Abstract
An in-depth molecular study of infectious bronchitis viruses (IBV) with particular interest in evolutionary aspects of IBV in Spain was carried out in the present study based on the S1 gene molecular characterization of twenty-six Spanish strains isolated over a fourteen-year period. Four genotypes were identified based on S1 gene sequence analyses and phylogenetic studies. A drastic virus population shift was demonstrated along time and the novel Italy 02 serotype was shown to have displaced the previous predominant serotype 4/91 in the field. Detailed analyses of synonymous to non-synonymous ratio of the S1 gene sequences of this new serotype Italy 02 suggested positive selection pressures might have contributed to the successful establishment of Italy 02 serotype in our country. In addition, differences on the fitness abilities of new emergent genotypes were indicated. Furthermore, intergenic sequences (IGs)-like motifs within S1 gene sequences of IBV isolates were suggested to enhance the recombination abilities of certain serotypes.
Keywords: Infectious bronchitis virus, Genotyping, S1 gene, Selection pressures, Phylogeny, Coronavirus, Recombination, Virus neutralization, Evolution, Sequencing
Introduction
Avian infectious bronchitis virus (IBV) is considered one of the major poultry pathogens, being probably endemic in all regions with intensive poultry production (Office, 2004). It is primarily a respiratory pathogen of domestic fowl, although strains can vary in pathogenicity and tissue tropism (Jones and Ambali, 1987, Jones and Jordan, 1970, Winterfield and Hitchner, 1962). IBV is a Group 3 member in the genus Coronavirus (Gonzalez et al., 2003) and its genome consists of a 27 Kb single-stranded positive-sense RNA molecule that encodes for four structural proteins. The nucleoprotein (N) is associated with viral genome to form the nucleocapsid, whereas the remaining structural proteins, the spike (S), small membrane (M), and envelope proteins (E), are inserted in the envelope surrounding the nucleocapsid. In addition, the genome also encodes for the replicase complex responsible for carrying out the unique discontinuous transcription process that leads to the generation of a nested set of six 3′ and 5′ coterminal subgenomic mRNAs (Stern and Kennedy, 1980, Stern and Sefton, 1982).
The continuous emergence of new IBV serotypes has complicated the design of appropriate control programs due to the antigenic variation and the low degree of cross protection observed among IBV serotypes and has pointed out the necessity for accurate techniques to diagnose and classify this viral agent (Cowen and Hitchner, 1975). Nucleotide sequencing and subsequent genetic analysis of the S1 protein gene sequences provides a fast and accurate method to classify and predict IBV serotype, but also a powerful instrument to monitor phylogenetic and epidemiological evolution of IBV subtypes (Adzhar et al., 1997, Cavanagh and Davis, 1986, Cavanagh et al., 1992b, Cavanagh et al., 1986, Kant et al., 1992, Koch et al., 1990, Lee and Jackwood, 2001, Liu et al., 2006, Mase et al., 2004, Moore et al., 1998, Wang and Huang, 2000). Mutation and recombination processes have been demonstrated to be involved in the genetic variation, and therefore in the evolution of IBV, leading to the emergence of new variant strains and giving rise to virus population diversity (Cavanagh et al., 1992a, Kottier et al., 1995, Lee and Jackwood, 2000, Wang et al., 1997). However, very distinct spatiotemporal and epidemiological behaviours are shown among these new strains. Whereas some emergent viruses rapidly spread to other geographic areas and become established, as happened with the 4/91 serotype in Europe or the Georgia serotype in North America, others remain restricted to the region of origin. More interestingly, hardly ever genotypes are exchanged among different continents (Kusters et al., 1987, Zanella et al., 2003). Nevertheless, factors that determine distinct spreading and fitness abilities of the new emergent strains are poorly understood. Hence, monitoring IBV subpopulation dynamics in a specific region over time may reveal molecular features that contributed to the different fitting and spreading abilities of IBV strains.
IBV has been diagnosed in Spain since the early seventies by virus isolation and serological techniques. In 2002, an epidemiological survey among the most important Spanish poultry companies pointed out IB as one of the main infectious diseases affecting farms and revealed the necessity of an in-depth study to determine the epidemiological situation and to improve the efficacy of vaccination and control programs against the disease. Furthermore, the availability of IBV strains isolated in Spain during the past 14 years provided a unique opportunity for a detailed genetic study to determine not only the prevalent IBV genotypes in our country but also the dynamics of viral subpopulation which is necessary for a deeply understanding of IBV evolution and epidemiology.
In this study, twenty-six IB viruses isolated in Spain from clinical outbreaks occurring between 1992 and 2005 were molecularly characterized by sequencing the whole S1 gene. To increase our insight into factors involved in IBV evolutionary process, specific tests to determine the presence of positive selection forces were carried out. Furthermore, several methods were implemented to identify putative recombination events among these isolates and establish the extent of recombination into IBV evolution. Finally, the antigenic relationships of the major genotypes identified in our country with reference strains were assessed by cross VN assays to ascertain the practical implications of molecular changes observed into virus immunogenicity.
Results
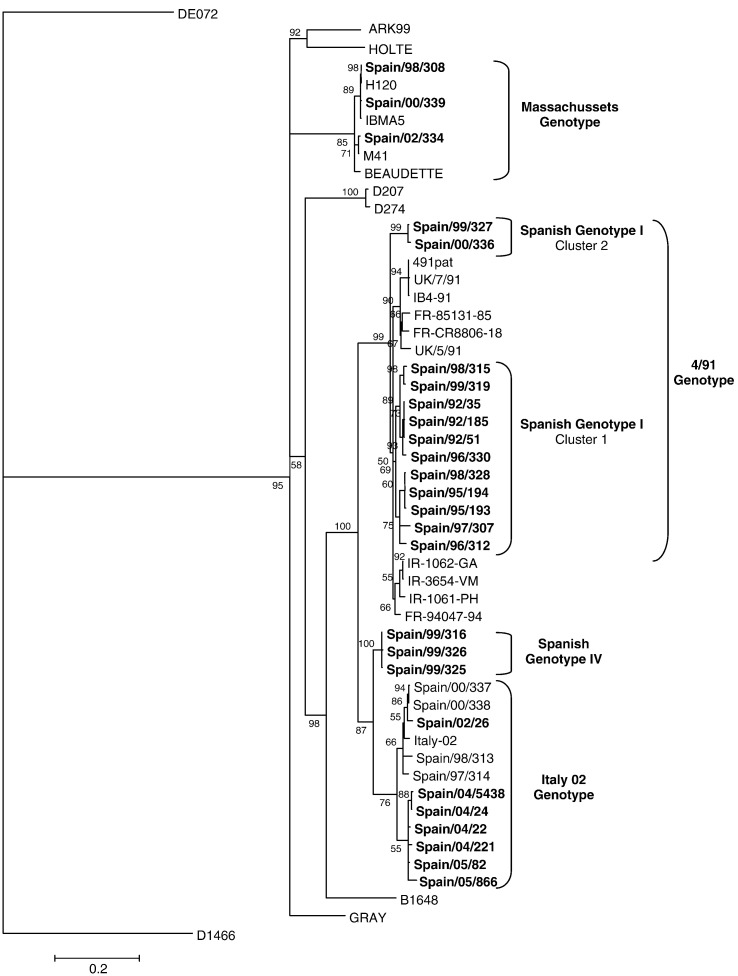
S1 gene phylogenetic and sequence analyses revealed that four different genotypes have been present in Spain over the last 14 years
Phylogenetic analysis based on both S1 gene nucleotide and amino acid deduced sequences of the twenty-six Spanish isolates and the reference IBV strains showed that Spanish isolates were separated into four distinct genetic groups or genotypes (Fig. 1 ). Genotype I included thirteen out of the twenty-six field isolates, isolated between 1992 and 2000, that were grouped with 4/91 reference isolates. Seven Spanish field isolates, from 1997 to 2005, were included in genotype II, and showed maximum nucleotide and amino acid identities with Italy 02 strains. Genotype III comprised three field viruses isolated between 1996 and 2000 that were grouped with isolates of the Massachusetts serotype. Particularly, Spain/98/308 and Spain/00/339 isolates were grouped with Massachusetts vaccine strains (H120) in the phylogenetic tree, and they are likely to be reisolations of vaccine strains used in the immunization of those flocks. Finally, Spanish genotype IV included 3 isolates that split in a unique genetic group in the phylogenetic analysis. All viruses included in genotype IV were isolated in 1999 and showed maximum nucleotide and amino acid identities with Italy 02 isolates (90.6–92% and 89–91.1% respectively).
Fig. 1.
Phylogenetic tree constructed by a maximum likelihood method (Tree-Puzzle 5.2). Nucleotide sequences of the complete S1 gene of Spanish field isolates investigated in this study (in bold letters) and reference IBV strains obtained from GenBank database are included in the comparison.
Genetic studies of Spanish isolates of 4/91 genotype (genotype I) showed the presence of two distinct genetic clusters
The 13 isolates assigned into the Spanish genotype I group shared maximum nucleotide and amino acid divergences of 7.3% and 10.3% respectively. Phylogenetic analyses comparing the complete S1 gene sequences of the 13 Spanish genotype I field isolates and all 4/91 S1 gene sequences previously published in the GenBank database, including British, French and Iranian isolates, revealed that Spanish isolates were clearly grouped in two separated genetic clusters identified as Spanish genotype I cluster 1 (including 11 isolates) and Spanish genotype I cluster 2 (including 2 isolates). Average nucleotide and amino acid identities shared by both clusters were 93.1% and 91.3% respectively. The minimum amino acid divergence observed between two isolates from the same year was 0.4% (between Spain/92/185 and Spain/92/51), whereas the maximum was 9.9% (between Spain/99/319 and Spain/99/327). Both cluster 1 and cluster 2 isolates showed maximum average nucleotide and amino acid identities with French isolate FR-94047-94. Comparison of 4/91 Spanish field isolates sequences revealed a common unique deletion at amino acid position 58 (located in HVR1) in 6 isolates included in cluster 1. Four isolates within this same group shared an amino acid deletion at position 340, located in HVR3. Both isolates included in cluster 2 showed 8 unique amino acid substitutions. In addition, isolate Spain/99/327 in cluster 2 showed 3 amino acid deletions at positions 55, 56 and 57 (located in HVR1). No cumulative changes were observed among 4/91 Spanish isolates.
Sequence analysis of Spanish isolates of Italy 02 genotype (Genotype II) clearly revealed presence of cumulative amino acid changes within S1 gene
A total of 11 Spanish field strains of Italy 02 genotype, seven from the present study and 4 previously described (Dolz et al., 2006), and the reference strain Italy 02 were included in the present study. Maximum nucleotide and amino acid divergence within Spanish Italy 02 isolates was 5.7% and 9.9%, respectively. The maximum amino acid distance between two viruses isolated the same year was 3.6% (Spain/04/5438 and Spain/04/221). No insertions or deletions were observed within S1 gene sequences of the Spanish Italy 02 isolates, except for the Spain/98/313 that showed two deletions at amino acid positions 139 and 140. Analysis of the sequences showed that 12 amino acid positions were cumulative changes that become fixed in a progressive manner through time of isolation.
Sequence analysis of Spanish genotype IV (Spain/99/325) showed a drastic shift on sequence homology when 5′ and 3′ ends of S1 gene were compared
S1 gene sequences of genotype IV isolates were aligned and compared with the remainder Spanish field isolates and a total of 90 mutated amino acid positions were observed in genotype IV isolates with respect to Spanish 4/91, Italy 02 and Massachusetts strains. Each mutated residue was compared with the residue present in the same amino acid position in the other 3 Spanish genotypes. Mutated positions were mostly shared with 4/91 and Italy 02 strains, but distinct sharing rates were observed between the 5′ and 3′ regions within the S1 gene. A total of 58 mutated residues were identified within the 200 amino acid 5′ terminal region. Only 1 (1.7%) out of the 58 mutated residues within this region was shared with 4/91 strains alone, whereas 33 (56.9%) were shared with Italy 02 isolates. The remaining positions were predominantly shared simultaneously with 4/91 and Italy 02 isolates. Very few unique mutations, 2 out of 58, were observed. When the 341 amino acid 3′ terminal region was analysed, 28 (87.5%) out of 32 identified mutated positions coincided with residues of 4/91 isolates, whereas only 1 (3.1%) coincided with Italy 02 isolates.
These observations correlated with distinct amino acid identities observed when both regions were compared separately among the three genotypes. Genotype IV had maximum amino acid identities with Italy 02 Spanish genotype (87.3%–95.4%) when the S1 200 Aa 5′ terminal region was compared, whereas amino acid identities in the same region with 4/91 Spanish genotype was much lower (68.8%–75.8%). On the contrary, when the 341 Aa 3′ end of the S1 gene was compared, genotype IV showed maximum amino acid identities with 4/91 Spanish isolates (92.9%–97.9%), and lower similarities with Italy 02 Spanish isolates (88.8%–91.2%).
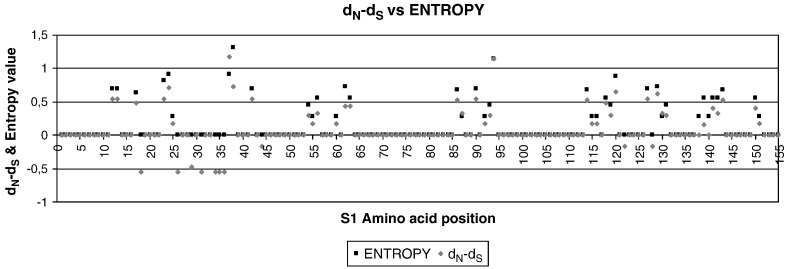
Positive selection pressure is detected in several amino acid positions within Italy 02 isolates, particularly within epitope region 2
The average dS/dN ratio within the 13 Spanish isolates of the 4/91 genotype was estimated to be 3.43, whereas within the Italy 02 Spanish isolates was 1.97. To identify regions or specific positions within the S1 gene under positive selection, the dN − dS difference per codon was calculated for the Spanish Italy 02 isolates. The resulting graphic of the dN − dS difference plotted against entropy values for the initial 150 amino acid positions of Spanish Italy 02 isolates is shown in Fig. 2 . Clearly, amino acid positions with positive dN − dS values also showed positive entropy values and were predominantly located within regions previously identified as virus-neutralizing and hemagglutination-inhibiting epitopes. Epitope 1 region contained 9 amino acid positions with positive dN − dS values, and five out of these 9 positions (amino acid residues 37, 42, 60, 62 and 63) were also fixed amino acid substitutions. Epitope 2 region contained 18 amino acid positions with positive dN − dS values, and five of them (residues 92, 129, 131, 141 and 143) became also fixed amino acid positions.
Fig. 2.
Entropy rates and non-synonymous and synonymous difference (dN − dS) plotted per each amino acid position for the first 150 amino acids of the S1 gene. Both values were calculated using an alignment that included all Spanish field isolates of Italy 02 genotype.
Putative recombination events have been detected in three out of the four IBV genotypes in Spain
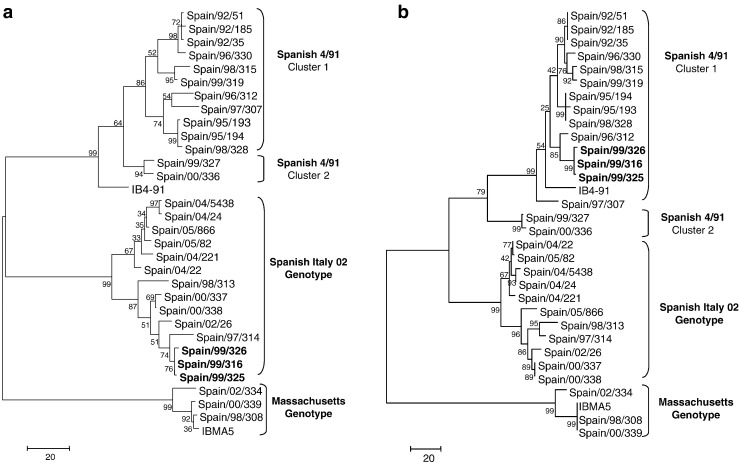
A total of seven recombination events, including 16 recombination breakpoints, were identified among Spanish IBV field isolates. One recombination event, beginning at nucleotide 702 and ending at nucleotide 1389, was identified in both isolates of Spanish 4/91 genotype cluster II by five methods. A total of five recombination events were detected within Spanish Italy 02 isolates, however just only one of the six events was common to the 11 isolates. It began at nucleotide position 583 and ended at position 930, and it was detected by 5 out of the six methods implemented. Finally, one recombination event, beginning at nucleotide position 281 and ending at position 586, was detected in all three isolates of Spanish genotype IV by 4 of the 6 methods used. Phylogenetic analysis performed separately with sequence region comprised within recombination event and the remainder sequence regions generated different tree topologies in all three recombination events described (Fig. 3 ).
Fig. 3.
Phylogenetic analysis of putative recombinant breakpoint located at position 586 of Italy 02 and Genotype IV isolates. All twenty-six Spanish IBV field isolates were included in the comparison. Trees were generated by the Maximum Parsimony method with 1000 bootstraps in the Mega 3 package. 3a. Phylogenetic tree obtained when the S1 region from nt 1 to 586 in the alignment was used. Italy 02 and Genotype IV (in bold letters) isolates were clustered together in the same phylogenetic group. 3b. Phylogenetic tree obtained when the S1 region from nt 586 to 1623 in the alignment was used. Genotype IV isolates (in bold letters) were grouped with Spanish 4/91 cluster 1 isolates, whereas Spanish Italy 02 isolates formed an own phylogenetic cluster.
Cross-over sites were not homogeneously distributed within S1 gene sequences. Eleven out of 16 breakpoints identified within S1 gene sequences of Spanish field isolates were concentrated in the half 5′ terminal region. Particularly two regions comprised between 100 and 200 nucleotide positions and 400 and 600 nucleotide positions concentrated 7 out of the 16 cross-over sites. Thus, detailed sequence analysis of these regions among all Spanish isolates was carried out to identify sequence motifs, particularly IBV intergenic sequences (IGs) CTTAACA and CTTAACAA, which might have facilitated RNA–RNA recombination. All Spanish isolates included within genotype IV and Italy 02 genotypes showed IGs CTTAACA twice within S1 gene, at nucleotide positions 370 and 445, with the exception of Spanish isolate Spain/05/21 that only showed it once at nucleotide position 445. Any of the Spanish isolates of 4/91 genotype neither Massachusetts genotype included this motif within S1 gene sequence. Nucleotide sequence CTTAACAA was only found in genotype IV isolates and two Italy 02 isolates, Spain/97/314 and Spain/98/313, between nucleotide positions 440 and 450. Any of the other Spanish isolates had this motif within S1 gene sequences.
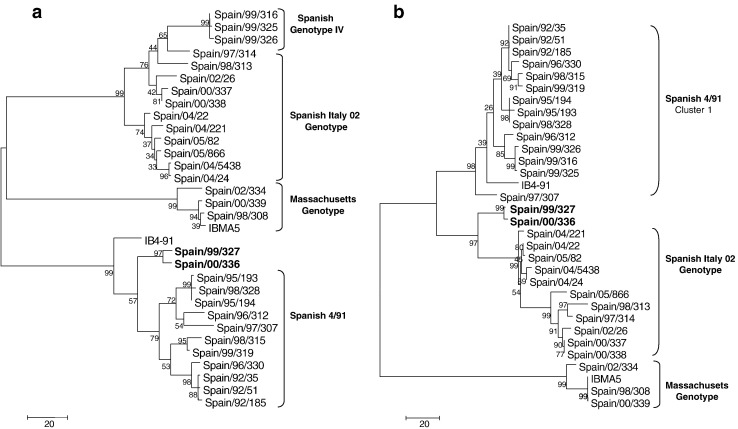
Antigenic relatedness studies of the field isolates Spain/99/319 and Spain/99/325
To further serologically characterize Spanish IBV isolates, two Spanish field isolates Spain/99/319 and Spain/99/325 included in genotype 1 cluster 1 and genotype 4 respectively were selected to assess VN assay Fig. 4 . Antigenic relatedness and neutralizing indexes are summarized in Table 2 .
Fig. 4.
Phylogenetic analysis of putative recombinant breakpoint located at position 700 of Spanish 4791 cluster 2 isolates. All twenty-six Spanish IBV field isolates were included in the comparison. Trees were generated by the Maximum Parsimony method with 1000 bootstraps in the Mega 3 package. 4a. Phylogenetic tree obtained when the S1 region from nt 1 to 700 in the alignment was used. Spanish 4/91 cluster 2 isolates (in bold letters) were grouped with the remaining Spanish 4/91 isolates. 4b. Phylogenetic tree obtained when the S1 region from nt 700 to 1623 in the alignment was used. Spanish 4/91 cluster 2 isolates (in bold letters) were clustered in an own phylogenetic branch closely related with Spanish Italy 02 isolates.
Table 2.
Neutralizing index and antigenic relatedness of Spanish field isolates Spain/99/319 (Genotype I) and Spain/99/325 (Genotype IV) field isolates and the main reference IBV European serotypes
| Virus 1 | Virus 2 | Neutralizing index virus 1a | Neutralizing index virus 2b | Antigenic relatedness (%) |
|---|---|---|---|---|
| Spain/99/325c | M41 | 0.6 | 0.6 | 23.2 |
| D274 | 0.84 | 0.6 | 33.0 | |
| D1466 | 0.6 | 0.6 | 24.3 | |
| 4/91 | 0.8 | 0.6 | 26.3 | |
| Spain/00/337e | 2.8 | 1.98 | 67.5 | |
| Spain/99/319 | 0.8 | 1.7 | 47.3 | |
| Spain/99/325 | 2.9 | 2.9 | 100.0 | |
| Spain/99/319d | M41 | 0.6 | 0.6 | 27.3 |
| D274 | 1.2 | 0.6 | 46.3 | |
| D1466 | 0.84 | 0.6 | 33.8 | |
| 4/91 | 3 | 0.6 | 59.8 | |
| Spain/00/337 | 1.4 | 1.2 | 43.6 | |
| Spain/99/325 | 1.7 | 0.8 | 47.3 | |
| Spain/99/319 | 2.1 | 2.1 | 100.0 |
Maximum antigenic relatedness obtained for each isolate is indicated in bold letters.
Neutralizing index obtained from antisera of virus 1 against virus 2.
Neatruazing index obtained from antisera of virus 2 against virus 1.
Spanish isolate representative from Genotype IV.
Spanish isolate representative from Genotype I (4/91 Spanish isolate).
Spanish field isolate previously demonstrated to belong to Italy 02 serotype (Dolz et al., 2006).
Discussion
Since it was first described in the early nineties in UK, 4/91 IBV serotype spread over many other countries and became one of the most predominant serotypes in Europe (Capua et al., 1999, Cavanagh et al., 2005, Cook et al., 1996, Gough et al., 1992, Meulemans et al., 2001). The present study shows 4/91 as one of the most predominant genotypes also in Spain and demonstrates it has been circulating in our country since 1992. Previous studies of the degree of variation exhibited within the S1 gene sequences of 4/91 serotype isolates reported maximum nucleotide and amino acid differences of 4% and 8% respectively (Cavanagh et al., 2005). In the present investigation, 4/91 Spanish isolates showed higher genetic divergences among them, and more interestingly they were split in two major phylogenetic clusters, suggesting that 4/91 genotype has gone through evident evolutive changes during these years in our country. Furthermore, cross VN results carried out with a field 4/91 Spanish isolate demonstrates that these unique evolutive processes contributed to greatly decrease the antigenic relatedness between the 4/91 Spanish field strains and the reference 4/91 serotype. Hence, these findings suggest that antigenic subtypes may be present in field and in the brink of arising as a new serotype, regardless of the level of genetic variation displayed.
It is particularly remarkable that two Spanish 4/91 strains isolated the same year practically showed the maximum divergence exhibited within one IBV genotype and furthermore, they were grouped in separate phylogenetic clusters. Although a very rapid evolutive rate from a common ancestor might have lead to a similar situation, no cumulative changes were observed among Spanish 4/91 isolates and unique amino acidic changes observed in one of these two groups were not shared by the other. Moreover, the average dS/dN ratio for the whole S1 gene was not suggestive of a high evolutive rate. Therefore, one possible explanation for this scenario would be that 4/91 strains isolated in Spain evolved from at least two distinct evolutive lineages probably originated in two different ancestors. Taking into account that both isolates assigned to Spanish 4/91 cluster 2 are the more recently isolated, a new reintroduction of 4/91 isolates in our country could be hypothesized as has been previously suspected in other countries (Adzhar et al., 1997). On the other hand, based on the results of recombination analyses carried out in this study, the possibility that recombination processes might have been involved in the rapid evolution of the Spanish 4/91 cluster 2 giving rise to a new lineage cannot be excluded.
Interestingly, the present study also shows an important shift in virus subpopulation dynamics along time with a clear replacement of predominant genotypes in our country. Whereas 4/91 was the predominant genotype in Spain in the nineties, the situation took a turn at the end of that decade when the new Italy 02 genotype appeared and displaced the previous predominant genotype. High sequence divergences and the presence of cumulative changes observed within Spanish Italy 02 isolates further support a fast evolutionary rate of this serotype in our country. A wide variety of causes could have been driving the observed virus subpopulation replacement. However, we found strong evidences of the positive selection of mutations within the S1 epitope regions of Italy 02 isolates which suggest a selection pressure-driven evolution of this genotype in Spain. Evidently, some cautionary remarks should be considered as our analyses were only focused on the S1 protein gene, but selection pressures could have occurred in other regions within IBV genome. Therefore, to accurately discern the evolutionary history of this genotypic shift, whole genome studies should be carried out.
At this point an intriguing question comes out regarding factors contributing to create a positive selection pressure. Our data suggests positive selected mutations of Italy 02 might have lead these new isolates escaping the immune system. Evidences of positive selection pressures acting over specific residues within antigenic sites have been previously reported for other viruses against which partial cross protection is induced after infection or vaccination, similarly to what happens with IBV (Grenfell et al., 2004, Haydon et al., 2001, Hurst, 2002). In this sense, it is noteworthy that at the late nineties, commercial vaccines against the 4/91 became available in Spain. Therefore, it is likely that vaccination implemented against 4/91 serotype was very successful in order to achieve an effective control of infections caused by this serotype in our country, based on the drastic reduction of 4/91 isolations from clinical cases. But on the other hand, it is tempting to speculate that the high immune pressure against 4/91 isolates might have provided an environment in which the novel variant genotype Italy 02 had higher fitness advantage contributing to a faster establishment and spreading of this serotype in Spain.
Another challenging aspect of IBV epidemiology in Spain is the emergence of the Genotype IV isolates and its relationship with Italy 02 genotype. Genotype IV was established as a new genotype based on the mosaic nature of its S1 gene. VN studies revealed Italy 02 and Genotype IV as being subtypes within a common serotype. However, the restricted epidemiological relevance of genotype IV, all three isolates included within this genotype were isolated in 1999 and in the same geographic region, clearly indicates distinct fitness abilities among IBV isolates. Therefore, this finding demonstrates that new unknown IBV isolates are emerging constantly, but only those with advantageous features within a particular environment become established, whereas others disappear and remain unidentified.
The extent of recombination events among IBV field isolates has been poorly investigated, even though recombination is a well-known feature of IBV (Cavanagh et al., 1992a, Kottier et al., 1995, Lee and Jackwood, 2000). Our data suggests that recombination events might have been frequently involved in the evolution of IBV rather than sporadically. Interestingly, most of the recombinant breakpoints identified among Spanish IBV field isolates were clustered in restricted regions within the 5′ half part of the S1 gene and involved Italy 02 isolates, suggesting the presence of sequence or structure motifs that could act as specific “hotspots”. RNA sequences and structures that promote viral RNA recombination have been demonstrated in several viruses (Carpenter et al., 1995, Nagy and Bujarski, 1997). Particularly, IGs that are required for transcription acting as a slow-down and detaching signal for the transcription–replication complex have been speculated to promote recombination exchange of whole genes in IBV (Brian and Spaan, 1997, Enjuanes et al., 2005, Lee and Jackwood, 2000). Based in our results, “IGs-like” motifs, that probably arose because random mutations, could have favoured recombination within specific sites in the S1 gene, increasing the potential of Italy 02 genotype to undergo recombination processes.
There have been few scientific studies covering the genetic and antigenic variation within IBV serotypes and dynamics along time of IBV genotypes in a specific region (Cavanagh et al., 2005, Lee and Jackwood, 2001, Liu et al., 2006, Mase et al., 2004). Herein, we present one of the most comprehensive studies of the genetic variation and evolution within a population of IBV isolates. Molecular analyses have revealed that vaccination, despite greatly contributes to the control of specific genotypes, may create and advantageous environment leading to successful spreading of new genotypes. Furthermore, our results indicate that recombination might have been frequently involved in the emergence and evolution of IBV.
Materials and methods
Viruses and antisera
Twenty-six field avian IBVs isolated between 1992 and 2005 in Spain were included in this study. Viruses were obtained from clinical cases submitted to Centre de Recerca en Sanitat Animal (CReSA), Centre de Sanitat Avícola de Catalunya i Aragó (CESAC) and Centro de Investigación en Sanidad Animal (CISA). Epidemiological information of these isolates is summarized in Table 1 . All field viruses were isolated in specific pathogen-free (SPF) embryonated eggs, acquired from Charles Rivers Spafas (North Franklin, USA). Two commercial vaccines currently licensed for use in Spain, IB 4/91 and IB MA5, were also included in the study. The reference IBV serotypes M41, D274, D1466 and 4/91 and antisera against them to be used in the virus neutralization (VN) assay were purchased from the Animal Health Center (Deventer, The Netherlands). The Spanish field virus Spain/00/337, also used in the VN assay, had been previously isolated and characterized as belonging to the Italy 02 serotype in our laboratory (Dolz et al., 2006). To obtain virus stocks for VN assays, allantoic fluid from field isolates Spain/99/319 and Spain/99/325 and reference strains was harvested, frozen at − 80 C and then titrated in SPF chicken embryos. Endpoint dilutions and titres were determined by the Reed and Muench method (Villegas, 1998). Monospecific antisera against field isolates were obtained following the procedure previously described (Dolz et al., 2006, Gelb and Jackwood, 1998).
Table 1.
Epidemiologic information of Spanish field IBV isolates included in the study
| IBV isolatea | Yearb | Location | Type of chicken | Age | Tissuec | Accession numbers |
|---|---|---|---|---|---|---|
| Spain/92/35 | 1992 | N/A | Broiler | N/A | Trachea | DQ386091 |
| Spain/92/51 | 1992 | N/A | Broiler | N/A | Trachea | DQ064801 |
| Spain/92/185 | 1992 | N/A | Broiler | N/A | Trachea | DQ386092 |
| Spain/95/193 | 1995 | N/A | Broiler | N/A | Trachea | DQ386093 |
| Spain/95/194 | 1995 | N/A | Broiler | N/A | Trachea | DQ064802 |
| Spain/96/312 | 1996 | Catalunya | Broiler | 55d | Trachea | DQk064803 |
| Spain/96/330 | 1996 | Galicia | Broiler | 32d | Trachea, kidney | DQ386094 |
| Spain/96/334 | 1996 | N/A | N/A | N/A | Trachea | DQ064804 |
| Spain/97/307 | 1997 | Catalunya | Broiler | 43d | Trachea | DQ064805 |
| Spain/98/308 | 1998 | Valencia | Slow growth | 35d | Trachea | DQ064807 |
| Spain/98/315 | 1998 | Valencia | Broiler | 35d | Trachea, kidney, cecal tonsil | DQ386095 |
| Spain/98/328 | 1998 | N/A | N/A | N/A | Trachea, kidney | DQ386096 |
| Spain/99/316 | 1999 | Center | Broiler | 23d | Kidney | DQ064809 |
| Spain/99/319 | 1999 | Catalunya | Slow growth | 38d | Trachea, cecal tonsil | DQ064810 |
| Spain/99/325 | 1999 | Center | Broiler | 40d | Kidney | DQ064811 |
| Spain/99/326 | 1999 | Center | Broiler | 40d | Kidney | DQ064812 |
| Spain/99/327 | 1999 | Catalunya | Broiler | 49d | Trachea | DQ386097 |
| Spain/00/336 | 2000 | Catalunya | Broiler | 44d | Trachea | DQ386098 |
| Spain/00/339 | 2000 | Catalunya | Broiler | 35d | Trachea | DQ064815 |
| Spain/02/26 | 2002 | Navarra | Broiler | 47d | N/A | DQ386099 |
| Spain/04/22 | 2004 | Catalunya | Broiler | 49d | Trachea | DQ386100 |
| Spain/04/24 | 2004 | Catalunya | Broiler | 36d | Trachea | DQ386101 |
| Spain/04/221 | 2004 | C. Mancha | N/A | N/A | N/A | DQ386103 |
| Spain/04/5438 | 2004 | Catalunya | Broiler | 49d | Trachea | DQ386105 |
| Spain/05/82 | 2005 | C. Mancha | N/A | N/A | N/A | DQ386104 |
| Spain/05/866 | 2005 | Murcia | Broiler | 41d | Trachea | DQ386102 |
Isolates are sorted by year of isolation and then by isolate number. The GenBank accession number of the obtained S1 genes sequences for each isolate is included.
N/A = Data not available.
Isolates have been named as proposed by Cavanagh (2002).
Year of isolation.
Tissues used for IBV isolation.
RT-PCR and nucleotide sequencing
RNA was extracted from 150 μl of allantoic fluid using Nucleospin RNA Virus Kit (Macherey-Nagel) according to manufacturer's instructions. The complete S1 gene was amplified with oligonucleotides S1Uni2+ and IBP1- as previously described (Adzhar et al., 1996). The 1800-bp RT-PCR products were purified by QIAquick gel extraction Kit and Minelute PCR purification Kit (Qiagen Inc.) following manufacturer's instructions. Sequencing of the whole S1 gene was carried out using a combination of eight internal primers previously published (Dolz et al., 2006). Some modifications were implemented to primers S1PR2+, S1PR2–, S1PR5+ and S1PR6+ because several mismatches were detected in some isolates, preventing correct sequencing. Sequences of modified primers were S1PR2+nou (5′ TAGCCTACTTTGTTAATGGTA 3′), S1PR2–nou (5′ GTACCATTAACAAAGTAGGCTA 3′), S1PR5+nou (5′ TAGATACTTCAGGAGCCATAGACA 3′) and S1PR6+nou (5′ CCATAGACATATTTGTTGTTC 3′). Purified RT-PCR products were sequenced by the dideoxy-mediated chain-termination method using ABI PRISM BigDye® Terminator v3.1 Cycle Sequencing Kit (PE Biosystems) as described by the manufacturer. Sequences were analyzed with an automated nucleic acid analyzer (ABI PRISM 3100 Avant; PE Biosystems).
Nucleotide and amino acid deduced sequence analyses
Assembly of sequence contigs and sequence analysis were performed with the Bioedit v7.0.5 package. The twenty-six IBV Spanish field isolates, the two vaccine strains, and the reference IBV strains downloaded from the GenBank database were first aligned using ClustalW included in the Bioedit v7.0.5. Phylogenetic trees were inferred using a maximum likelihood (ML) algorithm with the Tree-Puzzle 5.2 software (Schmidt et al., 2002). In all cases trees were estimated using the best fitting model of nucleotide substitution identified by Modeltest 3.8 (Posada and Crandall, 1998).
GenBank accession numbers
Nucleotide and amino acid deduced S1 sequences of reference IBV strains included in the comparisons were available in GenBank under the following accession numbers: Ark-99 (L10384); Beaudette (X02342); B1648 (X87238); De072 (U77298); D1466 (M21971); D207 (M21969); D274 (X15832); D3896 (X52084); FR-CR88061-88 (AJ618986); FR-85131-85 (AJ618985); FR-96047-94 (AJ818987); Gray (L14069); Holte (L18988); H120 (M21970); H52 (AF352315); Italy 02 (AJ457137); IR-1061-PH (AY544778); IR-1062-GA (AY544777); IR-3654-VM (AY544776); JMK (L14070); M41 (X04722); Spain/97/314 (DQ064806); Spain/98/313 (DQ064808); Spain/00/337 (DQ064813) and Spain/00/338 (DQ064814); UK/2/91 (Z83976); UK/3/91 (Z83977); UK/5/91 (Z83978); UK/7/91 (Z83975); UK/7/93 (Z83979); UK-1233-95 (AJ618984); 4/91 pathogenic (AF093794).
Selection pressure analysis on the S1 gene
Potential selective pressures along the S1 gene of major Spanish genotypes isolates were assessed first by calculating the average ratio of synonymous substitutions per synonymous site (dS) to non-synonymous substitutions per non-synonymous site (dN) (dS/dN). In addition, to detect specific residues within the S1 gene under strong selective pressure the subtraction of synonymous from non-synonymous nucleotide changes (dN − dS) per codon was calculated and plotted versus the amino acid sequence entropy, that is a measure of the lack of predictability at each position in the alignment and gives an idea of the amount of variability through a column in the alignment (Schneider and Stephens, 1990). Both, average dS/dN ratio and codon dN and dS were calculated with the SNAP web utility (http://hiv-web.lanl.gov/content/hiv-db/SNAP/WEBSNAP/SNAP.html). Amino acid entropy was calculated with BioEdit v7.0.5 software.
Recombination analysis
To determine whether recombination events had been involved in the evolution of Spanish IBV isolates, and to identify putative recombination breakpoints in the S1 gene, RDP v2.0 software was used (Martin et al., 2005). Only Spanish field isolates were included in the study because the objective was to investigate the extent of recombination events in field. All field Spanish isolates were aligned using ClustalW program implemented in BioEdit v7.0.5 package. Then, six methods implemented in the RDP v2.0 software package, including RDP, Bootscan, GENECONV, Maxchi, Chimaera and SiScan were employed to identify recombinant breakpoints (Gibbs et al., 2000, Padidam et al., 1999, Posada, 2002, Salminen et al., 1995). General settings used for all methods were as follows: window size = 20, highest acceptable P value = 0.001 and Bonferroni correction. Only recombination breakpoints detected by more than three methods were considered. Also, once potential recombination events were identified, sequence alignment was split at that point and phylogenetic analysis of the two regions was performed to evaluate conflicting evolutionary histories of both regions. Phylogenetic trees were constructed by using a maximum parsimony method with 1000 bootstraps (Kumar et al., 2004). Same methods and software have been previously used in recombination studies with SARS (Zhang et al., 2005).
Cross-VN analysis
To determine the antigenic relationships among the field Spanish isolates Spain/99/319 and Spain/99/325 and IBV reference strains M41, D274, 4/91, D1466 and Italy 02, reciprocal beta VN assays (diluted-serum constant-virus) were performed as described by Thayer and Beard (Thayer and Beard, 1998). Four-fold serial dilutions of each antiserum, and 100 EID50 0.1 ml− 1 of each virus were used. Antiserum, virus and negative controls were carried out in each assay. Endpoint titres were calculated by the Reed and Müench method (Villegas, 1998). The VN titres against homologous and heterologous virus were used to calculate the antigenic relatedness (r-values) by the formula of Archetti and Horsfall (Archetti and Horsfall, 1950).
References
- Adzhar A., Shaw K., Britton P., Cavanagh D. Universal oligonucleotides for the detection of infectious bronchitis virus by the polymerase chain reaction. Avian Pathol. 1996;25:817–836. doi: 10.1080/03079459608419184. [DOI] [PubMed] [Google Scholar]
- Adzhar A., Gough R.E., Haydon D., Shaw K., Britton P., Cavanagh D. Molecular analysis of the 793/B serotype of infectious bronchitis virus in Great Britain. Avian Pathol. 1997;26:625–640. doi: 10.1080/03079459708419239. [DOI] [PubMed] [Google Scholar]
- Archetti I., Horsfall F.L. Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum. J. Exptl. Med. 1950;92:441–462. doi: 10.1084/jem.92.5.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian A., Spaan W. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 1997;8:101–111. doi: 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capua I., Minta Z., Karpinska E., Mawditt K., Britton P., Cavanagh D., Gough R.E. Co-circulation of four types of infectious bronchitis virus (793/B, 624/I, B1648 and Massachusetts) Avian Pathol. 1999;28:587–592. doi: 10.1080/03079459994380. [DOI] [PubMed] [Google Scholar]
- Carpenter C.D., Oh J.W., Zhang C., Simon A.E. Involvement of a stem-loop structure in the location of junction sites in viral RNA recombination. J. Mol. Biol. 1995;245:608–622. doi: 10.1006/jmbi.1994.0050. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J. Gen. Virol. 1986;67(Pt 7):1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Darbyshire J.H., Peters R.W. Coronavirus IBV: virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus-neutralizing or haemagglutination-inhibiting antibody, or induce chicken tracheal protection. J. Gen. Virol. 1986;67(Pt 7):1435–1442. doi: 10.1099/0022-1317-67-7-1435. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Cook J.K.A. Infectious bronchitis virus: evidence for recombination within the Massachusetts serotype. Avian Pathol. 1992;21:401–408. doi: 10.1080/03079459208418858. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Cook J.K.A., Li D., Kant A., Koch G. Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992;21:33–43. doi: 10.1080/03079459208418816. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Picault J.P., Gough R., Hess M., Mawditt K., Britton P. Variation in the spike protein of the 793/B type of infectious bronchitis virus, in the field and during alternate passage in chickens and embryonated eggs. Avian Pathol. 2005;34:20–25. doi: 10.1080/03079450400025414. [DOI] [PubMed] [Google Scholar]
- Cook J.K., Orbell S.J., Woods M.A., Huggins M.B. A survey of the presence of a new infectious bronchitis virus designated 4/91 (793B) Vet. Rec. 1996;138:178–180. doi: 10.1136/vr.138.8.178. [DOI] [PubMed] [Google Scholar]
- Cowen B.S., Hitchner S.B. Serotyping of avian infectious bronchitis viruses by the virus-neutralization test. Avian Dis. 1975;19:583–595. [PubMed] [Google Scholar]
- Dolz R., Pujols J., Ordonez G., Porta R., Majo N. Antigenic and molecular characterization of isolates of the Italy 02 infectious bronchitis virus genotype. Avian Pathol. 2006;35:77–85. doi: 10.1080/03079450600597295. [DOI] [PubMed] [Google Scholar]
- Enjuanes L., Sola I., Alonso S., Escors D., Zuniga S. Coronavirus reverse genetics and development of vectors for gene expression. Curr. Top Microbiol. Immunol. 2005;287:161–197. doi: 10.1007/3-540-26765-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb J., Jr., Jackwood M.W. Infectious Bronchitis. In: Swayne D.E., Glisson J.R., Jackwood M.W., Pearson J.E., Reed W.M., editors. A Laboratory Manual for the Isolation and Identification of Avian Pathogens. The American Association of Avian Pathologists; Pennsylvania: 1998. pp. 169–174. [Google Scholar]
- Gibbs M.J., Armstrong J.S., Gibbs A.J. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics. 2000;16:573–582. doi: 10.1093/bioinformatics/16.7.573. [DOI] [PubMed] [Google Scholar]
- Gonzalez J.M., Gomez-Puertas P., Cavanagh D., Gorbalenya A.E., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough R.E., Randall C.J., Dagless M., Alexander D.J., Cox W.J., Pearson D. A 'new' strain of infectious bronchitis virus infecting domestic fowl in Great Britain. Vet. Rec. 1992;130:493–494. doi: 10.1136/vr.130.22.493. [DOI] [PubMed] [Google Scholar]
- Grenfell B.T., Pybus O.G., Gog J.R., Wood J.L., Daly J.M., Mumford J.A., Holmes E.C. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- Haydon D.T., Bastos A.D., Knowles N.J., Samuel A.R. Evidence for positive selection in foot-and-mouth disease virus capsid genes from field isolates. Genetics. 2001;157:7–15. doi: 10.1093/genetics/157.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst L.D. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18:486. doi: 10.1016/s0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- Jones R.C., Ambali A.G. Re-excretion of an enterotropic infectious bronchitis virus by hens at point of lay after experimental infection at day old. Vet. Rec. 1987;120:617–618. doi: 10.1136/vr.120.26.617. [DOI] [PubMed] [Google Scholar]
- Jones R.C., Jordan F.T.W. The exposure of day-old chicks to infectious bronchitis and the subsequent development of the oviduct. Vet. Rec. 1970;87:504–505. doi: 10.1136/vr.87.17.504. [DOI] [PubMed] [Google Scholar]
- Kant A., Koch G., van Roozelaar D.J., Kusters J.G., Poelwijk F.A., van der Zeijst B.A. Location of antigenic sites defined by neutralizing monoclonal antibodies on the S1 avian infectious bronchitis virus glycopolypeptide. J. Gen. Virol. 1992;73(Pt 3):591–596. doi: 10.1099/0022-1317-73-3-591. [DOI] [PubMed] [Google Scholar]
- Koch G., Hartog L., Kant A., van Roozelaar D.J. Antigenic domains on the peplomer protein of avian infectious bronchitis virus: correlation with biological functions. J. Gen. Virol. 1990;71(Pt 9):1929–1935. doi: 10.1099/0022-1317-71-9-1929. [DOI] [PubMed] [Google Scholar]
- Kottier S.A., Cavanagh D., Britton P. Experimental evidence of recombination in coronavirus infectious bronchitis virus. Virology. 1995;213:569–580. doi: 10.1006/viro.1995.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:2. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Kusters J.G., Niesters H.G., Bleumink-Pluym N.M., Davelaar F.G., Horzinek M.C., Van der Zeijst B.A. Molecular epidemiology of infectious bronchitis virus in The Netherlands. J. Gen. Virol. 1987;68(Pt 2):343–352. doi: 10.1099/0022-1317-68-2-343. [DOI] [PubMed] [Google Scholar]
- Lee C.W., Jackwood M.W. Evidence of genetic diversity generated by recombination among avian coronavirus IBV. Arch. Virol. 2000;145:2135–2148. doi: 10.1007/s007050070044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W., Jackwood M.W. Origin and evolution of Georgia 98 (GA98), a new serotype of avian infectious bronchitis virus. Virus Res. 2001;80:33–39. doi: 10.1016/s0168-1702(01)00345-8. [DOI] [PubMed] [Google Scholar]
- Liu S.W., Zhang Q.X., Chen J.D., Han Z.X., Liu X., Feng L., Shao Y.H., Rong J.G., Kong X.G., Tong G.Z. Genetic diversity of avian infectious bronchitis coronavirus strains isolated in China between 1995 and 2004. Arch Virol. 2006;151:1133–1148. doi: 10.1007/s00705-005-0695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.P., Williamson C., Posada D. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics. 2005;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
- Mase M., Tsukamoto K., Imai K., Yamaguchi S. Phylogenetic analysis of avian infectious bronchitis virus strains isolated in Japan. Arch. Virol. 2004;149:2069–2078. doi: 10.1007/s00705-004-0369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans G., Boschmans M., Decaesstecker M., Berg T.P.v.d., Denis P., Cavanagh D., van den Berg T.P. Epidemiology of infectious bronchitis virus in Belgian broilers: a retrospective study, 1986 to 1995. Avian Pathol. 2001;30:411–421. doi: 10.1080/03079450120066412. [DOI] [PubMed] [Google Scholar]
- Moore K.M., Bennett J.D., Seal B.S., Jackwood M.W. Sequence comparison of avian infectious bronchitis virus S1 glycoproteins of the Florida serotype and five variant isolates from Georgia and California. Virus Genes. 1998;17:63–83. doi: 10.1023/A:1008057118625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Bujarski J.J. Engineering of homologous recombination hotspots with AU-rich sequences in brome mosaic virus. J. Virol. 1997;71:3799–3810. doi: 10.1128/jvi.71.5.3799-3810.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Office e.d.i. Avian infectious bronchitis. In: OIE, editor. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Paris; 2004. Chapter 2.7.6. [Google Scholar]
- Padidam M., Sawyer S., Fauquet C.M. Possible emergence of new geminiviruses by frequent recombination. Virology. 1999;265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- Posada D. Evaluation of methods for detecting recombination from DNA sequences: empirical data. Mol. Biol. Evol. 2002;19:708–717. doi: 10.1093/oxfordjournals.molbev.a004129. [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Salminen M.O., Carr J.K., Burke D.S., McCutchan F.E. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.F., Kennedy S.I. Coronavirus multiplication strategy. II. Mapping the avian infectious bronchitis virus intracellular RNA species to the genome. J. Virol. 1980;36:440–449. doi: 10.1128/jvi.36.2.440-449.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.F., Sefton B.M. Coronavirus proteins: biogenesis of avian infectious bronchitis virus virion proteins. J. Virol. 1982;44:794–803. doi: 10.1128/jvi.44.3.794-803.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer S.G., Beard C.W. Serologic procedures. In: Swayne D.E., Glisson J.R., Jackwood M.W., Pearson J.E., Reed W.M., editors. A Laboratory Manual for the Isolation and Identification of Avian Pathogens. The American Association of Avian Pathologists; Pennsylvania: 1998. pp. 255–266. [Google Scholar]
- Villegas P. Titration of biological suspensions. In: Swayne D.E., Glisson J.R., Jackwood M.W., Pearson J.E., Reed W.M., editors. A Laboratory Manual for the Isolation and Identification of Avian Pathogens. The American Association of Avian Pathologists; Pennsylvania: 1998. pp. 248–254. [Google Scholar]
- Wang C.H., Huang Y.C. Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Arch. Virol. 2000;145:291–300. doi: 10.1007/s007050050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Xu Y., Collisson E.W. Experimental confirmation of recombination upstream of the S1 hypervariable region of infectious bronchitis virus. Virus Res. 1997;49:139–145. doi: 10.1016/S0168-1702(97)01466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterfield R.W., Hitchner S.B. Etiology of an infectious nephritis-nephrosis syndrome of chickens. Am. J. Vet. Res. 1962;23:1273–1279. [PubMed] [Google Scholar]
- Zanella A., Lavazza A., Marchi R., Moreno Martin A., Paganelli F. Avian infectious bronchitis: characterization of new isolates from Italy. Avian Dis. 2003;47:180–185. doi: 10.1637/0005-2086(2003)047[0180:AIBCON]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Zhang X.W., Yap Y.L., Danchin A. Testing the hypothesis of a recombinant origin of the SARS-associated coronavirus. Arch. Virol. 2005;150:1–20. doi: 10.1007/s00705-004-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]