Abstract
Using the complete genome sequences of 19 coronavirus genomes, we analyzed the codon usage bias, dinucleotide relative abundance and cytosine deamination in coronavirus genomes. Of the eight codons that contain CpG, six were markedly suppressed. The mean NNU/NNC ratio of the six amino acids using either NNC or NNU as codon is 3.262, suggesting cytosine deamination. Among the 16 dinucleotides, CpG was most markedly suppressed (mean relative abundance 0.509). No correlation was observed between CpG abundance and mean NNU/NNC ratio. Among the 19 coronaviruses, CoV-HKU1 showed the most extreme codon usage bias and extremely high NNU/NNC ratio of 8.835. Cytosine deamination and selection of CpG suppressed clones by the immune system are the two major independent biochemical and biological selective forces that shape codon usage bias in coronavirus genomes. The underlying mechanism for the extreme codon usage bias, cytosine deamination and G + C content in CoV-HKU1 warrants further studies.
Keywords: Coronavirus, Cytosine deamination, CpG suppression, Codon usage bias
Introduction
Codon usage bias is one of the most important indicators of the selective forces that shape genome evolution. In general, codon usage bias may be a result of mutation pressure and/or relative abundance of the corresponding acceptor tRNA molecules. For human RNA viruses, it has been observed in one study that codon usage bias was related to mutation pressure, G + C content, segmented nature of the genome and the route of transmission of the virus (Jenkins and Holmes, 2003). In other studies, it has been suggested that mutation pressure may result in bias in dinucleotide usage, such as CpG suppression, in small eukaryotic viruses (Karlin et al., 1994, Shackelton et al., 2006). Other factors, such as cytosine deamination, which results in C → U changes, have also been proposed to be responsible for shaping the G + C contents and GC skews of RNA viruses (Pyrc et al., 2004). Recently, it has been observed that codon usage is an important driving force in the evolution of astroviruses and small DNA viruses (Sewatanon et al., 2007, Van Hemert et al., 2007). Despite all these fragmented observations, no study has integrated the various factors and been able to explain the basis for codon usage bias in viruses successfully.
Coronaviruses are positive sense, single-stranded RNA (ssRNA) viruses found in a wide range of animals in which they can cause respiratory, enteric, hepatic and neurological diseases of varying severity. The sizes of the genomes of coronaviruses are about 30 kb, the largest among RNA viruses. Based on genotypic and serological characterization, coronaviruses were divided into three distinct groups (Brian and Baric, 2005, Lai and Cavanagh, 1997, Ziebuhr, 2004). As a result of the low fidelity of the RNA-dependent–RNA polymerases, the mutation rates of RNA virus genomes are high, in the order of 1 per 10,000 nucleotides replicated. Furthermore, the unique mechanism of viral replication has resulted in a high frequency of recombination in coronaviruses (Lai and Cavanagh, 1997, Woo et al., 2006b). Their tendency for recombination and high mutation rates have made their genomes highly plastic, allowed them to adapt to new hosts and ecological niches, and given them the potential to be good candidates for causing pandemics. These factors have made the study of coronavirus evolution particularly important, both biologically and for practical purposes (Grigoriev, 2004, Gu et al., 2004, Yap et al., 2003). However, the relative importance of the various selective forces that shape the codon usage bias in coronaviruses and their underlying biological and biochemical basis are still poorly understood.
The recent severe acute respiratory syndrome (SARS) epidemic, the discovery of SARS coronavirus (SARS-CoV) and identification of SARS-CoV-like viruses from Himalayan palm civets and a raccoon dog from wild live markets in China have led to a boost in interests in discovery of novel coronaviruses in both humans and animals (Guan et al., 2003, Marra et al., 2003, Peiris et al., 2003, Rota et al., 2003, Snijder et al., 2003, Woo et al., 2004). For human coronaviruses, in 2004, a novel group 1 human coronavirus, human coronavirus NL63 (HCoV-NL63), was reported (Fouchier et al., 2004, Van der Hoek et al., 2004); and in 2005, we described the discovery, complete genome sequence and molecular diversity of another novel group 2 human coronavirus, coronavirus HKU1 (CoV-HKU1) (Lau et al., 2006, Woo et al., 2005a, Woo et al., 2005b, Woo et al., 2005c, Woo et al., 2006b). As for animal coronaviruses, six group 1 (Poon et al., 2005, Tang et al., 2006, Woo et al., 2006a, Lau et al., 2007), six group 2, including bat SARS coronavirus, sable antelope coronavirus, giraffe coronavirus, and two new subgroups of group 2 coronaviruses (Lau et al., 2005, Li et al., 2005, Woo et al., 2006a, Woo et al., 2007), 11 group 3 (Cavanagh et al., 2002, East et al., 2004, Jonassen et al., 2005, Liu et al., 2005, Hasoksuz et al., 2007) coronaviruses, and two unclassified coronaviruses from Asian leopard cats and Chinese ferret badgers (Dong et al., 2007) have recently been described. Since the number of coronavirus species with complete genomes available has increased from 9 in 2003 to 19 in 2007, this has provided a golden opportunity to study genome evolution in coronaviruses.
In this study, we analyzed the codon usage bias, dinucleotide relative abundance, cytosine deamination in coronavirus genomes and the codon usage bias in the hosts of the various coronaviruses. The relative importance of the various forces in shaping the codon usage bias in the various coronaviruses and the extreme codon usage bias and cytosine deamination in CoV-HKU1 were also discussed.
Results
Codon usage in coronavirus genomes
The mean (S.D.) effective number of codons (Nc) of the 19 coronaviruses is 45.448 (4.207) (Table 1 ). The codon usage fractions in the 19 coronavirus genomes are shown in Table 2 . For all amino acids, the codon usage patterns of every individual coronavirus species are similar to the general codon usage patterns in coronaviruses. CoV-HKU1, HCoV-NL63, murine hepatitis virus (MHV) and bat coronavirus HKU5 (bat-CoV HKU5) are the four coronaviruses with relatively larger number of codons showing usage fractions outside the mean ± 2 S.D. usage fraction range of the corresponding codons, probably due to their relatively high (MHV and bat-CoV HKU5) or low (CoV-HKU1 and HCoV-NL63) G + C contents (Table 1, Table 2).
Table 1.
Coronavirus genomes used in the present study
| Coronavirus | Host | GenBank accession no. | Reference | Genome size (bases) | G + C content (%) | GC skew | Mononucleotide frequencies (%) |
Nc | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| G | A | U | C | ||||||||
| Group 1a | |||||||||||
| TGEV | Pig | NC_002306 | Almazan et al., 2000 | 28,586 | 37.5 | 0.097 | 20.6 | 29.5 | 32.9 | 17.0 | 44.737 |
| FIPV | Cat | AY994055 | Haijema et al., 2003 | 29,355 | 38.1 | 0.102 | 21.0 | 29.2 | 32.7 | 17.1 | 46.150 |
| PRCV | Pig | DQ811787 | Zhang et al., 2007 | 27,550 | 37.4 | 0.107 | 20.7 | 29.3 | 33.2 | 16.7 | 44.406 |
| Group 1b | |||||||||||
| HCoV-229E | Human | NC_002645 | Thiel et al., 2001 | 27,317 | 38.2 | 0.129 | 21.6 | 27.2 | 34.6 | 16.7 | 44.281 |
| HCoV-NL63 | Human | NC_005831 | van der Hoek et al., 2004 | 27,553 | 34.4 | 0.161 | 20.0 | 26.3 | 39.2 | 14.4 | 37.275 |
| PEDV | Pig | NC_003436 | Kocherhans et al., 2001 | 28,033 | 42.0 | 0.086 | 22.8 | 24.7 | 33.2 | 19.2 | 48.424 |
| BtCoV | Bat | DQ648858 | Tang et al., 2006 | 28,203 | 40.1 | 0.102 | 22.1 | 26.2 | 33.7 | 18.0 | 46.905 |
| Bat-CoV HKU2 | Bat | EF203064 | Lau et al., 2007 | 27,164 | 38.9 | 0.140 | 22.2 | 24.9 | 35.1 | 16.8 | 43.342 |
| Group 2a | |||||||||||
| HCoV-OC43 | Human | NC_005147 | Vijgen et al., 2005 | 30,738 | 36.8 | 0.176 | 21.7 | 27.6 | 35.6 | 15.2 | 43.791 |
| CoV-HKU1 | Human | NC_006577 | Woo et al., 2005b | 29,926 | 32.0 | 0.188 | 19.0 | 27.8 | 40.1 | 13.0 | 35.671 |
| BCoV | Cattle | NC_003045 | Chouljenko et al., 2001 | 31,028 | 37.1 | 0.174 | 21.8 | 27.4 | 35.5 | 15.3 | 43.856 |
| PHEV | Pig | NC_007732 | Vijgen et al., 2006 | 30,480 | 37.2 | 0.164 | 21.7 | 27.3 | 35.4 | 15.6 | 44.380 |
| MHV | Mouse | NC_001846 | Leparc-Goffart et al., 1997 | 31,357 | 41.7 | 0.142 | 23.9 | 26.0 | 32.3 | 17.9 | 51.237 |
| Group 2b | |||||||||||
| SARS-CoV | Human | NC_004718 | Marra et al., 2003 | 29,751 | 40.7 | 0.020 | 20.8 | 28.5 | 30.7 | 20.0 | 49.423 |
| Bat-SARS-CoV HKU3 | Bat | DQ022305 | Lau et al., 2005 | 29,728 | 41.1 | 0.027 | 21.1 | 28.4 | 30.5 | 20.0 | 49.882 |
| Group 2c | |||||||||||
| Bat-CoV HKU4 | Bat | EF065506 | Woo et al., 2006b | 30,286 | 37.8 | 0.093 | 20.7 | 27.6 | 34.6 | 17.1 | 44.585 |
| Bat-CoV HKU5 | Bat | EF065511 | Woo et al., 2006b | 30,488 | 42.9 | 0.004 | 21.6 | 26.6 | 30.4 | 21.4 | 53.230 |
| Group 2d | |||||||||||
| Bat-CoV HKU9 | Bat | EF065513 | Woo et al., 2006b | 29,114 | 41.0 | 0.138 | 23.3 | 25.3 | 33.7 | 17.7 | 46.162 |
| Group 3 | |||||||||||
| IBV | Chicken | NC_001451 | Boursnell et al., 1987 | 27,608 | 37.9 | 0.144 | 21.7 | 28.9 | 33.2 | 16.2 | 45.777 |
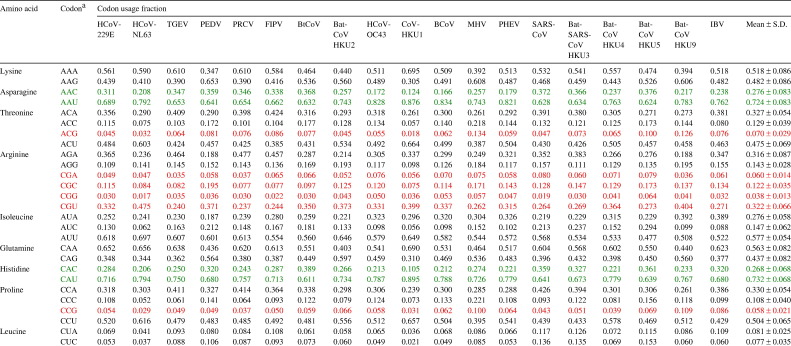
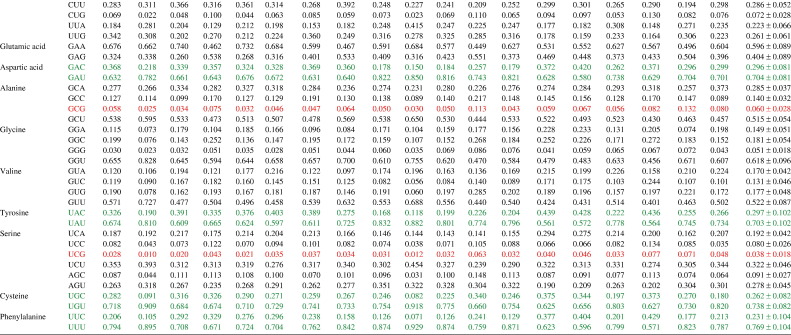
Table 2.
Codon usage fractions in coronaviruses
a Codons with CpG are in red and codons of amino acids that use either NNC or NNU as the codon are in green. (For interpretation of the references to colour in this table legend, the reader is referred to the web version of this article.)
To study the possible effect of CpG suppression on codon usage bias, the usage fractions of the eight codons that contain CpG (CCG, GCG, UCG, ACG, CGC, CGG, CGU and CGA) were analyzed. Of these eight codons, six [CCG (mean 0.058), GCG (mean 0.060), UCG (mean 0.038), ACG (mean 0.070), CGG (mean 0.038) and CGA (mean 0.060)] were markedly suppressed. CGC is slightly suppressed (mean 0.122) whereas CGU is over-represented (mean 0.322).
To study the possible effect of cytosine deamination on codon usage bias, codons of amino acids that can use C or U in the codons were analyzed. For all amino acids that only use either NNU or NNC as codon (asparagine, histidine, aspartic acid, tyrosine, cysteine and phenylalanine), all NNU are markedly over represented with usage fractions of more than 0.700, whereas the usage fractions of all NNC are less than 0.300. For amino acids that use NNU, NNC or other codons (threonine, isoleucine, proline, leucine, alanine, glycine, valine and serine), the usage fractions of all NNU are at least three times more than those of the corresponding NNC. For leucine, UUA (mean 0.223) is used much more frequently than CUA (mean 0.081), and UUG (mean 0.261) is used much more frequently than CUG (mean 0.072).
To study the possible effect of A ↔ G transition on codon usage bias, codons of amino acids that can use A or G in the codons were analyzed. For amino acids that use either NNA or NNG as codons (lysine, glutamine and glutamic acid) and those that use NNA, NNG or other codons but excluding those codons with CpG (arginine, glycine and valine), the usage fractions of NNA are often higher than those of NNG, but the differences between the usage fractions of NNA and NNG are not as marked as those between the usage fractions of NNU and NNC.
Codon usage in CoV-HKU1
Among all the 19 coronaviruses, CoV-HKU1 showed the most extreme codon usage bias. CoV-HKU1 is the only coronavirus that showed Nc outside the mean ± 2 S.D. range. CoV-HKU1 also possessed the lowest G + C content, highest GC skew, lowest percentages of G and C and highest percentage of U among all coronavirus genomes (Table 1). For the six amino acids that only use either NNU or NNC as codon (asparagine, histidine, aspartic acid, tyrosine, cysteine and phenylalanine), amino acids that use NNU, NNC or other codons (threonine, isoleucine, proline, leucine, alanine, glycine, valine and serine), and for leucine that use UNN or CNN as codon, the average (S.D.) ratio of the usage fractions of the codons with U to those with C is 9.66 (2.49) (Table 2). For amino acids that use either NNA or NNG as codons (lysine, glutamine and glutamic acid) and those that use NNA, NNG or other codons but excluding those codons with CpG (arginine, glycine and valine), the average (S.D.) ratio of the usage fractions of the codons with A to those with G is 2.72 (0.57) (Table 2).
Codon usage in hosts of coronaviruses
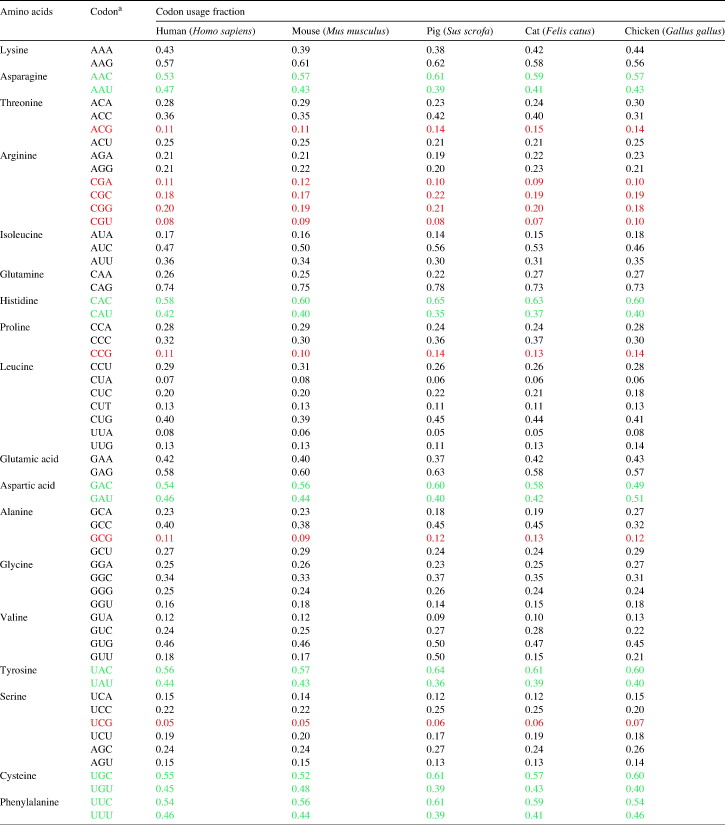
The codon usage fractions in the hosts of coronaviruses, including human, mouse, pig, cat and chicken, are shown in Table 3 . To study the possible effect of CpG suppression on codon usage bias, the usage fractions of the eight codons that contain CpG (CCG, GCG, UCG, ACG, CGC, CGG, CGU and CGA) were analyzed. Among these eight codons, six (CCG, GCG, UCG, ACG, CGU and CGA) were suppressed, of which five were also suppressed in the coronavirus genomes. To study the possible effect of C ↔ U transition and A ↔ G transition on codon usage bias, codons of amino acids that can use C or U and those of amino acids that can use A or G in the codons were analyzed. No pattern of difference was observed between the use of NNU and NNC and between the use of NNA and NNG.
Table 3.
Codon usage fractions in different hosts of coronaviruses
a Codons with CpG are in red and codons of amino acids that use either NNC or NNU as the codon are in green. (For interpretation of the references to colour in this table legend, the reader is referred to the web version of this article.)
Dinucleotide relative abundance in coronavirus genomes
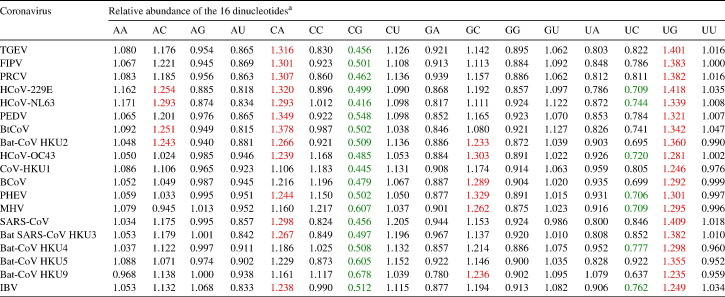
The relative abundance of the 16 dinucleotides in the 19 coronavirus genomes are shown in Table 4 . Among the 16 dinucleotides, the relative abundance of CpG showed the most marked deviation from the “normal range” (mean ± S.D. = 0.509 ± 0.063, 0.271 less than 0.78), with all 19 genomes showing CpG under-representation. In addition, the relative abundance of UpG and CpA also showed slight deviation from the “normal range” (mean ± S.D. = 1.331 ± 0.057 and 1.257 ± 0.070, respectively, both > 1.23), with all 19 and 13 genomes showing UpG and CpA over-representation, respectively.
Table 4.
Relative abundance of the 16 dinucleotides in the 19 coronavirus species with complete genomes available
a Numbers > 1.23 and < 0.78 are shown in red and green, respectively. (For interpretation of the references to colour in this table legend, the reader is referred to the web version of this article.)
Correlations between CpG suppression and cytosine deamination in coronaviruses
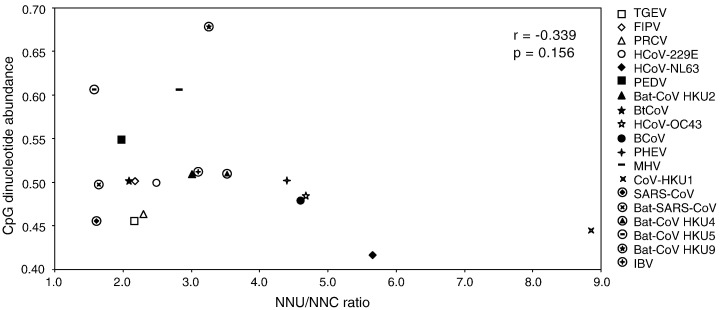
The relationship between CpG suppression and cytosine deamination in the 19 coronavirus genomes is shown in Fig. 1 . The mean (S.D.) of the NNU/NNC in the six amino acids that only use either NNC or NNU as the codons of the 19 coronavirus genomes is 3.262 (1.785). CoV-HKU1 showed extremely high NNU/NNC ratio of 8.835. No significant correlation was observed between CpG abundance and mean NNU/NNC ratio in the 19 coronavirus genomes (r = − 0.339, P = 0.156).
Fig. 1.
Correlation between CpG dinucleotide abundance and NNU/NNC ratio in the 19 coronavirus genomes.
Discussion
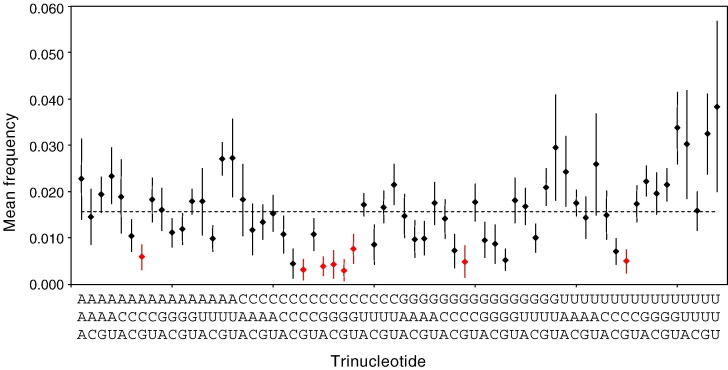
Marked CpG suppression is observed in all coronavirus genomes. The discovery of Toll-like receptors (TLRs) that recognize pathogen-associated molecular patterns and the downstream molecular pathways was one of the biggest advances in the understanding of vertebrate innate immunity in recent years. Among the TLR that recognize viral components, TLR3, 7, 8 and 9 detect viral nucleic acids (Bowie and Haga, 2005). It has been shown that TLR9 bound to CpG of double-stranded DNA and elicited the downstream inflammatory response, and administration of CpG oligodeoxynucleotides has been shown to protect mice from herpes simplex virus 2 infections (Ashkar et al., 2003, Lund et al., 2003). Furthermore, it has been shown that CpG is under-represented in the genomes of small DNA viruses, which could be related to their evasion of the host immune systems (Karlin et al., 1994, Shackelton et al., 2006). Although CpG suppression was also observed in RNA viruses, no known TLR has been shown to recognize CpG of ssRNA. However, recently it has been shown that ssRNA can stimulate human CD14+CD11c+ monocytes to produce large amounts of interleukin 12, but this activation of monocytes by CpG oligoribonucleotides was not mediated through TLR3, 7, 8 or 9 (Sugiyama et al., 2005). The results suggested that CpG oligoribonucleotides may stimulate monocytes through a novel mechanism distinct from previously known immunostimulatory nucleic acids. In the present study, we showed that the mean CpG relative abundance in the coronavirus genomes is markedly suppressed (Table 4). This concurs with the results observed in a study on di- and trinucleotide frequencies in nine coronaviruses 10 years ago (Tobler and Ackermann, 1998). The most logical way to avoid CpG is to mutate them to either UpG or CpA. This is in line with the observation that these two dinucleotides are over-represented in the coronavirus genomes, but their deviations from the upper limit of the “normal range” is not as remarkable as that of CpG from the lower limit of the “normal range”, as the CpG suppression pressure is equally shared by UpG and CpA over-representation. Interestingly, only CpG containing codons in the context of purine-CpG (ACG and GCG), pyrimidine-CpG (UCG and CCG) and CpG-purine (CGA and CGG) are suppressed (Table 2), whereas CpG-pyrimidine (CGU and CGC) are not. However, when trinucleotide frequencies were analyzed in the 19 coronavirus genomes, all the eight trinucleotides with CpG were suppressed (Fig. 2 ). This indicates that there is probably another force that has led to an increase use of CGU and CGC as codons for arginine, but this force does not act on trinucleotides over the whole genome in general. This force is probably unrelated to the relative abundance of the corresponding tRNA molecules in the hosts of the coronaviruses, as the pattern of bias in the hosts is not the same as that in the coronaviruses.
Fig. 2.
Mean frequencies of 64 trinucleotides in the 19 coronavirus genomes. The dots and the bars represent the mean frequencies and the 95% confidence intervals of the trinucleotides. The dotted line represents the frequency of each trinucleotide (1/64 = 0.015625) if the bases are distributed in random. The CpG containing trinucleotides are in red.
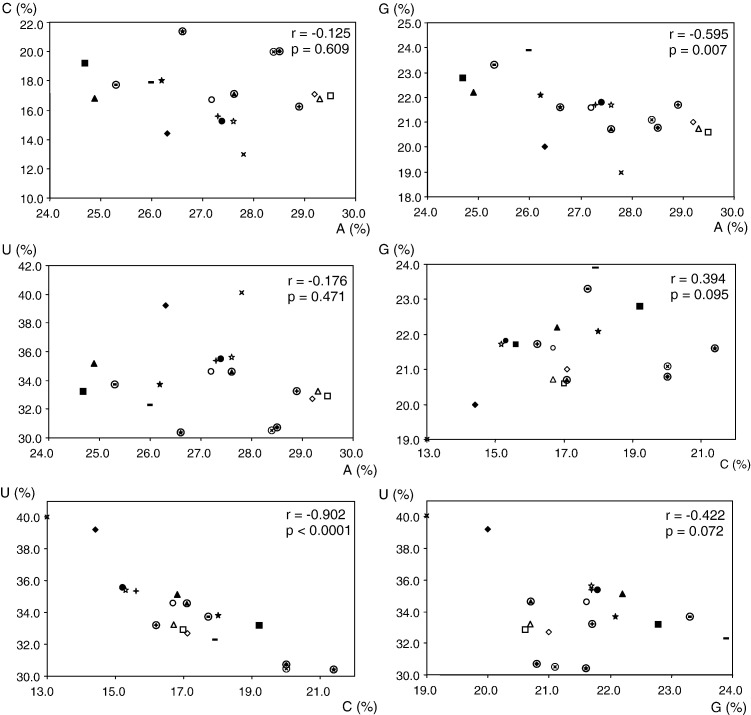
In addition to CpG suppression, marked cytosine deamination is also observed in all coronavirus genomes. Although it has been recognized that deamination of cytosine is a significant source of spontaneous mutations for a few decades (Duncan and Miller, 1980), DNA-cytosine deaminases, which are able to attack cytosines in single-stranded DNA, have only been discovered in the recent few years (Bransteitter et al., 2003, Sohail et al., 2003). The discovery of the ability to edit human immunodeficiency virus DNA, and subsequently RNA as well, by the human cytidine deaminase APOBEC3G has allowed the speculation that APOBEC-mediated cytosine deamination may contribute to the sequence variation of RNA viruses that replicate without any DNA intermediates (Bishop et al., 2004). GC skew, which reflects cytosine deamination, has been studied in various coronaviruses, and it has been shown that the GC skews of coronavirus genomes become less pronounced in the one third of the genome that encodes the structural proteins (Grigoriev, 2004, Pyrc et al., 2004). In the present study, using the six amino acids that are only encoded by NNU or NNC, hence excluding most other pressures that may affect the relative abundance of cytosine and uracil, we showed that all these NNU and NNC had usage fractions of > 0.700 and < 0.300, respectively (Table 2). In fact, for all codons that encode the same amino acid and with either C or U in any position, the usage fraction of the codon that uses U is invariably higher than the one that uses C in all coronaviruses. Furthermore, the percentage of C showed strong inverse relationships with the percentage of U in coronavirus genomes (r = − 0.902, P < 0.0001) (Fig. 3 ). All these suggest that cytosine deamination is an important biochemical force in shaping coronavirus evolution.
Fig. 3.
Correlations among mononucleotide frequencies in the 19 coronavirus genomes. The symbols for the various coronaviruses are the same as those used in Fig. 1.
Cytosine deamination and selection of CpG suppressed clones by the immune system are the two major independent biochemical and biological selective forces that shape codon usage bias in coronavirus genomes. Codon usage bias in coronaviruses is unrelated to the relative abundance of the corresponding tRNA molecules, as the patterns of bias in codon usage fractions in the hosts are not the same as those in the coronaviruses (Table 2, Table 3). Although others have tried to explain variations in codon usage in coronaviruses by compositional constraints (Gu et al., 2004), we think that both codon usage bias and nucleotide composition of the coronavirus genomes, which are apparently related to each other, are both results of other biological and biochemical selective forces, rather than nucleotide composition as a cause of codon usage bias. On the other hand, most of the codon usage bias in the coronaviruses can be easily explained by CpG suppression and cytosine deamination (Table 2). For asparagine, isoleucine, histidine, aspartic acid, glycine, valine, tyrosine, cysteine and phenylalanine, NNU are used more frequently than NNC because of cytosine deamination. For lysine, glutamine and glutamic acid, NNA are used slightly more frequently than NNG because of cytosine deamination in the minus strand during RNA replication. For threonine, ACG is suppressed because of CpG suppression and ACU is used more frequently than ACC because of cytosine deamination. For arginine, CGA and CGG are suppressed because of CpG suppression and CGU is used more frequently than CGC because of cytosine deamination. AGA is used more frequently than AGG and CGA is used more frequently than CGG because of cytosine deamination in the minus strand during RNA replication. For proline, CCG is suppressed because of CpG suppression and CCU is used more frequently than CCC because of cytosine deamination. For leucine, CUU is used more frequently than CUC, UUA is used more frequently than CUA, and UUG is used more frequently than CUG because of cytosine deamination. For alanine, GCG is suppressed because of CpG suppression and GCU is used more frequently than GCC because of cytosine deamination. For serine, UCG is suppressed because of CpG suppression and UCU is used more frequently than UCC while ACU is used more frequently than ACC because of cytosine deamination. In addition to showing that CpG suppression and cytosine deamination are probably the two most important biological/biochemical forces that shape codon usage bias, we also demonstrated that these two forces are independent (Fig. 1), although cytosine deamination and subsequent selection of CpG suppressed clones by the immune system may be one of the mechanisms that has led to the resultant CpG suppression. Furthermore, we speculate that the species-specific number of CpG containing codons may not simply be the result of mutation pressure to avoid CpG, but an equilibrium between the immune pressure and the required number of CpG containing codons to serve biological functions such as to maintain RNA structure stability. Such an additional factor could explain the mere correlation between the NNU/NNC ratio and CpG dinucleotide abundance.
The underlying mechanism for the extreme codon usage bias, cytosine deamination and G + C content in CoV-HKU1 is enigmatic. The contribution of cytosine deamination to genome evolution varies from very low to very high among the 19 coronavirus genomes. For bat-CoV HKU5, SARS-CoV and bat-SARS-CoV, the mean NNU/NNC ratios are less than 1.7 (Fig. 1). Codon usage bias in these coronaviruses is relatively mild (Nc of 53.23, 49.423 and 49.882, respectively; Table 1), and is mainly due to CpG suppression (Table 2). On the other hand, for CoV-HKU1, the mean NNU/NNC ratio is more than 8.8 (Fig. 1), which is likely a result of rapid cytosine deamination. Although the biochemical basis for this extreme cytosine deamination is not known, this is probably the explanation for the extremely strong codon usage bias in CoV-HKU1 (Nc of 35.671) and its lowest G + C content of 32% among all coronavirus genomes (Table 1).
Materials and methods
Coronavirus and host genomes
One genome sequence of each of the 19 coronavirus species with complete genome sequence available was downloaded from the GenBank database (Table 1). The genomes of the hosts of the coronaviruses, including those of human, mouse, pig, cat and chicken, were also downloaded.
Codon usage
Codon usage bias was calculated according to the method described by Wright (1990). Using this method, when only one codon is used for each amino acid, Nc for the virus would be 20, and when all codons are used equally, the Nc for the virus would be 61. The codon usage fraction of a particular codon in a genome is calculated by the ratio of the number of that codon to the number of the amino acid that codon and other synonymous codons encode for in the protein coding sequence of the genome. The method for calculating codon usage bias accounting for background nucleotide composition (Nc′) (Novembre, 2002) was not used because it had been proposed to suffer from methodology problems, although those problems did not affect the conclusions which had been drawn by using Nc of this study (Fuglsang, 2006).
Dinucleotide relative abundance in coronavirus genomes
The relative abundance of the dinucleotides in the coronavirus genomes was assessed using the method described by Karlin and Burge (1995). The odds ratio ρ xy = f xy/f x f y, where f x denotes the frequency of the nucleotide X and f xy the frequency of the dinucleotide XY, etc., for each dinucleotide were calculated. From data simulations and statistical theory, ρ xy ≤ 0.78 (extreme under-representation) or ρ xy ≥ 1.23 (extreme over-representation) occurs for sufficiently long (≥ 20 kb) random sequences with the probability at most 0.001 for virtually any base composition.
Correlations between CpG suppression and cytosine deamination in coronaviruses
To study possible correlations between CpG suppression and cytosine deamination in coronaviruses, the relative abundance of CpG and the mean ratio of NNC to NNU in the six amino acids (asparagine, histidine, aspartic acid, tyrosine, cysteine and phenylalanine) that only use either NNC or NNU as the codons (NNU/NNC ratio, representing contribution of cytosine deamination) were calculated for all 19 coronavirus genomes. Analysis of correlation between CpG deamination and NNU/NNC ratio was performed using Pearson's correlation (SPSS version 11.0).
Acknowledgments
We are grateful to the generous support of Mr. Hui Hoy and Mr. Hui Ming in the genomic sequencing platform. This work was partly supported by the Research Grant Council Grant; University Development Fund, Outstanding Young Researcher Award, HKU Special Research Achievement Award and The Croucher Senior Medical Research Fellowship, The University of Hong Kong; The Tung Wah Group of Hospitals Fund for Research in Infectious Diseases; the HKSAR Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau; and the Providence Foundation Limited in memory of the late Dr. Lui Hac Minh.
References
- Almazan F., Gonzalez J.M., Penzes Z., Izeta A., Calvo E., Plana-Duran J., Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkar A.A., Bauer S., Mitchell W.J., Vieira J., Rosenthal K.L. Local delivery of CpG oligodeoxynucleotides induces rapid changes in the genital mucosa and inhibits replication, but not entry, of herpes simplex virus type 2. J. Virol. 2003;77:8948–8956. doi: 10.1128/JVI.77.16.8948-8956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop K.N., Holmes R.K., Sheehy A.M., Malim M.H. APOBEC-mediated editing of viral RNA. Science. 2004;305:645. doi: 10.1126/science.1100658. [DOI] [PubMed] [Google Scholar]
- Boursnell M.E., Brown T.D., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Bowie A.G., Haga I.R. The role of Toll-like receptors in the host response to viruses. Mol. Immunol. 2005;42:859–867. doi: 10.1016/j.molimm.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Bransteitter R., Pham P., Scharff M.D., Goodman M.F. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Mawditt K., Welchman Dde B., Britton P., Gough R.E. Coronaviruses from pheasants (Phasianus colchicus) are genetically closely related to coronaviruses of domestic fowl (infectious bronchitis virus) and turkeys. Avian Pathol. 2002;31:81–93. doi: 10.1080/03079450120106651. [DOI] [PubMed] [Google Scholar]
- Chouljenko V.N., Lin X.Q., Storz J., Kousoulas K.G., Gorbalenya A.E. Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J. Gen. Virol. 2001;82:2927–2933. doi: 10.1099/0022-1317-82-12-2927. [DOI] [PubMed] [Google Scholar]
- Dong B.Q., Liu W., Fan X.H., Vijaykrishna D., Tang X.C., Gao F., Li L.F., Li G.J., Zhang J.X., Yang L.Q., Poon L.L., Zhang S.Y., Peiris J.S., Smith G.J., Chen H., Guan Y. Detection of a novel and highly divergent coronavirus from Asian leopard cats and Chinese ferret badgers in southern china. J. Virol. 2007;81:6920–6926. doi: 10.1128/JVI.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan B.K., Miller J.H. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- East M.L., Moestl K., Benetka V., Pitra C., Honer O.P., Wachter B., Hofer H. Coronavirus infection of spotted hyenas in the Serengeti ecosystem. Vet. Microbiol. 2004;102:1–9. doi: 10.1016/j.vetmic.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Hartwig N.G., Bestebroer T.M., Niemeyer B., de Jong J.C., Simon J.H., Osterhaus A.D. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang A. Accounting for background nucleotide composition when measuring codon usage bias: brilliant idea, difficult in practice. Mol. Biol. Evol. 2006;23:1345–1347. doi: 10.1093/molbev/msl009. [DOI] [PubMed] [Google Scholar]
- Grigoriev A. Mutational patterns correlate with genome organization in SARS and other coronaviruses. Trends Genet. 2004;20:131–135. doi: 10.1016/j.tig.2004.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Zhou T., Ma J., Sun X., Lu Z. Analysis of synonymous codon usage in SARS coronavirus and other virus in the Nidovirales. Virus Res. 2004;101:155–161. doi: 10.1016/j.virusres.2004.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 2003;77:4528–4538. doi: 10.1128/JVI.77.8.4528-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasoksuz M., Alekseev K., Vlasova A., Zhang X., Spiro D., Halpin R., Wang S., Ghedin E., Saif L.J. Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe. J, Virol. 2007;81:4981–4990. doi: 10.1128/JVI.02361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G.M., Holmes E.C. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res. 2003;92:1–7. doi: 10.1016/s0168-1702(02)00309-x. [DOI] [PubMed] [Google Scholar]
- Jonassen C.M., Kofstad T., Larsen I.L., Lovland A., Handeland K., Follestad A., Lillehaug A. Molecular identification and characterization of novel coronaviruses infecting graylag geese (Anser anser), feral pigeons (Columbia livia) and mallards (Anas platyrhynchos) J. Gen. Virol. 2005;86:1597–1607. doi: 10.1099/vir.0.80927-0. [DOI] [PubMed] [Google Scholar]
- Karlin S., Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- Karlin S., Doerfler W., Cardon L.R. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J. Virol. 1994;68:2889–2897. doi: 10.1128/jvi.68.5.2889-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocherhans R., Bridgen A., Ackermann M., Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. 2001;23:137–144. doi: 10.1023/A:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus. Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Yip C.C., Tse H., Tsoi H.W., Cheng V.C., Lee P., Tang B.S., Cheung C.H., Lee R.A., So L.Y., Lau Y.L., Chan K.H., Yuen K.Y. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J. Clin. Microbiol. 2006;44:2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Wang M., Lam C.S., Xu H., Guo R., Chan K.H., Zheng B.J., Yuen K.Y. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007 doi: 10.1016/j.virol.2007.06.009. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leparc-Goffart I., Hingley S.T., Chua M.M., Jiang X., Lavi E., Weiss S.R. Altered pathogenesis of a mutant of the murine coronavirus MHV-A59 is associated with a Q159L amino acid substitution in the spike protein. Virology. 1997;239:1–10. doi: 10.1006/viro.1997.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Liu S., Chen J., Chen J., Kong X., Shao Y., Han Z., Feng L., Cai X., Gu S., Liu M. Isolation of avian infectious bronchitis coronavirus from domestic peafowl (Pavo cristatus) and teal (Anas) J. Gen. Virol. 2005;86:719–725. doi: 10.1099/vir.0.80546-0. [DOI] [PubMed] [Google Scholar]
- Lund J., Sato A., Akira S., Medzhitov R., Iwasaki A. Toll-like receptor 9 mediated recognition of herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Novembre J.A. Accounting for background nucleotide composition when measuring codon usage bias. Mol. Biol. Evol. 2002;8:1390–1394. doi: 10.1093/oxfordjournals.molbev.a004201. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Chu D.K., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H., Lau S.K., Woo P.C., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S. Identification of a novel coronavirus in bats. J. Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Jebbink M.F., Berkhout B., van der Hoek L. Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J. 2004;17:1–7. doi: 10.1186/1743-422X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sewatanon J., Srichatrapimuk S., Auewarakul P. Compositional bias and size of genomes of human DNA viruses. Intervirology. 2007;50:123–132. doi: 10.1159/000098238. [DOI] [PubMed] [Google Scholar]
- Shackelton L.A., Parrish C.R., Holmes E.C. Evolutionary basis of codon usage and nucleotide composition bias in vertebrate DNA viruses. J. Mol. Evol. 2006;62:551–563. doi: 10.1007/s00239-005-0221-1. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail A., Klapacz J., Samaranayake M., Ullah A., Bhagwat A.S. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 2003;31:2990–2994. doi: 10.1093/nar/gkg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Gursel M., Takeshita F., Coban C., Conover J., Kaisho T., Akira S., Klinman D.M., Ishii K.J. CpG RNA: identification of novel single-stranded RNA that stimulates human CD14+CD11c+ monocytes. J. Immunol. 2005;174:2273–2279. doi: 10.4049/jimmunol.174.4.2273. [DOI] [PubMed] [Google Scholar]
- Tang X.C., Zhang J.X., Zhang S.Y., Wang P., Fan X.H., Li L.F., Li G., Dong B.Q., Liu W., Cheung C.L., Xu K.M., Song W.J., Vijaykrishna D., Poon L.L., Peiris J.S., Smith G.J., Chen H., Guan Y. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S.G. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- Tobler K., Ackermann M. Comparison of the di- and trinucleotide frequencies from the genomes of nine different coronaviruses. Adv. Exp. Med. Biol. 1998;440:801–804. doi: 10.1007/978-1-4615-5331-1_104. [DOI] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert F.J., Berkhout B., Lukashov V.V. Host-related nucleotide composition and codon usage as driving forces in the recent evolution of the Astroviridae. Virology. 2007;361:447–454. doi: 10.1016/j.virol.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moes E., Thoelen I., Wollants E., Lemey P., Vandamme A.M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Lemey P., Maes P., Van Reeth K., Nauwynck H., Pensaert M., Van Ranst M. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006;80:7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Tsoi H.W., Chan K.H., Wong B.H., Che X.Y., Tam V.K., Tam S.C., Cheng V.C., Hung I.F., Wong S.S., Zheng B.J., Guan Y., Yuen K.Y. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet. 2004;363:841–845. doi: 10.1016/S0140-6736(04)15729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Huang Y., Lau S.K., Tsoi H.W., Yuen K.Y. In silico analysis of ORF1ab in coronavirus HKU1 genome reveals a unique putative cleavage site of coronavirus HKU1 3C-like protease. Microbiol. Immunol. 2005;49:899–908. doi: 10.1111/j.1348-0421.2005.tb03681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Tsoi H.W., Huang Y., Poon R.W., Chu C.M., Lee R.A., Luk W.K., Wong G.K., Wong B.H., Cheng V.C., Tang B.S., Wu A.K., Yung R.W., Chen H., Guan Y., Chan K.H., Yuen K.Y. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J. Infect. Dis. 2005;192:1898–1907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Li K.S., Poon R.W., Wong B.H., Tsoi H.W., Yip B.C., Huang Y., Chan K.H., Yuen K.Y. Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Yip C.C., Huang Y., Tsoi H.W., Chan K.H., Yuen K.Y. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Wang M., Lau S.K., Xu H., Poon R.W., Guo R., Wong B.H., Gao K., Tsoi H.W., Huang Y., Li K.S., Lam C.S., Chan K.H., Zheng B.J., Yuen K.Y. Comparative analysis of 12 genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 2007;81:1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. The effective number of codons used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Yap Y.L., Zhang X.W., Danchin A. Relationship of SARS-CoV to other pathogenic RNA viruses explored by tetranucleotide usage profiling. BMC Bioinform. 2003;4:43. doi: 10.1186/1471-2105-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Hasoksuz M., Spiro D., Halpin R., Wang S., Stollar S., Janies D., Hadya N., Tang Y., Ghedin E., Saif L. Complete genomic sequences, a key residue in the spike protein and deletions in nonstructural protein 3b of US strains of the virulent and attenuated coronaviruses, transmissible gastroenteritis virus and porcine respiratory coronavirus. Virology. 2007;358:424–435. doi: 10.1016/j.virol.2006.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 2004;7:412–419. doi: 10.1016/j.mib.2004.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]