Abstract
Coronaviruses (CoV), including SARS and mouse hepatitis virus (MHV), are enveloped RNA viruses that induce formation of double-membrane vesicles (DMVs) and target their replication and transcription complexes (RTCs) on the DMV-limiting membranes. The DMV biogenesis has been connected with the early secretory pathway. CoV-induced DMVs, however, lack conventional endoplasmic reticulum (ER) or Golgi protein markers, leaving their membrane origins in question. We show that MHV co-opts the host cell machinery for COPII-independent vesicular ER export of a short-living regulator of ER-associated degradation (ERAD), EDEM1, to derive cellular membranes for replication. MHV infection causes accumulation of EDEM1 and OS-9, another short-living ER chaperone, in the DMVs. DMVs are coated with the nonlipidated LC3/Atg8 autophagy marker. Downregulation of LC3, but not inactivation of host cell autophagy, protects cells from CoV infection. Our study identifies the host cellular pathway hijacked for supplying CoV replication membranes and describes an autophagy-independent role for nonlipidated LC3-I.
Keywords: MICROBIO, CELLBIO
Highlights
► Coronavirus replicative membrane vesicles (DMVs) are associated with LC3-1 ► LC3-I, but not an intact autophagy machinery, is essential for CoV replication ► CoV hijacks ER-derived vesicles exporting short-lived ERAD regulators to generate DMVs
Introduction
As an early and crucial event upon infection, coronaviruses (CoV) such as the severe acute respiratory syndrome (SARS)-CoV and mouse hepatitis virus (MHV) induce the formation of double-membrane vesicles (DMVs) in host cells and target their RTCs on the limiting membranes of these structures (de Haan and Reggiori, 2008, Gosert et al., 2002, Miller and Krijnse-Locker, 2008, Salonen et al., 2005). A recent analysis of SARS-CoV- and MHV-infected cells by electron tomography has revealed that these DMVs are part of a reticular network of modified endoplasmic reticulum (ER) membranes and contain double-stranded RNA (dsRNA) in their interior (Knoops et al., 2008). The ER origin of the DMVs is supported by the finding that two nonstructural proteins (nsp3 and nsp4) with transmembrane segments that are part of the RTCs become N-glycosylated (Harcourt et al., 2004, Kanjanahaluethai et al., 2007, Oostra et al., 2008). Moreover, nsp4 localizes to the ER when separately expressed and moves to the DMVs upon viral infection (Oostra et al., 2007). Whereas previous work has shown that the early secretory pathway and CoV-induced DMV biogenesis are closely connected (Knoops et al., 2010, Oostra et al., 2007, Verheije et al., 2008), the lack of ER, ER-Golgi intermediate compartment (ERGIC), or Golgi protein markers in CoV-induced DMVs suggests that their biogenesis does not depend on the conventional routing of proteins through this transport pathway (Oostra et al., 2007, Snijder et al., 2006, Verheije et al., 2008). Thus, even though CoV must hijack ER-derived host cell membranes for replication, the precise origin of the DMV lipid bilayers, the host protein content, and the identity of the cellular factors essential for DMV formation remain mysterious (Knoops et al., 2008). The possible involvement of the autophagy machinery in the conversion of host membranes into DMVs has been reported. However, whereas Atg5, an essential component of the autophagy machinery (Mizushima et al., 2001, Yoshimori and Noda, 2008), has been shown to be dispensable for MHV replication (Zhao et al., 2007), contradictory immunofluorescence (IF) data report the presence (Prentice et al., 2004, Zhao et al., 2007) or the absence (de Haan and Reggiori, 2008, Snijder et al., 2006) of the autophagosome protein marker LC3/Atg8 on DMVs (de Haan and Reggiori, 2008). In this study, we have used MHV, the virus prototype for the CoV biology investigation, to unveil the origin of the virus-induced DMVs and to identify host cell factors essential for CoV replication.
In the ER, newly synthesized, unstructured polypeptides can attract the folding as well as the degradation machineries that are operating in the lumen of this organelle (Calì et al., 2008b, Hebert and Molinari, 2007). Under normal growth conditions, therefore, the activity of the ERAD machinery must be maintained low to avoid premature interruption of folding programs and favor attainment of the native structure over degradation of immature polypeptides (Calì et al., 2008a). Consistently, it has been reported that EDEM1, a crucial regulator of ERAD (Molinari et al., 2003, Oda et al., 2003), is selectively cleared from the ER to tune down the ERAD activity (Calì et al., 2008a). This regulation mechanism has been named ERAD tuning. It relies on the selective sorting of EDEM1 and probably other short-living ER chaperones in 200–800 nm vesicles called EDEMosomes. These vesicles, coated with nonlipidated LC3/Atg8 (LC3-I) (Calì et al., 2008a, Calì et al., 2008b), emerge from the ER through a COPII coat-independent mechanism (Zuber et al., 2007) and deliver their content to endosomal compartments for disposal in a series of poorly characterized events (Calì et al., 2008a, Calì et al., 2008b, Le Fourn et al., 2009, Zuber et al., 2007).
We have discovered that MHV exploits the pathway of EDEMosome formation to generate the DMVs required for viral replication. In doing so, MHV interferes with the degradation of EDEM1 and OS-9, another short-living chaperone that we identify here as a second EDEMosome cargo, by trapping them into the DMVs. Our data are consistent with the model in which DMVs are generated from the host ER (Knoops et al., 2008) and explain the absence of conventional ER resident chaperones in their interior. In addition, we show that, whereas the autophagy pathway is not essential for MHV infection, nonlipidated LC3-I coats CoV-induced DMVs and is essential for MHV replication.
Results and Discussion
MHV Infection Does Not Require an Intact Autophagy Machinery
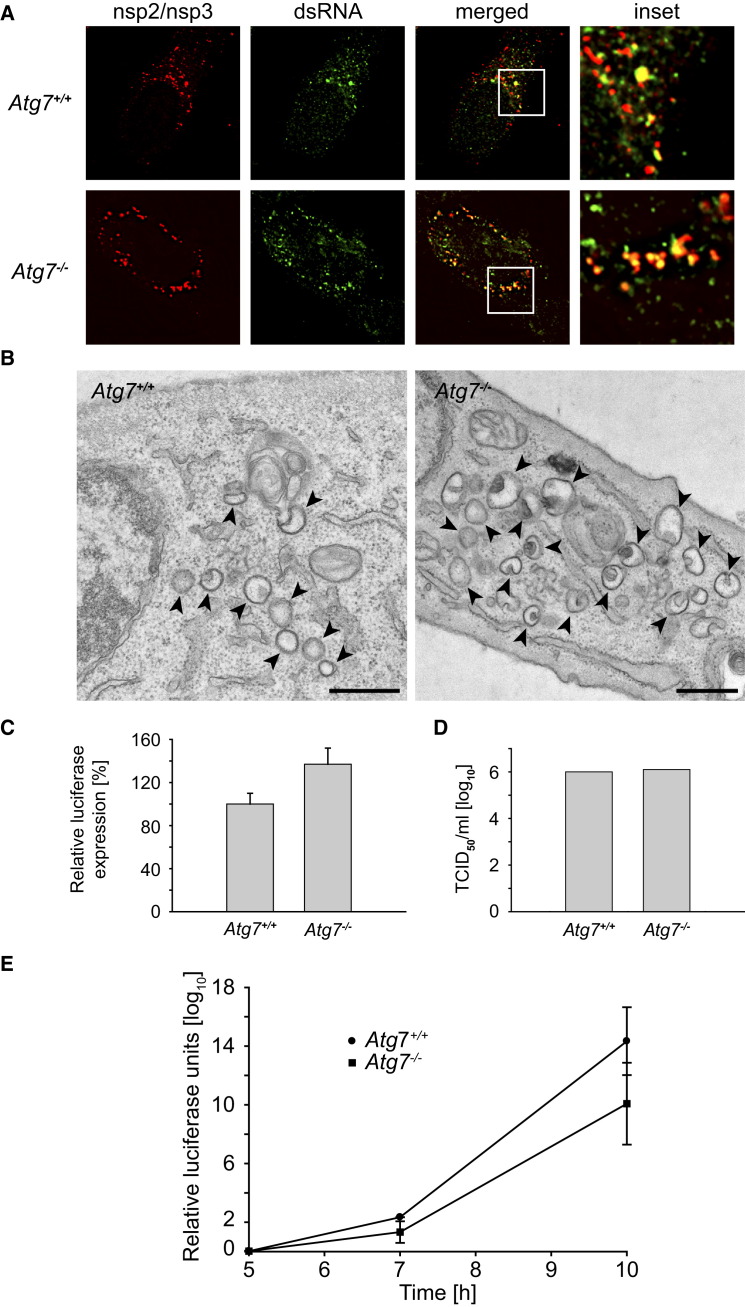
To conclusively establish whether autophagy is required for MHV infection, we assessed the consequences of deleting ATG7, a gene essential for autophagy (Komatsu et al., 2005), on DMV biogenesis. IF analysis of two RTC components, nsp2 and nsp3, in MHV-infected wild-type (Atg7 +/+) and ATG7 knockout mouse embryonic fibroblasts (Atg7−/− MEF) revealed that these two proteins were similarly distributed to numerous punctuate structures in both cell lines (Figure 1A). These puncta represent the virus-induced DMVs that contain viral dsRNA (colocalization in Figure 1A; Harcourt et al., 2004, Knoops et al., 2008). The presence in both Atg7 +/+ and Atg7−/− MEF of these double-membrane structures with a diameter of approximately 200–350 nm (Knoops et al., 2008, Stertz et al., 2007) was confirmed by conventional electron microscopy (Figure 1B). Moreover, cells with and without ATG7 were equally susceptible to MHV infection. This was established by assessing the virus replication using a recombinant luciferase-expressing MHV (Figure 1C) and by determining the titer of a virus stock on these cells by using the mean tissue culture infection dose (TCID50) test (Figure 1D). MHV replication in Atg7 +/+ and Atg7−/− MEF during the course of an infection was also very similar (Figure 1E). Thus, the conventional host cell autophagy is not required for formation of MHV-induced DMVs, nor for viral replication and production of viral progeny.
Figure 1.
MHV Replication, Assembly, and Release Is Autophagy Independent
(A) Atg7+/+ and Atg7−/− MEF were infected with MHV before being processed for IF at 7 hr postinfection (p.i.). The RTC components nsp2 and nsp3 colocalize with dsRNA to cytoplasmic puncta in both wild-type and knockout cells.
(B) DMVs have normal morphology in autophagy-deficient cells. Cells analyzed in (A) were also processed for EM. DMVs were absent in noninfected cells (data not shown). Arrowheads highlight DMVs. Scale bar, 500 nm.
(C) The Atg7+/+ and Atg7−/− MEF were inoculated with recombinant MHV-2aFLS, and luciferase expression was measured at 7 hr p.i. The graph shows the relative luciferase expression compared to Atg7+/+ cells after correction for cell viability from an experiment performed in triplicate.
(D) End point 10-fold dilutions of an MHV stock were titrated on Atg7+/+ and Atg7−/− MEF. Similar titers were observed indicating that ATG7 deletion does not affect MHV entry, replication, or assembly. Values presented in the graph are calculated and expressed as the log10 of TCID50 units per ml of supernatant, and the plotted values represent the average of two experiments.
(E) The experiment described in (C) was repeated in a time course manner, and luciferase expression was determined at 5, 7, and 10 hr p.i.
Error bars in (C–E) indicate the standard deviations between experiments.
Nonlipidated LC3/Atg8 Associates with CoV-Induced DMVs
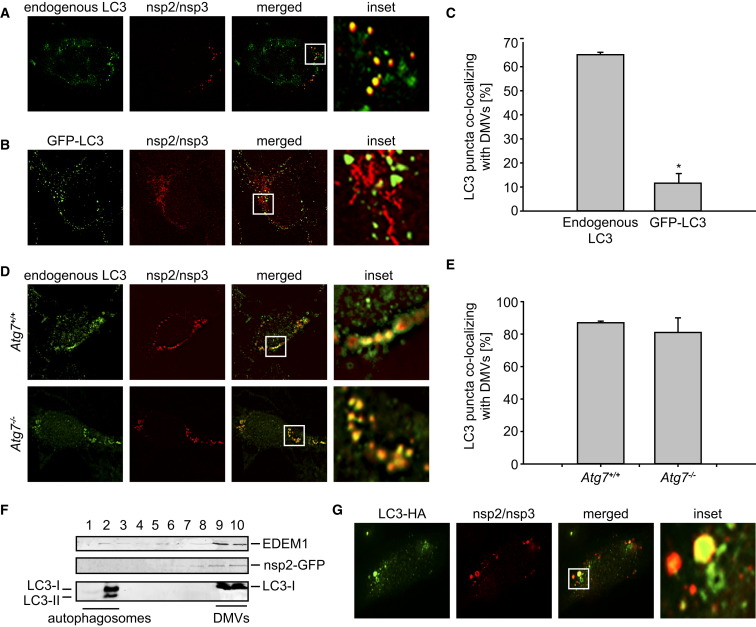
Because contrasting data have been published on the presence (Prentice et al., 2004, Zhao et al., 2007) or the absence (de Haan and Reggiori, 2008, Snijder et al., 2006) of LC3/Atg8 on CoV-induced DMVs, we examined this issue. Our analysis by IF showed that endogenous LC3 extensively colocalized with the DMV protein markers nsp2 and nsp3 (Figures 2A and 2C). This colocalization was observed during the entire course of the MHV infection (Figures S1A and S1B available online). In contrast, ectopically expressed GFP-LC3, a conventional protein marker for autophagosome membranes (Klionsky et al., 2008, Mizushima et al., 2004), did not colocalize with nsp2 and nsp3 (Figures 2B and 2C). These apparently conflicting data were confirmed in other cell lines (e.g., in HeLa cells; Figures S1C and S1D) and explain the contradictory data in the literature.
Figure 2.
Autophagy-Independent Recruitment of LC3-I onto MHV-Induced DMVs
(A and B) HEK293 cells stably transfected (B) or not (A) with a plasmid expressing GFP-LC3 were infected with MHV-Srec and processed for IF at 7 hr p.i.
(C) Summary statistics of the samples shown in (A) and (B) expressed as the percentage of LC3 or GFP-LC3 puncta colocalizing with the nsp2/nsp3 signals. Error bars represent the standard error of the mean percentage from counting 40 cells in three independent experiments. The asterisk indicates that the two samples are significantly different (tdf = 78 = 10.4; p < 0.00001).
(D) Atg7+/+ and Atg7−/− MEF were infected with MHV before being processed for IF at 7 hr p.i.
(E) Statistical analysis of the samples shown in (D) performed as described in (C).
(F) HeLa-CEACAM1a cells were infected with MHV-nsp2GFP for 7 hr before fractionating a cell extract on a continuous Optiprep gradient. Ten fractions were collected from the top to the bottom of the gradient and probed with antibodies against EDEM1, GFP, and LC3. The fractionation profile was confirmed by performing this experiment three times.
(G) HeLa-CEACAM1a cells were transiently transfected with a plasmid expressing C-terminally HA-tagged, nonlipidable LC3 before being infected with MHV. Cells were fixed at 7 hr p.i. and processed for IF. DMVs and LC3-HA were detected with antibodies against nps2/nsp3 and HA, respectively.
See also Figure S1.
LC3 is present in the cell predominantly in a cytoplasmic form (LC3-I) that, upon autophagy induction, is converted into an active lipidated form (LC3-II) by specific covalent linkage to the phosphatidylethanolamine present on autophagosomal membranes (Klionsky et al., 2008, Mizushima et al., 2004). The lipidation of LC3-I and the formation of LC3-II-coated autophagosomes require several proteins, including Atg7 (Komatsu et al., 2005). However, Atg7 was not necessary for the association of endogenous LC3 to DMVs (Figure 2D and 2E). Consequently, Atg7 and LC3 lipidation are not essential for the formation of LC3-positive DMVs. Analysis of the protein content in DMVs induced upon MHV-nsp2GFP infection of HeLa cells and separated on continuous density gradients as described (Calì et al., 2008a) showed the presence of LC3-I in the denser fractions containing the DMV protein marker nsp2-GFP (Figure 2F). These fractions were clearly separated from the lighter autophagosomes containing LC3-II that floated at the top of the gradient. Moreover, IF analyses revealed that ectopically expressed C-terminally HA-tagged, nonlipidable LC3 localizes on DMVs (Figure 2G). Taken all together, these data show that an intact host autophagy machinery is dispensable for the virus life cycle and that lipidation is not required for LC3 association with the DMV membranes.
Analogies between MHV-Induced DMVs and EDEMosomes
The ER origin of the MHV-induced DMVs (Knoops et al., 2008, Oostra et al., 2007), the absence of conventional ER markers in their membranes and lumen (Oostra et al., 2007, Snijder et al., 2006, Verheije et al., 2008), their association with LC3-I, and the fact that DMVs are stained with antibodies against endogenous LC3, but not with ectopically expressed GFP-LC3 (Figures 2, S1C, and S1D) are features reminiscent of those describing the EDEMosomes (Calì et al., 2008a). Significantly, the LC3-I coat distinguishes DMVs (this study) and EDEMosomes (Calì et al., 2008a; Figure S3D) from autophagosomes, which are associated with LC3-II and can be decorated with GFP-LC3 (Klionsky et al., 2008, Mizushima et al., 2004).
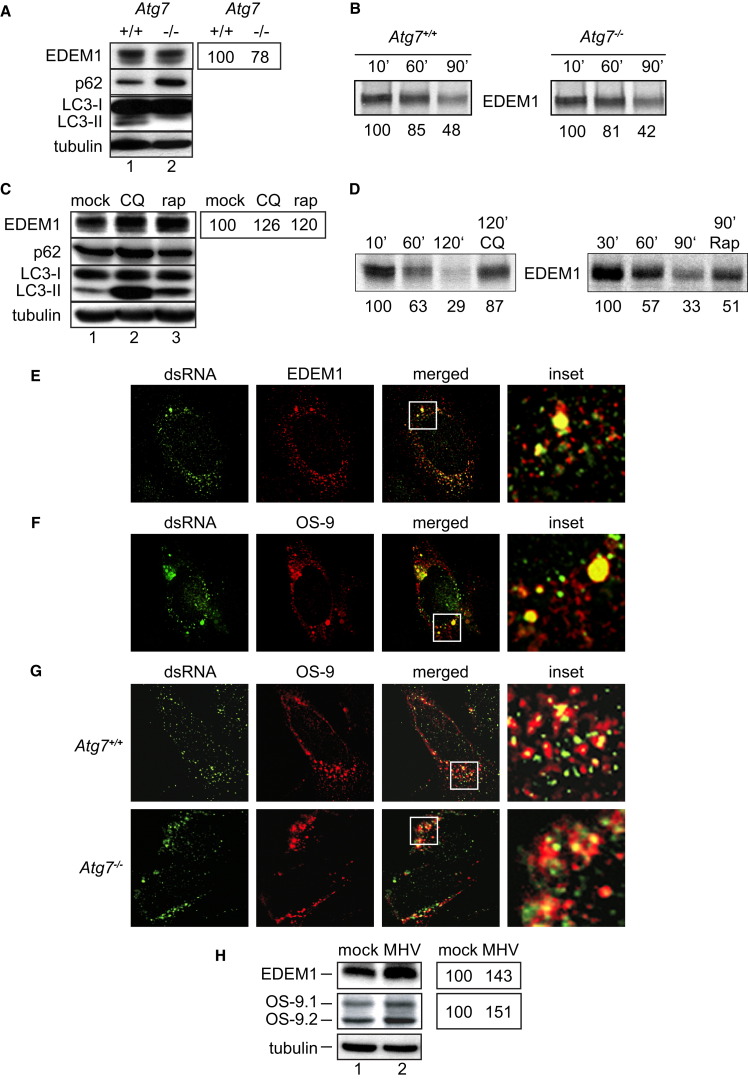
As in the case of formation of the ER-derived, CoV-induced DMVs, it is unclear whether an active autophagy machinery is required for formation of ER-derived EDEMosomes and/or for disposal of EDEM1 (Calì et al., 2008a versus Le Fourn et al., 2009). To better understand this, we compared variations in the intracellular levels of EDEM1 and of p62, a canonical substrate of autophagy (Bjørkøy et al., 2005), under conditions that either inactivate, e.g., ATG7 deletion (Figures 3A and 3B) or cell incubation with chloroquine (CQ) (Figures 3C and 3D) (Klionsky et al., 2008, Komatsu et al., 2005) or activate autophagy, e.g., rapamycin treatment (Figures 3C and 3D) (Klionsky et al., 2008). Deletion of ATG7 inhibits the p62 turnover (Waguri and Komatsu, 2009), thus substantially increasing the intracellular level of this autophagy substrate (Figure 3A). On the contrary, deletion of ATG7 did not result in substantial variations of the level of EDEM1 (Figure 3A), thus showing that Atg7 and conventional autophagy are dispensable for EDEM1 turnover. Consistently, when wild-type and Atg7−/− MEF were metabolically radiolabeled and chased for 10–90 min, the amount of residual endogenous EDEM1 decreased with similar kinetics in the two cell lines, confirming similar rate of disposal in the presence or absence of Atg7 (Figure 3B). Cell exposure to CQ inhibited the p62 degradation, resulting in higher levels of this protein, as expected for an autophagy substrate (Figure 3C; Bjørkøy et al., 2009). CQ delayed EDEM1 turnover and resulted in intracellular accumulation of EDEM1 (Figures 3C and 3D; Calì et al., 2008a). Finally, induction of autophagy with rapamycin reduced the intracellular levels of p62, as expected for an autophagy substrate (Figure 3C; Bjørkøy et al., 2009), but increased those of EDEM1 (Figure 3C) by delaying its turnover (Figure 3D). These results confirmed that, as reported above for CoV replication, the pathway regulating EDEM1 turnover is clearly distinct from autophagy. They also highlight another analogy between the ERAD tuning pathway and the CoV infection. Cell exposure to the autophagy-inducer rapamycin negatively affected both EDEM1 turnover (Figures 3C and 3D) and MHV replication, as shown by measuring the levels of both the N nucleocapsid levels and luciferase in cells infected with the recombinant luciferase-expressing MHV (Figure S2). All together, these observations led us to hypothesize that MHV hijacks the ERAD tuning machinery to co-opt cellular membranes for DMV generation.
Figure 3.
Components of the ERAD Tuning Pathway Are Associated with DMVs
(A) Cell extracts from Atg7+/+ (lane 1) and Atg7−/− (lane 2) were separated by SDS-PAGE and western blot membranes probed with antibodies against EDEM1, p62, LC3, and tubulin. Repetition of the analysis showed no significant differences in the EDEM1 level in Atg7+/+ versus Atg7−/− MEF. The percentages (right) indicate the relative EDEM1 levels in the knockout cells compared to wild-type cells and represent the average of two experiments.
(B) Atg7+/+ and Atg7−/− MEF were metabolically labeled and chased for the times indicated before lysis and EDEM1 immuno-isolation. The residual radiolabeled EDEM1 present in each lane was quantified and indicated below each band. Repetition of the analysis showed no significant differences in the EDEM1 turnover in Atg7+/+ versus Atg7−/− MEF.
(C) Atg7+/+ MEF were untreated or treated with 100 mM CQ or 1 mM rapamycin (rap) for 4 hr before preparation of cell extracts and analysis as in (A). The percentages (right) indicate the relative EDEM1 levels in drug-treated cells compared to mock-treated cells and represent the average of two experiments.
(D) Same as (B) to confirm, in a pulse-chase radiolabeling experiment, that CQ and rap delay EDEM1 turnover.
(E and F) HeLa cells were infected with MHV-Srec and processed for IF at 7 hr p.i. using antibodies against dsRNA and (E) EDEM1 or (F) OS-9.
(G) Atg7+/+ and Atg7−/− MEF infected with MHV-Srec were fixed at 7 hr p.i. and processed for IF using antibodies against OS-9 and dsRNA.
(H) Cell extracts were analyzed by western blot using antibodies against EDEM1 or OS-9. The percentages (right) indicate the relative EDEM1 and OS-9 levels in infected cells compared to control cells and represent the average of two experiments.
See also Figures S2, S3, and S4.
MHV Infection Interferes with ERAD Tuning and Results in Accumulation of ERAD Tuning Substrates in the Virus-Induced DMVs
As mentioned above, despite their ER origin (Knoops et al., 2008, Zuber et al., 2007), CoV-induced DMVs and EDEMosomes do not contain conventional ER chaperones or protein markers of the early compartments of the secretory pathway (Calì et al., 2008a, Oostra et al., 2007, Snijder et al., 2006, Stertz et al., 2007, Verheije et al., 2008). By IF analysis of MHV-infected wild-type and Atg7−/− knockout cells, we systematically assessed the presence in DMVs of several conventional protein markers of the early compartment of the secretory pathway, essentially confirming that none of them were in the virus-induced DMVs (Figure S3A and data not shown). Only two antibodies stained the virus-induced dsRNA- or nsp2/nsp3-positive compartments, namely those recognizing EDEM1 and OS-9 (Figures 3E and 3F). The presence of EDEM1 during the entire course of the infection (Figures S3B and S3C) confirmed our working model postulating that CoV hijack the ERAD tuning pathway to promote formation of their DMVs. Our model was further corroborated by the observation that cell infection with MHV substantially interfered with ERAD tuning, thus causing intracellular accumulation of EDEM1 and OS-9 (Figure 3H) due to the relocalization and confinement of these chaperones into the lasting MHV-induced DMVs (Figures 2F, 3E, 3F, S3B, and S3C). In noninfected cells, EDEM1 has a half-life of about 60 min (Figures 3B and 3D; Calì et al., 2008a). Because MHV infection induces a host translational shutoff (Raaben et al., 2007), the persistence of EDEM1 (and OS-9) for several hours confirms the defective clearance of this protein in infected cells (Figures 3H, S3B, and S3C). Entrapping of a fraction of cellular OS-9 in the DMVs was unexpected but significant. Like EDEM1, OS-9 is a regulator of protein disposal from the ER and it is expressed in two splice variants, OS-9.1 and OS-9.2 (Bernasconi et al., 2008, Christianson et al., 2008). Immunodetection of OS-9 in the MHV-induced DMVs (Figures 3F and 3G) led us to verify by subcellular fractionation whether OS-9 localizes to the same LC3-I-positive vesicular structures containing EDEM1 in noninfected cells (Calì et al., 2008a). Our analysis showed that, at steady state, about 20% of OS-9.1 and a smaller amount of OS-9.2 were indeed found in LC3-I-positive EDEMosomes, which sedimented in fractions 7–9 of a continuous Optiprep gradient, whereas other conventional ER chaperones were fully excluded from these fractions (Figure S3D; Calì et al., 2008a).
Despite their presence in DMVs, EDEM1 and OS-9 are not required for MHV replication. In fact, MHV infectivity was not affected by their knockdown, as measured by examining the synthesis of the viral structural N protein, the DMV formation by IF, and by assessing virus replication by either determining the TCID50 value of a virus stock on these cells or assaying the luciferase expression levels at different p.i. time points (Figure S4). Taken together, these results support the hypothesis that CoV hijack the ERAD tuning machinery to generate the replicative DMVs.
LC3-I Is Required for CoV Replication
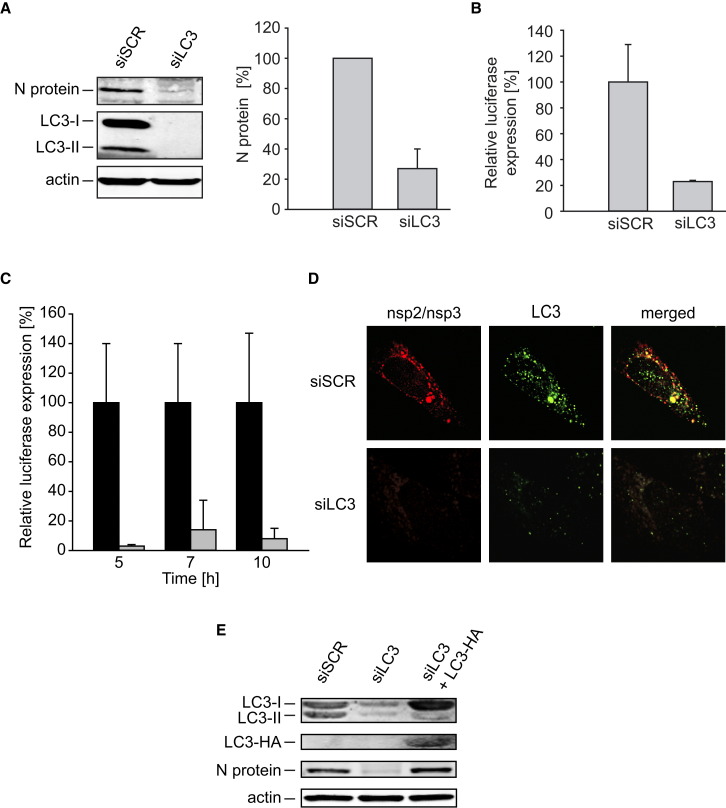
The association of LC3-I with MHV-induced DMVs is supported by data showing that these vesicles are decorated with anti-LC3 antibodies in cells lacking Atg7 and LC3-II (Figure 2D), they co-sediment with LC3-I, but not LC3-II, in density gradients (Figure 2F), and they colocalize with LC3-HA (Figure 2G). We therefore verified whether LC3 is required for viral replication even though the autophagy protein Atg7, which is required for its covalent membrane association, is dispensable (Figure 1). Efficient depletion of LC3A and LC3B was obtained by specific RNA interference (Figure 4A). LC3 downregulation protected cells from MHV infection as assessed by inhibition of the synthesis of the structural N protein (Figure 4A) and by a substantial decrease in luciferase levels measured in both single time point and time-course experiments (Figures 4B and 4C). The inhibition of MHV replication observed in LC3 knockdown cells was caused by a defect in DMV biogenesis, as no nsp2 and nsp3 signal was detected by IF (Figure 4D). The initial DMVs and RTCs generated upon MHV infection are necessary for the massive synthesis of extra nsp proteins, which, in turn, leads to the formation of a multitude of additional DMVs and RTCs. Consequently, a defect in DMV biogenesis results in a severe block of nsp production. Crucially, back transfection of LC3 knockdown cells with the plasmid-expressing C-terminally HA-tagged, nonlipidable LC3 restored MHV replication measured by monitoring N protein synthesis (Figure 4E).
Figure 4.
LC3-I Is Required for the Formation of DMVs
(A and B) HeLa-CEACAM1a cells were transfected with either siRNA directed against LC3A and LC3B (siLC3) or nontargeting (siSCR) siRNA. After 48 hr, cells were infected with MHV-2aFLS and analyzed at 7 hr p.i. This analysis has been repeated five times.
(A) Cell lysates were analyzed by western blot using antibodies against the N protein, LC3, and actin. The percentage indicates the relative N protein levels in LC3-depleted cells compared to control cells and represent the average of three experiments.
(B) Luciferase activity was measured as described in the Experimental Procedures, and error bars indicate the standard deviations for an experiment made in triplicate.
(C) The experiment described in (B) was repeated three times in a time course manner, and luciferase expression was determined at 5, 7, and 10 hr p.i. Error bars represent the standard deviations between experiments.
(D) Localization of nsp2/nsp3 and LC3 was examined by IF in cells treated as in (A) and (B).
(E) HeLa-CEACAM1a cells were transfected with either siRNA directed against LC3A and LC3B (siLC3) or nontargeting (siSCR) siRNA. After 24 hr, one of the samples was transfected with a plasmid expressing C-terminally HA-tagged, nonlipidable LC3, and MHV was inoculated at 48 hr. Cell lysates were prepared at 7 hr p.i. and analyzed by western blot using antibodies against the N protein, LC3, HA, and actin. Note that the higher levels of LC3-I in the siLC3+LC3-HA lane are due to LC3-HA, which run as LC3-I in the used SDS-PAGE gel.
During evolution, pathogens have developed molecular devices to exploit conserved cellular pathways to optimally infect, replicate, and/or leave host cells. Until now, no host protein has been directly implicated in CoV replication. The fact that both CoV-induced DMV formation and EDEM1/OS-9 turnover rely on analog mechanisms leads us to postulate that CoV hijack the ERAD tuning machinery for the generation of DMVs, which provide the membranous support for viral RTCs.
Based on our data, our current working model is that one or more CoV nsps do associate with a still elusive EDEMosome cargo receptor that normally mediates segregation and vesicular export from the ER of EDEM1 and OS-9. Accordingly to what is known about other vesicular transport pathways (Sato and Nakano, 2007), we postulate that the EDEMosome cargo receptor interacts with subunits of a cytosolic vesicle protein coat (nonlipidated LC3-I seems a good candidate). It is unlikely that the viral nsps recruit LC3-I at the cytosolic surface of DMVs because LC3-I is associated to EDEMosomes in uninfected cells (Calì et al., 2008a) and because we have not found direct interaction between nsp and LC3-I (unpublished data). Astonishingly, whereas EDEMosomes are transient structures that end their journey in late endosomes and/or lysosomes, MHV-induced DMVs are persistent cytoplasmic organelles (Ulasli et al., in press). Therefore, the presence of either nsps or other viral products inhibits the fusion of EDEMosomes/DMVs with the degradative compartments of the endosomal system. A main goal of future research is the identification of the elusive EDEMosome cargo receptor and the characterization of its involvement in CoV infection. It also remains to be established whether the ERAD tuning and CoV infection share mechanistic analogies with the recently characterized Atg5/Atg7-independent type of autophagy (Nishida et al., 2009). Importantly, in our study, we also show that blocking DMV formation by depleting LC3 severely impairs MHV infection. Consequently, future investigations on the involvement of viral proteins and ERAD tuning components, as well as the unconventional intervention of certain factors of the host cell autophagy machinery in the formation of CoV-induced DMVs, might provide valuable therapeutic targets.
Experimental Procedures
Cell Culture, Viruses, and Expression Plasmid
Cell culture, viruses, construction of the expression plasmid, and statistical analyses are described in the Supplemental Experimental Procedures.
Gene Silencing with siRNA
Sets of three different siRNA duplexes targeting different sites within the coding sequences of LC3A and LC3B were designed by and obtained from Applied Biosystems/Ambion (Nieuwerkerk a/d lJssel). One day after seeding, the HeLa-CEACAM1a cells were transfected with 40 nM of siRNA using Lipofectamine 2000. Experiments were conducted at 48 hr posttransfection. Antibodies against LC3 (NanoTools) were used to assess by western blot the depletion of the targeted gene.
Immunofluorescence Microscopy
Cells were grown and processed for IF on coverslides as described (Verheije et al., 2008). Fluorescence signals were visualized with a DeltaVision RT fluorescence microscope equipped with a CoolSnap camera (Applied Precision). Images were generated by collecting a stack of 20 pictures with focal planes 0.20 μm apart in order to cover the entire volume of the cell and by subsequently deconvolving them using the SoftWoRx software (Applied Precision). A single focal plane is shown at each time. Primary immunological reactions were carried out with polyclonal anti-EDEM1 (Sigma), anti-OS-9 (Novus Biologicals), anti-nsp2 and nsp3 (Schiller et al., 1998), and anti-Sec23 (Affinity Bioreagents) antisera and monoclonal anti-LC3 (NanoTools), anti-HA (a kind gift of G. Bu, Washington University), anti-dsRNA K1 (English and Scientific Consulting Bt.), anti-KDEL (Calbiochem) and anti-ERGIC53 (Alexis Biochemicals) antibodies. All experiments were repeated two or three times. Selected images show a representative fluorescence profile.
Western Blots
The cell extracts were prepared with lysis buffer (200 mM NaCl, 50 mM HEPES [pH 6.8], 2% CHAPS, and protease inhibitors) and boiled for 5 min in sample buffer. Proteins were separated on a SDS-PAGE gel and transferred onto a PVDF membrane before analysis with antibodies against EDEM1, OS-9, N protein (a gift of S. Siddell, University of Bristol), Erp57 (Soldà et al., 2006), calnexin (a gift of A. Helenius, ETH Zurich), p62 (Progen Biotechnik), mouse BiP (Stressgen), tubulin (Applied Biological Materials), and actin (MP Biomedicals). Alexa Fluor680-conjugated goat anti-rabbit or rabbit anti-mouse secondary antibodies (Molecular Probes) were used for the visualization of the immunoblots using an Odyssey system (Li-Cor Biosciences). Tubulin and actin represent the loading controls.
Miscellaneous Procedures
Electron microscopy, quantification of the virus replication using firefly luciferase-carrying virions MHV-2aFLS, measurement of the TCID50, subcellular fractionation on Optiprep gradients, and pulse-chase radiolabeling experiments followed by immunoprecipitations are described in Calì et al., 2008a, Slot and Geuze, 2007, Verheije et al., 2008.
Acknowledgments
The authors thank S. Baker, A. Helenius, K. Kirkegaard, M. Komatsu, S. Siddell, and S. Tooze for reagents; P. van Kerkhof and E. te Lintelo for technical advices; and I. Pen for guidance on the statistics. C.A.M.d.H. and F.R. are supported by the Utrecht University (High Potential grant). M.M. is supported by grants from the Foundation for Research on Neurodegenerative Diseases, the Fondazione San Salvatore, the Swiss National Center of Competence in Research on Neural Plasticity and Repair, the Swiss National Science Foundation, and ONELIFE Advisors SA.
Published: June 16, 2010
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at doi:10.1016/j.chom.2010.05.013.
Supplemental Information
References
- Bernasconi R., Pertel T., Luban J., Molinari M. A dual task for the Xbp1-responsive OS-9 variants in the mammalian endoplasmic reticulum: inhibiting secretion of misfolded protein conformers and enhancing their disposal. J. Biol. Chem. 2008;283:16446–16454. doi: 10.1074/jbc.M802272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Pankiv S., Øvervatn A., Brech A., Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- Calì T., Galli C., Olivari S., Molinari M. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem. Biophys. Res. Commun. 2008;371:405–410. doi: 10.1016/j.bbrc.2008.04.098. [DOI] [PubMed] [Google Scholar]
- Calì T., Vanoni O., Molinari M. The endoplasmic reticulum crossroads for newly synthesized polypeptide chains. Prog. Mol. Biol. Transl. Sci. 2008;83:135–179. doi: 10.1016/S0079-6603(08)00604-1. [DOI] [PubMed] [Google Scholar]
- Christianson J.C., Shaler T.A., Tyler R.E., Kopito R.R. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Reggiori F. Are nidoviruses hijacking the autophagy machinery? Autophagy. 2008;4:276–279. doi: 10.4161/auto.5241. [DOI] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert D.N., Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- Kanjanahaluethai A., Chen Z., Jukneliene D., Baker S.C. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology. 2007;361:391–401. doi: 10.1016/j.virol.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Swett-Tapia C., van den Worm S.H., Te Velthuis A.J., Koster A.J., Mommaas A.M., Snijder E.J., Kikkert M. Integrity of the early secretory pathway promotes, but is not required for, severe acute respiratory syndrome coronavirus RNA synthesis and virus-induced remodeling of endoplasmic reticulum membranes. J. Virol. 2010;84:833–846. doi: 10.1128/JVI.01826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Fourn V., Gaplovska-Kysela K., Guhl B., Santimaria R., Zuber C., Roth J. Basal autophagy is involved in the degradation of the ERAD component EDEM1. Cell. Mol. Life Sci. 2009;66:1434–1445. doi: 10.1007/s00018-009-9038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M., Calanca V., Galli C., Lucca P., Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Oda Y., Hosokawa N., Wada I., Nagata K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299:1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- Oostra M., te Lintelo E.G., Deijs M., Verheije M.H., Rottier P.J., de Haan C.A. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 2007;81:12323–12336. doi: 10.1128/JVI.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., Hagemeijer M.C., van Gent M., Bekker C.P., te Lintelo E.G., Rottier P.J., de Haan C.A. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 2008;82:12392–12405. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaben M., Groot Koerkamp M.J., Rottier P.J., de Haan C.A. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell. Microbiol. 2007;9:2218–2229. doi: 10.1111/j.1462-5822.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A., Ahola T., Kääriäinen L. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Nakano A. Mechanisms of COPII vesicle formation and protein sorting. FEBS Lett. 2007;581:2076–2082. doi: 10.1016/j.febslet.2007.01.091. [DOI] [PubMed] [Google Scholar]
- Schiller J.J., Kanjanahaluethai A., Baker S.C. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology. 1998;242:288–302. doi: 10.1006/viro.1997.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot J.W., Geuze H.J. Cryosectioning and immunolabeling. Nat. Protoc. 2007;2:2480–2491. doi: 10.1038/nprot.2007.365. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldà T., Garbi N., Hämmerling G.J., Molinari M. Consequences of ERp57 deletion on oxidative folding of obligate and facultative clients of the calnexin cycle. J. Biol. Chem. 2006;281:6219–6226. doi: 10.1074/jbc.M513595200. [DOI] [PubMed] [Google Scholar]
- Stertz S., Reichelt M., Spiegel M., Kuri T., Martínez-Sobrido L., García-Sastre A., Weber F., Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulasli M., Verheije M.H., de Haan C.A., Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 2010;12:844–886. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije M.H., Raaben M., Mari M., Te Lintelo E.G., Reggiori F., van Kuppeveld F.J., Rottier P.J., de Haan C.A. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 2008;4:e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waguri S., Komatsu M. Biochemical and morphological detection of inclusion bodies in autophagy-deficient mice. Methods Enzymol. 2009;453:181–196. doi: 10.1016/S0076-6879(08)04009-3. [DOI] [PubMed] [Google Scholar]
- Yoshimori T., Noda T. Toward unraveling membrane biogenesis in mammalian autophagy. Curr. Opin. Cell Biol. 2008;20:401–407. doi: 10.1016/j.ceb.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Thackray L.B., Miller B.C., Lynn T.M., Becker M.M., Ward E., Mizushima N.N., Denison M.R., Virgin H.W., 4th Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3:581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- Zuber C., Cormier J.H., Guhl B., Santimaria R., Hebert D.N., Roth J. EDEM1 reveals a quality control vesicular transport pathway out of the endoplasmic reticulum not involving the COPII exit sites. Proc. Natl. Acad. Sci. USA. 2007;104:4407–4412. doi: 10.1073/pnas.0700154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.