Abstract
Hepatitis C virus (HCV) causes chronic hepatitis, which often results in the development of liver cirrhosis and hepatocellular carcinoma (HCC) worldwide. In this study, we demonstrated that the non-structural protein NS3 of HCV enhances cyclooxygenase-2 (COX-2) gene promoter activity, COX-2 mRNA expression, COX-2 protein production, and prostaglandin E2 (PGE2) release in HepG2 cells in a concentration-dependent fashion. We also showed that transcription factor NF-κB is required for the activation of COX-2 regulated by NS3. In addition, multiple signaling pathways are involved cooperatively in the expression of COX-2 activated by the viral protein in a calcium-independent manner, which requires signaling components including JNK, ERK, and PKD2. A thorough investigation of mechanism involved in the activation of COX-2 regulated by HCV would provide insights into our understanding the processes of liver inflammatory response and hepatocellular carcinoma development caused by the viral infection and also into the development of novel therapeutics against HCV infection.
Keywords: Hepatitis C virus, NS3 protein, Cyclooxygenase-2, Gene regulation, Virus infection, Pathogenesis, Inflammation, PKC, JNK, ERK, Signaling pathway
Introduction
Hepatitis C virus (HCV) is a major pathogen that causes chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC) with 170 million people infected worldwide (Saito et al., 1990, Di Bishceglie, 1997). However, the exact mechanisms of HCV-associated pathogenesis and carcinogenesis are largely unknown.
HCV is a member of the flaviviridae family with a single-stranded and positive-sense RNA genome of approximately 9.6 kb in length (Choo et al., 1989, Rice et al., 1996). The viral RNA genome contains a single open reading frame flanked by un-translated regions (UTR) at both 5′ and 3′ ends. A large viral polyprotein precursor of about 3010 amino acids is translated and sequentially cleaved by cellular and viral proteases into at least 10 different proteins, including structural proteins (C, E1, E2, and p7) and non-structural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (Reed and Rice, 2000, Luo and Colacino, 2004). Replication of HCV RNA occurs in the cytoplasmic membrane-bound multiple-protein complex containing the HCV RNA and non-structural proteins NS3 to NS5B (Di Bishceglie, 1997, Luo and Colacino, 2004). Based on genome sequence similarity, HCV is grouped into six major genotypes and numerous subtypes (Tanaka et al., 1995).
NS3 is one of the viral serine proteases that is essential for virus replication and would be a good target for antiviral drugs. The serine protease is located in the N-terminal portion of NS3, which is responsible for cleavage at the NS3/4A, NS4A/4B, NS4B/5A, and NS5A/5B junctions. NS4A, a cofactor for NS3, stabilizes it to augment its serine protease activity and is essential for complete cleavage of the HCV polyprotein (Reed and Rice, 2000). The C-terminal portion of NS3 possesses the NTPase/helicase activity, which is essential for viral RNA replication (Kim et al., 1995).
In addition to its key role in the life cycle of HCV, NS3 is also involved in viral persistence and hepatocarcinogenesis. NS3 could transform NIH3T3 cells (Sakamuro et al., 1995) and rat fibroblasts (Zemel et al., 2001). NIH3T3 cells constitutively expressing truncated NS3 (aa 1–433) were more resistant to Actinomycin D-induced apoptosis (Fujita et al., 1996). NS3 could also block transforming growth factor-α/Smad3-mediated apoptosis (Cheng et al., 2004). In addition, NS3–NS4A complex suppresses beta interferon (IFN-β) induction by inhibiting retinoic acid-inducible gene I-mediated activation of IFN regulatory factor 3 to help establish persistent HCV infection (Foy et al., 2003, Breiman et al., 2005, Foy et al., 2005). Recently it was reported that NS3 could inhibit the function of tumor suppressor p53 in a sequence-dependent manner, which partly interprets the different degree of oncogenic capacity of different HCV isolates (Lin et al., 2006).
Cyclooxygenases (COX), including constitutively expressed COX-1 and inducible COX-2, are the rate-limiting enzymes for production of prostanoids (prostaglandins and thromboxanes) from arachidonic acid. COX-2 has been categorized as a cellular factor associated with inflammation (McAdam et al., 2000), cellular growth (Williams et al., 2000), prevention of apoptosis (McGinty et al., 2000), and tumorigenesis (Ambs et al., 1998). It has also reported that COX-2 induces angiogenesis and is essential for tumor growth (Tsujii et al., 1998). Increased prostaglandin production and enhanced release of angiogenic growth factor by COX-2 may induce neovascularization (Ambs et al., 1998, Williams et al., 2000).
A number of investigations have demonstrated that viral proteins can stimulate COX-2 expression. For examples, latent membrane protein 1 of Epstein–Barr virus (EBV) (Murono et al., 2001), X protein of hepatitis B virus (HBV) (Lara-Pezzi et al., 2002), gp120 and Tat proteins of human immunodeficiency virus (HIV) (Bagetta et al., 1998), and core and NS5A proteins of HCV (Nunez et al., 2004) have been reported to be able to activate COX-2 expression. We have previously demonstrated that nucleocapsid protein (Yan et al., 2006) and spike protein (Liu et al., 2006, Liu et al., 2007) of SARS-associated coronavirus (SARS-CoV) were able to activate COX-2 expression. However, the effects of other proteins of HCV on the regulation of COX-2 protein expression and inflammation are still unclear.
In this study, we investigated the roles of NS3 protein of HCV in the activation of COX-2 expression and the mechanisms involved in such regulation. The effects of NS3 on COX-2 gene promoter activity, COX-2 mRNA expression, COX-2 protein production, and PGE2 release are determined. The roles of transcription factor NF-κB and multiple signaling pathways in NS3-activated COX-2 expression are also investigated and discussed.
Results
NS3 of HCV activates COX-2 expression in HepG2 cells
It has been reported that HCV could induce the expression of cyclooxygenase-2 in both clinic samples and in cell culture system (Waris and Siddiqui, 2005) and that both core and NS5A of HCV are required for the induction of COX-2 expression (Nunez et al., 2004). Here we investigated whether the rest proteins of HCV play roles, if any, in the regulation of COX-2 expression. HepG2 cells were co-transfected with plasmid (pCOX-2-Luc) carrying the reporter fusion gene, in which luciferase gene is under the control of COX-2 promoter and plasmids pCMV-Core, pCMV-E1, pCMV-E2, pCMV-P7, pCMV-NS2, pCMV-NS3, pCMV-NS4A, pCMV-NS4B, pCMV-NS5A, and pCMV-NS5B, carrying each of HCV genes, core, E1, E2, P7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B, respectively. Results from luciferase activity assay showed that NS3 and Core proteins of HCV significantly enhanced the expression of luciferase, while the rest of HCV proteins tested in this study had no significant effect on the fusion gene expression in HepG2 cells (Fig. 1A).
Fig. 1.
Roles of HCV proteins in the activation of COX-2 promoter. (A) HepG2 cells were co-transfected with 0.2 μg reporter plasmid pCOX-2-Luc containing the luciferase gene under the control of COX-2 promoter and 0.4 μg plasmids, pCMV-flag, pCMV-flag-Core, pCMV-flag-E1, pCMV-flag-E2, pCMV-flag-P7, pCMV-flag-NS2, pCMV-flag-NS3, pCMV-flag-NS4A, pCMV-flag-NS4B, pCMV-flag-NS5A, and pCMV-flag-NS5B, carrying individual genes of HCV, respectively. Luciferase activity was measured as described in the Materials and methods section. Results are expressed as the mean ± sd of three independent experiments performed in triplicate and normalized by β-galactosidase assay. (B) Equal amount of pCMV-NS3 and pCMV-NS4A was transfected or co-transfected, respectively, into HepG2 cells with reporter plasmid pCOX-2-Luc. Results are expressed as the mean ± sd of three independent experiments performed in triplicate and normalized by β-galactosidase assay. ⁎, p < 0.05 versus pCMV-flag.
Considering the fact that NS4A protein always functions as a cofactor in NS3 protease function including cellular signaling pathways, we further checked whether NS4A plays a role in NS3-regulated COX-2 expression. Cells were co-transfected with plasmids carrying the NS3 gene and/or the NS4A gene, respectively, along with pCOX-2-Luc. Results from luciferase activity assay showed that the level of COX-2 promoter activity in the presence of NS3 was almost the same as that in the presence of both NS3 and NS4A (Fig. 1B). These results suggested that NS3 alone could activate the expression COX-2 promoter in HepG2 cells without the cooperation of NS4A.
NS3 activates COX-2 expression in a dose-dependent manner
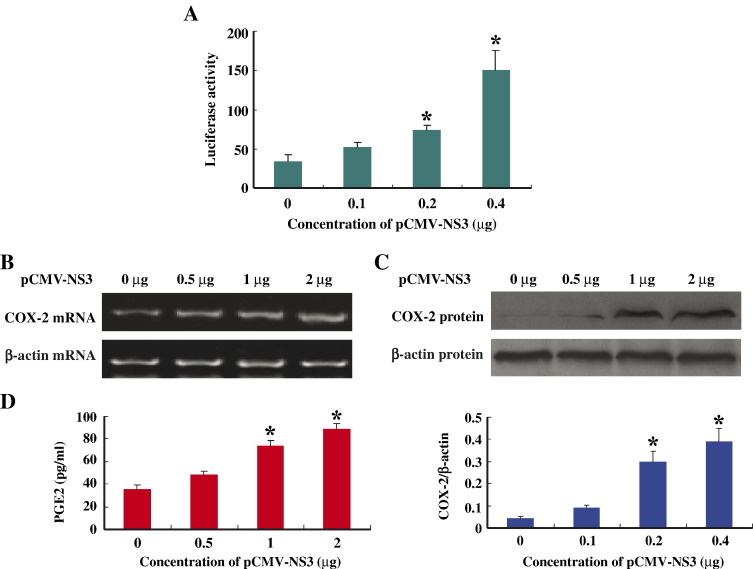
To further confirm the role of NS3 in the activation of COX-2 expression, we determined the effects of different concentrations of NS3 on the activation of COX-2 promoter. HepG2 cells were co-transfected with the report plasmid pCOX-2-luc and different concentrations of plasmid pCMV-NS3. Results from luciferase activity assay showed that the level of COX-2 promoter activity increased as the concentration of NS3 increased, indicting NS3 stimulates COX-2 promoter in a concentration-dependent fashion (Fig. 2A).
Fig. 2.
Effects of NS3 protein on the expression of COX-2 in HepG2 cells. (A) HepG2 cells were co-transfected with pCOX-2-Luc and different concentrations of pCMV-flag-NS3 as indicated. Luciferase activity was measured and results are expressed as the mean ± sd of three independent experiments performed in triplicate and normalized by β-galactosidase assay. ⁎, p < 0.05 versus pCMV-flag control. (B) HepG2 cells were transfected with different concentrations of pCMV-flag-NS3 as indicated. Forty-eight hours post-transfection, total RNA was isolated and used as templates for RT-PCR using COX-2-specific (top panel) or β-actin-specific primers (low panel). (C) Forty-eight hours post-transfection, proteins were prepared from transfected HepG2 cells and used for western blot analysis using antibody to COX-2 protein (top panel) or to β-actin protein (low panel). (D) Levels of prostaglandin E2 (PGE2) produced were measured from transfected HepG2 cells with the Biotrak Prostaglandin E2 Enzyme Immunoassay system. ⁎, p < 0.05 versus pCMV-flag control.
To determine the effects of NS3 on the activation of COX-2 mRNA expression, COX-2 protein production, and prostaglandin E2 (PGE2) generation, HepG2 cells were transfected with different concentrations of pCMV-NS3. Results from RT-PCR using COX-2-specific or β-actin-specific primers showed that the levels of COX-2 mRNA were increased as the concentration of pCMV-NS3 increased, but the levels of β-actin mRNA remained relatively constant (Fig. 2B). Results from western blot analysis using antibody to COX-2 protein or to β-actin protein revealed that the levels of COX-2 protein were increased as pCMV-NS3 concentration increased, while β-actin protein level was remained relatively unchanged (Fig. 2C). The production of PGE2 was assayed with the Biotrak Prostaglandin E2 Enzyme Immunoassay system and results indicated that the level of PGE2 was increased as the concentration of pCMV-NS3 increased (Fig. 2D). Taken together, these results demonstrated that NS3 protein of HCV induces COX-2 expression and PGE2 production in a dose-dependent manner in HepG2 cells.
NF-κB is required for the activation of COX-2 gene regulated by NS3
Activation of COX-2 gene relies on many consensus cis-regulatory elements on its promoter, including two NF-κB binding sites (Jijon et al., 2002) (Fig. 3A). To define the role of NF-κB recognition elements in the activation of COX-2 regulated by NS3, three mutants (Mut1, Mut2, and Mut3) of NF-κB binding site were generated by site specific mutagenesis (Fig. 3A). Reporter plasmids were constructed, in which the luciferase gene was under the control of Mut1, Mut2, and Mut3 promoter, respectively.
Fig. 3.
Effects of NF-κB on the activation of COX-2 regulated by NS3. (A) Diagrams of COX-2 core promoter structures, including wild-type COX-2 promoter (− 891/+ 9) and three NF-κB mutant promoters, Mut1 (− 447/− 438, ΔNF-κB-a), Mut2 (− 222/− 213, ΔNF-κB-b), and Mut3 (− 222/− 213, − 447/− 438, ΔNF-κB-a and -b). (B) HepG2 cells were co-transfected with 0.4 μg pCMV-flag-NS3 and 0.2 μg control plasmid or reporter plasmids, pCOX-2-Luc, pCOX-2-Mut1-Luc, pCOX-2-Mut2-Luc, and pCOX-2-Mut3-Luc, respectively. The inducted level of luciferase activity was compared with that of vector control. Data are expressed as mean ± sd of three independent experiments. ⁎, p < 0.05 versus control. †, p < 0.05 versus wild type group. (C) HepG2 cells were transfected with a reporter vector, in which the luciferase reporter gene is under the control of a NF-κB-binding sequence and plasmid pCMV-flag-NS3 at different concentrations as indicated. Luciferase activity was measured and data represent the mean ± sd of three independent experiments. ⁎, p < 0.05 versus control. (D) HepG2 cells were transfected with pCMV-flag or pCMV-flag-NS3 and then treated with or without 10 μM NF-κB inhibitor (MG132) for 24 h. Forty-eight hours post-transfection, total RNA was isolated and used as templates for RT-PCR using COX-2-specific (top panel) or β-actin-specific primers (low panel). (E) HepG2 cells were transfected with pCMV-flag or pCMV-flag-NS3 and then treated with or without 10 μM MG132 for 24 h. Forty-eight hours post-transfection, proteins were prepared and used for western blot analysis using antibody to COX-2 (top panel) or to β-actin (low panel). Blots were also normalized by measuring the amount of β-actin (Histogram). Bars are mean of optical density ratio of each group, ⁎, p < 0.05 versus vector control. †, p < 0.05 versus pCMV-NS3.
Plasmids carrying these mutant COX-2 promoters were then co-transfected with pCMV-NS3 into HepG2 cells, respectively. Results from luciferase assay showed that the two single mutations (Mut1 and Mut2) reduced the level of COX-2 promoter activation regulated by NS3 and the double mutation (Mut3) eliminated NS3-regulated COX-2 activation (Fig. 3B). These results suggested that NF-κB was involved in the regulation of COX-2 gene expression. In subsequent experiment, cells were transiently transfected with reporter plasmid, in which the expression of luciferase gene is under the control of NF-κB-binding element. Results showed that NS3 stimulated the NF-κB-binding element activity in a dose-dependent manner (Fig. 3C). This result demonstrated that NF-κB was sufficient for the activation of COX-2 expression regulated by NS3 protein.
NF-κB is a ubiquitously expressed transcription factor that regulates the induction of genes involved in cellular immune response and inflammatory function. To further confirm the involvement of NF-κB in the activation of COX-2 regulated by NS3, HepG2 cells were transfected with pCMV-NS3 and treated with or without NF-κB inhibitor, MG132. Results from RT-PCR analysis showed that MG132 significantly decreased the level of COX-2 mRNA activated by NS3 (Fig. 3D). Similar results were obtained when western blot analysis was carried out, which indicated that MG132 significantly reduced the level of COX-2 protein activated by NS3 (Fig. 3E). These results demonstrated that transcription factor NF-κB was required for the activation of COX-2 expression regulated by NS3 protein.
Multiple signal transduction pathways are involved in the activation of COX-2 expression regulated by NS3
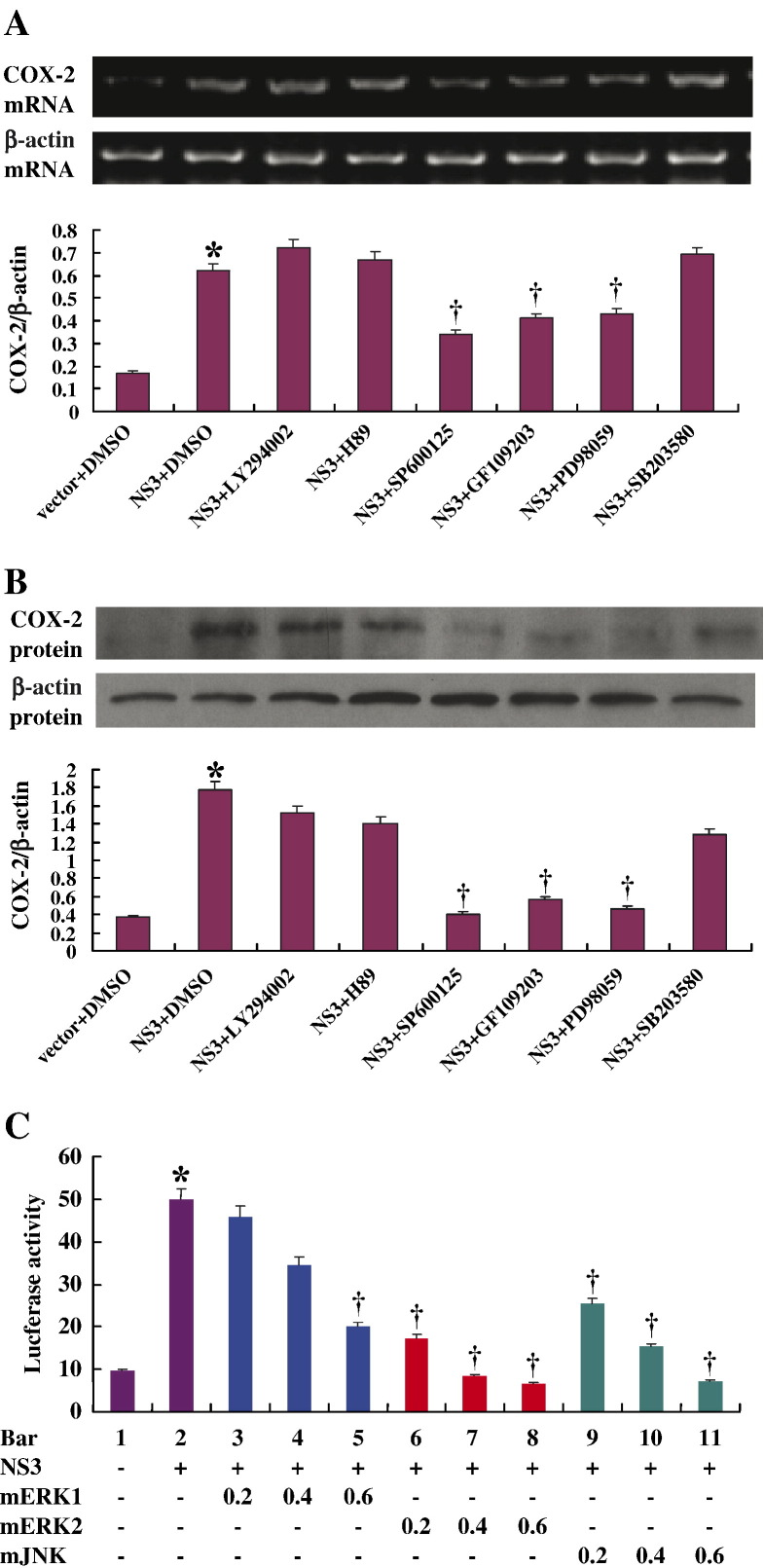
Given the critical roles of COX-2 in liver inflammation and carcinogenesis, it is important to understand the key signaling pathways involved in the regulation of COX-2 expression in human liver cell line. To delineate the signaling cascades involved, HepG2 cells were transfected with pCMV-NS3 and then treated with inhibitors of individual components of signaling pathways.
Results from RT-PCR analyses showed that the level of COX-2 mRNA activated by NS3 was suppressed by SP600125 (c-Jun inhibitor), GF109203 (PKC inhibitor), and PD98059 (MEK1/2 inhibitor), respectively, but not by LY294002 (PI3K inhibitor), H89 (protein kinase A inhibitor), or SB203580 (SAPK2α/β/p38α/β MAPK inhibitor) (Fig. 4A). Similar results were observed when western blot analysis was performed, which showed that the level of COX-2 protein expression regulated by NS3 was inhibited by SP600125, GF109203, and PD98059, respectively, but not by LY294002, H89, or SB203580 (Fig. 4B).
Fig. 4.
Involvement of signaling pathways in the activation of COX-2 expression regulated by NS3. (A) HepG2 cells were transfected with pCMV-flag-NS3 (2 μg/well of 6-well plate) and then treated with inhibitors of individual component of signaling pathways, 50 μM GF109203 (PKC inhibitor), 30 μM SP600125 (c-Jun N-terminal kinase inhibitor), 50 μM PD98059 (MEK1/2 inhibitor), 25 μM LY294002 (PI3K inhibitor), 10 μM H89 (protein kinase A inhibitor), and 10 μM SB203580 (SAPK2α/β/p38α/βMAPK inhibitor), respectively. Total RNA was isolated from treated cells and used as template for RT-PCR analyses using primers for COX-2 (top panel) or for β-actin (low panel). Blots were also normalized by measuring the amount of β-actin (Histogram). Bars are mean of optical density ratio of each group. ⁎, p < 0.05 versus vector control. †, p < 0.05 versus pCMV-NS3. (B) HepG2 cells were transfected with pCMV-flag-NS3 and treated with inhibitors, respectively. Proteins were prepared from transfected cells and used for western blot analyses using antibody to COX-2 (top panel) or to β-actin (low panel). Blots were also normalized by measuring the amount of β-actin (histogram). Bars are mean of optical density ratio of each group. ⁎, p < 0.05 versus vector control. †, p < 0.05 versus pCMV-NS3. (C) HepG2 cells were co-transfected with pCMV-flag-NS3 (0.4 μg/well of 24-well plate), pCOX-2-Luc (0.2 μg/well of 24-well plate), and dominant negative mutants of ERK1/2, JNK at different concentrations as indicated, or control vector. Luciferase activity was measured and data are expressed as mean ± sd of three independent experiments. ⁎, p < 0.05 versus vector control. †, p < 0.05 versus pCMV-NS3.
The roles of ERK and JNK in the activation of COX-2 mediated by NS3 protein were further determined by introducing three dominant kinase-inactive mutants, mERK1, mERK2, and mJNK, which can block corresponding kinase activities by competing with endogenous kinases (Werlen et al., 1998, Khoo et al., 2003). Cells were co-transfected with pCMV-NS3, pCOX-2-Luc reporter, and each of these three kinase mutants, respectively. Results from luciferase activity assays showed that all three mutants reduced COX-2 promoter activity in a dose-dependent manner, especially mERK2 and mJNK (Fig. 4C). Taken together, these data suggested that ERK, JNK, and PKC were involved in NS3-regulated COX-2 activation.
NS3 protein of HCV activates JNK and ERK signaling pathways
To evaluate the effects of NS3 protein of HCV on the activation of JNK and ERK signaling cascades, we used antibodies specific to the phosphorylated forms of the two mitogen-activated protein kinases ERK1/2 and JNK. Results from western blot and densitometry analyses showed that the levels of phosphorylation of both JNK (Fig. 5A) and ERK1/2 (Fig. 5B) were significantly increased when NS3 protein was present. These results suggested that NS3 plays a role in the activation of JNK and ERK signaling pathways through phosphorylation of these two protein kinases.
Fig. 5.

Roles of JNK and ERK signaling pathways in NS3-mediated COX-2 expression. HepG2 cells were transfected with 2 μg of pCMV-flag-NS3 or control plasmid. Proteins were prepared from transfected cells and the levels of phospho-JNK and phospho-ERK1/2 were determined by western blot with phospho-specific antibodies (top panels). Protein expression levels of non-phospho-JNK and non-phospho-ERK in cell lysates were also detected as controls (low panels). (A) Western blot results for phospho-JNK (top panel) and JNK (low panel) and blots normalized by measuring the amount of phospho-JNK/JNK (Histogram). Bars are mean of optical density ratio of each group. (B) Western blot results for phospho-ERK (top panel) and ERK (low panel) and blots normalized by measuring the amount of phospho-ERK/ERK (Histogram). Bars are mean of optical density ratio of each group. ⁎, p < 0.05 compared with control.
NS3-activated COX-2 expression is calcium-independent and involves PKD2 kinase
It is known that PKCs comprise a family of intracellular serine/threonine-specific kinases. Depending on the type of isoforms, activation of PKCs is typically initiated by Ca2+, lipid second messengers, and/or protein activators. To elucidate whether Ca2+ is involved in the expression of COX-2 regulated by NS3, HepG2 cells were co-transfected with pCMV-NS3 and pCOX-2-Luc and then treated with BAPAT/AM or EGTA at different concentrations. Results from luciferase assays showed that BAPAT/AM (Fig. 6A) and EGTA (Fig. 6B) had no significant effect on the activity of COX-2 promoter regulated by NS3, suggesting that Ca2+ was not involved in the induction of COX-2 regulated by NS3.
Fig. 6.
Effects of PKC signaling pathway on NS3-mediated COX-2 expression. HepG2 cells were co-transfected with 0.4 μg pCMV-flag-NS3 and 0.2 μg pCOX-2-Luc. Forty hours post-transfection, cells were treated with BAPAT/AM (A) or EGTA (B) at different concentrations as indicated for 8 h. Luciferase activity was measured and results are expressed as the mean ± sd of three independent experiments performed in triplicate and normalized by β-galactosidase assay. (C) HepG2 cells were co-transfected with 0.4 μg pCMV-flag-NS3 or 2 μg pCMV-flag and 1 μg plasmids expressing siRNA specific to PKCδ, PKCζ, PKCα, PKCβ1, PKD1, PKD2, and siCtrl, respectively. Proteins were prepared and used in western blot for detection of COX-2 (top panel) and β-actin protein (low panel). Western blots were normalized by measuring the amount of COX-2/β-actin and bars are mean of optical density ratio of each group (histogram). ⁎, p < 0.05 versus vector control. †, p < 0.05 versus pCMV-NS3. (D) Expression of PKD2 mRNA in the presence or absence of siRNA was determined by RT-PCR for PKD2 (top panel) or for β-actin (low panel). (E) Expression of PKCα mRNA in the presence or absence of siRNA was determined by RT-PCR for PKCα (top panel) or for β-actin (low panel). (F) Expression of PKCζ mRNA in the presence or absence of siRNA was determined by RT-PCR for PKCζ (top panel) or for β-actin (low panel). For better view, the results were shown in histograms and bars are mean of optical density ratio of each (lower panels). ⁎, p < 0.05 versus siCtrl.
We then extended our study to identify the specific isozymes of PKC involved in NS3-regulated activation of COX-2 using RNA interference (RNAi) approach. Small interfering RNA (siRNA) molecules that specifically knock down genes encoding for each PKC isozymes were used to study the function of PKC based on previous studies (Storz et al., 2004, Liu et al., 2007). HepG2 cells were transfected with pCMV-NS3 and then treated with siRNAs, including siPKCδ, siPKCζ, siPKCα, siPKCβ1, siPKD1, and siPKD2, respectively. Results from western blot analysis using antibody to COX-2 protein showed that siPKD2, but not other siRNAs tested, had a significant effect on the activation of COX-2 protein expression regulated by NS3 (Fig. 6C). These results suggested that PKD2 signaling pathway was involved in the activation of COX-2 regulated by NS3 protein.
Expression of PKD2 (Fig. 6D), PKCα (Fig. 6E), and PKCζ (Fig. 6F) mRNA was also determined before and after knock down by specific siRNA using RT-PCR analyses. Results showed that expression of different PKC isoforms was decreased significantly in the presence of PKC isoform-specific siRNAs (siPKCα, siPKCζ, and siPKD2), but siCtrl (a control siRNA containing an irrelevant sequence to PKCs and no homology to any human gene sequence) had no significant effect on the expression of PKCs (Figs. 6D, E, and E). These results indicated that various PKC isoform-specific siRNAs worked efficiently in the assays and that among PKCs, only PKD2 was involved in the activation of COX-2 regulated by NS3 protein.
Discussion
COX-2 was reported to be highly expressed in liver inflammation and carcinogenesis, thus representing a potential target for drug intervention during these processes (Rahman et al., 2001, Nunez et al., 2004, Aggarwal et al., 2006). Recent study provided evidence that COX-2 is over-expressed in the liver tissues of patients with chronic HCV infection, implying a positive correlation with the fibrotic stage of liver disease. Although the molecular basis for hepatocellular COX-2 expression in patients with chronic HCV infection remains to be defined, there is evidence that core and NS5A proteins of HCV can function as transcriptional transactivators for many cellular genes (Kato et al., 2000, Reyes, 2002, Nunez et al., 2004).
In the process of investigating the roles of additional proteins of HCV in COX-2 expression during HCV infection, we found that NS3 protein could induce COX-2 promoter activity, COX-2 mRNA expression, COX-2 protein production, and PGE2 generation in human liver cell line HepG2 in a concentration-dependent fashion. These results suggested that NS3 protein of HCV can also function as transcriptional transactivator for cellular genes.
NF-κB has been reported to mediate COX-2 activation induced by diverse stimulating agents. We demonstrated in this study that NF-κB is also essential for NS3 protein-induced COX-2 expression since mutations of binding sites for NF-κB and the inhibitor of NF-κB could eliminate NS3 function in the activation of COX-2.
Based on the findings that NS3 protein activates COX-2 expression and NF-κB is involved in such regulation, we further investigated the roles of different MAP kinases on the activation of COX-2 regulated by NS3. Our results demonstrated that ERK, JNK, and PKC were involved in the activation of COX-2 regulated by NS3 since inhibition of each of those kinases abolished the expression of COX-2 activated by NS3. We also observed that inhibitors of PI3K, protein kinase A, and SAPK2α/β/p38α/βMAPK had no effect on NS3-activated COX-2 expression. Using RNA interference approach, we further found that the induction of COX-2 by NS3 required the atypical isoform PKD2 since PKD2 specific siRNA inhibited COX-2 protein expression activated by NS3 and siRNA specific to other PKC isoforms had no effect on NS3-induced COX-2 expression.
Many viral proteins can induce multiple signaling pathways, which may crosstalk with each other or converge on common downstream effectors (Chen et al., 2003). In embryonic kidney cells, the latent membrane protein-1 of EBV induced COX-2 expression through MAPK, JNK/AP-1, and JAK/STAT signaling pathways and required the transcription factor NF-κB (Saunders et al., 2001).
Our results in this study suggested that the NS3 of HCV regulates multiple signaling pathways, such as ERK, JNK, and PKD2, to control the activity of transcription factor NF-κB, which in turn to activate the expression of COX-2 that may induce host inflammatory response and, perhaps, carcinogenesis. A thorough understanding of COX-2 gene expression regulated by the NS3 protein of HCV would provide insights into the development of novel therapeutics and into our understanding of liver inflammation and cancer development caused by HCV infection.
Materials and methods
Plasmids, antibodies, and reagents
SB203580, PD98059, LY294002, SP600125, GF109203, H89, EGTA, MG132, DTT, fura-2/acetoxymethyl ester (AM), and pluronic-F127 were purchased from Sigma Chemical Co (St. Louis, MO, USA). All protein kinase inhibitors were dissolved in DMSO and used at a final concentration of 50 μM for PD98059, 10 μM for SB203580, 50 μM for GF109203, 30 μM for SP600125, 25 μM for LY294002, 10 μM for H89, and 10 μM for MG132. Antibody against COX-2 was from Cayman Chemical Company (Ann Arbor, MI, USA). Antibodies specific for ERK, phospho-ERK, JNK, and phospho-JNK were purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA, USA).
Luciferase reporter vector (pGL3) containing a COX-2 promoter region (− 891/+ 9), its truncation mutants, and site specific mutants were constructed previously (Zhu et al., 2002). A NS3 construct was created by RT-polymerase chain reaction (RT-PCR) amplification of the open reading frame from HCV 2b. To create NS3-encoding vectors, the NS3 gene was amplified using the following primers: 5′-AAAGGATCCATGGCGCCCATCACG-3′ (sense) 5′-CCCGAATTCCTAAGTGACGACCTCTAGGTC-3′ (antisense), in which BamHI and EcoRI sites were introduced, respectively. The polymerase chain reaction (PCR) product was cloned into BamHI and EcoRI sites of pCMV-tag2B to generate plasmid pCMV-NS3, in which NS3 protein was tagged by FLAG. The resulting construct was confirmed by DNA sequencing. Mutants of ERK1 and ERK2 were gifts from Dr. Cobb (University of Texas Southwestern Medical Center) and a mutant of JNK was a gift from Dr. Karin (University of California at San Diego, San Diego, CA, USA). PKC siRNA vectors were constructed by ligating the corresponding pairs of oligonucleotide to pSilencer 2.0 (Ambion, Inc., Augustin, TX, USA) based on the sequences described previously (Storz et al., 2004).
Cell culture
The human hepatoma cell line HepG2 was grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate at 37 °C in a 5% CO2 incubator.
Transient transfection and luciferase reporter gene assays
Cells were plated at density of 4.0 × 105 cells per 24-well plate or 6-well plate and grown to the confluence reaching about 80% at the time of transfection. Cells were co-transfected with 0.4 μg of plasmid pCMV-NS3 together with 0.2 μg plasmid containing luciferase reporter and 0.2 μg of the pCMV-β-galactosidase plasmid, using Sofast™ transfection reagent (Xiamen Sunma Biotechnology Co. Ltd., China) according to the protocol provided by the manufacturer. In all co-transfection experiments, appropriate vector DNA was used to ensure similar DNA concentrations in all conditions. Twenty-four hours post-transfection, cells were serum-starved for another 24 h before being harvested. Harvested cells were lysed with luciferase assay cell culture lysis reagent (Promega, Madison, WI, USA). Cell lysates and luciferase assay substrate (Promega) were mixed and light intensity was detected by the luminometer (Turner T20/20). Assays were performed in triplicate and expressed as means ± sd. All transfections included a β-galactosidase expression vector to serve as an internal control.
Semiquantitative RT-PCR analysis
Total RNA was isolated using a TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse-transcribed with oligo(dT) primer. PCR was performed in 25 μl reactions with the primer pairs (20 μM) described in Table 1 . β-Actin specific primers were run in all reactions as an internal control. The PCR products were amplified for 28 cycles. The selected cycle number was chosen to stop the PCR reaction during its log phase to ensure availability of all reagents. Additional RT-PCR related information is summarized in Table 1. PCR products were analyzed by electrophoresis on 1% agarose gel containing ethidium bromide.
Table 1.
Summary of sequences of primers used for RT-PCR in this study
| Gene | Product size (bp) | Primer sequence (5′ to 3′) | Tm (°C) | Accession no. |
|---|---|---|---|---|
| F-COX-2 | 667 | TACAATGCTGACTATGGCTAC | 55 | NM_000963 |
| R-COX-2 | ACTGATGCGTGAAGTGCTG | |||
| F-β-actin | 561 | ATGATATCGCCGCGCTCG | 55 | NM_001101 |
| R-β-actin | CGCTCGGTGAGGATCTTCA | |||
| F-PKD2 | 723 | CACTGCCAATGCCACCTA | 56.7 | NM_016457 |
| R-PKD2 | ACTGAGCGGCGGAACGA | |||
| F-PKCα | 284 | GCCTATGGCGTCCTGT | 52.7 | NM_002737 |
| R-PKCα | TTGGGCTTGAATGGTG | |||
| F-PKCζ | 573 | CTCCTGGTGCGGTTGAA | 56 | NM_002744 |
| R-PKCζ | GTTGTCGGTGATGATGTCG |
Western blot analysis
Whole-cell lysates were prepared by lysing HepG2 with radioimmune precipitation buffer (50 mM Tris, pH7.5, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM sodium formate, 1 mM phenylmethylsulfonyl fluoride, and 10% cocktail protease inhibitor [Roche]). Cells were washed with ice-cold PBS, collected by centrifugation, and pellets were resuspended in RIPA buffer described above for 30 min on ice and vortexed for 10 s. Lysates were centrifuged at 12,000 rpm for 10 min. The supernatants were boiled for 5 min in SDS–polyacrylamide gel electrophoresis (PAGE) sample buffer. One hundred micrograms of cultured cell lysates was subjected to 12% SDS–PAGE gel. Gels were electroblotted onto a nitrocellulose membrane in 25 mM Tris, 192 mM glycine, and 20% methanol by electrophoresis. Nonspecific IgGs were blocked with 5% nonfat dried milk before being probed with antibodies used in this study. After an additional washing step with blocking buffer, immunoblots were visualized with the ECL detection system (Pierce, Rockford, IL, USA).
Intracellular PGE2 measurements
Subconfluent HepG2 cells were transiently transfected various amounts of pCMV-NS3. Forty-eight hours post-transfection, culture cells were washed thoroughly with cold phosphate-buffered saline, pH7.4, collected by centrifugation, and pellets were resuspended in PBS pH7.4 containing 0.01% Triton X-100, 0.01% EDTA, and 10% cocktail protease inhibitor (Roche). Cell membranes were broken to release intracellular PGE2. PGE2 levels were then assayed with the Biotrak Prostaglandin E2 Enzyme Immunoassay system (R&D Systems) according to the manufacturer's protocol.
Statistical analysis
All experiments were reproducible and were carried out in duplicate or quadruplicate. Each set of experiments was repeated at least three times with similar results, and a representative one is shown. The results are presented at the means ± sd. Student's t test for paired samples was used to determine statistical significance. Differences were considered statistically significant at a value of P ≤ 0.05. In all cases where comparative data are presented, the autoradiographic images originated from the same exposure of the same gel: in some cases, for clarity of presentation, lanes containing samples not germane to this study were removed.
Acknowledgments
This work was supported by research grants from the Major State Basic Research Development Program of China (973 program No. 2005CB522901), the National Natural Science Foundation of China (No. 30470087 and No. 30570070), and the Department of Science Technology of Hubei Province (No. 2005ABC003) to J. Wu and the National Basic Research Program of China (973 program No. 2007CB512803) to Y. Zhu.
Many thanks to Dr. Cobb from University of Texas Southwestern Medical Center for kindly providing the mutants of ERK1 and ERK2 and to Dr. Karin from University of California at San Diego for the gift of mutant of JNK.
References
- Aggarwal B.B., Shishodia S., Sandur S.K., Pandey M.K., Sethi G. Inflammation and cancer: how hot is the link? Biochem. Pharmacol. 2006;30:1605–1621. doi: 10.1016/j.bcp.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Ambs S., Merriam W.G., Bennett W.P., Felley-Bosco E., Ogunfusika M.O., Oser S.M., Klein S., Shields P.G., Billiar T.R., Harris C.C. Frequent nitric oxide synthase-2 expression in human colon adenomas: implication for tumor angiogenesis and colon cancer progression. Cancer Res. 1998;58:334–341. [PubMed] [Google Scholar]
- Bagetta G., Corasaniti M.T., Paoletti A.M., Berliocchi L., Nistico R., Giammarioli A.M., Malorni W., Finazzi-Agro A. HIV-1 gp120-induced apoptosis in the rat neocortex involves enhanced expression of cyclooxygenase type 2 (COX-2) Biochem. Biophys. Res. Commun. 1998;244:819–824. doi: 10.1006/bbrc.1998.8321. [DOI] [PubMed] [Google Scholar]
- Breiman A., Grandvaux N., Lin R., Ottone C., Akira S., Yoneyama M., Fujita T., Hiscott J., Meurs E.F. Inhibition of RIG-independent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKe. J. Virol. 2005;79:3969–3978. doi: 10.1128/JVI.79.7.3969-3978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Noueiry A., Ahlquist P. An alternate pathway for recruiting template RNA to the brome mosaic virus RNA replication complex. J. Virol. 2003;77:2568–2577. doi: 10.1128/JVI.77.4.2568-2577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P.L., Chang M.H., Chao C.H., Lee Y.H. Hepatitis C viral proteins interact with Smad3 and differentially regulate TGF-β/Smad3-mediated transcriptional activation. Oncogene. 2004;23:7821–7838. doi: 10.1038/sj.onc.1208066. [DOI] [PubMed] [Google Scholar]
- Choo Q.L., Kuo G., Weiner A.J., Overby L.R., Bradley D.W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Di Bishceglie A.M. Hepatitis C and hepatocellular carcinoma. Hepatology. 1997;26:34S–38S. doi: 10.1002/hep.510260706. [DOI] [PubMed] [Google Scholar]
- Foy E., Li K., Wang C., Sumpter R., Jr., Ikeda M., Lemon S.M., Gale M., Jr. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- Foy E., Li K., Sumpter R., Jr., Loo YM., Johnson C.L., Wang C., Fish P.M., Yoneyama M., Fujita T., Lemon S.M., Gale M., Jr. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Ishido S., Muramatsu S., Itoh M., Hotta H. Suppression of actinomycin D-induced apoptosis by the NS3 protein of hepatitis C virus. Biochem. Biophys. Res. Commun. 1996;229:825–831. doi: 10.1006/bbrc.1996.1887. [DOI] [PubMed] [Google Scholar]
- Jijon H.B., Panenka W.J., Madsen K.L., Parsons H.G. MAP kinases contribute to IL-8 secretion by intestinal epithelial cells via a posttranscriptional mechanism. Am. J. Physiol. 2002;283:C31–C34. doi: 10.1152/ajpcell.00113.2001. [DOI] [PubMed] [Google Scholar]
- Kato N., Yoshida H., Ono-Nita S., Kato J., Goto T., Otsuka M., Lan K., Matsushima K., Shiratori Y., Ornata M. Activation of intracellular signaling by hepatitis B and C viruses: C-viral core is the most potent signal inducer. Hepatology. 2000;32:405–412. doi: 10.1053/jhep.2000.9198. [DOI] [PubMed] [Google Scholar]
- Khoo S., Griffen S.C., Xia Y., Baer R.J., German M.S., Cobb M.H. Regulation of insulin gene transcription by ERK1 and ERK2 in pancreatic beta cells. J. Biol. Chem. 2003;278:32969–32977. doi: 10.1074/jbc.M301198200. [DOI] [PubMed] [Google Scholar]
- Kim D.W., Gwack Y., Han J.H., Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- Lara-Pezzi E., Gómez-Gaviro E.V., Gálvez B.G., Mira E., Iñiguez M.A., Martínez-A M.G., Arroyo A.G., López-Cabrera M. The hepatitis B virus X protein promotes tumor cell invasion by inducing membrane-type matrix metalloproteinase-1 and cyclooxygenase-2 expression. J. Clin. Invest. 2002;110:1831–1838. doi: 10.1172/JCI200215887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D., Motoko N.J., Motofumi T., Yuki N.T., Masanori I., Nobuyuki K., Kiyonao S., Hak H. NS3 protein of Hepatitis C virus associates with the tumor suppressor p53 and inhibits its function in an NS3 sequence-dependent manner. J. Gen. Viol. 2006;87:1703–1713. doi: 10.1099/vir.0.81735-0. [DOI] [PubMed] [Google Scholar]
- Liu M., Gu C., Wu J., Zhu Y. N-terminal of the spike protein of SARS-associated virus is essential for the induction of COX-2 expression in mammalian cells. Virus Genes. 2006;33:309–317. doi: 10.1007/s11262-005-0070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Yang Y., Gu C., Yue Y., Wu K.K., Wu J., Zhu Y. Spike protein of SARS-CoV stimulates cyclooxygenase-2 expression via both calcium-dependent and calcium-independent protein kinase C pathways. FASEB J. 2007;21:1586–1596. doi: 10.1096/fj.06-6589com. [DOI] [PubMed] [Google Scholar]
- Luo G., Colacino J.M. Molecular virology of hepatitis C virus. Hepat. Prev. Treat. 2004:67–85. [Google Scholar]
- McAdam B.F., Mardini I.A., Habib A., Burke A., Lawson J.A., Kapoor S., FitzGerald G.A. Effect of regulated expression of human cyclooxygenase isoforms on eicosanoid and isoeicosanoid production in inflammation. J. Clin. Invest. 2000;105:1473–1482. doi: 10.1172/JCI9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty A., Chang Y.W., Sorokin A., Bokemeyer D., Dunn M.J. Cyclooxygenase-2 expression inhibits trophic withdrawal apoptosis in nerve growth factor-differentiated PC12 cells. J. Biol. Chem. 2000;275:12095–12101. doi: 10.1074/jbc.275.16.12095. [DOI] [PubMed] [Google Scholar]
- Murono S., Inoue H., Tanabe T., Joab I., Yoshizaki T., Furukawa H., Pagano J.S. Induction of cyclooxygeanse-2 by Epstein–Barr virus latent membrane protein 1 is involved in vascular endothelial growth factor production in nasopharyngeal carcinoma cells. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6905–6910. doi: 10.1073/pnas.121016998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez O., Fernandez-Martinez A., Majano P.L., Apolinario A., Gomez-Gonzalo M., Benedicto I., Lopez-Cabrera M., Bosca L., Clemente G., Garcia-Monzon C., Martin-Sanz P. Increased intrahepatic cyclooxygenase 2, matrix metalloproteinase 2, and matrix metalloproteinase 9 expression is associated with progressive liver disease in chronic hepatitis C virus infection: role of viral core and NS5A proteins. Gut. 2004;53:1665–1672. doi: 10.1136/gut.2003.038364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M.A., Dhar D.K., Yamaguchi E., Maruyama S., Sato T., Hayashi H., Ono T., Yamanoi A., Kohno H., Nagasue N. Coexpression of inducible nitric oxide synthase and COX-2 in hepatocellular carcinoma and surrounding liver: possible involvement of COX-2 in the angiogenesis of hepatitis C virus-positive cases. Clin. Cancer Res. 2001;7:1325–1332. [PubMed] [Google Scholar]
- Reed K.E., Rice C.M. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- Reyes G.R. The nonstructural NS5A protein of hepatitis C virus: an expanding, multifunctional role in enhancing hepatitis C virus pathogenesis. J. Biomed. Sci. 2002;9:187–197. doi: 10.1007/BF02256065. [DOI] [PubMed] [Google Scholar]
- Rice C.M., Fields B.N., Knipe D.M., Howley P.M. Flaviviridae: the viruses and their replication. Fields Virol. 1996;1:931–960. [Google Scholar]
- Saito T., Miyamura T., Ohbayashi A., Harada H., Katayama T., Kikuchi S., Watanabe Y., Koi S., Onji M., Ohta Y., Choo Q., Houghton M., Kuo G. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. U. S. A. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamuro D., Furukawa T., Takegami T. Hepatitis C virus nonstructural protein NS3 transforms NIH3T3 cells. J. Virol. 1995;69:3893–3896. doi: 10.1128/jvi.69.6.3893-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders M.A., Sansores-Garcia L., Gilroy D.W., Wu K.K. Selective suppression of CCAAT enhancer-binding protein β binding and cyclooxygenase-2 promoter activity by sodium salicylate in quiescent human fibroblasts. J. Biol. Chem. 2001;276:18897–18900. doi: 10.1074/jbc.M011147200. [DOI] [PubMed] [Google Scholar]
- Storz P., Doppler H., Toker A. Protein kinase C selectively regulates protein kinase D-dependent activation of NF-κB in oxidative stress signaling. Mol. Cell. Biol. 2004;24:2614–2626. doi: 10.1128/MCB.24.7.2614-2626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kato N., Cho M.J., Shimotohno K. A novel sequence found at the 3′ terminus of the hepatitis C virus genome. Biochem. Biophys. Res. Commun. 1995;215:744–749. doi: 10.1006/bbrc.1995.2526. [DOI] [PubMed] [Google Scholar]
- Tsujii M., Kawano S., Tsuji S., Sawaoka H., Hori M., DuBois R.N. Cyclooxygenase regulates angiogenesis induced by colon cancer cells. Cell. 1998;93:705–716. doi: 10.1016/s0092-8674(00)81433-6. [DOI] [PubMed] [Google Scholar]
- Waris G., Siddiqui A. Hepatitis C virus stimulates the expression of cyclooxygenase-2 via oxidative stress: role of prostaglandin E2 in RNA replication. J. Virol. 2005;79:9725–9734. doi: 10.1128/JVI.79.15.9725-9734.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Werlen G., Jacinto E., Xia Y., Karin M. Calcineurin preferentially synergizes with PKC-theta to activate JNK and IL-2 promoter in T lymphocytes. EMBO J. 1998;17:3101–3111. doi: 10.1093/emboj/17.11.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.S., Tsujii M., Reese J., Dey S.K., DuBois R.N. Host cyclooxygenase-2 modulates carcinoma growth. J. Clin. Invest. 2000;105:1589–1594. doi: 10.1172/JCI9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Hao Q., Mu Y., Timani K.A., Ye L., Zhu Y., Wu J. Nucleocapsid protein of SARS-CoV activates the expression of cyclooxygenase-2 by binding directly to regulatory elements for nuclear factor kappa B and CCAAT/enhancer binding protein. Int. J. Biochem. Cell Biol. 2006;3:1417–1428. doi: 10.1016/j.biocel.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zemel R., Gerechet S., Greif H., Bachmatove L., Birk Y., Golan-Goldhrish A., Kunin M., Berdichevsky Y., Benhar I., Tur-Kaspa R. Cell transformation induced by hepatitis C virus NS3 serine protease. J. Viral. Hepatitis. 2001;8:96–102. doi: 10.1046/j.1365-2893.2001.00283.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Saunders M.A., Yeh H., Deng W., Wu K.K. Dynamic regulation of cyclooxygenase-2 promoter activity by isoforms of CCAAT/enhancer binding proteins. J. Biol. Chem. 2002;277:6923–6928. doi: 10.1074/jbc.M108075200. [DOI] [PubMed] [Google Scholar]