DDA 75–85%). | An ophthalmic delivery system with improved mechanical and mucoadhesive properties that could provide prolonged retention time for the treatment of ocular diseases. | Chitosan increased retention time fourfold in comparison with a conventional solution. Chitosan confers mucoadhesive force in a concentration-dependent manner. | Brazil | [446] | |
| Analgesic efficacy and safety of a novel intranasal morphine plus chitosan formulation compared to oral and intravenous route of morphine administration in a postsurgical dental pain model. | Active nasal sprays containing morphine (75 or 150 mg/mL) solubilized in an aqueous solution, combined with chitosan glutamate (5 mg/mL). | Efficacy and safety of intranasal morphine vs oral and intravenous morphine to provide postsurgical opioid analgesia for moderate-to-severe pain. | Single doses of intranasal morphine (7.5 mg and 15 mg) were safe and well tolerated and provide rapid analgesia to patients with acute moderate-to-severe postsurgical pain. The 15 mg dose of intranasal morphine showed an efficacy profile similar to the 7.5 mg dose of intravenous morphine. Both treatments demonstrated rapid onset of efficacy that persisted throughout the 6-h assessment period. | United States | [447] | |
| Long-term clinical outcome of percutaneous injection with holmium166-chitosan based chitosan complex (Milican) for the treatment of small hepatocellular carcinoma. | Holmium166-chitosan based complex (Milican) to retain the radioactive material within the tumor. | Long-term tumor response and safety of percutaneous holmium166-chitosan based complex injection therapy for small hepatocellular carcinoma as a local ablative treatment. | Among the 40 treated patients and during the long period follow-up, tumors recurred in 28 patients of which 24 recurred at another intrahepatic site. The 1-year and 2-year cumulative local recurrence rates were 18.5% and 34.9%, respectively. The survival rates at 1, 2, and 3 years were 87.2%, 71.8%, and 65.3%, respectively. | Korea | 2b | [448] |
| Induction of protective serum response (meningococcal bactericidal, diphtheria-neutralizing antibodies and mucosal immunoglobulin A) in volunteers by nasal insufflations of Neisseria meningitidis serogroup C polysaccharide–CRM197 conjugate vaccine mixed with chitosan. | Neisseria meningitidis serogroup C polysaccharide–CRM197 conjugate vaccine mixed with chitosan (formulation of chitosan glutamate G213). | Efficiency of nasal insufflations of the Neisseria meningitidis serogroup C polysaccharide–CRM197 conjugates vaccine mixed with chitosan. | A simple nasal insufflation of existing MCP–CRM197 conjugate vaccines in chitosan offers an inexpensive but effective needle-free prime and boost immunization against serogroup C N. meningitidis and diphtheria. | United Kingdom | 1 | [449] |
| Chitosan delivery systems for the treatment of oral mucositis: in vitro and in vivo studies. | Nystatin incorpored in to gel composed of chitosan at different molecular weights (MW: 272,000–1,400,000, DDA: 80–84%). | Efficiency of an occlusive bioadhesive system (chitosan based gel) for prophylaxis and/or treatment of oral mucositis in comparison with a commercial suspension. | Concentration of nystatin in saliva was higher with gels prepared with low molecularweight chitosan. Nystatin saliva levels above the minimum inhibitory concentration were maintained to 90 min. | Turkey | [450] | |
| Intranasal immunization with genetically detoxified diphtheria toxin induces T cell responses in humans: enhancement of Th2 responses and toxin-neutralizing antibodies by formulation with chitosan. | Powder formulation containing CRM197 with chitosan and mannitol. | Potential of intranasal booster immunization (CRM197 alone or with chitosan) to induce systemic T cell responses. | Th2 responses were enhanced following booster immunization with CRM197 and chitosan, which correlate with protective levels of toxin-neutralizing antibodies in intranasally boosted individuals. | Ireland | [451] |
5.4.2. Off target and side effects associated with chitosan-based nanoparticles
The success of nanoparticle-based gene therapy depends on its effectiveness and absence of serious side effects. Toxicity and off-target effects are critical factors that must be taken into consideration when designing gene delivery systems for in vivo use. Two types of off-target effects need to be avoided or minimized i) interference with non-targeted gene expression, and ii) undesired immune stimulation through recognition of nanoparticles or nanoparticle compounds by the innate system. Off-target effects associated with RNAi have attracted attention. Indeed RNAi can recognize and interfere with the expression of mRNAs that share partial homology with the target mRNA. Hence, the importance of carefully selecting siRNA sequences by using algorithms and by performing in vitro tests cannot be underestimated.
The effects of chitosan-based NA nanoparticles on the induction of immune responses have not been completely elucidated, with exceptions for gene therapy studies aimed at recombinant cytokine expression [389], [398] or immune response induction associated with antigen-specific immunization [342], [379], [399]. Lee et al. also showed that no significant amounts of pro-inflammatory cytokine (IL-6) were produced by human prostate cancer cells treated with chitosan-based Poly-siRNA Glycol nanoparticles [280]. Wiegand et al. showed that depending on incubation time and concentration, high Mn chitosans (120 kDa) compared to low Mn (5 kDa and 0.22 kDa) are more able to stimulate the release of inflammatory cytokines (Il-6 and Il-8) by HaCaT cells [367]. Huang et al. [309] suggested that cellular uptake of both chitosan as a soluble molecule and in a condensed nanoparticle were not associated with cytotoxicity. In fact, viability of A549 cells was generally not affected by soluble chitosan and nanoparticles at low concentrations but showed a progressive decline when exposed to increasing concentrations beyond 0.741 mg/ml. In vitro studies showed that Mn in the range 10–213 kDa did not affect cytotoxicity while decreasing the DDA of the polymer from 88% to 61% attenuated cytotoxicity. The latter supported the findings of Ekrami and Shen that cytotoxicity is directly related to nanoparticle surface charge density [400]. Wiegand, Winter et al. confirmed that high Mn chitosans (120 kDa) compared to low Mn (5–0.22 kDa) can negatively impact HaCat cell viability and proliferation in vitro [367]. In addition, Richardson, Kolbe et al. showed that chitosans of different Mn (< 5 kDa, 5–10 kDa and > 10 kDa) displayed little cytotoxicity against CCRF-CEM and L132 cells (IC50 > 1 mg/ml), with no hemolytic effects [95]. In vivo, the LD50 of chitosan in mice generally exceeds 16 g/kg [401].
5.4.3. Non-allergenic character of chitosan-based systems
Many investigations have been performed in order to elucidate a possible relationship between allergy and the presence of chitin in crustaceans. This is an important issue if chitin extracted from crustaceans is used for production of chitosan and chitosan-derivatives for biomedical applications. Muzzarelli revealed in his 2010 review some conceptual and methodological errors in different research reports where a hasty link was made between allergy and chitin [402]. Taking into consideration the contradictory findings of different research reports, Muzzarelli concluded that purified chitins and chitosans were free of proteins, fats and contaminants that could induce an allergic reaction and should be classified as non-allergenic chemicals. In addition, results from many studies have converged to a similar conclusion that chitin can stimulate the innate immune response by activating macrophages and other innate immune cells, leading to type 1 cytokines (IL-12, TNF-α, IL-18, IFN-γ) production which downregulate type 2 allergic immune responses [402], [403], [404], [405]. Strong et al. also showed that intranasal application of chitin microparticles is an effective treatment to reduce serum IgE, airway hyper-responsiveness and lung inflammation in mouse models of allergy [406]. The treatment not only increased Th1 cytokines IL-12, IFN-γ and TNF-α but also decreased IL-4 production, a cytokine involved in B cell production of allergen-specific IgE.
To date, no research group has reported an allergic response associated with the use of chitosan or chitosan-derivative based NA nanoparticles. However, there are many who have investigated the potential of chitosan-based DNA nanoparticles as DNA vaccine for allergy treatment [341], [379], [407]. Roy et al. showed that chitosan (Mn 390 kDa)-based pDNA nanoparticles (sized to 100–200 nm) coding for dominant peanut allergen gene (Arah2) and delivered orally can modify the immune system in mice and protect against food allergen-induced hypersensitivity [341]. The IgE and histamine levels as well the anaphylactic response were substantially lower in the vaccinated animals compared to those in the control group. Li et al. showed that an oral vaccine of chitosan (Mn 390 kDa; DDA 83.5%)-based pDNA nanoparticles (100–400 nm) coding for mite dust allergen Derp2 prevented subsequent sensitization of Th2 cell-regulated specific IgE responses by increasing IFN-γ and decreasing IL-4 in serum [379]. In this study, no immunostimulatory activity was reported when using chitosan alone. However, since Lee et al. [408] and Da Silva et al. [409] reported a size-dependant immunostimulation by chitin; it would be of interest to investigate the immunostimulatory potential of chitosan particles of different sizes.
5.4.4. Comparison with other delivery systems
Liposomes and lipoplexes as nonviral transfection vehicles for plasmid and RNAi pose toxicity concerns. The repeated administration, in vivo, of lipid-based delivery systems caused phospholipidosis [410]. Intravenous injection of stable NA-lipid particles (SNALP) has successfully targeted the liver to silence the apolipoprotein B (ApoB) gene in mice and nonhuman primates [357]. However, a significant 20-fold transient elevation in serum transaminases (aspartate transaminase, alanine transaminase) indicative of hepatocellular necrosis was identified at the effective dose. Liposomal formulations of NAs are known inducers of inflammatory cytokines including tumor necrosis factor-alpha, interferon-gamma, and interleukin-6 related to liver damage [411]. Polyethylene glycol (PEG) modification of liposomes (PEGylation), for the purpose of reducing their toxicity, was also found to elicit acute hypersensitivity after repeated dosing [412]. Similarly, the highly studied cationic family of synthetic polymers such as PEI possess high gene transfer efficiency but are associated with significant toxicity issues [6], [413] limiting their broad use in clinical trials. PEI cytotoxicity was characterized as a two-phase process where the polycation–cell interaction induces loss of cell membrane integrity and the induction of apoptosis. Insights into PEI toxicity highlight the importance of polycation/organelle interactions (i.e., mitochondria and lysosomes) on the induction of toxicity [414], [415]. In general, cationic polymers display less toxicity associated with cytokine induction and immune activation compared to their cationic lipid counterparts [15].
5.5. Future prospects
Chitosan and chitosan derivatives as nanocarriers for gene therapy face the traditional barriers such as susceptibility to degradation by nucleases of their cargo, low cellular membrane permeability, low solubility at physiological pH and lack of specificity. In order to increase translation from bench to bedside, efforts are focusing on the chemical and biological modification of chitosan in order to increase stability, specificity and endosomal escape. Stabilization against aggregation of nanocomplexes for in vivo delivery may be achieved by grafting a neutral polymer to chitosan in order to reduce the surface charge and increase the stability of chitosan–polynucleotide complexes. Koping-Hoggard et al. demonstrated that high levels of transfection may be achieved using low Mn chitosans [89]. Attempts to specifically target the cell receptors for DNA delivery have been performed using PEI grafted chitosan, galactosylated chitosan, galactosylated chitosan-graft-poly(vinylpyrrolidone) (PVP), trimethylated chitosan oligomers, N-dodecylated chitosan, deoxycholic acid modified chitosan and ligand attached chitosans [167], [179], [218], [222], [227], [416], [417]. Specific ligands on chitosan-based nanocomplexes should enhance their in vivo biodistribution/accumulation into the desired target tissues/cells. However systemic administration of these complexes is expected to achieve preferential uptake by desired target only if a suitable targeting moiety is appropriately incorporated into the formulation [152], [153], [155], [300], [371], [386], [398], [418], [419], [420].
Mohammadi et al. coupled a specific ligand (FAP-B) to chitosan–DNA nanoparticles via electrostatic attraction in order to target lung epithelial cells and showed that the transfection efficiency of Chitosan–DNA–FAP-B nanoparticles (size of 279 ± 27 nm) was about 10-fold higher than chitosan–DNA nanoparticles [419]. However, in vivo studies have yet to be performed to confirm nanoparticle efficiency. Luo et al. designed a new siRNA nanocarrier to target specifically lung cells by coupling salbutamol, a β2-adrenoceptor agonist, to guanidinylated chitosan [386]. This nanocarrier of 20–90 nm size remarkably enhanced the efficacy of gene silencing in vitro and in vivo when using transgenic mice for EGFP gene silencing. Therapeutic siRNA should be complexed to this new category of nanocarrier to confirm RNAi efficiency in vivo. Another innovative and promising approach that needs additional characterization was explored by Kumar et al. who demonstrated that a simple external magnetic field can help direct the nanoparticles to a specific site in vivo [421]. These magnetic nanoparticles were conjugated with plasmid DNA expressing EGFP and then coated with chitosan. These particles when injected into mice through the tail vein were directed to heart and kidney by means of external magnets. Also, chitosan modifications can be considered to enhance the functional delivery of the NA payload through an efficient endosomal escape of nanoparticles. Transfection efficiency was improved by modifying depolymerized trimethylated chitosans with a histidine moiety [154], where histidine can lead to the osmotic swelling and membrane disruption and eventually the vesicular escape of DNA. The imidazole group of histidine possesses a pK a near 6 thus absorbing protons and possess buffering capacity in the endosomal pH range (pH 5–6.5) [154], [422] possibly increasing the proton sponge effect.
6. Conclusion
In summary, chitosans have been investigated and developed to deliver NAs for nearly two decades and are now entering a stage of maturity where first results from human clinical trials are imminent. Many of the challenges associated with production and accurate analyses of specific chitosans have been overcome, resulting in the identification of specific systems that have high efficiency for protein expression from plasmid vectors and gene silencing using siRNA. The physicochemical and biological basis for the successful function of these systems has also been partly elucidated in vitro and to a lesser extent in vivo. Remaining challenges to effectively treat human disease include the production of sufficient quantities of highly characterized nanoparticles with features that satisfy regulatory requirements, and the matching of current capabilities in protein expression, gene silencing, targeting and safety profile to specific clinical indications.
Footnotes
This review is part of the Advanced Drug Delivery Reviews theme issue on “Polysaccharide-based systems in drug and gene delivery”.
References
- 1.Friedmann T., Robin R. Gene therapy for human genetic disease? Science. 1972;175:949–955. doi: 10.1126/science.175.4025.949. [DOI] [PubMed] [Google Scholar]
- 2.Dewey R.A., Morrissey G., Cowsill C.M., Stone D., Bolognani F., Dodd N.J.F., Southgate T.D., Klatzmann D., Lassmann H., Castro M.G., Lowenstein P.R. Chronic brain inflammation and persistent herpes simplex virus 1 thymidine kinase expression in survivors of syngeneic glioma treated by adenovirus-mediated gene therapy: Implications for clinical trials. Nat. Med. 1999;5:1256–1263. doi: 10.1038/15207. [DOI] [PubMed] [Google Scholar]
- 3.Fox J.L. Gene-therapy death prompts broad civil lawsuit. Nat. Biotechnol. 2000;18:1136. doi: 10.1038/81104. [DOI] [PubMed] [Google Scholar]
- 4.Niidome T., Huang L. Gene therapy progress and prospects: nonviral vectors. Gene Ther. 2002;9:1647–1652. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]
- 5.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.de Fougerolles A., Vornlocher H.P., Maraganore J., Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discovery. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulligan R., Howard B., Berg P. Synthesis of rabbit beta-globin in cultured monkey kidney cells following infection with a SV40 beta-globin recombinant genome. Nature. 1979;277:108–114. doi: 10.1038/277108a0. [DOI] [PubMed] [Google Scholar]
- 8.Filion D., Lavertu M., Buschmann M.D. Ionization and solubility of chitosan solutions related to thermosensitive chitosan/glycerol-phosphate systems. Biomacromolecules. 2007;8:3224–3234. doi: 10.1021/bm700520m. [DOI] [PubMed] [Google Scholar]
- 9.Lv H., Zhang S., Wang B., Cui S., Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 10.Boussif O., Lezoualc'h F., Zanta M.A., Mergny M.D., Scherman D., Demeneix B., Behr J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishii T., Okahata Y., Sato T. Mechanism of cell transfection with plasmid/chitosan complexes. Biochim. Biophys. Acta, Biomembr. 2001;1514:51–64. doi: 10.1016/s0005-2736(01)00362-5. [DOI] [PubMed] [Google Scholar]
- 12.Lee M., Nah J.W., Kwon Y., Koh J.J., Ko K.S., Kim S.W. Water-soluble and low molecular weight chitosan-based plasmid DNA delivery. Pharm. Res. 2001;18:427–431. doi: 10.1023/a:1011037807261. [DOI] [PubMed] [Google Scholar]
- 13.Issa M.M., Koping-Hoggard M., Tommeraas K., Varum K.M., Christensen B.E., Strand S.P., Artursson P. Targeted gene delivery with trisaccharide-substituted chitosan oligomers in vitro and after lung administration in vivo. J. Control. Release. 2006;115:103–112. doi: 10.1016/j.jconrel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 14.Howard K.A., Rahbek U.L., Liu X., Damgaard C.K., Glud S.Z., Andersen M.O., Hovgaard M.B., Schmitz A., Nyengaard J.R., Besenbacher F., Kjems J. RNA interference in vitro and in vivo using a novel chitosan/siRNA nanoparticle system. Mol. Ther. 2006;14:476–484. doi: 10.1016/j.ymthe.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Al-Dosari M.S., Gao X. Nonviral gene delivery: principle, limitations, and recent progress. AAPS J. 2009;11:671–681. doi: 10.1208/s12248-009-9143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Fuentes M., Alonso M.J. Chitosan-based drug nanocarriers: where do we stand? J. Control. Release. 2012;161:496–504. doi: 10.1016/j.jconrel.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 17.Kean T., Thanou M. Biodegradation, biodistribution and toxicity of chitosan. Adv. Drug Deliv. Rev. 2010;62:3–11. doi: 10.1016/j.addr.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Muzzarelli R.A.A. Chitins and chitosans for the repair of wounded skin, nerve, cartilage and bone. Carbohydr. Polym. 2009;76:167–182. [Google Scholar]
- 19.Hoemann C.D., Sun J., Legare A., McKee M.D., Buschmann M.D. Tissue engineering of cartilage using an injectable and adhesive chitosan-based cell-delivery vehicle. Osteoarthr. Cartil. 2005;13:318–329. doi: 10.1016/j.joca.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Chevrier A., Hoemann C.D., Sun J., Buschmann M.D. Chitosan-glycerol phosphate/blood implants increase cell recruitment, transient vascularization and subchondral bone remodeling in drilled cartilage defects. Osteoarthr. Cartil. 2007;15:316–327. doi: 10.1016/j.joca.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Chevrier A., Hoemann C.D., Sun J., Buschmann M.D. Temporal and spatial modulation of chondrogenic foci in subchondral microdrill holes by chitosan-glycerol phosphate/blood implants. Osteoarthr. Cartil. 2011;19:136–144. doi: 10.1016/j.joca.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 22.Hoemann C.D., Sun J., McKee M.D., Chevrier A., Rossomacha E., Rivard G.E., Hurtig M., Buschmann M.D. Chitosan-glycerol phosphate/blood implants elicit hyaline cartilage repair integrated with porous subchondral bone in microdrilled rabbit defects. Osteoarthr. Cartil. 2007;15:78–89. doi: 10.1016/j.joca.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 23.Iliescu M., Hoemann C.D., Shive M.S., Chenite A., Buschmann M.D. Ultrastructure of hybrid chitosan-glycerol phosphate blood clots by environmental scanning electron microscopy. Microsc. Res. Tech. 2008;71:236–247. doi: 10.1002/jemt.20545. [DOI] [PubMed] [Google Scholar]
- 24.Hoemann C.D., Chen G., Marchand C., Tran-Khanh N., Thibault M., Chevrier A., Sun J., Shive M.S., Fernandes M.J., Poubelle P.E., Centola M., El-Gabalawy H. Scaffold-guided subchondral bone repair: implication of neutrophils and alternatively activated arginase-1 + macrophages. Am. J. Sports Med. 2010;38:1845–1856. doi: 10.1177/0363546510369547. [DOI] [PubMed] [Google Scholar]
- 25.Piramal_Healthcare . EU market approval: Piramal Enterprises Presents Positive Results of Pivotal Phase III Clinical Trial for BST-CarGel® at the World Congress of the International Cartilage Repair Society (ICRS) Press Release; 2012. [Google Scholar]
- 26.Lavertu M., Methot S., Tran-Khanh N., Buschmann M.D. High efficiency gene transfer using chitosan/DNA nanoparticles with specific combinations of molecular weight and degree of deacetylation. Biomaterials. 2006;27:4815–4824. doi: 10.1016/j.biomaterials.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 27.Malmo J., Sorgard H., Varum K.M., Strand S.P. siRNA delivery with chitosan nanoparticles: Molecular properties favoring efficient gene silencing. J. Control. Release. 2012;158:261–268. doi: 10.1016/j.jconrel.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Strand S.P., Danielsen S., Christensen B.E., Varum K.M. Influence of chitosan structure on the formation and stability of DNA-chitosan polyelectrolyte complexes. Biomacromolecules. 2005;6:3357–3366. doi: 10.1021/bm0503726. [DOI] [PubMed] [Google Scholar]
- 29.Park J.H., Saravanakumar G., Kim K., Kwon I.C. Targeted delivery of low molecular drugs using chitosan and its derivatives. Adv. Drug Deliv. Rev. 2010;62:28–41. doi: 10.1016/j.addr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Rudzinski W.E., Aminabhavi T.M. Chitosan as a carrier for targeted delivery of small interfering RNA. Int. J. Pharm. 2010;399:1–11. doi: 10.1016/j.ijpharm.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 31.Mao S., Sun W., Kissel T. Chitosan-based formulations for delivery of DNA and siRNA. Adv. Drug Deliv. Rev. 2010;62:12–27. doi: 10.1016/j.addr.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Braconnot H. Recherches analytiques sur la nature des Champignons. Ann. Chim. 1811;79:265–304. [Google Scholar]
- 33.Rouget C. Des substances amylacées dans des animaux, spécialement des Articulés (chitine) Compte Rendus. 1859;48:792–795. [Google Scholar]
- 34.Islam M.S., Khan S., Tanaka M. Waste loading in shrimp and fish processing effluents: potential source of hazards to the coastal and nearshore environments. Mar. Pollut. Bull. 2004;49:103–110. doi: 10.1016/j.marpolbul.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 35.Beaney P., Lizardi-Mendoza J., Healy M. Comparison of chitins produced by chemical and bioprocessing methods. J. Chem. Technol. Biotechnol. 2005;80:145–150. [Google Scholar]
- 36.Brück W.M., Slater J.W., Carney B.F. In: Chitin, Chitosan, Oligosaccharides and Their Derivatives. Kim S.-K., editor. CRC Press; Boca Raton: 2011. Chitin and chitosan from marine organisms; pp. 11–23. [Google Scholar]
- 37.Kurita K. Chitin and chitosan: functional biopolymers from marine crustaceans. Mar. Biotechnol. 2006;8:203–226. doi: 10.1007/s10126-005-0097-5. [DOI] [PubMed] [Google Scholar]
- 38.Nwe N., Furuike T., Tamura H. In: Chitin, Chitosan, Oligosaccharides and Their Derivatives. Kim S.-K., editor. CRC Press; Boca Raton: 2011. Chitin and chitosan from terrestrial organisms; pp. 3–10. [Google Scholar]
- 39.Rødde R.H., Einbu A., Vårum K.M. A seasonal study of the chemical composition and chitin quality of shrimp shells obtained from northern shrimp (Pandalus borealis) Carbohydr. Polym. 2008;71:388–393. [Google Scholar]
- 40.Nemtsev S.V., Zueva O.Y., Khismatullin M.R., Albulov A.I., Varlamov V.P. Isolation of Chitin and Chitosan from honeybees. Appl. Biochem. Microbiol. 2004;40:39–43. [Google Scholar]
- 41.Paulino A.T., Simionato J.I., Garcia J.C., Nozaki J. Characterization of chitosan and chitin produced from silkworm crysalides. Carbohydr. Polym. 2006;64:98–103. [Google Scholar]
- 42.Knezevic-Jugovic Z., Petronijevic Z., Smelcerovic A. In: Chitin, Chitosan, Oligosaccharides and Their Derivatives. Kim S.-K., editor. CRC Press; Boca Raton: 2011. Chitin and chitosan from microorganisms; pp. 25–36. [Google Scholar]
- 43.Chatterjee S., Adhya M., Guha A.K., Chatterjee B.P. Chitosan from Mucor rouxii: production and physico-chemical characterization. Process. Biochem. 2005;40:395–400. [Google Scholar]
- 44.Stamford T.C.M., Stamford T.L.M., Stamford N.P., Neto B.d.B., Campos-Takaki G.M.d. Growth of Cunninghamella elegans UCP 542 and production of chitin and chitosan using yam bean medium. 2007;10:61–68. [Google Scholar]
- 45.Sietsma J.H., Wessels J.G.H. Solubility of (1→3)-β-D/(1→6)-β-D-glucan in fungal walls: importance of presumed linkage between glucan and chitin. J. Gen. Microbiol. 1981;125:209–212. doi: 10.1099/00221287-125-1-209. [DOI] [PubMed] [Google Scholar]
- 46.Muzzarelli R.A.A., Tanfani F., Scarpini G. Chelating, film-forming, and coagulating ability of the chitosan–glucan complex from Aspergillus niger industrial wastes. Biotechnol. Bioeng. 1980;22:885–896. doi: 10.1002/bit.260220412. [DOI] [PubMed] [Google Scholar]
- 47.Rinaudo M. Chitin and chitosan: properties and applications. Prog. Polym. Sci. 2006;31:603–632. [Google Scholar]
- 48.Rudall K.M. Chitin and its association with other molecules. J. Polym. Sci., Part C. 1969;28:83–102. [Google Scholar]
- 49.Gaill F., Persson J., Sugiyama J., Vuong R., Chanzy H. The chitin system in the tubes of deep sea hydrothermal vent worms. J. Struct. Biol. 1992;109:116–128. [Google Scholar]
- 50.Blackwell J., Parker K.D., Rudall K.M. Chitin in pogonophore tubes. J. Mar. Biol. Assoc. U. K. 1965;45:659–661. [Google Scholar]
- 51.Rudall K.M. Skeletal structure in insects. Biochem. Soc. Symp. 1965;25:83–92. [PubMed] [Google Scholar]
- 52.Roberts G.A.F. Macmillian; London: 1992. Chitin chemistry. [Google Scholar]
- 53.Atkins E. Conformations in polysaccharides and complex carbohydrates. J. Biosci. 1985;8:375–387. [Google Scholar]
- 54.Minke R., Blackwell J. The structure of α-chitin. J. Mol. Biol. 1978;120:167–181. doi: 10.1016/0022-2836(78)90063-3. [DOI] [PubMed] [Google Scholar]
- 55.Lotmar W., Picken L. A new crystallographic modification of chitin and its distribution. Cellular and Molecular Life Sciences. 1950;6:58–59. [Google Scholar]
- 56.Rudall K.M. The chitin/protein complexes of insect cuticles. Adv. Insect. Physiol. 1963;1:257–313. (J. W. L. Beament, J. E. Treherne, and V. B. Wigglesworth, editors. Academic) [Google Scholar]
- 57.Saito Y., Putaux J.L., Okano T., Gaill F., Chanzy H. Structural Aspects of the Swelling of β Chitin in HCl and its Conversion into α Chitin. Macromolecules. 1997;30:3867–3873. [Google Scholar]
- 58.Percot A., Viton C., Domard A. Optimization of chitin extraction from shrimp shells. Biomacromolecules. 2002;4:12–18. doi: 10.1021/bm025602k. [DOI] [PubMed] [Google Scholar]
- 59.Galed G., Diaz E., Goycoolea F.M., Heras A. Influence of N-deacetylation conditions on chitosan production from α-chitin. Nat. Prod. Commun. 2008;3:543–550. [Google Scholar]
- 60.Ottøy M.H., Vårum K.M., Smidsrød O. Compositional heterogeneity of heterogeneously deacetylated chitosans. Carbohydr. Polym. 1996;29:17–24. [Google Scholar]
- 61.G.I. Krepets, A.Y. Mikhailin, Method of chitin and chitosan production from chitin-containing raw materials, US Pat. 5,053,113.
- 62.Jo G.-H., Park R.-D., Jung W.-J. In: Chitin, Chitosan, Oligosaccharides and Their Derivatives. Kim S.-K., editor. CRC Press; Boca Raton: 2011. Enzymatic production of chitin from crustacean shell waste; pp. 37–45. [Google Scholar]
- 63.Pacheco N., Garnica-Gonzalez M.N., Gimeno M., Bárzana E., Trombotto S.P., David L., Shirai K. Structural Characterization of Chitin and Chitosan Obtained by Biological and Chemical Methods. Biomacromolecules. 2011;12:3285–3290. doi: 10.1021/bm200750t. [DOI] [PubMed] [Google Scholar]
- 64.Rao M.S., Stevens W.F. Chitin production by Lactobacillus fermentation of shrimp biowaste in a drum reactor and its chemical conversion to chitosan. J. Chem. Technol. Biotechnol. 2005;80:1080–1087. [Google Scholar]
- 65.Kumar M.N.V.R., Muzzarelli R.A.A., Muzzarelli C., Sashiwa H., Domb A.J. Chitosan chemistry and pharmaceutical perspectives. Chem. Rev. 2004;104:6017–6084. doi: 10.1021/cr030441b. [DOI] [PubMed] [Google Scholar]
- 66.Mao H.Q., Roy K., Troung-Le V.L., Janes K.A., Lin K.Y., Wang Y., August J.T., Leong K.W. Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J. Control. Release. 2001;70:399–421. doi: 10.1016/s0168-3659(00)00361-8. [DOI] [PubMed] [Google Scholar]
- 67.Kim T.-H., Jiang H.-L., Jere D., Park I.-K., Cho M.-H., Nah J.-W., Choi Y.-J., Akaike T., Cho C.-S. Chemical modification of chitosan as a gene carrier in vitro and in vivo. Prog. Polym. Sci. 2007;32:726–753. [Google Scholar]
- 68.Shigemasa Y., Saito K., Sashiwa H., Saimoto H. Enzymatic degradation of chitins and partially deacetylated chitins. Int. J. Biol. Macromol. 1994;16:43–49. doi: 10.1016/0141-8130(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 69.Aiba S. Studies on chitosan: 4. Lysozymic hydrolysis of partially N-acetylated chitosans. Int. J. Biol. Macromol. 1992;14:225–228. doi: 10.1016/s0141-8130(05)80032-7. [DOI] [PubMed] [Google Scholar]
- 70.Aiba S. Studies on chitosan: 3. Evidence for the presence of random and block copolymer structures in partially N-acetylated chitosans. Int. J. Biol. Macromol. 1991;13:40–44. doi: 10.1016/0141-8130(91)90008-i. [DOI] [PubMed] [Google Scholar]
- 71.Kurita K., Sannan T., Iwakura Y. Studies on chitin, 4 - Evidence for formation of block and random copolymers of N-Acetyl-D-glucosamine and D-glucosamine by hetero- and homogeneous hydrolyses. Makromol. Chem. 1977;178:3197–3202. [Google Scholar]
- 72.Vårum K.M., Antohonsen M.W., Grasdalen H., Smidsrød O. Determination of the degree of N-acetylation and the distribution of N-acetyl groups in partially N-deacetylated chitins (chitosans) by high-field n.m.r. spectroscopy. Carbohydr. Res. 1991;211:17–23. doi: 10.1016/0008-6215(91)84142-2. [DOI] [PubMed] [Google Scholar]
- 73.Vårum K.M., Anthonsen M.W., Grasdalen H., Smidsrød O. 13C-N.m.r. studies of the acetylation sequences in partially N-deacetylated chitins (chitosans) Carbohydr. Res. 1991;217:19–27. doi: 10.1016/0008-6215(91)84113-s. [DOI] [PubMed] [Google Scholar]
- 74.Lamarque G., Viton C., Domard A. Comparative Study of the first heterogeneous deacetylation of α- and β-chitins in a multistep process. Biomacromolecules. 2004;5:992–1001. doi: 10.1021/bm034498j. [DOI] [PubMed] [Google Scholar]
- 75.Lamarque G., Viton C., Domard A. Comparative study of the second and third heterogeneous deacetylations of α- and β-chitins in a multistep process. Biomacromolecules. 2004;5:1899–1907. doi: 10.1021/bm049780k. [DOI] [PubMed] [Google Scholar]
- 76.Lamarque G., Cretenet M., Viton C., Domard A. New route of deacetylation of α- and β-chitins by means of freeze−pump out−thaw cycles. Biomacromolecules. 2005;6:1380–1388. doi: 10.1021/bm049322b. [DOI] [PubMed] [Google Scholar]
- 77.Sannan T., Kurita K., Iwakura Y. Studies on chitin, 1 - Solubility change by alkaline treatment and film casting Die. Makromol. Chem. 1975;176:1191–1195. [Google Scholar]
- 78.Sannan T., Kurita K., Iwakura Y. Studies on chitin, 2 - Effect of deacetylation on solubility. Makromol. Chem. 1976;177:3589–3600. [Google Scholar]
- 79.Lavertu M., Darras V., Buschmann M.D. Kinetics and efficiency of chitosan reacetylation. Carbohydr. Polym. 2012;87:1192–1198. [Google Scholar]
- 80.Berger J., Reist M., Chenite A., Felt-Baeyens O., Mayer J.M., Gurny R. Pseudo-thermosetting chitosan hydrogels for biomedical application. Int. J. Pharm. 2005;288:17–25. doi: 10.1016/j.ijpharm.2004.07.036. [DOI] [PubMed] [Google Scholar]
- 81.Vachoud L., Zydowicz N., Domard A. Formation and characterisation of a physical chitin gel. Carbohydr. Res. 1997;302:169–177. [Google Scholar]
- 82.Qun G., Ajun W., Yong Z. Effect of reacetylation and degradation on the chemical and crystal structures of chitosan. J. Appl. Polym. Sci. 2007;104:2720–2728. [Google Scholar]
- 83.Zhao Y., Ju W.-T., Park R.-D. In: Chitin, Chitosan, Oligosaccharides and Their Derivatives. Kim S.-K., editor. CRC Press; Boca Raton: 2011. Enzymatic modifications of chitin and chitosan; pp. 185–192. [Google Scholar]
- 84.John M., Roehrig H., Schmidt J., Wieneke U., Schell J. Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc. Natl. Acad. Sci. U. S. A. 1993;90:625–629. doi: 10.1073/pnas.90.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vollmer W., Tomasz A. The pgdA gene encodes for a peptidoglycanN-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 2000;275:20496–20501. doi: 10.1074/jbc.M910189199. [DOI] [PubMed] [Google Scholar]
- 86.Tokuyasu K., Ono H., Ohnishi-Kameyama M., Hayashi K., Mori Y. Deacetylation of chitin oligosaccharides of dp 2–4 by chitin deacetylase from Colletotrichum lindemuthianum. Carbohydr. Res. 1997;303:353–358. doi: 10.1016/s0008-6215(97)00166-3. [DOI] [PubMed] [Google Scholar]
- 87.Aye K.N., Karuppuswamy R., Ahamed T., Stevens W.F. Peripheral enzymatic deacetylation of chitin and reprecipitated chitin particles. Bioresour. Technol. 2006;97:577–582. doi: 10.1016/j.biortech.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 88.Huang M., Fong C.-W., Khor E., Lim L.-Y. Transfection efficiency of chitosan vectors: effect of polymer molecular weight and degree of deacetylation. J. Control. Release. 2005;106:391–406. doi: 10.1016/j.jconrel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 89.Koping-Hoggard M., Varum K.M., Issa M., Danielsen S., Christensen B.E., Stokke B.T., Artursson P. Improved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomers. Gene Ther. 2004;11:1441–1452. doi: 10.1038/sj.gt.3302312. [DOI] [PubMed] [Google Scholar]
- 90.Falk M., Smith D.G., McLachlan J., McInnes A.G. Studies on chitan (β-(1→4)-linked 2-acetamido-2-deoxy-d-glucan) fibers of the diatom thalassiosira fluviatilis hustedt: ii. Proton magnetic resonance, infrared, and x-ray studies. Can. J. Chem. 1966;44:2269–2281. [Google Scholar]
- 91.Rupley J.A. The hydrolysis of chitin by concentrated hydrochloric acid and the preparation of low-molecular-weight substrates for lysozyme. Biochem. Biophys. Acta. 1964;83:245–255. doi: 10.1016/0926-6526(64)90001-1. [DOI] [PubMed] [Google Scholar]
- 92.Vårum K.M., Kristiansen Holme H., Izume M., Torger Stokke B., Smidsrød O. Determination of enzymatic hydrolysis specificity of partially N-acetylated chitosans. Biochim. Biophys. Acta, Gen. Subj. 1996;1291:5–15. doi: 10.1016/0304-4165(96)00038-4. [DOI] [PubMed] [Google Scholar]
- 93.Vårum K.M., Ottøy M.H., Smidsrød O. Acid hydrolysis of chitosans. Carbohydr. Polym. 2001;46:89–98. [Google Scholar]
- 94.Obaidat R., Al-Jbour N., Al-Sou’d K., Sweidan K., Al-Remawi M., Badwan A. Some physico-chemical properties of low molecular weight chitosans and their relationship to conformation in aqueous solution. J. Solution Chem. 2010;39:575–588. [Google Scholar]
- 95.Richardson S.C., Kolbe H.V., Duncan R. Potential of low molecular mass chitosan as a DNA delivery system: biocompatibility, body distribution and ability to complex and protect DNA. Int. J. Pharm. 1999;178:231–243. doi: 10.1016/s0378-5173(98)00378-0. [DOI] [PubMed] [Google Scholar]
- 96.Fukamizo T., Ohkawa T., Ikeda Y., Goto S. Specificity of chitosanase from Bacillus pumilus. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1994;1205:183–188. doi: 10.1016/0167-4838(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 97.Adachi W., Sakihama Y., Shimizu S., Sunami T., Fukazawa T., Suzuki M., Yatsunami R., Nakamura S., Takénaka A. Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17. J. Mol. Biol. 2004;343:785–795. doi: 10.1016/j.jmb.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 98.Horn S.J., Sikorski P., Cederkvist J.B., Vaaje-Kolstad G., Sørlie M., Synstad B., Vriend G., Vårum K.M., Eijsink V.G.H. Costs and benefits of processivity in enzymatic degradation of recalcitrant polysaccharides. Proc. Natl. Acad. Sci. 2006;103:18089–18094. doi: 10.1073/pnas.0608909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heggset E.B., Tuveng T.R., Hoell I.A., Liu Z., Eijsink V.G.H., Vårum K.M. Mode of action of a family 75 chitosanase from Streptomyces avermitilis. Biomacromolecules. 2012;13:1733–1741. doi: 10.1021/bm201521h. [DOI] [PubMed] [Google Scholar]
- 100.Allan G.G., Peyron M. In: Chitin and Chitosan. Skjak-Brack G., Anthonsen T., Sandford P., editors. Elsevier Science Publishers Ltd; London: 1989. The kinetics of the depolymerization of chitosan by nitrous acid; pp. 443–466. [Google Scholar]
- 101.Allan G.G., Peyron M. Molecular weight manipulation of chitosan. I. Kinetics of depolymerization by nitrous acid. Carbohydr. Res. 1995;277:257–272. doi: 10.1016/0008-6215(95)00207-a. [DOI] [PubMed] [Google Scholar]
- 102.Allan G.G., Peyron M. Molecular weight manipulation of chitosan. II. Prediction and control of extent of depolymerization by nitrous acid. Carbohydr. Res. 1995;277:273–282. doi: 10.1016/0008-6215(95)00207-a. [DOI] [PubMed] [Google Scholar]
- 103.Tommeraas K., Varum K.M., Christensen B.E., Smidsrod O. Preparation and characterisation of oligosaccharides produced by nitrous acid depolymerisation of chitosans. Carbohydr. Res. 2001;333:137–144. doi: 10.1016/s0008-6215(01)00130-6. [DOI] [PubMed] [Google Scholar]
- 104.Tømmeraas K., Köping-Höggård M., Vårum K.M., Christensen B.E., Artursson P., Smidsrød O. Preparation and characterisation of chitosans with oligosaccharide branches. Carbohydr. Res. 2002;337:2455–2462. doi: 10.1016/s0008-6215(02)00334-8. [DOI] [PubMed] [Google Scholar]
- 105.United States Pharmacopeial Convention NF vol. 29 Supplement n°2, "Chitosan", United States Pharmacopeial Convention, 2011, pp. 5361–5365.
- 106.ASTM Standard F2103 . Standard Guide for Characterization and Testing of Chitosan Salts as Starting Materials Intended for Use in Biomedical and Tissue-Engineered Medical Product Applications. ASTM International; West Conshohocken, PA: 2011. www.astm.org. [DOI] [Google Scholar]
- 107.Lavertu M., Xia Z., Serreqi A.N., Berrada M., Rodrigues A., Wang D., Buschmann M.D., Gupta A. A validated H-1 NMR method for the determination of the degree of deacetylation of chitosan. J. Pharm. Biomed. Anal. 2003;32:1149–1158. doi: 10.1016/s0731-7085(03)00155-9. [DOI] [PubMed] [Google Scholar]
- 108.Nordtveit R.J., Vårum K.M., Smidsrød O. Degradation of partially N-acetylated chitosans with hen egg white and human lysozyme. Carbohydr. Polym. 1996;29:163–167. [Google Scholar]
- 109.Jean M., Smaoui F., Lavertu M., Methot S., Bouhdoud L., Buschmann M.D., Merzouki A. Chitosan-plasmid nanoparticle formulations for IM and SC delivery of recombinant FGF-2 and PDGF-BB or generation of antibodies. Gene Ther. 2009;16:1097–1110. doi: 10.1038/gt.2009.60. [DOI] [PubMed] [Google Scholar]
- 110.Kasaai M.R. Various methods for determination of the degree of N-acetylation of chitin and chitosan: a review. J. Agric. Food Chem. 2009;57:1667–1676. doi: 10.1021/jf803001m. [DOI] [PubMed] [Google Scholar]
- 111.Muzzarelli R.A.A., Rocchetti R. Determination of the degree of acetylation of chitosans by first derivative ultraviolet spectrophotometry. Carbohydr. Polym. 1985;5:461–472. [Google Scholar]
- 112.Hirai A., Odani H., Nakajima A. Determination of degree of deacetylation of chitosan by proton NMR spectroscopy. Polym. Bull. (Berlin) 1991;26:87–94. [Google Scholar]
- 113.Tan S.C., Khor E., Tan T.K., Wong S.M. The degree of deacetylation of chitosan: advocating the first derivative UV-spectrophotometry method of determination. Talanta. 1998;45:713–719. doi: 10.1016/s0039-9140(97)00288-9. [DOI] [PubMed] [Google Scholar]
- 114.Mao S., Shuai X., Unger F., Wittmar M., Xie X., Kissel T. Synthesis, characterization and cytotoxicity of poly(ethylene glycol)-graft-trimethyl chitosan block copolymers. Biomaterials. 2005;26:6343–6356. doi: 10.1016/j.biomaterials.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 115.Weinhold M.X., Sauvageau J.C.M., Kumirska J., Thöming J. Studies on acetylation patterns of different chitosan preparations. Carbohydr. Polym. 2009;78:678–684. [Google Scholar]
- 116.ASTM Standard F2602 . Standard Test Method for Determining the Molar Mass of Chitosan and Chitosan Salts by Size Exclusion Chromatography with Multi angle Light Scattering Detection (SEC MALS) ASTM International; West Conshohocken, PA: 2008. 2008e1. www.astm.org. [DOI] [Google Scholar]
- 117.Beri R.G., Walker J., Resse E.T., Rollings J.E. Characterization of chitosans via coupled size-exclusion chromatography and multiple-angle laser light-scattering technique. Carbohydr. Res. 1993;238:11–26. [Google Scholar]
- 118.Christensen B.E., Myhr M.H., Aune O., Hagen S., Berge A., Ugelstad J. Macroporous, monodisperse particles and their application in aqueous size exclusion chromatography of high molecular weight polysaccharides. Carbohydr. Polym. 1996;29:217–223. [Google Scholar]
- 119.Wyatt P.J. Light scattering and the absolute characterization of macromolecules. Anal. Chim. Acta. 1993;272:1–40. [Google Scholar]
- 120.Wang W., Bo S.Q., Li S.Q., Qin W. Determination of the Mark-Houwink equation for chitosans with different degrees of deacetylation. Int. J. Biol. Macromol. 1991;13:281–285. doi: 10.1016/0141-8130(91)90027-r. [DOI] [PubMed] [Google Scholar]
- 121.Rinaudo M., Milas M., Dung P.L. Characterization of chitosan. Influence of ionic strength and degree of acetylation on chain expansion. Int. J. Biol. Macromol. 1993;15:281–285. doi: 10.1016/0141-8130(93)90027-j. [DOI] [PubMed] [Google Scholar]
- 122.Ottøy M.H., Vårum K.M., Christensen B.E., Anthonsen M.W., Smidsrød O. Preparative and analytical size-exclusion chromatography of chitosans. Carbohydr. Polym. 1996;31:253–261. [Google Scholar]
- 123.Berth G., Dautzenberg H., Peter M.G. Physico-chemical characterization of chitosans varying in degree of acetylation. Carbohydr. Polym. 1998;36:205–216. [Google Scholar]
- 124.Tsaih M.L., Chen R.H. Molecular weight determination of 83% degree of decetylation chitosan with non-Gaussian and wide range distribution by high-performance size exclusion chromatography and capillary viscometry. J. Appl. Polym. Sci. 1999;71:1905–1913. [Google Scholar]
- 125.Kasaai M.R., Arul J., Charlet G. Intrinsic viscosity–molecular weight relationship for chitosan. J. Polymer Sci., Part B: Polymer Phys. 2000;38:2591–2598. [Google Scholar]
- 126.Berth G., Dautzenberg H. The degree of acetylation of chitosans and its effect on the chain conformation in aqueous solution. Carbohydr. Polym. 2002;47:39–51. [Google Scholar]
- 127.Fee M., Errington N., Jumel K., Illum L., Smith A., Harding S.E. Correlation of SEC/MALLS with ultracentrifuge and viscometric data for chitosans. Eur. Biophys. J. 2003;32:457–464. doi: 10.1007/s00249-003-0317-8. [DOI] [PubMed] [Google Scholar]
- 128.Schatz C., Pichot C., Delair T., Viton C., Domard A. Static light scattering studies on chitosan solutions: from macromolecular chains to colloidal dispersions. Langmuir. 2003;19:9896–9903. [Google Scholar]
- 129.Schatz C., Viton C., Delair T., Pichot C., Domard A. Typical physicochemical behaviors of chitosan in aqueous solution. Biomacromolecules. 2003;4:641–648. doi: 10.1021/bm025724c. [DOI] [PubMed] [Google Scholar]
- 130.Sorlier P., Rochas C., Morfin I., Viton C., Domard A. Light scattering studies of the solution properties of chitosans of varying degrees of acetylation. Biomacromolecules. 2003;4:1034–1040. doi: 10.1021/bm034054n. [DOI] [PubMed] [Google Scholar]
- 131.Lamarque G., Lucas J.-M., Viton C., Domard A. Physicochemical behavior of homogeneous series of acetylated chitosans in aqueous solution: role of various structural parameters. Biomacromolecules. 2005;6:131–142. doi: 10.1021/bm0496357. [DOI] [PubMed] [Google Scholar]
- 132.Yanagisawa M., Kato Y., Yoshida Y., Isogai A. SEC-MALS study on aggregates of chitosan molecules in aqueous solvents: influence of residual N-acetyl groups. Carbohydr. Polym. 2006;66:192–198. [Google Scholar]
- 133.Christensen B.E., Vold I.M.N., Vårum K.M. Chain stiffness and extension of chitosans and periodate oxidised chitosans studied by size-exclusion chromatography combined with light scattering and viscosity detectors. Carbohydr. Polym. 2008;74:559–565. [Google Scholar]
- 134.Nguyen S., Winnik F.M., Buschmann M.D. Improved reproducibility in the determination of the molecular weight of chitosan by analytical size exclusion chromatography. Carbohydr. Polym. 2009;75:528–533. [Google Scholar]
- 135.Brugnerotto J., Desbrières J., Roberts G., Rinaudo M. Characterization of chitosan by steric exclusion chromatography. Polymer. 2001;42:9921–9927. [Google Scholar]
- 136.Anthonsen M.W., Varum K.M., Hermansson A.M., Smidsrod O., Brant D.A. Aggregates in acidic solutions of chitosans detected by static laser light scattering. Carbohydr. Polym. 1994;25:13–23. [Google Scholar]
- 137.Popa-Nita S., Alcouffe P., Rochas C., David L., Domard A. Continuum of structural organization from chitosan solutions to derived physical forms. Biomacromolecules. 2009;11:6–12. doi: 10.1021/bm9012138. [DOI] [PubMed] [Google Scholar]
- 138.Brugnerotto J., Lizardi J., Goycoolea F.M., Argüelles-Monal W., Desbrières J., Rinaudo M. An infrared investigation in relation with chitin and chitosan characterization. Polymer. 2001;42:3569–3580. [Google Scholar]
- 139.Domard A., Piron E. Recent approach of metal binding by chitosan and derivatives. Adv. Chitin Sci. 2000;4:295–301. [Google Scholar]
- 140.Zavodinsky V.G., Gnidenko A.A., Davidova V.N., Yermak I.M. Computer simulation of the interaction of bacterial endotoxin with polycation-chitozan, chemistry and computational simulation. Butlerov Commun. 2003;4:11–12. [Google Scholar]
- 141.S. Baker, W.P. Wiesmann, Methods of making a chitosan product having an ultra-low endotoxin concentration and the ultra-low endotoxin chitosan product derived therefrom and method of accurately determining inflammatory and anti-inflammatory cellular response to such materials, US Pat. US20080248508A1.
- 142.S. Baker, W.P. Wiesmann, R. Shannon, A chitosan product having an ultra-low endotoxin concentration and method of accurately determining inflammatory and anti-inflammatory cellular response to such materials, US Pat. US8119780B2.
- 143.Pillai C.K.S., Paul W., Sharma C.P. Chitin and chitosan polymers: chemistry, solubility and fiber formation. Prog. Polym. Sci. 2009;34:641–678. [Google Scholar]
- 144.K. Kiehm, B. Hauptmeier, P. Boderke, Chitosan beads and filler comprising such beads for cosmetic and medical applications, WO Pat. WO2011124380A1.
- 145.Polnok A., Borchard G., Verhoef J.C., Sarisuta N., Junginger H.E. Influence of methylation process on the degree of quaternization of N-trimethyl chitosan chloride. Eur. J. Pharm. Biopharm. 2004;57:77–83. doi: 10.1016/s0939-6411(03)00151-6. [DOI] [PubMed] [Google Scholar]
- 146.Rinaudo M., Dung P.L., Gey C., Milas M. Substituent distribution on O, N-carboxymethylchitosans by 1H and 13C n.m.r. Int. J. Biol. Macromol. 1992;14:122–128. doi: 10.1016/s0141-8130(05)80001-7. [DOI] [PubMed] [Google Scholar]
- 147.Tiera M.J., Qiu X.-P., Bechaouch S., Shi Q., Fernandes J.C., Winnik F.M. Synthesis and characterization of phosphorylcholine-substituted chitosans soluble in physiological pH conditions. Biomacromolecules. 2006;7:3151–3156. doi: 10.1021/bm060381u. [DOI] [PubMed] [Google Scholar]
- 148.Casé A.H., Picola I.P.D., Zaniquelli M.E.D., Fernandes J.C., Taboga S.R., Winnik F.M., Tiera M.J. Physicochemical characterization of nanoparticles formed between DNA and phosphorylcholine substituted chitosans. J. Colloid Interface Sci. 2009;336:125–133. doi: 10.1016/j.jcis.2009.02.069. [DOI] [PubMed] [Google Scholar]
- 149.Malmo J., Varum K.M., Strand S.P. Effect of chitosan chain architecture on gene delivery: comparison of self-branched and linear chitosans. Biomacromolecules. 2011;12:721–729. doi: 10.1021/bm1013525. [DOI] [PubMed] [Google Scholar]
- 150.Strand S.P., Issa M.M., Christensen B.E., Varum K.M., Artursson P. Tailoring of chitosans for gene delivery: novel self-branched glycosylated chitosan oligomers with improved functional properties. Biomacromolecules. 2008;9:3268–3276. doi: 10.1021/bm800832u. [DOI] [PubMed] [Google Scholar]
- 151.Germershaus O., Mao S., Sitterberg J., Bakowsky U., Kissel T. Gene delivery using chitosan, trimethyl chitosan or polyethylenglycol-graft-trimethyl chitosan block copolymers: establishment of structure-activity relationships in vitro. J. Control. Release. 2008;125:145–154. doi: 10.1016/j.jconrel.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 152.Morris V.B., Sharma C.P. Folate mediated in vitro targeting of depolymerised trimethylated chitosan having arginine functionality. J. Colloid Interface Sci. 2010;348:360–368. doi: 10.1016/j.jcis.2010.04.090. [DOI] [PubMed] [Google Scholar]
- 153.Park I.K., Park Y.H., Shin B.A., Choi E.S., Kim Y.R., Akaike T., Cho C.S. Galactosylated chitosan-graft-dextran as hepatocyte-targeting DNA carrier. J. Control. Release. 2000;69:97–108. doi: 10.1016/s0168-3659(00)00298-4. [DOI] [PubMed] [Google Scholar]
- 154.Morris V.B., Sharma C.P. Folate mediated histidine derivative of quaternised chitosan as a gene delivery vector. Int. J. Pharm. 2010;389:176–185. doi: 10.1016/j.ijpharm.2010.01.037. [DOI] [PubMed] [Google Scholar]
- 155.Park I.K., Kim T.H., Park Y.H., Shin B.A., Choi E.S., Chowdhury E.H., Akaike T., Cho C.S. Galactosylated chitosan-graft-poly(ethylene glycol) as hepatocyte-targeting DNA carrier. J. Control. Release. 2001;76:349–362. doi: 10.1016/s0168-3659(01)00448-5. [DOI] [PubMed] [Google Scholar]
- 156.Jeong Y.-I., Kim D.-G., Jang M.-K., Nah J.-W. Preparation and spectroscopic characterization of methoxy poly(ethylene glycol)-grafted water-soluble chitosan. Carbohydr. Res. 2008;343:282–289. doi: 10.1016/j.carres.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 157.Jiang H.-L., Kwon J.-T., Kim E.-M., Kim Y.-K., Arote R., Jere D., Jeong H.-J., Jang M.-K., Nah J.-W., Xu C.-X., Park I.-K., Cho M.-H., Cho C.-S. Galactosylated poly(ethylene glycol)-chitosan-graft-polyethylenimine as a gene carrier for hepatocyte-targeting. J. Control. Release. 2008;131:150–157. doi: 10.1016/j.jconrel.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 158.Satoh T., Nagasaki T., Sakairi N., Shinkai S. 6-Amino-6-deoxychitosan. Preparation and application as plasmid vector in COS-1 cells. Chem. Lett. 2004;33:340–341. [Google Scholar]
- 159.Satoh T., Kano H., Nakatani M., Sakairi N., Shinkai S., Nagasaki T. 6-Amino-6-deoxy-chitosan. Sequential chemical modifications at the C-6 positions of N-phthaloyl-chitosan and evaluation as a gene carrier. Carbohydr. Res. 2006;341:2406–2413. doi: 10.1016/j.carres.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 160.Strand S.P., Lelu S., Reitan N.K., Davies C.D., Artursson P., Varum K.M. Molecular design of chitosan gene delivery systems with an optimized balance between polyplex stability and polyplex unpacking. Biomaterials. 2010;31:975–987. doi: 10.1016/j.biomaterials.2009.09.102. [DOI] [PubMed] [Google Scholar]
- 161.Tømmeraas K., Strand S.P., Christensen B.E., Smidsrød O., Vårum K.M. Preparation and characterization of branched chitosans. Carbohydr. Polym. 2011;83:1558–1564. [Google Scholar]
- 162.Gao S., Chen J., Xu X., Ding Z., Yang Y.-H., Hua Z., Zhang J. Galactosylated low molecular weight chitosan as DNA carrier for hepatocyte-targeting. Int. J. Pharm. 2003;255:57–68. doi: 10.1016/s0378-5173(03)00082-6. [DOI] [PubMed] [Google Scholar]
- 163.Jiang H.L., Kwon J.T., Kim Y.K., Kim E.M., Arote R., Jeong H.J., Nah J.W., Choi Y.J., Akaike T., Cho M.H., Cho C.S. Galactosylated chitosan-graft-polyethylenimine as a gene carrier for hepatocyte targeting. Gene Ther. 2007;14:1389–1398. doi: 10.1038/sj.gt.3302997. [DOI] [PubMed] [Google Scholar]
- 164.Satoh T., Kakimoto S., Kano H., Nakatani M., Shinkai S., Nagasaki T. In vitro gene delivery to HepG2 cells using galactosylated 6-amino-6-deoxychitosan as a DNA carrier. Carbohydr. Res. 2007;342:1427–1433. doi: 10.1016/j.carres.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 165.Kim T.H., Ihm J.E., Choi Y.J., Nah J.W., Cho C.S. Efficient gene delivery by urocanic acid-modified chitosan. J. Control. Release. 2003;93:389–402. doi: 10.1016/j.jconrel.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 166.Moreira C., Oliveira H., Pires L.R., Simoes S., Barbosa M.A., Pego A.P. Improving chitosan-mediated gene transfer by the introduction of intracellular buffering moieties into the chitosan backbone. Acta Biomater. 2009;5:2995–3006. doi: 10.1016/j.actbio.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 167.Jiang H.L., Kim Y.K., Arote R., Nah J.W., Cho M.H., Choi Y.J., Akaike T., Cho C.S. Chitosan-graft-polyethylenimine as a gene carrier. J. Control. Release. 2007;117:273–280. doi: 10.1016/j.jconrel.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 168.Wong K., Sun G. Zhang, H. Dai, Y. Liu, He, K.W. Leong, PEI-g-chitosan, a Novel Gene Delivery System with Transfection Efficiency Comparable to Polyethylenimine in Vitro and after Liver Administration in Vivo. Bioconjugate Chem. 2005;17:152–158. doi: 10.1021/bc0501597. [DOI] [PubMed] [Google Scholar]
- 169.Lu B., Xu X.D., Zhang X.Z., Cheng S.X., Zhuo R.X. Low molecular weight polyethylenimine grafted N-maleated chitosan for gene delivery: properties and in vitro transfection studies. Biomacromolecules. 2008;9:2594–2600. doi: 10.1021/bm8004676. [DOI] [PubMed] [Google Scholar]
- 170.Lu B., Sun Y.-X., Li Y.-Q., Zhang X.-Z., Zhuo R.-X. N-Succinyl-chitosan grafted with low molecular weight polyethylenimine as a serum-resistant gene vector. Mol. Biosyst. 2009;5:629–637. doi: 10.1039/b822505b. [DOI] [PubMed] [Google Scholar]
- 171.Chang K.L., Higuchi Y., Kawakami S., Yamashita F., Hashida M. Development of lysine-histidine dendron modified chitosan for improving transfection efficiency in HEK293 cells. J. Control. Release. 2011;156:195–202. doi: 10.1016/j.jconrel.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 172.Xu Z., Wan X., Zhang W., Wang Z., Peng R., Tao F., Cai L., Li Y., Jiang Q., Gao R. Synthesis of biodegradable polycationic methoxy poly(ethylene glycol)-polyethylenimine-chitosan and its potential as gene carrier. Carbohydr. Polym. 2009;78:46–53. [Google Scholar]
- 173.Griffiths P.R., de Haseth J.A. John Wiley & Sons, Inc.; Hoboken, NJ: 2007. Fourier Transform Infrared Spectroscopy. [Google Scholar]
- 174.Darras V., Nelea M., Winnik F.M., Buschmann M.D. Chitosan modified with gadolinium diethylenetriaminepentaacetic acid for magnetic resonance imaging of DNA/chitosan nanoparticles. Carbohydr. Polym. 2010;80:1137–1146. [Google Scholar]
- 175.Tharanathan R.N., Kittur F.S. Chitin — the undisputed biomolecule of great potential. Crit. Rev. Food Sci. Nutr. 2003;43:61–87. doi: 10.1080/10408690390826455. [DOI] [PubMed] [Google Scholar]
- 176.Sieval A.B., Thanou M., Kotze´ A.F., Verhoef J.C., Brussee J., Junginger H.E. Preparation and NMR characterization of highly substitutedN-trimethyl chitosan chloride. Carbohydr. Polym. 1998;36:157–165. [Google Scholar]
- 177.Florea B.I., Thanou M., Geldof M., Meaney C., Junginger H.E., Borchard G. Proc. Int. Symp. Controlled Release Bioact. Mater., 27th. 2000. Modified chitosan oligosaccharides as transfection agents for gene therapy in cystic fibrosis; pp. 846–847. [Google Scholar]
- 178.Snyman D., Hamman J.H., Kotze J.S., Rollings J.E., Kotzé A.F. The relationship between the absolute molecular weight and the degree of quaternisation of N-trimethyl chitosan chloride. Carbohydr. Polym. 2002;50:145–150. [Google Scholar]
- 179.Thanou M., Florea B.I., Geldof M., Junginger H.E., Borchard G. Quaternized chitosan oligomers as novel gene delivery vectors in epithelial cell lines. Biomaterials. 2002;23:153–159. doi: 10.1016/s0142-9612(01)00090-4. [DOI] [PubMed] [Google Scholar]
- 180.Kean T., Roth S., Thanou M. Trimethylated chitosans as non-viral gene delivery vectors: cytotoxicity and transfection efficiency. J. Control. Release. 2005;103:643–653. doi: 10.1016/j.jconrel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 181.Loubaki E., Ourevitch M., Sicsic S. Chemical modification of chitosan by glycidyl trimethylammonium chloride. characterization of modified chitosan by 13C- and 1H-NMR spectroscopy. Eur. Polym. J. 1991;27:311–317. [Google Scholar]
- 182.Kim C.-H., Kim S.-Y., Choi K.-S. Synthesis and antibacterial activity of water-soluble chitin derivatives. Polym. Adv. Technol. 1997;8:319–325. [Google Scholar]
- 183.Faizuloev E., Marova A., Nikonova A., Volkova I., Gorshkova M., Izumrudov V. Water-soluble N-[(2-hydroxy-3-trimethylammonium)propyl]chitosan chloride as a nucleic acids vector for cell transfection. Carbohydr. Polym. 2012;89:1088–1094. doi: 10.1016/j.carbpol.2012.03.071. [DOI] [PubMed] [Google Scholar]
- 184.Hermanson G.T. Academic Press; Amsterdam: 2008. Bioconjugate Techniques. [Google Scholar]
- 185.Lee J.I., Ha K.-S., Yoo H.S. Quantum-dot-assisted fluorescence resonance energy transfer approach for intracellular trafficking of chitosan/DNA complex. Acta Biomater. 2008;4:791–798. doi: 10.1016/j.actbio.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 186.Tømmeraas K., Strand S.P., Tian W., Kenne L., Vårum K.M. Preparation and characterisation of fluorescent chitosans using 9-anthraldehyde as fluorophore. Carbohydr. Res. 2001;336:291–296. doi: 10.1016/s0008-6215(01)00275-0. [DOI] [PubMed] [Google Scholar]
- 187.Vold I.M.N., Christensen B.E. Periodate oxidation of chitosans with different chemical compositions. Carbohydr. Res. 2005;340:679–684. doi: 10.1016/j.carres.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 188.Ma O., Lavertu M., Sun J., Nguyen S., Buschmann M.D., Winnik F.M., Hoemann C.D. Precise derivatization of structurally distinct chitosans with rhodamine B isothiocyanate. Carbohydr. Polym. 2008;72:616–624. [Google Scholar]
- 189.Nishimura S., Kohgo O., Kurita K., Kuzuhara H. Chemospecific manipulations of a rigid polysaccharide: syntheses of novel chitosan derivatives with excellent solubility in common organic solvents by regioselective chemical modifications. Macromolecules. 1991;24:4745–4748. [Google Scholar]
- 190.Kurita K., Ikeda H., Yoshida Y., Shimojoh M., Harata M. Chemoselective protection of the amino groups of chitosan by controlled phthaloylation: facile preparation of a precursor useful for chemical modifications. Biomacromolecules. 2001;3:1–4. doi: 10.1021/bm0101163. [DOI] [PubMed] [Google Scholar]
- 191.Cai G., Jiang H., Tu K., Wang L., Zhu K. A facile route for regioselective conjugation of organo-soluble polymers onto chitosan. Macromol. Biosci. 2009;9:256–261. doi: 10.1002/mabi.200800153. [DOI] [PubMed] [Google Scholar]
- 192.M. Lavertu, S. Methot, M.D. Buschmann, Composition and Method for Efficient Delivery of Nucleic Acids to Cells Using Chitosan, Pat. WO2007059605–A1; EP1948810–A1; US2009075383–A1; CA2628313–A1.
- 193.Klausner E.A., Zhang Z., Chapman R.L., Multack R.F., Volin M.V. Ultrapure chitosan oligomers as carriers for corneal gene transfer. Biomaterials. 2010;31:1814–1820. doi: 10.1016/j.biomaterials.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 194.Dautzenberg H., Jaeger W. Effect of charge density on the formation and salt stability of polyelectrolyte complexes. Macromol. Chem. Phys. 2002;203:2095–2102. [Google Scholar]
- 195.Felgner P.L., Gadek T.R., Holm M., Roman R., Chan H.W., Wenz M., Northrop J.P., Ringold G.M., Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. U. S. A. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wu G.Y., Wu C.H. Receptor-mediated in vitro gene transformation by a soluble DNA carrier system. J. Biol. Chem. 1987;262:4429–4432. [PubMed] [Google Scholar]
- 197.Kabanov V.A., Zezin A.B. A new class of complex water-soluble polyelectrolytes. Macromol. Chem. Phys. Suppl. 1984;6:259–276. [Google Scholar]
- 198.Kabanov A.V., Kabanov V.A. DNA complexes with polycations for the delivery of genetic material into cells. Bioconjugate Chem. 1995;6:7–20. doi: 10.1021/bc00031a002. [DOI] [PubMed] [Google Scholar]
- 199.Philipp B., Dautzenberg H., Linow K.J., Koetz J., Dawydoff W. Polyelectrolyte complexes - recent developments and open problems. Prog. Polym. Sci. 1989;14:91–172. [Google Scholar]
- 200.Dautzenberg H. Polyelectrolyte complex formation in highly aggregating systems. 1. Effect of salt: Polyelectrolyte complex formation in the presence of NaCl. Macromolecules. 1997;30:7810–7815. [Google Scholar]
- 201.Dautzenberg H., Hartmann J., Grunewald S., Brand F. Stoichiometry and structure of polyelectrolyte complex particles in diluted solutions. Ber. Bunsen-Ges. Phys. Chem. Chem. Phys. 1996;100:1024–1032. [Google Scholar]
- 202.Thuenemann A.F., Mueller M., Dautzenberg H., Joanny J.-F., Loewen H. Polyelectrolyte complexes. Adv. Polym. Sci. 2004;166:113–171. [Google Scholar]
- 203.Tsuchida E., Osada Y., Sanada K. Interaction of poly(styrene sulfonate) with polycations carrying charges in the chain backbone. J. Polym. Sci., Part A-1. 1972;10:3397–3404. [Google Scholar]
- 204.Rungsardthong U., Ehtezazi T., Bailey L., Armes S.P., Garnett M.C., Stolnik S. Effect of polymer ionization on the interaction with DNA in nonviral gene delivery systems. Biomacromolecules. 2003;4:683–690. doi: 10.1021/bm025736y. [DOI] [PubMed] [Google Scholar]
- 205.Danielsen S., Varum K.M., Stokke B.T. Structural analysis of chitosan mediated DNA condensation by AFM: Influence of chitosan molecular parameters. Biomacromolecules. 2004;5:928–936. doi: 10.1021/bm034502r. [DOI] [PubMed] [Google Scholar]
- 206.Oupicky D., Konak C., Ulbrich K., Wolfert M.A., Seymour L.W. DNA delivery systems based on complexes of DNA with synthetic polycations and their copolymers. J. Control. Release. 2000;65:149–171. doi: 10.1016/s0168-3659(99)00249-7. [DOI] [PubMed] [Google Scholar]
- 207.Schatz C., Domard A., Viton C., Pichot C., Delair T. Versatile and efficient formation of colloids of biopolymer-based polyelectrolyte complexes. Biomacromolecules. 2004;5:1882–1892. doi: 10.1021/bm049786+. [DOI] [PubMed] [Google Scholar]
- 208.MacLaughlin F.C., Mumper R.J., Wang J., Tagliaferri J.M., Gill I., Hinchcliffe M., Rolland A.P. Chitosan and depolymerized chitosan oligomers as condensing carriers for in vivo plasmid delivery. J. Control. Release. 1998;56:259–272. doi: 10.1016/s0168-3659(98)00097-2. [DOI] [PubMed] [Google Scholar]
- 209.Jean M., Alameh M., De Jesus D., Thibault M., Lavertu M., Darras V., Nelea M., Buschmann M.D., Merzouki A. Chitosan-based therapeutic nanoparticles for combination gene therapy and gene silencing of in vitro cell lines relevant to type 2 diabetes. Eur. J. Pharm. Sci. 2012;45:138–149. doi: 10.1016/j.ejps.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 210.Howard K.A., Paludan S.R., Behlke M.A., Besenbacher F., Deleuran B., Kjems J. Chitosan/siRNA nanoparticle-mediated TNF-alpha knockdown in peritoneal macrophages for anti-inflammatory treatment in a murine arthritis model. Mol. Ther. 2009;17:162–168. doi: 10.1038/mt.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Katas H., Alpar H.O. Development and characterisation of chitosan nanoparticles for siRNA delivery. J. Control. Release. 2006;115:216–225. doi: 10.1016/j.jconrel.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 212.Gaspar V.M., Sousa F., Queiroz J.A., Correia I.J. Formulation of chitosan-TPP-pDNA nanocapsules for gene therapy applications. Nanotechnology. 2011;22 doi: 10.1088/0957-4484/22/1/015101. [DOI] [PubMed] [Google Scholar]
- 213.Csaba N., Koping-Hoggard M., Alonso M.J. Ionically crosslinked chitosan/tripolyphosphate nanoparticles for oligonucleotide and plasmid DNA delivery. Int. J. Pharm. 2009;382:205–214. doi: 10.1016/j.ijpharm.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 214.Ravina M., Cubillo E., Olmeda D., Novoa-Carballal R., Fernandez-Megia E., Riguera R., Sanchez A., Cano A., Alonso M.J. Hyaluronic Acid/Chitosan-g-Poly(ethylene glycol) Nanoparticles for Gene Therapy: An Application for pDNA and siRNA Delivery. Pharm. Res. 2010;27:2544–2555. doi: 10.1007/s11095-010-0263-y. [DOI] [PubMed] [Google Scholar]
- 215.Zeng P., Xu Y., Zeng C.H., Ren H., Peng M.L. Chitosan-modified poly(D, L-lactide-co-glycolide) nanospheres for plasmid DNA delivery and HBV gene-silencing. Int. J. Pharm. 2011;415:259–266. doi: 10.1016/j.ijpharm.2011.05.053. [DOI] [PubMed] [Google Scholar]
- 216.Kumar M., Bakowsky U., Lehr C.M. Preparation and characterization of cationic PLGA nanospheres as DNA carriers. Biomaterials. 2004;25:1771–1777. doi: 10.1016/j.biomaterials.2003.08.069. [DOI] [PubMed] [Google Scholar]
- 217.Lee K.Y., Kwon I.C., Kim Y.H., Jo W.H., Jeong S.Y. Preparation of chitosan self-aggregates as a gene delivery system. J. Control. Release. 1998;51:213–220. doi: 10.1016/s0168-3659(97)00173-9. [DOI] [PubMed] [Google Scholar]
- 218.Kim Y.H., Gihm S.H., Park C.R., Lee K.Y., Kim T.W., Kwon I.C., Chung H., Jeong S.Y. Structural characteristics of size-controlled self-aggregates of deoxycholic acid-modified chitosan and their application as a DNA delivery carrier. Bioconjugate Chem. 2001;12:932–938. doi: 10.1021/bc015510c. [DOI] [PubMed] [Google Scholar]
- 219.Huang M., Fong C.W., Khor E., Lim L.Y. Transfection efficiency of chitosan vectors: effect of polymer molecular weight and degree of deacetylation. J. Control. Release. 2005;106:391–406. doi: 10.1016/j.jconrel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 220.Koping-Hoggard M., Tubulekas I., Guan H., Edwards K., Nilsson M., Varum K.M., Artursson P. Chitosan as a nonviral gene delivery system. Structure–property relationships and characteristics compared with polyethylenimine in vitro and after lung administration in vivo. Gene Ther. 2001;8:1108–1121. doi: 10.1038/sj.gt.3301492. [DOI] [PubMed] [Google Scholar]
- 221.Nielsen E.J.B., Nielsen J.M., Becker D., Karlas A., Prakash H., Glud S.Z., Merrison J., Besenbacher F., Meyer T.F., Kjems J., Howard K.A. Pulmonary gene silencing in transgenic EGFP mice using aerosolised chitosan/siRNA nanoparticles. Pharm. Res. 2010;27:2520–2527. doi: 10.1007/s11095-010-0255-y. [DOI] [PubMed] [Google Scholar]
- 222.Sato T., Ishii T., Okahata Y. In vitro gene delivery mediated by chitosan. Effect of pH, serum, and molecular mass of chitosan on the transfection efficiency. Biomaterials. 2001;22:2075–2080. doi: 10.1016/s0142-9612(00)00385-9. [DOI] [PubMed] [Google Scholar]
- 223.Huang H.N., Li T.L., Chan Y.L., Chen C.L., Wu C.J. Transdermal immunization with low-pressure-gene-gun mediated chitosan-based DNA vaccines against Japanese encephalitis virus. Biomaterials. 2009;30:6017–6025. doi: 10.1016/j.biomaterials.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 224.Leong K.W., Mao H.Q., Truong-Le V.L., Roy K., Walsh S.M., August J.T. DNA-polycation nanospheres as non-viral gene delivery vehicles. J. Control. Release. 1998;53:183–193. doi: 10.1016/s0168-3659(97)00252-6. [DOI] [PubMed] [Google Scholar]
- 225.Romoren K., Pedersen S., Smistad G., Evensen O., Thu B.J. The influence of formulation variables on in vitro transfection efficiency and physicochemical properties of chitosan-based polyplexes. Int. J. Pharm. 2003;261:115–127. doi: 10.1016/s0378-5173(03)00301-6. [DOI] [PubMed] [Google Scholar]
- 226.Holzerny P., Ajdini B., Heusermann W., Bruno K., Schuleit M., Meinel L., Keller M. Biophysical properties of chitosan/siRNA polyplexes: Profiling the polymer/siRNA interactions and bioactivity. J. Control. Release. 2012;157:297–304. doi: 10.1016/j.jconrel.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 227.Erbacher P., Zou S., Bettinger T., Steffan A.M., Remy J.S. Chitosan-based vector/DNA complexes for gene delivery: biophysical characteristics and transfection ability. Pharm. Res. 1998;15:1332–1339. doi: 10.1023/a:1011981000671. [DOI] [PubMed] [Google Scholar]
- 228.Liu W.G., Sun S.J., Cao Z.Q., Xin Z., Yao K.D., Lu W.W., Luk K.D.K. An investigation on the physicochemical properties of chitosan/DNA polyelectrolyte complexes. Biomaterials. 2005;26:2705–2711. doi: 10.1016/j.biomaterials.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 229.Ma P.L., Buschmann M.D., Winnik F.M. Complete physicochemical characterization of DNA/chitosan complexes by multiple detection using asymmetrical flow field-flow fractionation. Anal. Chem. 2010;82:9636–9643. doi: 10.1021/ac100711j. [DOI] [PubMed] [Google Scholar]
- 230.Ma P.L., Buschmann M.D., Winnik F.M. One-step analysis of DNA/chitosan complexes by field-flow fractionation reveals particle size and free chitosan content. Biomacromolecules. 2010;11:549–554. doi: 10.1021/bm901345q. [DOI] [PubMed] [Google Scholar]
- 231.Xu L., Anchordoquy T. Drug delivery trends in clinical trials and translational medicine: challenges and opportunities in the delivery of nucleic acid-based therapeutics. J. Pharm. Sci. 2011;100:38–52. doi: 10.1002/jps.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bakeev K.N., Izumrudov V.A., Zezin A.B., Kabanov V.A. Kinetics and mechanism of the formation of polyelectrolyte complexes. Dokl. Akad. Nauk SSSR. 1988;299:1405–1408. [Google Scholar]
- 233.Kasper J.C., Schaffert D., Ogris M., Wagner E., Friess W. The establishment of an up-scaled micro-mixer method allows the standardized and reproducible preparation of well-defined plasmid/LPEI polyplexes. Eur. J. Pharm. Biopharm. 2011;77:182–185. doi: 10.1016/j.ejpb.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 234.Davies L.A., Nunez-Alonso G.A., Hebel H.L., Scheule R.K., Cheng S.H., Hyde S.C., Gill D.R. A novel mixing device for the reproducible generation of nonviral gene therapy formulations. Biotechniques. 2010;49:666–668. doi: 10.2144/000113498. [DOI] [PubMed] [Google Scholar]
- 235.Jean M., Alameh M., Buschmann M.D., Merzouki A. Effective and safe gene-based delivery of GLP-1 using chitosan/plasmid-DNA therapeutic nanocomplexes in an animal model of type 2 diabetes. Gene Ther. 2011;18:807–816. doi: 10.1038/gt.2011.25. [DOI] [PubMed] [Google Scholar]
- 236.Ankerfors C., Ondaral S., Wagberg L., Odberg L. Using jet mixing to prepare polyelectrolyte complexes: Complex properties and their interaction with silicon oxide surfaces. J. Colloid Interface Sci. 2010;351:88–95. doi: 10.1016/j.jcis.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 237.Anchordoquy T.J., Armstrong T.K., Molina M.D. Low molecular weight dextrans stabilize nonviral vectors during lyophilization at low osmolalities: concentrating suspensions by rehydration to reduced volumes. J. Pharm. Sci. 2005;94:1226–1236. doi: 10.1002/jps.20353. [DOI] [PubMed] [Google Scholar]
- 238.Miyata K., Kakizawa Y., Nishiyama N., Yamasaki Y., Watanabe T., Kohara M., Kataoka K. Freeze-dried formulations for in vivo gene delivery of PEGylated polyplex micelles with disulfide crosslinked cores to the liver. J. Control. Release. 2005;109:15–23. doi: 10.1016/j.jconrel.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 239.Cherng J.Y., Talsma H., Crommelin D.J., Hennink W.E. Long term stability of poly((2-dimethylamino)ethyl methacrylate)-based gene delivery systems. Pharm. Res. 1999;16:1417–1423. doi: 10.1023/a:1018907310472. [DOI] [PubMed] [Google Scholar]
- 240.Andersen M.O., Howard K.A., Paludan S.R., Besenbacher F., Kjems J. Delivery of siRNA from lyophilized polymeric surfaces. Biomaterials. 2008;29:506–512. doi: 10.1016/j.biomaterials.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 241.Kuriyel R., Fushijima M., Jung G.W. Advancements in membrane processes for pharmaceutical applications. CRC Press; 2009. pp. 409–426. [Google Scholar]
- 242.Dalwadi G., Sunderland V.B. Purification of PEGylated nanoparticles using tangential flow filtration (TFF) Drug Dev. Ind. Pharm. 2007;33:1030–1039. doi: 10.1080/03639040601180143. [DOI] [PubMed] [Google Scholar]
- 243.Dalwadi G., Benson H.A.E., Chen Y. Comparison of diafiltration and tangential flow filtration for purification of nanoparticle suspensions. Pharm. Res. 2005;22:2152–2162. doi: 10.1007/s11095-005-7781-z. [DOI] [PubMed] [Google Scholar]
- 244.Trefry J.C., Monahan J.L., Weaver K.M., Meyerhoefer A.J., Markopolous M.M., Arnold Z.S., Wooley D.P., Pavel I.E. Size selection and concentration of silver nanoparticles by tangential flow ultrafiltration for SERS-based biosensors. J. Am. Chem. Soc. 2010;132:10970–10972. doi: 10.1021/ja103809c. [DOI] [PubMed] [Google Scholar]
- 245.Limayem I., Charcosset C., Fessi H. Purification of nanoparticle suspensions by a concentration/diafiltration process. Sep. Purif. Technol. 2004;38:1–9. [Google Scholar]
- 246.Mumper R.J., Wang J., Claspell J.M., Rolland A.P. Vol. 22. 1995. Novel polymeric condensing carriers for gene delivery; pp. 178–179. (Proc. Intern. Symp. Control. Rel. Bioact. Mater.). [Google Scholar]
- 247.Prabha S., Zhou W.Z., Panyam J., Labhasetwar V. Size-dependency of nanoparticle-mediated gene transfection: studies with fractionated nanoparticles. Int. J. Pharm. 2002;244:105–115. doi: 10.1016/s0378-5173(02)00315-0. [DOI] [PubMed] [Google Scholar]
- 248.Kabanov A.V. Taking polycation gene delivery systems from in vitro to in vivo. Pharm. Sci. Technol. Today. 1999;2:365–372. doi: 10.1016/s1461-5347(99)00186-8. [DOI] [PubMed] [Google Scholar]
- 249.Lundqvist M., Stigler J., Elia G., Lynch I., Cedervall T., Dawson K.A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Bowman K., Sarkar R., Raut S., Leong K.W. Gene transfer to hemophilia A mice via oral delivery of FVIII-chitosan nanoparticles. J. Control. Release. 2008;132:252–259. doi: 10.1016/j.jconrel.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lou Y.L., Peng Y.S., Chen B.H., Wang L.F., Leong K.W. Poly(ethylene imine)-g-chitosan using EX-810 as a spacer for nonviral gene delivery vectors. J. Biomed. Mater. Res. Part A. 2009;88A:1058–1068. doi: 10.1002/jbm.a.31961. [DOI] [PubMed] [Google Scholar]
- 252.Filipe V., Hawe A., Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by nanosight for the measurement of nanoparticles and protein aggregates. Pharm. Res. 2010;27:796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Koping-Hoggard M., Mel'nikova Y.S., Varum K.M., Lindman B., Artursson P. Relationship between the physical shape and the efficiency of oligomeric chitosan as a gene delivery system in vitro and in vivo. Journal of Gene Medicine. 2003;5:130–141. doi: 10.1002/jgm.327. [DOI] [PubMed] [Google Scholar]
- 254.Mittnacht U., Hartmann H., Hein S., Oliveira H., Dong M., Pego A.P., Kjems J., Howard K.A., Schlosshauer B. Chitosan/siRNA nanoparticles biofunctionalize nerve implants and enable neurite outgrowth. Nano Lett. 2010;10:3933–3939. doi: 10.1021/nl1016909. [DOI] [PubMed] [Google Scholar]
- 255.Liu X., Howard K.A., Dong M., Andersen M.O., Rahbek U.L., Johnsen M.G., Hansen O.C., Besenbacher F., Kjems J. The influence of polymeric properties on chitosan/siRNA nanoparticle formulation and gene silencing. Biomaterials. 2007;28:1280–1288. doi: 10.1016/j.biomaterials.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 256.Ma P.L., Lavertu M., Winnik F.M., Buschmann M.D. New insights into chitosan-DNA interactions using isothermal titration microcalorimetry. Biomacromolecules. 2009;10:1490–1499. doi: 10.1021/bm900097s. [DOI] [PubMed] [Google Scholar]
- 257.Noh S.M., Park M.O., Shim G., Han S.E., Lee H.Y., Huh J.H., Kim M.S., Choi J.J., Kim K., Kwon I.C., Kim J.S., Baek K.H., Oh Y.K. Pegylated poly-L-arginine derivatives of chitosan for effective delivery of siRNA. J. Control. Release. 2010;145:159–164. doi: 10.1016/j.jconrel.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 258.Khai-Woon E., Chen H.Y., Lo Y.L., Huang S.J., Wang L.F. Succinated chitosan as a gene carrier for improved chitosan solubility and gene transfection. Nanomed. Nanotechnol. Biol. Med. 2011;7:174–183. doi: 10.1016/j.nano.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 259.Thibault M., Nimesh S., Lavertu M., Buschmann M.D. Intracellular trafficking and decondensation kinetics of chitosan-pDNA polyplexes. Mol. Ther. 2010;18:1787–1795. doi: 10.1038/mt.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Nimesh S., Thibault M.M., Lavertu M., Buschmann M.D. Enhanced gene delivery mediated by low molecular weight chitosan/DNA complexes: effect of pH and serum. Mol. Biotechnol. 2010;46:182–196. doi: 10.1007/s12033-010-9286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Erbacher P., Roche A.C., Monsigny M., Midoux P. Putative role of chloroquine in gene transfer into a human hepatoma cell line by DNA lactosylated polylysine complexes. Exp. Cell Res. 1996;225:186–194. doi: 10.1006/excr.1996.0169. [DOI] [PubMed] [Google Scholar]
- 262.Corsi K., Chellat F., Yahia L., Fernandes J.C. Mesenchymal stem cells, MG63 and HEK293 transfection using chitosan-DNA nanoparticles. Biomaterials. 2003;24:1255–1264. doi: 10.1016/s0142-9612(02)00507-0. [DOI] [PubMed] [Google Scholar]
- 263.Noguchi H., Yoshikawa K. Morphological variation in a collapsed single homopolymer chain. J. Chem. Phys. 1998;109:5070–5077. [Google Scholar]
- 264.Lu H.D., Zhao H.Q., Wang K., Lv L.L. Novel hyaluronic acid-chitosan nanoparticles as non-viral gene delivery vectors targeting osteoarthritis. Int. J. Pharm. 2011;420:358–365. doi: 10.1016/j.ijpharm.2011.08.046. [DOI] [PubMed] [Google Scholar]
- 265.Lu B., Wang C.F., Wu D.Q., Li C., Zhang X.Z., Zhuo R.X. Chitosan based oligoamine polymers: synthesis, characterization, and gene delivery. J. Control. Release. 2009;137:54–62. doi: 10.1016/j.jconrel.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 266.Li Z.T., Guo J., Zhang J.S., Zhao Y.P., Lv L., Ding C., Zhang X.Z. Chitosan-graft-polyethylenimine with improved properties as a potential gene vector. Carbohydr. Polym. 2010;80:254–259. [Google Scholar]
- 267.Ping Y., Liu C., Zhang Z., Liu K.L., Chen J., Li J. Chitosan-graft-(PEI-beta-cyclodextrin) copolymers and their supramolecular PEGylation for DNA and siRNA delivery. Biomaterials. 2011;32:8328–8341. doi: 10.1016/j.biomaterials.2011.07.038. [DOI] [PubMed] [Google Scholar]
- 268.Bloomfield V.A. Condensation of DNA by multivalent cations: considerations on mechanism. Biopolymers. 1991;31:1471–1481. doi: 10.1002/bip.360311305. [DOI] [PubMed] [Google Scholar]
- 269.Lee D.W., Yun K.S., Ban H.S., Choe W., Lee S.K., Lee K.Y. Preparation and characterization of chitosan/polyguluronate nanoparticles for siRNA delivery. J. Control. Release. 2009;139:146–152. doi: 10.1016/j.jconrel.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 270.Dehousse V., Garbacki N., Jaspart S., Castagne D., Piel G., Colige A., Evrard B. Comparison of chitosan/siRNA and trimethylchitosan/siRNA complexes behaviour in vitro. Int. J. Biol. Macromol. 2010;46:342–349. doi: 10.1016/j.ijbiomac.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 271.Katas H., Somavarapu S., Alpar H.O. Nanocarriers for siRNA based on cationic polymers: chitosan and TAT peptide. J. Pharm. Pharmacol. 2005;57:S54. [Google Scholar]
- 272.Alameh M., DeJesus D., Jean M., Darras V., Thibault M., Lavertu M., Buschmann M.D., Merzouki A. Low molecular weight chitosan nanoparticulate system at low N:P ratio for nontoxic polynucleotide delivery. Int. J. Nanomed. 2012;7:1399–1414. doi: 10.2147/IJN.S26571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Wahlund K.G., Giddings J.C. Properties of an asymmetrical flow field-flow fractionation channel having one permeable wall. Anal. Chem. 1987;59:1332–1339. doi: 10.1021/ac00136a016. [DOI] [PubMed] [Google Scholar]
- 274.Schimpf M. 2000. Field Flow Fractionation Handbook. [Google Scholar]
- 275.Richard I., Thibault M., De Crescenzo G., Buschmann M.D., Lavertu M. Ionization behavior of chitosan and chitosan-DNA polyplexes indicate that chitosan has a similar capability to induce a proton-sponge effect as PEI. Biomacromolecules. 2013;14:1732–1740. doi: 10.1021/bm4000713. [DOI] [PubMed] [Google Scholar]
- 276.Jiang X., Dai H., Leong K.W., Goh S.H., Mao H.Q., Yang Y.Y. Chitosan-g-PEG/DNA complexes deliver gene to the rat liver via intrabiliary and intraportal infusions. J. Gene Med. 2006;8:477–487. doi: 10.1002/jgm.868. [DOI] [PubMed] [Google Scholar]
- 277.Li C.X., Guo T.Y., Zhou D.Z., Hu Y.L., Zhou H., Wang S.F., Chen J.T., Zhang Z.P. A novel glutathione modified chitosan conjugate for efficient gene delivery. J. Control. Release. 2011;154:177–188. doi: 10.1016/j.jconrel.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 278.Yu Y.Y., Wang Z., Cai L., Wang G., Yang X., Wan X.P., Xu X.H., Li Y., Gao R. Synthesis and characterization of methoxy poly(ethylene glycol)-O-chitosan-polyethylenimine for gene delivery. Carbohydr. Polym. 2010;81:269–274. [Google Scholar]
- 279.Zhao X., Yin L.C., Ding J.Y., Tang C., Gu S.H., Yin C.H., Mao Y.M. Thiolated trimethyl chitosan nanocomplexes as gene carriers with high in vitro and in vivo transfection efficiency. J. Control. Release. 2010;144:46–54. doi: 10.1016/j.jconrel.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 280.Lee S.J., Huh M.S., Lee S.Y., Min S., Lee S., Koo H., Chu J.U., Lee K.E., Jeon H., Choi Y., Choi K., Byun Y., Jeong S.Y., Park K., Kim K., Kwon I.C. Tumor-Homing Poly-siRNA/Glycol Chitosan Self-Cross-Linked Nanoparticles for Systemic siRNA Delivery in Cancer Treatment. Angew Chem. Int. Ed. Engl. 2012;51:7203–7207. doi: 10.1002/anie.201201390. [DOI] [PubMed] [Google Scholar]
- 281.Ogris M., Brunner S., Schuller S., Kircheis R., Wagner E. PEGylated DNA/transferrin-PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 1999;6:595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- 282.Albanese A., Tang P.S., Chan W.C.W. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng. 2012;14:1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 283.Elsabahy M., Wooley K.L. Design of polymeric nanoparticles for biomedical delivery applications. Chem. Soc. Rev. 2012;41:2545–2561. doi: 10.1039/c2cs15327k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Allen C., Dos Santos N., Gallagher R., Chiu G.N.C., Shu Y., Li W.M., Johnstone S.A., Janoff A.S., Mayer L.D., Webb M.S., Bally M.B. Controlling the physical behavior and biological performance of liposome formulations through use of surface grafted poly(ethylene glycol) Biosci. Rep. 2002;22:225–250. doi: 10.1023/a:1020186505848. [DOI] [PubMed] [Google Scholar]
- 285.Allen T.M. Long-circulating (sterically stabilized) liposomes for targeted drug delivery. Trends Pharmacol. Sci. 1994;15:215–220. doi: 10.1016/0165-6147(94)90314-x. [DOI] [PubMed] [Google Scholar]
- 286.Zhang Y.Q., Chen J.J., Zhang Y.D., Pan Y.F., Zhao J.F., Ren L.F., Liao M.M., Hu Z.Y., Kong L., Wang J.W. A novel PEGylation of chitosan nanoparticles for gene delivery. Biotechnol. Appl. Biochem. 2007;46:197–204. doi: 10.1042/BA20060163. [DOI] [PubMed] [Google Scholar]
- 287.Merdan T., Kunath K., Petersen H., Bakowsky U., Voigt K.H., Kopecek J., Kissel T. PEGylation of poly(ethylene imine) affects stability of complexes with plasmid DNA under in vivo conditions in a dose-dependent manner after intravenous injection into mice. Bioconjugate Chem. 2005;16:785–792. doi: 10.1021/bc049743q. [DOI] [PubMed] [Google Scholar]
- 288.Lai E., van Zanten J.H. Monitoring DNA/poly-L-lysine polyplex formation with time-resolved multiangle laser light scattering. Biophys. J. 2001;80:864–873. doi: 10.1016/S0006-3495(01)76065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Welsh S., Kay S.A. Reporter gene expression for monitoring gene transfer. Curr. Opin. Biotechnol. 1997;8:617–622. doi: 10.1016/s0958-1669(97)80038-9. [DOI] [PubMed] [Google Scholar]
- 290.Caceres G., Xiao Y.Z.U., Jiao J.A., Zankina R., Aller A., Andreotti P. Imaging of luciferase and GFP-transfected human tumours in nude mice. Luminescence. 2003;18:218–223. doi: 10.1002/bio.729. [DOI] [PubMed] [Google Scholar]
- 291.R. Varma, S. Magdaleno, Appropriate positive and negative controls are always important in biological experiments, Life Technologies application note.
- 292.Venkatesh S., Smith T.J. Chitosan-membrane interactions and their probable role in chitosan-mediated transfection. Biotechnol. Appl. Biochem. 1998;27:265–267. [PubMed] [Google Scholar]
- 293.Koping-Hoggard M., Nilsson M., Edwards K., Artursson P. Vol. 25. 1998. Chitosan-DNA polyplex: a new efficient, biodegradable gene delivery system; pp. 368–369. (Proc. Int'l. Symp. Control. Rel. Bioact. Mater.). [Google Scholar]
- 294.Kiang T., Bright C., Cheung C.Y., Stayton P.S., Hoffman A.S., Leong K.W. Formulation of chitosan-DNA nanoparticles with poly(propyl acrylic acid) enhances gene expression. J. Biomater. Sci. Polym. Ed. 2004;15:1405–1421. doi: 10.1163/1568562042368112. [DOI] [PubMed] [Google Scholar]
- 295.Douglas K.L., Piecirillo C.A., Tabrizian M. Effects of alginate inclusion on the vector properties of chitosan-based nanoparticles. J. Control. Release. 2006;115:354–361. doi: 10.1016/j.jconrel.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 296.Thibault M., Astolfi M., Tran-Khanh N., Lavertu M., Darras V., Merzouki A., Buschmann M.D. Excess polycation mediates efficient chitosan-based gene transfer by promoting lysosomal release of the polyplexes. Biomaterials. 2011;32:4639–4646. doi: 10.1016/j.biomaterials.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 297.Application Note. 2010. Nanosight, visualisation, sizing and counting of fluorescent and fluorescently-labelled nanoparticles. [Google Scholar]
- 298.Braeckmans K., Buyens K., Bouquet W., Vervaet C., Joye P., De Vos F., Plawinski L., Doeuvre L., Angles-Cano E., Sanders N.N., Demeester J., De Smedt S.C. Sizing Nanomatter in Biological Fluids by Fluorescence Single Particle Tracking. Nano Lett. 2010;10:4435–4442. doi: 10.1021/nl103264u. [DOI] [PubMed] [Google Scholar]
- 299.van Gaal E.V.B., Spierenburg G., Hennink W.E., Crommelin D.J.A., Mastrobattista E. Flow cytometry for rapid size determination and sorting of nucleic acid containing nanoparticles in biological fluids. J. Control. Release. 2010;141:328–338. doi: 10.1016/j.jconrel.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 300.Park I.K., Ihm J.E., Park Y.H., Choi Y.J., Kim S.I., Kim W.J., Akaike T., Cho C.S. Galactosylated chitosan (GC)-graft-poly(vinyl pyrrolidone) (PVP) as hepatocyte-targeting DNA carrier. Preparation and physicochemical characterization of GC-graft-PVP/DNA complex (1) J Control Release. 2003;86:349–359. doi: 10.1016/s0168-3659(02)00365-6. [DOI] [PubMed] [Google Scholar]
- 301.Bieber T., Meissner W., Kostin S., Niemann A., Elsasser H.P. Intracellular route and transcriptional competence of polyethylenimine-DNA complexes. J. Control. Release. 2002;82:441–454. doi: 10.1016/s0168-3659(02)00129-3. [DOI] [PubMed] [Google Scholar]
- 302.Peng S.F., Tseng M.T., Ho Y.C., Wei M.C., Liao Z.X., Sung H.W. Mechanisms of cellular uptake and intracellular trafficking with chitosan/DNA/poly(gamma-glutamic acid) complexes as a gene delivery vector. Biomaterials. 2011;32:239–248. doi: 10.1016/j.biomaterials.2010.08.081. [DOI] [PubMed] [Google Scholar]
- 303.Jones N.A., Hill I.R.C., Stolnik S., Bignotti F., Davis S.S., Garnett M.C. Polymer chemical structure is a key determinant of physicochemical and colloidal properties of polymer-DNA complexes for gene delivery. Biochim. Biophys. Acta-Gene Struct. Expression. 2000;1517:1–18. doi: 10.1016/s0167-4781(00)00220-7. [DOI] [PubMed] [Google Scholar]
- 304.Rojanarata T., Opanasopit P., Techaarpornkul S., Ngawhirunpat T., Ruktanonchai U. Chitosan-Thiamine Pyrophosphate as a Novel Carrier for siRNA Delivery. Pharm. Res. 2008;25:2807–2814. doi: 10.1007/s11095-008-9648-6. [DOI] [PubMed] [Google Scholar]
- 305.Wedmore I., McManus J.G., Pusateri A.E., Holcomb J.B. A special report on the chitosan-based hemostatic dressing: Experience in current combat operations. J. Trauma-Injury Infect. Crit. Care. 2006;60:655–658. doi: 10.1097/01.ta.0000199392.91772.44. [DOI] [PubMed] [Google Scholar]
- 306.Varum K.M., Myhr M.M., Hjerde R.J.N., Smidsrod O. In vitro degradation rates of partially N-acetylated chitosans in human serum. Carbohydr. Res. 1997;299:99–101. doi: 10.1016/s0008-6215(96)00332-1. [DOI] [PubMed] [Google Scholar]
- 307.Boot R.G., Renkema G.H., Strijland A., van Zonneveld A.J., Aerts J.M. Cloning of a cDNA encoding chitotriosidase, a human chitinase produced by macrophages. J. Biol. Chem. 1995;270:26252–26256. doi: 10.1074/jbc.270.44.26252. [DOI] [PubMed] [Google Scholar]
- 308.Muzzarelli R.A.A. Human enzymatic activities related to the therapeutic administration of chitin derivatives. Cell. Mol. Life Sci. 1997;53:131–140. doi: 10.1007/PL00000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Huang M., Khor E., Lim L.Y. Uptake and cytotoxicity of chitosan molecules and nanoparticles: effects of molecular weight and degree of deacetylation. Pharm. Res. 2004;21:344–353. doi: 10.1023/b:pham.0000016249.52831.a5. [DOI] [PubMed] [Google Scholar]
- 310.Schipper N.G.M., Varum K.M., Artursson P. Chitosans as absorption enhancers for poorly absorbable drugs.1. Influence of molecular weight and degree of acetylation on drug transport across human intestinal epithelial (Caco-2) cells. Pharm. Res. 1996;13:1686–1692. doi: 10.1023/a:1016444808000. [DOI] [PubMed] [Google Scholar]
- 311.Gorzelanny C., Poeppelmann B., Pappelbaum K., Moerschbacher B.M., Schneider S.W. Human macrophage activation triggered by chitotriosidase-mediated chitin and chitosan degradation. Biomaterials. 2010;31:8556–8563. doi: 10.1016/j.biomaterials.2010.07.100. [DOI] [PubMed] [Google Scholar]
- 312.Guzman-Morales J., Ariganello M.B., Hammami I., Thibault M., Jolicoeur M., Hoemann C.D. Biodegradable chitosan particles induce chemokine release and negligible arginase-1 activity compared to IL-4 in murine bone marrow-derived macrophages. Biochem. Biophys. Res. Commun. 2011;405:538–544. doi: 10.1016/j.bbrc.2011.01.063. [DOI] [PubMed] [Google Scholar]
- 313.Fotakis G., Timbrell J.A. In vitro cytotoxicity assays: comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol. Lett. 2006;160:171–177. doi: 10.1016/j.toxlet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 314.Kai E., Ochiya T. A method for oral DNA delivery with N-acetylated chitosan. Pharm. Res. 2004;21:838–843. doi: 10.1023/b:pham.0000026437.32238.6f. [DOI] [PubMed] [Google Scholar]
- 315.Douglas K.L., Piccirillo C.A., Tabrizian M. Cell line-dependent internalization pathways and intracellular trafficking determine transfection efficiency of nanoparticle vectors. Eur. J. Pharm. Biopharm. 2008;68:676–687. doi: 10.1016/j.ejpb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 316.Turan K., Nagata K. Chitosan-DNA nanoparticles: the effect of cell type and hydrolysis of chitosan on in vitro DNA transfection. Pharm. Dev. Technol. 2006;11:503–512. doi: 10.1080/10837450600940873. [DOI] [PubMed] [Google Scholar]
- 317.Brunner S., Furtbauer E., Sauer T., Kursa M., Wagner E. Overcoming the nuclear barrier: cell cycle independent nonviral gene transfer with linear polyethylenimine or electroporation. Mol. Ther. 2002;5:80–86. doi: 10.1006/mthe.2001.0509. [DOI] [PubMed] [Google Scholar]
- 318.Won Y.Y., Sharma R., Konieczny S.F. Missing pieces in understanding the intracellular trafficking of polycation/DNA complexes. J. Control. Release. 2009;139:88–93. doi: 10.1016/j.jconrel.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Sonawane N.D., Szoka F.C., Jr., Verkman A.S. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J. Biol. Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 320.Chen H.H., Ho Y.P., Jiang X., Mao H.Q., Wang T.H., Leong K.W. Quantitative comparison of intracellular unpacking kinetics of polyplexes by a model constructed from quantum Dot-FRET. Mol. Ther. 2008;16:324–332. doi: 10.1038/sj.mt.6300392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Kiang T., Wen H., Lim H.W., Leong K.W. The effect of the degree of chitosan deacetylation on the efficiency of gene transfection. Biomaterials. 2004;25:5293–5301. doi: 10.1016/j.biomaterials.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 322.Steen M., Kirchberger T., Guse A.H. NAADP mobilizes calcium from the endoplasmic reticular Ca2 + store in T-lymphocytes. J. Biol. Chem. 2007;282:18864–18871. doi: 10.1074/jbc.M610925200. [DOI] [PubMed] [Google Scholar]
- 323.Itaka K., Harada A., Yamasaki Y., Nakamura K., Kawaguchi H., Kataoka K. In situ single cell observation by fluorescence resonance energy transfer reveals fast intra-cytoplasmic delivery and easy release of plasmid DNA complexed with linear polyethylenimine. J. Gene Med. 2004;6:76–84. doi: 10.1002/jgm.470. [DOI] [PubMed] [Google Scholar]
- 324.Kichler A., Leborgne C., Coeytaux E., Danos O. Polyethylenimine-mediated gene delivery: a mechanistic study. J. Gene Med. 2001;3:135–144. doi: 10.1002/jgm.173. [DOI] [PubMed] [Google Scholar]
- 325.Behr J.P. The proton sponge: a trick to enter cells the viruses did not exploit. Chimia. 1997;51:34–36. [Google Scholar]
- 326.Bowman K., Leong K.W. Chitosan nanoparticles for oral drug and gene delivery. Int. J. Nanomed. 2006;1:117–128. doi: 10.2147/nano.2006.1.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Forrest M.L., Gabrielson N., Pack D.W. Reduction of polyethylenimine buffering capacity enhances in-vitro gene delivery activity. Mol. Ther. 2004;9:S138. [Google Scholar]
- 328.Gabrielson N.P., Pack D.W. Acetylation of polyethylenimine enhances gene delivery via weakened polymer/DNA interactions. Biomacromolecules. 2006;7:2427–2435. doi: 10.1021/bm060300u. [DOI] [PubMed] [Google Scholar]
- 329.Godbey W.T., Barry M.A., Saggau P., Wu K.K., Mikos A.G. Poly(ethylenimine)-mediated transfection: a new paradigm for gene delivery. J. Biomed. Mater. Res. 2000;51:321–328. doi: 10.1002/1097-4636(20000905)51:3<321::aid-jbm5>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 330.Godbey W.T., Wu K.K., Mikos A.G. Poly(ethylenimine)-mediated gene delivery affects endothelial cell function and viability. Biomaterials. 2001;22:471–480. doi: 10.1016/s0142-9612(00)00203-9. [DOI] [PubMed] [Google Scholar]
- 331.Kim T.H., Kim S.I., Akaike T., Cho C.S. Synergistic effect of poly(ethylenimine) on the transfection efficiency of galactosylated chitosan/DNA complexes. J. Control. Release. 2005;105:354–366. doi: 10.1016/j.jconrel.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 332.Schaffer D.V., Fidelman N.A., Dan N., Lauffenburger D.A. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol. Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 333.Ho Y.P., Chen H.H., Leong K.W., Wang T.H. Evaluating the intracellular stability and unpacking of DNA nanocomplexes by quantum dots-FRET. J. Control. Release. 2006;116:83–89. doi: 10.1016/j.jconrel.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.LabatMoleur F., Steffan A.M., Brisson C., Perron H., Feugeas O., Furstenberger P., Oberling F., Brambilla E., Behr J.P. An electron microscopy study into the mechanism of gene transfer with lipopolyamines. Gene Ther. 1996;3:1010–1017. [PubMed] [Google Scholar]
- 335.Liang D.C., Liu W.G., Zuo A.J., Sun S.J., Cheng N., Guo G., Zhang J.Y., De Yao K. Pre-deliver chitosanase to cells: a novel strategy to improve gene expression by endocellular degradation-induced vector unpacking. Int. J. Pharm. 2006;314:63–71. doi: 10.1016/j.ijpharm.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 336.Hacein-Bey-Abina S., von Kalle C., Schmidt M., Le Deist F., Wulffraat N., McIntyre E., Radford I., Villeval J.L., Fraser C.C., Cavazzana-Calvo M., Fischer A. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 337.Check E. A tragic setback. Nature. 2002;420:116–118. doi: 10.1038/420116a. [DOI] [PubMed] [Google Scholar]
- 338.Gurunathan S., Klinman D.M., Seder R.A. DNA vaccines: immunology, application, and optimization*. Annu. Rev. Immunol. 2000;18:927–974. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- 339.Dodane V., Amin Khan M., Merwin J.R. Effect of chitosan on epithelial permeability and structure. Int. J. Pharm. 1999;182:21–32. doi: 10.1016/s0378-5173(99)00030-7. [DOI] [PubMed] [Google Scholar]
- 340.McNeela E.A., O'Connor D., Jabbal-Gill I., Illum L., Davis S.S., Pizza M., Peppoloni S., Rappuoli R., Mills K.H. A mucosal vaccine against diphtheria: formulation of cross reacting material (CRM(197)) of diphtheria toxin with chitosan enhances local and systemic antibody and Th2 responses following nasal delivery. Vaccine. 2000;19:1188–1198. doi: 10.1016/s0264-410x(00)00309-1. [DOI] [PubMed] [Google Scholar]
- 341.Roy K., Mao H.Q., Huang S.K., Leong K.W. Oral gene delivery with chitosan–DNA nanoparticles generates immunologic protection in a murine model of peanut allergy. Nat. Med. 1999;5:387–391. doi: 10.1038/7385. [DOI] [PubMed] [Google Scholar]
- 342.Khatri K., Goyal A.K., Gupta P.N., Mishra N., Vyas S.P. Plasmid DNA loaded chitosan nanoparticles for nasal mucosal immunization against hepatitis B. Int. J. Pharm. 2008;354:235–241. doi: 10.1016/j.ijpharm.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 343.Schirmbeck R., Reimann J. Revealing the potential of DNA-based vaccination: lessons learned from the hepatitis B virus surface antigen. Biol. Chem. 2001;382:543–552. doi: 10.1515/BC.2001.068. [DOI] [PubMed] [Google Scholar]
- 344.Bramwell V.W., Eyles J.E., Somavarapu S., Alpar H.O. Liposome/DNA complexes coated with biodegradable PLA improve immune responses to plasmid encoding hepatitis B surface antigen. Immunology. 2002;106:412–418. doi: 10.1046/j.1365-2567.2002.01448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Bos G.W., Kanellos T., Crommelin D.J., Hennink W.E., Howard C.R. Cationic polymers that enhance the performance of HbsAg DNA in vivo. Vaccine. 2004;23:460–469. doi: 10.1016/j.vaccine.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 346.Takashima A., Morita A. Dendritic cells in genetic immunization. J. Leukoc. Biol. 1999;66:350–356. doi: 10.1002/jlb.66.2.350. [DOI] [PubMed] [Google Scholar]
- 347.Shedlock D.J., Weiner D.B. DNA vaccination: antigen presentation and the induction of immunity. J. Leukoc. Biol. 2000;68:793–806. [PubMed] [Google Scholar]
- 348.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 349.Hohlfeld R., Engel A.G. The immunobiology of muscle. Immunol. Today. 1994;15:269–274. doi: 10.1016/0167-5699(94)90006-X. [DOI] [PubMed] [Google Scholar]
- 350.Otterlei M., Varum K.M., Ryan L., Espevik T. Characterization of binding and TNF-alpha-inducing ability of chitosans on monocytes: the involvement of CD14. Vaccine. 1994;12:825–832. doi: 10.1016/0264-410x(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 351.Chou T.C., Fu E., Shen E.C. Chitosan inhibits prostaglandin E2 formation and cyclooxygenase-2 induction in lipopolysaccharide-treated RAW 264.7 macrophages. Biochem. Biophys. Res. Commun. 2003;308:403–407. doi: 10.1016/s0006-291x(03)01407-4. [DOI] [PubMed] [Google Scholar]
- 352.Meyer K.B., Thompson M.M., Levy M.Y., Barron L.G., Szoka F.C., Jr. Intratracheal gene delivery to the mouse airway: characterization of plasmid DNA expression and pharmacokinetics. Gene Ther. 1995;2:450–460. [PubMed] [Google Scholar]
- 353.Tuomela M., Malm M., Wallen M., Stanescu I., Krohn K., Peterson P. Biodistribution and general safety of a naked DNA plasmid, GTU-MultiHIV, in a rat, using a quantitative PCR method. Vaccine. 2005;23:890–896. doi: 10.1016/j.vaccine.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 354.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 355.Bantounas I., Phylactou L.A., Uney J.B. RNA interference and the use of small interfering RNA to study gene function in mammalian systems. J. Mol. Endocrinol. 2004;33:545–557. doi: 10.1677/jme.1.01582. [DOI] [PubMed] [Google Scholar]
- 356.Castanotto D., Rossi J.J. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Zimmermann T.S., Lee A.C., Akinc A., Bramlage B., Bumcrot D., Fedoruk M.N., Harborth J., Heyes J.A., Jeffs L.B., John M., Judge A.D., Lam K., McClintock K., Nechev L.V., Palmer L.R., Racie T., Rohl I., Seiffert S., Shanmugam S., Sood V., Soutschek J., Toudjarska I., Wheat A.J., Yaworski E., Zedalis W., Koteliansky V., Manoharan M., Vornlocher H.P., MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 358.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 359.Whelan J. First clinical data on RNAi. Drug Discovery Today. 2005;10:1014–1015. doi: 10.1016/S1359-6446(05)03547-6. [DOI] [PubMed] [Google Scholar]
- 360.Corey D.R. Chemical modification: the key to clinical application of RNA interference? J. Clin. Invest. 2007;117:3615–3622. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Corey D.R. RNA learns from antisense. Nat. Chem. Biol. 2007;3:8–11. doi: 10.1038/nchembio0107-8. [DOI] [PubMed] [Google Scholar]
- 362.Stein C.A. Phosphorothioate antisense oligodeoxynucleotides: questions of specificity. Trends Biotechnol. 1996;14:147–149. doi: 10.1016/0167-7799(96)20006-X. [DOI] [PubMed] [Google Scholar]
- 363.Urban-Klein B., Werth S., Abuharbeid S., Czubayko F., Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 364.Salva E., Kabasakal L., Eren F., Ozkan N., Cakalagaoglu F., Akbuga J. Local delivery of chitosan/VEGF siRNA nanoplexes reduces angiogenesis and growth of breast cancer in vivo. Nucleic Acid Ther. 2012;22:40–48. doi: 10.1089/nat.2011.0312. [DOI] [PubMed] [Google Scholar]
- 365.Han H.D., Mora E.M., Roh J.W., Nishimura M., Lee S.J., Stone R.L., Bar-Eli M., Lopez-Berestein G., Sood A.K. Chitosan hydrogel for localized gene silencing. Cancer Biol. Ther. 2011;11:839–845. doi: 10.4161/cbt.11.9.15185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Kim H.S., Han H.D., Armaiz-Pena G.N., Stone R.L., Nam E.J., Lee J.W., Shahzad M.M., Nick A.M., Lee S.J., Roh J.W., Nishimura M., Mangala L.S., Bottsford-Miller J., Gallick G.E., Lopez-Berestein G., Sood A.K. Functional roles of Src and Fgr in ovarian carcinoma. Clin. Cancer Res. 2011;17:1713–1721. doi: 10.1158/1078-0432.CCR-10-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Wiegand C., Winter D., Hipler U.C. Molecular-weight-dependent toxic effects of chitosans on the human keratinocyte cell line HaCaT. Skin Pharmacol. Physiol. 2010;23:164–170. doi: 10.1159/000276996. [DOI] [PubMed] [Google Scholar]
- 368.Yang Y.M., Hu W., Wang X.D., Gu X.S. The controlling biodegradation of chitosan fibers by N-acetylation in vitro and in vivo. J. Mater. Sci. Mater. Med. 2007;18:2117–2121. doi: 10.1007/s10856-007-3013-x. [DOI] [PubMed] [Google Scholar]
- 369.Xu J., McCarthy S.P., Gross R.A., Kaplan D.L. Chitosan film acylation and effects on biodegradability. Macromolecules. 1996;29:3436–3440. [Google Scholar]
- 370.Hirano S., Tsuchida H., Nagao N. N-acetylation in chitosan and the rate of its enzymic hydrolysis. Biomaterials. 1989;10:574–576. doi: 10.1016/0142-9612(89)90066-5. [DOI] [PubMed] [Google Scholar]
- 371.Gao S., Dagnaes-Hansen F., Nielsen E.J., Wengel J., Besenbacher F., Howard K.A., Kjems J. The effect of chemical modification and nanoparticle formulation on stability and biodistribution of siRNA in mice. Mol. Ther. 2009;17:1225–1233. doi: 10.1038/mt.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Yan J., Yang L., Wang G., Xiao Y., Zhang B., Qi N. Biocompatibility evaluation of chitosan-based injectable hydrogels for the culturing mice mesenchymal stem cells in vitro. J. Biomater. Appl. 2010;24:625–637. doi: 10.1177/0885328208100536. [DOI] [PubMed] [Google Scholar]
- 373.Huh M.S., Lee S.Y., Park S., Lee S., Chung H., Choi Y., Oh Y.K., Park J.H., Jeong S.Y., Choi K., Kim K., Kwon I.C. Tumor-homing glycol chitosan/polyethylenimine nanoparticles for the systemic delivery of siRNA in tumor-bearing mice. J. Control. Release. 2010;144:134–143. doi: 10.1016/j.jconrel.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 374.Matsumura Y., Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 375.Yan C., Chen D., Gu J., Qin J. Nanoparticles of 5-fluorouracil (5-FU) loaded N-succinyl-chitosan (Suc-Chi) for cancer chemotherapy: preparation, characterization–in-vitro drug release and anti-tumour activity. J. Pharm. Pharmacol. 2006;58:1177–1181. doi: 10.1211/jpp.58.9.0003. [DOI] [PubMed] [Google Scholar]
- 376.Torchilin V. Tumor delivery of macromolecular drugs based on the EPR effect. Adv. Drug Deliv. Rev. 2011;63:131–135. doi: 10.1016/j.addr.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 377.Cheng M., Li Q., Wan T., Hong X., Chen H., He B., Cheng Z., Xu H., Ye T., Zha B., Wu J., Zhou R. Synthesis and efficient hepatocyte targeting of galactosylated chitosan as a gene carrier in vitro and in vivo. J. Biomed. Mater. Res. B Appl. Biomater. 2011;99:70–80. doi: 10.1002/jbm.b.31873. [DOI] [PubMed] [Google Scholar]
- 378.Guliyeva U., Oner F., Ozsoy S., Haziroglu R. Chitosan microparticles containing plasmid DNA as potential oral gene delivery system. Eur. J. Pharm. Biopharm. 2006;62:17–25. doi: 10.1016/j.ejpb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 379.Li G.P., Liu Z.G., Liao B., Zhong N.S. Induction of Th1-type immune response by chitosan nanoparticles containing plasmid DNA encoding house dust mite allergen Der p 2 for oral vaccination in mice. Cell. Mol. Immunol. 2009;6:45–50. doi: 10.1038/cmi.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Chen J., Yang W.L., Li G., Qian J., Xue J.L., Fu S.K., Lu D.R. Transfection of mEpo gene to intestinal epithelium in vivo mediated by oral delivery of chitosan-DNA nanoparticles. World J. Gastroenterol. 2004;10:112–116. doi: 10.3748/wjg.v10.i1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Sun C.J., Pan S.P., Xie Q.X., Xiao L.J. Preparation of chitosan-plasmid DNA nanoparticles encoding zona pellucida glycoprotein-3alpha and its expression in mouse. Mol. Reprod. Dev. 2004;68:182–188. doi: 10.1002/mrd.20058. [DOI] [PubMed] [Google Scholar]
- 382.Zheng F., Shi X.W., Yang G.F., Gong L.L., Yuan H.Y., Cui Y.J., Wang Y., Du Y.M., Li Y. Chitosan nanoparticle as gene therapy vector via gastrointestinal mucosa administration: results of an in vitro and in vivo study. Life Sci. 2007;80:388–396. doi: 10.1016/j.lfs.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 383.Hejazi R., Amiji M. Chitosan-based gastrointestinal delivery systems. J. Control. Release. 2003;89:151–165. doi: 10.1016/s0168-3659(03)00126-3. [DOI] [PubMed] [Google Scholar]
- 384.Tian J., Yu J., Sun X. Chitosan microspheres as candidate plasmid vaccine carrier for oral immunisation of Japanese flounder (Paralichthys olivaceus) Vet. Immunol. Immunopathol. 2008;126:220–229. doi: 10.1016/j.vetimm.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 385.Illum L., Jabbal-Gill I., Hinchcliffe M., Fisher A.N., Davis S.S. Chitosan as a novel nasal delivery system for vaccines. Adv. Drug Deliv. Rev. 2001;51:81–96. doi: 10.1016/s0169-409x(01)00171-5. [DOI] [PubMed] [Google Scholar]
- 386.Luo Y., Zhai X., Ma C., Sun P., Fu Z., Liu W., Xu J. An inhalable beta(2)-adrenoceptor ligand-directed guanidinylated chitosan carrier for targeted delivery of siRNA to lung. J. Control. Release. 2012;162:28–36. doi: 10.1016/j.jconrel.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 387.Kong X., Zhang W., Lockey R.F., Auais A., Piedimonte G., Mohapatra S.S. Respiratory syncytial virus infection in Fischer 344 rats is attenuated by short interfering RNA against the RSV-NS1 gene. Genet Vaccines Ther. 2007;5:4. doi: 10.1186/1479-0556-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Jiang H.L., Xu C.X., Kim Y.K., Arote R., Jere D., Lim H.T., Cho M.H., Cho C.S. The suppression of lung tumorigenesis by aerosol-delivered folate-chitosan-graft-polyethylenimine/Akt1 shRNA complexes through the Akt signaling pathway. Biomaterials. 2009;30:5844–5852. doi: 10.1016/j.biomaterials.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 389.Mandke R., Singh J. Cationic nanomicelles for delivery of plasmids encoding interleukin-4 and interleukin-10 for prevention of autoimmune diabetes in mice. Pharm. Res. 2012;29:883–897. doi: 10.1007/s11095-011-0616-1. [DOI] [PubMed] [Google Scholar]
- 390.Basarkar A., Singh J. Poly (lactide-co-glycolide)-polymethacrylate nanoparticles for intramuscular delivery of plasmid encoding interleukin-10 to prevent autoimmune diabetes in mice. Pharm. Res. 2009;26:72–81. doi: 10.1007/s11095-008-9710-4. [DOI] [PubMed] [Google Scholar]
- 391.Rosenberg S.A., Aebersold P., Cornetta K., Kasid A., Morgan R.A., Moen R., Karson E.M., Lotze M.T., Yang J.C., Topalian S.L., et al. Gene transfer into humans–immunotherapy of patients with advanced melanoma, using tumor-infiltrating lymphocytes modified by retroviral gene transduction. N. Engl. J. Med. 1990;323:570–578. doi: 10.1056/NEJM199008303230904. [DOI] [PubMed] [Google Scholar]
- 392.Gene therapy clinical trials worldwide. 2012. http://www.wiley.com/legacy/wileychi/genmed/clinical/
- 393.Noone P.G., Hohneker K.W., Zhou Z., Johnson L.G., Foy C., Gipson C., Jones K., Noah T.L., Leigh M.W., Schwartzbach C., Efthimiou J., Pearlman R., Boucher R.C., Knowles M.R. Safety and biological efficacy of a lipid-CFTR complex for gene transfer in the nasal epithelium of adult patients with cystic fibrosis. Mol. Ther. 2000;1:105–114. doi: 10.1006/mthe.1999.0009. [DOI] [PubMed] [Google Scholar]
- 394.Bedikian A.Y., Richards J., Kharkevitch D., Atkins M.B., Whitman E., Gonzalez R. A phase 2 study of high-dose Allovectin-7 in patients with advanced metastatic melanoma. Melanoma Res. 2010;20:218–226. doi: 10.1097/CMR.0b013e3283390711. [DOI] [PubMed] [Google Scholar]
- 395.Sidi A.A., Ohana P., Benjamin S., Shalev M., Ransom J.H., Lamm D., Hochberg A., Leibovitch I. Phase I/II marker lesion study of intravesical BC-819 DNA plasmid in H19 over expressing superficial bladder cancer refractory to bacillus Calmette-Guerin. J. Urol. 2008;180:2379–2383. doi: 10.1016/j.juro.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 396.Davis M.E., Zuckerman J.E., Choi C.H., Seligson D., Tolcher A., Alabi C.A., Yen Y., Heidel J.D., Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.El-Kamary S.S., Pasetti M.F., Mendelman P.M., Frey S.E., Bernstein D.I., Treanor J.J., Ferreira J., Chen W.H., Sublett R., Richardson C., Bargatze R.F., Sztein M.B., Tacket C.O. Adjuvanted intranasal Norwalk virus-like particle vaccine elicits antibodies and antibody-secreting cells that express homing receptors for mucosal and peripheral lymphoid tissues. J. Infect. Dis. 2010;202:1649–1658. doi: 10.1086/657087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Shi Q., Wang H., Tran C., Qiu X., Winnik F.M., Zhang X., Dai K., Benderdour M., Fernandes J.C. Hydrodynamic delivery of chitosan-folate-DNA nanoparticles in rats with adjuvant-induced arthritis. J. Biomed. Biotechnol. 2011;2011:148763. doi: 10.1155/2011/148763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Raghuwanshi D., Mishra V., Das D., Kaur K., Suresh M.R. Dendritic cell targeted chitosan nanoparticles for nasal DNA immunization against SARS CoV nucleocapsid protein. Mol. Pharm. 2012;9:946–956. doi: 10.1021/mp200553x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Ekrami H.M., Shen W.C. Carbamylation decreases the cytotoxicity but not the drug-carrier properties of polylysines. J. Drug Target. 1995;2:469–475. doi: 10.3109/10611869509015916. [DOI] [PubMed] [Google Scholar]
- 401.Lai W.F., Lin M.C. Nucleic acid delivery with chitosan and its derivatives. J. Control. Release. 2009;134:158–168. doi: 10.1016/j.jconrel.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 402.Muzzarelli R.A. Chitins and chitosans as immunoadjuvants and non-allergenic drug carriers. Mar. Drugs. 2010;8:292–312. doi: 10.3390/md8020292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Shibata Y., Foster L.A., Metzger W.J., Myrvik Q.N. Alveolar macrophage priming by intravenous administration of chitin particles, polymers of N-acetyl-D-glucosamine, in mice. Infect. Immun. 1997;65:1734–1741. doi: 10.1128/iai.65.5.1734-1741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Shibata Y., Foster L.A., Bradfield J.F., Myrvik Q.N. Oral administration of chitin down-regulates serum IgE levels and lung eosinophilia in the allergic mouse. J. Immunol. 2000;164:1314–1321. doi: 10.4049/jimmunol.164.3.1314. [DOI] [PubMed] [Google Scholar]
- 405.Shibata Y., Honda I., Justice J.P., Van Scott M.R., Nakamura R.M., Myrvik Q.N. Th1 adjuvant N-acetyl-D-glucosamine polymer up-regulates Th1 immunity but down-regulates Th2 immunity against a mycobacterial protein (MPB-59) in interleukin-10-knockout and wild-type mice. Infect. Immun. 2001;69:6123–6130. doi: 10.1128/IAI.69.10.6123-6130.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Strong P., Clark H., Reid K. Intranasal application of chitin microparticles down-regulates symptoms of allergic hypersensitivity to Dermatophagoides pteronyssinus and Aspergillus fumigatus in murine models of allergy. Clin. Exp. Allergy. 2002;32:1794–1800. doi: 10.1046/j.1365-2222.2002.01551.x. [DOI] [PubMed] [Google Scholar]
- 407.Chew J.L., Wolfowicz C.B., Mao H.Q., Leong K.W., Chua K.Y. Chitosan nanoparticles containing plasmid DNA encoding house dust mite allergen, Der p 1 for oral vaccination in mice. Vaccine. 2003;21:2720–2729. doi: 10.1016/s0264-410x(03)00228-7. [DOI] [PubMed] [Google Scholar]
- 408.Lee C.G., Da Silva C.A., Lee J.Y., Hartl D., Elias J.A. Chitin regulation of immune responses: an old molecule with new roles. Curr. Opin. Immunol. 2008;20:684–689. doi: 10.1016/j.coi.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Da Silva C.A., Chalouni C., Williams A., Hartl D., Lee C.G., Elias J.A. Chitin Is a Size-Dependent Regulator of Macrophage TNF and IL-10 Production. J. Immunol. 2009;182:3573–3582. doi: 10.4049/jimmunol.0802113. [DOI] [PubMed] [Google Scholar]
- 410.Reasor M.J., Hastings K.L., Ulrich R.G. Drug-induced phospholipidosis: issues and future directions. Expert Opin. Drug Saf. 2006;5:567–583. doi: 10.1517/14740338.5.4.567. [DOI] [PubMed] [Google Scholar]
- 411.Tousignant J.D., Gates A.L., Ingram L.A., Johnson C.L., Nietupski J.B., Cheng S.H., Eastman S.J., Scheule R.K. Comprehensive analysis of the acute toxicities induced by systemic administration of cationic lipid:plasmid DNA complexes in mice. Hum. Gene Ther. 2000;11:2493–2513. doi: 10.1089/10430340050207984. [DOI] [PubMed] [Google Scholar]
- 412.Ishida T., Ichihara M., Wang X., Yamamoto K., Kimura J., Majima E., Kiwada H. Injection of PEGylated liposomes in rats elicits PEG-specific IgM, which is responsible for rapid elimination of a second dose of PEGylated liposomes. J. Control. Release. 2006;112:15–25. doi: 10.1016/j.jconrel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 413.Boeckle S., von Gersdorff K., van der Piepen S., Culmsee C., Wagner E., Ogris M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J. Gene Med. 2004;6:1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 414.Moghimi S.M., Symonds P., Murray J.C., Hunter A.C., Debska G., Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol. Ther. 2005;11:990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 415.Hunter A.C., Moghimi S.M. Cationic carriers of genetic material and cell death: a mitochondrial tale. Biochim. Biophys. Acta. 2010;1797:1203–1209. doi: 10.1016/j.bbabio.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 416.Li F., Liu W.G., Yao K.D. Preparation of oxidized glucose-crosslinked N-alkylated chitosan membrane and in vitro studies of pH-sensitive drug delivery behaviour. Biomaterials. 2002;23:343–347. doi: 10.1016/s0142-9612(01)00111-9. [DOI] [PubMed] [Google Scholar]
- 417.Park I.K., Jiang H.L., Cook S.E., Cho M.H., Kim S.I., Jeong H.J., Akaike T., Cho C.S. Galactosylated chitosan (GC)-graft-poly(vinyl pyrrolidone) (PVP) as hepatocyte-targeting DNA carrier: in vitro transfection. Arch. Pharm. Res. 2004;27:1284–1289. doi: 10.1007/BF02975895. [DOI] [PubMed] [Google Scholar]
- 418.Lee D., Lockey R., Mohapatra S. Folate receptor-mediated cancer cell specific gene delivery using folic acid-conjugated oligochitosans. J. Nanosci. Nanotechnol. 2006;6:2860–2866. doi: 10.1166/jnn.2006.465. [DOI] [PubMed] [Google Scholar]
- 419.Mohammadi Z., Abolhassani M., Dorkoosh F.A., Hosseinkhani S., Gilani K., Amini T., Najafabadi A.R., Tehrani M.R. Preparation and evaluation of chitosan-DNA-FAP-B nanoparticles as a novel non-viral vector for gene delivery to the lung epithelial cells. Int. J. Pharm. 2011;409:307–313. doi: 10.1016/j.ijpharm.2011.02.043. [DOI] [PubMed] [Google Scholar]
- 420.Oliveira H., Pires L.R., Fernandez R., Martins M.C., Simoes S., Pego A.P. Chitosan-based gene delivery vectors targeted to the peripheral nervous system. J. Biomed. Mater. Res. A. 2010;95:801–810. doi: 10.1002/jbm.a.32874. [DOI] [PubMed] [Google Scholar]
- 421.Kumar A., Jena P.K., Behera S., Lockey R.F., Mohapatra S. Multifunctional magnetic nanoparticles for targeted delivery. Nanomedicine. 2010;6:64–69. doi: 10.1016/j.nano.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Midoux P., Monsigny M. Efficient gene transfer by histidylated polylysine/pDNA complexes. Bioconjugate Chem. 1999;10:406–411. doi: 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- 423.Terbojevich M., Cosani A., Focher B., Marsano E. High-performance gel-permeation chromatography of chitosan samples. Carbohydr. Res. 1993:301–314. [Google Scholar]
- 424.Terbojevich M., Cosani A., Focher B., Naggi A., Torri G. Chitosans from Euphausia superba. 1: Solution properties. Carbohydr. Polym. 1992;18:35–42. [Google Scholar]
- 425.Drogoz A., David L., Rochas C., Domard A., Delair T. Polyelectrolyte complexes from polysaccharides: Formation and stoichiometry monitoring. Langmuir. 2007;23:10950–10958. doi: 10.1021/la7008545. [DOI] [PubMed] [Google Scholar]
- 426.Iqbal M., Lin W., Jabbal-Gill I., Davis S.S., Steward M.W., Illum L. Nasal delivery of chitosan-DNA plasmid expressing epitopes of respiratory syncytial virus (RSV) induces protective CTL responses in BALB/c mice. Vaccine. 2003;21:1478–1485. doi: 10.1016/s0264-410x(02)00662-x. [DOI] [PubMed] [Google Scholar]
- 427.Zhao Q.Q., Hu Y.L., Zhou Y., Li N., Han M., Tang G.P., Qiu F., Tabata Y., Gao J.Q. Gene-carried hepatoma targeting complex induced high gene transfection efficiency with low toxicity and significant antitumor activity. Int. J. Nanomed. 2012;7:3191–3202. doi: 10.2147/IJN.S30909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Yang S.J., Chang S.M., Tsai K.C., Tsai H.M., Chen W.S., Shieh M.J. Enhancement of chitosan nanoparticle-facilitated gene transfection by ultrasound both in vitro and in vivo. J. Biomed. Mater. Res. B Appl. Biomater. 2012;100:1746–1754. doi: 10.1002/jbm.b.32741. [DOI] [PubMed] [Google Scholar]
- 429.Guo R., Xu S., Ma L., Huang A., Gao C. The healing of full-thickness burns treated by using plasmid DNA encoding VEGF-165 activated collagen-chitosan dermal equivalents. Biomaterials. 2011;32:1019–1031. doi: 10.1016/j.biomaterials.2010.08.087. [DOI] [PubMed] [Google Scholar]
- 430.Kievit F.M., Veiseh O., Bhattarai N., Fang C., Gunn J.W., Lee D., Ellenbogen R.G., Olson J.M., Zhang M. PEI-PEG-Chitosan Copolymer Coated Iron Oxide Nanoparticles for Safe Gene Delivery: synthesis, complexation, and transfection. Adv. Funct. Mater. 2009;19:2244–2251. doi: 10.1002/adfm.200801844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Dass C.R., Contreras K.G., Dunstan D.E., Choong P.F. Chitosan microparticles encapsulating PEDF plasmid demonstrate efficacy in an orthotopic metastatic model of osteosarcoma. Biomaterials. 2007;28:3026–3033. doi: 10.1016/j.biomaterials.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 432.Kim T.H., Jiang H.L., Nah J.W., Cho M.H., Akaike T., Cho C.S. Receptor-mediated gene delivery using chemically modified chitosan. Biomed. Mater. 2007;2:S95–100. doi: 10.1088/1748-6041/2/3/S02. [DOI] [PubMed] [Google Scholar]
- 433.Hussein Y.R., Sood A.K., Bandyopadhyay S., Albashiti B., Semaan A., Nahleh Z., Roh J., Han H.D., Lopez-Berestein G., Ali-Fehmi R. Clinical and biological relevance of enhancer of zeste homolog 2 in triple-negative breast cancer. Hum. Pathol. 2012;43:1638–1644. doi: 10.1016/j.humpath.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Steg A.D., Katre A.A., Goodman B., Han H.D., Nick A.M., Stone R.L., Coleman R.L., Alvarez R.D., Lopez-Berestein G., Sood A.K., Landen C.N. Targeting the notch ligand JAGGED1 in both tumor cells and stroma in ovarian cancer. Clin. Cancer Res. 2011;17:5674–5685. doi: 10.1158/1078-0432.CCR-11-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Lu C., Han H.D., Mangala L.S., Ali-Fehmi R., Newton C.S., Ozbun L., Armaiz-Pena G.N., Hu W., Stone R.L., Munkarah A., Ravoori M.K., Shahzad M.M., Lee J.W., Mora E., Langley R.R., Carroll A.R., Matsuo K., Spannuth W.A., Schmandt R., Jennings N.B., Goodman B.W., Jaffe R.B., Nick A.M., Kim H.S., Guven E.O., Chen Y.H., Li L.Y., Hsu M.C., Coleman R.L., Calin G.A., Denkbas E.B., Lim J.Y., Lee J.S., Kundra V., Birrer M.J., Hung M.C., Lopez-Berestein G., Sood A.K. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18:185–197. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Ghosn B., Singh A., Li M., Vlassov A.V., Burnett C., Puri N., Roy K. Efficient gene silencing in lungs and liver using imidazole-modified chitosan as a nanocarrier for small interfering RNA. Oligonucleotides. 2010;20:163–172. doi: 10.1089/oli.2010.0235. [DOI] [PubMed] [Google Scholar]
- 437.Han H.D., Mangala L.S., Lee J.W., Shahzad M.M., Kim H.S., Shen D., Nam E.J., Mora E.M., Stone R.L., Lu C., Lee S.J., Roh J.W., Nick A.M., Lopez-Berestein G., Sood A.K. Targeted gene silencing using RGD-labeled chitosan nanoparticles. Clin. Cancer Res. 2010;16:3910–3922. doi: 10.1158/1078-0432.CCR-10-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Cai Y.Q., Chen S.R., Han H.D., Sood A.K., Lopez-Berestein G., Pan H.L. Role of M2, M3, and M4 muscarinic receptor subtypes in the spinal cholinergic control of nociception revealed using siRNA in rats. J. Neurochem. 2009;111:1000–1010. doi: 10.1111/j.1471-4159.2009.06396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Feng S., Agoulnik I.U., Truong A., Li Z., Creighton C.J., Kaftanovskaya E.M., Pereira R., Han H.D., Lopez-Berestein G., Klonisch T., Ittmann M.M., Sood A.K., Agoulnik A.I. Suppression of relaxin receptor RXFP1 decreases prostate cancer growth and metastasis. Endocr. Relat. Cancer. 2010;17:1021–1033. doi: 10.1677/ERC-10-0073. [DOI] [PubMed] [Google Scholar]
- 440.de Martimprey H., Bertrand J.R., Fusco A., Santoro M., Couvreur P., Vauthier C., Malvy C. siRNA nanoformulation against the ret/PTC1 junction oncogene is efficient in an in vivo model of papillary thyroid carcinoma. Nucleic Acids Res. 2008;36:e2. doi: 10.1093/nar/gkm1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Pille J.Y., Li H., Blot E., Bertrand J.R., Pritchard L.L., Opolon P., Maksimenko A., Lu H., Vannier J.P., Soria J., Malvy C., Soria C. Intravenous delivery of anti-RhoA small interfering RNA loaded in nanoparticles of chitosan in mice: safety and efficacy in xenografted aggressive breast cancer. Hum. Gene Ther. 2006;17:1019–1026. doi: 10.1089/hum.2006.17.1019. [DOI] [PubMed] [Google Scholar]
- 442.Arbel J., Rozenbaum E., Reges O., Neuman Y., Levi A., Erel J., Haskia A.R., Caneti M., Sherf M., Mosseri M. USage of chitosan for Femoral (USF) haemostasis after percutaneous procedures: a comparative open label study. EuroIntervention. 2011;6:1104–1109. doi: 10.4244/EIJV6I9A192. [DOI] [PubMed] [Google Scholar]
- 443.U.N.I.o. Health ClinicalTrial.gov. 2013. http://clinicaltrials.gov
- 444.Malmquist J.P., Clemens S.C., Oien H.J., Wilson S.L. Hemostasis of oral surgery wounds with the HemCon Dental Dressing. Int. J. Oral Maxillofac. Surg. 2008;66:1177–1183. doi: 10.1016/j.joms.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 445.Jaffer S., Sampalis J.S. Efficacy and safety of chitosan HEP-40 in the management of hypercholesterolemia: a randomized, multicenter, placebo-controlled trial. Altern. Med. Rev. 2007;12:265–273. [PubMed] [Google Scholar]
- 446.Gratieri T., Gelfuso G.M., Rocha E.M., Sarmento V.H., de Freitas O., Lopez R.F. A poloxamer/chitosan in situ forming gel with prolonged retention time for ocular delivery. Eur. J. Pharm. Biopharm. 2010;75:186–193. doi: 10.1016/j.ejpb.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 447.Christensen K.S., Cohen A.E., Mermelstein F.H., Hamilton D.A., McNicol E., Babul N., Carr D.B. The analgesic efficacy and safety of a novel intranasal morphine formulation (morphine plus chitosan), immediate release oral morphine, intravenous morphine, and placebo in a postsurgical dental pain model. Anesth. Analg. 2008;107:2018–2024. doi: 10.1213/ane.0b013e318187b952. [DOI] [PubMed] [Google Scholar]
- 448.Kim J.K., Han K.H., Lee J.T., Paik Y.H., Ahn S.H., Lee J.D., Lee K.S., Chon C.Y., Moon Y.M. Long-term clinical outcome of phase IIb clinical trial of percutaneous injection with holmium-166/chitosan complex (Milican) for the treatment of small hepatocellular carcinoma. Clin. Cancer Res. 2006;12:543–548. doi: 10.1158/1078-0432.CCR-05-1730. [DOI] [PubMed] [Google Scholar]
- 449.Huo Z., Sinha R., McNeela E.A., Borrow R., Giemza R., Cosgrove C., Heath P.T., Mills K.H., Rappuoli R., Griffin G.E., Lewis D.J. Induction of protective serum meningococcal bactericidal and diphtheria-neutralizing antibodies and mucosal immunoglobulin A in volunteers by nasal insufflations of the Neisseria meningitidis serogroup C polysaccharide-CRM197 conjugate vaccine mixed with chitosan. Infect. Immun. 2005;73:8256–8265. doi: 10.1128/IAI.73.12.8256-8265.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Aksungur P., Sungur A., Unal S., Iskit A.B., Squier C.A., Senel S. Chitosan delivery systems for the treatment of oral mucositis: in vitro and in vivo studies. J. Control. Release. 2004;98:269–279. doi: 10.1016/j.jconrel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 451.McNeela E.A., Jabbal-Gill I., Illum L., Pizza M., Rappuoli R., Podda A., Lewis D.J., Mills K.H. Intranasal immunization with genetically detoxified diphtheria toxin induces T cell responses in humans: enhancement of Th2 responses and toxin-neutralizing antibodies by formulation with chitosan. Vaccine. 2004;22:909–914. doi: 10.1016/j.vaccine.2003.09.012. [DOI] [PubMed] [Google Scholar]