Abstract
Eight accessory proteins have been identified in severe acute respiratory syndrome-associated coronavirus (SARS-CoV). They are believed to play roles in the viral life cycle and may contribute to the pathogenesis and virulence. ORF9b as one of these accessory proteins is located in subgenomic mRNA9 and encodes a 98 amino acid protein. However, whether 9b protein is a structural component of SARS-CoV particles remains unknown. In this study, we demonstrate that 9b protein is translated from bicistronic mRNA9 via leaky ribosome scanning and it is incorporated into both virus-like particles (VLPs) and purified SARS-CoV virions. Further analysis shows that sufficient incorporation of 9b protein into VLPs is dependent upon the co-expression of E and M proteins, but not upon the presence of either S or N protein. Our data indicate that 9b protein of SARS-CoV is another virion-associated accessory protein. This finding will lead to a better understanding of the properties of the SARS-CoV 9b protein.
Keywords: SARS-CoV, ORF9b, Virion-associated protein
Introduction
Severe acute respiratory syndrome (SARS) which broke out in 2002–2003 has caused worldwide panic due to its high mortality in humans (Peiris et al., 2004). In 2003, SARS-associated coronavirus (SARS-CoV) was identified as the aetiological agent for this disease (Drosten et al., 2003, Ksiazek et al., 2003, Peiris et al., 2003). The genome of SARS-CoV is a plus-stranded single RNA which is 29,727 nucleotides in length excluding the polyadenylation tract at the 3′ end (Rota et al., 2003). Fourteen ORFs have been identified, which translate into two replicative polyproteins (pp1a and pp1ab), 4 structural proteins (S, E, M, N) and 8 accessory proteins (3a, 3b, 6, 7a, 7b, 8a, 8b, 9b) (Marra et al., 2003, Rota et al., 2003). The accessory proteins are unique to SARS-CoV, as they have little homology in amino acid sequence with accessory proteins of other coronaviruses. All current studies concerning accessory proteins of coronaviruses including SARS-CoV suggest that they are not essential for virus replication (de Haan et al., 2002, Yount et al., 2005), but do affect virus release, stability, pathogenesis, and finally contribute to the virulence (Weiss and Navas-Martin, 2005). Understanding the properties and functions of SARS-CoV specific accessory proteins may help to explain the differences in pathogenicity between SARS-CoV and other known coronaviruses.
Among the 8 accessory proteins of SARS-CoV, 3a protein was first found to be a structural component of SARS-CoV virions as it was incorporated into purified virions and virus-like particles (Ito et al., 2005, Shen et al., 2005). Our previous study also shows that 3a protein functions as an ion channel to facilitate virus release (Lu et al., 2006). In addition, ORF7a protein was also characterized as a virion-associated protein of SARS-CoV (Huang et al., 2006a), which induces apoptosis ((Kopecky-Bromberg et al., 2006, Tan et al., 2004) and arrests the cell cycle when over-expressed (Yuan et al., 2006). Subsequently, both SARS-CoV ORF6 and ORF7b proteins were determined to be incorporated into virus particles(Huang et al., 2007, Schaecher et al., 2007). ORF6 protein was further reported to function as an IFN antagonist and to accelerate replication of a related mouse virus (Kopecky-Bromberg et al., 2007, Tangudu et al., 2007). Four other accessory proteins have not been well investigated, and some of them may be included in virions and have important functions (Tan et al., 2006).
There are nine subgenomic mRNAs in SARS-CoV, four of which (mRNA3, 7, 8 and 9) are bicistronic producing two ORFs initiating at the first or additional downstream start codon (Thiel et al., 2003, Weiss and Navas-Martin, 2005). On bicistronic mRNA9, a 98 amino acid viral accessory protein, 9b, is encoded from a complete internal ORF within the N gene. The expression of ORF9b has been detected in infected cells and in clinical specimens (Chan et al., 2005). Anti-9b antibodies have also been found in the serum of SARS patients (Qiu et al., 2005). The crystal structure additionally indicates that 9b protein is an unusual membrane binding protein with a long hydrophobic lipid-binding tunnel. It was therefore proposed to be associated with intracellular vesicles and may have function in SARS-CoV assembly (Meier et al., 2006). However, further analysis of 9b protein is necessary for understanding its contribution in the viral life cycle.
In this study, we have examined the expression mechanism of 9b protein from mRNA9. We find that 9b protein is translated via a leaky ribosomal scanning mechanism. Furthermore, we provide the first evidence that 9b protein is present both in virus-like particles and in purified virions, and that the efficient incorporation of 9b protein is dependent upon the co-expression of E and M proteins. These data suggest that 9b protein is not only a viral accessory protein but also a structural component of SARS-CoV virions.
Results
Translation of ORF9b via leaky ribosomal scanning
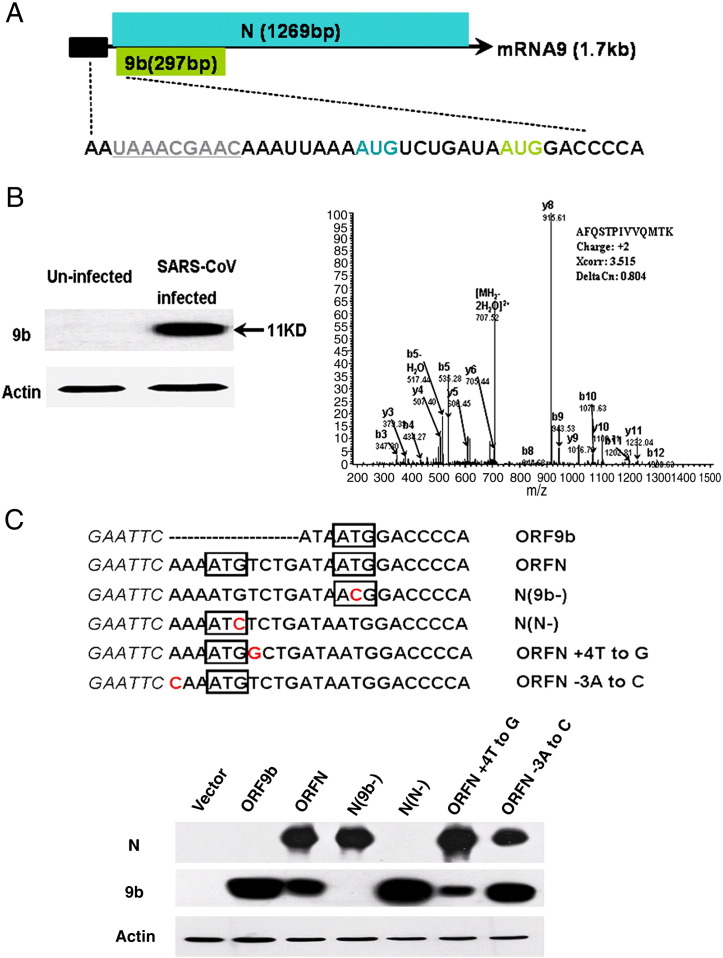
Our previous study using a proteomics approach has identified ORF3a as a viral protein of SARS-CoV (Zeng et al., 2004). Meanwhile, several peptides belonging to ORF9b protein had also been detected in the cytosol of SARS-CoV infected Vero E6 cells (Data not shown). To further investigate the properties of SARS-CoV 9b protein, purified recombinant 9b protein was used to produce both anti-9b polyclonal and monoclonal antibodies. The expression of 9b protein in SARS-CoV infected cells was confirmed by Western blot analysis with anti-9b monoclonal antibody. An 11 kDa protein (9b protein) was recognized in infected but not in uninfected cells (Fig. 1 B left). The specificity was further ensured by immunoprecipitation with anti-9b polyclonal antibody. The complex precipitated was resolved under gel electrophoresis and the corresponding band was submitted to mass spectrometry. A peptide: “AFQSTPIVVQMTK” representing amino acids 69–81 of the SARS-CoV 9b protein was identified (Fig. 1B right).
Fig. 1.
Expression of 9b protein from mRNA9. (A) SARS-CoV mRNA9 organization. ORFN, ORF9b, frameshift and transcription-regulating sequence (TRS), are shown. Black box represents RNA leading sequence. ORFN is highlighted in blue, while ORF9b is shown in an enlarged representation highlighted in green. TRS sequence is marked by underlines and in grey. Start codon of ORFN and 9b are shown in corresponding colors, adapted from (Meier et al., 2006, Thiel et al., 2003). (B) Left. 11 kDa 9b protein was detected in SARS infected FRhK-4 cells by anti-9b monoclonal antibody, but not in uninfected cells. β-actin was detected in both infected and uninfected cells. Right. 9b protein was immunoprecipitated by anti-9b polyclonal antibody from SARS infected FRhK-4 cell lysates in RIPA buffer, the mass spectrometry analysis of the corresponding gel slices detected a specific peptide that represents 9b protein. (C) Translation of ORF9b by leaky ribosomal scanning. Schematic diagram of cDNA constructs is shown (upper panel). Wild-type ORF9b sequence (ORF9b), wild-type N gene sequence (ORFN), a point mutation eliminating 9b initiation codon in ORFN sequence (N(9b−)), a point mutation eliminating N initiation codon (N(N−)), an optimal Kozak context around the ORFN initiation codon (ORFN + 4T to G), and a weaker Kozak context around the ORFN initiation codon (ORFN − 3A to C). 293T cells were transfected with plasmids encoding the indicated cDNAs and analyzed by Western blot with both anti-N and anti-9b monoclonal antibodies for each sample. β-actin was detected by actin specific polyclonal antibody.
As none of the previous studies has illustrated the expression mechanism of 9b protein, we then examined whether 9b protein was able to be translated from subgenomic mRNA9 alone and how it was translated. The 9b protein encoded by an internal ORF starts at the 18th nucleotide downstream of the transcription regulatory sequence of mRNA9 (Thiel et al., 2003). The start codon for ORF9b is very close to the start codon for ORFN, with only 10 nucleotides in between (Fig. 1A). To confirm that 9b protein can be translated from an mRNA corresponding to the SARS-CoV subgenomic RNA9, the sequence encoding ORFN which contains ORF9b was cloned into the eukaryotic expression vector pCAGGS and transfected into 293T cells. The results showed that 9b protein was able to be expressed in cells transfected with plasmids containing the N gene coding region (Fig. 1C ORFN), but that the expression level was relatively weaker compared with that of cells transfected with ORF9b alone (Fig. 1C ORF9b). When the ORF9b initiation codon was mutated from ATG to ACG (Fig. 1C N(9b−)), 9b protein was not expressed while the expression level of N protein showed little change. When the ORFN initiation codon was mutated from ATG to ATC (Fig. 1C N(N−)), N protein was not expressed while the expression level of 9b protein dramatically increased. These data not only demonstrated the expression of 9b protein from the N gene coding region, but also led us to further investigate the translation regulatory mechanism for 9b protein.
The downstream ORF of multicistronic mRNA is mainly translated by leaky scanning ribosomes or internal ribosome entry (Kozak, 1989, Thiel and Siddell, 1994, van Vliet et al., 2002). Analysis of the sequence flanking the initiation codon of ORFN and ORF9b showed that the initiation of the N gene represented a suboptimal Kozak context (A at − 3, T at + 4), while the initiation of the 9b gene indicated an optimal Kozak context (A at − 3, G at + 4) (Kozak, 1986). In addition, a lower expression level of 9b protein had been observed already in cells transfected with ORFN and then in cells transfected with ORF9b or N(N−). All of these findings make it reasonable to presume that the translation initiation of ORF9b is regulated by a leaky ribosomal scanning mechanism under which the downstream gene expression is affected by the translation efficiency of the upstream gene (Kozak, 1987, Kozak, 1989). To test this, a more ideal Kozak context was induced in ORFN (Fig. 1C ORFN + 4T to G). We detected an obviously decreased expression level of 9b protein as expected. Meanwhile, when the Kozak context of ORFN was changed to a much weaker one (Fig. 1C ORFN − 3A to C), an increased expression of 9b protein was observed. Together, these data suggest that ORF9b protein is translated via a leaky ribosomal scanning mechanism.
ORF9b protein is present in SARS-CoV VLPs
In order to examine whether 9b protein is incorporated into virus particles, we established a SARS-CoV VLP system in vitro to study the presence of 9b protein in virus-like particles.
To ensure that the release of 9b protein is associated with virus particles, we first examined whether 9b protein was able to be released from cells expressing 9b protein alone. Cells were transfected with pCAGGS-9b plasmid. Parental pCAGGS-3a or vector alone was also transfected as control. Western blot analysis of the cell lysates clearly showed that both 3a and 9b proteins were expressed (Fig. 2 ). The clarified culture supernatants from 9b and 3a-expressing cells and control cells were applied to the top of a 20% sucrose cushion for purification. Western blot analysis of the pellets demonstrated that 9b protein was not released into the culture medium of 9b-expressing cells, while 3a protein was released as previously described (Huang et al., 2006b). The abundant host protein, actin, was not detectable in the supernatants from any group of cells (Fig. 2).
Fig. 2.
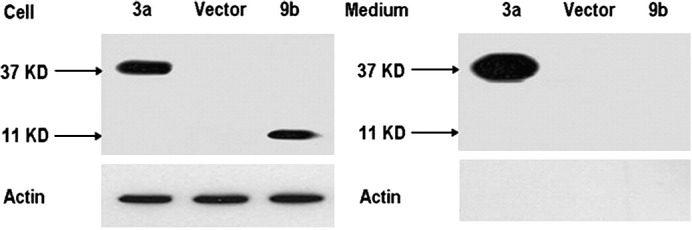
ORF9b protein is not released from 9b-expressing cells. 293T cells were transfected with pCAGGS-3a-HA (3a), pCAGGS (vector), or pCAGGS-9b (9b) independently. At 48 h post transfection, supernatants were clarified, applied to a 20% sucrose cushion, and centrifuged at 100,000 ×g for 3 h at 4 °C. The pellets were suspended in 1×SDS-PAGE loading buffer (Medium). Cell lysates were prepared with 1×SDS-PAGE loading buffer (Cell). Samples were subjected to Western blot analysis with anti-HA antibody to detect 3a protein (37 kDa), anti-9b antibody (11 kDa) to detect 9b protein and anti-actin antibody for internal control.
We then transfected pCAGGS-S, pCAGGS-E, pCAGGS-M, pCAGGS-N(9b−) and pCAGGS-9b simultaneously into 293T cells. At 48 h post transfection, the culture medium was collected and clarified. The VLPs purified with 20% sucrose were then added to the top of a 20–60% continuous sucrose gradient for fractionation. Twelve fractions were collected, and each was examined by Western blot analysis. The results showed that 9b protein was released from the VLP system, and exhibited in fractions 5 to 9 along with S, N, and M proteins (Fig. 3 ). The densities of these fractions were approximately between 1.12 g/ml and 1.20 g/ml. The greatest amount of 9b protein was detected in fraction 7 (density, 1.16 g/ml), which also contained the highest level of S, N and M proteins. The data suggest that 9b protein is incorporated into virus-like particles containing S, N, M and E proteins. E protein was not detectable in Western blot due to its low abundance in SARS-CoV VLPs (Huang et al., 2006a).
Fig. 3.
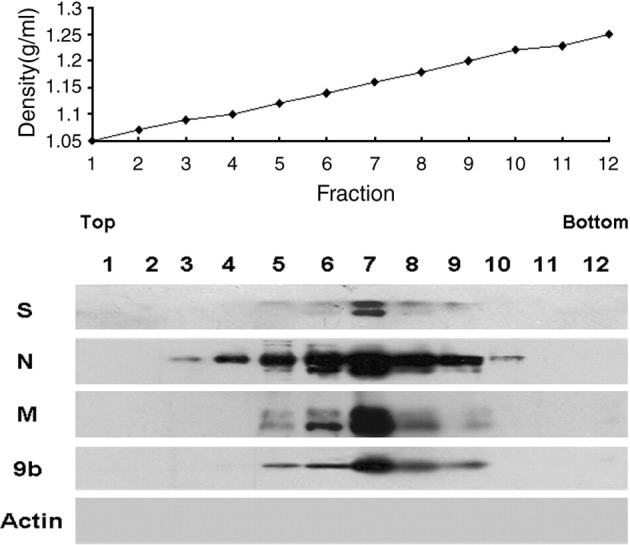
Presentation of 9b protein in SARS-CoV VLPs. Subconfluent 293T cells were transfected with 3 μg of pCAGGS-S, 3 μg of pCAGGS-N(9b−), 3 μg of pCAGGS-M, 3 μg of pCAGGS-E and 3 μg of pCAGGS-9b each. At 48 h post transfection, culture medium was harvested and clarified. The released VLPs were first pelleted by centrifugation through a 20% sucrose cushion, and then further purified over a 20–60% sucrose gradient. Twelve fractions were collected from top to bottom; each fraction concentrated by 20% sucrose ultracentrifugation was analyzed using Western blot analysis with anti-S monoclonal antibody (S), anti-N monoclonal antibody (N), anti-HA tag monoclonal antibody (M), and anti-9b monoclonal antibody (9b). β-actin was detected by actin specific polyclonal antibody (Actin). The density of each fraction was measured and is shown.
ORF9b protein is incorporated into purified SARS-CoV virions
To further confirm the potential association of 9b protein with SARS-CoV virions, purified SARS-CoV was prepared from the supernatant of virus-infected FRhK-4 cells. The medium was collected at 36 h post infection, at which time the cells remained intact and CPE had not appeared. Clarified supernatant was then subjected to sucrose gradient ultracentrifugation as described above and twelve fractions from top to bottom were collected and analyzed. The results showed that S, N, M and 9b proteins were present in fractions 7 (density, 1.16 g/ml) to 10 (density, 1.20 g/ml) at the same time (Fig. 4 ). The peak of the 9b protein was detected in fraction 8 along with the highest level of S, N and M proteins. The density of fraction 8 was approximately 1.18 g/ml, consistent with the density of SARS-CoV particles (Huang et al., 2006a, Ito et al., 2005, Schaecher et al., 2007). These data further suggest that 9b protein is incorporated into SARS-CoV virions.
Fig. 4.
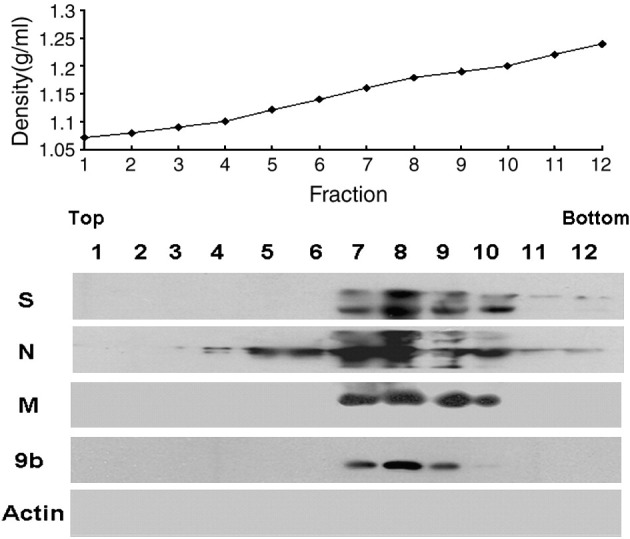
Incorporation of 9b protein into purified SARS-CoV virions. Clarified supernatant from SARS infected cells was subjected to 20% sucrose ultracentrifugation. The pellets resuspended in NTE buffer were further applied on a 20–60% sucrose gradient cushion. Twelve fractions were collected from top to bottom after ultracentrifugation, each fraction was condensed using 20% sucrose and subjected to Western blot analysis with anti-S monoclonal antibody (S), anti-N monoclonal antibody (N), anti-M polyclonal antibody (M) and anti-9b monoclonal antibody (9b). β-actin was detected by actin specific polyclonal antibody (Actin). The density of each fraction was measured and is shown.
Incorporation of 9b protein into VLPs is influenced by E and M proteins
As 9b protein is present in virions, it is advantageous to characterize the role of other SARS-CoV structural proteins in incorporation of 9b protein into VLPs. Cultures of 293T cells were transfected with the indicated plasmids, and pCAGGS vector was added to adjust the total amount of DNA to equivalent levels. As shown in Fig. 5 , in the pellets of the culture medium collected from cells co-transfected with pCAGGS-E and pCAGGS-9b, a low level of 9b protein was detected suggesting that 9b protein is able to be released when only E protein is available. A marked increase of 9b protein in culture medium was observed when M protein was co-expressed with 9b and E proteins. The amount of 9b protein in VLPs did not change significantly when S or N, or both of them were added. The results indicate that sufficient incorporation of 9b protein into SARS-CoV VLPs is dependent upon co-expression of E and M proteins but is not influenced by either S or N protein.
Fig. 5.
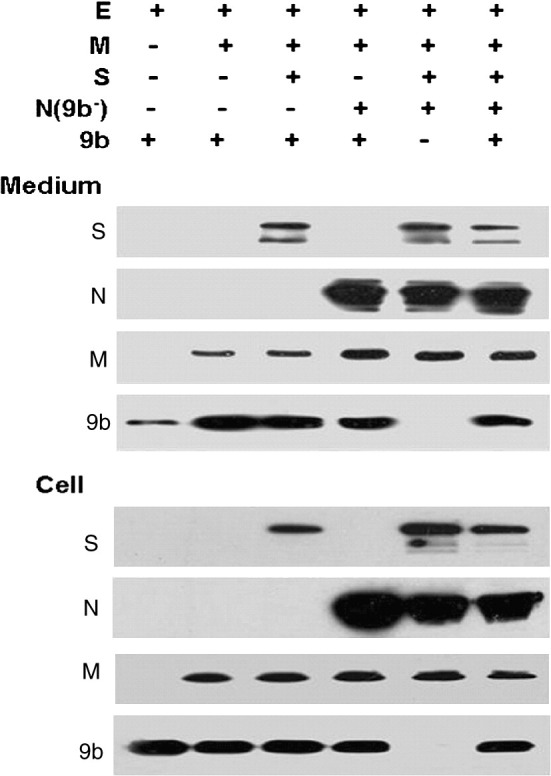
Release of SARS-CoV 9b protein in the presence of other viral structural proteins. 9b protein was co-expressed with E protein, M protein, S protein, and N protein in different combinations in 293T cells, as indicated. The quantity of each plasmid for this analysis was same, and total DNA levels were adjusted by adding pCAGGS. At 48 h post transfection, medium was harvested and pelleted through a 20% sucrose cushion. Medium (Medium) and cell lysates (Cell) were analyzed for S protein (S), N protein (N), M protein (M), and 9b protein (9b) by Western blotting. S, N, and 9b proteins were detected using indicated monoclonal antibodies, while M protein with HA tag was detected using anti-HA antibody.
The effect of 9b protein on VLP production was further analyzed. VLPs containing S, E, M and N proteins in 9b-expressing cells and non-expressing cells were examined and compared. Unfortunately, no obvious differences in the assembly of these proteins have been detected (Fig. 5).
Discussion
As a newly emerging coronavirus in the human, SARS-CoV has led to acute inflammation and a lethal syndrome in patients. Phylogenetic analysis showed that SARS-CoV had been classified as group 2b CoV distantly related to known group 2 CoV (Gorbalenya et al., 2004, Snijder et al., 2003). The functions of viral replicase and four basic structural proteins (S, E, M, and N) of SARS-CoV are similar to those of other coronaviruses. However, the accessory proteins of SARS-CoV show little homology to those of known coronaviruses. The specific properties of these accessory proteins may contribute to the differences in pathogenicity between SARS-CoV and other coronaviruses. In this report, we demonstrate that SARS-CoV 9b protein is another viral accessory protein incorporated into virus particles. Our finding makes 9b protein the fifth accessory protein of SARS-CoV to be found present in virions.
For efficient utilization of their limited genome, viruses frequently use bicistronic RNAs to produce alternative open reading frames. SARS-CoV ORF1b, ORF3b, ORF7b, ORF8b, and ORF9b are all transcribed via alternative open reading frames. In addition to ORF7b protein (Schaecher et al., 2007), ORF9b protein is another accessory protein that is proven to be translated via leaky ribosomal scanning. Other virus accessory proteins encoded by multicistronic mRNAs are translated either by leaky ribosomal scanning or by internal entry of the ribosome (Senanayake and Brian, 1997, Thiel and Siddell, 1994). The mechanisms must be identified individually.
After the first SARS-CoV accessory protein, 3a, had been identified as a structural component of virions, accessory proteins 7a, 7b, and 6 were subsequently determined to be incorporated into virions (Huang et al., 2006a, Huang et al., 2007, Ito et al., 2005, Schaecher et al., 2007, Shen et al., 2005). Here, our data further indicate that 9b protein is the fifth virion-associated accessory protein of SARS-CoV to be recognized. Analysis of VLPs and purified SARS-CoV particles showed that the highest level of 9b protein was detected in the fractions also abundant in S, N, and M proteins. While this property is almost equivalent among these accessory structural proteins, they do exhibited different biochemical characteristics. Some, such as 3a and 6 proteins, are released from expressing cells, while others, like 7a and 9b proteins, are not detectable in the supernatant of expressing cells. As a result, the release of the protein itself is seen not to be a determinant of incorporation. The localization of these proteins to intracellular vesicle components like Golgi and ER may help them to be incorporated through mechanisms bringing them close to the site of virus assembly. The mechanism of incorporating accessory proteins into virus particles is still not well understood and must be further studied. Furthermore, it will be important to address the necessity of incorporation of all these accessory proteins into virions.
Release of 9b protein was detected from cells co-expressing E and 9b proteins, but not from cells expressing 9b protein alone. This result was similar to what was found with 7a protein; the release of these two accessory proteins with E protein may be caused by their incorporation into putative E protein-containing vesicles (Maeda et al., 1999, Huang et al., 2006a). When M protein but not S or N protein was added, the release of 9b into the medium was increased markedly, indicating that E and M proteins are sufficient to allow incorporation of 9b protein into virus-like particles. This is consistent with former studies showing that E and M proteins could form smooth virus-like particles when co-expressed, while S and N were unnecessary for VLP formation (Hsieh et al., 2005, Mortola and Roy, 2004). As we do not have evidence for interactions between 9b and E or M proteins, we cannot determine whether the incorporation of 9b protein into VLPs generated by E and M co-expressing cells is caused by the physical associations among them. However, this mechanism requires further examination.
ORF9b on SARS-CoV bicistronic mRNA9 is a fully internal ORF in the N gene coding region. There are also coordinate proteins similar to SARS-CoV 9b which are so-called “Internal” or “I” proteins in other group II coronaviruses (Fischer et al., 1997, Lapps et al., 1987, Senanayake and Brian, 1997). In MHV, I protein is an accessory viral structural protein which can contribute to plaque morphology (Fischer et al., 1997). In our study, I protein of SARS-CoV (9b protein) was also shown to be a structural component of the virions. It is reasonable to propose that 9b protein of SARS-CoV may also play a role in the viral life cycle. However, a more thorough functional analysis of 9b protein still must be performed. Although a recent study with SARS-CoV accessory protein deletion mutants showed little effect on pathogenicity in a mouse model (Dediego et al., 2008), these accessory proteins, including 9b protein, may still play a supportive role in the viral life cycle in patients.
Materials and methods
Cell culture, transfection, and virus infection
293T, Vero E6 and FRhK-4 cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (Gibco) at 37 °C in a CO2 incubator. Lipofectamine (Invitrogen) was applied for transient transfection following the manufacturer's procedure. For SARS-CoV infection, FRhK-4 cells were inoculated with virus (GZ 50 Strain) as previously described (Zhong et al., 2003) at MOI of 5 for 1 h in medium without FBS. After 1 h, the cells were washed with medium and cultured with complete medium for the required time. All procedures with SARS-CoV infection were performed in a biosafety level-3 laboratory.
Plasmids
Viral RNA was extracted from filtered supernatant of virus-infected FRhK-4 cells using the RNesay Mini Kit (Qiagen) following the manufacturer's protocol. The cDNA was prepared by reverse transcription using M-MLV reverse transcriptase (Invitrogen) with oligonucleotide primers. S, E, M, N, 3a and 9b genes were amplified by PCR with specific primer using Pyrobest polymerase (Takara). S, N and 9b genes were cloned directly into pCAGGS vector under a powerful chicken β-actin promoter (kindly provided by Dr. Jun-ichi Miyazaki, Osaka University). The deletion of ORFN (N(N−)) was obtained by mutating the initiation codon of N gene from ATG to ATC, while the deletion of ORF9b (N(9b−)) in the N gene coding region was achieved by mutating the initiation codon of ORF9b from ATG to ACG. For the mutation of the KOZAK context, primers containing the mutated nucleotides (− 3A to C, + 4T to G) were synthesized to amplify the expected ORFs and then cloned into pCAGGS vector. E, M and 3a genes were first cloned into pcDNA-3′HA (Invitrogen), and then the genes together with HA tag at the 3′ end were amplified by PCR and subcloned into the pCAGGS vector. All plasmids were confirmed by sequence analysis.
Antibodies
The monoclonal and polyclonal antibodies against 9b protein were both produced by the Antibody Research Centre, Shanghai Institute of Biological Science. Mouse anti-S and mouse anti-N antibodies were also produced by the Antibody Research Centre, Shanghai Institute of Biological Science. Rabbit anti-M antibody (AP6008b) was purchased from Abgent. Mouse anti-HA monoclonal antibody was obtained from Covance and rabbit anti-actin from Sigma. HRP-conjugated anti-mouse IgG was purchased from Sigma and HRP-conjugated anti-rabbit IgG from Southern Biotech.
Mass spectrometry
Samples were treated as previously described (Zeng et al., 2004). Briefly, the infected cell lysates, which had been immunoprecipitated using 9b polyclonal antibody, were subjected to SDS-PAGE and the corresponding gel slices were excised and digested with trypsin. The digested peptides were analyzed by mass spectrometry (LCQ Deca XP Plus, Thermo Finnigan). Data analysis was carried out using a shot-gun approach.
Ultracentrifugation
For SARS-CoV purification, the supernatant containing the virus was inactivated with 1:2000 formaldehyde at 4 °C for 72 h as previously described (Qu et al., 2005). For the SARS-CoV virus-like particle system, the medium was collected 48 h post co-transfection for S, E, M, N and 9b. The inactivated virus and the medium from the VLP system were first purified by centrifugation at 1000 ×g for 15 min at 4 °C and then by ultracentrifugation over a 20% sucrose cushion at 100,000 ×g for 3 h in an SW41 ultracentrifuge rotor (Beckman). The pellets were resuspended in NTE buffer (100 mM NaCl, 10 mM Tris–HCl [pH7.5], 1 mM EDTA) and loaded onto a 20–60% continuous sucrose gradient for fractionation at 100,000 ×g over 12 h at 4 °C in a SW41 rotor (Hsieh et al., 2005, Mortola and Roy, 2004). Twelve fractions from top to bottom were collected and concentrated separately by ultracentrifugation at 100,000 ×g for 3 h. Each pellet was then dissolved in 1×SDS-PAGE loading buffer and analyzed by Western blot assay.
To determine whether 9b protein is released from cells expressing 9b protein, supernatants from cells transfected with ORF3a, ORF9b or empty vector were clarified by centrifugation at 1000 ×g for 15 min at 4 °C. This was followed by ultracentrifugation over 20% sucrose at 100,000 ×g for 3 h in a SW41 rotor. The pellet was dissolved in 1×SDS-PAGE loading buffer and submitted to Western blot analysis.
Immunoprecipitation and Western blot analysis
FRhK-4 cells were infected with SARS-CoV at 24 h post infection and the cell lysates were collected as described previously (Lu et al., 2006). Briefly, infected cells were lysed with solution containing 40 mM Tris (pH 8.3) and 0.5% NP-40 at 22 °C for 5 min. The supernatant was collected and subjected to immunoprecipitation and Western blot assay. Clarified supernatant was then incubated with anti-9b polyclonal antibody together with 5% BSA and protein A/G beads at 4 °C overnight. After immunoprecipitation, beads were washed 5 times with RIPA buffer; the complex was eluted and submitted to mass spectrometry analysis.
Expression of the SARS-CoV proteins in 293T cells was studied 24 h after transient transfection. The cells were lysed in SDS loading buffer and subjected to SDS-PAGE electrophoresis. Proteins were transferred to a nitrocellulose membrane and detected using corresponding antibodies.
Acknowledgments
We thank Prof. Vincent Deubel, Prof. Paul Zhou, Prof. Jing Zhong, Prof. Ke Lan, (Institute Pasteur of Shanghai, CAS, China), Prof. Shi-Shan Yuan (Shanghai Institute of Animal Parasitology, Chinese Academy of Agriculture Science), Prof. Xue-Liang Zhu (Shanghai Institute of Biological Science), Dr. Sheri Skinner (USA), Prof. Hao Shen (USA) and Prof. Hans Klenk (Marburg University, Germany) for reviewing the manuscript and for constructive suggestions. We thank Mr. Yi-Min Wang (Freiburg University, Germany) for helpful comments on this study. This work was supported by grants from the National Natural Science Foundation of China (30325018, 30530700, 30623003 and 30421005) and CAS project (KSCX1-YW-R-43), grants from the Technology Commission of Shanghai Municipality (07DZ22916, 064319034, 04DZ14902, 04DZ19108, 06DZ22032 and 04DZ19112), a grant from National 863 key project (2006AA02A27), a grant from the Sino-Germany center on SARS project (GZ238(202/11)), a grant from E-institutes of the Shanghai Universities Immunology Division and a grant from Li Kha Shing Foundation.
Contributor Information
Jia-Rui Wu, Email: wujr@sibs.ac.cn.
Bing Sun, Email: bsun@sibs.ac.cn.
References
- Chan W., Wu C., Chow S., Cheung T., To K., Leung W., Chan P., Lee K., Ng H., Au D., Lo A. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS) Mod. Pathol. 2005;18:1432–1439. doi: 10.1038/modpathol.3800439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dediego M., Pewe L., Alvarez E., Rejas M., Perlman S., Enjuanes L. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376:379–389. doi: 10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C., Masters P., Shen X., Weiss S., Rottier P. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R., Berger A., Burguiere A., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H., Osterhaus A., Schmitz H., Doerr H. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Fischer F., Peng D., Hingley S., Weiss S., Masters P. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J. Virol. 1997;71:996–1003. doi: 10.1128/jvi.71.2.996-1003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A., Snijder E., Spaan W. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 2004;78(15):7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P., Chang S., Huang C., Lee T., Hsiao C., Kou Y., Chen I., Chang C., Huang T., Chang M. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Ito N., Tseng C., Makino S. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 2006;80:7287–7294. doi: 10.1128/JVI.00414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Narayanan K., Ito N., Peters C., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is released in membranous structures from 3a protein-expressing cells and infected cells. J. Virol. 2006;80:210–217. doi: 10.1128/JVI.80.1.210-217.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Peters C., Makino S. Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells. J. Virol. 2007;81:5423–5426. doi: 10.1128/JVI.02307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N., Mossel E., Narayanan K., Popov V., Huang C., Inoue T., Peters C., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 2005;79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S., Martinez-Sobrido L., Palese P. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J. Virol. 2006;80:785–793. doi: 10.1128/JVI.80.2.785-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S., Martínez-Sobrido L., Frieman M., Baric R., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81(2):548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J. Cell. Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T., Erdman D., Goldsmith C., Zaki S., Peret T., Emery S., Tong S., Urbani C., Comer J., Lim W., Rollin P., Dowell S., Ling A., Humphrey C., Shieh W., Guarner J., Paddock C., Rota P., Fields B., DeRisi J., Yang J., Cox N., Hughes J., LeDuc J., Bellini W., Anderson L., Group S.W. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lapps W., Hogue B., Brian D. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology. 1987;157:47–57. doi: 10.1016/0042-6822(87)90312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Zheng B., Xu K., Schwarz W., Du L., Wong C., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, J., Maeda, A., and Makino, S. (1999). Release of coronavirus E protein in membrane vesicles from virus-infected cells and E protein-expressing cells Virology 263(2), 265–272. [DOI] [PMC free article] [PubMed]
- Marra M., Jones S., Astell C., Holt R., Brooks-Wilson A., Butterfield Y., Khattra J., Asano J., Barber S., Chan S., Cloutier A., Coughlin S., Freeman D., Girn N., Griffith O., Leach S., Mayo M., McDonald H., Montgomery S., Pandoh P., Petrescu A., Robertson A., Schein J., Siddiqui A., Smailus D., Stott J., Yang G., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G., Tyler S., Vogrig R., Ward D., Watson B., Brunham R., Krajden M., Petric M., Skowronski D., Upton C., Roper R. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Meier C., Aricescu A., Assenberg R., Aplin R., Gilbert R., Grimes J., Stuart D. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14:1157–1165. doi: 10.1016/j.str.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortola E., Roy P. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 2004;576:174–178. doi: 10.1016/j.febslet.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J., Lai S., Poon L., Guan Y., Yam L., Lim W., Nicholls J., Yee W., Yan W., Cheung M., Cheung V., Chan K., Tsang D., Yung R., Nq T., Yuen K., Group S.S. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J., Guan Y., Yuen K. Severe acute respiratory syndrome. Nature Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M., Shi Y., Guo Z., Chen Z., He R., Chen R., Zhou D., Dai E., Wang X., Si B., Song Y., Li J., Yang L., Wang J., Wang H., Pang X., Zhai J., Du Z., Liu Y., Zhang Y., Li L., Wang J., Sun B., Yang R. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect. 2005;7:882–889. doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu D., Zheng B., Yao X., Guan Y., Yuan Z., Zhong N., Lu L., Xie J., Wen Y. Intranasal immunization with inactivated SARS-CoV (SARS-associated coronavirus) induced local and serum antibodies in mice. Vaccine. 2005;23:924–931. doi: 10.1016/j.vaccine.2004.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P., Oberste M., Monroe S., Nix W., Campagnoli R., Icenogle J., Penaranda S., Bankamp B., Maher K., Chen M., Tong S., Tamin A., Lowe L., Frace M., DeRisi J., Chen Q., Wang D., Erdman D., Peret T., Burns C., Ksiazek T., Rollin P., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A., Drosten C., Pallansch M., Anderson L., Bellini W. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Schaecher S., Mackenzie J., Pekosz A. The ORF7b protein of SARS-CoV is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 2007;81:718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake S., Brian D. Bovine coronavirus I protein synthesis follows ribosomal scanning on the bicistronic N mRNA. Virus Res. 1997;48:101–105. doi: 10.1016/S0168-1702(96)01423-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Lin P., Chao Y., Zhang A., Yang X., Lim S., Hong W., Tan Y. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 2005;330:286–292. doi: 10.1016/j.bbrc.2005.02.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E., Bredenbeek P., Dobbe J., Thiel V., Ziebuhr J., Poon L., Guan Y., Rozanov M., Spaan W., Gorbalenya A. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Fielding B., Goh P., Shen S., Tan T., Lim S., Hong W. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase dependent pathway. J. Virol. 2004;78:14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Lim S., Hong W. Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus. Antiviral Research. 2006;72:78–88. doi: 10.1016/j.antiviral.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangudu C., Olivares H., Netland J., Perlman S., Gallagher T. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J. Virol. 2007;81:1220–1229. doi: 10.1128/JVI.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Siddell S. Internal ribosome entry in the coding region of murine hepatitis virus mRNA 5. J. Gen. Virol. 1994;75:3041–3046. doi: 10.1099/0022-1317-75-11-3041. [DOI] [PubMed] [Google Scholar]
- Thiel V., Ivanov K., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E., Rabenau H., Doerr H., Gorbalenya A., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- van Vliet A., Smits S., Rottier P., de Groot R. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 2002;21:6571–6580. doi: 10.1093/emboj/cdf635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Roberts R., Sims A., Deming D., Frieman M., Sparks J., Denison M., Davis N., Baric R. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Wu J., Shan Y., Yao Z., Dong B., Chen B., Zhao Z., Wang S., Chen J., Cong Y. SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology. 2006;346:74–85. doi: 10.1016/j.virol.2005.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R., Yang R., Shi M., Jiang M., Xie Y., Ruan H., Jiang X., Shi L., Zhou H., Zhang L., Wu X., Lin Y., Ji Y., Xiong L., Jin Y., Dai E., Wang X., Si B., Wang J., Wang H., Wang C., Gan Y., Li Y., Cao J., Zuo J., Shan S., Xie E., Chen S., Jiang Z., Zhang X., Wang Y., Pei G., Sun B., Wu J. Characterization of the 3a protein of SARS-associated coronavirus in infected vero E6 cells and SARS patients. J. Mol. Biol. 2004;341:271–279. doi: 10.1016/j.jmb.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N., Zheng B., Li Y., Poon, Xie Z., Chan K., Li P., Tan S., Chang Q., Xie J., Liu X., Xu J., Li D., Yuen K., Peiris, Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]