Summary
Developmental disorder Floating-Harbor Syndrome (FHS) is caused by heterozygous truncating mutations in SRCAP, a gene encoding a chromatin remodeler mediating incorporation of histone variant H2A.Z. Here, we demonstrate that FHS-associated mutations result in loss of SRCAP nuclear localization, alter neural crest gene programs in human in vitro models and Xenopus embryos, and cause craniofacial defects. These defects are mediated by one of two H2A.Z subtypes, H2A.Z.2, whose knockdown mimics and whose overexpression rescues the FHS phenotype. Selective rescue by H2A.Z.2 is conferred by one of the three amino acid differences between the H2A.Z subtypes, S38/T38. We further show that H2A.Z.1 and H2A.Z.2 genomic occupancy patterns are qualitatively similar, but quantitatively distinct, and that H2A.Z.2 incorporation at AT-rich enhancers and expression of their associated genes are both sensitized to SRCAP truncations. Altogether, our results illuminate the mechanism underlying a human syndrome and uncover selective functions of H2A.Z subtypes during development.
Keywords: human disorder, enhancers, neural crest, craniofacial, genetic mutation, histone variant, SRCAP, chromatin remodeler, development, epigenetics, H2A.Z
Graphical Abstract

Introduction
Floating-Harbor Syndrome (FHS) is a developmental disorder caused by heterozygous mutations in SRCAP, a broadly expressed ATP-dependent chromatin remodeler, which mediates the incorporation of a histone variant, H2A.Z, into nucleosomes (Hood et al. 2012; White et al. 2010; Nikkel et al. 2013). The mutation of SRCAP in FHS results in distinctive craniofacial features (Figure 1A), such as a triangular facial shape with a reduced jaw (often accompanied by cleft lip/palate) and a large nose as well as other abnormalities, including bone growth delay, gastrointestinal anomalies, high voice pitch and communicative language deficits, and decreased stature and weight during early childhood (Fryns et al. 1996; Patton et al. 1991; Hersh et al. 1998; Nikkel et al. 2013). All FHS cases described to date are associated with heterozygous truncating mutations in one allele of SRCAP, clustering just upstream of the C-terminal AT-hooks (Figure 1B and C) (Hood et al. 2012). The molecular and developmental mechanism of FHS has not been studied and it remains unclear how the loss of the C-terminal region affects SRCAP function and results in FHS.
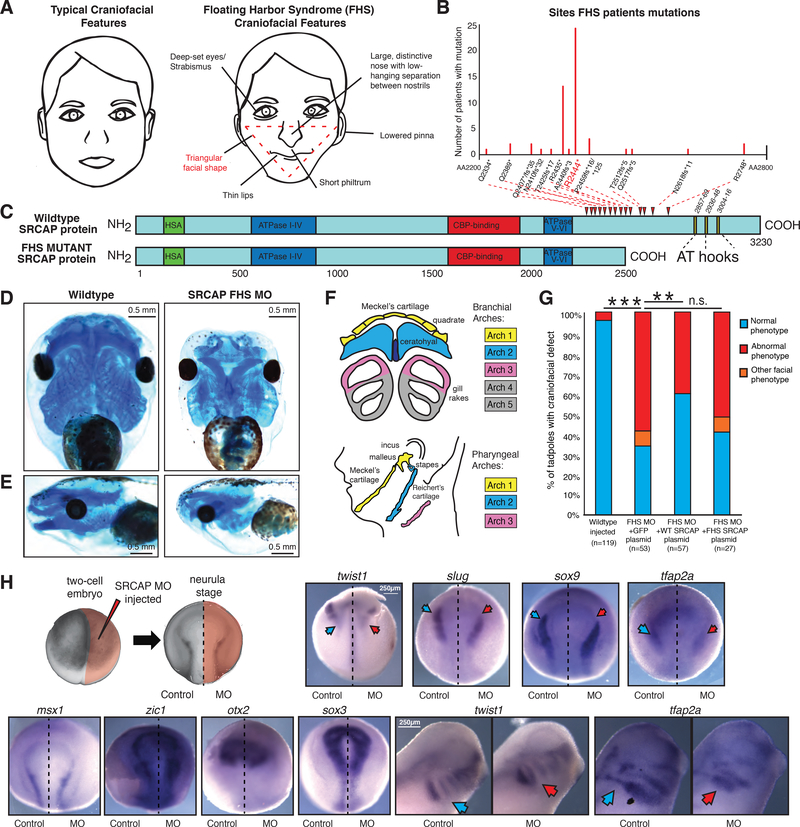
Figure 1. Recapitulating Floating-Harbor Syndrome in Xenopus laevis affects neural crest derived craniofacial structures.
(A) Graphical representation of typical (left) and Floating-Harbor Syndrome (right) craniofacial features. Characteristic triangular facial shape (most characteristic and diagnostic feature) demarcated in red.
(B) Plot of frequencies of SRCAP mutations in Floating-Harbor Syndrome probands. The x-axis goes from amino acid 2200 to 2800, and each mutation is denoted on this axis. The most frequent mutation R2444* is highlighted in red.
(C) Schematic of wild type and the FHS truncated SRCAP proteins. The hot spot for FHS truncating mutations is indicated by red arrowheads. Protein domains are annotated with HSA in green, ATPase in blue, CBP-binding in red, AT-hooks in yellow. The amino acid scale is below the schematic.
(D-E) Ventral (D) and side (E) view of X. laevis head with craniofacial cartilage stained with Alcian blue at stage 40, Wildtype (mock injected) and SRCAP FHS morphant (5.0 μM MO). 0.5 mm scale bar shown. Animals from n = 3 biologically independent experiments.
(F) Diagram of homology between branchial arch structures in X. laevis to pharyngeal arches of the developing human face, with key homologous structures highlighted in matching colors.
(G) Blinded quantification of rescue of characteristic craniofacial phenotype with co-injection of FHS MO and 200pg pB CAG GFP-FLAG, or pB CAG WT-SRCAP-GFP-FLAG, or pB CAG FHS-SRCAP-GFP-FLAG. Statistical test used was Fisher’s Exact Test (FET). FET p-value < 0.005 = **, FET p-value < 10e-5 = **, FET p-value > 0.05 = n.s. Animals from n=4 independent experiments.
(H) Diagram of injection set up at two-cell stage and of asymmetrical FHS SRCAP MO expression at neurula stage. In situ hybridization at neurula stage for neural crest specification genes twist1, slug, and sox9 (abnormal phenotype in 9/11, 5/5, 5/6 embryos respectively), for neural crest induction and specification gene tfap2a (abnormal phenotype in 11/12 embryos), for neural plate border maintenance genes zic1 and msx1 (abnormal phenotype in 1/10, 0/6 embryos respectively), for early neural patterning gene otx2 and for neural plate gene sox3 (abnormal phenotype in 0/5, 1/5 embryos respectively), with 5.0 μM FHS MO injected on right side only, control on left. Ventral side shown, with anterior at top and posterior at bottom. 250μm scale bar shown for neurula images. In situ hybridization at tailbud stage (stage 28), with each pair of images from same animal (control image flipped in vertical plane). In situ probes twist1 and tfap2a (abnormal phenotype in 8/10, 11/13 embryos respectively) visualize neural crest migration. 250μm scale bar shown for tailbud images. Blue arrows denote normal gene expression pattern, red arrows denote impact on expression for FHS morphant. Image brightness and color adjusted to optimize visualization.
The primary molecular function of SRCAP and its associated remodeling complex is to catalyze replacement of the canonical H2A-H2B dimer with the H2A.Z-H2B dimer (Liang et al. 2016; Ruhl et al. 2006; Wong et al. 2007; Hong et al. 2014; Zhou et al. 2016). The presence of histone variant H2A.Z (instead of canonical H2A) affects nucleosome dynamics at cis-regulatory regions such as enhancers and promoters and facilitates access of regulatory complexes to the genome during transcriptional activation, priming or repression (Calo & Wysocka 2013; Ku et al. 2012; Maze et al. 2014; Buschbeck & Hake 2017). During vertebrate evolution, the ancestral H2A.Z gene underwent a duplication event, resulting in two distinct paralogs, H2AFZ and H2AFV encoding subtypes H2A.Z.1 and H2A.Z.2, which are identical except for substitutions at three amino acid residues (Dryhurst et al. 2009). Although H2A.Z.1 and H2A.Z.2 are often considered collectively as H2A.Z, evidence suggests that the two subtypes may have specialized expression patterns and functions (Dryhurst et al. 2009; Eirín-López et al. 2009; Vardabasso et al. 2015; Vardabasso et al. 2016; Dunn et al. 2017; Maze et al. 2014). Ancestral H2A.Z-depositing chromatin remodeler SWR1 itself also underwent a duplication event in vertebrates, leading to emergence of two proteins capable of H2A.Z incorporation, p400 and SRCAP (Pünzeler et al. 2017; Bonisch & Hake 2012).
To understand the molecular and cellular underpinnings of FHS, we introduced disease-associated SRCAP truncations into Xenopus embryos and human embryonic stem cell derived CNCCs. We find that frogs expressing truncated SRCAP have a craniofacial phenotype that affects structures homologous to those malformed in humans with FHS, and consistent with dysfunction of cranial neural crest cells (CNCCs), the major cell type of origin for the developing face. The analysis of human CNCCs bearing heterozygous SRCAP truncations suggests that FHS is a consequence of the disruption of SRCAP nuclear localization and interaction with chromatin, and a resulting perturbation of neural crest gene expression programs. Surprisingly, gain and loss of function analyses in frog embryos revealed that the effects of SRCAP on craniofacial development are selectively mediated by one of the two H2A.Z subtypes present in vertebrates, H2A.Z.2. This specificity is not associated with tissue-restricted H2A.Z.2 expression, but instead can be explained by the relatively higher ratio of H2A.Z.2 to H2A.Z.1 at enhancers versus promoters, and sensitization of AT-rich enhancers and their associated genes (which include many neural crest migration and facial morphogenesis regulators) to SRCAP FHS truncations. Remarkably, a single divergent amino acid at position 38 endows H2A.Z.2 with the ability to rescue the FHS phenotypes, and this S38/T38 substitution has previously been implicated in mediating structural polymorphisms and in vivo differences in nucleosomal dynamics between H2A.Z.1 and H2A.Z.2 (Horikoshi et al. 2013). Thus, by elucidating molecular and cellular mechanisms underlying FHS, a poorly understood human syndrome, we have uncovered developmental, functional, and evolutionary specialization of the H2A.Z isoforms.
Results
SRCAP truncations perturb craniofacial cartilage development in X. laevis
To investigate how SRCAP truncations perturb morphogenesis and lead to FHS, we used the frog, Xenopus laevis. We designed a morpholino oligonucleotide (MO) that blocks the final splice-acceptor site of X. laevis SRCAP, leading to intron incorporation and C-terminal truncation of the SRCAP protein corresponding in size to FHS truncations in humans (Figure 1C, Fig. S1B–E). Injection of this MO into embryos at the two-cell stage resulted in reproducible craniofacial cartilage abnormalities in the derived morphant tadpoles, as compared to control injected embryos (Figure 1D). We found malformations in both Meckel’s and ceratohyal cartilages, a reduction in gill rake cartilage, and a change in the orientation of the eyes from facing laterally to facing ventrally. The resultant characteristic concave or triangular craniofacial shape (Figure 1E; quantifications of distinct craniofacial phenotypes are shown in Figure S1K) was further evident from optical projection tomography (OPT) of dissected cartilages (Figure S1H). The severity of craniofacial phenotypes was SRCAP MO dosage dependent (Figure S1G, S1L–M) with the characteristic ceratohyal deformation occurring at levels of MO where wild type full-length SRCAP is still present in the embryos, mimicking the human disease setting (Figure S1C).
Meckel’s cartilage and the ceratohyal cartilage are derived from the first and second branchial arches, structures homologous to the first and second pharyngeal arches, from which the lower face and jaw form in humans, whereas gill rakes have homology to the third and fourth pharyngeal arches, which contribute to formation of the hyoid bone and laryngeal thyroid cartilage, structures involved in the mechanics of speech (Figure 1F) (Jones & Smith 2008). Developmental defects in these structures may help to explain a number of additional FHS manifestations including hearing loss, other ear-nose-throat defects, dental problems, and voice changes. We noted that ~ 50% of the FHS morphant tadpoles had gut coiling defects (Figure S1I–J), which may be related to gut motility problems observed in a subset of FHS patients, and potentially result from enteric neural crest defects (Nikkel et al. 2013). For the rest of this study, however, we will focus on craniofacial aspects of the disease.
To validate the specificity of the observed defects we performed rescue experiments in which the FHS MO was co-injected with plasmids encoding GFP, WT-SRCAP, or FHS-SRCAP. Co-injection of FHS MO with WT SRCAP cDNA, but not with truncated FHS SRCAP or GFP cDNA resulted in significant rescue of the observed craniofacial phenotypes (Fig. 1G, S1N). To control for off-target effects, we designed a second splice-blocking MO that similarly promotes last intron inclusion and a decrease in full-length SRCAP protein (Fig. S1D, S1Q) which recapitulated the same craniofacial phenotypes (Figure S1O–P).
FHS morphant embryos show changes in neural crest gene expression
Our observed craniofacial phenotypes were reminiscent of manifestations of cranial neural crest defects previously described in Xenopus and other vertebrates (Calo et al. 2018; Dubey & Saint-Jeannet 2017; Devotta et al. 2016; Luo et al. 2003; Kong et al. 2014; Hu et al. 2014). To explore the possibility that observed cartilage malformations are a consequence of cranial neural crest defects, we asymmetrically injected one cell of two-cell stage embryos with the SRCAP MO and a fluorescent tracer and carried out RNA in situ hybridizations with selected markers (Figure 1H). At neurula stage, we found that genes associated with neural plate border territory specification (e.g. zic1, msx1), neural plate patterning (e.g, sox3) or neural ectoderm patterning (e.g. otx2) were not altered (Milet & Monsoro-Burq 2012; Pegoraro & Monsoro-Burq 2013; Bhat et al. 2013; Sargent 2006; Sinner et al. 2006). However, expression of tfap2a, a transcription factor that plays a role Xenopus neural crest induction and later specification was reduced on the MO injected side (Figure 1G) (Bosse 2010; Luo et al. 2003). Furthermore, we observed that expression of a number of genes implicated in neural crest specification, which act in the same signaling cascades with tfap2a, including twist1, slug, and sox9, was diminished on the MO-injected side, and these effects were especially evident in the anterior part of the neural plate border (Figure 1H) (Pegoraro & Monsoro-Burq 2013; Luo et al. 2003). At later stages (stage 28) when neural crest cells migrate into the branchial arches, we observed a reduction of twist1 and tfap2a staining in the ventral parts of the branchial arches on the MO-treated side, consistent with defective or delayed neural crest migration (Figure 1H) (Betancur et al. 2010; Lander et al. 2013). These results suggest that craniofacial malformations observed in FHS may originate from defects in neural crest specification and/or migration.
FHS mutations perturb nuclear localization of SRCAP
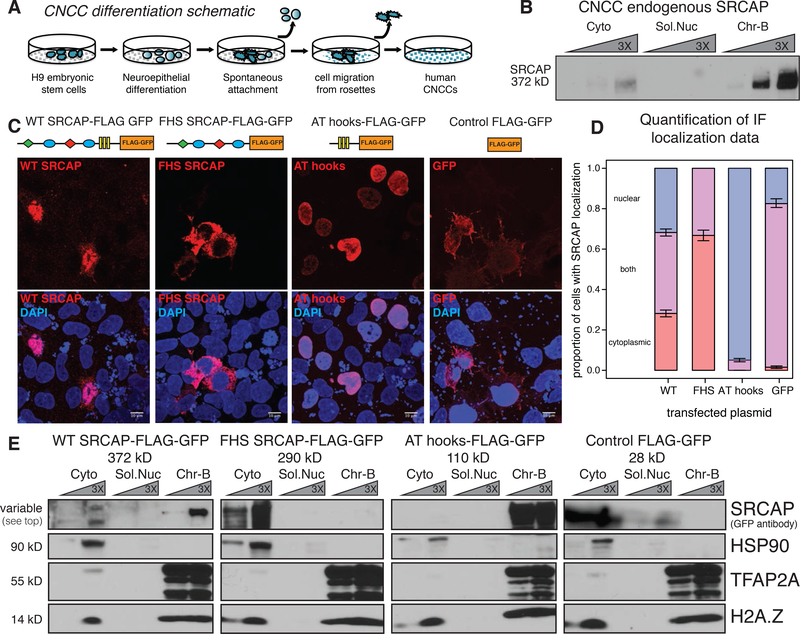
To study the molecular consequences of SRCAP truncations in the context of CNCCs, we turned to an in vitro differentiation model (Figure 2A) (Rada-Iglesias et al. 2012; Bajpai et al. 2010; Prescott et al. 2015; Calo et al. 2018). By biochemical fractionation, we first confirmed that in human CNCCs, endogenous wild type (WT) SRCAP is predominantly bound to chromatin (Figure 2B). Overexpression of GFP-Flag epitope-tagged versions of WT SRCAP, FHS SRCAP and the C-terminal AT hook domain in CNCCs revealed that overexpressed WT SRCAP was predominantly nuclear and chromatin-bound. By contrast, truncated FHS SRCAP, while stable, was largely excluded from the nucleus and was present in the cytoplasm (Figure 2C–E, Fig. S2A). Interestingly, the C-terminal AT hooks which are absent in FHS, were almost exclusively nuclear and chromatin-bound (Figure 2C–E, Fig. S2A). These observations were recapitulated in other cell lines, including human embryonic kidney cells (HEK293T), suggesting that the observed localization of SRCAP reflects the intrinsic properties of the protein domains rather than being determined by cell type specific features of CNCCs (Figure S2B–C). Analysis of the SRCAP sequence revealed that the SRCAP AT hooks are not only putative DNA binding domains, but also contain three strong nuclear localization signals that are absent upon truncation (Figure S2D–F) (Kosugi et al. 2009). This suggests that FHS mutations may perturb nuclear localization of SRCAP and that the AT-hooks, which are uniquely present in wild type vertebrate SRCAP among the SWR1-related proteins (Figure S1A), harbor signals important both for nuclear localization and chromatin association.
Figure 2. Nuclear localization and chromatin association defects of FHS SRCAP protein.
(A) Schematic of differentiation of hESC to CNCCs.
(B) Biochemical fractionation of human CNCCs with immunoblotting against endogenous SRCAP protein (Kerafast antibody). 1X, 3X, and 9X protein dilutions. Cyto – Cytoplasmic fraction, Sol.Nuc – soluble nuclear fraction, Chr-B- chromatin bound fraction. Predicted protein size on left.
(C) Confocal microscopy of anti-GFP immunofluorescence staining of CNCCs overexpressing FLAG-GFP tagged WT SRCAP, FHS mutant SRCAP, SRCAP AT hooks, and FLAG-GFP alone (DAPI – Blue; tagged protein – Red). Bottom panel shows merged image with DAPI staining. 10μm scale bar.
(D) Quantification of cellular localization of overexpression proteins with nucleus defined by DAPI signal; 95% confidence intervals from quasibinomial glm model.
(E) Biochemical fractionation of CNCCs overexpressing FLAG-GFP tagged WT SRCAP, FHS mutant SRCAP, SRCAP AT hooks, FLAG-GFP alone with GFP antibody for SRCAP or control proteins, HSP90 to mark cytoplasmic fraction (Cyto), TFAP2A and pan-H2A.Z to mark chromatin-bound fraction (Chr-B). 1X and 3X dilution of each sample.
See also Figure S2.
Using CRISPR/Cas9 genome editing to generate isogenic FHS hESC lines
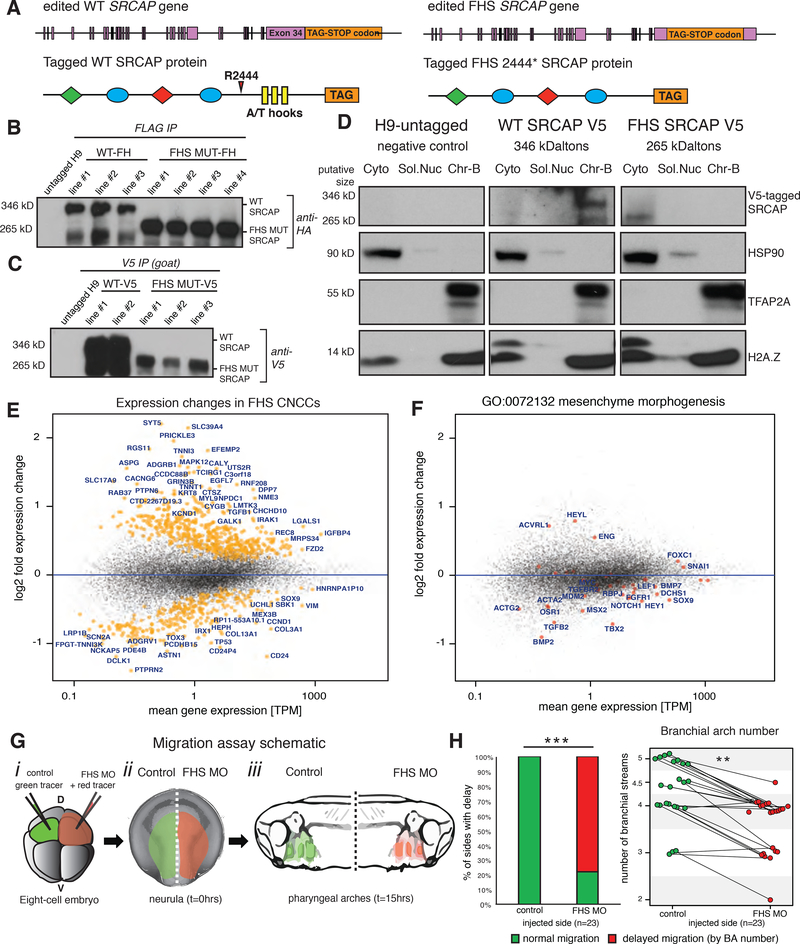
We investigated how the truncation of SRCAP affects neural crest cell function. To generate an in vitro disease model of FHS, we used CRISPR/Cas9 genome editing to introduce into hESCs the most common FHS nonsense mutation, R2444*, which is present in almost half of the known cases of the syndrome (Figure 1B) (Nikkel et al. 2013). Given that FHS mutations are heterozygous, we generated heterozygous H9 hESC lines with the R2444 mutation and, to facilitate subsequent analyses of the mutant protein, we fused a FLAG-HA tag or a V5-tag to the C-terminus of truncated SRCAP (Figure 3A). To obtain corresponding control reagents (and to exclude a possibility that the C-terminal tags, rather than the truncation, interfere with protein function), we also isolated heterozygous lines with tagged WT SRCAP (Figure 3A). The presence of tagged WT and FHS SRCAP proteins of the expected molecular weight was confirmed by immunoblotting of proteins enriched by immunoprecipitation (Figure 3B–C). Biochemical fractionation of CNCCs differentiated from the modified hESC lines revealed that endogenously tagged WT SRCAP is associated with chromatin, whereas tagged FHS SRCAP is primarily cytoplasmic, in agreement with our previous experiments with overexpressed proteins (Figure 3D). In the FHS mutant lines, we did not observe an exclusion of H2A.Z from chromatin, although a subtle increase of H2A.Z recovery in the soluble fractions was detectable (Figure 3D). Nonetheless, we did not expect a major loss of H2A.Z chromatin incorporation in the FHS cell lines because an intact copy of the WT SRCAP gene as well as SRCAP paralog p400 are both present and expressed in these CNCCs.
Figure 3. Engineered FHS SRCAP heterozygous human CNCCs show downregulation of critical migration and morphogenesis genes.
(A) CRISPR/Cas9 targeting strategy for endogenous tagging and/or truncation of SRCAP gene using homologous recombination with ultramers to add FLAG-HA or V5 tag.
(B) Validation of FLAG-HA tagged Wildtype SRCAP and FHS mutant CNCCs by whole cell immunoprecipitation for FLAG with immunoblotting against HA tag. Predicted protein size indicated on left.
(C) Validation of V5 tagged Wildtype SRCAP and FHS mutant CNCCs by whole cell immunoprecipitation for V5 with immunoblotting also against V5 tag. Predicted protein size indicated on left.
(D) Biochemical fractionation of CNCCs with untagged SRCAP (H9-untagged), endogenously V5-tagged WT SRCAP protein, endogenously V5-tagged FHS mutant SRCAP protein. Immunoblotting with mouse-V5 antibody. HSP90 in cytoplasmic fraction (Cyto), TFAP2A and pan-H2A.Z in chromatin-bound fraction (Chr-B). Predicted protein size on left.
(E) Gene expression changes between SRCAP WT and FHS mutant CRISPR/Cas9 lines determined by RNA-seq (FLAG-HA tagged lines WT1–3 and FHS MUT1–4). Significant changes at FDR<0.1 denoted in orange.
(F) Enrichment of genes involved in mesenchyme morphogenesis (GO term: 0072132) with gene denoted in red, gene names in blue.
(G) Diagram of migration assay schematic. (i) injection into one dorsal cell at four-cell stage with 250pg red fluorescent tracer mCherry mRNA and 5.0μM FHS MO #1, injection of 250pg green fluorescent tracer Kaede mRNA into both animal pole-dorsal cells at eight-cell stage. (ii-iii) embryos with red and green fluorescence analyzed at neurula stage (time=0) and tailbud stage (time=15 hours), respectively.
(H) Quantification of neural crest migratory delay. Statistical test used was Pearson’s chi-squared 2-sample test for equality of proportions with continuity correction. *** - p-value <2.2e-16. n=3 independent experiments. Normal migration in green, abnormal migration in red. Quantification of number of branchial arch streams, with lines matching number of arch streams from same embryo at time=15 hours. Statistical test used was two-sample Kolmogorov-Smirnov test. p-value = 0.003819. ** - p-value <0.005. Animals from n = 3 biologically independent experiments.
See also Figure S3.
Heterozygous FHS SRCAP mutation leads to perturbations of CNCC gene expression programs
We asked whether FHS-associated mutations in SRCAP lead to defects in CNCC formation, proliferation or molecular identity. FHS SRCAP hESC lines differentiated into CNCCs that were indistinguishable from CNCCs derived from either epitope-tagged WT SRCAP lines or unmodified hESC both morphologically and by staining with select neural crest cell markers (Figure S3A) (Rada-Iglesias et al. 2012). We further characterized the differentiation dynamics and cellular heterogeneity using a panel of four cluster of differentiation (CD) markers previously utilized for monitoring CNCC differentiations (Prescott et al. 2015). Again, we detected no major differences between CNCCs derived from FHS SRCAP and WT SRCAP cells (Figure S3B). Furthermore, while SRCAP and H2A.Z dysfunction has previously been associated with cell cycle changes, we found no difference in cell cycle profiles between WT and FHS SRCAP CNCCs (Figure S3E) (Vardabasso et al. 2015; A. J. Morrison & Shen 2009). These observations indicate that disease-mimicking heterozygous FHS mutations have no major effect on induction or proliferation of CNCCs.
To address whether CNCC gene expression programs are perturbed in FHS SRCAP cell lines, we performed polyA+ RNA-seq analysis, using CNCCs derived from two independent differentiation experiments using three WT SRCAP lines and four FHS SRCAP lines. We found significant changes in gene expression between FHS and WT SRCAP CNCCs, including downregulation of neural crest transcription factors SOX9, PAX7, MSX2, PITX1, BARX1, and HEY1, secreted proteins involved in inter-cellular communication (such as Semaphorins, ADAM metallopeptidases, SLIT family genes, NOTCH genes, and proto-cadherins), and genes associated with locomotion/cell motility and adhesion (Figure 3E). One of the most strongly downregulated genes encoded a surface protein, CD24, and we indeed confirmed that mutant CNCCs had decreased surface levels of CD24 (Figure S3F–G). Among the genes that are upregulated in the mutant cells, we noted a number of chromatin regulators (e.g. NACC2, RBBP7, HDAC10) and cell junction proteins (e.g. CDH8, CTTNB1, CTTND1/2). GO term analyses of the significantly affected transcripts revealed enrichment of mesenchyme morphogenesis and calcium dependent cell adhesion genes, and these transcripts were typically downregulated in the mutant cells, consistent with the postulated role of SRCAP as a positive regulator of transcription (Figure 3F, Figure S3C). A subset of genes associated with a migratory neural crest signature in the chick embryo (J. A. Morrison et al. 2017), were also downregulated in the mutant cells (Figure S3D). These findings indicate that FHS-associated SRCAP truncation leads to subtle, but broad and consistent gene expression defects, with pervasive downregulation of genes associated in cell migration, adhesion and mesenchyme morphogenesis.
Neural crest migration is delayed in FHS SRCAP morphant embryos
To test if SRCAP truncation affects neural crest migration in vivo, we coinjected the FHS MO and a red fluorescent tracer (mCherry) mRNA into one ventral cell at the 4-cell stage. Subsequently, a green tracer (unconverted kaede) mRNA was symmetrically injected into both ventral cells at the 8-cell stage to mark future migrating neural crest cells (Figure 3G). These embryos were developed and imaged at the neurula stage, when the neural crest is specified, and then at several additional timepoints, as the neural crest cells migrate to populate the branchial arches (Figure 3G). We quantified the number of migratory streams on the tracer-only versus FHS MO injected side of the tadpoles. In the controls, the number of migratory streams ranged from three streams (at the initial stages of branchial arch migration) to five streams (at the later stages of migration). Within this range, there were also embryos in the stages of migration when migratory streams split into two distinct migratory streams (quantified as 4.5). We observed that fewer migratory streams were consistently detected on the FHS MO injected side as compared to the control side at all examined stages (Figure 3H and S3I). These observations suggest that SRCAP truncation may cause defects in neural crest specification which subsequently delays neural crest migration, or alternatively, that neural crest migration is directly perturbed. Whichever the case, these migratory defects are likely to be responsible, at least in part, for the craniofacial manifestations of the syndrome.
Knockdown of histone variant H2A.Z.2 but not H2A.Z.1 phenocopies FHS SRCAP craniofacial abnormalities
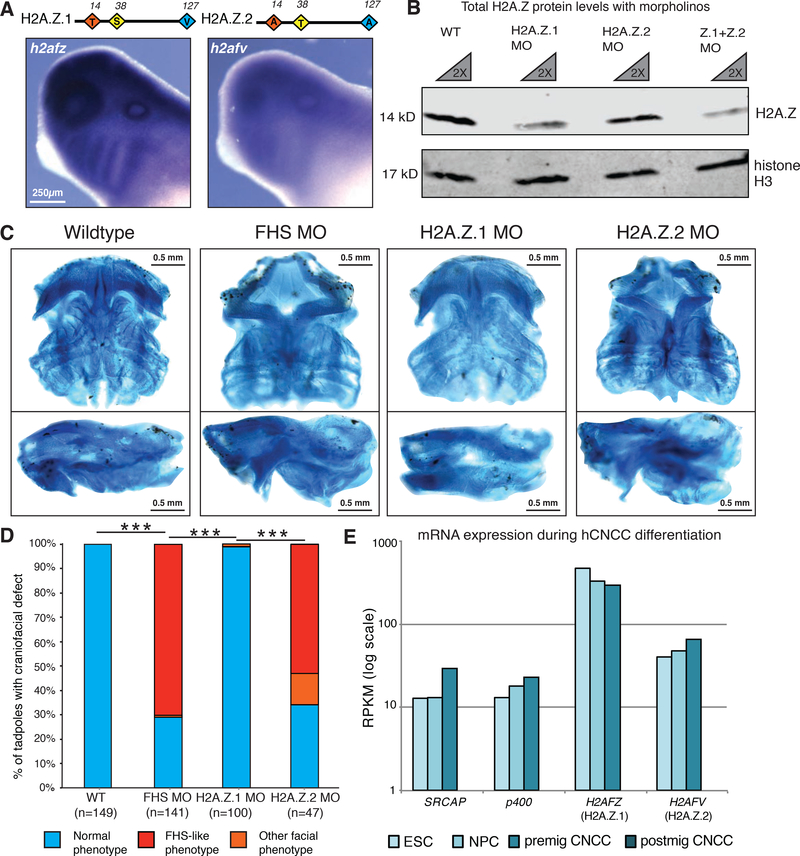
Since H2A.Z incorporation is the main molecular function of SRCAP, we asked whether downregulation of H2A.Z in X. laevis can phenocopy SRCAP truncation. Vertebrates have evolved two H2A.Z genes, H2AFZ (encoding H2A.Z.1) and H2AFV (encoding H2A.Z.2), whose protein products are identical except for three divergent amino acids (Figure 4A). We found that like srcap, both genes are broadly expressed in the frog, including in the branchial arches, though we detected consistently higher signals for the h2afz mRNA probe than for the h2afv mRNA probe (Figure 4A, S4A). To differentiate between the H2A.Z isoforms, we designed subtype-selective splice-blocking MOs. Given that no antibodies exist that can distinguish between endogenous H2A.Z.1 and H2A.Z.2, we assayed H2A.Z protein levels in bulk, and were able to detect a decrease in total H2A.Z protein levels upon injection of either the H2A.Z.1 or H2A.Z.2-specific MOs. Co-injection of both MOs resulted in a more significant reduction of H2A.Z levels (Figure 4B, quantified in Figure S4B). Notably, H2A.Z.1 MO had a larger effect on total H2A.Z protein levels, in agreement with an overall higher expression of its mRNA in the frog. We used RT-PCR to confirm that injection of each MO leads to an intron inclusion at the intended, isoform-specific exon-intron junction, that there are no off-target effects of either H2A.Z MO on the transcript for the other H2A.Z subtype, and that both transcripts were targeted when the two MOs were co-injected (Figure S4C).
Figure 4. Knockdown of H2A.Z.2 phenocopies the craniofacial features of FHS morphant frogs.
(A) Schematic of H2A.Z.1 and H2A.Z.2 proteins. Red, yellow, blue diamonds denote three amino acids divergent between H2A.Z.1 and H2A.Z.2. In situ hybridization staining for h2afz and h2afv mRNA at tailbud stage in wildtype X. laevis tadpoles. 250μm scale bar shown.
(B) Western blot of cellular extract from dissected X. laevis at tailbud stage, with wildtype and 2.5 μM H2AFZ MO and 2.5 μM H2AFV MO samples used. Antibodies against total-H2A.Z and total histone H3 (loading control). 1X and 2X dilution of each sample. Imaged and quantified on LI-COR Odyssey (see supplemental figure).
(C) Ventral and side views of dissected X. laevis cartilage stained with Alcian blue at stage 40, Wildtype (mock injected), SRCAP FHS morphant (SRCAP truncation) (5 μM), H2A.Z.1 morpholino (2.5 μM morpholino), H2A.Z.2 morpholino (2.5 μM morpholino). 0.5 mm scale bar shown. Animals from n = 3 biologically independent experiments.
(D) Blinded quantification of characteristic craniofacial phenotype. Statistical test was Pearson’s chi-squared 2-sample test for equality of proportions with continuity correction. *** - p-value <2.2e-16. Animals from n=3 independent experiments.
(E) mRNA expression of indicated genes (measured by RPKM) from hESCs, neural precursor cells (NPCs), pre-migratory neural crest cells (premig NCCs), and post-migratory neural crest cells (postmig NCCs) (Rada-Iglesias et al. 2012).
See also Figure S4.
We injected various doses of each MO (2.5μM, 5.0 μM, etc.) to the two-cell stage frog embryos. At high doses (≥5μM), H2A.Z.1 MO injection led to an early developmental arrest, resembling the early embryonic lethality seen in H2A.Z.1 knockout mice (Faast et al. 2001). At lower doses (2.5μM) the embryos displayed a significant decrease in H2A.Z levels but low penetrance phenotypes, characterized by either no observable craniofacial and growth defects in majority of embryos, or by very severe head development defects and growth impairment (Figure 4C, Figure S4D,H). However, these morphants failed to phenocopy the FHS phenotype observed when SRCAP is truncated. By contrast, in H2A.Z.2 morphants, we observed no early embryonic lethality at either low or high MO dose. Instead, we saw a craniofacial cartilage phenotype that was similar to that observed when SRCAP is truncated in X. laevis (Figure 4C, S4D–F). For both H2A.Z subtype MOs, when injected at a dose that resulted in either a severe developmental defect (e.g. H2A.Z.1 MO at 5μM) or a craniofacial cartilage defect (e.g. H2A.Z.2 MO at 2.5μM), we were able to substantially rescue the phenotypes by co-injection of respective subtype mRNAs (Figure S4H–J).
To compare craniofacial phenotypes between the various morphants, we conducted a blinded analysis and found that the penetrance and nature of the cartilage defects in H2A.Z.2 morphants were similar to those seen in the FHS SRCAP morphants, while the control or H2A.Z.1 MO injected tadpoles (2.5μM) showed no/minimal craniofacial defects; the differences between the two H2A.Z subtypes were highly statistically significant (Figure 4D; S4G, note that for these measurements we could only consider a subset of H2A.Z.1 morphants in which head formation occurred). Optical projection tomography (OPT) again revealed morphological similarity between the cartilage dissected from the FHS SRCAP and H2A.Z.2 morphants (Movie S1). Additionally, we observed that the H2A.Z.2 MO-injected tadpoles were more heavily pigmented compared to the H2A.Z.1 MO-injected tadpoles (Figure S4D). Our observations suggest that: (i) malformations associated with FHS are likely caused by defects in H2A.Z deposition during embryogenesis, and (ii) deficiency of H2A.Z.2 phenocopies the effects of the SRCAP FHS mutation, indicating that H2A.Z.2 may be selectively mediating the craniofacial manifestations in FHS.
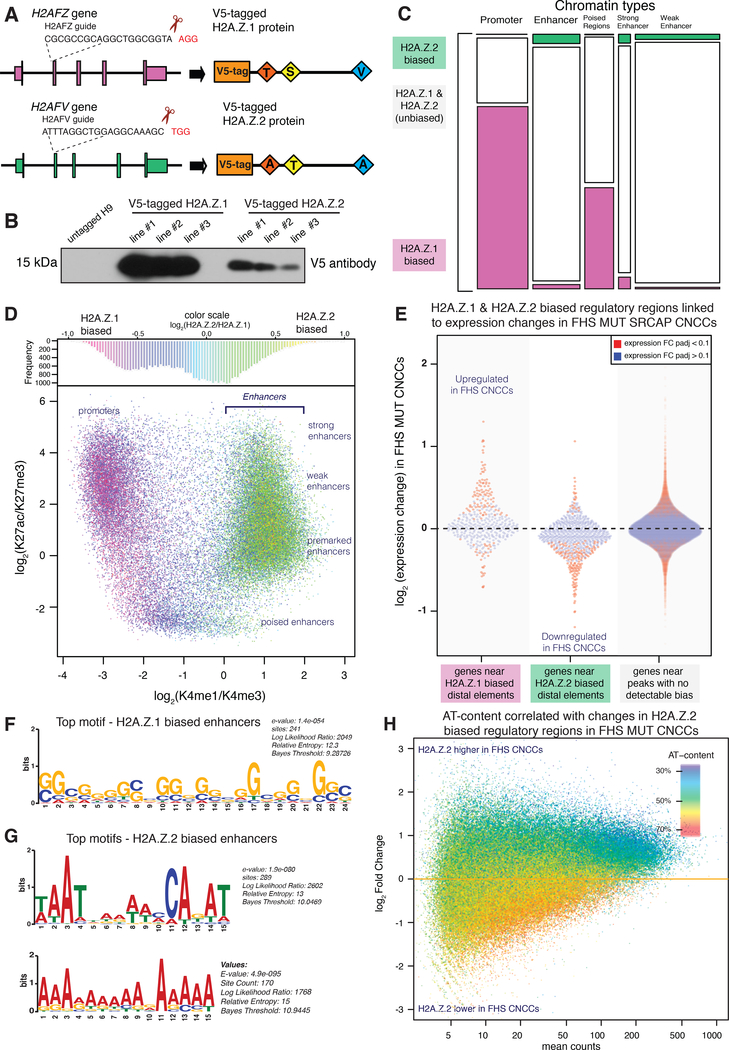
Endogenous tagging of H2A.Z.1 and H2A.Z.2 uncovers qualitatively similar, but quantitatively distinct genomic incorporation patterns
To explore the molecular consequences of H2A.Z.2 depletion, we returned to our human CNCC differentiation model. First, we found that although absolute H2A.Z.1 mRNA levels were generally substantially higher (between 4-fold and 10-fold) than H2A.Z.2 mRNA levels throughout hESC differentiation to CNCC, H2A.Z.2 and SRCAP mRNA levels progressively increased, while those of H2A.Z.1 mRNA decreased throughout the course of differentiation (Figure 4E). To compare genomic occupancy patterns of H2A.Z.1 and H2A.Z.2 in the absence of antibodies that can distinguish the two subtypes, we used CRISPR/Cas9 genome editing to generate hESC lines in which the N-terminus of either H2A.Z.1 or H2A.Z.2 was tagged with V5 (Figure 5A). We verified that V5-tagged H2A.Z was detectible by Western blot and localized to the nucleus (Figure 5B, S5A). Of note, V5-tagged H2A.Z.1 was expressed at approximately five-fold higher levels than tagged H2A.Z.2, a difference consistent with the relative mRNA expression levels of the endogenous genes in human CNCCs (Figure 4E).
Figure 5. H2A.Z.2-biased regions are found at AT-rich enhancers near genes downregulated in FHS.
(A) CRISPR/Cas9 targeting strategy in hESCs for endogenously V5-tagging N-terminus of H2A.Z.1 and H2A.Z.2 by homologous recombination. H2A.Z.1 in magenta, H2A.Z.2 in green.
(B) Immunoblot against V5 tag for H2A.Z.1 lines 1–3 and H2A.Z.2 lines 1–3, untargeted H9s as negative control.
(C) H2A.Z sites classified into five categories with k-means algorithm based on chromatin modifications and into H2A.Z.1-biased sites (green) or H2A.Z.2-biased sites (purple) based on V5-tag ChIP-seq data.
(D) Distribution of H2A.Z.1/H2A.Z.2 ratio over CNCC regulatory element landscape. Upper panel: histogram of H2A.Z.1/H2A.Z.2 ratio distribution. Lower panel: scatter plot of regulatory regions, colored by H2A.Z.1/H2A.Z.2 ratio as in upper panel. x-axis: log ratio of H3K4me1 to H3K4me3 differentiating promoters from enhancer-like elements; y-axis: log ratio of H3K27ac to H3K27me3 reflecting region transcriptional activity and Polycomb silencing, respectively.
(E) Genes in proximity of H2A.Z.2-biased regulatory regions downregulated in FHS CNCCs. 1D scatterplot of gene expression changes between WT and FHS SRCAP CNCCs for genes in proximity to H2A.Z.1-biased or H2A.Z.2-biased promoter-distal ChIP-seq peaks or genes with only unbiased elements in vicinity. Genes with RNA-seq differential expression analysis FDR<0.1 colored in red, others in blue.
(F) Motif associated by MEME with H2A.Z.1 biased enhancers, using identified CNCC enhancers as background model (Bailey et al. 2009).
(G) Primary and secondary motifs associated by MEME with H2A.Z.2 biased enhancers, using identified CNCC enhancers as background model (Bailey et al. 2009).
(H) Association of changes in H2A.Z.2 incorporation for WT and FHS SRCAP CNCCs with DNA AT-content. Plot of H2A.Z.2 incorporation in FHS CNCCs compared to WT CNCCs with a range of AT-content from 30% (blue) to 50% (green) to 70% (red). X-axis is mean counts, normalized reads for each site with adjusted mean as base factor.
See also Figure S5.
We performed anti-V5 ChIP-seq using CNCCs derived from three independent V5-tagged H2A.Z.1 and H2A.Z.2 lines (Figure S5B). Given that H2A.Z is known to broadly occupy enhancers and promoters, we used our previously described epigenomic annotations of putative cis-regulatory elements in CNCCs (Rada-Iglesias et al. 2012; Prescott et al. 2015) to characterize relative H2A.Z subtype enrichments at these regions. Remarkably, this analysis revealed that while H2A.Z.1 and H2A.Z.2 incorporation patterns are qualitatively highly similar (with the highest enrichment levels at promoters and broad association with active and premarked enhancers), they are quantitatively distinct. Specifically, active promoters and H3K27me3 marked regulatory regions typically showed higher ratio of H2A.Z.1 vs H2A.Z.2 (although this may be simply a consequence of higher overall H2A.Z.1 expression in CNCCs) (Figure 5C–D, S5B–D). By contrast, enrichments of H2A.Z.1 and H2A.Z.2 at both active and premarked enhancers were typically comparable, with a subset of enhancers characterized by a higher ratio of H2A.Z.2, despite its five-fold lower protein levels (Figure 5C–D, S5B–D). Consistent with these observations, when we compared H2A.Z.1 and H2A.Z.2-biased regions, we found that the majority of H2A.Z.1-biased regions were located within 5kb of a TSS while the majority of H2A.Z.2 peaks were located farther than 5kb of a TSS (Figure S5E). Thus, the two H2A.Z subtypes show quantitative differences in genomic incorporation patterns.
H2A.Z.2-biased enhancers are associated with genes downregulated in FHS mutant CNCCs
We investigated whether H2A.Z.2-enriched enhancers were near genes that were dysregulated in our heterozygous FHS SRCAP CNCCs. Towards that end, we identified genes that were proximal to H2A.Z.2-biased enhancers, H2A.Z.1-biased enhancers, or those containing comparable enrichment of the two subtypes (unbiased). This third gene set included the majority of the transcripts expressed in CNCCs. We found that genes proximal to H2A.Z.2 enriched enhancers were more likely to be downregulated in FHS SRCAP CNCCs, while such downregulation was not observed for the genes associated with enhancers with no detectable H2A.Z subtype bias or for those enriched for H2A.Z.1 (Wilcoxon-Mann-Whitney test: p-value <0.0001; Figure 5E). In fact, genes associated with H2A.Z.1 biased enhancers were more likely to be expressed at higher levels in FHS mutant cells compared to the wildtype cells (Figure S5F). Thus, genes near enhancers that have a relatively high enrichment of H2A.Z.2 appear to be more sensitive to partial loss of SRCAP function caused by heterozygous truncations, resulting in their preferential downregulation.
Gene ontology analysis of these H2A.Z.2 enriched enhancers using the Genomic Regions Enrichment of Annotations (GREAT) tool (McLean et al. 2010) revealed a number of relevant terms, including chemotaxis and migration, cranial skeleton morphogenesis, as well as several human phenotypes associated with malformations present in FHS (i.e. tooth number, nose morphology, ear malformation, and distal/middle symphalangism of the fifth finger). No such enrichments were detected at H2A.Z.1-enriched enhancers (Figure S5G–H). Notably, distinct sequence motifs were overrepresented at H2A.Z.2 biased enhancers relative to H2A.Z.1 bound enhancers, with the former being characterized by AT-richness and the latter by the GC-richness (Figure 5G–H) (Bailey et al. 2009). This finding is intriguing, because the major domain lost from SRCAP upon FHS mutation are three AT hooks, which not only endow SRCAP with nuclear localization signals, but also have autonomous ability to bind chromatin (Figure 2E).
FHS mutation leads to decreased H2A.Z.2 incorporation selectively at AT-rich enhancers
To test the consequences of FHS mutation on H2A.Z.2 incorporation in CNCCs, we used one of the previously targeted V5-tagged H2A.Z.2 hESC lines and performed further editing to introduce a FLAG-HA tag either along with the FHS 2444* mutation or at the endogenous C-terminus of SRCAP (Figure S5I–J). We differentiated these matched WT SRCAP/V5 H2A.Z.2 and FHS SRCAP/V5 H2A.Z.2 lines to CNCCs in quadruplicate and carried out ChIP-seq with the V5 antibody to map H2A.Z.2 enrichment. We found that the regulatory elements that showed diminished H2A.Z.2 incorporation in the FHS cells corresponded to the highly AT-rich distal regions, which were generally non-overlapping with TSS (Figure 5H, S5K). This selective loss of H2A.Z.2 incorporation at AT-rich enhancer regions suggests the importance of the SRCAP AT-hooks in directing the specificity of H2A.Z genomic incorporation.
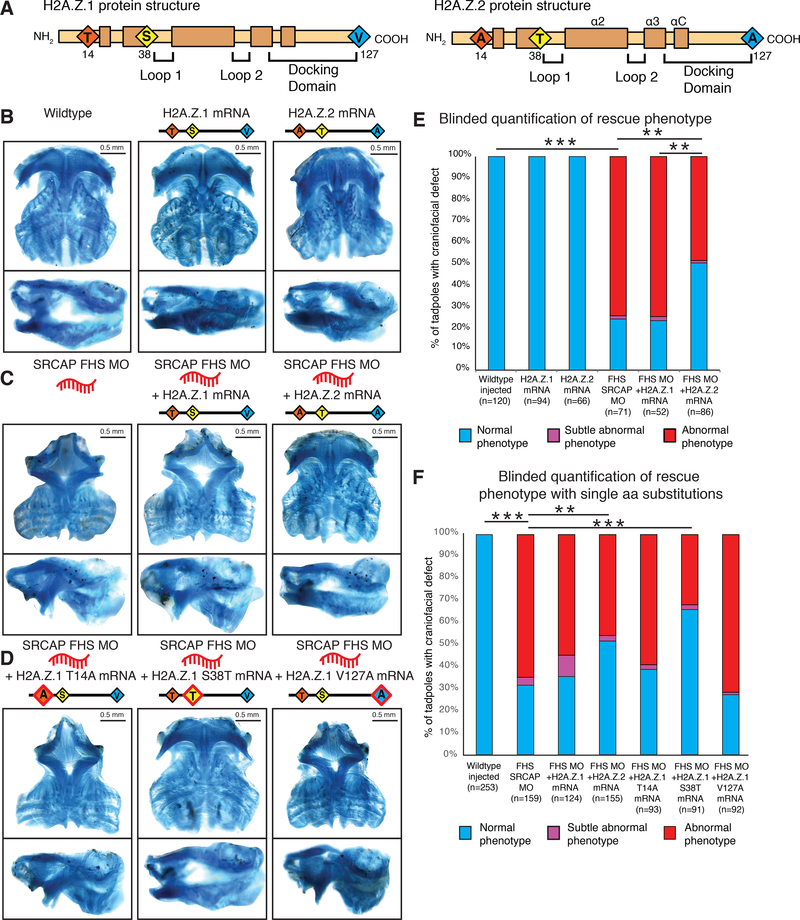
Overexpression of H2A.Z.2 but not H2A.Z.1 rescues craniofacial abnormalities seen in FHS SRCAP morphant in X. laevis
Our combined results from the in vitro FHS model and Xenopus embryos suggest that FHS manifestations arise from defects in H2A.Z.2 incorporation caused by a dosage deficiency (and loss of AT-rich region specificity) of SRCAP. We hypothesized that in the context of partial loss of SRCAP activity, supplying higher levels of H2A.Z might to a certain extent overcome these deficits, resulting in a phenotypic rescue. To test this hypothesis, we combined SRCAP MO injection with overexpression of mRNA encoding H2A.Z.2 or H2A.Z.1 (see Figure 6A for depiction of three amino acid residues that differ between the two isoforms). After injecting two-cell stage frog embryos with truncating FHS SRCAP MO #1 (as described in Fig.1) with or without HA-tagged H2A.Z.1 or H2A.Z.2 mRNA, we analyzed the cartilage of resultant tadpoles. In the absence of the SRCAP MO, we observed that the overexpression of H2A.Z.1 or H2A.Z.2 mRNA alone had no significant effect on craniofacial development (Figure 6B, S6A). However, co-injection with H2A.Z.2 mRNA, but not with H2A.Z.1 mRNA partially rescued the ceratohyal and Meckel’s cartilage defects observed in FHS SRCAP morphants (Pearson’s chi-squared 2-sample test: p<0.0005), leading to a relative normalization of the defects associated with FHS (Figure 6C, 6E, S6A). Furthermore, the cranial cartilage size is significantly increased in the H2A.Z.2 co-injected morphants, as compared to either the MO alone or the MO with H2A.Z.1 (Tukey HSD, p-value<0.005), albeit not to the full extent observed in control embryos (Figure S6D). These results suggest that by specifically increasing the levels of H2A.Z.2 in the context of FHS, it is possible to rescue the downstream effects of the SRCAP mutation.
Figure 6. H2A.Z.2 but not H2A.Z.1 can rescue FHS in vivo due to a single amino acid substitution.
(A) Schematic of H2A.Z.1 and H2A.Z.2 protein domains. Red, yellow, blue diamonds denote three amino acids divergent between H2A.Z.1 and H2A.Z.2. Structural domains including loop 1, loop 2 and docking domain in brackets and alpha helices are indicated.
(B-D) Ventral and lateral views of dissected X. laevis cartilage Alcian blue stained at stage 40, and injected as schematically indicated above each image. 0.5 mm scale bar shown. All FHS MO #1 injected at 5.0 μM, all mRNA at 2.5ng each.
(B) wildtype (mock injected), H2A.Z.1 mRNA overexpression and H2A.Z.2 mRNA overexpression.
(C) SRCAP FHS MO #1, injected alone or with H2A.Z.1 mRNA or H2A.Z.2 mRNA.
(D) SRCAP FHS MO #1, injected with H2A.Z.1 T14A mRNA, or H2A.Z.1 S38T mRNA, or H2A.Z.1 V127A mRNA.
(E) Blinded quantification of characteristic craniofacial phenotype for FHS rescue with H2AZ.1 and H2AZ.2 mRNA. Statistical test was Fisher’s Exact Test (FET). FET p-value < 0.005 = **, FET p-value < 10e-5 = **. Animals from n = 3 biologically independent experiments.
(F) Blinded quantification of characteristic craniofacial phenotype for FHS rescue with H2AZ subtypes and H2AZ.1 single amino acid substitution mRNA, includes samples from Fig. 6E. Statistical test was Pearson’s chi-squared 2-sample test for equality of proportions with continuity correction. FET p-value < 0.005 = **, FET p-value < 10e-5 = ***. Animals from n = 3 biologically independent experiments.
See also Figure S6.
Single conservative amino acid substitution of H2A.Z.1 S38T confers capacity to rescue the FHS craniofacial phenotypes
To establish which of the three amino acid changes confer upon H2A.Z.2 the ability to rescue the effects of SRCAP truncations, we mutated each of the divergent H2A.Z.1 residues to the corresponding H2A.Z.2 residues (T14A, S38T, or V127A, respectively), then injected these HA-tagged H2A.Z mutant mRNAs with the FHS SRCAP MO and analyzed the resultant craniofacial phenotypes. Although injection of each mRNA led to a comparable increase in total H2A.Z protein levels (Figure S6C), only overexpression of H2A.Z.1 S38T mRNA could significantly rescue the FHS SRCAP MO phenotype, while T14A or V127A mRNAs had a relatively minor effect (Figure 6D, 6F, S6B). Rescue with H2A.Z.1 S38T mRNA resulted in a size increase in the ceratohyal cartilage area and in the eye-to-eye distance that was statistically significant, and such increase was not seen for either the H2A.Z.1 T14A or V127A mRNAs (Tukey HSD, p-value<0.005) (Figure S6E). These findings indicate that a single, conservative amino acid change S/T38 endows H2A.Z.2 with ability to rescue the FHS phenotypes, although we cannot exclude a possibility that the other two substitutions may provide a minor contribution. We discuss below a model that may explain functional and developmental specialization of the H2A.Z paralogs.
Discussion
In this study, we illuminate the molecular mechanism underlying FHS, a poorly understood human craniofacial syndrome caused by mutations in the chromatin remodeler SRCAP. We find that syndrome-associated mutations perturb nuclear localization and chromatin association of the truncated protein. The AT-hooks region, which is a unique structural feature of SRCAP within the SWR1 family of remodelers (Fig. S1A), has the autonomous ability to translocate to the nucleus and to associate with chromatin. Our data suggest that the AT hook region supplies WT SRCAP not only with nuclear localization signal(s), but also contributes to its nucleosome binding properties, likely via AT-rich DNA recognition, and these features are lost in the FHS-associated truncations.
FHS mutations are heterozygous in affected patients leaving the other SRCAP allele intact. Notably, we found no evidence for dimerization of WT and FHS SRCAP in FHS heterozygous cells, suggesting that the syndrome is caused by a dosage deficiency of SRCAP, although we cannot exclude potential dominant effects as well. FHS-associated impairment of SRCAP activity manifests itself preferentially at AT-rich enhancers and can be partially overcome by supplying an excess of H2A.Z.2. AT-rich enhancers also exhibit the highest ratio of H2A.Z.2 to H2A.Z.1, and thus may be relatively more dependent on the incorporation of the former subtype for their activation, despite the H2A.Z.1 being present at higher levels in CNCCs. In contrast, most promoters and a small subset of enhancers with relatively higher H2A.Z.1/H2A.Z.2 ratio show high GC-content. These observations suggest that underlying sequence properties can influence H2A.Z subtype incorporation by heretofore unknown mechanisms that could be mediated by the distinct inherent dynamics/allosteric properties of the H2A.Z-containing nucleosomes themselves, or by the divergent targeting or sequence preferences of the remodelers and chaperones involved in their deposition.
Perhaps our most surprising observation is that overexpression of H2A.Z.2 but not H2A.Z.1 partially rescues the SRCAP deficiency that underlies FHS, and that a single amino acid difference (S/T38) is responsible for this feature of H2A.Z.2. These findings indicate that H2A.Z.2 and H2A.Z.1 have evolutionarily divergent roles during some aspects of development, despite the high amino acid sequence similarities between the two paralogs and their overlapping expression patterns. It is intriguing that vertebrates have evolved the two histone variants H2A.Z.1 and H2A.Z.2, as well as two different chromatin remodeling complexes SRCAP and p400, and that these duplication events occurred around the same period of evolution that the neural crest is thought to have emerged (Dryhurst et al. 2009; Obri et al. 2014). Given our results, we propose that in rare reported patient cases with FHS-like features and without SRCAP mutations (Goff et al. 2012), genetic testing for H2A.Z.2 mutations should be performed.
How does a single amino acid change, the substitution of H2A.Z.1 residue S38 to T38, which is both conservative and occurs within a structured histone domain, render H2A.Z.2 able to rescue the FHS craniofacial phenotype in Xenopus? Neither amino acid residue 38 nor the other two amino acid residues that differ between H2A.Z.1 and H2A.Z.2 appear to directly affect the association of H2A.Z with SRCAP in biochemical and structural studies (Horikoshi et al. 2013; Horikoshi et al. 2016; Hong et al. 2014) (Figure S7A). Instead, the S38T substitution appears to alter the conformation of the adjacent L1 loop (Horikoshi et al. 2013) (Figure S7B). Interestingly, the divergent L1 loop properties and L1-L1 contacts within the nucleosome have been shown to confer differential stability and dynamics to distinct H2A variant nucleosomes, including canonical H2A, H2A.Z, H2A.X, macroH2A or H2A.W, and have been proposed to regulate allosteric networks within the nucleosome (Osakabe et al. 2018; Chakravarthy & Luger 2006; Bowerman & Wereszczynski 2016). Consistent with the idea that S38T substitution and associated L1 loop polymorphisms may contribute to the regulation of nucleosome mobility, the two H2A.Z subtypes differ in their FRAP dynamics, with H2A.Z.1 showing faster and H2A.Z.2 showing slower recovery after photobleaching (Horikoshi et al. 2013). Moreover, H2A.Z.1 S38T mutation decreases whereas H2A.Z.2 T38S mutation increases the respective subtype mobility in FRAP (Horikoshi et al. 2013). Thus, subfunctionalization of the two H2A.Z subtypes based on divergence of just a single amino acid residue (S/T38) appears to alter both nucleosomal mobility and sensitivity to a diminished activity of the SRCAP remodeler, though it remains to be elucidated how these two properties are related.
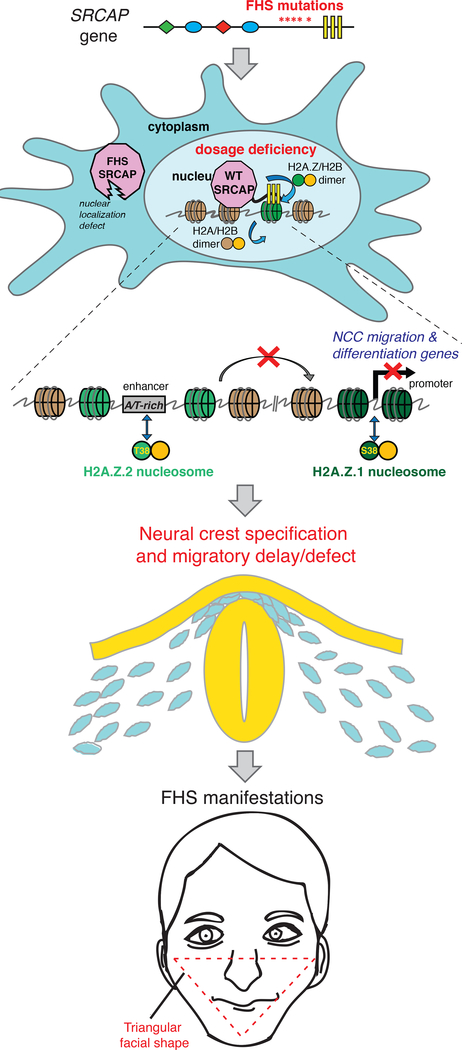
In summary, we propose a mechanistic model underlying FHS and H2A.Z subtype specialization (Figure 7). In this model, heterozygous truncations of SRCAP eliminate nucleosome-binding AT-hooks, leading to the decreased dose of SRCAP activity on chromatin. This decrease in H2A.Z-remodeling activity preferentially affects H2A.Z.2 incorporation into nucleosomes through a mechanism dependent on H2A.Z amino acid residue S38, which when substituted with a threonine confers a structural change in H2A.Z that results in in vivo differences in nucleosomal exchange. AT-rich enhancers that have relatively higher levels of H2A.Z.2 are sensitized to the diminished activity of SRCAP, resulting in preferential downregulation of their associated genes. Many of these genes have functions in neural crest migration and facial morphogenesis. Consequently, neural crest migration and specification is perturbed, resulting in craniofacial anomalies associated with FHS. Thus, a single, conservative amino acid substitution in a broadly expressed histone variant can affect dynamic properties of the nucleosome and lead to differential sensitivity to the dosage of the remodeler, resulting in selective effects on gene expression and mediating a developmental disorder with highly tissue-specific manifestations.
Figure 7. Proposed model of Floating-Harbor Syndrome and H2A.Z subtype specialization.
In FHS, heterozygous SRCAP mutation truncates the protein prior to DNA-binding AT-hooks, causing loss of SRCAP from nucleus and chromatin. With a diminished dose of functional SRCAP present in FHS, nuclear H2A.Z-remodeling activity is reduced. Genomic incorporation pattern of the two H2A.Z subtypes is qualitatively similar, but is biased towards promoters for H2A.Z.1 and AT-rich enhancers for H2A.Z.2. In FHS CNCCs, H2A.Z.2 is preferentially lost from AT-rich enhancers, and associated genes are downregulated. These sensitized regions regulate genes important for CNCC migration and differentiation. FHS patients have craniofacial anomalies related to defects in these developmental processes.
See also Figure S7.
STAR Methods
Lead Contact and Materials Availability
Further information and requests for cell lines, plasmids, and resources should be directed to and will be fulfilled by the Lead Contact Joanna Wysocka (wysocka@stanford.edu). DNA constructs and other research reagents generated by the authors will be distributed upon request to other research investigators under a Material Transfer Agreement.
Experimental Model and Subject Details
ESC culture and CRSIPR/Cas9 genetic engineering
H9 human embryonic stem cells (WA09 from WiCell) were grown on 6-well plates coated with growth-factor-reduced matrigel (BD Biosciences) and were cultured in feeder-free, serum-free medium mTESR-1 (StemCell Technologies). For routine maintenance, the ESCs were passaged at ~1:10 ratio with ReLeSR (StemCell Technologies) one time every 6 days. For passaging, cells were incubated in ReLeSR for 30 seconds, which was then removed and the plate placed in the incubator for 6–8 minutes. mTESR was then added back to the cells, and the plate was gently tapped to detach them from the plate and the cells were then replated on fresh matrigel-coated plates.
To generate the CRISPR/Cas9 genetically engineered lines described in the paper, H9 ESCs were split and cultured in mTESR with 2.5μM Rock inhibitor (Thermo Fisher). The next day, the cells were transfected in mTESR without antibiotics with 2.5μM Rock inhibitor using FuGENE 6 transfection reagent (Promega) at a 1:6 DNA to FuGENE ratio with 3μg of the guide RNA (in a pX330-U6-Chimeric_BB-CBh-hSpCas9 plasmid (Addgene plasmid # 42230 (Cong & F. Zhang 2015)),1.5μg of a 200 bp oligodeoxynucleotide repair template, plus 1μg pb EF1alpha GFP Ubc puro expressing plasmid, and 500ng transposase (System Biosciences) (see Supplemental Table 1 for guide sequences). Cells were then cultured in mTESR with Rock inhibitors until confluent, when they were split ~1:3. Puromycin (Invivogen) selection was started at 0.25mg/ml in mTESR media. Cells were grown in puromycin containing media until all cells in a control well (no selection resistance) had died. Selected wells were allowed to propagate for 7–10 day until clear colonies had grown up and those clones were handpicked and replated. Each handpicked clone was allowed to grow and was then split for DNA isolation (DirectPCR Lysis Reagent, Viagen) and propagation or freezing. Once DNA was isolated from each clone, the region of interest was amplified by PCR and digested with expected restriction site or was amplified by PCR with a primer within the incorporated tag (Supplemental Table 1). Positive clones were Sanger sequenced and clones with the correct mutation and/or tag were selected for validation by immunoprecipitation and/or western blot.
HEK293T cells
Human embryonic kidney cells, HEK293T from America Type Culture Collection (ATCC), were maintained on 10cm2 tissue culture plates in D-MEM high-glucose media (Hyclone), with 10% FBS, 1× NEAA, and 1x Glutamax, supplemented with antibiotics unless otherwise indicated. HEK293T cells were passaged approximately every three days with trypsin.
Xenopus animal model
Adult wildtype male and female frogs (Xenopus laevis) of reproductive age were purchased from Nasco (see Key Resource Table for detailed source information), maintained in a Stanford University facility under the care of Stanford veterinary staff, and were used to obtain embryos by in vitro fertilization. Adult frogs were housed in 12-hour light-dark cycles, were fed three days per week, and were maintained in water at temperature of 16–21°C and at pH 6.5–8.5. Adult female frogs were primed by injection of 800 units of human chorionic gonadotropin (Sigma-Aldrich) into the hindleg approximately 12 to 16 hours prior to egg collection. Adult male frogs were euthanized by injection of anesthetic Tricaine-S (2g/L MS222 in H2O) using IACUC approved methods and the testes were extracted and stored in 1× MMR (100mM NaCl, 2mM KCl, 1mM MgSO4, 2mM CaCl2, 5mM Hepes pH 7.8, 100μM EDTA) for purposes of in vitro fertilization. Eggs were gently squeezed from the ovulating females and were fertilized with sperm from the extracted testes in 0.1× MMR. The fertilized eggs were allowed to develop at 19°C for 30–45 minutes, then were dejellied in 2% cysteine solution (pH 7.8), and were returned to 0.1× MMR at room temperature until they began to divide (~1 hour 45 minutes after fertilization). For all injections, dividing embryos were transferred to Ficoll injection solution (4% (w/v) Ficoll in 0.3× MMR) during the injections and for 1 hour post injection, at which time they were returned to and maintained in 0.1× MMR at 19°C. In vitro fertilization methods were previously published (Sive et al. 2000). All animal experiments performed in this study were conducted in accordance with national guidelines and with protocol approval (Protocol 14405) from the IACUC at Stanford University.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit polyclonal anti-SRCAP - WB | Kerafast | ESL103 |
| goat polyclonal anti-SRCAP - WB | Abcam | ab99408 |
| mouse monoclonal (SV5-Pk1) anti-V5 - WB, IF | Abcam | ab27671 |
| rabbit polyclonal anti-V5 - ChIP | Abcam | ab9116 |
| goat polyclonal anti-V5 (agarose) - IP | Abcam | ab1229 |
| mouse monoclonal anti-FLAG (M2) (agarose)-IP | Sigma-Aldrich | A2220 |
| rabbit polyclonal anti-HA - WB | Abcam | ab9110 |
| mouse monoclonal (HA.C5) anti-HA -WB | Abcam | ab18181 |
| mouse monoclonal (3B5) anti-TFAP2A - WB, IF | Santa Cruz | sc-12726 |
| mouse monoclonal (G-6) anti-NR2F1/COUP-TFI - IF | Santa Cruz | sc-74560 |
| rabbit polyclonal anti-p75 - IF | Promega | G3231 |
| rabbit polyclonal anti-GFP - WB, IF | Abcam | ab290 |
| rabbit polyclonal anti-H2A.Z (pan H2A.Z) - WB, IF | Abcam | ab4174 |
| Rabbit mAb anti-HSP90 (C45G5) (HRP Conjugate) - WB | Cell Signaling | 79641S |
| rabbit polyclonal anti-Histone H3 - WB | Abcam | ab1791 |
| mouse monoclonal anti-Histone H3 (mAbcam 10799) - WB | Abcam | ab10799 |
| rabbit polyclonal anti-CBP (A-22) - WB | Santa Cruz | SC-369 |
| rabbit monoclonal anti-HA | Cell Signaling | C29F4 |
| CD266 (FN14)-PE - FC | Miltenyi | 130-104-329 |
| CD105-VioBlue - FC | Miltenyi | 130-099-667 |
| CD10-APC - FC | Miltenyi | 130-100-082 |
| CD266 (FN14)-PE - FC | Miltenyi | 130-104-329 |
| CD99-APC-Vio770 - FC | Miltenyi | 130-104-367 |
| CD24-PerCP-Vio700 - FC | Miltenyi | 130-112-849 |
| Anti-Digoxigenin-AP, Fab fragments from sheep - ISH | Sigma-Aldrich | 11093274910 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Neurobasal medium | Life Technologies | 21103049 |
| Gem21 NeuroPlex™Supplement With Vitamin A | Gemini Bio-Products | 400-160 |
| N2 NeuroPlex™ Supplement | Gemini Bio-Products | 400-163 |
| HyClone™ Classical Liquid Media: DMEM/F12, 1:1; w/L-Glutamine, w/o HEPES; 500mL | Fisher Scientific | SH3027101 |
| Bovine Insulin Powder | Gemini Bio-Products | 700-112P |
| Pepro-Tech Inc HUMAN EGF 1MG | Fisher Scientific | 50-813-060 |
| Recombinant Human FGF basic (154 a.a.) | PeproTech 100 | 100-18B-1mg |
| Bovine Serum Albumin (BSA) Powder - Serum Replacement Grade | Gemini Bio-Products | 700-104P |
| ChIRON 99021 | Selleck | CHIR99021 |
| GlutaMAX Supplement (100X) | Life Technologies | 35050061 |
| Antibiotic-Antimycotic (100X) | Life Technologies | 15240062 |
| accutase | EMD Millipore | SF006 |
| fibronectin | Fisher Scientific | FC01010MG |
| BMP2 | Peprotech | 120-02 |
| collagenase | Thermo Fisher | 17104019 |
| Gibco KO DMEM | Gibco™ | 10829018 |
| Matrigel | Corning | CB40234A |
| Nuclease micrococcal from Staphylococcus aureus | Sigma-Aldrich | N3755 |
| BM Purple AP Substrate | Sigma-Aldrich | 11442074001 |
| Alcian Blue 8GX | Sigma-Aldrich | A5268 |
| cOmplete™, EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich | 11873580001 |
| Super piggyBac Transposase expression vector | System Biosciences | PB210PA-1 |
| Chorionic gonadotropin human | Sigma-Aldrich | CG10-10VL |
| Tricaine-S (MS-222) | Pentair Aquatic Eco-Systems | TRS1 |
| Critical Commercial Assays | ||
| Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry | ThermoFisher | C10634 |
| FxCycle Violet Stain | ThermoFisher | F10347 |
| MEGAscript kit T7 kit | ThermoFisher | AM1334 |
| MEGAscript kit SP6 kit | ThermoFisher | AM1330 |
| MessageMachine T7 Ultra kit | ThermoFisher | AMB13455 |
| PureLink™ RNA Mini Kit | ThermoFisher | 12183018A |
| Megaclear Transcription Clean-Up kit | ThermoFisher | AM1908 |
| NEBNext® Multiplex Oligos for Illumina | New England Biolabs | E7335S |
| Amersham ECL Prime Western Blotting | GE Lifesciences | RPN2232 |
| Agencourt AMPure XP beads | Beckman Coulter Diagnostics | A63880 |
| Digoxigenin-11-UTP | Sigma-Aldrich | 3359247910 |
| DIG RNA Labeling Kit (SP6/T7) | Sigma-Aldrich | 11175025910 |
| VECTASHIELD® Antifade Mounting Medium with DAPI | Vector Laboratories | H-1200 |
| Trizol | Invitrogen | 15596026 |
| DirectPCR Lysis Reagent | Viagen | 301-C |
| Dynabeads oligo(dT)25 | ThermoFisher | 61002 |
| Dynabeads Protein G | ThermoFisher | 10004D |
| Fugene HD | Promega | E2311 |
| Deposited Data | ||
| RNA-seq and ChIP-seq data | This paper | GEO accession # GSE134532 |
| Experimental Models: Cell Lines | ||
| H9 embryonic stem cell line | WiCell Research Institute | WA09 |
| HEK293T | ATCC | #CRL-3216 |
| H9 - WT SRCAP C-terminal FLAG-HA tag line #1 | This study | N/A |
| H9 - WT SRCAP C-terminal FLAG-HA tag line #2 | This study | N/A |
| H9 - WT SRCAP C-terminal FLAG-HA tag line #3 | This study | N/A |
| H9 - FHS R2444 MUT SRCAP C-terminal FLAG-HA tag line #1 | This study | N/A |
| H9 - FHS R2444 MUT SRCAP C-terminal FLAG-HA tag line #2 | This study | N/A |
| H9 - FHS R2444 MUT SRCAP C-terminal FLAG-HA tag line #3 | This study | N/A |
| H9 - FHS R2444 MUT SRCAP C-terminal FLAG-HA tag line #4 | This study | N/A |
| H9 - WT SRCAP C-terminal V5 tag line #1 | This study | N/A |
| H9 - WT SRCAP C-terminal V5 tag line #2 | This study | N/A |
| H9 - FHS R2444 MUT SRCAP C-terminal V5 tag line #1 | This study | N/A |
| H9 - FHS R2444 MUT SRCAP C-terminal V5 tag line #2 | This study | N/A |
| H9 - FHS R2444 MUT SRCAP C-terminal V5 tag line #3 | This study | N/A |
| H9 - WT H2AZ.1 N-terminal V5 tag line #1 | This study | N/A |
| H9 - WT H2AZ.1 N-terminal V5 tag line #2 | This study | N/A |
| H9 - WT H2AZ.1 N-terminal V5 tag line #3 | This study | N/A |
| H9 - WT H2AZ.2 N-terminal V5 tag line #1 | This study | N/A |
| H9 - WT H2AZ.2 N-terminal V5 tag line #2 | This study | N/A |
| H9 - WT H2AZ.2 N-terminal V5 tag line #3 | This study | N/A |
| H9 - WT H2AZ.2 N-terminal V5 tag + WT SRCAP C-terminal FLAG-HA tag line | This study | N/A |
| H9 - WT H2AZ.1 N-terminal V5 tag + FHS R2444 MUT SRCAP C-terminal FLAG-HA tag line | This study | N/A |
| Experimental Models: Organisms/Strains | ||
| Male Xenopus laevis, Pigmented, Wildtype | NASCO | LM00715MX |
| Oocyte Positive Female Xenopus laevis, Pigmented, Wildtype | NASCO | LM00531 |
| Oligonucleotides | ||
| Morpholino - FHS SRCAP MO: CCTTAAGTACAAAAGGTGTAGGGC | Gene-Tools, LLC | N/A |
| FHS SRCAP MO #2: ACTTGCTCCTTAAGTACAAAAGGTG | Gene-Tools, LLC | N/A |
| Morpholino - H2AFZ MO #1: GCACTACAAAAATGTACTTACCACC | Gene-Tools, LLC | N/A |
| Morpholino - H2AFZ MO #2: ACCACAAAAGTCACTTACCACCACC | Gene-Tools, LLC | N/A |
| Morpholino - H2AFV MO: ACAGGGAACTAAGAGGTACAAAGGA | Gene-Tools, LLC | N/A |
| Primers for RT-PCR, CRISPR-Cas9, in situ probes; see Table SI | This paper | N/A |
| Recombinant DNA | ||
| pcDNA3.1+ CMV/T7 promoter N-terminal-HA tag H2A.Z.1 | This paper | N/A |
| pcDNA3.1+ CMV/T7 promoter N-terminal-HA tag H2A.Z.2 | This paper | N/A |
| pcDNA3.1+ CMV/T7 promoter N-terminal-HA tag H2A.Z.1 T14A | This paper | N/A |
| pcDNA3.1+ CMV/T7 promoter N-terminal-HA tag H2A.Z.1 S38T | This paper | N/A |
| pcDNA3.1+ CMV/T7 promoter N-terminal-HA tag H2A.Z.1 V127A | This paper | N/A |
| pB CAG WT-SRCAP-eGFP-FLAG pGK Blast | This paper | N/A |
| pB CAG FHS-truncated-SRCAP-eGFP-FLAG pGK Blast | This paper | N/A |
| pB CAG AT-hooks-eGFP-FLAG pGK Blast | This paper | N/A |
| pB CAG eGFP-FLAG pGK Blast | This paper | N/A |
| pB EF1alpha GFP puro | Gift from Ed Grow | N/A |
| pB EF1alpha mCherry | Gift from Hannah Long | N/A |
| pCS2+ kaede | Bajpai et al. 2010 | N/A |
| pCS2+-membrane-mCherry | Megason 2009 | Addgene #53750 |
| pX330 FHS 2444* SRCAP gRNA | Cong et al. 2013 | Addgene #42230 |
| pX330 C-terminal SRCAP gRNA | Cong et al. 2013 | Addgene #42230 |
| pX330 N-terminal H2AFZ gRNA | Cong et al. 2013 | Addgene #42230 |
| pX330 N-terminal H2AFV gRNA | Cong et al. 2013 | Addgene #42230 |
| Software and Algorithms | ||
| Cutadapt | Martin et al. 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MEME | Bailey et al. 2009 | http://meme-suite.org/tools/meme |
| NLS Mapper | Kosugi et al. 2009 | http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi |
| GREAT | McLean et al. 2010 | http://great.stanford.edu/public/html/ |
| topGO (Bioconductor) | Alexa A et al. 2016 | http://bioconductor.org/packages/topGO |
| igv | Broad Institute and Regents of the University of California Robinson et al. 2011. | http://www.broadinstitute.org/igv |
| The R package for Statistical Computing | R Core Team, 2013 | https://www.r-project.org/ |
| Samtools (v 1.4) | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Macs2 (v2.1.120160309) | Zhang et al., 2008 | https://github.com/taoliu/MACS |
| DESeq2 | Love et al., 2014 | http://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| BEDtools | Quinlan et al. 2010 | https://github.com/arq5x/bedtools2 |
| hisat2 | Kim et al. 2011 | https://ccb.jhu.edu/software/hisat2/index.shtml |
| featureCounts (subread package) | Liao et al. 2014 | http://subread.sourceforge.net/ |
| SVA (Bioconductor) | Leek et al. 2012 | http://bioconductor.org/packages/release/bioc/html/sva.html |
| The PyMOL Molecular Graphics System (v2.0) | Schrödinger, LLC. | https://pymol.org/2 |
| FlowJo | FLOWJO LLC | https://www.flowjo.com/ |
| FIJI | Schindelin et al. 2012 | https://imagej.net/Fiji |
| BD FACSAria II | BD Biosciences | http://www.bdbiosciences.com/cn/home |
| NextSeq 500; The NextSeq System Suite software; NCS v2.0 | Illumina | ftp://webdata2:webdata2@ussd-ftp.illumina.com/downloads/Software/nextseq/nextseq-system-suite-v2-0-1.zip |
| Bioanalyzer | Agilent | https://www.genomics.agilent.com/en/Bioanalyzer-System/2100-Expert-Software/?cid=AG-PT-106&tabId=AG-PR-1002 |
| MATLAB (v.2014b) | The Mathworks, Inc. 1994–2018 | https://www.mathworks.com/products/matlab.html |
| 3D Slicer | Federov et al. 2012 | http://www.slicer.org |
| Other | ||
| QImaging MicroPublisher camera | QImaging | N/A |
| Schneider Xenoplan 1:1 telecentric objective | Schneider | N/A |
| Leica imaging stereoscope | Leica | N/A |
| Leica TSC SP2 confocal microscope | Leica | N/A |
| Bioruptor Plus B01020001 | Diagenode | N/A |
| Covaris sonicator E220 | Covaris | N/A |
| Amersham Imager 680 | Amersham | N/A |
| LI-COR Odyssey CLX | LI-COR Biosciences | N/A |
Method Details
CNCC derivation and culture
Embryonic stem cell lines were differentiated into cranial neural crest cells as described (Rada-Iglesias et al. 2012; Prescott et al. 2015). hESCs were grown to confluence and colonies were detached from the plate with 2mg/ml collagenase (Invitrogen). Cells were then washed with PBS and manually dissociated into clusters of 100–200 cells by pipetting. The cells were then resuspended in NCC differentiation media and transferred into a petri dish. NCC differentiation media includes - 1:1 Neurobasal medium/D-MEM F-12 medium (Invitrogen), 0.5× B-27 supplement with Vitamin A (50× stock, Invitrogen), 0.5× N-2 supplement (100× stock, Invitrogen), 20 ng/ml bFGF (Peprotech), 20 ng/ml EGF (Sigma-Aldrich), 5 μg/ml bovine insulin (Sigma-Aldrich) and 1× Glutamax-I supplement (100× stock, Invitrogen). Neural spheres were cultured for 4 days with media changed daily and spheres separated from debris by gentle centrifugation. Then neural spheres were allowed to sit for 3 days so sphere attachment would begin to occur. Media was then changed daily to allow neural crest cells to migrate out of the neural rosettes for ~4 days. Neural crest cells were then dissociated with accutase and passaged onto 7.5μg/ml fibronectin coated plates, removing any spheres. These cells were moved into Early Maintenance Media, which includes: 1:1 Neurobasal medium/D-MEM F-12 medium (Invitrogen), 0.5× B-27 supplement with Vitamin A (50× stock, Invitrogen), 0.5× N-2 supplement (100× stock, Invitrogen), 20 ng/ml bFGF (Peprotech), 20 ng/ml EGF (Sigma-Aldrich), 1 mg/ml bovine serum albumin, serum replacement grade (Gemini Bio-Products # 700–104P) and 1× Glutamax-I supplement (100× stock, Invitrogen). Neural crest-derived cells were cultured on fibronectin plates, passaging every ~3 days with accutase. After 2 additional passages, media was changed to BMP/ChIR media, which increased cell proliferation and decreased migration, and included the components from early maintenance media plus 3μM ChIRON 99021 (Selleck, CHIR-99021) and 50pg/ml BMP2 (Peprotech).
Overexpression experiments (NCCs and 293T)
HEK293T cells were maintained as previously described and CNCCs were maintained in BMP/ChIR media, but media for transfection experiments did not include antibiotics. The overexpression plasmids used were: pB CAG WT-SRCAP-eGFP-FLAG pGK Blast, pB CAG FHS-truncated-SRCAP-eGFP-FLAG pGK Blast, pB CAG AT-hooks-eGFP-FLAG pGK Blast, and pB CAG eGFP-FLAG pGK Blast (pB = piggybac). CNCCs were split 2 hours prior to transfection, HEK293T cells were split the day before. 2.5 μg of DNA was transfected for each construct in 1:3 ratio of DNA to FuGENE 6 (Promega) for passage 4 CNCCs into each well and 5.0 μg of DNA was transfected with Lipofectamine 2000 (ThermoFisher) into each well of HEK293T cells, with further details according to each reagent’s published protocol. Media was changed 2 hours after transfection and cells were allowed to grow for 1–3 days until expression could be observed under an epifluorescence microscope at which time they were harvested for downstream experiments.
Immunoprecipitation
Whole cell lysate was extracted with cold extraction buffer (300 mM NaCl, 100 mM Tris pH 8, 0.2 mM EDTA, 0.1% NP40, 10% glycerol) with protease inhibitors and lysed for 30 minutes at 4°C then centrifuged to remove debris. Samples were quantified with Bradford reagent. 10% of sample was saved as input and the rest of the sample was added to either washed FLAG M2 agarose beads (20μl bead slurry; Sigma Aldrich) or with V5 agarose beads (20μl bead slurry; Abcam ab1229). Samples were allowed to bind to antibodies for 2 hours at 4ºC and were then washed four times with extraction buffer without glycerol. IPs and inputs were resuspended in Laemmli buffer and boiled. Samples were separated on Tris-glycine PAGE gels, then transferred in high-molecular weight transfer buffer (39mM glycine, 48mM Tris base, 20% methanol, 0.1% SDS), blocked with 6% milk in PBST, and immunoblotted with HA antibody (1:2000 dilution; Abcam 9110) or V5 (1:2000 dilution; Abcam 27671), probed with the appropriate secondary antibody, and using high-sensitivity chemiluminescence reagent (GE Lifesciences). Imaging of immunoblots was done with a film developer or Amersham Imager 680 (indicated in figure legends).
Cell cycle staining and flow cytometry
Staining for cell cycle analysis was done with Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry reagents (ThermoFisher) and staining for DNA content with FxCycle Violet Stain (ThermoFisher) according to the manufacturer’s protocol. Flow cytometry was used to determine the number of cells in G1, G2/M and S phases of the cell cycle (AriaII SORP and BD Fortessa) and cell numbers for each cell cycle phase were assessed via gate statistics in FlowJo.
Chromatin Immunoprecipitation and preparation of ChIP-seq libraries
Approximately 7.5 million cells were fixed with 1% methanol-free formaldehyde for 5–10 minutes, and quenched with a final concentration of 0.125M glycine. Samples were resuspended in Lysis buffer 1 (50 mM Hepes-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) with protease inhibitors (Sigma-Aldrich), rotated (10 min at 4°C), centrifuged (1350 × g at 4°C). Then pellets were resuspended in Lysis buffer 2 (10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) with protease inhibitors (Sigma-Aldrich), rotated (10 min at room temperate), and centrifuged (1350 × g at 4°C). Next, pellets were resuspended in Lysis buffer 3 (10 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-Deoxycholate, 0.5% N-lauroylsarcosine) with protease inhibitors (Sigma-Aldrich). The resuspended chromatin was sheared using a Covaris sonicator to produce fragments of approximately 500–2000bp. Triton X-100 (to 1% final concentration) was added to the fragmented chromatin, which was immunoprecipitated overnight at 4°C with 5μg antibody (V5 antibody, rabbit polyclonal, Abcam 15828). Protein G Dynabeads (ThermoFisher) were first blocked with Block solution (0.5% BSA (w/v) in 1× PBS) and then added to chromatin to select for antibody-bound chromatin during a 4–6 hour incubation. After washing the beads with RIPA wash buffer (50 mM Hepes-KOH pH 7.5, 500 mM LiCl, 1 mM EDTA, 1% NP-40, 0.7% Na-Deoxycholate). Crosslinking was reversed in Elution buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS) at 65°C overnight (12–16 hours). RNase A (0.2 mg/mL) was added and samples were incubated at 37°C for 2 hours to digest RNA, and Proteinase K (0.2 mg/mL) was added and samples were incubated at 55°C for 2 hours to digest protein, and the DNA was purified by phenol-chloroform extraction. 10–30ng of DNA were used for library preparation with end repair, A-tailing, and adaptor ligation (New England Biosciences). The libraries were then size selected for a size of 220–500 bps by separation by gel electrophoresis, and gel extraction. The libraries were then indexed using NEBNext Multiplex Oligos for Illumina kit (Cat# E7335S) and 12–15 amplification cycles for ChIP-seq. Adaptors were depleted via size selection on a gel or with Ampure beads. Library quality and quantity was assessed by Bioanalyzer, multiplexed ten to twelve samples per lane for next-gen sequencing by single-end, 75bp reads on a NEXTseq platform or paired-end, 75bp reads on the HiSeq 4000 platform.
RNA isolation and preparation of RNA-seq libraries
Total RNA was extracted from at least 1×106 CNCCs at passage 5 on fibronectin from two independent differentiations using Trizol reagent (Invitrogen). 10μg of total RNA underwent two rounds of polyA tail purification with Dynal oligo(dT) beads (Invitrogen). The mRNA was then fragmented with 10X Fragmentation Buffer (Ambion) for exactly 5 minutes and the fragmented mRNA was purified. First strand cDNA synthesis was performed with Random Hexamer Primers (Invitrogen) and SuperScript II enzyme (Invitrogen). Next, second strand cDNA synthesis was performed with RNase H (Invitrogen) and DNA Pol I (Invitrogen), and cDNA was purified with a QIAquick column (Qiagen). 30ng of cDNA was used for library preparation, with end repair, A-tailing, and adaptor ligation (New England Biosciences). The double stranded cDNA was then prepared for sequencing using NEBNext Multiplex Oligos for Illumina kit (Cat# E7335S) and 10–12 amplification cycles for RNA-seq. Adaptors were depleted via a size selection with magnetic beads (Agencourt XP). Library quality and quantity was assessed by Bioanalyzer and multiplexed four to six samples per lane for single-end, 75bp reads on Illumina HiSeq 2500 or NEXTseq platform.
Immunocytochemistry
CNCCs were plated onto coverslips coated with 10μg/ml fibronectin. Cells were fixed in fresh 4% paraformaldehyde in PBS for 10–15 minutes at room temperature. Coverslips were then washed 3 times with PBS for 5 minutes and permeabilized (if not cell surface marker) with 0.1% Triton in PBS for 5 minutes. Slides were then incubated in block solution (1–3% BSA and 0.1% TritonX-100 in PBS) for 1 hour to overnight and then incubated with primary antibody for 1 hour to overnight depending on the antibody, washed 3 times with PBS for 5 minutes, and incubated in secondary antibody (1:500 dilution) for 1 hour in dark chamber. Slides were mounted with Vectashield with DAPI (Vector Laboratories) and imaged on a confocal microscope (Leica TSC SP2). Antibodies used were: TFAP2A (1:50 dilution; Santa Cruz, sc-12726), p75 (1:200 dilution; Promega, G3231), NR2F1 (1:50 dilution; Santa Cruz, SC-74560X), or GFP (1:500; Abcam, ab290).
Surface marker flow cytometry
Five “cluster of differentiation” markers previously identified in our laboratory were used to assess the ability of WT SRCAP and FHS MUT SRCAP lines to make neural crest cells (Prescott et al. 2015). These markers were CD10 (MME), CD99, CD105 (ENG), CD266 (TNFRSF12A) and CD271 (NGFR, p75) (Miltenyi Biotec). Cells were stained with surface marker antibodies in FACS buffer (5% FBS, 5mM EDTA, 0.1% sodium azide in PBS) and surface expression of each marker was determined using flow cytometry (AriaII SORP and BD Fortessa). All antibodies used (5μL each per sample) for surface marker analysis: CD10-APC, CD24 PerCP-Vio700, CD99-APC-Vio770, CD105-VioBlue, CD266-PE, CD271-PE-Vio770 (Miltenyi).
Chromatin fractionation
Cells were lysed in buffer A (10mM HEPES pH 7.9, 10mM KCl, 1.5mM MgCl2, 0.34M sucrose, 10% glycerol, 5mM EDTA) with protease inhibitors to extract the cytoplasm. The soluble nuclear fraction was then extracted with No-salt Buffer (3mM EDTA, 0.5mM EGTA) with protease inhibitors. The chromatin bound fraction was then resuspended in buffer A without EDTA (10mM HEPES pH 7.9, 10mM KCl, 1.5mM MgCl2, 0.34M sucrose, 10% glycerol) with protease inhibitors (Sigma-Aldrich), plus 20mM CaCl2. The chromatin bound fraction was digested with micrococcal nuclease (1 Sigma u/uL) for 2 minutes. Reaction was stopped by the addition of EGTA to a final concentration 1mM. Each fraction was then clarified by high-speed sonication. These fractions were resuspended in Laemmli buffer and boiled. Then samples were separated on a Tris-glycine PAGE gel, then in high-molecular weight transfer buffer (39mM glycine, 48mM Tris base, 20% methanol, 0.1% SDS), blocked with 6% milk in PBST, and immunoblotted with antibodies including GFP (1:1000 dilution; Abcam, ab290), SRCAP (1:1000 dilution; Kerafast), V5 (1:2000 dilution; Abcam 27671), HSP90 (1:1000 dilution; CST C45G5), TFAP2A (1:500 dilution; Santa Cruz 12726), H2AZ (1:1000 dilution; Abcam 4174), CREBBP (1:500 dilution; Santa Cruz SC-583) probed with the appropriate secondary antibody (Thermo Fisher), and developed using high-sensitivity chemiluminescence reagent (GE Lifesciences). The biochemical fractionation procedure and microccocal nuclease treatment was previously described in more detail (Wysocka et al. 2001; Méndez & Stillman 2000). Immunoblots were imaged using X-ray film.
Xenopus laevis embryology and Morpholino experiments
Xenopus laevis embryos were staged based on Nieuwkoop and Faber staging series.
Embryos were injected at the two-cell stage, with 5nl of morpholino and/or mRNA injected into each blastomere at a concentration determined for each morpholino. Morpholinos were designed with experts at Gene Tools (Eisen 2008; Morcos 2007). The doses are as follows: 5μM FHS SRCAP morpholino (5’-CCTTAAGTACAAAAGGTGTAGGGC-3’), 10μM FHS SRCAP morpholino #2 (5’-ACTTGCTCCTTAAGTACAAAAGGTG-3’), 2.5μM H2A.Z.2 morpholino (5’-ACAGGGAACTAAGAGGTACAAAGGA-3’), 2.5μM each of two H2A.Z.1 morpholinos (5’-ACCACAAAAGTCACTTACCACCACC-3’ and 5’-GCACTACAAAAATGTACTTACCACC-3’ to recognize both X.laevis genes). Morpholino concentrations were determined in a dose titration curve for each morpholino independently and were subsequently all injected in the same batches (FHS SRCAP MO #2 was injected separately). At these dosages, embryos were able to develop through Nieuwkoop and Faber late tadpole stages (stage 40–45), at which time they were fixed and the cartilage was stained with Alcian blue stain (20% (w/v) Alcian Blue 8GX, 70% (v/v) ethanol, 30% (v/v) glacial acetic acid), and imaged (Jones & Smith 2008). A trained observer blindly scored the images for presence of the observed craniofacial abnormalities, specifically the identified characteristic phenotype (see quantification and statistical analysis section for further details). Whole mount images of Alcian stained tadpoles were taken using transmissive and incident light on Leica imaging stereoscope (LEICA M165 FC/DFC7000 T). Craniofacial cartilage was also dissected and imaged on the Leica imaging stereoscope. Samples with significant amounts of pigment (H2A.Z.2 dose titration) were bleached (3% hydrogen peroxide in 1% KOH) using a lightbox for visualization purposes.
Optical projection tomography of Xenopus laevis craniofacial cartilage
Alcian blue stained specimens were embedded in 1% agarose, agarose blocks were dehydrated with methanol and cleared with BABB solution (1:2 v/v benzyl alcohol:benzyl benzoate). Specimens were mounted on custom build motorized rotation stage with 5 degrees of freedom adjustment of the rotation axis and immersed in BABB bath. Transmitted light projections at 0.36 degree intervals, 1000 per specimen were collected with QImaging MicroPublisher camera outfitted with Schneider Xenoplan 1:1 telecentric objective.
H2A.Z.1 and H2A.Z.2 mRNA cloning and FHS SRCAP X.laevis rescue experiments
H2A.Z.1 and H2A.Z.2 were cloned into a pcDNA3.1+ vector with an N-terminal HA tag. Mutant forms of H2A.Z.1 (T14A, S38T, V127A) were synthesized in geneBlocks by IDT, cloned into PGEM-T, then transferred into pcDNA3.1+ with an N-terminal HA-tag. PCR primers including the coding region and the T7 promoter were designed and a product was amplified. 5’ capped mRNA was synthesized and a polyA tail was added using the MessageMachine T7 Ultra kit (Ambion), and the mRNA was cleaned up with a MegaClear kit (Ambion). To test the capacity of WT and FHS SRCAP to rescue the FHS morphant phenotype, 5.0 μM FHS MO was co-injected with 200pg of GFP-FLAG expressing plasmid, WT SRCAP-GFP-FLAG, or FHS SRCAP-GFP-FLAG (pB plasmids previously described above in section ‘Overexpression experiments’). The embryos were assessed for GFP expression and then allowed to develop For rescuing H2A.Z.1 and/or H2A.Z.2, 5.0μM H2A.Z.1 MOs (to more consistently get the severe phenotype) was co-injected with 2.5ng of H2A.Z.1 mRNA, and 2.5μM H2A.Z.2 MO was co-injected with 2.5ng of H2A.Z.2 mRNA. For rescuing the FHS SRCAP craniofacial defects, 2.5 ng/embryo of H2A.Z.1 or H2A.Z.2 mRNA (or mutant H2A.Z.1 T14A, S38T, V127A) were co-injected with 5μM of the FHS SRCAP morpholino. For all experiments in which craniofacial cartilage was visualized and imaged, embryos were able to develop through Nieuwkoop and Faber late tadpole stages (stage 40–45), at which time they were fixed and the cartilage was stained with Alcian blue stain (20% (w/v) Alcian Blue 8GX, 70% (v/v) ethanol, 30% (v/v) glacial acetic acid) and were imaged for a trained observer to blindly score for presence of the characteristic craniofacial abnormalities or for rescue.
X. laevis kaede migration assay
Embryos were allowed to develop to the four-cell stage and then one of the two dorsal cells was injected with 250pg mCherry mRNA and 5μM FHS SRCAP morpholino. At the eight-cell stage, both dorsal animal cells from the same embryos were injected with 250pg kaede mRNA. Starting at stage 16, each embryo was placed in a separate well of a 24 well plate and imaged at 0 hours, 5 hours and 15 hours on a Leica imaging stereoscope (LEICA M165 FC/DFC7000 T).
Knockdown validations by protein analysis and cDNA validation
Embryos were allowed to develop to Nieuwkoop and Faber stages 14 (neurula stage for mRNA overexpression) or 22–26 (tailbud stage for MO only) and non-embryonic tissue was dissected away. For validation by western blot, two to five tadpoles were used per experiment to obtain enough protein. Whole cell lysate was extracted with cold extraction buffer (300 mM NaCl, 100 mM Tris pH 8, 0.2 mM EDTA, 0.1% NP40, 10% glycerol) with protease inhibitors for 30 minutes at 4°C then centrifuged to remove debris. The samples were then sonicated for 20 minutes (30 seconds on, 30 seconds off per cycle) at 4°C and then centrifuged again to reduce genomic DNA contamination. The resulting lysates were quantified with Bradford reagent and then resuspended in Laemmli buffer and boiled. Then samples were separated on Tris-glycine PAGE gels, then transferred in High-Molecular Weight Transfer Buffer, blocked with 6% milk in PBST, and immunoblotted with SRCAP C-terminal antibody (1:1000 dilution; Kerafast), SRCAP N-terminal antibody (1:500 dilution; Abcam ab99408), H2A.Z antibody (1:1000 dilution; Abcam ab4174 or 1:1000 dilution; Active Motif 39113, as indicated in figure), HA antibody (1:1000 dilution; Abcam ab18181), alpha-tubulin antibody (1:500 dilution; Sigma-Aldrich T9026), H3 antibody (1:1000 dilution; Abcam ab1791 for chemiluminescence or 1:1000 dilution; Abcam ab10799 for LI-COR imaging) probed with the appropriate secondary antibody, and developed using high-sensitivity chemiluminescence reagent (GE Lifesciences) or quantitatively using a LI-COR Odyssey imager. For validation by cDNA analysis, 5–10 embryos were used per experiment. Total RNA was extracted using Trizol reagent (Invitrogen) and first-strand cDNA was synthesized with SuperScript reverse transcriptase. Analytical PCR was then performed using Taq polymerase with appropriate primer sets (see STAR Methods for primers) and separated on an agarose gel.
In situ hybridizations
In situ probe templates were amplified from Xenopus laevis cDNA and cloned into a p-GEM-T vector (Promega). The antisense transcript was synthesized using the appropriate T7 or SP6 polymerase using MEGAscript kit (ThermoFisher) with 10X DIG UTP RNA labeling mix (Sigma-Aldrich) or using DIG RNA labeling Kit (Sigma-Aldrich #11175025910). In situ probe sequences are reported in Supplemental Table 1. X. laevis embryos were injected asymmetrically with 5μM of the FHS SRCAP MO and 500pg kaede mRNA, then targeted embryos were selected at early neurula stage and sorted into left-injected or right-injected groups based on expression of the fluorescent marker for separate processing. Once sorted, embryos were collected at either Nieuwkoop and Faber stage 14 (neurula stage) or stage 26–28 (tailbud stage) and were fixed in MEMFA (0.1 M MOPS pH 7.4, 2 mM EGTA, 1 mM MgSO4, 3.7% (v/v) formaldehyde) and stored in 100% methanol at −20°C. For in situ hybridization, embryos were rehydrated, washed in PBST (2 times, 5 min) and bleached in a light box for 1–2 hours (bleaching solution: H2O, 1xSSC, 1% hydrogen peroxide, 5% formamide, made fresh in this order). After washing, embryos were treated with 10μg/ml Proteinase K in PBST for 5 minutes, and acetylated in 0.1M triethanolamine with 25μl acetic anhydride per 10 ml volume (2 times, 5 min incubation each), then washed and refixed for 20 min with 4% PFA. Embryos were prehybridized in HYB buffer (50% formamide, 5XSSC, 1mg/ml Torula RNA, 100μg/ml heparin, 1X Denhart’s, 0.1% Tween-20, 0.1% CHAPS (w/v), 10mM EDTA) for 6 hours at 60°C, then replaced with 1μg/ml probe in HYB buffer and hybridized overnight at 60°C. Embryos were washed with HYB buffer at 60°C (1 time, 3 min), then 2XSSC Buffer at 60°C (2 times, 3 minutes; 3 times 20 min), then 2x SSC with 20 μg/ml RNase A for 30 min at 37°C, then 2XSSC Buffer (1 time, 10 min, RT), then 0.2XSSC Buffer (2 times, 30 min, 60°C), then Maleic Acid Buffer (MAB) pH 7.5 (2 times, 15 min, RT), and then MAB + 2% BMB Blocking Reagent (Sigma-Aldrich) (1 hour, RT). Embryos were incubated in MAB with 2% BMB Blocking Reagent and 1/3000 dilution of the anti-digoxygenin AP antibody (Sigma-Aldrich) overnight at 4°C. Embryos were then washed in MAB (5 times, >1 hour, RT), then washed in Alkaline Phosphatase buffer (5M NaCl, 1M Tris pH 9.5, 1M MgCl2 with 0.005 M levamisol and 1% Tween freshly added) (2 times, 5 min). Embryos were developed gradually in BM Purple (Sigma-Aldrich) in the dark until signal appeared. The developing reaction was stopped by washing with MAB and embryos were refixed in 4% PFA for 1 hour at RT, washed with 2XSCC then dehydrated in 100% methanol for storage. Protocol adapted from previously published protocols (Sive et al. 2000; Monsoro-Burq 2007). Embryos were imaged using transmissive and incident light on Leica imaging stereoscope (LEICA M165 FC/DFC7000 T) and images were visualized in FIJI.
Quantification and Statistical Analysis
ChIP-seq analysis
ChIP-seq reads were trimmed with cutadapt and aligned with bowtie2 to hg19 reference genome as previously described (Rada-Iglesias et al. 2010; Prescott et al. 2015). ChIP peaks were called in MACS2 with callpeak using --broad setting with untagged line ChIP as control. To obtain a statistically robust set of H2A.Z.1 and H2A.Z.2 biased sites, read counts over macs2 called peaks together with large set of random decoy sites was calculated with bedtools coverage and differential count analysis was performed with DESseq2 platform. Differential ChIP peak analysis between statistically robust peaks from previous steps was performed with DESeq2. Peaks were classified based on a chromatin modification dataset (Prescott et al. 2015) with k-means algorithm (R). Bigwig files for genome browser visualization were generated with MACS2 and visualized in IGV. Gene ontology term analysis for enhancers were performed using GREAT association rules (McLean et al., 2010). Motif analysis for ChIP peaks was performed in MEME for identified H2A.Z.1 or H2A.Z.2 biased enhancers, with identified CNCC enhancers as background model (Bailey et al. 2009). CNCC enhancer dataset utilized was previously published (Prescott et al. 2015).
RNA-seq analysis and enhancer association
RNA-seq reads were aligned with hisat2 and tabulated using featureCounts (subread package) against GENCODE 24 gene models, differential analysis from three wildtype biological replicates and four mutant biological replicates from two differentiations was performed with DESeq2. Unaccounted variation between replicates and batches were removed using SVA. Genes with expression changes were associated with enhancers by proximity to TSS and statistical analysis was done in R using the Wilcoxon-Mann-Whitney test. Gene ontology term analysis conducted for RNA-seq data was performed in topGO. CNCC differentiation expression data was previously published (Rada-Iglesias et al. 2012).
Cell cycle analysis and quantification
After determining the number of cells in G1, G2/M and S phases of the cell cycle by flow cytometry, cell counts for each cell cycle phase was obtained through gate statistics in FlowJo. Statistical analysis was performed in R with quasi-binomial logistic regression model in glm.
Surface marker analysis
The five “cluster of differentiation” markers - CD10, CD99, CD105, CD266 and CD271– that had been previously identified in our laboratory (Prescott et al. 2015) were assessed via surface expression by flow cytometry and analyzed in FlowJo (AriaII SORP and BD Fortessa). Cell counts were obtained through gate statistics in FlowJo and statistical analysis was performed in R using multivariate ANOVA.
Quantification of protein levels
For immunoblots imaged quantitatively using a LI-COR Odyssey imager, signal intensity of each band was quantified in LI-COR Image Studio or in FIJI (as indicated in figure legend). Relative signal intensity of protein of interest versus loading control was calculated and graphed in R.
Immunofluorescence localization quantification and statistical analysis
Images were taken with 20X objective on a confocal microscope (Leica TSC SP2) and transfected cells were identified and categorized based on location of GFP signal compared to DAPI signal in FIJI. Categories were defined as: nuclear, cytoplasmic, or both, with the nucleus defined by a mask of the DAPI signal. Cell counts indicated in figure legend. Quantification, graphical representation, and statistical analysis using 95% confidence intervals from quasibinomial glm model were generated in R. Sample size for experiments were: WT SRCAP (n=714), FHS SRCAP (n=325), AT hooks (n=694), GFP only control (n=389).
NLS analysis
Amino acid sequence for wildtype SRCAP and FHS 2444* SRCAP were input into NLS Mapper which calculates levels of NLS activity specific for importin αβ pathway (Kosugi et al. 2009). Both monopartite and bipartite sequences are reported.
PyMOL analysis
Published H2A.Z.1 and H2A.Z.2 structures were obtained from PDB (3WA9 and 3WAA respectively), aligned and RMSD predicted using align command. The loop 1 residue structure is displayed and regions were demarcated using PyMOL GUI tools. Additional information displayed was extracted from previous published datasets (Horikoshi et al. 2013; Horikoshi et al. 2016; Hong et al. 2014).
Xenopus laevis image quantification and statistical analysis
For the following experiments in Xenopus laevis embryos, sample size and number of independent biological experiments is indicated in the relevant figure and/or figure legend:
Blind analysis of craniofacial phenotype
For each experiment, a blinded dataset was generated of Alcian blue-stained Xenopus embryo craniofacial cartilage. Each image in the dataset was given a random number and presented in a random order. The blinded observer was trained to categorize the craniofacial phenotype for each Xenopus embryo with a training set (consisting of images not included within any dataset). Categories used (as indicated in figures) were: normal, abnormal, subtle abnormal, other craniofacial phenotype, or unknown. The trained, blinded observer went through the dataset and categorized the craniofacial phenotype for each image (characterization of “unknown” was not included in the quantifications). The resulting quantifications were graphed and statistically analyzed in R using Fisher’s Exact Test (FET) and Pearson’s chi-squared 2-sample test for equality of proportions with continuity correction as indicated in the figure legends.
In situ hybridization phenotype
Images were taken for each probe set of in situ hybridized embryos (asymmetrically injected with SRCAP MO #1). For each embryo, the MO injected side was compared to the uninjected (control) side. The expression pattern for the gene was characterized as normal or abnormal with MO injection. For each in situ phenotype, a representative image is shown in the figure and the sample size and phenotypic distribution are indicated in the figure legend (Figure 1H).
Gut looping phenotype
Images of wildtype and FHS MO #1 injected embryos were characterized as displaying normal or abnormal looping (see Figure S1 for example phenotype) and the resulting quantifications were graphed and statistically analyzed in R using Pearson’s chi-squared 2-sample test for equality of proportions with continuity correction.
Xenopus laevis morphometry analysis
Quantifications of craniofacial dimensions, as described in Figure S1H, were done in Leica software and in FIJI. Tadpoles were imaged with a calibrated objective on LEICA imaging stereoscope (LEICA M165 FC/DFC7000 T). The calibrated images were used to perform measurements of total length (only animals with an intact tail were included for this measurement), interocular distance, and cartilage measurements described and shown in Figure with FIJI v2 using standard tools (straight line tool for interocular distance, freehand line tools for other length measurements, freehand selection tool for area measurements, and angle tool to determine angle) followed by measurement analysis. These measurements were imported into R, the plots were generated using boxplot function based on injected morpholino type and then statistical analysis was performed using lm, wilcox, anova and TukeyHSD, and aov functions, as indicated in the figure legend. Each type of statistical analysis is indicated in the relevant figure legend.
Kaede migration assay
Kaede migration assay images were taken at time 15 hours with mCherry visible on one side (morphant side) and kaede (GFP) visible on one or both sides. These images were used to score neural crest migration in control side and morphant sides. The number of branchial arches in these images were counted on the control and morphant sides, were quantified and the statistical analysis was conducted in R using the following statistical tests as indicated in figure legend: Pearson’s chi-squared 2-sample test for equality of proportions with continuity correction and two-sample Kolmogorov-Smirnov test.
Optical projection tomography analysis
With data collected with OPT camera, the 3D reconstruction was performed with inverse Radon transform (iradon) in MATLAB. Reconstructed volumetric data (surface models) were visualized with Slicer.
Data and Code Availability
The accession number for the RNA and ChIP sequencing data reported in this paper is NCBI GEO: GSE134532.
Supplementary Material
Supplemental Movie 1, Related to Figure 1.
3-dimensional optical projection tomography image of dissected cartilage stained with Alcian blue. Wildtype craniofacial cartilage (blue), FHS SRCAP MO craniofacial cartilage (bright green), H2A.Z.2 MO craniofacial cartilage (aquamarine).
Supplemental Table 1, Related to Figure STAR Methods.
Primers used for CRISPR-Cas9 guides and validations, Xenopus laevis RT-PCR, and to produce in situ probes.
Identifying change in the localization of FHS mutant SRCAP protein by overexpression in CNCCs and HEK293T cells, Related to Figure 2.
(A) Live cell imaging using epifluorescence microscope in CNCCs for overexpressed FLAG-GFP tagged Wildtype SRCAP, FHS mutant SRCAP, SRCAP AT hooks, FLAG-GFP alone, showing GFP signal. Scale bars shown is 100 μm.
(B) Confocal microscopy of immunofluorescence staining in HEK293T cells for overexpressed FLAG-GFP tagged Wildtype SRCAP and FHS mutant SRCAP with DAPI is in blue and SRCAP-tagged protein is in red. Immunofluorescence staining with primary antibody against GFP tag, DAPI to denote nucleus. Scale bars shown are 10μm.
(C) Chromatin fractionation in HEK293T cells for overexpressed FLAG-GFP tagged Wildtype SRCAP and FHS mutant SRCAP. Cyto – Cytoplasmic fraction, Sol.Nuc – soluble nuclear fraction, Chr-B- chromatin bound fraction. GFP primary antibody for SRCAP proteins, CREBBP in chromatin bound fraction (Chr-B) and cytoplasmic fraction (Cyto), total histone H3 and pan-H2A.Z in the chromatin-bound fraction (Chr-B).
(D) Nuclear localization signal analysis using NLS Mapper (Kosugi et al. 2009). Full protein amino acid sequence with nuclear localization signals in red, AT-hooks of SRCAP highlighted in yellow.
(E) Predicted monopartite and bipartite NLSs for Wildtype SRCAP, with NLSs lost upon SRCAP truncation in red. Score represents relative strength of NLS.
(F) Nuclear localization signal analysis for FHS MUT SRCAP 2444* in NLS Mapper (Kosugi et al. 2009). Truncated protein amino acid sequence with nuclear localization signals are in red.
In vivo recapitulation of SRCAP FHS truncation leads to a characteristic craniofacial phenotype that is phenocopied by epistatic gene H2A.Z.2, Related to Figure 1.
(A) Comparison of SRCAP orthologs. Protein domains are annotated with HSA in green, ATPase in blue, CBP-binding in red, AT-hooks in yellow, and SANT domain in purple. Protein name and relevant organism are indicated.
(B) Morpholino strategy for generating FHS truncated SRCAP mRNA, with domains defined as in (A). Splice blocking by morpholino denoted by bar-headed line at target region.
(C) Western blot of cellular extract from dissected X. laevis at tailbud stage, with wildtype and 5.0 μM FHS SRCAP MO samples used. Antibodies against C-terminal SRCAP (short and long exposures), N-terminal SRCAP (showing wildtype and truncated SRCAP), and total histone H3 (loading control). 1X and 2X dilution of each sample.
(D) RT-PCR showing successful targeting of final intron-exon junction with FHS SRCAP MO #1 at two concentrations (5.0μM, 20μM) and FHS SRCAP MO #2 (10μM). Primers designed to span exons, with expected products at (i) ~126 bp. (ii) FHS product with intron incorporated expected to be 844bp. Bands indicated with blue and red arrows, respectively.
(E) Diagram of MO targeting and expected protein product based on Sanger sequencing results from RT-PCR products from (i) wildtype (126 bp band) and (ii) FHS morphant (844 bp band) (from Fig. S1D).
(F) Ventral and lateral views of dissected X. laevis cartilage stained with Alcian blue at stage 40, Wildtype (water injected) and SRCAP FHS MO #1 (SRCAP truncation) (5.0 μM). 0.5 mm scale bar shown. Animals from n >3 biologically independent experiments.
(G) Ventral view of FHS dose titration with X. laevis cartilage stained with Alcian blue at stage 40. Wildtype (water injected), SRCAP FHS morphant (SRCAP truncation with FHS MO #1) at 0.1 μM, 1.0 μM, 5.0 μM, 10.0 μM, and 20.0 μM. 0.5 mm scale bar shown.
(H) Surface models from 3D Optical projection tomography images of dissected cartilage from Wildtype (blue) and FHS SRCAP MO #1 (green) with ventral views. 3D reconstruction produced using inverse Radon transform in MATLAB and visualized in Slicer.
(I) Images of SRCAP gut looping in wildtype and in FHS MO #1 (5.0 μM) injected morphants, with example diagrams of typical and atypical looping patterns observed on right. 0.5 mm scale bar shown.
(J) Quantification of SRCAP gut looping defect. Normal counter-clockwise gut looping is indicated in green, abnormal gut looping (typically disorganization of loops, definitively no coiling) in red. Statistical test was Pearson's chi-squared 2-sample test for equality of proportions with continuity correction. *** - p-value <2.2e-16. Animals from n=4 independent experiments.
(K) Quantitative analysis of craniofacial phenotype due to FHS truncation. Wildtype in light blue, FHS truncated in light green. At top are diagrams of features measured. Nose to tail length in red (p-value not significant), distance between eyes in pink (p-value =8.719e-12, angle between Meckel’s cartilage and ceratohyal cartilage in green (p-value < 2.12e-16), area of ceratohyal cartilage in blue (p-value < 5.046e-12), area of gillrake cartilage in orange (p-value = 1.477e-10), area of entire craniofacial cartilage in yellow (p-value = 0.03523). Statistical analysis by Wilcoxon-Mann Whitney test, n.s. - p-value > 0.05, * - p-value < 0.05, *** - p-value < 0.0005. Further details of how measurements were made can be found in STAR Methods section.
(L) Ventral view of dissected X. laevis cartilage from wildtype embryos and embryos asymmetrically injected with 10 μM of FHS SRCAP MO #1 (injected side shown on the right) stained with Alcian blue at Nieuwkoop and Faber stages 40 and 46. 0.5 mm scale bar shown.
(M) Wildtype and SRCAP FHS morphant (5.0 μM) with Alcian staining of Meckel’s cartilage at 12x magnification. Wildtype and SRCAP FHS morphant (5.0 μM) with Alcian staining of Meckel’s cartilage and ceratohyal cartilage. 250 μm scale bars shown.
(N) Ventral view of representative tadpoles for SRCAP rescue experiment. Showing wildtype, and co-injection of FHS MO and 200pg pB CAG GFP-FLAG, or pB CAG WT-SRCAP-GFP-FLAG, or pB CAG FHS-SRCAP-GFP-FLAG. Injection schematic above each image and below normal craniofacial phenotype is indicated in green, abnormal craniofacial phenotype is indicated in red. 0.5 mm scale bar shown.
(O) Ventral view of representative tadpoles of wildtype frogs and frogs injected with 10 μM FHS MO #2. Injection schematic indicated above each image and below normal craniofacial phenotype is indicated in green, abnormal craniofacial phenotype is indicated in red. 0.5 mm scale bar shown.
(P) Blinded quantification of characteristic craniofacial phenotype caused by FHS MO#2. Statistical test used was Fisher’s Exact Test (FET). FET p-value < 10e-5 = ***. Animals from n=3 independent experiments.
(Q) Western blot of cellular extract from dissected X. laevis at tailbud stage, with wildtype and 10 μM FHS MO #2 samples. Antibodies against C-terminal SRCAP and total H2A.Z (loading control). 1X and 2X dilution of each sample. Imaged on LI-COR Odyssey.
Characterizing molecular and cellular phenotypes in FHS mutant cranial neural crest cells, Related to Figure 3.
(A) Confocal microscopy of immunofluorescence staining in WT and FHS MUT SRCAP CNCCs against canonical NCC markers NR2F1 (red), TFAP2A (orange), and p75 (green) with DAPI nuclear stain (blue). 10μm scale bars shown.
(B) Fluorescence activated cell surface marker analysis of WT (WT-V5 lines #1-#3) and FHS MUT (FHS-V5 lines #1-#4) NCCs, with markers CD271 PE-Cy7, CD266 PE, CD105 PacificBlue, CD24 PerCP-Cy5, and CD10 APC. All are previously characterized markers for normal neural crest cells.
(C) Enrichment of genes involved in calcium-dependent cell-cell adhesion (GO:0016339) with gene enrichment in red and associated gene names in blue.
(D) Enrichment of migration genes enriched in HHSt11cluster 1 (J. A. Morrison et al. 2017) that are affected in FHS MUT SRCAP CNCCs with gene enrichment in red and associated gene names in blue.
(E) Cell cycle analysis of WT (WT-V5 lines #1-#3) and FHS (FHS-V5 lines #1-#4) NCCs for two distinct differentiations with cell cycle phases identified using Click-iT and FxCycle DNA staining with flow cytometry analysis. No significant differences detect by binomial regression. Confidence intervals form binomial regression (glm).
(F) CD24 flow cytometry analysis of WT and FHS MUT NCCs. CD24 by CD10 in one experiment, with three WT and three FHS MUT lines differentiated to CNCCs.
(G) Fraction of CD24-low CNCCs compared between WT (orange) and FHS MUT (blue) over n=3 independent differentiations. Statistical analysis was analysis of covariance test with p=0.00003.
(H) Imaging time course from migration assay (see Fig 3H) showing embryos selected for correct fluorescence at neurula stage (time=0 hours), migratory stage (time=5 hours) and tailbud stage (time=15 hours). 250μm scale bar shown. White line for each branchial arch and diagram on right shows number and patterning of branchial arches counted for quantifications.
Characterization of histone variant H2A.Z subtype H2A.Z.2 dysfunction in X.laevis, Related to Figure 4.
(A) In situ hybridization staining for srcap expression at tailbud stage in wildtype X. laevis tadpoles. 250μm scale bar shown.
(B) Quantification of H2A.Z protein levels from immunoblot images taken with LI-COR Odyssey. For each sample, H2A.Z signal was normalized to signal from 1X dilution of H3 (loading control). Normalization of both 1X and 2X dilution for each sample shown.
(C) RT-PCR showing specific H2A.Z.1 and H2A.Z.2 mRNA targeting. One h2afz primer set and one h2afv primer set was used for RT-PCR. WT – mRNA from control injected embryos; H2A.Z.1 MO– mRNA from H2A.Z.1 MO injected embryos; H2A.Z.2 MO– mRNA from H2A.Z.2 MO injected embryos; Z.1 MO + Z.2 MO - mRNA from embryros co-injected with H2A.Z.1 MO and H2A.Z.2 MO. Expected PCR sizes without targeting are indicated by blue arrows (h2afz – 229bp; h2afv – 200bp), splicing defects affecting PCR product size due to morpholino targeting are indicated by red arrows.
(D) Ventral view of tadpoles stained with Alcian blue at stage 40, Wildtype (water injected), SRCAP FHS morphant (SRCAP truncation) (5 μM), H2A.Z.1 morpholino (2.5 μM morpholino), H2A.Z.2 morpholino (2.5 μM morpholino). Scale bar is 0.5 mm for each image. Animals from n = 3 biologically independent experiments.
(E) Ventral view of H2A.Z.2 dose titration with X.laevis cartilage stained with Alcian blue at stage 40. Wildtype (water injected), H2A.Z.2 morpholino at 1.0 μM, H2A.Z.2 morpholino at 2.5 μM, H2A.Z.2 morpholino at 5.0 μM. Scale bar is 0.5 mm for each image.
(F) H2A.Z.1 (2.5 μM) and H2A.Z.2 morphant (2.5 μM) with Alcian staining of ceratohyal cartilage at 12x magnification. 250μm scale bars shown.
(G) Quantitative analysis of craniofacial phenotype for all morpholinos. Normal phenotypes in light blue, abnormal phenotypes in light green. Nose to tail length in red (p-value not significant), distance between eyes in pink, ceratohyal cartilage area in blue. Statistical analysis was ANOVA followed by Tukey HSD test, n.s. - p-value > 0.05, * - p-value < 0.05, ** - p-value < 0.005, *** - p-value < 0.0005.
(H) Ventral view of tadpoles stained with Alcian blue at stage 40, wildtype, H2A.Z.1 MO (5.0 μM), H2A.Z.1 MO (5.0 μM) with HA-tagged H2A.Z.1 mRNA overexpression, H2A.Z.2 MO (2.5 μM), H2A.Z.2 MO (2.5 μM) with HA-tagged H2A.Z.2 mRNA overexpression, and H2A.Z.1 MO (5.0 μM) with H2A.Z.2 MO (2.5 μM). All mRNA injected at 2.5ng each. 0.5 mm scale bar indicated for each image. Animals from n = 2 biologically independent experiments (except H2A.Z.1 MO with H2A.Z.2 MO, as only one experiment had any survival).
(I) Blinded quantification of rescue capacity for developmental defects with for H2A.Z.1 MO and H2A.Z.2 MO with overexpression of H2A.Z.2 or H2A.Z.2 mRNA, respectively. Statistical test was Fisher’s Exact Test (FET). FET p-value < 0.05 = *, FET p-value < 0.005 =**, FET p-value < 0.00005 = ****. n=2 independent experiments.
(J) Western blot of cellular extract from dissected X. laevis at neurula stage, with wildtype, H2A.Z.1 MO (5.0 μM), H2A.Z.1 MO (5.0 μM) with HA-tagged H2A.Z.1 mRNA overexpression, H2A.Z.2 MO (2.5 μM), H2A.Z.2 MO (2.5 μM) with HA-tagged H2A.Z.2 mRNA overexpression, and H2A.Z.1 MO (5.0 μM) with H2A.Z.2 MO (2.5 μM) samples. All mRNA injected at 2.5ng each. Antibodies against HA-tag (overexpressed HA-tagged H2A.Z.1 or H2A.Z.2) (mouse HA) and alpha tubulin (loading control). 1X and 2X dilution of each sample. Imaged on LI-COR Odyssey.
FHS SRCAP defects can be rescued by overexpression of histone variant H2A.Z.2 due to a single subtype-specific amino acid difference, Related to Figure 6.
(A) Ventral view of tadpoles stained with Alcian blue at stage 40, wildtype, FHS MO #1, HA-tagged H2A.Z.1 mRNA overexpression, HA-tagged H2A.Z.2 mRNA overexpression, FHS MO #1 with HA-tagged H2A.Z.1 mRNA overexpression, and FHS MO #1 HA-tagged H2A.Z.2 mRNA overexpression. All FHS MO #1 at 5.0 μM, all mRNA at 2.5ng each. 0.5 mm scale bar indicated for each image. Animals from n = 3 biologically independent experiments.
(B) Ventral view of tadpoles stained with Alcian blue at stage 40, wildtype, FHS MO #1 with HA-tagged H2A.Z.1 T14A mRNA overexpression FHS MO #1 with HA-tagged H2A.Z.1 S38T mRNA overexpression, and FHS MO #1 HA-tagged H2A.Z.1 V127A mRNA overexpression. All FHS MO #1 at 5.0 μM, all mRNA at 2.5ng each. 0.5 mm scale bar indicated for each image. Animals from n = 3 biologically independent experiments.
(C) Western blot of cellular extract from dissected X. laevis at neurula stage, with wildtype, FHS MO #1, FHS MO #1 with HA-tagged H2A.Z.1 mRNA overexpression, FHS MO #1 with HA-tagged H2A.Z.2 mRNA overexpression, FHS MO #1 with HA-tagged H2A.Z.1 T14A mRNA overexpression, FHS MO #1 with HA-tagged H2A.Z.1 S38T mRNA overexpression, and FHS MO #1 HA-tagged H2A.Z.1 V127A mRNA overexpression samples. Antibodies against total H2A.Z (endogenous H2A.Z and overexpressed HA-tagged H2A.Z.1 or H2A.Z.2 or mutants) (rabbit) and H3 (mouse, loading control). 1X and 2X dilution of each sample. Imaged on LI-COR Odyssey, changes in signal intensity of total H2A.Z in relation to control H3 signal shown below blots with quantifications done in ImageJ.
(D) Quantitative analysis of craniofacial phenotype due to FHS truncation upon rescue by overexpression with H2A.Z.1 and H2A.Z.2 mRNA. Normal range in green, abnormal range in red, partial rescue in light green. Nose to tail length in red (p-value not significant), distance between eyes in pink, and area of ceratohyal cartilage in blue. Statistical analysis ANOVA followed by Tukey HSD test, n.s. - p-value > 0.05, * - p-value < 0.05, ** - p-value < 0.005, *** - p-value < 0.0005.
(E) Quantitative analysis of craniofacial phenotype due to FHS truncation upon rescue by overexpression with H2A.Z.1 and H2A.Z.2 mRNA and H2A.Z.1 single amino acid mutants (T14A, S38T, V127A). Normal range in green, abnormal range in red, partial rescue in light green. Nose to tail length in red (p-value not significant), distance between eyes in pink, and area of ceratohyal cartilage in blue. Statistical analysis was ANOVA followed by Tukey HSD test, n.s. - p-value > 0.05, * - p-value < 0.05, ** - p-value < 0.005, *** - p-value < 0.0005.
Structural differences between H2A.Z-subtypes H2A.Z.1 and H2A.Z.2, Related to Figure 7.
(A) Alignments of crystal structures of H2A.Z.1 (purple/pink) and H2A.Z.2 (green/yellow) (Horikoshi et al. 2013; Horikoshi et al. 2016). Amino acid differences A14/T14, S38/T38, and V127/A127 denoted with arrows. Predicted SRCAP binding site denoted with gray arrows (Hong et al. 2014). RMSD calculated in Pymol.
(B) Main and side chain structures of H2A.Z.1 (pink) and H2A.Z.2 (yellow) loop 1. Amino acid difference S38/T38 denoted with arrows (Horikoshi et al. 2013; Horikoshi et al. 2016).
FHS SRCAP defects are in part due to histone variant H2A.Z.2 dysfunction, Related to Figure 5.
(A) Confocal microscopy of immunofluorescence staining in CNCCs with H2A.Z.1 or H2A.Z.2 endogenously V5- tagged; DAPI in blue, V5 (mouse antibody) in green. 10μm scale bar denoted.
(B) ChIP-seq traces for V5-tagged H2A.Z.1 (lines 1–3 overlaid, shown in gray) and V5-tagged H2A.Z.2 (lines #1-#3 overlaid, shown in gray) at the SOX9 gene. Alignment to hg19, window at chr17: 69,940,012–70,184,231. H2A.Z.1 biased peaks shaded in purple, H2A.Z.2 biased peaks shaded in green, unbiased peaks shaded in gray.
(C) Identification of statistically significant H2A.Z.1 and H2A.Z.2 biased peaks. Plot of H2A.Z.1 ChIP enrichment versus H2A.Z.2 ChIP enrichment with identification of significant Z.1 sites (red), significant Z.2 sites (green), and not significant sites (grey). X-axis is mean counts, which are normalized reads for each site with an adjusted mean for a base factor.
(D) Association of H2A.Z.1 and H2A.Z.2 biased peaks with regulatory elements. Plot of H2A.Z.1 ChIP enrichment versus H2A.Z.2 ChIP enrichment with identification of promoters (red), enhancers (blue), and other regulatory sites (grey). X-axis is mean counts, which are normalized reads for each site with an adjusted mean for a base factor.
(E) All H2A.Z.1 and H2A.Z.2 biased peaks GREAT analysis - x-axis is distance to TSS in kb, y-axis is number of region-gene associations. TSS is indicated on graph (McLean et al. 2010).
(F) All H2A.Z.1 and H2A.Z.2 biased enhancers, as defined by chromatin modifications (Fig.4E) - GREAT analysis - x-axis is distance to TSS in kb, y-axis is number of region-gene associations. TSS is indicated on graph. Background set all identified enhancer regions from CNCCs (McLean et al. 2010).
(G) GO biological processes terms for H2A.Z.1 and H2A.Z.2 biased enhancers with description, binomial rank, p-value, and FDR (McLean et al. 2010). Processes relevant to FHS and neural crest development in red.
(H)GO phenotypes (human and mouse) terms for H2A.Z.1 and H2A.Z.2 biased enhancers with description, binomial rank, p-value, and FDR (McLean et al. 2010). Phenotypes relevant to FHS symptoms in red.
(I) CRISPR/Cas9 targeting strategy in hESCs for retargeting the V5-tagged H2A.Z.2 line with endogenous tagging and/or truncation of SRCAP gene using homologous recombination with ultramers to add FLAG-HA tag. SRCAP and H2A.Z.2 protein domains annotated as previously described.
(J) Validation of FLAG-HA tagged Wildtype SRCAP and FHS mutant CNCCs by whole cell immunoprecipitation for FLAG with immunoblotting against HA tag. Immunoblot against input sample for V5 tag, endogenous SRCAP protein, and HSP90 as a loading control, with untargeted H9s as negative control.
(K) Association of statistically significant changes in H2A.Z.2 incorporation between WT and FHS MUT CNCCs with regulatory elements. Plot of H2A.Z.1 ChIP enrichment versus H2A.Z.2 ChIP enrichment with identification of TSSs/promoters (red), enhancers and other non-TSS sites (blue). X-axis is mean counts, which are normalized reads for each site with an adjusted mean for a base factor.
Acknowledgements
We thank Drs. D. Kingsley, M. Fuller, S. Prescott and Wysocka lab members for comments on the manuscript. We thank Dr. Shu-Yu Wu for his work on RT-PCR experiments, and C. Jensen for help with the migration assays. We thank Drs. C. Reid and J. Chang for their advice on in situ hybridizations. This work was supported by: NIH R35 GM131757, Stinehart-Reed and Howard Hughes Medical Institute grants to JW, NSF Graduate Research Fellowship Program (NSF GRFP) and Ruth L. Kirschstein NRSA F31 from NIDCR (1F31DE025534) awards to RSG, and Wellcome Trust, Sir Henry Wellcome Postdoctoral Fellowship (106051/Z/14/Z) to HKL.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Alexa A & Rahnenfuhrer J, 2018. topGO: Enrichment Analysis for Gene Ontology. R package version 2.34.0. [Google Scholar]
- Bailey TL et al. , 2009. MEME SUITE: tools for motif discovery and searching. Nucleic acids research, 37(Web Server issue), pp.W202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai R et al. , 2010. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature, 463(7283), pp.958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M & Sauka-Spengler T, 2010. Assembling Neural Crest Regulatory Circuits into a Gene Regulatory Network. Annual Review of Cell and Developmental Biology, 26(1), pp.581–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat N, Kwon H-J & Riley BB, 2013. A gene network that coordinates preplacodal competence and neural crest specification in zebrafish. Developmental Biology, 373(1), pp.107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonisch C & Hake SB, 2012. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic acids research, 40(21), pp.10719–10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse KM, 2010. A Framework Gene Regulatory Network Controlling Neural Crest Cell Diversification. [Google Scholar]
- Buschbeck M & Hake SB, 2017. Variants of core histones and their roles in cell fate decisions, development and cancer. Nature Reviews Molecular Cell Biology, 18(5), pp.299–314. [DOI] [PubMed] [Google Scholar]
- Calo E & Wysocka J, 2013. Modification of enhancer chromatin: what, how, and why? Molecular Cell, 49(5), pp.825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E et al. , 2018. Tissue-selective effects of nucleolar stress and rDNA damage in developmental disorders. Nature, 554(7690), pp.112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L & Zhang F, 2015. Genome Engineering Using CRISPR-Cas9 System. Methods in molecular biology (Clifton, N.J.), 1239(Chapter 10), pp.197–217. [DOI] [PubMed] [Google Scholar]
- Devotta A et al. , 2016. Sf3b4-depleted Xenopus embryos: A model to study the pathogenesis of craniofacial defects in Nager syndrome. Developmental Biology, 415(2), pp.371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryhurst D et al. , 2009. Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates. BMC biology, 7(1), p.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey A & Saint-Jeannet J-P, 2017. Modeling human craniofacial disorders in Xenopus. Current pathobiology reports, 5(1), pp.79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CJ et al. , 2017. Histone Hypervariants H2A.Z.1 and H2A.Z.2 Play Independent and Context-Specific Roles in Neuronal Activity-Induced Transcription of Arc/Arg3.1 and Other Immediate Early Genes. eNeuro, 4(4), pp.ENEURO.0040–17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirín-López JM et al. , 2009. The evolutionary differentiation of two histone H2A.Z variants in chordates (H2A.Z-1 and H2A.Z-2) is mediated by a stepwise mutation process that affects three amino acid residues. BMC evolutionary biology, 9(1), p.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J, 2008. Controlling morpholino experiments: don’t stop making antisense. Development. [DOI] [PubMed] [Google Scholar]
- Faast R et al. , 2001. Histone variant H2A.Z is required for early mammalian development. Current Biology, 11(15), pp.1183–1187. [DOI] [PubMed] [Google Scholar]
- Fedorov A et al. , 2012. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magnetic resonance imaging, 30(9), pp.1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryns J-P et al. , 1996. The Floating-Harbor syndrome: two affected siblings in a family. Clinical genetics, 50(4), pp.217–219. [DOI] [PubMed] [Google Scholar]
- Goff CL et al. , 2012. Not All Floating-Harbor Syndrome Cases are Due to Mutations in Exon 34 of SRCAP. Human mutation, 34(1), pp.88–92. [DOI] [PubMed] [Google Scholar]
- Hersh JH et al. , 1998. Changing phenotype in Floating-Harbor syndrome. American Journal of Medical Genetics Part A, 76(1), pp.58–61. [PubMed] [Google Scholar]
- Hong J et al. , 2014. The catalytic subunit of the SWR1 remodeler is a histone chaperone for the H2A.Z-H2B dimer. Molecular Cell, 53(3), pp.498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RL, Lines MA, Nikkel SM, Schwartzentruber J, Beaulieu C, Nowaczyk MJM, Allanson J, Kim CA, Wieczorek D, Moilanen JS, Lacombe D, Gillessen-Kaesbach G, Whiteford ML, Quaio CRDC, Gomy I, Bertola DR, Albrecht B, Platzer K, McGillivray G, Zou R, McLeod DR, Chudley AE, Chodirker BN, Marcadier J, Majewski J, Bulman DE, White SM, Boycott KM & Consortium18, F.C., 2012. REPOR TMutations in SRCAP, Encoding SNF2-Related CREBBP Activator Protein, Cause Floating-Harbor Syndrome. The American Journal of Human Genetics, 90(2), pp.308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi N et al. , 2016. Crystal structures of heterotypic nucleosomes containing histones H2A.Z and H2A. Open biology, 6(6), p.160127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi N et al. , 2013. Structural polymorphism in the L1 loop regions of human H2A.Z.1 and H2A.Z.2. Acta crystallographica. Section D, Biological crystallography, 69(Pt 12), pp.2431–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N, Strobl-Mazzulla PH & Bronner ME, 2014. Epigenetic regulation in neural crest development. Developmental Biology, 396(2), pp.159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM & Smith JC, 2008. An overview of Xenopus development. Methods in molecular biology (Clifton, N.J.), 461, pp.385–394. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B & Salzberg SL, 2015. HISAT: a fast spliced aligner with low memory requirements. Nature Methods, 12(4), pp.357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y et al. , 2014. Neural crest development and craniofacial morphogenesis is coordinated by nitric oxide and histone acetylation. Chemistry & biology, 21(4), pp.488–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S et al. , 2009. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proceedings of the National Academy of Sciences, 106(25), pp.10171–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M et al. , 2012. H2A.Z landscapes and dual modifications inpluripotent and multipotent stem cells underliecomplex genome regulatory functions. Genome Biology, 13(10), p.R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander R et al. , 2013. Interactions between Twist and other core epithelial-mesenchymal transition factors are controlled by GSK3-mediated phosphorylation. Nature communications, 4, p.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B & Salzberg SL, 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods, 9(4), pp.357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT et al. , 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics (Oxford, England), 28(6), pp.882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England), 25(16), pp.2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X et al. , 2016. Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1. Nature Structural & Molecular Biology, 23(4), pp.317–323. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK & Shi W, 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England), 30(7), pp.923–930. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W & Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12), p.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T et al. , 2003. Induction of neural crest in Xenopus by transcription factor AP2alpha. Proceedings of the National Academy of Sciences of the United States of America, 100(2), pp.532–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal; Vol 17, No 1: Next Generation Sequencing Data Analysis DO - 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- MathWorks, Inc, 1996. MATLAB : the language of technical computing : computation, visualization, programming : installation guide for UNIX version 5, Natwick : Math Works Inc., 1996. [Google Scholar]
- Maze I et al. , 2014. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nature Reviews Genetics, 15(4), pp.259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY et al. , 2010. GREAT improves functional interpretation of cis-regulatory regions. Nature Biotechnology, 28(5), pp.495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megason SG, 2009. In toto imaging of embryogenesis with confocal time-lapse microscopy. Methods Mol Biol. 2009; 546:317–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez J & Stillman B, 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Molecular and cellular biology, 20(22), pp.8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milet C & Monsoro-Burq AH, 2012. Neural crest induction at the neural plate border in vertebrates. Developmental Biology, 366(1), pp.22–33. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq AH, 2007. A rapid protocol for whole-mount in situ hybridization on Xenopus embryos. CSH protocols, 2007, p.pdb.prot4809. [DOI] [PubMed] [Google Scholar]
- Morcos PA, 2007. Achieving targeted and quantifiable alteration of mRNA splicing with Morpholino oligos. Biochemical and biophysical research communications, 358(2), pp.521–527. [DOI] [PubMed] [Google Scholar]
- Morrison AJ & Shen X, 2009. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nature Reviews Molecular Cell Biology, 10(6), pp.373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JA et al. , 2017. Single-cell transcriptome analysis of avian neural crest migration reveals signatures of invasion and molecular transitions. eLife, 6, p.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikkel SM et al. , 2013. The phenotype of floating-harbor syndrome: clinical characterization of 52 individuals with mutations in exon 34 of SRCAP. Orphanet journal of rare diseases, 8(1), p.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obri A et al. , 2014. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature, 505(7485), pp.648–653. [DOI] [PubMed] [Google Scholar]
- Patton MA et al. , 1991. Floating-Harbor syndrome. Journal of medical genetics, 28(3), pp.201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro C & Monsoro-Burq AH, 2013. Signaling and transcriptional regulation in neural crest specification and migration: lessons from xenopus embryos. Wiley interdisciplinary reviews. Developmental biology, 2(2), pp.247–259. [DOI] [PubMed] [Google Scholar]
- Prescott SL et al. , 2015. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell, 163(1), pp.68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pünzeler S et al. , 2017. Multivalent binding of PWWP2A to H2A.Z regulates mitosis and neural crest differentiation. The EMBO journal, 36(15), pp.2263–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR & Hall IM, 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics (Oxford, England), 26(6), pp.841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A et al. , 2010. A unique chromatin signature uncovers early developmental enhancers in humans. Nature, 470(7333), pp.279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A et al. , 2012. Epigenomic Annotation of Enhancers Predicts Transcriptional Regulators of Human Neural Crest. Cell Stem Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT et al. , 2011. Integrative genomics viewer. Nature Biotechnology, 29(1), pp.24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl DD et al. , 2006. Purification of a Human SRCAP Complex That Remodels Chromatin by Incorporating the Histone Variant H2A.Z into Nucleosomes. Biochemistry, 45(17), pp.5671–5677. [DOI] [PubMed] [Google Scholar]
- Sargent TD, 2006. Transcriptional regulation at the neural plate border. Advances in experimental medicine and biology, 589, pp.32–44. [DOI] [PubMed] [Google Scholar]
- Schindelin J et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods, 9(7), pp.676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger,, LLC, 2015. The PyMOL Molecular Graphics System, Version 1.8.
- Sinner D et al. , 2006. Global analysis of the transcriptional network controlling Xenopus endoderm formation. Development, 133(10), pp.1955–1966. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM; Early Development of Xenopus Laevis: A Laboratory Manual; Cold Spring Harbor Laboratory Press; New York, 2000; pp. 249–297. [Google Scholar]
- Vardabasso C et al. , 2015. Histone Variant H2A.Z.2 Mediates Proliferation and Drug Sensitivity of Malignant Melanoma. Molecular Cell, 59(1), pp.75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardabasso C, Hake SB & Bernstein E, 2016. Histone variant H2A.Z.2: A novel driver of melanoma progression. Molecular & cellular oncology, 3(2), p.e1073417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SM et al. , 2010. The phenotype of Floating-Harbor syndrome in 10 patients. American Journal of Medical Genetics Part A, 152A(4), pp.821–829. [DOI] [PubMed] [Google Scholar]
- Wong MM, Cox LK & Chrivia JC, 2007. The Chromatin Remodeling Protein, SRCAP, Is Critical for Deposition of the Histone Variant H2A.Z at Promoters. Journal of Biological Chemistry, 282(36), pp.26132–26139. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Reilly PT & Herr W, 2001. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Molecular and cellular biology, 21(11), pp.3820–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y et al. , 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biology, 9(9), p.R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CY et al. , 2016. Mechanisms of ATP-Dependent Chromatin Remodeling Motors. Annual review of biophysics, 45(1), pp.153–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Movie 1, Related to Figure 1.
3-dimensional optical projection tomography image of dissected cartilage stained with Alcian blue. Wildtype craniofacial cartilage (blue), FHS SRCAP MO craniofacial cartilage (bright green), H2A.Z.2 MO craniofacial cartilage (aquamarine).
Supplemental Table 1, Related to Figure STAR Methods.
Primers used for CRISPR-Cas9 guides and validations, Xenopus laevis RT-PCR, and to produce in situ probes.
Identifying change in the localization of FHS mutant SRCAP protein by overexpression in CNCCs and HEK293T cells, Related to Figure 2.
(A) Live cell imaging using epifluorescence microscope in CNCCs for overexpressed FLAG-GFP tagged Wildtype SRCAP, FHS mutant SRCAP, SRCAP AT hooks, FLAG-GFP alone, showing GFP signal. Scale bars shown is 100 μm.
(B) Confocal microscopy of immunofluorescence staining in HEK293T cells for overexpressed FLAG-GFP tagged Wildtype SRCAP and FHS mutant SRCAP with DAPI is in blue and SRCAP-tagged protein is in red. Immunofluorescence staining with primary antibody against GFP tag, DAPI to denote nucleus. Scale bars shown are 10μm.
(C) Chromatin fractionation in HEK293T cells for overexpressed FLAG-GFP tagged Wildtype SRCAP and FHS mutant SRCAP. Cyto – Cytoplasmic fraction, Sol.Nuc – soluble nuclear fraction, Chr-B- chromatin bound fraction. GFP primary antibody for SRCAP proteins, CREBBP in chromatin bound fraction (Chr-B) and cytoplasmic fraction (Cyto), total histone H3 and pan-H2A.Z in the chromatin-bound fraction (Chr-B).
(D) Nuclear localization signal analysis using NLS Mapper (Kosugi et al. 2009). Full protein amino acid sequence with nuclear localization signals in red, AT-hooks of SRCAP highlighted in yellow.
(E) Predicted monopartite and bipartite NLSs for Wildtype SRCAP, with NLSs lost upon SRCAP truncation in red. Score represents relative strength of NLS.
(F) Nuclear localization signal analysis for FHS MUT SRCAP 2444* in NLS Mapper (Kosugi et al. 2009). Truncated protein amino acid sequence with nuclear localization signals are in red.
In vivo recapitulation of SRCAP FHS truncation leads to a characteristic craniofacial phenotype that is phenocopied by epistatic gene H2A.Z.2, Related to Figure 1.
(A) Comparison of SRCAP orthologs. Protein domains are annotated with HSA in green, ATPase in blue, CBP-binding in red, AT-hooks in yellow, and SANT domain in purple. Protein name and relevant organism are indicated.
(B) Morpholino strategy for generating FHS truncated SRCAP mRNA, with domains defined as in (A). Splice blocking by morpholino denoted by bar-headed line at target region.
(C) Western blot of cellular extract from dissected X. laevis at tailbud stage, with wildtype and 5.0 μM FHS SRCAP MO samples used. Antibodies against C-terminal SRCAP (short and long exposures), N-terminal SRCAP (showing wildtype and truncated SRCAP), and total histone H3 (loading control). 1X and 2X dilution of each sample.
(D) RT-PCR showing successful targeting of final intron-exon junction with FHS SRCAP MO #1 at two concentrations (5.0μM, 20μM) and FHS SRCAP MO #2 (10μM). Primers designed to span exons, with expected products at (i) ~126 bp. (ii) FHS product with intron incorporated expected to be 844bp. Bands indicated with blue and red arrows, respectively.
(E) Diagram of MO targeting and expected protein product based on Sanger sequencing results from RT-PCR products from (i) wildtype (126 bp band) and (ii) FHS morphant (844 bp band) (from Fig. S1D).
(F) Ventral and lateral views of dissected X. laevis cartilage stained with Alcian blue at stage 40, Wildtype (water injected) and SRCAP FHS MO #1 (SRCAP truncation) (5.0 μM). 0.5 mm scale bar shown. Animals from n >3 biologically independent experiments.
(G) Ventral view of FHS dose titration with X. laevis cartilage stained with Alcian blue at stage 40. Wildtype (water injected), SRCAP FHS morphant (SRCAP truncation with FHS MO #1) at 0.1 μM, 1.0 μM, 5.0 μM, 10.0 μM, and 20.0 μM. 0.5 mm scale bar shown.
(H) Surface models from 3D Optical projection tomography images of dissected cartilage from Wildtype (blue) and FHS SRCAP MO #1 (green) with ventral views. 3D reconstruction produced using inverse Radon transform in MATLAB and visualized in Slicer.
(I) Images of SRCAP gut looping in wildtype and in FHS MO #1 (5.0 μM) injected morphants, with example diagrams of typical and atypical looping patterns observed on right. 0.5 mm scale bar shown.
(J) Quantification of SRCAP gut looping defect. Normal counter-clockwise gut looping is indicated in green, abnormal gut looping (typically disorganization of loops, definitively no coiling) in red. Statistical test was Pearson's chi-squared 2-sample test for equality of proportions with continuity correction. *** - p-value <2.2e-16. Animals from n=4 independent experiments.
(K) Quantitative analysis of craniofacial phenotype due to FHS truncation. Wildtype in light blue, FHS truncated in light green. At top are diagrams of features measured. Nose to tail length in red (p-value not significant), distance between eyes in pink (p-value =8.719e-12, angle between Meckel’s cartilage and ceratohyal cartilage in green (p-value < 2.12e-16), area of ceratohyal cartilage in blue (p-value < 5.046e-12), area of gillrake cartilage in orange (p-value = 1.477e-10), area of entire craniofacial cartilage in yellow (p-value = 0.03523). Statistical analysis by Wilcoxon-Mann Whitney test, n.s. - p-value > 0.05, * - p-value < 0.05, *** - p-value < 0.0005. Further details of how measurements were made can be found in STAR Methods section.
(L) Ventral view of dissected X. laevis cartilage from wildtype embryos and embryos asymmetrically injected with 10 μM of FHS SRCAP MO #1 (injected side shown on the right) stained with Alcian blue at Nieuwkoop and Faber stages 40 and 46. 0.5 mm scale bar shown.
(M) Wildtype and SRCAP FHS morphant (5.0 μM) with Alcian staining of Meckel’s cartilage at 12x magnification. Wildtype and SRCAP FHS morphant (5.0 μM) with Alcian staining of Meckel’s cartilage and ceratohyal cartilage. 250 μm scale bars shown.
(N) Ventral view of representative tadpoles for SRCAP rescue experiment. Showing wildtype, and co-injection of FHS MO and 200pg pB CAG GFP-FLAG, or pB CAG WT-SRCAP-GFP-FLAG, or pB CAG FHS-SRCAP-GFP-FLAG. Injection schematic above each image and below normal craniofacial phenotype is indicated in green, abnormal craniofacial phenotype is indicated in red. 0.5 mm scale bar shown.
(O) Ventral view of representative tadpoles of wildtype frogs and frogs injected with 10 μM FHS MO #2. Injection schematic indicated above each image and below normal craniofacial phenotype is indicated in green, abnormal craniofacial phenotype is indicated in red. 0.5 mm scale bar shown.
(P) Blinded quantification of characteristic craniofacial phenotype caused by FHS MO#2. Statistical test used was Fisher’s Exact Test (FET). FET p-value < 10e-5 = ***. Animals from n=3 independent experiments.
(Q) Western blot of cellular extract from dissected X. laevis at tailbud stage, with wildtype and 10 μM FHS MO #2 samples. Antibodies against C-terminal SRCAP and total H2A.Z (loading control). 1X and 2X dilution of each sample. Imaged on LI-COR Odyssey.
Characterizing molecular and cellular phenotypes in FHS mutant cranial neural crest cells, Related to Figure 3.
(A) Confocal microscopy of immunofluorescence staining in WT and FHS MUT SRCAP CNCCs against canonical NCC markers NR2F1 (red), TFAP2A (orange), and p75 (green) with DAPI nuclear stain (blue). 10μm scale bars shown.
(B) Fluorescence activated cell surface marker analysis of WT (WT-V5 lines #1-#3) and FHS MUT (FHS-V5 lines #1-#4) NCCs, with markers CD271 PE-Cy7, CD266 PE, CD105 PacificBlue, CD24 PerCP-Cy5, and CD10 APC. All are previously characterized markers for normal neural crest cells.
(C) Enrichment of genes involved in calcium-dependent cell-cell adhesion (GO:0016339) with gene enrichment in red and associated gene names in blue.
(D) Enrichment of migration genes enriched in HHSt11cluster 1 (J. A. Morrison et al. 2017) that are affected in FHS MUT SRCAP CNCCs with gene enrichment in red and associated gene names in blue.
(E) Cell cycle analysis of WT (WT-V5 lines #1-#3) and FHS (FHS-V5 lines #1-#4) NCCs for two distinct differentiations with cell cycle phases identified using Click-iT and FxCycle DNA staining with flow cytometry analysis. No significant differences detect by binomial regression. Confidence intervals form binomial regression (glm).
(F) CD24 flow cytometry analysis of WT and FHS MUT NCCs. CD24 by CD10 in one experiment, with three WT and three FHS MUT lines differentiated to CNCCs.
(G) Fraction of CD24-low CNCCs compared between WT (orange) and FHS MUT (blue) over n=3 independent differentiations. Statistical analysis was analysis of covariance test with p=0.00003.
(H) Imaging time course from migration assay (see Fig 3H) showing embryos selected for correct fluorescence at neurula stage (time=0 hours), migratory stage (time=5 hours) and tailbud stage (time=15 hours). 250μm scale bar shown. White line for each branchial arch and diagram on right shows number and patterning of branchial arches counted for quantifications.
Characterization of histone variant H2A.Z subtype H2A.Z.2 dysfunction in X.laevis, Related to Figure 4.
(A) In situ hybridization staining for srcap expression at tailbud stage in wildtype X. laevis tadpoles. 250μm scale bar shown.
(B) Quantification of H2A.Z protein levels from immunoblot images taken with LI-COR Odyssey. For each sample, H2A.Z signal was normalized to signal from 1X dilution of H3 (loading control). Normalization of both 1X and 2X dilution for each sample shown.
(C) RT-PCR showing specific H2A.Z.1 and H2A.Z.2 mRNA targeting. One h2afz primer set and one h2afv primer set was used for RT-PCR. WT – mRNA from control injected embryos; H2A.Z.1 MO– mRNA from H2A.Z.1 MO injected embryos; H2A.Z.2 MO– mRNA from H2A.Z.2 MO injected embryos; Z.1 MO + Z.2 MO - mRNA from embryros co-injected with H2A.Z.1 MO and H2A.Z.2 MO. Expected PCR sizes without targeting are indicated by blue arrows (h2afz – 229bp; h2afv – 200bp), splicing defects affecting PCR product size due to morpholino targeting are indicated by red arrows.
(D) Ventral view of tadpoles stained with Alcian blue at stage 40, Wildtype (water injected), SRCAP FHS morphant (SRCAP truncation) (5 μM), H2A.Z.1 morpholino (2.5 μM morpholino), H2A.Z.2 morpholino (2.5 μM morpholino). Scale bar is 0.5 mm for each image. Animals from n = 3 biologically independent experiments.
(E) Ventral view of H2A.Z.2 dose titration with X.laevis cartilage stained with Alcian blue at stage 40. Wildtype (water injected), H2A.Z.2 morpholino at 1.0 μM, H2A.Z.2 morpholino at 2.5 μM, H2A.Z.2 morpholino at 5.0 μM. Scale bar is 0.5 mm for each image.
(F) H2A.Z.1 (2.5 μM) and H2A.Z.2 morphant (2.5 μM) with Alcian staining of ceratohyal cartilage at 12x magnification. 250μm scale bars shown.
(G) Quantitative analysis of craniofacial phenotype for all morpholinos. Normal phenotypes in light blue, abnormal phenotypes in light green. Nose to tail length in red (p-value not significant), distance between eyes in pink, ceratohyal cartilage area in blue. Statistical analysis was ANOVA followed by Tukey HSD test, n.s. - p-value > 0.05, * - p-value < 0.05, ** - p-value < 0.005, *** - p-value < 0.0005.
(H) Ventral view of tadpoles stained with Alcian blue at stage 40, wildtype, H2A.Z.1 MO (5.0 μM), H2A.Z.1 MO (5.0 μM) with HA-tagged H2A.Z.1 mRNA overexpression, H2A.Z.2 MO (2.5 μM), H2A.Z.2 MO (2.5 μM) with HA-tagged H2A.Z.2 mRNA overexpression, and H2A.Z.1 MO (5.0 μM) with H2A.Z.2 MO (2.5 μM). All mRNA injected at 2.5ng each. 0.5 mm scale bar indicated for each image. Animals from n = 2 biologically independent experiments (except H2A.Z.1 MO with H2A.Z.2 MO, as only one experiment had any survival).
(I) Blinded quantification of rescue capacity for developmental defects with for H2A.Z.1 MO and H2A.Z.2 MO with overexpression of H2A.Z.2 or H2A.Z.2 mRNA, respectively. Statistical test was Fisher’s Exact Test (FET). FET p-value < 0.05 = *, FET p-value < 0.005 =**, FET p-value < 0.00005 = ****. n=2 independent experiments.
(J) Western blot of cellular extract from dissected X. laevis at neurula stage, with wildtype, H2A.Z.1 MO (5.0 μM), H2A.Z.1 MO (5.0 μM) with HA-tagged H2A.Z.1 mRNA overexpression, H2A.Z.2 MO (2.5 μM), H2A.Z.2 MO (2.5 μM) with HA-tagged H2A.Z.2 mRNA overexpression, and H2A.Z.1 MO (5.0 μM) with H2A.Z.2 MO (2.5 μM) samples. All mRNA injected at 2.5ng each. Antibodies against HA-tag (overexpressed HA-tagged H2A.Z.1 or H2A.Z.2) (mouse HA) and alpha tubulin (loading control). 1X and 2X dilution of each sample. Imaged on LI-COR Odyssey.
FHS SRCAP defects can be rescued by overexpression of histone variant H2A.Z.2 due to a single subtype-specific amino acid difference, Related to Figure 6.
(A) Ventral view of tadpoles stained with Alcian blue at stage 40, wildtype, FHS MO #1, HA-tagged H2A.Z.1 mRNA overexpression, HA-tagged H2A.Z.2 mRNA overexpression, FHS MO #1 with HA-tagged H2A.Z.1 mRNA overexpression, and FHS MO #1 HA-tagged H2A.Z.2 mRNA overexpression. All FHS MO #1 at 5.0 μM, all mRNA at 2.5ng each. 0.5 mm scale bar indicated for each image. Animals from n = 3 biologically independent experiments.
(B) Ventral view of tadpoles stained with Alcian blue at stage 40, wildtype, FHS MO #1 with HA-tagged H2A.Z.1 T14A mRNA overexpression FHS MO #1 with HA-tagged H2A.Z.1 S38T mRNA overexpression, and FHS MO #1 HA-tagged H2A.Z.1 V127A mRNA overexpression. All FHS MO #1 at 5.0 μM, all mRNA at 2.5ng each. 0.5 mm scale bar indicated for each image. Animals from n = 3 biologically independent experiments.
(C) Western blot of cellular extract from dissected X. laevis at neurula stage, with wildtype, FHS MO #1, FHS MO #1 with HA-tagged H2A.Z.1 mRNA overexpression, FHS MO #1 with HA-tagged H2A.Z.2 mRNA overexpression, FHS MO #1 with HA-tagged H2A.Z.1 T14A mRNA overexpression, FHS MO #1 with HA-tagged H2A.Z.1 S38T mRNA overexpression, and FHS MO #1 HA-tagged H2A.Z.1 V127A mRNA overexpression samples. Antibodies against total H2A.Z (endogenous H2A.Z and overexpressed HA-tagged H2A.Z.1 or H2A.Z.2 or mutants) (rabbit) and H3 (mouse, loading control). 1X and 2X dilution of each sample. Imaged on LI-COR Odyssey, changes in signal intensity of total H2A.Z in relation to control H3 signal shown below blots with quantifications done in ImageJ.
(D) Quantitative analysis of craniofacial phenotype due to FHS truncation upon rescue by overexpression with H2A.Z.1 and H2A.Z.2 mRNA. Normal range in green, abnormal range in red, partial rescue in light green. Nose to tail length in red (p-value not significant), distance between eyes in pink, and area of ceratohyal cartilage in blue. Statistical analysis ANOVA followed by Tukey HSD test, n.s. - p-value > 0.05, * - p-value < 0.05, ** - p-value < 0.005, *** - p-value < 0.0005.
(E) Quantitative analysis of craniofacial phenotype due to FHS truncation upon rescue by overexpression with H2A.Z.1 and H2A.Z.2 mRNA and H2A.Z.1 single amino acid mutants (T14A, S38T, V127A). Normal range in green, abnormal range in red, partial rescue in light green. Nose to tail length in red (p-value not significant), distance between eyes in pink, and area of ceratohyal cartilage in blue. Statistical analysis was ANOVA followed by Tukey HSD test, n.s. - p-value > 0.05, * - p-value < 0.05, ** - p-value < 0.005, *** - p-value < 0.0005.
Structural differences between H2A.Z-subtypes H2A.Z.1 and H2A.Z.2, Related to Figure 7.
(A) Alignments of crystal structures of H2A.Z.1 (purple/pink) and H2A.Z.2 (green/yellow) (Horikoshi et al. 2013; Horikoshi et al. 2016). Amino acid differences A14/T14, S38/T38, and V127/A127 denoted with arrows. Predicted SRCAP binding site denoted with gray arrows (Hong et al. 2014). RMSD calculated in Pymol.
(B) Main and side chain structures of H2A.Z.1 (pink) and H2A.Z.2 (yellow) loop 1. Amino acid difference S38/T38 denoted with arrows (Horikoshi et al. 2013; Horikoshi et al. 2016).
FHS SRCAP defects are in part due to histone variant H2A.Z.2 dysfunction, Related to Figure 5.
(A) Confocal microscopy of immunofluorescence staining in CNCCs with H2A.Z.1 or H2A.Z.2 endogenously V5- tagged; DAPI in blue, V5 (mouse antibody) in green. 10μm scale bar denoted.
(B) ChIP-seq traces for V5-tagged H2A.Z.1 (lines 1–3 overlaid, shown in gray) and V5-tagged H2A.Z.2 (lines #1-#3 overlaid, shown in gray) at the SOX9 gene. Alignment to hg19, window at chr17: 69,940,012–70,184,231. H2A.Z.1 biased peaks shaded in purple, H2A.Z.2 biased peaks shaded in green, unbiased peaks shaded in gray.
(C) Identification of statistically significant H2A.Z.1 and H2A.Z.2 biased peaks. Plot of H2A.Z.1 ChIP enrichment versus H2A.Z.2 ChIP enrichment with identification of significant Z.1 sites (red), significant Z.2 sites (green), and not significant sites (grey). X-axis is mean counts, which are normalized reads for each site with an adjusted mean for a base factor.
(D) Association of H2A.Z.1 and H2A.Z.2 biased peaks with regulatory elements. Plot of H2A.Z.1 ChIP enrichment versus H2A.Z.2 ChIP enrichment with identification of promoters (red), enhancers (blue), and other regulatory sites (grey). X-axis is mean counts, which are normalized reads for each site with an adjusted mean for a base factor.
(E) All H2A.Z.1 and H2A.Z.2 biased peaks GREAT analysis - x-axis is distance to TSS in kb, y-axis is number of region-gene associations. TSS is indicated on graph (McLean et al. 2010).
(F) All H2A.Z.1 and H2A.Z.2 biased enhancers, as defined by chromatin modifications (Fig.4E) - GREAT analysis - x-axis is distance to TSS in kb, y-axis is number of region-gene associations. TSS is indicated on graph. Background set all identified enhancer regions from CNCCs (McLean et al. 2010).
(G) GO biological processes terms for H2A.Z.1 and H2A.Z.2 biased enhancers with description, binomial rank, p-value, and FDR (McLean et al. 2010). Processes relevant to FHS and neural crest development in red.
(H)GO phenotypes (human and mouse) terms for H2A.Z.1 and H2A.Z.2 biased enhancers with description, binomial rank, p-value, and FDR (McLean et al. 2010). Phenotypes relevant to FHS symptoms in red.
(I) CRISPR/Cas9 targeting strategy in hESCs for retargeting the V5-tagged H2A.Z.2 line with endogenous tagging and/or truncation of SRCAP gene using homologous recombination with ultramers to add FLAG-HA tag. SRCAP and H2A.Z.2 protein domains annotated as previously described.
(J) Validation of FLAG-HA tagged Wildtype SRCAP and FHS mutant CNCCs by whole cell immunoprecipitation for FLAG with immunoblotting against HA tag. Immunoblot against input sample for V5 tag, endogenous SRCAP protein, and HSP90 as a loading control, with untargeted H9s as negative control.
(K) Association of statistically significant changes in H2A.Z.2 incorporation between WT and FHS MUT CNCCs with regulatory elements. Plot of H2A.Z.1 ChIP enrichment versus H2A.Z.2 ChIP enrichment with identification of TSSs/promoters (red), enhancers and other non-TSS sites (blue). X-axis is mean counts, which are normalized reads for each site with an adjusted mean for a base factor.
Data Availability Statement
The accession number for the RNA and ChIP sequencing data reported in this paper is NCBI GEO: GSE134532.