Conspectus
Metal ions can be beneficial or toxic, depending on their identity, oxidation state, and concentration. Therefore, being able to detect and quantify different types of metals using portable sensors or in situ imaging agents is important for better environmental monitoring, medical in vitro diagnostics, and imaging of biological systems. While numerous metal ions of different oxidation states are present in the environment and biological systems, only a limited number of them can be detected effectively using current methods. In this account, we summarize research results from the authors’ group that overcome this limitation by developing a novel class of activity-based sensors based on metal-dependent DNAzymes, which are DNA molecules with enzymatic activity. First, we have developed an in vitro selection method to obtain DNAzymes from a large DNA library of up to 1015 sequences that can carry out cleavage of an oligonucleotide substrate only in the presence of a specific metal ion with high selectivity. Negative selection steps can further be used to improve the selectivity against potentially competing targets by removing sequences that recognize the competing metal ions. Second, we have developed a patented catalytic beacon method to transform the metal-dependent DNAzyme cleavage reaction into a turn-on fluorescent signal by attaching a fluorophore and quenchers to the DNAzyme complex. Taking advantage of the difference in melting temperatures of DNA hybridization before and after metal ion-dependent cleavage of the DNAzyme substrate, the fluorophore on the DNA cleavage product can be released from its quenchers to create a turn-on fluorescent signal. Because DNAzymes are easy to conjugate with other signaling moieties, such as gold nanoparticles, lanthanide-doped upconversion nanoparticles, electrochemical agents, and gadolinium complexes, these DNAzymes can also readily be converted into colorimetric sensors, upconversion luminescence sensors, electrochemical sensors, or magnetic resonance contrast agents. In addition to describing recent progress in developing and applying these metal ion sensors for environmental monitoring, point-of-care diagnostics, cellular imaging, and in vivo imaging in zebrafish, we have summarized major advantages of this class of activity-based sensors. In addition to advantages common to most activity-based sensors, such as enzymatic turnovers that allow for signal amplification and using initial rates instead of absolute signals for quantification to avoid interferences from sample matrices, the DNAzyme-based sensors allow for in vitro selection to expand the method to almost any metal ion under a variety of conditions, negative selection to improve selectivity against competing targets, and re-selection of DNAzymes and combination of active and inactive variants to fine-tune the dynamic range of detection. The use of melting temperature differences to separate target binding from signaling moieties in the catalytic beacon method allows using different fluorophores and nanomaterials to extend the versatility and modularity of this sensing platform. Furthermore, sensing and imaging artifacts can be minimized by using an inactive mutant DNAzyme as a negative control, while spatiotemporal control of sensing/imaging can be achieved using optical, photothermal, and endogenous orthogonal caging methods. Finally, current challenges, opportunities, and future perspectives for DNAzymes as activity-based sensors are also discussed.
Graphical Abstract
1. Introduction
Activity-based sensors are an important class of probes in bioanalytical chemistry, in which the signal readout is produced by a chemical reaction catalyzed by the sensor only in the presence of the analyte, instead of relying on a signal readout based on analyte binding through the traditional lock-and-key mode of molecular recognition.1 A major class of activity-based sensors can trigger reactions with multiple turnovers from binding of a single target, which can produce significant signal amplification. Because of this amplification, some targets that may be too low in concentration to be detectable using binding-based sensors can be detected with these multi-turnover reactivity-based sensors. These multi-turnover sensors are found in both biological systems and commercial products. For instance, many biological events, such as protease activity, are sensed by multi-turnover enzymatic reactions,2 while one of the most commercially successful sensors is glucose detection using a glucose meter, whose core technology relies on enzymatic activity such as glucose oxidase. Furthermore, another major advantage of activity-based sensors is that there are in general two types of signal readings that can be taken to correlate with analyte concentration: a single reading at a determined “end timepoint” or a reaction rate calculated from multiple readings at timepoint intervals. While a single endpoint reading is more convenient and often sufficient, calculation of a reaction rate based on previously established reaction kinetics3 is often more reliable and less affected by fluctuations or other signal interferences that can be present in biological systems, such as differences in pH, temperature, and sensor loading. This mode of detection can be especially useful for targets that may interfere with the signal transduction, such as paramagnetic ions which can have high fluorescence quenching effects. Based on these advantages, activity-based sensors can be effective in many sensing applications.
While many activity-based sensors use small molecules or proteins, DNAzymes, also called deoxyribozymes, have emerged as a new class of activity-based sensors,4–6 as they are DNA molecules capable of catalyzing a specific reaction. Although there are no known naturally occurring DNAzymes, synthetic DNAzymes were first identified in the lab nearly thirty years ago to catalyze the cleavage of a DNA substrate strand at a single ribonucleotide incorporated in the middle of the strand.7 Since then, DNAzymes have been identified to catalyze many other reactions, such as DNA ligation and porphyrin metalation;8,9 however, since almost all DNAzyme-based sensors developed so far have been based on RNA cleavage, we will focus our discussion on these RNA-cleaving DNAzymes.
DNAzymes are typically discovered in the lab through a process called in vitro selection, which starts with an initial pool of 1014-1015 oligonucleotides carrying different random sequences and employs a selection strategy to separate out oligonucleotides with a desired function (Figure 1A).4–6,9,10 In the case of RNA cleavage, one strategy uses solid supports, such as magnetic or agarose beads, onto which the DNA library is immobilized, to collect the active DNAzymes that have been cleaved and detached from the solid phase support. Another method uses gel electrophoresis to separate the active DNAzymes, which have undergone cleavage and so are shorter in length, from the longer inactive DNAzymes which have not undergone cleavage. To obtain active sequences, multiple rounds of selection are carried out with amplification steps in between using Polymerase Chain Reaction (PCR) to regenerate the pool. Over these selection rounds, the selection pool gradually increases in the proportion of active DNAzyme sequences. When the pool reaches a high enough activity for the desired function, it can be sequenced to identify the best individual oligonucleotide sequences that carry out the function.
Figure 1. In Vitro Selection of Metal-Specific DNAzymes as Activity-Based Sensors.
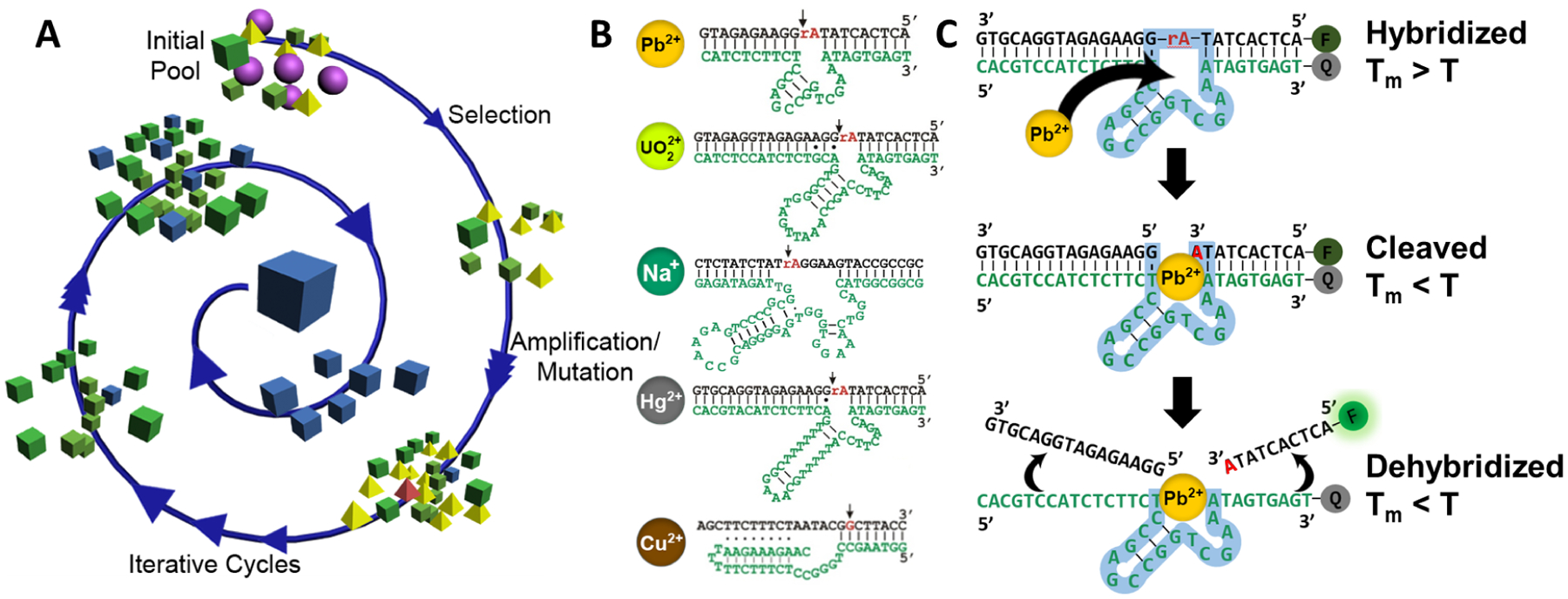
A) DNAzymes are obtained through in vitro selection in which a pool of 1014-1015 random DNA sequences is subjected to increasing selection pressure to select out sequences with the desired function over multiple selection rounds. This process can obtain DNAzymes that catalyze a reaction only in the presence of a specific target with high activity and selectivity. B) Predicted secondary structure of metal-dependent DNAzymes specific for Pb2+, UO22+, Na+, Hg2+, and Cu2+. C) Schematic of the catalytic beacon sensing strategy to produce a fluorescent turn-on signal in the presence of the metal ion target based on cleavage and subsequent dehybridization of a fluorophore-labeled substrate strand. The catalytic loop sequences are highlighted with light blue shadow.
2. DNAzymes as Activity-Based Sensors
Given the limited number of functional groups that are available for DNAzymes composed of the four canonical nucleotides, their catalytic activities generally require metal ions as cofactors. Therefore, we have adopted the in vitro selection as a way to obtain DNAzymes with selectivity toward different metal ions by performing the selection in the presence of a metal ion of interest. Furthermore, DNAzymes with improved performance can be obtained by increasing the stringency of the selection, such as using shorter reaction times and lower metal concentrations in the later rounds of selection (Figure 1A).4–6 Once these metal-dependent RNA-cleaving DNAzymes are obtained (Figure 1B), they can be converted into activity-based sensors for these metal ions using the “catalytic beacon” approach (Figure 1C), which was pioneered by our group. The approach is based on the difference of melting temperature (Tm) of the annealing, or hybridization, of the enzyme strand to the substrate strand, before and after the metal-dependent cleavage of the substrate strand by the DNAzyme. Using the fluorescent 8–17 DNAzyme sensor for Zn2+ and Pb2+ detection as an example (Figure 1C), the Tm between the substrate and enzyme strands of this DNAzyme sensor is higher than the ambient temperature (T), keeping the two strands hybridized and the fluorophore and quenchers in close proximity for effective quenching. In the presence of the target, Pb2+, the enzyme strand catalyzes the cleavage of the substrate strand to generate two DNA fragments, each of which has a much lower Tm than the full-length substrate had to the enzyme strand. This Tm change results in dehybridization of the fluorophore-labeled fragment from the DNAzyme duplex. The increased distance between fluorophore and quencher results in a fluorescence turn-on signal. To minimize background fluorescence from incomplete hybridization, we further improved upon the original catalytic beacon system by placing a second quencher on the other end of the substrate strand, resulting in >10-fold improved signal-to-noise ratio.11
Almost all DNAzymes share a similar secondary structure with two binding arms surrounding the cleavage site (Figure 1B), which allow for binding to the substrate sequence and are used as PCR primers during the selection, and it has been shown that the sequence identity or length of the arms are not involved in metal-binding or activity. Because of this property, a DNAzyme can be adapted to work at various detection temperatures and conditions by simply changing the binding arm sequencing, as demonstrated by the creation of a Pb2+-dependent DNAzyme sensor that is invariant to temperature from 4°C – 30°C.12 Furthermore, the catalytic beacon design can be amended to produce any type of sensor that can generate a signal based on a distance-dependent two-component readout strategy, such as for fluorescent, colorimetric, magnetic resonance, and photoacoustic sensors.
2.1. DNAzymes as Activity-Based Sensors for Environmental Monitoring
The increasing attention to environmental pollution has initiated the development of sensors for rapid, on-site, and real-time environmental monitoring of pollutants, especially heavy metal ions.13 However, the detection of a specific heavy metal within the complex matrix of environmental samples can remain challenging, especially when coupled with the desire to use portable, on-site devices in real-time. Many heavy metal detection methods, such as Inductively Coupled Plasma Mass Spectrometry (ICP-MS), must be used off-site and so can take days to obtain the results. To address these issues, a new generation of environmental sensors must be designed with high sensitivity and selectivity for specific metal ions and with signal outputs that can be read with small, portable instruments, such as a fluorometer or colorimeter. While many sensors based on other types of molecules have been reported, very few of them have been shown to be effective toward sensing heavy metal ions under real-world conditions. To address this limitation, we have designed DNAzymes fluorescent sensors that can be detected using a portable fluorimeter using the catalytic beacon approach described in Section 2 (Figure 1c). The invention of catalytic beacons is a milestone in the development of DNAzyme activity-based sensors for environmental monitoring of many different metal ions, including Pb2+,3 Cu2+,14,15 UO2+,16 Hg2+,17 Ag+,18 Ca2+,19 Tl3+,20 Cd2+,21 Cr3+,22 and Cr4+,22 with impressive sensitivity and selectivity (Table 1, Figure 1b). Some of these sensors have also been commercialized by ANDalyze Inc. for the detection of heavy metal ions using an integrated device that consists of consumable fluorescent sensor cartridges and a hand-held fluorimeter. More importantly, these products have successfully received US EPA ETV validation in 2014, and are currently being used worldwide, including in a school district in St Paul, MN,23 for the on-site detection of lead in drinking water. Furthermore, fluorescent DNAzyme sensors have also been attached to gold surfaces24 and within gold-coated nanocapillary array membranes,25 which allow for sensor regeneration and long-term dry storage, as well for potential future applications within microfluidic devices.
Table 1.
DNAzyme-Based Activity Sensors for Metal Ions using Catalytic Beacons
| Metal ions | Detection Range | LOD | Selectivity | EPA value | Ref. |
|---|---|---|---|---|---|
| Pb2+ | 10 nM – 4 μM | 1.0 nM | >80 fold | 75 nM | 3 |
| Cu2+ | 35 nM – 20 μM | 35 nM | >2,000 fold | 20 μM | 14 |
| 5 nM – 200 nM | 1.6 nM | N. A. | 15 | ||
| UO22+ | 0 –2 μM | 45 pM | >1,000,000 fold | 126 nM | 16 |
| Hg2+ | 0 – 500 nM | 2.4 nM | >100,000 fold | 10 nM | 17 |
| Ag+ | 0 – 500 nM | 21.8 nM | >3,000 fold | 3.2 μg/L | 18 |
| Ca2+ | 0 – 500 nM | 17 μM | >6,000 fold | N. A. | 19 |
| Tl3+ | 0 – 500 nM | 1.5 nM | >10,000 fold | 10 nM | 20 |
| Cd2+ | 0 – 500 nM | 1.1 nM | >100,000 fold | 45 nM | 21 |
| Cr3+ | 0 – 20 μM | 70 nM | >100 fold | 11 μM | 22 |
| Cr4+ | 0 – 10 μM | 140 nM | N. A. | 300 nM | 22 |
N.A. = Not Applicable
While portable fluorimeters provide very accurate results, they are relatively expensive, making them inefficient for public use at home. To make the DNAzyme sensors more accessible, we have also developed colorimetric sensors based on the color change produced from aggregation and disaggregation of gold nanoparticles (AuNPs, Figure 2b). A DNA substrate containing the RNA cleavage site is hybridized to DNA strands conjugated to the AuNPs, serving as a linker to bring individual AuNPs together in an aggregated state, resulting in a blue color, due to surface plasmon effects of AuNPs. In the presence of a metal ion, the DNAzyme will cleave the DNA substrate and thus the linkage between the AuNPs, allowing for disaggregation of the AuNPs, resulting in a red color. In addition to the above labeled method, a label-free colorimetric sensor with even higher sensitivity has also been demonstrated (Figure 2d).26,27 To make the on-site detection even easier, we have developed an easy-to-use dipstick test using non-cross-linked AuNP-DNAzyme conjugates, which allows for one-step colorimetric detection of Pb2+ extracted from paints,28 providing a promising approach for environmental monitoring at home or in low-resource settings (Figure 2e). In addition to the aforementioned sensing formats, a smart MRI contrast agent with switchable relaxivity in response to metal ions was developed based on conjugation of a DNAzyme with a gadolinium complex and streptavidin (Figure 2g). The binding of a metal ion to its DNAzyme could cleave and dissociate the Gd(III)-DOTA-labeled substrate strand from the large streptavidin labeled enzyme strand, resulting in a decrease of the T1 correlation time for a detectable change of MRI signal.29 Furthermore, the Plaxco group has developed DNAzyme-based electrochemical sensors by utilizing methylene blue (MB)-modified DNAzymes covalently attached to a gold surface (Figure 2c). Before cleavage, the relatively rigid DNAzyme structure prevents close contact of MB to the gold surface, however following cleavage, the MB-modified DNAzyme retains a more flexible, mostly single-stranded structure which allows for close contact of MB with the gold surface to transfer electrons from MB to the surface, producing the electrochemical signal.30
Figure 2. Timeline of In Vitro DNAzyme Sensors for Environmental Monitoring and Medical Diagnostics.
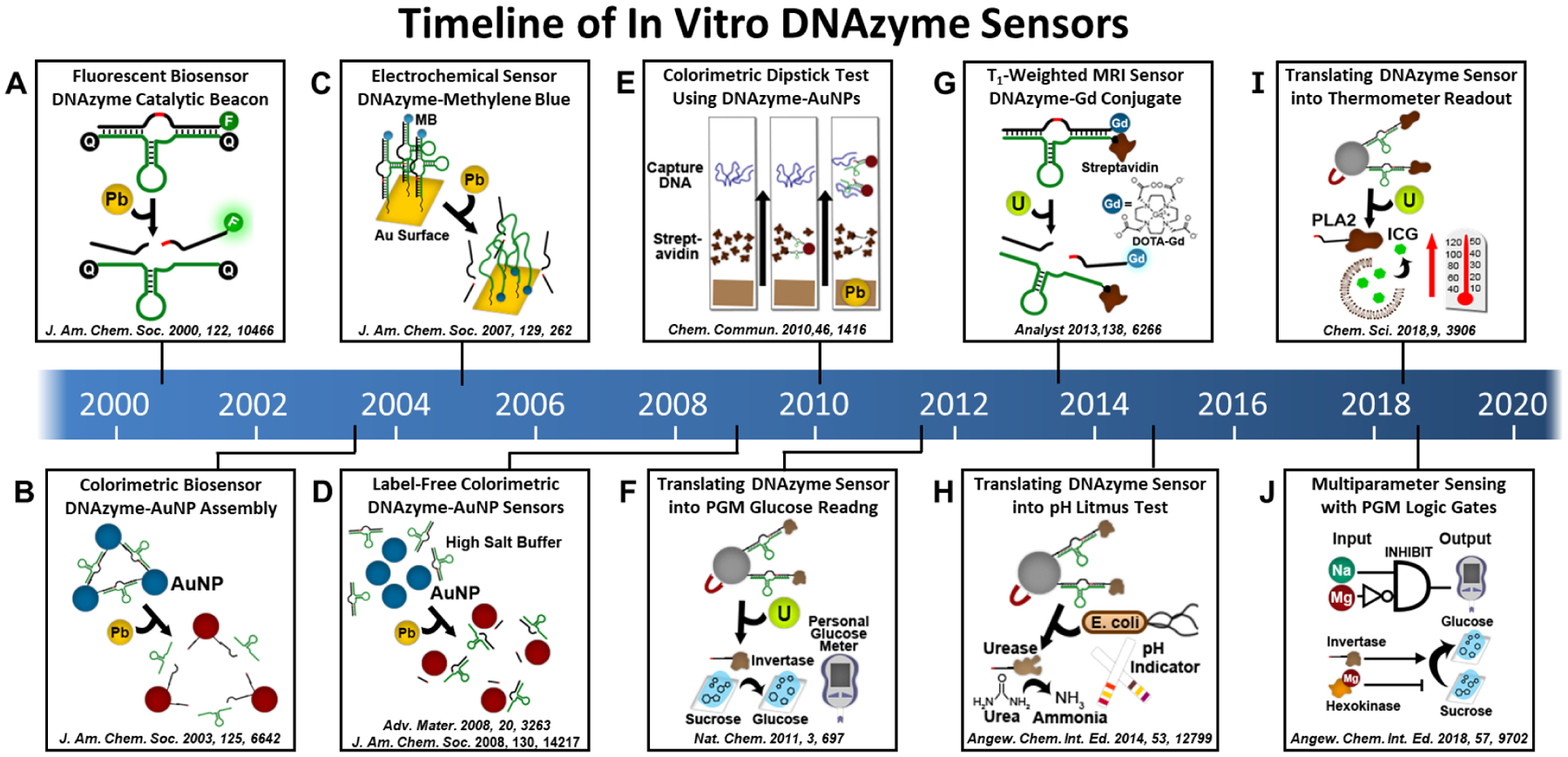
Timeline of the major milestones for the use of metal-dependent DNAzymes as activity-based sensors with many different sensing modalities, including fluorescent, colorimetric, electrochemical, and magnetic resonance imaging (MRI) sensors. DNAzyme activity-based sensors have also been applied to produce signals that can be detected by commercial point-of-care devices, such as a personal glucose meter, a pH meter, and a thermometer.
2.2. DNAzymes as Activity-Based Sensors for In Vitro Medical Diagnostics
In addition to environmental monitoring, DNAzyme activity-based sensors have also been applied for in vitro medical diagnostics. Among these sensors, point-of-care (POC) sensors that allow rapid, on-site, and low-cost detection are highly desirable. While a number of POC devices have been reported, including those attached to smartphones, very few are commercially available, because most of them are bulky, unfamiliar to use, and take millions of dollars to translate results in the academic labs into commercial products due to costs of scale-up manufacturing. To overcome these limitations, we and others have taken advantage of existing widely available meters, including personal glucose meters (PGMs),31–33 pH meters,34 and thermometers,35 and adapted them to measure a wide range of targets. By using small portable meters whose assay formats are familiar to millions of users around the world, including FDA-approved PGMs that attach to smartphones, our technology makes it easier for the market to adopt the sensors. Furthermore, using existing meters for detection can minimize the developmental costs of scale-up manufacturing for new meters. A major challenge in this endeavor is to translate target recognition events into a readily measurable signal without modifying these meters. To meet this challenge, we are the first to demonstrate the integration of DNAzyme-based target recognition with protein enzymes such as invertase, which can catalyze the specific reactions required to generate a change of glucose concentration that is detectable with a PGM. For instance, by linking a DNA-invertase conjugate with the substrate (39S) of the UO22+-dependent DNAzyme (39E), we have demonstrated a new sensor system in which the presence of UO22+ can be coupled to generation of glucose detectable with a PGM (Figure 2f).31 To facilitate portable multiparameter sensing in complex biological samples, we further demonstrated a new biocomputing platform that integrates PGMs with logic capability using metal-specific DNAzymes and protein enzymes as building blocks (Figure 2j).33 A YES gate for sodium ions using a Na+-specific DNAzyme (NaA43E) and its substrate (NaA43S) was first developed and applied for early evaluation of clinical parameters of hyponatremia and hypernatremia by binary reading. In addition, the Boolean INHIBIT logic gate for Na+ and Mg2+ was constructed by combining a Na+-specific DNAzyme with Mg2+-dependent hexokinase, using glucose as a signal readout, which showed potential application for the clinical analysis of hypermagnesemia. Additionally, a pH signal which can be measured with pH litmus paper has been developed by Yingfu Li’s lab for detection of bacteria using urease, which converts urea to ammonia, in conjunction with their bacteria-dependent DNAzymes (Figure 2h).34 In addition to these glucose and pH readouts, we have further taken advantage of the excellent photothermal properties of indocyanine green (ICG)-encapsulated liposomes and target-responsive DNAzymes conjugated to phospholipase A2 (PLA2) to provide a quantitative link between the concentration of a target and the temperature readout of a thermometer for portable sensing of UO22+ (Figure 2i).35 This sensor is based on the target-induced release of DNA-PLA2 conjugates from a DNAzyme-modified magnetic bead, which catalyzes the hydrolysis of liposomes to release the ICG dye and subsequently generates a temperature increase in solution via NIR-laser irradiation.
2.3. DNAzymes as Activity-Based Sensors for Cellular Imaging
Because of their high biocompatibility and ability to detect biologically relevant metal ions in complex matrices in vitro, DNAzymes have also been used in cellular metal ion imaging applications as activity-based fluorescent sensors using the catalytic beacon approach (Figure 3).36,37 The first intracellular application of DNAzymes as sensors was reported by the author’s lab by conjugating the uranyl-specific 39E DNAzyme with AuNPs, which enabled delivery into cultured cells by endocytosis with localization in lysosomes (Figure 3a).38 The AuNPs provide additional beneficial effects by protecting the DNAzyme ends to slow down their exonuclease-based degradation in cells and by acting as a highly efficient quencher for the fluorophore before metal-dependent sensor activation and release of the fluorophore from the AuNP. Following this work, the intracellular detection of other types of metal ions has been reported using different metal-selective DNAzymes, including for Zn2+,39 Na+,40 Cu2+,41 Mg2+,42 and Pb2+.43 Additionally, other delivery methods have been used for delivery to other subcellular locations, including Lipofectamine,39 cationic polypeptides,40 and membrane insertion,42 as reviewed elsewhere.44 Another major advancement of this field has been the development of photocaging groups, which allow the activity-based DNAzyme sensors to be “caged” or inactivated to avoid unwanted sensor activation before imaging (Figure 3b, 3d, 3f, 3h).39 The sensors can then be activated with spatiotemporal control, such as with 365 nm light irradiation, as described in more detail in Section 3.7.
Figure 3. Timeline of Intracellular and In Vivo DNAzyme Sensors.
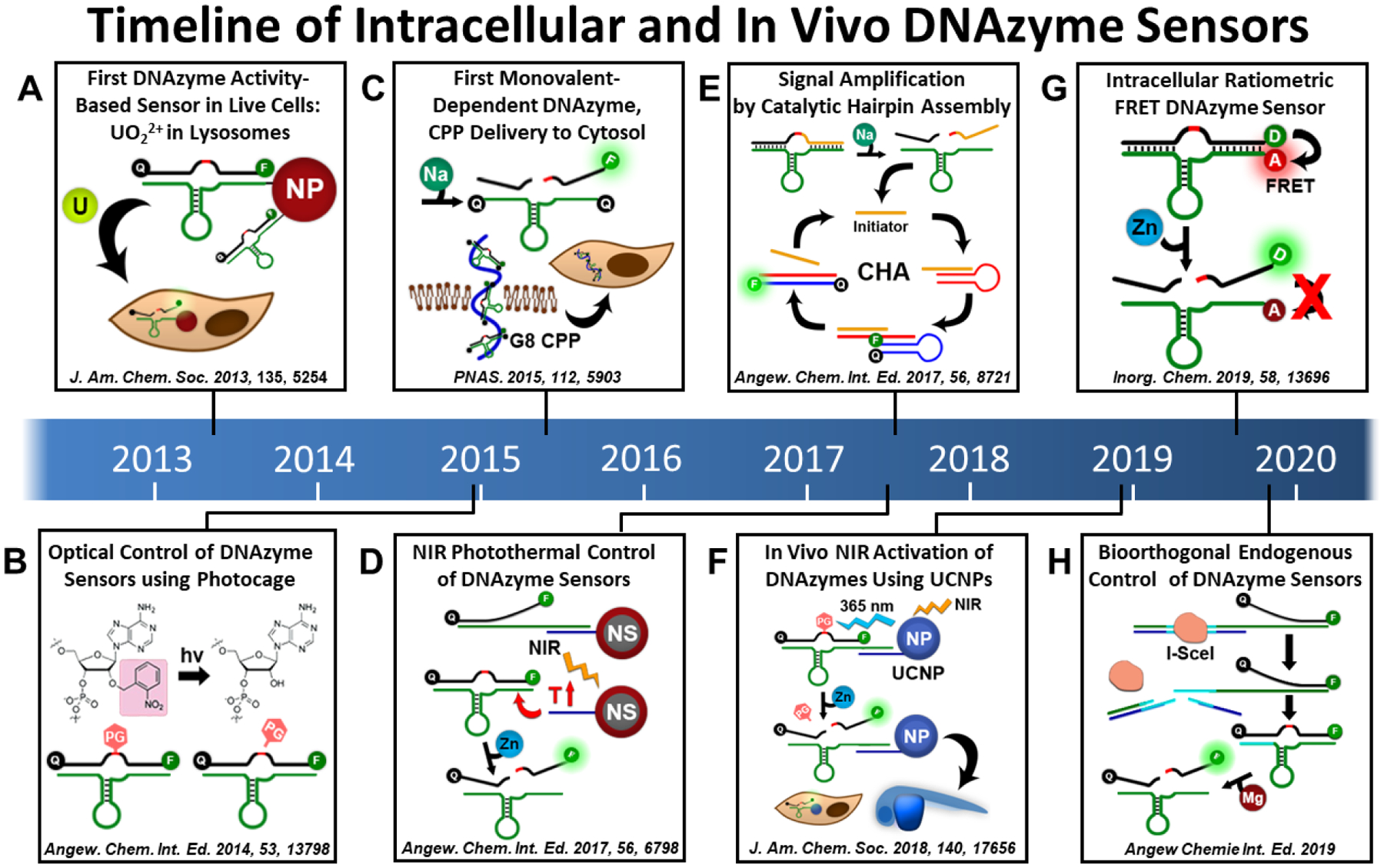
Timeline of the major milestones for use of metal-dependent DNAzyme fluorescent sensors for live cell imaging and in vivo imaging in zebrafish. These advancements include the use of different transfection agents for different subcellular delivery of the sensors, different photocaging strategies to activate the DNAzymes only after delivery with spatiotemporal control, amplification strategies to improve sensitivity for the detection of endogenous metal ion concentrations, and delivery into zebrafish embryos for in vivo sensing of Zn2+ concentrations.
2.4. DNAzymes as Activity-Based Sensors for In Vivo Imaging in Zebrafish
Recently, we have also applied DNAzyme sensors for imaging in live zebrafish by conjugating photocaged DNAzymes onto lanthanide-doped upconversion nanoparticles (UCNPs, Figure 3f).45 These UCNPs can be excited with deep tissue penetrating NIR 980 nm light to emit light of much lower wavelength. These UCNPs can thus be used as a decaging agent to produce the 365 nm light required by the previously developed photocaging methods, which are crucial for the application of DNAzyme sensors in living systems. Using this strategy, we developed a platform that can be used to image the Zn2+ distribution with spatiotemporal control, thereby giving insights into the dynamic Zn2+ ion distributions in intracellular and in vivo zebrafish embryo models.
3. Advantages of DNAzymes as Activity-Based Sensors
In addition to the general advantages of activity-based sensors as described in the Introduction, DNAzymes have several key advantages over other types of activity-based sensors, which are mostly small molecule- or protein-based. The main advantage of DNAzyme-based sensors comes from their similar secondary structures and reaction mechanisms, which makes them a relatively generalized platform such that techniques developed for one DNAzyme sensor can usually be readily applied to most other DNAzyme sensors, as described in more detail in Section 3.5. As DNAzyme sensors consist mainly of nucleic acids, they can make use of delivery methods developed for other nucleic acid-based systems. DNAzyme sensors are also relatively stable both in vitro and within biological systems, especially in comparison to protein-based sensors, and their stability can be further enhanced by various commercialized DNA modification methods (see Section 4.1). Furthermore, it should be noted that though there are some reported sensors based on ribozymes, which will share many of the same properties and advantages of DNAzyme-based sensors, because RNA is inherently less stable than DNA, their application as sensors, especially in biological systems, is limited based on their relatively rapid degradation through cellular pathways or self-hydrolysis through their 2′-OH. Applications of ribozymes have been mostly limited to detection and degradation of specific RNA sequences, rather than detection of non-nucleic acid targets like metal ions. Below, we outline in detail additional advantages specific to DNAzyme-based sensors.
3.1. Using In Vitro Selection to Obtain DNAzymes for a Broad Range of Metal Ions and Conditions
As mentioned briefly in Section 2, a major advantage of DNAzyme-based sensors is the ability to obtain DNAzymes that are selective for almost any metal ion by carrying out in vitro selection in the presence of the metal ion of interest. Since there are numerous metal ions in the periodic table and many of these metal ions have different oxidation states, it is extremely challenging to use rational design to create a receptor that recognizes only one target and not the others, because the difference between different targets is often quite subtle. This difficulty in using rational design to obtain metal ion sensors contributes to the lack of effective sensors for many metal ions, despite the first metal sensor (for Ca2+) being reported more than 30 years ago. In contrast, combinatorial screening or selections do not require prior knowledge of specific metal binding and allow sensors to be obtained from a library that are selective for almost any target. In comparison to combinatorial screening of small organic molecules or peptides/proteins, in vitro selection of DNAzymes can sample a much larger library (≈1015 vs. 104), can continually enrich the pool with the “winner” DNAzyme sequences using PCR amplification over multiple selection rounds, and can introduce mutations using error-prone PCR to explore even more sequences than the initial selection pool, mimicking natural evolution (Figure 1a). What makes in vitro selection of DNAzymes so unique is that the genotype (i.e., the DNAzyme sequence) and phenotype (i.e., its function, such as cleavage) is combined within the same DNA molecule, so that it is easier to identify and amplify the “winners”. Furthermore, unlike antibody generation, in vitro selection can be carried out under a variety of conditions such as different pHs,46 temperatures,47 and buffer compositions,48,49 making it possible to customize the sensors for different applications, such as contaminated water for environmental monitoring, blood or urine for POC diagnostics, and cellular extracts for cell imaging.
3.2. Using Negative Selection to Improve Selectivity Against Competing Targets
Another advantage of in vitro selection over rational design is the ability to use negative or counter selection to increase the selectivity of the DNAzyme over similar competing targets. Most rational approaches start with design of one receptor and, when the receptor lacks enough selectivity over competing targets, the entire process of design, synthesis, and testing must be repeated again, often multiple times, and may still have unsatisfactory selectivity at the end. In vitro selection starts with a large library and thus can afford to use negative selection steps to remove the population of sequences that recognize competing targets. These negative selection steps can be performed directly prior or subsequent to positive selection steps. This method is especially useful for detection of metal ions, as selectivity over similar competing metal ions can often be a challenge for rationally designed sensors, such as for metals of the same charge (e.g., Na+ over Li+, K+, and NH4+) or for different oxidation states of the same metal (e.g., Fe2+ over Fe3+). For instance, selectivity against Pb2+ activity of DNAzymes has been a major challenge of the field, since Pb2+ has relatively fast ribonucleotide cleavage catalysis by itself. However, by using negative selection, Co2+-reactive DNAzymes were able to be obtained with more than four-fold selectivity for Co2+ over Pb2+, as well as some selectivity over Zn2+. The Co2+ DNAzymes obtained without negative selection had more activity for both Pb2+ and Zn2+ than Co2+, indicating a significant enhancement of Co2+ selectivity from the negative selections.50
3.3. Using Selection and Re-selection to Fine-Tune Affinities, Sensitivity, and Dynamic Ranges
In addition to improving selectivity using negative selection, in vitro selection can fine-tune target-binding affinity and the DNAzyme activity rate by customizing each step of the selection, such as incubating the selection mixture in the presence of different concentrations of the target for different amounts of time. Often this technique is performed with higher concentrations of the targets and longer incubation times first, with the target concentration and incubation time slowly decreasing over subsequent rounds of selection to increase the selection pressure. In this way, higher affinity and faster DNAzymes can be enriched in later rounds of selection. If in vitro selection does not result in desirable affinity or activity, a re-selection can be performed. Re-selections use a known DNAzyme with some initial affinity and activity as a pool template, instead of using a completely randomized pool, and incorporate partial randomization at each oligonucleotide position with a small percentage chance (up to 15%) of mutating the original nucleotide. This is especially useful for DNAzymes as studies have shown that just a few nucleotide changes can make a difference in DNAzyme activity under different conditions.51 Re-selection has been used to improve the activity of DNAzymes by more than 500-fold.52
Though in general it is often desirable to obtain the highest affinity possible, for practical applications it is often desirable to be able to fine-tune affinity, such as for different sensing applications that require different limits of detection, which is especially applicable to environmental sensing. This can be accomplished by using different pHs, as DNAzyme catalysis often involves general acid-base chemistry which is pH dependent,26 or by using different ratios of active and inactive DNAzymes,53 since inactive sequences can still bind metal ions without subsequent signal transduction. This technique will effectively decrease the apparent affinity of the sensors with increasing ratios of inactive DNAzymes, allowing a broad range of affinities to be used from the same DNAzyme.
3.4. Separating Target Binding from the Signaling Moiety to Allow Facile Signal Transductions
While it is difficult to obtain receptors to recognize targets selectively, it may be even harder to transform this recognition into detectable signals without compromising the recognition, because adding signaling moieties such as fluorophores and chromophores close to the target binding site can often abolish or decrease the target recognition, while moving the signaling moieties away from the binding site can result in minimal signal transduction. Because the DNAzyme catalytic beacon design separates target binding from the signaling moieties and relies on only dehybridization of the cleaved substrate strand to generate the signal, the signaling moieties can be changed without affecting the DNAzyme activity. It is also relatively easy to attach these moieties onto the DNAzyme either through solid phase oligonucleotide synthesis or various chemical conjugation techniques. This advantage allows any fluorophore to be used with optimal properties (e.g., wavelength, lifetime, quantum yield, etc.) for the desired application. It also addresses a major issue in the field of metal ion sensing: the lack of fluorescent sensors for paramagnetic metal ions, which are known to abolish fluorescent signals through contact quenching with fluorophores. The separation of the metal-binding site from the signaling moiety overcomes this limitation, making it possible for DNAzyme sensors to sense paramagnetic metal ions.14
3.5. Using Different Nanomaterials or Other Signaling Moieties to Extend the Versatility and Modularity of the Sensing Platform
The separation of the signaling moiety from target binding allows for the use of different fluorophores, which is especially useful for multiplexed sensing, such that different DNAzymes can use different fluorophores for the simultaneous detection of different analytes. Similarly, different signaling moieties can also be incorporated easily. In general, DNAzymes can be used with any sensing method that can generate signal differences from bringing two moieties closer together or farther apart. For instance, in addition to organic fluorophores, as was first demonstrated for the catalytic beacon design (Figure 1c), quantum dots can also be attached to the DNAzymes in place of the fluorophore.54 Colorimetric sensors have also been developed, as described in Section 2.1, by attaching AuNPs to DNAzymes, which are blue when aggregated closely together but turn red upon disaggregation via substrate cleavage (Figure 2b, 2d).53 Other signaling moieties have also been developed along similar principles, including various electrochemical sensors (Figure 2c),30 sensors based on glucose meters (Figure 2f, 2j),31 and MRI sensors (Figure 2g).29
3.6. Overcoming Sensing/Imaging Artifacts with Inactive DNAzymes as Negative Controls
Activity-based sensors that rely on separation or cleavage reactions to generate a signal, like DNAzymes, are often vulnerable to artifacts that result from degradation of the sensing reagents, rather than from reaction with the target, especially in complex samples, such as contaminated water, blood, or cells. To overcome these artifacts, DNAzyme activity-based sensors have a distinct advantage over other types of activity-based sensors, because they can make use of an “inactive” DNAzyme as a negative control that has almost the same sequence, except that a conserved nucleotide critical to DNAzyme activity is mutated to render the DNAzyme inactive in the presence of the target. Furthermore, a non-cleavable substrate negative control can also be used by replacing the ribonucleotide in the scissile position with the corresponding deoxyribonucleotide. Using these negative controls in parallel with the active DNAzyme allows artifacts due to other effects, such as degradation of the sensors, to be ruled out and as such is always used by our group for verification of DNAzyme sensors in biological systems.
3.7. Spatiotemporal Control of Sensing and Imaging with Caging/Decaging Strategies
Targets such as metal ions display different concentrations in different cellular and biological locations, and these concentrations will also often fluctuate with time or based on certain biological events. Therefore, sensing or imaging of these targets with spatiotemporal control is a very important feature that few activity-based sensors can currently offer. To address this issue, we have developed three photoactivatable methods to provide spatiotemporal control of DNAzyme sensors.
The first is a photocaging method in which the 2′-OH at the adenosine ribonucleotide of the substrate strand is replaced by a 2′-O-nitrobenzyl group, thereby inhibiting metal-dependent RNA cleavage during delivery (Figure 3b).39 Prior to imaging, irradiation with 365 nm light will release the nitrobenzyl modification, restoring the 2′-OH and re-activating the DNAzyme, which can be accomplished with high spatiotemporal control. Since 365 nm is close to UV light that can damage cells, we have further improved upon this system by using upconversion nanoparticles that can upconvert 980 nm near-IR (NIR) light irradiation into 365 nm light to decage the nitrobenzyl group, making it possible to image Zn2+ in zebrafish with minimal UV-vis damage, while also having significantly increased light penetration depth (Figure 3f).45
The second is a photothermal-caging method in which the binding arm of the DNAzyme strand is hybridized to a short DNA oligonucleotide that is conjugated to a gold nanoshell (AuNS) (Figure 3d).55 The short strand prevents the DNAzyme strand from binding to its substrate strand, thus inhibiting its metal-dependent activity. Upon NIR illumination, the AuNS produces a localized temperature increase, which is just enough to dehybridize the short strand, allowing the DNAzyme to bind to the substrate strand to form an active DNAzyme construct for target ion detection.
In addition to these two exogenous decaging methods, two different endogenous decaging methods have also recently been developed, which rely on endogenous moieties within the cellular system to decage the DNAzyme sensor. These systems rely on either reactive oxygen species (H2O2 or HClO)56 or a homing endonuclease (Sce-I)57 to decage the DNAzyme sensors upon cellular delivery (Figure 3h).
3.8. Suitability for Detection of Labile Metal Ions in Biological Systems
Though RNA-cleaving DNAzymes have been selected for different types of targets, including bacteria,9,34 they are especially well-suited for reaction in the presence of different metal ions. Metal ions have been shown to play a role, either directly or indirectly, in the acid-base catalytic mechanism of DNAzymes,58 as well as in the proper folding of DNAzymes.59 Due to both of these potential contributions to act as cofactors for the DNAzyme reaction, DNAzymes with high specificity for a particular metal ion have been obtained for many different metal ions (Table 1), including monovalents,40 divalents,50 and trivalents.22 Furthermore, when DNAzyme-based sensors are used in biological systems for metal detection, they are able to detect specific labile metal pools, or the species of a certain metal that are loosely associated with other molecules and are not tightly bound to proteins, rather than the total amount of a certain metal. Though total metal levels within biological systems can be useful to measure, the labile metal pools are often more informative of metal homeostasis or misregulation and so they are an important emerging area of study that DNAzyme-based probes can help to elucidate.
4. Current Challenges and Opportunities for DNAzymes as Activity-Based Sensors
Though DNAzyme activity-based sensors have many advantages, as outlined in Section 3, there are many aspects of DNAzymes that remain as challenges and potential hurdles to their application as sensors. In this section, we outline several of these challenges as well as the ways in which they are being addressed to design the most effective DNAzyme-based sensors.
4.1. Stability of DNAzymes for Sensing and Imaging
Even though DNA-based sensors are significantly more stable than their RNA- or protein-based counterparts, they are generally less stable than organic and polymer-based sensors. Most DNAzyme sensors are relatively short (<100 nucleotides) and are stable at room temperature in aqueous solutions for many weeks and in dry powder for several years. Even though the substrate does contain a single ribonucleotide, it is significantly more stable than all-RNA sequences, and when the DNAzyme is stored as a dry powder, it has a shelf life of more than 9 months.60 Since most sensing can finish in 2–15 minutes when the dry DNAzyme is dissolved in a solution, the DNAzyme sensor stability is not an issue for in vitro tests. In cellular and in vivo systems, while the required time for DNAzyme sensor delivery and imaging can be many hours or even days, the main barriers to stability are nucleases. The exonuclease-based degradation can be minimized by conjugating signaling moieties such as fluorophores and quenchers at the terminal sites of DNAzymes, or by using other modifications such as phosphorothioate bonds and inverted nucleotides. Additionally, because DNAzymes form stable 3D structures like globular proteins,58 they are often harder to be recognized by both exo- and endonucleases than linear DNA helices. If the stability is an issue, negative selection (see Section 3.2) can be used to obtain DNAzymes that are stable against cellular extract or human serum. In any case, negative controls using inactive DNAzymes (see Section 3.6) should always be used when stability is a concern to make sure that the cleavage is due to DNAzyme activity rather than DNA degradation via other processes.
4.2. Sensitivity to Temperature Variations
Since the catalytic beacon method relies on the change in melting temperature before and after the metal-dependent cleavage, the performance of the DNAzyme sensors can be affected by variations of the ambient temperature. While this is not a major issue for cellular and in vivo imaging, because the physiological temperature varies little, sensing for environmental monitoring and medical diagnostics can be affected. To address this issue, we have introduced mismatches in the binding arms, which do not affect the DNAzyme activity and metal selectivity but can tune the temperature range in which the DNAzyme sensors will work.12 When combining two different DNAzymes that work at high and low temperatures, we have produced a DNAzyme sensing system that is resistant to temperature variation over the range of 4°C – 30°C.
4.3. Delivery into Cells for Intracellular Imaging
Since DNAzymes are highly-negatively charged and thus are not directly uptaken by most cell types, an effective delivery method for intracellular imaging with DNAzymes is crucial, both to increase the DNAzyme stability and to allow for sufficient uptake and precise localization in cells. To address these issues, a variety of delivery methods have been developed, which can be grouped into a few broad categories.44,61 Metal nanoparticles are typically used for delivery to endosomes or lysosomes as they are mostly uptaken via endocytosis. With this method, the DNAzymes are typically covalently attached to the nanoparticles through various types of attachment chemistry. Similar to metal nanoparticles, DNA nanostructures such as DNA dendrimers or DNA tetrahedrons have also been used by incorporating the DNAzyme enzyme or substrate sequences directly into the DNA strands of the nanostructures. These systems will also typically be uptaken via endocytosis and so deliver to endosomes/lysosomes, but do not require any of the above attachment methods. For delivery to subcellular locations outside of lysosomes, cationic molecules, such as cationic polymers, peptides, or lipid aggregates (micelles/vesicles), are often used to mask the negative charges of the DNAzymes and can associate together through electrostatic interactions without having to directly conjugate them together. These transfection agents enable their cargo to cross membranes and subsequently allow for endosomal escape to deliver to the cytosol. One common commercial reagent is Lipofectamine, which can be used for either cytosolic or nuclear localization. Of note, another major advantage of DNAzyme-based sensors is that they can take advantage of various transfection agents developed for other oligonucleotide agents, such as antisense DNA or siRNA. Furthermore, while the current focus of the field has been on transfection to mammalian cells, delivery to other cell types such as bacterial, yeast, or plant cells, as well as for in vivo systems and more complex cell cultures, will require further development of new transfection methods.
4.4. Detection of Endogenous Metal Ions Through Enzyme-Free Amplification Methods
Since the concentration of targets can vary widely in cells, some targets may not be readily detectable using DNAzyme-based sensors if their concentration is lower than the sensor detection limit. To meet this challenge, we have designed an enzyme-free amplification method62 by using the cleaved substrate strand of a DNAzyme sensor as an initiator to trigger catalytic hairpin assembly (CHA), resulting in almost 10-fold higher sensitivity in comparison with the non-amplification method,40 and thereby allowing detection of endogenous Na+ levels in cells under physiological conditions (Figure 3e). CHA and other isothermal, enzyme-free amplification methods, as reviewed elsewhere,44 allow for signal amplification without the need of co-transfecting protein enzymes (usually DNA polymerases, such as for rolling circle amplification), which allows for more efficient and less perturbing sensor systems, while still enabling signal amplification for significantly improved sensitivity.
4.5. Quantitative Detection of Metal Ions in Biological Systems with Ratiometric Sensors
Quantitative detection of endogenous metal ions in biological systems, as opposed to more qualitative detection from the first generation of DNAzyme-based sensors, also remains a significant challenge. One major issue is the inability to know the precise amount of sensor delivered to the system, due to variation in DNA delivery efficiency, which can be addressed by using ratiometric sensors. Ratiometric sensors rely on taking the signal from two distinct signaling moieties, and so is much less dependent on sensor concentration. We have recently developed a ratiometric fluorescence probe based on FRET (Förster Resonance Energy Transfer) by replacing the catalytic beacon fluorophore and quencher with a FRET donor and acceptor, which allowed for improved intracellular detection of Zn2+ (Figure 3g).44 However, obtaining truly quantitative detection in biological systems will also require careful calibration of the sensors within the biological environment, such as in a buffer or cell lysate that mimics the ionic strength, molecular components, molecular crowding, and other conditions of biological systems.
4.6. Temporal Resolution and DNAzyme Reaction Time
Although caging groups, as discussed in Section 3.7, can allow for both spatio- and temporal activation of DNAzyme sensors, the temporal resolution, or the speed at which a signal readout can be obtained, is still limited by both the DNAzyme reaction rate as well as the amount of time required for decaging. The former can be improved by further reselection to obtain faster DNAzymes, while the latter can be addressed by developing new and faster decaging methods. However, it is likely that there will always be some delay between DNAzyme activation and the signal readout, which could potentially limit the types of biological events that can be monitored. Furthermore, as is the case with all activity-based sensors, DNAzyme sensors are essentially irreversible and thus changes in the metal concentrations within a single sample, especially decreases, cannot be detected since the sensors cannot be “turned off” after activation. This problem can be mitigated by using identical samples with a different time-point for each sample, but this approach may not be suitable for all applications. It is possible that many DNAzyme-based activity sensors can be made reversible by using a substrate that can be regenerated after a reaction, such as through ligation of the cleaved fragments or extension of the longer fragment to restore the released shorter fragment.63 Alternatively, some DNAzymes can be converted into aptamers which produce a signal upon binding to a metal, without catalyzing a reaction, as has been demonstrated for Ag+ 64 and Na+,65 effectively creating a binding-based sensor.
5. Conclusions and Perspectives
Since the first publication of a DNAzyme-based metal sensor in 2000, our group has been developing DNAzymes as a novel class of activity-based metal ion sensors and imaging agents for both in vitro and in vivo applications. In this account, we have reviewed recent progress of sensing in environmental monitoring and medical diagnostics, as well as of imaging in living cells and animals such as zebrafish. Given the progress made in the past 19 years, we have summarized the major advantages and current challenges of our DNAzyme sensor platform and conclude by outlining further opportunities and future applications for DNAzyme-based sensors and imaging agents. Despite the advantages and progress made to date, this field is still an active area of research to further develop and maximize the potentials of DNAzymes as sensors and imaging agents.
5.1. Better Understanding of Structure and Mechanism
While many DNAzymes have been obtained through in vitro selection, many of which are highly selective for metal ions (e.g., the uranyl-dependent DNAzyme has > 1-million-fold selectivity against other metal ions), we still know very little about the structural features and reaction mechanisms responsible for such selectivity and activity, with most of these studies being limited to the 8–17 DNAzyme,66–68 including the recently published crystal structure of the 8–17 DNAzyme, which is only the second DNAzyme structure so far obtained.58 Understanding the structural features responsible for metal-dependent binding49,69 may explain the key contributing factors in DNAzyme selectivity and provide possible strategies for rational design of DNAzymes with even higher selectivity. Exploring the mechanisms of how DNAzymes catalyze their reactions could help develop sensors with faster response times and lower detection limits.
5.2. Broadening DNAzyme Activity-Based Sensing Using Modified Nucleotides and Aptazymes
Most DNAzymes that have been selected so far use the four canonical nucleobases. The limited number of functional groups in the four canonical nucleobases (i.e., primary/secondary amines and carbonyls) can potentially restrict the sensitivity and selectivity of DNAzyme sensors, as well as the targets that they can detect, which are mostly positively charged metal ions or biomolecules. To overcome this limitation, non-canonical nucleotides with more functional groups, such as those containing alkylthio, imidazole, and aromatic groups, can be introduced into DNAzymes during selection steps in order to find DNAzymes that can bind metal ions or other targets more strongly. Another under-explored area of research is to couple DNAzymes with aptamers, called aptazymes, so that the binding of targets by the aptamers can either inhibit or enhance the activity of the DNAzymes. Both of these techniques can greatly expand the number of DNAzymes developed in the coming years for new targets.
5.3. Alternative Sensing Mechanisms for Different Applications
Although DNAzyme-based sensors have been applied for in vivo zebrafish models, the signal detection still relies on the transparency of early fish larvae and is limited when applied to imaging in nontransparent living animals. This limitation can be overcome by using NIR-based modalities, since NIR has much deeper penetration in biological tissues, such as with NIR fluorophores/quenchers70 or with the photoacoustic imaging technique, which has already been used with DNA aptamer sensors in vivo.71 Furthermore, chemi- and bioluminescence sensors have advantages in their signal-to-noise ratio and response dynamics, as well as not requiring an excitation light source that can often be costly and cytotoxic, which may be valuable for generating novel DNAzyme-based sensors for environmental monitoring and in vivo imaging. Other non-intensity-based sensing mechanisms focusing on a change in fluorescence anisotropy or lifetime are also valuable new techniques for future sensor development.
5.4. Subcellular Targeting
To obtain deeper insight into biological functions, it is important to image metal ions and other targets in different subcellular locations, because most molecules are not evenly distributed inside cells and reside in different organelles and compartments to enable different functions. The visualization of subcellular dynamics of metal ions is one of the next frontiers not only for DNAzyme-based sensing, but also for all activity-based sensing research. Since DNAzymes can be readily conjugated to various subcellular targeting molecules or nanostructures for subcellular imaging, a whole array of new research in metal homeostasis and trafficking in subcellular locations can be explored in future.
5.5. Concluding Remarks
In view of the current interest in understanding the biological functions of metal ions and metabolites in biochemical and biomedical research, the continued development of molecular imaging tools will be of great importance to studies across a huge range of investigations in physiology and pathology. Even though DNAzymes have been developed as sensors in a much shorter time and by a much smaller number of researchers than for other types of sensors, they have emerged as one of the most powerful and versatile types of sensors that will continue to make a major impact on both scientific research and societal well-being.
Acknowledgements
We wish to thank the Lu group members who have contributed over the years to the many studies described here, and the U.S. National Institutes of Health (GM124316 and MH110975) for financial support.
Biography
Biographical Information
Ryan Lake is a graduate student in the Department of Chemistry at the University of Illinois at Urbana-Champaign. He received his B.A. degree in Biochemistry from Earlham College in 2014. His current research interests include the application of DNAzymes as intracellular sensors and the discovery and application of novel functional nucleic acids.
Zhenglin Yang is a graduate student in the Department of Biochemistry at the University of Illinois at Urbana-Champaign. He received his B.A. degree in Life Sciences from Peking University in 2011 and his M.Sc. degree in Molecular Genetics from the University of Toronto in 2015. His research interests are in biomedical theranostics by selecting and engineering functional nucleic acids.
JingJing Zhang is an Associate Professor in the School of Chemistry and Chemical Engineering at Nanjing University. He received his Ph.D. degree in Chemistry from Nanjing University in 2010 with Prof. Jun-jie Zhu. After two years of postdoctoral research at Nanjing University, he moved to Prof. Yi Lu’s group at UIUC as a postdoctoral research associate before joining Nanjing University in 2019. His research interests focus on point-of-care diagnostics, synthetic biosensors, and in vivo photoacoustic imaging.
Yi Lu is the Jay and Ann Schenck Professor in the Department of Chemistry at the University of Illinois at Urbana–Champaign. He received his B.S. degree from Peking University in 1986 and his Ph.D. degree from UCLA in 1992 under Dr. Joan S. Valentine. After two years of postdoctoral research in Dr. Harry B. Gray’s group at Caltech, he started his own independent career at UIUC in 1994. His group interests are in bioinorganic, biomaterial, and bioanalytical chemistry.
References
- (1).Aron AT; Ramos-Torres KM; Cotruvo JA; Chang CJ Recognition- and Reactivity-Based Fluorescent Probes for Studying Transition Metal Signaling in Living Systems. Acc. Chem. Res 2015, 48, 2434–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Sanman LE; Bogyo M Activity-Based Profiling of Proteases. Annu. Rev. Biochem 2014, 83, 249–273. [DOI] [PubMed] [Google Scholar]
- (3).Li J; Lu Y A Highly Sensitive and Selective Catalytic DNA Biosensor for Lead Ions. J. Am. Chem. Soc 2000, 122, 10466–10467. [Google Scholar]
- (4).Liu J; Cao Z; Lu Y Functional Nucleic Acid Sensors. Chem. Rev 2009, 109, 1948–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Xiang Y; Lu Y DNA as Sensors and Imaging Agents for Metal Ions. Inorg. Chem 2014, 53, 1925–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zhou W; Saran R; Liu J Metal Sensing by DNA. Chem. Rev 2017, 117, 8272–8325. [DOI] [PubMed] [Google Scholar]
- (7).Breaker RR; Joyce GF A DNA Enzyme That Cleaves RNA. Chem. Biol 1994, 1, 223–229. [DOI] [PubMed] [Google Scholar]
- (8).Silverman SK Catalytic DNA: Scope, Applications, and Biochemistry of Deoxyribozymes. Trends Biochem. Sci 2016, 41, 595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Liu M; Chang D; Li Y Discovery and Biosensing Applications of Diverse RNA-Cleaving DNAzymes. Acc. Chem. Res 2017, 50, 2273–2283. [DOI] [PubMed] [Google Scholar]
- (10).McGhee CE; Lake RJ; Lu Y Preparation of MetalloDNAzymes In Artificial Metalloenzymes and MetalloDNAzymes in Catalysis; Wiley-Blackwell, 2018; pp 41–68. [Google Scholar]
- (11).Liu J; Lu Y Improving Fluorescent DNAzyme Biosensors by Combining Inter- and Intramolecular Quenchers. Anal. Chem 2003, 75, 6666–6672. [DOI] [PubMed] [Google Scholar]
- (12).Nagraj N; Liu J; Sterling S; Wu J; Lu Y DNAzyme Catalytic Beacon Sensors That Resist Temperature-Dependent Variations. Chem. Commun 2009, 4103–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Zhang J; Cheng F; Li J; Zhu JJ; Lu Y Fluorescent Nanoprobes for Sensing and Imaging of Metal Ions: Recent Advances and Future Perspectives. Nano Today 2016, 11, 309–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Liu J; Lu Y A DNAzyme Catalytic Beacon Sensor for Paramagnetic Cu2+ Ions in Aqueous Solution with High Sensitivity and Selectivity. J. Am. Chem. Soc 2007, 129, 9838–9839. [DOI] [PubMed] [Google Scholar]
- (15).Huang P-JJ; Liu J An Ultrasensitive Light-up Cu2+ Biosensor Using a New DNAzyme Cleaving a Phosphorothioate-Modified Substrate. Anal. Chem 2016, 88, 3341–3347. [DOI] [PubMed] [Google Scholar]
- (16).Liu J; Brown AK; Meng X; Cropek DM; Istok JD; Watson DB; Lu Y A Catalytic Beacon Sensor for Uranium with Parts-per-Trillion Sensitivity and Millionfold Selectivity. Proc. Natl. Acad. Sci 2007, 104, 2056–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Liu J; Lu Y Rational Design of “Turn-On” Allosteric DNAzyme Catalytic Beacons for Aqueous Mercury Ions with Ultrahigh Sensitivity and Selectivity. Angew. Chem.-Int. Ed 2007, 46, 7587–7590. [DOI] [PubMed] [Google Scholar]
- (18).Saran R; Liu J A Silver DNAzyme. Anal. Chem 2016, 88, 4014–4020. [DOI] [PubMed] [Google Scholar]
- (19).Zhou W; Saran R; Huang P-JJ; Ding J; Liu J An Exceptionally Selective DNA Cooperatively Binding Two Ca2+ Ions. ChemBioChem 2017, 18, 518–522. [DOI] [PubMed] [Google Scholar]
- (20).Huang P-JJ; Vazin M; Liu J Desulfurization Activated Phosphorothioate DNAzyme for the Detection of Thallium. Anal. Chem 2015, 87, 10443–10449. [DOI] [PubMed] [Google Scholar]
- (21).Huang P-JJ; Liu J Rational Evolution of Cd2+-Specific DNAzymes with Phosphorothioate Modified Cleavage Junction and Cd2+ Sensing. Nucleic Acids Res. 2015, 43, 6125–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhou W; Vazin M; Yu T; Ding J; Liu J In Vitro Selection of Chromium-Dependent DNAzymes for Sensing Chromium(III) and Chromium(VI). Chem. - Eur. J 2016, 22, 9835–9840. [DOI] [PubMed] [Google Scholar]
- (23).St Paul Public School Lead in Water Testing https://www.youtube.com/watch?v=UdWxFsLsKOc (accessed Aug 5, 2019).
- (24).Swearingen CB; Wernette DP; Cropek DM; Lu Y; Sweedler JV; Bohn PW Immobilization of a Catalytic DNA Molecular Beacon on Au for Pb(II) Detection. Anal. Chem 2005, 77, 442–448. [DOI] [PubMed] [Google Scholar]
- (25).Wernette DP; Swearingen CB; Cropek DM; Lu Y; Sweedler JV; Bohn PW Incorporation of a DNAzyme into Au-Coated Nanocapillary Array Membranes with an Internal Standard for Pb(II) Sensing. Analyst 2006, 131, 41–47. [DOI] [PubMed] [Google Scholar]
- (26).Wang Z; Lee JH; Lu Y Label-Free Colorimetric Detection of Lead Ions with a Nanomolar Detection Limit and Tunable Dynamic Range by Using Gold Nanoparticles and DNAzyme. Adv. Mater 2008, 20, 3263–3267. [Google Scholar]
- (27).Lee JH; Wang ZD; Liu JW; Lu Y Highly Sensitive and Selective Colorimetric Sensors for Uranyl (UO22+): Development and Comparison of Labeled and Label-Free DNAzyme-Gold Nanoparticle Systems. J. Am. Chem. Soc 2008, 130, 14217–14226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Mazumdar D; Liu JW; Lu G; Zhou JZ; Lu Y Easy-to-Use Dipstick Tests for Detection of Lead in Paints Using Non-Cross-Linked Gold Nanoparticle-DNAzyme Conjugates. Chem. Commun 2010, 46, 1416–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Xu W; Xing H; Lu Y A Smart T1-Weighted MRI Contrast Agent for Uranyl Cations Based on a DNAzyme-Gadolinium Conjugate. Analyst 2013, 138, 6266–6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Xiao Y; Rowe AA; Plaxco KW Electrochemical Detection of Parts-Per-Billion Lead via an Electrode-Bound DNAzyme Assembly. J. Am. Chem. Soc 2007, 129, 262–263. [DOI] [PubMed] [Google Scholar]
- (31).Xiang Y; Lu Y Using Personal Glucose Meters and Functional DNA Sensors to Quantify a Variety of Analytical Targets. Nat. Chem 2011, 3, 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Xiang Y; Lu Y An Invasive DNA Approach toward a General Method for Portable Quantification of Metal Ions Using a Personal Glucose Meter. Chem. Commun 2012, 49, 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhang J; Lu Y Biocomputing for Portable, Resettable, and Quantitative Point-of-Care Diagnostics: Making the Glucose Meter a Logic-Gate Responsive Device for Measuring Many Clinically Relevant Targets. Angew. Chem. Int. Ed 2018, 57, 9702–9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tram K; Kanda P; Salena BJ; Huan S; Li Y Translating Bacterial Detection by DNAzymes into a Litmus Test. Angew. Chem. Int. Ed 2014, 53, 12799–12802. [DOI] [PubMed] [Google Scholar]
- (35).Zhang J; Xing H; Lu Y Translating Molecular Detections into a Simple Temperature Test Using a Target-Responsive Smart Thermometer. Chem. Sci 2018, 9, 3906–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).McGhee CE; Loh KY; Lu Y DNAzyme Sensors for Detection of Metal Ions in the Environment and Imaging Them in Living Cells. Curr. Opin. Biotechnol 2017, 45, 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Zhang J; Lan T; Lu Y Molecular Engineering of Functional Nucleic Acid Nanomaterials toward In Vivo Applications. Adv. Healthc. Mater 2019, 8, 1801158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Wu P; Hwang K; Lan T; Lu Y A DNAzyme-Gold Nanoparticle Probe for Uranyl Ion in Living Cells. J. Am. Chem. Soc 2013, 135, 5254–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Hwang K; Wu P; Kim T; Lei L; Tian S; Wang Y; Lu Y Photocaged DNAzymes as a General Method for Sensing Metal Ions in Living Cells. Angew. Chem. Int. Ed 2014, 53, 13798–13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Torabi S-F; Wu P; McGhee CE; Chen L; Hwang K; Zheng N; Cheng J; Lu Y In Vitro Selection of a Sodium-Specific DNAzyme and Its Application in Intracellular Sensing. Proc. Natl. Acad. Sci 2015, 112, 5903–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Li L; Feng J; Fan Y; Tang B Simultaneous Imaging of Zn2+ and Cu2+ in Living Cells Based on DNAzyme Modified Gold Nanoparticle. Anal. Chem 2015, 87, 4829–4835. [DOI] [PubMed] [Google Scholar]
- (42).Qiu L; Zhang T; Jiang J; Wu C; Zhu G; You M; Chen X; Zhang L; Cui C; Yu R; Tan W Cell Membrane-Anchored Biosensors for Real-Time Monitoring of the Cellular Microenvironment. J. Am. Chem. Soc 2014, 136, 13090–13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Zhang L; Huang H; Xu N; Yin Q Functionalization of Cationic Poly(p-Phenylene Ethynylene) with Dendritic Polyethylene Enables Efficient DNAzyme Delivery for Imaging Pb2+ in Living Cells. J. Mater. Chem. B 2014, 2, 4935–4942. [DOI] [PubMed] [Google Scholar]
- (44).Hwang K; Mou Q; Lake RJ; Xiong M; Holland B; Lu Y Metal-Dependent DNAzymes for the Quantitative Detection of Metal Ions in Living Cells: Recent Progress, Current Challenges, and Latest Results on FRET Ratiometric Sensors. Inorg. Chem 2019, 58, 13696–13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Yang Z; Loh KY; Chu Y-T; Feng R; Satyavolu NSR; Xiong M; Nakamata Huynh SM; Hwang K; Li L; Xing H; Zhang X; Chemla YR; Gruebele M; Lu Y Optical Control of Metal Ion Probes in Cells and Zebrafish Using Highly Selective DNAzymes Conjugated to Upconversion Nanoparticles. J. Am. Chem. Soc 2018, 140, 17656–17665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Liu Z; Mei SHJ; Brennan JD; Li Y Assemblage of Signaling DNA Enzymes with Intriguing Metal-Ion Specificities and PH Dependences. J. Am. Chem. Soc 2003, 125, 7539–7545. [DOI] [PubMed] [Google Scholar]
- (47).Nelson KE; Bruesehoff PJ; Lu Y In Vitro Selection of High Temperature Zn2+-Dependent DNAzymes. J. Mol. Evol 2005, 61, 216–225. [DOI] [PubMed] [Google Scholar]
- (48).Zhou W; Saran R; Chen Q; Ding J; Liu J A New Na+-Dependent RNA-Cleaving DNAzyme with over 1000-Fold Rate Acceleration by Ethanol. ChemBioChem 2016, 17, 159–163. [DOI] [PubMed] [Google Scholar]
- (49).Torabi S-F; Lu Y Identification of the Same Na+-Specific DNAzyme Motif from Two In Vitro Selections Under Different Conditions. J. Mol. Evol 2015, 81, 225–234. [DOI] [PubMed] [Google Scholar]
- (50).Bruesehoff P; Li J; Augustine AJ; Lu Y Improving Metal Ion Specificity during in Vitro Selection of Catalytic DNA. Comb. Chem. High Throughput Screen 2002, 5, 327–335. [DOI] [PubMed] [Google Scholar]
- (51).Nelson KE; Ihms HE; Mazumdar D; Bruesehoff PJ; Lu Y The Importance of Peripheral Sequences in Determining the Metal Selectivity of an in Vitro-Selected Co2+-Dependent DNAzyme. ChemBioChem 2012, 13, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Chan L; Tram K; Gysbers R; Gu J; Li Y Sequence Mutation and Structural Alteration Transform a Noncatalytic DNA Sequence into an Efficient RNA-Cleaving DNAzyme. J. Mol. Evol 2015, 81, 245–253. [DOI] [PubMed] [Google Scholar]
- (53).Liu J; Lu Y A Colorimetric Lead Biosensor Using DNAzyme-Directed Assembly of Gold Nanoparticles. J. Am. Chem. Soc 2003, 125, 6642–6643. [DOI] [PubMed] [Google Scholar]
- (54).Wu C-S; Khaing Oo MK; Fan X Highly Sensitive Multiplexed Heavy Metal Detection Using Quantum-Dot-Labeled DNAzymes. ACS Nano 2010, 4, 5897–5904. [DOI] [PubMed] [Google Scholar]
- (55).Wang W; Satyavolu NSR; Wu Z; Zhang J-R; Zhu J-J; Lu Y Near-Infrared Photothermally Activated DNAzyme-Gold Nanoshells for Imaging Metal Ions in Living Cells. Angew. Chem. Int. Ed 2017, 56, 6798–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Xiao L; Gu C; Xiang Y Orthogonal Activation of RNA-Cleaving DNAzymes in Live Cells by Reactive Oxygen Species. Angew. Chem. Int. Ed 2019, 58, 14167–14172. [DOI] [PubMed] [Google Scholar]
- (57).Lin Y; Yang Z; Lake RJ; Zheng C; Lu Y Enzyme-Mediated Endogenous and Bioorthogonal Control of a DNAzyme Fluorescent Sensor for Imaging Metal Ions in Living Cells. Angew. Chem. Int. Ed 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Liu H; Yu X; Chen Y; Zhang J; Wu B; Zheng L; Haruehanroengra P; Wang R; Li S; Lin J; Li J; Sheng J; Huang Z; Ma J; Gan J Crystal Structure of an RNA-Cleaving DNAzyme. Nat. Commun 2017, 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Brown AK; Li J; Pavot CMB; Lu Y A Lead-Dependent DNAzyme with a Two-Step Mechanism. Biochemistry 2003, 42, 7152–7161. [DOI] [PubMed] [Google Scholar]
- (60).ANDalyze, Inc. Home Page. http://www.andalyze.com (accessed Apr 25, 2019).
- (61).Fan H; Zhang X; Lu Y Recent Advances in DNAzyme-Based Gene Silencing. Sci. China Chem 2017, 60, 591–601. [Google Scholar]
- (62).Wu Z; Fan H; Satyavolu NSR; Wang W; Lake R; Jiang J-H; Lu Y Imaging Endogenous Metal Ions in Living Cells Using a DNAzyme-Catalytic Hairpin Assembly Probe. Angew. Chem. Int. Ed 2017, 56, 8721–8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Fan H; Bai H; Liu Q; Xing H; Zhang X-B; Tan W Monitoring Telomerase Activity in Living Cells with High Sensitivity Using Cascade Amplification Reaction-Based Nanoprobe. Anal. Chem 2019, 91, 13143–13151. [DOI] [PubMed] [Google Scholar]
- (64).Saran R; Kleinke K; Zhou W; Yu T; Liu J A Silver-Specific DNAzyme with a New Silver Aptamer and Salt-Promoted Activity. Biochemistry 2017, 56, 1955–1962. [DOI] [PubMed] [Google Scholar]
- (65).Zhou W; Ding J; Liu J A Highly Specific Sodium Aptamer Probed by 2-Aminopurine for Robust Na+ Sensing. Nucleic Acids Res. 2016, 44, 10377–10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Cepeda-Plaza M; McGhee CE; Lu Y Evidence of a General Acid-Base Catalysis Mechanism in the 8–17 DNAzyme. Biochemistry 2018, 57, 1517–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Kim HK; Rasnik I; Liu JW; Ha TJ; Lu Y Dissecting Metal Ion-Dependent Folding and Catalysis of a Single DNAzyme. Nat. Chem. Biol 2007, 3, 763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Mazumdar D; Nagraj N; Kim H-K; Meng X; Brown AK; Sun Q; Li W; Lu Y Activity, Folding and Z-DNA Formation of the 8–17 DNAzyme in the Presence of Monovalent Ions. J. Am. Chem. Soc 2009, 131, 5506–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Hwang K; Hosseinzadeh P; Lu Y Biochemical and Biophysical Understanding of Metal Ion Selectivity of DNAzymes. Inorganica Chim. Acta 2016, 452, 12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Yang C; Yin X; Huan S-Y; Chen L; Hu X-X; Xiong M-Y; Chen K; Zhang X-B Two-Photon DNAzyme-Gold Nanoparticle Probe for Imaging Intracellular Metal Ions. Anal. Chem 2018, 90, 3118–3123. [DOI] [PubMed] [Google Scholar]
- (71).Zhang J; Smaga LP; Satyavolu NSR; Chan J; Lu Y DNA Aptamer-Based Activatable Probes for Photoacoustic Imaging in Living Mice. J. Am. Chem. Soc 2017, 139, 17225–17228. [DOI] [PMC free article] [PubMed] [Google Scholar]


