Abstract
Like vaccines, biologic proteins can be very immunogenic for reasons including route of administration, dose frequency and the underlying antigenicity of the therapeutic protein. Because the impact of immunogenicity can be quite severe, regulatory agencies are developing risk-based guidelines for immunogenicity screening. T cell epitopes are at the root of the immunogenicity issue. Through their presentation to T cells, they activate the process of anti-drug antibody development. Preclinical screening for T cell epitopes can be performed in silico, followed by in vitro and in vivo validation. Importantly, screening for immunogenicity is complicated by the discovery of regulatory T cell epitopes, which suggests that immunogenicity testing must now take regulatory T cells into consideration. In this review, we address the application of computational tools for preclinical immunogenicity assessment, the implication of the discovery of regulatory T cell epitopes, and experimental validation of those assessments.
Keywords: Antigen, MHC, Antibodies, Anti-drug antibody, Antigen presentation, Immunoinformatics, T lymphocyte antigens, T effector cells, T regulatory cells, Tolerance
1. Introduction
The biotechnology revolution has made great strides in recent years: drug developers are producing novel therapeutic proteins, including monoclonal antibodies (mAbs) and antibody-like protein scaffolds. However, in the push to deliver novel biologics to the market, developers have on occasion overlooked factors that may contribute to protein immunogenicity, with sometimes disastrous results. Depending on the therapeutic context, autologous or human-like proteins have proven to be surprisingly immunogenic, suggesting that assumptions about immune tolerance also require careful consideration in biologics design.
Fortunately, years of thorough study of the parameters influencing vaccine immunogenicity and efficacy now allow parallels to be drawn for protein therapeutics. Factors such as delivery route and vehicle, formulation, dosing, innate immune system activation, and the ability of the protein to interface with the humoral (B cell) and cellular (T cell) immune systems, may all influence the potential immunogenicity of proteins to some degree, whether these proteins are administered for therapeutic purposes (as in enzymes, monoclonals, replacement proteins) or as vaccines [1].
Both cellular and humoral immune responses may result from therapeutic protein administration, similar to the effect of vaccines. Anti-drug antibodies (ADA) may neutralize the therapeutic effects of a drug and/or alter its pharmacokinetics. T cells are certainly involved in this immune response when IgG class ADA are observed, because antibody isotype switching is a key function of T-dependent antigens [2]. Serious adverse events can be provoked if the ADA cross-react with a critical autologous protein. Two examples of undesirable ADA responses include autoimmune thrombocytopenia (ITP) following exposure to recombinant thrombopoietin [3], and pure red cell aplasia, which was associated with a particular formulation of erythropoietin (Eprex) [4]. Since the effects of immunogenicity can be quite severe, regulatory agencies are developing risk-based guidelines for immunogenicity screening [5].
T cell epitope content is one of the factors that contributes to antigenicity. The binding strength of T cell epitopes to major histocompatibility complex (MHC or HLA) molecules is a key determinant in T cell epitope immunogenicity. This allows the epitopes with higher binding affinities to be more likely to be displayed on the surface of the cell (in the context of MHC molecules) where they are recognized by their corresponding T cell receptor (TCR) [6]. Thus, the fate of a protein drug may be determined by its constituent 9-mers and their corresponding HLA binding affinities. Fortunately, after years of development and validation, T cell epitopes can now be predicted relatively accurately using in silico tools. As a result, immunoinformatics tools are now being applied to triage protein therapeutics into higher risk and lower risk categories prior to clinical development based on their T cell epitope content. Therefore, finding peptide sequences that may trigger or alternatively suppress a T cell response may be the most important step to be taken in the pre-clinical development of a protein drug.
One approach to measuring the potential immunogenicity of a protein is to parse protein sequences into overlapping 9-mer peptide frames, and to search for potential T cell epitopes by estimating the HLA binding potential of these short sequences, in silico, to eight common class II HLA alleles that “cover” the majority of human genetic backgrounds [7]. A critical step in this process is to use an algorithm that normalizes HLA allele-specific scores in order to be able to compare scores of any 9-mer across multiple HLA alleles [8]. Approaches that do not normalize across alleles are highly likely to over-predict for selected HLA, thereby skewing the analysis and reducing its accuracy. The potential immunogenicity of a protein can then be represented as a “sum of the scores” for the overlapping 9-mer peptide frames, and the potential for any single peptide sequence to be immunogenic can be estimated from its cumulative HLA binding score.
Applying this approach to both biologics (usually autologous proteins such as monoclonals) and foreign proteins, we have discovered that the in silico signature, or binding score, of a T cell epitope tends to be within one to two deviations from the mean of a set of random protein sequences. This allows relatively accurate prediction of potential T cell epitopes. However, knowledge of potential HLA binding does not translate into knowledge of the type of T cell response that may ensue. Potentially, effector (inflammatory) or regulatory (suppressive) T cell immunity may be activated. This complicates the process of screening for immunogenicity. The presence of T cell epitopes may contribute to immunogenicity, but is not in itself sufficient. The controlling factor appears to be the “context” in which a T cell epitope is recognized by a T cell. If signals evoke release of inflammatory cytokines, resulting in expression of co-stimulatory factors by antigen-presenting cells, an effector immune response is triggered. In the absence of such signals, and the presence of others (such as an abundance of IL-2, or other regulatory cytokines such as TGFβ), a T cell may be deviated towards a regulatory or tolerogenic response. Thus, for protein biologics, a T cell epitope may be friend (regulatory) or foe (effector) depending on the context of the response (Fig. 1 ). This is an important observation, as it turns out. Regulatory T cell epitopes may be a key feature of biologic drugs, but their existence and their role in the immune response to biologics is not yet well understood.
Fig. 1.
The same T cell epitope may trigger an effector immune response, characterized by inflammatory cytokines, up regulation of B cell activity, and development of anti-drug antibodies, or a regulatory T cell response characterized by suppressor cytokines, decreased cellular proliferation, and decreased antibody secretion. The “context” of the immune response is the most important factor that determines the T cell phenotype.
1.1. Immunogenicity factors: foreignness, T cell epitope content, “danger signal”
Tolerance to self-proteins is an integral aspect of the immune system, without which immune response would be severely hampered or dangerous to “self”. Despite the importance of tolerance to the regulation of auto-reactive immune responses, it is quite clear that tolerance can be overcome, and that T cell epitopes contribute to the induction of immune response to autologous proteins. There are many parallels between the development of autoimmune disease and immunogenicity of therapeutic proteins, including the reduction of regulatory T cell (Treg) immune responses and induction of T effector responses [9]. Deviation of the immune response away from tolerance towards immunogenicity probably involves B cells, effector T cells, and danger signals such as toll-like receptor ligands.
Since tolerance is considered the normal state of immune response to autologous proteins, the development of ADA to recombinant autologous proteins can be regarded as a “breach” of tolerance. Auto-reactive B cells are found in normal individuals, but such cells are rarely stimulated to produce antibodies by circulating levels of native proteins. The presence of low levels of circulating antibodies against autologous proteins is a well-known phenomenon in drug development, but these auto-antibodies rarely interfere with the drug. In contrast, the presence of cell responses to autologous proteins are a critical determinant of autoimmune disease; they also contribute to the development of higher levels of anti-self antibodies by providing cytokine “help” to B cells [10].
How does immune response to autologous proteins occur? In theory, T and B cell receptors to “self” are eliminated in the course of immune maturation in the thymus (T cells) and the bone marrow (B cells). However, there is evidence that auto-reactive T cells are present in the peripheral circulation. Thymic deletion of self-reactive T cells is imperfect. 25–40% of auto-reactive T cells escape into the periphery because of low avidity [11], [12], absence of thymic expression, inefficient expression or inefficient processing of T cell epitopes [13]. This has been documented in diabetes [14], rheumatoid arthritis [15] and multiple sclerosis [16]. Tolerance to self can be overcome if a danger signal [17] is present, or if cross-reactivity between self epitopes and epitopes of pathogens triggers an effector immune response against self (e.g. human coronavirus [18] and lyme disease antigen [19]). The importance of danger signals to the development of T effector responses to autologous proteins is worth bearing in mind, since many protein therapeutics are produced using methods that may incorporate such signals at levels that are below the usual limits of detection. Another factor that may play a role in the induction of autoimmunity is known as “heterologous immunity” or T cell response to epitopes that cross-react with self [20]. Unfortunately, the role of heterologous immunity in the development of ADA to protein therapeutics remains unexplored at present, even though it is likely to be a contributing factor.
In the context of B cell response, the development of antibodies (including auto-antibodies) can be T-independent or T-dependent. Although T cell independent (Ti) antibody development is often noted as a source of antibodies to protein therapeutics, Ti activation of B cells generally does not lead to antibody affinity maturation or to B cell memory. More commonly, T cell dependent (Td) activation of B cells is triggered, leading to a robust antibody response, affinity maturation, isotype switching, and the development of B cell memory. The induction of IgG class ADA, whether to chimeric antibodies, human antibodies, recombinant proteins, or proteins of non-human origin, normally implies that some of the sequences in the protein triggered a T cell response that led to a Td B cell response, and thus, isotype switching [21].
The source of a danger signal may be quite different for vaccines and biologic proteins. Vaccine adjuvants contain innate immune system triggers such as Complete Freund's Adjuvant and CpG oligodeoxynucleotides. These adjuvants are examples of toll-like receptor (TLR) ligands. TLRs are pattern recognition receptors expressed by antigen-presenting cells that recognize structurally conserved molecular patterns in microbes such as LPS, bacterial DNA and certain bacterial polysaccharides [22]. Protein therapeutics, as a rule, are highly purified proteins that do not contain danger signals, but the process of manufacturing may lead to introduction of TLR ligands, or, alternatively, some components of the formulation may degrade in the course of drug processing and formulation, leading to the formation of a danger signal (see, for example, the discussion on Eprex below).
In summary, three critical factors contribute to immune response to protein therapeutics. First, B cells must recognize protein antigens in their native form using the B cell receptor (membrane bound immunoglobulin). Second, B cells or other antigen-presenting cells (APC) must process and present T cell epitopes derived from the protein therapeutic, triggering a T cell response. Third, a danger signal is required to create the proper “context” for triggering a T cell response. Regardless of the initial trigger, immune response to protein drugs, whether antibodies or recombinant human proteins such as FVIII, erythropoietin, and myozyme, is due to the combined activities of T and B cells that recognize antigen and the presence of a danger signal “trigger”.
1.2. Types of T cell responses (effector and regulatory)
While T cells begin on a path to maturation in the thymus, their fate is only reached in the periphery. That process begins when naïve T cells engage epitope-loaded MHC at the surface of antigen-presenting cells through the T cell receptor. A series of molecular events ensues, resulting in differential expression of functional and phenotypic markers that make it possible to experimentally distinguish one T cell type from another [23]. Stimulatory and inhibitory responses are mediated by effector and regulatory T cells, respectively. In general, effector T cells produce an inflammatory response to clear infections, whereas regulatory T cells generate a suppressive response that promotes immunological tolerance.
1.2.1. T helper effector cells
Effector T cells fall into two broad classes: T helper cells (Th) and cytotoxic T lymphocytes (CTLs). Phenotypically, these populations are distinguished by cell surface proteins; Th cells express CD4 and CTLs CD8. Th cells orchestrate events critical to both the humoral and cellular arms of the adaptive immune system. They prime naïve CTL and B cell responses, generate secondary responses and contribute to the induction and sustainability of immune memory [24]. T helper (CD4+) T cells are critically important instigators of ADA response. The T cell receptor (TCR) assures immune response specificity through interaction with epitope-loaded MHC on antigen-presenting cells (APCs). TCRs are as pleiomorphic as B cell receptors (immunoglobulin). In the case of CD4+ T cells, the TCR recognizes MHC class II molecules that are expressed by dendritic cells, macrophages or B cells.
Anti-drug immune responses commence with ingestion of the protein drug by APCs such as dendritic cells or B cells. Foreign antigens are taken up by APCs into endocytic vesicles where proteases degrade proteins into peptides. Endosomes then fuse with MHC class II-containing vesicles (MIIC vesicles) enabling peptides to bind to MHC molecules. In a concentration-dependent manner, and aided by HLA-DM, peptides compete for binding in the MHC groove. Peptide–MHC complexes are transported to the cell surface for presentation to T cells.
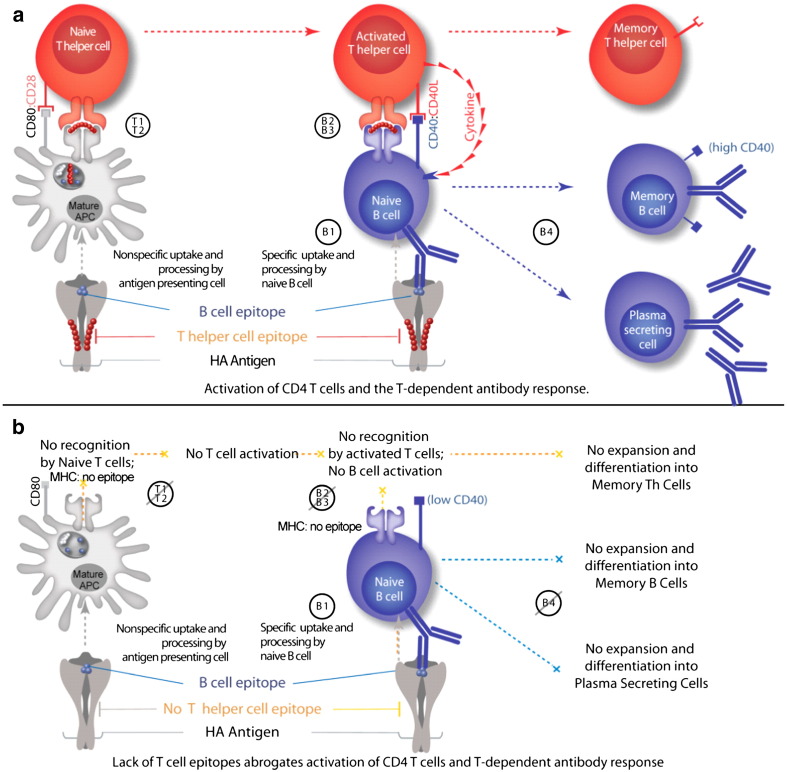
Naïve CD4+ T cells that have a TCR that binds to the MHC class II–epitope complex are activated with co-stimulatory interaction between CD80/CD86 and CD28 (signals T1 and T2 in Fig. 2a). TCR activation switches on downstream signaling pathways in T cells that lead to secretion of IL-2. This cytokine, in turn, stimulates further intracellular signaling by binding in an autocrine fashion to the IL-2 receptor, which leads to T cell proliferation [25]. Activated CD4+ Th cells secrete a wide range of cytokines, including interferon-gamma (IFN-γ), IL-4 and IL-17, the hallmark cytokines of the major Th lineages Th1, Th2 and Th17, respectively [26].
Fig. 2.
a and b. Stimulation of T helper (Th) cells by antigens involves first activation by interleukin 1 (IL-1) (T1) and then presentation of the antigen at the surface of antigen-presenting cells (usually macrophages, dendritic cells or B cells) (T2) in association with class II major histocompatibility complex molecules (MHC II); for extrinsic (foreign) proteins this requires initial capture of the protein, followed by denaturation and/or degradation so as to associate the molecule or fragments with MHC II. T cells stimulated in this way express receptors for interleukin 2 (IL-2), and secrete various molecules, including factors which stimulate B cells to divide and/or secrete Ig, interferon-gamma and IL-2. (B1) In turn, IL-2 causes proliferation of Th and cytotoxic/suppressor T cells. Encounter of the same antigen by a B cell results in processing and presentation of the epitope at the B cell surface in the context of MHC (B1). Co-stimulatory factors such as B7 interact with the memory B cell (B2), resulting in the secretion of cytokines by the T cell (B3). These cytokines allow the B cell to mature and become a dedicated IgG-producing (isotype switched) memory B cell (B4).
Although commitment to a particular T cell subset is not necessarily fixed and is currently the subject of intense investigation, the function of the classic Th1, Th2, Th17 and Treg subsets is well-described. Th1 cells activate macrophages and aid in the expansion of CTLs. Th2 cells aid in the adaptive humoral response by triggering B cells to produce antibodies (signals B2 and B3 in Fig. 2a). T cells that secrete IL-17, the third and newest major Th lineage, are pro-inflammatory and important in pathogen defense [27]. Interestingly, Th17 cells can be induced to become regulatory T cells, further underscoring the flexibility of CD4+ T cell subsets (and their T cell epitope-specific immune responses) to exhibit different phenotypes, depending on the context of the immune response [28]. T helper cells that secrete IL-2 play an important role in the development of B memory cells. In the absence of T help (and IL-2), B cells do not expand, affinity mature, or isotype switch [29] (Fig. 2a, b).
Dysfunctional or inappropriate responses elicited by T cells can give rise to a number of disease conditions. For example, inappropriate responses associated with Th2 cells give rise to allergic diseases, and inappropriate responses associated with Th1 or Th17 cells may result in autoimmune diseases [25]. Again, the T cell response is not fundamentally defined by the epitope; rather, the phenotype is determined by the environment in which the epitope is presented in to the T cell.
1.2.2. Regulatory T cells
Regulatory or “suppressor” T cells are key players in the complex set of interactions that regulate immune responses to ensure tolerance to self. There are two types of tolerance: central and peripheral. Central T cell tolerance occurs during thymic development, when T cells that have high affinity TCR for autologous epitopes are deleted. B cells that can recognize self-proteins may similarly be deleted in the bone marrow. In addition, peripheral tolerance mechanisms exist to control autoimmunity and prevent auto-reactivity after lymphocytes have exited the thymus or bone marrow. According to current thinking, auto-reactive T cells with moderate affinity may escape deletion and be converted to function as ‘natural’ regulatory T cells (nTreg) to suppress autoimmunity [30]. Tolerance in the periphery is characterized by plasticity of T cell lineage commitment, where mature T cells are converted to a Treg phenotype upon activation through their T cell receptor in the presence of IL-10 and TGF-β. The role of these ‘adaptive’ Treg (aTreg) cells may be to dampen effector immune responses either to control a vigorous pro-inflammatory immune reaction to infection or possibly to facilitate co-existence with beneficial symbiotic bacteria and viruses [31], [32]. Tregs exhibit a CD4+CD25+FoxP3+ phenotype; these proteins are important to maintaining tolerance as attenuated CD25 and FoxP3 expression is associated with autoimmune disease [33], [34].
The major function of regulatory T cells is to regulate T cell-mediated immunity and suppress auto-immunogenic T cells that were not destroyed during their development in the thymus. This is extremely important to help prevent autoimmune diseases. Tregs are promoted by the transforming growth factor-beta (TGF-β) by leading to the induction of FoxP3 expression, which in turn renders the T cell suppressive function [35]. Tregs secrete specific cytokines that have a suppressing effect on effector response. IL-2 is a cytokine that is important for T cell proliferation. It contributes to signal transduction in Tregs and the maintenance of homeostasis and competitive fitness of natural Treg cells [35]. Stimulating CD4+ cells with antigen in vitro in the presence of TGF-β has led to the induction of FoxP3 expression. However, following re-stimulation with antigen in the absence of exogenous TGF-β, most cells seem to lose the FoxP3 expression [35].
IL-35 is a cytokine produced only by regulatory T cells, which plays a role in maximizing suppressive activity of immune cells [36]. Conversely, IL-4 and IFN-γ have been shown to play repressive roles in FoxP3 transcriptional activity [35], which drives T cell fate determination away from the Treg lineage.
Studies in animal models of autoimmunity have also shown an increased frequency or severity of autoimmunity in the absence of Tregs [37], [38] and that transfer of Treg cells is sufficient to protect from or reverse autoimmunity. Also, antigen-specific inducible T regulatory cells that express FoxP3 stably may generate transplantation tolerance, preventing or aiding autoimmunity, allergy and inflammation [32].
1.3. Definition of T cell epitopes
While B cells and antibodies generally recognize conformational epitopes from surface proteins, T cells recognize linear epitopes derived from proteins that are processed by APCs. As described above, antigenic (and autologous) proteins are broken down by proteolytic enzymes in the APC. During this process very large numbers of peptide fragments are produced. Any one of these fragments could be a T cell epitope, but only about 2% of all the fragments generated have the right amino acid side chains that allow them to bind in the MHC binding groove and be presented on the surface of the APC. One of the critical determinants of immunogenicity is the strength of T cell epitope binding to MHC molecules [39]. Peptides binding with higher affinity to MHC are more likely to be displayed on the cell surface where they can be recognized by TCRs. Both T effectors and Treg have TCR that bind to MHC–peptide complexes; as far as can be determined, it is the nature of the T cell (Treg or T effector) that determines the overall effect of the T cell responses; the T cell epitope sequence and its affinity for MHC does not appear to differ between T cell types.
1.4. Immunoinformatic identification of T cell epitopes
Over the last decade, researchers have developed a number of computer algorithms that map the locations of MHC class I- and class II-restricted T cell epitopes within proteins of various origins. Some in silico methods include frequency analysis, support-vector machines, hidden Markov models, and neural networks. For recent reviews of T cell epitope-mapping tools, see Sette [40], Brusic [41] and De Groot [42] among others. What these tools have in common is an ability to quickly screen large volumes of protein sequences for putative T cell epitopes. This preliminary screen reduces the search space dramatically, typically by at least 20-fold. MHC class II prediction methods are useful for the evaluation of Td immunogenicity, as MHC class II-restricted T cells provide help to B cells, leading to the development of ADA.
Such in silico predictions of T helper epitopes have already been successfully applied to the design of vaccines [43], [44] and to the selection of epitopes in studies of autoimmunity [45]. The authors use the EpiMatrix system, which has been validated following more than a decade of use for in vitro and in vivo studies (e.g. [46], [47], [48], [49], [50]). EpiMatrix employs HLA class I and class II “pocket profiles” that describe HLA pocket binding coefficients, and applies these coefficients to the prediction of overlapping 9- and 10-mer peptide epitopes. Each frame-by-allele evaluation is considered a single “assessment.” All EpiMatrix assessment scores (Z-scores) equal to or above 1.64 are defined as “Hits”; that is to say potentially immunogenic and worthy of further consideration. A sample analysis is provided in Fig. 3 below.
Fig. 3.
The influenza HA peptide 306–318 is an epitope known to be promiscuously immunogenic. It has high Z-scores for all 8 alleles in EpiMatrix. The ranking of the EpiMatrix scores (by color) is shown in the Z-score legend. Peptides that have predicted Z-scores above 1.64 (approximately the top 5% of all 9-mers derived from any given protein) have a significant chance of binding to MHC molecules and scores above 2.32 (approximately the top 1%) are highly likely to bind to MHC molecules. A cluster score is calculated by summing the EpiMatrix scores for each of the eight class II alleles. Cluster scores higher than 10 are considered to be significant. The influenza HA peptide shown here has a cluster score of 18. The band-like pattern (EpiBar) showing “hits” for potential binding for all eight class II alleles is characteristic of promiscuous epitopes.
The EpiMatrix report provided in Fig. 3 illustrates an important concept about T cell epitopes — those that are the most immunogenic tend to have more than one MHC or HLA binding motif. Another way to say this is that the best T cell epitopes tend to contain “clusters” of HLA binding motifs and clustering is highly correlated with immunogenicity. We have found that T cell epitope “clusters” range from 9 to roughly 25 amino acids in length and, considering their affinity to multiple alleles and across multiple frames, can contain anywhere from 4 to 40 binding motifs. A cluster score is calculated by summing the EpiMatrix scores for each of the eight class II alleles. Cluster scores higher than 10 are considered to be significant. Further, we have noticed that many of the most reactive T cell epitope clusters are concentrated in a single 9-mer frame, a feature we refer to as an EpiBar. As shown in Fig. 3, an EpiBar is a single 9-mer frame which is predicted to bind at least four different HLA alleles. Sequences that contain EpiBars include influenza hemagglutinin 307–319 (cluster score of 18), tetanus toxin 825–850 (cluster score of 16), and GAD65 557–567 (cluster score of 19). The significance of promiscuous binding is underscored by its association with immunodominance as illustrated for several proteins including tetanus toxin [51], influenza hemagglutinin [52], hepatitis B virus nucleocapsid antigen [53], hepatitis C virus nonstructural protein 3 [54] and HIV proteins gag and nef [55].
To consider the overall immunogenic potential of a whole protein, EpiMatrix Z-scores are summed for all “Hits” and adjusted for the length of the protein. By testing a large number of randomly generated protein sequences, an expected value of zero can be determined (the average score for a random protein sequence) and all other protein sequences can be compared to that standard (Fig. 4 ). Proteins scoring in the top quartile of the distribution (roughly speaking all proteins scoring above 20) contain an unusually high number of potential T cell epitopes. This method is described in a number of publications, most recently in De Groot et al. Clinical Immunology 2009 [56]. Importantly, the presence of one or more dominant T cell epitope clusters can enable significant immune responses to otherwise low-scoring proteins. Therefore, any evaluation of immunogenic potential should consider not only overall potential but also regional potentials.
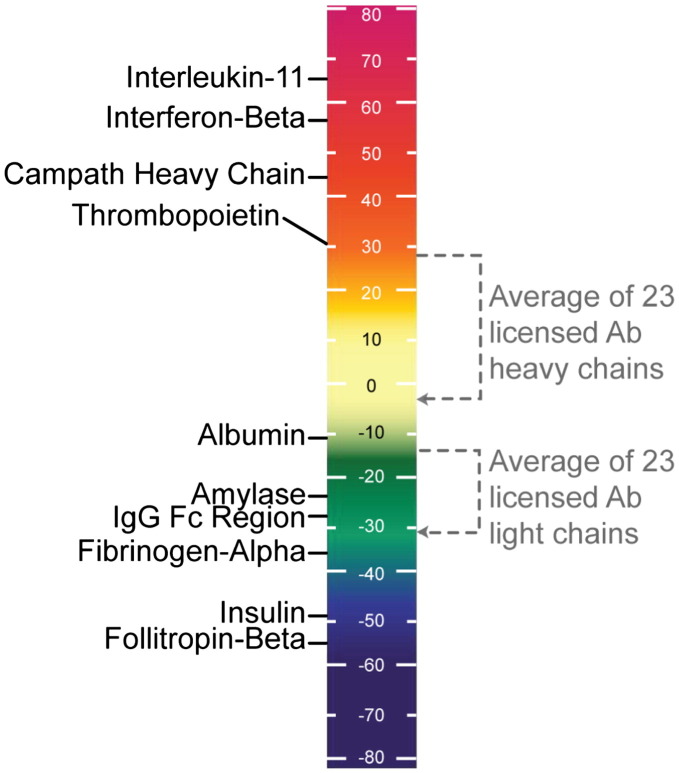
Fig. 4.
Summary of the immunogenicity “scale” findings for selected autologous proteins shows how antibody sequences rank, compared to standard controls. EpiMatrix protein immunogenicity scores higher than 20 are considered to be potentially immunogenic. Note that the low-scoring proteins on the lower left side of the scale are known to engender little to no immunogenicity while the higher scoring proteins on the upper left side of the scale are all known immunogens. For monoclonal antibodies, we adjust the antibody scores for the presence of pre-defined regulatory T cell epitopes [6] as we have evidence that the presence of these epitopes decreases the overall immunogenicity of antibodies in the clinic. Discovering these peptide sequences and identifying putative T cell epitopes may potentially be the most important aspect for protein therapeutic development.
1.4.1. T cell epitope prediction: a retrospective analysis
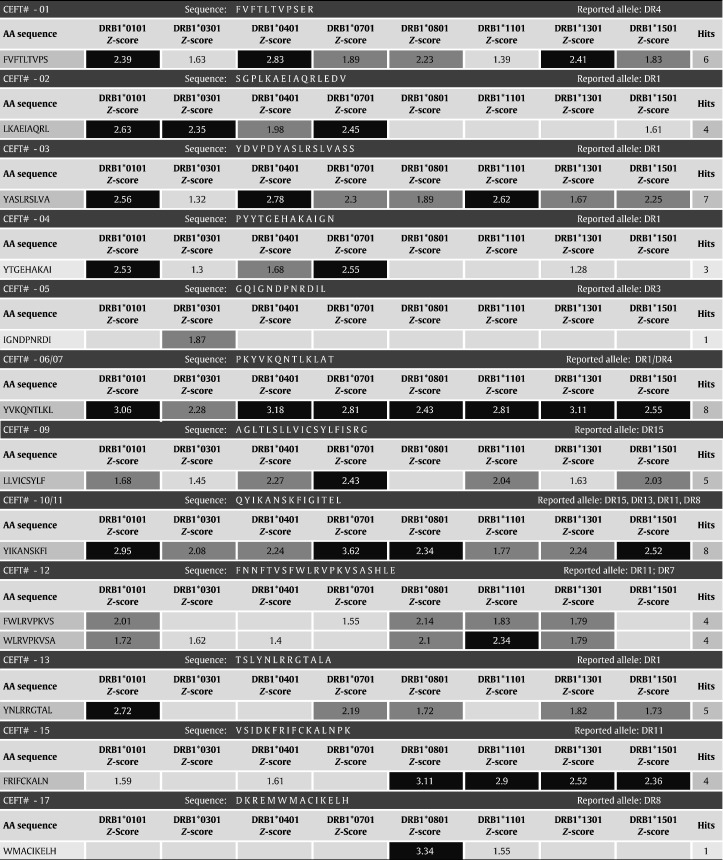
T cell epitope-mapping tools such as EpiMatrix [57] can be highly accurate predictors of immune response. So as to illustrate the predictive capabilities of EpiMatrix, we have recently analyzed a set of highly immunogenic peptide epitopes that are included in a commercial reagent (CEFT produced by PANATecs) commonly used as a positive control in immunological tests such as ELISPOT or cytokine flow cytometry. As shown in Table 1 , we analyzed all 23 epitopes in the CEFT reagent, which had previously been identified as promiscuous epitopes [58]. This peptide pool contains eight HLA Class II-restricted peptides from influenza virus, eight from Epstein-Barr virus, six from tetanus toxin and one from the human cytomegalovirus (Table 1). The CEFT reagent is immunogenic as measured by IFN-γ release (in ELISpot) or proliferation (in CFSE assays) [58]. It contains peptides restricted by the eight archetypal Class II alleles used for epitope prediction with EpiMatrix (DRB1⁎0101, DRB1⁎0301, DRB1⁎0401, DRB1⁎0701, DRB1⁎0801, DRB1⁎1101, DRB1⁎1301, and DRB1⁎1501).
Table 1.
EpiMatrix analysis of the CEFT peptide pool.
CEFT peptides receiving a high score (> 2.32) are indicated in black; peptides receiving a moderate score (1.64–2.31) are dark grey; and peptides scoring in the top 10% of scores are in light grey. Three (3) of the twenty-three (23) CEFT peptides included in the reagent (CEFT#18, 20, and 21) are not shown above because HLA do not belong to the “standard” set of eight archetypal alleles (DRB3⁎0201, DRB1⁎1001, and DQ2). CEFT# 19 was given a score in the top 10% for its specified allele of DRB1⁎0301, which would normally not be interpreted as a significant binder for DR3, yet an EpiBar is present within the sequence, illustrating its potential to be a strong candidate as a promiscuous epitope.
CEFT peptide sequences were evaluated using the EpiMatrix panel of Class II predictive matrices based on estimated probabilities of MHC binding. The resulting Z-scores again fall on a common scale that can be directly compared across HLA alleles (Table 1). Nearly all of these well-known T cell epitopes have clusters of immunogenic potential and EpiBars. The correspondence with EpiMatrix prediction is very high: seventeen (17) of the twenty peptides included in the CEFT reagent, or 85%, were correctly predicted to bind to each of their specified alleles, using EpiMatrix. The negative predictive value of EpiMatrix (NPV) was calculated to 94%.
As illustrated by the CEFT analysis, based on the “immunogenicity score” of a peptide sequence [42], it is possible to make an informed decision about the likelihood that the peptide sequence will provoke an immune response. As shown in the next section, we have used the EpiMatrix immunogenicity scale to prospectively predict the clinical immunogenicity of a novel “peptibody” and a bioengineered autologous protein [59], [60].
1.4.2. T cell epitope prediction: a prospective analysis
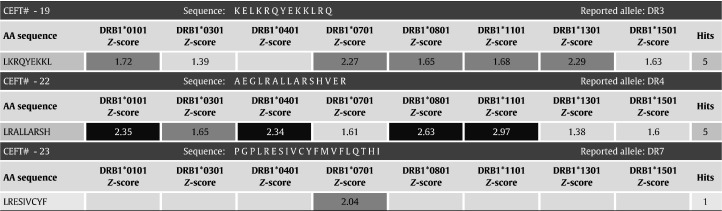
In silico screening along with ex vivo immunogenicity testing has been used to evaluate protein therapeutics in preclinical development. Two examples of correctly predicted clinical immunogenicity protein therapeutics include FPX and GDNF, which were subsequently published [3], [59]. FPX is a recombinant fusion protein consisting of two identical, biologically active peptides linked to human Fc fragment. EpiMatrix predicted a strong signal for immunogenicity within the 14-amino acid carboxy-terminal region of the peptide portion of FPX. With administration of FPX in 76 healthy human subjects, 37% developed antibodies after a single injection. A memory T cell response against the carboxy-terminus of the peptide was observed in antibody positive subjects but not in antibody negative subjects. The predicted promiscuity of the predicted T cell epitope(s) was confirmed by representation of all common HLA alleles in antibody positive subjects. HLA-haplotype DRB1⁎0701/1501 was predicted to be associated with the highest T cell and antibody response; this was confirmed in T cell and antibody assays. Tatarewicz and Moxness also used EpiMatrix to screen GDNF, a protein therapeutic that was subsequently proven to be immunogenic in clinical trials [46]. The protein contains epitope clusters that rank as high as well-known immunogenic epitopes on the EpiMatrix immunogenicity scale (Fig. 5 ) [46].
Fig. 5.
This figure shows the EpiMatrix immunogenicity scale to prospectively predict clinical immunogenicity. It compares novel protein sequences to proteins of known varying immunogenic potential. The new protein therapeutic GDNF was about to be used as treatment for Parkinson's disease or amyotrophic lateral sclerosis (ALS). In EpiMatrix analysis, however, it scores as high as published highly immunogenic peptides from influenza hemagglutinin and tetanus toxin. This might be the cause for the observed side effects during its clinical use as therapeutic.
Several other publications have also linked T cell epitopes, immune response and immunogenicity. For example, Barbosa et al. [61] confirmed the role of T cells in the immune response to Betaseron by linking ADA to HLA-DR type. In addition to the two published studies [60], [46], in silico assessments have been predictive of clinical immunogenicity in at least two unpublished studies (Vibha Jawa personal communication [62]).
2. T cell epitope contribution to immunogenicity to foreign antigens
2.1. Presence of T cell epitopes correlates with antibody response to foreign proteins
T help and cytotoxic T cell response are required to enhance the efficacy of vaccines against viruses. The importance of T cell help is nowhere more obvious than for influenza vaccines. Conventional influenza vaccines are designed to stimulate a protective antibody response to hemagglutinin (HA). Even though T cell help is required for high specific IgG antibody titers against the HA antigen [63], antibody titers are usually relatively weak and do not last very long. Induction of CD4+ T cell response to influenza Th cell epitopes improves antibody response against B cell epitopes and generates long-term memory [64], [65]. The live-attenuated influenza vaccine (LAIV) is much more immunogenic, suggesting that better-conserved T cell epitopes present in the internal flu proteins that are present in the LAIV [66] contribute to the immunogenicity of the vaccine. Therefore, the presence of T cell epitopes is critically important for the induction of B cell response, and particularly B cell memory, against foreign antigens.
2.2. Epitope density contributes to enhanced immunogenicity of foreign proteins
We have found that T cell epitopes containing a higher concentration of putative MHC binding motifs per amino acid tend to be more immunogenic. Presumably, this is due to the ability of these epitopes to be effectively processed by APCs and presented on a range of different MHC molecules (since more than one binding motif is present), thereby triggering a range of CD4+ T cell clones, leading to a synergistic effect. We have numerous examples from our vaccine research. For example, the EpiMatrix epitope-mapping algorithm was used to identify highly promiscuous T cell epitopes within the predicted secreted proteins from the F. tularensis tularensis genome as well as from known expressed proteins. Peptide epitopes were tested in ELISpot assays using blood from human subjects that had recovered from F. tularensis tularensis infection. ELISpot assays showed positive IFN-gamma responses to 21 of 25 individual Class II peptides and to peptide pools, in most human subjects [67]. Although differences in the HLA restriction of the molecules contributes to subject variability, these MHC binding motif-rich epitopes are generally highly immunogenic, leading to a 93% overall positive response rate in our vaccine programs.
2.3. Absence of T cell epitopes corresponds to lower immunogenicity of foreign proteins
The inability of a specific HLA molecule to present epitopes from a given vaccine antigen is well known to be a cause of vaccine failure. For example, Celis et al. reported that a significant number of HBsAg-reactive T cells from various HBV-immune individuals recognize a determinant localized near the amino terminus of HBsAg [68], [69] and individuals who cannot present the T cell epitopes in this region are unable to mount a protective immune response following vaccination [70], [71]. In this example, the lack of a T cell epitope to provide help to the B cells led to a lower antibody response.
2.4. Abrogation of T cell epitopes diminishes immunogenicity
Lack of T help and the consequent abrogation of B cell response contribute to the idea that deliberate removal of T cell epitopes might reduce immunogenicity [72]. One of the first deimmunized proteins was “Sakstar” or stapylokinase [73]. Modification or removal of the specific amino acids that contribute to HLA binding led to a reduction in the potential of epitope to stimulate T cell response. Similarly, a number of epitope-abrogation studies have been performed using FVIII. Jones et al. identified a 15-mer sequence in human FVIII that bound strongly to DRB1⁎0401, ⁎1101, and ⁎1501, moderately to ⁎0701, weakly to ⁎0101, and not at all to ⁎0301 and ⁎1301 in HLA Class II binding assays. They then modified the sequence of this epitope to reduce its potential to bind to HLA. The modified peptide did not bind to any allele and was less immunogenic in vitro [74], [75].
3. The regulatory T cell epitope wrinkle
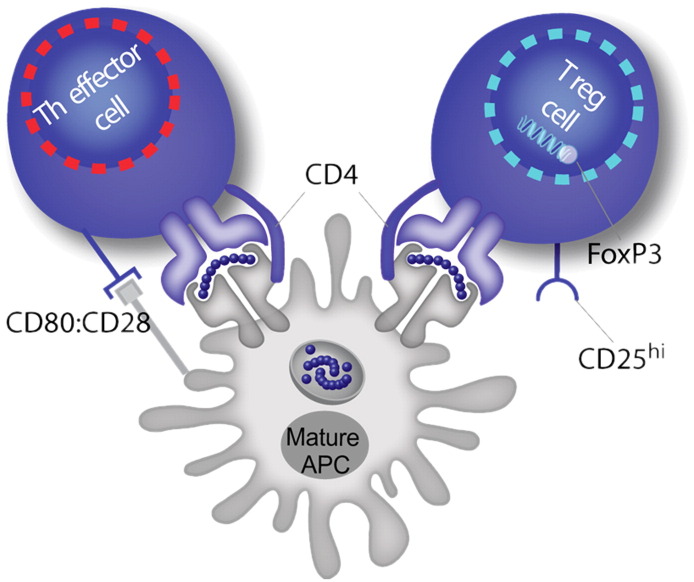
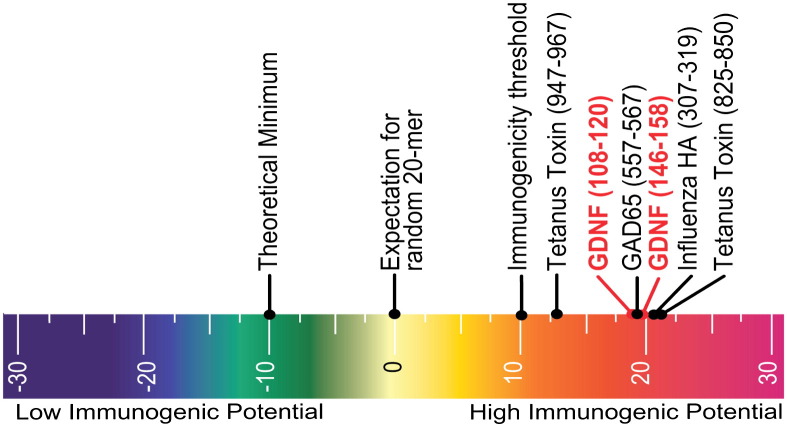
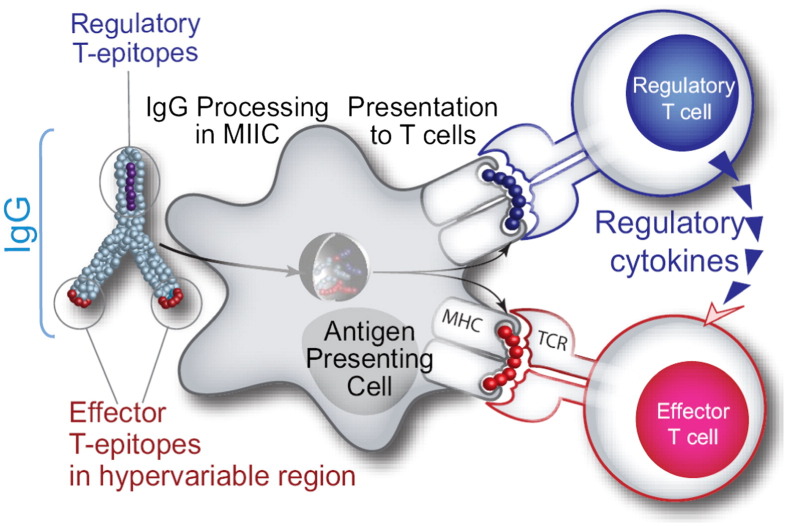
Using EpiMatrix, we screened human antibody sequences for HLA binding sequences. We discovered seven epitope clusters conserved in human antibodies, each containing at least one EpiBar. These sequences are derived from the Fc region and from the framework regions of variable domains. Because EpiMatrix successfully identifies regions which code for T cell activation, conservation of these epitopes suggests that they play an important immunological role. Typically, we screen proteins for immunogenicity but we realized that these putative binders may in fact be tolerizing. In other words, these EpiMatrix results can be interpreted to mean that antibody sequences are presented to T cells to stimulate Tregs rather than effector T cells. These regulatory T cell epitopes (Tregitopes), when co-incubated with a target antigen, induce bystander tolerance, which could explain the relatively lower immunogenicity of humanized antibodies. Indeed, we found that the immunogenicity of therapeutic monoclonal antibodies is correlated to their Tregitope content (R-square 0.7, p = .002), a finding of direct relevance to the biologics industry [6]. Fig. 6 illustrates the hypothesized mechanism of Tregitope-mediated immunosuppression. A major implication of this concept is that other autologous proteins may also contain regulatory T cell epitopes. As therapeutics, these proteins will interface with the immune system and their regulatory and effector epitopes will be presented to T cells. Therefore, regulatory T cell epitopes must also be accounted for when addressing protein immunogenicity.
Fig. 6.
We have discovered conserved T cell epitopes in IgG that engage natural regulatory T cells. We hypothesize that antibody-derived Treg epitopes (dark blue epitope on the top) activate regulatory T cells, which leads to suppression of effector T cells that recognize effector epitopes (red epitope on the bottom), like those of IgG hypervariable regions to which central tolerance does not exist. Whether this suppression is mediated by regulatory cytokines alone or by contact-dependent signaling, or both, has yet to be determined [95]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Current Applications of T cell epitope mapping to biologics development
4.1. Screening for T cell epitopes and de-immunization
The use of cytokines, growth factors, and monoclonal antibodies in clinical settings is often associated with the development of antibodies directed against therapeutic proteins. Therefore, diminishing the immunogenicity of therapeutic proteins without hindering their function may improve clinical outcomes [72]. Epitope mapping and evaluation of the potential for immunogenicity on an “immunogenicity scale” is one approach; the same tools can be used to identify clusters of epitopes for removal. As described previously, reduction in T cell epitope content has been associated with reduced immunogenicity. This can be accomplished by substitution of key amino acids in the T cell epitope sequences which abrogate binding to HLA and thereby attenuate epitope potential to trigger a T cell response. Altered T cell epitopes no longer bind to HLA. Therefore, uncovering the immunogenic regions and specific amino acid residues involved in binding to HLA can allow protein therapeutics developers to abrogate T-dependent antibody responses, reducing the immunogenicity of their candidate drug. Such epitope-modifications are easily evaluated in vitro and in vivo prior to release of the protein therapeutic for clinical development. This method of reducing immunogenicity, also known as de-immunization, is described in detail in [72].
4.2. Consideration for regulatory T cell epitopes
Although T cells that are auto-reactive are said to be deleted in thymic development, some may escape deletion and cause prospective epitopes in autologous proteins to trigger T cells. However, not all T cell epitope clusters can be considered to be potentially immuno-stimulatory. As discussed above, some T cells specific for autologous proteins escape thymic deletion and become natural regulatory T cells (Tregs); they appear to serve as regulators or suppressors of auto-reactive immune responses [30]. Therefore, epitopes that induce Treg responses might be useful to down-regulate immune responses in the context of protein therapeutics and autoimmune disease. Tregs have been shown to suppress auto-antibodies and thus they may also play a role in the suppression of ADA [9], [76]. We have developed a Treg epitope (Tregitope) adjusted EpiMatrix score for monoclonal antibodies to predict anti-therapeutic antibody response by deducting the scores of Tregitopes from the EpiMatrix raw score. In our experience adjusted scores are better correlated to the observed clinical immune response. This scoring protocol enables categorization of antibodies into four groups: 1) low Tregitope and high T effector (neo epitope) content; 2) low Tregitope and low neo epitope content; 3) high Tregitope and low neo epitope or 4) high Tregitope content and high neo epitope content (Table 2 ).
Table 2.
Analysis of anti-therapeutic antibody responses in relation to neo epitope and Tregitope content.
| Monoclonal antibodies | High Tregitope content (%) | Low Tregitope content (%) |
|---|---|---|
| Low neo epitope content | Nuvion (0) | Synagis (1) |
| High neo epitope content | Humira (12) | Rituxan (27) |
Antibodies with low epitope content and antibodies which contain only regulatory epitopes, such as Nuvion and Synagis, would be less likely to induce T cell-mediated immune responses than antibodies containing high numbers of effector epitopes and low numbers of regulatory epitopes, such as Rituxan. These predictions appear to correlate with the observed immunogenicity of these drugs in the clinic [77]. In some cases high numbers of regulatory epitopes can silence immune responses to antibodies containing high numbers of effector epitopes, such as may be the case with Humira (Table 2).
To address immunogenicity concerns related to low Tregitope content in antibodies, targeted sequence modifications may be made by site-directed mutagenesis to enrich for Tregitopes. Alternatively, when structure/function considerations preclude the mutagenesis approach, Tregitopes may be engineered at the carboxy-terminus of the heavy and/or light chains, far from the antigen binding region. This approach could also be adopted to reduce immunogenicity of other classes of protein therapeutics.
4.3. Alternative methods for reducing immunogenicity
Other means of improving the safety and efficacy of biologics by reducing their immunogenicity include humanization and PEGylation. Following FDA approval of the first therapeutic monoclonal antibodies (mAb), many were found to elicit anti-drug antibodies in immune-competent patients [78], [79], [80], [81]. For example, OKT3, an immunosuppressive drug used to treat transplant rejection and the first monoclonal antibody approved for human use, elicited antibodies in 86% of patients [82]. Not surprisingly, in hindsight, the explanation for immunogenicity was patient response to foreign (mouse) sequences. To improve the next generation of mAbs, constant domain sequences of antibody light and heavy chains were replaced by human constant regions to produce chimeric antibodies. This approach met with mixed success because chimeric antibodies still raised human anti-mouse antibodies against the mouse variable regions. While an improvement over fully murine monoclonals, chimeric antibodies were shown to raise varying immune responses depending on their target and their indication. For example, Rituximab, a chimeric anti-CD20 antibody, elicited no immune response from B cell chronic lymphocytic leukemia patients [83], [84] but was immunogenic in 27% of Sjogren's syndrome and 65% of systemic lupus erythomatosus patients [85], [86]. Later, monoclonal antibodies were further humanized by grafting mouse CDR regions onto a human scaffold so that the only mouse sequences to remain were those that encode antigen specificity. Remarkably, some of these antibodies are still immunogenic. For example, Campath-1H (Alemtuzumab), a humanized monoclonal antibody that targets human CD52 expressed in many lymphoid neoplasms, has been shown to be immunogenic in > 60% of rheumatoid arthritis patients [87], [88], [89]. Indeed, a completely human antibody sequence also does not guarantee immune stealth. 17% of rheumatoid arthritis and Crohn's disease patients receiving Humira (Adalimumab), a tumor necrosis factor alpha (TNF-α) inhibitor, developed anti-drug antibodies [90], [91]. The immunogenicity of this antibody has been linked to the presence of five to seven key T helper epitopes as defined by a number of different laboratories (Fiona Harding, Philip Stas, Matt Baker, personal communication).
PEGylation, the conjugation of polyethylene glycol (PEG) to a biologic, has also been used to reduce immunogenicity. PEGylation leads to an increase in protein solubility and hydrodynamic size, allowing a therapeutic protein to evade renal clearance and increase its circulatory time in the body. The large size of the PEG molecules may interfere with antibody binding [92]. PEGylation has been used successfully to minimize the immunogenicity of therapeutic enzymes such as arginase, asparaginase and purine nucleoside phosphorylase, because it blocks antibody binding but does not prevent the diffusion of small molecule substrates [93], [94].
5. Summary
Integrating T cell epitope-mapping tools into scientific research saves time and promises to increase the safety and efficacy of protein therapeutics. As with all bioinformatics tools, in vitro and in vivo confirmations of in silico observations are necessary; validated T cell epitope-mapping tools can then be used with more confidence to begin new scientific explorations [95].
A number of therapeutic protein developers have already incorporated in silico, ex vivo and in vivo preclinical immunogenicity screening protocols into their product development strategy. Mapping epitopes that are contained in recombinant autologous and therapeutic proteins may allow their identification and subsequent modulation, thereby reducing the chance that a biologic will induce a T cell-mediated immune response. T cell epitopes should not all be considered dangerous — in some cases, T cell epitopes can be associated with a regulatory T cell response. Regulatory T cells have the potential to help develop and maintain tolerance. Enhanced use of T cell epitope-mapping tools may improve our ability to identify T cell epitope “friends” (Treg epitopes) and “foes” (effector epitopes) in the context of protein therapeutics and autoimmunity and enable us to harness these important mediators of immune response. Effective differentiation between T cell epitope friends and foes will facilitate development of less immunogenic therapeutics to smooth progress towards improved human health outcomes.
Acknowledgements
The authors wish to thank Julie McMurry for providing the beautiful illustrations for this manuscript. The concept of epitope density emerged from discussions with Gene Koren and Paul Knopf, whose contributions to the immunogenicity scale are hereby gratefully acknowledged. The authors wish to acknowledge Ryan Tassone and Elizabeth McClaine for editorial support. William Martin and Anne S. De Groot are senior officers and majority shareholders at EpiVax, a privately-owned immunotherapeutics company located in Providence RI. These authors acknowledge that there is a potential conflict of interest related to their relationship with EpiVax and attest that the work contained in this research report is free of any bias that might be associated with the commercial goals of the company.
Footnotes
This review is part of the Advanced Drug Delivery Reviews theme issue on “Optimizing the Future for Biotechnology Therapies, the Key Role of Protein Engineering”.
References
- 1.De Groot A.S., Scott D.W. Immunogenicity of protein therapeutics. Trends. Immunol. 2007;28:482–490. doi: 10.1016/j.it.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 2.Zubler R.H. Naive and memory B cells in T-cell-dependent and T-independent responses. Springer. Semin. Immunopathol. 2001;23:405–419. doi: 10.1007/s281-001-8167-7. [DOI] [PubMed] [Google Scholar]
- 3.Koren E., Zuckerman L.A., Mire-Sluis A.R. Immune responses to therapeutic proteins in humans— clinical significance, assessment and prediction. Curr. Pharm. Biotechnol. 2002;3:349–360. doi: 10.2174/1389201023378175. [DOI] [PubMed] [Google Scholar]
- 4.Casadevall N., Nataf J., Viron B., Kolta A., Kiladjian J.J., Martin-Dupont P., Michaud P., Papo T., Ugo V., Teyssandier I., Varet B., Mayeux P. Pure red-cell aplasia and antierythropoietin antibodies in patients treated with recombinant erythropoietin. N. Engl. J. Med. 2002;346:469–475. doi: 10.1056/NEJMoa011931. [DOI] [PubMed] [Google Scholar]
- 5.Koren E., Smith H.W., Shores E., Shankar G., Finco-Kent D., Rup B., Barrett Y.C., Devanarayan V., Gorovits B., Gupta S., Parish T., Quarmby V., Moxness M., Swanson S.J., Taniguchi G., Zuckerman L.A., Stebbins C.C., Mire-Sluis A. Recommendations on risk-based strategies for detection and characterization of antibodies against biotechnology products. J. Immunol. Methods. 2008;333:1–9. doi: 10.1016/j.jim.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 6.De Groot A.S., Martin W. Reducing risk, improving outcomes: bioengineering less immunogenic protein therapeutics. Clin. Immunol. 2009;131:189–201. doi: 10.1016/j.clim.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Southwood S., Sidney J., Kondo A., del Guercio M.F., Appella E., Hoffman S., Kubo R.T., Chesnut R.W., Grey H.M., Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- 8.De Groot A.S., Rayner J., Martin W. Modelling the immunogenicity of therapeutic proteins using T cell epitope mapping. Dev. Biol. (Basel) 2003;112:71–80. [PubMed] [Google Scholar]
- 9.Reveille J.D. The genetic basis of autoantibody production. Autoimmun. Rev. 2006;5:389–398. doi: 10.1016/j.autrev.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Jang E., Cho S.H., Park H., Paik D.J., Kim J.M., Youn J. A positive feedback loop of IL-21 signaling provoked by homeostatic CD4+CD25− T cell expansion is essential for the development of arthritis in autoimmune K/BxN mice. J. Immunol. 2009;182:4649–4656. doi: 10.4049/jimmunol.0804350. [DOI] [PubMed] [Google Scholar]
- 11.Liu G.Y., Fairchild P.J., Smith R.M., Prowle J.R., Kioussis D., Wraith D.C. Low avidity recognition of self-antigen by T cells permits escape from central tolerance. Immunity. 1995;3:407–415. doi: 10.1016/1074-7613(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 12.Bouneaud C., Kourilsky P., Bousso P. Impact of negative selection on the T cell repertoire reactive to a self-peptide: a large fraction of T cell clones escapes clonal deletion. Immunity. 2000;13:829–840. doi: 10.1016/s1074-7613(00)00080-7. [DOI] [PubMed] [Google Scholar]
- 13.Kyewski B., Derbinski J. Self-representation in the thymus: an extended view. Nat. Rev., Immunol. 2004;4:688–698. doi: 10.1038/nri1436. [DOI] [PubMed] [Google Scholar]
- 14.Knip M., Siljander H. Autoimmune mechanisms in type 1 diabetes. Autoimmun. Rev. 2008;7:550–557. doi: 10.1016/j.autrev.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 15.VanderBorght A., Geusens P., Raus J., Stinissen P. The autoimmune pathogenesis of rheumatoid arthritis: role of autoreactive T cells and new immunotherapies. Semin. Arthritis Rheum. 2001;31:160–175. doi: 10.1053/sarh.2001.27736. [DOI] [PubMed] [Google Scholar]
- 16.McFarland H.F., Martin R. Multiple sclerosis: a complicated picture of autoimmunity. Nat. Immunol. 2007;8:913–919. doi: 10.1038/ni1507. [DOI] [PubMed] [Google Scholar]
- 17.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 18.Boucher A., Desforges M., Duquette P., Talbot P.J. Long-term human coronavirus-myelin cross-reactive T-cell clones derived from multiple sclerosis patients. Clin. Immunol. 2007;123:258–267. doi: 10.1016/j.clim.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steere A.C., Klitz W., Drouin E.E., Falk B.A., Kwok W.W., Nepom G.T., Baxter-Lowe L.A. Antibiotic-refractory Lyme arthritis is associated with HLA-DR molecules that bind a Borrelia burgdorferi peptide. J. Exp. Med. 2006;203:961–971. doi: 10.1084/jem.20052471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page K.R., Scott A.L., Manabe Y.C. The expanding realm of heterologous immunity: friend or foe? Cell. Microbiol. 2006;8:185–196. doi: 10.1111/j.1462-5822.2005.00653.x. [DOI] [PubMed] [Google Scholar]
- 21.Zubler R.H. Naive and memory B cells in T-cell-dependent and T-independent responses, Springer. Semin. Immunopathol. 2001;23:405–419. doi: 10.1007/s281-001-8167-7. [DOI] [PubMed] [Google Scholar]
- 22.Miyake K. Innate immune sensing of pathogens and danger signals by cell surface Toll-like receptors. Semin. Immunol. 2007;19:3–10. doi: 10.1016/j.smim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Purcell A.W., McCluskey J., Rossjohn J. More than one reason to rethink the use of peptides in vaccine design. Nat. Rev., Drug Discov. 2007;6:404–414. doi: 10.1038/nrd2224. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi H., Kuwana M., Amagai M. A single helper T cell clone is sufficient to commit polyclonal naive B cells to produce pathogenic IgG in experimental pemphigus vulgaris. J. Immunol. 2009;182:1740–1745. doi: 10.4049/jimmunol.182.3.1740. [DOI] [PubMed] [Google Scholar]
- 25.Wilson C.B., Rowell E., Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat. Rev., Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 26.Lohr J., Knoechel B., Caretto D., Abbas A.K. Balance of Th1 and Th17 effector and peripheral regulatory T cells. Microbes Infect. 2009 doi: 10.1016/j.micinf.2009.04.012. [Electronic publication ahead of Print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghilardi N., Ouyang W. Targeting the development and effector functions of TH17 cells. Semin. Immunol. 2007;19:383–393. doi: 10.1016/j.smim.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 28.Osorio F., LeibundGut-Landmann S., Lochner M., Lahl K., Sparwasser T., Eberl G., Reis e Sousa C. DC activated via dectin-1 convert Treg into IL-17 producers. Eur. J. Immunol. 2008;38:3274–3281. doi: 10.1002/eji.200838950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dörner T., Radbruch A. Antibodies and B cell memory in viral immunity. Immunity. 2007;27:384–392. doi: 10.1016/j.immuni.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Bluestone J.A., Abbas A.K. Natural versus adaptive regulatory T cells. Nat. Rev., Immunol. 2003;3:253–257. doi: 10.1038/nri1032. [DOI] [PubMed] [Google Scholar]
- 31.Bresson D., von Herrath M. Resuscitating adaptive Tregs with combination therapies? Novartis Found. Symp. 2008;292:50–60. doi: 10.1002/9780470697405.ch5. (discussion 60-7, 122-9, 202-3) [DOI] [PubMed] [Google Scholar]
- 32.Reiner S.L. Decision making during the conception and career of CD4+ T cells. Nat. Rev., Immunol. 2009;9:81–82. doi: 10.1038/nri2490. [DOI] [PubMed] [Google Scholar]
- 33.Wan Y.Y., Flavell R.A. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 34.Caudy A.A., Reddy S.T., Chatila T., Atkinson J.P., Verbsky J.W. CD25 deficiency causes an immune dysregulation, polyendocrinopathy, enteropathy, X-linked-like syndrome, and defective IL-10 expression from CD4 lymphocytes. J. Allergy Clin. Immunol. 2007;119:482–487. doi: 10.1016/j.jaci.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 35.Huehn J., Polansky J.K., Hamann A. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat. Rev., Immunol. 2009;9:83–89. doi: 10.1038/nri2474. [DOI] [PubMed] [Google Scholar]
- 36.Collison L.W., Workman C.J., Kuo T.T., Boyd K., Wang Y., Vignali K.M., Cross R., Sehy D., Blumber R.S., Vignali D.A. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450:556–559. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 37.Lahl K., Loddenkemper C., Drouin C., Freyer J., Arnason J., Eberl G., Hamann A., Wagner H., Huehn J. T. Sparwasser. Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J. Exp. Med. 2007;204:57–63. doi: 10.1084/jem.20061852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smyk-Pearson S.K., Bakke A.C., Held P.K., Wildin R.S. Rescue of the autoimmune scurfy mouse by partial bone marrow transplantation or by injection with T-enriched splenocytes. Clin. Exp. Immunol. 2003;133:193–199. doi: 10.1046/j.1365-2249.2003.02217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lazarski C.A., Chaves F.A., Jenks S.A., Wu S., Richards K.A., Weaver J.M., Sant A.J. The kinetic stability of MHC class II peptide complexes is a key parameter that dictates immunodominance. Immunity. 2005;23:29–40. doi: 10.1016/j.immuni.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Q., Wang P., Kim Y., Haste-Andersen P., Beaver J., Bourne P.E., Bui H.H., Buus S., Frankild S., Greenbaum J., Lund O., Lundegaard C., Nielsen M., Ponomarenko J., Sette A., Zhu Z., Peters B. Immune epitope database analysis resource (IEDB-AR) Nucleic Acids Res. 2008;36:W513–W518. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brusic V., Bajic B., Petrovsky N. Computational methods for prediction of T-cell epitopes—a framework for modelling, testing, and applications. Methods. 2004;34:436–443. doi: 10.1016/j.ymeth.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 42.De Groot A.S., Moise L. Prediction of immunogenicity for therapeutic proteins: state of the art. Curr. Opin. Drug Discov. Dev. 2007;10:332–334. [PubMed] [Google Scholar]
- 43.Ahlers J.D., Belyakov I.M., Thomas E.K., Berzofsky J.A. High-affinity T helper epitope induces complementary helper and APC polarization, increased CTL, and protection against viral infection. J. Clin. Invest. 2001;108:1677–1685. doi: 10.1172/JCI13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Groot A.S., Sbai H., Saint-Aubin C., McMurry J.A., Martin W. Immuno-informatics: mining genomes for vaccine components. Immunol. Cell Biol. 2002;8:255–269. doi: 10.1046/j.1440-1711.2002.01092.x. [DOI] [PubMed] [Google Scholar]
- 45.Inaba H., Martin W., De Groot A.S., Qin S., De Groot L.J. Thyrotropin receptor epitopes and their relation to histocompatibility leukocyte antigen-DR molecules in Graves' disease. J. Clin. Endocrinol. Metab. 2006;91:2286–2294. doi: 10.1210/jc.2005-2537. [DOI] [PubMed] [Google Scholar]
- 46.De Groot A.S., Jesdale B.M., Szu E., Schafer J.R., Chicz R.M., Deocampo G. An interactive web site providing major histocompatibility ligand predictions: application to HIV research. AIDS Res. Hum. Retrovir. 1997;13:529–531. doi: 10.1089/aid.1997.13.529. [DOI] [PubMed] [Google Scholar]
- 47.Bond K.B., Sriwanthana B., Hodge T.W., De Groot A.S., Mastro T.D., Young N.L., Promadej N., Altman J.D., Limpakarnjanarat K., McNicholl J.M. An HLA-directed molecular and bioinformatics approach identifies new HLA-A11 HIV-1 subtype E cytotoxic T lymphocyte epitopes in HIV-1-infected Thais. AIDS Res. Hum. Retrovir. 2001;17:703–717. doi: 10.1089/088922201750236988. [DOI] [PubMed] [Google Scholar]
- 48.McMurry J.A., Sbai H., Gennaro M.L., Carter E.J., Martin W., De Groot A.S. Analyzing Mycobacterium tuberculosis proteomes for candidate vaccine epitopes. Tuberculosis (Edinb.) 2005;85:95–105. doi: 10.1016/j.tube.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 49.Dong Y., Demaria S., Sun X., Santori F.R., Jesdale B.M., De Groot A.S., Rom W.N., Bushkin Y. HLA-A2-restricted CD8+-cytotoxic-T-cell responses to novel epitopes in Mycobacterium tuberculosis superoxide dismutase, alanine dehydrogenase, and glutamine synthetase. Infect. Immun. 2004;72:2412–2415. doi: 10.1128/IAI.72.4.2412-2415.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koita O.A., Dabitao D., Mahamadou I., Tall M., Dao S., Tounkara A., Guiteye H., Noumsi C., Thiero O., Kone M., Rivera D., McMurry J.A., Martin W., De Groot A.S. Confirmation of immunogenic consensus sequence HIV-1 T-cell epitopes in Bamako, Mali and Providence, Rhode Island. Hum. Vaccin. 2006;2:119–128. doi: 10.4161/hv.2869. [DOI] [PubMed] [Google Scholar]
- 51.Panina-Bordignon P., Tan A., Termijtelen A., Demotz S., Corradin G., Lanzavecchia A. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur. J. Immunol. 1989;19:2237–2242. doi: 10.1002/eji.1830191209. [DOI] [PubMed] [Google Scholar]
- 52.Roche P.A., Cresswell P. High-affinity binding of an influenza hemagglutinin-derived peptide to purified HLA-DR. J. Immunol. 1990;144:1849–1856. [PubMed] [Google Scholar]
- 53.Ferrari C., Bertoletti A., Penna A., Cavalli A., Valli A., Missale G., Pilli M., Fowler P., Giuberti T., Chisari F.V. Identification of immunodominant T cell epitopes of the hepatitis B virus nucleocapsid antigen. J. Clin. Invest. 1991;88:214–222. doi: 10.1172/JCI115280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diepolder H.M., Gerlach J.T., Zachoval R., Hoffmann R.M., Jung M.C., Wierenga E.A., Scholz S., Santantonio T., Houghton M., Southwood S., Sette A., Pape G.R. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J. Virol. 1997;71:6011–6019. doi: 10.1128/jvi.71.8.6011-6019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaufmann D.E., Bailey P.M., Sidney J., Wagner B., Norris PJ., Johnston M.N., Cosimi L.A., Addo M.M., Lichterfeld M., Altfeld M., Frahm N., Brander C., Sette A., Walker B.D., Rosenberg E.S. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J. Virol. 2004;78:4463–4477. doi: 10.1128/JVI.78.9.4463-4477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.L. Moise, M. Ardito, J. Desrosiers, J. Schriewer, M. Buller, S. Frey, S.F. Gregory, S.F. Moss, J. Lee, H. Kornfeld, W. Martin, A.S. De Groot, Immunome-derived Epitope-driven Vaccines (ID-EDV) Protect against Viral or Bacterial Challenge in Humanized Mice. Vaccine (in press) (Electronic publication ahead of print).
- 57.Roberts C.G.P., Meister G.E., Jesdale B.M., Lieberman J., Berzofsky J.A., De Groot A.S. Prediction of HIV peptide epitopes by a novel algorithm. AIDS Res. Hum. Retrovir. 1996;12:593–610. doi: 10.1089/aid.1996.12.593. [DOI] [PubMed] [Google Scholar]
- 58.PT-PA-CEFT-001 Data Sheet. Axxora.com. <http://www.axxora.com/?content=open.php%3FPID%3DPT-PA-CEFT-001>. 2009 May.
- 59.Tatarewicz S.M., Wei X., Gupta S., Masterman D., Swanson S.J., Moxness M.S. Development of a maturing T-cell-mediated immune response in patients with idiopathic Parkinson's disease receiving r-metHuGDNF via continuous intraputaminal infusion. J. Clin. Immunol. 2007;27:620–627. doi: 10.1007/s10875-007-9117-8. [DOI] [PubMed] [Google Scholar]
- 60.Koren E., De Groot A.S., Jawa V., Beck K.D., Boone T., Rivera D., Li L., Mytych D., Koscec M., Weeraratne D., Swanson S., Martin W. Clinical validation of the “in silico” prediction of immunogenicity of a human recombinant therapeutic protein. Clin. Immunol. 2007;124:26–32. doi: 10.1016/j.clim.2007.03.544. [DOI] [PubMed] [Google Scholar]
- 61.Barbosa M.D., Vielmetter J., Chu S., Smith D.D., Jacinto J. Clinical link between MHC class II haplotype and interferon-beta (IFN-beta) immunogenicity. Clin. Immunol. 2006;118:42–50. doi: 10.1016/j.clim.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 62.Jawa V. Clinical Validation of in silico Prediction of Immunogenicity of a Recombinant Therapeutic Protein, Presentation at the 2nd Protein Therapeutics Discovery and Development Conference, Sept 7–8, 2008, San Diego CA. 2008. link to abstract available on line at: http://www.gtcbio.com/userAgenda.aspx?id=128. [Google Scholar]
- 63.Kamperschroer C., Dibble J.P., Meents D.L., Schwartzberg P.L., Swain S.L. SAP is required for Th cell function and for immunity to influenza. J. Immunol. 2006;177:5317–5327. doi: 10.4049/jimmunol.177.8.5317. [DOI] [PubMed] [Google Scholar]
- 64.McMurry J.A., Johansson B.E., De Groot A.S. A call to cellular and humoral arms: enlisting cognate T cell help to develop broad-spectrum vaccines against influenza A. Hum. Vaccin. 2008;4:148–157. doi: 10.4161/hv.4.2.5169. [DOI] [PubMed] [Google Scholar]
- 65.Galli G., Medini D., Borgogni E., Zedda L., Bardelli M., Malzone C., Nuti S., Tavarini S., Sammicheli C., Hilbert A.K., Brauer V., Banzhoff A., Rappuoli R., Del Giudice G., Castellino F. Adjuvanted H5N1 vaccine induces early CD4+ T cell response that predicts long-term persistence of protective antibody levels. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3877–3882. doi: 10.1073/pnas.0813390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chaloupka I., Schuler A., Marschall M., Meier-Ewert H. Comparative analysis of six European influenza vaccines. Eur. J. Clin. Microbiol. Infect. Dis. 1996;15:121–127. doi: 10.1007/BF01591484. [DOI] [PubMed] [Google Scholar]
- 67.McMurry J.A., Gregory S.H., Moise L., Rivera D., Buus S., De Groot A.S. Diversity of Francisella tularensis Schu4 antigens recognized by T lymphocytes after natural infections in humans: identification of candidate epitopes for inclusion in a rationally designed tularemia vaccine. Vaccine. 2007;25:3179–3191. doi: 10.1016/j.vaccine.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 68.Celis E., Ou D., Otvos L., Jr. Recognition of hepatitis B surface antigen by human T lymphocytes. Proliferative and cytotoxic responses to a major antigenic determinant defined by synthetic peptides. J. Immunol. 1998;140:1808–1815. [PubMed] [Google Scholar]
- 69.Min W.P., Kamikawaji N., Mineta M., Tana T., Kashiwagi S., Sasazuki T. Identification of an epitope for T-cells correlated with antibody response to hepatitis B surface antigen in vaccinated humans. Hum. Immunol. 1996;46:93–99. doi: 10.1016/0198-8859(96)00009-2. [DOI] [PubMed] [Google Scholar]
- 70.Zuckerman J.N. Nonresponse to hepatitis B vaccines and the kinetics of anti-HBs production. J. Med. Virol. 1996;50:283–288. doi: 10.1002/(SICI)1096-9071(199612)50:4<283::AID-JMV1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 71.McDermott A.B., Cohen S.B.A., Zuckerman J.N., Madrigal J.A. Human leukocyte antigens influence the immune response to a pre-S/S hepatitis B vaccine. Vaccine. 1999;17:330–339. doi: 10.1016/s0264-410x(98)00203-5. [DOI] [PubMed] [Google Scholar]
- 72.De Groot A.S., Knopf P., Martin W. De-immunization of therapeutic proteins by T-cell epitope modification. Dev. Biol. (Basel) 2005;122:171–194. [PubMed] [Google Scholar]
- 73.Collen D., Bernaerts R., Declerck P., De Cock F., Demarsin E., Jenné S., Laroche Y., Lijnen H.R., Silence K., Verstreken M. Recombinant staphylokinase variants with altered immunoreactivity. I: Construction and characterization. Circulation. 1996;94:197–206. doi: 10.1161/01.cir.94.2.197. [DOI] [PubMed] [Google Scholar]
- 74.Jones T.D., Phillips W.J., Smith B.J., Bamford C.A., Nayee P.D., Baglin T.P., Gaston J.S., Baker M.P. Identification and removal of a promiscuous CD4+ T cell epitope from the C1 domain of factor VIII. J. Thromb. Haemost. 2005;3:991–1000. doi: 10.1111/j.1538-7836.2005.01309.x. [DOI] [PubMed] [Google Scholar]
- 75.Gilles J.G., Lavend'homme R., Peerlinck K., Jacquemin M.G., Hoylaerts M., Jorieux S., Mazurier C., Vermylen J., Saint-Remy J.M. Some factor VIII (FVIII) inhibitors recognise a FVIII epitope(s) that is present only on FVIII-vWF complexes. Thromb. Haemost. 1999;82:40–45. [PubMed] [Google Scholar]
- 76.S. Hai, J.A. McMurry, P. Knopf, W. Martin, A.S. De Groot, Immunogenicity screening using in silico methods: Correlation between T-cell epitope content and clinical immunogenicity of monoclonal antibodies. In Therapeutic Antibodies: from Theory to Practice, Zhiqiang An editor. John Wiley and Sons, in press, Scheduled for publication 2009.
- 77.Hwang W.Y., Foote J. Immunogenicity of engineered antibodies. Methods. 2005;36:3–10. doi: 10.1016/j.ymeth.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 78.McIntyre J.A., Kincade M., Higgins N.G. Detection of IGA anti-OKT3 antibodies in OKT3-treated transplant recipients. Transplantation. 1996;61:1465–1469. doi: 10.1097/00007890-199605270-00009. [DOI] [PubMed] [Google Scholar]
- 79.Uckun F.M., Messinger Y., Chen C.L., O'Neill K., Myers D.E., Goldman F., Hurvitz C., Casper J.T., Levine A. Treatment of therapy-refractory B-lineage acute lymphoblastic leukemia with an apoptosis-inducing CD19-directed tyrosine kinase inhibitor. Clin. Cancer. Res. 1999;5:3906–3913. [PubMed] [Google Scholar]
- 80.Kaminski M.S., Zelenetz A.D., Press O.W., Saleh M., Leonard J., Fehrenbacher L., Lister T.A., Stagg R.J., Tidmarsh G.F., Kroll S., Wahl R.L., Knox S.J., Vose J.M. Pivotal study of iodine I 131 tositumomab for chemotherapy-refractory low-grade or transformed low-grade B-cell non-Hodgkin's lymphomas. J. Clin. Oncol. 2001;19:3918–3928. doi: 10.1200/JCO.2001.19.19.3918. [DOI] [PubMed] [Google Scholar]
- 81.Stroomer J.W., Roos J.C., Sproll M., Quak J.J., Heider K.H., Wilhelm B.J., Castelijns J.A., Meyer R., Kwakkelstein M.O., Snow G.B., Adolf G.R., van Dongen G.A. Safety and biodistribution of 99mTechnetium-labeled anti-CD44v6 monoclonal antibody BIWA 1 in head and neck cancer patients. Clin. Cancer Res. 2000;6:3046–3055. [PubMed] [Google Scholar]
- 82.McIntyre J.A., Kincade M., Higgins N.G. Detection of IGA anti-OKT3 antibodies in OKT3-treated transplant recipients. Transplantation. 1996;61:1465–1469. doi: 10.1097/00007890-199605270-00009. [DOI] [PubMed] [Google Scholar]
- 83.Davis T.A., Grillo-López A.J., White C.A., McLaughlin P., Czuczman M.S., Link B.K., Maloney D.G., Weaver R.L., Rosenberg J., Levy R. Rituximab anti-CD20 monoclonal antibody therapy in non-Hodgkin's lymphoma: safety and efficacy of re-treatment. J. Clin. Oncol. 2000;18:3135–3143. doi: 10.1200/JCO.2000.18.17.3135. [DOI] [PubMed] [Google Scholar]
- 84.Piro L.D., White C.A., Grillo-López A.J., Janakiraman N., Saven A., Beck T.M., Varns C., Shuey S., Czuczman M., Lynch J.W., Kolitz J.E., Jain V. Extended Rituximab (anti-CD20 monoclonal antibody) therapy for relapsed or refractory low-grade or follicular non-Hodgkin's lymphoma. Ann. Oncol. 1999;10:655–661. doi: 10.1023/a:1008389119525. [DOI] [PubMed] [Google Scholar]
- 85.Pijpe J., van Imhoff G.W., Spijkervet F.K., Roodenburg J.L., Wolbink G.J., Mansour K., Vissink A., Kallenberg C.G., Bootsma H. Rituximab treatment in patients with primary Sjögren's syndrome: an open-label phase II study. Arthritis Rheum. 2005;52:2740–2750. doi: 10.1002/art.21260. [DOI] [PubMed] [Google Scholar]
- 86.Looney R.J., Anolik J.H., Campbell D., Felgar R.E., Young F., Arend L.J., Sloand J.A., Rosenblatt J., Sanz I. B cell depletion as a novel treatment for systemic lupus erythematosus: a phase I/II dose-escalation trial of rituximab. Arthritis Rheum. 2004;50:2580–2589. doi: 10.1002/art.20430. [DOI] [PubMed] [Google Scholar]
- 87.Weinblatt M.E., Maddison P.J., Bulpitt K.J., Hazleman B.L., Urowitz M.B., Sturrock R.D., Coblyn J.S., Maier A.L., Spreen W.R, Manna V.K. CAMPATH-1H, a humanized monoclonal antibody, in refractory rheumatoid arthritis. An intravenous dose-escalation study. Arthritis Rheum. 1995;38:1589–1594. doi: 10.1002/art.1780381110. [DOI] [PubMed] [Google Scholar]
- 88.Isaacs J.D., Watts R.A., Hazleman B.L., Hale G., Keogan M.T., Cobbold S.P., Waldmann H. Humanised monoclonal antibody therapy for rheumatoid arthritis. Lancet. 1992;340:748–752. doi: 10.1016/0140-6736(92)92294-p. [DOI] [PubMed] [Google Scholar]
- 89.Reiff A. A review of Campath in autoimmune disease: biologic therapy in the gray zone between immunosuppression and immunoablation. Hematology. 2005;10:79–93. doi: 10.1080/10245330400026139. [DOI] [PubMed] [Google Scholar]
- 90.Radstake T.R., Svenson M., Eijsbouts A.M., van den Hoogen F.H., Enevold C., van Riel P.L., Bendtzen K. Formation of antibodies against infliximab and adalimumab strongly correlates with functional drug levels and clinical responses in rheumatoid arthritis. Ann. Rheum. Dis. 2008 doi: 10.1136/ard.2008.092833. [DOI] [PubMed] [Google Scholar]
- 91.Bartelds G.M., Wijbrandts C.A., Nurmohamed M.T., Stapel S., Lems W.F., Aarden L., Dijkmans B.A., Tak P.P., Wolbink G.J. Clinical response to adalimumab: relationship to anti-adalimumab antibodies and serum adalimumab concentrations in rheumatoid arthritis. Ann. Rheum. Dis. 2007;66:921–926. doi: 10.1136/ard.2006.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Harris J.M., Martin N.E., Modi M. Pegylation: a novel process for modifying pharmacokinetics. Clin. Pharmacokinet. 2001;40:539–551. doi: 10.2165/00003088-200140070-00005. [DOI] [PubMed] [Google Scholar]
- 93.Savoca K.V., Abuchowski A., van Es T., Davis F.F., Palczuk N.C. Preparation of a non-immunogenic arginase by the covalent attachment of polyethylene glycol. Biochim. Biophys. Acta. 1979;578:47–53. doi: 10.1016/0005-2795(79)90111-9. [DOI] [PubMed] [Google Scholar]
- 94.Hershfield M.S., Chaffee S., Koro-Johnson L., Mary A., Smith A.A., Short S.A. Use of site-directed mutagenesis to enhance the epitope-shielding effect of covalent modification of proteins with polyethylene glycol. Proc. Natl. Acad. Sci. U. S. A. 1991;88:7185–7189. doi: 10.1073/pnas.88.16.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.DeGroot A.S. Immunomics: discovering new targets for vaccines and therapeutics. Drug Discov. Today. 2006;11:203–209. doi: 10.1016/S1359-6446(05)03720-7. [DOI] [PubMed] [Google Scholar]