ABSTRACT
Introduction: Clinical management and identification of respiratory diseases has become more rapid and increasingly specific due to widespread use of PCR(polymerase chain reaction) multiplex technologies. Although significantly improving clinical diagnosis, multiplexed PCR assays could have a greater impact on local and global disease surveillance. The authors wish to propose methods of evaluating respiratory multiplex assays to maximize diagnostic yields specifically for surveillance efforts.
Areas covered: The authors review multiplexed assays and critically assess what barriers have limited these assays for disease surveillance and how these barriers might be addressed. The manuscript focuses specifically on the case study of using multiplexed assays for surveillance of respiratory pathogens. The authors also provide a method of validation of specific surveillance measures.
Expert commentary: Current commercially available respiratory multiplex PCR assays are widely used for clinical diagnosis; however, specific barriers have limited their use for surveillance. Key barriers include differences in testing phase requirements and diagnostic performance evaluation. In this work the authors clarify phase testing requirements and introduce unique diagnostic performance measures that simplify the use of these assays on a per target basis for disease surveillance.
KEYWORDS: Multiplex respiratory assays, surveillance, respiratory pathogens, sensitivity, specificity, diagnostic odds ratio, ROC curves, Youden Index
1. Introduction
Respiratory multiplex platforms and assays significantly improve treatment of respiratory infections and reduce associated complications [1]. There are four major FDA-approved respiratory multiplex assays: Luminex NxTAG Respiratory Pathogen Panel (RPP), Nanosphere Verigene Respiratory Panel (RP) Flex, BioFire Film Array Respiratory Panel (RP), and eSensor Respiratory Viral Panel (RVP). Because treatment and clinical management of respiratory tract infections (RTIs) depend on the etiological agent, the ability to rapidly characterize respiratory pathogens is extremely important. Multiplex methods are particularly valuable in the case of RTIs, allowing clinicians to simultaneously test for a wide range of possible pathogens, as opposed of relying on numerous individuals tests. Correct diagnosis of RTIs is also critical to antibiotic stewardship, since roughly 1 in 3 antibiotics prescribed are unnecessary and most of these unneeded prescriptions are for respiratory infections [2]. Another challenge is diagnosing RTI’s across geographic regions where respiratory disease prevalence may be seasonal or region-specific. A broad spectrum assay screening a variety of targets would identify changes in seasonal trends significantly impacting regional clinical decisions throughout the year. RSV(respiratory syncytial virus) infections, for example, while seasonal, affect different parts of the United States at different times of the year with outbreaks varying within regions and between communities [3]. Multiplex methods also help resolve genotype variation. Recognizing genotypic differences helps in diagnosis by determining the course of clinical treatments. For instance, adamantine-resistant A/H3N2 strains have circulated globally for the past decade and oseltamivir-resistant seasonal A/H1N1 have circulated globally since 2007 [4]. During the 2016–2017 influenza season, the predominant strain was A/H3N2, compared to a year earlier when H1N1pdm09 was prevalent. These trends call to attention of the need to not only identify, but also characterize respiratory pathogens if possible to circumvent ineffective treatments. The benefits of multiplex assays have resulted in an increasing number of hospital and clinical laboratories adopting these methods strictly for clinical use [5]. Multiplex technologies can help aide clinical decisions by simultaneously screening for a variety of respiratory pathogens often detecting specific genetic differences increasing the value of these methods for diagnostic purposes. For example, in children and older adults, influenza and RSV are primary medical burdens that require distinct treatment, thus resolving the cause of infection is critical [6,7]. For surveillance, limited studies have shown the epidemiological use of these technologies on control and prevention; however, when used, respiratory multiplex assays tremendously benefit public health. For example, despite not being included in the first-generation Luminex xTAG RVP (released in 2008), results could still be used as a rule-out method for 2009 H1N1pdm09 strain [8]. The use of the Luminex xTAG RVP allowed for rapid diagnosis and better surveillance during the outbreak. Similarly, during the 2014 EV-D68 outbreak, both the eSensor RVP and the FilmArray RP were used as reliable diagnostic tests in surveillance algorithms to distinguish between Rhinovirus and Enterovirus-D68 [9,10] mitigating the further spread of disease.
Multiplexed diagnostic assays have the potential to revolutionize disease surveillance; however, these technologies have not had as great of impact on control and prevention efforts as compared to healthcare utilities because surveillance needs are fundamentally different from clinical diagnostic priorities. Whereas diagnostic assays are used to identify disease etiology and appropriate treatment for infected individuals, disease surveillance is conducted at a population level to identify trends and disease burden. Despite the substantial benefits (e.g. high efficiency, rapid turnaround time, high throughput, screening for a multitude of targets) of multiplexed assays to enhance surveillance efforts, no comprehensive review has assessed how multiplex assays can meet surveillance needs or whether future studies should focus on specific laboratory needs when performing surveillance and outbreak testing. Here, we review the primary barrier to using multiplexed assays for RTIs’ surveillance. By focusing on the limitation of current respiratory multiplex assays for surveillance, we are able to highlight alternative methods to evaluate performance of new multiplexed assays to better meet the needs of programs conducting surveillance.
2. Current FDA-approved multiplexed assays for RTIs
In many clinical laboratories, the use of culture and serology has been greatly reduced (if not altogether replaced) due to the availability of nucleic acid amplification tests (NAATs). NAATs dramatically reduce turnaround time allowing rapid response for control and prevention and for confirming suspect cases in hours instead of days and weeks. NAATs have also shown greater sensitivity (Sen) (defined as the ability of a diagnostic test to correctly identify individuals with disease) and specificity (Spe) (defined as the ability of a diagnostic test to correctly identify individuals without disease) above conventional methods for all public health relevant targets. NAATs are sufficient for making clinical decisions and unlike traditional methods remove subjective interpretation that can differ from one technician to the next. The epidemiological use of NAATs such as RT-PCR allows understanding of possible transmission in a population, but, also important, gives information for defining an appropriate case definition. Despite the widespread use of NAATs, only a few multiplex methods are available that not only detect respiratory pathogens, but are also FDA-approved.
2.1. Luminex NxTAG RPP
In 2008, the Luminex Corporation (Austin, Texas) unveiled the xTAG RVPv1, a high complexity high throughput bead-based qualitative PCR assay capable of detecting a variety of respiratory pathogens [11]. The xTAG RVPv1 assay was the first FDA-approved broad respiratory panel and is extensively used in clinics and public health laboratories (PHLs). Utilizing a bead hybridization fluid array, the most current version of the panel is the Luminex NxTAG RPP that can screen 20 total targets from nasopharyngeal specimens (NPS): RSV type A and B, influenza A variants, influenza A H1 and H3, influenza B, parainfluenza 1, 2, 3, and 4, human metapneumovirus (hMPV), adenovirus, rhinovrus/enterovirus, coronavirus type HKU1, NL63, 229E, OC43, human bocavirus, and bacterial targets, C. pneumoniae and M. pneumoniae. Results of up to 96 samples can be obtained in a single working day (3 h post-extraction).
2.2. Verigene Nanosphere RP Flex
The Verigene system (now manufactured by Luminex) is a moderate complexity, customizable throughput assay that detects a variety of pathogens not found in other major FDA-approved respiratory multiplex panel such as a Bordetella spp. group. The Verigene platform consists of a reader and a separate processing unit capable of sample-to-results in about 2 h with minimal hands-on time and is fully automated [12]. The most recent panel offered on the Verigene system is the Respiratory Pathogens Flex (RP Flex), which received FDA approval in 2015. The panel screens against both viruses and bacteria highlighting the Bordetella spp. bacterial group. The RP Flex is the first multiplex respiratory test to allow customization by user’s reporting preference allowing a ‘pay for what you need’ option. Five reporting blocks that may be released to users are: Block 1: adenovirus and hMPV; Block 2: influenza A, A/H1, A/H3, and influenza B; Block 3: parainfluenza 1–4 and rhinovirus; Block 4: RSV A and B; and Block 5: B. pertussis, B. holmesii, and B. parapertussis/B.bronchiseptica. While onboard PCR multiplexing is performed for all targets, only those results preselected are paid for and reported. Endemically circulating viruses may be targeted in specific populations while budgeting for targets of interest. For example, Block 1 and Block 4 may be of great interest for laboratories serving senior or pediatric populations as outbreaks in RSV and adenovirus are common in these sub-populations [7,13,14]. Additionally, during flu season, Block 2 would give a great profile on circulating influenza viruses. For diagnostic laboratories, surveillance and population-based trend data are highly valued and therefore the flex option would most likely be bypassed in favor of a fixed report for all targets.
2.3. GenMark eSensor RVP
GenMark Diagnostics, Inc. (Carlsbad, California) received FDA approval for the eSensor RVP, a high complexity high-throughput assay in 2012 [15]. The assay runs on the GenMark’s XT-8 instrument modular system which can integrate from one to three analyzers processing up to eight samples each screening against 14 viral respiratory pathogens. The initial workflow begins with 60 min of prep time followed by a 3.5 h once placed into the system. The respiratory viral panel itself contains several Influenza targets, RSV subtypes, parainfluenza types, and hMPV. Additionally, the panel only reports rhinovirus instead of a combined rhinovirus/enterovirus result; however, cross-reactivity with enterovirus D68 (EV-D68) has been reported [9]. The panel also is the only multiplex panel to specifically detect adenovirus types (B/E and C).
2.4. FilmArray RP
The FilmArray RP is an FDA-approved (2011) low-throughput, moderate/low complexity, multiplex nested PCR assay automated from extraction to result [16]. The assay is able to identify and characterize 17 viral as well as three bacterial pathogens manufactured by BioFire Diagnostics (Salt Lake City, Utah). The 17 viral targets include: adenovirus, coronavirus HKU1, NL63, 229E and OC43, hMPV, rhinovrus/enterovirus, influenza A, influenza A subtype H1, H3, and (H1N1)pdm09, influenza B, parainfluenza 1,2,3, and 4, and RSV. The three bacterial targets include: B. pertussis, Chlamydophila pneumoniae, and Mycoplasma pneumoniae. A major benefit of the FilmArray RP is the minimal hands-on operator time (2–4 min/sample). The system includes onboard automated nucleic acid extraction directly from NPS samples. The time-to-result is about 1 h. A drawback of the FilmArray RP system is that only one sample can be processed at a time. Thus, in a single 8-h workday, a total of 6 to 8 samples may be processed. The FilmArray 2.0 System received FDA clearance in early 2015. The updated model could include up to eight modular units sequentially linked to one PC allowing higher throughput. In early 2016, BioFire Diagnostics (now a subsidiary of BioMerieux) received FDA clearance for an expanded instrument system, the FilmArray Torch. The newer platform is comprised of scalable modules that may include up to a 12-module system capable of screening up to ~90 samples per 8-h workday. The newer platform follows the same technology and allows the processing of the same RP assay.
3. Analytical requirement differences of surveillance and clinical diagnosis
The analytical requirements for surveillance of respiratory pathogens differ from those of clinical diagnosis (Table 1). In the clinical setting, rapid identification is a top priority so that the appropriate treatments can be identified and delivered. By contrast, for surveillance purposes, identification is also critical but for resolving etiologies of outbreaks, understanding epidemiological trends, and aiding disease control and prevention. Here, we review the testing requirement differences for the three phases (pre-analytical, analytical, and post-analytical) of clinical and surveillance testing.
Table 1.
Analytical requirements for multiplex diagnostic assays for diagnosis and surveillance of respiratory pathogens.
| Clinical diagnosis of respiratory pathogens |
Surveillance of respiratory pathogens and outbreak control |
|||
|---|---|---|---|---|
| Characteristics | Critical attributes for assays | Characteristics | Critical attributes for assays | |
| Pre-analytical | Samples processed individually or in small batches; number of samples varies depending on size of laboratory and prevalence of disease. | Small labs (or labs in low prevalence areas) require low-cost individual tests; larger labs (or labs in high prevalence areas) may do better with moderate to high-throughput assays. | Periodically, large numbers of samples are screened to determine etiology and origin of outbreak. | Potential to process large number of samples rapidly at minimal cost to support surge testing. |
| Typically collect specimens from individuals with clinical symptoms, who are more likely to have a high viral or bacterial load. | Per sample cost, low turnaround time and ability to differentiate between viral and bacterial respiratory pathogens (e.g. viruses versus bacterial) more important than sensitivity or specificity. | Because syndromic surveillance is performed for identification of causative agents and possible risk factors, labs often test asymptomatic individuals in addition to symptomatic ones. | Critical that assays are both sensitive and specific. | |
| Likely to have standard specimen type with minimal variation for a given pathogen. | FDA-approval is required for standardization in clinical decision-making; closed assay systems (i.e. not modifiable) are ideal preventing operator error and contamination. | Because remnant samples may be used and collected specimen types may vary, sample matrices are highly variable, which may limit interpretation on FDA-approved assays. | FDA-approval is required for clinical decision-making; however, laboratories tailor and validate assays for new sample types and emerging pathogens depending on need. | |
| Post-analytical | Treatment and patient care decisions are based on results. | Need to accurately distinguish between pathogens which have similar symptoms but different treatments. | Results used both for individual clinical management and for understanding population-level trends. | Need to accurately distinguish between pathogens which have similar symptoms but different treatments for clinical care AND need phylogenetic information to identify epidemiological trends/identify sources of outbreaks. |
3.1. Pre-analytical and analytical requirements
The primary differences between the pre-analytical testing requirements for disease surveillance and clinical diagnosis begins during specimen collection. Because healthcare facilities only collect specimens from individuals exhibiting RTI symptoms, the likelihood of identifying the source of infection is high [17]. This selective specimen collection by clinical labs minimizes unnecessary testing, which in turn reduces cost and use of medical services. For laboratories performing surveillance-based testing, specimens are collected from both symptomatic and asymptomatic individuals. Comparing the test results for symptomatic and healthy individuals provide an evidence base for epidemiological associations. Critically, active surveillance studies have reported that as many as 44% of viral respiratory infections may go unreported due to lack of symptoms [18]. Thus, monitoring asymptomatic individuals can potentially mitigate further spread of infection by detecting asymptomatic carriers of respiratory disease.
Another difference between clinical and surveillance testing occurs during specimen processing. All four of the current major FDA-approved multiplex assays (FilmArray RP, Nanosphere RP Flex, Luminex NxTAG RVP, and GenMark eSensor RVP) have received FDA approval for the testing of NPS only. Despite several proof-of-principal studies [19–22] that have shown comparable diagnostic Sen and Spe when using other samples types (e.g. sputum, bronchoalveolar lavage fluids, bronchial washes, anterior nares swabs, throat swabs, and tracheal aspirates), processing of other specimen types and collection methods requires additional independent studies by the testing laboratory especially if results are used to make a clinical decision. Independent studies include analytical validation studies of lab-developed tests (LDTs) that establish comparable precision, accuracy, reference range, and reportable range. Laboratories are also required to assess additional assay performance characteristics (e.g. analytical specificity and analytical sensitivity and linearity) [23]. For surveillance testing, similar independent studies are required especially when results are reported to healthcare agencies for clinical purposes. However, because surveillance testing is used to establish disease trends and reveal etiology, a variety of sample types are often collected and processed since microbial load is unknown and may be higher in one type than another [24]. To report these results, surveillance labs are also required to perform additional independent validation studies or report results as Research Use Only or Investigational Use Only. The required additional validation of other sample types likely limits the adoption of multiplexed assays for surveillance purpose.
Finally, there are differences between the number of samples typically processed for clinical diagnostic and surveillance work. Because effective clinical care depends on rapid turnaround time, clinical labs are more likely to process samples individually or in smaller batches. When identifying the source of an outbreak, rapid results are critical for control and prevention also requiring small batch testing. However, for routine surveillance whose aim is to trend population data, samples can be tested in large batches to minimize costs. However, large batch testing is often not possible for some multiplex assays due to the inherent design by manufacturers. For instance, both the FilmArray RP and the Nanosphere RP Flex can only process one sample at a time, while the Luminex NxTAG RPP and the eSensor RVP can process multiple samples at one time. It should be noted that if needed, both the FilmArray RP and the Nanosphere RP Flex allow for modular expansion to test more than one sample at a time but even then there are limitations to the number of samples that can be processed at one time. Thus, the need for surveillance labs to perform large batch testing has not been met which likely has slowed the adoption of respiratory multiplex assays by surveillance labs.
3.2. Post-analytical requirements
There are also differences when considering whether an assay will be used for either clinical diagnosis or surveillance at the post-analytical phase of testing. The original intended use of the major FDA-approved respiratory multiplex assays was to provide rapid results to aide clinical management [11,15,16,25]. As a result, since the commercial release of these assays, studies have mostly reported the post-analytical use of respiratory multiplex results in clinical settings [1,26]. However, post-analytical interpretation of results varies depending on how testing results are used. For example, in healthcare settings, results from multiplex testing are used to help manage patient care. However, for surveillance testing, results are used to trend population data, identify the causative agent of an outbreak, and confirm cases. These slight differences are significant and must be resolved.
4. Validity of performance measures
Differences during the pre- and post-analytical phases of testing help define the critical attributes needed in multiplex assays when used for either clinical testing or surveillance (Table 1). A major attribute is diagnostic validity, which measures the accuracy and usefulness of an assay [27]. For clinical use, the basic measures to assess diagnostic validity are Sen and Spe. However, despite the universal use of these measures in assessing assay performance, they are not without limitations [28]. While Sen and Spe are summarized population parameters used to confirm the presence or absence of disease, conceptually, Sen and Spe are not appropriate on a case-by-case basis (as disease progression differs from one patient to another). More critically, Sen and Spe are independent of disease prevalence, which changes how an assay may perform [29,30]. Thus, appropriate clinical decision-making using these population parameters requires more parametric inputs to aid healthcare decisions [31]. The interpretation of Sen and Spe in the clinical area is incomplete requiring more scrutiny and explanation for patient management [32].
A relatively small number of studies have directly compared the diagnostic performance between the major FDA-approved multiplex respiratory assays currently available. These studies have mostly concluded similar diagnostic performance (Table 2) based on comparisons between Sen, Spe, and Cohen’s Kappa, κ [33]. Popwitch et al. presented the most complete comparison of more than two FDA-approved multiplex respiratory assays [34]. The study included an analytical comparison of the BioFire FilmArray RP, GenMark eSensor RVP, Luminex xTAGv1, and Luminex RVP FAST using retrospective samples collected in general clinical setting screened by each panel. The eSensor RVP reported the highest Sen (100%) for nearly all targets (rhinovirus/enterovirus, 90.7% the lone exception). Low Sen for adenovirus was a weakness for most assays: FilmArray RP, 57.1%; xTAG RVPv1, 74.3%; xTAG RVP FAST, 82.9%;and eSensor RVP, 100%. Detection of Influenza B was also difficult: xTAG RVP FAST, 45.5%; FilmArray RP, 77.3%; xTAG RVPv1, 95.5%; and eSensor RVP, 100%. When assessing age-specific assay performance, only the eSensor RVP failed to show any statistically significant difference across age groups. The xTAG RVP FAST and FilmArray RP showed higher Sen for adults (≥18) than children (≤5) and teens (<18). Other comparative studies have similarly evaluated diagnostic performance based on Sen and Spe comparing the FDA-approved multiplex respiratory tests to culture, serology, and LDTs. The consensus shows that for all comparable targets, multiplex assays (based on Sen and Spe) are superior to traditional methods and relatively the same across each multiplex assay with few target exceptions (see Table 3).
Table 2.
Comparison of sensitivity (Sen), specificity (Spe), and diagnostic odds ratio (DOR, with 95%CI) of FDA-approved respiratory panels, based on data reported by manufacturers to the FDA as part of the approval processes for the assays. For the purposes of surveillance studies, assays with larger DORs have better accuracy for the pathogen(s) of interest.
| FilmArray RP |
Nanosphere RP Flex |
eSensor RVP |
Luminex NxTAG RPP |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sen | Spe | DOR | 95% CI | Sen | Spe | DOR | 95% CI | Sen | Spe | DOR | 95% CI | Sen | Spe | DOR | 95% CI | |
| Adeno | 95% | 97% | 547.59 | 192–1561.3* | 86% | 97% | 213.2 | 2.8–469.9* | n.d. | n.d. | n.d. | n.d. | 100% | 98% | 61,438.5 | 52,533.3–71,853.3* |
| Adeno B/E | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 100% | 99% | 110,001 | 46,915.4–257,915.7* | n.d. | n.d. | n.d. | n.d. |
| Adeno C | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 100% | 97% | 28,383.35294 | 11,911.8–67,631.5* | n.d. | n.d. | n.d. | n.d. |
| CoHKU1 | 100% | 99% | 123,876 | 49,003–313,149.2 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 93% | 100% | 6529.2 | 533.9–79,836.2* |
| CoNL63 | 100% | 99% | 165,501 | 82,619–331,528.6 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 95% | 99% | 3435.8 | 1469.6–8032.4* |
| Co229E | 92% | 100% | 3671.68 | 395.8–34,059.4* | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 100% | 99% | 165,501 | 129,533.6–211,455.4* |
| CoOC43 | 81% | 100% | 1371.94 | 426.1–4417.5* | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 97% | 100% | 7532.3 | 798.6–71,039.91* |
| hMPV | 97% | 100% | 7171.37 | 1496.1–34,374.1* | 100% | 100% | 332,001 | 5.7–778,167.9* | 100% | 100% | 498,501 | 121,426.9–2,046,525* | 94% | 99% | 1665.9 | 1183.1–2345.6* |
| Rhino/Entero | 98% | 94% | 561 | 295.3–1065.6* | 82% | 97% | 151.5 | 3.7–219.2* | 89% | 96% | 203.5166192 | 46.3–895.1* | 95% | 96% | 527.7 | 442.2–629.8* |
| InfA | 100% | 100% | 998,001 | 295.3–1065.6* | 98% | 99% | 9579.4 | 1.4–74,460.2* | 96% | 95% | 488.1794872 | 190.7–1249.8* | 95% | 98% | 911.8 | 745.3–1115.5 |
| InfA/H1 | 100% | 100% | 998,001 | 319,033.3–3,121,950* | 98% | 100% | 14,773.7 | 1.3–125,258.6* | 97% | 100% | 29,273.72727 | 3983.8–215,106.9* | 100% | 99% | 110,001 | 90,760.01–133,321.1* |
| InfA/H3 | 100% | 100% | 998,001 | 374,104–2,662,377* | 100% | 100% | 498,501 | 4.6–1,530,067* | 100% | 97% | 37,424.07692 | 23,683.6–59,161.3* | 99% | 98% | 2789.4 | 1374.7–5659.9* |
| InfA/H1-2009 | 100% | 100% | 998,001 | 790,354.4–1,260,202* | n.d. | n.d. | n.d. | n.d. | 100% | 99% | 65,601 | 36,662.8–117,380.4* | n.d. | n.d. | n.d. | n.d. |
| InfB | 100% | 100% | 998,001 | 22,630.6–4,394,843* | 98% | 100% | 12,201 | 1.4–9445.6* | 93% | 98% | 665.4736842 | 239.3–1850.4* | 96% | 99% | 3082.2 | 1603.4–5924.7* |
| Para1 | 100% | 100% | 998,001 | 126,362.2–7,882,150* | 100% | 100% | 8991 | 1.8–55,995.1* | 100% | 100% | 998,001 | 111,447.8–8,936,976* | 100% | 100% | 998,001 | 94,900.4–10,495,281* |
| Para2 | 98% | 100% | 9277.19 | 1136.6–75,719* | 100% | 100% | 11,975.0 | 1.1–123,475.6* | 83% | 100% | 2436.294118 | 189.0–31,397.8* | 50% | 100% | 999 | 2.78–357,781.9* |
| Para3 | 96% | 99% | 3599.48 | 765.4–16,927.5* | 82% | 100% | 4677.1 | 1.9–25,206.3* | 94% | 98% | 677.4922623 | 226.6–2025.2* | 95% | 99% | 2459.3 | 279.6–21,630.8* |
| Para4 | 100% | 100% | 248,751 | 92,525.8–668,754.6* | 79% | 100% | 1900.0 | 2.2–7629.9* | n.d. | n.d. | n.d. | n.d. | 60% | 100% | 298.5 | 48.7–1828.4* |
| RSV | 99% | 98% | 9579.43 | 1287.9–71,249.8* | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| RSVA | n.d. | n.d. | n.d. | n.d. | 100% | 100% | 998,001 | 4.4–3,408,002* | 100% | 95% | 17,850.0566 | 12,343.6–25,812.9* | 100% | 99% | 141,715.3 | 120,945.7–166,051.6* |
| RSVB | n.d. | n.d. | n.d. | n.d. | 100% | 99% | 82,251 | 4.6–221,854.5* | 100% | 96% | 23,366.85366 | 14,391.4–37,939.9* | 99% | 99% | 10,878.8 | 5528–7-21,405.9* |
| Bocavirus | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 96% | 99% | 2407.6 | 289.3–20,032.8* |
| B.pert | 67% | 100% | 2001 | 90.2–44,392.2* | 100% | 100% | 998,001 | 3.3–4,702,441* | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| B. para/bronch | n.d. | n.d. | n.d. | 720.4–49,759.4* | 100% | 100% | 998,001 | 1.5–11,010,489* | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| B. holmesii | n.d. | n.d. | n.d. | n.d. | 100% | 100% | 998,001 | 2.2–7,088,148* | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| C.pneumo | 100% | 100% | 998,001 | 116,527.2–8,547,411 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | – | 100% | 0 | n.a. |
| M.pneumo | 96% | 100% | 7580.37 | 851.6–67,473.8* | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 78% | 100% | 3501 | 372.4–32,916.5* |
DOR: Diagnostic Odds Ratio calculated as LR+/LR– per Glas et al. FilmArray RP: InfluA/H1 Sen not reported. Luminex xTAP RPP: C.pneumo site testing results for discrepant specimens were available or reported.
n.d.: target is not included as a target in the panel.
As reported in the FDA-decision summary, Sen was reported as 0%; thus, DOR was not able to be calculated.
95% CI: Calculated as LN(DOR) ± 1.96 (√(1/TP + 1/TN + 1/FP + 1/FN)),*p ≤ 0.05.
When reported, instead of using Sen and Spe at 100%, we used 99.9% in our calculations to prevent either a DOR = undefined,
Table 3.
List of all direct comparative studies of the major FDA-approved multiplex respiratory assays.
| Study | Comparison | Cohen’s Kappa, κ (95% CI) | Interpretation of κ [33,35] |
|---|---|---|---|
| Lee et al. (2017) [36] | Luminex NxTAG RPP: Luminex xTAG RVPv2 | 0.85 (0.757–0.932) | Strong |
| Chen et al. (2016) [37] | Luminex NxTAG RPP: FilmArray RP | 0.92 (0.90–0.94) | Almost perfect |
| Hwang et al. (2014) [38] | xTAG RVP: Nanosphere RV+ | 0.908a | Almost perfect |
| Popowitch, et al. (2013) [34] | FilmArray RP: eSensor RVP: Luminex xTAG RVPv1: Luminex xTAG RVP | Not reported | n/a |
| Babady et al. (2012) [21] | Luminex xTAG RVP: FilmArray RP | 0.685b | Moderate |
| Rand et al. (2011) [49] | FilmArray RP: Luminex xTAG RVP | 0.91 (0.85–0.97) | Almost perfect |
| Pabbaraju et al. (2011) [39] | Luminex xTAG RVP: Luminex xTAG RVP FAST | 0.548–1.00c | Weak to almost perfect |
aStudy only included comparison of RSV, influenza A and B.
bNo 95% CI reported.
cCalculated κ for all targets individually. Low end kappa coefficient is for influenza B only.
4.1. Diagnostic validity for surveillance use
While comparison of Sen and Spe potentially holds much value for clinical diagnoses and helps understand the critical attributes for multiplex assays, surveillance requires a broader epidemiological diagnostic approach. Diagnostic measures that capture strength and direction of association, and help epidemiologists strategize control and prevention efforts would be useful. Measures such as the DOR, ROC curves, and the Youden Index (J) may serve as good alternatives to Sen and Spe to fill this need.
DOR is interpreted as the ratio of the odds of a positive test in the presence of disease to the odds of positive test in the absence of disease. The higher the DOR the better discriminatory power of a test [40]. DOR can be calculated as
| (1) |
or equivalently:
| (2) |
DOR differentiates between those in the population with disease and those without disease based on the likelihood of a positive test result or equivalently calculated from the Sen and Spe for a specific assay. As a measure, the DOR shows the overall strength and epidemiological importance of association between test result and disease [40,41]. For instance, a larger DOR would describe a very strong association and conversely, a low DOR would describe a weaker association. A higher DOR translates to better discriminatory test performance to identify patients with disease [41]. The reported Sen and Spe from the 510(k) FDA approval summary reports of the major FDA-approved respiratory multiplex assays are summarized in Table 3, along with the DOR for each assay and for all targets available per assay. The DORs for all of the FDA-approved multiplex respiratory assays are high, which dictates that each of these assays has good discriminatory power. The strength of association for each of the assays varies depending on the target (i.e. pathogen of interest). As a result, which assay is the ‘best’ depends on the target being studied. The eSensor RVP shows greater diagnostic performance than other multiplex assays (i.e. the FilmArray RP and the Luminex NxTAG RPP) when confirming outbreak cases of adenovirus (specifically types B/C and E), common in the United States as recently as 2013 [42]. By contrast, the FilmArray RP performs better than all other comparable assays for all influenza types (A/H1, A/H3, A/H1-2009, and B). The Nanosphere RP Flex performs better than all other comparable assays for Bordetella bacterial targets (i.e. B. pertussis, B. parapertussis/B. bronchiseptica, and B. holmesii). In summary, the best assay among targets that overlap across all assays is the FilmArray RP. However, not all targets overlap as many assays have a variety of respiratory pathogens targeted by the panel, thus the best performing assay needs to be evaluated at a per target basis. ROC curves provide an alternative approach for evaluating diagnostic performance providing epidemiological insight and guidance in differentiating the usefulness of respiratory multiplex assays. ROC curves are simple visualizations that allow comparison of classifiers (i.e. tests) by plotting the True Positive Rate (TRP = Sen) against the False Positive Rate (FPR = 1–Spe) [43]. For RTIs, FPs represent a significant burden to healthcare systems especially if infection is in a vulnerable subpopulation or in individuals where coinfection exacerbate an already existing condition. For surveillance, FPs represent cases of misallocated resources, where efforts are required to treat a non-existing condition in hopes of mitigating the further spread of disease in subpopulations. If comparing routine clinical care to routine surveillance, FPs are a much greater burden to the healthcare industry, especially if the test is either expensive or if follow-up is invasive or expensive. Within ROC space, a point (FPR, TPR) on the left-hand side denotes a classifier that exhibits better performance over random chance (Figure 1). Better performing classifiers are always preferred. The discriminatory line separates ‘liberal’ and ‘conservative’ classifiers. Any point in the liberal space represents a classifier that will likely identify all the TP despite a weak signal and at the expense of having a high FPR. Conversely, any point in the conservative space will identify all the TPs in the presence of a strong signal at the expense of a low FPR. During an outbreak from a novel respiratory variant (e.g. influenza, coronavirus [44]), a high FPR is acceptable especially if we are assured that by identifying all the TPs, appropriate control and prevention could mitigate the spread of infection. Additionally, because milder forms of RTI go unreported [45] and not every respiratory infection is detected (for many reasons) true prevalence and incidence is difficult to estimate. A high FPR respiratory multiplex test could potentially minimize the error of not knowing true prevalence estimates by including false positives in the total number of positives observed. While may not be ideal for correcting true prevalence estimates, a high FPR assay could help in approaching the true burden of disease. Finally, from the ROC space plot analysis, clinical decisions benefit from the use of conservative diagnostic tests; however, for routine surveillance, liberal diagnostic tests would be more effective. Using ROC space provides a valuable method for comparing the major FDA-approved respiratory multiplex assays for either clinical diagnostic and surveillance use.
Figure 1.
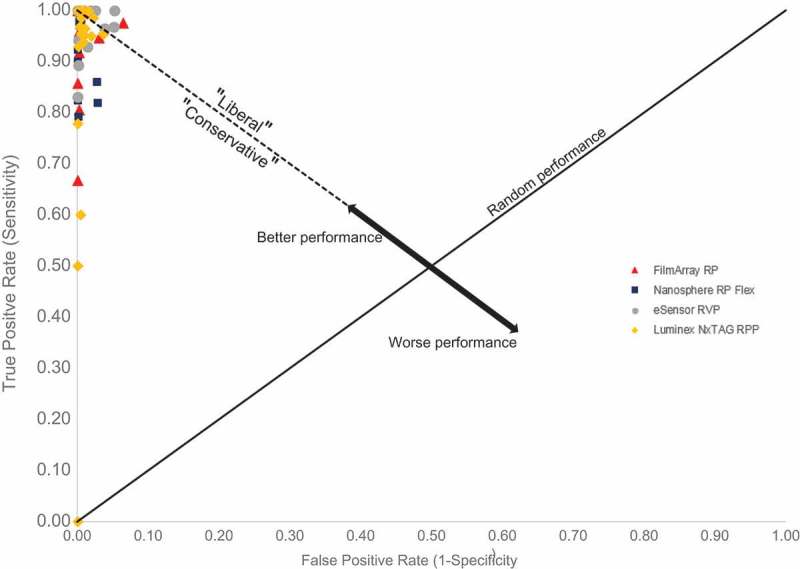
ROC space of major FDA-approved respiratory panels showing ‘Liberal’ and ‘Conservative’ calls from discrimination. Better performing assays lie on the left of random chance; worse performing assays lie on the right. Outlier at (0,0) is for C. pneumo sample tested by Luminex NxTAG RPP during clinical prospective studies; sample was discrepant with follow-up clinical site testing unknown.
Upon closer inspection of the ROC space plot (Figure 2), important similarities and differences arise between the FilmArray RP, Nanosphere RP Flex, Luminex NxTAG RPP, and the eSensor RVP. While all of these assays are better performers than random chance, a slight majority of target calls lie on the conservative side of discrimination suggesting that when considering assays as whole performers, few false positives will be reported as most calls will correctly identify positive infection. Every assay has calls throughout the ROC space suggesting that, despite an overall liberal or conservative classification of any assay, these assays could still be used for either patient care or surveillance. The ROC space plot also reveals that the most conservative multiplex assay is the Luminex NxTAG RPP which suggests the assay would be most beneficial over other assays when used for clinical diagnostic purposes. The most liberal test is the eSensor RVP, which would serve well for those performing surveillance work. From the ROC space plot, we can also make per target assay specific recommendations. The FilmArray RP has the lowest FPR across all targets except when liberal calling rhinovirus/enterovirus, RSV and CoHKU1, and conservative calling adenovirus infections. Thus, in an outbreak of rhinovirus/enterovirus, RSV or CoHKU1, the FilmArray RP would perform much better than other assays. The Nanosphere RP Flex also has a low FPR, but unlike the FilmArray RP, hardly makes any liberal calls except for RSV B suggesting a greater clinical application than others. For highly relevant clinical and public health targets (e.g. Influenza A, A/H1/A/H3, A/H1-2009, and influenza B), the FilmArray RP is nearly perfect by detecting relatively few false positives when correctly identifying positive infection. For pandemic potential targets such as coronaviruses, the FilmArray RP is again an excellent diagnostic test that has both a higher Sen and lower FPR comparable to all other assays with the sole exception of CoNL63, detected slightly better on the Luminex NxTAG RPP. Only two assays screen for Bordetella spp.: the FilmArray RP and the Nanosphere RP Flex. Among these two assays, the Nanosphere RP Flex is a near perfect assay by correctly calling positive Bordetella spp. infections. The worst performing targets across all assays were conservative calls for parainfluenza 2 (Luminex NxTAG RPP), parainfluenza 4 (Luminex NxTAG RPP), and B. pertussis (FilmArray RP). The use of ROC plots to assess graphically the diagnostic value for respiratory multiplex assays adds tremendous value where each target could be evaluated independently of other targets and in comparison to the same target across each multiplex assay.
Figure 2.
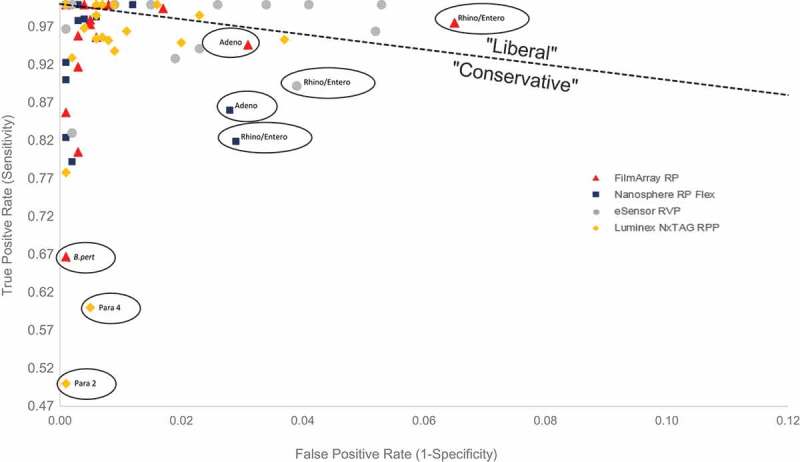
Figure 1 with x and y-axis adjusted (minus C. pneumo for Luminex NxTAG RPP) to view targets details in ROC space. Outliers circled and identified. C. pneumo target for Luminex NxTAG RPP omitted (True Positive Rate = 0.0).
Another useful epidemiologic measure often presented in conjunction with ROC curves is the Youden Index (J) [46]. J is a statistical summary measure that (similar to DOR) incorporates both Sen and Spe. J is interpreted as diagnostic test maximum effectiveness representing the optimal cutoff threshold where the maximum difference between TPR and FPR exists [47]. Essentially, J is a point (Sen, 1–Spe) in ROC space that defines where a diagnostic test will be able to correctly identify the most positives and the fewest false positives. J has a value between 0 and 1. J graphically represents the farthest point in ROC space away from random performance. J is expressed as
| (3) |
While J is not dependent on disease prevalence, it does require a dichotomous test result which may not always be applicable depending by target [48]. For instance, characterizing influenza A strains on the FilmArray RP gives one of three results, ‘Detected,’ ‘Not Detected,’ and ‘Equivocal.’ Detection of RSV on the same panel is either ‘Detected’ or ‘Not Detected.’ Similarly, the Luminex xTAG RVP provides ‘Equivocal’ results for some targets but not all [49]. For evaluating respiratory multiplex assays, J provides a simple measure of diagnostic effectiveness. Upon calculating J for each of the respiratory multiplex assays, a heat map (Figure 3) is presented that allows a visual comparison of diagnostic performance that could be used for either patient care or surveillance. The lighter the color, the closer J = 1 which is interpreted as a target that is detected much better on a respective assay. Grey boxes indicate targets not screened in respective assays. The heat map is particularly useful for identifying which assay is best suited to confirm coinfections and respond rapidly in clinical diagnosis and surveillance. For example, RSV and rhinovirus coinfections have been associated with severe pediatric disease [50]. First, the better performing assay to detect rhinovirus is identified (per the heat map, either the Luminex NxTAG RPP or the FilmArray RP). Next, consideration of the need to confirm either RSV or RSV A and RSV B is done. While the FilmArray RP can only confirm RSV, the Luminex NxTAG RPP confirms either RSV A or RSV B. With knowledge that RSV A have been implicated in more severe complications than RSV B, the Luminex NxTAG RPP would be the best suitable assay to confirm coinfections of RSV and rhinovirus. For surveillance, this same method of identifying the best performing assay of coinfection using a similar heat map could be applied to reduce the probability of having a large outbreak [51].
Figure 3.
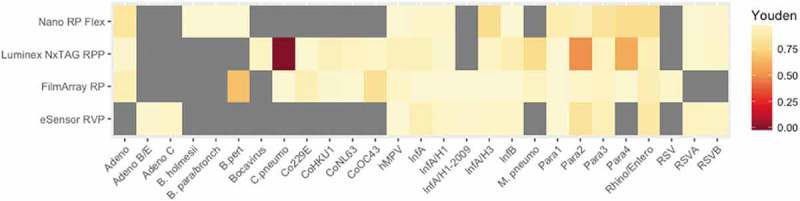
Heatmap illustrating Youden Index (J) for all FDA-approved respiratory multiplex assay targets. Boxes in gray denote not detected targets. No 95% CI calculated per the assumption that some of the index values calculated are close to 0 or 1. Per the FDA decision summary, only one C. pneumo target for the Luminex NxTAG RPP was tested during clinical prospective studies with discrepant site testing unknown.
Collectively, the use of the DOR, ROC space plots, and J allow a unique method for evaluating FDA respiratory multiplex assays for surveillance use. Each measure provides unique information that adds to surveillance strategies for aiding diagnosis and mitigating the spread of disease. The DOR allows control and prevention programs to identify strength and direction of association between test result and presence or absence of disease. ROC space plots allow differentiation between better and worse performing assays on a per target basis. And J is a supplemental measure that is most useful to minimizing the number of false positives and false negatives. From an overall evaluation based on these measures of all FDA-approved multiplex respiratory assays, the BioFire FilmArray RP is the recommended platform for surveillance use against respiratory pathogens. However, when evaluating surveillance of specific targets, such as Bordetella spp or RSV A, the recommended platform for use is the Nanosphere Verigene RP Flex. For surveillance detection of untypeable influenza A, the BioFire FilmArray RP is recommended, and for confirming adenovirus infection, the GenMark eSensor RVP is recommended. Based on ROC space plots, the Luminex NxTAG RVP is most suitable for clinical diagnoses for a majority of targets. These recommendations are based on the current formulations and reported FDA summary reports of each of these assays and would change upon release of newer versions of these same panels.
5. Validation of surveillance measures
If DORs are correlative to Sen and Spe (i.e. a high Sen or low Spe corresponding to a high DOR or low DOR, respectively), then DORs provide no supportive information and serve no practical need since Sen and Spe could be used as direct proxies. At best, these additional measures would help epidemiologists understand the performance of multiplex assays since odds ratio is a terminology that is well known within the field. However, if the correlation between DORs and Sen and Spe are different enough, then DORs must be treated as essential measures that reveal unique interpretation and use of multiplex assays. When comparing DORs to Sen, the correlation coefficients (R2) for each FDA-approved multiplex assay range from 0.29 to 0.57 suggesting poor correlation despite a positive association overall (Figure 4(a)). Similarly, when comparing DORs to Spe, the correlation coefficients for each assay range from 0.09 to 0.46 also suggesting very poor positive correlation (Figure 4(b)). As a standard, when R2 > 0, a positive correlation exists and any R2 near 0.95 and 1.0 is considered highly correlative. Despite the overall low positive correlation for each assay, interpretation for how each assay performs by target is different. There are instances when a high DOR highly correlates to a high Sen and conversely a low DOR to a low Sen. The same trend is observed for Spe. However, high correlation is target specific for each assay and the low R2 suggests that Sen and Spe should not be used as epidemiological measures when assessing how respiratory multiplex assays perform. Overall, DORs are separate and unique measures that add significant epidemiological value in assessing the use of multiplex assays.
Figure 4.
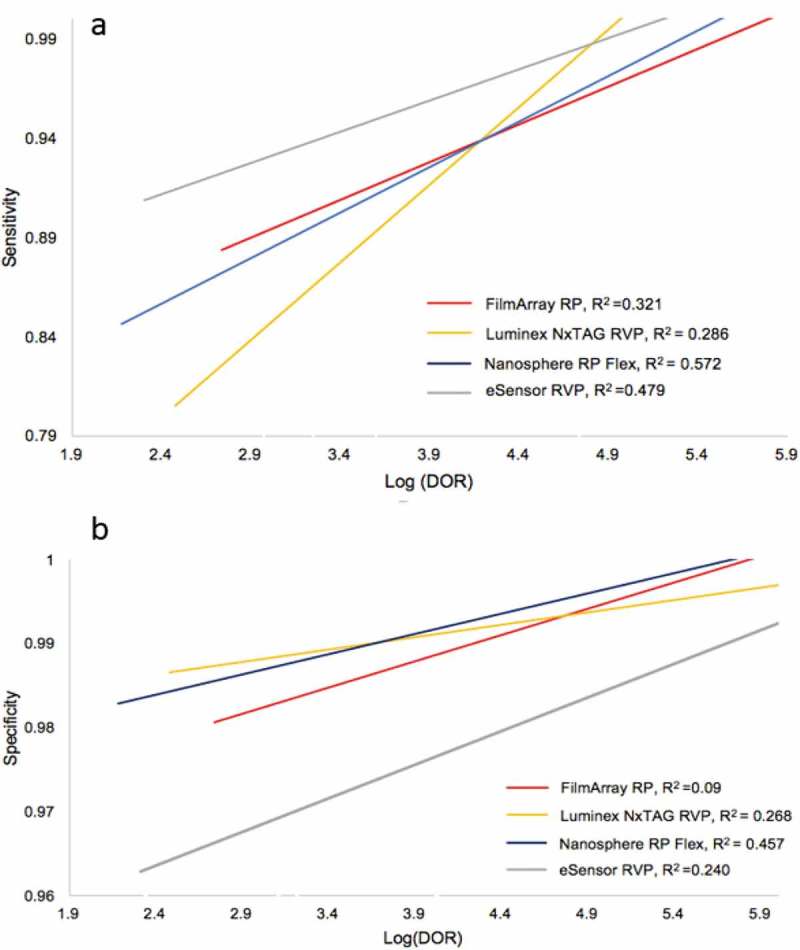
Correlation of Sensitivity and Log(DOR) (a) and Specificity and Log(DOR) (b) for all FDA-approved respiratory multiplex assays. Linear association is indicated by colored lines representing best fit across all targets for each of the four major FDA-approved respiratory multiplex assays. Correlation coefficient (R2) is given for each assay.
6. Limitations of current multiplexed assays for surveillance of respiratory pathogens
Despite the tremendous potential benefits of respiratory multiplex technologies for surveillance, these assays are not without limitations. One of the important aspects in designing PCR-based assays is primer selection. All of the FDA-approved multiplex respiratory technologies presented contain primer pools and probes corresponding to common respiratory targets. Optimized primers yield reliable qualitative results for all the targets in each individual panel. However, a universal challenge for all manufactures is selecting appropriate genomic target sequences to optimize hybridization and thereby minimizing unwanted nonspecific interactions which could lead to diagnostic errors (i.e. FP and FN). Two targets have challenged manufacturers: adenovirus and rhinovirus/enterovirus. Early assay releases of the Luminex xTAG RVP and the BioFire FilmArray RP reported poor adenovirus detection and resulted in reduced sensitivity, when presented in clinical and contrived samples [19,52,53]. Current versions of the FilmArray RP and the xTAG RVP now include updated primers aligned against the complete coding sequences of all adenovirus serotypes. The updated FilmArray RP panel has shown diagnostic improvement over the previous version [54]. eSensor RVP is the only multiplex assay capable of detecting subgroups B/E and C. For surveillance, assay choice depends on surveillance population needs. The increasing DOR order of the major FDA-approved respiratory multiplex panels as designed are: Nanosphere RP Flex, FilmArray RP, and Luminex NxTAG RPP.
Other problematic targets for respiratory multiplex testing are rhinoviruses and enteroviruses. Due to genetic homology, these pathogens lack resolution when detected on either the Luminex NxTAG RVP or FilmArray RP and as a result, when detected are simply reported as either rhinovirus or rhinovirus/enterovirus without differentiation. The primer similarity of rhinoviruses and enteroviruses increases the risk of cross-amplification and interference during multiplex testing. Per the product insert, Nanosphere RP Flex has observed cross-reactivity of rhinovirus/enterovirus primers with Human poliovirus 2 and 3, coxsackievirus A24, and EV-D68. When samples are called ‘positive’ for rhinovirus/enterovirus, laboratories should reflex to either culture or sequencing to further resolve identification. Detecting and follow-up testing for rhinovirus/enterovirus samples has become an important concern due to a nationwide outbreak of EV-D68 across the United States [55]. Symptoms included hallmark cold symptoms (fever, runny nose, and muscle aches) as well as difficulty breathing. A recent study has since described the association of EV-D68 infection and acute flaccid myelitis (related to acute flaccid paralysis) [56] highlighting the importance of good resolution between rhinovirus and enterovirus for all respiratory multiplex assays. A similar primer cross-reactivity issue is present for coronavirus OC43 and HKU1 viruses in the FilmArray RP assay. Another limitation of respiratory multiplex assays is that precise quantification of infection cannot be determined as all of the assays presented are FDA-approved qualitative test unable to determine viral or bacterial load, which (if available) may be helpful during surveillance in identifying an appropriate case definition.
Additionally, costs of respiratory multiplex testing are a significant limitation that must be justified in both clinical care settings and in surveillance efforts. While the use of respiratory multiplex assays is costlier than traditional methods, not every program can justify the use of these platforms without establishing the cost–benefit to patient- and population-based outcomes. However, the use of respiratory multiplex assays can lead to overall cost savings in all settings. The use of xTAG RVPv1 over a combination of serology and culture resulted in a cost savings of approximately $530,000 for four hospitals screening pediatric patients [57]. The xTAG RVPv1 also improved laboratory efficiency as well as reduce laboratory costs by increasing workflow [58]. Similarly, implementation of the FilmArray RP improved treatment stewardship for influenza A and influenza B, dramatically improving patient care [59]. The relative costs of multiplex respiratory assays have been previously compared and show increasing costs on the order of: FilmArray RP > eSensor RVP > xTAG RVPv1 [34,59]. The Nanosphere Verigene RP Flex should be considered the most economically feasible FDA-approved respiratory multiplex assay on the market as laboratories are only charged on preselected targets while results of remaining targets are hidden. Additional costs to consider include throughput and turnaround time. Overall, a cost benefit analysis is necessary for every laboratory and program considering implementing a respiratory multiplex test.
7. Conclusion
The current landscape of respiratory multiplex testing includes a variety of technologies that have revolutionized clinical practice; however, due to specific barriers, these technologies could impact surveillance efforts even more. Despite numerous studies showing real-world application, full adoption of these methods for routine surveillance testing and outbreak response is limited in large part due to how these technologies are evaluated. When compared to culture and serology, all of the FDA-approved assays presented in this report provide equivalent Sen and Spe, measures that have traditionally been markers for diagnostic performance. However, better measures of diagnostic performance such as DORs, ROC space plots, and the Youden Index make assay use and result interpretation clearer and much more effective for surveillance use. These measures allow comparisons on a per-target basis, providing a much more specific use of these technologies for surveillance and allow epidemiological strength of associations and direction to be determined. Additionally, these measures are not restrictive to multiplex respiratory platforms, but could also be critically used for other classes of multiplex assays and panels (e.g. enteric panels, blood culture, meningitis). The next few years will see the evolution of respiratory multiplex technologies in more rapid, more efficient, easier to use (and easier to interpret) panels capable of screening a broader range of pathogens. The challenge bestowed on laboratories is evaluating how these newer technologies can meet public health demand.
8. Expert commentary
Respiratory multiplex assays have greatly impacted patient care by reducing turnaround time, increasing throughput, and offering greater diagnostic validity over conventional methods. Sen and Spe are highly valued in determining better performing assays for clinical application. However, these same measures have limited use for surveillance since evaluating the impact of multiplex assays on population-based outcomes requires measures that capture epidemiological associations and express the performance of these assays on a population-based level. Measures that express broader epidemiological impact of respiratory multiplex assays would be useful to mitigate disease transmission, help control and prevention efforts, and identify better performing assays in populations. In light of this need, DORs, ROC space plots, and the Youden Index are plausible measures that should be used. These non-traditional evaluation measures establish not only strength and direction of association between test result and presence of disease, but also allow communicable control programs to identify better performing assays in populations, which would greatly aid outbreak response. Further, for manufacturers, these evaluation measures create opportunity to reveal panel targets and panels that perhaps should be reformulated if recommended for surveillance use.
The implication for evaluating multiplex assays for surveillance efforts instead of clinical use reaches beyond respiratory disease. While RTIs are used as a case study in this report, multiplex assays exist for screening a variety of pathogens, such as antibiotic-resistant organisms. KPC-producing carbapenem-resistant Enterobacteriaceae (CRE) has been reported in every state as of December 2017 [60]. Healthcare setting morality rates for CRE infection may be as high as 50% for some groups [61]. Currently, the Vergiene Gram-Negative Blood Culture Test, a multiplex molecular assay, identifies gram-negative bacilli from blood culture and also a variety of resistance mechanisms including KPC, OXA, IMP, and VIM. Also commercially available is the Cepheid Xpert Carba-R, a multiplex assay to identify and differentiate carbapenemase gene families. Performance evaluation of these assays and others for surveillance would distinguish better performing assays from weaker tests greatly aiding epidemiological strategies in curbing the spread of antibiotic resistance and helping lower the burden of disease.
9. Five-year view
Within five years, while multiplex respiratory assays become more rapid, more comprehensive and more automated, next-generation sequencing (NGS) platforms and assays will dominate the diagnostic market for surveillance purposes. NGS technologies are a powerful high-throughput tool capable of a variety of different healthcare and non-healthcare applications [62,63]. The introduction of NGS has already began a shift in public health practice. As a diagnostic method, multiplex NGS has been shown to confirm viral infections as detected by a PCR-based multiplex assay [64]. Similarly, Graf et al. showed the reliability of metagenomics (a specific NGS-type of analysis) as a comparable method to the eSensor RVP not just in agreement but also in detecting additional pathogens [65]. For surveillance, NGS provides detailed genomic information that aides in identifying resistance, virulence, and sequences changes over time [63,66–70]. One of the biggest challenges of applying NGS toward surveillance is the need of FDA regulatory standards and methods. NGS presents a unique regulatory challenge that requires evaluation of not just performance, but also of the big data pipelines used in determining characterization and phylogenetic relationships. However, the benefits of NGS in providing detailed omic information that could significantly improve surveillance greatly outweighs the regulatory and big data challenges.
Key issues
Advances in multiplex PCR instrument systems have replaced most traditional methods for detecting respiratory pathogens
Within the past decade, several multiplex platforms such as the Luminex NxTAG RPP, Verigene RP Flex, eSensor RVP, and the FilmArray RP have acquired FDA approval for use in public health and clinical laboratories. FDA-approved respiratory multiplex assays have successfully demonstrated public health benefits in a variety of patient populations.
From a clinical standpoint, respiratory multiplex assays have been evaluated for use and need by comparing Sen and Spe.
From a surveillance perspective, respiratory multiplex assays should be evaluated based on epidemiologically relevant and interpretable measures such as the DOR, ROC space plots, and the Youden Index.
DORs reveal direction and strength of association between test result and presence or absence of disease.
ROC space plots and the Youden Index are great tools for comparing diagnostic assays on a per target basis which can aide outbreak response.
Comparison of DORs and Sen and Spe show a low positive correlation suggesting DORs are unique measures that add valuable information regarding performance of multiplex assays.
Limitations of the use of multiplex assays include primer selection and cost.
Surveillance measures can be used to evaluate a variety of multiplex assays used to confirm and rule-out disease such as antibiotic-resistant organisms.
Funding Statement
This paper was not funded.
Declaration of Interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer Disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Subramony A, Zachariah P, Krones A, et al. Impact of multiplex polymerase chain reaction testing for respiratory pathogens on healthcare resource utilization for pediatric inpatients. J Pediatr. 2016;173:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fleming-Dutra KE, Hersh AL, Shapiro DJ.. Prevalence of inappropriate antibiotic prescriptions among us ambulatory care visits, 2010-2011. JAMA. 2016. May 3;315(17):1864–1873. [DOI] [PubMed] [Google Scholar]
- 3.CDC RSV Census regional trends updated 2017; cited 2016 February4 Available from: https://www.cdc.gov/surveillance/nrevss/rsv/region.html.
- 4.Hayden FG, De Jong MD.. Emerging influenza antiviral resistance threats. J Infect Dis. 2011. January 1;203(1):6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su S, Fry AM, Kirley PD. Survey of influenza and other respiratory viruses diagnostic testing in US hospitals, 2012-2013. Influenza Other Respir Viruses. 2016. March;10(2):86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong KK, Jain S, Blanton L, et al. Influenza-associated pediatric deaths in the United States, 2004-2012. Pediatrics. 2013. November;132(5):796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Foundation for Infectious Diseases Respiratory syncytial virus in older adults: a hidden annual epidemic. Cited 2018 April30 Available from: http://www.nfid.org/publications/reports/rsv-report.pdf.
- 8.Ginocchio CC, St George K. Likelihood that an unsubtypeable influenza A virus result obtained with the Luminex xTAG respiratory virus panel is indicative of infection with novel A/H1N1 (swine-like) influenza virus. J Clin Microbiol. 2009. July;47(7):2347–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Revealed that untypeable influenza A results on the Luminex xTAG RVP could be interpreted as indicative for the presence of the 2009 pandemic strain.
- 9.McAllister SC, Schleiss MR, Arbefeville S, et al. Epidemic 2014 enterovirus d68 cross-reacts with human rhinovirus on a respiratory molecular diagnostic platform. PLoS One. 2015;10(3):e0118529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibib DR, Matushek SM, Beavis KG, et al. BioFire FilmArray respiratory panel for detection of enterovirus d68. J Clin Microbiol. 2016. February;54(2):457–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahony J, Chong S, Merante F, et al. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid micobead-based assay. J Clin Microbiol. 2007. September;45(9):2965–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jannetto PJ, Buchan BW, Vaughan KA, et al. Real-time detection of influenza A, influenza B, and respiratory syncytial virus A and B in respiratory specimens by use of nanoparticle probes. J Clin Microbiol. 2010. November;48(11):3997–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.French CE, McKenzie BC, Cooper C, et al. Risk of nosocomial respiratory syncytial virus infection and effectiveness of control measures to prevent transmission events: a systematic review. Influenza Other Respir Viruses. 2016. Jul;10(4):268–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghebremedhin B. Human adenovirus: viral pathogen with increasing importance. Eur J Microbiol Immunol (Bp). 2014. March;4(1):26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierce VM, Hodinka RL. Comparison of the GenMark Diagnostics eSensor respiratory viral panel to real-time PCR for detection of respiratory viruses in children. J Clin Microbiol. 2012. November;50(11):3458–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poritz MA, Blaschke AJ, Byington CL, et al. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One. 2011;6(10):e26047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen RR, Wieringa J, Koekkoek SM, et al. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J Clin Microbiol. 2011. July;49(7):2631–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau LL, Cowling BJ, Fang VJ, et al. Viral shedding and clinical illness in naturally acquired influenza virus infections. J Infect Dis. 2010. May 15;201(10):1509–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruggiero P, McMillen T, Tang YW, et al. Evaluation of the BioFire FilmArray respiratory panel and the GenMark eSensor respiratory viral panel on lower respiratory tract specimens. J Clin Microbiol. 2014. January;52(1):288–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byington CL, Ampofo K, Stockmann C, et al. Community surveillance of respiratory viruses among families in the utah better identification of germs-longitudinal viral epidemiology (BIG-LoVE) study. Clin Infect Dis. 2015. October 15;61(8):1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babady NE, Mead P, Stiles J, et al. Comparison of the Luminex xTAG RVP Fast Assay and the Idaho Technology FilmArray RP Assay for detection of respiratory viruses in pediatric patients at a cancer hospital. J Clin Microbiol. 2012. July;50(7):2282–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song Cho G, Moon BY, Lee BJ, et al. High rates of detection of respiratory viruses in the nasal washes and muscosae of patients with chronic rhinosinusitis. J Clin Microbiol. 2013. March;51(3):979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halling KC, Schrijver I, Persons DL. Test verification and validation for molecular diagnostic assays. Arch Pathol Lab Med. 2012. January;136(1):11–13. [DOI] [PubMed] [Google Scholar]
- 24.Lieberman D, Shleyfer E, Castel H, et al. Nasopharyngeal versus oropharyngeal sampling for isolation of potential respiratory pathogens in adults. J Clin Microbiol. 2006. February;44(2):525–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jannetto PJ, Buchan BW, Vaughan KA. Real-time detection of influenza A, influenza B, and respiratory syncytial virus A and B in respiratory specimens by use of nanoparticle probes. J Clin Microbiol. 2010. November;48(11):3997–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchand-Austin A, Farrell DJ, Jamieson FB, et al. Respiratory infection in institutions during early stages of pandemic (H1N1) 2009, Canada. Emerg Infect Dis. 2009. December;15(12):2001–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feighner JP, Herbstein J. Diagnostic validity In: Last CG, Hersen M, editors. Issues in diagnostic research. Boston (MA): Springer; 1987. [Google Scholar]
- 28.Parikh R, Parikh S, Arun E, et al. Likelihood ratios: clinical application in day-to-day practice. Indian J Ophthalmol. 2009. May-Jun;57(3):217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenner H, Gefeller O. Variation of sensitivity, specificity, likelihood ratios and predictive values with disease prevalence. Stat Med. 1997. May 15;16(9):981–991. [DOI] [PubMed] [Google Scholar]
- 30.Moons KG, Van Es GA, Deckers JW. Limitations of sensitivity, specificity, likelihood ratio, and Bayes’ theorem in assessing diagnostic probabilities: a clinical example. Epidemiology. 1997. January;8(1):12–17. [DOI] [PubMed] [Google Scholar]; •• Describes the limitations of common diagnostic performance measures.
- 31.Idrovo AJ. Three criteria for ecological fallacy. Environ Health Perspect. 2011. August;119(8):a332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akobeng AK. Understanding diagnostic tests 1: sensitivity, specificity and predictive values. Acta Paediatr. 2007. March;96(3):338–341. [DOI] [PubMed] [Google Scholar]
- 33.McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb). 2012. October;22(3):276–282. [PMC free article] [PubMed] [Google Scholar]
- 34.Popowitch EB, O’Neill SS, Miller MB. Comparison of the BioFire FilmArray RP, GenMark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP fast multiplex assays for detection of respiratory viruses. J Clin Microbiol. 2013. May;51(5):1528–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sim J, Wright CC. The kappa statistic in reliability studies: use, interpretation, and sample size requirements. Phys Ther. 2005. March;85(3):257–268. [PubMed] [Google Scholar]
- 36.Lee CK, Lee HK, Ng CWS, et al. Comparison of Luminex NxTAG Respiratory Pathogen Panel and xTAG Respiratory Viral Panel FAST Version 2 for the detection of respiratory viruses. Ann Lab Med. 2017. May;37(3):267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen JH, Lam HY, Yip CC, et al. Clinical evaluation of the new high-throughput Luminex NxTAG Respiratory Pathogen Panel Assay for multiplex respiratory pathogen detection. J Clin Micrbiol. 2016. July;54(7):1820–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang SM, Lim MS, Han M, et al. Comparison of xTAG respiratory virus panel and Verigene Respiratory Virus Plus for detecting influenza virus and respiratory syncytial virus. J Clin Lab Anal. 2015. March;29(2):116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pabbaraju K, Tokaryk KL, Wong S, et al. Comparison of the Luminex xTAG Respiratory Viral Panel with in-house nucleic acid amplification tests for diagnosis of respiratory virus infections. J Clin Microbiol. 2008. September;46(9):3056–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simundic AM. Measures of diagnostic accuracy: basic definitions. EJIFCC. 2009. January;19(4):203–211. [PMC free article] [PubMed] [Google Scholar]
- 41.Glas AS, Lijmer JG, Prins MH, et al. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003. November;56(11):1129–1135. [DOI] [PubMed] [Google Scholar]
- 42.Scott MK, Chommanard C, Lu X, et al. Human adenovirus associated with severe respiratory infection, Oregon, USA, 2013-2014. Emerg Infect Dis. 2016. June;22(6):1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fawcett T. An introduction to ROC analysis. Pattern Recogn Lett. 2006;27:861–874. [Google Scholar]
- 44.Coleman CM, Frieman MB. Coronaviruses: important emerging human pathogens. J Virol. 2014. May;88(10):5209–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.WHO Morbidity rate for children aged 0-4 years due to acute respiratory illness In: Children’s environmental health indicators. cited 2016 May5 Available from: http://www.who.int/ceh/indicators/ARImorbidity0_4.pdf?ua=1 [Google Scholar]
- 46.Youden WJ. Index for rating diagnostic tests. Cancer. 1950. January;3(1):32–35. [DOI] [PubMed] [Google Scholar]
- 47.Ruopp MD, Perkins NJ, Whitcomb BW, et al. Youden index and optimal cut-point estimated from observations affected by a lower limit of detection. Biom J. 2008. June;50(3):419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou XH, McClish DK, Obuchowski NA. Combined measures of sensitivity and specificity In: Statistical methods in diagnostic medicine. New York (NY): John Wiley & Sons; 2009. p. 23–24. [Google Scholar]
- 49.Rand KH, Rampersaud H, Houchk HJ. Comparison of two multiplex methods for detection of respiratory viruses: FilmArray RP and xTAG RVP. J Clin Microbiol. 2011. July;49(7):2449–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Costa LF, Queiroz DA, Lopes Da Silveira H, et al. Human rhinovirus and disease severity in children. Pediatrics. 2014. February;133(2):e312–e321. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez JP, Ghanbarnejad F, Eguiluz VM. Risk of coinfection outbreaks in temporal networks: a case study of a hospital contact network Front. Phys., 2017. Oct; 5(46). [Google Scholar]
- 52.Esposito S, Scala A, Bianchini S, et al. Identification of human adenovirus in respiratory samples with Luminex Respiratory Virus Panel Fast V2 assay and real-time polymerase chain reaction. Int J Mol Scie. 2016. February 26;17(3):297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faden H, Wynn RJ, Campagna L, et al. Outbreak of adenovirus type 30 in a neonatal intensive care unit. J Pediatr. 2005. April;146(4):523–527. [DOI] [PubMed] [Google Scholar]
- 54.Song E, Wang H, Salamon D, et al. Performance characteristics of FilmArray Respiratory Panel v1.7 for detection of adenovirus in a large cohort of pediatric nasopharyngeal samples: one test may not fit all. J Clin Microbiol. 2016. June;54(6):1479–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.CDC Enterovirus D68. updated 2016; cited 2016 May9 Available from: https://www.cdc.gov/non-polio-enterovirus/about/ev-d68.html.
- 56.Aliabadi N, Messacar K, Pastula DM, et al. Enterovirus D68 infection in children with acute flaccid myelitis, Colorado, USA, 2014. Emerg Infect Dis. 2016. August;22(8):1387–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mahony JB, Blackhouse G, Babwah J, et al. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J Clin Microbiol. 2009. September;47(9):2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dundas NE, Ziadie MS, Revell PA, et al. A lean laboratory: operational simplicity and cost effectiveness of the Luminex xTAG Respiratory Viral Panel. J Mol Diagn. 2011. March;13(2):175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu M, Qin X, Astion ML, et al. Implementing of FilmArray Respiratory Viral Panel in a core laboratory improves testing turnaround time and patient care. Am J Clin Pathol. 2013. January;139(1):118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.CDC Healthcare-associate infections: tracking CRE. updated 2018 February27; cited 2018 April30 Available from: https://www.cdc.gov/hai/organisms/cre/trackingcre.html
- 61.CDC, Jacob JT, Klein E, et al. Vital signs: carbapenem-resistant Enterobacteriaceae. MMWR. 2013. March 8;62(09):165–170. [PMC free article] [PubMed] [Google Scholar]
- 62.Struelens MJ, Brisse S. From molecular to genomic epidemiology: transforming surveillance and control of infectious diseases. Euro Surveill. 2013;18(4):20386. [DOI] [PubMed] [Google Scholar]
- 63.Koser CU, Ellington MJ, Cartwright EJ, et al. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 2012;8(8):e1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zoll J, Rahamat-Langendoen J, Ahout I, et al. Direct multiplexed whole genome sequencing of respiratory tract samples reveals full viral genomic information. J Clin Microbiol. 2015;66:6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Graf EH, Simmon KE, Tardif KD, et al. Unbiased detection of respiratory virus by use of RNA sequencing-based metagenomics: a systematic comparison to a commercial PCR panel. J Clin Microbiol. 2016. April;54(4):1000–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGinnis J, Laplante J, Shudt M, et al. Next generation sequencing for whole genome analysis and surveillance of influenza A viruses. J Clin Virol. 2016;79:44–50. [DOI] [PubMed] [Google Scholar]
- 67.Dallman TJ, Byrne L, Launders N, et al. The utility and public health implications of PCR and whole genome sequencing for the detection and investigation of an outbreak of Shig toxin-producing Escherichia coli serogroup O26:H11. Epidemiol Infect. 2015. June;143(8):1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carriço JA, Sabat AJ, Friedrich AW, et al. Bioinformatics in bacterial molecular epidemiology and public health: databases, tools and the next-generation sequencing revolution. Euro Surveill. 2013;18(4):1-9. [DOI] [PubMed] [Google Scholar]
- 69.Koser CU, Ellington MJ, Peacock SJ. Whole genome sequencing to control antimicrobial resistance. Trends Genet. 2014. September;30(9):401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aanensen DM, Feil EJ, Holden MT, et al. Whole-genome sequencing for routine pathogen surveillance in public health: a population snapshot of invasive Staphylococcus aureus in Europe. MBio. 2016. May 5;7(3):1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]


