Abstract
Apart from bat-SARS-CoV, we have identified a novel group 1 coronavirus, bat-CoV HKU2, in Rhinolophus sinicus (Chinese horseshoe bats). Since it has been suggested that the receptor-binding motif (RBM) of SARS-CoV may have been acquired from a group 1 coronavirus, we conducted a surveillance study and identified bat-SARS-CoV and bat-CoV HKU2 in 8.7% and 7.5% respectively of R. sinicus in Hong Kong and Guangdong. Complete genome sequencing of four strains of bat-CoV HKU2 revealed the smallest coronavirus genome (27164 nucleotides) and a unique spike protein evolutionarily distinct from the rest of the genome. This spike protein, sharing similar deletions with other group 2 coronaviruses in its C-terminus, also contained a 15-amino acid peptide homologous to a corresponding peptide within the RBM of spike protein of SARS-CoV, which was absent in other coronaviruses except bat-SARS-CoV. These suggest a common evolutionary origin in the spike protein of bat-CoV HKU2, bat-SARS-CoV, and SARS-CoV.
Keywords: Chinese horseshoe bats, Coronavirus, HKU2, SARS, Novel, Receptor, Origin
Introduction
Coronaviruses can infect a wide variety of animals in which they can cause respiratory, enteric, hepatic and neurological diseases of varying severity. Based on genotypic and serological characteristics, coronaviruses were classified into three distinct groups (Brian and Baric, 2005, Lai and Cavanagh, 1997, Ziebuhr, 2004). As a result of the unique mechanism of viral replication, coronaviruses have a high frequency of recombination (Lai and Cavanagh, 1997). Such a high recombination rate, coupled with the infidelity of the polymerases of RNA viruses, may allow them to adapt to new hosts and ecological niches (Herrewegh et al., 1998, Woo et al., 2006c).
The severe acute respiratory syndrome (SARS) epidemic in 2003, caused by a novel coronavirus, SARS coronavirus (SARS-CoV), has aroused interests in the discovery of novel coronaviruses in both humans and animals (Guan et al., 2003, Marra et al., 2003, Peiris et al., 2003, Rota et al., 2003, Woo et al., 2004). Before that, only 19 (two human, 13 mammalian and four avian) coronaviruses were known. After the epidemic, two novel human coronaviruses, human coronavirus NL63 (HCoV-NL63), a group 1 coronavirus, and coronavirus HKU1 (CoV-HKU1), a group 2 coronavirus, have been discovered (Fouchier et al., 2004, Lau et al., 2006, van der Hoek et al., 2004, Woo et al., 2005a, Woo et al., 2005b). In the recent two years, at least 10 previously unrecognized coronaviruses from bats were also described in Hong Kong and mainland China (Lau et al., 2005, Li et al., 2005b, Poon et al., 2005, Tang et al., 2006, Woo et al., 2006a, Woo et al., 2006d), suggesting that bats play an important role in the ecology and evolution of coronaviruses.
Although the identification of SARS-CoV-like viruses in Himalayan palm civets and raccoon dogs in live-animal markets in southern China suggested that wild animals could be the origin of SARS (Guan et al., 2003), the absence of related viruses in wild civets in extensive surveillance studies and the rapid evolution of SARS-CoV genomes in market civets suggested that these caged animals were likely only intermediate hosts and there is a yet unidentified natural reservoir for SARS-CoV (Li et al., 2005a, Song et al., 2005, Tu et al., 2004, Yang et al., 2005). Recently, we have described the discovery of a SARS-CoV-like virus, bat SARS coronavirus (bat-SARS-CoV), in Chinese horseshoe bats in Hong Kong (Lau et al., 2005). Similar viruses have also been found in other species of horseshoe bats in mainland China (Li et al., 2005b), suggesting that horseshoe bats are reservoir of SARS-CoV-like viruses. However, genome sequence comparison of SARS-CoV-like coronaviruses from horseshoe bats and human/civet SARS-CoV showed that they shared only 88–92% nucleotide identities. More importantly, the amino acid sequence identities between the spike (S) proteins of bat and human/civet viruses were only 78–80% (Lau et al., 2005, Li et al., 2005b, Ren et al., 2006). Therefore, events such as mutation and/or recombination would have occurred during the evolution of these SARS-CoV-like viruses before the possible emergence of direct progenitors of SARS-CoV capable of infecting palm civets and subsequently humans.
In a recent report on angiotensin-converting enzyme 2 (ACE2)–S protein interactions of SARS-CoV, it was suggested that the receptor-binding motif (RBM) of SARS-CoV may have been acquired from a group 1 virus related to HCoV-NL63 (Li et al., 2006). Interestingly, a novel group 1 coronavirus, bat coronavirus HKU2 (bat-CoV HKU2), was identified in Chinese horseshoe bats in addition to bat-SARS-CoV in our previous surveillance studies (Lau et al., 2005, Woo et al., 2006d). To better understand the epidemiology and evolution of bat-CoV HKU2 and explore possible recombination events between this group 1 coronavirus and bat-SARS-CoV that could have led to the emergence of SARS-CoV, we conducted an extensive surveillance for coronaviruses in Chinese horseshoe bats in Hong Kong and Guangdong, the province in southern China where the SARS epidemic originated, over a 2-year period. Four complete genomes of bat-CoV HKU2, three from Hong Kong and one from Guangdong, were also sequenced and analyzed. Comparison of bat-CoV HKU2 genomes with other coronavirus genomes revealed a spike protein distinct from the spike proteins of other group 1 coronaviruses, with a peptide homologous to a segment of the RBM of the S protein of SARS-CoV.
Results
Coronavirus surveillance in Chinese horseshoe bats
A total of 770 respiratory and alimentary specimens from 348 and 64 Chinese horseshoe bats were obtained in Hong Kong and in the Guangdong province in Southern China, respectively. RT-PCR for a 440-bp fragment in the RdRp genes of coronaviruses was positive in alimentary specimens from 58 (16.7%) of the 348 bats from Hong Kong, and from 8 (12.5%) of the 64 bats from Guandong. None of the respiratory specimens was positive. Sequencing results suggested the presence of two different coronaviruses among the 64 positive bats. Of the 58 positive bats from Hong Kong, the sequences of 29 samples possessed ≥ 99% nucleotide identities to bat-CoV HKU2 (GenBank accession no. DQ249235), while those of the other 29 samples possessed ≥ 99% nucleotide identities to bat-SARS-CoV (GenBank accession no. DQ022305) (Lau et al., 2005, Woo et al., 2006d). The bats positive for bat-CoV HKU2 and bat-SARS-CoV were from nine of the 18 sampling locations in Hong Kong, with bats from three locations harboring both viruses (Fig. 1 ). Of the eight positive bats from Guangdong, the sequences of six alimentary samples possessed 97–98% nucleotide identities to bat-CoV HKU2, while that of one possessed 98% nucleotide identities to bat-SARS-CoV. The remaining positive sample contained both bat-CoV HKU2 and bat-SARS-CoV with 98% nucleotide identities. Attempts to stably passage bat-CoV HKU2 in cell lines were unsuccessful.
Fig. 1.
Map showing locations of sampling in Hong Kong. Dark circles represent locations positive for bat-CoV HKU2, squares represent locations positive for bat-SARS-CoV, and triangles represent locations positive for both bat-CoV HKU2 and bat-SARS-CoV. Blank circles represent locations negative for bat-SARS-CoV and bat-CoV HKU2. Location A was where bat-CoV HKU2/HK/33/2004 was found, location B was where bat-CoV HKU2/HK/298/2004 was found, and location C was where bat-CoV HKU2/HK/46/2006 was found.
Characterization of bat-CoV HKU2 genomes
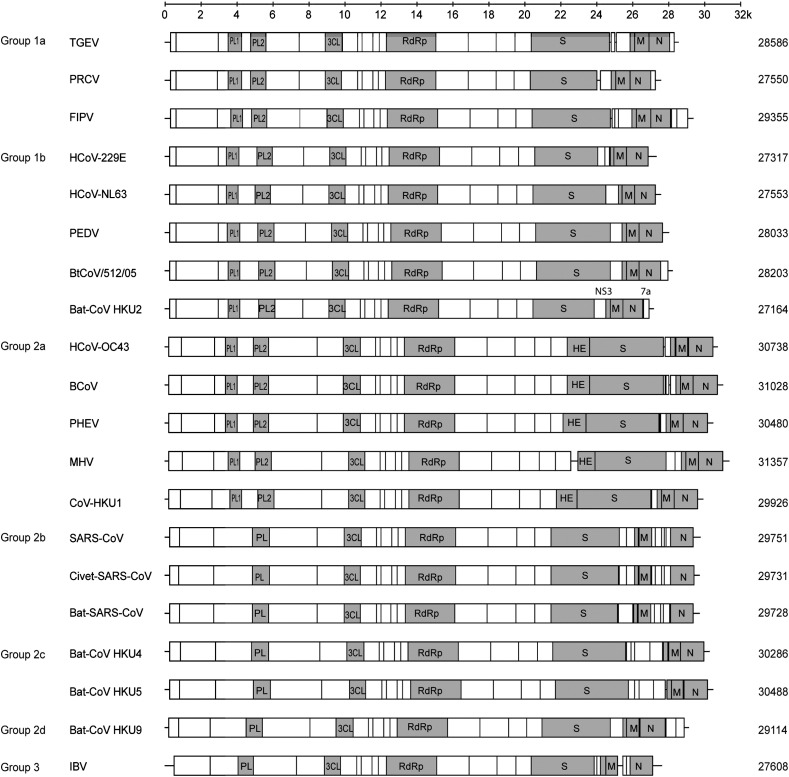
Complete genome sequence data of four strains of bat-CoV HKU2 were obtained by assembly of the sequences of the RT-PCR products obtained directly from four individual specimens collected at different time and places. Three strains were obtained from Hong Kong (bat-CoV HKU2/HK/33/2004, bat-CoV HKU2/HK/298/2004 and bat-CoV HKU2/HK/46/2006) (Fig. 1), while one was obtained from Guangdong (bat-CoV HKU2/GD/430/2006). Their genomes were 27,164-nucleotide, polyadenylated RNA, the smallest genome size among all coronaviruses with genome sequences available (Table 1 and Fig. 2 ). The G + C content was 39% (Table 1). The four strains share the same genome structures and were highly similar in their nucleotide sequence. The three Hong Kong strains were more closely related to each other with 99.9% overall nucleotide identities, while that from Guangdong had 98.5% nucleotide identities with the three Hong Kong strains. Their genome organization was similar to other coronaviruses (Table 2 and Fig. 2). Bat-CoV HKU2 possessed the putative transcription regulatory sequence (TRS) motif, 5′-AACUAAA-3′, at the 3′ end of the leader sequence and precedes each ORF (Table 2). This TRS has also been shown to be the TRS for HCoV-NL63 (Pyrc et al., 2004), whereas a shorter sequence, 5″-CUAAAC-3′, was found to be the TRS for other group 1 coronaviruses such as TGEV and FIPV (Dye and Siddell, 2005, Hiscox et al., 1995).
Table 1.
Comparison of genomic features of bat-CoV HKU2 and other coronaviruses and amino acid identities between the predicted 3CLpro, RdRp, Hel, S, E, M, and N proteins of bat-CoV HKU2 and the corresponding proteins of other coronaviruses
| Coronavirusesa | Genome |
Features |
Pairwise amino acid identity (%) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Size (bases) | G + C content | 3CLpro | RdRp | Hel | S | E | M | N | |
| Group 1a | |||||||||
| TGEV | 28586 | 0.38 | 63.2 | 75.8 | 77.5 | 21.9 | 28.9 | 47.3 | 40.3 |
| FIPV | 29355 | 0.38 | 61.3 | 75.9 | 77.6 | 22.3 | 28.9 | 45.5 | 41.6 |
| PRCV | 27550 | 0.37 | 62.9 | 75.6 | 77.3 | 26.0 | 30.1 | 47.5 | 40.0 |
| Group 1b | |||||||||
| HCoV-229E | 27317 | 0.38 | 64.2 | 81.2 | 81.1 | 27.0 | 51.3 | 56.5 | 44.5 |
| HCoV-NL63 | 27553 | 0.34 | 64.0 | 79.4 | 81.6 | 25.7 | 50.0 | 59.6 | 46.8 |
| PEDV | 28033 | 0.42 | 65.2 | 78.5 | 78.4 | 24.0 | 46.2 | 64.8 | 39.6 |
| BtCoV/512/2005 | 28203 | 0.40 | 62.3 | 77.8 | 78.2 | 25.9 | 50.0 | 61.3 | 46.9 |
| Bat-CoV HKU6 | NAb | NA | NA | 77.9 | 78.2 | NA | NA | NA | NA |
| Bat-CoV HKU7 | NA | NA | NA | 82.7 | 80.6 | NA | NA | NA | 46.3 |
| Bat-CoV HKU8 | NA | NA | NA | 80.7 | 81.6 | NA | NA | NA | 48.7 |
| Bat-CoV 1A (partial CDS) | NA | NA | NA | 80.0 | NA | NA | NA | NA | 50.5 |
| Bat-CoV 1B (partial CDS) | NA | NA | NA | 80.0 | NA | NA | NA | NA | 50.2 |
| Bat-CoV HKU2 | 27164 | 0.39 | – | – | – | – | – | – | – |
| Group 2a | |||||||||
| CoV-HKU1 | 29926 | 0.32 | 45.5 | 56.6 | 53.6 | 24.5 | 30.4 | 34.3 | 27.8 |
| HCoV-OC43 | 30738 | 0.37 | 44.2 | 57.4 | 54.5 | 24.1 | 32.3 | 36.6 | 24.7 |
| MHV | 31357 | 0.42 | 46.9 | 56.3 | 54.2 | 24.8 | 28.6 | 36.5 | 26.9 |
| BCoV | 31028 | 0.37 | 44.2 | 57.3 | 54.6 | 23.4 | 32.3 | 36.3 | 23.9 |
| PHEV | 30480 | 0.37 | 43.9 | 57.2 | 54.5 | 24.5 | 33.3 | 35.7 | 24.7 |
| Group 2b | |||||||||
| SARS-CoV | 29751 | 0.41 | 43.5 | 59.9 | 61.3 | 25.6 | 27.3 | 30.7 | 22.4 |
| Bat-SARS-CoV HKU3 | 29728 | 0.41 | 43.8 | 59.8 | 60.8 | 25.9 | 27.3 | 31.2 | 22.4 |
| Group 2c | |||||||||
| Bat-CoV HKU4 | 30286 | 0.38 | 47.7 | 59.4 | 61.8 | 23.6 | 22.6 | 33.9 | 26.3 |
| Bat-CoV HKU5 | 30488 | 0.43 | 46.6 | 58.7 | 61.8 | 23.5 | 25.3 | 30.6 | 26.4 |
| Group 2d | |||||||||
| Bat-CoV HKU9 | 29114 | 0.41 | 45.6 | 58.0 | 61.8 | 25.5 | 23.8 | 34.2 | 19.3 |
| Group 3 | |||||||||
| IBV | 27608 | 0.38 | 40.1 | 58.5 | 56.1 | 24.2 | 18.9 | 22.7 | 22.1 |
TGEV, porcine transmissible gastroenteritis virus; FIPV, feline infectious peritonitis virus; PRCV, porcine respiratory coronavirus; HCoV-229E, human coronavirus 229E; HCoV-NL63, human coronavirus NL63; PEDV, porcine epidemic diarrhea virus; CoV-HKU1, coronavirus HKU1; HCoV-OC43, human coronavirus OC43; MHV, murine hepatitis virus; BCoV, bovine coronavirus; PHEV, porcine hemagglutinating encephalomyelitis virus; SARS-CoV, SARS coronavirus; bat-SARS-CoV HKU3, bat SARS coronavirus HKU3; IBV, infectious bronchitis virus.
NA, data not available for analysis.
Fig. 2.
Genome organizations of bat-CoV HKU2 compared to representative coronaviruses from each group. The conserved functional domains ORF 1ab and the structural proteins are represented by gray boxes. The genome sizes (bp) are shown on the right.
Table 2.
Coding potential and putative transcription regulatory sequences of bat-CoV HKU2
| Coronaviruses | ORFs | Start-end (nucleotide position) | No. of nucleotides | No. of amino acids | Frame | Putative TRS |
|
|---|---|---|---|---|---|---|---|
| Nucleotide position in genome | TRS sequence | ||||||
| Bat-CoV HKU2 | 1ab | 297–20,479 (shift at 12,446) | 20,183 | 6727 | +3/+2 | 122 | AACUAAAC(167)AUG |
| S | 20,476–23,862 | 3387 | 1128 | +1 | 20,470 | AACUAAAUG | |
| NS3 | 23,862–24,551 | 690 | 229 | +3 | 23,817 | AACUAAAC(37)AUG | |
| E | 24,532–24,759 | 228 | 75 | +1 | 24,523 | AACUAAAC(1)AUG | |
| M | 24,768–25,457 | 690 | 229 | +3 | 24,754 | AACUAAAC(6)AUG | |
| N | 25,469–26,596 | 1128 | 375 | +2 | 25,452 | AACUAAAC(9)AUG | |
| NS7a | 26,608–26,907 | 300 | 99 | +1 | 26,600 | AACUAAACAUG | |
Similar to other coronaviruses, the replicase ORF1ab encodes a number of putative proteins, including nsp3 [which contains the putative papain-like protease (PLpro)], nsp5 [putative chymotrypsin-like protease (3CLpro)], nsp12 (putative RdRp), nsp13 [putative helicase (Hel)], which are produced by proteolytic cleavage by PLpro and 3CLpro at specific sites (Woo et al., 2005c). Similar to other group 1 coronaviruses, the genome of bat-CoV HKU2 has two putative PLpro, which are homologous to PL1pro and PL2pro of other group 1 coronaviruses (Fig. 2).
One ORF, which encodes a putative 229-amino acid nonstructural protein, NS3, was observed between the S and E genes. This NS3 possessed 42% amino acid identities to the NS3 of HCoV-NL63, 37% identities to that of BtCoV/512/05, 36% identities to that of PEDV, and 29% identities to the NS3b of TGEV. No functional domains were identified by PFAM and InterProScan. TMHMM analysis showed three putative transmembrane domains in NS3 of bat-CoV HKU2 (residues 38–60, 81–103, and 118–140).
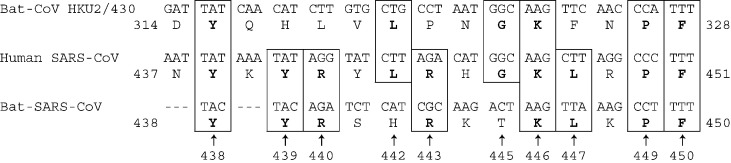
The most striking feature of bat-CoV HKU2 genome was observed in its S protein which possessed the shortest amino acid sequence (1128 amino acid residues) among the S proteins of all coronaviruses, as a result of deletions in the N-terminal region (Supplementary Fig. 1). It had ≤ 27% amino acid identities to the S proteins of all known coronaviruses, as opposed to other genes which showed higher amino acid identities to the corresponding genes in other group 1 coronaviruses (especially group 1b) than to group 2 and group 3 coronaviruses (Table 1). When the S protein of bat-CoV HKU2 is aligned with the S protein of other group 1 coronaviruses, many of the amino acid residues conserved among and specific to group 1b coronaviruses were not found; whereas residues conserved among all coronaviruses, especially those in the C-terminal region, were identified (Supplementary Fig. 1). In fact, the N-terminal region of the S protein of bat-CoV HKU2 possessed very low amino acid identities to the corresponding regions in any group of coronaviruses, which was due to both amino acid substitutions and deletions. Despite this, a short peptide consisting of 15 amino acids (residues 314 to 328) was found to be homologous to a corresponding peptide within the RBM in the S1 domain of SARS-CoV (residues 437 to 451) (Fig. 3 ). A similar peptide was also observed in bat-SARS-CoV, but not in any other known coronaviruses, suggesting that it is specific to SARS-CoV, bat-SARS-CoV and bat-CoV HKU2, with a common origin. Of the 15 amino acids within this homologous peptide, six (tyrosine 438, leucine 442, glycine 445, lysine 446, proline 449, and phenylalanine 450) were conserved between SARS-CoV and bat-CoV HKU2, with four using identical codons. Of these six amino acid residues, only four (tyrosine 438, lysine 446, proline 449, and phenylalaine) were found in bat-SARS-CoV, with two using identical codons. On the other hand, four additional amino acid residues (tyrosine 439, arginine 440, arginine 443, and leucine 447), not found in bat-CoV HKU2, were conserved between SARS-CoV and bat-SARS-CoV, though with different codon usage. In contrast to a previous study which suggested that the extended receptor-binding domain of HCoV-NL63 includes a stretch of residues with weak homology to the RBM of SARS-CoV (unpublished observations, Li et al., 2006), we and another group of researchers did not identify any significant homology between the spike protein of the two coronaviruses (Hofmann et al., 2006). When compared to the S proteins of other group 1 coronaviruses and SARS-CoV, large deletions were observed in the S protein of bat-CoV HKU2 in the region corresponding to the RBM of SARS-CoV. Since the amino acid sequence of the S protein of bat-SARS-CoV also differed significantly from that of SARS-CoV in this region, it is likely that this is a site of frequent mutation and/or recombination among coronaviruses in Chinese horseshoe bats. This highly variable region within the S protein of bat-CoV HKU2 and bat-SARS-CoV may have been important for host receptor adaptation. Although the overall amino acid identities of the S protein of bat-CoV HKU2 were equally low when compared to the S proteins of all three groups of coronaviruses, the S protein of bat-CoV HKU2 shares the two conserved regions of deletions both of 14 amino acids among group 2 coronaviruses in its C-terminus (Supplementary Fig. 1). This suggests that this segment of the S protein of bat-CoV HKU2 may have co-evolved with the corresponding regions in group 2 coronaviruses. Nevertheless, the receptor for bat-CoV HKU2 remains to be determined. Aminopeptidase N (CD13) has been shown to be the receptor for many group 1 coronaviruses including HCoV-229E, canine coronavirus, FIPV, PEDV, and TGEV (Delmas et al., 1992, Yeager et al., 1992). As for group 2 coronaviruses, carcinoembryonic antigen-cell adhesion molecule 1 (CEACAM1) was identified as the receptor for murine hepatitis virus (MHV), while sialic acids were found to be the receptor for bovine coronavirus (BCoV) and human coronavirus OC43 (HCoV-OC43) (Krempl et al., 1995, Williams et al., 1991). However, human ACE2 (hACE2) have been shown to be the receptor for both SARS-CoV, a group 2 coronavirus, and HCoV-NL63, a group 1 coronavirus, although the two viruses utilize different binding sites for receptor recognition (Hofmann et al., 2005, Li et al., 2003). The S protein of bat-CoV HKU2 does not exhibit significant homology to the known receptor binding domains of HCoV-229E, HCoV-NL63, or MHV (Bonavia et al., 2003, Hofmann et al., 2006, Kubo et al., 1994). Further experiments are required to delineate the receptor for bat-CoV HKU2.
Fig. 3.
A short stretch of peptide within the RBM of S protein of SARS-CoV with homology to the corresponding region in the S of bat-CoV HKU2 and bat-SARS-CoV. The conserved amino acids are in bold and boxed.
At the 3′ end of the genome after the N gene, there is one ORF that encodes a 99-amino acid nonstructural protein, NS7a. BLAST search revealed no amino acid similarities between this putative nonstructural protein and other known proteins and no functional domain was identified by PFAM and InterProScan. TMHMM analysis showed two putative transmembrane domains in NS7a (residues 4–26 and 59–81). Previously, FIPV and TGEV, both group 1a coronavirus, were the only coronaviruses known to possess genes downstream of N (Fig. 1). It has been suggested that the two genes downstream of N in FIPV may be important for virulence (Haijema et al., 2004, Olsen, 1993). In TGEV, the gene downstream of N has been suggested to play a role in membrane association of replication complexes or assembly of the virus (Tung et al., 1992). In our recent report on the discovery of bat coronavirus HKU9, a novel bat coronavirus belonging to group 2d coronaviruses, two ORFs downstream to N were also found (Woo et al., 2006a). In another group 1b coronavirus recently identified from bats in China, BtCoV/512/05, an ORF downstream to N was also identified (Tang et al., 2006). These suggest that ORFs downstream to N can be present in coronaviruses other than group 1a and may be more prevalent among bat coronaviruses. Further experiments will delineate the function of such ORFs in bat coronaviruses.
Phylogenetic analyses
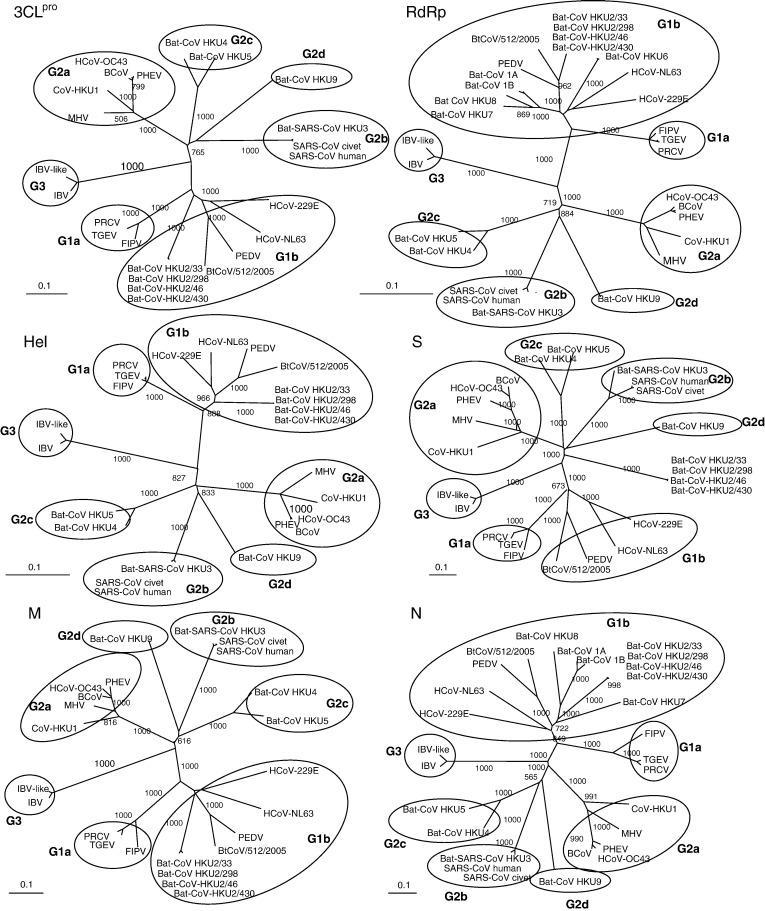
The phylogenetic trees constructed using the amino acid sequences of the 3CLpro, RdRp, Hel, S, M, and N of bat-CoV HKU2 and other coronaviruses are shown in Fig. 4 and the corresponding pairwise amino acid identities are shown in Table 1. As shown in all six trees, the four strains of bat-CoV HKU2 were clustered together, reflecting their high sequence similarities. For all the genes except S, bat-CoV HKU2 formed a distinct branch that clustered with other group 1 coronaviruses. This is supported by the higher amino acid identities to the corresponding genes in other group 1 coronaviruses (especially group 1b) than to those of group 2 and group 3 coronaviruses (Table 1). However, for the S gene, bat-CoV HKU2 formed a branch distinct from the three groups of known coronaviruses. The same tree topology was obtained when using the maximum likelihood method and Bayesian approach (data not shown). This finding is in line with results obtained from pairwise amino acid comparisons, which showed that the S of bat-CoV HKU2 possessed equally low amino acid identities (≤ 27%) to the S of all three groups of coronaviruses (Table 1).
Fig. 4.
Phylogenetic analysis of 3CLpro, RdRp, Hel, S, M, and N of bat-CoV HKU2. The trees were constructed by neighbor joining method using Kimura's two-parameter correction and bootstrap values calculated from 1000 trees. 306, 949, 609, 1758, 270, and 586 amino acid positions in 3CLpro, RdRp, Hel, S, M, and N, respectively, were included in the analysis. The scale bar indicates the estimated number of substitutions per 10 amino acids. HCoV-229E, human coronavirus 229E; PEDV, porcine epidemic diarrhea virus; TGEV, porcine transmissible gastroenteritis virus; FIPV, feline infectious peritonitis virus; HCoV-NL63, human coronavirus NL63; CoV-HKU1, coronavirus HKU1; HCoV-OC43, human coronavirus OC43; MHV, murine hepatitis virus; BCoV, bovine coronavirus; PHEV, porcine hemagglutinating encephalomyelitis virus; IBV, infectious bronchitis virus; SARS-CoV, SARS coronavirus; bat-SARS-CoV HKU3, bat-SARS-like coronavirus HKU3; bat-CoV HKU4, bat coronavirus HKU4; bat-CoV HKU5, bat coronavirus HKU5; bat-CoV HKU9, bat coronavirus HKU9.
Recombination analysis
To evaluate if segments of the SARS-CoV genome have arisen as a result of recombination between bat-SARS-CoV and bat-CoV HKU2, a sliding window analysis was conducted. No statistical support for recombination was obtained, which may be due to the high sequence divergence between the bat-SARS-CoV and bat-CoV HKU2 genomes.
Estimation of synonymous and non-synonymous substitution rates
The Ka/Ks ratios for the various coding regions in bat-CoV HKU2 are shown in Table 3 . Higher Ka/Ks ratios were observed within ORF1ab, especially nsp3 (which encodes the putative PLpro domains), nsp5 (which encodes the putative 3CLpro), and nsp14 (which encodes the helicase), whereas the ratios appeared to be lower among the structural genes. Notably, the Ka/Ks ratio for the S of bat-CoV HKU2 is only 0.03, suggesting that this gene is unlikely undergoing rapid evolution under positive selection.
Table 3.
Estimation of non-synonymous substitution and synonymous rates in the genomes of bat-CoV HKU2
| Coding regions | Ka/Ks |
|---|---|
| Bat-CoV HKU2 | |
| nsp1 | 0.197 |
| nsp2 | 0.105 |
| nsp3 | 0.470 |
| nsp4 | 0.059 |
| nsp5 | 0.320 |
| nsp6 | 0.133 |
| nsp7 | Ka = 0, Ks = 0.01925 |
| nsp8 | 0.855 |
| nsp9 | Ka = 0, Ks = 0.00864 |
| nsp10 | Ka = 0, Ks = 0 |
| nsp11 | Ka = 0, Ks = 0 |
| nsp12 | 0.037 |
| nsp13 | 0.027 |
| nsp14 | 0.338 |
| nsp15 | 0.178 |
| nsp16 | Ka = 0.00071, Ks = 0 |
| S | 0.030 |
| NS3 | 0.194 |
| E | 0.098 |
| M | 0.148 |
| N | 0.076 |
| NS7a | Ka = 0, Ks = 0.01847 |
Discussion
In this study, bat-CoV HKU2 was found among 29 (8.3%) of 348 Chinese horseshoe bats from Hong Kong and 7 (10.9%) of 64 bats from Guangdong. All bats infected with bat-CoV HKU2 appeared healthy. The finding that bat-CoV HKU2 can only be detected in alimentary specimens suggests that it possesses enteric tropism. The genomes of the four strains of bat-CoV HKU2 being sequenced were highly similar, with conserved nucleotide and amino acid sequences in most of their genes (Fig. 4). Traditionally, coronaviruses have been classified into groups 1, 2, and 3. Based on a comprehensive comparative analysis of the genomes of the various groups of coronaviruses, coronaviruses can be classified into group 1 (subgroups 1a and 1b), group 2 (subgroups 2a, 2b, 2c, and 2d) and group 3 (Woo et al., 2006a), with SARS-CoV being classified as group 2b coronaviruses (Eickmann et al., 2003, Snijder et al., 2003). Comparative amino acid sequence analysis showed that the predicted proteins in bat-CoV HKU2, except the S protein, were most similar to subgroup 1b of group 1 coronaviruses than to other groups of coronaviruses (Table 1). Based on phylogenetic analysis of the 3CLpro, RdRp, Hel, M, and N genes, the four strains of bat-CoV HKU2 formed a distinct branch within subgroup 1b of group 1 coronaviruses. They also possessed genomic features most similar to other members within this subgroup (Fig. 2). The genomes of group 1a coronaviruses encode two to three nonstructural proteins between S and E, whereas most group 1b coronaviruses encode only one such protein, except HCoV-229E which encodes two (Thiel et al., 2001). The genome organization of bat-CoV HKU2, most similar to Bt/CoV/512/05, a recently reported bat coronavirus from Scotophilus kuhlii in China, contains a small ORF downstream to the N gene, which is not observed in other group 1b coronaviruses. These results support that bat-CoV HKU2 represents a novel member within subgroup 1b of group 1 coronaviruses.
The S protein of bat-CoV HKU2 possesses several unique features. First, it represents the shortest S protein among the S proteins of known coronaviruses, as a result of substantial deletions especially in the N-terminal region corresponding to the RBM of SARS-CoV. These deletions within the S protein were also largely responsible for the smallest coronavirus genome observed among all coronaviruses. Second, although comparative genome analysis strongly suggests that bat-CoV HKU2 belonged to group 1b coronaviruses, its S protein is not closely related to the S proteins of any known coronaviruses. The S proteins of coronaviruses, being responsible for receptor binding and host species adaptation, are known to be one of the most variable regions within coronavirus genomes. Nevertheless, S proteins of coronaviruses within the same group or subgroup are more closely related among themselves than to members from a different group or subgroup, as shown in the same cluster upon phylogenetic analysis (Fig. 4). As demonstrated in a previous study, the within-group amino acid similarities of the S proteins of coronaviruses ranged from 59 to 91% while between-group similarities were from 22 to 36% (Tang et al., 2006). In particular, the within-group similarity of the S proteins of group 1 coronavirus was found to be 59%. In contrast, the S protein of bat-CoV HKU2 possessed ≤ 27% amino acid identities to the S proteins of any known coronaviruses and formed a distinct branch away from the three groups of coronaviruses on phylogenetic analysis, suggesting that this gene had a very different phylogenetic position and hence evolutionary history as compared to other regions within the genome of bat-CoV HKU2. This virus would have either acquired this unique S protein from a yet unidentified coronavirus through recombination, or undergone rapid evolution in its S protein because of strong selective pressure. Since the Ka/Ks ratio for the S gene of bat-CoV HKU2 was found to be low when using the four strains collected from different sites and dates (Table 3), the latter hypothesis would be less supported. Moreover, further analysis revealed a unique short peptide with significant homology to a corresponding peptide within the RBM of SARS-CoV, which was not seen in any other coronaviruses except bat-SARS-CoV. The C-terminus of the S protein of bat-CoV HKU2 also contained regions of deletions conserved among group 2 coronaviruses. Therefore, the S protein of bat-CoV HKU2 is likely to share a common origin with other group 2 coronaviruses, especially group 2b coronaviruses, although bat-CoV HKU2 belongs to group 1 coronaviruses. This suggests that the S of bat-CoV HKU2 could have been acquired from a group 2 or related coronavirus by recombination. Although recombination between different groups of coronaviruses has not been reported previously, targeted recombination between MHV and it has been proposed that recombination may have occurred between influenza C virus and coronavirus (Luytjes et al., 1988). Since the hemagglutinin esterase (HE), a unique protein only found in group 2 but not in group 1 or 3 coronaviruses, shared 30% amino acid homology to the hemagglutinin protein of influenza C virus, it was suggested that the HE of group 2 coronaviruses could have been acquired from influenza C virus by their ancestor through recombination. The present data suggest that the S protein of bat-CoV HKU2, bat-SARS-CoV, and SARS-CoV could have originated from an unknown ancestor coronavirus and was thereafter separately evolved, with the 15-amino acid homologous region being left-in molecular signatures. Further studies are required to elucidate the possible common ancestor virus and its host species.
Although it remains to be determined if bats are reservoir for the direct precursor of SARS-CoV, Chinese horseshoe bats are a potential mixing vessel for the generation of new coronavirus variants. Apart from bat-CoV HKU2, bat-SARS-CoV was also found among 29 (8.3%) Chinese horseshoe bats from Hong Kong and 2 (3.1%) bats from Guangdong in the present study. Coinfection by both bat-CoV HKU2 and bat-SARS-CoV was also found in one bat from China. In our previous study, bat-CoV HKU2 was also detected in a bat positive for antibodies against bat-SARS-CoV (Lau et al., 2005). Recombination, a characteristic feature of coronaviruses, has been observed between both different strains of the same coronavirus species and different species of coronaviruses. Recombination between different strains of coronaviruses was first recognized in MHV, which has been utilized as a valuable molecular tool in the generation of mutants by targeted RNA recombination (Keck et al., 1988). Similar phenomenon was subsequently demonstrated in other coronaviruses such as infectious bronchitis virus, a group 3 coronavirus and between MHV and BCoV, both being group 2 coronaviruses (Kottier et al., 1995, Lavi et al., 1998). Recently, by complete genome analysis of 22 strains of CoV-HKU1, we have also documented for the first time natural recombination events in a human coronavirus giving rise to at least three different genotypes (Woo et al., 2006c). Recombination between two different species of coronavirus, feline coronavirus type I and canine coronavirus, has also been suggested to be responsible for generation of feline coronavirus type II (Herrewegh et al., 1998). Although the existing data did not provide enough evidence for recombination between bat-CoV HKU2 and bat-SARS-CoV in the generation of SARS-CoV, their co-infection of the same bat species would allow ample opportunities for recombination and emergence of other SARS-CoV-like viruses capable of inter-species transmission.
The role of bats in the evolution and ecology of coronaviruses is yet to be explored. The existence of coronaviruses in bats was unknown until after the SARS epidemic when we first identified a novel group 1 coronavirus and bat-SARS-CoV from bats in Hong Kong (Lau et al., 2005, Poon et al., 2005). An astonishing diversity of coronaviruses was subsequently found among the bat population in Hong Kong and other parts of China (Li et al., 2005b, Tang et al., 2006, Woo et al., 2006a, Woo et al., 2006d). Since bats are commonly found and served in wild animal markets and restaurants in Guangdong (Woo et al., 2006b), and given their species diversity, roosting behavior, and migrating ability, these animals could well be the source for emergence of zoonotic epidemics like SARS. In a previous study, it has been suggested that there was species-specific host restriction of coronavirus in bats, with most coronaviruses from a single bat species clustered together (Tang et al., 2006). However, there is evidence that one bat species can be infected by more than one coronavirus species, and more than one bat species can be infected by the same coronavirus. The consistent detection of bat-CoV-HKU2 and bat-SARS-CoV in Chinese horseshoe bats over the 2-year study period from both Hong Kong and Guangdong suggested that this bat species is an established reservoir for both viruses which belonged to two different groups. Chinese horseshoe bat, under the family Rhinolophidae, is a common insectivorous species found in Hong Kong and China. Apart from Rhinolophus sinicus, R. ferrumequinum, another horseshoe bat species found in China, has also been found to harbor both group 1 and group 2 coronaviruses (Tang et al., 2006). Therefore, it is likely that bats, especially members of Rhinolophidae, can be infected by both group 1 and group 2 coronaviruses, a situation similar to humans who can be infected by group 1 (HCoV-229E and HCoV-NL63) and group 2 (SARS-CoV, HCoV-OC43, and CoV-HKU1) coronaviruses. As for the infection of more than one bat species by the same coronavirus, SARS-CoV-like viruses have been detected in at least three different species of Rhinolophidae in China (Li et al., 2005b). More extensive surveillance for coronaviruses in different species of horseshoe bats would shed light on the role of this bat family in the ecology and evolution of coronaviruses.
Materials and methods
Sample collection
Chinese horseshoe bats (R. sinicus) were captured from various locations in Hong Kong and in the Guangdong province of Southern China over a 2-year period (April 2004 to April 2006). Their respiratory and alimentary specimens were collected using procedures described previously (Lau et al., 2005, Yob et al., 2001). All specimens were placed in viral transport medium before transportation to the laboratory for RNA extraction.
RNA extraction
Viral RNA was extracted from the respiratory and alimentary specimens using QIAamp Viral RNA Mini Kit (QIAgen, Hilden, Germany). The RNA was eluted in 50 μl of AVE buffer and was used as the template for RT-PCR.
RT-PCR for coronaviruses and DNA sequencing
Coronavirus screening was performed by amplifying a 440-bp fragment of the RNA-dependent RNA polymerase (RdRp) gene of coronaviruses using conserved primers (5′-GGTTGGGACTATCCTAAGTGTGA-3′ and 5′-CCATCATCAGATAGAATCATCATA-3′) designed by multiple alignments of the nucleotide sequences of available RdRp genes of known coronaviruses (Woo et al., 2005a). Reverse transcription was performed using the SuperScript III kit (Invitrogen, San Diego, CA, USA). The PCR mixture (25 μl) contained cDNA, PCR buffer (10 mM Tris–HCl pH 8.3, 50 mM KCl, 3 mM MgCl2, and 0.01% gelatin), 200 μM of each dNTPs, and 1.0 U Taq polymerase (Applied Biosystem, Foster City, CA, USA). The mixtures were amplified in 60 cycles of 94 °C for 1 min, 48 °C for 1 min, and 72 °C for 1 min and a final extension at 72 °C for 10 min in an automated thermal cycler (Applied Biosystem, Foster City, CA, USA). Standard precautions were taken to avoid PCR contamination and no false-positive was observed in negative controls.
The PCR products were gel-purified using the QIAquick gel extraction kit (QIAgen, Hilden, Germany). Both strands of the PCR products were sequenced twice with an ABI Prism 3700 DNA Analyzer (Applied Biosystems, Foster City, CA, USA), using the two PCR primers. The sequences of the PCR products were compared with known sequences of the RdRp genes of coronaviruses in the GenBank database.
Viral culture
Three of the samples positive for bat-CoV HKU2 were cultured in LLC-Mk2 (rhesus monkey kidney), MRC-5 (human lung fibroblast), FRhK-4 (rhesus monkey kidney), Huh-7.5 (human hepatoma), Vero E6 (African green monkey kidney), HRT-18 (colorectal adenocarcinoma) cell lines and primary kidney epithelium and lung fibroblast cells derived from a Chinese horseshoe bat.
Complete genome sequencing of bat-CoV HKU2
Four complete genomes of bat-CoV HKU2 detected in the present study were amplified and sequenced using the RNA extracted from the alimentary specimens as templates. The RNA was converted to cDNA by a combined random-priming and oligo(dT) priming strategy. As the initial results revealed that they were group 1 coronaviruses, the cDNA was amplified by degenerate primers designed by multiple alignment of the genomes of human coronavirus 229E (HCoV-229E) (GenBank accession no. NC_002645), porcine epidemic diarrhea virus (PEDV) (GenBank accession no. NC_003436), porcine transmissible gastroenteritis virus (TGEV) (GenBank accession no. NC_002306), feline infectious peritonitis virus (FIPV) (GenBank accession no. AY994055), and HCoV-NL63 (GenBank accession no. NC_005831), and additional primers covering the original degenerate primer sites were designed from the results of the first and subsequent rounds of sequencing. These primer sequences are available on request. The 5′ ends of the viral genomes were confirmed by rapid amplification of cDNA ends using the 5′/3′ RACE kit (Roche, Germany). Sequences were assembled and manually edited to produce final sequences of the viral genomes.
Genome analysis
The nucleotide sequences of the genomes and the deduced amino acid sequences of the open reading frames (ORFs) were compared to those of other coronaviruses. Phylogenetic tree construction was performed using neighbor joining method with ClustalX 1.83. Protein family analysis was performed using PFAM and InterProScan (Apweiler et al., 2001, Bateman et al., 2002). Prediction of transmembrane domains was performed using TMHMM (Sonnhammer et al., 1998).
Estimation of synonymous and non-synonymous substitution rates
The number of synonymous substitutions per synonymous site, Ks, and the number of non-synonymous substitutions per non-synonymous site, Ka, for each coding region were calculated using the Nei-Gojobori method (Jukes-Cantor) in MEGA 3.1 (Kumar et al., 2004). Six pairwise comparisons on the four strains of bat-CoV HKU2 were performed for each coding region.
Recombination analysis
Sliding window analysis was used to detect possible recombination, using a nucleotide alignment of the genome sequences of the four strains of bat-CoV HKU2 and bat-SARS-CoV (GenBank accession no. DQ022305) generated by ClustalX version 1.83 and edited manually. Bootscan analysis was performed using Simplot version 3.5.1 (Lole et al., 1999) (F84 model; window size, 1000 bp; step, 200 bp) with the genome sequence of SARS-CoV (GenBank accession no. NC_004718) as a query.
Nucleotide sequence accession numbers
The nucleotide sequences of the four genomes of bat-CoV HKU2 have been lodged within the GenBank sequence database under accession no. EF203064 to EF203067.
Acknowledgments
We thank Director Stella Hung, Sin-Pang Lau, Chik-Chuen Lay, and Ping-Man So [HKSAR Department of Agriculture, Fisheries, and Conservation (AFCD)] and Hong Kong Police Force for facilitation and support; Chung-Tong Shek, Cynthia S. M. Chan, and Tak-yin Lau from AFCD for their excellent technical assistance; and Dr. King-Shun Lo (Laboratory Animal Unit) and Dr. Cassius Chan for collection of animal specimens.
We are grateful to the generous support of Mr. Hui Hoy and Mr. Hui Ming in the genomic sequencing platform. This work is partly supported by the Research Grant Council Grant; University Development Fund and Outstanding Young Researcher Award, The University of Hong Kong; The Tung Wah Group of Hospitals Fund for Research in Infectious Diseases; the HKSAR Research Fund for the Control of Infectious Diseases (04050232) of the Health, Welfare and Food Bureau; and the Providence Foundation Limited in memory of the late Dr. Lui Hac Minh.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.virol.2007.06.009.
Appendix A. Supplementary data
Multiple alignments of S proteins of bat-CoV HKU2, other group 1 coronaviruses, human and civet SARS-CoV and bat-SARS-CoV. Amino acid residues fully conserved across all coronaviruses are marked with ‘⁎’; strongly conserved residues are marked with ‘:’; and weakly conserved residues are marked with ‘.’. RBM of SARS-CoV, and RBDs of HCoV-229E and HCoV-NL63 were shaded in grey. Residues within the S of SARS-CoV that directly contact hACE2 are in white letters. The cysteines that contribute to ACE2 binding in SARS-CoV are bolded. The short stretch of homologous peptide between bat-CoV HKU2 and SARS-CoV within the RBM of SARS-CoV is boxed. Large deletions within the S of bat-CoV HKU2 were shaded in dark grey. The two regions of deletions in the C-terminal conserved between bat-CoV HKU2 and group 2 coronaviruses are boxed and shaded.
References
- Apweiler R., Attwood T.K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D., Durbin R., Falquet L., Fleischmann W., Gouzy J., Hermjakob H., Hulo N., Jonassen I., Kahn D., Kanapin A., Karavidopoulou Y., Lopez R., Marx B., Mulder N.J., Oinn T.M., Pagni M., Servant F., Sigrist C.J., Zdobnov E.M. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S.R., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavia A., Zelus B.D., Wentworth D.E., Talbot P.J., Holmes K.V. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 2003;77:2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., L'Haridon R., Vogel L.K., Sjostrom H., Noren O., Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye C., Siddell S.G. Genomic RNA sequence of Feline coronavirus strain FIPV WSU-79/1146. J. Gen. Virol. 2005;86:2249–2253. doi: 10.1099/vir.0.80985-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickmann M., Becker S., Klenk H.D., Doerr H.W., Stadler K., Censini S., Guidotti S., Masignani V., Scarselli M., Mora M., Donati C., Han J.H., Song H.C., Abrignani S., Covacci A., Rappuoli R. Phylogeny of the SARS coronavirus. Science. 2003;302:1504–1505. doi: 10.1126/science.302.5650.1504b. [DOI] [PubMed] [Google Scholar]
- Fouchier R.A., Hartwig N.G., Bestebroer T.M., Niemeyer B., de Jong J.C., Simon J.H., Osterhaus A.D. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 2004;78:3863–3871. doi: 10.1128/JVI.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A., Smeenk I., Horzinek M.C., Rottier P.J., de Groot R.J. Feline coronavirus type II strains 79–1683 and 79–1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A., Mawditt K.L., Cavanagh D., Britton P. Investigation of the control of coronavirus subgenomic mRNA transcription by using T7-generated negative-sense RNA transcripts. J. Virol. 1995;69:6219–6227. doi: 10.1128/jvi.69.10.6219-6227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Simmons G., Rennekamp A.J., Chaipan C., Gramberg T., Heck E., Geier M., Wegele A., Marzi A., Bates P., Pohlmann S. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 2006;80:8639–8652. doi: 10.1128/JVI.00560-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck J.G., Matsushima G.K., Makino S., Fleming J.O., Vannier D.M., Stohlman S.A., Lai M.M. In vivo RNA-RNA recombination of coronavirus in mouse brain. J. Virol. 1988;62:1810–1813. doi: 10.1128/jvi.62.5.1810-1813.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottier S.A., Cavanagh D., Britton P. Experimental evidence of recombination in coronavirus infectious bronchitis virus. Virology. 1995;213:569–580. doi: 10.1006/viro.1995.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempl C., Schultze B., Herrler G. Analysis of cellular receptors for human coronavirus OC43. Adv. Exp. Med. Biol. 1995;380:371–374. doi: 10.1007/978-1-4615-1899-0_60. [DOI] [PubMed] [Google Scholar]
- Kubo H., Yamada Y.K., Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 1994;68:5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Yip C.C., Tse H., Tsoi H.W., Cheng V.C., Lee P., Tang B.S., Cheung C.H., Lee R.A., So L.Y., Lau Y.L., Chan K.H., Yuen K.Y. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J. Clin. Microbiol. 2006;44:2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi E., Haluskey J.A., Masters P.S. The pathogenesis of MHV nucleocapsid gene chimeric viruses. Adv. Exp. Med. Biol. 1998;440:537–541. doi: 10.1007/978-1-4615-5331-1_69. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N., Murakami A., He Y., Marasco W.A., Guan Y., Choe H., Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li W., Wong S.K., Li F., Kuhn J.H., Huang I.C., Choe H., Farzan M. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J. Virol. 2006;80:4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W., Bredenbeek P.J., Noten A.F., Horzinek M.C., Spaan W.J. Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Olsen C.W. A review of feline infectious peritonitis virus: molecular biology, immunopathogenesis, clinical aspects, and vaccination. Vet. Microbiol. 1993;36:1–37. doi: 10.1016/0378-1135(93)90126-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Chu D.K., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H., Lau S.K., Woo P.C., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S. Identification of a novel coronavirus in bats. J. Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Jebbink M.F., Berkhout B., van der Hoek L. Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J. 2004;1:7. doi: 10.1186/1743-422X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Li W., Yu M., Hao P., Zhang Y., Zhou P., Zhang S., Zhao G., Zhong Y., Wang S., Wang L.F., Shi Z. Full-length genome sequences of two SARS-like coronaviruses in horseshoe bats and genetic variation analysis. J. Gen. Virol. 2006;87:3355–3359. doi: 10.1099/vir.0.82220-0. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu X.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L., von Heijne G., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Tang X.C., Zhang J.X., Zhang S.Y., Wang P., Fan X.H., Li L.F., Li G., Dong B.Q., Liu W., Cheung C.L., Xu K.M., Song W.J., Vijaykrishna D., Poon L.L., Peiris J.S., Smith G.J., Chen H., Guan Y. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S.G. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- Tu C., Crameri G., Kong X., Chen J., Sun Y., Yu M., Xiang H., Xia X., Liu S., Ren T., Yu Y., Eaton B.T., Xuan H., Wang L.F. Antibodies to SARS coronavirus in civets. Emerg. Infect. Dis. 2004;10:2244–2248. doi: 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung F.Y., Abraham S., Sethna M., Hung S.L., Sethna P., Hogue B.G., Brian D.A. The 9-kDa hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology. 1992;186:676–683. doi: 10.1016/0042-6822(92)90034-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.K., Jiang G.S., Holmes K.V. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 1991;88:5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Tsoi H.W., Chan K.H., Wong B.H., Che X.Y., Tam V.K., Tam S.C., Cheng V.C., Hung I.F., Wong S.S., Zheng B.J., Guan Y., Yuen K.Y. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet. 2004;363:841–845. doi: 10.1016/S0140-6736(04)15729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Tsoi H.W., Huang Y., Poon R.W., Chu C.M., Lee R.A., Luk W.K., Wong G.K., Wong B.H., Cheng V.C., Tang B.S., Wu A.K., Yung R.W., Chen H., Guan Y., Chan K.H., Yuen K.Y. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J. Infect. Dis. 2005;192:1898–1907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Huang Y., Lau S.K., Tsoi H.W., Yuen K.Y. In silico analysis of ORF1ab in coronavirus HKU1 genome reveals a unique putative cleavage site of coronavirus HKU1 3C-like protease. Microbiol. Immunol. 2005;49:899–908. doi: 10.1111/j.1348-0421.2005.tb03681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Wang M., Lau S.K., Xu H., Poon R.W., Guo R., Wong B.H., Gao K., Tsoi H.W., Huang Y., Li K.S., Lam C.S., Chan K.H., Zheng B.J., Yuen K.Y. Comparative analysis of 12 genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 2006;81:1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Yuen K.Y. Infectious diseases emerging from Chinese wet-markets: zoonotic origins of severe respiratory viral infections. Curr. Opin. Infect. Dis. 2006;19:401–407. doi: 10.1097/01.qco.0000244043.08264.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Yip C.C., Huang Y., Tsoi H.W., Chan K.H., Yuen K.Y. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Li K.S., Poon R.W., Wong B.H., Tsoi H.W., Yip B.C., Huang Y., Chan K.H., Yuen K.Y. Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.Y., Werner H.C., Kong W.P., Leung K., Traggiai E., Lanzavecchia A., Nabel G.J. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc. Natl. Acad. Sci. U.S.A. 2005;102:797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yob J.M., Field H., Rashdi A.M., Morrissy C., van der Heide B., Rota P., bin Adzhar A., White J., Daniels P., Jamaluddin A., Ksiazek T. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 2001;7:439–441. doi: 10.3201/eid0703.010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 2004;7:412–419. doi: 10.1016/j.mib.2004.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple alignments of S proteins of bat-CoV HKU2, other group 1 coronaviruses, human and civet SARS-CoV and bat-SARS-CoV. Amino acid residues fully conserved across all coronaviruses are marked with ‘⁎’; strongly conserved residues are marked with ‘:’; and weakly conserved residues are marked with ‘.’. RBM of SARS-CoV, and RBDs of HCoV-229E and HCoV-NL63 were shaded in grey. Residues within the S of SARS-CoV that directly contact hACE2 are in white letters. The cysteines that contribute to ACE2 binding in SARS-CoV are bolded. The short stretch of homologous peptide between bat-CoV HKU2 and SARS-CoV within the RBM of SARS-CoV is boxed. Large deletions within the S of bat-CoV HKU2 were shaded in dark grey. The two regions of deletions in the C-terminal conserved between bat-CoV HKU2 and group 2 coronaviruses are boxed and shaded.