Abstract
Post-translational modification of proteins and peptides by ubiquitin, a highly evolutionarily conserved 76 residue protein, and ubiquitin-like modifiers has emerged as a major regulatory mechanism in various cellular activities. Eukaryotic viruses are known to modulate protein ubiquitination to their advantage in various ways. At the same time, the evidence for the importance of deubiquitination as a viral target also is growing. This review centers on known viral interactions with protein deubiquitination, on viral enzymes for which deubiquitinating activities were recently demonstrated, and on the roles of viral ubiquitin-like sequences.
Keywords: β-Catenin, Deubiquitinating enzymes, Epstein–Barr nuclear antigen 1, Herpes simplex virus regulatory protein ICP0, Ubiquitin, Ubiquitin-specific protease 7
Introduction
Post-translational modification of proteins and peptides by ubiquitin (Ub), a highly evolutionarily conserved 76 residue protein, and ubiquitin-like modifiers (Ubls) (Fig. 1 ) has emerged as a major regulatory mechanism in various cellular activities including signal transduction, transcription, membrane protein trafficking, nuclear transport, autophagy, and immune responses (d'Azzo et al., 2005, Haglund and Dikic, 2005, Welchman et al., 2005). Protein modifications by Ub and Ubls, such as Nedd8, ISG15, and SUMO, modulate protein–protein interactions (Kerscher et al., 2006), while ubiquitin-like Atg8 homologs become lipidated and attached to cellular membranes (Tanida et al., 2004). Ub and most Ubls are produced as precursor proteins, and only carboxy-terminal processing after recognition sequence motifs by deubiquitinating enzymes (DUBs) generates the active modifiers (Amerik and Hochstrasser, 2004). Conjugations of Ub and Ubls to their targets relies on analogous enzymatic cascades comprising the sequential action of three enzymes (Passmore and Barford, 2004): a modifier activating enzyme (E1), one of several modifier carrier enzymes (E2s), and a member of the large and diverse group of modifier-target ligases (E3s), which chiefly determine target specificity. The enzymatic trio transfers the carboxy terminal glycine of Ub to the epsilon-NH2 group of an internal lysine residue of the target protein, or less often to its terminal amino group. In contrast to the known Ubl modifications, Ub can further be assembled into polymeric chains (polyubiquitination). One out of five internal lysine residues at position 6, 11, 29, 48, and 63 of Ub, but mostly lysine-48 or lysine-63, is used for the attachment of additional Ub units.
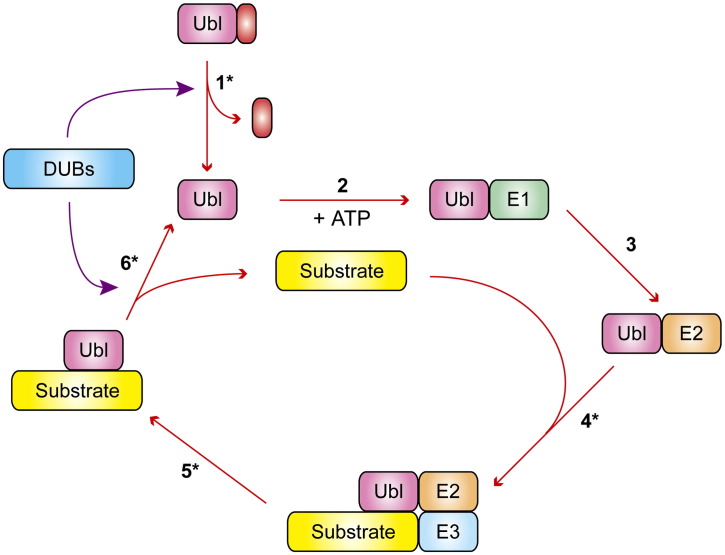
Fig. 1.
General schematic for Ubl conjugation and deconjugation and points of viral interference. Proteolytic maturation of Ubl (including Ub) precursor proteins by DUBs (step 1) exposes a new C-terminus in the modifier, which is then activated by E1 in an ATP-dependent manner (step 2) and next transferred to the E2 (step 3). An E3 generally recognizes the target protein (step 4) and facilitates the ligation of the Ubl to it (step 5), altering its protein interaction repertory and thereby its function. Polyubiquitination as a particular example of Ubl modification of a protein can lead to its proteasomal degradation. Ubl deconjugation by DUBs reversely regulates target protein function and, additionally, replenishes the pool of free Ubl (step 6). Points of viral interference are marked (*). Viruses are known to intervene with the Ub conjugation pathway (at steps 4 and 5, see text), but also target Ubl deconjugation (step 6), inclusively referred to as deubiquitination, which is the principal theme of this review. Viruses may also interfere with Ubl precursor processing (step 1), as suggested by the ability of some viral DUBs to process the ISG15 precursor protein (see text).
While attachment of at least four lysine-48 linked Ub molecules usually promotes the degradation of a protein by the Ub-proteasome system (UPS), the most important machinery for the degradation of cytoplasmic and nuclear proteins, chain formation via other lysine residues, or conjugation of individual Ub molecules mediates largely non-proteolytic functions of Ub (Ciechanover, 2006). Ub and Ubl modifications are reversed through the isopeptidase activities of DUBs (Fig. 1), with most studied DUBs deconjugating only a small number of targets (Nijman et al., 2005). In fact, deubiquitination, a term used here inclusive of Ub and Ubl deconjugation, is an emerging regulatory process in signaling pathways, chromatin structure, endocytosis, and apoptosis (Nijman et al., 2005) and is important for physiological activities including neuronal function, development, and immunity (Evans, 2005).
Viruses of eukaryotes are known to take advantage of protein ubiquitination in various ways. Entry or release of different viruses, for example, was shown to depend on the proteasome or on certain cellular E3s (Banks et al., 2003, Bieniasz, 2006, Ros and Kempf, 2004, Yu and Lai, 2005). What is more, many viruses manipulate protein ubiquitination in order to overcome host cell defense mechanisms, including apoptosis, the type 1 interferon (IFN) response, and major histocompatibility complex (MHC) class 1 antigen presentation. To this end, numerous viruses encode proteins that redirect cellular E3s of the UPS to proteins with antiviral activity (Fig. 1, step 4), including for example the tumor suppressor protein p53 (induction of apoptosis) and the signal transducers and activators of transcription (IFN response). Alternatively, some viruses express their own E3s (Fig. 1, step5), which commit cellular defense proteins, such as p53 or MHC class 1 molecules, to degradation. Such viral strategies were reviewed recently by Shackelford and Pagano (2005), Gao and Luo (2006), and Barry and Früh (2006) and are not considered here in detail.
Different from ubiquitination, only few examples for the targeting of protein deubiquitination (Fig. 1, step 6) by viruses have been described to date. They include the potential recruitment of DUBs for the stabilization of β-catenin in Epstein–Barr virus (EBV)-infected B cells (Ovaa et al., 2004, Shackelford et al., 2003), and the specific targeting of the cellular DUB ubiquitin-specific protease 7 (USP7) by the Epstein–Barr nuclear antigen 1 (EBNA1) and the herpes simplex virus type 1 (HSV-1) regulatory protein ICP0 (Everett et al., 1997, Holowaty and Frappier, 2004). Despite the compelling biochemical evidence for the specificity of these two interactions, the importance of viral targeting of USP7 remains vague. The possibility that modulation of deubiquitination is, nevertheless, a more common viral strategy has gained support by the recent in vitro demonstration of deubiquitinating activities for three viral enzymes: the adenovirus protease adenain (Balakirev et al., 2002), the papain-like protease (PLpro) of severe acute respiratory syndrome coronavirus (SARS-CoV) (Barretto et al., 2005, Lindner et al., 2005), and a protease domain contained in the N-terminal fragment of the large tegument protein UL36 (UL36USP) from several herpesviruses, namely, HSV-1, EBV, and mouse and human cytomegalovirus (MCMV and HCMV) (Kattenhorn et al., 2005, Schlieker et al., 2005, Wang et al., 2006). However, the roles of these deubiquitinating activities during virus infection remain elusive. Here, the known viral interactions with protein deubiquitination are reviewed, and potential roles of viral DUBs are considered.
Deubiquitinating enzymes
The majority of the DUBs from all kingdoms of life (excluding archaea), including the known and predicted viral enzymes, represent cysteine protease homologs, with the remainder forming a separate family of metalloproteases (Rawlings et al., 2004). At least six structural classes (families) of cysteine protease DUBs have been identified: the ubiquitin-specific protease (USP), autophagin (ATG), ubiquitin C-terminal hydrolase (UCH), ovarian tumor-related protease (OTU), Josephin-domain protease (JD), and ubiquitin-like protein-specific protease (ULP) families (Amerik and Hochstrasser, 2004, Nijman et al., 2005, Sulea et al., 2006). They all feature structural variations of the papain fold and display canonical papain-like spatial arrangement of catalytic centers. Many are further characterized by additional variable amino- and/or carboxy-terminal sequences, for which a growing number of structures and functions, including localization, substrate recognition, and/or activation of catalytic activity, are being reported (Nijman et al., 2005, Reyes-Turcu et al., 2006, Sulea et al., 2006).
Despite recent advances in the understanding of DUB structure–function relationships (Amerik and Hochstrasser, 2004, Nijman et al., 2005, Sulea et al., 2006), inference of biological importance for uncharacterized DUBs from sequence information alone remains challenging. The human genome encodes around 90 cysteine protease DUBs and 12 metalloprotease DUBs (Rawlings et al., 2004). The number of catalytically active human cysteine protease DUBs, inclusive of putative alternative splice isoforms, amounts to over 100. In order to further the functional characterization of DUBs on a large scale, Ploegh and co-workers synthesized specific probes for the proteomic profiling of cysteine protease DUB activities (Borodovsky et al., 2002). They used an intein-based chemical ligation method to modify the free C-terminus of a hemagglutinin (HA)-tagged Ub with a series of thiol-reactive groups, such as vinylmethylsulfone (VME) or bromoethylamine. After incubation of cellular lysates with some of these active site-directed suicide substrates and subsequent anti-HA immunoprecipitation, tandem mass spectrometry facilitated the simultaneous identification of multiple DUBs and some associated proteins (Borodovsky et al., 2002). HAUbVME exhibited the broadest reactivity, and, in the following, was used to detect active cysteine protease DUBs and associated proteins in EBV-infected B cells (Ovaa et al., 2004, Shackelford et al., 2003), and human papillomavirus (HPV)-infected cervical carcinoma cells as well as HPV E6/E7 immortalized keratinocytes (Rolén et al., 2006).
β-Catenin stabilization in Epstein–Barr virus infection
In a study by Ovaa et al. (2004), lymphoblastoid transformation of freshly isolated B cells by in vitro EBV infection led to an increase in the activities of six cellular DUBs, identified as UCH37, UCH-L3, UCH-L1, USP7, USP9X, and USP15, at different times postinfection. Interestingly, USP9X was previously shown to interact with the bifunctional armadillo repeat protein β-catenin in vitro as well as in cultured epithelial cells and was able to stabilize β-catenin in these cells (Taya et al., 1999). β-Catenin is further known to be stabilized in latency type III EBV-transformed B cells (see below).
β-Catenin is a component of cell-cell adherens junctions. At the same time, a low-level cytoplasmic pool of this protein functions in canonical Wnt signaling (reviewed by Brembeck et al., 2006). In the absence of Wnt signaling, cytoplasmic β-catenin is continuously phosphorylated by casein kinase followed by glycogen synthase kinase 3-β, which together with the two scaffold proteins axin and the adenomatosis polyposis coli tumor-suppressor protein constitute the so-called degradation complex. These consecutive phosphorylations events initiate the polyubiquitination of β-catenin by a Ub ligase complex, known as Skp1/cullin/F box proteinβ-TrCP (SCFβ-TrCP) (Liu et al., 2001, and references therein), and its rapid proteasomal degradation. Binding of the Wnt ligand to its cell surface receptors is thought to trigger a series of phosphorylation events that result amongst others in the degradation of axin, causing disassembly of the above-mentioned degradation complex and consequently the stabilization of β-catenin which then accumulates in the nucleus. Here, β-catenin acts as a coactivator of Wnt target genes that regulate cellular proliferation and differentiation during animal development and tissue homeostasis (Städeli et al., 2006).
β-Catenin stabilization in latency type III EBV-transformed B cells occurs by mechanisms that involve EBV latent membrane proteins 1 (LMP1) and 2A (LMP2A) (Hayward et al., 2006). This does, however, not necessarily entail its nuclear accumulation, indicating that effects of EBV induced β-catenin stabilization are dependent on the cellular context and in lymphoid cells may be distinct from Wnt activation (Morrison et al., 2004). Lately, Pagano and coworkers identified LMP1 as a transcriptional down-regulator of another Ub ligase complex that targets β-catenin for proteasomal degradation and is called seven in absentia homolog-1 (Jang et al., 2005). In any event, the observed up-regulation of DUBs in EBV transformed B cells by Ovaa et al. (2004), together with the almost simultaneous demonstration that β-catenin in the type III latently infected B cell line Sav III exists in a complex with active, yet unidentified DUBs by Shackelford et al. (2003), strengthens the possibility that DUBs contribute to EBV induced β-catenin stabilization.
Viral targeting of ubiquitin-specific protease 7
USP7 is an evolutionary conserved mammalian DUB of the USP family, which was originally identified by its ability to bind to two different herpesviral proteins, namely, EBNA1 of EBV, and the HSV-1 regulatory protein ICP0 for which it has also been coined herpes virus-associated ubiquitin-specific protease (HAUSP) (Everett et al., 1997, Holowaty and Frappier, 2004). USP7 displays debranching activity against lysine-48 linked polyubiquitin chains and was shown to play an important part in the dynamic regulation of nuclear p53 turnover (Brooks and Gu, 2006, Cheon and Baek, 2006).
Ubiquitin-specific protease 7 and the p53-MDM2 pathway
In unstressed cells, p53 is constitutively polyubiquitinated, which leads to its proteasomal degradation, keeping p53 levels low (Gomez-Lazaro et al., 2004). p53 levels dramatically increase upon various types of stress, including viral infection, triggering either growth arrest or apoptosis. Virus mediated gain of p53 ubiquitinating activity as a means of apoptosis avoidance was already mentioned above. Expression of USP7, the target protein of EBNA1 and ICP0, however, effectively promotes increased levels of p53 by antagonizing proteasomal degradation of p53 through deubiquitination (Li et al., 2004). As may be expected, partial reduction of USP7 levels by RNA interference (RNAi) was shown to destabilize p53 (Li et al., 2004). With the collaboration of the adapter protein Daxx (death domain associated protein) (Tang et al., 2006), USP7 also deubiquitinates the mouse double minute 2 (MDM2) oncogene, one of several E3s that mediate p53 proteasomal degradation. This counteracts autoubiquitination of MDM2 and its proteasomal degradation (Li et al., 2004). In fact, a genetic knockout of USP7 caused depletion of MDM2 and, despite the aforementioned rescue effect of USP7 on p53, effectively stabilized p53 (Cummins et al., 2004, Li et al., 2004). Brooks and Gu (2006) have recently proposed that the predominant role of MDM2 during cellular stress is to ubiquitinate p53, and to keep its levels in check in order to maintain growth arrest while avoiding default execution of the apoptotic program and thereby to afford cell survival in the event of successful cellular repair and overcoming of the original challenge. In view of these activities, USP7 appears to be well-positioned to relay signals that regulate the p53-MDM2 pathway (Fig. 2 ). But it also becomes clear that the possible reduction of p53 levels through viral inhibition of USP7 would likely need to be well-balanced in order not to achieve the opposite. Recent structure–functional studies on the EBV protein EBNA1 have brought to light how this protein may accomplish this and are outlined in the following. The role of the ICP0-USP7 interaction in p53 metabolism during HSV-1 infection has as yet proven complex to delineated (see below).
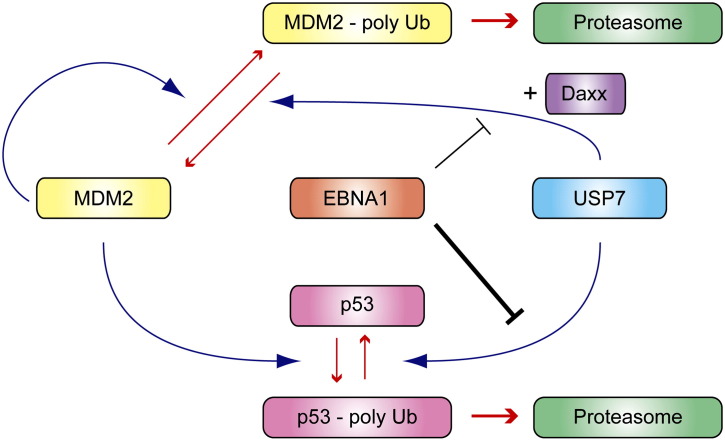
Fig. 2.
Model for regulation of the p53-MDM2 pathway by USP7 and interference by EBNA1. The E3 enzyme MDM2 catalyses both autoubiquitination and ubiquitination of p53 leading to proteasomal degradation in both cases. Deubiquitination by USP7 stabilizes p53 and MDM2, with the adaptor protein Daxx directing USP7 to MDM2. Partial reduction of USP7 activity by RNAi destabilizes p53 through reduced deubiquitination. Contrarily, genetic knockout of USP7 increasingly commits MDM2 to proteasomal degradation thus leading to an important reduction in p53 ubiquitination and, indirectly, effective p53 stabilization. The EBV protein EBNA1, MDM2, and p53 compete for same binding site on USP7, with affinities decreasing in this order. Hence, inhibition of USP7 by EBNA1 may exhibit selectivity and contribute twofold to p53 destabilization. By blocking the deubiquitination of p53 more efficiently than the deubiquitination of MDM2, it may allow for sufficient levels of MDM2 that maintain p53 ubiquitination.
Epstein–Barr nuclear antigen 1
For a long time, EBNA1 has been known as a regulator of both transcription and replication of the EBV genome, as well as being required for the segregation of EBV genomes with chromosomes during mitosis (Frappier, 2004, Wang and Sugden, 2005). EBNA1 is essential for viral persistence, promotes cellular immortalization and is found consistently expressed in EBV-associated human malignancies including Burkitt's lymphoma, Hodgkin's lymphoma, and nasopharyngeal carcinoma. Functional studies by Saridakis et al. (2005) have indicated that an interaction of transfected EBNA1 with USP7 fosters cellular degradation of p53, presumably by preventing p53 deubiquitination through USP7, and confers apoptosis resistance to UV-irradiated cells. Structural analyses from the laboratories of Shi and Frappier have recently provided insight into the molecular mechanism by which EBNA1 modulates p53 turnover and likely contributes to the anti-apoptotic and survival factor function of EBNA1 in vivo. Crystal structures for short EBNA1, p53, and MDM2 peptides, respectively, in complex with the N-terminal tumor necrosis factor-receptor associated factor (TRAF)-like domain of USP7 show that these three proteins use a consensus tetrapeptide recognition sequence to engage in structurally conserved contacts with the same surface groove of the TRAF-like domain of USP7 (Hu et al., 2006, Saridakis et al., 2005, Sheng et al., 2006). The buried contact surface areas and extent of directed interactions, as well as the measured binding affinities of corresponding peptides to the USP7 TRAF-like domain increase in the order of p53 < MDM2 < EBNA1 (Hu et al., 2006, Sheng et al., 2006). It appears that the competitive binding to USP7 of EBNA1 versus p53 and MDM2 prevents deubiquitination of the lowest affinity binder (p53) sufficiently to reduce cellular p53 levels, while it permits deubiquitination of the intermediate binder (MDM2) to continue, avoiding MDM2 depletion and p53 stabilization, as observed in the USP7 knockout (Fig. 2). Otherwise, i.e., in case MDM2 levels also dramatically drop in the presence of EBNA1, one of the remaining E3s for p53 (Brooks and Gu, 2006) may play an important role in keeping p53 levels low in EBV-infected cells.
Herpes simplex virus regulatory protein ICP0
The immediate–early HSV-1 gene product ICP0 is required for efficient initiation of lytic infection by stimulating the reactivation of quiescent viral genomes (Hagglund and Roizman, 2004). ICP0 functions as an E3 whose putative in vivo targets include p53. Early in HSV-1 infection, ICP0 associates with nuclear domain 10 (ND10), nuclear substructures that are found juxtaposed to the genomes of many DNA viruses including herpesviruses and adenoviruses (Everett, 2006). ND10 have been implicated in DNA repair, the IFN response, and the regulation of p53 activity (Everett, 2006, Takahashi et al., 2004) and might be preferred sites of transcription and replication of DNA viral genomes (Ching et al., 2005). ICP0 induces the proteasomal degradation of the ND10 organizing promyelocytic leukemia (PML) protein and promotes rapid dispersal of ND10 (Everett, 2006). This has been proposed to alleviate PML protein mediated anti-HSV-1 effects of IFN (Chee et al., 2003). Recent results by Everett et al. (2006), obtained by the use of RNAi to reduce PML levels, indeed argue for the contribution of PML to a cellular antiviral repression mechanism that is countered by ICP0.
Everett and coworkers (1997) had previously demonstrated that ICP0 increases the proportion of USP7 localized to ND10. Contrary to EBNA1, p53, and MDM2, the binding of ICP0 to USP7 was mapped to a domain of unknown structure, located C-terminally to the catalytic core domain of USP7 (Holowaty et al., 2003). Possible effects of ICP0 on p53 metabolism were shown not to depend on the interaction of ICP0 with USP7 but on the particular cell type under investigation (Boutell and Everett, 2004). Instead, the significance of ICP0 binding to USP7 possibly lies in the ability of USP7 to counteract autoubiquitination of ICP0 and to protect it from proteasomal degradation (Canning et al., 2004). Although USP7 is conversely ubiquitinated and marked for proteasomal degradation by ICP0, the biological net effect of the reciprocal activities between ICP0 and USP7 is thought to be the stabilization of ICP0 early during HSV-1 lytic infection or reactivation from latency, when ICP0 levels are low (Boutell et al., 2005). Interestingly, the aforementioned USP7-MDM2 adaptor protein Daxx (Tang et al., 2006) is a major ND10 component (Everett et al., 2006), raising the question whether it also has an adaptor function for USP7 in this nuclear compartment.
Viral deubiquitinating activities
Deubiquitinating activities for viral enzymes have directly been demonstrated for the adenovirus protease adenain, SARS-CoV PLpro, and herpesviral UL36USP. Structural aspects of these specificities have been reviewed recently by Sulea et al. (2006). In the following, potential roles for these confirmed viral DUBs are considered against the background of their established functions and properties.
Adenovirus protease adenain
During adenovirus infection, adenain is first made in an essentially inactive form of 23 kDa which localizes to both the cytoplasm and the nucleus (reviewed by Mangel et al., 2003). In the nucleus, binding to viral DNA partially activates the enzyme inside nascent virions, allowing it to cleave an eleven amino acid peptide, named pVIc, from the precursor of the viral DNA binding capsid protein VI (pVI). pVIc binding fully activates adenain, and the peptidic cofactor becomes eventually disulphide-linked to the enzyme. Activated adenain is thought to subsequently complete the proteolytic maturation of altogether six virus capsid precursor proteins inside the virion. The stepwise activation of adenain by viral DNA and pVIc prevents precursor protein cleavage before virion assembly and the generation of immature capsids. Maturation of the capsid proteins is important for their ability to promote low-pH activated endosomal lysis and cytoplasmic entry of viral capsids during the next infection cycle (Cotten and Weber, 1995).
Cytoplasmic adenain is believed to contribute to cell lysis and release of virions by the cleavage of cytoskeletal proteins, including cytokeratin 18 and actin. In contrast to nuclear adenain, the cytoplasmic form has no access to either of the two capsid bound viral cofactors, viral DNA and pVIc. Instead, the C-terminal sequence of actin, which is highly homologous to pVIc, efficiently replaces pVIc acting as a cellular cofactor (Brown and Mangel, 2004). Within the extracellular virus, adenain resides in an oxidized and dormant state, but becomes activated again up on infection and re-entry into a reducing cellular environment. Adenain eventually participates in the final steps of the viral uncoating program, i.e., dissociation of the viral DNA from the capsid at the nuclear pore complex, by the digestion of pVI (Greber et al., 1996).
Using a biotinylated form of the specific DUB inhibitor Ub-aldehyde as a probe, Balakirev et al. (2002) retrieved adenain from a lysate of adenovirus-infected HeLa cells. They showed that the enzyme accounted for a time-dependent increase in global and, especially, nuclear deubiquitinating activity in late phase adenovirus-infected cells compared to mock-infected cells, as judged by the decline of the Ub conjugate pool analyzed by Western blotting with anti-Ub antibodies (Balakirev et al., 2002). Additionally, the authors overexpressed in the same cell line hexahistidine-tagged Ub together with either adenain or an autocleavable fusion of the enzyme to its activating peptide cofactor pVIc, followed by enrichment of Ub conjugates from cell lysates over a metal affinity resin. Anti-Ub Western blotting of the resulting fractions demonstrated, again, an overall reduction in Ub conjugates with both versions of the transfected enzyme. Enzymatic in vitro assays with recombinant purified adenain and substrate proteins, and chemically synthesized pVIc peptide showed that adenain, indeed, exhibits debranching activity against lysine-48 linked polyubiquitin and could also process the ISG15 precursor protein, but not a fusion of the yeast SUMO homolog Smt3 to the green fluorescent protein. This specificity, seemingly, conflicts with the structural classification of adenain as a member of the ULP family of deSUMOylating enzymes (Balakirev et al., 2002) but agrees well with the molecular binding site features of the enzyme (reviewed by Sulea et al., 2006).
The described prevalence of adenain-dependent deubiquitination of nuclear proteins in infected cells (Balakirev et al., 2002) is in accordance with the presence of adenain in this subcellular compartment, where it is involved in virion maturation. At the same time, however, activation of nuclear adenain is thought to occur only inside the nascent virion particle, with about 70 adenain-pVIc complexes remaining incorporated per particle (Mangel et al., 2003). While this may suggest that virion resident adenain deubiquitinates capsid proteins, no change in the overall pattern of the anti-Ub staining in comparison to nuclei of mock-infected cells was observed (Balakirev et al., 2002), indicating that the overall decline in nuclear Ub conjugates, for the most part, reflected deconjugation of cellular proteins. This could mean that activated adenain partially escapes from nascent virions and acts on proteins in the surrounding nucleoplasm (the possibility that nuclear adenain is activated in a virion independent manner is considered below).
As already mentioned, besides HSV-1, adenoviruses belong to the DNA viruses whose genomes associate with ND10. Interestingly, ND10 in turn are associated with nuclear aggresomes, sites that recruit chaperones, Ub, and proteasomes and that may be specialized in protein degradation (reviewed by Wileman, 2006). Moreover, ND10 have been proposed to present passageways for proteins, including viral proteins that are destined for proteasomal degradation in the proximity of ND10 (Bailey and O'Hare, 2005, Hay, 2005). In this light, it is tempting to speculate that deubiquitination by adenain safeguards the delivery of viral proteins to nascent capsids by protecting them from proteasomal degradation at close-by aggresomes, much as cellular USP7 is thought to stabilize ICP0 of HSV-1 (Boutell et al., 2005).
It is also noteworthy that the plasmid mediated overexpression of adenain in HeLa cells, both with and without its peptidic cofactor, resulted in an overall loss of cellular Ub conjugates (Balakirev et al., 2002). The strong dependence of adenain catalytic activity on cofactor complex formation and the ability of the C-terminal sequence of the traditionally cytoplasmic protein actin to act as a cofactor (Brown and Mangel, 2004) may suggest that predominantly cytoplasmic adenain activity was detected in this experiment. Alternatively, a notable fraction of adenain may have entered the nucleus together with actin derived peptide cofactor. Actin itself has also been identified in the nucleus as a component of protein complexes active in various aspects of gene transcription (Grummt, 2006, Miralles and Visa, 2006). Actin may therefore also be involved in a virion independent stimulation of adenain activity in the nucleus.
Severe acute respiratory syndrome coronavirus papain-like protease
After entry of the coronaviral single-stranded positive-sense RNA genome into the cytoplasm, the viral replicase gene is translated directly from it. Two coronaviral proteases, 3C-like protease (3CLpro) and PLpro, are part of the replicase polyprotein, the precursor of the altogether sixteen non-structural proteins (nsps) that form the viral RNA replication complex (Snijder et al., 2003, Thiel et al., 2003). 3CLpro, on the one hand, is contained in nsp5, and after autocleavage, releases all downstream replicase subunits. PLpro, on the other hand, originally referred to a domain of around 24 kDa within nsp3, whose boundaries are defined by homology to the papain-fold (Herold et al., 1999). PLpro processes the amino-proximal nsps (Harcourt et al., 2004). In accordance with previous structural bioinformatics prediction (Sulea et al., 2005), the crystal structure of SARS-CoV PLpro recently established the enzyme's membership in the USP family of DUBs (Ratia et al., 2006). Additionally, it revealed an unexpected Ub-like domain at the N-terminus of the PLpro catalytic core domain. Ub-like domains are defined by a common β-grasp three-dimensional structure (Kiel and Serrano, 2006). Within the multidomain nsp3, PLpro is further preceded by an acidic sequence forming the N-terminus of nsp3, a macro-domain with ADP-ribose-1″-phosphatase activity, which is potentially involved in viral RNA modification (Saikatendu et al., 2005), and a so-called SARS unique domain (Snijder et al., 2003). PLpro is followed by a hydrophobic domain with putative transmembrane regions (Harcourt et al., 2004). PLpro cotranslationally liberates nsp1 to 3 in this order (Harcourt et al., 2004). All three cleavage products become part of the replication complex, which is found bound to double-membrane vesicles that are characterized by autophagosome markers (Prentice et al., 2004). A more recent ultrastructural study, however, points to the endoplasmic reticulum as the direct origin of the membranes associated with SARS-CoV replication complex, including nsp3 (Snijder et al., 2006).
SARS-CoV PLpro, which now refers to the enzyme's catalytic core domain plus the N-terminal Ub-like domain, was recombinantly expressed and purified (Barretto et al., 2005, Lindner et al., 2005). As predicted based on the similarity of its catalytic core domain to the corresponding domain of USP7 (Sulea et al., 2005), SARS-CoV PLpro displays DUB activity. Specifically, it debranches lysine-48 polyubiquitin chains, very efficiently hydrolyzes the general DUB substrate Ub-7-amino-4-methylcoumarin (Ub-AMC) (Barretto et al., 2005, Lindner et al., 2005), and exhibits ISG15 precursor processing activity (Lindner et al., 2005). Although this proves the enzyme's proficiency as a DUB in vitro, it is not clear whether SARS-CoV PLpro can gain access to potential deubiquitination targets, other than replicase polyprotein sequences themselves, during its synthesis as part of the replicase polyprotein, cotranslational autoprocessing, and incorporation into the membrane bound replication complex. It is noteworthy that the synthesis of both negative- and plus-strand coronavirus RNA requires ongoing viral protein production (Kim et al., 1995, Perlman et al., 1986, Sawicki and Sawicki, 1986), and it is conceivable that concomitant deubiquitination by SARS-CoV PLpro protects replicase subunits against proteasomal degradation.
Herpesvirus UL36USP
Prompted by the known interaction of ICP0 with cellular USP7, Ploegh and coworkers made use of the active-site directed probe HAUbVME, mentioned above, to monitor DUB activity in lysates of primary fibroblasts infected with HSV-1 (Kattenhorn et al., 2005). A major HAUb-adduct corresponding to an ∼ 47-kDa protein occurred late in infection and persisted. The protein was identified as N-terminal fragment of the essential large (3164 amino acid residues) tegument protein UL36 (also called VP1/2 or ICP1/2). Further labeling attempts with similar Ubl probes, and in vitro enzymatic assays using a 533 residues recombinant N-terminal fragment of UL36, established that the enzyme, baptized UL36USP, is specific for Ub, but exhibits relatively low catalytic efficiency. Recombinant HSV-1 UL36USP disassembled lysine-48 but not lysine-63 polyubiquitin chains which may implicate the enzyme in protein stabilization (Kattenhorn et al., 2005).
Sequence alignment of UL36 homologs from α-, β-, and γ-herpesvirus genomes identified a putative cysteine box around the HAUbVME reactive cysteine residue, and a histidine box located around 130 amino acids further downstream (Kattenhorn et al., 2005). Although none of the UL36USP sequences shows similarity to any known DUB, the conservation of cysteine box and histidine box motifs is characteristic of cysteine protease DUB families (Amerik and Hochstrasser, 2004). In addition to HSV-1 (α-subfamily), analogous labeling experiments confirmed the presence of DUB activity and specificity for Ub in recombinant UL36USP from EBV (γ-subfamily), and MCMV and HCMV (β-subfamily) (Schlieker et al., 2005, Wang et al., 2006). The sequence of the recombinant UL36USP variant examined for EBV extended only little beyond the histidine box motif, demonstrating that an N-terminal UL36 fragment of just under 22 kDa carries the UL36USP specificity (Schlieker et al., 2005).
Wang et al. (2006) detected HAUbVME labeling of full-length UL36 in HCMV, simian CMV (SCMV) and HSV-1-infected fibroblasts. They also reported the detection of the 47-kDa fragment of UL36 for HSV-1 previously described by Kattenhorn et al. (2005) but ascribed its occurrence to uncontrolled proteolysis (Wang et al., 2006). The authors went ahead to confirm catalytic activity in isolated wild-type and mutant HCMV extracellular particles. Mutation of either the putative catalytic cysteine or histidine residue abolished catalytic activity, identifying UL36USP as the sole source of DUB activity in the virion. For additional mutants of several conserved cysteine box and histidine box residues, the spread and development of cytopathic effects as well as virus yields in virus-infected cells were wild-type like. Only the respective cysteine and histidine mutants gave notably lower virus yields and delayed the development of cytopathic effects. However, the cysteine mutation caused no apparent changes during cell infection as judged by electron microscopy. Overall, this indicates that UL36USP is important for optimal HCMV replication but is not essential in cell culture (Wang et al., 2006).
So far, we can only speculate on possible roles for UL36USP considering the known implications of the large tegument protein UL36, which according to earlier studies shows equal cytoplasmic and nuclear distribution (McNabb and Courtney, 1992). UL36 was demonstrated to interact with a region of the viral α sequence, which is required for cleavage and packaging of the viral genome in the nucleus (Chou and Roizman, 1989) into icosahedral capsids (see Pomeranz et al., 2005, for a review of the α-herpesvirus life cycle). Studies of α-herpesvirus-infected cells using microscopic techniques coupled with protein labeling methods have led to two different theories that aim at explaining how nucleocapsids exit the nucleus, obtain the viral tegument, and acquire an envelop. They imply different pathways for the capsid attachment of UL36, which is thought to form the innermost, capsid-proximal layer of the tegument. According to the more prevalent of the two theories (reviewed by Mettenleiter, 2004, Mettenleiter et al., 2006), viral capsids reach the cytoplasm by successive envelopment and de-envelopment at the inner and outer membrane of the nuclear envelope, respectively. Primary attachment of UL36 in the cytoplasm then affords further assembly of inner tegument proteins. The inner tegument subsequently coalesces with the outer tegument, which assembles independently at future sites of budding into the exocytic pathway, likely into the trans-Golgi network (Mettenleiter, 2006). The alternative theory (Leuzinger et al., 2005, Wild et al., 2005) assumes a dual pathway for nuclear egress of capsids. Capsids may either undergo nuclear envelopment followed by intraluminal transport to the Golgi, or they may leave the nucleus directly through dilated nuclear pores and afterwards bud from the cytoplasm into compartments of the exocytic pathway. Here, the deposition of tegument is thought to occur during the budding at the inner nuclear membrane or at cytoplasmic membranes (Leuzinger et al., 2005, Wild et al., 2005), hence presumably involving nuclear or cytoplasmic UL36, respectively. It has however to be noted that UL36 has not yet been detected as a component of virions inside the lumen of the nuclear envelope (Mettenleiter and Minson, 2006). Wherever herpesvirus tegumentation occurs, tegument proteins are known to engage in many complex protein–protein interactions (reviewed by Mettenleiter, 2006). Yeast two-hybrid analyses suggest that UL36 uses a domain downstream of UL36USP to directly bind to UL37, another inner tegument protein (Klupp et al., 2002, Vittone et al., 2005). Fusion of Golgi derived vesicles with the plasma membrane eventually releases infectious virions from the cell.
After infection by fusion of the viral envelope with the cell membrane, UL36 initially stays associated with the capsid. UL36 is in fact emerging as a main candidate among herpesviral proteins that may dock incoming capsids to the dynein motor during transport along microtubules (MTs) to the nuclear pores (Granzow et al., 2005) and during retrograd axonal transport (Antinone et al., 2006, Luxton et al., 2005). In this regard, it is interesting to note that the minus end-directed transport of misfolded proteins to the microtubule-organizing center during aggresome formation was shown to require the interaction of the dynein motor with the cellular DUB ataxin-3 (Burnett and Pittman, 2005). As UL36USP, ataxin-3 debranches lysine-48 polyubiquitin chains, but possible targets of this activity during retrograd transport are unknown. Last of all, a temperature sensitive mutation in HSV-1 UL36 impeded the release of viral DNA from the capsid into the nucleus at the nuclear pore (Batterson et al., 1983).
Taken together, during herpesvirus infection UL36 reportedly localizes to the nucleus, the cytoplasm, and the lumen of the nuclear envelope as well as the exocytic pathway where it is eventually part of the infectious virion, whose yield is reduced by mutational inactivation of UL36USP. It remains speculative at this point whether deubiquitination of viral or cellular proteins by its UL36USP domain plays part in any of the processes that have been associated with the presence of UL36 in these locations, i.e., nucleocapsid formation, tegumentation, budding, viral egress from and entry into cells, MT-dependent capsid transport, and nuclear entry of the viral genome.
Viral ubiquitin and ubiquitin-like sequences
The genomes of some viruses encode Ub or Ub-like sequences. The baculovirus Autographa californica nucleopolyhedrovirus, for example, expresses a viral Ub precursor protein during the late phase of infection (Guarino, 1990). Although non-essential for replication in cell culture, the gene is required for optimal virus production (Reilly and Guarino, 1996). The baculoviral Ub is functional in protein conjugation in vitro but inhibits the formation of more extended lysine-48 linked polyubiquitin chains necessary for proteasomal targeting (Haas et al., 1996). The authors speculated that this protects otherwise short-lived viral proteins from proteasomal degradation.
Viruses also express Ub or Ub-like sequences as part of multidomain polyproteins. Intriguingly, Ub and Ubls including NEDD8, SUMO, and Atg8 homologs represent the most frequent inserts in the polyprotein of several strains of bovine viral diarrhea virus (BVDV), where their presence is associated with a viral cytopathogenic phenotype (Baroth et al., 2000, Meyers et al., 1998, Qi et al., 1998, Tautz et al., 1993). In fact, these inserts function as polyprotein processing signals that allow cellular proteases, presumably DUBs, to process the polyprotein at positions corresponding to the precursors processing sites of the respective cellular Ub or Ubl sequences. In the case of an Atg8 sequence insert in the BVDV isolate JaCP, the processing enzyme was indeed identified as a specific cellular DUB, namely, autophagin-1 (ATG4B) (Fricke et al., 2004).
Compared to Ub and Ubls, the already mentioned N-terminal domain of SARS-CoV PLpro (Ratia et al., 2006) for example represents a different but not less common type of Ub-like domain (Kiel and Serrano, 2006). Here, the lack of a C-terminal recognition sequence motif precludes proteolytic processing by DUBs and subsequent conjugation to other molecules. Nevertheless, such intrinsic Ub-like domains also serve in mediating protein–protein interactions in a variety of multidomain proteins (see Kiel and Serrano, 2006, and references therein). These include players of the UPS, such as specialized Ub receptors (Elsasser and Finley, 2005), and last but not least DUBs (Nijman et al., 2005; Zhu and Sulea, unpublished data). The domain arrangement in SARS-CoV PLpro is in fact reminiscent of USP14 and its homologs, where the Ub-like domain likewise precedes the catalytic core domain. USP14 and its yeast homolog Ubp6 both bind to the regulatory subunit of the proteasome via their Ub-like domain which greatly stimulates their catalytic activities (Hu et al., 2005, Schmidt et al., 2005). Comparison of the crystal structures for the free and Ub bound catalytic core domain of USP14 suggests that the enzyme is activated by conformational translocation of two enzyme surface loops, which block access of the Ub C-terminus to the active site in the free enzyme (Hu et al., 2005). Proteasome binding may promote this activation step. USP14 was further shown to debranch lysine-48 linked polyubiquitin chains from the distal end (Hu et al., 2005). USP14/Ubp6 is thought to prevent the translocation of Ub from incoming substrates into the inner core particle of the proteasome and to contribute to the homeostasis of the cellular pool of free Ub (Schmidt et al., 2005). It is tempting to speculate that the Ub-like domain of SARS-CoV PLpro may similarly anchor the enzyme to a larger protein complex. There is, however, no indication from the available crystal structure of the free enzyme, like for USP14, that its catalytic activity may require activation (Ratia et al., 2006).
Conclusions
The binding of ICP0 and EBNA1 to different domains of the cellular DUB USP7 contributes to the maintenance of a productive life cycle for HSV-1 and the establishment of latency for EBV, respectively, representing the only well established examples of viral interference with deubiquitination so far. Interestingly, however, infection by EBV, HCMV, and HPV modulates the activities of several cellular DUBs (Ovaa et al., 2004, Rolén et al., 2006, Wang et al., 2006) the significance of which remains to be established. DUBs, such as USP9X, for instance, might be involved in β-catenin stabilization in latency type III EBV-transformed B cells.
It will further be important to establish the roles of viral deubiquitinating activities. Like USP7, the three recently confirmed viral DUBs, adenain, SARS-CoV PLpro, and herpesvirus UL36USP, all exhibit lysine-48 linked polyubiquitin debranching activities. It is conceivable that viruses avail themselves of this activity in order to stabilize viral gene products or host cell proteins whose proteasomal degradation promotes anti-viral responses such as IκB (Evans, 2005). The ability of adenain and SARS-CoV PLpro to additionally cleave the ISG15 precursor protein has led to the speculation that these enzymes mimic cellular USP18 (Sulea et al., 2006), a DUB with delSGylating activity and a negative regulator of the IFN response (Dao and Zhang, 2005). Recent data by Zhang and coworkers (Malakhova et al., 2006) revealed, however, that USP18 attenuates JAK-STAT signaling, and thereby the type 1 IFN response, in a non-enzymatic manner, i.e., by directly competing with JAK1 for binding to the IFNAR2 subunit of the type 1 IFN receptor. As for SARS-CoV PLpro, the importance of the deubiquitinating activity of USP18 remains to be determined.
Table 1 summarizes the demonstrated and potential roles in viral infection of both cellular and viral DUBs discussed thus far. The review of the possible subcellular sites of action for the three confirmed viral DUBs presented here suggests that their deubiquitinating activities could, similar to the polyprotein processing activities of adenain and SARS-CoV PLpro, also contribute to more basic viral needs such as viral genome replication and packaging, or viral egress and entry. Similar to adenain (Greber et al., 1996), UL36USP may for example exhibit digestive activity during the release of viral DNA from the capsid, explaining the prevention of this step in a temperature sensitive mutant of HSV-1 UL36 (Batterson et al., 1983).
Table 1.
Demonstrated and potential roles of DUBs in virus infection
| DUB | Roles | References |
|---|---|---|
| Demonstrated rolesa | ||
| Adenovirus protease adenain | Release of its own activating peptidic cofactor, maturation of capsid precursor proteins, promotion of cell lysis by cleavage of cytokeratin 18 and actin, support of viral uncoating, and DNA release by capsid protein digestion at the nuclear pore | Mangel et al. (2003) |
| SARS-CoV PLpro | Processing of nsp1 to 3 from the viral replicase polyprotein | Harcourt et al. (2004) |
| Potential rolesb | ||
| USP9X | Stabilization of β-catenin in EBV-infected B cells | Shackelford et al. (2003) |
| USP7 | Impaired stabilization of p53 in the presence of EBNA1 of EBV leading to apoptosis avoidance Stabilization of ICP0 of HSV-1 supporting reactivation of quiescent viral genomes and lytic infection | Hu et al., 2006, Saridakis et al., 2005, Sheng et al., 2006, Boutell et al., 2005, Canning et al., 2004 |
| Adenovirus protease adenain | Stabilization of virion proteins, negative regulation of the IFN responsec | Balakirev et al. (2002) |
| SARS-CoV PLpro | Stabilization of viral polyprotein sequences, negative regulation of the IFN responsec | Lindner et al. (2005); Barretto et al. (2005) |
| Herpesvirus UL36USP | Stabilization of viral proteins | Kattenhorn et al. (2005) |
Among the viral enzymes with deubiquitinating activity, proteolytic roles during virus infection have only been demonstrated for the adenovirus protease adenain and SARS-CoV PLpro. In both cases, however, they involve hydrolysis of polypeptides at regular peptide bonds by these enzymes and not isopeptide bond cleavage.
DUBs have been proposed to benefit viral infection by stabilizing viral gene products or, selectively, cellular proteins by protecting them from proteasomal degradation.
The delSGylating activities of adenain and SARS-CoV PLpro may mimic USP18, a demonstrated negative regulator of the interferon response (Dao and Zhang, 2005). Yet, an isopeptidase independent mechanism for this function of USP18 was discovered recently (Malakhova et al., 2006), and the significance of its de-ISGylating activity remains unclear.
The example of UL36USP of HCMV has shown that infection of cell cultures with mutant viruses does not necessarily cause a tangible phenotype. For adenain and SARS-CoV PLpro it may prove difficult to dissect their essential functions in polyprotein cleavage from the significance of their DUB activities. For SARS-CoV, the actual importance of the nsp1/2, nsp2/3, and nsp3/4 cleavages carried out by the PLpro is still not clear. Nevertheless, in cell culture infection nsp1 appears to suppress host gene expression by promoting host mRNA degradation (Kamitani et al., 2006), and nsp2, although dispensable, was shown to be required for optimal virus replication (Graham et al., 2005). In order to, however, more comprehensively study potential roles of cellular and viral DUBs in virus pathogenicity, e.g., through the modulation of the host antiviral immune responses, animal models of viral infection are needed (cf. Arrode and Davrinche, 2003, Cantin et al., 1999, Cinatl et al., 2005, Jogler et al., 2006).
The acquisition of Ub and Ubl sequences by strains of BVDV as polyprotein processing signals is intriguing and suggests a possible scenario for the evolutionary origin of viral DUBs. In bovine cells, the essential processing of the Atg8 insert in the BVDV isolate JaCP was demonstrated to be carried out by the DUB ATG4B. This processing step also occurred when an Atg8 containing BVDV polyprotein sequence was recombinantly expressed in avian, fish, and insect cells, as well as in a rabbit reticulocyte lysate (Fricke et al., 2004). The utilization of a Ub or Ubl sequence as polyprotein processing signal may present an advantageous viral strategy because a newly infected host cell is very likely to already express a DUB that can perform the processing reaction. It is tempting to speculate that Ub or Ubl sequence containing viruses originally have happened to insert sequences of cellular DUBs into their genomes as functional protein processing enzymes, thereby becoming independent of the cellular enzymes. As a fitness advantage, this could have enhanced the viral host cell spectrum.
Acknowledgments
The comments on the text by Traian Sulea and Robert Ménard are gratefully acknowledged. This is NRCC publication no. 47552.
References
- Amerik A.Y., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Antinone S.E., Shubeita G.T., Coller K.E., Lee J.I., Haverlock-Moyns S., Gross S.P., Smith G.A. The herpesvirus capsid surface protein, VP26, and the majority of the tegument proteins are dispensable for capsid transport toward the nucleus. J. Virol. 2006;80:5494–5498. doi: 10.1128/JVI.00026-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrode G., Davrinche C. Dendritic cells and HCMV cross-presentation. Curr. Top. Microbiol. Immunol. 2003;276:277–294. doi: 10.1007/978-3-662-06508-2_13. [DOI] [PubMed] [Google Scholar]
- Bailey D., O'Hare P. Comparison of the SUMO1 and ubiquitin conjugation pathways during the inhibition of proteasome activity with evidence of SUMO1 recycling. Biochem. J. 2005;392:271–281. doi: 10.1042/BJ20050873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakirev M.Y., Jaquinod M., Haas A.L., Chroboczek J. Deubiquitinating function of adenovirus proteinase. J. Virol. 2002;76:6323–6331. doi: 10.1128/JVI.76.12.6323-6331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks L., Pim D., Thomas M. Viruses and the 26S proteasome: hacking into destruction. Trends Biochem. Sci. 2003;28:452–459. doi: 10.1016/S0968-0004(03)00141-5. [DOI] [PubMed] [Google Scholar]
- Baroth M., Orlich M., Thiel H.J., Becher P. Insertion of cellular NEDD8 coding sequences in a pestivirus. Virology. 2000;278:456–466. doi: 10.1006/viro.2000.0644. [DOI] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry M., Früh K. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci. STKE. 2006:e21. doi: 10.1126/stke.3352006pe21. [DOI] [PubMed] [Google Scholar]
- Batterson W., Furlong D., Roizman B. Molecular genetics of herpes simplex virus: VIII. further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz P.D. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K.D., Ploegh H.L., Kessler B.M. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- Boutell C., Everett R.D. Herpes simplex virus type 1 infection induces the stabilization of p53 in a USP7- and ATM-independent manner. J. Virol. 2004;78:8068–8077. doi: 10.1128/JVI.78.15.8068-8077.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutell C., Canning M., Orr A., Everett R.D. Reciprocal activities between herpes simplex virus type 1 regulatory protein ICP0, a ubiquitin E3 ligase, and ubiquitin-specific protease USP7. J. Virol. 2005;79:12342–12354. doi: 10.1128/JVI.79.19.12342-12354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembeck F.H., Rosario M., Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of β-catenin. Curr. Opin. Genet. Dev. 2006;16:51–59. doi: 10.1016/j.gde.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Brooks C.L., Gu W. p53 ubiquitination: Mdm2 and beyond. Mol. Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.T., Mangel W.F. Interaction of actin and its 11-amino acid C-terminal peptide as cofactors with the adenovirus proteinase. FEBS Lett. 2004;563:213–218. doi: 10.1016/S0014-5793(04)00285-6. [DOI] [PubMed] [Google Scholar]
- Burnett B.G., Pittman R.N. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4330–4335. doi: 10.1073/pnas.0407252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canning M., Boutell C., Parkinson J., Everett R.D. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J. Biol. Chem. 2004;279:38160–38168. doi: 10.1074/jbc.M402885200. [DOI] [PubMed] [Google Scholar]
- Cantin E., Tanamachi B., Openshaw H. Role for gamma interferon in control of herpes simplex virus type 1 reactivation. J. Virol. 1999;73:3418–3423. doi: 10.1128/jvi.73.4.3418-3423.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee A.V., Lopez P., Pandolfi P.P., Roizman B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 2003;77:7101–7105. doi: 10.1128/JVI.77.12.7101-7105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon K.W., Baek K.H. HAUSP as a therapeutic target for hematopoietic tumors. Int. J. Oncol. 2006;28:1209–1215. [PubMed] [Google Scholar]
- Ching R.W., Dellaire G., Eskiw C.H., Bazett-Jones D.P. PML bodies: a meeting place for genomic loci? J. Cell Sci. 2005;118:847–854. doi: 10.1242/jcs.01700. [DOI] [PubMed] [Google Scholar]
- Chou J., Roizman B. Characterization of DNA sequence-common and sequence-specific proteins binding to cis-acting sites for cleavage of the terminal a sequence of the herpes simplex virus 1 genome. J. Virol. 1989;63:1059–1068. doi: 10.1128/jvi.63.3.1059-1068.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin proteolytic system: from a vague idea, through basic mechanisms, and onto human diseases and drug targeting. Neurology. 2006;66:S7–S19. doi: 10.1212/01.wnl.0000192261.02023.b8. [DOI] [PubMed] [Google Scholar]
- Cinatl J., Michaelis M., Hoever G., Preiser W., Doerr H.W. Development of antiviral therapy for severe acute respiratory syndrome. Antiviral Res. 2005;66:81–97. doi: 10.1016/j.antiviral.2005.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Weber J.M. The adenovirus protease is required for virus entry into host cells. Virology. 1995;213:494–502. doi: 10.1006/viro.1995.0022. [DOI] [PubMed] [Google Scholar]
- Cummins J.M., Rago C., Kohli M., Kinzler K.W., Lengauer C., Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1. doi: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- Dao C.T., Zhang D.E. ISG15: a ubiquitin-like enigma. Front. Biosci. 2005;10:2701–2722. doi: 10.2741/1730. [DOI] [PubMed] [Google Scholar]
- d'Azzo A., Bongiovanni A., Nastasi T. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic. 2005;6:429–441. doi: 10.1111/j.1600-0854.2005.00294.x. [DOI] [PubMed] [Google Scholar]
- Elsasser S., Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Evans P.C. Regulation of pro-inflammatory signalling networks by ubiquitin: identification of novel targets for anti-inflammatory drugs. Expert Rev. Mol. Med. 2005;7:1–19. doi: 10.1017/S1462399405009415. [DOI] [PubMed] [Google Scholar]
- Everett R.D. Interactions between DNA viruses, ND10 and the DNA damage response. Cell. Microbiol. 2006;8:365–374. doi: 10.1111/j.1462-5822.2005.00677.x. [DOI] [PubMed] [Google Scholar]
- Everett R.D., Meredith M., Orr A., Cross A., Kathoria M., Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R.D., Rechter S., Papior P., Tavalai N., Stamminger T., Orr A. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 2006;80:7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier L. Viral plasmids in mammalian cells. In: Funnell B.E., Phillips G.J., editors. Plasmid Biology. ASM Press; Washington, DC: 2004. pp. 325–339. [Google Scholar]
- Fricke J., Voss C., Thumm M., Meyers G. Processing of a pestivirus protein by a cellular protease specific for light chain 3 of microtubule-associated proteins. J. Virol. 2004;78:5900–5912. doi: 10.1128/JVI.78.11.5900-5912.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Luo H. The ubiquitin–proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 2006;84:5–14. doi: 10.1139/y05-144. [DOI] [PubMed] [Google Scholar]
- Gomez-Lazaro M., Fernandez-Gomez F.J., Jordan J. p53: twenty five years understanding the mechanism of genome protection. J. Physiol. Biochem. 2004;60:287–307. doi: 10.1007/BF03167075. [DOI] [PubMed] [Google Scholar]
- Graham R.L., Sims A.C., Brockway S.M., Baric R.S., Denison M.R. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 2005;79:13399–13411. doi: 10.1128/JVI.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzow H., Klupp B.G., Mettenleiter T.C. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 2005;79:3200–3205. doi: 10.1128/JVI.79.5.3200-3205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber U.F., Webster P., Weber J., Helenius A. The role of the adenovirus protease on virus entry into cells. EMBO J. 1996;15:1766–1777. [PMC free article] [PubMed] [Google Scholar]
- Grummt I. Actin and myosin as transcription factors. Curr. Opin. Genet. Dev. 2006;16:191–196. doi: 10.1016/j.gde.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Guarino L.A. Identification of a viral gene encoding a ubiquitin-like protein. Proc. Natl. Acad. Sci. U.S.A. 1990;87:409–413. doi: 10.1073/pnas.87.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A.L., Katzung D.J., Reback P.M., Guarino L.A. Functional characterization of the ubiquitin variant encoded by the baculovirus Autographa californica. Biochemistry. 1996;35:5385–5394. doi: 10.1021/bi9524981. [DOI] [PubMed] [Google Scholar]
- Hagglund R., Roizman B. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 2004;78:2169–2178. doi: 10.1128/JVI.78.5.2169-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K., Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay R.T. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hayward S.D., Liu J., Fujimuro M. Notch and Wnt signaling: mimicry and manipulation by gamma herpesviruses. Sci. STKE. 2006:re4. doi: 10.1126/stke.3352006re4. [DOI] [PubMed] [Google Scholar]
- Herold J., Siddell S.G., Gorbalenya A.E. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold. J. Biol. Chem. 1999;274:14918–14925. doi: 10.1074/jbc.274.21.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowaty M.N., Frappier L. HAUSP/USP7 as an Epstein–Barr virus target. Biochem. Soc. Trans. 2004;32:731–732. doi: 10.1042/BST0320731. [DOI] [PubMed] [Google Scholar]
- Holowaty M.N., Sheng Y., Nguyen T., Arrowsmith C., Frappier L. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J. Biol. Chem. 2003;278:47753–47761. doi: 10.1074/jbc.M307200200. [DOI] [PubMed] [Google Scholar]
- Hu M., Li P., Sing L., Jeffrey P., Chenova T., Wilkinson K.D., Cohen R.E., Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Gu L., Li M., Jeffrey P.D., Gu W., Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 Pathway. PLoS Biol. 2006;4:e27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K.L., Shackelford J., Seo S.Y., Pagano J.S. Up-regulation of β-catenin by a viral oncogene correlates with inhibition of the seven in absentia homolog 1 in B lymphoma cells. Proc. Natl. Acad. Sci. U. S. A. 2006;102:18431–18436. doi: 10.1073/pnas.0504054102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogler C., Hoffmann D., Theegarten D., Grunwald T., Uberla K., Wildner O. Replication properties of human adenovirus in vivo and in cultures of primary cells from different animal species. J. Virol. 2006;80:3549–3558. doi: 10.1128/JVI.80.7.3549-3558.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattenhorn L.M., Korbel G.A., Kessler B.M., Spooner E., Ploegh H.L. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that Is conserved across the family Herpesviridae. Mol. Cell. 2005;19:547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Kerscher O., Felberbaum R., Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Kiel C., Serrano L. The ubiquitin domain superfold: structure-based sequence alignments and characterization of binding epitopes. J. Mol. Biol. 2006;355:821–844. doi: 10.1016/j.jmb.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Kim J.C., Spence R.A., Currier P.F., Lu X., Denison M.R. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology. 1995;208:1–8. doi: 10.1006/viro.1995.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klupp B.G., Fuchs W., Granzow H., Nixdorf R., Mettenleiter T.C. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 2002;76:3065–3071. doi: 10.1128/JVI.76.6.3065-3071.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuzinger H., Ziegler U., Schraner E.M., Fraefel C., Glauser D.L., Heid I., Ackermann M., Mueller M., Wild P. Herpes simplex virus 1 envelopment follows two diverse pathways. J. Virol. 2005;79:13047–13059. doi: 10.1128/JVI.79.20.13047-13059.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Brooks C.L., Kon N., Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Menard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Stevens J., Rote C.A., Yost H.J., Hu Y., Neufeld K.L., White R.L., Matsunami N. Siah-1 mediates a novel β-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol. Cell. 2001;7:927–936. doi: 10.1016/s1097-2765(01)00241-6. [DOI] [PubMed] [Google Scholar]
- Luxton G.W.G., Haverlock S., Coller K.E., Antinone S.E., Pincetic A., Smith G.A. From the Cover: targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5832–5837. doi: 10.1073/pnas.0500803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakhova O.A., Kim K.I., Luo J.K., Zou W., Kumar K.G., Fuchs S.Y., Shuai K., Zhang D.E. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangel W.F., Baniecki M.L., McGrath W.J. Specific interactions of the adenovirus proteinase with the viral DNA, an 11-amino-acid viral peptide, and the cellular protein actin. Cell. Mol. Life Sci. 2003;60:2347–2355. doi: 10.1007/s00018-003-2318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb D.S., Courtney R.J. Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J. Virol. 1992;66:7581–7584. doi: 10.1128/jvi.66.12.7581-7584.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter T.C. Budding events in herpesvirus morphogenesis. Virus Res. 2004;106:167–180. doi: 10.1016/j.virusres.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Mettenleiter T.C. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 2006;113:163–169. doi: 10.1016/j.vetmic.2005.11.040. [DOI] [PubMed] [Google Scholar]
- Mettenleiter T.C., Minson T. Egress of alphaherpesviruses. J. Virol. 2006;80:1610–1611. doi: 10.1128/JVI.80.3.1610-1612.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter T.C., Klupp B.G., Granzow H. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 2006;9:423–429. doi: 10.1016/j.mib.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Meyers G., Stoll D., Gunn M. Insertion of a sequence encoding light chain 3 of microtubule-associated proteins 1A and 1B in a pestivirus genome: connection with virus cytopathogenicity and induction of lethal disease in cattle. J. Virol. 1998;72:4139–4148. doi: 10.1128/jvi.72.5.4139-4148.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F., Visa N. Actin in transcription and transcription regulation. Curr. Opin. Cell Biol. 2006;18:261–266. doi: 10.1016/j.ceb.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Morrison J.A., Gulley M.L., Pathmanathan R., Raab-Traub N. Differential signaling pathways are activated in the Epstein–Barr virus-associated malignancies nasopharyngeal carcinoma and Hodgkin lymphoma. Cancer Res. 2004;64:5251–5260. doi: 10.1158/0008-5472.CAN-04-0538. [DOI] [PubMed] [Google Scholar]
- Nijman S.M.B., Luna-Vargas M.P.A., Velds A., Brummelkamp T.R., Dirac A.M.G., Sixma T.K., Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Ovaa H., Kessler B.M., Rolen U., Galardy P.J., Ploegh H.L., Masucci M.G. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2253–2258. doi: 10.1073/pnas.0308411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore L.A., Barford D. Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem. J. 2004;379:513–525. doi: 10.1042/BJ20040198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Ries D., Bolger E., Chang L.J., Stoltzfus C.M. MHV nucleocapsid synthesis in the presence of cycloheximide and accumulation of negative strand MHV RNA. Virus Res. 1986;6:261–272. doi: 10.1016/0168-1702(86)90074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz L.E., Reynolds A.E., Hengartner C.J. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 2005;69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Ridpath J.F., Berry E.S. Insertion of a bovine SMT3B gene in NS4B and duplication of NS3 in a bovine viral diarrhea virus genome correlate with the cytopathogenicity of the virus. Virus Res. 1998;57:1–9. doi: 10.1016/s0168-1702(98)00073-2. [DOI] [PubMed] [Google Scholar]
- Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N.D., Tolle D.P., Barrett A.J. MEROPS: the peptidase database. Nucleic Acids Res. 2004;32:D160–D164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly L.M., Guarino L.A. The viral ubiquitin gene of Autographa californica nuclear polyhedrosis virus is not essential for viral replication. Virology. 1996;218:243–247. doi: 10.1006/viro.1996.0185. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Horton J.R., Mullally J.E., Heroux A., Cheng X., Wilkinson K.D. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Rolén U., Kobzeva V., Gasparjan N., Ovaa H., Winberg G., Kisseljov F., Masucci M.G. Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines. Mol. Carcinog. 2006;45:260–269. doi: 10.1002/mc.20177. [DOI] [PubMed] [Google Scholar]
- Ros C., Kempf C. The ubiquitin-proteasome machinery is essential for nuclear translocation of incoming minute virus of mice. Virology. 2004;324:350–360. doi: 10.1016/j.virol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K., Velasquez J., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Structural basis of severe acute respiratory syndrome coronavirus ADP-Ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saridakis V., Sheng Y., Sarkari F., Holowaty M.N., Shire K., Nguyen T., Zhang R.G., Liao J., Lee W., Edwards A.M. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein–Barr nuclear antigen 1: implications for EBV-mediated immortalization. Mol. Cell. 2005;18:25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J. Virol. 1986;57:328–334. doi: 10.1128/jvi.57.1.328-334.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C., Korbel G.A., Kattenhorn L.M., Ploegh H.L. A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae. J. Virol. 2005;79:15582–15585. doi: 10.1128/JVI.79.24.15582-15585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Hanna J., Elsasser S., Finley D. Proteasome-associated proteins: regulation of a proteolytic machine. Biol. Chem. 2005;386:725–737. doi: 10.1515/BC.2005.085. [DOI] [PubMed] [Google Scholar]
- Shackelford J., Pagano J.S. Targeting of host–cell ubiquitin pathways by viruses. Essays Biochem. 2005;41:139–156. doi: 10.1042/EB0410139. [DOI] [PubMed] [Google Scholar]
- Shackelford J., Maier C., Pagano J.S. Epstein–Barr virus activates {beta}-catenin in type III latently infected B lymphocyte lines: association with deubiquitinating enzymes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15572–15576. doi: 10.1073/pnas.2636947100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y., Saridakis V., Sarkari F., Duan S., Wu T., Arrowsmith C.H., Frappier L. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat. Struct. Mol. Biol. 2006;13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J.M., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Städeli R., Hoffmans R., Basler K. Transcription under the control of nuclear Arm/[beta]-catenin. Curr. Biol. 2006;16:R378–R385. doi: 10.1016/j.cub.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Sulea T., Lindner H.A., Purisima E.O., Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 2005;79:4550–4551. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulea T., Lindner H.A., Menard R. Structural aspects of recently discovered viral deubiquitinating activities. Biol. Chem. 2006;387:853–862. doi: 10.1515/BC.2006.108. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Lallemand-Breitenbach V., Zhu J., de The H. PML nuclear bodies and apoptosis. Oncogene. 2004;23:2819–2824. doi: 10.1038/sj.onc.1207533. [DOI] [PubMed] [Google Scholar]
- Tang J., Qu L.K., Zhang J., Wang W., Michaelson J.S., Degenhardt Y.Y., El-Deiry W.S., Yang X. Critical role for Daxx in regulating Mdm2. Nat. Cell Biol. 2006;8:855–862. doi: 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- Tanida I., Ueno T., Kominami E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz N., Meyers G., Thiel H.J. Processing of poly-ubiquitin in the polyprotein of an RNA virus. Virology. 1993;197:74–85. doi: 10.1006/viro.1993.1568. [DOI] [PubMed] [Google Scholar]
- Taya S., Yamamoto T., Kanai-Azuma M., Wood S.A., Kaibuchi K. The deubiquitinating enzyme Fam interacts with and stabilizes beta-catenin. Genes Cells. 1999;4:757–767. doi: 10.1046/j.1365-2443.1999.00297.x. [DOI] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Vittone V., Diefenbach E., Triffett D., Douglas M.W., Cunningham A.L., Diefenbach R.J. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 2005;79:9566–9571. doi: 10.1128/JVI.79.15.9566-9571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Sugden B. Origins of bidirectional replication of Epstein–Barr virus: models for understanding mammalian origins of DNA synthesis. J. Cell. Biochem. 2005;94:247–256. doi: 10.1002/jcb.20324. [DOI] [PubMed] [Google Scholar]
- Wang J., Loveland A.N., Kattenhorn L.M., Ploegh H.L., Gibson W. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 2006;80:6003–6012. doi: 10.1128/JVI.00401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchman R.L., Gordon C., Mayer R.J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev., Mol. Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Wild P., Engels M., Senn C., Tobler K., Ziegler U., Schraner E.M., Loepfe E., Ackermann M., Mueller M., Walther P. Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J. Virol. 2005;79:1071–1083. doi: 10.1128/JVI.79.2.1071-1083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wileman T. Aggresomes and autophagy generate sites for virus replication. Science. 2006;312:875–878. doi: 10.1126/science.1126766. [DOI] [PubMed] [Google Scholar]
- Yu G.Y., Lai M.M.C. The ubiquitin-proteasome system facilitates the transfer of murine coronavirus from endosome to cytoplasm during virus entry. J. Virol. 2005;79:644–648. doi: 10.1128/JVI.79.1.644-648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]