Abstract
Background
Saliva evaluation could be a possible alternative to blood and/or tissue analyses, for researching specific molecules associated to the presence of systemic diseases and malignancies. The present systematic review has been designed in order to answer to the question “are there significant associations between specific salivary biomarkers and diagnosis of systemic diseases or malignancies?”.
Material and Methods
The Preferred Reporting Item for Systematic Reviews and Meta-analysis (PRISMA) statement was used to guide the review. The combinations of “saliva” and “systemic diseases” or “diagnosis” or “biomarkers” or “cancers” or “carcinoma” or “tumors”, were used to search Medline, Scopus and Web of Science databases. Endpoint of research has been set at May 2019. Studies were classified into 3 groups according to the type of disease investigated for diagnosis: 1) malignant tumors; 2) neurologic diseases and 3) inflammatory/metabolic/cardiovascular diseases. Assessment of quality has been assigned according to a series of questions proposed by the National Institute of Health. Level of evidence was assessed using the categories proposed in the Oxford Center for Evidence-Based medicine (CEMB) levels for diagnosis (2011).
Results
Seventy-nine studies met the inclusion and exclusion criteria. Fifty-one (64%) investigated malignant tumors, 14 (17.5%) neurologic and 14 (18.5%) inflammatory/cardiovascular/metabolic diseases. Among studies investigating malignant tumors, 12 (23.5%) were scored as “good” and 11 of these reported statistically significant associations between salivary molecules and pathology. Two and 5 studies were found to have a good quality, among those evaluating the association between salivary biomarkers and neurologic and inflammatory/metabolic/cardiovascular diseases, respectively.
Conclusions
The present systematic review confirms the existence of some “good” quality evidence to support the role of peculiar salivary biomarkers for diagnosis of systemic diseases (e.g. lung cancer and EGFR).
Key words:Salivary diagnostics, biomarkers, systemic diseases, malignant tumors, early diagnosis.
Introduction
Currently, one of the most relevant targets of medicine and healthcare is early diagnosis. Detecting a disease at an early stage may improve the possibility of success of treatment, prevent complications and enhance prognosis and quality of life (1).
The concept of “point-of-care diagnosis” includes a field of investigation that explores technologies allowing patients and health providers to gain actionable medical information rapidly and conveniently (1).
The term “precision medicine” refers to the uses of molecular profiles, genomic, transcriptomic, proteomic and metabolomic, to adapt a personalized therapeutic strategy for peculiar patients: a. in the right moment, b. to determine the predisposition to diseases, and c. to provide timely and targeted prevention (2).
The combination of such profiles and the identification of biomarkers is leading to the development of new technologies, based on easy and non-invasive methods to collect diagnostic human specimens, possibly with a high specificity and sensitivity and customized on single patient.
In the last ten years, research has focused on the use of biomarkers in a previously poorly investigated human fluid: saliva.
Saliva is a fluid constantly produced by salivary glands and It has a complex molecular composition. Saliva is abundantly delivered in the oral cavity, its collection being simple and non-invasive. Moreover, transportation and storing are easy. For such reasons, saliva evaluation could be considered as a possible alternative to blood and/or tissue analyses, for researching specific molecules (DNA, RNA, proteins and metabolites) associated to the presence of systemic diseases and malignancies (3).
The present systematic review has been designed in order to answer to the question “are there significant associations between specific salivary biomarkers and diagnosis of systemic diseases or malignancies?”, formulated according to the “Patient-Intervention-Comparison-Outcome” (PICO) worksheet.
Material and Methods
The Preferred Reporting Item for Systematic Reviews and Meta-analysis (PRISMA) statement was used to guide this systematic review (98).
- Search strategy
The combinations of “saliva” and “systemic diseases” or “diagnosis” or “biomarkers” or “cancers” or “carcinoma” or “tumors”, have been used for searching Medline, Scopus and Web of Science databases. Only English literature was searched. We considered articles published after 2000 (endpoint of research has been set at May 2019). Periodic screening of the databases was performed, between September 2017 and May 2019.
- Inclusion and exclusion criteria
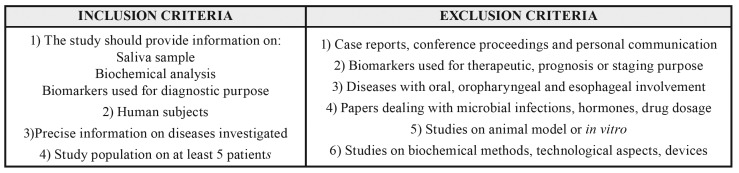
Only papers reporting details on salivary sampling and biochemical analysis were included. Papers selected were primarily focused on the use of saliva for diagnostic purposes. We only included studies performed on humans, detailing the disease of patients and providing precise information on diagnosis. Studies reporting data on at least 5 patients were included.
Case reports, conference proceedings and personal communication were excluded.
Studies dealing with biomarkers evaluated for therapy, prognosis or staging of systemic diseases were not included.
We excluded researches specifically investigating salivary biomarkers in patients with systemic diseases with oral, oropharyngeal and esophageal involvement. Such a choice was taken in order to exclude presence of pathologies in the proximity of the site of saliva collection, thus avoiding confounding factors (e.g. contamination with peripheral molecules not originally presents in the salivary secrete).
Papers dealing with systemic microbial infections, hormones, drug dosage, were further excluded.
We excluded studies specifically reporting on biochemical methods, technological aspects, devices used or proposed for saliva evaluation or detection of specific molecules.
The criteria are summarized in Table 1.
Table 1. Inclusion and exclusion criteria.
- Data extraction
Titles and abstracts were screened by two independent investigators. Equivocal titles/abstracts were included for full-text evaluation.
Reviews of literature addressing the topic of salivary biomarkers and diagnosis of systemic diseases and malignant tumors were carefully read and all references were screened in order to include papers possibly not selected through the entry terms used within the databases. Other relevant literature was identified from the reference lists of the retrieved articles.
Information extracted from each study were summarized in an Excel Table and they included title, citation date (authors, publication year), pathology investigated, type of biomarkers, device used to analyze the sample, results and presence of statistically significant association.
- Data analysis
Studies were assessed for overlapping series of patients on the basis of the recruitment Centre and period. Wherever multiple studies reported the same set of data in fully detecTable overlapping series of patients, only the most recent or the most complete series was included in the review.
Studies were classified into 3 groups according to the type of disease investigated for diagnosis: 1) malignant tumors; 2) neurologic diseases and 3) inflammatory/metabolic/cardiovascular diseases.
- Quality assessment and critical appraisal
Assessment of quality has been assigned according to a series of questions proposed by the National Institute of Health (NIH) for each typology of study (controlled intervention studies, systematic reviews and meta-analysis, observational cohort and cross-sectional studies, case control studies, before-after studies with no control group, case series studies) (4).
Critical appraisal has been summarized through assignation of a value ranging from 0 to 100% to each of the study selected, based on the percentage of “yes” choices on the overall number of answers given. Furthermore, the number of patients enrolled in each study was taken into consideration.
Studies with a percentage of quality ranging from 80 to 100% were defined as “good”. Studies with a quality ranging between 50 and 80% were defined as “fair” and studies scoring less than 50% in quality were defined as “poor”.
Level of evidence was assessed using the categories proposed in the Oxford Center for Evidence-Based medicine (CEMB) levels for diagnosis [2011] (5).
Disagreement were resolved by discussion between the reviewers.
Results
The systematic literature search provided 79 studies which met the inclusion and exclusion criteria.
Seventy-five papers were case-control studies and 4 case-series studies.
Fifty-one (64%) papers were focused on malignant tumors, 14 (17.5%) papers on neurologic diseases and 14 (18.5%) on inflammatory/cardiovascular/metabolic diseases (Table 2, Table 3, Table 4).
Table 2. Papers on salivary biomarkers for diagnosis of malignant tumors.
Table 3. Papers on salivary biomarkers for diagnosis of neurologic diseases.
Table 4. Papers on salivary biomarkers for diagnosis of inflammatory/cardiovascular/metabolic diseases.
Molecules investigated were DNA, RNA, proteins, metabolites, microbiota and combination of these.
Results of search strategy are summarized in Fig. 1.
Figure 1.
Search strategy.
Table 2 cont. Papers on salivary biomarkers for diagnosis of malignant tumors.
- Critical appraisal of the selected papers
Following the NIH guidelines modified according to the methodology of the present systematic review, 19 (24%) studies were scored as “good”, 45 (57%) were “fair” and 15 (19%) had “poor” quality.
The most frequently encountered risk of bias (ROBs) were the absence of concurrent controls (71 papers), the lack of sample size justification (70 papers), the lack of randomization (70 papers) and absence of report of blinding exposure assessors (68 papers).
- Level of evidence
Application of the Oxford CEMB guidelines highlighted that all of the selected papers have a low level of evidence (4 on 5, the fifth level being the lowest), because of their case control or case series design.
- Malignant tumors
Breast cancer was the most investigated disease (18 papers), followed by lung cancer (10 papers), gastric cancer (7 papers) and pancreatic cancer (6 papers). Other cancers included leukemia, prostate, ovarian, colorectal, pancreatobiliary and hepatocellular cancer.
Molecules most frequently investigated were proteins and RNA.
Twelve studies (23.5%) were scored as “good” and 11 of these reported statistically significant associations between molecules searched and pathology (6-17).
The twelve studies are summarized in Table 5.
Table 5. Good quality studies on salivary biomarkers for diagnosis of malignant tumors.
- Neurologic diseases
Fourteen papers were focused on neurologic diseases. Among these, 7 papers investigated Alzheimer’s disease, 6 investigated Parkinson’s diseases and one multiple sclerosis.
Two papers were scored as “good”. One study searched DJ-1 proteins in Parkinson’s patients (not statistically significant association) (18). The other one analyzed salivary metabolites in patient with Alzheimer’s disease and found a statistically significant association between disease and biomarkers (19).
Details on such papers are reported in Table 6.
Table 6. Good quality studies on salivary biomarkers for diagnosis of neurologic disease.
- Inflammatory/cardiovascular/metabolic diseases
Fourteen papers were included in this category, the pathologies investigated being diabetes, myocardial infarction, rheumatoid arthritis, gastric diseases, celiac disease and coronary syndrome.
Myocardial infarction was the most frequently studied disease (7 papers, 50%).
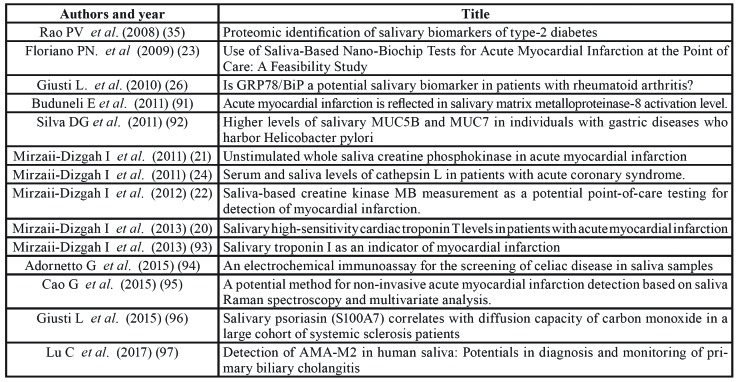
“Good” quality articles were 5, four focused on myocardial infarction and one on coronary syndrome. All studies searched for proteins and 5 of these reported significant results. (20-24)
Papers are summarized in Table 7.
Table 7. Good quality studies on salivary biomarkers for diagnosis of inflammatory/cardiovascular/metabolic diseases.
- Biochemical technologies
It goes beyond the aim of the present systematic review to critically discuss the biochemical mehods utilized in the studies included.
Almost all the studies on genetic molecules used Polymerase Chain Reaction (PCR) technique.
The studies on proteins used mainly Enzyme-Linked Immunosorbent Assays (ELISA) or Mass Spectrometry (MS). Few studies used Surface Enhanced Raman Scattering (SERS) and Luminex.
For searching metabolites, the preferred technique was MS.
Discussion
The present systematic review has highlighted an increased scientific interest toward the use of salivary biomarkers for diagnosis of systemic diseases and malignant tumors. Even limiting the field of interest only to diagnosis and applying strict inclusion and exclusion criteria, the number of papers appears considerable.
It is worthy to mention here that among the 79 papers included, only 12 studies were conducted before 2010, the rest being published after such year (12,23,25-34).
The most investigated salivary molecules are proteins (43 studies), followed by metabolites (15 studies) and RNA (12 studies). Surprisingly, the less studied salivary biomarkers are those based on DNA (3 studies) and microbiota (2 studies) analysis, despite their popularity for other aims (e.g ancestry investigations, biocompatibility for transplant, forensic analysis, dietary implications) (35,36).
In recent years, there has been a shifting of interest in the typology of molecules investigated. In fact, even if proteins remained predominant, there has been an increase of researches dealing with salivary metabolites (16 papers in 2018 and 2 papers in the first half of 2019) (37,38).
Most of the studies included in the present systematic review (69 out of 79 – 87%) showed statistically significant correlations between one or more biomarkers and specific pathologies. Such results would, in general, indicate the possibility to use peculiar salivary molecules for early diagnosis of diseases. However, critical appraisal and quality assessment highlighted that most of these studies did not satisfy a relevant percentage the items suggested by the NIH formats. As a matter of facts, only 19 studies received a score indicating “good” quality. Also, the level of evidence of all of the examined studies appears quite low.
One of the most frequently encountered ROBs was the lack of sample size justification. Calculation of a sample size is a fundamental step for creating reliable researches. Groups of patients too small have little chance of meeting the study objectives (39). Therefore, particularly for studies on salivary biomarkers it seems very important to report the justification of the population size. On the other hand, it should be taken into account that several studies included in the present review were pilot studies (27,40). For such a typology of research, it is usually difficult to provide a statistical reliable sample size justification, on the basis of the absence of background data in the literature (41). It is opinion of the Authors of the present systematic review that, the described ROB may largely depend on the fact that many of the studies took into consideration many variables at the same time (e.g. panel or combinations of very different biomolecules, biomarkers evaluated for the first time in saliva, patients with diseases at different stages), thus making more or less impossible to calculate a reliable sample size.
The second most frequently encountered ROBs involved the absence of blinding of exposure assessor. Blinding is important to remove bias that could influence the way the data is processed. The two major biases that can be controlled using blinding are the performance bias (differences that occur due to knowledge of intervention allocation, in either the researcher or the participant that cause differences in the care received) and the ascertainment bias (when data for a study or analysis is collected, surveyed, screened, or recorded, such that some members of the intended population are less likely to be included than others) (42). In the studies evaluated, the ROBs “absence of blindness” was induced by the fact that it was not specified if the biochemical analyst was or was not unaware of the provenance of the specimen (e.g. case or control group).
The most investigated disease in the present review was breast cancer (18 out of 79 studies), the most common malignant tumor among women (25% off all females tumors), with approximately 1.7 million new cases diagnosed every year (43). Fourteen studies (78%) reported statistically significant association between the presence of breast tumor and finding of one or more markers in patients saliva. Molecules such C-erbB2, CA 15-3, Cathepsin D, sialic acid and P53, EGF, VEGF and the CEA, seem to be promising salivary markers possibly very useful either for diagnosis of breast carcinoma and for follow-up of patients after treatment. According to the quality assessment tool adopted here, only one study dealing with breast cancer and salivary biomarkers obtained a “good” quality score, its results being apparently very robust (6). However, such a study investigated a panel of proteins, detected trough the SERS technology, which are not yet completely characterized and identified (6). Therefore, the utility of these proteins is currently somewhat questionable and their role should be confirmed in further studies.
The second most studied malignant tumor, according to the present systematic review is lung carcinoma (10 studies). Lung cancer is the most common cancer in humans (11.6% of all malignant tumours) and the leading cause of death for malignancy (18.4%) (44). All of the researches on patients with lung carcinomas provided statistically significant results to the association between salivary molecules and the pulmonary malignancy. Specifically, molecules identified included a mutation of the EGFR gene, 5 mRNA (CCNI, EGFR, FGF 19, FRS 2, GREB1), several bacteria (e.g. Sphingomonas, Blastomonas, Acinetobacter, Streptococcus) and proteins such as the calprotectin, alkaline phosphatase, cytokines, AZGP1 and haptoglobine (HP). All the reported molecules showed a good statistical association with diagnosis of lung carcinomas at different stages of developement. The “good” quality studies on salivary biomarkers and lung cancer are those demonstrating an association with EGFR, the 5 mRNA, microbiota and cytokines (8-11). According to the results of the present review they can be considered already reliable markers.
Data on association between salivary biomarkers and diagnosis of malignant tumors are available also for gastric and pancreatic cancer. Gastric cancer affects approximately one million individuals per year worldwide, having a mortality rate of approximately (1.033.701 new cases in 2018 and 782.685 death in 2018, in the world) (44). It is often detected late because up to 80 % of patients are asymptomatic during the early phases of disease (45). Similarly, pancreatic carcinoma is insidious, very aggressive and in most cases diagnosed at a very late stage, being associated to a very poor prognosis (46).
With regard to gastric cancer 7 studies were included in the present review (15-17,25,47-49). All of these reported statistically significant results. The “good” quality studies were three and they were focused on salivary bacteria (Prevotella, Leptotrichia, Rothia, Aggregatibacter, Campylobacter, Megasphaera, Granulicatella), RNA (SPINK7, PPL, SEMA4B, miR140-5p, miR301a) and some lectins.
All of the studies on pancreatic cancer demonstrated a significant association between the salivary molecules and the disease. Two of these were also scored as “good” after quality assessment. Molecules reported in such analysis were derived from transcriptomics (4mRNA (KRAS, MBD3L2, ACRV1, DPM1), and 2 miRNA (miR-3679-5p and miR-940)).
It is worthy to mention here that both for gastric and pancreatic cancer, the possibility of early diagnosis through salivary diagnostics, not based on the subjective radiographical images interpretation, could potentially contribute to prevent most of the deaths related to such cancers (3).
Particularly in the field of oncology, an easy and non-invasive method based on salivary biomarkers, may hypothetically constantly monitor and screen saliva, thus detecting very recurrences very (50).
Studies on neurological disorders are focused on Parkinson’s and Alzheimer's diseases. It is interesting to highlight that the use of salivary biomarkers in patients with neurological pathologies has gained a great interest in the last couple of years (8 out 14 articles published in 2017-18). Such an increasing of interest might be explained taking into consideration that the diagnosis of these diseases are essentially clinical. The identification of objective features (including biomolecules within body fluid) seems therefore of paramount importance for diagnosis, monitoring of disease progression and management as well as the development of novel therapeutic interventions (51).
Among studies on inflammatory, cardiovascular and metabolic diseases those reporting significant results are focused on myocardial infarction and rheumatoid arthritis (20-24,26).
Research on myocardial infarction searched C-reactive protein, myoglobin, myeloperoxidase, creatine phosphokinase, creatin kinase MB, cardiac troponin T and cathepsin-L, all reporting statistically significant result. In one of these studies the use a saliva-based nanochip was proposed (23). Such a new technology, based on nanotechnologies and innovative materials, is apparently very promising, deserving further researches.
Rheumatoid arthritis is sometime difficult to diagnose, especially in early stages, because of the variability of the symptoms and the absence of specific markers (52). Early diagnosis of the disease usually improves the success of treatment and could possibly reduce the quantity of drugs administered. In the study on included in the present review, Authors identified particularly one salivary protein (GRP78/BiP) which was significantly associated to the disease (p<0.001, 83.3% sensitivity and 95% specificity) (26).
A limit of the present systematic review is the lack of a quantitative analysis. The heterogeneity of diseases evaluated, their stage at diagnosis, the extremely wide range of molecules investigated as well as the differences in procedures for saliva collection, handling, storing and processing, makes it impossible to draw reliable pooled results or to perform a meta-analysis. On the other hand, the results of the present qualitative analysis can provide useful information in the field of salivary diagnostics. Particularly, the findings reported here can be the background for further studies which possibly might take into account the ROBs highlighted in the qualitative analysis of papers. Future studies might be useful to confirm and improve the potentiality of salivary analysis techniques (in terms of sensitivity and specificity) as well as to develop new, smaller, patient-friendly devices possibly with affordable costs.
Diffusion and ready availability of a panel of sensors for detecting salivary biomarkers associated to systemic diseases could have a strong impact on public healthcare and economy. The use of saliva for analysis might replace the use of blood, with a possible economic impact based on the easier and non-invasive method for collection (50). Such a perspective, may well lead to a higher commercial availability of screening assays (53) and possibly bring to the development of analytic tools directly administered in the dental or general physician office.
The use of salivary biomarkers for diagnosis of systemic diseases (“salivary diagnostics”) is gaining increasing interest.
The present systematic review confirms the existence of some “good” quality evidence to support the role of peculiar salivary biomarkers for diagnosis of systemic diseases (e.g. lung cancer and EGFR). However, it seems necessary to encourage further researches for improving the sensitivity and specificity of salivary diagnostics analysis.
The perspective of realizing a reliable “lab-on-a-chip” for diagnosis and follow-up of systemic diseases and/or malignant tumors through saliva evaluation seems an attainable target of modern medicine.
Acknowledgments
Conflicts of interest None declared.
Funding None declared.
References
- 1.Aro K, Wei F, Wong DT, Tu M. Saliva Liquid Biopsy for Point-of-Care Applications. Front Public Health. 2017;5:77. doi: 10.3389/fpubh.2017.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yager P, Domingo GJ, Gerdes J. Point-of-Care Diagnostics for Global Health. Annu Rev Biomed Eng. 2008;10:107–44. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- 3.Kaczor-Urbanowicz KE, MartinCarreras-Presas C, Aro K, Tu M, Garcia-Godoy F, Wong DT. Saliva diagnostics - Current views and directions. Exp Biol Med. 2017;242:459–72. doi: 10.1177/1535370216681550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools.
- 5.Phillips B. Oxford Centre for Evidence-based Medicine-Levels of Evidence. Retrieved from http://www.cebm.net/oxford-centre-evidence-based-medicine-levels-evidence-march-2009/
- 6.Feng S, Huang S, Lin D, Chen G, Xu Y, Li Y. Surface-enhanced Raman spectroscopy of saliva proteins for the noninvasive differentiation of benign and malignant breast tumors. Int J Nanomedicine. 2015;10:537–47. doi: 10.2147/IJN.S71811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laidi F, Bouziane A, Zaoui F. Salivary expression of soluble HER2 in breast cancer patients with positive and negative HER2 status. Onco Targets Ther. 2014;7:1285–89. doi: 10.2147/OTT.S64230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Xiao H, Zhou H, Santiago S, Lee JM, Garon EB. Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell Mol Life Sci. 2012;69:3341–50. doi: 10.1007/s00018-012-1027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koizumi T, Shetty V, Yamaguchi M. Salivary cytokine panel indicative of non-small cell lung cancer. J Int Med Res. 2018;46:3570–82. doi: 10.1177/0300060518775563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei F, Lin CC, Joon A et al. Noninvasive saliva-based EGFR gene mutation detection in patients with lung cancer. Am J Respir Crit Care Med. 2014;190:1117–26. doi: 10.1164/rccm.201406-1003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, Mu X, Wang Y, Zhu D, Zhang J, Liang C. Dysbiosis of the Salivary Microbiome Is Associated With Non-smoking Female Lung Cancer and Correlated With Immunocytochemistry Markers. Front Oncol. 2018;8:520. doi: 10.3389/fonc.2018.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Farrell JJ, Zhou H, Elashoff D, Akin D, Park NH. Salivary Transcriptomic Biomarkers for Detection of Resectable Pancreatic Cancer. Gastroenterology. 2010;138:949–57. doi: 10.1053/j.gastro.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Z, Yin X, Gong B, et al. Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer. Cancer Prev Res. 2015;8:165–3. doi: 10.1158/1940-6207.CAPR-14-0192. [DOI] [PubMed] [Google Scholar]
- 14.Holten-Andersen L, Christensen IJ, Jensen SB, Reibel J, Laurberg S, Nauntofte B. Saliva and plasma TIMP-1 in patients with colorectal cancer: a prospective study. Scand J Gastroenterol. 2012;47:1234–41. doi: 10.3109/00365521.2012.711855. [DOI] [PubMed] [Google Scholar]
- 15.Shu J, Yu H, Li X, Zhang D, Liu X, Du H. Salivary glycopatterns as potential biomarkers for diagnosis of gastric cancer. Oncotarget. 2017;8:35718–27. doi: 10.18632/oncotarget.16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun JH, Li XL, Yin J, Li YH, Hou BX, Zhang Z. A screening method for gastric cancer by oral microbiome detection. Oncol Rep. 2018;39:2217–24. doi: 10.3892/or.2018.6286. [DOI] [PubMed] [Google Scholar]
- 17.Li F, Yoshizawa JM, Kim KM, Kanjanapangka J, Grogan TR, Wang X. Discovery and Validation of Salivary Extracellular RNA Biomarkers for Noninvasive Detection of Gastric Cancer. Clin Chem. 2018;64:1513–21. doi: 10.1373/clinchem.2018.290569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masters JM, Noyce AJ, Warner TT, Giovannoni G, Proctor GB. Elevated salivary protein in Parkinson's disease and salivary DJ-1 as a potential marker of disease severity. Parkinsonism relat disord. 2015;21:1251–5. doi: 10.1016/j.parkreldis.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 19.Ahmadi-Motamayel F, Goodarzi MT, Tarazi S, Vahabian M. Evaluation of salivary acetylcholinesterase and pseudocholinesterase in patients with Alzheimer's disease: A case-control study. Spec Care Dentist. 2019;39:39–44. doi: 10.1111/scd.12342. [DOI] [PubMed] [Google Scholar]
- 20.Mirzaii-Dizgah I, Riahi E. Salivary high-sensitivity cardiac troponin T levels in patients with acute myocardial infarction. Oral Dis. 2013;19:180–4. doi: 10.1111/j.1601-0825.2012.01968.x. [DOI] [PubMed] [Google Scholar]
- 21.Mirzaii-Dizgah I, Jafari-Sabet M. Unstimulated whole saliva creatine phosphokinase in acute myocardial infarction. Oral Dis. 2011;17:597–600. doi: 10.1111/j.1601-0825.2011.01817.x. [DOI] [PubMed] [Google Scholar]
- 22.Mirzaii-Dizgah I, Hejazi SF, Riahi E, Salehi MM. Saliva-based creatine kinase MB measurement as a potential point-of-care testing for detection of myocardial infarction. Clin Oral Investig. 2012;16:775–9. doi: 10.1007/s00784-011-0578-z. [DOI] [PubMed] [Google Scholar]
- 23.Floriano PN, Christodoulides N, Miller CS, Ebersole JL, Spertus J, Rose BG. Use of Saliva-Based Nano-Biochip Tests for Acute Myocardial Infarction at the Point of Care: A Feasibility Study. Clin chem. 2009;55:1530–8. doi: 10.1373/clinchem.2008.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirzaii-Dizgah I, Riahi E. Serum and Saliva Levels of Cathepsin L in Patients with Acute Coronary Syndrome. J Contemp Dent Pract. 2011;12:114–9. doi: 10.5005/jp-journals-10024-1019. [DOI] [PubMed] [Google Scholar]
- 25.Wu ZZ, Wang JG, Zhang XL. Diagnostic model of saliva protein finger print analysis of patients with gastric cancer. World journal of gastroenterology. 2009;15:865–70. doi: 10.3748/wjg.15.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giusti L, Baldini C, Ciregia F, Giannaccini G, Giacomelli C, De Feo F. Is GRP78/BiP a potential salivary biomarker in patients with rheumatoid arthritis? Proteomics Clinical Appl. 2010;4:315–24. doi: 10.1002/prca.200900082. [DOI] [PubMed] [Google Scholar]
- 27.Streckfus C, Bigler L, Tucci M, Thigpen JT. A preliminary study of CA15-3, c-erbB-2, epidermal growth factor receptor, cathepsin-D, and p53 in saliva among women with breast carcinoma. Cancer Invest. 2000;18:101–9. doi: 10.3109/07357900009038240. [DOI] [PubMed] [Google Scholar]
- 28.Streckfus C, Bigler L. The use of soluble, salivary c-erbB-2 for the detection and post-operative follow-up of breast cancer in women: the results of a five-year translational research study. Adv Dent Res. 2005;18:17–24. doi: 10.1177/154407370501800105. [DOI] [PubMed] [Google Scholar]
- 29.Turan T, Demir S, Aybek H, Atahan O, Tuncay OL, Aybek Z. Free and Total Prostate-Specific Antigen Levels in Saliva and the Comparison with Serum Levels in Men. Eur Urol. 2000;38:550–4. doi: 10.1159/000020354. [DOI] [PubMed] [Google Scholar]
- 30.Streckfus CF, Bigler LR, Zwick M. The use of surface-enhanced laser desorption/ionization time-of-flight mass spectrometry to detect putative breast cancer markers in saliva: A feasibility study. J Oral Pathol Med. 2006;35:292–300. doi: 10.1111/j.1600-0714.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 31.Brooks MN, Wang J, Li Y, Zhang R, Elashoff D, Wong DT. Salivary protein factors are elevated in breast cancer patients. Mol Med Rep. 2008;1:375–8. [PMC free article] [PubMed] [Google Scholar]
- 32.Agha-Hosseini F, Mirzaii-Dizgah I, Rahimi A. Correlation of serum and salivary CA15-3 levels in patients with breast cancer. Med Oral Patol Oral Cir Bucal. 2009;14:521–4. doi: 10.4317/medoral.14.e521. [DOI] [PubMed] [Google Scholar]
- 33.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao PV, Reddy AP, Lu X, Dasari S, Krishnaprasad A, Biggs E et al. Proteomic identification of salivary biomarkers of type-2 diabetes. J Proteome Res. 2009;8:239–45. doi: 10.1021/pr8003776. [DOI] [PubMed] [Google Scholar]
- 35.Pandeshwar P, Das R. Role of oral fluids in DNA investigations. J Forensic Leg Med. 2014;22:45–50. doi: 10.1016/j.jflm.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Valentini A, Miquel C, Nawaz MA, Bellemain E, Coissac E, Pompanon F et al. New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: the trnL approach. Mol Ecol Resour. 2009;9:51–60. doi: 10.1111/j.1755-0998.2008.02352.x. [DOI] [PubMed] [Google Scholar]
- 37.Vivacqua G, Suppa A, Mancinelli R, Belvisi D, Fabbrini A, Costanzo M. Salivary alpha-synuclein in the diagnosis of Parkinson's disease and Progressive Supranuclear Palsy. Parkinsonism Relat Disord. 2019;63:143–8. doi: 10.1016/j.parkreldis.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 38.Marksteiner J, Oberacher H, Humpel C. Acyl-Alkyl Phosphatidlycholines are Decreased in Saliva of Patients with Alzheimer's Disease as Identified by Targeted Metabolomics. J Alzheimers Dis. 2019;68:583–89. doi: 10.3233/JAD-181278. [DOI] [PubMed] [Google Scholar]
- 39.Julious SA. Sample sizes for clinical trials with normal data. Stat Med. 2004;23:1921–86. doi: 10.1002/sim.1783. [DOI] [PubMed] [Google Scholar]
- 40.Pu D, Liang H, Wei F et al. Evaluation of a novel saliva-based epidermal growth factor receptor mutation detection for lung cancer: A pilot study. Thorac Cancer. 2016;7:428–36. doi: 10.1111/1759-7714.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Julious SA. Sample size of 12 per group rule of thumb for a pilot study. Pharmaceut Statist. 2005;4:287–91. [Google Scholar]
- 42.Faggion CM Jr. Evaluating the Risk of Bias of a Study. J Evid Based Dent Pract. 2015;15:164–70. doi: 10.1016/j.jebdp.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 44.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 45.Layke JC, Lopez PP. Gastric Cancer: Diagnosis and Treatment Options. Am Fam Physician. 2004;69:1133–40. [PubMed] [Google Scholar]
- 46.McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24:4846–61. doi: 10.3748/wjg.v24.i43.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y, Cheng S, Zhang A, Song J, Chang J, Wang K. Salivary Analysis Based on Surface Enhanced Raman Scattering Sensors Distinguishes Early and Advanced Gastric Cancer Patients from Healthy Persons. J Biomed Nanotechnol. 2018;14:1773–84. doi: 10.1166/jbn.2018.2621. [DOI] [PubMed] [Google Scholar]
- 48.Shu J, Yu H, Du H, Zhang J, Zhang K, Li X. Identification of N- and O-linked glycans recognized by AAL in saliva of patients with atrophic gastritis and gastric cancer. Cancer Biomark. 2018;22:1–13. doi: 10.3233/CBM-171087. [DOI] [PubMed] [Google Scholar]
- 49.Xiao H, Zhang Y, Kim Y, Kim S, Kim JJ, Kim KM. Differential Proteomic Analysis of Human Saliva using Tandem Mass Tags Quantification for Gastric Cancer Detection. Sci Rep. 2016;6:22165. doi: 10.1038/srep22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee YH, Wong DT. Saliva: an emerging biofluid for early detection of diseases. Am J Dent. 2009;22:241–8. [PMC free article] [PubMed] [Google Scholar]
- 51.Hong Z, Shi M, Chung KA, Quinn JF, Peskind ER, Galasko D. DJ-1 and α-synuclein in human cerebrospinal fluid as biomarkers of Parkinson's disease. Brain. 2010;133:713–26. doi: 10.1093/brain/awq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott DL, Wolfe F, Huizinga TWJ. Rheumatoid arthritis. Lancet. 2010;376:1094–108. doi: 10.1016/S0140-6736(10)60826-4. [DOI] [PubMed] [Google Scholar]
- 53.Javaid MA, Ahmed AS, Durand R, Tran SD. Saliva as a diagnostic tool for oral and systemic diseases. J Oral Biol Craniofac Res. 2016;6:67–76. doi: 10.1016/j.jobcr.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao H, Zhang L, Zhou H, Lee JM, Garon EB, Wong DT. Proteomic analysis of human saliva from lung cancer patients using two-dimensional difference gel electrophoresis and mass spectrometry. Mol Cell Proteomics. 2012;11:M111. doi: 10.1074/mcp.M111.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee YH, Kim JH, Zhou H, Kim BW, Wong DT. Salivary transcriptomic biomarkers for detection of ovarian cancer: for serous papillary adenocarcinoma. J Mol Med. 2012;90:427–34. doi: 10.1007/s00109-011-0829-0. [DOI] [PubMed] [Google Scholar]
- 56.Zhang L, Xiao H, Karlan S, Zhou H, Gross J, Elashoff D. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS One. 2010;5:e15573. doi: 10.1371/journal.pone.0015573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Yang T, Lin J. Spectral analysis of human saliva for detection of lung cancer using surface-enhanced Raman spectroscopy. J Biomed Opt. 2012;17:037003. doi: 10.1117/1.JBO.17.3.037003. [DOI] [PubMed] [Google Scholar]
- 58.De Abreu Pereira D, Areias VR, Franco MF, Benitez MC, do Nascimento CM, de Azevedo CM. Measurement of HER2 in saliva of women in risk of breast cancer. Pathol Oncol Res. 2013;19:509–13. doi: 10.1007/s12253-013-9610-8. [DOI] [PubMed] [Google Scholar]
- 59.Chen D, Song N, Ni R, Zhao J, Hu J, Lu Q. Saliva as a sampling source for the detection of leukemic fusion transcripts. J Transl Med. 2014;12:321. doi: 10.1186/s12967-014-0321-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng F, Wang Z, Huang Y, Duan Y, Wang X. Investigation of salivary free amino acid profile for early diagnosis of breast cancer with ultra performance liquid chromatography-mass spectrometry. Clin Chim Acta. 2015;447:23–31. doi: 10.1016/j.cca.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 61.Humeau M, Vignolle-Vidoni A, Sicard F, Martins F, Bournet B, Buscail L. Salivary MicroRNA in Pancreatic Cancer Patients. PLoS One. 2015;10:e0130996. doi: 10.1371/journal.pone.0130996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan X, Yang M, Liu J, Gao R, Hu J, Li J. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res. 2015;5:3111–22. [PMC free article] [PubMed] [Google Scholar]
- 63.Delmonico L, Moreira Ados S, Franco MF, Esteves EB, Scherrer L, Gallo CV. CDKN2A (p14(ARF)/p16(INK4a)) and ATM promoter methylation in patients with impalpable breast lesions. Hum Pathol. 2015;46:1540–7. doi: 10.1016/j.humpath.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 64.Takayama T, Tsutsui H, Shimizu I, Toyama T, Yoshimoto N, Endo Y. Diagnostic approach to breast cancer patients based on target metabolomics in saliva by liquid chromatography with tandem mass spectrometry. Clin Chim Acta. 2016;452:18–26. doi: 10.1016/j.cca.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 65.Torres PJ, Fletcher EM, Gibbons SM, Bouvet M, Doran KS, Kelley ST. Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ. 2015;3:e1373. doi: 10.7717/peerj.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wood N, Streckfus CF. The Expression of Lung Resistance Protein in Saliva: A Novel Prognostic Indicator Protein for Carcinoma of the Breast. Cancer Invest. 2015;33:510–5. doi: 10.3109/07357907.2015.1081920. [DOI] [PubMed] [Google Scholar]
- 67.Tsutsui H, Mochizuki T, Inoue K, Toyama T, Yoshimoto N, Endo Y. High-throughput LC-MS/MS based simultaneous determination of polyamines including N-acetylated forms in human saliva and the diagnostic approach to breast cancer patients. Anal Chem. 2013;85:11835–42. doi: 10.1021/ac402526c. [DOI] [PubMed] [Google Scholar]
- 68.Xie Z, Chen X, Li J, Guo Y, Li H, Pan X. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget. 2016;7:25408–19. doi: 10.18632/oncotarget.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhong L, Cheng F, Lu X, Duan Y, Wang X. Untargeted saliva metabonomics study of breast cancer based on ultra performance liquid chromatography coupled to mass spectrometry with HILIC and RPLC separations. Talanta. 2016;158:351–60. doi: 10.1016/j.talanta.2016.04.049. [DOI] [PubMed] [Google Scholar]
- 70.Machida T, Tomofuji T, Maruyama T, Yoneda T, Ekuni D, Azuma T et al. miR‑1246 and miR‑4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol Rep. 2016;36:2375–81. doi: 10.3892/or.2016.5021. [DOI] [PubMed] [Google Scholar]
- 71.Liu HJ, Guo YY, Li DJ. Predicting novel salivary biomarkers for the detection of pancreatic cancer using biological feature-based classification. Pathol Res Pract. 2017;213:394–99. doi: 10.1016/j.prp.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 72.Bel'skaya L. The activity of metabolic enzymes in the saliva of lung cancer patients. Natl J Physiol Pharm Pharmacol. 2017;7:646–53. [Google Scholar]
- 73.Sun Y, Liu S, Qiao Z, Shang Z, Xia Z, Niu X. Systematic comparison of exosomal proteomes from human saliva and serum for the detection of lung cancer. Anal Chim Acta. 2017;982:84–95. doi: 10.1016/j.aca.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 74.Laidi F, Bouziane A, Errachid A, Zaoui F. Usefulness of Salivary and Serum Auto-antibodies Against Tumor Biomarkers HER2 and MUC1 in Breast Cancer Screening. Asian Pac J Cancer Prev. 2016;17:335–9. doi: 10.7314/apjcp.2016.17.1.335. [DOI] [PubMed] [Google Scholar]
- 75.Hernández-Arteaga A, de Jesús Zermeño Nava J, Kolosovas-Machuca ES, Velázquez-Salazar J, et al. Diagnosis of breast cancer by analysis of sialic acid concentrations in human saliva by surface-enhanced Raman spectroscopy of silver nanoparticles. Nano Res. 2017;10:3662–70. [Google Scholar]
- 76.Cavaco C, Pereira JAM, Taunk K, Taware R, Rapole S, Nagarajaram H. Screening of salivary volatiles for putative breast cancer discrimination: an exploratory study involving geographically distant populations. Anal Bioanal Chem. 2018;410:4459–68. doi: 10.1007/s00216-018-1103-x. [DOI] [PubMed] [Google Scholar]
- 77.Zermeño-Nava JJ, Martínez-Martínez MU, Rámirez-de-Ávila AL, Hernández-Arteaga AC, García-Valdivieso MG, Hernández-Cedillo A. Determination of sialic acid in saliva by means of surface-enhanced Raman spectroscopy as a marker in adnexal mass patients: ovarian cancer vs benign cases. J Ovarian Res. 2018;11:61. doi: 10.1186/s13048-018-0433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tajmul M, Parween F, Singh L, Mathur SR, Sharma JB, Kumar S. Identification and validation of salivary proteomic signatures for non-invasive detection of ovarian cancer. Int J Biol Macromol. 2018;108:503–14. doi: 10.1016/j.ijbiomac.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 79.Zhang J, Zhong Y, Zhang P, Du H, Shu J, Liu X. Identification of abnormal fucosylated-glycans recognized by LTL in saliva of HBV-induced chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Glycobiology. 2019;29:242–59. doi: 10.1093/glycob/cwy108. [DOI] [PubMed] [Google Scholar]
- 80.Xie Z, Zhou F, Yang Y, Li L, Lei Y, Lin X. Lnc-PCDH9-13:1 Is a Hypersensitive and Specific Biomarker for Early Hepatocellular Carcinoma. EBioMedicine. 2018;33:57–67. doi: 10.1016/j.ebiom.2018.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Devic I, Hwang H, Edgar JS, Izutsu K, Presland R, Pan C. Salivary α-synuclein and DJ-1: potential biomarkers for Parkinson's disease. Brain. 2011;134:e178. doi: 10.1093/brain/awr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsuruoka M, Hara J, Hirayama A, Sugimoto M, Soga T, Shankle WR. Capillary electrophoresis-mass spectrometry-based metabolome analysis of serum and saliva from neurodegenerative dementia patients. Electrophoresis. 2013;34:2865–72. doi: 10.1002/elps.201300019. [DOI] [PubMed] [Google Scholar]
- 83.Al-Nimer MS, Mshatat SF, Abdulla HI. Saliva α-Synuclein and A High Extinction Coefficient Protein: A Novel Approach in Assessment Biomarkers of Parkinson's Disease. N Am J Med Sci. 2014;6:633–7. doi: 10.4103/1947-2714.147980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liang Q, Zhang T and Jiang Y, et al. Metabolomics-Based Screening of Salivary Biomarkers for Early Diagnosis of Alzheimer's Disease. RSC Adv. 2015;5:96074–9. [Google Scholar]
- 85.Liang Q, Liu H, Li X, et al. High-throughput metabolomics analysis discovers salivary biomarkers for predicting mild cognitive impairment and Alzheimer's disease. RSC Adv. 2016;6:75499–504. [Google Scholar]
- 86.Yilmaz A, Geddes T, Han B, Bahado-Singh RO, Wilson GD, Imam K. Diagnostic Biomarkers of Alzheimer's Disease as Identified in Saliva using 1H NMR-Based Metabolomics. Alzheimers Dis. 2017;58:355–9. doi: 10.3233/JAD-161226. [DOI] [PubMed] [Google Scholar]
- 87.Ashton NJ, Ide M, Schöll M, Blennow K, Lovestone S, Hye A. No association of salivary total tau concentration with Alzheimer's disease. Neurobiol Aging. 2018;70:125–7. doi: 10.1016/j.neurobiolaging.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 88.Cao Z, Wu Y, Liu G, Jiang Y, Wang X, Wang Z. α-Synuclein in salivary extracellular vesicles as a potential biomarker of Parkinson's disease. Neurosci Lett. 2019;696:114–20. doi: 10.1016/j.neulet.2018.12.030. [DOI] [PubMed] [Google Scholar]
- 89.Song W, Kothari V, Velly AM, Cressatti M, Liberman A, Gornitsky M. Evaluation of salivary heme oxygenase-1 as a potential biomarker of early Parkinson's disease. Mov Disord. 2018;33:583–91. doi: 10.1002/mds.27328. [DOI] [PubMed] [Google Scholar]
- 90.Manconi B, Liori B, Cabras T, Vincenzoni F, Iavarone F, Lorefice L. Top-down proteomic profiling of human saliva in multiple sclerosis patients. J Proteomics. 2018;187:212–22. doi: 10.1016/j.jprot.2018.07.019. [DOI] [PubMed] [Google Scholar]
- 91.Buduneli E, Mäntylä P, Emingil G, Tervahartiala T, Pussinen P, Barış N. Acute myocardial infarction is reflected in salivary matrix metalloproteinase-8 activation level. J Periodontol. 2011;82:716–25. doi: 10.1902/jop.2010.100492. [DOI] [PubMed] [Google Scholar]
- 92.Silva DG, Stevens RH, Macedo JM, Hirata R, Pinto AC, Alves LM. Higher levels of salivary MUC5B and MUC7 in individuals with gastric diseases who harbor Helicobacter pylori. Arch Oral Biol. 2009;54:86–90. doi: 10.1016/j.archoralbio.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 93.Mirzaii-Dizgah I, Riahi E. Salivary troponin I as an indicator of myocardial infarction. Indian J Med Res. 2013;138:861–5. [PMC free article] [PubMed] [Google Scholar]
- 94.Adornetto G, Fabiani L, Volpe G, De Stefano A, Martini S, Nenna R. An electrochemical immunoassay for the screening of celiac disease in saliva samples. Anal Bioanal Chem. 2015;407:7189–96. doi: 10.1007/s00216-015-8884-y. [DOI] [PubMed] [Google Scholar]
- 95.Gang Cao, Maowen Chen, Yuanxiang Chen, Zufang Huang, Jinyong Lin, Jia Lin. A potential method for non-invasive acute myocardial infarction detection based on saliva Raman spectroscopy and multivariate analysis. Laser Phys. 2015;12:125702. [Google Scholar]
- 96.Giusti L, Sernissi F, Donadio E, et al. Salivary psoriasin (S100A7) correlates with diffusion capacity of carbon monoxide in a large cohort of systemic sclerosis patients. J Transl Med. 2016;14:262. doi: 10.1186/s12967-016-1023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu C, Hou X, Li M , Wang L, Zeng P, Jia H, et al Detection of AMA-M2 in human saliva: Potentials in diagnosis and monitoring of primary biliary cholangitis. Sci Rep. 2017;7:796. doi: 10.1038/s41598-017-00906-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]