Abstract
Autophagy is a cellular response against stresses which include the infection of viruses and bacteria. We unravel that Dengue virus-2 (DV2) can trigger autophagic process in various infected cell lines demonstrated by GFP-LC3 dot formation and increased LC3-II formation. Autophagosome formation was also observed under the transmission electron microscope. DV2-induced autophagy further enhances the titers of extracellular and intracellular viruses indicating that autophagy can promote viral replication in the infected cells. Moreover, our data show that ATG5 protein is required to execute DV2-induced autophagy. All together, we are the first to demonstrate that DV can activate autophagic machinery that is favorable for viral replication.
Keywords: Dengue virus, Autophagy, Autophagosome, Viral replication
Introduction
Dengue virus (DV) is a positive-single strand RNA virus that belongs to the family of Flaviviridae. The four DV serotypes are transmitted by mosquitoes. In most cases, the symptoms of dengue fever (DF) are self-limited. However, in a small proportion of people the disease progresses to the severe hemorrhagic manifestations of dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS).
Autophagy is characterized by the accumulation of autophagic vacuoles and regulated processes of degradation and recycling of cellular constituents, which are important in organelle turnover and the bioenergetics management of starvation. As well, autophagy plays important roles in cell growth, development, and disease pathology.
During autophagy, portions of the cytoplasm or small organelles are sequestered into double-membrane vesicles called autophagosomes. Autophagosomes ultimately fuse with lysosomes to generate single-membrane vesicles termed autophagolysosomes, in which the contents are subsequently degraded.
Several Atg proteins have been implicated in autophagosome formation. The Atg5 and Atg12 are required to recruit other proteins to the autophagosomal membrane and cytosol to form the autophagic vacuole (Nemoto et al., 2003). LC3 is the mammalian equivalent of yeast Atg 8. The precursor form of LC3 is post-modified into two forms, LC3-I and LC3-II, which are localized in the cytosol (LC3-I) and in autophagosomal membranes (LC3-II). LC3-II can be used to estimate the abundance of autophagosomes before they are destroyed through fusion with lysosomes (Kabeya et al., 2000, Mizushima et al., 2001). Similarly, GFP-LC3 fusion protein redistributes from a diffusive state to a vascular aggregative pattern when autophagosomes are formed (Kabeya et al., 2000, Mizushima et al., 2001). Transmission electron microscopy (TEM) has been used to detect autophagosome formation, while GFP-LC3 dot formation and expression levels of LC3-II have been used to measure the level of autophagy and the autophagosome formation (Kabeya et al., 2000, Mizushima et al., 2001).
Autophagy not only influences tumor formation, but also plays a role in infectious disease. Macrophages can eliminate Legionella pneumophila infection through cholesterol- or lipid-raft-rich induction of autophagy (Amer and Swanson, 2005). Conversely, autophagy in pathogen infected non-phagocytes may elevate pathogen replication and infection (Gutierrez et al., 2005). Infection of human cells with poliovirus and rhinovirus induces the formation of double-membrane cytoplasm vesicles and autophagosome. Autophagosomes are used as the sites of viral RNA replication and these double-membrane structures provide the site for viral RNA replication, possibly enabling the non-lytic release of cytoplasma contents including progeny virions from poliovirus and rhinovirus infected cells (Jackson et al., 2005). Furthermore, autophagosome is required for the formation of double membrane-bound coronavirus replication complexes, with the formation of the double membrane vesicles significantly enhancing viral replication efficiency (Prentice et al., 2004). Not only RNA virus (poliovirus etc.) but also DNA virus (Epstein-Barr virus etc.) infection can induce autophagic machinery, and whether the activation of autophagic machinery can enhance viral replication (poliovirus and mouse hepatitis virus etc.) or not (vaccinia virus and herpes simplex virus type 1 etc.) depend on the type of viruses (Alexander et al., 2007, Jackson et al., 2005, Paludan et al., 2005, Prentice et al., 2004, Wileman, 2006, Zhang et al., 2006). In this report, we demonstrate that DV infection can activate the machinery of autophagy and cause autophagosome formation, which promotes DV replication in an ATG5 dependent manner.
Results
GFP-LC3 dot formation in DV2-infected cells
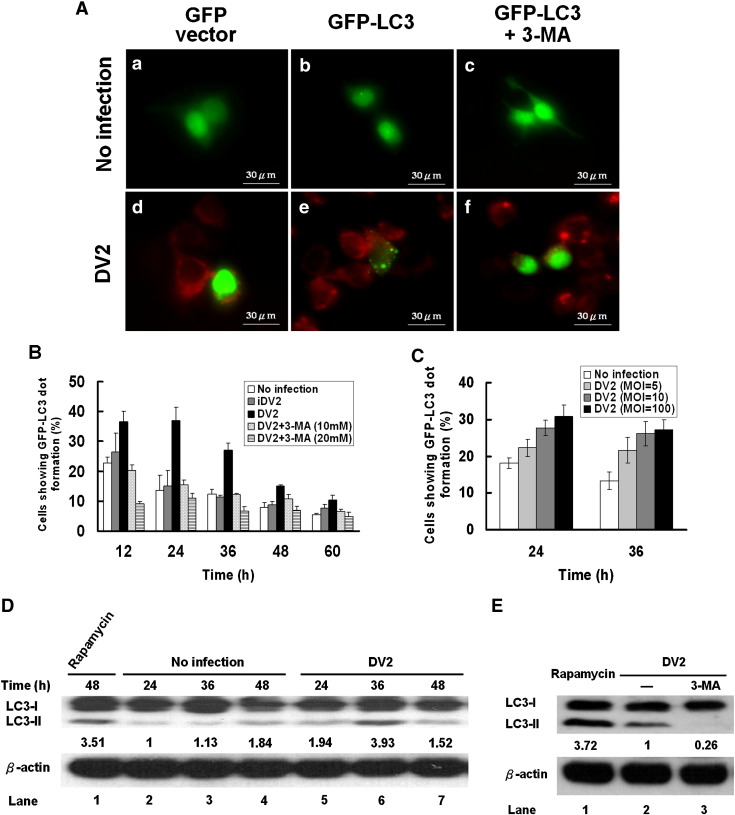
Because GFP-LC3 fusion protein can bind autophagosomes, the green fluorescent LC3 dot formation in the cell represents autophagosome formation. To clarify whether DV2 infection can activate the machinery of autophagy and induce the formation of autophagosome, GFP-LC3 dot formation during DV2 infection was investigated. Under the fluorescence microscope, the existence of DV2 was demonstrated by labeling the cells with DV2 anti-prM antibody (Red). DV2 infection induced the dot formation of GFP-LC3 fusion protein in the virus infected cells (Red background) (Fig. 1A-e), compared to the cells harboring GFP-LC3 without DV2 infection (Fig. 1A-b). Rapamycin and 3-methyladenine (3-MA) were used to clarify autophagosome formation during DV infection. The cytotoxicity of these two drugs at various dosages was tested and no toxicity was defected (Supplementary Fig. 1). Therefore, the optimal dosages of rapamycin (50 nM) and 3-MA (10 mM) were used in this study. The degree of GFP-LC3 dot formation was decreased by treatment of DV2-infected cells with 3-MA (an inhibitor of autophagy) (Fig. 1A-f). The cells treated with 3-MA alone or the cells transfected with GFP vector regardless DV2 infection or not could not induce GFP-LC3 dot formation (Figs. 1A-a, A-c, A-d). The cells treated with heat-inactivated virus (iDV2) also could not induce GFP-LC3 dot formation (data not shown). Similarly, GFP-LC3 dot formation was evidently induced in the positive control cells (cells exposed to the autophagosome inducers rapamycin; data not shown). Our data demonstrated that DV2 infection can trigger autophagic machinery and induce autophagosome formation.
Fig. 1.
GFP-LC3 dot formation and LC3-II expression level in Huh7 cells infected with DV2. (A) Huh7 cells were transfected with the plasmid either expressing GFP alone or GFP-LC3 fusion protein as indicated. The cells infected with DV2 or uninfected with or without 3-MA treatment for 36 h were labeled with anti-DV2-prM antibody. (B) Quantitative presentation of the percentage of GFP-LC3 dot formation cells in the total GFP expressing cells after various treatments. (C) Quantitative presentation of the percentage of GFP-LC3 dot formation cells with different DV titers. (D) The levels of LC3-II expression in the cells with or without DV2 infection at indicated times were detected by Western blotting. Quantification of the intensity of each band is listed under each band. β-actin was used as an internal control, and rapamycin treatment was a positive control. (E) The expression levels of LC3-II in DV2 infected cells and in the presence or absence of 10 mM 3-MA were detected by Western blotting. Rapamycin (50 nM) treated cells were used as the positive control. Each data point is representative of the results of three independent experiments.
The time course and the dosage effect of DV2 infection on the percentage of the cells with increased GFP-LC3 dot formation are shown quantitatively. Briefly, after pEGFPC1-LC3 plasmid DNA transfection, the cells expressing green fluorescence and the cells showing GFP-LC3 dot formation were counted, individually. The percentage of GFP-LC3 dot formation was calculated as: number of GFP-LC3 dot formation cells/number of total GFP expressing cells. Fig. 1B shows that the percentage of the cells with increased GFP-LC3 dot formation. The difference of the percentage of GFP-LC3 dot formation cells between DV2 infected (DV2) and noninfected groups (No infection) reached the plateau at 24 h and 36 h p.i. and then declined, indicating the greatest viral induced GFP-LC3 dot formation is at these time points (Fig. 1B). The GFP-LC3 dot formation was autophagy related, which was demonstrated by 3-MA suppression in a dose dependent manner. Moreover, DV2 induced GFP-LC3 dot formation correlated with increasing MOIs of DV2 either at 24 or at 36 h p.i. (Fig. 1C). DV2-induced GFP-LC3 dot formation was also detected in BHK and MEF cells (data not shown), suggesting that DV2-induced autophagy is a general event. Our data demonstrate that DV2 can transiently induce LC3 dot formation in a virus titer dependent manner in infected cells.
Increased formation of LC3 type II in DV2-infected Huh7 cells
In quiescent cells, LC3-I as a major form of LC3 evenly distributes in the cytoplasm. Upon exposure to various stresses which provoke autophagy, LC3-II formation was induced. It then sticks onto the autophagosomes. Because the level of LC3-II or the ratio of LC3-II/LC3-I expression has been used to represent the level of autophagy, Fig. 1D shows the expression levels of LC3-I and LC3-II in DV2-infected Huh7 cells at 24, 36 and 48 h p.i. The level of LC3-II was evidently increased at 36 h p.i. and declined at 48 h p.i. in DV2-infected cells compared to the uninfected cells (Fig. 1D, lanes 3 and 4 vs. lanes 6 and 7). Accordingly, DV2-induced LC3-II expression in Huh7 cells was suppressed by 3-MA treatment (Fig. 1E, lane 2 vs. 3). Rapamycin treatment of Huh7 cells dramatically increased LC3-II expression was used as a positive control (Fig. 1D, lane 1; Fig. 1E, lane 1). The expression levels of LC3-II in heat inactivated virus-infected cells compared to uninfected control showed no significant difference (data not shown). Our data showed that DV2 induced overexpression of LC3-II reached the maximum at 36 h p.i. The decline of LC3-II expression at 48 h is caused by dissociation of LC3-II from autolysosomes followed by LC3 recycling. Moreover, the LC3-II formation was further enhanced (0.58 to 1.35 folds) in the DV2 infected cells while the inhibitor of microtubule disrupting agent vinblastine was added to block the fusion of autophagosomes with lysosomes (Supplementary Fig. 2), suggesting that the autophagy flux is very large (Hoyvik et al., 1986). Collectively, our data indicate that in infected cells LC3-II can be induced by DV2 indicating the occurrence of autophagy.
Co-localization of LAMP1 and LC3 in DV2-infected Huh7 cells
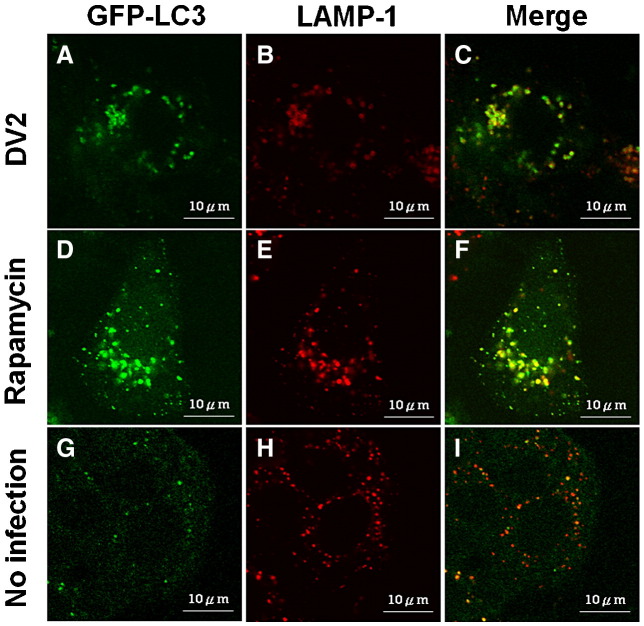
LAMP1, a lysosomal protein and a marker of endosomes and lysosomes, can be found at the early stage of autophagolysosome formation during autophagy. It is also used as an indicator of autophagolysosome formation. LAMP1 and LC3 do not distribute in the same region in non-autophagic cells. LC3 co-localizes with LAMP1 during the maturation of autolysosomes has been reported (Jackson et al., 2005). Co-localization of LAMP1 and LC3 was evidently increased in DV2-infected cells under the confocal microscope (Fig. 2C). LAMP1 and LC3 colocalization was also detected in the cells treated with rapamycin to induce autophagosome formation (Fig. 2F). Colocalization of LAMP1 and LC3 was seldom seen in uninfected Huh7 cells (Fig. 2I) and in the infected cell with 3-MA treatment (data not shown). In summary, colocalization of LC3 and LAMP1 observed in DV2-infected cells support our notion that DV infection can trigger autophagic process and induce the formation of autophagolysosomes.
Fig. 2.
Co-localization of GFP-LC3 and LAMP1 proteins in Huh7 cells with and without DV2 infection or with rapamycin treatment. Huh7 cells were transfected with pEGFPC1-LC3 for 12 h. (A to C) Huh7 cells were infected with DV2 for 36 h. (D to F) Huh7 cells were treated with rapamycin (50 nM) for 36 h as a positive control. (G to I) The Huh7 cells without DV2 infection and rapamycin treatment were used as the negative control. The cells were then treated with LAMP1 monoclonal antibody for 2 h and observed under the confocal microscope. LC3: green. LAMP1: Red.
Autophagosome was detected in DV2-infected Huh7 cells and wild type MEF cells
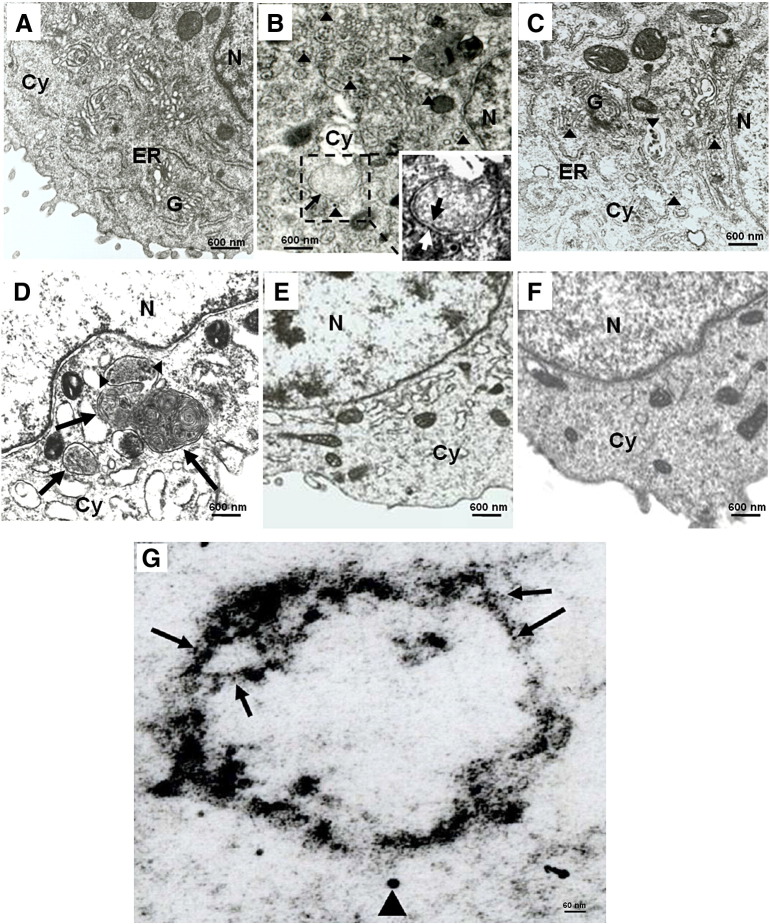
To further confirm that autophagy could be induced by DV2 infection, double membrane autophagosome-like vesicle in DV2 infected cells was observed under TEM. In DV2-infected Huh7 cells, the double-membrane vesicle defined as autophagosome vesicle was found in DV2 infected Huh7 cells (Fig. 3B, arrow). The inlet shows enlargement of the autophagosome in the dotted-black-box (Fig. 3B, white-arrow: outer membrane, black-arrow: inner membrane). Similar double-membrane vesicle (autophagosome vesicle) was rarely seen in uninfected cells (Fig. 3A) or infected cells in which autophagy process was blocked by 3-MA (Fig. 3C). Moreover, DV2-like particles between 20–40 nm were seen in the cytoplasm (Figs. 3B and C, arrowhead). DV and accompanied autophagosome formation were also detected in human monocytes (data not shown). In contrast, uninfected Huh7 cells, no dengue viral particles were detected (Fig. 3A). Moreover, autophagosome vesicles were not detected in the cells exposed to iDV2 (data not shown). Consistently, we detected the DV2-induced LC3 aggregates around the double membraned autophagosome by immuno-gold labeling of LC3 under TEM. We also detected DV2-like particle in DV2 infected Huh7 cells (Fig. 3G and Supplementary Fig. 3). We reveal that dengue virus particle, LC3 aggregate and double membrane autophagosome exist in the DV2 infected Huh7 cells, indicating DV can trigger LC3 dot formation and formation of autophagosome-like vesicles.
Fig. 3.
Detection of autophagosome in DV2-infected Huh7 and mouse MEF cells under TEM. The cells were fixed at 36 h post DV2 infection and observed under TEM. (A) Normal Huh7 cells without infection and treatment; (B) Huh7 cells were infected with DV2. The dot-line box encompassing the autophagosome was further enlarged in the inlet to show the outer- and inner-membrane (white-arrow: outer-membrane and black-arrow: inner-membrane). (C) Huh7 cells were infected with DV2 plus 10 mM 3-MA treatment. (D) ATG5+/+ MEF cells were infected with DV2. (E) ATG5−/− MEF cells were infected with DV2. (F) ATG5+/+ MEF cells were infected with DV2 plus 3-MA treatment. Arrowheads: virus-like particle. Arrow: autophagosome vesicle. Cy: cytoplasm; ER: endoplasmic reticulum; G: Golgi apparatus; N: nucleus. (G) Huh7 cells infected with DV2 for 36 h were fixed and stained with anti-LC3 antibody, and visualized by a secondary antibody coupled to 5-nm gold particles. The arrow indicates gold particles representing LC3, and the arrowhead indicates virus-particle (20–40 nm).
ATG5 is an initiator of autophagosome formation during autophagy progression (Kuma et al., 2004). To clarify whether DV2-induced autophagosome formation is through ATG5 regulatory system related process, wild-type and ATG5 gene knockout MEF cells infected with DV2 for 36 h were examined for double membrane autophagosome formation. Initially, we demonstrated that DV2 indeed could infect and replicate in both wild-type and ATG5 knockout MEF cells by immunofluorescence analysis using anti-DV2-NS1 antibody (data not shown). In DV2-infected MEF cells, only a few DV2 particles were seen in the region with autophagosome formation compared to DV2-infected Huh7 cells. This is probably caused by low infection as well as replication rate in this cell line (Fig. 4E). After DV2 infection, autophagosome vesicles and virus like particles were detected in wild-type MEF cells (Fig. 3D, arrow: autophagosome; arrowhead: virus like particles) but not in ATG5 knockout MEF cells (Fig. 3E). The double membrane autophagosome formation was suppressed while the wild-type ATG5 MEF cells were treated with the autophagy inhibitor 3-MA (Fig. 3F). Our ultra structure photographs indicate that DV2 can induce autophagosome formation. It is a general event and is mediated by the ATG5 regulatory system.
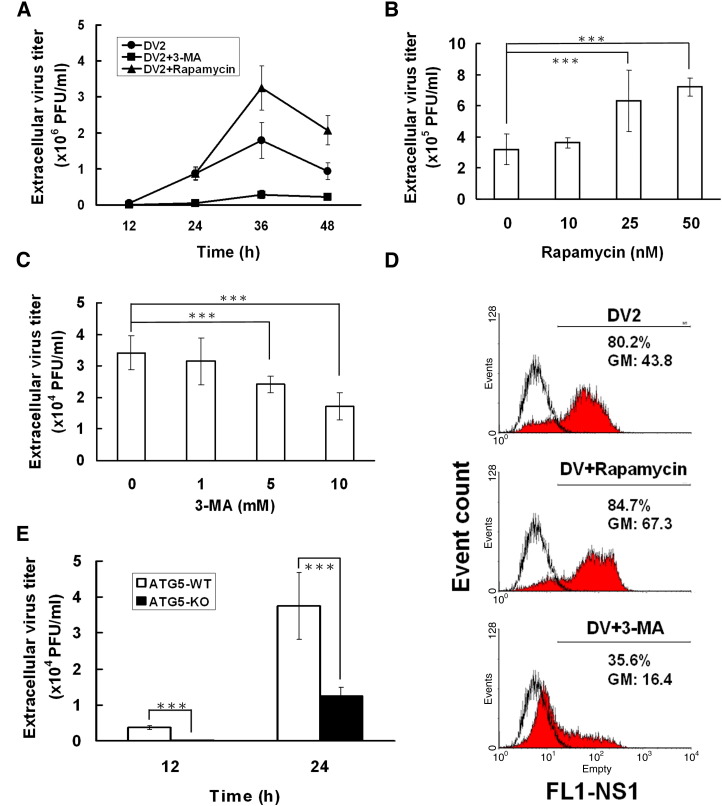
Fig. 4.
Determination of DV2 titer in Huh7 and MEF cells by plaque assay and flow-cytometry. (A) Extracellular viral titers in DV2 infected Huh7 cells with or without 3-MA or rapamycin were determined by plaque assay at the indicated times. Moreover, the extracellular viral titers in DV2 infected Huh7 cells combined with various dosages of Rapamycin (B) or 3-MA (C) were determined by plaque assay at 36 h p.i. (D) The intracellular DV-NS1 expression levels in DV2 infected Huh7 cells were determined using anti-DV-NS1 antibody labeling followed by flow-cytometry analysis. FL1-NS1: intensity of NS1 fluorescence; Event count: cell number; GM: Mean fluorescence of NS1 expressing cells. (E) DV2 titers in ATG5 wild-type and ATG5 knockout MEF cells were measured by plaque assay at indicated times. Each data point is representative of the results of three independent experiments. ATG5-WT: wild-type MEF cells; ATG5-KO: ATG5 knockout MEF cells; ⁎⁎⁎: means p < 0.05.
DV2-induced autophagic machinery can enhance viral replication
Induction of autophagy pathways can increase the replication of poliovirus and coronavirus (Jackson et al., 2005, Prentice et al., 2004). Therefore, it is intriguing to clarify the effect of DV2-induced autophagy on DV2 replication. After DV2 infection of Huh7 cells, extracellular viral titer from the culture supernatant and intracellular viral titers from the infected cells were determined by plaque assay and flow cytometry, respectively. The extracellular DV2 titer increased in DV2 infected Huh7 cells along with the increase of infection time from 12 to 36 h p.i., and then declined at 48 h p.i. (Fig. 4A). The replication rate reached the maximum at 36 h p.i. DV2 replication was evidently suppressed while autophagy was blocked by 3-MA from 24 h p.i. to 48 h p.i. (Fig. 4A). In DV2 infected cells, rapamycin was added to further induce autophagy, accordingly the DV2 titer was further increased at 36 and 48 h p.i. In addition, the extracellular DV2 titer increased in DV2 infected Huh7 cells was rapamycin dosage dependent (Fig. 4B). In contrast, 3-MA suppressed DV2 titer in a dosage dependent manner as well (Fig. 4C). However, DV2 titer was only suppressed about 50% at highest concentration of 3-MA treatments (10 mM), indicating that autophagic machinery is not the only mechanism for DV2 replication.
The DV non-structure protein 1 (NS1) plays a positive role in viral RNA replication, the expression level of NS1 can be used to represent the level of the intracellular DV2 replication (Mackenzie et al., 1996). Therefore, the level of DV2-NS1 protein in the infected Huh7 cells was labeled by anti-DV2-NS1 antibody and evaluated by flow cytometry. The mean fluorescence of the DV2-NS1 expression in DV2-infected Huh7 cells was approximately 43.8 (Fig. 4D, DV vs. No infection; GM). Rapamycin treatment of DV2 infected cells increased DV2-NS1 expression to 67.3. In contrast, treatment of DV2 infected cells with 3-MA suppressed DV2-NS1 expression to 16.4 (Fig. 4D). Taken together, our plaque assay and flow cytometry data demonstrate that both extracellular and intracellular DV titer can be induced by DV induced autophagy. The increase of intracellular virus titer indicates the increase of viral replication.
To further verify that DV2 induced autophagy can promote its replication, ATG5 knockout (ATG5-KO) MEF cells were used. The ATG5 protein is essential for the formation of autophagosomal membrane; the ATG12/ATG5 conjugation system was revealed in yeast and confirmed in mammalian cells (Mizushima et al., 2001, Nemoto et al., 2003). The extracellular DV2 virus titers after infection of wild-type and ATG5 knockout MEF cells for 12 and 24 h were evaluated by plaque assay. Fig. 4E shows that the extracellular DV2 titer from wild-type MEF cells was about 3-fold higher than that from ATG5 knockout cells at 24 h p.i. Our data indicate that ATG5 could not only control autophagosome formation but also affect autophagy related DV2 replication.
Discussion
The present study offers compelling evidence that DV2 infection could trigger autophagy machinery, induce autophagosome formation and promote viral replication in various cell lines. Moreover, ATG5 is involved in the activation of autophagic machinery, autophagosome formation and promotion of DV2 replication (which is different with coronavirus and vaccinia virus).
Autophagy can be induced in RNA and DNA viruses infected cells, however the detailed mechanisms are still elusive (Jackson et al., 2005, Nakashima et al., 2006, Prentice et al., 2004). Currently, three possible mechanisms participating in virus-induced autophagy have been reported. Firstly, double stranded RNA may induce autophagosome formation through reducing PKR activity and down-regulation of mTOR (Talloczy et al., 2002). Secondly, virus infection can induce autophagosome formation through ER stress (Yorimitsu and Klionsky, 2007). Thirdly, mTOR positively regulates cellular translation (Wullschleger et al., 2006). Several viral proteins can block cellular transcription and translation through suppression of mTOR activity, which may also induce autophagosome formation (Kato et al., 2002, Kopecky-Bromberg et al., 2006). However, what mechanism participates in DV infection induce autophagic machinery activation remaining unknown. Our unpublished data unravel the activities of mTOR and p70S6K (the down-stream effector of mTOR) were suppressed by DV2 infection. The suppression was further enhanced or reversed by 3-MA indicating that mTOR is involved in DV2 induced autophagy. DV infection can induce ER stress has been reported (Yu et al., 2006). ER stress, as has been induced autophagy in DV infection also deserves exploration.
Higher viral load correlates with the severity of dengue disease including DHF and DSS (Lei et al., 2001). Dendritic cells, monocytes, macrophages, endothelial cells, and liver cells are the natural target cells of the virus (Green and Rothman, 2006). According to the antibody-dependent enhancement hypothesis, anti-DV antibody binding with DV of different serotypes enhances viral entry into the target cells in an Fc receptor dependent or independent manner (Huang et al., 2006, Kliks et al., 1989). If the secondary infection of DVs is a different serotype, antibody-dependent enhancement may occur, leading to an increased virus titer in the host, promoting progression of the disease from mild DF to severe DHF/DSS. In this study, we unravel that DV2 can induce autophagy activation and autophagy activation can sustain more DV2 replication. Whether viral load induced by antibody-dependent enhancement is via an increased autophagy in DV-infected cells is noteworthy for exploration.
Autophagosome formation can increase poliovirus load, suggesting that autophagic machinery plays a favorable role in viral replication (Jackson et al., 2005). In this study, the increase of viral load was correlated with the increase of GFP-LC3 dot formation and LC3-II formation (Fig. 1D). Furthermore, both intracellular and extracellular viral loads are increased in the presence of the autophagy inducer rapamycin (Figs. 4B and D) and are reduced in the presence of the autophagy inhibitor 3-MA (Figs. 4C and D). And the difference in the viral loads between autophagic and non-autophagic cells is not due to the difference in infection efficiency because the levels of DV2-E protein in these two cell lines detected by immuno-fluorescence analysis were identical (data not shown). In summary, our results clearly indicate that DV2-induced autophagic machinery is favorable for DV2 replication. However, when autophagy is blocked by 3-MA or in ATG5-KO MEF cells, DV2 replication continues but at a reduced level, indicating that autophagic machinery may play a promoting role but not an essential role in DV2 replication. This view is consistent with a previous report (Jackson et al., 2005), which suggested that DV2 may utilize the autophagic machinery for virus replication. Both of the extracellular and intracellular DV titers can be increased by DV2 induced autophagy (Figs. 4A and D), indicating that autophagic machinery cannot only assist in conveying DV progeny from autophagosome to outside of the infected cells but also promote the virus replication inside the cells. If that is the case, whether autophagosome is the site for DV replication and what is the underlying mechanism requiring further exploration as well. Differently, ATG5 and/or Beclin-1 are not required for vaccinia virus and coronavirus replication have been reported, indicating that different viruses may utilize different signaling pathway to trigger autophagic machinery and affect viral replication (Wileman, 2006, Zhang et al., 2006, Zhao et al., 2007).
In addition, autophagic machinery can block innate antiviral immune responses (IFN-beta) has been reported (Jounai et al., 2007). Therefore, whether the increase of the viral titer was caused by down-regulation of IFN-beta through autophagy activation should be investigated.
Materials and methods
Cell line and virus
Huh7 (human hepatoma cell line) cells were cultured in DMEM, (Gibco, USA) supplemented with 10% FBS (Trace BioSciences, Australia) at 37 °C in a 5% CO2 incubator. Wild type mouse embryo-fibroblast (MEF) and ATG5−/− MEF cells were grown under the same conditions. Cells were infected with DV2 strain PL0146 isolated from Taiwan. The virus was routinely maintained in C6/36 cells. For virus infection, cells were adsorbed with DV2 at a multiplicity of infection (MOI) of 10 at 37 °C for 2 h. The cells were then washed three times with PBS and incubated at 37 °C in DMEM.
Plaque assay
BHK-21 cells were plated into 12-well plates (2 × 102 cells/well) and cultured in DMEM (GIBCO) under CO2-enriched conditions as described above. After adsorption with a serially-diluted virus solution for 2 h, the solution was replaced with fresh DMEM containing 2% FBS and 0.5% methyl cellulose (Sigma-Aldrich, USA). Five days postinfection (p.i.), the medium was removed, the cells were fixed, and stained with the crystal violet solution consisting of 1% crystal violet, 0.64% NaCl, and 2% formalin.
Transmission electron microscopy
Thirty-six hours after infection, Huh7 cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer containing 4% sucrose, 1 mM MgCl2, and 1 mM CaCl2, and post-fixed in 1% osmium tetroxide. The cells were further dehydrated with ethanol and embedded with LR White (Agar Scientific, UK). Ultrathin sections were stained with uranyl acetate and lead citrate, and then observed using a Hitachi 7000 transmission electron microscope (TEM; Hitachi, Tokyo, Japan). For immuno-gold labeling, ultra-thin sections were pretreated with 10% H2O2 for 10 min at room-temperature (RT). After treatment for 30 min at RT with blocking buffer (0.8% BSA in PBS), the ultrathin sections were exposed overnight at 4 °C with a 1:10 dilution in blocking buffer of anti-LC3 antibody (Abgent, San Diego, CA, USA). Sections were then exposed for 90 min at RT to similarly-diluted goat anti-rabbit IgG conjugated with 5-nm gold particles (British Biocell International, Cardiff, UK). Finally, ulttrathin sections were stained with uranyl acetate and lead citrate, and were examined under the TEM.
Immunofluorescence
Cells cultured in 10-cm culture dishes were transfected with 20 μg of pEGFPC1-LC3 or pEGFPC1. The dot formation of GFP-LC3 was detected under a fluorescence microscope (TCS SP2, Leica, USA) after drug treatment and/or DV2 infection. After transient transfection and DV infection, all the cells expressing GFP were counted (total cells). Among them, the cell containing ≧ 5 GFP-LC3 dots formation is defined as autophagy positive cells. Therefore, the % of the cells showing significant dots formation is = no. of autophagic cells/no. of total cells expressing GFP.
DV2 antigens and lysosome-associated membrane proteins-1 (LAMP1) expression in DV2-infected or drug-treated cells were detected by indirect immunofluorescence labeling. To examine the expression of prM protein of DV2 in the presence of GFP-LC3, the infected cells were fixed and immuno-labeled with anti-DV2-prM monoclonal antibodies (gifts from Dr. Huan-Yao Lei). Rhodamine-conjugated goat anti-mouse IgG monoclonal antibody (Santa Cruz Biotechnology, USA) was applied as the secondary antibody. Non-specific fluorescence was assessed using the secondary antibody. Samples were investigated using laser confocal scanning microscopy (FB1000, Olympus, Japan). Anti-LAMP1 monoclonal-antibody (Santa Cruz Biotechnology, USA) was used to detect the expression of LAMP1 using the aforementioned protocol.
Western blot analysis
Cells were cultured and infected with DV2 in 10-cm culture dishes. The whole cellular extract was subjected to the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) at various time post-infection (p.i.), and the separated proteins were electrically transferred to a PVDF membrane (Millipore Corporation, USA). The membrane was blocked with the sequential additions of the primary (anti-LC3 Ab; Abcam, USA; anti-β-actin Ab; Santa Cruz Biotechnology, USA) and secondary antibodies, incubation with enhanced chemiluminescence (ECL) solution for 1 min, and exposure to on XAR film (Eastman Kodak, NY).
Flow cytometry
Cells grown in a 6 cm plate were treated with drugs (3-MA or rapamycin) and/or infected with DV2. The cells were harvested and fixed with 3.7% paraformaldehyde after resuspended in PBS at 36 h p.i. After three PBS washes, cells were exposed to blocking buffer and stained with anti-DV2-NS1 antibody (gifts from Dr. Huan-Yao Lei). Green fluorescent protein (GFP)-conjugated goat anti-mouse IgG monoclonal antibody (Invitrogen, USA) was used as the secondary antibody after PBS washing. Cells were assayed using a FACScan fluorescence-activated cell sorter (BD Biosciences, USA).
Statistical analysis
Data are presented as the mean ± SE. Differences between the test and control groups were analyzed using the Student's t test. Significance was set at P < 0.05.
Acknowledgments
We thank Dr. N. Mizushima and Dr. T. Yoshimori for providing the GFP-LC3 plasmid and ATG5 wild-type as well as ATG5 knockout MEF cells. This research was supported by grants NHRI-CN-CL9303P, NHRI-CN-CL9601S from the National Health Research Institute, and NSC 96-2628-B-006-003-MY3 from National Science Council, Taiwan, R.O.C.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.virol.2008.02.016.
Appendix A. Supplementary data
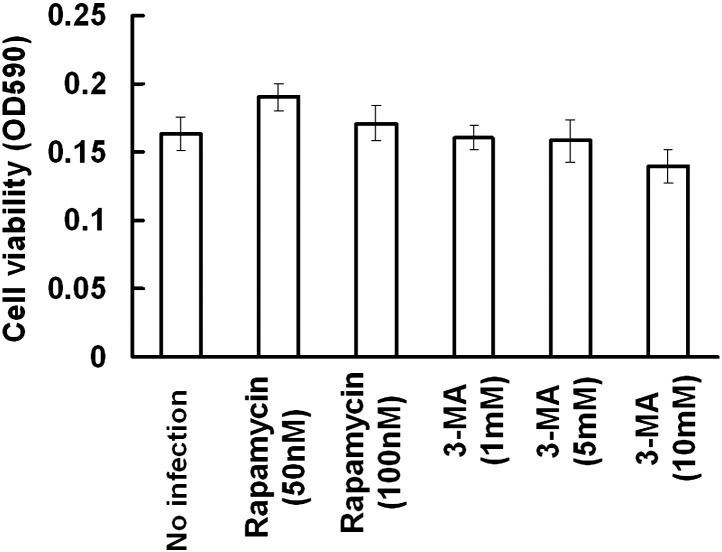
Supplementary Fig 1.
The effect of Rapamycin and 3-MA on Huh7 cells. Huh7 cells were treated with various dosages of Rapamycin or 3-MA for 36 h. Cell variability after drug treatment was determined by MTT assay.
Supplementary Fig 2.
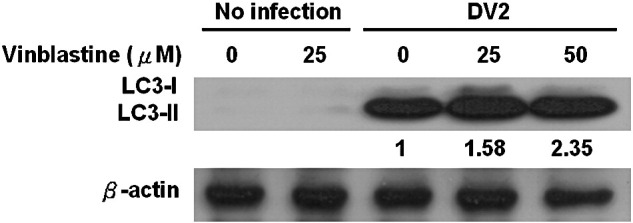
The expression levels of LC3 in Huh7 cells after drug treatment to block autophagolysosome formation. Huh7 cells were lysed and protein was analyzed for the levels of LC3-II by Western blotting. The expression levels of LC3-II in DV2 infected cells and in the presence or absence of drug vinblastine were detected by Western blotting. Noninfected and nontreated cells were used as the negative controls.
Supplementary Fig 3.
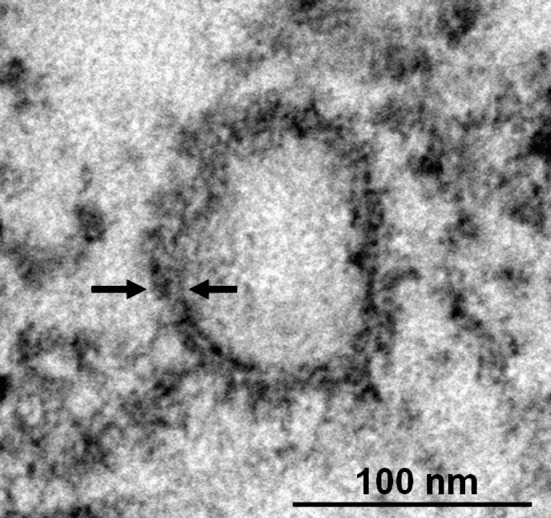
Detection of coclocalization of LC3 and autophagosome in the DV2 infected Huh7 cells. Huh7 cells infected with DV2 for 36 h were fixed and stained with anti-LC3 antibody, and visualized by a secondary antibody coupled with HRP. The arrow indicates LC3 labeled region surrounding the double-membrane vesicle.
References
- Alexander D.E., Ward S.L., Mizushima N., Levine B., Leib D.A. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J. Virol. 2007;81:12128–12134. doi: 10.1128/JVI.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer A.O., Swanson M.S. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell. Microbiol. 2005;7:765–778. doi: 10.1111/j.1462-5822.2005.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S., Rothman A. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr. Opin. Infect. Dis. 2006;19:429–436. doi: 10.1097/01.qco.0000244047.31135.fa. [DOI] [PubMed] [Google Scholar]
- Gutierrez M.G., Vazquez C.L., Munafo D.B., Zoppino F.C., Beron W., Rabinovitch M., Colombo M.I. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell. Microbiol. 2005;7:981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- Hoyvik H., Gordon P.B., Seglen P.O. Use of a hydrolysable probe, [14C]lactose, to distinguish between pre-lysosomal and lysosomal steps in the autophagic pathway. Exp. Cell Res. 1986;166:1–14. doi: 10.1016/0014-4827(86)90503-3. [DOI] [PubMed] [Google Scholar]
- Huang K.J., Yang Y.C., Lin Y.S., Huang J.H., Liu H.S., Yeh T.M., Chen S.H., Liu C.C., Lei H.Y. The dual-specific binding of dengue virus and target cells for the antibody-dependent enhancement of dengue virus infection. J. Immunol. 2006;176:2825–2832. doi: 10.4049/jimmunol.176.5.2825. [DOI] [PubMed] [Google Scholar]
- Jackson W.T., Giddings T.H., Jr., Taylor M.P., Mulinyawe S., Rabinovitch M., Kopito R.R., Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jounai N., Takeshita F., Kobiyama K., Sawano A., Miyawaki A., Xin K.Q., Ishii K.J., Kawai T., Akira S., Suzuki K., Okuda K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J., Kato N., Yoshida H., Ono-Nita S.K., Shiratori Y., Omata M. Hepatitis C virus NS4A and NS4B proteins suppress translation in vivo. J. Med. Virol. 2002;66:187–199. doi: 10.1002/jmv.2129. [DOI] [PubMed] [Google Scholar]
- Kliks S.C., Nisalak A., Brandt W.E., Wahl L., Burke D.S. Antibody-dependent enhancement of dengue virus growth in human monocytes as a risk factor for dengue hemorrhagic fever. Am. J. Trop. Med. Hyg. 1989;40:444–451. doi: 10.4269/ajtmh.1989.40.444. [DOI] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Palese P. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J. Virol. 2006;80:785–793. doi: 10.1128/JVI.80.2.785-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Lei H.Y., Yeh T.M., Liu H.S., Lin Y.S., Chen S.H., Liu C.C. Immunopathogenesis of dengue virus infection. J. Biomed. Sci. 2001;8:377–388. doi: 10.1007/BF02255946. [DOI] [PubMed] [Google Scholar]
- Mackenzie J.M., Jones M.K., Young P.R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A., Tanaka N., Tamai K., Kyuuma M., Ishikawa Y., Sato H., Yoshimori T., Saito S., Sugamura K. Survival of parvovirus B19-infected cells by cellular autophagy. Virology. 2006;349:254–263. doi: 10.1016/j.virol.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Nemoto T., Tanida I., Tanida-Miyake E., Minematsu-Ikeguchi N., Yokota M., Ohsumi M., Ueno T., Kominami E. The mouse APG10 homologue, an E2-like enzyme for Apg12p conjugation, facilitates MAP-LC3 modification. J. Biol. Chem. 2003;278:39517–39526. doi: 10.1074/jbc.m300550200. [DOI] [PubMed] [Google Scholar]
- Paludan C., Schmid D., Landthaler M., Vockerodt M., Kube D., Tuschl T., Munz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talloczy Z., Jiang W., Virgin H.W.T., Leib D.A., Scheuner D., Kaufman R.J., Eskelinen E.L., Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wileman T. Aggresomes and autophagy generate sites for virus replication. Science. 2006;312:875–878. doi: 10.1126/science.1126766. [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D.J. Endoplasmic reticulum stress: a new pathway to induce autophagy. Autophagy. 2007;3:160–162. doi: 10.4161/auto.3653. [DOI] [PubMed] [Google Scholar]
- Yu C.Y., Hsu Y.W., Liao C.L., Lin Y.L. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J. Virol. 2006;80:11868–11880. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Monken C.E., Zhang Y., Lenard J., Mizushima N., Lattime E.C., Jin S. Cellular autophagy machinery is not required for vaccinia virus replication and maturation. Autophagy. 2006;2:91–95. doi: 10.4161/auto.2.2.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Thackray L.B., Miller B.C., Lynn T.M., Becker M.M., Ward E., Mizushima N.N., Denison M.R., Virgin H.W.t. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3:581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]