Abstract
Severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) caused a severe outbreak in several regions of the world in 2003. The SARS-CoV genome is predicted to contain 14 functional open reading frames (ORFs). The first ORF (1a and 1b) encodes a large polyprotein that is cleaved into nonstructural proteins (nsp). The other ORFs encode for four structural proteins (spike, membrane, nucleocapsid and envelope) as well as eight SARS-CoV-specific accessory proteins (3a, 3b, 6, 7a, 7b, 8a, 8b and 9b). In this report we have cloned the predicted nsp8 gene and the ORF6 gene of the SARS-CoV and studied their abilities to interact with each other. We expressed the two proteins as fusion proteins in the yeast two-hybrid system to demonstrate protein–protein interactions and tested the same using a yeast genetic cross. Further the strength of the interaction was measured by challenging growth of the positive interaction clones on increasing gradients of 2-amino trizole. The interaction was then verified by expressing both proteins separately in-vitro in a coupled-transcription translation system and by coimmunoprecipitation in mammalian cells. Finally, colocalization experiments were performed in SARS-CoV infected Vero E6 mammalian cells to confirm the nsp8–ORF6 interaction. To the best of our knowledge, this is the first report of the interaction between a SARS-CoV accessory protein and nsp8 and our findings suggest that ORF6 protein may play a role in virus replication.
Abbreviations: AD, activation domain; BD, binding domain; 3-AT, 3-amino trizole; N, nucleocapsid
Keywords: SARS coronavirus, Protein–protein interaction, Yeast two-hybrid system, Non structural protein, Accessory protein, Immunoprecipitation
Introduction
Severe acute respiratory syndrome (SARS) is a newly emerging infectious disease. The etiologic agent of SARS has been identified as a novel coronavirus, namely SARS-associated coronavirus (SARS-CoV) (Ksiazek et al., 2003, Marra et al., 2003, Peiris et al., 2003, Rota et al., 2003). As of June 30th, 2003, 8447 probable SARS cases including 811 deaths were reported by the World Health Organization (WHO) from 32 countries or regions worldwide. The significant morbidity and mortality, and the potential for re-emergence, makes SARS-CoV a continued worldwide public health threat.
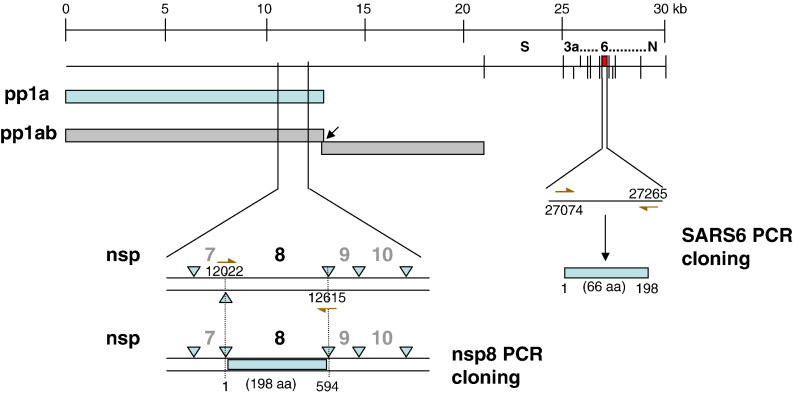
Like other known coronaviruses, SARS-CoV is an enveloped, plus-strand RNA virus that features the largest RNA genomes currently known. The SARS viral genome comprises approximately 30,000 nucleotides, which are organized into 14 functional open reading frames (ORFs) (Rota et al., 2003, Thiel et al., 2003). Analysis of the nucleotide sequence of this novel coronavirus revealed a similar pattern of gene organization typical of coronaviruses (Marra et al., 2003, Rota et al., 2003). Two large, 5′-terminal overlapping ORFs, 1a and 1b of the replicase gene encode two polyproteins, pp1a and pp1ab (Fig. 1 ) (Rota et al., 2003, Thiel et al., 2003). Like other coronaviruses, the nascent SARS-CoV replicase polyproteins are processed by virus-encoded proteinases, and in this case, two proteinases have been identified: a 3C-like proteinase (3CLpro) and a papain-like proteinase (PLP) (Denison et al., 1992, Snijder et al., 2003, Ziebuhr, 2004). The processed end products of pp1a are designated nonstructural proteins (nsp) 1 to nsp11 and those of pp1ab are designated nsp1 to nsp16. Cleavage by the viral main protease, 3CLpro results in generating the nsp8 protein which is currently of undesignated function. The nsp8 protein has been shown to associate with several other nsps and to colocalize with these nsps in cytoplasmic complexes that are important for viral RNA synthesis (Prentice et al., 2004a, Prentice et al., 2004b, Sutton et al., 2004, Zhai et al., 2005). The +RNA coronavirus usually encode a non-structural protein (nsp12 for SARS-CoV) that includes an RdRp domain, conserved in all RNA viruses. The SARS-CoV uniquely encodes a second RdRp residing in nsp8, responsible for initiation of the synthesis of complementary oligonucleotides smaller than 6 residues long, at relatively low fidelity. These nsp8 RdRp produced primers are postulated to be utilized by the primer-dependent nsp12 RdRp for replication (Imbert et al., 2006).
Fig. 1.
Cloning of the putative nsp8 and ORF6 genes from the SARS CoV genome. The full-length nsp8 (198 amino acids) and ORF6 (66 amino acids) genes of the SARS coronavirus (Tor2 isolate) were PCR-amplified from a genomic construct of clone NC_004718 and cloned into the pCR-XL-TOPO vector (Invitrogen) as described in Table 1. The scale on top shows genomic region.
The remaining 12 ORFs encode the four structural proteins, spike (S), membrane (M), nucleocapsid (N) and envelope (E), and eight accessory proteins (3a, 3b, 6, 7a, 7b, 8a, 8b and 9b). These accessory proteins are postulated to be non-essential in tissue culture but may provide a selective advantage in the infected host. The ORF3a protein has been studied to some details (Tan et al., 2006) and has been shown to be expressed in infected cells (Tan et al., 2004c, Yu et al., 2004, Zeng et al., 2004). ORF3a protein has been shown to bind the spike protein (Zeng et al., 2004, Tan et al., 2004c) and recent work demonstrated that it is a novel structural protein (Ito et al., 2005). The ORF7a protein was also detected in infected cells and has been found to localize in the ER-Golgi intermediate compartments where coronaviruses are known to assemble (Fielding et al., 2004, Nelson et al., 2005). In addition, ORF3a, ORF3b and ORF7a proteins have been shown to induce apoptosis (Tan et al., 2004a, Law et al., 2005, Yuan et al., 2005). Two recent papers on the ORF6 accessory protein also demonstrated its expression during infection and have showed that it may be important for viral pathogenesis (Geng et al., 2005, Pewe et al., 2005). Most recently the ORF6 protein has been shown to accelerate replication of a related mouse virus, a property that may contribute towards its increased in vivo virulence (Tangudu et al., 2007).
In this study, the nsp8 and ORF6 proteins were tested for interaction in a yeast cellular environment using the yeast two-hybrid system. These interactions were further verified by in-vitro coimmunoprecipitation experiments using proteins expressed by a coupled-transcription/translation system or in mammalian cells by lipid-mediated transfection of DNA. Finally, the nsp8–ORF6 interaction was confirmed by colocalization of the two proteins in SARS-CoV infected mammalian cells (Vero E6). This unique SARS-CoV accessory protein 6 interacting with a non-structural protein, nsp8, suggests that ORF6 may have a role in SARS virus replication.
Results and discussion
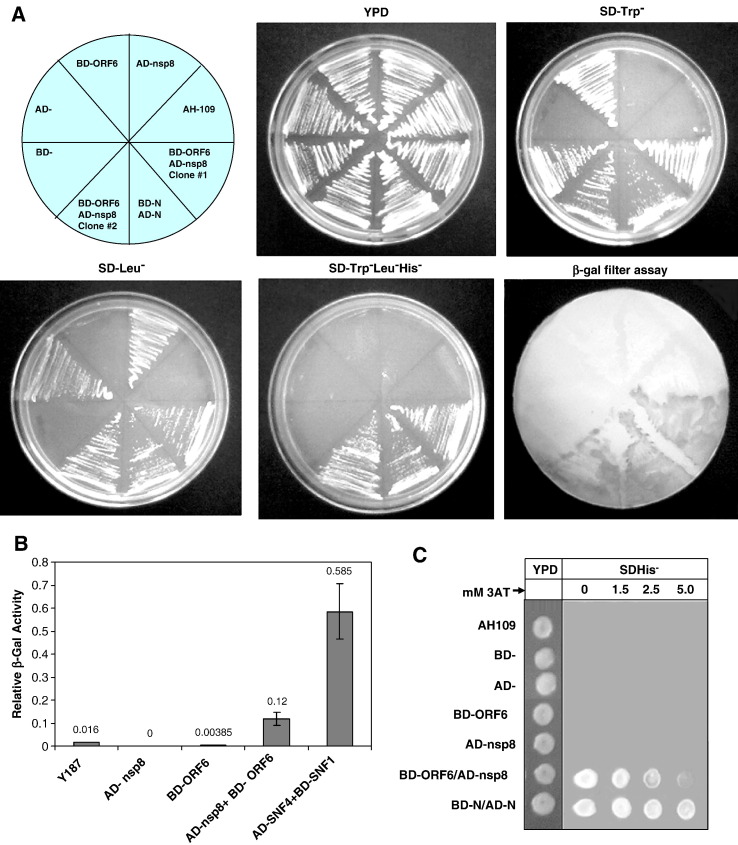
The nsp8 protein of the SARS coronavirus was cloned from the Tor2 Singapore isolate and expressed using the yeast two-hybrid vectors resulting in an N-terminal in-frame fusion protein with the GAL4 activation domain (AD). Similarly the ORF6 protein was PCR cloned and expressed in yeast two-hybrid vectors resulting in an N-terminal in-frame fusion protein with the GAL4 DNA binding domain (BD). In order to check for correct reading-frame constructs, the final plasmids were sequenced and checked for in-vitro protein expression. Saccharomyces cerevisiae AH109 cells were transformed with single plasmids, or co-transformed with the GAL4 BD- and AD-vectors containing SARS Co-V ORF6 and/or nsp8, respectively. The AH109 host strain containing pAS2-N and pACT2-N were used as positive controls (Surjit et al., 2004). AH109 contains integrated copies of both HIS3 and lacZ reporter genes under the control of GAL4 binding sites. The results of the two-hybrid assay are shown in Fig. 2A. Single transformants used in this assay were yeast (AH109) cells containing BD-ORF6, and host cells containing AD-nsp8, also yeast cells containing only the BD- and only the AD-vectors, were also used as negative controls. All these transformants and co-transformants grew well on the unrestricted YPD plate (non-selective media). Also, the untransformed host cells were plated as a negative control. Single transformants containing the BD-by itself or as a fusion with the ORF6 protein, showed growth on the synthetic dextrose Trp− plate (SDTrp−). Correspondingly, single transformants containing the AD- by itself or as a fusion with nsp8 showed growth on the synthetic dextrose Leu− plate (SDLeu−). The co-transformants were similarly plated on YPD and synthetic dextrose medium lacking Trp or Leu or Trp, Leu and upon positive growth on these was subsequently plated on His− medium (SD Trp−Leu−His−) to test for His prototrophy. Growth of the co-transformants, containing the BD-ORF6 and AD-nsp8 constructs, in both SDTrp− and SDLeu− plates simply showed that both plasmids were present in the transformed cells. Growth of these clones on the SD Trp−Leu−His− media showed that the transcription of the HIS3 gene was turned on by the reconstitution of the GAL4 transactivator due to a specific ORF6–nsp8 interaction. Colonies were transferred on to nitrocellulose filters and a β-galactosidase filter assay was performed as described in methods. The co-transformants containing both the BD-ORF6 and AD-nsp8 constructs along with all positive and negative controls were used in this assay. Results obtained from the β-galactosidase filter assay were in agreement with the results obtained from the SDHis− growth experiments.
Fig. 2.
Yeast two-hybrid results showing that nsp8 and ORF6 proteins interact with each other. (A) AD-nsp8 fusion protein and BD-ORF6 fusion protein tested as single and co-transformants on various synthetic growth media lacking specific amino-acids. YPD media shows uninhibited growth of all transformants and host strain. SDTrp− and SD-Leu− selective media have Tryptophan and Leucine missing, respectively. SD Trp−Leu−His− media are triple dropout plates lacking Tryptophan, Leucine and Histidine. Growth on the SD Trp−Leu−His− plate and blue color (shown here in shades of grey) on the β-gal filter assay represents a positive interaction. BD-N:AD-N combination was used as a positive control in these yeast two-hybrid experiments (Surjit et al., 2004). (B) Liquid β-galactosidase assay results of the nsp8–ORF6 protein–protein interactions. Single transformants and co-transformants were analyzed in a liquid β-galactosidase assay and compared with each other. Values given are in arbitrary units. The numbers above each bar represent the mean from five independent transformants. Y187 corresponds to the untransformed host strain. Transformants with more than one plasmid are separated by a slash. Positive controls used in this assay are denoted by BD-SNF1/AD-SNF4 (Harper et al., 1993). (C) Measurement of strength of the nsp8–ORF6 interactions on an increasing 3-AT gradient. Activation of the HIS3 reporter was determined for the host AH109 strain, single transformants (BD, AD, BD-ORF6 and AD-nsp8), and co-transformants (BD-ORF6/AD-nsp8 and BD-N/AD-N). Hundred-fold serial dilutions of all of the above-mentioned log-phase cultures were plated on YPD (left) followed by SD-His-plus 50 mM 3-AT in increasing concentrations (0 mM to 5 mM 3-AT).
The liquid β-galactosidase assay was conducted and the activity determined using the substrate chlorophenol red-β-D-galactopyranoside (CPRG) as described in methods. The host strain alone, along with single transformants containing either AD-nsp8 or BD-ORF6 and co-transformants containing AD-/BD- without a fusion protein was tested. Negative controls (host untransformed cells) showed almost no liquid β-galactosidase activity. BD-SNF1/AD-SNF4 was the positive control whereas the clones containing BD-ORF6/AD-nsp8 were the test samples in this experiment (Fig. 2B). Relative enzymatic activity was determined in five independent transformants from each group. Our results from this assay indicate a moderate strength protein–protein interaction between the AD-nsp8 and BD-ORF6 proteins. This observation was further confirmed by the use of a 3-AT gradient on which the His+ phenotype was tested (Fig. 2C). 3-AT is known to be a competitive inhibitor of HIS3 protein, thus enhancing the stringency of selection, it reduces the background signal by inhibiting the product of the HIS3 reporter. This result clearly showed that the interaction of nsp8 with ORF6 was not as strong as the N–N dimerization positive control interaction. On plates with increasing concentrations of 3-AT, the growth ability of the His+ colonies diminished and was almost undetectable at 5 mM 3-AT concentrations.
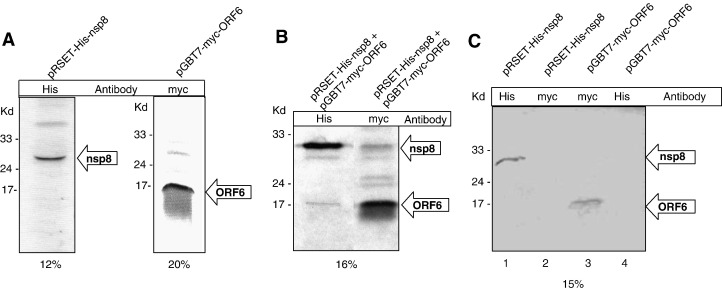
To further confirm these interactions we performed a coimmunoprecipitation assay using proteins expressed by a cell-free coupled transcription–translation system. The rabbit reticulocyte lysates expressing His6-nsp8 and myc-ORF6 proteins separately were tested for expression (Fig. 3A). For detection of the expressed proteins, the lysates were immunoprecipitated using the respective antibodies i.e. anti-His antibody was used for detection of the nsp8 protein and anti-myc antibody was used for the detection of the ORF6 protein. When the two cell lysates were mixed in equal proportions, one of the two antibodies was added and subsequently pulled out using Protein A Sepharose beads, we observed the interacting protein to be present in the gel as well. This procedure was repeated conversely to show the other corresponding antibody pull out the interacting protein partner as well (Fig. 3B). When anti-His antibody was used to pull out the nsp8 protein from the mixture of proteins, it also pulled out the ORF6 protein. Similarly, when anti-myc antibody was used to pull out the ORF6 protein, the nsp8 protein was also observed to be pulled out as an interaction partner of ORF6. In a control experiment we observed no ORF6 protein binding directly to the anti-His antibody Sepharose-A complex and similarly observed no binding of the nsp8 protein to the anti-myc antibody Separose-A complex (Fig. 3C). These results clearly showed that the two proteins nsp8 and ORF6 interacted with each other.
Fig. 3.
Confirmation of the nsp8–ORF6 interaction using an in-vitro cell-free coupled transcription/translation coimmunoprecipitation assay. (A) The His6-tagged nsp8 and myc-tagged ORF6 proteins were produced by coupled transcription–translation in the presence of 35S-Met and detected using their corresponding antibodies (Anti-His and anti-myc, respectively). Both nsp8 and ORF6 proteins were visible and corresponded to their correct molecular sizes on 12% and 20% SDS-PAGE, respectively. (B) The 35S-Met-His-nsp8 and 35S-Met-myc-ORF6 proteins were detected by autoradiography. The nsp8–ORF6 complex was detected by both anti-His and anti-myc antibodies on 16% SDS-PAGE. When nsp8 was probed for using anti-His antibody, the ORF6 protein was visible and when ORF6 protein was probed using myc antibody, the nsp8 protein was visible. (C) In a control reaction, the 35S-Met-His6-nsp8 and 35S-Met-myc-ORF6 proteins showed no cross-reactivity with the other antibody used in the pull-down experiments. Also this control experiment shows that the proteins do not non-specifically bind to the beads used in the results shown in panel B. Lane 1 shows the nsp8 protein immunoprecipitated by anti-His, lane 2 does not show nsp8 protein band when immunoprecipitated by anti-myc. Similarly, lane 3 shows myc-tagged ORF6 protein immunoprecipitated by anti-myc and lane 4 does not show ORF6 protein where it was immunoprecipitated using anti-His.
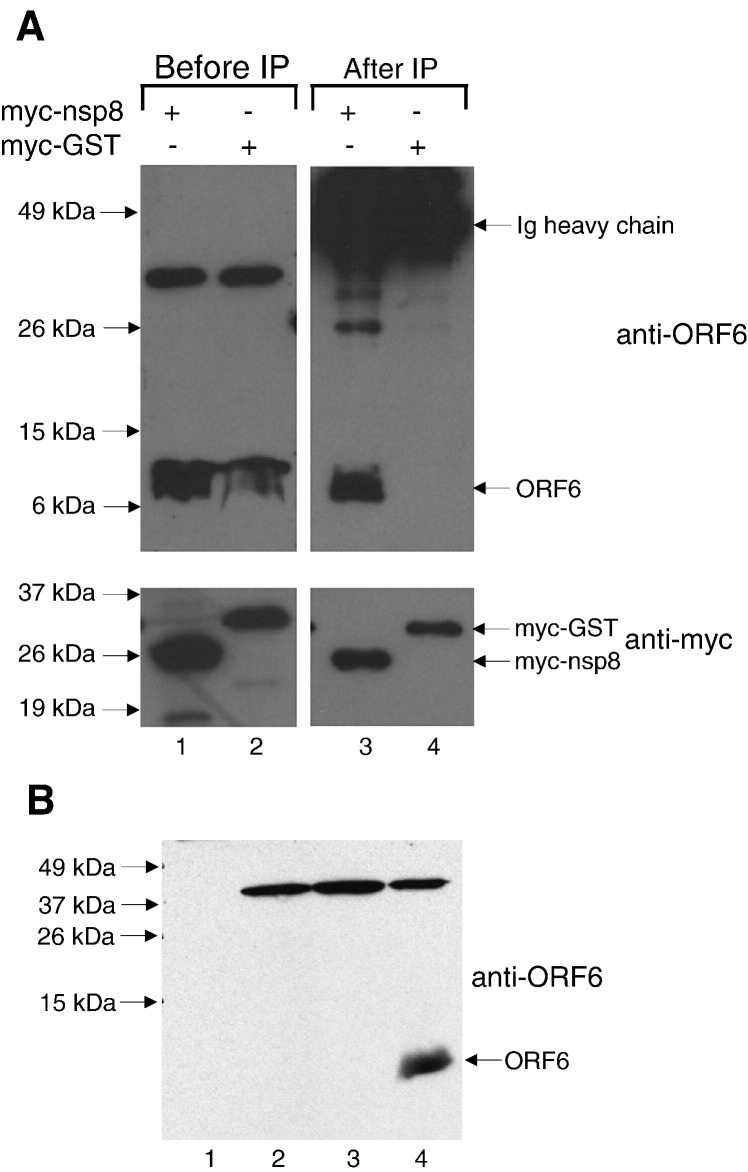
In order to verify the interaction between nsp8 and ORF6 in mammalian cells, co-immunoprecipitation experiments were performed using lysates obtained from Vero E6 cells expressing transiently transfected myc-tagged nsp8 and untagged ORF6 proteins carried by a replicative recombinant vaccinia virus. Myc-polyclonal antibody conjugated to protein A Sepharose beads was used to pull-down myc-nsp8 or myc-GST (negative control), and any ORF6 protein that co-immunoprecipitated was detected using a rabbit polyclonal antibody raised against a peptide of ORF6 (Abgent). This antibody specifically detected the ORF6 protein (∼ 7 kDa) in cells that were infected with ORF6 recombinant vaccinia virus (Fig. 4 , lanes 1 and 2). As shown in Fig. 4, ORF6 was co-immunoprecipitated by myc-nsp8 (lane 3) but not by myc-GST (lane 4), showing that the ORF6 and nsp8 proteins can interact in Vero E6 cells.
Fig. 4.
(A) Cell lysates were obtained from Vero E6 cells expressing ORF6 and myc-nsp8 or ORF6 and myc-GST (negative control) and subjected to Western blot analysis with either anti-ORF6 polyclonal (upper panel, lanes 1 and 2) or anti-myc monoclonal antibodies (lower panel, lanes 1 and 2). Equal amounts of lysates were subjected to immunoprecipitation using anti-myc polyclonal antibodies and protein A beads. The immuno-complexes were subjected to Western blot with either anti-ORF6 polyclonal (upper panel, lanes 3 and 4) or anti-myc monoclonal antibodies (lower panel, lanes 3 and 4). (B) Equal amounts of total cellular protein from Vero E6 cells either uninfected (lane 1) or infected with wildtype Dryvax (lane 2), a recombinant vaccinia virus expressing the SARS-CoV S protein (lane 3), or the recombinant vaccinia virus expressing the ORF6 protein used in this study (lane 4) were subjected to Western blot and probed with anti-ORF6 rabbit polyclonal antibody. This demonstrates that the ∼ 37 kDa band seen in lanes 1 and 2 of panel A is a cross-reacting protein that is probably a vaccinia virus protein or an up-regulated cellular protein which is detected by the rabbit anti-ORF6 antibody used in this study.
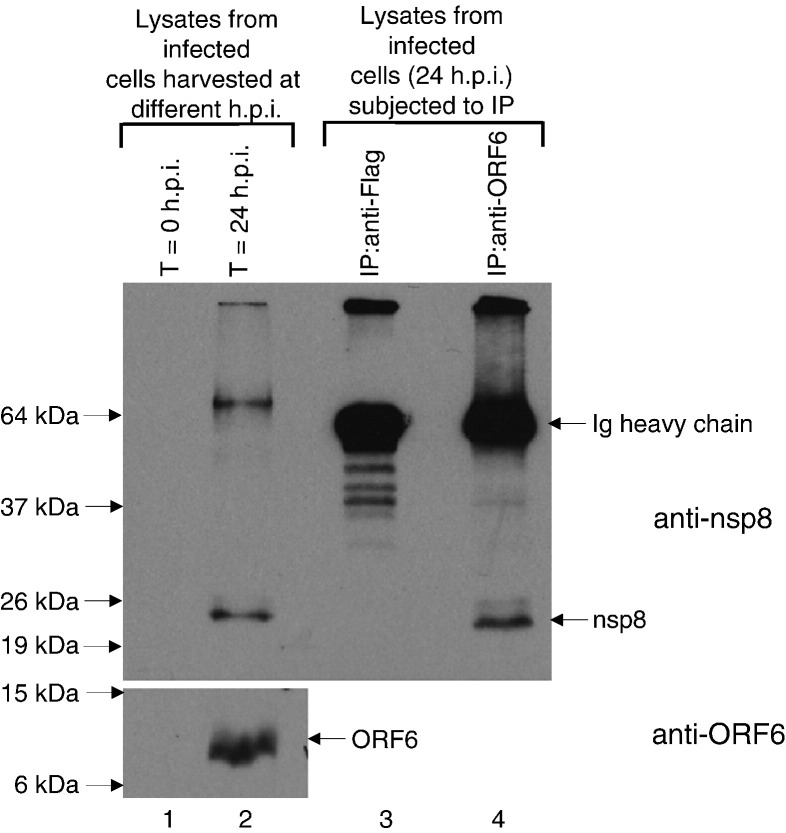
In order to confirm that the interaction between nsp8 and ORF6 occurs during SARS-CoV infection, co-immunoprecipitation experiments were performed with lysates obtained from SARS-CoV infected cells. Infection of Vero E6 cells with an isolate of SARS-CoV (strain HKU 39849) was carried out in a physical containment level 4 (PC4) laboratory as previously described (Kaye et al., 2006). The cells were harvested at 0 or 24 h post-infection (h.p.i.) and the lysates were subjected to Western blot analysis to determine the expression of nsp8 and ORF6 proteins (Fig. 5 ). Two specific proteins in the cells at 24 h.p.i. were detected with purified supernatant from hybridomas producing monoclonal antibodies against the nsp8 protein (see Materials and methods) and these were not found in the cells harvested at 0 h.p.i. (upper panel, lanes 1 and 2). The protein that migrated at ∼ 24 kDa matched the predicted molecular mass of nsp8 while the slower migrating protein (∼ 65 kDa) may be an aggregated form of nsp8. As expected, the ORF6 protein was also detected at 24 h.p.i. and not at 0 h.p.i. (lower panel, lanes 1 and 2). Cell lysates obtained at 24 h.p.i. were then subjected to immunoprecipitation with either an irrelevant antibody (rabbit anti-Flag polyclonal antibody, lane 3) or rabbit anti-ORF6 polyclonal antibody (lane 4) and protein-A agarose beads. Western blot analyses showed that nsp8 protein (∼ 24 kDa) was co-immunoprecipitated when anti-ORF6 antibody was used but not when an irrelevant antibody was used, indicating that the nsp8 interacts specifically with ORF6 in SARS-CoV infected cells (upper panel, lanes 3 and 4).
Fig. 5.
Lysates were obtained from SARS-CoV infected Vero E6 cells harvested at 0 or 24 h post infection (h.p.i.) and Western blot analyses were performed to determine the expression of SARS-CoV proteins using mouse anti-nsp8 monoclonal antibody (upper panel, lanes 1 and 2) and rabbit anti-ORF6 polyclonal antibody (lower panel, lanes 1 and 2). Cell lysates obtained at 24 h.p.i. were then subjected to immunoprecipitation using anti-Flag (irrelevant antibody as negative control) or anti-ORF6 rabbit polyclonal antibodies and protein A beads. The immuno-complexes were subjected to Western blot analysis with mouse anti-nsp8 monoclonal antibody (upper panel, lanes 3 and 4).
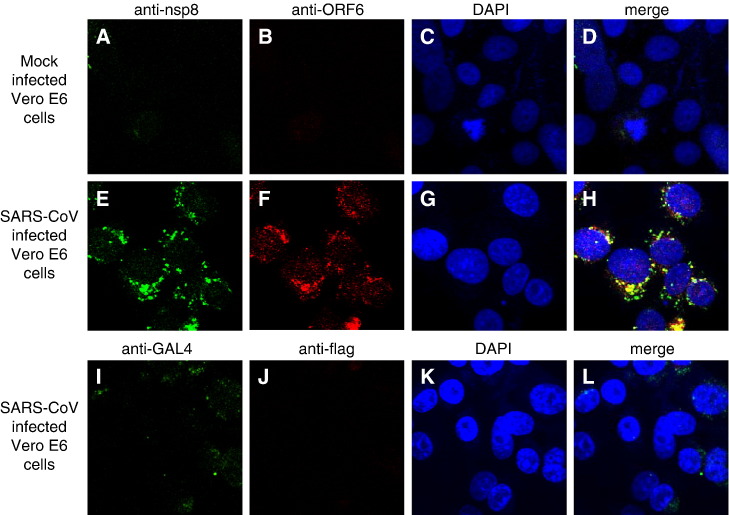
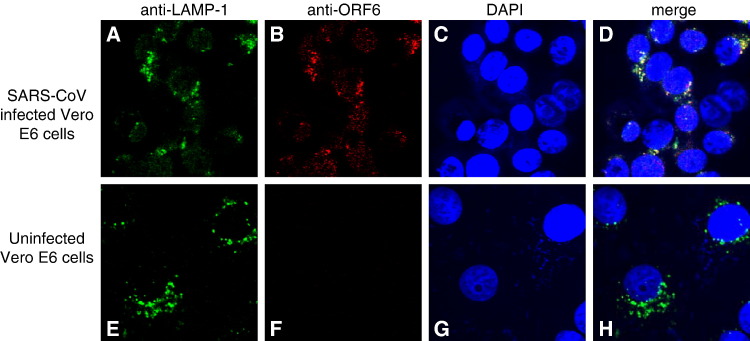
Indirect immunofluorescence analysis of ORF6 expression in SARS-CoV-infected cells (Fig. 6 , middle panel) showed that, consistent with previous publications (Geng et al., 2005, Pewe et al., 2005), ORF6 was found in the cytoplasm. Most of the ORF6 protein seen in infected cells were observed in a punctate staining pattern consistent with a vesicle-associated intracellular distribution. Using the rabbit polyclonal antibody against ORF6 (Abgent) and purified supernatant from hybridomas producing monoclonal antibodies against the nsp8 protein (see Materials and methods), cells infected with an isolate of SARS-CoV (strain HKU39849) were examined by indirect immunofluorescence to determine if the two proteins colocalized, as their interaction would suggest. The extent of colocalization seen between the two proteins was significant, both localizing to the same set of punctate structures (Fig. 6, middle panel, D). The punctate distribution of nsp8 has been previously described (Prentice et al., 2004a) and the colocalization of ORF6 protein with nsp8 suggests that ORF6 could be associated with the SARS-CoV replicase complex, localized to a subset of the vesicular trafficking complex in mammalian cells. These ORF6-containing vesicular structures did not colocalize with AP-1 and LAMP-2 (data not shown), which are present in early and late endosomes respectively. However, the ORF6 protein does colocalize with LAMP-1 (Fig. 7 ), which is a lysosomal marker. LAMP-1 has been used as a marker for characteristic double-membrane vesicles seen in infections by other viruses (Jackson et al., 2005) and it has been shown that at least one other coronavirus induces these double-membrane vesicles, presumably as a site for the assembly of virus replication complexes (Prentice et al., 2004b). Given this, the ORF6 protein might localize to these vesicles which have been suggested to be derived from the cellular autophagosomal machinery, but further study will be required. A recent structural study of the nsp8 protein has suggested a role for the nsp8–nsp7 hexadecamer in binding dsRNA intermediates in the viral replication cycle (Zhai et al., 2005), further lending impetus to such a study.
Fig. 6.
Indirect immunofluorescence used to study cellular localization of nsp8 and ORF6 in SARS-CoV infected Vero E6 cells. (Upper panel) Mouse anti-nsp8 (A) and rabbit anti-ORF6 (B) antibodies were used against uninfected cells to determine if the antibodies showed any unspecific staining. Nuclei are represented by means of a DAPI stain (C). (Middle panel) Vero E6 cells infected with SARS-CoV were examined by indirect immunofluorescence using the same antibodies. The expression of nsp8 is represented by FITC staining (E), while the expression of ORF6 is represented by Rhodamine staining (F). Nuclei are represented by means of a DAPI stain (G). The merged image showed that the nsp8 and ORF6 proteins colocalize to punctuate structures in the cytoplasm (H). (Lower panel) Irrelevant antibodies (mouse anti-GAL4TA (I) and rabbit anti-flag (J)) were used to probe SARS-CoV infected cells at the same final concentrations as their counterparts in the middle panel to show the specificity of the antibodies used in detecting the ORF6 and nsp8 proteins.
Fig. 7.
Indirect immunofluorescence was used to probe for LAMP-1 (A, E) and ORF6 (B, F) staining in SARS-CoV infected cells (upper panel) and uninfected Vero E6 cells (lower panel). Nuclei are represented by means of a DAPI stain (C, G). The merged image showed that there is colocalization between the LAMP-1 protein and ORF6 proteins in SARS-CoV infected cells (D, upper panel).
This is the first report describing the interaction between the ORF6 accessory protein and the nsp8 protein. We demonstrated the strength and specificity of this interaction by three independent methods, i.e. by yeast-2-hybrid, co-immunoprecipitation using in-vitro translated proteins in a cell-free system and co-immunoprecipitation using lysates from Vero E6 cells. Interestingly, two previous publications on ORF6 have shed some light on its possible function in viral pathogenesis. Geng and co-workers detected ORF6 protein in lung and intestine of SARS patients and showed that it could stimulate cellular DNA synthesis when overexpressed (Geng et al., 2005). Pewe et al. (2005) showed that when SARS-CoV ORF gene was introduced into an attenuated murine coronavirus, the recombinant virus grew faster in cell culture and also exhibited enhanced lethality of infection in mice. In contrast, Yount and co-workers reported that the SARS-CoV virus with ORF6 deleted has the same viral replication kinetics as the wild-type virus in cell-culture and mice (Yount et al., 2005). Hence, ORF6 is non-essential for viral replication in these models although it cannot rule out that ORF6 (and/or the other accessory proteins of SARS-CoV) contributes to the virulence and pathogenesis of SARS coronavirus infection in its natural host. Further experiments in more robust animal models, like non-human primates that demonstrate clinical disease upon SARS-CoV infection, will be needed to determine the actual function of ORF6 during SARS infection in vivo. Since our data shows that ORF6 interacts with nsp8 which is part of the replicase complex (Prentice et al., 2004a, Prentice et al., 2004b), it will be important in future studies to determine if the interaction between ORF6 and nsp8 can modulate the viral synthesis process and to determine how ORF6 modulates viral pathogenesis in these animal models.
Materials and methods
Growth media, yeast strains and plasmid constructs
All strains, plasmids and plasmid constructs used in this study are described in Table 1 . The full-length nsp8 and ORF6 genes of the SARS coronavirus (Tor2 isolate) were PCR-amplified from a genomic construct of clone NC_004718 (Fig. 1), and cloned into the pCR-XL-TOPO vector (Invitrogen). The full-length nsp8 and ORF6 genes were subjected to DNA sequencing and the inserts were verified against the corresponding region of the SARS coronavirus complete genome NC_004718. The nsp8 gene was excised from the pCR-XL-TOPO-nsp8 construct using the restriction enzymes NcoI and EcoRI, and ligated into the yeast two-hybrid Activation Domain vector, pACT2 to generate an N-terminal in-frame fusion with the GAL4 activation domain (AD). The ORF6 gene was excised from the pCR-XL-TOPO-ORF6 construct using the restriction enzymes EcoRI and PstI, and cloned into the yeast two-hybrid DNA Binding Domain (BD) vector pGBKT7 in fusion with the GAL4 DNA-binding domain to express an N-terminal fusion protein. Similarly the NcoI and EcoRI sites were used to clone the nsp8 gene into the pRSET vector to give the construct pRSET-nsp8. All DNA manipulations were performed as described by Sambrook et al. (1989). All constructs were verified by restriction digestion and DNA sequencing.
Table 1.
Yeast strains, plasmids and recombinant plasmid constructs used in this study
| Strain/plasmid/construct | Genotype/description/reference |
|---|---|
| Strains | |
| AH109 | MATatrp1-901 his3 leu2-3, 112 ura3-52 ade2 gal4 gal80URA3∷GAL-lacZ LYS2∷GAL-HIS3 |
| Y187 | MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, 112, gal4Δ, met-, gal80Δ, URA3∷GAL1UAS- GAL1TATA-lacZ |
| Plasmids | |
| pGADT7/pACT2 | GAL4 AD vector [GAL4(768–881)]; LEU2, 2 μm, Ampr |
| pGBKT7/pAS2 | GAL4 DNA-BD vector [GAL4(1–147)]; TRP1, 2 μm, Ampr |
| Constructs | |
| pACT2-nsp8 | pCR-XL-TOPO-nsp8-easy-nsp8 digested with NcoI and EcoRI, fragment ligated into pACT2 |
| pGBKT7-ORF6 | pCR-XL-TOPO-ORF6 digested with NcoI and EcoRI, fragment ligated into pGBKT7 |
| pRSET-nsp8 | pGEMT-nsp8 digested with NcoI and EcoRI, fragment ligated into pRSET |
Yeast two-hybrid techniques
The GAL4-based two-hybrid system, kindly provided by Dr. Stephen Elledge (Harper et al., 1993), containing pGBKT7 (DNA-binding domain vector) and pACT2 (activation domain vector), together with the yeast reporter strain S. cerevisiae AH109 (trp1-901 his3 leu2-3, 112 ura3-52 ade2 gal4 gal80URA3∷GAL-lacZ LYS2∷GAL-HIS3) were employed. The host strain containing the Nucleocapsid protein (N) of SARS virus fusion constructs pACT2-N and pAS2-N, shown previously to form a homodimer (Surjit et al., 2004) was used as a positive control. The AH109 host contains integrated copies of both HIS3 and lacZ reporter genes under the control of GAL4 binding sites. The AH109 yeast strain was transformed with the appropriate plasmids, using the lithium acetate procedure and grown on SD plates in the absence of Trptophane (Trp) and Leucine (Leu); (SDTrp− and SDLeu−). Protein interaction was tested on SD plates without Leu, Trp and Histidine (SDLeu− Trp− His−). After 3 days at 30 °C, individual colonies were streaked out and tested for liquid and filter-lift β-galactosidase activity, 5 mM 3-amino-1,2,3-trizole assay (3AT) and the liquid β-galactosidase assay (Ober et al., 2002, Peiris et al., 2003, Pewe et al., 2005). The filter β-galactosidase assay, a parameter directly reflecting the strength of protein–protein interactions, was performed by streaking doubly transformed yeast colonies onto filter paper and allowing them to grow for 2 days on selection medium. Yeast was permeabilized by freezing yeast-impregnated filters in liquid nitrogen, and thawing at room temperature. This filter was placed over a second filter that was pre-soaked in Z buffer (pH 7.0) containing 10 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) and 0.27% β-mercaptoethanol. Filters were left for 18 h to develop a blue color, which indicated a positive protein–protein interaction. The liquid β-galactosidase activity was determined using the substrate CPRG as described previously (Tyagi et al., 2001, Tyagi et al., 2002). Relative enzymatic activity was determined in five independent transformants. Data for quantitative assays were collected for yeast cell number and are the mean ± S.E.M. of triplicate assays. Appropriate positive/negative controls and buffer blanks were used. The yeast Y187 host strain containing BD-1111 and AD-1112 were used as positive controls for these assays.
In-vitro cell-free coupled transcription/translation binding analysis
The 35[S]-methionine labeled full-length pRSET-His6-nsp8 protein (198 amino acids nsp8 with N-terminal His6-tag) and radiolabelled 35[S]-methionine full-length ORF6 accessory protein (66 amino acids with an N-terminal myc tag), were expressed in separate reactions using a coupled in-vitro transcription–translation system (TNT coupled reticulocyte lysate system; Promega) as per manufacturer's instructions. 10 μl of the His6-nsp8 labeled protein lysate was mixed with 10 μl of myc-ORF6 protein lysate. This mixture was incubated on ice for 2 h and the primary antibody (either anti-His or anti-myc) was added to the mixture. Subsequently the mixture was incubated at 4 °C for 1 overnight. Sepharose-A beads were added to the mixture and incubated at 4 °C with gentle shaking for 1 h. The beads were washed three times with PBS, resuspended in 10 μl of SDS-PAGE loading buffer and boiled for 5 min to dissociate the bound proteins and detected by autoradiography. Appropriate control reactions were performed to validate the data.
Mammalian cell cultures
The CV-1 (African Green monkey kidney) cell-line was a kind gift from Baxter Vaccines, Orth/Donau, Austria. The Vero E6 cell-line was purchased from the American Type Culture Collection (Manassas, VA, USA). All cells were cultured at 37 °C in 5% CO2 in DMEM containing 1 g/l glucose, 2 mM l-glutamine, 1.5 g/l sodium bicarbonate, 0.1 mM non-essential amino acids, 0.1 mg/ml streptomycin and 100 U penicillin, and 10% FBS (HyClone, Utah, USA).
Generation of ORF6 recombinant replicating vaccinia virus
ORF6 recombinant replicating vaccinia virus was generated using the system developed by Baxter Vaccine (Orth/Donau, Austria). The vector, cell-lines and viruses used here were kind gifts from Baxter Vaccine. ORF6 was amplified from cDNA prepared from SARS-CoV infected cells as previously described (Tan et al., 2004b) and the pair of primers used is ORF6_forward (5′-GCAAGCTTATGTTTCATCTTGTTGA-3′) and ORF6_reverse (5′-CCGGCGGCCGCTTATGGATAATCTAACACC-3′). The amplicon was digested with HindIII and NotI and cloned into the compatible sites of the pDD4-mh5 vector to create plasmid pDD4-mh5/ORF6. The pDD4-mh5 vector can be used to generate growth-competent recombinants expressing foreign genes in the D4 locus via a rescue method as previously described (Holzer and Falkner, 1997). Essentially, the parental virus lacking the D4 gene is replication negative in the usual permissive host lines and only the reintegration of D4 into the virus genome allows for growth in these lines. This is in contrast to classical drug-based screening approaches which have previously been well established in the generation of vaccinia recombinants.
CV-1 cells were grown to 70–80% confluence in 60 mm dishes. Confluent monolayers were washed once with PBS, followed by a two washes with 1 ml DME containing no FBS or antibiotics. The culture medium was removed and overlaid with 0.2 ml defective vaccinia virus (dVVL) (∼ multiplicity of infection is 1 pfu/cell) diluted in 0.6 ml DMEM for 1 h at 37 °C in CO2, with shaking every 15 min. dVVL is derived from the vaccinia virus Lister strain (ATCC VR-862; Ober et al., 2002, and was titered in the complementing cell line RK44.20; Holzer and Falkner, 1997). The supernatant was removed, replaced with 2 ml DMEM containing no FBS or antibiotics and cells were transfected with 5 μg pDD4-mh5/ORF6 using lipofectamine reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. After 3 days, advanced cytopathic effect was evident and the cells were harvested and subjected to Western blot analysis (see below) for determining ORF6 expression. Alternatively, the cells and culture supernatant were harvested together and then subjected to three rounds of freeze-thawing and clarified by centrifugation at 1500 rpm for 5 min. The supernatant, which constitute the P1 viral stock, was used to infect fresh CV-1 cells after which the P2 viral stock was harvested in the same manner and stored at − 80 °C before use. The P2 viral stock was titered in CV-1 cells. This approach of dominant host range selection provides a stringent and time-saving method for obtaining vaccinia recombinants carrying foreign genes and has been described previously (Holzer et al., 1998). The recombinant vaccinia virus expressing the SARS-CoV S protein was previously generated using the same methods (Lip et al., 2006).
Expression of viral proteins in mammalian cells and co-immuoprecipitation experiments
The nsp8 gene was PCR amplified from the pRSET-nsp8 plasmid with the following pair of primers: nsp8_forward (5′-CGGGATCCGGCACCATGGCTATTGCTTCAG-3′) and nsp8_reverse (5′-CCGCTCGAGTCACTGTAGTTTAACAGCTG-3′). The amplicon was digested with BamHI and XhoI and cloned into compatible sites in the mammalian expression pXJ40myc vector which contains a myc-tag at the N-terminus for ease of detection. Typically, ∼ 1 × 106 cells Vero E6 were plated on 6 cm dish and allowed to attach overnight and then infected with ORF6 recombinant vaccinia virus at a multiplicity of infection of 1. Infection media was replaced 1 h post-infection and the cells were transfected with 1 ug of pXJ40myc-nsp8 using lipofectamine reagent as described above. The cells were harvested ∼ 16 h later and lysed in IP buffer (50 mM Tris (pH 8.0), 150 mM NaCl, 0.5% NP40, 0.5% deoxycholic acid, 0.005% SDS) and used in co-immunoprecipitation experiments as previously described (Tan et al., 2004c). Briefly, the lysate was incubated with an anti-myc polyclonal antibody (A14, Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4 °C, followed by adsorption onto a 50 μl suspension of protein A-sepharose beads (Roche Molecular Biochemicals). Beads were then washed 3 times with cold IP buffer and subjected to Western blot analysis as previously described. Primary antibodies used here were anti-myc monoclonal (9E10, Santa Cruz Biotechnology) and rabbit anti-ORF6 (PUP3, Abgent, San Diego, CA, USA).
For co-immunoprecipitation experiments performed with SARS-CoV infected Vero E6 cells, Vero E6 cells were plated in 25 cm2 flasks and infected with an isolate of SARS-CoV (strain HKU39849) as previously described (Kaye et al., 2006). The cells were harvest at 0 or 24 h.p.i. and the lysates were then subjected to Western blot analysis and co-immunoprecipitation experiments as described above. Anti-nsp8 monoclonal (described below) and rabbit anti-ORF6 antibodies were used for Western blot while rabbit anti-flag polyclonal (Sigma) or rabbit anti-ORF6 antibodies were used for immunoprecipitation.
Immunofluorescence analysis
Indirect immunofluorescence was performed using SARS-CoV infected Vero E6 cells as previously described (Tan et al., 2004c). Briefly, Vero E6 cells were plated onto 4-well chamber slides (Lab-Tek) and infected with an isolate of SARS-CoV (strain HKU39849) as previously described (Kaye et al., 2006). 42 h post-infection, chamber slides were washed twice in phosphate-buffered saline (PBS); cells were then fixed and permeabilized with methanol for 5 min and subsequently subjected to gamma-irradiation to neutralize infectivity of the virus. Cells were then refixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X100. Blocking was done using PBS with 1% bovine serum albumin (Sigma) and each chamber was incubated with relevant and irrelevant control antibodies before being washed with PBS + 1% BSA several times. Chamber slides were then incubated using FITC-conjugated goat anti-mouse and rhodamine-conjugated goat anti-rabbit secondary antibodies (1:200; Santa Cruz) before being washed again and mounted using glass coverslips and a mixture of Fluorsave mounting medium (Calbiochem) and Vectashield mounting medium with DAPI (Vector Laboratories). Imaging was done with an Olympus Fluoview upright confocal microscope (Olympus).
Antibodies for detecting endogenous AP-1 and the FLAG epitope were purchased from Sigma and antibodies against LAMP-1 and LAMP-2 were purchased from Abcam plc. The anti-GAL4TA antibody was purchased from Santa Cruz. Monoclonal antibody against the nsp8 protein was prepared as follows: bacterially expressed glutathione S-transferase (GST)-nsp8 fusion protein was used to immunize BALB/c mice as previously described (Fielding et al., 2004). The spleen was excised from a mouse that showed strong antibody response and hybridoma fusion was performed to generate hybridomas producing monoclonal anti-nsp8 antibodies as previously described (Lip et al., 2006). All procedures on the use of laboratory animals were performed by trained personnel in accordance with the regulations and guidelines of the National Advisory Committee for Laboratory Animal Research (NACLAR), Singapore. The culture supernatants from several of these hybridoma clones were pooled and purified using a HiTrap protein G HP column (Amersham) and verified by immunofluoresence assay to be specific to the nsp8 protein.
The mouse monoclonal primary antibodies (mouse anti-nsp8 and anti-GAL4TA) were used at concentrations of 0.2 mg/ml and the rabbit polyclonal antibodies (anti-ORF6 and anti-flag) were used at concentrations of 0.0025 mg/ml. Identical dilutions of secondary antibodies were used for all samples.
Acknowledgments
This work was supported by internal funds from the International Centre for Genetic Engineering and Biotechnology (India), Institute of Molecular and Cell Biology (Agency for Science, Technology and Research (A⁎STAR), Singapore), and Microbiology Department, National University of Singapore and a research grant from the Department of Biotechnology, Government of India. We thank Baxter Vaccine (Orth/Donau, Austria) for sharing their proprietary vaccinia virus expression system, and personnel at the Biological Resource Centre (Agency for Science, Technology and Research (A⁎STAR), Singapore), Monoclonal Antibody Unit (Institute of Molecular and Cell Biology, Singapore) and PC4 laboratory (Victorian Infectious Diseases Reference Laboratory, Australia) for technical assistance.
References
- Denison M.R., Zoltick P.W., Hughes S.A., Giangreco B., Olson A.L., Perlman S., Leibowitz J.L., Weiss S.R. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology. 1992;189(1):274–284. doi: 10.1016/0042-6822(92)90703-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding B.C., Tan Y.J., Shuo S., Tan T.H., Ooi E.E., Lim S.G., Hong W., Goh P.Y. Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome coronavirus. J. Virol. 2004;78(14):7311–7318. doi: 10.1128/JVI.78.14.7311-7318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H., Liu Y.M., Chan W.S., Lo A.W., Au D.M., Waye M.M., Ho Y.Y. The putative protein 6 of the severe acute respiratory syndrome-associated coronavirus: expression and functional characterization. FEBS Lett. 2005;579(30):6763–6768. doi: 10.1016/j.febslet.2005.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W., Adami G.R., Wei N., Keyomarsi K., Elledge S.J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75(4):805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Holzer G.W., Falkner F.G. Construction of a vaccinia virus deficient in the essential DNA repair enzyme uracil DNA glycosylase by a complementing cell line. J. Virol. 1997;71(7):4997–5002. doi: 10.1128/jvi.71.7.4997-5002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer G.W., Gritschenberger W., Mayrhofer J.A., Wieser V., Dorner F., Falkner F.G. Dominant host range selection of vaccinia recombinants by rescue of an essential gene. Virology. 1998;249(1):160–166. doi: 10.1006/viro.1998.9307. [DOI] [PubMed] [Google Scholar]
- Imbert I., Guillemot J.C., Bourhis J.M., Bussetta C., Coutard B., Egloff M.P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25(20):4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C., Inoue T., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 2005;79(5):3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson W.T., Ginddings T.H., J.r., Taylor M.P., Mulinyawe S., Rabinovitch M., Kopito R.R., Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biology. 2005;3(5):e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye M., Druce J., Tran T., Kostecki R., Chibo D., Morris J., Catton M., Birch C. SARS-associated coronavirus replication in cell lines. Emerg. Infect. Dis. 2006;12(1):128–133. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Law P.T., Wong C.H., Au T.C., Chuck C.P., Kong S.K., Chan P.K., To K.F., Lo A.W., Chan J.Y., Suen Y.K., Chan H.Y., Fung K.P., Waye M.M., Sung J.J., Lo Y.M., Tsui S.K. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 2005;86(Pt. 7):1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- Lip K.M., Shen S., Yang X., Keng C.T., Zhang A., Oh H.L., Li Z.H., Hwang L.A., Chou C.F., Fielding B.C., Tan T.H., Mayrhofer J., Falkner F.G., Fu J., Lim S.G., Hong W., Tan Y.J. Monoclonal antibodies targeting the HR2 domain and the region immediately upstream of the HR2 of the S protein neutralize in vitro infection of severe acute respiratory syndrome coronavirus. J. Virol. 2006;80:941–950. doi: 10.1128/JVI.80.2.941-950.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Nelson C.A., Pekosz A., Lee C.A., Diamond M.S., Fremont D.H. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure. 2005;13(1):75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober B.T., Bruhl P., Schmidt M., Wieser V., Gritschenberger W., Coulibaly S., Savidis-Dacho H., Gerencer M., Falkner F.G. Immunogenicity and safety of defective vaccinia virus lister: comparison with modified vaccinia virus Ankara. J. Virol. 2002;76(15):7713–7723. doi: 10.1128/JVI.76.15.7713-7723.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pewe L., Zhou H., Netland J., Tangudu C., Olivares H., Shi L., Look D., Gallagher T., Perlman S. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 2005;79(17):11335–11342. doi: 10.1128/JVI.79.17.11335-11342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78(18):9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279(11):10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. 2nd Ed. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Kumar P., Chow V.T., Lal S.K. The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun. 2004;317(4):1030–1036. doi: 10.1016/j.bbrc.2004.03.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L.L., Diprose J., Alderton D., Walsh M., Grimes J.M., Stuart D.I. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12(2):341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Fielding B.C., Goh P.Y., Shen S., Tan T.H., Lim S.G., Hong W. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 2004;78(24):14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Goh P.Y., Fielding B.C., Shen S., Chou C.F., Fu J.L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E., Lim S.G., Hong W. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11(2):362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Teng E., Shen S., Tan T.H., Goh P.Y., Fielding B.C., Ooi E.E., Tan H.C., Lim S.G., Hong W. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78(13):6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Lim S.G., Hong W. Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus. Antivir. Res. 2006;72(2):78–88. doi: 10.1016/j.antiviral.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangudu C., Olivares H., Netland J., Perlman S., Gallagher T. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J. Virol. 2007;81(3):1220–1229. doi: 10.1128/JVI.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84(Pt. 9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Tyagi S., Jameel S., Lal S.K. Self-association and mapping of the interaction domain of hepatitis E virus ORF3 protein. J. Virol. 2001;75(5):2493–2498. doi: 10.1128/JVI.75.5.2493-2498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S., Salier J.P., Lal S.K. The liver-specific human alpha(1)-microglobulin/bikunin precursor (AMBP) is capable of self-association. Arch. Biochem. Biophys. 2002;399(1):66–72. doi: 10.1006/abbi.2001.2745. [DOI] [PubMed] [Google Scholar]
- Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J., Denison M.R., Davis N., Baric R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79(23):14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.J., Chen Y.C., Hsiao C.H., Kuo T.C., Chang S.C., Lu C.Y., Wei W.C., Lee C.H., Huang L.M., Chang M.F., Ho H.N., Lee F.J. Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett. 2004;565(1–3):111–116. doi: 10.1016/j.febslet.2004.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Shan Y., Zhao Z., Chen J., Cong Y. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol. J. 2005;2:66. doi: 10.1186/1743-422X-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R., Yang R.F., Shi M.D., Jiang M.R., Xie Y.H., Ruan H.Q., Jiang X.S., Shi L., Zhou H., Zhang L., Wu X.D., Lin Y., Ji Y.Y., Xiong L., Jin Y., Dai E.H., Wang X.Y., Si B.Y., Wang J., Wang H.X., Wang C.E., Gan Y.H., Li Y.C., Cao J.T., Zuo J.P., Shan S.F., Xie E., Chen S.H., Jiang Z.Q., Zhang X., Wang Y., Pei G., Sun B., Wu J.R. Characterization of the 3a protein of SARS-associated coronavirus in infected vero E6 cells and SARS patients. J. Mol. Biol. 2004;341(1):271–279. doi: 10.1016/j.jmb.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12(11):980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 2004;7(4):412–419. doi: 10.1016/j.mib.2004.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]