Abstract
Background
Matrix Gla protein (MGP) is a secreted protein contributed to the immunomodulatory functions of mesenchymal stromal cells. Microarray profiling found a significantly higher expression level of the extracellular matrix gene MGP in patients with ulcerative colitis (UC). However, little is known about the role of MGP in UC and its upstream signaling regulation. This study aimed to identify the expression of MGP in UC and its upstream regulator mechanism.
Methods
Colonic mucosa biopsies were obtained from patients with UC and healthy controls. DNA microarray profiling was used to explore underlying genes correlating with UC development. Mice were fed with water containing different concentrations of dextran sodium sulfate (DSS) to induce an experimental colitis model. Colonic tissues were collected and evaluated using immunohistochemistry, immunoblot, real-time polymerase chain reaction, and chromatin immunoprecipitation assay. Bioinformatics analysis was performed to identify candidate MGP gene-promoter sequence and transcription-initiation sites. Luciferase-reporter gene assay was conducted to examine the potential transcription factor of MGP gene expression.
Results
The expression of MGP was significantly increased in colonic tissues from UC patients and DSS-induced colitis models, and was positively correlated with disease severity. Bioinformatics analysis showed a conserved binding site for Egr-1 in the upstream region of human MGP gene. The significantly higher level of Egr-1 gene expression was found in UC patients than in healthy controls. The activity of luciferase was significantly enhanced in the Egr-1 expression plasmid co-transfected group than in the control group and was further inhibited when co-transfected with the Egr-1 binding-site mutated MGP promoter.
Conclusions
Up-regulated expression of MGP was found in UC patients and DSS-induced colitis. The expression of MGP can be regulated by Egr-1.
Keywords: ulcerative colitis, matrix Gla protein, Egr-1
Introduction
Inflammatory bowel disease (IBD) is a chronic non-specific intestinal inflammatory disease mainly including Crohn’s disease (CD) and ulcerative colitis (UC). These disorders lead to a low quality of life in patients because of frequent fever, abdominal pain, diarrhea, and a variety of complications [1]. The etiology of IBD is not well understood; however, multiple studies have revealed that genetic and environmental factors involved in the pathogenesis of IBD [2]. The alteration of genes might play an important role in the progress of IBD [3]. So far, more than 300 differentially expressed genes have been identified in IBD, which were mainly involved in the regulation of inflammation and immunity, as well as the maintenance of the intestinal epithelial-barrier function [4, 5].
Gene-expression profiling in disease can uncover the underlying gene changes contributing to the IBD and promote the identification of targets [6]. A meta-analysis combining genome-wide association studies have identified 163 loci associated with IBD in more than 75,000 patients and control individuals of European descent [7, 8]. Of these loci, 110 are at risk for both CD and UC phenotypes, while 30 and 23 loci were unique to CD and UC, respectively [7, 8].
In this study, we first performed DNA microarray analysis to assess the gene expression in UC patients with severe symptoms. Among those differentially expressed genes, we focused on extracellular matrix gene matrix Gla protein (MGP). We examined the protein expression of MGP in colon tissue in UC patients and in a dextran sodium sulfate (DSS)-induced colitis model. In addition, to understand whether MGP contributed to the pathogenesis of IBD, the upstream signaling regulation of MGP was explored in this study.
Materials and methods
Patients
A total of 50 patients with UC who underwent colonoscopy were enrolled in this study. All patients were diagnosed by clinical characteristics, endoscopy, and pathology. Disease severity in these patients was determined clinically by the modified Mayo scoring system [9]. Control tissues were from a healthy population. This study was approved by the Institutional Review Board of Peking Union Medical College Hospital.
DNA microarray profiling
Colonic mucosa biopsies were obtained from the ascending colon of patients with severe UC (n = 3) and healthy subjects (n = 3) undergoing screening colonoscopies in the Department of Gastroenterology at Peking Union Medical College Hospital. The frozen samples were homogenized, and the total RNA was extracted. Samples for the DNA-profiling studies were processed by Agilent microarrays (Agilent Technologies Inc., China) according to the company’s standard procedures.
Microarray data analysis
The microarray image analysis was performed using Agilent G4450AA feature extraction software 9.5, Agilent scan control software, and Agilent GeneSpring software GX9.0. Statistical analysis was performed using an unpaired t-test with a 5% false-discovery rate GeneSpring GX v9.0 or later (Agilent Technologies) to generate significant gene lists. Hierarchical clustering was calculated using Gene Cluster software v3.0. Clustering was performed on both samples and genes, using the fold-change cutoff of 1.5 and the single linkage rule. Individual clusters were further analysed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) 6.8 to determine common and unique functional pathways. A functional annotation clustering tool was used by choosing the default option. The threshold value of the Enrichment Score was set at above 1.0, thereby avoiding the loss of valuable information. The genes were classified as the significantly over-represented biological process group.
Experimental colitis mice model induced by DSS
Male C57BL/6 mice (6–8 weeks old) from Beijing Wei Tong Li Hua Laboratory Animal Technology Co., Ltd) were fed with a standard laboratory diet and kept in an air-conditioned room with a day/night cycle. The mice were fed with water containing different concentrations of DSS for 5 days. The parameters of body-weight loss, diarrhea, and survival were recorded daily for 9 days. Colons were collected from the cecum to the anus after 9 days from the beginning of DSS feeding and the colon length was measured. Colons were evaluated for disease activity index and macroscopic damage as described previously [10]. RNA and protein were extracted.
Immunohistochemistry
Paraffin-embedded colon tissue sections were de-paraffinized and rehydrated through dimethylbenzene and grade concentrations of ethanol solutions. After retrieval of the antigen and blocking endogenous peroxidase activity, the sections were incubated with primary antibody (anti-MGP, 1:500, Abcam ab192396) at 4°C overnight followed by Polymer Detection System reagents (Zhong Shan Jin Qiao company, Beijing, China, PV-9000) and DAB (Zhong Shan Jin Qiao company, Beijing, China company, No ZLI-9018). The slides were analysed by two experienced pathologists in a blinded fashion. The positively stained mucosal cells were counted in 10 randomized fields (×400) with a light microscope (Olympus, Melville, NY) and the average positive rate was taken.
RNA isolation and real-time PCR
Total RNA was isolated from colon tissue of UC patients and healthy controls using Trizol reagent and then treated with DNase I (Thermo Scientific, EN0521) to eliminate possible DNA contamination. RNA was quantified by optical density (A260) and stored at –80°C until use. cDNA was prepared using a Reverse Transcription system (Promega Applied BiosystemsTM, 4359659). Real-time quantitative PCR was performed using a SYBR premix Ex Taq (TaKaRa, Dalian, China) according to the manufacturer’s recommendations. Ninety-six-well plates were used on a BIO-RAD IQ 5 thermocycler. Cycling conditions were as follows: an initial step at 95°C of 5 minutes for enzyme activation, followed by 40 cycles alternating between 10 seconds at 95°C, 30 seconds at 60°C, and a final dissociation step. Obtained Ct values were normalized against â-actin. The relative gene-expression level was determined by using the delta-delta Ct method. The primers used in real-time PCR were as follows: MGP (forward 5'-CGCCTTAGCGGTAGTAACTTTGTG-3′; reverse 5′-CAGGCTTAGAGCGTTCTCGGAT-3′), â-actin (forward 5′-CATGTACGTTGCTATCCAGGC-3′; reverse 5'-CTCCTTAATGTCACGCACGAT-3′).
Western blotting
Samples for Western blotting were prepared using a radio immunoprecipitation assay lysis buffer (Applygen, C1053, Beijing, China) containing proteinase inhibitors. After 20 minutes of incubation on ice, the protein mixture was extracted by centrifuging the sample at 13,000 rpm for 20 minutes. A bicinchoninic acid assay was used to determine the protein concentration before samples were diluted into 2 mg/mL with sodium dodecyl sulfate loading buffer (Auragene, # P003B, Changsha, China). For immunoblot assay, equal amounts of protein from each sample were electrophoresed on a 10% sulfate-polyacrylamide gel and transferred onto a nitrocellulose membrane. Five percent milk-blocked membranes were then incubated with primary antibody in 5% BSA-TBST overnight at 4°C, then washed twice by TBST and incubated with a secondary antibody. After a second wash, results were detected using an Aura ECL chemiluminescence kit (Thermoscientific, No. 32109).
Bioinformatics analysis of the regulatory region and transcriptional factor-binding sites at the 5’ end of the human MGP gene
The first base of the initiation codon ATG starting translating from the MGP gene was marked +1. MGP mRNA and 5’ upstream 2,000-bp sequences aligned with different species were analysed to find the evolutionary conserved regions (ECRs) by an online software ECR browser (http://ecrbrowser.dcode.org/). The evolutional conserved region (chr12: 15038606–15039196) located at 5’ upstream of MGP was predicated as the promoter active region.
Online software NNPP (http://www.fruitfly.org/seq_tools/promoter.html) and Promoter 2.0 (http://www.cbs.dtu.dk/services/Promoter/) were then applied for the prediction of the candidate MGP gene-promoter sequence and transcription-initiation sites. The putative transcriptional factor-binding sites were searched in the upstream region of the human MGP gene by online software TRAP (http://trap.molgen.mpg.de/cgi-bin/trap_form.cgi).
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using a SimpleChIP® Plus Sonication Chromatin IP Kit #56383 from Cell Signaling Technology following the manufacturer’s protocol. In brief, the colon tissues from 3% DSS-induced colitis mice and control mice were cross-linked with 1% formaldehyde for 10 minutes at room temperature, followed by adding glycine to end the cross-linking. The cross-linked chromatin was sonicated to yield fragments of 200–1,000 bp. Ten micrograms of soluble chromatin per sample were aliquoted and diluted with ChIP buffer and 2% of the diluted chromatin for each sample was saved as input. Antibodies against Egr-1 (Santa Cruz Biotechnology sc101033) or IgG were added to the diluted chromatin of each sample for immunoprecipitation overnight at 4°C. Immunocomplexes were captured on the ChIP-specific protein G-magnetic beads, and washed sequentially with a low-salt wash buffer three times and a high-salt wash buffer for the final wash. After washing, the immunocomplexes were eluted by incubation and vortex with 200 ul of elution buffer for 15 minutes at 25°C and then reverse cross-linking for 3 hours at 65°C. The DNA fragments were purified using a DNA purification column in the kit according to the manufacturer’s protocol. The immunoprecipitated DNA samples were analysed by real-time PCR using the MGP-specific ChIP primers: forward 5′-GGGTTGGCACTGAACTAGCA-3′; reverse 5′-CTTCCTCTGTGGGCTTTTGC-3′.
Luciferase-reporter gene assay
The putative promoter region (DNA fragment from position −626 to +164 relative to the transcriptional start site) of the human MGP gene was amplified from the human genome and inserted into the pGL3-basic vector. The sequences and orientations of the MGP promoter fragment inside the pGL3-basic were confirmed by sequencing. The site-directed mutagenesis was achieved using the PCR-overlapping method.
Each pGL3-basic construct (e.g. pGL3-basic-MGP promoter, or pGL3-Egr1 binding-site mutated MGP promoter) and internal control vector pRL-TK were co-transfected with either Egr-1 gene-expression plasmid or empty plasmid pGL3-basic into 293 T cells to test the effect of Egr-1 on MGP promoter-driven luciferase activity. After 30 hours, luciferase-reporter gene activity was detected following dual luciferase assay kit steps. The culture media were discarded and the cells were then washed twice with cold PBS. Then, 100 µl of passive lysis buffer was added to each well and the cells were placed on a shaker shaking slowly for 15–20 minutes at room temperature. After repeated freezing and thawing, cell lysates were transferred to a 0.5-ml centrifuge tube for a brief centrifugation of 10 seconds. After mixing 20 µl of cell lysate with 100 µl of fluorescence luciferase substrate (LAR II), the luminescence value of the liquid scintillation luciferase was measured. The reaction-terminated liquid (100 µl Stop & Glo Reagent) was added and mixed to measure the internal standard Renilla luciferase using a liquid scintillation counter. The relative luciferase activity was measured as firefly luciferase activity normalized by internal Renilla luciferase activity. Each recombinant plasmid and control plasmid was transfected and detected three times.
Results
Extracellular matrix and cell adhesion were the top two gene ontology (GO) terms of our interests based on GO-enrichment analysis of differentially expressed genes involved in UC development
To explore the underlying genes correlating with UC development, gene-expression profiling through DNA microarray was performed in severe UC patients (n = 3) and healthy controls (n = 3). As shown in the heatmap (Figure 1), 510 up-regulated genes and 328 down-regulated genes, with at least 1.5-fold differences in mRNA expression, were identified in UC patients compared to healthy controls. The result of hierarchical clustering clearly classified UC patients and healthy controls into two different groups. It was demonstrated that the transgene expression of UC patients had essential differences compared with healthy controls. GO-enrichment analysis of up-regulated/down-regulated genes showed that extracellular matrix and cell adhesion were the top two GO terms of our interest (Table 1). The top 10 up-regulated/down-regulated genes in UC patients are listed in Table 2. The MGP gene, the extracellular matrix structural constituent, was the fifth highest, with an 8.91-fold increase. Therefore, we speculated that MGP may play an important role in the pathogenesis of UC.
Figure 1.
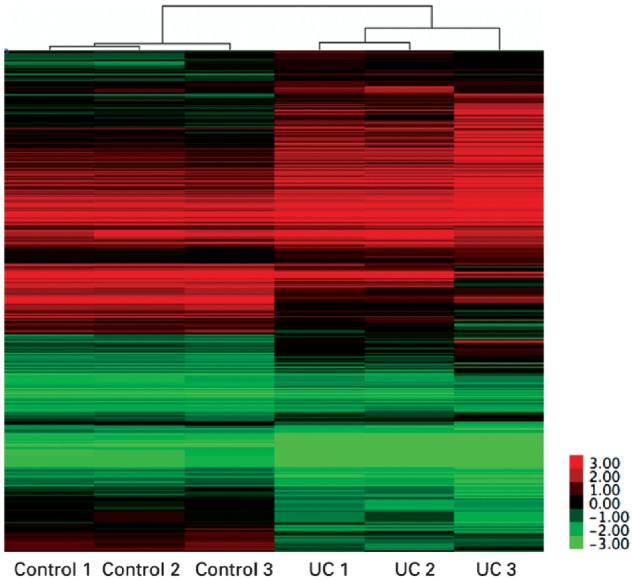
Hierarchical clustering of expression profiles of ulcerative colitis (UC) patients and healthy people (Control). Clustering was based on 510 up-regulated genes and 328 down-regulated genes (fold change >1.5). Colors represent expression levels of each individual gene (Log 2 fold-changes).
Table 1.
Top 10 biological process GO terms of up-regulated/down-regulated pathway genes in patients with ulcerative colitis
| Term | Count (%) | P-value |
|---|---|---|
| Up-regulated pathway genes | ||
| Positive regulation of transcription from RNA polymerase II promoter | 69 (0.0718) | 1.35E-08 |
| Signal transduction | 66 (0.0686) | 4.31E-05 |
| Negative regulation of transcription from RNA polymerase II promoter | 49 (0.051) | 6.17E-06 |
| Extracellular matrix organization | 43 (0.0447) | 3.38E-22 |
| Cell adhesion | 42 (0.0437) | 1.60E-08 |
| Positive regulation of transcription, DNA-templated | 35 (0.0364) | 1.68E-04 |
| Negative regulation of apoptotic process | 34 (0.0354) | 3.51E-05 |
| Negative regulation of transcription, DNA-templated | 33 (0.0343) | 4.41E-04 |
| Angiogenesis | 32 (0.0333) | 2.37E-11 |
| Response to drug | 30 (0.0312) | 5.47E-07 |
| Down-regulated pathway genes | ||
| Oxidation–reduction process | 15 (0.0515) | 6.00E-03 |
| Inflammatory response | 9 (0.0309) | 5.83E-02 |
| Immune response | 9 (0.0309) | 9.46E-02 |
| Transport | 8 (0.0275) | 9.01E-02 |
| Fatty acid beta-oxidation | 7 (0.0240) | 8.22E-06 |
| Nucleosome assembly | 7 (0.0240) | 2.05E-03 |
| Defense response to virus | 7 (0.0240) | 1.01E-02 |
| Transmembrane transport | 7 (0.0240) | 5.44E-02 |
| Metabolic process | 6 (0.0206) | 3.90E-02 |
| Negative regulation of viral genome replication | 5 (0.0172) | 9.56E-04 |
Table 2.
Top 10 up-regulated/down-regulated genes in UC patients
| Gene symbol | Description | Fold change | GO molecular function term |
|---|---|---|---|
| Up-regulated genes | |||
| EGR1 | Early growth response 1 | 18.33 | Sequence-specific DNA binding transcription factor activity |
| FOS | FBJ murine osteosarcoma viral oncogene homolog | 14.48 | Sequence-specific DNA binding transcription factor activity |
| ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif 1 | 9.71 | Metalloendopeptidase activity |
| OGN | Osteoglycin | 9.57 | Protein binding//growth factor activity |
| MGP | Matrix Gla protein | 8.91 | Extracellular matrix structural constituent |
| CYR61 | Cysteine-rich, angiogenic inducer, 61 | 8.9 | Integrin binding |
| SFRP2 | Secreted frizzled-related protein 2 | 7.85 | Fibronectin binding |
| CTGF | Connective-tissue growth factor | 7.58 | Fibronectin binding |
| DES | Desmin | 7.48 | Structural constituent of cytoskeleton |
| ACTG2 | Actin, gamma 2, smooth muscle, enteric | 6.82 | ATP binding |
| Down-regulated genes | |||
| AQP8 | Aquaporin 8 | 0.14 | Water-channel activity |
| FDCSP | Follicular dendritic cell secreted protein | 0.18 | |
| CR2 | Complement component (3d/Epstein Barr virus) receptor 2 | 0.22 | Complement receptor activity |
| SLC38A4 | Solute carrier family 38, member 4 | 0.23 | Amino-acid transmembrane transporter activity |
| SLC9A3 | Solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 | 0.33 | Sodium:hydrogen antiporter activity |
| HAVCR1 | Hepatitis A virus cellular receptor 1 | 0.34 | Receptor activity |
| BMP5 | Bone morphogenetic protein 5 | 0.34 | Cytokine activity//growth factor activity//BMP receptor binding |
| MS4A1 | Membrane-spanning 4-domains, subfamily A, member 1 | 0.34 | Epidermal growth factor receptor binding |
| GUCA2B | Guanylate cyclase activator 2B (uroguanylin) | 0.36 | Calcium-sensitive guanylate cyclase activator activity |
| CXCL13 | Chemokine (C-X-C motif) ligand 13 | 0.39 | Chemokine activity |
The expression of MGP was increased in colonic tissues of UC patients and DSS-induced UC mice, and correlated with the severity of the disease
We verified the role of MGP in 50 UC and 17 healthy subjects. According to the modified Mayo scoring system, 50 UC subjects were divided into four groups: remission group (score ≤2 and no single score >1, n = 7), mild group (score 3–5, n = 13), moderate group (score 6–10, n = 17), and severe group (score 11–12, n = 13). Healthy subjects served as the control group. Demographic data and clinical features are shown in Table 3. The age, sex, clinical classification, and Montreal classification at the baseline did not show significant difference among remission, mild, moderate, and severe groups in UC patients.
Table 3.
Demographic characteristics of ulcerative colitis (UC) patients and healthy controls
| Characteristics | Controls (n = 17) | UC patients |
||||
|---|---|---|---|---|---|---|
| Remission (n = 7) | Mild (n = 13) | Moderate (n = 17) | Severe (n = 13) | P-value | ||
| Age, years (mean ± SD) | 47.1 ± 9.4 | 41.6 ± 14.9 | 42.2 ± 14.3 | 39.0 ± 11.7 | 47.8 ± 11.4 | 0.22a |
| Female gender, n (%) | 8 (47.1) | 5 (71.4) | 5 (38.5) | 8 (47.1) | 4 (30.8) | 0.51b |
| Age of onset, years (mean ± SD) | – | 37.4 ± 15.2 | 35.5 ± 12.3 | 33.3 ± 9.2 | 40.2 ± 10.4 | 0.41a |
aComplete random analysis of variance.
bChi-square test.
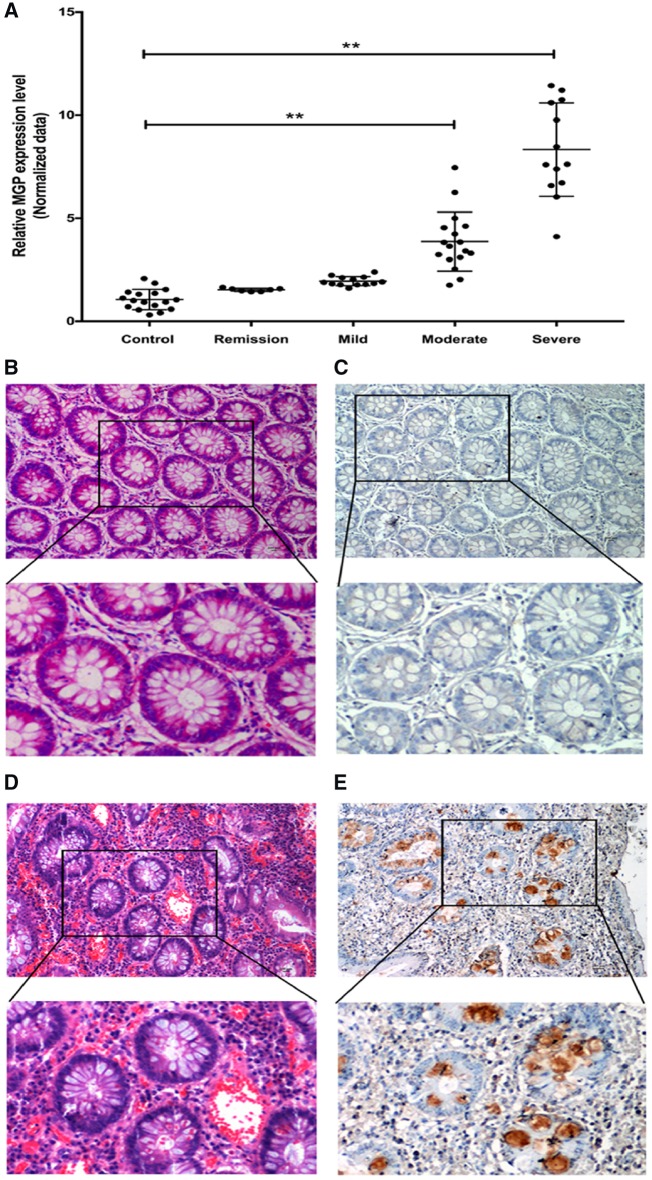
As shown in Figure 2A, the average MGP mRNA level in UC patients gradually increased with the severity of the disease. The MGP mRNA level was significantly higher in the moderate group (relative fold change: 3.87 ± 0.43) and severe group (relative fold change: 8.33 ± 0.47) compared to the control group. However, no significant changes in the MGP mRNA level were found in the remission and mild groups compared to the control group. Immunohistochemistry (IHC) was also performed to detect MGP protein in the colonic mucosa of severe UC patients (n = 9) and healthy controls (n = 6). In the control group, colonic mucosal MGP staining was weak or no obvious MGP staining was found (Figure 2B and C). However, MGP staining was positive, and located in the cytoplasm and extracellular matrix of colonic mucosal epithelial cells and goblet cells in severe UC patients (Figure 2D and E).
Figure 2.
Expression of MGP in colonic mucosa of ulcerative colitis patients and healthy controls based on quantitative RT–PCR and immunohistochemistry (IHC). (A) Relative MGP-expression profile, examined by quantitative RT–PCR, in the control, remission, mild, moderate, and severe groups, respectively. Asterisks (**) denote a significant difference between the healthy control and UC patient samples (P ≤ 0.01). (B) H&E and (C) IHC stains of MGP in colonic mucosa of healthy controls (upper: magnification ×100; lower: magnification ×200). (D) H&E and (E) IHC stains of MGP in colonic mucosa of UC patients (upper: magnification ×100; lower: magnification ×200).
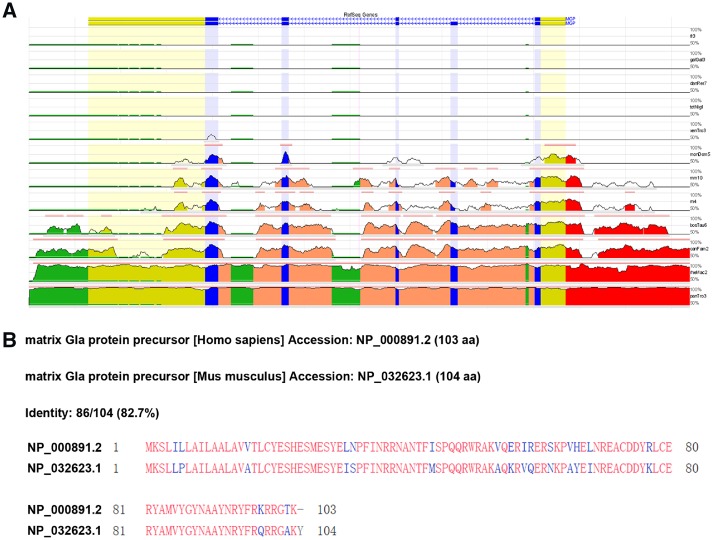
Moreover, the MGP mRNA, protein sequence, and 5’ upstream 2,000-bp sequences aligned with different species were analysed to find the ECRs using an online software ECR browser (http://ecrbrowser.dcode.org/). As a result, we found that the MGP gene-coding region and protein sequence were conserved across the different species including human and mouse (Figure 3). An evolutional conserved region between human and mouse (human hg19 chr12: 15038606–15039196, mouse mm10: chr6: 136872291–136876911) located at 5’ upstream of the MGP was also found and predicated as a putative promoter active region.
Figure 3.
Conservation analysis of human MGP gene mRNA, protein sequence, and upstream regulatory region. (A) Conservation analysis of human MGP gene mRNA and upstream regulatory region with other vertebrates. The analysis software evolutionary conserved regions (ECRs) were used. The ECR browser is a dynamic graphical interface that allows users to visualize and analyse ECRs in genomes of sequenced species. The x-axis represents the positions of bases in the genomes. The y-axis represents the percentage (%) identity between the bases of the aligned genomes at a particular position. The base genome is hg19, human and the selected reference genomes are as follows: fr3, fugu; galGaI3, chicken; danRer7, zebrafish; tetNig1, tetraodon; xenTro2, frog; monDom4, opossum; mm9, mouse; rn4, rat; bosTau6, cow; camFam2, dog; rheMac2, rhesus macaque; panTro3, chimpanzee. (B) Protein-sequence alignment of human and mouse MGP using NCBI online software ECR browser (http://ecrbrowser.dcode.org/).
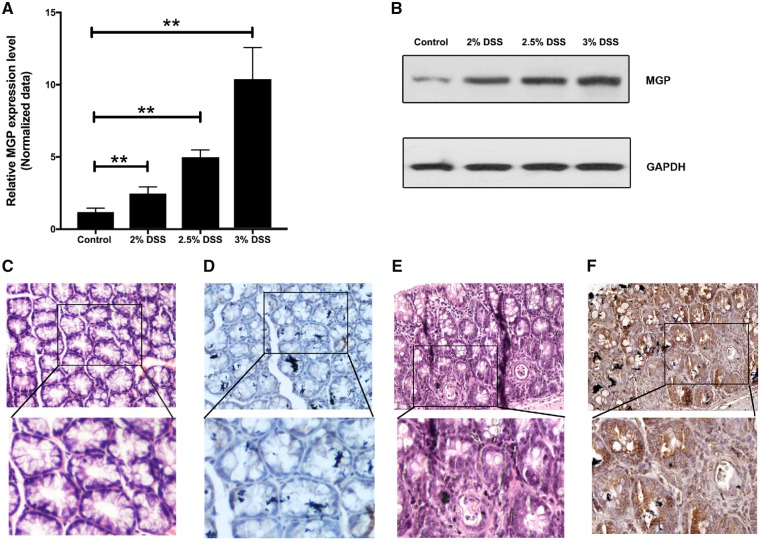
The above information indicates that MGP function and regulation can be studied in a mouse model. We then generated mouse colitis models with serial concentrations of DSS (2%, 2.5%, and 3%) and used these models to detect the MGP-expression profile. The relative fold change of the MGP mRNA level was 2.21 ± 0.06 in the 2% DSS group (n = 5), 4.91 ± 1.10 in the 2.5% DSS group (n = 9), and 10.7 ± 0.56 in the 3% DSS group (n = 6), respectively, compared with the control group (n = 5) (Figure 4A). Consistently, the levels of MGP protein in the DSS colitis mice were also obviously increased with the increase in DSS concentration (Figure 4B) as compared with that in the control group. IHC was also performed to detect MGP protein in the colon tissue of DSS colitis mice and control mice. Similar staining patterns with the colonic mucosa of UC patients were also observed in the colon tissue of DSS colitis mice (Figure 4C–F).
Figure 4.
Expression of MGP in colonic tissue of dextran sodium sulfate (DSS) model mice and normal controls based on quantitative RT–PCR, Western blot, and immunohistochemistry (IHC). Relative MGP mRNA (A) and protein levels (B) in colonic tissue of DSS model mice treated with different concentrations of DSS and controls. Control group (n = 5), 2% DSS group (n = 5), 2.5% DSS group (n = 9), and 3% DSS group (n = 6). The data were represented with mean ± standard error. Asterisks (**) denote a significant difference between the control and different DSS groups (P ≤ 0.01). (C) H&E and (D) IHC stains of MGP in colonic mucosa of normal controls (upper: magnification ×100; lower: magnification ×200). (E) H&E and (F) IHC stain of MGP in colonic mucosa of 2.5% colitis model mice (upper: magnification ×100; lower: magnification ×200).
The increase in MGP gene expression was regulated by Egr-1
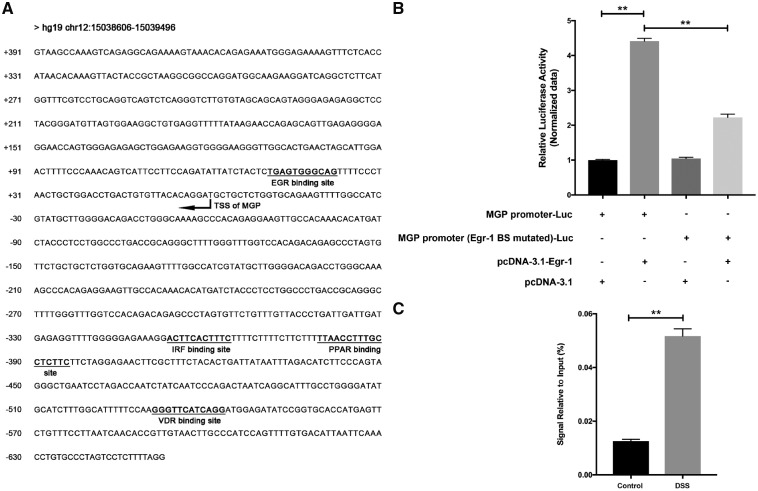
To identify the upstream regulator of MGP gene expression, the putative transcriptional factor-binding sites were searched in the upstream region of the human MGP gene using online software TRAP (http://trap.molgen.mpg.de/cgi-bin/trap_form.cgi). As a result, we found a conserved binding site for Egr-1, VDR, PPAR, etc. in the upstream region of the human MGP gene (Figure 5A). Gene-expression microarray results showed that the Egr-1 gene has significantly higher expression in UC patients than in healthy controls (18.33-fold). Therefore, Egr-1 was selected to be further validated.
Figure 5.
Promoter and upstream regulator analysis of MGP gene expression. (A) Part of human MGP gene mRNA and upstream region (hg19 chr12: 15038606–15039496) was selected for transcriptional factor-binding site analysis. Transcription start site of MGP was marked based on MGP mRNA sequence. (B) Relative luciferase activity was measured on different combinations of luciferase-reporter gene construct and Egr-1 expression plasmid or empty control (with: +, without: –). Asterisks (**) denote a significant difference between the control and different treatment groups (P ≤ 0.01). (C) Chromatin immunoprecipitation (ChIP) analysis of Egr-1 binding on MGP promoter in dextran sodium sulfate (DSS) induced colitis model mice. Asterisks (**) denote a significant difference between the control and DSS groups (P ≤ 0.01).
The putative promoter region (DNA fragment from position −626 to +164 relative to the transcriptional start site) of the human MGP gene was amplified from the human genome by PCR and inserted into the pGL3-basic plasmid. The luciferase-reporter gene plasmid with the MGP promoter (MGP promoter-luc) was co-transfected with the Egr-1 expression plasmid (Egr-1 group) or negative control plasmid (control group) to examine the potential effect of Egr-1 on the MGP promoter-driven luciferase expression. The results showed that the activity of luciferase in the Egr-1 group was 4.41-fold higher than that of the control group (P < 0.001). As a comparison, when the luciferase-reporter gene plasmid with the Egr-1 binding-site mutated MGP promoter (Egr-1 mutated MGP promoter-luc) was co-transfected with the Egr-1 expression plasmid, the enhancement of Egr-1 to the activity of the luciferase was decreased. These results suggest that transcription factor Egr-1 can enhance the promoter activity of the MGP gene (Figure 5B).
In order to further testify whether Egr-1 can bind to the promoter region of MGP, ChIP assay was performed using DSS-induced colitis mice. The results of ChIP assay showed that the enrichment of Egr-1 on the MGP promoter region in DSS-induced colitis mouse was significantly higher than that in the control group (P < 0.01, Figure 5C). These results confirmed the binding of transcription factor Egr-1 to the MGP promoter.
Discussion
The present study explored the gene-expression profiling from UC patients to provide clinically relevant special genes and pathways for UC, of which the extracellular matrix and cell adhesion were the top two GO terms that captured our interest. Among the differently expressed genes, the level of the extracellular matrix gene MGP expression was 8.91 times higher in UC patients than in the healthy controls. Although these results suggested the possibly important role of MGP in UC, we need to further expand the sample-size verification. Therefore, the following verification experiments identified the MGP as influencing the protein induced in UC and DSS colitis models. The severity of UC patients correlated positively with the higher mRNA expression of MGP. As in UC patients, DSS-induced colitis had a similar trend in the increase in MGP mRNA expression by different concentrations of DSS and the severity of colonic inflammation. MGP localized in the epithelium cells and global cells in UC patients and animal models. In addition, we found Egr-1, serving as an upstream protein, can regulate the transcription of MGP.
The pathogenesis of IBD is not yet fully understood. Multiple factors may be involved in the pathogenesis, including genetics and environmental, immunological, and intestinal microflora. mRNA analysis by microarrays can investigate simultaneously thousands of transcripts and genes. In general, studies showed that the changed genes in IBD were mainly involved in the immune response, cell adhesion, barrier integrity, and tissue remodeling [11]. The results presented here were from an mRNA array analysis of a very well-controlled material of a colonic mucosal sample from severe active UC and healthy controls. In this study, there were 510 up-regulated genes and 328 down-regulated genes, with at least 1.5-fold differences in mRNA expression. The related pathways were involved in transcription regulation, signal transduction, extracellular matrix organization, cell adhesion, negative regulation of the apoptotic process, and angiogenesis.
Recently, there has been an increasing interest in the role of the extracellular matrix and cell adhesion [12]. One study showed that MGP might have a protective role in the therapy of IBD [13]. In this study, we found that the extracellular matrix gene MGP-expression level was significantly higher in UC patients. MGP is an insoluble, small protein, with a molecular weight of only 14.7 kD, which belongs to a tissue-derived vitamin K-dependent protein family. MGP is a secreted protein and has a loci in the promoter sequence binding with vitamin D receptor [14]. Shiraishi et al. showed that vitamin K has a protective effect against DSS colitis, which is associated with IL6 down-regulation [15]. Several studies have revealed that the vitamin K status appeared lower in CD patients [16]. Therefore, MGP as a protein associated with vitamin K should be given much more attention.
Most of the research on MGP has been associated with physiological angiogenesis, tissue mineralization, and vascular calcification [17–19]. It was reported that MGP might be synthesized in the majority of human immune system cells involved in innate or adaptive immune responses [20]. Therefore, MGP might act as a mediator linking inflammation and calcification events as a secreted protein [21, 22]. Mesenchymal stromal cells (MSCs)-secreted MGP could ameliorate the clinical and histopathological severity of colonic inflammation, with an obvious inhibiting action on the number of T cells and degree of cytokine production in peritoneal lavage fluid and colon tissues of colitis mice [11]. Down-regulation of MGP expression significantly weakened this curative effect [13]. However, the expression and influence of MGP on patients with IBD are still to be elucidated.
In this study, we found higher mRNA and protein expression of MGP in patients with UC and DSS mice models, which indicated that MGP might be involved in the pathogenesis of IBD. MGP mRNA expression was significantly correlated with inflammatory markers. Bioinformatics analysis indicated that the putative transcription binding site of MGP was Egr-1. Moreover, the mRNA-expression microarray in UC patients showed that Egr-1 gene was significantly more highly (18.33-fold) expressed than in healthy controls. Electrophoretic mobility shift assay along with ChIP assay confirmed a specific Egr-1 overlapping site spanning in the MGP minimal promoter. The regulation of MGP transcription by Egr-1 has been shown to play a critical role in the expression of MGP. It was reported that Egr-1 regulated epithelial-barrier disruption in human intestinal epithelial cells [23].
One of the limitations of this study was the small sample size of the microarray analysis. We selected three representative samples that had the most severe clinical symptoms for microarray analysis at first. We then verified our findings from microarrays for altered gene expression with larger samples (50 samples). Although these results suggested the possibly important role of MGP in UC, we need to expand the sample size for further verification. Another limitation of this study was that, although we found the phenomena of higher MGP expression in UC, the function of the MGP gene in UC is still unknown. Based on the characteristics of MGP with being synthesized and secreted in the extracellular matrix and the characteristics of affecting immune cells, we speculate that MGP plays a protective role in the pathogenesis of IBD. Further study is certainly required to test our hypothesis.
Another fact is that MGP has a loci in the promoter sequence binding with vitamin D receptors [14]. Vitamin D-receptor signaling plays an important role in the process of intestinal inflammation [24]. Several studies have validated the protective effect of vitamin D-receptor signaling in the destruction of intestinal epithelial-barrier function [24, 25]. Moreover, although a recent study showed that the abundant MSC-derived MGP participated in the therapeutic mechanisms for MSCs treating CD [13], the expression and effect of MGP on CD patients remain to be clarified. Our future studies should be focused on investigating the relationship between MGP and vitamin D-receptor signaling in IBD, and also MGP expression and effects on CD.
In summary, here we reported the expression of MGP in UC patients and DSS mice, and the upstream regulator mechanism. These results will help us to understand and uncover the bio-functions of the MGP gene involved in the pathogenesis of IBD.
Authors’ contributions
X.Y.D. performed the majority of the experiments and drafted the manuscript. M.X.W. offered help during the experiments. H.M.Z. participated in the collection of the human material and clinical data. H.L. served as a scientific adviser. J.M.Q. and H.Y. conceived of the experiments, and reviewed and approved the final version of the manuscript.
Funding
This study was supported by the National Nature Science Foundation of China [No. 81570505], Health Research & Special Projects Grant of China [No. 201002020], Ministry of Science and Technology of China [No. 2015CB943203], and CAMS Innovation Fund for Medical Sciences [No. 2016-I2M-3–001].
Acknowledgements
The authors would like to thank Dr Hong-Ying Wang for her consistent instruction, and we would like to express our gratitude to Dr Zhen-Ya Li, Zhi-Gang Wang, and Ya-Shu Zhu for their contribution to the data analysis.
Conflicts of interest
None declared.
References
- 1. Navaneethan U, Zhu X, Lourdusamy D. et al. Colorectal cancer resection rates in patients with inflammatory bowel disease: a population-based study. Gastroenterol Rep (Oxf) 2018;6:263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Knights D, Lassen KG, Xavier RJ.. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut 2013;62:1505–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Horvath B, Liu G, Wu X. et al. Overexpression of p53 predicts colorectal neoplasia risk in patients with inflammatory bowel disease and mucosa changes indefinite for dysplasia. Gastroenterol Rep (Oxf) 2015;3:344–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eguchi R, Karim MB, Hu P. et al. An integrative network-based approach to identify novel disease genes and pathways: a case study in the context of inflammatory bowel disease. BMC Bioinformatics 2018;19:264.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xie D, Zhang Y, Qu H.. Crucial genes of inflammatory bowel diseases explored by gene expression profiling analysis. Scand J Gastroenterol 2018;53:685–91. [DOI] [PubMed] [Google Scholar]
- 6. Ahuja V, Subodh S, Tuteja A. et al. Genome-wide gene expression analysis for target genes to differentiate patients with intestinal tuberculosis and Crohn's disease and discriminative value of FOXP3 mRNA expression. Gastroenterol Rep 2016;4:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mcgovern DP, Kugathasan S, Cho JH.. Genetics of inflammatory bowel diseases. Gastroenterology 2015;149:1163–76.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jostins L, Ripke S, Weersma RK. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D'Haens G, Sandlborn WJ, Feagan BG. et al. A review of activity indices and efficacy end points for clinical trials of medical therapy in adults with ulcerative colitis. Gastroenterology 2007;132:763–86. [DOI] [PubMed] [Google Scholar]
- 10. Zhao H, Zhang H, Wu H. et al. Protective role of 1, 25(OH)2 vitamin D3 in the mucosal injury and epithelial barrier disruption in DSS-induced acute colitis in mice. BMC Gastroenterol 2012;12:57.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu JZ, van Sommeren S, Huang H. et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nishimoto SK, Nishimoto M.. Matrix Gla protein binds to fibronectin and enhances cell attachment and spreading on fibronectin. Int J Cell Biol 2014;2014:807013.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng Y, Liao Y, Huang W. et al. Mesenchymal stromal cells-derived matrix Gla protein contribute to the alleviation of experimental colitis. Cell Death Dis 2018;9:691.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cancela L, Hsieh CL, Francke U. et al. Molecular structure, chromosome assignment, and promoter organization of the human matrix Gla protein gene. J Biol Chem 1990;265:15040–8. [PubMed] [Google Scholar]
- 15. Shiraishi E, Iijima H, Shinzaki S. et al. Vitamin K deficiency leads to exacerbation of murine dextran sulfate sodium-induced colitis. J Gastroenterol 2016;51:346–56. [DOI] [PubMed] [Google Scholar]
- 16. Fabisiak N, Fabisiak A, Watala C. et al. Fat-soluble vitamin deficiencies and inflammatory bowel disease: systematic review and meta-analysis. J Clin Gastroenterol 2017;51:878–89. [DOI] [PubMed] [Google Scholar]
- 17. Barrett H, O’Keeffe M, Kavanagh E. et al. Is Matrix Gla protein associated with vascular calcification? A systematic review. Nutrients 2018;10:E415.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khoshniat S, Bourgine A, Julien M. et al. Phosphate-dependent stimulation of MGP and OPN expression in osteoblasts via the ERK1/2 pathway is modulated by calcium. Bone 2011;48:894–902. [DOI] [PubMed] [Google Scholar]
- 19. Luo G, Ducy P, Mckee MD. et al. Spontaneous calcification of arteries and cartilage in mice lacking matrix GLA protein. Nature 1997;386:78–81. [DOI] [PubMed] [Google Scholar]
- 20. New SE, Goettsch C, Aikawa M. et al. Macrophage-derived matrix vesicles: an alternative novel mechanism for microcalcification in atherosclerotic plaques. Circ Res 2013;113:72–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Viegas CSB, Costa RM, Santos L. et al. Gla-rich protein function as an anti-inflammatory agent in monocytes/macrophages: implications for calcification-related chronic inflammatory diseases. PLoS One 2017;12:e0177829.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rios-Arce ND, Collins FL, Schepper JD. et al. Epithelial barrier function in gut-bone signaling. Adv Exp Med Biol 2017;1033:151–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Choi HJ, Kim J, Park SH. et al. Pro-inflammatory NF-kappaB and early growth response gene 1 regulate epithelial barrier disruption by food additive carrageenan in human intestinal epithelial cells. Toxicol Lett 2012;211:289–95. [DOI] [PubMed] [Google Scholar]
- 24. Mousa A, Misso M, Teede H. et al. Effect of vitamin D supplementation on inflammation: protocol for a systematic review. BMJ Open 2016;6:e010804.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu W, Chen Y, Golan MA. et al. Intestinal epithelial vitamin D receptor signaling inhibits experimental colitis. J Clin Invest 2013;123:3983–96. [DOI] [PMC free article] [PubMed] [Google Scholar]