Abstract
Positive-strand RNA viruses are known to rearrange cellular membranes to facilitate viral genome replication. The biogenesis and three-dimensional organization of these membranes and the link between replication and virus assembly sites is not fully clear. Using electron microscopy, we find Dengue virus (DENV)-induced vesicles, convoluted membranes, and virus particles to be endoplasmic reticulum (ER)-derived, and we detect double-stranded RNA, a presumed marker of RNA replication, inside virus-induced vesicles. Electron tomography (ET) shows DENV-induced membrane structures to be part of one ER-derived network. Furthermore, ET reveals vesicle pores that could enable release of newly synthesized viral RNA and reveals budding of DENV particles on ER membranes directly apposed to vesicle pores. Thus, DENV modifies ER membrane structure to promote replication and efficient encapsidation of the genome into progeny virus. This architecture of DENV replication and assembly sites could explain the coordination of distinct steps of the flavivirus replication cycle.
Keywords: MICROBIO, CELLBIO
Introduction
Dengue virus (DENV) is the causative agent of dengue fever, the most important arboviral disease of humans. Between 50 and 100 million cases are reported annually, and up to 500,000 people are admitted to hospitals with dengue hemorrhagic fever and dengue shock syndrome (Gubler, 2002, Halstead, 1988, Halstead, 2007).
DENV is a flavivirus: a genus that comprises over 70 viruses, many of which are arthropod borne. Flavivirus particles contain a lipid envelope and an inner nucleocapsid composed of capsid protein and a single-stranded, positive-sense RNA genome (Cleaves and Dubin, 1979). During infection, the viral RNA is translated into a polyprotein, which is co- and posttranslationally cleaved by cellular and viral proteases into the structural proteins C, prM, and E and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (reviewed in Bartenschlager and Miller, 2008, Murray et al., 2008). The structural capsid (C), membrane (M), and envelope (E) proteins are components of extracellular mature virus particles; NS proteins are expressed within the infected host cell but are not detectable in virions. They are thought to be involved in DENV genome replication, which occurs in close association with cellular membranes. NS3 and its cofactor, NS2B, are involved in processing of the viral polyprotein. NS3 also has RNA helicase and nucleotide triphosphatase activity. NS5 has RNA-dependent RNA polymerase activity and is involved in capping of progeny viral RNA genomes. The role of NS1 in RNA replication is not fully understood. Little is known about functions of the hydrophobic proteins NS2A, NS4A, and NS4B, but several studies suggest that they may anchor the viral replicase proteins to cellular membranes (Chambers et al., 1989) and contribute to virion assembly (Kummerer and Rice, 2002, Leung et al., 2008) and to inhibition of IFNα/β response (Muñoz-Jordan et al., 2003). Finally, NS4A potentially plays a role in induction of membrane alterations, assumed to serve as a scaffold for the formation of the viral replicase complex (Miller et al., 2007, Roosendaal et al., 2006).
Replication of flaviviruses occurs in close association with virus-induced intracellular membrane structures that wrap around the RNA amplification machinery, the so-called replication complex (RC), containing viral proteins, viral RNA, and probably host cell factors (Mackenzie, 2005, Miller and Krijnse-Locker, 2008, Salonen et al., 2005). RCs may promote efficient viral replication by anchoring the replication machinery in membrane compartments or by shielding it off from host defense mechanisms (Netherton et al., 2007, Novoa et al., 2005). Electron microscopy (EM) showed that DENV induces membrane alterations of various morphologies, including convoluted membranes (CM) and vesicle packets (VPs) (Grief et al., 1997). Immuno-EM studies indicated that VP and CM may represent the site of DENV replication and RNA translation/polyprotein processing, respectively (Mackenzie et al., 1996). Light microscopy studies showed that NS proteins localize to cytosolic, virus-induced structures (Kapoor et al., 1995, Miller et al., 2006, Miller et al., 2007, Umareddy et al., 2006) also positive for double-stranded RNA (dsRNA).
While immuno-EM analysis of infected cells is an important tool for localizing viral and cellular proteins in thin sections, electron tomography (ET) is currently the most suitable method to analyze complex three-dimensional (membrane) structures. It relies on the reconstruction of 3D images of specimens 4–8 times thicker than in thin-section transmission EM (TEM) and reveals the 3D architecture of intracellular structures at high resolution (Baumeister, 2005, Frey et al., 2006, McIntosh et al., 2005). It has been successfully employed to determine the architecture of virus-induced membrane alterations (Knoops et al., 2008, Kopek et al., 2007) or the organization of other large intracellular structures (Hoog et al., 2007).
In this study, we have combined immuno-EM, thin-section TEM, and ET to determine the origin and structure of DENV-induced membranes. We have also used ET of DENV-infected samples to unravel the 3D architecture of virus-induced membrane compartments, including the presumed sites of viral replication and assembly. From our results, we establish a biologically relevant model of the possible topological link between flavivirus replication and assembly in infected cells.
Results
Immunofluorescence Microscopy Reveals Presumed DENV Replication Sites
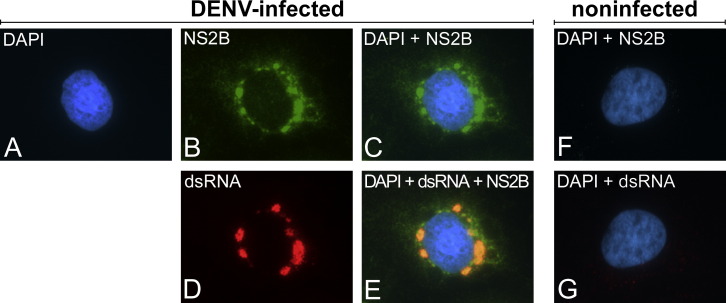
A subset of DENV NS proteins was previously shown to localize to dot-like cytoplasmic structures (Mackenzie et al., 1996, Miller et al., 2006, Miller et al., 2007). We extended this observation by raising rabbit antisera monospecific for NS1, NS2B, NS3, NS4A, NS4B, and NS5 that were tested for light microscopy analysis of DENV-infected cells ( Figures 1, S1, and S2). All antibodies displayed a distinct labeling pattern that was not seen in uninfected cells (Figure S1). This pattern, exemplified by the NS2B-specific labeling, consisted of cytoplasmic dot-like structures in the perinuclear area of the cell (Figure 1). They colocalized with dsRNA, a generally accepted marker for the (presumed) intermediates during DENV RNA replication (Figures 1D and 1E). An exception was NS5, which in agreement with previous reports (Buckley et al., 1992, Kapoor et al., 1995, Miller et al., 2006) localized predominantly to the nucleus of infected cells. However, biochemical fractionation experiments show that up to 30% of NS5 is in the cytosol and about 70% in the nuclear fraction (data not shown), arguing that cytosolic NS5 is poorly accessible to the used antibodies (but still detectable; see below). Thus, DENV NS proteins localize to conspicuous cytosolic structures that likely correspond to the site of viral replication.
Figure 1.
Colocalization of DENV NS2B with Double-Stranded RNA in Infected Cells
(A–G) Huh-7 cells infected with DENV2 (A–E) or none-infected (F and G) were fixed 24 hr postinfection and labeled with a monospecific polyclonal rabbit antiserum to DENV NS2B and a monospecific mouse monoclonal antibody against dsRNA. Nuclear DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole dihydrochloride).
EM Analysis of DENV-Infected Cells Reveals a Complex Set of Membranes
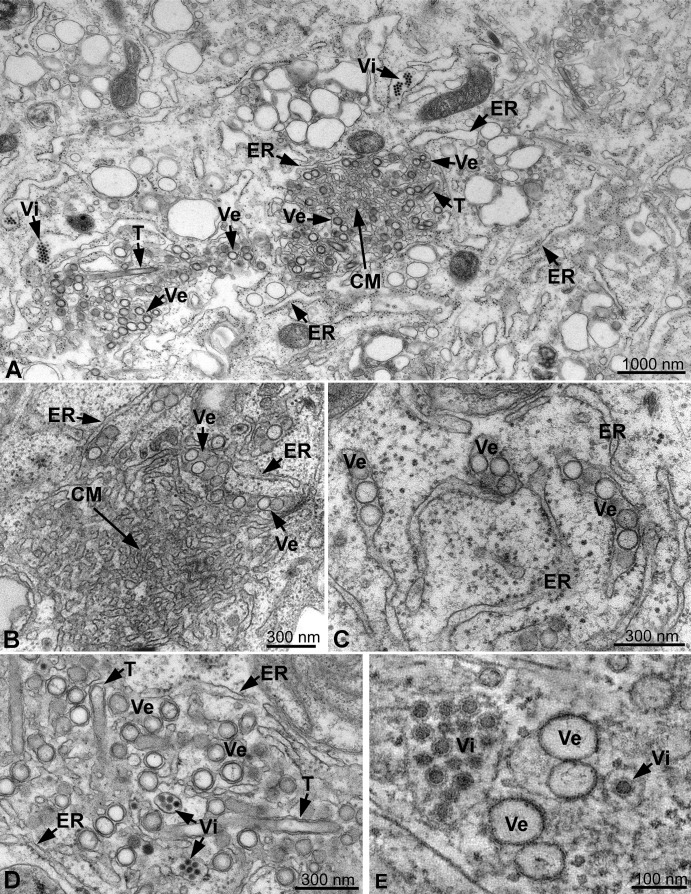
TEM of resin-embedded infected cells fixed at 24 hr postinfection revealed a complex collection of convoluted and vesicular membrane structures ( Figure 2A). CM, a term that we use for consistency with reports on other flaviviruses, were usually found in the center of large structures. They were surrounded by spherical vesicles (Figure 2B) that appeared as 80–90 nm single-membrane vesicles located within the lumen of the rough ER (Figure 2C) or as double-membrane vesicles (DMVs) (Figure 2D) and were closely associated with cisternae reminiscent of the rough ER. Occasionally, double-membrane tubes were seen between the vesicles (Figures 2A and 2D), but their sporadic appearance suggests that they are not absolutely required for DENV replication. Of note, the way the 80–90 nm vesicles appeared (DMVs or single-membrane vesicles within the ER lumen) depended on sample preparation protocol (Figures S3A–S3C).
Figure 2.
Ultrastructure of DENV-Induced Membrane Alterations
(A) Thin-section TEM images of DENV-infected, resin-embedded Huh-7 cells fixed at 24 hr postinfection are shown in (A)–(D). Overview of the virus-induced structures is shown in (A); virus-induced vesicles (Ve) and occasionally membrane tubes (T) accumulate around stacks of convoluted membranes (CMs). Ve and CM are intimately associated with the ER. Clusters of virus particles (Vi) reside in the periphery of virus-induced membrane structures and are aligned in stacks in the lumen of dilated ER cisternae.
(B) Arrays of CM and associated Ve, surrounded by ER membranes.
(C) Higher magnification view of Ve in the lumen of the ER.
(D) Tight apposition of inner vesicle (Ve) or tube (T) membrane and outer ER membrane gives the impression of “free” DMVs or tubes next to ER cisternae containing virus particles.
(E) Projection of eight consecutive 1 nm tomogram slices of a high-pressure-frozen/freeze-substituted DENV-infected cell volume analyzed by ET. Stacked and single virus particles (Vi) are surrounded by a membrane that displays a spiky pattern resembling viral membrane proteins. Vi are inside membrane cisternae, which are tightly associated with the virus-induced vesicles (Ve).
Virus particles were seen in the ER lumen, typically in dilated ER cisternae located at the periphery of the vesicles and CM (Figure 2A), and were often arranged in regular arrays (Figures 2A, 2D, and 2E). The particles had an outer diameter of about 45 nm and were composed of an electron-dense central core and a membranous layer that displayed the regularity of a spike-like surface protein, strongly suggesting that the particles represent enveloped virions (see below).
In conclusion, the dot-like staining pattern seen by light microscopy most likely corresponds to several distinct membrane structures: one- and two-membraned vesicles, CM, and virions, all closely associated with cisternae reminiscent of the rough ER.
Localization of Viral Proteins and DsRNA to DENV-Induced Membrane Structures
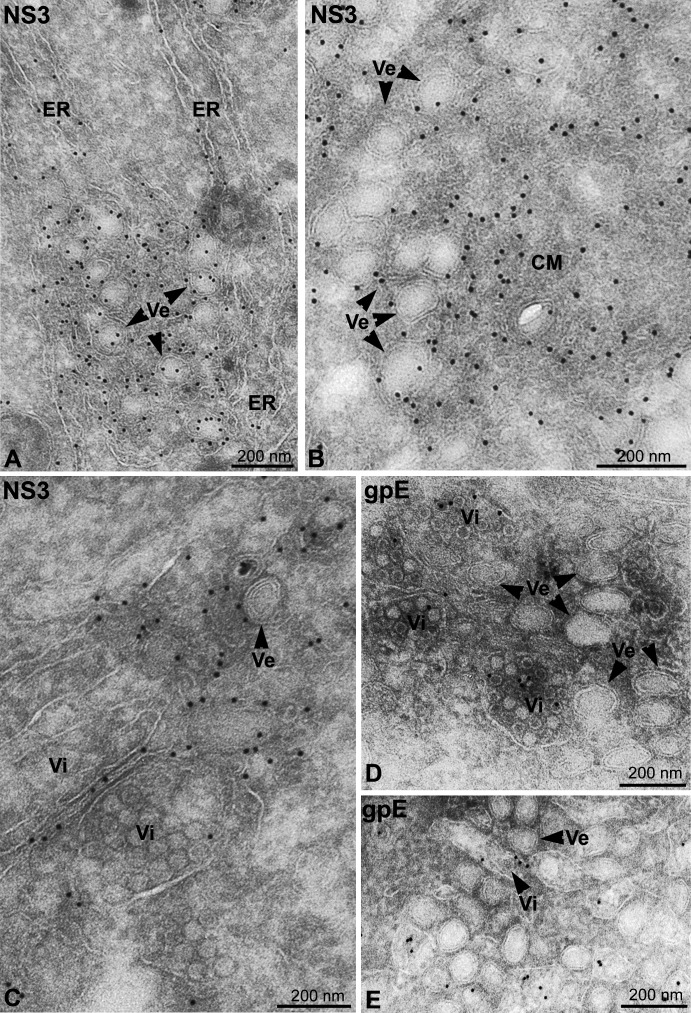
We used immuno-EM to localize viral proteins and RNA to the membrane structures. NS3-specific antiserum specifically labeled DMVs, into which ER cisternae extended ( Figure 3A), and CM (Figure 3B). Since NS3 has helicase activity that is essential for RNA replication (Matusan et al., 2001) the localization of NS3 suggests that it is an integral part of the viral RC. Dilated ER cisternae packed with particles of ∼45 nm were readily seen at the periphery of labeled structures (Figure 3C). These particles were not labeled with the NS3-specific antiserum (Figure 3C) but with a monoclonal antibody to the DENV E glycoprotein, confirming that they were newly assembled virions (Figure 3D). Whereas the anti-E antibody also labeled the ER (data not shown), it did not significantly label the vesicles or the CM (Figures 3D and 3E). The antibody also revealed single virus particles in the lumen of ER cisternae that were typically located between the vesicles rather than at their periphery (Figure 3E; see below).
Figure 3.
EM Localization of NS3 and E in DENV-Infected Cells
(A–E) DENV-infected Huh-7 cells were fixed 24 hr postinfection, and thawed cryosections were labeled with antibodies specific for NS3 (A–C) or the glycoprotein E (D and E). Vesicles (Ve, arrowheads) into which cisternae of the ER (indicated) extend are shown in (A). The NS3-specific antibody significantly labels both Ve (arrowheads) and the CMs in (B). In (C), NS3-specific antibody labels a Ve (arrowhead), ER membranes, and the limiting membrane of a dilated ER cisterna containing virions (Vi). Dilated cisternae filled with Vi that are labeled with anti-E in (D), whereas the Ve (arrowheads) are not labeled. The E-specific antibody labels a single virion (Vi, arrowhead) within the lumen of an ER cisterna that is located within a site of Ve accumulation in (E). Note that the Ve appear as double-membraned spheres in thawed cryosections.
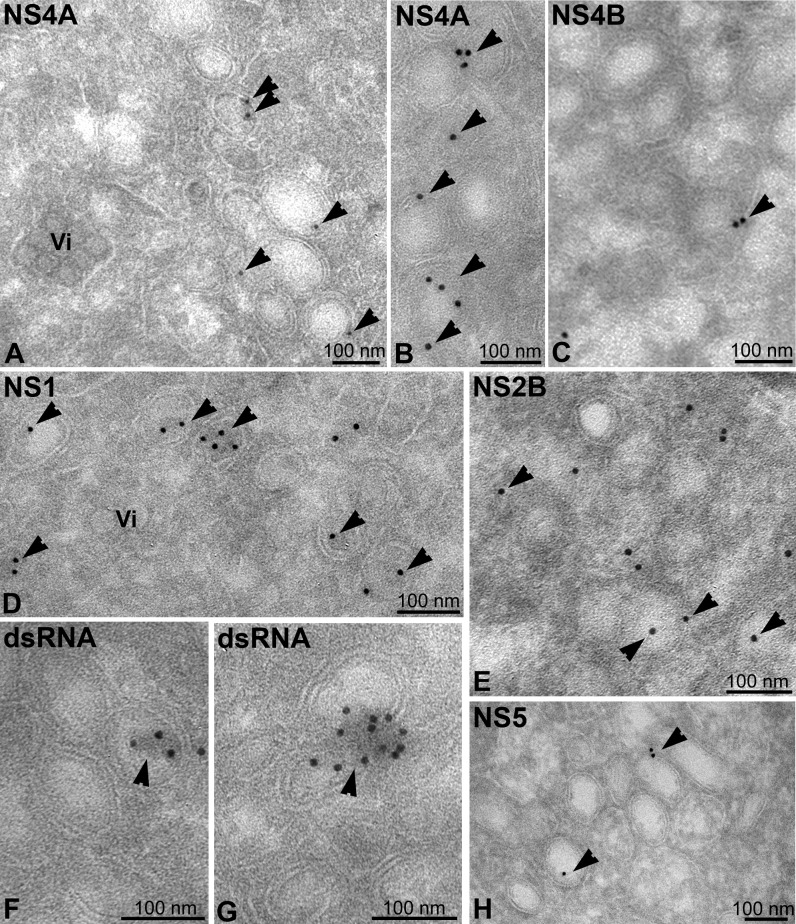
Antibodies against other NS proteins (NS4A, NS4B, NS1, NS2B, and NS5) also showed specific labeling of the vesicles, although weaker than for NS3 ( Figures 4A–4E and 4H, arrowheads). While this suggested that the CM labeled with NS3 only, a lower affinity of the antibodies or poor accessibility of the other NS proteins in the CM cannot be excluded. DsRNA was found as a discrete electron-dense structure inside (Figure 4F) or on the cytosolic surface of a subset of vesicles (Figure 4G), suggesting that dsRNA is present only in some of the vesicles at a given time point. The electron density detected along with the immuno gold labeling most likely is due to uranyl acetate that was used to contrast the labeled cryosections, although we cannot exclude the possibility that its formation was linked to the use of the dsRNA-specific antibody. However, uranyl acetate binds to phosphate groups present in nucleic acids, supporting the notion that the dsRNA-specific antibody indeed detected viral RNA.
Figure 4.
EM Localization of NS Proteins and DsRNA in DENV-Infected Cells
(A–H) DENV-infected Huh-7 cells were fixed 24 hr postinfection, and thawed cryosections were labeled with antibodies against NS4A (A and B), NS4B (C), NS1 (D), NS2B (E), dsRNA, the presumed intermediate produced during DENV replication (F and G), or NS5 (H). All images show DENV-induced vesicles. All NS antibodies specifically label vesicles (arrowheads). Note that the virions (Vi) in (A) are not labeled with anti-NS4A and that the dsRNA-specific antibody (F and G) labels a discrete electron-dense structure located within (F) or outside (G) the vesicles.
In summary, immuno-EM confirmed that the membrane structures seen in resin-embedded cells were induced by DENV infection and contained all NS proteins. Moreover, the internal location of the NS proteins, as well as of dsRNA, the presumed intermediate of RNA replication, implied that viral RNA synthesis occurred in the vesicles.
Virus-Induced Membranes Are ER Derived
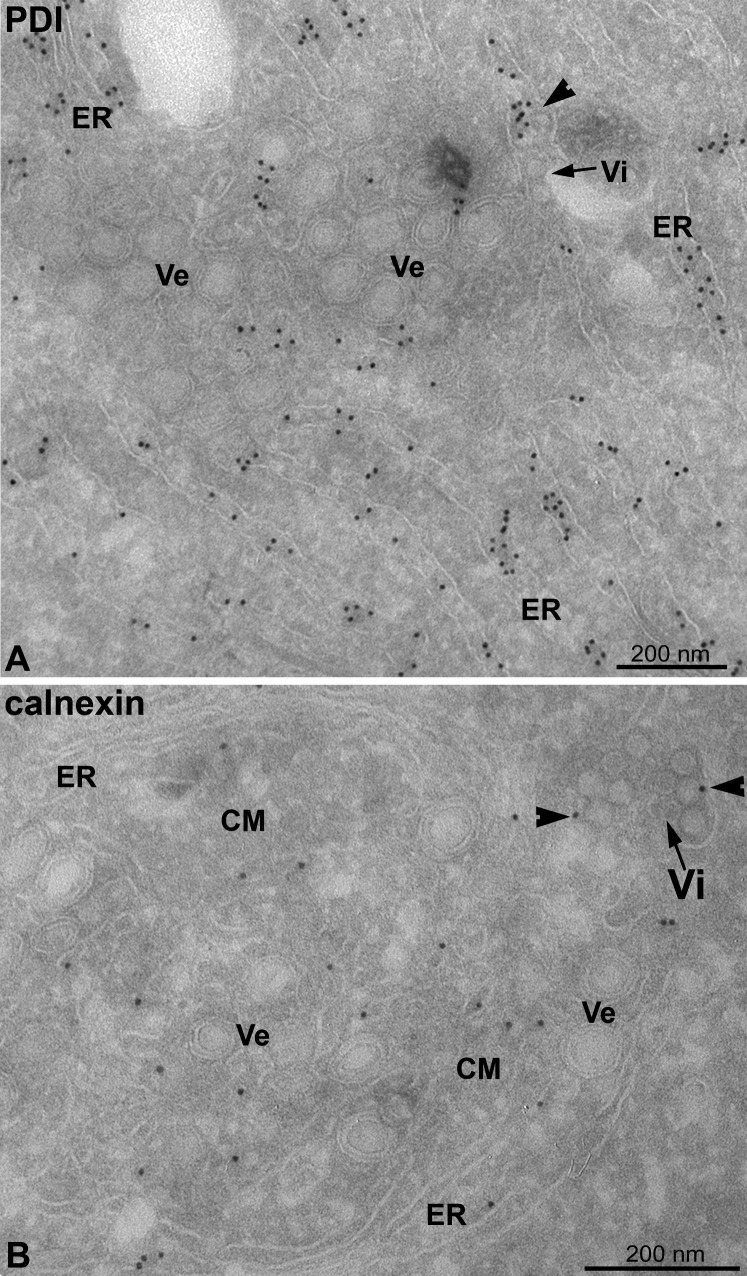
We used immuno-EM with antibodies to cellular marker proteins to determine the cellular origin of virally modified membranes. Marker proteins of late endosomes/lysosomes (lamp-1), Golgi complex (GM130, p230), and COPI and COPII transport machineries involved in vesicular trafficking (β′-COP and sec13) did not label the virus-induced membrane structures (Figure S4 and Table S1). In contrast, vesicles, CM, and the cisternae found in their close proximity were clearly and specifically labeled for the ER markers PDI and calnexin ( Figures 5A, 5B, and S4). The PDI labeling of the vesicles was relatively low compared to the ER labeling, suggesting that this protein may be partially excluded from these virus-induced membrane structures. Calnexin, in contrast, seemed enriched on the CM compared to the ER, while it was less abundant in vesicles. Finally, the limiting membrane of the dilated virus-containing cisternae and their lumen was labeled for calnexin (Figure 5B, arrowheads) and PDI (Figure 5A, arrowheads), respectively, defining this compartment as ER-derived. Thus, the vesicles, CM, and virus-harboring cisternae are derived from the ER. This conclusion has further support from our finding by immunofluorescence microscopy that virus-induced structures contain Syntaxin17, a marker for smooth ER (Figure S4 and Table S1).
Figure 5.
Virally Modified Membranes Are Derived from the ER
(A and B) DENV-infected Huh-7 cells were fixed 24 hr postinfection, and thawed cryosections were labeled with PDI- (A) or calnexin-specific (B) antibodies. Arrowheads indicate anti-PDI or -calnexin labeling; arrows indicate virions (Vi) in the lumen of the ER. Note the ER cisternae in (A) that are abundantly labeled with PDI and that extend into the membrane structure containing vesicles (Ve) and CMs. Whereas PDI shows low but specific labeling of the vesicles in (A), calnexin specifically labels the CMs in (B).
Electron Tomography Reveals a Network of Virally Modified Membranes and Vesicles Containing Single Pores
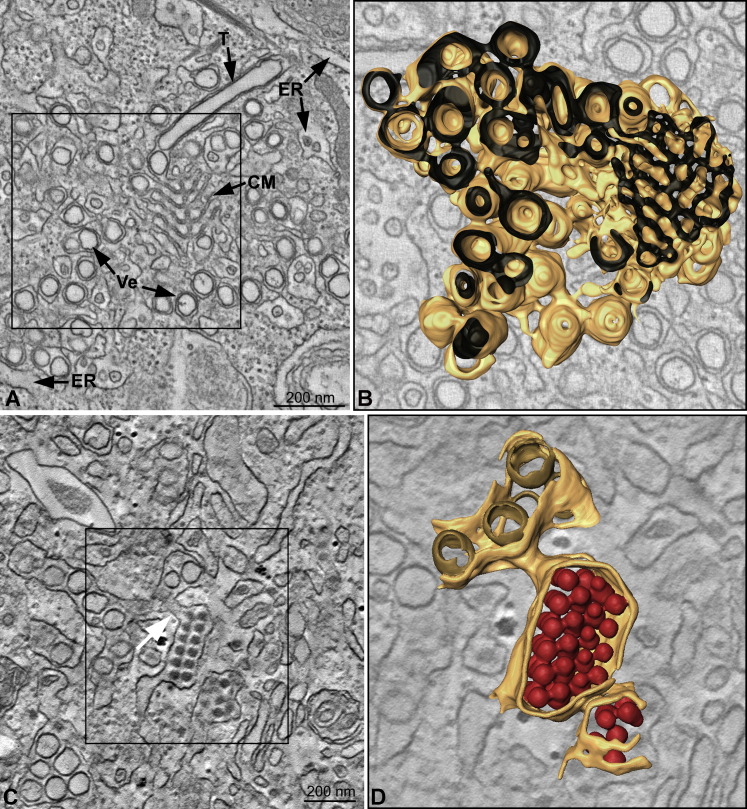
Our thin-section EM analysis prompted us to study the 3D relationship between vesicles, CM, and ER membranes as well as the architecture of inner and outer membranes of DENV-induced vesicles by ET. Tomograms of cellular volumes containing virally modified membranes revealed that CM, ER, and the outer membrane of vesicles are part of one continuous membrane network ( Figure 6A and Movie S1). Whereas in 2D images, the vesicles sometimes appeared as “free” DMVs, ET showed that they were interconnected via their outer membrane. Three-dimensional surface rendering (Figure 6B and Movie S1) illustrates that the CM, DENV-induced vesicles, and tubes are covered by one continuous surface and are thus part of a network of interconnected membranes derived from the ER.
Figure 6.
DENV-Induced Membrane Structures and ER Form a Continuous Intracellular Membrane Network
(A) Huh-7 cells fixed at 26.5 hr postinfection were embedded in epoxy resin and analyzed by ET. Slice through a tomogram shows DENV-induced CMs, vesicles (Ve), and tubes (T) that form a network of interconnected membranes in continuity with the ER. Image represents an ∼9.5 nm thick slice (Z binning of eight 1.18 nm slices).
(B) Three-dimensional surface model of the membranes in the boxed area in (A). The outer (cytosolic) face of the continuous membrane network is depicted in yellow; the ER lumen is dark.
(C) Three nanometer slice through a tomogram (Z binning of three 0.998 nm slices). Stacked virus particles are seen in ER cisternae that are directly connected to virus-induced vesicles (white arrow).
(D) Three-dimensional surface model of the virus-induced structures in the boxed area in (C) showing the continuity of virus-and vesicle-containing ER cisternae. ER membranes are depicted in yellow, inner vesicle membranes in light brown, and virus particles in red.
Virus particles accumulated either as densely packed stacks in dilated cisternae of the rough ER (Figures 6C and 6D and Movie S2) or occurred as single enveloped virions within the lumen of the ER and the nuclear envelope (black arrowheads in Figures 7F, 7H, and 7I and Movie S2). Virus-filled ER cisternae were always found in close proximity to the DENV-induced vesicles and often directly connected to the ER that contained vesicles (Figures 6C and 6D and Movie S2, white arrow). Since the intralumenal virions have a membrane envelope (Figures 2E, 3D, and 3E), our data clearly indicate that they have budded from ER membranes, presumably in close proximity to the DENV-induced vesicles. Additional single virions were seen inside vesicles located in the periphery of the Golgi complex and in coated-membrane structures (Figures 7F and 7H and Movie S3, black arrowheads), strongly suggesting that individual virions are transported to the Golgi complex by secretory vesicles from peripheral ER cisternae, where they accumulate initially.
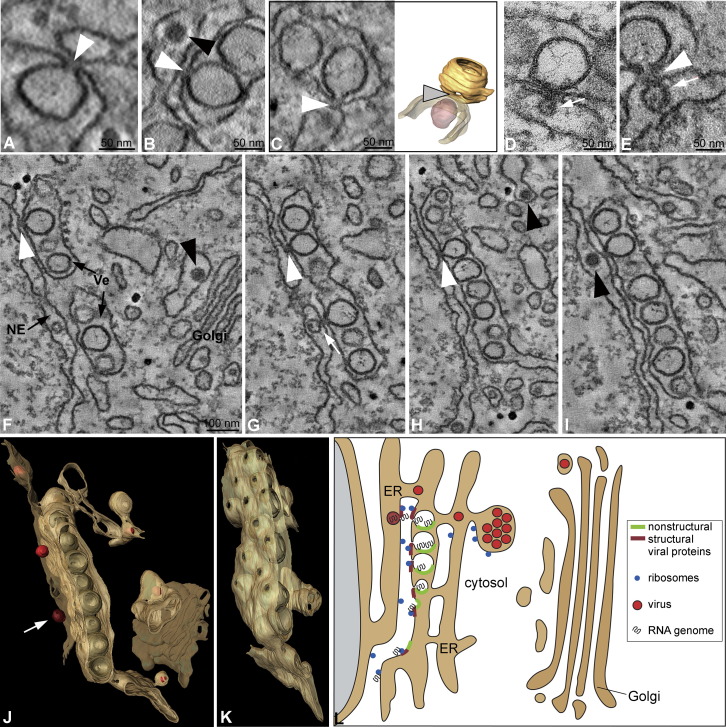
Figure 7.
Characterization of DENV-Induced Vesicles and Viral Budding Sites and a Model of Their Possible Relation
(A–C) Single slices of tomograms showing DENV-induced vesicles as invaginations of the ER membrane (white arrowheads). Note the diffuse electron density at the cytosolic face of the vesicle openings and the tight apposition of vesicle openings to the opposing ER/NE containing a virus particle (black arrowhead in [B]). The right panel in (C) shows a 3D surface rendering of the ER/vesicle continuity (yellow) and the tightly apposed ER (semitransparent) containing a virus particle (red).
(D) Sixty nanometer section of the same sample as in (C), showing a vesicle in the ER and a putative virus budding event into the ER lumen located opposite the vesicle pore (arrow).
(E) Single slice of a tomogram showing a vesicle in the ER, its pore (white arrowhead), and a putative budding site (arrow) opposite the vesicle pore.
(F–I) Four slices through a tomogram, each ∼2 nm thick. Note the continuity of vesicle and ER membranes (white arrowheads) and the tight apposition of vesicle openings to the opposing NE. Virus particles (black arrowheads) are seen in the NE, in the ER, and in a cisterna close to the Golgi stack.
(J) Three-dimensional surface rendering of ER and Golgi membranes (yellow), outer NE membrane (semitransparent), DENV-induced vesicles in the ER (light brown), virus particles (red), and a putative virus budding site (arrow) of the tomogram shown in (F–I).
(K) The vesicle-containing ER segment shown in (J), rotated by 90° around the y axis, highlighting the vesicle openings.
(L) Model of the relation between DENV replication, assembly, and virion release. Upon infection, the viral genome associates with the rough ER (ribosomes in blue), and the viral polyprotein, composed of the structural proteins (red) and the NS proteins (green), is synthesized on rough ER membranes. NS4A, together with other viral and perhaps cellular proteins, induces invaginations of the ER membrane, leading to the formation of vesicles that are connected to the cytosol via a pore. Inside these invaginations, RNA replication occurs. Viral capsid protein associates with progeny RNA genomes liberated through the pore-like structure into the cytosol. Virus budding occurs through the ER membrane located in close proximity to or opposite of the vesicles. Individual virions travel toward distal sites of the ER lumen, where they collect in dilated ER cisternae. They are transported, likely as individual virions, via secretory vesicles to the Golgi complex, where virion maturation occurs. Samples (A–I) were prepared as in Figure 6 and analyzed by ET, with the exception of (D), which was analyzed by TEM.
Analysis of DENV-induced vesicle size revealed an inner vesicle diameter of approximately 87.5 nm (± 8.5 nm, n = 149) and thus a vesicle volume of about 3.5 × 10−4 μm3. Virus particles had an average outer diameter of 45.1 nm (± 2.7 nm, n = 56). Close inspection of the vesicles in 3D revealed that the inner vesicle membrane was often continuous with the outer ER membrane, connecting the lumen to the cytosol via a pore-like opening. Thus, the vesicles appeared as invaginations of the ER rather than as sealed single-membrane vesicles (Figures 7A–7C and 7E, white arrowheads). The membrane invaginations were seen irrespective of the sample preparation protocol (Figures S3A′–S3C′, white arrowheads), but their visibility depended on the orientation of membranes perpendicular to the x-y plane of tomographic slices and was frequently hampered by a diffuse electron density at the cytosolic side of the vesicle pore (Figures 7A, 7B, and 7E). Upon Gaussian filtering of the 3D images, 58.2% (± 7.1%, n = 264) of the vesicles that were fully included in the volumes of ten different tomograms were found to display a pore and to be continuous with ER membranes (Figure S3). Each vesicle contained only one such pore, further suggesting that their inner membrane is an invagination of the ER membrane. Consequently, the lumen of most if not all of these virus-induced vesicles is topologically identical to and open toward the cytosol. Analysis of pores that were readily visible without filtering revealed that they were almost round and had an average maximum width of 11.2 nm (± 1.6 nm, n = 27). Thus, both the topology of the vesicle interior and the immunolabeling results are consistent with the conclusion that the vesicles are the site of viral RNA replication. Likewise, the size of the pore would allow release of viral RNA genomes to be used for RNA translation and virus assembly.
Virus Assembly Occurs in Close Proximity to DENV-Induced Vesicles
Close inspection of vesicle pores revealed that they were often just opposite a tightly apposed ER cisterna (Figures 7A–7C and 7E, white arrowhead) or nuclear envelope (NE) membrane (Figures 7F–7H, white arrowhead). The cytosolic space between the pores and opposed membranes was frequently filled with a diffuse electron-dense material (most obvious in Figures 7A, 7B, 7F, and 7H, white arrowheads). The ER/NE lumen directly opposite the vesicle pores often displayed small membrane invaginations (Figures 7D, 7E, and 7G and Movie S3, small arrows), which had a slightly bigger diameter (about 60 nm) than virions and contained a distinct electron density, indicating that they represented virus budding sites (Figures 7D, 7E, and 7G, small arrows). These putative budding sites were always found in close proximity to single enveloped virus particles inside the ER lumen (Figures 7B and 7I and Movie S3, black arrowheads), further suggesting that virus budding occurs on ER membranes in close proximity to the vesicle “pores.” Three-dimensional surface rendering of the ER and its associated membranes (Figure 7J and Movie S4) illustrates the above findings: a portion of the ER lumen (yellow) encloses numerous densely packed vesicles (light brown) and is tightly apposed to another ER/NE membrane (yellow, semi-transparent; only the outer NE membrane is displayed). This opposed ER/NE lumen contains single virus particles (red) and, in addition, displays a putative virus budding site (small arrow). Additional virus particles are found in more peripheral segments of the ER that are oriented toward the Golgi stack or in single vesicles in the periphery of the Golgi complex. Vesicle openings that connect the vesicle lumen with the cytosol are highlighted in Figure 7K. In this tomogram, the vesicle “pores” were almost exclusively oriented toward the nucleus, although in other tomograms taken in more peripheral parts of the cell, the vesicle openings were found more randomly distributed toward adjacent ER membranes.
Thus, ET has revealed that the virally modified membranes are tightly interconnected and that virus budding occurs on membranes that are distinct but tightly apposed to membranes most likely involved in viral replication. This topological link may ensure efficient production and delivery of viral RNA for the assembly of infectious virus progeny.
Discussion
In the present study, we identified distinct membrane structures, including CM and vesicles in DENV-infected cells, in agreement with earlier reports (Barth, 1992, Mackenzie et al., 1996, Ng et al., 1994, Westaway et al., 1997b) and extended the analysis using different EM techniques, including ET, to study the 3D architecture of virus-induced membrane rearrangements potentially involved in DENV replication. Induction of such membrane structures that most likely serve as a scaffold for the assembly of viral RCs has been described for virtually all groups of positive-strand RNA viruses, but their architecture and origin differ among various virus groups (reviewed in Salonen et al., 2005). The positive immunolabeling for PDI and calnexin (immuno-EM) as well as CLIMP63 (Miller et al., 2006) shows that the DENV-induced membrane structures are derived from the rough ER. Immunofluorescence-based localization of the smooth ER marker Syntaxin17 (Steegmaier et al., 2000) to the membrane structures also implicates smooth ER in the formation of virus-induced compartments. By using ET, we could unambiguously show that all these membrane structures are interconnected and part of a single ER-derived endomembrane system, providing a platform for the transport of viral proteins and genomes between RNA replication vesicles, ribosome-containing membranes (RNA translation), and virus assembly sites. By TEM, the vesicles to which all of the tested NS proteins localized appeared as “free” DMVs or as single vesicles in the ER lumen. We show that their appearance depends on the sample preparation protocol, a fact that should be considered when analyzing the ultrastructure of membrane rearrangements induced by other RNA viruses. Three-dimensional analysis of the vesicles reveals that they are invaginations of the ER membrane. Thus, the inside of the vesicles is connected to the cytosol and probably is the site of viral RNA replication, as indicated by the labeling for NS proteins and dsRNA. This result is reminiscent of an earlier report on DENV, in which colocalization of NS1 and dsRNA to VPs and cytoplasmic vacuoles was found in infected Vero cells and insect cells (Mackenzie et al., 1996). Our results do not, however, support the model by Uchil and Satchidanandam (2003), who proposed that flavivirus replication vesicles consist of a closed inner vesicle surrounded by an outer membrane that is continuous with CM and the ER. In contrast, we show that the interior of the vesicle is connected to the surrounding cytosol by a pore, which only became visible by use of ET. The pore may regulate import of factors required for RNA replication as well as export of newly synthesized genomes to be used for translation or virus assembly (see below).
The most striking observation from ET was the identification of virus budding sites in close proximity to the pores of replication vesicles. It has been assumed for a long time that flaviviruses acquire their envelope by budding into the ER lumen, and here we provide a direct visualization, in 3D, of this process. Our results not only reveal the DENV budding sites, but also allow us to draw a model of how the topology of DENV-induced membrane rearrangements may contribute to the different steps of the replication cycle (Figure 7L): upon infection, genomic viral RNA associates with ribosomes at the rough ER, where translation occurs. The resulting polyprotein is cleaved co- and posttranslationally, forming a membrane-associated RC. Whether polyprotein processing occurs in CM as proposed for Kunjin virus (KUNV) remains to be determined. Invaginations of the ER membrane are induced, presumably by NS4A, in conjunction with other viral and cellular factors (Miller et al., 2007) and give rise to membranous vesicles that are connected to the cytosol via a pore. We assume that the structural proteins core, E, and prM are excluded from these vesicles, an assumption that is supported by our immunolabeling showing NS proteins on one set of membranes and E on another set. Based on the dsRNA labeling, Bromo-UTP labeling of nascent DENV RNA (S.M. and R.B., unpublished data), and biochemical studies (Mackenzie et al., 1996, Miller et al., 2006, Miller et al., 2007, Uchil and Satchidanandam, 2003), RNA synthesis likely occurs within the lumen of the vesicles. Nucleotides and other factors required for replication may enter the vesicle via the pore that also would allow exit of the newly synthesized RNA genome to the cytosol, where it is used for translation or particle assembly. Nucleocapsid formation and virus budding into the ER lumen occur in close proximity of such RNA exit sites. Although virus particles appear to accumulate in dilated ER cisternae, single virus particles contained in vesicles were often found at the cis-Golgi, arguing that single DENV particles are transported to the Golgi, where prM is cleaved by the cellular furin protease (Shapiro et al., 1997). The release of mature virus particles most likely occurs through the constitutive secretory pathway.
Vesicle formation is probably induced by NS4A, as suggested by recent studies with DENV and KUNV (Miller et al., 2007, Roosendaal et al., 2006). DENV NS4A appears to contain a central peripheral membrane domain that intercalates into the lumenal leaflet of the ER membrane (Miller et al., 2007). NS4A oligomers would dilate the lumenal leaflet, resulting in membrane invagination toward the ER lumen, a topology that fits our observation. While this would explain how the vesicles are generated, we do not know how a pore of distinct size is formed and why the vesicles do not pinch off. One possibility is that the required host cell machinery is not available at these sites. Further studies will be required to clarify the molecular composition and biogenesis of the pores.
Colocalization studies performed, especially with the poliovirus, have suggested a role for autophagy in the biogenesis of viral RCs as well as virus assembly and release (reviewed in Miller and Krijnse-Locker, 2008). Lee and colleague have recently provided evidence that DENV infection enhances autophagolysosome formation and that inhibition of the autophagic host cell machinery by phosphatidylinositol-3′-kinase (PI3KC3) inhibitor 3-methyladenine reduces DENV titers (Lee et al., 2008). The effects are moderate, however, arguing that this pathway may contribute to DENV replication to only a minor extent. Whereas the double-membrane phenotype seen in our TEM resembles the membrane dynamics during autophagy, a role for this cellular process is unlikely, at least under the conditions used here. Hallmarks of autophagy include membrane wrapping, leading to a double-membrane compartment involved in lysosomal degradation. As shown unequivocally by our ET data, the DENV-induced vesicles are invaginations rather than sealed double-membrane vesicles, and our immunolabeling experiments that exclude lamp-1 from these vesicles do not support an involvement of lysosomes. We note that Lee and colleagues used a different DENV strain that may be more virulent than the New Guinea C (NGC) strain used in our study, which could account for the discrepant results.
Analysis of the size of DENV-induced vesicles revealed an inner vesicle diameter of approximately 87.5 nm (± 8.5 nm, n = 149) and thus a vesicle volume of about 3.5 × 10−4 μm3. Assuming a volume of 0.655 nm3 per hydrated nucleotide, as estimated from the crystal structure of dsRNA (Johnson and Rueckert, 1997), and taking into account the genome length of the DENV NGC strain (about 10,700 nucleotides), a vesicle may harbor up to 50 single-stranded RNA molecules. However, our EM studies reveal that the vesicles are rather electron lucent, indicating that they contain only few RNA molecules.
Whereas the role of the vesicles as the sites of RNA replication is rather well established, the role of the CM in DENV replication is much less clear. In the case of KUNV, CMs are supposed to be the site of polyprotein processing, as deduced from the immunolabeling for NS2B and NS3 and the absence of NS1 and NS4B (Westaway et al., 1997a, Westaway et al., 1997b). However, a similar role of CM for DENV remains to be demonstrated. Morphologically, the CM resembled smooth ER membranes, consistent with our finding that ribosomes were not detectable in their central region. In addition, the smooth ER marker Syntaxin17 was detected in DENV-induced membrane compartments. We speculate that CM might represent a storage site for proteins and lipids involved in DENV replication that can be recruited to vesicles upon demand. The fact that the CMs are continuous with the vesicles and contain NS3 would be consistent with this proposal.
While this study was ongoing, two groups reported the 3D structure of “replication vesicles” of other positive-strand RNA viruses, the SARS-Coronavirus (SARS-CoV) and the flock house virus (FHV) (Knoops et al., 2008, Kopek et al., 2007). In the case of FHV (Kopek et al., 2007), newly synthesized viral RNA and the sole replication factor, protein A, were found to colocalize in invaginations of the outer mitochondrial membrane. These spherules have a diameter of about 50 nm and are connected to the cytosol via a necked 10 nm channel. It is assumed that this pore allows exchange of factors required for RNA replication as well as release of progeny RNA. Formation of the spherule is induced by protein A oligomers that appear to bend membranes in a process similar to viral matrix proteins. Kopek and coworkers have also calculated that one spherule of FHV contains, on average, only three RNA replication intermediates. These results reveal a surprising parallel to the DENV-induced vesicles, supporting the assumption of an evolutionary conserved mechanism.
In the case of SARS-CoV, a more complex picture emerged from the ET analyses (Knoops et al., 2008). DsRNA, a presumed intermediate of replication, was detected exclusively in DMVs, whereas antibodies targeting replicase subunits labeled most intensively a reticulovesicular network of modified ER membranes containing DMVs. This observation is reminiscent of the complex ER-derived network described here in DENV-infected cells. Surprisingly, however, the inner membrane layers of the SARS-CoV DMVs were sealed, whereas neck-like connections were detected between the outer layers of different DMVs. This topology raises the challenging questions of how such large (200–300 nm diameter) sealed vesicles form and how viral RNA is released to gain access to ribosomes and the structural proteins. While the underlying mechanisms are not known, evidence was obtained that the membrane network involved in RNA replication is continuous with membranes involved in virus assembly (Knoops et al., 2008), arguing that these two processes are tightly coupled.
An apparently different strategy appears to be used by poliovirus. Several pioneering studies performed by the laboratory of Kurt Bienz and Denise Egger suggest that poliovirus induces ER-derived membranous vesicle clusters in a COPII-dependent manner (Egger et al., 2000, Rust et al., 2001). RNA replication is thought to occur on the cytosolic surface of these ER-derived vesicles, primarily at sites where vesicles cluster, whereas RNA translation may take place primarily at the more distal sites of these structures. However, in poliovirus-infected cells, membranous structures with two lipid bilayers have also been detected that may be induced by a process analogous to the formation of autophagic vacuoles (Schlegel et al., 1996). These results illustrate the different strategies positive-strand RNA viruses have developed to achieve efficient RNA replication.
The data described here for DENV may have implications for our understanding of the biogenesis and architecture of the RC of other members of the Flaviviridae family, most notably hepatitis C virus (HCV). In cells either containing a persistently replicating subgenome or infected with HCV, intensive structural rearrangements, designated the membranous web, were found (Gosert et al., 2003, Miller and Krijnse-Locker, 2008). However, the induced membrane alterations are much more heterogenous, with irregular assemblies of membranous vesicles that vary in size and profound alterations of presumably ER-derived membranes. At variance with flaviviruses, the membranous web of HCV contains interspersed lipid droplets that play a major role in the assembly of infectious virus particles (Boulant et al., 2007, Miyanari et al., 2007, Shavinskaya et al., 2007). These droplets appear to be tightly linked to the replication sites, but due to the high complexity of these membrane structures, a detailed analysis will be very difficult. In this respect, the 3D model developed here for DENV may serve as a template to unravel the architecture of the HCV replication and assembly compartments.
In conclusion, we have resolved the 3D structures of flavivirus-induced membrane rearrangements. Our results provide a possible explanation for the spatial coupling of the different steps of the DENV replication cycle. The next obvious step is to integrate available information about the structures and functions of the viral and cellular factors involved in the biogenesis of these replication factories, but also to decipher the temporal relationship orchestrating the steps, from uncoating of the viral genome up to the assembly of infectious progeny virus. We can assume that these studies will provide new insight into fundamental cell biological processes, but also define targets for the development of preventive and therapeutic strategies to combat this insidious disease.
Experimental Procedures
Cells and Viruses
The human hepatoma cell line Huh-7 (Nakabayashi et al., 1982) was maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen; Karlsruhe, Germany) supplemented with 2 mM L-glutamine, nonessential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum (FCS) (DMEM complete). Cells were infected with the DENV 2 NGC (Progen Biotechnik GmbH; Heidelberg, Germany) at a multiplicity of infection of 2–4. Virus was diluted in DMEM containing 2% FCS, and cells were incubated for 4 hr at 37°C / 5% CO2 with occasional rocking. After 4 hr, the inoculum was replaced by complete DMEM, and cells were incubated for the indicated times.
Antisera and Immunofluorescence Microscopy
Primary antibodies used were: rabbit polyclonal anti-calnexin and mouse monoclonal anti-protein disulfide isomerase (both from Stressgen; Victoria, BC, Canada), lamp-1 (The Developmental Studies Hybridoma Bank; Iowa City, IA), mouse monoclonal anti-p230 (BD Biosciences; Heidelberg, Germany), mouse monoclonal anti-Arf1 antibody (Abcam; Cambridge, MA), mouse monoclonal anti-GOS28 (Acris; Herford, Germany), and mouse monoclonal anti-dsRNA (English & Scientific Consulting; Szirak, Hungary). The monoclonal antibody to E was a kind gift of Dr. Marie Flamand (Pasteur Institute; Paris). The following people kindly provided us with antibodies to cellular markers: human anti-sec-13 was from Wanjin Hong (IMB; Singapore), anti-β′-COP was from Jeremy Simpson (EMBL; Heidelberg, Germany), anti-GM130 was from Antonella de Matteis (Mario Negri Sud; Santa Maria Imbargo, Italy), and goat anti-Syntaxin17 was from R. Sheller (Howard Hughes Medical Institute, Stanford University School of Medicine; Stanford, CA). The production of anti-DENV NS3, NS4A, NS4B, and NS5 polyclonal antibodies in rabbits was described previously (Miller et al., 2006, Miller et al., 2007). Secondary goat antibodies used in immunofluorescence microscopy were conjugated to Alexa Fluor 546 and 488 (Molecular Probes; Karlsruhe, Germany). For western blot analyses, a secondary rabbit antibody coupled to horseradish-peroxidase (Sigma; Deisenhofen, Germany) was used.
Details of the protocol for immunofluorescence microscopy are provided in the Supplemental Data.
Plasmid Constructs and Expression of DENV prM, NS1-, and NS2B-GST Fusion Proteins and Generation of Antisera
Construction of expression plasmids and production of recombinant antigens for immunization of rabbits is described in the Supplemental Data.
Electron Microscopy and Tomography
Cells were infected for 24 or 26.5 hr as above, fixed, and processed for immuno-EM as previously described (Miller et al., 2007). Sectioning was done with a Leica EM UC6 Microtome, FC6 Cryochamber (Leica Microsystems; Wetzlar, Germany), and a diamond knife (Drukker; Hanau, Germany). Immunocytochemical labeling of thawed cryosections was performed essentially as described (Griffiths et al., 1983).
For conventional TEM, cells were infected as above and fixed with 2.5% glutaraldehyde in cacodylate buffer (100 mM sodium cacodylate, 50 mM KCl, 2.5 mM CaCl2 [pH 7.2]) for 30–60 min at room temperature. Fixed cells were high-pressure frozen/freeze substituted with acetone/0.1% osmium tetroxide as described (Buser and Walther, 2008) (Figure 2E) or postfixed with 1% osmium tetroxide for 1 hr on ice and dehydrated in a graded acetone series at room temperature (all other figures). Samples were embedded in epoxy resin (Roth; Karlsruhe, Germany) according to the manufacturer's instructions. Sections were obtained with a Leica Ultracut UCT Microtome and diamond knife, counterstained with lead citrate, and examined with a Philips BioTwin CM120 TEM (120 kV) or a Zeiss 10C TEM (80 kV). For ET, 250–300 nm thick sections were prepared as described elsewhere (Hoog et al., 2007). Unstained grids were placed in a high-tilt holder (Model 2020; Fischione Instruments; Corporate Circle, PA), and digital images were recorded as single- (Figure 6C) or dual-axis (all other tomograms) tilt series (FEI 4k Eagle camera; binning factor 2, binned pixel size 0.592 nm (Figures 7F–7I), 0.998 nm (Figure 6C), or 1.18 nm (Figure 6A) on the specimen level) over a −60°–60° tilt range (increment 1°) at a defocus of −0.2 μm on an FEI Tecnai TF30 microscope operated at 300 kV. Tomograms were reconstructed using the IMOD software package (version 3.12.20) (Kremer et al., 1996). The Amira Visualization Package (version 5.2.0, Visage Imaging) was used for 3D measurements, surface rendering, and Gaussian image filtering (3 × 3 × 3 kernel). All 3D surface models were generated from unfiltered tomograms by manually masking areas of interest, thresholding, and smoothing labels, followed by manually filling gaps and additional smoothing when necessary to render the surfaces more continuous. Tomograms were denoised using a nonlinear anisotropic diffusion algorithm for presentation in Movies S1–S3.
Acknowledgments
We thank Ulrike Herian, Uta Haselmann, Anja Habermann, and Eberhard Schmid for excellent technical assistance; Matthias Weiss for help with RNA volume calculation; Progen Biotechnik GmbH for providing the DENV2 NGC isolate; Andrew Davidson (School of Medical Sciences; Bristol, UK) for the gift of the DENV2 NGC-encoding plasmid; and all those colleagues who kindly provided antibodies essential to carry out this study. We are very thankful to the Electron Microscopy Core Facility (EMCF) at EMBL Heidelberg for providing access to their equipment, expertise, and technical support. This work was supported by the German Research Council (SFB 638, Teilprojekt A5 to R.B.), the EU (DENCO project, contract number 517708 to R.B.), the CellNetworks Cluster of Excellence, and the Wellcome Trust (program grant H5RCYV0 to S.D.F.).
Published: April 22, 2009
Footnotes
Supplemental Data include Supplemental Experimental Procedures, four figures, one table, and four movies and can be found online at http://www.cell.com/cell-host-microbe/supplemental/S1931-3128(09)00098-5.
Supplemental Data
Animation through the central portion of a Z series of 1.18 nm thick digital slices (total thickness ∼170 nm) of a dual-axis tomogram (corresponding to Figures 6A and 6B), reconstructed from an ∼250 nm thick section of a DENV-infected Huh-7 cell. Note the direct continuity of CM, vesicles, and surrounding ER membranes.
Animation through a Z series of 1.996 nm thick digital slices (total thickness ∼198 nm) of a single-axis tomogram (corresponding to Figure 6C), reconstructed from an ∼250 nm thick section of a DENV-infected Huh-7 cell. Colored overlay shows a 3D surface model of the virus- and vesicle-containing ER cisternae. ER membranes are depicted in yellow, inner vesicle membranes in light brown, and virus particles in red. Note the continuity of membranes of virus- and vesicle-containing ER cisternae (white arrow).
Animation through the central portion of Z series of 1.78 nm thick digital slices (total thickness ∼178 nm) of a dual-axis tomogram (corresponding to Figures 7F–7I), reconstructed from an ∼250 nm thick section of a DENV-infected Huh-7 cell. Colored overlay shows a 3D surface model of the virus-induced membranes. ER and Golgi membranes are depicted in yellow, inner vesicle membranes in light brown, and virus particles in red. Note the continuity of the ER and vesicle membranes and the putative budding profiles on the NE membrane opposite of the pores. Single virus particles are located within the NE and virions within vesicles near the Golgi stack.
Three-dimensional surface rendering of ER and Golgi membranes (yellow), virus-induced vesicles (light brown), and virus particles (red) of the reconstructed 3D volume displayed in Figures 7F–7I and Movie S3. Note the vesicle pores directly connecting the vesicle lumen to the cytosol.
References
- Bartenschlager R., Miller S. Molecular aspects of Dengue virus replication. Future Microbiol. 2008;3:155–165. doi: 10.2217/17460913.3.2.155. [DOI] [PubMed] [Google Scholar]
- Barth O.M. Replication of dengue viruses in mosquito cell cultures—a model from ultrastructural observations. Mem. Inst. Oswaldo Cruz. 1992;87:565–574. doi: 10.1590/s0074-02761992000400017. [DOI] [PubMed] [Google Scholar]
- Baumeister W. A voyage to the inner space of cells. Protein Sci. 2005;14:257–269. doi: 10.1110/ps.041148605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S., Targett-Adams P., McLauchlan J. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 2007;88:2204–2213. doi: 10.1099/vir.0.82898-0. [DOI] [PubMed] [Google Scholar]
- Buckley A., Gaidamovich S., Turchinskaya A., Gould E.A. Monoclonal antibodies identify the NS5 yellow fever virus non-structural protein in the nuclei of infected cells. J. Gen. Virol. 1992;73:1125–1130. doi: 10.1099/0022-1317-73-5-1125. [DOI] [PubMed] [Google Scholar]
- Buser C., Walther P. Freeze-substitution: the addition of water to polar solvents enhances the retention of structure and acts at temperatures around −60 degrees C. J. Microsc. 2008;230:268–277. doi: 10.1111/j.1365-2818.2008.01984.x. [DOI] [PubMed] [Google Scholar]
- Chambers T.J., McCourt D.W., Rice C.M. Yellow fever virus proteins NS2A, NS2B, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology. 1989;169:100–109. doi: 10.1016/0042-6822(89)90045-7. [DOI] [PubMed] [Google Scholar]
- Cleaves G.R., Dubin D.T. Methylation status of intracellular dengue type 2 40 S RNA. Virology. 1979;96:159–165. doi: 10.1016/0042-6822(79)90181-8. [DOI] [PubMed] [Google Scholar]
- Egger D., Teterina N., Ehrenfeld E., Bienz K. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 2000;74:6570–6580. doi: 10.1128/jvi.74.14.6570-6580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey T.G., Perkins G.A., Ellisman M.H. Electron tomography of membrane-bound cellular organelles. Annu. Rev. Biophys. Biomol. Struct. 2006;35:199–224. doi: 10.1146/annurev.biophys.35.040405.102039. [DOI] [PubMed] [Google Scholar]
- Gosert R., Egger D., Lohmann V., Bartenschlager R., Blum H.E., Bienz K., Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grief C., Galler R., Cortes L.M., Barth O.M. Intracellular localisation of dengue-2 RNA in mosquito cell culture using electron microscopic in situ hybridisation. Arch. Virol. 1997;142:2347–2357. doi: 10.1007/s007050050247. [DOI] [PubMed] [Google Scholar]
- Griffiths G., Simons K., Warren G., Tokuyasu K.T. Immunoelectron microscopy using thin, frozen sections: application to studies of the intracellular transport of Semliki Forest virus spike glycoproteins. Methods Enzymol. 1983;96:466–485. doi: 10.1016/s0076-6879(83)96041-x. [DOI] [PubMed] [Google Scholar]
- Gubler D.J. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10:100–103. doi: 10.1016/s0966-842x(01)02288-0. [DOI] [PubMed] [Google Scholar]
- Halstead S.B. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- Halstead S.B. Dengue. Lancet. 2007;370:1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- Hoog J.L., Schwartz C., Noon A.T., O'Toole E.T., Mastronarde D.N., McIntosh J.R., Antony C. Organization of interphase microtubules in fission yeast analyzed by electron tomography. Dev. Cell. 2007;12:349–361. doi: 10.1016/j.devcel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- Johnson J.E., Rueckert R.R. In: Structural Biology of Viruses. Chiu W., Burnett R.M., Garcea R., editors. Oxford University Press; New York: 1997. Packing and release of the viral genome; pp. 269–287. [Google Scholar]
- Kapoor M., Zhang L., Ramachandra M., Kusukawa J., Ebner K.E., Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J. Biol. Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Van den Worm S., Zevenhoven-Dobbe J., Van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:1957–1974. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopek B.G., Perkins G., Miller D.J., Ellisman M.H., Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kummerer B.M., Rice C.M. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 2002;76:4773–4784. doi: 10.1128/JVI.76.10.4773-4784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.R., Lei H.Y., Liu M.T., Wang J.R., Chen S.H., Jiang-Shieh Y.F., Lin Y.S., Yeh T.M., Liu C.C., Liu H.S. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374:240–248. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J.Y., Pijlman G.P., Kondratieva N., Hyde J., Mackenzie J.M., Khromykh A.A. The role of nonstructural protein NS2A in flavivirus assembly. J. Virol. 2008;82:4731–4741. doi: 10.1128/JVI.00002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J. Wrapping things up about virus RNA replication. Traffic. 2005;6:967–977. doi: 10.1111/j.1600-0854.2005.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J.M., Jones M.K., Young P.R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- Matusan A.E., Pryor M.J., Davidson A.D., Wright P.J. Mutagenesis of the Dengue virus type 2 NS3 protein within and outside helicase motifs: effects on enzyme activity and virus replication. J. Virol. 2001;75:9633–9643. doi: 10.1128/JVI.75.20.9633-9643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh R., Nicastro D., Mastronarde D. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 2005;15:43–51. doi: 10.1016/j.tcb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Miller S., Sparacio S., Bartenschlager R. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 2006;281:8854–8863. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- Miller S., Kastner S., Krijnse-Locker J., Buhler S., Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 2007;282:8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- Muñoz-Jordan J.L., Sánchez-Burgos G.G., Laurent-Rolle M., García-Sastre A. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA. 2003;100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C.L., Jones C.T., Rice C.M. Architects of assembly: roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat. Rev. Microbiol. 2008;6:699–708. doi: 10.1038/nrmicro1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi H., Taketa K., Miyano K., Yamane T., Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- Netherton C., Moffat K., Brooks E., Wileman T. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv. Virus Res. 2007;70:101–182. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M.L., Yeong F.M., Tan S.H. Cryosubstitution technique reveals new morphology of flavivirus-induced structures. J. Virol. Methods. 1994;49:305–314. doi: 10.1016/0166-0934(94)90145-7. [DOI] [PubMed] [Google Scholar]
- Novoa R.R., Calderita G., Arranz R., Fontana J., Granzow H., Risco C. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. 2005;97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosendaal J., Westaway E.G., Khromykh A., Mackenzie J.M. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and golgi trafficking of the NS4A protein. J. Virol. 2006;80:4623–4632. doi: 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust R.C., Landmann L., Gosert R., Tang B.L., Hong W., Hauri H.P., Egger D., Bienz K. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 2001;75:9808–9818. doi: 10.1128/JVI.75.20.9808-9818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A., Ahola T., Kaariainen L. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A., Giddings T.H., Jr., Ladinsky M.S., Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J., Sciaky N., Lee J., Bosshart H., Angeletti R.H., Bonifacino J.S. Localization of endogenous furin in cultured cell lines. J. Histochem. Cytochem. 1997;45:3–12. doi: 10.1177/002215549704500102. [DOI] [PubMed] [Google Scholar]
- Shavinskaya A., Boulant S., Penin F., McLauchlan J., Bartenschlager R. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 2007;282:37158–37169. doi: 10.1074/jbc.M707329200. [DOI] [PubMed] [Google Scholar]
- Steegmaier M., Oorschot V., Klumperman J., Scheller R.H. Syntaxin 17 is abundant in steroidogenic cells and implicated in smooth endoplasmic reticulum membrane dynamics. Mol. Biol. Cell. 2000;11:2719–2731. doi: 10.1091/mbc.11.8.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchil P.D., Satchidanandam V. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 2003;278:24388–24398. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- Umareddy I., Chao A., Sampath A., Gu F., Vasudevan S.G. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J. Gen. Virol. 2006;87:2605–2614. doi: 10.1099/vir.0.81844-0. [DOI] [PubMed] [Google Scholar]
- Westaway E.G., Khromykh A.A., Kenney M.T., Mackenzie J.M., Jones M.K. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology. 1997;234:31–41. doi: 10.1006/viro.1997.8629. [DOI] [PubMed] [Google Scholar]
- Westaway E.G., Mackenzie J.M., Kenney M.T., Jones M.K., Khromykh A.A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Animation through the central portion of a Z series of 1.18 nm thick digital slices (total thickness ∼170 nm) of a dual-axis tomogram (corresponding to Figures 6A and 6B), reconstructed from an ∼250 nm thick section of a DENV-infected Huh-7 cell. Note the direct continuity of CM, vesicles, and surrounding ER membranes.
Animation through a Z series of 1.996 nm thick digital slices (total thickness ∼198 nm) of a single-axis tomogram (corresponding to Figure 6C), reconstructed from an ∼250 nm thick section of a DENV-infected Huh-7 cell. Colored overlay shows a 3D surface model of the virus- and vesicle-containing ER cisternae. ER membranes are depicted in yellow, inner vesicle membranes in light brown, and virus particles in red. Note the continuity of membranes of virus- and vesicle-containing ER cisternae (white arrow).
Animation through the central portion of Z series of 1.78 nm thick digital slices (total thickness ∼178 nm) of a dual-axis tomogram (corresponding to Figures 7F–7I), reconstructed from an ∼250 nm thick section of a DENV-infected Huh-7 cell. Colored overlay shows a 3D surface model of the virus-induced membranes. ER and Golgi membranes are depicted in yellow, inner vesicle membranes in light brown, and virus particles in red. Note the continuity of the ER and vesicle membranes and the putative budding profiles on the NE membrane opposite of the pores. Single virus particles are located within the NE and virions within vesicles near the Golgi stack.
Three-dimensional surface rendering of ER and Golgi membranes (yellow), virus-induced vesicles (light brown), and virus particles (red) of the reconstructed 3D volume displayed in Figures 7F–7I and Movie S3. Note the vesicle pores directly connecting the vesicle lumen to the cytosol.