Abstract
New methods to identify trace amount of infectious pathogens rapidly, accurately and with high sensitivity are in constant demand to prevent epidemics and loss of lives. Early detection of these pathogens to prevent, treat and contain the spread of infections is crucial. Therefore, there is a need and urgency for sensitive, specific, accurate, easy-to-use diagnostic tests. Versatile biofunctionalized engineered nanomaterials are proving to be promising in meeting these needs in diagnosing the pathogens in food, blood and clinical samples. The unique optical and magnetic properties of the nanoscale materials have been put to use for the diagnostics. In this review, we focus on the developments of the fluorescent nanoparticles, metallic nanostructures and superparamagnetic nanoparticles for bioimaging and detection of infectious microorganisms. The various nanodiagnostic assays developed to image, detect and capture infectious virus and bacteria in solutions, food or biological samples in vitro and in vivo are presented and their relevance to developing countries is discussed.
Abbreviations: WHO, World Health Organization; ELISA, Enzyme Linked Immuno Sorbent Assay; PCR, Polymerase Chain Reaction; NPs, nanoparticles; Qdots, quantum dots; RSV, Respiratory Syncytial Virus; FITC, Fluorescein Isothiocyanate; Zn-DPA, Zn (II)-dipicolylamine; HBV, hepatitis B virus; HCV, hepatitis C virus; Qdot-B, Qdot-barcodes; HIV, human immunodeficiency virus; FSNPS, fluorescent silica nanoparticles; FRET, Förster resonance energy transfer; FAM-SE, (5-carboxy-fluorescein succinimidyl ester); ROX-SE, (6-carboxy-X-rhodamine, succinimidyl ester); R6G-SE, (5-carboxyrhodamine 6G, succinimidyl ester); TMR-SE, (carboxytetramethylrhodamine, succinimidyl ester); OsBpy, Tris (2, 2′bipyridyl) osmium bis (hexafluorophosphate); RuBpy, Tris(bipyridine) ruthenium (II) dichloride; FNP-IIFM, fluorescent nanoparticle-based indirect immunofluorescence microscopy; Eu III, Europium; CaDPA, calcium dipicolinate; LOD, limit of detection; SEC1, staphylococcal enterotoxin C1; CT, cholera toxin; PA, anthrax protective agent; CCMV, cow pea chlorotic mottle virus; MRI, Magnetic Resonance Imaging; SpA, Protein A; Gd-DOTA, Gadolinium-1,4,7,10-tetraazacyclododecane tetraacetic acid; ICP-MS, inductively coupled plasma mass spectrometry; SPR, surface plasmon resonance; Au NP, gold nanoparticle; HSV-2, Herpes simplex Virus type 2; HSV-1, Herpes simplex Virus type 1; RLS, Resonance Light Scattering; ss, single stranded; HRS, Hyper-Rayleigh scattering; ds, double stranded; TEM, transmission electron microscopy; H. pyroli, Helicobacter pyroli; SERS, surface enhanced Raman scattering; SMCC, Succinimidyl-4-(N-Maleimidomethyl)Cyclohexane-1-Carboxylate; Bg, Bacillus globigii; Ova, Ovalbumin; CFU, colony forming unit; ATP, adenosine triphosphate; IR, Infra Red; SQUID, Superconducting Quantum Interference Device; MNP, magnetic nanoparticles; MALDI-MS, Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry; POA, adopted pigeon ovalbumin; MGNP, magnetic glycol nanoparticles; SPIO, superparamagnetic iron oxide; MRS, magnetic relaxation sensors; NMR, Nuclear Magnetic Resonance
Keywords: Fluorescent nanoparticles, Multiplexing, Viral imaging, Bacterial detection, Surface plasmon resonance, Colorimetric assay, Magnetic nanosensors, Immunomagnetic separation
1. Introduction
Infectious disease is a clinically evident disease resulting from the presence of pathogenic agent which can either be a pathogenic virus or bacteria or fungi or a parasite. These diseases are also called communicable diseases due to their ability to get transmitted from one person to another (malaria, tuberculosis) and also sometimes from one species to another (flu, influenza). Infectious diseases can be broadly classified as: a) known diseases which are persistently there (e.g., dengue, malaria, tuberculosis); b) new, previously unknown diseases (e.g., severe acute respiratory syndrome); and c) diseases which threaten to increase in the near future (e.g., avian influenza). These diseases possess a great threat as more than half of the deaths occurring worldwide can be attributed to these diseases, especially in developing countries.
According to the statistics, cholera, a disease endemic in the Indian subcontinent, Russia and sub-Saharan Africa, is spread mostly through contaminated drinking water and unsanitary conditions. An estimated of 200,000 cases are reported by the World Health Organization (WHO) annually. Recent years have seen dengue outbreaks all over Asia and Africa, with a WHO estimate of 50 million cases of dengue fever appear each year. Several influenza epidemics in the 20th century caused millions of death worldwide, including the worst epidemic; the Spanish influenza outbreak that killed more than 500,000 people in 1918. Today influenza is less of a public health threat, though it continues to be a serious disease that affects many people. Malaria almost eradicated 30 years ago, is on the rise again, affecting more than 500 million people annually, causing between 1 and 3 million deaths. Measles has seen a drastic reduction in countries where a vaccine is already available, but it is still prevalent in developing countries. Rotavirus the most common cause of viral gastroenteritis worldwide kills more than 600,000 children year each, mostly in developing countries. Tuberculosis causes nearly 2 million deaths every year, and WHO estimates that nearly 1 million people will be infected between 2000 and 2020 if more effective procedures are not adopted. Typhoid fever causes and estimated 600,000 deaths annually, out of 12–17 million cases [1]. Because of the threat posed by these infectious diseases, the speedy detection has a major impact on the success of strategies for disease zoning, control or eradication.
Diagnosis of an infectious disease involves the detection of the infecting agent, either directly or indirectly. The following are common methods that are currently used for the diagnosis of infectious diseases and these methods are either used independently or in combination with each other.
-
1.
Microbial culture: This is the most common method where the pathogenic agent is allowed to grow in the specific growth medium and then detected based on the size, shape, color of the colony formed and the changes brought by their growth.
-
2.
Microscopy: The samples taken directly from the patients (sputum, blood, urine, stool, etc.) can be viewed under the microscope for the detection and identification of the pathogenic agent. Microscopy can also be used in combination with biochemical staining and/or fluorescently labeled antibodies. Both optical microscopes and fluorescence microscopes are used for imaging pathogenic agent.
-
3.
Biochemical tests: These tests are capable of identifying the target pathogenic agent based on the detection of metabolic or enzymatic products characteristic of that agent. Acids, alcohols and gases are usually detected in these tests.
-
4.
Immunoassays: These are the most widely used tests that are based on the detection of specific binding event of an antibody to its antigen. The antigen–antibody complex is detected by labeling either the antigen or the antibody. The methods for labeling includes enzyme (Enzyme Linked Immuno Sorbent Assay, ELISA), radioisotopes (radio-immunoassays), magnetic labels (magnetic immunoassays), fluorescent labels (fluoroimmunoassays) or colloidal gold (lateral-flow assays).
-
5.
Molecular diagnostics: This method is based on changes in genomic and proteomic expression patterns due to infection by pathogenic agent. Polymerase Chain Reaction (PCR) method is usually used to amplify the target DNA or RNA for reliable detection.
Rapid, accurate and early detection of trace amounts of pathogenic bacteria and viruses is important and critical for clinical diagnosis and prevention. The major drawback is that most of these methods are laborious and time consuming as they involve the pathogen amplification step. Current methods for bacteria detection require microorganism to grow into visible colonies, which take 1–5 days. Methods that rely upon fluorescent molecular probes such as molecular beacons (a single-stranded DNA probe with hairpin-like structure; one end of the strand is attached to a fluorescent dye and the other end to a fluorescent quencher), suffer from severe photo-bleaching of the fluorescent dye that limits long-term analysis. Moreover, these probes are not highly sensitive for early detection. Similar limitations are observed using fluorescent protein based probes. Another drawback is that some of these methods do not provide the information whether or not a pathogen is infectious (strain specific).
In developing countries, most deaths from infectious diseases occur among children and young adults leading to the loss of healthy and productive life. Thus, better and widely available diagnostic methods are required to control infectious diseases in developing countries. Unfortunately, some of the commonly used methods for the diagnosis of infectious diseases are not quite cost-effective for wide implementation in developing countries. With the limited resources available in these countries, development of highly cost-effective and easy-to-perform diagnostic tests would be attractive and must receive highest priority [2]. In this context, nanodiagnostics have strong potential in introducing reliable, rapid, cost-effective, easy-to-use diagnostic methods that could benefit developing countries in the near future.
Nanodiagnostics involve the use of nanotechnology in clinical diagnosis to meet the demands for increased sensitivity and early detection in less time. The large surface area of nanomaterials enables attachment of large number of target-specific molecules of interest for ultra-sensitive detection. With such capability, diagnosis at the molecular and single cell level is possible. Because of high sensitivity, nanotechnology enables detection of a few microorganisms or target molecular analytes specific to pathogens. Conventional methods are limited to achieve this ultra-sensitivity. In addition, unique properties of nanomaterials could allow rapid (as short as few minutes) and real-time detection of the pathogens. Moreover, relatively small sample volumes are required for the tests. All these attractive features would have positive impact on the cost-effectiveness [3] for implementing nanotechnology for diagnosis of infectious diseases, especially in developing countries.
Till date, nanoparticles (NPs) of various types have been primarily studied and have shown great promise for nanodiagnostics of infectious diseases. Nanoparticle technology based on fluorescent NPs (e.g. dye-loaded NPs, quantum dots (Qdots)), magnetic NPs and metallic NPs (e.g. gold and silver NPs) has been successfully used to image, track and detect various infectious microorganisms [3], [4], [5]. In this review article, we focus on the developments (Table 1 ) of these NPs for bioimaging and detection of various infectious microorganisms in relevance to developing countries.
Table 1.
Summary of the NPs studied in the diagnosis of infectious pathogens.
| Pathogen | Nanomaterial | Recognition | Detection method | Efficiency/detection limit | Reference |
|---|---|---|---|---|---|
| E. coli O157:H7 | Qdots | Biotinylated antibody | Fluorescence microscopy | 2 orders more sensitive than conventional dyes | [13] |
| Qdots and magnetic NPs | Antibody | Fluorometry | 100 times more sensitive than FITC | [11] | |
| Qdots | Fim-H mannose-specific lectin | Fluorometry | 104 bacteria/ml | [17] | |
| Dye-doped silica NPs | Antibody | Plate counting/flow cytometry | 1–400 E. coli within 20 min | [25] | |
| Surface plating | 1.6 × 101 to 7.2 × 107 CFU/ml | [64] | |||
| Magnetic NPs | Antibody | IR spectroscopy | 104–105 CFU/ml | [65] | |
| ATP bioluminescence | 20 CFU/ml | [66] | |||
| Au NPs | Antibody | Microscope and visual | 10 ng | [53] | |
| B. anthracis | Eu doped silica NPs | Ca dipicolinate | Fluorescence spectra | 0.2 nM in 2 min | [27] |
| Eu doped polystyrene NPs | Antibody | Fluoroimmunoassay | 0.01–100 ng/ml | [32] | |
| M. tuberculosis | Dye-doped silica NPs | Antibody | Fluorescence microscopy | Amplified signal within 4 h | [26] |
| Au NPs | Oligonucleotide | UV–Vis spectroscopy | Visual detection | [45] | |
| Listeria monocytogenes | Magnetic NPs | Antibody | PCR | 226 CFU/.5 ml | [81] |
| M. pneumoniae | Ag nanorod array | SERS | – | [56] | |
| S. saprophyticus | Magnetic NPs | vancomycin | MALDI-MS | 7 × 104 CFU/ml in urine | [72] |
| S. aureus | Magnetic NPs | Vancomycin | MALDI-MS | 7 × 104 CFU/ml in urine | [72] |
| CCMV | Antibody | Test strip | Visual detection | [35] | |
| Au NPs | Antibody | I.C. Assay Test device | 100% Sensitivity | [43] | |
| Cholera toxin | Liposomes | Gangliosides | Fluoroimmunoassay | 1 nM | [29] |
| Au NPs | Thiolated lactose | Test strip | 10 fg/ml in 20 min | [30] | |
| Visual and UV–Vis | 10 min | [46] | |||
| Salmonella | Au NPs | Antibody | Test strip | Red dots appearance in 2 h | [44] |
| Salmonella enteritidis | Au and magnetic NPs | DNA assay | Fluorescence | 1 ng/ml | [69] |
| H. pylori | Au NPs | Antibody | SEM | 10 ng | [53] |
| Gram-negative bacteria | Magnetic NPs | Vancomycin | Fluorescence microscopy | 4 CFU/ml | [74] |
| SEC1 | Dye-doped Silica NPs | Antibody | Fluorescence microscopy | – | [28] |
| Staphylococcus | Magnetic NPs | Antibody | TEM | 10 CFU/ml in human blood | [75] |
| E. faecalis, S. epidermidis | Magnetic NPs | Vancomycin | Plate counting | – | [70] |
| P fimbriated E. coli | Magnetic NPs | Pigeon ovalbumin | MALDI-MS | ∼ 9.6 × 104 CFU/0.5 ml | [76] |
| Multiplexed | |||||
| E. coli, Salmonella | Qdots | Antibody | Fluorescence spectroscopy | 104 CFU/ml | [12] |
| Cholera toxin, ricin, Shinga-like toxin1, staphylococcal enterotoxin B | Qdots | Antibody | Fluroimmunoassay | In ng/ml quantity | [14] |
| E. coli, Salmonella, S. aureus | Dye-doped silica NPs | Antibody | Luminophore immunoassay | within 20 min | [23] |
| Virus | |||||
| RSV | Qdots | Antibody | Color change | Single step/short time | [8] |
| Ag nanorod array | SERS | – | [56] | ||
| Rotavirus | Ag nanorod array | – | SERS | – | [56] |
| Influenza virus | Liposomes | Sialic acid on glycoproteins | Colorimetry | 1 HAUs | [31] |
| HBV | Au NPs/Ag staining | Protein-A antibody | Protein chip assay | Color appears in 15 min | [49] |
| NLO properties | 60 pM | [51] | |||
| HSV-2 | Au NPs | Antibodies | I.C. assay | 100% Sensitivity | [42] |
| HIV-1 DNA | Au NPs | Probe DNA | NLO properties | 100 pM | [52] |
| HIV | Ag nanorod array | – | SERS | – | [56] |
| H5N1 | Magnetic NPs | Antibody | MRI | 5 viral particles in 10 µl | [78] |
| SQUID | 5 pg/ml | [80] | |||
| Adenovirus | Eu doped polystyrene NPs | Antibody | Fluoroimmunoassay | 800 fold more sensitive than conventional immunoassays | [33] |
| Multiplexed | |||||
| HBV, HCV, HIV | Qdots | Antibody | Fluorescence | 100 µl sample/1 h/50 times more sensitive | [19] |
| Au nanowire barcoded | Thiolated DNA | Fluorescence imaging | [58] | ||
| Coated with silica | |||||
| HBV, HCV | Au NPs/Ag staining | Protein A | Protein chip assay | Visual detection | [48], [49] |
| Protozoan parasite | |||||
| Giardia lamblia | Au NPs/Ag staining | Antibody | UV–Vis spectroscopy | 1.088 × 103 cells/ml | [50] |
| P. falciparum | Eu doped polystyrene NPs | Antibody | Fluoroimmunoassay | – | [34] |
I.C. = Immunochromatographic; HAUS = hemagglutinating units.
2. Surface modifications and bioconjugation of nanoparticles
Nanoparticles in their as synthesized form are often not appropriate for biological applications because of their surface characteristics. Surface modification is usually required for improving their aqueous dispersibility and biocompatibility, and obtaining appropriate surface functional groups for bioconjugation purposes. Surface attachment of biorecognition elements such as proteins, antibodies, DNA, etc. to NPs is usually carried out using conventional bioconjugate chemistries. Biomolecules can be attached to NPs through direct linkage by either physical adsorption or covalent coupling. In physical adsorption, hydrophobic and electrostatic interactions between biomolecules and the NPs dominate over the interaction among NPs. Limitations of physical adsorption are: (i) less control on the number of attached biomolecules per NPs, (ii) orientation of biomolecules, and (iii) desorption of biomolecules. Covalent coupling is preferred to avoid these limitations. Covalent coupling of the biomolecules to NPs is usually done after modifying the surface of NPs with functional group(s) of interest, for example, sulfide, amine or carboxyl groups. In addition to providing site for biomolecule conjugation, the functional groups also play crucial role in controlling colloidal stability of the NPs.
3. Fluorescent nanoparticles
Fluorescent NPs are considered as a new class of photostable highly-sensitive fluorescent tags for labeling various biological specimens such as cells and tissue samples. These fluorescent tags serve as new tools for biologists to perform sensitive bioimaging and sensing studies in real time. Fluorescent Qdots, dye-loaded silica and polystyrene NPs have been successfully used for imaging and sensing of various infectious diseases as discussed below.
3.1. Quantum dots
Qdots are semiconductor nanocrystals, typically in the size range between 1 nm and 10 nm, composed of groups II–VI (e.g., CdSe) or II–V (e.g., InP) elements of the periodic table. Qdots are extremely bright, photostable and possess high quantum yield. These properties are suitable for real-time sensitive imaging and sensing applications. Qdots exhibit size and composition tunable fluorescence properties that are suitable for multiplexed imaging using single excitation wavelength. Following studies demonstrate the potential of Qdots for imaging and detection of infectious diseases.
Respiratory Syncytial Virus (RSV) is a negative sense, single-stranded RNA virus of the family Paramyxoviridae and is a leading cause of lower respiratory tract infections. It has two characteristic surface proteins: F protein (fusion protein of the virus) and G protein (attachment protein of the virus); and it gets its name from the F protein which causes the cell membranes on nearby cells to merge, forming syncytia. A joint study carried out by Tripp and Nie groups demonstrated the suitability of using dual color Qdots for real-time detection of single virus particles and viral protein expression [6]. Their study was based on the principles of microcapillary flow cytometry and single molecule detection for high sensitivity that involved two types of fluorescent NPs: (i) 40-nm carboxyl-modified fluorescent NPs, (named, G NPs, 505/515; R NPs, 488/685) and (ii) streptavidin-coated Qdots 488/605. For the RSV detection, 40 nm NPs were coupled to either anti-RSV F protein or anti-RSV G protein monoclonal antibodies and allowed to interact with RSV. The photons produced by the laser excitation of the fluorophores were detected in the real time. This system operates on the concept that if two NPs are free to move in a solution, photons generated by them will arrive at the detector at different times unless they are bound to the same target. Hence, this can be used to detect target molecules at low concentrations with high sensitivity and discriminate between aggregate particles. Furthermore, a relative amount of surface F protein expression can be determined using this Qdot nanotechnology.
In another study, Nie group demonstrated real-time detection of individual molecules of genes, proteins and virus particles in a microfluidic channel using color-coded NP probes and two-color co-localization imaging [7]. For this study, two bioconjugated NPs (green and red emitting) were designed to recognize the same target molecule at two different sites. For RSV detection, the F protein antibody was conjugated to red emitting NPs and the G protein antibody was conjugated to green antibody. The sandwich-type binding brings two color-coded NPs together to form a NP pair with the RSV (wild type) for which both G and F proteins are expressed on the virus surface. Upon binding, the green–red pair moves in the solution as a single complex. When excited by a laser beam (488 nm excitation), the pair emits green and red fluorescence light simultaneously (i.e. time correlation). Real-time coincidence analysis of single photon events confirmed the presence of RSV in the sample. Tripp et al. demonstrated rapid and specific detection of RSV both in vitro and in vivo [8] using monoclonal antibody anti-F protein conjugated to 585 nm and 540 nm emitting cadmium telluride Qdots (RSV-NPs). These RSV-NPs were used also to quantitate RSV titers in a single-step fashion, thereby shortening the detection time needed to quantitate virus titers by plaque assay. These RSV-NPs were able to detect RSV plaques 2 days earlier than compared with conventional immunostaining, hence, indicating that the single-step RSV-NPs procedure is more sensitive in detecting limiting levels of RSV.
Wright group reported the use of streptavidin-coated Qdots (605 nm for F protein and 525 nm for G protein) to identify the presence of RSV infection over time [9]. Western blot analysis of the infection using an established sensitive chemiluminescent protocol, used as an alternative method of detection, could not detect the RSV F protein under similar conditions, for at least 96 h following infection, clearly providing the evidence of advantage of Qdots over such conventional method for low limit detection.
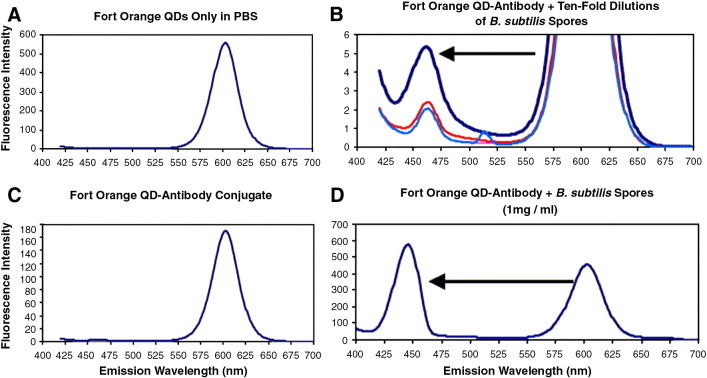
For bacterial detection, Dwarakanath et al. reported a method for possible quantitation of bacteria based on emission wavelength shifts of Qdots on binding with bacteria [10]. The test was done for Salmonella typhimurium, E. coli and B. subtilis spores using CdSe/ZnS Qdots. Both antibody-Qdot and DNA aptamer-Qdot conjugates exhibited dramatic blue shift of at least 140 nm upon binding to the bacterial surface. The authors attributed this shift to the changes in the chemical environment and physical deformation of the Qdot conjugate upon binding to bacteria. The blue shift peak grows upon conjugation to antibodies/aptamers and subsequent binding to bacterial surfaces. With increase in concentration of the bacteria, the intensity of this peak increases while the intensity of the major emission peak of the Qdot either diminishes or disappears as shown in Fig. 1 .
Fig. 1.
Fluorescence emission spectra of Fort Orange Qdots before conjugation to IgG antibody (A), after conjugation (B), and after binding increasing amounts of B. subtilis variant niger spores (C,D).
Adapted from reference [10] with permission from Elsevier.
Su and Li demonstrated the use of magnetic beads coated with anti-E. coli O157 antibodies to selectively target bacteria and biotin-conjugated anti-E. coli antibodies to form sandwich immune complexes for magnetic separation of bacteria which was followed by labeling with streptavidin-coated Qdots for measuring the cell concentration [11]. The detection limit was at least 100 times lower than that of Fluorescein Isothiocyanate (FITC) based method and the detection time was less than 2 h. In another study this group reported the simultaneous detection of E. coli O157:H7 and S. typhimurium using semiconductor Qdots with different emission wavelengths (525 nm and 705 nm) [12]. The target bacteria were isolated from the sample mixture using specific antibody coated magnetic beads. The bead–cell complexes were reacted with Qdots to form bead–cell-Qdots. The fluorescence intensities at 525 nm and 705 nm emission wavelengths were then measured for quantitative detection. This method has the capability of simultaneously detecting multiple bacterial species within a couple of hours. The detection time is much shorter than PCR based methods for simultaneous detection of pathogens.
Krauss group used CdSe/ZnS streptavidin-coated Qdots to detect individual pathogenic E. coli O157:H7 in phosphate buffer saline solution [13]. Biotinylated anti-E. coli O157:H7 recognized streptavidin-coated Qdots via well known avidin–biotin binding. Once treated, Qdot labeled antibody selectively targeted pathogenic E. coli O157:H7 over common lab strain E. coli DH5α. This assay exhibited 2 orders of magnitude more sensitivity than using an organic dye with minimal non-specific binding between the Qdots and the bacterial cells.
Sensitive and simultaneous detection of multiple toxins is challenging using conventional fluorescent dyes. Mattoussi group demonstrated that photostable and highly-sensitive Qdots were suitable for multiplexed sandwich fluoroimmunoassays for the detection of cholera toxin, ricin, shinga-like toxin1 and staphylococcal enterotoxin B simultaneously in single wells of a microtiter plate [14]. Initially the assay performance for the detection of each toxin was examined separately with a single color CdSe/ZnS Qdot. The authors then selected Qdots emitting at 510 nm, 555 nm, 590 nm and 610 nm for the conjugation of each of the four different toxin specific antibodies so that the respective toxin can be detected by means of a characteristic color of the Qdot. The overall fluorescence signal obtained from four different Qdots selective to four different toxins was analyzed quantitatively using a simple deconvolution scheme.
In a similar study Zhao et al. reported a method for simultaneous detection of food-borne pathogenic E. coli O157:H7, S. typhimurium and Shigella flexneri bacteria [15]. The method involved silica coated magnetic NP beads for the enrichment of bacteria followed by detection using fluorescent Qdot labels. Pathogen specific antibodies were surface immobilized onto amine functionalized silica coated magnetic NP beads using glutaraldehyde coupling chemistry. Once treated with the specific pathogenic bacteria, a “bead–cell” complex was formed. Qdots with three different emission wavelengths (620, 560 and 520 nm) were conjugated to anti-S. typhimurium antibody, anti-S. flexneri antibody, and anti-E. coli O157:H7 antibody, respectively. The “bead–cell” complexes were then labeled with antibody-conjugated Qdots, allowing selective and specific detection of these pathogenic bacteria using a fluorescence microscopic setup.
Sensitive and selective staining of bacterial mutants using Qdot labels was demonstrated by Smith's group. This principle of detection is based on selective targeting affinity of Zn(II)-dipicolylamine (Zn-DPA) coordination complex to phospholipids on the bacterial cell surface of specific strain. In their assay, streptavidin coated red-emitting (655 nm) CdSe/ZnS Qdots were treated with biotinylated Zn-DPA. Among three different E. coli strains (JM83, UTI89 and AO16), only JM83 strain (rough strain) was stained with the Qdots–Zn-DPA conjugate [16].
Mukhopadhyay et al. demonstrated the use of fluorescent mannose conjugated CdS Qdots to detect E. coli at levels as low as 104 bacteria/ml of sample. These Qdots induce E. coli aggregation and the aggregation process is dependent on the mannose-specific lectin present on the cell surface [17]. Detection of E. coli was specific as there was no aggregation with an E. coli strain defective in the Fim-H lectin or with galactose coated Qdots. Edgar et al. reported a rapid and simple method for detecting bacteria susceptible to specific phages, particularly the ones that are slow growing, e.g., Mycobacterium, or highly infectious ones, e.g., Bacillus anthracis [18]. This method combines in vivo biotinylation of engineered host-specific bacteriophage and conjugation of phage to streptavidin-coated Qdots. This method allowed specific and sensitive (∼ 100 fold amplification of signal over background) detection of E. coli (as low as 10 cells/ml of experimental sample) within an hour time.
In a major breakthrough Chan group has created a diagnostic system based on Qdots, capable of multiplexed, high throughput analysis of infectious agents in human serum samples, with minimal cross reactivity, low sample volume (less than 100 μl), rapidity (less than 1 h), and 50 times greater sensitivity than currently available FDA-approved methods [19]. As a proof of concept three targets were taken (hepatitis B virus, HBV; hepatitis C virus, HCV; and human immunodeficiency virus, HIV) and three different Qdot-barcodes (QdotB) with different wavelengths (QdotB1—570 nm; QdotB2—615 nm, QdotB3—570 and 615 nm). These Qdot-Bs were then incubated in human serum sample spiked with antibodies for the corresponding viral antigens, forming sandwich assay complexes which provided the fluorescence signal peak. This assay protocol can also be integrated into microfluidic chip to minimize human involvement in the entire detection process and according to the authors the sensitivity can also be further enhanced. Because of the use of Qdots for preparing barcodes, this device can be further developed into a portable handheld point-of-care diagnostic system as a major advance in detecting, monitoring, treating and preventing infectious disease spread in developed and developing countries.
Qdots have been well studied to label various bacteria and viruses. They have shown superior sensitivity and stability than traditional organic fluorophores. Moreover, broad absorption spectra and narrow emission spectra allow for simultaneous excitation and detection, respectively of multi color Qdots. Also they have additional advantage in the case of low antigen concentration or low affinity/avidity of antibodies for the antigen and their stability allows for the real-time study of biological processes.
3.2. Silica nanoparticles
Fluorescent silica NPs (FSNPs) received immense interest in bioimaging and biosensing in recent years due to their several attractive features. Firstly, silica NP matrix is capable of encapsulating agents such as organic and metallorganic fluorescent dyes, Qdots, metal ions and metal NPs without any noticeable alteration of their physical, chemical characteristics. Secondly, silica NP matrix is optically transparent allowing excitation and emission light to pass through it [20]. Thirdly, hydroxylated silica NP surface can be easily modified using various silane based reagents, polymeric coating etc. to improve biocompatibility and aqueous dispersibility. Surface modification often requires facilitating bioconjugation. Fourthly, silica NPs are hydrophilic and easily dispersed in aqueous solution. Lastly, silica NPs exhibit good mechanical and chemical stability [21], [22]. Due to these features, FSNPs of various kinds have been fabricated for imaging and sensing of infectious diseases as discussed below.
Tan group has developed triple dye-doped FRET (Förster resonance energy transfer, also known as Fluorescence Energy Transfer, a process in which there is energy transfer between two chromophores, in proximity, typically less than 10 nm) silica NPs for multiplexed bacteria monitoring [23]. By varying the ratio of the three dyes encapsulated in the NPs, excitation with a single wavelength led to emission of unique colors. The dyes used were amine reactive dyes, FAM-SE (5-carboxy-fluorescein succinimidyl ester), ROX-SE (6-carboxy-X-rhodamine, succinimidyl ester), R6G-SE (5-carboxyrhodamine 6G, succinimidyl ester), and TMR-SE (carboxytetramethylrhodamine, succinimidyl ester).These NPs were conjugated to monoclonal antibodies specific for pathogenic E. coli, S. typhimurium and Staphylococcus aureus, and then incubated with small concentrations of the bacteria. Simultaneous and sensitive detection of the multiple bacterial targets was achieved.
Previously, this group reported dual fluorophore-loaded silica NPs for multiple target detection [24]. The two luminophores, Tris(2,2′bipyridyl)osmium bis (hexafluorophosphate) (OsBpy) and Tris(bipyridine)ruthenium(II) dichloride (RuBpy) were entrapped simultaneously inside the silica nanoparticles and excited using a single wavelength. These NPS exhibited high signal amplification and photostability. A simple system using avidin–biotin was applied to immunoassay for demonstrating the use of these NPs for multiplexed signaling. Tan group also used single dye-loaded (RuBpy) bioconjugated NPs for in situ pathogen quantification down to a single bacterium within 20 min [25]. Each of the ∼ 60 nm size single dye-loaded NP encapsulated about 10,000 metallorganic RuBpy dye molecules. The antibody-conjugated NPs could readily and specifically identify E. coli O157:H7, through antibody–antigen interaction and recognition. Furthermore, since this method does not require an amplification or enrichment step, the bacterial detection can be adapted to real-time detection.
Tan group also developed a method for rapid detection of Mycobacterium tuberculosis [26]. In this method an anti-M. tuberculosis antibody was used as primary antibody to recognize M. tuberculosis, and then an antibody binding protein labeled with RuBpy doped silica NPs was used to generate fluorescent signal for examination. The detection was done with the help of fluorescent nanoparticle-based indirect immunofluorescence microscopy (FNP-IIFM). With this method, M. tuberculosis was detected in bacterial mixture as well as in spiked sputum. The assay was done within 4 h, including sample pre-treatment.
In a study by Lu et al., Europium (Eu III)-based fluorescence NP sensor for rapid and ultra-sensitive detection of B. anthracis spores in aqueous solution was developed [27]. B. anthracis spores can be identified through the detection of calcium dipicolinate (CaDPA), a unique biomarker for bacillus spores. The Eu III-complex (sensing moiety) grafted onto the surface of dye-doped silica NPs senses CaDPA. Thus, a NP sensor with optimal geometry was produced, while offering the greatest possible surface area for sensing B. anthracis spores. With this sensor, only in 2 min a limit of detection (LOD) of 0.2 nM CaDPA could be achieved, which is approximately six orders of magnitude lower than an infectious dosage of the spores.
Based on RuBpy doped amine modified fluorescent silica NPs as labels, Zhang proposed a highly-sensitive fluoroimmunoassay for the determination of staphylococcal enterotoxin C1 (SEC1) [28]. This method uses anti-SEC1 coated NPs for detection which is possible in food samples and enables fluorescence microscopy imaging for the determination of SEC1.
In summary, luminescent silica NPs demonstrate the advantages of multiplexing capability, greater sensitivity, photostability and ease of functionalization, over regular fluorescent dyes. Each silica NP can encapsulate tens of thousands of fluorescent dye molecules, providing highly amplified and reproducible signal. These advanced optical features make silica NPs attractive for bioimaging and detection as well as real-time tracking and monitoring of infectious diseases.
3.3. Liposomes
Liposomes are small vesicles consisting of one or more concentric lipid bilayers surrounding aqueous compartments. Particle size and physicochemical characteristics of liposomes can be manipulated for specific applications.
Gangliosides are expressed on cell surface and serve as natural receptors for bacterial and viral toxins. Therefore, for detection of toxins, methods have been developed based on gangliosides targeting. Singh et al. engineered and fabricated GT1b or GM1 ganglioside bearing liposomes (∼ 120–130 nm) for detecting three bacterial toxins tetanus, botulinum and cholera [29]. Their study demonstrated that these engineered liposomes were able to mimic cells and recognize target toxins, thus forming a basis for toxin detection. These liposomes were labeled with fluorescent markers (rhodamine dyes) followed by a sandwich fluoroimmunoassay on antibody coated microtiter plates. Through this assay, they reported detection of toxins as low as 1 nM concentration.
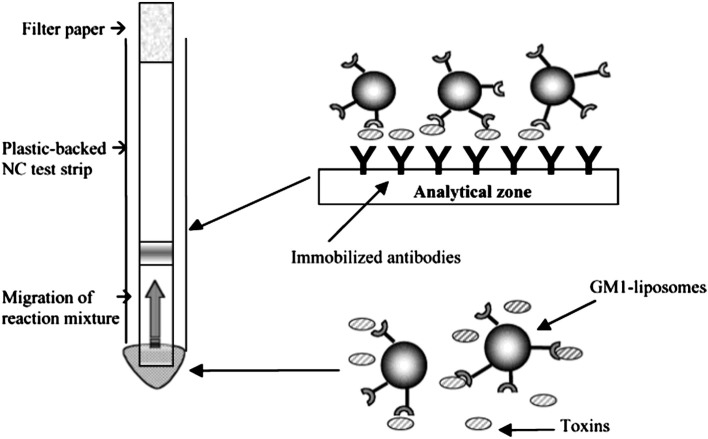
In a similar study, Durst group developed a sensitive bioassay for cholera toxin (CT) detection using GM1 gangliosides containing liposomes (∼ 200 nm size) [30]. In their sandwich assay, CT was detected as a colored band on the nitrocellulose membrane strip, where CT bound to liposomes can be captured by immobilized antibodies as shown in Fig. 2 . The limit of detection was found to be 10 fg/ml and the assay could be completed in 20 min.
Fig. 2.
Test strip assay format. CT in the reaction mixture binds to the gangliosides on the liposome surface. The CT–GM1-liposome complex migrates through the nitrocellulose test strip by capillary action until it reaches the analytical zone, where toxins in the complex are captured by immobilized antibodies. This binding zone is shown as the dark band on the test strip.
Adapted from reference [30] with permission from American Chemical Society.
In another study Charych group designed sialic acid conjugated liposomes for colorimetric detection of influenza viruses. These liposomes mimic cell surface molecular recognition and signal transduction. The virus recognition is based on binding of sialic acid to hemagglutinin lectin present on virus surface. Upon binding to influenza virus, liposome particles changed color from blue to pink/orange. This color change was visually detectable and was quantified spectroscopically [31].
All these experimental evidences demonstrate that liposomes can be used for detection of infectious agents and perform equal or even better than conventional methods.
3.4. Polystyrene nanoparticles
Polystyrene nanoparticles were used as a carrier for Eu(III) ions that serve as fluorescent reporter. Eu(III) has unique photophysical characteristics (such as sharp line-like emission peaks, longer lifetime, large stokes shift etc.) that are useful for sensitive detection of biological targets including infectious agents. Hewlett group developed an immunoassay based on polystyrene NPs loaded with Eu (III) β-diketone chelates for the detection of anthrax protective agent (PA) [32]. The Eu (III) loaded polymeric NPs could be directly integrated to ELISA platform and detection could be performed using a fluorescence plate reader. Their study involved plasma samples spiked with PA with record dynamic detection range of 0.01 to 100 ng/ml and the assay sensitivity was approximately 100 fold more than conventional ELISA. Moreover, their assay was able to successfully detect PA in all 11 samples whereas traditional ELISA could detect only 4 out of 11 samples, showing 100% success for sample detection capability. In another study based on Eu (III) chelate loaded polystyrene NPs, Härmä group developed an immunoassay for adenovirus [33]. The assay is based on direct antigen detection via sandwich forming immunoreactions using these NPs and high affinity monoclonal antibodies. The assay performed on purified adenovirus of serotype 2 and nasopharyngeal aspirates revealed high detection sensitivity of three orders more (800 fold) when compared to conventional immunoassays.
A rapid test for malaria diagnosis based on agglutination of sensitive polystyrene particles was developed by Udomsangpetch group [34] to overcome limitations associated with the high cost of currently available rapid diagnostic tests. This test involves in vitro aggregation of NPs with antigen or antibody-called latex-antigen (or antibody) conjugates, in the presence of malaria specific antibody (or antigen). The assay was successfully evaluated for P. falciparum at an outpatient malaria clinic (Mae Sod, Thailand). Authors claim the test to be quickest and easiest to be performed in a field.
3.5. Virus particles
S. aureus is a bacterium that can cause a wide range of human diseases from minor skin infections like boils and abscesses to more serious illnesses such as septicaemia, pneumonia and endocarditis. S. aureus is diagnosed by a standard method of culturing an isolate of the bacteria from blood agar plate followed by a latex test to identify the bacteria.
Multifunctional virus particles conjugated to imaging agents could serve as a probe for detecting biofilm forming S. aureus. Douglas group demonstrated that cow pea chlorotic mottle virus (CCMV, 28 nm size) could be labeled with fluorescent and Magnetic Resonance Image (MRI) contrast agents [35]. These virus particles were loaded with imaging contrast agents (fluorophore and MRI contrast agent) and targeted to a biofilm of S. aureus bacteria [35]. The targeting strategy was based on protein–ligand interaction that consisted of streptavidin sandwiched between biotinylated CCMV and biotinylated anti-SpA monoclonal antibody. The antibody was bound to the protein A (SpA) that is expressed on the bacterial cell surface. The biotinylated CCMV was tagged with fluorescein, a fluorescent dye and the targeting was confirmed by flow cytometry analysis and epifluorescence microscopy. By means of fluorescence imaging, the depth of penetration of the CCMV particles into S. aureus biofilm was observed and the density of binding was found to be exceptionally high. The CCMV particles were assessed for its capacity to deliver MRI contrast agent Gadolinium-1,4,7,10-tetraazacyclododecane tetraacetic acid (Gd-DOTA) to S. aureus cells. The concentration of Gd in the cell was analyzed by inductively coupled plasma mass spectrometry (ICP-MS) which indicated a value of 1.8 × 105 Gd atoms per bacterial cell. These results demonstrate the usefulness of virus particles for both diagnosis and possible treatment of biofilm forming bacterial infections.
In summary, fluorescent nanoparticles such as quantum dots, dye-doped silica nanoparticles, liposomes, polystyrene nanoparticles and virus nanoparticles have been used to demonstrate their applicability for imaging and sensing of various pathogens. Qdots and dye-doped silica NPs based assays have demonstrated multiplexing capabilities for the simultaneous detection of infectious diseases. However, these assays may prove to be unaffordable in low resources settings as sophisticated instruments such as flow cytometry, fluorescence microscopy are used for the detection of the diseases. These assays can be adopted if the detecting methods are made affordable and simpler. For developing countries, simple assays such as the liposome based strip test for the detection of cholera toxin, polystyrene NPs based test for malaria are ideal. Other applicable inexpensive techniques could be Qdot assisted detection of infectious diseases on an immunoassay platform which can be achieved on a lateral-flow based test device that would use light emitting diodes as excitation source. Also handheld microfluidic devices using Qdot-barcodes can be further modified for use in low resource settings.
4. Metal nanostructures
Metal nanoparticles typically gold and silver, when excited with electromagnetic radiation, produce intense absorption due to the collective oscillations of surface electrons. The frequency of oscillation of these electrons is in resonance with the incident light. This resonance frequency or the surface plasmon band lies in the visible part of the electromagnetic spectrum giving rise to intense colors from solutions of colloidal metal particles [36]. This optical property known as surface plasmon resonance (SPR) can be measured by absorption and scattering spectroscopic techniques. Molecules adsorbed onto the metal surface can induce changes in the electron density on the surface causing a shift in the SPR absorption band, i.e., a change in the color. This is the basis for the application of gold and silver NPs as sensitive sensors in diagnostics [37], [38]. The SPR property is affected by the particle size, shape, material and the distance between particles (i.e., monodisperse and aggregated NPs have different properties) [37]. The SPR absorption has an absorption coefficient several orders of magnitude larger than strongly absorbing dyes [39]. The resonance light scattering property of the metal NPs has also been widely applied to detect biological species that form assemblies/aggregates with the NPs. These assemblies or aggregated species when excited by wavelength of light close to the region of their absorption bands, enhanced resonance light scattering signals are generated. Such signals can be measured spectroscopically or even by mere visualization.
4.1. Gold nanoparticles
The potential of nanomaterials in biodiagnostic applications was first recognized by Mirkin and coworkers in 1996 when they used gold NP-based assay to detect DNA. The color of the gold NP (Au NP) solution changed from red to blue upon the analyte-directed aggregation [40], [41]. This is a consequence of surface plasmon particle interaction and aggregate scattering properties.
4.1.1. Colorimetric assay
The SPR application in diagnosing infectious diseases has been based on antibody or antigen detection and DNA hybridization. The colorimetric change is a simple and inexpensive way of diagnosing disease and provides a sensitive test for the presence of target analytes.
Herpes simplex virus type 2 (HSV-2) is a common human pathogen that causes a variety of infections. Laboratory-based methods such as Western blotting, immunoblotting, and ELISA are the current testing methods for HSV-2 infection. Laderman et al. developed and evaluated lateral-flow immunochromatographic assay device for rapid screening of serum and whole blood samples for the HSV-2 infection so that patient treatment can be fast [42]. A dual-antigen directed sandwich assay was applied for the detection of anti-HSV IgG to HSV-2. The device consists of nitrocellulose membrane test strip immobilized with separate antigens for Herpes simplex virus type 1 (HSV1) (gG1, pre-absorption line) and the detection zone HSV2 (gG2, named as test line) and another detection zone anti-human IgG (named as control line) at different positions. A mixture of Au NPs bound to anti-human IgG and sample was passed along the test strip. A buffer facilitates migration of sample along the test strip. The sample came in contact sequentially with the pre-absorption line, HSV2 test line and the control line. The presence of a HSV-2 infection could be detected in 15–20 min by the appearance of a pink color both in the test and control lines. A pink control line alone indicates a negative test for HSV-2.
Another immunochromatographic assay test device was developed for rapid detection of the S. aureus in patient specimen [43]. Protein A, a specific product of the bacteria that is secreted into the medium during the growth phase of the bacteria, serves as a biomarker. The assay consisted of a sandwich format where the target antigen was recognized by the anti-protein-A antibodies, one immobilized on the test strip nitrocellulose membrane while the other conjugated to the 20 nm Au NPs. The antibody-conjugated Au NPs recognized the protein-A antigen and were further captured by the antibody immobilized on the test strip. This positive result was generated by a colorimetric response in the test and control lines. Appearance of color in the control line alone confirmed negative results. The analytical sensitivity of the device assay was similar to the conventional immunoassays. The advantages include obtaining results in 5 min, requiring no sophisticated instrumentations and long shelf-life at room temperature. Fang et al. demonstrated another Au NP immunoassay readout technique for easy screening of Salmonella [44]. The test was conducted on a nitrocellulose membrane (called the colony-print) onto which bacterial colonies are transferred from an agar surface incubated with stool specimens. The colony-print was then treated with anti-Salmonella antibody coated Au NPs. The coated NPs recognized the Salmonella species bacteria and formed red dots on the blotted membrane in 2 h. The selectivity of this method is 99.1% without the need for any equipment including a microscope.
Au NP-based colorimetric assay was reported for direct and fast detection of M. tuberculosis DNA in clinical samples [45]. This method required an initial round of PCR for enrichment of DNA sample. The Au NPs were conjugated to the thiolated oligonucleotide specific to the mycobacteria. The assay was performed by mixing the nanoprobe with clinical sample containing target DNA. The nanoprobe solution was red in color due to the SPR effect. At high NaCl concentration, the nanoprobe aggregates in the absence of a complementary DNA sequence which causes the solution to turn purple. When the nanoprobe undergoes hybridization with the complementary DNA from M. tuberculosis, no aggregation occurs and the solution remains red in color.
Russell and coworkers have found that Au NPs are very effective detectors of cholera toxin secreted by the bacteria vibrio cholera which causes acute infection of the intestine [46]. Their bioassay comprised of 16 nm Au NPs coated with thiolated lactose where the –SH group anchors the lactose to the Au NP surface. The thiolated lactose derivative plays the role of molecular recognition ligand and mimics the GM1 ganglioside—the receptor to which cholera toxin binds in the small intestine. The lactose–NP solution has a surface plasmon band centered at 520 nm and appears red in color. When the cholera toxin binds to the lactose derivative, aggregation of the NPs is induced. The aggregation causes a broadening and a red shift of the surface plasmon absorption band such that the NP solution appears deep purple color. The bioassay could detect the toxin within 10 min and provide quantitative toxin concentration data. The lactose-stabilized NPs were stable on freeze drying and could be resuspended in water to detect the cholera toxin in an electrolyte solution that mimicked the watery stool that is typical of cholera patients to test the selectivity for the toxin. This colorimetric bioassay provides a rapid, simple and colorimetric tool for the measurement of the infectious bacterial cholera toxin.
Ag staining of Au NPs is a simple, time-saving, sensitive and inexpensive method of signal amplification. In this method, metallic Ag is deposited onto Au NPs where the latter serve as nucleation sites. Black, highly intense color of metallic Ag drastically enhances visual contrast. Taton et al. reported this method for the detection of target DNA with the sensitivity that exceeds two orders of magnitude to that of fluorescence based detection system [47]. The advantage of this method is that it does not require any sophisticated instrument for signal detection. Using this signal amplification method, a protein chip assay was developed for simultaneous detection of HBV and HCV [48], [49]. In this protein chip assay, antigens of HBV and HCV were immobilized on a glass slide (called as chip) to capture HBV and HCV in human sera. Au NPs of 15 ± 2 nm size labeled with staphylococcal protein A was mixed with the sample. The Au NPs were used as an indicator and silver staining was applied to amplify the detection signals, producing a black image on the chip. Thus, by seeing the color change, the pathogen gene could be diagnosed.
Detection of intestinal pathogen (e.g. Giardia lambia cysts) that causes infectious gastroenteritis is usually done using microscopic diagnoses, real-time PCR and ELISA methods. However, these methods are time and labor consuming. Sim et al. have applied Ag staining onto Au NPs based signal amplification method to amplify the immunoassay signal for the detection of G. lambia cysts [50] . In a typical procedure, antibody functionalized Au NPs of 25 nm size were used to capture the target pathogen cells. The captured cells were collected and then separated by centrifuge filters (of 0.45 µm size while the pathogen size is 6–10 µm). The Au NP seeds attached to the cells were dispersed in the growth solution (gold solution) wherein the gold seeds acted as nuclei and catalyzed the deposition of Ag metal atoms resulting in signal enhancement that was measured by the UV–Vis spectrophotometer.
4.1.2. Resonance Light Scattering (RLS) application
HCV is a single-stranded (ss) RNA that is responsible for chronic infectious liver disease. ELISA method is the widely used method of diagnosing HCV antibodies. But ELISA will not be able to detect the infection at an early stage as the antibodies against HCV antigens will not be produced then. Also, detectable antibodies could be absent in HCV infection that may be present in immunocompromised patients. The need for a direct RNA based test to detect the presence of HCV at an early stage of infection has motivated Ray and coworkers for screening and quantifying sequence-specific HCV RNA using non-linear optical properties of Au NPs by Hyper-Rayleigh scattering (HRS) technique [51]. The assay is based on the fact that double and single-stranded oligonucleotides have different electrostatic properties. A fluorescent Raman dye rhodamine 6G tagged ssRNA was adsorbed onto the Au NPs. Fluorescence quenching of the dye and the enhancement of resonant Raman scattering from the dye were observed. Upon binding to the target RNA, duplex structure of the double stranded (ds) RNA is formed. Electrostatic repulsion between the ds RNA and the NP causes the ds RNA not to adsorb onto the Au NPs and the fluorescence of the dye persists. As soon as the ds RNA is separated from the Au NPs, a second effect is observed which is the aggregation of the Au NPs. This aggregation is evidenced by transmission electron microscopy (TEM) and further confirmed by colorimetric studies. The aggregation causes increase in size resulting in increase in HRS intensity thereby confirming the detection of HCV virus RNA. Ray et al. used a similar HRS assay with Au nanorods for sensing sequence-specific HIV-1 virus DNA [52].
The RLS property of Au NPs was applied to develop a sensitive heterogeneous immunoassay using a microfluidic platform for the detection of the gastrointestinal microbial pathogens Helicobacter pyroli (H. pyroli) and E. coli O157:H7 [53]. A Microfluidic platform uses micro liter volumes in channels with sub-millimeter dimensions [54]. It aims to reduce the analysis time of the immunoassay and requires low amounts of the sample and the reagent making it inexpensive. The RLS imaging technique was applied to measure the light scattering of aggregation of the Au NPs induced by the binding of the antibody functionalized NP to the microbial pathogens. The detection was evidenced by golden brown color in the microchannels (of ∼ 1.5 µl volume) visualized under a dark-field microscope and the images were captured by a digital camera. The assay was capable of detecting the H. pyroli and E. coli O157:H7 antigens at quantities as low as 10 ng. The microchannel immunoassay could be stored up to eight months with no loss in signal intensity and did not require sophisticated instruments to detect the NPs in microchannels.
In summary, the development of Au NPs in diagnosis of infectious diseases is valuable. The SPR properties of Au NPs offer major advantages such as rapid, label-free analysis and applications in portable miniature devices.
4.2. Silver nanoparticles
Silver nanoparticles have been used in surface enhanced Raman scattering (SERS) based detection of target analytes. Because of high sensitivity, this technique has been used in single molecule detection. The sensitivity of detection relies upon the close proximity of the analytes to the NP surface. The following studies demonstrate the application of this technique for pathogen detection.
Naja group demonstrated application of SERS technique using E. coli model bacteria. The technique involved protein-A modified silver NPs and E. coli specific antibodies. Binding of E. coli-antibody-conjugated to protein-A modified NP produced SERS spectra of bacteria. The SERS spectral fingerprint exhibited superior molecular selectivity as compared to the corresponding bulk Raman vibrational signatures, demonstrating the potential of silver NPs in SERS [55].
Driskell et al. showed that SERS active silver nanorod array could be used to detect and differentiate the molecular fingerprints of several important human pathogens, particularly RSV, HIV, rotavirus, and the bacterium Mycoplasma pneumoniae [56]. Their method can be used to classify viruses at the strain level.
In summary, colorimetric immunoassay based on Au NPs is promising for the diagnosis of infectious diseases. It has also been demonstrated that the sensitivity of this assay could be further improved by Ag staining. The immunoassay itself could be adapted to a lateral-flow based test device in a highly cost-effective manner. Thus, application of Au NPs based nanodiagnostic technique for infectious diseases is quite feasible in a constrained setting environment. However, further research on Au NPs based immunoassay is necessary for improving specificity and sensitivity.
4.3. Metallic nanowires
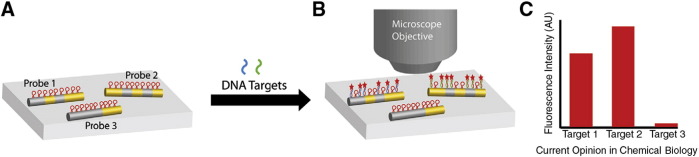
In addition to NPs, nanowire based detection strategies have been developed for multiplexed bioanalysis and the detection is based on either optical or electrical readouts [57]. Barcoded metallic nanowires, synthesized by template electrodeposition of multiple metal segments such as silver, gold, etc. have been studied for multiplexed protein and nucleic acid detection (codes 0 and 1 in the nanowire indicate Au and Ag segments respectively) (Fig. 3 ).
Fig. 3.
Barcoded metallic nanowires for multiplexed biodetection. (A) Nanowires with different patterns of Au and Ag segments are functionalized with different molecular beacon probe sequences, which are nonfluorescent in the absence of target strands. When a mixture of target molecules, in this case complementary to probes 1 and 2, is incubated with the mixture of barcoded nanowires, (B) some nanowire-bound probes become fluorescent because of complementary target binding. Reflectance and fluorescence microscope images are acquired for the identification of nanowires and quantification of fluorescence, respectively, and (C) mean fluorescence intensities are calculated from populations of individual nanowires of each barcode pattern, to quantify the amount of each target present.
Adapted from reference [57] with permission from Elsevier.
A sandwich DNA hybridization assay was performed on the silica coated Au/Ag nanowires for the detection of HIV, HBV and HCV specific oligonucleotide sequences [58]. The thiolated probe DNA was attached to the silica coating of nanowires using a bifunctional crosslinker, sulfo-SMCC (Succinimidyl-4-(N-Maleimidomethyl)Cyclohexane-1-Carboxylate) which links an amine to a thiol. Each of the pathogenic probe DNAs was attached to a particular pattern of nanowire. The probe nanowires were combined together and allowed to interact with the target DNA for hybridization followed by the addition of fluorescently labeled DNA. The nanowire of a given pattern appears fluorescent only if it has the correct target DNA sequence. An optical microscope was used for the fluorescence and reflectance imaging. The Au/Ag reflectance ratios from optical reflectance images were acquired and the nanowire pattern identification was done using software. When only the HCV target was present on a nanowire pattern 0110, it was visible in the fluorescence image and similarly when HBV target was present, the nanowire pattern 0011 shows fluorescence. It was observed that the silica coating improved the fluorescence intensity compared to uncoated nanowires.
In another study, Au/Ag/Ni barcoded nanowires demonstrated the multiplexed detection of three antigens that are bio-threat stimulants. They used Bacillus globigii (Bg) spores to simulate B. anthracis and other bacterial species; RNA MS2 bacteriophage to simulate Variola (virus for smallpox) and other pathogenic viruses; and ovalbumin (Ova) protein to simulate protein toxins such as ricin or botulinum toxin. The sensitivities were found to be 1 × 105 colony forming or plaque forming units, and 5 ng/mg for the protein ovalbumin [59]. An optical reflectance image could be processed to identify the strip pattern while fluorescence images report information on the degree of binding between the antibody-conjugated nanowires and a fluorophore-tagged antigen target.
Metallic barcode nanowire based DNA detection assay is highly promising. Specifically, multiplexed detection and electrical readout capabilities are attractive. However, large-scale synthesis of high-quality barcode nanowires is challenging. Furthermore, complexity in target detection that uses both fluorescence and reflectance imaging modalities will limit the application of this technique in constrained setting environments.
5. Magnetic nanoparticles
Magnetic nanoparticles have been demonstrated to have exciting and promising applications in medical diagnostics and therapy [60], [61] as well as immunoassay based diagnostics [62]. In the immunoassay diagnostics, the functionalized magnetic particle bound to a biomolecule recognition unit is used as a label instead of enzymes or fluorescent material to capture the target analyte. Using a magnet, the target–magnetic particle complex can be separated for further analysis.
Majority of the food-borne disease outbreaks have been linked to Campylobacter, E. coli O157:H7, Salmonella and Listeria monocytogenes [63]. Iron oxide NPs, such as magnetite Fe3O4 or Fe2O3 have been well studied with great interest in isolating these pathogens from food, blood, etc. using an external magnet and quantify them with various analytical techniques (Table 1).
Varshney et al. used streptavidin-coated magnetic NPs and biotin labeled antibodies to detect E. coli O157:H7 [64]. These functionalized NPs were mixed with inoculated ground beef samples and the NP–antibody–E. coli O157:147 complexes were separated from the food matrix by magnetic separation, washed and the bacteria were quantified by surface plating. About 94% of the bacteria could be captured ranging from 1.6 × 101 to 7.2 × 107 CFU/ml (CFU—colony forming unit) in 15 min without any enrichment. The magnetic NPs had the advantages of higher capture efficiency and minimal sample preparation, and mechanical mixing was not needed compared to microbeads based immunomagnetic separation.
In another study, lactic acid coated magnetic (Fe3O4) NPs were functionalized with the antibodies of E. coli O157:147 or S. typhimurium using water carbodiimide chemistry. These were used to capture and separate the E. coli or S. typhimurium in inoculated spinach and milk as well as from a mixture of other bacterial species [65]. The pathogen NPs complex was confirmed in less than 30 min with Infra Red (IR) spectroscopy that presented characteristic spectroscopic finger printing of these pathogens with a detection limit of 104–105 CFU/ml. To enable this protocol as a point-of-detection technology, the binding and detection of the NPs to the pathogen were applied to a portable mid-IR spectrometer.
Cheng et al. have determined the E. coli number (20 CFU/ml) in a milk sample by biotin functionalized amine Fe3O4 NPs using the adenosine triphosphate (ATP) bioluminescence method in about 1 h [66].
L. monocytogenes, another food-borne pathogen was detected in spiked milk sample by magnetic NP-based immune-magnetic separation combined with real-time PCR. The detection limit was found to be 226 CFU/0.5 ml [67]. The isolation of the pathogen from the food matrix removes the interference of the food constituents to carry out PCR amplification. In the absence of magnetic separation, there was no amplification of the L. monocytogenes DNA when extracted from the milk sample with a concentration of even 107 CFU/ml. In another study for detecting L. monocytogenes, Grossman et al. demonstrated the use of Superconducting Quantum Interference Device (SQUID) [68]. γ-Fe2O3 of 50 nm size coupled to L. monocytogenes antibodies was added to an aqueous solution of the bacteria and the particles were allowed to bind to the target. A pulsed magnetic field was applied to align the magnetic dipole moments. Upon removal of the magnetic field, the magnetic relaxation signal of the NP–bacterial complex was measured. The basis of the detection is that unbound NPs relax too quickly to be detected while particles bound to the large bacteria were able to rotate and relax slowly. The detection limit in a 20 μl sample volume was (5.6 ± 1.1) × 106 of the cells. This assay had the advantage of doing away with the step of washing away of the unbound particles.
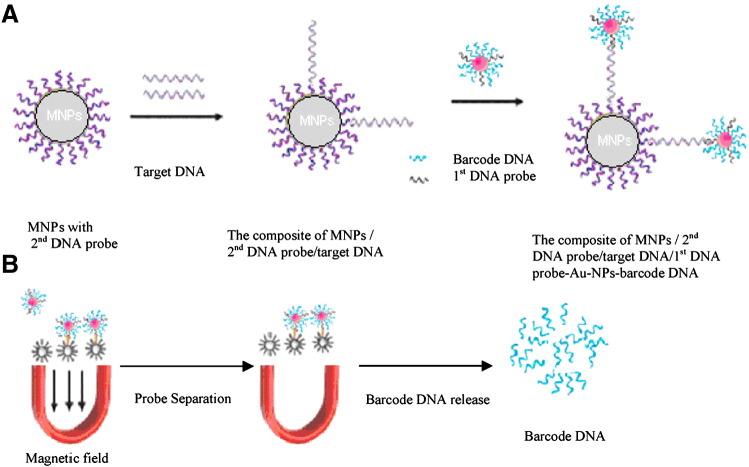
Zhang et al. developed a fluorescent bio-barcoded DNA assay for the rapid detection of the Salmonella enteritidis gene, based on two NPs: magnetic NPs (MNP) and gold NPs (Au NP) [69]. The Au NPs are conjugated with the 1st target-specific DNA probe which can recognize the target gene, and a fluorescein-labeled barcode DNA. The MNPs were coated with the 2nd target-specific DNA probe. Upon mixing with the target gene, a sandwich structure is formed followed by magnetic separation of the sandwich structure (Fig. 4 ). The barcode DNA from the Au NPs is released by heating the mixture and the released barcode DNA is measured by fluorescence with a detection limit of 1 ng/ml. The Au NPs here contribute to signal amplification by carrying a large number of barcode DNA per DNA probe binding event and the MNPs act as separator and pre-concentrator.
Fig. 4.
Schematic of the bio-barcode assay: (A) formation of MNP-2nd DNA probe/target DNA/1st DNA probe-Au NPs-barcode DNA; (B) barcode DNA separation and release.
Adapted from reference [69] with permission from Elsevier.
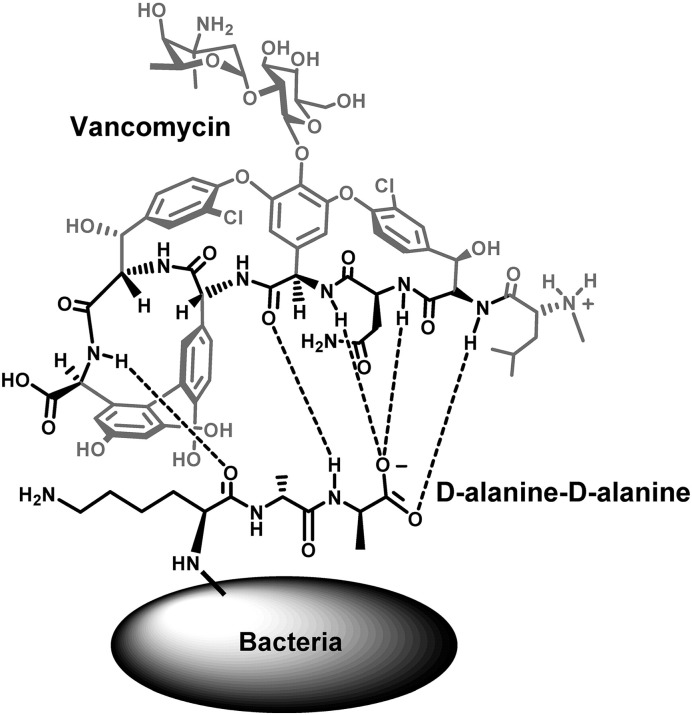
Other studies have demonstrated the applications of small molecule probes as promising alternatives to antibodies for use in NP-based magnetic capture assays. Vancomycin is one such small molecule, a glycopeptide antibiotic that interacts with a broad range of Gram-positive bacteria by binding to D-Ala–D-Ala units of the bacterial cell wall through the formation of hydrogen bonds [70] (Fig. 5 ).
Fig. 5.
Representation of the vancomycin-d-alanyl–d-alanine interaction responsible for mediating the interaction between the NPs and the bacteria. The critical components for the strong H-bonding interaction both on the vancomycin molecule (the heptapeptide backbone) and the d-alanyl–d-alanine dipeptide exposed from the bacterial surface are highlighted.
Adapted from reference [70] with permission from American Chemical Society.
The vancomycin functionalized magnetic NPs for pathogen detection have been reviewed by Gu et al. [71]. Vancomycin can be bound to the magnetic NP surface by activating the –COOH group of vancomycin followed by reaction with the amine groups on the surface of the iron oxide NPs. Another method is to react carbodiimide activated carboxyl functionalized magnetic NPs to the primary amine group of the vancomycin [72]. The vancomycin conjugated iron oxide NPs were used as probes to selectively trap S. Saprophyticus (a pathogen that commonly infects the urinary tract of young women) and S. aureus bacteria from urine sample using a magnetic field. The isolated cells were then characterized by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry (MALDI-MS) to differentiate microorganism species based on their characteristic mass spectra [72]. The magnetic separation effectively reduced the interference of protein and metabolite signals in the mass spectra as vancomycin has high specificity to the bacterial cell walls. Vancomycin conjugated MNPs were able to isolate pathogens at concentrations of 7 × 104 CFU/ml from the urine sample. The same pathogens when captured by IgG antibody-modified magnetite (Fe3O4) NPs could be isolated at 3 × 107 CFU/ml in a urine sample indicating the higher sensitivity of vancomycin conjugated magnetic NPs [73].
Xu group studied thiolated vancomycin coated FePt NPs for capturing bacteria at ultra-low concentrations of 4 CFU/ml within 1 h using scanning electron microscopy [62], [74]. In another study by the same group, magnetic property was combined along with fluorescence to detect bacterial pathogens in human blood samples within 2 h with a sensitivity as low as 10 CFU/ml [62], [75]. The magnetic component consisted of the magnetic FePt NPs of 10 nm size which were coated with thiolated vancomycin. The fluorescent property arises from the conjugation of fluorescein amine and vancomycin. The coagulase-negative Staphylococcus bacteria contaminated platelet was mixed and shaken with the vancomycin conjugated magnetic NPs followed by magnetic isolation of the bacteria–NP conjugate. The isolated bacteria was then mixed with the fluorescent vancomycin and analyzed by a fluorescence microscope.
Different strategies to anchor vancomycin on silica coated iron oxide NP surface and its influence on capturing a wide range of pathogenic bacteria were studied by Kell et al. [70]. They attached vancomycin to the modified NP surface utilizing its functional groups –COOH, –NH2 as well as placing a spacer between the NP and vancomycin probe. These various anchoring strategies resulted in a change in the orientation/architecture of the vancomycin probe on the NP surface which had a critical influence on the efficient capture of pathogenic bacteria such as E. faecalis, S. epidermidis etc. The magnetically isolated bacteria were determined via plate counting of the NP–pathogen conjugates. The NP size as well as the surface coverage of vancomycin on the NP also influenced the capture of the bacteria.
Liu et al. in their study to detect E. coli expressing P fimbriae (a most common source for urinary tract infection), adopted pigeon ovalbumin (POA) bound Fe3O4 coated Al2O3 NPs as affinity probes for these bacteria [76]. The POA is a phosphorylated protein with high levels of galactose units for which the P fimbriae E. coli has high affinity. To immobilize the POA onto the Fe3O4 NPs, coating of aluminum oxide was carried out under microwave heating to facilitate the phosphate–alumina complex. The POA functionalized magnetic NPs could target selectively the P fimbriated E. coli from sample solutions and was characterized by MALDI-MS with a detection limit of ∼ 9.6 × 104 CFU/0.5 ml.
Other examples of small molecule probes alternative to antibody include carbohydrates. Silica coated magnetite NPs functionalized with carbohydrate d-mannose was studied for the detection of bacteria E. coli [77]. The carbohydrate provides anchor to the bacteria for attachment to the mammalian cell which subsequently results in the infection. The carbohydrate coated NP known as magnetic glyco NP (MGNP) could detect E. coli in 5 min. About 88% of the bacteria could be removed from the media by magnetic separation due to the high surface to volume ratio of the MGNPs because of the higher contact surface area. Also, the MGNPs were studied to differentiate E. coli strains based on the fact that several bacteria bind to the same carbohydrate with different affinities and this would help in distinguishing pathogen strains in clinical applications.
For detecting viruses, Weissleder group developed biocompatible superparamagnetic iron oxide (SPIO) nanosensors that act as magnetic relaxation sensors (MRS) as an alternative method of detecting biomolecules by MRI [78], [79]. The nanosensor was studied for the detection of HSV-1 virus and adenovirus in serum samples. The sensor comprised of amino-dextran coated SPIO NPs onto which antibodies of the virus were conjugated to the NPs. The antibody-conjugated probe NPs were incubated with serum and the clustering of NPs as a result of the probe–target conjugation resulted in a corresponding change in MRI signal. The virus particles could be detected at low concentrations of 5 viral particles in 10 µl without the need of PCR amplification. This method utilizes common NMR (Nuclear Magnetic Resonance) and MRI instrumentation.
The similar concept of clustering of viral target induced magnetic NPs was applied for diagnosing avian influenza H5N1 virus at ultra-low concentration of about 5 pg/ml using SQUID (Superconducting Quantum Interference Device) [80]. The clustered MNPs are formed as a result of the interaction of the viral pathogen with the antibodies on the MNPs. The response of these larger clustered magnetic NPs to the external AC magnetic field is much less than that of the original, individual MNPs.
Engineered MNPs provide a fast, efficient and convenient method for the separation and enrichment of low count of pathogen from large volumes of sample. These MNPs display high capture efficiencies due to high surface to volume ratio and faster reaction kinetics. Thus the use of MNPs for direct pathogen isolation is more favorable over PCR based technique. Detection of pathogen that involves highly-expensive equipment such as MALDI-MS, MRI and SQUID based detection systems are not practical to implement in developing countries. However, MR based spectroscopic technique could be used in the near future provided the MR instrument is miniaturized and made cost-effective.
6. Conclusions
In this paper, we reviewed recent innovative developments on engineered nanomaterials with unique optical and magnetic properties and reported their promising applications in various nanodiagnostic assays for the detection of trace amount of infectious pathogens (Table 1). The nanodiagnostic assays that have been studied include the immunofluorescent, colorimetric, immunomagnetic separation as well as combination of these assays. The working principle of these assays to image, detect and capture infectious virus and bacteria in solutions or biological samples was discussed along with pros and cons of these assays. Detection methods such as flow cytometry, spectroscopy (absorption, Raman spectroscopy, fluorescence, IR etc.), microscopy and visualization by naked eye that were used in these assays were discussed. Key features of nanodiagnostic assays were identified that include sensitivity, multiplexing capabilities and reduced sample volume. These features could provide excellent results for in vitro diagnostics of infectious diseases and could be further extended to in vivo diagnosis. Yet another attractive advantage is the application of nanodiagnostic assays into portable, easy to perform and read out diagnostic tools that are cost-effective.
These nanotechnology innovations applied to diagnostics are promising with immense humanitarian benefits. However, when applying these innovations to developing countries or low resource settings, the user requirement needs understanding [82]. A diagnostic test requirement would be based on the infectious disease type as well as the different levels of health care facilities such as the community/rural health facility, the district/town hospital and the urban hospital. In developing countries, rural patients can often be misdiagnosed due to lack of access to diagnostic tests such as the diagnosis of malaria instead of HIV [83]. A lateral-flow based diagnostic testing or a test strip membrane would be very beneficial as by mere visualization of the test, the presence of an infection can be determined rapidly. This would prevent spreading of the infection, false diagnosis and resistance to the medication as in the case of bacterial infections. A quantitative test may not be of much use for a community or rural health worker while it may be more informative to a doctor at a hospital or clinic. But these quantitative tests often are expensive, require specialized equipments and highly trained technicians to carry out the analysis. Researchers have used specialized equipments such as flow cytometry, SQUID, spectroscopic techniques, TEM, confocal microscopy etc., to analyze trace amount of the pathogen. Some of these equipments may be acquired by a few urban hospitals but the cost factor and access to such facilities are not at reach to a large number of people in developing countries.
Researchers and device developers would have to keep in mind affordable portable diagnostic tests that can generate results in a few minutes and that can handle mass screening in the developing countries. Some examples of such achievements are [84], a nanotechnology based TB diagnostic kit, designed by the Central Scientific Instruments Organization of India and currently in the clinical trial phase, does not require skilled technicians for use and offers efficiency, portability, user-friendliness and availability for use for as little as 30 rupees (less than US$1). Furthermore, a group at RMIT University, in Australia, is conducting research into the application of novel tethered nanoparticles as low-cost, color based assays for TB diagnosis and at the Chidicon Medical Center in Nigeria, researchers are studying nanoscale copolymer assemblies for diagnostic imaging and therapeutic management of infectious diseases.
In conclusion, nanotechnology has strong potential for diagnosing infectious diseases and this review demonstrates various assays for this application. Yet, there exist challenges associated with successful implementation of these in developing countries. This area of research is still at infancy. Government agencies would play a major role in establishing cross-country research initiatives that would connect scientists and device developers from both developed and developing countries.
Acknowledgement
This work has been partly supported by the NSF (grant #s CBET-63016011 and NIRT EEC-0506560) and NIH grant # 2P01HL059412-11A1.
Footnotes
This review is part of the Advanced Drug Delivery Reviews theme issue on “Nanotechnology Solutions for Infectious Diseases in Developing Nations”.
Contributor Information
Padmavathy Tallury, Email: ptallury@mail.ucf.edu.
Swadeshmukul Santra, Email: ssantra@mail.ucf.edu.
References
- 1.http://www.infoplease.com/ipa/A0903696.
- 2.Mabey D., Peeling R.W., Ustianowski A., Perkins M.D. Diagnostics for the developing world. Nat. Rev. Microbiol. 2004;2:231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- 3.Jain K.K. Nanotechnology in clinical laboratory diagnostics. Clin. Chim. Acta. 2005;358:37–54. doi: 10.1016/j.cccn.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Azzazy H.M.E., Mansour M.M.H., Kazmierczak S.C. Nanodiagnostics: a new frontier for clinical laboratory medicine. Clin. Chem. 2006;52:1238–1246. doi: 10.1373/clinchem.2006.066654. [DOI] [PubMed] [Google Scholar]
- 5.Luo P.G., Stutzenberger F.J. vol. 63. Academic Press; 2008. Nanotechnology in the detection and control of microorganisms; pp. 145–172. (Advances in Applied Microbiology). [DOI] [PubMed] [Google Scholar]
- 6.Agrawal A., Tripp R.A., Anderson L.J., Nie S. Real-time detection of virus particles and viral protein expression with two-color nanoparticle probes. J. Virol. 2005;79:8625–8628. doi: 10.1128/JVI.79.13.8625-8628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agrawal A., Zhang C., Byassee T., Tripp R.A., Nie S. Counting single native biomolecules and intact viruses with color-coded nanoparticles. Anal. Chem. 2006;78:1061–1070. doi: 10.1021/ac051801t. [DOI] [PubMed] [Google Scholar]
- 8.Tripp R.A., Alvarez R., Anderson B., Jones L., Weeks C., Chen W. Bioconjugated nanoparticle detection of respiratory syncytial virus infection. Int. J. Nanomed. 2007;2:117–124. doi: 10.2147/nano.2007.2.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentzen E.L., House F., Utley T.J., Crowe J.E., Wright D.W. Progression of respiratory syncytial virus infection monitored by fluorescent quantum dot probes. Nano Lett. 2005;5:591–595. doi: 10.1021/nl048073u. [DOI] [PubMed] [Google Scholar]
- 10.Dwarakanath S., Bruno J.G., Shastry A., Phillips T., John A., Kumar A., Stephenson L.D. Quantum dot–antibody and aptamer conjugates shift fluorescence upon binding bacteria. Biochem. Biophys. Res. Commun. 2004;325:739–743. doi: 10.1016/j.bbrc.2004.10.099. [DOI] [PubMed] [Google Scholar]
- 11.Su X.L., Li Y. Quantum dot biolabeling coupled with immunomagnetic separation for detection of Escherichia coli O157: H7. Anal. Chem. 2004;76:4806–4810. doi: 10.1021/ac049442+. [DOI] [PubMed] [Google Scholar]
- 12.Yang L., Li Y. Simultaneous detection of Escherichia coli O157:H7 and Salmonella typhimurium using quantum dots as fluorescence labels. Analyst. 2006;131:394–401. doi: 10.1039/b510888h. [DOI] [PubMed] [Google Scholar]
- 13.Hahn M.A., Tabb J.S., Krauss T.D. Detection of single bacterial pathogens with semiconductor quantum dots. Anal. Chem. 2005;77:4861–4869. doi: 10.1021/ac050641i. [DOI] [PubMed] [Google Scholar]
- 14.Goldman E.R., Clapp A.R., Anderson G.P., Uyeda H.T., Mauro J.M., Medintz I.L., Mattoussi H. Multiplexed toxin analysis using four colors of quantum dot fluororeagents. Anal. Chem. 2004;76:684–688. doi: 10.1021/ac035083r. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y., Ye M., Chao Q., Jia N., Ge Y., Shen H. Simultaneous detection of multifood-borne pathogenic bacteria based on functionalized quantum dots coupled with immunomagnetic separation in food samples. J. Agric. Food Chem. 2009;57:517–524. doi: 10.1021/jf802817y. [DOI] [PubMed] [Google Scholar]
- 16.Leevy W.M., Lambert T.N., Johnson J.R., Morris J., Smith B.D. Quantum dot probes for bacteria distinguish Escherichia coli mutants and permit in vivo imaging. Chem. Commun. 2008:2331–2333. doi: 10.1039/b803590c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukhopadhyay B., Martins M.B., Karamanska R., Russell D.A., Field R.A. Bacterial detection using carbohydrate-functionalised CdS quantum dots: a model study exploiting E. coli recognition of mannosides. Tetrahedron Lett. 2009;50:886–889. [Google Scholar]
- 18.Edgar R., McKinstry M., Hwang J., Oppenheim A.B., Fekete R.A., Giulian G., Merril C., Nagashima K., Adhya S. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc. Natl Acad. Sci. USA. 2006;103:4841–4845. doi: 10.1073/pnas.0601211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klostranec J.M., Xiang Q., Farcas G.A., Lee J.A., Rhee A., Lafferty E.I., Perrault S.D., Kain K.C., Chan W.C.W. Convergence of quantum dot barcodes with microfluidics and signal processing for multiplexed high-throughput infectious disease diagnostics. Nano Lett. 2007;7:2812–2818. doi: 10.1021/nl071415m. [DOI] [PubMed] [Google Scholar]
- 20.Tallury P., Payton K., Santra S. Silica-based multimodal/multifunctional nanoparticles for bioimaging and biosensing applications. Nanomedicine. 2008;3:579–592. doi: 10.2217/17435889.3.4.579. [DOI] [PubMed] [Google Scholar]
- 21.Wang L., Zhao W., Tan W. Bioconjugated silica nanoparticles: development and applications. Nano Res. 2008;1:99–115. [Google Scholar]
- 22.Wang L., Wang K.M., Santra S., Zhao X.J., Hilliard L.R., Smith J.E., Wu J.R., Tan W.H. Watching silica nanoparticles glow in the biological world. Anal. Chem. 2006;78:646–654. [Google Scholar]
- 23.Wang L., Zhao W.J., O'Donoghue M.B., Tan W.H. Fluorescent nanoparticles for multiplexed bacteria monitoring. Bioconjug. Chem. 2007;18:297–301. doi: 10.1021/bc060255n. [DOI] [PubMed] [Google Scholar]
- 24.Wang L., Yang C., Tan W. Dual-luminophore-doped silica nanoparticles for multiplexed signaling. Nano Lett. 2005;5:37–43. doi: 10.1021/nl048417g. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X.J., Hilliard L.R., Mechery S.J., Wang Y.P., Bagwe R.P., Jin S.G., Tan W.H. A rapid bioassay for single bacterial cell quantitation using bioconjugated nanoparticles. Proc. Natl Acad. Sci. USA. 2004;101:15027–15032. doi: 10.1073/pnas.0404806101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin D.L., He X.X., Wang K.M., Zhao X.J.J., Tan W.H., Chen J.Y. Fluorescent nanoparticle-based indirect immunofluorescence microscopy for detection of Mycobacterium tuberculosis. J. Biomed. Biotechnol. 2007:9. doi: 10.1155/2007/89364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelong Ai B.Z.L.L. Europium-based fluorescence nanoparticle sensor for rapid and ultrasensitive detection of an anthrax biomarker13. Angew. Chem., Int. Ed. 2009;48:304–308. doi: 10.1002/anie.200804231. [DOI] [PubMed] [Google Scholar]
- 28.Hun X., Zhang Z.J. A novel sensitive staphylococcal enterotoxin C, fluoroimmunoassay based on functionalized fluorescent core–shell nanoparticle labels. Food Chem. 2007;105:1623–1629. [Google Scholar]
- 29.Singh A.K., Harrison S.H., Schoeniger J.S. Gangliosides as receptors for biological toxins: development of sensitive fluoroimmunoassays using ganglioside-bearing liposomes. Anal. Chem. 2000;72:6019–6024. doi: 10.1021/ac000846l. [DOI] [PubMed] [Google Scholar]
- 30.Ahn-Yoon S., DeCory T.R., Baeumner A.J., Durst R.A. Ganglioside–liposome immunoassay for the ultrasensitive detection of cholera toxin. Anal. Chem. 2003;75:2256–2261. doi: 10.1021/ac026428t. [DOI] [PubMed] [Google Scholar]
- 31.Reichert A., Nagy J.O., Spevak W., Charych D. Polydiacetylene liposomes functionalized with sialic acid bind and colorimetrically detect influenza virus. J. Am. Chem. Soc. 1995;117:829–830. [Google Scholar]
- 32.Tang S., Moayeri M., Chen Z., Harma H., Zhao J., Hu H., Purcell R.H., Leppla S.H., Hewlett I.K. Detection of anthrax toxin by an ultrasensitive immunoassay using Europium nanoparticles. Clin. Vaccine Immunol. 2009;16:408–413. doi: 10.1128/CVI.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valanne A., Huopalahti S., Soukka T., Vainionpää R., Lövgren T., Härmä H. A sensitive adenovirus immunoassay as a model for using nanoparticle label technology in virus diagnostics. J. Clin. Virol. 2005;33:217–223. doi: 10.1016/j.jcv.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Polpanich D., Tangboriboonrat P., Elaissari A., Udomsangpetch R. Detection of malaria infection via latex agglutination assay. Anal. Chem. 2007;79:4690–4695. doi: 10.1021/ac070502w. [DOI] [PubMed] [Google Scholar]
- 35.Suci P.A., Berglund D.L., Liepold L., Brumfield S., Pitts B., Davison W., Oltrogge L., Hoyt K.O., Codd S., Stewart P.S., Young M., Douglas T. High-density targeting of a viral multifunctional nanoplatform to a pathogenic biofilm-forming bacterium. Chem. Biol. 2007;14:387–398. doi: 10.1016/j.chembiol.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Liz-Marzan L.M. Tailoring surface plasmons through the morphology and assembly of metal nanoparticles. Langmuir. 2006;22:32–41. doi: 10.1021/la0513353. [DOI] [PubMed] [Google Scholar]
- 37.Eustis S., El-Sayed M.A. Why gold nanoparticles are more precious than pretty gold: noble metal surface plasmon resonance and its enhancement of the radiative and nonradiative properties of nanocrystals of different shapes. Chem. Soc. Rev. 2006;35:209–217. doi: 10.1039/b514191e. [DOI] [PubMed] [Google Scholar]
- 38.Sperling R.A., Gil P.R., Zhang F., Zanella M., Parak W.J. Biological applications of gold nanoparticles. Chem. Soc. Rev. 2008;37:1896–1908. doi: 10.1039/b712170a. [DOI] [PubMed] [Google Scholar]
- 39.Yguerabide J., Yguerabide E.E. Light-scattering submicroscopic particles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications — II. Experimental characterization. Anal. Biochem. 1998;262:157–176. doi: 10.1006/abio.1998.2760. [DOI] [PubMed] [Google Scholar]
- 40.Mirkin C.A., Letsinger R.L., Mucic R.C., Storhoff J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 41.Rosi N.L., Mirkin C.A. Nanostructures in biodiagnostics. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 42.Laderman E.I., Whitworth E., Dumaual E., Jones M., Hudak A., Hogrefe W., Carney J., Groen J. Rapid, sensitive, and specific lateral-flow immunochromatographic point-of-care device for detection of Herpes simplex virus type 2-specific immunoglobulin G antibodies in serum and whole blood. Clin. Vaccine Immunol. 2008;15:159–163. doi: 10.1128/CVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang S.-H. Gold nanoparticle-based immunochromatographic test for identification of Staphylococcus aureus from clinical specimens. Clin. Chim. Acta. 2006;373:139–143. doi: 10.1016/j.cca.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 44.Fang S.B., Tseng W.Y., Lee H.-C., Tsai C.-K., Huang J.-T., Hou S.-Y. Identification of Salmonella using colony-print and detection with antibody-coated gold nanoparticles. J. Microbiol. Methods. 2009;77:225–228. doi: 10.1016/j.mimet.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Baptista P.V., Koziol-Montewka M., Paluch-Oles J., Doria G., Franco R. Gold-nanoparticle–probe-based assay for rapid and direct detection of Mycobacterium tuberculosis DNA in clinical samples. Clin. Chem. 2006;52:1433–1434. doi: 10.1373/clinchem.2005.065391. [DOI] [PubMed] [Google Scholar]
- 46.Schofield C.L., Field R.A., Russell D.A. Glyconanoparticles for the colorimetric detection of cholera toxin. Anal. Chem. 2007;79:1356–1361. doi: 10.1021/ac061462j. [DOI] [PubMed] [Google Scholar]
- 47.Taton T.A., Mirkin C.A., Letsinger R.L. Scanometric DNA array detection with nanoparticle probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y.F., Pang D.W., Zhang Z.L., Zheng H.Z., Cao J.P., Shen J.T. Visual gene diagnosis of HBV and HCV based on nanoparticle probe amplification and silver staining enhancement. J. Med. Virol. 2003;70:205–211. doi: 10.1002/jmv.10379. [DOI] [PubMed] [Google Scholar]
- 49.Duan L.L., Wang Y.F., Li S.S.C., Wan Z.X., Zhai J.X. Rapid and simultaneous detection of human hepatitis B virus and hepatitis C virus antibodies based on a protein chip assay using nano-gold immunological amplification and silver staining method. BMC Infec. Dis. 2005;5 doi: 10.1186/1471-2334-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X.X., Cao C., Han S.J., Sim S.J. Detection of pathogen based on the catalytic growth of gold nanocrystals. Water Res. 2009;43:1425–1431. doi: 10.1016/j.watres.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 51.Griffin J., Singh A.K., Senapati D., Lee E., Gaylor K., Jones-Boone J., Ray P.C. Sequence-specific HCV RNA quantification using the size-dependent nonlinear optical properties of gold nanoparticles. Small. 2009;5:839–845. doi: 10.1002/smll.200801334. [DOI] [PubMed] [Google Scholar]
- 52.Darbha G.K., Rai U.S., Singh A.K., Ray P.C. Gold-nanorod-based sensing of sequence specific HIV-1 virus DNA by using hyper-Rayleigh scattering spectroscopy. Chem. Eur. J. 2008;14:3896–3903. doi: 10.1002/chem.200701850. [DOI] [PubMed] [Google Scholar]
- 53.Lin F.Y.H., Sabri M., Alirezaie J., Li D., Sherman P.M. Development of a nanoparticle-labeled microfluidic immunoassay for detection of pathogenic microorganisms. Clin. Diagn. Lab. Immunol. 2005;12:418–425. doi: 10.1128/CDLI.12.3.418-425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossier J.S., Perruche B., Morier P., Vollet C., Heini D., Michel P., Reymond F. Microfluidic based immunoassay. NanoBiotechnology. 2005;1:311–312. [Google Scholar]
- 55.Naja G., Bouvrette P., Hrapovic S., Luong J.H.T. Raman-based detection of bacteria using silver nanoparticles conjugated with antibodies. Analyst. 2007;132:679–686. doi: 10.1039/b701160a. [DOI] [PubMed] [Google Scholar]
- 56.Driskell J.D., Shanmukh S., Yong-Jun L., Hennigan S., Jones L., Yi-Ping Z., Dluhy R.A., Krause D.C., Tripp R.A. Infectious agent detection with SERS-active silver nanorod arrays prepared by oblique angle deposition. IEEE Sens. J. 2008;8:863–870. [Google Scholar]
- 57.He B., Morrow T.J., Keating C.D. Nanowire sensors for multiplexed detection of biomolecules. Curr. Opin. Chem. Biol. 2008;12:522–528. doi: 10.1016/j.cbpa.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sioss J.A., Stoermer R.L., Sha M.Y., Keating C.D. Silica-coated, Au/Ag striped nanowires for bioanalysis. Langmuir. 2007;23:11334–11341. doi: 10.1021/la7019846. [DOI] [PubMed] [Google Scholar]
- 59.Tok J.B.H., Chuang F.Y.S., Kao M.C., Rose K.A., Pannu S.S., Sha M.Y., Chakarova G., Penn S.G., Dougherty G.M. Metallic striped nanowires as multiplexed immunoassay platforms for pathogen detection. Angew. Chem., Int. Ed. 2006;45:6900–6904. doi: 10.1002/anie.200601104. [DOI] [PubMed] [Google Scholar]
- 60.Duguet E., Vasseur S., Mornet S., Devoisselle J.M. Magnetic nanoparticles and their applications in medicine. Nanomedicine. 2006;1:157–168. doi: 10.2217/17435889.1.2.157. [DOI] [PubMed] [Google Scholar]
- 61.Ito A., Shinkai M., Honda H., Kobayashi T. Medical application of functionalized magnetic nanoparticles. J. Biosci. Bioeng. 2005;100:1–11. doi: 10.1263/jbb.100.1. [DOI] [PubMed] [Google Scholar]
- 62.Gu H., Xu K., Xu C., Xu B. Biofunctional magnetic nanoparticles for protein separation and pathogen detection. Chem. Commun. 2006:941–949. doi: 10.1039/b514130c. [DOI] [PubMed] [Google Scholar]
- 63.Yang H., Li H., Jiang X. Detection of foodborne pathogens using bioconjugated nanomaterials. Microfluid. Nanofluid. 2008;5:571–583. [Google Scholar]
- 64.Varshney M., Yang L.J., Su X.L., Li Y.B. Magnetic nanoparticle–antibody conjugates for the separation of Escherichia coli 0157: H7 in ground beef. J. Food Prot. 2005;68:1804–1811. doi: 10.4315/0362-028x-68.9.1804. [DOI] [PubMed] [Google Scholar]
- 65.Ravindranath S.P., Mauer L.J., Deb-Roy C., Irudayaraj J. Biofunctionalized magnetic nanoparticle integrated mid-infrared pathogen sensor for food matrixes. Anal. Chem. 2009;81:2840–2846. doi: 10.1021/ac802158y. [DOI] [PubMed] [Google Scholar]
- 66.Cheng Y., Liu Y., Huang J., Li K., Zhang W., Xian Y., Jin L. Combining biofunctional magnetic nanoparticles and ATP bioluminescence for rapid detection of Escherichia coli. Talanta. 2009;77:1332–1336. doi: 10.1016/j.talanta.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 67.Yang H., Qu L., Wimbrow A.N., Jiang X., Sun Y. Rapid detection of Listeria monocytogenes by nanoparticle-based immunomagnetic separation and real-time PCR. Int. J. Food Microbiol. 2007;118:132–138. doi: 10.1016/j.ijfoodmicro.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 68.Grossman H.L., Myers W.R., Vreeland V.J., Bruehl R., Alper M.D., Bertozzi C.R., Clarke J. Detection of bacteria in suspension by using a superconducting quantum interference device. Proc. Natl Acad. Sci. USA. 2004;101:129–134. doi: 10.1073/pnas.0307128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang D., Carr D.J., Alocilja E.C. Fluorescent bio-barcode DNA assay for the detection of Salmonella enterica serovar Enteritidis. Biosens. Bioelectron. 2009;24:1377–1381. doi: 10.1016/j.bios.2008.07.081. [DOI] [PubMed] [Google Scholar]
- 70.Kell A.J., Stewart G., Ryan S., Peytavi R., Boissinot M., Huletsky A., Bergeron M.G., Simard B. Vancomycin-modified nanoparticles for efficient targeting and preconcentration of Gram-positive and Gram-negative bacteria. ACS Nano. 2008;2:1777–1788. doi: 10.1021/nn700183g. [DOI] [PubMed] [Google Scholar]
- 71.Gu H.W., Xu K.M., Xu C.J., Xu B. Biofunctional magnetic nanoparticles for protein separation and pathogen detection. Chem. Commun. 2006:941–949. doi: 10.1039/b514130c. [DOI] [PubMed] [Google Scholar]
- 72.Lin Y.S., Tsai P.J., Weng M.F., Chen Y.C. Affinity capture using vancomycin-bound magnetic nanoparticles for the MALDI-MS analysis of bacteria. Anal. Chem. 2005;77:1753–1760. doi: 10.1021/ac048990k. [DOI] [PubMed] [Google Scholar]
- 73.Ho K.C., Tsai P.J., Lin Y.S., Chen Y.C. Using biofunctionalized nanoparticles to probe pathogenic bacteria. Anal. Chem. 2004;76:7162–7168. doi: 10.1021/ac048688b. [DOI] [PubMed] [Google Scholar]
- 74.Gu H.W., Ho P.L., Tsang K.W.T., Yu C.W., Xu B. Using biofunctional magnetic nanoparticles to capture Gram-negative bacteria at an ultra-low concentration. Chem. Commun. 2003:1966–1967. doi: 10.1039/b305421g. [DOI] [PubMed] [Google Scholar]
- 75.Gao J., Li L.H., Ho P.L., Mak G.C., Gu H.W., Xu B. Combining fluorescent probes and biofunctional magnetic nanoparticles for rapid detection of bacteria in human blood. Adv. Mater. 2006;18:3145–3148. [Google Scholar]
- 76.Liu J.C., Tsai P.J., Lee Y.C., Chen Y.C. Affinity capture of uropathogenic Escherichia coli using pigeon ovalbumin-bound Fe3O4@Al2O3 magnetic nanoparticles. Anal. Chem. 2008;80:5425–5432. doi: 10.1021/ac800487v. [DOI] [PubMed] [Google Scholar]
- 77.El-Boubbou K., Gruden C., Huang X. Magnetic glyco-nanoparticles: a unique tool for rapid pathogen detection, decontamination, and strain differentiation. J. Am. Chem. Soc. 2007;129:13392. doi: 10.1021/ja076086e. [DOI] [PubMed] [Google Scholar]
- 78.Perez J.M., Josephson L., O'Loughlin T., Hogemann D., Weissleder R. Magnetic relaxation switches capable of sensing molecular interactions. Nat. Biotechnol. 2002;20:816–820. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
- 79.Josephson L., Perez J.M., Weissleder R. Magnetic nanosensors for the detection of oligonucleotide sequences. Angew. Chem., Int. Ed. 2001;40:3204–3206. doi: 10.1002/1521-3773(20010903)40:17<3204::AID-ANIE3204>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 80.Yang S.Y., Chieh J.J., Wang W.C., Yu C.Y., Lan C.B., Chen J.H., Horng H.E., Hong C.-Y., Yang H.C., Huang W. Ultra-highly sensitive and wash-free bio-detection of H5N1 virus by immunomagnetic reduction assays. J. Virol. Methods. 2008;153:250–252. doi: 10.1016/j.jviromet.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 81.Yang H., Qu L.W., Lin Y., Jiang X.P., Sun Y.P. Detection of Listeria monocytogenes in biofilms using immunonanoparticles. J. Biomed. Nanotechnol. 2007;3:131–138. [Google Scholar]
- 82.Yager P., Domingo G.J., Gerdes J. Point-of-care diagnostics for global health. Ann. Rev. Biomed. Eng. 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- 83.Goho A. Diagnosing the Developing World. Sci. News. 2004;165:108. [Google Scholar]
- 84.Maclurcan D.C. Nanotechnology and developing countries: part 1: what possibilities? J. Nanotechnology Online. 2005;1:1–10. [Google Scholar]