Abstract
Aims: The dysregulation and essential role of WNTs in glioma have been widely implicated. However, there is a paucity of literature on the expression status of all the 19 WNTs in glioma. Our study was aimed to evaluate the expression and prognostic values of the 19 WNTs in glioma. Methods: mRNA expression and clinical data were retrieved from the Cancer Genome Atlas (TCGA) database, Chinese Glioma Genome Atlas (CGGA), GTEx and ONCOMINE databases. The 50 frequent neighbor genes of WNT5A and WNT10B were shown with PPI network, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. Results: We found that the mRNA expression of WNT5A was significantly higher in glioma; however, the WNT10B expression was significantly lower in glioma. Furthermore, the expression of WNT5A and WNT10B was associated with the clinicopathology of glioma. The survival analysis revealed that the higher expressions of WNT5A and WNT16 were associated poor overall survival (OS) in patients with glioma. Conversely, overexpression of WNT3, WNT5B, and WNT10B was associated with better OS. Finally, Go and KEGG analysis revealed WNT5A was associated with multiple signal translations, and crucial oncogenes (EGFR and MDM2) and 2 important tumor suppressors (PTEN and IKN4a/ARF) were found closely correlated with WNT5A in glioma. Conclusion: Among 19WNTs, WNT5A can serve as a candidate to diagnose and therapy glioma, while WNT10B might be valuable for anti-glioma research. The presumed direction was provided to explore the relation of WNTs signal and multiple pathways in glioma.
Keywords: bioinformatics, expression, glioma, prognosis, WNT
Introduction
Glioma represents the most frequent primary intracranial neoplasms and is characterized by aggressive growth and poor prognosis [1]. The WHO classification divides glioma originating from a glial cell into 4 (I–IV) grades based on the malignant degree [1]. Glioblastoma (GBM; WHO grade IV), which constitutes 54.9% of all gliomas, is the most aggressive and highly invasive type of glioma. For the patients with GBM, the median overall survival is merely 12–15 months and the 5-year survival rate remains less than 5% [2,3]. As the heterogeneity represents a major challenge in precise diagnosis and therapy of glioma [4], the molecular signatures of glioma are urgent to be explored for diagnosis and therapy [5].
In the hundreds of previous studies, 8 members of 19 WNTs, WNT1, 2, 2B, 3A, 5A, 6, 7A and 7B, were showed being related with glioma development [4,6–9]. Nowadays, increasing evidences report that WNTs play essential role in glioma progress [2]. WNT1 has been suggested to be overexpressed with a paradoxical role in glioma [10,11]. WNT2 knockdown in glioma cells significantly suppressed growth and associated with the decrease of PI3K/p-AKT expression in vitro and in vivo [12]. Furthermore, WNT1 and WNT3A were found to increase glioma-derived stem-like cells chemo-resistance, proliferation and migration [11]. In addition, WNT5A was found to induce rapid growth and migration of glioma and associated with the presence of tumor-associated microglia [8,9,13,14]. WNT6 expression was shown to increase the features of glioma aggressiveness [6]. WNT7A and WNT7B was shown to regulate glioma-vascular interactions [7]. Collectively, these pieces of evidence indicate that targeting WNT signaling might be an effective therapeutic strategy against glioma.
Nineteen WNTs of Homo sapiens are numbered in the order of discovery: WNT1, 2, 2B, 3, 3A, 4, 5A, 5B, 6, 7A, 7B, 8A, 8B, 9A, 9B, 10A, 10B, 11 and 16 [15,16]. WNT signaling cascade was an evolutionarily conserved pathway [17]. Abnormality of WNT signaling cascade has been found in many cancers. WNTs are known to function via three WNT signaling pathways, including the canonical WNT/β-catenin pathway, and the two non-canonical pathways such as WNT/PCP and WNT/Ca2+ pathways. Accumulating evidences indicate that 11 members of WNT1, 2, 2B, 3, 3A, 7A, 7B, 9A, 9B, 10A and 10B are predominant ligands of canonical pathway [11,18–23]; WNT7A and WNT7B are also the ligands of WNT/PCP pathway [24–26]. WNT5A and 5B are mainly considered as the ligands of WNT/Ca2+pathway and take part in the canonical pathway as well [20,27]. Besides, six WNT members, WNT4, 6, 8A, 8B, 11 and 16, remain largely unknown for which specific pathway they function [21,28,29]. However, which WNT of 19 WNT family members are the most potential to be biomarkers of glioma and the way they take part in glioma progress are still not clear.
In the present study, we extensively used bioinformatic approaches to analyze the expression profiles and evaluate the prognostic significance of all the 19 members of the WNT family in glioma with publicly available data from TCGA, CGGA, CCLE and other databases. The PPI network, GO and KEGG analyses were performed to analyze the biofunction and molecular mechanism of several essential WNTs in glioma. Therefore, we could find out the most reliable prognostic markers and therapeutic strategies against glioma in 19 WNTs.
Materials and methods
ONCOMINE database
ONCOMINE (www.oncomine.org) is currently the world’s largest online data-mining platform including data of over 20 cancers from TCGA and GEO databases [30]. The transcriptional mRNA expression data of 19 WNTs across different cancers along with corresponding normal tissues was retrieved from ONCOMINE database. The P-value was set up at 0.01.
Broad Institute Cancer Cell Line Encyclopedia (CCLE) database
The CCLE (https://www.broadinstitute.org/ccle) database was used to retrieve the mRNA expression of WNTs in various cancer cell lines, and the data are verified and illustrated as box plots [31]. In addition, the mRNA expression of 19 WNTs was compared in multiple well-known glioma cell lines.
The Cancer Genome Atlas (TCGA) database and Chinese Glioma Genome Atlas (CGGA) database
The TCGA database is a comprehensive and coordinated project, including information about sequencing and pathological data of more than 30 types of human tumors. Clinicopathological parameters and WNTs’ mRNA expression from 665 patients with glioma (GBM:156, LGG:509) were downloaded for our study. The CGGA database is a free used platform to investigate glioma from Chinese cohorts including over 600 glioma samples of patients. The mRNA microarray of 301 patients and matched clinical data were retrieved in the present study. The clinicopathological correlation and prognostic value of WNTs’ mRNA expression in glioma were accepted only when the results are statistically significant in both databases.
Gene Expression Profiling Interactive Analysis (GEPIA)
In our study, the mRNA expression data of 163 GBM samples and 518 LGG samples were retrieved from TCGA database, and the mRNA expression data of 207 NB samples was retrieved form GTEx database. For analysis, we utilized GEPIA to compare mRNA expression of WNTs between glioma and NB tissues. Furthermore, we performed the survival analysis of 19 WNTs in patients with glioma [32].
c-BioPortal
c-BioPortal (www.cbioportal.org) is an online open-access website to easily explore, visualize and analyze the multidimensional genomic data of cancers. Following the c-BioPortal’s online instruction, 50 frequent neighbor genes of WNT5A and WNT10B were determined as summarized in Table 2.
Table 2. Neighboring genes of WNT5A and WNT10B.
| Gene | Neighbor genes |
|---|---|
| WNT5A | AP2A1, AP2A2, AP2M1, AP2S1, ARHGEF6, CALM3, CER1, CSNK1G2, CTNNB1, DCC, DOCK1, DVL1, DVL3, EDNRA, EFNB2, EFNB3, ELMO1, EPHB4, FLNA, GNA11, GNA15, GNB3, GNB4, GNG11, GNG7, GNG8, GNGT1, HRAS, ILK, KRAS, LRP6, MAPK1, MYC, NOS1, NOS3, PIK3CA, PIK3CD, PIK3CG, PIK3R1, PORCN, PRKCA, PRKCD, PRKCG, PTK2, RHOG, SAG, SFRP2, SPHK1, VAV1, VAV2 |
| WNT10B | ARR3, CDK8, CEBPA, CER1, CREBBP, CSNK1A1L, CSNK1D, CSNK1E, CSNK1G2, CTNNB1, DVL1, DVL3, EBF1, EP300, GNA11, GNA14, GNA15, GNB1, GNB2, GNB3, GNB4, GNG12, GNG13, GNG7, GNG8, HELZ2, LRP6, MED10, MED12, MED13, MED13L, MED14, MED15, MED16, MED20, MED21, MED22, MED25, MED26, MED27, MED29, MED30, MED4, MED7 PORCN, PPARG, RXRA, SAG, SFRP2, TGFB1 |
PPI network and module analysis
STRING (https://string-db.org/) was used to make PPI network of WNT5A/10B and 50 frequent neighbor genes. Then, Cytoscape was applied to examine the potential correlation between these genes. In addition, the MCODE app and BONGO app in Cytoscape were used to check modules of the PPI network (degree cutoff = 2, max. Depth = 100, k-core = 2, and node score cutoff = 0.2).
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis
GO analysis predicted the functions of WNT5A and 10B and in glioma on the basis of three aspects, including molecular function, biological processes and biological pathway. KEGG analysis showed the important role of WNT5A, visualized with the online website DAVID (https://david.ncifcrf.gov/summary.jsp).
Statistical methods
IMB SPSS 22.0 and GraphPad Prism 7.0 software were used to analyze statistical data and plot the graphs. All values were expressed as means ± standard deviations (SD). P < 0.05 was considered to be statistically significant. A Student’s t-test was used to assess the differences between two groups. Different grade of malignant glioma was analyzed by one-way analysis of variance (ANOVA) followed by Dunnett test. The Kaplan–Meier method was used to plot survival curves and compared using the log-rank test.
Results
mRNA expression of WNTs in glioma compared with NB tissue
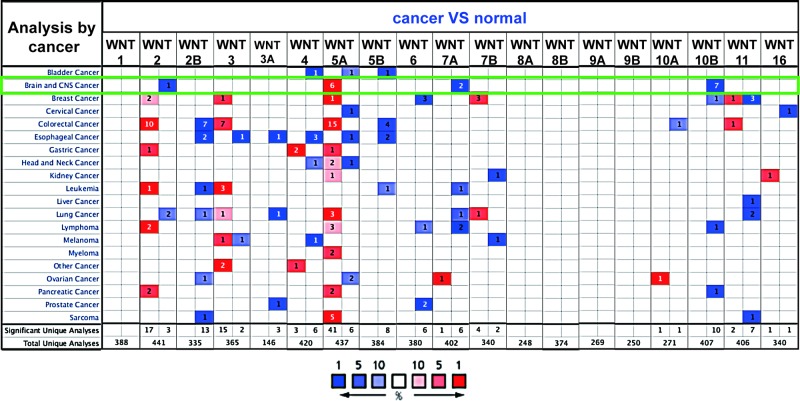
Thus far, 19 WNT ligands have been identified in Homo sapiens. By analyzing the ONCOMINE database, the expression profiles of WNTs were analyzed across 20 cancers and compared with corresponding normal tissues. The higher level of WNT5A mRNA expression is elevated in most cancers. For CNS tumor, higher expression of WNT5A, lower expression of WNT7A and lower expression of WNT10B were found in glioma compared with NB tissues (Figure 1).
Figure 1. Transcriptional expression of 19 WNTs in different cancer types using the ONCOMINE database. mRNA overexpression (red) or down-regulation (blue).
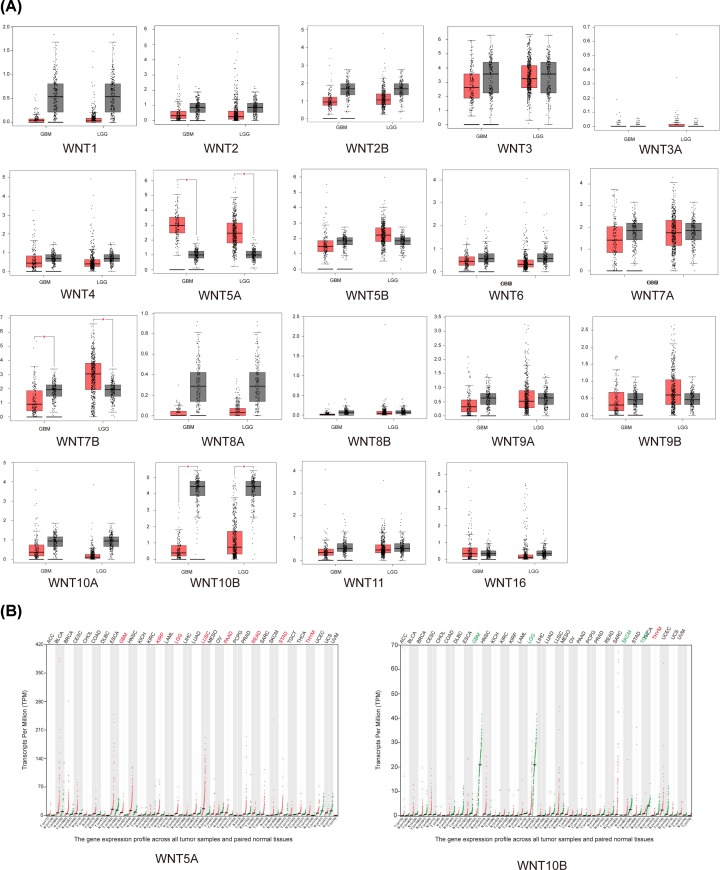
Glioblastoma (GBM; WHO grade IV) was the most malignant of all glioma, the others were low grade glioma (LGG; WHO grade I–III). We analyzed 19 WNTs expression in GBM and LGG compared with NB tissues using GEPIA website. The result suggested that only higher expression of WNT5A and lower expression of WNT10B were detected with statistical significance (Figure 2A). In multiple human tumors, significantly higher mRNA expression of WNT5A and lower mRNA expression of WNT10B in GBM and LGG were also shown (Figure 2B).
Figure 2. mRNA expression of 19 WNT genes in glioma including GBM and LGG compared with NB tissue using GEPIA.
(A) Statistically significant higher expression of WNT5A and lower expression of WNT10B in both GBM and LGG compared with NB (GBM: n= 163, LGG: 518, NB: n= 207) analyzed with TCGA and GTEx datasets (P <0.05). (B) Higher expression of WNT5A and lower expression of WNT10B in GBM and LGG compared with other human tumors.
mRNA expression of WNTs in glioma cell lines
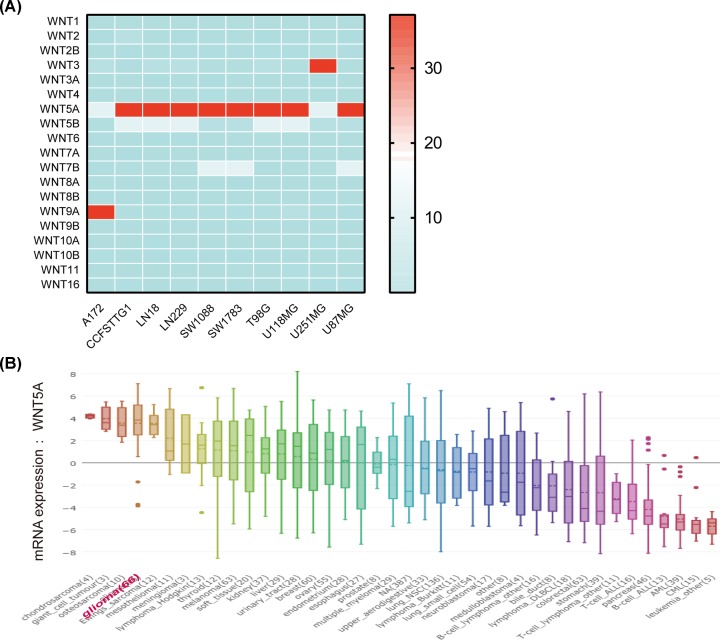
Then, we analyzed the mRNA expression of WNTs in glioma cell lines by assembling the CCLE database. We expanded the procedure of detailed annotation in preclinical human glioma models. WNT5A was differentially higher expressed in glioma cell lines among 19 WNTs, implying that WNT5A might play distinct functions in glioma (Figure 3A). CCLE analysis was also revealed that WNT5A mRNA expression was significantly up-regulated in glioma compared with other human cancers (Figure 3B).
Figure 3. The expression of WNTs in glioma cell lines using CCLE database.
(A) Among 19 WNTs, higher expression of WNT5A in multiple widely used glioma cell lines. (B) Significantly higher expression of WNT5A in glioma compared with other cancer cell lines.
The prognostic values of WNTs in glioma
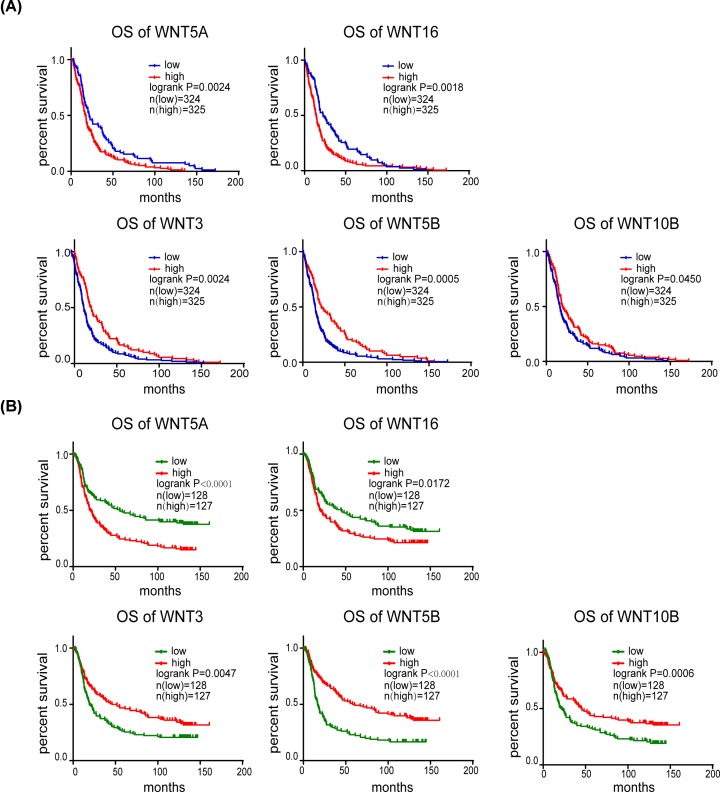
To investigate the prognostics significance of WNTs in glioma, we further analyzed mRNA expression data of WNTs with clinical data using Kaplan–Meier survival analysis. Higher mRNA levels of WNT5A and WNT16 were significantly associated with poor OS in patients with glioma (Figure 4A,B). Conversely, the increased mRNA expression levels of WNT3, WNT5B and WNT10B were correlated with better OS in patients with glioma (Figure 4A,B).
Figure 4. The prognostic value of the mRNA level of WNTs in patients with glioma analyzed with TCGA and CGGA databases.
(A) The prognostic value of the mRNA level of WNTS in patients with glioma analyzed using the TCGA database. (B) The prognostic value of the mRNA level of WNTS in patients with glioma analyzed using the CGGA database.
Clinicopathological correlation of WNTs mRNA expression in patients with glioma
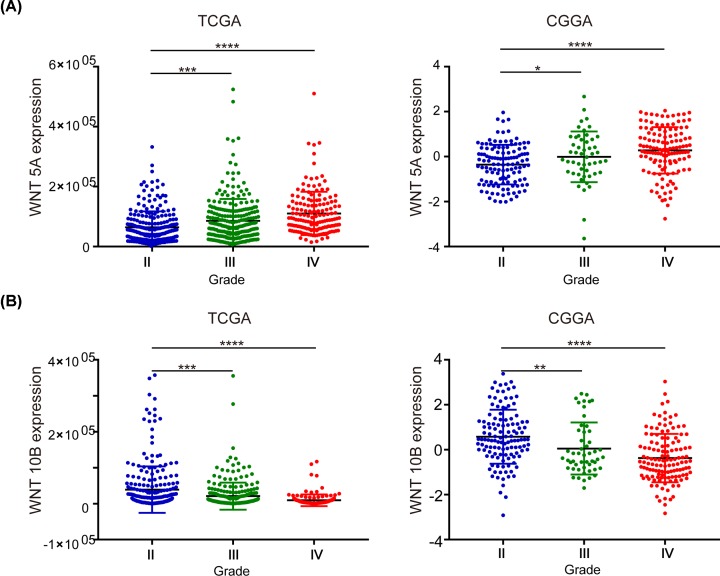
Analyzing the mRNA data of 19 WNTs from TCGA and CGGA database, we found that WNT5A mRNA expression level was significantly correlated with the grade of glioma in both TCGA and CGGA databases (Figure 5A); however, WNT10B expression was inversely correlated with the grade of glioma (Figure 5B). Furthermore, the WNTs mRNA expression in glioma of detailed histological classification compared with NB tissues was analyzed with ONCOMINE database. Consistently, higher expression of WNT5A in GBM, Anaplastic Astrocytoma and anaplastic oligodendroglioma was observed; while lower expression of WNT 10B in GBM, oligodendroglioma, astrocytoma and anaplastic astrocytoma was detected. Moreover, WNT7A was also showed lower expressed in astrocytoma than in NB tissues (Table 1).
Figure 5. Correlation between the expression levels of WNT5A and WNT10B mRNA and tumor grade in patients with glioma.
(A) Correlation between mRNA expression of WNT5A and tumor grade in patients with glioma as analyzed using CGGA and TCGA data. (B) Correlation between mRNA expression of WNT10B and tumor grade in patients with glioma as analyzed using CGGA and TCGA data (*P <0.05, **P <0.01, ***P <0.0001, ****P <0.0001).
Table 1. Significant differences in the transcription level of WNTs expression between different types of glioma and normal brain tissue using the ONCOMINE database.
| Database type | Tumor type | P-value | t-test | Fold change | |
|---|---|---|---|---|---|
| WNT5A | Sun brain | Glioblastoma vs. Normal | 3.35 ×10−23 | 12.796 | 2.577 |
| Anaplastic Astrocytoma vs. Normal | 9.86 × 10−7 | 5.963 | 2.250 | ||
| TCGA | Brain Glioblastoma vs. Normal | 2.58 × 10−14 | 24.091 | 4.336 | |
| Lee Brain | Glioblastoma vs. Normal | 1.08 × 10−9 | 24.264 | 14.423 | |
| Bredel Brain 2 | Glioblastoma vs. Normal | 6.70 × 10−8 | 8.799 | 3.768 | |
| French Brain | Anaplastic Oligodendroglioma vs. Normal | 5.16 × 10−7 | 6.287 | 3.127 | |
| WNT7A | Rickman Brain | Astrocytoma vs. Normal | 1.09 × 10−7 | -6.673 | -9.341 |
| WNT10B | Bredel Brain 2 | Glioblastoma vs. Normal | 1.03 × 10−12 | -11.601 | -6.374 |
| Sun Brain | Glioblastoma vs. Normal | 2.49 × 10−27 | -15.662 | -9.563 | |
| Oligodendroglioma vs. Normal | 1.25 × 10−16 | -10.636 | -5.914 | ||
| Diffuse Astrocytoma vs. Normal | 3.97 × 10−5 | -6.781 | -4.025 | ||
| Anaplastic Astrocytoma vs. Normal | 7.94 × 10−8 | -7.364 | -6.300 | ||
| Rickman Brain | Astrocytoma vs. Normal | 3.15 × 10−5 | -7.069 | -2.892 | |
| TCGA Brain | Brain Glioblastoma vs. Normal | 1.86 × 10−9 | -20.916 | -7.016 |
Differences in transcriptional expression were compared by Students’ t-test. Cut-off of P-value and fold-change were as follows: P-value: 0.01, fold-change: 1.5, gene rank: 10%.
Predicted functions and pathways of WNT5A and WNT10B in patients with glioma
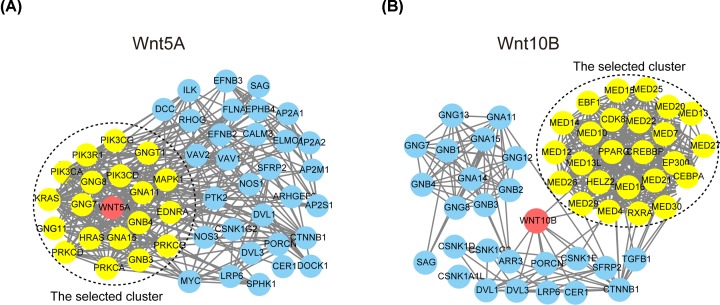
As WNT5A and WNT10B were the most correlated with glioma prognosis and clinicopathology, we analyzed the 50 most frequently altered neighbor genes of WNT5A and WNT10B in glioma (Table 2) and constructed integrated networks using String (Figure 6A,B). Then, we applied MCODE and BINGO Apps in Cytoscape for further analysis. Results showed the cluster of WNT5A was correlated with signaling pathway and the cluster of WNT10B was correlated with RNA polymerase II transcription mediator activity (Figure 6A,B).
Figure 6. The PPI network complex of WNT5A/10B and the 50 most frequently altered neighbor genes analyzed with String and Cytoscape.
(A) The Module analysis of WNT5A and the 50 most frequently altered neighbor genes. (B) The Module analysis of WNT10B and the 50 most frequently altered neighbor genes. The nodes meant proteins; the edges meant the interaction of proteins. Module analysis via Cytoscape software (degree cutoff = 2, node score cutoff = 0.2, k-core = 2, and max. Depth = 100)
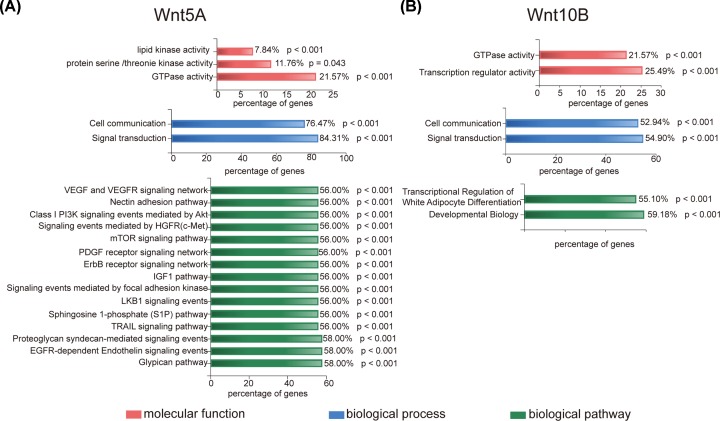
The GO enrichment analysis was then performed to determine the function of WNT5A and WNT10B with neighbor genes in glioma using DAVID. Go analysis predicted that WNT5A regulated molecular functions including GTPase activity, biological processes such as signal translation, and biological pathways such as EGFR-dependent signaling events in glioma (Figure 7A). In addition, GO analysis predicted that WNT10B regulated molecular functions including GTPase activity, biological processes such as signal translation, and biological pathways such as development biology in glioma (Figure 7B).
Figure 7. GO enrichment analysis of three main functions of WNT5A and WNT10B, including molecular function, biological process and biological pathway.
(A) GO enrichment analysis predicted three main functions of WNT5A. (B) GO enrichment analysis predicted three main functions of WNT10B.
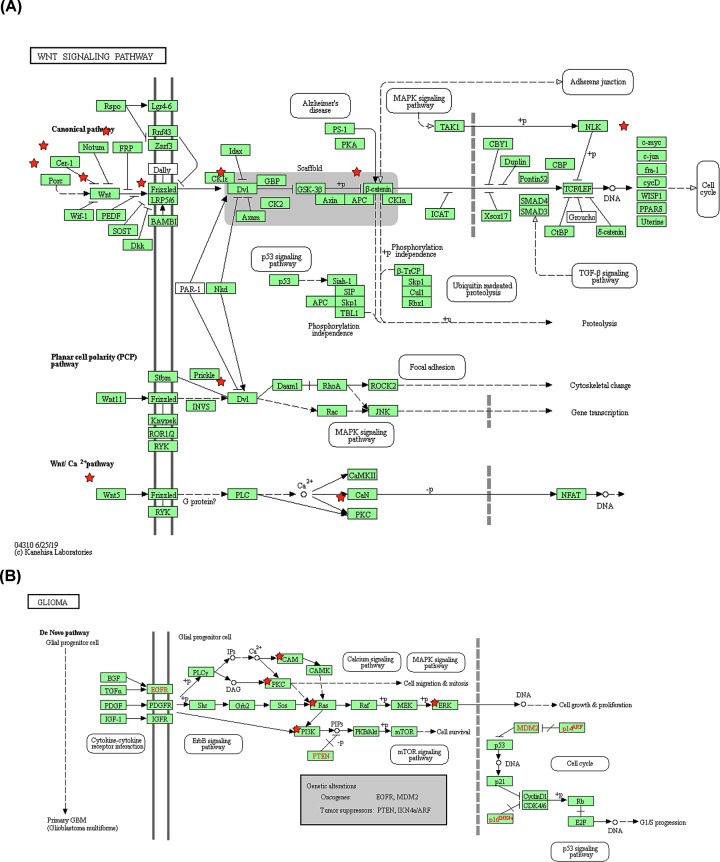
Finally, KEGG pathway enrichment analysis revealed WNT5A and neighbor genes were essential in the occurrence glioma. WNT5A played role in glioma by taking part in WNT/Ca2+pathway and the canonical pathway, however, WNT10B was only ligands of canonical pathway (Figure 8A). In the de Novo pathway of glioma occurrence, two crucial oncogenes (EGFR and MDM2) and two important tumor suppressors (PTEN and IKN4a/ARF) were found to be closely correlated with WNT5A in glioma (Figure 8B).
Figure 8. KEGG pathway enrichment analysis of WNT5A/10B and the 50 most frequently altered neighbor genes in glioma analyzed with DAVID.
(A) Three WNT signaling pathways in glioma that WNT5A and WNT10B took part in. (B) The associated pathways of WNT5A and the 50 neighbor genes in glioma occurrence and progress.
Discussion
Historically, WNTs were found to regulate many processes in embryonic development, physiology and homeostasis [33,34]. In 1982, for the first time, Nusse and Varmus described WNT-1 (Int-1) as an oncogene in mouse mammary tumors [35]. Up to now, increasing evidences suggested that defective WNT signaling is a causative factor in various cancers including breast cancer [36–38], lung cancer [39], pancreas cancer [40], colorectal cancer [41], hepatocellular carcinoma [42] and as well as glioma [6,8,14]. Despite the fact that some members of WNTs have been confirmed to play crucial roles in glioma, our study is first to investigate expression patterns, prognostic significance, and potential function of all the 19 WNT family members in glioma. We presumed that the findings of this study might contribute to the study of heterogeneity in glioma, and improve the accuracy of prognosis and treatment response and may aid in individualized therapy of patients with glioma. In our study, we found not all WNTs worked equally in glioma. WNT5A was seemed to be a crucial tumor promotor in glioma; WNT10B was seemed to a tumor suppressor in glioma; and other WNTs were not detected statistically different expression and distinct function in glioma.
For WNT5A, significantly higher mRNA expression of WNT5A in glioma was observed compared with NB. The mRNA levels of WNT5A was positively associated with the WHO grade of glioma. The survival analysis revealed that the higher mRNA expression levels of WNT5A was significantly associated with shorter OS in patients with glioma. Among the 19 WNTs, WNT5A is the most commonly studied in glioma, particularly GBM. These results are consistent with earlier studies, WNT5A has indeed been shown overexpression in GBM, and required to maintain the proliferative capability, infiltrative ability, and stem-cell-like characteristics of GBM [8,9,43]. Reis et al. reported elevated expression of WNT5A in glioma comparing with normal tissue [3]. Binda et al. also found the correlation between poor prognosis and WNT5A expression [44]. Hu et al. revealed higher WNT5A expression in recurrent GBM compared with primary GBMs, and pointed out that epigenetic activation of WNT5A triggered stem-cell like GBM invasive growth and differentiation [8]. MicroRNAs (miRNAs) including miR-30a and miR-129-5p has been documented to function as tumor suppressors by targeting WNT5A [45,46].
For WNT10B, in contrast with the results of WNT5B, significant lower mRNA expression of WNT10B in glioma was observed compared with NB. Furthermore, we verified that the mRNA expression of WNT10B was inversely proportional to the WHO grade of glioma. The survival analysis revealed that the higher mRNA expression of WNT10B was associated with better OS of patients. Till date, researches about WNT10B in glioma is few. Only Tayrac et al. reported the under-expression of WNT10B in glioblastoma [47]. According to our study, we suggested a new point that WNT10B might function as a glioma suppressor.
To understand the function and molecular mechanism of WNT5A and WNT10B in glioma, we performed PPI network, GO and KEGG analyses. First, the cluster of WNT5A was correlated with signaling pathway and the cluster of WNT10B was correlated with RNA polymerase II transcription mediator activity in PPI network. Then, GO analysis predicted that WNT5A regulated molecular functions including GTPase activity, biological processes such as signal translation, and biological pathways such as EGFR-dependent signaling events in glioma; differently, WNT10B regulated molecular functions including GTPase activity, biological processes such as signal translation, and biological pathways such as development biology in glioma. Finally, KEGG pathway analysis showed that WNT5A is primarily through the WNT/Ca+ pathway, also activates the canonical pathway [20,48–50] and WNT10B has been only reported in the canonical WNT signaling pathway [51–54]. KEGG pathway analysis also showed that the neighbor genes of WNT5A were widely distributed in multiple pathways in the de novo pathways of glioma occurrence, in which crucial oncogenes (EGFR and MDM2) and 2 important tumor suppressors (PTEN and IKN4a/ARF) were closely correlated with WNT5A.
Despite the down-regulation of WNT10B expression in glioma, the most important molecule in the classical pathway, β-catenin is up-regulated in glioma and also reported accumulation in the nucleus of glioma [55,56]. Classical WNT pathway activation may be associated with an increase of WNT5A, but there is no literature reporting WNT5A is the main factor of classic classical pathway activation in glioma. We can just guess WNT5A may cause classic pathways activated in glioma. Furthermore, it is very important to explore WNT5A promoting glioma development through the WNT/Ca+ pathway in glioma more clearly [57]. Our analysis with TCGA and CGGA data showed positive correlation between the expression levels of WNT5A and NFAT subtypes, which are important downstream transcription factors of the non-classical pathway (Supplementary Figure S1). Lastly, the interaction of WNT pathway with many other signaling pathways makes the mechanism of action of WNT pathway more complex [58,59], and our results in Figure 8B provided a direction to explore that.
In the present study, we also revealed that increased mRNA expressions of WNT16 was associated with shorter OS in patients with glioma, while the increased mRNA expressions of WNT3 and WNT5B were associated with longer OS. However, there was not enough evidence for their different expression between glioma and NB tissues.
Although our study provided important insights into the prognostic and research values of WNT5A and WNT10B in glioma patients at the mRNA level, there still are some limitations to the present study. On the one hand, all the data analyzed in our study retrieved from the online databases were RNA expression data, thus, a larger quantity of protein data such as immunohistochemistry is required to validate our findings. On the other hand, though we performed analysis to predict the mechanism and biofunction of WNT5A and WNT10B in glioma, a remarkable amount of work is urgent to investigate the distinct role of WNTs in glioma.
Conclusion
We systemically investigated the expression profiles and prognostic significance of all the 19 members of the WNT family in glioma. Our results indicated that mRNA expressions of WNT5A and WNT10B were significantly differentially regulated in glioma compared with NB tissues, and associated with pathology and grade of glioma. The study also indicated that WNT5A can serve as a candidate to diagnose and therapy glioma, while WNT10B might be valuable for anti-glioma research. Our data also provided presumed directions to explore interactions in canonical WNT pathway, non-canonical WNT pathway and multiple other signal pathways of interaction in glioma occurrence and development.
Supplementary Material
Abbreviations
- CGGA
Chinese Glioma Genome Atlas
- GBM
glioblastoma
- GEPIA
Gene Expression Profiling Interactive Analysis
- GO
gene ontology
- GTEx
Genotype‐Tissue Expression
- KEGG
Kyoto encyclopedia of genes and genomes
- LGG
low grade glioma
- NB
normal brain
- OS
over survival
- TCGA
cancer genome atlas
- WNT
Wingless / Integrated
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This study was supported by the National Nature Science Fund of China [grant number 81872064]; Outstanding Youths Development Scheme of Nanfang Hospital, Southern Medical University [grant number 2016J008]; Natural Science Fund of Tibet Autonomous Region, China [grant number XZ2017ZR-ZYZ27]. The funders had no role in study design, data collection, data analysis, decision to publish, or preparation of the manuscript.
Author Contribution
Anqi Xu and Ye Song made main contribution to concept and design. The first author Anqi Xu also made contribution to the manuscript preparation, manuscript editing and data analysis. Corresponding author Ye Song also provided funding and took part in data curation. The other authors, Huiping Yang, Kunjie Gao, Zhengming Zhan and Zibin Song made contribution to data analysis, statistical analysis and manuscript review. Tengyue Huang helped analyze and interpret data and optimize figures. All authors have read the final manuscript and approve to publish.
References
- 1.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K. et al. (2016) The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. (Berl) 131, 803–820 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 2.McCord M., Mukouyama Y.S., Gilbert M.R. and Jackson S. (2017) Targeting WNT Signaling for Multifaceted Glioblastoma Therapy. Front. Cell. Neurosc. 11, 318 10.3389/fncel.2017.00318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reis M., Czupalla C.J., Ziegler N., Devraj K., Zinke J., Seidel S. et al. (2012) Endothelial Wnt/beta-catenin signaling inhibits glioma angiogenesis and normalizes tumor blood vessels by inducing PDGF-B expression. J. Exp. Med. 209, 1611–1627 10.1084/jem.20111580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neftel C., Laffy J., Filbin M.G., Hara T., Shore M.E., Rahme G.J. et al. (2019) An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 178, 835e21–849e21 10.1016/j.cell.2019.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song Y., Luo Q., Long H., Hu Z., Que T., Zhang X. et al. (2014) Alpha-enolase as a potential cancer prognostic marker promotes cell growth, migration, and invasion in glioma. Mol. Cancer 13, 65 10.1186/1476-4598-13-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goncalves C.S., Vieira de Castro J., Pojo M., Martins E.P., Queiros S., Chautard E. et al. (2018) WNT6 is a novel oncogenic prognostic biomarker in human glioblastoma. Theranostics 8, 4805–4823 10.7150/thno.25025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griveau A., Seano G., Shelton S.J., Kupp R., Jahangiri A., Obernier K. et al. (2018) A Glial Signature and Wnt7 Signaling Regulate Glioma-Vascular Interactions and Tumor Microenvironment. Cancer Cell 33, 874e7–889e7 10.1016/j.ccell.2018.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu B., Wang Q., Wang Y.A., Hua S., Sauve C.G., Ong D. et al. (2016) Epigenetic Activation of WNT5A Drives Glioblastoma Stem Cell Differentiation and Invasive Growth. Cell 167, 1281e18–1295e18 10.1016/j.cell.2016.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J.M., Jun E.S., Jung J.S., Suh S.Y., Han J.Y., Kim J.Y. et al. (2007) Role of Wnt5a in the proliferation of human glioblastoma cells. Cancer Lett. 257, 172–181 10.1016/j.canlet.2007.07.011 [DOI] [PubMed] [Google Scholar]
- 10.Zhang B., Shen C., Ge F., Ma T. and Zhang Z. (2017) Epigenetically controlled Six3 expression regulates glioblastoma cell proliferation and invasion alongside modulating the activation levels of WNT pathway members. J. Neurooncol. 133, 509–518 10.1007/s11060-017-2476-y [DOI] [PubMed] [Google Scholar]
- 11.Kaur N., Chettiar S., Rathod S., Rath P., Muzumdar D., Shaikh M.L. et al. (2013) Wnt3a mediated activation of Wnt/beta-catenin signaling promotes tumor progression in glioblastoma. Mol. Cell. Neurosci. 54, 44–57 10.1016/j.mcn.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 12.Pu P., Zhang Z., Kang C., Jiang R., Jia Z., Wang G. et al. (2009) Downregulation of Wnt2 and beta-catenin by siRNA suppresses malignant glioma cell growth. Cancer Gene Ther. 16, 351–361 10.1038/cgt.2008.78 [DOI] [PubMed] [Google Scholar]
- 13.Kamino M., Kishida M., Kibe T., Ikoma K., Iijima M., Hirano H. et al. (2011) Wnt-5a signaling is correlated with infiltrative activity in human glioma by inducing cellular migration and MMP-2. Cancer Sci. 102, 540–548 10.1111/j.1349-7006.2010.01815.x [DOI] [PubMed] [Google Scholar]
- 14.Dijksterhuis J.P., Arthofer E., Marinescu V.D., Nelander S., Uhlen M., Ponten F. et al. (2015) High levels of WNT-5A in human glioma correlate with increased presence of tumor-associated microglia/monocytes. Exp. Cell Res. 339, 280–288 10.1016/j.yexcr.2015.10.022 [DOI] [PubMed] [Google Scholar]
- 15.Bredel M., Bredel C., Juric D., Harsh G.R., Vogel H., Recht L.D. et al. (2005) Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 65, 8679–8689 10.1158/0008-5472.CAN-05-1204 [DOI] [PubMed] [Google Scholar]
- 16.Denysenko T., Annovazzi L., Cassoni P., Melcarne A., Mellai M. and Schiffer D. (2016) WNT/beta-catenin Signaling Pathway and Downstream Modulators in Low- and High-grade Glioma. Cancer Genomics Proteomics 13, 31–45 [PubMed] [Google Scholar]
- 17.Baril M., Es-Saad S., Chatel-Chaix L., Fink K., Pham T., Raymond V.A. et al. (2013) Genome-wide RNAi screen reveals a new role of a WNT/CTNNB1 signaling pathway as negative regulator of virus-induced innate immune responses. PLoS Pathog. 9, e1003416 10.1371/journal.ppat.1003416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He W., Kang Y.S., Dai C. and Liu Y. (2011) Blockade of Wnt/beta-catenin signaling by paricalcitol ameliorates proteinuria and kidney injury. J. Am. Soc. Nephrol. 22, 90–103 10.1681/ASN.2009121236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doubravska L., Krausova M., Gradl D., Vojtechova M., Tumova L., Lukas J. et al. (2011) Fatty acid modification of Wnt1 and Wnt3a at serine is prerequisite for lipidation at cysteine and is essential for Wnt signalling. Cell. Signal. 23, 837–848 10.1016/j.cellsig.2011.01.007 [DOI] [PubMed] [Google Scholar]
- 20.Kremenevskaja N., von Wasielewski R., Rao A.S., Schofl C., Andersson T. and Brabant G. (2005) Wnt-5a has tumor suppressor activity in thyroid carcinoma. Oncogene 24, 2144–2154 10.1038/sj.onc.1208370 [DOI] [PubMed] [Google Scholar]
- 21.Nygren M.K., Dosen-Dahl G., Stubberud H., Walchli S., Munthe E. and Rian E. (2009) beta-catenin is involved in N-cadherin-dependent adhesion, but not in canonical Wnt signaling in E2A-PBX1-positive B acute lymphoblastic leukemia cells. Exp. Hematol. 37, 225–233 10.1016/j.exphem.2008.10.007 [DOI] [PubMed] [Google Scholar]
- 22.Sousa K.M., Villaescusa J.C., Cajanek L., Ondr J.K., Castelo-Branco G., Hofstra W. et al. (2010) Wnt2 regulates progenitor proliferation in the developing ventral midbrain. J. Biol. Chem. 285, 7246–7253 10.1074/jbc.M109.079822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J. and Shackleford G.M. (1996) Murine Wnt10a and Wnt10b: cloning and expression in developing limbs, face and skin of embryos and in adults. Oncogene 13, 1537–1544 [PubMed] [Google Scholar]
- 24.Eubelen M., Bostaille N., Cabochette P., Gauquier A., Tebabi P., Dumitru A.C. et al. (2018) A molecular mechanism for Wnt ligand-specific signaling. Science 361, 1126–1178 [DOI] [PubMed] [Google Scholar]
- 25.von Toerne C., Schmidt C., Adams J., Kiss E., Bedke J., Porubsky S. et al. (2009) Wnt pathway regulation in chronic renal allograft damage. Am. J. Transpl. 9, 2223–2239 10.1111/j.1600-6143.2009.02762.x [DOI] [PubMed] [Google Scholar]
- 26.Delaunay D., Cortay V., Patti D., Knoblauch K. and Dehay C. (2014) Mitotic spindle asymmetry: a Wnt/PCP-regulated mechanism generating asymmetrical division in cortical precursors. Cell Rep. 6, 400–414 10.1016/j.celrep.2013.12.026 [DOI] [PubMed] [Google Scholar]
- 27.Wang S.H., Chang J.S., Hsiao J.R., Yen Y.C., Jiang S.S., Liu S.H. et al. (2017) Tumour cell-derived WNT5B modulates in vitro lymphangiogenesis via induction of partial endothelial-mesenchymal transition of lymphatic endothelial cells. Oncogene 36, 1503–1515 10.1038/onc.2016.317 [DOI] [PubMed] [Google Scholar]
- 28.Ono M., Yin P., Navarro A., Moravek M.B., Coon J.S.T., Druschitz S.A. et al. (2013) Paracrine activation of WNT/beta-catenin pathway in uterine leiomyoma stem cells promotes tumor growth. PNAS 110, 17053–17058 10.1073/pnas.1313650110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitoh T., Mine T. and Katoh M. (2002) Up-regulation of WNT8B mRNA in human gastric cancer. Int. J. Oncol. 20, 343–348 [PubMed] [Google Scholar]
- 30.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D. et al. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia (New York, NY) 6, 1–6 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III et al. (2019) Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 10.1038/s41586-019-1186-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Z., Li C., Kang B., Gao G., Li C. and Zhang Z. (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mihara E., Hirai H., Yamamoto H., Tamura-Kawakami K., Matano M., Kikuchi A. et al. (2016) Active and water-soluble form of lipidated Wnt protein is maintained by a serum glycoprotein afamin/alpha-albumin. eLife. 5, 7554–11621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Won J.Y., Jang W.Y., Lee H.R., Park S.Y., Kim W.Y., Park J.H. et al. (2017) Novel missense loss-of-function mutations of WNT1 in an autosomal recessive Osteogenesis imperfecta patient. Eur. J. Med. Genet. 60, 411–415 10.1016/j.ejmg.2017.05.002 [DOI] [PubMed] [Google Scholar]
- 35.Nusse R. and Varmus H. (2012) Three decades of Wnts: a personal perspective on how a scientific field developed. EMBO J. 31, 2670–2684 10.1038/emboj.2012.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luga V., Zhang L., Viloria-Petit A.M., Ogunjimi A.A., Inanlou M.R., Chiu E. et al. (2012) Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 151, 1542–1556 10.1016/j.cell.2012.11.024 [DOI] [PubMed] [Google Scholar]
- 37.Wend P., Runke S., Wend K., Anchondo B., Yesayan M., Jardon M. et al. (2013) WNT10B/beta-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO Mol. Med. 5, 264–279 10.1002/emmm.201201320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pohl S.G., Brook N., Agostino M., Arfuso F., Kumar A.P. and Dharmarajan A. (2017) Wnt signaling in triple-negative breast cancer. Oncogenesis 6, e310 10.1038/oncsis.2017.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tennis M., Van Scoyk M. and Winn R.A. (2007) Role of the wnt signaling pathway and lung cancer. J. Thoracic Oncol. Intern. Assoc. Study Lung Cancer 2, 889–892 10.1097/JTO.0b013e318153fdb1 [DOI] [PubMed] [Google Scholar]
- 40.Hou P., Ma X., Zhang Q., Wu C.J., Liao W., Li J. et al. (2019) USP21 deubiquitinase promotes pancreas cancer cell stemness via Wnt pathway activation. Genes Dev. 33, 1361–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q., Lai Q., He C., Fang Y., Yan Q., Zhang Y. et al. (2019) RUNX1 promotes tumour metastasis by activating the Wnt/beta-catenin signalling pathway and EMT in colorectal cancer. J. Exper. Clin. Cancer Res. 38, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui Y., Wu X., Lin C., Zhang X., Ye L., Ren L. et al. (2019) AKIP1 promotes early recurrence of hepatocellular carcinoma through activating the Wnt/beta-catenin/CBP signaling pathway. Oncogene 38, 5516–5529 10.1038/s41388-019-0807-5 [DOI] [PubMed] [Google Scholar]
- 43.Pulvirenti T., Van Der Heijden M., Droms L.A., Huse J.T., Tabar V. and Hall A. (2011) Dishevelled 2 signaling promotes self-renewal and tumorigenicity in human gliomas. Cancer Res. 71, 7280–7290 10.1158/0008-5472.CAN-11-1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Binda E., Visioli A., Giani F., Trivieri N., Palumbo O., Restelli S. et al. (2017) Wnt5a Drives an Invasive Phenotype in Human Glioblastoma Stem-like Cells. Cancer Res. 77, 996–1007 10.1158/0008-5472.CAN-16-1693 [DOI] [PubMed] [Google Scholar]
- 45.Zeng A., Yin J., Li Y., Li R., Wang Z., Zhou X. et al. (2018) miR-129-5p targets Wnt5a to block PKC/ERK/NF-kappaB and JNK pathways in glioblastoma. Cell Death. Dis. 9, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y., Wu Z., Li L. and Xie M. (2017) miR-30a inhibits glioma progression and stem celllike properties by repression of Wnt5a. Oncol. Rep. 38, 1156–1162 10.3892/or.2017.5728 [DOI] [PubMed] [Google Scholar]
- 47.de Tayrac M., Etcheverry A., Aubry M., Saikali S., Hamlat A., Quillien V. et al. (2009) Integrative genome-wide analysis reveals a robust genomic glioblastoma signature associated with copy number driving changes in gene expression. Genes Chromosomes Cancer 48, 55–68 [DOI] [PubMed] [Google Scholar]
- 48.Asem M.S., Buechler S., Wates R.B., Miller D.L. and Stack M.S.. Wnt5a Signaling in Cancer. LID - 10.3390/cancers8090079 [doi] LID - E79 [pii]. (2072-6694 (Print)) [Google Scholar]
- 49.Kuhl M., Sheldahl L.C., Malbon C.C. and Moon R.T. (2000) Ca(2+)/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J. Biol. Chem. 275, 12701–12711 10.1074/jbc.275.17.12701 [DOI] [PubMed] [Google Scholar]
- 50.Bauer M., Benard J., Gaasterland T., Willert K. and Cappellen D. (2013) WNT5A encodes two isoforms with distinct functions in cancers. PLoS One 8, e80526 10.1371/journal.pone.0080526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wend P., Wend K., Krum S.A. and Miranda-Carboni G.A. (2012) The role of WNT10B in physiology and disease. Acta Physiologica 204, 34–51 10.1111/j.1748-1716.2011.02296.x [DOI] [PubMed] [Google Scholar]
- 52.Lee J.G. and Heur M. (2015) WNT10B enhances proliferation through beta-catenin and RAC1 GTPase in human corneal endothelial cells. J. Biol. Chem. 290, 26752–26764 10.1074/jbc.M115.677245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshikawa H., Matsubara K., Zhou X., Okamura S., Kubo T., Murase Y. et al. (2007) WNT10B functional dualism: beta-catenin/Tcf-dependent growth promotion or independent suppression with deregulated expression in cancer. Mol. Biol. Cell 18, 4292–4303 10.1091/mbc.e06-10-0889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou J., Gao Y.H., Zhu B.Y., Shao J.L., Ma H.P., Xian C.J. et al. (2019) Sinusoidal Electromagnetic Fields Increase Peak Bone Mass in Rats by Activating Wnt10b/beta-Catenin in Primary Cilia of Osteoblasts. J. Bone Mineral Res. 34, 1336–1351 10.1002/jbmr.3704 [DOI] [PubMed] [Google Scholar]
- 55.Zhang J., Cai H., Sun L., Zhan P., Chen M., Zhang F. et al. (2018) LGR5, a novel functional glioma stem cell marker, promotes EMT by activating the Wnt/beta-catenin pathway and predicts poor survival of glioma patients. J. Exp. Clinical Cancer Res. 37, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siebert G.K. (1986) The fixed torsion bridge. Phillip J. Restaur. Zahnmed. 3, 321–323 [PubMed] [Google Scholar]
- 57.Ahsan R., Baisiwala S. and Ahmed A.U. (2017) Rogue one: another faction of the Wnt empire implicated in assisting GBM progression. Translational Cancer Res. 6, S321–S327 10.21037/tcr.2017.03.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nusse R. and Clevers H. (2017) Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 169, 985–999 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 59.Semenov M.V., Habas R., Macdonald B.T. and He X. (2007) SnapShot: Noncanonical Wnt Signaling Pathways. Cell 131, 1378 10.1016/j.cell.2007.12.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.