Abstract
Store-operated calcium (Ca2+) entry (SOCE) occurs through a widely distributed family of ion channels activated by the loss of Ca2+ from the endoplasmic reticulum (ER). The best understood of these is the Ca2+ release-activated Ca2+ (CRAC) channel, which is notable for its unique activation mechanism as well as its many essential physiological functions and the diverse pathologies that result from dysregulation. In response to ER Ca2+ depletion, CRAC channels are formed through a diffusion trap mechanism at ER–plasma membrane (PM) junctions, where the ER Ca2+-sensing stromal interaction molecule (STIM) proteins bind and activate hexamers of Orai pore-forming proteins to trigger Ca2+ entry. Cell biological studies are clarifying the architecture of ER–PM junctions, their roles in Ca2+ and lipid transport, and functional interactions with cytoskeletal proteins. Molecular structures of STIM and Orai have inspired a multitude of mutagenesis and electrophysiological studies that reveal potential mechanisms for how STIM is toggled between inactive and active states, how it binds and activates Orai, and the importance of STIM-binding stoichiometry for opening the channel and establishing its signature characteristics of extremely high Ca2+ selectivity and low Ca2+ conductance.
OVERVIEW OF STORE-OPERATED CALCIUM ENTRY
One of the major pathways for Ca2+ influx across the plasma membrane (PM) of cells is store-operated Ca2+ entry (SOCE), so called because it is triggered by stimuli that reduce the level of Ca2+ in the endoplasmic reticulum (ER), also known as the major cellular “Ca2+ store” (Putney 1986). Initially described in electrically nonexcitable cells, SOCE is now known to operate in practically all cells, including excitable cells such as neurons and muscle. At a cellular level, SOCE controls a wide variety of processes, including gene expression, secretion, motility, and maintaining a high Ca2+ concentration in the ER. Its physiological importance is underscored by numerous pathologies caused by gain- or loss-of-function. In humans, loss-of-function leads to severe combined immunodeficiency (SCID), autoimmunity, myopathy, and ectodermal dysplasia, whereas gain-of-function causes York and Stormorken syndromes (characterized by thrombocytopenia, bleeding diathesis, miosis, and tubular aggregate myopathy) (Lacruz and Feske 2015). The dire consequences of dysregulation show the importance of precisely controlling SOCE to drive a variety of essential physiological functions.
The prototypical store-operated channel, the Ca2+ release-activated Ca2+ (CRAC) channel, was first described through electrophysiological studies of mast cells and T lymphocytes (Lewis and Cahalan 1989; Hoth and Penner 1992, 1993; Zweifach and Lewis 1993). These and later studies revealed the distinguishing features of the CRAC channel, including activation by store depletion, an extremely high (∼2000:1) selectivity for Ca2+ over Na+, extremely low single-channel Ca2+ conductance (∼20 fS), and several types of Ca2+-dependent feedback regulation (for review, see Prakriya and Lewis 2015).
The breakthrough for pursuing the mechanisms underlying CRAC channel function was the identification of stromal interaction molecule (STIM) (Liou et al. 2005; Roos et al. 2005) and Orai proteins (Feske et al. 2006; Vig et al. 2006b; Zhang et al. 2006). Mammals express two STIM proteins, STIM1 and STIM2, and three Orai channels, Orai1, 2, and 3; STIM1 and Orai1 form the basis for the CRAC channel as originally described by electrophysiology and are the focus of this review. CRAC channels become activated through a peculiar mechanism based on protein relocalization (Fig. 1). In unstimulated (resting) cells with replete Ca2+ stores, STIM and Orai are mostly free to diffuse throughout the ER and PMs, respectively. Ligands bound to cell-surface receptors activate phospholipase C to generate inositol 1,4,5-trisphosphate (IP3) from phosphatidylinositol 4,5-bisphosphate (PIP2) in the PM. The ensuing release of ER Ca2+ through IP3 receptors reduces luminal [Ca2+]ER below its resting level of ∼400 µm. This is sensed by STIM, triggering its activation and relocalization to ER–PM junctions, which are specialized membrane contact sites where the ER comes to within 10–25 nm of the PM. At these sites, STIM1 binds to Orai1, trapping it and opening the CRAC channel to trigger Ca2+ entry (Fig. 1B). The process is reversible: After the cessation of the stimulus, Ca2+ is pumped back into the ER, STIM deactivates and unbinds from Orai, the channels close, and both proteins diffuse away from the junctions. Since the initial identification of STIM and Orai proteins as the foundation of SOCE, much progress has been made in deciphering the molecular and cellular mechanisms underlying this unusual choreography. As a result, we are beginning to achieve a deeper understanding of the intricate ways in which SOCE is regulated.
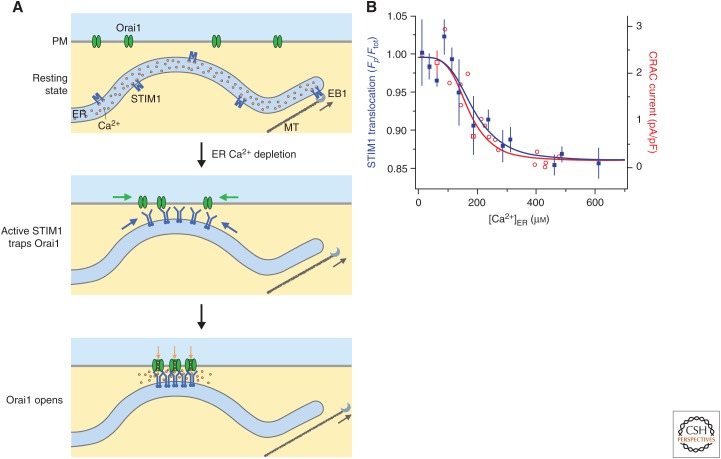
Figure 1.
Cellular choreography of store-operated calcium channels. (A) (Top) In resting cells with full endoplasmic reticulum (ER) Ca2+ stores, STIM1 and Orai1 diffuse freely in the ER and plasma membrane (PM), respectively. STIM1 also binds to EB1 at the tips of growing microtubules (MTs), which extends ER tubules toward the cell periphery. (Middle) After store depletion, STIM1 is activated, detaches from EB1, and accumulates at ER–PM junctions through interactions with negatively charged inositol phospholipids in the PM. Orai1 diffusing in the PM is trapped through binding to the CAD/SOAR domain of STIM1. (Bottom) Binding of STIM1 to the six Orai1 subunits of the Ca2+ release-activated Ca2+ (CRAC) channel opens the channel to allow local Ca2+ entry. (B) STIM1 translocation to ER–PM junctions (blue, measured by the ratio of labeled STIM1 at the periphery to the total STIM1 in the cell, Fp/Ftot) and CRAC channel current density (red) show a similar steep dependence on ER Ca2+ concentration ([Ca2+]ER). (Panel B is from Luik et al. 2008; adapted, with permission, from HHS Public Access and Nature Publishing.)
ACTIVATION OF STIM BY STORE DEPLETION
STIM proteins serve two primary functions in SOCE—to sense ER Ca2+ depletion and activate Orai channels. These type I dimeric ER membrane proteins contain a number of specialized domains to carry out these tasks (Soboloff et al. 2012; Prakriya and Lewis 2015). On the luminal side, there are (1) a canonical EF-hand that binds Ca2+, (2) a non-Ca2+-binding, noncanonical EF-hand, and (3) a sterile α motif (SAM) domain, collectively known as the EF-SAM domain. The cytosolic domain contains (1) a putative coiled-coil domain 1 (CC1), (2) the CRAC activation domain (CAD) or STIM-Orai-activating region (SOAR) comprising coiled-coil domains CC2 and CC3, and (3) additional domains involved in inactivation (IDSTIM) and EB1 binding, and a polybasic domain (PBD) at the carboxyl terminus (Fig. 2A). The CAD/SOAR domain (Fig. 2B) binds to Orai1 to activate it (Kawasaki et al. 2009; Park et al. 2009; Yuan et al. 2009), such that regulation of SOCE is achieved primarily by controlling the access of the CAD/SOAR to Orai1. Ca2+ release from the luminal domain sets in motion the entire sequence leading to SOCE, so the proper regulation of SOCE depends critically on precise switching between the inactive and active states of STIM.
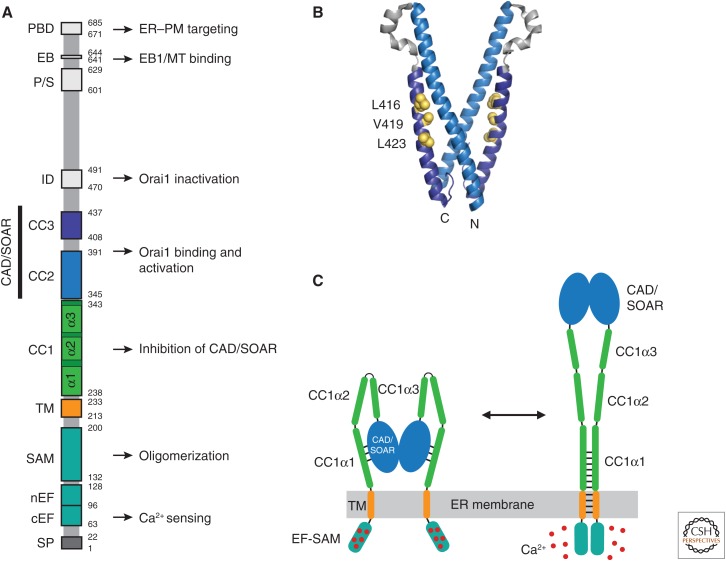
Figure 2.
STIM1 functional domains and a model for activation. (A) Domain organization of STIM1. Functional domains include the signal peptide (SP), canonical EF-hand (cEF), noncanonical EF-hand (nEF), sterile α motif (SAM), transmembrane (TM) domain, putative coiled-coil domains 1–3 (CC1–3), Ca2+ release-activated Ca2+ (CRAC) activation domain (CAD, aa 342–448) or STIM-Orai-activating region (SOAR, aa 344–442), inactivation domain (ID), proline/serine-rich domain (P/S), EB1-binding (EB) domain, and the polybasic domain (PBD). Known functions are indicated. (B) Crystal structure of the dimeric CAD/SOAR (3TEQ.pdb) (Yang et al. 2012). Amino- and carboxy-terminal ends of the front subunit and selected residues implicated in interactions with CC1 (L416, V419, L423) are indicated in yellow. (C) A coiled-coil model for STIM1 activation by ER Ca2+ depletion (regions downstream from CAD/SOAR omitted for clarity). In the resting state (left), the luminal EF-SAM domains are bound to 5–6 Ca2+ ions and separated, which allows CC1α1 to bind to CAD/SOAR and sequester it close to the endoplasmic reticulum (ER) membrane. Following store depletion (right), Ca2+ release triggers dimerization of the EF-SAM domain, bringing the TM domains together to form a coiled-coil. This rearrangement favors dissociation of CC1α1 from CAD/SOAR and extends the coiled-coil beyond the ER membrane to move CAD/SOAR toward the plasma membrane (PM).
Although the EF-hand of STIM1 binds only a single Ca2+ ion (Stathopulos et al. 2006), recent evidence shows that the luminal domain binds a total of 5–6 Ca2+ per subunit of the STIM1 dimer (Gudlur et al. 2018; Stathopulos and Ikura 2019). The locations of the additional Ca2+-binding sites have not been determined, but they are closely coupled to the EF-hand, and release from both is required for STIM1 activation. The overall affinity (∼200 µm) is consistent with the K1/2 for STIM1 accumulation at ER–PM junctions, and the number of Ca2+-binding sites may help explain the high cooperativity of this process (Hill coefficient of 4–8) (Fig. 1B; Brandman et al. 2007; Luik et al. 2008).
In the Ca2+-bound state, the luminal EF-SAM regions in the dimer are held apart; these domains are critical for preventing spontaneous activation of STIM1 (Li et al. 2007; Korzeniowski et al. 2017). This configuration allows the cytosolic CC1 domain to associate with CAD/SOAR to sequester it near the ER membrane (Fig. 2C). This coiled-coil “clamp,” originally proposed by Korzeniowski et al. (2010), has been shown elegantly by the ability of a mutant STIM1 truncated at the end of CC1 or its first helical segment CC1α1 to capture soluble CAD/SOAR in cells and release it on store depletion (Ma et al. 2015). Of the three helical segments predicted to form CC1, CC1α1 appears most critical (Fahrner et al. 2014; Ma et al. 2015). CC1α3 also contributes, perhaps by enhancing CC1α1 interactions with CAD/SOAR (Yang et al. 2012; Ma et al. 2015). Mutations of residues in CC1α1 and CC3 of CAD/SOAR prevent binding of CC1 to CAD/SOAR (Muik et al. 2011; Fahrner et al. 2014; Ma et al. 2015), leading to the suggestion that L416, V419, and L423 in CC3 pair with L258 and L261 in CC1α1 to form a coiled-coil interface (Ma et al. 2015), although this has not yet been directly shown (Fig. 2B).
Shortly following store depletion and the consequent release of Ca2+ from STIM1, Förster resonance energy transfer (FRET) between labeled amino termini of STIM1 increases, suggesting that formation of higher-order STIM1 oligomers is the initiating step in activation of SOCE (Liou et al. 2007; Covington et al. 2010). This is supported by studies showing that artificial cross-linking of STIM1 luminal domains leads to puncta formation and Orai1 activation in cells with full Ca2+ stores (Luik et al. 2008). More recent work suggests a reinterpretation of these results. A similar FRET increase occurs with a truncated STIM1 (luminal, transmembrane [TM], and CC1 domains only) attached to a dimerizing GrpE protein, suggesting that the initiating step may instead be movements within the STIM1 dimer leading to increased proximity and/or reorientation of the luminal domains (Fig. 2C; Zhou et al. 2013; Ma et al. 2015; Gudlur et al. 2018). Interestingly, the CC3 region of CAD/SOAR has a propensity to dimerize (Muik et al. 2009), perhaps enabling formation of higher-order oligomers of STIM1 dimers if it is released from its interactions with CC2 (Stathopulos et al. 2013). An important future goal is to determine the oligomeric state of STIM1 during the various phases of its activation, accumulation at ER–PM junctions, and interactions with Orai.
The rearrangement of the luminal domains after store depletion triggers an extension of the cytosolic domains (Muik et al. 2011) leading to STIM1 activation (Fig. 2C). Recent findings suggest how these conformational changes may be coupled. Association of the EF-SAM domains changes the arrangement of the TM domains (Ma et al. 2015), which may promote formation of a coiled-coil structure (Hirve et al. 2018). The subsequent release of CC1α from CAD/SOAR then triggers an extension of the TM coiled-coil to include the membrane-proximal part of CC1α1 (Hirve et al. 2018). Interestingly, this region of CC1α1 contains an imperfect heptad repeat and several sentinel residues that may temper the ability to form a coiled-coil and prevent spontaneous activation of STIM1 (Hirve et al. 2018). This is potentially critical for the precise regulation of STIM1 activity, to avoid the consequences of unregulated activation of Ca2+ entry.
STIM2 is highly homologous to STIM1 but differs in key respects that determine its function. Most obvious is its reduced Ca2+ affinity (∼500 µm) (Zheng et al. 2011), which prelocalizes it to ER–PM junctions and enables it to respond to small changes in [Ca2+]ER near the resting level and maintain ER Ca2+ homeostasis important for protein folding and signaling functions of the ER (Brandman et al. 2007). Interestingly, STIM2 has been reported to form heterodimers with STIM1 to help recruit STIM1 and facilitate its activation at junctions (Ong et al. 2015). In addition, alternative splicing of STIM2 converts STIM2 to an inhibitor of SOCE, which may play a role during development as RNA splicing programs are engaged (Miederer et al. 2015; Rana et al. 2015). A developmental role has also been ascribed to Orai2, which reduces SOCE in naïve T cells by forming heteromers with Orai1; in this case, the loss of Orai2 as naïve cells differentiate into effector T cells enhances T-cell function during later stages of the immune response (Vaeth et al. 2017). By increasing the number of distinct functional forms of STIM and Orai proteins, alternative splicing and heteromerization significantly expand their potential range of functions in shaping SOCE.
HOW STIM AND Orai FIND EACH OTHER
Self-organization is one of the defining features of the SOCE mechanism. The activation of STIM1 by Ca2+ store depletion acts as a switch to trigger all subsequent events, including colocalization of STIM1 and Orai1 at ER–PM junctions and opening of the CRAC channel. Thus, activation of STIM1 independently of [Ca2+]ER, either by mutation of the EF-hand (D76A mutation) or by in situ cross-linking of STIM1 proteins drives CRAC channel activation in cells with full Ca2+ stores (Liou et al. 2005; Zhang et al. 2005; Luik et al. 2008).
The basis of this self-organizing behavior is a diffusion trap. Both STIM1 and Orai1 move by diffusion in the ER and PM, respectively (Liou et al. 2007; Covington et al. 2010; Wu et al. 2014). Following store depletion, extension of the cytosolic domain of STIM1 enables the PBD at the carboxyl terminus to reach across the 10–25 nm gap of the ER–PM junctions. There, it interacts with negatively charged phospholipids, in particular PIP2, and acts to concentrate STIM1 proteins at ER–PM junctions (Ercan et al. 2009; Korzeniowski et al. 2009; Walsh et al. 2010). The RAS-association domain family member RASSF4 appears to enhance SOCE by maintaining PIP2 levels in the PM and stabilizing ER–PM junctions (Chen et al. 2017). Although the PBD is not absolutely necessary for SOCE in overexpression studies, its ability to precluster STIM1 increases the speed and efficiency with which STIM1 can bind, trap, and activate Orai1 (Li et al. 2007; Zheng et al. 2018). Importantly, preclustering by the PBD may be more critical at endogenous protein levels, in which individual STIM1 dimers may be too sparse to effectively engage Orai. STIM proteins in some species, for example, Drosophila, lack a PBD, raising questions of whether other proteins may serve a clustering function. In Drosophila, such a role could be played by IP3 receptors, which cluster at ER–PM junctions and are required for STIM-Orai accumulation and SOCE in response to store depletion (Chakraborty et al. 2016).
Only a minimal interaction with STIM1 (∼1 dimer per channel) is sufficient to trap Orai1, although it is not enough to open the channel (Hoover and Lewis 2011). Although puncta appear to be static based on the fluorescence of STIM1-Orai1 in depleted cells, the trap is actually highly dynamic; escape of photoactivated green fluorescent protein (PAGFP)-STIM1 or PAGFP-Orai1 from puncta suggests that the proteins cycle through a series of bound and unbound states at junctions, and exchange with surrounding pools with half times of 50–100 sec (Wu et al. 2014). A recycling pool of Orai1 in intracellular vesicles is also trapped by STIM1 binding at junctions, leading to its enrichment in the PM (Hodeify et al. 2015, 2018).
ER–PM Junctions
ER–PM junctions are one example of a large family of membrane contact sites between the ER and other organelles (mitochondria, endosomes, lysosomes, peroxisomes, and lipid droplets), which have become widely recognized as sites of lipid and Ca2+ transfer and communication within cellular compartments (Phillips and Voeltz 2016; Saheki and De Camilli 2017). ER–PM junctions typically cover only a few percent of the cell surface and have dimensions of 100–200 nm with the ER and PM separated by a gap of 10–25 nm (Wu et al. 2006; Orci et al. 2009; Hsieh et al. 2017), although overexpression of STIM1 can greatly increase their size by enhancing adhesion of ER to the PM (Várnai et al. 2007; Orci et al. 2009). ER–PM junctions are long-lived although dynamic. STIM forms puncta at the same locations after repeated bouts of depletion, implying a lifetime on a minutes time scale (Liou et al. 2005). However, quantitation of junctions by EM indicate that while junctions pre-exist in resting cells, a proportion of new contacts are formed in response to store depletion (Wu et al. 2006), implying that luminal [Ca2+]ER influences their formation or stability. The identities of tethering proteins that control the formation and maintenance of junctions are of great interest. ER proteins like the extended synaptotagmins (E-Syt1-3) (Giordano et al. 2013) and GRAMD2A (Besprozvannaya et al. 2018) have been identified as tethering proteins at junctions where STIM1 accumulates, although their role is still not fully understood, as knockouts only partially reduce or do not affect SOCE. This implies some level of redundancy in the tethering process, with more proteins likely to be discovered. STIM proteins themselves are candidates. Although overexpression of STIM1 has the ability to greatly enlarge the spatial extent of ER–PM junctions, its role at endogenous levels needs further study. SOCE and lipid homeostasis at junctions are interconnected; local increases in Ca2+ entering through CRAC channels trigger accumulation of E-Syt1, which brings the two membranes closer together and enhances phosphatidylinositol (PI) transfer to the PM through recruitment of the PI transfer protein Nir2 (Kim et al. 2015; Chang and Liou 2016). In this way, Ca2+-dependent accumulation of E-Syt1 may act to maintain PIP2 levels in local domains while transporting diacylglycerol to the ER to prevent its accumulation in the PM (Saheki et al. 2016).
Roles of Cytoskeletal Proteins
The cytoskeleton interacts with the SOCE machinery in several ways. STIM1 binds to the tips of growing microtubules (MTs) through a specific interaction with EB1, an MT tip-binding protein (Fig. 1A; Grigoriev et al. 2008). Although hitchhiking on MTs has the potential to move STIM quickly through the cell, EB1 knockdown or inhibiting MT dynamics with taxol does not affect the amplitude of SOCE. These observations indicate that MT association is not required to deliver STIM1 to junctions, although it may serve to extend new ER tubules toward the cell periphery where they may form new junctions (Grigoriev et al. 2008). Recent evidence suggests that binding to MTs may actually temper the ability of STIM1 to diffuse to the ER–PM junction to interact with Orai (Chang et al. 2018). Interestingly, store depletion releases STIM1 from MTs (Sampieri et al. 2009; Alvarez-Barrientos et al. 2013). The release can also be triggered by phosphorylation of STIM1 by ERK at sites that inhibit association with EB1 (Alvarez-Barrientos et al. 2013), revealing potential cross talk between mitogen-activated protein kinase (MAPK) and Ca2+-signaling pathways.
Septins also influence the degree to which STIM and Orai accumulate and engage at ER–PM junctions. Septins 2, 4, and 5 facilitate SOCE by preventing preclustering of Orai1 in resting cells, by promoting the recruitment of STIM1 to ER–PM junctions and by stabilizing STIM-Orai interactions after store depletion (Sharma et al. 2013). Septins in various combinations are known to form filaments and rings, and precisely how they facilitate SOCE is not fully understood but may involve their ability to reorganize PIP2 domains in the vicinity of ER–PM junctions (Sharma et al. 2013). Similar effects have been reported for homologs in Drosophila, in which septin 7 also acts to suppress constitutive activity of Orai (Deb et al. 2016).
Cortical actin surrounds the perimeter of ER–PM junctions, and contributes to their stability and spatial distribution. Disassembly of actin with latrunculin B or cytochalasin D has been shown to increase junctional mobility and reduce their abundance, at least in part through promoting spontaneous fusion of neighboring junctions (Luik et al. 2006; Hsieh et al. 2017). Interestingly, actin disassembly interferes with PIP2 homeostasis at ER–PM junctions without significantly impacting SOCE mediated by STIM and Orai (Hsieh et al. 2017). In addition, the ER membrane protein PERK (widely studied for its role in the ER stress response) also affects the initiation and/or expansion of ER–PM junctions through its control of cortical actin polymerization and localization. On store depletion, PERK dimerizes and binds the actin-binding protein filamin-A to remodel cortical actin; without PERK, junction formation is inhibited by accumulation of cortical actin at the PM, which presumably prevents the close approach of the ER to the PM (van Vliet et al. 2017). Together, these studies identify intriguing links between actin remodeling and Ca2+-signaling pathways that deserve further investigation.
THE STOICHIOMETRY AND STRUCTURE OF THE CRAC CHANNEL
The stoichiometry of the CRAC channel has long been a subject of debate (Yen and Lewis 2019). Early evidence based largely on photobleaching of Orai-GFP chimeras and tandem tetrameric Orai concatemers supported a tetrameric stoichiometry (Ji et al. 2008; Mignen et al. 2008). Other studies suggested that it is trafficked to the PM as a dimer but forms tetramers after contact with STIM1 following store depletion (Penna et al. 2008; Demuro et al. 2011). The crystal structure of Drosophila Orai (dOrai) was therefore surprising in revealing a hexameric structure (Hou et al. 2012). Functional studies of hexameric Orai1 concatemers subsequently supported this conclusion (Cai et al. 2016; Yen et al. 2016). Intrinsic pore properties of hexameric concatemers (unitary conductance, Ca2+-block affinity, and ion selectivity) closely match those of native CRAC channels or channels made from monomeric Orai1 cDNA, strongly affirming the hexameric pore geometry of native CRAC channels (Yen et al. 2016). How to account for the results of earlier studies using GFP photobleaching and tetrameric Orai1 concatemers remains an open question (Yen and Lewis 2019).
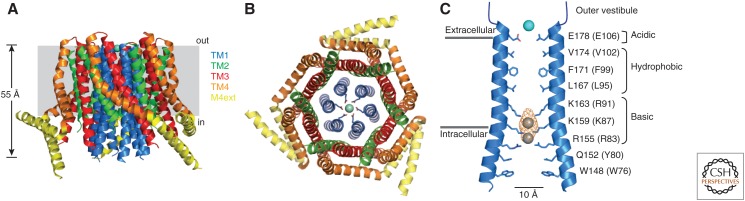
The hexameric crystal structure of dOrai comprises a narrow central pore lined entirely by six TM1 domains, surrounded by a shell of interlocking TM2 and TM3 helices, and peripheral TM4 domains that interact with the lipid bilayer and STIM (Fig. 3). The ion conduction pathway is formed by several domains in series (Fig. 3C). On the exterior side is a cluster of acidic residues (D110/D112/D114) that serves to concentrate divalent cations (Frischauf et al. 2015) near the selectivity filter that is formed by a ring of E106 side chains at the extracellular mouth of the pore (Prakriya et al. 2006; Vig et al. 2006a; Yeromin et al. 2006). Directly below the selectivity filter is a hydrophobic region lined by V102/F99/L95 and a highly charged basic region with three rings of K/R side chains (R91/K87/R83), both of which have been implicated in gating (see below). Extension of the pore beyond the basic region contains residues important for Ca2+-dependent inactivation (Y80, W76). Overall, the pore is quite narrow, accounting at least in part for the extremely low conductance of the channel (∼20 fS) (Zweifach and Lewis 1993; Prakriya and Lewis 2006). The structure has had an enormous impact in the field by providing a platform for studying the physical basis of signature CRAC channel properties like ion selectivity and conductance, as well as the gating mechanisms of activation and inactivation.
Figure 3.
The closed-channel structure of Drosophila Orai (dOrai). (A) Side view of the crystal structure of dOrai, showing the arrangement of the transmembrane (TM) domains of the subunits (4HKR.pdb). (Panel A created based on data in Hou et al. 2012.) (B) Top view of dOrai, showing the six TM1 subunits arranged around a central pore, an interlocking cage of TM2/TM3 subunits, and peripheral TM4 subunits with three paired coiled-coil interactions of the cytoplasmic M4ext helices. (C) Side view of TM1 and amino-terminal extension helices from two opposed subunits in the crystal structure, showing pore-lining residues and functional pore domains. The corresponding human Orai1 residues are shown in parentheses. A Ba2+ ion is shown in blue above E178. The anion density with Fe atoms modeled into the structure are shown in yellow and gray in the inner pore.
HOW STIM BINDS TO ORAI
STIM binding to Orai is direct (Park et al. 2009) and sufficient to open the channel in vitro (Zhou et al. 2010; Gudlur et al. 2014), although other proteins may modulate binding in cells (Prakriya and Lewis 2015). Binding of STIM to the carboxyl terminus of Orai (the cytosolic extension of the M4 helix, or M4ext) is well established, based on findings that mutagenesis or deletion of the M4ext inhibits Orai1 activation and clustering at ER–PM junctions as well as binding of CAD/SOAR to Orai1 carboxy-terminal fragments (for review, see Prakriya and Lewis 2015). Studies of human Orai1 found no evidence for binding to the 2–3 loop, but this occurs in Caenorhabditis elegans, suggesting a different mode of activation in this species (Kim et al. 2018). Whether the amino terminus of Orai1 binds STIM1 in the context of full-length Orai1 is still an open question. The amino terminus (amino acids 73–91) is required for Orai1 activation, amino-terminal fragments bind STIM1 fragments in vitro (Park et al. 2009; Zhou et al. 2010; Gudlur et al. 2014), and STIM–Orai FRET is reduced by deletion or mutations in the amino terminus (McNally et al. 2013; Palty and Isacoff 2016). However, binding of STIM1 to full-length Orai1 has not been directly shown, and a weakened interaction with amino-terminal Orai1 mutants would in any case be expected because the amino terminus is needed for opening, and opening is energetically coupled to carboxy-terminal binding. In fact, a STIM-independent role for the amino terminus in gating has recently been shown for constitutively active Orai1 mutants in the absence of STIM1 (Zhou et al. 2016; Yeung et al. 2018). Although these results do not rule out a direct interaction of STIM1 or accessory proteins with the amino terminus, they support the idea that the amino terminus interacts with other Orai1 domains such as TM3 or the 2–3 loop to regulate opening (Fahrner et al. 2018).
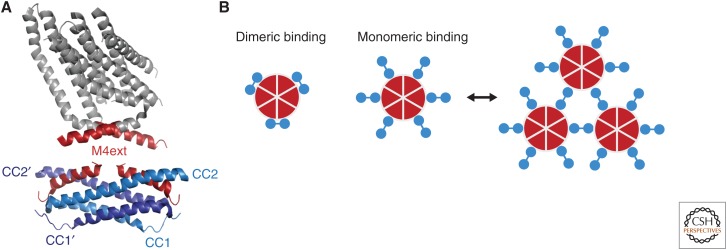
The CAD/SOAR domain of STIM1 by itself is able to activate Orai1, and a crystal structure reveals a dimeric “V” shape, with CC2 and CC3 regions connected by a loop of two short helices (Fig. 2B; Yang et al. 2012). Several models have emerged to explain how the CAD/SOAR domain may bind to Orai1. A dimeric binding mechanism has been proposed in which the two CAD/SOAR domains of a single STIM1 dimer simultaneously engage two adjacent M4ext helices. The model is based on a nuclear magnetic resonance (NMR) spectroscopy solution structure in which a pair of CAD/SOAR fragments (comprising CC1α3 and CC2) form a pair of binding pockets that bind two oppositely oriented M4ext peptides (Fig. 4A; Stathopulos et al. 2013). The model is supported to some extent by the surprising finding that an Orai subunit with the “nonbinding” L273D mutation actually enhances STIM1 binding when present next to a wild-type (WT) subunit, which could occur if binding of one STIM subunit by the WT M4ext increases the avidity of the second STIM subunit to the neighboring L273D M4ext (Yen and Lewis 2018). However, there is as yet no direct evidence for simultaneous occupancy of adjacent M4exts using functionally competent fragments or full-length proteins.
Figure 4.
Dimeric and monomeric models of STIM1-Orai1 binding. (A) A dimeric binding model, showing two subunits from the crystal structure of dOrai (top), with the cytoplasmic M4ext regions highlighted in red (4HKR.pdb) (Hou et al. 2012). The nuclear magnetic resonance (NMR) spectroscopy structure below shows two corresponding human Orai1 M4ext fragments (aa 272–292) bound to a dimer of CC1–CC2 STIM1 fragments (aa 312–383; 2MAK.pdb) (Stathopulos et al. 2013). (B) Schematic views of dimeric and monomeric binding models in which three or six STIM1 dimers (blue) are bound to the six Orai1 subunits (red), respectively. Monomeric binding may allow STIM1 dimers to create cross-linked arrays of channels (right).
Evidence also exists for a monomeric binding mechanism, in which only one subunit from each STIM1 dimer binds to a single M4ext, with the second one remaining free (or binding to another channel) (Fig. 4B). In support of this model, a dimeric CAD/SOAR concatemer with one subunit rendered binding-incompetent by a F394H mutation binds and activates Orai1 as well as WT CAD/SOAR dimer (Zhou et al. 2015). Although the details of the binding interface remain to be worked out, the model appears consistent with observations that STIM1 can cross-link Orai1 channels; for example, expression of CAD/SOAR creates Orai1 puncta in cells (Zhou et al. 2018), cross-links individual Orai1 channels (Park et al. 2009), and slows their diffusion (Park et al. 2009; Zhou et al. 2018). Cross-linking is also consistent with EM observations that Orai1 particles show a preferred spacing of ∼15 Å (Perni et al. 2015), approximating the expected distance between two channels connected by a CAD/SOAR dimer. Other evidence suggests that channel activity may be enhanced by cross-linking (Zhou et al. 2018). Because these studies were all conducted with overexpressed proteins, it will be important to test whether cross-linking also occurs at endogenous levels and, if so, how it affects activity.
A third model has been proposed in which initial STIM1 binding partially activates the channel, triggering a conformational change that creates a higher affinity STIM-binding site that fully activates (Palty et al. 2017). This model is based on observations that STIM1 mutants that cannot bind to WT channels can bind to, and enhance, the opening of Orai1 channels that have been partially pre-activated by mutations or attachment of CAD/SOAR monomers.
Currently, there is no clear conclusion regarding which binding mode best describes STIM1 binding in vivo. A structure of a constitutively open mutant channel (dOrai H206A) displays straightened M4ext helices that are unlikely to bind STIM1 in the way depicted in the Stathopulos et al. NMR structure (Hou et al. 2018), but it is possible that the mutant channel does not fully mimic the STIM-Orai complexed state. It is also conceivable that the different models represent steps on the way to the STIM-bound open state, as implied by a sequential binding scheme. One problem is that the support for each model is indirect or based on fragments separated from the protein environment. A structure for the complete STIM-Orai complex will ultimately be needed to determine the intricacies of STIM-Orai association and allow more definitive interpretations of the current models.
HOW STIM BINDING AFFECTS ORAI FUNCTION
STIM1 binding affects Orai1 in multiple ways. In addition to activation and inactivation gating, the STIM:Orai ratio has a strong impact on ion selectivity as well (Scrimgeour et al. 2009; McNally et al. 2012). Many constitutively active Orai1 mutants display reduced Ca2+ selectivity, which is restored by the addition of STIM1 (McNally et al. 2012; Yamashita et al. 2017; Yeung et al. 2018), and the selectivity of WT channels also increases with the degree of STIM1 binding (McNally et al. 2012; Yen and Lewis 2018). These observations indicate that STIM1 binding modifies the pore structure, most likely at multiple locations.
Activation and the Orai Channel Gate
Orai1 activation is a highly nonlinear function of the number of sites bound by STIM1. In experiments on hexameric Orai1 concatemers, inhibiting STIM1 binding to just a single subunit of the channel reduces the open probability to <10% of the fully occupied WT channel, and dramatically alters the permeation properties of the open channel, tripling the unitary conductance while reducing Ca2+-binding affinity to the selectivity filter and the ability to discriminate between Cs+ and Na+ in the absence of Ca2+ (Yen and Lewis 2018). These results imply that under physiological conditions, all six subunits of the native CRAC channel bind STIM1 to open effectively and display the signature CRAC channel properties of extremely high Ca2+-selectivity and low single-channel conductance. A corollary is that as SOCE develops after store depletion, channels likely spend very little time incompletely bound by STIM, thereby avoiding substantial Na+ influx that would be energetically costly and reduce the driving force for Ca2+ entry through membrane depolarization. This raises questions as to how full binding to the native channel is promoted at low endogenous protein levels. Several factors may contribute, including preclustering of STIM1 at ER–PM junctions, cooperative binding to Orai1 possibly including cross-linking of multiple channels, and enhancement of binding through the action of accessory proteins such as CRACR2A (Srikanth et al. 2010b), junctate (Srikanth et al. 2012), and STIMATE (Jing et al. 2015).
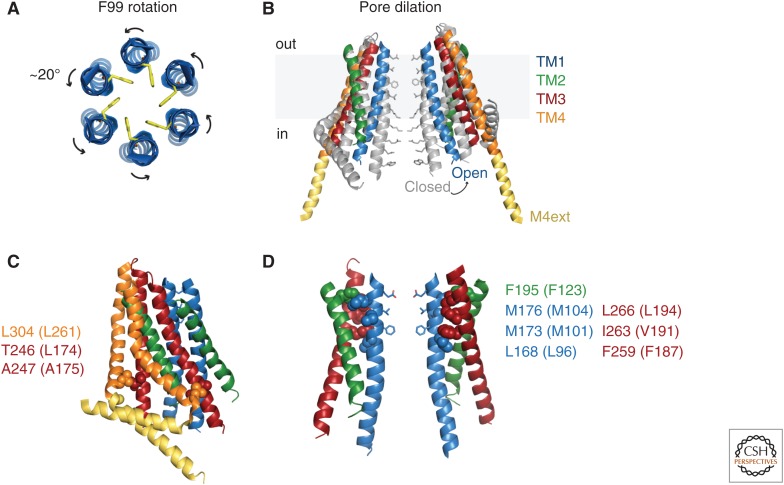
Two regions of the pore have been identified as potential gates. A hydrophobic section near the extracellular side of TM1, dominated by pore-lining side chains of V102 and F99, may operate by controlling the entry of water and cations (McNally et al. 2012; Gudlur et al. 2014). In support of this hydrophobic gate, Yamashita et al. found that the cysteine accessibilities at the F99 and G98 positions change in opposite directions on STIM1 binding, suggesting a ∼20° counterclockwise rotation, which would swing the hydrophobic F99s out of the pore lumen (Fig. 5A; Yamashita et al. 2017). Molecular dynamics (MD) simulations showed spontaneous rotations at this location and illustrate how water entry would then support Ca2+ permeation. A spontaneously active H134A Orai1 mutant also displayed rotation of F99 and slight pore widening at this site (Yeung et al. 2018). Interestingly, the pore rotation model may help to explain the ability of STIM1 binding to establish high Ca2+ selectivity (Yamashita et al. 2017). Because E106 is only two helical turns above F99, it is possible it rotates as well to form the high-affinity Ca2+-binding site that precludes Na+ permeation.
Figure 5.
Models for Orai1 activation gating. (A) Pore rotation model. Top view of six TM1 helices showing opening of the hydrophobic gate through rotation of F99 side chains out of the pore lumen (Yamashita et al. 2017). (B) Pore dilation model. Side view of two opposing dOrai subunits, showing the closed structure (gray) superimposed on the constitutively open H206A structure (6BBF.pdb). (Panel B created from data in Hou et al. 2018.) (C) Interactions between hydrophobic residues in TM4 (orange) and TM3 (red) in a pair of dOrai subunits. Residues are indicated for dOrai (human Orai1 equivalents in parentheses). Mutations at these sites open Orai1 in the absence of STIM1, indicating a role in keeping the channel closed, and possibly in channel opening by STIM1 (Zhou et al. 2016). (D) A hydrophobic cluster between TM1 and TM2/TM3 is essential for transmitting gating forces to open the pore. Space-filling side chains are shown for TM1 (blue), TM2 (green), and TM3 (red). The cluster resides opposite the hydrophobic gate (pore side chains of V102 and F99 shown below E106) and may contribute to the rotation of F99 depicted in A.
The more central basic region comprising R91, K87, and R83 may also gate Ca2+ conduction by imposing an electrostatic barrier or by binding anions (Fig. 3C; Zhang et al. 2011; Hou et al. 2012). Thus far, it has not been possible to obtain a structure of the STIM-bound open channel, but Hou et al. succeeded in deriving a low-resolution crystal structure of a Ca2+-selective constitutively active dOrai H206A mutant (equivalent to Orai1 H134A) that revealed a dramatic dilation of the basic pore region (∼10 Å at the level of R91) along with a straightening of the M4ext helices (Fig. 5B; Hou et al. 2018). The crossed M4ext helices in the original closed WT dOrai structure present a steric barrier to pore dilation, suggesting that they act as latches to prevent spontaneous opening. This has led to a proposal in which the M4exts straighten first to allow STIM1 to bind and drive pore dilation to relieve the block by the basic gate (Hou et al. 2018).
A third proposal incorporates aspects of both the hydrophobic and basic gates. Based on MD simulations of the open Orai1 H134A mutant, Frischauf et al. (2017) reported a slight dilation of the hydrophobic section and a 1–2 Å dilation of the basic region. In addition, cysteine cross-linking and MD simulations suggest that R91 side chains rotate out of the pore to promote permeation. Unlike the study of Yamashita et al., F99 rotation was not observed, although a different force field, solvent, and membrane conditions in the MD simulations makes it difficult to directly compare the two studies.
More work is needed to fully understand the influence of STIM1 binding on pore gating and selectivity. Because most of the studies of the open channel have relied on a mutant (human Orai1 H134A/dOrai H206A) that may not fully mimic the STIM-bound open state, a structure of Orai1 bound to STIM1 will be needed to fully resolve this issue. This important goal has been difficult to achieve presumably because of the flexibility and low-affinity interactions between STIM1 and Orai1.
From STIM Binding to Orai Activation
A central question is how binding of STIM1 to the carboxyl termini of Orai at the channel periphery acts allosterically to open the central pore gate(s). In the absence of an open-channel structure of the STIM-Orai complex, insights about transmission of the gating “signal” have been limited to functional studies of a series of Orai1 mutants. More than 20 mutations in all four TM domains have been found to open the channel in the absence of STIM1 (Srikanth et al. 2011; Zhang et al. 2011; McNally et al. 2012; Palty et al. 2015; Zhou et al. 2016; Frischauf et al. 2017; Yeung et al. 2018; Bulla et al. 2019), identifying a large number of helical interactions that serve to maintain the closed resting state. Although removal of even a single “brake” is enough to allow opening, most of these mutant channels lack the characteristic Ca2+ selectivity of the CRAC channel. STIM binding to the mutants generally increases their activity and restores their selectivity, suggesting that multiple open conformations are possible when a brake is lost, and that STIM binding exerts a more controlled effect on the pore geometry. A combination of mutagenesis and structural studies are beginning to identify a potential pathway for the gating signal. The TM4 helix, which bends within the membrane at P245, is likely to straighten based on activation caused by P245 mutations (Palty et al. 2015) and the straightened M4ext helices in the constitutively active dOrai H206A channel (Hou et al. 2018). This may interrupt critical hydrophobic interactions between TM4 (261LV262) and TM3 (174LA175), consistent with the activating effects of mutations at these sites (Fig. 5C; Zhou et al. 2016). How this initiates movement of the TM2/TM3 cage is unclear. However, it is tempting to speculate that a prominent hydrophobic patch at the interface between TM1 and TM2/3 could act as a gear to translate motion of TM2/TM3 into rotation of the adjacent hydrophobic gate in TM1, based on the ability of single alanine substitutions within the patch to effectively abrogate channel opening by STIM1 (Fig. 5D; Yeung et al. 2018). Identification of additional Orai1 mutations that release or create new brakes may be useful in tracing the path of the activation signal.
Ca2+-Dependent Inactivation
Like many other Ca2+-permeable channels, CRAC channels are regulated through feedback inhibition by Ca2+. Ca2+ entering through CRAC channels causes rapid inactivation on a tens of milliseconds time scale (Hoth and Penner 1993; Zweifach and Lewis 1995; Fierro and Parekh 1999). The target Ca2+-binding site, estimated to lie within several nanometers of the inner mouth of the pore (Zweifach and Lewis 1995), is yet to be identified. An acidic region of the STIM1 cytosolic domain (475DDVDDMDEE483; termed IDSTIM) is essential for Ca2+-dependent inactivation (CDI) (Derler et al. 2009; Lee et al. 2009; Mullins et al. 2009). Although the isolated IDSTIM domain binds Ca2+, this has yet to be functionally connected to the CDI process (Mullins et al. 2009). Calmodulin (CaM) was an early candidate for the CDI Ca2+ sensor, based on binding of CaM to amino-terminal Orai1 peptides through Orai1 residues W76 and Y80 that are critical for CDI (Mullins et al. 2009; Liu et al. 2012). However, the subsequent structure of dOrai (Hou et al. 2012) showed that W76 and Y80 face the inside of the pore, without sufficient room for CaM binding, and further experiments showed that a Ca2+-binding-deficient CaM mutant failed to affect CDI (Mullins et al. 2016). Thus, current evidence argues that CaM is not the sensor, but that W76 interacts functionally with IDSTIM to support CDI (Mullins and Lewis 2016). A possible role of hydrophobic residues in the Orai1 2–3 loop has also been implicated (Srikanth et al. 2010a), but how it relates to IDSTIM and the inner pore domain needs to be explored.
SUMMARY AND PERSPECTIVE
The complex choreography of SOCE is now starting to be understood at a mechanistic and structural level, but as expected each advance brings with it additional questions. Release of Ca2+ from the luminal domain of STIM1 initiates rearrangements of the EF-SAM, TM, and CC1 domains to release CAD/SOAR and extend it toward the PM to interact with Orai1. Many details of the mechanism are still missing, such as a complete structural model of STIM1 and the critical intermediates as it transitions from inactive to active conformations. The diffusion trap model for STIM-Orai accumulation at ER–PM junctions is now established, and cytoskeletal and other proteins have been identified that influence their association, but much remains to be learned about how they operate, as well as the complete set of tethering proteins that control the plasticity of junction formation and signaling pathways localized at junctions. A structural basis for Orai function is emerging, and the hexameric channel appears to require full occupancy by STIM1 to effectively open and display the pore characteristics of the native CRAC channel. However, the precise ways in which STIM interacts with Orai is not entirely clear; for example, does STIM bind to the Orai amino terminus, does it bind as a monomer or a dimer, and could it bind in multiple stages to different sites? There is evidence for hydrophobic and basic gates in the pore; could they both operate to control conduction, and how is the energy from STIM binding transmitted allosterically to control each one? For fast CDI, more needs to be learned about the identity of the Ca2+ sensors, the interactions of IDSTIM with the channel, and how this promotes channel closure. Definitive answers to many of these questions will begin with the elucidation of structures for the STIM-Orai complex in various functional states. The long-range goal is to understand how STIM and Orai operate at cellular and molecular scales, to understand their roles in physiology and pathophysiology, and to further the ultimate aim of achieving ways of perturbing function or correcting defects to treat SOCE-related disease states.
ACKNOWLEDGMENTS
The author is grateful to Ruoyi Qiu and Michelle Yen for assistance with figures and to the National Institutes of Health (NIH) for support (R37GM45374).
Footnotes
Editors: Geert Bultynck, Martin D. Bootman, Michael J. Berridge, and Grace E. Stutzmann
Additional Perspectives on Calcium Signaling available at www.cshperspectives.org
REFERENCES
- Alvarez-Barrientos A, Martin-Romero FJ, Tomas-Martin P, Lopez-Guerrero AM, Pozo-Guisado E, Casas-Rua V. 2013. Phosphorylation of STIM1 at ERK1/2 target sites regulates interaction with the microtubule plus-end binding protein EB1. J Cell Sci 126: 3170–3180. 10.1242/jcs.125054 [DOI] [PubMed] [Google Scholar]
- Besprozvannaya M, Dickson E, Li H, Ginburg KS, Bers DM, Auwerx J, Nunnari J. 2018. GRAM domain proteins specialize functionally distinct ER–PM contact sites in human cells. eLife 7: e31019 10.7554/eLife.31019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Liou J, Park W, Meyer T. 2007. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell 131: 1327–1339. 10.1016/j.cell.2007.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulla M, Gyimesi G, Kim JH, Bhardwaj R, Hediger MA, Frieden M, Demaurex N. 2019. ORAI1 channel gating and selectivity is differentially altered by natural mutations in the first or third transmembrane domain. J Physiol 597: 561–582. 10.1113/JP277079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Zhou Y, Nwokonko RM, Loktionova NA, Wang X, Xin P, Trebak M, Wang Y, Gill DL. 2016. The Orai1 store-operated calcium channel functions as a hexamer. J Biol Chem 291: 25764–25775. 10.1074/jbc.M116.758813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Deb BK, Chorna T, Konieczny V, Taylor CW, Hasan G. 2016. Mutant IP3 receptors attenuate store-operated Ca2+ entry by destabilizing STIM–Orai interactions in Drosophila neurons. J Cell Sci 129: 3903–3910. 10.1242/jcs.191585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CL, Liou J. 2016. Homeostatic regulation of the PI(4,5)P2-Ca2+ signaling system at ER–PM junctions. Biochim Biophys Acta 1861: 862–873. 10.1016/j.bbalip.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CL, Chen YJ, Quintanilla CG, Hsieh TS, Liou J. 2018. EB1 binding restricts STIM1 translocation to ER–PM junctions and regulates store-operated Ca2+ entry. J Cell Biol 217: 2047–2058. 10.1083/jcb.201711151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Chang CL, Lee WR, Liou J. 2017. RASSF4 controls SOCE and ER–PM junctions through regulation of PI(4,5)P2. J Cell Biol 216: 2011–2025. 10.1083/jcb.201606047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington ED, Wu MM, Lewis RS. 2010. Essential role for the CRAC activation domain in store-dependent oligomerization of STIM1. Mol Biol Cell 21: 1897–1907. 10.1091/mbc.e10-02-0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb BK, Pathak T, Hasan G. 2016. Store-independent modulation of Ca2+ entry through Orai by Septin 7. Nat Commun 7: 11751 10.1038/ncomms11751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuro A, Penna A, Safrina O, Yeromin A V, Amcheslavsky A, Cahalan MD, Parker I. 2011. Subunit stoichiometry of human Orai1 and Orai3 channels in closed and open states. Proc Natl Acad Sci 108: 17832–17837. 10.1073/pnas.1114814108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derler I, Fahrner M, Muik M, Lackner B, Schindl R, Groschner K, Romanin C. 2009. A Ca2+ release-activated Ca2+ (CRAC) modulatory domain (CMD) within STIM1 mediates fast Ca2+-dependent inactivation of ORAI1 channels. J Biol Chem 284: 24933–24938. 10.1074/jbc.C109.024083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan E, Momburg F, Engel U, Temmerman K, Nickel W, Seedorf M. 2009. A conserved, lipid-mediated sorting mechanism of yeast Ist2 and mammalian STIM proteins to the peripheral ER. Traffic 10: 1802–1818. 10.1111/j.1600-0854.2009.00995.x [DOI] [PubMed] [Google Scholar]
- Fahrner M, Muik M, Schindl R, Butorac C, Stathopulos P, Zheng L, Jardin I, Ikura M, Romanin C. 2014. A coiled-coil clamp controls both conformation and clustering of stromal interaction molecule 1 (STIM1). J Biol Chem 289: 33231–33244. 10.1074/jbc.M114.610022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrner M, Pandey SK, Muik M, Traxler L, Butorac C, Stadlbauer M, Zayats V, Krizova A, Plenk P, Frischauf I, et al. 2018. Communication between N terminus and loop2 tunes Orai activation. J Biol Chem 293: 1271–1285. 10.1074/jbc.M117.812693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A. 2006. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 441: 179–185. 10.1038/nature04702 [DOI] [PubMed] [Google Scholar]
- Fierro L, Parekh AB. 1999. Fast calcium-dependent inactivation of calcium release-activated calcium current (CRAC) in RBL-1 cells. J Membr Biol 168: 9–17. 10.1007/s002329900493 [DOI] [PubMed] [Google Scholar]
- Frischauf I, Zayats V, Deix M, Hochreiter A, Jardin I, Muik M, Lackner B, Svobodova B, Pammer T, Litvinukova M, et al. 2015. A calcium-accumulating region, CAR, in the channel Orai1 enhances Ca2+ permeation and SOCE-induced gene transcription. Sci Signal 8: ra131–ra131. 10.1126/scisignal.aab1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf I, Litviňuková M, Schober R, Zayats V, Svobodová B, Bonhenry D, Lunz V, Cappello S, Tociu L, Reha D, et al. 2017. Transmembrane helix connectivity in Orai1 controls two gates for calcium-dependent transcription. Sci Signal 10: eaao0358 10.1126/scisignal.aao0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P. 2013. PI(4,5)P2-dependent and Ca2+-regulated ER–PM interactions mediated by the extended synaptotagmins. Cell 153: 1494–1509. 10.1016/j.cell.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I, Gouveia SM, van der Vaart B, Demmers J, Smyth JT, Honnappa S, Splinter D, Steinmetz MO, Putney JW, Hoogenraad CC, et al. 2008. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr Biol 18: 177–182. 10.1016/j.cub.2007.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudlur A, Quintana A, Zhou Y, Hirve N, Mahapatra S, Hogan PG. 2014. STIM1 triggers a gating rearrangement at the extracellular mouth of the ORAI1 channel. Nat Commun 5: 5164 10.1038/ncomms6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudlur A, Zeraik AE, Hirve N, Rajanikanth V, Bobkov AA, Ma G, Zheng S, Wang Y, Zhou Y, Komives EA, et al. 2018. Calcium sensing by the STIM1 ER-luminal domain. Nat Commun 9: 4536 10.1038/s41467-018-06816-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirve N, Rajanikanth V, Hogan PG, Gudlur A. 2018. Coiled-coil formation conveys a STIM1 signal from ER lumen to cytoplasm. Cell Rep 22: 72–83. 10.1016/j.celrep.2017.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodeify R, Selvaraj S, Wen J, Arredouani A, Hubrack S, Dib M, Al-Thani SN, McGraw T, Machaca K. 2015. A STIM1-dependent “trafficking trap” mechanism regulates Orai1 plasma membrane residence and Ca2+ influx levels. J Cell Sci 128: 3143–3154. 10.1242/jcs.172320 [DOI] [PubMed] [Google Scholar]
- Hodeify R, Nandakumar M, Own M, Courjaret RJ, Graumann J, Hubrack SZ, Machaca K. 2018. The CCT chaperonin is a novel regulator of Ca2+ signaling through modulation of Orai1 trafficking. Sci Adv 4: eaau1935 10.1126/sciadv.aau1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover PJ, Lewis RS. 2011. Stoichiometric requirements for trapping and gating of Ca2+ release-activated Ca2+ (CRAC) channels by stromal interaction molecule 1 (STIM1). Proc Natl Acad Sci 108: 13299–13304. 10.1073/pnas.1101664108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth M, Penner R. 1992. Depletion of intracellular calcium stores activates a calcium current in mast cells. Nature 355: 353–356. 10.1038/355353a0 [DOI] [PubMed] [Google Scholar]
- Hoth M, Penner R. 1993. Calcium release-activated calcium current in rat mast cells. J Physiol 465: 359–386. 10.1113/jphysiol.1993.sp019681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Pedi L, Diver MM, Long SB. 2012. Crystal structure of the calcium release-activated calcium channel Orai. Science 338: 1308–1313. 10.1126/science.1228757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Burstein SR, Long SB. 2018. Structures reveal opening of the store-operated calcium channel Orai. eLife 7: e36758 10.7554/eLife.36758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TS, Chen YJ, Chang CL, Lee WR, Liou J. 2017. Cortical actin contributes to spatial organization of ER–PM junctions. Mol Biol Cell 28: 3171–3180. 10.1091/mbc.e17-06-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Xu P, Li Z, Lu J, Liu L, Zhan Y, Chen Y, Hille B, Xu T, Chen L. 2008. Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proc Natl Acad Sci 105: 13668–13673. 10.1073/pnas.0806499105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing J, He L, Sun A, Quintana A, Ding Y, Ma G, Tan P, Liang X, Zheng X, Chen L, et al. 2015. Proteomic mapping of ER–PM junctions identifies STIMATE as a regulator of Ca2+ influx. Nat Cell Biol 17: 1339–1347. 10.1038/ncb3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Lange I, Feske S. 2009. A minimal regulatory domain in the C terminus of STIM1 binds to and activates ORAI1 CRAC channels. Biochem Biophys Res Commun 385: 49–54. 10.1016/j.bbrc.2009.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Guzman-Hernandez ML, Wisniewski E, Balla T. 2015. Phosphatidylinositol-phosphatidic acid exchange by Nir2 at ER–PM contact sites maintains phosphoinositide signaling competence. Dev Cell 33: 549–561. 10.1016/j.devcel.2015.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KM, Wijerathne T, Hur JH, Kang UJ, Kim IH, Kweon YC, Lee AR, Jeong SJ, Lee SK, Lee YY, et al. 2018. Distinct gating mechanism of SOC channel involving STIM-Orai coupling and an intramolecular interaction of Orai in Caenorhabditis elegans. Proc Natl Acad Sci 115: E4623–E4632. 10.1073/pnas.1714986115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniowski M, Popovic M, Szentpetery Z, Varnai P, Stojilkovic S, Balla T. 2009. Dependence of STIM1/Orai1-mediated calcium entry on plasma membrane phosphoinositides. J Biol Chem 284: 21027–21035. 10.1074/jbc.M109.012252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniowski MK, Manjarrés IM, Varnai P, Balla T. 2010. Activation of STIM1-Orai1 involves an intramolecular switching mechanism. Sci Signal 3: ra82 10.1126/scisignal.2001122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniowski MK, Wisniewski E, Baird B, Holowka DA, Balla T. 2017. Molecular anatomy of the early events in STIM1 activation—Oligomerization or conformational change? J Cell Sci 130: 2821–2832. 10.1242/jcs.205583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacruz RS, Feske S. 2015. Diseases caused by mutations in ORAI1 and STIM1. Ann NY Acad Sci 1356: 45–79. 10.1111/nyas.12938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Yuan JP, Zeng W, So I, Worley PF, Muallem S. 2009. Molecular determinants of fast Ca2+-dependent inactivation and gating of the Orai channels. Proc Natl Acad Sci 106: 14687–14692. 10.1073/pnas.0904664106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RS, Cahalan MD. 1989. Mitogen-induced oscillations of cytosolic Ca2+ and transmembrane Ca2+ current in human leukemic T cells. Cell Regul 1: 99–112. 10.1091/mbc.1.1.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lu J, Xu P, Xie X, Chen L, Xu T. 2007. Mapping the interacting domains of STIM1 and Orai1 in Ca2+ release-activated Ca2+ channel activation. J Biol Chem 282: 29448–29456. 10.1074/jbc.M703573200 [DOI] [PubMed] [Google Scholar]
- Liou J, Kim ML, Do HW, Jones JT, Myers JW, Ferrell JE, Meyer T. 2005. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol 15: 1235–1241. 10.1016/j.cub.2005.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J, Fivaz M, Inoue T, Meyer T. 2007. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci 104: 9301–9306. 10.1073/pnas.0702866104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zheng X, Mueller GA, Sobhany M, Derose EF, Zhang Y, London RE, Birnbaumer L. 2012. Crystal structure of calmodulin binding domain of Orai1 in complex with Ca2+/calmodulin displays a unique binding mode. J Biol Chem 287: 43030–43041. 10.1074/jbc.M112.380964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luik RM, Wu MM, Buchanan J, Lewis RS. 2006. The elementary unit of store-operated Ca2+ entry: Local activation of CRAC channels by STIM1 at ER–plasma membrane junctions. J Cell Biol 174: 815–825. 10.1083/jcb.200604015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luik RM, Wang B, Prakriya M, Wu MM, Lewis RS. 2008. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature 454: 538–542. 10.1038/nature07065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G, Wei M, He L, Liu C, Wu B, Zhang SL, Jing J, Liang X, Senes A, Tan P, et al. 2015. Inside-out Ca2+ signalling prompted by STIM1 conformational switch. Nat Commun 6: 7826 10.1038/ncomms8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally BA, Somasundaram A, Yamashita M, Prakriya M. 2012. Gated regulation of CRAC channel ion selectivity by STIM1. Nature 482: 241–245. 10.1038/nature10752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally BA, Somasundaram A, Jairaman A, Yamashita M, Prakriya M. 2013. The C- and N-terminal STIM1 binding sites on Orai1 are required for both trapping and gating CRAC channels. J Physiol 591: 2833–2850. 10.1113/jphysiol.2012.250456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miederer AM, Alansary D, Schwär G, Lee PH, Jung M, Helms V, Niemeyer BA. 2015. A STIM2 splice variant negatively regulates store-operated calcium entry. Nat Commun 6: 6899 10.1038/ncomms7899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignen O, Thompson JL, Shuttleworth TJ. 2008. Orai1 subunit stoichiometry of the mammalian CRAC channel pore. J Physiol 586: 419–425. 10.1113/jphysiol.2007.147249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik M, Fahrner M, Derler I, Schindl R, Bergsmann J, Frischauf I, Groschner K, Romanin C. 2009. A cytosolic homomerization and a modulatory domain within STIM1 C terminus determine coupling to ORAI1 channels. J Biol Chem 284: 8421–8426. 10.1074/jbc.C800229200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik M, Fahrner M, Schindl R, Stathopulos P, Frischauf I, Derler I, Plenk P, Lackner B, Groschner K, Ikura M, et al. 2011. STIM1 couples to ORAI1 via an intramolecular transition into an extended conformation. EMBO J 30: 1678–1689. 10.1038/emboj.2011.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins FM, Lewis RS. 2016. The inactivation domain of STIM1 is functionally coupled with the Orai1 pore to enable Ca2+-dependent inactivation. J Gen Physiol 147: 153–164. 10.1085/jgp.201511438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins FM, Park CY, Dolmetsch RE, Lewis RS. 2009. STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc Natl Acad Sci 106: 15495–15500. 10.1073/pnas.0906781106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins FM, Yen M, Lewis RS. 2016. Orai1 pore residues control CRAC channel inactivation independently of calmodulin. J Gen Physiol 147: 137–152. 10.1085/jgp.201511437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong HL, de Souza LB, Zheng C, Cheng KT, Liu X, Goldsmith CM, Feske S, Ambudkar IS. 2015. STIM2 enhances receptor-stimulated Ca2+ signaling by promoting recruitment of STIM1 to the endoplasmic reticulum-plasma membrane junctions. Sci Signal 8: ra3 10.1126/scisignal.2005748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Le Coadic M, Shen WW, Demaurex N, Cosson P. 2009. STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc Natl Acad Sci 106: 19358–19362. 10.1073/pnas.0911280106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palty R, Isacoff EY. 2016. Cooperative binding of stromal interaction molecule 1 (STIM1) to the N and C termini of calcium release-activated calcium modulator 1 (Orai1). J Biol Chem 291: 334–341. 10.1074/jbc.M115.685289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palty R, Stanley C, Isacoff EY. 2015. Critical role for Orai1 C-terminal domain and TM4 in CRAC channel gating. Cell Res 25: 963–980. 10.1038/cr.2015.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palty R, Fu Z, Isacoff EY. 2017. Sequential steps of CRAC channel activation. Cell Rep 19: 1929–1939. 10.1016/j.celrep.2017.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Hoover PJ, Mullins FM, Bachhawat P, Covington ED, Raunser S, Walz T, Garcia KC, Dolmetsch RE, Lewis RS. 2009. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell 136: 876–890. 10.1016/j.cell.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penna A, Demuro A, Yeromin A V, Zhang SL, Safrina O, Parker I, Cahalan MD. 2008. The CRAC channel consists of a tetramer formed by Stim-induced dimerization of Orai dimers. Nature 456: 116–120. 10.1038/nature07338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perni S, Dynes JL, Yeromin A V, Cahalan MD, Franzini-Armstrong C. 2015. Nanoscale patterning of STIM1 and Orai1 during store-operated Ca2+ entry. Proc Natl Acad Sci 112: E5533–E5542. 10.1073/pnas.1515606112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, Voeltz GK. 2016. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol 17: 69–82. 10.1038/nrm.2015.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M, Lewis RS. 2006. Regulation of CRAC channel activity by recruitment of silent channels to a high open-probability gating mode. J Gen Physiol 128: 373–386. 10.1085/jgp.200609588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M, Lewis RS. 2015. Store-operated calcium channels. Physiol Rev 95: 1383–1436. 10.1152/physrev.00020.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M, Feske S, Gwack Y, Srikanth S, Rao A, Hogan P. 2006. Orai1 is an essential pore subunit of the CRAC channel. Nature 443: 230–233. 10.1038/nature05122 [DOI] [PubMed] [Google Scholar]
- Putney JW. 1986. A model for receptor-regulated calcium entry. Cell Calcium 7: 1–12. 10.1016/0143-4160(86)90026-6 [DOI] [PubMed] [Google Scholar]
- Rana A, Yen M, Sadaghiani AM, Malmersjö S, Park CY, Dolmetsch RE, Lewis RS. 2015. Alternative splicing converts STIM2 from an activator to an inhibitor of store-operated calcium channels. J Cell Biol 209: 653–670. 10.1083/jcb.201412060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J, DiGregorio PJ, Yeromin A V, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, et al. 2005. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol 169: 435–445. 10.1083/jcb.200502019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki Y, De Camilli P. 2017. Endoplasmic reticulum-plasma membrane contact sites. Annu Rev Biochem 86: 659–684. 10.1146/annurev-biochem-061516-044932 [DOI] [PubMed] [Google Scholar]
- Saheki Y, Bian X, Schauder CM, Sawaki Y, Surma MA, Klose C, Pincet F, Reinisch KM, De Camilli P. 2016. Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat Cell Biol 18: 504–515. 10.1038/ncb3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampieri A, Zepeda A, Asanov A, Vaca L. 2009. Visualizing the store-operated channel complex assembly in real time: Identification of SERCA2 as a new member. Cell Calcium 45: 439–446. 10.1016/j.ceca.2009.02.010 [DOI] [PubMed] [Google Scholar]
- Scrimgeour N, Litjens T, Ma L, Barritt GJ, Rychkov GY. 2009. Properties of Orai1 mediated store-operated current depend on the expression levels of STIM1 and Orai1 proteins. J Physiol 587: 2903–2918. 10.1113/jphysiol.2009.170662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Quintana A, Findlay GM, Mettlen M, Baust B, Jain M, Nilsson R, Rao A, Hogan PG. 2013. An siRNA screen for NFAT activation identifies septins as coordinators of store-operated Ca2+ entry. Nature 499: 238–242. 10.1038/nature12229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboloff J, Rothberg BS, Madesh M, Gill DL. 2012. STIM proteins: Dynamic calcium signal transducers. Nat Rev Mol Cell Biol 13: 549–565. 10.1038/nrm3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S, Jung HJ, Ribalet B, Gwack Y. 2010a. The intracellular loop of Orai1 plays a central role in fast inactivation of Ca2+ release-activated Ca2+ channels. J Biol Chem 285: 5066–5075. 10.1074/jbc.M109.072736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S, Jung HJ, Kim KD, Souda P, Whitelegge J, Gwack Y. 2010b. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat Cell Biol 12: 436–446. 10.1038/ncb2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S, Yee MKW, Gwack Y, Ribalet B. 2011. The third transmembrane segment of Orai1 protein modulates Ca2+ release-activated Ca2+ (CRAC) channel gating and permeation properties. J Biol Chem 286: 35318–35328. 10.1074/jbc.M111.265884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S, Jew M, Kim KD, Yee MK, Abramson J, Gwack Y. 2012. Junctate is a Ca2+-sensing structural component of Orai1 and stromal interaction molecule 1 (STIM1). Proc Natl Acad Sci 109: 8682–8687. 10.1073/pnas.1200667109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopulos PB, Ikura M. 2019. Does stromal interaction molecule-1 have five senses? Cell Calcium 77: 79–80. 10.1016/j.ceca.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Stathopulos P, Li G, Plevin M, Ames J, Ikura M. 2006. Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region: An initiation mechanism for capacitive Ca2+ entry. J Biol Chem 281: 35855–35862. 10.1074/jbc.M608247200 [DOI] [PubMed] [Google Scholar]
- Stathopulos PB, Schindl R, Fahrner M, Zheng L, Gasmi-Seabrook GM, Muik M, Romanin C, Ikura M. 2013. STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry. Nat Commun 4: 2963 10.1038/ncomms3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaeth M, Yang J, Yamashita M, Zee I, Eckstein M, Knosp C, Kaufmann U, Karoly Jani P, Lacruz RS, Flockerzi V, et al. 2017. ORAI2 modulates store-operated calcium entry and T cell-mediated immunity. Nat Commun 8: 14714 10.1038/ncomms14714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet AR, Giordano F, Gerlo S, Segura I, Van Eygen S, Molenberghs G, Rocha S, Houcine A, Derua R, Verfaillie T, et al. 2017. The ER stress sensor PERK coordinates ER-plasma membrane contact site formation through interaction with filamin-A and F-actin remodeling. Mol Cell 65: 885–899.e6. 10.1016/j.molcel.2017.01.020 [DOI] [PubMed] [Google Scholar]
- Várnai P, Tóth B, Tóth DJ, Hunyady L, Balla T. 2007. Visualization and manipulation of plasma membrane–endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 complex. J Biol Chem 282: 29678–29690. 10.1074/jbc.M704339200 [DOI] [PubMed] [Google Scholar]
- Vig M, Beck A, Billingsley J, Lis A, Parvez S, Peinelt C, Koomoa D, Soboloff J, Gill D, Fleig A, et al. 2006a. CRACM1 multimers form the ion-selective pore of the CRAC channel. Curr Biol 16: 2073–2079. 10.1016/j.cub.2006.08.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, et al. 2006b. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 312: 1220–1223. 10.1126/science.1127883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CM, Chvanov M, Haynes LP, Petersen OH, Tepikin A V, Burgoyne RD. 2010. Role of phosphoinositides in STIM1 dynamics and store-operated calcium entry. Biochem J 425: 159–168. 10.1042/BJ20090884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MM, Buchanan J, Luik RM, Lewis RS. 2006. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol 174: 803–813. 10.1083/jcb.200604014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MM, Covington ED, Lewis RS. 2014. Single-molecule analysis of diffusion and trapping of STIM1 and Orai1 at endoplasmic reticulum-plasma membrane junctions. Mol Biol Cell 25: 3672–3685. 10.1091/mbc.e14-06-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Yeung PSW, Ing CE, McNally BA, Pomès R, Prakriya M. 2017. STIM1 activates CRAC channels through rotation of the pore helix to open a hydrophobic gate. Nat Commun 8: 14512 10.1038/ncomms14512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Jin H, Cai X, Li S, Shen Y. 2012. Structural and mechanistic insights into the activation of Stromal interaction molecule 1 (STIM1). Proc Natl Acad Sci 109: 5657–5662. 10.1073/pnas.1118947109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen M, Lewis RS. 2018. Physiological CRAC channel activation and pore properties require STIM1 binding to all six Orai1 subunits. J Gen Physiol 150: 1373–1385. 10.1085/jgp.201711985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen M, Lewis RS. 2019. Numbers count: How STIM and Orai stoichiometry affect store-operated calcium entry. Cell Calcium 79: 35–43. 10.1016/j.ceca.2019.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen M, Lokteva LA, Lewis RS. 2016. Functional analysis of Orai1 concatemers supports a hexameric stoichiometry for the CRAC channel. Biophys J 111: 1897–1907. 10.1016/j.bpj.2016.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeromin A, Zhang S, Jiang W, Yu Y, Safrina O, Cahalan M. 2006. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature 443: 226–229. 10.1038/nature05108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung PS-W, Yamashita M, Ing CE, Pomès R, Freymann DM, Prakriya M. 2018. Mapping the functional anatomy of Orai1 transmembrane domains for CRAC channel gating. Proc Natl Acad Sci 115: E5193–E5202. 10.1073/pnas.1718373115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Zeng W, Dorwart M, Choi Y, Worley P, Muallem S. 2009. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat Cell Biol 11: 337–343. 10.1038/ncb1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Yu Y, Roos J, Kozak J, Deerinck T, Ellisman M, Stauderman K, Cahalan M. 2005. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature 437: 902–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SL, Yeromin A V, Zhang XHF, Yu Y, Safrina O, Penna A, Roos J, Stauderman KA, Cahalan MD. 2006. Genome-wide RNAi screen of Ca2+ influx identifies genes that regulate Ca2+ release-activated Ca2+ channel activity. Proc Natl Acad Sci 103: 9357–9362. 10.1073/pnas.0603161103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SL, Yeromin A V, Hu J, Amcheslavsky A, Zheng H, Cahalan MD. 2011. Mutations in Orai1 transmembrane segment 1 cause STIM1-independent activation of Orai1 channels at glycine 98 and channel closure at arginine 91. Proc Natl Acad Sci 108: 17838–17843. 10.1073/pnas.1114821108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Stathopulos PB, Schindl R, Li GY, Romanin C, Ikura M. 2011. Auto-inhibitory role of the EF-SAM domain of STIM proteins in store-operated calcium entry. Proc Natl Acad Sci 108: 1337–1342. 10.1073/pnas.1015125108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Zhou L, Ma G, Zhang T, Liu J, Li J, Nguyen NT, Zhang X, Li W, Nwokonko R, et al. 2018. Calcium store refilling and STIM activation in STIM- and Orai-deficient cell lines. Pflugers Arch 470: 1555–1567. 10.1007/s00424-018-2165-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Meraner P, Kwon HT, Machnes D, Masatsugu O, Zimmer J, Huang Y, Stura A, Rao A, Hogan PG. 2010. STIM1 gates the store-operated calcium channel ORAI1 in vitro. Nat Struct Mol Biol 17: 112–116. 10.1038/nsmb.1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Srinivasan P, Razavi S, Seymour S, Meraner P, Gudlur A, Stathopulos PB, Ikura M, Rao A, Hogan PG. 2013. Initial activation of STIM1, the regulator of store-operated calcium entry. Nat Struct Mol Biol 20: 973–981. 10.1038/nsmb.2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang X, Wang X, Loktionova NA, Cai X, Nwokonko RM, Vrana E, Wang Y, Rothberg BS, Gill DL. 2015. STIM1 dimers undergo unimolecular coupling to activate Orai1 channels. Nat Commun 6: 8395 10.1038/ncomms9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Cai X, Loktionova NA, Wang X, Nwokonko RM, Wang X, Wang Y, Rothberg BS, Trebak M, Gill DL. 2016. The STIM1-binding site nexus remotely controls Orai1 channel gating. Nat Commun 7: 13725 10.1038/ncomms13725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Nwokonko RM, Cai X, Loktionova NA, Abdulqadir R, Xin P, Niemeyer BA, Wang Y, Trebak M, Gill DL. 2018. Cross-linking of Orai1 channels by STIM proteins. Proc Natl Acad Sci 115: E3398–E3407. 10.1073/pnas.1720810115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifach A, Lewis RS. 1993. Mitogen-regulated Ca2+ current of T lymphocytes is activated by depletion of intracellular Ca2+ stores. Proc Natl Acad Sci 90: 6295–6299. 10.1073/pnas.90.13.6295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifach A, Lewis RS. 1995. Rapid inactivation of depletion-activated calcium current (ICRAC) due to local calcium feedback. J Gen Physiol 105: 209–226. 10.1085/jgp.105.2.209 [DOI] [PMC free article] [PubMed] [Google Scholar]