Abstract
In this study, we analyzed the replication and budding sites of severe acute respiratory syndrome coronavirus (SARS-CoV) at early time points of infection. We detected cytoplasmic accumulations containing the viral nucleocapsid protein, viral RNA and the non-structural protein nsp3. Using EM techniques, we found that these putative viral replication sites were associated with characteristic membrane tubules and double membrane vesicles that most probably originated from ER cisternae. In addition to its presence at the replication sites, N also accumulated in the Golgi region and colocalized with the viral spike protein. Immuno-EM revealed that budding occurred at membranes of the ERGIC (ER–Golgi intermediate compartment) and the Golgi region as early as 3 h post infection, demonstrating that SARS-CoV replicates surprisingly fast. Our data suggest that SARS-CoV establishes replication complexes at ER-derived membranes. Later on, viral nucleocapsids have to be transported to the budding sites in the Golgi region where the viral glycoproteins accumulate and particle formation occurs.
Keywords: SARS-coronavirus, Replication, Budding, Double membrane vesicles
Introduction
SARS-coronavirus (SARS-CoV) is the etiologic agent of severe acute respiratory syndrome (SARS) (Drosten et al., 2003, Fouchier et al., 2003, Ksiazek et al., 2003, Kuiken et al., 2003, Peiris et al., 2003). The virus belongs to the family of Coronaviridae which are enveloped viruses with a positive-sense RNA genome of about 30,000 nt length. Coronaviruses share a common set of genes, encoding non-structural proteins (nsps) involved in RNA replication (ORF 1a/1ab) and structural proteins such as the nucleocapsid protein N, the membrane protein M, the envelope protein E and the spike protein S. In addition, there are several open reading frames with undefined or poorly characterized functions, some of which also encode structural proteins (Holmes, 1996, Navas-Martin and Weiss, 2004).
The Coronaviridae family consists of three phylogenetic groups. Human pathogens are found in group I (229E and NL63) and in group II (OC43 and HKU1). SARS-CoV constitutes either a new phylogenetic group (Eickmann et al., 2003) or an early split-off of group II (Snijder et al., 2003). Mouse hepatitis virus (MHV), a member of group II, is the prototype coronavirus used to study the cell biological aspects of replication and assembly. For MHV it was found that ORF 1a/1ab gene products and the N protein colocalize with the viral RNA in cytoplasmic complexes. These replication sites are associated with intracellular membranes of characteristic shape. The formation of double membrane vesicles (DMVs) was observed specifically in infected cells (van der Meer et al., 1999, Gosert et al., 2002, Brockway et al., 2003, Prentice et al., 2004a). However, the origin of the membranes has not been clearly identified. It has been suggested that MHV uses components of the cellular autophagy machinery to form DMVs (Prentice et al., 2004a). Autophagy is normally involved in the turnover of long-lived proteins and damaged organelles (Stromhaug and Klionsky, 2001) and is part of the innate immune response (Levine, 2005). Recently, it has been shown that the autophagy pathway can be exploited by intracellular pathogens, including members of the Picorna- and Coronaviridae, to facilitate their replication (Prentice et al., 2004a, Jackson et al., 2005). Assembly and budding of new MHV particles take place in the pre-Golgi compartment also called the ERGIC (ER–Golgi intermediate compartment) (Tooze et al., 1984, Klumperman et al., 1994). Therefore, it was speculated that viral genomic RNA and some viral proteins, including nucleocapsid, have to translocate from the RNA replication sites to the budding compartment at the ERGIC (Bost et al., 2001). By contrast, other components of the replication complex, such as the RNA-dependent RNA polymerase, stay associated with the viral replication sites throughout the infection cycle (Brockway et al., 2003).
For SARS-CoV, the intracellular sites of replication and budding have not been fully elucidated. With respect to viral particle formation, it has been described that SARS-CoV budding occurs at a pre-Golgi compartment as well as at the outer nuclear membrane 3–5 days post infection (p.i.) (Goldsmith et al., 2004), whereas in another study virus particles budding into the lumen of structures which resembled swollen Golgi cisternae were detected already at 5 h p.i. (Ng et al., 2003).
Antibodies directed against the ORF 1a/1ab proteins of SARS-CoV were used to analyze the intracellular localization of the viral replication sites. As described for other coronaviruses, the non-structural proteins accumulated in discrete cytoplasmic complexes (Harcourt et al., 2004, Ivanov et al., 2004, Prentice et al., 2004b). In addition, SARS-CoV non-structural proteins colocalized with LC3, a marker protein for autophagosomes, reminiscent of the replication complexes of MHV (Kabeya et al., 2000, Prentice et al., 2004a, Prentice et al., 2004b). However, ultrastructural evidence supporting this conclusion has not been provided. Moreover, it was demonstrated that the complexes containing the non-structural proteins colocalized with newly synthesized RNA on virus induced DMVs (Harcourt et al., 2004). While we were preparing our manuscript, Snijder et al. published a study on the ultrastructure of the SARS-CoV replication sites (Snijder et al., 2006). They show colocalization of different SARS-CoV non-structural proteins (nsp) at 9 h p.i. and the appearance of putative viral replication sites associated with DMVs from 6 h p.i. on. Interestingly, they observed ER membranes in vicinity to the DMVs and found SARS-CoV nsps present at ER membranes. However, direct evidence for the ER origin of the DMVs was not provided.
The aim of our study was to further characterize both the RNA replication site and the budding compartment of SARS-CoV. We focused in our studies on early time points of infection, reasoning that early in infection the ultrastructural architecture of the cell might be better preserved. Using immunofluorescence analysis and in situ hybridization, we could show that at early time points after infection the viral nucleocapsid protein (SARS-CoV N) accumulates in cytoplasmic complexes together with viral genomic RNA and nsp3. We then investigated the ultrastructure of these putative replication sites. Standard EM revealed the presence of DMVs and characteristic membrane tubules that protruded from ribosome-studded ER cisternae already at 3 h p.i. Cryo-immuno-EM (iEM) confirmed that the sites double positive for N and nsp3 labeling were associated with membrane tubules that originated from ER cisternae and probably represent precursors of DMVs. The viral glycoprotein SARS-CoV S, by contrast, was detected exclusively in the Golgi area and was clearly distinct from the replication sites. Further EM studies revealed that budding of SARS-CoV particles takes place at the ERGIC and Golgi region. Therefore, we conclude that genome replication and budding of SARS-CoV occur on separate membrane-associated sites in the cytoplasm: ER membranes are most likely used for the formation of replication sites, whereas budding takes place at membranes in the ERGIC and Golgi region. Strikingly, SARS-CoV runs through its replication cycle very rapidly as budding was observed already at 3 h p.i.
Results
Intracellular distribution of SARS-CoV N, SARS-CoV nsp3 and SARS-CoV S early in infection
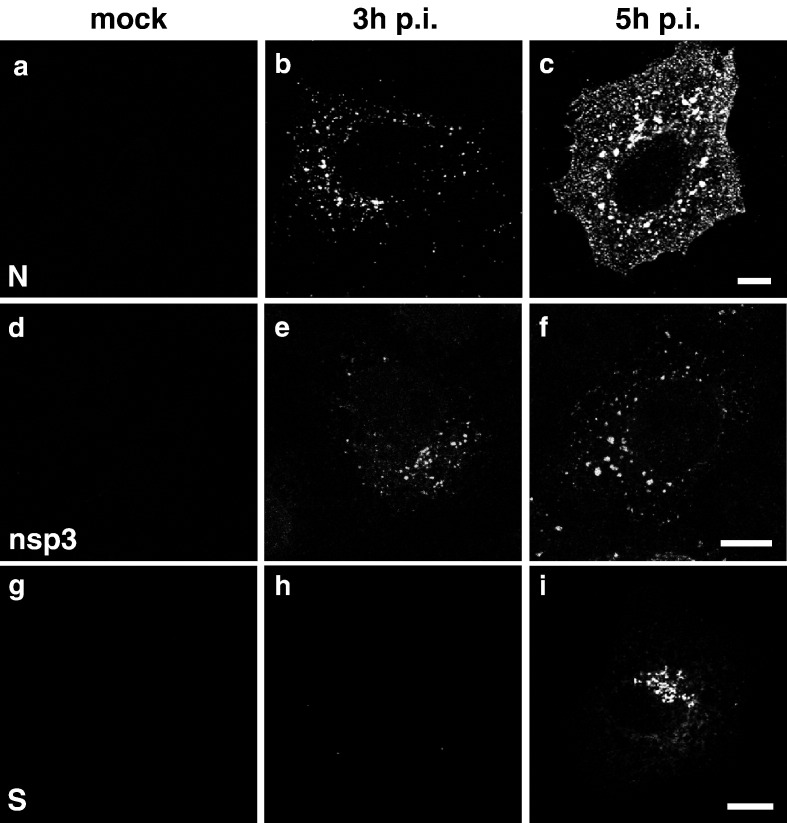
In a first set of experiments we analyzed the intracellular localization of the SARS-CoV proteins N, nsp3 and S. The nucleocapsid (N) protein of coronaviruses is involved in replication and transcription of the genome, but it is also a structural component of the viral particle (Almazan et al., 2004, Schelle et al., 2005). SARS-CoV nsp3 is a non-structural protein which functions as a papain-like accessory proteinase (PLpro) (Thiel et al., 2003, Harcourt et al., 2004). In addition, ADP-ribose-1″-mono-phosphatase activity has been described for nsp3 (Putics et al., 2005), and it has been shown that nsp3 is part of the viral replication complex (Harcourt et al., 2004, Snijder et al., 2006). The spike protein (S) is a structural protein present in the membrane of viral particles. We generated specific antibodies, a polyclonal rabbit serum and a mouse monoclonal antibody, both specific for SARS-CoV N, and a monoclonal mouse antibody against SARS-CoV S to determine the intracellular localization of N and S during the course of infection. In addition, we monitored nsp3 localization using an antibody described by Harcourt et al. (2004). Vero cells were mock-infected or infected with SARS-CoV for 3 h or 5 h, permeabilized and analyzed by immunofluorescence. Anti-N staining revealed that as early as 3 h p.i., SARS-CoV N was detectable in small cytoplasmic foci (Fig. 1b). At 5 h p.i. these foci had grown to large accumulations and perinuclear enrichment of N became visible (Fig. 1c). Staining against nsp3 resulted in a similar pattern at 3 h p.i. (Fig. 1e), but was different from the N distribution at 5 h (Fig. 1f): Nsp3 was enriched in small cytoplasmic foci but no perinuclear accumulation was observed. When cells were stained for SARS-CoV S, we observed a different distribution: SARS-CoV S became first detectable at 5 h p.i. and was found exclusively in the perinuclear area (Fig. 1i).
Fig. 1.
Distribution of SARS-CoV N, SARS-CoV nsp3 and SARS-CoV S at early time points of infection. Vero cells were either mock-infected (a, d, g) or infected with SARS-CoV for 3 h (b, e, h) or 5 h (c, f, i) with an MOI of 10. Cells were fixed, permeabilized and analyzed by immunofluorescence using antibodies directed against SARS-CoV N (a–c), SARS-CoV nsp3 (d–f) or SARS-CoV S protein (g–i). Scale bars represent 10 μm.
Cytoplasmic accumulations of SARS-CoV N colocalize with nsp3 and viral genomic RNA; perinuclear N colocalizes with SARS-CoV S and a Golgi marker
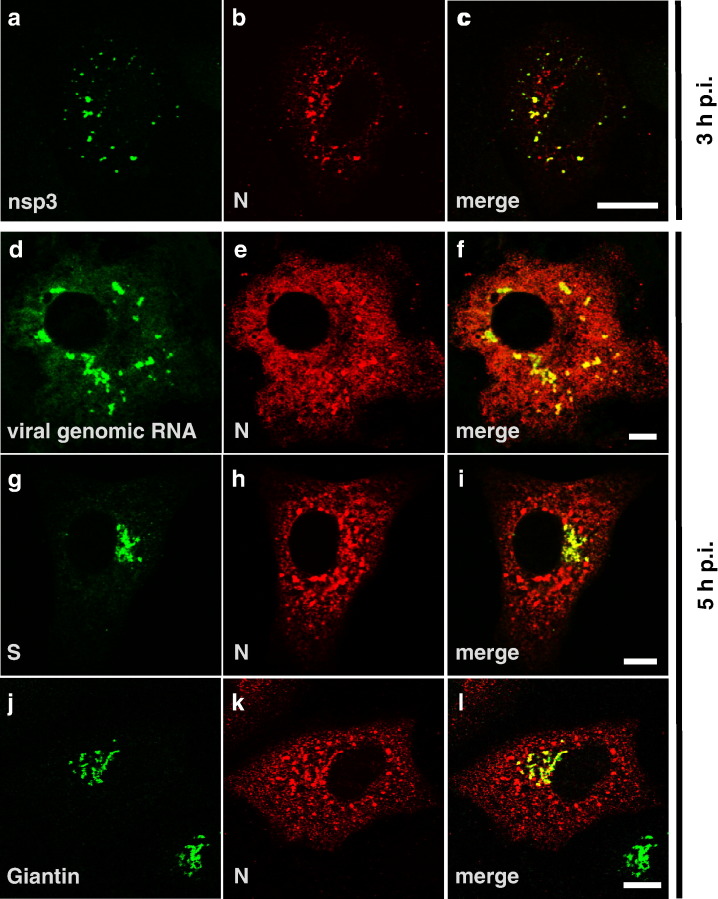
Since the intracellular staining patterns of SARS-CoV N and nsp3 were very similar at 3 h p.i., we double-stained for SARS-CoV nsp3 and SARS-CoV N (Figs. 2a–c). We found a clear colocalization of nsp3 and N in cytoplasmic foci at 3 h p.i. Another study had shown that nsp3 colocalized with newly synthesized viral RNA in SARS-CoV-infected cells and it was suggested that the sites of nsp3 and RNA accumulation represented viral replication sites (Harcourt et al., 2004). Given the role of N in coronavirus replication (Almazan et al., 2004, Schelle et al., 2005), we investigated whether cytoplasmic N foci also contained viral RNA. To this aim, Vero cells were infected with SARS-CoV, fixed and analyzed by in situ hybridization combined with immunofluorescence staining of the viral N protein. For the detection of viral RNA, an FITC-labeled RNA probe which hybridizes to the SARS-CoV nsp1-ORF was used. The nsp1-ORF is located at the 5′ end of the viral genome and is therefore only present on the full-length genomic RNA of SARS-CoV. Thus, our probe of negative polarity should detect full-length viral RNA of positive polarity. When the probe was tested on mock-infected cells, no signal was detected, confirming the specificity of the RNA probe (data not shown). In SARS-CoV-infected cells, at 5 h p.i. clear signals for viral RNA were detected (Fig. 2d), whereas at earlier time points viral RNA-specific labeling could not be observed (data not shown). When the cells were examined in parallel for expression of the viral N protein (Fig. 2e), it turned out that accumulations of the N protein colocalized with sites of strong viral RNA signals (Fig. 2f). Additional diffuse cytoplasmic N staining that did not colocalize with viral RNA was also evident, most likely representing newly synthesized free N protein. It should be mentioned that the distribution of N in Fig. 2e was slightly different than in Figs. 2h and k: more diffuse N staining throughout the cytoplasm was visible in Fig. 2e. This might be explained by the different fixation procedure used for in situ hybridization. Next, we compared the signals for the viral nucleoprotein N and the viral glycoprotein S at 5 h p.i. by double-immunostaining since we had observed perinuclear accumulation of both proteins. Indeed, colocalization of SARS-CoV N with SARS-CoV S was observed in the perinuclear area, whereas the N signals in the peripheral dots were clearly distinct from the S glycoprotein-positive area (Figs. 2g–i). When we stained for the Golgi marker protein Giantin in parallel, we found a clear colocalization of the perinuclear N and S containing accumulations with Giantin (Figs. 2j–l and data not shown). In contrast, staining for nsp3 resulted in no overlap with staining either for S or the Golgi marker (data not shown).
Fig. 2.
Cytoplasmic accumulations of SARS-CoV N colocalize with nsp3 and viral genomic RNA; perinuclear N colocalizes with S and a Golgi marker. (a–c) Vero cells were infected with SARS-CoV (MOI of 10) for 3 h, fixed and stained with specific antibodies for SARS-CoV nsp3 (a, green) and N (b, red) in parallel. (d–f) Vero cells were infected with SARS-CoV (MOI of 10) for 5 h, fixed and in situ hybridization was performed with a FITC-labeled RNA probe of negative polarity directed against the nsp1-ORF (d, green). After in situ hybridization, immunofluorescence staining was performed with an antibody specific for SARS-CoV N (e, red). (g–l) Vero cells were infected with SARS-CoV (MOI of 10) for 5 h, fixed, permeabilized and stained with antibodies specific for SARS-CoV S or Giantin, a marker for cis-Golgi, (g, j, green) and SARS-CoV N (h, k, red). Areas of colocalization appear in yellow in the merged images (c, f, i, l). Scale bars represent 10 μm.
In summary, our immunofluorescence studies have shown that as early as 3 h p.i., cytoplasmic accumulations of SARS-CoV N which colocalize with SARS-CoV nsp3 and viral genomic RNA are formed in infected cells. These cytoplasmic foci most probably represent viral replication sites, a hypothesis which is supported by the results of two other studies (Harcourt et al., 2004, Snijder et al., 2006). From 5 h p.i. on, N staining also became visible in the perinuclear area colocalizing with S and a Golgi marker whereas nsp3 was clearly excluded from these sites. This suggested that the N- and S-double-positive areas represent assembly and budding sites.
Budding of SARS-CoV takes place in the Golgi region already at 3 h p.i
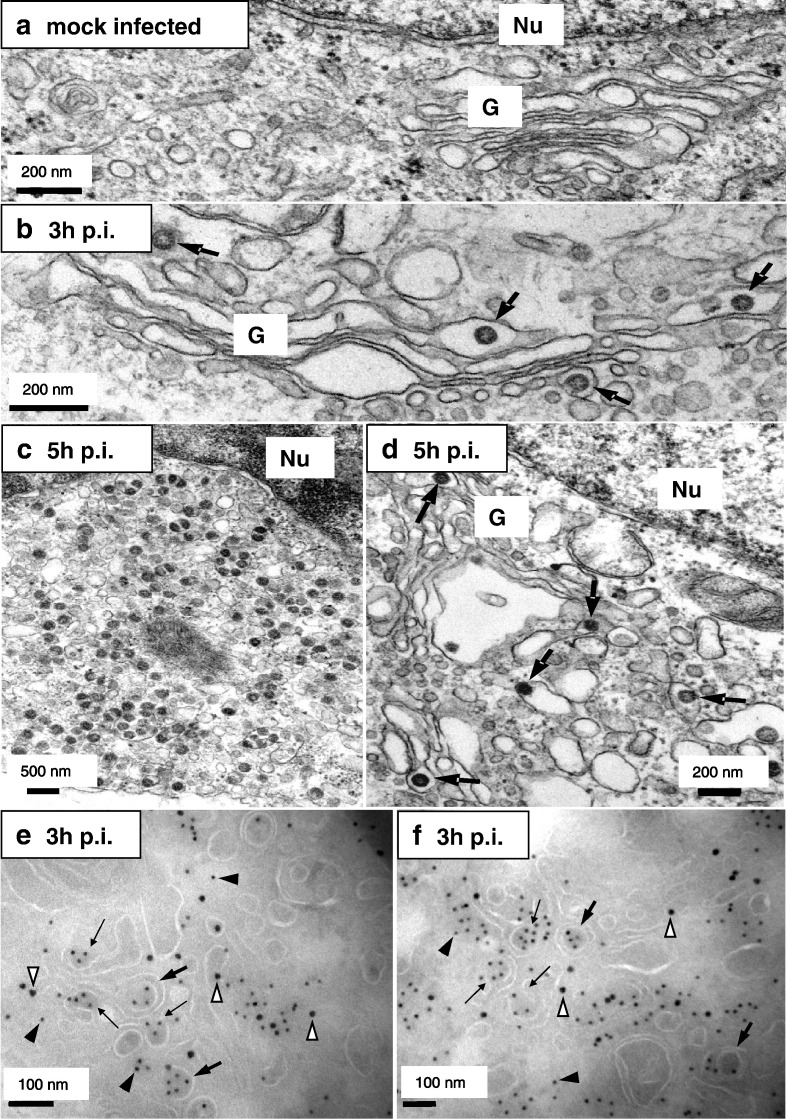
To analyze budding of SARS-CoV, we performed standard EM, which is well suitable for the detection of virus particles. Surprisingly, we found viral particles already at 3 h p.i. in Golgi cisternae and vesicles close to the Golgi (Fig. 3b, large arrows), whereas no viral structures could be detected in uninfected cells (Fig. 3a). At 5 h p.i. many particles were found throughout the cell (Figs. 3c, d). The budding and presence of numerous SARS-CoV particles in Golgi cisternae clearly indicated that they were newly assembled and did not represent virus inoculum. As an additional control we also infected Vero cells with UV-irradiated SARS-CoV. In contrast to infection with native virus, viral particles were not detectable in these samples (data not shown). For other coronaviruses it was shown that budding takes place in the ERGIC (Tooze et al., 1984, Klumperman et al., 1994). To gain insights into the assembly and budding sites of SARS-CoV, we performed immuno-EM (iEM) studies. Unfortunately, the antibody specific for S was not suitable for iEM. We therefore used an N-specific antibody to label viral budding profiles and particles. Additionally, cryosections were labeled with an antibody specific for β′COP-I, a well established marker for the ERGIC and Golgi region (Lowe and Kreis, 1995). We found a clear colocalization of N (Figs. 3e, f, filled arrowheads) with the β′COP-I marker (Figs. 3e, f, open arrowheads) indicating accumulation of N in the ERGIC and Golgi region. Again, as early as 3 h p.i. we observed partially enveloped N-containing structures of approximately the size of virions, most likely representing budding particles (Figs. 3e, f, small arrows) as well as completely budded particles (Figs. 3e, f, large arrows). This strongly suggests that budding of SARS-CoV particles takes place in the ERGIC and Golgi area. It should be noted that we observed SARS-CoV particles by EM at a time point when we could not observe SARS-CoV N and SARS-CoV S accumulation in the Golgi by immunofluorescence staining. This may be explained by the much higher sensitivity of EM which enabled us to detect newly formed SARS-CoV particles at the ultrastructural level even at 3 h p.i.
Fig. 3.
Budding of SARS-CoV occurs in the ERGIC and Golgi area already at 3 h p.i. (a–d) Vero cells were mock-infected (a) or infected with SARS-CoV (MOI of 10) for 3 h (b) or 5 h (c, d), fixed and analyzed by standard EM. The nuclei are marked with “Nu” and the Golgi with “G”. Viral particles are highlighted with arrows (b–d). Scale bars represent 200 or 500 nm as indicated. (e, f) Vero cells were infected with SARS-CoV (MOI of 10) for 3 h, fixed and analyzed by cryo-iEM with antibodies directed against SARS-CoV N (10 nm gold, filled arrowheads) and β′COP-I as a marker for the ERGIC and Golgi area (15 nm gold, open arrowheads). Budding profiles are marked with small arrows, large arrows point to viral particles. Scale bars represent 100 nm.
Ultrastructural analysis of the putative SARS-CoV replication sites
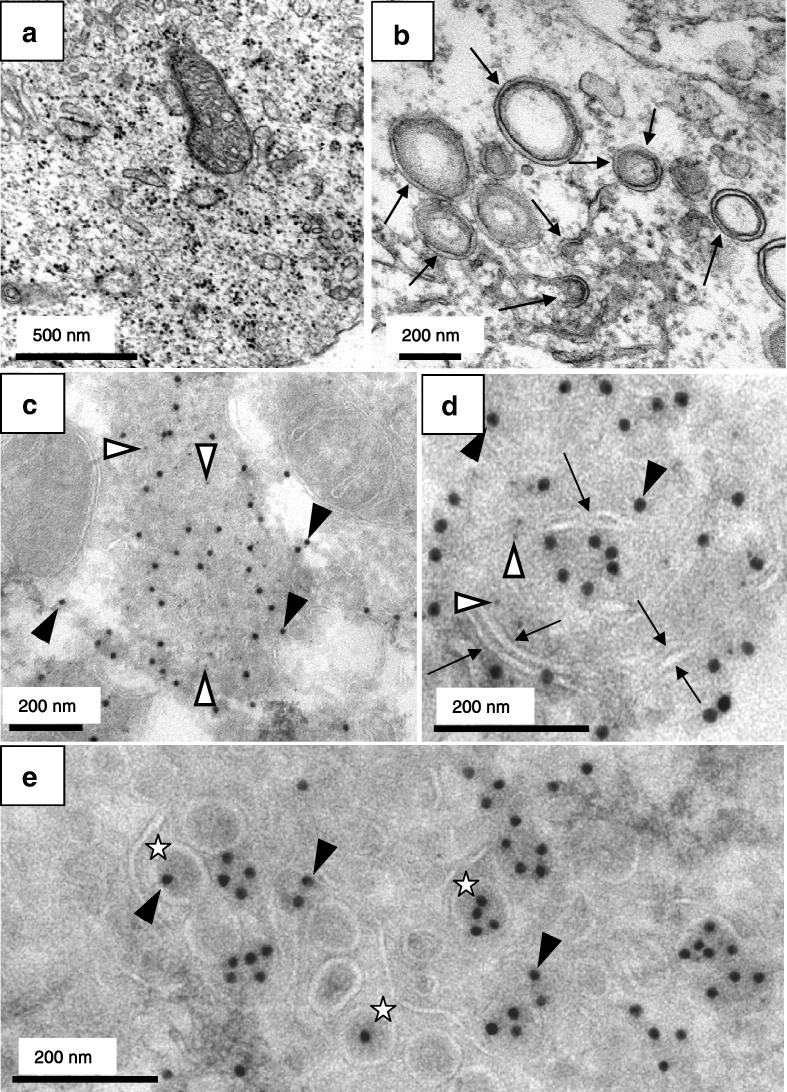
Our immunofluorescence studies had suggested that viral replication sites were formed already at 3 h p.i. To confirm this, we analyzed cells infected with SARS-CoV by standard EM. Harcourt et al. had described characteristic DMVs in SARS-CoV-infected cells representing viral replication sites (Harcourt et al., 2004). Indeed, we readily found DMVs in infected cells at 3 h p.i. (Fig. 4b) but not in mock-infected cells (Fig. 4a). The vesicles had a size of about 200–400 nm and the distance between the two opposing membranes was about 15 nm. Next, we performed cryo-iEM to further characterize viral replication sites. Thawed cryosections of cells infected with SARS-CoV for 3 h were labeled with antibodies specific for SARS-CoV N or nsp3. The specificity of both antibodies in cryo-iEM was controlled with mock-infected cells where almost no signal was detected (data not shown). N and nsp3 were visible in cytoplasmic foci (Figs. 4c, d) which were clearly distinct from the Golgi-associated budding sites and most probably correspond to the putative replication sites identified by immunofluorescence and in situ hybridization (Fig. 2). As shown above (Fig. 3), the N-specific labeling was also associated with budding viral particles in the Golgi area. In this region almost no nsp3 labeling was found indicating the absence of replication sites from the Golgi region (Fig. 4e). Most interestingly, we observed characteristic membrane tubules of about 15 nm in diameter within the N/nsp3 accumulations (marked with arrows in Fig. 4d) that were not found in mock-infected cells. As we frequently found ER cisternae in close proximity to the N/nsp3 double positive sites, we postulated that the specific membrane tubules were derived from ER cisternae. In line with this hypothesis we found enrichment of an ER marker protein at the N/nsp3 double positive sites in immunofluorescence studies (data not shown).
Fig. 4.
Ultrastructural analysis of the putative SARS-CoV replication sites. (a, b) Vero cells were mock-infected (a) or infected with SARS-CoV (MOI of 10) for 3 h (b), fixed and analyzed by standard EM. Double membrane vesicles characteristic for SARS-CoV replication sites are highlighted with arrows. (c–e) Vero cells were infected with SARS-CoV (MOI of 10) for 3 h, fixed and analyzed by cryo-iEM. Cells were labeled with antibodies directed against SARS-CoV N (15 nm gold, filled arrowheads) and SARS-CoV nsp3 (5 nm gold, open arrowheads). Areas of colocalization of N and nsp3 are shown in panels c and d. Membrane tubules specific for these sites are marked with arrows. Sites where viral particles (marked with stars) were present only contained N labeling but almost no nsp3 labeling (e). Scale bars represent 200 or 500 nm as indicated.
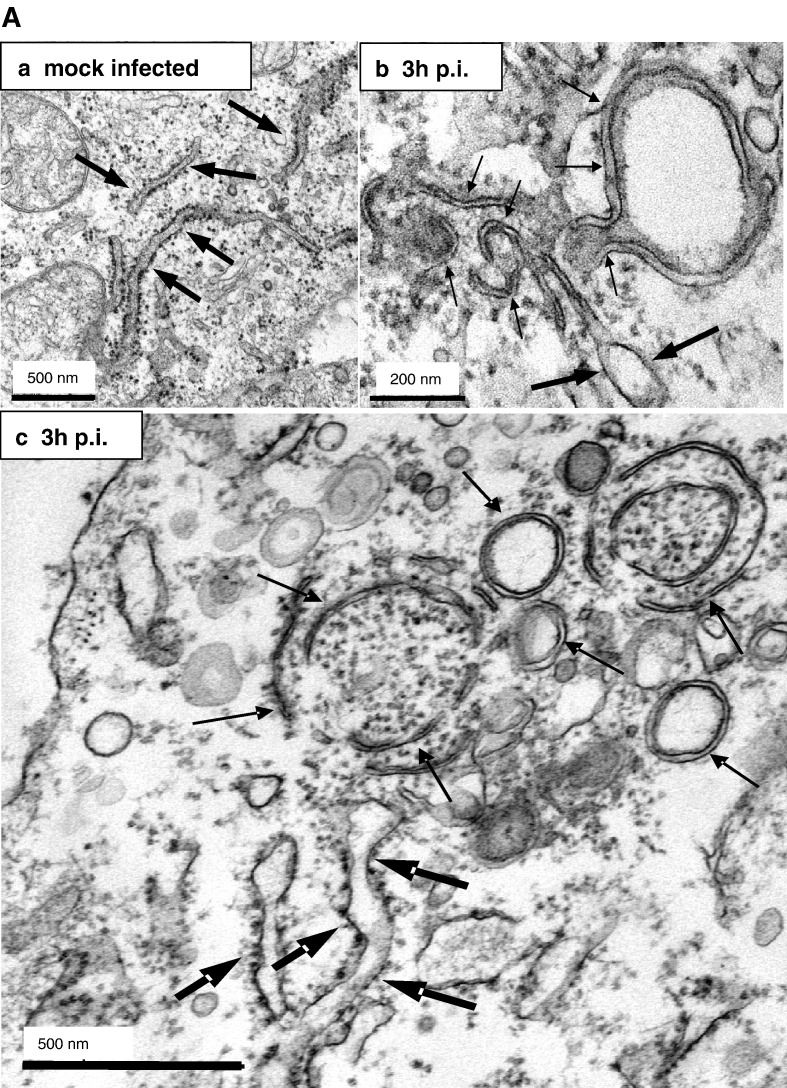
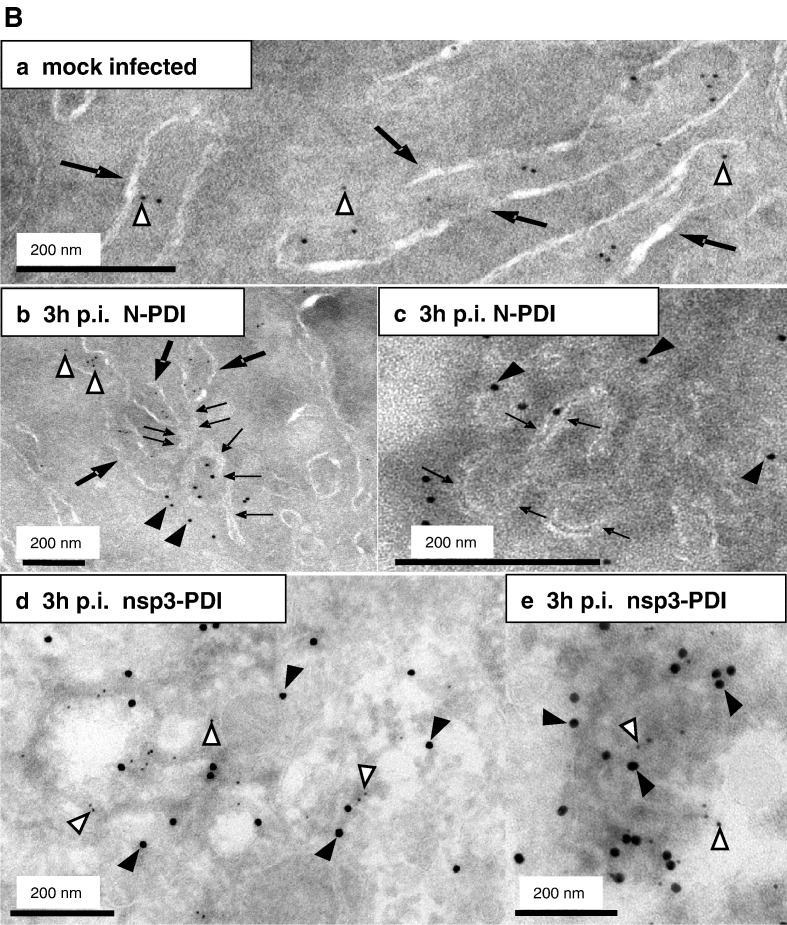
Membrane tubules within the SARS-CoV replication sites are derived from ER membranes
In order to further analyze the association of viral replication sites with ER membranes, we performed an ultrastructural analysis of both non-infected and infected cells using a combination of immuno- and standard EM techniques. At 3 h after infection, cells were fixed and processed for either standard embedding in epoxy-resin (Fig. 5A) or cryosectioning (Fig. 5B). First, we analyzed conventionally embedded control and infected cells, reasoning that in these preparations double membrane vesicles and ER membranes might be well preserved. As shown in Fig. 4b, DMVs were readily found in SARS-CoV-infected cells at 3 h p.i. (Figs. 5Ab, c) but were only rarely detected in mock-infected cells (Fig. 5Aa). Furthermore, thin membrane tubules that protruded from ribosome-studded ER cisternae were also observed (Figs. 5Ab, c). These structures were of about 10 to 20 nm in diameter and were closely associated with the DMVs. The consistent finding of concentrically arranged, curved or nearly closed tubules together with double membrane vesicles strongly suggests that the latter structures are derived from tubular ER-protrusions. To confirm ER origin of the characteristic membrane tubules, we next performed cryo-iEM. Thawed cryosections were labeled with antibodies specific for SARS-CoV N and protein disulfide isomerase (PDI), a well established marker for the ER that labels specifically the lumen of ER cisternae. The specificity of the double-immunogold labeling was controlled with sections of mock-infected cells (Fig. 5Ba). The lumen of the ER cisternae (Fig. 5B, large arrows) was specifically labeled with the anti-PDI antibody (Fig. 5B, open arrowheads) in both control cells (Fig. 5Ba) and infected cells (Fig. 5Bb). Incubation of sections with the anti-N antibody did hardly lead to any immunogold labeling in mock-infected but clear signals in infected cells, confirming its specificity and excluding any crossreaction with the anti-PDI antibody (Fig. 5B, filled arrowheads). The membrane tubules associated with the N-specific signal had essentially the same dimensions as the specific tubules observed in conventionally embedded sections (compare Figs. 5A and B) and indeed originated from PDI-positive ER cisternae (Fig. 5Bb, small arrows). When sections were labeled with an nsp3-specific antibody in combination with the anti-PDI antibody, both labels were found clustering together at the edge of extracted sites in the cytoplasm which probably correspond to DMVs (Figs. 5Bd, e). Although in this case the specific membrane tubules were not as clearly visible as in Figs. 5Bb and c, the nsp3–PDI labeling confirmed the association of the replication sites with the ER. Therefore, one can summarize that the putative viral RNA replication sites comprise distinct ER-derived membrane protrusions that specifically occur in infected cells and were never observed in control cells. In ultrathin sections (60–70 nm) that merely allow a two-dimensional view of three-dimensional structures, these protrusions appeared as tubule-like structures of about 10–20 nm (Figs. 5Bb, c, small arrows). It is very likely that these structures in fact represent flat ER-derived double membrane sheets when seen in three dimensions. This assumption is supported by the lack of vesicular structures of the same diameter that would represent cross-sectioned tubules. These ER-derived protrusions were strongly curved, suggesting that they are precursors of DMVs formed at the viral replication sites.
Fig. 5.
Membrane tubules within the SARS-CoV replication sites are derived from ER membranes. A) Vero cells were mock-infected (a) or infected with SARS-CoV (MOI of 10) for 3 h (b, c), fixed and analyzed by standard EM. ER cisternae are marked with large arrows, whereas the double membrane vesicles and the membrane tubules characteristic for the putative replication sites are highlighted with small arrows. Scale bars represent 200 nm or 500 nm as indicated. B) Vero cells were mock-infected (a) or infected with SARS-CoV (MOI of 10) for 3 h (b–e), fixed and analyzed by cryo-iEM. Cells were labeled with antibodies directed against SARS-CoV N (a–c, 10 nm gold, filled arrowheads) or SARS-CoV nsp3 (d–e, 15 nm gold, filled arrowheads) and PDI as a marker for ER (a–e, 5 nm gold, open arrowheads). Panel B c and e show putative replication complexes in a higher magnification. PDI labeling surrounded the N assembly in c but is not visible in this cutout. ER cisternae are marked with large arrows (a, b) and small arrows delineate membrane tubules characteristic for the putative replication sites (b, c). Scale bars represent 200 nm.
Additionally, we compared the localization of the autophagosomal marker LC3 with the distribution of N in SARS-CoV-infected cells by immunofluorescence analysis, as it has been suggested that autophagy might play a role in the formation of the viral replication sites (Prentice et al., 2004b). Using an LC3-specific antiserum, we found partial colocalization of N and LC3 at 3 h and 5 h p.i. (Supplementary Figs. 1a–f). However, when we used a plasmid encoding GFP-LC3 (Mizushima et al., 2004) we could not observe any colocalization of N and LC3 (Supplementary Figs. 1g–i) or nsp3 and LC3 (data not shown). Thus, further studies are required to determine the role of autophagy in the formation of SARS-CoV replication sites.
Discussion
In this study, we investigated cell biological aspects of the replication cycle of SARS-CoV by analyzing the intracellular localization of viral proteins. We found cytoplasmic accumulations of SARS-CoV N and nsp3 as early as 3 h p.i. and colocalization of N with viral genomic RNA at 5 h p.i. Cytoplasmic accumulations of the non-structural protein nsp3 which colocalized with other non-structural proteins and newly synthesized RNA have been described before, and it was suggested that these accumulations are the SARS-CoV replication sites (Harcourt et al., 2004, Snijder et al., 2006). Since we found colocalization of N with nsp3 as well as with viral genomic RNA, this strongly suggests that the N assemblies represent the replication sites of SARS-CoV. Interestingly, we also found colocalization of these putative replication sites with a marker protein for the ER, and indeed ultrastructural analysis proved that the N assemblies were associated with distinct membrane protrusions that are continuous with and thus most probably originate from ER cisternae. Therefore, we hypothesize that SARS-CoV recruits membranes from the ER to build the sites of viral replication. Recently, Prentice et al. described cytoplasmic foci in SARS-CoV-infected cells that were positive for proteins encoded by ORF 1a/1ab and also for the autophagosomal marker LC3 (Prentice et al., 2004b). In analogy to MHV, it was suggested that these foci are RNA replication complexes associated with membranes of autophagosomes. In the case of MHV it was shown that autophagy is induced by infection and plays an important role for viral replication (Prentice et al., 2004a). We also compared the localization of LC3 with the N accumulations by using an LC3-specific antibody and found colocalization of N and LC3 to some extent. However, using a plasmid encoding GFP-LC3 we got a different result: no overlap of the N- and LC3-specific staining was detected confirming a recent study by Snijder et al. (2006). Thus, the role of autophagy in the formation of SARS-CoV replication sites still needs to be elucidated. During the process of autophagy, cytoplasmic material is engulfed by membranes that are thought to be derived from the ER or formed de novo, leading to the formation of DMVs (Klionsky and Emr, 2000, Kirkegaard et al., 2004). Indeed, drastic membrane rearrangements, especially DMVs, were described to be associated with the replication complexes of the coronavirus MHV (Gosert et al., 2002) and the arterivirus equine arteritis virus (Pedersen et al., 1999). Accordingly, we also found formation of DMVs in SARS-CoV-infected cells in line with two recent studies (Goldsmith et al., 2004, Snijder et al., 2006). We provide morphological evidence that the ER-derived membrane protrusions associated with the putative replication complexes are the precursors of the DMVs. Therefore, we conclude that the formation of DMVs in SARS-CoV-infected cells occurs by curving and fusion of ER-derived double membrane sheets. This is consistent with the protrusion-and-detachment model for the formation of DMVs in arterivirus-infected cells (Pedersen et al., 1999).
Earlier studies demonstrated the appearance of viral replicase components at 4 h p.i. in SARS-CoV-infected cells (Harcourt et al., 2004) and release of viral particles from infected cells at 5 h p.i. (Ng et al., 2003), already suggesting that SARS-CoV is a very fast replicating virus. Indeed, using immunofluorescence we found colocalization of N and S with a Golgi marker indicating budding events from 5 h p.i. on. Strikingly, using EM we observed newly formed SARS-CoV particles in the Golgi even at 3 h p.i. Importantly, budding profiles and viral particles were strongly labeled with the N-specific antibody, but nsp3 labeling was only rarely associated with the budding sites. This confirms our immunofluorescence data which had shown that nsp3 stays associated with the replication sites and was absent from S accumulations. We detected budding of viral particles into β′COP-I-positive membranes of the ERGIC and Golgi area, which is in line with the finding of another study that SARS-CoV particles bud into membranes of the pre-Golgi compartment (Goldsmith et al., 2004). However, in the same study additional particle formation at the outer nuclear membrane was observed at 3 days p.i., an effect which might occur late in infection when the infected cell contains very high amounts of viral proteins and viral genomic RNA. In another study, budding into vacuoles, supposed to be derived from the Golgi, was described (Ng et al., 2003). Indeed, we detected similar vacuoles containing multiple particles from 7 h p.i. on (data not shown) and also observed some budding profiles at these vacuoles. However, we found that the Golgi area is the main budding site of SARS-CoV early in infection. Budding is probably a very rapid process, and the newly formed particles are directly transported to the cis-Golgi. This would explain why we see a prominent accumulation of SARS-CoV N and S in the cis-Golgi with immunofluorescence analysis.
In summary, our results support the following model for the replication cycle of SARS-CoV: early in infection, viral replication complexes are established at ER-derived membranes that are transformed into characteristic DMVs by drastic membrane rearrangements. The possible involvement of autophagy as well as the role of single viral proteins in this process has to be addressed in future studies. Later on, viral genomic RNA and N protein translocate to the budding sites at the ERGIC and Golgi region, where the viral glycoproteins are located and budding of new particles occurs. Intriguingly, SARS-CoV passes through these different steps of the replication cycle very fast as we observed the final step, budding, already at 3 h p.i.
Materials and methods
Cells and viruses
African green monkey kidney (VeroE6) cells were maintained in DMEM supplemented with 10% fetal calf serum and antibiotics. The FFM-1 strain of SARS-CoV was kindly provided by Stephan Becker, Robert-Koch-Institut, Berlin, Germany (Drosten et al., 2003).
Antibodies and plasmids
For detection of SARS-CoV N, a rabbit polyclonal antiserum (Spiegel et al., 2005) and a mouse monoclonal antibody were used. To generate the mouse monoclonal antibody, mice were immunized with bacterially expressed recombinant SARS-CoV N (Spiegel et al., 2005). Briefly, 100 μg of purified SARS-CoV N dissolved in complete Freund's adjuvant was used to immunize BALB/c mice. Two weeks after immunization, mice were boosted with the same dose and 2 weeks later, splenocytes from one mouse were fused with SP2 cells to generate hybridomas. Hybridoma supernatants were screened on 96-well plates coated with the bacterially expressed SARS-CoV N, and specific monoclonal antibodies were selected. The specificity of all monoclonal antibodies was confirmed by immunofluorescence and Western blot analyses using cells transfected with a SARS-CoV N expression plasmid and in SARS-CoV-infected cells. In this study, we used the monoclonal antibody 9C10, one of the selected clones. We also generated a mouse monoclonal antibody against SARS-CoV S. To this aim, the S cDNA sequence (Urbani strain) was first cloned into the expression vector pCAGGS by using EcoRI and XhoI restriction endonucleases. Reverse transcriptase PCR (RT-PCR) was done by using RNA from SARS-CoV-infected cells, kindly provided by Dr. Paragas (USAMRIID, Fort Detrick). SARS-CoV S cDNA was cloned into pRB21 to generate a recombinant vaccinia virus expressing SARS-CoV S (rVV-S) as described (Blasco and Moss, 1995). To generate monoclonal antibodies, one mouse was immunized with 106 pfu of the rVV-S. Two weeks after rVV-S immunization, the mouse was boosted with recombinant SARS-CoV S fragments produced in bacteria. Purified GST-S fragment F2 (aa 258 to 572) and GST-S fragment F3 (aa 492 to 856) were administrated intravenously. Two weeks after boosting, splenocytes were fused with SP2 cells, and hybridomas were screened for their ability to generate antibodies which specifically recognize Vero cells transfected with the pCAGGS-SARS-CoV S expression vector. One of the selected SARS-CoV S specific monoclonal antibodies, 5E5G10, was used in this study. To detect SARS-CoV nsp3, we used a polyclonal rabbit serum (Harcourt et al., 2004), kindly provided by S. Baker, Loyola University of Chicago, Illinois. A monoclonal mouse anti-Giantin antibody to stain the cis-Golgi (Linstedt and Hauri, 1993) was kindly provided by H.P. Hauri, Biozentrum Basel, Switzerland. The polyclonal rabbit anti-PDI antibody was purchased from Santa Cruz Biotechnology (California, U.S.) and a polyclonal rabbit antiserum directed against β′COP-I was provided by J. Simpson, EMBL, Heidelberg, Germany (Lowe and Kreis, 1995). A polyclonal rabbit antiserum directed against LC3 (Kabeya et al., 2000) and the plasmid pEGFP-LC3 (Mizushima et al., 2004) which were both used to label autophagosomes were gifts from T. Yoshimori, National Institute of Genetics, Shizuoka, Japan.
Infection
Vero cells were incubated with SARS-CoV in DMEM with 2% fetal calf serum for 1 h. Subsequently, the inoculum was washed off and cells were incubated with DMEM containing 10% fetal calf serum. Infection experiments were performed under biosafety level 3 conditions.
Immunofluorescence analysis
Vero cells grown on glass cover slips were infected with SARS-CoV at an MOI of 10. When studying the localization of GFP-LC3, cells were transfected with 0.5 μg plasmid per well of a 6-well-plate using Metafectene (Biontex, Munich, Germany) according to the manufacturer's protocol 24 h before infection. At the time points indicated, cells were fixed with 3% paraformaldehyde and then permeabilized with 0.5% Triton-X-100 in phosphate-buffered saline (PBS). Cells were stained for viral proteins and intracellular compartments using specific primary antibodies and subsequently fluorophore (Cy2, Cy3)-conjugated donkey secondary antibodies (Dianova, Hamburg, Germany). Immunostained cells were analyzed with a Leica (Heidelberg, Germany) TCSSP2 confocal laser scanning microscope.
In situ hybridization
cDNA encoding the nsp1-ORF (nt 265–801 of the SARS-CoV genome) of SARS-CoV was cloned into the pcDNA 3.1 TOPO TA cloning vector (Invitrogen) using the following primers: 5′tctagaatggagagccttgttcttggtgtcaacgagaaaa3′ and 5′ctcgagtcatccattgagctcacgagtgagtt3′. For in vitro transcription, the cDNA was cloned into pBSK(I) using BamHI and EcoRV restriction enzymes. The plasmid was linearized, and a riboprobe of negative polarity was prepared by in vitro transcription with T7 RNA polymerase in the presence of fluorescein isothiocyanate (FITC)-labeled UTP (Roche, Mannheim, Germany). To detect viral RNA in infected cells, Vero cells grown on cover slips were infected with SARS-CoV using an MOI of 10 and fixed with 3% paraformaldehyde at 5 h p.i. In situ hybridization and subsequent immunofluorescence staining were performed as described (Bolten et al., 1998, Egger et al., 1999).
Electron microscopy (EM)
The samples were either processed for standard electron microscopy (standard EM) (Luft, 1961) or cryo-immuno-EM (cryo-iEM). In brief, for standard EM cells were fixed in phosphate-buffered 1% glutaraldehyde, pH 7.2 and postfixed in 1% osmium tetroxide. The samples were then dehydrated in a graded ethanol series and propylene oxide and finally embedded in epoxy-resin EMbed-812 (EMS, Electron Microscopy Sciences). Ultrathin sections (60 nm) were cut on an Ultracut S ultramicrotome (Leica), mounted on Formvar- and carbon-coated grids and counterstained with 1% aqueous uranyl acetate and 0.2% lead citrate. Processing for cryo-iEM and immunogold labeling were performed essentially as described before (Reichelt et al., 2004). Samples were fixed in phosphate-buffered 0.2% glutaraldehyde, pH 7.2 and 2% paraformaldehyde, embedded in 10% gelatin and infiltrated with 2.3 M sucrose overnight. Sample blocks were mounted on cryoultramicrotomy pins and frozen in liquid nitrogen. Ultrathin sections (70 nm) were cut on an Ultracut Cryoultramicrotome (Leica) at − 125 °C, picked up with a mixture (1:1) of 2% methylcellulose and 2.3 M sucrose and transferred to Formvar- and carbon-coated grids. Immunogold double-labeling was performed essentially as described (Slot et al., 1991). First, the specificity of the antibodies used (anti-β′COP I, anti-PDI, anti-nsp3, anti-N) was controlled by single-immunogold labeling. To detect the primary antibodies we used Protein A conjugated with colloidal gold (PA–gold) of different sizes (5 nm, 10 nm or 15 nm). Double-labeling was performed by incubating with the first antibody and detection with PA–gold, then fixation with 1% GA for 5 min, quenching and incubation with the second antibody and detection with PA–gold of a different size. Finally, sections were counterstained with 0.5% uranyl acetate in 2% methylcellulose for 10 min on ice and grids were looped out to dry. Samples were analyzed with a Philips 400 electron microscope or with a JEOL 1230 TEM at 80 kV, and digital photographs were taken using a Gatan Multiscan 791 digital camera.
Acknowledgments
We thank S. Becker for providing the FFM-1 strain of SARS-CoV, S. Baker, H-P. Hauri, J. Simpson and T. Yoshimori for antibodies and plasmids, J. Paragas for SARS-CoV RNA and J. Krijnse-Locker, G. Griffith (EMBL, Heidelberg) and John Perrino (Stanford, CSIF) for training and advice concerning electron microscopy and K. Kirkegaard for supporting this collaboration and for helpful discussion. Furthermore, we want to acknowledge B. Moss for providing us with the pRB21 plasmid as well as the vRB21 vaccinia strain to generate recombinant vaccinia virus, as well as A. Pritsker at Mount Sinai Hybridoma Center for generating the monoclonal antibody and R. Cádagan for excellent technical assistance. We thank O. Haller and P. Staeheli for helpful discussions and critical comments on the manuscript. This work was supported by grants from the Sino-German Center for Research Promotion (FW) and the Deutsche Forschungsgemeinschaft (FW) as well as by NIH grants (AG-S) and the Dean's fellowship from Stanford University (MR).
This work was conducted by Silke Stertz in partial fulfillment of the requirements for a Ph.D. degree from the Faculty of Biology of the University of Freiburg, Germany.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.virol.2006.11.027.
Appendix A. Supplementary data
Localization of the putative SARS-CoV replication sites compared to the autophagy marker protein LC3. (a–f) Vero cells were mock-infected (a) or infected with SARS-CoV (MOI of 10) for 3 h (b, d–f) or 5 h (c), fixed and stained with specific antibodies for SARS-CoV N (red) and LC3 (green) in parallel. In a–c, the merged images of the double immunostainings are shown. In d–f, a cutout of the cell shown in b is depicted at higher magnification. To better demonstrate the observed partial colocalization (arrows), the N staining (d), the LC3 signal (e) and the merged image (f) are shown separately in this area. (g–i) Vero cells were transfected with an EGFP-LC3-encoding plasmid. Twenty-four hours later, the cells were mock-infected (g) or infected with SARS-CoV (MOI of 10) for 3 h (h) or 5 h (i), fixed and stained for SARS-CoV N (red). Scale bars represent 10 μm.
References
- Almazan F., Galan C., Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004;78(22):12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco R., Moss B. Selection of recombinant vaccinia viruses on the basis of plaque formation. Gene. 1995;158(2):157–162. doi: 10.1016/0378-1119(95)00149-z. [DOI] [PubMed] [Google Scholar]
- Bolten R., Egger D., Gosert R., Schaub G., Landmann L., Bienz K. Intracellular localization of poliovirus plus- and minus-strand RNA visualized by strand-specific fluorescent in situ hybridisation. J. Virol. 1998;72:8578–8585. doi: 10.1128/jvi.72.11.8578-8585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost A.G., Prentice E., Denison M.R. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology. 2001;285(1):21–29. doi: 10.1006/viro.2001.0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway S.M., Clay C.T., Lu X.T., Denison M.R. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 2003;77(19):10515–10527. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Egger D., Bolten R., Rahner C., Bienz K. Fluorochrome-labeled RNA as a sensitive, strand-specific probe for direct fluorescence in situ hybridization. Histochem. Cell Biol. 1999;111(4):319–324. doi: 10.1007/s004180050363. [DOI] [PubMed] [Google Scholar]
- Eickmann M., Becker S., Klenk H.D., Doerr H.W., Stadler K., Censini S., Guidotti S., Masignani V., Scarselli M., Mora M., Donati C., Han J.H., Song H.C., Abrignani S., Covacci A., Rappuoli R. Phylogeny of the SARS coronavirus. Science. 2003;302(5650):1504–1505. doi: 10.1126/science.302.5650.1504b. [DOI] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423(6937):240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith C.S., Tatti K.M., Ksiazek T.G., Rollin P.E., Comer J.A., Lee W.W., Rota P.A., Bankamp B., Bellini W.J., Zaki S.R. Ultrastructural characterization of SARS coronavirus. Emerg. Infect Dis. 2004;10(2):320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76(8):3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78(24):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, K., 1996. Coronaviridaae: The Viruses and their Replication. F.V.F.B., Ed. Philadelphia: Lippincott-Raven, Philadelphia: Lippincott-Raven.
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78(11):5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson W.T., Giddings T.H., Jr., Taylor M.P., Mulinyawe S., Rabinovitch M., Kopito R.R., Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3(5):e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Taylor M.P., Jackson W.T. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev., Microbiol. 2004;2(4):301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Emr S.D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J., Rottier P.J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994;68(10):6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.K., Tam J.S., Zambon M.C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J.C., Stohr K., Peiris J.S., Osterhaus A.D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120(2):159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Linstedt A.D., Hauri H.P. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol. Biol. Cell. 1993;4(7):679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M., Kreis T.E. In vitro assembly and disassembly of coatomer. J. Biol. Chem. 1995;270(52):31364–31371. doi: 10.1074/jbc.270.52.31364. [DOI] [PubMed] [Google Scholar]
- Luft J.H. Improvements in epoxy resin embedding methods. J. Biophys. Biochem. Cytol. 1961;9:409–414. doi: 10.1083/jcb.9.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Martin S.R., Weiss S. Coronavirus replication and pathogenesis: implications for the recent outbreak of severe acute respiratory syndrome (SARS), and the challenge for vaccine development. J. NeuroVirol. 2004;10(2):75–85. doi: 10.1080/13550280490280292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M.L., Tan S.H., See E.E., Ooi E.E., Ling A.E. Proliferative growth of SARS coronavirus in Vero E6 cells. J. Gen. Virol. 2003;84(Pt. 12):3291–3303. doi: 10.1099/vir.0.19505-0. [DOI] [PubMed] [Google Scholar]
- Pedersen K.W., van der Meer Y., Roos N., Snijder E.J. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 1999;73(3):2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279(11):10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78(18):9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Filipowicz W., Hall J., Gorbalenya A.E., Ziebuhr J. ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 2005;79(20):12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt M., Stertz S., Krijnse-Locker J., Haller O., Kochs G. Missorting of LaCrosse virus nucleocapsid protein by the interferon-induced MxA GTPase involves smooth ER membranes. Traffic. 2004;5(10):772–784. doi: 10.1111/j.1600-0854.2004.00219.x. [DOI] [PubMed] [Google Scholar]
- Schelle B., Karl N., Ludewig B., Siddell S.G., Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 2005;79(11):6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot J.W., Geuze H.J., Gigengack S., Lienhard G.E., James D.E. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell Biol. 1991;113(1):123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80(12):5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M., Pichlmair A., Martinez-Sobrido L., Cros J., Garcia-Sastre A., Haller O., Weber F. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 2005;79(4):2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug P.E., Klionsky D.J. Approaching the molecular mechanism of autophagy. Traffic. 2001;2(8):524–531. doi: 10.1034/j.1600-0854.2001.20802.x. [DOI] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84(Pt. 9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Tooze J., Tooze S., Warren G. Replication of coronavirus MHV-A59 in sac-cells: determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 1984;33(2):281–293. [PubMed] [Google Scholar]
- van der Meer Y., Snijder E.J., Dobbe J.C., Schleich S., Denison M.R., Spaan W.J., Locker J.K. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 1999;73(9):7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Localization of the putative SARS-CoV replication sites compared to the autophagy marker protein LC3. (a–f) Vero cells were mock-infected (a) or infected with SARS-CoV (MOI of 10) for 3 h (b, d–f) or 5 h (c), fixed and stained with specific antibodies for SARS-CoV N (red) and LC3 (green) in parallel. In a–c, the merged images of the double immunostainings are shown. In d–f, a cutout of the cell shown in b is depicted at higher magnification. To better demonstrate the observed partial colocalization (arrows), the N staining (d), the LC3 signal (e) and the merged image (f) are shown separately in this area. (g–i) Vero cells were transfected with an EGFP-LC3-encoding plasmid. Twenty-four hours later, the cells were mock-infected (g) or infected with SARS-CoV (MOI of 10) for 3 h (h) or 5 h (i), fixed and stained for SARS-CoV N (red). Scale bars represent 10 μm.