Abstract
We have previously demonstrated that over-expression of spike protein (S) of severe acute respiratory syndrome coronavirus (SARS-CoV) or its C-terminal subunit (S2) is sufficient to induce apoptosis in vitro. To further investigate the possible roles of S2 in SARS-CoV-induced apoptosis and pathogenesis of SARS, we characterized the host expression profiles induced upon S2 over-expression in Vero E6 cells by oligonucleotide microarray analysis. Possible activation of mitochondrial apoptotic pathway in S2 expressing cells was suggested, as evidenced by the up-regulation of cytochrome c and down-regulation of the Bcl-2 family anti-apoptotic members. Inhibition of Bcl-2-related anti-apoptotic pathway was further supported by the diminution of S2-induced apoptosis in Vero E6 cells over-expressing Bcl-xL. In addition, modulation of CCN E2 and CDKN 1A implied the possible control of cell cycle arrest at G1/S phase. This study is expected to extend our understanding on the pathogenesis of SARS at a molecular level.
Keywords: Severe acute respiratory syndrome coronavirus, Spike protein, Apoptosis, Proliferation, Microarray
Introduction
Severe acute respiratory syndrome (SARS) is a highly contagious disease caused by a member of the Coronaviridae family named SARS Coronavirus (SARS-CoV) (Drosten et al., 2003, Ksiazek et al., 2003, Peiris, 2003). The disease was first documented during late 2002 and the outbreak was effectively controlled by mid-2003 through rigorous quarantine measures (Zhong et al., 2003). However, the ability of the virus in infecting multiple cell types (Hattermann et al., 2005, Kaye, 2006) and animals (Martina et al., 2003) implied the risk of viral circulation in animal reservoirs and hence the re-emergence of the virus in human population. Understanding the molecular mechanisms of viral pathogenesis may provide important information for rational design of antiviral drugs, which may be important to combat against another possible SARS epidemic. The viral–host factors contributing to the pathogenesis of SARS-CoV are incompletely understood. In addition to respiratory tract illness, lymphoid depletion in spleen and lymph nodes is a common clinical manifestation in fatal cases of SARS (Wong et al., 2003). Cytokine overdrive and apoptosis induced by the virus were suggested to contribute to such lymphoid depletion (Wong et al., 2003). These observations, together with the fact that SARS-CoV is capable to induce apoptosis in the viral susceptible Vero E6 cell line (Yan et al., 2004), highlighted the important role of apoptosis in the pathogenesis of SARS.
The SARS-CoV spike protein (S) is a class I fusion protein. Cellular entry of the virus was demonstrated to be mediated by S through the receptor binding domain located in the N-terminal subunit (S1) (Dimitrov, 2004, Hofmann and Pohlmann, 2004) and the fusion peptide in the C-terminal subunit (S2) (Dimitrov, 2004, Hofmann and Pohlmann, 2004). Recent studies also suggested the presence of neutralizing domains in S2 (Duan et al., 2005, Keng et al., 2005, Zeng et al., 2006, Zhang et al., 2004). We have previously demonstrated that the SARS-CoV S and S2, but not S1 nor other structural proteins, including envelope (E), membrane (M) and nucleocapsid (N), can induce apoptosis in Vero E6 cells (Chow et al., 2005). Other recent studies showed that the E (Yang et al., 2005), M (Lai et al., 2006, Zhao et al., 2006), N (Surjit et al., 2004, Zhao et al., 2006), ORF 3a (Law et al., 2005), 3b (Yuan et al., 2005) and 7a (Tan et al., 2004) could trigger apoptosis in different cell lines or under specific culture conditions. As S is abundantly expressed during the virus replication cycle, and that the silencing of S expression exhibits an inhibition of cytopathic effect induced by SARS-CoV infection (He et al., 2006), we sought to elucidate the molecular mechanisms of apoptosis mediated by S2 in Vero E6 cells.
In depicting the biochemistry of SARS-CoV infections, several microarray studies identified the differential expression of genes related to apoptosis, cell cycle and stress response in Vero (Leong et al., 2005), PBMC (Reghunathan et al., 2005, Yu et al., 2005) and Huh-7 cells (Tang et al., 2005). Other in vitro studies indicated that infection of SARS-CoV involves the phosphorylation of p38 MAPK (Mizutani et al., 2004b), down-regulation of Bcl-2 (Bordi et al., 2006, Ren et al., 2005), up-regulation of Bax and activation of caspase 3 (Ren et al., 2005). However, few efforts were put in trying to identify the role of individual proteins of SARS-CoV in these biochemical pathways. In this regard, using an adenoviral system for gene delivery, we studied the changes of expression profiles in serially sampled Vero E6 cells over-expressing S2 through microarray technology. Based on our results, we proposed the alternations of several physiological pathways that may be involved in S2-induced apoptosis, including the activation of intrinsic apoptotic pathway, inhibition of NFκB-downstream members and MAPK cascade members. Moreover, a number of genes critically involved in cell cycle control were also found to be differentially regulated.
Results and discussion
Apoptosis induced by SARS-CoV S2 over-expression
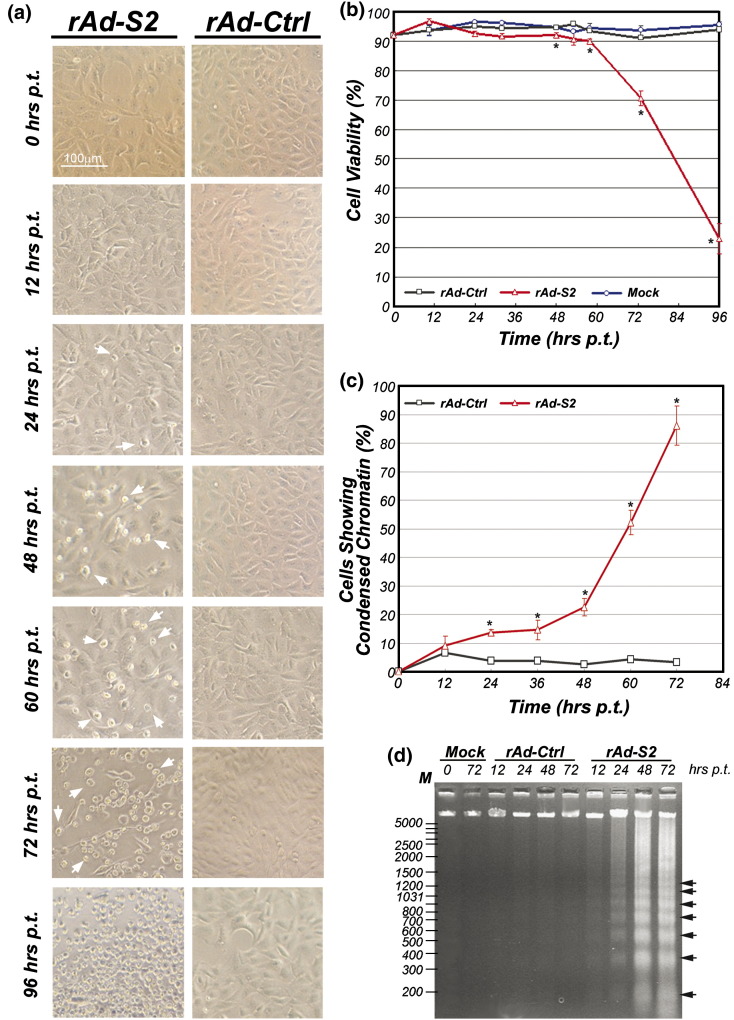
To determine the appropriate time points for microarray analysis, we first characterized the time-dependent induction of apoptosis by S2 over-expression mediated by recombinant adenoviruses (rAd). Starting from 24 h (h) post-transductions (p.t.), cytopathic effects, including abnormal cell morphology and shrinkage, were already observed in rAd-S2 but not rAd-Ctrl-transduced Vero E6 cells (Fig. 1a). Results of cell viability assay and chromatin condensation assay agreed well with the above observations (Figs. 1b and c). Cell viability of rAd-S2-transduced cells dropped to the minimum at 96 h p.t. (Fig. 1b), and the percentage of rAd-S2-transduced cells undergoing apoptosis, as shown by chromatin condensation assay (Fig. 1c) and internucleosomal DNA cleavage analysis (Fig. 1d), increased rapidly and was peaked at 72 h p.t., which is the last time-point of the assays. According to these apoptotic signatures, which agree well with previous reports (Chow et al., 2005, Jamshidi-Parsian et al., 2005), four time points at 12, 24, 48 and 72 h p.t., were chosen for the microarray analysis.
Fig. 1.
Time course study of S2-induced apoptosis in Vero E6 cells. (a) Morphology of Vero E6 cells under light microscope (40×) after rAd-Ctrl or rAd-S2 transductions from 0 to 96 h p.t. in 12- to 24-h intervals. Representative figures of three independent experiments are shown. (b) Cell viability along the time course is estimated quantitatively with Trypan blue exclusion assay. (c) The percentage of rAd-Ctrl or rAd-S2-transduced cells showing chromatin condensation was counted under fluorescence microscope after Hoechst 33342 staining. For both panels b and c, average of three independent experiments is shown with standard error of the mean (SEM). (d) Agarose gel electrophoresis showing the characteristic DNA laddering pattern resulting from internucleosomal DNA cleavage in rAd-S2-transduced cells in four selected time points. Three micrograms of low molecular weight DNA was loaded into each well. The ladders with 200 bp increments are indicated with arrowheads. The result is the representative of three independent experiments. ⁎p < 0.01 when compared with cells transduced by rAd-Ctrl under the same condition.
The global gene expression profile in Vero E6 cells over-expressing SARS-CoV S2
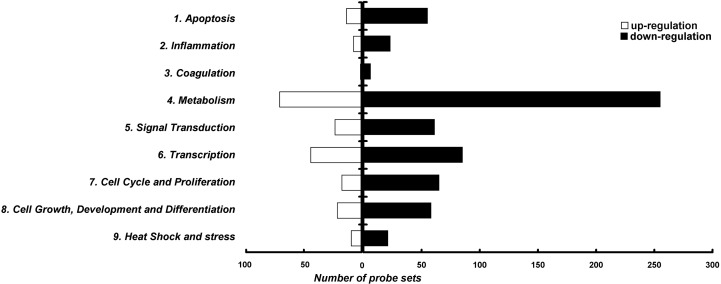
A total of 1702 probe sets (3.2% of total probe sets) in the array have exhibited a significant up- or down-regulation (at least two-fold changes) in at least one of the time points upon S2 over-expression. Only 33 of these differentially hybridized probe sets showed both up- and down-regulation across the four time points, while the rest of them showed either only up (403 probe sets)- or down-regulated expression (1266 probe sets) during the course of the experiment. Gene ontology study classified 829 of the 1702 differential hybridized probe sets into 9 groups with known functions (Fig. 2 ) according to a defined classification (Tang et al., 2005). The relatively large number of unclassified probe sets may be due to the less characterized rhesus macaque genome, with unclassified probe sets and hypothetical proteins contributed to 33.6% and 7.1% of total probe sets respectively. Among the differentially hybridized probe sets with known function, 68 and 82 probe sets (representing 49 and 68 different genes, respectively), are related to apoptosis and cell cycle/proliferation respectively. The above data are summarized in Table 1 . Changes of genes expression related to other cellular pathways in S2 expressing cells can be found in our Web page (http://evolution.hku.hk/publications/SARS_microarray.htm).
Fig. 2.
Functional classification of differentially expressed probe sets. Probe sets that were differentially expressed by more than two-folds were classified into 11 groups manually, with 9 groups of known functions and two groups representing hypothetical/miscellaneous proteins and unknown proteins. The number of probe sets with known functions showing up- and down-regulations is shown with empty and filled bars respectively. The first group represents the probe sets related to apoptosis while probe sets regulating cell cycle and proliferation are grouped into group 7.
Table 1.
Signal log ratios (log2 scale) on the fold change of gene expression in S2-expressing Vero E6 cells (only genes highly related to apoptosis and cell cycle and proliferation are shown)
| Categories and subcategories |
Probe set ID |
Gene symbol |
Description |
Log 2 ratio (sample/control)a (h p.i.) |
|||
|---|---|---|---|---|---|---|---|
| 12 |
24 |
48 |
72 |
||||
| Apoptosis | |||||||
| Anti-apoptosis | |||||||
| MmugDNA.28580.1.S1_at | ANXA4 | Annexin A4 | − 0.39 | − 0.28 | − 0.45 | − 1.01 | |
| MmugDNA.34272.1.S1_s_at | API5 | Apoptosis inhibitor 5 | − 1.44 | − 0.29 | − 0.36 | − 1.08 | |
| MmugDNA.29603.1.S1_at | BCL2 | B-cell CLL/lymphoma 2 | 0.40 | − 0.49 | − 1.24 | − 1.78 | |
| MmugDNA.12133.1.S1_at | BCLXL | BCL2-like 1 (BCL2L1) | − 1.56 | − 0.23 | − 0.77 | − 1.92 | |
| MmugDNA.1928.1.S1_at | CTNNA1 | Catenin (cadherin-associated protein), alpha 1, 102 kDa | − 0.50 | − 0.08 | − 0.62 | − 1.22 | |
| MmugDNA.39222.1.S1_at | CFL1 | Cofilin 1 (non-muscle) | − 1.11 | − 0.12 | − 0.53 | − 1.44 | |
| MmugDNA.35450.1.S1_at | CSTB | Cystatin B (stefin B) | − 1.03 | − 0.97 | − 0.85 | 0.01 | |
| MmugDNA.21273.1.S1_at | HSPA5 | Heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) | 0.15 | 0.39 | 1.88 | 2.15 | |
| MmugDNA.725.1.S1_at | HIPK3 | Homeodomain interacting protein kinase 3 | − 2.13 | − 0.17 | − 0.17 | − 1.82 | |
| MmugDNA.29385.1.S1_at | MIRN21 | MicroRNA 21 | − 0.65 | − 0.22 | − 0.79 | − 1.12 | |
| MmugDNA.38922.1.S1_at | MCL1 | Myeloid cell leukemia sequence 1 (BCL2-related) | − 0.95 | − 0.07 | − 0.48 | − 1.09 | |
| MmugDNA.11909.1.S1_s_at | ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | − 1.35 | − 0.17 | − 1.17 | − 1.01 | |
| MmugDNA.34988.1.S1_at | TXNDC | Thioredoxin domain containing | − 1.40 | 0.02 | − 0.05 | − 1.11 | |
| MmugDNA.40138.1.S1_at | TNFAIP3 | Tumor necrosis factor, alpha-induced protein 3 | 0.57 | 0.45 | 0.72 | 1.19 | |
| MmugDNA.32694.1.S1_at | TNFAIP8 | Tumor necrosis factor, alpha-induced protein 8 | − 0.18 | − 0.69 | − 0.95 | − 1.46 | |
| MmugDNA.30167.1.S1_at | TRA1 | Tumor rejection antigen (gp96) 1 | − 0.24 | 0.42 | 1.41 | 0.93 | |
| MmugDNA.6621.1.S1_at | TRA1 | Tumor rejection antigen (gp96) 1 | 0.16 | 0.42 | 1.41 | 1.14 | |
| MmugDNA.3345.1.S1_at | VEGF | Vascular endothelial growth factor | − 0.94 | − 0.48 | − 0.05 | − 1.05 | |
| Pro-apoptosis | |||||||
| MmugDNA.6382.1.S1_at | BIRC3 | Baculoviral IAP repeat-containing 3 | − 0.33 | − 0.37 | − 0.40 | − 1.05 | |
| MmugDNA.30317.1.S1_at | BIRC4 | Baculoviral IAP repeat-containing 4 | 1.47 | 0.28 | 0.53 | 1.22 | |
| MmugDNA.2328.1.S1_at | BAG2 | BCL2-associated athanogene 2 | 1.12 | 0.10 | − 0.12 | 0.65 | |
| MmugDNA.4144.1.S1_s_at | BCL2L11 | BCL2-like 11 (apoptosis facilitator) | − 1.61 | − 0.49 | − 0.40 | − 0.85 | |
| MmugDNA.23713.1.S1_at | BOMB | BH3-only member B protein | − 0.99 | − 0.28 | − 0.57 | −1.23 | |
| MmugDNA.41236.1.S1_at | CASP2 | Caspase 2, apoptosis-related cysteine peptidase (neural precursor cell expressed, developmentally down-regulated 2) | − 0.72 | 0.35 | − 0.21 | − 1.40 | |
| MmugDNA.13586.1.S1_at | CASP8 | Caspase 8, apoptosis-related cysteine peptidase | − 1.49 | − 0.45 | − 0.62 | − 1.38 | |
| MmugDNA.15712.1.S1_x_at | CTSB | Cathepsin B | 0.13 | 0.02 | − 0.07 | 1.08 | |
| MmugDNA.19990.1.S1_s_at | CD99 | CD99 antigen | − 1.08 | − 0.24 | − 0.25 | − 1.28 | |
| MmugDNA.15069.1.S1_at | CYCS | Cytochrome c, somatic | 1.50 | 0.58 | 0.40 | 1.63 | |
| MmugDNA.39077.1.S1_at | DAPK3 | Death-associated protein kinase 3 | − 1.19 | − 0.68 | − 0.47 | − 0.37 | |
| MmugDNA.29093.1.S1_at | DOC1 | Down-regulated in ovarian cancer 1 | − 0.13 | − 0.31 | − 0.79 | − 1.53 | |
| MmugDNA.24700.1.S1_at | ESRRBL1 | Estrogen-related receptor beta like 1 | − 1.25 | − 0.32 | − 0.67 | − 1.51 | |
| MmugDNA.7450.1.S1_at | FAF1 | Fas (TNFRSF6) associated factor 1 | − 0.40 | − 0.60 | − 0.48 | − 1.08 | |
| MmugDNA.34508.1.S1_at | HIP1 | Huntingtin interacting protein 1 | 1.10 | − 0.02 | 0.11 | 0.47 | |
| MmugDNA.38424.1.S1_at | IHPK2 | Inositol hexaphosphate kinase 2 | − 0.23 | − 0.50 | − 0.60 | − 1.18 | |
| MmugDNA.22620.1.S1_at | LGALS7 | Lectin, galactoside-binding, soluble, 7 (galectin 7) | − 0.10 | − 0.19 | − 0.53 | − 1.20 | |
| MmugDNA.21644.1.S1_at | MAPK1 | Mitogen-activated protein kinase 1 | − 1.41 | − 0.32 | − 0.27 | − 1.42 | |
| MmugDNA.834.1.S1_at | RPL4 | Mitogen-activated protein kinase kinase kinase 13 | − 1.75 | − 0.46 | − 0.41 | − 1.44 | |
| MmugDNA.19461.1.S1_at | WIG1 | p53 target zinc finger protein | − 1.76 | − 0.73 | − 0.70 | − 1.94 | |
| MmugDNA.22348.1.S1_at | PSEN1 | Presenilin 1 (Alzheimer disease 3) | − 1.00 | − 0.33 | 0.12 | − 0.61 | |
| MmugDNA.5589.1.S1_at | PURB | Purine-rich element binding protein B | − 0.90 | − 0.94 | − 1.14 | − 1.09 | |
| MmugDNA.21841.1.S1_s_at | RHOB | Ras homolog gene family, member B | − 1.44 | − 0.63 | − 0.86 | − 1.23 | |
| MmugDNA.25692.1.S1_at | STK3 | Serine/threonine kinase 3 (STE20 homolog, yeast) | − 0.90 | − 0.15 | − 0.35 | − 1.02 | |
| MmugDNA.39549.1.S1_at | STK4 | Serine/threonine kinase 4 | − 1.89 | − 0.40 | − 0.61 | − 1.29 | |
| MmugDNA.7204.1.S1_at | SGK | Serum/glucocorticoid regulated kinase | 0.20 | 0.38 | 0.88 | 1.38 | |
| MmugDNA.40778.1.S1_at | SH3MD2 | SH3 multiple domains 2 | 1.00 | − 0.01 | 0.12 | 0.16 | |
| MmugDNA.37821.1.S1_at | TNFRSF19 | Tumor necrosis factor receptor superfamily, member 19 | − 1.47 | − 0.97 | − 0.84 | − 2.01 | |
| MmugDNA.39342.1.S1_s_at | TNFRSF21 | Tumor necrosis factor receptor superfamily, member 21 | − 0.42 | − 0.26 | − 0.74 | − 1.17 | |
| MmugDNA.34645.1.S1_s_at | ZFP36L1 | Zinc finger protein 36, C3H type-like 1 | − 0.16 | − 0.41 | − 0.73 | − 1.05 | |
| DNA repair | |||||||
| MmugDNA.12810.1.S1_at | BRCA2 | Breast cancer 2, early onset | − 1.18 | − 0.52 | 0.26 | − 0.17 | |
| Cell cycle and proliferation | |||||||
| MmugDNA.880.1.S1_at | ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | − 0.04 | − 0.32 | − 1.36 | − 2.22 | |
| MmugDNA.18873.1.S1_at | APBB2 | Amyloid beta (A4) precursor protein-binding, family B, member 2 (Fe65-like) | − 0.85 | − 0.33 | − 0.54 | − 1.02 | |
| MmugDNA.40593.1.S1_at | ANAPC5 | Anaphase promoting complex subunit 5 | − 1.21 | − 0.18 | − 0.36 | − 1.03 | |
| MmugDNA.25197.1.S1_at | ASPM | Asp (abnormal spindle)-like, microcephaly associated (Drosophila) | 1.14 | − 0.23 | − 0.16 | 0.64 | |
| MmugDNA.27494.1.S1_at | BTBD7 | BTB (POZ) domain containing 7 | − 1.60 | − 0.32 | − 0.77 | − 1.95 | |
| MmugDNA.40767.1.S1_at | BUB3 | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | − 1.02 | − 0.24 | − 0.41 | − 0.94 | |
| MmugDNA.24430.1.S1_at | CDC25A | Cell division cycle 25A | 0.46 | 0.83 | 1.08 | 1.55 | |
| MmugDNA.8073.1.S1_at | CDC25C | Cell division cycle 25C | − 1.00 | − 0.86 | − 0.46 | − 1.49 | |
| MmugDNA.7114.1.S1_at | CDCA2 | Cell division cycle associated 2 | − 0.54 | − 0.83 | − 1.44 | − 0.18 | |
| MmugDNA.15060.1.S1_at | CEP1 | Centrosomal protein 1 | 1.01 | − 0.02 | − 0.21 | 0.54 | |
| MmugDNA.27098.1.S1_at | CCND1 | Cyclin D1 | 1.04 | 0.56 | 0.20 | 0.76 | |
| MmugDNA.8564.1.S1_at | CCNE2 | Cyclin E2 | − 1.34 | 0.22 | 0.25 | − 0.82 | |
| MmugDNA.38294.1.S1_at | CCNG2 | Cyclin G2 | − 1.56 | − 0.63 | 0.01 | − 2.04 | |
| MmugDNA.14485.1.S1_at | CDK6 | Cyclin-dependent kinase 6 | − 1.23 | − 0.66 | − 0.91 | − 1.46 | |
| MmugDNA.12064.1.S1_at | CDKN2B | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | − 1.13 | − 0.30 | − 0.04 | − 1.12 | |
| MmugDNA.31081.1.S1_at | DAZAP2 | DAZ associated protein 2 | − 1.27 | − 0.55 | − 1.06 | − 1.85 | |
| MmugDNA.15195.1.S1_at | DBC1 | Deleted in bladder cancer 1 | 0.06 | − 0.35 | − 1.26 | − 0.91 | |
| MmugDNA.36289.1.S1_s_at | DLEU2 /// BCMSUNL | Deleted in lymphocytic leukemia, 2 /// BCMS upstream neighbor-like | 0.32 | 0.45 | 0.74 | 1.31 | |
| MmugDNA.16818.1.S1_at | DAB2 | Disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | − 0.11 | − 0.14 | − 0.23 | − 1.01 | |
| MmugDNA.32685.1.S1_at | DDR2 | Discoidin domain receptor family, member 2 | 0.66 | 0.17 | 0.36 | 1.52 | |
| MmugDNA.28062.1.S1_s_at | DLG1 | Discs, large homolog 1 (Drosophila) | − 0.14 | − 0.48 | − 0.39 | − 1.08 | |
| MmugDNA.32495.1.S1_at | DDIT3 | DNA-damage-inducible transcript 3 | 0.11 | 1.30 | 0.87 | 1.32 | |
| MmugDNA.24043.1.S1_at | DUSP1 | Dual specificity phosphatase 1 | − 1.04 | − 0.10 | 0.30 | − 0.04 | |
| MmugDNA.23886.1.S1_at | DNCH1 | Dynein, cytoplasmic, heavy polypeptide 1 | 0.66 | 0.73 | 0.35 | 1.12 | |
| MmugDNA.13015.1.S1_at | EML4 | Echinoderm microtubule associated protein like 4 | 1.25 | 0.32 | 0.32 | 1.20 | |
| MmugDNA.38400.1.S1_at | ENTH | Enthoprotin | − 1.06 | − 0.36 | − 0.37 | − 1.10 | |
| MmugDNA.31099.1.S1_s_at | EPLIN | Epithelial protein lost in neoplasm beta | − 0.78 | − 0.75 | − 1.34 | − 1.40 | |
| MmugDNA.35735.1.S1_at | EPB41L4A | Erythrocyte membrane protein band 4.1 like 4A | − 0.68 | − 0.41 | − 0.75 | − 1.43 | |
| MmugDNA.29681.1.S1_at | FBXO45 | F-box protein 45 | 0.91 | 0.26 | 0.40 | 1.07 | |
| MmugDNA.25606.1.S1_s_at | FUS | Fusion (involved in t(12;16) in malignant liposarcoma) | − 2.62 | − 0.90 | − 1.42 | − 1.31 | |
| MmugDNA.7304.1.S1_s_at | GSPT1 | G1 to S phase transition 1 | − 0.37 | − 0.31 | − 0.87 | − 1.50e | |
| MmugDNA.33793.1.S1_at | GTSE1 | G-2 and S-phase expressed 1 | 1.01 | − 0.45 | − 0.19 | 0.08 | |
| MmugDNA.33680.1.S1_at | GSN | Gelsolin (amyloidosis, Finnish type) | − 1.14 | 0.00 | − 0.16 | − 0.89 | |
| MmugDNA.38529.1.S1_at | ENPEP | Glutamyl aminopeptidase (aminopeptidase A) | − 0.60 | − 0.43 | − 0.25 | − 1.08 | |
| MmugDNA.25216.1.S1_at | GPC4 | Glypican 4 | − 0.43 | − 0.59 | − 1.08 | − 1.42 | |
| MmugDNA.41267.1.S1_at | GAS1 | Growth arrest- specific 1 | − 0.16 | 0.13 | − 0.27 | − 1.40 | |
| MmugDNA.23210.1.S1_x_at | IGF2 | Insulin-like growth factor 2 (somatomedin A) | 0.32 | 0.14 | − 0.89 | − 1.02 | |
| MmugDNA.2176.1.S1_at | ISG20 | Interferon stimulated exonuclease gene 20 kDa | 0.09 | 0.19 | 0.43 | 1.07 | |
| MmugDNA.18344.1.S1_at | JAG1 | Jagged 1 (Alagille syndrome) | − 0.10 | − 0.82 | − 1.04 | − 1.07 | |
| MmugDNA.30627.1.S1_at | JUB | Jub, ajuba homolog (Xenopus laevis) | − 0.68 | − 0.06 | − 0.27 | − 1.09 | |
| MmugDNA.21869.1.S1_at | KIF22 | Kinesin family member 22 | − 1.13 | − 0.37 | − 0.31 | − 1.16 | |
| MmugDNA.17868.1.S1_at | MAD2L1 | MAD2 mitotic arrest deficient-like 1 (yeast) | − 1.17 | − 0.40 | − 0.22 | − 0.89 | |
| MmugDNA.11043.1.S1_s_at | MAP7 | Microtubule-associated protein 7 | − 0.34 | − 0.59 | − 0.58 | − 1.19 | |
| MmugDNA.38080.1.S1_at | MPHOSPH9 | M-phase phosphoprotein 9 | − 0.87 | − 0.77 | − 0.61 | − 1.50 | |
| MmugDNA.40010.1.S1_at | NF2 | Neurofibromin 2 (bilateral acoustic neuroma) | − 0.40 | 0.02 | − 0.08 | − 1.13 | |
| MmugDNA.11581.1.S1_at | NEK9 | NIMA (never in mitosis gene a)-related kinase 9 | − 1.36 | − 0.42 | − 0.71 | − 1.71 | |
| MmugDNA.25696.1.S1_at | NEK2 | NIMA (never in mitosis gene a)-related kinase 2 | − 1.53 | − 0.80 | − 0.44 | − 1.37 | |
| MmugDNA.15458.1.S1_at | NOTCH2 | Notch homolog 2 (Drosophila) | − 0.96 | − 0.30 | − 0.40 | − 1.14 | |
| MmugDNA.31795.1.S1_at | NUCKS1 | Nuclear casein kinase and cyclin-dependent kinase substrate 1 | − 1.19 | − 0.82 | − 1.07 | − 0.43 | |
| MmugDNA.18184.1.S1_at | PAK3 | P21 (CDKN1A)-activated kinase 3 | 1.40 | 0.09 | 0.17 | 1.52 | |
| MmugDNA.6179.1.S1_at | PARD6B | Par-6 partitioning defective 6 homolog beta (C. elegans) | − 1.09 | − 0.32 | − 0.55 | − 0.95 | |
| MmugDNA.28892.1.S1_at | PPIG | Peptidylprolyl isomerase G (cyclophilin G) | 1.14 | 0.00 | − 0.02 | 0.51 | |
| Cell cycle and proliferation | |||||||
| MmugDNA.30055.1.S1_at | PAFAH1B1 | Platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45 kDa | − 1.03 | − 0.40 | − 0.51 | − 1.19 | |
| MmugDNA.27441.1.S1_at | PPP2R1B | Protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), beta isoform | − 1.02 | − 0.37 | − 0.47 | − 0.79 | |
| MmugDNA.3729.1.S1_at | RIF1 | RAP1 interacting factor homolog (yeast) | − 1.76 | − 0.49 | − 0.70 | − 1.64 | |
| MmugDNA.23090.1.S1_at | RERG | RAS-like, estrogen-regulated, growth inhibitor | − 0.67 | − 0.43 | − 1.40 | − 2.26 | |
| MmugDNA.24655.1.S1_s_at | RBBP6 | Retinoblastoma binding protein 6 | 0.77 | 0.33 | 0.27 | 1.01 | |
| MmugDNA.1893.1.S1_at | RNGTT | RNA guanylyltransferase and 5′-phosphatase | − 1.06 | − 0.50 | − 0.51 | − 1.27 | |
| MmugDNA.43216.1.S1_at | S100A6 | S100 calcium binding protein A6 (calcyclin) | − 0.20 | − 0.25 | − 0.30 | − 1.15 | |
| MmugDNA.18017.1.S1_at | SEP2 | Septin 2 | − 2.05 | − 0.63 | − 0.90 | − 2.10 | |
| MmugDNA.7350.1.S1_s_at | SSR1 | Signal sequence receptor, alpha (translocon-associated protein alpha) | − 1.12 | − 0.04 | 0.23 | − 0.20 | |
| MmugDNA.29506.1.S1_at | SMC5L1 | SMC5 structural maintenance of chromosomes 5-like 1 (yeast) | 0.81 | 0.27 | 0.06 | 1.17 | |
| MmugDNA.10117.1.S1_s_at | SKP2 | S-phase kinase-associated protein 2 (p45) | − 1.68 | − 0.61 | − 0.65 | − 1.02 | |
| MmugDNA.23385.1.S1_at | TSPAN2 | Tetraspanin 2 | − 0.57 | − 0.51 | − 0.70 | − 1.07 | |
| MmugDNA.39770.1.S1_at | TSPAN31 | Tetraspanin 31 | − 0.80 | − 0.40 | − 0.24 | − 1.19 | |
| MmugDNA.20960.1.S1_at | TGFA | Transforming growth factor, alpha | − 1.50 | − 0.17 | − 0.44 | − 1.39 | |
| MmugDNA.35789.1.S1_at | TUSC2 | Tumor suppressor candidate 2 | − 0.71 | − 0.11 | − 0.22 | − 1.03 | |
| MmugDNA.40025.1.S1_at | UHMK1 | U2AF homology motif (UHM) kinase 1 | − 1.38 | − 0.36 | − 0.60 | − 1.80 | |
The bolded numbers represent genes with more than two-fold up-regulation, while the italicized numbers represent genes with more than two-fold down-regulation.
Differential gene expression confirmed by real-time PCR
To verify the result of the microarray analysis, relative expression levels of selected genes were assayed with real-time quantitative PCR. The relative quantification by real-time PCR suggested the creditability of the array analysis (Table 2 ). It is noted that the scale of expression level detected by these two systems may vary, possibly due to the variation of probing regions, sensitivity of assays, as well as the saturation of fluorescence signals in array analysis.
Table 2.
Comparison of the gene expression levels in S2-expressing Vero E6 cells analyzed with microarray and real-time PCR
| Gene name | Microarraya |
Real timeb |
||||||
|---|---|---|---|---|---|---|---|---|
| 12 | 24 | 48 | 72 | 12 | 24 | 48 | 72 | |
| ATF1 | − 1.47 | − 0.47 | − 0.50 | − 1.24 | − 1.13 | − 0.15 | − 0.10 | − 1.63 |
| Bcl−xL | − 1.56 | − 0.23 | − 0.77 | − 1.92 | − 0.87 | − 0.58 | − 0.22 | − 1.73 |
| CDK6 | − 1.23 | − 0.66 | − 0.91 | − 1.46 | − 0.50 | − 0.08 | −1.00 | − 1.98 |
| CYCS | 1.50 | 0.58 | 0.40 | 1.63 | 1.03 | 0.62 | 0.45 | 1.02 |
| GRP78 | 0.15 | 0.39 | 1.88 | 2.15 | − 0.26 | 0.67 | 1.44 | 1.84 |
| GRP94 | 0.16 | 0.42 | 1.41 | 1.15 | 0.41 | 0.23 | 0.96 | 1.21 |
| IGF2 | 0.32 | 0.14 | − 0.89 | − 1.02 | 0.65 | 0.58 | − 1.83 | − 2.28 |
| IkBkB | − 1.10 | − 0.56 | − 0.58 | − 0.76 | − 1.02 | 0.32 | − 0.13 | − 0.55 |
| JunD | − 0.27 | 0.01 | 0.22 | − 1.00 | − 0.85 | −0.32 | 0.17 | − 1.12 |
| MAPK1 | − 1.41 | − 0.32 | − 0.27 | − 1.42 | − 1.22 | − 0.83 | 0.52 | − 2.80 |
For both assays, bolded numbers represent genes with more than two-fold up-regulation, while italicizec numbers represent genes with more than two-fold down-regulation.
For each gene, only the probe set with the largest variation is shown.
The average expression level of triplicate experiments is shown.
Transcriptional inactivation of extrinsic apoptotic pathway
Apoptosis is initiated through two interrelated pathways, one involving the engagement of TNF family of death receptors, namely the extrinsic pathway, and the other involving Bax-mediated release of cytochrome c (CYCS) from mitochondria, namely the intrinsic pathway. In the extrinsic pathway, engagement of death receptors have been shown to trigger the activation of MAPK/JNK, caspase 8 and caspase 3, which leads to apoptosis (Baetu and Hiscott, 2002). Based on our microarray data, expression of a number of genes involved in the extrinsic pathways of apoptosis was down-regulated. TRAIL, which is a ligand of death receptor 4/5 and a potent apoptotic inducer, and FAF1, which binds to FAS antigen and initiates apoptosis, were both found to be down-regulated upon S2 over-expression. These observations suggest a possible suppression of TRAIL-induced and FasL-induced apoptotic pathways. A downstream target of these two pathways, caspase 8, was also down-regulated at 48 and 72 h p.t. Although the down-regulation of HIPK3, a gene that activates the Fas apoptotic pathway (Curtin and Cotter, 2004), was observed, the possibility that the effect of HIPK3 as a feedback mechanism cannot be excluded. Inhibition of apoptosis is a common strategy employed by viruses to evade host defense mechanism that limits viral replication by triggering the apoptosis of the infected cells (Hiscott et al., 2001). From this, the blockage of extrinsic apoptotic pathway by S2 may represent a viral defense mechanism to avoid premature cell death before the completion of viral replication.
Transcriptional activation of intrinsic apoptotic pathway
In our study, several lines of evidence suggest the triggering of intrinsic apoptotic pathway in response to S2 expression. From the microarray data, the expression of Mdm2, a protein that mediates the degradation of pro-apoptotic tumor suppressor protein p53 (Momand et al., 1992), was showed to be down-regulated. The down-regulation of Mdm2 thus suggests the possible increase of the activity of a pro-apoptotic Bcl-2 family member, Bax, via p53 (Miyashita and Reed, 1995). Bax accelerates the opening of the mitochondrial porin channel and mediates the release of CYCS (Narita et al., 1998). Indeed, as indicated in our data, the expression of S2 did up-regulate the transcription of CYCS, the leakage of CYCS is thus expected to be increased, which enables the formation of apoptosome and the subsequent activation of the downstream effector caspase cascade, and ultimately leads to apoptosis (Cain et al., 2002). Collectively, these data suggest that the intrinsic apoptotic pathway may possibly be responsible for the S2-induced apoptosis in Vero E6 cells.
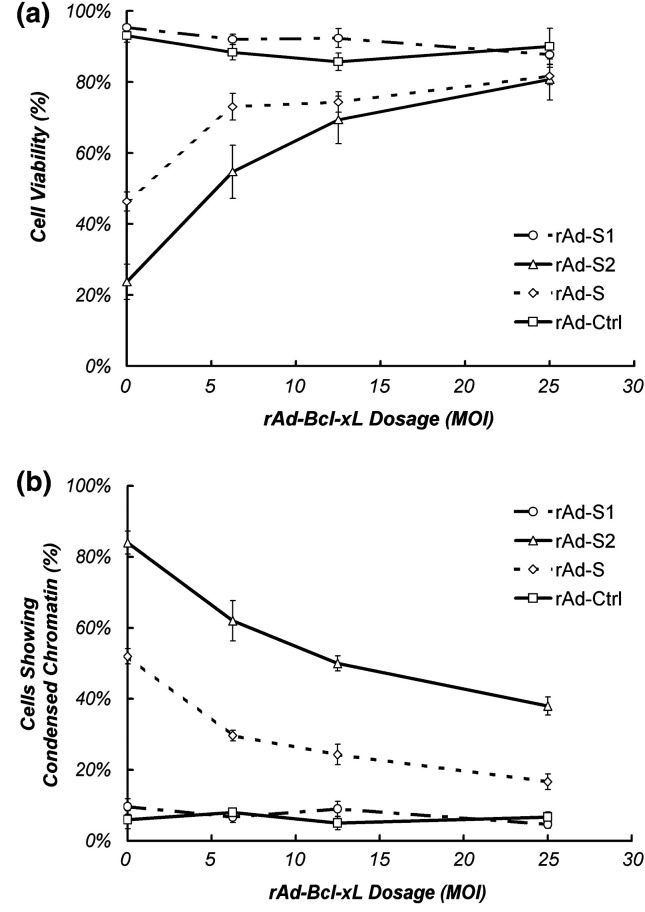
In SARS-CoV-infected cells, down-regulation of Bcl-2 and up-regulation of Bax has been reported (Ren et al., 2005). These, together with the capacity of Bcl-2 to overcome the apoptotic effects of SARS-CoV (Bordi et al., 2006), imply the involvement of the intrinsic apoptotic pathway. In our array data, despite the fact that a pro-apoptotic member of the Bcl-2 family, Bim (Marani et al., 2002), was down-regulated, a number of anti-apoptotic Bcl-2 family members, including Mcl-1, Bcl-xL and Bcl-2 (Gross et al., 1999, Reed, 1996), were also found to be down-regulated. These anti-apoptotic members are known to inhibit the release of CYCS from mitochondria to cytosol by blocking the mitochondrial porin channel (Shimizu et al., 1999). To further elucidate the possible inhibitory role of the anti-apoptotic Bcl-2 protein members in S2-induced apoptosis, we over-expressed Bcl-xL in Vero E6 cells through rAd transduction. As shown in Fig. 3 , over-expression of Bcl-xL significantly reduced the extent of apoptosis in Vero E6 cells induced by both S2 and S. In contrast, co-transduction of rAd-Ctrl with rAd-S2 or rAd-S has no significant effect on the level of apoptosis mediated by S nor S2 (data not shown). Besides the important role of Bcl-xL in mitochondrial apoptosis, it is noted that over-expression of Bcl-xL may inhibit other mitochondria-independent apoptotic pathways like phosphorylation of JNK (Srivastava et al., 1999).
Fig. 3.
Bcl-xL blocks S2-induced apoptosis in Vero E6 cells. Different dosage of rAd-Bcl-xL was co-infected with 50 MOI of rAd-S, -S1, -S2 or rAd-Ctrl and the effect on cell death and apoptosis induced by the rAds were assayed through (a) Trypan blue exclusion assay and (b) Hoechst 33342 staining at day 5 and day 3 p.t. respectively. Both figures indicated the average of three independent experiments with SEM.
Induction of endoplasmic reticulum stress responses
The expression of full length spike protein has been demonstrated to induce specific ER stress and unfolded protein response (UPR) while other viral proteins like M, N and E do not (Chan et al., 2006). Previous reports showed that ER stress may trigger 3 branches of UPR including activating transcription factor 6 (ATF6), inositol-requiring enzyme 1 (IRE1) and PKR-like ER kinase (PERK)-related signaling pathways (Schroder and Kaufman, 2005). Chan et al. (2006) demonstrated the ER stress induced by spike protein does not trigger ATF6 and IRE1-related signaling pathways while GRP78 and GRP94, indicators for activation of PERK-related signaling pathways (Schroder and Kaufman, 2005), are up-regulated. Interestingly, our array data on the expression of these genes, as well as the real-time PCR analysis of GRP78 and GRP94 expression (Table 2), are consistent with these findings, suggesting a possible role of the S2 subunit in the specific ER stress induced by S. Although GRP78/94-induced UPR takes part in alleviating ER stress and promoting anti-apoptotic signaling (Ma and Hendershot, 2004), a prolonged ER stress may induce mitochondrial apoptosis by releasing calcium ion and down-regulating Bcl-2 (Breckenridge et al., 2003). Coincidentally, Bcl-2 was found to be down-regulated in this study (Table 1). ER stress may also activate caspase 4, which is a homologue to murine caspase 12 (Hitomi et al., 2004), to induce mitochondria-independent apoptotic response. However, due to the fact that murine caspase 12 is triggered through IRE1 activation (Yoneda et al., 2001) and that the over-expression of spike protein does not activate IRE1 (Chan et al., 2006), it is anticipated that caspase 4 may not be activated by SARS-CoV spike protein. Taken together, since a similar transcriptional response was observed by the expression of S of both SARS-CoV and MHV but not other viral proteins (Chan et al., 2006, Versteeg et al., 2007), we proposed that the potential ER stress observed in our assay is specific to S2 expression.
Inactivation of the NFκB pathway
The transcription factor NFκB is a key regulator in cell proliferation, protecting cells from apoptosis under most circumstances and accelerating apoptosis in the others (Piva et al., 2006). Our data suggest a possible inhibition of NFκB activities, as supported by the regulation of several targets and regulators of NFκB. In particular, IκBκB, which is responsible for the activation of NFB (Scheidereit, 2006), was down-regulated at the early time-point upon S2 expression. In addition, at both early (12 h p.t.) and late (72 h p.t.) time points, a negative regulator of NFκB, the TNFAIP3 (Wertz et al., 2004), was up-regulated, while positive regulators, including ECT2, FKBP1A, TRAIL, TMEM9B and TRIM38 (Matsuda et al., 2003), were down-regulated. NFκB-dependent decrease of the expression level of anti-apoptotic factors like Bcl-2 (Catz and Johnson, 2001), Bcl-xL (Chen et al., 2000), Mcl-1 (Henson et al., 2003), A1/Bfl-1 and TRAF-1 (Akari et al., 2001) has been demonstrated.
Down-regulation of the MAPK pathway
The MAPK-dependent signaling cascade is known to be important for regulating apoptosis, cell differentiation and proliferation. Potential suppression of the MAPK pathways is suggested to be mediated by the down-regulation of its key members, p38 MAPK (MAPK1) and p38 MAPKα (MAPK14), and the key targets, including ATF1 (Tan et al., 1996), CREB1 (Tan et al., 1996), JunD (Stocco et al., 2002) and Sp1 (Moon et al., 2006). While their transcriptional profiles have not been investigated in SARS-CoV-infected cells, based on our microarray results, we anticipate a conceivable role of S2 in down-regulating these genes at the transcription level. Suppression of MAPK cascade by S2 may imply various downstream responses as suggested by its overall regulation observed in other cell types. Consequential down-regulations of ATF1, CREB1, JunD and Sp1 observed in S2-expressing Vero E6 cells suggested the trend towards cell death. It is noted that the p38 MAPK, CREB, MSK-1, ERK1/2 and ATF1 (Mizutani et al., 2004b), and PI3k/Akt-dependent pathways (Mizutani et al., 2004a) were activated in SARS-CoV-infected Vero E6 cells, although their role in S2-mediated apoptosis has not yet been defined. Indeed, the activation of these proteins may possibly due to the presence of other viral proteins or the double-stranded RNA intermediates of SARS-CoV through different machineries (Mizutani et al., 2004b).
Transcriptional regulation of genes involved in cell cycle control
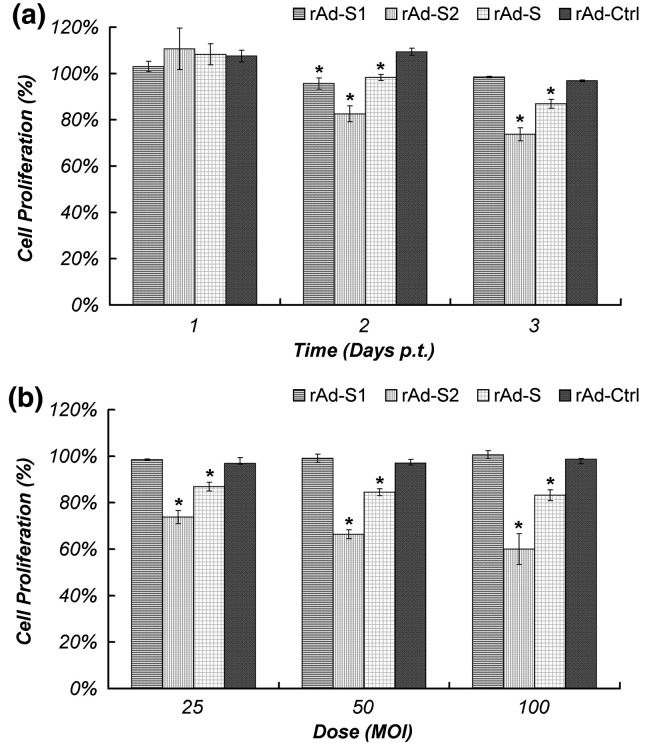
Cell cycle progression is mainly governed by proteins including cyclins, cyclin-dependent kinase (CDK) and CDK inhibitors, which cooperatively control the phosphorylation of retinoblastoma (Rb) (Sherr, 1996). The cyclin E2 (CCN E2)/CDK6 complex is important for the progression of cell cycle in the G1/S phase. While IGF2 acts upstream of the complex (Zhang et al., 1999), the activity of CTNN B1 is regulated by the complex through binding (Park et al., 2004). MDM2 is a negative regulator of p53 and it has been demonstrated to inhibit the G1 arrest (Chen et al., 1996). The reduced MDM2 expression was expected to activate the inhibitor of CDKN 1A (p21, Cip1) via p53 (Waldman et al., 1995). In this study, transcriptional regulations of a number of cyclins, CDKs and their inhibitors were detected. Cyclin D1 (CCN D1) was found to be up-regulated, while the expressions of IGF2, CCN E2, CDK6 and CTNN B1 were all suppressed upon S2 over-expression. CDKN 2B, a CDK inhibitor involved in negative regulation of cell proliferation, was also found to be down-regulated (Table 1). Thus, our data suggest the possibility of cell cycle arrest in G1/S phase by S2 through modulating G1 controlling cyclins. While the regulation of cell cycle mediators leading to cell proliferation inhibition is a common phenomenon in virus infection cycle, we observed, as described above, an inhibition of cell proliferation in cells transduced with rAd-S2 (Fig. 4 ). Further study to confirm the S2-induced arrest at G1/S phase will be indicative to understand the role of S2 in the pathogenesis of SARS-CoV.
Fig. 4.
S2-mediated inhibition of cell proliferation in Vero E6 cells. Vero E6 cells were transduced with the indicated rAds and their proliferations were estimated with MTT cell proliferation assay and are expressed as percentage of proliferation of mock-infected cells. Time- and dose-dependent anti-proliferation effect of S2 was shown in panels a and b, respectively. The time-dependent effect was performed with rAd of 25 MOI and cells in the dose-dependent experiment were collected at 3 days p.t. Results shown are the average of three independent experiments with SEM. ⁎p < 0.01 when compared with cells transduced by rAd-Ctrl under the same condition.
It has been shown that SARS-CoV can induce cell proliferation arrest (Mizutani et al., 2006). Recent studies suggested the induction of G0/G1 phase arrest by SARS-CoV ORF 3b and 7a (Yuan et al., 2005, Yuan et al., 2006) and S phase arrest by N protein (Surjit et al., 2006). While the phase in which cell cycle is arrested in SARS-CoV-infected cells remained to be determined, the inhibition of G1 phase progression in mouse hepatitis virus (MHV) p28 protein-transduced (Chen et al., 2004) and in IBV-infected (Dove et al., 2006) cells with increase viral protein productions suggested the possible role of potential G1 arrest in rAd-S2-transduced cells. This in turn plays a role in the lytic replication cycle, driving the infected cells to apoptosis showing cytopathic effect.
Overall conclusive remarks
In this study, we investigated the possible modulation of the host cellular factors by S2 of SARS-CoV, which may be important in mediating cell death and cell proliferation arrest. We observed a suppression of extrinsic apoptotic pathway and an activation of intrinsic pathway. Together with the down-regulation of genes involved in the MAPK and NFB signaling pathway, these changes are believed to be responsible for the S2-induced apoptotic cell death. Modulation of expression of G1 cyclins also suggests the capacity of inducing G1 cell cycle arrest by S2. These data collectively elucidate the possible role of S2 in disease pathogenesis and viral replication. In summary, our investigation unraveled the candidate physiological pathways involved in S2-induced apoptosis at a molecular level, providing a foundation for researchers to design experiments based on testable hypothesis targeting individual genes. This information is expected to shed light on the molecular mechanisms of the pathogenesis of SARS, which is important for the development of antiviral therapy.
Materials and methods
Cell cultures
HEK293-derived AD-293 cells (Stratagene) used to propagate recombinant adenoviruses (rAds), and the African green monkey kidney cell line Vero E6 cells (CRL-1586, American Type Culture Collection) used for adenoviral transduction and microarray analysis were cultured and maintained as described (Chow et al., 2005).
Preparation of recombinant adenoviruses and transduction of Vero E6 cells
Recombinant adenoviruses expressing S (rAd-S), S1 (rAd-S1) and S2 (rAd-S2), as well as a control rAd with no transgene (rAd-Ctrl), were constructed, propagated, purified and titrated as previously described (Chow et al., 2005). For all adenoviral transductions in this study, monolayer of Vero E6 cells was infected with minimal volume of purified rAds at a multiplicity of infections (MOI) of 100 unless otherwise specified. At 2 h p.t., minimum essential medium with eagle's salts (EMEM; Gibco-BRL) was added to the transduced cells at a final concentration of 1% heat-inactivated fetal bovine serum (FBS; Gibco-BRL) and 1% antibiotics–antimycotic (Gibco-BRL).
Assessment of apoptosis induced by S2 expression
Cell viability, nuclear morphology and the extent of DNA fragmentation of rAd-S2 and rAd-Ctrl-transduced Vero E6 cells were investigated at the indicated time intervals using Trypan blue exclusion assay, Hoechst 33342 staining (Sigma) and genomic DNA laddering assay, respectively. All three assays were performed in triplicate as previously described (Chow et al., 2005).
RNA sample preparation and microarray analysis
To identify the differentially expressed transcripts induced by S2 expression, the global gene expression profiles of rAd-Ctrl and rAd-S2-transduced Vero E6 cells at 12, 24, 48 and 72 h p.t. were compared using GeneChip® Rhesus Macaque Genome Array (Affymetrix). The microarray was spotted with 52,865 probe sets referring to more than 47,000 Macaca mulatta transcripts. For both rAd-Ctrl and rAd-S2-transduced cells, total RNA were extracted from approximately 5 × 106 cells at each of the 4 indicated time points using RNeasy Mini Kit (Qiagen) according to manufacturer's instructions. The integrity of the extracted RNA was confirmed by electrophoresis in a 1% denaturing agarose gel and the aliquots were stored at − 80 °C until further use. Fifteen micrograms of the extracted RNA from each sample at each of the time points was used in microarray analysis. Standard sample preparation using the one cycle target labeling system, slide hybridization and scanning was performed in the Institute of Systems Biology (ISB; Seattle, USA) according to manufacturer's instructions. The intensity value was then processed with RMAExpress for background adjustment, quartile normalization and summarization with the Web-based SEBAMS system. The physiological functions of the transcripts were annotated according to the original annotation of the array, iHOP database (Robert Hoffmann) and NetAffy database (Affymetrix).
Real-time quantitative PCR
Real-time quantitative PCR analysis was performed to validate the transcript expression pattern detected in the microarray for selected transcripts (n = 6) using specific primers (Table 3 ). First strand cDNA was synthesized from 2 μg of the extracted RNA at each of the time point using MMLV-reverse transcriptase (Promega) according to manufacturer's instructions. Real-time PCR analysis was then performed with the EvaGreen real-time PCR system (Biotium Inc.) and the iCycler real-time PCR machine (Bio-Rad) according to the manufacturers' instructions. The amplification cycle included initial denaturation at 94 °C 10 min and 45 cycles of (94 °C 10 s, 55 °C 10 s, 72 °C 10 s, 80 °C 7 s), followed by 76 cycles of (+ 0.5 °C per s) from 56 °C for melting curve analysis. Amplification of the specific genes was repeated in triplicate. The relative expression (RE) of each gene in rAd-transduced cells were calculated relative to that of β-actin (REβ-actin) using the ΔCt method, i.e. REβ-actin = 2ΔCt, while ΔCt = (Ctβ-actin − Cttarget gene). The normalized results were represented as the number of fold changes when compared to the rAd-Ctrl in each time point, which was calculated as log 2 [REβ-actin (rAd-S2 transduced)] log2 [REβ-actin (rAd-Ctrl transduced)]− 1.
Table 3.
List of primers for quantitative real-time PCR analysis
| Primer name | Primer sequence (5′-3′) | Product size | |
|---|---|---|---|
| 1 | β-Actin_F | ATCGTGCGTGACATTAAGGAG | 179 |
| β-Actin_R | AGGAAGGAAGGCTGGAAGAG | ||
| 2 | ATF1_F | CAGGCACAGATGGAGTACAG | 131 |
| ATF1_R | CTGATTGCTGGGCACAAGTA | ||
| 3 | Bcl-xL_F | TTGAACAGGTAGTGAATGAACTC | 139 |
| Bcl-xL_R | AAGCTGCGATCCGACTCACC | ||
| 4 | CDK6_F | CCGAAGTCTTGCTCCAGTC | 124 |
| CDK6_R | CCTAGTTGATCGACATCTGAA | ||
| 5 | CYCS_F | TCCACATGGCTGTCAAGAA | 113 |
| CYCS_R | CACGACGCCCAGTTATCTA | ||
| 6 | GRP78_F | GGCAACTGGCTGAAAGGT | 92 |
| GRP78_R | GGCAGTGCAGCAGAGGTC | ||
| 7 | GRP94_F | CGCTTCGGTCAGGATATC | 151 |
| GRP94_R | CTGTCGTGTCTTCTGTTGTCT | ||
| 8 | IGF2_F | GTCCAGCAATCGGAAGTGA | 125 |
| IGF2_R | GGAAGTGGAACCGAGAGATT | ||
| 9 | IkBkB_F | GCAGCAAGGAGAACAGAGG | 152 |
| IkBkB_R | CGACGGTCACTGTGTACTTCT | ||
| 10 | JunD_F | CCTCAAGAGTCAGAACACGGA | 98 |
| JunD_R | CGCTGTTGACGTGGCTGA | ||
| 11 | MAPK1_F | CATCGCCGAAGCACCATT | 108 |
| MAPK1_R | CTGTATCCTGGCTGGAATCT |
MTT cell proliferation assay
Cell proliferation in rAd-transduced Vero E6 cells was evaluated in a time- and dose-dependent manner using MTT Cell Proliferation Kit (Roche) according to manufacturer's instructions. Briefly, 1 × 104 Vero E6 cells were seeded in each well of a 96-well tissue culture plate (Falcon) 1 day before rAd transduction at the indicated MOIs. To assess the extent of cell proliferation, MTT labeling reagent was added in the indicated time points at a final concentration of 0.5 mg ml− 1 and the plate was incubated at 37 °C in dark for 4 h before the addition of solubilization solution. The plate was then allowed to stand overnight before the measurement of absorbance at 595 nm.
Bcl-xL co-transduction assay
Recombinant adenovirus carrying Bcl-xL (rAd-Bcl-xL) was cloned, propagated and purified in the same way of other rAds as described, whereas the Bcl-xL transgene was cloned from the cDNA of Vero E6 cells. The expression of Bcl-xL in rAd-Bcl-xL-transduced Vero E6 cells was confirmed by Western blot targeting the transgene's V5 tag (data not shown). Ten thousand cells were seeded 1 day before transduction. Serial diluted rAd-Bcl-xL was then added to the cells 30 min before the transduction of other candidate rAds at 50 MOI. After an incubation for another 1.5 h, EMEM was added to each well to achieve a final concentration of 1% FBS.
Acknowledgment
This work was supported by HWF CR/3/6/3921/03 research grant funded by the Hong Kong Government.
Contributor Information
Yin-Shan Yeung, Email: ysyeung@graduate.hku.hk.
Chi-Wai Yip, Email: h0024004@hkusua.hku.hk.
Chung-Chau Hon, Email: h9826299@hkusua.hku.hk.
Ken Y.C. Chow, Email: chow@pasteur.fr.
Iris C.M. Ma, Email: h0105962@hkusua.hku.hk.
Fanya Zeng, Email: fzeng@hkucc.hku.hk.
Frederick C.C. Leung, Email: fcleung@hkucc.hku.hk.
References
- Akari H., Bour S., Kao S., Adachi A., Strebel K. The human immunodeficiency virus type 1 accessory protein Vpu induces apoptosis by suppressing the nuclear factor kappaB-dependent expression of antiapoptotic factors. J. Exp. Med. 2001;194(9):1299–1311. doi: 10.1084/jem.194.9.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetu T.M., Hiscott J. On the TRAIL to apoptosis. Cytokine Growth Factor Rev. 2002;13(3):199–207. doi: 10.1016/s1359-6101(02)00006-0. [DOI] [PubMed] [Google Scholar]
- Bordi L., Castilletti C., Falasca L., Ciccosanti F., Calcaterra S., Rozera G., Di Caro A., Zaniratti S., Rinaldi A., Ippolito G., Piacentini M., Capobianchi M.R. Bcl-2 inhibits the caspase-dependent apoptosis induced by SARS-CoV without affecting virus replication kinetics. Arch. Virol. 2006;151(2):369–377. doi: 10.1007/s00705-005-0632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge D.G., Germain M., Mathai J.P., Nguyen M., Shore G.C. Regulation of apoptosis by endoplasmic reticulum pathways. Oncogene. 2003;22(53):8608–8618. doi: 10.1038/sj.onc.1207108. [DOI] [PubMed] [Google Scholar]
- Cain K., Bratton S.B., Cohen G.M. The Apaf-1 apoptosome: a large caspase-activating complex. Biochimie. 2002;84(2–3):203–214. doi: 10.1016/s0300-9084(02)01376-7. [DOI] [PubMed] [Google Scholar]
- Catz S.D., Johnson J.L. Transcriptional regulation of bcl-2 by nuclear factor kappa B and its significance in prostate cancer. Oncogene. 2001;20(50):7342–7351. doi: 10.1038/sj.onc.1204926. [DOI] [PubMed] [Google Scholar]
- Chan C.P., Siu K.L., Chin K.T., Yuen K.Y., Zheng B., Jin D.Y. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2006;80(18):9279–9287. doi: 10.1128/JVI.00659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wu X., Lin J., Levine A.J. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol. Cell. Biol. 1996;16(5):2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Edelstein L.C., Gelinas C. The Rel/NF-kappaB family directly activates expression of the apoptosis inhibitor Bcl-x(L) Mol. Cell. Biol. 2000;20(8):2687–2695. doi: 10.1128/mcb.20.8.2687-2695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.J., Sugiyama K., Kubo H., Huang C., Makino S. Murine coronavirus nonstructural protein p28 arrests cell cycle in G0/G1 phase. J. Virol. 2004;78(19):10410–10419. doi: 10.1128/JVI.78.19.10410-10419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K.Y., Yeung Y.S., Hon C.C., Zeng F., Law K.M., Leung F.C. Adenovirus-mediated expression of the C-terminal domain of SARS-CoV spike protein is sufficient to induce apoptosis in Vero E6 cells. FEBS Lett. 2005;579(30):6699–6704. doi: 10.1016/j.febslet.2005.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin J.F., Cotter T.G. JNK regulates HIPK3 expression and promotes resistance to Fas-mediated apoptosis in DU 145 prostate carcinoma cells. J. Biol. Chem. 2004;279(17):17090–17100. doi: 10.1074/jbc.M307629200. [DOI] [PubMed] [Google Scholar]
- Dimitrov D.S. Virus entry: molecular mechanisms and biomedical applications. Nat. Rev., Microbiol. 2004;2(2):109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove B., Brooks G., Bicknell K., Wurm T., Hiscox J.A. Cell cycle perturbations induced by infection with the coronavirus infectious bronchitis virus and their effect on virus replication. J. Virol. 2006;80(8):4147–4156. doi: 10.1128/JVI.80.8.4147-4156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A., Cinatl M., Eickmann J., Escriou M., Grywna N., Kramme K., Manuguerra S., Muller C., Rickerts S., Sturmer V., Vieth M., Klenk S., Osterhaus H.D., Schmitz A.D., Doerr H. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Duan J., Yan X., Guo X., Cao W., Han W., Qi C., Feng J., Yang D., Gao G., Jin G. A human SARS-CoV neutralizing antibody against epitope on S2 protein. Biochem. Biophys. Res. Commun. 2005;333(1):186–193. doi: 10.1016/j.bbrc.2005.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross A., McDonnell J.M., Korsmeyer S.J. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999;13(15):1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- Hattermann K., Muller M.A., Nitsche A., Wendt S., Donoso Mantke O., Niedrig M. Susceptibility of different eukaryotic cell lines to SARS-coronavirus. Arch. Virol. 2005;150(5):1023–1031. doi: 10.1007/s00705-004-0461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M.L., Zheng B.J., Chen Y., Wong K.L., Huang J.D., Lin M.C., Peng Y., Yuen K.Y., Sung J.J., Kung H.F. Kinetics and synergistic effects of siRNAs targeting structural and replicase genes of SARS-associated coronavirus. FEBS Lett. 2006;580(10):2414–2420. doi: 10.1016/j.febslet.2006.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson E.S., Gibson E.M., Villanueva J., Bristow N.A., Haney N., Gibson S.B. Increased expression of Mcl-1 is responsible for the blockage of TRAIL-induced apoptosis mediated by EGF/ErbB1 signaling pathway. J. Cell. Biochem. 2003;89(6):1177–1192. doi: 10.1002/jcb.10597. [DOI] [PubMed] [Google Scholar]
- Hiscott J., Kwon H., Genin P. Hostile takeovers: viral appropriation of the NF-kappaB pathway. J. Clin. Invest. 2001;107(2):143–151. doi: 10.1172/JCI11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi J., Katayama T., Eguchi Y., Kudo T., Taniguchi M., Koyama Y., Manabe T., Yamagishi S., Bando Y., Imaizumi K., Tsujimoto Y., Tohyama M. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. J. Cell Biol. 2004;165(3):347–356. doi: 10.1083/jcb.200310015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pohlmann S. Cellular entry of the SARS coronavirus. Trends Microbiol. 2004;12(10):466–472. doi: 10.1016/j.tim.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshidi-Parsian A., Dong Y., Zheng X., Zhou H.S., Zacharias W., McMasters K.M. Gene expression profiling of E2F-1-induced apoptosis. Gene. 2005;344:67–77. doi: 10.1016/j.gene.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Kaye M. SARS-associated coronavirus replication in cell lines. Emerg. Infect. Dis. 2006;12(1):128–133. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keng C.T., Zhang A., Shen S., Lip K.M., Fielding B.C., Tan T.H., Chou C.F., Loh C.B., Wang S., Fu J., Yang X., Lim S.G., Hong W., Tan Y.J. Amino acids 1055 to 1192 in the S2 region of severe acute respiratory syndrome coronavirus S protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. J. Virol. 2005;79(6):3289–3296. doi: 10.1128/JVI.79.6.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J., Cox Y., Hughes N., LeDuc J.M., Bellini J.W., Anderson W.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lai C.W., Chan Z.R., Yang D.G., Lo W.H., Lai Y.K., Chang M.D., Hu Y.C. Accelerated induction of apoptosis in insect cells by baculovirus-expressed SARS-CoV membrane protein. FEBS Lett. 2006;580(16):3829–3834. doi: 10.1016/j.febslet.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law P.T., Wong C.H., Au T.C., Chuck C.P., Kong S.K., Chan P.K., To K.F., Lo A.W., Chan J.Y., Suen Y.K., Chan H.Y., Fung K.P., Waye M.M., Sung J.J., Lo Y.M., Tsui S.K. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 2005;86(Pt 7):1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- Leong W.F., Tan H.C., Ooi E.E., Koh D.R., Chow V.T. Microarray and real-time RT-PCR analyses of differential human gene expression patterns induced by severe acute respiratory syndrome (SARS) coronavirus infection of Vero cells. Microbes Infect. 2005;7(2):248–259. doi: 10.1016/j.micinf.2004.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Hendershot L.M. The role of the unfolded protein response in tumour development: friend or foe? Nat. Rev., Cancer. 2004;4(12):966–977. doi: 10.1038/nrc1505. [DOI] [PubMed] [Google Scholar]
- Marani M., Tenev T., Hancock D., Downward J., Lemoine N.R. Identification of novel isoforms of the BH3 domain protein Bim which directly activate Bax to trigger apoptosis. Mol. Cell. Biol. 2002;22(11):3577–3589. doi: 10.1128/MCB.22.11.3577-3589.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G., Peiris J.S., Lim W., Osterhaus A.D. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425(6961):915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda A., Suzuki Y., Honda G., Muramatsu S., Matsuzaki O., Nagano Y., Doi T., Shimotohno K., Harada T., Nishida E., Hayashi H., Sugano S. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene. 2003;22(21):3307–3318. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- Miyashita T., Reed J.C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80(2):293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- Mizutani T., Fukushi S., Saijo M., Kurane I., Morikawa S. Importance of Akt signaling pathway for apoptosis in SARS-CoV-infected Vero E6 cells. Virology. 2004;327(2):169–174. doi: 10.1016/j.virol.2004.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T., Fukushi S., Saijo M., Kurane I., Morikawa S. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem. Biophys. Res. Commun. 2004;319(4):1228–1234. doi: 10.1016/j.bbrc.2004.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T., Fukushi S., Iizuka D., Inanami O., Kuwabara M., Takashima H., Yanagawa H., Saijo M., Kurane I., Morikawa S. Inhibition of cell proliferation by SARS-CoV infection in Vero E6 cells. FEMS Immunol. Med. Microbiol. 2006;46(2):236–243. doi: 10.1111/j.1574-695X.2005.00028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J., Zambetti G.P., Olson D.C., George D., Levine A.J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69(7):1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- Moon S.K., Choi Y.H., Kim C.H., Choi W.S. p38MAPK mediates benzyl isothiocyanate-induced p21WAF1 expression in vascular smooth muscle cells via the regulation of Sp1. Biochem. Biophys. Res. Commun. 2006;350(3):662–668. doi: 10.1016/j.bbrc.2006.09.092. [DOI] [PubMed] [Google Scholar]
- Narita M., Shimizu S., Ito T., Chittenden T., Lutz R.J., Matsuda H., Tsujimoto Y. Bax interacts with the permeability transition pore to induce permeability transition and cytochrome c release in isolated mitochondria. Proc. Natl. Acad. Sci. U. S. A. 1998;95(25):14681–14686. doi: 10.1073/pnas.95.25.14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.S., Kim S.I., Lee M.S., Youn C.Y., Kim D.J., Jho E.H., Song W.K. Modulation of beta-catenin phosphorylation/degradation by cyclin-dependent kinase 2. J. Biol. Chem. 2004;279(19):19592–19599. doi: 10.1074/jbc.M314208200. [DOI] [PubMed] [Google Scholar]
- Peiris J.S. Severe acute respiratory syndrome (SARS) J. Clin. Virol. 2003;28(3):245–247. doi: 10.1016/j.jcv.2003.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piva R., Belardo G., Santoro M.G. NF-kappaB: a stress-regulated switch for cell survival. Antioxid. Redox Signal. 2006;8(3–4):478–486. doi: 10.1089/ars.2006.8.478. [DOI] [PubMed] [Google Scholar]
- Reed J.C. Mechanisms of Bcl-2 family protein function and dysfunction in health and disease. Behring.-Inst.-Mitt. 1996;(97):72–100. [PubMed] [Google Scholar]
- Reghunathan R., Jayapal M., Hsu L.Y., Chng H.H., Tai D., Leung B.P., Melendez A.J. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6:2. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L., Yang R., Guo L., Qu J., Wang J., Hung T. Apoptosis induced by the SARS-associated coronavirus in Vero cells is replication-dependent and involves caspase. DNA Cell Biol. 2005;24(8):496–502. doi: 10.1089/dna.2005.24.496. [DOI] [PubMed] [Google Scholar]
- Scheidereit C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 2006;25(51):6685–6705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- Schroder M., Kaufman R.J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. Cancer cell cycles. Science. 1996;274(5293):1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Shimizu S., Narita M., Tsujimoto Y. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature. 1999;399(6735):483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- Srivastava R.K., Sollott S.J., Khan L., Hansford R., Lakatta E.G., Longo D.L. Bcl-2 and Bcl-X(L) block thapsigargin-induced nitric oxide generation, c-Jun NH(2)-terminal kinase activity, and apoptosis. Mol. Cell. Biol. 1999;19(8):5659–5674. doi: 10.1128/mcb.19.8.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocco C.O., Lau L.F., Gibori G. A calcium/calmodulin-dependent activation of ERK1/2 mediates JunD phosphorylation and induction of nur77 and 20alpha-hsd genes by prostaglandin F2alpha in ovarian cells. J. Biol. Chem. 2002;277(5):3293–3302. doi: 10.1074/jbc.M110936200. [DOI] [PubMed] [Google Scholar]
- Surjit M., Liu B., Jameel S., Chow V.T., Lal S.K. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 2004;383(Pt 1):13–18. doi: 10.1042/BJ20040984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Chow V.T., Lal S.K. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 2006;281(16):10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Rouse J., Zhang A., Cariati S., Cohen P., Comb M.J. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 1996;15(17):4629–4642. [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Fielding B.C., Goh P.Y., Shen S., Tan T.H., Lim S.G., Hong W. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 2004;78(24):14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B.S., Chan K.H., Cheng V.C., Woo P.C., Lau S.K., Lam C.C., Chan T.L., Wu A.K., Hung I.F., Leung S.Y., Yuen K.Y. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J. Virol. 2005;79(10):6180–6193. doi: 10.1128/JVI.79.10.6180-6193.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg G.A., van de Nes P.S., Bredenbeek P.J., Spaan W.J. The 638 coronavirus spike protein induces ER stress and upregulation of intracellular 639 chemokine mRNA concentrations. J. Virol. 2007;81(20):10981–10990. doi: 10.1128/JVI.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman T., Kinzler K.W., Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55(22):5187–5190. [PubMed] [Google Scholar]
- Wertz I.E., O'Rourke K.M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D.L., Ma A., Koonin E.V., Dixit V.M. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430(7000):694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Wong R.S., Wu A., To K.F., Lee N., Lam C.W., Wong C.K., Chan P.K., Ng M.H., Yu L.M., Hui D.S., Tam J.S., Cheng G., Sung J.J. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. 2003;326(7403):1358–1362. doi: 10.1136/bmj.326.7403.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Xiao G., Zhang J., Hu Y., Yuan F., Cole D.K., Zheng C., Gao G.F. SARS coronavirus induces apoptosis in Vero E6 cells. J. Med. Virol. 2004;73(3):323–331. doi: 10.1002/jmv.20094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Xiong Z., Zhang S., Yan Y., Nguyen J., Ng B., Lu H., Brendese J., Yang F., Wang H., Yang X.F. Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem. J. 2005;392(Pt 1):135–143. doi: 10.1042/BJ20050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T., Imaizumi K., Oono K., Yui D., Gomi F., Katayama T., Tohyama M. Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J. Biol. Chem. 2001;276(17):13935–13940. doi: 10.1074/jbc.M010677200. [DOI] [PubMed] [Google Scholar]
- Yu S.Y., Hu Y.W., Liu X.Y., Xiong W., Zhou Z.T., Yuan Z.H. Gene expression profiles in peripheral blood mononuclear cells of SARS patients. World J. Gastroenterol. 2005;11(32):5037–5043. doi: 10.3748/wjg.v11.i32.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Shan Y., Zhao Z., Chen J., Cong Y. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol. J. 2005;2:66. doi: 10.1186/1743-422X-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Wu J., Shan Y., Yao Z., Dong B., Chen B., Zhao Z., Wang S., Chen J., Cong Y. SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology. 2006;346(1):74–85. doi: 10.1016/j.virol.2005.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F., Hon C.C., Yip C.W., Law K.M., Yeung Y.S., Chan K.H., Malik Peiris J.S., Leung F.C. Quantitative comparison of the efficiency of antibodies against S1 and S2 subunit of SARS coronavirus spike protein in virus neutralization and blocking of receptor binding: implications for the functional roles of S2 subunit. FEBS Lett. 2006;580(24):5612–5620. doi: 10.1016/j.febslet.2006.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wang G., Li J., Nie Y., Shi X., Lian G., Wang W., Yin X., Zhao Y., Qu X., Ding M., Deng H. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J. Virol. 2004;78(13):6938–6945. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Kim M., Choi Y.H., Goemans B., Yeung C., Hu Z., Zhan S., Seth P., Helman L.J. Diminished G1 checkpoint after gamma-irradiation and altered cell cycle regulation by insulin-like growth factor II overexpression. J. Biol. Chem. 1999;274(19):13118–13126. doi: 10.1074/jbc.274.19.13118. [DOI] [PubMed] [Google Scholar]
- Zhao G., Shi S.Q., Yang Y., Peng J.P. M and N proteins of SARS coronavirus induce apoptosis in HPF cells. Cell Biol. Toxicol. 2006;22(5):313–322. doi: 10.1007/s10565-006-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N.S., Zheng B.J., Li Y.M., Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362(9393):1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]