Abstract
Penaeus monodon shrimp collected from across the Indo-Pacific region during 1997–2004 were screened for the presence of yellow head-related viruses. Phylogenetic analyses of amplified ORF1b gene segments identified at least six distinct genetic lineages (genotypes). Genotype 1 (YHV) was detected only in shrimp with yellow head disease. Genotype 2 (GAV) was detected in diseased shrimp with the less severe condition described as mid-crop mortality syndrome and in healthy shrimp from Australia, Thailand and Vietnam. Other genotypes occurred commonly in healthy shrimp. Sequence comparisons of structural protein genes (ORF2 and ORF3), intergenic regions (IGRs) and the long 3′-UTR supported the delineation of genotypes and identified both conserved and variant structural features. In putative transcription regulating sequences (TRSs) encompassing the sub-genomic mRNA 5′-termini, a core motif (5′-GUCAAUUACAAC-3′) is absolutely conserved. A small (83 nt) open reading frame (ORF4) in the 3′-UTR of GAV is variously truncated in all other genotypes and a TRS-like element preceding ORF4 is invariably corrupted by a A > G/U substitution in the central core motif (5′-UU(G/U)CAAC-3′). The data support previous evidence that ORF4 is a non-functional gene under construction or deconstruction. The 3′-UTRs also contain predicted 3′-terminal hairpin-loop structures that are preserved in all genotypes by compensatory nucleotide substitutions, suggesting a role in polymerase recognition for minus-strand RNA synthesis.
Keywords: Yellow head virus, Gill-associated virus, Phylogeny, Replicase gene, Genotypic variation, Genotypes
Introduction
Yellow head disease (YHD) emerged in 1990 in farmed black tiger shrimp (Penaeus monodon) in Thailand (Limsuwan, 1991). It is a devastating disease that can cause total crop losses within a few days following the appearance of gross signs (Chantanachookin et al., 1993). The causative agent, yellow head virus (YHV), is a bacilliform, enveloped, (+) ssRNA virus classified in the genus Okavirus, family Roniviridae within the order Nidovirales (Walker et al., 2005). A related nidovirus, gill-associated virus (GAV), occurs commonly as a chronic infection in healthy wild and farmed P. monodon in eastern Australia (Spann et al., 1995, Cowley et al., 2000a, Walker et al., 2001). GAV has been associated with a disease described as mid-crop mortality syndrome (MCMS) in which mortalities progressively accumulate from the mid-late juvenile stage onwards (Spann et al., 1997, Walker et al., 2001). A variant YHV genotype has also been detected in healthy P. monodon broodstock from Thailand (Soowannayan et al., 2003) and YHV or related viruses have been reported in P. monodon and Penaeus japonicus farmed in Taiwan (Wang et al., 1996, Wang and Chang, 2000). However, these viruses were not associated with typical YHD and their relationships to YHV or GAV remain unconfirmed. There are also reports of YHD or YHV in P. monodon in several other countries in the Asian region including the Philippines, India, Indonesia, Sri Lanka, Malaysia, Vietnam and China (Walker et al., 2001) but these have rarely been confirmed by laboratory analysis.
Like other nidoviruses, the polyadenylated, (+) RNA genomes of YHV (26,662 nt) and GAV (26,235 nt) are expressed from a nested set of genomic and subgenomic mRNAs (Cowley and Walker, 2002, Cowley et al., 2002a, Sittidilokratna et al., 2008). The genomes are organized similarly (5′-ORF1a/ORF1b-ORF2-ORF3-(ORF4)-polyA-3′). ORF1a encodes a long polyprotein (pp1a) containing 3C-like and papain-like proteases required for auto-processing, and overlaps ORF1b which encodes a ‘SDD’ RNA-dependent RNA polymerase, helicase, metal-ion-binding, exonuclease, uridylate-specific endoribonuclease and ribose-2′-O-methyl transferase domains (Cowley et al., 2000b, Ziebuhr et al., 2003, Sittidilokratna et al., 2002, Sittidilokratna et al., 2008). A -1 ribosomal frame-shift element at the ORF1a/ORF1b overlap facilitates translation of the complete pp1ab polyprotein (Cowley et al., 2002a). ORF2 encodes the nucleoprotein (p20) and ORF3 encodes a polyprotein that is processed to generate two envelope glycoproteins (gp116 and gp64) and a small N-terminal triple-membrane-spanning (TMS) fragment of unknown function (Cowley and Walker, 2002, Jitrapakdee et al., 2003, Cowley et al., 2004a, Sittidilokratna et al., 2006). ORF4 is a short open reading frame that, although possibly expressed in GAV (Cowley and Walker, 2008), is severely truncated in YHV and unlikely to encode a functional protein (Sittidilokratna et al., 2008). Intergenic regions (IGRs) upstream of ORF2 and ORF3 contain conserved transcription regulatory sequences (TRSs) that facilitate transcription of two sub-genomic mRNAs (sgmRNAs) which, together with the genome-length mRNA, form a 3′-coterminal nested set characteristic of nidoviruses (Cowley et al., 2002a, Sittidilokratna et al., 2008). The genome sequences of YHV and GAV are ∼ 79% identical, with amino acid sequence identity ranging from ∼ 73% in gp116 to ∼ 84% in pp1ab (Sittidilokratna et al., 2008). Based on levels of sequence divergence, YHV and GAV have been regarded as closely related variants that are likely to represent geographic topotypes in a larger complex (Cowley et al., 1999, Walker et al., 2001). Currently, they are each classified as the species Gill-associated virus (Walker et al., 2005).
This paper reports the analysis of tissue samples from healthy and diseased P. monodon collected from several countries in the Indo-Pacific region. Analysis of partial ORF1b sequences identified at least six distinct genetic lineages (genotypes) of yellow head-related viruses of which only YHV (genotype 1) was detected in shrimp from YHD-outbreak ponds. Sequencing of the structural gene region of representatives of genotypes 3, 4 and 5 identified significant diversity, particularly in IGRs and the N-terminal gp116 region, but preservation of TRS-like elements and a 3′-terminal hairpin structure likely to be involved in negative-strand RNA synthesis.
Results
A new genotype (genotype 3) detected in healthy P. monodon broodstock and postlarvae
In an initial study, a semi-nested RT-PCR targeting a 279 nt ORF1b sequence located just downstream of helicase domain motif VI was used to detect YHV-related viruses in 38 apparently healthy P. monodon broodstock sampled from commercial hatcheries in Central Thailand in March 2000. PCR products were amplified from 25 (66%) of the broodstock (Fig. 1A). Nucleotide sequences were determined for six of these in addition to PCR products amplified from juvenile shrimp from YHD outbreaks in Thailand and Taiwan and healthy broodstock and postlarvae from hatcheries in Australia, Vietnam and Sarawak in Malaysia (Table 1 ). Using 25 sequences, ClustalX was used to align the 231 nt segment bounded by the nested PCR primers (equivalent to the sequence A17492 to A17722 in the GAV reference strain; GenBank AF227196). An unrooted neighbour-joining phylogenetic tree generated from this alignment segregated the sequences into three clusters (Fig. 1B). One cluster (genotype 1) included the YHV reference strain (GenBank AY052786) and all eight viruses from juvenile shrimp from YHD outbreaks. Whilst slight nucleotide variation occurred between some of these viruses, the sequences of one Thai (THA-00-D11) and three of the four Taiwanese viruses (TWN-00-D1, TWN-00-D2, TWN-00-D3) were identical. The second cluster (genotype 2) included the GAV reference strain, the two other Australian viruses from either healthy or MCMS-affected shrimp, and one of the five Vietnamese viruses from healthy postlarvae (VNM-01-H65). The third phylogenetic cluster (genotype 3), which has not been described previously, comprised all viruses from healthy broodstock from Thailand and Sarawak and the other four Vietnamese viruses detected in healthy postlarvae. The Vietnamese viruses appeared to form a sub-group within genotype 3.
Fig. 1.
(A) 1% agarose-TAE gels illustrating RT-nested PCR products amplified from 16 of 19 healthy P. monodon broodstock (lanes 1–19) that were collected from central Thailand in March 2002. A semi-nested one-step RT-PCR and primers 2s12, 2a12 and 2a43 were used to amplify products of 280 and 575 bp. A DNA ladder (lane M) and positive control (YHV) reaction (lane +) are indicated. (B) Unrooted, neighbour-joining phylogenetic tree constructed from a ClustalX multiple alignment of a 231 nt sequence in the ORF1b replicase gene obtained for 25 viruses including the reference strains of YHV (THA-98-Ref) and GAV (AUS-96-Ref). The samples were collected from Thailand, Taiwan, Australia, Vietnam and Malaysia in 2000 and 2001. The sources of viruses and their codes are described in Table 1. Bootstrap values shown at key branch nodes were determined for 1000 replicates. Horizontal branch lengths indicate phylogenetic distance calculated as the number of nucleotide substitutions per total nucleotide residues. The clustering of viruses into three discrete genetic lineages (genotypes 1 to 3) is indicated.
Table 1.
Samples of shrimp (Penaeus monodon) collected from sites in the Indo-Pacific region
| Sample codea | Collection date | Sample origin | Life stage | Tissues type | Health status | Genotype |
|---|---|---|---|---|---|---|
| AUS-97-MCMS1 | 1997 | Queensland, Australia | Sub-adult | LO | MCMS | 2 (GAV) |
| AUS-97-MCMS2 | 24-04-1997 | Queensland, Australia | Adult | gill | MCMS | 2 (GAV) |
| AUS-97-MCMS3 | 1997 | Queensland, Australia | Sub-adult | LO | MCMS | 2 (GAV) |
| AUS-00-H1 | 2000 | Queensland, Australia | Sub-adult | LO | Healthy | 2 (GAV) |
| AUS-00-H2 | 2000 | Queensland, Australia | Sub-adult | LO | Healthy | 2 (GAV) |
| AUS-00-HL4 | 16-04-2000 | Queensland, Australia | Adult | LO | Healthy | 2 (GAV) |
| AUS-00-HL5 | 2000 | Queensland, Australia | Sub-adult | LO | Healthy | 2 (GAV) |
| AUS-00-HL11 | 2000 | Queensland, Australia | Sub-adult | LO | Healthy | 2 (GAV) |
| IDN-04-H4 | 23-01-2004 | Indonesia | Sub-adult | Muscle | Healthy | 3 |
| IDN-04-H7 | 23-01-2004 | Indonesia | Sub-adult | Muscle | Healthy | 3 |
| IDN-04-H10 | 23-02-2004 | Indonesia | Sub-adult | Muscle | Healthy | 3 |
| IDN-04-H11 | 10-02-2004 | Indonesia | Sub-adult | Muscle + pleopod | Healthy | 3 |
| IND-02-H5 | 15-06-2002 | Nellore, India | Pl14 | Whole | Healthy | 4 |
| IND-02-H7 | 15-06-2002 | Nellore, India | Pl11 | Whole | Healthy | 4 |
| IND-02-H9 | 15-06-2002 | Nellore, India | Pl15 | Whole | Healthy | 4 |
| MOZ-04-H1 | 13-01-2004 | Mozambique | Brooder | Gill | Healthy | 3 |
| MOZ-04-H6 | 13-01-2004 | Mozambique | Brooder | Gill | Healthy | 6 |
| MOZ-04-H8 | 13-01-2004 | Mozambique | Brooder | Gill | Healthy | 6 |
| MOZ-04-H9 | 13-01-2004 | Mozambique | Brooder | Gill | Healthy | 6 |
| MOZ-04-H11 | 13-01-2004 | Mozambique | Brooder | Gill | Healthy | 6 |
| MOZ-04-H12 | 13-01-2004 | Mozambique | Brooder | Gill | Healthy | 6 |
| MYS-01-H1 | 2001 | Sarawak, Malaysia | Brooder | LO + pleopod | Healthy | 3 |
| MYS-03-H1 | 06-06-2003 | Malaysia | Sub-adult | Pleopod | Healthy | 3 |
| MYS-03-H2 | 06-06-2003 | Malaysia | Sub-adult | Pleopod | Healthy | 3 |
| MYS-03-H3 | 06-06-2003 | Malaysia | Sub-adult | Pleopod | Healthy | 3 |
| MYS-03-H4 | 06-06-2003 | Malaysia | Sub-adult | Pleopod | Healthy | 5 |
| PHL-03-H8 | 02-10-2003 | Iloilo, Philippines | Pl12 | Whole | Healthy | 5 |
| THA-00-H3 | 2000 | Thailand | Brooder | Pleopod | Healthy | 3 |
| THA-00-H7 | 2000 | Thailand | Brooder | Pleopod | Healthy | 3 |
| THA-00-H8 | 2000 | Thailand | Brooder | Pleopod | Healthy | 3 |
| THA-00-H10 | 2000 | Thailand | Brooder | Pleopod | Healthy | 3 |
| THA-00-H11 | 2000 | Thailand | Brooder | Pleopod | Healthy | 3 |
| THA-00-H12 | 2000 | Thailand | Brooder | Pleopod | Healthy | 3 |
| THA-00-H19 | 2000 | Thailand | Brooder | Pleopod | Healthy | 3 |
| THA-00-D1 | 2000 | Thailand | Sub-adult | Pleopod | Diseased | 1 (YHV) |
| THA-00-D9 | 2000 | Thailand | Sub-adult | Pleopod | Diseased | 1 (YHV) |
| THA-00-D11 | 2000 | Thailand | Sub-adult | Pleopod | Diseased | 1 (YHV) |
| THA-00-D12 | 2000 | Thailand | Sub-adult | Pleopod | Diseased | 1 (YHV) |
| THA-00-DRH | 2000 | Thailand | Sub-adult | Half head | Diseased | 1 (YHV) |
| THA-03-D1 | 2003 | Nankorn, Thailand | Juvenile | Gill | Diseased | 1 (YHV) |
| THA-03-D2 | 2003 | Nankorn, Thailand | Juvenile | Gill | Diseased | 1 (YHV) |
| THA-03-D3 | 2003 | Nankorn, Thailand | Juvenile | Gill | Diseased | 1 (YHV) |
| THA-01-D4 | 2001 | Nakorn Pathom, Thailand | Juvenile | Gill | Diseased | 1 (YHV) |
| THA-01-D8 | 2001 | Nakorn Pathom, Thailand | Juvenile | Gill | Diseased | 1 (YHV) |
| THA-01-D9 | 2001 | Nakorn Pathom, Thailand | Juvenile | Gill | Diseased | 1 (YHV) |
| THA-01-D10 | 2001 | Nakorn Pathom, Thailand | Juvenile | Gill | Diseased | 1 (YHV) |
| THA-03-D29 | 2003 | Chachoengsao, Thailand | Juvenile | Gill | Diseased | 1 (YHV) |
| THA-03-D30 | 2003 | Chachoengsao, Thailand | Juvenile | Gill | Diseased | 1 (YHV) |
| THA-03-D33 | 2003 | Rachaburi, Thailand | Juvenile | Gill | Diseased | 1 (YHV) |
| THA-02-D34 | 2002 | Thailand | Juvenile | Gill | Diseased | 1 (YHV) |
| THA-04-H20 | 28-03-2004 | Supanburi, Thailand | Pl20 | Whole | Healthy | 2 (GAV) |
| THA-03-HA | 2003 | Thailand | Adult | Gill | Healthy | 2 (GAV) |
| THA-03-HB | 2003 | Thailand | Adult | Gill | Healthy | 2 (GAV) |
| THA-03-HG | 2003 | Thailand | Adult | Gill | Healthy | 2 (GAV) |
| THA-04-HK | 16-04-2004 | Prathum Thani, Thailand | Adult | Gill | Healthy | 2 (GAV) |
| THA-03-DB1 | 27-10-2003 | Thailand | Brooder | Gill + pleopod | Diseased | 1 (YHV) |
| THA-03-HB3 | 30-09-2003 | Thailand | Brooder | Gill + pleopod | Healthy | 2 (GAV) |
| THA-03-SG21 | 2003 | Thailand | Sub-adult | Gill | Slow growth | 5 |
| TWN-00-D1 | 2000 | Taiwan | Sub-adult | Gill | Diseased | 1 (YHV) |
| TWN-00-D2 | 2000 | Taiwan | Sub-adult | Gill | Diseased | 1 (YHV) |
| TWN-00-D3 | 2000 | Taiwan | Sub-adult | Gill | Diseased | 1 (YHV) |
| TWN-00-D4 | 2000 | Taiwan | Sub-adult | Gill | Diseased | 1 (YHV) |
| TWN-03-H9 | 05-07-2003 | Taiwan | Juvenile | Pleopod | Healthy | 3 |
| TWN-03-H11 | 05-07-2003 | Taiwan | Juvenile | Pleopod | Healthy | 3 |
| VNM-01-H41 | 2001 | Vietnam | Pl | Whole | Healthy | 3 |
| VNM-01-H42 | 2001 | Vietnam | Pl | Whole | Healthy | 3 |
| VNM-01-H2 | 2001 | Vietnam | Pl | Whole | Healthy | 3 |
| VNM-01-H82 | 2001 | Vietnam | Pl | Whole | Healthy | 3 |
| VNM-01-H65 | 2001 | Vietnam | Pl | Whole | Healthy | 2 (GAV) |
| VNM-02-H81 | 02-02-2002 | Vietnam | Pl | Whole | Healthy | 3 |
| VNM-02-H258 | 01-02-2002 | Nha Trang, Vietnam | Pl12 | Whole | Healthy | 3 |
| VNM-02-H278 | 06-03-2002 | Hon Chong, Vietnam | Pl12 | Whole | Healthy | 3 |
| VNM-02-H264 | 03-03-2002 | Ca Na, Vietnam | Pl12 | Whole | Healthy | 3 |
| VNM-01-H77 | 31-12-2001 | Phan Thiet, Vietnam | Pl | Whole | Healthy | 2 (GAV) |
| VNM-02-H64 | 18-01-2002 | Nha Trang, Vietnam | Pl13 | Whole | Healthy | 2 (GAV) |
| VNM-02-H93 | 04-01-2002 | Hon Khoai, Vietnam | Pl10 | Whole | Healthy | 3 |
| VNM-02-H6 | 04-01-2002 | Ngoc Hien, Vietnam | Pl12 | Whole | Healthy | 2 (GAV) |
| VNM-02-H5 | 01-04-2002 | Vietnam | Sub-adult | Muscle | Healthy | 3 |
| VNM-02-H70 | 04-01-2002 | Hon Khoai, Vietnam | Pl10 | Whole | Healthy | 3 |
Shrimp sample code assignment: country of origin-year of collection-health status-identification number/code. Abbreviations: healthy (H), diseased (D), mid-crop mortality syndrome (MCMS), slow growth (SG), lymphoid organ (LO), postlarvae (PL).
Other genotypes detected in P. monodon from various Indo-Pacific regions
A more extensive study was conducted using a larger set of P. monodon sampled over a wider geographic range. A new RT-nested PCR designed to amplify a 722 nt ORF1b gene segment encompassing the semi-nested RT-PCR sequence was used in initial analyses. This test was further modified to utilize degenerate primer pairs YH30-F2/R2 and YHV31-F2/R2 to better accommodate sequence variations among the known genotypes and was applied to ∼ 200 P. monodon sampled between 1997 and 2004 from Indonesia, the Philippines, Taiwan, Vietnam, Thailand, Malaysia, India, Sri Lanka, Mozambique and Australia (Table 1). Samples included whole postlarvae as well as gill, lymphoid organ, hepatopancreas or whole head tissues of juvenile farmed shrimp or adult broodstock from hatcheries. Most samples were from healthy shrimp but some originated from shrimp either displaying typical signs of YHD or that were moribund and collected from MCMS-affected ponds in Australia.
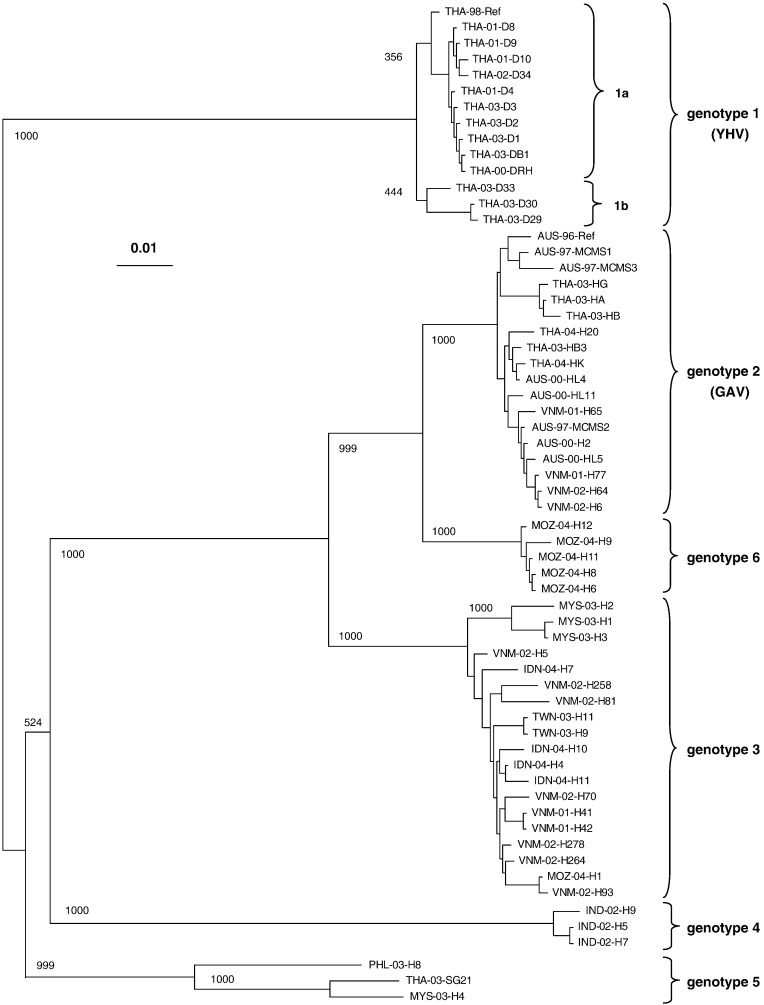
In total, 57 of the ∼ 200 samples generated nested PCR amplicon yields suitable for sequencing. Sequences of these 57 viruses, together with YHV and GAV reference strain sequences, were aligned using ClustalX. An unrooted neighbour-joining tree generated from the alignment of the 688–671 nt segment (equivalent to the GAV reference strain sequence G17259 to A17929) segregated the viruses into six major clusters well supported by bootstrap values (> 70%) (Fig. 2 ). The clustering of viruses in genotypes 1, 2 and 3 substantiated initial findings with the 231 nt sequence in that all 13 viruses clustering with YHV (genotype 1) originated from juvenile YHD-affected shrimp and all seven viruses from healthy and diseased Australian shrimp clustered with GAV (genotype 2). Ten Thai and Vietnamese viruses from healthy shrimp also clustered with GAV. The 17 genotype 3 viruses displayed the widest geographic distribution, being identified in healthy broodstock and postlarvae from hatcheries in Thailand, Vietnam, Taiwan, Indonesia, Malaysia and Mozambique. The three viruses from Malaysian shrimp also appeared to form a subcluster in genotype 3.
Fig. 2.
Unrooted, neighbour-joining phylogenetic tree constructed from a ClustalX multiple alignment of a 668–671 nt sequence in the ORF1b replicase gene obtained for 57 viruses including the reference strains of YHV (THA-98-Ref) and GAV (AUS-96-Ref). The samples were collected from throughout the Indo-Pacific region from 1997–2004. The sources of viruses and their codes are described in Table 1. Bootstrap values shown at key branch nodes were determined for 1000 replicates. Horizontal branch lengths indicate phylogenetic distance calculated as the number of nucleotide substitutions per total nucleotide residues. The clustering of viruses into the six discrete genetic lineages (genotypes 1 to 6) is indicated.
In addition to these genotypes, the tree delineated three new clusters. One lineage (genotype 4) was clearly separated from all other genotypes and comprised viruses detected in three healthy postlarvae batches sampled from hatcheries in Nellore, India. Another lineage (genotype 6) was most closely related to genotype 2 (GAV) and comprised viruses detected in five of the six broodstock from Mozambique. The other Mozambique virus clustered in genotype 3. However, as Mozambique shrimp were supplied from a commercial breeding facility in Malaysia, it is possible this sample was mislabeled. Another lineage (genotype 5) comprised viruses derived from a healthy sub-adult from Malaysia (MYS-03-H4), a sub-adult from Thailand displaying slower than normal growth (THA-03-SG21), and a batch of healthy postlarvae sampled from the Philippines (PHL-03-H8). The relationships among the three genotype 5 viruses were more disparate than viruses clustering within the other lineages. Pair-wise alignments indicated that THA-03-SG21 and MYS-03-H4 share 97.3% sequence identity and that the levels of identity of these two viruses to PHL-03-H8 (92.8% and 93.4%) was less than that distinguishing viruses clustered in genotypes 2 and 6 (∼ 96.5%) and comparable to that distinguishing viruses clustered in genotypes 2 and 3 (∼ 93.3%). PHL-03-H8 was the only YH-related virus detected among 18 P. monodon sampled from the Philippines and it is likely that genotype 5 will resolve into a two separate lineages once more viruses are analyzed. Phylogenetic analysis using the maximum parsimony method clustered the viruses similarly into six genotypes (data not shown).
Translation of the 57 amplified ORF1b gene fragments revealed 35 unique 223 aa sequences that extended from the start of the pp1ab helicase domain motif V to a locus 173 aa downstream of motif VI (Cowley et al., 2000b, Sittidilokratna et al., 2002). An unrooted phylogenetic tree was generated from a ClustalX alignment of the sequences using the neighbour-joining distance matrix. The viruses clustered into the same phylogenetic lineages as defined by the nucleotide sequences, except that the three genotype 6 viruses merged with viruses clustering in genotype 2 (data not shown).
Partial genome sequence analysis of genotype 3, 4 and 5 viruses
Partial genomic sequences of representatives of genotypes 3 (VNM-02-H93), 4 (IND-02-H9) and 5 (THA-03-SG21) were compiled from three to five fragments amplified by RT-nested PCR, RT-semi-nested PCR and 3′-polyA-anchored RT-PCR. For genotypes 3 and 4, the ∼ 6.15 kb sequence extended from IGR1 to the 3′-polyA tail and thus encompassed ORF2 (N protein) and ORF3 (glycoproteins) and the genome 3′-UTR. Due to the poor quality of tissue available to extract RNA, no sequence was obtained for the central ORF3 region or the near 3′-terminal region of genotype 5. Comparisons with genomic sequences of the reference strains of YHV (genotype 1) and GAV (genotype 2) identified high levels of overall nucleotide sequence identity between the genotypes and a similar genome organization (5′…IGR1-ORF2-IGR2-ORF3-IGR3-ORF4-UTR-polyA-3′).
Nucleoprotein gene
ORF2 gene amino acid sequence lengths (144 aa) were identical for GAV and the genotype 3, 4 and 5 viruses and slightly longer for YHV (146 aa). Sequence identity varied between 84.0% (GAV and YHV) and 94.4% (GAV and genotype 3), with most variability, including the two amino acid insertions in YHV, occurring in N- and C-terminal regions (Fig. 3A). Phylogenetic analysis using the N gene sequences (Fig. 3B) confirmed the relationships determined using the ORF1b sequence, with genotypes 2 and 3 and genotypes 4 and 5 clustering on separate branches and genotype 1 (YHV) being more distantly related.
Fig. 3.
(A) ClustalX multiple alignment of the deduced amino acid sequences of the nucleocapsid proteins encoded in ORF2 of the reference genotype 1 (THA-98-Ref) and genotype 2 (AUS-96-Ref) isolates and representatives of genotypes 3 (VNM-02-H93), 4 (IND-02-H9) and 5 (THA-03-SG21). The more variable sequences in the N- and C-terminal regions are shaded. (B) Neighbour-joining tree constructed from a Clustal X multiple alignment of the nucleotide sequence of the nucleocapsid protein genes. Bootstrap values were calculated for 1000 replicate analyses and horizontal branch lengths are proportional to phylogenetic distances.
Glycoprotein gene
In YHV and GAV, ORF3 encodes a polyprotein (pp3) that is cleaved to generate envelope glycoproteins gp116 and gp64, and an N-terminal TMS fragment that has yet to be identified in virions or infected cells (Jitrapakdee et al., 2003, Cowley and Walker, 2002). The pp3 sequences of the genotype 3 (1642 aa) and genotype 4 viruses (1638 aa) were similar in length to GAV (1640 aa) and somewhat shorter than YHV (1666 aa). A ClustalX alignment of the pp3 sequences indicated overall identity to vary between 75.4% (genotypes 1 and 4) and 86.7% (genotypes 2 and 4). Identity was lower in gp116 (71.3%–83.4%) than gp64 (81.8%–92.4%). Of the four genotypes, YHV was most distantly related in all three polypeptides generated by pp3 processing. All 50 cysteine residues located in the predicted ectodomains were perfectly conserved and, of the 25 likely disulphide bridges, two occurred in the TMS fragment, 12 in gp116, and 11 in gp64. Predicted N-linked glycosylation sites are less well conserved amongst the genotypes. Both sites in the GAV TMS ectodomain also exist in genotypes 3 and 4 but only one is present in YHV. In the gp116 ectodomain, GAV contains eight potential sites. YHV contains seven of which six are present in GAV (Cowley and Walker, 2002, Jitrapakdee et al., 2003). All seven potential sites in genotype 3 are shared with GAV as are six of the seven sites in genotype 4, the other being unique. Five of the potential sites in gp116 are conserved across all four genotypes. In the gp64 ectodomain, all four potential N-linked glycosylation sites are conserved across the genotypes.
TMHMM hydropathy profiles of the pp3 sequences of genotypes 3 and 4 predicted six transmembrane (TM) helices with ectodomains between helices (TM1–TM2, TM3–TM4, and TM5–TM6) and a membrane topology identical to that predicted previously for YHV and GAV (Fig. 4 ). SignalP3.0 analysis of each pp3 predicted two signal peptidase type I-like cleavage sites immediately downstream of TM3 and TM5. The TM3 cleavage sites of genotypes 3 (AFA228↓ST), genotype 4 (SFA228↓KE) and genotype 5 (AFS228↓QE) correspond to those used in YHV (AFA228↓TI) and GAV (TFA228↓KE) to generate the gp116 N-terminus (Cowley and Walker, 2002, Jitrapakdee et al., 2003). The TM5 cleavage site generates the gp116 C-terminus and the gp64 N-terminus. Although this site is highly conserved in YHV and GAV (ASA1127↓LA), substitutions were observed upstream of the point of cleavage in genotype 3 (ATA1104↓LA) and genotype 4 (VSS1099↓LA). As indicated, there is marked variability in the amino acid sequence immediately downstream of the TM3 cleavage site (Fig. 4). Although all seven cysteine residues are preserved, there are differences in the number and location of N-linked glycosylation sites and, in YHV, there is a large sequence deletion at the gp116 N-terminus.
Fig. 4.
A ClustalX multiple alignment of amino acid sequences spanning the N-terminal gp116 region of the ORF3 gene of the reference genotype 1 (THA-98-Ref) and genotype 2 (AUS-96-Ref) isolates, and representatives of genotypes 3 (VNM-02-H93), 4 (IND-02-H9) and 5 (THA-03-SG21). The site of proteolytic cleavage of the YHV pp3 polyprotein is indicated. Absolutely conserved (⁎) and similar (: or .) amino acids are indicated according to the similarity groups defined in ClustalX. Conserved cysteine and residues are indicated (+). Potential N-linked glycosylation sites are indicated in bold face and underlined in the alignment and denoted ( ) on the illustration. Six predicted transmembrane-spanning domains (TM1–TM6) are numbered.
) on the illustration. Six predicted transmembrane-spanning domains (TM1–TM6) are numbered.
Intergenic regions, ORF4 and 3′-UTR
The IGRs upstream of ORF2 (IGR1) and ORF3 (IGR2) and the ∼ 650 nt 3′-terminal region downstream of ORF3 were aligned using ClustalX and compared. As observed previously (Cowley et al., 2002a, Sittidilokratna et al., 2002), IGR1 is significantly longer in YHV (352 nt) than in GAV (93 nt) but each contains a highly conserved motif predicted to function as a TRS. In GAV, the 5′-terminus of sub-genomic (sg) mRNA1 is located within the TRS and the terminal seven nucleotides (5′-AACACCU-) are preserved in YHV (Cowley et al., 2002a, Sittidilokratna et al., 2008). As shown in Figure 5A, IGR1 lengths in the genotype 3 (91 nt), 4 (112 nt) and 5 (104 nt) viruses are similar to GAV and contain a highly conserved block of 25 nt including a 13 nt motif (5′-GGUCAAUUACAAC-3′) encompassing the sgmRNA1 5′-terminus that is conserved across all five genotypes. IGR2 sequences (54–57 nt) are highly conserved across all genotypes. In the most conserved block of 24 nt (5′-AUU(G/U)GUCAAUUACAACC(U/A)AAAUUUU-3′), substitutions occur at only two positions, one in genotype 4 (G/U) and another in genotypes 3 and 4 (U/A) (Fig. 6A). WebLogo presentation of ClustalX alignments indicated that the sequence 5′-GUCAAUUACAACxxAxxUU-3′, encompassing the 5′-termini of sgmRNA1 and 2 of GAV, is invariant in all five genotypes (Fig. 5B).
Fig. 5.
(A) Conserved sequences in the intergenic regions upstream of ORF2, ORF3 and ORF4 in the reference genotype 1 (THA-98-Ref) and genotype 2 (AUS-96-Ref) isolates and representatives of genotypes 3 (VNM-02-H93), 4 (IND-02-H9) and 5 (THA-03-SG21). The initiation and termination codons of flanking ORFs are indicated in bold face and underlined. The position in each sequence corresponding to the 5′-terminal adenosine of GAV sgmRNA1 and sgmRNA2 is shaded. The highly conserved 14 nt sequence surrounding the terminal adenosine is underlined, as is the region upstream of ORF4 in which the sequence is partially conserved. Nucleotide substitutions (uridine) in the conserved regions are shaded in black. Identical nucleotides in all aligned sequences are indicated (⁎). (B) WebLogo presentation of nucleotides conserved and less conserved between genotypes 1, 2, 3, 4 and 5 in the TRS elements in IGR1 and in IGR2 as well as between the TRS elements of both IGRs. The cDNA rather than RNA sequence was analysed. (C) Multiple alignment of the putative genotype 2 (GAV) ORF4 gene amino acid sequence with the comparable truncated sequences of genotypes 1, 3, 4 and 5. Amino acids shared in all 5 genotypes are indicated (⁎).
Fig. 6.
Conserved RNA secondary structures predicted using MFOLD to form in the 3′-UTR sequence of genotypes 1, 2, 3 and 4 downstream of the putative ORF4 gene to the genome 3′-polyA tail. Helices 1 to 4 are indicated and the positions of all nucleotide differences that occurred amongst the four genotypes, including compensatory changes that preserved base-pairing in helix 2 and that maintained helix 4 in genotype 1, are indicated (▶).
In GAV, the 3′-terminal region downstream of ORF3 contains a TRS-like element followed by a short ORF4 predicted to encode an unidentified 83 aa (9.2 kDa) polypeptide (Cowley and Walker, 2002). In YHV, the TRS-like sequence is highly conserved but ORF4 is severely truncated (20 aa) (Sittidilokratna et al., 2008). As shown in Figure 5A, the 3′-terminal regions of genotype 3, 4 and 5 viruses also contain TRS-like elements immediately upstream of truncated ORF4 sequences. This region is highly conserved across all five genotypes, with a conserved block of 22 nt sequence encompassing the TRS-like element (5′-UAGCUA(U/)UU(G/U)CAACCUUAUUCU-A-3′) containing only a U insertion in genotype 3 and a G/U substitution in YHV. This sequence includes 9 of 15 nt that were invariant and two others highly dominant in the WebLogo analysis of IGR1 and IGR2 (Fig. 5B). However, in the putative IGR3 of all genotypes, the conserved the 5′-A nucleotide of the sgmRNA terminal sequence motif (5′-ACAAC) is corrupted by a (G/U) substitution (Fig. 6A).
Due to multiple insertions and deletions, ORF4 in YHV (20 aa) (Sittidilokratna et al., 2008) and genotypes 3 (36 aa), 4 (36 aa) and 5 (37 aa) is severely truncated relative to GAV (83 aa). Nevertheless, a ClustalX alignment of the ORF4 sequences indicated that 16 of 20 aa in the N-terminal region upstream of the insertion/deletions were conserved across all five genotypes (Fig. 5C).
A ClustalX alignment of available sequences for YHV, GAV and genotypes 3 and 4 indicated that the 128–131 nt 3′-UTR extending from the GAV ORF4 termination codon to the polyA tail is extremely conserved (data not shown). MFOLD analysis showed the sequences formed similar RNA hairpin structures stabilized by four helices (ΔG = − 32.4 to − 41.3 kcal/mol) (Fig. 6). Significantly, compared to YHV, the integrity of helix 2 in GAV and genotypes 3 and 4 was preserved by compensatory nucleotide changes in either one or both strands (C:G > U:A, G:C > A:U and U:G > C:G). Both strands of YHV helix 4 possessed compensatory C:G > U:A changes and the leading strand possessed a CC > AU change allowing the formation of additional A:U and U:A base pairs that extended the helix. Moreover, a two nucleotide (UA) insertion maintained a hairpin loop size (6 nt) in YHV comparable to the loops of GAV (7 nt) and genotypes 3 (7 nt) and 4 (6 nt). The conformity of these low energy RNA secondary structures suggests that they may be important for the viral polymerase binding and initiation of anti-genomic RNA synthesis.
Discussion
Yellow head virus is a major pathogen of farmed P. monodon shrimp and is listed by the World Organisation for Animal Health as causing a notifiable disease (OIE, 2007). This paper provides evidence for the existence of at least six distinct genetic lineages of YHV distributed across most of the natural geographic range of P. monodon from Africa, through South and Southeast Asia, to eastern Australia. Viruses assigned to genotypes other that genotype 1 (YHV) or 2 (GAV) were detected exclusively as low level infections in apparently healthy shrimp. All viruses clustering with the 1998 Chachoengsao (Thailand) reference YHV isolate (genotype 1) occurred in shrimp displaying typical signs of YHD sampled from pond outbreaks in Thailand and Taiwan between 2000 and 2003. Some genotype 2 (GAV) viruses were detected in moribund Australian shrimp sampled from MCMS-affected ponds. However, whilst similar in histopathology to YHD, MCMS is far less severe, developing less rapidly and rarely causing mass mortalities (Owens, 1997, Spann et al., 1997). GAV is also far less virulent (∼ 106-fold by LD50) than YHV in experimental bioassays (Sittidilokratna and Walker, unpublished) and occurs at a high prevalence in healthy wild and farmed P. monodon along eastern Australia (Spann et al., 1995, Cowley et al., 2000a, Walker et al., 2001) due to its likely maintenance in a natural cycle by vertical transmission (Cowley et al., 2002b, Cowley and Walker, 2008). Although one virus (THA-03-SG21) assigned to genotype 5 was detected in a shrimp affected by a condition of unknown aetiology described as monodon slow growth syndrome (MSGS) (Chayaburakul et al., 2004), there is no evidence to suggest its causal association with this condition for which at least one other virus (Laem Singh virus) has been implicated (Sritunyalucksana et al., 2006).
The nucleotide sequence comparisons of an ORF1b gene region overlapping the highly conserved helicase domain delineated at least six genotypes, although the genotype 6 viruses were not distinguishable from genotype 2 (GAV) viruses when deduced amino acid sequences were compared. Extensive surveys have failed to provide evidence of genotypes other than genotype 2 in eastern Australian P. monodon. It appears, therefore, that GAV is an endemic natural infection of this tiger shrimp population and that other genotypes have not been introduced to Australia because of a prohibition on the importation of live crustaceans (Doyle et al., 1996). Many genotype 2 viruses found at lower prevalence in healthy P. monodon sampled from hatcheries in Thailand and Vietnam showed no clear segregation from Australian viruses, suggesting their possible recent introduction through the export of eastern Australian broodstock. The exclusive clustering of Indian viruses in genotype 4 and Mozambique viruses (with one exception) in genotype 6 may also reflect their isolation from the active trade in P. monodon broodstock and seed that occurs throughout East and South-East Asia. It has been hypothesized that, prior to the advent of aquaculture, efficient vertical transmission allowed individual genotypes to co-evolve with penaeid shrimp host populations in geographic isolation (Cowley et al., 2002b, Cowley and Walker, 2008). The similarity of Australian genotype 2 and Mozambique genotype 6 viruses is intriguing and mirrors the close relationship between Australian and African strains of infectious hypodermal and haematopoietic necrosis virus (IHHNV) infecting P. monodon (Cowley et al., 2004b). Although there has been no recorded traffic in shrimp to between Australia and Africa for aquaculture purposes, the translocation of viruses via shipping routes cannot be excluded.
The 3′-terminal genome regions of YHV, GAV and representative genotype 3, 4 and 5 viruses were examined to establish the extent to which the structural protein genes have diverged, the relevance of the putative ORF4 gene, the conservation of TRS elements in the IGRs and whether RNA secondary structures potentially important for polymerase recognition might exist in the 3′-UTR. Sequence relationships in the structural protein genes were largely consistent with genotype assignments based on the phylogeny of the ORF1b gene sequence and supported the view that YHV is more distant from each of the other genotypes. The YHV nucleoprotein encoded by ORF2 is 2 aa longer and sequence variations, which occur primarily near the N- and C-termini, include substitutions in the binding site of monoclonal antibody (MAb) Y20 which has been used for differential detection of YHV and GAV (Sithigorngul et al., 2000, Sithigorngul et al., 2002, Sithigorngul et al., 2007, Soowannayan et al., 2003, Sittidilokratna et al., 2006). MAb Y20 should also detect genotypes 3, 4 and 5 as their sequences are identical to GAV in this region. In contrast, the binding site for MAbs Y19 and YII4 (IVPDPSL) is preserved across all five genotypes and appears to be a useful group-specific epitope (Sittidilokratna et al., 2006).
In ORF3, YHV is also most divergent in terms of the length of the expressed polyprotein (pp3) and sequence identity of each of the three glycoproteins (gp116, gp64 and the TMS protein) that are released by proteolytic processing (Jitrapakdee et al., 2003). Sequence divergence is most significant in gp116 due to insertions of several amino acid blocks at its N-terminus, the longest of which occurs immediately downstream of the TM3 domain signal peptidase type I cleavage site. It is not yet clear how these insertions might affect the structure of gp116 but it is conceivable they may be responsible for the high virulence of YHV. The efficiency of proteolytic cleavage of virion surface proteins influences virulence and pathogenicity in several diverse groups of RNA viruses (Nagai et al., 1976, Nagai and Klenk, 1977, Rubin and Fields, 1980, Toyoda et al., 1987, Gorman et al., 1988, Scolaro et al., 1990, Gotto and Kawaoka, 1998). Persistent YHV infection in some palemonid shrimp species in the absence of disease has been associated with suppressed gp116 expression relative to the p20 nucleoprotein (Longyant et al., 2005), and it is possible this is also due to aberrant pp3 processing. Variations in sequence at the critical cleavage site at the N-terminus of gp116 may modulate the availability and turnover of mature envelope glycoproteins with consequences for virion maturation, release from infected cells, and progression to chronic persistent infection or acute infection and disease.
Sequences of the putative TRS elements within IGR1 and IGR2 were highly conserved across all genotypes, emphasizing the critical importance of a core 12 nt sequence encompassing the 5′-termini of GAV sgmRNAs (Cowley et al., 2002a). It has been postulated that the ronivirus TRS elements direct the attenuation of polymerase extension during minus-strand RNA synthesis and that the anti-TRS component of minus-strand templates might directly promote subsequent polymerase recognition and transcription of the corresponding sgmRNAs (Pasternak et al., 2006). Significantly, neither this core 12 nt block nor the absolutely conserved adenosine residue at sgmRNA1 and sgmRNA2 5′-termini are preserved in the TRS-like element in the untranslated sequence immediately upstream of ORF4. The absence of these features appears to preclude both efficient attenuation of minus-strand synthesis and the initiation of sgmRNA transcription, and this consistent with the absence of abundant sgmRNA initiating at this site in GAV or YHV (Cowley et al., 2002a, Sittidilokratna et al., 2008). Although there is some evidence of low-level expression of ORF4 protein in GAV (Cowley and Walker, 2008), this is thought to occur either by internal initiation from sgmRNA2 or from variable-length RNAs transcribed in low abundance from sites in the upstream region of the 3′-UTR (J.A. Cowley et al., unpublished data). Nevertheless, the severe truncation of ORF4 in YHV and genotypes 2, 3, and 5 suggests it is unlikely to generate a functional product. It is not clear whether the ORF4 gene is under construction or deconstruction but the existence of the conserved TRS-like sequences upstream of ORF4 and the high level of sequence homology within ORF4 and downstream of the point of truncation indicate that the 3′-region of the ronivirus genome is a genetic resource in active evolutionary transition.
MFOLD analysis of the 3′-UTR sequences of YHV, GAV and genotypes 3 and 4 identified a highly conserved, thermodynamically stable RNA hairpin with four helices, the first of which (helix 1) included the first adenosine residue of the poly-A tail. Helix 1 was absolutely conserved and the importance of helices 2 and 4 in maintaining the structure was supported by the occurrence of compensatory nucleotide changes to maintain base-pairing in different genotypes. Only one nucleotide change, which slightly relaxed the stability of long helix 3, occurred in three of the four genotypes. The 3′-UTR of other positive-strand RNA viruses, including coronaviruses and arteriviruses, contains sequence and/or structural elements critical to polymerase recognition and minus-strand genomic RNA synthesis (Buck, 1996, Dreher, 1999). In the coronavirus, mouse hepatitis virus (MHV), a 55 nt 3′-terminal sequence including the poly-A tail is the minimal signal needed to initiate minus-strand RNA synthesis (Lin et al., 1994). In MHV and other subgroup II coronaviruses, this region forms an RNA pseudoknot, and an upstream bulged RNA hairpin structure and sequence elements that bind four host proteins also appear to be involved in minus-strand RNA synthesis (Williams et al., 1995, Williams et al., 1999, Hsue and Masters, 1997, Hsue and Masters, 1998, Nanda et al., 2004). Stem–loop structures in the 3′-terminal region of arteriviruses also act as signals for the viral polymerase and host protein recognition and are needed to initiate minus-strand synthesis (Verheije et al., 2002, Maines et al., 2005, Beerens and Snijder, 2006). The highly conserved nature of the ronivirus 3′-UTR RNA structure suggests it may also act as a polymerase recognition signal for minus-strand RNA synthesis.
Materials and methods
Shrimp samples
Between 1997 and 2004, postlarvae and tissues of juvenile and adult P. monodon were sampled from hatcheries and farms in India, Taiwan, Malaysia, Vietnam, Indonesia, Thailand and the Philippines. Tissue from Mozambique shrimp was provided via a commercial breeding facility in Malaysia. Pools of postlarvae as well as gill tissue, pleopods or whole heads were collected into alcohol preservative (80% ethanol, 20% glycerol) and stored at ambient temperature until processed. Fresh tissue collected from Australian P. monodon was snap frozen on dry ice and stored at − 80 °C. Details of the origin and nature of the P. monodon samples are listed in Table 1.
RNA isolation
Total RNA was isolated from preserved shrimp tissues as soon as possible after receipt. Pools of 10–15 postlarvae or 10–50 mg tissue were processed using TRIzol reagent (Invitrogen). RNA was re-suspended in 20–40 μl RNase-free sterile distilled water, quantified by spectrophotometry (A260 nm) and stored at − 80 °C.
RT-PCR amplification of ORF1b gene fragments for phylogenetic analysis
Several RT-PCR methods were used to amplify ORF1b gene segments. All PCR primer sequences are described in Table 2 . In initial analyses, a semi-nested one-step RT-PCR employing sense primer 2s12 and antisense primers 2a12 and 2a43 was used to amplify 575 bp and 279 bp products, the larger product being amplified only when viral RNA levels were sufficiently high. An RT-nested PCR used subsequently to amplify longer (1346 bp and 719 bp) products employed primers 2s38M and 2a43M for PCR and primers 2s113 and 2a114 for nested PCR. After new variants were identified, 8-fold degenerate consensus primers were designed to broaden PCR specificity. The target sites of the reverse PCR primer (YH30-R2) and both nested PCR primers (YH31-F2 and YH31-R2) were not changed but the forward PCR primer (YH30-F2) was moved to a more conserved genome region, reducing the amplicon length to 1004 bp.
Table 2.
PCR primer sequences
| PCR Primera | 5′-3′ Sequence |
|---|---|
| ORF1b gene fragments for phylogenetic comparisons | |
| 2s12 | CGCTTCCAATGTATCTGCATGCACC |
| 2a12 | GTGTGAACACCTTCTTGGCTTCCT |
| 2a43 | GAGATGATTTGATTCTTGAATTTCTG |
| 2s38M | CATTGCCGTCCTTGCTAGCTC |
| 2a43M | AGATGATTTGATCTTGAATTTCT |
| 2s113 | AGATCCATGCAATTTGGGAATCATC |
| 2a114 | TTTGGTACGGATGTTGGTGAGGA |
| YH30-F2 | CTACCAYTCAAACATCATCAAYAAYCA |
| YH30-R2 | GAGATGATYTGRTKCTTGAATTTCTG |
| YH31-F2 | CARATCCATGCMATYTGGGAATCATC |
| YH31-R2 | TTTGGTACGGATGTTGGTGAGGA |
| ORF3 gene 5′- and 3′-termini of genotypes 3 and 4 | |
| GAV217F | CGTAACAMRGCARGCYTAGCTATGCA |
| GAV219F | GGTGACTTCTGCAATAACAACGCATGGATG |
| GAV224R | TAKAGAATRTATTTGAGRATRTAATCCCA |
| GAV225R | GCCACCGAGTGARAAAGARGTGAAGGTDCC |
| GAV228R | TGATCAACAGTGACRTTWACCATKTTHT |
| Central ORF3 gene region of genotypes 3 and 4 | |
| GAV237F | CGCCATAGTATCGGAAAACAAATTCTT |
| GAV238R | GCGAATAGCCTGTTGGATTGTTGCA |
| GAV239F | GACTGGAGCATACAAGTTCAAGAGC |
| GAV240R | GGCCTGCTGAATGGTTGCAAAGTTT |
| GAV246R | GCTCCTAATGGGTCGTAACTTCTTACG |
| GAV247R | GGAGTCACCRTCRTGRTAYTCYTC |
| GAV251F | ATGAGACAATTCCAACGCCCAGTG |
| GAV252R | ACGGCTTGTGATGGTAGAGATGAT |
| Genomic 3′-termini of genotypes 3, 4 and 5 | |
| Uni-P | GCCGGAGCTCTGCAGAATTC |
| GAV196R | GGTGGCTGKAGCAGATATGATGCAA |
| GAV197F | CGTGGGGCCGAGTCATCTGCCTT |
| GAV198F | TCCGACACCATTGTKGGYGCTGCAGG |
| GAV199F | ACTGATTCTCTYAAYACTTTCGGCGC |
| ORF2 plus IGR1 and IGR2 of genotypes 3 and 4 | |
| ORF2-4F | ATTGACAAYCCACAYAARTTYAARATG |
| ORF2-5R | ACAAGAGGAAGATCAGAAATATACC |
| ORF2-4F | ATTGACAAYCCACAYAARTTYAARATG |
| ORF2-6R | TAGCAAATTGTAGTGCTTTGCCATG |
| DNA spanning ORF2, IGR1 and IGR2 of genotype 5 | |
| Geno5-F1-P | ACAGGCAAGAGTCATCTCGTCGAA |
| Geno5-R2-N | TGTCACTCCCCACGTTCTCTGGA |
Mixed nucleotide abbreviations: R = A/G, Y = C/T, M = A/C, K = G/T, W = A/T, D = A/G/T, H = A/C/T.
Primer codes: forward (F), reverse (R), sense (s), antisense (a).
Semi-nested RT-PCRs (15 μl) contained 0.2 x MMLV-RT Buffer (Epicentre Biotechnologies), 0.5× Elongase Buffer A and 0.5× Elongase Buffer B (Invitrogen), 0.8 mM dithiothreitol, 5% DMSO, 0.8 mM each dNTP, 20 U RNasin ribonuclease inhibitor (Promega), 2.5 U MMLV RT (Epicentre Biotechnologies), 0.45 U Elongase (Invitrogen), 0.2 U DyNAzyme II (Finnzymes), 0.5 μmol each primer (2s12, 2a12 and 2a43) and 100 ng RNA. Reactions were incubated at 48 °C/45 min, 94 °C/60 s followed by 45 cycles of 94 °C/20 s, 55 °C/30 s, 62 °C/20 s and 70 °C/120 s and then at 70 °C/5 min and 25 °C/60 s.
In the RT-nested PCR, cDNA was synthesized in a 20 μl reaction containing 1–3 μg total RNA, 50 ng random hexamer primers (Promega), 1 mM each dNTP and either 100 U Superscript II or III reverse transcriptase (Invitrogen) as described by the manufacturer. In the PCR, 1 μl cDNA (equivalent to 50–150 ng RNA) was amplified in a 25 μl reaction containing 1 x Taq DNA polymerase buffer (10 mM Tris–HCl pH 9.0, 50 mM KCl, 0.1% Triton X-100), 1.5 mM MgCl2, 17.5 pmol each of primers 2s38M and 2a43M, 200 μM each dNTP and 1.25 U Taq DNA polymerase (Promega). The reaction was incubated at 94 °C/1 min followed by 35 cycles of 94 °C/30 s, 56 °C/30 s, 72 °C/90 s and then at 72 °C/7 min. A 2 μl aliquot of PCR was added nested PCR (25 μl final) prepared as above but containing primers 2s113 and 2a114. PCR cycling conditions were the same except that the extension time was reduced to 45 s.
The consensus RT-nested PCR used 1 μl random-primed cDNA and primers YH30-F2 and YH30-R2 in the PCR, with primers YH31-F2 and YH31-R2 used in the nested PCR. The reaction conditions were as described except for the use of a 58°C annealing temperature and a 60 s extension time in the PCR. An iCycler (BioRad) thermal cycler was used in all PCRs.
3′-polyA-anchored PCR to amplify 3′-terminal genome sequences
Based on sequences available for the reference YHV (GenBank AF540644) and GAV (GenBank AF039647) isolates, degenerate PCR primers were designed to target a C-terminal region of ORF3 (GAV199F) and regions downstream in the ORF3-ORF4 IGR (GAV198F and GAV197F). A degenerate reverse primer (GAV196R) was targeted to a sequence just upstream of the 3′-polyA tail. In the 3′-polyA-anchored PCR method (Cowley and Walker, 2002), cDNA synthesized using the Uni-dT16-A/C/G primer was amplified by PCR using primers GAV199F and Uni-P. This PCR amplified low amounts of a 1044 bp DNA product for the genotype 3 (VNM-02-H93) and 4 (IND-02-H9) viruses but none was detected for the genotype 5 (THA-03-SG21) virus. Using these PCR products as templates, semi-nested (primers GAV197F and Uni-P) and nested PCRs (primers GAV198F and GAV196R) were used to amplify 507 bp and 744 bp products, respectively, which overlapped and included the 3′-polyA tail. As these PCRs did not amplify products for genotype 5, a region from ORF3 to a position downstream of ORF4 in GAV was amplified by semi-nested PCR using random-primed cDNA and primers GAV198F and GAV196R for PCR (∼ 0.7 kb product) and primers GAV197F and GAV196R for semi-nested PCR (∼ 0.4 kb bp product). Primer sequences are described in Table 2.
RT-PCR amplification of ORF3
For genotypes 3 and 4, 5′- and 3′-terminal fragments of the ∼ 4.9 kb ORF3 gene were amplified by RT-PCR using random-primed cDNA and the PCR primer pairs GAV217F/GAV228R (∼ 1.3 kb product) and GAV219F/GAV224R (∼ 0.7 bp product), respectively. A semi-nested PCR employing the primer pair GAV219F/GAV225R (∼ 0.7 bp product) was used to obtain better yields of the 3′-terminal fragment. Reverse primers GAV224R and GAV225R targeted sequences of the 3′-polyA-anchored PCR products that were relatively conserved amongst genotypes 1, 2, 3 and 4. These and subsequent PCRs to generate long fragments of the ORF3 gene employed the Expand Long Template PCR system (Roche) with Buffer 3 adjusted to contain 3.0 mM MgCl2 according to the manufacturer's instructions.
Amplification of the central ORF3 region of genotypes 3 and 4 was attempted by RT-PCR using random-primed cDNA and the primer pairs GAV239F/GAV240R (∼ 3.5 kb product) and GAV237F/GAV238R (∼ 3.1 kb product), respectively, designed to sequences determined for the 5′- and 3′-terminal fragments. As no product was amplified for genotype 3, the PCR was repeated using primer GAV239F in combination with degenerate reverse primer GAV247R designed to a sequence relatively conserved in genotypes 1 and 2. This PCR used similar reaction and cycling conditions and amplified the expected ∼ 1.1 kb DNA product. Using the sequence of this fragment, primer GAV251F was designed and a semi-nested PCR employing this primer and primers GAV252R and GAV246R was used to amplify a ∼ 2.6 kb product spanning the remaining ORF3 gene sequence. Primer sequences are described in Table 2.
RT-PCR amplification of ORF2 and flanking IGR1 and IGR2 sequences
The ORF2 gene and flanking IGR1 and IGR2 sequences of genotypes 3 and 4 were amplified by RT-semi-nested PCR using random-primed cDNA and degenerate sense primers targeting a ORF1b gene (ORF2-4F) sequence relatively conserved between genotypes 1 and 2 and primers targeting ORF3 gene sequences (ORF2-5R and ORF2-6R) more highly conserved amongst genotypes 1, 2, 3, and 4. PCR using the Expand Long Template PCR system (Roche) generated poor ∼ 0.8 kb amplicon yields. Semi-nested PCR amplified good yields of the expected ∼ 0.75 kb products, in addition to others, and these were gel purified prior to sequence analysis. The ORF2 gene and IGR1 and IGR2 sequences of genotype 5 (THA-03-SG21) were derived from a cloned ∼ 3.2 kb DNA product amplified using primers targeting sequences within the ORF1b (Geno5-F1-P) and ORF3 genes (Geno5-R2-N) and the Superscript II One-Step RT-PCR system (Invitrogen). Primer sequences are described in Table 2.
Nucleotide sequence analysis
Prior to being sequenced in both orientations, DNA products amplified by PCR were either purified directly using QIAquick columns (QIAGEN), with or without prior resolution in 0.8% agarose-TAE gels. Sequencing employed PCR primers listed in Table 2 and additional sequence-specific primers for long PCR products. Sequencing reactions used 3.3 μM primer and ABI Prism BigDye Terminator V3.0 or V3.1 reagent and were analyzed at the Australian Genome Research Facility. Sequence chromatograms were edited using SeqEd 1.0.3 (ABI). Deduced amino acid sequences were generated using MacVector 7.0 software. All nucleotide sequences have been deposited in Genbank under accession numbers EU784957-EU785043.
Phylogenetic analysis
Multiple sequence alignments were generated using ClustalX Version 1.82 software (Thompson et al., 1997). Unrooted neighbour-joining trees, constructed using the distance matrix described by Saitou and Nei (1987), were generated using the PHYLIP output of ClustalX. The robustness of neighbour-joining trees was evaluated by bootstrap analysis of a 1000 replicates. Trees were presented graphically using NJ-plot and drawn using TreeView Version 1.6.6 (Page, 1996).
Acknowledgments
The authors would like to thank Dr Nusra Sittidilokratna and Prof. Boonsirm Withychumnarnkul of CENTEX Shrimp (Thailand), Tran Hoa and Dang Oanh of Cantho University (Vietnam), Dr Celia Lavilla-Torres of SEAFDEC (Philippines), Prof. Chu-Fang Lo of National Taiwan University, Mr Su Chen of Farming Intelligene Technology Corporation (Taiwan), and Dr KV Rajendran of CSIRO (Australia) for generously supplying RNA and preserved shrimp tissue samples. This research and the PhD undertaken by PKM Wijegoonawardane were funded, in part, by the Asian Development Bank via the National Aquatic Resources Research and Development Agency of Sri Lanka, and by the Food and Agricultural Organisation (FAO) of the United Nations.
References
- Beerens N., Snijder E.J. RNA signals in the 3′-terminus of the genome of Equine arteritis virus are required for viral RNA synthesis. J. Gen. Virol. 2006;87:1977–1983. doi: 10.1099/vir.0.81750-0. [DOI] [PubMed] [Google Scholar]
- Buck K.W. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantanachookin C., Boonyaratpalin S., Kasornchandra J., Direkbusaratana S., Ekpanithanpong U., Supamataya K., Sriurairatana S., Flegel T.W. Histology and untrastructure reveal a new granulosis-like virus in Penaeus monodon affected by yellow-head disease. Dis. Aquat. Org. 1993;17:145–157. [Google Scholar]
- Chayaburakul K., Nash G., Pratanpipat P., Sriurairatana S., Withyachumnarnkul B. Multiple pathogens found in growth-retarded black tiger shrimp Penaeus monodon cultivated in Thailand. Dis. Aquat. Org. 2004;60:89–96. doi: 10.3354/dao060089. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Walker P.J. The complete sequence of gill-associated virus of Penaeus monodon prawns indicates a gene organisation unique among nidoviruses. Arch. Virol. 2002;147:1977–1987. doi: 10.1007/s00705-002-0847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley J.A., Walker P.J. Molecular biology and pathogenesis of roniviruses. In: Perlman S., Gallagher T., Snijder E.J., editors. Nidoviruses. ASM Press; Washington D.C.: 2008. pp. 361–377. [Google Scholar]
- Cowley J.A., Dimmock C.M., Wongteerasupaya C., Boonsaeng V., Panyim S., Walker P.J. Yellow head virus from Thailand and gill-associated virus from Australia are closely related but distinct prawn viruses. Dis. Aquat. Org. 1999;36:153–157. doi: 10.3354/dao036153. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Spann K.M., Walker P.J. Detection of Australian gill-associated virus (GAV) and lymphoid organ virus (LOV) of Penaeus monodon by RT-nested PCR. Dis. Aquat. Org. 2000;39:159–167. doi: 10.3354/dao039159. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Spann K.M., Walker P.J. Gill-associated virus Penaeus monodon prawns: an invertebrate nidovirus with ORF1a and ORF1b gene related arteri- and coronaviruses. J. Gen. Virol. 2000;81:1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Walker P.J. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic RNAs that do not possess 5′-leader sequences. J. Gen. Virol. 2002;83:927–935. doi: 10.1099/0022-1317-83-4-927. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Hall M.R., Cadogan L.C., Spann K.M., Walker P.J. Vertical transmission of covert gill-associated virus (GAV) infections in Penaeus monodon. Dis. Aquat. Org. 2002;50:95–104. doi: 10.3354/dao050095. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Cadogan L.C., Spann K.M., Sittidilokratna N., Walker P.J. The gene encoding the nucleocapsid protein of gill-associated nidovirus of Penaeus monodon prawns is located upstream of the glycoprotein gene. J. Virol. 2004;78:8935–8941. doi: 10.1128/JVI.78.16.8935-8941.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley J.A., Rajendran K.V., McCulloch R.J., Walker P.J. Australasian Aquaculture 2004, Book of Abstracts. World Aquaculture Society; Baton Rouge: 2004. The detection of infectious hypodermal and haematopoietic necrosis virus (IHHNV) in Australian Penaeus monodon and its potential evolutionary origin; p. 107. [Google Scholar]
- Doyle K.A., Beers P.T., Wilson D.W. Quarantine of aquatic animals in Australia. Rev. Sci. Tech. Off. Int. Epiz. 1996;15:659–673. doi: 10.20506/rst.15.2.951. [DOI] [PubMed] [Google Scholar]
- Dreher T.W. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 1999;37:151–174. doi: 10.1146/annurev.phyto.37.1.151. [DOI] [PubMed] [Google Scholar]
- Gorman J.J., Nestorowicz A., Mitchell S.J., Corino G.L., Selleck P.W. Characterization of the sites of proteolytic activation of Newcastle disease virus membrane glycoprotein precursors. J. Biol. Chem. 1988;263:12522–12531. [PubMed] [Google Scholar]
- Gotto H., Kawaoka Y. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc. Natl. Acad. Sci. (USA) 1998;95:10224–10228. doi: 10.1073/pnas.95.17.10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsue B., Masters P.S. A bulged stem–loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsue B., Masters P.S. An essential secondary structure in the 3′ untranslated region of the mouse hepatitis virus genome. Adv. Exp. Med. Biol. 1998;440:297–302. doi: 10.1007/978-1-4615-5331-1_39. [DOI] [PubMed] [Google Scholar]
- Jitrapakdee S., Unajak S., Sittidilokratna N., Hodgson R.A.J., Cowley J.A., Walker P.J., Panyim S., Boonsaeng V. Identification and analysis of gp116 and gp64 structural glycoproteins of yellow head nidovirus of Penaeus monodon shrimp. J. Gen. Virol. 2003;84:863–873. doi: 10.1099/vir.0.18811-0. [DOI] [PubMed] [Google Scholar]
- Limsuwan C. Tamsetakit Co. Ltd.; Bangkok (in Thai): 1991. Handbook for Cultivation of Black Tiger Prawns. [Google Scholar]
- Lin Y.J., Liao C.L., Lai M.M. Identification of the cis-acting signal for minus strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J. Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longyant S., Sithigorngul P., Chaivisuthangkura P., Rukpratanporn S., Sithigorngul W., Menasveta P. Differences in susceptibility of palaemonid shrimp species to yellow head virus (YHV) infection. Dis. Aquat. Org. 2005;64:5–12. doi: 10.3354/dao064005. [DOI] [PubMed] [Google Scholar]
- Maines T.R., Young M., Dinh N.N., Brinton M.A. Two cellular proteins that interact with a stem loop in the simian hemorrhagic fever virus 3′(+)NCR RNA. Virus Res. 2005;109:109–124. doi: 10.1016/j.virusres.2004.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y., Klenk H.D. Activation of precursors to both glycoproteins of Newcastle disease virus by proteolytic cleavage. Virology. 1977;77:125–134. doi: 10.1016/0042-6822(77)90412-3. [DOI] [PubMed] [Google Scholar]
- Nagai Y., Klenk H.D., Rott R. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology. 1976;72:494–508. doi: 10.1016/0042-6822(76)90178-1. [DOI] [PubMed] [Google Scholar]
- Nanda S.K., Johnson R.F., Liu Q., Leibowitz J.L. Mitochondrial HSP70, HSP40, and HSP60 bind to the 3′-untranslated region of the murine hepatitis virus genome. Arch. Virol. 2004;149:93–111. doi: 10.1007/s00705-003-0196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE . OIE Aquatic Animal Health Code. 10th edn. World Organization for Animal Health; Paris: 2007. Yellowhead disease; pp. 180–185. [Google Scholar]
- Owens L. Special topic review: The history of the emergence of viruses in Australian prawn aquaculture. World J. Microbiol. Biotech. 1997;13:427–431. [Google Scholar]
- Page R.D.M. Tree View: An application to display phylogenetic trees on personal computers. Bioinformatics. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pasternak A.O., Spaan W.J.M., Snijder E.J. Nidovirus transcription: how to make sense…? J. Gen. Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- Rubin D.H., Fields B.N. Molecular basis of reovirus virulence. Role of the M2 gene. J. Exp. Med. 1980;152:853–868. doi: 10.1084/jem.152.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Scolaro L.A., Mersich S.E., Damonte E.B. A mouse attenuated mutant of Junin virus with an altered envelope glycoprotein. Arch. Virol. 1990;111:257–262. doi: 10.1007/BF01311059. [DOI] [PubMed] [Google Scholar]
- Sithigorngul P., Chauychuwong P., Sithigorngul W., Longyant S., Chaivisuthangkura P., Menasveta P. Development of a monoclonal antibody specific to yellow head virus (YHV) from Penaeus monodon. Dis. Aquat. Org. 2000;42:27–34. doi: 10.3354/dao042027. [DOI] [PubMed] [Google Scholar]
- Sithigorngul P., Rukpratanporn S., Longyant S., Chaivisuthangkura P., Sithigorngul W., Menasveta P. Monoclonal antibodies specific to yellow-head virus (YHV) of Penaeus monodon. Dis. Aquat. Org. 2002;49:71–76. doi: 10.3354/dao049071. [DOI] [PubMed] [Google Scholar]
- Sithigorngul W., Rukpratanporn S., Sittidilokratna N., Pecharaburanin N., Longyant S., Chaivisuthangkura P., Sithigorngul P. A convenient immunochromatographic test strip for rapid diagnosis of yellow head virus infection in shrimp. J. Virol. Meth. 2007;140:193–199. doi: 10.1016/j.jviromet.2006.11.034. [DOI] [PubMed] [Google Scholar]
- Sittidilokratna N., Hodgson R.A.J., Cowley J.A., Jitrapakdee S., Boonsaeng V., Panyim S., Walker P.J. Complete ORF1b-gene sequence indicates yellow head virus is an invertebrate nidovirus. Dis. Aquat. Org. 2002;50:87–93. doi: 10.3354/dao050087. [DOI] [PubMed] [Google Scholar]
- Sittidilokratna N., Dangtip S., Cowley J.A., Walker P.J. RNA transcription analysis and completion of the genome sequence of yellow head nidovirus. Virus Res. 2008;136:157–165. doi: 10.1016/j.virusres.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittidilokratna N., Petchampai N., Boonsaeng V., Walker P.J. Structural and antigenic analysis of the yellow head virus nucleocapsids protein. Virus Res. 2006;116:21–29. doi: 10.1016/j.virusres.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soowannayan C., Flegel T.W., Sithigorngul P., Slater J., Hyatt A., Cameri S., Wise T., Crane M.St.J., Cowley J.A., McCulloch R., Walker P.J. Detection and differentiation of yellow head complex viruses using monoclonal antibodies. Dis. Aquat. Org. 2003;57:193–200. doi: 10.3354/dao057193. [DOI] [PubMed] [Google Scholar]
- Spann K.M., Cowley J.A., Walker P.J., Lester R.J.G. Gill-associated virus from cultured Penaeus monodon from Queensland, Australia. Dis. Aquat. Org. 1997;31:169–179. [Google Scholar]
- Spann K.M., Vickers J.E., Lester R.J.G. Lymphoid organ virus of Penaeus monodon from Australia. Dis. Aquat. Org. 1995;23:127–134. [Google Scholar]
- Sritunyalucksana K., Apisawetakan S., Boon-Nat A., Withyachumnarnkul B., Flegel T.W. A new RNA virus found in black tiger shrimp Penaeus monodon from Thailand. Virus Res. 2006;118:31–38. doi: 10.1016/j.virusres.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda T., Sakaguchi T., Imai K., Inocencio N.M., Gotoh B., Hamaguchi M., Nagai Y. Structural comparison of the cleavage-activation site of the fusion glycoprotein between virulent and avirulent strains of Newcastle disease virus. Virology. 1987;158:242–247. doi: 10.1016/0042-6822(87)90261-3. [DOI] [PubMed] [Google Scholar]
- Verheije M.H., Olsthoorn R.C., Kroese M.V., Rottier P.J., Meulenberg J.J. Kissing interaction between 3' noncoding and coding sequences is essential for porcine arterivirus RNA replication. J. Virol. 2002;76:1521–1526. doi: 10.1128/JVI.76.3.1521-1526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker P.J., Bonami J.R., Boonsaeng V., Chang P.S., Cowley J.A., Enjuanes L., Flegel T.W., Lightner D.V., Loh P.C., Snijder E.J., Tang K. Roniviridae. In: Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy, VIIIth Report of the ICTV. Elsevier/Academic Press; London: 2005. pp. 973–977. [Google Scholar]
- Walker P.J., Cowley J.A., Spann K.M., Hodgson R.A.J., Hall M.R., Withychumnarnkul B. Yellow head complex viruses: transmission cycles and topographical distribution in the Asia-Pacific region. In: Browdy C.L., Jory D.E., editors. The New Wave: Proceedings of the Special Session on Sustainable Shrimp Culture, Aquaculture 2001. The World Aquaculture Society; Baton Rouge: 2001. pp. 227–237. [Google Scholar]
- Wang Y.C., Chang P.S. Yellow head virus infection in the giant tiger prawn Penaeus monodon cultured in Taiwan. Fish Pathol. 2000;35:1–10. [Google Scholar]
- Wang C.S., Tang K.F.J., Kou G.H., Chen S.N. Yellow head disease-like virus infection in the Kuruma shrimp Penaeus japonicus cultured in Taiwan. Fish Pathol. 1996;31:177–182. [Google Scholar]
- Williams G.D., Chang R.Y., Brian D.A. Evidence for a pseudoknot in the 3' untranslated region of the bovine coronavirus genome. Adv. Exp. Med. Biol. 1995;380:511–514. doi: 10.1007/978-1-4615-1899-0_81. [DOI] [PubMed] [Google Scholar]
- Williams G.D., Chang R.-Y., Brian D.A. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 1999;73:8349–8355. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Bayer S., Cowley J.A., Gorbalenya A.E. The 3C-like proteinase of an invertebrate nidovirus links coronavirus and potyvirus homologs. J. Virol. 2003;77:1415–1426. doi: 10.1128/JVI.77.2.1415-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]