Abstract
Non-coding RNAs are RNA molecules that are not translated into the protein, making up the vast majority of the human genome. Long non-coding RNAs (lncRNA) are in the RNA group that has longer than 200 nucleotides, and non-protein coding transcripts. In recent years, the potential has attracted considerable attention as new important biological regulators. LncRNAs play a critical role in regulating the activity and localization of proteins, processing the production of small RNAs, and processing other RNAs. They are also involved in cell differentiation, cell cycle, proliferation, apoptosis, migration and invasion by modulation of gene expression. Abnormal expression of LncRNAs has an important role in the function of oncogenes and tumor suppressor genes. Recently, there has been an increasing number of studies on the tumorigenic effects of specific lncRNAs in the initiation and progression of cancer. In this review, general information about lncRNAs is provided, including the biological importance of lncRNAs in cancer diseases and their potential development in therapeutic applications.
Keywords: Cancer, long noncoding RNA, oncogene, tumor suppressor
In a recent study on the human genome showed that sequenced more than 90% of the DNA are readily transcribed and only a few of them are active transcripts (2%) are protein encoded RNAs transcribed from DNA but not translated into proteins, which are called as non-coding RNAs (ncRNAs) [1]. Studies that have been conducted up to date show that the proportion of non-coding RNAs in some functions of biological systems is higher than that of coding RNAs. In this regard, the most non-coding RNAs (99%) play a role in primer RNA transcripts. Moreover, the ratio of non-coding RNA to RNA varieties that produce poly-A tailed mature RNA is to be known as 80–90%. Approximately 60% of the protein-coding genes are targeted by non-coding RNAs and one or more homologous antisense IncRNA is found in about 70% of the genes encoding the protein [2]. Long non-coding RNAs have more than 200 nucleotides. Non-coding RNAs can be classified according to their functions (Fig. 1) [3]. They have been found to be involved in the physiological and pathological processes of human diseases, including cancer, which is thought to be transcriptional “noises” [4]. Long non-coding RNAs play a role in regulating miRNA synthesis, directing chromatin-modifying structures to gene regions, managing transcriptional programs [5]. In Figure 2, some of the long non-coding RNAs that are present in the biologic functions are shown.
FIGURE 1.
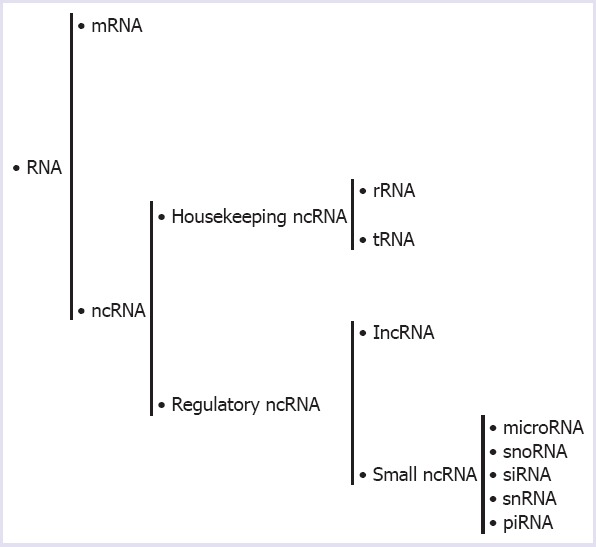
Classification of Non-coding RNAs. They were classified into housekeeping and regulatory RNAs according to their functions. Housekeeping ncRNAs consist of carrier RNA and ribosomal RNA. On the other hand, regulatory RNAs contain small non-coding RNAs (sncRNAs) and long non-coding RNAs (lncRNAs). Small ncRNAs that vary in a base range of approximately 15–40; microRNAs (miRNA), small nucleolar RNAs (snoRNAs), small inhibitory RNAs (siRNAs), small nuclear RNAs (snRNAs), and RNAs interacting with PIWI (piRNAs).
FIGURE 2.
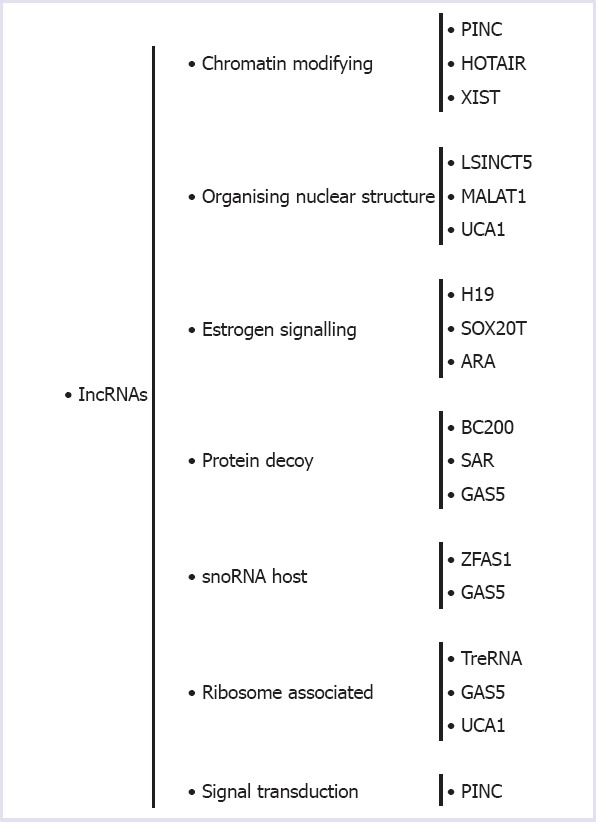
Overview of the molecular functions of IncRNAs.
The Role of Long Non-Coding RNAs in Cancer Formation
So far, 8,179 (60.31%) lncRNAs have been found to play a crucial role in at least one type of cancer formation [6]. Approximately, 15% up and 11.18% down-regulated lncRNAs have been shown in seven types of cancer, including gastric, lung, prostate, breast, pancreatic, hepatocellular and ovarian cancer types [7–10]. lncRNA dysregulation can be affected by somatic copy number alterations (SCNAs). SCNAs have been identified in many cancer types, and 36.27% of the lncRNA gene copy numbers have been found a significant correlation with RNA expression levels [11].
Lung squamous cell carcinoma and ovarian cancer have stronger RNA-SCNA correlations than fewer SCNAs cancer types, such as acute myeloid leukemia and prostate adenocarcinoma [11, 12]. LncRNAs regulate gene expression and genomic functions in the nucleus. They play an active role in the transcription and chromatin-related epigenetic mechanisms by affecting chromatin structure and modifications. It has been reported that lncRNAs modulate chromatin by covalently interacting with enzymatic complexes and controling ATP dependent chromatin remodeling complexes that may change the nucleosome spacing [13].
RNA expressions have been found to have higher specificity for lncRNAs encoded in primary tumor tissues, biopsies after metastatic progression, cancer cell lines, and adjacent normal tissues. LncRNAs have an important role in gene regulation because only 25–40% of the protein-coding genes in genomic sequences occur to have overlapping antisense transcription [14, 15].
Mechanisms of lncRNAs in Gene Regulation
LncRNAs regulate gene expression and protein synthesis, but the mechanisms underlying these activities are unclear [16], which remained under-researched. The gene regulation can be cis (close to the transcribed lncRNA) or trans (far from the transcriptional region) when the effects of chromatin modulation are gene-specific [17]. LncRNAs have been shown to play an important role in the basic processes of gene regulation, such as chromatin modification, transcription and post-translation of proteins, localization-silencing of genes through endogenous siRNA production and microRNA modulation. It has been understood that the levels of aberrant expression of lncRNAs have significant a role in all stages of cancer development, including the onset, progression and metastasis of cancer [18]. Several lncRNAs have tumor suppressor or oncogenic properties in more than one type of cancer [19, 20].
Tumor Suppressor and Oncogenic Long Non-Coding RNAs
LncRNAs play a crucial role in developing cancer. Expression of lncRNAs in tumors is significantly regulated by epigenetic and genomic modifications; hence, these lncRNAs can control both protein-coding and non-coding genes and interact with known other cancer genes [21, 22]. Long non-coding RNAs have different features that act like tumor suppressors, oncogenes, or in their therapeutic potentials [23–25]. Their complicated structures and ability to be involved in multicomponent complexes are thought to be the main cause of these differences. It is shown that IncRNAs, which vary in gene expression specific to a tissue or cell type, play both tumor suppressor and oncogenic roles in various types of cancer [26].
In recent studies, H19, which was initially reported to be a mutated tumor suppressor gene, showed abnormal expression in many types of cancer. H19 has a role of activation in breast, lung, cervical, esophagus and bladder tumors [18]. H19 induces cell growth and proliferation in the bladder and hepatocellular cancers [27]. Overexpression of XIST lncRNA is associated with tumor malignancies in several cancers. It has been demonstrated that XIST suppresses KLN2 expression in non-small cell lung cancers. In another study, it was disclosed that XIST might be an oncogenic IncRNA that promotes miR-140/mIR-124 expression from microRNAs and promotes gene expression. XIST plays a role in the cell cycle through miR-140/miR-124, thereby promoting growth in pancreatic carcinoma [28].
The high-level transcription of KCNQTOT1 IncRNA has been detected in colorectal cancers, but its functional role is still unknown [29]. It has been shown that IncRNA ANRIL may have an effect on tumorigenesis in various types of cancer. Tumor size was also found high in patients with high-level ANRIL expression in osteosarcoma (OS) tissues. This indicates that patients’ survival times reduced and there is a positive correlation between high ANRIL expression and poor prognosis of OS (bone tumor) patients [30]. Additionally, several studies have indicated that an increase in the lncRNA ANRIL enhances the growth of nasopharyngeal carcinoma growth and hence, tumor formation [31]. Other studies reported HOTAIR to act as a trans-acting lncRNA and to be associated with breast cancer, lung cancer and gastric cancer [32]. In breast cancer, HOTAIR competes with BRCA1 protein. It was reported that mir-7 and mir-34a regulate the HOTAIR expression level, which interacts with mir-141 and mir-34a to reinforce cancer development [33]. Overexpression of HOTAIR induces invasion and metastasis of breast cancer cells. [34]. It was reported that overexpression of HOTAIR might cause metastasis to spread to more areas and move to different organs, promoting the proliferation of cancer cells in tissues in gastric cancer [35].
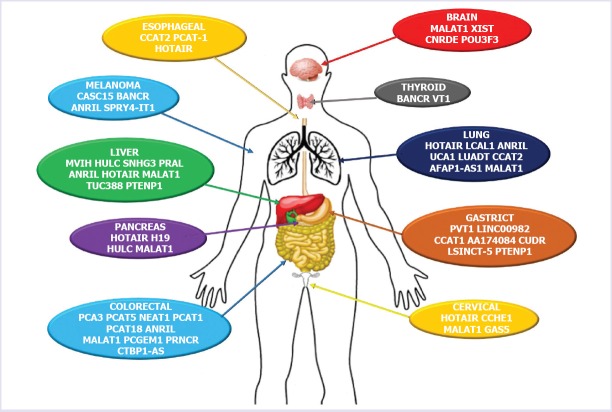
It has been described that UCA1 exerts oncogenic functions in many types of cancer [36]. Overexpression of UCA1 has been reported in bladder carcinoma [37], tongue squamous cell carcinoma [38], ovarian cancer [39], and melanoma [40]. Studies have shown that long non-coding RNA GAS5 affects the miR-32-5p/PTEN axis and prevents cancer cells from developing and metastasizing into different tissues [41]. It has been demonstrated that miR-32-5p plays a tumor regulatory role. It may play a role in the development of pancreatic carcinoma (PC) because of its overexpression in PC tissues. In another study, it was disclosed that overexpression of GAS5 might inhibit invasion, migration, and proliferation of colorectal cancer (CRC) HT-29 cells. In the same study, it was also shown that colorectal cancer cells were induced by overexpression of GAS5 in apoptosis [42]. Figure 3 also shows the types of IncRNA that play a role in some types of cancer.
FIGURE 3.
Differentially expressed tissue-specific long non-coding RNAs.
Therapeutic Approaches towards Long-coding RNAs in the Treatment of Cancer
Up to now, regarding protein-coding genes peptides, proteins, antibodies and small RNAs have been known as therapeutic agents. In recent studies, long non-coding RNAs have emerged as promising drug targets [43, 44].
As a result of the interaction of structural functions and complex structures of RNA genes with multiple factors, there can be more complex interactions. Consequently, the functional effects of targeting lncRNA should be analyzed in the light of a systematic approach rather than a singly based target. Defining lncRNAs, which are involved in cellular processes that contribute oncogenesis, tumor suppressor, or tumorigenesis, will provide new therapeutic developments in cancer that targets lncRNA [45].
pHLIP-PNA targets solid tumors, and it was reported that lncRNA is a new therapeutic marker for in vivo cancer. It was shown that 91H, BCAR4, HULC, MALAT-1, TUGl, UCA1 are oncogenic, Loc285194, MEG3 are tumor suppressor, and LINC00161 and ODRUL are drug dependent lncRNAs. They may have potential therapeutic targets in osteosarcoma [46].
In vitro studies hold promising results for understanding the functions of lncRNA. In vivo validation of lncRNA, which are supposed to be used as therapeutic targets, is challenging and needs to be optimized. Gene editing, such as TALEN or CRISPR/Cas9 methodologies, is thought to enable evaluation of lncRNA functions in details [34, 47, 48]. LncRNAs can specifically interact with chromatin-modifying complexes, DNA, mRNA, ncRNA, and proteins to change cellular physiology, may show dysregulation in diseases like cancer and may form a basis for therapeutic interruption [49]. To improve treatment efficiency, an integrated approach is required to unify lncRNA with other proteins and genes. Advanced studies targeting lncRNA will provide ggolden opportunities for the development of new therapeutic methods to cure cancers and other diseases.
Conclusion
Many studies showed that LncRNAs are expressed in different types of cancer play and they have an important role in the pathogenesis of diseases. Recent studies have shown that lncRNAs are fundamental building blocks of cellular differentiation, cell progenitor organogenesis and tissue homeostasis. lncRNAs can act as a tumor suppressor or an oncogene by interacting with the promoter or enhancer region of a gene to modulate the gene expression. In some cases, lncRNAs can also regulate a protein’s function. Studies conducted to elucidate the functions of lncRNA in cancer will increase our understanding of molecular mechanisms of this disease better. As a result, LncRNAs can be considered as new important therapeutic targets for the treatment and diagnosis of diseases.
Footnotes
Conflict of Interest: No conflict of interest was declared by the authors.
Financial Disclosure: The authors declared that this study has received no financial support.
Authorship Contributions: Concept – EG, MHA, SK, MG, TMO; Design – EG, MHA, SK, MG, TMO; Supervision – EG, BY; Analysis and/or interpretation – MHA, SK, MG; Literature review – TMO, SK, EG; Writing – TMO, BY, SK, MG, MHA, EG; Critical review – EG, BY, TMO.
REFERENCES
- 1.Li X, Wang Z. The role of noncoding RNA in thyroid cancer. Gland Surg. 2012;1:146–50. doi: 10.3978/j.issn.2227-684X.2012.10.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su Y, Wu H, Pavlosky A, Zou LL, Deng X, Zhang ZX, et al. Regulatory non-coding RNA:new instruments in the orchestration of cell death. Cell Death Dis. 2016;7:e2333. doi: 10.1038/cddis.2016.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei MM, Zhou GB. Long Non-coding RNAs and Their Roles in Non-small-cell Lung Cancer. Genomics Proteomics Bioinformatics. 2016;14:280–8. doi: 10.1016/j.gpb.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ping G, Xiong W, Zhang L, Li Y, Zhang Y, Zhao Y. Silencing long noncoding RNA PVT1 inhibits tumorigenesis and cisplatin resistance of colorectal cancer. Am J Transl Res. 2018;10:138–49. [PMC free article] [PubMed] [Google Scholar]
- 5.Hansji H, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Front Genet. 2014;5:379. doi: 10.3389/fgene.2014.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE:the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misawa A, Takayama KI, Inoue S. Long non-coding RNAs and prostate cancer. Cancer Sci. 2017;108:2107–14. doi: 10.1111/cas.13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malhotra A, Jain M, Prakash H, Vasquez KM, Jain A. The regulatory roles of long non-coding RNAs in the development of chemoresistance in breast cancer. Oncotarget. 2017;8:110671–84. doi: 10.18632/oncotarget.22577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–63. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huo X, Han S, Wu G, Latchoumanin O, Zhou G, Hebbard L, et al. Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma:implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol Cancer. 2017;16:165. doi: 10.1186/s12943-017-0734-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529–40. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balbin OA, Malik R, Dhanasekaran SM, Prensner JR, Cao X, Wu YM, et al. The landscape of antisense gene expression in human cancers. Genome Res. 2015;25:1068–79. doi: 10.1101/gr.180596.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, et al. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–20. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parasramka MA, Maji S, Matsuda A, Yan IK, Patel T. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol Ther. 2016;161:67–78. doi: 10.1016/j.pharmthera.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi D, Zhang C, Liu X. Long noncoding RNAs in cervical cancer. J Cancer Res Ther. 2018;14:745–53. doi: 10.4103/jcrt.JCRT_669_17. [DOI] [PubMed] [Google Scholar]
- 18.Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget. 2014;5:10976–96. doi: 10.18632/oncotarget.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yarmishyn AA, Kurochkin IV. Long noncoding RNAs:a potential novel class of cancer biomarkers. Front Genet. 2015;6:145. doi: 10.3389/fgene.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauptman N, Glavač D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655–69. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geisler S, Lojek L, Khalil AM, Baker KE, Coller J. Decapping of long noncoding RNAs regulates inducible genes. Mol Cell. 2012;45:279–91. doi: 10.1016/j.molcel.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng Q, Ren M, Li Y, Song X. LncRNA-RMRP Acts as an Oncogene in Lung Cancer. PLoS One. 2016;11:e0164845. doi: 10.1371/journal.pone.0164845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanoglu A, Balta AZ, Berber U, Ozdemir Y, Emirzeoglu L, Sayilir A, et al. MicroRNA expression profile in patients with stage II colorectal cancer:a Turkish referral center study. Asian Pac J Cancer Prev. 2015;16:1851–5. doi: 10.7314/apjcp.2015.16.5.1851. [DOI] [PubMed] [Google Scholar]
- 25.Shenouda SK, Alahari SK. MicroRNA function in cancer:oncogene or a tumor suppressor?Cancer Metastasis Rev. 2009;28:369–78. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 26.Inamura K. Major Tumor Suppressor and Oncogenic Non-Coding RNAs:Clinical Relevance in Lung Cancer. Cells. 2017;6:pii:E12. doi: 10.3390/cells6020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang S, Gong X, Zhang G, Huang G, Lu Y, Li Y. The lncRNA XIST interacts with miR-140/miR-124/iASPP axis to promote pancreatic carcinoma growth. Oncotarget. 2017;8:113701–18. doi: 10.18632/oncotarget.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sunamura N, Ohira T, Kataoka M, Inaoka D, Tanabe H, Nakayama Y, et al. Regulation of functional KCNQ1OT1 lncRNA by β-catenin. Sci Rep. 2016;6:20690. doi: 10.1038/srep20690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu G, Liu G, Yuan D, Dai J, Cui Y, Tang X. Long non-coding RNA ANRIL is associated with a poor prognosis of osteosarcoma and promotes tumorigenesis via PI3K/Akt pathway. J Bone Oncol. 2018;11:51–5. doi: 10.1016/j.jbo.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu JH, Tang JM, Li J, Li XW. Upregulation of SOX2-activated lncRNA ANRIL promotes nasopharyngeal carcinoma cell growth. Sci Rep. 2018;8:3333. doi: 10.1038/s41598-018-21708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin Y, Guo W, Li N, Fu F, Lin S, Wang C. Polymorphisms of long non-coding RNA HOTAIR with breast cancer susceptibility and clinical outcomes for a southeast Chinese Han population. Oncotarget. 2017;9:3677–89. doi: 10.18632/oncotarget.23343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pawłowska E, Szczepanska J, Blasiak J. The Long Noncoding RNA HOTAIR in Breast Cancer:Does Autophagy Play a Role? Int J Mol Sci. 2017;18:jpii:E2317. doi: 10.3390/ijms18112317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen WM, Chen WD, Jiang XM, Jia XF, Wang HM, Zhang QJ, et al. HOX transcript antisense intergenic RNA represses E-cadherin expression by binding to EZH2 in gastric cancer. World J Gastroenterol. 2017;23:6100–10. doi: 10.3748/wjg.v23.i33.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nasrollahzadeh-Khakiani M, Emadi-Baygi M, Nikpour P. Augmented expression levels of lncRNAs ecCEBPA and UCA1 in gastric cancer tissues and their clinical significance. Iran J Basic Med Sci. 2017;20:1149–58. doi: 10.22038/IJBMS.2017.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12:4851–8. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 38.Fang Z, Wu L, Wang L, Yang Y, Meng Y, Yang H. Increased expression of the long non-coding RNA UCA1 in tongue squamous cell carcinomas:a possible correlation with cancer metastasis. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;117:89–95. doi: 10.1016/j.oooo.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Liu SP, Yang JX, Cao DY, Shen K. Identification of differentially expressed long non-coding RNAs in human ovarian cancer cells with different metastatic potentials. Cancer Biol Med. 2013;10:138–41. doi: 10.7497/j.issn.2095-3941.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian Y, Zhang X, Hao Y, Fang Z, He Y. Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma Res. 2014;24:335–41. doi: 10.1097/CMR.0000000000000080. [DOI] [PubMed] [Google Scholar]
- 41.Gao ZQ, Wang JF, Chen DH, Ma XS, Wu Y, Tang Z, et al. Long non-coding RNA GAS5 suppresses pancreatic cancer metastasis through modulating miR-32-5p/PTEN axis. Cell Biosci. 2017;7:66. doi: 10.1186/s13578-017-0192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Wang Y, Zhang CG, Xiao Xiao HJ, Hu Hou JM, et al. Effect of long non-coding RNA Gas5 on proliferation, migration, invasion and apoptosis of colorectal cancer HT-29 cell line. Cancer Cell Int. 2018;18:4. doi: 10.1186/s12935-017-0478-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang J, Zhuo H, Zhang X, Jiang R, Ji J, Deng L, et al. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis. 2014;5:e1549. doi: 10.1038/cddis.2014.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amit D, Hochberg A. Development of targeted therapy for bladder cancer mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. J Transl Med. 2010;8:134. doi: 10.1186/1479-5876-8-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura T, Aikata H, Takahashi S, Takahashi I, Nishibuchi I, Doi Y, et al. Stereotactic body radiotherapy for patients with small hepatocellular carcinoma ineligible for resection or ablation therapies. Hepatol Res. 2015;45:378–86. doi: 10.1111/hepr.12359. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Dou P, Liu T, He S. Application of Long Noncoding RNAs in Osteosarcoma:Biomarkers and Therapeutic Targets. Cell Physiol Biochem. 2017;42:1407–19. doi: 10.1159/000479205. [DOI] [PubMed] [Google Scholar]
- 47.Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–45. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 48.Hainer SJ, Charsar BA, Cohen SB, Martens JA. Identification of Mutant Versions of the Spt16 Histone Chaperone That Are Defective for Transcription-Coupled Nucleosome Occupancy in Saccharomyces cerevisiae. G3 (Bethesda) 2012;2:555–67. doi: 10.1534/g3.112.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics Proteomics Bioinformatics. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]