Abstract
Localisation of both viral and cellular proteins to the nucleolus is determined by a variety of factors including nucleolar localisation signals (NoLSs), but how these signals operate is not clearly understood. The nucleolar trafficking of wild type viral proteins and chimeric proteins, which contain altered NoLSs, were compared to investigate the role of NoLSs in dynamic nucleolar trafficking. Three viral proteins from diverse viruses were selected which localised to the nucleolus; the coronavirus infectious bronchitis virus nucleocapsid (N) protein, the herpesvirus saimiri ORF57 protein and the HIV-1 Rev protein. The chimeric proteins were N protein and ORF57 protein which had their own NoLS replaced with those from ORF57 and Rev proteins, respectively. By analysing the sub-cellular localisation and trafficking of these viral proteins and their chimeras within and between nucleoli using confocal microscopy and photo-bleaching we show that NoLSs are responsible for different nucleolar localisations and trafficking rates.
Keywords: Nucleolus, Nucleolar trafficking, FRAP, FLIP, Herpesvirus, Coronavirus, HIV
Introduction
The nucleolus is a sub-nuclear compartment involved in many processes crucial to the efficient functioning of a eukaryotic cell (Carmo-Fonseca et al., 2000, Lam et al., 2005). These include, but are not limited to, ribosomal RNA synthesis (Grummt, 2003), modulation of cell growth (Hernandez-Verdun and Roussel, 2003) and response to cell stress (Rubbi and Milner, 2003). It is composed of over 700 proteins (Leung et al., 2006) which can be grouped into separate classes depending on their role in the nucleolus and wider cell function (Leung et al., 2003, Leung and Lamond, 2003).
The nucleolus is formed from a myriad of protein:protein and protein:nucleic acid interactions (Carmo-Fonseca, 2002, Louvet et al., 2005). Several features can be identified in the nucleolus corresponding to its central role in RNA synthesis including fibrillar centres (FCs), dense fibrillar components (DFCs), and a granular compartment (GC) (Louvet et al., 2005). As with other sub-nuclear structures, although the nucleolus appears to be stable, when viewed as fixed cell preparations for fluorescence or under the electron microscope, nucleolar proteins are undergoing continuous exchange with the nucleoplasm (Andersen et al., 2005, Hernandez-Verdun, 2006, Hernandez-Verdun et al., 2002, Lamond and Sleeman, 2003, Phair and Misteli, 2000). Therefore the structure of the nucleolus has been proposed to be maintained by the equilibrium between binding and release of sequestered proteins (Misteli, 2001). As such to account for protein localisation to discrete compartments within the nucleus including the nucleolus, a scanning mechanism has been proposed (Misteli, 2001) in which proteins diffuse through the nucleoplasm until they find their target, rather than requiring specific targeting signals per se (Misteli, 2001).
DNA, retro and RNA virus proteins and in some cases viral genomes traffic to and from the nucleolus (Hiscox, 2002 , 2007). For example, in the DNA virus herpesvirus saimiri (HVS), nucleolar localization of the viral ORF57 protein was shown to be required for the nuclear-cytoplasmic trafficking of intron-less viral RNAs (Boyne and Whitehouse, 2006, Goodwin et al., 1999, Williams et al., 2005). A number of retroviral proteins, for example from human immunodeficiency virus (HIV-1), localise to the nucleolus including the Rev (Cochrane et al., 1990, Perkins et al., 1989) and Tat proteins (Kuppuswamy et al., 1989, Ruben et al., 1989). One of the functions of Rev protein within the life cycle of HIV is trafficking of intron-containing viral RNA from the nucleus to the cytoplasm (Pollard and Malim, 1998). Multimerisation of Rev is crucial to this function (Stauber et al., 1998), and this step is predominately thought to occur in the nucleolus (Daelemans et al., 2004). Indeed the interaction of Rev with viral RNA can be disrupted through the use of nucleolar Rev binding elements (Michienzi et al., 2006), HIV-1 RNA can be cleaved in the nucleolus using ribozymes (Michienzi et al., 2000) and likewise small nucleolar RNA-TAR decoy RNAs can be used to sequester Tat (Michienzi et al., 2002). All of these approaches have illustrated the essential role of the nucleolus in virus biology as well as providing potential genetic therapeutic strategies against HIV-1 infection (Rossi et al., 2007). Despite the cytoplasm being the primary site of viral RNA synthesis, many positive strand RNA virus proteins have been shown to localise to the nucleolus (Hiscox, 2003 Hiscox, 2007, Rowland and Yoo, 2003, Taliansky and Robinson, 2003, Weidman et al., 2003), and these are generally viral capsid/nucleocapsid proteins whose function is in binding to viral RNA. For example, many coronavirus and the closely related arterivirus nucleocapsid (N) proteins have been shown to localise to the nucleolus (Hiscox et al., 2001, Rowland et al., 1999, Tijms et al., 2002, Wurm et al., 2001). Mutation of the nuclear/nucleolar localisation signal (NLS/NoLS) in the arterivirus porcine reproductive and respiratory syndrome virus (PRRSV) N protein in the context of recombinant virus infected cells resulted in cytoplasmic localisation of the protein and lower viral titers when compared to wild type virus (Lee et al., 2006). Upon passage in swine, NLS/NoLS revertant mutants were observed (Lee et al., 2006) pointing to a role for the nucleolus/nucleus in the virus life cycle (Pei et al., 2008). In the positive strand plant RNA groundnut rosette virus the nucleolus and interaction with nucleolar proteins has been shown to be essential for systemic infection (Kim et al., 2007a , 2007b). In negative strand RNA viruses the nucleoprotein and non-structural protein 1 (NS1) protein in certain strains of influenza virus has also been shown to localise to the nucleolus (Melen et al., 2007, Ozawa et al., 2007), and NS1 protein has been shown to interact with nucleolin (Murayama et al., 2007).
How viral and cellular proteins traffic to the nucleolus and what determines their sub-nucleolar localisation is not clearly understood, but NoLSs are thought to be involved (Carmo-Fonseca et al., 2000). How these operate to direct proteins to the nucleolus is generally not well understood. To investigate the role of NoLSs in dynamic trafficking and directing sub-nucleolar localisation, three viral proteins were selected from a DNA, RNA and retrovirus, which had defined but different NoLSs. The avian coronavirus, infectious bronchitis virus N protein, a phosphorylated viral RNA binding protein (Chen et al., 2005, Spencer et al., 2008, Spencer and Hiscox, 2006), localises to both the cytoplasm and the nucleolus in both infected and cells over-expressing N protein (Chen et al., 2002, Hiscox et al., 2001, Wurm et al., 2001). N protein can be predominately found within the DFC and GC (Dove et al., 2006, Reed et al., 2006) and contains an eight amino acid motif (WRRQARFK) which is necessary and sufficient for directing nucleolar localisation (Reed et al., 2006). N protein also contains a defined nuclear export signal (Reed et al., 2007). HVS ORF57 protein localises predominately to the nucleus, nucleolus and nuclear speckles (Goodwin et al., 1999, Williams et al., 2005) and contains a bi-partite NoLS which is composed of two different but functional NLSs, a classical pat4 motif (KRPR) and a second 10 amino acid motif (RRPSRPFRKP) (Boyne and Whitehouse, 2006). HIV-1 Rev protein, localises to the nucleolus in the DFCs and GC (Dundr et al., 1995) and contains a 16 amino acid motif (RQARRNRRRRWRERQRQ) which directs nucleolar localisation (Cochrane et al., 1990, Kubota et al., 1989). All of these signals have been shown to be necessary (i.e. if they are deleted/mutated in the context of wild type protein then nucleolar localisation is abolished) and sufficient (they will retarget an exogenous protein to the nucleolus) for protein localisation to the nucleolus (Cochrane et al., 1990, Goodwin and Whitehouse, 2001, Reed et al., 2006).
In this study nucleolar localisation signals, nucleolar retention signals, nucleolar accumulation signals are all referred to as NoLSs. By analysing the trafficking of these three viral proteins we demonstrated that each protein had different nucleolar trafficking rates due to their alternative NoLSs. This was confirmed by swapping the NoLSs between viral proteins and thereby altering their trafficking dynamics and localisation.
Results and discussion
Nucleolar localisation of viral proteins
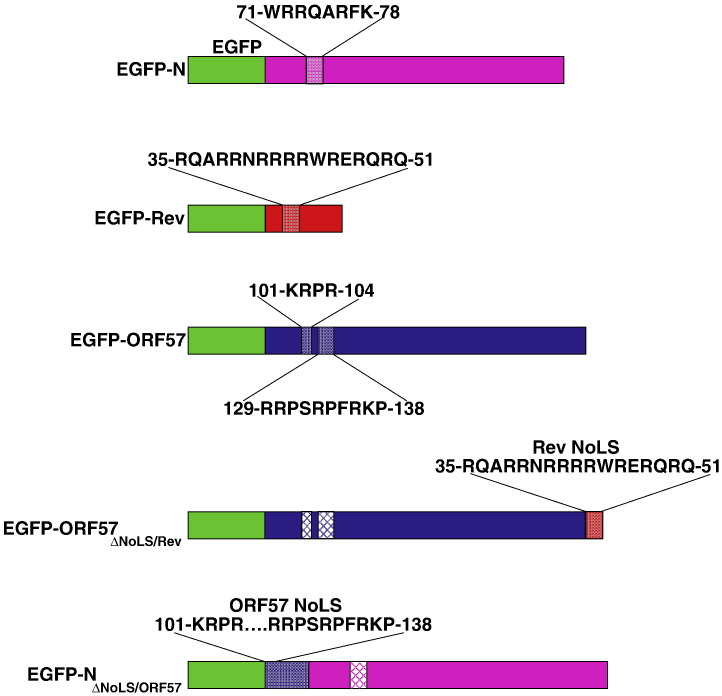
To investigate the role of viral NoLSs in localisation and dynamic nucleolar trafficking, a series of expression plasmids were utilised which led to the production of wild type viral proteins tagged to EGFP (EGFP-N, EGFP-Rev and EGFP-ORF57) (Fig. 1 ), and chimeric proteins in which the function of the wild type NoLS had been disabled and replaced with a heterologous NoLS from another viral protein. These chimeric proteins were generated from EGFP-N protein in which its own NoLS had been disrupted (Reed et al., 2006) and replaced with the corresponding NoLS from ORF57 (generating protein EGFP-NΔNoLS/ORF57) and the EGFP-ORF57 protein in which its NoLS had been disrupted and replaced the HIV-1 Rev protein NoLS (generating protein EGFP-ORF57ΔNoLS/Rev) (Boyne and Whitehouse, 2006) (Fig. 1). Fluorescent fusion tagged proteins were used in this study to allow live imaging, dynamic trafficking, to avoid fixation artefacts of sub-cellular localisation and also difficulties associated with using anti-bodies to detect proteins in the nucleolus (Sheval et al., 2005.
Fig. 1.
Diagrammatic representation of the EGFP-viral wild type and chimeric fusion proteins used in this study. The position and amino acid of each NoLS is indicated and in the chimeric fusion proteins the inactivated NoLS is denoted in crosshatch. IBV N protein, Rev protein and ORF57 protein are shown in pink, red and blue, respectively. EGFP is denoted in green.
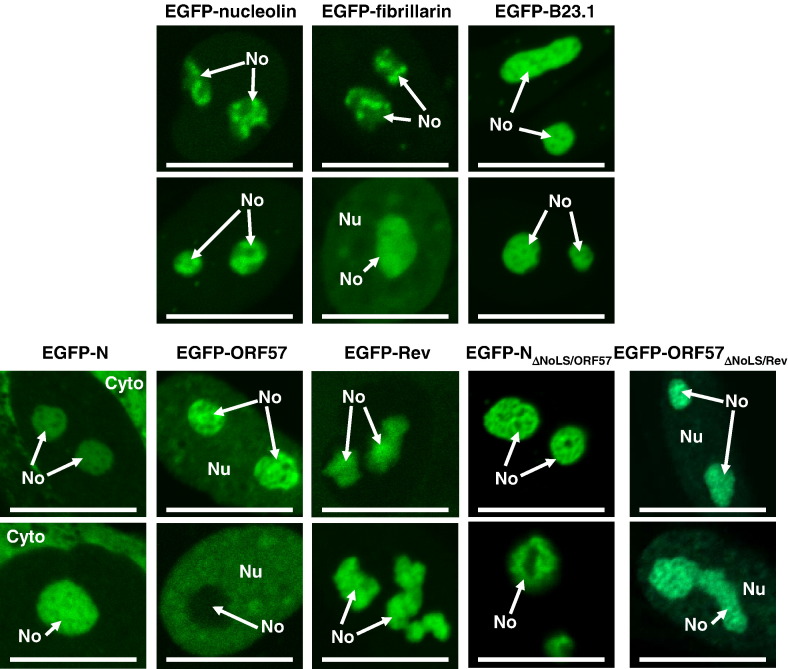
To provide markers for the nucleolus and for comparison in the dynamic trafficking analysis described below, Vero cells were transfected with expression plasmids which led to the production of fluorescent tagged cellular nucleolar proteins, nucleolin (EGFP-nucleolin), fibrillarin (EGFP-fibrillarin) and one of the two splice variants of B23 (Chang and Olson, 1989); B23.1 (EGFP-B23.1). EGFP-nucleolin localised predominately to the nucleolus (with some nuclear localisation) and was located mainly within the GC, but was excluded from FCs (Fig. 2 ). EGFP-fibrillarin had a globular type nucleolar localisation and localised principally to the DFCs with some nuclear accumulation (Fig. 2). EGFP-B23.1 localised mainly to the DFCs in the nucleolus but did not accumulate in the FCs (Fig. 2). This is similar to previously published data on the nucleolar localisation of these proteins (Chen and Huang, 2001, Dundr et al., 2000).
Fig. 2.
Sub-cellular localisation of EGFP-nucleolin, EGFP-fibrillarin and EGFP-B23.1 and EGFP-N, EGFP-Rev, EGFP-ORF57, EGFP-NΔNoLS/ORF57 and EGFP-ORF57ΔNoLS/Rev in Vero cells as determined by live cell confocal microscopy. The width of the marker is 15 μm and the image encompasses one nucleus. The nucleolus, nucleus and cytoplasm are indicated as No, Nu and Cyto, respectively.
The data indicated that in Vero cells EGFP-N protein localised either to the cytoplasm or the cytoplasm and nucleolus with a small amount of nuclear localisation (two examples of the latter are shown) (Fig. 2). EGFP-ORF57 protein localised predominately to the nucleolus with a variable distribution in the nucleus, and occasionally did not localise to the nucleolus (Fig. 2). EGFP-Rev protein localised predominately to the nucleolus (Fig. 2). Whilst EGFP-N protein and EGFP-Rev protein had a nucleolar distribution which was similar to EGFP-B23.1, localising to the DFCs, EGFP-ORF57 did not have a nucleolar localisation pattern which corresponded to the marker proteins, and could not be assigned either to the FC, DFC or GC. Previously, Rev protein has been reported to localise to the DFC and GC and to co-localise with B23.1 (Dundr et al., 1995). Interestingly over-expression of the viral and chimeric viral proteins led to different nucleolar morphologies suggesting that these proteins may alter the structure of the nucleolus. Certainly in both herpes-simplex-virus-1 (Lymberopoulos and Pearson, 2007, Bertrand and Pearson, 2008) and IBV infected cells (Dove et al. 2006) nucleolar morphology can alter.
In both cases the chimeric proteins also localised to the nucleolus in Vero cells, with the same apparent distribution as the protein from which the NoLS had been derived (Fig. 2). EGFP-NΔNoLS/ORF57 localised predominately to the nucleolus and in some cases had distinct possible nuclear speckles which are thought to co-localise with splicing factors (which has previously been observed for ORF57; Cooper et al., 1999), rather than the nucleolus and cytoplasm observed with EGFP-N protein. EGFP-ORF57ΔNoLS/Rev preferentially accumulated in the nucleolus over the nucleus and in some cases had an exclusively nucleolar distribution, similar to EGFP-Rev protein. This data suggests that viral NoLSs can be substituted between viral proteins whilst still maintaining the phenotype of nucleolar localisation. However, the heterologous NoLS may alter the localisation within the nucleolus.
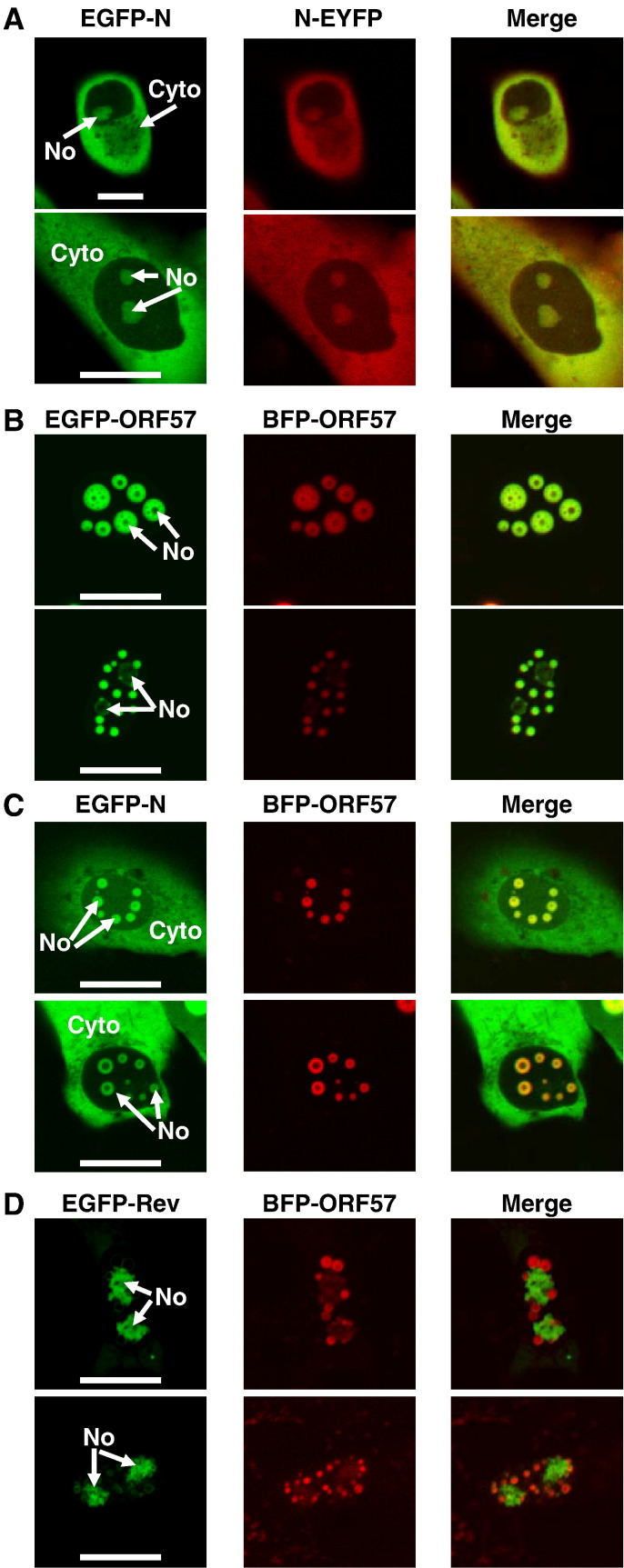
To confirm that the chimeric proteins were able to localise to the nucleolus and potentially had different localisation patterns to the parental proteins, these proteins were expressed in Vero cells which also expressed the appropriate parental proteins from which the NoLS had been derived. Given that all of the fluorescent proteins used were EGFP-tagged, two other wild type fluorescent labelled proteins were generated, EYFP-N and BFP-ORF57, to use in conjunction with the EGFP-wild type and chimeric proteins. Note that in all images both EYFP-N protein and BFP-ORF57 protein were false coloured red post image capture.
First to confirm that both EYFP-N protein and BFP-ORF57 protein had identical localisation to EGFP-N protein and EGFP-ORF57 protein, respectively, and thus could be used when comparing the localisation of the chimeric proteins, these proteins were co-expressed in Vero cells. Both EGFP-N protein and EYFP-N protein co-localised to the cytoplasm and the nucleolus as determined by the merged image (Fig. 3A). EGFP-ORF57 protein and BFP-ORF57 protein co-localised as determined by the merged image (Fig. 3B). In both cases variable nuclear localisation was observed and two examples of this are shown in which the proteins appear to be mainly localised to the nucleolus, or localised to the nucleolus and other sub-nuclear structures (Fig. 3B). Co-expression of EGFP-N and BFP-ORF57 indicated that whilst EGFP-N localised in the cytoplasm and the nucleolus, BFP-ORF57 localised in the nucleolus only (Fig. 3C) as would be predicted based upon their expression profiles when expressed in isolation. In cells expressing EGFP-Rev protein and BFP-ORF57 protein (Fig. 3D), EGFP-Rev protein localised predominately to the nucleolus and BFP-ORF57 predominately to other nuclear structures, with a weaker localisation in the nucleolus. This suggested that when co-expressed, EGFP-Rev protein may occupy the nucleolus in preference to BFP-ORF57 protein; this may be related to trafficking dynamics and/or relative affinity of interaction with components of the nucleolus.
Fig. 3.
Live cell confocal imaging analysis of the sub-cellular localisation of (A) EGFP-N and EYFP-N (false coloured red) proteins, (B) EFGP-ORF57 and BFP-ORF57 (false coloured red) proteins, (C) EGFP-N and BFP-ORF57 (false coloured red) proteins, and (D) EGFP-Rev and BFP-ORF57 (false coloured red) proteins, in Vero cells. Merged images are also presented. The width of the marker is 10 μm. The nucleolus and cytoplasm are indicated as No and Cyto, respectively.
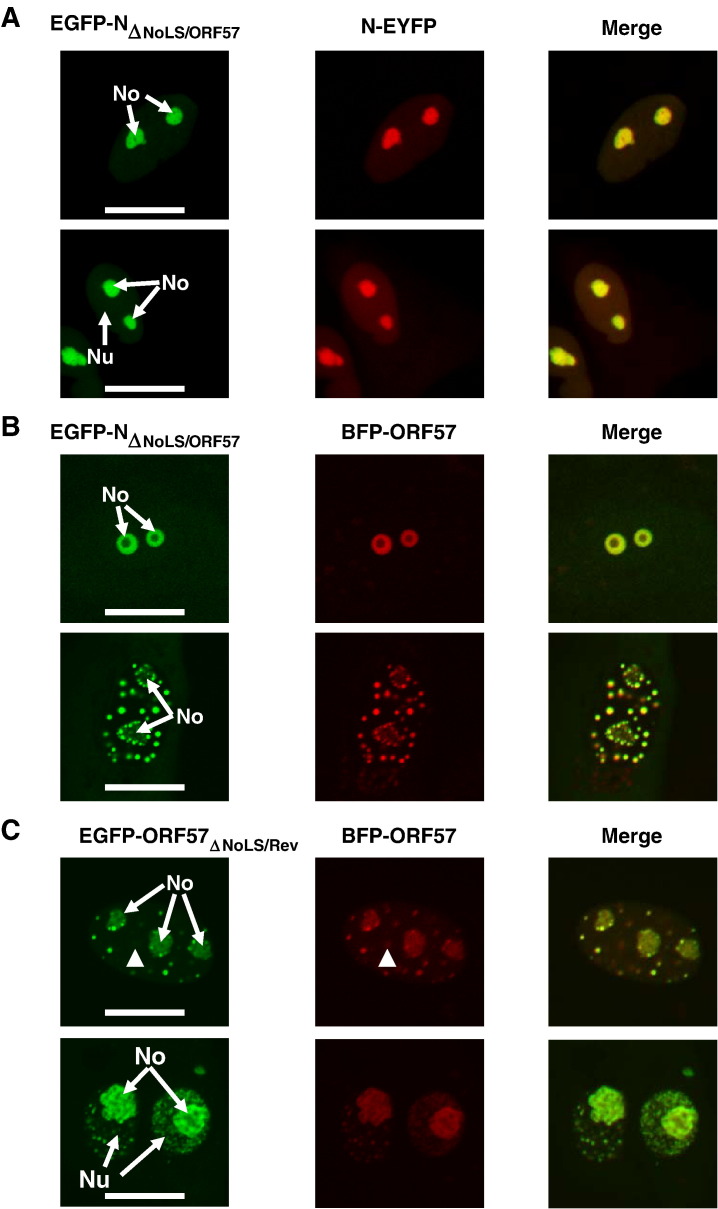
The sub-cellular localisation of the chimeric proteins was then compared to the wild type protein from which the NoLS was derived. We predicted that if the identified NoLS was the sole determinant of nucleolar localisation then in cells co-expressing EGFP-NΔNoLS/ORF57 protein and EYFP-N protein, EGFP-NΔNoLS/ORF57 protein would localise to the nucleolus and EYFP-N protein would localise to the cytoplasm and the nucleolus. When co-expressed in Vero cells, EGFP-NΔNoLS/ORF57 protein had the same distribution as when expressed in isolation (Fig. 4A). However, EYFP-N protein had a localisation pattern that was predominately nucleolar/nuclear (Fig. 4A), similar to EGFP-NΔNoLS/ORF57 protein. A minority of the protein still localised to the cytoplasm but was below the threshold of detection when the fluorescence in the nucleolus was captured in the linear range, indicating a localisation pattern intermediate between fluorescent tagged wild type N protein and EGFP-NΔNoLS/ORF57 protein. This result may indicate that EGFP-NΔNoLS/ORF57 protein and EYFP-N protein dimerised and that the ORF57 NoLS was the dominant localisation signal operating in the protein complex. Certainly purified IBV N protein can form multimers which are linked by disulphide bridge formation (Chen et al., 2003 , 2005).
Fig. 4.
Live cell confocal imaging analysis of the sub-cellular localisation of (A) EGFP-NΔNoLS/ORF57 and EYFP-N (false coloured red) proteins, (B) EGFP-NΔNoLS/ORF57 and BFP-ORF57 (false coloured red) proteins, (C) EGFP-ORF57ΔNoLS/Rev and BFP-ORF57 (false coloured red) proteins in Vero cells (arrows indicate one point of difference between samples, others are visible in the merged image). The width of the marker is 10 μm. The nucleolus and nucleus are indicated as No and Nu, respectively.
As would be predicted, co-expression of EGFP-NΔNoLS/ORF57 protein and BFP-ORF57 protein showed identical co-localisation indicating that the ORF57 NoLS directed both proteins to the same sub-nuclear structures (Fig. 4B). Co-expression of EGFP-ORF57ΔNoLS/Rev and BFP-ORF57 indicated that in general both proteins co-localised (Fig. 4C) (although there were minor differences, arrowed and visible in merged image), despite the parental EGFP-Rev protein and BFP-ORF57 protein having different localisation patterns (Fig. 3D). Similar, to the self-association of N protein, data indicated that ORF57 protein can also self-associate (Malik and Clements, 2004) which may therefore account for the co-localisation of EGFP-ORF57ΔNoLS/Rev and BFP-ORF57, and again indicating that one NoLS might be dominant over another.
Although Vero cells are permissive for infection with IBV, and thus may be considered an appropriate model for studying the sub-cellular localisation of N protein, we also investigated the sub-cellular localisation of EGFP-N, EGFP-Rev and EGFP-ORF57, EGFP-NΔNoLS/ORF57 and EGFP-ORF57ΔNoLS/Rev and also the cellular nucleolar marker proteins in cell lines related to the host species of the appropriate viral protein. This included; the avian cell line Doug Foster 1 (DF-1) which is permissive for IBV infection, the Owl monkey kidney (OMK) cell which supports the replication of HVS and the human HeLa cell line which is permissive for HIV-1 infection (provided the appropriate receptor is exogenously expressed) (Supplementary data). In all cases EGFP-nucleolin, EGFP-fibrillarin and EGFP-B23.1 localised to the nucleolus. In DF-1 and Vero cells EGFP-N protein localised to either to the cytoplasm or the cytoplasm and the nucleolus with approximately a 50% frequency for each phenotype, in line with that previously observed for this cell type (Cawood et al., 2007). In approximately 90% of OMK and HeLa cells EGFP-N protein localised to the cytoplasm only, while in approximately 10% of these cells localisation was both cytoplasmic and nucleolar. In contrast, in all four cell types (e.g. Vero, DF-1, OMK and HeLa cells) EGFP-ORF57 displayed variable nuclear localisation; nucleolar, nuclear and nucleolar, nuclear and small structures (possibly speckles), or nuclear only. EGFP-Rev protein localised to the nucleolus in all cell types examined. In all four cell types EGFP-NΔNoLS/ORF57 localised predominately to the nucleolus, with some cells also showing localisation to small nuclear structures. In all four cell types, EGFP-ORF57ΔNoLS/Rev localised to the nucleolus. Vero cells were then selected for subsequent dynamic trafficking analysis to standardise data capture.
Viral proteins traffic with different rates in the nucleolus
To compare the dynamic trafficking of these viral proteins within the nucleolus fluorescence recovery after photo-bleaching (FRAP) and fluorescence loss in photo-bleaching (FLIP) were utilised. In the FRAP experiments a defined portion of the nucleolus was photo-bleached and the relative ability of each protein to refill this area was compared. In the FLIP experiments, cells were imaged which contained two nucleoli, one of which was continuously photo-bleached, in order to investigate protein trafficking between the two. (Note that complete photo-bleaching of the cell over the time course of the trafficking experiments described below indicated that no significant de novo synthesis of fluorescent labelled proteins occurred).
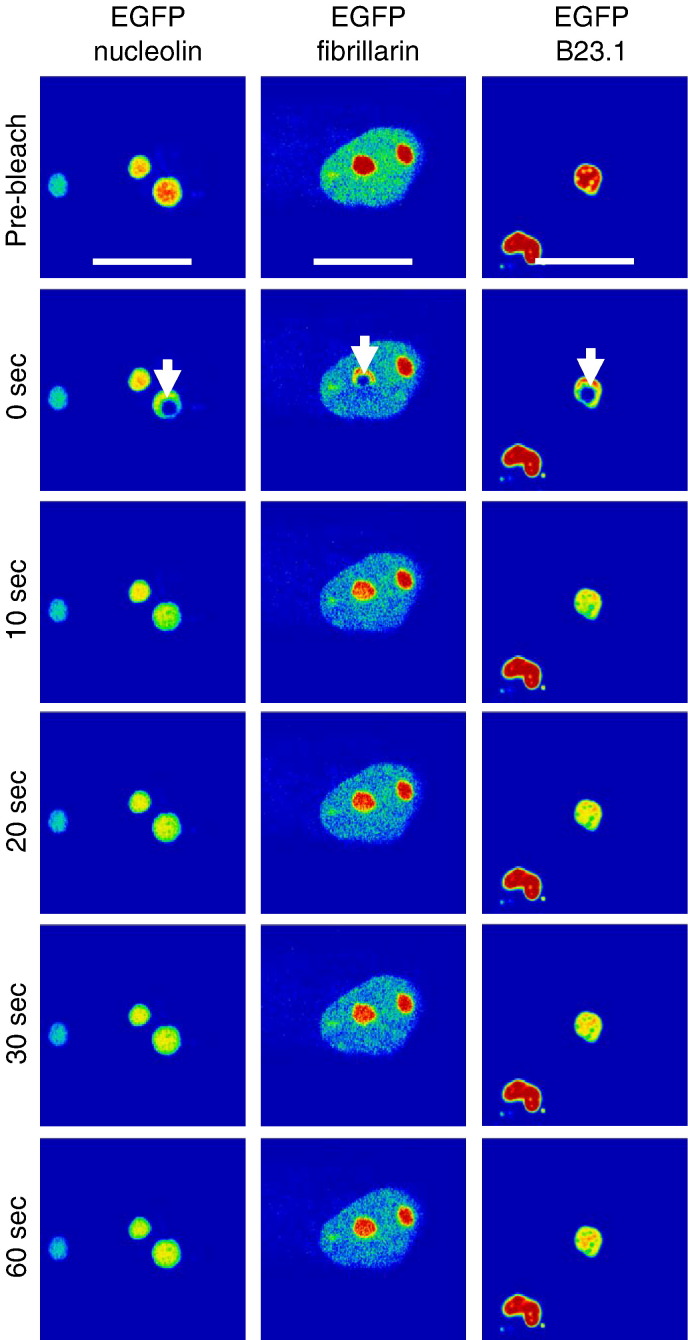
Although other studies have investigated the dynamic trafficking of nucleolin, fibrillarin and B23.1 under various metabolic conditions and cell types (Andersen et al., 2005, Angelier et al., 2005, Dundr et al., 2000, Phair and Misteli, 2000, You et al., 2008), these predominately nucleolar proteins were used in this analysis to compare to the viral proteins and also to provide a reference to previous work. In general the trafficking of nucleolar proteins can be classified into two broad criteria — fast and slow, depending on their function (Hernandez-Verdun, 2006). In this study 24 h post-transfection a defined region of the nucleolus was photo-bleached using FRAP and the time taken for the nucleolar marker proteins (Fig. 5 ), viral proteins and chimeric viral proteins (Fig. 6 ) to refill this photo-bleached area was determined (Fig. 7 ). The data indicated that EGFP-nucleolin (t 1/2 = 2.93 ± 1.28 s) and EGFP-fibrillarin (t 1/2 = 2.13 ± 0.41 s) had recovery rates that were not significantly different from each other (P = 0.22, significant difference P ≤ 0.05). Whereas the recovery rate of EGFP-B23.1 (t 1/2 = 7.69 ± 3.68 s) was significantly different from both nucleolin and fibrillarin (P = 0.02 and 0.01, respectively; significant difference P ≤ 0.05). However, recovery of these proteins was rapid, in line with previous observations (Chen and Huang, 2001, Dundr et al., 2000, Phair and Misteli, 2000). Proteins which are considered as having slower trafficking times, such as the ribosomal subunit proteins S5 and L9, generally give t 1/2 values of approximately 72 s (Chen and Huang, 2001).
Fig. 5.
FRAP analysis of EGFP-nucleolin, EGFP-fibrillarin and EGFP-B23.1. Images were sampled pre-bleach and post-bleach five times a second for 60 s. EGFP-fusion proteins are false coloured to show their concentration using the ‘rainbow’ feature on the Zeiss LSM imaging software, where red is high intensity and blue is low intensity. The experiment was repeated five times and a representative image from each cell series is shown for pre-bleach, immediately post-bleach (0 s), 10, 20, 30 and 60 s post-bleach. The bleach area is denoted in the immediate post-bleach image (0 s), and the width of the marker is 10 μm.
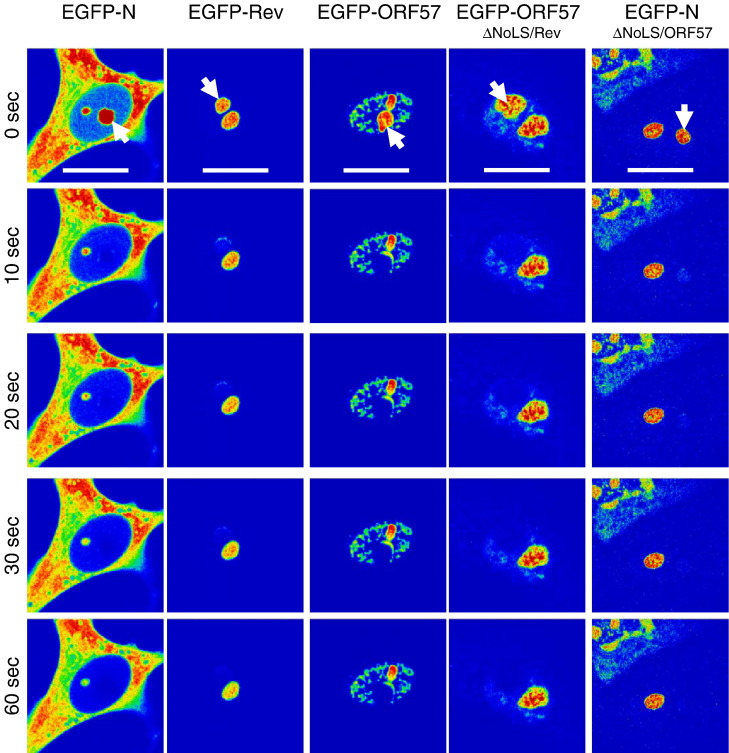
Fig. 6.
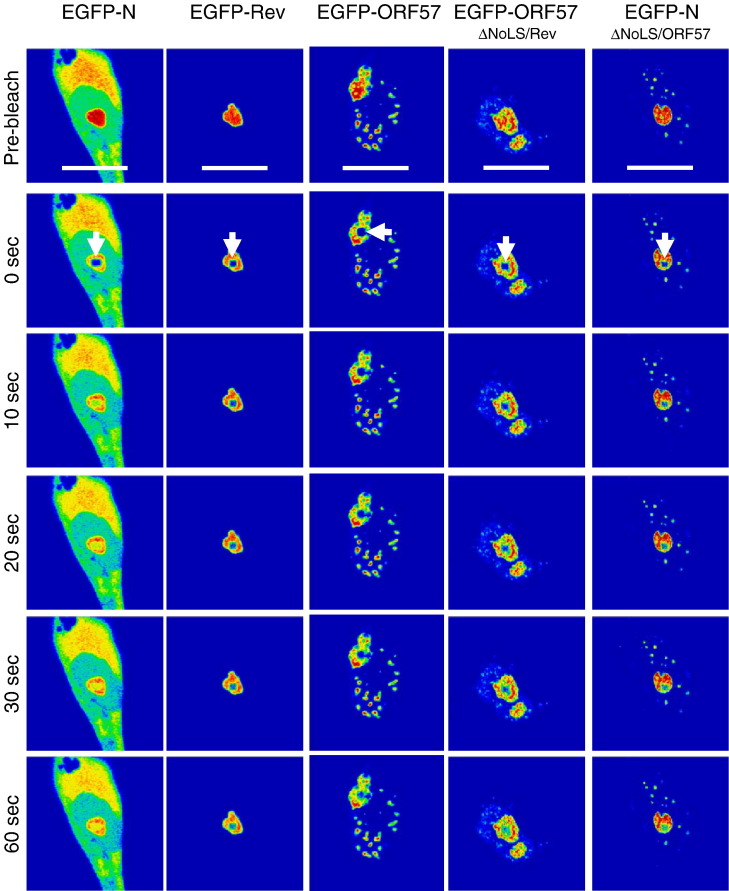
FRAP analysis of EGFP, EGFP-N, EGFP-Rev, EGFP-ORF57, EGFP-ORF57ΔNoLS/Rev and EGFP-NΔNoLS/ORF57. Images were sampled pre-bleach and post-bleach five times a second for 60 s. EGFP-fusion proteins are false coloured to show their concentration using the ‘rainbow’ feature on the Zeiss LSM imaging software, where red is high intensity and blue is low intensity. The experiment was repeated five times and a representative image from each cell series is shown for pre-bleach, immediately post-bleach (0 s), 10, 20, 30 and 60 s post-bleach. The bleach area in the nucleolus is denoted in the immediate post-bleach image (0 s), and the width of the marker is 15 μm.
Fig. 7.
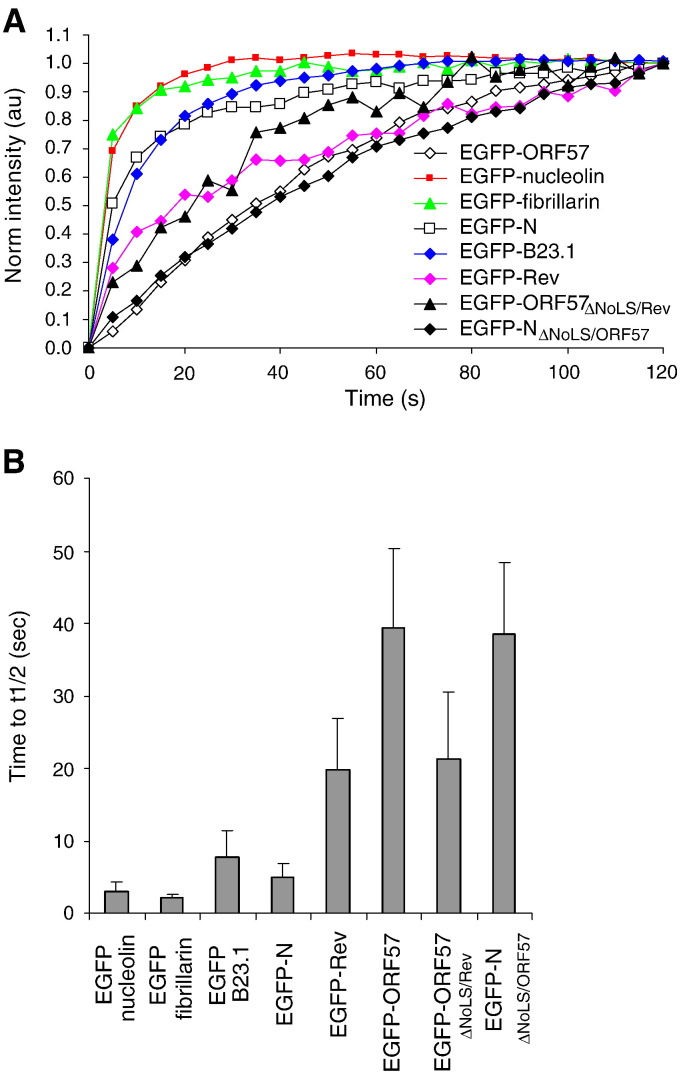
(A). FRAP analysis was performed on cells transfected with EGFP-tagged cellular and viral proteins. The recovery curves are shown over a period of 2 min following a photo-bleach period. Data was normalized following the bleach period so that the initial post-bleach was set at zero, and the final intensity level was set at one. (B). Quantitative analysis showing the relative half life recovery in seconds of GFP tagged cellular and viral proteins within the nucleolus of transfected Vero cells.
Analysis of the trafficking of viral proteins indicated that the recovery of EGFP-N protein (t 1/2 = 5.03 ± 1.95 s) was not significantly different from EGFP-nucleolin and EGFP-B23.1 (P = 0.06 and 0.11, respectively; significant difference P ≤ 0.05), but was significantly different from EGFP-fibrillarin (P = 0.01, significant difference P ≤ 0.05). However, the rate of recovery and hence an indication of the trafficking of EGFP-N protein within the nucleolus, is clearly comparable to the three nucleolar marker proteins used in this study. Therefore it can be considered as fast, at least compared to the recovery times previously described for nucleolin, fibrillarin and B23.1 (Chen and Huang, 2001, Dundr et al., 2000). In contrast, the recovery of EGFP-Rev protein (t 1/2 = 24.21 ± 3.15 s) and EGFP-ORF57 (t 1/2 = 42.97 ± 7.29 s) protein were significantly slower than the former four proteins e.g. compared to EGFP-N protein (P < 0.00 for both, significant difference P ≤ 0.05) and also from each other (P < 0.00, significant difference P ≤ 0.05) and can be considered as slow. This is in line with previous observations for the trafficking of Rev protein within the nucleolus (Daelemans et al., 2004).
The recovery rates observed with the trafficking of viral proteins in the nucleolus may not be attributed to size dependent diffusion within the nucleolus, in that a smaller protein may be predicted to traffic quicker within the nucleolus. For example, the predicted molecular weights of N, ORF57 and Rev monomeric proteins are approximately 45, 48 and 15 kDa, respectively (excluding the EGFP tag), yet the trafficking of EGFP-Rev protein is slower than EGFP-N protein, while the trafficking of EGFP-N protein is faster than the largest protein, EGFP-ORF57. This analysis is somewhat complicated in that all three proteins have been reported to form multimers (Chen et al., 2003, Daelemans et al., 2004, Malik and Clements, 2004). However, taken together our data suggests that similar to cellular nucleolar proteins, different viral proteins (from diverse viruses) can display different nucleolar trafficking profiles. Indeed, although conducted in different cell lines, the PRRSV N protein has faster recovery rates than the IBV N protein and nucleolin, fibrillarin and B23.1 (You et al., 2008).
Dynamic trafficking of viral proteins within the nucleolus is determined by the viral NoLS
Interestingly, both the chimeric EGFP-ORF57ΔNoLS/Rev protein (t 1/2 = 38.50 ± 9.78 s) and EGFP-NΔNoLS/ORF57 protein (t 1/2 = 19.11 ± 9.23 s) had recovery times that were not significantly different from the proteins from which their nucleolar targeting signals were derived (P = 0.28 and 0.44, respectively; significant difference P ≤ 0.05). In contrast, the recovery of these proteins within the nucleolus was significantly different from the parental EGFP-tagged wild type proteins e.g. EGFP-ORF57ΔNoLS/Rev protein compared to EGFP-ORF57, P = 0.01 (significant difference P ≤ 0.05) and EGFP-NΔNoLS/ORF57 protein compared to EGFP-N, P < 0.00 (significant difference P ≤ 0.05). This data suggested that the heterologous NoLS alone could determine dynamic nucleolar trafficking of a protein.
This was further demonstrated when the diffusion coefficient (D) value of EGFP-nucleolin, EGFP-fibrillarin, EGFP-B23.1, EGFP-N, EGFP-Rev, EGFP-ORF57, EGFP-ORF57ΔNoLS/Rev and EGFP-NΔNoLS/ORF57 proteins were calculated from the recovery curves (Phair and Misteli, 2000). This value is the rate at which a protein repopulates a photo-bleached area (Table 1 ). This data indicated that the chimeric viral proteins had similar D values corresponding to the wild type protein from which their NoLS was derived. For comparison EGFP-labelled PRRSV N protein was shown to have a D value of 0.839 μm2/s (You et al., 2008). This is approximately six times more mobile than EGFP-labelled IBV N protein, and again indicated that viral proteins could have variable mobility within the nucleolus.
Table 1.
| Expressed protein | Diffusion coefficient (D) |
|---|---|
| EGFP-nucleolin | 0.257 ± 0.143 μm2/s |
| EGFP-fibrillarin | 0.299 ± 0.05 μm2/s |
| EGFP-B23.1 | 0.097 ± 0.043 μm2/s |
| EGFP-N protein | 0.138 ± 0.047 μm2/s |
| EGFP-Rev protein | 0.039 ± 0.027 μm2/s |
| EGFP-ORF57 protein | 0.017 ± 0.006 μm2/s |
| EGFP-ORF57ΔNoLS/Rev | 0.037 ± 0.013 μm2/s |
| EGFP-NΔNoLS/ORF57 | 0.017 ± 0.003 μm2/s |
The nucleolus is a dynamic structure and recent evidence suggests it is continuously exchanging nucleolar proteins with the nucleoplasm (Matthews and Olson, 2006). To investigate if this occurred with viral proteins and whether the NoLS was involved in regulating this exchange, we performed FLIP experiments on cells with two nucleoli which contained the fluorescently tagged viral protein. In this assay, a selected nucleolus was continually imaged and photo-bleached for a period of 3 min and the fluorescence intensity of the non-bleached nucleolus was assessed. Rapid loss of fluorescence in the non-bleached nucleolus is suggestive of a dynamic interchange of the tagged protein between the nucleoplasm and nucleolus. No reduction in fluorescence intensity would suggest a static protein. No significant bleaching was observed in neighbouring nuclei in all of the experiments (data not shown).
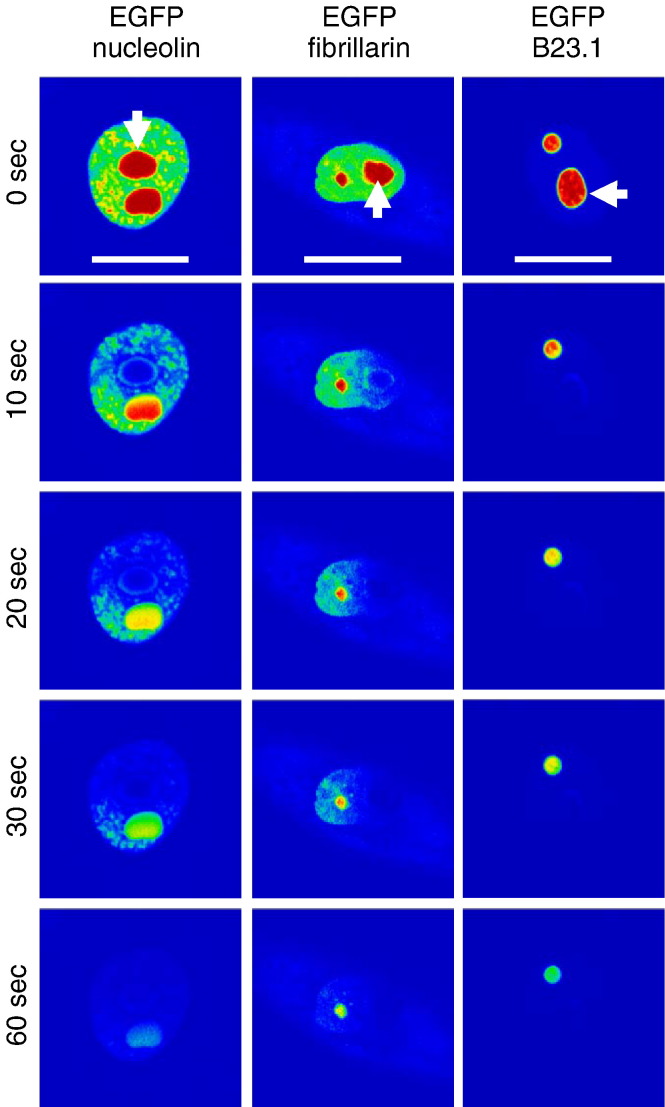
Comparison of the relative trafficking between two nucleoli using FLIP in general reflected the mobility data obtained in the FRAP experiments. The three cellular nucleolar proteins examined were rapidly lost from the non-bleached nucleolus, with greatly reduced signal at 60 s (Fig. 8 ). This is similar to the data of Phair and Misteli (2000), who found that if one nucleolus was continuously photo-bleached then this resulted in the complete loss of nucleolin, fibrillarin and B23.1 from the other. Our data indicated that there was no general correlation between rate of loss and whether the protein had a nucleolar/nuclear rather than an exclusively nuclear distribution (e.g. EGFP-nucleolin versus EGFP-B23.1). Of the viral proteins examined, loss of fluorescence from the unbleached nucleolus was not related to the molecular weight of the protein, with EGFP-ORF57 having a slower amount of loss compared to EGFP-Rev and EGFP-N, the latter of which had the fastest amount of loss (Fig. 9 ). A possible explanation for this observation may be due to the presence of a pool of the viral protein in the nucleoplasm which is more readily drawn on by the photo-bleached nucleoli, as observed with EGFP-nucleolin and EGFP-fibrillarin. Again, the chimeric proteins had loss rates which were similar to the protein from which the NoLS was derived. EGFP-NΔNoLS/ORF57 protein was retained in the non-photo-bleached nucleolus similar to EGFP-ORF57 and EGFP-ORF57ΔNoLS/Rev protein had a greater loss than EGFP-ORF57.
Fig. 8.
FLIP analysis of EGFP-nucleolin, EGFP-fibrillarin and EGFP-B23.1. One nucleolus was continuously photo-bleached (indicated by an arrow) and images were sampled at 0, 10, 20, 30 and 60 s. EGFP-fusion proteins are false coloured to show their concentration using the ‘rainbow’ feature on the Zeiss LSM imaging software, where red is high intensity and blue is low intensity. The experiment was repeated five times and a representative image from each cell series is shown. The width of the marker is 10 μm.
Fig. 9.
FLIP analysis of EGFP-N, EGFP-Rev, EGFP-ORF57, EGFP-ORF57ΔNoLS/Rev and EGFP-NΔNoLS/ORF57. One nucleolus was continuously photo-bleached (indicated by an arrow) and images were sampled at 0, 10, 20, 30 and 60 s. EGFP-fusion proteins are false coloured to show their concentration using the ‘rainbow’ feature on the Zeiss LSM imaging software, where red is high intensity and blue is low intensity. The experiment was repeated five times and a representative image from each cell series is shown. The width of the marker is 10 μm.
The data presented in this study suggested that viral NoLSs could have a dual role in determining the localisation and trafficking of a protein within the nucleolus. This is supported by the use of chimeric viral proteins which have their own NoLS mutated and a heterologous NoLS inserted. These chimeric proteins have an altered localisation pattern and trafficking rate which overrides the wild type phenotype and would appear to be dependent on the heterologous NoLS. Interestingly substitution of the ORF57 NoLS with the Rev Protein NoLS restored the viral RNA trafficking function of ORF57 protein (Boyne and Whitehouse, 2006).
Given that trafficking of the viral and chimeric proteins may not be related to their molecular weight, the different NoLSs may be interacting with different cellular partners to promote differential trafficking and localisation of a protein within the nucleolus. Within the nucleolus the two obvious candidates are interactions with nucleolar proteins and/or RNA. Previous data has indicated that N protein can interact with nucleolin and fibrillarin (Chen et al., 2002, Dove et al., 2006, Reed et al., 2006). Indeed the trafficking profile of N protein, in this study is similar to nucleolin and fibrillarin. This suggests a protein:protein interaction with N protein and a cellular nucleolar protein may be responsible for its trafficking. However, Rev protein has been shown to co-localise and interact with B23.1 (Dundr et al., 1995, Li, 1997, Marasco et al., 1994, Miyazaki et al., 1996) although the trafficking profiles are different. Similarly ORF57 can also associate with viral RNA and other cellular protein which are found in the nucleolus (Boyne et al., 2008, Williams et al., 2005). Likewise, although PRRSV N protein has been shown to interact with fibrillarin (Yoo et al., 2003) the mobility of the two proteins within the nucleolus is different (EGFP labelled PRRSV N protein had a D value of 0.839 μm2/s and EGFP-labelled fibrillarin had a D value of 0.278 μm2/s) (You et al., 2008).
The specific trafficking dynamics of virus proteins which localise to the nucleolus may be important for function. Certainly, abolishing the nucleolar localisation of viral proteins through mutation of specific trafficking signals is detrimental for virus biology (Huang et al., 2001, Kim et al., 2007a , 2004 Kim et al., 2007b, Lee et al., 2006 Lee et al., 1998, Pei et al., 2008). Disrupting the efficiency of nucleolar localisation of viral proteins through the use of chimeric or mutated NoLSs may therefore be a way of attenuating virus replication, whether as part of an antiviral strategy or for the design of recombinant growth attenuated virus and potential vaccine candidates.
Materials and methods
Cell culture
Vero, DF-1, HeLa and OMK cells were grown at 37°C with 5% CO2 in Dulbecco's modified Eagles media (DMEM) supplemented with 10% foetal calf serum and penicillin/streptomycin. 33 mm glass based tissue culture dishes were seeded with 2 × 105 cells 24 h prior to transfection.
Expression plasmids
The construction and use of pEGFP, pEGFP-nucleolin, pEGFP-fibrillarin, pEGFP-B23.1, pEGFP-N, pEGFP-ORF57, pBFP-ORF57, pEGFP-HIV-Rev and EGFP-ORF57ΔNoRS/Rev have been described previously (Boyne and Whitehouse, 2006, Goodwin et al., 1999, Goodwin and Whitehouse, 2001, Reed et al., 2006, You et al., 2005). pEGFP-NΔNoLS/ORF57 was constructed by amplifying the ORF 57 nucleolar targeting signal using PCR with forward primer 5′-AAAGAATTCAAGCGACCCCGTATCAGC-3′ and reverse primer 5′-AAAGAATTCTGGCTTTCTGAAAGGCCT-3′, and sub-cloned into TOPO TA vector pCR2.1. The ORF57 nucleolar targeting signal was isolated using incorporated EcoRI restriction sites and inserted into EcoRI digested pEGFP-NΔNoLS (in which the IBV N protein NoLS had been disabled and therefore the protein did not localise to the nucleolus; Reed et al., 2006). This produced an expression construct where the ORF 57 NoLS was C-terminal of EGFP and N-terminal of IBV N protein. pEYFP-N was constructed by using pEGFP-N as the template for PCR. Primers were designed to introduce a XhoI restriction site 5′ of the construct (EdC1IBVN-fwd CCGCTCGAGATATGGCAAGCGGTAAAGCAGCTGGA) and a SalI site 3′ of the construct (EdN1IBVN-rev CGGGTCGACATTCAAAGTTCATTCTCTCCTAGAGCTGC). PCR products were then purified and sub-cloned into pCR-Blunt (Invitrogen) as per the manufacturer's instructions. DNA was digested with XhoI and SalI (New England Biolabs) before being ligated into pEYFP-N1 (Clontech), which, when expressed, produced full-length N protein fused to the N-terminus of EYFP. The orientation of all inserts was confirmed by sequencing and correct expression by Western blot (data not shown).
Confocal imaging
Confocal images were captured on an LSM510 META microscope (Carl Zeiss Ltd., Germany) equipped with a 40× and 63×, NA 1.4, oil immersion lens. Pinholes were set to allow optical sections of 1 μm to be acquired. EGFP was excited with the 488 nm argon laser line running at 2% and emission was collected through a LP505 filter. All fluorescence was measured in the linear range as the detector is a photomultiplier, and the range indicator was utilised to ensure no saturated pixels were obtained on image capture. Images were averaged four times.
FRAP microscopy and data analysis
Cells were plated onto glass based 33 mm culture dishes, transfected and imaged 24 h later on an inverted LSM 510 META confocal microscope (Carl Zeiss, Herts, UK). Cells were maintained at 37 °C with a heated stage throughout the experiments. For imaging cell culture medium was exchanged for CO2 independent medium (Gibco) to maintain cell homeostasis throughout the experiments. All images were captured using a 63× objective and a digital zoom factor of 4 within the software. EGFP was excited with the 488 nm laser line delivered from a 30 mW argon laser running at 6.1 A and 1% power output; these settings were established as causing no residual background bleaching of the sample with the appropriate controls.
Photo-bleaching was performed on a defined area of 12 pixels squared, which equated to 20.16 μm2 area, within the nucleolus with the 488 nm laser line at 100% power output for 100 iterations, bleaching took approximately 1.2 s. Five images were collected prior to the bleach and images were collected continually for up to 120 s subsequently. Images are presented up to 60 s. Recovery of fluorescence was detailed using the ROI Mean module of the LSM510 software and 12-bit data sets were exported into Microsoft Excel for analysis.
Data from 5–7 experiments were normalized so that the first time point following the bleach period was 0 and the final intensity measurement was 1 using the previously published equation I t = (X t − Y) / (Z − Y), where I is the intensity at time t, Y is the intensity immediately after the photo-bleach and Z in the final intensity measurement (Stenoien et al., 2002). This allowed the time to half final recovery (t 1/2) to be easily determined from the resultant graphical data. Mean t 1/2 are presented with standard deviations determined using Microsoft Excel.
Diffusion coefficient (D) values were calculated from the half life recovery (t 1/2) using the following diffusion equation D = (w 2 / 4t 1/2) × 0.88 where w is the width of the bleach area, approx 1.68 μm, and a constant factor of 0.88 was used for a Gaussian beam profile (Axelrod et al., 1976). Data from 5 experiments were used per construct and standard deviation was calculated in the usual manner using Microsoft Excel.
FLIP microscopy
Transfected Vero cells were imaged in glass base dishes as outlined above. Imaging and photo-bleaching were performed with the same laser settings as detailed in the FRAP microscopy. In each FLIP experiment a single cell nucleus was imaged 5 times followed by a period of photo-bleaching for a total time of 3 min. Photo-bleaching was performed on one nucleolus within the cell for 50 iterations (mean bleach time 2.1 s). To ensure cell viability after laser power submission, DIC images were taken of the cells before and after FLIP analysis during preliminary experiments to establish photo-bleaching conditions.
Acknowledgments
This work was funded by the award of a BBSRC DTG studentship to EE, a BBSRC project grant BBSB03416 to JAH and GB, BBSRC Committee Studentship BBSSP200310434 to JAH, and BBSRC project grant BBSB03475 and Yorkshire Cancer Research pump prime grant to AW and JAH. AW is the recipient of a Leverhulme Research Fellowship. The confocal microscope facility was funded by the Wellcome Trust and SRIF. The authors would like to thank Dr. John Rossi (Beckman Research Institute) for his generous donation of the HIV-1 Rev EGFP expression construct used in this study.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.virol.2008.05.032.
Appendix A.
Sub-cellular localisation of EGFP, EGFP-nucleolin, EGFP-fibrillarin and EGFP-B23.1 and EGFP-N, EGFP-Rev, EGFP-ORF57, EGFP-NΔNoLS/ORF57 and EGFP-ORF57ΔNoLS/Rev in DF-1 cells as determined by live cell confocal microscopy. The width of the marker is 15 μm. The nucleolus, nucleus and cytoplasm are indicated as No, Nu and Cyto, respectively.
Sub-cellular localisation of EGFP, EGFP-nucleolin, EGFP-fibrillarin and EGFP-B23.1 and EGFP-N, EGFP-Rev, EGFP-ORF57, EGFP-NΔNoLS/ORF57 and EGFP-ORF57ΔNoLS/Rev in OMK cells as determined by live cell confocal microscopy. The width of the marker is 15 μm. The nucleolus, nucleus and cytoplasm are indicated as No, Nu and Cyto, respectively.
Sub-cellular localisation of EGFP, EGFP-nucleolin, EGFP-fibrillarin and EGFP-B23.1 and EGFP-N, EGFP-Rev, EGFP-ORF57, EGFP-NΔNoLS/ORF57 and EGFP-ORF57ΔNoLS/Rev in HeLa cells as determined by live cell confocal microscopy. The width of the marker is 15 μm. The nucleolus, nucleus and cytoplasm are indicated as No, Nu and Cyto, respectively.
References
- Andersen J.S., Lam Y.W., Leung A.K., Ong S.E., Lyon C.E., Lamond A.I., Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Angelier N., Tramier M., Louvet E., Coppey-Moisan M., Savino T.M., De Mey J.R., Hernandez-Verdun D. Tracking the interactions of rRNA processing proteins during nucleolar assembly in living cells. Mol. Biol. Cell. 2005;16:2862–2871. doi: 10.1091/mbc.E05-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D., Koppel D.E., Schlessinger J., Elson E., Webb W.W. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand L., Pearson A. The conserved N-terminal domain of herpes simplex virus 1 UL24 protein is sufficient to induce the spatial redistribution of nucleolin. J. Gen. Virol. 2008;89:1142–1151. doi: 10.1099/vir.0.83573-0. [DOI] [PubMed] [Google Scholar]
- Boyne J.R., Colgan K.J., Whitehouse A. Herpesvirus saimiri ORF57: a post-transcriptional regulatory protein. Front. Biosci. 2008;13:2928–2938. doi: 10.2741/2898. [DOI] [PubMed] [Google Scholar]
- Boyne J.R., Whitehouse A. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. USA. 2006;103:15190–15195. doi: 10.1073/pnas.0604890103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M. The contribution of nuclear compartmentalization to gene regulation. Cell. 2002;108:513–521. doi: 10.1016/s0092-8674(02)00650-5. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M., Mendes-Soares L., Campos I. To be or not to be in the nucleolus. Nature Cell Biol. 2000;2:E107–E112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- Cawood R., Harrison S.M., Dove B.K., Reed M.L., Hiscox J.A. Cell cycle dependent localisation of the coronavirus nucleocapsid protein. Cell Cycle. 2007;6:863–867. doi: 10.4161/cc.6.7.4032. [DOI] [PubMed] [Google Scholar]
- Chang J.H., Olson M.O. A single gene codes for two forms of rat nucleolar protein B23 mRNA. J. Biol. Chem. 1989;264:11732–11737. [PubMed] [Google Scholar]
- Chen D., Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell. Biol. 2001;153:169–176. doi: 10.1083/jcb.153.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Coote B., Attree S., Hiscox J.A. Evaluation of a nucleoprotein-based enzyme-linked immunosorbent assay for the detection of antibodies against infectious bronchitis virus. Avian Path. 2003;32:519–526. doi: 10.1080/0307945031000154125. [DOI] [PubMed] [Google Scholar]
- Chen H., Gill A., Dove B.K., Emmett S.R., Kemp F.C., Ritchie M.A., Dee M., Hiscox J.A. Mass spectroscopic characterisation of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding using surface plasmon resonance. J. Virol. 2005;79:1164–1179. doi: 10.1128/JVI.79.2.1164-1179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wurm T., Britton P., Brooks G., Hiscox J.A. Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J. Virol. 2002;76:5233–5250. doi: 10.1128/JVI.76.10.5233-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane A.W., Perkins A., Rosen C.A. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J. Virol. 1990;64:881–885. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M., Goodwin D.J., Hall K.T., Stevenson A.J., Meredith D.M., Markham A.F., Whitehouse A. The gene product encoded by ORF 57 of herpesvirus saimiri regulates the redistribution of the splicing factor SC-35. J. Gen. Virol. 1999;80:1311–1316. doi: 10.1099/0022-1317-80-5-1311. [DOI] [PubMed] [Google Scholar]
- Daelemans D., Costes S.V., Cho E.H., Erwin-Cohen R.A., Lockett S., Pavlakis G.N. In vivo HIV-1 Rev multimerization in the nucleolus and cytoplasm identified by fluorescence resonance energy transfer. J. Biol. Chem. 2004;279:50167–50175. doi: 10.1074/jbc.M407713200. [DOI] [PubMed] [Google Scholar]
- Dove B.K., You J.-H., Reed M.L., Emmett S.R., Brooks G., Hiscox J.A. Changes in nucleolar architecture and protein profile during coronavirus infection. Cellular Microbiol. 2006;8:1147–1157. doi: 10.1111/j.1462-5822.2006.00698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Leno G.H., Hammarskjold M.L., Rekosh D., Helga-Maria C., Olson M.O. The roles of nucleolar structure and function in the subcellular location of the HIV-1 Rev protein. J. Cell. Sci. 1995;108:2811–2823. doi: 10.1242/jcs.108.8.2811. [DOI] [PubMed] [Google Scholar]
- Dundr M., Misteli T., Olson M.O. The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 2000;150:433–446. doi: 10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin D.J., Hall K.T., Stevenson A.J., Markham A.F., Whitehouse A. The open reading frame 57 gene product of herpesvirus saimiri shuttles between the nucleus and cytoplasm and is involved in viral RNA nuclear export. J. Virol. 1999;73:10519–10524. doi: 10.1128/jvi.73.12.10519-10524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin D.J., Whitehouse A. A gamma-2 herpesvirus nucleocytoplasmic shuttle protein interacts with importin alpha 1 and alpha 5. J. Biol. Chem. 2001;276:19905–19912. doi: 10.1074/jbc.M009513200. [DOI] [PubMed] [Google Scholar]
- Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D. Nucleolus: from structure to dynamics. Histochem. Cell Biol. 2006;125:127–137. doi: 10.1007/s00418-005-0046-4. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D., Roussel P. Regulators of nucleolar functions. Prog. Cell Cycle Res. 2003;5:301–308. [PubMed] [Google Scholar]
- Hernandez-Verdun D., Roussel P., Gebrane-Younes J. Emerging concepts of nucleolar assembly. J. Cell. Sci. 2002;115:2265–2270. doi: 10.1242/jcs.115.11.2265. [DOI] [PubMed] [Google Scholar]
- Hiscox J.A. Brief review: the nucleolus — a gateway to viral infection? Arch. Virol. 2002;147:1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 2003;95:13–22. doi: 10.1016/S0168-1702(03)00160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A. RNA viruses: hijacking the dynamic nucleolus. Nature Rev. Microbiol. 2007;5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A., Wurm T., Wilson L., Cavanagh D., Britton P., Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2001;75:506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.H., Yung B.Y., Syu W.J., Lee Y.H. The nucleolar phosphoprotein B23 interacts with hepatitis delta antigens and modulates the hepatitis delta virus RNA replication. J. Biol. Chem. 2001;276:25166–25175. doi: 10.1074/jbc.M010087200. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Macfarlane S., Kalinina N.O., Rakitina D.V., Ryabov E.V., Gillespie T., Haupt S., Brown J.W., Taliansky M. Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc. Natl. Acad. Sci. USA. 2007;104:11115–11120. doi: 10.1073/pnas.0704632104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Ryabov E.V., Kalinina N.O., Rakitina D.V., Gillespie T., MacFarlane S., Haupt S., Brown J.W., Taliansky M. Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 2007;26:2169–2179. doi: 10.1038/sj.emboj.7601674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Ryabov E.V., Brown J.W., Taliansky M. Involvement of the nucleolus in plant virus systemic infection. Biochem. Soc. Trans. 2004;32:557–560. doi: 10.1042/BST0320557. [DOI] [PubMed] [Google Scholar]
- Kubota S., Siomi H., Satoh T., Endo S., Maki M., Hatanaka M. Functional similarity of HIV-I rev and HTLV-I rex proteins: identification of a new nucleolar-targeting signal in rev protein. Biochem. Biophys. Res. Commun. 1989;162:963–970. doi: 10.1016/0006-291x(89)90767-5. [DOI] [PubMed] [Google Scholar]
- Kuppuswamy M., Subramanian T., Srinivasan A., Chinnadurai G. Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nuc. Acid. Res. 1989;17:3551–3561. doi: 10.1093/nar/17.9.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam Y.W., Trinkle-Mulcahy L., Lamond A.I. The nucleolus. J. Cell Sci. 2005;118:1335–1337. doi: 10.1242/jcs.01736. [DOI] [PubMed] [Google Scholar]
- Lamond A.I., Sleeman J.E. Nuclear substructure and dynamics. Current Biol. 2003;13:R825. doi: 10.1016/j.cub.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Lee C., Hodgins D., Calvert J.G., Welch S.K., Jolie R., Yoo D. Mutations within the nuclear localization signal of the porcine reproductive and respiratory syndrome virus nucleocapsid protein attenuate virus replication. Virology. 2006;346:238–250. doi: 10.1016/j.virol.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.H., Chang S.C., Chen C.J., Chang M.F. The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J. Biol. Chem. 1998;273:7650–7656. doi: 10.1074/jbc.273.13.7650. [DOI] [PubMed] [Google Scholar]
- Leung A.K., Andersen J.S., Mann M., Lamond A.I. Bioinformatic analysis of the nucleolus. Biochem J. 2003;376:553–569. doi: 10.1042/BJ20031169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K., Lamond A.I. The dynamics of the nucleolus. Crit. Rev. Eukaryot. Gene Expr. 2003;13:39–54. doi: 10.1615/critreveukaryotgeneexpr.v13.i1.40. [DOI] [PubMed] [Google Scholar]
- Leung A.K., Trinkle-Mulcahy L., Lam Y.W., Andersen J.S., Mann M., Lamond A.I. NOPdb: Nucleolar Proteome Database. Nuc. Acid. Res. 2006;34:D218–D220. doi: 10.1093/nar/gkj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.P. Protein B23 is an important human factor for the nucleolar localization of the human immunodeficiency virus protein Tat. J. Virol. 1997;71:4098–4102. doi: 10.1128/jvi.71.5.4098-4102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvet E., Junera H.R., Le Panse S., Hernandez-Verdun D. Dynamics and compartmentation of the nucleolar processing machinery. Exp. Cell Res. 2005;304:457–470. doi: 10.1016/j.yexcr.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Lymberopoulos M.H., Pearson A. Involvement of UL24 in herpes-simplex-virus-1-induced dispersal of nucleolin. Virology. 2007;363:397–409. doi: 10.1016/j.virol.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Malik P., Clements J.B. Protein kinase CK2 phosphorylation regulates the interaction of Kaposi's sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K. Nuc. Acid. Res. 2004;32:5553–5569. doi: 10.1093/nar/gkh876. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Marasco W.A., Szilvay A.M., Kalland K.H., Helland D.G., Reyes H.M., Walter R.J. Spatial association of HIV-1 tat protein and the nucleolar transport protein B23 in stably transfected Jurkat T-cells. Arch. Virol. 1994;139:133–154. doi: 10.1007/BF01309460. [DOI] [PubMed] [Google Scholar]
- Matthews D.A., Olson M.O. What's new in the nucleolus? EMBO Reports. 2006;7:1–4. doi: 10.1038/sj.embor.7400786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melen K., Kinnunen L., Fagerlund R., Ikonen N., Twu K.Y., Krug R.M., Julkunen I. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J. Virol. 2007;81:5995–6006. doi: 10.1128/JVI.01714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michienzi A., Cagnon L., Bahner I., Rossi J.J. Ribozyme-mediated inhibition of HIV 1 suggests nucleolar trafficking of HIV-1 RNA. Proc. Natl. Acad. Sci. U S A. 2000;97:8955–8960. doi: 10.1073/pnas.97.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michienzi A., De Angelis F.G., Bozzoni I., Rossi J.J. A nucleolar localizing Rev binding element inhibits HIV replication. AIDS Res. Ther. 2006;3:13. doi: 10.1186/1742-6405-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michienzi A., Li S., Zaia J.A., Rossi J.J. A nucleolar TAR decoy inhibitor of HIV-1 replication. Proc. Natl. Acad. Sci. USA. 2002;99:14047–14052. doi: 10.1073/pnas.212229599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Protein dynamics: implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y., Nosaka T., Hatanaka M. The post-transcriptional regulator Rev of HIV: implications for its interaction with the nucleolar protein B23. Biochimie. 1996;78:1081–1086. doi: 10.1016/s0300-9084(97)86733-8. [DOI] [PubMed] [Google Scholar]
- Murayama R., Harada Y., Shibata T., Kuroda K., Hayakawa S., Shimizu K., Tanaka T. Influenza A virus non-structural protein 1 (NS1) interacts with cellular multifunctional protein nucleolin during infection. Biochem. Biophys. Res. Commun. 2007;362:880–885. doi: 10.1016/j.bbrc.2007.08.091. [DOI] [PubMed] [Google Scholar]
- Ozawa M., Fujii K., Muramoto Y., Yamada S., Yamayoshi S., Takada A., Goto H., Horimoto T., Kawaoka Y. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J. Virol. 2007;81:30–41. doi: 10.1128/JVI.01434-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y., Hodgins D.C., Lee C., Calvert J.G., Welch S.K., Jolie R., Keith M., Yoo D. Functional mapping of the porcine reproductive and respiratory syndrome virus capsid protein nuclear localization signal and its pathogenic association. Virus Res. 2008;135:107–114. doi: 10.1016/j.virusres.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Perkins A., Cochrane A.W., Ruben S.M., Rosen C.A. Structural and functional characterization of the human immunodeficiency virus rev protein. J. Acquir. Immune Defic. Syndr. 1989;2:256–263. [PubMed] [Google Scholar]
- Phair R.D., Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Pollard V.W., Malim M.H. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- Reed M.L., Dove B.K., Jackson R.M., Collins R., Brooks G., Hiscox J.A. Delineation and modelling of a nucleolar retention signal in the coronavirus nucleocapsid protein. Traffic. 2006;7:833–848. doi: 10.1111/j.1600-0854.2006.00424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed M.L., Howell G., Harrison S.M., Spencer K.A., Hiscox J.A. Characterization of the nuclear export signal in the coronavirus infectious bronchitis virus nucleocapsid protein. J. Virol. 2007;81:4298–4304. doi: 10.1128/JVI.02239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J.J., June C.H., Kohn D.B. Genetic therapies against HIV. Nat. Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland R.R., Kerwin R., Kuckleburg C., Sperlich A., Benfield D.A. The localisation of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 1999;64:1–12. doi: 10.1016/s0168-1702(99)00048-9. [DOI] [PubMed] [Google Scholar]
- Rowland R.R.R., Yoo D. Nucleolar-cytoplasmic shuttling of PRRSV nucleocapsid protein: a simple case of molecular mimicry or the complex regulation by nuclear import, nucleolar localization and nuclear export signal sequences. Virus Res. 2003;95:23–33. doi: 10.1016/S0168-1702(03)00161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi C.P., Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben S., Perkins A., Purcell R., Joung K., Sia R., Burghoff R., Haseltine W.A., Rosen C.A. Structural and functional characterization of human immunodeficiency virus tat protein. J Virol. 1989;63:1–8. doi: 10.1128/jvi.63.1.1-8.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheval E.V., Polzikov M.A., Olson M.O.J., Zatsepina O.V. A higher concentration of an antigen within the nucleolus may prevent its proper recognition by specific antibodies. Eur. J. Histochem. 2005;49:117–123. [PubMed] [Google Scholar]
- Spencer K.A., Dee M., Britton P., Hiscox J.A. Role of phosphorylation clusters in the biology of the coronavirus infectious bronchitis virus nucleocapsid protein. Virology. 2008;370:373–381. doi: 10.1016/j.virol.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer K.A., Hiscox J.A. Characterisation of the RNA binding properties of the coronavirus infectious bronchitis virus nucleocapsid protein amino-terminal region. FEBS Lett. 2006;580:5993–5998. doi: 10.1016/j.febslet.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber R.H., Afonina E., Gulnik S., Erickson J., Pavlakis G.N. Analysis of intracellular trafficking and interactions of cytoplasmic HIV-1 Rev mutants in living cells. Virology. 1998;251:38–48. doi: 10.1006/viro.1998.9295. [DOI] [PubMed] [Google Scholar]
- Stenoien D.L., Mielke M., Mancini M.A. Intranuclear ataxin1 inclusions contain both fast- and slow-exchanging components. Nat. Cell Biol. 2002;4:806–810. doi: 10.1038/ncb859. [DOI] [PubMed] [Google Scholar]
- Taliansky M.E., Robinson D.J. Molecular biology of umbraviruses: phantom warriors. J. Gen. Virol. 2003;84:1951–1960. doi: 10.1099/vir.0.19219-0. [DOI] [PubMed] [Google Scholar]
- Tijms M.A., van der Meer Y., Snijder E.J. Nuclear localization of non-structural protein 1 and nucleocapsid protein of equine arteritis virus. J. Gen. Virol. 2002;83:795–800. doi: 10.1099/0022-1317-83-4-795. [DOI] [PubMed] [Google Scholar]
- Weidman M.K., Sharma R., Raychaudhuri S., Kundu P., Tsai W., Dasgupta A. The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res. 2003;95:75–85. doi: 10.1016/s0168-1702(03)00164-3. [DOI] [PubMed] [Google Scholar]
- Williams B.J., Boyne J.R., Goodwin D.J., Roaden L., Hautbergue G.M., Wilson S.A., Whitehouse A. The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA through the cellular mRNA export pathway. Biochem. J. 2005;387:295–308. doi: 10.1042/BJ20041223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm T., Chen H., Britton P., Brooks G., Hiscox J.A. Localisation to the nucleolus is a common feature of coronavirus nucleoproteins and the protein may disrupt host cell division. J. Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo D., Wootton S.K., Li G., Song C., Rowland R.R. Colocalization and interaction of the porcine arterivirus nucleocapsid protein with the small nucleolar RNA-associated protein fibrillarin. J. Virol. 2003;77:12173–12183. doi: 10.1128/JVI.77.22.12173-12183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J.H., Dove B.K., Enjuanes L., DeDiego M.L., Alvarez E., Howell G., Heinen P., Zambon M., Hiscox J.A. Sub-cellular localisation of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Gen. Virol. 2005;86:3303–3310. doi: 10.1099/vir.0.81076-0. [DOI] [PubMed] [Google Scholar]
- You J.H., Howell G., Pattnaik A.K., Osorio F.A., Hiscox J.A. A model for the dynamic nuclear/nucleolar/cytoplasmic trafficking of the porcine reproductive and respiratory syndrome virus (PRRSV) nucleocapsid protein based on live cell imaging. Virology. 2008;378:34–47. doi: 10.1016/j.virol.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sub-cellular localisation of EGFP, EGFP-nucleolin, EGFP-fibrillarin and EGFP-B23.1 and EGFP-N, EGFP-Rev, EGFP-ORF57, EGFP-NΔNoLS/ORF57 and EGFP-ORF57ΔNoLS/Rev in DF-1 cells as determined by live cell confocal microscopy. The width of the marker is 15 μm. The nucleolus, nucleus and cytoplasm are indicated as No, Nu and Cyto, respectively.
Sub-cellular localisation of EGFP, EGFP-nucleolin, EGFP-fibrillarin and EGFP-B23.1 and EGFP-N, EGFP-Rev, EGFP-ORF57, EGFP-NΔNoLS/ORF57 and EGFP-ORF57ΔNoLS/Rev in OMK cells as determined by live cell confocal microscopy. The width of the marker is 15 μm. The nucleolus, nucleus and cytoplasm are indicated as No, Nu and Cyto, respectively.
Sub-cellular localisation of EGFP, EGFP-nucleolin, EGFP-fibrillarin and EGFP-B23.1 and EGFP-N, EGFP-Rev, EGFP-ORF57, EGFP-NΔNoLS/ORF57 and EGFP-ORF57ΔNoLS/Rev in HeLa cells as determined by live cell confocal microscopy. The width of the marker is 15 μm. The nucleolus, nucleus and cytoplasm are indicated as No, Nu and Cyto, respectively.