Abstract
The receptor-binding domain (RBD) on spike protein of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) is the main region interacting with the viral receptor-ACE2 and is a useful target for induction of neutralizing antibodies against SARS-CoV infection. Here we generated two monoclonal antibodies (mAbs), targeting RBD, with marked virus neutralizing activity. The mAbs recognize a new conformational epitope which consists of several discontinuous peptides (aa. 343–367, 373–390 and 411–428) and is spatially located neighboring the receptor-binding motif (RPM) region of the RBD. Importantly, W423 and N424 residues are essential for mAb recognition and are highly conserved among 107 different strains of SARS, indicating that the residues are the most critical in the epitope which is a novel potential target for therapeutic mAbs. A human–mouse chimeric antibody, based upon the original murine mAb, was also constructed and shown to possess good neutralizing activity and high affinity.
Abbreviations: SARS, severe acute respiratory syndrome; SARS-CoV, severe acute respiratory syndrome-associated coronavirus; mAb/pAb, monoclonal/polyclonal antibody; RBD, receptor-binding domain; RBD-Fc, receptor-binding domain linked with a human IgG Fc; RPM, receptor-binding motif; ACE2, angiotensin-converting enzyme 2; CPE, cytopathic effect; pfu, plague forming unit.
Keywords: SARS-CoV, Receptor-binding domain, Neutralization, Monoclonal antibody, Epitope, Chimeric
Introduction
Severe acute respiratory syndrome (SARS), which spread widely in 2002 to 2004 in China and all over the world, resulted in a great loss of life and property. This fatal disease was caused by a newly identified coronavirus: severe acute respiratory syndrome-associated coronavirus (SARS-CoV) (Rota et al., 2003). SARS-CoV is a new kind of coronavirus that is significantly different from previously known ones (Stadler et al., 2003). It is a single-stranded positive-sense RNA virus, which contains four structural proteins embedded on the surface of the viral particle (Stadler et al., 2003). Among them, it has been demonstrated that spike protein (S protein) is the important functional protein that mediates entry of the virus into susceptible cells. The S protein of SARS-CoV is a type I transmembrane glycoprotein that consists of 1255 amino acids. It has two functional domains: S1 (aa. 1–680) and S2 (aa. 681–1255) as predicted by sequence alignment with S proteins of other coronaviruses (Rota et al., 2003, Holmes, 2003).
The S1 domain mediates viral attachment to the cellular receptor ACE2 (Li et al., 2003). The functional portion within the S1 domain, which directly interacts with ACE2, has been identified as the receptor-binding domain (RBD) (Wong et al., 2004). It is located between aa. 318–510 and has been proven to be a major neutralizing determinant (Wong et al., 2004, He et al., 2004a). It has been demonstrated that RBD can induce highly potent neutralizing antibodies. Therefore, it can be considered an excellent candidate for development of a subunit vaccine against SARS (He et al., 2004a). Neutralizing antibodies targeting RBD can be generated by direct immunization of mice with inactivated SARS virus or RBD protein (He et al., 2004a, He et al., 2004b).
Identification of regions containing crucial amino acids within the RBD is essential for assuring recognition of neutralizing antibodies. Such identification could help to increase the understanding of the neutralizing mechanism and to aid discovery of potential drug targets.
So far, it has been reported that the RBD contains several linear and conformational epitopes, among which, most of the neutralizing epitopes are conformational ones (He et al., 2005). There are also some specific fragments or crucial amino acids within the RBD region that are related to neutralizing activity (Chakraborti et al., 2005, Yi et al., 2005). Recently the crystal structure of the RBD and ACE2 complex was clearly demonstrated and a receptor-binding motif (RPM aa. 424–494) within the RBD was characterized as the important region that directly contacts ACE2 (Li et al., 2005).
Although the studies mentioned above brought us much information, neutralizing epitopes have still not been completely characterized. In particular, detailed information concerning conformational neutralizing epitopes and critical amino acids is limited. Moreover, recent studies have shown that the RPM is an important region recognized by neutralizing monoclonal antibodies (Prabakaran et al., 2006, Hwang et al., 2006). However, information on neutralizing epitopes existing outside the RPM was still incomplete.
In this study, we generate two neutralizing monoclonal antibodies, termed S-9-11 and N-176-15, against the RBD. They exhibit good neutralizing activity in a syncytia inhibition assay, a pseudovirus infection assay and a neutralization assay in SARS virus infected cells. A mapping experiment indicates that both mAbs recognize several discontinuous peptides (aa. 343–367, 373–390 and 411–428) and that binding activity is conformation-dependent. Mutations of W423 and N424 completely abrogate binding activity of the RBD to the neutralizing monoclonal antibodies. It was further revealed that W423 and N424 residues within a 411–428 peptide are located adjoining the receptor-binding motif (RBM) of the RBD, an important region holding the RBD–ACE2 interaction surface. W423 and N424 are very stable among 107 different strains of SARS (Song et al., 2005), indicating that the mAbs would possibly possess a broad neutralizing spectrum. A human–mouse chimeric antibody was thus constructed based upon the N-176-15 mAb. It retained a high affinity (KD = 1.6 × 10− 10 M) as compared to the original murine mAb (KD = 2.8 × 10− 9 M). In addition, it maintained binding specificity and neutralizing activity.
The present work describes discovery of a new conformational epitope in the RBD located outside the RPM and formed by several discontinuous peptides with conserved amino acids W423 and N424 as critical mAb binding sites. These findings will be helpful in development of therapeutic antibodies against SARS-CoV.
Results
Vaccinia virus induces high titers of antibodies against RBD
In order to produce a useful antigen that mimics natural Spike protein on the particle surface of SARS-CoV, the full length S gene was inserted into a vaccinia virus expression system (Chen et al., 2005). To confirm that the expressed virus worked well, this constructed virus was used to immunize mice and subsequent neutralizing activity of resulting antisera was tested by a syncytia formation experiment. Once neutralizing activity in the sera was established, the appropriate mouse was selected for raising mAbs. To generate neutralizing mAbs with high efficiency, the region covering the receptor-binding domain (RBD) of the S gene was fused with the human IgG Fc fragment and utilized as a screening antigen.
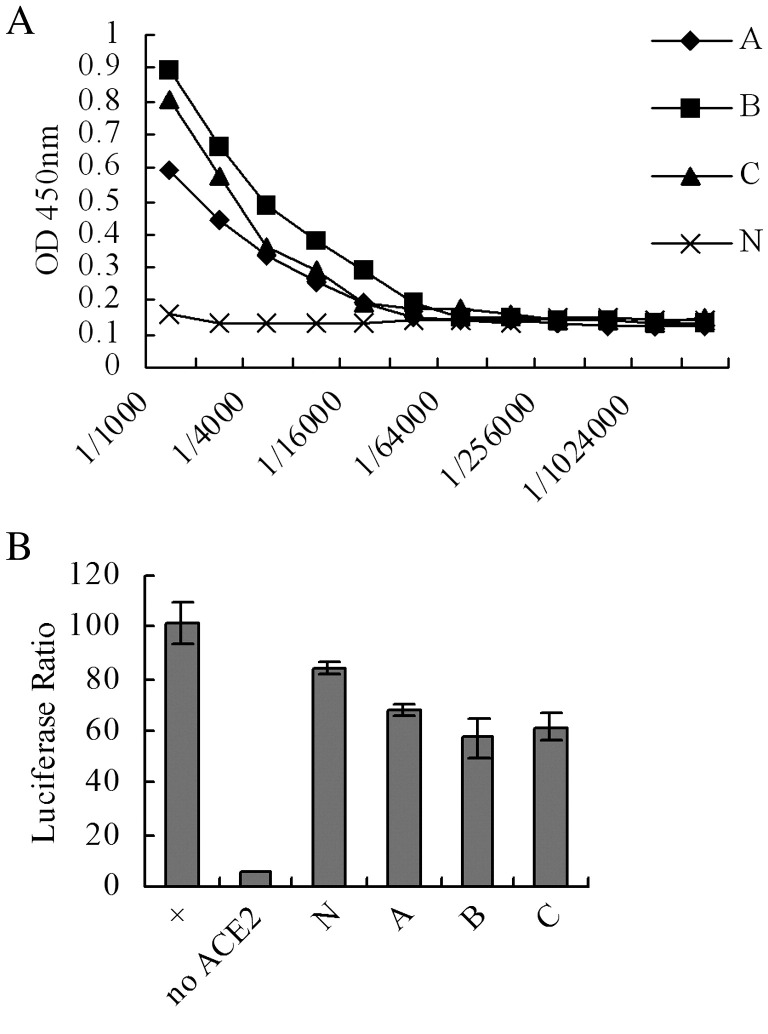
Experiments showed that mice immunized with the vaccinia virus developed a remarkable antibody response against RBD protein (Fig. 1A). The antibody titer of the serum was significantly enhanced following booster immunization (data not shown). One week after the second immunization, antiserum was collected and tested for neutralizing activity using the syncytia inhibition assay. The results show that vaccinia virus immunization induces a considerable concentration of neutralizing antibodies blocking the binding activity between S protein and ACE2 (Fig. 1B), suggesting that the vaccinia virus antigen could trigger significant neutralizing antibody responses targeting the RBD region.
Fig. 1.
RBD-specific neutralizing Ab was induced in mice. (A) A vaccinia virus which contains the full length of the spike gene was used to immunize mice. Titers of the RBD-specific Ab were tested using ELISA in sera collected 1 week after the second immunization. ‘A’, ‘B’ and ‘C’: three individual immunized mice, ‘N’: mouse without immunization. (B) Neutralizing activity was measured by syncytia inhibition assay. Immunized mouse sera collected 1 week after the second booster immunization was screened by syncytia inhibition assay. ‘No ACE2’: HEK293T cells without ACE2 plasmid transfection. ‘+’: positive control in which HEK293T cells were transfected with ACE2 plasmid and no antiserum added. Neutralizing effects of normal mouse sera (N) and immunized mouse sera (A, B and C) were measured in groups of ACE2 transfected HEK293T cells. Mouse whose serum bears highest neutralizing activity was selected for raising mAbs.
mAbs possess high affinity and significant neutralizing activity
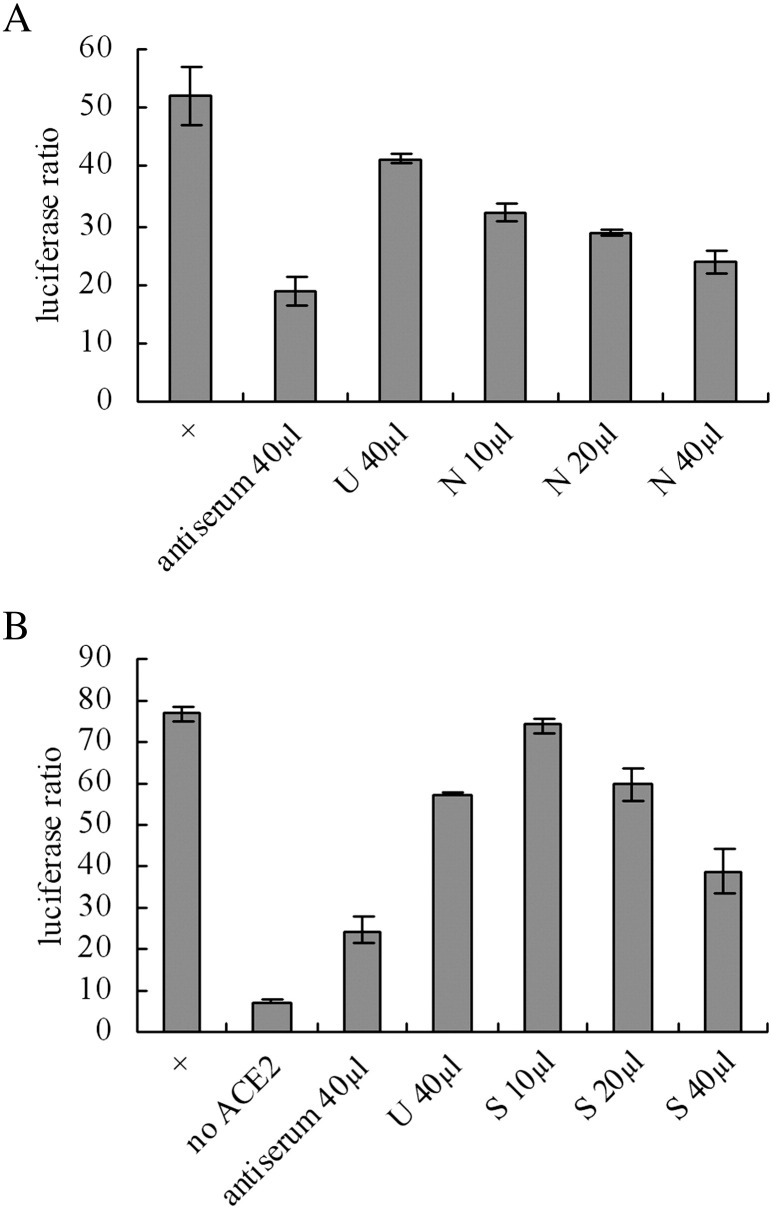
After selection, hybridoma clones reacting to the RBD domain were selected (data not shown), from which two clones exhibiting neutralizing activity confirmed by syncytia formation inhibition assay (Figs. 2A and B), were chosen and termed S-9-11 and N-176-15. When the isotypes were analyzed, S-9-11 was found to be IgG 2a and N-176-15 to be IgG 1, both belonging to the IgG, к class. Ascites produced with the mAbs possessed a very high titer indicated in their binding to the RBD in ELISA, in which their dilution rates reached 1:128,000. These data demonstrate that the mAbs raised in these studies are of very high affinity and exhibit a potential inhibitory effect in blocking the interaction of SARS-CoV Spike protein with ACE2.
Fig. 2.
Neutralizing activity of mAbs (A: N-176-15 and B: S-9-11) at different doses was determined by syncytia inhibition assay. The method is the same as that described in Fig. 1. ‘+’: no mAb or antiserum. ‘No ACE2’: HEK293T cells without ACE2 plasmid transfection. ‘U’: An anti-mouse CD4 mAb selected as an unrelated control. ‘S’: mAb S-9-11, ‘N’: mAb N-176-15, the volume of mAb ascites added into each well (with medium of 1 ml) was also marked.
Neutralizing effect of mAbs is confirmed in pseudotype assay and SARS virus infected cells
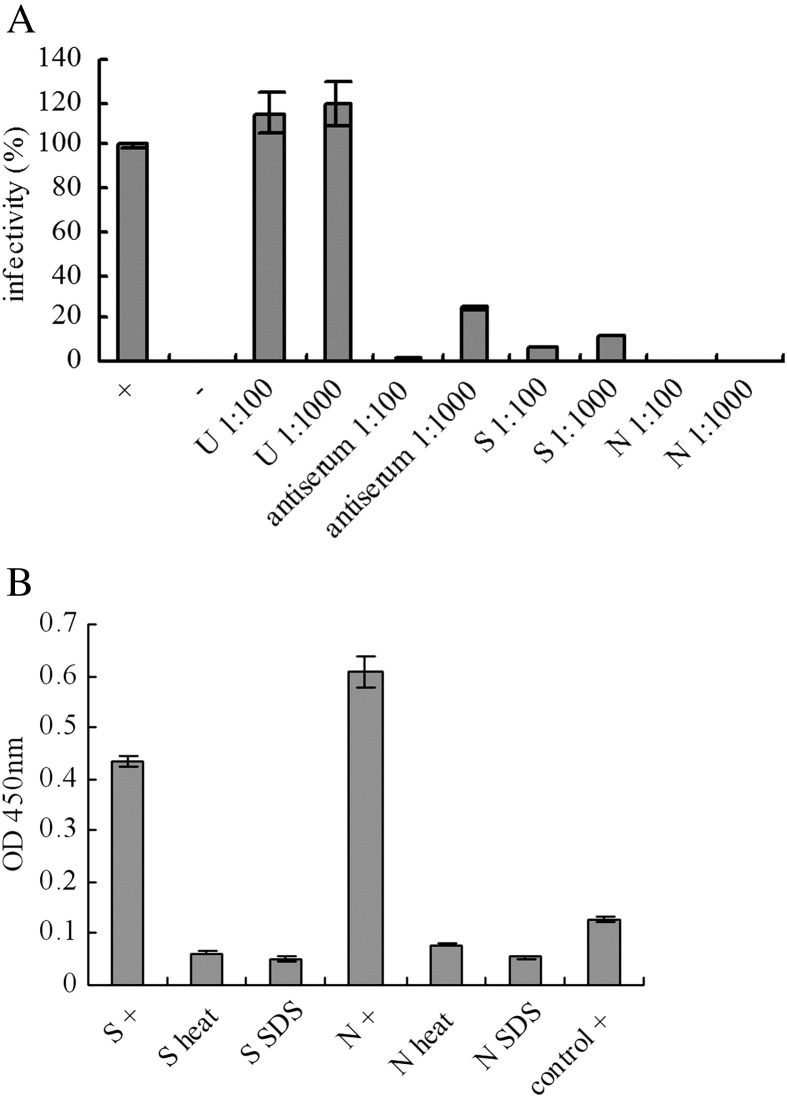
Based upon the primary data generated from syncytia inhibition experiments, it was suggested that the S-9-11 and N-176-15 mAbs were likely to possess potential neutralizing activity. In order to further confirm this notion, two methods were selected to evaluate neutralizing activity. Initially, a pseudotype virus assay was utilized. Results indicated a significant inhibition of pseudovirus infection achieved by use of S-9-11 and N-176-15 mAbs (Fig. 3A). The experiments revealed that the mAbs could block the infection of SARS pseudovirus into target Vero E6 cells through interference with S protein and ACE2 interaction. Secondly, we evaluated neutralizing activity of the mAbs in SARS virus-infected FRhK-4 cells. A serial 2-fold dilution (beginning at 1:80) was carried out to arrive at the highest dilution rate allowing a complete suppression of CPE induced by the virus in at least 5 of 10 wells. When mAb S-9-11 ascites was tested, it showed a marked neutralizing effect upon SARS virus infection with a dilution titer at 1:5000. In additional experiments, mAb N-176-15 ascites showed a similar neutralizing effect (data not shown). The results strongly indicate that both S-9-11 and N-176-15 mAbs are potentially useful candidates for therapeutic use against SARS infection.
Fig. 3.
(A) Neutralizing activity of mAbs (S-9-11 and N-176-15) was further confirmed by pseudovirus infection assay. ‘+’: no Ab, ‘−’: no pseudovirus infection. The dilution of the mAb ascites and antiserum is 1:100 and 1:1000. ‘S’: S-9-11 ascites, ‘N’: N-176-15 ascites, ‘U’: an unrelated mAb ascites (anti mouse CD4). Infectivity: ‘+’ is calculated as 100% and ‘−’ as zero. (B) A denatured antigen ELISA used to determine whether the epitope is linear or conformational. RBD-Fc denatured by heat or SDS/β-mercaptoethanol was coated on 96-well micro-plates. ‘S’: mAb S-9-11, ‘N’: mAb N-176-15. ‘+’: RBD-Fc without denaturation. ‘Control’: an unrelated mAb.
Mapping epitopes recognized by neutralizing mAbs
Based upon our primary data suggesting high affinity and marked neutralizing activity for S-9-11 and N-176-15 mAbs as compared to other mAbs (data not shown), it was of interest to map epitopes recognized by the mAbs. To do so, we first carried out an ELISA experiment to determine whether epitopes recognized by the mAbs might be linear or conformational. Results showed that after RBD-Fc protein was denatured by heat or by SDS/β-mercaptoethanol, binding activity of mAbs with the antigen was completely lost (Fig. 3B), indicating that the epitope is conformation-dependent.
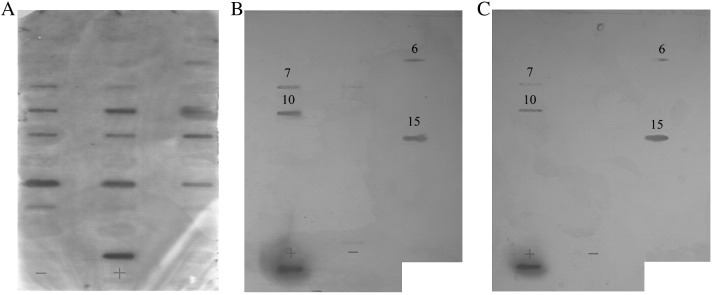
Since the RBD domain is located at amino acid residues 318 to 510 as recognized by the mAbs, a series of synthetic overlapping peptides (27 total) covering the entire RBD sequence were selected (Table 1 ) and experiments were performed to identify specific regions or amino acids that are crucial to the binding of the mAbs with the RBD. Initially, the synthetic peptides were coated on plates and their reactivity with the mAbs tested by ELISA. Unexpectedly, the reactivity was undetectable (data not shown). Subsequently, a dot-blot experiment was employed. To guarantee its success, we first tested the overlapping peptides with our laboratory-prepared rabbit anti-Spike S1 polyclonal antibodies recognizing S1 protein from amino acid residues 1 to 678 (Wu et al., 2004). Results showed that the anti-S1 polyclonal antibodies clearly recognized several peptides with different binding intensities, which depended upon their different antigenicities (Fig. 4A). As expected, the pAb did not react with HA peptide (negative control). The data demonstrated the utility of this system. From this experiment, we were able to identify three major antigenic regions: peptides no. 6–7 (aa. 343–367), no. 10–15 (aa. 373–428), and no. 19–22 (aa. 442–478) (Fig. 4A). We then tested the binding activity of the mAbs with these peptides. Results showed that both mAbs were able to bind the same peptides: no.6, 7, 10 and 15, which represented amino acid residues 343–367, 373–390 and 411–428 (Table 1, Figs. 4B and C). The no. 8 peptide position also demonstrated a mild binding signal but was too weak to be considered recognizable. It was also shown that the two mAbs (S-9-11 and N-176-15) recognized the same peptides indicating they share the same epitope. Although the mAbs recognized a conformational epitope, we were still able to use dot-blot analysis to detect peptide reactivity and to analyze epitopes recognized by the mAbs within the whole RBD region.
Table 1.
Overlapping peptides covering the RBD region
| 1 | VVPSGDVVRFPNITNL | 10 | KLNDLCFSNVYADSFVVK | 19 | YLRHGKLRPFERDISNV |
| 2 | VVRFPNITNLCPFGEVF | 11 | NVYADSFVVKGDDVRQIA | 20 | RPFERDISNVPFSPDGK |
| 3 | TNLCPFGEVFNATKFPSV | 12 | VKGDDVRQIAPGQTGVIA | 21 | SNVPFSPDGKPCTPPAL |
| 4 | VFNATKFPSVYAWERKKI | 13 | IAPGQTGVIADYNYKL | 22 | DGKPCTPPALNCYWPL |
| 5 | SVYAWERKKISNCVADY | 14 | GVIADYNYKLPDDFMGCV | 23 | PPALNCYWPLNDYGFY |
| 6 | KKISNCVADYSVLYNSTF | 15 | KLPDDFMGCVLAWNTRNI | 24 | YWPLNDYGFYTTTGIGY |
| 7 | DYSVLYNSTFFSTFKCY | 16 | CVLAWNTRNIDATSTGNY | 25 | GFYTTTGIGYQPYRVVVL |
| 8 | STFFSTFKCYGVSATKL | 17 | NIDATSTGNYNYKYRYLR | 26 | GYQPYRVVVLSFELLNA |
| 9 | KCYGVSATKLNDLCFSNV | 18 | NYNYKYRYLRHGKLRPF | 27 | VVLSFELLNAPATVCGPK |
Amino acid residue sequences of overlapping peptides synthesized to cover the entire RBD region. The total peptide number is 27.
Fig. 4.
Potential epitopes were mapped within the RBD domain. (A) Dot-blot assay for rabbit anti-S1 polyclonal antibodies. ‘+’: RBD-Fc, ‘−’: an unrelated HA peptide. (B) Dot-blot assay of mAb N-176-15. (C) Dot-blot assay of mAb S-9-11.
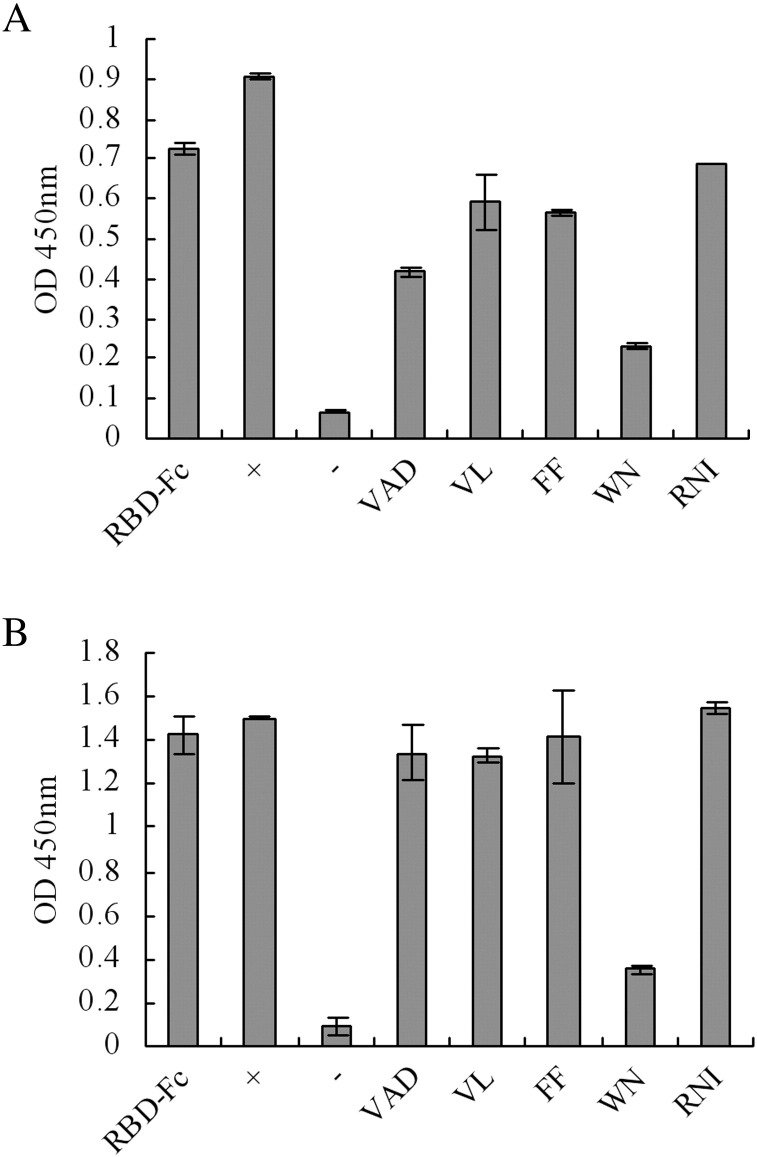
We then constructed a computer-based model of the variable regions of mAb N-176-15 to predict its interaction pattern with the RBD. Based upon the analysis, several clusters of amino acid residues from the RBD were selected (Fig. 5 ). Combining the dot-blot experiment and the computer prediction, we predicted that five sites (VAD, VL, FF, WN, RNI) might be important for mAb binding (Fig. 5). A site-directed mutation experiment was then carried out to test whether the putative sites could influence their binding. We mutated the amino acids of the five sites into alanines (Table 2 ), generated mutated RBD-Fc with transient transfection and determined their binding activity with the mAbs using a sandwich ELISA (Fig. 6 ). The results show that the W423 and N424 mutated RBD-Fc lost significant ability to bind with the mAbs, indicating that these two amino acids are crucial for the binding of mAbs N-176-15 and S-9-11 with the RBD.
Fig. 5.
Potential crucial amino acids for binding of mAb with RBD are predicted. Underlined letters represent amino acids recognized by the mAbs in dot-blot experiments. Capital letters represent amino acids that were predicted by computer model to be potentially important for the binding of the mAb with the RBD. Combining the results of the two methods, amino acids VAD, VL, FF, WN and RNI were selected to be the most important amino acids determined in a site-directed mutagenesis experiment.
Table 2.
Primers for site-directed mutagenesis; construction of RBD-Fc expressing plasmids
| aa. residues mutated | Primer sequences |
|---|---|
| FF → AA | SP: TAC AAC TCA ACA GCT GCT TCA ACC TTT AAG |
| AS: CTT AAA GGT TGA AGC AGC TGT TGA GTT GTA | |
| WN → AA | SP: TGT GTC CTT GCT GCT GCT ACT AGG AAC ATT |
| AS: AAT GTT CCT AGT AGC AGC AGC AAG GAC ACA | |
| VL → AA | SP: GCT GAT TAC TCT GCT GCT TAC AAC TCA ACA |
| AS: TGT TGA GTT GTA AGC AGC AGA GTA ATC AGC | |
| VAD → AAA | SP: AAA ATT TCT AAT TGT GCT GCT GCT TAC TCT GTG CTC TAC |
| AS: GTA GAG CAC AGA GTA AGC AGC AGC ACA ATT AGA AAT TTT | |
| RNI → AAA | SP: CTT GCT TGG AAT ACT GCT GCT GCT GAT GCT ACT TCA ACT |
| AS: AGT TGA AGT AGC ATC AGCAGC AGC AGT ATT CCA AGC AAG |
The candidate amino acid residues were mutated to alanines within the primers designed for mutation. SP: sense primer. AS: anti-sense primer.
Fig. 6.
Identification of potentially important amino acids by site-directed mutagenesis for the binding of the mAbs with the RBD. ELISA was used to measure the binding activity of the mAbs with the RBD or with mutated RBD generated from supernatants of HEK293T cells transfected with RBD-Fc plasmid or mutated RBD-Fc plasmids. ‘+’: supernatant of RBD-Fc plasmid transfection, ‘RBD-Fc’: purified RBD-Fc protein dissolved in distilled water at a concentration of 20 μg/ml, ‘−’: supernatant of cells without transfection. ‘VAD’ etc.: five mutated RBD-Fc proteins (selected amino acids mutated to alanine) expressed in cell culture supernatants. (A) Reaction measured by mAb S-9-11. (B) Reaction measured by mAb N-176-15.
Finally, we determined the degree of variation of these two amino acid residues in different SARS strains. We chose 107 SARS spike genes, among which 14 are from palm civet (4 from 2003, other 2004) and 93 from human (4 from 2004, 11 from early 2003, 17 from middle 2003, 61 from late 2003) (Song et al., 2005). The W423 and N424 are very conserved in all SARS S gene sequences without any mutation. The results indicate that the mAbs would probably have a broad viral neutralizing spectrum and that this epitope is highly conserved among different SARS strains.
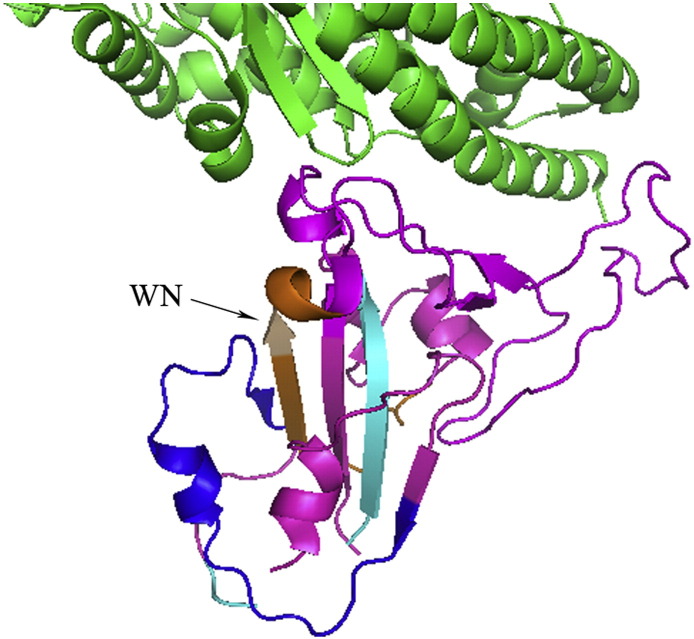
The structure of the RBD offered a direct and more precise profile of this newly identified neutralizing epitope. Peptides identified in dot-blot analysis formed on the RBD a spatial structure, which localized immediately beside the RPM region (Fig. 7 ). Interestingly, N424 was the first amino acid residue determined for the RPM (Li et al., 2005). Although this neutralizing epitope was not well localized on the surface that directly contacts ACE2, antibodies were able to bind this region and block the attachment of ACE2 due to spatial hindrance. This explains the neutralization mechanism of the two mAbs at a molecular level.
Fig. 7.
Spatial location of the epitope predicted using a computer-analysis model. Green: ACE2, Magenta: RBD. The location of binding peptides and amino acids W423, N424 are marked as blue: aa. 343–367, cyan: aa. 373–390, orange: aa. 411–428, wheat: W423 and N424.
A chimeric antibody shows a potential blocking effect on SARS-CoV infection
We constructed a chimeric antibody based upon the N-176-15 clone, and retaining the affinity, specificity and neutralizing activity of the murine mAb. As the data shows, the chimeric antibody recognizes the same peptides as does the murine antibody in the dot-blot assay, indicating that the chimeric nature of the antibody does not change its binding specificity (Fig. 8 ). A SPR experiment was carried out to determine the binding affinity of the chimeric antibody with the RBD. The data indicates that both the chimeric antibody and the murine mAb show a very high binding affinity with the RBD, their KDs being 1.6 × 10− 10 M and 2.0 × 10− 9 M respectively. This high affinity explains the strong neutralizing effect of the mAb. There is no doubt that the constructed chimeric antibody completely preserves the binding affinity of the murine mAb. In addition, we tested the neutralizing activity of the chimeric antibody in the pseudovirus infection system. The results reveal that the chimeric antibody has a similar function to the murine one in completely blocking entry of the pseudovirus into Vero E6 cells at a concentration of 1 μg/ml (data not shown). All data indicate that we have successfully constructed a SARS chimeric neutralizing antibody with great potential for therapeutic use against SARS infection.
Fig. 8.
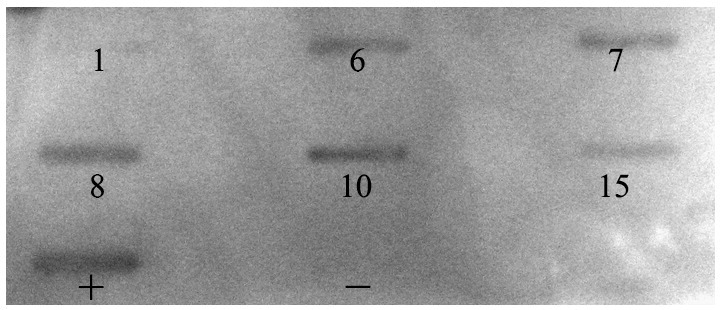
Chimeric antibody recognition of the peptides recognized by N-176-15 tested in dot-blot. ‘+’: RBD-Fc, ‘−’: an unrelated HA peptide.
Discussion
From this study, it is demonstrated that the receptor-binding domain of SARS-CoV plays a crucial role in the process of viral infection and host immune response, especially as a dominant antigen in inducing neutralizing antibody development.
Information concerning epitopes on the RBD has been reported. It has been revealed that the RBD contains various linear and conformational epitopes and that most of the neutralizing epitopes are conformational ones (He et al., 2005). Although the neutralizing antibodies show a higher affinity, the spatial profiles of these conformational epitopes had not been well understood. There have also been some studies on functionally important amino acids or segments of the RBD. It was reported that the deletion of a positively charged region (422–463) on the RBD protein abolished the capacity of the S protein to induce neutralizing antibodies (Yi et al., 2005). It was also reported that a single amino acid substitution (R441A) in the full-length S DNA vaccine failed to induce neutralizing antibodies or to abolish viral entry (Yi et al., 2005). These results indicated that particular portions of and amino acids on the RBD are crucial for retaining the RBD's normal function. However, whether critical amino acids or segments exist in conformational neutralizing epitopes was still not known. The crystal structure of the RBD–ACE2 complex has been identified, introducing detailed information concerning RBD structure and function (Li et al., 2005). The structure revealed that the RBD could be further divided into 2 separate subdomains. One is the RBD core and the other is the RBD loop (RPM) (aa. 424–494). The RBD loop is the region that directly contacts the ACD2 molecule. The RBD core, on the other hand, makes contact with accessory proteins (Li et al., 2005). The epitope recognized by a humanized neutralizing monoclonal antibody, 80R, has been mapped and its crucial amino acid residues on the RBD loop region binding to the neutralizing mAb has been determined (Sui et al., 2004, Sui et al., 2005). However, little evidence or description was reported regarding epitopes, especially conformational ones, located outside the RBD loop region.
Currently an important task is to characterize neutralizing epitopes, to clarify their spatial profile and to determine important regions or amino acids involved in neutralizing antibody recognition. Because most neutralizing epitopes are conformational ones, it is more meaningful to develop a suitable strategy by studying their structure and then identifying the critical amino acids in a given antigen.
In this study, we report, for the first time, a conformational neutralizing epitope localized outside the RPM region, indicating that neutralizing antibodies do not bind only the surface of RBD–ACE2 interaction but also bind other sites. Ultimately, three peptides (aa. 343–367, 373–390 and 411–428) are seen to form a conformational epitope; within them the amino acids W423 and N424 in the 411–428 peptide, are a central point for mAb recognition and for eliciting neutralizing antibodies.
In this research process, it is notable that a peptide-related dot-blot method was used to analyze conformational epitopes. The results indicate that a neutralizing mAb that recognizes a natural-conformation-dependent antigen can bind in mild affinity with peptides that could only be detected by the dot-blot method.
Many clones of neutralizing monoclonal antibodies with the target of S protein have been raised so far. A variety of methods have been utilized; for example, the traditional PEG cell fusion method and the phage display method (Sui et al., 2004, Chou et al., 2005; and Brink et al., 2005). Antigens for immunization include inactivated whole virus, recombinant S protein and RBD-Fc protein (He et al., 2004b, Chou et al., 2005; and Zhou et al., 2004). Here we used a modified recombinant vaccinia virus Ankara with SARS S protein on the surface as the antigen with which to immunize mice. The results show that this vaccinia virus was able to elicit high titers of neutralizing antibodies (Chen et al., 2005). Therefore, it appears to be an ideal immunizing antigen for generation of monoclonal antibodies as well as a possible vaccine candidate. For screening of hybridoma clones, we used mammalian-cell expressed RBD-Fc protein, whose natural conformation is well preserved. With this strategy, we successfully obtained two hybridoma clones generating monoclonal antibodies to target the RBD region of the S protein with high affinity and good neutralizing effect.
For the evaluation of neutralizing effects, we utilized two different systems, one a syncytia inhibition assay, and the other a pseudovirus infection assay. These procedures allowed us to evaluate the neutralizing effects of antibodies or peptides conveniently in vitro.
Since the W423 and N424 amino acids in the neutralizing epitope constitute a conserved site and the mAb against the site thus constitutes a potential therapeutic mAb, we constructed a chimeric antibody based upon the murine monoclonal antibody N-176-15 clone. The chimeric antibody is shown to have the same specificity as the original murine one and retains its neutralizing effect. Therefore, our work demonstrates the feasibility of constructing neutralizing chimeric antibodies with potential therapeutic value that might be further developed into SARS therapeutic drugs.
In summary, mAbs recognizing a new conformational epitope were raised, the epitope consisting of several discontinuous peptides (aa. 343–367, 373–390 and 411–428) and spatially located near the receptor-binding motif (RPM) region of the RBD. The W423 and N424 residues in the 411–428 peptide are essential for mAb recognition and are highly conserved among 107 different strains of SARS, indicating that the residues are the most critical in the epitope and constitute a novel potential target for inducing a broad response from neutralizing mAbs. A human–mouse chimeric antibody was also constructed with good neutralizing activity and high affinity based upon the original murine mAb. The characteristics demonstrated for such a mAb demonstrate a notable potential for use in prevention of SARS infection.
Materials and methods
Animals
Balb/c mice were purchased from Shanghai Laboratory Animal center, Chinese Academy of Sciences. Animals were kept in conventional conditions and were handled in compliance with Chinese Academy of Sciences guidelines for Animal Care and Use.
Antigen for immunization
Recombinant modified vaccinia virus Ankara expressing the S protein of SARS-CoV was used as the antigen for immunization (Chen et al., 2005). This modified SARS vaccine showed excellent potential to induce neutralizing antibodies that target primarily the RBD region. Mammalian-cell expressed RBD-Fc serves as a natural form of protein of the RBD region (He et al., 2005) and was used as an antigen for screening hybridoma clones.
Immunization of mice and generation of monoclonal antibodies
Six-week old female Balb/c mice were used for immunization. For each injection, the quantity of antigen used per mouse was 5 × 107 pfu and an intramuscular multi-site injection method was adopted. Boosting was carried out 4 times, each at 3-week intervals. Serum was collected 3 days after the second immunization and was stored at − 20 °C. Three days after the last boost, mouse spleen cells were harvested and fused with myeloma cell lines NS-1 and SP2/0 using 50% PEG (w/v) and were distributed in 96-well plates. Culture supernatants from each well were screened using ELISA for antibody reactivity against the RBD. Positive wells were then selected using a limiting dilution method. After three cycles of cloning, hybridoma clones were established. Monoclonal antibody isotypes were identified using a mouse sub-isotyping kit (Bio-Rad USA). Monoclonal antibodies were produced from mouse ascites by injection of 106 hybridoma cells per mouse intraperitoneally and were purified by Protein G column chromatography.
ELISA
RBD-Fc protein dissolved in 0.1 M carbonate buffer (pH 9.6) was coated on 96-well micro-titer plates (10 μg/ml, 50 μl/well) at 4 °C overnight. After blocking with phosphate-buffered saline (PBS) containing 10% bovine serum and 0.1% Tween 20 at 37 °C for 2 h, the plates were incubated with culture supernatants or diluted antiserum at 37 °C for 2 h. Bound antibodies were detected with HRP-conjugated goat anti-mouse Ig antibody (Santa Cruz USA). Tetramethylbenzidine (TMB) was used as substrate (Sigma USA), and the absorbance was measured using a micro-titer plate autoreader (Thermo USA) at 450 nm. In a sandwich ELISA, protein G purified S-9-11 or N-176-15 mAbs were coated on 96-well micro-titer plates. After blocking, supernatant containing RBD-Fc or its mutated form was placed into the plates; an anti-human IgG (Fc specific) HRP-conjugated antibody (Sigma USA) was used as detecting antibody in a detailed protocol as described above. In an RBD denatured ELISA, RBD-Fc protein and RBD-Fc protein denatured with heat (80 °C10 min) or 5%SDS + 5%β-mercaptoethanol were coated on ELISA 96-well micro-titer plates and incubated at 4 °C overnight in a detailed protocol as described as above.
Syncytia inhibition assay
This assay was described in our previous article (Lu et al., 2005). In brief, we co-transfected HEK293T cells (approximately 50% confluent in 60 mm dishes) with plasmids pCDNA3.1-ACE2 and pUHD 15-1(SV40), while another group of HEK293T cells (approximately 50% confluent in 60 mm dishes) were co-transfected with pCDNA3.1-spike-Ig and pUHD 10-3(Luc). A control plasmid, pRellina, was also utilized as an internal reference. Twenty-four hours after transfection, the two groups of cells were trypsinized, re-suspended in DMEM/10% FBS and adjusted to a proper final concentration. The antibody ascites or antiserum obtained during mAb generation was added to the spike protein group of transfected cells at serial doses of 10, 20 or 40 μl per well and incubated for 0.5 h at 37 °C. Subsequently, the two groups of cells were mixed together (cell number 1:1) in 48-well plates with a total cell number of 4 × 105, a volume of 1 ml per well and were incubated at 37 °C. Twenty-four hours later, medium was aspirated and cells were lysed with passive lysis buffer (Promega USA). Luciferase intensity was measured using a luminometer TD-20/20 (Promega USA).
Pseudovirus infection assay
HEK293T cells were plated at approximately 50% confluence in 6-well plates. Twenty-four hours later, plasmids coding SARS spike protein and pNL4-3 luc Env− Vpr− were co-transfected using lipofectamine (Invitrogen USA) to generate an HIV backbone pseudovirus with SARS spike protein embedded on the surface (Chen et al., 2005). Forty-eight hours later, supernatant was collected from the cells using low speed centrifugation (3000 rpm, 5 min), aliquoted and frozen at − 140 °C. Vero E6 cells were plated as target cells before infection in 24-well plates at approximately 50% confluence. During infection, medium was aspirated and 100 μl pseudovirus (supernatants) mixed with 900 μl 12% FBS/DMEM added per well simultaneously with or without purified mAb, ascites or antiserum at 37 °C for incubation. Twenty-four hours later, another 1 ml of 12%FBS/DMEM was added to each well. Seventy-two hours after infection, medium was aspirated, cells were washed with PBS twice, lysed with passive lysis buffer (Promega USA) at room temperature, and stored at − 80 °C or analyzed immediately for luciferase activity.
SARS virus neutralization assay
Micro-titer plates were used in the neutralization assay. Serial two-fold dilutions (1:80 to 1:10,240) of mAb were separately mixed with 100 TCID50 of virus (GZ50), incubated at 37 °C for 1 h and then added to FRhK-4 cells. A virus back-titration (virus in serial two-fold dilution with medium), virus positive control (100 TCID50) and negative cell controls with medium in parallel with the neutralization test were included in the assay. Each dilution of serum or virus control was repeated in 10 wells. Results were observed daily and CPE endpoints were read and recorded up to 3 days after virus inoculation. The TCID50 was calculated using the Reed–Muench method. The titer of the neutralization antibody was determined based upon the highest dilution of the mAb at which the mAb could completely suppress CPE induced by the virus in at least 5 of 10 wells. All virus experiments were carried out in a bio-safety P3 laboratory at the University of Hong Kong.
Dot-blot analysis
This assay was performed to identify binding epitopes of the monoclonal antibodies. Antigens used were overlapping peptides covering the entire RBD domain (aa. 318–510). For each peptide, 5 μg diluted in 50 μl PBS was blotted on PVDF membrane, which was then washed five times in TTBS. The membrane was blocked in 3% BSA (in TTBS) at 37 °C for 2 h. Diluted monoclonal antibody or antiserum in 3% BSA (in TTBS) was then added and incubated at 4 °C overnight. Subsequently, the membrane was incubated with a secondary antibody (goat anti-mouse/rabbit HRP conjugated antibody (Santa Cruz USA)) at 37 °C for 1 h. The blots were developed using ECL western blotting substrate reagents (Pierce USA).
Bioinformatic analysis
A computer-based simulation of the structure of the N-176-15 VH/VL region and construction of the interaction model of this structure with the RBD domain was carried out to determine specific amino acids critical for binding activity. We first cloned the variable region of the N-176-15 mAb. With the sequence and PDB database, a structure model was constructed; the binding model of the putative structure with the already-known RBD structure was simulated and potentially crucial amino acids were predicted.
Site-directed mutagenesis
A PCR method was used to construct mutated forms of the RBD protein. The mutated amino acids were constructed utilizing PCR primers. After PCR, mutated forms of the RBD gene were obtained and subsequently cloned into a pCDNA3.1 vector with a human IgG Fc tag gene. The mutated amino acids and the specific PCR primers are included in Table 2. A transient mammalian protein expression system was used to express the RBD protein and its mutated form. Briefly, the RBD-Fc expression plasmid was transiently transfected using lipofectamine (Invitrogen USA) into HEK293T cells, plated 24 h before in 6-well plates. Forty-eight hours later, cells were harvested and lysed with 0.5 ml RIPA buffer per well. Supernatants were collected after centrifugation at 12 000 rpm for 10 min. Using the sandwich ELISA assay described above, it was possible to successfully detect mAb binding activity to RBD or to its mutated form in cell lysis supernatants.
Chimeric antibody construction and characterization
To construct a mouse–human chimeric IgG, total RNA was isolated from N-176-15 hybridoma cells using TRI Reagent (Sigma USA). The variable domain gene was obtained by a 5′RACE polymerase chain reaction (5′RACE PCR), reverse transcription and gel purification. Subsequently, the gene was inserted into a pGEM-T Vector (Promega USA) and sequenced (Sangon China). The variable region gene including signal sequence was constructed in a WS3 plasmid containing a human kappa constant domain and a human CH1–CH2–CH3 domain. The plasmid was then transfected into CHO-dhfr− cell lines using lipofectamine reagent (Invitrogen USA) to construct chimeric-antibody-secreting cells. Clones were screened using ELISA to detect chimeric antibody in culture supernatant and were established using a limiting dilution method. After establishment of stably transfected clones, a dot-blot procedure was used to measure the binding specificity of the chimeric antibody and a SPR (Biacore USA) assay was employed to characterize its affinity. A pseudovirus infection assay was used to determine neutralizing activity of the chimeric antibody.
Statistical analyses and reproducibility
Experiments were repeated at least twice, and usually three or more times. Figures show data compiled from several experiments, or from a representative experiment, as specified. Results represent the mean ± SD where applicable. Statistical analyses for parametric data were performed by independent t-test.
Disclosures
The authors declare that there's no financial conflict of interest.
Acknowledgments
We thank Dr. Vincent Deubel and Dr. Sheri Skinner for reviewing the manuscript and for helpful comments. This work was supported by grants from the National Natural Science Foundation of China (30325018, 30530700, 30623003 and 30421005), the CAS project (KSCX1-YW-R-43), a grant from the 863 key project (2006AA02A247), grants from the Technology Commission of Shanghai Municipality (04DZ14902, 04DZ19108, 06DZ22032 and 04DZ19112), and a grant from the E-institutes of Shanghai Universities Immunology Division.
References
- Chakraborti S., Prabakaran P., Xiao X., Dimitrov D.S. The SARS coronavirus S glycoprotein receptor binding domain: fine mapping and functional characterization. Virology Journal. 2005;2:73. doi: 10.1186/1743-422X-2-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhang L., Qin C., Ba L., Yi C.E., Zhang F., Wei Q., He T., Yu W., Yu J., Gao H., Tu X., Gettie A., Farzan M., Yuen K.y., Ho D.D. Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J. Virol. 2005;79:2678–2688. doi: 10.1128/JVI.79.5.2678-2688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.h.W., Wang S., Sakhatskyy P.V., Mboudoudjeck I., Lawrence J.M., Huang S., Coley S., Yang B., Li J., Zhu Q., Lu S. Epitope mapping and biological function analysis of antibodies produced by immunization of mice with an inactivated Chinese isolate of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) Virology. 2005;334:134–143. doi: 10.1016/j.virol.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324:773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Siddiqui P., Jiang S. Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem. Biophys. Res. Commun. 2004;325:445–452. doi: 10.1016/j.bbrc.2004.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Lu H., Siddiqui P., Zhou Y., Jiang S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J. Immunol. 2005;174:4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- Holmes K.V. SARS-associated coronavirus. N. Engl. J. Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- Hwang W.C., Lin Y., Santelli E., Sui J., Jaroszewski L., Stec B., Farzan M., Marasco W.A., Liddington R.C. Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J. Biol. Chem. 2006;281:34610–34616. doi: 10.1074/jbc.M603275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Lu W., Wu X.D., De Shi M., Yang R.F., Heb Y.Y., Bian C., Shi T.L., Yang S., Zhu X.L., Jiang W.H., Li Y.X., Yan L.C., Ji Y.Y., Lin Y., Lin G.M., Tian L., Wang J., Wang H.X., Xie Y.H., Pei G., Wua J.R., Sun B. Synthetic peptides derived from SARS coronavirus S protein with diagnostic and therapeutic potential. FEBS Lett. 2005;579:2130–2136. doi: 10.1016/j.febslet.2005.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhry V., Xiao X., Ji X., Dimitrov D.S. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.h., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C.T., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Günther S., Osterhaus A.D.M.E., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Wang Chern S.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu X.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., Rappuoli R. SARS-beginning to understand a new virus. Nat. Rev. Micro. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S.t.C., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Roberts A., Matthews L.J., Murakami A., Vogel L., Wong S.K., Subbarao K., Farzan M., Marasco W.A. Evaluation of human monoclonal antibody 80R for immunoprophylaxis of severe acute respiratory syndrome by an animal study, epitope mapping, and analysis of spike variants. J. Virol. 2005;79:5900–5906. doi: 10.1128/JVI.79.10.5900-5906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink E.N., ter Meulen J., Cox F., Jongeneelen M.A.C., Thijsse A., Throsby M., Marissen W.E., Rood P.M.L., Bakker A.B.H., Gelderblom H.R., Martina B.E., Osterhaus A.D.M.E., Preiser W., Doerr H.W., de Kruif J., Goudsmit J. Molecular and biological characterization of human monoclonal antibodies binding to the spike and nucleocapsid proteins of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:1635–1644. doi: 10.1128/JVI.79.3.1635-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino-acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.D., Shang B., Yang R.F., Yu H., Ma Z.H., Shen X., Ji Y.Y., Lin Y., Wu Y.D., Lin G.M., Tian L., Gan X.Q., Yang S., Jiang W.H., Dai E.H., Wang X.Y., Jiang H.L., Xie Y.H., Zhu X.L., Pei G., Li L., Wu J.R., Sun B. The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells. Cell Res. 2004;14:400–406. doi: 10.1038/sj.cr.7290240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C.E., Ba L., Zhang L., Ho D.D., Chen Z. Single amino acid substitutions in the severe acute respiratory syndrome coronavirus spike glycoprotein determine viral entry and immunogenicity of a major neutralizing domain. J. Virol. 2005;79:11638–11646. doi: 10.1128/JVI.79.18.11638-11646.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Wang H., Luo D., Rowe T., Wang Z., Hogan R.J., Qiu S., Bunzel R.J., Huang G., Mishra V., Voss T.G., Kimberly R., Luo M. An exposed domain in the severe acute respiratory syndrome coronavirus spike protein induces neutralizing antibodies. J. Virol. 2004;78:7217–7226. doi: 10.1128/JVI.78.13.7217-7226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]