Abstract
The complete genome sequence of the first equine coronavirus (ECoV) isolate, NC99 strain was accomplished by directly sequencing 11 overlapping fragments which were RT–PCR amplified from viral RNA. The ECoV genome is 30,992 nucleotides in length, excluding the polyA tail. Analysis of the sequence identified 11 open reading frames which encode two replicase polyproteins, five structural proteins (hemagglutinin esterase, spike, envelope, membrane, and nucleocapsid) and four accessory proteins (NS2, p4.7, p12.7, and I). The two replicase polyproteins are predicted to be proteolytically processed by three virus-encoded proteases into 16 non-structural proteins (nsp1–16). The ECoV nsp3 protein had considerable amino acid deletions and insertions compared to the nsp3 proteins of bovine coronavirus, human coronavirus OC43, and porcine hemagglutinating encephalomyelitis virus, three group 2 coronaviruses phylogenetically most closely related to ECoV. The structure of subgenomic mRNAs was analyzed by Northern blot analysis and sequencing of the leader–body junction in each sg mRNA.
Keywords: Equine coronavirus, Entire genome, Subgenomic RNA, Transcription regulatory sequence, Non-structural protein 3
Introduction
Coronaviruses are mainly associated with respiratory and gastrointestinal disease in humans (Drosten et al., 2003, Holmes, 2001, Ksiazek et al., 2003, Peiris et al., 2003, van der Hoek et al., 2004, Woo et al., 2005) and respiratory, enteric, neurological, or hepatic disease in animals (Holmes, 2001). Coronaviruses have also been isolated from bats, poultry and other birds (Cavanagh, 2005, Chu et al., 2006, Poon et al., 2005, Ren et al., 2006). On the basis of antigenic and genetic analyses, coronaviruses are divided into three groups (Gonzalez et al., 2003, Gorbalenya et al., 2004, Snijder et al., 2003). Group 1 viruses include human coronaviruses 229E (HCoV-229E) and NL63 (HCoV-NL63), canine coronavirus (CCoV), feline coronavirus (FCoV), porcine transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), and bat coronavirus. Group 2 viruses are subdivided into group 2a which includes murine hepatitis virus (MHV), human coronaviruses OC43 (HCoV-OC43) and HKU1 (HCoV-HKU1), bovine coronavirus (BCoV), porcine hemagglutinating encephalomyelitis virus (PHEV), and rat coronavirus (RCov), and group 2b which includes SARS-coronavirus (SARS-CoV). Group 3 viruses include avian viruses, such as avian infectious bronchitis virus (IBV), and turkey coronavirus (TCoV).
Members of the family Coronaviridae are enveloped, positive-stranded RNA viruses with exceptionally large, polycistronic genomes (27–32 kb). The 5′-proximal two-thirds of the genome comprises two open reading frames (ORFs), ORF1a and ORF1b, which encode the replicase polyproteins (pp) 1a and pp1ab (Ziebuhr, 2005). Expression of the pp1ab requires a − 1 ribosomal frameshift during translation of the genomic RNA (Brierley et al., 1987). The two replicase polyproteins are processed extensively by two or three viral proteases encoded by ORF1a to generate up to 16 end-products termed nonstructural proteins (nsp) 1 to 16 and multiple processing intermediates (Ziebuhr, 2005, Ziebuhr et al., 2000). The N-proximal region of the polyproteins is processed by one or two papain-like proteases (PLpro), whereas the central and C-proximal region is processed by the viral main protease, 3C-like protease (3CLpro) (Ziebuhr, 2005, Ziebuhr et al., 2000). The 3′-proximal one-third of the genome encodes structural proteins and various accessory proteins. Genes encoding the four structural proteins present in all coronaviruses occur in the 5′ to 3′ order as spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins (Brian and Baric, 2005, Lai et al., 2006). Some coronaviruses contain an additional structural protein, the hemagglutinin–esterase (HE) protein which is located upstream of the S protein gene (Lai et al., 2006). In contrast to the replicase proteins which are directly translated from the genomic RNA, coronavirus structural and accessory proteins are expressed from a nested set of 3′ co-terminal subgenomic (sg) mRNAs that also possess a common 5′ leader sequence derived from the 5′ end of the genome (Pasternak et al., 2006, Sawicki et al., 2007). The common 5′ leader is fused to the 3′ body segments through a mechanism that is presumed to involve discontinuous minus strand RNA synthesis to produce subgenome-length templates for subgenomic mRNA synthesis, with the transcription regulatory sequence (TRS) elements determining the fusion sites of leader and body segments (see recent review of Pasternak et al., 2006, Sawicki et al., 2007 for details).
Equine coronavirus (ECoV) was first isolated from feces of a diarrheic foal in 1999 (ECoV-NC99) in North Carolina, USA (Guy et al., 2000). Little is known about ECoV and its clinical significance. Molecular characterization of ECoV and development of diagnostic and prophylactic reagents necessitate sequencing of ECoV. In this study, we determined the full-length nucleotide sequence of the ECoV-NC99 strain of equine coronavirus. The viral genome and proteome were analyzed and the predicted features of ECoV nonstructural, structural, and accessory proteins were compared to those of other coronaviruses. Synthesis of sg mRNAs in ECoV-infected cells was analyzed by Northern blotting. The leader–body junction sequence in each sg mRNA was determined and the exact position of TRS used for synthesis of each sg mRNA was mapped on the genome. The evolutionary relationship between ECoV and other phylogenetically closely related group 2a coronaviruses was explored.
Results and discussion
ECoV genome sequence analysis
We report here the full-length genomic sequence of the first ECoV isolate, the NC99 strain, and this is also the first reported complete genome sequence of ECoV. The nucleotide sequence was determined by directly sequencing 11 overlapping cDNA fragments which were RT–PCR amplified from viral RNA. The ECoV-NC99 genome comprises 30,992 nucleotides (nt), excluding the 3′ poly (A) tail, and has a GC content of 37.2%. The nucleotide sequence data have been deposited in GenBank under accession number EF446615.
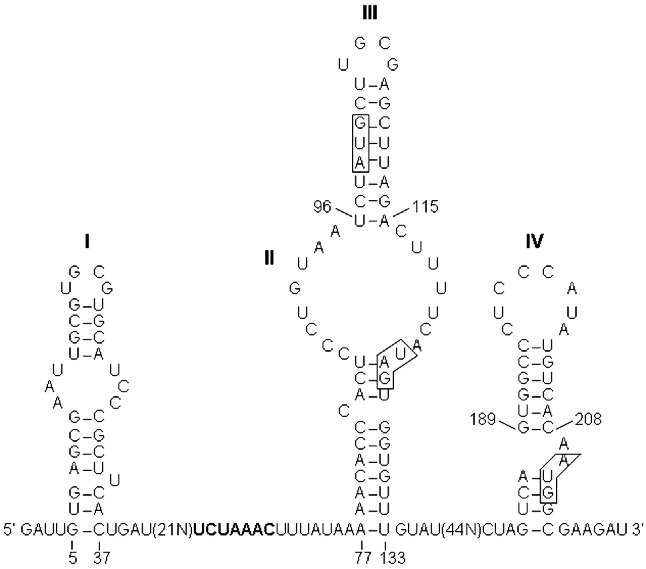
Both 5′ and 3′ ends of the ECoV genome contain short untranslated regions (UTR). The 5′ UTR comprises 209 nt (1–209) and includes a potential short internal ORF of 8 codons (nt 99–125). Four stem–loop structures (I, II, III, and IV) were identified in the 5′ UTR and a short stretch of nucleotides that are part of the ORF1a (see Supplementary Fig. S1). The bulged stem–loop III (96–115) and IV (189–208) closely resemble the stem–loop III and IV that have been identified as replication signaling elements in bovine coronavirus and other group 2 coronaviruses (Raman and Brian, 2005, Raman et al., 2003, Wu et al., 2003). The 3′ UTR of the ECoV genome comprises 289 nt (30,704–30,992) and contains a putative bulged stem–loop structure (nt 30,703–30,770) and a putative pseudoknot structure (30,766–30,819) (see Supplementary Fig. S2). Similar putative bulged stem–loop structure and pseudoknot structure have been identified in murine hepatitis virus and other group 2 coronaviruses; these have been shown to be essential for viral replication (Goebel et al., 2004a, Goebel et al., 2004b, Hsue and Masters, 1997, Hsue et al., 2000, Williams et al., 1999).
Analysis of the ECoV-NC99 genome reveals 11 potential ORFs (1a, 1b, 2–8, 9a and 9b) as shown in Fig. 1 and Table 1 . The ORFs 1a and 1b encode the replicase polyproteins pp1a and pp1ab. The ORFs 2–8, 9a and 9b encode structural and accessory proteins NS2, HE, S, p4.7, p12.7, E, M, N, and I, respectively.
Fig. 1.
Schematic diagrams of ECoV genome organization. The ECoV entire genome organization is depicted (middle). The 5′ leader, ORFs 1a and 1b encoding replicase polyproteins are shown, with the ribosomal frameshift site indicated. Structural and accessory proteins are also indicated: NS2 protein (encoded by ORF2), hemagglutinin esterase (HE, ORF3), spike protein (S, ORF4), p4.7 protein (ORF5), p12.7 protein (ORF6), envelope protein (E, ORF7), membrane protein (M, ORF8), nucleocapsid protein (N, ORF9a), and I protein (ORF9b). Predicted cleavage products (nsp1–nsp16) of the replicase polyproteins are depicted (Bottom). Arrows represent sites in the corresponding replicase polyproteins that are cleaved by papain-like proteases (white arrows) or the 3C-like cysteine protease (black arrows). A number of putative functional domains predicted in the ECoV pp1a and pp1ab are indicated. PL1, papain-like proteinase 1 (aa 1059–1275); PL2, papain-like proteinase 2 (aa 1570–1867); X, X-domain which contains adenosine diphosphate-ribose 1ʺ-phosphatase (ADRP) (aa 1276–1435); TM, transmembrane domain; 3CL, 3C-like proteinase; RdRp, RNA-dependent RNA polymerase; Z, zinc-binding domain; HEL, helicase domain; ExoN, exonuclease; N, nidoviral uridylate-specific endoribonuclease (NendoU); MT, 2′-O-ribose methyltransferase (2′-O-MT). Domains Ac (aa 846–1058) and Y (aa 2310–2796) are described by Ziebuhr et al. (2001). The spike protein (1363 amino acids) of ECoV is represented by a black line (Top). The N-terminal signal peptide (amino acid residues 1–14 or 17), the heptad repeat 1 (HR1, amino acid residues 991–1902), the heptad repeat 2 (HR2, amino acid residues 1259–1304), the transmembrane domain (amino acid residues 1308–1330), and the cytoplasmic domain (amino acid residues 1331–1363) are depicted. A potential cleavage recognition sequence (RRQRR) at residues 764–768 and the predicted cleavage site between residues 768 and 769 are indicated. The generated cleavage products S1 and S2 subunits are depicted. The positions of the receptor-binding domain on the S1 subunit and the fusion peptide on the S2 subunit are currently unknown.
Table 1.
Coding potential of the ECoV-NC99 genome sequence
| ORF | Encoded protein | Nucleotide position in the genome | No. of nucleotides | No. of amino acids (aa) | mRNA used for expressiona |
|---|---|---|---|---|---|
| 5′ Leader | 1–64 | 64 | |||
| 5′ UTR | 1–209 | 209 | |||
| ORF1a | pp1a | 210–13,499 | 13,290 | 4429 | 1 |
| ORF1a/b | pp1ab | 210–21,595 | 21,386 | 7128 | 1 |
| ORF2 | NS2 | 21,610–22,446 | 837 | 278 | 2 |
| ORF3 | HE | 22,458–23,729 | 1272 | 423 | 3 |
| ORF4 | S | 23,744–27,835 | 4092 | 1363 | 4 |
| ORF5 | p4.7 | 27,825–27,947 | 123 | 40 | 5 |
| ORF6 | p12.7 | 28,076–28,405 | 330 | 109 | 6 |
| ORF7 | E | 28,392–28,646 | 255 | 84 | 7 |
| ORF8 | M | 28,661–29,353 | 693 | 230 | 8 |
| ORF9a | N | 29,363–30,703 | 1341 | 446 | 9 |
| ORF9b | I | 29,424–30,044 | 621 | 206 | 9 |
| 3′ UTR | 30,704–30,992 | 289 |
The mRNA used for expression of each protein is derived from the Northern blotting analysis and the comparison with other group 2a coronaviruses. See the text for details.
The replicase ORF1a (nt 210–13,499) and replicase ORF1b (13,478–21,595) occupy 21.4 kb (69%) of the ECoV-NC99 genome. The translation of ORF1a generates a precursor pp1a of 4,429 amino acids. Similar to other coronaviruses, translation of ORF1b involves a − 1 ribosomal frameshift, generating a 7128-amino acid pp1ab. The ribosomal frameshift is assumed to be directed by two signals in the ORF1a/1b overlapping region: a slippery sequence 5′UUUAAAC3′ (nt 13,472–13,478) and a predicted downstream RNA pseudoknot structure (nt 13,484–13,559) (see Supplementary Fig. S3). The pp1a and pp1ab proteins are predicted to be proteolytically processed by viral-encoded proteases into 16 non-structural proteins (nsp1–16, Table 2 ) required for viral replication and transcription. By comparison to other coronaviruses, a number of putative functional domains are predicted in the ECoV pp1a and pp1ab and these are summarized in Fig. 1 and Table 2 (Gorbalenya et al., 1991, Gorbalenya et al., 2006, Snijder et al., 2003, Ziebuhr, 2005, Ziebuhr et al., 2001). Enzymatic activities of nsp3, nsp5, nsp12, nsp13, nsp14 and nsp15 have been experimentally confirmed for some coronaviruses (Barretto et al., 2005, Cheng et al., 2005, Guarino et al., 2005, Heusipp et al., 1997, Ivanov et al., 2004a, Ivanov et al., 2004b, Ivanov and Ziebuhr, 2004, Lindner et al., 2005, Minskaia et al., 2006, Putics et al., 2005, Putics et al., 2006, Seybert et al., 2000, Seybert et al., 2005, Tanner et al., 2003, Ziebuhr, 2005, Ziebuhr et al., 2001). The 3CLpro (catalytic residues His-3333 and Cys-3437) is predicted to cleave the C-terminal half of the ECoV pp1a and the ORF1b-encoded part of pp1ab. The putative PL1pro (catalytic residues Cys-1078 and His-1229) and PL2pro (catalytic residues Cys-1675 and His-1832) are predicted to process the N-proximal regions of the ECoV pp1a (Fig. 1 and Table 2).
Table 2.
Predicted end-products of proteolytic processing of the ECoV replicase polyproteins pp1a and pp1ab
| Cleavage product | Nucleotide positiona | Polyprotein | Position in pp1a/pp1ab (aa) | Length (aa) | Putative funcitional domain(s)b | Putative proteases predicted to release protein from polyproteins |
|---|---|---|---|---|---|---|
| nsp1 | 210–941 | pp1a/pp1ab | 1Met-Gly244 | 244 | PL1pro | |
| nsp2 | 942–2744 | pp1a/pp1ab | 245Val-Ala845 | 601 | PL1pro | |
| nsp3 | 2745–8597 | pp1a/pp1ab | 846Gly-Gly2796 | 1951 | Ac, PL1pro, ADRP, PL2pro, TM1, Y | PL2pro |
| nsp4 | 8598–10,085 | pp1a/pp1ab | 2797Ala-Gln3292 | 496 | TM2 | PL2pro + 3CLpro |
| nsp5 | 10,086–10,994 | pp1a/pp1ab | 3293Ser-Gln3595 | 303 | 3CLpro | 3CLpro |
| nsp6 | 10,995–11,855 | pp1a/pp1ab | 3596Ser-Gln3882 | 287 | TM3 | 3CLpro |
| nsp7 | 11,856–12,122 | pp1a/pp1ab | 3883Ser-Gln3971 | 89 | Part of RNA binding hexadecameric supercomplex | 3CLpro |
| nsp8 | 12,123–12,713 | pp1a/pp1ab | 3972Ala-Gln4168 | 197 | Part of RNA binding hexadecameric supercomplex | 3CLpro |
| nsp9 | 12,714–13,043 | pp1a/pp1ab | 4169Asn-Gln4278 | 110 | ssRNA-binding protein | 3CLpro |
| nsp10 | 13,044–13,454 | pp1a/pp1ab | 4279Ala-Gln4415 | 137 | 2 zinc fingers | 3CLpro |
| nsp11 | 13,455–13,496 | pp1a | 4416Ser-Ser4429 | 14 | 3CLpro | |
| nsp12 | 13,455–16,237 | pp1ab | 4416Ser-Gln5343 | 928 | RdRp | 3CLpro |
| nsp13 | 16,238–18,034 | pp1ab | 5344Ser-Gln5942 | 599 | ZBD, HEL | 3CLpro |
| nsp14 | 18,035–19,597 | pp1ab | 5943Cys-Gln6463 | 521 | Exonuclease (ExoN) | 3CLpro |
| nsp15 | 19,598–20,695 | pp1ab | 6464Ser-Gln6829 | 366 | NendoU | 3CLpro |
| nsp16 | 20,696–21,592 | pp1ab | 6830Ala-Ile7128 | 299 | 2′-O-MT | 3CLpro |
Domains Ac and Y are described by Ziebuhr et al. (2001).
Nucleotide position means the location of the nucleotides encoding corresponding proteins in the entire genome of equine coronavirus-NC99 strain.
PL1pro, papain-like proteinase 1; PL2pro, papain-like proteinase 2; ADRP, adenosine diphosphate-ribose 1ʺ-phosphatase (formerly known as ‘X-domain’); 3CLpro, 3C-like proteinase; TM, transmembrane domain; GFL, growth factor-like domain; RdRp, RNA-dependent RNA polymerase; ZBD, zinc-binding domain; HEL, helicase domain; NendoU, nidoviral uridylate-specific endoribonuclease; 2′-O-MT, 2′-O-ribose methyltransferase.
The most striking differences between the ECoV replicase and other group 2 coronaviruses replicases were identified in nsp3. The ECoV nsp3 protein has 3 aa deletions and 55 aa insertions compared to the nsp3 proteins of BCoV, HCoV-OC43, and PHEV, three viruses phylogenetically most closely related to ECoV. These insertions and deletions are clustered at two regions: the Ac domain and the region between the PL2pro and the Y domain. The functional significance of these insertions and deletions is unknown as yet; however, the functions of PL1pro, PL2pro, and ADRP are not anticipated to be affected since insertions and deletions are not located in the functional domains of these enzymes (Fig. 1).
ORF2 (nt 21,610–22,446) of ECoV-NC99 encodes the predicted NS2 protein with 278 amino acids. The NS2 of ECoV has 67%, 67%, and 45% amino acid identity with the respective NS2 proteins of BCoV, HCoV-OC43, and PHEV. The lower amino acid identity with PHEV may be attributable to the fact that PHEV has a truncated NS2 protein (Vijgen et al., 2006). Sequence analysis revealed that the ECoV NS2 protein contains a domain (aa 46–135) with similarity to the putative cyclic phosphodiesterase (CPD, Martzen et al., 1999). The CPD domain has also been identified in the NS2 proteins of other group 2a coronaviruses as well as in the 3′end of the pp1a protein of toroviruses (Gorbalenya et al., 2006, Snijder et al., 1991, Snijder et al., 2003). The NS2 of ECoV was predicted to contain 9 potential phosphorylation sites. The NS2 of ECoV does not contain a signal peptide and is a non-secretory protein. The function of the NS2 protein in coronaviruses has not been studied in detail. It is known that the NS2 gene is non-essential for MHV replication in transformed cells (Schwarz et al., 1990). However, a recent study showed that a point mutation in the NS2 of MHV led to its attenuation in mice in spite of its wild-type replication in tissue culture (Sperry et al., 2005).
ORF3 (nt 22,458–23,729) of ECoV-NC99 encodes the predicted HE protein containing 423 amino acids. Nine potential N-glycosylation sites were predicted. SignalP analysis revealed a signal peptide probability of 0.802 with a potential cleavage site between residues 17 and 18. It was predicted that the N-terminal 390 amino acids are located outside the cell surface or viral envelope with a transmembrane helix at amino acids 391–413 and an internal domain at amino acids 414–423. The putative active site for esterase activity, FGDS (Kienzle et al., 1990), is present at amino acids 36–39 of the HE protein in ECoV.
ORF4 (nt 23,744–27,835) of ECoV-NC99 encodes the predicted spike (S) protein containing 1363 amino acids. Eighteen potential N-glycosylation sites were predicted. An N-terminal signal peptide was identified with a potential cleavage site between amino acids 14 and 15 predicted by SignalP-NN or between amino acids 17 and 18 predicted by SignalP-HMM. The ECoV S protein was predicted to be a typical type I membrane protein with the N-terminal 1307 residues exposed on the outside of the cell surface or virus particle, a transmembrane domain near the C terminus (residues 1308–1330), followed by a cytoplasmic tail (residues 1331–1363). Following multiple alignments with the S proteins of other group 2a coronaviruses, a potential cleavage recognition sequence (RRQRR) was identified at residues 764–768 which would predict a cleavage between amino acids 768 and 769, separating the ECoV S protein into S1 and S2 subunits (Fig. 1). The ECoV S1 subunit is expected to contain a receptor-binding domain whose position has not yet been determined. The S2 subunit is predicted to mediate membrane fusion. Two heptad repeat (HR) regions, which are conserved in position and sequence among the three groups of coronaviruses and play important roles in membrane fusion (see reviews of Eckert and Kim, 2001, Hernandez et al., 1996), were identified in the ECoV S2 subunit (HR1: aa 991–1092; HR2: aa 1259–1304) (Fig. 1). The ECoV S2 subunit is anticipated to possess a fusion peptide whose position is yet unknown. Some coronavirus S proteins have been shown to contain important neutralization epitopes (Godet et al., 1994, Kubo et al., 1994, Yoo et al., 1991) and mutations in the S protein have been associated with altered viral antigenicity and pathogenicity (Ballesteros et al., 1997, Bernard and Laude, 1995, Dalziel et al., 1986, Gallagher and Buchmeier, 2001, Leparc-Goffart et al., 1997). Whether the S protein of ECoV has such properties remains to be determined.
ORF5 (nt 27,825–27,947) of ECoV-NC99 is predicted to encode a hypothetical protein of 40 amino acids with an estimated molecular weight of 4.7 kDa (termed p4.7 protein). It was predicted to be a non-secretory protein and did not contain any transmembrane helix. This protein is not closely matched to any known protein based on a search using BLASTP, PSI-BLAST, or FASTA programs.
ORF6 (nt 28,076–28,405) of ECoV-NC99 is predicted to encode a protein of 109 amino acids corresponding to the BCoV 12.7 kDa non-structural protein (p12.7). This ORF overlaps by 15 nucleotides with the ORF7 that encodes the E protein. No signal peptide or any transmembrane helix was present. No N-glycosylation site was found.
ORF7 (nt 28,392–28,646) of ECoV-NC99 encodes the predicted E protein containing 84 amino acids. No N-glycosylation site was identified. It was predicted to contain a signal anchor (probability 0.999). One transmembrane domain was predicted at residues 18–36 by TMpred analysis or at residues 15–37 by TMHMM analysis. Both programs predicted the N-terminus of the protein to be external to the cell surface or viral envelope. In the case of other coronaviruses, there is increasing evidence that the E protein together with the M protein is instrumental in viral assembly and budding; the cytoplasmic tails of both proteins have an important interactive role in this process (Corse and Machamer, 2000, Corse and Machamer, 2002, Corse and Machamer, 2003, Vennema et al., 1996).
ORF8 (nt 28,661–29,353) of ECoV-NC99 encodes the predicted M protein containing 230 amino acids. It was predicted to contain a signal anchor (probability 0.947). Three transmembrane domains were predicted to be present at positions 25–46, 57–78, and 81–102 by TMpred analysis or at positions 25–44, 49–71, and 81–103 by TMHMM analysis. The N-terminal 24 amino acid residues were predicted to be outside and the C-terminal 127 or 128-amino acid hydrophilic domain was predicted to be inside the virus. One potential N-glycosylation site was predicted at position 26 (NFS). The presence of potential O-glycosylation sites was predicted at the extreme N-terminus of the M protein (MSSTPTPAPGYT). Whether these sites are glycosylated or not needs to be experimentally verified. Previous studies have shown that the M protein of group 1 and 3 coronaviruses (e.g. TGEV and IBV) are N-glycosylated, whereas the M protein of group 2 coronavirus MHV is only O-glycosylated (de Haan et al., 2002, Lai et al., 2006). The M protein is the most abundant envelope component and plays a key role in coronavirus assembly by interacting with the E, S, N and HE proteins (Bosch et al., 2005, de Haan and Rottier, 2005).
ORF9a (nt 29,363–30,703) of ECoV-NC99 encodes the predicted N protein containing 446 amino acids. It was predicted to contain 36 potential phosphorylation sites. No signal peptide or any transmembrane helix was present. The N protein of coronaviruses has been shown to be multifunctional, e.g. interaction with the viral RNA genome to form a viral nucleocapsid, interaction with the M protein, and the ability for self-association (Masters, 1992, Narayanan et al., 2000, Narayanan et al., 2003). Recently it has also been reported that the N protein may play a role in coronavirus replication (Almazan et al., 2004, Schelle et al., 2005).
ORF9b (nt 29,424–30,044) of ECoV-NC99 encodes a hypothetical protein (I) containing 206 amino acids within ORF9a which encodes the N protein. It was predicted to contain 10 potential phosphorylation sites. No signal peptide or any transmembrane helix was present. In the case of MHV, expression of the protein I has been detected in virus-infected cells but this protein is nonessential for viral replication and viral production (Fischer et al., 1997).
Northern blot analysis of ECoV genomic and subgenomic mRNAs
It is generally accepted that the replicase proteins are directly synthesized from the coronavirus genome, whereas the structural and accessory proteins are expressed from a nested set of subgenomic mRNAs. However, the number of sg mRNAs and the characteristics and expression pattern of the proteins they encode (e.g. a sg mRNA may sometimes express multiple proteins) varies for each virus. In order to investigate ECoV sg mRNA synthesis, Northern blot analysis was performed to evaluate the synthesis of genomic and subgenomic RNAs in ECoV-infected cells. A digoxigenin-labeled RNA probe complementary to the 3′ end (nt 30,660–30,946) of the ECoV genome was used for a Northern blot hybridization analysis. As shown in Fig. 2 , nine mRNAs were detected in ECoV-infected HRT-18G cells at 72 h p.i. Absence of such mRNAs in mock-infected cells confirms that these mRNAs are ECoV-specific. According to the estimated sizes of the mRNAs, it is reasonable to assume that sg mRNAs 2–8 express the NS2, HE, S, p4.7, p12.7, E, and M proteins, respectively and that mRNA 9 expresses the N protein and probably the I protein as well.
Fig. 2.
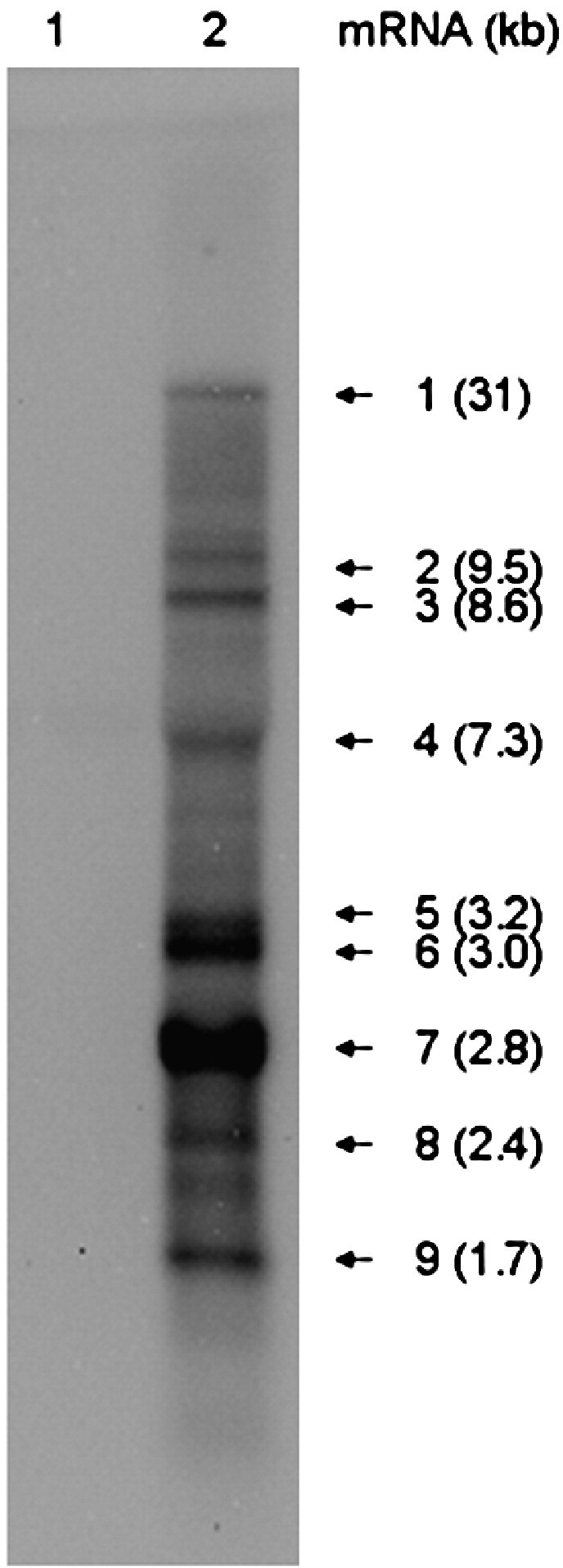
Northern blot analysis of intracellular RNA isolated from ECoV-infected HRT-18G cells. A DIG-labeled probe which was complementary to the 3′ end (nt 30,660–30,946) of ECoV genome was used to detect the genomic and subgenomic mRNAs in ECoV-infected (lane 2) and mock-infected (lane 1) HRT-18G cells at 72 h p.i.
Determination of leader–body junction sequences of sg mRNAs
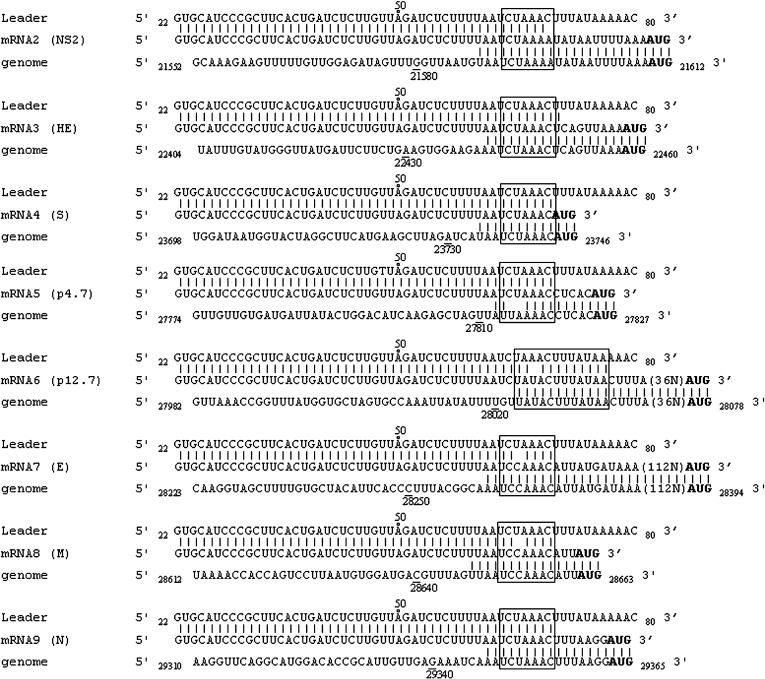
There is a general agreement that the TRS elements determine the fusion sites of the 5′ leader and the 3′ body segments in coronavirus sg mRNAs. In order to determine the precise location of the leader and body TRSs used for ECoV sg mRNA synthesis, the leader–body junction and flanking sequences of each ECoV sg mRNA were determined using sg mRNA-specific RT–PCRs (see Table 3 and Materials and methods for details). The sg mRNA sequences were aligned to the leader and corresponding ‘body’ genomes as shown in Fig. 3 . Analysis of the leader–body junction sequences revealed that the core sequence of the TRS motifs is 5′UCUAAAC3′. The leader TRS (5′UCUAAAC3′) and the body TRS (5′UCUAAAC3′) used for synthesizing HE mRNA, S mRNA, and N mRNA exactly match each other. There is one mismatch between the leader TRS and the body TRS (5′UCUAAAA3′) used for generating the mRNA of the NS2 protein. There is also one mismatch between the leader TRS and the body TRS (5′UCCAAAC3′) used for generating E mRNA and M mRNA. There are two mismatches between the leader TRS and the body TRS (5′UUAAAAC3′) used for generating the mRNA of the p4.7 protein. Interestingly, in the case of the mRNA of the p12.7 protein, the leader and the body segment is joined at the unusual consensus variant 5′UAAACUUUAUAA3′. Previously it has been shown that the mRNA of the p12.7 protein of BCoV also utilizes an unusual consensus variant for joining the leader and body segment (Hofmann et al., 1993). From the sequence data, we conclude that the ECoV common leader on sg mRNAs is the first 64 nucleotides of the ECoV genome.
Table 3.
Oligonucleotide primers used for RT–PCR amplification of the leader–body junction of sg mRNAs
| Primer ID | Position | Sequence (5′–3′) | Use |
|---|---|---|---|
| 22813N | 22,792–22,813 | GCGTTATCACCAGAAGCGGTGC | Reverse transcription for mRNA2 (NS2) and reverse primer for mRNA3 (HE) PCR |
| 25095N | 25,076–25,095 | CGCCTATTCCAGGCAGAAGG | Reverse transcription for mRNA3 (HE) and mRNA4 (S) |
| 29101N | 29,078–29,101 | GGCAGTAAGAGTATGATGGTCCTC | Reverse transcription for mRNA5 (p4.7), mRNA6 (p12.7) and mRNA7 (E) |
| 30945N | 30,921–30,945 | CTGGGTGGTAACTTAACATGCTGGC | Reverse transcription for mRNA8 (M) and mRNA9 (N) |
| 1P | 1–21 | GATTGTGAGCGAATTGCGTGC | Forward primer for all sg mRNA PCR |
| 21982N | 21,958–21,982 | GACGGGACTGACCAACTACACAACC | Reverse primer for mRNA2 (NS2) PCR |
| 24283N | 24,262–24,283 | GCGTGGTGACCCAATACCACTG | Reverse primer for mRNA4 (S) PCR |
| 28100N | 28,078–28,100 | TCCTCTCAGGTCTCCAGATGTCC | Reverse primer for mRNA5 (p4.7) PCR |
| 28334N | 28,312–28,334 | CAGCCTCCTCTATAGTATTGGCG | Reverse primer for mRNA6 (p12.7) PCR |
| 28641N | 28,617–28,641 | CGTCATCCACATTAAGGACTGGTGG | Reverse primer for mRNA7 (E) PCR |
| 29016N | 28,992–29,016 | GGGTTGAAACTCCACCAACTACCAG | Reverse primer for mRNA8 (M) PCR |
| 29710N | 29,691–29,710 | GCGTTGATTGCCATCGGCTG | Reverse primer for mRNA9 (N) PCR |
Fig. 3.
ECoV sg mRNA leader–body junction and flanking sequences. The sg mRNA sequences are shown in alignment with the leader and the genome sequences. The genomic positions of the nucleotides in the leader and genome sequences are indicated. The start codon AUG in each sg mRNA is depicted in bold. Boxed regions are the putative TRS used for each sg mRNA synthesis. The 36N and 112N in the parenthesis mean that 36 and 112 nucleotides at that region are not shown. Homologous nucleotides between the leader and the mRNA or between the mRNA and the genome are indicated with connecting lines.
Phylogenetic analysis of ECoV
Phylogenetic analyses of ECoV and other coronaviruses were performed based on the amino acid sequences of replicase polyprotein pp1a, the ORF1b-encoded part of the pp1ab, S, E, M, and N. Phylogenetic analysis clustered coronaviruses into three major groups (G1, G2a, and G3) irrespective of the gene used for analysis (Fig. 4 ). The SARS-CoV forms a separate branch and is classified as subgroup 2b (G2b) as suggested previously (Gorbalenya et al., 2004, Snijder et al., 2003). Phylogenetic analysis clearly demonstrated that ECoV falls into the cluster of group 2a coronaviruses and is most closely related to BCoV, HCoV-OC43, and PHEV.
Fig. 4.
Phylogenetic analysis of the amino acid sequences of replicase polyprotein pp1a, the ORF1b-encoded part of the pp1ab, spike (S), envelope (E), membrane (M), and nucleocapsid (N) of ECoV-NC99. Multiple amino acid sequence alignments were carried out by using ClustalX 1.83 and the unrooted neighbor-joining trees were constructed using PAUP 4.0b10. Bootstrap analysis was carried out on 1000 replicate data sets. CCoV, canine coronavirus (GenBank accession number D13096); TGEV, porcine transmissible gastroenteritis virus Purdue (NC_002306); FCoV, feline coronavirus (NC_007025); HCoV-NL63, human coronavirus NL63 (NC_005831); HCoV-229E, human coronavirus 229E (NC_002645); PEDV, porcine epidemic diarrhea virus CV777 (NC_003436); BCoV, bovine coronavirus ENT (NC_003045); HCoV-OC43, human coronavirus OC43 strain VR759 (NC_005147); PHEV, porcine hemagglutinating encephalomyelitis virus VW572 (DQ011855); MHV, murine hepatitis viruses A59 (NC_001846) and JHM (NC_006852); SDAV, rat sialodacryoadenitis coronavirus (AF207551); HCoV-HKU1, coronavirus HKU1 (NC_006577); SARS-CoV, SARS coronavirus Tor2 (NC_004718); IBV, avian infectious bronchitis virus Beaudette (NC_001451).
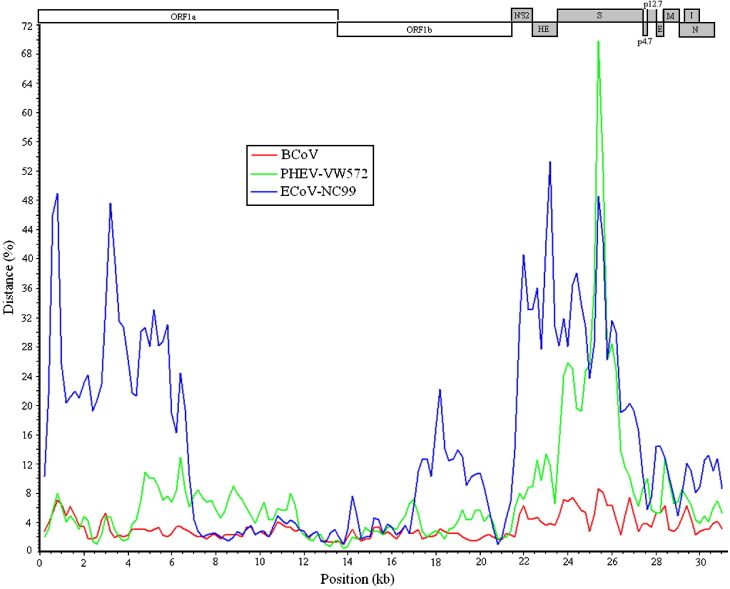
To further explore the possible evolutionary relationships among ECoV, BCoV, HCoV-OC43, and PHEV, the genetic distances of ECoV, BCoV, and PHEV to HCoV-OC43 were determined over the entire genome using the SimPlot analysis (Lole et al., 1999). As shown in Fig. 5 , the BCoV strains and HCoV-OC43 had lowest genetic distances over the complete genome; the genetic distance between PHEV and HCoV-OC43 was similar to the distance between BCoV and HCoV-OC43 in most regions of the genome with exception of the spike gene where the genetic distance of PHEV to HCoV-OC43 was significantly greater than the distance of BCoV to HCoV-OC43; the genetic distance of ECoV to HCoV-OC43 was significantly greater than the distance of either BCoV or PHEV to HCoV-OC43 in the regions of the first half of ORF1a, the central part of ORF1b, NS2 and HE genes; the genetic distance with respect to the spike gene between ECoV and HCoV-OC43 was similar to the distance between PHEV and HCoV-OC43 but greatly higher than the distance between BCoV and HCoV-OC43. The genetic distances of BCoV and PHEV to HCoV-OC43 observed in this study are consistent with previously reported findings (Vijgen et al., 2005, Vijgen et al., 2006). Vijgen et al., 2006, Vijgen et al., 2005 concluded that PHEV diverged from the common ancestor before BCoV and HCoV-OC43. Our analysis suggested that ECoV had diverged earlier than PHEV from a common ancestor. In summary, ECoV had emerged earlier than PHEV, BCoV, and HCoV-OC43, notwithstanding the fact that ECoV was not isolated until 1999 from a diarrheic foal in USA.
Fig. 5.
Genetic distance between ECoV, BCoV, PHEV and HCoV-OC43. The average genetic distances were calculated over the entire genome using the SimPlot program with a sliding window size of 400 bp and a step size of 200 bp. Each curve represents a comparison of the sequence data of ECoV-NC99, the BCoV strains, and PHEV-VW572 to the reference sequence data of the HCoV-OC43 ATCC strain VR759 (NC_005147). The sequence data of the BCoV strains used for comparison are the 50% consensus sequence of six BCoV strains: BCoV-ENT (NC_003045), BCoV-Alpaca (DQ915164), BCoV-DB2 (DQ811784), BCoV-Mebus (U00735), BCoV-Quebec (AF220295), and BCoV-LUN (AF391542). The linear representation of the ECoV-NC99 genome was shown at the top of the diagram.
Conclusion
In this study, we have determined the first complete genome sequence of ECoV and provided the first comprehensive analysis of the ECoV genome. Completion of the genome sequence of ECoV will contribute to our understanding of this virus at the molecular level and also enrich the database of coronaviruses. The sequence data are expected to aid in the development of diagnostic and prophylactic reagents. The sequence data of ECoV-NC99 will also help identify and characterize other ECoV isolates and enhance our understanding of the molecular epidemiology of coronavirus. Neonatal enterocolitis is an economically significant disease for horse breeders. Further studies are needed to determine the prevalence of ECoV infection in equine populations and the relative role of ECoV as a cause of enteric disease in horses.
Materials and methods
Cells and virus
The human rectal tumor cell line HRT-18G (American Type Culture Collection [ATCC, CRL-11663]) was grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 4 mM l-glutamine, 5% fetal bovine serum, and penicillin/streptomycin at 37 °C in the presence of 5% CO2. The equine coronavirus-NC99 (Guy et al., 2000) was propagated once in HRT-18G cells to produce the working virus stocks.
Isolation of viral RNA, RT–PCR amplification and sequencing
The complete genome of ECoV was determined by sequencing 11 overlapping RT–PCR products encompassing the entire genome (nt 1–3615; nt 3446–5458; nt 4953–6600; nt 5497–9678; nt 9347–13,021; nt 12,451–15,736; nt 15,425–19,307; nt 19,039–22,812; nt 22,566–26,390; nt 26,065–29,662; and nt 29,363–30,992). Viral RNA was isolated from ECoV stocks using the QIAamp viral RNA mini kit (Qiagen). Viral RNA was first reverse transcribed with AccuScript reverse transcriptase (Stratagene) following the manufacturer's instructions. Then, PCR amplification was performed with proof-reading PfuUltra high-fidelity DNA polymerase (Stratagene) in a volume of 50 μl: 5 μl PfuUltra PCR buffer (10×), 1.0 μl dNTP mix (10 mM each), 1 μl of each primer (20 μM), 2 μl cDNA template, 1 μl PfuUltra DNA polymerase, and 39.0 μl nuclease-free water. The reaction mixtures were incubated at 95 °C for 2 min, followed by 35 cycles of amplification at 95 °C for 45 s, 50–53 °C for 45 s, and 72 °C for 4.5 min, with a final incubation at 72 °C for 10 min. The PCR products were gel-purified using QIAquick gel extraction kit (Qiagen). Both sense and anti-sense strands were sequenced using the Applied Biosystems Big Dye Terminator V3.0 sequencing chemistry on ABI 3730 DNA sequencers (Davis Sequencing Center). Partial genomic sequence (9487 nucleotides) of ECoV had been previously determined by two groups (Guy et al., 2000, GenBank accession number AF251144; Wu et al., 2003, AF523846 and AF523850. H.Y. Wu, J.S. Guy, and D.A. Brian, unpublished data, AY316300). These regions were re-sequenced in this study. To determine the remaining genomic sequence of ECoV-NC99, initial RT–PCR and sequencing primers were designed based on multiple alignments of the genomes of BCoV (GenBank accession number NC_003045), HCoV-OC43 (NC_005147), PHEV (DQ011855), and MHV (NC_001846); additional primers were designed based on the results of the first and subsequent rounds of sequencing. All of the primer sequences are attached in the Supplementary Table S1.
DNA and protein sequence analysis
The nucleotide sequences were assembled and manually edited using CodonCode Aligner version 1.5.2 to produce the complete sequence of the viral genome. ORF analysis was performed using Vector NTI Advance 10 (Invitrogen). RNA secondary structures of 5′ and 3′ UTRs and the ribosomal frameshift signals were predicted using the MFOLD program with the default parameter settings (Mathews et al., 1999, Zuker, 2003). Potential 3C-like protease cleavage sites were predicted using the NetCorona 1.0 server (Kiemer et al., 2004). Prediction of signal peptides and their cleavage sites was conducted using SignalP 3.0 server (Nielsen et al., 1997). Potential N-glycosylation sites, O-glycosylation sites, and phosphorylation sites were predicted using NetNGlyc, NetOGlyc, and NetPhos, respectively (Blom et al., 1999, Julenius et al., 2005). Prediction of transmembrane domains was performed using TMpred (Hofmann and Stoffel, 1993) and TMHMM server 2.0 (Sonnhammer et al., 1998). Protein similarity searches were performed using BLASTP version 2.2.16, PSI-BLAST against the Protein Data Bank (PDB) (Altschul et al., 1997, Schaffer et al., 2001) and FASTA version 34.26 against the uniprot protein database with the default parameter settings (Pearson and Lipman, 1988). Pairwise amino acid comparison was performed using EMBOSS Pairwise Alignment Algorithms with the default parameter settings (http://www.ebi.ac.uk/emboss/align). Multiple sequence alignments were performed using ClustalX version 1.83 (Thompson et al., 1997). Phylogenetic analysis and unrooted neighbor-joining trees were carried out using PAUP version 4.0b10 with the default parameter settings. Bootstrap analysis was carried out on 1000 replicate data sets. The genetic distance between genomes was determined using the SimPlot version 3.5.1 (Lole et al., 1999).
Analysis of viral RNA by Northern blotting
One anti-sense RNA probe base pairing to the 3′ end of the ECoV genome (nt 30,660–30,946) was developed to evaluate the synthesis of genomic and subgenomic RNAs in ECoV-infected cells by Northern blotting. The ECoV RNA was amplified using two primer pairs (forward primer 30660P: 5′AGCAGATGGATGATCCCCTC3′; reverse primer 30946N: 5′ACTGGGTGGTAACTTAACATGCTG3′) and the QIAgen One-step RT–PCR kit (Qiagen). The gel-purified RT–PCR products were cloned into a linearized plasmid vector with overhanging 3′ T residues (pDrive Cloning Vector, Qiagen). The authenticity and orientation of the insert was determined by sequencing both strands of DNA with M13 reverse and forward primers. Plasmid DNA was linearized with BamHI (Roche), phenol/chloroform extracted, ethanol precipitated, and resuspended in nuclease-free water. A digoxigenin (DIG)-labeled RNA probe was prepared using the DIG RNA labeling kit (Roche) according to the manufacturer's instructions.
Intracellular RNA was extracted at 72 h p.i. from ECoV-infected HRT-18G cells using the RNAqueous-4PCR kit (Ambion). Northern hybridization with the DIG-labeled RNA probe was carried out following the protocols that had been previously described for equine arteritis virus (Balasuriya et al., 2004).
Determination of the leader–body junction sequence
The leader–body junction sites of all ECoV sg mRNAs were RT–PCR amplified and sequenced. Briefly, intracellular RNA was extracted from ECoV-infected HRT-18G cells using the RNAqueous-4PCR kit (Ambion). Reverse transcription was carried out with an RT primer located downstream to the body TRS region in a sg mRNA (Table 3) using SuperscriptIII reverse transcriptase (Invitrogen) following the manufacturer's instructions. Due to the nested nature of sg mRNAs, such an RT primer also binds to the corresponding positions in all larger viral mRNAs, including the genomic RNA. Subsequently, cDNA was PCR amplified with a forward primer (1P) located in the leader sequence and a reverse primer located just upstream of the RT primer in the body of the mRNA (Table 3). Amplification was performed in a volume of 50 μl: 5 μl PfuTurbo PCR buffer (10×), 0.4 μl dNTP mix (25 mM each), 1 μl of each primer (20 μM), 2 μl cDNA template, 1 μl PfuTurbo® DNA polymerase, and 39.6 μl nuclease-free water. The reaction mixtures were incubated at 95 °C for 2 min, followed by 35 cycles at 95 °C for 45 s, 50–56 °C for 45 s, and 72 °C for 3 min, with a final incubation at 72 °C for 10 min. RT–PCR products corresponding to each mRNA species could be distinguished by size differences on agarose gel. PCR products were gel-purified and sequenced to obtain the leader–body junction sequences for each sg mRNA.
Nucleotide sequence accession number
The nucleotide sequence of ECoV was deposited in GenBank under the accession number EF446615.
Acknowledgments
This work was partly supported by funds from Fort Dodge Animal Health and Kentucky Agricultural Experiment Station, College of Agriculture, University of Kentucky.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.virol.2007.06.035.
Appendix A. Supplementary data
Oligonucleotide primers used for RT–PCR amplification and sequencing of the entire genome of equine coronavirus NC99*.
Supplementary Fig. S1.
Model of the predicted secondary structure of the 5′ UTR of the ECoV-NC99 genome. The secondary structure was predicted using the MFOLD program with the default parameter settings and five structures were generated. The free energy of the structure shown here is dG = − 48.41. The other four predicted structures are all in agreement with this structure in terms of step–loop structures I, III, and IV. However, two structures agree and two other structures disagree with the step–loop II described in the structure shown here. The leader-associated transcription regulatory sequence UCUAAAC (nt 63 to 69) is shown in bold. The start and stop codons for the intra-5′ UTR short ORF (nt 99 to 125) and the start codon for ORF1a are boxed. The bulged stem–loop III (nt 96–115) and IV (189–208) strikingly resemble the stem–loop III and IV identified in bovine coronavirus and other group 2 coronaviruses (Raman and Brian, 2005, Raman et al., 2003, Wu et al., 2003).
Supplementary Fig. S2.
Model of the predicted secondary structure of the 3′ UTR of the ECoV-NC99 genome. The secondary structure was predicted using the MFOLD program with the default parameter settings. Only one structure was generated with a free energy dG = − 37. The shown region (nt 30,701 to 30,824) in the 3′ UTR is predicted to contain a bulged stem–loop structure (nt 30,703–30,770) and a pseudoknot structure (nt 30,766–30,819). The stop codon of the nucleocapsid gene is bolded. Similar putative bulged stem–loop structure and pseudoknot structure have been identified in murine hepatitis virus and other group 2 coronaviruses and have been shown to be essential for viral replication (Goebel et al., 2004a, Goebel et al., 2004b, Hsue and Masters, 1997, Hsue et al., 2000, Williams et al., 1999).
Supplementary Fig. S3.
Model showing the ECoV ORF1a/1b frameshift-directing signals. The secondary structure was predicted using the MFOLD program with the default parameter settings and four structures were generated. The free energy of the structure shown here is dG = − 29.2. The other three predicted structures are in general agreement with the structure shown here. The slippery sequence 5′UUUAAAC3′ (nt 13,472–13,478, indicated in bold) and a downstream predicted pseudoknotted structure (nt 13,484–13,559) in the ORF1a/1b overlapping region are assumed to direct the − 1 ribosomal frameshift. The stop codon for the ORF1a is boxed.
References
- Almazan F., Galan C., Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004;78(22):12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasuriya U.B., Hedges J.F., Smalley V.L., Navarrette A., McCollum W.H., Timoney P.J., Snijder E.J., MacLachlan N.J. Genetic characterization of equine arteritis virus during persistent infection of stallions. J. Gen. Virol. 2004;85(Pt. 2):379–390. doi: 10.1099/vir.0.19545-0. [DOI] [PubMed] [Google Scholar]
- Ballesteros M.L., Sanchez C.M., Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227(2):378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79(24):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard S., Laude H. Site-specific alteration of transmissible gastroenteritis virus spike protein results in markedly reduced pathogenicity. J. Gen. Virol. 1995;76(Pt. 9):2235–2241. doi: 10.1099/0022-1317-76-9-2235. [DOI] [PubMed] [Google Scholar]
- Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294(5):1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., de Haan C.A., Smits S.L., Rottier P.J. Spike protein assembly into the coronavirion: exploring the limits of its sequence requirements. Virology. 2005;334(2):306–318. doi: 10.1016/j.virol.2005.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Boursnell M.E., Binns M.M., Bilimoria B., Blok V.C., Brown T.D., Inglis S.C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34(6):439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Cheng A., Zhang W., Xie Y., Jiang W., Arnold E., Sarafianos S.G., Ding J. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology. 2005;335(2):165–176. doi: 10.1016/j.virol.2005.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Poon L.L., Chan K.H., Chen H., Guan Y., Yuen K.Y., Peiris J.S. Coronaviruses in bent-winged bats (Miniopterus spp.) J. Gen. Virol. 2006;87(Pt. 9):2461–2466. doi: 10.1099/vir.0.82203-0. [DOI] [PubMed] [Google Scholar]
- Corse E., Machamer C.E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 2000;74(9):4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse E., Machamer C.E. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 2002;76(3):1273–1284. doi: 10.1128/JVI.76.3.1273-1284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse E., Machamer C.E. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology. 2003;312(1):25–34. doi: 10.1016/S0042-6822(03)00175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel R.G., Lampert P.W., Talbot P.J., Buchmeier M.J. Site-specific alteration of murine hepatitis virus type 4 peplomer glycoprotein E2 results in reduced neurovirulence. J. Virol. 1986;59(2):463–471. doi: 10.1128/jvi.59.2.463-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Rottier P.J. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., de Wit M., Kuo L., Montalto C., Masters P.S., Weiss S.R., Rottier P.J. O-Glycosylation of the mouse hepatitis coronavirus membrane protein. Virus Res. 2002;82(1–2):77–81. doi: 10.1016/S0168-1702(01)00390-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Fischer F., Peng D., Hingley S.T., Weiss S.R., Masters P.S. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J. Virol. 1997;71(2):996–1003. doi: 10.1128/jvi.71.2.996-1003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279(2):371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet M., Grosclaude J., Delmas B., Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 1994;68(12):8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S.J., Hsue B., Dombrowski T.F., Masters P.S. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2004;78(2):669–682. doi: 10.1128/JVI.78.2.669-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S.J., Taylor J., Masters P.S. The 3′ cis-acting genomic replication element of the severe acute respiratory syndrome coronavirus can function in the murine coronavirus genome. J. Virol. 2004;78(14):7846–7851. doi: 10.1128/JVI.78.14.7846-7851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J.M., Gomez-Puertas P., Cavanagh D., Gorbalenya A.E., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003;148(11):2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Lai M.M. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 1991;288(1–2):201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Snijder E.J., Spaan W.J. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 2004;78(15):7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino L.A., Bhardwaj K., Dong W., Sun J., Holzenburg A., Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 2005;353(5):1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J.S., Breslin J.J., Breuhaus B., Vivrette S., Smith L.G. Characterization of a coronavirus isolated from a diarrheic foal. J. Clin. Microbiol. 2000;38(12):4523–4526. doi: 10.1128/jcm.38.12.4523-4526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez L.D., Hoffman L.R., Wolfsberg T.G., White J.M. Virus–cell and cell–cell fusion. Annu. Rev. Cell Dev. Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- Heusipp G., Harms U., Siddell S.G., Ziebuhr J. Identification of an ATPase activity associated with a 71-kilodalton polypeptide encoded in gene 1 of the human coronavirus 229E. J. Virol. 1997;71(7):5631–5634. doi: 10.1128/jvi.71.7.5631-5634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K., Stoffel W. TMbase – a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Hofmann M.A., Chang R.Y., Ku S., Brian D.A. Leader–mRNA junction sequences are unique for each subgenomic mRNA species in the bovine coronavirus and remain so throughout persistent infection. Virology. 1993;196(1):163–171. doi: 10.1006/viro.1993.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V. Coronaviruses. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Straus S.E., editors. Fields Virology. 4th ed. Lippincott Williams and Wilkins; Philadelphia: 2001. pp. 1187–1203. [Google Scholar]
- Hsue B., Masters P.S. A bulged stem–loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 1997;71(10):7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsue B., Hartshorne T., Masters P.S. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2000;74(15):6911–6921. doi: 10.1128/jvi.74.15.6911-6921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 2004;78(14):7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. U.S.A. 2004;101(34):12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78(11):5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julenius K., Molgaard A., Gupta R., Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15(2):153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- Kiemer L., Lund O., Brunak S., Blom N. Coronavirus 3CLpro proteinase cleavage sites: possible relevance to SARS virus pathology. BMC Bioinformatics. 2004;5:72. doi: 10.1186/1471-2105-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienzle T.E., Abraham S., Hogue B.G., Brian D.A. Structure and orientation of expressed bovine coronavirus hemagglutinin–esterase protein. J. Virol. 1990;64(4):1834–1838. doi: 10.1128/jvi.64.4.1834-1838.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kubo H., Yamada Y.K., Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 1994;68(9):5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Perlman S., Anderson L.J. Coronaviridae. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., editors. Fields Virology. 5th ed. Lippincott Williams and Wilkins; Philadelphia: 2006. pp. 1305–1335. [Google Scholar]
- Leparc-Goffart I., Hingley S.T., Chua M.M., Jiang X., Lavi E., Weiss S.R. Altered pathogenesis of a mutant of the murine coronavirus MHV-A59 is associated with a Q159L amino acid substitution in the spike protein. Virology. 1997;239(1):1–10. doi: 10.1006/viro.1997.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Menard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79(24):15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73(1):152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martzen M.R., McCraith S.M., Spinelli S.L., Torres F.M., Fields S., Grayhack E.J., Phizicky E.M. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286(5442):1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- Masters P.S. Localization of an RNA-binding domain in the nucleocapsid protein of the coronavirus mouse hepatitis virus. Arch. Virol. 1992;125(1–4):141–160. doi: 10.1007/BF01309634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288(5):911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2006;103(13):5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74(17):8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Kim K.H., Makino S. Characterization of N protein self-association in coronavirus ribonucleoprotein complexes. Virus Res. 2003;98(2):131–140. doi: 10.1016/j.virusres.2003.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht J., Brunak S., von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10(1):1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Pasternak A.O., Spaan W.J., Snijder E.J. Nidovirus transcription: how to make sense…? J. Gen. Virol. 2006;87(Pt. 6):1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U.S.A. 1988;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Chu D.K., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H., Lau S.K., Woo P.C., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S. Identification of a novel coronavirus in bats. J. Virol. 2005;79(4):2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Filipowicz W., Hall J., Gorbalenya A.E., Ziebuhr J. ADP-ribose-1ʺ-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 2005;79(20):12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Gorbalenya A.E., Ziebuhr J. Identification of protease and ADP-ribose 1ʺ-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J. Gen. Virol. 2006;87(Pt. 3):651–656. doi: 10.1099/vir.0.81596-0. [DOI] [PubMed] [Google Scholar]
- Raman S., Brian D.A. Stem–loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2005;79(19):12,434–12,446. doi: 10.1128/JVI.79.19.12434-12446.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S., Bouma P., Williams G.D., Brian D.A. Stem–loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2003;77(12):6720–6730. doi: 10.1128/JVI.77.12.6720-6730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Li W., Yu M., Hao P., Zhang Y., Zhou P., Zhang S., Zhao G., Zhong Y., Wang S., Wang L.F., Shi Z. Full-length genome sequences of two SARS-like coronaviruses in horseshoe bats and genetic variation analysis. J. Gen. Virol. 2006;87(Pt. 11):3355–3359. doi: 10.1099/vir.0.82220-0. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81(1):20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer A.A., Aravind L., Madden T.L., Shavirin S., Spouge J.L., Wolf Y.I., Koonin E.V., Altschul S.F. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29(14):2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelle B., Karl N., Ludewig B., Siddell S.G., Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 2005;79(11):6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz B., Routledge E., Siddell S.G. Murine coronavirus nonstructural protein ns2 is not essential for virus replication in transformed cells. J. Virol. 1990;64(10):4784–4791. doi: 10.1128/jvi.64.10.4784-4791.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., Hegyi A., Siddell S.G., Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 2000;6(7):1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., Posthuma C.C., van Dinten L.C., Snijder E.J., Gorbalenya A.E., Ziebuhr J. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J. Virol. 2005;79(2):696–704. doi: 10.1128/JVI.79.2.696-704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., den Boon J.A., Horzinek M.C., Spaan W.J. Comparison of the genome organization of toro- and coronaviruses: evidence for two nonhomologous RNA recombination events during Berne virus evolution. Virology. 1991;180(1):448–452. doi: 10.1016/0042-6822(91)90056-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L., von Heijne G., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Sperry S.M., Kazi L., Graham R.L., Baric R.S., Weiss S.R., Denison M.R. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 2005;79(6):3391–3400. doi: 10.1128/JVI.79.6.3391-3400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner J.A., Watt R.M., Chai Y.B., Lu L.Y., Lin M.C., Peiris J.S., Poon L.L., Kung H.F., Huang J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278(41):39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15(8):2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moes E., Thoelen I., Wollants E., Lemey P., Vandamme A.M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79(3):1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Lemey P., Maes P., Van Reeth K., Nauwynck H., Pensaert M., Van Ranst M. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006;80(14):7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G.D., Chang R.Y., Brian D.A. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 1999;73(10):8349–8355. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79(2):884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y., Guy J.S., Yoo D., Vlasak R., Urbach E., Brian D.A. Common RNA replication signals exist among group 2 coronaviruses: evidence for in vivo recombination between animal and human coronavirus molecules. Virology. 2003;315(1):174–183. doi: 10.1016/S0042-6822(03)00511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo D.W., Parker M.D., Song J., Cox G.J., Deregt D., Babiuk L.A. Structural analysis of the conformational domains involved in neutralization of bovine coronavirus using deletion mutants of the spike glycoprotein S1 subunit expressed by recombinant baculoviruses. Virology. 1991;183(1):91–98. doi: 10.1016/0042-6822(91)90121-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Curr. Top Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81(Pt. 4):853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J., Thiel V., Gorbalenya A.E. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 2001;276(35):33220–33232. doi: 10.1074/jbc.M104097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotide primers used for RT–PCR amplification and sequencing of the entire genome of equine coronavirus NC99*.